
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell. Neurosci. , 06 December 2019
Sec. Cellular Neuropathology
Volume 13 - 2019 | https://doi.org/10.3389/fncel.2019.00545
This article is part of the Research Topic Interactions of the Nervous System with Bacteria View all 14 articles
Human gastrointestinal (GI)-tract microbiome-derived lipopolysaccharide (LPS): (i) has been recently shown to target, accumulate within, and eventually encapsulate neuronal nuclei of the human central nervous system (CNS) in Alzheimer’s disease (AD) brain; and (ii) this action appears to impede and restrict the outward flow of genetic information from neuronal nuclei. It has previously been shown that in LPS-encased neuronal nuclei in AD brain there is a specific disruption in the output and expression of two AD-relevant, neuron-specific markers encoding the cytoskeletal neurofilament light (NF-L) chain protein and the synaptic phosphoprotein synapsin-1 (SYN1) involved in the regulation of neurotransmitter release. The biophysical mechanisms involved in the facilitation of the targeting of LPS to neuronal cells and nuclei and eventual nuclear envelopment and functional disruption are not entirely clear. In this “Perspectives article” we discuss current advances, and consider future directions in this research area, and provide novel evidence in human neuronal-glial (HNG) cells in primary culture that the co-incubation of LPS with amyloid-beta 42 (Aβ42) peptide facilitates the association of LPS with neuronal cells. These findings: (i) support a novel pathogenic role for Aβ42 peptides in neurons via the formation of pores across the nuclear membrane and/or a significant biophysical disruption of the neuronal nuclear envelope; and (ii) advance the concept that the Aβ42 peptide-facilitated entry of LPS into brain neurons, accession of neuronal nuclei, and down-regulation of neuron-specific components such as NF-L and SYN1 may contribute significantly to neuropathological deficits as are characteristically observed in AD-affected brain.
The highest known density of microorganisms anywhere in the biosphere is in the human GI-tract microbiome at about ~1011 microorganisms per gram of GI-tract content (Angelucci et al., 2019; Castillo-Álvarez and Marzo-Sola, 2019; Fox et al., 2019). This vast number represents a remarkably complex and highly dynamic source of microbes; of the approximate ~1,800 different microbial phyla that make up the GI-tract microbiome, the overwhelming majority are facultative anaerobic bacteria with archaea, fungi, microbial eukaryotes, protozoa, viruses, and other microbes making up the remainder (Bhattacharjee and Lukiw, 2013; Fox et al., 2019; Tierney et al., 2019). One major species of bacteria in the human GI-tract microbiome, about ~100-fold more abundant than Escherichia coli in certain GI-tract regions is Bacteroides fragilis, an obligate, anaerobic, non-spore forming Gram-negative, rod-shaped enterotoxigenic bacterium. B. fragilis: (i) generates a remarkable array of highly neurotoxic exudates; and (ii) produces a particularly virulent, pro-inflammatory LPS glycolipid subtype (BF-LPS) that accumulates in Alzheimer’s disease (AD) brain (Sears, 2009; Fathi and Wu, 2016; Lukiw, 2016a,b; Wexler and Goodman, 2017; Zhao et al., 2017a,b,c; Allen et al., 2019). Besides BF-LPS, B. fragilis-derived neurotoxins include small non-coding RNA (sncRNA), bacterial amyloids, endo-, exo-, and enterotoxins such as fragilysin, and truncated LPS molecules known as lipooligosaccharides (LOS). These neurotoxins have recently been shown to be capable of transversing normally restrictive gastrointestinal (GI) tract and blood-brain barriers (BBBs) in transgenic murine models of AD (Varatharaj and Galea, 2017; Sweeney et al., 2018; Tulkens et al., 2018; Barton et al., 2019; Erdö and Krajcsi, 2019; Panza et al., 2019; Sweeney and Lowary, 2019). Both the GI-tract and BBB may become weakened with aging or following surgery, disease or trauma (Sweeney et al., 2018; Sweeney and Lowary, 2019). For example, BF-LPS and the B. fragilis-derived enterotoxin fragilysin very effectively disrupt cell-cell adhesion, in part by E-cadherin cleavage and/or the action of LPS binding protein and Toll-like receptor 4 (TLR4), and subsequent LPS internalization, followed by translocation of neurotoxins into the systemic circulation, past the BBB and on into the parenchyma of the brain [Wu et al., 1998; Holton, 2008; Tsukamoto et al., 2018; Barton et al., 2019; Jeon et al., 2019; Lukiw, 2019 (submitted)].
Multiple, independent research laboratories have reported: (i) the association of LPS and microbial-derived amyloid with AD brain (Zhao and Lukiw, 2015; Zhao et al., 2015); (ii) the remarkable affinity of specific LPS isoforms with AD brain parenchyma (Lukiw, 2016a,b); (iii) that Gram-negative bacterial molecules associate with AD neuropathology (Zhan et al., 2016); (iv) that microbiome-derived E.coli LPS and B. fragilis LPS associate the hippocampal CA1 region of AD brain (Zhao et al., 2017a,b,c); (v) of LPS accumulation within neocortical neurons of the AD brain that impair transcriptional output (Zhao et al., 2017a,b,c); (vi) that there is a strong association of LPS with neuronal nuclei and the specific LPS-mediated impairment of expression of the neurofilament light (NF-L) chain gene expression (Lukiw et al., 2018); (vii) of LPS association with the amyloid plaques, neurons and oligodendrocytes in AD brain (Zhan et al., 2018); and (viii) a significantly reduced expression of the AD-relevant synaptic components such as synapsin-1 (SYN1) in LPS-treated human neuronal-glial (HNG) cells in primary culture (Zhao et al., 2019). Most recently, it has been shown that LPS has a very strong affinity for, and association with, the neuronal nuclear envelope of the HNG cells in primary culture. This is also observed in the superior temporal lobe neocortex (Brodmann area A22; Wernicke’s area) and the hippocampal CA1 region of AD-affected brain (Lukiw, 2016a,b; Zhao et al., 2017a,b,c, 2019; Lukiw et al., 2018; Ticinesi et al., 2019; Figure 1). Interestingly in moderate-to-late-stage AD LPS totally encapsulates neuronal nuclei in the AD brain with the subsequent restriction in the output of genetic information from those neuronal nuclei (Lukiw et al., 2018; Zhao and Lukiw, 2018a,b; Zhao et al., 2019). Interestingly, gene expression profiling showed a long-lasting deficit in neuron- and synaptic-specific gene expression and signaling in the hippocampus and neocortex of both transgenic murine models for AD and in patients with mild cognitive impairment or AD (Colangelo et al., 2002; Counts et al., 2014; Jaber et al., 2019; Parra-Damas and Saura, 2019).
