- 1Fraunhofer Institute for Molecular Biology and Applied Ecology IME, Aachen, Germany
- 2Institute for Molecular Biotechnology, Worringerweg 1, RWTH Aachen University, Aachen, Germany
Immunoglobulin subclass IgG1 is bound and neutralized effectively by Staphylococcus aureus protein A, allowing the bacterium to evade the host’s adaptive immune response. In contrast, the IgG3 subclass is not bound by protein A and can be used to treat S. aureus infections, including drug-resistant strains such as methicillin-resistant Staphylococcus aureus (MRSA). However, the yields of recombinant IgG3 are generally low because this subclass is prone to degradation, and recovery is hindered by the inability to use protein A as an affinity ligand for antibody purification. Here, we investigated plants (Nicotiana spp.) as an alternative to microbes and mammalian cell cultures for the production of an IgG3 antibody specific for the S. aureus alpha toxin. We targeted recombinant IgG3 to different subcellular compartments and tested different chromatography conditions to improve recovery and purification. Finally, we tested the antigen-binding capacity of the purified antibodies. The highest IgG3 levels in planta (>130 mg kg−1 wet biomass) were achieved by targeting the endoplasmic reticulum or apoplast. Although the purity of IgG3 exceeded 95% following protein G chromatography, product recovery requires further improvement. Importantly, the binding affinity of the purified antibodies was in the nanomolar range and thus comparable to previous studies using murine hybridoma cells as the production system.
Introduction
Antibiotic-resistant bacteria are increasingly recognized as a major threat to human health (Conlon et al., 2013). Such bacteria are predicted to become more prevalent due to the continual evolution of resistance and the lack of new antibiotics with novel mechanisms of action in the development pipeline (Guo et al., 2020). Resistance becomes more likely if bacteria are exposed to antibiotics below the minimal inhibitory concentration (MIC) in humans or animals, for example, due to low-level tissue distribution, as observed for vancomycin (Micek, 2007; Wistrand-Yuen et al., 2018). Antibiotics may also persist in the environment at such concentrations (Szymańska et al., 2019; Alduina, 2020), reflecting their indiscriminate use in humans and livestock followed by excretion (King et al., 2019). One of the most prevalent antibiotic-resistant pathogens is methicillin-resistant Staphylococcus aureus (MRSA) (Hibbitts and O’Leary, 2018), a Gram-positive species that causes 10 times as many infections as all multidrug-resistant Gram-negative pathogens combined (Craft et al., 2019) [∼120,000 per year in the US (Kourtis et al., 2019)]. Penicillin-resistant S. aureus emerged in the mid-1940s (DeLeo and Chambers, 2009), followed by MRSA in the 1960s and vancomycin-resistant S. aureus in the early 2000s (De Lencastre et al., 2007). Strict policies for the use of antibiotics initially reduced the frequency of MRSA in hospitals, but these strains started to emerge in the community in the 1990s (De Lencastre et al., 2007).
Although ∼30% of the human population is colonized asymptomatic by S. aureus (Craft et al., 2019), this opportunistic pathogen can cause severe infections once it gains access to the body, for example, through surgical wounds (Lowy, 1998). Once inside the body, a vast arsenal of virulence factors promotes S. aureus pathogenicity, including toxins, immunomodulators and exoenzymes (Oogai et al., 2011). For example, proteases and cytotoxins such as the pore-forming 33-kDa alpha toxin induce the release of nutrients from host tissues, supporting S. aureus growth and dissemination (Dinges et al., 2000). The pathogen can also evade host immune defenses by expressing the cell-surface virulence factor protein A, which binds to conserved immunoglobulin G (IgG) domains, thus preventing phagocytosis by macrophages (Atkins et al., 2008; Falugi et al., 2013). Given the lack of an effective vaccine against multidrug-resistant S. aureus strains and development of antibiotic resistances resulting from antibiotic release in the environment (Gothwal and Shashidhar, 2015; Miller et al., 2019), alternative strategies for the treatment of S. aureus infections are urgently needed.
Antibody-based therapies targeting S. aureus and its toxins have been deployed successfully to fight this pathogen (Shopsin et al., 2016). The alpha toxin is a promising target (Hua et al., 2014) because it is conserved in many S. aureus strains and is toxic to a broad range of mammalian cells (Dinges et al., 2000; Hua et al., 2014). The production of antibodies in plants is a promising strategy to meet the growing demand for diagnostics and therapeutics (Schillberg et al., 2005; Capell et al., 2020) because high biomass yields of ∼500,000 kg ha−1 y−1 (Huebbers and Buyel, 2021) can be combined with high yields of recombinant antibodies per unit biomass [∼2 g kg−1 (Zischewski et al., 2016)] to favor the broad application of antibodies against S. aureus. However, IgG3 subclass antibodies are preferable for this purpose because all other IgG subclasses are recognized by S. aureus protein A (Nezlin and Nezlin, 1998). Importantly, although the inability of protein A to bind IgG3 is an asset for therapeutic activity, it prevents the use of protein A as an affinity ligand for antibody purification (Hober et al., 2007) and detection by surface plasmon resonance (SPR) spectroscopy using off-the-shelf sensor chips. Alternative ligands for the purification and quantitation of IgG3 are therefore required, and protein G from Streptococcus spp. group C/G is a promising candidate (Higgins et al., 1995). Whereas protein A binds the antibody Fc region mainly via hydrophobic interactions, protein G binds the same region mostly via charged and polar interactions (Nezlin and Nezlin, 1998). Therefore, elution from protein G resins typically requires high rather than low salt concentrations to disrupt ionic interactions (Hey and Zhang, 2012).
In addition to the purification/detection challenges described above, IgG3 is more susceptible than IgG1 to proteolysis and aggregation (Saito et al., 2019), which limits the accumulation of recombinant IgG3 in higher eukaryotes (Mandal et al., 2016; Saito et al., 2019; Kallolimath et al., 2020). Therefore, we systematically investigated the suitability of plants (Nicotiana spp.) as expression systems to produce native IgG3 and domain exchange variants directed against S. aureus alpha toxin and assessed the suitability of protein G for the purification and high-throughput quantification of recombinant IgG3 antibodies produced in plants.
Materials and Methods
Cloning of Expression Constructs
Constructs (Supplementary Figures S1A, B) encoding the native human heavy chain of IgG3 (Uniprot ID P01860) or IgG1 [M12 (Gaur, 2004)] combined with a human lambda light chain were codon optimized for Nicotiana benthamiana and synthesized by GeneArt (Thermo Fisher Scientific, Darmstadt, Germany). N-terminal NcoI and C-terminal XhoI sites were used for subcloning in pTRAc expression vectors featuring the tobacco mosaic virus omega prime sequence (Omega), the tobacco etch virus leader sequence (TL), or the 5′ untranslated region (5′ UTR) of the Petroselinum hortense chalcone synthase gene (CHS) (Gengenbach et al., 2020). These constructs also included the N-terminal leader peptide of the antibody mAb24 heavy chain (LPH) targeting the secretory pathway (Fischer et al., 1999) and a C-terminal SEKDEL sequence for retrieval to the endoplasmic reticulum (ER) (Munro and Pelham, 1987). Additional vectors promoting antibody accumulation in the apoplast, vacuole, and plastids (Figure 1A) were generated from these initial constructs (Supplementary Figures S1A, B) by deleting the SEKDEL sequence (apoplast) and replacing the initial signal sequences with a KISIA signal sequence (vacuole) (Ocampo et al., 2016) or a rbcs-CTP signal sequence (plastids) (Maclean et al., 2007) using PCR primers with overhangs (Supplementary Table S1) in combination with restriction enzyme treatment or Gibson assembly (Gibson et al., 2009).
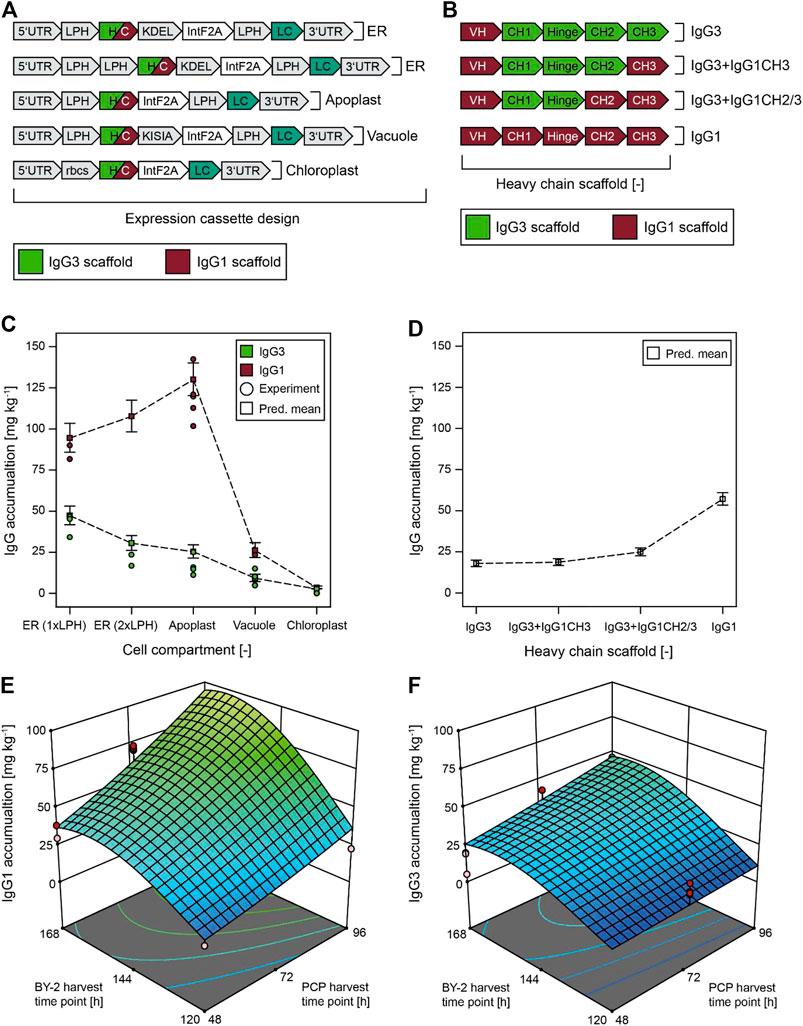
FIGURE 1. Models predicting the accumulation of IgG3 variants and the IgG1 control in BY-2 PCPs depending on the compartment, the heavy chain scaffold, and the BY-2/PCP harvest time point (I-optimal design with 216 runs). (A) Schematic of IgG3 mRNA design for different subcellular compartments in PCPs and differentiated plants. (B) IgG variants investigated for product accumulation in different subcellular compartments. (C) Compartment-specific accumulation of native IgG3 and the IgG1 control in the ER, apoplast, vacuole, and chloroplast. (D) IgG3 variant-specific accumulation as an average over all compartments tested (see Supplementary Figure S3 for individual accumulation levels). BY-2 cultivation and PCP harvest times were 196 and 96 h respectively in both (C, D). Error bars in (C, D) represent least significant difference (LSD) bars around the predicted means (non-overlapping LSD bars indicate that predictions at a given factor combination are significantly different). (E, F) Models predicting IgG1 (E) or IgG3 (F) accumulation in PCPs in the ER, dependent on the BY-2 and PCP harvest time points. Dots in (C, E, F) represent experimental data. CH–constant heavy chain antibody domain, ER–endoplasmic reticulum, HC–antibody heavy chain, LC–antibody light chain, LPH–N-terminal leader peptide of the antibody mAb24 heavy chain, PCP–plant cell pack, VH–variable heavy chain antibody domain.
Domain exchange variants (Figure 1B) were generated by amplifying IgG1 domains from the initial M12 construct by PCR (Supplementary Table S1) and introducing them into all the IgG3 constructs by Gibson assembly for expression in different compartments. The identity of the resulting constructs (Supplementary Table S2) was confirmed by DNA sequencing (Fraunhofer IME in-house sequencing service) using the primers listed in Supplementary Table S1. Constructs were assembled and propagated in Escherichia coli strains Dh5α (restriction cloning) or Top10 (Gibson assembly) and introduced into competent Agrobacterium tumefaciens (Rhizobium radiobacter) GV3101:pMP90RK cells by electroporation (2400 V, 25 μF, and 200 Ω) as previously described (Buyel et al., 2013). Restriction enzymes, Q5 high fidelity polymerase, T4 DNA ligase and Gibson Assembly Mastermix (all from New England Biolabs, Ipswich, MA, United States) were used according to the manufacturer’s recommendations. All primers were synthesized by Eurofins (Eurofins Genomics, Ebersberg, Germany).
