- 1Department of Civil and Environmental Engineering, University of Wisconsin-Madison, Madison, WI, United States
- 2Great Lakes Bioenergy Research Center, Madison, WI, United States
- 3Wisconsin Energy Institute, University of Wisconsin-Madison, Madison, WI, United States
- 4Department of Wood Science, University of British Columbia, Vancouver, BC, Canada
- 5Joint BioEnergy Institute, Emeryville, CA, United States
- 6Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 7Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, United States
- 8Department of Bacteriology, University of Wisconsin-Madison, Madison, WI, United States
The economic and environmental sustainability of lignocellulosic biomass biorefineries is predicated on generating biofuels and bioproducts from cell-wall polysaccharide and lignin polymers. Historical efforts in plant genetic engineering have focused on the development of strategies that facilitate biomass deconstruction, with more recently efforts including the synthesis of high-value chemicals in planta. One such genetic modification is the expression of the bacterial quinate and shikimate utilization B (qsuB) gene that increases the accumulation of protocatechuic acid in lignocellulosic biomass. Herein, we evaluated the effectiveness of an alkaline pretreatment process to extract phenolics directly from wild-type and QsuB-transgenic lines of Arabidopsis, poplar, and sorghum, and then upgrade them to the polyester precursor 2-pyrone-4,6-dicarboxylic acid (PDC) with an engineered strain of Novosphingobium aromaticivorans. Protocatechuic acid extracted from all QsuB transgenic lines was found to be mostly in the glycosylated form. Glycosylated protocatechuic acid and other plant-derived phenolics were effectively metabolized by N. aromaticivorans, and PDC production was greatest using extracts from an Arabidopsis QsuB transgenic line (∼5% w/w), followed by QsuB sorghum (∼1.1% w/w), and QsuB poplar (∼0.4% w/w) lines. The comparison of PDC production from wild-type and QsuB transgenic lines of Arabidopsis, poplar, and sorghum demonstrates the utility of a mild alkaline pretreatment to liberate phenolics from plant biomass that are either naturally present or that accumulate as a consequence of genetic engineering strategies. All QsuB transgenic lines outperformed their wild-type counterparts with respect to observed PDC yields. In addition, microbial funneling to PDC was effective even when most of the protocatechuic acid extracted was in glycosylated form, clearly demonstrating that this bacterium can metabolize these aromatic conjugates. These findings illustrate the benefits of combining plant and microbial engineering for bioproduct formation from phenolics in lignocellulosic biorefineries.
1 Introduction
The world’s dependence on fossil fuels, a non-renewable resource with negative global environmental impacts, has reinforced the importance of sustainability when converting renewable feedstocks into fuels and bioproducts. Lignocellulosic biomass has garnered significant attention as a potential feedstock to provide precursors for generating chemicals currently made from non-renewable resources (Corma et al., 2007; Isikgor and Becer, 2015). Lignocellulosic biomass is primarily composed of plant cell wall polysaccharide (cellulose and hemicelluloses) and phenolic (lignin) polymers (Xu et al., 2013). Multiple strategies for conversion of polysaccharide polymers into biofuels have been developed (Alvira et al., 2010; Kim et al., 2016), and there is increasing recognition that, although challenging due to recalcitrance and complexity of the lignin polymer (Labeeuw et al., 2015), the economic and environmental sustainability of lignocellulosic biomass biorefineries will depend on producing chemicals from the phenolic fraction of the biomass (Linger et al., 2014; Narron et al., 2016; Gall et al., 2017).
A research frontier to increase the efficiency of biofuel and bioproduct production from lignocellulosic biomass is the genetic modification of energy crops to facilitate deconstruction and recovery of the polysaccharide and lignin polymers (Chen and Dixon, 2007; Simmons et al., 2010; Mahon and Mansfield, 2019; de Vries et al., 2021a). Plant genetic engineering has been directed towards the modification of several key cell wall constituents, including pectins (Lionetti et al., 2010), hemicelluloses (Park et al., 2004), cellulose (Coleman et al., 2009; Fan et al., 2020), and lignin (Eudes et al., 2012; Mansfield et al., 2012; Bonawitz and Chapple, 2013; Ralph et al., 2019). More specifically, in an attempt to engineer plants with more easily deconstructed lignin that could produce higher monoaromatic yields, genetic modifications have aimed at reducing the formation of C–C interunit bonds during lignin biosynthesis by elevating syringyl levels (Stewart et al., 2009), incorporating ester-linkages into the lignin backbone (Grabber et al., 2008; Ralph, 2010; Wilkerson et al., 2014), or introducing alternative monomers into lignification (Eudes et al., 2015; Smith et al., 2015; de Vries et al., 2021b; Mahon et al., 2021). Transgenic plants with added chemically-labile ester linkages have shown improved digestibility under milder conditions (Zhou et al., 2017; Bhalla et al., 2018), higher accumulation of soluble aromatics after mild alkaline pretreatment (Sibout et al., 2016) and, in some cases, formation of glycosylated phenolics that are easily extracted with aqueous methanol solvents (Siebert et al., 1996; Eudes et al., 2012; Wu et al., 2017; Unda et al., 2022).
The viability of using transgenic plants in lignocellulosic biorefineries will depend on the ability to use mild and cost-effective biomass deconstruction techniques, and the effective utilization of all major cell wall polymers for the production of biofuels and bioproducts. As lignin composition and content have been the primary targets to genetically improve biomass deconstruction and offer major sources of readily available phenolics in the plant cell wall, we were interested in evaluating and comparing the effectiveness of using mild alkaline pretreatment (Kim et al., 2016) on different biomass sources as a method to extract phenolic compounds for microbial conversion to a valuable bioproduct, while leaving the major polysaccharides and residual lignin available for further valorization. For this study, we selected transgenic plants modified to express a bacterial quinate and shikimate utilization B (qsuB) gene (Teramoto et al., 2009), which encodes a 3-dehydroshikimate dehydratase (QsuB) that catalyzes the conversion of 3-dehydroshikimate into protocatechuate (3,4-dihydroxybenzoate). Expression of this gene in Arabidopsis (Eudes et al., 2015) or tobacco plants (Wu et al., 2017) results in the accumulation of conjugated forms of protocatechuate (Lin and Eudes, 2020), from which free protocatechuic acid (PCA) can be recovered by acid hydrolysis (Wu et al., 2017). QsuB transgenics have also been reported in energy crops such as poplar (Unda et al., 2022), sorghum (Lin and Eudes, 2020; Tian et al., 2022), and switchgrass (Hao et al., 2021). For microbial conversion of aromatics, we employed an engineered strain of the α-proteobacterium Novosphingobium aromaticivorans DSM 12444 (Perez et al., 2019; Perez et al., 2022) which has the capacity to funnel a variety of aromatic monomers into 2-pyrone-4,6-dicarboxylic acid (PDC), a potential polyester and adhesive precursor (Hishida et al., 2009; Shikinaka et al., 2018). We selected PDC as a target product for microbial funneling because it is an intermediate at the intersection of converging pathways for degrading different types of phenolics that can be found in deconstructed lignin (Perez et al., 2019; Perez et al., 2022).
2 Results
To evaluate whether the expression of qsuB in plants influenced the microbial funneling of aromatics to PDC by N. aromaticivorans strain PDC (Perez et al., 2019), we extracted phenolics from wild-type (WT) and transgenic biomass samples of Arabidopsis, poplar, and sorghum using a mild alkaline pretreatment process (see Section 4). This procedure cleaves ester bonds and liberates soluble metabolites to generate an alkaline pretreatment liquor (APL) rich in phenolics that is, after pH adjustment, suitable for use as an organic growth substrate in microbial funneling experiments.
