
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Neurosci. , 04 October 2021
Sec. Non-Neuronal Cells
Volume 15 - 2021 | https://doi.org/10.3389/fncel.2021.717209
This article is part of the Research Topic The Function of Schwann Cells in Peripheral Nervous System View all 11 articles
 Bo He1,2*†
Bo He1,2*† Vincent Pang2†
Vincent Pang2† Xiangxia Liu2,3†
Xiangxia Liu2,3† Shuqia Xu2
Shuqia Xu2 Yi Zhang2
Yi Zhang2 David Djuanda2
David Djuanda2 Guanggeng Wu2
Guanggeng Wu2 Yangbin Xu1*
Yangbin Xu1* Zhaowei Zhu1*
Zhaowei Zhu1*To preliminarily explore the primary changes in the expression of genes involved in peripheral nerve processes, namely, regeneration, angiogenesis, and the immune response, and to identify important molecular therapeutic targets, 45 Sprague-Dawley (SD) rats were randomly divided into a control group and an injury group. In the injury group, tissue samples were collected at 4 and 7 days after the injury for next-generation sequencing (NGS) analysis combined with gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and Venn diagram construction to identify the differentially expressed mRNAs (DEmRNAs) associated with regeneration, angiogenesis, and the immune response of the nerve. The expression of genes in the distal and proximal ends of the injured nerve after injury was analyzed by qRT-PCR. NGS revealed that compared with the control group, the injury group had 4020 DEmRNAs 4 days after injury and 3278 DEmRNAs 7 days after injury. A bioinformatics analysis showed that C-C chemokine receptor type 5 (CCR5), Thy1 cell surface antigen (Thy1), Notch homolog 1 (Notch1), and semaphorin 4A (Sema4A) were all associated with regeneration, angiogenesis, and the immune response of the nerve at both 4 and 7 days after injury, but qPCR revealed no significant difference in the expression of Thy1 (P = 0.29) or Sema4A (P = 0.82) in the proximal end, whereas a significant difference was observed in CCR5 and Notch1 (P < 0.05). The trend in the Notch1 change was basically consistent with the RNA-seq result after injury, which implied its indispensable role during endothelial cell proliferation and migration, macrophage recruitment, and neurotrophic factor secretion.
Peripheral nerve regeneration is a complex process. Active Schwann cells (SCs) play an important role in nerve repair process. They not only allow the myelin sheath debris of the axon that disintegrates due to Wallerian degeneration to be cleared quickly but also attract immunocytes migrating to the injured site; this allows the regenerating axons to grow smoothly into the bands of Büngner and eventually improves the efficacy of neurological functional recovery (Bozkurt et al., 2009). We also found in our research that efficacy of peripheral nerve defect repair is related to the blood supply within the graft (Zhu et al., 2017). Vascularized nerve grafting maintains the continuity of the blood supply during nerve transplantation, ensures the activity of SCs, and leads to continuous neurotrophic factor secretion. Due to local ischemia, SCs become necrotic and replaced by fibroblasts, which results in an excessively low concentration of neurotrophic factors secreted by local SCs and slow remyelination of regenerating axons; this is unfavorable for the ingrowth and maturation of regenerating axons (He et al., 2014). Many angiogenic factors are involved in the repair process. Among them, endothelial growth factor (VEGF) and angiopoietin (Ang) are the two most important proangiogenic factors, as they act synergistically in angiogenesis. Studies have shown that VEGF promotes axonal growth through angiogenesis (Ebenezer et al., 2012). In a previous study involving in vivo experiments in rats, we found that COMP-Ang1, an angiogenic factor, was able to promote early angiogenesis in nerve repair scaffolds and the proliferation of SCs in peripheral nerves (Qiu et al., 2015). Besides, immune response provided significant assistance, especially during the early phrase after nerve injury. The myelin sheath debris is phagocytosed and removed by macrophages, which are brought in by the blood supply, and local SCs. At the same time, immune cells also secrete some cytokines that promote proximal nerve fiber growth to form growth cones (Lindborg et al., 2018).
Although axonogenesis, angiogenesis and the immune response are three different processes, they have numerous interactions after nerve injury (Cattin et al., 2015). Many cytokines and genes are involved in the mutual regulation of regeneration, angiogenesis, and the immune response in the peripheral nerve, but the specific mechanism remains unclear. To further study the molecular mechanisms of regeneration, angiogenesis, and the immune response in the nerve after peripheral nerve injury, this study aimed to perform transcriptome analysis of injured nerves in a rat model of sciatic nerve crush injury induced by clamping. We preliminarily explored the primary changes in gene expression that are involved in regeneration, angiogenesis, and the immune response of the peripheral nerve and provide a scientific basis for more effective treatment of peripheral nerve injury by identifying important molecular therapeutic targets.
Forty-Five healthy male Sprague-Dawley (SD) rats weighing approximately 150–200 g were chosen and randomly separated into three groups as follows: control, 4-day injury, and 7-day injury groups (15 rats per group). This study was approved by the Experimental Animal Administration Committee of Sun Yat-sen University. Efforts were taken to minimize animal suffering during the experiment. The study design is presented in Figure 1.
Experimental rats (injured for 4 days and injured for 7 days) were anesthetized by intraperitoneal injection of 10% chloral hydrate (0.3 ml/100 g) (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) and were subjected to sciatic nerve crush injury according to a previously published procedure (Zhu et al., 2017). Under aseptic conditions, the skin of the left leg was cut parallel to the femur, and the sciatic nerve was exposed by splitting the superficial gluteus muscle. The middle segment of the left sciatic nerve was clamped with hemostatic forceps for approximately 30 s. Subsequently, they were released, and the wound was closed, after which the animals were kept in a quiet and comfortable environment with a temperature between 20 and 25°C and humidity between 50 and 65%, and enough food and water were ensured. We also offered light for 14 h to maintain the normal circadian rhythm of each animal. The condition of the animals and the wounds were observed regularly.
Three rats were randomly selected from each group. In the 4-day and 7-day injury groups, the nerve was completely cut at the lesion site at each selected observation time, and samples were obtained 1 cm proximal to the lesion and 1 cm distal to the lesion. The nerve samples were fixed in 4% paraformaldehyde at 4°C for 24 h. Subsequently, the nerve was placed in 20% sucrose for 24 h for the first round of dehydration, which was followed by placement in 30% sucrose for 24 h for the second round of dehydration. Ten-micrometer-thick sections of the middle segment of the nerve were cut using a cryostat and placed on a coverslip; immunofluorescence staining was then performed with a rabbit anti-mouse NF-200 antibody (Sigma-Aldrich, Tokyo, Japan, 1:400), a CD31 antibody (Sigma-Aldrich, Tokyo, Japan, 1:400), and a FITC-conjugated goat anti-rabbit IgG secondary antibody (TCS Biologicals, Botolph Clayton, United Kingdom; 1:200). Antibodies were diluted in PBS containing 3% rat serum, 3% goat serum, and 0.02% sodium azide (Sigma-Aldrich, Tokyo, Japan) to reduce background staining.
For image study, photos were taken under the same condition as previously described (Zhu et al., 2015). For the determination of NF200 and CD31 expression, the integrated optical density (IOD) of positive staining was assessed using Image-Pro Plus. For each animal and for each stain, calculations were made on three randomly chosen sections, and the results were averaged.
Two rats were randomly selected from each group, and the injured nerves were harvested as samples for next-generation RNA sequencing (NGS). All sequencing programs were performed by Shanghai Biotechnology Corporation (Shanghai, China).
During transcriptome sequencing, to enable comparability of gene expression levels among different genes and different samples, the reads were converted into FPKM (fragments per kilobase of transcript per million mapped reads) to normalize the gene expression level (Mortazavi et al., 2008). We first applied Stringtie (version: 1.3.0) (Pertea et al., 2015; Pertea et al., 2016) to count the number of fragments for each gene after HisAT2 alignment, and then we used the TMM (trimmed mean of M values) (Robinson and Oshlack, 2010) method for normalization; finally, we used Perl script to calculate the FPKM value for each gene.
EdgeR (Robinson et al., 2010) was used to perform the differential gene analysis between samples. After the P-value was derived, multiple hypothesis testing correction was performed, and the threshold of the p-value was determined by controlling the FDR (false discovery rate) (Benjamini et al., 2001); the corrected p-value is the q-value. Moreover, we calculated the fold differential expression based on the FPKM values, namely, the fold change. Differential gene screening conditions were set at a q-value ≤ 0.05 and a fold change ≥ 2. Both gene ontology (GO) analysis1 and KEGG pathway analysis2 were performed to determine the roles of the differentially expressed mRNAs. Fisher’s exact test was used to test the significance of GO terms and KEGG pathway identifier enrichment in the differentially expressed gene list. RNA sequencing and bioinformatics analyses were carried out by Shanghai Biotechnology Corporation (Shanghai, China).
