- Department of Microbiology, Sikkim University, Gangtok, India
Over-exploitation and energy security concerns of the diminishing fossil fuels is a challenge to the present global economy. Further, the negative impact of greenhouse gases released using conventional fuels has led to the need for searching for alternative biofuel sources with biomass in the form of lignocellulose coming up as among the potent candidates. The entrapped carbon source of the lignocellulose has multiple applications other than biofuel generation under the biorefinery approach. However, the major bottleneck in using lignocellulose for biofuel production is its recalcitrant nature. Carbohydrate Active Enzymes (CAZymes) are enzymes that are employed for the disintegration and consumption of lignocellulose biomass as the carbon source for the production of biofuels and bio-derivatives. However, the cost of enzyme production and their stability and catalytic efficiency under stressed conditions is a concern that hinders large-scale biofuel production and utilization. Search for novel CAZymes with superior activity and stability under industrial condition has become a major research focus in this area considering the fact that the most conventional CAZymes has low commercial viability. The gut of plant-eating herbivores and other organisms is a potential source of CAZyme with high efficiency. The review explores the potential of the gut microbiome of various organisms in the production of an efficient CAZyme system and the challenges in using the biofuels produced through this approach as an alternative to conventional biofuels.
1 Introduction
Microorganism with ubiquitous nature and high genetic diversity has evolved over millions of years to adapt to and form a unique ecological niche in different ecosystems on earth. A different group of microbes including bacteria, fungi, protozoa, viruses, etc. interacts together to form a complex microbial community termed microbiota which, along with their entire genetic content, is termed microbiome (Dekaboruah et al., 2020). These microbiomes are found in different environments such as soil, deep oceans, hot springs, etc. They are also found to be present in different living forms such as humans, insects, plants, animals, etc., and consist of all commensal, symbiotic, and pathogenic microbes living in association and interacting with their host organism (Wagg et al., 2019).
In recent years, various significant interests have evolved in the gut microbiome of different life forms since the microbiota is involved in the nutritional and metabolic activities that directly influence the health of the host. Earlier studies focused majorly on the composition of these microbiomes but the functional aspect of these microbiomes and their influence on the host health is analysed currently (Antwis et al., 2017; Coyte and Rakoff-Nahoum, 2019). Microorganisms in microbiomes interact with their host and among themselves to form synergistic relationships. It also determines the composition and influences not only where they reside but it also effects the physiology of their hosts (Foo et al., 2017; Contijoch et al., 2019). One such example is the gut microbiome of different organisms with various plant parts and polysaccharides as their nutritional source such as termites, ruminants, and other herbivores. The microorganisms are essential for the utilization of the saccharides trapped in this recalcitrant carbon source since the host lacks the enzymes required for their metabolism (Chettri and Verma, 2022a). Here the active microbial breakdown of dietary plant polysaccharides takes place and the gut of ruminants consists of plant polysaccharides, where 70% of the volume of the gut is devoted to microbial fermentation (Flint, 2020).
The gut microbiota from these organisms has become an interestingly attractive source of enzymes responsible for the degradation of lignocellulose also called the Carbohydrate Active Enzymes (CAZymes) (Wardman et al., 2022). The CAZymes have been divided into six major classes i.e., Glycosyl hydrolase (GHs), Glycosyl transferase (GTs), Polysaccharide Lyases (PLs), Carbohydrate Esterases (CEs) and Auxiliary Activities (AAs) which are involved in the hydrolysis, formation, and modification of glycosidic bonds and are essential for carbohydrate metabolism (Chettri et al., 2020). These CAZymes are of high industrial value and are also essential pertaining to the concept of sustainable development (Jakeer et al., 2020). High energy demand, emission of harmful pollutants from conventional fossil fuel and their limited availability has sparked research interest in alternate energy source in the form of biofuel which is generated using different carbon source with lignocellulose being considered the most abundant, economic and environmentally friendly option (Zhou et al., 2021).
Lignocellulose is the most copious renewable biomass on earth, and it is also considered a carbon-neutral source that decreases carbon dioxide emission and atmospheric pollution. It is the major component of different waste streams from different industries, agriculture, forestry and municipalities. Lignocellulose is composed of the following components: Cellulose, hemicelluloses and lignin which form a highly complex structure that is recalcitrant to degradation (Zhu et al., 2020). The CAZymes from various sources play a crucial role in the hydrolysis of the sugar polysaccharides in the lignocellulose for biofuel production. To increase the commercial viability of lignocellulose as an alternate energy source an efficient CAZyme system is required for which different ecosystems in nature are mined (Wertz and Béchade, 2020). Since the gut microbiome is such a prominent source and several studies indicate the potential of lignocellulose (Calusinska et al., 2020; Peng et al., 2021; Huang et al., 2022; Xiao et al., 2022), the present review examines the gut microbiome of herbivores and insects from this perspective. Previous review articles were limited to a specific part of organisms such as insects (Jang and Kikuchi, 2020; Bhujbal et al., 2021), or herbivores (Ozbayram et al., 2020; Chettri and Verma, 2022b). We searched the literature for prominent CAZymes from the guts of various organisms in the animal kingdom where a clear relationship between diet, gut flora composition, and CAZyme was found.
2 Gut microbiome as a source of carbohydrate active enzymes
The gut of herbivorous animals is mainly the source of microorganisms that exhibit hydrolytic activity, allowing them to convert complex feed material into readily absorbable monomers. The gut of herbivorous animals, especially ruminants, is a rich source of microorganisms capable of producing carbohydrate-active enzymes (CAZymes) as they consume plant fibers and lignocellulosic plants. The gut microbiome of carnivores, on the other hand, specializes in the breakdown of proteins as an energy source and does not have CAZyme-producing organisms. Omnivores, such as humans, have certain bacteria in their gut that produce CAZymes because humans consume plants directly or indirectly.
2.1 Gut microbiome of ruminants
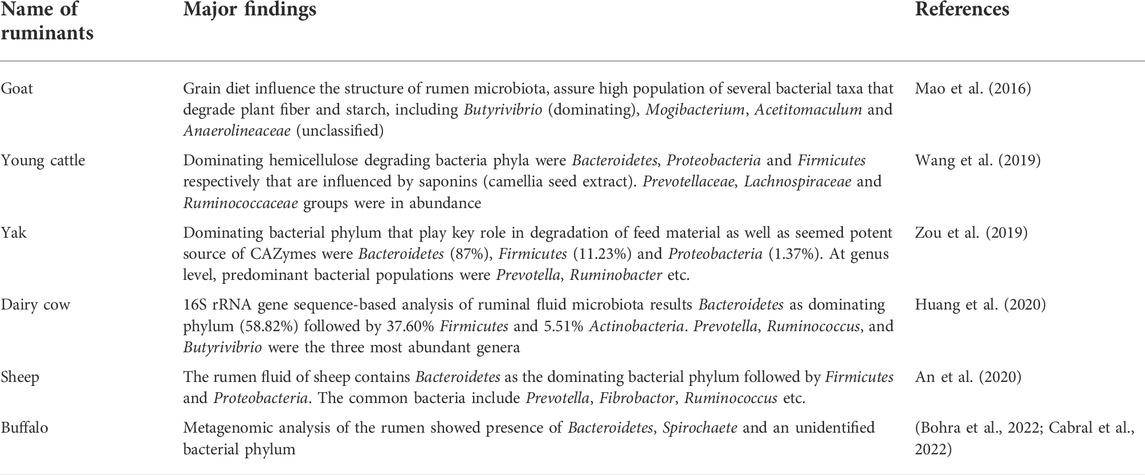
The rumen is the most important organ of ruminants with numerous and diverse microflora. There are many types of microorganisms belonging to different species (Zou et al., 2018). The rumen functions as an anaerobic digestive chamber that maintains a relatively constant pH of 6–7. It is inhabited by more than 5,000 species of microorganisms, 95% of which are bacteria while the remaining 3%–4% belong to archaea, protozoa, anaerobic fungi, viruses, and bacteriophages (Cholewińska et al., 2021). These microorganisms break down forage fibers (generally cellulose and hemicellulose) through enzymatic reactions in the rumen into monomeric compounds that can be easily digested and absorbed by the body (Chen H et al., 2021). The main function of the rumen microflora is to convert plant food into energy. The resulting products are volatile fatty acids (VFAs) - mainly propionate and butyrate - and proteins. These are the main source of energy for the animals. This process has a direct impact on the physiological parameters of the animals, including production rates (De Nardi et al., 2016). Studies have shown that many important factors influence the abundance and composition of the rumen microbiota, for example, individual differences between animals, diet composition, and fineness-to-roughness ratio (Cholewińska et al., 2021).
The rumen of adult ruminants consists of many bacterial species involved in the breakdown of cellulose and lignin. Some common microorganisms are Fibrobacter succinogenes, Ruminococcus albus, Ruminococcus flavefaciens, and Prevotella ruminicola (Liu et al., 2020), of which F. succinogenes is the most abundant and significant in the degradation of plant cellulose (Sha et al., 2020). The common microbiota of ruminants in the rumen and surrounding gut is summarized in Table 1.
