
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 22 February 2022
Sec. Cellular Neurophysiology
Volume 16 - 2022 | https://doi.org/10.3389/fncel.2022.811493
This article is part of the Research Topic New Horizons in Cellular Optogenetics View all 7 articles
Cells reside in a dynamic microenvironment that presents them with regulatory signals that vary in time, space, and amplitude. The cell, in turn, interprets these signals and accordingly initiates downstream processes including cell proliferation, differentiation, migration, and self-organization. Conventional approaches to perturb and investigate signaling pathways (e.g., agonist/antagonist addition, overexpression, silencing, knockouts) are often binary perturbations that do not offer precise control over signaling levels, and/or provide limited spatial or temporal control. In contrast, optogenetics leverages light-sensitive proteins to control cellular signaling dynamics and target gene expression and, by virtue of precise hardware control over illumination, offers the capacity to interrogate how spatiotemporally varying signals modulate gene regulatory networks and cellular behaviors. Recent studies have employed various optogenetic systems in stem cell, embryonic, and somatic cell patterning studies, which have addressed fundamental questions of how cell-cell communication, subcellular protein localization, and signal integration affect cell fate. Other efforts have explored how alteration of signaling dynamics may contribute to neurological diseases and have in the process created physiologically relevant models that could inform new therapeutic strategies. In this review, we focus on emerging applications within the expanding field of optogenetics to study gene regulation, cell signaling, neurodevelopment, and neurological disorders, and we comment on current limitations and future directions for the growth of the field.
Cellular signaling is mediated by highly dynamic and intertwined pathways that control crucial cell behaviors. During organismal development, these signaling processes are spatially and dynamically regulated to orchestrate cell expansion, cell differentiation, and tissue morphogenesis. Likewise, complex signaling pathways enable and protect adult organismal function. Investigating these complex mechanisms has required the development of tools to perturb their activity.
Conventional chemical perturbations can offer some levels of control but have limited capability in their spatial and temporal resolution (Webster et al., 1988; Gossen et al., 1995; Metzger et al., 1995; Rivera et al., 1996; Stanton et al., 2018). As an alternative approach, optogenetics offers the potential for unprecedented precise temporal, spatial, and dosage control of signaling. Specifically, the spatial precision can be achieved on the order of ~micrometers (μm), and the temporal precision can be achieved at ~ms (<1 ms; Shemesh et al., 2017). Optogenetics, which combines genetic and optical tools for the precise perturbation of target cellular processes, initially applied naturally derived photosensitive opsins to study and control the activity of individual neurons and by extension investigate brain circuity. In particular, opsins, derived from various microbial species, were re-purposed for mammalian cell expression and upon illumination at specific wavelengths unleashed ions flow across cell membranes to activate or inhibit target neurons (Zhang et al., 2006; Deisseroth, 2015), as extensively reviewed in recent literature (Looger, 2011; Zhang et al., 2011). This review will focus instead on newly developed light-responsive systems and optics hardware that have enabled applications in new areas such as control of protein activity, signaling dynamics, up- or down- regulation of gene expression, subcellular localization, and other applications. We will discuss how the high spatiotemporal precision of optogenetics has led to novel insights into cellular signaling networks and future trends in the field of optogenetics for a variety of applications in basic science and therapeutics.
In recent years, optogenetic tools have extended to many classes of proteins containing light-responsive domains (summarized in Table 1) discovered in plants, bacteria, invertebrates, etc. Their photoreceptor domains exhibit light-inducible properties such as reversible protein-protein interactions, oligomerization, and conformational changes, which can be harnessed to engineer photoswitches for regulating gene activation, repression, protein localization, and other functions (Figure 1).
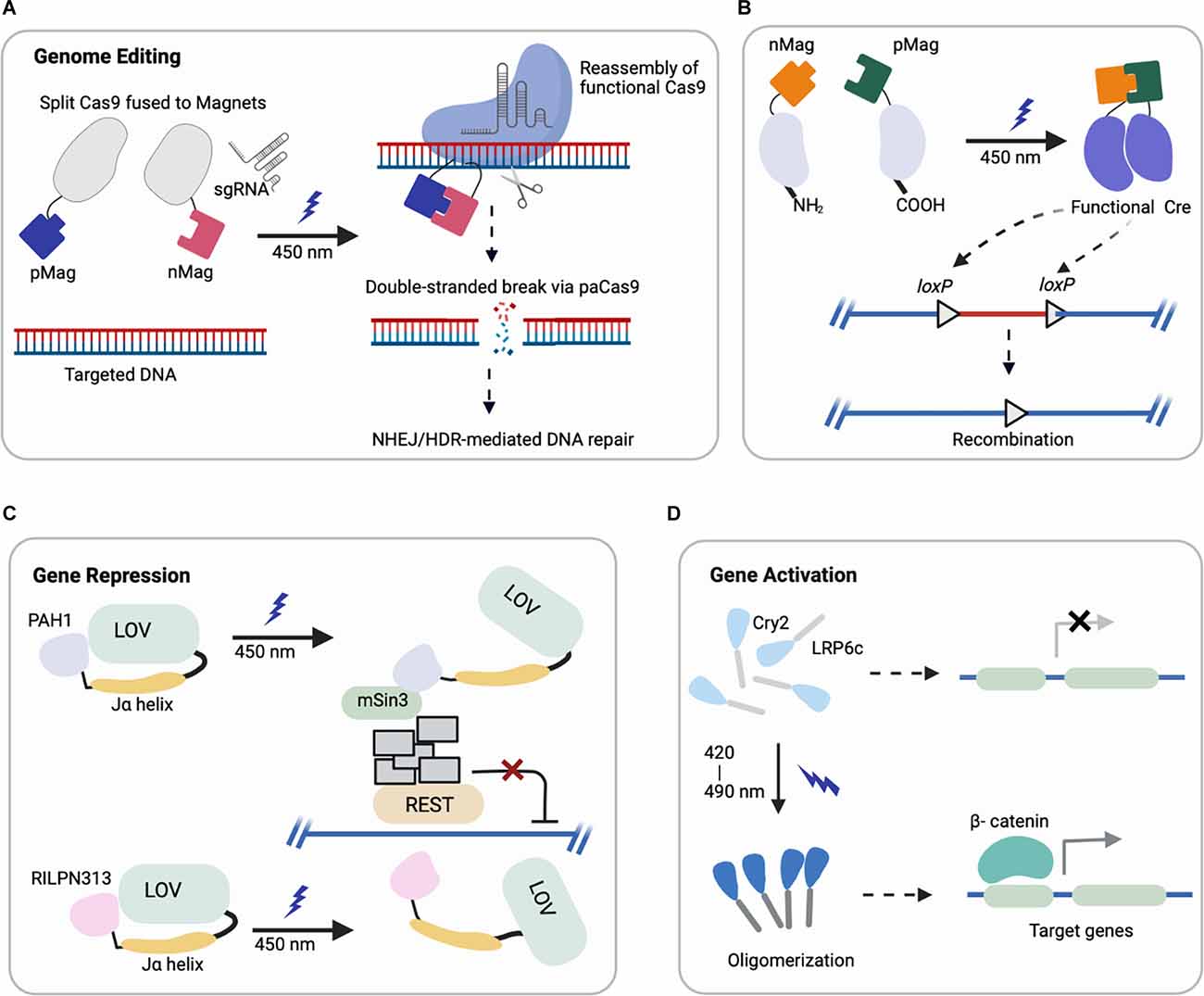
Figure 1. Representative schematics of four major optogenetic-based gene regulations using different systems (created with Biorender). (A,B) Light-induced uncaging of affinity domains (positive pMag) and (negative nMag) results in dimerization of attached domains and reassembly of (A) functional Cas9 for indel mutation and (B) split Cre recombinase for DNA recombination. (C) Upon light-induction, the Jα helix unfolds from LOV2 core to uncage a fused protein (PAH1, RILPN313) that inhibits the binding and activity of targeted transcription factor (REST). (D) Cry2 clusters an attached protein (LRP6c) in response to light that activates the downstream target genes, e.g., Wnt signaling.
