- 1School of Medicine, Yunnan University, Yunnan, China
- 2NHC Key Laboratory of Drug Addiction Medicine, Kunming Medical University, Kunming, China
- 3International Research Center for Regenerative Medicine, BOAO International Hospital, Qionghai, China
- 4High School Attached to Shanghai Normal University, Shanghai, China
Methamphetamine (METH) addiction and withdrawal cause serious harm to both the immune system and nervous system. However, the pathogenesis remains largely unknown. Herein, we investigated the peripheral cytokines and exosomal transcriptome regulatory networks in the patients with METH use disorders (MUDs) undergoing withdrawal. Twenty-seven cytokines were simultaneously assessed in 51 subjects, including 22 at the acute withdrawal (AW) stage and 29 at the protracted withdrawal (PW) stage, and 31 age and gender-matched healthy controls (HCs). Compared to the HCs, significantly decreased levels of interleukin (IL)-1β, IL-9, IL-15, Basic FGF, and MIP1a, increased levels of IL-1rα, IL-6, Eotaxin IP-10, VEGF, and RANTES were identified in AW. These disturbances were mostly or partly restored to the baseline in PW. However, the cytokines IL-6, IL-7, and IL-12p70 were consistently increased even after one year of withdrawal. Besides, a significant decrease in CD3+T and CD4+T cell numbers was observed in AW, and the diminishment was restored to baseline in PW. Comparatively, there were no statistically significant changes in CD8+T, NK, and B cells. Furthermore, the exosomal mRNAs and long non-coding RNAs (lncRNA) were profiled, and the lncRNA-miRNA-mRNA networks were constructed and associated with METH AW and PW stages. Notably, the chemokine signaling was remarkably upregulated during AW. By contrast, the differentially expressed mRNAs/lincRNAs were significantly enriched in neurodegeneration-related diseases. Taken together, a group of METH withdrawal-related cytokines and exosomal mRNA/lncRNA regulatory networks were obtained, which provides a useful experimental and theoretical basis for further understanding of the pathogenesis of the withdrawal symptoms in MUDs.
Introduction
Methamphetamine (METH) is one of the most common and highly addictive psychostimulants that cause severe damage to a wide spectrum of organ systems (Darke et al., 2008; Glasner-Edwards et al., 2010). In addition to those METH induced neuropsychiatric and neurodegenerative symptoms such as depression, anxiety, and/or cognitive impairments (Hall et al., 1996; McKetin et al., 2008), methamphetamine use disorders (MUDs) could also give rise to considerable problems associated with immunoglobins and cytokine/chemokine signaling pathways, which may contribute to spread of devastating infectious diseases (Potula et al., 2010; Harms et al., 2012) and thereafter influence the recovery from neurodegeneration defects (Loftis and Huckans, 2013). To date, despite the astounding efforts into the knowledge base of addiction medicine, the underlying molecular mechanism and functional interplay between immune response and neurodegeneration remain largely lacking (Zhang et al., 2021; Zhou et al., 2021).
The development of omics-based high-throughput technologies nowadays allows for a simultaneous determination of a large number of potential biomarkers and their correlations to the disease progressions (Hoshino et al., 2020). For example, the cytokine multiplex assay allows researchers to evaluate the concentrations of multiple cytokines of interest in a limited sample volume (Lynch et al., 2014). Likewise, exosomes are tiny endosomal-derived membrane micro-vesicles that contain a large amount of regulatory information and may participate in a series of basic biological processes (Beatriz et al., 2021; Qiu et al., 2021). Compared to those diluted nucleotide pieces in the peripheral blood, the exosomal content is much more stable. Therefore, the value of analyzing exosome-encapsulated DNA/RNA has been recognized (Cheng et al., 2015; Ramirez et al., 2018), and the exosomes in liquid biopsy are commonly utilized to develop diagnostic tools in clinical applications (Grieco et al., 2021; Masoumipour et al., 2021; Zani-Ruttenstock et al., 2021). Among these, studies have shown that exosomes could be used for predicting Alzheimer’s disease (AD) and evaluating the severity of intellectual disability in patients with Parkinson’s disease and dementia (Cheng et al., 2015; Stuendl et al., 2016). As for MUDs, the disease management would take advantage of having adequate peripheral biomarkers and gain greater insight into immune response in patients undergoing METH withdrawal.
We had already identified those differentially expressed long non-coding RNAs (lncRNAs) and mRNAs in the plasma exosomes of heroin addicts, which are specifically associated with heroin withdrawal stages and the irregularity of both adaptive and innate immunity (Zhang et al., 2021). The present study was designed to investigate the pattern of METH-withdrawal altered immune responses, and to unveil the function of circulating exosome capsulated molecules in patients with MUDs. Our present approach aims to uncover the mechanism associated with the peripheral exosomal biomarkers and transcriptome regulatory network in developing withdrawal symptoms.
Materials and Methods
Study participants and ethics statement
In order to analyze the dynamics of the cytokines in MUDs, we recruited 82 male participants in the Kunming Drug Rehabilitation Center between January 2018 and October 2019, including 51 MUDs patients and 31 healthy control subjects. For exosomal RNA sequencing analysis, 20 MUDs consisting of 10 subjects each at 7 to 14-day and 1-year stages of METH withdrawal, and 10 healthy individuals were recruited and previously described in Chen et al. (2021) and Yu et al. (2021). All participants provided written informed consent before enrollment. The recruitment procedures and protocols were approved by the Research Ethics Committee of the First Affiliated Hospital of Kunming Medical University (2018-L-42). The drug use history of patients with MUDs was obtained by self-report and verified with caregivers. Samples were collected as previously described (Zhang et al., 2021).
The inclusion criteria were as follows: (1) patients were diagnosed with methamphetamine dependence, (2) patients were between the ages of 20 and 55. The exclusion criteria were defined as: (1) patients with medical or neurological disease or trauma affecting their central nervous system, (2) patients who have been reported to have a history of HIV or other infectious diseases, (3) patients who had severe endocrine, cardiovascular, or a history of loss of consciousness for more than 30 min.
Determination of peripheral cytokine levels in human plasma
Fresh whole blood samples were collected from the study participants using a 10 ml EDTA-2Na vacuum tubes. The blood samples were centrifuged at 1,500 g for 15 min, and the plasma was transferred into a new tube. The plasma was centrifuged at 20,000 g at 4°C for 15 min to remove platelets. Purified plasma was harvested, aliquoted, and stored at −80°C until assay.
The Luminex Human Cytokine Assay was performed using the Luminex Human Magnetic Assay Kit (R&D Systems, MN, USA) according to the manufacturer’s instructions. Standard curves were generated by Bio-plex Manager software to determine unknown sample concentrations. The detection kit includes 27 cytokines, including bFGF, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17A, IP-10, IFN-γ, MCP-1, MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, and VEGF.
Immune cell subsets
Fresh whole blood samples were analyzed with flow cytometry. For flow cytometry, we followed the protocol using 50 μl of whole blood and added 5 μl of monoclonal antibody, then incubated for 15 min at room temperature in the dark. Added 800 μl BD 1× lysis solution to each sample and incubated for 10 min at room temperature in the dark. The sample was centrifuged at 500 g for 5 min, and the pellet was collected and resuspended with 300 μl of FACS Flow, and analyzed with BD FACS Canto II cytometer.
The antibody used in this study was BD MultitestTM 6-color TBNK Reagent (Cat# 644611), including CD3-FITC/CD4-PE-Cy7/CD8-APC-Cy7/CD16&CD56-PE/CD19-APC/CD45-PerCP-Cy5.5 (BD, United States). The absolute cell counts were calculated by multiplying the percentage of immune cell subsets by the concentration of total lymphocytes present in the peripheral blood.
