
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 12 August 2022
Sec. Non-Neuronal Cells
Volume 16 - 2022 | https://doi.org/10.3389/fncel.2022.932572
This article is part of the Research Topic The Role of Glial Cells in Neurodegenerative Diseases View all 4 articles
In recent years, glial cells have been acknowledged as key players in the pathogenesis of Alzheimer’s disease (AD), a neurodegenerative condition in which an accumulation of intracellular neurofibrillary tangles and extracellular fibrillar amyloid beta is notably observed in the central nervous system. Genome-wide association studies have shown, both in microglia and astrocytes, an increase in gene variants associated with a higher risk of developing late-onset AD. Microglia, the resident innate immune cells of the brain, and astrocytes, glial cells crucial for vascular integrity and neuronal support, both agglomerate near amyloid beta plaques and dystrophic neurites where they participate in the elimination of these harmful parenchymal elements. However, their role in AD pathogenesis has been challenging to resolve due to the highly heterogeneous nature of these cell populations, i.e., their molecular, morphological, and ultrastructural diversity, together with their ever-changing responsiveness and functions throughout the pathological course of AD. With the recent expansions in the field of glial heterogeneity through innovative advances in state-of-the-art microscopy and -omics techniques, novel concepts and questions arose, notably pertaining to how the diverse microglial and astrocytic states interact with each other and with the AD hallmarks, and how their concerted efforts/actions impact the progression of the disease. In this review, we discuss the recent advances and findings on the topic of glial heterogeneity, particularly focusing on the relationships of these cells with AD hallmarks (e.g., amyloid beta plaques, neurofibrillary tangles, synaptic loss, and dystrophic neurites) in murine models of AD pathology and post-mortem brain samples of patients with AD.
Alzheimer’s disease (AD), the most common form of dementia, is a neurodegenerative disease notably associated with severe synaptic loss and brain atrophy, clinically resulting in cognitive decline (Terry et al., 1991; Spires-Jones and Hyman, 2014; Pini et al., 2016; Halliday, 2017; Marino et al., 2019). The progressive impairment of learning and memory, among other cognitive functions, that characterizes AD is neuropathologically associated with intracellular neurofibrillary tangles composed of hyperphosphorylated tau protein and extracellular plaque deposition of amyloid beta (Aß) (DeTure and Dickson, 2019). Several key brain regions (e.g., hippocampus, dentate gyrus, entorhinal cortex, prefrontal cortex) involved in these functions are more vulnerable or prone to the various AD hallmarks aforementioned (Halliday, 2017; Mrdjen et al., 2019; Leng et al., 2021). However, while the presence of these abnormal features is a strong indicator of AD, the preserved cognition of some individuals with these AD hallmarks suggests that different mechanisms, apart from the pathological markers of AD, might be at play (Jack et al., 2016; Zolochevska and Taglialatela, 2016). Indeed, previous hypotheses regarding the pathogenesis of AD primarily comprise the amyloid cascade, which places the neurotoxic accumulation of the Aß protein at the center of the disease process (Hardy and Higgins, 1992; Ricciarelli and Fedele, 2017). Recent amendments to this hypothesis include mitochondrial dysfunction (Chakravorty et al., 2019), cellular senescence (Saez-Atienzar and Masliah, 2020; Walton et al., 2020; Guerrero et al., 2021) and vascular dysfunction (Govindpani et al., 2019). All these emerging hypotheses have highlighted the impact of glial cells (e.g., astrocytes, microglia) on AD pathology (Liu et al., 2018; Kaur et al., 2019; Hashemiaghdam and Mroczek, 2020).
Microglia first appear in the yolk sac where they egress to the central nervous system (CNS) around embryonic day 9.5 in mice (Ginhoux et al., 2010) and the 4–5th week of gestation in humans (Andjelkovic et al., 1998; Monier et al., 2006; Verney et al., 2010). From synaptic pruning and plasticity to surveillance of the parenchyma and removal of cellular debris, microglia exert crucial functions necessary for the proper development and maintenance of the CNS homeostasis throughout life (Davalos et al., 2005; Nimmerjahn et al., 2005; Paolicelli et al., 2011; Schafer et al., 2012). While understanding the role of microglia in the pathogenesis of AD has gained more traction in recent years with the advances in single-cell(sc)-omic techniques, it is still unclear if and/or how these cells can be modulated to help prevent and/or treat this neurodegenerative disease (Šimončičová et al., 2022). To study the mechanisms underlying this neurodegenerative disease, murine models of AD pathology, genetically altered to induce human AD hallmarks, are most commonly used (Jankowsky and Zheng, 2017). A summary of all the murine models of AD pathology mentioned in this Review is provided in Table 1. Studies from Sosna et al., and Spangenberg et al., have shown that long-term treatment starting early during the disease (at 1.5–2 months of age) with an inhibitor of colony-stimulating factor 1 receptor (CSF1R), notably crucial for microglial survival, was able to reduce the number of Aß plaques in the 5xFAD model [Swedish, Florida and London mutation in the amyloid precursor protein (APP) with M146L and L286V mutations in the humanized presenilin 1 (PSEN1) (Oakley et al., 2006)] (Sosna et al., 2018; Spangenberg et al., 2019). Oppositely, introducing a CSF1R inhibitor later on at 10 months of age did not detectably alter Aß load in the same 5xFAD model (Spangenberg et al., 2016) or in 12-month-old APP-PS1 mice [Swedish mutation in APP and humanized PSEN1 (Jankowsky et al., 2004; Radde et al., 2006)] (Unger et al., 2018), emphasizing the differential roles exerted by microglia across AD pathology progression (Casali et al., 2020). In addition, the functional spatio-temporal heterogeneity of microglia, observed even in non-pathological conditions and throughout the lifespan (Hammond et al., 2019; Zheng et al., 2021), confers an additional challenge alongside the diversity of their population comprised of different states [e.g., dark microglia, disease-associated microglia (DAM), neurodegenerative phenotype (MGnD)] observed in various mouse models of AD pathology and human post-mortem samples (Bisht et al., 2016b; Keren-Shaul et al., 2017; Krasemann et al., 2017).
In the last 5 years, the heterogeneous nature of astrocytes was also brought to the forefront of the dementia field with several studies underlining a myriad of astrocytic states in both health and disease (e.g., disease-associated astrocytes) (Liddelow et al., 2017; Habib et al., 2020; Escartin et al., 2021). Astrocytes were shown to play a key part in neuronal support; from sustaining metabolic needs by providing lactate to neurons according to the astrocyte-neuron lactate shuttle hypothesis, to recycling exocytotoxic glutamate into glutamine to be taken up by pre-synaptic axon terminals (Tsacopoulos and Magistretti, 1996; Pellerin et al., 2007; Verkhratsky and Nedergaard, 2018). In both patients with AD and mouse models of AD pathology, astrocytes were shown to interact with fibrillar Aß (Nagele et al., 2004) and dystrophic neurites found nearby Aß plaques (Gomez-Arboledas et al., 2018). For instance, ablation of astrocytes and proliferative astrocytes in organotypic 5xFAD brain culture slices and 9-month-old APP23/GFAP-TK mice [APP23 is a mouse model overexpressing a human APP with a Swedish mutation (Sturchler-Pierrat et al., 1997) crossed with glial fibrillary acidic protein (GFAP) thymidine kinase (TK) mice], respectively, both showed increased levels of Aß, indicating a crucial role for astrocytes in the resolution of AD pathology (Katsouri et al., 2020; Davis et al., 2021). Similar to microglia, astrocytes display a spatio-temporal heterogeneity during aging and in various pathological conditions (Lana et al., 2020). For instance, depending on their proximity to Aß plaques, astrocytes present varying morphologies (atrophy, hypertrophy) and molecular signatures in post-mortem brains of patients with AD and mouse models of AD pathology (Schitine et al., 2015). However, further investigations are required to fully understand the functional impact of their heterogeneous nature on the onset and progression of AD pathology.
Understanding the complex relationships of heterogeneous glial cells with their micro-environment will be crucial to unravel not only the mechanisms underlying AD, but also help to determine their responses to various potential therapeutic targets. Therefore, in this review, we will highlight the different microglial and astrocytic states observed in both mouse models of AD pathology and human post-mortem samples, discussing specifically how this heterogeneity affects their interaction with each other and the hallmarks of AD.
Microglia aggregate near Aß plaques where they partially initiate and maintain a highly dynamic phagocytic and (neuro)inflammatory response (Manchikalapudi et al., 2019; Shukla et al., 2019; von Saucken et al., 2020; Grubman et al., 2021). While recent studies have helped elucidate the roles of microglia in the pathogenesis of AD, their functions throughout the course of the disease are still a topic boasting a variety of opinions. As much as microglia are able to take up Aß (Hickman et al., 2008; Sebastian Monasor et al., 2020), indicating their ability to initially inhibit plaque growth, they are ultimately not as effective at removing existing deposits later on in the disease process (Sebastian Monasor et al., 2020; Zhang G. et al., 2021). In addition, extensive literature highlights the impaired or altered functions that these cells exhibit throughout the progression of the disease (Prokop et al., 2013; Gyoneva et al., 2016; Kaur et al., 2019; Sebastian Monasor et al., 2020). How these changes in function may relate to morphological, ultrastructural, and transcriptomic states will be further discussed below.
Microglia survey the parenchyma through their ramified and highly motile processes. This morphological state, termed ramified or surveying microglia, can rapidly shift in response to micro-environmental changes (Davalos et al., 2005; Nimmerjahn et al., 2005). Ameboid microglia represent one of these morphological shifts and are observed notably after acute brain injury and in conditions where phagocytosis of debris or pathogens takes place (Giulian, 1987; Savage et al., 2019). This ameboid state has a large and round cell body which at first possess thin, almost invisible, processes that later on retract, which is thought to aid in microglia’s ability to migrate across the brain (Lu et al., 2011; Parakalan et al., 2012; Leyh et al., 2021). In AD pathology, the interaction between microglia and Aß can drive this morphological state. Curiously, morphological alterations linked to microglia’s close association with Aß plaques in the neocortex of 5-month-old male and female TgCRND8wt/tg; CXC3CR1GFP/wt mice [mouse model with Swedish and Indiana mutations in APP (Chishti et al., 2001)] include a decrease in ramification, surface area, cell volume, number of processes and junctions (Plescher et al., 2018), an atrophy unlike the increased cell soma size traditionally associated with ameboid microglia (Leyh et al., 2021). This ameboid morphology has also been suggested to reflect microglial proliferation observed nearby Aß plaques in the post-mortem hippocampus of patients with AD (Marlatt et al., 2014).
Other morphological states include hypertrophic microglia–large cell bodies with short and thick processes–which have been identified in various pathological mouse models (e.g., traumatic brain injury, AD pathology), along with human AD cases (Walker and Lue, 2015; Zanier et al., 2015; Romero-Molina et al., 2018; Ohm et al., 2020). Indeed, this state is conserved in human post-mortem brain samples, being described in the hippocampus of aged individuals with AD (mean age of 77 and Braak stage IV-VI) (Bachstetter et al., 2015). Another state termed dystrophic microglia (also referred to as senescent in the literature) was uncovered in aging and notably near tau pathology (neurofibrillary degeneration) in AD human brains (Streit et al., 2004, 2009, 2020). These cells were additionally found in the middle temporal gyrus of healthy individuals (Swanson et al., 2020) and in the dorsal hippocampus of aged (7.5 years old) male tree shrews that developed hyperphosphorylated tau (Rodriguez-Callejas et al., 2020). Characteristic features of these cells include spherical swellings in their tortuous processes, notable accumulation of lipofuscin deposits (Streit et al., 2004, 2009, 2020; Lopes et al., 2008), and immunohistoreactivity to L-ferritin, a marker of cellular senescence (Lopes et al., 2008; Swanson et al., 2020). However, recent studies suggest that this microglial morphology could be linked to the pH of the brain rather than AD pathology as it was shown that lowering the pH of the brain was associated with an increased abundance of dystrophic microglia (Paasila et al., 2019). While hypoxia is associated with low pH, this study investigated the effect of changing pH without altering the oxygen content (Paasila et al., 2019).
Ultrastructural heterogeneity, including microglia’s diverse relationships with Aß and dystrophic neurites, can be observed in AD pathology using electron microscopy (EM) (Stalder et al., 1999; El Hajj et al., 2019). EM is a powerful imaging tool allowing to demystify the presence and functional state of various organelles within cells and their interactions with the micro-environment, but also the topographical heterogeneity of microglia. For instance, using scanning EM, Dyne et al., investigated the ultrastructural morphology (cell body shape, processes length, surface topology, aspect ratio) of human C20 cells [immortalized human microglial cell line from resected adult human brain samples] that were treated for 24 h with Aß. These cells showed a diverse topography based on their surface (designated as smooth, blebbed, ruffled or pitted depending on the number and size of their cavities) and displayed enlarged pores compared to an anti-inflammatory condition [interleukin (IL)-4 treatment for 24 h], a change hypothesized to be associated with phagocytosis (Dyne et al., 2021).
