
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 13 March 2025
Sec. Virus and Host
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1563440
Objective: As one of the major human oncogenic viruses, Kaposi’s Sarcoma-associated Herpesvirus (KSHV) is closely related to several cancers such as Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL). KSHV can infect a broad tropism of human primary cells in vitro and in vivo. Embryonic stem cell-like pluripotent stem cells can be generated by the simultaneous introduction of several factors, into somatic cells, yielding induced pluripotent stem (iPS) cells. However, it remains unclear whether human induced pluripotent stem cells (hiPSCs) are permissive to KSHV and how this oncogenic virus infection may affect cellular gene profile.
Methods: In the current study, we examined whether hiPSCs were permissive to KSHV infection. The flow cytometry was used to assess the impacts of KSHV infection on hiPSCs viability and apoptosis. The Illumina RNA-Sequencing was used to determine cellular gene profile changed in KSHV-infected hiPSCs and lytically induced cells.
Results: We report that KSHV successfully establishes latent infection in hiPSCs, which can be completely induced to lytic reactivation and release infectious virions. KSHV de novo infection arrests the growth of hiPSCs through inducing cell apoptosis. Transcriptomic analysis revealed significant changes in global cellular gene expression in KSHV-infected hiPSCs as well as lytically induced cells.
Conclusion: Our findings demonstrate hiPSCs as a powerful tool to explore the potential impacts of KSHV infection on stem cell functions and virus pathogenesis in stem cell differentiated cells.
Kaposi’s Sarcoma-associated Herpesvirus (KSHV) was first discovered in Kaposi’s sarcoma (KS) lesion from an AIDS patient, and this virus serves as the causative agent of KS, primary effusion lymphoma (PEL), and multicentric Castleman disease (MCD) (Mesri et al., 2014). Recent studies have linked KSHV to the KSHV inflammatory cytokine syndrome (KICS) and osteosarcoma (Polizzotto et al., 2012; Chen et al., 2021). Like other herpesviruses, KSHV life cycle comprises two main phases: latency and lytic replication. During latency, only limited viral latent genes and viral microRNAs are expressed. Once being stimulated to the lytic phase, a wide array of viral genes is expressed, culminating in the assembly of virion particles and finally the release of mature progeny viruses to infect new cells (Avey et al., 2015). KSHV infects a variety of in vivo target cells, such as endothelial cells, B cells, monocytes, epithelial cells, and keratinocytes. KSHV also infects a diversity of in vitro target cells and establishes in vitro latency in many of these cell types (Chakraborty et al., 2012). Mesenchymal stem cells (MSCs) exhibit the potential to differentiate into a variety of mesodermal cell lineages, such as adipocytes, chondrocytes, osteocytes, and myocytes (Ankrum et al., 2014). Interestingly, KSHV have been found to infect human primary MSCs derived from diverse organs, including bone marrow, adipose tissue, dental pulp, gingiva tissue, and exfoliated deciduous teeth (Lee et al., 2016). Also, KSHV can efficiently infect, transform, and reprogram rat primary MSCs into KS-like tumor cells (Jones et al., 2012).
Takahashi and Yamanaka first time demonstrated induction of pluripotent stem cells (iPSCs) from mouse embryonic or adult fibroblasts by introducing four factors, Oct3/4, Sox2, c-Myc, and Klf4, under embryonic stem cell culture conditions (Takahashi and Yamanaka, 2006). This groundbreaking discovery offers significant potential in disciplines such as biology, pathophysiology, and cellular regenerative medicine. As a result, iPSCs derived differentiated cells (e.g., dendritic cells and macrophages) have become a pioneering avenue for cell therapy research (Senju et al., 2011; Wu et al., 2024). However, it remains unclear whether human iPSCs (hiPSCs) are permissive to KSHV and how this oncogenic virus infection may affect cellular gene profile. These interesting questions will be addressed in the current study using the in vitro culture system, next-generation sequencing (NGS) analysis and different functional assays.
KSHV+ PEL cell line BCBL-1 was kindly provided by Dr. Dean Kedes (University of Virginia) and cultured in RPMI 1640 media (Gibco) with supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine, 0.05 mM β-mercaptoethanol, and 0.02% (wt/vol) sodium bicarbonate. For KSHV infection experiments, BCBL-1 cells were incubated with 0.6 mM valproic acid for 4-6 days, then the virions was purified from the culture supernatant by using the ultracentrifugation at 20,000 × g for 3 h, 4°C. The viral pellets were resuspended in 1/100 of the original volume with cell culture medium. The infectious titers of virus were determined as described previously (Qin et al., 2010). Human iSLK.219 cells (Dox-inducible SLK carrying rKSHV.219) are latently infected with a recombinant rKSHV.219 virus and contain a doxycycline (Dox)-inducible RTA (Myoung and Ganem, 2011). The rKSHV.219 virus expresses both the red fluorescent protein (RFP) under the control of KSHV lytic PAN promoter and the green fluorescent protein (GFP) under the control of the elongation factor 1 promoter (EF-1α) (Vieira and O’Hearn, 2004). HEK293T (Human embryonic kidney 293T) cells were purchased from American Type Culture Collection (ATCC) and cultured as recommended by the manufacturer. The hiPSCs were derived originally from wild type adult skin fibroblasts, which were induced to pluripotent stem cells in Dr. Bruce Conklin’s laboratory (Miyaoka et al., 2014) and confirmed by the staining of NANOG, one of stem cell markers expression (Figure 1). These cells were cultivated in mTeSR1 Basal medium supplemented with 5x Supplements (Stemcell Technologies) and 1% penicillin and streptomycin (Gibco) on 1:60 growth-factor-reduced Matrigel Matrix coated dish or plates.
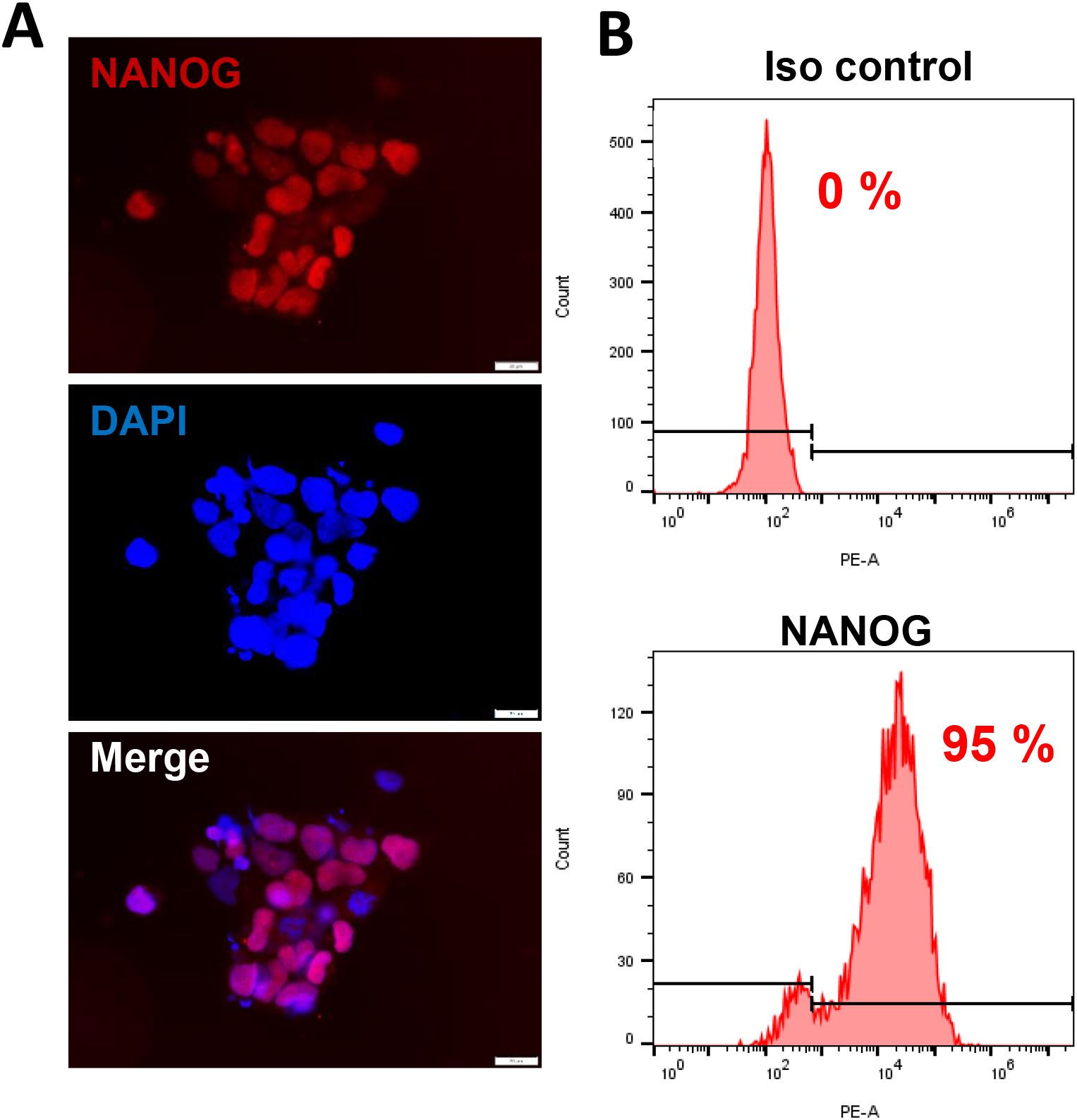
Figure 1. The culture of human induced pluripotent stem cells (hiPSCs). (A, B) The hiPSCs were cultured in mTeSR1 medium with supplements (Stemcell Technologies). The expression of stem cell marker, NANOG, was detected by using immunofluorescence and flow cytometry (isotype Ab as a negative control), respectively. Scale bar, 20 μm.
