
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 07 February 2017
Sec. Cellular Neuropathology
Volume 11 - 2017 | https://doi.org/10.3389/fncel.2017.00023
 Martyna Podobinska1
Martyna Podobinska1 Ilona Szablowska-Gadomska2
Ilona Szablowska-Gadomska2 Justyna Augustyniak1
Justyna Augustyniak1 Ioanna Sandvig3
Ioanna Sandvig3 Axel Sandvig3
Axel Sandvig3 Leonora Buzanska1*
Leonora Buzanska1*The coordinated development of the nervous system requires fidelity in the expression of specific genes determining the different neural cell phenotypes. Stem cell fate decisions during neurodevelopment are strictly correlated with their epigenetic status. The epigenetic regulatory processes, such as DNA methylation and histone modifications discussed in this review article, may impact both neural stem cell (NSC) self-renewal and differentiation and thus play an important role in neurodevelopment. At the same time, stem cell decisions regarding fate commitment and differentiation are highly dependent on the temporospatial expression of specific genes contingent on the developmental stage of the nervous system. An interplay between the above, as well as basic cell processes, such as transcription regulation, DNA replication, cell cycle regulation and DNA repair therefore determine the accuracy and function of neuronal connections. This may significantly impact embryonic health and development as well as cognitive processes such as neuroplasticity and memory formation later in the adult.
The development of the nervous system is regulated by a multitude of intracellular molecular and cellular signals interacting with the extracellular microenvironment in a temporal and spatial manner. These signals induce expression of genes involved in lineage commitment, differentiation, maturation, migration and cell survival. In addition, silencing of genes responsible for the maintenance of stem cells in a pluripotent state and for cell fate decisions are important hallmarks of neurodevelopment (Lilja et al., 2012). Neural stem cells (NSCs) are able to proliferate and self-renew and also give rise to all neural lineage cells in the central nervous system (CNS): neurons, astrocytes and oligodendrocytes (Altman, 1962; Gage, 2000).
After development is completed, NSCs are still present in the adult brain throughout its entire lifetime in two neurogenic niches: the subventricular zone (SVZ) lining the lateral ventricles, and the subgranular zone (SGZ), which is part of the dentate gyrus (DG) of the hippocampus (Eriksson et al., 1998). It is well documented in pioneering (Hsieh and Gage, 2004) and more recent studies that epigenetic alteration plays a crucial role in the maintenance of the resident NSC multipotent state as well as in their differentiation process (Mohamed Ariff et al., 2012).
The term “epigenetics” was coined by Conrad Waddington (1905–1975) in 1942 to describe interactions between genotype and the environment, which together shape the expressed characteristic traits of an organism, defined as the phenotype (Waddington, 1942). Different definitions of the term epigenetics exist and a clear consensus is still lacking. The name epi- (Greek: ɛπí- over, above, outer)-genetics refers to exogenous dynamic mechanisms which change gene expression without changing the DNA sequence. Epigenetic changes can however be inherited through subsequent cell divisions. Each cell type has a unique gene expression profile (transcriptome) and also a distinctive chromatin signature (Sha and Boyer, 2009). Interestingly, epigenetic changes are dependent on environmental factors. It has been documented that maternal behavior during pregnancy or prenatal stress may lead to epigenetic alterations in the offspring (Wu et al., 2004; Morgan and Bale, 2011).
Epigenetic mechanisms that control changes in gene expression levels can be divided into three main groups: (i) DNA methylation; (ii) histone and chromatin modifications, including histone variants; and (iii) non-coding RNAs (ncRNAs) such as long non-coding RNA (lncRNA) and small non-coding RNAs (sncRNAs), including microRNAs (miRNAs); and small interfering RNAs (siRNAs; Jenuwein and Allis, 2001; Sun et al., 2008; Mohamed Ariff et al., 2012; Fenoglio et al., 2013).
This review article will focus on the regulation of DNA methylation and two types of histone modification: methylation and acetylation and their relevance to stem cell fate decisions in neurodevelopmental processes. The epigenetic events are considered to act in a switch-like mode (Hoffmann et al., 2015), however the potential of the cell to undertake developmental decisions, stemness/lineage commitment and further differentiation is highly dependent on the activity of the genes typical for the defined stage of development. A schematic correlation of neurodevelopmental hierarchy of stem cells, along with their epigenetic status is presented on Figure 1. Developmental genes are referred to as being at an activated (on), repressed (off) or bivalent (poised) stage. This is linked to the pattern of DNA and histone permissive and inhibitory epigenetic marks, which are further discussed in this review article.
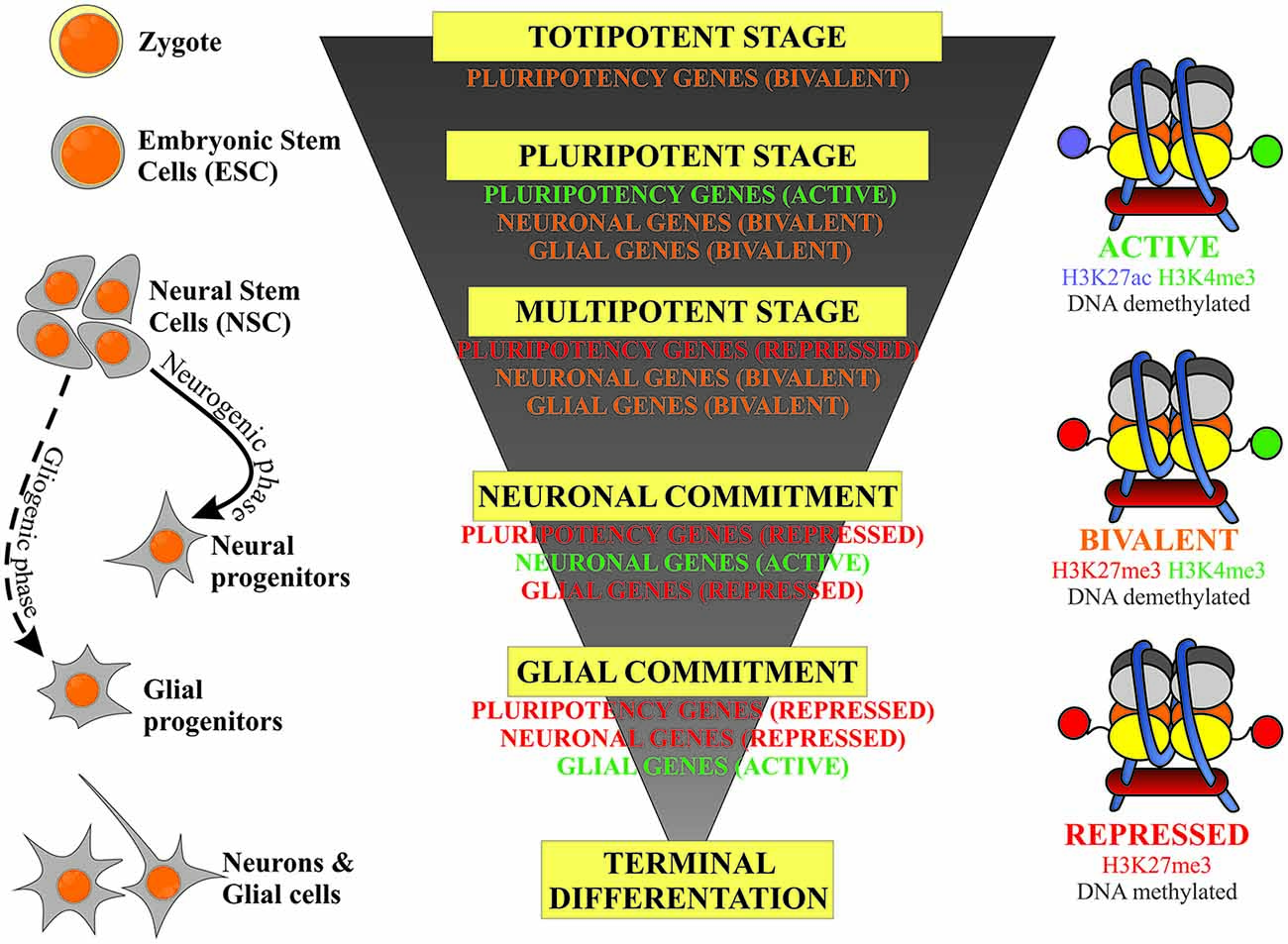
Figure 1. Schematic representation of the correlation of the stem cells’ developmental hierarchy with their epigenetic status based on the balance between inhibitory (DNA and H3K27 methylation) and permissive (H3K4 methylation) epigenetic marks.
DNA methylation is a crucial process in embryogenesis and normal development and one of the most frequently investigated epigenetic modifications. It is involved in regulation of gene expression (Keshet et al., 1986), X-chromosome inactivation (Heard et al., 1997), genomic imprinting (Li et al., 1993; Denomme and Mann, 2013), regulation of chromatin structure (Bernstein et al., 2007), transposon silencing (Walsh et al., 1998) and control of telomere length (Gonzalo et al., 2006). Methylation of cytosine at the C5 position is catalyzed by various DNA methyltransferases (DNMTs), which catalyze the transfer of a methyl group (-CH3), derived from the S-adenosylmethionine (SAM; methyl donor) onto cytosine and, as a result, lead to formation of 5-methylcytosine (5mC; Cheng et al., 1993). There is also an alternative form of DNA methylation; Kriaucionis and Heintz (2009) discovered 5-hydroxymethylcytosine (5hmC), an unusual DNA nucleotide that occurs frequently in the mouse brain.
The DNA methylation pattern is established during embryonic development by de novo DNMTs (DNMT3A and DNMT3B supported by DNMT3L), while maintenance of this pattern is linked with activity of the DNMT1 (Razin and Szyf, 1984; Li and Zhao, 2008). Lister et al. (2009) suggest that DNMT3A may be responsible for catalyzing cytosine methylation at non-5′-C-phosphate-G-3′(CpG) sites.
