- 1Department of Microbiology/Biotechnology, Interfaculty Institute of Microbiology and Infection Medicine, Faculty of Science, University of Tübingen, Tübingen, Germany
- 2German Center for Infection Research (DZIF), Partner Site Tübingen, Tübingen, Germany
- 3Cluster of Excellence “Controlling Microbes to Fight Infections” (CMFI), University of Tübingen, Tübingen, Germany
- 4Department Bioresources for Bioeconomy and Health Research, Leibniz Institute DSMZ -German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany
- 5Braunschweig Integrated Centre of Systems Biology (BRICS), Braunschweig, Germany
- 6Institut für Mikrobiologie, Technische Universität Braunschweig, Braunschweig, Germany
Lysolipin I is a halogenated, polycyclic xanthone natural product belonging to the polyketide class of antibiotics, naturally produced by Streptomyces violaceoniger TÜ96 and Streptomyces tendae TÜ4042. The biosynthesis is encoded on a 43 kb-spanning biosynthetic gene cluster. Heterologous expression of the gene cluster has been established in previous work by using the cosmid 4H04, which was transferred to Streptomyces albus. In the current study, we demonstrate the optimization of production yields of therapeutically interesting lysolipin derivatives with extended activity against Gram-negatives and less cytotoxic bioactivities, respectively, by using mutated heterologous S. albus producer strains. Production yields were significantly increased by adapting cultivation conditions as well as by inactivating the transcriptional repressor gene llpRI, which lead to increased and consistent lysolipin (derivatives) production. Furthermore, cultivation of a S. albus 4H04∆llpOI mutant strain in bromide-containing fermentation medium resulted in the production of a new brominated lysolipin derivative (C28H20BrNO9).
Introduction
Lysolipin I is a polycyclic xanthone natural product belonging to the polyketide class of antibiotics (Figure 1A), which is naturally produced by Streptomyces violaceoniger TÜ96 and Streptomyces tendae TÜ4042 (Drautz et al., 1975a; Lopez et al., 2010). It shows an extraordinarily potent antibacterial activity, with a minimum inhibitory concentration (MIC) in the low nanomolar range (less than 10 nM) against Gram-negative microorganisms such as Proteus or Pseudomonas, as well as Gram-positive microorganisms like Bacillus, Staphylococcus, Corynebacterium (Drautz et al., 1975a). Own recent data show also low nanomolar activity against vancomycin-resistant enterococci (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) as well as anaerobe microorganisms such as Helicobacter pylori. Interestingly, the Gram-negative E. coli and Salmonella typhimurium were resistant if their cell wall was intact, but susceptible in case of permeation-enhanced strains (Drautz et al., 1975a). The mechanisms of action of the lysolipins bactericidal activity has not been fully elucidated yet, although the compounds are suspected to target the cell envelope, possibly interfering with peptidoglycan synthesis by binding to lipid-bound murein precursors (Drautz et al., 1975a; Drautz et al., 1975b). The usability of lysolipin as antibacterial agent is limited due to the fact that it also shows cytotoxicity (Lopez et al., 2010). First experiments have shown that structural modifications of the lysolipin core structure can differentially influence antibacterial activity and cytotoxicity (Wohlleben et al., 2005; WO/2006/032232). Modification of compounds can in principle be achieved either by chemical means or by genetic modification of the producer strains. A total chemical synthesis of lysolipin has not been established so far, however, a modular chemical synthesis