- 1Environmental Management Research Institute (EMRI), National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Japan
- 2School of Life Science and Technology, Tokyo Institute of Technology, Yokohama, Japan
Bioluminescence (BL) is an amazing optical readout that has great potential to be utilized in various bioassays and molecular imaging, but also has some unique defects in practical applications. The recent innovative research on BL has enriched the available repertories of the toolbox. While these efforts greatly diversified the users’ choices in the applications, the wide choices on the contrary do not promise their successful applications to bioassays. This is mainly due to complexity-driven confusion with the diversity and the lack of accurate knowledge on the advantages and disadvantages of BL. This review is intended to showcase the advantages and disadvantages of BL, and serve as a searchlight to find directions for future studies. We hope that this review provides instant references for readers on BL and leads them to properly understand the “bright” and “dark” sides of BL to narrow down their choices in their applications.
GRAPHICAL ABSTRACT | Bioluminescence from the bright and dark sides (The image was designed to contrast the “bright” and “dark” sides of bioluminescence as the theme of this review).
1 Bioluminescence from the “bright” and “dark” sides
There are many light-emitting organisms in nature. More than 2000 species of fireflies are known on land (Kumar et al., 2019) and the number is updated every year. Such light-emitting organisms are rather rare on land but extremely common in the oceans. Such light-emitting organisms are found in oceans at all depths with the greatest numbers found in the upper 1,000 m of the vast open ocean (Widder, 2001). This light emission from living organisms is especially called bioluminescence (BL). This BL is a cold light-emitting event by oxidation of a small molecular weight substrate called luciferin, catalyzed by an enzyme named luciferase inside living organisms. The representative luciferases and luciferins were detailed in Table 1 and Figure 1.
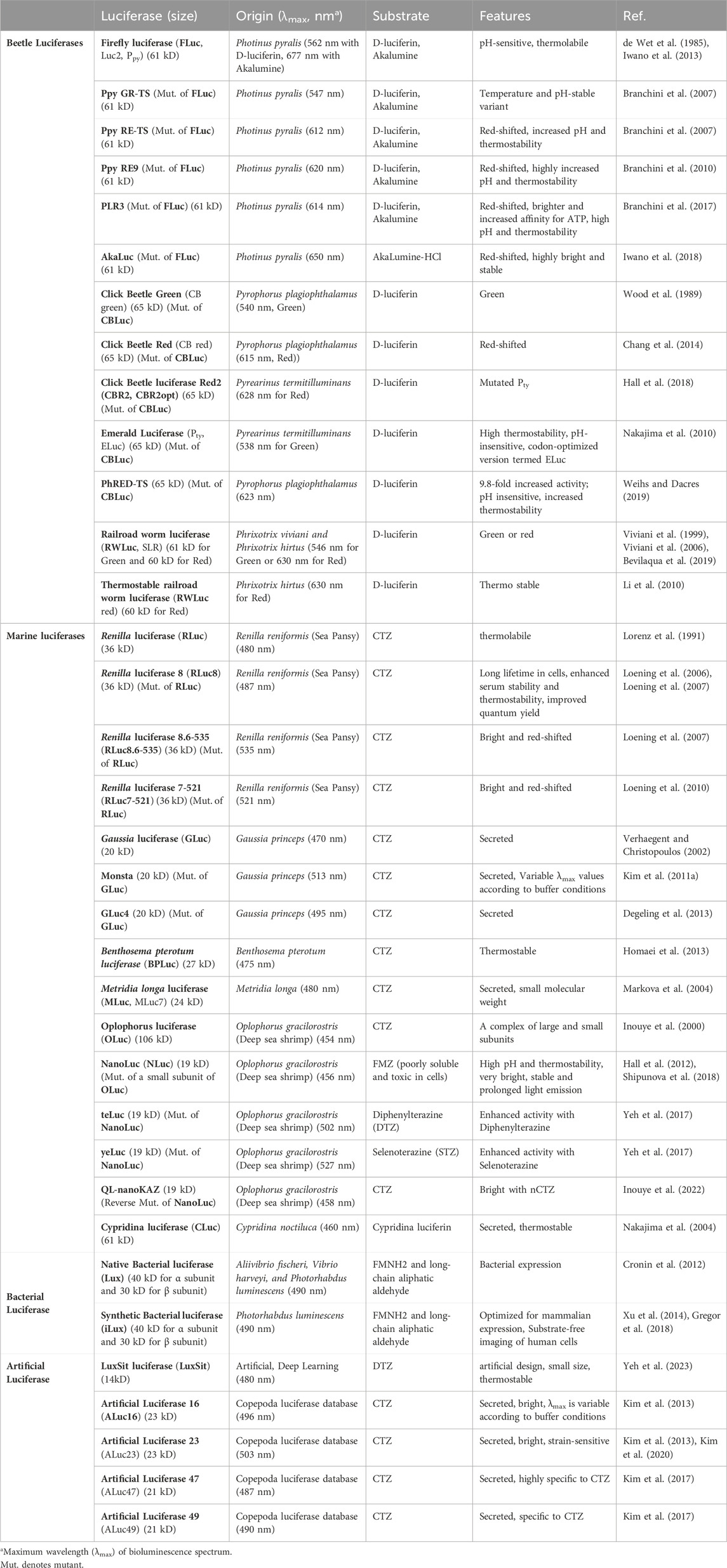
Table 1. Optical properties of representative luciferases and their variants. This table was reproduced and updated from Kim et al. (Kim and Paulmurugan, 2021) with permission from Springer Nature.
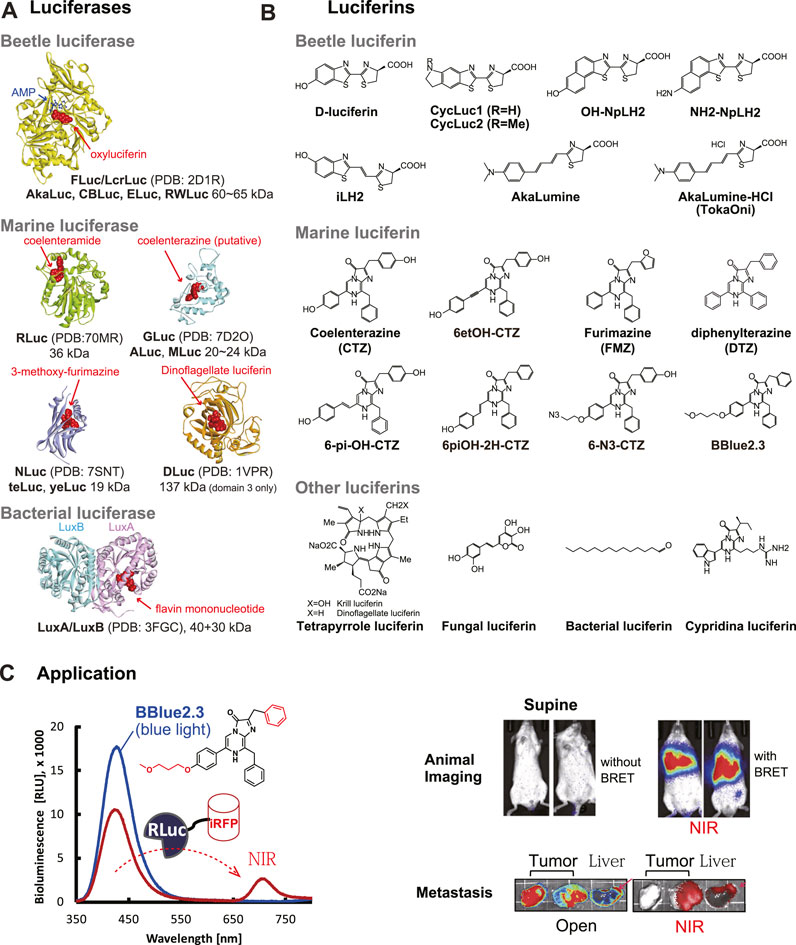
Figure 1. (A) The 3D structures of beetle and marine luciferases. The substrates in the active sites were highlighted with red balls. The 3D structures were obtained from the following references: FLuc/LcrLuc (2D1R) (Nakatsu et al., 2006), RLuc (7OMR) (Schenkmayerova et al., 2023), GLuc (7D2O) (Wu et al., 2020), NLuc (7SNT) (Nemergut et al., 2023), DLuc (1VPR) (Schultz et al., 2005), and LuxA/LuxB (3FGC) (Campbell et al., 2009). (B) Natural and synthetic luciferins for beetle and marine luciferases. (C) Mouse imaging with a CTZ analog BBlue2.3. BBlue2.3 emits strong blue light in the presence of RLuc. The blue light can excite the adjacent near-infrared fluorescent protein (iRFP) and is transferred to NIR at ca. 720 nm, because iRFP has a soret band at around 380 nm. This NIR light is useful for a deep tissue imaging of living mice. The optical images were reproduced from Kim et al. (Nishihara et al., 2019) with permission from Ivyspring under CC BY-NC 4.0.
Because BL shows obvious contrasts in the advantages and drawbacks, many of recent studies have been directed to tackle the drawbacks of BL, while capitalizing the advantages: e.g., 1) development of new luciferases and synthesis of novel luciferins; 2) protein engineering to create genetically encoded probes with luciferases; and 3) integration of all the ingredients including the probes into new imaging systems to address molecular events in living organisms. These three domains are closely related in the technical progression. Luciferases and luciferins with excellent optical properties enable us to design new optical probes with a novel strategy. The new bioluminescent probes are expected to facilitate better BL imaging (BLI) of molecular events of interest in live cells and animal models.
This review is intended to showcase the advantages and disadvantages of BL, and serve as a searchlight to find directions for future studies.
1.1 The “bright” side of bioluminescence
As BL has many distinctive advantages over the other optical readouts, it has been broadly used in various bioassays and molecular imaging modalities. The distinctive benefits of BLI are summarized as follow: (Loening et al., 2006; Novobilsky and Hoglund, 2020):
(i) High sensitivity in bioassay and molecular imaging. BL can occur in the presence of a minimal amount of luciferase and its specific substrate luciferin. The corresponding assays have a potential to be very sensitive in a dark chamber (de Wet et al., 1987). Even single cell can be imaged in living mouse models (Iwano et al., 2018).
(ii) The low background and high signal-to-noise (S/N) ratio. BLI systems can drastically suppress the background intensities before BL initiation. Thanks to the absence of inherent background luminescence in mammalian cells and animal models, BLI systems generally allow high S/N ratios.
(iii) High quantum yield (QY). The luciferin-luciferase reactions generally have a high quantum yield (QY), compared to the other chemiluminescence, and consequently a significant amount of light is generated for each chemical reaction that occurs. For example, the QY of firefly luciferase (FLuc) was reported to be ca. 41% (Ando et al., 2008), and the QYs of Cypridina and Aqeuorin were known to be ca. 30% and 16%, respectively (Shimomura, 2006). Although highly variable according to the luciferases, these QYs of BL are generally higher than those of other chemiluminescence (CL) such as 0.09% of Luminol (Ando et al., 2007).
(iv) Wider dynamic range of signals. BLI systems generally have wider dynamic ranges, compared to other modalities such as FLI. Although the dynamic ranges are manifold according to the imaging systems, they generally appear ranging from three to eight orders (Fan and Wood, 2007; Branchini et al., 2018). Recently advanced BLI systems with high sensitivity can determine even smaller number of cells in the dynamic range. For example, a new BLI system was previously highlighted with a simultaneous determination of BL cells ranging from 101 to 2 × 104 cells on a microplate (Nishihara et al., 2019). However, it is also true that the wide dynamic range depends on a number of parameters including the imaging system and the luciferin substrate concentration.
(v) No requirement of external light source. BLI does not require external light source. Because of this virtue, BLI does not cause light toxicity in living organisms and does not necessitate a complex measuring instrumentation equipped with optical filters to discern the signal from the background. This feature eventually contributes to low background and high signal-to-noise (S/N) ratio (Li Q. et al., 2019).
(vi) Quantitative imaging modality. BLI is considered as more quantitative than fluorescence-based imaging modality. Fluorescence (FL) is influenced by laser power for excitation, but BLI is not influenced by such an external light source. In a well-controlled experimental setup, BL intensities are proportional to the cell numbers. Thanks to the linearity, even the cell numbers are countable based on the BL intensities (Iwano et al., 2018; Nishihara et al., 2019).
(vii) Versatility in various imaging systems as an optical readout. BL systems are applicable to various bioassay modalities and organisms from bacteria to animals. For example, a luciferase can be embedded in various bioassays and nicely work as an optical reporter, which include bioluminescence resonance energy transfer (BRET), reporter-gene assay (RGA), protein-fragment complementation assay (PCA), and molecular strain probe systems.
(viii) Quick signal development. The BL initiation is easy and immediate without any pre-treatment nor complex instrumental build-up. The luciferase reporters immediately work after expression, whereas fluorescent proteins necessitate a maturation time (oxidation) of the fluorophore and a warming up time of the instrumentation (Laser) before measurement.
(ix) Low costs compared with other imaging modalities. BL is a low-cost imaging modality, compared with the other established ones such as PET, SPECT, and MRI (Massoud and Gambhir, 2003). BL is easily determined by a low-cost instrument, and the reagents are relatively cheap.
(x) Suitability in deep tissue imaging. BLI is suitable for a few milli- and centimeter order of deep tissue imaging (Massoud and Gambhir, 2003). Because of this virtue, it is generally useful for small animal imaging.
(xi) Biocompatible and non-radioactive feature. BL systems are originally derived from living organisms. Hence, all the ingredients for BL development are biocompatible and not harmful for living subjects.
(xii) Low possible false-positive signals. The luciferase-luciferin reactions are based on enzymatic specificity. Hence, the BL-based bioassays are less disturbed by false-positive signals. In contrast, FL-based assays are more likely disturbed by fluorescent ingredients in biological samples. BL-based bioassays are considered more quantitative and straightforward in the process of the data.
(xiii) Suitability for high-throughput screening of chemicals. BL-based biosensors are suitable for high-throughput screening for chemical biology and drug discovery applications due to their ability to maintain sensitivity, signal strength, and biological fidelity in automated systems (Azad et al., 2021).
(xiv) Secretory nature of some marine luciferases. Some marine luciferases from copepods are naturally secreted into the extracellular compartments of animal cells or the blood stream of animal models after expression (Takenaka et al., 2012). The feature enables us to easily and repeatedly measure the reporter levels in the culture medium or blood samples of animals without any pre-treatment step for separating reporters from the cell debris.
1.2 The “dark” side of bioluminescence
Conversely, the disadvantages of BL imaging and analysis may be summarized as follows:
(i) Low optical intensity of BL, compared with FL. BLI exerts relatively low photon emission, compared with the other imaging modality such as fluorescence imaging (FLI), although it generally exerts high QYs. For example, Emerald luciferase (ELuc) from click beetle generates as low as 1.17 × 104 photons/sec/cell with D-luciferin (Niwa et al., 2023). NanoLuc (engineered from deep sea shrimp luciferase) and FLuc emit 3.72 × 10−18 Watts/molecule (i.e., 8.6 photons/sec/molecule) and 7.25 × 10−20 Watts/molecule (i.e., 0.2 photons/sec/molecule), respectively (Klein et al., 2023). Because of the low photon emission, BLI is generally measured in a group of cells, and requires a long light acquisition time typically from seconds to minutes. Because of this feature, BLI is disadvantageous for monitoring temporal dynamics of molecular events in living organisms.