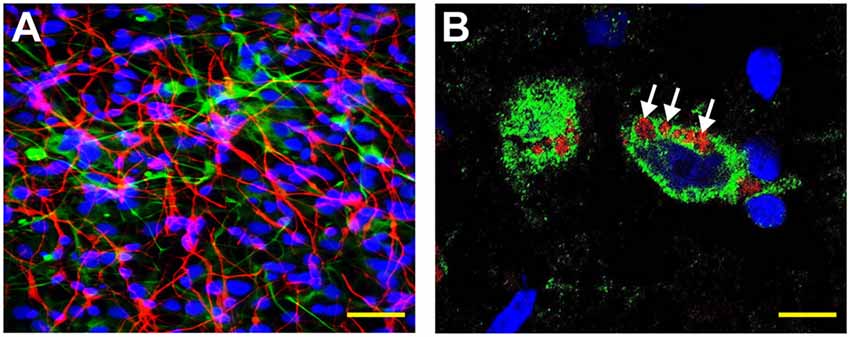
Figure 1. Human neuronal-glial (HNG) cells (transplantation grade) in primary co-culture were used to study the dynamics of amyloid-beta 42 (Aβ42) peptide-mediated entry of lipopolysaccharide (LPS) into neurons (Bhattacharjee and Lukiw, 2013; Zhan et al., 2018; Zhao and Lukiw, 2018a,b; Zhao et al., 2019). (A) HNG cells are a primary co-culture of neuronal [β-tubulin III (βTUBIII)-stained; red; λmax = 690 nm] and glial (GFAP-stained; green; λmax = 520 nm) human brain cells; HNG cells are also stained for nuclei (DAPI-stained; blue; λmax = 470 nm); cells shown are ~2 weeks in culture; HNG cells are about ~60% neurons (red) and about ~40% astroglial (green) at ~65% confluence; human primary neuronal and glial “support” cell co-cultures are utilized, because human neuronal cells do not culture well by themselves (Cui et al., 2010; Zhao et al., 2017c); HNG cells were exposed to 50 nM LPS for 36 h in the presence or absence of 10 nM Aβ42 peptides; other LPS concentrations at similar times displayed analogous trends; yellow scale bar (lower right) ~50 μm. (B) Affinity of LPS for the neuronal nuclear envelope (white arrows); LPS (red; λmax = 690 nm); β-tubulin III (βTUBIII)-stained (green; λmax = 520 nm) and nuclei (blue; λmax = 470 nm) stained HNG cells; white arrows indicate punctate and perinuclear clustering of LPS and LPS affinity for the nuclear envelope as has been previously reported (Hill and Lukiw, 2015; Zhan et al., 2016, 2018; Yang and Chiu, 2017; Zhao et al., 2017a,b); yellow scale bar (lower right) = 20 μm.
Recent evidence shows that LPS-type glycolipids not only have an unusually high affinity for neuronal nuclear membranes but also that amyloid-β 42 (Aβ42) peptides significantly facilitate LPS access and translocation into human neuronal cells and across neuronal nuclear envelopes in HNG cells in primary co-culture (Figures 2A–L). There are several possible explanations for the facilitation of LPS entry into, and their association with, neuronal membranes and neuronal nuclei by Aβ42 peptides or Aβ42 oligomers and these include:
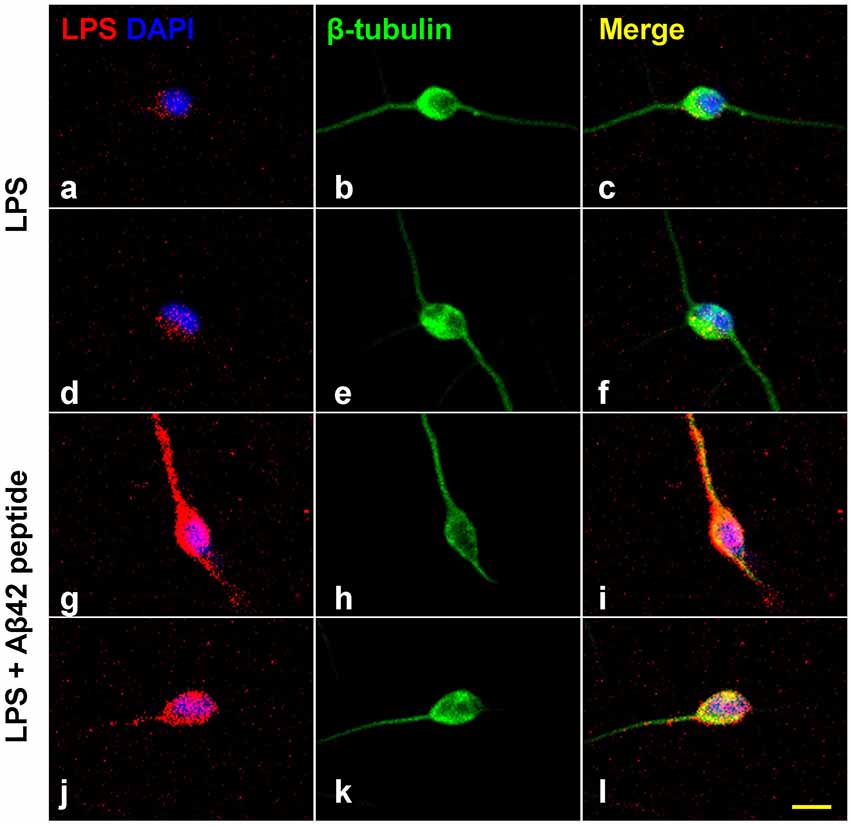
Figure 2. Increased affinity of LPS for neuronal nuclei in the presence of Aβ42 peptide. Panels (A–L) show LPS-neuronal interactions in the presence or absence of Aβ42 peptide; (A–F) in the absence of Aβ42 peptide and (G–L) in the presence of Aβ42 peptide. LPS preferentially associates with human neuronal nuclei both in Alzheimer’s disease (AD) and in LPS-addition experiments (Zhao et al., 2017a,b,c; Zhao and Lukiw, 2018a,b). Panels (A,D) show LPS (red) affinity for a polar region of a single DAPI-stained neuronal nucleus (blue). Panels (B,E) show single neuronal nucleus stained with neuron-specific β-tubulin III (green). Panels (C,F) show merged stain indicating LPS affinity for the polar region involving a single DAPI-stained neuronal nucleus. Panels (G,J) show the presence of Aβ42 significantly increases the affinity of LPS for single DAPI-stained neuronal nucleus. Panels (H,K) show single neuronal nucleus stained with neuron-specific β-tubulin III (green). Panels (I,L) show merged stain indicating LPS affinity for the neurite and soma of a single DAPI-stained neuronal nuclei. The results suggest that LPS is stimulated to associate with DAPI-stained neuronal nuclei in the presence of the hydrophobic Aβ42 peptide; neither Aβ40 peptide or β-actin showed comparable “association” effects (Zhao et al., 2017a,b,c; Lukiw et al., 2018); yellow scale bar (lower right) = 50 μm.
Amyloid peptides (both Aβ40 and Aβ42) form a remarkable number of heterotypic structures and configurations under pathological conditions. These include: (i) the self-assembly and deposition of multiple types of fibrillar and globular structures and polymorphic assemblies both in solution and on membrane surfaces; and (ii) the formation of heterogeneous ionic pores spanning the lipid bilayer that is linked to the pathogenicity of these molecules. These later findings are supported by multiple independent reports regarding the capability of Aβ42 peptides [2 hydrophobic amino acid residues (isoleucine and alanine) longer (at the C-terminal) than Aβ40] to form up to 2.4-nm diameter pores through lipid bilayer membranes (Lashuel et al., 2002; Connelly et al., 2012; Sciacca et al., 2012; Ullah et al., 2015; Di Scala et al., 2016; Jang et al., 2016; Davidson, 2019; Hicks et al., 2019; Nguyen et al., 2019; Sun et al., 2019; Österlund et al., 2019). Interestingly, the slightly longer and more hydrophobic Aβ42-based peptide assemblies in oligomeric preparations have been observed to form voltage-independent, non-selective ion channels in contrast to Aβ40 peptide-based oligomers, fibers, and monomers which do not generally support pore structure formation (Bode et al., 2017, 2019; Nguyen et al., 2019). Although LPS is intrinsically heterogeneous and over time tends to form aggregates of ~1–4 mDa or greater, smaller LOS or LPS monomers in the range of ~50 to ~100 kDa appear to have little difficulty in transversing ~2.4 nm diameter pores to reach their final destination within the nucleoplasm (Zimmer et al., 1988; Millipore Sigma; Lipopolysaccharides1).
