Recent studies have shown that SIGLEC8, a member of the CD33-related Siglec receptor family, is highly expressed in brain microglia (Olah et al., 2018; Estus et al., 2019; Gonzalez-Gil et al., 2022). A recent study by Gonzalez-Gil et al. demonstrated that SIGLEC8 binds to large proteoglycans, including the receptor protein tyrosine phosphatase RPTPζ carrying O-linked terminally sialylated keratan sulfate chains. These chains are produced by the sialyltransferase ST3GAL4 and the keratan sulfate sulfotransferase CHST1 (Gonzalez-Gil et al., 2022). By examining the Human Central Nervous System Cell Type Expression Correlates website (Kelley et al., 2018), I confirmed SIGLEC8 expression in microglia and at lower levels in oligodendrocytes (Figure 1A). I was also able to ask where SIGLEC8 was likely to signal by looking at co-expression of the receptor, ligand and glycosylation enzymes in the brain. While the highest level of SIGLEC8 transcript was seen in the frontal cortex, it was also detected in the parietal cortex and the cerebellum. PTPRZ1 was expressed in multiple cortical lobes and in the cerebellum, the sialyltransferase ST3GAL4 followed a similar expression pattern, and CHST1 expression was observed in both the frontal cortex and cerebellum (Figures 1B, C). These data when taken together, suggest SIGLEC8 may primarily signal by interacting with RPTPζ decorated with terminally sialylated keratan sulfate on cerebellar astrocytes.
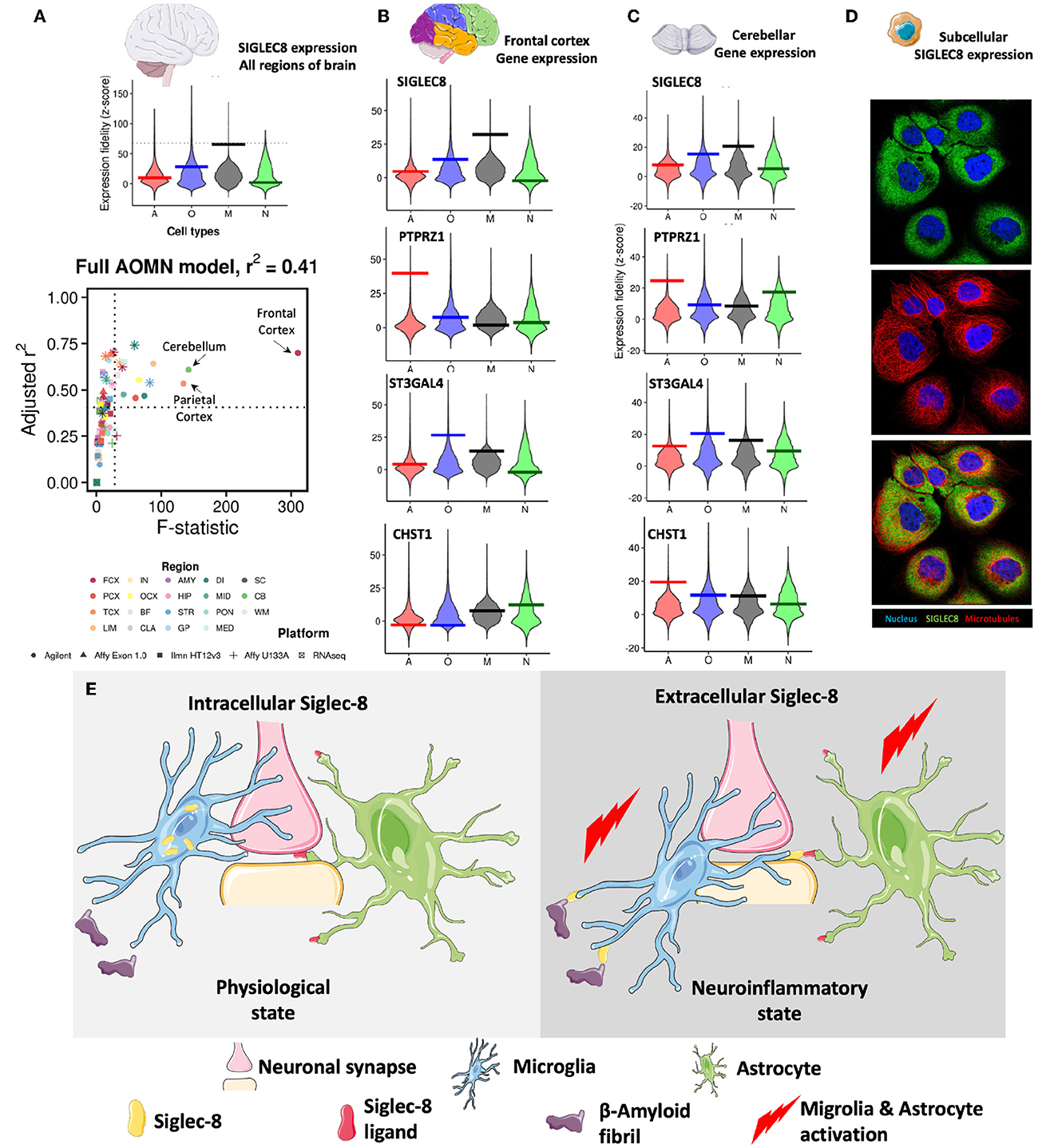
Figure 1. (A) SIGLEC8 is expressed by microglia and its expression is highest in the frontal cortex, parietal cortex, and the cerebellum. Genome-wide distributions of expression fidelity for astrocytes (A), oligodendrocytes (O), microglia (M), and neurons (N) over all analyzed samples are shown. The horizontal line denotes the expression fidelity of the query gene for each cell type. The dashed horizontal line denotes the threshold above which all fidelity scores had Confidence = 100. Lower panel shows modeling results for expression levels of the SIGLEC8 as a function of variation in the inferred abundance of individual cell types. Simple linear regression is used to predict gene expression levels as a function of estimated neuron, astrocyte, oligodendrocyte, or