
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Cell. Infect. Microbiol. , 06 March 2025
Sec. Clinical Microbiology
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1581237
This article is part of the Research Topic Developing Therapeutics for Antimicrobial Resistant Pathogens: Volume II View all 8 articles
 Raja Veerapandian1*
Raja Veerapandian1* Parveez Ahamed Abdul Azees2
Parveez Ahamed Abdul Azees2 Thiruselvam Viswanathan3
Thiruselvam Viswanathan3 Bennett Tochukwu Amaechi2*
Bennett Tochukwu Amaechi2* Govindsamy Vediyappan4,5*
Govindsamy Vediyappan4,5*Editorial on the Research Topic
Developing therapeutics for antimicrobial resistant pathogens: volume II
Antimicrobial resistance (AMR) is one of the most pressing global public health challenges, with bacterial AMR alone directly causing 1.27 million deaths worldwide in 2019 and contributing to 4.95 million deaths, excluding those caused by AMR in viruses, fungi, and parasites (Antimicrobial Resistance, 2022; WHO, 2023). Despite the growing threat of AMR, the development of new therapeutics has stalled, with no clinical candidates close to reaching the market (WHO, 2022). In response to this growing threat, we launched a Research Topic in 2021 titled “Developing Therapeutics for Antimicrobial-Resistant Pathogens” (Veerapandian et al., 2022). Building on the success of Volume I, we initiated Volume II in 2024. The latest Research Topic features seven manuscripts presenting diverse perspectives on combating AMR pathogens.
Carbapenem-resistant Klebsiella pneumoniae is the major public health problem with high mortality rates (Semet et al., 2024). A comprehensive understanding of its virulence mechanisms and effective therapeutic strategies is critically needed. Tu et al. elucidated the molecular characteristics and pathogenic mechanisms of hypervirulent carbapenem-resistant Klebsiella pneumoniae (CR-hvKP) isolated from a human patient sample. Whole-genome sequencing identified three plasmids: Plasmid 1, a pLVPK-like virulence plasmid carrying multiple virulence genes like rmpA, rmpA2, iroB, iucA, and terB; Plasmid 2, which contains transposable elements such as IS15 and IS26; and Plasmid 3, a resistance plasmid harboring the blaKPC-3 carbapenem resistance gene. Mouse virulence assays with CR-hvKP demonstrated a high mortality rate and a significant increase in proinflammatory cytokine levels, including IL-1β, IL-6, and TNF-α. Interesting article from Aluisio et al. examined the use of cell-free supernatant (CFS) from Lactobacillus gasseri against carbapenem-resistant K. pneumoniae. Whole genome sequencing of the L. gasseri 1A-TV strain revealed two bacteriocin biosynthetic gene clusters (BBGCs) namely BBGC1 which encodes two class IIc bacteriocins, including a gassericin A-like bacteriocin, while BBGC2 harbors the class III bacteriocin helveticin J. Notably, 1A-TV CFS effectively inhibited the growth of all K. pneumoniae isolates tested. Nuclear magnetic resonance (NMR) analysis of the L. gasseri 1A-TV strain CFS identified and quantified various metabolites involved in carbohydrate and amino acid metabolism, along with organic acids such as ethanol, lactate, acetate, and succinate.
Use of Antimicrobial peptides (AMPs) are one of the promising alternative approaches against AMR pathogens (Xuan et al., 2023; Jeyarajan et al., 2024). Although resistance to AMPs remains relatively low, the potential for resistance development has been reported by multiple mechanism employed by pathogens (Band and Weiss, 2015; Duperthuy, 2020). Hernández-García et al. examined resistance mechanisms associated with Murepavadin, a novel peptidomimetic antibiotic that specifically targets Pseudomonas aeruginosa LptD. A genome-wide association study (GWAS) indicated that high resistance to Murepavadin is likely linked to mutations in lpxL1 and lpxL2, resulting in reduced hexa-acylated lipid A, which corresponds to lower inflammatory induction and increased susceptibility to host-derived AMPs.
Combination therapy is an effective approach used recently to treat various bacterial and fungal infections (Basavegowda and Baek, 2022; Bonincontro et al., 2023). Zhao et al. demonstrated that aloe plant extracts containing aloe emodin, emodin, and rhein exhibit strain-specific antibacterial effects against polymyxin-resistant Acinetobacter baumannii. Interestingly, combination therapy synergistically restored the sensitivity of resistant A. baumannii strains to polymyxins. Moreover, this drug combination showed low cytotoxicity, promoted wound healing, and reduced bacterial burden in mice infected with a polymyxin-resistant A. baumannii strain.