EM imaging was also employed to investigate ultrastructural microglial alterations in 14-month-old APP-PS1 male mice where microglia were shown to possess increased markers of cellular stress [e.g., endoplasmic reticulum (ER) dilation], a phenomenon which was exacerbated in microglia associated with dystrophic neurites and Aß (El Hajj et al., 2019). Microglial ultrastructural heterogeneity in AD pathology is also highlighted by the presence of dark microglia, a state associated with numerous markers of cellular stress and advanced/tertiary lysosomal organelles (Bisht et al., 2016b; St-Pierre et al., 2019, 2020). While this state is rarely present in adulthood during normal physiological conditions, it is notably more abundant in pathological conditions, including in 14-month-old APP-PS1 male mice (Bisht et al., 2016b). There are notable differences between dark microglia and their non-dark (or more typical) counterparts, which includes markers of oxidative stress (e.g., a condensed, electron-dense cyto- and nucleoplasm, altered mitochondria, dilated ER) and a remodeled nuclear chromatin pattern (Bisht et al., 2016a,b; St-Pierre et al., 2019, 2020). Dark microglia possess highly ramified, dark processes which frequently interact with axon terminals and dendritic spines (Bisht et al., 2016a); suggesting that they likely play a role in the synaptic dysfunction and loss observed in AD.
Genome-wide association studies uncovered numerous genes associated with a higher risk of developing late-onset AD [reviewed in Bertram and Tanzi (2009)]. One of these genes, triggering receptor expressed on myeloid cells 2 (trem2), was shown to be crucial for the appearance of various microglial states, including the DAM reported in male and female 5xFAD mice at 3, 6, and 8 months of age (Keren-Shaul et al., 2017). The DAM transcriptomic state is associated with a TREM2-dependent decrease of homeostatic genes (e.g., p2ry12, tmem119) and increase of disease signature genes (e.g., clec7a, spp1, itgax) (Keren-Shaul et al., 2017). The necessity of TREM2 for the emergence of DAM was also confirmed by independent studies in which 7-month-old male and female 5xFAD mice were crossed with either homozygous or heterozygous TREM2-/- mice and injected intracerebrally with tau. The latter resulted in a reduced expression of DAM genes in the 5xFAD × TREM2+/- mice compared to 5xFAD × TREM2+/- mice, which was further exacerbated by the whole-brain knock-out of TREM2, when examined in both the cortex and hippocampus of these mice (Delizannis et al., 2021).
DAM were suggested to restrict themselves/reside primarily nearby Aß plaques–making it an important area of interest for therapeutical targeting (Keren-Shaul et al., 2017). Since the initial discovery by Keren-Shaul et al., this particular microglial signature has been uncovered in several other mouse models of AD pathology, including in 9-month-old homozygous APP-KI male and female mice [intercrossing of the heterogenous APPNL–F–G-KI mice, a mouse model with a Swedish, Iberian and Artic mutation in APP (Saito et al., 2014)] (Swartzlander et al., 2018). A summary of the overlap between DAM and other microglial states mentioned in this review is provided in Figure 1. In addition, DAM genes were observed notably in 6-month-old male TgCRND8 mice using RNA sequencing on laser capture microdissected non- and plaque-associated microglia; the DAM signature was largely restricted to nearby Aß plaques, highlighting the necessity of Aß plaques to induce this unique state (Rothman et al., 2018). Of note, while primary mouse microglial cells treated 12 h with fibrillar Aß recapitulated the DAM signature, cells treated for 12 h with oligomeric Aß did not, stressing the importance of an amyloid conformation for this particular microglial response (McFarland et al., 2021). Tau, in conjuncture with Aß, was also shown to drive the expression of the DAM signature in 7-month-old 5xFAD × P301S mice injected stereotactically with tau [P301S is mouse model expressing microtubule associated protein tau (MAPT) with a human PS301 mutation (Allen et al., 2002; Bellucci et al., 2004; Yoshiyama et al., 2007)] (Lodder et al., 2021) and 13-month-old APP-PS1 female mice both with and without the P301L mutation that induces the production of hyperphosphorylated tau (Natunen et al., 2020). An increased expression of DAM genes (e.g., clec7a, cd68, trem2) was also shown in tau pathology mouse models; e.g., in the hippocampus of 9–12-month-old P301S male and female mice (Romero-Molina et al., 2018) and in the cerebral cortex of 7-month-old rTg4510 mice [model expressing P301L mutation in MAPT (Ramsden et al., 2005; Santacruz et al., 2005)] (Sobue et al., 2021).
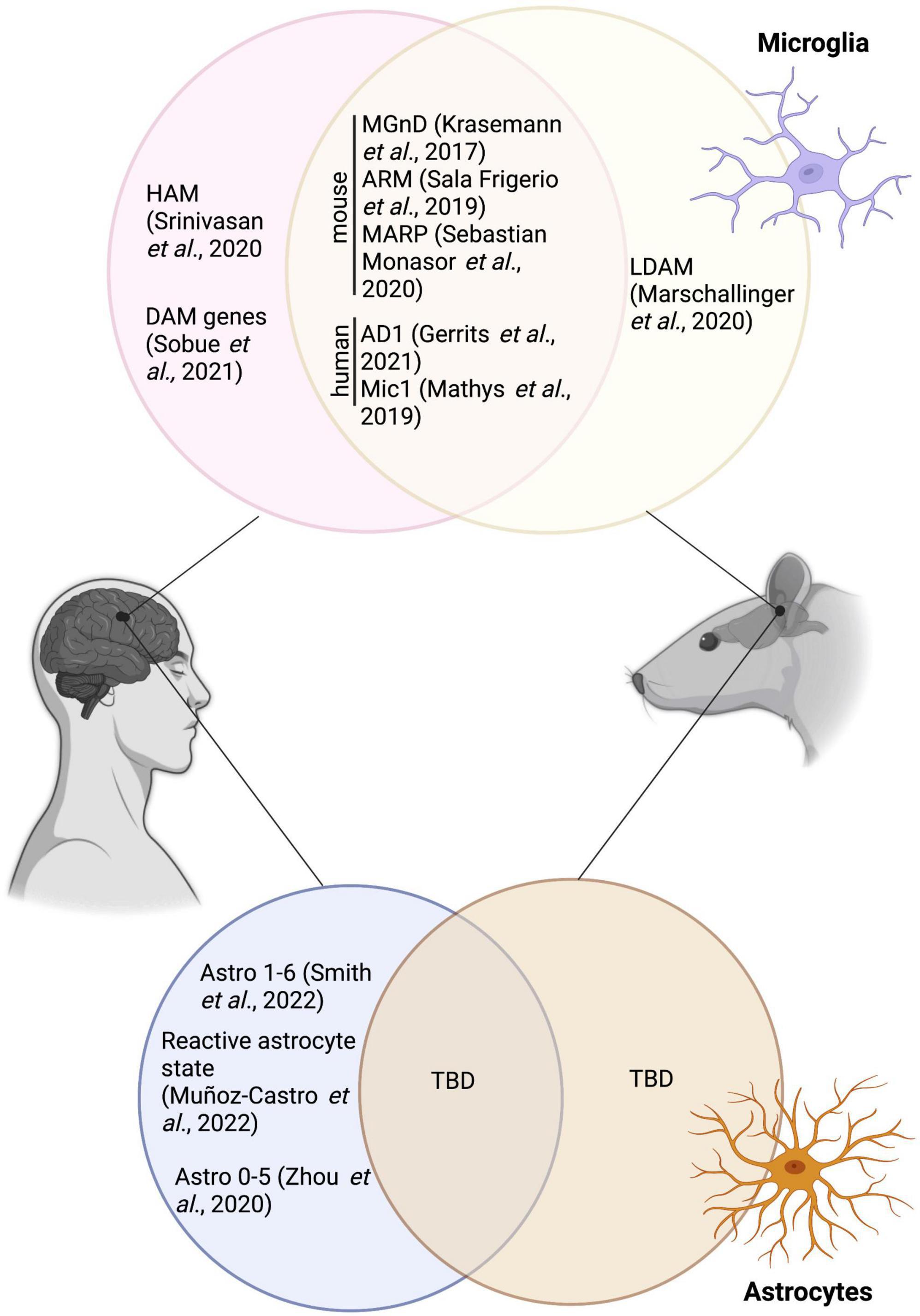
Figure 1. Overlap between the various microglial and astrocytic signatures described in human and mice, and the disease associated microglia and disease-associated astrocytes. HAM, human AD microglia; MGnD, neurodegenerative disease phenotype; ARM, activated-response microglia; MARP, microglial Aß response proteins; AD1, Alzheimer’s disease 1; LDAM, lipid droplet-accumulating microglia; TBD, to be determined. Figure was created using Biorender.
Further investigation identified various substates of DAM emphasizing the vast heterogeneity observed even within individual microglial states. These include, for instance, a pro- or anti-inflammatory substate in 6–8 month-old female 5xFAD mice (Rangaraju et al., 2018), and a senescent substate (as shown by telomere shortening, increased beta-galactosidase activity and changes in the expression of genes such as serpine1, cdkn1/2a and casp8 associated with senescence) in 4, 6, 10 and 12/13 month-old APP-PS1 male and female mice (Hu et al., 2021). In addition to this intra-state diversity reported for the DAM signature, both the abundance and gene enrichment scores were found to be affected by the utilized mouse strain (Yang et al., 2021), further stressing the myriad of factors capable of affecting microglial heterogeneity.
Moreover, the DAM signature transcriptomically overlaps with the activated-response microglia (ARM), another microglial state identified in the cortex and hippocampus of 3, 6, 12 and 21-month-old APPNL–G–F and C57BL/6 male and female mice, the latter which increases with age and pathology (Sala Frigerio et al., 2019). This signature seems to be strongly driven, however, by Aß as shown by the drastic increase of the hippocampal ARM population in 10/11-month-old vs 4-month-old APP-PS1 mice, and was also mildly increased in a tau pathology mouse model (Thy-Tau22 mice) [mouse model with G272V and P301S mutations in MAPT (Schindowski et al., 2006)] at 4 vs 10/11 months of age (Sierksma et al., 2020). Fibrillar Aß was further associated with an age-related progressive alteration of microglial signatures, reflected through changes in microglial Aß response proteins (MARPs), which partially overlap with the DAM signature and show impaired phagocytosis in two models of amyloid deposition: in 1, 3, 6 as well as 12 month-old APP-PS1 and APPNL–G–F mice, both male and female (Sebastian Monasor et al., 2020). Other microglial states reported in AD pathology include the MGnD, which displays a down-regulation of homeostatic genes (e.g., p2ry12, tmem119, fcrls) and an up-regulation of genes such as apoe, clec7a, and trem2, linked to the phagocytosis of apoptotic neurons and dystrophic neurites in 24-month-old male and female APP-PS1 mice (Krasemann et al., 2017). It was also reported that the MGnD state presents an increased secretion of extracellular vesicles containing tau in 6-month-old male and female APPNL–G–F mice injected with adeno-associated virus (AAV)-P301L-tau, highlighting this state’s potential to propagate tau (Clayton et al., 2021).
While partial or overlapping DAM signatures were uncovered in numerous mouse models of AD pathology, the particular DAM signature did not correlate with the lipid droplet-accumulating microglia (LDAM), a state associated with increased production of reactive oxygen species (ROS) and impaired phagocytosis, in the hippocampus of 20-month-old C57BL/6 and GRN-/- male mice, a model of frontotemporal dementia (Marschallinger et al., 2020). Recent studies have, however, revealed a conservation of the DAM signature in brains of patients with AD in comparison to healthy individuals. Indeed, using single-nucleus (sn) RNA-seq, Gerrits et al., identified two microglial profiles: AD1–composed of three subclusters enriched for some DAM genes (e.g., LPL, ITGAX, SPP1) (higher abundance found in patients with AD) and AD2–three regrouped microglial subclusters enriched notably in homeostatic genes (higher abundance in healthy control patients) (Gerrits et al., 2021). A reduced expression of P2RY12, a marker traditionally linked to homeostatic microglia and downregulated in mouse disease-associated microglial signatures [DAM (Keren-Shaul et al., 2017), MGnD (Krasemann et al., 2017)], and dark microglia (Bisht et al., 2016b) was observed in frontal and temporal cortices of human post-mortem AD brains compared to control individuals (Maeda et al., 2021). While some studies found an overlap between the mouse DAM and human microglial signatures in the post-mortem prefrontal cortex of patients with AD (Mathys et al., 2019), others have instead shown that this distinct population was not conserved. Indeed, human AD microglia (HAM) in the frontal and temporal cortex do not possess the same profile described in mouse models of AD pathology, but are present in another neurodegenerative condition (post-mortem brains samples of patients with multiple sclerosis) (Srinivasan et al., 2020). A lack of DAM in human was also reported by Sobue et al., in the precuneus of individuals with AD (Sobue et al., 2021). A summary of microglial states identified transcriptomically in AD pathology is provided in Table 2.
