- 1Department of Physiology and Neurobiology, University of Connecticut, Storrs, CT, United States
- 2Connecticut Institute for the Brain and Cognitive Sciences, University of Connecticut, Storrs, CT, United States
- 3Institute for Systems Genomics, University of Connecticut, Storrs, CT, United States
Insects detect odorants using two large families of heteromeric receptors, the Odorant Receptors (ORs) and Ionotropic Receptors (IRs). Most OR and IR genes encode odorant-binding “tuning” subunits, whereas four (Orco, Ir8a, Ir25a, and Ir76b) encode co-receptor subunits required for receptor function. Olfactory neurons are thought to degenerate in the absence of Orco in ants and bees, and limited data suggest this may happen to some olfactory neurons in Drosophila fruit flies as well. Here, we thoroughly examined the role of co-receptors on olfactory neuron survival in Drosophila. Leveraging knowledge that olfactory neuron classes are defined by the expression of different tuning receptors, we used tuning receptor expression in antennal transcriptomes as a proxy for the survival of distinct olfactory neuron classes. Consistent with olfactory neuron degeneration, expression of many OR-family tuning receptors is decreased in Orco mutants relative to controls, and transcript loss is progressive with age. The effects of Orco are highly receptor-dependent, with expression of some receptor transcripts nearly eliminated and others unaffected. Surprisingly, further studies revealed that olfactory neuron classes with reduced tuning receptor expression generally survive in Orco mutant flies. Furthermore, there is little apoptosis or neuronal loss in the antenna of these flies. We went on to investigate the effects of IR family co-receptor mutants using similar approaches and found that expression of IR tuning receptors is decreased in the absence of Ir8a and Ir25a, but not Ir76b. As in Orco mutants, Ir8a-dependent olfactory neurons mostly endure despite near-absent expression of associated tuning receptors. Finally, we used differential expression analysis to identify other antennal genes whose expression is changed in IR and OR co-receptor mutants. Taken together, our data indicate that odorant co-receptors are necessary for maintaining expression of many tuning receptors at the mRNA level. Further, most Drosophila olfactory neurons persist in OR and IR co-receptor mutants, suggesting that the impact of co-receptors on neuronal survival may vary across insect species.
Introduction
Insects use olfaction to navigate their environments, contributing to recognition of food, toxins, and mates. Odorants are detected by their peripheral olfactory organs, the antennae and maxillary palps. Each appendage is densely covered in cuticular hairs known as sensilla, which encompass the dendrites of olfactory neurons. In Drosophila melanogaster, the antenna and maxillary palp have ∼1,200 and ∼120 olfactory neurons, respectively, with 1–4 olfactory neurons per sensillum (Stocker, 1994). The ∼45 olfactory neuron classes in Drosophila express different odorant receptors, which determine the range of odorants that the neurons sense. Axons from each class of olfactory neurons project to a singular glomerulus in the antennal lobe, the site where olfactory information is first processed in the brain (Gomez-Diaz et al., 2018).
Environmental odors contain an enormous variety of molecular structures, underlying the need for numerous receptors to encode these stimuli. Insect genomes contain two sizeable families of insect odorant receptors, the Odorant Receptors (ORs) and Ionotropic Receptors (IRs) (Robertson, 2019; Supplementary Figure 1). Most IRs respond best to carboxylic acids and amines, whereas the ORs respond to esters, alcohols, ketones, and aromatics as well as long-chain hydrocarbon pheromones (Gomez-Diaz et al., 2018; Yan et al., 2020; Ni, 2021). Some members of the IR family alternatively function as taste, humidity, or temperature receptors (Ni, 2021). Both ORs and IRs form tetrameric odorant-gated cation channels (Sato et al., 2008; Smart et al., 2008; Abuin et al., 2011; Butterwick et al., 2018). Each OR subunit has seven transmembrane domains, with a hydrophobic ligand-binding pocket located in the transmembrane region (Butterwick et al., 2018; Del Marmol et al., 2021). The IRs are distantly related to the glutamate receptor family, with each subunit containing an extracellular ligand-binding domain, three transmembrane domains, and a re-entrant pore loop (Abuin et al., 2011, 2019).
The ORs form heteromeric ion channels comprised of an obligate subunit, Odorant Receptor Co-Receptor (Orco), and one of many potential odorant-binding tuning subunits, “OrX”, that define the odors to which the receptor can respond (Larsson et al., 2004; Neuhaus et al., 2005; Benton et al., 2006; Hallem et al., 2006; Sato et al., 2008). OrX subunits produce only minimal odor responses when expressed in heterologous cells in the absence of Orco, and odor sensitivity of OrX-expressing olfactory neurons is abolished in Orco mutants (Larsson et al., 2004; Neuhaus et al., 2005; Sato et al., 2008; Smart et al., 2008). In the absence of Orco in vivo, OrX protein is unstable and mostly unable to traffic to the ciliated neuronal dendrites (Larsson et al., 2004; Benton et al., 2006). The Orco co-receptor is highly conserved across insect species, whereas the tuning receptors are highly variable and evolve to detect odorants in different ecological niches (Jones et al., 2005; Robertson, 2019).
Similarly, IR receptors are heteromers comprised of one of many odorant-binding tuning subunits, “IrX”, and one or more co-receptors, Ir8a, Ir25a, and Ir76b (Abuin et al., 2011; Ni, 2021; Vulpe and Menuz, 2021). As with the OR family, the formation of functional IR receptors requires co-receptors in heterologous cells and in vivo (Abuin et al., 2011, 2019). Studies in Drosophila indicate that neuronal responses to carboxylic acids are lost in Ir8a mutants, whereas amine responses are lost in either Ir25a or Ir76b mutants (Abuin et al., 2011; Vulpe and Menuz, 2021). Ir8a and Ir25a contain an amino-terminal domain (ATD) predicted to affect receptor assembly, and an extracellular “co-receptor extra loop” (CREL) domain involved in ciliary trafficking of receptors (Croset et al., 2010; Abuin et al., 2011, 2019). Ir25a and Ir8a co-receptors are well conserved across insects (Croset et al., 2010; Rytz et al., 2013). In contrast, the Ir76b co-receptor does not contain the ATD or CREL domains and is less well-conserved (Croset et al., 2010).
Recent data suggest that the impact of Orco goes beyond odorant receptor protein function and localization to also play a neurodevelopmental role. Orco mutation in ants, honeybees, and Manduca moths induces a decrease in the volume and number of antennal lobe glomeruli (Trible et al., 2017; Yan et al., 2017; Fandino et al., 2019; Chen et al., 2021). This neurodevelopmental defect is associated with a loss of olfactory neurons in the antenna in ants and reduced antennal expression of most OR tuning receptors in honeybees (Trible et al., 2017; Yan et al., 2017; Chen et al., 2021). In contrast, mutation of Orco does not affect the gross anatomy or volume of the antennal lobe in Drosophila, the species with the best characterized olfactory system (Larsson et al., 2004; Chiang et al., 2009; Trible et al., 2017). There are only limited data on whether Orco impacts olfactory neuron survival in the Drosophila antenna. Olfactory neurons were reported to have generally normal morphology, and loss of olfactory neurons may be less prevalent than in other species (Larsson et al., 2004). For example, the number of antennal Or47b-expressing olfactory neurons is only ∼25% reduced at 14 days post-eclosion (DPE), and there is no loss of Or22a-expressing neurons at 6 DPE (Chiang et al., 2009; Hueston et al., 2016).
However, one study reported a widespread and progressive retraction of olfactory neuron axons in Drosophila Orco mutants, consistent with the initial stages of neuronal degeneration (Chiang et al., 2009). Further, a recent study found that olfactory neurons in the Drosophila maxillary palp progressively degenerate in Orco mutants, with more than 60% lost by 15 days post-eclosion (Task and Potter, 2021). This degradation differentially affects palp olfactory neuron classes, with nearly complete loss of Or46a-expressing neurons but no loss of Or42b-expressing neurons. Similarly, the expression of some OR tuning receptors in the antenna is decreased at 4 DPE in the absence of Orco (Jafari et al., 2021). Considering that olfactory neuron loss may be progressive with age and/or class-dependent, antennal olfactory neuron degeneration may have been overlooked previously.
Here, we set out to systematically and comprehensively examine the role of odorant co-receptors on the survival of antennal olfactory neurons in Drosophila. We use antennal RNASeq to identify OR and IR tuning subunits whose expression is lost in co-receptor mutants, as an indicator of olfactory neuron classes that may have degenerated. We then used transgenic reporters and lineage tracing to determine the fate of multiple specific olfactory neuron classes. Together, our data reveal that loss of odorant co-receptors leads to a decrease in mRNA expression of many tuning receptors, but few antennal olfactory neurons degenerate as a consequence of this insult.
Materials and methods
Fly stocks
Co-receptor mutant lines were obtained from the Bloomington Drosophila Stock Center: #23130 Orco2, #41744 Ir8a1, #51309 Ir76b1, and #41737 Ir25a2. Each of these mutations was outcrossed for at least ten generations into wCS, our standard genetic background that is a Cantonized w1118 line (Koh et al., 2014). Such flies also served as the control flies (WT) for RNASeq experiments. We also obtained driver and reporter lines from the Bloomington Drosophila Stock Center: #9946 Or13a-GAL4, #9947 Or19a-GAL4, #24614 Or49b-GAL4, #23125 Or82a-GAL4, #23131 Or83c-GAL4, #41732 Ir64a-GAL4, #32219 10XUAS-IVS-mCD8:RFP, and #28280 UAS-G-TRACE (UAS-RedStinger,UAS-FLP.D,UBI-p63E(FRT.STOP)Stinger). The Ir75a-GAL4 line was reported previously (Vulpe et al., 2021). Specific genotypes of fly lines used in this study are provided in Supplementary Table 1.
RNA isolation and sequencing
Antennae were dissected from flies either 0–2 DPE (“1 DPE”), 4–9 DPE (“7 DPE”), or 16–20 DPE (“20 DPE”), with approximately equal numbers of males and females. Samples were collected from wCS (WT), Ir25a2, Ir8a1, Ir76b1, Orco2 and Ir25a2;Ir76b1 flies at the time points specified in the text. WT samples of the same age were compared to Orco2 flies in Figure 1 and to IR co-receptor mutants in Figure 5. We isolated RNA using a protocol described previously (Mohapatra and Menuz, 2019). In brief, flies were dipped in liquid nitrogen, and antennae were manually dissected into 1.5 mL Eppendorf tubes in a liquid nitrogen bath. Antennae from 150 to 200 flies were dissected for each sample. Antennae were homogenized using disposable RNAse free micropestles (USA Scientific) and QIAshredder columns (Qiagen). RNA was extracted using an RNAeasy Micro Kit (Qiagen) and digested with DNAse from an iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad). The Center for Genome Innovation at the University of Connecticut carried out quality control on the RNA samples (∼0.5 μg each) and generated libraries with the TruSeq Stranded mRNA library kit (Illumina). Sequencing produced ∼10.3–30.3 million paired-end reads per sample.