microglia abundance in all human CNS regional datasets containing the query gene. Adjusted R2 values and t-values are shown for each cell type (labeled by color, with shapes denoting technology platforms). (B) Expression data of SIGLEC8, PTPRZ1, ST3GAL4, and CHST1 in the frontal cortex. (C) Expression data of SIGLEC8, PTPRZ1, ST3GAL4, and CHST1 in the cerebellum [data for (A–C) obtained from (http://oldhamlab.ctec.ucsf.edu)] (D) SIGLEC8 is localized to the subcellular compartment. Immunofluorescence images of the A-431 cell line stained with the HPA012556 SIGLEC8 antibody (https://www.proteinatlas.org/ENSG00000105366-SIGLEC8/subcellular#img, Human Protein Atlas https://www.proteinatlas.org) (E) Hypothetical effects of intracellular and extracellular expression of SIGLEC-8 Region FCX, frontal cortex; PCX, parietal cortex; TCX, temporal cortex; LIM, limbic cortex; IN, insular cortex; OCX, occipital cortex; BF, basal forebrain; CLA, claustrum; AMY, amygdala; HIP, hippocampus; STR, striatum; GP, globus pallidus; DI, diencephalon; MID, midbrain; PON, pons; MED, medulla; SC, spinal cord; CB, cerebellum; WM, white matter. Some representative images were obtained from smart.servier.com.
Surprisingly, when I investigated SIGLEC8 localization in the cell, I found evidence for intracellular expression (Figure 1D). While this observation warrants detailed molecular studies, it suggests that SIGLEC8 localization might be regulated in a similar fashion as the CD33 form (CD33m) that protects from Late Onset Alzheimer's Disease (LOAD) (Hollingworth et al., 2011) and it corroborates the hypothesis that Siglec receptors might exhibit intracellular functions. In fact, the evidence that the LOAD-protective CD33 allele results in the expression of a protein unable to bind its natural ligand led to the suggestion that protection from disease was driven by a loss-of-function mechanism (Bradshaw et al., 2013; Griciuc et al., 2013). Parallel studies (Saha et al., 2019) have indicated that CD33m retention in intracellular peroxisomes prevented it from signaling (Siddiqui et al., 2017; Saha et al., 2019), adding credit to the loss-of-function hypothesis. However, the recent discovery of a non-functional CD33 allele not associated with LOAD has challenged this view.