To generate APL from WT and transgenic samples, biomass was first milled to create a fine powder that was then processed under alkaline conditions to release easily recoverable aromatics with minimal impact on the lignin polymer backbone. The concentration of free phenolics in the resulting APL was measured by liquid chromatography-mass spectrometry (LC-MS/MS) (Supplementary Table S1). In addition, to quantify the total concentration of phenolics, including those that may be present as glycosylated conjugates (Lin and Eudes, 2020; Unda et al., 2022), aliquots of APL were subjected to a mild acid hydrolysis and the resulting concentrations of free phenolics were then quantified (Supplementary Table S2).
2.1 Quantification of phenolics in APL from WT and QsuB arabidopsis
The APL produced from WT Arabidopsis (Figure 1A) contained traces of p-hydroxybenzoic acid, p-hydroxybenzaldehyde, and p-coumaric acid. After acid hydrolysis of the APL, vanillic acid was also detected at low concentrations (0.05 mM). These results are consistent with prior observations that Arabidopsis does not inherently have large amounts of phenolics that acylate the cell wall polymers, nor significant amounts of soluble phenolics that can be extracted following alkaline pretreatment (Smith et al., 2015).
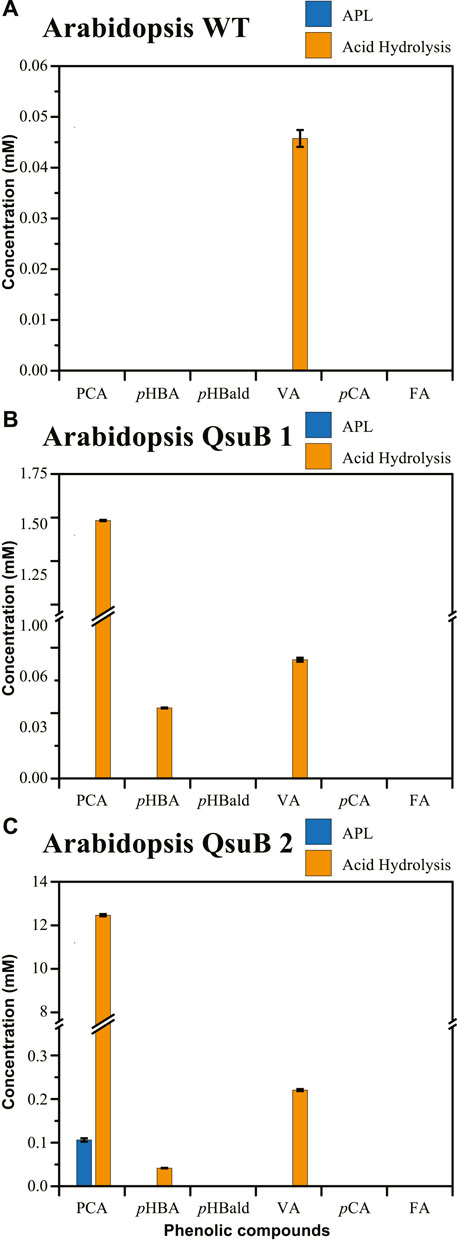
FIGURE 1. Concentration of phenolics present in APL (blue bars) and APL subjected to acid hydrolysis (orange bars) for (A) wild-type Arabidopsis, (B) Arabidopsis QsuB line 1, (C) Arabidopsis QsuB line 2. Abbreviations: protocatechuic acid (PCA), p-hydroxybenzoic acid (pHBA), p-hydroxybenzaldehyde (pHBald), vanillic acid (VA), p-coumaric acid (pCA), ferulic acid (FA). Figures 1B,C have a y-axis break to show a range of concentrations.
Two different Arabidopsis QsuB transgenic lines were examined by the same method. The QsuB line 1 (Figure 1B) has been shown to accumulate conjugated PCA (Eudes et al., 2015). The APL generated from this line showed a similar phenolic profile as the WT Arabidopsis, with traces of various phenolics (Supplementary Table S2). However, elevated concentrations of PCA (1.49 mM) were measured after acid hydrolysis of the APL from QsuB line 1, in agreement with a previous report that this line contains large amounts of monosaccharide-conjugated PCA (Eudes et al., 2015). Also, low concentrations of p-hydroxybenzoic acid (0.03 mM) and vanillic acid (0.06 mM) were detected after acid hydrolysis (Supplementary Table S2). A second QsuB line was designed to express a feedback-insensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (AroG*) in addition to QsuB in order to enhance carbon flux toward in planta PCA production through the shikimate pathway (see Materials and Methods). This line (Figure 1C, QsuB line 2) showed low amounts of free PCA (0.11 mM) and traces of other phenolics in the APL (Supplementary Table S2), but after acid hydrolysis of the APL a significantly higher concentration of PCA (12.46 mM) was detected, indicating an improvement in PCA accumulation in this transgenic line enabled by the co-expression of qsuB with aroG*. In the Arabidopsis QsuB line 2, vanillic acid (0.22 mM) and p-hydroxybenzoic acid (0.04 mM) were also detected after acid hydrolysis of the APL, in agreement with the presence of several monosaccharide-conjugated aromatics (Supplementary Figure S1).
Notably, vanillic acid was only detected after acid treatment of the APL from WT and QsuB transgenic lines (Supplementary Table S2), suggesting that there was a small amount of vanillic acid phenolic glycoside liberated by alkaline pretreatment. Moreover, the amount of vanillic acid released after acid treatment was markedly higher for the QsuB line 2 than for QsuB line 1. This suggests that the gene stacking strategy employed in QsuB line 2, compared to QsuB line 1, improved the metabolic flux towards PCA production, resulting in not only an increased level of conjugated PCA in the plant biomass, but also the accumulation of conjugated vanillic acid.
2.2 Quantification of phenolics in APL from WT and QsuB poplar
We also analyzed the phenolics present in APL samples from WT (Figure 2A) and QsuB poplars (Figures 2B,C). In the APL from WT poplar, we detected p-hydroxybenzoic acid (0.46 mM), low concentrations of ferulic acid (0.05 mM), and traces of p-coumaric acid (Supplementary Table S2). After acid hydrolysis, neither ferulic acid nor p-coumaric acid was detected and the concentration of p-hydroxybenzoic acid did not substantially change, indicating that conjugated forms of these acids are not present in the APL of WT poplar wood (Supplementary Table S2). These results are consistent with the known presence of p-hydroxybenzoate esters bound to lignin in Populus hardwoods (Smith, 1955; Ralph, 2010). We also prepared similar samples from two QsuB transgenic lines, which are QsuB-poplar lines 1 and 15 in Unda et al. (2022). Only traces of PCA were detected in the APL derived from either line. However, following acid hydrolysis, PCA was detected in both QsuB poplar lines (0.83 mM and 0.58 mM, respectively), implying that most of the PCA accumulation in the poplar QsuB lines was in the form of glycosylated or otherwise conjugated PCA that was extractable under alkaline conditions (Unda et al., 2022).