An interesting fact is that in mammals the DNA of mature sperm and egg is highly methylated compared with somatic cells. However, the largest extent of this methylation disappears during the preimplantation developmental stage (genome wide demethylation). After implantation, the embryo undergoes a de novo DNA methylation process (Reik et al., 2001; Denomme and Mann, 2013). Furthermore, Dnmt1 or Dnmt3a and Dnmt3b knockout mice die during mid-gestation (Li et al., 1992; Okano et al., 1999; Ueda et al., 2006). There are two mechanisms for the regulation of gene expression by DNA methylation: (i) DNA methylation prevents the binding of certain transcription factors, which blocks transcription; and (ii) methylated CpG dinucleotides are recognized by a family of proteins containing methyl-CpG-binding domain (MBD) such as MBD1 and methyl CpG binding protein 2 (MeCP2), which actively block their binding sites and are consequently responsible for gene inactivation (Li and Zhao, 2008; Sun et al., 2008). MBD-proteins can block transcription alone or in cooperation with enzymes responsible for histone modification (Nan et al., 1998). Interestingly, recent studies have shown that actively transcribed genes have high levels of gene body methylation (Lister et al., 2009; Wu et al., 2010).
Traditionally DNA methylation was considered as stable, irreversible epigenetic modification. However, studies conducted in recent years revealed methylation changes in postnatal brains (Guo et al., 2011a).
It has been proposed that the demethylation process may take place in two different ways; either active or passive. The passive way is associated with suppression of DNMT1, while active demethylation occurs enzymatically and is catalyzed by TET1—tet methylcytosine dioxygenase 1 (5mC hydroxylase) (Wu and Zhang, 2010; Guo et al., 2011b) involved in oxidative reaction of 5mC to 5hmC transition.
The other mechanism, which is involved in active DNA demethylation is associated with growth arrest and DNA-damage-inducible alpha (GADD45A) protein. GADD45 proteins are involved in many processes associated with cellular response to stress. They have been implicated in cell-cycle control, DNA repair and suppression of cell growth (Kaufmann et al., 2011). High GADD45A expression is associated with global DNA demethylation (Barreto et al., 2007). Gadd45 genes mediate the repair-based DNA demethylation and may occur by three mechanisms as described (Teperek-Tkacz et al., 2011; Niehrs and Schäfer, 2012). Some authors have suggested that DNA methylation patterns may be useful epigenetic markers in characterizing different types of pluripotent and differentiated cells (Watanabe et al., 2012). The basis for their assumptions is the fact that there are different patterns of DNA methylation in pluripotent stem cells, during their differentiation and in terminally differentiated cells. It has been demonstrated that the DNA methylation profile undergoes dynamic changes during the process of cell differentiation, particularly in HCP promoters (promoters with high CpG content; Meissner et al., 2008).
Dnmt1, Dnmt3a and Dnmt3b are expressed during CNS development and differentiation (Feng et al., 2007; Hutnick et al., 2009). As indicated above, DNA methylation has been shown to play an important role during embryonic development but is also significant in the formation of the nervous system as well as for NSC differentiation (Stroud et al., 2011; Szulwach et al., 2011; Wu et al., 2011; Chen et al., 2013, 2014). Alterations in DNA methylation during CNS development are consistent with the regulation of chromatin-modifying enzymes occurring during embryonic development (Gapp et al., 2014). For example, DNMT1 and DNMT3 maintain DNA methylation and regulate the division of neural progenitor cells and mature neurons as well as play an important role in adult neurogenesis and synaptic plasticity of mature neurons. Furthermore, Dnmt1 and Dnmt3a deletion in mouse forebrain excitatory neurons caused a decrease in DNA methylation, abnormal neuron size and dysregulation in expression of genes important for synaptic plasticity (class I MHC and STAT1). These changes resulted in dysfunction in long-term plasticity in the hippocampal CA1 region, and caused learning and memory deficits (Feng et al., 2010). It was also shown that lack of DNMT1 results in perturbation of resident populations of neural stem/precursor cells and post-mitotic neurons. Specifically, the differences were, respectively, in terms of reduced numbers of neurons and a reduction of cortical and hippocampal volumes, and a negative effect on neuronal survival (Noguchi et al., 2015). Moreover Noguchi et al. (2016) showed that presence of DNMT1 is essential for hippocampal DG development, given that its lack in early development leads to poor granule cell layer in the adult DG. It was also revealed that during neurogenesis DNMT1 plays a pivotal role in the regulation of neural differentiation. Such control of NSC differentiation also serves the purpose of preventing astrocytes from premature differentiation (Noguchi et al., 2016).
DNMT3A influences mouse NSC maturation and differentiation. During neurogenesis, DNMT3A is expressed in embryonic neural precursor cells (NPCs) within the SVZ and in postmitotic CNS neurons as well as in oligodendrocytes (Feng et al., 2005). In order to understand the role of DNMT3A in NSC differentiation and proliferation, Wu et al. (2012) used NSC derived from DNMT3A-deficient mouse embryonic stem cells (ESCs). These Dnmt3a−/− NSCs were globally hypomethylated and exhibited an increased proliferation rate compared to control NSCs, which indicates that DNMT3A plays an important role in mouse NSC proliferation. Moreover, the same authors observed altered timing of NSC differentiation. In early passages (P3), Dnmt3a−/− NSCs exhibited precocious differentiation towards both astrocytes and oligodendrocytes, while neuronal differentiation was not impaired. Furthermore, loss of DNMT3A led to an increased number of both astrocytes and oligodendrocytes (Wu et al., 2012).
During development, neural precursors in the SVZ give rise to neurons and, subsequently, astrocytes and oligodendrocytes. This developmental, hierarchical transition is closely associated with epigenetic remodeling of many genes. The principal role in astrogliogenesis is played by Glial Fibrillary Acidic Protein (GFAP) transcription, which is activated by developmental demethylation of its promotor. Since the process of GFAP activation is triggered by STAT3 transcription factor, the methylation of the CpG at the STAT3 binding site in the GFAP promotor represses gliogenesis (Takizawa et al., 2001; Feng et al., 2007; Namihira et al., 2009). These data focused attention to DNA methylation as a critical determinant of astrocyte differentiation in the fetal brain.
Recent studies have shown that during the perinatal period, the male and female mouse brains differ in DNMT enzyme activity in the highly sexually dimorphic preoptic area (POA). Exposure to gonadal steroids leads to a decrease in DNMT activity, which results in a reduction of DNA methylation (i.e., a reduction of fully methylated CpG sites) and activation of masculinizing genes. Moreover, DNMT3A inhibition caused brain masculinization, which resulted in male copulative behavior. These findings indicated that brain feminization is an active process maintained by higher levels DNA methylation (Nugent et al., 2015).
The family of Gadd45 genes is another important player in CNS development. Candal et al. (2004) have shown that Gadd45gamma is involved in the development of medaka fish brain, specifically in cell fate decisions, by affecting cell cycle exit. Further research demonstrated that, during mouse neural development, mainly two of Gadd45 genes, Gadd45a and Gadd45g, are expressed with slight temporospatial differences. High Gadd45a expression is present in the neural tube during its closure, in the dorsal root and cranial ganglia VII-X, and in the olfactory epithelium. Expression of Gadd45g is also observed in the neural tube, dorsal root and cranial ganglia VII-X, and olfactory epithelium, but also in neural precursors, forebrain, midbrain and hindbrain (Kaufmann et al., 2011).
Furthermore, recent studies have shown that Gadd45b expression (but not Gadd45a, or Gadd45g) and DNA methylation in the developing rodent amygdala are different in individuals of different sexes (Kigar et al., 2016).
Many studies have shown that epigenetic regulation affects activity-dependent mature brain functions. Activity-dependent neurogenesis in the adult hippocampus is one of the mechanisms involved in neural plasticity. However, the specific mechanisms of action of this phenomenon are poorly understood. Ma et al. (2009) discovered that Gadd45b is needed for activity-induced DNA demethylation of some of the gene promoters involved in adult neurogenesis. Region-specific DNA demethylation, performed by GADD45B, enabled expression of paracrine neurogenic niche factors (BDNF, FGF-1, FGF-2) in mature neurons, which induced activity-dependent adult neurogenesis (Ma et al., 2009).
Zhou and his group have shown that DNA methylation was programmed along with neural tube development by the pattern of appearance of DNA 5mC and 5hmC epigenetic modifications (Zhou, 2012). They further investigated differences between 5mC and 5hmC methylation sites in the brain (Chen et al., 2014). They discovered that 5hmC is located in alternate regions to 5mC, appears at different times during neurodevelopment and is connected with different MBD proteins. The spatiotemporal pattern of 5mC and 5hmC DNA methylation sites is closely associated with the state of differentiation of NSCs in the neural tube. Appearance of 5hmC is always preceded by the presence of 5mC. It was suggested that the surge of 5mC is connected with priming of NPCs for differentiation. Additionally, Stroud et al. (2011) have shown that the timing of 5hmC appearance is closely related with initiation of neuroepithelial cell differentiation. 5mC and 5hmC reveal different binding partners. While 5mC preferably binds to MBD1 and MeCP2, 5hmC is associated with MBD3. Moreover, negative correlation between the amount of 5hmC and MeCP2 levels is observed. 5mC is mostly distributed in the promoter regions of the genes and is co-localized with repressive histone marks, H3K9me3 and H3K27me3, while 5hmC is localized in the gene body, downstream from the transcription start site (TSS) and is co-localized with the H3K4me2 histone mark, which is a marker of active chromatin (Stroud et al., 2011). Szulwach et al. (2011) examined 5hmC occurrence in mouse cerebellum and hippocampus during neurodevelopment. They found a correlation in the spatial distribution between 5hmC and NeuN-positive neuronal cells in the granule cell layer in developing and adult cerebellum, wherein the amount of 5hmC increased in the latter. In calbindin-positive Purkinje cells the amount of 5hmC was increased in both developing and adult cerebellum, compared to NeuN-positive cells. However, the progenitor cells in the external granule layer, which are a highly proliferative fraction, lack 5hmC. In the hippocampus, these authors observed very low levels or lack of 5hmC in immature (NeuN-negative) neurons. The amount of 5hmC clearly increased in NeuN-positive cells in the DG granule cell layer in both developing and adult mice. Together, these data suggest that 5hmC clearly appears in mature neuronal cells, which implies a role in neuronal development (Szulwach et al., 2011). It is thus suggested that 5hmC is associated with euchromatin and plays an important role in maintaining the activity of genes characteristic for specific NSC subtypes (Stroud et al., 2011; Szulwach et al., 2011; Wu et al., 2011; Chen et al., 2014).