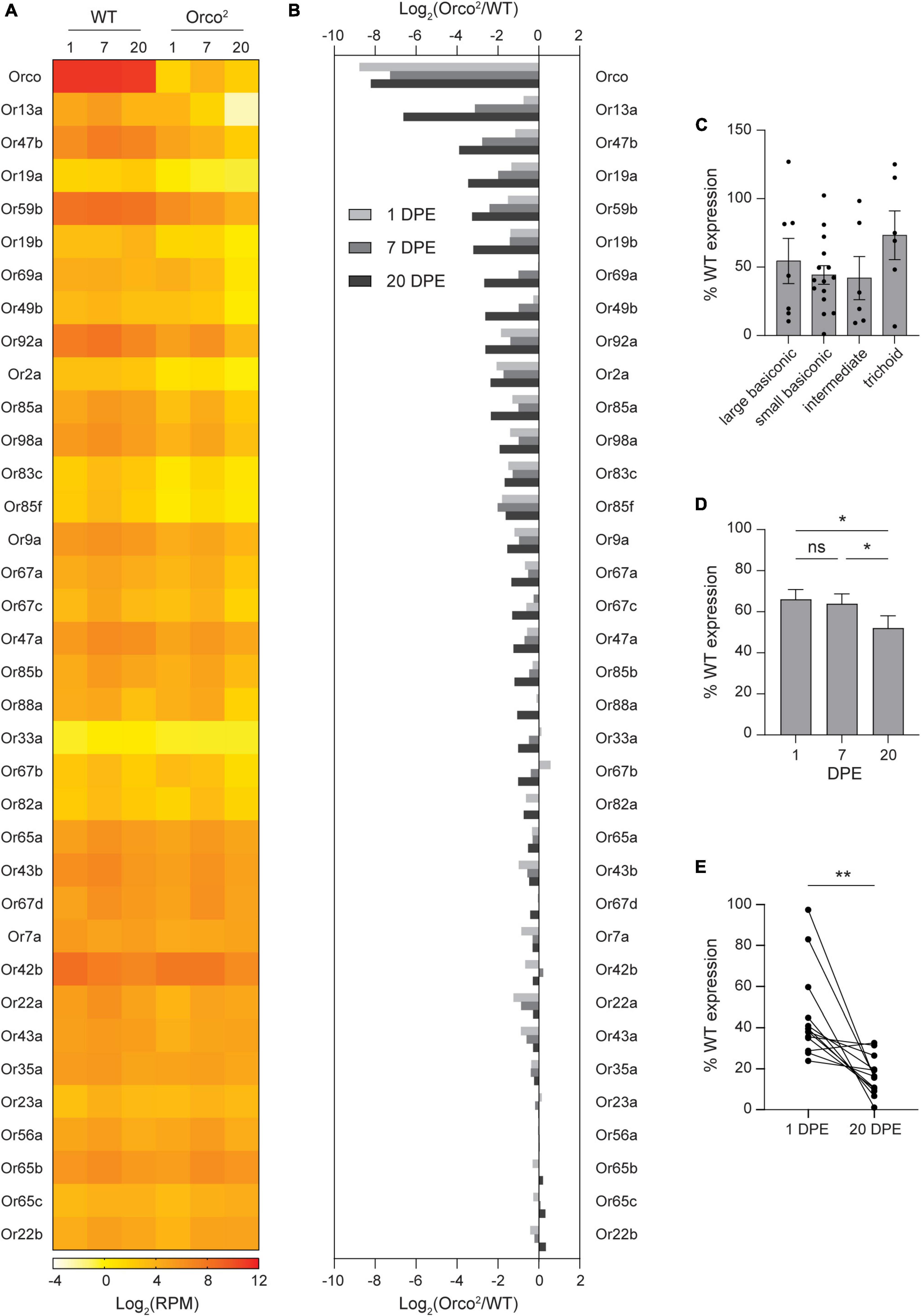
Figure 1. Expression of select OR tuning receptors is progressively lost in Orco2 flies. (A) Heat map showing the average antennal expression in Log2(RPM) for each of the known antennal OR genes at 1, 7, and 20 DPE in WT and Orco2 flies. (B) Bar graph reporting the Log2 of the expression ratio between Orco2 and WT flies for members of the OR family. ORs are arranged in descending order of the Log2 expression ratio at 20 DPE. (C) Expression of antennal OR tuning receptors in Orco2 flies as a percentage of expression in WT flies at 20 DPE. (D) Average expression of antennal OR tuning receptors in Orco2 flies as a percentage of expression in WT flies at different ages. (E) Graph showing expression of each of the 13 strongly reduced OR tuning receptors at 1 and 20 DPE in Orco2 flies as a percentage of expression in WT flies. Statistical significance is presented as *p < 0.05 and **p < 0.01. Other comparisons were not significant (p > 0.05).
RNASeq quantification and differential expression analysis
Raw reads were stored in BaseSpace Sequence Hub (Illumina) and downloaded to the University of Connecticut High Performance Computing cluster. Trimmed reads were generated using Sickle version 1.33 with the default parameters (Joshi and Fass, 2011). The Drosophila gene annotation file and reference genome Release 6.46 were downloaded from NCBI (dos Santos et al., 2014; Gramates et al., 2022). Trimmed reads were aligned to the reference genome with mitochondrial genes removed using STAR with the default parameters (Dobin et al., 2013). Mapped reads generated by STAR are available under BioProject accession number PRJNA735732 at the NCBI Sequence Read Archive (SRA). Raw counts of reads mapping uniquely to each gene were generated using HTSeq (Anders et al., 2015). HTSeq-count was used with “intersection-non-empty” and “non-unique-none” modes to handle reads mapping to more than one feature. To analyze expression of Ir76b on an exon-by-exon basis, we fed the aligned reads from STAR into DEXSeq and ran the program with the default parameters to estimate the RPM for each exon (Anders et al., 2012; Reyes et al., 2013). Three independent biological replicates were analyzed for most genotypes and ages. Because samples were not included for analysis if they were contaminated with other body parts (ex. heads), we utilized only two Ir76b1 7 DPE and Orco2 20 DPE samples. Further analysis was carried out on 1, 7, and 20 DPE samples using the raw gene counts from HTSeq-count with the EdgeR v3.40.2 package in R studio v4.2.1 (Robinson et al., 2010). First, the Reads per Million Mapped Reads (RPM) value for each gene in each sample was calculated using EdgeR (Dataset S1, “RPM”). These values were used for all figures, except in Figure 5A. In that figure, a small value (0.05) was added to each RPM value when calculating the Log2(RPM) values to avoid undefined values resulting from zero expression of Ir8a in Ir8a1 mutants.
For differential gene expression analysis on 7 DPE samples, a general linearized model in EdgeR was first used to identify differentially expressed (DE) genes over all samples (Dataset S1, “GLM”). To identify the specific samples in which the genes were differentially expressed, an EdgeR ExactTest was run between each co-receptor mutant and WT in a pair-wise fashion (Dataset S1, “ExactTest Orco vs WT”, “ExactTest Ir8a vs WT”, “ExactTest Ir25a vs WT”, “ExactTest Ir76b vs WT”, and “ExactTest Ir25a;Ir76b vs WT”). We identified DE genes for each co-receptor mutant as those with FDR < 0.01 in both the GLM and ExactTest and with a > 2-fold change in expression (Dataset S1, “all DE genes”). Finally, we identified 206 protein-coding DE genes that were differentially expressed in at least one genotype (Dataset S1, “206 protein encoding DE genes”).
DAVID with Knowledgebase v2023q2 was used for functional annotation of the 206 DE genes (Dataset S1, “206 protein encoding DE genes”) (Sherman et al., 2022). We used DAVID Functional Annotation Clustering to determine the types of functions that are statistically enriched among the DE genes in each co-receptor mutant relative to WT, with separate analyses run for upregulated and downregulated DE genes. We considered terms significant if they had a Benjamini and Hochberg adjusted p-value of <0.01. We used FlyBase (Release 6.54) to update the FlyBase Gene ID numbers and Gene Symbols for each gene in Dataset S1 “206 protein encoding DE genes” (Gramates et al., 2022).
Antennal whole-mount imaging
Fly crosses were used to generate experimental flies with either the WT or co-receptor background. For OrX > RFP and OrX > G-TRACE experiments, antennae were dissected from male and female flies either 1–2 DPE (“1 DPE”) or 17–22 DPE (“20 DPE”). For Ir75a > RFP and Ir64a > G-TRACE, antennae were dissected from 7 to 9 DPE (“7 DPE”) male flies. Dissected antennae were washed in 1X PBS, and then mounted in VectaShield (Vector Labs). Whole-mounted antennae were imaged on a Nikon A1R confocal microscope in the University of Connecticut Advanced Light Microscopy Facility. RFP-labeled antennae were imaged within 4 h after dissection, and G-TRACE antennae were imaged within 45 min after dissection, due to the weak expression of nuclear EGFP. Stacks of images (0.5 μm z-step) were acquired with a 40x oil-immersion objective. The imaging gain and offset were set the same for each compared pair of fly lines (same driver, reporter, and age, but either mutant or WT background). Images were analyzed with ImageJ/Fiji software using the Cell Counter tool (Schindelin et al., 2012).
Antennal immunohistochemistry
Immunohistochemistry was carried out on male and female flies. Flies were 17–21 DPE for anti-elav, anti-cleaved-caspase-3 and anti-cleaved-Dcp-1 staining and 6-8 DPE for anti-Ir64a staining. Approximately 4–5 flies of each sex were anesthetized, placed in an alignment collar, encased with OCT (Tissue-Tek) in a silicone mold, and frozen over dry ice. Blocks containing ∼8–10 aligned heads were snapped off and stored in 1.5 mL microcentrifuge tubes at −80°C until sectioning. A Leica CM3050S cryostat was used to collect 20 μm thick tissue sections at −20°C. All further steps were at room temperature, unless noted. Tissue sections were fixed for 10 min in a solution of PBS containing 4% paraformaldehyde. Sections were washed 3 × 5 min with PBS, permeabilized with PBS containing 0.2% TritonX-100 (PBST) for 30 min and blocked with PBST containing 1% Bovine Serum Albumin (BSA) for 30 min. The primary antibodies were diluted into 1% BSA-PBST, and 200 μL of the antibody solution was applied to each slide under a bridged coverslip. Primary antibodies were rabbit anti-cleaved-caspase-3 (1:500, RRID:AB_2341188), rabbit anti-cleaved-Dcp-1 (1:500, RRID:AB_2721060), rat anti-elav (1:10, RRID:AB_528218), and rabbit anti-Ir64a antibody (1:100, RRID:AB_2566854). Tissue sections were incubated overnight at 4°C, before being washed 3 × 10 min in PBST. Then, sections were incubated for 2 h in the dark with 200 μL of the secondary antibody solution composed of the appropriate secondary antibody diluted 1:500 into 1% BSA-PBST. Secondary antibodies were goat anti-rabbit Alexa Fluor 568 (RRID:AB_143157) for the caspase antibodies, goat anti-rat Alexa Fluor 488 (RRID:AB_2534074) for anti-elav, and goat anti-rabbit Alexa Fluor Plus 555 (RRID:AB_2633281) for anti-Ir64a. Slides were then washed 3 × 5 min, air dried for 30 min in the dark, and mounted in VectaShield (Vector Labs).