One important system involves light, oxygen, and voltage domains (LOV), isolated from plant photosensor phototropin 1 (phot1). Phot1 molecule has two LOV domains, with the second domain (LOV2) primarily involved in kinase activation (Christie et al., 1999, 2012; Wu et al., 2009). In particular, Avena sativa phot1 LOV2 (AsLOV2) has a tightly associated C-terminal Jα helix in a folded state, but upon exposure to blue light, the bound flavin mononucleotide (FMN) chromophore undergoes a conformational change to displace the Jα helix from the protein core, leading to uncaging of an effector kinase domain (Harper et al., 2003). By engineering fusions between other signaling effectors to LOV domains, this light-induced conformational change can be used to unleash signaling activity.
In another system, cryptochrome 2 (Cry2), derived from the plant A. thaliana, has a photolyase homology region (PHR) that interacts with a partner CIB1 in a blue light responsive manner, due to a flavin adenine dinucleotide (FAD) bound to Cry2 as a chromophore cofactor (Kennedy et al., 2010). In one optogenetic design, the Cry2PHR domain can dimerize with CIB1 lacking the basic helix-loop-helix domain (CIBN) within seconds of exposure. In addition to dimerization, Cry2PHR can also be harnessed for self-oligomerization with blue light activation (Bugaj et al., 2013, 2015). Specifically, the detection of protein clusters can be achieved with ~10 s, and dissociation occurs with a half-life ~5.5 min.
In a common system for red light activation, phytochrome (Phy), derived from Arabidopsis thaliana, contains two structural domains (Quail, 2010). The N-terminal photosensory domain binds to a tetrapyrrole chromophore, such as phycocyanobilin (PCB), and can be triggered by red light (650 nm) to photoisomerize and bind to phytochrome-interacting factors (PIFs) to modulate downstream pathways (Quail, 2002). This Phy/PIF dimerization has been harnessed to stimulate protein-protein interactions in mammalian cells (Levskaya et al., 2009). More recently, a near-infrared-light (~750 nm) responsive dimerization of bacterial phytochrome (BphP1) with its partner PpsR2 system has reportedly been harnessed for optogenetics (Kaberniuk et al., 2016). In addition to the deep tissue penetration of red- and far-red light, an advantage of Phy/PIF and Bphp1/PpsR2 systems is their reversibility and speed. Specifically, the dissociation of these protein pairs can be induced with lights of different wavelengths, enabling studies to perturb dynamics of signaling networks with a fast on/off rate.
Magnets, an improved system that incorporates positive and negative amino acids into the Vivid (VVD) N-terminal helix, also enables heterodimerization with electrostatic attraction upon uncaging of the protein (Kawano et al., 2015). VVD contains LOV domain that can be excited with blue light illumination to uncage the effector domain (Zoltowski et al., 2007).
Other dimerization systems such as ultraviolet B (UVB) and constitutively photomorphogenic 1 (COP1) offer an alternative excitation wavelength (~280–315 nm). UV response locus 8 (UVR8) can translocate and bind to COP1 in the presence of UVB (Favory et al., 2009), liberating COP1-bound transcription factors in the nucleus to modulate downstream signaling in an irreversible manner (Cloix et al., 2012; Wu et al., 2012).
To activate the variety of photosensory domains that are being harnessed to control signaling pathways and gene expression, the complementary development of optics technologies for illumination is also needed. Current optical stimulation devices have been developed for both in vitro and in vivo models. In one in vitro system, an array of light-emitting diodes (LEDs) can be constructed for large surface illumination in a uniform manner, though spatial resolution can be limited in this design (Tucker et al., 2014). To improve the spatial control of illumination, other systems such as digital micromirror device (DMD) microscopes (Allen, 2017) and light activation at variable amplitudes (LAVA) boards (Repina et al., 2020) have been developed. DMD-based systems offer single-cell resolution with dynamic, user-defined patterns, though with low throughput. By comparison, LAVA boards enable dynamically controlled illumination of multi-well plates for extended culture times, with the option of using masks for spatial patterning with ~100 μm resolution. Other types of devices are summarized in Table 2 (Gerhardt et al., 2016; Bugaj and Lim, 2019). Current in vivo hardware primarily relies on optical fiber or implantable LEDs sensors passed through or mounted to the skull of an animal to target cell populations in a spatially restricted manner. Such neural interfaces can generally achieve a temporal precision at ~ ms with a spatial precision ~ hundreds of micrometers (Aravanis et al., 2007; Kim et al., 2013).
Optogenetics has been extensively applied for the spatiotemporal modulation of gene expression, including genome editing, gene activation, and repression.
(i) Genome Editing: CRISPR/Cas9 (clustered, regularly interspaced, short palindromic repeats, CRISPR-associated protein 9) can be engineered for targeted genome modification in mammalian cells at specific DNA loci, which enables the causal dissection of gene functions (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013). Optogenetic control of targeted genome editing can facilitate our improved understanding of gene networks. The first reported photoactivatable Cas9 (paCas9) was based on the dimerization system called Magnets (Kawano et al., 2015). In this work, the Cas9 nucleases were split into two parts fused to photoinducible dimerizing domains (positive Magnet and negative Magnet), which assemble and reconstitute Cas9 activity upon blue light illumination (Nihongaki et al., 2015a). This photoactivatable Cas9 cleaved a targeted endogenous genomic locus and induced indel mutation by nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in a light-dependent fashion (Figure 1A). Using multiple sgRNAs targeting human genes EMX1 and VEGFA, the light-induced Cas9 indel mutations in both genes simultaneously. Though the reported study focused on HEK293T cells, EMX1 is ubiquitously expressed in neural stem cells and guides differentiation of layer-specific neuronal phenotypes (Schuurmans and Guillemot, 2002), such that this system could be further harnessed for investigating corticogenesis or other neurodevelopmental processes.
Another study adapted a photoactivatable Cre (PA-Cre) system for site-specific genome recombination, where low intensity or short pulsed illumination enabled efficient activation with high spatial and temporal precision in vivo (Kawano et al., 2016). In particular, Cre recombinase was split (CreN59/CreC60) into two fragments and fused to components of the Magnet system for heterodimerization under blue light (Figure 1B). This design yielded a substantial increase in DNA recombination compared to a prior design split system (Kennedy et al., 2010). Recently, an improved version of this PA-Cre system was reported to enhance efficiency and reduce dark background recombination when targeting active neurons in the murine dorsal raphe nucleus (Morikawa et al., 2020). Successful recombination was observed under blue light condition for mouse brains in vivo and in neural cells in vitro, demonstrating the promise of this system for future applications in studying neural pathways in mouse models. Other groups have applied analogous systems with different recombinases to modulate excitatory neurons and parvalbumin interneurons in the mouse brain, which can potentially be combined with PA-Cre for dual locus genome editing (Li et al., 2020).