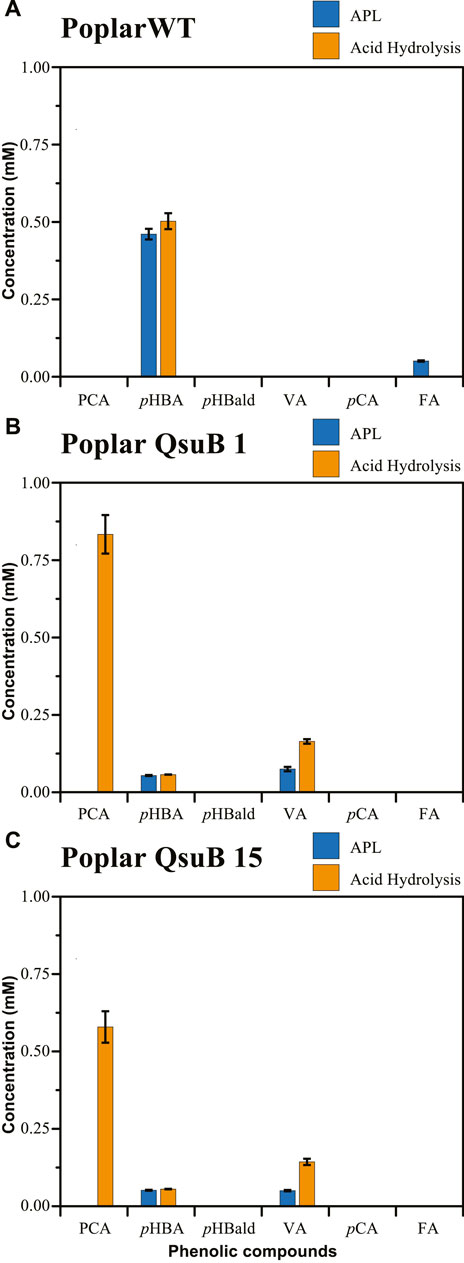
FIGURE 2. Concentration of phenolics present in APL (blue bars) and APL subjected to acid hydrolysis (orange bars) for (A) wild-type poplar, (B) poplar QsuB line 1, (C) poplar QsuB line 15. Abbreviations: protocatechuic acid (PCA), p-hydroxybenzoic acid (pHBA), p-hydroxybenzaldehyde (pHBald), vanillic acid (VA), p-coumaric acid (pCA), ferulic acid (FA).
We also found that the APL from both poplar QsuB lines had 89% less p-hydroxybenzoic acid (0.05 mM) than the corresponding WT poplar (0.46 mM), in agreement with the observations of Unda et al. (2022). The biosynthesis of p-hydroxybenzoic acid in poplar requires precursors that are derived from the shikimate pathway (i.e., phenylalanine, cinnamic acid, and p-coumaric acid) (Terashima et al., 1975). The activity of QsuB is predicted to convert 3-dehydroshikimate into protocatechuate in this pathway, presumably reducing the carbon flow towards the production of p-hydroxybenzoic acid precursors. However, the precise metabolic reason for this tradeoff is unknown.
We also observed differences in the concentrations of vanillic acid between WT and the QsuB 1 and 15 transgenic poplar lines (Figure 2). Free vanillic acid was only detected in the APL of the QsuB 1 and 15 transgenic poplar lines (0.08 mM and 0.05 mM, respectively). Furthermore, vanillic acid concentrations increased after acid hydrolysis of the APL from the QsuB 1 and 15 transgenic poplar lines (Figures 2B,C), suggesting the accumulation of conjugated vanillic acid, presumably in a glycosylated form, as a result of 3-O-methylation of PCA, as proposed by Unda et al. (2022).
2.3 Quantification of phenolics in APL from WT and QsuB sorghum
We also compared the phenolics present in APL of WT and one line of QsuB transgenic sorghum (Tian et al., 2022) (Figure 3). In sorghum, hydroxycinnamates such as p-coumaric and ferulic acids normally acylate lignin as well as arabinosyl subunits of arabinoxylans in the plant cell wall (Harris and Hartley, 1976; Hatfield et al., 1999; Karlen et al., 2016). As predicted, these phenolics were extracted with alkaline pretreatment and found in high concentrations (p-coumaric acid at 1.22 mM in WT and 1.69 mM in the QsuB line; ferulic acid at 0.61 mM in WT and 0.75 mM in the QsuB line) in the APLs of WT and the transgenic sorghum line. p-Hydroxybenzaldehyde was also detected, in low concentrations, in both APLs (Figure 3) and a low concentration of PCA was found (0.10 mM) only in the APL of the QsuB transgenic line (Figure 3B). The soluble PCA concentration increased (0.96 mM) when the APL from the transgenic sorghum was subjected to acid hydrolysis, again suggesting that, as observed with QsuB-expressing Arabidopsis and poplar, most of the PCA that accumulated in the QsuB transgenic sorghum was in the form of conjugated PCA (Figure 3B).
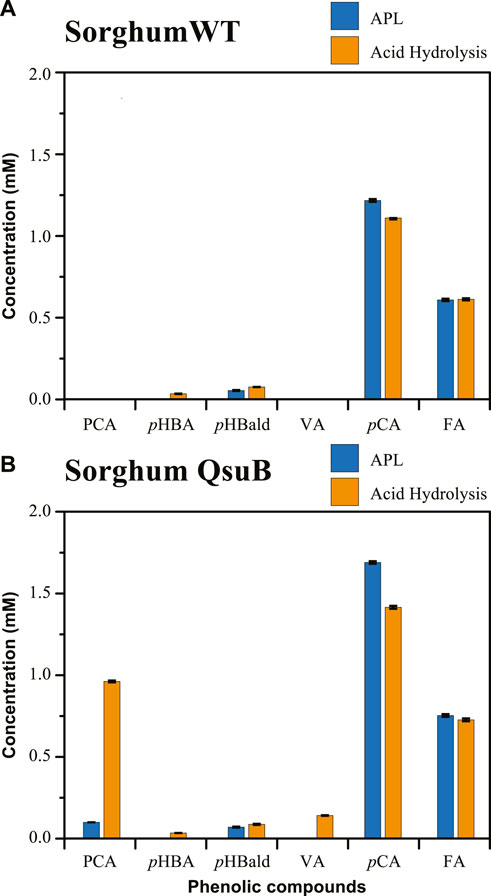
FIGURE 3. Concentration of phenolics present in APL (blue bars) and APL subjected to acid hydrolysis (orange bars) for (A) wild-type sorghum, and (B) QsuB sorghum. Abbreviations: protocatechuic acid (PCA), p-hydroxybenzoic acid (pHBA), p-hydroxybenzaldehyde (pHBald), vanillic acid (VA), p-coumaric acid (pCA), ferulic acid (FA).
In addition, vanillic acid was detected in acid-hydrolyzed APL from the QsuB transgenic line (0.14 mM), supporting the hypothesis that expression of the qsuB gene in sorghum may also result in accumulation of glycosylated vanillic acid, as observed in the Arabidopsis QsuB line 2 and the QsuB transgenic poplar lines, and supported by the identification of glycosylated forms of vanillic acid in poplar (Unda et al., 2022). Traces of p-hydroxybenzoic acid were detected in the APL from WT and QsuB sorghum, and low concentrations of this compound were measured after acid hydrolysis of the APL (Supplementary Table S2).
In summary, the differences in ester groups in the lignin of Arabidopsis, poplar, and sorghum result in characteristically different contents of phenolics in the APLs obtained from these plants. In general, the phenolic concentrations in WT and QsuB lines, with and without an acid hydrolysis step, show that 1) most of the PCA available in QsuB transgenic plants is found as conjugated PCA, which is likely glycosylated PCA as has been hypothesized for Arabidopsis, poplar, and tobacco plants (Lin and Eudes, 2020; Unda et al., 2022); and 2) glycosylated forms of vanillic acid appear to also be extracted from QsuB transgenics by alkaline pretreatment.
2.4 Microbial funneling of APL phenolics to PDC by N. aromaticivorans
Having established that aromatics in the APL are present as free and conjugated phenolics, we evaluated the possibility of microbially converting these aromatics to higher-value products. For this, we tested the production of PDC from APL using the engineered N. aromaticivorans PDC strain that funnels a variety of plant-derived phenolics to PDC (Perez et al., 2019; Linz et al., 2021; Perez et al., 2022).