There have been very few mechanistic studies exploring Siglec-8 in neurodegeneration. One that stands out mainly utilizes in vivo models of neurodegeneration utilizing Siglec-F—is the mouse paralog of Siglec-8 (Siddiqui et al., 2019)—and human cell line models to elucidate effects of Siglec-8 expression (Morshed et al., 2020). The investigators first identified a common response involving Siglec-F and Inpp5d likely linked to microglia (Morshed et al., 2020). They also explored RNASeq data and showed that aging-associated microglia had higher expression levels of Siglec-8 (Morshed et al., 2020). They also showed by immunofluorescence imaging, that Iba1+/MHC-II+ microglia in patients with LOAD had
the highest level of Siglec-8 expression (Morshed et al., 2020). Similar changes were noted in Siglec-F expression levels in mouse microglia during aging and neurodegeneration (Morshed et al., 2020). Additionally, they show Siglec-F and Siglec-8 induce cell death in a similar ITIM-dependent manner (Morshed et al., 2020). Lastly, the authors investigated the in vitro effects of IFNγ and TGFβ upregulate the expression of Siglec-8 in induced pluripotent stem cell-derived microglia (Morshed et al., 2020). The authors go on to suggest that targeting pathways downstream of Siglec-8/F may reduce neuroinflammation due to chronic receptor activation (Morshed et al., 2020).
It must be noted here that several variants of SIGLEC8 have been identified, and some of these (rs36498, rs10409962, and rs11672925) have been linked to asthma (Gao et al., 2010; Angata, 2014). It remains to be seen what the effect of these variants are on microglial Siglec-8 expression and function and if these are similar to the CD33 variants. Taking all these data together, I opine that intracellular Siglec receptors might instead have distinct roles from cell surface Siglecs, and they might modulate cell activation or protect from disease. Briefly, I suggest two distinct states of Siglec-8 expression exist (Figure 1E). One is an intercellular pool that does not interact with either sialylated β-amyloid proteins/prions (Srivastava et al., 2015) or astrocytes displaying sialylated keratin sulfate chains. These promote normal microglial phagocytic function and astocyte quiescence. On the other hand, Siglec-8 when expressed on the surface of microglia it binds to its ligands on the β-amyloid proteins and astrocytes leading to activation of these cell types, leading down neurodegenerative pathways.
Of course, it may be that Siglec-8 expression is simply modulated by aging processes, and not genetics. In this opinion piece, I also did not consider Siglec-8's other important ligand-−6'-sulfo sialyl Lewis X (O'Sullivan et al., 2018)—and its expression in the brain. It may be that these interactions are all working in concert in promoting neurodegenerative processes! These unaddressed issues are likely to be better understood when additional mechanistic experiments of Siglec-8 and its role in the brain are carried out. Future studies that address the potential roles of Siglecs retained within the cell will be particularly useful in understanding Alzheimer's disease and other cerebellar disorders like age-related cerebellar atrophy.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Angata, T. (2014). Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology 24, 785–793. doi: 10.1093/glycob/cwu043
Bradshaw, E. M., Chibnik, L. B., Keenan, B. T., Ottoboni, L., Raj, T., Tang, A., et al. (2013). CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 16, 848–850. doi: 10.1038/nn.3435