Stained antennal sections were imaged on a Nikon A1R confocal microscope. Stacks of images (0.5 μm z-step) were acquired with a 40x oil-immersion objective. ImageJ/Fiji software was used to analyze antennal histology. For anti-cleaved-caspase-3, anti-cleaved-Dcp-1, and anti-elav staining, an eleven-slice volume from each antenna was selected in the middle of the stack. All labeled cells in the volume were counted for anti-cleaved-caspase-3 and anti-cleaved-Dcp-1 quantification. Due to the density of elav staining, the number of elav+ cells in the eleven-slice volume was quantified in the region under a ∼1100 μm2 rectangle placed over the distal region of the antenna. For quantification of Ir64a+ neurons, only antennal sections in which the third chamber of the sacculus was visible with DIC were used because this region contains Ir64a+ neurons (Ai et al., 2010). Here, 20 slice volumes centered on the sacculus were used for quantification, and all cells in the volume were counted.
Statistical analysis
Data were analyzed using GraphPad Prism 10. Bar graphs depict the mean ± SEM overlaid with the individual data points. Student’s t-tests or a paired t-test were used to compare two conditions as described in the text. A one-way ANOVA or repeated measures ANOVA followed by Tukey’s post hoc test was used for comparisons of multiple genotypes, as noted in the text. Differences were considered significant if p < 0.05. Values are given as the mean ± SEM.
Results
Expression of select tuning OrXs is progressively reduced in Orco2 mutants
We reasoned that any classes of olfactory neurons that degenerate in the absence of Orco can be revealed by a loss of mRNA expression for their associated OR tuning receptors. Transcriptomic profiling can be used to quantitatively compare tuning receptor expression at multiple time points on all antennal olfactory neuron classes in parallel. We therefore used RNA Sequencing to compare antennal expression of OR tuning receptors in wild-type and Orco2 mutants, in which Orco expression is abolished. The Orco2 line was backcrossed for ten generations to our control white-eyed Canton-S flies (WT) to minimize genetic variability between lines. Antennal RNA harvested from the WT and Orco2 flies at 1, 7, and 20 DPE was subjected to next-generation transcriptional profiling, and receptor expression was quantified with HTSeq (Dataset S1, “RPM”). As expected, Orco expression is nearly eliminated in Orco2 flies (< 1% of WT at all ages) (Figures 1A, B). Remaining Orco expression is most likely derived from the exons that remain in the Orco2 flies because the mutation replaces only the first five transmembrane domains with a mini-white marker, and a smaller downstream portion of the gene remains in the genome.
We examined the expression of the 35 known antennal OR tuning receptors that are expressed in the antenna. We found that the loss of Orco leads to reduced OR tuning receptor expression (Figures 1A, B), with average OrX expression 52% of WT levels at 20 DPE. In contrast, expression of IR tuning receptors is maintained in Orco2 antenna, with average IrX expression 104% of WT levels at 20 DPE (Supplementary Figure 2).
The average loss of OR tuning receptors obscures a wide range of individual receptor variability. The greatest reduction is seen for Or13a, whose expression in Orco2 mutants is 1% of WT at 20 DPE (Figures 1A, B). Other tuning receptors with less than 1/3rd of WT level expression are Or47b (7%), Or19a (9%), Or59b (10%), Or19b (11%), Or69a (16%), Or49b (16%), Or92a (16%), Or2a (19%), Or85a (20%), Or98a (26%), Or83c (31%), and Or85f (33%). On the other extreme, expression of 10 tuning receptors remains at >80% of WT level at 20 DPE: Or7a, Or42b, Or22a, Or43a, Or35a, Or23a, Or56a, Or65b, Or65c, and Or22b. The identity of the tuning receptors whose expression is strongly Orco-dependent is not explained by sensillar morphology. The 13 tuning receptors with strongly reduced expression are found in all morphological classes of Orco-expressing sensilla: large and small basiconic, intermediate, and trichoid sensilla. Likewise, there is no difference in average tuning receptor loss by sensilla morphology (one-way ANOVA p > 0.05) (Figure 1C).
Loss of OR tuning receptor expression is generally progressive. On average, OR tuning receptor expression in Orco2 mutants relative to that in WT flies declines mildly with age (repeated measures one-way ANOVA followed by Tukey’s post hoc test p < 0.05, n = 35 receptors) (Figure 1D). The progressive nature of tuning receptor loss is clearer when considering only those receptors with strongly reduced expression at 20 DPE. For example, 60% of Or13a expression remained at 1 DPE, 11% at 7 DPE, and only 1% at 20 DPE (Figures 1A, B). The average expression in Orco2 mutants of the 13 most strongly reduced tuning receptors declined from 46% of WT at 1 DPE to 17% of WT at 20 DPE (paired t-test, p < 0.01, n = 13 receptors) (Figure 1E).
Reduced OrX expression is rarely associated with olfactory neuron degeneration
We wondered whether the loss of OrX expression was due to the degeneration of antennal olfactory neurons in Orco2 mutants, similar to what was previously observed in the maxillary palp in Drosophila and the antenna in ants (Trible et al., 2017; Task and Potter, 2021). To investigate the effects of Orco on the survival of specific classes of antennal olfactory neurons, we drove UAS-mCD8:RFP expression using validated OrX-GAL4 drivers in WT flies and Orco2 mutants and quantified the number of labeled olfactory neurons in whole mount antennae. We first examined neurons that express Or13a, the tuning receptor with the greatest reduction in expression in Orco2 mutants at 20 DPE. Examination of Or13a > RFP antennae revealed a ∼65% reduction in the number of Or13a+ olfactory neurons in Orco2 flies at 20 DPE (Student’s t-test p < 0.0001, n = 25 antenna each) (Figures 2A, B). The loss of Or13a+ neurons is progressive, as there is no significant difference in the number of neurons at 1 DPE (Student’s t-test p > 0.05, n = 25 antenna each).
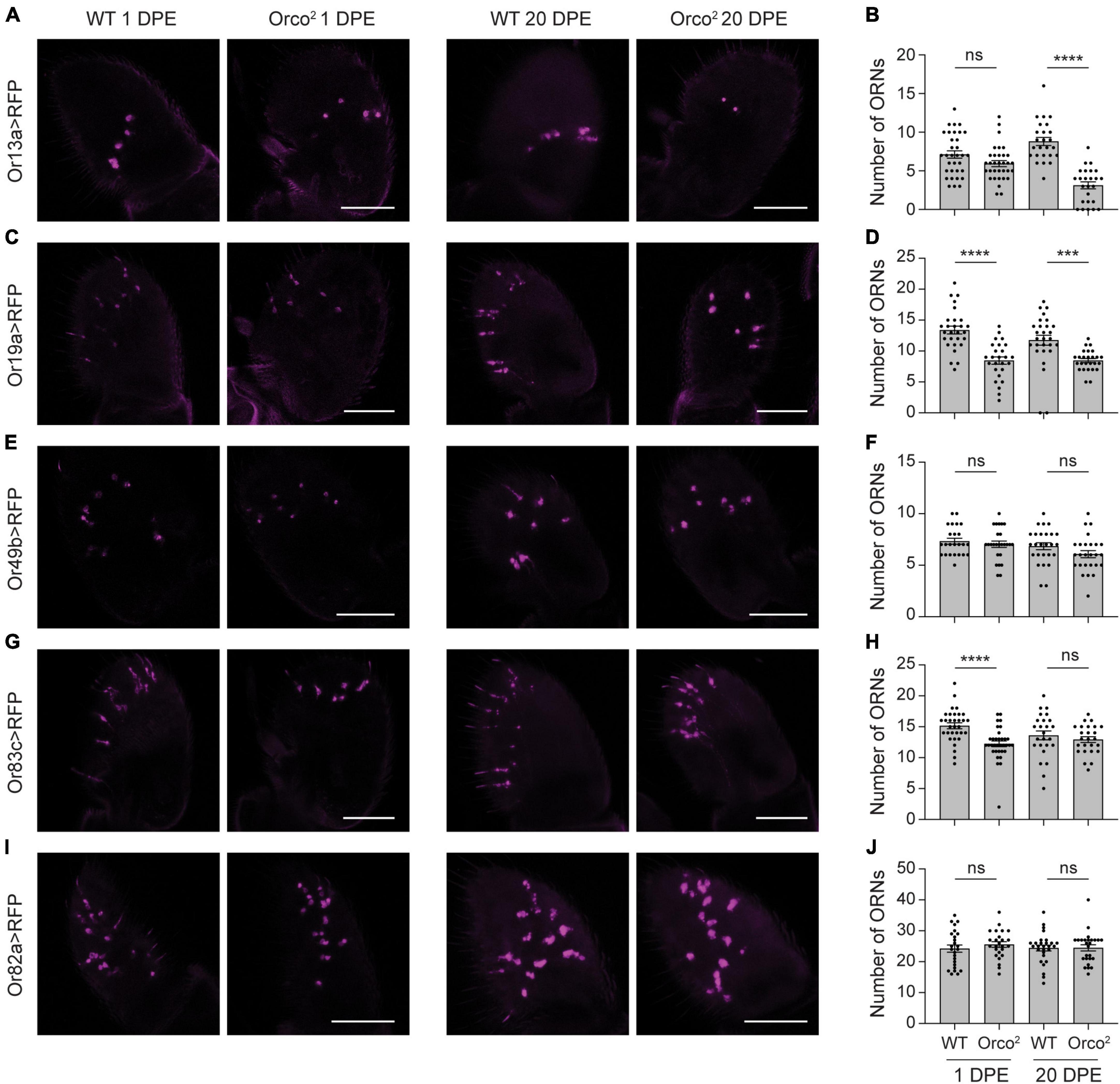
Figure 2. Reductions in OrX expression are sometimes associated with mild loss of associated olfactory neurons. (A) Whole-mount images of antenna taken from 1 and 20 DPE flies in which UAS-mCD8:RFP is driven by Or13a-GAL4 in a WT or Orco2 mutant background. Scale bar, 50 μm. (B) Quantification of the number of olfactory receptor neurons (ORNs) detected in each antenna. Bar graph depicts the mean ± SEM overlaid with the individual data points. (C–J) Similar but using the Or19a-GAL4 (C,D), Or49b-GAL4 (E,F), Or83c-GAL4 (G,H), and Or82a-GAL4 (I,J) drivers. Statistical significance is presented as ***p < 0.001, ****p < 0.0001, and ns p > 0.05.
We also examined neurons that express Or19a, the receptor with the third greatest reduction in expression in Orco2 flies. There is a ∼30% loss of Or19a+ olfactory neurons at both 1 and 20 DPE (Student’s t-tests p < 0.0001 at 1 DPE and p < 0.001 at 20 DPE, n = 26–28 antenna each) (Figures 2C, D). We next looked at Or49b+ and Or83c+ neurons, as expression of both receptors is strongly reduced in Orco2 flies. We found that the number of Or49b+ neurons is unchanged in the absence of Orco (Student’s t-tests p > 0.05 at 1 and 20 DPE, n = 24–26 antenna each) (Figures 2E, F). Although we detected slightly fewer (∼20%) Or83c+ olfactory neurons at 1 DPE in Orco2 flies compared to controls, there is no difference in number at 20 DPE (Student’s t-tests p < 0.0001 at 1 DPE and p > 0.05 at 20 DPE, n = 25–34 antenna each) (Figures 2G, H). Finally, we examined Or82a+ olfactory neurons as a representative for receptors with a more moderate loss of receptor expression (Or82a: 40% decreased versus WT at 20 DPE). Here, we found no significant difference in the number of olfactory neurons in WT and Orco2 flies, both at 1 and 20 DPE (Student’s t-tests p > 0.05 at 1 and 20 DPE, n = 25–28 antenna each) (Figures 2I, J). There appears to be a non-linear relationship between reductions in tuning receptor expression and loss of olfactory neurons in Orco2 mutants at 20 DPE (Supplementary Figure 3), with substantial (> 25%) reductions in olfactory neuron number only observed with >90% reductions in tuning receptor expression.