(ii) Necessity of the specific substrates. BLI necessitates a specific substrate as the “luminophore” and optionally needs cofactors such as Mg2+ and ATP. Because of this necessity, an excess amount of the substrate should be exogenously delivered for cell-based studies or injected into the blood stream of animals. Deficiency of the substrates or the cofactors can cause false-negative consequences.
(iii) The optical performance driven by the chemical property of the substrates. The substrate as a luminophore is the center of the optical properties of BL. The chemical properties determine the overall optical performance including the color and intensity.
For example, the hydrophilicity and π-electron conjugation of the functional groups of the substrate collectively determine the plasma membrane (PM) permeability, color, and optical intensity (Nishihara et al., 2019). If the functional groups are highly hydrophobic, the water solubility is poor and causes trouble in the injection of the substrate solutions into an animal.
(iv) Secretory nature of some marine luciferases. Some marine luciferases such as Metridia longa luciferase (MLuc) and Gaussia princeps luciferase (GLuc) from copepods have secretory nature, which can be a reason of the reporter loss in mammalian cells. This nature is generally suppressed by tagging with an endoplasmic reticulum (ER) retention signal like a C-terminal KDEL sequence. This ER retention can cause a new problem of the substrate supply. It is because the substrate has to pass through the two membrane barriers to reach out their ER-retained luciferases of live cells. To bypass this problem, secretory luciferases have been displayed on the PM of live cells to ease the substrate diffusion into the PM (Santos et al., 2009).
(v) The optical intensity influenced by reaction conditions. Because the BL is an outcome of an enzymatic reaction, any factors affecting enzyme reactions potentially influence the BL intensity and color. It includes pH, ion levels, temperature, buffer species, etc. This feature may be the reason that weakens the quantitative feature of BL. Considering every luciferase and luciferin has its optimal reaction conditions, one needs to optimize the assay conditions for the best optical performance.
(vi) Poor spatial resolution. BLI systems provide a less spatial resolution modality than other imaging modalities such as FLI. The spatial images of cells are generally smeared and dim, whereas FLI is much more sharp, vivid, and clear.
(vii) Potential cross-leakage of BL signals in multi-reporter systems. Because of the broad bandwidths of luciferases, the BL spectra are prone to be overlapped in multi-reporter systems. This feature causes an optical cross-leakage between the BL signals of multi-reporter systems. Optical filters may be useful for minimizing the BL signal contamination.
(viii) Potential interferences of chemicals in samples. Because BL depends on enzymatic reactions of luciferases, BL emission can be interfered by various small chemicals in samples. Such inhibitors include linear, planar compounds including a benzothiazole, benzoxazole, or benzimidiazole core. It is because the benzothiazoles bind to the D-luciferin pocket of FLuc as blockers (Thorne et al., 2012). Resveratrol and compounds containing m-carboxylate group are also known to inhibit FLuc activities (Bakhtiarova et al., 2006). RLuc is also inhibited by compounds containing an aryl sulfonamide core (Ho et al., 2013). It is also known that cAMP-dependent protein kinase (PKA) inhibitor H89 works as an inhibitor of RLuc (Herbst et al., 2009). These inhibitory feature of some chemicals in samples works false-negatively and is a unique disadvantage of BL assay systems.
2 Beetle and marine luciferases
2.1 Outline of luciferases
There are many light-emitting organisms in nature. From the natural sources, many luciferases emitting cold light have been established. The chemical study on the light-emitting mechanism did not begin until the early 20th century. The luciferin–luciferase reaction of fireflies was first demonstrated by Harvey (1917). FLuc was partially purified and characterized for the first time by McElroy et al. in 1955, and then crystallized by Green et al. in 1956 (Shimomura, 2006). The beetle luciferases are considered to have evolved from the ancestral fatty acyl-CoA synthetase (ACS), an enzyme, which is present in all insects (Adams and Miller, 2020). The specifications of luciferases were briefly summarized in Table 1 and especially important ones were highlighted in this section and in Figure 1A. The corresponding luciferins from natural resources were briefly summarized in Figure 1B. A typical example of animal imaging with an CTZ analog substrate was shown in Figure 1C.
Because the luciferases from natural resources are limited in the color variety in the relatively shorter wavelength, a lot of efforts have been directed to extend the color palette to the red and infrared side (650–950 nm) in order to achieve the minimized light attenuation and tissue absorption of the optical signal. The red- and blue-shifted BL has been achieved through mutagenesis of luciferases, together with synthesis of the specific substrates with different luminophore structures (Iwano et al., 2013; Mezzanotte et al., 2017; Iwano et al., 2018). The specific mutation sites were depicted in Figure 2.
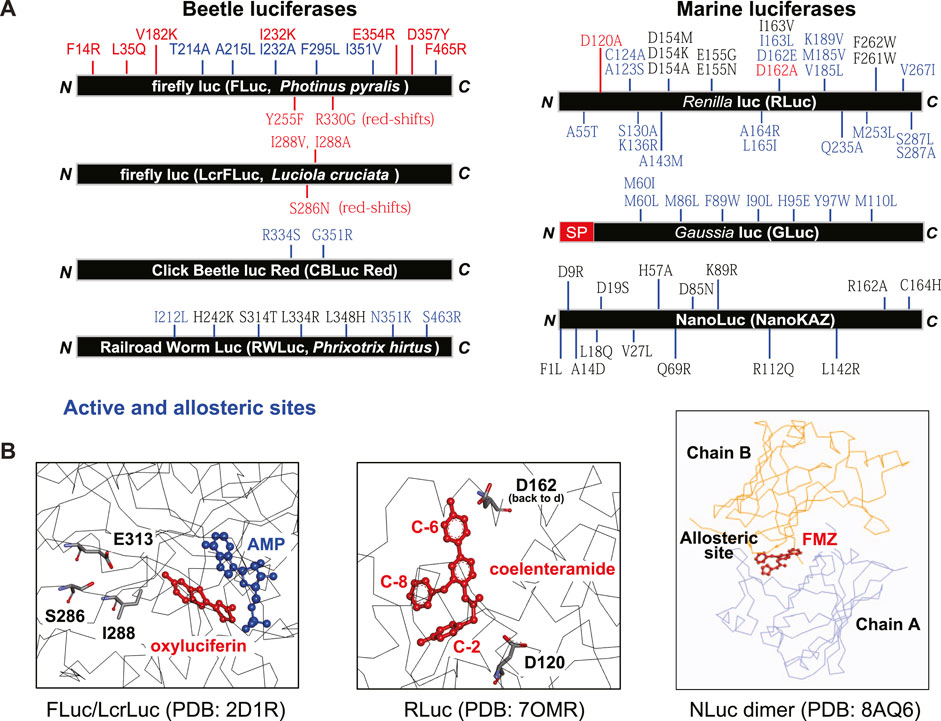
Figure 2. (A) Summarized map showing the mutation sites of various luciferases for improving the optical performances. FLuc (Japanese Genji-Botaru, Luciola cruciate) (Nakatsu et al., 2006), Railroad worm luciferase (Li et al., 2010; Bevilaqua et al., 2019), Click beetle luciferase red (Hall et al., 2018), Renilla luciferase (Loening et al., 2006; Loening et al., 2007; Woo and von Arnim, 2008; Loening et al., 2010), Gaussia luciferase (Welsh et al., 2009; Kim et al., 2011a), and NanoLuc (Yeh et al., 2017; Inouye et al., 2022; Nemergut et al., 2023). (B) The substrate-bound active and allosteric sites. Known, effective mutation sites in the active sites were highlighted by stick structures (left and center), and the FMZ-bound allosteric site between chains A and B is shown in the right. The 3D structure information was obtained from the following references: FLuc/LcrLuc (2D1R) (Nakatsu et al., 2006), RLuc (7OMR) (Schenkmayerova et al., 2023), NLuc (8AQ6) (Nemergut et al., 2023).
Among beetle luciferases, firefly luciferase (FLuc) derived from North American Photinus pyralis (Ppy) has been exceptionally well investigated and has been widely utilized in various BLI systems (Adams and Miller, 2020). The native luciferases have been modified by random and site-directed mutagenesis for improving the optical properties including the intensity, stability, color, and spectra. Because the crystal structures of Ppy FLuc and Japanese Luciola cruciata (Lcr, Genji-botaru) FLuc were reported (Conti et al., 1996; Nakatsu et al., 2006), their active sites have been a target of the mutagenesis for improving the optical properties including red-shifted BL. The mutation sites in FLuc share consensus sites with the other beetle luciferases such as click beetle and railroad worm luciferases (Figure 2) (Nakatsu et al., 2006; Li et al., 2010).
One of the most successful examples is the red-shifted BL with the pair of Akaluc (luciferase) and Akalumine (luciferin). A bright mutant named Akaluc through random mutagenesis of FLuc was established, which has 28 amino acid mutations across the sequence (Iwano et al., 2013). Akaluc shows improved thermostability and robust BL with the maximal emission peak at 650 nm with the substrate Akalumine as an optimized variant of D-luciferin.
Marine luciferases derived from marine organisms such as sea pansy and copepods, catalyze the oxidation of their specific substrates to produce BL. Unlike the beetle luciferase, the marine luciferases require only molecular oxygen (O2) for the reaction, but do not necessitate cofactors such as ATP and Mg2+. The marine luciferase includes the sea pansy Renilla reniformis (Lorenz et al., 1991), the copepod G. princeps (Verhaegent and Christopoulos, 2002) and M. longa (Markova et al., 2004), the ostracod Cypridina noctiluca (61 kDa) (Nakajima et al., 2004), the dinoflagellate Pyrocystis lunula (Lecuyer et al., 1979), and the deep sea shrimp, Oplophorus gracilirostris (Inouye et al., 2000). GLuc and MLuc were cloned from copepods G. princeps and M. longa, respectively, and successfully utilized in BLI of mammalian cells (Santos et al., 2009). A few years later, Takenaka et al. established 25 kinds of new copepod luciferases including M. pacifica luciferases (MpLuc1, and 2) (Takenaka et al., 2008; Takenaka et al., 2012).
The marine luciferases can be divided into secretory and non-secretory ones. RLuc and its variants are the representative non-secreted luciferases, whereas many of the other marine luciferases such as copepod luciferases (GLuc) and M, longa luciferase (MLuc)) are innate secretory ones.
RLuc was originally established by Karkhanis et al. (Shimomura, 2006). Many RLuc variants were created by mutagenesis based on “consensus sequence-driven mutagenesis strategy (CSMS)” (Loening et al., 2006). This strategy was developed on the premise that frequently occurring amino acids at a given position allow a larger thermostabilizing effect than less frequent amino acids. This approach allowed them to find potential mutagenesis sites from the aligned sequences of the same lineage of luciferases. Thanks to the strategy, Loening et al. established various red-shifted variants of RLuc and applied them to animal models (Loening et al., 2007; Loening et al., 2010). The mutants were named RLuc8, RLuc8.6, RLuc8.6-535, RLuc8.6-535SG, etc.
Kim et al., expended the concept of the CSMS to artificially construct whole amino acid sequences (named Artificial luciferases (ALuc)), whose identities are distinctive from any existing luciferases (Kim et al., 2013; Kim et al., 2017). The ALucs have been made through extracting and linking frequently occurring amino acids from an alignment of many existing copepod luciferases in public databases, where copepod luciferases were selected because they are the smallest ones among luciferases with two-repeated catalytic domains like a mirror image, besides the variable N-terminal domain including the secretion signal (Inouye and Sahara, 2008). This effort to create new ALucs is ongoing to date (Kim et al., 2023c).
Another marine luciferase from deep sea shrimp O. gracilirostris (OLuc) was engineered to develop the brightest NanoLuc. OLuc consists of two 35 kDa subunits and two 19 kDa subunits (Shimomura et al., 1978). NanoLuc (or NLuc) is established through random and direct mutagenesis of the smaller luciferase unit of OLuc (19 kDa). Although it utilizes native coelenterazine (CTZ) as the substrate, a CTZ analog named furimazine (FMZ) boosts the BL intensity by 25-fold (Hall et al., 2012). The red-shifted BL of NanoLuc was further achieved through two pathways: one is by mutagenesis of NanoLuc and the other is by organic synthesis of the specific substrates. Yeh et al. (2017) synthesized a series of FMZ analogs exerting red-shifted BL with NanoLuc. They further established bright and red-shifted variants of NanoLuc named teLuc and yeLuc. The maximal intensities of teLuc and yeLuc were found to be 502 nm and 527 nm, respectively (Yeh et al., 2017). The allosteric behavior of NanoLuc also has been investigated using experimental and computational techniques for further engineering (Nemergut et al., 2023).
2.2 The merits and demerits of beetle luciferases
Beetle luciferases derived from insects necessitate the cofactors, ATP and Mg2+, besides the specific substrate and molecular oxygen (O2) for BL emission. Beetle luciferases oxidize the substrate luciferins and lead them to the excited state. The excited intermediates emit photons when they relax to the ground state. Many researchers capitalize on this mechanism to modify the luciferase and/or luciferin scaffolds for improving the optical intensity and color variety of BL.
These beetle luciferases provide the following exclusive advantages:
(i) Red-shifted BL spectra. The BL spectra of beetle luciferases are generally witnessed at the wavelength region longer than those of marine luciferases. This feature minimizes the light attenuation in physiological samples and animal models.
(ii) BL signal stability. Beetle luciferases generally develop stable BL signals. This feature positively works for the fidelity of the bioassays, compared to marine luciferases implementing busting and rapidly decaying BL signals.
(iii) Low autoluminescence. The possibility of autoluminescence of beetle luciferase systems is much lower than that of marine luciferase systems. Because beetle luciferase-luciferin reaction necessitates ATP and Mg2+, besides O2, the threshold for BL emission is considered high.
The beetle luciferases also have the following disadvantages:
(i) Large molecular weights. Beetle luciferases are generally large in the molecular weights, compared to marine luciferases. For example, the molecular weight (Mw) of 62 kDa of FLuc is approximately two to three fold larger than those of marine luciferases such as RLuc (36 kDa) and NanoLuc (19 kDa) (Auld and Inglese, 2016). Because of this bulkiness, beetle luciferases as a reporter can cause steric hinderance to the host probes in bioassay systems.
(ii) Dimmer BL intensity. The absolute BL intensities of beetle luciferases are considered generally dimmer than those of marine luciferases. For example, NanoLuc/FMZ pair was reported much brighter than FLuc/D-luciferin pair in human embryonic kidney (HEK) 293T cells (Yeg et al., red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging). Even in RGAs, NanoLuc and GLuc were reported to implement better BL intensity and signal-to-background ratios than FLuc in RGAs (Neefjes et al., 2021). Humanized GLuc is also extremely brighter than humanized RLuc and FLuc (Tannous et al., 2005).