The most recent studies, using single cell immuno-identification and cell-specific quantitative methylation assays, revealed cell-wide DNA de-methylation and subsequent re-methylation of Purkinje Neurons in the developing cerebellum (Zhou et al., 2016). This process involved both 5mC and 5hmC. This is the first time when such global de-methylation has been observed beyond totipotent stages of development.
The correlation of different MBD proteins with the activated euchromatin and inactive heterochromatin sites (respectively MBD1 and MBD3) corresponds to an earlier observation by Zhao et al. (2003) that the absence of MBD1 causes reduced neuronal differentiation and increased genomic instability in NSCs and that adult Mbd1−/− mice exhibit deregulation in the DG of the hippocampus during adult neurogenesis.
TET (10 to 11 translocation) TET1, TET2 and TET3 proteins are dioxygenases that catalyze the conversion of the modified genomic base 5mC into 5hmC and are involved in further oxidative DNA demethylation process (Tahiliani et al., 2009). Thus, TET occupancy at gene promoters is negatively correlated with levels of DNA methylation.
The active participation of TET proteins in neural fate specification has been the subject of intense investigation given that in mammals it occurs abundantly in ESCs and neurons (Wu and Zhang, 2011; Li et al., 2014). Two different lines of evidence confirmed that non-promotor based 5hmC methylation sites are involved in maintaining active chromatin states of neurogenic genes (Wu et al., 2010; Zhang et al., 2013). It was first documented that DNMT3A dependent non-proximal promoter methylation triggers expression of neurogenic genes by functionally antagonizing Polycomb repression (Wu et al., 2010). In addition Zhang et al. (2013) have shown TET1 associated regulation of neural progenitor proliferation in the developing cortex as well as in a population of adult NSCs.
The precise and dynamic pattern of 5mC and 5hmC distribution within chromatin as the specific sites of DNA methylation located either in gene promoters or within the gene body also raise a more general question regarding the role of global methylation/demethylation and their link with the activity of the specific developmental genes. The mechanism underlying the widely accepted observation that DNA 5mC methylation at proximal promoters at CpG islands facilitates silencing of cell type–specific genes, while such lineage restriction is attenuated when 5hmC methylation occurs in non-proximal promoters at euchromatic DNA in of transcriptionally permissive (poised) developmental genes, remains to be elucidated.
The structure of the nucleosome and its post-translational modifications (PTMs) are presented in Figure 2. Nucleosomes are the basic units of chromatin structure, consisting of DNA and histones. DNA with a length of 146 bp is wrapped in 1.67 left-handed superhelical turns around an octameric histone core which comprises a (H3–H4)2 tetramer and two H2A–H2B dimers. Nucleosomes are joined by 10–50-bp-long stretches of unwrapped linker DNA and linker histone (H1), which together are involved in chromatin compaction (Luger et al., 1997; Richmond and Davey, 2003).
Histones are small alkaline proteins consisting of a globular C-terminal domain and an N-terminal tail, which is positively charged and extends outwards from the nucleosome (Jenuwein and Allis, 2001). Histone tails are highly basic due to numerous lysine and arginine residues and play an important role in the structural stability of the nucleosome and in chromatin compaction. Regulation of the structure of nucleosomes is possible due to internucleosomal interactions between N-terminal tails and DNA or other parts of histones (Arya and Schlick, 2009). There are also sites of numerous PTMs, such as acetylation, phosphorylation, methylation, ubiquitination, SUMOylation, ADP-ribosylation, deimination, proline isomerization, crotonylation and citrullination. Both chromatin structure and histone modifications affect the availability of nuclear factors presented to the DNA. PTM can alter chromatin organization and regulate different processes, such as DNA replication, chromatin assembly after replication, transcription or DNA repair processes (Hanks et al., 1983; Shiio and Eisenman, 2003; Cuthbert et al., 2004; Nelson et al., 2006; Shogren-Knaak et al., 2006; Kouzarides, 2007; Ismail et al., 2010; Biswas et al., 2011; Martinez-Zamudio and Ha, 2012; Rothbart and Strahl, 2014). Jenuwein and Allis (2001) proposed the “Histone Code Hypothesis”, suggesting that different combinations of histone modifications may have different outcomes and can interact with each other in a synergistic or antagonistic way. According to this hypothesis, modification marks on the histone tails should be present and can be recognized and thus provide binding sites for effector proteins (Jenuwein and Allis, 2001).
Three main classes of enzymes involved in the PTMs of histones can be distinguished according to their function: “writers”, “readers” and “erasers”. Writers are enzymes that catalyze the addition of methyl, acetyl or other chemical groups to histone tails, for example Polycomb Repressive Complex 2 (PRC2), SUV39H, DOT1L, with histone methyltransferase (HMT) activity. Readers are proteins, that are able to recognize and bind to specific modifications, for example, PRC1, which reads methylation of H3K27 caused by PRC2 and is subsequently involved in histone ubiquitinization, or chromodomain helicase DNA binding protein 1 (CHD1). Erasers are enzymes such as histone deacetylases (HDACs), or lysine specific demethylase (LSD; KDM6B), which are involved in the removal of modifications (Goldberg et al., 2007; Weng et al., 2012). Each modification may have several writers, readers and erasers (Kouzarides, 2007).
Over 580 histone regulators from eight model organisms classified into distinct families have been collected in a large database1 (Xu et al., 2017). Examples of some compounds belonging to these three groups are presented in Figure 3.
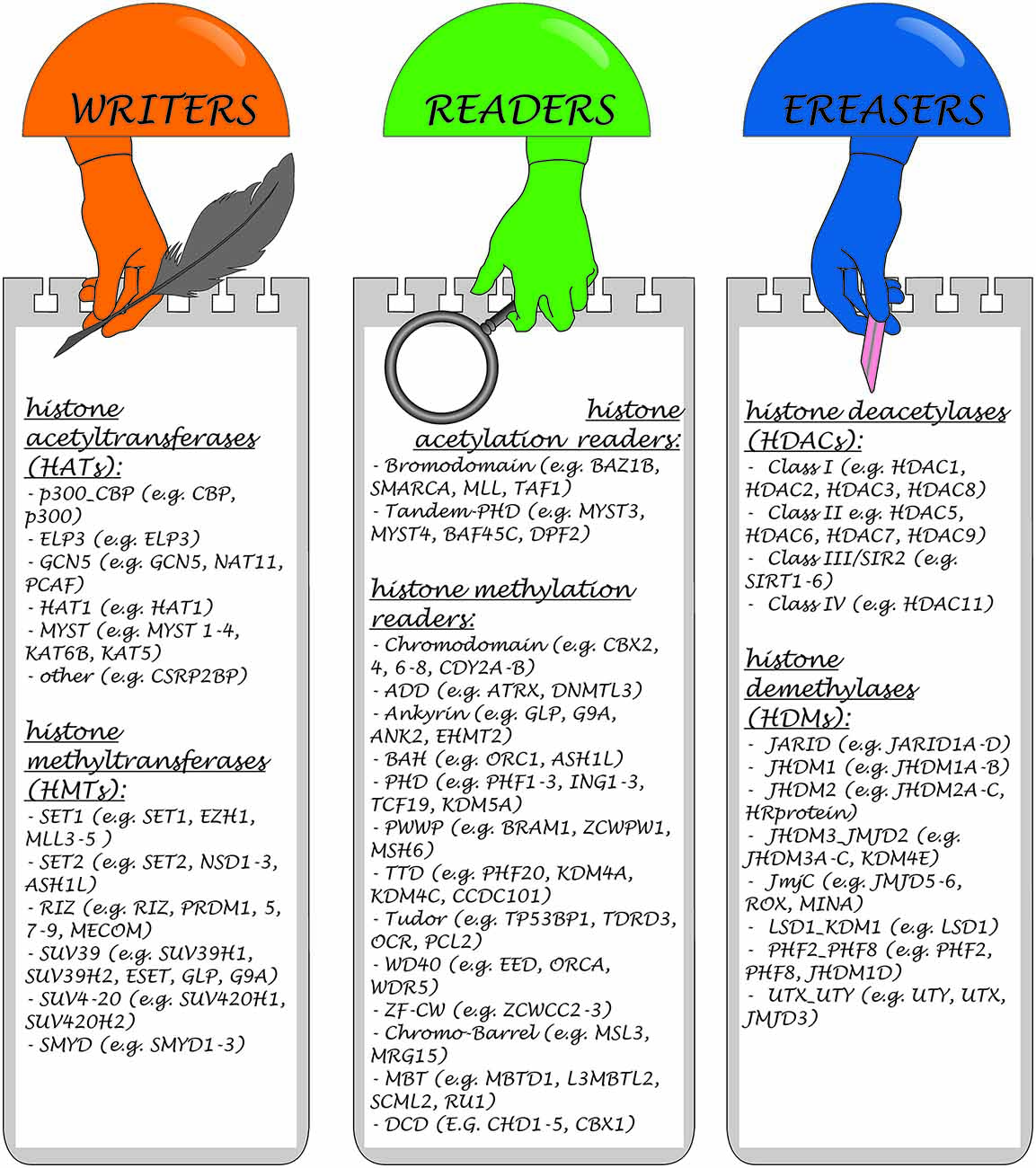
Figure 3. Classification of enzymes involved in the histone PTMs and representative members of each class.