As recently visualized by atomic force, transmission electron microscopy, mobility-mass spectrometry and liquid surface X-ray scattering there is a remarkable influence of Aβ monomers, short fibrillar Aβ oligomers, globular non-fibrillar Aβ oligomers and full-length Aβ fibrils on lipid bilayer membrane integrity and stability (Bode et al., 2019; Nguyen et al., 2019; Österlund et al., 2019; Vander Zanden et al., 2019). Abundant evidence indicates an Aβ oligomeric fibril-induced reorganization of membrane lipid packing and the induction of membrane destabilization and lipid disorganization by globular non-fibrillar Aβ oligomers (Di Lorenzo et al., 2019; Vander Zanden et al., 2019). Scanning electron microscopy (SEM) and thioflavin-T fluorescence assay have revealed: (i) that LPS and/or LPS-binding protein (LBP) have strong disruptive effects on the structural and biophysical organization of Aβ peptides and amyloidogenesis in Parkinson’s disease (Montagne et al., 2017; Pretorius et al., 2018); and (ii) that LPS strongly induces NF-kB signaling, inflammatory responses, neuroinflammation, the generation of Aβ42 peptides and amyloidogenesis in transgenic murine models of AD (Gu et al., 2018; Jeon et al., 2019; Sheppard et al., 2019). Conversely, as evidenced by atomic force and electron microscopy imaging, short fibrillar Aβ42 oligomers appear to have a profound detergent-like, highly-localized, solubilizing effect on lipid membrane bilayers and this may predispose to hydrophobic interaction with LPS already present in the parenchyma of AD brain (Bode et al., 2019).
Used for highly regulated nucleocytoplasmic transport, the nuclear envelope of typical neuronal cells contain about ~10,000 nuclear pore complexes/transporters (significantly more than the ~3,000 nuclear pores of a typical eukaryotic cell), and each ~110 MDa nuclear pore complex (NPC) consists of about ~1,000 nucleoporin proteins (Cooper, 2000; Kabachinski and Schwartz, 2015; Davidson, 2019; Lin and Hoelz, 2019; Sun et al., 2019). The affinity of LPS for any NPC component or any nucleoporin protein is not well understood and is an understudied area of both the neurobiology, microbiology and neuropathology of the human central nervous system (CNS). The perinuclear accumulation of LPS and LPS-mediated envelopment of human neuronal nuclei (Zhao et al., 2017a,b,c), and the restriction of the outflow of neuron-specific information, such as those mRNAs encoding the neuron-specific neurofilament light (NF-L) chain protein and SYN1 (Zhao et al., 2019), underscore the novel pathogenic potential of LPS in supporting dysfunction in neuronal cytoarchitecture and the capacity for efficient inter-neuronal signaling by disrupting SYN1 availability and hence synaptic integrity. In addition, LPS strongly associates with amyloid plaques (Zhan et al., 2018) and perinuclear LPS, and encasement of neuronal nuclei by LPS may also contribute to the biophysical blockage of exit of mRNA through the NPC into the cytoplasm in AD brain (Zhao et al., 2019). Interestingly, using stable isotope labeling of amino acids in cell culture and quantitative proteomics, it has recently been shown that the interactome of the 695 amino acid beta-amyloid precursor protein βAPP695, which is the direct precursor to Aβ42 peptide, interacts strongly with the NPC and nucleoporin proteins in neuronal cells (Andrew et al., 2019). This suggests some novel roles for both βAPP695 and Aβ42 peptide in both NPC function and amyloid peptide processing and generation. The unique phospholipid composition of the inner nuclear neuronal membrane (that encases the genome) and the outer neuronal nuclear membrane that together form the nuclear envelope, their extremely high ratio of phospholipid to cholesterol, the biophysics of nuclear lipid membrane remodeling and lipid raft formation may predispose the neuronal nuclear envelope to the potential interaction between amyloid peptides and LPS, and with NPC nucleoporin proteins.
Lipopolysaccharide (LPS) is a type of prokaryotic glycoconjugate-glycolipid comprised of three major domains: (i) an “O” antigen consisting of an “O polysaccharide”; (ii) a “core” polysaccharide domain (the innermost hydrophilic domain of the three regions of LPS); and (iii) a hydrophobic “lipid A” domain. The “core” polysaccharide domain contains an oligosaccharide covalently attached directly to the “lipid A” moiety and commonly contains sugars such as heptose, 3-deoxy-D-mannooctulosonic acid as well as non-carbohydrate components that include phosphate, amino acids linkages and ethanolamine components characteristic of each Gram-negative bacterial genus and species (Whitfield and Trent, 2014; Tulkens et al., 2018). Both the 50–100 kDa LPS monomer (especially the “lipid A” domain responsible for much of the toxicity of Gram-negative bacteria) and the 4.5 kDa Aβ42 peptide monomer are highly hydrophobic and this alone may favor their mutual interaction with the lipid bilayer of neuronal membranes (National Institutes of Health, PubChem, 2019). As mentioned earlier, Aβ42-based oligomers are highly disruptive toward lipid bilayer membranes, but whether the chaotropic actions of LPS and Aβ42 peptide are additive or synergistic is unknown as their specific interactions are currently not well understood.
While Aβ42 peptides clearly support LPS entry into neurons it is not clear why neuronal membranes are specifically targeted for these actions and/or why other cellular plasma membranes (lipid bilayers) are not preferred or involved to a lesser extent; perhaps this has something to do with the unique neuronal membrane proteolipid composition and/or the electrical activity of these cell types or other unique neuronal features including Aβ42 peptide-mediated pore formation (see above; Nguyen et al., 2019; Österlund et al., 2019). While LPS has been shown to completely envelope neuronal nuclei in the superior temporal lobe neocortex (Brodmann area A22) in both aging, and especially in AD brain, there is emerging evidence that the end result of this biophysical occlusion of nuclear pores is the restriction of outflow of genetic information, i.e., messenger RNA (mRNA) through these nuclear pores, however some other pathogenic mechanism may be involved (Clement et al., 2016; Lukiw et al., 2018; Zhao et al., 2019; Cornelison et al., 2019). Very recently it has been demonstrated that there is an LPS-induced translocation of cytosolic NF-κB into the cell nucleus (Bagaev et al., 2019) and LPS-induced neuronal hypertrophy (Tellez-Merlo et al., 2019) but these potentially pathogenic and neuroinflammatory outcomes require further investigation. It would be very useful to know at what point an induced disruption along the gut-brain axis and LPS-signaling pathways might be beneficial in the clinical management of AD.