Exosomal RNA sequencing and identification of differentially expressed lncRNA and mRNA
The exosomes from peripheral blood were isolated as previously described (Chen et al., 2021). The Exosupur® columns (Echobiotech, China) were used to purify 2 ml of 0.8 μm-filtered plasma samples followed by elution of exosomes with PBS. The eluted fractions were concentrated to 200 μl using Amicon® Ultra spin filters with a molecular weight cut-off of 100 kDa (Merck, Germany). Total RNAs were then extracted from the purified exosomes using the miRNeasy® Mini kit according to the manufacturer’s protocol, then qualified with Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., CA, USA).
For RNA sequencing library construction, 1.5 μg RNA per sample was used as input material. The rRNA was removed using the Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI, USA). The sequencing libraries were generated using NEBNext UltraTM Directional RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s instructions. Raw reads of RNA-seq data were filtered using fast QC, then aligned to the GRCh38 human genome assembly using HISAT2 (Pertea et al., 2016). Annotations of mRNA and lncRNA in the human genome were retrieved from the GENCODE (v.25). The mRNAs and lncRNAs were quantified and analyzed using DESeq2 R package (Love et al., 2014) and StringTie 1.3.1 (Pertea et al., 2016), respectively.
Functional and pathway enrichment
Gene Ontology (GO) and KEGG pathways enrichment analyses were performed in DAVID website1 that are significantly enriched compared to the entire human genome background (Huang da et al., 2009; Sherman et al., 2022).
Prediction of miRNA/lncRNA targets and lncRNA-miRNA-mRNA regulatory network construction
The targets of DE miRNAs were analyzed by miRTarBase, a database containing targets validated experimentally (Huang et al., 2020). To summarize the overlap between predicted mRNA targets and DE mRNAs, Venn diagrams were constructed by FunRich 3.1 software. LncRNAs that could regulate miRNA expression were predicted by DIANA-LncBase v32 (Karagkouni et al., 2020). The lncRNA-miRNA-mRNA regulatory network was constructed according to the prediction results of lncRNA targets and miRNA targets, and the interaction between lncRNA, miRNA, and mRNA was displayed. Cytoscape software was used to plot the lncRNA-miRNA-mRNA regulatory network.
Data availability and data analysis
All sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE172306. Kruskal–Wallis test or analysis of variance (ANOVA) test followed by a post-hoc test (Bonferroni’s t-test) was used to test for differences in continuous variables. Comparison between groups was performed using a student’s t-test or a Chi-square analysis as appropriate. The Pearson correlation was used for correlation analyses.
Results
Clinical characteristics
To characterize the immune responses in patients undergoing METH withdrawal, we recruited 51 male patients with MUDs, of which 22 and 29 subjects were approximately 7 to 14 days (acute withdrawal stage, AW) and 1 year (protracted withdrawal stage, PW) after the initiation of the abstinence from METH, respectively, as well as 31 age-matched healthy male volunteers (healthy controls, HC). The main clinical characteristics of the study participants were summarized in Table 1. There were no significant differences for any variables, including age, substance-use history, and education level, between the two groups of MUDs and HCs.
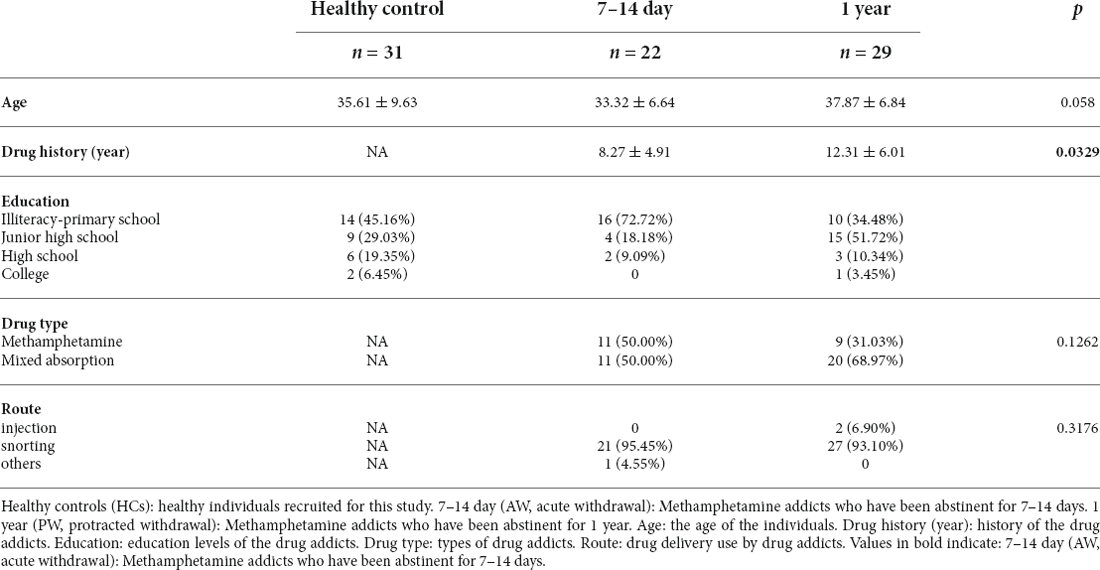
Table 1. Demographic and clinical characteristics of methamphetamine addicts (n = 51) and health controls (n = 31).
Alterations in plasma cytokine levels and immune cell subsets in patients undergoing METH withdrawal
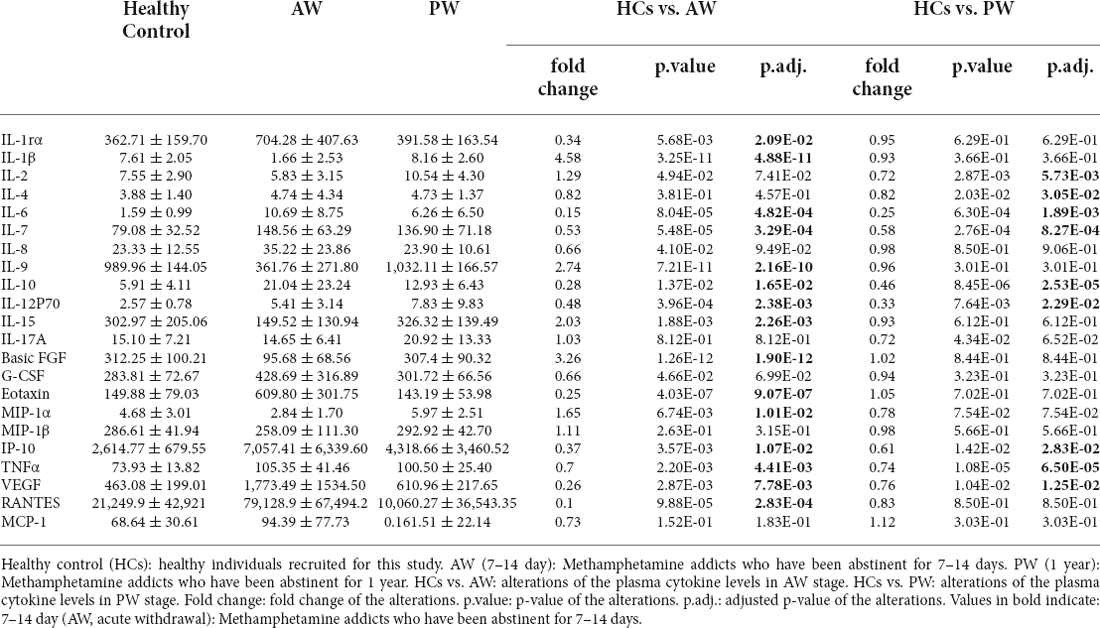
The alterations of 27 cytokines in the plasma associated with METH withdrawal were determined using Luminex Human Cytokine 27-plex assay (see “Materials and methods” Section). IFN-γ and GM-CSF were excluded from analysis in the dataset because these two fell below the lower level of quantitation in >20% of samples. The concentrations of IL-8, IL-17A, G-CSF, MIP-1β, and MCP-1 in the plasma of MUD patients at both AW and PW were not significantly different from those in the HCs. In the AW subjects, compared with those in the HCs, significantly lower levels of IL-1β (p = 4.88E-11), IL-9 (p = 2.16E-10), IL-15 (p = 2.26E-03), Basic FGF (p = 1.90E-12), and MIP-1α (p = 1.01E-2) were observed (Table 2). Synchronously, there were remarkable increases in the levels of IL-1rα (p = 2.09E-02), IL-6 (p = 4.82E-04), IL-10 (p = 1.65E-02), IL-12P70 (p = 2.38E-03), Eotaxin (p = 9.07E-07), IP-10 (p = 1.07E-02), VEGF (p = 7.78E-03) and TANTES (p = 2.83E-04) in the AW subjects than in the HCs (Table 2). Compared with those in the HCs, significantly higher levels of IL-2 (p = 5.73E-03), IL-4 (p = 3.05E-02), IL-6 (p = 1.89E-03), IL-7 (p = 8.27E-04), IL-10 (p = 8.27E-04), IL-12P70 (p = 2.29E-02), IP-10 (p = 2.83E-02), TNFα (p = 6.50E-05), and VEGF (p = 1.25E-02; Table 2) were found in the PW subjects.