Cells were seeded in eight-well chamber slides (Nunc) prior to virus infection, then fixed with 4% PFA and stained with a first antibody recognizing NANOG (Cell Signaling) or LANA (Advanced Biotechnologies), followed by a secondary antibody conjugated to Texas Red (Invitrogen) and DAPI. Fluorescence signal was measured using the Olympus IX83 microscope (Olympus).
Flow cytometry was used for the quantitative assessment of apoptosis with the FITC-Annexin V/propidium iodide (PI) Apoptosis Detection Kit I (BD Pharmingen) on a FACS Calibur 4-color flow cytometer (BD Bioscience). The detection of stem cell markers expression in hiPSCs using flow cytometry has been described in our previous study (Qin et al., 2011).
Total RNA was isolated using the RNeasy Mini kit (Qiagen), and cDNA was synthesized using a SuperScript III First-Strand Synthesis SuperMix Kit (Invitrogen). Primers used for amplification of target genes were listed in Supplementary Table S1. Amplification was carried out using an iCycler IQ Real-Time PCR Detection System, and cycle threshold (Ct) values were tabulated in duplicate for each gene of interest in each experiment. “No template” (water) controls were used to ensure minimal background contamination. Using mean Ct values tabulated for each gene, and paired Ct values for β-actin or GAPDH as a loading control, fold changes for experimental groups relative to assigned controls were calculated using automated iQ5 2.0 software (Bio-rad).
RNA-Sequencing of triplicate samples was performed by BGI Americas Corporation using their unique DNBSEQ sequencing technology. The completed RNA-Sequencing data was submitted to NCBI Sequence Read Archive (SRA# PRJNA1186964). Raw sequencing reads were analyzed using the RSEM software (version 1.3.0; human GRCh38 genome sequence and annotation) and gene expression was quantified as previously described (Kheir et al., 2019). The EBSeq software was utilized to call differentially expressed genes that were statistically significant using a false discovery rate (FDR) less than 0.05. Differentially expressed genes between cells with or without virus infection were used as input for the GO_enrichment and KEGG pathways analyses.
For RNA interference (RNAi) assays, FZD10 On-Target plus SMART pool small interfering RNA (siRNA; Dharmacon) or negative control siRNA, were delivered using the DharmaFECT Transfection Reagent as recommended by the manufacturer.
Significance for differences between experimental and control groups (with no fewer than 3 experiments per group performed) was determined using the two-tailed Student’s t-test (Excel 2016), and p values < 0.05 or < 0.01 were considered significant or highly significant, respectively.
We first tested whether hiPSCs cells (Figure 1) are permissive to KSHV infection. As shown in Figure 2, the immunofluorescence results showed that either wild type KSHV or rKSHV.219 successfully established latent infection in hiPSCs (the infection rate > 80%). We next tested whether rKSHV.219 infected cells could be induced to lytic reactivation by different chemical inducers such valproic acid (VA), sodium butyrate (NaB) and 12-O-tetradecanoyl-phorbol-13-acetate (TPA). Our RT-qPCR and immunofluorescence assays results showed NaB plus TPA treatment having the best effects on lytic reactivation when compared to VA alone treatment (Figures 3A, B). When we collected the viral supernatants to infect naïve HEK293T cells, we found that NaB plus TPA induction produced the most infectious virions through detection of intracellular viral copies by using qPCR (Figure 3C).
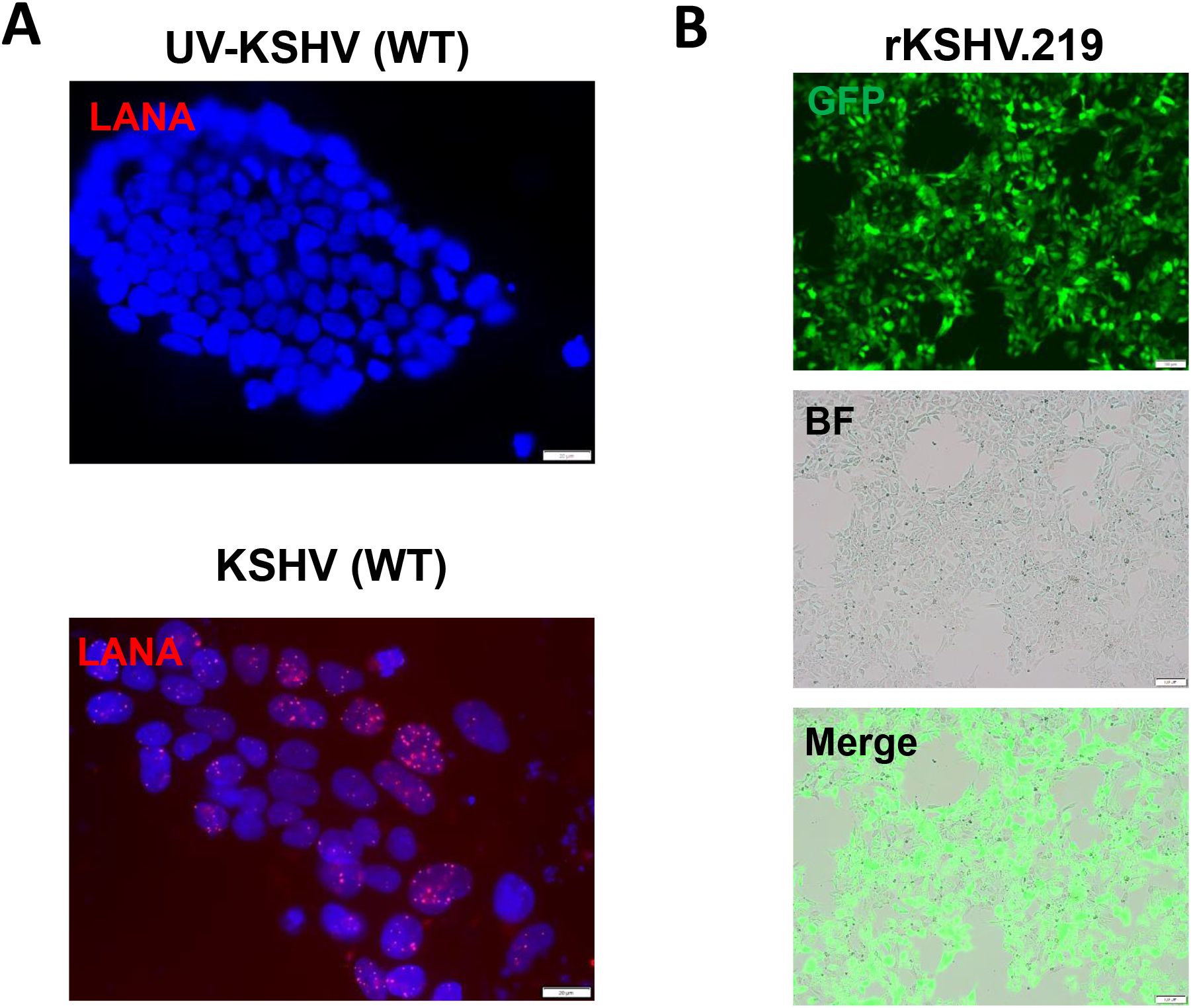
Figure 2. KSHV de novo infection of hiPSCs. (A, B) The hiPSCs were infected by either wild type (WT) KSHV or rKSHV.219 (MOI~3), respectively, the images were acquired by using fluorescence microscope (LANA staining for WT virus) at 72 h p.i. Scale bar: 100 μm. The UV-inactivated virus was used as a negative control.