Histone acetylation is the most well-defined histone modification and is linked with the lysine residues on the N-terminal tails of histones H3 (Lys9, Lys14, Lys18, Lys23) and H4 (Lys5, Lys8, Lys12, Lys16). During the acetylation process, acetyl groups from Acetyl-Coenzyme A are transferred to the lysine residues of the histone N termini. This changes the charge and decreases the electrostatic interactions between the positively charged histones and the negatively charged DNA. Furthermore, acetylation of key lysine residues of the core histones H3 and H4 inhibits the association of linker histone—H1, which promotes further chromatin decondensation. This process makes the chromatin available to transcription factors through its relaxation (Ridsdale et al., 1990; Lilja et al., 2012). Histone deacetylation, on the other hand, causes chromatin condensation, preventing binding of transcription factors to their target sequences in gene promoters of NSCs resulting in gene suppression (Hsieh and Gage, 2004).
Potentially active euchromatin and inactive heterochromatin exhibit different acetylation patterns. Euchromatin is normally acetylated at lysine 5, 8 12 and 16 on histone H4, whereas in heterochromatin these sites are hypoacetylated (O’Neill and Turner, 1995). The level of histone acetylation is a dynamic process and depends on two groups of enzymes; histone acetyltransferases (HAT), which are capable of activating acetylation, and HDACs removing this modification, i.e., HDAC activity. So far, eighteen HDACs have been identified in mammals. Based on domain organization and sequence homology with yeast, HDACs can be grouped into four classes (Table 1). It has been shown that each class has a different expression pattern. For example, class I HDACs 1, 2, 3, 8 are the most commonly localized in the nucleus of all cells. On the other hand, HDAC4, 5, 7 and 9 (class II) are found in both the nucleus and cytoplasm and exhibit more tissue-specific expression, suggesting that they are involved in cell differentiation and tissue specification processes (Haigis and Sinclair, 2010; Lian et al., 2012; New et al., 2012).
In mammals, HDACs from class I and II contain two catalytic domains or form dimers. This suggests that some proteins require two active centers within the HDACs complex (Zhang et al., 2006). So far, several complexes containing HDAC1/HDAC2 have been identified.
One of them, the SIN3/HDAC corepressor complex, is a multiprotein complex implicated in the regulation of key biological processes such as cell cycle regulation, differentiation, protein stabilization and destabilization, transcriptional regulation, cellular senescence or energy metabolism by deacetylation of histones H3 and H4. The SIN3/HDAC corepressor complex is comprised of several proteins, including SIN3 Associated Proteins 30 (SAP30), SAP18, HDAC1, HDAC2, retinoblastoma binding protein 4 (RBAP4), RBAP7, SDS3, MeCP2 and Rb-binding protein (RBP1). The modular structure of the SIN3/HDAC corepressor complex enables different outcomes depending on the associated proteins (Silverstein and Ekwall, 2005; Grzenda et al., 2009; Kadamb et al., 2013). Furthermore, the SIN3/HDAC may also act as a scaffold, enabling access for chromatin remodelers such as histone lysine methylases and demethylases or transcription factors. In this manner, SIN3/HDAC may influence chromatin remodeling processes and transcription regulation (Silverstein and Ekwall, 2005; Grzenda et al., 2009; Kadamb et al., 2013).
Another complex containing HDAC1/HDAC2, the nucleosome remodeling and deacetylase complex (NuRD), can act both as nucleosome remodeler and HDAC (Xue et al., 1998). Additionally, the NuRD complex can play an important role in the regulation of gene transcription, DNA repair or maintenance of genome stability. The NuRD is widely considered to be a transcriptional co-repressor complex, however some studies indicate that it may also have a role in the activation of gene expression (Shimbo et al., 2013). The NuRD complex is formed by several subunits, inter alia, chromodomain helicase DNA-binding proteins (CHD3, CHD4, CHD5), histone deacetylases (HDAC1 and HDAC2), histone binding proteins (RBAP48 and RBAP46), MBD2, MBD3 and metastasis associated gene 1, 2 and 3 (MTA1, MTA2, MTA3) proteins (Allen et al., 2013). It has been suggested that heterogeneity of the subunits of this complex and different combinations of proteins in these subunits provide unique functional properties (Bowen et al., 2004).
At least two types of complexes containing histones deacetylases, namely Co-REST and N-COR, have been shown to be involved in the neural commitment and differentiation of stem cells.
CoREST (co-repressor for element-1-silencing transcription factor) complex, regulates neuronal gene expression and is abundantly present in mammalian ESCs, neural progenitors and differentiated non-neuronal cells. Furthermore, CoREST is a platform which recruits different epigenetic factors such as MeCP2, HDAC1/2, LSD1, BHC80 and BRAF35 (Shi et al., 2004; Ballas et al., 2005).
In mammalian genomes there are three genes coding the RCOR protein and which form three types of CoREST complexes: CoREST1, CoREST2 and CoREST3. Recent studies have shown that each of them exhibits different activity and function. CoREST3 mostly resembles the CoREST1, however it has weaker transcriptional repressive activity. CoREST2 exhibits lower HDAC activity and is not inhibited by HDACs inhibitors (e.g., SAHA or TSA) like CoREST1 and CoREST3 complexes (Barrios et al., 2014).
The fourth example of the complex containing HDACs is the nuclear receptor co-repressor (N-CoR)/silencing mediator of retinoid and thyroid hormone receptor (SMRT) complex, comprised of several proteins, including HDAC3, transducin β-like protein 1 (TBL1)/TBL1-related protein 1 (TBLR1) and G-protein pathway suppressor 2 (GPS2). It is involved in the regulation of the differentiation of NSCs and in mouse forebrain development (Jepsen et al., 2007).
It has been demonstrated that some of the above-mentioned proteins and complexes play an important role during NSC differentiation and CNS development.
For example, SKI (transcriptional regulator, present in progenitor cells and young neurons) level changes during cell development, while its lack leads to reduced progenitor cell number and direct differentiation into neurons. The impact of SKI on neurogenesis may be associated with its ability to negatively regulate the transforming growth factor β (TGFβ) and bone morphogenetic protein (BMP) signaling pathways by binding to SMAD protein complexes. It is postulated that through its influence on the TGFβ pathway, SKI regulates the balance between proliferation and differentiation of NPC during cortical development.
Moreover, SKI interactions with SIN3/HDAC corepressor complex play an important role during cortical development. In was demonstrated that SKI is not universally expressed in whole brain but is present only in proliferating progenitor cells in the ventricular zone and in subtypes of differentiated projection neurons of the cortical plate (Luo, 2003; Baranek and Atanasoski, 2012; Baranek et al., 2012).
It was also shown that MBD3 is an important player during embryonic development. Mbd3−/− ESCs remain in the pluripotent state and are not able to commit to developmental lineages (Kaji et al., 2006). Furthermore, NuRD can block reprogramming of somatic cells into pluripotent stem cells (Luo, 2003), however, in the presence of NANOG, MBD3/NuRD induces NSC pluripotency (dos Santos et al., 2014). Such results indicate a context-dependent manner of MBD3/NuRD role in facilitating pluripotency and stemness.
CoREST2 complex has been shown to regulate ESC proliferation and pluripotency (Yang et al., 2011), while CoREST1 complex plays a significant role during neuronal differentiation. All CoREST complexes are expressed in the adult rat brain, both in neuronal and glial cells, however during neuronal differentiation CoREST1 and CoREST2 are downregulated, while CoREST3 expression remains unchanged (Sáez et al., 2015).
It has been shown that CoREST complex stops further differentiation of neuronal progenitors by repressing neuronal differentiation genes. The CoREST complex, which has HDAC activity, interacts with an HMT and this leads to H3K4 methylation. The simultaneous histone deacetylation and H3K4 methylation keep neuronal differentiation genes in a poised state, allowing their further activation. Moreover, in adult not-neuronal cells, CoREST complex in cooperation with an H3K4-specific histone LSD1 and H3K9-specific HMT leads to stable inhibition of neural gene expression (Shi et al., 2004; Ballas et al., 2005). Interestingly, it has been shown that CoREST apart from its key role in neuronal differentiation also influences glial subtype specification and oligodendrocyte lineage maturation. This is attributed to the influence of CoREST complex on the expression of glial genes, thus suggesting that CoREST may be involved in the regulation of astrocyte and oligodendrocyte function, including modulation of synaptic plasticity and axonal myelination (Abrajano et al., 2009). Rodenas-Ruano et al. (2012) demonstrated that in rat hippocampal neurons, expression of genes that encode proteins which build subunits of NMDA receptors is also regulated by REST-dependent DNA methylation. On this basis, these authors suggest that REST is an important player involved in experience-dependent activation of genes involved in synaptic plasticity (Rodenas-Ruano et al., 2012). It has also been shown that one of the phenotypes of Huntington’s Disease is correlated with transcriptional misregulations and abnormal cellular distributions of REST in neurons (Zuccato et al., 2007).
The CoREST complex together with the LSD1, and the HDAC1/2 are the main components of the LSD1-CoREST-HDAC (LCH) transcriptional repressor complexes. Recent studies revealed that in mammals there are four forms of LCH, which is associated with alternative splicing isoforms of LSD1 (LSD1, LSD1-2A, LSD1-8A, LSD1-2A/8A). The LSD1-8A and LSD1-2A/8A isotypes have been associated with neuronal differentiation. Moreover, it was shown that different isoforms are expressed during different stages of neuronal development. At the beginning of the neuronal differentiation of rat cortical neurons all LSD1 isoforms are expressed, however the most abundant ones are LSD1-2A and LSD. In the subsequent days of differentiation, increasing expression of LSD1-8A was observed. It was also observed that early-stage silencing of the LSD1-8A isoform causes a delay in neurite maturation, which indicates a role in the regulation of the proper timing of neurite maturation (Zibetti et al., 2010).