After a period of withdrawal to PW, the abnormal cytokines (e.g., IL-1rα, IL-1β, IL-9, IL-15, Basic FGF, Eotaxin, MIP-1α, and TANTES) could be restored to the normal range, however, several cytokines, IL-6, IL-7, IL-10, IL-12P70, IP-10, TNFα, and VEGF could only be restored partially after one year of withdrawal. Besides, it was worth noting that the levels of IL-2 and IL-4 were normal range in AW, but dysregulated in the PW stage in patients with MUDs.
Furthermore, the immune cell subsets in the patients of MUDs during both AW and PW stages were analyzed (Figure 1 and Supplementary Figure S1). The absolute counts of CD3+T (p = 0.0004) and CD3+CD4+T (p = 0.0001) cells from the AW group were significantly lower than those of the HCs. A similar tendency was also observed for CD3+CD8+T cells, although there was no statistical difference. Considering that CD3+T cells are composed of CD3+CD4+T and CD3+CD8+T, these results suggest the tendency of CD3+T is dependent on CD3+CD4+T. In addition, the absolute counts of CD3+CD4+T were not fully recovered even one year after initiation of METH abstinence.
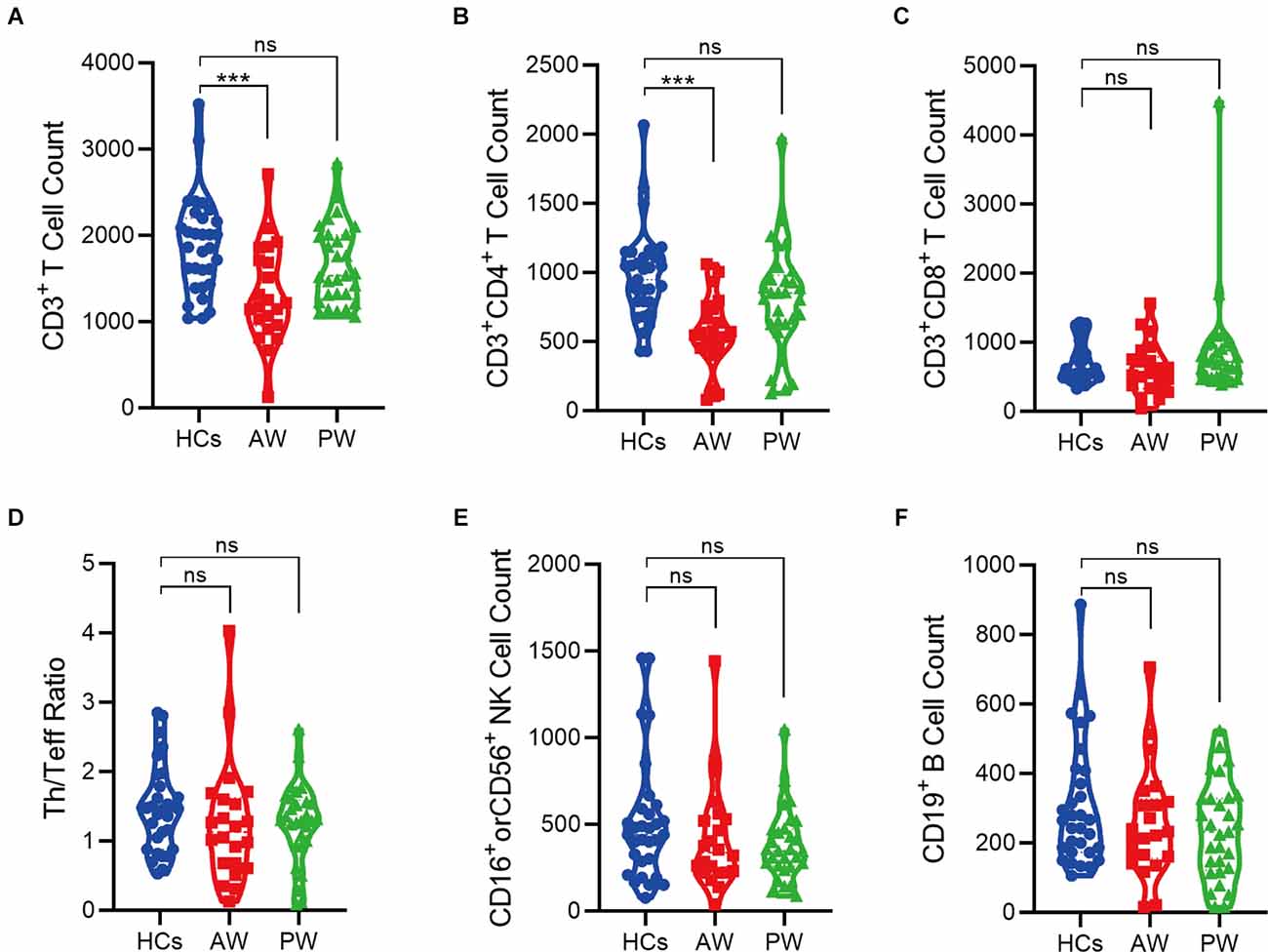
Figure 1. Alterations of peripheral immune cell subsets in patients undergoing methamphetamine (METH) withdrawal. (A–F) Total T cell count (CD3+) (A), Th cell count (CD3+CD4+) (B), Teff cell count (CD3+CD8+) (C), Th/Teff ratio (CD3+CD4+/ CD3+CD8+) (D), NK cell count (CD16+ or CD56+) (E), and B cell count (CD19+) (F) in the MUDs patients during withdrawal. HCs, healthy controls; AW, acute withdrawal; PW, protracted withdrawal. ***p < 0.001; ns, no significant difference.
Differential expression of exosomal mRNAs and lncRNAs at acute METH withdrawal stage
To investigate the plasma exosomal mRNA/lncRNA expression changes associated with METH withdrawal, we additionally recruited 20 MUDs currently undergoing withdrawal (10 AW subjects and 10 PW subjects), and then compared their transcriptome with 10 HCs (Figure 2A). The clinical information of these study participants as well as the isolation and validation of exosomes from plasma samples were previously described (Yu et al., 2021). The population’s basic information variables, such as age, BMI, substance-use history, and education level were well balanced between the two groups of MUDs and the HCs (Supplementary Table S1).