In these experiments, we observed that labeling of some olfactory neurons is weaker in Orco2 mutants compared to WT flies (Figure 2), even if the number of cells did not change. This could be a consequence of using OrX-GAL4 lines to label different classes of olfactory neurons. If OR tuning receptor mRNA expression is reduced, then OrX-GAL4 expression may also be decreased. Likewise, RFP expression tended to be somewhat stronger at 20 DPE than 1 DPE, consistent with the known increase in odorant receptor expression to adult levels during the first few days post-eclosion (Jafari et al., 2021). Together, this may explain how there can be fewer Or83c+ cells in Orco2 mutants at 1 DPE, but no change at 20 DPE. It also suggests that the loss of Or13a+ and Or19a+ olfactory neurons in Orco2 mutants at 20 DPE may be overestimated.
To attempt to overcome this issue, we utilized the G-TRACE lineage labeling technique (Evans et al., 2009). In this approach, real time expression of GAL4 labels cells with nuclear marker RedStinger, and cells that have ever expressed the GAL4 are labeled with nuclear EGFP using a FLP recombinase-FRT approach. By using G-TRACE, we hypothesized that we could detect ORNs at 20 DPE, as long as they transiently express the OrX-GAL4 at a sufficient level earlier in development. The benefit of this system is that FLP recombination only needs to occur once to allow for permanent GFP-marking of ORNs, even if OrX-GAL4 expression (and therefore RedStinger expression) is very weak at the 20 DPE time point.
We first used Or13a-GAL4 to drive the G-TRACE system. We found a ∼60% reduction in the number of red Or13a+ cells in Orco2 mutants compared to WT flies at 20 DPE (Student’s t-test p < 0.0001, n = 17–18 antenna each) (Figures 3A, B). No RFP+ cells were seen in the absence of the GAL4 driver, but these control flies do have a small number of GFP+ cells, suggesting low-level FLP recombination in the absence of a GAL4 driver. The number of GFP+ RFP– cells is similar in all conditions (one-way ANOVA p > 0.05, n = 17–18 antenna each) (Figures 3A, B). Accordingly, there is a significant reduction in the total number of Or13a+ cells in Orco2 antennae compared to WT flies (one-way ANOVA followed by Tukey’s post hoc test p < 0.01, n = 17–18 antenna each). Together, our G-TRACE data are consistent with the partial loss of Or13a+ ORNs observed with the simpler RFP reporter experiments (Figures 2A, B).
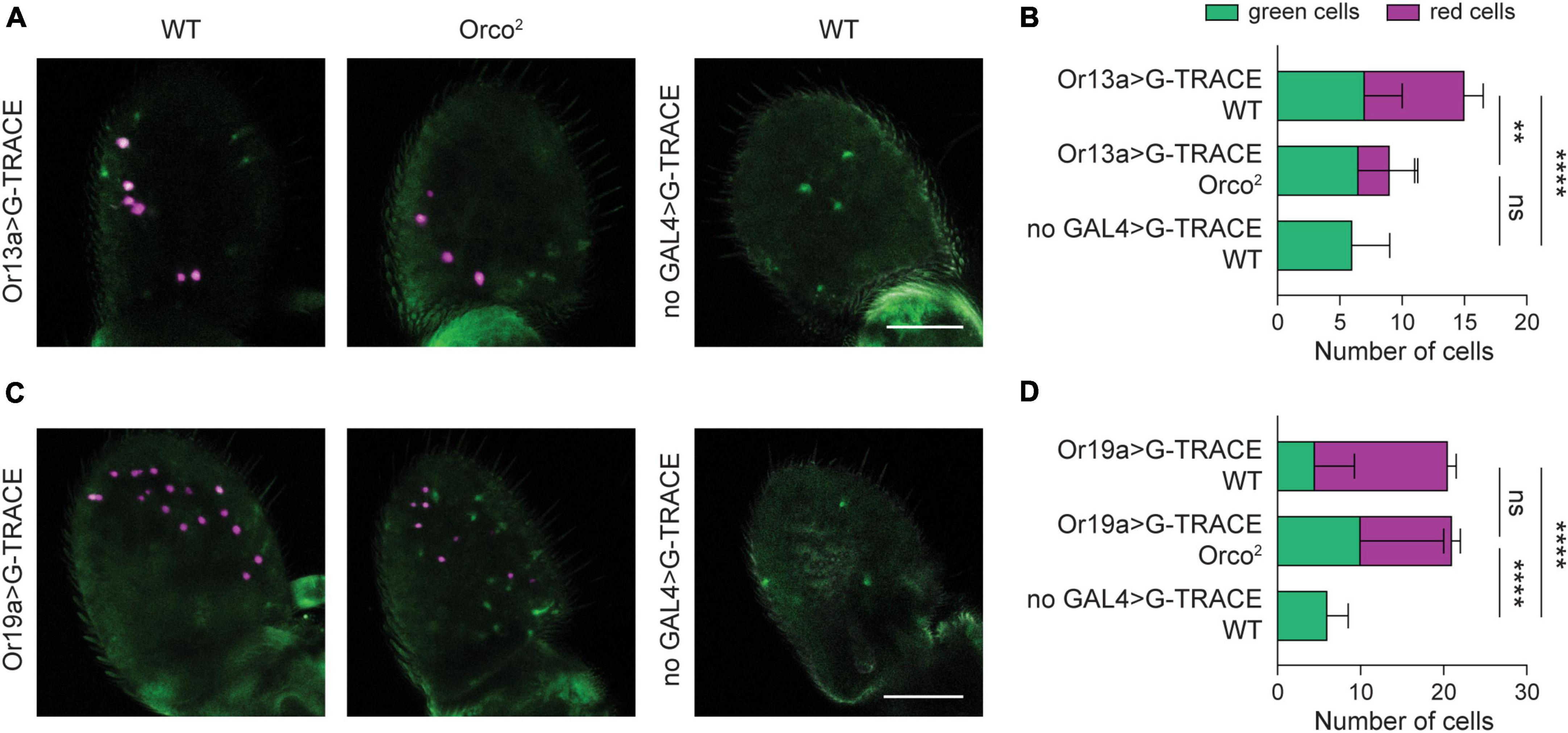
Figure 3. Retention of Or19a+ olfactory neurons despite severe loss of Or19a mRNA expression. (A) Whole-mount images of antenna taken from 20 DPE flies in which the G-TRACE cell lineage tracing system transgenes are driven by Or13a-GAL4 in a WT or Orco2 mutant background. The right panel shows a control WT antenna in which the GAL4 is absent. Scale bar, 50 μm. (B) Quantification of the average number of GFP+ RFP– cells (green cells) and RFP+ cells (red cells) per antenna in each genotype. Bar graph depicts the mean ± SEM. (C,D) Similar but using the Or19-GAL4 driver. Statistical significance for comparison of the total number of cells detected is presented as **p < 0.01, ****p < 0.0001, and ns p > 0.05.
We next used Or19a-GAL4 to drive the G-TRACE system (Figures 3C, D). We found a ∼36% decrease in the number of RFP+ cells in Orco2 flies compared to WT controls (Student’s t-test p < 0.0001, n = 18–19 antenna each), consistent with the loss observed earlier (Figure 2D). However, this loss is concomitant with an increase in the number of GFP+ RFP– cells (one-way ANOVA followed by Tukey’s post hoc test p < 0.01 vs WT and p < 0.05 vs no-GAL4 control, n = 17–19 antenna each). As a result, the total number of labeled Or19a+ cells is unchanged in Orco2 mutants (one-way ANOVA followed by Tukey’s post hoc test p > 0.05, n = 17–19 antenna each). Together, this suggests that reduced Or19a-GAL4 expression in Orco2 mutants causes some Or19a neurons to appear RFP–, and that Or19a+ olfactory neurons persist in Orco2 mutants despite > 90% loss of Or19a mRNA expression.
Loss of Orco does not cause widespread olfactory neuron degeneration
Our data using markers for individual olfactory neuron classes suggest that most antennal olfactory neurons do not degenerate in the absence of Orco, unlike those in the maxillary palp. However, we only examined a subset of the many olfactory neuron classes. To more comprehensively investigate whether cell death occurs in Orco2 flies, we used histology to look for signs of apoptosis. In flies, cleaved effector caspases serve as cell death markers (Vasudevan and Ryoo, 2016). There are four effector caspases in Drosophila, of which two (Drice/caspase-3 and Dcp-1) are expressed at >1 RPM in Orco2 flies at 20 DPE in the antenna; their expression level was similar to that in WT flies (Dataset S1, “RPM”). To determine if the caspase proteins were activated by cleavage, we stained antennal sections with anti-cleaved-caspase antibodies. On average, we found less than one cell labeled with anti-cleaved-caspase-3 in antenna from either WT or Orco2 flies (Student’s t-test p > 0.05, n = 6 antenna each) (Figures 4A, B). Similarly, we detected few cells labeled with anti-cleaved-Dcp-1 in WT and Orco2 antennae, although there was an increase in the absence of Orco (WT: 0.2 ± 0.2 and Orco2: 3.3 ± 0.9 cells, Student’s t-test p < 0.01, n = 6 antenna each) (Figures 4C, D).