Infiltration of Plants and Plant Cell Packs With A. tumefaciens
Nicotiana tabacum and N. benthamiana plants used for transient expression were cultivated on stone-wool blocks in a phytotron for 7 weeks as previously described (Menzel, 2018). N. tabacum Bright Yellow 2 (BY-2) cells were cultivated in continuous 5-L suspension cultures [100 g L−1 cell wet mass, packed cell volume 30–40% v v−1 (Holland et al., 2010)] or shake flasks, and were concentrated to a cell wet mass of 200 g L−1 for the preparation of plant cell packs (PCPs) by sedimentation as previously described (Gengenbach et al., 2020). Shake flask BY-2 cells were cultivated in 0.25-L glass flasks containing 0.05 L medium of supplemented Murashige and Skoog (MS) major and minor salts mixture [4.3 g L−1 MS medium (M0221, Duchefa, Haarlem, Netherlands), supplemented with 0.1 g L−1myo-inositol, 0.001 g L−1 thiamine, 0.0002 g L−1 2,4-dichlorophenoxyacetic acid, 0.2 g L−1 potassium phosphate and 0.1 g L−1 Pluronic L61] supplemented with 30 g L−1 sucrose or 30 g L−1 glucose as the carbon source. Shake flask cultures were inoculated with 2·108 cells L−1 and cultivated at 26°C and 160 rpm for 5–7 days as required by the Design of Experiments (DoE) settings.
The A. tumefaciens pre-cultures [PAM4 medium (Houdelet et al., 2017) containing 50 mg L−1 carbenicillin, 25 mg L−1 kanamycin and 25 mg L−1 rifampicin] were inoculated from glycerol stocks to an OD600nm of 0.04 and cultivated at 28°C for 24 h, shaking at 160 rpm in baffled 100-ml glass flasks (infiltration of differentiated plants) or at 1,000 rpm in 96-deepwell plates (infiltration of PCPs). The main cultures (500 ml in Fernbach flasks for the infiltration of differentiated plants, or 500 µL well−1 in 96-deepwell plates for the infiltration of PCPs) were inoculated from the pre-cultures to an OD600nm of 0.1 and incubated for 24 h using the same medium and cultivation settings as described above.
We used an automated protocol to generate PCPs from BY-2 cells followed by infiltration with A. tumefaciens as previously described (Gengenbach et al., 2020). We used 100 µL infiltration solution per PCP [0.5 g L−1 MS micro and macro salts (M0221, Duchefa), 50.0 g L−1 (146 mM) sucrose, 2.0 g L−1 (10 mM) glucose monohydrate, 0.0392 g L−1 (0.2 mM) acetosyringone, and 2.928 g L−1 (15 mM) 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.6] and an infiltration OD600nm of 0.4. Infiltrated PCPs were incubated at 26°C and 80% relative humidity for 48–96 h over a water reservoir (Gengenbach et al., 2020).
The infiltration solution for differentiated plants was prepared by adjusting A. tumefaciens cultures with 2 × infiltration buffer and deionized water to an OD600nm of 0.5 [final concentration of 0.5 g L−1 MS micro and macro salts (M0221, Duchefa) and 0.0392 g L−1 (0.2 mM) acetosyringone, pH 5.6]. The stem and leaves of whole plants were submerged in infiltration solution and infiltrated by applying a vacuum of 0.01 MPa (100 mbar) and releasing it after 1 min. Infiltrated plants were incubated in an inverted position at 22.2 ± 1.5°C, using 16-h light cycles with a light intensity of ∼50 μmol s−1 m−2 supplied by eight Osram Lumilux 58W/830 warm white and eight Osram 58W/840 cool white lamps (Osram, Munich, Germany). Plants were watered every 1.5 h for 7.7 s using MRS-10122 nozzles (Micro Rain Systems, Altenburg, Germany) with a water flow rate of 149 ± 5 ml min−1 (relative humidity 30% before and 80% after spray pulse) and were harvested after 5 days as previously described (Menzel et al., 2016). Vir gene expression was induced through acetosyringone in Agrobacterium infiltration suspensions by incubation for 1 h before infiltration of plants and PCPs. The purity of all media components was ≥99.5% and/or in compliance with the requirements of the European Pharmacopoeia (Ph. Eur.).
Protein Extraction From BY-2 Cells and Differentiated Plants
Plants were extracted using a 3 v m−1 ratio of extraction buffer (50 mM sodium phosphate, 10 mM sodium bisulfite, 500 mM sodium chloride, pH 8.0) in a blender (Koninklijke Philips, Amsterdam, Netherlands) for 3 × 30 s with 30-s breaks between mixing cycles as previously described (Buyel et al., 2014). Extracts intended only for analysis were clarified by centrifugation (twice at 16,000 × g, for 20 min at 4°C) and supernatants were stored at −20°C before analysis. Extracts intended for purification were clarified by passing through a 22-cm2 PDH4 depth filter composed of K700 and KS50 filter layers with ∼10 and ∼0.6 μm nominal retention ratings, respectively (Pall, Dreieich, Germany), at a flow rate of 12 ml min−1, and a Sartopore Capsule 0.20-μm filter (Sartorius-Stedim, Göttingen, Germany) as previously described (Opdensteinen et al., 2018).
Each 60-mg PCP was extracted with the same buffer ratio used for differentiated plants. The PCPs were transferred to 1.2-ml collection microtube strips (Qiagen, Hilden, Germany) containing a single 3-mm steel bead per microtube, and were extracted in a MM300 bead mill (Retsch, Han, Germany) at 28 Hz for 2 × 3 min (Gengenbach et al., 2020). Extracts were clarified by centrifugation (5,100 × g for 8 min at 4°C) and supernatants were stored at −20°C. The purity of all chemicals used to prepare extraction buffers was ≥99.5%, except sodium bisulfite (≥97%).
Antibody Purification Using Protein G Spin Columns
Miniaturized NAb protein G spin columns containing 0.2 ml resuspendable resin (Thermo Fisher Scientific, Waltham, MA, United States) were used to test IgG3 purification from plant extracts. The columns were equilibrated twice with 400 µL equilibration buffer [2.2 g L−1 (12.3 mM) disodium hydrogen phosphate, 1.2 g L−1 (7.7 mM) sodium dihydrogen phosphate, 8.8 g L−1 (150 mM) sodium chloride, pH 7.2]. We then loaded 500 µL of plant extract, which had been passed through a 0.2-µm filter. The columns were then incubated with washing buffer using conductivities of 5–50 mS cm−1 and pH values of 4.0–8.0 as required by the DoE settings. Citrate (0.02 M) was used as the buffering agent for wash buffers in the pH range 4.0–5.0, whereas phosphate (0.02 M) was used for buffers in the pH range 6.0–8.0. Elution [7.5 g L−1 (100 mM) glycine] was carried out at conductivities of 5–25 mS cm−1 and pH values of 2.0–3.0 as required by the DoE settings. The conductivity of the wash and elution buffers was adjusted as required by adding sodium chloride.
All wash and elution steps were carried out using 3 × 400 µL of the appropriate buffers. Wash and elution fractions were immediately mixed with neutralization buffer [121.1 g L−1 (1,000 mM) tris(hydroxymethyl)aminomethane (Tris), pH 9.0] using a 15% (v v−1) ratio for pH 2.0, a 5% (v v−1) ratio for pH 3.0–4.0 and a 2.5% (v v−1) ratio for pH 5.0–6.0. The resin was distributed in the columns for each chromatography step by end-over-end mixing using an Intelli RM-2L mixer (LTF Labortechnik, Wasserburg, Germany) with a speed of 15 rpm and a contact time of 2 min. The purity of all chemicals used to prepare small-scale chromatography buffers was ≥99.5% pro analysis (pa).
Laboratory-Scale Protein G Chromatography
An Äkta pure FPLC system and HiTrap Protein G HP column (both from GE Healthcare, Chicago, IL, United States) with a column volume (CV) of 1 ml were used for laboratory-scale protein G chromatography. The column was equilibrated with 5 CVs of equilibration buffer (composition as stated above) before loading 100 CVs of N. benthamiana extract, which had been passed through a 0.2-µm filter. The column was then washed with 5 CVs of equilibration buffer and five CVs of wash buffer [2.2 g L−1 (12.3 mM) disodium hydrogen phosphate, 2.2 g L−1 (7.7 mM) sodium dihydrogen phosphate, pH 8.0]. Fractions were eluted in 6 CVs of elution buffer [7.5 g L−1 (100 mM) glycine, pH 2.0] using sodium chloride to adjust the conductivity to 25 or 50 mS cm−1. Elution fractions were immediately neutralized using a 15% (v v−1) ratio of neutralization buffer [121.1 g L−1 (1,000 mM) Tris, pH 9.0]. A flow rate of 1 ml min−1 and a contact time of 2 min were used for all chromatography steps. The purity of all chemicals used to prepare small-scale chromatography buffers was ≥99.5% pro analysis (pa).
Statistical Design of Experiments
Design‐Expert v13 (Stat‐Ease, Minneapolis, MN, United States) was used to set up and analyze all models described herein. The effect of the cellular compartment (ER, single and duplicated signal sequence directing the target protein to the secretory pathway, apoplast, vacuole, and plastids), IgG scaffold (IgG1, IgG3, IgG3 with IgG1 CH3 or IgG1 CH2 and CH3), BY-2 cultivation time (120–168 h) and PCP incubation time (48–96 h) on antibody accumulation and integrity were investigated using an I‐optimal split-plot response surface design (PCP harvest time as a hard-to-change factor) with 216 runs.
The effects of the wash buffer (pH 4.0–8.0, conductivity 5–50 mS cm−1) and elution buffer (pH 2.0–3.0, conductivity 5–25 mS cm−1) composition on IgG3 purity and recovery after purification from plant extracts by protein G chromatography were investigated using a D-optimal response surface design with 30 runs conducted in three blocks. The model accuracy was assessed by comparing its predictions for the 80% or 95% confidence intervals with experimental data from verification runs not included in the model.
Sample Analysis
Samples from IgG3 expression and purification were separated on 4–12% Bis-Tris polyacrylamide gels before staining with SimplyBlue SafeStain (Thermo Fisher Scientific), silver staining as previously described (Arfi et al., 2016), or western blotting. Silver-stained lithium dodecylsulfate (LDS) gels were used to assess the antibody purity after chromatography by densiometric analysis using AIDA Image Analyzer (Raytest, Straubenhardt, Germany) as previously described (Menzel et al., 2016). The peak area for bands corresponding to antibodies and the total peak area of all bands [i.e., stained host cell proteins (HCPs) and antibodies] in a sample were used to estimate the antibody purity. Western blots were prepared as previously described (Kastilan et al., 2017) using alkaline phosphatase (AP)-conjugated goat anti-human IgG HC + LC (Jackson ImmunoResearch, West Grove, PA, United States) at a concentration of 0.12 mg L−1. The peak area corresponding to intact antibody chains and the total peak area were measured as above and the antibody integrity was calculated as the corresponding ratio. Western blots not intended for quantification were incubated with a mixture of AP-conjugated goat anti-human IgG HC + LC and AP-conjugated goat anti-human Ig λ chain (Merck, Darmstadt, Germany) at concentrations of 0.15 and 0.12 mg L−1, respectively.
Protein G Immobilization
A Sierra SPR 2/4 (Bruker Daltonics SPR, Hamburg, Germany) or Biacore T200 (GE Healthcare, Uppsala, Sweden) were used for SPR experiments. Recombinant E. coli-derived protein G (Thermo Fisher Scientific) was coupled to a CM5 sensor chip (Biacore T200) or a high capacity amine sensor (Sierra SPR 2/4) by ethyl-3-diaminopropyl-carbodiimide/N-hydroxylsuccinimide (EDC/NHS) chemistry (Johnsson et al., 1991). Briefly, the sensor surface was conditioned by alternating exposure to 30 mM hydrochloric acid and 25 mM sodium hydroxide followed by activation with a freshly prepared mixture of 0.48 M EDC and 0.10 M NHS for 10 min. The protein G solution (200 mg L−1 in 10 mM sodium acetate, pH 4.0) was then injected over the activated surface for 15 min before inactivation by exposure to 1.0 M ethanolamine for 10 min. A reference surface was functionalized in a similar manner, omitting the injection of the protein G solution. Protein G immobilization levels (200 mg L−1 in 10 mM sodium acetate, pH 4.0) were typically between 5,000 and 6,000 resonance units (RU), indicating efficient surface functionalization. Functionalized sensor surfaces were conditioned with several 30-s injections of 30 mM hydrochloric acid before each assay. Protein G-coated sensors were stored in sterile PBS at 4°C.