For these experiments, we employed aliquots of APL from each biomass type, pH-adjusted to 7.0 with hydrochloric acid, and supplemented with glucose and ammonium sulfate to provide another organic carbon and nitrogen source to support growth of the N. aromaticivorans PDC strain (Perez et al., 2019). Each of these cultures was incubated for 48 h, and the concentrations of PDC and phenolics in the culture medium at the end of the incubation were quantified.
Figure 4 shows the extracellular PDC concentration after 48 h of incubation, in comparison to the concentration of phenolics at the start of the incubation period (all values normalized to the grams of plant biomass treated). To account for the presence of free and conjugated phenolics, the total concentration of aromatics was calculated by adding the highest concentration of each phenolic in samples before and after acid treatment (Figures 1–3). In addition, the initial phenolic concentrations were adjusted to consider the dilution of the APL material brought about by mixing the APL with the pre-grown microbial culture. This analysis showed that all N. aromaticivorans cultures produced PDC when supplemented with the different APL samples that contained aromatics. It also indicated that the final extracellular concentration of PDC was dependent on the concentration of total phenolics present in the APL (Table 1).
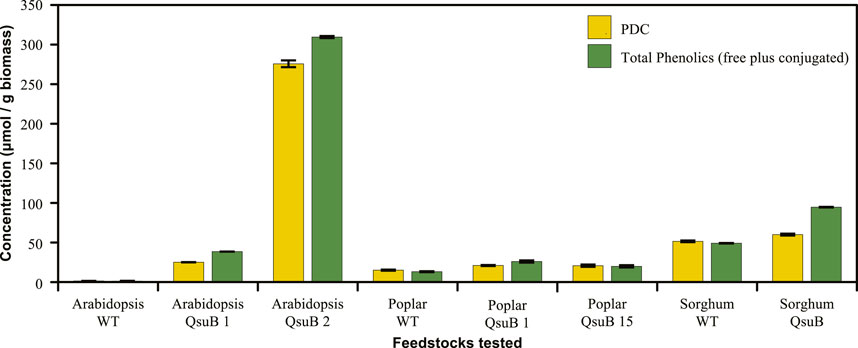
FIGURE 4. Comparison between the total normalized concentration of phenolics (free plus conjugated) and the concentration of 2-pyrone-4,6-dicarboxylic acid (PDC) produced after 48 h incubation with the N. aromaticivorans PDC strain.
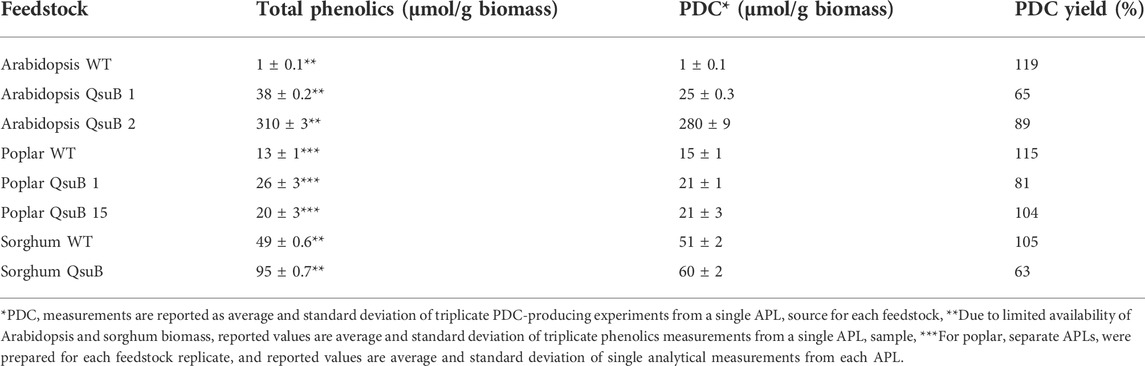
TABLE 1. Total phenolics at beginning of microbial incubations, normalized by dry weight of biomass treated, and PDC produced, both normalized by biomass treated and as yield from total phenolics.
Adding APL from WT Arabidopsis to the N. aromaticivorans PDC strain resulted in negligible PDC accumulation (Table 1), which was likely due to the low initial concentration of phenolics in this sample. When using APL from the QsuB transgenic Arabidopsis lines, the amount of PDC produced was commensurate with the initial concentration of aromatics, which is related to the presence of PCA conjugates in the APL from these plants (Figure 1). Furthermore, there was a difference in observed PDC yields for the two Arabidopsis transgenic lines (Table 1), with N. aromaticivorans PDC strain converting 65% of the total phenolics in the APL to PDC from Arabidopsis QsuB line 1, and 89% from Arabidopsis QsuB line 2. This difference is related to the presence of AroG* in QsuB line 2, although the precise reason for this difference in PDC yield remains unknown.
In N. aromaticivorans cultures provided with APL from WT poplar, the PDC accumulation from aromatics in this sample was close to stoichiometric (Table 1). From N. aromaticivorans cultures supplemented with APL from poplar QsuB transgenics, nearly stoichiometric amounts of PDC were produced from the QsuB line 15, whereas an ∼81% yield of PDC was observed in the analogous sample prepared from QsuB line 1. When sorghum APL was added to N. aromaticivorans cultures, again near-stoichiometric PDC production was observed from the aromatics derived from WT plants, and a 63% yield for the QsuB line tested (Table 1).
When an analysis of free and conjugated phenolics in the media at the end of the microbial growth experiments was performed, we only detected traces of PCA in the cultures fed APL from the Arabidopsis QsuB line 2. This suggests that the N. aromaticivorans PDC strain metabolized the aromatics in the APL during the incubation period and suggests that the bacteria were able to cleave conjugated phenolics, metabolize the resulting free phenolics, and convert most of them into PDC.
When comparing the APLs of WT with the APLs of the transgenic plants, the overall PDC production was higher in the QsuB lines for all three treated plant materials (Figure 4). In Arabidopsis, PDC production was 25 μmol/g biomass when using APL from Arabidopsis QsuB line 1 and ∼10-fold higher (280 μmol/g biomass) when the N. aromaticivorans PDC strain was provided with APL from the Arabidopsis QsuB line 2. When provided with APL from poplar, the PDC production by the N. aromaticivorans PDC strain increased 40% in both poplar QsuB lines compared to a sample from WT poplar. When using APL from sorghum, PDC production by the N. aromaticivorans PDC strain increased by 18% compared to that found when using material from WT plants.
3 Discussion
In the context of a lignocellulosic biomass industry that produces biofuels and bioproducts, genetic modifications of lignin in energy crops can serve two purposes. First, reducing lignin content and making the lignin polymer more easily deconstructed facilitates the recovery of carbohydrates that can be used for biofuel production (Grabber et al., 2008; Wilkerson et al., 2014; Eudes et al., 2015). Second, modifying the lignin to add pendent groups that are easily cleaved can increase the type of valuable chemicals that could be recovered from plant biomass (Siebert et al., 1996; Wu et al., 2017; Mahon et al., 2021), provided that separating these products from other extracted materials is economically feasible.
The QsuB transgenic lines tested here were constructed with the purpose of imbuing plants with the ability to accumulate PCA, which is not a typical pendent group in lignin of wild-type plants (Eudes et al., 2015; Tian et al., 2022; Unda et al., 2022). These transgenic plants show growth phenotypes compared to controls (Eudes et al., 2015; Tian et al., 2022; Unda et al., 2022), and therefore, for future larger scale studies, it will be important to assess the agronomic performance of the engineered crops under natural environments.