Bioactive compounds from the marine ecosystem remain largely unexplored. Metabolites from the marine algae is reported for its antimicrobial activity (Pérez et al., 2016; Makhlof et al., 2024). Sangavi et al. explored the anticariogenic potential of the methanolic extract of Padina boergesenii (MEPB). MEPB effectively inhibited Streptococcus mutans biofilms without affecting bacterial viability while also impairing key virulence factors such as adherence, acid production, acid tolerance, glucan synthesis, cell surface hydrophobicity, and extracellular DNA (eDNA) production. Its non-toxic nature was confirmed using human buccal epithelial cells. Additionally, GC-MS/MS analysis identified palmitic acid, myristic acid, and stearic acid as the major active constituents of MEPB and the further biofilm inhibition assays with these compounds demonstrated significant antibiofilm activity, with palmitic acid reducing biofilm formation by 85%, myristic acid by 72%, and stearic acid by 83%.
Candida species are known to form polymicrobial biofilms with various bacteria, including Streptococcus species, Staphylococcus species, and Escherichia coli, leading to severe clinical outcomes and challenges to treatment (Veerapandian and Vediyappan, 2019; Pohl, 2022). Shaik et al. investigated various phthalimide derivatives, including N-butylphthalimide (NBP), N-methylphthalimide (NMP), N-aminophthalimide (NAP), N-hydroxymethylphthalimide (NHP), N-carbethoxyphthalimide (NCP), and N-(2-butynyl)phthalimide (N2BP), for their antifungal activity against Candida albicans and Candida parapsilosis. These compounds effectively inhibited biofilm formation, hyphal development, and cell aggregation. Notably, NBP significantly downregulated key hyphal- and biofilm-associated genes (ECE1, HWP1, and UME6) upon treatment. Furthermore, NBP demonstrated broad-spectrum antibiofilm activity by inhibiting biofilm formation of bacterial pathogens such as Escherichia coli, Staphylococcus epidermidis, Staphylococcus aureus, and Vibrio parahaemolyticus. Additionally, NBP effectively disrupted polymicrobial biofilms composed of S. epidermidis and C. albicans. Veerapandian et al. established a mouse model of oropharyngeal candidiasis (OPC) and demonstrated that oral infection with Candida albicans can result in its dissemination under immunosuppressed conditions, even in the absence of antibiotics or chemotherapeutic agents. The authors observed alterations in the gut microbiome that favored the proliferation of Enterococcus species, C. albicans, and potentially other pathogenic bacteria in immunosuppressed mice, which may have exacerbated mucosal damage and facilitated their dissemination.