strategy towards the production of halogenated xanthone molecules has been reported recently (Meringdal et al., 2022), which can now serve as a basis for further compound modification. Compound derivatisation of lysolipin by genetic engineering has previously been reported (Wohlleben et al., 2005; WO/2006/032232). Prerequisite for compound derivatisation by genetic engineering is the knowledge of the biosynthetic pathway and the corresponding genes of the producer strains, which has been described for lysolipin biosynthesis by Lopez et al., 2010. However, since production yields of lysolipin were low in the natural producer strains, the biosynthetic gene cluster (BGC), consisting of 44 genes spanning a region of ∼43 kb (Figure 1B) was heterologously expressed in Streptomyces albus J1074 as expression host, yielding a stable production of 50 mg/L lysolipin under non-optimized conditions (Wohlleben et al., 2005; WO/2006/032232). Another possibility to increase production yields is genetic manipulation of cluster-situated transcriptional regulatory genes. In the lysolipin BGC, five genes (llpR1-llpRV) are present, which code for different types of transcriptional regulators (Lopez et al., 2010). LlpRI resembles a transcriptional regulator of the PadR type, which acts as a transcriptional repressor (Rohrer, 2017). LlpRII and LlpRIII show sequence similarities to transcriptional activators of the TenA family. LlpRIV has strong similarity to Streptomyces antibiotic regulatory proteins (SARPs), known to acts as activators of antibiotic biosynthesis (Rohrer, 2017), whereas LlpRV shows sequence similarity to DNA binding proteins from Streptomyces coelicolor A3 (2) and contains a helix-turn-helix motif (Rohrer, 2017). The involvement of all individual llpR regulatory genes has been demonstrated by mutagenesis in previous work, where deletion of llpRI in S. tendae TÜ 4042 led to significantly increased lysolipin production (>300%; ∼0.5 g/L) (Rohrer, 2017) (Supplementary Figure S1). In addition, two genes have been suggested to be involved in conferring resistance against lysolipin, which are llpN encoding a potential efflux protein and llpG encoding a potential glycosyl transferase (Lopez et al., 2010). Heterologous expression of llpN in the lysolipin-sensitive S. albus strains led to significantly higher tolerance against lysolipin (up to 500 μg/mL lysolipin) (Unsin, 2008), whereas involvement of llpG in conferring resistance to lysolipin has not been demonstrated experimentally. Furthermore, lysolipin was derivatized applying genetic engineering on genes encoding tailoring enzymes of lysolipin biosynthesis. A special structural feature of lysolipin is a xanthone mojety, which is highly decorated by post-polyketide synthase enzymatic reactions catalyzed e.g. by oxygenases, methyltransferases, hydroxylases, a halogenase etc. encoded by the respective modification genes located on the lysolipin BGC (Lopez et al., 2010; Hofeditz et al., 2018). In previous work it has been shown that the deletion of llpOI encoding a putative FAD-dependent monooxygenase resulted in the production of a lysolipin derivative CBS40 with improved antibacterial activity against Escherichia coli, whereas deletion of llpOIV, encoding a putative cytochrome P450 hydroxylase, and llpMVI, encoding a putative O-methyltransferase led to lysolipin derivatives CBS68 and a mixture of CBS70 and CBS72, respectively, with less cytotoxicity values (patent WO/2007/079715, Combinature Biopharm, Berlin, Germany). Thus, first experiments have proven that structural modifications of the lysolipin core structure is feasible and can differentially influence antibacterial activity and cytotoxicity.