Estus, S., Shaw, B. C., Devanney, N., Katsumata, Y., Press, E. E., Fardo, D. W., et al. (2019). Evaluation of CD33 as a genetic risk factor for Alzheimer's disease. Acta Neuropathol. 138, 187–199. doi: 10.1007/s00401-019-02000-4
Gao, P. S., Shimizu, K., Grant, A. V., Rafaels, N., Zhou, L. F., Hudson, S. A., et al. (2010). Polymorphisms in the sialic acid-binding immunoglobulin-like lectin-8 (Siglec-8) gene are associated with susceptibility to asthma. Eur. J. Hum. Genet. 18, 713–719. doi: 10.1038/ejhg.2009.239
Gonzalez-Gil, A., Porell, R. N., Fernandes, S. M., Maenpaa, E., Li, T. A., Li, T., et al. (2022). Human brain sialoglycan ligand for CD33, a microglial inhibitory Siglec implicated in Alzheimer's disease. J. Biol. Chem. 298, 12246. doi: 10.1016/j.jbc.2022.101960
Griciuc, A., Serrano-Pozo, A., Parrado, A. R., Lesinski, A. N., Asselin, C. N., Mullin, K., et al. (2013). Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78, 631–643. doi: 10.1016/j.neuron.2013.04.014
Hollingworth, P., Harold, D., Sims, R., Gerrish, A., Lambert, J. C., Carrasquillo, M. M., et al. (2011). Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 43, 429–435. doi: 10.1038/ng.803
Kelley, K. W., Nakao-Inoue, H., Molofsky, A. V., and Oldham, M. C. (2018). Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184. doi: 10.1038/s41593-018-0216-z
Morshed, N., Ralvenius, W. T., Nott, A., Watson, L. A., Rodriguez, F. H., Akay, L. A., et al. (2020). LaRocque L, Mastroeni D. Phosphoproteomics identifies microglial Siglec-F inflammatory response during neurodegeneration. Mol. Syst. Biol. 16, e9819. doi: 10.15252/msb.20209819
Olah, M., Patrick, E., Villani, A. C., Xu, J., White, C. C., Ryan, K. J., et al. (2018). A transcriptomic atlas of aged human microglia. Nat. Commun. 9, 539. doi: 10.1038/s41467-018-02926-5
O'Sullivan, J. A., Carroll, D. J., Cao, Y., Salicru, A. N., and Bochner, B. S. (2018). Leveraging Siglec-8 endocytic mechanisms to kill human eosinophils and malignant mast cells. J. Allergy Clin. Immunol. 141, 1774–1785. doi: 10.1016/j.jaci.2017.06.028
Saha, S., Siddiqui, S. S., Khan, N., Verhagen, A., Jiang, W., Springer, S., et al. (2019). Controversies about the subcellular localization and mechanisms of action of the Alzheimer's disease-protective CD33 splice variant. Acta Neuropathol. 138, 671–672. doi: 10.1007/s00401-019-02065-1
Siddiqui, S. S., Matar, R., Merheb, M., Hodeify, R., Vazhappilly, C. G., Marton, J., et al. (2019). Siglecs in brain function and neurological disorders. Cells. 8, 1125. doi: 10.3390/cells8101125
Siddiqui, S. S., Springer, S. A., Verhagen, A., Sundaramurthy, V., Alisson-Silva, F., Jiang, W., et al. (2017). The Alzheimer's disease–protective CD33 splice variant mediates adaptive loss of function via diversion to an intracellular pool. J. Biol. Chem. 292, 15312–15320. doi: 10.1074/jbc.M117.799346
Keywords: SIGLEC, Alzheheimer's disease, CD33, SIGLEC8, sialic acid
Citation: Dhar C (2023) Does SIGLEC8 localize to the subcellular compartment like the Alzheimer's disease protective CD33 splice variant?. Front. Cell. Neurosci. 17:1124150. doi: 10.3389/fncel.2023.1124150
Received: 15 December 2022; Accepted: 28 March 2023;
Published: 13 April 2023.
Edited by:
Ertugrul Kilic, Istanbul Medipol University, TürkiyeReviewed by:
Burak Yulug, Alanya Alaaddin Keykubat University, TürkiyeCopyright © 2023 Dhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chirag Dhar, Y2hpcmFnLmRoYXJAdmVubi5iaW8=
 Chirag Dhar
Chirag Dhar