There exists the intriguing and enigmatic possibility, as has been suggested for other major organ groups, that the human brain and/or CNS might have its own, as yet poorly characterized microbiome (Bhattacharjee and Lukiw, 2013; Hill et al., 2014a,b; Köhler et al., 2016; Emery et al., 2017; Zhao et al., 2017b,c; Roberts et al., 2018; Zhao and Lukiw, 2018a,b; Zhou and Bian, 2018; Javan et al., 2019; Mazmanian, 2019). Currently, it is understood that microbes can enter the brain and CNS through the BBB which becomes leaky via physical damage, disease and/or aging, and/or via nerves that innervate both the brain and the gut (Roberts et al., 2018; Javan et al., 2019). Very recent studies indicate the presence of bacteria in the human and mouse brain at the BBB under noninfectious or non-traumatic conditions. Microbes have been identified via morphological criteria and ultrastructural imaging analysis with high bacterial counts found in the human hippocampus and prefrontal cortex, but low bacterial counts in other brain anatomical regions such as the striatum (Roberts et al., 2018; Javan et al., 2019). Significantly increased bacterial populations have been observed in association with neurological deterioration in AD brain tissues compared with controls (Emery et al., 2017). Other supportive studies come from investigations involving the human thanatomicrobiome—the microbiome of death—that reflects the post-mortem microbial changes which vary by organ and as a function of time and temperature (Zhao et al., 2017a,b,c; Zhou and Bian, 2018; Javan et al., 2019). Further support for the idea that a microbiome may already be present in the brain arises when considering that microbes (or microbial-derived neurotoxins) in the GI tract would need to travel a significant distance to reach the brain compartments post-mortem vs. the extremely rapid proliferation of bacteria in the brain shortly after death. This might contribute locally to the presence of bacterial-derived neurotoxins such as LPS and other microbial-derived molecules in brain tissues (Hill et al., 2014a,b; Lukiw, 2016a,b; Zhan et al., 2016; Emery et al., 2017). Put another way, post-mortem microscopic examination of the post-mortem brain routinely detects bacteria far more rapidly than the bio-physiological capability of microbes to transit from the GI-tract across the systemic circulation into the brain or other CNS compartments (Roberts et al., 2018; Javan et al., 2019).
The remarkable affinity of the glycoconjugate LPS, the major component of the outer membrane of Gram-negative bacteria such as B. fragilis and E.coli, for the neuronal nuclear envelope in human brain cells and tissues was first described just ~2 years ago (Zhao et al., 2017a,b,c; Zhan et al., 2018). The mean abundance of GI-tract-sourced LPS can be increased by either an up-regulation in the biosynthesis of LPS itself or via an increase in the number of Gram-negative bacteria capable of generating and releasing LPS in part through the process of dysbiosis. Because both the de novo induction of LPS and bacterial division times of ~15–20 min are relatively rapid microbiological-pathological events, it seems also that the production of LPS can be a rapidly undertaken and perhaps even exponential biological event (Raetz and Whitfield, 2002; Whitfield and Trent, 2014; Sweeney and Lowary, 2019). Interestingly, growth rates in the GI-tract microbiome for Gram-negative bacteria have been shown to be dependent on ingested dietary fiber, and the relative proportion of B. fragilis in the GI tract can for example decrease 2–3-fold after a fiber-laden meal while a high-fat-cholesterol (HFC) meal has the opposite effect (Heinritz et al., 2016; Huang and Liu, 2019). Indeed, dietary modification by increasing both soluble and/or insoluble fiber intake has been shown to decrease the abundance of B. fragilis in the GI-tract microbiome on a time-scale of hours-to-days after the ingestion of the fiber-enriched meal itself (Simpson and Campbell, 2015; Chen et al., 2017; Dhillon et al., 2019; Huang and Liu, 2019; Parada Venegas et al., 2019). Dietary manipulation, probiotics and prebiotic supplementation and increased ingestion of fiber is one research area urgently requiring more study, because the management of diet could yield real and more effective therapies for both the treatment of neurodegeneration and malignancy (Rios-Covian et al., 2017; Poeker et al., 2018; Dhillon et al., 2019; Huang and Liu, 2019). It should be mentioned that under realistic physiological conditions the ~1,800 phyla of bacteria of the GI-tract microbiome are together most likely capable of generating an extremely complex neurotoxic cocktail of exudates, and at this point in time we are analyzing just a very small number of GI-tract derived neurotoxins from a vast neurotoxic pool of huge abundance and bewildering biological complexity (Hicks et al., 2019; Tierney et al., 2019).
Over the last few years, the GI-tract microbiome-brain axis has emerged as a focus of increasing interest in the establishment of a neurophysiological and neurobiological basis for age-related, developmental, neurodegenerative, neuroinflammatory and psychiatric disease. The microorganisms which constitute the human GI-tract microbiome have potential to secrete some of the most neurotoxic and inflammation-inducing substances known, including bacterial glycolipid lipopolysaccharide (LPS) from abundant, anaerobic, GI-tract resident Gram-negative bacteria (Whitfield and Trent, 2014; Batista et al., 2019; Patrick et al., 2019; Ticinesi et al., 2019). As a particularly abundant commensal, non-motile, non-spore forming obligatory anaerobic, Gram-negative bacillus of the human GI-tract microbiome, Bacteroides fragilis (B. fragilis), releases an intensely pro-inflammatory species of LPS (BF-LPS), amongst the most pro-inflammatory substances known, that in HNG cells in primary culture induces the pro-inflammatory transcription factor NF-kB (p50/p65) complex (Lukiw, 2016a,b; Zhao and Lukiw, 2018a,b; Batista et al., 2019; Sweeney and Lowary, 2019). LPS translocation into the nucleoplasm and access to neuronal nuclei are greatly facilitated in the presence of Aβ42 peptides (Figures 1, 2). LPS-triggered NF-kB (p50/p65) up-regulation is associated with: (i) the induction of pro-inflammatory, pathogenic microRNA-regulated gene expression programs in the AD brain; these microRNAs have multiple NF-kB (p50/p65) recognition features in their immediate promoters (Pogue and Lukiw, 2018); and (ii) multiple independent laboratories have provided evidence that GI-tract derived glycolipids such as LPS associated with the pro-inflammatory, cytoarchitectural and/or synaptic neuropathology of AD brain and transgenic murine models of AD (Bhattacharjee and Lukiw, 2013; Hill and Lukiw, 2015; Zhan et al., 2016, 2018; Lukiw et al., 2018; Zhao et al., 2019). Many of these noxious biopolymers are potent enterotoxins which can neutralize cadherins and other cell-cell adhesion molecules, inducing leakage through the GI-tract epithelial barrier, which normally is largely impermeable, allowing neurotoxin access to the systemic circulation and subsequent translocation across the BBB (Leshchyns’ka and Sytnyk, 2016; Sweeney et al., 2018; Jeon et al., 2019; Sweeney and Lowary, 2019). Clinically, the detection of these GI-tract microbiome-derived neurotoxins in blood serum may be of prodromal, prognostic and/or diagnostic value as biomarkers for the onset and/or propagation of neurological disease or malignancy; or of forensic value in the determination of temporal aspects of the post-mortem interval (Li and Yu, 2017; Zhou and Bian, 2018).