IgG3 Quantification by SPR Spectroscopy
IgG3 was quantified using a Sierra SPR 2/4 or Biacore T200. All measurements were conducted at 25°C and at a flow rate of 30 μL min−1 using HBS-EP running buffer [10 mM HEPES, 150 mM sodium chloride, 3 mM EDTA, 0.05% (v v−1) polysorbate 20, pH 7.4]. Samples were diluted in HBS-EP running buffer until the signals were in the liner range (Rademacher et al., 2008), typically 1:40 or 1:80, and were injected for 3 min. IgG3 concentrations in samples (csample) were calculated based on the resonance signal using Eq. 1, with plant-derived IgG1 monoclonal antibody (mAb) 2G12 as a standard (Ma et al., 2015):
where RUsample is the reference-corrected resonance signal of the sample, RUstd is the reference-corrected resonance signal of the 2G12 standard, Mm,std and Mm,sample are the molar masses of the standard and sample, and oligostd and oligosample describe the oligomerization state of standard and sample. The slope obtained from a dilution series of standards with a known concentration is shown as cstd RUstd−1, and dilsample is the sample dilution. To ensure the accuracy of IgG3 quantification, the RU signal from samples containing IgG3 was corrected using the RU signal recorded with wild-type extract diluted in a similar manner. Protein G-functionalized sensor surfaces were regenerated with a 30-s pulse of 30 mM hydrochloric acid (Ramessar et al., 2008).
Antigen-Binding Assays
Antigen-binding assays were performed with protein G-purified IgG3 expressed in the ER or apoplast and recombinant alpha toxin produced in E. coli (Abcam, Cambridge, United Kingdom) on a Biacore T200 at 25°C, using HBS-EP running buffer at a flowrate of 30 μL min−1. Briefly, IgG3 was injected over a protein G-coated CM5 sensor at a concentration of 7.35 mM (ER IgG3) or 10.62 mM (apoplast IgG3) in HBS-EP. Following IgG3 capture, we injected alpha toxin at concentrations in the range 400–0.024 nM in HBS-EP. The alpha toxin (∼33 kDa) was dialyzed against HBS-EP at 4°C for ∼16 h to remove glycerol (initial concentration 20% v v−1) from the formulation. The protein G sensor surface was regenerated after every antibody and alpha toxin injection using a 30-s pulse of 30 mM hydrochloric acid. The association rate constant (ka), the dissociation rate constant (kd), and the equilibrium dissociation constant between antibodies and alpha toxin (KD) were derived from the signal measured with different alpha toxin concentrations using the Biacore T200 evaluation software (GE Healthcare). Because the same KD (Eq. 2) can be calculated from different combinations of kd and ka values, we also used the latter to compare our IgG3 antibodies with the literature.
Statistical Methods
Two-sided two-sample t-tests were used to compare mAb degradation and mAb accumulation levels with a significance level of α = 0.05, using Shapiro-Wilk to test for normality and the F-test to test for equal variances. Welch’s t-test was used for non-normal or heteroscedastic data. The non-parametric Mann-Whitney U-test was used to compare mAb accumulation levels if data were non-normally distributed, using a significance level of α = 0.05. Tests were conducted using OriginPro 2015 (OriginLab, Northampton, MA, United States). A p-value ≤0.05 was regarded as significant. The significance of differences between mean model predictions (Figures 1C, D and Supplementary Figures S2C–E) was assessed by a least significant difference (LSD) test using a 95% confidence interval. LSD bars were calculated using Design-Expert v13.
Results and Discussion
Construct Design Rationale and Assessment of Functionality
IgG3 heavy chains are more susceptible to degradation than other subtypes, particularly the hinge region (Saito et al., 2019), and unassembled heavy and light chain monomers are in general more exposed to proteolysis than assembled mAbs (Gardner et al., 1993; Chen et al., 2016; Zischewski et al., 2016). To minimize the latter issue, we attempted to support mAb assembly with a theoretically optimal 1:1 ratio (Lin et al., 2018) of heavy and light chains by creating single-cassette expression vectors using an F2A linker peptide (Supplementary Figure S1) (Ryan et al., 1991). However, given that similar mRNA levels for each chain do not necessarily translate to similar protein levels (e.g., due to differences in translation, folding or degradation), the effect of excess IgG3 heavy and light chain monomers should be investigated more carefully in the future (Schlatter et al., 2005).
Because the F2A linker can generate a non-native N-terminus on the downstream (light chain) protein, which would attract regulatory scrutiny, we used a refined self-excising linker composed of an intein and F2A peptide (IntF2A) yielding native heavy and light chain sequences (Zhang et al., 2017). Deviating from the literature, the 58-amino-acid F2A peptide in the IntF2A linker was replaced with a 22-amino-acid F2A peptide (Supplementary Figure S1) to reduce potential ribosomal frameshifting (Zhang et al., 2017). The absence of unwanted proteinase recognition sites in the modified IntF2A linker was confirmed using PROSPER (Song et al., 2012).
In addition to a native human IgG3 heavy chain (Supplementary Figure S1), we also used the IgG1 control M12, which is known to accumulate to levels of ∼1 g kg−1 fresh mass in N. benthamiana (Zischewski et al., 2016). Both mAbs contained an identical antigen-binding site consisting of the M12 variable domain (Gaur, 2004) fitted with complementarity-determining regions (CDRs) directed against S. aureus alpha toxin (Sellman et al., 2018) to reduce potential differences in accumulation associated with the variable region in N. benthamiana (Zischewski et al., 2016) and to facilitate the analysis of constant domain stability.
We next assessed the functionality of the IgG3 and IgG1 constructs (Supplementary Figures S1A, B) by transient expression in PCPs, testing different 5′ UTRs and BY-2 cell cultivation conditions. We found that the IgG3 construct produced bands of ∼65 and ∼25 kDa whereas the IgG1 construct produced bands of ∼55 and ∼25 kDa (Supplementary Figure S1C), as anticipated based on previous reports (Kallolimath et al., 2020). The apparent mass of the IgG1 produced from a single expression cassette using the IntF2A linker matched the apparent mass of an IgG1 produced using separate expression cassettes for the heavy and light chains (Ma et al., 2015), confirming that the polypeptides were correctly processed.
As anticipated, the apparent mass of the IgG3 heavy chain was greater than that of the IgG1 counterpart, due to the elongated hinge region (Supplementary Figure S1C). However, the difference between the IgG1 heavy chain hinge region (15 amino acids) and its IgG3 counterpart (62 amino acids) was ∼10 kDa rather than the ∼6 kDa predicted from the amino acid sequence. This was attributed to the presence of additional glycosylation sites in the IgG3 heavy chain (Plomp et al., 2015; De Haan et al., 2019), further contributing to the difference in apparent molecular mass compared to IgG1 (Ma et al., 1994). For example, even ER-retained mAbs are modified in tobacco with high-mannose-type N-linked glycans featuring 6–9 mannose residues (Gomord et al., 2004), which have a mass of ∼1.4–1.9 kDa (Guo et al., 2021). Consequently, a mass increase of ∼2 kDa would be anticipated solely based on the additional N-glycosylation site in the IgG3 CH3 region compared to IgG1 (De Haan et al., 2019). Three additional O-glycosylation sites are also present in the IgG3 hinge region (but not in the IgG1 hinge region) although these should not be modified in ER-retained mAbs (Fernandez-del-Carmen et al., 2013).
IgG1 and IgG3 accumulation levels were not affected by the choice of 5′ UTR (Supplementary Figure S1C), but only ∼11% of the IgG1 was degraded compared to ∼38% for IgG3, a significant difference (two-sided, two-sample t-test, p<0.001, n = 6, α = 0.05). We detected distinct degradation products for each mAb, including a ∼45 kDa product for IgG3 that matched earlier reports (Navarre et al., 2012). In plants, IgG3 heavy chain degradation was previously attributed to the activity of serine proteases (Navarre et al., 2012) as well as the lack of protective O-linked glycans on the IgG3 hinge region (Kallolimath et al., 2020). This highlights the importance of subcellular targeting, because protease activity and glycosylation are compartment-specific (Hehle et al., 2011). The lower stability of IgG3 compared to IgG1 agreed with previous studies reporting >10-fold differences in the accumulation of these subclasses (Kallolimath et al., 2020).
Interestingly, BY-2 cells cultivated in a stirred-tank reactor accumulated less of the product (16.02 ± 1.40 mg kg−1, n = 6) than the cells cultivated in shake flasks (20.69 ± 3.46 mg kg−1, n = 6). The use of glucose instead of sucrose as a carbon source more than doubled the initial mAb levels in PCPs derived from BY-2 cells in shake flasks (34.94 ± 3.71 mg kg−1, n = 6), indicating that the optimization of media and cultivation conditions can help to improve recombinant protein expression levels in BY-2 cells. These differences in mAb accumulation were significant when comparing stirred-tank and shake-flask BY-2 cells (two-sided, two-sample t-test, p<0.017, n = 6, α = 0.05) and sucrose vs glucose as a carbon source (two-sided, two-sample t-test, p<0.001, n = 6, α = 0.05).
Expression Screening in PCPs
Next, we systematically investigated the effect of different BY-2 and PCP harvest time points as well as different target compartments and IgG scaffolds on IgG3 accumulation and product integrity using a DoE approach. Whereas harvesting the BY-2 cells earlier increased mAb integrity (Supplementary Figure S2, Supplementary Table S3), incubating the PCPs for 96 h rather than 48 h reduced degradation, but the overall impact of both parameters was small (∼5% change in integrity). In contrast, targeting the antibodies to particular subcellular compartments increased the integrity by >30%, with the highest integrity for both mAbs observed in the ER, followed by the apoplast and vacuole. In agreement with our data, retention in the ER has been shown to promote the folding and stability of antibody-derived proteins (Conrad and Fiedler, 1998).
The different 5′ UTRs again had no major impact on IgG3 or IgG1 levels (Figure 1A and Supplementary Figure S3) and we used the CHS 5′ UTR for all subsequent experiments. However, the subcellular compartment had a substantial effect on product accumulation, with higher IgG3 levels observed in the ER compared to the apoplast and vacuole, reaching ∼50 mg kg−1 wet biomass in the ER. IgG1 levels reached ∼150 mg kg−1 in the apoplast, correlating with the greater integrity discussed above. This sequence of compartments was consistent with previous reports of mAb degradation in the apoplast by acidic proteases that are not found in the ER (Hehle et al., 2011). Our data were also consistent with previous reports concerning single-chain fragment variable (scFv) antibodies (Ram et al., 2002), but contrasted with the >10-fold higher accumulation of a mouse IgG1 in the central vacuole compared to the secretory pathway (Ocampo et al., 2016), indicating that vacuolar proteases may selectively target human and/or IgG3-specific sequences. Our antibodies did not accumulate in the chloroplast, and we speculate that this may reflect the insufficient translocation of the light chain following cleavage of the IntF2A linker. Others have observed similar issues for bicistronic expression vectors featuring an F2A peptide (De Felipe et al., 2003) and therefore included individual signal sequences for all segments of F2A-separated polypeptides such as antibodies (Ho et al., 2013), which we plan to investigate in the future. In this context, we used a duplicated signal sequence at the start of the open reading frame to investigate whether the accumulation of IgG3 was limited by the inefficient translocation of the heavy chain to the ER (Supplementary Figure S1B). Our reasoning was that a failure to secrete both antibody chains would interfere with efficient assembly in the same manner as unbalanced expression. Although the duplicate signal peptide sequence did not appear to have a substantial effect on product accumulation in PCPs (Supplementary Figure S3), it had a relevant impact on the accumulation of antibodies in intact plants (see below).
We found that the quantity of IgG3 directed to the secretory pathway was about half that of the corresponding IgG1. Others have observed a >10-fold difference between the secretion of IgG3 (using the barley α-amylase signal peptide) and IgG1 (Kallolimath et al., 2020). We predicted that the self-cleaving linker would favor mAb assembly with a theoretically optimal 1:1 ratio of heavy and light chain, thus helping to limit the degradation of monomeric IgG3, which is more severe than the degradation of IgG1 due to the elongated, protease-sensitive hinge region (Kallolimath et al., 2020). We then investigated IgG3 domain variants in a DoE to identify which sections of the antibody may be limiting IgG3 accumulation compared to IgG1. We found that replacing the IgG3 CH3 or CH3 and CH2 domains with IgG1 counterparts increased the absolute accumulation of IgG3 to 75% of the donor IgG1 level in the ideal compartments (ER and apoplast), reaching a maximum of 140 mg kg−1 (Supplementary Figure S3). However, the IgG3 levels remained below 30% relative to IgG1 levels when averaged over all compartments (Figure 1D). These results support previous studies showing that the IgG3 CH3 and CH2 domains limit mAb accumulation by promoting aggregation, but other factors such as low stability in an acidic environment may also play a role (Saito et al., 2019). Higher mAb levels (up to ∼180 mg kg−1 IgG1) compared to the initial set of experiments (up to ∼150 mg kg−1 IgG1) were attributed to variation between different BY-2 batches, which we will investigate in more detail the future. We did not exchange the IgG3 hinge region and CH1 domain (e.g., while maintaining an IgG3 CH2 and/or CH3 domain) because the CH2 and CH3 domains have the greatest effect on IgG3 stability (Saito et al., 2019). A potential disadvantage of simultaneously replacing the CH2 and CH3 domains of IgG3 with IgG1 counterparts is the restoration of protein A binding to the CH2-CH3 interface, which is therapeutically undesirable (Rispens et al., 2014). However, engineering individual amino acids such as R435 can help to overcome this issue (Shah et al., 2017) while maintaining desirable IgG3 features, including a flexible hinge region and effector functions such as complement activation or antibody-dependent cell cytotoxicity (Damelang et al., 2019).