(ii) Gene Activation: Optogenetic control of gene expression has been achieved with several different approaches, and a typical strategy combines a photosensory module with a split transcription factor that can be reassembled upon light illumination. The first reported optogenetic system for regulating gene expression, published in 2002 (Shimizu-Sato et al., 2002), was based on the fusion of the Phy-Gal4-DNA binding domain and a PIF3-Gal4-activation domain to induce downstream gene activation.
Engineering DNA-binding became far easier with the advent CRISPR/Cas9 genome editing, and this capability can be harnessed for other applications. For example, Zhang and colleagues (Konermann et al., 2013) developed light-inducible transcriptional effectors (LITEs) that enable direct optical control of mammalian cell gene expression. Cry2 systems rely upon photoactivated dimerization with its interacting partner CIB1 or the self-oligomerization of the Cry2-PHR. In this study, a customizable DNA-binding domain was fused to the light-sensitive module of Cry2, and upon blue light illumination, its dimerization partner CIB1, fused to a desired effector domain, became recruited to a target locus to modulate gene expression. This approach enabled independent, multiplexed genomic anchors to target a range of genomic loci. More recently, to improve the targeting of multiple sequences in the same gene simultaneously for robust activation, Cry2 was fused to the VP64 transactivation domain while its interacting partner CIB1 was fused to dCas9 (Jinek et al., 2012), the catalytically inactive form of Cas9. With multiple gRNAs co-transfected into mammalian cells to direct the binding of dCas9 to the targeted promoter, transcription could be induced robustly in the presence of blue light. The advantage of such a system compared to the conventional chemical-based strategy in gene activation is reversibility by the removal of light illumination, enabling dynamic patterns of gene modulation (Polstein and Gersbach, 2015). While transcriptional upregulation has been achieved in multiple studies, the magnitude of upregulation can be too low to induce an effective biological response (Nihongaki et al., 2015a, b). To address this shortcoming, a modified system incorporating synergistic activation mediators has been developed (Nihongaki et al., 2017). In this system, a single gRNA is attached to MS2 binding sequences to recruit the MS2 coat protein fused with two activators (p65 and heat shock factor 1) onto the same dCas9 protein, resulting in a 50–100-fold higher activation (ASCL1) than the previous constructs in HEK293T cells.
Researchers have applied this system to target NEUROD1 expression to induce neuronal differentiation in human iPSCs. Using the modified gene activation system, the upregulation was improved by 860-fold after 1 d of blue light illumination, and activated cells stained positively for the neuronal marker β-III tubulin (Tuj1) after 4 d of illumination. Another research has induced functional neuronal differentiation using a far-red light-mediated system, which offers low phototoxicity and deep tissue penetration (Shao et al., 2018).
(iii) Gene Repression: In addition to activating target gene expression, multiple systems have been developed to repress transcription. For example, repressive histone modifiers have been fused to light-sensitive proteins to suppress Grm2 and Neurog2 gene expression in primary neurons and Neuro2a cells with light illumination (Konermann et al., 2013). Another example harnessed a small peptide degron. Specifically, a small peptide degron RRRGN was fused to the C-terminus of the photoresponsive Jα helix of LOV2 domain. The LOV2 domain contains a flavin-binding core domain, and blue light illumination induces a side-chain rotation of the conserved flavin-interacting residue followed by dissociation and unfolding of the C-terminal Jα helix (Shcherbakova et al., 2015). This dissociation from the LOV core domain under blue light induction exposed the degron, which subsequently stimulated protein degradation (Bonger et al., 2014). Another study further engineered a system to include an additional Cry2-based system together with the LOV-domain, such that the blue light can simultaneously block new mRNA transcription while inducing the degradation of existing proteins (Pathak et al., 2017), showed a further reduction in targeting protein level compared to the single LOV2-system reported previously.
Researchers have also leveraged this LOV2 conformation change to engineer chimeric proteins to inhibit the repressor element (RE) 1-silencing transcription factor (REST) in primary neurons (Paonessa et al., 2016). REST is a transcriptional repressor that is normally expressed at low levels by mature neurons, but in various brain pathologies becomes up regulated (Mandel et al., 2011; Pozzi et al., 2013). A minimal interacting REST sequence was fused to the LOV2 C-terminus. On blue light illumination, LOV2 unfolded and freed the C-terminal domain to interact with and inhibit the binding of REST (Figure 1C). With effective light-medicated REST inhibition, neurons showed increased brain-derived neurotrophic factor transcription and boosted Na+ currents and neuronal firing (Paonessa et al., 2016). Future investigation can apply these modulating systems in vivo for brain pathologies associated with REST overexpression or hyperactivity (Zuccato et al., 2003; Lu et al., 2014).
Optogenetic tools generally allow modulation of the single gene expression using a single light color system; however, recent studies have highlighted the possibility for multiplexed control of several genes to regulate biological processes (Redchuk et al., 2018), using near infrared (NIR) and blue light controlled systems with minimal spectral crosstalk. In addition, cell lines that include more than one optogenetic system (UVR8, PhyB/PIF, BphP1/PpsR2, etc.) activated by distinct wavelengths can also be generated, enabling downstream exploration of combinatorial or orthogonal signaling pathways (Müller et al., 2014). Other method includes constructing a light-responsive promoter library, which can be induced with pulsatile signaling light and enable multi-gene regulation at a fixed expression ratio (Benzinger and Khammash, 2018).
While these optogenetic tools can modulate gene expressions in targeted cells, recent studies have noted some potential limitations of current systems. These include the chances of toxicity from high intensity blue light illumination, toxicity resulting from high expression levels of Cas activator domain (Ewen-Campen et al., 2017; Casas-Mollano et al., 2020), activity noise, and variation of gene expressions associated with split-component systems (Guinn and Balázsi, 2019). Remedies include using pulses in place of continuous irradiation (Wang et al., 2012), a weaker promoter for Cas constructs (Jia et al., 2018), and negative feedback gene circuit regulation, respectively. As an example of the latter, a recent study demonstrated a five-fold noise reduction by incorporating negative feedback regulation gene circuits (i.e., TetR repressor fused with a Tet-inhibiting peptide; Guinn and Balázsi, 2019). Such synthetic circuits can also be multiplexed orthogonally to probe multigene expressions in contribution of cell phenotypes (Szenk et al., 2020) and buffering gene dosage variation across cell populations for a more homogeneous gene expression control (Yang et al., 2021).
While optogenetics has not yet been broadly applied to study development, there is considerable potential. For example, the nervous system is a highly ordered, complex, and tightly regulated system central to organismal survival and function. This system develops through the expansion and differentiation of various neural stem and progenitor cells, processes that are tightly orchestrated by extracellular ligand signals. These ligands bind to receptor tyrosine kinases (RTKs; Schlessinger, 2014), adhesion, G-protein-coupled (Rosenbaum et al., 2009), and other receptors to initiate signal transduction cascades and trigger downstream cellular responses such as growth, differentiation, migration, and survival. Protein ligands or synthetic agonists have been broadly useful for probing cellular function and malfunction in vitro and in vivo. However, such signal activation has poor spatial and temporal control, limited by slow diffusion and convection as well as technical limitations in signal presentation and temporal activation. In contrast, optogenetic control of cellular signaling is an exciting approach for spatiotemporal control of signaling (Repina et al., 2017). These approaches thereby offer a valuable toolbox to investigate the complex spatiotemporal role of major signaling pathways in neurodevelopment and adult neurogenesis.