We defined axonogenesis, angiogenesis, and immune response by GO categories, whose item numbers were 0007409, 0001525, and 0006955, respectively. The upregulated differentially expressed mRNAs (DEmRNAs) related to these three items were analyzed, and a Venn diagram was generated by web tools3.
Ten rats were randomly selected from each group. In the 4-day and 7-day injury groups, the nerve was completely transected at the lesion site, and samples were obtained 1 cm proximal to the lesion and 1 cm distal to the lesion. Quantitative analysis of the targeted mRNA expression was performed using real-time RT-PCR as previously described (Djuanda et al., 2021). The RNA concentration and purity were detected using a NanoDrop 2000, and RNA with an excessively high concentration was diluted appropriately to a final concentration of 200 ng/μl. One microgram of each specimen was used for RNA reverse transcription using the RevertAid First Strand cDNA Synthesis Kit (Thermo, MA, United States). An appropriate amount of cDNA was amplified using FastStart Universal SYBR Green Master Mix (Roche, Basel, Switzerland) in a fluorescence quantitative PCR machine (StepOne Plus, Thermo, MA, United States). For the specific procedure, please refer to the product manual. This experiment was repeated three times for each specimen. GAPDH expression was used to normalize the expression of mRNAs, and information about the genes to be tested and their primers is shown in Table 1. The specificity of real-time PCR was confirmed via routine agarose gel electrophoresis and melting-curve analysis. The 2–ΔΔ Ct method was used to calculate relative gene expression. GAPDH served as the reference gene.
Network software built by Cytoscape (version: 3.7.14) with the GeneMania plug-in (version: 3.5.15) was used to analyze the interactions between genes sorted by the Venn diagram. Rattus norvegicus was chosen, and the parameter was set to the top 20 related genes using automatic weight. Through the score ranking of each gene, the network of genes interacting with the sorted genes was obtained.
The RT-PCR data are presented as the mean ± SD as describedpreviously (Liu et al., 2018). SPSS 16.0 software (SPSS, Chicago, IL, United States) was used to analyze quantitative RT-PCR results. Statistical analysis was performed with Kruskal–Wallis test to determine the significant differences between the groups. α < 0.05 was considered statistically significant.
The sciatic nerves of SD rats in the control group and those of SD rats at 4 and 7 days after injury were obtained to examine the expression of the nerve axon marker NF-200 and the vascular endothelial cell marker CD31. The results showed that NF200 expression was negatively correlated with time and that its expression gradually changed from a line-like distribution to a punctate scattered distribution, as observed by microscopy; however, CD31 expression was positively correlated with time, and multiple small luminal structures were observed inside the nerve at day 7 (see Figure 2). Additionally, we found that compared to control group, the injury group had a decrease in NF200-positive fibers after injury, and on day 7, most NF200-positive staining had disappeared. These phenomena suggested that Wallerian degeneration of sciatic nerves after injury was ongoing and that our crush models were established successfully. In contrast to the transection models, the crush models still had some NF200 + axon fibers after injury, which were evident by immunofluorescence staining at day 7.
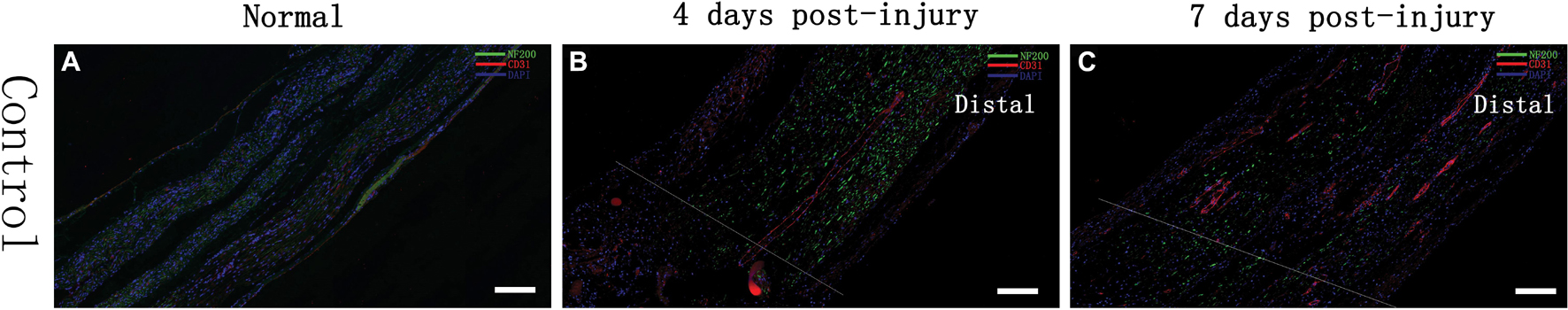
Figure 2. Immunofluorescence assessment of the sciatic nerve in SD rats Changes in the expression of an axon-associated protein (NF-200) and a vascular endothelial cell protein (CD31) in the early stage of a contusion injury (4 days, 7 days) compared with controls. The scale bar represents 200 μm. (A) Normal rat sciatic nerve; (B) 4 days after injury, rat sciatic nerve injury and beyond; (C) 7 days after injury, rat sciatic nerve injury and beyond.
Analysis of IOD demonstrated that the expression of NF200 was higher in control group (12262 ± 2863), and decreased gradually after injury in day 4 (7478 ± 1603) and day 7 (4061 ± 2933). Same trend appeared in CD31 expression, which showed highest in control group (12043 ± 2849), then decrease in day 4 (1374 ± 848) and day 7 (3595 ± 2011).
We used NGS to analyze the expression of genes in the transcriptome at 4 and 7 days after clamp-induced sciatic nerve crush injury in SD rats, and the distribution of differentially expressed mRNAs (DEmRNAs) is shown in heat maps (Figure 3). In all, 30,957 mRNAs were detected by RNA-seq and were screened according to the criteria of a fold change ≥ 2.0 and q < 0.05. Compared with the control rats, rats examined 4 days after clamp-induced crush injury had 4020 DEmRNAs, with the expression of 2500 being upregulated and 1520 being downregulated. Four days after injury, 137 mRNAs were activated in the experimental group, and of these, the most obvious change was in Hmga2 (FPKM = 0 vs. 16.42, q < 0.05). Sixty-four genes were silenced, and the greatest change was seen in fgf4 (FPKM = 4.00 vs. 0, q < 0.05). The top 10 DEmRNAs that were activated or silenced are shown in Table 2. In addition, among the mRNAs that were expressed in both the control and experimental groups, the mRNA with the most significant increase was Tyrp1 (log2FC = 9.75, q < 0.05), whereas the one with the largest decrease was Sec14l5 (log2FC = −11.16, q < 0.05). The top 10 DEmRNAs that were the most significantly upregulated or downregulated are shown in Table 3. Seven days after injury, there were 3278 DEmRNAs; expression of 1977 mRNAs was upregulated, and expression of 1301 mRNAs were downregulated. Among them, 88 mRNAs were activated, and the most obvious change was seen in Apoc1 (FPKM = 0 vs. 5.96, P < 0.05); 35 mRNAs were silenced, and the most obvious change was seen in Hapln2 (FPKM = 4.26 vs. 0, P < 0.05). The top 10 DEmRNAs that were activated or silenced are shown in Table 4. In addition, among the mRNAs that were expressed in both the control and the experimental groups, the mRNA with the largest increase was tyrosinase-related protein 1 (Tyrp1) (log2FC = 10.92, P < 0.05), whereas the one with the largest decrease was Rn60_10_0890.5 (log2FC = −8.63, P < 0.05). The top 10 DEmRNAs that were the most significantly upregulated or downregulated are shown in Table 5.