Lastly, obligate anaerobic bacteria such as Bacteroides fragilis make up the largest proportion of Gram-negative microbes in the human GI-tract microbiome. In a recent study of 2,100 human donors, the most recent estimate is that all together microbial constituents of this microbiome harbor at least 22.3 million non-redundant prokaryotic genes in contrast to the 26.6 thousand protein-encoding transcripts of the human genome (Venter et al., 2001; Tierney et al., 2019). Hence, GI-tract microbial genes outnumber host genes by about 840-to-1, which represents staggering genetic complexity (Fields et al., 1994; Venter et al., 2001; Tierney et al., 2019). With this comes a GI-tract microbial proteome of remarkable proportion and speciation that includes highly neurotoxic and pro-inflammatory exudates such as LPS (Hicks et al., 2019; Roy Sarkar and Banerjee, 2019). It is tempting to speculate that we are just scratching the surface of our understanding of the potential impact of these prokaryotic GI-tract microbiome-derived genes and their extruded neurotoxic molecules on our own host gene signaling and expression systems which are likely to have a tremendous impact and relevance to both human health and disease.
All datasets generated for this study are included in the article.
The animal study was reviewed and approved by Louisiana State University (LSU) Ethics Committee, Louisiana State University, New Orleans, LA, USA.
WLi, WLu, TB and YZ performed and analyzed all experiments. WLu wrote the article.
The research on microRNAs, ethnobiology, botanical neurotoxins, pro-inflammatory and pathogenic signaling in the Lukiw laboratory involving the microbiome, the innate-immune response, amyloidogenesis, synaptogenesis, and neuro-inflammation in AD, prion and in other human neurological- and plant-viroid-based diseases was supported through an unrestricted grant to the LSU Eye Center from Research to Prevent Blindness (RPB); the Louisiana Biotechnology Research Network (LBRN) and National Institutes of Health (NIH) grants NEI EY006311, NIA AG18031, and NIA AG038834 (WLu). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging, the National Center for Research Resources, or the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research review presented as a “Frontiers Perspectives” article was presented in part at the Vavilov Institute of General Genetics Autumn 2018 Seminar Series ( ) in Moscow, Russia, October 2018, at the Society for Neuroscience (SFN) Annual Meeting, Chicago, IL, USA, November 2019. Sincere thanks are extended to Drs. L. Cong, F. Culicchia, C. Eicken, K. Navel, A.I. Pogue, W. Poon, E. Head, and the late Drs. J.M. Hill and P.N. Alexandrov for helpful discussions in this research area, for short postmortem interval (PMI) human brain and retinal tissues or extracts, for initial bioinformatics and data interpretation, and to A.I. Pogue and D. Guillot for expert technical assistance and medical artwork. We would like to further thank the following brain and tissue banks for access to high-quality post-mortem tissues and valuable analytical advice: the National Institute of Neurological Disorders and Stroke (NINDS), Bethesda, MD, USA; the Oregon Health Sciences University, Portland OR, USA; the Southern Eye Bank, Metairie LA, USA; the University of California (UCI) MIND Institute, Irvine, CA, USA; and the many neuropathologists, physicians, and researchers in the US and Canada who have provided high quality, short PMI human brain tissue fractions for scientific analysis.
) in Moscow, Russia, October 2018, at the Society for Neuroscience (SFN) Annual Meeting, Chicago, IL, USA, November 2019. Sincere thanks are extended to Drs. L. Cong, F. Culicchia, C. Eicken, K. Navel, A.I. Pogue, W. Poon, E. Head, and the late Drs. J.M. Hill and P.N. Alexandrov for helpful discussions in this research area, for short postmortem interval (PMI) human brain and retinal tissues or extracts, for initial bioinformatics and data interpretation, and to A.I. Pogue and D. Guillot for expert technical assistance and medical artwork. We would like to further thank the following brain and tissue banks for access to high-quality post-mortem tissues and valuable analytical advice: the National Institute of Neurological Disorders and Stroke (NINDS), Bethesda, MD, USA; the Oregon Health Sciences University, Portland OR, USA; the Southern Eye Bank, Metairie LA, USA; the University of California (UCI) MIND Institute, Irvine, CA, USA; and the many neuropathologists, physicians, and researchers in the US and Canada who have provided high quality, short PMI human brain tissue fractions for scientific analysis.
Allen, J., Hao, S., Sears, C. L., and Timp, W. (2019). Epigenetic changes induced by Bacteroides fragilis toxin. Infect Immun. 6:e00447–18. doi: 10.1128/IAI.00447-18
Andrew, R. J., Fisher, K., Heesom, K. J., Kellett, K. A. B., and Hooper, N. M. (2019). Quantitative interaction proteomics reveals differences in the interactomes of amyloid precursor protein isoforms. J. Neurochem. 149, 399–412. doi: 10.1111/jnc.14666
Angelucci, F., Cechova, K., Amlerova, J., and Hort, J. (2019). Antibiotics, gut microbiota, and Alzheimer’s disease. J. Neuroinflammation 16:108. doi: 10.1186/s12974-019-1494-4
Bagaev, A. V., Garaeva, A. Y., Lebedeva, E. S., Pichugin, A. V., Ataullakhanov, R. I., and Ataullakhanov, F. I. (2019). Elevated pre-activation basal level of nuclear NF-κB in native macrophages accelerates LPS-induced translocation of cytosolic NF-κB into the cell nucleus. Sci. Rep. 9:4563. doi: 10.1038/s41598-018-36052-5
Barton, S. M., Janve, V. A., McClure, R., Anderson, A., Matsubara, J. A., Gore, J. C., et al. (2019). Lipopolysaccharide induced opening of the blood brain barrier on aging 5XFAD mouse model. J. Alzheimers Dis. 67, 503–513. doi: 10.3233/jad-180755
Batista, C. R. A., Gomes, G. F., Candelario-Jalil, E., Fiebich, B. L., and de Oliveira, A. C. P. (2019). Lipopolysaccharide-induced neuroinflammation as a bridge to understand neurodegeneration. Int. J. Mol. Sci. 20:E2293. doi: 10.3390/ijms20092293
Bhattacharjee, S., and Lukiw, W. J. (2013). Alzheimer’s disease and the microbiome. Front. Cell. Neurosci. 7:153. doi: 10.3389/fncel.2013.00153
Bode, D. C., Baker, M. D., and Viles, J. H. (2017). Ion channel formation by amyloid-β42 oligomers but not amyloid-β40 in cellular membranes. J. Biol. Chem. 292, 1404–1413. doi: 10.1074/jbc.M116.762526
Bode, D. C., Freeley, M., Nield, J., Palma, M., and Viles, J. H. (2019). Amyloid-β oligomers have a profound detergent-like effect on lipid membrane bilayers, imaged by atomic force and electron microscopy. J. Biol. Chem. 294, 7566–7572. doi: 10.1074/jbc.AC118.007195
Castillo-Álvarez, F., and Marzo-Sola, M. E. (2019). Role of the gut microbiota in the development of various neurological diseases. Neurologia doi: 10.1016/j.nrl.2019.03.017 [Epub ahead of print].