Following gastrulation, precise spatiotemporal presentation of morphogen signals regionally patterns the neural plate, neural groove, and neural tube and thereby the full central nervous system (Nikolopoulou et al., 2017). More specifically, the induction of the neuroectoderm and patterning of both the dorsal-ventral and anterior-posterior axis of the central nervous systems rely on spatiotemporal activation of Wnt, BMP, SHH, FGF, and retinoic acid (RA; Kiecker and Lumsden, 2012). However, challenges with ectopically modulating morphogen signals with temporal and spatial control have classically limited the generation of accurate in vitro models of neurodevelopment (Kelava and Lancaster, 2016).
Optogenetic activation of signaling cascades typically relies on a genetically encoded light responsive dimerizing/oligomerizing protein fused to a signaling effector to mimic the effect of upstream ligand binding (Kramer et al., 2021), and it can do so with spatiotemporal control. In particular, several optogenetic systems controlling morphogen signals relevant to development have recently been reported, leveraging Cry2 (Kennedy et al., 2010; Bugaj et al., 2013). One of the first examples of leveraging Cry2 oligomerization to control cellular signaling was optogenetic control of canonical Wnt activation. Our optoWnt system involves fusion of the intracellular portion of the canonical Wnt co-receptor LRP6 to the PHR domain of Cry2 (Figure 1D), and we have reported optogenetic activation of Wnt signaling in both adult neural stem cells (NSCs; Bugaj et al., 2013) and human embryonic stem cells (hESCs; Repina et al., 2020). Other emerging and relevant morphogen signals to neurodevelopment include optoFGFR1(Kim et al., 2014), optoTGF (Li et al., 2018), optoBMP (Humphreys et al., 2020), and optoBrn2 (Sokolik et al., 2015), which have clear applications in modeling neurodevelopment and neurogenesis if translated to either pluripotent stem cells or adult neural stem cells.
optoWnt has been recently deployed to investigate the temporal role of β-catenin activity dynamics on adult neurogenesis (Rosenbloom et al., 2020). It was determined that sustained optoWnt activation promoted proliferation and differentiation, while transient or disrupted activation induced cellular apoptosis. This discrepancy may explain the cell death observed during adult neurogenesis, indicating a potential inherent mechanism that corrects for deviations in differentiation cues.
In studies outside the nervous system, optogenetic control over Ras/ERK signaling in Drosophila, relying on the association of Phy-PIF for the membrane localization of SOS proteins (Son of Sevenless), revealed the role of this pathway in promoting endodermal differentiation at the cost of mesodermal fate during a critical time window (McFann et al., 2021). Specifically, Ras signaling was controlled using a specific region of SOS with catalytic activity (SOScat), which activates Ras signal when recruited to the plasma membrane (optoSOS). This was accomplished via PIF-SOScat fusion and a membrane PhyB (Toettcher et al., 2013). In hESCs, mixed optoWnt activation in a subpopulation of cells has been linked to mesendoderm specification and migration from a non-Wnt stimulated population, providing insight into gastrulation and early ectoderm specification (Repina et al., 2020). In an alternative application, investigating the temporal dynamics of optoBrn2 in hESC reveals a positive-feedback mechanism, where pluripotency networks do not respond to optoBrn2 signals below a certain magnitude or duration threshold (Sokolik et al., 2015). This result may indicate how hESCs are able to distinguish signal vs. stochastic fluctuations during development. Finally, optogenetics can function as a precision perturbation tool in developmental biology (Krueger et al., 2019). In vivo application include decoding signaling dynamics during Xenopus development, where light-mediated Raf/MEK/ERK activation, accomplished by membrane localization of Raf1 via CRY2-CIBN association, was demonstrated to induce a tail-like structure after germ layer formation (Krishnamurthy et al., 2016, 2017), and optoWnt activation results in axis duplication (Krishnamurthy et al., 2021).
Overall, optogenetic toolkits represent a powerful and exciting approach to understand and probe the mechanisms underlying neurodevelopment and neurogenesis. In the future, optogenetic control of morphogen signals may better inform the spatiotemporal signal dynamics governing neurulation, neural tube patterning, and neural differentiation and enable more physiologically relevant in vitro models of these stages.
Beyond early-stage work in neurodevelopment, recent optogenetic systems provide novel approaches to investigate the mechanisms behind cellular survival, function, and synaptic communication in the adult nervous system. One important class of cellular signaling pathways in the nervous system is the tropomyosin-related kinase (Trk) family of RTKs, as they are known to be important for neuronal survival, neurite outgrowth, synaptic function, and synaptic plasticity (Chang et al., 2014). Additionally, Trk signaling cascades are implicated in neurodegenerative disorders, including Alzheimer’s, chronic pain, inflammation, and cancer (Amatu et al., 2019). Upon binding to the respective neurotrophins, members of the Trk family, namely A, B, and C, activate several major signal transduction pathways, including the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated protein kinase (ERK) kinase (MEK), phosphatidylinositol 3-kinase (PI3K)/Akt, and phospholipase Cγ1 (PLCγ1)/Ca2+ pathways.
Recently, several such pathways have been placed under light control using Cry2, enabling investigations into mechanisms underlying cellular behavior in the nervous system. For example, an optogenetic TrkA (optoTrkA) system (Duan et al., 2018; Khamo et al., 2019), involving the fusion of the intracellular domain of TrkA to the Cry2-CIB1 dimerizing system, successfully stimulated nerve growth factor (NGF)/TrkA signaling in PC12 cells. Under light stimulation, the group observed both neurite outgrowth and survival of dorsal root ganglion nerves, similar to the effects of exogenous NGF. Another group recently reported that a fusion of the PHR region of Cry2 to the TrkB receptor (optoTrkB) induced robust activation of canonical TrkB signaling (Chang et al., 2014). They demonstrated that sustained light illumination resulted in upregulated extracellular signal related kinase (ERK) activity, neurite outgrowth, and filopodia formation, which mimicked brain-derived neurotrophic factor (BDNF) ligand signaling.
Following similar approaches, Cry2-based optogenetic toolkits for additional signaling cascades have also been recently reported, including optoRaf (Su et al., 2020), optoRhoA (Bugaj et al., 2013), optoCdc42 (Bugaj et al., 2013), optoEphB2 (Locke et al., 2017), and light sensitive myosin motors (Zhang et al., 2021). Overall, these newly developed optogenetic toolkits enable the spatiotemporal profiling of signal activation in vitro and enable applications in understanding the signaling dynamics underlying key neurotrophin cascades and cellular function in the nervous system.
In addition to Cry2-based optogenetic systems, the LOV domain from phototropin (Christie et al., 2012) has been harnessed in neurons. For example, in photoactivable Rac1 (Wu et al., 2009) the LOV domain sterically occluded the binding of Rac1 to its effector. Upon light illumination, a conformational change induced unwinding of the Jα helix from the LOV domain and thereby released the inhibition of Rac1 signaling. The researchers thereby demonstrated that selectively activating Rac1 signaling could induce cell motility and control the direction of cell movement, enabling analysis of Rac1 regulation in RhoA in cell mobility. Rac1 is a key GTPase that regulates actin cytoskeletal dynamics (Raftopoulou and Hall, 2004), and in dendritic spines, Rac1 specifically regulates the spine size via rearrangement of actin cytoskeleton (Hayashi-Takagi et al., 2010; Yuste, 2011). Previous work has implied the important role of structural plasticity of spines in synaptic transmission for learning-related activities (Hayashi-Takagi et al., 2015). Given the role of Rac1 in neurites and neurons, modulating Rac1 activity using precise and reversible optogenetic tool may offer novel insights into neuronal function and health (Penzes et al., 2011; Hayashi-Takagi, 2017).