(iii) Requirement of cofactors. Beetle luciferases necessitate cofactors, ATP and Mg2+, besides the specific substrate D-luciferin and O2. In contrast, marine luciferases need the specific substrate D-luciferin and O2 only for BL emission. Any deficiency of the cofactors or uneven biodistribution can implement false-negative results.
2.3 The merits and demerits of marine luciferases
(i) Strong BL intensity. It is generally accepted that the BL intensities of marine luciferases such as NanoLuc and GLuc variants are generally brighter than those of beetle luciferases.
(ii) Small molecular weights. Marine luciferases are generally smaller than beetle luciferases in the size. In contrast of the 62 kDa of FLuc, marine luciferases are ranging from 15.7 to 20 kDa in the molecular weights (Auld and Inglese, 2016). This compactness exerts less steric hinderance to the host molecule after labeling.
(iii) Simplicity in the light emitting mechanism. Marine luciferases follow a relatively simple mechanism to generate BL: The reaction is initiated by the binding of O2 at the C-2 position of the CTZ, giving peroxide. The peroxide then forms a four-membered ring “dioxetanone.” The dioxetanone promptly decomposes as a result of the splitting of two binds in a concerted fashion, producing CO2 and the amide anion of coelenteramide in its excited state. The excited state of the amide anion emits light when the energy level falls to the ground state (Shimomura, 2006). In contrast, beetle luciferases follow complex steps to emit BL in the presence of ATP and Mg2+. It indicates that the BL emission steps are more likely to be influenced by other factors and ingredients.
Marine luciferases have the following common demerits as optical readouts in bioassays and molecular imaging.
(i) BL emission in a shorter wavelength range. Marine luciferases generally emit BL at the shorter wavelength region. The shorter wavelengths suffer from severe light attenuation in the tissue of living subjects. For example, a luciferase from Oplophorus gracilorostris (deep sea shrimp) is a typical marine luciferase emitting blue BL (462 nm) through oxidizing CTZ in the presence of O2. In mammalian tissues, hemoglobin impairs the BL transmission below 600 nm.
(ii) Limited color palette. Marine luciferases have a limited range of the available color palette, especially deficiency in red-shifted colors above 550 nm (Weihs and Dacres, 2019). This is especially disadvantageous in multi-reporter systems.
(iii) Bursting but rapid decay of the bioluminescence. Marine luciferases generally exert bursting BL emissions with the substrate. However, the BL quickly decays, too. Because of this nature, the measurement requires a luminometer equipped with an injector to measure the transient peak BL.
(iv) Autoluminescence. Because marine luciferases require only O2, besides the substrate, the threshold of BL emission is relatively low, compared to that of beetle luciferases. Even in the absence of the marine luciferases, the representative substrates, CTZ and its analogs, are prone to be decomposed in physiological samples and exert relatively high autoluminescence (Nishihara et al., 2019). This feature makes sample handling difficult and increase error bars in bioassays.
(v) Potential influence of multivalent cations. Among luciferases, copepod luciferases are especially sensitive to multivalent cations (Kim et al., 2015). An excess amount of multivalent cations can enhance or inhibit the activities of copepod luciferases, because copepod luciferases have unique EF-hand-like structures, which are known as the binding site of Ca2+ (Kim and Izumi, 2014).
(vi) Poor folding efficiency in mammalian cells. Some marine luciferases have problems with the folding efficiency in mammalian cells and require special consideration on it such as codon optimization and folding locations. Copepod luciferases especially contain rich disulfide bonds to stabilize the structure, and thus are prone to be misfolded in the cytosol of mammalian cells.
3 Luciferins
Substrate is a key component for glowing BL with luciferases. The role of substrates was previously explained as “luminophore,” because it is the energy source and oxidized by luciferases to generate BL. The luciferases recruit the “luminophore” substrate from the aqua phase, whereas fluorescent proteins embed the “fluorophore” inside the molecular scaffold. It is interesting to note that the chemical structure of the common substrate of marine luciferases, CTZ, is similar to the fluorophore of fluorescent proteins (65SYG67), sharing the same imidazolone ring structure (Kim et al., 2012).
While luciferases are extremely diverse, luciferins are surprisingly simple to be categorized, because a limited number of specific luciferins (to date only 12) are shared by many luciferases. Thus, BL systems can be surprisingly simplified into a few groups according to the substrate luciferins. Among them, D-luciferin and CTZ occupy the major portion.
A recent trend in luciferin studies is to append functionalities to the natural luciferins using organic synthesis. The authors classified and explained general luciferins in the “non-activatable luciferin” section, while more functional luciferins, e.g., containing on-off switches were summarized below in the “activatable substrates” section. The chemical structures were depicted in Figure 3.
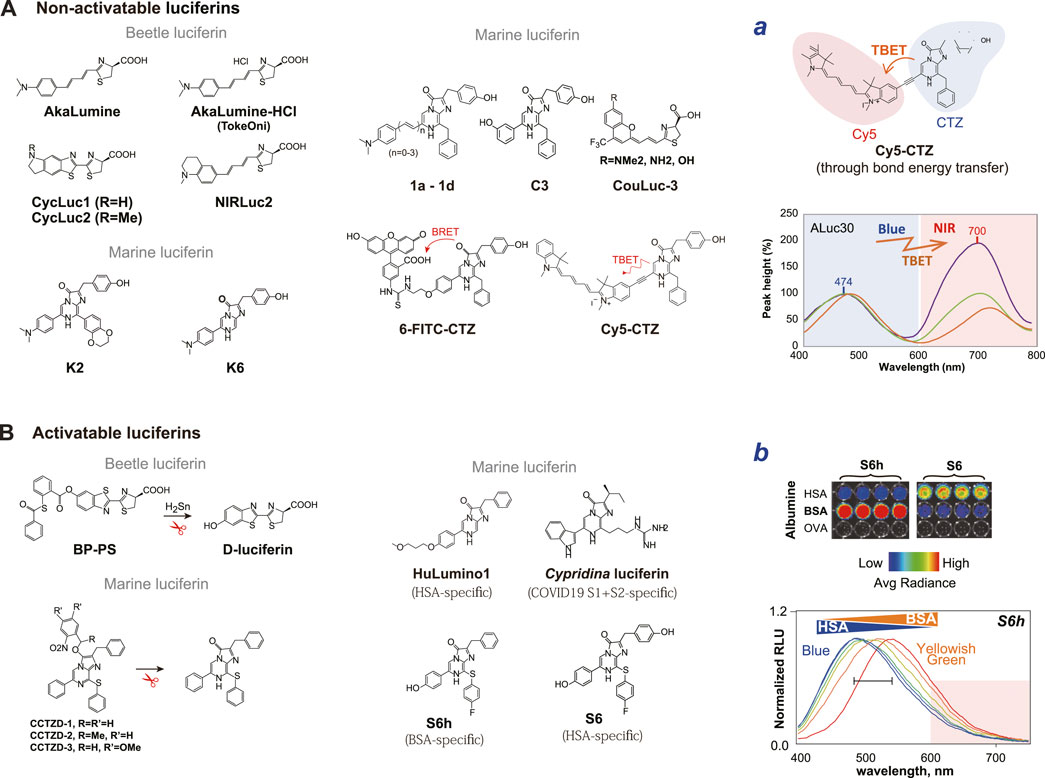
Figure 3. (A) The chemical structures of non-activatable luciferins. The non-activatable substrates are functional but cannot actively recognize analytes. The right panel (Inset a) exemplifies the optical spectra of Cy5-CTZ. The optical images were reproduced from Kim et al. (Kim et al., 2023d) with permission from Springer. (B) The chemical structures of activatable luciferins. The activatable luciferins can determine analytes and report BL. The right panel (Inset b) exemplifies albumin-specific light emission of S6h and S6. The color generated by S6h is variable according to the ratios of the bovine and human serum albumins. The optical images were reproduced from Kim et al. (Kim S.-B. et al., 2023) with permission from MDPI. The specific substrate names are referred from the following references: AkaLumine (Iwano et al., 2013); AkaLumine-HCl (TokeOni) (Iwano et al., 2013); CycLuc1 and 2 (Li Q. et al., 2019); NIRLuc2 (Ikeda et al., 2020); K2, K5, and K6 (Kamiya et al., 2022b); 1a-1d (Tamaki et al., 2021); C3 (Kamiya et al., 2022a); 6-FITC-CTZ (Nishihara et al., 2018); Cy5-CTZ (Abe et al., 2019); BP-PS (caged luciferin) (Li J. B. et al., 2019); CCTZD-1, -2, -3 (caged CTZ analog) (Zhang et al., 2018); S6 and S6h (Kim S.-B. et al., 2023; Kamiya et al., 2023); HuLumino1 (Nishihara et al., 2020); and Cypridian luciferin (Nishihara et al., 2024).
3.1 Non-activatable luciferins
Luciferin is an essential component for BL reaction as the energy source. Luciferins are supplied by the biosynthesis in living organisms, although most of the mechanisms are unknown. While a myriad number of light-emitting species exist in nature, their luciferin repertoires are astonishingly small. The representative ones include 1) beetle luciferin (D-luciferin), 2) coelenterazine (CTZ), 3) tetrapyrrole luciferin, 4) fungal luciferin, 5) bacterial luciferin, and 6) Cypridina luciferin (Fleiss and Sarkisyan, 2019) (Figure 1). The disadvantages of the natural luciferins may be explained as follows:
Disadvantages of D-luciferin. D-luciferin as the common substrate for beetle luciferases is unevenly biodistributed in mice. After i.v. injection, D-luciferin immediately shows high uptake in the kidney and liver. Later, it is predominantly accumulated in the bladder, but minimally diffused to the brain (Berger et al., 2008).
Disadvantages of CTZ. The common substrate for the marine luciferin, CTZ, has relatively poor solubility in hydrophilic solvents and chemically unstable property.
The principal role of these natural luciferins is to luminesce with luciferases, and do not have additional functionalities such as analyte sensing and conditional color variation. In this review, we introduce a series of substrates that have intentional chemical modification of the native substrates, which are named “non-activatable luciferin.” This review focuses on the D-luciferin and CTZ because they are the most used and representative substrates of beetle and marine luciferases (Figure 3).
D-luciferin consists of a benzothiazole moiety attached to a thiazole carboxylic acid moiety (Oba et al., 2013). These two moieties have been modified for better optical properties such as red-shifted BL and the history proves that such modified D-luciferins are tolerated by beetle luciferases.
Iwano et al. (2013) synthesized an analog named Akalumine to generate NIR BL by extending the π-conjugation length between the benzothiazole and thiazole moieties, and established the red-shifted BLI system with a FLuc mutant Akaluc. They accomplished a diverse hue of visible and NIR BL, whose longest emission maximum was observed at 675 nm.
One important modification of D-luciferin was achieved through alkylation and cyclic alkylamino modification of the hydroxy group (-OH) (Otto-Duessel et al., 2006). The alkyl and aminoluciferins are further modified by bulky fluorescent dyes for red-shifted emission of BL (Mezzanotte et al., 2013). A cyclic alkylamino modification of the OH group of Akalumine was also conducted by Ikeda et al. (2020).
Prescher et al. rationally designed a unique class of NIR-emitting coumarin luciferin analogs specifically recognizing a mutant luciferase and emitting NIR BL (Yao et al., 2021; Love et al., 2023). These types of luciferins are suitable for multiplexed imaging of molecular events in animal modalities.
CTZ is the luciferin of a wide variety of marine luciferases including RLuc and GLuc, and is another modified tripeptide that is biosynthesized from one phenylalanine and two tyrosine residues (Shimomura, 2006). This CTZ-based imaging system naturally emits BL in blue in the presence of O2. Modification of the C-2, C-6, and C-8 positions of the CTZ backbone has been actively studied by several research groups to exert 1) luciferase specificity, 2) red-shifted BL, and 3) enhanced BL. These modified CTZ analogs are known to be tolerated by marine luciferases.
High selectivity to RLuc is accomplished by introducing a double bond at the C-6 position, besides the elimination of OH group at the C-2 position (Nishihara et al., 2017). It is also known that an ethynylation at the C6 position enhances the overall optical intensities of the analogs with ALucs (Nishihara et al., 2017). Kamiya et al. (2022b) also reported that elimination of the functional group at the C-8 position, named K5 and K6, is useful for the RLuc specificity, whereas the bulkiness at the C-8 position, named K2, is driven to ALuc specificity.
Red-shifted BL is obtained through conjugating fluorescent dyes to the C-2 or C-6 position of the backbone of native CTZ (Nishihara et al., 2018). For example, a CTZ analog named Cy5-CTZ was prepared by conjugating Cyanine-5 (Cy5) dye to CTZ through an acetylene linker. The acetylene linker enables through-bond energy transfer (TBET) between the energy donor CTZ and the energy acceptor Cy5. This novel CTZ analog is intrinsically fluorescent and emits NIR-shifted luminescence in the Cy5 channel upon reacting with appropriate luciferases, RLuc and ALuc (Abe et al., 2019; Kim et al., 2023d).
Even blue-shifted CTZ analogs can be utilized for NIR BLI in animal models through a large stock-shift of NIR fluorescent proteins (iRFPs) (Nishihara et al., 2019). It was accomplished through combining the blue-shifted CTZ analog with RLuc8.6SG-linked iRFPs.
One of the most desirable functionalities of luciferin may be red-shifted BL, which can minimize the light attenuation in physiological samples or in the tissues of living subjects. Abe et al. (2019) recently synthesized a CTZ analog that emits NIR BL. The CTZ analog named Cy5-CTZ was prepared by conjugating Cy5 dye to CTZ through an acetylene linker. The acetylene linker enables TBET between the energy donor CTZ and the energy acceptor Cy5. This novel derivative is intrinsically fluorescent and emits NIR-shifted luminescence in the Cy5 channel upon reacting with an appropriate luciferase, RLuc. The authors demonstrated that Cy5-CTZ is optically stable in physiological samples, rapidly permeabilizes through the plasma membrane, and luminesces NIR BL.
3.2 Activatable luciferins
This review defines “activatable luciferins” as chemically modified luciferins for exerting special abilities such as responding to a specific stimulator or recognizing a target protein, and emitting optical signal(s), together with the original role as the energy source for light emission.
A prototypical “photoactivatable” luciferin should embed a conceptional on-off switch in response to a specific stimulator or a molecular event. For this purpose, the functional groups of luciferins have been chemically modified.
Caged luciferins are typical photoactivatable means, where the substrate itself is protected by a cage and released by a specific signal or molecular event. These caged luciferins commonly carry a bulky group that blocks the interaction of luciferin backbone with the enzyme. The cages are removed by specific wavelengths of UV irradiation in animal models (Zhang et al., 2018), a stimulator (analyte) including oxygen species (Kojima et al., 2015), copper ions (Heffern et al., 2016; O'Sullivan et al., 2022), calcium ion (Tian et al., 2022), and labile iron levels (Aron et al., 2017).
The balance of sulfur redox plays an important role in maintaining homeostasis of the living subjects. The D-luciferin backbone was modified to visualize endogenous hydrogen polysulfides (H2Sn) and applied to animal models (Li J. B. et al., 2019). Likewise, some active substrates that is sensitive to reactive nitrogen species (RNS) have also been synthesized (Li et al., 2018).