Transfer to Differentiated Plants
Having identified ideal conditions for the expression of IgG3 in PCPs, we investigated their transferability to differentiated N. benthamiana and N. tabacum plants, which are the anticipated production platforms (Capell et al., 2020; Buyel et al., 2017). In terms of subcellular targeting, IgG3 accumulation in N. benthamiana and N. tabacum plants matched the results in PCPs (Figure 2C). The highest IgG3 accumulation levels (∼50 mg kg−1) were achieved by targeting the ER, followed by the apoplast, the vacuole and the chloroplast. Interestingly, the construct with a duplicated signal peptide directing the IgG3 heavy chain to the secretory pathway showed significantly higher accumulation levels (Mann-Whitney U-test, p = 0.013, α = 0.05, n = 6) compared to a construct featuring a single signal sequence in differentiated plants (Figure 2C). This may indicate that, in contrast to PCPs, the efficiency of heavy chain trafficking limits IgG3 expression in differentiated plants as previously shown for hybrid mAbs representing different immunoglobulin classes (Frigerio et al., 2000). The IgG3 degradation products in differentiated plants (Figure 2B) resembled those detected in PCPs (Supplementary Figure S1C), regardless of the carbon source (sucrose or glucose) or cultivation setting (shake flask or stirred tank) used for BY-2 cultivation. These results indicate that PCPs are suitable as a screening tool, at least for IgG3, because the accumulation and integrity results transferred well to differentiated plants. In addition, the co-infiltration of intact plants with A. tumefaciens strains carrying a p19 silencing suppressor construct and the IgG3 construct (1:1 ratio, combined infiltration OD600nm = 1.0) increased IgG3 accumulation from ∼50 mg kg−1 to >130 mg kg−1 (Figure 2C). Further improvement could be achieved by specific optimization, such as varying the infiltration ratio or combining p19 co-infiltration with domain exchange variants, which was beyond the scope of this study.
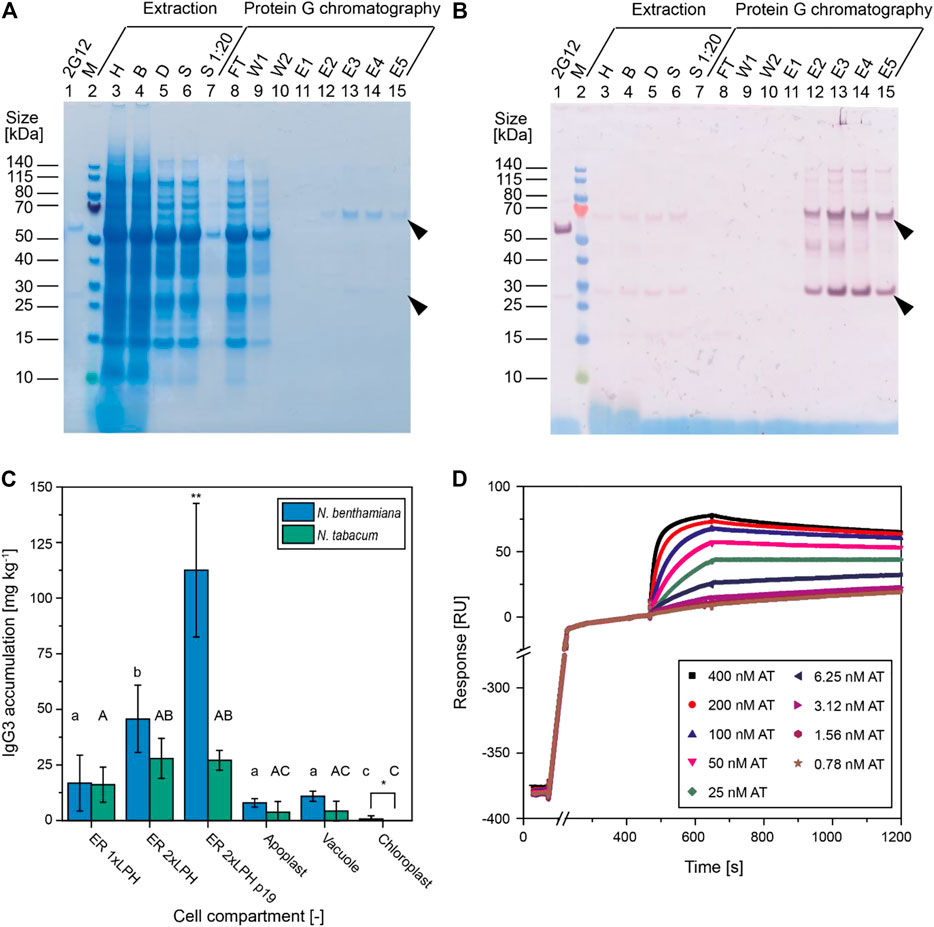
FIGURE 2. Expression of IgG3 mAbs in Nicotiana spp., laboratory-scale purification, and assessment of functionality. (A, B) Samples from IgG3 purification were analyzed on a Coomassie stained LDS gel (A) and by western blot (B) using AP-labeled goat anti-human IgG and goat anti-human Ig λ antibodies. (C) IgG3 accumulation levels in differentiated N. benthamiana and N. tabacum plants as measured by SPR. IgG3 accumulation levels correspond to constructs featuring a CHS 5′ UTR (p19 = co-infiltration with p19 silencing suppressor, 2 × LPH - construct with duplicated LPH leader sequence). Data are means ± standard deviations from 3–6 plants. Significant (alpha 0.05, p-value ≤0.05, two-sided, two-sample Welch’s t-test) differences between compartments using the same species are denoted by letters (lower case for N. benthamiana, upper case for N. tabacum) whereas differences between species in the same compartment are denoted by * and a ** indicates a significant difference to all other data. (D) Purified IgG3 antibodies were captured on a protein G-coated SPR sensor and 14 different concentrations (0.024–400 nM) of recombinant alpha toxin were injected. Data in D are raw resonance units without correction for baseline drift, recorded on a Biacore T200 with ER-derived IgG3. Alpha toxin concentrations <0.780 nM are omitted for clarity. Black block arrows indicate the expected molecular mass of heavy and light chains. H–homogenate, B–bag filtrate, D–depth filtrate, S–sterile (0.2 µm) filtrate, FT–flow-through, W1–running buffer wash, W2–pH 8.0, 5 mS cm−1 wash, E1–5–protein G eluate, AT–S. aureus alpha toxin, ER–endoplasmic reticulum, LPH–N-terminal leader peptide of the antibody mAb24 heavy chain.
IgG3 Purification From Plant Extracts by Protein G Chromatography
Minimal interaction between IgG3 and protein A is desirable for MRSA treatment and diagnosis (Kota et al., 2020), but this excludes IgG3 purification by protein A chromatography, which is the industry standard for mAbs (Loghem et al., 1982). Protein L cannot be used in place of protein A because protein L binds kappa light chains rather than the lambda light chains used here. However, protein G binds to a broader range of antibodies than protein A, including all four human IgG subclasses (Nezlin and Nezlin, 1998). We therefore used the Streptococcus spp. group C/G cell wall-derived defense protein G (Gronenborn et al., 1991) to purify IgG3 from plant extracts in small-scale spin columns.
In a DoE approach, we found that the pH and conductivity of the wash and elution buffers significantly affected IgG3 purity, with the highest purities (close to 100%) achieved with a low wash buffer pH (4.0), a low elution buffer pH (2.0), and a low wash buffer conductivity (5 mS cm−1) combined with a high elution buffer conductivity (25 mS cm−1) (Figure 3, Supplementary Table S4). The purity of IgG3 in the elution fractions was lower when using a high wash buffer conductivity, which we attributed to the elimination of the charged and polar contacts (Hey and Zhang, 2012) primarily responsible for IgG3–protein G interactions in the Fc region (Nezlin and Nezlin, 1998). In contrast, hydrophobic interactions dominate antibody binding to protein A (Nezlin and Nezlin, 1998). A high wash buffer pH (8.0) moderately reduced the elution purity of IgG3 compared to pH 4.0, probably reflecting the weaker interaction between IgG3 and protein G under these conditions. This agreed with earlier studies reporting an optimal pH of 4.0–5.0 for protein G binding, in contrast to pH 8.0 for protein A binding (Nezlin and Nezlin, 1998). Alkaline conditions (pH 11.0) have also been reported for the elution of recombinant IgG1 from protein G (Tkaczyk et al., 2012). We did not consider such conditions because many protein-based affinity chromatography resins are sensitive to alkaline pH (Gülich et al., 2002). For example, protein G denatures at pH 11.5 (Goward et al., 1991), so elution at pH 11.0 would substantially reduce the resin lifetime and thus increase costs.
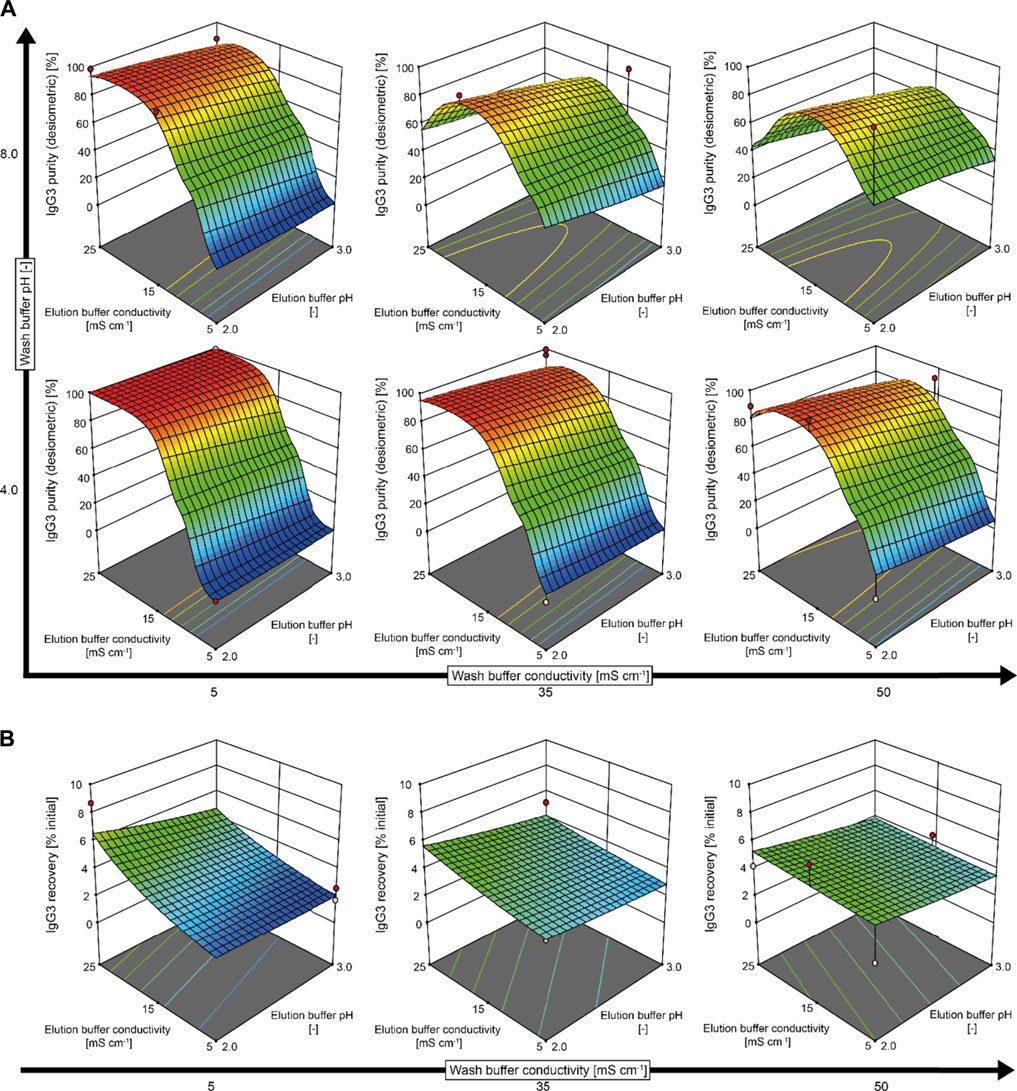
FIGURE 3. Models predicting the effects of the wash and elution buffer pH and conductivity on IgG3 purity and recovery after protein G chromatography (D-optimal design with 30 runs). (A) IgG3 purities after protein G chromatography were assessed by densiometric analysis of silver-stained LDS gels using the software AIDA. (B) IgG3 recoveries were derived from IgG3 concentrations measured in the feed and elution fractions using a protein G-coated SPR sensor. Miniaturized spin columns containing resuspendable protein G resin were used for purification. A contact time of 2 min was used for all chromatography steps. Red dots represent experimental data. Note: the model surface appears distorted around 15 mS cm−1 because there are additional levels in that region.