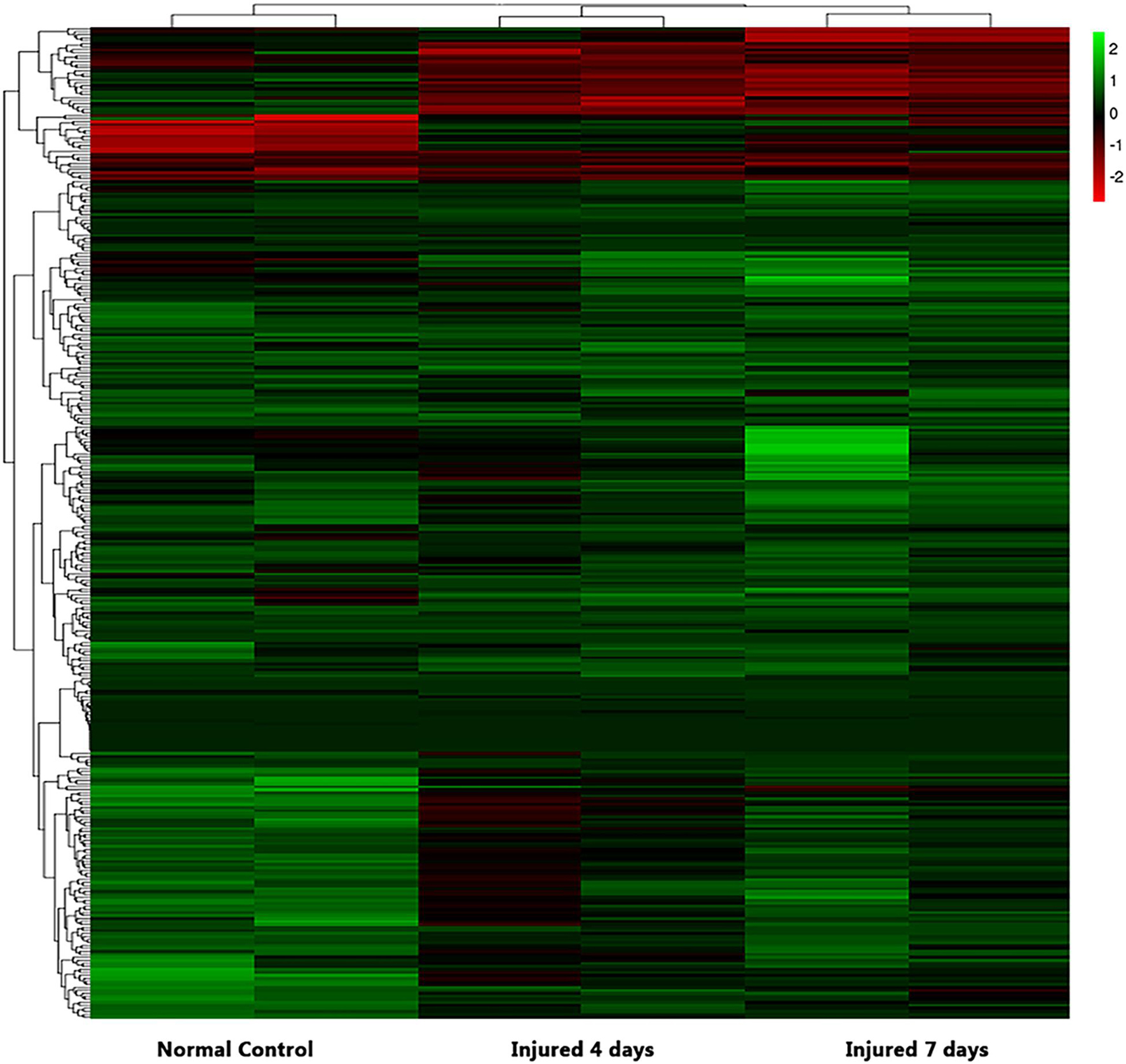
Figure 3. Heatmap of differential mRNA expression after RNA sequencing at different times (4 and 7 days after injury) soon after sciatic nerve contusion in SD rats. Black indicates no significant fold change in mRNA expression between the groups, green indicates increased expression, and red indicates decreased expression.
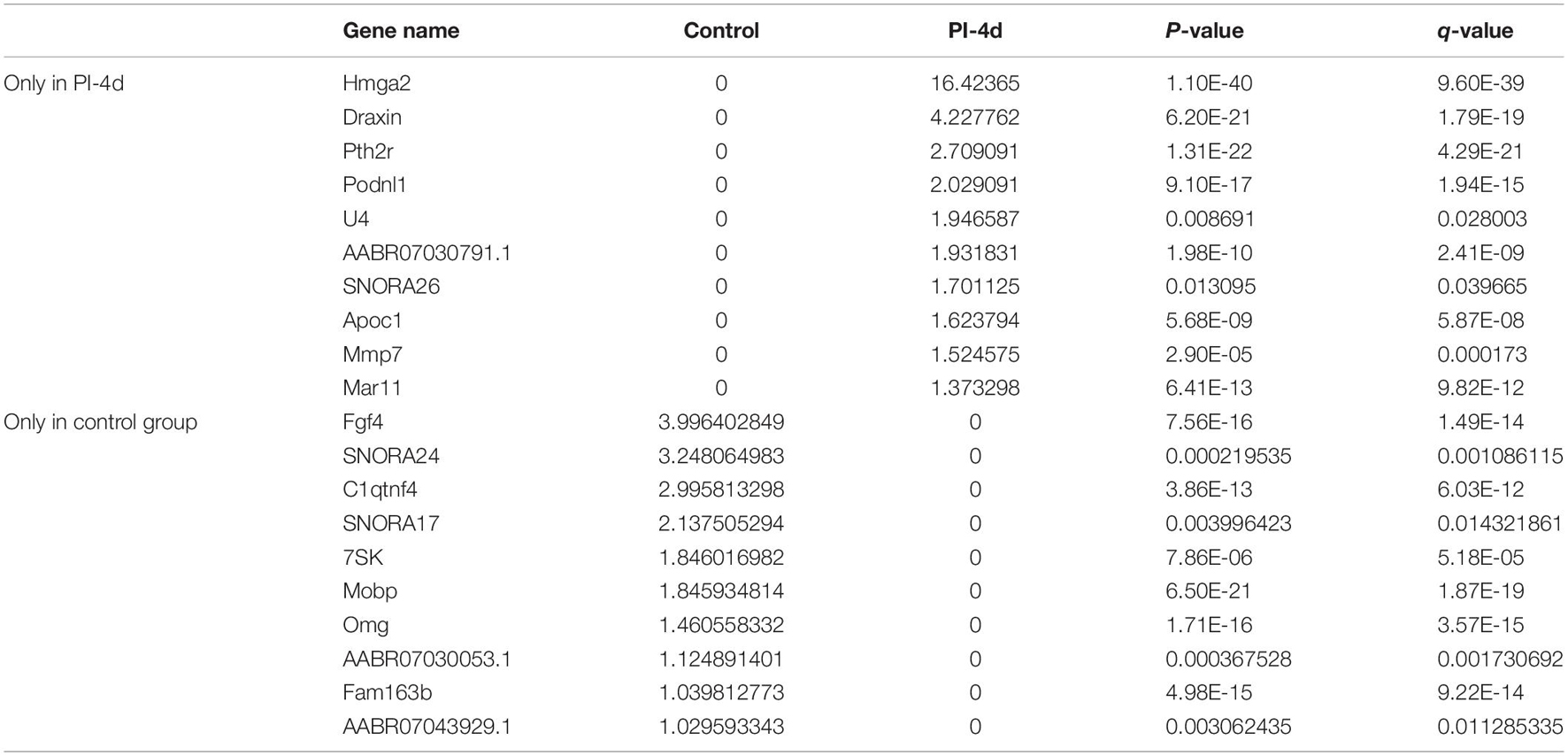
Table 2. Top 10 DEmRNAs only expressed 4 days after crush injury (Post-Injury 4 days, PI-4 d) or only expressed in normal sciatic nerves.
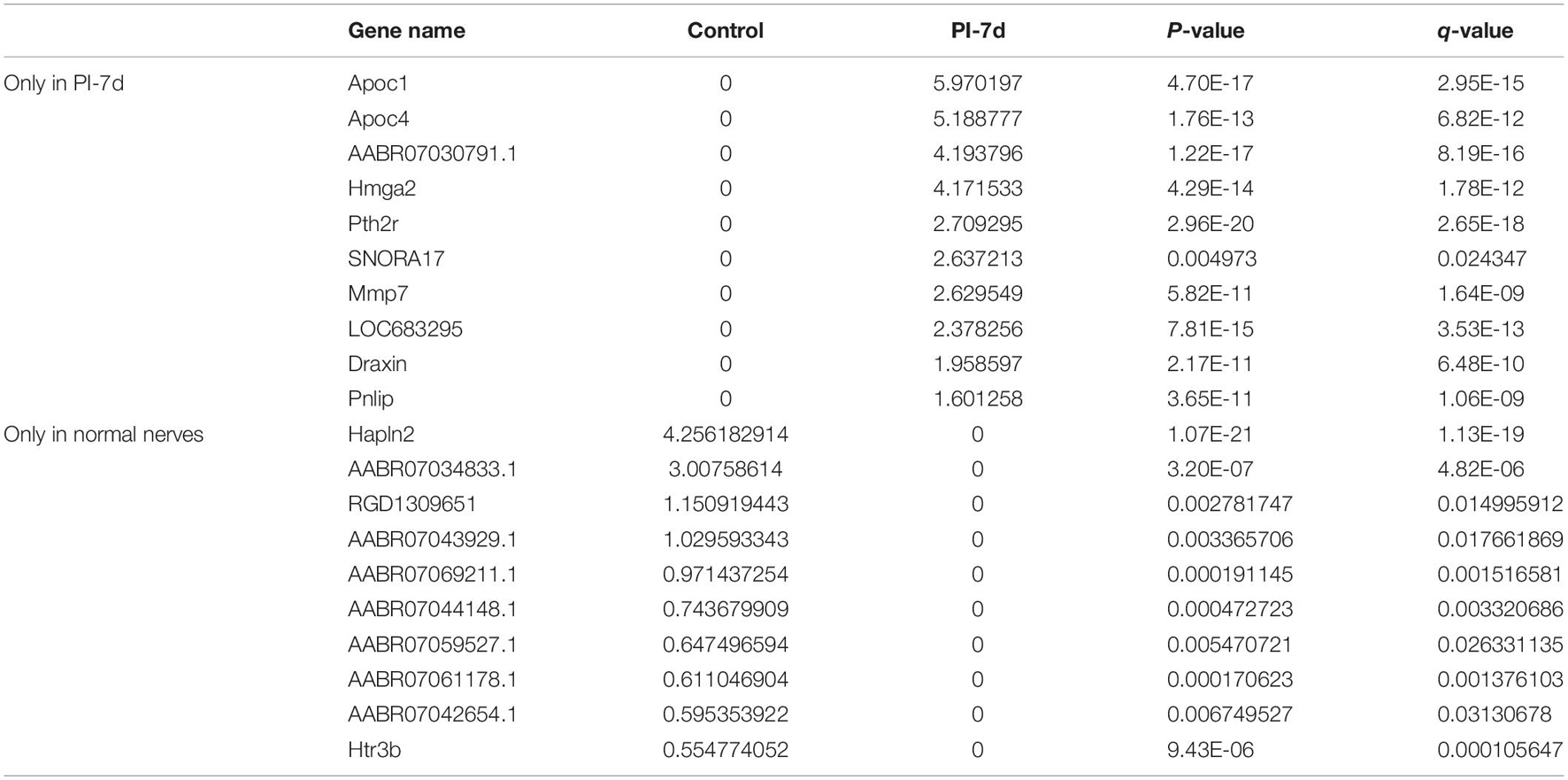
Table 4. Top 10 DEmRNAs only expressed 7 days after crush injury (Post-Injury 7 days, PI-7d) or only expressed in normal sciatic nerves.