Some CTZ analogs with special functional groups can specifically recognize serum albumins from different sources. These CTZ analogs bind the drug binding sites I and/or II of serum albumins and generate luminescence signals, where the albumins work like pseudo-luciferase. Based on this concept, Nishihara et al. reported a CTZ indicator for imaging human serum albumin (HSA) (Nishihara et al., 2020). Kim et al. (2023a) also developed albumin indicators, S6 and S6h, for sensing HSA and bovine serum albumin (BSA). It was also reported that Cypridina luciferin quantifies the spike protein levels of COVID-19 (SARS-CoV-2) acting as a pseudo-luciferase (Nishihara et al., 2024).
The optical properties of activatable luciferins may be summarized as follows:
(i) Minimized autoluminescence. Caged luciferin is another important category of D-luciferin analogs. Caged luciferin cannot luminesce until an enzymatic cleavage event occurs and thus can minimize the autoluminescence. β-Galactoside-linked D-luciferin was previously introduced, which is only active in case its galactoside moiety is removed by the co-expressed β-galactosidase (Porterfield et al., 2015).
(ii) Low BL intensity. Activatable substrates are chemically modified with bulky groups for implementing an on-off switch in the backbone. The modified substrates generally weaken the BL intensities, compared to the original ones.
4 Bioluminescence imaging (BLI) systems
The ingredients, luciferins and luciferases, have been systemized for bioassays and molecular imaging.
The BLI systems have been divided into “static” and “activatable” groups (Reumann et al., 2010). Kim et al. (2011b) also categorized the BLI systems into two major groups: 1) a family of genetic and transcriptional probes (GTPs), and 2) a family of non-transcriptional and activatable probes (NAPs) (Figure 4).
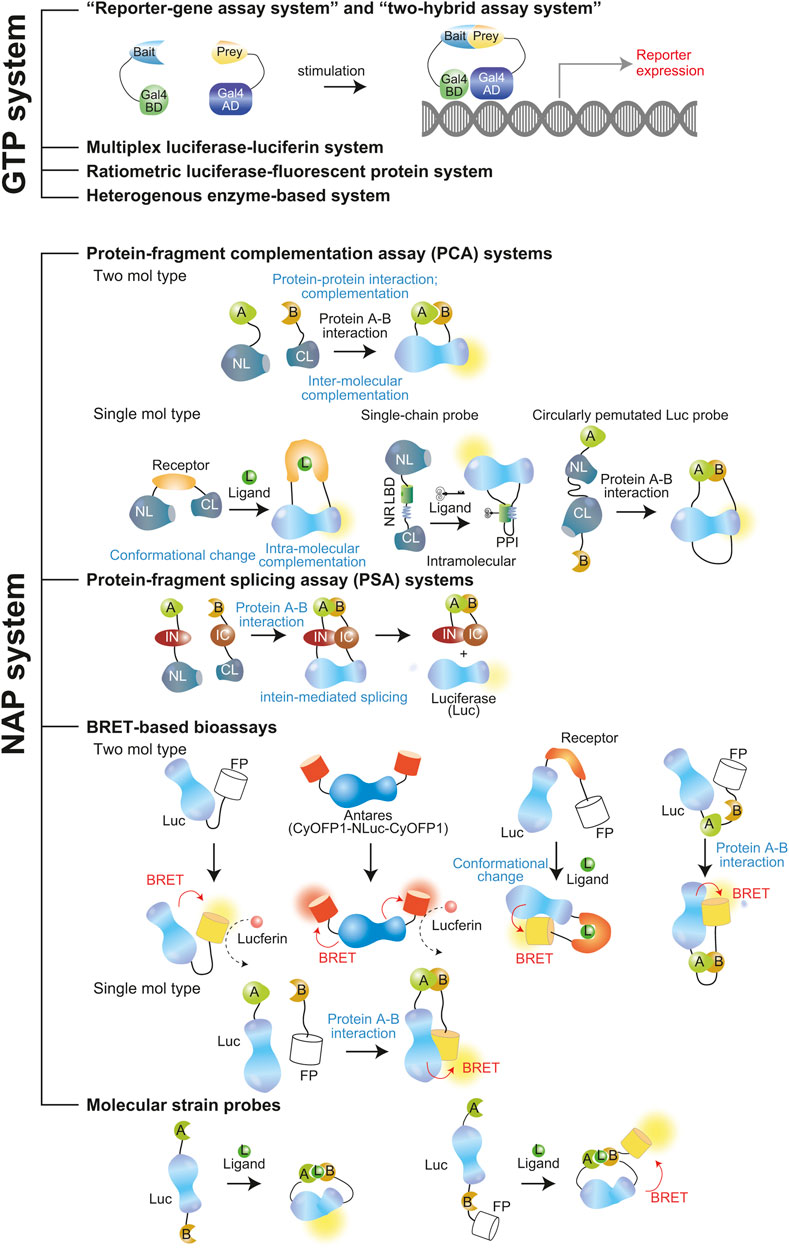
Figure 4. Schematic illustration of GTP and NAP systems. The molecular probes were referred to as the following names: Reporter gene assay system and two-hybrid assay system (Fetchko and Stagljar, 2004; Neefjes et al., 2021), Protein-fragment complementation assay (single- and two-chain types) (Paulmurugan and Gambhir, 2006; Kim et al., 2019a), Protein-fragment splicing assay (Ozawa et al., 2001a; Kim et al., 2004), BRET-based bioassays (Yeh et al., 2017; Nishihara et al., 2019), Molecular strain probes (Kim et al., 2016; Bae Kim et al., 2020).
Typical feature of GTP comprises transcription of the luciferase in the imaging process. This family of probes generally requires a long ligand-stimulation time until sufficient accumulation of the reporter protein is reached. This is mainly controlled at the promoter level. An RGA and a two-hybrid assay are grouped in this family. On the other hand, NAPs are expressed beforehand and pre-localized in adequate intracellular compartments of interest. The probe is ready to develop BL upon stimulation of a signal. Therefore, the NAPs intrinsically respond quickly to signals and generally are expected to have higher S/N ratio than GTPs. PCA, protein-fragment splicing assay (PSA), and single-chain probes, are a few which are categorized under NAPs.
The NAPs can be further categorized into split-reporter imaging systems and split-free imaging systems. The details are explained in the following sections.
4.1 Genetic and transcriptional probe (GTP) systems
Luciferases as an optical reporter can be genetically engineered to construct various optical probes. GTP systems typically comprise a transcription step of the luciferase as a reporter in the imaging process. Therefore, the on-off switch of this type of probe systems depends on the gene transcription machinery in live cells or living subjects. These imaging probes conceptionally embed a full-length luciferase, termed “split-free reporter imaging system”.
The most broadly utilized GTP systems are “reporter-gene assay system” and “two-hybrid assay system.” These systems commonly make use of an expression vector encoding a specific promoter sequence linked to the coding region that regulates the transcription of the reporter luciferase. A ligand-activated transfection factor binds to the promoter sequence and triggers expression of the reporter luciferase accumulation in cells which can be measured at various time points. The high-throughput mammalian two-hybrid applications, facilitate reporter genes use in large-scale interactome mapping and drug discovery projects.
The GTP systems can comprise multiple reporters and be utilized in various molecular imaging aspects as an expanded tool box of bioluminescent imaging. They may be conceptionally summarized into three categories:
(i) Orthogonal multiplex luciferase–luciferin system. These GTP systems let multiple luciferases coexist in the same context. They oxidize completely distinctive luciferins without cross-talks and thus one can easily be distinguished for each BL signal (Chan et al., 2012). Recently, a triple reporter gene assay system was developed based on a humanized mushroom luciferase, besides NanoLuc and FLuc (Mujawar et al., 2023). Kim et al. also developed a multiplex reporter system through combining two luciferases with each luciferase-specific luciferins, which was newly synthesized (Nishihara et al., 2017). Similar orthogonal luciferase–luciferin systems have been developed by Jones et al., who synthesized many luciferins to find positive binding pairs with each luciferase (Jones et al., 2017). These systems allow researchers distinguish the signal from each other.
(ii) Ratiometric luciferase-fluorescent protein system. One of the most direct and intuitive methods is to consecutively link a couple of fluorescent and bioluminescent reporters. A pH-sensitive BL reporter system named “pHLuc” is an excellent example for such reporter system (Ong et al., 2020). pHLuc enables us to ratiometrically determine pH levels in living cells and animal models through a linkage of pH-sensitive green fluorescent protein (GFP) with pH-stable full-length luciferases, Antares and NanoLuc have been established. pHLuc successfully visualized variance in the level of acidosis across the tumor.
(iii) Heterogenous multiplex imaging system. This unique system consists of a luciferase–luciferin system combined with other reporter enzymes like β-galactosidase (Martin et al., 1996). and constitutes a multiple reporter system. FLuc and β-galactosidase activities are determined with the same aliquot of cell lysates. Therefore, one can reduce manual labor and increase experimental accuracy.
The advantages of GTPs may be summarized as follows:
(i) Superior BL intensities. GTPs make use of full-length luciferases as an optical readout and thus are generally strong in the BL intensities. In contrast, molecular probes carrying dissected luciferase fragments for a temporal loss and conditional recovery of the activities such as PCA and PSA are generally poor in the absolute BL intensities. It is because this method inevitably hampers the intrinsic enzymatic property of the luciferase and actually recovers merely 0.5%–5% of the original luciferase activity (Paulmurugan and Gambhir, 2005; Kim et al., 2007c).
(ii) Simple molecular designs. Bioassay probes using full length luciferases as an optical readout generally have simple molecular designs. Conversely, molecular probes for PCAs generally require a sophisticated probe design and a tedious optimization step for deciding a suitable splitting site in the luciferase.
The disadvantages of GTPs may be summarized as follows:
(i) Poor S/N ratios. Bioassay systems using a full-length luciferase as an optical readout generally have a poor S/N ratio because they do not have strict on-off switches. For example, the basal physiological activities implement slow accumulation of the reporter protein in reporter gene assays, elevating the background signals.
(ii) Long stimulation time. The expression of a reporter gene requires a long stimulation time for the reporter accumulation to obtain sufficient S/N ratios. Hence, monitoring temporal dynamics of target proteins is limited.
(iii) Representing a limited kind of molecular events. Reporter-gene assays using a full-length luciferase make use of the expression machinery in the nucleus of cells as the on-off switch. Therefore, the assay is suitable for determining the molecular events implementing the protein expression inside the cells.
4.2 Non-transcriptional and activatable probe (NAP) systems
NAP systems express their probes beforehand and pre-localized in adequate intracellular compartments of interest. The probe itself embeds a conceptual on-off switch and is ready to develop an optical signal upon any stimulation of analytes. NAP systems are best for studies of the spatial and temporal dynamics of protein pairs.
The typical examples of NAP systems include 1) PCA systems, 2) PSA systems, 3) BRET-based Bioassays, and 4) molecular strain probes.
PCA and PSA probes occupy the main body of NAPs. While PCAs and PSAs have many merits as bioassays, it is tedious and time-consuming to determine the optimal dissection sites of each luciferase for the design of the probes. To address this issue, Kim et al. previously suggested a hydrophilicity search of the amino acid sequence using the scale of Kyte and Doolittle (Swiss Institute of Bioinformatics, SIB) (Kyte and Doolittle, 1982). Kim (2012) suggested that this search on a remarkably hydrophilic region empirically narrow down the potential dissection site region for PCA and PSA. The approach is empirically confirmed with the proven, successful dissection sites from Kim et al. and other researchers (Figure 5; Table 2) (Luker et al., 2004; Paulmurugan and Gambhir, 2007). This approach is explained as a remarkably hydrophilic interface in the middle of the sequence should be exposed to the aqueous phase and preferably accessible by the substrate in the aqueous phase. Hence, we can easily exert temporal inactivation of the luciferases by dissection at the hydrophilic region and the exposed hydrophilic sites are preferably survived in the context of animal cells after expression.
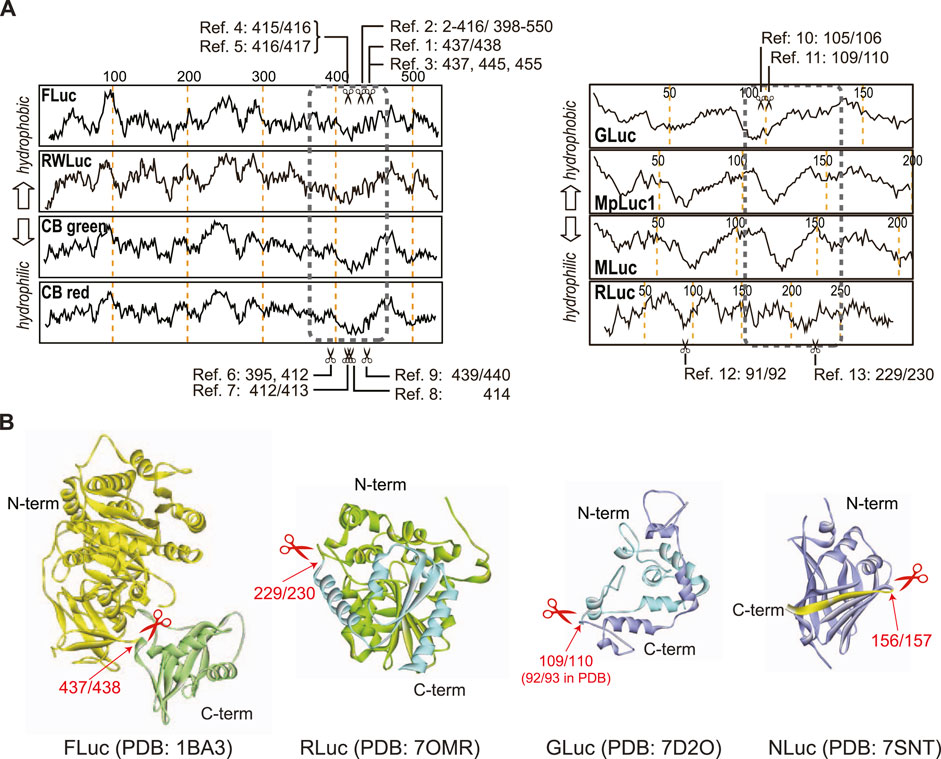
Figure 5. (A) Hydrophilicity map of amino acids and representative dissection sites (scissors marks), that is reported in literatures. Each reference number refers to the paired citation: Ref. 1 (Ozawa et al., 2001a), Ref. 2 (Luker et al., 2004), Ref. 3 (Paulmurugan and Gambhir, 2005), Ref. 4 (Kim et al., 2007a), Ref. 5 (Kim et al., 2007b), Ref. 6 (Villalobos et al., 2010), Ref. 7 (Kim et al., 2008), Ref. 8 (Hida et al., 2009), Ref. 9 (Kim et al., 2007c), Ref. 10 (Kim et al., 2009b), Ref. 11 (Remy and Michnick, 2006), Ref. 12 (Kaihara and Umezawa, 2008), and Ref. 13 (Paulmurugan and Gambhir, 2003). (B) The suitable dissection sites of beetle and marine luciferases. The 3D structures of FLuc, RLuc, Gluc, and NanoLuc were obtained from the reference structures, 1BA3 (Franks et al., 1998), 7OMR (Schenkmayerova et al., 2023), 7D2O (Wu et al., 2020), and 7SNT (Nemergut et al., 2023), respectively. The arrows indicate the dissection sites in the luciferase structures.