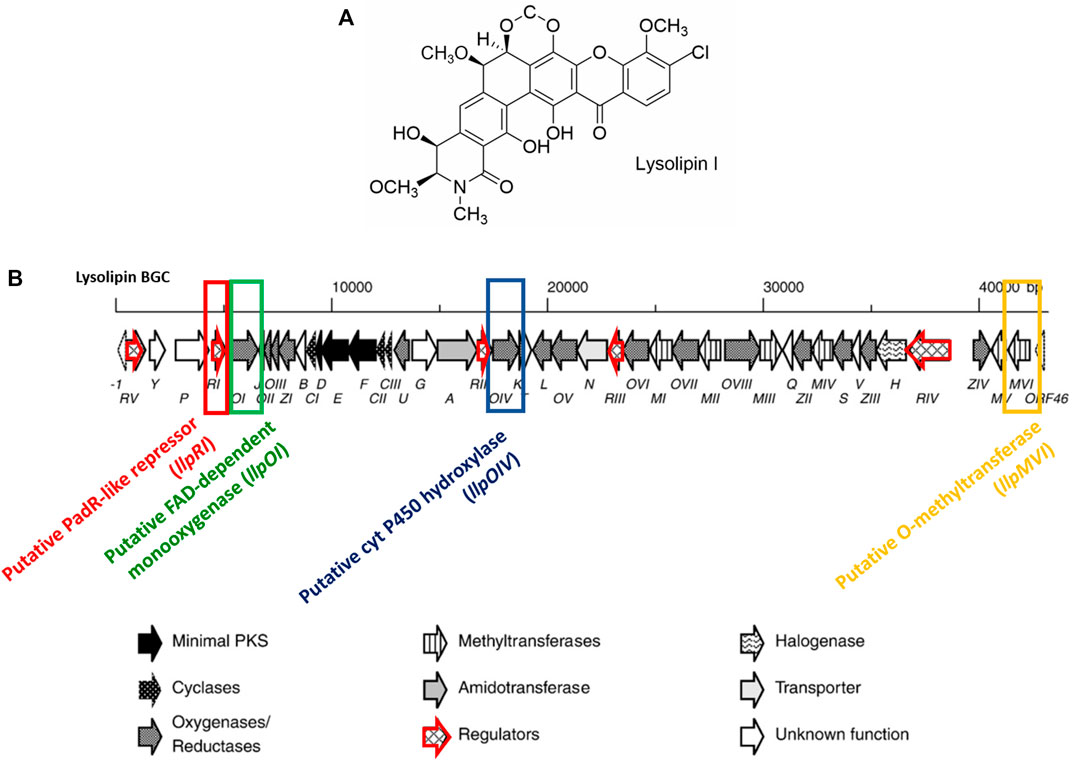
Figure 1. Chemical structure of lysolipin I (A) and encoding lysolipin BGC (B). Genes targeted for genetic knockout studies are highlighted by color: llpRI (red), llpOI (green), llpOIV (blue), and llpMVI (yellow). Each gene is presented by an arrow and filling patterns are indicated at the bottom of the figure according to Lopez et al., 2010. Regulatory genes are additionally highlighted with a red frame.
In the current study, we aimed to optimize lysolipin production yields in the heterologous expression host S. albus and generate new lysolipin derivatives based on the already known lysolipin lead structures.
Results and discussion
To define the optimal conditions for lysolipin production, the heterologous host strain S. albus 4H04 (Wohlleben et al., 2005; patent WO/2006/032232) expressing the lysolipin BGC was grown in 14 different cultivation media. For this purpose, S. albus 4H04 was cultivated in 100 mL of R5 medium (Handayani et al., 2021) with 100 μg/mL apramycin in a 500-mL Erlenmeyer flasks (with steel springs) on an orbital shaker (180 rpm) at 28°C as a preculture. After 3 days of cultivation, 10 mL of preculture were transferred to 100 mL main culture medium NL200, NL300, NL330, NL400, NL500, NL550, NL800, SGG, TDG, ISP2, ISP3, MS, R5 (Handayani et al., 2021; Wex et al., 2021), or E1 (Hardter et al., 2012), respectively. Cell cultures were grown for 7 days and extracted as reported previously by adjusting the pH to 3, followed by extraction with ethyl acetate (1:1), concentrating samples in vacuo, and then re-dissolving in methanol (Lopez et al., 2010). The methanolic extracts were analyzed for lysolipin production by HPLC-DAD-MS analysis as reported previously using a calibration curve with pure lysolipin I as a standard (Lopez et al., 2010). Thereby it was found that NL800 and E1 (Supplementary Table S1) were the best lysolipin production media, yielding 1.1 mg/mL and 0.6 mg/mL of substance, respectively (Figure 2; only six best production media are shown). Regarding the NL800-derived sample, this was >60% more lysolipin in comparison to the previously reported maximal lysolipin production of ∼0.5 g/L of the genetically manipulated native lysolipin S. tendae producer strain. However, due to the presence of 10 g/L glycerol in NL800, which was detrimental for subsequent chemical analysis and purification of the compound, E1 was selected as suitable production medium for lysolipin in the further work steps. To further increase lysolipin production, the transcriptional repressor gene llpRI was targeted for gene inactivation in the S. albus 4H04 strain. The llpRI mutant was constructed with the REDirect cloning strategy (Gust et al., 2003) as described by Zhang et al., 2018 by using primers listed in Supplementary Table S2. Thereby a llpRI disruption cassette with a chloramphenicol (cam) resistance marker and artificial MunI restriction sites was introduced into E. coli BW25113:pKD20 harboring the 4H04 cosmid (Supplementary Figure S2). The llpRI gene of cosmid 4H04 was deleted via λ-RED-driven recombination yielding 4H04 with an integrated llpRI disruption cassette (4H04∆llpRIcam). The 4H04∆llpRIcam construct was isolated from E. coli and digested with MunI. The resulting large (∼40 kb) fragment was purified from an agarose gel and religated to yield construct 4H04∆llpRI. The 4H04∆llpRI construct was introduced into S. albus J1074 by conjugation with E. coli ET12567/pUB307 (Flett et al., 2006) as described in Kieser et al., 2000. This resulted in the llpRI knockout strain S. albus 4H04∆llpRI. HPLC-MS analyses with extract samples of S. albus 4H04∆llpRI showed that lysolipin I production (peak with retention time (RT) ∼11.9 min) is increased 3-fold in comparison to the reference sample S. albus 4H04 (Figure 3 A vs. B), proving that LlpRI is acting as transcriptional repressor of lysolipin biosynthesis. A PadR-like DNA binding motif ATGT-8N-ACAT (Kaval et al., 2015) was identified in the intergenic region between llpRY and llpP, which indicates the binding site for LlpRI. The consistent and strong lysolipin production set the basis to genetically engineer the heterologous host strain to produce therapeutically promising lysolipin derivatives. For that, we specifically focused on optimizing the production of the lysolipin derivatives CBS40, CBS68, and CBS70+72 due to their interesting bioactive properties as outlined in the introduction (Supplementary Table S3). For this purpose, the genes llpOI, llpOIV, and llpMVI were knocked out individually, as the respective mutant strains were expected to produce CBS40, CBS68, and CBS70+72 as lysolipin derivatives, respectively. As the original in-frame deletion strains were no longer available, these mutants were newly constructed exactly as described in patent WO/2007/079715. Thereby, strain S. albus 4H04∆llpOI (≙ CB3919LlpOI_A) contained a mutation of gene llpOI encoding the FAD-dependent monooxygenase gene LlpOI, S. albus 4H04∆llpOIV (≙ CB3919LlpOIV) harbored a mutation of gene llpOIV encoding the cytochrome P450 hydroxylase LlpOIV, whereas strain S. albus 4H04∆llpMVI (≙ CB3919LlpMVI_A) contained a mutation of gene llpMVI, encoding the methyltransferase LlpMVI. HPLC-MS analysis was carried out with extract samples of the mutant cultures in comparison to extracts from the lysolipin producing S. albus 4H04 reference strain. In the CBS terminology lysolipin I is designated as CBS42 with a sum formula of C29H24ClNO11 (patent WO/2007/079715). HPLC-MS analysis of extract samples from S. albus 4H04∆llpOI revealed a peak with a RT of ∼12.4 min and a mass of m/z = 550 [M+H]+, which correlates to the compound features of CBS40 (C28H20CINO9) (Figure 4A) (for comparisons see (patent WO/2007/079715). Samples from S. albus 4H04∆llpOIV showed a peak at RT 12.1 min and a mass of m/z = 522.08 [M+H]+, correlating to the compound CBS68 (C27H20CINO8) (Figure 4B), respectively, whereas samples from S. albus 4H04∆llpMVI revealed a specific peak at RT 10.6 min for which two prominent masses were identified at m/z = 508 [M+H]+ and m/z = 538 [M+H]+, which match with the lysolipin derivatives CBS70 (C26H18ClNO8) and CBS72 (C27H20CINO9), respectively (Figure 4C) (for comparisons see patent WO/2007/079715). Thus, the production of the respective lysolipin derivatives was confirmed.
