- Department of Microbiology and Parasitology, Faculty of Biology - Aquatic One Health Research Center (ARCUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain
Introduction: The World Health Organization has identified multi-drug resistant Klebsiella pneumoniae strains as the highest priority in 2024. Understanding the regulatory routes of virulence features is crucial for the development of novel anti-virulence strategies. SdiA, a LuxR-like quorum sensing (QS) receptor that responds to N-acyl-homoserine lactones (AHLs), is involved in the regulation of virulence traits in some Gram-negative bacteria. The function of this receptor in the virulence of K. pneumoniae remains uncertain. The objective of the present study was to elucidate the function of SdiA in K. pneumoniae biofilm formation and virulence.
Methods: To this end, a genetic knockout of sdiA was conducted, and virulence-related phenotypic studies were performed following AHL provision.
Results and Discussion: The results demonstrate that sdiA deficiency increases susceptibility to phage infection and human serum resistance, and promotes biofilm maturation and cell filamentation, although no effect on virulence was observed in vivo in the Galleria mellonella infection model. On the other hand, C6-HSL promoted sdiA-dependent biofilm maturation, capsule production and serum resistance while reducing virulence against G. mellonella in the absence of sdiA. The addition of C6-HSL did not affect phage susceptibility. The results of this study demonstrate that AHLs and SdiA exert a dual influence on virulence phenotypes, operating both independently and hierarchically. These findings provide new insights into the virulence of K. pneumoniae and its regulation by SdiA.
1 Introduction
The emergence of MDR bacteria represents a significant public health concern, given the lack of effective treatment options (Tacconelli et al., 2018). K. pneumoniae has been classified by the WHO as a maximum priority pathogen for the development of new antimicrobial strategies in 2024, with an increasing incidence of convergent strains with MDR and hypervirulence traits (González- et al., 2021; WHO, 2024).
A comprehensive understanding of the factors that regulate virulence traits is essential for the development of novel anti-virulence therapies. Among these, blocking QS systems may play an important role in controlling virulence in many MDR pathogens. QS systems are responsible for regulating gene expression in bacterial populations in accordance with cell density through the production of autoinducing molecules, which act as signals (Wang et al., 2020). In Gram-negative bacteria, these are AHLs, which are often synthesised by LuxI-type synthases and recognised by LuxR-type receptors (Papenfort and Bassler, 2016). In the case of some species of Enterobacteriaceae, such as Escherichia coli, Salmonella spp. and Klebsiella pneumoniae, a putative LuxI synthase is absent, yet an orphan LuxR receptor (SdiA) is present and it has been shown that this receptor is capable of detecting AHLs produced by other bacteria (Sabag-Daigle et al., 2015; Michael et al., 2001; Ahmer, 2004; Sabag-Daigle and Ahmer, 2012; Cao et al., 2022; Mayer et al., 2023), among other ligands (Janssens et al., 2007; Styles et al., 2020).
The regulatory role of SdiA in virulence has been widely investigated in E. coli and Salmonella (Ahmer, 2004; Mayer et al., 2023; Schwieters and Ahmer, 2024). In E. coli, it has been shown that SdiA has a promoting effect on survival in the gastrointestinal tract through acid tolerance upregulating gad expression (Sabag-Daigle et al., 2012), and the promotion of resistance to quinolones through expression of AcrAB efflux pump (Rahmati et al., 2002). However, these observations were made only in the context of sdiA being overexpressed, and no such phenotypes were observed when sdiA was in its native position on the chromosome (Dyszel et al., 2010b). Conversely, SdiA has also been demonstrated to exert a repressive effect on other phenotypes, including motility and adhesion, through repression of fliC (flagella) and fimA (fimbriae) expression (Mayer et al., 2023). Furthermore, SdiA has been proposed as a repressor of biofilm formation, as sdiA-lacking strains show higher biofilm forming ability through uvrY repression (Suzuki et al., 2002). In addition, a reduction in biofilm formation (Lee et al., 2009) and an increased phage sensitivity (Ghosh et al., 2009) has been observed in E. coli following the addition of AHL in an sdiA-dependent manner. Nevertheless, there are also studies that argue that SdiA does not affect biofilm formation in this species (Sabag-Daigle et al., 2012). However, the majority of laboratory strains of E. coli are low biofilm formers, which may result in an underestimation of the impact of QS on this phenotype (Król et al., 2019). Regarding Salmonella, SdiA has shown to regulate two loci in an AHL-dependent activation manner: srgE and rck (Smith and Ahmer, 2003; Schwieters and Ahmer, 2024). The first one, srgE, codifies an effector secreted by the type III secretor system involved in virulence (Habyarimana et al., 2014); and the second one, rck, is involved in adhesion to eukaryotic cells and resistance to complement killing in human serum (Ahmer et al., 1998; Michael et al., 2001; Mambu et al., 2017), suggesting a role of SdiA in pathogenesis. Another study has highlighted the significance of SdiA in Salmonella adhesion and biofilm formation, as adhesion to eukaryotic cells and biofilm formation was reduced in the mutant strain irrespective of the presence of AHL (Askoura et al., 2021). However, the contribution of SdiA to the pathogenesis of Salmonella remains unclear, as SdiA AHL-activated strains exhibit no greater advantage than sdiA-deficient strains in the gut environment (Smith et al., 2008; Dyszel et al., 2010a; Schwieters and Ahmer, 2024). Furthermore, sdiA expression levels in Salmonella can exhibit considerable fluctuations in biofilm cells, contingent on the culture medium and the duration of the incubation period (Wang et al., 2016).
The function of SdiA has also been the subject of investigation in the bacterium Enterobacter cloacae. Research has indicated that the inactivation of sdiA has a positive effect on biofilm formation and adhesion (Shankar et al., 2013), and another study has observed AHL-dependent induced SdiA-regulation of copper transport and type VI secretion system in this species (Sabag-Daigle et al., 2015). Furthermore, a study conducted with a sdiA-lacking Cronobacter sakazakii strain showed increased expression of capsule and lipopolysaccharide (LPS) synthesis genes (Cao et al., 2022).
The role of QS signalling in K. pneumoniae has only been the subject of a limited number of studies, and there is considerable inconsistency in the literature regarding AHL-regulated QS. For instance, some authors have proposed that K. pneumoniae is devoid of the luxI homologues for synthesis of AHLs (Subramoni and Venturi, 2009; Pacheco et al., 2021). However, other researchers have reported the production of AHLs in K. pneumoniae strains (Wang et al., 2006; Ngeow et al., 2013; Hosny and Fadel, 2021). With regard to SdiA, the only study found in the literature conducted by Pacheco et al. (2021) proposed that SdiA functions as a repressor of biofilm formation and fimbriae expression in K. pneumoniae. To conduct the study, the authors employed a sdiA transposon-based insertion mutant of the K. pneumoniae ATCC 10031 collection strain and examined the impact of N-octanoyl-L-homoserine lactone (C8-HSL) as an exogenous AHL on a microtiter-based biofilm formation cultivation model. In this study, the authors observed that the biofilm-repressing effect of the AHL was sdiA-dependent. However, a preliminary work carried out in our laboratory with several clinical strains about the effect of different AHLs on biofilm formation revealed no effect of C8-HSL and high variability on strain response (unpublished results). Therefore, a deeper understanding is needed in order to develop new anti-virulence strategies.
The aim of this study is to deeper our knowledge about the function of SdiA and AHL supplementation in virulence-related traits and biofilm formation of K. pneumoniae. The most active AHL was selected for evaluation of virulence phenotypes, including biofilm formation, capsular synthesis, serum resistance, and phage sensitivity, plus virulence assessment in vivo in Galleria mellonella. The K. pneumoniae strain selected for this study was KLEB-33, a multiresistant, hypermucoviscous and hyperbiofilm-forming clinical strain (Smith and Ahmer, 2003) that harbours several virulence genes that are characteristic of hypervirulent K. pneumoniae strains (Russo and Marr, 2019). The genetic and phenotypic characteristics of KLEB-33 render it an optimal model for the study of the emerging convergent K. pneumoniae strains, as the majority of the QS studies to date have been performed in collection strains. For comparative purposes, the aforementioned phenotypes were also studied in a non-virulent, low biofilm forming, and non-MDR ATCC 13883T strain. The results of this study demonstrate that C6-HSL is the most effective AHLs on promoting biofilm formation. Additionally, C6-HSL and SdiA exert a dual influence on virulence phenotypes, operating both independently and hierarchically. The experiments conducted have facilitated a more profound comprehension of the QS mechanisms in K. pneumoniae.