Clustering analysis of DEmRNAs can help to identify the function of unknown transcripts or the unknown function of known transcripts by gathering similar expression patterns or similar genes to a class (Yao et al., 2012). The results showed that at 4 days after injury, DEmRNAs were enriched mainly in mitotic chromosome condensation [Rich factor (RF) = 5.52, q < 0.05], protein binding involved in cell-cell adhesion (RF = 5.52, q < 0.05), and condensed chromosome outer kinetochore (RF = 5.06, q < 0.05). However, 7 days after injury, DEmRNAs were enriched mainly in the G protein-coupled receptor signaling pathway involved in heart processes (RF = 6.14, q < 0.05), condensed chromosome outer kinetochores (RF = 5.62, q < 0.05), and MHC class II protein complexes (RF = 5.62, q < 0.05). The GO enrichment diagram and the distribution of GO entries enriched in the top 10 are shown in Figures 4A1,B1 and Table 6.
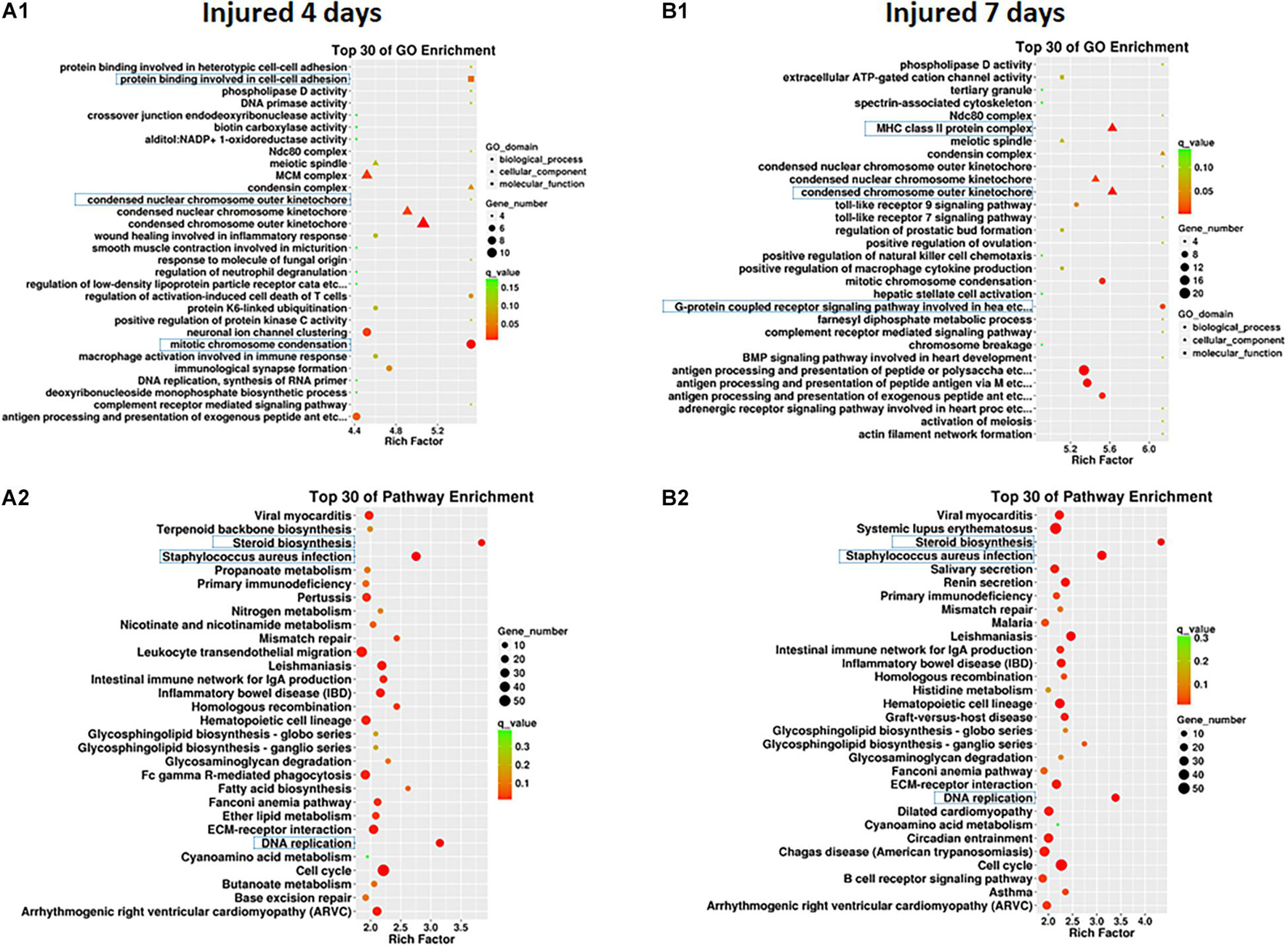
Figure 4. GO enrichment map (A1,B1) and KEGG pathway enrichment map (A2,B2) of differentially expressed mRNAs in SD rats 4 and 7 days after sciatic nerve injury compared with rats without injury during the early stage after sciatic nerve contusion. (A1) GO gene enrichment map at 4 days after injury; (B1) GO gene enrichment map at 7 days after injury. The different shapes in the figure represent different categories, with circles representing biological processes, triangles representing cellular components, and squares representing molecular functions. A larger shape area indicates a greater number of DEmRNAs in that category. The significant differences in the distribution of DEmRNAs in different subcategories between the two groups are indicated by different colors, where green indicates q-values > 0.15, yellow indicates that q-values are between 0.10 and 0.15, orange indicates that q-values are between 0.05 and 0.10, and red indicates q-values < 0.05. The test criterion was set at 0.05. (A2) KEGG gene enrichment map at 4 days after injury; (B2) KEGG gene enrichment map at 7 days after injury. A larger graphical area in the figure indicates a higher number of DEmRNAs in that channel. The significant differences in the distribution of DEmRNAs in different channels between the two groups are indicated by different colors, where green indicates q-values > 0.3, yellow indicates that q-values are between 0.2 and 0.3, orange indicates that q-values are between 0.1 and 0.2, and red indicates q-values < 0.1. The test criterion was 0.1.

Table 6. The classification corresponding to the top 10 DEmRNAs at 4 and 7 days after sciatic nerve crush injury in SD rats in the GO analysis.
Pathway enrichments was performed by means of KEGG analysis to determine the pathways in which DEmRNAs are involved during the early stage after injury to understand the molecular interactions after injury. The results showed that the top three pathways enriched were steroid biosynthesis (3.84 vs. 4.33, q < 0.05), DNA replication (3.15 vs. 3.39, q < 0.05), and Staphylococcus aureus infection (2.75 vs. 3.11, q < 0.05) at both 4 and 7 days after injury. The KEGG enrichment diagram and the distribution of the top 10 GO entries enriched are shown in Figures 4A2,B2 and Table 7.