As any biomass pretreatment employed to recover PCA from the plant biomass would likely also remove naturally occurring pendent groups such as p-hydroxybenzoic acid from poplar, or p-coumaric and ferulic acids from sorghum, we investigated the concept of microbial funneling to convert the resulting mixture of aromatics to a single product, in this case, the dicarboxylic acid PDC. This approach both avoids the need for separating PCA from other aromatics and takes advantage of the naturally occurring pendent groups that are also easily released by plant biomass. Thus, the observed PDC yields should be influenced by the effectiveness of accumulating PCA in the transgenic lines and by the presence of pendent groups in the plant’s lignin.
The measurement of phenolics in the APL (Supplementary Table S2) and the corresponding product yields when the engineered N. aromaticivorans PDC strain was used to produce PDC from the phenolics in the APL (Figure 4) provided several important observations. First, the majority of the PCA in the APL from all QsuB transgenic lines was in the form of aromatic conjugates. This is deduced from the measurement of higher PCA concentrations when the APL from transgenics was subjected to acid hydrolysis (Supplementary Table S2). This observation suggests that in the QsuB transgenic lines of three different plants, most of the PCA was not incorporated into the lignin as a pendent group; instead, it accumulated as glycosylated PCA, presumably in vacuoles where these conjugates are reported to be stored (Terashima et al., 1975; Hatfield et al., 1999; de Vries et al., 2021a). Second, because glycosylated PCA was effectively converted to PDC by N. aromaticivorans (Figure 4), it can be concluded that in the context of microbial funneling by this bacterium, glycosylated conjugates are an appropriate reservoir for accumulation of phenolics likely because it contains enzymes that cleave the conjugates to release free PCA. The identity of these catabolic enzymes is currently unknown, and it is also unknown whether other microorganisms that are being investigated for microbial funneling (Beckham et al., 2016) express a similar enzymatic function. Additional biochemical and genomic studies are needed to identify these proposed enzymes, analyze their substrate specificities, and determine whether they are widespread among different microorganisms that are being evaluated for producing chemicals from biomass aromatics. Third, unrealized in this research, but potentially important in the context of microbial funneling of phenolics, is the fact that the cleavage of glycosyl-conjugated phenolics releases a sugar molecule that can be used as a growth substrate for the microbial funneling microorganisms. The engineering of PDC-producing N. aromaticivorans strains relies on blocking the natural pathways for the degradation of phenolics so that all aromatic compounds are channeled to PDC (Perez et al., 2019; Perez et al., 2021). This implies that aromatic compounds are not growth substrates for the engineered strain, so other organic carbon sources need to be supplemented to the growth media. In this study, cultures were provided glucose as the growth substrate. If a large fraction of phenolics accumulates in the plant in the form of glycosylated conjugates, the need for the addition of glucose or another carbon source may be reduced or obviated completely. In addition, although not investigated here, other organic substances extracted into the APL, such as acetate, could potentially serve as additional growth substrates for N. aromaticivorans.
To compare PDC yields from the different biomass sources tested, we calculated the bioproduct mass yield based on the amount of plant material employed for the APL production, as this metric accounts for potential inefficiencies associated with the extraction of phenolics from the biomass. Thus, we determined the mass of PDC produced per gram of biomass (Figure 5). This analysis shows that the PDC yields increased when using APL from the QsuB transgenic lines on a weight-by-weight basis compared to using APL from WT plants. Furthermore, the highest PDC production yield by N. aromaticivorans was obtained from growth in the presence of APL samples from the Arabidopsis QsuB line 2, in which ∼5% of the biomass was converted to PDC. This higher yield can be attributed to the substantial accumulation of PCA as conjugates in Arabidopsis QsuB line 2 by the co-expression of aroG* and qsuB, and the nearly stoichiometric yield of PDC from the phenolics in APL (Table 1).
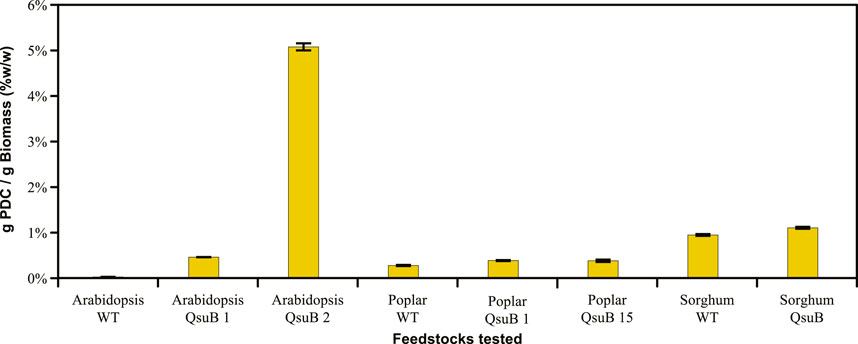
FIGURE 5. Comparison of 2-pyrone-4,6-dicarboxylic acid (PDC) yields among wild-type and QsuB lines of Arabidopsis, poplar, and sorghum, normalized per dry weight of biomass treated.
Sorghum APL had the next highest PDC yields (after Arabidopsis QsuB line 2) per unit of biomass, with 0.9% from the WT and 1.1% from the QsuB transgenic (Figure 5). The small increase in PDC yield when using APL from the QsuB sorghum line indicates that most of the PDC produced by N. aromaticivorans likely comes from the high concentrations of p-coumaric and ferulic acids that are normally found in the APL derived from this plant (Figure 3), as the PDC production in WT sorghum could be explained by the phenolics measured in the APL (Figure 4). Poplar had the lowest PDC yields, on a weight-per-weight basis (Figure 5), of the plants tested when APL was provided to the N. aromaticivorans PDC strain. In poplar samples, 0.27% of the biomass was converted to PDC when APL from WT plants was added to the N. aromaticivorans PDC strain, compared to 0.39% when cells were provided with APL from the QsuB transgenic lines. It is known that p-hydroxybenzoic acid is the main phenolic in the APL from WT poplar, whereas PCA was the main phenolic in the APL from the QsuB transgenic lines. Most of the PDC production when cells are provided with APL from the QsuB transgenic poplar is therefore attributed to PCA, with a potentially higher PDC production offset by the reduced levels of p-hydroxybenzoic acid in the APL from this transgenic poplar line.
It is important to note that the method used to generate the APLs from all the plants tested was not intended to break down the lignin polymer, so the remaining aromatics present in the polymer backbone would remain available to generate additional streams of phenolic compounds for conversion to valuable products after lignin depolymerization. It is also possible that the alkaline pretreatment method could be optimized to extract more phenolics from the biomass. For instance, in the case of poplar, Unda et al. reported a total PCA accumulation in the QsuB line 15 of 5.07 mg per gram of biomass (Unda et al., 2022). If all the PCA is extracted, and assuming a stoichiometric yield of PDC from PCA, the potential PDC production by N. aromaticivorans from the PCA in this transgenic line would be 6.1 mg PDC/g biomass (0.61% w/w), which is ∼1.6 times greater than the PDC yield of 0.39% observed in this work.
In this regard, a recent study of PDC production from lignin that was depolymerized by hydrogenolysis reported PDC yields from WT poplar and sorghum of 0.8% w/w and 0.6% w/w, respectively (Perez et al., 2022). Moreover, in planta production of PDC in engineered Arabidopsis showed 3% w/w accumulation in the biomass (Lin et al., 2021), which is lower than the ∼5% yield that was obtained from the Arabidopsis QsuB line 2 (Figure 5). Thus, to increase the yields of PDC that could be obtained from biomass, it may be possible to develop other transgenics with stacked traits that take advantage of in planta PDC production plus increased phenolics accumulation, such as in the QsuB transgenics.