The recovery of IgG3 increased with decreasing elution buffer pH and increasing elution buffer conductivity (Figure 3B, Supplementary Table S4). As for purity, IgG3 recovery increased as the wash buffer conductivity was reduced, probably by preventing the partial elution of the product from the protein G resin. The highest IgG3 recovery using the miniaturized spin columns (∼10%) was achieved under the same conditions that optimized IgG3 purity. The low recovery was surprising because we did not detect IgG3 in the protein G chromatography wash or flow-through fractions. Considering the susceptibility of IgG3 to acidic conditions (Saito et al., 2019), the strongly acidic elution pH of 2.0–3.0 probably induced antibody denaturation and aggregation (Scheffel and Hober, 2021), thus hindering the quantification of IgG3 in the elution fractions. Further optimization of the protein G chromatography elution step will therefore be necessary to develop an economically viable process. Magnesium chloride or arginine can facilitate mAb elution under gentle conditions such as neutral pH (Tsumoto et al., 2007). For example, a 1.25 M magnesium chloride elution step has been applied successfully to elute plant-derived mAbs from an affinity resin (Rühl et al., 2018). Because protein G interactions with antibody Fc domains are mainly governed by charged and polar contacts, magnesium chloride concentrations ≤1.0 M will probably suffice for elution, whereas up to 4.0 M magnesium chloride is recommended for the elution of mAbs from protein A at neutral pH (Tsumoto et al., 2007). Alternatively, mAb-specific purification strategies can be used, such as epitope-based ligands (Rühl et al., 2018).
We next transferred the optimal purification conditions from spin columns to laboratory-scale 1.0-ml packed-bed protein G columns. As discussed for the spin columns, we achieved IgG3 purities >90% after a single purification step (Figures 3A,B, Supplementary Figure S5A). The remaining impurities were product related, i.e., degradation products of the antibody heavy chain, as they were larger than the antibody light chain, as indicated by western blotting (Figure 2B). Such degradation products can be removed by additional chromatography steps such as ion-exchange chromatography, as recently used to separate mAbs produced in Pichia pastoris from their degradation products (Purcell et al., 2017). IgG3 recovery increased from ∼10 to ∼19% (50 mS cm−1 elution buffer) in the packed-bed columns (Table 1, Supplementary Figure S5B). Although IgG3 antibodies were, once again, not detected in the wash or flow-through fractions (Figures 2A,B), the protein G chromatography step had the lowest recovery along the entire IgG3 purification process (Table 1). Optimizing this step, for example, by combining modified depth filtration with the gentle magnesium chloride elution method discussed above, should be the focus of future research.

TABLE 1. Overview of per step recoveries and total IgG3 recoveries during IgG3 purification from N. benthamiana extracts. Data are means ± standard deviations from n = 4 (extraction to sterile filtration) or n = 2 (protein G chromatography) purification processes.
Assessment of mAb Functionality
IgG3 expressed in the ER or apoplast and subsequently purified using protein G was able to bind the S. aureus alpha toxin (Figure 2D). The ER-derived IgG3 had a higher association rate (ka) and lower dissociation rate (kd) and thus achieved overall stronger alpha toxin binding (KD) than its counterpart purified from the apoplast (Table 2). This may reflect the ability of serine proteases in the apoplast to damage IgG3, interfering with antigen binding (Navarre et al., 2012; Hehle et al., 2011). Targeting IgG3 to the ER resulted in higher accumulation levels and greater affinity for the antigen (Table 2), making this the preferred compartment for IgG3 production using our Nicotiana platforms. However, the plant-derived antibodies showed a ∼5-fold lower affinity (KD) for their antigen compared to mAbs derived from B-cell murine hybridomas (Tkaczyk et al., 2012). The major difference was the lower association rate (ka) compared to earlier reports, whereas the dissociation rate (kd) was similar (Table 2). Given that many plant-derived mAbs perform as well as their counterparts produced in mammalian cells (Rattanapisit et al., 2019; Sack et al., 2007), and considering the similar dissociation rate of our IgG3 and others discussed in the literature (Table 2), we speculate that the differences in activity were related to the antigen batch (Yang et al., 2016). On one hand, we removed glycerol from the alpha toxin samples by overnight dialysis because 20% (v v−1) glycerol interfered with the refractive index change during SPR spectroscopy (Quinn et al., 2000), potentially masking the signal from antigen binding. This step has not been reported for the murine hybridoma-derived mAbs used in the literature. On the other hand, when comparing suppliers, we found differences in the alpha toxin amino acid sequence, which can affect mAb binding. A fair comparison of the antibodies produced in plants and animal cells would therefore require parallel measurements using identical antigens and reagent batches as previously described for SPR experiments (Katsamba et al., 2006; Yang et al., 2016).

TABLE 2. Comparison of empirical kinetic parameters determined for our plant-derived IgG3 antibodies directed against S. aureus alpha toxin with values presented in the literature. Kinetic parameters were calculated using the Biacore T200 evaluation software. KD = equilibrium dissociation constant, ka = association rate constant, kd = dissociation rate constant.
High-Throughput Quantification of IgG3 Directly in Clarified Plant Extracts
High-throughput assays based on SPR can speed up the development of new pharmaceuticals (Myszka and Rich, 2000). Protein A-coated SPR sensors have been used successfully to quantify mAbs in clarified plant extracts without prior purification (Sack et al., 2015). However, due to the lack of interaction with protein A, alternative ligands are necessary to detect IgG3. Here, we established a high-throughput compatible assay using EDC/NHS chemistry for the covalent attachment of protein G to SPR sensors, allowing the rapid quantification of IgG3 in plant extracts (Johnsson et al., 1991; Rademacher et al., 2008).
A protein G-coated SPR sensor showed minimal nonspecific binding (∼20 RU) when we injected a clarified wild-type N. benthamiana extract diluted 1:40 in HBS-EP running buffer (Figure 4A). In contrast, the clarified and similarly diluted extract of N. benthamiana plants transiently expressing IgG3 generated a strong signal on the same sensor (∼275 RU), confirming the successful binding of the plant-derived IgG3 antibodies to the functionalized sensor surface even in crude extracts (Figure 4B). The low nonspecific binding of the wild-type extract probably reflected the presence of host cell impurities, in agreement with the high selectivity of protein G for IgG3 during chromatographic purification (Figures 2A,B). We used injections of 30 mM hydrochloric acid for 30 s to regenerate the sensor, which can be challenging due to fouling of the SPR sensors with proteins and other host cell impurities (Goode et al., 2015). However, the resonance signal of the functionalized surface and the control surface returned to baseline after regeneration with hydrochloric acid, indicating that specifically bound IgG3 and non-specifically bound host cell impurities were successfully removed (Figure 4C). Repeated injection and regeneration cycles resulted in a capacity loss of 5.1% (1.0 mg L−1 2G12 standard) after 30 injections of sample and hydrochloric acid (Figure 4D). For comparison, protein A-based sensors allow >120 regeneration cycles before a similar capacity loss occurs (Quinn et al., 2000), whereas antibody-based sensors often fail after 10 regeneration cycles (Quinn et al., 2000; Puttharugsa et al., 2011). These data indicate that the protein G surface remains functional after regeneration and can be used continuously, which is desirable given the high cost of SPR sensor chips (Chatelier et al., 1995). Furthermore, the capacity loss can probably be slowed by replacing the acidic elution buffer with a magnesium chloride solution, as suggested above for the purification of IgG3 by chromatography. Protein G-coated sensors are therefore suitable for the repeated high-throughput quantification of IgG3, even in crude samples. Protein G-coated SPR sensors can also facilitate multi-step assays. For example, IgG3 capture by protein G followed by the injection of antigen to assess antibody binding kinetics ensures a controlled antibody orientation relative to the sensor surface, and thus the analysis of affinity instead of avidity (Bae et al., 2005). This may not be possible following chemical immobilization, where the antibody orientation is random and antigen binding may be sterically blocked (Bae et al., 2005).
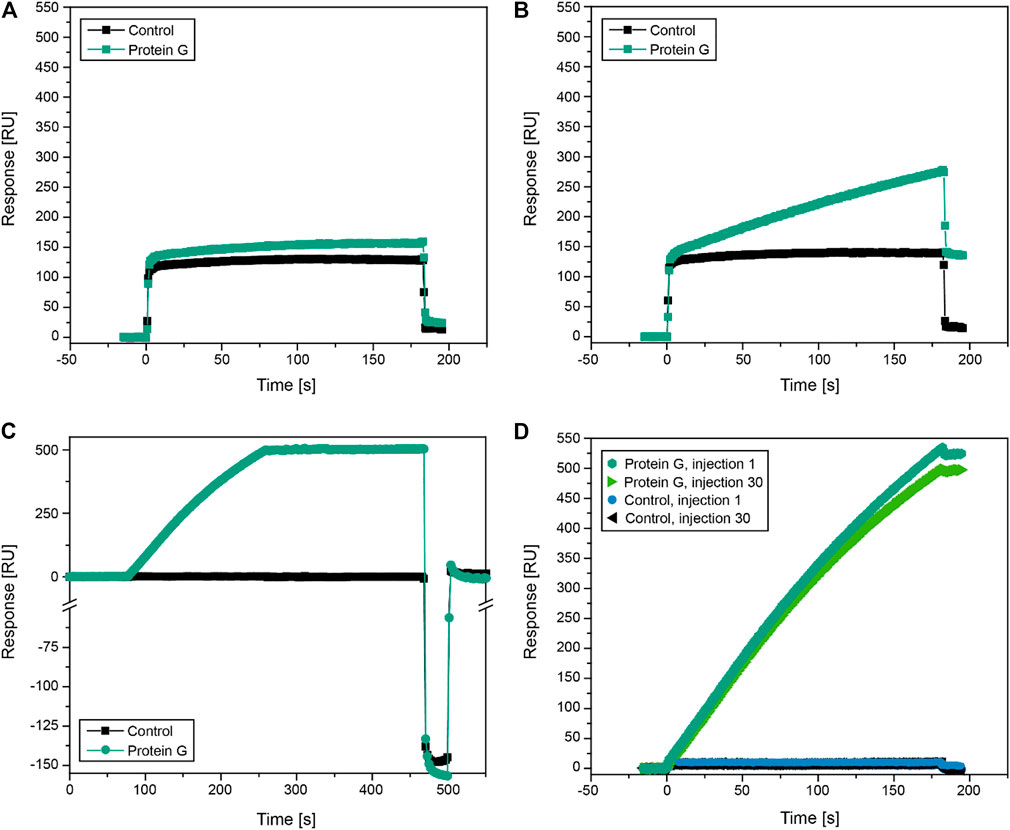
FIGURE 4. Test of protein G-coated sensors for the high-throughput quantification of recombinant IgG3 in plant extracts by SPR. (A, B) Protein G-coated sensors were challenged with clarified wild-type N. benthamiana extract (A) and clarified extract from N. benthamiana plants expressing IgG3 (B), both diluted 1:40 in HBS-EP running buffer. (C) Regeneration of protein G-coated sensor by elution of captured mAbs (2G12) with a 30-s injection of 30 mM hydrochloric acid. (D) Assessment of protein G sensor capacity loss over time by re-measuring samples with an identical mAb concentration of 1.0 mg L−1 (2G12). Data were recorded on a Sierra SPR2/4 instrument.