Chen, T., Long, W., Zhang, C., Liu, S., Zhao, L., and Hamaker, B. R. (2017). Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci. Rep. 7:2594. doi: 10.1038/s41598-017-02995-4
Clement, C., Hill, J. M., Dua, P., Culicchia, F., and Lukiw, W. J. (2016). Analysis of RNA from Alzheimer’s disease post-mortem brain tissues. Mol. Neurobiol. 53, 1322–1328. doi: 10.1007/s12035-015-9105-6
Colangelo, V., Schurr, J., Ball, M. J., Pelaez, R. P., Bazan, N. G., and Lukiw, W. J. (2002). Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J. Neurosci. Res. 70, 462–473. doi: 10.1002/jnr.10351
Connelly, L., Jang, H., Arce, F. T., Capone, R., Kotler, S. A., Ramachandran, S., et al. (2012). Atomic force microscopy and MD simulations reveal pore-like structures of all-D-enantiomer of Alzheimer’s β-amyloid peptide: relevance to the ion channel mechanism of AD pathology. J. Phys. Chem. B 116, 1728–1735. doi: 10.1021/jp2108126
Cooper, G. M. (2000). The Nuclear Envelope and Traffic Between the Nucleus and Cytoplasm; The Cell: A Molecular Approach. 2nd Edn. Sunderland, MA: Sinauer Associates. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK9927. Accessed November 5, 2019.
Cornelison, G. L., Levy, S. A., Jenson, T., and Frost, B. (2019). Tau-induced nuclear envelope invagination causes a toxic accumulation of mRNA in Drosophila. Aging Cell 18:e12847. doi: 10.1111/acel.12847
Counts, S. E., Alldred, M. J., Che, S., Ginsberg, S. D., and Mufson, E. J. (2014). Synaptic gene dysregulation within hippocampal CA1 pyramidal neurons in mild cognitive impairment. Neuropharmacology 79, 172–179. doi: 10.1016/j.neuropharm.2013.10.018
Cui, J. G., Li, Y. Y., Zhao, Y., Bhattacharjee, S., and Lukiw, W. J. (2010). Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-κB in stressed human astroglial cells and in Alzheimer disease. J. Biol. Chem. 285, 38951–38960. doi: 10.1074/jbc.M110.178848
Davidson, M. W., and Florida State University. (2019). Nuclear Pores. Available online at: https://micro.magnet.fsu.edu/cells/nucleus/nuclearpores.html. Accessed November 5, 2019.
Dhillon, J., Li, Z., and Ortiz, R. M. (2019). Almond snacking for 8 wk increases α-diversity of the gastrointestinal microbiome and decreases Bacteroides fragilis abundance compared with an isocaloric snack in college freshmen. Curr. Dev. Nutr. 3:nzz079. doi: 10.1093/cdn/nzz079
Di Lorenzo, F., De Castro, C., Silipo, A., and Molinaro, A. (2019). Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 43, 257–272. doi: 10.1093/femsre/fuz002
Di Scala, C., Yahi, N., Boutemeur, S., Flores, A., Rodriguez, L., Chahinian, H., et al. (2016). Common molecular mechanism of amyloid pore formation by Alzheimer’s β-amyloid peptide and α-synuclein. Sci. Rep. 6:28781. doi: 10.1038/srep28781
Emery, D. C., Shoemark, D. K., Batstone, T. E., Waterfall, C. M., Coghill, J. A., Cerajewska, T. L., et al. (2017). 16S rRNA next generation sequencing analysis shows bacteria in Alzheimer’s post-mortem brain. Front. Aging Neurosci. 9:195. doi: 10.3389/fnagi.2017.00195
Erdö, F., and Krajcsi, P. (2019). Age-related functional and expressional changes in efflux pathways at the blood-brain barrier. Front. Aging Neurosci. 11:196. doi: 10.3389/fnagi.2019.00196
Fathi, P., and Wu, S. (2016). Isolation, detection, and characterization of enterotoxigenic Bacteroides fragils in clinical samples. Open Microbiol. J. 10, 57–63. doi: 10.2174/1874285801610010057
Fields, C., Adams, M. D., White, O., and Venter, J. C. (1994). How many genes in the human genome? Nat. Genet. 7, 345–346. doi: 10.1038/ng0794-345
Fox, M., Knorr, D. A., and Haptonstall, K. M. (2019). Alzheimer’s disease and symbiotic microbiota: an evolutionary medicine perspective. Ann. N Y Acad. Sci. 1449, 3–24. doi: 10.1111/nyas.14129
Gu, S. M., Lee, H. P., Ham, Y. W., Son, D. J., Kim, H. Y., Oh, K. W., et al. (2018). Piperlongumine improves lipopolysaccharide-induced amyloidogenesis by suppressing NF-kB pathway. Neuromolecular Med. 20, 312–327. doi: 10.1007/s12017-018-8495-9
Heinritz, S. N., Weiss, E., Eklund, M., Aumiller, T., Heyer, C. M., Messner, S., et al. (2016). Impact of a high-fat or high-fiber diet on intestinal microbiota and metabolic markers in a pig model. Nutrients 8:E317. doi: 10.3390/nu8050317
Hicks, M., Bartha, I., di Iulio, J., Venter, J. C., and Telenti, A. (2019). Functional characterization of 3D protein structures informed by human genetic diversity. Proc. Natl. Acad. Sci. U S A 116, 8960–8965. doi: 10.1073/pnas.1820813116
Hill, J. M., Bhattacharjee, S., Pogue, A. I., and Lukiw, W. J. (2014a). The gastrointestinal tract microbiome and potential link to Alzheimer’s disease. Front. Neurol. 5:43. doi: 10.3389/fneur.2014.00043
Hill, J. M., Clement, C., Pogue, A. I., Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2014b). Pathogenic microbes, the microbiome, and Alzheimer’s disease (AD). Front. Aging Neurosci. 6:127. doi: 10.3389/fnagi.2014.00127
Hill, J. M., and Lukiw, W. J. (2015). Microbial-generated amyloids and Alzheimer’s disease (AD). Front. Aging Neurosci. 7:9. doi: 10.3389/fnagi.2015.00009
Holton, J. (2008). Enterotoxigenic Bacteroides fragilis. Curr. Infect. Dis. Rep. 10, 99–104. doi: 10.1007/s11908-008-0018-7
Huang, P., and Liu, Y. (2019). A reasonable diet promotes balance of intestinal microbiota: prevention of precolorectal cancer. Biomed. Res. Int. 2019:3405278. doi: 10.1155/2019/3405278
Jaber, V. R., Zhao, Y., Sharfman, N. M., Li, W., and Lukiw, W. J. (2019). Addressing Alzheimer’s disease (AD) neuropathology using anti-microRNA (AM) strategies. Mol. Neurobiol. 56, 8101–8108. doi: 10.1007/s12035-019-1632-0