In conclusion, this study of WT and QsuB transgenic lines of Arabidopsis, poplar, and sorghum demonstrates the utility of a mild alkaline pretreatment to extract plant biomass phenolics that can be metabolized and funneled to PDC by an engineered N. aromaticivorans strain. All QsuB transgenic lines outperformed their WT counterparts with respect to observed PDC yields. In addition, microbial funneling was effective even when most of the PCA extracted into the APL was in glycosylated form, suggesting that this bacterium can metabolize these aromatic conjugates. The results of these studies also suggest other strategies to engineer plants and microbes for lignin valorization in lignocellulosic biorefineries.
4 Materials and methods
4.1 Biomass characterization
The Arabidopsis line C4H:qsuB-1 (Eudes et al., 2015) was used for this study (QsuB line 1). The Arabidopsis QsuB line 2 was obtained by introducing a DNA construct containing both the pAtC4H::qsuB cassette used for qsuB expression (as in QsuB line 1) and a sequence encoding a plastid-targeted feedback-insensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli [AroG*, with the asterisk denoting that it is the feedback-insensitive version of AroG, as described in Lin et al. (2021)] preceded by the promoter of the CesA8 (cellulose synthase 8) gene from poplar (PtCesA8, Wilkerson et al., 2014) for aroG* expression (pPtCesA8::aroG cassette) [Lin et al. (2021) unpublished]. Arabidopsis lines were cultivated in growth chambers until the fully mature senesced stage and stems were harvested for subsequent work as previously described (Eudes et al., 2015).
The QsuB sorghum line pSbUbi::qsuB #22 described in Tian et al. (2022) was used for this study. It contains a codon-optimized sequence encoding a plastid-targeted version of QsuB under the control of the promoter of a sorghum polyubiquitin gene for constitutive qsuB expression. Plants were grown at the UC Berkeley South greenhouse Oxford facility between June and November 2019 with a minimum temperature set at 22°C. Transgenic seeds in the second generation (T2) and wild-type segregant seeds were germinated directly on soil (Sunshine mix #4, Sun Gro, Agawam, MA) in one-gallon pots and plants were grown until seeds reached the black layer stage (i.e., full physiological maturity). One tablespoon of Osmocote Plus 15-9-12 was added to the soil biweekly until the flowering stage. Watering was terminated at the end of the growing period and pots containing plants were allowed to dry for another three weeks. Stems were harvested and further dried in an oven at 50°C for five days before ball-milling.
The pTKan-pC4H::schl::qsuB expression construct described previously by Eudes et al. (2015) was transferred into Agrobacterium tumefaciens EHA105 for transformation of Populus alba × grandidentata (P39) hybrid. Transgenic trees were grown in two-gallon pots containing perennial soil (50% peat, 25% fine bark and 25% pumice; pH 6.0), in the University of British Columbia (Vancouver, B.C.) campus greenhouse where they were maintained on flood-tables with supplemental lighting (16-h days) and watered daily with fertilized water. Trees were harvested after 5 months of growth; stems were debarked and air-dried for several weeks.
4.2 Alkaline pretreatment process
The biomass was milled (Retsch MM400) in a 50 ml stainless steel jar with one 15 mm stainless steel ball-bearing, shaking at 30 Hz for 1 min–5 min to achieve a fine powder. Samples of milled biomass (∼2 g) were mixed with sodium hydroxide (1% NaOH, 20 ml) in sealed 125-ml Erlenmeyer flasks, then treated for 90 min at 90°C in an oil bath. Immediately after, the container was placed on ice for 10 min. Subsequently, the biomass and aqueous phase were separated by centrifugation (4,300 rcf for 15 min) and the supernatant was recovered. The biomass solids were rinsed three times by adding distilled deionized water (20 ml, 15 ml, and 15 ml), centrifuging, and then the supernatant was recovered. The supernatants recovered in the centrifugation and rinsing steps were combined and adjusted to pH 7.0 using 1 M hydrochloric acid. The solution was then subjected to a high-efficiency centrifugation (48,400 rcf, for 1 h at 4°C) to remove any additional insoluble material. The supernatant solution constituted the alkaline pretreatment liquor (APL) used in further experiments.
4.3 Acid hydrolysis of APL
APL (200 µl) was mixed with 10 µl of 0.2 M NaOH and 10 µl of 1.26 mg/ml o-anisic acid. The mixture was evaporated to ∼20 µl, then 300 µl of 1 M HCl was added. The solution was heated to 95°C for 3 h. Subsequently, 800 µl of ethyl acetate was added and thoroughly mixed at room temperature. The supernatant was recovered, separated, and evaporated. The pellet was dissolved in 200 µl of 80% aqueous methanol.
4.4 PDC production by N. aromaticivorans
A strain of Novosphingobium aromaticivorans DSM12444 engineered to accumulate PDC (N. aromaticivorans PDC strain) when provided with aromatic phenolics was employed (Perez et al., 2019). The strain has deletions in ligI (Saro_2819 or SARO_RS14300 in the most recent annotation), desC (Saro_2864 or SARO_RS14525), and desD (Saro_2865 or SARO_RS14530) that interrupt the central metabolic pathway for degradation of phenolics, allowing it to extracellularly accumulate the intermediate metabolite PDC. For PDC production using N. aromaticivorans strain PDC, the APL was supplemented with glucose and ammonium sulfate, to reach final concentrations of 1 g/L each, and filtered using 0.2 µm sterile PES membrane filters. Then, 10-ml aliquots of this solution were inoculated with 2-ml aliquots of an N. aromaticivorans strain PDC culture that had been grown overnight in standard mineral base media (Perez et al., 2022) supplemented with 3 g/L glucose and incubated in an environmental growth chamber at 30°C using a shaker at 200 rpm. The culture was incubated for 48 h; bacterial growth was monitored, and 1-ml samples were collected periodically and centrifuged at 3,300 rcf for 6 min, the supernatant was recovered and filtered, and samples were stored at −18°C until further analysis.
4.5 Analysis of extracellular metabolites
Quantitative analysis of phenolics was accomplished by using a Shimadzu triple-quadrupole liquid chromatography-mass spectrometer (LC-MS, Nexera XR HPLC-8045 MS/MS). The mobile phase was a binary gradient that consisted of a mixture of water containing 0.1% formic acid (solvent A) and methanol (solvent B). The stationary phase was a Kinetex F5 column (Phenomenex, 2.6 μm pore size, 2.1 mm ID, 150 mm length, P/N: 00F-4723-AN). All compounds were detected by multiple-reaction-monitoring (MRM) and quantified using the strongest MRM transition (Supplementary Table S1).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
GU contributed to conceptualization, performed experiments, analyzed data, and wrote the manuscript; JP contributed to conceptualization, performed experiments, analyzed data, and edited the manuscript; FU contributed to conceptualization, provided poplar biomass, analyzed data, and edited the manuscript; C-YL provided Arabidopsis and sorghum biomass, analyzed data, and edited the manuscript; CS contributed to conceptualization, analyzed data, and edited the manuscript; SK contributed to conceptualization, performed experiments, analyzed data, and edited the manuscript; SM contributed to conceptualization, provided poplar biomass, analyzed data, and edited the manuscript; AE contributed to conceptualization, provided Arabidopsis and sorghum biomass, analyzed data, and edited the manuscript; JR contributed to conceptualization, analyzed data, and edited the manuscript; TD contributed to conceptualization, directed study, analyzed data, and had major contribution writing the manuscript; DN contributed to conceptualization, directed study, analyzed data, and had major contribution writing the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Great Lakes Bioenergy Research Center, United States. Department of Energy, Office of Science, Office of Biological and Environmental Research (grant number DE-SC0018409), and by the Joint BioEnergy Institute, United States. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract between Lawrence Berkeley National Laboratory and the United States. Department of Energy (grant number DE-AC02-05CH11231).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2022.1036084/full#supplementary-material.