Finally, optogenetic control of cellular signaling pathways not only offers exciting new tools to probe biological questions in vitro, but it also presents a potential novel perturbation in vivo to study mechanisms of neuroregeneration and disease in model organisms. Optogenetic domains have been harnessed for the recruitment of ligands to a specified localization to induce Ca2+ signaling in vivo. In particular, the PHR domain of Cry2 was fused to STIM1, and light inducible oligomerized STIM1 translocated to the plasma membrane and bound endogenous calcium release-activated calcium (CRAC) channels to trigger Ca2+ influx into the cell (Kyung et al., 2015). Using the OptoSTIM1 system, expressed in the hippocampus of living mice, researchers observed an increase of contextual fear memory in the light-illuminated group, suggesting potential mechanisms of Ca2+ dependent memory formation. Finally, a recent study demonstrated optoAKT and optoRAF (for the control of ERK signaling) in Drosophila enhanced axon regeneration and proper axon guidance in the peripheral nervous system (PNS) as well as axon regrowth and functional recovery in the CNS (Wang et al., 2020). In this example, optogenetics may provide a potential strategy in the intervention of neural degeneration.
Overall, optogenetic control of protein activity is a powerful toolkit to probe the spatiotemporal role of signaling dynamics with high precision and throughput. As new optogenetic systems emerge, future applications in understanding neurodevelopment and function may benefit from in vitro and in vivo optogenetic approaches.
Beyond studying signaling pathways and advancing understanding of complex cellular networks in neurodevelopment and physiological brain functions, optogenetics has also been extended to medical therapeutics and for modulation of cell behaviors in the context of understanding of disease pathologies. The need to develop efficient therapies for neurodegenerative diseases is urgent, given the increasing percentages of the population living longer. One of the reasons for the healthcare burden of neurological disorders is the lack of effective targeted treatments. The simultaneous development of genome engineering and optogenetics has the potential for interrogating mechanistically the pathology involved in genetic-related brain disorders and possibly offering more targeted therapies for clinical applications.
For example, stable, optogenetically-modified mammalian cells could be implanted for relevant clinical conditions (Oggu et al., 2017). Given the deeper tissue penetrance and low toxicity of red or NIR, recently developed red light systems developed for high tissue penetration, transgene, or signal induction with controlled localization, timing, and dosage is increasingly possible in the adult brain. For example, the implantation of wireless, minimally invasive illumination devices can enable optogenetic modulation of neural systems in large organisms (Montgomery et al., 2015; Park et al., 2015). Some previous studies have also shown that light can be efficiently delivered by a helmet-type configuration applied to the brain (Lanzafame et al., 2014). Using engineered stem cells, one can also introduce a therapeutic target gene with a light inducible expression system, which then can be spatiotemporally controlled for producing or inhibiting specific responses (Pomeroy et al., 2017). For example, human embryonic stem cell-mesencephalic dopaminergic neurons have been optogenetically controlled in a model of Parkinson’s disease using a fiber-optic cannula, which exploited optogenetics as an on-off switch for neuronal activity, allowing this function of the cells to be tested independently of other possible functions (Steinbeck et al., 2015). While the study demonstrated the utility of optogenetics to dissect the mechanism underlying graft function, the proof-of-concept of using engrafted, engineered stem cells with optogenetic control is also promising for potential clinical applications of optogenetic cell-based therapies in neurological disorders.
However, there are also some potential limitations in the translation of optogenetic systems to clinical usages, including: (1) the gene cassette size that can be packaged in vectors for transfection; (2) safe and efficient delivery vehicles that can target the cell or tissue of interest (though engineering of viral delivery systems such as engineered adeno-associated viruses can help accelerate progress); (3) potential inflammation and immunogenicity caused by optogenetic tools; (4) limited red-light or NIR light sensitive proteins available with high tissue penetration and low phototoxicity, especially in wavelengths ranges not currently well-explored; (5) potential long-term effects of permanent introduction of non-human proteins with optical properties; and (6) a lack of appropriate illumination devices that present narrower-wavelengths for mitigating surrounding tissue phototoxicity (Wojtovich and Foster, 2014), or high spatial illumination resolution that can circumvent subcellular protein diffusion problems.
To overcome some of these limitations and further improve the light-responsive protein properties, systematic protein engineering processes such as directed evolution (Packer and Liu, 2015; Wu et al., 2019) can be applied to engineer the next generation of optogenetic systems with enhanced efficiency/specificity (Herwig et al., 2017; Bedbrook et al., 2019). Some characteristics to consider can include: (a) specificity in response to an activating wavelength, with low or no background activity under the dark condition, (b) rapid and greater sensitivity to low levels of activation light to allow for longer exposures at low illumination intensity, (c) reduced size of protein components to facilitate delivery.
In summary, state-of-the-art optogenetic systems are enabling numerous discoveries and insights into fundamental functions of cellular signaling networks. With the substantial acceleration in the range of applications, the role of optogenetics will broadly enable the examination of how genes, proteins, cells, and cellular connections modulate local and global network activity to develop complex tissue structures and encode behaviors. Such fundamental knowledge gained from optogenetic research will also potentially enable clinical therapeutics.
DZ and DS substantially contributed to the conception and design of the article and interpreting the relevant literature. DZ wrote the first draft of the manuscript. HJ and JC wrote sections of the manuscript. DS revised the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
This work was supported by NIH R01NS074831 (to DS), a Chan-Zuckerberg Biohub award (DS, DZ), the Tang Scholars program (JC), and the National Science Foundation Graduate Research Fellowship Program (HJ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cartoon graphics created with BioRender.com.