Furthermore, histone acetylation plays an important role in both maintenance of ESC pluripotency and neural differentiation. The histone acetylation level is mostly regulated by distinct HDAC members, which are expressed in specific patterns at different neurodevelopmental stages. Qiao et al. (2015) discovered that H3K9Ac occurs in a very specific manner during hESC neural differentiation in vitro. They demonstrated that during the first 4 days of hESC differentiation, the H3K9Ac level decreased. This is related to reduced acetylation at several pluripotency genes. Shortly afterwards, in day 4–8, H3K9Ac levels increased along with neural differentiation. Furthermore, they showed that the use of HDAC inhibitors (HDACi) during the first 4 days of differentiation promoted pluripotency and inhibited neural commitment (Qiao et al., 2015).
As previously mentioned, PTMs include histone acetylation, modulate chromatin accessibility and gene expression. Histone acetylation affects recruitment of chromatin modifier enzymes, such as bromodomain-containing proteins (BRD). It was shown that BRDs play a role in ESC self-renewal and development. For example, both BRD2 and BRD4 mutants are embryonically lethal. BRD2 null mice embryos exhibit abnormalities in neural tube development (Shang et al., 2009). Horne et al. (2015) demonstrated that BRD4 binds at the NANOG promoter and maintains its expression and is thus critical for the maintenance of ESC pluripotency.
Different HDACs are expressed at multiple developmental stages. For example, HDAC1 is highly expressed in mice NSCs and glia, while HDAC2 expression is present in neural progenitors and in post-mitotic neurons. Moreover, HDAC2 expression is absent in most of fully differentiated glia, suggesting that sequential expression of specific HDACs is necessary for the differentiation of neuronal and glial subtypes (MacDonald and Roskams, 2008). Recent studies suggest that HDAC inhibition influences neurogenesis in the mouse DG by disturbing the ratio of the neural progenitor types (Foti et al., 2013). Similarly, HDAC interferes with self-renewal in the SVZ (Zhou et al., 2011; Foti et al., 2013).
Histone methylation is the process whereby methyl groups are transferred to lysine (K) or arginine (R) residues. Histone methylation is a reversible process and it is catalyzed by either HMTs or histone demethylases (HDMs). There are two major types of HMTs; arginine-specific, catalyzing methylation of arginine residue on histone H3 and H4, and lysine-specific. HMTs, which catalyze lysine methylation, are subdivided into SET domain-containing (e.g., SUV39H1, SUV39H2) and non-SET domain-containing (e.g., DOT1L; Rea et al., 2000; Feng et al., 2002).
There are currently two known groups of histone LSDs: a flavin adenine dinucleotide (FAD)-dependent amine oxidase, and a Fe(II) and α-ketoglutarate-dependent dioxygenase. The first group is represented by LSD1 (Lysine-specific demethylase 1) and the second by the Jumonji domain-containing proteins (JmjC), which are classified into seven subgroups (e.g., lysine-specific HDM 5B, KDM5B; Shi et al., 2004; Tsukada et al., 2006; Li et al., 2014).
Histone methylation may have different outcomes depending on site (H3 K: 4, 9, 27, 36, 79; H4 K30 or R3) and type of methylation (mono-, di-, or tri-methylation—me1, me2 or me3). Methylation of lysine at sites 4, 36 and 79 on histone 3 (H3K4, H3K36, H3K79) correlates with transcriptional activity and is most commonly linked to Trithorax Group (TrxG) proteins, whereas methylation of lysine 9 and 27 on the same histone (H3K9, H3K27) as well as lysine 20 on histone 4 (H4K20) is typically associated with gene repression and Polycomb Group (PcG) proteins (Bernstein et al., 2006; Schuettengruber et al., 2007).
For example, in housekeeping genes of ESCs there are marks characteristic for transcription initiation, H3K4me3, while at developmental genes there are marks typical for active (H3K4me3) and repressive (H3K27me3) genes, while such double modified genes are generally silent (Bernstein et al., 2006). The specific state of the chromatin, which presents marks of activation and inactivation, is described as the “bivalent” state (“poised state”; Boheler, 2009).
In vertebrates, the PcG proteins comprise a class of transcriptional repressors that are found in multi-subunit complexes. There are two types of Polycomb repressive complexes: PRC1, which is involved in the maintenance of gene repression, and PRC2, which is associated with initiation of gene repression (Rajasekhar and Begemann, 2007). Both complexes catalyze histone covalent modifications and dynamically influence cell lineage commitment (Schuettengruber and Cavalli, 2009).
Regulation of developmental activity and pluripotency genes is linked with activity of the PcG, which binds to the methylated lysine in position 27 on histone 3 and silencing genes (Scheper and Copray, 2009). Maherali et al. (2007) suggested that changes in inactivation marks and PcG play a more significant role for the reprogramming process than methylation on lysine 4 of histone 3. Interestingly, during cell differentiation repressive marks (H3K9me3 and H3K27me3), which in ESCs cover around 4% of the genes, expand to 12%–16% (Hawkins et al., 2010).
During CNS development, the differentiation of each type of progenitor populations is dependent on the activity of various extracellular signals which facilitate access of transcription factors to promoters of specific glial or neuronal differentiation genes. During neural progenitor differentiation into astrocytes, H3K9 methylation of the GFAP promoter is replaced by H3K4 methylation. It was demonstrated that during this process the fibroblast growth factor 2 (FGF2) causes inhibition of H3K9 methylation and induces H3K4 methylation, which enables binding of STAT/CBP complex to the STAT binding site at the promoter of GFAP and results in gene activation (Song and Ghosh, 2004). Furthermore, recent studies suggest that histone lysine methylation levels are abnormally regulated in the aged hippocampus, as the baseline resting levels for tri-methylation of H3K4me3 and acetylation of H3K9, K14Ac were found to be different in aged rats with memory deficits compared to those of young adults and failed to increase following object-learning activity. However, environmental enrichment promoted object learning in the aged animals suggesting that histone lysine methylation is implicated in the epigenetic benefits of the environment on memory function (Morse et al., 2015).
The EZH2 (enhancer of Zeste homolog 2)—HMT, which is a part of PRC2, represses gene transcription by triple methylation of lysine 27 (H3K27me3). The PRCs have been widely studied in mouse ESCs showing that these complexes play an important role in maintaining pluripotency and regulating cell fate and identity (de Napoles et al., 2004; Leeb and Wutz, 2007; Pereira C. F. et al., 2010). Furthermore, growing evidence suggests that the PRCs also play an important role in different aspects of mammalian development, including the regulation of tissue specific stem and progenitor cells (Leeb and Wutz, 2010; Laugesen and Helin, 2014).
It has been reported that EZH2, PcG protein, the catalytic subunit of its PRC2 complex, keeps a balance between cortical progenitor cell self-renewal and differentiation and affects the course of neurogenesis (Pereira J. D. et al., 2010). During NSC differentiation into neurons, expression of EZH2 decreases, while it is absent when NSC differentiate into astrocytes and remains at a high level during NSC commitment towards the oligodendrocytic lineage (Sher et al., 2008). EZH2 was shown to negatively regulate neuronal induction during mesenchymal stem cell differentiation by binding to the promoter region and suppressing expression of PIP5K1C (protein present in synapses; Yu et al., 2011). Conversely, EZH2 is involved in suppression of GFAP activation, by binding to its promotor site. This is mediated by CHD4 acting as a EZH2 partner, where the dissociation of CHD4 from the PcG is crucial for the onset of gliogenesis (Sparmann et al., 2013). This suggests a specific role of EHZ2/CHD4 interaction in PcG repressive complex in preventing the premature onset of gliogenesis in maintaining the proper developmental balance in different glial populations.
Furthermore, recent studies have shown that during early mouse development redistribution of H3K27me3 conducted by EZH2 takes place and that it coincides with genome-wide accumulation of H3K9me2, which is mediated by the G9A enzyme. This histone H3 lysine 9 dimethylation was found to be crucial for proper development in post-implantation stage as it regulates the expression of genes involved in proliferation and cell fate decisions, such as Cd3e, Cdkn1a, Asz1, Rhox5, Pou5f1. Embryos that lacked G9A, which adds methyl groups at specific developmentally linked genes and causes their silencing, exhibited prematurely activated and inappropriate development. Moreover, G9A enzyme was found to be important in the mechanisms responsible for turning off gene expression enhancers (Zylicz et al., 2015).
Sex-determining region Y-related HMG box 2 (SOX 2) is an NSC and NPC marker important for their self-renewal and adult neurogenesis (Graham et al., 2003; Favaro et al., 2009). SOX2 was shown to be actively involved in epigenetic regulation of neural development by binding to the promotors of genes involved in neural differentiation, thus limiting EZH2 binding and preventing PRC2 activity. It was also shown that SOX2 is involved in regulating the expression of genes important during neuronal development, such as neurogenin 2 (Ngn2), neurogenic differentiation 1 (NeuroD1), brain-derived neurotrophic factor (Bdnf) and growth arrest-DNA-damage-inducible beta (Gadd45b). This suggests that SOX2 promotes an open chromatin state, thus enabling proneural and early neurogenic gene activation (Amador-Arjona et al., 2015).
Recent evidence suggests that another chromatin component, Zuotin-related factor 1 (ZRF1), may change developmental gene expression by displacing the PRC1 complex from chromatin and is thus involved in cell fate decisions. Moreover, ZRF1 plays a role in transcriptional regulation and cooperates with deubiquitinase enzyme (Richly et al., 2010). Taken together, these studies demonstrate the importance of histone methylation status and appropriate level of enzymes for the proper development, function and maintenance of the nervous system.
Different histone modifications may interact with DNA methylation and alter the expression of individual genes. Laurent et al. (2010) suggested that the level of DNA methylation is related to local chromatin conformation. Their findings indicated that there is a strong negative correlation between the level of DNA methylation and histone marks of active transcription (H3K4me3). Interaction between DNA methylation and histone modification is associated with the occurrence of one of two families of methylated CpG DNA binding proteins (MBD or BTB/POZ family; Rose and Klose, 2014).