2 Materials and methods
2.1 Bacterial strains and culture conditions
This study employed the K. pneumoniae ATCC 13883T and KLEB-33 strains. KLEB-33 is a MDR hyper-biofilm-forming clinical strain harbouring hypervirulence genes used as convergent-model strain (Silva-Bea et al., 2024a). This strain was obtained from a previous study (Silva-Bea et al., 2024a) approved by the Institutional Ethics Committee (CEImPA 03/2018). The present study did not require to be reviewed or approved by an ethics committee. ATCC 13883T is a low-biofilm forming non-MDR nor hypervirulent collection strain used for comparative purposes. The strains were routinely grown at 37 °C/200 rpm on 5 mL Lysogeny broth (LB) or LB agar (1.5% w/v). LB broth was supplemented with glucose 0.4% when required. Antibiotics were added when required, and synthetic AHL signals dissolved in acetonitrile at a concentration of 10 mg/mL, and comprising acyl chains with a carbon length of 4 to 18, including the oxo- substituted AHLs oxo-C4-HSL and oxo-C6-HSL, were added at a final concentration of 5, 2 or 0.2 µM, as required. An equal amount of solvent was added to the control cultures in all experiments involving AHL addition.
2.2 Construction of sdiA mutants
The sdiA gene was deleted from the ATCC 13883T and KLEB-33 strains using the CRISPR/Cas9-based system with the pCas9KP-Apr and pSGKP-Km plasmids, as previously described (Wang et al., 2018). The sequences of the single-guide RNA (sgRNA) spacer, the single-stranded DNA (ssDNA) sequences employed for allelic knockout, and the primers used for mutation confirmation are shown in Supplementary Table S1. Completely removal of the gene was confirmed by Sanger sequencing.
Planktonic growth was assessed in 15 mL LB cultures, plus the effect of AHL addition. Briefly, cultures inoculated at an initial absorbance at 600 nm of 0.01 (Abs600 nm) were incubated at 200 rpm, 37 °C/24 h, with growth measured at 1, 2, 4, 6, 8, 10 and 24 h, in triplicate. The lineal relationship between Colony Forming Units (CFUs) and Abs600 nm was confirmed using the Miles and Misra method (Miles et al., 1938), with the experiments being repeated twice.
2.3 Biofilm cultivation
K. pneumoniae KLEB-33 biofilms were cultivated in LB or LB+Glucose (0.4%) using the active attachment (AA) method as previously described (Silva-Bea et al., 2024b). Briefly, biofilms were grown for 24 h/37 °C in 12-well plates (VWR, 734-2778) in aerobiosis using a custom-made aluminium lid with glass coverslips (18x18 mm) attached as a substrate. Bacteria were inoculated at final Abs600 nm of 0.05. The culture media and treatment were replaced at 12 h to facilitate the growth of adherent cells. The Rolling Biofilm Bioreactor (RBB) cultivation method (Romero et al., 2022) was also used to promote biofilm growth and maturation of KLEB-33 and ATCC 13883T strains of K. pneumoniae. In this system bacteria were inoculated at a final Abs600 nm of 0.01 and incubated at 37 °C/72 h, with media and treatment changes every 24 h. The biofilm biomass was quantified by staining with crystal violet (CV) (0.04%) and measuring the absorbance at Abs590 nm after washing the coverslips with 33% acetic acid (Exterkate et al., 2010).
RBB biofilms were stained with Syto9 (ThermoFisher S34854) and subsequently imaged by confocal laser scanning microscopy (CLSM) (Leica Stellaris 8) to quantify biofilm height and coverage at 24, 48 and 72 h. Furthermore, 24 h biofilms were also examined for bacterial filamentation. To examine the composition and structure of biofilms, 48 h samples were also stained with YOYO™-1 iodide (ThermoFisher Y3601), Concanavalin A conjugated with Alexa Fluor® 594 (ThermoFisher C11253) and lipophilic FM™ 4-64 (ThermoFisher, F34653) fluorescent dyes to stain biofilm extracellular DNA (eDNA), extracellular polysaccharides, and cell membranes, respectively. A total of six fields were collected per sample. Images were subsequently analysed using ImageJ (v1.54) and Leica Application Suite X Office (v1.4.6.28433).
2.4 Quorum quenching activity
Quorum quenching (QQ) activity was evaluated in accordance with the methodology previously described (Parga et al., 2023). Briefly, 500 µL of the filtrated (0.22 µm) supernatant and pellet (resuspended in PBS pH 6.5) portions of 15 mL 24 h LB culture samples were exposed to C6-HSL (10 µM) for 6, 12, 24 and 48 h. pH of samples was adjusted when necessary to 6.5 to prevent the spontaneous opening of the lactone ring. PBS pH 6.5 with C6-HSL (10 µM) was used as negative control. After incubation, 100 µL of each sample was added to wells prepared in soft LB agar plates (0.8%) with the biosensor Chromobacterium subtsugae CV026, and incubated at 30 °C/24 h. Pellet samples were filtrated to avoid contamination of biosensor. Absence of production of violacein by the biosensor was indicative of positive QQ activity. Biosensor was routinely grown in LB broth supplemented with kanamycin (25 µg/mL).
2.5 Percoll density gradient centrifugation and capsule staining
Percoll density gradient centrifugation was employed to quantify strain capsule expression, in accordance with the methodology described (Dorman et al., 2018). Bacteria were adjusted to Abs590 nm = 1, collected from overnight cultures by centrifugation and resuspended in 2 mL PBS. The bacterial suspension was added to the top of a Percoll density gradient comprising 80, 60, 40 and 20% solutions in PBS to separate the bacterial fractions after centrifugation (2600 g) at 4 °C/20 min (9x acceleration; 1x deceleration). The distance between the bottom of the tube and the cell layer was measured. Capsule staining was also conducted using the Maneval method (Hughes and Smith, 2007).
2.6 Human serum sensitivity
The susceptibility to human serum was assessed as previously described (Dorman et al., 2018; Lv et al., 2022). Briefly, the bacterial inoculum was adjusted to an Abs600 nm of 1 in PBS, and 100 µL of bacterial suspension was added to 200 µL of pre-warmed (37 °C) human serum (Merk, S7023). Mixture was incubated at 37 °C/2 h. Colony-forming units per millilitre (CFU/mL) were determined on LB agar plates.
2.7 Phage susceptibility
Phage susceptibility was evaluated using the specific lytic phage Webervirus kpv33d1 (Sonia Rey et al., unpublished) as previously described (Xie et al., 2018). Briefly, the bacterial inoculum was grown in LB to an Abs600 nm of 0.5, diluted 1/5, and 10 µL were added to each well containing 90 µL of pre-diluted phage, resulting in a final Abs600 nm of 0.01 (approximately 106 CFU/mL). Phage at 1010 Plate Forming Units (PFU)/mL was serially diluted 1/10 to up to 9 times in 90 µL LB in 96-well U-bottom plates, from a Multiplicity Of Infection (MOI) of 103 to 10-5. A gas-permeable membrane (Breathe-Easy®, Z380059) was applied, and the plate was incubated at 37 °C/24 h, with Abs600 nm readings of the cultures every 30 min.
2.8 Galleria mellonella infection model
The virulence was evaluated using the G. mellonella survival assay, as previously described (Insua et al., 2013; Gato et al., 2020). Briefly, 15 larvae (300 - 400 mg body weight) were injected with 10 μL of a suspension containing 103, 105, or 107 CFUs in PBS. Larvae injected with an equal volume of sterile PBS or PBS plus C6-HSL (5 µM) were used as controls. Larvae were incubated at 37 °C in the dark and mortality was monitored every 24 h for up to 3 days.