Allen, J. (2017). Application of patterned illumination using a DMD for optogenetic control of signaling. Nat. Methods 14:1114. doi: 10.1038/nmeth.f.402
Amatu, A., Sartore-Bianchi, A., Bencardino, K., Pizzutilo, E. G., Tosi, F., and Siena, S. (2019). Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 30, viii5–viii15. doi: 10.1093/annonc/mdz383
Aravanis, A. M., Wang, L. P., Zhang, F., Meltzer, L. A., Mogri, M. Z., Schneider, M. B., et al. (2007). An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J. Neural Eng. 4, S143–S156. doi: 10.1088/1741-2560/4/3/S02
Bedbrook, C. N., Yang, K. K., Robinson, J. E., Mackey, E. D., Gradinaru, V., and Arnold, F. H. (2019). Machine learning-guided channelrhodopsin engineering enables minimally invasive optogenetics. Nat. Methods 16, 1176–1184. doi: 10.1038/s41592-019-0583-8
Benzinger, D., and Khammash, M. (2018). Pulsatile inputs achieve tunable attenuation of gene expression variability and graded multi-gene regulation. Nat. Commun. 9:3521. doi: 10.1038/s41467-018-05882-2
Bonger, K. M., Rakhit, R., Payumo, A. Y., Chen, J. K., and Wandless, T. J. (2014). General method for regulating protein stability with light. ACS Chem. Biol. 9, 111–115. doi: 10.1021/cb400755b
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S., and Schaffer, D. V. (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252. doi: 10.1038/nmeth.2360
Bugaj, L. J., and Lim, W. A. (2019). High-throughput multicolor optogenetics in microwell plates. Nat. Protoc. 14, 2205–2228. doi: 10.1038/s41596-019-0178-y
Bugaj, L. J., Spelke, D. P., Mesuda, C. K., Varedi, M., Kane, R. S., and Schaffer, D. V. (2015). Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat. Commun. 6:6898. doi: 10.1038/ncomms7898
Casas-Mollano, J. A., Zinselmeier, M. H., Erickson, S. E., and Smanski, M. J. (2020). CRISPR-Cas activators for engineering gene expression in higher eukaryotes. CRISPR J. 3, 350–364. doi: 10.1089/crispr.2020.0064
Chang, K. Y., Woo, D., Jung, H., Lee, S., Kim, S., Won, J., et al. (2014). Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat. Commun. 5:4057. doi: 10.1038/ncomms5057
Christie, J. M., Gawthorne, J., Young, G., Fraser, N. J., and Roe, A. J. (2012). LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol. Plant. 5, 533–544. doi: 10.1093/mp/sss020
Christie, J. M., Salomon, M., Nozue, K., Wada, M., and Briggs, W. R. (1999). LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. U S A 96, 8779–8783. doi: 10.1073/pnas.96.15.8779
Cloix, C., Kaiserli, E., Heilmann, M., Baxter, K. J., Brown, B. A., O’Hara, A., et al. (2012). C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc. Natl. Acad. Sci. U S A 109, 16366–16370. doi: 10.1073/pnas.1210898109
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Deisseroth, K. (2015). Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225. doi: 10.1038/nn.4091
Duan, L., Hope, J. M., Guo, S., Ong, Q., François, A., Kaplan, L., et al. (2018). Optical activation of TrkA signaling. ACS Synth. Biol. 7, 1685–1693. doi: 10.1021/acssynbio.8b00126
Ewen-Campen, B., Yang-Zhou, D., Fernandes, V. R., González, D. P., Liu, L. P., Tao, R., et al. (2017). Optimized strategy for in vivo Cas9-activation in Drosophila. Proc. Natl. Acad. Sci. U S A 114, 9409–9414. doi: 10.1073/pnas.1707635114
Favory, J. J., Stec, A., Gruber, H., Rizzini, L., Oravecz, A., Funk, M., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in arabidopsis. EMBO J 28, 591–601. doi: 10.1038/emboj.2009.4
Gerhardt, K. P., Olson, E. J., Castillo-Hair, S. M., Hartsough, L. A., Landry, B. P., Ekness, F., et al. (2016). An open-hardware platform for optogenetics and photobiology. Sci. Rep. 6:35363. doi: 10.1038/srep35363
Gossen, M., Freundlieb, S., Bender, G., Müller, G., Hillen, W., and Bujard, H. (1995). Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769. doi: 10.1126/science.7792603
Guinn, M. T., and Balázsi, G. (2019). Noise-reducing optogenetic negative-feedback gene circuits in human cells. Nucleic Acids Res. 47, 7703–7714. doi: 10.1093/nar/gkz556
Harper, S. M., Neil, L. C., and Gardner, K. H. (2003). Structural basis of a phototropin light switch. Science 301, 1541–1544. doi: 10.1126/science.1086810
Hayashi-Takagi, A. (2017). Synapse pathology and translational applications for schizophrenia. Neurosci. Res. 114, 3–8. doi: 10.1016/j.neures.2016.09.001
Hayashi-Takagi, A., Takaki, M., Graziane, N., Seshadri, S., Murdoch, H., Dunlop, A. J., et al. (2010). Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat. Neurosci. 13, 327–332. doi: 10.1038/nn.2487
Hayashi-Takagi, A., Yagishita, S., Nakamura, M., Shirai, F., Wu, Y. I., Loshbaugh, A. L., et al. (2015). Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature 525, 333–338. doi: 10.1038/nature15257
Herwig, L., Rice, A. J., Bedbrook, C. N., Zhang, R. K., Lignell, A., Cahn, J. K. B., et al. (2017). Directed evolution of a bright near-infrared fluorescent rhodopsin using a synthetic chromophore. Cell Chem. Biol. 24, 415–425. doi: 10.1016/j.chembiol.2017.02.008
Humphreys, P. A., Woods, S., Smith, C. A., Bates, N., Cain, S. A., Lucas, R., et al. (2020). Optogenetic control of the BMP signaling pathway. ACS Synth. Biol. 9, 3067–3078. doi: 10.1021/acssynbio.0c00315
Jia, Y., Xu, R. G., Ren, X., Ewen-Campen, B., Rajakumar, R., Zirin, J., et al. (2018). Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc. Natl. Acad. Sci. U S A 115, 4719–4724. doi: 10.1073/pnas.1800677115
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-Guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jinek, M., East, A., Cheng, A., Lin, S., Ma, E., and Doudna, J. (2013). RNA-programmed genome editing in human cells. eLife 2:e00471. doi: 10.7554/eLife.00471
Kaberniuk, A. A., Shemetov, A. A., and Verkhusha, V. V. (2016). A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat. Methods 13, 591–597. doi: 10.1038/nmeth.3864
Kawano, F., Okazaki, R., Yazawa, M., and Sato, M. (2016). A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 12, 1059–1064. doi: 10.1038/nchembio.2205
Kawano, F., Suzuki, H., Furuya, A., and Sato, M. (2015). Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 6:6256. doi: 10.1038/ncomms7256
Kelava, I., and Lancaster, M. A. (2016). Stem cell models of human brain development. Cell Stem Cell 18, 736–748. doi: 10.1016/j.stem.2016.05.022
Kennedy, M. J., Hughes, R. M., Peteya, L. A., Schwartz, J. W., Ehlers, M. D., and Tucker, C. L. (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. doi: 10.1038/nmeth.1524
Khamo, J. S., Krishnamurthy, V. V., Chen, Q., Diao, J., and Zhang, K. (2019). Optogenetic delineation of receptor tyrosine kinase subcircuits in PC12 cell differentiation. Cell Chem. Biol. 26, 400–410.e3. doi: 10.1016/j.chembiol.2018.11.004
Kiecker, C., and Lumsden, A. (2012). The role of organizers in patterning the nervous system. Annu. Rev. Neurosci. 35, 347–367. doi: 10.1146/annurev-neuro-062111-150543
Kim, N., Kim, J. M., Lee, M., Kim, C. Y., Chang, K. Y., and Heo, W. D. (2014). Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem. Biol. 21, 903–912. doi: 10.1016/j.chembiol.2014.05.013