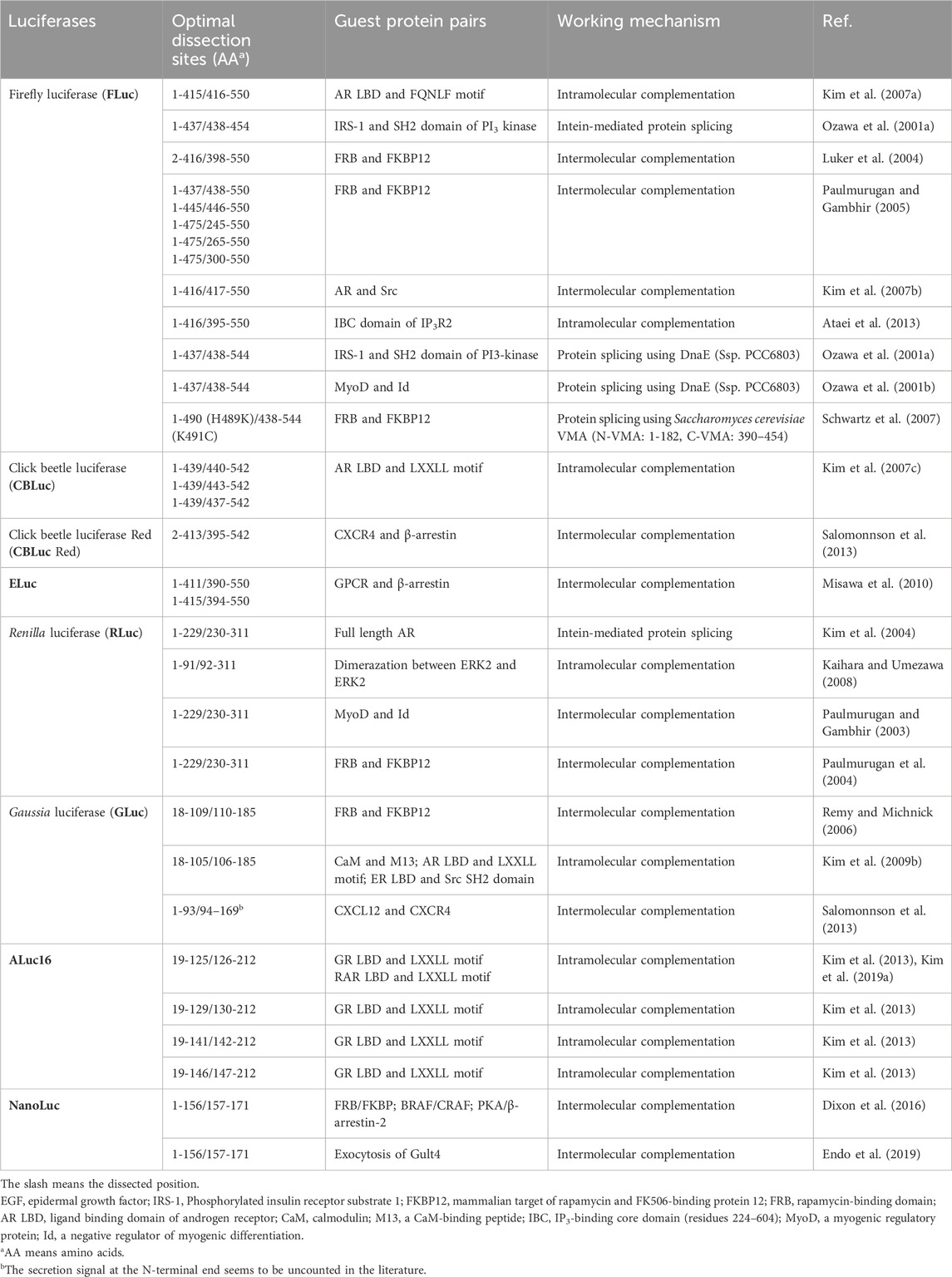
Table 2. Optimal dissection sites of luciferases for construction of bioluminescent probes and molecular imaging. This table was reproduced and updated from Kim et al. (Kim and Paulmurugan, 2021) with permission from Springer Nature.
The common merits of NAP systems may be summarized as follows:
(i) Rapid responses. NAP systems are pre-expressed in cells and ready to respond to analytes. Upon stimulation, PCA probes completed the optical signal development within few minutes after ligand stimulation (Kim et al., 2007a; Taneoka et al., 2009; Hattori et al., 2013). This near real-time feature is advantageous for repeatedly imaging the dynamics of the molecular events in cells. This is a greatly rapid time frame, considering that reporter-gene assays require overnight incubation until the reporter is accumulated enough to be determined by luminometers.
(ii) High S/N ratios. The NAP systems are generally expected to have higher S/N ratio than GTPs. NAPs contain the on-off switches inside the probe and suppress the basal false positive signals.
(iii) Reversibility. Not always, but many NAP systems are conceptually reversible and thus allow for the detection of kinetic constants and equilibrium aspects of the molecular events of interest in living cells.
The common demerits of NAP systems may be summarized as follows:
(i) Poor BL intensities. Not always, but many NAP probes are designed to suppress the BL intensities upon construction of the on-off switch. For example, PCAs carry dissected luciferase fragments for a temporal loss and conditional recovery of luciferase activities. However, this methodology inevitably hampers the intrinsic enzymatic property of the luciferase and recovers merely 0.5%–5% of the original luciferase activity (Paulmurugan and Gambhir, 2005; Kim et al., 2007c).
Similarly, molecular strain probes are designed to cause steric hindrance to the substrate access in the basal conditions by fusing protein pairs of interest at the N- and C-terminal ends of the embedded luciferase with minimal linkers. The steric hindrance is relieved upon occurrence of the intramolecular protein–protein interaction (PPI). This suppression-driven working mechanism explains why the overall optical signals are generally weaker than those of GTPs.
(ii) Complex molecular designs. NAPs generally require complex molecular designs for containing their unique on-off switches. For example, PCA- and PSA-based probe systems proceed with a sophisticated probe design and a tedious optimization step. This optimization includes deciding the overall layout of the components and a suitable dissection site inside the luciferase reporter.
(iii) Irreversible and potentially high background intensity of PSAs. All the PSAs and some of PCAs are irreversible in the reaction mechanisms. PSAs especially proceed with spontaneous splicing reactions by time and thus the background intensities are generally drifted by time. This feature worsens the S/N ratios.
4.2.1 Protein-fragment complementation assay (PCA) systems
PCAs provide a unique experimental strategy for quantitatively imaging the dynamics of PPIs in animal cells. A luciferase as a reporter is genetically dissected into N- and C-terminal fragments and thus it temporarily loses its enzymatic activity. The N- and C-terminal fragments are then genetically fused to a pair of proteins of interest. If the two proteins interact, the adjacent reporter fragments are approximated together and their activity is reconstituted (Figure 5) (Remy and Michnick, 2006; Li P. et al., 2019).
A frontier study on conditional association of protein fragments was demonstrated in 1994 with split-ubiquitin (Johnsson and Varshavsky, 1994). The complementation of split-fluorescent protein was first examined in Escherichia coli by Ghosh et al. (2000). Calmodulin (CaM)‒M13 binding was examined in mammalian cells with fragmented yellow fluorescent protein (YFP) in 2001 (Nagai et al., 2001).
Michnick et al. (2010) also pioneered the development of PCAs: They conducted a whole cell-wide mapping of PPIs in yeast using luciferase fragments in 2008 (Tarassov et al., 2008) and 2010 (Levy et al., 2010). They also presented detailed protocols for large-scale analysis of PPIs with the survival selection of dihydrofolate reductase (DHFR) reporter PCA in 2010. The known examples of PCAs are summarized in Table 2.
The above-mentioned PCA systems are based on an association between two independently expressed fusion proteins. In contrast, Paulmurugan and Gambhir (2006) and Kim et al. (2007a) independently developed single-chain probes for sensing estrogenicity and androgenicity of ligands, respectively, where all the components required for a ligand sensing and light emission are integrated into a single molecular backbone.
4.2.2 Protein-fragment splicing assay (PSA) systems
PSAs have been introduced for determining PPI in animal cells. PSAs make use of protein ligase intein-extein fragments that enable us to provoke spontaneous protein splicing between two fusion proteins without energy. In the probe design, split-fragments of a luciferase are genetically linked to a protein pair of interest via dissected intein fragments, where the split-luciferases act as the extein. The split-luciferase temporarily loses its enzymatic activity. Upon approximation of the split-fragments, the inteins trigger protein splicing, which is a self-catalyzed excision of the intein and ligation (reconstitution) of the flanking split-luciferase (Gimble, 1998). The known examples of PSAs are summarized in Table 2.
PSA was pioneered by Ozawa et al. using both a split-GFP in 2000 (Ozawa et al., 2000) and split- FLuc in 2001 (Ozawa et al., 2001a). BLI of PPIs in living mice was first reported by Paulmurugan et al., where both strategies of protein complementation and intein-mediated protein splicing were comparatively examined (Paulmurugan et al., 2002).
4.2.3 Bioluminescence resonance energy transfer (BRET) -based bioassays
Bioluminescence resonance energy transfer (BRET) is an emerging assay strategy for determining PPIs in animal cells. The BRET-based bioassays are based on the nonradiative energy transfer from a bioluminescent donor (e.g., RLuc) and a fluorescent protein acceptor. The donor and acceptor are tandemly fused to a pair of proteins of interest, respectively. In case that the proteins are approximated at a distance of less than 100 Å, the BRET signal enhances. BRET allows sensitive analysis of small spatial changes of proteins. It is used to interrogate PPIs or conformation changes of proteins of interest. As this review cannot recognize all the BRET systems in literature, we briefly describe the main ones below.
The first generation BRET assay, called “BRET1” used RLuc and an enhanced YFP (EYFP) as donor–acceptor combination, which yielded a spectral separation of nearly 50 nm (Xu et al., 1999). To date, many versions of BRET systems have been reported with various combinations of FP and luciferases: for example, 1) systems named from BRET1 to BRET9 (Otsuji et al., 2004; Dragulescu-Andrasi et al., 2011; Li et al., 2013; Machleidt et al., 2015; Sun et al., 2016; Hiblot et al., 2017; Bae Kim et al., 2020), 2) “NanoBRET” and “eNanoBRET” using NanoLuc as the energy donor (Dale et al., 2019), 3) “Nano-lantern” (Saito et al., 2012), 4) “Antares” using an orange fluorescent protein (CYOFP1) and NanoLuc (Chu et al., 2016), (v) NIR-BRET based on a −300 nm blue-to-near infrared shift of the emission (Nishihara et al., 2019), and a Venus-engineered bacterial luciferase (heterodimer of luxA and luxB) for BRET for enhanced BL intensities (Kaku et al., 2021).
RLuc and its derivatives have been widely used in most of the BRET systems as the energy donor. Recently, the other marine luciferases such as GLuc, NanoLuc, ALuc, and Cypridina luciferase (CLuc) have also been used as energy donors in the BRET-based assays. Alternatively, modification of the substrates was also targeted for creating unique NIR-BRET systems (Nishihara et al., 2019).
The unique merits of BRET may be explained as follows: Different from FRET, BRET does not require outer light source and has low background intensities and relatively high S/N ratios. Because of this virtue, BRET does not suffer from phototoxicity, photobleaching, and autofluorescence (Weihs et al., 2020). BRET is also an appropriate means to examine PPIs, protein folding, and protease activities because the BRET-permissive distance of less than 10 nm is very similar to the dimensions of biological macromolecular protein complexes.
4.2.4 Molecular strain probes
Kim et al. (2009a) previously demonstrated a unique non-transcriptional assay system based on molecular tension of a luciferase artificially appended by PPI. For the basic probe design, a full-length luciferase was sandwiched between two-component proteins of interest with the minimal length of flexible linkers. Upon stimulation of a ligand, the two-component proteins are approximated PPI and append a molecular tension to the sandwiched luciferase. Kim et al. (2009a) found that tensed luciferases can enhance the BL intensity. To date, they examined various luciferases and found that RLuc8, ALuc23, and ALuc49 are suitable for developing the molecular tension probes. For example, the full-length RLuc8 is sandwiched between the ligand-binding domain of estrogen receptor α (ER LBD) and the SH2 domain of Src. This fusion protein elevates the optical intensities ligand-dependently. Likewise, full-length ALuc23 or ALuc49 was sandwiched between FKBP-rapamycin binding protein (FRB) and FK506-binding protein (FKBP), and showed excellent sensorial performance as a bioluminescent probe (Kim et al., 2016; Kim et al., 2023b). The imaging system works even in a cell-free condition (Kim et al., 2019b) and in the application to BRET system (Bae Kim et al., 2020).
5 Summary
BL is an excellent optical readout that has been harnessed in various bioassays and molecular imaging. The recent innovative research greatly diversified the repertories of the toolbox. Considering the potential choices of users, it is valuable to properly understand the merits and demerits of current BL-driven bioassays and molecular imaging systems.
Because we previously reported two major reviews on BL studies in Anal. Sci. and Anal. Chem. (Ozawa et al., 2013; Kim and Paulmurugan, 2021), we are also intended to update recent research articles in this review. While this review initially showcases BL resources (luciferase and luciferin) and their applications to BL-driven bioassays, it also highlights both the excellence and inferiority of respective BL-based bioassays and molecular imaging modalities to provide readers with balanced views. The “bright” side of BL was discussed in detail, which includes the long dynamic range, low background intensity, high S/N ratios, biocompatibility, simplicity in the measurement system, low cost, and versatility in the assay design. In parallel, we introduced the “dark” side as disadvantages of BL such as low optical intensity, necessity of the specific substrates, poor spatial resolution, and potential signal cross-leakage owing to broad bandwidths.
It is expected that future efforts are devoted to basic material studies, in addition to its parallel application in tracking biological events of cells in vitro and in vivo. Similarly, it is also expected that many BL studies exert a wide variety of colors palettes and NIR BL systems. The direct merits of these developments could be that one can construct multiplex bioassay systems using multiple optical readouts, while the use of red and NIR BL can enhance imaging applications in physiological samples and in living subjects.
The other important research tendency in BL studies is the convergence and heterogenicity: BRET is a good example of such a convergence between BL and FL. Likewise, BL can be adapted to various heterogenous materials like nanoparticles and fluorescent dyes. The genius convergence is fully open to future analytical chemists.
Taken through great efforts, BL should be settled to be truly quantitative, highly sensitive, and versatile optical readouts. Such great breakthroughs in BL will be achieved from ideas inspired by understanding of nature, computer modeling, and even simple imagination.