A greater fraction of enzymes targeted against oligosaccharides is associated with bacteria present in the animal’s digestive system, including β-glucuronidase, β-D-mannosidase, β-xylosidase, β-1,3-xylosidase, galactan 1, α-L-arabinofuranosidase, β-D-galactosidase, exo-β-glucosaminidase, arabinanase, xylanase, and 3-β-galactosidase (Sha et al., 2020).
2.2 Gut microbiome of other vertebrates
The gastrointestinal tract (GI) of vertebrates is a special environment that is composed of interactions that are ecologically conserved as well as an evolutionary progression between the host and resident microflora (Youngblut et al., 2019). However, the gut microflora varies greatly among animals, as the morphology of the GI tract differs among species (Siddiqui et al., 2022).
Approximately 90% of the fish gut microbiota is composed of Proteobacteria, Firmicutes, and Bacteroidetes. Some common bacterial genera are Bacillus, Staphylococcus, Lactobacillus, Actinobacteria, etc. (Clements et al., 2014). In fish, hydrolysis of β-glycosidic bonds of non-starch polysaccharides (derived from plant feeds) by CAZymes is scarce or absent. However, in vitro studies on several carbohydrate-active Bacillus species isolated from the hindgut of herbivorous fish showed cellulase and xylanase activity. This observation was supported by the presence of genes encoding specific extracellular CAZymes that help fish digest the non-starch polysaccharides of plant feeds (Serra et al., 2019).
The digestive tract of birds’ GI harbors a high density of microbial communities, up to 1011 CFU/g in the hindgut. As in the case of other vertebrates, the avian microbiota GI is dominated by Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria (Waite and Taylor, 2015). The dominant bacterial genera are Enterococcus, Escherichia, Bacillus, etc. (Siddiqui et al., 2022). A South American bird, the hoatzin (Opisthocomus hoazin), has unique nutritional characteristics. Unlike other birds, it carried out foregut fermentation in leaves and plants. As a result, its digestive strategy is more similar to ruminants than birds. Their forestomach microbiota is dominated by Firmicutes, Bacteroidetes, and Proteobacteria (Godoy-Vitorino et al., 2012). This may be an important source for isolating CAZyme-producing bacteria.
According to studies, only ∼17% of vertebrate taxa are known in reptiles (Keenan et al., 2013). The most abundant phyla in the reptile gut are Proteobacteria, Bacteroidetes, Firmicutes, and Fusobacteria (Zhang et al., 2019). Some common bacterial genera are Pseudomonas, Staphylococcus, Enterobacter, Aeromonas, etc. (Siddiqui et al., 2022), while there is little information on the gut microbiome of amphibians. As in humans and mice, Bacteroidetes, Firmicutes, and Proteobacteria are the main components of the gut microbiome of amphibians. Some Flavobacteria are also observed (Colombo et al., 2015). There is no documentation supporting the presence of CAZyme-producing bacteria in reptiles and amphibians, as they are mostly carnivorous animals.
According to studies, only about 10% of vertebrate taxa are known to exist in mammals, particularly humans and mice (Keenan et al., 2013). The gut of mammals contains about 1,014 microorganisms (>1 kg). For example, human gut bacteria include predominantly species of the following genera: Escherichia, Enterobacter, Clostridium, Klebsiella, Lactobacillus, Bacteroides, Butyrivibrio, Bifidobacterium, Prevotella, etc. various yeasts and other microorganisms may also be present (Nicholson et al., 2005). Most CAZymes are produced by members of the Bacteroides sp., Butyrivibrio fbrisolvens, Clostridium hathewayi, Enterobacter cloacea, Escherichia coli, Klebsiella pneumoniae, Prevotella copri, Prevotella salivae, etc. (Ghosh and Pramanik, 2021).
In addition to ruminants, CAZymes have also been studied in various mammals. For example, in an adult elephant, the members of the phylum Proteobacteria show a maximum CAZyme abundance of 91%, followed by Actinobacteria and Bacteroidetes each with a value of 4.6% and 2.5% respectively (Jakeer et al., 2020). The pseudo-ruminant dromedary camels have a rumen microbiota which is handy compared to ruminants. The bacterial community for CAZymes was Prevotella (23%), Ruminococcus (1%), Bacteroides (4%), Butyrivibrio (1%), and Fibrobacter (4%), (Hinsu et al., 2021). Researchers had identified an endonuclease with cellulase activity in a bacterial strain isolated from the intestine of a bamboo rat; a rich CAZyme family was also detected in the appendix microbiome. In lignocellulosic diets, the dominant microbial families in the gut include Muribaculaceae, Lachnospiraceae, Ruminococcaceae, and Desulfovibrionaceae (Xiao et al., 2022).
2.3 Gut microbiome of insects
Insects are the most diverse group of animals in different ecological niches. It is estimated that the insect gut contains microorganisms 10 times higher than their cells and their microbial genome is also 100 times their genes (Krishnan et al., 2014). The insect gut is inhabited by diverse microorganisms including bacteria (most abundant), fungi, archaea, protozoa, and viruses (Munoz-Benavent et al., 2021). The most common and abundant bacteria include Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Tenericutes (Brune and Dietrich, 2015).
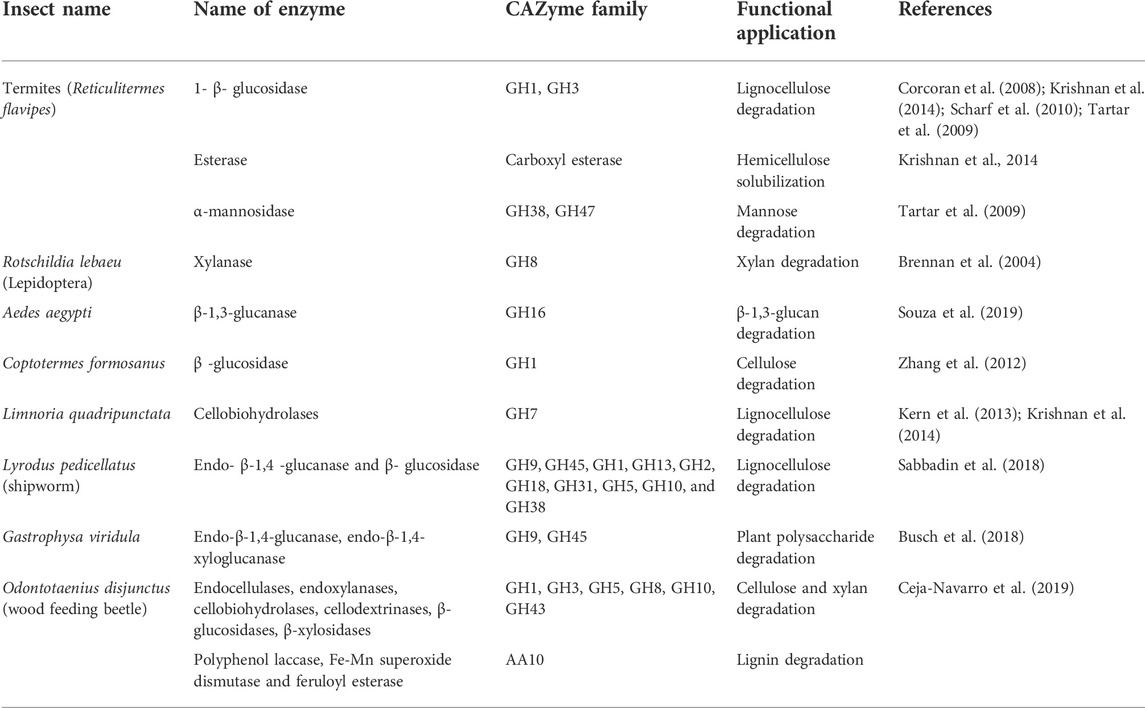
Termites are one of the most successful groups of organisms that can degrade woody materials. Therefore, their gut is a potential source of CAZyme-producing bacteria (Krishnan et al., 2014). Wood-eating termites are generally higher termites in which Spirochaetes and Fibrobacteres are the dominant bacteria responsible for digesting lignocellulose. In lower termites, lignocellulose is degraded by symbiotic flagellates (Munoz-Benavent et al., 2021). Shipworms are a good example of wood-feeding organisms that contain multiple endosymbiotic bacteria capable of producing a large number of carbohydrate-active enzymes (CAZymes).
3 Industrial production of carbohydrate active enzymes
The commercial enzymes production, including CAZymes, can be classified into two phases, upstream and downstream processes. The upstream process involves the selection of a suitable microorganism/strain, followed by strain improvement and process development in order to optimize growth and enzyme production conditions (Zeng et al., 2020). The different substrates, fermentation conditions, and environmental factors such as pH, temperature, etc. are considered to maximize enzyme production. The optimized conditions will then be applied to flask/laboratory scale production, followed by a pilot plant that will eventually be scaled up to an industrial-scale fermenter. Novel approaches to fermenter design and mathematical modelling will be used for the upstream processes (Wehrs et al., 2019; Tarafdar et al., 2021).