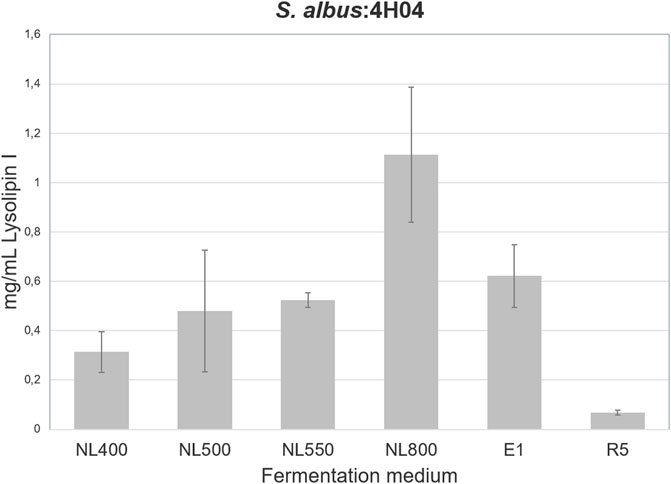
Figure 2. Lysolipin I production in six different cultivation media after cultivation for 7 days at 28°C. Results are shown from three independent biological replicates.
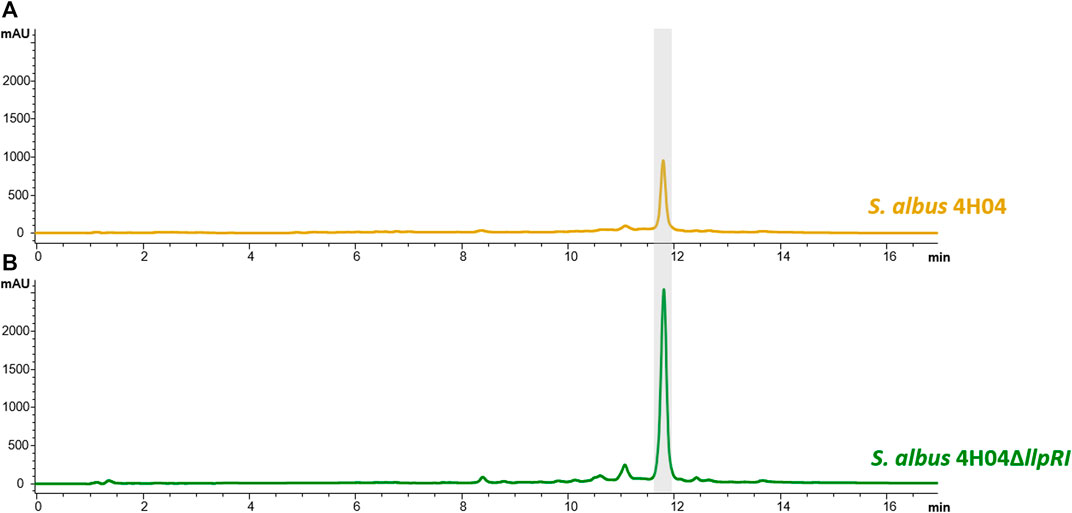
Figure 3. HPLC spectrum of extract samples from S. albus 4H04 (A) and S. albus 4H04∆llpRI (B) at 365 nm with lysolipin-specific peaks at RT 11.9 min. Samples were obtained from cultures grown for 7 days at 28°C.