Conclusion
We systematically tested the effects of different BY-2/PCP cultivation conditions, subcellular compartments, different 5′ UTRs and IgG scaffolds on the accumulation and integrity of IgG3 in PCPs (Saito et al., 2019; Kallolimath et al., 2020). Our data indicate that a self-cleaving linker and the careful selection of an expression compartment can help to reduce IgG3 degradation in plants. The ER and apoplast were the most suitable compartments for recombinant IgG3 production, reaching accumulation levels of up to ∼50 mg kg−1 (native IgG3) or 130 mg kg−1 (domain exchange variants or following co-infiltration with p19) in the ER in PCPs and differentiated N. benthamiana plants. The similar accumulation and degradation patterns between intact plants and PCPs indicated that the latter are effective as a screening system and results can be readily translated to large-scale production.
The plant-derived IgG3 antibodies bound to the S. aureus alpha toxin, indicating that transient expression in Nicotiana spp. can be used to supply functional IgG3 mAbs in as little as 3 days (PCPs) or 5 days (differentiated plants). Previous domain exchange experiments with IgG1 increased accumulation by up to 8-fold (to 2 g kg−1) (Zischewski et al., 2016), suggesting that further improvements may also be possible for IgG3, for example, by combining BY-2 medium optimization, exchanging IgG3 domains with IgG1, or co-infiltration with p19. Accordingly, plant-derived IgG3 may help to meet the growing demand for diagnostics and therapeutics (Schillberg et al., 2005; Capell et al., 2020), especially for MRSA infections (Miller et al., 2019), which are increasingly difficult to treat with antibiotics. Whereas highly pure IgG3 was recovered after protein G chromatography, the recovery was poor and both the protein G chromatography and depth filtration steps require further optimization, including the use of alternative eluents such as magnesium chloride (Hey and Zhang, 2012; Rühl et al., 2018). The rapid SPR screening assay we developed will facilitate the analysis of samples during such optimization projects.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
SM and PO conducted experiments. PO, SM, and JB designed and evaluated experiments. PO wrote the manuscript. JB corrected and revised the manuscript. JB secured the funding for the project.
Funding
This work was funded in part by the Fraunhofer-Gesellschaft Internal Programs under grant no. Attract 125-600164 and the state of North-Rhine-Westphalia under the Leistungszentrum grant no. 423 “Networked, adaptive production.” The work was also supported by the Deutsche Forschungsgemeinschaft (DFG) in the framework of the Research Training Group “Tumor-targeted Drug Delivery” grant 331065168.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank Dr. Holger Spiegel for his support during kinetic measurements with IgG3 antibodies and Dr. Richard M. Twyman for editorial assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2021.737010/full#supplementary-material
Supplementary Figure S1 | Design of base constructs for IgG1 and IgG3 retained in the ER and assessment of construct functionality by transient expression in BY-2 PCPs. (A, B) IgG1 (A) and IgG3 (B) expression cassettes featuring a self-cleaving linker composed of an intein and an F2A peptide (Zhang et al., 2017) facilitating single-promoter transcription and stoichiometric translation of the mAb heavy and light chains. Deviating from the literature, a short F2A peptide (22 amino acids) was used. The CDRs directed against S. aureus alpha toxin (Sellman et al., 2018) were introduced into the variable domains of the M12 antibody (Gaur, 2004). (C) Investigation of IgG1 and IgG3 integrity by western blotting using an AP-labeled polyclonal goat anti-human IgG. BY-2 cells were cultivated for 168 h and PCPs were incubated for 72 h. Black block arrows indicate the expected molecular mass of the heavy and light chains. CHS–Petroselinum hortense chalcone synthase gene 5′ UTR, omega–omega prime sequence from tobacco mosaic virus, TL–tobacco etch virus leader sequence, WT–wild type PCP extract, S1 to S4–IgG1 standard (2G12, Fraunhofer IME) at concentrations of 14.0, 7.0, 5.0, and 2.5 mg L−1. CH – constant heavy chain antibody domain, CL – constant light chain antibody domain, ER – endoplasmic reticulum, LPH – N-terminal leader peptide of the antibody mAb24 heavy chain, PCP – plant cell pack, VH – variable heavy chain antibody domain, VL – variable light chain antibody domain.
Supplementary Figure S2 | Predictive model for compartment-specific IgG1 and IgG3 antibody integrity. (A–D) IgG3 integrity dependent on plant cell (pack) harvest times averaged over all compartments (A) and per compartment at 168 h BY-2 and 96 h PCP harvest (C) IgG1 integrity dependent on plant cell (pack) harvest times (B) and compartment at 168 h BY-2 and 96 h PCP harvest (D). (E) Effect of heavy chain scaffold (Figure 1B) on mAb integrity. Numbers inside the plot in panels (A) and (B) represent mAb iso-integrity lines. The mAb integrity was assessed by densitometric analysis of silver-stained LDS gels using AIDA software. Error bars in (C–E) represent least significant difference (LSD) bars around the model predicted means (dots). CH – constant heavy chain antibody domain, LPH – N-terminal leader peptide of the antibody mAb24 heavy chain, PCP – plant cell pack.
Supplementary Figure S3 | Accumulation and integrity of IgG1 and IgG3 variants in BY-2 PCPs. (A–D) Accumulation of IgG3 (A), IgG3 variant with IgG1 CH3 domain (B), IgG3 variant featuring IgG1 CH2 and CH3 domains (C) and IgG1 (D) dependent on the subcellular compartment and 5′ UTR. Even though silver-stained LDS gels indicated the presence of ∼10% intact mAbs when targeting the chloroplasts (Supplementary Figure S2), these mAbs did not bind to a protein G-coated SPR sensor, indicating that chloroplast-derived mAbs were not functional. Error bars indicate the standard deviation (n = 3 PCPs). BY-2 cells were cultivated for 168 h and PCPs were incubated for 96 h. (E) Integrity of ER-targeted IgG1 control and IgG3 variants detected with AP-labeled polyclonal goat anti-human IgG and goat anti-human Ig λ antibodies on a western blot. Black block arrows indicate the expected molecular mass of heavy and light chains. CHS – Petroselinum hortense chalcone synthase gene 5′ UTR, LPH – leader peptide of the antibody mAb24 heavy chain, omega – omega prime sequence from tobacco mosaic virus, TL – tobacco etch virus leader sequence. CH – constant heavy chain antibody domain, PCP – plant cell pack.
Supplementary Figure S4 | Comparison of model predictions (80% or 95% confidence interval, CI) and verification runs for mAb accumulation and integrity. (A) SPR measurement of IgG accumulation dependent on subcellular compartment and IgG scaffold. (B) Antibody integrities evaluated by densitometric analysis of silver-stained LDS gels dependent on the subcellular compartment and IgG scaffold. The broad confidence interval for IgG accumulation originated from substantial batch-to-batch variation during expression in PCPs. The data correspond to constructs featuring a CHS 5′ UTR. BY-2 cells were cultivated for 168 h and PCPs were incubated for 96 h. Apo – apoplast targeting, CH – constant heavy chain antibody domain, ERH – endoplasmic reticulum targeting with hexa-His-tag, LPH – N-terminal leader peptide of the antibody mAb24 heavy chain, PCP – plant cell pack, Vac – vacuolar targeting.
Supplementary Figure S5 | Comparison of model predictions (95% confidence interval, CI, orange lines) and verification runs (green diamonds at 25 mS cm−1; blue diamonds at 50 mS cm−1) for IgG3 purity and recovery after purification by protein G chromatography. The wash step was performed at pH 8.0 and 5.0 mS cm−1 using a contact time of 2 min in all experiments. Elution buffer conductivities up to 25 mS cm−1 (always pH 2.0) were used to establish the descriptive models, which cannot be used for predictions at higher conductivities, i.e. at 50 mS cm−1 used to test extreme conditions (blue diamonds). Independent of conductivity, model predictions and verification run performance matched well for purity, the model under-predicted the recovery, most likely due to scale-effects.
Supplementary Table S1 | Primers used to generate expression vectors for target protein expression in the apoplast, vacuole and plastids, as well as IgG3 domain exchange variants, starting from two IgG1/3 base constructs (Supplementary Figure S1).
Supplementary Table S2 | Expression vectors used to assess IgG3 production in Nicotiana spp.
Supplementary Table S3 | Model factors with a significant influence on IgG accumulation and integrity during transient expression in BY-2 PCPs as identified by restricted maximum likelihood analysis.
Supplementary Table S4 | Model factors with a significant influence on IgG purity and recovery after protein G chromatography as identified by analysis of variance (ANOVA).
Abbreviations
F2A, self-cleaving peptide derived from foot-and-mouth disease virus; Fab, antigen-binding fragment; Fc, crystallizable fragment; IgG, immunoglobulin G; Int, intein; mAb, monoclonal antibody; MRSA, methicillin-resistant Staphylococcus aureus; MIC, minimal inhibitory concentration; SPR, surface plasmon resonance.
References
Arfi, Z. A., Hellwig, S., Drossard, J., Fischer, R., and Buyel, J. F. (2016). Polyclonal Antibodies for Specific Detection of Tobacco Host Cell Proteins Can Be Efficiently Generated Following RuBisCO Depletion and the Removal of Endotoxins. Biotechnol. J. 11, 507–518. doi:10.1002/biot.201500271
Atkins, K. L., Burman, J. D., Chamberlain, E. S., Cooper, J. E., Poutrel, B., Bagby, S., et al. (2008). S. aureus IgG-Binding Proteins SpA and Sbi: Host Specificity and Mechanisms of Immune Complex Formation. Mol. Immunol. 45, 1600–1611. doi:10.1016/j.molimm.2007.10.021
Bae, Y. M., Oh, B.-K., Lee, W., Lee, W. H., and Choi, J.-W. (2005). Study on Orientation of Immunoglobulin G on Protein G Layer. Biosens. Bioelectron. 21, 103–110. doi:10.1016/j.bios.2004.09.003
Buyel, J. F., Gruchow, H. M., Boes, A., and Fischer, R. (2014). Rational Design of a Host Cell Protein Heat Precipitation Step Simplifies the Subsequent Purification of Recombinant Proteins from Tobacco. Biochem. Eng. J. 88, 162–170. doi:10.1016/j.bej.2014.04.015
Buyel, J. F., Kaever, T., Buyel, J. J., and Fischer, R. (2013). Predictive Models for the Accumulation of a Fluorescent Marker Protein in Tobacco Leaves According to the Promoter/5′UTR Combination. Biotechnol. Bioeng. 110, 471–482. doi:10.1002/bit.24715
Buyel, J. F., Twyman, R. M., and Fischer, R. (2017). Very-large-scale Production of Antibodies in Plants: the Biologization of Manufacturing. Biotechnol. Adv. 35, 458–465. doi:10.1016/j.biotechadv.2017.03.011
Capell, T., Twyman, R. M., Armario-Najera, V., Ma, J. K.-C., Schillberg, S., and Christou, P. (2020). Potential Applications of Plant Biotechnology against SARS-CoV-2. Trends Plant Sci. 25, 635–643. doi:10.1016/j.tplants.2020.04.009
Chatelier, R. C., Gengenbach, T. R., Griesser, H. J., Brighamburke, M., and Oshannessy, D. J. (1995). A General Method to Recondition and Reuse Biacore Sensor Chips Fouled with Covalently Immobilized Protein/peptide. Anal. Biochem. 229, 112–118. doi:10.1006/abio.1995.1386
Chen, L., Yang, X., Luo, D., and Yu, W. (2016). Efficient Production of a Bioactive Bevacizumab Monoclonal Antibody Using the 2A Self-Cleavage Peptide in Transgenic rice Callus. Front. Plant Sci. 7, 1156. doi:10.3389/fpls.2016.01156
Conlon, B. P., Nakayasu, E. S., Fleck, L. E., LaFleur, M. D., Isabella, V. M., Coleman, K., et al. (2013). Activated ClpP Kills Persisters and Eradicates a Chronic Biofilm Infection. Nature 503, 365–370. doi:10.1038/nature12790
Conrad, U., and Fiedler, U. (1998). Compartment-specific Accumulation of Recombinant Immunoglobulins in Plant Cells: an Essential Tool for Antibody Production and Immunomodulation of Physiological Functions and Pathogen Activity. Plant Mol. Biol. 38, 101–109. doi:10.1007/978-94-011-5298-3_5
Craft, K. M., Nguyen, J. M., Berg, L. J., and Townsend, S. D. (2019). Methicillin-resistant Staphylococcus aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. Med. Chem. Commun. 10, 1231–1241. doi:10.1039/c9md00044e
Damelang, T., Rogerson, S. J., Kent, S. J., and Chung, A. W. (2019). Role of IgG3 in Infectious Diseases. Trends Immunol. 40, 197–211. doi:10.1016/j.it.2019.01.005
De Felipe, P., Hughes, L. E., Ryan, M. D., and Brown, J. D. (2003). Co-translational, Intraribosomal Cleavage of Polypeptides by the Foot-And-Mouth Disease Virus 2A Peptide. J. Biol. Chem. 278, 11441–11448. doi:10.1074/jbc.m211644200
De Haan, N., Falck, D., and Wuhrer, M. (2019). Monitoring of Immunoglobulin N- and O-Glycosylation in Health and Disease. Glycobiology 30, 226–240. doi:10.1093/glycob/cwz048
De Lencastre, H., Oliveira, D., and Tomasz, A. (2007). Antibiotic Resistant Staphylococcus aureus: a Paradigm of Adaptive Power. Curr. Opin. Microbiol. 10, 428–435. doi:10.1016/j.mib.2007.08.003
DeLeo, F. R., and Chambers, H. F. (2009). Reemergence of Antibiotic-Resistant Staphylococcus aureus in the Genomics Era. J. Clin. Invest. 119, 2464–2474. doi:10.1172/jci38226
Dinges, M. M., Orwin, P. M., and Schlievert, P. M. (2000). Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13, 16–34. doi:10.1128/cmr.13.1.16
Falugi, F., Kim, H. K., Missiakas, D. M., Schneewind, O., and Gilmore, M. S. (2013). Role of Protein A in the Evasion of Host Adaptive Immune Responses by Staphylococcus aureus. mBio 4, e00575–13. doi:10.1128/mBio.00575-13
Fernandez-del-Carmen, A., Juárez, P., Presa, S., Granell, A., and Orzáez, D. (2013). Recombinant Jacalin-like Plant Lectins Are Produced at High Levels in Nicotiana Benthamiana and Retain Agglutination Activity and Sugar Specificity. J. Biotechnol. 163, 391–400. doi:10.1016/j.jbiotec.2012.11.017
Fischer, R., Schumann, D., Zimmermann, S., Drossard, J., Sack, M., and Schillberg, S. (1999). Expression and Characterization of Bispecific Single-Chain Fv Fragments Produced in Transgenic Plants. Eur. J. Biochem. 262, 810–816. doi:10.1046/j.1432-1327.1999.00435.x
Frigerio, L., Vine, N. D., Pedrazzini, E., Hein, M. B., Wang, F., Ma, J. K.-C., et al. (2000). Assembly, Secretion, and Vacuolar Delivery of a Hybrid Immunoglobulin in Plants. Plant Physiol. 123, 1483–1494. doi:10.1104/pp.123.4.1483
Gardner, A. M., Aviel, S., and Argon, Y. (1993). Rapid Degradation of an Unassembled Immunoglobulin Light Chain Is Mediated by a Serine Protease and Occurs in a Pre-golgi Compartment. J. Biol. Chem. 268, 25940–25947. doi:10.1016/s0021-9258(19)74477-9
Gaur, R. K. (2004). Structural Study of Human Antibody Fragments with Specificity for Mucin-1 Antigen. Bibliothek der RWTH Aachen.