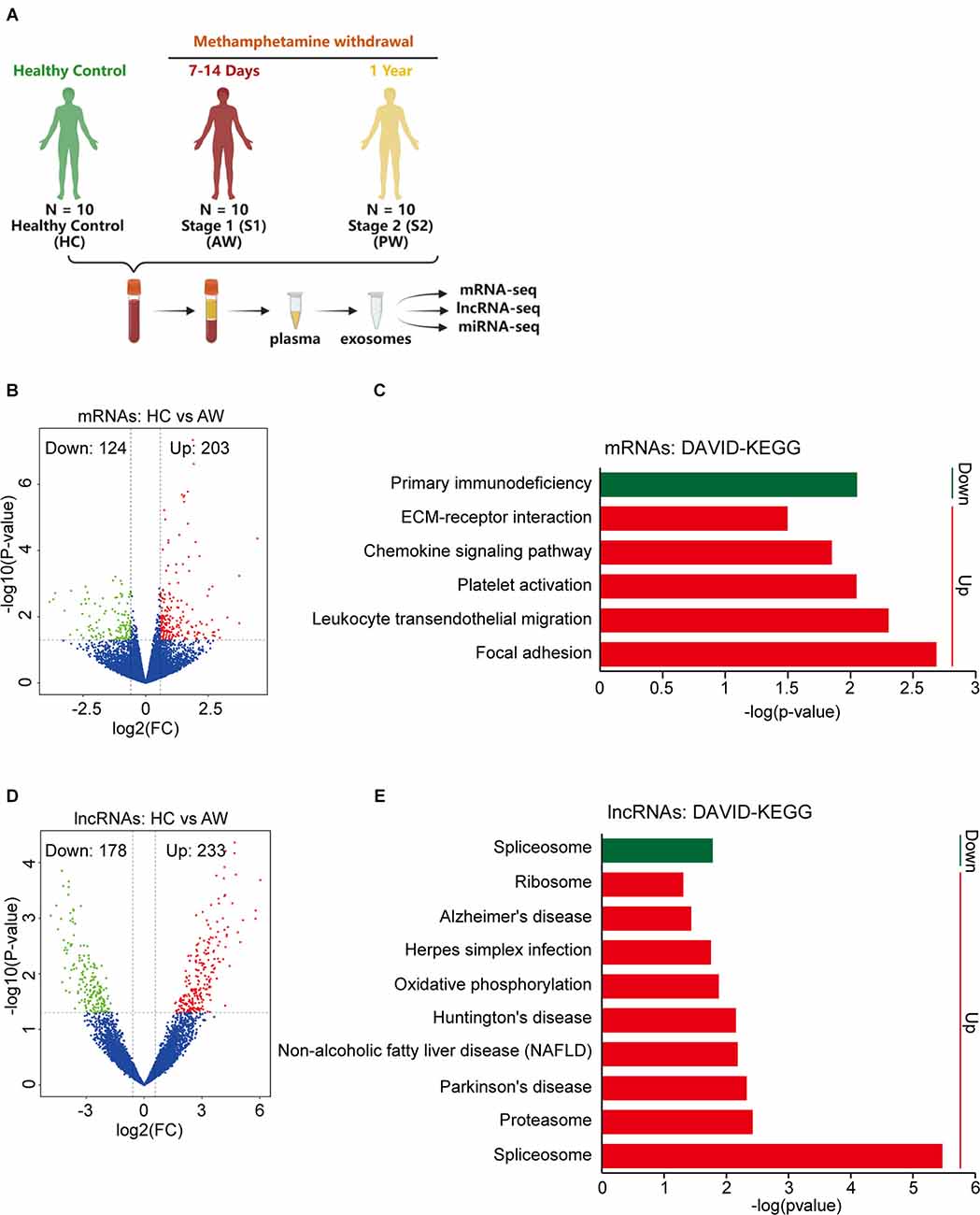
Figure 2. Differential expression of exosomal mRNAs and lncRNAs at acute METH withdrawal (AW) stage. (A) Sample collection and high-throughput sequencing processing schedule for exosomal transcriptome alterations in MUD patients. (B) Volcano plot showed the different expression (DE) mRNAs at the AW stage. (C) DAVID-KEGG analysis of DE mRNAs at AW stage. (D) Volcano plot showed the DE lncRNAs at AW stage. (E) DAVID-KEGG analysis of DE lncRNAs at AW stage. Green represents downregulated genes; Red represents upregulated genes. mRNA, messenger RNA.
The expression profiles of exosomal mRNA/lncRNA in the peripheral blood were determined by a high-throughput sequencing technique. We first compared the HC group vs. the AW group (S1), to identify the mRNAs/lncRNAs that were specifically responsible for the acute METH withdrawal stage. At the mRNA level, a total of 327 differentially expressed (DE) mRNAs, of which 124 downregulated and 203 upregulated were associated with the AW stage (Figure 2B). DAVID GO term analysis of these overlapped DE mRNAs showed that the platelets were persistently impaired in the AW stage. Genes involved in the biological process terms including platelet degranulation (15 genes, p < 0.0001), platelet activation (five genes, p = 0.0008), and platelet aggregation (six genes, p = 0.007) were significantly upregulated (Supplementary Table S2). Furthermore, KEGG pathway analysis indicated that these DE mRNAs were significantly enriched in the Chemokine signaling pathway (Increased, p = 0.014), Leukocyte transendothelial migration (Increased, p = 0.005), Platelet activation (Increased, p = 0.009), and Primary immunodeficiency (Decreased, p = 0.009; Figure 2C and Supplementary Table S2).
Next, we compared the lncRNA expression profiles between the HC group vs. the AW group. Using the threshold (|log2 (fold change)| > 0.5, p < 0.05), a total of 411 DE lncRNAs (233 downregulated and 178 upregulated lncRNAs) were identified to associate with METH AW (Figure 2D). GO and KEGG analyses of these DE lncRNAs suggested that the neurodegeneration diseases-related lncRNAs were significantly upregulated in the AW group, including Parkinson’s disease (p = 0.005), Huntington’s disease (p = 0.007) and Alzheimer’s disease (p = 0.04; Figure 2E and Supplementary Table S3). Overall, these results suggest that both the nervous system and the immune systems were significantly damaged during METH AW.
Differential expression of exosomal mRNAs and lncRNAs during the protracted METH withdrawal
To determine the health impact of METH dependence after a long-term withdrawal, we further compared the exosomal mRNA/lncRNA profiles between the HC group vs. the PW group. Notably, a total of 518 mRNAs were identified differentially expressed, in which 227 mRNAs were upregulated, and 291 mRNAs were downregulated at the PW stage (Figure 3A). These upregulated mRNAs were significantly enriched in the Phosphatidylinositol signaling system (p = 0.001), Pathways in cancer (p = 0.003), Glucagon signaling pathway (p = 0.007) etc., whereas the downregulated genes were significantly enriched in the Proteasome (p < 0.001), Protein export (p = 0.001), Oxidative phosphorylation (p = 0.009) and Parkinson’s disease (p = 0.01; Figure 3B and Supplementary Table S3). Compared to the HC group, a total of 188 upregulated and 337 downregulated lncRNAs were identified in the PW group (Figure 3C). Interestingly, the exosomal DE lncRNA at PW also exhibited unique features. DAVID KEGG term analysis suggested that these upregulated lncRNAs were remarkably associated with neurodegenerative diseases, such as Parkinson’s disease (p = 0.01), Alzheimer’s disease (p = 0.027), Huntington’s disease (p = 0.047; Figure 3D and Supplementary Table S3). These results indicated that unlike, METH AW stage, the key difference in exosomal genes/transcripts for patients undergoing protracted METH withdrawal is more likely associated with neurological diseases.