Author contributions
SB-K: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. TF: Data curation, Formal Analysis, Methodology, Resources, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI grants to SB-K: numbers 20K21851, 21H04948, and 23KK0101.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, M., Nishihara, R., Ikeda, Y., Nakajima, T., Sato, M., Iwasawa, N., et al. (2019). Near-infrared bioluminescence imaging with a through-bond energy transfer cassette. Chembiochem 20 (15), 1919–1923. doi:10.1002/cbic.201900149
Adams, S. T., and Miller, S. C. (2020). Enzymatic promiscuity and the evolution of bioluminescence. The FEBS J. 287 (7), 1369–1380. doi:10.1111/febs.15176
Ando, Y., Niwa, K., Yamada, N., Enomoto, T., Irie, T., Kubota, H., et al. (2008). Firefly bioluminescence quantum yield and colour change by pH-sensitive green emission. Nat. Photonics 2 (1), 44–47. doi:10.1038/nphoton.2007.251
Ando, Y., Niwa, K., Yamada, N., Irie, T., Enomoto, T., Kubota, H., et al. (2007). Development of a quantitative bio/chemiluminescence spectrometer determining quantum yields: Re-examination of the aqueous luminol chemiluminescence standard. Photochem. Photobiol. 83 (5), 1205–1210. doi:10.1111/j.1751-1097.2007.00140.x
Aron, A. T., Heffern, M. C., Lonergan, Z. R., Vander Wal, M. N., Blank, B. R., Spangler, B., et al. (2017). In vivo bioluminescence imaging of labile iron accumulation in a murine model of Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. U. S. A. 114 (48), 12669–12674. doi:10.1073/pnas.1708747114
Ataei, F., Torkzadeh-Mahani, M., and Hosseinkhani, S. (2013). A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens. Bioelectron. 41, 642–648. doi:10.1016/j.bios.2012.09.037
Auld, D. S., and Inglese, J. (2016). Interferences with luciferase reporter enzymes. Bethesda (MD): Eli Lilly & Company.
Azad, T., Janse van Rensburg, H. J., Morgan, J., Rezaei, R., Crupi, M. J. F., Chen, R., et al. (2021). Luciferase-based biosensors in the era of the COVID-19 pandemic. ACS Nanosci. Au 1 (1), 15–37. doi:10.1021/acsnanoscienceau.1c00009
Bae Kim, S., Fujii, R., Natarajan, A., Massoud, T. F., and Paulmurugan, R. (2020). Ligand-activated BRET9 imaging for measuring protein-protein interactions in living mice. Chem. Commun. 56 (2), 281–284. doi:10.1039/c9cc07634d
Bakhtiarova, A., Taslimi, P., Elliman, S. J., Kosinski, P. A., Hubbard, B., Kavana, M., et al. (2006). Resveratrol inhibits firefly luciferase. Biochem. biophysical Res. Commun. 351 (2), 481–484. doi:10.1016/j.bbrc.2006.10.057
Berger, F., Paulmurugan, R., Bhaumik, S., and Gambhir, S. S. (2008). Uptake kinetics and biodistribution of 14C-D-luciferin--a radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: impact on bioluminescence based reporter gene imaging. Eur. J. Nucl. Med. Mol. imaging 35 (12), 2275–2285. doi:10.1007/s00259-008-0870-6
Bevilaqua, V. R., Matsuhashi, T., Oliveira, G., Oliveira, P. S. L., Hirano, T., and Viviani, V. R. (2019). Phrixotrix luciferase and 6'-aminoluciferins reveal a larger luciferin phenolate binding site and provide novel far-red combinations for bioimaging purposes. Sci. Rep. 9 (1), 8998. doi:10.1038/s41598-019-44534-3
Branchini, B. R., Ablamsky, D. M., Davis, A. L., Southworth, T. L., Butler, B., Fan, F., et al. (2010). Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal. Biochem. 396 (2), 290–297. doi:10.1016/j.ab.2009.09.009
Branchini, B. R., Ablamsky, D. M., Murtiashaw, M. H., Uzasci, L., Fraga, H., and Southworth, T. L. (2007). Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal. Biochem. 361 (2), 253–262. doi:10.1016/j.ab.2006.10.043
Branchini, B. R., Southworth, T. L., Fontaine, D. M., Kohrt, D., Florentine, C. M., and Grossel, M. J. (2018). A firefly luciferase dual color bioluminescence reporter assay using two substrates to simultaneously monitor two gene expression events. Sci. Rep. 8 (1), 5990. doi:10.1038/s41598-018-24278-2
Branchini, B. R., Southworth, T. L., Fontaine, D. M., Kohrt, D., Welcome, F. S., Florentine, C. M., et al. (2017). Red-emitting chimeric firefly luciferase for in vivo imaging in low ATP cellular environments. Anal. Biochem. 534, 36–39. doi:10.1016/j.ab.2017.07.001
Campbell, Z. T., Weichsel, A., Montfort, W. R., and Baldwin, T. O. (2009). Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the β subunit. Biochemistry 48 (26), 6085–6094. doi:10.1021/bi900003t
Chan, C. T., Reeves, R. E., Geller, R., Yaghoubi, S. S., Hoehne, A., Solow-Cordero, D. E., et al. (2012). Discovery and validation of small-molecule heat-shock protein 90 inhibitors through multimodality molecular imaging in living subjects. Proc. Natl. Acad. Sci. U. S. A. 109 (37), E2476–E2485. doi:10.1073/pnas.1205459109
Chang, M., Anttonen, K. P., Cirillo, S. L. G., Francis, K. P., and Cirillo, J. D. (2014). Real-time bioluminescence imaging of mixed mycobacterial infections. Plos One 9 (9), e108341. doi:10.1371/journal.pone.0108341
Chu, J., Oh, Y., Sens, A., Ataie, N., Dana, H., Macklin, J. J., et al. (2016). A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 34 (7), 760–767. doi:10.1038/nbt.3550
Conti, E., Franks, N. P., and Brick, P. (1996). Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4 (3), 287–298. doi:10.1016/s0969-2126(96)00033-0
Cronin, M., Akin, A. R., Collins, S. A., Meganck, J., Kim, J. B., Baban, C. K., et al. (2012). High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. Plos One 7 (1), e30940. doi:10.1371/journal.pone.0030940
Dale, N. C., Johnstone, E. K. M., White, C. W., and Pfleger, K. D. G. (2019). NanoBRET: the bright future of proximity-based assays. Front. Bioeng. Biotechnol. 7, 56. doi:10.3389/fbioe.2019.00056
Degeling, M. H., Bovenberg, M. S. S., Lewandrowski, G. K., de Gooijer, M. C., Vleggeert-Lankamp, C. L. A., Tannous, M., et al. (2013). Directed molecular evolution reveals Gaussia luciferase variants with enhanced light output stability. Anal. Chem. 85 (5), 3006–3012. doi:10.1021/ac4003134
de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., and Subramani, S. (1987). Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7 (2), 725–737. doi:10.1128/mcb.7.2.725-737.1987
de Wet, J. R., Wood, K. V., Helinski, D. R., and DeLuca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 82 (23), 7870–7873. doi:10.1073/pnas.82.23.7870
Dixon, A. S., Schwinn, M. K., Hall, M. P., Zimmerman, K., Otto, P., Lubben, T. H., et al. (2016). NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11 (2), 400–408. doi:10.1021/acschembio.5b00753
Dragulescu-Andrasi, A., Chan, C. T., De, A., Massoud, T. F., and Gambhir, S. S. (2011). Bioluminescence resonance energy transfer (BRET) imaging of protein–protein interactions within deep tissues of living subjects. Proc. Natl. Acad. Sci. U.S.A. 108 (29), 12060–12065. doi:10.1073/pnas.1100923108
Endo, M., Miyasaki, M., Li, Q. J., Kawamura, G., and Ozawa, T. (2019). A detection method for GLUT4 exocytosis based on spontaneous split luciferase complementation. Anal. Sci. 35 (8), 835–838. doi:10.2116/analsci.19c003
Fan, F., and Wood, K. V. (2007). Bioluminescent assays for high-throughput screening. ASSAY Drug Dev. Technol. 5 (1), 127–136. doi:10.1089/adt.2006.053
Fetchko, M., and Stagljar, I. (2004). Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods (Duluth) 32 (4), 349–362. doi:10.1016/j.ymeth.2003.10.010
Fleiss, A., and Sarkisyan, K. S. (2019). A brief review of bioluminescent systems (2019). Curr. Genet. 65 (4), 877–882. doi:10.1007/s00294-019-00951-5
Franks, N. P., Jenkins, A., Conti, E., Lieb, W. R., and Brick, P. (1998). Structural basis for the inhibition of firefly luciferase by a general anesthetic. Biophysical J. 75 (5), 2205–2211. doi:10.1016/S0006-3495(98)77664-7
Ghosh, I., Hamilton, A. D., and Regan, L. (2000). Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J. Am. Chem. Soc. 122 (23), 5658–5659. doi:10.1021/ja994421w
Gimble, F. S. (1998). Putting protein splicing to work. Chem. & Biol. 5 (10), R251–R256. doi:10.1016/s1074-5521(98)90109-0
Gregor, C., Gwosch, K. C., Sahl, S. J., and Hell, S. W. (2018). Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc. Natl. Acad. Sci. U. S. A. 115 (5), 962–967. doi:10.1073/pnas.1715946115
Hall, M. P., Unch, J., Binkowski, B. F., Valley, M. P., Butler, B. L., Wood, M. G., et al. (2012). Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7 (11), 1848–1857. doi:10.1021/cb3002478
Hall, M. P., Woodroofe, C. C., Wood, M. G., Que, I., van't Root, M., Ridwan, Y., et al. (2018). Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun. 9, 132. doi:10.1038/s41467-017-02542-9
Harvey, E. N. (1917). What substance is the source of the light in the firefly? Science 46 (1184), 241–243. doi:10.1126/science.46.1184.241
Hattori, M., Haga, S., Takakura, H., Ozaki, M., and Ozawa, T. (2013). Sustained accurate recording of intracellular acidification in living tissues with a photo-controllable bioluminescent protein. Proc. Natl. Acad. Sci. U. S. A. 110 (23), 9332–9337. doi:10.1073/pnas.1304056110
Heffern, M. C., Park, H. M., Au-Yeung, H. Y., Van de Bittner, G. C., Ackerman, C. M., Stahl, A., et al. (2016). In vivo bioluminescence imaging reveals copper deficiency in a murine model of nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. U. S. A. 113 (50), 14219–14224. doi:10.1073/pnas.1613628113
Herbst, K. J., Allen, M. D., and Zhang, J. (2009). The cAMP-dependent protein kinase inhibitor H-89 attenuates the bioluminescence signal produced by Renilla Luciferase. PloS one 4 (5), e5642. doi:10.1371/journal.pone.0005642
Hiblot, J., Yu, Q. L. Y., Sabbadini, M. D. B., Reymond, L., Xue, L., Schena, A., et al. (2017). Luciferases with tunable emission wavelengths. Angew. Chem. Int. Ed. 56 (46), 14556–14560. doi:10.1002/anie.201708277
Hida, N., Awais, M., Takeuchi, M., Ueno, N., Tashiro, M., Takagi, C., et al. (2009). High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLos One 4 (6), e5868. doi:10.1371/journal.pone.0005868
Ho, P. I., Yue, K., Pandey, P., Breault, L., Harbinski, F., McBride, A. J., et al. (2013). Reporter enzyme inhibitor study to aid assembly of orthogonal reporter gene assays. ACS Chem. Biol. 8 (5), 1009–1017. doi:10.1021/cb3007264
Homaei, A. A., Mymandi, A. B., Sariri, R., Kamrani, E., Stevanato, R., Etezad, S. M., et al. (2013). Purification and characterization of a novel thermostable luciferase from Benthosema pterotum. J. Photochem. Photobiol. B Biol. 125, 131–136. doi:10.1016/j.jphotobiol.2013.05.015
Ikeda, Y., Nomoto, T., Hiruta, Y., Nishiyama, N., and Citterio, D. (2020). Ring-fused firefly luciferins: expanded palette of near-infrared emitting bioluminescent substrates. Anal. Chem. 92 (6), 4235–4243. doi:10.1021/acs.analchem.9b04562
Inouye, S., and Sahara, Y. (2008). Identification of two catalytic domains in a luciferase secreted by the copepod Gaussia princeps. Biochem. Biophysical Res. Commun. 365 (1), 96–101. doi:10.1016/j.bbrc.2007.10.152
Inouye, S., Sato, J.-I., Sahara-Miura, Y., Tomabechi, Y., Sumida, Y., Sekine, S.-I., et al. (2022). Reverse mutants of the catalytic 19 kDa mutant protein (nanoKAZ/nanoLuc) from Oplophorus luciferase with coelenterazine as preferred substrate. PloS one 17 (9), e0272992. doi:10.1371/journal.pone.0272992
Inouye, S., Watanabe, K., Nakamura, H., and Shimomura, O. (2000). Secretional luciferase of the luminous shrimp Oplophorus gracilirostris: cDNA cloning of a novel imidazopyrazinone luciferase. FEBS Lett. 481 (1), 19–25. doi:10.1016/s0014-5793(00)01963-3
Iwano, S., Obata, R., Miura, C., Kiyama, M., Hama, K., Nakamura, M., et al. (2013). Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron 69 (19), 3847–3856. doi:10.1016/j.tet.2013.03.050
Iwano, S., Sugiyama, M., Hama, H., Watakabe, A., Hasegawa, N., Kuchimaru, T., et al. (2018). Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 359 (6378), 935–939. doi:10.1126/science.aaq1067
Johnsson, N., and Varshavsky, A. (1994). Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91 (22), 10340–10344. doi:10.1073/pnas.91.22.10340
Jones, K. A., Porterfield, W. B., Rathbun, C. M., McCutcheon, D. C., Paley, M. A., and Prescher, J. A. (2017). Orthogonal luciferase-luciferin pairs for bioluminescence imaging. J. Am. Chem. Soc. 139 (6), 2351–2358. doi:10.1021/jacs.6b11737
Kaihara, A., and Umezawa, Y. (2008). Genetically encoded bioluminescent indicator for ERK2 dimer in living cells. Chem. – Asian J. 3 (1), 38–45. doi:10.1002/asia.200700186
Kaku, T., Sugiura, K., Entani, T., Osabe, K., and Nagai, T. (2021). Enhanced brightness of bacterial luciferase by bioluminescence resonance energy transfer. Sci. Rep. 11 (1), 14994. doi:10.1038/s41598-021-94551-4
Kamiya, G., Kitada, N., Furuta, T., Hirano, T., Maki, S., and Kim, S. B. (2022a). C-series coelenterazine-driven bioluminescence signature imaging. Int. J. Mol. Sci. 23 (21), 13047. doi:10.3390/ijms232113047