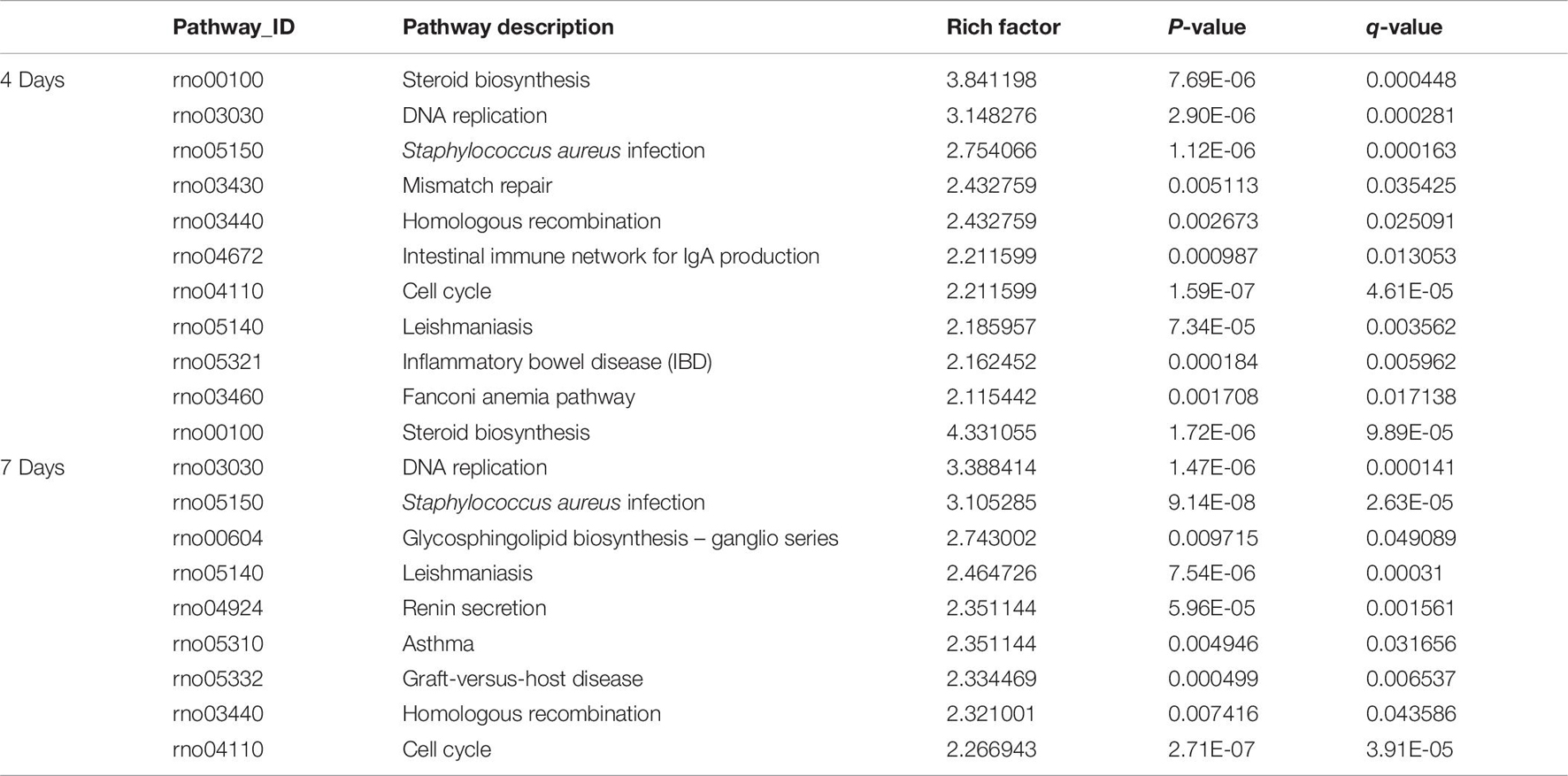
Table 7. The classification corresponding to the top 10 DEmRNAs at 4 and 7 days after sciatic nerve crush injury in SD rats in the KEGG analysis.
At 4 days after injury, 6 DEmRNAs were found to be involved in both nerve regeneration and angiogenesis: semaphorin 3E (Sema3e), fibroblast growth factor 8 (Fgf8), neural cell adhesion molecule (Ncam), ephrin type-B receptor 3 (Ephb3), Sonic hedgehog (Shh), and TNF receptor superfamily member 12A (Tnfrsf12a). At 7 days after injury, increased expression in only Sema3e, Ephb3, Shh, and Tnfrsf12a was maintained (P < 0.05). At 4 and 7 days after injury, 2 DEmRNAs were found to be involved in both regeneration and the immune response of the nerve: nerve growth factor receptor (Ngfr) and Sema7a (P < 0.05).
A Venn diagram was used to perform the three-intersection analysis of the DEmRNA-associated signaling pathways of regeneration, angiogenesis, and the immune response of the nerve, and the intersection of the upregulated DEmRNAs at 4 and 7 days after injury is shown in Figure 5. The results showed that the intersecting mRNAs of regeneration, angiogenesis, and the immune response of the nerve at 4 and 7 days after injury included C-C chemokine receptor type 5 (Ccr5), Thy1 cell surface antigen (Thy1), Notch homolog 1 (Notch1), and Sema4A, and the expression levels of all but Thy1 were significantly increased compared with the control group (P < 0.05).
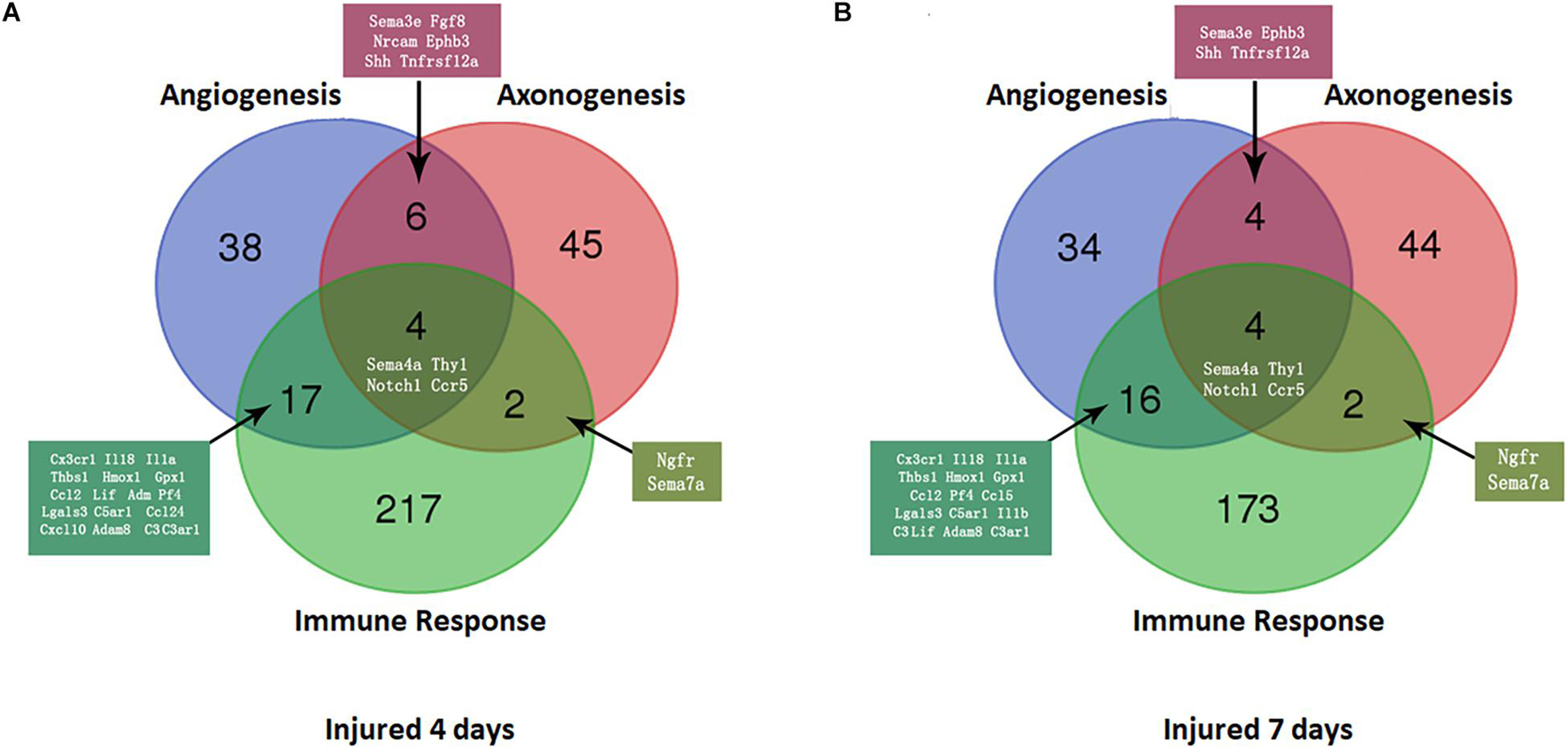
Figure 5. Venn diagram of mRNAs with increased expression at the intersection of angiogenesis, axonal regeneration, and the immune response at different time points. (A) 4 days after sciatic nerve crush injury; (B) 7 days after sciatic nerve crush injury. The number and names of intersecting genes are indicated in the figure.