Downstream processing includes recovery of the enzyme, concentration, and purification of the enzyme for various applications. Depending on the type of enzyme and the application, different techniques such as chromatography, phase separation, and, depending on the location of the enzyme, cell lysis are used (Yan et al., 2018; Cortes, 2020). The purified CAZyme is then used for enzymatic hydrolysis of lignocellulose to produce simple sugar molecules, which are then further used for biofuel production.
A wider range of natural polysaccharide-based substrates from the agro-industrial sector, e.g., lignocellulose, is available for biotransformation into industrially relevant and high-value products such as “biofuel” (Arevalo-Gallegos et al., 2017). CAZymes are used in biofuel production to convert this recalcitrant polysaccharide into fermentable carbohydrate monomers (Tingley et al., 2021). They offer a sustainable alternative to surmount the drawbacks linked with conventional chemical catalysts (Saini et al., 2022). Bacteria and anaerobic fungi are considered promising for lignocellulosic biofuel production due to their natural diversity of CAZymes (Saye et al., 2021). Thus, rumen isolates and their CAZymes provide an efficient system for the degradation of lignocellulose for biofuel production (Patel et al., 2020).
4 Lignocellulose structure
Lignocellulose is a very complex and heterogeneous substance consisting of two main biopolymers, i.e., holocellulose and lignin. The holocellulose in turn comprises two different biopolymers, namely cellulose and hemicellulose (Smith, 2019). The ratio of these components can be different among species, species, and even within an individual plant depending on age, growth stage, and environmental parameters (Bajpai, 2016).
4.1 Cellulose structure
Cellulose is the predominant component of the lignocellulosic matrix and endows mechanical strength on the plant cell wall. Some bacterial, algal, and tunicate species are also known to produce cellulose (Seddiqi et al., 2021). It is an unbranched, linear homopolysaccharide composed of glucopyranose units linked by β-1,4-glycosidic bonds, with each monomer unit twisted 180° relative to its neighbours. The recurring unit of this biopolymer i.e. cellobiose consists of two adjacent glucose units linked by glucosidic bonds (C-O-C) at positions C1 and C4 (Chen P et al., 2021). The length of the cellulose chain depends on the degree of polymerization (DP), which can vary widely, from about 20 in synthetic cellulose to about 10,000 or more in cotton, plant fibres, and bacterial cellulose (Fang et al., 2020). The presence of two types of hydrogen bonds between the OH group of C3 and the oxygen atom of the pyranose ring within the same molecule and the oxygen of the hydroxymethyl group at the C6 position of a molecule and the oxygen atom of the glycosidic bond of the adjacent molecule gives cellulose fibrils a high axial rigidity. This crystal forms a fervent structure that cannot be penetrated by water or enzymes, making them insoluble in water. These intermolecular hydrogen bonds and van der Waals interactions promote the parallel stacking of multiple long-chain cellulose polymers, which form elemental fibrils that further group into microfibrils (Gupta et al., 2019). Microfibrils are further aggregated into cellulose fibres (Bajpai 2016). Within cellulose fibrils, a large percentage of cellulose is arranged in highly ordered crystalline forms, while a small portion of disordered cellulose chains exist in amorphous form (Gupta et al., 2019). Crystalline cellulose exists in several different allomorphs (cellulose I, II, III, and IV), of which cellulose I is the thermodynamically metastable and cellulose II has maximum stability (Baghaei and Skrifvars, 2020).
4.2 Hemicellulose structure
Unlike cellulose, hemicellulose is a heterogeneous polymer of 5-C sugars (arabinose, xylose, and rhamnose), 6-C sugars (galactose, mannose, and glucose), and uronic acids (D-galacturonic acid, 4-O-methylglucuronic acid, and D-glucuronic acid) (Ayeni et al., 2015). These monosaccharides are arranged in either homopolymeric or heteropolymeric configurations with small side branches linked by β-(1,4) and β-(1,3) glycosidic linkages (Bajpai, 2016). The backbone of hemicelluloses is arranged in different compositions depending on biomass type. Agricultural biomass such as straw, grasses, and hardwood hemicelluloses are mainly composed of xylan, while glucomannan is the major component in softwood hemicelluloses (Bajpai, 2016). In addition to a common 1,4-linked β-D-xylopyranose units backbone, they may comprise acetic acid, arabinose, ferulic acid, glucuronic acid or its 4-O-methyl ether, and p-coumaric acid (Qaseem et al., 2021).
4.3 Lignin structure
Lignin is one of the most abundant terrestrial biopolymers, surpassed only by cellulose and accounting for a significant proportion of the lignocellulosic biomass. It is an amorphous heteropolymer that is not composed of carbohydrates and has a complex structure resulting from the oxidative coupling reaction between three monolignols (phenylpropane unit): p-coumaryl alcohol, sinapyl alcohol, and coniferyl alcohol. In the lignin polymer, these modified phenylpropanoid monomers are recognized as p-hydroxyphenyl (H-lignin), syringyl (S-lignin), and guaicyl (G-lignin) units. Alkyl-alkyl, alkyl-aryl, and aryl-aryl ether linkages are the primary linkages used for the polymerization of phenolic monomers (Ralph et al., 2019).
4.4 Lignocellulose degradation
The complex protective barrier of lignin and hemicellulose surrounding cellulose along with the crystalline structure of cellulose makes it difficult to break down lignocellulose into its components. Various chemical, physicochemical, physical, and biological pretreatment methods play an important role in depolymerizing lignocellulosic biomass into its major components. Biological pretreatment by microorganisms is a cost-effective and environmentally friendly approach (Kumar et al., 2020).
The various enzymes produced by these organisms target the lignocellulosic components to degrade them. Enzymes involved in lignin polymerization are broadly classified into two classes based on their mode of action: Lignin-modifying enzymes (LMEs) and lignin-degrading auxiliary enzymes (LDAs). LMEs include lignin peroxidase, manganese peroxidase, versatile peroxidase, laccases, and dye-degrading peroxidase. Rather than degrading lignin themselves, LMEs assist LDAs in the degradation process. Glyoxal oxidase, glucose dehydrogenase, aryl alcohol oxidase, cellobiose dehydrogenase, and pyranose-2-oxidase are lignin-degrading auxiliaries involved in various steps of lignin degradation (Sharma et al., 2020). The complex structural heterogeneity and chemical diversity of hemicellulose require the synergistic activity of a wide range of enzymes for its effective hydrolysis. The cellulase enzyme system includes three soluble extracellular enzymes: 1, 4-β-endoglucanase (EC 3.2.1.4), 1, 4-β-exoglucanase (cellobiohydrolase) (EC 3.2.1.91), and β-glucosidase (β-D-glucoside glucohydrolase or cellobiase) (EC 3.2.1.21) (Nandy et al., 2021). Absolute hydrolysis of cellulose to glucose is only possible through the synergy of the three enzymes mentioned above or thorough mineralization to H2O and CO2 (Popovic et al., 2019).
The isolates from the rumen possess these CAZymes, which have been explored by several researchers for the degradation of lignocellulose (Table 2).