Kamiya, G., Kitada, N., Furuta, T., Hirano, T., Maki, S. A., and Kim, S. B. (2023). S-series coelenterazine-driven combinatorial bioluminescence imaging systems for mammalian cells. Int. J. Mol. Sci. 24 (2), 1420. doi:10.3390/ijms24021420
Kamiya, G., Kitada, N., Maki, S., and Kim, S. B. (2022b). Multiplex quadruple bioluminescent assay system. Sci. Rep. 12 (1), 17485. doi:10.1038/s41598-022-20468-1
Kim, S. B. (2012). Labor-effective manipulation of marine and beetle luciferases for bioassays. Protein Eng. Des. Sel. 25 (6), 261–269. doi:10.1093/protein/gzs016
Kim, S. B., Awais, M., Sato, M., Umezawa, Y., and Tao, H. (2007a). Integrated molecule-format bioluminescent probe for visualizing androgenicity of ligands based on the intramolecular association of androgen receptor with its recognition peptide. Anal. Chem. 79 (5), 1874–1880. doi:10.1021/ac061934u
Kim, S. B., Fujii, R., Nishihara, R., Bose, R. J. C., Citterio, D., Suzuki, K., et al. (2019a). Molecular imaging of retinoic acids in live cells using single-chain bioluminescence probes. ACS Comb. Sci. 21 (6), 473–481. doi:10.1021/acscombsci.9b00035
Kim, S. B., Furuta, T., Kamiya, G., Kitada, N., Paulmurugan, R., and Maki, S. A. (2023b). Bright molecular strain probe templates for reporting protein-protein interactions. Sensors 23 (7), 3498. doi:10.3390/s23073498
Kim, S. B., Furuta, T., Kitada, N., and Maki, S. A. (2023c). Creation of artificial luciferase 60s from sequential insights and their applications to bioassays. Sensors 23 (14), 6376. doi:10.3390/s23146376
Kim, S. B., Furuta, T., Ohmuro-Matsuyama, Y., Kitada, N., Nishihara, R., and Maki, S. A. (2023d). Bioluminescent imaging systems boosting near-infrared signals in mammalian cells. Photochem Photobiol. Sci. 22 (6), 1267–1278. doi:10.1007/s43630-023-00367-8
Kim, S. B., Hattori, M., and Ozawa, T. (2012). Intelligent design of nano-scale molecular imaging agents. Int. J. Mol. Sci. 13 (12), 16986–17005. doi:10.3390/ijms131216986
Kim, S. B., and Izumi, H. (2014). Functional artificial luciferases as an optical readout for bioassays. Biochem. Biophysical Res. Commun. 448 (4), 418–423. doi:10.1016/j.bbrc.2014.04.128
Kim, S.-B., Kamiya, G., Furuta, T., Kitada, N., and Maki, S. A. (2023a). Coelenterazine indicators for the specific imaging of human and bovine serum albumins. Sensors 23 (13), 6020. doi:10.3390/s23136020
Kim, S. B., Kanno, A., Ozawa, T., Tao, H., and Umezawa, Y. (2007b). Nongenomic activity of ligands in the association of androgen receptor with Src. ACS Chem. Biol. 2 (7), 484–492. doi:10.1021/cb7000439
Kim, S. B., Miller, S., Suzuki, N., Senda, T., Nishihara, R., and Suzuki, K. (2015). Cation-driven optical properties of artificial luciferases. Anal. Sci. 31 (10), 955–960. doi:10.2116/analsci.31.955
Kim, S. B., Nishihara, R., Citterio, D., and Suzuki, K. (2016). Genetically encoded molecular tension probe for tracing protein - protein interactions in mammalian cells. Bioconjugate Chem. 27 (2), 354–362. doi:10.1021/acs.bioconjchem.5b00421
Kim, S. B., Nishihara, R., Citterio, D., and Suzuki, K. (2017). Fabrication of a new lineage of artificial luciferases from natural luciferase pools. ACS Comb. Sci. 19 (9), 594–599. doi:10.1021/acscombsci.7b00081
Kim, S. B., Nishihara, R., Fujii, R., Paulmurugan, R., Citterio, D., and Suzuki, K. (2019b). In vitro determination of rapamycin-triggered FKBP-FRB interactions using a molecular tension probe. Anal. Sci. 35 (1), 71–78. doi:10.2116/analsci.18sdp08
Kim, S. B., Otani, Y., Umezawa, Y., and Tao, H. (2007c). Bioluminescent indicator for determining protein-protein interactions using intramolecular complementation of split click beetle luciferase. Anal. Chem. 79 (13), 4820–4826. doi:10.1021/ac0621571
Kim, S. B., Ozawa, T., Watanabe, S., and Umezawa, Y. (2004). High-throughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc. Natl. Acad. Sci. U. S. A. 101 (32), 11542–11547. doi:10.1073/pnas.0401722101
Kim, S. B., and Paulmurugan, R. (2021). Bioluminescent imaging systems for assay developments. Anal. Sci. 37 (2), 233–247. doi:10.2116/analsci.20R003
Kim, S. B., Sato, M., and Tao, H. (2009a). Molecular tension-indexed bioluminescent probe for determining Protein−Protein interactions. Bioconjugate Chem. 20, 2324–2330. doi:10.1021/bc900330w
Kim, S. B., Sato, M., and Tao, H. (2009b). Split Gaussia luciferase-based bioluminescence template for tracing protein dynamics in living cells. Anal. Chem. 81 (1), 67–74. doi:10.1021/ac801658y
Kim, S. B., Suzuki, H., Sato, M., and Tao, H. (2011a). Superluminescent variants of marine luciferases for bioassays. Anal. Chem. 83 (22), 8732–8740. doi:10.1021/ac2021882
Kim, S. B., Takenaka, Y., and Torimura, M. (2011b). A bioluminescent probe for salivary cortisol. Bioconjugate Chem. 22 (9), 1835–1841. doi:10.1021/bc200220k
Kim, S. B., Torimura, M., and Tao, H. (2013). Creation of artificial luciferases for bioassays. Bioconjugate Chem. 24 (12), 2067–2075. doi:10.1021/bc400411h
Kim, S. B., Umezawa, Y., Kanno, K. A., and Tao, H. (2008). An integrated-molecule-format multicolor probe for monitoring multiple activities of a bioactive small molecule. ACS Chem. Biol. 3 (6), 359–372. doi:10.1021/cb800004s
Klein, M. A., Lazarev, S., Gervasi, C., Cowan, C., Machleidt, T., and Friedman Ohana, R. (2023). Luciferase calibrants enable absolute quantitation of bioluminescence power. ACS Meas. Sci. au 3 (6), 496–503. doi:10.1021/acsmeasuresciau.3c00036
Kojima, R., Takakura, H., Kamiya, M., Kobayashi, E., Komatsu, T., Ueno, T., et al. (2015). Development of a sensitive bioluminogenic probe for imaging highly reactive oxygen species in living rats. Angew. Chem. Int. Ed. 54 (49), 14768–14771. doi:10.1002/anie.201507530
Kumar, S. N., Fred, A. L., Kumar, H. A., and Varghese, P. S. (2019). Firefly optimization based improved fuzzy clustering for CT/MR image segmentation. Cham, Switzerland: Springer Nature.
Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157 (1), 105–132. doi:10.1016/0022-2836(82)90515-0
Lecuyer, B., Arrio, B., Fresneau, C., and Volfin, P. (1979). Dinoflagellate luciferases - purification of luciferases from gonyaulax-polyedra, pyrocystis-lunula, and pyrocystis-fusiformis. Archives Biochem. Biophysics 196 (2), 371–384. doi:10.1016/0003-9861(79)90288-1
Levy, E. D., Landry, C. R., and Michnick, S. W. (2010). Signaling through cooperation. Science 328 (5981), 983–984. doi:10.1126/science.1190993
Li, F. Y., Yu, J. P., Zhang, Z. P., Cui, Z. Q., Wang, D. B., Wei, H. P., et al. (2013). Use of hGluc/tdTomato pair for sensitive BRET sensing of protease with high solution media tolerance. Talanta 109, 141–146. doi:10.1016/j.talanta.2013.02.007
Li, J. B., Chen, L. L., Wang, Q. Q., Liu, H. W., Hu, X. X., Yuan, L., et al. (2018). A bioluminescent probe for imaging endogenous peroxynitrite in living cells and mice. Anal. Chem. 90 (6), 4167–4173. doi:10.1021/acs.analchem.8b00198
Li, J. B., Wang, Q. Q., Liu, H. W., Yuan, L., and Zhang, X. B. (2019a). A bioluminescent probe for imaging endogenous hydrogen polysulfides in live cells and a murine model of bacterial infection. Chem. Commun. 55 (31), 4487–4490. doi:10.1039/c9cc01699f
Li, P., Wang, L., and Di, L. J. (2019b). Applications of protein fragment complementation assays for analyzing biomolecular interactions and biochemical networks in living cells. J. Proteome Res. 18 (8), 2987–2998. doi:10.1021/acs.jproteome.9b00154
Li, Q., Zeng, J. F., Miao, Q. Q., and Gao, M. Y. (2019c). Self-illuminating agents for deep-tissue optical imaging. Front. Bioeng. Biotechnol. 7, 326. doi:10.3389/fbioe.2019.00326
Li, X. Y., Nakajima, Y., Niwa, K., Viviani, V. R., and Ohmiya, Y. (2010). Enhanced red-emitting railroad worm luciferase for bioassays and bioimaging. Protein Sci. 19 (1), 26–33. doi:10.1002/pro.279
Loening, A. M., Dragulescu-Andrasi, A., and Gambhir, S. S. (2010). A red-shifted Renilla luciferase for transient reporter-gene expression. Nat. Methods 7 (1), 5–6. doi:10.1038/nmeth0110-05
Loening, A. M., Fenn, T. D., Wu, A. M., and Gambhir, S. S. (2006). Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 19 (9), 391–400. doi:10.1093/protein/gzl023
Loening, A. M., Wu, A. M., and Gambhir, S. S. (2007). Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods 4 (8), 641–643. doi:10.1038/nmeth1070
Lorenz, W. W., McCann, R. O., Longiaru, M., and Cormier, M. J. (1991). Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc. Natl. Acad. Sci. U. S. A. 88 (10), 4438–4442. doi:10.1073/pnas.88.10.4438
Love, A. C., Caldwell, D. R., Kolbaba-Kartchner, B., Townsend, K. M., Halbers, L. P., Yao, Z., et al. (2023). Red-shifted coumarin luciferins for improved bioluminescence imaging. J. Am. Chem. Soc. 145 (6), 3335–3345. doi:10.1021/jacs.2c07220
Luker, K. E., Smith, M. C., Luker, G. D., Gammon, S. T., Piwnica-Worms, H., and Piwnica-Worms, D. (2004). Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. U. S. A. 101 (33), 12288–12293. doi:10.1073/pnas.0404041101
Machleidt, T., Woodroofe, C. C., Schwinn, M. K., Mendez, J., Robers, M. B., Zimmerman, K., et al. (2015). NanoBRET-A novel BRET platform for the analysis of protein-protein interactions. ACS Chem. Biol. 10 (8), 1797–1804. doi:10.1021/acschembio.5b00143
Markova, S. V., Golz, S., Frank, L. A., Kalthof, B., and Vysotski, E. S. (2004). Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa. J. Biol. Chem. 279 (5), 3212–3217. doi:10.1074/jbc.m309639200
Martin, C. S., Wight, P. A., Dobretsova, A., and Bronstein, I. (1996). Dual luminescence-based reporter gene assay for luciferase andβ-galactosidase. Biotechniques 21 (3), 520–524. doi:10.2144/96213pf01
Massoud, T. F., and Gambhir, S. S. (2003). Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes & Dev. 17 (5), 545–580. doi:10.1101/gad.1047403
Mezzanotte, L., Aswendt, M., Tennstaedt, A., Hoeben, R., Hoehn, M., and Lowik, C. (2013). Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast Media & Mol. Imaging 8 (6), 505–513. doi:10.1002/cmmi.1549
Mezzanotte, L., van 't Root, M., Karatas, H., Goun, E. A., and Lowik, C. W. G. M. (2017). In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol. 35 (7), 640–652. doi:10.1016/j.tibtech.2017.03.012
Michnick, S. W., Ear, P. H., Landry, C., Malleshaiah, M. K., and Messier, V. (2010). A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein protein interactions in living cells. Methods Enzym. 470, 335–368. doi:10.1016/S0076-6879(10)70014-8
Misawa, N., Kafi, A. K. M., Hattori, M., Miura, K., Masuda, K., and Ozawa, T. (2010). Rapid and high-sensitivity cell-based assays of protein-protein interactions using split click beetle luciferase complementation: an approach to the study of G-protein-coupled receptors. Anal. Chem. 82 (6), 2552–2560. doi:10.1021/ac100104q
Mujawar, A., Phadte, P., Palkina, K. A., Markina, N. M., Mohammad, A., Thakur, B. L., et al. (2023). Triple reporter assay: a non-overlapping luciferase assay for the measurement of complex macromolecular regulation in cancer cells using a new mushroom luciferase-luciferin pair. Sensors 23 (17), 7313. doi:10.3390/s23177313
Nagai, T., Sawano, A., Park, E. S., and Miyawaki, A. (2001). Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc. Natl. Acad. Sci. U. S. A. 98 (6), 3197–3202. doi:10.1073/pnas.051636098
Nakajima, Y., Kobayashi, K., Yamagishi, K., Enomoto, T., and Ohmiya, Y. (2004). CDNA cloning and characterization of a secreted luciferase from the luminous Japanese ostracod, Cypridina noctiluca. Biosci. Biotechnol. Biochem. 68 (3), 565–570. doi:10.1271/bbb.68.565
Nakajima, Y., Yamazaki, T., Nishii, S., Noguchi, T., Hoshino, H., Niwa, K., et al. (2010). Enhanced beetle luciferase for high-resolution bioluminescence imaging. Plos One 5 (3), e10011. doi:10.1371/journal.pone.0010011
Nakatsu, T., Ichiyama, S., Hiratake, J., Saldanha, A., Kobashi, N., Sakata, K., et al. (2006). Structural basis for the spectral difference in luciferase bioluminescence. Nature 440 (7082), 372–376. doi:10.1038/nature04542
Neefjes, M., Housmans, B. A. C., van den Akker, G. G. H., van Rhijn, L. W., Welting, T. J. M., and van der Kraan, P. M. (2021). Reporter gene comparison demonstrates interference of complex body fluids with secreted luciferase activity. Sci. Rep. 11 (1), 1359. doi:10.1038/s41598-020-80451-6
Nemergut, M., Pluskal, D., Horackova, J., Sustrova, T., Tulis, J., Barta, T., et al. (2023). Illuminating the mechanism and allosteric behavior of NanoLuc luciferase. Nat. Commun. 14 (1), 7864. doi:10.1038/s41467-023-43403-y
Nishihara, R., Abe, M., Nishiyama, S., Citterio, D., Suzuki, K., and Kim, S. B. (2017). Luciferase-specific coelenterazine analogues for optical contamination-free bioassays. Sci. Rep. 7, 908. doi:10.1038/s41598-017-00955-6
Nishihara, R., Dokainish, H. M., Kihara, Y., Ashiba, H., Sugita, Y., and Kurita, R. (2024). Pseudo-luciferase activity of the SARS-CoV-2 spike protein for Cypridina luciferin luciferin. ACS Central Sci. 10(2), 283–290. doi:10.1021/acscentsci.3c00887