The DNA methylation program as well as the methylation of histone codes are implicated in neural tube closure during embryonic development (Zhou, 2012).The link of this process to the neuronal differentiation was documented by the appearance of expression of neuronal marks such as NESTIN, CRABP (Cellular retinoic acid binding protein), NEU-N (Neuronal Nuclei) and MAP2 (microtubule-associated protein2).
Furthermore, acetylation of histones, together with the methylation status of DNA play an important role in the regulation of differentiation of developmental processes by regulation of tissue-specific genes. Both types of promoters (high and low-CpG-density promoters) in ESCs are rich in regions of unmethylated CpGs. However, when ESCs were differentiated into NPCs, HCP loss of the H3K4me3 mark caused an increase in the DNA methylation level. Meissner et al. (2008) discovered that in ESCs HCP of housekeeping genes are associated with transcription initiation mark on histone 3 (H3K4me3) and as a consequence, they become highly expressed. In contrast, HCP of developmental genes are marked by both H3K4me3 and H3K27me3 (repressive mark), which results in their silencing. Moreover, promoters of housekeeping genes as well as those of developmental genes are tagged with the H3K4me2 mark, which provides an open chromatin conformation.
The epigenetic regulatory mechanisms of DNA methylation and histone modification are widely involved in reprogramming, maintaining stemness and NSC differentiation. Epigenome modifications accompany developmental processes from the stage of the first embryonic division through stem cell commitment into defined cell subtypes and determination of their topographic distribution in the adult brain. Furthermore, these mechanisms are implicated in basic cellular processes, such as transcription regulation, DNA replication, cell cycle regulation and DNA repair. Their failure may thus result in severe neurodevelopmental pathologies as well as neurodegenerative diseases. Loss of specific enzymes responsible for deacetylation is associated with embryo lethality, which confirms their importance in embryonic development. Methylation and acetylation are further implicated in cognitive processes such as learning/plasticity and memory formation. The elucidation of these mechanisms in the context of epigenetic modifications may therefore be instrumental for our ability to understand and influence damage and repair processes in the developing as well as adult CNS.
LB and MP: substantial contributions to the conception or design of the work; drafting the work and revising the work critically for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the work. IS-G: substantial contributions to the conception or design of the work; drafting the work; final approval of the version to be published; agreement to be accountable for all aspects of the work. JA, IS and AS: substantial contributions to the conception or design of the work; revising the work critically for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer NNK and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
This work was supported by the National Centre for Research and Development grant No. Strategmed 1/234261/2/NCBR/2014 and Mossakowski Medical Research Centre statutory funds. IS and AS would like to acknowledge financial support from the Liaison Committee between the Central Norway Regional Health Authority (HMN) and the Norwegian University of Science and Technology (NTNU).
Abrajano, J. J., Qureshi, I. A., Gokhan, S., Zheng, D., Bergman, A., and Mehler, M. F. (2009). Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS One 4:e7665. doi: 10.1371/journal.pone.0007665
Allen, H. F., Wade, P. A., and Kutateladze, T. G. (2013). The NuRD architecture. Cell. Mol. Life Sci. 70, 3513–3524. doi: 10.1007/s00018-012-1256-2
Altman, J. (1962). Are new neurons formed in the brains of adult mammals? Science 135, 1127–1128. doi: 10.1126/science.135.3509.1127
Amador-Arjona, A., Cimadamore, F., Huang, C. T., Wright, R., Lewis, S., Gage, F. H., et al. (2015). SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. U S A 112, E1936–E1945. doi: 10.1073/pnas.1421480112
Arya, G., and Schlick, T. (2009). A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A 113, 4045–4059. doi: 10.1021/jp810375d
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657. doi: 10.1016/j.cell.2005.03.013
Baranek, C., and Atanasoski, S. (2012). Modulating epigenetic mechanisms: the diverse functions of Ski during cortical development. Epigenetics 7, 676–679. doi: 10.4161/epi.20590
Baranek, C., Dittrich, M., Parthasarathy, S., Bonnon, C. G., Britanova, O., Lanshakov, D., et al. (2012). Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc. Natl. Acad. Sci. U S A 109, 3546–3551. doi: 10.1073/pnas.1108718109
Barreto, G., Schäfer, A., Marhold, J., Stach, D., Swaminathan, S. K., Handa, V., et al. (2007). Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671–675. doi: 10.1038/nature05515
Barrios, Á. P., Gómez, A. V., Sáez, J. E., Ciossani, G., Toffolo, E., Battaglioli, E., et al. (2014). Differential properties of transcriptional complexes formed by the CoREST family. Mol. Cell. Biol. 34, 2760–2770. doi: 10.1128/mcb.00083-14
Bernstein, B. E., Meissner, A., and Lander, E. S. (2007). The mammalian epigenome. Cell 128, 669–681. doi: 10.1016/j.cell.2007.01.033
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Biswas, M., Voltz, K., Smith, J. C., and Langowski, J. (2011). Role of histone tails in structural stability of the nucleosome. PLoS Comput. Biol. 7:e1002279. doi: 10.1371/journal.pcbi.1002279
Boheler, K. R. (2009). Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J. Cell. Physiol. 221, 10–17. doi: 10.1002/jcp.21866
Bowen, N. J., Fujita, N., Kajita, M., and Wade, P. A. (2004). Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta 1677, 52–57. doi: 10.1016/j.bbaexp.2003.10.010
Candal, E., Thermes, V., Joly, J. S., and Bourrat, F. (2004). Medaka as a model system for the characterisation of cell cycle regulators: a functional analysis of Ol-Gadd45gamma during early embryogenesis. Mech. Dev. 121, 945–958. doi: 10.1016/j.mod.2004.03.007
Chen, Y., Damayanti, N. P., Irudayaraj, J., Dunn, K., and Zhou, F. C. (2014). Diversity of two forms of DNA methylation in the brain. Front. Genet. 5:46. doi: 10.3389/fgene.2014.00046
Chen, Y., Ozturk, N. C., and Zhou, F. C. (2013). DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS One 8:e60503. doi: 10.1371/journal.pone.0060503
Cheng, X., Kumar, S., Posfai, J., Pflugrath, J. W., and Roberts, R. J. (1993). Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell 74, 299–307. doi: 10.1016/0092-8674(93)90421-L
Cuthbert, G. L., Daujat, S., Snowden, A. W., Erdjument-Bromage, H., Hagiwara, T., Yamada, M., et al. (2004). Histone deimination antagonizes arginine methylation. Cell 118, 545–553. doi: 10.1016/j.cell.2004.08.020
Denomme, M. M., and Mann, M. R. (2013). Maternal control of genomic imprint maintenance. Reprod. Biomed. Online 27, 629–636. doi: 10.1016/j.rbmo.2013.06.004
Eriksson, P. S., Perfilieva, E., Björk-Eriksson, T., Alborn, A. M., Nordborg, C., Peterson, D. A., et al. (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317. doi: 10.1038/3305
Favaro, R., Valotta, M., Ferri, A. L., Latorre, E., Mariani, J., Giachino, C., et al. (2009). Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 12, 1248–1256. doi: 10.1038/nn.2397
Feng, J., Chang, H., Li, E., and Fan, G. (2005). Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79, 734–746. doi: 10.1002/jnr.20404
Feng, J., Fouse, S. D., and Fan, G. (2007). Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 61, 58R–63R. doi: 10.1203/pdr.0b013e3180457635
Feng, J., Zhou, Y., Campbell, S. L., Le, T., Li, E., Sweatt, J. D., et al. (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430. doi: 10.1038/nn.2514
Feng, Q., Wang, H., Ng, H. H., Erdjument-Bromage, H., Tempst, P., Struhl, K., et al. (2002). Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058. doi: 10.1016/s0960-9822(02)00901-6
Fenoglio, C., Ridolfi, E., Galimberti, D., and Scarpini, E. (2013). An emerging role for long non-coding RNA dysregulation in neurological disorders. Int. J. Mol. Sci. 14, 20427–20442. doi: 10.3390/ijms141020427
Foti, S. B., Chou, A., Moll, A. D., and Roskams, A. J. (2013). HDAC inhibitors dysregulate neural stem cell activity in the postnatal mouse brain. Int. J. Dev. Neurosci. 31, 434–447. doi: 10.1016/j.ijdevneu.2013.03.008
Gage, F. H. (2000). Mammalian neural stem cells. Science 287, 1433–1438. doi: 10.1126/science.287.5457.1433
Gapp, K., Woldemichael, B. T., Bohacek, J., and Mansuy, I. M. (2014). Epigenetic regulation in neurodevelopmental and neurodegenerative diseases. Neuroscience 264, 99–111. doi: 10.1016/j.neuroscience.2012.11.040
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638. doi: 10.1016/j.cell.2007.02.006
Gonzalo, S., Jaco, I., Fraga, M. F., Chen, T., Li, E., Esteller, M., et al. (2006). DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8, 416–424. doi: 10.1038/ncb1386
Graham, V., Khudyakov, J., Ellis, P., and Pevny, L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765. doi: 10.1016/s0896-6273(03)00497-5
Grzenda, A., Lomberk, G., Zhang, J. S., and Urrutia, R. (2009). Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789, 443–450. doi: 10.1016/j.bbagrm.2009.05.007
Guo, J. U., Ma, D. K., Mo, H., Ball, M. P., Jang, M. H., Bonaguidi, M. A., et al. (2011a). Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351. doi: 10.1038/nn.2900
Guo, J. U., Su, Y., Zhong, C., Ming, G. L., and Song, H. (2011b). Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434. doi: 10.1016/j.cell.2011.03.022
Haigis, M. C., and Sinclair, D. A. (2010). Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295. doi: 10.1146/annurev.pathol.4.110807.092250