4.5 Production of biofuels
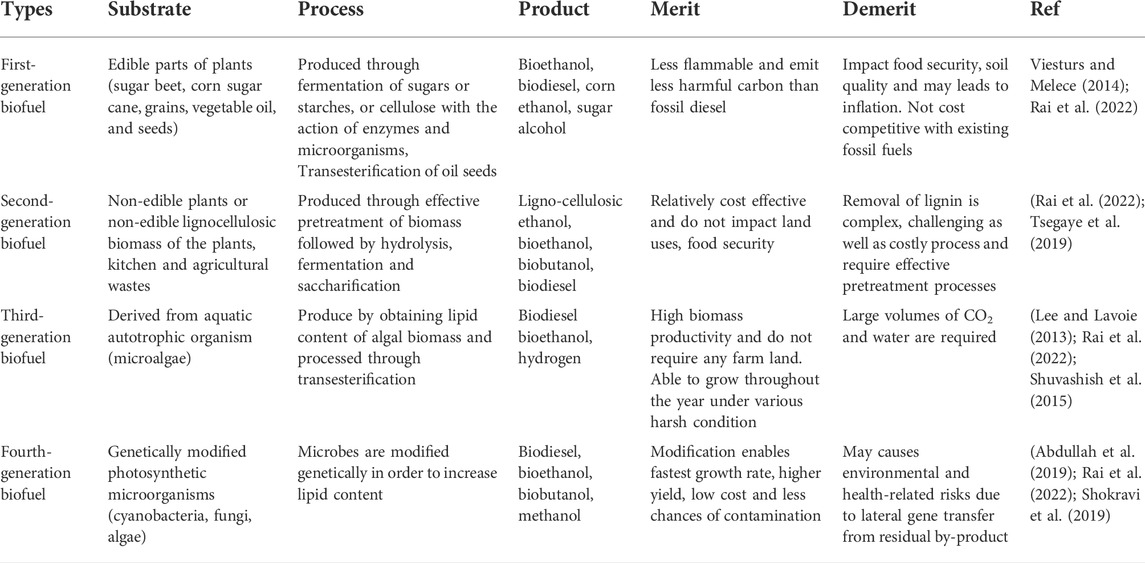
In recent decades, biofuel production has been explored worldwide as a sustainable substitute to fossil fuels because it is cost-effective, eco-friendly, and has an abundant substrate. Based on substrate source and the technical aspect of production, biofuels have been categorised into four generations (Rai et al., 2022) (Table 3). However, to date, more than 90% of bioethanol produced worldwide is derived from edible crops such as sugarcane, beets, corn, etc., which ultimately affects food security and leads to inflation (Choudhary et al., 2020). The United States and Brazil rank first and second, respectively, in the production of bioethanol and biodiesel (Sajid et al., 2021). In 2019, the U.S. recorded bioethanol production of 15.8 billion gallons, representing about 54% of global production. The second largest producer, Brazil, recorded bioethanol production of 35.6 billion gallons from sugarcane and corn, which accounted for 30% of global production during the same period. In 2019, the US recorded biodiesel production of 2.5 billion gallons, followed by Brazil with 5.8 billion litres (Sajid et al., 2021). Lignocellulosic biomass includes non-food energy feedstocks used as an alternative to cane sugar or corn-starch for biofuel production and is referred to as second-generation biofuel (Choudhary et al., 2020; Rai et al., 2022). However, the presence of lignin is a barrier to the expansion of biofuel production and commercialization (Tran et al., 2019). Therefore, pretreatment of biomass is required to remove the lignin component and increase the accessibility of cellulose and hemicellulose for hydrolysis and fermentation by increasing the porosity and surface area of the substrates (Tsegaye et al., 2019). Currently, there are various pretreatment methods for lignocellulosic biomass, which can be broadly classified into physical, chemical, physiochemical, and biological pretreatments. Physical pretreatment methods include biomass size reduction, extrusion, microwave irradiation, and ultrasonic treatment (Tayyab et al., 2018). Chemical pretreatment includes acid pretreatment, alkaline pretreatment, ozonolysis pretreatment, and organic solvent pretreatment (Rahardjo et al., 2021). Ammonia fiber explosion (AFEX) pretreatment, liquid hot water pretreatment, steam explosion pretreatment, and supercritical CO2 pretreatment fall under physiochemical pretreatment (Tran et al., 2019; Rahardjo et al., 2021). Nowadays, biological pretreatment with microorganisms is gaining a lot of attention due to its energy efficiency, environmental friendliness, and the fact that no or very few unwanted by-products are generated (Rahardjo et al., 2021). However, biological pretreatment is quite expensive and requires more time (Tayyab et al., 2018; Rahardjo et al., 2021).
The ability of the gut microbiome to produce CAZymes is also a focus of interest worldwide to find an optimal microbial candidate with different enzymatic mechanisms for the depolymerization of lignocellulose. The ability of gut microbes to degrade lignocellulose was discussed in the previous section. The resulting hydrolysis products, which include simple saccharides, can be used for biofuel production (Rabee et al., 2022) evaluated the higher ethanol production associated with the hydrolysis of barley straw by camel and sheep rumen juice containing Ruminococcus, Saccharofermentans, and Butyrivibrio. The physicochemical properties of biodiesel produced by Meyerozyma caribbica SSA1654, a manganese-dependent peroxidase-producing oleaginous strain isolated from the guts of the wood-eating termites Coptotermes formosanus and Reticulitermes chinenesis were thoroughly investigated by (Ali et al., 2021). In this study, CAZyme activity (β-glucosidase, xylanase) was also determined along with a coupled pathway of dye degradation for biodiesel production (Li et al., 2020). reported the presence of 136 genes of CAZyme from five subfamilies in B. velezensis LC1, a symbiotic bacterium isolated from the intestine of Cyrtotrachelus buqueti. This report also provides crucial insight into the potent ability of B. velezensis to convert lignocellulosic biomass to ethanol (Ben Guerrero et al., 2015). described the endoglucanase and xylanase activities of Cohnella, Paenibacillus, Acinetobacter, and Klebsiella bacteria associated with the gut digestome of N. aquilinus and C. fulviceps termites suitable for bioethanol production (Deka et al., 2013). conducted a batch simultaneous saccharification and fermentation experiment using the enzyme cellulase isolated from B. subtilis along with Z. mobilis and wild grass as the substrate to determine the different pretreatment methods by determining the ethanol yield coefficient (g ethanol/g substrate) (Corcoran et al., 2008). conducted a study on a two-step process of hydrolysis and fermentation to produce ethanol using sugarcane bagasse by microorganisms found in the rumen fluids of dairy cows.
5 Assessment of the feasibility of the application of biofuels
Although biofuels have several advantages compared to fossil fuels, economic and environmental viability are the most important issues that need to be critically analysed in order for biofuel to replace conventional fuels. The life cycle analysis of these biofuels provides information on this aspect of biofuels and helps in policy-making for sustainable production and application of biofuels. In addition, the technological aspect of biofuel application also needs to be reviewed.
5.1 Life cycle analysis
It is generally assumed that biofuels emit fewer pollutants during their life cycle compared to fossil fuels. However, the environmental impact of the energy crops used and the type of lignocellulosic treatment method used must also be examined to determine the overall impact of biofuels. Since there are limited land resources available for energy crop production, in addition to the need for already scarce freshwater, threats to food security and reduction in crop nutrient content are major concerns for biofuel production (Joshi et al., 2017). Therefore, for large-scale production of biofuels, a systematic assessment of their benefits is essential. For this purpose, LCA is used, which provides an overview of the impacts of a production system by collecting data on inputs and outputs and assessing their impact on the environment (Bilal and Iqbal, 2020) (Figure 1).
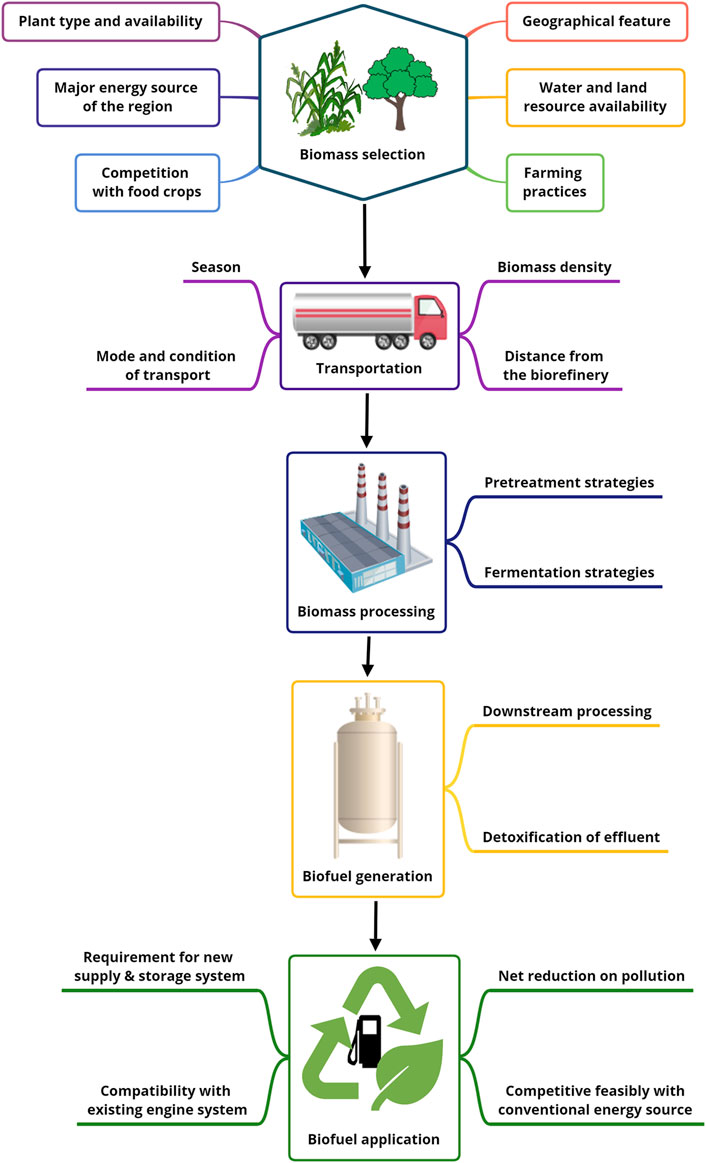
FIGURE 1. Schematic representation of the various points of Life cycle analysis for biofuel production from biomass.
In LCA, the goal is first defined along with the system boundary (stage of agriculture, biofuel production, etc.), then the inputs and outputs are listed, and then the impact of the system on predefined environmental processes such as ozone depletion, global warming, etc. Is quantified, and finally, the result is interpreted (Morales et al., 2015). The LCA provides an understanding of the impacts of biomass conversion techniques to create awareness among researchers, industry, and policymakers to develop strategies for sustainable biofuel production (Standardization, 2006). Both the economic and environmental impacts of biofuels are evaluated at LCA, using net energy production, overall reductions in pollutant production throughout the cycle, social and economic feasibility, and food security impacts as criteria (Weldu and Assefa, 2016; Mayer et al., 2020).