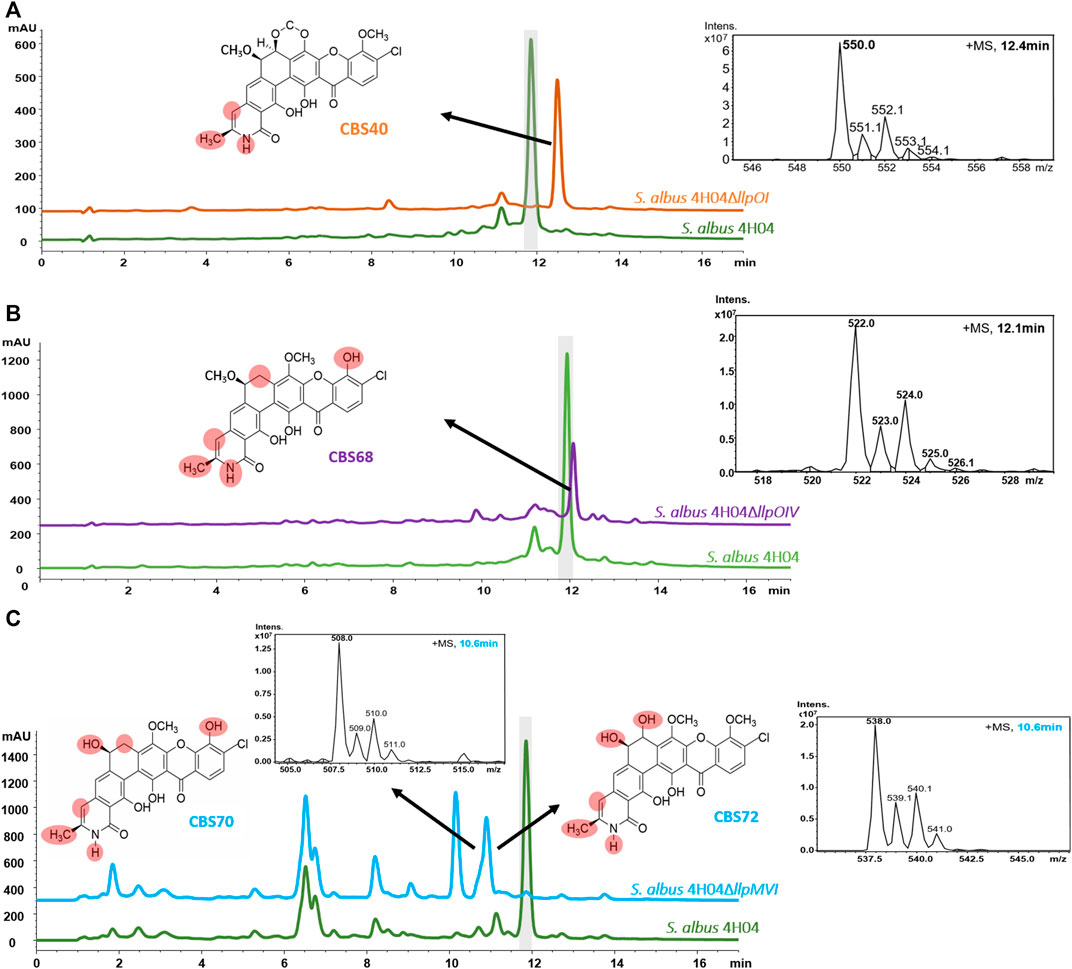
Figure 4. HPLC-MS chromatograms of extract samples from (A) S. albus 4H04∆llpOI (orange), (B) S. albus 4H04∆llpOIV (purple), and (C) S. albus 4H04∆llpMVI (blue) in comparison to HPLC chromatograms from S. albus 4H04 (green). Samples were obtained from cultures grown for 7 days at 28°C. Peaks related to lysolipin derivatives are indicated by arrows. Furthermore, MS spectra (black) and chemical structures of lysolipin derivatives are shown. Differences to the chemical structure of lysolipin I are highlighted by light red circles.