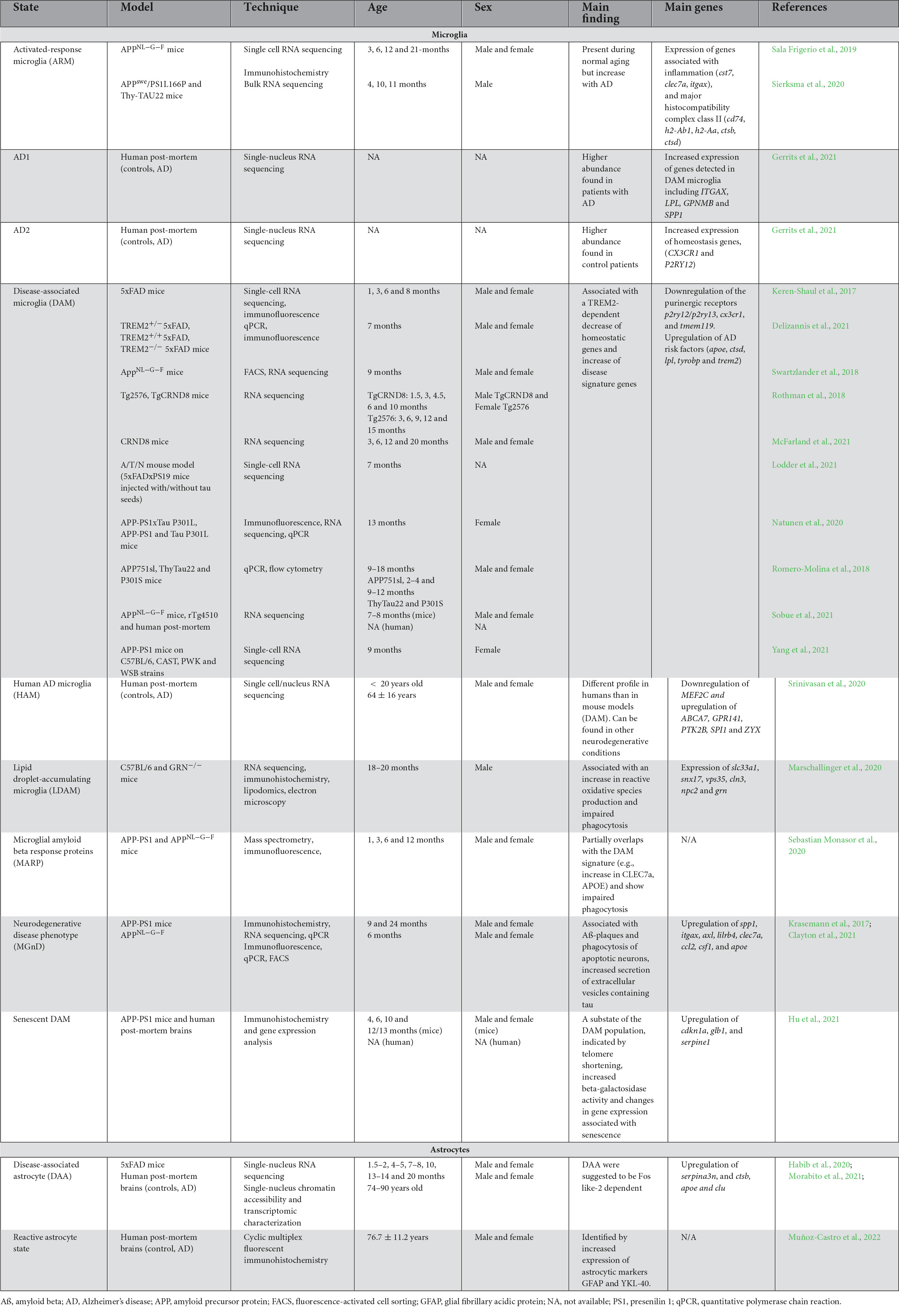
Table 2. Summary of microglial and astrocytic states observed in mouse models of AD pathology and human post-mortem brains of patients with AD.
While the most commonly used models of AD pathology at this point continue to be non-primates [reviewed in Jankowsky and Zheng (2017)], there are notable gaps between these microglia and the ones studied in human post-mortem samples. The challenge in translating results across species could in part result from the difficulty in removing intact microglial cells from post-mortem brain samples and the artifacts associated with long post-mortem intervals (Srinivasan et al., 2020), as well as a lack of murine models fully replicating the intricate features of the disease (Boche and Gordon, 2021). Of note, non-human primate models of AD, such as the common marmoset, have shown incredible similarities to human Aß pathology (Latimer et al., 2019; Li et al., 2019). Nonetheless, non-primate models of AD pathology have provided invaluable insights into microglial heterogeneity that highlight the complexity of microglia as a glial and immune cell type. An area of increasing interest is the design of specific radiotracers to study inflammation within the CNS through positron emission tomography (PET) [reviewed by Narayanaswami et al. (2018)]. This non-invasive, in vivo technique has been used previously to investigate microglial activity in conjunction with the (neuro)inflammation marker, translocator protein 18-kDa (TSPO) expressed on the outer membrane of mitochondria (Ghadery et al., 2019; Zhang L. et al., 2021). It is important to note that TSPO is not specific to microglia, being also expressed by other CNS cells (e.g., astrocytes) (Zhang L. et al., 2021). In addition, TPSO’s expression was shown to vary between rodents and human, where under pro-inflammatory conditions (treatment with lipopolysaccharide), TSPO gene expression increased in primary microglial cells from postnatal day 0–5 C57BL/6 mice contrary to adult microglia isolated from the temporal lobe of patients with epilepsy (Owen et al., 2017). Although there are many shortcomings to microglial PET analysis, further advances could lead to the development of specific radiotracers to study in vivo microglial heterogeneity in humans.
“Astrocyte reactivity” commonly refers to the plasticity or remodeling these cells undergo when responding to micro-environmental changes during pathological conditions (Escartin et al., 2021). Considered to represent a spectrum of varying states beginning in the developmental stages (Schitine et al., 2015; Clarke et al., 2021), astrocytes are often categorized based on their function and/or outcome on the brain (e.g., neuroprotective vs neurotoxic) (Liddelow et al., 2017; Miller, 2018; Matias et al., 2019; Westergard and Rothstein, 2020). The heterogenous nature of these glial cells ranges from varying morphology, ultrastructure to -omic signatures, to name a few. Micro-environmental changes, in both murine models of AD pathology and in human post-mortem brains of patients with AD, were also shown to drive this heterogeneity. For instance, astrocytes’ close interaction with AD hallmarks (e.g., dystrophic neurites, Aß plaques) can alter their morphology, notably resulting in hypertrophic and atrophic phenotypes (Zhou et al., 2019) as well as their gene and protein expression [e.g., GFAP, serpina3n, aquaporin 4 (AQP4), intercellular adhesion molecule 1 (ICAM-1)] (Akiyama et al., 1993; Orre et al., 2014; Habib et al., 2020; Spanos and Liddelow, 2020; Morabito et al., 2021).
On a morphological aspect, astrocytes are distinguished by their numerous branches that can further ramify, giving them a star-like appearance in light microscopy [for a detailed review on protoplasmic astrocytes morphology, see Zhou et al. (2019)] (Şovrea and Boşca, 2013). Similar to microglial spatial heterogeneity, astrocytes from different brain regions (e.g., cortical layers I-VI, hippocampus) possess diverse morphologies associated with specific transcriptomic clusters (e.g., larger and more arborized cells in the hippocampus CA3 vs smaller ones in the cortical layers 2–4) (Batiuk et al., 2020). In AD pathology, numerous studies denoted a change in astrocytic morphology associated with atrophy, including in human post-mortem brains of late-Braak stages (Verkhratsky et al., 2019), and in mouse models of AD pathology (Olabarria et al., 2010; Yeh et al., 2011; Kulijewicz-Nawrot et al., 2012; Diniz et al., 2017). This atrophy was particularly evident in 3xTg male mice [Swedish mutation in APP, P301S mutation in MAPT and M146 mutation in PSEN1 (Oddo et al., 2003)], among the medial prefrontal cortex (mPFC) at 3, 9, 12 and 18 months of age (Kulijewicz-Nawrot et al., 2012), the dentate gyrus at 6, 12 and 18 months of age (Olabarria et al., 2010) and the entorhinal cortex at 1, 3, 6, 9 and 12 months of age (Yeh et al., 2011). In addition, intracerebroventricular injections of oligomeric Aß in the hippocampus CA1 stratum radiatum of 3-month-old Swiss male mice were associated with an atrophy of the astrocytic cytoskeleton resulting in a decreased number of processes and reduced cell area (Diniz et al., 2017).
Other studies have shown an increased cytoskeleton hypertrophy or increased complexity of astrocytes near Aß plaques using GFAP immunolabeling in the hippocampus of 6, 12, and 18-month-old TgF344 male and female rats [Swedish mutation in APP and humanized PSEN1 (Cohen et al., 2013)] (Mampay et al., 2020), in the dentate gyrus and hippocampus CA1 of 18-month-old 3xTg male mice (Olabarria et al., 2010) and in the cerebral cortex of 3, 6 and 12-month-old APP-PS1 mice (increase in volume and process length) (Li et al., 2020). A sex-specific increase in the abundance of hypertrophic astrocytes in 7–9 month-old APP-PS1 female compared to male mice was observed in the outer molecular layer of the dentate gyrus (Richetin et al., 2017). These differences highlight the significant impact of the micro-environment, notably AD hallmarks, alongside the effect of sex, age, and model, on the morphological differences observed across studies. As aforementioned, an atrophy and/or hypertrophy of astrocytes has been shown in AD pathology, however, the variety of regions, models, species, ages, and analytic techniques makes it challenging to correlate morphological heterogeneity in human AD cases. For instance, in the subiculum of human post-mortem brain samples of early- and late-onset AD compared to healthy individuals, 3D reconstruction of astrocytes using GFAP immunolabeling showed no morphological differences (volume, cell area) (Taipa et al., 2018).
As shown with microglial studies, EM was used by numerous research teams to investigate the ultrastructure and function of astrocytes in AD pathology. Indeed, Lin et al., demonstrated the presence of tau filaments within astrocytes in the spinal cord white matter of a JPNL3 [expressing the tau mutation P301L (Lewis et al., 2000)] mouse model of tauopathy (Lin et al., 2003). Others found astrocytic engulfment of dystrophic neurites near Aß plaques in the hippocampus of 6 and 12-month-old APP-PS1 male mice and in the medial temporal lobe of patients with AD, further clarifying the phagocytic role of astrocytes that appears conserved across species (Gomez-Arboledas et al., 2018). Astrocytic processes were also shown to pierce inside the amyloid core of a large mature plaque where they fragmented the latter, underlining the intimate relationship between these two elements (Wegiel et al., 2000). EM was also pivotal in shedding light on certain astrocytic dysfunctions in AD, including mitochondrial ultrastructural alterations (Baloyannis, 2019) and the autophagic functions in vitro of APOE3 and APOE4 astrocytic cells from the cerebral cortices of postnatal day 1–3 mice (Simonovitch et al., 2016). Changes in the expression of the water channel AQP4 between perivascular astrocytic end-feet and plaque-associated astrocytes was also uncovered in 5xFAD and tg-ArcSwe [Swedish and Artic mutation in APP (Lord et al., 2006)] mice using this imaging technique (Yang et al., 2011, 2017; Smith et al., 2019). Despite EM’s extensive applications, few studies have explored the ultrastructural diversity and heterogeneity of astrocytes in AD pathology using this approach. Therefore, the field would greatly benefit from in-depth EM studies investigating astrocytes, similar to what was uncovered in microglia from mouse model of AD pathology (i.e., dark microglia) (Bisht et al., 2016b) and in post-mortem brain samples of individuals with AD (El Hajj et al., 2019) and schizophrenia (i.e., dystrophic-like microglia) (Uranova et al., 2018).
Many recent efforts to uncover astrocytic heterogeneity have employed -omics techniques to characterize the varying astrocytic states observed in AD pathology (see Table 2 for a summary of astrocytic molecular signatures in AD). Like microglia, astrocytic regional differences can already be appreciated at steady-state, prior to dishomeostasis driven by micro-environmental changes (John Lin et al., 2017). Indeed, scRNA-seq investigations have identified 5 astrocytic subclusters heterogeneously dispersed in both the cortex and the hippocampus of 2-month-old C57BL/6 mice (Batiuk et al., 2020). For instance, cortical cluster 1, which associated with a mature astrocytic signature [high expression of gfap and angiotensinogen (agt)], was located in the subpial layer while cluster 5, an intermediate between mature and immature astrocytes (genes linked notably with mitosis and energy metabolism), was found in cortical layers 2/3 and 5 (Batiuk et al., 2020). With the objective of determining heterogeneity based on transcriptional signatures, astrocytes were studied using snRNA-seq in the hippocampus and cortex of 7-month-old 5xFAD male and female mice (Habib et al., 2020). In this study, 6 transcriptional states were distinguished by their uniquely up- and down-regulated genes, including the disease-associated cluster, which expressed genes associated with APP processing and Aß production (e.g., serpina3n, ctsb) and clearance (e.g., apoe, clu), as well as inflammatory signaling (Habib et al., 2020). In later studies, the disease-associated astrocytic signature was suggested to be dependent on Fos like-2 (FOSL2) (Morabito et al., 2021), a subunit of the activator protein 1 (AP-1) transcription factor (Lee et al., 2021).