Jang, H., Arce, F. T., Lee, J., Gillman, A. L., Ramachandran, S., Kagan, B. L., et al. (2016). Computational methods for structural and functional studies of Alzheimer’s amyloid ion channels. Methods Mol. Biol. 1345, 251–268. doi: 10.1007/978-1-4939-2978-8_16
Javan, G. T., Finley, S. J., Tuomisto, S., Hal, L. A., Benbow, M. E., and Mills, D. (2019). An interdisciplinary review of the thanatomicrobiome in human decomposition. Forensic Sci. Med. Pathol. 15, 75–83. doi: 10.1007/s12024-018-0061-0
Jeon, J. I., Ko, S. H., and Kim, J. M. (2019). Intestinal epithelial cells exposed to Bacteroides fragilis enterotoxin regulates NF-κB activation and inflammatory responses through β-catenin expression. Infect. Immun. 87:e00312-19. doi: 10.1128/iai.00312-19
Kabachinski, G., and Schwartz, T. U. (2015). The nuclear pore complex-structure and function at a glance. J. Cell Sci. 128, 423–429. doi: 10.1242/jcs.083246
Köhler, C. A., Maes, M., Slyepchenko, A., Berk, M., Solmi, M., Lanctôt, K. L., et al. (2016). The gut-brain axis, including the microbiome, leaky gut and bacterial translocation: mechanisms and pathophysiological role in Alzheimer’s disease. Curr. Pharm. Des. 22, 6152–6166. doi: 10.2174/1381612822666160907093807
Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T., and Lansbury, P. T. Jr. (2002). Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418:291. doi: 10.1038/418291a
Leshchyns’ka, I., and Sytnyk, V. (2016). Synaptic cell adhesion molecules in Alzheimer’s disease. Neural Plast. 2016:6427537. doi: 10.1155/2016/6427537
Lin, D. H., and Hoelz, A. (2019). The structure of the nuclear pore complex (an update). Annu. Rev. Biochem. 88, 725–783. doi: 10.1146/annurev-biochem-062917-011901
Li, D., and Yu, F. (2017). Peripheral inflammatory biomarkers and cognitive decline in older adults with and without Alzheimer’s disease: a systematic review. J. Gerontol. Nurs. 43, 53–60. doi: 10.3928/00989134-20170519-01
Lukiw, W. J. (2016a). Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer’s disease. Front. Microbiol. 7:1544. doi: 10.3389/fmicb.2016.01544
Lukiw, W. J. (2016b). The microbiome, microbial-generated pro-inflammatory neurotoxins, and Alzheimer’s disease. J. Sport Health Sci. 5, 393–396. doi: 10.1016/j.jshs.2016.08.008
Lukiw, W. J., Cong, L., Jaber, V., and Zhao, Y. (2018). Microbiome-derived lipopolysaccharide (LPS) selectively inhibits neurofilament light chain (NF-L) Gene expression in human neuronal-glial (HNG) cells in primary culture. Front. Neurosci. 12:896. doi: 10.3389/fnins.2018.00896
Mazmanian, S. (2019). Bacteria and Brains. Available online at: https://www.genengnews.com/insights/bacteria-and-brains-an-interview-with-microbiome-expert-sarkis-mazmanian/. Accessed November 5 2019.
Montagne, A., Zhao, Z., and Zlokovic, B. V. (2017). Alzheimer’s disease: a matter of blood-brain barrier dysfunction? J. Exp. Med. 214, 3151–3169. doi: 10.1084/jem.20171406
National Institutes of Health, US National Library of Medicine; National Center for Biotechnology Information; PubChem. (2019). Available online at: https://pubchem.ncbi.nlm.nih.gov/compound/Human-beta-amyloid-peptide 1–42. Accessed November 5, 2019.
Nguyen, P. H., Campanera, J. M., Ngo, S. T., Loquet, A., and Derreumaux, P. (2019). Tetrameric Aβ40 and Aβ42 β-barrel structures by extensive atomistic simulations. i. in a bilayer mimicking a neuronal membrane. J. Phys. Chem. B 123, 3643–3648. doi: 10.1021/acs.jpcb.9b01206
Österlund, N., Moons, R., Ilag, L. L., Sobott, F., and Gräslund, A. (2019). Native ion mobility-mass spectrometry reveals the formation of β-barrel shaped amyloid-β hexamers in a membrane-mimicking environment. J. Am. Chem. Soc. 141, 10440–10450. doi: 10.1021/jacs.9b04596
Panza, F., Lozupone, M., Solfrizzi, V., Watling, M., and Imbimbo, B. P. (2019). Time to test antibacterial therapy in Alzheimer’s disease. Brain 142, 2905–2929. doi: 10.1093/brain/awz244
Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Parra-Damas, A., and Saura, C. A. (2019). Synapse-to-nucleus signaling in neurodegenerative and neuropsychiatric disorders. Biol. Psychiatry 86, 87–96. doi: 10.1016/j.biopsych.2019.01.006
Patrick, K. L., Bell, S. L., Weindel, C. G., and Watson, R. O. (2019). Exploring the “Multiple-hit Hypothesis” of neurodegenerative disease: bacterial infection comes up to bat. Front. Cell. Infect. Microbiol. 9:138. doi: 10.3389/fcimb.2019.00138
Poeker, S. A., Geirnaert, A., Berchtold, L., Greppi, A., Krych, L., Steinert, R. E., et al. (2018). Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model. Sci. Rep. 8:4318. doi: 10.1038/s41598-018-22438-y
Pogue, A. I., and Lukiw, W. J. (2018). Up-regulated pro-inflammatory microRNAs (miRNAs) in Alzheimer’s disease (AD) and age-related macular degeneration (AMD). Cell. Mol. Neurobiol. 38, 1021–1031. doi: 10.1007/s10571-017-0572-3
Pretorius, E., Page, M. J., Mbotwe, S., and Kell, D. B. (2018). Lipopolysaccharide-binding protein (LBP) can reverse the amyloid state of fibrin seen or induced in Parkinson’s disease. PLoS One 13:e0192121. doi: 10.1371/journal.pone.0192121
Raetz, C. R., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Rios-Covian, D., Salazar, N., Gueimonde, M., and de Los Reyes-Gavilan, C. G. (2017). Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Front. Microbiol. 8:376. doi: 10.3389/fmicb.2017.00376
Roberts, R. C., Farmer, C. B., and Walker, C. K. (2018). “The human brain microbiome; there are bacteria in our brains! Session 594-Neuroimmunology: Regulating Systems,” in Poster at the 594.08/YY23 Neuroscience Meeting Planner, San Diego, CA: Society for Neuroscience. Available online at: https://www.abstractsonline.com/pp8//4649/presentation/32057.