References
Alvira, P., Tomas-Pejo, E., Ballesteros, M., and Negro, M. J. (2010). Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: A review. Bioresour. Technol. 101 (13), 4851–4861. doi:10.1016/j.biortech.2009.11.093
Beckham, G. T., Johnson, C. W., Karp, E. M., Salvachua, D., and Vardon, D. R. (2016). Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 42, 40–53. doi:10.1016/j.copbio.2016.02.030
Bhalla, A., Bansal, N., Pattathil, S., Li, M. Y., Shen, W., Particka, C. A., et al. (2018). Engineered lignin in poplar biomass facilitates Cu-catalyzed alkaline-oxidative pretreatment. ACS Sustain. Chem. Eng. 6 (3), 2932–2941. doi:10.1021/acssuschemeng.7b02067
Bonawitz, N. D., and Chapple, C. (2013). Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 24 (2), 336–343. doi:10.1016/j.copbio.2012.11.004
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25 (7), 759–761. doi:10.1038/nbt1316
Coleman, H. D., Yan, J., and Mansfield, S. D. (2009). Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. U. S. A. 106 (31), 13118–13123. doi:10.1073/pnas.0900188106
Corma, A., Iborra, S., and Velty, A. (2007). Chemical routes for the transformation of biomass into chemicals. Chem. Rev. 107 (6), 2411–2502. doi:10.1021/cr050989d
de Vries, L., Guevara-Rozo, S., Cho, M., Liu, L. Y., Renneckar, S., and Mansfield, S. D. (2021a). Tailoring renewable materials via plant biotechnology. Biotechnol. Biofuels 14 (1), 167. doi:10.1186/s13068-021-02010-z
de Vries, L., MacKay, H. A., Smith, R. A., Mottiar, Y., Karlen, S. D., Unda, F., et al. (2021b). pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar. Plant Physiol. 188 (2), 1014–1027. doi:10.1093/plphys/kiab546
Eudes, A., George, A., Mukerjee, P., Kim, J. S., Pollet, B., Benke, P. I., et al. (2012). Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol. J. 10 (5), 609–620. doi:10.1111/j.1467-7652.2012.00692.x
Eudes, A., Sathitsuksanoh, N., Baidoo, E. E. K., George, A., Liang, Y., Yang, F., et al. (2015). Expression of a bacterial 3-dehydroshikimate dehydratase reduces lignin content and improves biomass saccharification efficiency. Plant Biotechnol. J. 13 (9), 1241–1250. doi:10.1111/pbi.12310
Fan, C. F., Wang, G. Y., Wu, L. M., Liu, P., Huang, J. F., Jin, X. H., et al. (2020). Distinct cellulose and callose accumulation for enhanced bioethanol production and biotic stress resistance in OsSUS3 transgenic rice. Carbohydr. Polym. 232, 115448. doi:10.1016/j.carbpol.2019.115448
Gall, D. L., Ralph, J., Donohue, T. J., and Noguera, D. R. (2017). Biochemical transformation of lignin for deriving valued commodities from lignocellulose. Curr. Opin. Biotechnol. 45, 120–126. doi:10.1016/j.copbio.2017.02.015
Grabber, J. H., Hatfield, R. D., Lu, F. C., and Ralph, J. (2008). Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9 (9), 2510–2516. doi:10.1021/bm800528f
Hao, Z. Y., Yogiswara, S., Wei, T., Benites, V. T., Sinha, A., Wang, G., et al. (2021). Expression of a bacterial 3-dehydroshikimate dehydratase (QsuB) reduces lignin and improves biomass saccharification efficiency in switchgrass (Panicum virgatum L.). BMC Plant Biol. 21 (1), 56. doi:10.1186/s12870-021-02842-9
Harris, P. J., and Hartley, R. D. (1976). Detection of bound ferulic acid in cell walls of the Gramineae by ultraviolet fluorescence microscopy. Nature 259 (5543), 508–510. doi:10.1038/259508a0
Hatfield, R. D., Wilson, J. R., and Mertens, D. R. (1999).Composition of cell walls isolated from cell types of grain sorghum stems, J. Sci. Food Agric., 79, 8912–9899. doi:10.1002/(SICI)1097-0010(19990501)79:6<891:AID-JSFA304>3.0
Hishida, M., Shikinaka, K., Katayama, Y., Kajita, S., Masai, E., Nakamura, M., et al. (2009). Polyesters of 2-pyrone-4, 6-dicarboxylic acid (PDC) as bio-based plastics exhibiting strong adhering properties. Polym. J. 41 (4), 297–302. doi:10.1295/polymj.PJ2008291
Isikgor, F. H., and Becer, C. R. (2015). Lignocellulosic biomass: A sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 6 (25), 4497–4559. doi:10.1039/c5py00263j
Karlen, S. D., Zhang, C. C., Peck, M. L., Smith, R. A., Padmakshan, D., Helmich, K. E., et al. (2016). Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2 (10), e1600393. doi:10.1126/sciadv.1600393
Kim, J. S., Lee, Y. Y., and Kim, T. H. (2016). A review on alkaline pretreatment technology for bioconversion of lignocellulosic biomass. Bioresour. Technol. 199, 42–48. doi:10.1016/j.biortech.2015.08.085
Labeeuw, L., Martone, P. T., Boucher, Y., and Case, R. J. (2015). Ancient origin of the biosynthesis of lignin precursors. Biol. Direct 10 (1), 23. doi:10.1186/s13062-015-0052-y
Lin, C. Y., and Eudes, A. (2020). Strategies for the production of biochemicals in bioenergy crops. Biotechnol. Biofuels 13 (1), 71. doi:10.1186/s13068-020-01707-x
Lin, C. Y., Vuu, K. M., Amer, B., Shih, P. M., Baidoo, E. E. K., Scheller, H. V., et al. (2021). In-planta production of the biodegradable polyester precursor 2-pyrone-4, 6-dicarboxylic acid (PDC): Stacking reduced biomass recalcitrance with value-added co-product. Metab. Eng. 66, 148–156. doi:10.1016/j.ymben.2021.04.011
Linger, J. G., Vardon, D. R., Guarnieri, M. T., Karp, E. M., Hunsinger, G. B., Franden, M. A., et al. (2014). Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. U. S. A. 111 (33), 12013–12018. doi:10.1073/pnas.1410657111
Linz, A. M., Ma, Y., Perez, J. M., Myers, K. S., Kontur, W. S., Noguera, D. R., et al. (2021). Aromatic dimer dehydrogenases from Novosphingobium aromaticivorans reduce monoaromatic diketones. Appl. Environ. Microbiol. 87 (24), 01742211–e201721. doi:10.1128/AEM.01742-21
Lionetti, V., Francocci, F., Ferrari, S., Volpi, C., Bellincampi, D., Galletti, R., et al. (2010). Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. U. S. A. 107 (2), 616–621. doi:10.1073/pnas.0907549107
Mahon, E. L., de Vries, L., Jang, S.-K., Middar, S., Kim, H., Unda, F., et al. (2021). Exogenous chalcone synthase expression in developing poplar xylem incorporates naringenin into lignins. Plant Physiol. 188 (2), 984–996. doi:10.1093/plphys/kiab499
Mahon, E. L., and Mansfield, S. D. (2019). Tailor-made trees: Engineering lignin for ease of processing and tomorrow's bioeconomy. Curr. Opin. Biotechnol. 56, 147–155. doi:10.1016/j.copbio.2018.10.014