Gengenbach, B. B., Opdensteinen, P., and Buyel, J. F. (2020). Robot Cookies - Plant Cell Packs as an Automated High-Throughput Screening Platform Based on Transient Expression. Front. Bioeng. Biotechnol. 8, 393. doi:10.3389/fbioe.2020.00393
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 6, 343–345. doi:10.1038/nmeth.1318
Gomord, V. r., Sourrouille, C., Fitchette, A.-C., Bardor, M., Pagny, S., Lerouge, P., et al. (2004). Production and Glycosylation of Plant-Made Pharmaceuticals: the Antibodies as a challenge. Plant Biotechnol. J. 2, 83–100. doi:10.1111/j.1467-7652.2004.00062.x
Goode, J. A., Rushworth, J. V. H., and Millner, P. A. (2015). Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 31, 6267–6276. doi:10.1021/la503533g
Gothwal, R., and Shashidhar, T. (2015). Antibiotic Pollution in the Environment: A Review. Clean. Soil Air Water 43, 479–489. doi:10.1002/clen.201300989
Goward, C. R., Irons, L. I., Murphy, J. P., and Atkinson, T. (1991). The Secondary Structure of Protein G′, a Robust Molecule. Biochem. J. 274, 503–507. doi:10.1042/bj2740503
Gronenborn, A., Filpula, D., Essig, N., Achari, A., Whitlow, M., Wingfield, P., et al. (1991). A Novel, Highly Stable Fold of the Immunoglobulin Binding Domain of Streptococcal Protein G. Science 253, 657–661. doi:10.1126/science.1871600
Gülich, S., Linhult, M., Ståhl, S., and Hober, S. (2002). Engineering Streptococcal Protein G for Increased Alkaline Stability. Protein Eng. Des. Selection 15, 835–842. doi:10.1093/protein/15.10.835
Guo, J., Tu, H., Rao B, M., Chillara, A. K., Chang, E., and Atouf, F. (2021). More Comprehensive Standards for Monitoring Glycosylation. Anal. Biochem. 612, 113896. doi:10.1016/j.ab.2020.113896
Guo, Y., Song, G., Sun, M., Wang, J., and Wang, Y. (2020). Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front Cel Infect Microbiol. 10, 107. doi:10.3389/fcimb.2020.00107
Hehle, V. K., Paul, M. J., Drake, P. M., Ma, J. K., and van Dolleweerd, C. J. (2011). Antibody Degradation in Tobacco Plants: a Predominantly Apoplastic Process. BMC Biotechnol. 11, 128. doi:10.1186/1472-6750-11-128
Hey, C., and Zhang, C. (2012). Process Development for Antibody Purification from Tobacco by Protein A Affinity Chromatography. Chem. Eng. Technol. 35, 142–148. doi:10.1002/ceat.201100259
Hibbitts, A., and O’Leary, C. (2018). Emerging Nanomedicine Therapies to Counter the Rise of Methicillin-Resistant Staphylococcus aureus. Materials 11, 321. doi:10.3390/ma11020321
Higgins, D. A., Cromie, R. L., Liu, S. S., Magor, K. E., and Warr, G. W. (1995). Purification of Duck Immunoglobulins: an Evaluation of Protein A and Protein G Affinity Chromatography. Vet. Immunol. Immunopathology 44, 169–180. doi:10.1016/0165-2427(93)05300-4
Ho, S. C. L., Bardor, M., Li, B., Lee, J. J., Song, Z., Tong, Y. W., et al. (2013). Comparison of Internal Ribosome Entry Site (IRES) and furin-2A (F2A) for Monoclonal Antibody Expression Level and Quality in CHO Cells. PLOS ONE 8, e63247. doi:10.1371/journal.pone.0063247
Hober, S., Nord, K., and Linhult, M. (2007). Protein A Chromatography for Antibody Purification. J. Chromatogr. B. 848, 40–47. doi:10.1016/j.jchromb.2006.09.030
Holland, T., Sack, M., Rademacher, T., Schmale, K., Altmann, F., Stadlmann, J., et al. (2010). Optimal Nitrogen Supply as a Key to Increased and Sustained Production of a Monoclonal Full-Size Antibody in BY-2 Suspension Culture. Biotechnol. Bioeng. 107, 278–289. doi:10.1002/bit.22800
Houdelet, M., Galinski, A., Holland, T., Wenzel, K., Schillberg, S., and Buyel, J. F. (2017). Animal Component-freeAgrobacterium Tumefacienscultivation media for Better GMP-Compliance Increases Biomass Yield and Pharmaceutical Protein Expression inNicotiana Benthamiana. Biotechnol. J. 12, 1600721. doi:10.1002/biot.201600721
Hua, L., Hilliard, J. J., Shi, Y., Tkaczyk, C., Cheng, L. I., Yu, X., et al. (2014). Assessment of an Anti-alpha-toxin Monoclonal Antibody for Prevention and Treatment of Staphylococcus Aureus-Induced Pneumonia. Antimicrob. Agents Chemother. 58, 1108–1117. doi:10.1128/aac.02190-13
Huebbers, J. W., and Buyel, J. F. (2021). On the Verge of the Market - Plant Factories for the Automated and Standardized Production of Biopharmaceuticals. Biotechnol. Adv. 46, 107681. doi:10.1016/j.biotechadv.2020.107681
Johnsson, B., Löfås, S., and Lindquist, G. (1991). Immobilization of Proteins to a Carboxymethyldextran-Modified Gold Surface for Biospecific Interaction Analysis in Surface Plasmon Resonance Sensors. Anal. Biochem. 198, 268–277. doi:10.1016/0003-2697(91)90424-r
Kallolimath, S., Hackl, T., Gahn, R., Grünwald-Gruber, C., Zich, W., Kogelmann, B., et al. (2020). Expression Profiling and Glycan Engineering of IgG Subclass 1-4 in Nicotiana Benthamiana. Front. Bioeng. Biotechnol. 8, 825. doi:10.3389/fbioe.2020.00825
Kastilan, R., Boes, A., Spiegel, H., Voepel, N., Chudobová, I., Hellwig, S., et al. (2017). Improvement of a Fermentation Process for the Production of Two PfAMA1-DiCo-Based Malaria Vaccine Candidates in Pichia pastoris. Sci. Rep. 7, 11991. doi:10.1038/s41598-017-11819-4
Katsamba, P. S., Navratilova, I., Calderon-Cacia, M., Fan, L., Thornton, K., Zhu, M., et al. (2006). Kinetic Analysis of a High-Affinity Antibody/antigen Interaction Performed by Multiple Biacore Users. Anal. Biochem. 352, 208–221. doi:10.1016/j.ab.2006.01.034
King, L. M., Bartoces, M., Fleming-Dutra, K. E., Roberts, R. M., and Hicks, L. A. (2019). Changes in US Outpatient Antibiotic Prescriptions from 2011-2016. Clin. Infect. Dis. 70, 370–377. doi:10.1093/cid/ciz225
Kota, R. K., Reddy, P. N., and Sreerama, K. (2020). Application of IgY Antibodies against Staphylococcal Protein A (SpA) of Staphylococcus aureus for Detection and Prophylactic Functions. Appl. Microbiol. Biotechnol. 104, 9387–9398. doi:10.1007/s00253-020-10912-5
Kourtis, A. P., Hatfield, K., Baggs, J., Mu, Y., See, I., Epson, E., et al. (2019). Vital Signs:Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-SusceptibleStaphylococcus aureusBloodstream Infections - United States. MMWR Morb. Mortal. Wkly. Rep. 68, 214–219. doi:10.15585/mmwr.mm6809e1
Lin, Y., Hung, C. Y., Bhattacharya, C., Nichols, S., Rahimuddin, H., Kittur, F. S., et al. (2018). An Effective Way of Producing Fully Assembled Antibody in Transgenic Tobacco Plants by Linking Heavy and Light Chains via a Self-Cleaving 2A Peptide. Front. Plant Sci. 9, 1379. doi:10.3389/fpls.2018.01379
Loghem, E., Frangione, B., Recht, B., and Franklin, E. C. (1982). Staphylococcal Protein A and Human IgG Subclasses and Allotypes. Scand. J. Immunol. 15, 275–278. doi:10.1111/j.1365-3083.1982.tb00649.x
Lowy, F. D. (1998). Staphylococcus aureusInfections. N. Engl. J. Med. 339, 520–532. doi:10.1056/nejm199808203390806
Ma, J. K.-C., Drossard, J., Lewis, D., Altmann, F., Boyle, J., Christou, P., et al. (2015). Regulatory Approval and a First-In-Human Phase I Clinical Trial of a Monoclonal Antibody Produced in Transgenic Tobacco Plants. Plant Biotechnol. J. 13, 1106–1120. doi:10.1111/pbi.12416
Ma, J. K.-C., Lehner, T., Stabila, P., Fux, C. I., and Hiatt, A. (1994). Assembly of Monoclonal Antibodies with IgG1 and IgA Heavy Chain Domains in Transgenic Tobacco Plants. Eur. J. Immunol. 24, 131–138. doi:10.1002/eji.1830240120
Maclean, J., Koekemoer, M., Olivier, A. J., Stewart, D., Hitzeroth, I. I., Rademacher, T., et al. (2007). Optimization of Human Papillomavirus Type 16 (HPV-16) L1 Expression in Plants: Comparison of the Suitability of Different HPV-16 L1 Gene Variants and Different Cell-Compartment Localization. J. Gen. Virol. 88, 1460–1469. doi:10.1099/vir.0.82718-0
Mandal, M. K., Ahvari, H., Schillberg, S., and Schiermeyer, A. (2016). Tackling Unwanted Proteolysis in Plant Production Hosts Used for Molecular Farming. Front. Plant Sci. 7, 267. doi:10.3389/fpls.2016.00267
Menzel, S., Holland, T., Boes, A., Spiegel, H., Bolzenius, J., Fischer, R., et al. (2016). Optimized Blanching Reduces the Host Cell Protein Content and Substantially Enhances the Recovery and Stability of Two Plant-Derived Malaria Vaccine Candidates. Front. Plant Sci. 7, 159. doi:10.3389/fpls.2016.00159
Menzel, S. (2018). Downstream Processing of Malaria Vaccine Candidates and Modeling of Chromatography. Bibliothek der RWTH Aachen.