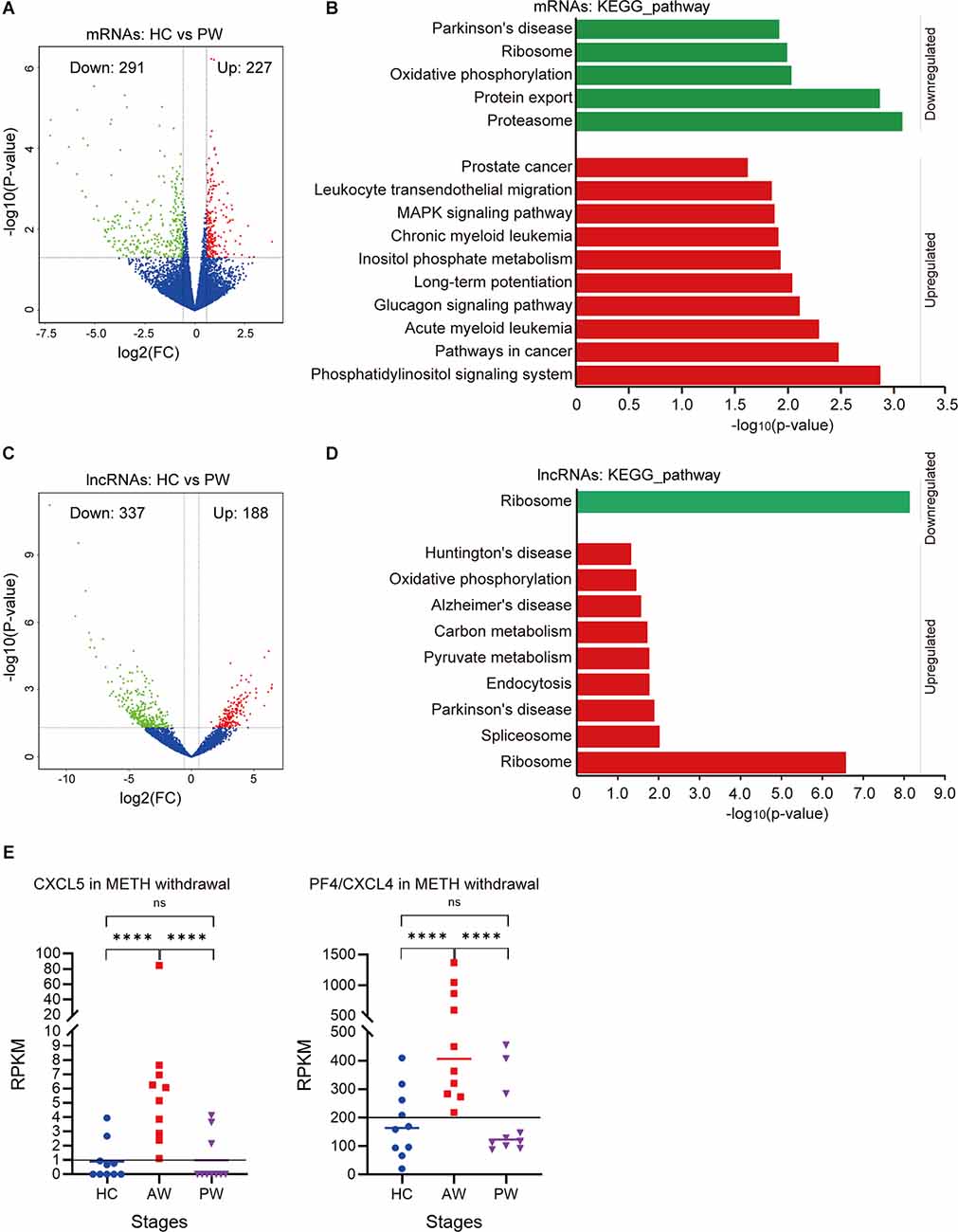
Figure 3. Potential expression profile differences at the AW stage and protracted METH withdrawal (PW) stage. (A) Volcano plot showed the DE mRNAs at the PW stage. (B) DAVID-KEGG analysis of DE mRNAs at the PW stage. (C) Volcano plot showed the DE lncRNAs at PW stage. (D) DAVID-KEGG analysis of DE lncRNAs at the PW stage. Green represents downregulated genes; Red represents upregulated genes. lncRNA, long noncoding RNA. (E) Potential biomarkers (CXCL5, CXCL4) in the AW stage exosomal transcriptome. ****p < 0.0001; ns, no significant difference.
It is noteworthy that the chemokine signaling was remarkably upregulated in the acute METH withdrawal stage (Supplementary Table S2). Further investigation of these DE mRNAs/lncRNAs during the entire withdrawal showed that the expression of CXCL5 and CXCL4 (PF4) was only upregulated in acute METH withdrawal but could be recovered in the protracted withdrawal (Figure 3E). Hence, the expression levels of CXCL5 and CXCL4 in plasma exosomes could be used as potential biomarkers for the METH withdrawal stage.
Construction of lncRNA-miRNA-mRNA regulatory network associated with METH AW and PW
The DE miRNA has been previously identified in Chen et al. (2021) and Yu et al. (2021). The mRNAs targeted by miRNAs were predicted using miRanda algorithm according to the miRNA-mRNA binding data. For the AW stage, there were 53 miRNA-mRNA target pairs in total were obtained removing duplicates which consisted of 22 miRNAs and 38 mRNAs (Supplementary Table S4). Three significant miRNAs, hsa-miR-152,3p, hsa-miR-1255b-5p, and hsa-miR-744,5p had the most target mRNAs. Next, the lncRNAs regulated by miRNAs were analyzed by miRanda algorithm. Totally, only three miRNA-lncRNA regulatory pairs were identified including three miRNAs and three lncRNAs (Supplementary Table S4). Based on the identified regulatory pairs of miRNA-mRNA and miRNA-lncRNA, a lncRNA-miRNA-mRNA network was constructed, consisting of 22 miRNAs, 8lncRNAs, and 38 mRNAs (Figure 4A). Meanwhile, the lncRNA-miRNA-mRNA network for the METH PW stage was constructed using the same strategy, consisting of 24 miRNAs, 25lncRNAs, and 50 mRNAs (Figure 4B and Supplementary Table S4). Of these miRNA/lncRNA targeted mRNAs, 35 were upregulated, and 15 were downregulated. Four significant miRNAs, hsa-miR-27a-3p, hsa-miR-338,3p, hsa-miR-370,3p, and hsa-miR-7,5p had the most target mRNAs (Supplementary Table S4).

Figure 4. LncRNA-miRNA-mRNA regulatory network associated with AW and PW stage. (A) The network related to the AW stage consists of 22 miRNAs, eight lncRNAs, and 38 mRNAs. (B) The network related to the PW stage consists of 24 miRNAs, 25 lncRNAs, and 50 mRNAs. Quadrilateral represents lncRNAs, triangle represents miRNAs, and ellipse represents mRNAs.
Discussion
Asa famous psychostimulant drug, METH abuse results in negative consequences, and it is on the rise worldwide (Prakash et al., 2017). Over two decades ago, Jerrells et al. (1989) suggested that withdrawal from alcohol and other substance leads to increased hypothalamic-pituitary-adrenal (HPA) axis reactivity and an acute inflammatory response in the. It is critical because increased acute inflammation may accelerate neurodegeneration and cognitive disability and eventually contribute to high relapse rates and impaired daily functioning. Therefore, to better understand the mechanism associated with developing withdrawal symptoms, it is essential to identify the withdrawal stage-related molecular signatures.