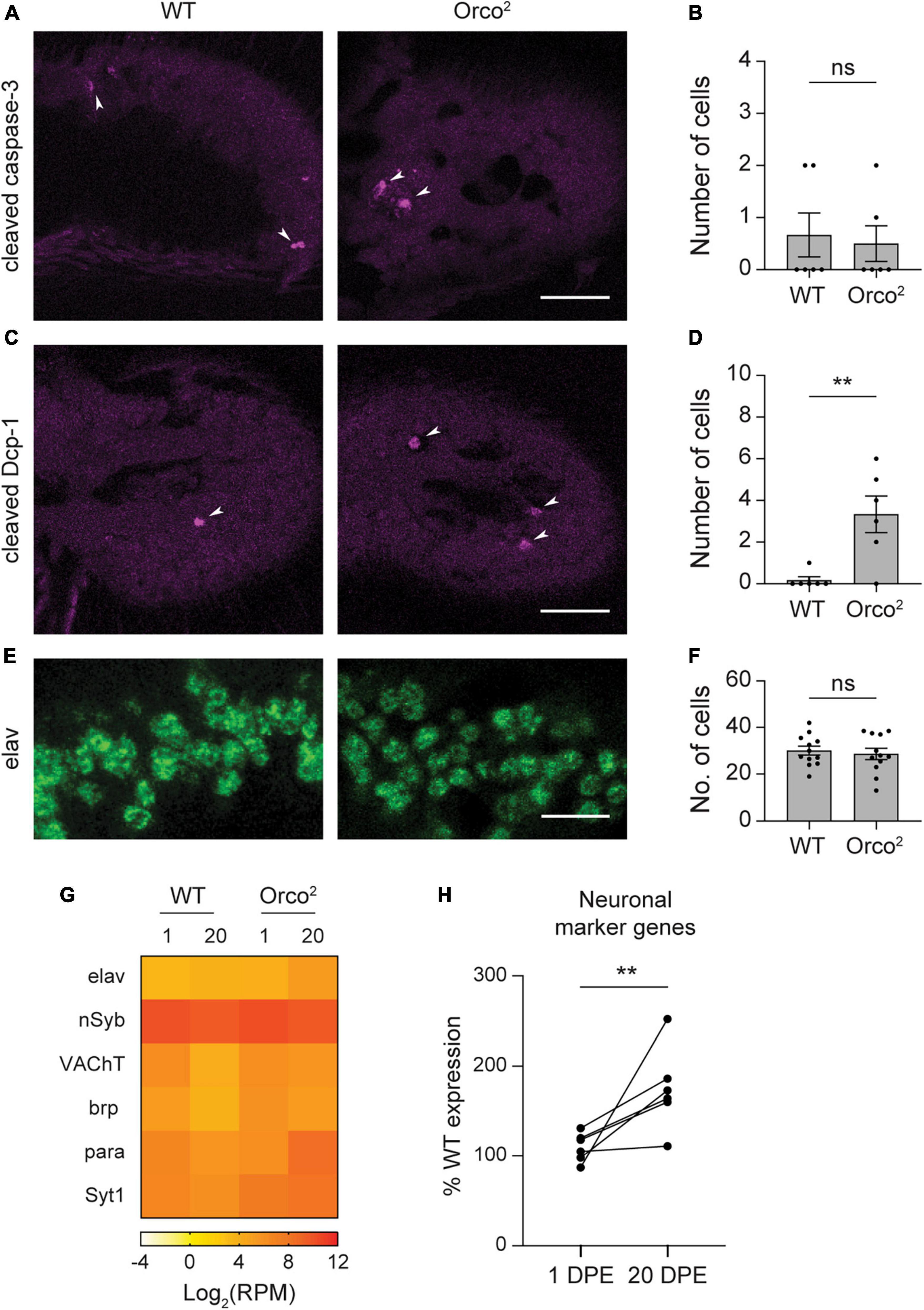
Figure 4. Loss of Orco does not cause widespread olfactory neuron degeneration. (A) Antennal sections from WT or Orco2 flies stained with anti-cleaved-caspase-3. White arrowheads point to labeled cells. Scale bar, 25 μm. (B) Quantification of the number of labeled cells detected in each antenna. Bar graph depicts the mean ± SEM overlaid with the individual data points. (C,D) Similar for staining with anti-cleaved-Dcp-1. (E,F) Similar for staining with anti-elav. The image covers the same areas as the rectangle used for quantification through an eleven-slice volume. Scale bar, 10 μm. (G) Heat map showing the average antennal expression in Log2(RPM) for neuronal marker genes at 1 and 20 DPE in WT and Orco2 flies. (H) Graph showing expression of each of the neuronal marker genes at 1 and 20 DPE in Orco2 flies as a percentage of expression in WT flies. Statistical significance is presented as **p < 0.01 and ns p > 0.05.
To more directly determine if there is a loss of neurons in the Orco2 mutants, we also compared the number of antennal cells labeled with the neuronal marker elav (Figures 4E, F). To do this, we stained antennal sections with anti-elav and compared the number of cells within a fixed volume. We found a similar number of elav-positive cells in WT and Orco2 antennae (WT: 30.0 ± 1.9 and Orco2: 28.6 ± 2.4 cells, Student’s t-test p > 0.05, n = 12 antenna each). Consistent with this finding, expression of elav transcripts was not reduced in Orco2 flies in our RNASeq data. Interestingly, although elav and other neuronal markers nSyb, VACht, brp, para, and Syt1 generally had unchanged expression in Orco2 mutants at 1 DPE, their expression was often higher at 20 DPE (paired t-test, p < 0.05) (Figures 4G, H). Together, these data suggest that the loss of OR tuning receptor expression in Orco2 mutants is independent of neuron degeneration, which does not broadly occur in Orco2 flies.
Loss of IR co-receptors affects expression of associated IR tuning receptors
Does expression of IR tuning receptor mRNA also rely on expression of their associated co-receptors? Previous electrophysiology experiments have shown that amine-sensitive IR tuning receptor responses are selectively eliminated in Ir76b, Ir25a, and Ir25a;Ir76b mutants, whereas acid-sensitive IR tuning receptor responses are selectively eliminated in Ir8a mutants (Abuin et al., 2011; Vulpe and Menuz, 2021). We therefore examined IR tuning receptor expression using antennal RNASeq on outcrossed IR family co-receptor mutants at 7 DPE, a time point at which average OR tuning receptor expression in the absence of Orco is only ∼60% of that in control flies.
As expected, expression of Ir8a and Ir25a is nearly eliminated in the Ir8a1 and Ir25a2 mutants, respectively (Figure 5A). Surprisingly, expression of Ir76b transcripts decreased only ∼40% in Ir76b1 mutants (Figure 5A). Closer examination revealed that expression of the first three exons is mostly lacking (Figure 5B), in agreement with the loss of receptor function in this line (Vulpe and Menuz, 2021).
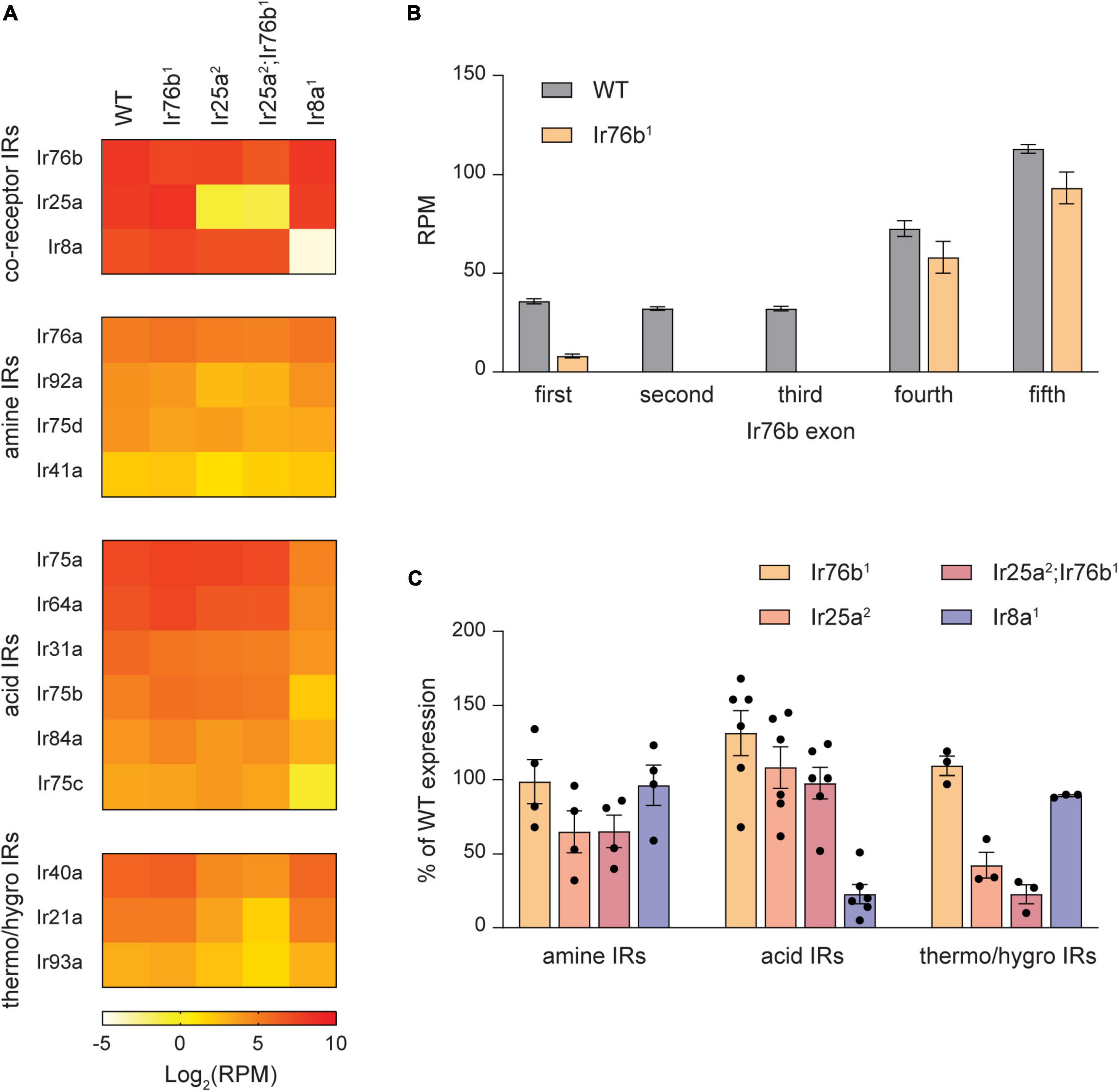
Figure 5. Expression of many IR tuning receptors depends on their IR co-receptor. (A) Heat map showing the average antennal expression in Log2(RPM) for detected antennal IR genes at 7 DPE in WT, Ir76b1, Ir25a2, Ir25a2;Ir76b1, and Ir8a1 flies. The IR genes are grouped by their type: co-receptors, Ir25a- and Ir76b-dependent amine-sensitive tuning receptors, Ir8a-dependent acid-sensitive receptors, and Ir25a-dependent thermosensitive and/or hygrosensitive receptors. (B) The expression level in RPM for each exon of the Ir76b gene in WT flies and Ir76b1 mutants. (C) Expression of amine-sensitive, acid-sensitive and thermo/hygrosensitive IR tuning receptors as a percentage of expression in WT flies in different IR co-receptor mutants.
Interestingly, we found that expression of most amine-sensitive IR tuning receptors is reduced in Ir25a2 mutants, but not Ir76b1 mutants (Figures 5A, C). On average, expression of amine-sensitive IR tuning receptors in Ir25a2 mutants is ∼65% of WT flies, with large variation by individual receptor. This reduction is not increased in Ir25a2;Ir76b1 double mutants. Expression of amine-sensitive IrX receptors is generally unaffected by the loss of the acid co-receptor Ir8a (Figures 5A, C). In contrast, expression of all acid-sensitive receptors is consistently and strongly reduced in Ir8a1 antennae, but unaffected by the loss of Ir25a or Ir76b (Figures 5A, C). We also noted that expression of Ir40a, Ir21a, and Ir93a, which form functional receptors with Ir25a in antennal hygrosensory and thermosensory neurons (Ni, 2021), depends on Ir25a expression. As expected, OR tuning receptor expression is mostly unaffected by the loss of IrX co-receptors (Supplementary Figure 4). Together, our data indicate that expression of many IR tuning receptors relies on expression of their specific IR co-receptors, like the reliance of OR tuning receptor expression on Orco.