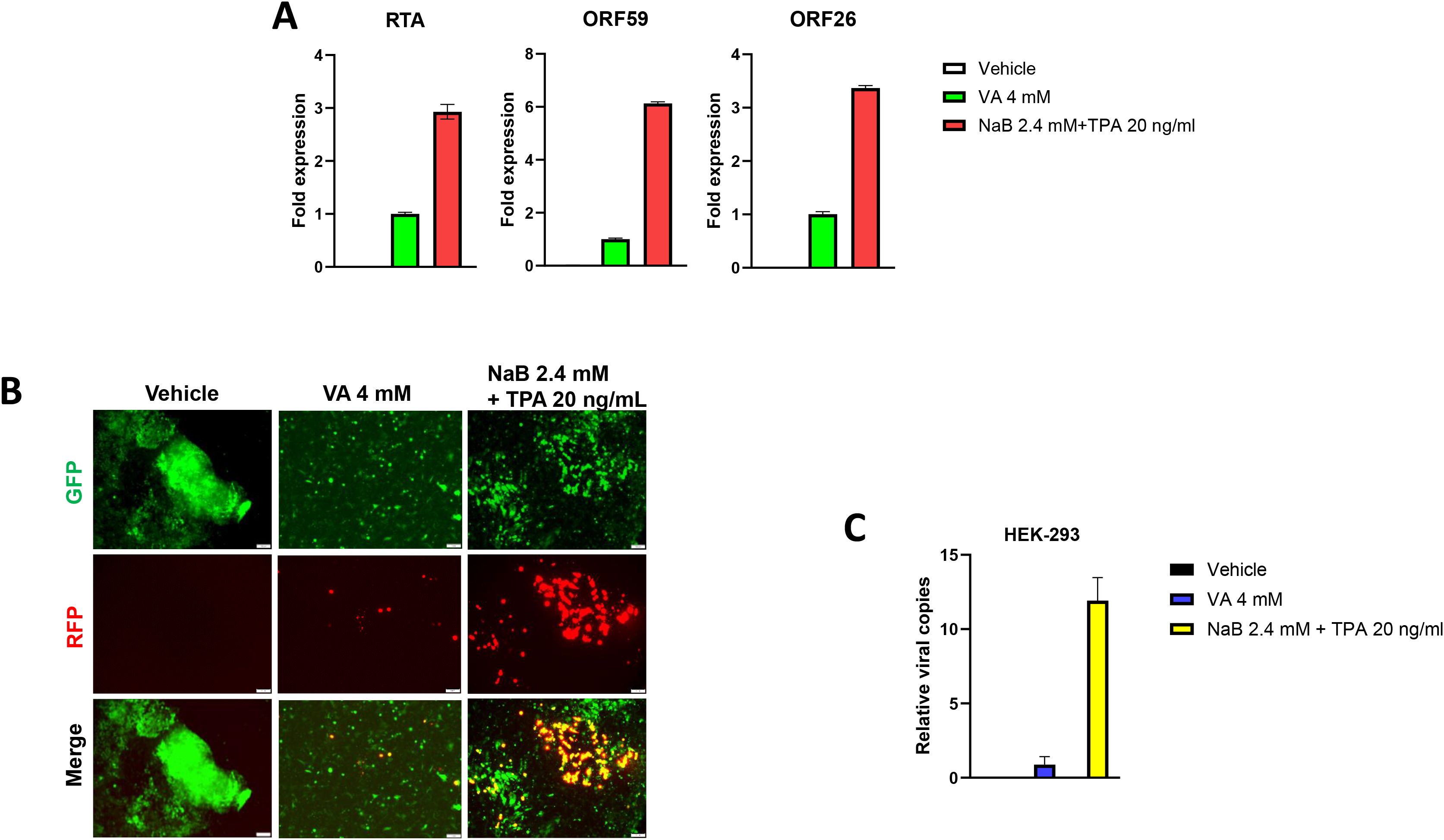
Figure 3. Successful induction of KSHV lytic reactivation and virions production from latently infected hiPSCs. (A, B) The hiPSCs were first infected by rKSHV.219 (MOI~3) for 72 h, then treated by different chemical inducers for another 72 h. The gene expression was detected and compared by using RT-qPCR (vs the VA 4 mM group), and the images were acquired by using fluorescence microscope. (C) The viral supernatants from (A) were collected to infect naïve HEK293T cells, and intracellular viral copies were detected and compared by using qPCR at 48 h p.i. The Error bars represent S.D. for 3 independent experiments. VA, valproic acid; NaB, sodium butyrate; TPA, 12-O-tetradecanoyl-phorbol-13-acetate.
Heparan sulfate (HS) represents the major cellular receptor for KSHV entry into host cells (Birkmann et al., 2001), although it remains unclear whether HS is responsible for KSHV entry into hiPSCs. Here we found that pretreatment of rKSHV.219 virions by Heparin, the competitor of HS, almost completely blocked viral entry into hiPSCs by using qPCR and immunofluorescence assays, respectively (Figure 4). We observed similar results when we used wild type KSHV virions (Supplementary Figure S1), although other alternative receptors or pathways are probably existed for KSHV entry into hiPSCs.
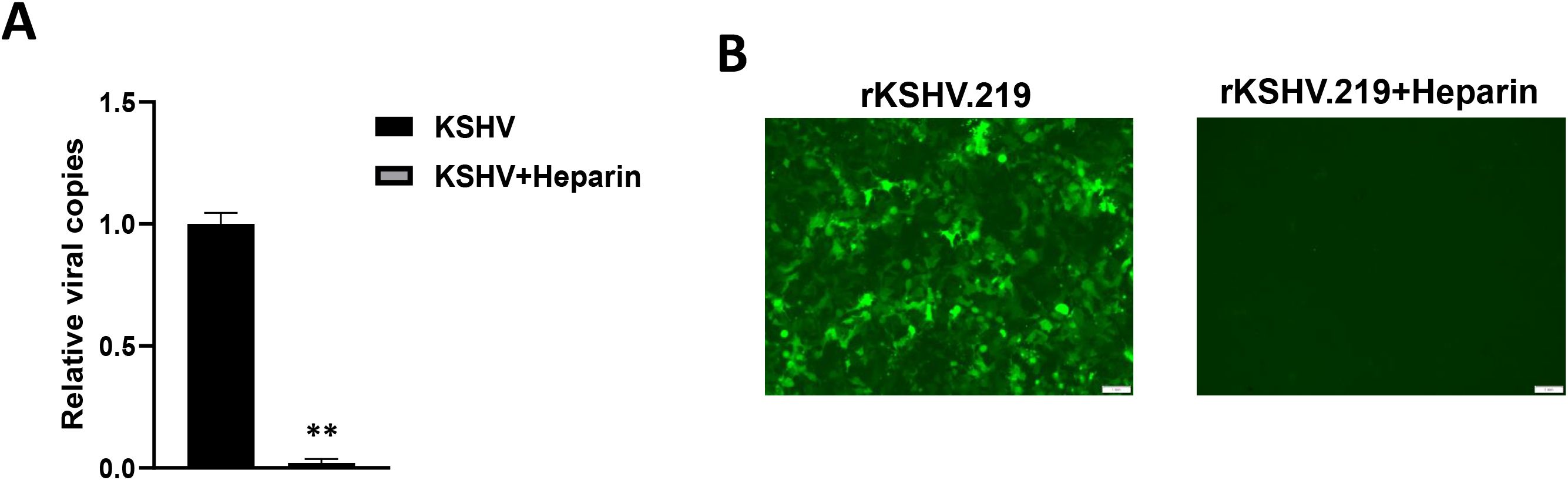
Figure 4. Heparan sulfate is the major cellular receptor responsible for KSHV entry into hiPSCs. (A) The rKSHV.219 virions were first treated with 0.5 mg/mL heparin (the competitor for heparan sulfate) for 1 h at 4°C, then the cells were infected with purified virions (MOI~3) for 2 h at 37°C. After that, cells were trypsinized and washed to remove extracellular virions. Total cellular DNA was prepared and the internalized viral copies were measured by qPCR. Error bars represent S.D. for 3 independent experiments, **p<0.01. (B) The images were acquired by using fluorescence microscope at 48 h p.i.