Nishihara, R., Hoshino, E., Kakudate, Y., Kishigami, S., Iwasawa, N., Sasaki, S., et al. (2018). Azide- and dye-conjugated coelenterazine analogues for a multiplex molecular imaging platform. Bioconjugate Chem. 29 (6), 1922–1931. doi:10.1021/acs.bioconjchem.8b00188
Nishihara, R., Niwa, K., Tomita, T., and Kurita, R. (2020). Coelenterazine analogue with human serum albumin-specific bioluminescence. Bioconjugate Chem. 31 (12), 2679–2684. doi:10.1021/acs.bioconjchem.0c00536
Nishihara, R., Paulmurugan, R., Nakajima, T., Yamamoto, E., Natarajan, A., Afjei, R., et al. (2019). Highly bright and stable NIR-BRET with blue-shifted coelenterazine derivatives for deep-tissue imaging of molecular events in vivo. Theranostics 9 (9), 2646–2661. doi:10.7150/thno.32219
Niwa, K., Kubota, H., Enomoto, T., Ichino, Y., and Ohmiya, Y. (2023). Quantitative analysis of bioluminescence optical signal. Biosensors 13 (2), 223. doi:10.3390/bios13020223
Novobilsky, A., and Hoglund, J. (2020). Small animal in vivo imaging of parasitic infections: a systematic review. Exp. Parasitol. 214, 107905. doi:10.1016/j.exppara.2020.107905
Oba, Y., Yoshida, N., Kanie, S., Ojika, M., and Inouye, S. (2013). Biosynthesis of firefly luciferin in adult lantern: decarboxylation of ʟ-cysteine is a key step for benzothiazole ring formation in firefly luciferin synthesis. Plos One 8 (12), e84023. doi:10.1371/journal.pone.0084023
Ong, T. T., Ang, Z., Verma, R., Koean, R., Tam, J. K. C., and Ding, J. L. (2020). pHLuc, a ratiometric luminescent reporter for in vivo monitoring of tumor acidosis. Front. Bioeng. Biotechnol. 8, 412. doi:10.3389/fbioe.2020.00412
O'Sullivan, J. J., Medici, V., and Heffern, M. C. (2022). A caged imidazopyrazinone for selective bioluminescence detection of labile extracellular copper(ii). Chem. Sci. 13 (15), 4352–4363. doi:10.1039/d1sc07177g
Otsuji, T., Okuda-Ashitaka, E., Kojima, S., Akiyama, H., Ito, S., and Ohmiya, Y. (2004). Monitoring for dynamic biological processing by intramolecular bioluminescence resonance energy transfer system using secreted luciferase. Anal. Biochem. 329 (2), 230–237. doi:10.1016/j.ab.2004.03.010
Otto-Duessel, M., Khankaldyyan, V., Gonzalez-Gomez, I., Jensen, M. C., Laug, W. E., and Rosol, M. (2006). In vivo testing of Renilla luciferase substrate analogs in an orthotopic murine model of human glioblastoma. Mol. Imaging 5 (2), 7290200600006–64. doi:10.2310/7290.2006.00006
Ozawa, T., Kaihara, A., Sato, M., Tachihara, K., and Umezawa, Y. (2001a). Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal. Chem. 73(11), 2516–2521. doi:10.1021/Ac0013296
Ozawa, T., Nogami, S., Sato, M., Ohya, Y., and Umezawa, Y. (2000). A fluorescent indicator for detecting protein-protein interactions in vivo based on protein splicing. Anal. Chem. 72 (21), 5151–5157. doi:10.1021/ac000617z
Ozawa, T., Takeuchi, M., Kaihara, A., Sato, M., and Umezawa, Y. (2001b). Protein splicing-based reconstitution of split green fluorescent protein for monitoring protein-protein interactions in bacteria: improved sensitivity and reduced screening time. Anal. Chem. 73 (24), 5866–5874. doi:10.1021/ac010717k
Ozawa, T., Yoshimura, H., and Kim, S. B. (2013). Advances in fluorescence and bioluminescence imaging. Anal. Chem. 85 (2), 590–609. doi:10.1021/ac3031724
Paulmurugan, R., and Gambhir, S. S. (2003). Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 75 (7), 1584–1589. doi:10.1021/ac020731c
Paulmurugan, R., and Gambhir, S. S. (2005). Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal. Chem. 77 (5), 1295–1302. doi:10.1021/ac0484777
Paulmurugan, R., and Gambhir, S. S. (2006). An intramolecular folding sensor for imaging estrogen receptor-ligand interactions. Proc. Natl. Acad. Sci. U. S. A. 103 (43), 15883–15888. doi:10.1073/pnas.0607385103
Paulmurugan, R., and Gambhir, S. S. (2007). Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal. Chem. 79 (6), 2346–2353. doi:10.1021/ac062053q
Paulmurugan, R., Massoud, T. F., Huang, J., and Gambhir, S. S. (2004). Molecular imaging of drug-modulated protein-protein interactions in living subjects. Cancer Res. 64 (6), 2113–2119. doi:10.1158/0008-5472.can-03-2972
Paulmurugan, R., Umezawa, Y., and Gambhir, S. S. (2002). Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U. S. A. 99 (24), 15608–15613. doi:10.1073/pnas.242594299
Porterfield, W. B., Jones, K. A., McCutcheon, D. C., and Prescher, J. A. (2015). A "caged" luciferin for imaging cell-cell contacts. J. Am. Chem. Soc. 137 (27), 8656–8659. doi:10.1021/jacs.5b02774
Remy, I., and Michnick, S. W. (2006). A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3 (12), 977–979. doi:10.1038/nmeth979
Reumann, M. K., Weiser, M. C., and Mayer-Kuckuk, P. (2010). Musculoskeletal molecular imaging: a comprehensive overview. Trends Biotechnol. 28 (2), 93–101. doi:10.1016/j.tibtech.2009.11.004
Saito, K., Chang, Y. F., Horikawa, K., Hatsugai, N., Higuchi, Y., Hashida, M., et al. (2012). Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 3, 1262. doi:10.1038/ncomms2248
Salomonnson, E., Stacer, A. C., Ehrlich, A., Luker, K. E., and Luker, G. D. (2013). Imaging CXCL12-CXCR4 signaling in ovarian cancer therapy. Plos One 8 (1), e51500. doi:10.1371/journal.pone.0051500
Santos, E. B., Yeh, R., Lee, J., Nikhamin, Y., Punzalan, B., Punzalan, B., et al. (2009). Sensitive in vivo imaging of T cells using a membrane-bound Gaussia princeps luciferase. Nat. Med. 15 (3), 338–344. doi:10.1038/nm.1930
Schenkmayerova, A., Toul, M., Pluskal, D., Baatallah, R., Gagnot, G., Pinto, G. P., et al. (2023). Catalytic mechanism for Renilla-type luciferases. Nat. Catal. 6 (1), 23–38. doi:10.1038/s41929-022-00895-z
Schultz, L. W., Liu, L. Y., Cegielski, M., and Hastings, J. W. (2005). Crystal structure of a pH-regulated luciferase catalyzing the bioluminescent oxidation of an open tetrapyrrole. Proc. Natl. Acad. Sci. U. S. A. 102 (5), 1378–1383. doi:10.1073/pnas.0409335102
Schwartz, E. C., Saez, L., Young, M. W., and Muir, T. W. (2007). Post-translational enzyme activation in an animal via optimized conditional protein splicing. Nat. Chem. Biol. 3 (1), 50–54. doi:10.1038/nchembio832
Shimomura, O. (2006). Bioluminescence: chemical principles and methods. Biolumin. Chem. Princ. Methods, 1–471. doi:10.1142/6102
Shimomura, O., Masugi, T., Johnson, F. H., and Haneda, Y. (1978). Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry 17 (6), 994–998. doi:10.1021/bi00599a008
Shipunova, V. O., Shilova, O. N., Shramova, E. I., Deyev, S. M., and Proshkina, G. M. (2018). A highly specific substrate for NanoLUC luciferase furimazine is toxic in vitro and in vivo. Russ. J. Bioorg. Chem. 44 (2), 225–228. doi:10.1134/S1068162018020085
Sun, S. H., Yang, X. B., Wang, Y., and Shen, X. H. (2016). In vivo analysis of protein-protein interactions with bioluminescence resonance energy transfer (BRET): progress and prospects. Int. J. Mol. Sci. 17 (10), 1704. doi:10.3390/ijms17101704
Takenaka, Y., Masuda, H., Yamaguchi, A., Nishikawa, S., Shigeri, Y., Yoshida, Y., et al. (2008). Two forms of secreted and thermostable luciferases from the marine copepod crustacean, Metridia pacifica. Gene 425 (1-2), 28–35. doi:10.1016/j.gene.2008.07.041
Takenaka, Y., Yamaguchi, A., Tsuruoka, N., Torimura, M., Gojobori, T., and Shigeri, Y. (2012). Evolution of bioluminescence in marine planktonic copepods. Mol. Biol. Evol. 29 (6), 1669–1681. doi:10.1093/molbev/mss009
Tamaki, S., Kitada, N., Kiyama, M., Fujii, R., Hirano, T., Kim, S. B., et al. (2021). Color-tunable bioluminescence imaging portfolio for cell imaging. Sci. Rep. 11 (1), 2219. doi:10.1038/s41598-021-81430-1
Taneoka, A., Sakaguchi-Mikami, A., Yamazaki, T., Tsugawa, W., and Sode, K. (2009). The construction of a glucose-sensing luciferase. Biosens. Bioelectron. 25 (1), 76–81. doi:10.1016/j.bios.2009.06.004
Tannous, B. A., Kim, D. E., Fernandez, J. L., Weissleder, R., and Breakefield, X. O. (2005). Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11 (3), 435–443. doi:10.1016/j.ymthe.2004.10.016
Tarassov, K., Messier, V., Landry, C. R., Radinovic, S., Molina, M. M., Shames, I., et al. (2008). An in vivo map of the yeast protein interactome. Science 320 (5882), 1465–1470. doi:10.1126/science.1153878
Thorne, N., Shen, M., Lea, W. A., Simeonov, A., Lovell, S., Auld, D. S., et al. (2012). Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem. & Biol. 19 (8), 1060–1072. doi:10.1016/j.chembiol.2012.07.015
Tian, X., Zhang, Y., Li, X., Xiong, Y., Wu, T., and Ai, H.-W. (2022). A luciferase prosubstrate and a red bioluminescent calcium indicator for imaging neuronal activity in mice. Nat. Commun. 13 (1), 3967. doi:10.1038/s41467-022-31673-x
Verhaegen, M., and Christopoulos, T. K. (2002). Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal. Chem. 74 (17), 4378–4385. doi:10.1021/ac025742k
Villalobos, V., Naik, S., Bruinsma, M., Dothager, R. S., Pan, M. H., Samrakandi, M., et al. (2010). Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem. & Biol. 17 (9), 1018–1029. doi:10.1016/j.chembiol.2010.06.018
Viviani, V. R., Arnoldi, F. G. C., Venkatesh, B., Neto, A. J. S., Ogawa, F. G. T., Oehlmeyer, A. T. L., et al. (2006). Active-site properties of Phrixotrix railroad worm green and red bioluminescence-eliciting luciferases. The J. Biochem. 140 (4), 467–474. doi:10.1093/Jb/Mvj190
Viviani, V. R., Bechara, E. J. H., and Ohmiya, Y. (1999). Cloning, sequence analysis, and expression of active Phrixothrix railroad-worms luciferases: relationship between bioluminescence spectra and primary structures. Biochemistry 38 (26), 8271–8279. doi:10.1021/bi9900830
Weihs, F., and Dacres, H. (2019). Red-shifted bioluminescence Resonance Energy Transfer: improved tools and materials for analytical in vivo approaches. TrAC Trends Anal. Chem. 116, 61–73. doi:10.1016/j.trac.2019.04.011
Weihs, F., Peh, A., and Dacres, H. (2020). A red-shifted Bioluminescence Resonance Energy Transfer (BRET) biosensing system for rapid measurement of plasmin activity in human plasma. Anal. Chim. Acta 1102, 99–108. doi:10.1016/j.aca.2019.12.044
Welsh, J. P., Patel, K. G., Manthiram, K., and Swartz, J. R. (2009). Multiply mutated Gaussia luciferases provide prolonged and intense bioluminescence. Biochem. Biophysical Res. Commun. 389 (4), 563–568. doi:10.1016/j.bbrc.2009.09.006
Widder, E. A. (2001). Marine bioluminescence. Why do so many animals in the open ocean make light? Biosci. Explain. 1, 1–9.
Woo, J., and von Arnim, A. G. (2008). Mutational optimization of the coelenterazine-dependent luciferase from Renilla. Plant Methods 4, 23. doi:10.1186/1746-4811-4-23
Wood, K. V., Lam, Y. A., Seliger, H. H., and McElroy, W. D. (1989). Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science 244 (4905), 700–702. doi:10.1126/science.2655091
Wu, N., Kobayashi, N., Tsuda, K., Unzai, S., Saotome, T., Kuroda, Y., et al. (2020). Solution structure of Gaussia Luciferase with five disulfide bonds and identification of a putative coelenterazine binding cavity by heteronuclear NMR. Sci. Rep. 10 (1), 20069. doi:10.1038/s41598-020-76486-4
Xu, T. T., Ripp, S., Sayler, G. S., and Close, D. M. (2014). Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. Plos One 9 (5), e96347. doi:10.1371/journal.pone.0096347
Xu, Y., Piston, D. W., and Johnson, C. H. (1999). A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. U. S. A. 96 (1), 151–156. doi:10.1073/pnas.96.1.151
Yao, Z., Caldwell, D. R., Love, A. C., Kolbaba-Kartchner, B., Mills, J. H., Schnermann, M. J., et al. (2021). Coumarin luciferins and mutant luciferases for robust multi-component bioluminescence imaging. Chem. Sci. 12 (35), 11684–11691. doi:10.1039/d1sc03114g
Yeh, A. H., Norn, C., Kipnis, Y., Tischer, D., Pellock, S. J., Evans, D., et al. (2023). De novo design of luciferases using deep learning. Nature 614 (7949), 774–780. doi:10.1038/s41586-023-05696-3
Yeh, H. W., Karmach, O., Ji, A., Carter, D., Martins-Green, M. M., and Ai, H. W. (2017). Red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 14 (10), 971–974. doi:10.1038/Nmeth.4400
Keywords: bioluminescence (BL), bioluminescence imaging (BLI), luciferase, luciferin, D-luciferin, coelenterazine (CTZ), bioassay
Citation: Kim S-B and Furuta T (2024) Bioluminescence from the bright and dark sides. Front. Chem. Biol 3:1459397. doi: 10.3389/fchbi.2024.1459397
Received: 04 July 2024; Accepted: 07 August 2024;
Published: 19 August 2024.
Edited by:
Tae-Jin Kim, Pusan National University, Republic of KoreaReviewed by:
Mingxing Ouyang, Changzhou University, ChinaKabir H. Biswas, Hamad bin Khalifa University, Qatar
Copyright © 2024 Kim and Furuta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung-Bae Kim, a2ltdS1zYkBhaXN0LmdvLmpw
 Sung-Bae Kim
Sung-Bae Kim Tadaomi Furuta
Tadaomi Furuta