Ir8a-dependent acid-sensitive olfactory neurons mostly persist in Ir8a1 flies
Given the pronounced reduction in Ir8a-dependent acid-sensitive tuning receptor expression in the absence of Ir8a (∼75%), we investigated whether this is due to the degeneration of the olfactory neurons that express these receptors. We first investigated Ir75a+ olfactory neurons at 7 DPE, as Ir75a transcript expression is only 18% of WT flies in Ir8a1 mutants at this age. Using Ir75a-GAL4 to drive UAS-mCD8:RFP we found no change in the number of neurons in Ir8a1 antenna (Student’s t-test p > 0.05, n = 24–26 antenna each) (Figures 6A, B).
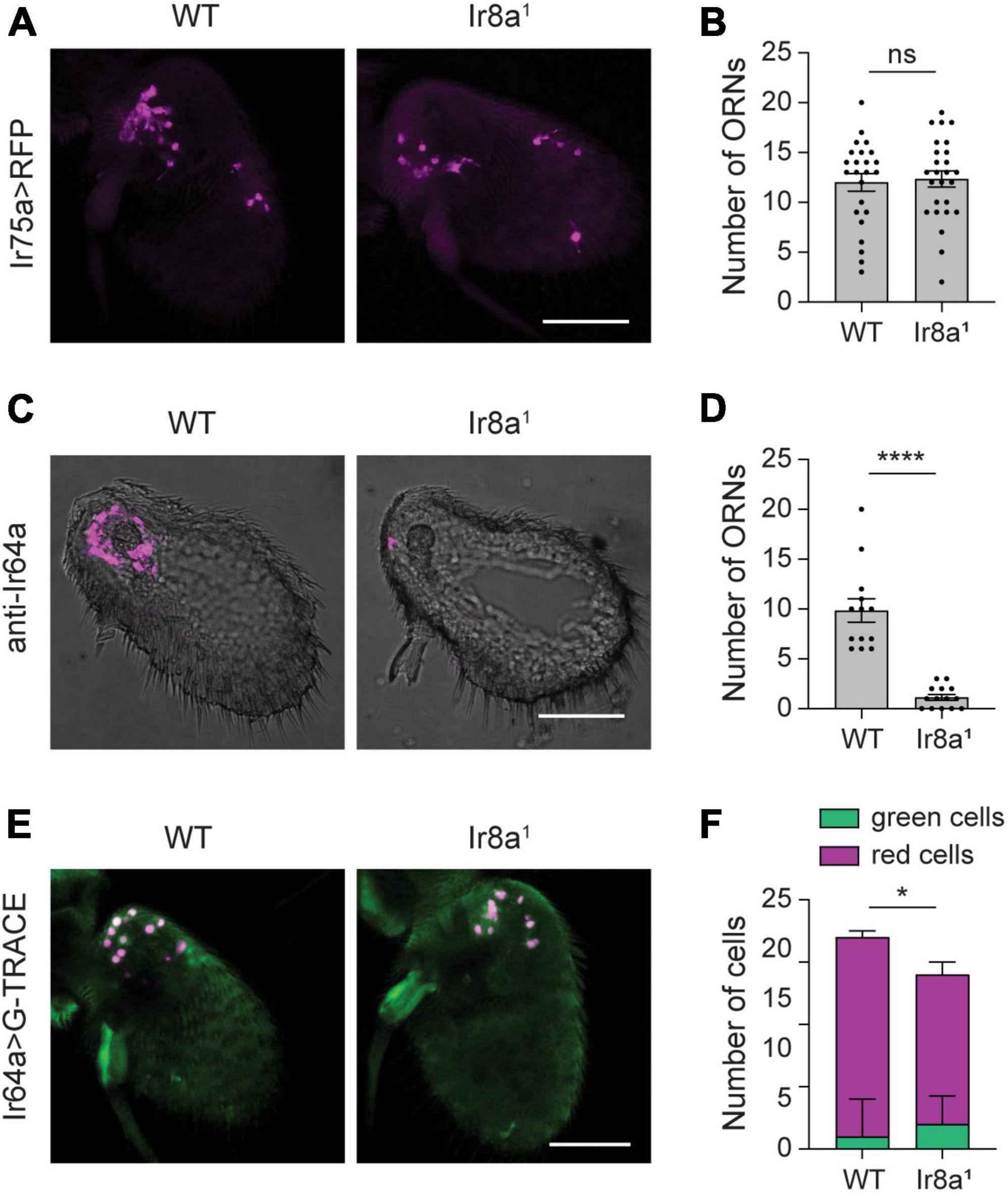
Figure 6. Little to no loss of Ir8a-dependent ORNs in Ir8a1 flies. (A) Whole-mount images of antenna taken from flies in which UAS-mCD8:RFP is driven by Ir75a-GAL4 in a WT or Ir8a1 mutant background. (B) Quantification of the number of olfactory receptor neurons (ORNs) detected in each antenna. (C) Antennal section from WT or Ir8a1 flies stained with anti-Ir64a. (D) Quantification of the number of ORNs detected in each antennal section. (E) Whole-mount images of antenna taken flies in which the G-TRACE cell lineage tracing system transgenes are driven by Ir64a-GAL4 in a WT or Ir8a1 mutant background. (F) Quantification of the average number of GFP+ RFP– cells (green cells) and RFP+ cells (red cells) per antenna in each genotype. Scale bars, 50 μm. Bar graphs depict the mean ± SEM. Those in panels (B,D) are overlaid with the individual data points. Statistical significance is presented as *p < 0.05, ****p < 0.0001, and ns p > 0.05.
We next examined Ir64a, a tuning receptor with 80% loss of transcript expression in Ir8a1 mutants. Here, we took advantage of a validated anti-Ir64a antibody to use immunohistochemistry to quantify the number of Ir64a-expressing neurons. We found that the number of olfactory neurons that express Ir64a protein is sharply lower (89%) in Ir8a1 mutants compared to controls (Student’s t-test p < 0.0001, n = 13–14 antennal sections each) (Figures 6C, D). However, there could be an apparent loss of neurons if loss of Ir64a protein expression makes it difficult to detect weakly labeled neurons. Therefore, we used Ir64a-GAL4 to drive the G-TRACE system to mark Ir64a+ olfactory neurons. There is a small (24%), but significant, loss of RFP+ neurons in Ir8a1 antenna (Student’s t-test p < 0.001, n = 17–18 antenna each) (Figures 6E, F). This is not accompanied by a significant increase in the number of GFP+ RFP– cells (Student’s t-test p > 0.05, n = 17–18 antenna each), but rather by a ∼18% loss of total labeled cells (Student’s t-test p < 0.05, n = 17–18 antenna each) (Figures 6E, F). Thus, loss of IR tuning receptor mRNA and protein expression is much greater than the loss of olfactory neurons in Ir8a1 mutants, like what is observed in Orco2 mutants.
Changes in gene expression associated with loss of odorant co-receptors
Is there a broader reorganization of gene expression in the absence of odorant co-receptors? To address this question, we used EdgeR to identify genes whose expression is differentially expressed in the antennae of co-receptor mutant flies compared to the genetic control line, focusing on those genes with at least a two-fold difference in expression. We detected a total of 206 DE protein-coding genes across all genotypes, with 33 to 159 DE genes per genotype (Figure 7A). There are roughly even numbers of upregulated and downregulated genes in each genotype. Many genes (113 of 206) are differentially expressed in at least two genotypes. For 111 of these genes, the direction of change (upregulation or downregulation) relative to control flies is the same in all genotypes in which it is differentially expressed.
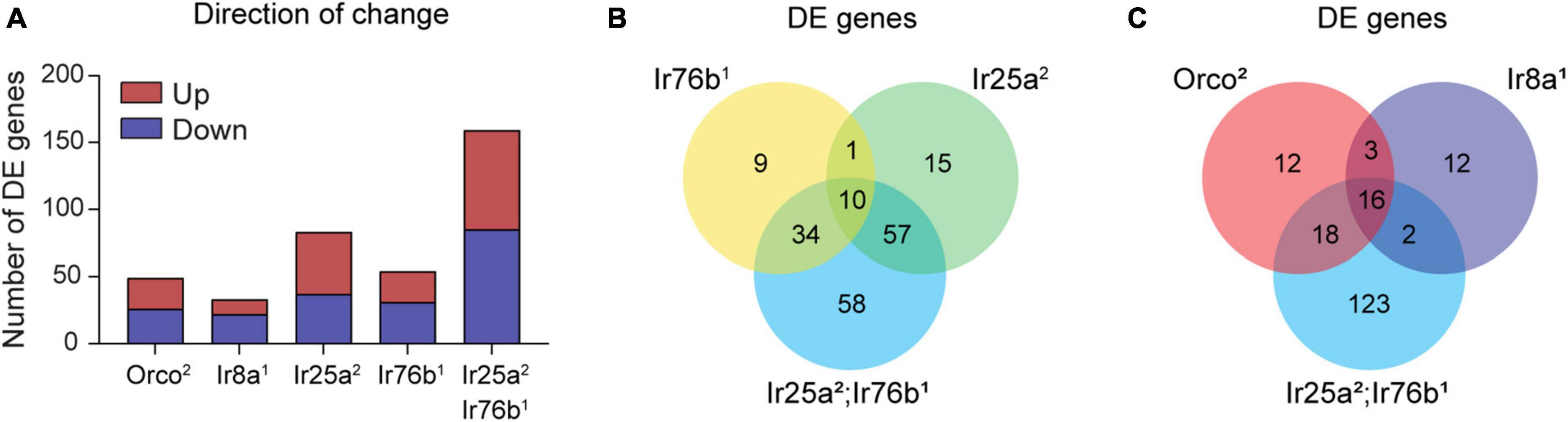
Figure 7. Differentially expressed genes are detected in co-receptor mutants. (A) Bar graph depicting the total number of DE genes identified in Orco2, Ir8a1, Ir25a2, Ir76b1, and Ir25a2;Ir76b1 mutants. Upregulated and downregulated genes in mutant flies relative to WT are shown in maroon and navy, respectively. (B,C) Venn diagrams showing the overlap between DE genes detected in the different co-receptor mutant lines.
We first examined the DE genes from flies lacking co-receptors for amine-sensitive IrX receptors (Figure 7B). Interestingly, the 54 DE genes in Ir76b1 flies are mostly distinct from the 83 genes in Ir25a2 flies, despite their similar role as co-receptors. This is aligned with our earlier finding that amine tuning receptor transcript expression depends on Ir25a, but not Ir76b (Figure 5C). It may also reflect the involvement of Ir25a, but not Ir76b, in antennal receptors mediating humidity and temperature sensing (Enjin et al., 2016; Knecht et al., 2016). Most genes (∼81%) that are differentially expressed in either Ir25a2 or Ir76b1 flies are also differentially expressed in the Ir25a2;Ir76b1 double mutants. However, approximately one-third of DE genes in the double mutants are not differentially expressed in either Ir25a2 or Ir76b1 flies (Figure 7B). This may indicate that there is some level of functional redundancy between Ir25a and Ir76b in their impact on antennal olfactory neuron function and gene transcription.