There are multiple therapeutic strategies available to combat AMR; however, a deeper understanding of its underlying mechanisms is still needed. The recent application of omics technologies has allowed researchers to simultaneously examine multiple layers of molecular activity (Chen et al., 2023). While omics data provide valuable insights into molecular changes following infection, their impact would be significantly greater if these technologies were also applied to therapeutic treatment groups for comparison. This approach would help uncover the molecular mechanisms responsible for the protective effects of therapeutics against diseases (Yow et al., 2022).
Additionally, microbiome studies will play a crucial role in advancing newly developed drugs to the next stage. A key concern when developing new drugs is their potential impact on the host’s normal flora (Patangia et al., 2022). Integrating microbiome analysis with assessments of a drug’s antimicrobial efficacy would provide a more comprehensive understanding of its overall effects.
In summary, we believe that the research articles featured in this Research Topic enhance our understanding of therapeutics for combating antimicrobial-resistant (AMR) pathogens and suggest future directions for addressing AMR challenges.
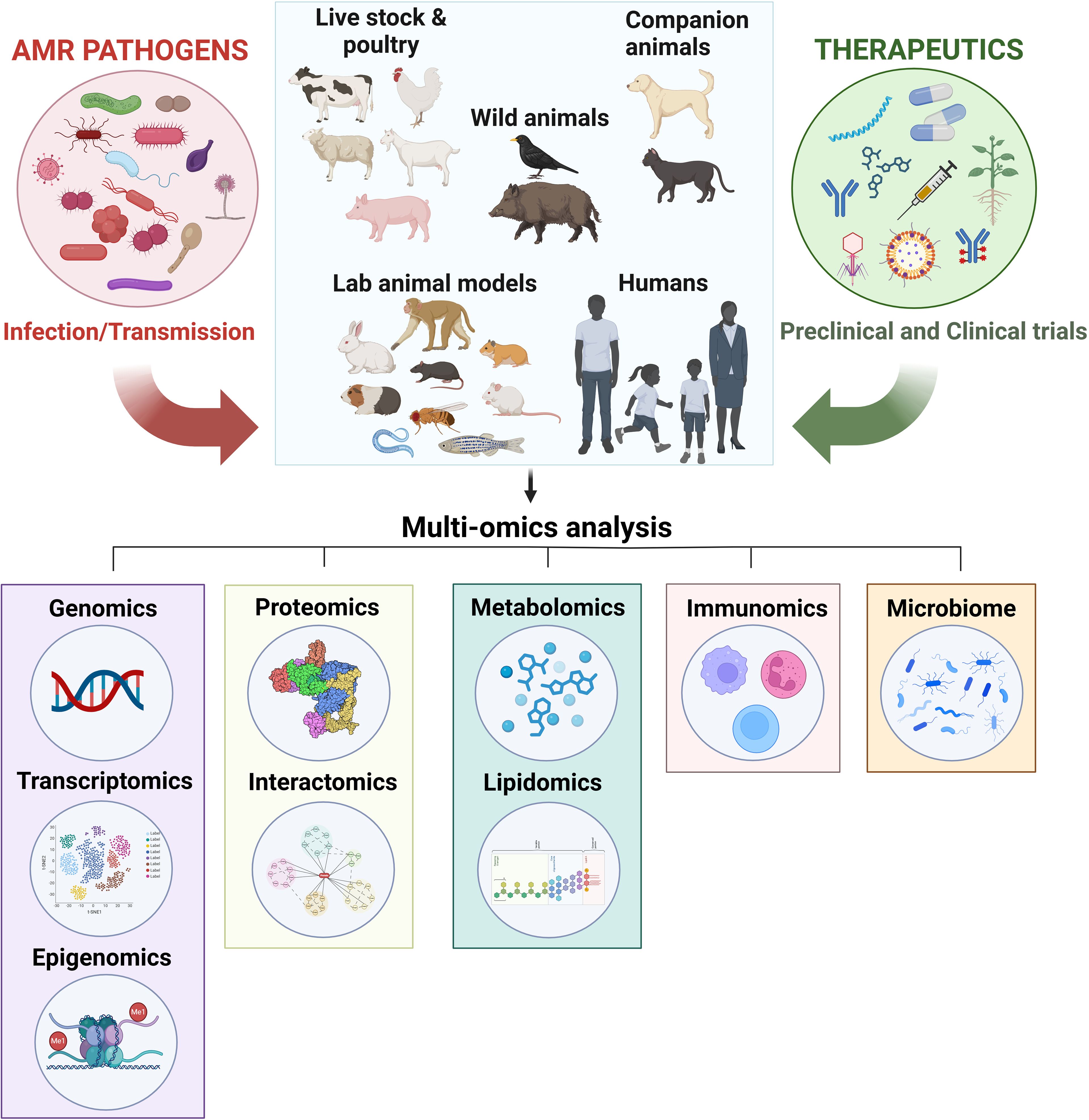
Figure 1. Utilizing omics and microbiome technologies to uncover the molecular mechanisms of therapeutics against AMR. Antimicrobial-resistant (AMR) pathogens, including bacteria, fungi, viruses, and parasites, can naturally infect humans and various animals, such as livestock (e.g., chickens, cattle, goats, sheep, and swine), companion animals (e.g., dogs and cats), and wildlife (e.g., birds and wild boars), leading to disease. Additionally, laboratory animals, including non-human primates, mice, rats, hamsters, guinea pigs, rabbits, zebrafish, Caenorhabditis elegans, and Drosophila melanogaster, are intentionally infected for research purposes. To combat these AMR pathogens, various therapeutic strategies are employed for treating naturally infected humans and animals, as well as for research on artificially infected laboratory animals. These strategies include plant-derived antimicrobials, antimicrobial peptides, nanomaterials, phage therapy, small compound libraries, drug repurposing, vaccines, antibody, and antibiotic-antibody conjugates. Multi-omics approaches, such as genomics (including transcriptomics and epigenomics), proteomics (including interactomics), metabolomics (including lipidomics), immunomics, and microbiome analysis, can be utilized to elucidate the mechanisms of these therapeutics, accelerating the discovery of effective treatment options.
RV: Writing – original draft, Writing – review & editing, Conceptualization. PA: Writing – review & editing. TV: Writing – review & editing. BA: Writing – review & editing, Conceptualization. GV: Writing – review & editing, Conceptualization.