2.9 Statistical analysis
Statistical analyses were conducted using GraphPad Prism 8.3.0. First, a Shapiro-Wilk test was used to ascertain whether the data exhibited a normal distribution. If normal, an analysis of variance (ANOVA) or a Student’s t-test was conducted. Alternatively, a Kruskal-Wallis or a Mann-Whitney test was performed, depending on whether there were more than two groups or only two groups, respectively. The significance values indicated by asterisks in the graphs presented in this paper are as follows: * = p<0.05; ** = p<0.005; *** = p<0.0005; and **** = p<0.00005.
3 Results and discussion
3.1 C6-HSL exhibited the greatest effect in promoting biofilm formation in AA cultivation system
Several reports indicate that SdiA can be activated by different AHLs in E. coli and Salmonella (Michael et al., 2001; Ahmer, 2004; Janssens et al., 2007; Panchal et al., 2024). It is therefore necessary to perform a screening of different AHLs in a robust and repeatable biofilm cultivation method to elucidate the function of SdiA in biofilm formation following AHL addition in K. pneumoniae. To this end, the effect of three short-chain homoserine lactones (C4-HSL, C6-HSL and C8-HSL), two oxo- substituted short-chain homoserine lactones (oxo-C4-HSL and oxo-C6-HSL) and five long-chain homoserine lactones (C10-HSL, C12-HSL, C14-HSL, C16-HSL and C18-HSL) on the AA biofilm cultivation system in the hyper-biofilm-forming strain KLEB-33 was investigated. The AA system has been already demonstrated to be a reliable and repeatable system for biofilm studies in this strain (Silva-Bea et al., 2024b).
The results showed that in the clinical strain KLEB-33 the AHLs C4-HSL, oxo-C4 and C6-HSL at 5 µM exhibited a substantial impact on biofilm formation in the absence of glucose (Figure 1A; Supplementary Figure S1A), with C12-HSL and C14-HSL also exhibiting a significant effect, albeit with a considerably lower magnitude than that observed for the short-chain AHLs (Figure 1B). In the presence of glucose 0.4%, oxo-C6-HSL also had a significant effect on this phenotype (Supplementary Figure S1A). However, none of the tested AHLs exhibited an effect on biofilm formation at 2 µM (Supplementary Figure S1B), although it should be noted that other phenotypes further examined in this study could respond to this lower AHL concentration. The influence of AHL provision on ATCC 13883T strain was also assessed in the AA cultivation system (Supplementary Figure S1C). However, its inherent low biofilm-forming capacity hindered the detection of significant differences in this biofilm model system. Following these results, C6-HSL in LB without glucose was selected for the subsequent experiments, as it exhibited the most pronounced contrast in comparison with the control cultures.
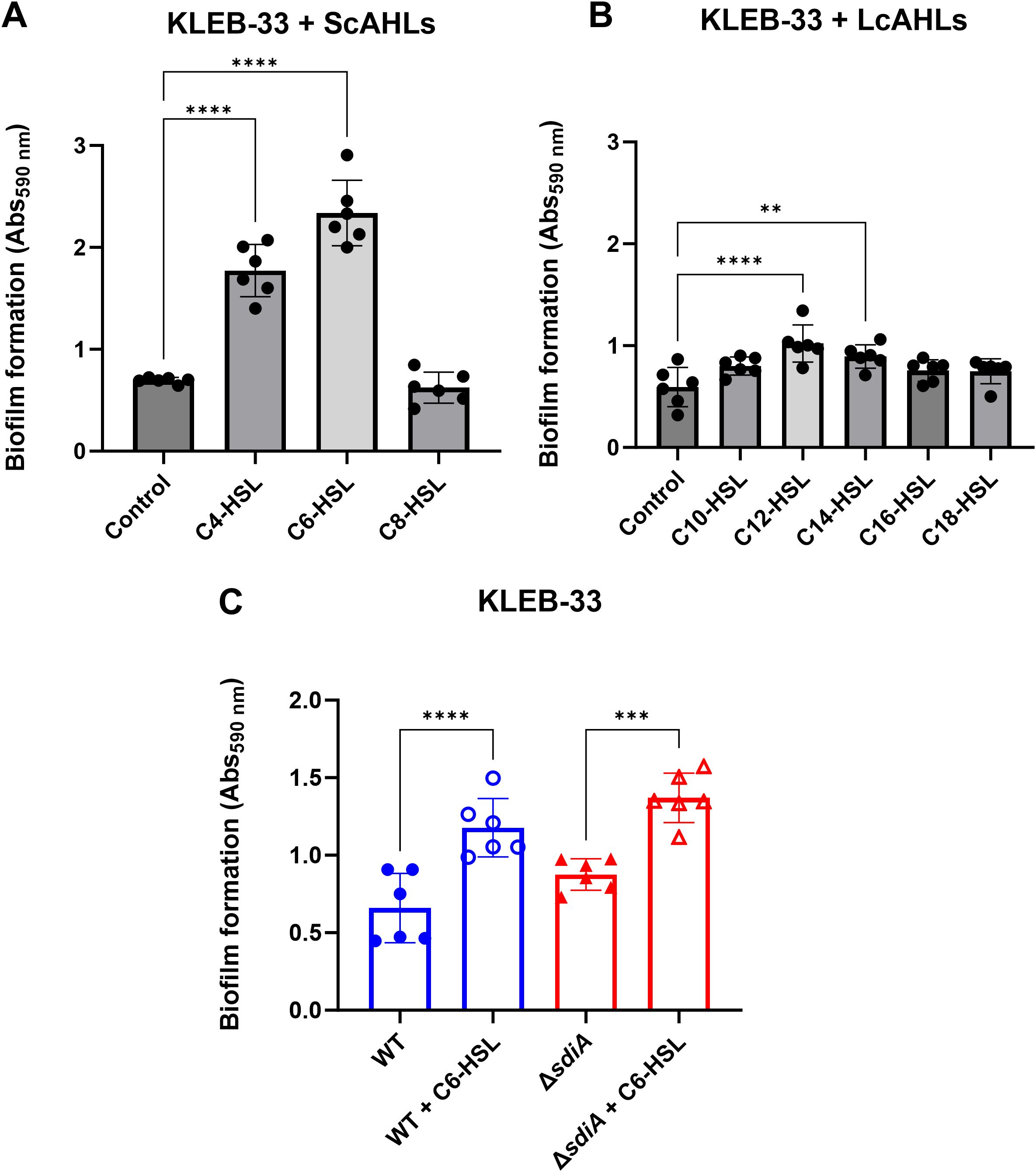
Figure 1. Effect of AHL addition (5 µM) on biofilm formation by K. pneumoniae KLEB-33 on glass coverslips in the Active Attachment (AA) biofilm model. The quantification of biofilm formation was conducted using CV staining, which was subsequently dissolved with 33% acetic acid and the absorbance measured at 590 nm (Abs590 nm). No effect was observed at 2 µM in previous experiments (Supplementary Figure S1). The impact of each AHL individually at 5 µM was examined for short-chain AHLs (ScAHLs) (A) and long-chain AHLs (LcAHLs) (B) in comparison to the solvent control. The repeatability of the C6-HSL effect was validated in subsequent experiments conducted on wild-type and sdiA-deficient KLEB-33 strains (C). All the experiments were conducted in duplicate (N = 3).
In contrast with observations made by Pacheco et al. (2021), our findings did not indicate a strong promoting effect on biofilm formation of C8-HSL. This suggests a high variability on the response to AHLs among different strains. Given the significant biofilm-promoting impact of C6-HSL observed in our experiments, we sought to ascertain whether this effect could be attributed to its direct interaction with SdiA. To this end, a ΔsdiA strain was constructed in KLEB-33 and was cultivated under C6-HSL supplementation. A significant increase in biofilm formation was also observed in the sdiA-lacking strain in response to C6-HSL addition (Figure 1C), indicating that the biofilm-promoting effect of C6-HSL was independent of sdiA. Moreover, despite SdiA being described in the literature as a biofilm repressor (Pacheco et al., 2021; Mayer et al., 2023), in the AA biofilm cultivation system the experiments revealed only a slight, non-significant increase in biofilm formation on the ΔsdiA in comparison to its wild-type. Growth was monitored with/without AHL supplementation in shaken cultures to ascertain that sdiA deficiency and/or C6-HSL addition does not affect growth (Supplementary Figure S2).