Mansfield, S. D., Kang, K. Y., and Chapple, C. (2012). Designed for deconstruction - poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 194 (1), 91–101. doi:10.1111/j.1469-8137.2011.04031.x
Narron, R. H., Kim, H., Chang, H. M., Jameel, H., and Park, S. (2016). Biomass pretreatments capable of enabling lignin valorization in a biorefinery process. Curr. Opin. Biotechnol. 38, 39–46. doi:10.1016/j.copbio.2015.12.018
Park, Y. W., Baba, K., Furuta, Y., Iida, I., Sameshima, K., Arai, M., et al. (2004). Enhancement of growth and cellulose accumulation by overexpression of xyloglucanase in poplar. FEBS Lett. 564 (1-2), 183–187. doi:10.1016/s0014-5793(04)00346-1
Perez, J. M., Kontur, W. S., Alherech, M., Coplien, J., Karlen, S. D., Stahl, S. S., et al. (2019). Funneling aromatic products of chemically depolymerized lignin into 2-pyrone-4-6-dicarboxylic acid with Novosphingobium aromaticivorans. Green Chem. 21 (6), 1340–1350. doi:10.1039/c8gc03504k
Perez, J. M., Kontur, W. S., Gehl, C., Gille, D. M., Ma, Y., Niles, A. V., et al. (2021). Redundancy in aromatic O-demethylation and ring opening reactions in Novosphingobium aromaticivorans and their impact in the metabolism of plant derived phenolics. Appl. Environ. Microbiol. 87 (8), 027944-e2820. doi:10.1128/aem.02794-20
Perez, J., Sener, C., Misra, S., Umana, G., Coplien, J., Haak, D., et al. (2022). Integrating lignin depolymerization with microbial funneling processes using agronomically relevant feedstocks. Green Chem. 24, 2795–2811. doi:10.1039/d1gc03592d
Ralph, J. (2010). Hydroxycinnamates in lignification. Phytochem. Rev. 9 (1), 65–83. doi:10.1007/s11101-009-9141-9
Ralph, J., Lapierre, C., and Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi:10.1016/j.copbio.2019.02.019
Shikinaka, K., Otsuka, Y., Nakamura, M., Masai, E., and Katayama, Y. (2018). Utilization of lignocellulosic biomass via novel sustainable process. J. Oleo Sci. 67 (9), 1059–1070. doi:10.5650/jos.ess18075
Sibout, R., Le Bris, P., Legee, F., Cezard, L., Renault, H., and Lapierre, C. (2016). Structural redesigning Arabidopsis lignins into alkali-soluble lignins through the expression of p-Coumaroyl-CoA: Monolignol Transferase PMT. Plant Physiol. 170 (3), 1358–1366. doi:10.1104/pp.15.01877
Siebert, M., Sommer, S., Li, S. M., Wang, Z. X., Severin, K., and Heide, L. (1996). Genetic engineering of plant secondary metabolism. Accumulation of 4-hydroxybenzoate glucosides as a result of the expression of the bacterial ubiC gene in tobacco. Plant Physiol. 112 (2), 811–819. doi:10.1104/pp.112.2.811
Simmons, B. A., Logue, D., and Ralph, J. (2010). Advances in modifying lignin for enhanced biofuel production. Curr. Opin. Plant Biol. 13 (3), 312–319. doi:10.1016/j.pbi.2010.03.001
Smith, D. C. C. (1955). p-Hydroxybenzoate groups in the lignin of aspen (Populus tremula). J. Chem. Soc. 1955, 2347–2351. doi:10.1039/JR9550002347
Smith, R. A., Gonzales-Vigil, E., Karlen, S. D., Park, J. Y., Lu, F. C., Wilkerson, C. G., et al. (2015). Engineering monolignol p-coumarate conjugates into poplar and Arabidopsis lignins. Plant Physiol. 169 (4), 2992–3001. doi:10.1104/pp.15.00815
Stewart, J. J., Akiyama, T., Chapple, C., Ralph, J., and Mansfield, S. D. (2009). The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 150 (2), 621–635. doi:10.1104/pp.109.137059
Teramoto, H., Inui, M., and Yukawa, H. (2009). Regulation of expression of genes involved in quinate and shikimate utilization in Corynebacterium glutamicum. Appl. Environ. Microbiol. 75 (11), 3461–3468. doi:10.1128/aem.00163-09
Terashima, N., Mori, I., and Kanda, T. (1975). Biosynthesis of p-hydroxybenzoic acid in poplar lignin. Phytochemistry 14 (9), 1991–1992. doi:10.1016/0031-9422(75)83111-6
Tian, Y., Yang, M., Lin, C.-Y., Park, J.-H., Wu, C.-Y., Kakumanu, R., et al. (2022). Expression of dehydroshikimate dehydratase in sorghum improves biomass yield, accumulation of protocatechuate, and biorefinery economics. ACS Sustain. Chem. Eng. 10 (38), 12520–12528. doi:10.1021/acssuschemeng.2c01160
Unda, F., Mottiar, Y., Mahon, E. L., Karlen, S. D., Kim, K. H., Loque, D., et al. (2022). A new approach to zip-lignin: 3, 4-dihydroxybenzoate is compatible with lignification. New Phytol. 235 (1), 234–246. doi:10.1111/nph.18136
Wilkerson, C. G., Mansfield, S. D., Lu, F., Withers, S., Park, J. Y., Karlen, S. D., et al. (2014). Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344 (6179), 90–93. doi:10.1126/science.1250161
Wu, W. H., Dutta, T., Varman, A. M., Eudes, A., Manalansan, B., Loque, D., et al. (2017). Lignin valorization: Two hybrid biochemical routes for the conversion of polymeric lignin into value-added chemicals. Sci. Rep. 7, 8420. doi:10.1038/s41598-017-07895-1
Xu, F., Yu, J., Tesso, T., Dowell, F., and Wang, D. (2013). Qualitative and quantitative analysis of lignocellulosic biomass using infrared techniques: A mini-review. Appl. Energy 104, 801–809. doi:10.1016/j.apenergy.2012.12.019
Keywords: QsuB, lignin, lignin valorization, value-added products, 2-pyrone-4,6-dicarboxylic acid (PDC), QsuB transgenics, aromatic compounds
Citation: Umana GE, Perez JM, Unda F, Lin C-Y, Sener C, Karlen SD, Mansfield SD, Eudes A, Ralph J, Donohue TJ and Noguera DR (2022) Biological funneling of phenolics from transgenic plants engineered to express the bacterial 3-dehydroshikimate dehydratase (qsuB) gene. Front. Chem. Eng. 4:1036084. doi: 10.3389/fceng.2022.1036084
Received: 03 September 2022; Accepted: 06 October 2022;
Published: 19 October 2022.
Edited by:
Florent Allais, AgroParisTech Institut des Sciences et Industries du Vivant et de L’environnement, FranceReviewed by:
David B. Hodge, Montana State University, United StatesDinesh Christendat, University of Toronto, Canada
Copyright © 2022 Umana, Perez, Unda, Lin, Sener, Karlen, Mansfield, Eudes, Ralph, Donohue and Noguera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel R. Noguera, bm9ndWVyYUBlbmdyLndpc2MuZWR1
 German E. Umana
German E. Umana Jose M. Perez
Jose M. Perez Faride Unda
Faride Unda Chien-Yuan Lin5,6
Chien-Yuan Lin5,6 Canan Sener
Canan Sener Steven D. Karlen
Steven D. Karlen Shawn D. Mansfield
Shawn D. Mansfield Aymerick Eudes
Aymerick Eudes John Ralph
John Ralph Timothy J. Donohue
Timothy J. Donohue Daniel R. Noguera
Daniel R. Noguera