Numerous studies with different lignocellulosic biomasses have been investigated for biofuel production and the associated risk according to different LCA models. The first approach to LCA for lignocellulosic biofuel production is the study of lignocellulosic briquettes and pellets, followed by different pre-treatment and fermentation strategies. A study was conducted with lignocellulosic biomass in different parts of Latin America to investigate its impact on freshwater eutrophication, cumulative energy demand, land use and soil acidification, and water consumption, apart from global warming. Among the different samples, Chilean pellets were the most efficient in terms of global warming with a −68.7 g CO2 equivalent, while Brazilian charcoal briquettes had the least impact with 103 g CO2 equivalent per functional unit. However, when looking at the overall impact, urban forest waste was the most efficient, while charcoal briquettes and pellets from palm branches were the worst (Silva et al., 2022).
The LCA study conducted by (Prasad et al., 2016) of the different pretreatment methods for lignocellulose showed that out of the four methods selected, i.e. organosolv (OS), liquid hot water (LHW), dilute acid (DA) and steam explosion (SE), LHW was the most efficient as twice as much sugar was released and CO2 emissions were significantly reduced although water consumption was higher. DA proved to be the least efficient, considering that a treatment temperature of 60°C and a duration of 12 h are required, which are maintained using electricity, resulting in an increased environmental impact.
In addition, LCA has found that the impact of various factors is case and region-specific and depends on geographical conditions such as water availability, farming practices, major energy sources, etc. (Murali and Shastri, 2022). In the case of sugarcane bagasse, the strategy of dilute acid pre-treatment followed by simultaneous saccharification and fermentation was found to be the most efficient method of ethanol production with higher yield and low biomass requirement and CO2 generation of 0.132 kg eq. per MJ. It was also found that the major step associated with CO2 release is the steam generation, which accounts for about 85%. It is proposed to replace natural gas to reduce the amount of CO2 released to 0.019 kg eq. per MJ. The amount of water required for the entire process was 232–672 L for each litre of ethanol produced, most of which came from agricultural water. The ammonia fibre explosion (AFEX) pretreatment method had the largest impact. The main reason is the large demand for ammonia and the related recycling problem (Murali and Shastri, 2022).
5.2 Economic and technological impacts
In addition to assessing the environmental impacts of biofuels, it is also important to consider the economic and technological limitations of their application. The first major hurdle is the technical and economic aspect of the type of biomass, collection, storage, processing, and related logistics. The mode of transportation, seasonal change-related variation in the biomass with increased volume: density ratio, and the distance of the field from the factory all contribute to the total transportation cost. The need for specific equipment to pre-treat the biomass, the cost of the enzymes required, the generation of unwanted by-products, the down streaming of the biofuel, and the neutralization of toxic substances in the effluent also have a direct impact on biofuel production costs (Balan, 2014; Mungodla et al., 2019).
Once biofuel is produced, the other major hurdle is its transportation, distribution, and compatibility with current engine systems. Biofuels require different systems to handle the fuel than the existing ones, so new pipelines and storage facilities need to be constructed (Kesieme et al., 2019). Although pipelines are the most efficient way of transporting fuel among the different transport models, it is observed that biofuels lead to pipeline corrosion (Gabetta, 2021). Although many countries have started using bioethanol as a fuel because it is compatible with gasoline, has a high-octane rating, and increases vehicle performance, ethanol has a lower energy value compared to gasoline, which increases fuel costs (Masum et al., 2015). The biofuel can also have a negative impact on engine durability, where the biofuel can precipitate, clog filters, or it can leak and dilute lubricating oils, damaging the engine. In addition, the effects of biofuels on other engine system components, such as fuel injectors, are not fully known (Joshi et al., 2017).
Since the currently available engine system has been developed over the years to achieve maximum efficiency concerning conventional fuels, biofuels are not fully compatible with them. The strategy for processing biofuels to increase their compatibility with these engine systems has a drastic impact on overall costs and pollutant emissions. Therefore, further development of engine technology may be an alternative to this. However, there has been increased interest in developing hybrid engines for vehicles that can run on both conventional fuels and biofuels (Bergthorson and Thomson, 2015).
6 Conclusion
Scientific developments in biological research related to microbial culture techniques have provided new insights into the microbiology and composition of the microbiome of many organisms that have come a long way. A major advance has been the study of the microbiome of various organisms, analyzing the diversity, function, importance, and conditions associated with dysbiosis. The gut microbiome of ruminants, termites, and other organisms that feed on plants consists of microbes with an efficient CAZyme system to utilize lignocellulose as a nutrient source. In the highly selective environment of these ecosystems, microorganisms with an efficient CAZyme system have evolved and have been studied with great success for the degradation of lignocellulose. Current trends show an increasing research interest in the gut of various organisms with the isolation and utilization of CAZymes for biofuel production. These enzymes, along with other strategies, have high potential as a future energy source while reducing waste management and pollution.
7 Challenges and future prospect
Although much of the current energy supply still comes from fossil fuels, lignocellulose will become an important substitute in the near future. CAZymes from the gut of herbivorous organisms may provide a revolutionary and environmentally friendly approach to this recalcitrant carbon source. However, a major challenge in isolating these novel microbes and CAZymes is the limitation associated with culture-dependent methods: Not all isolates can be grown on plates. In addition, most gut microbes are strict anaerobes, so more complex equipment is required for their growth and maintenance. In addition, the composition of the host microflora depends on other factors such as age, nutrition, health status, etc. The limited knowledge about the nutritional and environmental factors required for the growth and CAZyme production of gut microflora can be elucidated by culture-independent techniques such as metagenomics, transcriptomics, proteomics, and so on. Technological advances in high-throughput screening, computational studies and mathematical modelling, novel genetic and protein engineering techniques, etc., are now being used to maximize the potential of CAZyme for the gut microflora.
Author contributions
AV conceived of the presented idea. DC, SN, and UK wrote the manuscript. AV provided critical feedback, supervised the study, and helped shape the review.
Acknowledgments
The authors would like to thank the Department of Microbiology, Sikkim University, for providing the computational infrastructure and central library facilities for procuring references and plagiarism analysis (Ouriginal: Plagiarism Detection Software).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdullah, B., Muhammad, S. A. F. a. S., Shokravi, Z., Ismail, S., Kassim, K. A., Mahmood, A. N., et al. (2019). Fourth generation biofuel: A review on risks and mitigation strategies. Renew. Sustain. Energy Rev. 107, 37–50. doi:10.1016/j.rser.2019.02.018
Ali, S. S., Al-Tohamy, R., Koutra, E., Kornaros, M., Khalil, M., Elsamahy, T., et al. (2021). Coupling azo dye degradation and biodiesel production by manganese-dependent peroxidase producing oleaginous yeasts isolated from wood-feeding termite gut symbionts. Biotechnol. Biofuels 14 (1), 61–25. doi:10.1186/s13068-021-01906-0
An, X., Zhang, L., Luo, J., Zhao, S., and Jiao, T. (2020). Effects of oat hay content in diets on nutrient metabolism and the rumen microflora in sheep. Animals 10 (12), 2341. doi:10.3390/ani10122341
Antwis, R. E., Griffiths, S. M., Harrison, X. A., Aranega-Bou, P., Arce, A., Bettridge, A. S., et al. (2017). Fifty important research questions in microbial ecology. FEMS Microbiol. Ecol. 93 (5). doi:10.1093/femsec/fix044
Arevalo-Gallegos, A., Ahmad, Z., Asgher, M., Parra-Saldivar, R., and Iqbal, H. M. (2017). Lignocellulose: A sustainable material to produce value-added products with a zero waste approach—a review. Int. J. Biol. Macromol. 99, 308–318. doi:10.1016/j.ijbiomac.2017.02.097
Ayeni, A. O., Adeeyo, O. A., Oresegun, O. M., and Oladimeji, T. E. (2015). Compositional analysis of lignocellulosic materials: Evaluation of an economically viable method suitable for woody and non-woody biomass. Am. J. Eng. Res. 4 (4), 14–19.
Baghaei, B., and Skrifvars, M. (2020). All-cellulose composites: A review of recent studies on structure, properties and applications. Molecules 25 (12), 2836. doi:10.3390/molecules25122836
Bajpai, P. (2016). “Pretreatment of lignocellulosic biomass,” in Pretreatment of lignocellulosic biomass for biofuel production (Springer), 17–70.