Hanks, S. K., Rodriguez, L. V., and Rao, P. N. (1983). Relationship between histone phosphorylation and premature chromosome condensation. Exp. Cell Res. 148, 293–302. doi: 10.1016/0014-4827(83)90153-2
Hawkins, R. D., Hon, G. C., Lee, L. K., Ngo, Q., Lister, R., Pelizzola, M., et al. (2010). Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491. doi: 10.1016/j.stem.2010.03.018
Heard, E., Clerc, P., and Avner, P. (1997). X-chromosome inactivation in mammals. Annu. Rev. Genet. 31, 571–610. doi: 10.1146/annurev.genet.31.1.571
Hoffmann, A., Zimmermann, C. A., and Spengler, D. (2015). Molecular epigenetic switches in neurodevelopment in health and disease. Front. Behav. Neurosci. 9:120. doi: 10.3389/fnbeh.2015.00120
Horne, G. A., Stewart, H. J., Dickson, J., Knapp, S., Ramsahoye, B., and Chevassut, T. (2015). Nanog requires BRD4 to maintain murine embryonic stem cell pluripotency and is suppressed by bromodomain inhibitor JQ1 together with Lefty1. Stem Cells Dev. 24, 879–891. doi: 10.1089/scd.2014.0302
Hsieh, J., and Gage, F. H. (2004). Epigenetic control of neural stem cell fate. Curr. Opin. Genet. Dev. 14, 461–469. doi: 10.1016/j.gde.2004.07.006
Hutnick, L. K., Golshani, P., Namihira, M., Xue, Z., Matynia, A., Yang, X. W., et al. (2009). DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum. Mol. Genet. 18, 2875–2888. doi: 10.1093/hmg/ddp222
Ismail, I. H., Andrin, C., McDonald, D., and Hendzel, M. J. (2010). BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191, 45–60. doi: 10.1083/jcb.201003034
Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Science 293, 1074–1080. doi: 10.1126/science.1063127
Jepsen, K., Solum, D., Zhou, T., McEvilly, R. J., Kim, H. J., Glass, C. K., et al. (2007). SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450, 415–419. doi: 10.1038/nature06270
Kadamb, R., Mittal, S., Bansal, N., Batra, H., and Saluja, D. (2013). Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 92, 237–246. doi: 10.1016/j.ejcb.2013.09.001
Kaji, K., Caballero, I. M., MacLeod, R., Nichols, J., Wilson, V. A., and Hendrich, B. (2006). The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8, 285–292. doi: 10.1038/ncb1372
Kaufmann, L. T., Gierl, M. S., and Niehrs, C. (2011). Gadd45a, Gadd45b and Gadd45g expression during mouse embryonic development. Gene Expr. Patterns 11, 465–470. doi: 10.1016/j.gep.2011.07.005
Keshet, I., Lieman-Hurwitz, J., and Cedar, H. (1986). DNA methylation affects the formation of active chromatin. Cell 44, 535–543. doi: 10.1016/0092-8674(86)90263-1
Kigar, S. L., Chang, L., Hayne, M. R., Karls, N. T., and Auger, A. P. (2016). Sex differences in Gadd45b expression and methylation in the developing rodent amygdala. Brain Res. 1642, 461–466. doi: 10.1016/j.brainres.2016.04.031
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Kriaucionis, S., and Heintz, N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. doi: 10.1126/science.1169786
Laugesen, A., and Helin, K. (2014). Chromatin repressive complexes in stem cells, development and cancer. Cell Stem Cell 14, 735–751. doi: 10.1016/j.stem.2014.05.006
Laurent, L., Wong, E., Li, G., Huynh, T., Tsirigos, A., Ong, C. T., et al. (2010). Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320–331. doi: 10.1101/gr.101907.109
Leeb, M., and Wutz, A. (2007). Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 178, 219–229. doi: 10.1083/jcb.200612127
Leeb, M., and Wutz, A. (2010). Polycomb complexes–Genes make sense of host defense. Cell Cycle 9, 2692–2693. doi: 10.4161/cc.9.14.12455
Li, E., Beard, C., and Jaenisch, R. (1993). Role for DNA methylation in genomic imprinting. Nature 366, 362–365. doi: 10.1038/366362a0
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi: 10.1016/0092-8674(92)90611-f
Li, X., Liu, L., Yang, S., Song, N., Zhou, X., Gao, J., et al. (2014). Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. U S A 111, 7096–7101. doi: 10.1073/pnas.1324036111
Li, X., and Zhao, X. (2008). Epigenetic regulation of mammalian stem cells. Stem Cells Dev. 17, 1043–1052. doi: 10.1089/scd.2008.0036
Lian, Z. R., Xu, Y. F., Wang, X. B., Gong, J. P., and Liu, Z. J. (2012). Suppression of histone deacetylase 11 promotes expression of IL-10 in kupffer cells and induces tolerance following orthotopic liver transplantation in rats. J. Surg. Res. 174, 359–368. doi: 10.1016/j.jss.2010.12.035
Lilja, T., Heldring, N., and Hermanson, O. (2012). Like a rolling histone: epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochim. Biophys. Acta 1830, 2354–2360. doi: 10.1016/j.bbagen.2012.08.011
Lister, R., Pelizzola, M., Dowen, R. H., Hawkins, R. D., Hon, G., Tonti-Filippini, J., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322. doi: 10.1038/nature08514
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. doi: 10.1038/38444
Luo, K. (2003). Negative regulation of BMP signaling by the ski oncoprotein. J. Bone Joint Surg. Am. 85-A, 39–43. doi: 10.2106/00004623-200300003-00008
Ma, D. K., Jang, M. H., Guo, J. U., Kitabatake, Y., Chang, M. L., Pow-Anpongkul, N., et al. (2009). Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077. doi: 10.1126/science.1166859
MacDonald, J. L., and Roskams, A. J. (2008). Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. 237, 2256–2267. doi: 10.1002/dvdy.21626
Maherali, N., Sridharan, R., Xie, W., Utikal, J., Eminli, S., Arnold, K., et al. (2007). Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70. doi: 10.1016/j.stem.2007.05.014
Martinez-Zamudio, R., and Ha, H. C. (2012). Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol. Cell. Biol. 32, 2490–2502. doi: 10.1128/MCB.06667-11
Meissner, A., Mikkelsen, T. S., Gu, H., Wernig, M., Hanna, J., Sivachenko, A., et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770. doi: 10.1038/nature07107
Mohamed Ariff, I., Mitra, A., and Basu, A. (2012). Epigenetic regulation of self-renewal and fate determination in neural stem cells. J. Neurosci. Res. 90, 529–539. doi: 10.1002/jnr.22804
Morgan, C. P., and Bale, T. L. (2011). Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J. Neurosci. 31, 11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011
Morse, S. J., Butler, A. A., Davis, R. L., Soller, I. J., and Farah, D. L. (2015). Environmental enrichment reverses histone methylation changes in the aged hippocampus and restores age-related memory deficits. Biology (Basel) 4, 298–313. doi: 10.3390/biology4020298
Namihira, M., Kohyama, J., Semi, K., Sanosaka, T., Deneen, B., Taga, T., et al. (2009). Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell 16, 245–255. doi: 10.1016/j.devcel.2008.12.014
Nan, X., Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N., et al. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. doi: 10.1038/30764
de Napoles, M., Mermoud, J. E., Wakao, R., Tang, Y. A., Endoh, M., Appanah, R., et al. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676. doi: 10.1016/j.devcel.2004.10.005
Nelson, C. J., Santos-Rosa, H., and Kouzarides, T. (2006). Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126, 905–916. doi: 10.1016/j.cell.2006.07.026
New, M., Olzscha, H., and La Thangue, N. B. (2012). HDAC inhibitor-based therapies: can we interpret the code? Mol. Oncol. 6, 637–656. doi: 10.1016/j.molonc.2012.09.003
Niehrs, C., and Schäfer, A. (2012). Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 22, 220–227. doi: 10.1016/j.tcb.2012.01.002
Noguchi, H., Kimura, A., Murao, N., Matsuda, T., Namihira, M., and Nakashima, K. (2015). Expression of DNMT1 in neural stem/precursor cells is critical for survival of newly generated neurons in the adult hippocampus. Neurosci. Res. 95, 1–11. doi: 10.1016/j.neures.2015.01.014
Noguchi, H., Murao, N., Kimura, A., Matsuda, T., Namihira, M., and Nakashima, K. (2016). DNA methyltransferase 1 is indispensable for development of the hippocampal dentate gyrus. J. Neurosci. 36, 6050–6068. doi: 10.1523/JNEUROSCI.0512-16.2016
Nugent, B. M., Wright, C. L., Shetty, A. C., Hodes, G. E., Lenz, K. M., Mahurkar, A., et al. (2015). Brain feminization requires active repression of masculinization via DNA methylation. Nat. Neurosci. 18, 690–697. doi: 10.1038/nn.3988
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi: 10.1016/s0092-8674(00)81656-6
O’Neill, L. P., and Turner, B. M. (1995). Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 14, 3946–3957.
Pereira, C. F., Piccolo, F. M., Tsubouchi, T., Sauer, S., Ryan, N. K., Bruno, L., et al. (2010). ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6, 547–556. doi: 10.1016/j.stem.2010.04.013
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M. W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U S A 107, 15957–15962. doi: 10.1073/pnas.1002530107
Qiao, Y., Wang, R., Yang, X., Tang, K., and Jing, N. (2015). Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J. Biol. Chem. 290, 2508–2520. doi: 10.1074/jbc.M114.603761
Rajasekhar, V. K., and Begemann, M. (2007). Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells 25, 2498–2510. doi: 10.1634/stemcells.2006-0608
Razin, A., and Szyf, M. (1984). DNA methylation patterns. Formation and function. Biochim. Biophys. Acta 782, 331–342. doi: 10.1007/978-1-4613-8519-6_7
Rea, S., Eisenhaber, F., O’Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. doi: 10.1038/35020506
Reik, W., Dean, W., and Walter, J. (2001). Epigenetic reprogramming in mammalian development. Science 293, 1089–1093. doi: 10.1126/science.1063443
Richly, H., Rocha-Viegas, L., Ribeiro, J. D., Demajo, S., Gundem, G., Lopez-Bigas, N., et al. (2010). Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468, 1124–1128. doi: 10.1038/nature09574
Richmond, T. J., and Davey, C. A. (2003). The structure of DNA in the nucleosome core. Nature 423, 145–150. doi: 10.1038/nature01595
Ridsdale, J. A., Hendzel, M. J., Delcuve, G. P., and Davie, J. R. (1990). Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J. Biol. Chem. 265, 5150–5156.