To date, in vitro and animal experiments have reported that METH and other substances modulate the function of immune cells, for instance, phagocytosis, chemotaxis, cytokine response, and activity. In particular, METH affects cellular immunity by promoting T cell apoptosis (Iwasa et al., 1996). In the present study, we provide evidence that the CD3+T cells were diminished in the AW stage, indicating that METH dependence interferes with the development of cellular immunity once again (Hernandez-Santini et al., 2021). Harms et al. reported that chronic administration of METH results in a significant decrease in activated T lymphocyte lineage, including both CD4+ and CD8+T cells (Saito et al., 2006; Harms et al., 2012; Mata et al., 2015; Hernandez-Santini et al., 2021). Our results showed a lower number of CD4+T but CD8+T cells in the patients undergoing the AW stage. In line with previous findings, these results suggest that decreased number of CD4+ helper T cells could diminish the ability to defend against virus infection, such as HIV, making substances more susceptible to HIV infection. However, we didn’t observe a lower number of CD8+T cells, probably due to the small number of study participants enrolled. In addition, the immunosuppressive effects of METH were shown in animal models, including decreased NK activity and proliferation rate in splenic lymphocytes (Yu et al., 2002; Akiyama et al., 2008). It has also been reported that METH administration could promote B cell infiltration in the spleen and lung in mice (Mitha et al., 2021). However, we observed no significant change in the number of NK and B cells in patients undergoing METH withdrawal. There is a decreasing trend in B cell numbers in the PW stage. However, these results require further validation in a large group of patient samples.
METH administration in animal models causes microglial activation in key brain regions and contributes to neurodegenerative symptoms (Bowyer and Ali, 2006; Sekine et al., 2008). Notably, IL-1β and TNF-α have been reported to inhibit long-term potentiation by damaging circuit and neuronal plasticity (Cunningham et al., 1996; Butler et al., 2004). Chemokines MCP-1, MIP-1α, and MIP-1β have been shown to play a significant role in neurogenesis and maintain brain function and influence inflammatory processes. Multiple lines of studies in humans have shown neuropsychiatric functions such as mood and cognition may be influenced by cytokines and other immune molecules. Our present data showed that some of the METH acute withdrawal induced cytokine abnormalities such as IL-1β and MIP-1α could be restored to normal at the PW stage. However, the TNFα level of the AW stage was altered and could be only partially restored even a year after the initiation of abstinence. Collectively, these findings support the notion that METH may exert its adverse effects on neuropsychiatric functioning via impairing neuroinflammatory regulation.
Recently, exosomes have been identified as a new facet of inter-cellular communications, particularly in neurodegenerative disease studies (Buzas et al., 2014; Thompson et al., 2016). We performed exosomal RNA sequencing and revealed significant changes in mRNA and lncRNA expression profiles at both AW and PW stages, respectively. Interestingly, those specific signaling pathways that are associated with METH withdrawal progression mainly consisted of immune-related chemokine mRNAs and neurological disease-related lncRNAs. Some identified transcripts were previously reported. For example, the genetic variance in UBQLN1 has been recognized as candidate genes for Alzheimer’s disease (Bertram et al., 2005). Another gene ADD3 was discovered by the Psychiatric Genomics Consortium Bipolar Disorder Working Group with a large-scale meta-analysis (Charney et al., 2017). It is worth noting that unlike those reversible changes from the immune system, persistent alterations in the neurological disease-associated lncRNAs were revealed, suggesting that key lncRNAs may play central regulatory roles in mediating severe long-term CNS defects in METH addiction and withdrawal (Aliperti et al., 2021). Our group and others have shown that most immune parameters tested in human opioid abusers are suppressed following withdrawal and recovery time to baseline varies in studies (Eisenstein et al., 2006). The recovery of the immune system after one year of abstinence may be associated with the renewal of immune cells in the body. Regrettably, the hypothesis has not been experimentally determined in patients with MUDs. This novel view and hypothesis updated the previous study of METH and provided a new way to study the effect of METH on the regulatory elements of immunity.
As noted in the literature, numerous experimental models have been applied to explore the knowledge regarding the impartments in the nerve system and the immune system from METH exposure and withdrawal (Loftis and Janowsky, 2014; Potula et al., 2018; Miller et al., 2021). Because the exosomes could cross the blood-brain-barrier (BBB) into the bloodstream, making it possible to extract CNS-derived bioactive substances, the peripheral exosomes have been thought to be promising minimally invasive biomarkers and therapeutic targets for neuronal diseases (Gao et al., 2021; Pineles et al., 2022), multiple lines of evidence indicate that the circulating exosomes might act as the bridge for intercellular communications between neuronal cells (Meng et al., 2020), while contents of these exosomes were closely related to METH addiction status in rats (Li et al., 2018). Notably, screening of all DEmRNAs/lncRNAs during withdrawal found that the CXCL5 and CXCL4 (PF4) were specifically upregulated in the blood exosomes of patients with acute METH withdrawal. Hence, the expression levels of CXCL5 and CXCL4 in blood exosomes may be clinically used as novel biomarkers for the acute METH withdrawal stage. This discovery was in line with the previous finding that CXCL5 was reported remarkably upregulated in the METH-induced astrocyte activation in patients of MUDs at 3 months of withdrawal (Bortell et al., 2017). All the above data suggest that the contents of the peripheral exosomes were clinically valuable biomarkers that could predict or correlate with MUD trajectories and treatment responses to EMTH withdrawal symptoms.
The present study has several strengths. First, the characteristics of recruited patients in the immune cohort and exosome cohorts were systematically analyzed. Second, we divided MUD patients into AW and PW stages of METH withdrawal and identified novel biomarkers. Third, the biomarkers identified were in plasma exosomes rather than in the CNS, so diagnosis was relatively easy. Finally, this study provided in-human evidence that the immune and nervous systems were significantly disrupted in METH AW, whereas the immune systems could e partially restored in the PW stage. The study also has some limitations. The date on dynamic immune and exosomal miRNA/mRNA/lncRNA regulatory networks of MUDs were generated from two independent cohorts, so correlation analysis was not possible. The sample size was relatively small. It is assumed that studies with relatively large sample sizes should obtain more accurate results. The dynamic mRNAs and lncRNAs in peripheral blood exosomes required further studies to determine their origin.
Overall, the present study evaluated the responses of the immune systems and the transcriptomic profiles of the peripheral blood exosomes in patients with MUDs, providing a group of METH withdrawal-related cytokines and exosomal miRNA/mRNA/lncRNA regulatory networks. However additional studies remain needed to determine the further molecular mechanisms underlying the affected immune and nervous systems during METH withdrawal.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE172306.
Ethics Statement
The studies involving human participants were reviewed and approved by Research Ethics Committee of the First Affiliated Hospital of Kunming Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
KW and JY designed the study. ZZ, YM, FC, and GD performed the experiments. FC, PX, and ZX recruited the patients and collected the clinic data. HW, ZZ, and JY analyzed the data, performed statistical analyses, and wrote the manuscript. HW, ZZ, GD, and JY interpreted and discussed the data with all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81860100, 31860306, 81870458), Science and Technology Department of Yunnan Province (Grant No. 2018DH006, 2018NS0086, 202001AV070010, 2020DAMRA-005, 202002AA100007), Yunnan Province and the Education Department of Yunnan Province (Grant No. 2019J1226), Yunnan Province Clinical Research Center (Grant No. 2019ZF012), Fund of Central Public Welfare Research Institute (Grant No. 2019PT310003), and Yunling Scholar (Grant No. YLXL20170002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2022.961131/full#supplementary-material.
Supplementary Figure S1 | FACS analysis of (A) CD3+T, (B) CD3+CD4+T, (C) CD3+CD8+T, (D) CD16+ and CD56+NK, and (E) CD19+B cells in peripheral blood.
Supplementary Table S1 | DE mRNAs and lncRNAs between HC and MA groups (AW, PW).
Supplementary Table S2 | DAVID_GO and DAVID_KEGG analyses of DE mRNAs and lncRNAs in the AW group.
Supplementary Table S3 | DAVID_GO and DAVID_KEGG analyses of DE mRNAs and lncRNAs in the PW group.
Supplementary Table S4 | Networks of miRNA-mRNA target pairs at the AW and the PW stage.