We next compared the DE genes in Orco2 flies and Ir8a1 flies with the DE genes in Ir25a2;Ir76b1 double mutants, as the latter includes most DE genes in Ir25a2 or Ir76b1 single mutants. We found that ∼77% of genes that are DE in Ir25a2;Ir76b1 double mutants are not DE in either Orco2 flies or Ir8a1 flies. There is a core group of ∼16 genes that are DE in each of these genotypes relative to the control flies (Figure 7C). For 15/16 of the genes, the direction of change (upregulation or downregulation in the mutants) is consistent across all three mutant genotypes. One of these genes is white (w), whose upregulation relative to the w- WT flies is expected because the Orco2, Ir8a1 and Ir25a2 mutations contain a mini-white marker that restores w function. The other 15 shared DE genes have disparate functions, ranging from dehydrogenase enzymes to a tRNA-synthetase to a TRP channel (Dataset S1, “206 protein encoding DE genes”). The transcription of these genes may generally be affected by the loss of odorant co-receptors or olfactory signaling. Alternatively, their differential expression may be due to the absence of w in the control flies, or due to slight variations in the genetic background of the control line that developed after out-crossing. Other than the 16 shared DE genes, there is little overlap between the Ir8a1 DE genes and DE genes in either Orco2 or Ir25a2;Ir76b1 flies. However, more than half of the Orco2 DE genes are also DE in the Ir25a2;Ir76b1 double mutants.
We annotated the predicted functions of the 206 DE protein-encoding genes using DAVID (Dataset S1, “206 protein encoding DE genes”) (Sherman et al., 2022). We found that 16 are members of the OR and IR family. Other DE genes have a variety of predicted functions, including ion channels, transporters, receptors, kinases, and metabolic enzymes. We used DAVID Functional Annotation Clustering to determine what types of functions are statistically enriched among the DE genes in each genotype. There is only one significant term cluster associated with the DE genes downregulated in Orco2 flies, “Drosophila Olfactory Receptor”, consisting of OR tuning receptors. Similarly, the only significant term cluster among DE genes downregulated in Ir8a1 flies related to “Ionotropic Glutamate Receptor”, the IR tuning receptors. No term clusters are enriched among the upregulated DE genes in the Ir8a1 or Orco2 flies, nor among up- or downregulated genes in Ir76b1 flies.
The upregulated genes in both Ir25a2 and Ir25a2;Ir76b1 flies are enriched for genes associated with “Innate Immunity”. These included several members of the Bomanin family of small, secreted peptides that can be released into the hemolymph upon microbial infection. The downregulated genes in Ir25a2 flies are enriched for genes associated with the ligand “Heme”, including members of the cytochrome p450 family such as Cyp4d21, Cyp4p2, Cyp6a17, and Cyp6g2. This family is also nearly significantly enriched among the downregulated DE genes in Ir25a2;Ir76b1 flies (adjusted p-value 0.084). Cyps are thought to function as Odorant Degrading Enzymes (ODEs) within the antenna, and their transcription can be upregulated by their substrates such as odorants (Baldwin et al., 2021). However, it is unclear why they are downregulated in the absence of Ir25a, an effect not seen in other co-receptor mutants. The term “Ion Channel” is also enriched amongst the downregulated DE genes in Ir25a2 flies. In addition to members of the IR family, these include the TRP channel TrpA1 and two members of the DEG/ENaC family of sodium channels ppk14, and ppk25. However, this term is not enriched among the downregulated genes in Ir25a2;Ir76b1 double-mutants. Taken together, our term enrichment analysis suggests that the major transcriptional effect seen in co-receptor mutants is on associated tuning receptors, and that large-scale transcriptional remodeling does not occur.
Discussion
Previous work has suggested that Drosophila olfactory receptor neurons degenerate in Orco mutants due to a loss of neuronal activity, as evidenced by axonal degeneration and retraction during the first week post-eclosion (Chiang et al., 2009). Loss of olfactory neuron firing activity is thought to activate Gsk-3β kinase, a master regulator affecting neuronal morphology and synapse formation (Chiang et al., 2009; Aimino et al., 2023; Lai et al., 2023). Likewise, expression of the neuronal marker elav is strongly decreased in the maxillary palp in Orco mutants, suggesting widespread loss of olfactory neurons (Task and Potter, 2021). In contrast, our transcriptomic data suggest that the antennal transcriptome is not broadly remodeled in the absence of Orco, and that the most prominent category of differentially expressed genes are OR tuning receptors. Furthermore, there is no evidence of widespread apoptosis or loss of elav+ cells in Orco2 flies. Thus, our data show that the absence of Orco does not lead to substantial loss of olfactory neurons in the antenna, at least through the first 3 weeks post-eclosion.
We also addressed whether selected classes of olfactory neurons are lost in Orco mutants. This possibility was suggested by the highly variable impact of Orco2 mutation on expression of different OR tuning receptors in the antennal transcriptome at 20 DPE; there is >90% loss of Or13a, Or47b, and Or19a on one extreme, and 10 receptors with <20% reduction on the other. Our initial studies using OrX-GAL4 lines to label neurons with an RFP reporter suggested that there are ∼65% fewer Or13a+ neurons and ∼30% fewer Or19a+ neurons in Orco2 flies. This is consistent with the ∼30% loss of Or47b+ neurons previously reported in Orco mutants (Hueston et al., 2016). For each of these three tuning receptors, the loss of OrX mRNA expression is much greater than the loss of neurons would suggest. We therefore explored the possibility that there is only an apparent loss of neurons due to transient or weak OrX-GAL4 expression, which may parallel loss of OrX mRNA expression. Using the G-TRACE lineage tracing system, we found that Orco2 flies have no loss of Or19a+ neurons, although there is a bonafide loss of Or13+ neurons. Considered together with our data showing no loss of three additional olfactory neuron classes and previous work showing no loss of Or22a+ olfactory neurons, our data suggest that widespread loss of antennal odorant receptor expression is mostly independent of olfactory neuron degeneration in the absence of Orco. This conclusion could be further validated with ORN-class specific markers whose expression is independent of Orco expression; however, to our knowledge such markers have not been reported.
We found a similar dependence of IR tuning receptor mRNA expression on their associated co-receptors, Ir25a and Ir8a. This effect is particularly pronounced for the six Ir8a-dependent acid-sensitive receptors, whose average expression is decreased ∼75% in Ir8a1 flies. Like Orco-dependent olfactory neurons, our data suggest that there is little degeneration of Ir8a-dependent neurons in the absence of Ir8a. The loss of mRNA expression of the four Ir25a-dependent amine-sensitive receptors is weaker and more variable in Ir25a2 flies. Loss of Ir25a also led to moderately reduced expression of hygrosensory and thermosensory IrX receptors, which also form functional receptors with Ir25a (Enjin et al., 2016; Knecht et al., 2016). Intriguingly, no loss of amine-sensitive IrX receptors was seen in the absence of Ir76b, although this co-receptor is required along with Ir25a for the formation of functional amine-sensitive receptors (Vulpe and Menuz, 2021). Ir25a and Ir76b do not have a functionally redundant role in maintaining the transcriptional expression of amine-sensitive IrXs, as elimination of both co-receptors did not lead to a further reduction in their expression than the loss of Ir25a alone.
The mechanism underlying the loss of odorant tuning receptor expression in co-receptor mutants is unclear. Lowered olfactory neuron activity induces a loss of synapses in the antennal lobe and mimics the effects of Orco mutation on axon degeneration (Chiang et al., 2009; Aimino et al., 2023). However, this activity-dependent mechanism cannot explain the highly variable effects of Orco on OrX mRNA expression because that the odor-induced activity in all OrX-dependent neurons depends on Orco. Likewise, it cannot explain why maxillary palp Or46a+ ORNs are nearly eliminated in Orco2 mutants, whereas Or42a+ ORNs are unaffected (Task and Potter, 2021). An activity-dependent mechanism is also inconsistent with the loss of amine-sensitive IrX receptor expression in the absence of Ir25a, but not Ir76b, given the absence of odor-induced activity in amine-sensitive neurons in the absence of either Ir25a and Ir76b (Vulpe and Menuz, 2021). Therefore, additional factors must underlie the differential susceptibility of tuning receptors to transcript loss in the absence of family-specific co-receptors. Recent work has revealed repressive transcriptional interactions between some odorant tuning receptors in insects, one of many mechanisms that support the expression of a singular tuning receptor in a particular olfactory neuron class (Jafari et al., 2021; Mika and Benton, 2021; Mika et al., 2021; Maguire et al., 2022). It will be interesting to determine if there are also transcriptional interactions between odorant tuning receptors and their co-receptors.
Our results suggest that the role of Orco in neurodevelopment is species-specific, unlike its role in neurophysiology. Whereas Orco mutagenesis in two species of ants leads to a massive loss of olfactory neurons in the periphery (Trible et al., 2017; Yan et al., 2017), we found that this does not occur in Drosophila. It was recently reported that Orco mutagenesis does not lead to antennal neuron loss in hoverflies, nor does it affect expression of most OR tuning receptors in Helicoverpa moths (Fan et al., 2022; Wu et al., 2023). Whether the widespread reduction in OR tuning receptor expression in honeybees correlates with olfactory neuron degeneration will need to be investigated (Chen et al., 2021). Orco mutations have been generated in a wide variety of insects, including mosquitos, locusts, moths, and others (DeGennaro et al., 2013; Koutroumpa et al., 2016; Li et al., 2016; Yang et al., 2016; Liu et al., 2017, 2023; Fandino et al., 2019; Sun et al., 2020). It will be interesting to use these organisms as tools to examine the species-specificity of Orco-dependent olfactory neuron survival.
Data availability statement
The datasets presented in this study can be found at the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA735732.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
TL: Conceptualization, Formal analysis, Investigation, Methodology, Writing – review and editing. PM: Conceptualization, Formal analysis, Investigation, Methodology, Writing – review and editing. SB: Formal analysis, Investigation, Writing – review and editing. KM: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by NIH award R35GM133209 to KM.
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank Dr. Greg Suh for sharing the anti-Ir64a antibody. We appreciate assistance from Eleanor LoConte with outcrossing genetic mutants, Ben Groce and Thendral Nagarajan with antennal dissections, and Hope Sylva with initial whole-mount imaging trials. We thank Dr. Anastasios Tzingounis for discussions and comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2024.1390557/full#supplementary-material
References
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60.
Abuin, L., Prieto-Godino, L. L., Pan, H., Gutierrez, C., Huang, L., Jin, R., et al. (2019). In vivo assembly and trafficking of olfactory Ionotropic Receptors. BMC Biol. 17:34. doi: 10.1186/s12915-019-0651-7
Ai, M., Min, S., Grosjean, Y., Leblanc, C., Bell, R., Benton, R., et al. (2010). Acid sensing by the Drosophila olfactory system. Nature 468, 691–695.
Aimino, M. A., DePew, A. T., Restrepo, L., and Mosca, T. J. (2023). Synaptic development in diverse olfactory neuron classes uses distinct temporal and activity-related programs. J. Neurosci. 43, 28–55. doi: 10.1523/JNEUROSCI.0884-22.2022
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Anders, S., Reyes, A., and Huber, W. (2012). Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017. doi: 10.1101/gr.133744.111
Baldwin, S. R., Mohapatra, P., Nagalla, M., Sindvani, R., Amaya, D., Dickson, H. A., et al. (2021). Identification and characterization of CYPs induced in the Drosophila antenna by exposure to a plant odorant. Sci. Rep. 11:20530. doi: 10.1038/s41598-021-99910-9
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Butterwick, J. A., Del Marmol, J., Kim, K. H., Kahlson, M. A., Rogow, J. A., Walz, T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452.