As for microglial studies examining the translation of disease-associated states from mouse to human, conservation of the astrocytic disease-associated gene signature across species is still considered controversial in AD. A summary of the overlap between disease-associated astrocytes and other astrocytic states mentioned in this review is provided in Figure 1. For instance, Smith et al., uncovered 6 astrocytic subclusters, located in the entorhinal and somatosensory cortex from aged non-diseased control individuals (Braak 0-II) and patients with AD (Braak III-VI), using snRNA-seq. These subclusters were enriched for genes associated with diverse astrocytic roles and pathways from homeostatic functions (e.g., neurotransmitter uptake) to immune responses (Smith et al., 2022). The disease-associated signature was not attributed to a unique cluster, highlighting differences in astrocytic populations between species (Smith et al., 2022). Other astrocytic signatures found in the post-mortem human brain of patients with AD vs age-matched controls include a reactive astrocytic state, denoted notably by a high expression of the astrocytic proteins GFAP and chitinase 3-like 1 (chi3l1 or YKL-40), which was observed in the temporal association cortex using cyclic multiplex fluorescent immunohistochemistry combined with spectral clustering analysis (Muñoz-Castro et al., 2022). Human astrocytic heterogeneity in the post-mortem human AD brain was also described by Zhou et al. using snRNA-seq analysis in prefrontal cortex samples which showed a down-regulation of genes associated with the metabolic coordination between neurons and astrocytes [e.g., superoxide dismutase 2 (sod2), hypoxia inducible lipid droplet associated (hilpda), fatty acid binding protein 5 (fabp5)] (Zhou et al., 2020). While the aforementioned studies have helped to enlighten the heterogeneity of astrocytes in humans and mouse models of AD pathology, further investigation is required to fully unravel the puzzle behind the functions of the various astrocytic states observed in AD.
The evidence for the involvement of both diverse microglial and astrocytic states in the etiopathology of AD has been outlined above. In situ, microglia and astrocytes, as well as other cell types found throughout the CNS, exist in a pool of cellular signals, constantly exchanging information notably regarding the micro-environmental status (Fakhoury, 2018; Jha et al., 2019). Every step the field takes toward understanding these cellular and molecular interactions is another step closer to fully understanding the pathogenesis of AD. Within the broad categories of “microglia” and “astrocytes” lays even further complexity with recent studies demonstrating distinct states of both microglia (Bisht et al., 2016b; Keren-Shaul et al., 2017; Krasemann et al., 2017; Rangaraju et al., 2018; Hashemiaghdam and Mroczek, 2020; Nguyen et al., 2020; Xu et al., 2021; Yang et al., 2021) and astrocytes (Cunningham et al., 2019; Batiuk et al., 2020; Sofroniew, 2020). Some of the key questions that remain: How do these heterogeneous glial cells interact with each other? How do their interactions participate in the pathogenesis of AD?
The prominent role that both glial cells play in the pathogenesis of neurodegenerative diseases [through mechanisms such as blood-brain-barrier decay, extracellular matrix alterations, and production of ROS, for example (Serrano-Pozo et al., 2013; Acosta et al., 2017; Cragnolini et al., 2019; Gleichman and Carmichael, 2020; Liddelow et al., 2020)] also emphasizes the importance of studying inter-glial communication. This bidirectional communication was demonstrated to occur through released cytokines, metabolites, and neurotransmitters, among other mediators [reviewed in Jha et al. (2019)].
Because of the demonstrated roles of astrocytes and microglia in CNS inflammatory processes (Fakhoury, 2018), a comprehensive analysis of inflammatory signaling between glial cells reveals possible mechanisms for AD pathogenesis. Through the analysis of microglial inflammatory signaling molecules on astrocytes via a number of transgenic murine models, it has been demonstrated that the beneficial and/or harmful effects of astrocytes can be regulated by microglia and their subsequent cytokine secretions [tumor necrotic factor-alpha, complement component c1q and IL-1 alpha, for instance; (Liddelow et al., 2017; Shinozaki et al., 2017; Jha et al., 2019; Kim and Son, 2021)]. However, this relationship is not unidirectional, rather, the various microglial states and functions are heavily influenced by astrocytic signaling—particularly through plasminogen activator inhibitor-1 (Jeon et al., 2012), orosomucoid-2 (Jo et al., 2017), lipocalin-1 (Jha et al., 2015, 2019), pentraxin related protein-3 (Jeon et al., 2010), AQP4 (Smith et al., 2019) and trophic factors (Rocha et al., 2012; Norden et al., 2014) to name a few. For example, depletion of astrocytic lipid-binding protein, APOE, was shown to decrease tau-associated neurodegeneration and decrease microglial phagocytosis (Wang et al., 2021), and research is frequently emphasizing the role of APOE-TREM2 pathways as a means of microglial-astrocyte communication, particularly in modulating microglial homeostasis (Yeh et al., 2016; Krasemann et al., 2017; Taylor et al., 2020; Zhou et al., 2020; Mahan et al., 2022; Smith et al., 2022). Further, it has been demonstrated in 7-month-old APP/TTA (tetracycline transactivator) male mice that microglia-astrocyte communication through the complement system, specifically the complement C3, was involved in Aß pathology (Lian et al., 2016). Inhibition of this pathway, specifically via a signal transduction and activator of transcription-3 (STAT3) inhibitor, can mitigate tau pathology and inflammatory markers in the brain (excluding cerebellum) of PS19 tau transgenic mice (Litvinchuk et al., 2018). Another promising avenue of research pertain to toll-like receptors (TLRs) that many studies have implicated in the pathogenesis of AD (Liu et al., 2012; Li L. et al., 2021). Specifically, there is evidence that extracellular Aß and alpha-synuclein activate and upregulate TLR-2 and TLR-4, subsequently increasing a production of inflammatory mediators which interact with surrounding microglia and neuronal cells (Hughes et al., 2020; Li L. et al., 2021). Overall, TLRs, APOE, and C3 are molecules at the intersection of microglia-astrocyte communication which have demonstrated prominent roles in AD and are worth further investigation.
There is a substantial difficulty in deciphering the independent mechanisms of astrocytes and microglia in the pathogenesis of AD—making the investigation of their interactions and combined effects further complex. As described above, one promising methodology that is currently being utilized to investigate cellular heterogeneity and even to measure potential drug candidates for AD is multidimensional sc/sn-RNA-seq (Hammond et al., 2019; Batiuk et al., 2020; Olah et al., 2020; Xu et al., 2021; Smith et al., 2022). While a myriad of astrocytic and microglial states have been uncovered in recent years, various considerations must be taken into account when interpreting the transcriptomic heterogeneity of glial cells. Indeed, it is still unknown if the many microglial and astrocytic states observed are the results of AD pathology itself or technical artifacts. For instance, visualization of scRNA-seq and its interpretation can be distorted when using t-distributed stochastic neighbor embedding (t-SNE), which does not provide information on cellular heterogeneity based on neighboring cells, but rather on gene set. This difference can result in the visual misrepresentation of dense clusters when interpreting the data (Narayan et al., 2021). In addition, a myriad of technical and biological difficulties in scRNA-seq have been recently reported, from batch effect to incorrectly reported data and differing statistical approaches (Tung et al., 2017; Hicks et al., 2018; Squair et al., 2021), as well as artifacts caused by cell preparation (Nguyen et al., 2018). These variations highlight the challenges encountered when using the scRNA-seq technique to accurately study glial heterogeneity as well as the need to complement sequencing experiments with in situ techniques. In addition, with the expending popularity of this technique, various protocols were established (e.g., Smart-seq, Smart-seq2, MARS-seq) with differing results obtained in terms of gene detections and sensitivity, making it difficult to compare results across transcriptomic studies from different research groups (Ziegenhain et al., 2017). In isolated microglia, it has been shown that enzymatic digestion of the brain to generate single-cell samples affected their gene expression, underlining the sensitivity of this cell type to environmental changes and the need for careful processing (Marsh et al., 2022).
Moreover, further limitations pertaining to the varying states generated from sc- and snRNA-seq have to be taken into account. While snRNA-seq has gained popularity over single-cell due to its possible usage on frozen samples (Lake et al., 2016; Hu et al., 2017; Bakken et al., 2018; Thrupp et al., 2020), it remains controversial if this technique is suitable for glial cell investigation. Some studies have shown similar transcriptomic results in 2-month-old C57BL/6 male mice and in the superior frontal gyrus of human post-mortem brains when comparing fresh vs frozen samples as well as sc- vs sn- techniques (Gerrits et al., 2020). However, others have shown that snRNA-seq did not properly detect microglia in the temporal cortices of human brains (Thrupp et al., 2020), which would provide a possible explanation for the lack of translation regarding the mouse glial states (e.g., DAM) in human samples. Another reason behind the lack of gene conservation measured across species could arise from the cells themselves as differences between the transcriptomes of human and mouse astrocytes, for instance (e.g., responses to oxidative stress and hypoxia, metabolic differences), were reported (Li J. et al., 2021).
The abundant research described throughout this review, focusing on heterogeneity within the field of glia, does not paint a simple picture of isolated roles, singular functions, and straightforward interactions. Instead, with every new discovery attributing a discrete function to certain cytokines, lipoproteins, enzymes, or receptors, or with the imaging of a new functional state for each glial cell, the whole picture becomes further complex. The exact role of each cellular states is still in discovery, with their location, time-point, genes, and macro- and micro-environment all appearing as critical variables in determining cellular expression. As further research is conducted, the specialization of our analyses (e.g., pertaining to receptor expression, mitochondrial ultrastructure, cytoplasmic density, cytokine release, for example) of these cells will become increasingly important to fully understand the function and involvement in disease-states. Further, as techniques in the fields of imaging and -omics continue to develop, it is likely that the number of distinct categories will grow further as well. As described above, even within a particular glial state, such as DAM (Keren-Shaul et al., 2017; Rangaraju et al., 2018; Hu et al., 2021), there is immense diversity observed. Therefore, integrating different experimental approaches into mutual understanding is essential to the future of this field. With all this in mind, glial heterogeneity opens the idea that, while they may be categorized in simple terms, microglia and astrocytes account for a large number of distinct cell types and functional states and that these cellular states may act in accordance with each other but also, perhaps, in opposition. These various possibilities are important to keep in mind as these cellular states are investigated in health and in neurological disorders such as AD. As both the fields of microglia and astrocytes continue their respective progress, communication between these fields and concurrent research of glial communication is critical to providing discoveries in the search for therapeutic novelty in AD. Lastly, there is always a need for further examination in humans as those findings will inform preclinical experiments and help overcome translational barriers in AD research.
M-KS-P, SL, and JV wrote the manuscript. M-KS-P and M-ÈT conceptualized and edited the review. M-KS-P designed the figure. All authors approved the submitted manuscript.
M-KS-P was supported by doctoral training awards from the Canadian Institutes of Health Research (CIHR) and Fonds de recherche du Québec—Santé. JV holds a CIHR master’s training award. M-ÈT holds a Canada Research Chair (Tier 2) in Neurobiology of Aging and Cognition.