Ben Guerrero, E., Arneodo, J., Bombarda Campanha, R., Abrão de Oliveira, P., Veneziano Labate, M. T., Regiani Cataldi, T., et al. (2015). Prospection and evaluation of (Hemi) cellulolytic enzymes using untreated and pretreated biomasses in two argentinean native termites. Plos one 10 (8), e0136573. doi:10.1371/journal.pone.0136573
Bergthorson, J. M., and Thomson, M. J. (2015). A review of the combustion and emissions properties of advanced transportation biofuels and their impact on existing and future engines. Renew. Sustain. Energy Rev. 42, 1393–1417. doi:10.1016/j.rser.2014.10.034
Bhujbal, S. K., Kumar, M., Vijay, V. K., Kumar, V., and Ghosh, P. (2021). Potential of termite gut microbiota for biomethanation of lignocellulosic wastes: Current status and future perspectives. Rev. Environ. Sci. Biotechnol. 20 (2), 419–438. doi:10.1007/s11157-021-09576-y
Bilal, M., and Iqbal, H. (2020). Recent advancements in the life cycle analysis of lignocellulosic biomass. Curr. Sustain. Renew. Energy Rep. 7 (3), 100–107. doi:10.1007/s40518-020-00153-5
Bohra, V., Tikariha, H., Purohit, H. J., and Dafale, N. A. (2022). Unique pool of carbohydrate-degrading enzymes in novel bacteria assembled from cow and buffalo rumen metagenomes. Appl. Microbiol. Biotechnol. 106 (12), 4643–4654. doi:10.1007/s00253-022-12020-y
Brennan, Y., Callen, W. N., Christoffersen, L., Dupree, P., Goubet, F., Healey, S., et al. (2004). Unusual microbial xylanases from insect guts. Appl. Environ. Microbiol. 70 (6), 3609–3617. doi:10.1128/aem.70.6.3609-3617.2004
Brune, A., and Dietrich, C. (2015). The gut microbiota of termites: Digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 69, 145–166. doi:10.1146/annurev-micro-092412-155715
Busch, A., Kunert, G., Wielsch, N., and Pauchet, Y. (2018). Cellulose degradation in Gastrophysa viridula (Coleoptera: Chrysomelidae): Functional characterization of two CAZymes belonging to glycoside hydrolase family 45 reveals a novel enzymatic activity. Insect Mol. Biol. 27 (5), 633–650. doi:10.1111/imb.12500
Cabral, L., Persinoti, G. F., Paixão, D. A., Martins, M. P., Morais, M. A., Chinaglia, M., et al. (2022). Gut microbiome of the largest living rodent harbors unprecedented enzymatic systems to degrade plant polysaccharides. Nat. Commun. 13 (1), 629–716. doi:10.1038/s41467-022-28310-y
Calusinska, M., Marynowska, M., Bertucci, M., Untereiner, B., Klimek, D., Goux, X., et al. (2020). Integrative omics analysis of the termite gut system adaptation to Miscanthus diet identifies lignocellulose degradation enzymes. Commun. Biol. 3 (1), 275–312. doi:10.1038/s42003-020-1004-3
Ceja-Navarro, J. A., Karaoz, U., Bill, M., Hao, Z., White, R. A., Arellano, A., et al. (2019). Gut anatomical properties and microbial functional assembly promote lignocellulose deconstruction and colony subsistence of a wood-feeding beetle. Nat. Microbiol. 4 (5), 864–875. doi:10.1038/s41564-019-0384-y
Chen, H, H., Wang, C., Huasai, S., and Chen, A. (2021). Effects of dietary forage to concentrate ratio on nutrient digestibility, ruminal fermentation and rumen bacterial composition in Angus cows. Sci. Rep. 11 (1), 17023–17111. doi:10.1038/s41598-021-96580-5
Chen, P, P., Shrotri, A., and Fukuoka, A. (2021). Synthesis of cello-oligosaccharides by depolymerization of cellulose: A review. Appl. Catal. A General 621, 118177. doi:10.1016/j.apcata.2021.118177
Chettri, D., and Verma, A. K. (2022a). Importance of CAZyme producing microbial flora in the herbivore gut. J. Biotechnol. Bioresearch. Amsterdam, NL: Elsevier 3 (5), 000573.
Chettri, D., and Verma, A. K. (2022b). Importance of CAZyme producing microbial flora in the herbivore gut. New York, NY: Crimson.
Chettri, D., Verma, A. K., and Verma, A. K. (2020). Innovations in CAZyme gene diversity and its modification for biorefinery applications. Biotechnol. Rep. 28, e00525. doi:10.1016/j.btre.2020.e00525
Cholewińska, P., Górniak, W., and Wojnarowski, K. (2021). Impact of selected environmental factors on microbiome of the digestive tract of ruminants. BMC Vet. Res. 17 (1), 25–10. doi:10.1186/s12917-021-02742-y
Choudhary, M., Joshi, S., Singh, P., and Srivastava, N. (2020). “Biofuel production from lignocellulosic biomass: Introduction and metabolic engineering for fermentation scale-up,” in Genetic and metabolic engineering for improved biofuel production from lignocellulosic biomass (Elsevier), 1
Clements, K. D., Angert, E. R., Montgomery, W. L., and Choat, J. H. (2014). Intestinal microbiota in fishes: what's known and what's not. Amsterdam, NL: Wiley Online Library.
Colombo, B. M., Scalvenzi, T., Benlamara, S., and Pollet, N. (2015). Microbiota and mucosal immunity in amphibians. Front. Immunol. New Jersey, NJ: Hoboken 6, 111. doi:10.3389/fimmu.2015.00111
Contijoch, E. J., Britton, G. J., Yang, C., Mogno, I., Li, Z., Ng, R., et al. (2019). Gut microbiota density influences host physiology and is shaped by host and microbial factors. Elife 8, e40553. doi:10.7554/eLife.40553
Corcoran, B. A., Henry, J. C., Rice, R. R., Rismani-Yazdi, H. D., and Christy, A. D. (2008). Cellulosic ethanol from sugarcane bagasse using rumen microorganisms. 2008 Providence: Rhode Island
Coyte, K. Z., and Rakoff-Nahoum, S. (2019). Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29 (11), R538–R544. doi:10.1016/j.cub.2019.04.017
De Nardi, R., Marchesini, G., Li, S., Khafipour, E., Plaizier, K. J., Gianesella, M., et al. (2016). Metagenomic analysis of rumen microbial population in dairy heifers fed a high grain diet supplemented with dicarboxylic acids or polyphenols. BMC Vet. Res. 12 (1), 29–9. doi:10.1186/s12917-016-0653-4
Deka, D., Das, S. P., Sahoo, N., Das, D., Jawed, M., Goyal, D., et al. (2013). Enhanced cellulase production from Bacillus subtilis by optimizing physical parameters for bioethanol production. Hindawi, NY: International Scholarly Research Notices.