Kim, T.-I., McCall, J. G., Jung, Y. H., Huang, X., Siuda, E. R., Li, Y., et al. (2013). Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science 340, 211–216. doi: 10.1126/science.1232437
Konermann, S., Brigham, M. D., Trevino, A. E., Hsu, P. D., Heidenreich, M., Le, C., et al. (2013). Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476. doi: 10.1038/nature12466
Kramer, M. M., Lataster, L., Weber, W., and Radziwill, G. (2021). Optogenetic approaches for the spatiotemporal control of signal transduction pathways. Int. J. Mol. Sci. 22:5300. doi: 10.3390/ijms22105300
Krishnamurthy, V. V., Hwang, H., Fu, J., Yang, J., and Zhang, K. (2021). Optogenetic control of the canonical Wnt signaling pathway during Xenopus laevis embryonic development. J. Mol. Biol. 433:167050. doi: 10.1016/j.jmb.2021.167050
Krishnamurthy, V. V., Khamo, J. S., Mei, W., Turgeon, A. J., Ashraf, H. M., Mondal, P., et al. (2016). Reversible optogenetic control of kinase activity during differentiation and embryonic development. Development 143, 4085–4094. doi: 10.1242/dev.140889
Krishnamurthy, V. V., Turgeon, A. J., Khamo, J. S., Mondal, P., Sharum, S. R., Mei, W., et al. (2017). Light-mediated reversible modulation of the mitogen-activated protein kinase pathway during cell differentiation and Xenopus embryonic development. J. Vis. Exp. doi: 10.3791/55823
Krueger, D., Izquierdo, E., Viswanathan, R., Hartmann, J., Pallares Cartes, C., and De Renzis, S. (2019). Principles and applications of optogenetics in developmental biology. Development 146:dev175067. doi: 10.1242/dev.175067
Kyung, T., Lee, S., Kim, J. E., Cho, T., Park, H., Jeong, Y.-M., et al. (2015). Optogenetic control of endogenous Ca2+ channels in vivo. Nat. Biotechnol. 33, 1092–1096. doi: 10.1038/nbt.3350
Lanzafame, R. J., Blanche, R. R., Chiacchierini, R. P., Kazmirek, E. R., and Sklar, J. A. (2014). The growth of human scalp hair in females using visible red light laser and LED sources. Lasers Surg. Med. 46, 601–607. doi: 10.1002/lsm.22277
Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. doi: 10.1038/nature08446
Li, Y., Lee, M., Kim, N., Wu, G., Deng, D., Kim, J. M., et al. (2018). Spatiotemporal control of TGF-β signaling with light. ACS Synth. Biol. 7, 443–451. doi: 10.1021/acssynbio.7b00225
Li, H., Zhang, Q., Gu, Y., Wu, Y., Wang, Y., Wang, L., et al. (2020). Efficient photoactivatable Dre recombinase for cell type-specific spatiotemporal control of genome engineering in the mouse. Proc. Natl. Acad. Sci. U S A 117:33426. doi: 10.1073/pnas.2003991117
Locke, C., Machida, K., Tucker, C. L., Wu, Y., and Yu, J. (2017). Optogenetic activation of EphB2 receptor in dendrites induced actin polymerization by activating Arg kinase. Biol. Open. 6, 1820–1830. doi: 10.1242/bio.029900
Looger, L. L. (2011). Running in reverse: rhodopsins sense voltage. Nat. Methods 9, 43–44. doi: 10.1038/nmeth.1817
Lu, T., Aron, L., Zullo, J., Pan, Y., Kim, H., Chen, Y., et al. (2014). REST and stress resistance in ageing and Alzheimer’s disease. Nature 507, 448–454. doi: 10.1038/nature13163
Müller, K., Engesser, R., Timmer, J., Zurbriggen, M. D., and Weber, W. (2014). Orthogonal optogenetic triple-gene control in Mammalian cells. ACS Synth. Biol. 3, 796–801. doi: 10.1021/sb500305v
Mali, P., Yang, L., Esvelt Kevin, M., Aach, J., Guell, M., DiCarlo James, E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Mandel, G., Fiondella, C. G., Covey, M. V., Lu, D. D., Loturco, J. J., and Ballas, N. (2011). Repressor element 1 silencing transcription factor (REST) controls radial migration and temporal neuronal specification during neocortical development. Proc. Natl. Acad. Sci. U S A 108, 16789–16794. doi: 10.1073/pnas.1113486108
McFann, S., Dutta, S., Toettcher, J. E., and Shvartsman, S. Y. (2021). Temporal integration of inductive cues on the way to gastrulation. Proc. Natl. Acad. Sci. U S A 118:e2102691118. doi: 10.1073/pnas.2102691118
Metzger, D., Clifford, J., Chiba, H., and Chambon, P. (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. U S A 92, 6991–6995. doi: 10.1073/pnas.92.15.6991
Montgomery, K. L., Yeh, A. J., Ho, J. S., Tsao, V., Mohan Iyer, S., Grosenick, L., et al. (2015). Wirelessly powered, fully internal optogenetics for brain, spinal and peripheral circuits in mice. Nat. Methods 12, 969–974. doi: 10.1038/nmeth.3536
Morikawa, K., Furuhashi, K., de Sena-Tomas, C., Garcia-Garcia, A. L., Bekdash, R., Klein, A. D., et al. (2020). Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat. Commun. 11:2141. doi: 10.1038/s41467-020-16030-0
Nihongaki, Y., Furuhata, Y., Otabe, T., Hasegawa, S., Yoshimoto, K., and Sato, M. (2017). CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 14, 963–966. doi: 10.1038/nmeth.4430
Nihongaki, Y., Kawano, F., Nakajima, T., and Sato, M. (2015a). Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760. doi: 10.1038/nbt.3245
Nihongaki, Y., Yamamoto, S., Kawano, F., Suzuki, H., and Sato, M. (2015b). CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 22, 169–174. doi: 10.1016/j.chembiol.2014.12.011
Nikolopoulou, E., Galea, G. L., Rolo, A., Greene, N. D., and Copp, A. J. (2017). Neural tube closure: cellular, molecular and biomechanical mechanisms. Development 144, 552–566. doi: 10.1242/dev.145904
Oggu, G. S., Sasikumar, S., Reddy, N., Ella, K. K. R., Rao, C. M., Bokara, K. K., et al. (2017). Gene delivery approaches for mesenchymal stem cell therapy: strategies to increase efficiency and specificity. Stem Cell Rev. Rep. 13, 725–740. doi: 10.1007/s12015-017-9760-2
Packer, M. S., and Liu, D. R. (2015). Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394. doi: 10.1038/nrg3927
Paonessa, F., Criscuolo, S., Sacchetti, S., Amoroso, D., Scarongella, H., Pecoraro Bisogni, F., et al. (2016). Regulation of neural gene transcription by optogenetic inhibition of the RE1-silencing transcription factor. Proc. Natl. Acad. Sci. U S A 113, E91–E100. doi: 10.1073/pnas.1507355112
Park, S. I., Brenner, D. S., Shin, G., Morgan, C. D., Copits, B. A., Chung, H. U., et al. (2015). Soft, stretchable, fully implantable miniaturized optoelectronic systems for wireless optogenetics. Nat. Biotechnol. 33, 1280–1286. doi: 10.1038/nbt.3415
Pathak, G. P., Spiltoir, J. I., Höglund, C., Polstein, L. R., Heine-Koskinen, S., Gersbach, C. A., et al. (2017). Bidirectional approaches for optogenetic regulation of gene expression in mammalian cells using Arabidopsis cryptochrome 2. Nucleic Acids Res. 45:e167. doi: 10.1093/nar/gkx260
Penzes, P., Cahill, M. E., Jones, K. A., VanLeeuwen, J. E., and Woolfrey, K. M. (2011). Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293. doi: 10.1038/nn.2741
Polstein, L. R., and Gersbach, C. A. (2015). A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 11, 198–200. doi: 10.1038/nchembio.1753
Pomeroy, J. E., Nguyen, H. X., Hoffman, B. D., and Bursac, N. (2017). Genetically encoded photoactuators and photosensors for characterization and manipulation of pluripotent stem cells. Theranostics 7, 3539–3558. doi: 10.7150/thno.20593
Pozzi, D., Lignani, G., Ferrea, E., Contestabile, A., Paonessa, F., D’Alessandro, R., et al. (2013). REST/NRSF-mediated intrinsic homeostasis protects neuronal networks from hyperexcitability. EMBO J. 32, 2994–3007. doi: 10.1038/emboj.2013.231