We would like to acknowledge that the University of Victoria is located on the traditional territory of the lək̓ʷəŋən people and that the Songhees, Esquimalt, and WSÁNEĆ people have historical relationships to this land.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acosta, C., Anderson, H. D., and Anderson, C. M. (2017). Astrocyte dysfunction in Alzheimer disease. J. Neurosci. Res. 95, 2430–2447. doi: 10.1002/jnr.24075
Akiyama, H., Kawamata, T., Yamada, T., Tooyama, I., Ishii, T., and McGeer, P. L. (1993). Expression of intercellular adhesion molecule (ICAM)-1 by a subset of astrocytes in Alzheimer disease and some other degenerative neurological disorders. Acta Neuropathol. 85, 628–634. doi: 10.1007/BF00334673
Allen, B., Ingram, E., Takao, M., Smith, M. J., Jakes, R., Virdee, K., et al. (2002). Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J. Neurosci. 22, 9340–9351. doi: 10.1523/JNEUROSCI.22-21-09340.2002
Andjelkovic, A. V., Nikolic, B., Pachter, J. S., and Zecevic, N. (1998). Macrophages/microglial cells in human central nervous system during development: an immunohistochemical study. Brain Res. 814, 13–25. doi: 10.1016/s0006-8993(98)00830-0
Bachstetter, A. D., Van Eldik, L. J., Schmitt, F. A., Neltner, J. H., Ighodaro, E. T., Webster, S. J., et al. (2015). Disease-related microglia heterogeneity in the hippocampus of Alzheimer’s disease, dementia with Lewy bodies, and hippocampal sclerosis of aging. Acta Neuropathol. Commun. 3:32. doi: 10.1186/s40478-015-0209-z
Bakken, T. E., Hodge, R. D., Miller, J. A., Yao, Z., Nguyen, T. N., Aevermann, B., et al. (2018). Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 13:e0209648. doi: 10.1371/journal.pone.0209648
Baloyannis, S. J. (2019). ““Mitochondria and Alzheimer’s Disease: An Electron Microscopy Study,”,” in Redirecting Alzheimer Strategy, ed. D. Larrivee (Rijeka: IntechOpen), doi: 10.5772/intechopen.84881
Batiuk, M. Y., Martirosyan, A., Wahis, J., de Vin, F., Marneffe, C., Kusserow, C., et al. (2020). Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11:1220. doi: 10.1038/s41467-019-14198-8
Bellucci, A., Westwood, A. J., Ingram, E., Casamenti, F., Goedert, M., and Spillantini, M. G. (2004). Induction of inflammatory mediators and microglial activation in mice transgenic for mutant human P301S tau protein. Am. J. Pathol. 165, 1643–1652. doi: 10.1016/S0002-9440(10)63421-9
Bertram, L., and Tanzi, R. E. (2009). Genome-wide association studies in Alzheimer’s disease. Hum. Mol. Genet. 18, R137–R145. doi: 10.1093/hmg/ddp406
Bisht, K., Sharma, K. P., Lecours, C., Gabriela Sánchez, M., El Hajj, H., Milior, G., et al. (2016b). Dark microglia: a new phenotype predominantly associated with pathological states. Glia 64, 826–839. doi: 10.1002/glia.22966
Bisht, K., Sharma, K., Lacoste, B., and Tremblay, M. -È (2016a). Dark microglia: why are they dark? Commun. Integr. Biol. 9:e1230575. doi: 10.1080/19420889.2016.1230575
Boche, D., and Gordon, M. N. (2021). Diversity of transcriptomic microglial phenotypes in aging and Alzheimer’s disease. Alzheimers Dement. 18, 360–376. doi: 10.1002/alz.12389
Casali, B. T., MacPherson, K. P., Reed-Geaghan, E. G., and Landreth, G. E. (2020). Microglia depletion rapidly and reversibly alters amyloid pathology by modification of plaque compaction and morphologies. Neurobiol. Dis. 142:104956. doi: 10.1016/j.nbd.2020.104956
Chakravorty, A., Jetto, C. T., and Manjithaya, R. (2019). Dysfunctional Mitochondria and Mitophagy as Drivers of Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 11:311. doi: 10.3389/fnagi.2019.00311
Chishti, M. A., Yang, D. S., Janus, C., Phinney, A. L., Horne, P., Pearson, J., et al. (2001). Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J. Biol. Chem. 276, 21562–21570. doi: 10.1074/jbc.M100710200
Clarke, B. E., Taha, D. M., Tyzack, G. E., and Patani, R. (2021). Regionally encoded functional heterogeneity of astrocytes in health and disease: a perspective. Glia 69, 20–27. doi: 10.1002/glia.23877
Clayton, K., Delpech, J. C., Herron, S., Iwahara, N., Ericsson, M., Saito, T., et al. (2021). Plaque associated microglia hyper-secrete extracellular vesicles and accelerate tau propagation in a humanized APP mouse model. Mol. Neurodegener. 16:18. doi: 10.1186/s13024-021-00440-9
Cohen, R. M., Rezai-Zadeh, K., Weitz, T. M., Rentsendorj, A., Gate, D., Spivak, I., et al. (2013). A transgenic Alzheimer rat with plaques, tau pathology, behavioral impairment, oligomeric aβ, and frank neuronal loss. J. Neurosci. 33, 6245–6256. doi: 10.1523/JNEUROSCI.3672-12.2013
Cragnolini, A. B., Lampitella, G., Virtuoso, A., Viscovo, I., Panetsos, F., Papa, M., et al. (2019). Regional brain susceptibility to neurodegeneration: what is the role of glial cells? Neural Regen. Res. 15, 838–842. doi: 10.4103/1673-5374.268897
Cunningham, C., Dunne, A., and Lopez-Rodriguez, A. B. (2019). Astrocytes: heterogeneous and Dynamic Phenotypes in Neurodegeneration and Innate Immunity. Neuroscientist 25, 455–474. doi: 10.1177/1073858418809941
Davalos, D., Grutzendler, J., Yang, G., Kim, J. V., Zuo, Y., Jung, S., et al. (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758. doi: 10.1038/nn1472
Davis, N., Mota, B. C., Stead, L., Palmer, E. O. C., Lombardero, L., Rodríguez-Puertas, R., et al. (2021). Pharmacological ablation of astrocytes reduces Aβ degradation and synaptic connectivity in an ex vivo model of Alzheimer’s disease. J. Neuroinflamm. 18:73. doi: 10.1186/s12974-021-02117-y
Delizannis, A. T., Nonneman, A., Tsering, W., De Bondt, A., Van den Wyngaert, I., Zhang, B., et al. (2021). Effects of microglial depletion and TREM2 deficiency on Aβ plaque burden and neuritic plaque tau pathology in 5XFAD mice. Acta Neuropathol. Commun. 9:150. doi: 10.1186/s40478-021-01251-1
DeTure, M. A., and Dickson, D. W. (2019). The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 14:32. doi: 10.1186/s13024-019-0333-5
Diniz, L. P., Tortelli, V., Matias, I., Morgado, J., Araujo, A. P. B., Melo, H. M., et al. (2017). Astrocyte Transforming Growth Factor Beta 1 Protects Synapses against Aβ Oligomers in Alzheimer’s Disease Model. J. Neurosci. 37, 6797–6809. doi: 10.1523/JNEUROSCI.3351-16.2017
Dyne, E., Cawood, M., Suzelis, M., Russell, R., and Kim, M.-H. (2021). Ultrastructural analysis of the morphological phenotypes of microglia associated with neuroinflammatory cues. J. Comp. Neurol. 530, 1263–1275. doi: 10.1002/cne.25274
El Hajj, H., Savage, J. C., Bisht, K., Parent, M., Vallières, L., Rivest, S., et al. (2019). Ultrastructural evidence of microglial heterogeneity in Alzheimer’s disease amyloid pathology. J. Neuroinflamm. 16:87. doi: 10.1186/s12974-019-1473-9
Escartin, C., Galea, E., Lakatos, A., O’Callaghan, J. P., Petzold, G. C., Serrano-Pozo, A., et al. (2021). Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325. doi: 10.1038/s41593-020-00783-4
Fakhoury, M. (2018). Microglia and Astrocytes in Alzheimer’s Disease: implications for Therapy. Curr. Neuropharmacol. 16, 508–518. doi: 10.2174/1570159X15666170720095240
Gerrits, E., Brouwer, N., Kooistra, S. M., Woodbury, M. E., Vermeiren, Y., Lambourne, M., et al. (2021). Distinct amyloid-β and tau-associated microglia profiles in Alzheimer’s disease. Acta Neuropathol. 141, 681–696. doi: 10.1007/s00401-021-02263-w
Gerrits, E., Heng, Y., Boddeke, E. W. G. M., and Eggen, B. J. L. (2020). Transcriptional profiling of microglia; current state of the art and future perspectives. Glia 68, 740–755. doi: 10.1002/glia.23767
Ghadery, C., Best, L. A., Pavese, N., Tai, Y. F., and Strafella, A. P. (2019). PET Evaluation of Microglial Activation in Non-neurodegenerative Brain Diseases. Curr. Neurol. Neurosci. Rep. 19:38. doi: 10.1007/s11910-019-0951-x
Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. doi: 10.1126/science.1194637
Giulian, D. (1987). Ameboid microglia as effectors of inflammation in the central nervous system. J. Neurosci. Res. 18, 132–133. doi: 10.1002/jnr.490180123
Gleichman, A. J., and Carmichael, S. T. (2020). Glia in neurodegeneration: drivers of disease or along for the ride? Neurobiol. Dis. 142:104957. doi: 10.1016/j.nbd.2020.104957
Gomez-Arboledas, A., Davila, J. C., Sanchez-Mejias, E., Navarro, V., Nuñez-Diaz, C., Sanchez-Varo, R., et al. (2018). Phagocytic clearance of presynaptic dystrophies by reactive astrocytes in Alzheimer’s disease. Glia 66, 637–653. doi: 10.1002/glia.23270
Govindpani, K., McNamara, L. G., Smith, N. R., Vinnakota, C., Waldvogel, H. J., Faull, R. L., et al. (2019). Vascular Dysfunction in Alzheimer’s Disease: a Prelude to the Pathological Process or a Consequence of It? J. Clin. Med. 8:651. doi: 10.3390/jcm8050651
Grubman, A., Choo, X. Y., Chew, G., Ouyang, J. F., Sun, G., Croft, N. P., et al. (2021). Transcriptional signature in microglia associated with Aβ plaque phagocytosis. Nat. Commun. 12:3015. doi: 10.1038/s41467-021-23111-1
Guerrero, A., De Strooper, B., and Arancibia-Cárcamo, I. L. (2021). Cellular senescence at the crossroads of inflammation and Alzheimer’s disease. Trends Neurosci. 44, 714–727. doi: 10.1016/j.tins.2021.06.007
Gyoneva, S., Swanger, S. A., Zhang, J., Weinshenker, D., and Traynelis, S. F. (2016). Altered motility of plaque-associated microglia in a model of Alzheimer’s disease. Neuroscience 330, 410–420. doi: 10.1016/j.neuroscience.2016.05.061
Habib, N., McCabe, C., Medina, S., Varshavsky, M., Kitsberg, D., Dvir-Szternfeld, R., et al. (2020). Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706. doi: 10.1038/s41593-020-0624-8
Halliday, G. (2017). Pathology and hippocampal atrophy in Alzheimer’s disease. Lancet Neurol. 16, 862–864. doi: 10.1016/S1474-4422(17)30343-5
Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e6. doi: 10.1016/j.immuni.2018.11.004
Hardy, J. A., and Higgins, G. A. (1992). Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185. doi: 10.1126/science.1566067
Hashemiaghdam, A., and Mroczek, M. (2020). Microglia heterogeneity and neurodegeneration: the emerging paradigm of the role of immunity in Alzheimer’s disease. J. Neuroimmunol. 341:577185. doi: 10.1016/j.jneuroim.2020.577185
Hickman, S. E., Allison, E. K., and Khoury, J. E. (2008). Microglial Dysfunction and Defective β-Amyloid Clearance Pathways in Aging Alzheimer’s Disease Mice. J. Neurosci. 28:8354. doi: 10.1523/JNEUROSCI.0616-08.2008
Hicks, S. C., Townes, F. W., Teng, M., and Irizarry, R. A. (2018). Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 19, 562–578. doi: 10.1093/biostatistics/kxx053
Hu, P., Fabyanic, E., Kwon, D., Tang, S., Zhou, Z., and Wu, H. (2017). Dissecting cell-type composition and activity-dependent transcriptional state in mammalian brains by massively parallel single-nucleus RNA-Seq. Mol. Cell 68, 1006-1015.e7. doi: 10.1016/j.molcel.2017.11.017
Hu, Y., Fryatt, G. L., Ghorbani, M., Obst, J., Menassa, D. A., Martin-Estebane, M., et al. (2021). Replicative senescence dictates the emergence of disease-associated microglia and contributes to Aβ pathology. Cell Rep. 35:109228. doi: 10.1016/j.celrep.2021.109228
Hughes, C., Choi, M. L., Yi, J.-H., Kim, S.-C., Drews, A., George-Hyslop, P. S., et al. (2020). Beta amyloid aggregates induce sensitised TLR4 signalling causing long-term potentiation deficit and rat neuronal cell death. Commun. Biol. 3:79. doi: 10.1038/s42003-020-0792-9