Dekaboruah, E., Suryavanshi, M. V., Chettri, D., and Verma, A. K. (2020). Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol 202 (8), 2147–2167. doi:10.1007/s00203-020-01931-x
Fang, Z., Li, B., Liu, Y., Zhu, J., Li, G., Hou, G., et al. (2020). Critical role of degree of polymerization of cellulose in super-strong nanocellulose films. Matter 2 (4), 1000–1014. doi:10.1016/j.matt.2020.01.016
Flint, H. J. (2020). “The gut microbiome: Essential symbionts or unwelcome guests?,” in Why gut microbes matter: Understanding our microbiome. Editor H. J. Flint. Manhattan, NY: Springer International Publishing, 9–14. doi:10.1007/978-3-030-43246-1_2
Foo, J. L., Ling, H., Lee, Y. S., and Chang, M. W. (2017). Microbiome engineering: Current applications and its future. Biotechnol. J. 12 (3), 1600099. doi:10.1002/biot.201600099
Gabetta, G. (2021). “A tentative summary of corrosion issues in pipelines transporting hydrocarbons,” in Degradation assessment and failure prevention of pipeline systems. Manhattan, NY: Springer, 77
Godoy-Vitorino, F., Goldfarb, K. C., Karaoz, U., Leal, S., Garcia-Amado, M. A., Hugenholtz, P., et al. (2012). Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J. 6 (3), 531–541. doi:10.1038/ismej.2011.131
Gupta, P. K., Raghunath, S. S., Prasanna, D. V., Venkat, P., Shree, V., Chithananthan, C., et al. (2019). An update on overview of cellulose, its structure and applications. Cellulose 201 (9), 84727. doi:10.5772/intechopen.84727
Hinsu, A. T., Tulsani, N. J., Panchal, K. J., Pandit, R. J., Jyotsana, B., Dafale, N. A., et al. (2021). Characterizing rumen microbiota and CAZyme profile of Indian dromedary camel (Camelus dromedarius) in response to different roughages. Sci. Rep. 11 (1), 9400–9414. doi:10.1038/s41598-021-88943-9
Huang, J., Weng, L., Zhang, X., Long, K., An, X., Bao, J., et al. (2022). Trypoxylus dichotomus gut bacteria provides an effective system for bamboo lignocellulose degradation. Microbiol. Spectr. 10 (5), e0214722. doi:10.1128/spectrum.02147-22
Huang, S., Ji, S., Yan, H., Hao, Y., Zhang, J., Wang, Y., et al. (2020). The day‐to‐day stability of the ruminal and fecal microbiota in lactating dairy cows. Microbiologyopen 9 (5), e990. doi:10.1002/mbo3.990
Jakeer, S., Varma, M., Sharma, J., Mattoo, F., Gupta, D., Singh, J., et al. (2020). Metagenomic analysis of the fecal microbiome of an adult elephant reveals the diversity of CAZymes related to lignocellulosic biomass degradation. Symbiosis 81 (3), 209–222. doi:10.1007/s13199-020-00695-8
Jang, S., and Kikuchi, Y. (2020). Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 41, 33–39. doi:10.1016/j.cois.2020.06.004
Joshi, G., Pandey, J. K., Rana, S., and Rawat, D. S. (2017). Challenges and opportunities for the application of biofuel. Renew. Sustain. Energy Rev. 79, 850–866. doi:10.1016/j.rser.2017.05.185
Keenan, S. W., Engel, A. S., and Elsey, R. M. (2013). The alligator gut microbiome and implications for archosaur symbioses. Sci. Rep. 3 (1), 2877–7. doi:10.1038/srep02877
Kesieme, U., Pazouki, K., Murphy, A., and Chrysanthou, A. (2019). Biofuel as an alternative shipping fuel: Technological, environmental and economic assessment. Sustain. Energy Fuels 3 (4), 899–909. doi:10.1039/c8se00466h
Krishnan, M., Bharathiraja, C., Pandiarajan, J., Prasanna, V. A., Rajendhran, J., and Gunasekaran, P. (2014). Insect gut microbiome–An unexploited reserve for biotechnological application. Asian Pac. J. Trop. Biomed. 4, S16–S21. doi:10.12980/apjtb.4.2014c95
Kumar, A., Kumar, J., and Bhaskar, T. (2020). Utilization of lignin: A sustainable and eco-friendly approach. J. Energy Inst. 93 (1), 235–271. doi:10.1016/j.joei.2019.03.005
Lee, R. A., and Lavoie, J.-M. (2013). From first-to third-generation biofuels: Challenges of producing a commodity from a biomass of increasing complexity. Anim. Front. 3 (2), 6–11. doi:10.2527/af.2013-0010
Li, Y., Lei, L., Zheng, L., Xiao, X., Tang, H., and Luo, C. (2020). Genome sequencing of gut symbiotic Bacillus velezensis LC1 for bioethanol production from bamboo shoots. Biotechnol. Biofuels 13 (1), 34–12. doi:10.1186/s13068-020-1671-9
Liu, H., Hu, L., Han, X., Zhao, N., Xu, T., Ma, L., et al. (2020). Tibetan sheep adapt to plant phenology in alpine meadows by changing rumen microbial community structure and function. Front. Microbiol. 11, 587558. doi:10.3389/fmicb.2020.587558
Mao, S. Y., Huo, W. J., and Zhu, W. Y. (2016). Microbiome–metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18 (2), 525–541. doi:10.1111/1462-2920.12724
Masum, B., Masjuki, H. H., Kalam, M., Palash, S., and Habibullah, M. (2015). Effect of alcohol–gasoline blends optimization on fuel properties, performance and emissions of a SI engine. J. Clean. Prod. 86, 230–237. doi:10.1016/j.jclepro.2014.08.032
Mayer, F. D., Brondani, M., Carrillo, M. C. V., Hoffmann, R., and Lora, E. E. S. (2020). Revisiting energy efficiency, renewability, and sustainability indicators in biofuels life cycle: Analysis and standardization proposal. J. Clean. Prod. 252, 119850. doi:10.1016/j.jclepro.2019.119850
Morales, M., Quintero, J., Conejeros, R., and Aroca, G. (2015). Life cycle assessment of lignocellulosic bioethanol: Environmental impacts and energy balance. Renew. Sustain. Energy Rev. 42, 1349–1361. doi:10.1016/j.rser.2014.10.097
Mungodla, S. G., Linganiso, L. Z., Mlambo, S., and Motaung, T. (2019). Economic and technical feasibility studies: Technologies for second generation biofuels. J. Eng. Des. Technol. 17 (4), 670–704. doi:10.1108/jedt-07-2018-0111
Munoz-Benavent, M., Perez-Cobas, A. E., Garcia-Ferris, C., Moya, A., and Latorre, A. (2021). Insects’ potential: Understanding the functional role of their gut microbiome. J. Pharm. Biomed. Analysis 194, 113787. doi:10.1016/j.jpba.2020.113787
Murali, G., and Shastri, Y. (2022). Life-cycle assessment-based comparison of different lignocellulosic ethanol production routes. Biofuels 13 (2), 237–247. doi:10.1080/17597269.2019.1670465
Nandy, G., Chakraborti, M., Shee, A., Aditya, G., and Acharya, K. (2021). Gut microbiota from lower groups of animals: An upcoming source for cellulolytic enzymes with industrial potentials. Biointerface Res. Appl. Chem. 11, 13614. doi:10.33263/BRIAC115.1361413637
Nicholson, J. K., Holmes, E., and Wilson, I. D. (2005). Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3 (5), 431–438. doi:10.1038/nrmicro1152
Ozbayram, E. G., Kleinsteuber, S., and Nikolausz, M. (2020). Biotechnological utilization of animal gut microbiota for valorization of lignocellulosic biomass. Appl. Microbiol. Biotechnol. 104 (2), 489–508. doi:10.1007/s00253-019-10239-w
Patel, M., Patel, H. M., and Dave, S. (2020). Determination of bioethanol production potential from lignocellulosic biomass using novel Cel-5m isolated from cow rumen metagenome. Int. J. Biol. Macromol. 153, 1099–1106. doi:10.1016/j.ijbiomac.2019.10.240
Peng, X., Wilken, S. E., Lankiewicz, T. S., Gilmore, S. P., Brown, J. L., Henske, J. K., et al. (2021). Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat. Microbiol. 6 (4), 499–511. doi:10.1038/s41564-020-00861-0
Popovic, M., Woodfield, B. F., and Hansen, L. D. (2019). Thermodynamics of hydrolysis of cellulose to glucose from 0 to 100 C: cellulosic biofuel applications and climate change implications. J. Chem. Thermodyn. 128, 244–250. doi:10.1016/j.jct.2018.08.006
Prasad, A., Sotenko, M., Blenkinsopp, T., and Coles, S. R. (2016). Life cycle assessment of lignocellulosic biomass pretreatment methods in biofuel production. Int. J. Life Cycle Assess. 21 (1), 44–50. doi:10.1007/s11367-015-0985-5
Qaseem, M. F., Shaheen, H., and Wu, A.-M. (2021). Cell wall hemicellulose for sustainable industrial utilization. Renew. Sustain. Energy Rev. 144, 110996. doi:10.1016/j.rser.2021.110996
Rabee, A. E., Sayed Alahl, A. A., Lamara, M., and Ishaq, S. L. (2022). Fibrolytic rumen bacteria of camel and sheep and their applications in the bioconversion of barley straw to soluble sugars for biofuel production. Plos one 17 (1), e0262304. doi:10.1371/journal.pone.0262304
Rahardjo, A., Azmi, R., Muharja, M., Aparamarta, H., and Widjaja, A. (2021). in Pretreatment of tropical lignocellulosic biomass for industrial biofuel production: A review. IOP conference series: Materials science and engineering. Amsterdam, NL: Elsevier.
Rai, A. K., Al Makishah, N. H., Wen, Z., Gupta, G., Pandit, S., and Prasad, R. (2022). Recent developments in lignocellulosic biofuels, a renewable source of bioenergy. Fermentation 8 (4), 161. doi:10.3390/fermentation8040161
Ralph, J., Lapierre, C., and Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi:10.1016/j.copbio.2019.02.019
Sabbadin, F., Pesante, G., Elias, L., Besser, K., Li, Y., Steele-King, C., et al. (2018). Uncovering the molecular mechanisms of lignocellulose digestion in shipworms. Biotechnol. Biofuels 11 (1), 59–14. doi:10.1186/s13068-018-1058-3
Saini, J. K., Kaur, A., and Mathur, A. (2022). Strategies to enhance enzymatic hydrolysis of lignocellulosic biomass for biorefinery applications: A review. Bioresource technology, 127517.
Sajid, Z., da Silva, M. A. B., and Danial, S. N. (2021). Historical analysis of the role of governance systems in the sustainable development of biofuels in Brazil and the United States of America (USA). Sustainability 13 (12), 6881. doi:10.3390/su13126881
Saye, L. M., Navaratna, T. A., Chong, J. P., O’Malley, M. A., Theodorou, M. K., and Reilly, M. (2021). The anaerobic fungi: Challenges and opportunities for industrial lignocellulosic biofuel production. Microorganisms 9 (4), 694. doi:10.3390/microorganisms9040694
Seddiqi, H., Oliaei, E., Honarkar, H., Jin, J., Geonzon, L. C., Bacabac, R. G., et al. (2021). Cellulose and its derivatives: Towards biomedical applications. Cellulose 28 (4), 1893–1931. doi:10.1007/s10570-020-03674-w
Serra, C. R., Almeida, E. M., Guerreiro, I., Santos, R., Merrifield, D. L., Tavares, F., et al. (2019). Selection of carbohydrate-active probiotics from the gut of carnivorous fish fed plant-based diets. Sci. Rep. 9 (1), 6384–6415. doi:10.1038/s41598-019-42716-7
Sha, Y., Hu, J., Shi, B., Dingkao, R., Wang, J., Li, S., et al. (2020). Characteristics and functions of the rumen microbial community of Cattle-Yak at different ages. BioMed Res. Int. 2020, 1–9. doi:10.1155/2020/3482692
Sharma, S., Sharma, A., Mulla, S. I., Pant, D., Sharma, T., and Kumar, A. (2020). “Lignin as potent industrial biopolymer: An introduction,” in Lignin (Springer), 1–15.