It is noteworthy that the concentration of C6-HSL at which a significant impact on biofilm formation was observed is higher than the physiological AHL concentrations typically encountered in QS signalling species (Milton et al., 2001). Nevertheless, similar concentrations in the micromolar range have been used to elicit a biological effect in K. pneumoniae (Høyland-Kroghsbo et al., 2013; Gopu et al., 2016; Pacheco et al., 2021) and other bacterial species (Michael et al., 2001; Dyszel et al., 2010b) for the study of the role of SdiA. In the case of K. pneumoniae, the presence of AHL-degrading enzymes has previously been described (Chan, 2013), and the presence of QQ activity against C6-HSL was corroborated in the culture media of the strains used in this study (Supplementary Table S2), therefore, the high concentrations required to observe a phenotypic response could be also due to the partial inactivation of the AHLs. Nevertheless, and due to the high AHL concentration required to elicit a response, we cannot fully disregard the possibility that these molecules are acting through non-specific mechanisms, for example, by interacting with the cellular membranes, as reported previously for long-chain, oxo-substituted AHLs (Davis et al., 2010; Gahan et al., 2021).
3.2 The formation and maturation of K. pneumoniae biofilms are influenced by SdiA and C6-HSL in an opposing and hierarchically organised manner
In order to evaluate the impact of sdiA mutation and C6-HSL supplementation on biofilm structure and maturation, we also employed the rolling biofilm bioreactor (RBB) system (Romero et al., 2022). This system permits the cultivation of biofilms over extended periods and the generation of highly matured biofilms with high reproducibility, even in strains with low biofilm-forming capabilities, such as ATCC 13883T. Even though little differences were observed in the early stages of biofilm formation, the results obtained with the RBB system demonstrated that the KLEB-33 ΔsdiA strain exhibited a higher biofilm maturation in comparison to the wild-type strain. This was evidenced by the earlier formation of mushroom-like structures in the ΔsdiA strain after 48 hours of incubation and a higher biofilm biomass (Figure 2). Our observations in the RBB cultivation system are in accordance with a role of SdiA as a biofilm-repressor, as previously described in the literature (Sabag-Daigle et al., 2015; Pacheco et al., 2021; Mayer et al., 2023; Schwieters and Ahmer, 2024). Nevertheless, no notable differences were observed between the wild-type and ΔsdiA strains at 24 hours of incubation, as happened in the AA cultivation system, as the 24-hour biofilms had not yet reached a stage of development sufficient to manifest such differences. Indeed, comparable levels of biofilm formation were recorded in the AA and RBB cultivation systems for both strains following a 24-hour incubation period (Figures 1, 2). On the contrary, no significant increase in biofilm formation was observed in the ATCC 13883T ΔsdiA strain. This finding may again be attributed to the inherent lower biofilm formation ability of the strain.
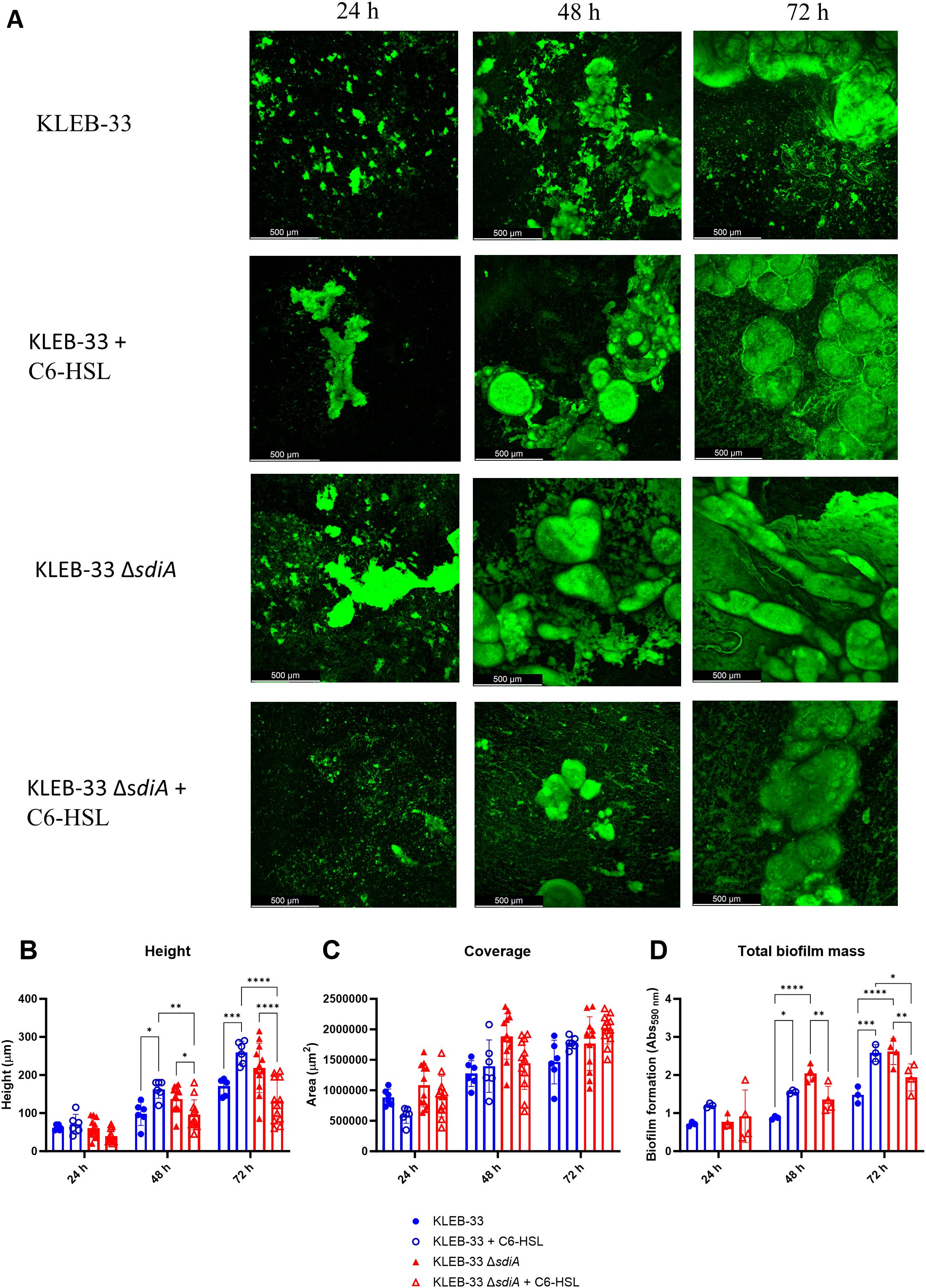
Figure 2. Impact of sdiA mutation and AHL addition (5 µM) on biofilm formation in K. pneumoniae KLEB-33. (A) Representative CLSM images (N = 3) of biofilms obtained after 24, 48 and 72 hours of incubation in the RBB cultivation system and staining with Syto9 fluorescent dye. (B, C) Quantification of height and biofilm coverage from confocal images using ImageJ (v1.54) image analysis software. (D) Crystal Violet quantification (N = 3) of biofilm biomass of wild-type and ΔsdiA K. pneumoniae KLEB-33 strains after treatment with C6-HSL (5 µM) or the solvent control (acetonitrile). The experiment was repeated twice.