In a time-course assay, we found that KSHV infection caused growth arrest of hiPSCs when compared to non-infected cells (Figure 5A). Using the DAPI staining, we observed obvious nuclear condensation or fragmentation only in KSHV-infected cells (Figure 5B). Our flow cytometry analysis further confirmed that KSHV infection caused increased cell apoptosis in hiPSCs (Figures 5C, D). We also examined the impacts of KSHV infection on the expression of stem cell markers from hiPSCs by flow cytometry analysis. Our results showed that the expression of NANOG and OCT-4 was significantly downregulated (~50% of reduction) in KSHV-infected hiPSCs (Figure 6). As a control, NANOG and OCT-4 are almost not expressed on BCBL-1 cells (Supplementary Figure S2).
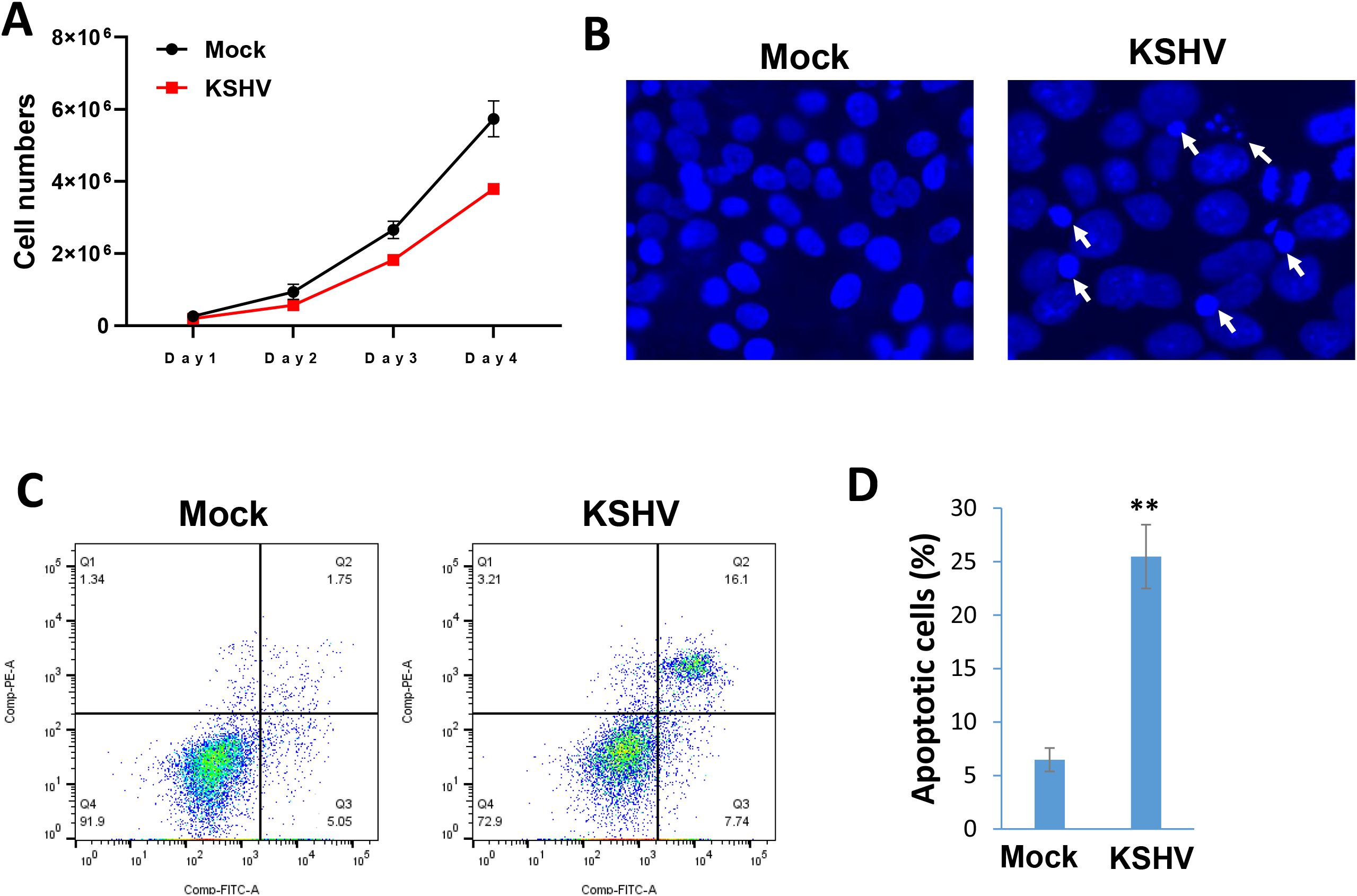
Figure 5. KSHV infection arrests the growth of hiPSCs through inducing cell apoptosis. (A) The hiPSCs-CMs were infected by KSHV, then the live cell numbers were calculated at indicated time points using Countess 3 Automated Cell Counter (Thermo Fisher). (B) Cells were stained by DAPI, then the images were acquired by using fluorescence microscope at 72 h p.i. The visualization of nuclear condensation or fragmentation was indicated by arrows. (C, D) Cell apoptosis was measured by Annexin V-PI staining and flow cytometry analysis. Error bars represent S.D. for 3 independent experiments, **p<0.01.
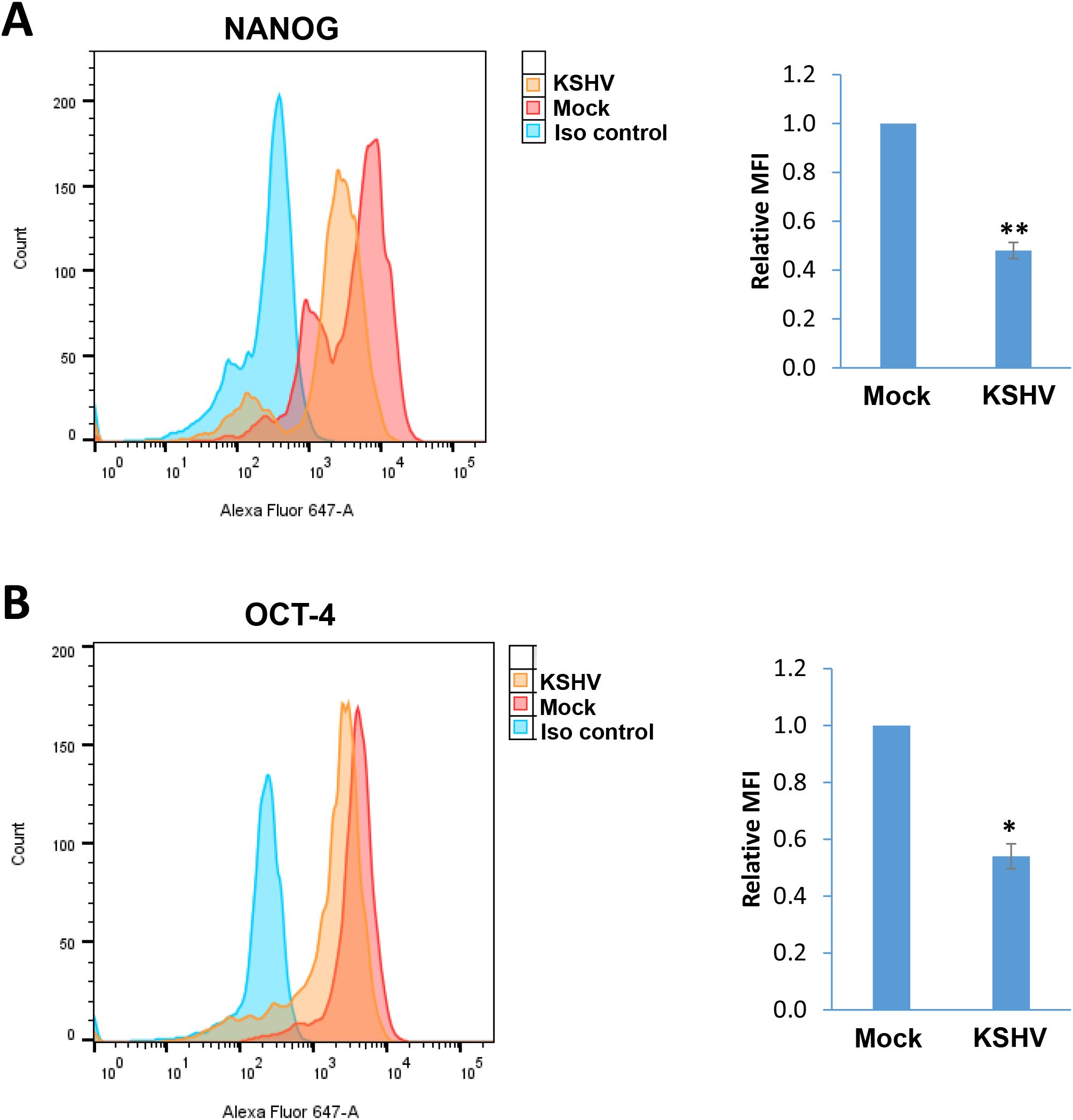
Figure 6. The impacts on stem cell markers expression by KSHV infection of hiPSCs. (A, B) Flow cytometry analyses were used to quantify NANGO and OCT-4 expression in mock- or KSHV-infected hiPSCs. Mean fluorescence intensity (MFI), reflecting the expression of each protein for the same number of cells in each condition was calculated using FlowJo software. Error bars represent S.D. for 3 independent experiments, *p<0.05, **p<0.01.