Shokravi, Z., Shokravi, H., Aziz, M. A., and Shokravi, H. (2019). The fourth-generation biofuel: A systematic review on nearly two decades of research from 2008 to 2019. Boca Raton, Florida: Fossil Free Fuels, 213
Shuvashish, B., Richa, S., Richa, A., Kumar, S. N., Madhulika, S., and Sachin, K. (2015). Scope of algae as third generation biofuels. Front. Bioeng. Biotechnol. 2, 90. doi:10.3389/fbioe.2014.00090
Siddiqui, R., Soopramanien, M., Alharbi, A. M., Alfahemi, H., and Khan, N. A. (2022). Novel sources of bioactive molecules: Gut microbiome of species routinely exposed to microorganisms. Veterinary Sci. 9 (8), 380. doi:10.3390/vetsci9080380
Silva, D., Filleti, R., Musule, R., Matheus, T., and Freire, F. (2022). A systematic review and life cycle assessment of biomass pellets and briquettes production in Latin America. Renew. Sustain. Energy Rev. 157, 112042. doi:10.1016/j.rser.2021.112042
Smith, M. D. (2019). “An abbreviated historical and structural introduction to lignocellulose,” in Understanding lignocellulose: Synergistic computational and analytic methods (Washington, DC: ACS Publications), 1
Souza, R. S., Gama, M. d. V. F., Schama, R., Lima, J. B. P., Diaz-Albiter, H. M., and Genta, F. A. (2019). Biochemical and functional characterization of glycoside hydrolase family 16 genes in Aedes aegypti larvae: Identification of the major digestive β-1, 3-glucanase. Front. Physiol. 10, 122. doi:10.3389/fphys.2019.00122
Standardization, I. O. f. (2006). Environmental management: Life cycle assessment; requirements and guidelines, 14044. Switzerland: ISO Geneva.
Tarafdar, A., Sirohi, R., Gaur, V. K., Kumar, S., Sharma, P., Varjani, S., et al. (2021). Engineering interventions in enzyme production: Lab to industrial scale. Bioresour. Technol. 326, 124771. doi:10.1016/j.biortech.2021.124771
Tartar, A., Wheeler, M. M., Zhou, X., Coy, M. R., Boucias, D. G., and Scharf, M. E. (2009). Parallel metatranscriptome analyses of host and symbiont gene expression in the gut of the termite Reticulitermes flavipes. Biotechnol. Biofuels 2 (1), 25–19. doi:10.1186/1754-6834-2-25
Tayyab, M., Noman, A., Islam, W., Waheed, S., Arafat, Y., Ali, F., et al. (2018). Bioethanol production from lignocellulosic biomass by environment-friendly pretreatment methods: A review. Appl. Ecol. Environ. Res. 16 (1), 225–249. doi:10.15666/aeer/1601_225249
Tingley, J. P., Low, K. E., Xing, X., and Abbott, D. W. (2021). Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues. Biotechnol. Biofuels 14 (1), 16–19. doi:10.1186/s13068-020-01869-8
Tran, T. T. A., Le, T. K. P., Mai, T. P., and Nguyen, D. Q. (2019). Bioethanol production from lignocellulosic biomass. Alcohol fuels-current technologies and future prospect. London, UK: Intechopen.
Tsegaye, B., Balomajumder, C., and Roy, P. (2019). Microbial delignification and hydrolysis of lignocellulosic biomass to enhance biofuel production: An overview and future prospect. Bull. Natl. Res. Cent. 43 (1), 51–16. doi:10.1186/s42269-019-0094-x
Viesturs, D., and Melece, L. (2014). Advantages and disadvantages of biofuels: Observations in Latvia. Jelgava, Latvia: Latvia University of Agriculture.
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10 (1), 4841–4910. doi:10.1038/s41467-019-12798-y
Wang, B., Ma, M. P., Diao, Q. Y., and Tu, Y. (2019). Saponin-induced shifts in the rumen microbiome and metabolome of young cattle. Front. Microbiol. 10, 356. doi:10.3389/fmicb.2019.00356
Wardman, J. F., Bains, R. K., Rahfeld, P., and Withers, S. G. (2022). Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20 (9), 542–556. doi:10.1038/s41579-022-00712-1
Wehrs, M., Tanjore, D., Eng, T., Lievense, J., Pray, T. R., and Mukhopadhyay, A. (2019). Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 27 (6), 524–537. doi:10.1016/j.tim.2019.01.006
Weldu, Y. W., and Assefa, G. (2016). Evaluating the environmental sustainability of biomass-based energy strategy: Using an impact matrix framework. Environ. Impact Assess. Rev. 60, 75–82. doi:10.1016/j.eiar.2016.05.005
Wertz, J. T., and Béchade, B. (2020). “Chapter Three - symbiont-mediated degradation of dietary carbon sources in social herbivorous insects,” in Advances in insect physiology. Editors K. M. Oliver, and J. A. Russell, 58, 63–109. doi:10.1016/bs.aiip.2020.04.001
Xiao, K., Liang, X., Lu, H., Li, X., Zhang, Z., Lu, X., et al. (2022). Adaptation of gut microbiome and host metabolic systems to lignocellulosic degradation in bamboo rats. ISME J. 16, 1980–1992. doi:10.1038/s41396-022-01247-2
Yan, J.-K., Wang, Y.-Y., Qiu, W.-Y., Ma, H., Wang, Z.-B., and Wu, J.-Y. (2018). Three-phase partitioning as an elegant and versatile platform applied to nonchromatographic bioseparation processes. Crit. Rev. food Sci. Nutr. 58 (14), 2416–2431. doi:10.1080/10408398.2017.1327418
Youngblut, N. D., Reischer, G. H., Walters, W., Schuster, N., Walzer, C., Stalder, G., et al. (2019). Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat. Commun. 10 (1), 2200–2215. doi:10.1038/s41467-019-10191-3
Zeng, W., Guo, L., Xu, S., Chen, J., and Zhou, J. (2020). High-throughput screening technology in industrial biotechnology. TRENDS Biotechnol. 38 (8), 888–906. doi:10.1016/j.tibtech.2020.01.001
Zhang, B., Ren, J., Yang, D., Liu, S., and Gong, X. (2019). Comparative analysis and characterization of the gut microbiota of four farmed snakes from southern China. PeerJ 7, e6658. doi:10.7717/peerj.6658
Zhang, D., Allen, A. B., and Lax, A. R. (2012). Functional analyses of the digestive β-glucosidase of Formosan subterranean termites (Coptotermes formosanus). J. insect physiology 58 (1), 205–210. doi:10.1016/j.jinsphys.2011.11.014
Zhou, Z., Liu, D., and Zhao, X. (2021). Conversion of lignocellulose to biofuels and chemicals via sugar platform: An updated review on chemistry and mechanisms of acid hydrolysis of lignocellulose. Renew. Sustain. Energy Rev. 146, 111169. doi:10.1016/j.rser.2021.111169
Zhu, P., Abdelaziz, O. Y., Hulteberg, C. P., and Riisager, A. (2020). New synthetic approaches to biofuels from lignocellulosic biomass. Curr. Opin. Green Sustain. Chem. 21, 16–21. doi:10.1016/j.cogsc.2019.08.005
Keywords: CAZyme, biofuel, biorefineries, ruminants, lignocellulose, rumen
Citation: Chettri D, Nad S, Konar U and Verma AK (2022) CAZyme from gut microbiome for efficient lignocellulose degradation and biofuel production. Front. Chem. Eng. 4:1054242. doi: 10.3389/fceng.2022.1054242
Received: 26 September 2022; Accepted: 24 October 2022;
Published: 07 November 2022.
Edited by:
Muhammad Sajid, Yibin University, ChinaReviewed by:
Sabeela Beevi Ummalyma, Institute of Bioresources and Sustainable Development (IBSD), IndiaClaudia Gutiérrez-Antonio, Universidad Autónoma de Querétaro, Mexico
Copyright © 2022 Chettri, Nad, Konar and Verma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anil Kumar Verma, YWt2ZXJtYUBjdXMuYWMuaW4=
 Dixita Chettri
Dixita Chettri Susmita Nad
Susmita Nad Ujjal Konar
Ujjal Konar Anil Kumar Verma
Anil Kumar Verma