The addition of C6-HSL (5 μM) also promoted the maturation of the biofilm in the KLEB-33 and ATCC 13883T wild-type strains after 48 hours of incubation (Figure 2; Supplementary Figure S3). However, in the roller biofilm bioreactor no such promotion effect was observed when AHL was added to the ΔsdiA for both strains studied, and these observations were corroborated by the quantification of the total biofilm biomass and thickness (Supplementary Figure S3, Figure 2). These results show that in the wild-type strain, SdiA functions as a repressor of genes involved in biofilm maturation, as previously reported (Pacheco et al., 2021; Mayer et al., 2023). However, this effect is negated in the presence of AHL, resulting in a biofilm maturation-promoting effect only when sdiA is present. The observed phenotype is only evident in the absence of sdiA, suggesting a hierarchical regulatory relationship between SdiA and C6-HSL.
Additionally, a significant increase in cell filamentation was observed in biofilms of the KLEB-33 ΔsdiA, a finding that aligns with previous observations (Pacheco et al., 2021) (Figure 3; Supplementary Figure S4). The formation of filamented bacteria has been documented in the literature as a mechanism that contributes to the persistence and colonisation of surfaces (Abell-King et al., 2022). Therefore, the elevated filamentation rates observed in the ΔsdiA strain are consistent with its enhanced capacity for biofilm formation (Figure 2). However, no differences were observed following C6-HSL supplementation, indicating that cell filamentation is dependent on SdiA, yet not triggered by AHL signalling. The AHL-independent effect of sdiA has been reported before (Sabag-Daigle et al., 2015; Dyszel et al., 2010b; Schwieters and Ahmer, 2024).
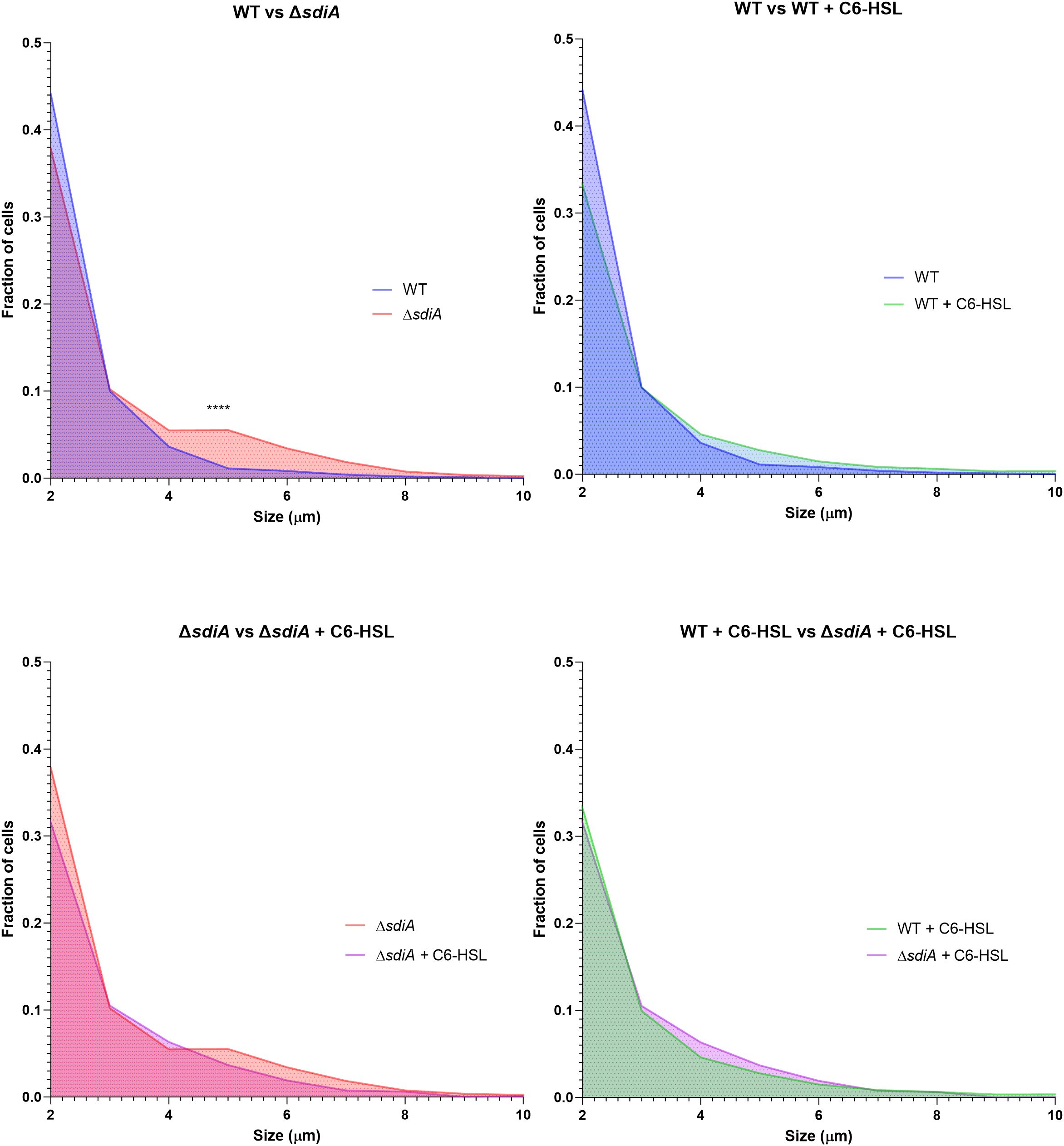
Figure 3. Distribution of cell size fractions in biofilms of K. pneumoniae KLEB-33 wild-type (WT) and ΔsdiA strains cultivated in the RBB system for 24 hours with supplementation of C6-HSL (5 µM) or the solvent control (acetonitrile). Cell sizes were measured using the ImageJ software (version 1.54). A total of 12 images were analysed per condition and sample (N = 3). The experiment was repeated twice.
In light of the observed alterations in biofilm maturation and structure following sdiA mutation and AHL supplementation, we sought to investigate whether sdiA deficiency or AHL signalling could influence the biofilm matrix composition in K. pneumoniae, as previously described for other pathogens (Das and Manefield, 2012; Yang and Lan, 2016). To this end, 48-hour RBB biofilms of KLEB-33 were fluorescently stained with YOYO-1 (eDNA), FM 4-64 (cell membranes) and Concanavalin A conjugated with AlexaFluor594 (ConA, extracellular polysaccharides). The results demonstrated a significant increase in fluorescence intensity for both the YOYO-1 and FM 4-64 signals (Figure 4) in the same conditions where enhanced biofilm formation was recorded (Figure 2: WT + C6-HSL and ΔsdiA). However, the ConA staining did not yield a strong fluorescence signal, which could suggest that eDNA may be the principal component of the biofilm matrix (Supplementary Figure S5). The function of eDNA as a crucial component of the biofilm matrix, facilitating the development of biofilms through the formation of adhesive “webs” (Supplementary Figure S5) that enhance cell cohesion and adhesion, has been previously postulated in a variety of bacteria (Mann et al., 2009; Campoccia et al., 2021; Romero et al., 2022). However, no clear relationship could be identified between SdiA or AHL addition and eDNA. This suggests that the regulation of this matrix component is not dependent on these regulatory systems. Moreover, and although other researchers have reported elevated polysaccharide production in a sdiA-deficient C. sakazakii strain (Cao et al., 2022), no changes in exopolysaccharide abundance were observed in CLSM biofilms images (Figure 4).