Chen, Z., Traniello, I. M., Rana, S., Cash-Ahmed, A. C., Sankey, A. L., Yang, C., et al. (2021). Neurodevelopmental and transcriptomic effects of CRISPR/Cas9-induced somatic orco mutation in honey bees. J. Neurogenet. 35, 320–332. doi: 10.1080/01677063.2021.1887173
Chiang, A., Priya, R., Ramaswami, M., Vijayraghavan, K., and Rodrigues, V. (2009). Neuronal activity and Wnt signaling act through Gsk3-beta to regulate axonal integrity in mature Drosophila olfactory sensory neurons. Development 136, 1273–1282. doi: 10.1242/dev.031377
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
DeGennaro, M., McBride, C. S., Seeholzer, L., Nakagawa, T., Dennis, E. J., Goldman, C., et al. (2013). orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498, 487–491. doi: 10.1038/nature12206
Del Marmol, J., Yedlin, M. A., and Ruta, V. (2021). The structural basis of odorant recognition in insect olfactory receptors. Nature 597, 126–131. doi: 10.1038/s41586-021-03794-8
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
dos Santos, G., Schroeder, A. J., Goodman, J. L., Strelets, V. B., Crosby, M. A., Thurmond, J., et al. (2014). FlyBase: Introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43, D690–D697. doi: 10.1093/nar/gku1099
Enjin, A., Zaharieva, E. E., Frank, D. D., Mansourian, S., Suh, G. S., Gallio, M., et al. (2016). Humidity sensing in Drosophila. Curr. Biol. 26, 1352–1358.
Evans, C. J., Olson, J. M., Ngo, K. T., Kim, E., Lee, N. E., Kuoy, E., et al. (2009). G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 6, 603–605. doi: 10.1038/nmeth.1356
Fan, X. B., Mo, B. T., Li, G. C., Huang, L. Q., Guo, H., Gong, X. L., et al. (2022). Mutagenesis of the odorant receptor co-receptor (Orco) reveals severe olfactory defects in the crop pest moth Helicoverpa armigera. BMC Biol. 20:214. doi: 10.1186/s12915-022-01411-2
Fandino, R. A., Haverkamp, A., Bisch-Knaden, S., Zhang, J., Bucks, S., Nguyen, T. A. T., et al. (2019). Mutagenesis of odorant coreceptor Orco fully disrupts foraging but not oviposition behaviors in the hawkmoth Manduca sexta. Proc. Natl. Acad. Sci. U. S. A. 116, 15677–15685. doi: 10.1073/pnas.1902089116
Gomez-Diaz, C., Martin, F., Garcia-Fernandez, J. M., and Alcorta, E. (2018). The two main olfactory receptor families in Drosophila, ORs and IRs: A comparative approach. Front. Cell Neurosci. 12:253. doi: 10.3389/fncel.2018.00253
Gramates, L. S., Agapite, J., Attrill, H., Calvi, B. R., Crosby, M. A., Dos Santos, G., et al. (2022). FlyBase: A guided tour of highlighted features. Genetics 220:iyac035. doi: 10.1093/genetics/iyac035
Hallem, E. A., Dahanukar, A., and Carlson, J. R. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135. doi: 10.1146/annurev.ento.51.051705.113646
Hueston, C. E., Olsen, D., Li, Q., Okuwa, S., Peng, B., Wu, J., et al. (2016). Chromatin modulatory proteins and olfactory receptor signaling in the refinement and maintenance of fruitless expression in olfactory receptor neurons. PLoS Biol. 14:e1002443. doi: 10.1371/journal.pbio.1002443
Jafari, S., Henriksson, J., Yan, H., and Alenius, M. (2021). Stress and odorant receptor feedback during a critical period after hatching regulates olfactory sensory neuron differentiation in Drosophila. PLoS Biol. 19:e3001101. doi: 10.1371/journal.pbio.3001101
Jones, W. D., Nguyen, T. A., Kloss, B., Lee, K. J., and Vosshall, L. B. (2005). Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 15, R119–R121. doi: 10.1016/j.cub.2005.02.007
Joshi, N. A., and Fass, J. N. (2011). Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ files. 1.33 ed.). Available online at: https://github.com/najoshi/sickle.
Knecht, Z. A., Silbering, A. F., Ni, L., Klein, M., Budelli, G., Bell, R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. Elife 5:e17879. doi: 10.7554/eLife.17879
Koh, T. W., He, Z., Gorur-Shandilya, S., Menuz, K., Larter, N. K., Stewart, S., et al. (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. doi: 10.1016/j.neuron.2014.07.012
Koutroumpa, F. A., Monsempes, C., Francois, M. C., de Cian, A., Royer, C., Concordet, J. P., et al. (2016). Heritable genome editing with CRISPR/Cas9 induces anosmia in a crop pest moth. Sci. Rep. 6:29620. doi: 10.1038/srep29620
Lai, S., Wang, P., Gong, J., and Zhang, S. (2023). New insights into the role of GSK-3beta in the brain: From neurodegenerative disease to tumorigenesis. PeerJ 11:e16635. doi: 10.7717/peerj.16635
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Li, Y., Zhang, J., Chen, D., Yang, P., Jiang, F., Wang, X., et al. (2016). CRISPR/Cas9 in locusts: Successful establishment of an olfactory deficiency line by targeting the mutagenesis of an odorant receptor co-receptor (Orco). Insect. Biochem. Mol. Biol. 79, 27–35. doi: 10.1016/j.ibmb.2016.10.003
Liu, Q., Liu, W., Zeng, B., Wang, G., Hao, D., and Huang, Y. (2017). Deletion of the Bombyx mori odorant receptor co-receptor (BmOrco) impairs olfactory sensitivity in silkworms. Insect. Biochem Mol. Biol. 86, 58–67. doi: 10.1016/j.ibmb.2017.05.007
Liu, X. Y., Yan, R., Chen, S. J., Zhang, J. L., and Xu, H. J. (2023). Orco mutagenesis causes deficiencies in olfactory sensitivity and fertility in the migratory brown planthopper, Nilaparvata lugens. Pest Manag. Sci. 79, 1030–1039. doi: 10.1002/ps.7286
Maguire, S. E., Afify, A., Goff, L. A., and Potter, C. J. (2022). Odorant-receptor-mediated regulation of chemosensory gene expression in the malaria mosquito Anopheles gambiae. Cell Rep. 38:110494. doi: 10.1016/j.celrep.2022.110494
Mika, K., and Benton, R. (2021). Olfactory receptor gene regulation in insects: Multiple mechanisms for singular expression. Front. Neurosci. 15:738088. doi: 10.3389/fnins.2021.738088
Mika, K., Cruchet, S., Chai, P. C., Prieto-Godino, L. L., Auer, T. O., Pradervand, S., et al. (2021). Olfactory receptor-dependent receptor repression in Drosophila. Sci. Adv. 7:eabe3745. doi: 10.1126/sciadv.abe3745
Mohapatra, P., and Menuz, K. (2019). Molecular profiling of the Drosophila antenna reveals conserved genes underlying olfaction in insects. G3 9, 3753–3771. doi: 10.1534/g3.119.400669
Neuhaus, E. M., Gisselmann, G., Zhang, W., Dooley, R., Stortkuhl, K., and Hatt, H. (2005). Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 8, 15–17. doi: 10.1038/nn1371
Ni, L. (2021). The structure and function of ionotropic receptors in Drosophila. Front. Mol. Neurosci. 13:638839. doi: 10.3389/fnmol.2020.638839
Reyes, A., Anders, S., Weatheritt, R. J., Gibson, T. J., Steinmetz, L. M., and Huber, W. (2013). Drift and conservation of differential exon usage across tissues in primate species. Proc. Natl. Acad. Sci. U. S. A. 110, 15377–15382. doi: 10.1073/pnas.1307202110
Robertson, H. M. (2019). Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 64, 227–242.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140.
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect. Biochem. Mol. Biol. 43, 888–897. doi: 10.1016/j.ibmb.2013.02.007
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi: 10.1093/nar/gkac194
Smart, R., Kiely, A., Beale, M., Vargas, E., Carraher, C., Kralicek, A. V., et al. (2008). Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect. Biochem. Mol. Biol. 38, 770–780. doi: 10.1016/j.ibmb.2008.05.002
Stocker, R. F. (1994). The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 275, 3–26. doi: 10.1007/BF00305372
Sun, H., Liu, F., Ye, Z., Baker, A., and Zwiebel, L. J. (2020). Mutagenesis of the orco odorant receptor co-receptor impairs olfactory function in the malaria vector Anopheles coluzzii. Insect. Biochem. Mol. Biol. 127:103497. doi: 10.1016/j.ibmb.2020.103497
Task, D., and Potter, C. J. (2021). Rapid degeneration of Drosophila olfactory neurons in Orco mutant maxillary palps. Micropubl. Biol. 2021:10.17912/micropub.biology.000398
Trible, W., Olivos-Cisneros, L., McKenzie, S. K., Saragosti, J., Chang, N.-C., Matthews, B. J., et al. (2017). Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 170, 727–735. doi: 10.1016/j.cell.2017.07.001
Vasudevan, D., and Ryoo, H. D. (2016). Detection of cell death in Drosophila tissues. Methods Mol. Biol. 1419, 131–144. doi: 10.1007/978-1-4939-3581-9_11
Vulpe, A., Kim, H. S., Ballou, S., Wu, S. T., Grabe, V., Nava Gonzales, C., et al. (2021). An ammonium transporter is a non-canonical olfactory receptor for ammonia. Curr. Biol. 31, 3382–3390. doi: 10.1016/j.cub.2021.05.025
Vulpe, A., and Menuz, K. (2021). Ir76b is a Co-receptor for amine responses in Drosophila olfactory neurons. Front. Cell Neurosci. 15:759238. doi: 10.3389/fncel.2021.759238
Wu, J. N., Cai, C. X., Liu, W. B., Ai, D., Cao, S., Wang, B., et al. (2023). Mutagenesis of odorant receptor coreceptor orco reveals the odorant-detected behavior of the predator Eupeodes corollae. Int. J. Mol. Sci. 24, 17284. doi: 10.3390/ijms242417284
Yan, H., Jafari, S., Pask, G., Zhou, X., Reinberg, D., and Desplan, C. (2020). Evolution, developmental expression and function of odorant receptors in insects. J Exp Biol 223(Pt Suppl. 1):jeb208215. doi: 10.1242/jeb.208215
Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., et al. (2017). An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170, 736–747. doi: 10.1016/j.cell.2017.06.051
Keywords: olfaction, receptor, transcriptome, Drosophila, olfactory sensory neuron, antenna
Citation: Long T, Mohapatra P, Ballou S and Menuz K (2024) Odorant receptor co-receptors affect expression of tuning receptors in Drosophila. Front. Cell. Neurosci. 18:1390557. doi: 10.3389/fncel.2024.1390557
Received: 23 February 2024; Accepted: 01 May 2024;
Published: 20 May 2024.
Edited by:
Tanja Angela Godenschwege, Florida Atlantic University, United StatesReviewed by:
Timothy Mosca, Thomas Jefferson University, United StatesCarolina Gomez-Diaz, University of Oviedo, Spain
Copyright © 2024 Long, Mohapatra, Ballou and Menuz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karen Menuz, a2FyZW4ubWVudXpAdWNvbm4uZWR1
†These authors have contributed equally to this work and share first authorship
 Teng Long1†
Teng Long1†