As shown in Figure 6, In the proximal ends of the injured nerve between the experimental group and the control group, the qPCR results revealed no significant difference in the expression of Thy1 (P > 0.05) or Sema4A (P > 0.05), whereas a significant difference was observed in CCR5 and Notch1 (P < 0.05); however, the trends in CCR5 was not completely consistent with the RNA-seq results. At 4 days after injury, the expression of CCR5 was significantly reduced (FC = 0.66 ± 0.02, P < 0.01), gradually increased and then remained lower than that of the control group at day 7 (FC = 0.80 ± 0.03, P < 0.01). The trend in the Notch1 change was basically consistent with the RNA-seq result after injury. In the proximal ends of injured nerves, Notch1 expression showed significant changes 4 days after the operation and continued to increase at day 7.
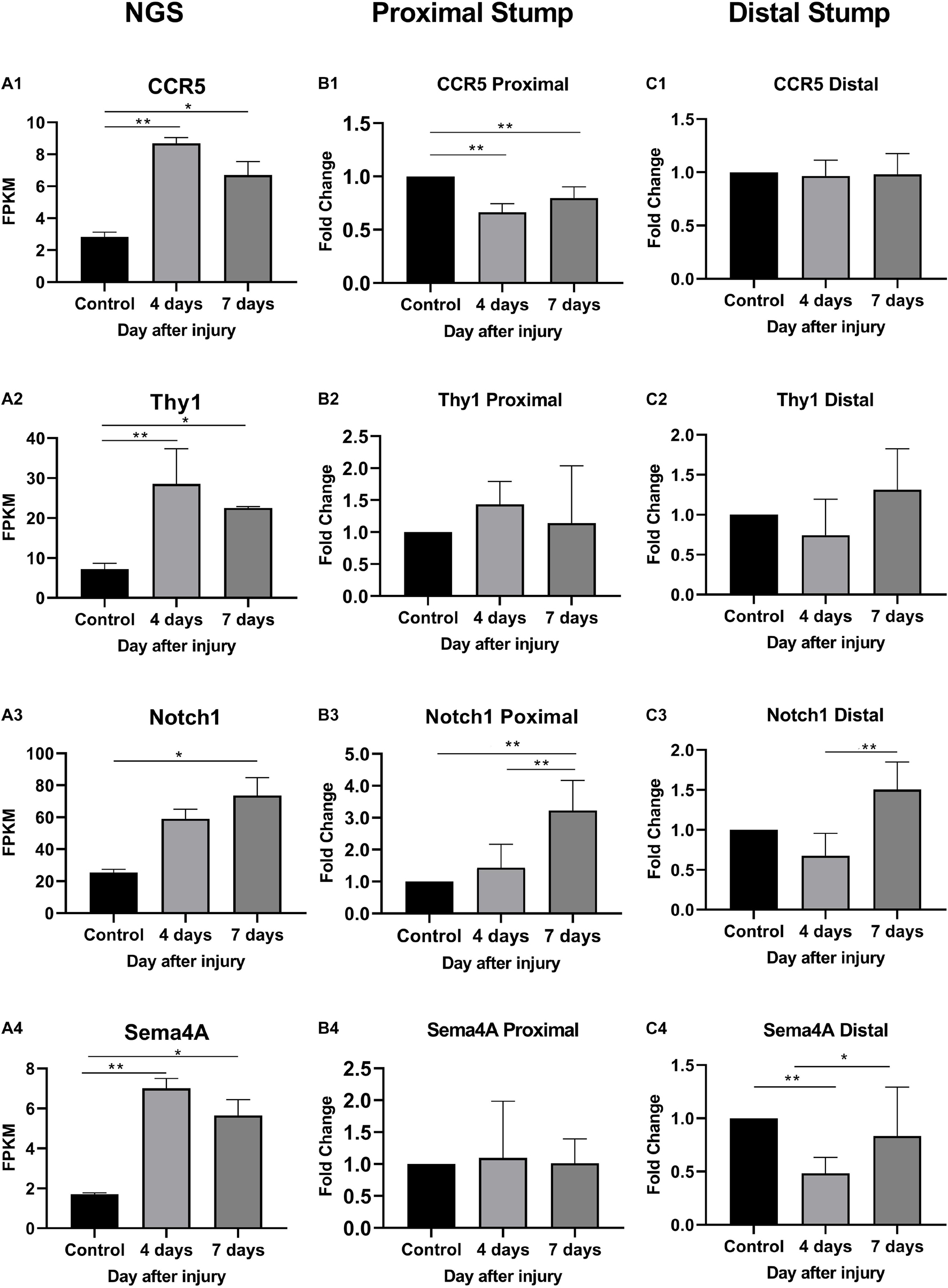
Figure 6. Expression levels of DEmRNAs common to angiogenesis, axonal regeneration and immune response. (A) NGS results; (B) qRT-PCR expression of genes in proximal part after injury; (C) qRT-PCR expression of genes in distal part after injury. (A1–C1) CCR5 expression; (A2–C2) Thy1 expression; (A3–C3) Notch1 expression; (A4–C4) Sema4a expression. * denotes P < 0.05 compared with the control group, and ** denotes P < 0.01 compared with the control group.
In distal ends of the injured nerves, the results of the three-intersection analyses in the control group, the 4-day injury group, and the 7-day injury group showed no significant difference in the expression of CCR5 (P > 0.05), but significant differences were observed in the expression of Thy1, Notch1, and Sema4A between the groups (P < 0.05). Compared with that in the control group, the expression of Thy1 was not significantly different in the day-4 (FC = 0.74 ± 0.16) or day-7 injury group (FC = 1.31 ± 0.18, P > 0.05), but Thy1 expression was significantly higher at day 7 than at day 4 (P < 0.05). Four days after injury, Notch1 expression was significantly lower than that in the control group (FC = 0.68 ± 0.10, P < 0.01); however, Notch1 expression then gradually increased and became significantly higher than that in the control group by day 7 (P < 0.01). Four days after injury, Sema4A expression was significantly reduced compared with that in the control group (FC = 0.48 ± 0.05, P < 0.01), and its expression gradually increased thereafter (FC = 0.86 ± 0.16, P > 0.05).
The gene network of the mutual intersections among the 21 genes with the highest correlation with Ccr5, Thy1, Notch1, and Sema4A is shown in Figure 7 and Supplementary Tables 1, 2. The results revealed a total of 47 links, of which co-expression accounted for 42.55%, predicted links accounted for 29.79%, and physical interactions accounted for 8.51%. Higher scores of the nodes indicate better correlation with the screened genes. The three nodes with the highest scores were X-linked inhibitor of apoptosis (Xiap, score: 0.012), F-box and WD repeat domain containing 7 (Fbxw7, score: 0.009) and Jagged 2 (Jag2, score: 0.008), and 8 were associated with the Notch signaling pathway.
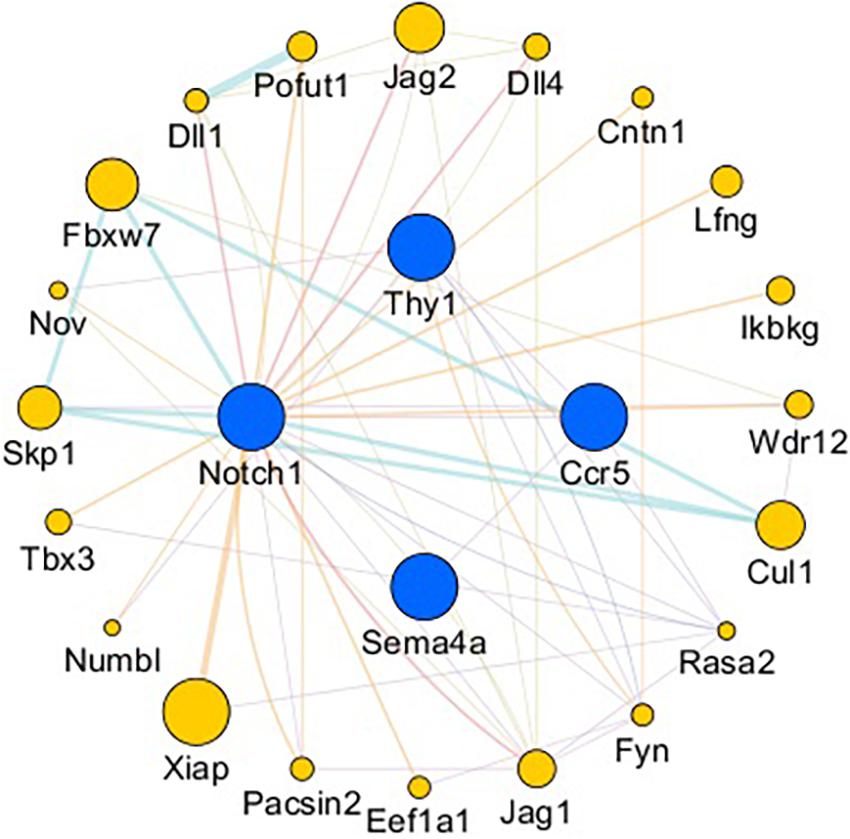
Figure 7. Gene interaction network. Blue nodes: Genes related to axonogenesis, angiogenesis, and the immune response. Yellow nodes: Genes that interacted with CCR5, Thy1, Notch1, and Sema4A. The node size is based on the score. The larger the node is, the higher the score will be.