Quail, P. H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. doi: 10.1038/nrm728
Raftopoulou, M., and Hall, A. (2004). Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23–32. doi: 10.1016/j.ydbio.2003.06.003
Redchuk, T. A., Kaberniuk, A. A., and Verkhusha, V. V. (2018). Near-infrared light-controlled systems for gene transcription regulation, protein targeting and spectral multiplexing. Nat. Protoc. 13, 1121–1136. doi: 10.1038/nprot.2018.022
Repina, N. A., McClave, T., Johnson, H. J., Bao, X., Kane, R. S., and Schaffer, D. V. (2020). Engineered illumination devices for optogenetic control of cellular signaling dynamics. Cell Rep. 31:107737. doi: 10.1016/j.celrep.2020.107737
Repina, N. A., Rosenbloom, A., Mukherjee, A., Schaffer, D. V., and Kane, R. S. (2017). At light speed: advances in optogenetic systems for regulating cell signaling and behavior. Annu. Rev. Chem. Biomol. Eng. 8, 13–39. doi: 10.1146/annurev-chembioeng-060816-101254
Rivera, V. M., Clackson, T., Natesan, S., Pollock, R., Amara, J. F., Keenan, T., et al. (1996). A humanized system for pharmacologic control of gene expression. Nat. Med. 2, 1028–1032. doi: 10.1038/nm0996-1028
Rosenbaum, D. M., Rasmussen, S. G. F., and Kobilka, B. K. (2009). The structure and function of G-protein-coupled receptors. Nature 459, 356–363. doi: 10.1038/nature08144
Rosenbloom, A. B., Tarczyński, M., Lam, N., Kane, R. S., Bugaj, L. J., and Schaffer, D. V. (2020). β-Catenin signaling dynamics regulate cell fate in differentiating neural stem cells. Proc. Natl. Acad. Sci. U S A 117, 28828–28837. doi: 10.1073/pnas.2008509117
Schlessinger, J. (2014). Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb Perspect Biol. 6:a008912. doi: 10.1101/cshperspect.a008912
Schuurmans, C., and Guillemot, F. (2002). Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr. Opin. Neurobiol. 12, 26–34. doi: 10.1016/s0959-4388(02)00286-6
Shao, J., Wang, M., Yu, G., Zhu, S., Yu, Y., Heng, B. C., et al. (2018). Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. U S A 115:E6722. doi: 10.1073/pnas.1802448115
Shcherbakova, D. M., Shemetov, A. A., Kaberniuk, A. A., and Verkhusha, V. V. (2015). Natural photoreceptors as a source of fluorescent proteins, biosensors and optogenetic tools. Annu. Rev. Biochem. 84, 519–550. doi: 10.1146/annurev-biochem-060614-034411
Shemesh, O. A., Tanese, D., Zampini, V., Linghu, C., Piatkevich, K., Ronzitti, E., et al. (2017). Temporally precise single-cell-resolution optogenetics. Nat. Neurosci. 20, 1796–1806. doi: 10.1038/s41593-017-0018-8
Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044. doi: 10.1038/nbt734
Sokolik, C., Liu, Y., Bauer, D., McPherson, J., Broeker, M., Heimberg, G., et al. (2015). Transcription factor competition allows embryonic stem cells to distinguish authentic signals from noise. Cell Syst. 1, 117–129. doi: 10.1016/j.cels.2015.08.001
Stanton, B. Z., Chory, E. J., and Crabtree, G. R. (2018). Chemically induced proximity in biology and medicine. Science 359:eaao5902. doi: 10.1126/science.aao5902
Steinbeck, J. A., Choi, S. J., Mrejeru, A., Ganat, Y., Deisseroth, K., Sulzer, D., et al. (2015). Optogenetics enables functional analysis of human embryonic stem cell-derived grafts in a Parkinson’s disease model. Nat. Biotechnol. 33, 204–209. doi: 10.1038/nbt.3124
Su, Y., Huang, X., Huang, Z., Huang, T., Li, T., Fan, H., et al. (2020). Early but not delayed optogenetic RAF activation promotes astrocytogenesis in mouse neural progenitors. J. Mol. Biol. 432, 4358–4368. doi: 10.1016/j.jmb.2020.06.020
Szenk, M., Yim, T., and Balázsi, G. (2020). Multiplexed gene expression tuning with orthogonal synthetic gene circuits. ACS Synth. Biol. 9, 930–939. doi: 10.1021/acssynbio.9b00534
Toettcher, J. E., Weiner, O. D., and Lim, W. A. (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. doi: 10.1016/j.cell.2013.11.004
Tucker, C. L., Vrana, J. D., and Kennedy, M. J. (2014). Tools for controlling protein interactions using light. Curr. Protoc. Cell Biol. 64:17.6.120. doi: 10.1002/0471143030.cb1716s64
Wang, X., Chen, X., and Yang, Y. (2012). Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266–269. doi: 10.1038/nmeth.1892
Wang, Q., Fan, H., Li, F., Skeeters, S. S., Krishnamurthy, V. V., Song, Y., et al. (2020). Optical control of ERK and AKT signaling promotes axon regeneration and functional recovery of PNS and CNS in Drosophila. eLife 9:e57395. doi: 10.7554/eLife.57395
Webster, N. J., Green, S., Jin, J. R., and Chambon, P. (1988). The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 54, 199–207. doi: 10.1016/0092-8674(88)90552-1
Wojtovich, A. P., and Foster, T. H. (2014). Optogenetic control of ROS production. Redox Biol. 2, 368–376. doi: 10.1016/j.redox.2014.01.019
Wu, Y. I., Frey, D., Lungu, O. I., Jaehrig, A., Schlichting, I., Kuhlman, B., et al. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108. doi: 10.1038/nature08241
Wu, D., Hu, Q., Yan, Z., Chen, W., Yan, C., Huang, X., et al. (2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484, 214–219. doi: 10.1038/nature10931
Wu, Z., Kan, S. B. J., Lewis, R. D., Wittmann, B. J., and Arnold, F. H. (2019). Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl. Acad. Sci. U S A 116:8852. doi: 10.1073/pnas.1901979116
Yang, J., Lee, J., Land, M. A., Lai, S., Igoshin, O. A., and St-Pierre, F. (2021). A synthetic circuit for buffering gene dosage variation between individual mammalian cells. Nat. Commun. 12:4132. doi: 10.1038/s41467-021-23889-0
Zhang, Z., Denans, N., Liu, Y., Zhulyn, O., Rosenblatt, H. D., Wernig, M., et al. (2021). Optogenetic manipulation of cellular communication using engineered myosin motors. Nat. Cell Biol. 23, 198–208. doi: 10.1038/s41556-020-00625-2
Zhang, F., Vierock, J., Yizhar, O., Fenno, L. E., Tsunoda, S., Kianianmomeni, A., et al. (2011). The microbial opsin family of optogenetic tools. Cell 147, 1446–1457. doi: 10.1016/j.cell.2011.12.004
Zhang, F., Wang, L. P., Boyden, E. S., and Deisseroth, K. (2006). Channelrhodopsin-2 and optical control of excitable cells. Nat. Methods 3, 785–792. doi: 10.1038/nmeth936
Zoltowski, B. D., Schwerdtfeger, C., Widom, J., Loros, J. J., Bilwes, A. M., Dunlap, J. C., et al. (2007). Conformational switching in the fungal light sensor vivid. Science 316, 1054–1057. doi: 10.1126/science.1137128
Keywords: optogenetics, cell signaling, spatiotemporal, gene regulation, neuroscience
Citation: Zhu D, Johnson HJ, Chen J and Schaffer DV (2022) Optogenetic Application to Investigating Cell Behavior and Neurological Disease. Front. Cell. Neurosci. 16:811493. doi: 10.3389/fncel.2022.811493
Received: 08 November 2021; Accepted: 04 January 2022;
Published: 22 February 2022.
Edited by:
Suneel Kateriya, Jawaharlal Nehru University, IndiaReviewed by:
Gabor Balazsi, Stony Brook University, United StatesCopyright © 2022 Zhu, Johnson, Chen and Schaffer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David V. Schaffer, c2NoYWZmZXJAYmVya2VsZXkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.