To optimize production yields of the bioactively interesting lysolipin derivatives, we further modified the strains by genetic engineering attempts and additionally inactivated the llpRI gene in those mutants. Here, we focused on the S. albus 4H04∆llpOI and S. albus 4H04∆llpMVI mutant strains, which each produced a broad-spectrum lysolipin derivative (CBS40) or lysolipin derivatives with reduced cytotoxicity (CBS70 and CBS72), respectively (Supplementary Table S3). Mutant construction was performed by the REDirect cloning strategy as reported above by using the pGUS/∆llpRI deletion construct, which was each introduced into S. albus 4H04∆llpOI and S. albus 4H04∆llpMVI by conjugation with E. coli ET12567/pUZ8002, resulting in the strains S. albus 4H04∆llpOI∆llpRI and S. albus 4H04∆llpMVI∆llpRI, respectively. The mutant strains were cultivated under lysolipin production conditions and extract samples were analyzed by HPLC-MS. Comparative analyses revealed that lysolipin derivative production was increased 2.5-fold in both, the S. albus 4H04∆llpOI∆llpRI and S. albus 4H04∆llpMVI∆llpRI strain in comparison to the respective reference strains S. albus 4H04∆llpOI and S. albus 4H04∆llpMVI, respectively (Supplementary Figures S3, S4 A vs. B, respectively). Thus, genetic engineering of the heterologous S. albus 4H04 expression strains successfully enabled optimization of lysolipin derivative production.
One compound feature of the lysolipins is a chlorine residue at the xanthone substructure of the molecule, which is attached by a cluster-encoded halogenase LlpH. To extend the spectrum of compound derivatives, we attempted to exchange the chlorine atom of the lysolipins against a bromine. For this purpose, we cultivated the S. albus 4H04 strain in E1 medium with 5 g NaBr instead of NaCl. HPLC-MS analysis revealed a peak at RT 12.1 and a specific mass of m/z = 642 [M+H]+, which correlates to a brominated lysolipin I (CBS49). Furthermore, a peak at RT 10.7 min was detected of which the mass pattern fit to a dechloro-lysolipin I (CBS44), in addition to a reduced production amount of lysolipin I (Supplementary Figure S5A). NaBr feeding was also carried out with the S. albus 4H04∆llpOI mutant strains, which led to the production of a dechlorinated version of CBS40, designated as CBS48 (C28H21NO9), which has been described before as a product from S. albus 4H04∆llpOI (patent WO/2007/079715), as well as a new brominated version of CBS40 (RT 12.5 min, of m/z = 594 [M+H]+) with a sum formula of C28H20BrNO9 (Supplementary Figure S5B).
In this way, numerous lysolipin derivatives could be produced by genetic engineering of the S. albus 4H04 heterologous expression strain, whereby lysolipin derivative production was significantly increased by inactivating the repressor gene llpRI and a new halogenated lysolipin derivative could be obtained due to the cultivation of the S. albus 4H04∆llpOI strain in bromide-containing fermentation medium. Altogether, these data hold potential for future drug development attempts with the lysolipin compound.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
HR: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing–review and editing. SR: Formal Analysis, Investigation, Visualization, Writing–review and editing. AK: Formal Analysis, Methodology, Writing–review and editing. WW: Funding acquisition, Project administration, Resources, Writing–review and editing. YM: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by a grant provided by the German Center for Infection Research (DZIF) TTU 09.912. We acknowledge support from the Open Access Publication Fund of the University of Tübingen.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchbi.2024.1445095/full#supplementary-material
References
Drautz, H., Keller-Schierlein, W., and Zähner, H. (1975a). Metabolic products of microorganisms, 149. Lysolipin I, a new antibiotic from Streptomyces violaceoniger. Arch. Microbiol. 106, 175–190. doi:10.1007/BF00446521
Drautz, H., Keller-Schierlein, W., and Zähner, H. (1975b). ein neuerHemmstoff der bakteriellen Zellwandsynthese. Path Microbiol. doi:10.1159/000162762
Flett, F., Mersinias, V., and Smith, C. P. (2006). High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155, 223–229. doi:10.1111/j.1574-6968.1997.tb13882.x
Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100, 1541–1546. doi:10.1073/pnas.0337542100
Handayani, I., Saad, H., Ratnakomala, S., Lisdiyanti, P., Kusharyoto, W., Krause, J., et al. (2021). Mining Indonesian microbial biodiversity for Novel natural compounds by a combined genome mining and molecular networking approach. Mar. Drugs 19, 316. doi:10.3390/md19060316
Hardter, U., Luzhetska, M., Ebeling, S., and Bechthold, A. (2012). Ethanol production in actinomycetes after expression of synthetic adhB and pdc. Open Biotechnol. J. 6, 13–16. doi:10.2174/1874070701206010013
Hofeditz, T., Unsin, C. E., Wiese, J., Imhoff, J. F., Wohlleben, W., Grond, S., et al. (2018). Lysoquinone-TH1, a new polyphenolic tridecaketide produced by expressing the lysolipin minimal PKS II in Streptomyces albus. Antibiotics 7, 53. doi:10.3390/antibiotics7030053
Kaval, K. G., Hahn, B., Tusamda, N., Albrecht, D., and Halbedel, S. (2015). The PadR-like transcriptional regulator LftR ensures efficient invasion of Listeria monocytogenes into human host cells. Front. Microbiol. 6, 772. doi:10.3389/fmicb.2015.00772
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces genetics. Norwich Research Park: John Innes Centre. Colney, Norwich NR4 7UH.