Micek, S. T. (2007). Alternatives to Vancomycin for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections. Clin. Infect. Dis. 45, S184–S190. doi:10.1086/519471
Miller, L. S., Fowler, V. G., Shukla, S. K., Rose, W. E., and Proctor, R. A. (2019). Development of a Vaccine against Staphylococcus aureus Invasive Infections: Evidence Based on Human Immunity, Genetics and Bacterial Evasion Mechanisms. FEMS Microbiol. Rev. 44, 123–153. doi:10.1093/femsre/fuz030
Munro, S., and Pelham, H. R. B. (1987). A C-Terminal Signal Prevents Secretion of Luminal ER Proteins. Cell 48, 899–907. doi:10.1016/0092-8674(87)90086-9
Myszka, D. G., and Rich, R. L. (2000). Implementing Surface Plasmon Resonance Biosensors in Drug Discovery. Pharm. Sci. Tech. Today 3, 310–317. doi:10.1016/s1461-5347(00)00288-1
Navarre, C., De Muynck, B., Alves, G., Vertommen, D., Magy, B., and Boutry, M. (2012). Identification, Gene Cloning and Expression of Serine Proteases in the Extracellular Medium of Nicotiana Tabacum Cells. Plant Cel Rep. 31, 1959–1968. doi:10.1007/s00299-012-1308-y
Nezlin, R. (1998). “Interactions outside the Antigen-Combining Site,” in The Immunoglobulins. Editor R. Nezlin (New York: Academic Press), 219–cp1. doi:10.1016/b978-012517970-6/50006-0
Ocampo, C. G., Lareu, J. F., Marin Viegas, V. S., Mangano, S., Loos, A., Steinkellner, H., et al. (2016). Vacuolar Targeting of Recombinant Antibodies in Nicotiana Benthamiana. Plant Biotechnol. J. 14, 2265–2275. doi:10.1111/pbi.12580
Oogai, Y., Matsuo, M., Hashimoto, M., Kato, F., Sugai, M., and Komatsuzawa, H. (2011). Expression of Virulence Factors by Staphylococcus aureus Grown in Serum. Appl. Environ. Microbiol. 77, 8097–8105. doi:10.1128/aem.05316-11
Opdensteinen, P., Clodt, J. I., Müschen, C. R., Filiz, V., and Buyel, J. F. (2018). A Combined Ultrafiltration/diafiltration Step Facilitates the Purification of Cyanovirin-N from Transgenic Tobacco Extracts. Front. Bioeng. Biotechnol. 6, 206. doi:10.3389/fbioe.2018.00206
Plomp, R., Dekkers, G., Rombouts, Y., Visser, R., Koeleman, C. A. M., Kammeijer, G. S. M., et al. (2015). Hinge-region O-Glycosylation of Human Immunoglobulin G3 (IgG3). Mol. Cell Proteomics 14, 1373–1384. doi:10.1074/mcp.m114.047381
Purcell, O., Patrick Opdensteinen, W. C., Lowenhaupt, K., Brown, A., Hermann, M., Cao, J., et al. (2017). Production of Functional Anti-ebola Antibodies in Glycoengineered Pichia pastoris. ACS Synth. Biol. 6 (12), 2183–2190. doi:10.1021/acssynbio.7b00234
Puttharugsa, C., Wangkam, T., Huangkamhang, N., Gajanandana, O., Himananto, O., Sutapun, B., et al. (2011). Development of Surface Plasmon Resonance Imaging for Detection of Acidovorax Avenae Subsp. Citrulli (Aac) Using Specific Monoclonal Antibody. Biosens. Bioelectron. 26, 2341–2346. doi:10.1016/j.bios.2010.10.007
Quinn, J. G., O'Neill, S., Doyle, A., McAtamney, C., Diamond, D., MacCraith, B. D., et al. (2000). Development and Application of Surface Plasmon Resonance-Based Biosensors for the Detection of Cell-Ligand Interactions. Anal. Biochem. 281, 135–143. doi:10.1006/abio.2000.4564
Rademacher, T., Sack, M., Arcalis, E., Stadlmann, J., Balzer, S., Altmann, F., et al. (2008). Recombinant Antibody 2G12 Produced in maize Endosperm Efficiently Neutralizes HIV-1 and Contains Predominantly Single-GlcNAc N-Glycans. Plant Biotechnol. J 6, 189–201. doi:10.1111/j.1467-7652.2007.00306.x
Ram, N., Ayala, M., Lorenzo, D., Palenzuela, D., Herrera, L., Doreste, V., et al. (2002). Expression of a Single-Chain Fv Antibody Fragment Specific for the Hepatitis B Surface Antigen in Transgenic Tobacco Plants. Transgenic Res. 11, 61–64. doi:10.1023/a:1013967705337
Ramessar, K., Rademacher, T., Sack, M., Stadlmann, J., Platis, D., Stiegler, G., et al. (2008). Cost-effective Production of a Vaginal Protein Microbicide to Prevent HIV Transmission. Proc. Natl. Acad. Sci. 105, 3727–3732. doi:10.1073/pnas.0708841104
Rattanapisit, K., Phakham, T., Buranapraditkun, S., Siriwattananon, K., Boonkrai, C., Pisitkun, T., et al. (2019). Structural and In Vitro Functional Analyses of Novel Plant-Produced Anti-human PD1 Antibody. Sci. Rep. 9, 15205. doi:10.1038/s41598-019-51656-1
Rispens, T., and Vidarsson, G. (2014). “Human IgG Subclasses,” in Antibody Fc. Editors M. E. Ackerman, and F. Nimmerjahn (Boston: Academic Press), 159–177. doi:10.1016/b978-0-12-394802-1.00009-1
Rühl, C., Knödler, M., Opdensteinen, P., and Buyel, J. F. (2018). A Linear Epitope Coupled to DsRed Provides an Affinity Ligand for the Capture of Monoclonal Antibodies. J. Chromatogr. A 1571, 55–64. doi:10.1016/j.chroma.2018.08.014
Ryan, M. D., King, A. M. Q., and Thomas, G. P. (1991). Cleavage of Foot-And-Mouth Disease Virus Polyprotein Is Mediated by Residues Located within a 19 Amino Acid Sequence. J. Gen. Virol. 72, 2727–2732. doi:10.1099/0022-1317-72-11-2727
Sack, M., Paetz, A., Kunert, R., Bomble, M., Hesse, F., Stiegler, G., et al. (2007). Functional Analysis of the Broadly Neutralizing Human anti‐HIV‐1 Antibody 2F5 Produced in Transgenic BY‐2 Suspension Cultures. FASEB j. 21, 1655–1664. doi:10.1096/fj.06-5863com
Sack, M., Rademacher, T., Spiegel, H., Boes, A., Hellwig, S., Drossard, J., et al. (2015). From Gene to Harvest: Insights into Upstream Process Development for the GMP Production of a Monoclonal Antibody in Transgenic Tobacco Plants. Plant Biotechnol. J. 13, 1094–1105. doi:10.1111/pbi.12438
Saito, S., Namisaki, H., Hiraishi, K., Takahashi, N., and Iida, S. (2019). A Stable Engineered Human IgG3 Antibody with Decreased Aggregation during Antibody Expression and Low pH Stress. Protein Sci. 28, 900–909. doi:10.1002/pro.3598
Scheffel, J., and Hober, S. (2021). Highly Selective Protein A Resin Allows for Mild Sodium Chloride-Mediated Elution of Antibodies. J. Chromatogr. A 1637, 461843. doi:10.1016/j.chroma.2020.461843
Schillberg, S., Twyman, R. M., and Fischer, R. (2005). Opportunities for Recombinant Antigen and Antibody Expression in Transgenic Plants-Technology Assessment. Vaccine 23, 1764–1769. doi:10.1016/j.vaccine.2004.11.002
Schlatter, S., Stansfield, S. H., Dinnis, D. M., Racher, A. J., Birch, J. R., and James, D. C. (2005). On the Optimal Ratio of Heavy to Light Chain Genes for Efficient Recombinant Antibody Production by CHO Cells. Biotechnol. Prog. 21, 122–133. doi:10.1021/bp049780w
Sellman, B., Tkaczyk, C., Hua, L., Chowdhury, P., and Varkey, R. (2018). Antibodies that Specifically Bind Staphylococcus aureus Alpha Toxin and Methods of Use (EP-2673373-B1). USA: MedImmune LLC. Available at: https://pubchem.ncbi.nlm.nih.gov/patent/EP-2673373-B1.
Shah, I. S., Lovell, S., Mehzabeen, N., Battaile, K. P., and Tolbert, T. J. (2017). Structural Characterization of the Man5 Glycoform of Human IgG3 Fc. Mol. Immunol. 92, 28–37. doi:10.1016/j.molimm.2017.10.001
Shopsin, B., Kaveri, S. V., and Bayry, J. (2016). Tackling Difficult Staphylococcus aureus Infections: Antibodies Show the Way. Cell Host & Microbe 20, 555–557. doi:10.1016/j.chom.2016.10.018
Song, J., Tan, H., Perry, A. J., Akutsu, T., Webb, G. I., Whisstock, J. C., et al. (2012). PROSPER: An Integrated Feature-Based Tool for Predicting Protease Substrate Cleavage Sites. PLOS ONE 7, e50300. doi:10.1371/journal.pone.0050300
Szymańska, U., Wiergowski, M., Sołtyszewski, I., Kuzemko, J., et al. (2019). Presence of Antibiotics in the Aquatic Environment in Europe and Their Analytical Monitoring: Recent Trends and Perspectives. Microchemical J. 147, 729–740. doi:10.1016/j.microc.2019.04.003
Tkaczyk, C., Hua, L., Varkey, R., Shi, Y., Dettinger, L., Woods, R., et al. (2012). Identification of Anti-alpha Toxin Monoclonal Antibodies that Reduce the Severity of Staphylococcus aureus Dermonecrosis and Exhibit a Correlation between Affinity and Potency. Clin. Vaccin. Immunol. 19, 377–385. doi:10.1128/cvi.05589-11
Tsumoto, K., Ejima, D., Senczuk, A. M., Kita, Y., and Arakawa, T. (2007). Effects of Salts on Protein-Surface Interactions: Applications for Column Chromatography. J. Pharm. Sci. 96, 1677–1690. doi:10.1002/jps.20821
Wistrand-Yuen, E., Knopp, M., Hjort, K., Koskiniemi, S., Berg, O. G., and Andersson, D. I. (2018). Evolution of High-Level Resistance during Low-Level Antibiotic Exposure. Nat. Commun. 9, 1599. doi:10.1038/s41467-018-04059-1
Yang, D., Singh, A., Wu, H., and Kroe-Barrett, R. (2016). Comparison of Biosensor Platforms in the Evaluation of High Affinity Antibody-Antigen Binding Kinetics. Anal. Biochem. 508, 78–96. doi:10.1016/j.ab.2016.06.024
Zhang, B., Rapolu, M., Kumar, S., Gupta, M., Liang, Z., Han, Z., et al. (2017). Coordinated Protein Co‐expression in Plants by Harnessing the Synergy between an Intein and a Viral 2A Peptide. Plant Biotechnol. J. 15, 718–728. doi:10.1111/pbi.12670
Keywords: affinity chromatography, alpha toxin, biopharmaceuticals, MRSA, protein G, plant molecular farming, plant cell packs, subcellular compartments
Citation: Opdensteinen P, Meyer S and Buyel JF (2021) Nicotiana spp. for the Expression and Purification of Functional IgG3 Antibodies Directed Against the Staphylococcus aureus Alpha Toxin. Front. Chem. Eng. 3:737010. doi: 10.3389/fceng.2021.737010
Received: 06 July 2021; Accepted: 06 September 2021;
Published: 05 October 2021.
Edited by:
Karla Mayolo-Deloisa, Monterrey Institute of Technology and Higher Education (ITESM), MexicoReviewed by:
Rüstem Keçili, Anadolu University, TurkeyXiaobin Jiang, Dalian University of Technology, China
Marie-Claire Goulet, Laval University, Canada
Copyright © 2021 Opdensteinen, Meyer and Buyel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. F. Buyel, johannes.buyel@rwth-aachen.de
†ORCID: J. F. Buyel, orcid.org/0000-0003-2361-143X
 P. Opdensteinen
P. Opdensteinen