To determine the global cellular changes caused by KSHV infection, we compared the gene profiles of mock- and KSHV-infected hiPSCs, using RNA-Sequencing analyses. The volcano plots showed the scattering of genes which were significantly upregulated (1365 transcripts) or downregulated (1811 transcripts) in KSHV-infected hiPSCs (Figure 7A). The top 20 significantly upregulated or downregulated genes in KSHV-infected hiPSCs were listed in Table 1. The GO_enrichment (Biological process module) analysis of these significantly changed genes identified several major functional categories potentially involved, such as cytoskeleton organization and functions, regulation of RNA splicing, DNA repair, nucleobase biosynthetic process, etc (Figure 7B). The KEGG analysis indicated that KSHV infection affected pathways important for MAPK, Relaxin, Hippo and Wnt signaling in hiPSCs (Figure 7C). Among them, the Hippo and Wnt signaling pathways play important roles in stem cells functions and development (Zhao et al., 2024; Yu et al., 2024). The bioinformatics analysis predicted that the changed candidates (42 in Hippo pathway and 41 in Wnt pathway, respectively) showed a close protein-protein interaction network and potential functional association (Supplementary Figure S3).
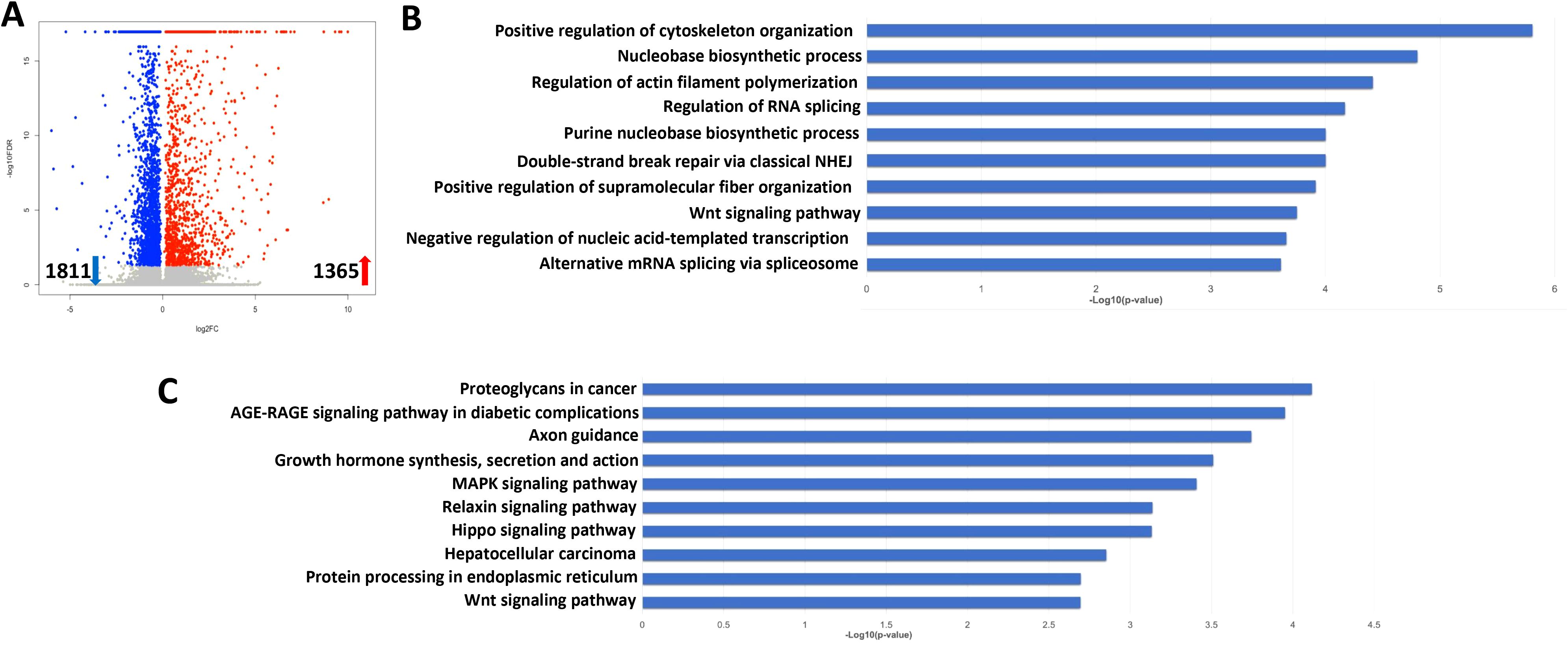
Figure 7. Transcriptome analysis of KSHV latently infected hiPSCs. (A) RNA-Sequencing was used to investigate changes in the transcriptome from rKSHV.219 infected cells (72 h p.i.) when compared to mock cells. The significantly altered genes (FDR<0.05) were shown in the Volcano plot panel. (B, C) The GO_enrichment (Biological process module) and KEGG pathway analysis of the significantly changed genes in rKSHV.219 infected cells.
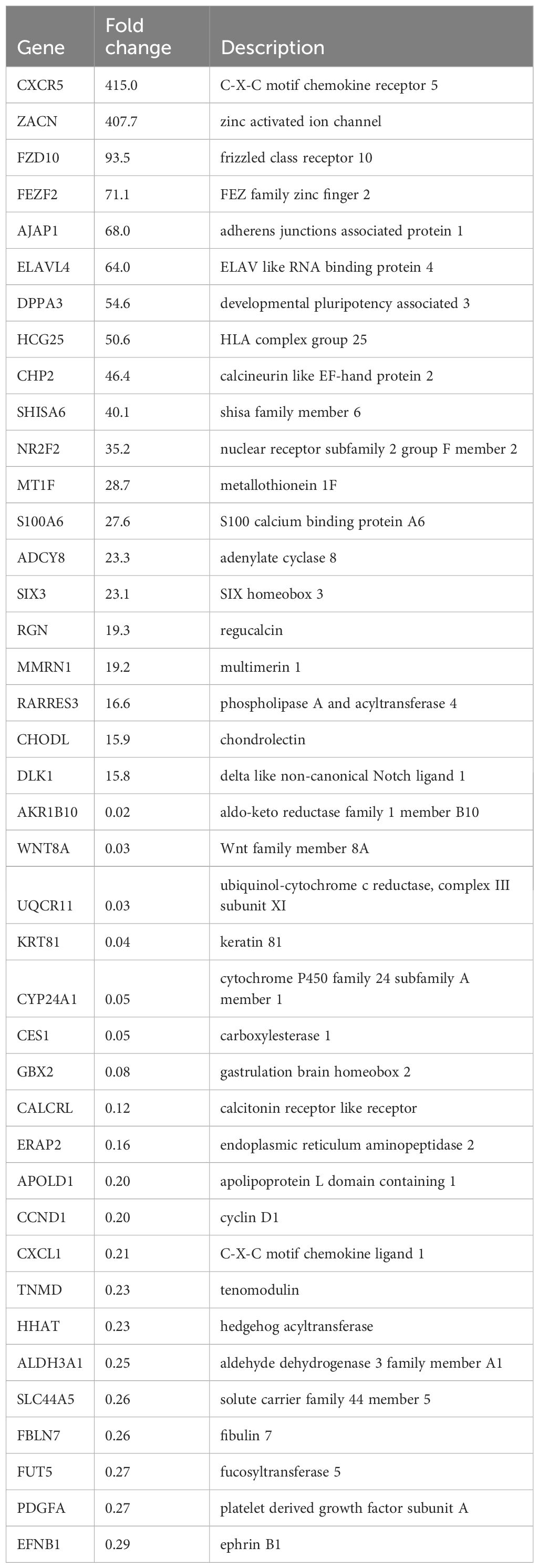
Table 1. The top 20 candidate genes upregulated or downregulated within KSHV latently infected hiPSCs.