Axonogenesis, angiogenesis and the immune response have numerous interactions after nerve injury (Cattin et al., 2015). In order to better verified cell clusters involved in the nerve injury and repair process, Kalinski et al. (2020) performed single-cell RNA sequencing in mice sciatic nerves after crush injury models. After being injured for 3 days, 24 different cell clusters were identified. The most prominently featured are immune cells, especially innate immune cells (Itgam/CD11b+). Other abundantly featured cell types include mesenchymal progenitor cells, Schwann cells and nerve vasculature cells. The early pathophysiological changes after peripheral nerve injury are reflected in Wallerian degeneration of the distal end and part of the proximal end of the nerve. The myelin sheath debris is phagocytosed and removed by macrophages, which are brought in by the blood supply, and local SCs. As we found in our research that at the very beginning after injury, NF-200 positive fibers were fragmented and finally disappeared, while CD31 positive vasculature was immediately established. At the same time, cells also secrete some cytokines that promote proximal nerve fiber growth to form growth cones. Only when the proximal regenerating axons pass through the site of injury, regenerate distally, and innervate the target organ is the process of nerve regeneration ultimately completed (Sayad Fathi and Zaminy, 2017). During this process, a series of signaling pathways regulate the local microenvironment inside the nerve until nerve regeneration is completed (Kato et al., 2013; Liu et al., 2015, 2016; Figure 8).
In the adult mammalian peripheral nervous system, injured axons maintain the capacity to regenerate spontaneously and provide the possibility for functional recovery post-peripheral nerve injury (Dickson, 2002; Abe and Cavalli, 2008). Peripheral nerve regeneration is accompanied by activation of various complicated molecular pathways and cellular events, which are motivated by differentially expressed genes and significantly changed pathways postinjury. Li et al. (2013) previously profiled global mRNA expression changes in proximal nerve segments, focusing on dynamic changes in the crucial biological processes, and they noted the time-dependent expression of key regulatory genes after sciatic nerve transection. The peripheral nerve injury response leads to the upregulation of various genes required for initiating, guiding, and sustaining axonal growth. Guo et al. (2018) tried to identify specific genes related to the immune response and axon regeneration after peripheral nerve injury. They found six genes, namely, Alcam, Nrp1, Nrp2, Rac1, Creb1, and Runx3, in rat models of sciatic nerve transection, and these genes were found to be partially localized to the axons, especially in the regenerative axons (Guo et al., 2018). Injury to peripheral nerves can activate innate and acquired immune responses. Of note, the success or failure of the regeneration process has been interfered with by the interaction and overlap mechanisms of the nervous and immune systems after nerve injury (Gaudet et al., 2011; Rotshenker, 2011; Xanthos and Sandkühler, 2014). We suggested that regeneration, angiogenesis, and the immune response of the nerve interact and affect each other, and the results of their interaction directly affect the functional recovery of target organs after nerve repair (Yao et al., 2012; Cattin et al., 2015; Guo et al., 2018). Therefore, in this study, we investigated the transcriptome of SD rats in a model of sciatic nerve crush injury induced by clamping and combined the results of our GO and KEGG analyses to identify four genes related to nerve regeneration, angiogenesis, and the immune response: CCR5, Thy1, Notch1, and Sema4A. Among them, only change of Notch1 was basically consistent with the RNA-seq result after injury, which may be responsible for coordinating this process and promote peripheral nerve regeneration.
The DLL4/Notch1/Akt pathway is principally related to angiogenesis because it regulates the proliferation and migration of ECs in both normal conditions and in malignancies (Jones and Li, 2007; Sun et al., 2018; Tian et al., 2018). Inhibition of the Notch1 pathway suppresses ECs proliferation (Sun et al., 2018; Tian et al., 2018). It has also been reported that VEGF triggers DLL4 expression in tip cells to promote migration and activation of stalk cell proliferation via the Notch pathway (Jones and Li, 2007; Hasan et al., 2017; Park et al., 2018). Additionally, components of the Notch1 pathway are highly expressed in arteries but not in veins (Park et al., 2018), where it limits proliferation to prevent an excessively branched vasculature. Notch signaling can activate macrophage mobilization (Bai et al., 2018; Hossain et al., 2018) and promote neural stem and progenitor cell proliferation (Wang et al., 2018). On the one hand, enhancing arterial ECs proliferation and tube formation via the Notch1 pathway is important in angiogenesis during regeneration. On the other hand, the Notch1 pathway modulates artery formation by controlling ECs proliferation. The combination of the effects on immunology and neural cells suggests the importance of Notch1 in neural regeneration after damage. In the current study, we found that Notch1 expression showed significant change at both ends between 4 and 7 days after crush injury. The trend in the Notch1 change was basically consistent with the RNA-seq result after injury. In the proximal ends of injured nerves, Notch1 expression showed significant changes 4 days after the operation and continued to increase at day 7. But in the distal end, 4 days after injury, Notch1 expression was significantly lower than that in the control group, then gradually increased and became significantly higher than that in the control group by day 7 (P < 0.01). Furthermore, when we investigated interactions with related genes, we found eight important genes were associated with the Notch signaling pathway, most of which were indispensable in regeneration process of axons or vasculature.
Gene expression of CCR5, Thy1, and Sema4A showed either no changes or changes in the opposite direction, which implied that these three genes didn’t not pass validation. However, it might be because of sites of samples were different between RNA-seq and qPCR analysis.
After peripheral nerve injury, CCR5, Thy1, Notch1, and Sema4A could be identified by bioinformatics analysis and transcriptional gene screening and found associated with regeneration, angiogenesis, and the immune response. Among them, the Notch1 change was basically consistent with the RNA-seq result after injury, which may be responsible for coordinating this process and promote peripheral nerve regeneration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE162548.
The animal study was reviewed and approved by Experimental Animal Administration Committee of Sun Yat-sen University.
BH, ZZ, and YX designed the study. VP established the animal models. XL, SX, and YZ analysed and interpreted the experimental data. DD and GW were responsible for the bioinformatics analysis. ZZ and VP generated the graphs. YX, ZZ, and BH edited and revised the manuscript. All authors read and approved the final manuscript.
This research was supported by the Natural Science Foundation of Guangdong Province (2021A1515010471) and the National Natural Science Foundation of China (81901024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2021.717209/full#supplementary-material
SD rats, Sprague-Dawley rats; SCs, Schwann cells; NGS, next-generation sequencing; FPKM, fragments per kilobase of transcript per million mapped reads; TMM, trimmed mean of M values; FDR, false discovery rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEmRNAs, differentially expressed mRNAs; FC, fold change; RF, rich factor; DRG, dorsal root ganglion; ECs, endothelial cells; VEGF, vascular endothelial growth factor; ECM, extracellular matrix; ACTM, acellular tissue matrix; Ang, angiopoietin; FGF, fibroblast growth factor; IOD, integrated optical density.