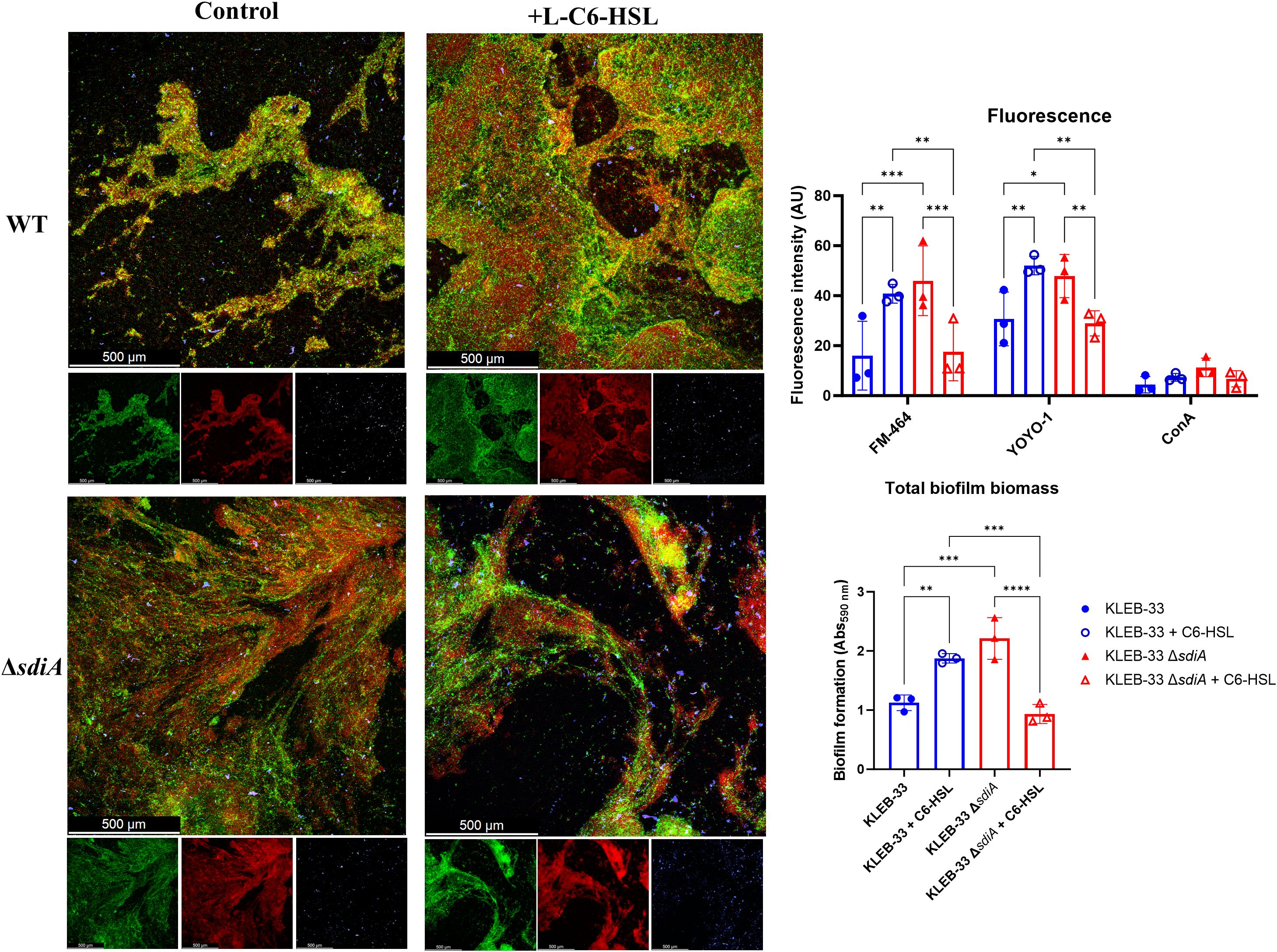
Figure 4. Representative CLSM (Leica Stellaris 8) images of RBB 48 hours biofilms of WT and ΔsdiA K. pneumoniae KLEB-33 treated with C6-HSL (5 µM) or the solvent control (acetonitrile). Biofilms were stained with YOYO-1 (eDNA, green), FM-464 (cell membranes, red), and Concanavalin A conjugated with AlexaFluor594 (exopolysaccharide, blue). A total of three fields were recorded in CLSM for each biofilm sample. Repeatability of the experiments was corroborated with total biomass quantification (N = 3) with CV staining (Abs590nm). Quantification of fluorescence intensity (AU: arbitrary units) was performed with ImageJ (v1.54).
3.3 QS-induced changes in biofilm formation have no correlation with capsule production in K. pneumoniae
To further examine the mechanism underlying the alterations in biofilm formation observed, we postulate that the supplementation of C6-HSL or the mutation of sdiA (which is associated with high biofilm formation) may be related to a reduction in capsule production. This is based on the findings of other researchers who have demonstrated that high-capsule-producing bacteria are more likely to be low biofilm formers, as the capsule polysaccharides have been shown to interfere with adhesion (Nunez et al., 2023). Consequently, a semi-quantitative analysis of the capsule production was conducted using Percoll density gradient centrifugation. This method enables the macroscopic differentiation of high and low capsule-producing bacteria based on their flotation characteristics (Dorman et al., 2018). Additionally, the capsules of the strains under study were subjected to staining using the Maneval’s method (Hughes and Smith, 2007). The results revealed differences between KLEB-33 and ATCC 13883T strains, as a thicker band (Figure 5) and capsule (Supplementary Figure S6) was observed in the collection strain. However, no appreciable differences in capsule production between the wild-type and the ΔsdiA for both strains studied were observed (Supplementary Figure S6; Figure 5). Regarding the addition of C6-HSL, a significant increase in capsule production was recorded in the wild-type strain of ATCC 13883T, whereas no effect was observed in KLEB-33 floatability (Figure 5), which is likely attributable to its relatively lower capsule production compared to the ATCC 13883T strain (Supplementary Figure S6, Figure 5). In fact, the low capsule production of KLEB-33 is consistent with its higher biofilm formation compared with ATCC 13883T strain, a low biofilm-forming and high capsule-producing strain (Silva-Bea et al., 2024b). However, it seems that the C6-HSL induced capsule production does not affect biofilm formation, as we observed higher biofilm formation with AHL supplementation.
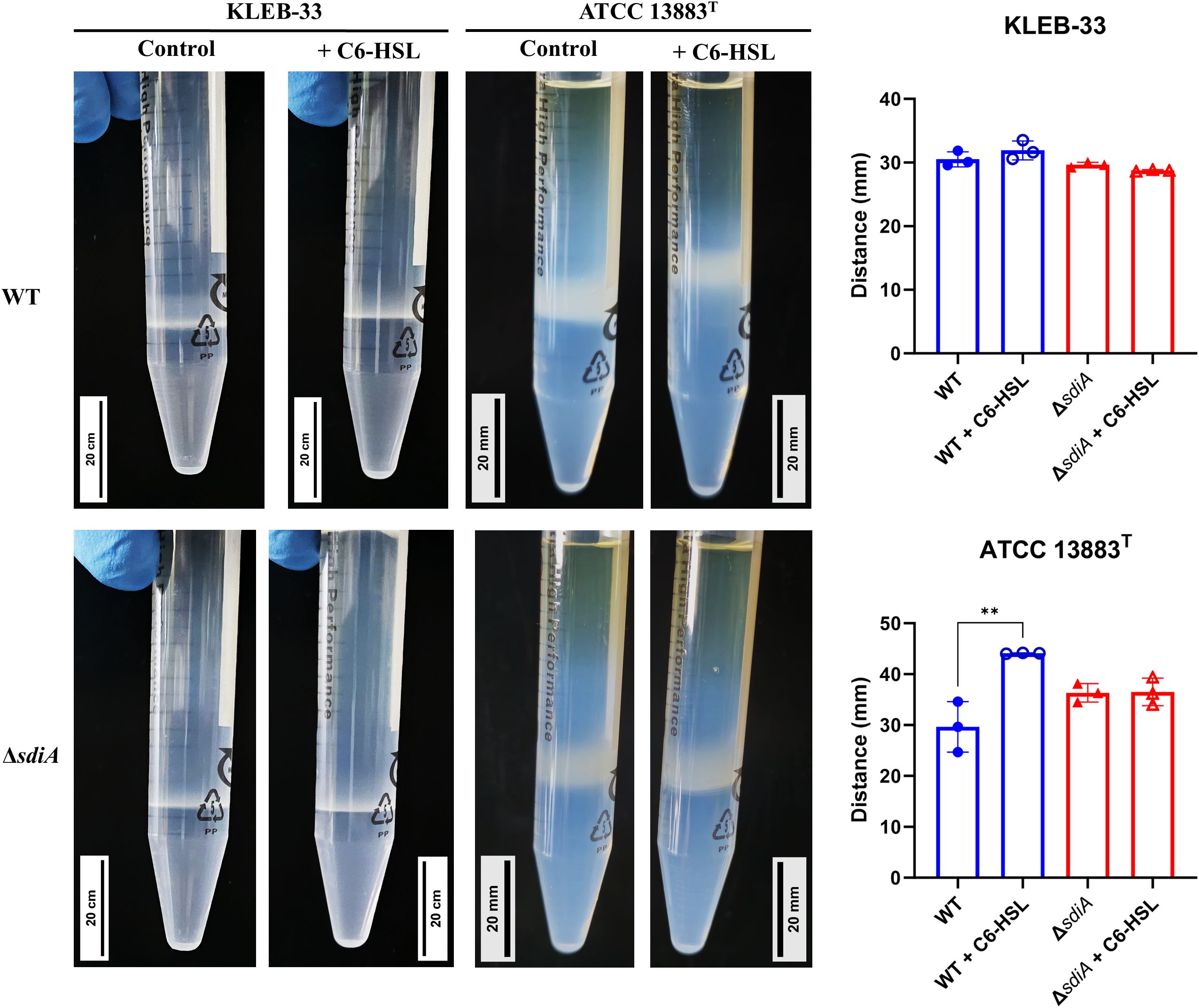
Figure 5. Percoll density gradient analysis conducted on K. pneumoniae KLEB-33 and ATCC 13883T wild-type (WT) and ΔsdiA strains with and without the addition of AHL (5 µM). Representative images are presented in left side, while histograms of the obtained measurements for bacterial cell layer height (N = 3) are shown in the right side. Experiment was repeated twice. An equal amount of solvent was added to the control cultures.