One of upregulated candidate genes identified from RNA-Sequencing analyses, frizzled class receptor 10 (FZD10), is a member of the frizzled gene family. Members of this family encode 7-transmembrane domain proteins that are receptors for the Wnt signaling protein ligands. Most frizzled receptors including FZD10 are coupled to the β-Catenin canonical signaling pathway (Katoh and Katoh, 2017). Our RT-qPCR results further confirmed that FZD10 transcriptional level was dramatically increased from KSHV-infected hiPSCs (Figure 8A). We then silenced FZD10 with RNAi, and found that it could significantly reduce the level of β-Catenin transcription (Figure 8B).
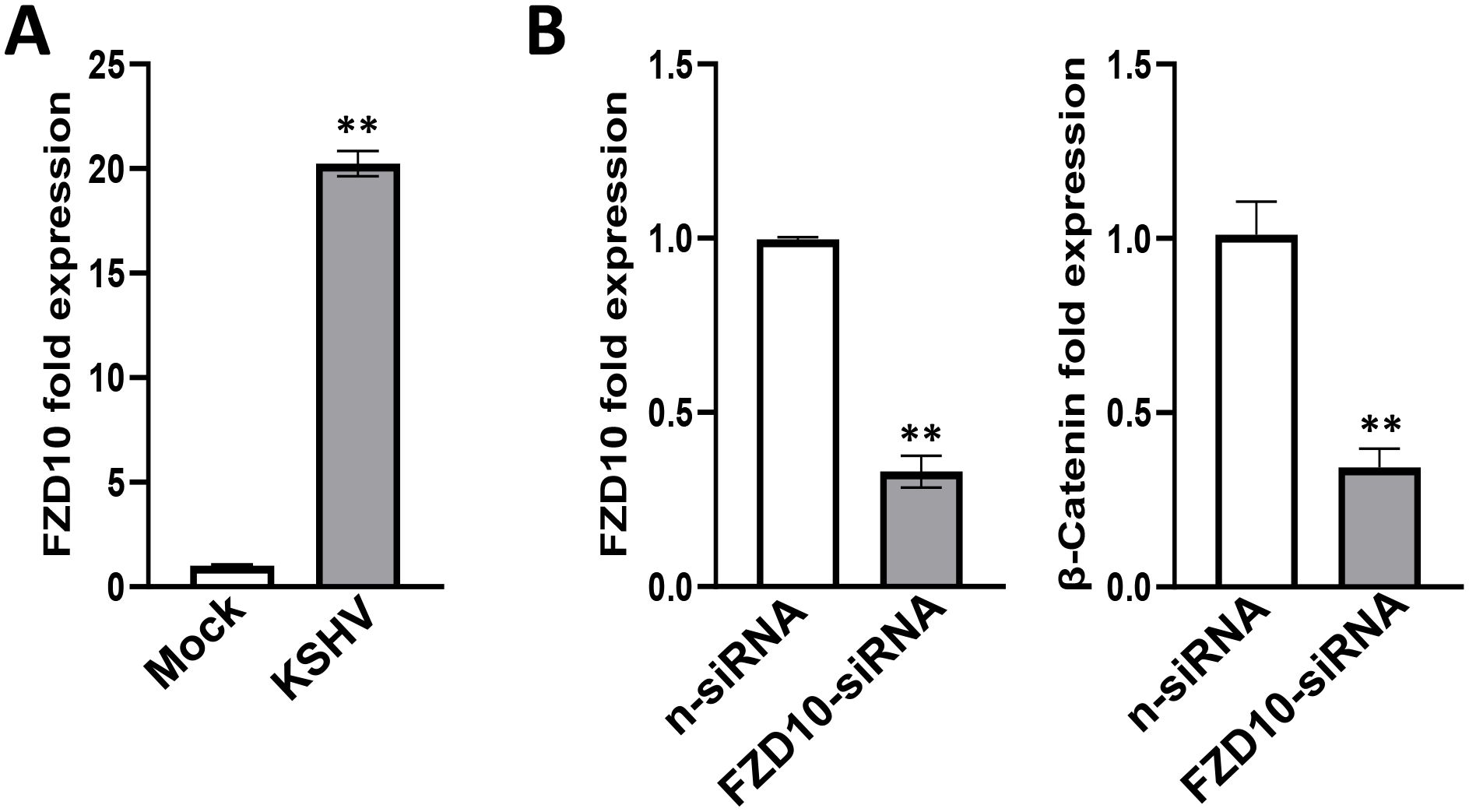
Figure 8. Functional validation of FZD10 candidate gene in KSHV-infected hiPSCs. (A) The hiPSCs were first infected by rKSHV.219 (MOI~3) for 72 h, then gene expression was detected and compared by using RT-qPCR. (B) The hiPSCs were first transfected with FZD10-siRNA or non-target control siRNA (n-siRNA) for 48 h, then infected by rKSHV.219. Gene expression was detected and compared by using RT-qPCR. Error bars represent S.D. for 3 independent experiments, **p<0.01.
We further explored the gene profiles changed in lytically induced KSHV-infected hiPSCs by NaB plus TPA. The volcano plots showed the scattering of genes which were significantly upregulated (3928 transcripts) or downregulated (2856 transcripts) in lytically induced cells (Figure 9A). The top 20 significantly upregulated or downregulated genes in lytically induced cells were listed in Table 2. The GO_enrichment analysis of these significantly changed genes identified several major functional categories, including regulation of cell migration/mobility, axonogenesis, regulation of cell proliferation, etc (Figure 9B). The KEGG analysis indicated that induction of lytic reactivation affected pathways important for focal adhesion, steroid biosynthesis, cellular signaling such as Wnt, TNF and MAPK (Figure 9C).