Abe, N., and Cavalli, V. (2008). Nerve injury signaling. Curr. Opin. Neurobiol. 18, 276–83. doi: 10.1016/j.conb.2008.06.005
Bai, X., Zhang, J., Cao, M., Han, S., Liu, Y., Wang, K., et al. (2018). MicroRNA-146a protects against LPS-induced organ damage by inhibiting Notch1 in macrophage. Int. Immunopharmacol. 63, 220–226. doi: 10.1016/j.intimp.2018.07.040
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N., and Golani, I. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284. doi: 10.1016/S0166-4328(01)00297-2
Bozkurt, A., Deumens, R., Beckmann, C., Olde Damink, L., Schugner, F., Heschel, I., et al. (2009). In vitro cell alignment obtained with a Schwann cell enriched microstructured nerve guide with longitudinal guidance channels. Biomaterials 30, 169–179. doi: 10.1016/j.biomaterials.2008.09.017
Cattin, A. L., Burden, J. J., Van Emmenis, L., Mackenzie, F. E., Hoving, J. J., Garcia Calavia, N., et al. (2015). Macrophage-Induced blood vessels guide schwann cell-mediated regeneration of peripheral nerves. Cell 162, 1127–1139. doi: 10.1016/j.cell.2015.07.021
Dickson, B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959–1964. doi: 10.1126/science.1072165
Djuanda, D., He, B., Liu, X., Xu, S., Zhang, Y., Xu, Y., et al. (2021). Comprehensive analysis of age-related changes in lipid metabolism and myelin sheath formation in sciatic nerves. J. Mol. Neurosci. [Online ahead of print] doi: 10.1007/s12031-020-01768-5
Ebenezer, G. J., McArthur, J. C., Polydefkis, M., Dorsey, J. L., O’Donnell, R., Hauer, P., et al. (2012). SIV-induced impairment of neurovascular repair: a potential role for VEGF. J. Neurovirol. 18, 222–230. doi: 10.1007/s13365-012-0102-5
Gaudet, A. D., Popovich, P. G., and Ramer, M. S. (2011). Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J. Neuroinflammation, 8:110. doi: 10.1186/1742-2094-8-110
Gorin, C., Rochefort, G. Y., Bascetin, R., Ying, H., Lesieur, J., Sadoine, J., et al. (2016). Priming dental pulp stem cells with fibroblast growth Factor-2 increases angiogenesis of implanted tissue-engineered constructs through hepatocyte growth factor and vascular endothelial growth factor secretion. Stem. Cells Transl. Med. 5, 392–404. doi: 10.5966/sctm.2015-0166
Guo, Q., Zhu, H., Wang, H., Zhang, P., Wang, S., Sun, Z., et al. (2018). Transcriptomic landscapes of immune response and axonal regeneration by integrative analysis of molecular pathways and interactive networks post-sciatic nerve transection. Front. Neurosci. 12:457. doi: 10.3389/fnins.2018.00457
Hasan, S. S., Tsaryk, R., Lange, M., Wisniewski, L., Moore, J. C., Lawson, N. D., et al. (2017). Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat. Cell Biol. 19, 928–940. doi: 10.1038/ncb3574
He, B., Zhu, Z., Zhu, Q., Zhou, X., Zheng, C., Li, P., et al. (2014). Factors predicting sensory and motor recovery after the repair of upper limb peripheral nerve injuries. Neural. Regen. Res. 9, 661–672. doi: 10.4103/1673-5374.130094
Hossain, F., Majumder, S., Ucar, D. A., Rodriguez, P. C., Golde, T. E., Minter, L. M., et al. (2018). Notch signaling in myeloid cells as a regulator of tumor immune responses. Front. Immunol. 9:1288. doi: 10.3389/fimmu.2018.01288
Jones, C. A., and Li, D. Y. (2007). Common cues regulate neural and vascular patterning. Curr. Opin. Genet. Dev. 17, 332–336. doi: 10.1016/j.gde.2007.07.004
Kalinski, A. L., Yoon, C., Huffman, L. D., Duncker, P. C., Kohen, R., Passino, R., et al. (2020). Analysis of the immune response to sciatic nerve injury identifies efferocytosis as a key mechanism of nerve debridement. eLife 9:e60223. doi: 10.7554/eLife.60223
Kato, S., Matsukawa, T., Koriyama, Y., Sugitani, K., and Ogai, K. (2013). A molecular mechanism of optic nerve regeneration in fish: the retinoid signaling pathway. Prog. Retin. Eye Res. 37, 13–30. doi: 10.1016/j.preteyeres.2013.07.004
Konofaos, P., and Ver Halen, J. P. (2013). Nerve repair by means of tubulization: past, present, future. J. Reconstr. Microsurg. 29, 149–164. doi: 10.1055/s-0032-1333316
Kumar, V., Sachan, T., Natrajan, M., and Sharma, A. (2014). High resolution structural changes of Schwann cell and endothelial cells in peripheral nerves across leprosy spectrum. Ultrastruct. Pathol. 38, 86–92. doi: 10.3109/01913123.2013.870273
Li, S., Liu, Q., Wang, Y., Gu, Y., Liu, D., Wang, C., et al. (2013). Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One, 8:e57000. doi: 10.1371/journal.pone.0057000
Lindborg, J. A., Niemi, J. P., Howarth, M. A., Liu, K. W., Moore, C. Z., Mahajan, D., et al. (2018). Molecular and cellular identification of the immune response in peripheral ganglia following nerve injury. J. Neuroinflamm. 15:192. doi: 10.1186/s12974-018-1222-5
Liu, D., Liang, X., and Zhang, H. (2016). Effects of high glucose on cell viability and differentiation in primary cultured schwann cells: potential role of ERK signaling pathway. Neurochem. Res. 41, 1281–1290. doi: 10.1007/s11064-015-1824-6
Liu, J., Wang, H. W., Liu, F., and Wang, X. F. (2015). Antenatal taurine improves neuronal regeneration in fetal rats with intrauterine growth restriction by inhibiting the Rho-ROCK signal pathway. Metab. Brain Dis. 30, 67–73. doi: 10.1007/s11011-014-9572-x
Liu, J. H., Tang, Q., Liu, X. X., Qi, J., Zeng, R. X., Zhu, Z. W., et al. (2018). Analysis of transcriptome sequencing of sciatic nerves in Sprague-Dawley rats of different ages. Neural. Regen. Res. 13, 2182–2190. doi: 10.4103/1673-5374.241469
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Park, J. K., Lee, T. W., Do, E. K., Moon, H. J., and Kim, J. H. (2018). Role of Notch1 in the arterial specification and angiogenic potential of mouse embryonic stem cell-derived endothelial cells. Stem Cell Res. Ther. 9:197. doi: 10.1186/s13287-018-0945-7
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Qiu, L., He, B., Hu, J., Zhu, Z., Liu, X., and Zhu, J. (2015). Cartilage oligomeric matrix protein Angiopoeitin-1 provides benefits during nerve regeneration in vivo and in vitro. Ann. Biomed. Eng. 43, 2924–2940.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140.
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25.
Roth, L., Koncina, E., Satkauskas, S., Cremel, G., Aunis, D., and Bagnard, D. (2009). The many faces of semaphorins: from development to pathology. Cell. Mol. Life Sci. 66, 649–666.
Rotshenker, S. (2011). Wallerian degeneration: the innate-immune response to traumatic nerve injury. J. Neuroinflammation 8:109. doi: 10.1186/1742-2094-8-109
Sayad Fathi, S., and Zaminy, A. (2017). Stem cell therapy for nerve injury. World J. Stem. Cells 9, 144–151.
Sun, B., Dong, C., Lei, H., Gong, Y., Li, M., Zhang, Y., et al. (2018). Propranolol inhibits proliferation and invasion of hemangioma-derived endothelial cells by suppressing the DLL4/Notch1/Akt pathway. Chem. Biol. Interact. 294, 28–33.
Tessier-Lavigne, M., and Goodman, C. S. (1996). The molecular biology of axon guidance. Science 274, 1123–1133.
Tian, D., Zeng, X., Wang, W., Wang, Z., Zhang, Y., and Wang, Y. (2018). Protective effect of rapamycin on endothelial-to-mesenchymal transition in HUVECs through the Notch signaling pathway. Vascul, Pharmacol. 113, 20–26.
Wang, R., Zhao, C., Li, J., Li, Y., Liu, Y., Dong, H., et al. (2018). Notch1 promotes mouse spinal neural stem and progenitor cells proliferation via p-p38-pax6 induced cyclin D1 activation. Exp. Cell Res. 373, 80–90.
Xanthos, D. N., and Sandkühler J. (2014). Neurogenic neuroinflammation: inflammatory CNS reactions in response to neuronal activity. Nat. Rev. Neurosci. 15, 43–53. doi: 10.1038/nrn3617
Yao, D., Li, M., Shen, D., Ding, F., Lu, S., Zhao, Q., et al. (2012). Gene expression profiling of the rat sciatic nerve in early Wallerian degeneration after injury. Neural. Regen. Res. 7, 1285–1292.
Zhou, Y., Gunput, R. A., and Pasterkamp, R. J. (2008). Semaphorin signaling: progress made and promises ahead. Trends Biochem. Sci. 33, 161–170.
Zhu, Z., Huang, Y., Zou, X., Zheng, C., Liu, J., Qiu, L., et al. (2017). The vascularization pattern of acellular nerve allografts after nerve repair in Sprague-Dawley rats. Neurol. Res. 39, 1014–1021.
Keywords: peripheral nerve injury, SD rats, transcriptome, bioinformatic, immune response
Citation: He B, Pang V, Liu X, Xu S, Zhang Y, Djuanda D, Wu G, Xu Y and Zhu Z (2021) Interactions Among Nerve Regeneration, Angiogenesis, and the Immune Response Immediately After Sciatic Nerve Crush Injury in Sprague-Dawley Rats. Front. Cell. Neurosci. 15:717209. doi: 10.3389/fncel.2021.717209
Received: 30 May 2021; Accepted: 08 September 2021;
Published: 04 October 2021.
Edited by:
Maria Dolores Ganfornina, University of Valladolid, SpainReviewed by:
Ashutosh Kumar, National Institute of Pharmaceutical Education and Research (NIPER-Kolkata), IndiaCopyright © 2021 He, Pang, Liu, Xu, Zhang, Djuanda, Wu, Xu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaowei Zhu, bm10bWVtb2lyQGFsaXl1bi5jb20=; Yangbin Xu, eHV5YW5nYmluQGhvdG1haWwuY29t; Bo He, aGVib2RvY0BhbGl5dW4uY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.