Lopez, P., Hornung, A., Welzel, K., Unsin, C., Wohlleben, W., Weber, T., et al. (2010). Isolation of the lysolipin gene cluster of Streptomyces tendae Tü 4042. Gene 461, 5–14. doi:10.1016/j.gene.2010.03.016
Meringdal, J. W., Kilian, A., Li, W. C., Heinemann, M. J. B., Rausch, M., Schneider, T., et al. (2022). Modular synthesis of halogenated xanthones by a divergent coupling strategy. J. Org. Chem. 87, 9375–9383. doi:10.1021/acs.joc.2c01157
Rohrer, S. (2017). Genetische und physiologische Untersuchung der Lysolipin-Produzenten Streptomyces tendae Tü 4042 und Streptomyces violaceoniger Tü 96. Diss. Univ. Tübingen. (Ger).
Unsin, C. E.-M. (2008). Genetische und biochemische Analyse der Lysolipin-Biosynthese und -Resistenz in Streptomyces tendae Tü 4042. Diss. Univ. Tübingen. (Ger).
Wex, K. W., Saur, J. S., Handel, F., Ortlieb, N., Mokeev, V., Kulik, A., et al. (2021). Bioreporters for direct mode of action-guided screening of antibiotic producer strains. Cell Chem. Biol. 15 (21), S2451–S9456. 00107-0. doi:10.1016/j.chembiol.2021.02.022
Wohlleben, W., Pelzer, S., Lopez, P., and Hornung, A. (2005). Novel lysolipin biosynthesis gene cluster. WO/2006/032232. Available at: https://patents.google.com/patent/WO2006032232A2/en?oq=WO%2f2006%2f032232.
Keywords: actinomycetes, Streptomyces, antibiotic, polyketides, lysolipin, genetic engineering
Citation: Robertsen H, Rohrer S, Kulik A, Wohlleben W and Mast Y (2024) Generation of lysolipin derivatives by genetic engineering. Front. Chem. Biol 3:1445095. doi: 10.3389/fchbi.2024.1445095
Received: 06 June 2024; Accepted: 16 July 2024;
Published: 31 July 2024.
Edited by:
Oleksandr S. Yushchuk, University of Insubria, ItalyReviewed by:
Patrick Caffrey, University College Dublin, IrelandMarie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2024 Robertsen, Rohrer, Kulik, Wohlleben and Mast. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yvonne Mast, eXZvbm5lLm1hc3RAZHNtei5kZQ==; Wolfgang Wohlleben, d29sZmdhbmcud29obGxlYmVuQGJpb3RlY2gudW5pLXR1ZWJpbmdlbi5kZQ==
 Helene Robertsen1,2
Helene Robertsen1,2 Andreas Kulik
Andreas Kulik Wolfgang Wohlleben
Wolfgang Wohlleben Yvonne Mast
Yvonne Mast