The authors express their gratitude to Dr. Mahalakshmi Vijayaraghavan from Texas Tech University Health Sciences Center El Paso (TTUHSC-EP) for her assistance in generating Figure 1. The figure was created using BioRender (https://BioRender.com/e51z049), and we appreciate TTUHSC El Paso for providing user access to the platform. Additionally, we extend our thanks to the authors of the contributed articles, the peer reviewers, and the Editorial Office of Frontiers in Cellular and Infection Microbiology for their valuable contributions to this Research Topic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Antimicrobial Resistance, C. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Band, V. I., Weiss, D. S. (2015). Mechanisms of antimicrobial peptide resistance in gram-negative bacteria. Antibiotics (Basel) 4, 18–41. doi: 10.3390/antibiotics4010018
Basavegowda, N., Baek, K. H. (2022). Combination strategies of different antimicrobials: an efficient and alternative tool for pathogen inactivation. Biomedicines 10, 2219. doi: 10.3390/biomedicines10092219
Bonincontro, G., Scuderi, S. A., Marino, A., Simonetti, G. (2023). Synergistic Effect of Plant Compounds in Combination with Conventional Antimicrobials against Biofilm of Staphylococcus aureus, Pseudomonas aeruginosa, and Candida spp. Pharm. (Basel) 16, 1531. doi: 10.3390/ph16111531
Chen, C., Wang, J., Pan, D., Wang, X., Xu, Y., Yan, J., et al. (2023). Applications of multi-omics analysis in human diseases. MedComm (2020) 4, e315. doi: 10.1002/mco2.v4.4
Duperthuy, M. (2020). Antimicrobial peptides: virulence and resistance modulation in gram-negative bacteria. Microorganisms 8 (2), 280. doi: 10.3390/microorganisms8020280
Jeyarajan, S., Ranjith, S., Veerapandian, R., Natarajaseenivasan, K., Chidambaram, P., Kumarasamy, A. (2024). Antibiofilm Activity of Epinecidin-1 and Its Variants Against Drug-Resistant Candida krusei and Candida tropicalis Isolates from Vaginal Candidiasis Patients. Infect. Dis. Rep. 16, 1214–1229. doi: 10.3390/idr16060096
Makhlof, M. E. M., El-Sheekh, M. M., El-Sayed, A. I. M. (2024). In vitro antibiofilm, antibacterial, antioxidant, and antitumor activities of the brown alga Padina pavonica biomass extract. Int. J. Environ. Health Res. 34, 1861–1878. doi: 10.1080/09603123.2023.2165045
Patangia, D. V., Anthony Ryan, C., Dempsey, E., Paul Ross, R., Stanton, C. (2022). Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 11, e1260. doi: 10.1002/mbo3.v11.1
Pérez, M. J., Falqué, E., Domínguez, H. (2016). Antimicrobial action of compounds from marine seaweed. Mar. Drugs 14 (3), 52. doi: 10.3390/md14030052
Pohl, C. H. (2022). Recent advances and opportunities in the study of candida albicans polymicrobial biofilms. Front. Cell Infect. Microbiol. 12, 836379. doi: 10.3389/fcimb.2022.836379
Semet, C., Efe, K., Akalın, H., İşçimen, R., Girgin, N. K., Özakın, C., et al. (2024). Outcome of carbapenem or colistin resistant Klebsiella pneumoniae bacteremia in the intensive care unit. Sci. Rep. 14, 25805. doi: 10.1038/s41598-024-73786-x
Veerapandian, R., Abdul Azees, P. A., Viswanathan, T., Amaechi, B. T., Vediyappan, G. (2022). Editorial: Developing therapeutics for antimicrobial resistant pathogens. Front. Cell Infect. Microbiol. 12, 1083501. doi: 10.3389/fcimb.2022.1083501
Veerapandian, R., Vediyappan, G. (2019). Gymnemic Acids Inhibit Adhesive Nanofibrillar Mediated Streptococcus gordonii-Candida albicans Mono-Species and Dual-Species Biofilms. Front. Microbiol. 10, 2328. doi: 10.3389/fmicb.2019.02328
WHO (2022). Lack of innovation set to undermine antibiotic performance and health gains. Available online at: https://www.who.int/news/item/22-06-2022-22-06-2022-lack-of-innovation-set-to-undermine-antibiotic-performance-and-health-gains (Accesed February 2, 2025).
WHO (2023). Antimicrobial resistance. Available online at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accesed February 2, 2025).
Xuan, J., Feng, W., Wang, J., Wang, R., Zhang, B., Bo, L., et al. (2023). Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Update 68, 100954. doi: 10.1016/j.drup.2023.100954
Keywords: drugs, anti-virulence drugs, anti-biofilm drugs, repurposed drugs, antibiotic-antibody combination therapy, nanotechnology, in silico drug design
Citation: Veerapandian R, Abdul Azees PA, Viswanathan T, Amaechi BT and Vediyappan G (2025) Editorial: Developing therapeutics for antimicrobial resistant pathogens: volume II. Front. Cell. Infect. Microbiol. 15:1581237. doi: 10.3389/fcimb.2025.1581237
Received: 21 February 2025; Accepted: 24 February 2025;
Published: 06 March 2025.
Edited and Reviewed by:
Nahed Ismail, University of Illinois Chicago, United StatesCopyright © 2025 Veerapandian, Abdul Azees, Viswanathan, Amaechi and Vediyappan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raja Veerapandian, ci52ZWVyYXBhbmRpYW5AdHR1aHNjLmVkdQ==; Bennett Tochukwu Amaechi, YW1hZWNoaUB1dGhzY3NhLmVkdQ==; Govindsamy Vediyappan, Z3ZlZGl5YXBAa3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.