Roy Sarkar, S., and Banerjee, S. (2019). Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 328, 98–104. doi: 10.1016/j.jneuroim.2019.01.004
Sciacca, M. F., Kotler, S. A., Brender, J. R., Chen, J., Lee, D. K., and Ramamoorthy, A. (2012). Two-step mechanism of membrane disruption by Aβ through membrane fragmentation and pore formation. Biophys. J. 103, 702–710. doi: 10.1016/j.bpj.2012.06.045
Sears, C. L. (2009). Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin. Microbiol. Rev. 22, 349–369. doi: 10.1128/CMR.00053-08
Sheppard, O., Coleman, M. P., and Durrant, C. S. (2019). Lipopolysaccharide-induced neuroinflammation induces presynaptic disruption through a direct action on brain tissue involving microglia-derived interleukin 1β. J. Neuroinflammation 16:106. doi: 10.1186/s12974-019-1490-8
Simpson, H. L., and Campbell, B. J. (2015). Review article: dietary fibre-microbiota interactions. Aliment. Pharmacol. Ther. 42, 158–179. doi: 10.1111/apt.13248
Sun, J., Shi, Y., and Yildirim, E. (2019). The nuclear pore complex in cell type-specific chromatin structure and gene regulation. Trends Genet. 35, 579–588. doi: 10.1016/j.tig.2019.05.006
Sweeney, R. P., and Lowary, T. L. (2019). New insights into lipopolysaccharide assembly and export. Curr. Opin. Chem. Biol. 53, 37–43. doi: 10.1016/j.cbpa.2019.07.004
Sweeney, M. D., Sagare, A. P., and Zlokovic, B. V. (2018). Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133–150. doi: 10.1038/nrneurol.2017.188
Tellez-Merlo, G., Morales-Medina, J. C., Camacho-Ábrego, I., Juárez-Díaz, I., Aguilar-Alonso, P., de la Cruz, F., et al. (2019). Prenatal immune challenge induces behavioral deficits, neuronal remodeling and increases brain nitric oxide and zinc levels in the male rat offspring. Neuroscience 406, 594–605. doi: 10.1016/j.neuroscience.2019.02.018
Ticinesi, A., Tana, C., and Nouvenne, A. (2019). The intestinal microbiome and its relevance for functionality in older persons. Curr. Opin. Clin. Nutr. Metab. Care 22, 4–12. doi: 10.1097/MCO.0000000000000521
Tierney, B. T., Yang, Z., Luber, J. M., Beaudin, M., Wibowo, M. C., Baek, C., et al. (2019). The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe 26, 283.e8–295.e8. doi: 10.1016/j.chom.2019.07.008
Tsukamoto, H., Takeuchi, S., Kubota, K., Kobayashi, Y., Kozakai, S., Ukai, I., et al. (2018). Lipopolysaccharide (LPS)-binding protein stimulates CD14-dependent Toll-like receptor 4 internalization and LPS-induced TBK1-IKKε-IRF3 axis activation. J. Biol. Chem. 293, 10186–10201. doi: 10.1074/jbc.M117.796631
Tulkens, J., Vergauwen, G., Van Deun, J., Geeurickx, E., Dhondt, B., Lippens, L., et al. (2018). Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut doi: 10.1158/1538-7445.sabcs18-1489 [Epub ahead of print].
Ullah, G., Demuro, A., Parker, I., and Pearson, J. E. (2015). Analyzing and modeling the kinetics of amyloid β pores associated with Alzheimer’s disease pathology. PLoS One 9:e0137357. doi: 10.1371/journal.pone.0137357
Vander Zanden, C. M., Wampler, L., Bowers, I., Watkins, E. B., Majewski, J., and Chi, E. Y. (2019). Fibrillar and nonfibrillar amyloid β structures drive two modes of membrane-mediated toxicity. Langmuir doi: 10.1021/acs.langmuir.9b02484 [Epub ahead of print].
Varatharaj, A., and Galea, I. (2017). The blood-brain barrier in systemic inflammation. Brain Behav. Immun. 60, 1–12. doi: 10.1016/j.bbi.2016.03.010
Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., et al. (2001). The sequence of the human genome. Science 291, 1304–1351. doi: 10.1126/science.1058040
Wexler, A. G., and Goodman, A. L. (2017). An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2:17026. doi: 10.1038/nmicrobiol.2017.26
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi: 10.1146/annurev-biochem-060713-035600
Wu, S., Lim, K. C., Huang, J., Saidi, R. F., and Sears, C. L. (1998). Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. U S A 95, 14979–14984. doi: 10.1073/pnas.95.25.14979
Yang, N. J., and Chiu, I. M. (2017). Bacterial signaling to the nervous system through toxins and metabolites. J. Mol. Biol. 429, 587–605. doi: 10.1016/j.jmb.2016.12.023
Zhan, X., Stamova, B., Jin, L. W., DeCarli, C., Phinney, B., and Sharp, F. R. (2016). Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 87, 2324–2332. doi: 10.1212/WNL.0000000000003391
Zhan, X., Stamova, B., and Sharp, F. R. (2018). Lipopolysaccharide associates with amyloid plaques, neurons and oligodendrocytes in Alzheimer’s disease brain: a review. Front. Aging Neurosci. 10:42. doi: 10.3389/fnagi.2018.00042
Zhao, Y., Cong, L., Jaber, V., and Lukiw, W. J. (2017a). Microbiome-derived lipopolysaccharide enriched in the perinuclear region of Alzheimer’s disease brain. Front. Immunol. 8:1064. doi: 10.3389/fimmu.2017.01064
Zhao, Y., Cong, L., and Lukiw, W. J. (2017b). Lipopolysaccharide (LPS) accumulates in neocortical neurons of Alzheimer’s disease (AD) brain and impairs transcription in human neuronal-glial primary co-cultures. Front. Aging Neurosci. 9:407. doi: 10.3389/fnagi.2017.00407
Zhao, Y., Jaber, V., and Lukiw, W. J. (2017c). Secretory products of the human GI tract microbiome and their potential impact on Alzheimer’s disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus. Front. Cell. Infect. Microbiol. 7:318. doi: 10.3389/fcimb.2017.00318
Zhao, Y., Dua, P., and Lukiw, W. J. (2015). Microbial sources of amyloid and relevance to amyloidogenesis and Alzheimer’s disease (AD). J. Alzheimers Dis. Parkinsonism 5:177. doi: 10.4172/2161-0460.1000177
Zhao, Y., and Lukiw, W. J. (2015). Microbiome-generated amyloid and potential impact on amyloidogenesis in Alzheimer’s disease (AD). J. Nat. Sci. 7:e138.
Zhao, Y., and Lukiw, W. J. (2018a). Bacteroidetes neurotoxins and inflammatory neurodegeneration. Mol. Neurobiol. 55, 9100–9107. doi: 10.1007/s12035-018-1015-y
Zhao, Y., and Lukiw, W. J. (2018b). Microbiome-mediated upregulation of microRNA-146a in sporadic Alzheimer’s disease. Front. Neurol. 9:145. doi: 10.3389/fneur.2018.00145
Zhao, Y., Sharfman, N. M., Jaber, V. R., and Lukiw, W. J. (2019). Down-regulation of essential synaptic components by GI-tract microbiome-derived lipopolysaccharide (LPS) in LPS-treated human neuronal-glial (HNG) cells in primary culture: relevance to Alzheimer’s disease (AD). Front. Cell. Neurosci. 13:314. doi: 10.3389/fncel.2019.00314
Zhou, W., and Bian, Y. (2018). Thanatomicrobiome composition profiling as a tool for forensic investigation. Forensic Sci. Res. 3, 105–110. doi: 10.1080/20961790.2018.1466430
Keywords: Alzheimer’s disease (AD), brain microbiome, dysbiosis, gastrointestinal (GI) tract, lipopolysaccharide (LPS), neurofilament light (NF-L), synapsin-1 (SYN1), the thanato-microbiome (the post-mortem microbiome)
Citation: Lukiw WJ, Li W, Bond T and Zhao Y (2019) Facilitation of Gastrointestinal (GI) Tract Microbiome-Derived Lipopolysaccharide (LPS) Entry Into Human Neurons by Amyloid Beta-42 (Aβ42) Peptide. Front. Cell. Neurosci. 13:545. doi: 10.3389/fncel.2019.00545
Received: 29 September 2019; Accepted: 22 November 2019;
Published: 06 December 2019.
Edited by:
Elisa L. Hill-Yardin, RMIT University, AustraliaReviewed by:
Stuart Douglas Portbury, University of Melbourne, AustraliaCopyright © 2019 Lukiw, Li, Bond and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walter J. Lukiw, d2x1a2l3QGxzdWhzYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.