Jack, C. R., Bennett, D. A., Blennow, K., Carrillo, M. C., Feldman, H. H., Frisoni, G. B., et al. (2016). A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 87, 539–547. doi: 10.1212/WNL.0000000000002923
Jankowsky, J. L., Fadale, D. J., Anderson, J., Xu, G. M., Gonzales, V., Jenkins, N. A., et al. (2004). Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 13, 159–170. doi: 10.1093/hmg/ddh019
Jankowsky, J. L., and Zheng, H. (2017). Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol. Neurodegener. 12:89. doi: 10.1186/s13024-017-0231-7
Jeon, H., Kim, J.-H., Kim, J.-H., Lee, W.-H., Lee, M.-S., and Suk, K. (2012). Plasminogen activator inhibitor type 1 regulates microglial motility and phagocytic activity. J. Neuroinflamm. 9:149. doi: 10.1186/1742-2094-9-149
Jeon, H., Lee, S., Lee, W.-H., and Suk, K. (2010). Analysis of glial secretome: the long pentraxin PTX3 modulates phagocytic activity of microglia. J. Neuroimmunol. 229, 63–72. doi: 10.1016/j.jneuroim.2010.07.001
Jha, M. K., Jo, M., Kim, J.-H., and Suk, K. (2019). Microglia-Astrocyte Crosstalk: an Intimate Molecular Conversation. Neuroscientist 25, 227–240. doi: 10.1177/1073858418783959
Jha, M. K., Lee, S., Park, D. H., Kook, H., Park, K.-G., Lee, I.-K., et al. (2015). Diverse functional roles of lipocalin-2 in the central nervous system. Neurosci. Biobehav. Rev. 49, 135–156. doi: 10.1016/j.neubiorev.2014.12.006
Jo, M., Kim, J.-H., Song, G. J., Seo, M., Hwang, E. M., and Suk, K. (2017). Astrocytic Orosomucoid-2 Modulates Microglial Activation and Neuroinflammation. J. Neurosci. 37, 2878–2894. doi: 10.1523/JNEUROSCI.2534-16.2017
John Lin, C.-C., Yu, K., Hatcher, A., Huang, T.-W., Lee, H. K., Carlson, J., et al. (2017). Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405. doi: 10.1038/nn.4493
Katsouri, L., Birch, A. M., Renziehausen, A. W. J., Zach, C., Aman, Y., Steeds, H., et al. (2020). Ablation of reactive astrocytes exacerbates disease pathology in a model of Alzheimer’s disease. Glia 68, 1017–1030. doi: 10.1002/glia.23759
Kaur, D., Sharma, V., and Deshmukh, R. (2019). Activation of microglia and astrocytes: a roadway to neuroinflammation and Alzheimer’s disease. Inflammopharmacology 27, 663–677. doi: 10.1007/s10787-019-00580-x
Keren-Shaul, H., Spinrad, A., Weiner, A., Matcovitch-Natan, O., Dvir-Szternfeld, R., Ulland, T. K., et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276-1290.e17. doi: 10.1016/j.cell.2017.05.018
Kim, S., and Son, Y. (2021). Astrocytes Stimulate Microglial Proliferation and M2 Polarization In Vitro through Crosstalk between Astrocytes and Microglia. Int. J. Mol. Sci. 22:8800. doi: 10.3390/ijms22168800
Krasemann, S., Madore, C., Cialic, R., Baufeld, C., Calcagno, N., El Fatimy, R., et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566.e–581.e. doi: 10.1016/j.immuni.2017.08.008
Kulijewicz-Nawrot, M., Verkhratsky, A., Chvátal, A., Syková, E., and Rodríguez, J. J. (2012). Astrocytic cytoskeletal atrophy in the medial prefrontal cortex of a triple transgenic mouse model of Alzheimer’s disease. J. Anatomy 221, 252–262. doi: 10.1111/j.1469-7580.2012.01536.x
Lake, B. B., Ai, R., Kaeser, G. E., Salathia, N. S., Yung, Y. C., Liu, R., et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. doi: 10.1126/science.aaf1204
Lana, D., Ugolini, F., and Giovannini, M. G. (2020). Space-Dependent Glia-Neuron Interplay in the Hippocampus of Transgenic Models of β-Amyloid Deposition. Int. J. Mol. Sci. 21:9441. doi: 10.3390/ijms21249441
Latimer, C. S., Shively, C. A., Keene, C. D., Jorgensen, M. J., Andrews, R. N., Register, T. C., et al. (2019). A nonhuman primate model of early Alzheimer’s disease pathologic change: implications for disease pathogenesis. Alzheimer’s Dement. 15, 93–105. doi: 10.1016/j.jalz.2018.06.3057
Lee, T.-S., Li, A. Y., Rapuano, A., Mantis, J., Eid, T., Seyfried, T. N., et al. (2021). Gene expression in the epileptic (EL) mouse hippocampus. Neurobiol. Dis. 147:105152. doi: 10.1016/j.nbd.2020.105152
Leng, K., Li, E., Eser, R., Piergies, A., Sit, R., Tan, M., et al. (2021). Molecular characterization of selectively vulnerable neurons in Alzheimer’s Disease. Nat. Neurosci. 24, 276–287. doi: 10.1038/s41593-020-00764-7
Lewis, J., McGowan, E., Rockwood, J., Melrose, H., Nacharaju, P., Van Slegtenhorst, M., et al. (2000). Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 25, 402–405. doi: 10.1038/78078
Leyh, J., Paeschke, S., Mages, B., Michalski, D., Nowicki, M., Bechmann, I., et al. (2021). Classification of Microglial Morphological Phenotypes Using Machine Learning. Front. Cell. Neurosci. 15:241. doi: 10.3389/fncel.2021.701673
Li, H.-W., Zhang, L., and Qin, C. (2019). Current state of research on non-human primate models of Alzheimer’s disease. Anim. Models Exp. Med. 2, 227–238. doi: 10.1002/ame2.12092
Li, J., Pan, L., Pembroke, W. G., Rexach, J. E., Godoy, M. I., Condro, M. C., et al. (2021). Conservation and divergence of vulnerability and responses to stressors between human and mouse astrocytes. Nat. Commun. 12:3958. doi: 10.1038/s41467-021-24232-3
Li, L., Acioglu, C., Heary, R. F., and Elkabes, S. (2021). Role of astroglial toll-like receptors (TLRs) in central nervous system infections, injury and neurodegenerative diseases. Brain Behav. Immun. 91, 740–755. doi: 10.1016/j.bbi.2020.10.007
Li, K.-Y., Gong, P.-F., Li, J.-T., Xu, N.-J., and Qin, S. (2020). Morphological and molecular alterations of reactive astrocytes without proliferation in cerebral cortex of an APP/PS1 transgenic mouse model and Alzheimer’s patients. Glia 68, 2361–2376. doi: 10.1002/glia.23845
Lian, H., Litvinchuk, A., Chiang, A. C.-A., Aithmitti, N., Jankowsky, J. L., and Zheng, H. (2016). Astrocyte-Microglia Cross Talk through Complement Activation Modulates Amyloid Pathology in Mouse Models of Alzheimer’s Disease. J. Neurosci. 36, 577–589. doi: 10.1523/JNEUROSCI.2117-15.2016
Liddelow, S. A., Guttenplan, K. A., Clarke, L. E., Bennett, F. C., Bohlen, C. J., Schirmer, L., et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. doi: 10.1038/nature21029
Liddelow, S. A., Marsh, S. E., and Stevens, B. (2020). Microglia and Astrocytes in Disease: dynamic Duo or Partners in Crime? Trends Immunol. 41, 820–835. doi: 10.1016/j.it.2020.07.006
Lin, W.-L., Lewis, J., Yen, S.-H., Hutton, M., and Dickson, D. W. (2003). Filamentous Tau in Oligodendrocytes and Astrocytes of Transgenic Mice Expressing the Human Tau Isoform with the P301L Mutation. Am. J. Pathol. 162, 213–218. doi: 10.1016/S0002-9440(10)63812-6
Litvinchuk, A., Wan, Y.-W., Swartzlander, D. B., Chen, F., Cole, A., Propson, N. E., et al. (2018). Complement C3aR Inactivation Attenuates Tau Pathology and Reverses an Immune Network Deregulated in Tauopathy Models and Alzheimer’s Disease. Neuron 100, 1337.e–1353.e. doi: 10.1016/j.neuron.2018.10.031
Liu, C.-Y., Yang, Y., Ju, W.-N., Wang, X., and Zhang, H.-L. (2018). Emerging Roles of Astrocytes in Neuro-Vascular Unit and the Tripartite Synapse With Emphasis on Reactive Gliosis in the Context of Alzheimer’s Disease. Front. Cell Neurosci. 12:193. doi: 10.3389/fncel.2018.00193
Liu, S., Liu, Y., Hao, W., Wolf, L., Kiliaan, A. J., Penke, B., et al. (2012). TLR2 is a primary receptor for Alzheimer’s amyloid β peptide to trigger neuroinflammatory activation. J. Immunol. 188, 1098–1107. doi: 10.4049/jimmunol.1101121
Lodder, C., Scheyltjens, I., Stancu, I. C., Botella Lucena, P., Gutiérrez, de Ravé, M., et al. (2021). CSF1R inhibition rescues tau pathology and neurodegeneration in an A/T/N model with combined AD pathologies, while preserving plaque associated microglia. Acta Neuropathol. Commun. 9:108. doi: 10.1186/s40478-021-01204-8
Lopes, K. O., Sparks, D. L., and Streit, W. J. (2008). Microglial dystrophy in the aged and Alzheimer’s disease brain is associated with ferritin immunoreactivity. Glia 56, 1048–1060. doi: 10.1002/glia.20678
Lord, A., Kalimo, H., Eckman, C., Zhang, X.-Q., Lannfelt, L., and Nilsson, L. N. G. (2006). The Arctic Alzheimer mutation facilitates early intraneuronal Abeta aggregation and senile plaque formation in transgenic mice. Neurobiol. Aging 27, 67–77. doi: 10.1016/j.neurobiolaging.2004.12.007
Lu, S.-M., Tremblay, M. -È, King, I. L., Qi, J., Reynolds, H. M., Marker, D. F., et al. (2011). HIV-1 Tat-induced microgliosis and synaptic damage via interactions between peripheral and central myeloid cells. PLoS One 6:e23915. doi: 10.1371/journal.pone.0023915
Maeda, J., Minamihisamatsu, T., Shimojo, M., Zhou, X., Ono, M., Matsuba, Y., et al. (2021). Distinct microglial response against Alzheimer’s amyloid and tau pathologies characterized by P2Y12 receptor. Brain Commun. 3:fcab011. doi: 10.1093/braincomms/fcab011
Mahan, T. E., Wang, C., Bao, X., Choudhury, A., Ulrich, J. D., and Holtzman, D. M. (2022). Selective reduction of astrocyte apoE3 and apoE4 strongly reduces Aβ accumulation and plaque-related pathology in a mouse model of amyloidosis. Mol. Neurodegener. 17:13. doi: 10.1186/s13024-022-00516-0
Mampay, M., Velasco-Estevez, M., Rolle, S. O., Chaney, A. M., Boutin, H., Dev, K. K., et al. (2020). Spatiotemporal immunolocalisation of REST in the brain of healthy ageing and Alzheimer’s disease rats. FEBS Open Bio. 11, 146–163. doi: 10.1002/2211-5463.13036
Manchikalapudi, A. L., Chilakala, R. R., Kalia, K., and Sunkaria, A. (2019). Evaluating the Role of Microglial Cells in Clearance of Aβ from Alzheimer’s Brain. ACS Chem. Neurosci. 10, 1149–1156. doi: 10.1021/acschemneuro.8b00627
Marino, S., Bonanno, L., Lo Buono, V., Ciurleo, R., Corallo, F., Morabito, R., et al. (2019). Longitudinal analysis of brain atrophy in Alzheimer’s disease and frontotemporal dementia. J. Int. Med. Res. 47, 5019–5027. doi: 10.1177/0300060519830830
Marlatt, M. W., Bauer, J., Aronica, E., van Haastert, E. S., Hoozemans, J. J. M., Joels, M., et al. (2014). Proliferation in the Alzheimer hippocampus is due to microglia, not astroglia, and occurs at sites of amyloid deposition. Neural Plast. 2014:693851. doi: 10.1155/2014/693851
Marschallinger, J., Iram, T., Zardeneta, M., Lee, S. E., Lehallier, B., Haney, M. S., et al. (2020). Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208. doi: 10.1038/s41593-019-0566-1
Marsh, S. E., Walker, A. J., Kamath, T., Dissing-Olesen, L., Hammond, T. R., de Soysa, T. Y., et al. (2022). Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 25, 306–316. doi: 10.1038/s41593-022-01022-8
Mathys, H., Davila-Velderrain, J., Peng, Z., Gao, F., Mohammadi, S., Young, J. Z., et al. (2019). Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337. doi: 10.1038/s41586-019-1195-2
Matias, I., Morgado, J., and Gomes, F. C. A. (2019). Astrocyte Heterogeneity: impact to Brain Aging and Disease. Front. Aging Neurosci. 11:59. doi: 10.3389/fnagi.2019.00059
McFarland, K. N., Ceballos, C., Rosario, A., Ladd, T., Moore, B., Golde, G., et al. (2021). Microglia show differential transcriptomic response to Aβ peptide aggregates ex vivo and in vivo. Life Sci. Alliance 4:e202101108. doi: 10.26508/lsa.202101108
Miller, S. J. (2018). Astrocyte Heterogeneity in the Adult Central Nervous System. Front. Cell Neurosci. 12:401. doi: 10.3389/fncel.2018.00401