Rodenas-Ruano, A., Chavez, A. E., Cossio, M. J., Castillo, P. E., and Zukin, R. S. (2012). REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat. Neurosci. 15, 1382–1390. doi: 10.1038/nn.3214
Rose, N. R., and Klose, R. J. (2014). Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta 1839, 1362–1372. doi: 10.1016/j.bbagrm.2014.02.007
Rothbart, S. B., and Strahl, B. D. (2014). Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839, 627–643. doi: 10.1016/j.bbagrm.2014.03.001
Sáez, J. E., Gómez, A. V., Barrios, Á. P., Parada, G. E., Galdames, L., González, M., et al. (2015). Decreased expression of CoREST1 and CoREST2 together with LSD1 and HDAC1/2 during neuronal differentiation. PLoS One 10:e0131760. doi: 10.1371/journal.pone.0131760
dos Santos, R. L., Tosti, L., Radzisheuskaya, A., Caballero, I. M., Kaji, K., Hendrich, B., et al. (2014). MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell 15, 102–110. doi: 10.1016/j.stem.2014.04.019
Scheper, W., and Copray, S. (2009). The molecular mechanism of induced pluripotency: a two-stage switch. Stem Cell Rev. 5, 204–223. doi: 10.1007/s12015-009-9077-x
Schuettengruber, B., and Cavalli, G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542. doi: 10.1242/dev.033902
Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B., and Cavalli, G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745. doi: 10.1016/j.cell.2007.02.009
Sha, K., and Boyer, L. A. (2009). The chromatin signature of pluripotent cells. StemBook, ed. The Stem Cell Research Community, StemBook. Available online at: http://www.stembook.org/node/585
Shang, E., Wang, X., Wen, D., Greenberg, D. A., and Wolgemuth, D. J. (2009). Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 238, 908–917. doi: 10.1002/dvdy.21911
Sher, F., Rössler, R., Brouwer, N., Balasubramaniyan, V., Boddeke, E., and Copray, S. (2008). Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells 26, 2875–2883. doi: 10.1634/stemcells.2008-0121
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Shiio, Y., and Eisenman, R. N. (2003). Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U S A 100, 13225–13230. doi: 10.1073/pnas.1735528100
Shimbo, T., Du, Y., Grimm, S. A., Dhasarathy, A., Mav, D., Shah, R. R., et al. (2013). MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 9:e1004028. doi: 10.1371/journal.pgen.1004028
Shogren-Knaak, M., Ishii, H., Sun, J. M., Pazin, M. J., Davie, J. R., and Peterson, C. L. (2006). Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. doi: 10.1126/science.1124000
Silverstein, R. A., and Ekwall, K. (2005). Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17. doi: 10.1007/s00294-004-0541-5
Song, M. R., and Ghosh, A. (2004). FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 7, 229–235. doi: 10.1038/nn1192
Sparmann, A., Xie, Y., Verhoeven, E., Vermeulen, M., Lancini, C., Gargiulo, G., et al. (2013). The chromodomain helicase Chd4 is required for Polycomb-mediated inhibition of astroglial differentiation. EMBO J. 32, 1598–1612. doi: 10.1038/emboj.2013.93
Stroud, H., Feng, S., Morey Kinney, S., Pradhan, S., and Jacobsen, S. E. (2011). 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 12:R54. doi: 10.1186/gb-2011-12-6-r54
Sun, G. Q., Zhao, C., and Shi, Y. (2008). Epigenetic control of neural stem cell self-renewal and specification. Adv. Biomed. Res. 1, 69–82. doi: 10.1007/978-1-4020-8502-4_4
Szulwach, K. E., Li, X., Li, Y., Song, C. X., Wu, H., Dai, Q., et al. (2011). 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616. doi: 10.1038/nn.2959
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yanagisawa, M., et al. (2001). DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1, 749–758. doi: 10.1016/s1534-5807(01)00101-0
Teperek-Tkacz, M., Pasque, V., Gentsch, G., and Ferguson-Smith, A. C. (2011). Epigenetic reprogramming: is deamination key to active DNA demethylation? Reproduction 142, 621–632. doi: 10.1530/REP-11-0148
Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi: 10.1038/nature04433
Ueda, Y., Okano, M., Williams, C., Chen, T., Georgopoulos, K., and Li, E. (2006). Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development 133, 1183–1192. doi: 10.1242/dev.02293
Walsh, C. P., Chaillet, J. R., and Bestor, T. H. (1998). Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117. doi: 10.1038/2413
Watanabe, A., Yamada, Y., and Yamanaka, S. (2012). Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120292. doi: 10.1098/rstb.2012.0292
Weng, M. K., Zimmer, B., Pöltl, D., Broeg, M. P., Ivanova, V., Gaspar, J. A., et al. (2012). Extensive transcriptional regulation of chromatin modifiers during human neurodevelopment. PLoS One 7:e36708. doi: 10.1371/journal.pone.0036708
Wu, G., Bazer, F. W., Cudd, T. A., Meininger, C. J., and Spencer, T. E. (2004). Maternal nutrition and fetal development. J. Nutr. 134, 2169–2172.
Wu, H., Coskun, V., Tao, J., Xie, W., Ge, W., Yoshikawa, K., et al. (2010). Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–448. doi: 10.1126/science.1190485
Wu, H., D’Alessio, A. C., Ito, S., Wang, Z., Cui, K., Zhao, K., et al. (2011). Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684. doi: 10.1101/gad.2036011
Wu, Z., Huang, K., Yu, J., Le, T., Namihira, M., Liu, Y., et al. (2012). Dnmt3a regulates both proliferation and differentiation of mouse neural stem cells. J. Neurosci. Res. 90, 1883–1891. doi: 10.1002/jnr.23077
Wu, S. C., and Zhang, Y. (2010). Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620. doi: 10.1038/nrm2950
Wu, H., and Zhang, Y. (2011). Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452. doi: 10.1101/gad.179184.111
Xu, Y., Zhang, S., Lin, S., Guo, Y., Deng, W., Zhang, Y., et al. (2017). WERAM: a database of writers, erasers and readers of histone acetylation and methylation in eukaryotes. Nucleic Acids Res. 45, D264–D270. doi: 10.1093/nar/gkw1011
Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Côté, J., and Wang, W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861. doi: 10.1016/s1097-2765(00)80299-3
Yang, P., Wang, Y., Chen, J., Li, H., Kang, L., Zhang, Y., et al. (2011). RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem Cells 29, 791–801. doi: 10.1002/stem.634
Yu, Y. L., Chou, R. H., Chen, L. T., Shyu, W. C., Hsieh, S. C., Wu, C. S., et al. (2011). EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1C-dependent calcium signaling. J. Biol. Chem. 286, 9657–9667. doi: 10.1074/jbc.m110.185124
Zhang, R. R., Cui, Q. Y., Murai, K., Lim, Y. C., Smith, Z. D., Jin, S., et al. (2013). Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245. doi: 10.1016/j.stem.2013.05.006
Zhang, Y., Gilquin, B., Khochbin, S., and Matthias, P. (2006). Two catalytic domains are required for protein deacetylation. J. Biol. Chem. 281, 2401–2404. doi: 10.1074/jbc.c500241200
Zhao, X., Ueba, T., Christie, B. R., Barkho, B., McConnell, M. J., Nakashima, K., et al. (2003). Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. U S A 100, 6777–6782. doi: 10.1073/pnas.1131928100
Zhou, F. C. (2012). DNA methylation program during development. Front. Biol. (Beijing) 7, 485–494. doi: 10.1007/s11515-012-9246-1
Zhou, Q., Dalgard, C. L., Wynder, C., and Doughty, M. L. (2011). Histone deacetylase inhibitors SAHA and sodium butyrate block G1-to-S cell cycle progression in neurosphere formation by adult subventricular cells. BMC Neurosci. 12:50. doi: 10.1186/1471-2202-12-50
Zhou, F. C., Resendiz, M., Lo, C. L., and Chen, Y. (2016). Cell-wide DNA de-methylation and re-methylation of purkinje neurons in the developing cerebellum. PLoS One 11:e0162063. doi: 10.1371/journal.pone.0162063
Zibetti, C., Adamo, A., Binda, C., Forneris, F., Toffolo, E., Verpelli, C., et al. (2010). Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J. Neurosci. 30, 2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010
Zuccato, C., Belyaev, N., Conforti, P., Ooi, L., Tartari, M., Papadimou, E., et al. (2007). Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J. Neurosci. 27, 6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007
Keywords: epigenetics, methylation, acetylation, stem cells, neurodevelopment, neuroplasticity
Citation: Podobinska M, Szablowska-Gadomska I, Augustyniak J, Sandvig I, Sandvig A and Buzanska L (2017) Epigenetic Modulation of Stem Cells in Neurodevelopment: The Role of Methylation and Acetylation. Front. Cell. Neurosci. 11:23. doi: 10.3389/fncel.2017.00023
Received: 07 October 2016; Accepted: 23 January 2017;
Published: 07 February 2017.
Edited by:
Dirk Schubert, Radboud UMC, NetherlandsReviewed by:
Takumi Takizawa, Gunma University, JapanCopyright © 2017 Podobinska, Szablowska-Gadomska, Augustyniak, Sandvig, Sandvig and Buzanska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution and reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonora Buzanska, YnV6YW5za2FAaW1kaWsucGFuLnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.