References
Akiyama, K., Isao, T., Ide, S., Ishikawa, M., and Saito, A. (2008). mRNA expression of the Nurr1 and NGFI-B nuclear receptor families following acute and chronic administration of methamphetamine. Prog. Neuropsychopharmacol. Biol. Psychiatry 32, 1957–1966. doi: 10.1016/j.pnpbp.2008.09.021
Aliperti, V., Skonieczna, J., and Cerase, A. (2021). Long non-coding RNA (lncRNA) roles in cell biology, neurodevelopment and neurological disorders. Noncoding RNA 7:36. doi: 10.3390/ncrna7020036
Beatriz, M., Vilaca, R., and Lopes, C. (2021). Exosomes: innocent bystanders or critical culprits in neurodegenerative diseases. Front. Cell Dev. Biol. 9:635104. doi: 10.3389/fcell.2021.635104
Bertram, L., Hiltunen, M., Parkinson, M., Ingelsson, M., Lange, C., Ramasamy, K., et al. (2005). Family-based association between Alzheimer’s disease and variants in UBQLN1. N. Engl. J. Med. 352, 884–894. doi: 10.1056/NEJMoa042765
Bortell, N., Basova, L., Semenova, S., Fox, H. S., Ravasi, T., Marcondes, M. C., et al. (2017). Astrocyte-specific overexpressed gene signatures in response to methamphetamine exposure in vitro. J. Neuroinflammation 14:49. doi: 10.1186/s12974-017-0825-6
Bowyer, J. F., and Ali, S. (2006). High doses of methamphetamine that cause disruption of the blood-brain barrier in limbic regions produce extensive neuronal degeneration in mouse hippocampus. Synapse 60, 521–532. doi: 10.1002/syn.20324
Butler, M. P., O’Connor, J. J., and Moynagh, P. N. (2004). Dissection of tumor-necrosis factor-alpha inhibition of long-term potentiation (LTP) reveals a p38 mitogen-activated protein kinase-dependent mechanism which maps to early-but not late-phase LTP. Neuroscience 124, 319–326. doi: 10.1016/j.neuroscience.2003.11.040
Buzas, E. I., Gyorgy, B., Nagy, G., Falus, A., and Gay, S. (2014). Emerging role of extracellular vesicles in inflammatory diseases. Nat. Rev. Rheumatol. 10, 356–364. doi: 10.1038/nrrheum.2014.19
Charney, A. W., Ruderfer, D. M., Stahl, E. A., Moran, J. L., Chambert, K., Belliveau, R. A., et al. (2017). Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl. Psychiatry 7:e993. doi: 10.1038/tp.2016.242
Chen, F., Zou, L., Dai, Y., Sun, J., Chen, C., Zhang, Y., et al. (2021). Prognostic plasma exosomal microRNA biomarkers in patients with substance use disorders presenting comorbid with anxiety and depression. Sci. Rep. 11:6271. doi: 10.1038/s41598-021-84501-5
Cheng, L., Doecke, J. D., Sharples, R. A., Villemagne, V. L., Fowler, C. J., Rembach, A., et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20, 1188–1196. doi: 10.1038/mp.2014.127
Cunningham, A. J., Murray, C. A., O’Neill, L. A., Lynch, M. A., and O’Connor, J. J. (1996). Interleukin-1 βbeta (IL-1 β ) and tumour necrosis factor (TNF) inhibit long-term potentiation in the rat dentate gyrus in vitro. Neurosci. Lett. 203, 17–20. doi: 10.1016/0304-3940(95)12252-4
Darke, S., Kaye, S., McKetin, R., and Duflou, J. (2008). Major physical and psychological harms of methamphetamine use. Drug Alcohol Rev. 2, 253–262. doi: 10.1080/09595230801923702
Eisenstein, T. K., Rahim, R. T., Feng, P., Thingalaya, N. K., and Meissler, J. J. (2006). Effects of opioid tolerance and withdrawal on the immune system. J. Neuroimmune Pharmacol. 1, 237–249. doi: 10.1007/s11481-006-9019-1
Gao, Y., Ma, H., Lv, C., Lan, F., Wang, Y., Deng, Y., et al. (2021). Exosomes and exosomal microRNA in non-targeted radiation bystander and abscopal effects in the central nervous system. Cancer Lett. 499, 73–84. doi: 10.1016/j.canlet.2020.10.049
Glasner-Edwards, S., Mooney, L. J., Marinelli-Casey, P., Hillhouse, M., Ang, A., Rawson, R., et al. (2010). Anxiety disorders among methamphetamine dependent adults: association with post-treatment functioning. Am. J. Addict. 19, 385–390. doi: 10.1111/j.1521-0391.2010.00061.x
Grieco, G. E., Fignani, D., Formichi, C., Nigi, L., Licata, G., Maccora, C., et al. (2021). Extracellular vesicles in immune system regulation and type 1 diabetes: cell-to-cell communication mediators, disease biomarkers and promising therapeutic tools. Front. Immunol. 12:682948. doi: 10.3389/fimmu.2021.682948
Hall, W., Hando, J., Darke, S., and Ross, J. (1996). Psychological morbidity and route of administration among amphetamine users in Sydney, Australia. Addiction 91, 81–87. doi: 10.1046/j.1360-0443.1996.9118110.x
Harms, R., Morsey, B., Boyer, C. W., Fox, H. S., and Sarvetnick, N. (2012). Methamphetamine administration targets multiple immune subsets and induces phenotypic alterations suggestive of immunosuppression. PLoS One 7:e49897. doi: 10.1371/journal.pone.0049897
Hernandez-Santini, A. C., Mitha, A. N., Chow, D., Hamed, M. F., Gucwa, A. L., Vaval, V., et al. (2021). Methamphetamine facilitates pulmonary and splenic tissue injury and reduces T cell infiltration in C57BL/6 mice after antigenic challenge. Sci. Rep. 11:8207. doi: 10.1038/s41598-021-87728-4
Hoshino, A., Kim, H. S., Bojmar, L., Gyan, K. E., Cioffi, M., Hernandez, J., et al. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 182, 1044–1061. e18. doi: 10.1016/j.cell.2020.07.009
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Huang, H. Y., Lin, Y. C., Li, J., Huang, K. Y., Shrestha, S., Hong, H. C., et al. (2020). miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic. Acids Res. 48, D148–D154. doi: 10.1093/nar/gkz896
Iwasa, M., Maeno, Y., Inoue, H., Koyama, H., and Matoba, R. (1996). Induction of apoptotic cell death in rat thymus and spleen after a bolus injection of methamphetamine. Int. J. Legal Med. 109, 23–28. doi: 10.1007/BF01369597
Jerrells, T. R., Peritt, D., Marietta, C., and Eckardt, M. J. (1989). Mechanisms of suppression of cellular immunity induced by ethanol. Alcohol. Clin. Exp. Res. 13, 490–493. doi: 10.1111/j.1530-0277.1989.tb00363.x
Karagkouni, D., Paraskevopoulou, M. D., Tastsoglou, S., Skoufos, G., Karavangeli, A., Pierros, V., et al. (2020). DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic. Acids Res. 48, D101–D110. doi: 10.1093/nar/gkz1036
Li, H. C., Lin, Y. B., Li, C., Luo, C. H., Zhou, Y. T., Ou, J. Y., et al. (2018). Expression of miRNAs in serum exosomes versus hippocampus in methamphetamine-induced rats and intervention of rhynchophylline. Evid. Based Complement. Alternat. Med. 2018:8025062. doi: 10.1155/2018/8025062
Loftis, J. M., and Huckans, M. (2013). Substance use disorders: psychoneuroimmunological mechanisms and new targets for therapy. Pharmacol. Ther. 139, 289–300. doi: 10.1016/j.pharmthera.2013.04.011
Loftis, J. M., and Janowsky, A. (2014). Neuroimmune basis of methamphetamine toxicity. Int. Rev. Neurobiol. 118, 165–197. doi: 10.1016/B978-0-12-801284-0.00007-5