Monier, A., Evrard, P., Gressens, P., and Verney, C. (2006). Distribution and differentiation of microglia in the human encephalon during the first two trimesters of gestation. J. Comp. Neurol. 499, 565–582. doi: 10.1002/cne.21123
Morabito, S., Miyoshi, E., Michael, N., Shahin, S., Martini, A. C., Head, E., et al. (2021). Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat. Genet. 53, 1143–1155. doi: 10.1038/s41588-021-00894-z
Mrdjen, D., Fox, E. J., Bukhari, S. A., Montine, K. S., Bendall, S. C., and Montine, T. J. (2019). The basis of cellular and regional vulnerability in Alzheimer’s disease. Acta Neuropathol. 138, 729–749. doi: 10.1007/s00401-019-02054-4
Muñoz-Castro, C., Noori, A., Magdamo, C. G., Li, Z., Marks, J. D., Frosch, M. P., et al. (2022). Cyclic multiplex fluorescent immunohistochemistry and machine learning reveal distinct states of astrocytes and microglia in normal aging and Alzheimer’s disease. J. Neuroinflamm. 19:30. doi: 10.1186/s12974-022-02383-4
Nagele, R. G., Wegiel, J., Venkataraman, V., Imaki, H., Wang, K.-C., and Wegiel, J. (2004). Contribution of glial cells to the development of amyloid plaques in Alzheimer’s disease. Neurobiol. Aging 25, 663–674. doi: 10.1016/j.neurobiolaging.2004.01.007
Narayan, A., Berger, B., and Cho, H. (2021). Assessing single-cell transcriptomic variability through density-preserving data visualization. Nat. Biotechnol. 39, 765–774. doi: 10.1038/s41587-020-00801-7
Narayanaswami, V., Dahl, K., Bernard-Gauthier, V., Josephson, L., Cumming, P., and Vasdev, N. (2018). Emerging PET Radiotracers and Targets for Imaging of Neuroinflammation in Neurodegenerative Diseases: outlook Beyond TSPO. Mol. Imaging 17:1536012118792317. doi: 10.1177/1536012118792317
Natunen, T., Martiskainen, H., Marttinen, M., Gabbouj, S., Koivisto, H., Kemppainen, S., et al. (2020). Diabetic phenotype in mouse and humans reduces the number of microglia around β-amyloid plaques. Mol. Neurodegener. 15:66. doi: 10.1186/s13024-020-00415-2
Nguyen, A. T., Wang, K., Hu, G., Wang, X., Miao, Z., Azevedo, J. A., et al. (2020). APOE and TREM2 regulate amyloid-responsive microglia in Alzheimer’s disease. Acta Neuropathol. 140, 477–493. doi: 10.1007/s00401-020-02200-3
Nguyen, Q. H., Pervolarakis, N., Nee, K., and Kessenbrock, K. (2018). Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front. Cell Dev. Biol. 6:108. doi: 10.3389/fcell.2018.00108
Nimmerjahn, A., Kirchhoff, F., and Helmchen, F. (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. doi: 10.1126/science.1110647
Norden, D. M., Fenn, A. M., Dugan, A., and Godbout, J. P. (2014). TGFβ produced by IL-10 redirected astrocytes attenuates microglial activation. Glia 62, 881–895. doi: 10.1002/glia.22647
Oakley, H., Cole, S. L., Logan, S., Maus, E., Shao, P., Craft, J., et al. (2006). Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006
Oddo, S., Caccamo, A., Shepherd, J. D., Murphy, M. P., Golde, T. E., Kayed, R., et al. (2003). Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron 39, 409–421. doi: 10.1016/s0896-6273(03)00434-3
Ohm, D. T., Fought, A. J., Martersteck, A., Coventry, C., Sridhar, J., Gefen, T., et al. (2020). Accumulation of neurofibrillary tangles and activated microglia is associated with lower neuron densities in the aphasic variant of Alzheimer’s disease. Brain Pathol. 31, 189–204. doi: 10.1111/bpa.12902
Olabarria, M., Noristani, H. N., Verkhratsky, A., and Rodríguez, J. J. (2010). Concomitant astroglial atrophy and astrogliosis in a triple transgenic animal model of Alzheimer’s disease. Glia 58, 831–838. doi: 10.1002/glia.20967
Olah, M., Menon, V., Habib, N., Taga, M. F., Ma, Y., Yung, C. J., et al. (2020). Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 11:6129. doi: 10.1038/s41467-020-19737-2
Orre, M., Kamphuis, W., Osborn, L. M., Jansen, A. H. P., Kooijman, L., Bossers, K., et al. (2014). Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol. Aging 35, 2746–2760. doi: 10.1016/j.neurobiolaging.2014.06.004
Owen, D. R., Narayan, N., Wells, L., Healy, L., Smyth, E., Rabiner, E. A., et al. (2017). Pro-inflammatory activation of primary microglia and macrophages increases 18 kDa translocator protein expression in rodents but not humans. J. Cereb. Blood Flow Metab. 37, 2679–2690. doi: 10.1177/0271678X17710182
Paasila, P. J., Davies, D. S., Kril, J. J., Goldsbury, C., and Sutherland, G. T. (2019). The relationship between the morphological subtypes of microglia and Alzheimer’s disease neuropathology. Brain Pathol. 29, 726–740. doi: 10.1111/bpa.12717
Paolicelli, R. C., Bolasco, G., Pagani, F., Maggi, L., Scianni, M., Panzanelli, P., et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. doi: 10.1126/science.1202529
Parakalan, R., Jiang, B., Nimmi, B., Janani, M., Jayapal, M., Lu, J., et al. (2012). Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC Neurosci. 13:64. doi: 10.1186/1471-2202-13-64
Pellerin, L., Bouzier-Sore, A.-K., Aubert, A., Serres, S., Merle, M., Costalat, R., et al. (2007). Activity-dependent regulation of energy metabolism by astrocytes: an update. Glia 55, 1251–1262. doi: 10.1002/glia.20528
Pini, L., Pievani, M., Bocchetta, M., Altomare, D., Bosco, P., Cavedo, E., et al. (2016). Brain atrophy in Alzheimer’s Disease and aging. Ageing Res. Rev. 30, 25–48. doi: 10.1016/j.arr.2016.01.002
Plescher, M., Seifert, G., Hansen, J. N., Bedner, P., Steinhäuser, C., and Halle, A. (2018). Plaque-dependent morphological and electrophysiological heterogeneity of microglia in an Alzheimer’s disease mouse model. Glia 66, 1464–1480. doi: 10.1002/glia.23318
Prokop, S., Miller, K. R., and Heppner, F. L. (2013). Microglia actions in Alzheimer’s disease. Acta Neuropathol. 126, 461–477. doi: 10.1007/s00401-013-1182-x
Radde, R., Bolmont, T., Kaeser, S. A., Coomaraswamy, J., Lindau, D., Stoltze, L., et al. (2006). Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7, 940–946. doi: 10.1038/sj.embor.7400784
Ramsden, M., Kotilinek, L., Forster, C., Paulson, J., McGowan, E., SantaCruz, K., et al. (2005). Age-dependent neurofibrillary tangle formation, neuron loss, and memory impairment in a mouse model of human tauopathy (P301L). J. Neurosci. 25, 10637–10647. doi: 10.1523/JNEUROSCI.3279-05.2005
Rangaraju, S., Dammer, E. B., Raza, S. A., Rathakrishnan, P., Xiao, H., Gao, T., et al. (2018). Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol. Neurodegener. 13:24. doi: 10.1186/s13024-018-0254-8
Ricciarelli, R., and Fedele, E. (2017). The Amyloid Cascade Hypothesis in Alzheimer’s Disease: it’s Time to Change Our Mind. Curr. Neuropharmacol. 15, 926–935. doi: 10.2174/1570159X15666170116143743
Richetin, K., Petsophonsakul, P., Roybon, L., Guiard, B. P., and Rampon, C. (2017). Differential alteration of hippocampal function and plasticity in females and males of the APPxPS1 mouse model of Alzheimer’s disease. Neurobiol. Aging 57, 220–231. doi: 10.1016/j.neurobiolaging.2017.05.025
Rocha, S. M., Cristovão, A. C., Campos, F. L., Fonseca, C. P., and Baltazar, G. (2012). Astrocyte-derived GDNF is a potent inhibitor of microglial activation. Neurobiol. Dis. 47, 407–415. doi: 10.1016/j.nbd.2012.04.014
Rodriguez-Callejas, J. D., Fuchs, E., and Perez-Cruz, C. (2020). Increased oxidative stress, hyperphosphorylation of tau, and dystrophic microglia in the hippocampus of aged Tupaia belangeri. Glia 68, 1775–1793. doi: 10.1002/glia.23804
Romero-Molina, C., Navarro, V., Sanchez-Varo, R., Jimenez, S., Fernandez-Valenzuela, J. J., Sanchez-Mico, M. V., et al. (2018). Distinct Microglial Responses in Two Transgenic Murine Models of TAU Pathology. Front. Cell Neurosci. 12:421. doi: 10.3389/fncel.2018.00421
Rothman, S. M., Tanis, K. Q., Gandhi, P., Malkov, V., Marcus, J., Pearson, M., et al. (2018). Human Alzheimer’s disease gene expression signatures and immune profile in APP mouse models: a discrete transcriptomic view of Aβ plaque pathology. J. Neuroinflamm. 15:256. doi: 10.1186/s12974-018-1265-7
Saez-Atienzar, S., and Masliah, E. (2020). Cellular senescence and Alzheimer disease: the egg and the chicken scenario. Nat. Rev. Neurosci. 21, 433–444. doi: 10.1038/s41583-020-0325-z
Saito, T., Matsuba, Y., Mihira, N., Takano, J., Nilsson, P., Itohara, S., et al. (2014). Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci. 17, 661–663. doi: 10.1038/nn.3697
Sala Frigerio, C., Wolfs, L., Fattorelli, N., Thrupp, N., Voytyuk, I., Schmidt, I., et al. (2019). The Major Risk Factors for Alzheimer’s Disease: age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cell Rep. 27, 1293-1306.e6. doi: 10.1016/j.celrep.2019.03.099
Santacruz, K., Lewis, J., Spires, T., Paulson, J., Kotilinek, L., Ingelsson, M., et al. (2005). Tau suppression in a neurodegenerative mouse model improves memory function. Science 309, 476–481. doi: 10.1126/science.1113694
Savage, J. C., Carrier, M., and Tremblay, M. -È (2019). Morphology of Microglia Across Contexts of Health and Disease. Methods Mol. Biol. 2034, 13–26. doi: 10.1007/978-1-4939-9658-2_2
Schafer, D. P., Lehrman, E. K., Kautzman, A. G., Koyama, R., Mardinly, A. R., Yamasaki, R., et al. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. doi: 10.1016/j.neuron.2012.03.026
Schindowski, K., Bretteville, A., Leroy, K., Bégard, S., Brion, J.-P., Hamdane, M., et al. (2006). Alzheimer’s disease-like tau neuropathology leads to memory deficits and loss of functional synapses in a novel mutated tau transgenic mouse without any motor deficits. Am. J. Pathol. 169, 599–616. doi: 10.2353/ajpath.2006.060002
Schitine, C., Nogaroli, L., Costa, M. R., and Hedin-Pereira, C. (2015). Astrocyte heterogeneity in the brain: from development to disease. Front. Cell Neurosci. 9:76. doi: 10.3389/fncel.2015.00076
Sebastian Monasor, L., Müller, S. A., Colombo, A. V., Tanrioever, G., König, J., Roth, S., et al. (2020). Fibrillar Aβ triggers microglial proteome alterations and dysfunction in Alzheimer mouse models. Elife 9:e54083. doi: 10.7554/eLife.54083
Serrano-Pozo, A., Muzikansky, A., Gómez-Isla, T., Growdon, J. H., Betensky, R. A., Frosch, M. P., et al. (2013). Differential relationships of reactive astrocytes and microglia to fibrillar amyloid deposits in Alzheimer disease. J. Neuropathol. Exp. Neurol. 72, 462–471. doi: 10.1097/NEN.0b013e3182933788
Shinozaki, Y., Shibata, K., Yoshida, K., Shigetomi, E., Gachet, C., Ikenaka, K., et al. (2017). Transformation of Astrocytes to a Neuroprotective Phenotype by Microglia via P2Y1 Receptor Downregulation. Cell Rep. 19, 1151–1164. doi: 10.1016/j.celrep.2017.04.047
Shukla, A. K., McIntyre, L. L., Marsh, S. E., Schneider, C. A., Hoover, E. M., Walsh, C. M., et al. (2019). CD11a expression distinguishes infiltrating myeloid cells from plaque-associated microglia in Alzheimer’s disease. Glia 67, 844–856. doi: 10.1002/glia.23575
Sierksma, A., Lu, A., Mancuso, R., Fattorelli, N., Thrupp, N., Salta, E., et al. (2020). Novel Alzheimer risk genes determine the microglia response to amyloid-β but not to TAU pathology. EMBO Mol. Med. 12:e10606. doi: 10.15252/emmm.201910606
Šimončičová, E., Gonçalves de Andrade, E., Vecchiarelli, H. A., Awogbindin, I. O., Delage, I., and Tremblay, M. -È (2022). Present and future of microglial pharmacology. Trends Pharmacol. Sci. ahead of print] doi: 10.1016/j.tips.2021.11.006.