3.4 Serum complement killing in K. pneumoniae is inversely affected by SdiA and AHL supplementation
Although the Percoll method and Maneval’s staining did not reveal macroscopic differences in capsule production following sdiA mutation, we sought to identify a phenotype that could depend on capsule production and be sufficiently sensitive to detect differences in capsule biosynthesis. To this end, we conducted human serum survival assays as this method is employed by some authors as an indirect means of assessing capsule production (Lv et al., 2022). This is because capsule biosynthesis has been associated with serum and complement-killing resistance (González- et al., 2021). The results of our experiments showed that the sdiA knockout strains exhibited significantly elevated serum sensitivity in both strains under study (Figure 6), thereby indicating that SdiA plays a role in this process.
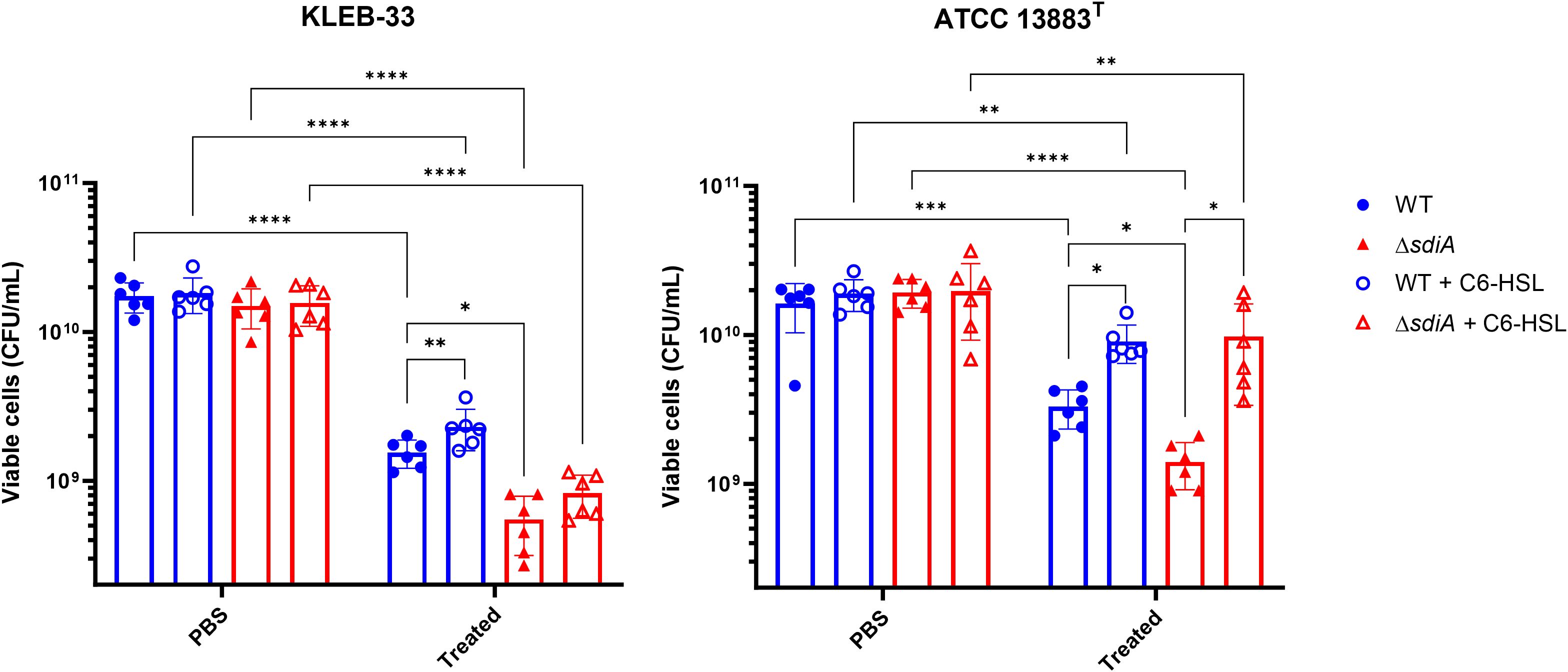
Figure 6. Serum resistance assays of K. pneumoniae KLEB-33 (left) and ATCC 13883T (right) wild-type (WT) and ΔsdiA strains following exposure to human serum for 2 hours, plus the effect of C6-HSL supplementation (5 µM). The control samples represent non-serum-treated bacteria in PBS. The experiment was repeated twice with N = 3. An equal amount of solvent was added to the control cultures.
The reduced serum resistance exhibited by the ΔsdiA strains may be linked to modifications in capsule production, which could also account for the increased biofilm formation. In fact, KLEB-33 has shown higher serum sensitivity than ATCC 13883T strain (Figure 6), showing a correlation with low capsule production and high biofilm formation, as described in the literature (Lv et al., 2022; Nunez et al., 2023). However, an alternative hypothesis is that an SdiA deficiency in K. pneumoniae may lead to alterations in other components of the cell surface that affect serum resistance. For instance, a number of studies indicates that LPS upregulation contributes to higher serum and phage sensitivity (González- et al., 2021; Majkowska-Skrobek et al., 2021; Tang et al., 2023). Moreover, the diminished serum resistance observed in the ΔsdiA strains is consistent with the findings of previous studies conducted in Salmonella spp (Lindsay and Ahmer, 2005). Additionally, type-1 fimbriae were found to be overexpressed in a sdiA-deficient strain of K. pneumoniae (Pacheco et al., 2021), and its overexpression has been linked to detrimental effects on complement survival in E. coli (Huja et al., 2014). Conversely, the addition of C6-HSL increased serum resistance independently of the presence of SdiA (Figure 6). Therefore, our experiments further support that C6-HSL supplementation promotes capsule production, a factor related with complement killing reduction (Dorman et al., 2018), as evidenced by the observation of enhanced serum resistance and a heightened band in Percoll experiments in the ATCC 13883T strain.