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lynch, H. E., Sanchez, A. M., D’Souza, M. P., Rountree, W., Denny, T. N., Kalos, M., et al. (2014). Development and implementation of a proficiency testing program for Luminex bead-based cytokine assays. J. Immunol. Methods 409, 62–71. doi: 10.1016/j.jim.2014.04.011
Masoumipour, M., Abbaspanah, B., and Mousavi, S. H. (2021). Extracellular vesicles: regenerative medicine prospect in hematological malignancies. Cell Biol. Int. 45, 2031–2044. doi: 10.1002/cbin.11660
Mata, M. M., Napier, T. C., Graves, S. M., Mahmood, F., Raeisi, S., Baum, L. L., et al. (2015). Methamphetamine decreases CD4 T cell frequency and alters pro-inflammatory cytokine production in a model of drug abuse. Eur. J. Pharmacol. 752, 26–33. doi: 10.1016/j.ejphar.2015.02.002
McKetin, R., Ross, J., Kelly, E., Baker, A., Lee, N., Lubman, D. I., et al. (2008). Characteristics and harms associated with injecting versus smoking methamphetamine among methamphetamine treatment entrants. Drug Alcohol Rev. 27, 277–285. doi: 10.1080/09595230801919486
Meng, Y., Ding, J., Li, C., Fan, H., He, Y., Qiu, P., et al. (2020). Transfer of pathological alpha-synuclein from neurons to astrocytes via exosomes causes inflammatory responses after METH exposure. Toxicol. Lett. 331, 188–199. doi: 10.1016/j.toxlet.2020.06.016
Miller, D. R., Bu, M., Gopinath, A., Martinez, L. R., and Khoshbouei, H. (2021). Methamphetamine dysregulation of the central nervous system and peripheral immunity. J. Pharmacol. Exp. Ther. 379, 372–385. doi: 10.1124/jpet.121.000767
Mitha, A. N., Chow, D., Vaval, V., Guerrero, P., Rivera-Rodriguez, D. E., Martinez, L. R., et al. (2021). Methamphetamine compromises the adaptive B cell-mediated immunity to antigenic challenge in C57BL/6 mice. Front. Toxicol. 3:629451. doi: 10.3389/ftox.2021.629451
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pineles, B., Mani, A., Sura, L., Rossignol, C., Albayram, M., Weiss, M. D., et al. (2022). Neuronal exosome proteins: novel biomarkers for predicting neonatal response to therapeutic hypothermia. Arch. Dis. Child. Fetal Neonatal Ed. 107, 60–64. doi: 10.1136/archdischild-2020-321096
Potula, R., Haldar, B., Cenna, J. M., Sriram, U., and Fan, S. (2018). Methamphetamine alters T cell cycle entry and progression: role in immune dysfunction. Cell Death Discov. 4:44. doi: 10.1038/s41420-018-0045-6
Potula, R., Hawkins, B. J., Cenna, J. M., Fan, S., Dykstra, H., Ramirez, S. H., et al. (2010). Methamphetamine causes mitrochondrial oxidative damage in human T lymphocytes leading to functional impairment. J. Immunol. 185, 2867–2876. doi: 10.4049/jimmunol.0903691
Prakash, M. D., Tangalakis, K., Antonipillai, J., Stojanovs.ka, L., Nurgali, K., Apostolopoulos, V., et al. (2017). Methamphetamine: effects on the brain, gut and immune system. Pharmacol. Res. 120, 60–67. doi: 10.1016/j.phrs.2017.03.009
Qiu, P., Zhou, J., Zhang, J., Dong, Y., and Liu, Y. (2021). Exosome: the regulator of the immune system in sepsis. Front. Pharmacol. 12:671164. doi: 10.3389/fphar.2021.671164
Ramirez, S. H., Andrews, A. M., Paul, D., and Pachter, J. S. (2018). Extracellular vesicles: mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 15:19. doi: 10.1186/s12987-018-0104-7
Saito, M., Yamaguchi, T., Kawata, T., Ito, H., Kanai, T., Terada, M., et al. (2006). Effects of methamphetamine on cortisone concentration, NK cell activity and mitogen response of T-lymphocytes in female cynomolgus monkeys. Exp. Anim. 55, 477–481. doi: 10.1538/expanim.55.477
Sekine, Y., Ouchi, Y., Sugihara, G., Takei, N., Yoshikawa, E., Nakamura, K., et al. (2008). Methamphetamine causes microglial activation in the brains of human abusers. J. Neurosci. 28, 5756–5761. doi: 10.1523/JNEUROSCI.1179-08.2008
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). Functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic. Acids Res. 50, W216–W221. doi: 10.1093/nar/gkac194
Stuendl, A., Kunadt, M., Kruse, N., Bartels, C., Moebius, W., Danzer, K. M., et al. (2016). Induction of alpha-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies. Brain 139, 481–494. doi: 10.1093/brain/awv346
Thompson, A. G., Gray, E., Heman-Ackah, S. M., Mager, I., Talbot, K., Andaloussi, S. E., et al. (2016). Extracellular vesicles in neurodegenerative disease - pathogenesis to biomarkers. Nat. Rev. Neurol. 12, 346–357. doi: 10.1038/nrneurol.2016.68
Yu, J., Cadet, J. L., and Angulo, J. A. (2002). Neurokinin-1 (NK-1) receptor antagonists abrogate methamphetamine-induced striatal dopaminergic neurotoxicity in the murine brain. J. Neurochem. 83, 613–622. doi: 10.1046/j.1471-4159.2002.01155.x
Yu, J., Chen, F., Xu, Y., Shi, K., Zhang, Z., Peng, Q., et al. (2021). Dynamics of neurotransmitter and extracellular vesicle-derived microRNA landscapes during heroin and methamphetamine withdrawal. medRxiv [Preprint]. doi: 10.1101/2021.04.19.21255653
Zani-Ruttenstock, E., Antounians, L., Khalaj, K., Figueira, R. L., and Zani, A. (2021). The role of exosomes in the treatment, prevention, diagnosis and pathogenesis of COVID-19. Eur J. Pediatr. Surg. 31, 326–334. doi: 10.1055/s-0041-1731294
Zhang, Z., Wu, H., Peng, Q., Xie, Z., Chen, F., Ma, Y., et al. (2021). Integration of molecular inflammatory interactome analyses reveals dynamics of circulating cytokines and extracellular vesicle long non-coding RNAs and mRNAs in heroin addicts during acute and protracted withdrawal. Front. Immunol. 12:730300. doi: 10.3389/fimmu.2021.730300
Keywords: methamphetamine, exosome, transcriptome, regulatory network, immune response
Citation: Wu H, Zhang Z, Ma Y, Chen F, Xiong P, Xie Z, Ding G, Yu J and Wang K (2022) Dynamic immune and exosome transcriptomic responses in patients undergoing psychostimulant methamphetamine withdrawal. Front. Cell. Neurosci. 16:961131. doi: 10.3389/fncel.2022.961131
Received: 04 June 2022; Accepted: 29 August 2022;
Published: 27 September 2022.
Edited by:
Xiaohuan Xia, Tongji Hospital Affiliated to Tongji University, ChinaReviewed by:
Xiao Han, Fudan University, ChinaHelioswilton Sales-Campos, Universidade Federal de Goiás, Brazil
Copyright © 2022 Wu, Zhang, Ma, Chen, Xiong, Xie, Ding, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juehua Yu, anVlaHVheXVAZ21haWwuY29t https://orcid.org/0000-0002-1661-0503; Kunhua Wang, a3VuaHVhd2FuZzFAMTYzLmNvbQ==
† These authors have contributed equally to this work
 Hongjin Wu
Hongjin Wu Zunyue Zhang
Zunyue Zhang Yuru Ma
Yuru Ma Fengrong Chen
Fengrong Chen Pu Xiong2
Pu Xiong2 Zhenrong Xie
Zhenrong Xie Guo Ding
Guo Ding Juehua Yu
Juehua Yu Kunhua Wang
Kunhua Wang