Simonovitch, S., Schmukler, E., Bespalko, A., Iram, T., Frenkel, D., Holtzman, D. M., et al. (2016). Impaired Autophagy in APOE4 Astrocytes. J. Alzheimers Dis. 51, 915–927. doi: 10.3233/JAD-151101
Smith, A. J., Duan, T., and Verkman, A. S. (2019). Aquaporin-4 reduces neuropathology in a mouse model of Alzheimer’s disease by remodeling peri-plaque astrocyte structure. Acta Neuropathol. Commun. 7:74. doi: 10.1186/s40478-019-0728-0
Smith, A. M., Davey, K., Tsartsalis, S., Khozoie, C., Fancy, N., Tang, S. S., et al. (2022). Diverse human astrocyte and microglial transcriptional responses to Alzheimer’s pathology. Acta Neuropathol. 143, 75–91. doi: 10.1007/s00401-021-02372-6
Sobue, A., Komine, O., Hara, Y., Endo, F., Mizoguchi, H., Watanabe, S., et al. (2021). Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol. Commun. 9:1. doi: 10.1186/s40478-020-01099-x
Sofroniew, M. V. (2020). Astrocyte Reactivity: subtypes, States, and Functions in CNS Innate Immunity. Trends Immunol. 41, 758–770. doi: 10.1016/j.it.2020.07.004
Sosna, J., Philipp, S., Albay, R., Reyes-Ruiz, J. M., Baglietto-Vargas, D., LaFerla, F. M., et al. (2018). Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer’s disease. Mol. Neurodegener. 13:11. doi: 10.1186/s13024-018-0244-x
Şovrea, A. S., and Boşca, A. B. (2013). Astrocytes reassessment - an evolving concept part one: embryology, biology, morphology and reactivity. J. Mol. Psychiatry 1:18. doi: 10.1186/2049-9256-1-18
Spangenberg, E., Severson, P. L., Hohsfield, L. A., Crapser, J., Zhang, J., Burton, E. A., et al. (2019). Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10:3758. doi: 10.1038/s41467-019-11674-z
Spangenberg, E. E., Lee, R. J., Najafi, A. R., Rice, R. A., Elmore, M. R. P., Blurton-Jones, M., et al. (2016). Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain 139, 1265–1281. doi: 10.1093/brain/aww016
Spanos, F., and Liddelow, S. A. (2020). An Overview of Astrocyte Responses in Genetically Induced Alzheimer’s Disease Mouse Models. Cells 9:2415. doi: 10.3390/cells9112415
Spires-Jones, T. L., and Hyman, B. T. (2014). The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron 82, 756–771. doi: 10.1016/j.neuron.2014.05.004
Squair, J. W., Gautier, M., Kathe, C., Anderson, M. A., James, N. D., Hutson, T. H., et al. (2021). Confronting false discoveries in single-cell differential expression. Nat. Commun. 12:5692. doi: 10.1038/s41467-021-25960-2
Srinivasan, K., Friedman, B. A., Etxeberria, A., Huntley, M. A., van der Brug, M. P., Foreman, O., et al. (2020). Alzheimer’s Patient Microglia Exhibit Enhanced Aging and Unique Transcriptional Activation. Cell Rep. 31:107843. doi: 10.1016/j.celrep.2020.107843
Stalder, M., Phinney, A., Probst, A., Sommer, B., Staufenbiel, M., and Jucker, M. (1999). Association of microglia with amyloid plaques in brains of APP23 transgenic mice. Am. J. Pathol. 154, 1673–1684. doi: 10.1016/S0002-9440(10)65423-5
St-Pierre, M.-K., Bordeleau, M., and Tremblay, M. È (2019). Visualizing Dark Microglia. Methods Mol. Biol. 2034, 97–110. doi: 10.1007/978-1-4939-9658-2_8
St-Pierre, M.-K., Šimončičová, E., Bögi, E., and Tremblay, M. È (2020). Shedding Light on the Dark Side of the Microglia. ASN Neuro. 12:1759091420925335. doi: 10.1177/1759091420925335
Streit, W. J., Braak, H., Xue, Q.-S., and Bechmann, I. (2009). Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer’s disease. Acta Neuropathol. 118, 475–485. doi: 10.1007/s00401-009-0556-6
Streit, W. J., Khoshbouei, H., and Bechmann, I. (2020). Dystrophic microglia in late-onset Alzheimer’s disease. Glia 68, 845–854. doi: 10.1002/glia.23782
Streit, W. J., Sammons, N. W., Kuhns, A. J., and Sparks, D. L. (2004). Dystrophic microglia in the aging human brain. Glia 45, 208–212. doi: 10.1002/glia.10319
Sturchler-Pierrat, C., Abramowski, D., Duke, M., Wiederhold, K.-H., Mistl, C., Rothacher, S., et al. (1997). Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc. Natl. Acad. Sci. U.S.A 94, 13287–13292.
Swanson, M. E. V., Murray, H. C., Ryan, B., Faull, R. L. M., Dragunow, M., and Curtis, M. A. (2020). Quantitative immunohistochemical analysis of myeloid cell marker expression in human cortex captures microglia heterogeneity with anatomical context. Sci. Rep. 10:11693. doi: 10.1038/s41598-020-68086-z
Swartzlander, D. B., Propson, N. E., Roy, E. R., Saito, T., Saido, T., Wang, B., et al. (2018). Concurrent cell type-specific isolation and profiling of mouse brains in inflammation and Alzheimer’s disease. JCI Insight 3:121109. doi: 10.1172/jci.insight.121109
Taipa, R., Ferreira, V., Brochado, P., Robinson, A., Reis, I., Marques, F., et al. (2018). Inflammatory pathology markers (activated microglia and reactive astrocytes) in early and late onset Alzheimer disease: a post mortem study. Neuropathol. Appl. Neurobiol. 44, 298–313. doi: 10.1111/nan.12445
Taylor, X., Cisternas, P., You, Y., You, Y., Xiang, S., Marambio, Y., et al. (2020). A1 reactive astrocytes and a loss of TREM2 are associated with an early stage of pathology in a mouse model of cerebral amyloid angiopathy. J. Neuroinflamm. 17:223. doi: 10.1186/s12974-020-01900-7
Terry, R. D., Masliah, E., Salmon, D. P., Butters, N., DeTeresa, R., Hill, R., et al. (1991). Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 30, 572–580. doi: 10.1002/ana.410300410
Thrupp, N., Sala Frigerio, C., Wolfs, L., Skene, N. G., Fattorelli, N., Poovathingal, S., et al. (2020). Single-Nucleus RNA-Seq Is Not Suitable for Detection of Microglial Activation Genes in Humans. Cell Rep. 32:108189. doi: 10.1016/j.celrep.2020.108189
Tsacopoulos, M., and Magistretti, P. J. (1996). Metabolic coupling between glia and neurons. J. Neurosci. 16, 877–885.
Tung, P.-Y., Blischak, J. D., Hsiao, C. J., Knowles, D. A., Burnett, J. E., Pritchard, J. K., et al. (2017). Batch effects and the effective design of single-cell gene expression studies. Sci. Rep. 7:39921. doi: 10.1038/srep39921
Unger, M. S., Schernthaner, P., Marschallinger, J., Mrowetz, H., and Aigner, L. (2018). Microglia prevent peripheral immune cell invasion and promote an anti-inflammatory environment in the brain of APP-PS1 transgenic mice. J. Neuroinflamm. 15:274. doi: 10.1186/s12974-018-1304-4
Uranova, N. A., Vikhreva, O. V., Rakhmanova, V. I., and Orlovskaya, D. D. (2018). Ultrastructural pathology of oligodendrocytes adjacent to microglia in prefrontal white matter in schizophrenia. NPJ Schizophr. 4:26. doi: 10.1038/s41537-018-0068-2
Verkhratsky, A., and Nedergaard, M. (2018). Physiology of Astroglia. Physiol. Rev. 98, 239–389. doi: 10.1152/physrev.00042.2016
Verkhratsky, A., Rodrigues, J. J., Pivoriunas, A., Zorec, R., and Semyanov, A. (2019). Astroglial atrophy in Alzheimer’s disease. Pflugers Arch. 471, 1247–1261. doi: 10.1007/s00424-019-02310-2
Verney, C., Monier, A., Fallet-Bianco, C., and Gressens, P. (2010). Early microglial colonization of the human forebrain and possible involvement in periventricular white-matter injury of preterm infants. J. Anat. 217, 436–448. doi: 10.1111/j.1469-7580.2010.01245.x
von Saucken, V. E., Jay, T. R., and Landreth, G. E. (2020). The effect of amyloid on microglia-neuron interactions before plaque onset occurs independently of TREM2 in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 145:105072. doi: 10.1016/j.nbd.2020.105072
Walker, D. G., and Lue, L.-F. (2015). Immune phenotypes of microglia in human neurodegenerative disease: challenges to detecting microglial polarization in human brains. Alzheimers Res. Ther. 7:56. doi: 10.1186/s13195-015-0139-9
Walton, C. C., Begelman, D., Nguyen, W., and Andersen, J. K. (2020). Senescence as an Amyloid Cascade: the Amyloid Senescence Hypothesis. Front. Cell Neurosci. 14:129. doi: 10.3389/fncel.2020.00129
Wang, C., Xiong, M., Gratuze, M., Bao, X., Shi, Y., Andhey, P. S., et al. (2021). Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia. Neuron 109, 1657-1674.e7. doi: 10.1016/j.neuron.2021.03.024
Wegiel, J., Wang, K. C., Tarnawski, M., and Lach, B. (2000). Microglia cells are the driving force in fibrillar plaque formation, whereas astrocytes are a leading factor in plague degradation. Acta Neuropathol. 100, 356–364. doi: 10.1007/s004010000199
Westergard, T., and Rothstein, J. D. (2020). Astrocyte Diversity: current Insights and Future Directions. Neurochem. Res. 45, 1298–1305. doi: 10.1007/s11064-020-02959-7
Xu, J., Zhang, P., Huang, Y., Zhou, Y., Hou, Y., Bekris, L. M., et al. (2021). Multimodal single-cell/nucleus RNA sequencing data analysis uncovers molecular networks between disease-associated microglia and astrocytes with implications for drug repurposing in Alzheimer’s disease. Genom. Res. 31, 1900–1912. doi: 10.1101/gr.272484.120
Yang, H. S., Onos, K. D., Choi, K., Keezer, K. J., Skelly, D. A., Carter, G. W., et al. (2021). Natural genetic variation determines microglia heterogeneity in wild-derived mouse models of Alzheimer’s disease. Cell Rep. 34:108739. doi: 10.1016/j.celrep.2021.108739
Yang, J., Lunde, L. K., Nuntagij, P., Oguchi, T., Camassa, L. M. A., Nilsson, L. N. G., et al. (2011). Loss of astrocyte polarization in the tg-ArcSwe mouse model of Alzheimer’s disease. J. Alzheimers Dis. 27, 711–722. doi: 10.3233/JAD-2011-110725
Yang, J., Zhang, R., Shi, C., Mao, C., Yang, Z., Suo, Z., et al. (2017). AQP4 Association with Amyloid Deposition and Astrocyte Pathology in the Tg-ArcSwe Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 57, 157–169. doi: 10.3233/JAD-160957
Yeh, C.-Y., Vadhwana, B., Verkhratsky, A., and Rodríguez, J. J. (2011). Early Astrocytic Atrophy in the Entorhinal Cortex of a Triple Transgenic Animal Model of Alzheimer’s Disease. ASN Neuro. 3:AN20110025. doi: 10.1042/AN20110025
Yeh, F. L., Wang, Y., Tom, I., Gonzalez, L. C., and Sheng, M. (2016). TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron 91, 328–340. doi: 10.1016/j.neuron.2016.06.015
Yoshiyama, Y., Higuchi, M., Zhang, B., Huang, S.-M., Iwata, N., Saido, T. C., et al. (2007). Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351. doi: 10.1016/j.neuron.2007.01.010
Zanier, E. R., Fumagalli, S., Perego, C., Pischiutta, F., and De Simoni, M.-G. (2015). Shape descriptors of the “never resting” microglia in three different acute brain injury models in mice. Intensive Care Med. Exp. 3:39. doi: 10.1186/s40635-015-0039-0
Zhang, G., Wang, Z., Hu, H., Zhao, M., and Sun, L. (2021). Microglia in Alzheimer’s Disease: a Target for Therapeutic Intervention. Front. Cell Neurosci. 15:749587. doi: 10.3389/fncel.2021.749587
Zhang, L., Hu, K., Shao, T., Hou, L., Zhang, S., Ye, W., et al. (2021). Recent developments on PET radiotracers for TSPO and their applications in neuroimaging. Acta Pharm. Sin. B 11, 373–393. doi: 10.1016/j.apsb.2020.08.006
Zheng, J., Ru, W., Adolacion, J. R., Spurgat, M. S., Liu, X., Yuan, S., et al. (2021). Single-cell RNA-seq analysis reveals compartment-specific heterogeneity and plasticity of microglia. iScience 24:102186. doi: 10.1016/j.isci.2021.102186
Zhou, B., Zuo, Y.-X., and Jiang, R.-T. (2019). Astrocyte morphology: diversity, plasticity, and role in neurological diseases. CNS Neurosci. Ther. 25, 665–673. doi: 10.1111/cns.13123
Zhou, Y., Song, W. M., Andhey, P. S., Swain, A., Levy, T., Miller, K. R., et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142. doi: 10.1038/s41591-019-0695-9
Ziegenhain, C., Vieth, B., Parekh, S., Reinius, B., Guillaumet-Adkins, A., Smets, M., et al. (2017). Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 65, 631–643.e4. doi: 10.1016/j.molcel.2017.01.023
Keywords: Alzheimer’s disease, heterogeneity, microglia, astrocyte, murine model, human brain samples
Citation: St-Pierre M-K, VanderZwaag J, Loewen S and Tremblay M-È (2022) All roads lead to heterogeneity: The complex involvement of astrocytes and microglia in the pathogenesis of Alzheimer’s disease. Front. Cell. Neurosci. 16:932572. doi: 10.3389/fncel.2022.932572
Received: 29 April 2022; Accepted: 11 July 2022;
Published: 12 August 2022.
Edited by:
Nora Bengoa-Vergniory, University of Oxford, United KingdomReviewed by:
Maria Weinert, Imperial College London, United KingdomCopyright © 2022 St-Pierre, VanderZwaag, Loewen and Tremblay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie-Ève Tremblay, ZXZldHJlbWJsYXlAdXZpYy5jYQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.