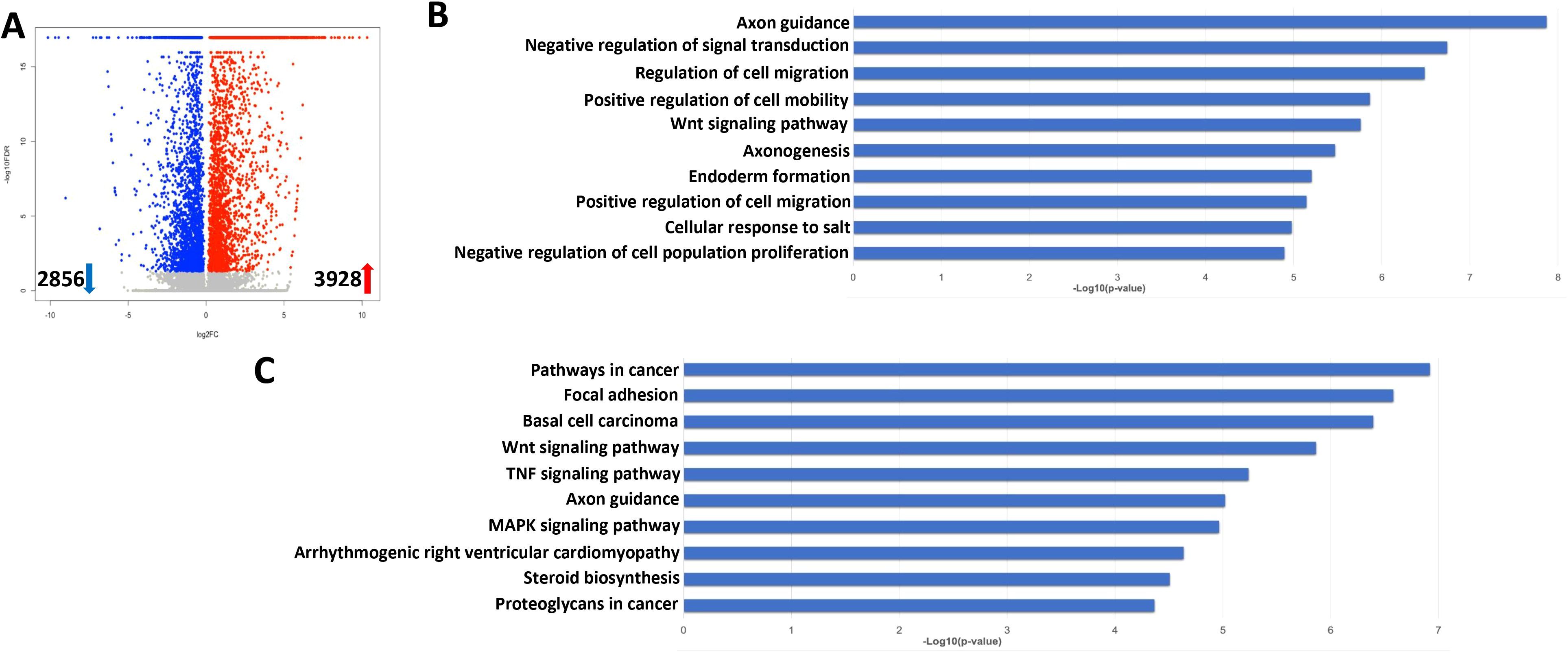
Figure 9. Transcriptome analysis of lytically induced KSHV infected hiPSCs. (A) RNA-Sequencing was used to investigate changes in the transcriptome from NaB/TPA induced rKSHV.219 infected cells (72 h) when compared to non-induced cells. The significantly altered genes (FDR<0.05) were shown in the Volcano plot panel. (B, C) The GO_enrichment (Biological process module) and KEGG pathway analysis of the significantly changed genes from infected cells.
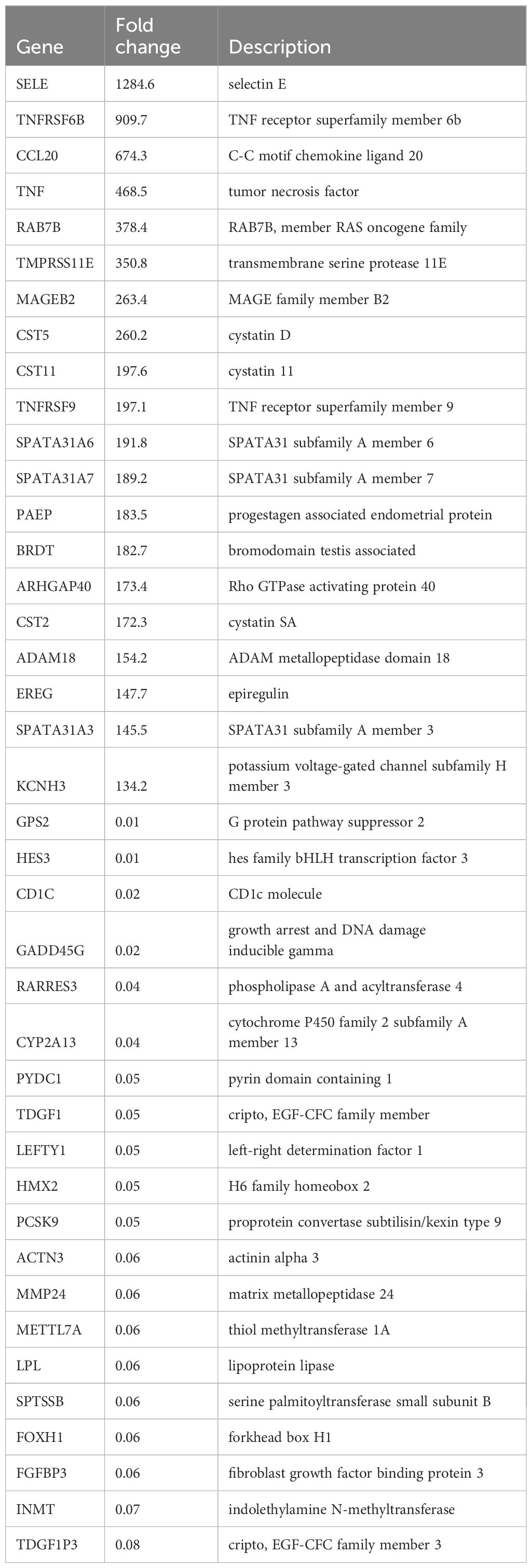
Table 2. The top 20 candidate genes upregulated or downregulated within lytically induced KSHV-infected hiPSCs.
As mentioned above, the new technology allows us to generate hiPSCs without human embryo, which can overcome the issues of histoincompatibility and ethical problems associated with the use human embryonic stem cells (Senju et al., 2011). Since KSHV can infect a broad tropism of human primary cells, some of them are not easily isolated from the donors or no commercial cell lines available. Thus, the hiPSCs may represent a powerful tool to study KSHV infection and pathogenesis in different types of differentiated cells. For instance, recent data indicate that KSHV infection is potentially associated with some heart diseases such as dilated cardiomyopathy (DCM) (Zhao et al., 2023; Huang et al., 2024; Hsueh et al., 2006). Zhao and colleague reported that increased KSHV seropositivity and quantitative titers were found in the patients with DCM compared with the non-DCM group. The risk of the individual end point of death from cardiovascular causes or heart transplantation was increased among DCM patients with the KSHV DNA seropositivity during follow-up (Zhao et al., 2023). As we know, primary isolated cardiomyocytes are very difficult to culture in vitro, so the hiPSCs induced cardiomyocytes will be a better option for such research.
In the current study, we found that the hiPSCs were permissive to KSHV infection including being completely induced to lytic reactivation and releasing infectious virions. Also, we identified heparan sulfate as the major cellular receptor for KSHV entry into hiPSCs. Interestingly, one recent study reported Neuropilin 1 as an entry receptor for KSHV infection of MSCs through TGFBR1/2-mediated micropinocytosis (Lu et al., 2023). In that study, the authors used different types of oral MSCs, including periodontal ligament stem cells (PDLSC), dental pulp stem cells (DPSC), and gingiva/mucosa-derived mesenchymal stem cells (GMSC). These data together with ours suggest that KSHV may use different receptors to entry into stem cells with varied origins.
Using the hiPSCs, we can also study the impacts of KSHV infection on these stem cells differentiation. Previous study found that KSHV infection of oral MSCs promoted multilineage differentiation and mesenchymal-to-endothelial transition (MEndT) (Li et al., 2018). Here, our transcriptomic analysis indicated that KSHV infection affected several cellular signaling pathways important for stem cells differentiation, such as Hippo and Wnt pathways. Previous study reported that KSHV-encoded G-protein coupled receptor (vGPCR) protein was able to inhibit the Hippo pathway kinases Lats1/2, promoting the activation of YAP/TAZ, cell proliferation and tumorigenesis (Liu et al., 2015). Interestingly, the vGPCR protein was found to activate the canonical Wnt/β-catenin signaling activities, which was dependent on the PI3K/Akt pathway (Angelova et al., 2014). Another viral protein, latency-associated nuclear antigen (LANA), was also found to regulate the activities of Wnt signaling pathway (Kusano and Eizuru, 2010). However, the molecular mechanisms of KSHV manipulating these signaling pathways to modulate hiPSCs differentiation and other functions still require further investigation. Notably, there is a close relationship between the Hippo pathway and the canonical Wnt pathway, and their crosstalk may play a role in some human diseases (Sileo et al., 2022).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRA# PRJNA1186964.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
JF: Writing – original draft. ZL: Writing – review & editing. HZ: Writing – review & editing. LD: Writing – original draft, Writing – review & editing. ZQ: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by NIH R03DE031978, R21AI186566, and the Arkansas Bioscience Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000. ZL was supported by a National Cancer Institute grant R01CA261258, a National Institute of General Medical Sciences COBRE grant P20GM121288, a U.S.-Japan Cooperative Medical Sciences Program Collaborative Award from the National Institute of Allergy and Infectious Diseases and CRDF Global (grant number DAA3-19-65602-1), a Ladies Leukemia League research grant, a Tulane school of medicine faculty research pilot grant, and a Carol Lavin Bernick faculty grant. HZ received support from NIGMS (R35GM151226). Funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1563440/full#supplementary-material
Angelova, M., Ferris, M., Swan, K. F., McFerrin, H. E., Pridjian, G., Morris, C. A., et al. (2014). Kaposi’s sarcoma-associated herpesvirus G-protein coupled receptor activates the canonical Wnt/beta-catenin signaling pathway. Virol. J. 11, 218. doi: 10.1186/s12985-014-0218-8
Ankrum, J. A., Ong, J. F., Karp, J. M. (2014). Mesenchymal stem cells: immune evasive, not immune privileged. Nat. Biotechnol. 32, 252–260. doi: 10.1038/nbt.2816
Avey, D., Brewers, B., Zhu, F. (2015). Recent advances in the study of Kaposi’s sarcoma-associated herpesvirus replication and pathogenesis. Virol. Sin. 30, 130–145. doi: 10.1007/s12250-015-3595-2
Birkmann, A., Mahr, K., Ensser, A., Yaguboglu, S., Titgemeyer, F., Fleckenstein, B., et al. (2001). Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75, 11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001
Chakraborty, S., Veettil, M. V., Chandran, B. (2012). Kaposi’s sarcoma associated herpesvirus entry into target cells. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00006
Chen, Q., Chen, J., Li, Y., Liu, D., Zeng, Y., Tian, Z., et al. (2021). Kaposi’s sarcoma herpesvirus is associated with osteosarcoma in Xinjiang populations. Proc. Natl. Acad. Sci. U.S.A. 118. doi: 10.1073/pnas.2016653118
Hsueh, S. C., Chung, M. T., Fang, R., Hsiung, M. C., Young, M. S., Lu, H. F. (2006). Primary cardiac lymphoma. J. Chin. Med. Assoc. 69, 169–174. doi: 10.1016/S1726-4901(09)70200-X
Huang, X., Huang, X., Li, Y., Li, L., Liao, J., Huang, H., et al. (2024). Kaposi’s sarcoma-associated herpesvirus infection and its association with all-cause and cardiovascular mortality in the general adults: A prospective cohort study. J. Med. Virol. 96, e29953. doi: 10.1002/jmv.29953
Jones, T., Ye, F., Bedolla, R., Huang, Y., Meng, J., Qian, L., et al. (2012). Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Invest. 122, 1076–1081. doi: 10.1172/JCI58530
Katoh, M., Katoh, M. (2017). Molecular genetics and targeted therapy of WNT-related human diseases (Review). Int. J. Mol. Med. 40, 587–606. doi: 10.3892/ijmm.2017.3071
Kheir, F., Zhao, M., Strong, M. J., Yu, Y., Nanbo, A., Flemington, E. K., et al. (2019). Detection of epstein-barr virus infection in non-small cell lung cancer. Cancers (Basel) 11:759–783; . doi: 10.3390/cancers11060759
Kusano, S., Eizuru, Y. (2010). Human I-mfa domain proteins specifically interact with KSHV LANA and affect its regulation of Wnt signaling-dependent transcription. Biochem. Biophys. Res. Commun. 396, 608–613. doi: 10.1016/j.bbrc.2010.04.111
Lee, M. S., Yuan, H., Jeon, H., Zhu, Y., Yoo, S., Shi, S., et al. (2016). Human mesenchymal stem cells of diverse origins support persistent infection with kaposi’s sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. MBio 7, e02109–e02115. doi: 10.1128/mBio.02109-15
Li, Y., Zhong, C., Liu, D., Yu, W., Chen, W., Wang, Y., et al. (2018). Evidence for kaposi sarcoma originating from mesenchymal stem cell through KSHV-induced mesenchymal-to-endothelial transition. Cancer Res. 78, 230–245. doi: 10.1158/0008-5472.CAN-17-1961
Liu, G., Yu, F. X., Kim, Y. C., Meng, Z., Naipauer, J., Looney, D. J., et al. (2015). Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene 34, 3536–3546. doi: 10.1038/onc.2014.281
Lu, Z. Z., Sun, C., Zhang, X., Peng, Y., Wang, Y., Zeng, Y., et al. (2023). Neuropilin 1 is an entry receptor for KSHV infection of mesenchymal stem cell through TGFBR1/2-mediated macropinocytosis. Sci. Adv. 9, eadg1778. doi: 10.1126/sciadv.adg1778
Mesri, E. A., Feitelson, M. A., Munger, K. (2014). Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15, 266–282. doi: 10.1016/j.chom.2014.02.011
Miyaoka, Y., Chan, A. H., Judge, L. M., Yoo, J., Huang, M., Nguyen, T. D., et al. (2014). Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 11, 291–293. doi: 10.1038/nmeth.2840
Myoung, J., Ganem, D. (2011). Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174, 12–21. doi: 10.1016/j.jviromet.2011.03.012
Polizzotto, M. N., Uldrick, T. S., Hu, D., Yarchoan, R. (2012). Clinical manifestations of kaposi sarcoma herpesvirus lytic activation: multicentric castleman disease (KSHV-MCD) and the KSHV inflammatory cytokine syndrome. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00073
Qin, Z., Dai, L., Bratoeva, M., Slomiany, M. G., Toole, B. P., Parsons, C. (2011). Cooperative roles for emmprin and LYVE-1 in the regulation of chemoresistance for primary effusion lymphoma. Leukemia 25, 1598–1609. doi: 10.1038/leu.2011.144
Qin, Z., Freitas, E., Sullivan, R., Mohan, S., Bacelieri, R., Branch, D., et al. (2010). Upregulation of xCT by KSHV-encoded microRNAs facilitates KSHV dissemination and persistence in an environment of oxidative stress. PloS Pathog. 6, e1000742. doi: 10.1371/journal.ppat.1000742
Senju, S., Haruta, M., Matsumura, K., Matsunaga, Y., Fukushima, S., Ikeda, T., et al. (2011). Generation of dendritic cells and macrophages from human induced pluripotent stem cells aiming at cell therapy. Gene Ther. 18, 874–883. doi: 10.1038/gt.2011.22
Sileo, P., Simonin, C., Melnyk, P., Chartier-Harlin, M. C., Cotelle, P. (2022). Crosstalk between the hippo pathway and the wnt pathway in huntington’s disease and other neurodegenerative disorders. Cells 11:3631–3651. doi: 10.3390/cells11223631
Takahashi, K., Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Vieira, J., O’Hearn, P. M. (2004). Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 325, 225–240. doi: 10.1016/j.virol.2004.03.049
Wu, Z., Su, Y., Li, J., Liu, X., Liu, Y., Zhao, L., et al. (2024). Induced pluripotent stem cell-derived mesenchymal stem cells: whether they can become new stars of cell therapy. Stem Cell Res. Ther. 15, 367. doi: 10.1186/s13287-024-03968-x
Yu, M., Qin, K., Fan, J., Zhao, G., Zhao, P., Zeng, W., et al. (2024). The evolving roles of Wnt signaling in stem cell proliferation and differentiation, the development of human diseases, and therapeutic opportunities. Genes Dis. 11, 101026. doi: 10.1016/j.gendis.2023.04.042
Zhao, Y., Li, H., Du, H., Yin, Z., He, M., Fan, J., et al. (2023). A Kaposi’s sarcoma-associated herpes virus-encoded microRNA contributes to dilated cardiomyopathy. Signal Transduct Target Ther. 8, 226. doi: 10.1038/s41392-023-01434-3
Keywords: KSHV, hiPSCs, stem cell, herpesvirus, infection
Citation: Fan J, Lin Z, Zhang H, Dai L and Qin Z (2025) Infection of human induced pluripotent stem cells by an oncogenic herpesvirus. Front. Cell. Infect. Microbiol. 15:1563440. doi: 10.3389/fcimb.2025.1563440
Received: 20 January 2025; Accepted: 25 February 2025;
Published: 13 March 2025.
Edited by:
Haitao Guo, University of Pittsburgh, United StatesReviewed by:
Jian Zhu, The Ohio State University, United StatesCopyright © 2025 Fan, Lin, Zhang, Dai and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Dai, bGRhaUB1YW1zLmVkdQ==; Zhiqiang Qin, enFpbkB1YW1zLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.