3.5 SdiA deficiency increases phage sensitivity in K. pneumoniae
The quantification of capsule production and complement-killing resistance experiments suggested that SdiA may affect cell surface components. Furthermore, a study conducted in E. coli demonstrated that SdiA plays a role in bacteriophage sensitivity through an AHL-dependent mechanism (Ghosh et al., 2009). Accordingly, an experiment was conducted to investigate whether modifications of the cell surface resulting from QS signalling could influence the susceptibility to phage infection. The infectivity of Webervirus kpv33d1, a high lytic wastewater-derived phage isolated against KLEB-33 strain (Sonia-Rey et al., unpublished), was tested on the wild type and genetically modified strains lacking the sdiA gene with or without AHL supplementation. Different levels of phage multiplicity of infection (MOI) were used for KLEB-33 (0.00001 to 0.1) and ATCC 13883T (1 to 100) strains due to differences in phage specificity (Figure 7). The deletion of the sdiA gene resulted in higher phage susceptibility in comparison to the wild type strain in the presence of a phage MOI of 0.001 and 10 in the KLEB-33 and ATCC 13883T strains, respectively. The addition of C6-HSL at concentrations of 5, 2 and 0.2 µM did not result in any observable effect on phage susceptibility in KLEB-33 (Supplementary Figure S7). These findings are consistent with those previously observed in E. coli (Ghosh et al., 2009) and also align with the results of serum resistance experiments, as evidenced by the heightened sensitivity of the ΔsdiA strains compared to the parental strains. The observation of higher serum and phage sensitivity of ΔsdiA may be also related to the higher filamentation rates observed (Figure 3), with higher cell surface being exposed to complement killing and phage attachment. However, the fact that C6-HSL addition has no effect on phage infection provides further support for a regulatory role of SdiA independent of C6-HSL.
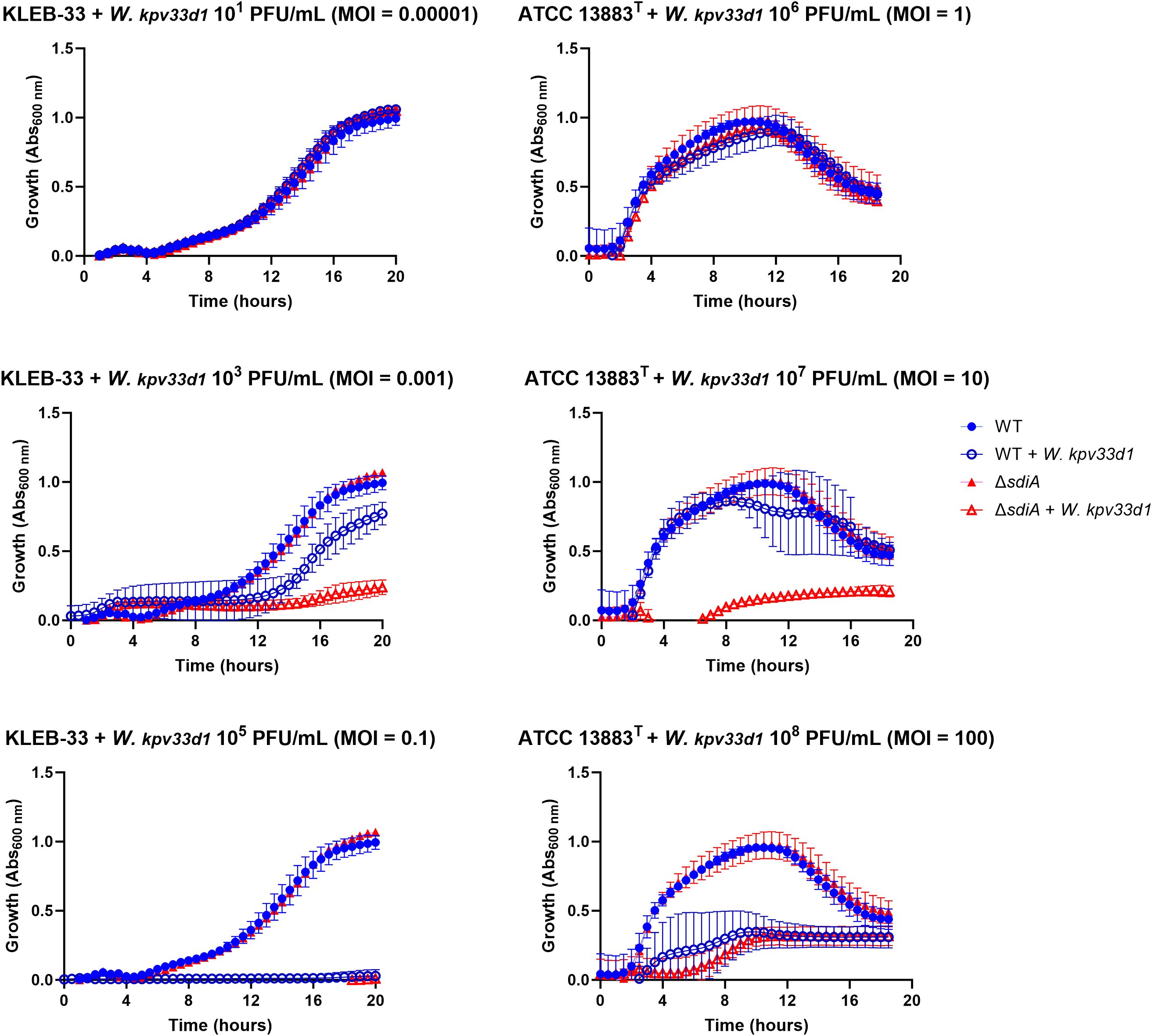
Figure 7. Susceptibility assays of K. pneumoniae KLEB-33 and ATCC 13883T wild-type (WT) and ΔsdiA strains (106 CFU/mL) to Webervirus kpv33d1 phage. A dose-response assay was conducted using 96-well microtiter plates and varying MOI values. Experiments were performed twice for each strain separately (N = 3). No effect of C6-HSL addition (5 µM) was observed (Supplementary Figure S7).
3.6 C6-HSL signalling increases Galleria mellonella survival after infection with sdiA-deficient K. pneumoniae
In order to test how the mutation of sdiA or C6-HSL supplementation could affect virulence in vivo, we examined whether strains lacking SdiA could exhibit higher mortality rates in the G. mellonella infection model. As a result, despite the mortality rates recorded daily being consistently higher in the mutant, no statistically significant differences were observed between the wild-type and ΔsdiA strains for both KLEB-33 and ATCC 13883T (Figure 8A). Regarding the addition of C6-HSL, a markedly reduced virulence was observed in the KLEB-33 ΔsdiA strain at a dose of 105 CFU in the presence of AHL in comparison with the absence of AHL supplementation (Figure 8B). However, C6-HSL had no effect on virulence in either of the wild-type strains. Again, these results seem to indicate a separate regulatory pathway for SdiA and AHLs. The effect of decreased virulence observed following AHL treatment of the sdiA mutant strain was not replicated in ATCC 13883T. This may be attributed to the higher mortality rates observed in G. mellonella infected with this strain in comparison with KLEB-33, which demonstrated significantly lower mortality rates despite being a hyper biofilm-forming strain harbouring additional virulence genes (Silva-Bea et al., 2024a). However, the G. mellonella model proved inadequate for differentiating between classic and hypervirulent K. pneumoniae strains, and thus may not be a reliable indicator of virulence in murine models or in humans in certain cases (Russo and MacDonald, 2020). Additionally, a recent report showed unexpectedly lower virulence in convergent K. pneumoniae strains (Kochan et al., 2023). The notable reduction in virulence observed in KLEB-33 ΔsdiA supplemented with C6-HSL is consistent with the lower biofilm formation observed in RBB biofilms. The biofilm formation levels of ΔsdiA supplemented with C6-HSL observed in RBB experiments are very similar to WT levels (Figure 2). However, the WT strain did not show low virulence in G. mellonella model. A study performed in Salmonella has already reported depletion of adherence ability to HeLa cells in the presence of C6-HSL only in the ΔsdiA (Askoura et al., 2021). Therefore, we speculate that C6-HSL may control the expression of virulence and biofilm-formation genes independently and in opposition to SdiA, following a hierarchical regulation. This would explain why the biofilm and virulence repressive effect of C6-HSL could only be observed when SdiA was absent. However, though potentially statistically significant with a higher number of animals, there is no data to suggest that the in vivo survival phenotypes are linked to any of the in vitro virulence assays tested.
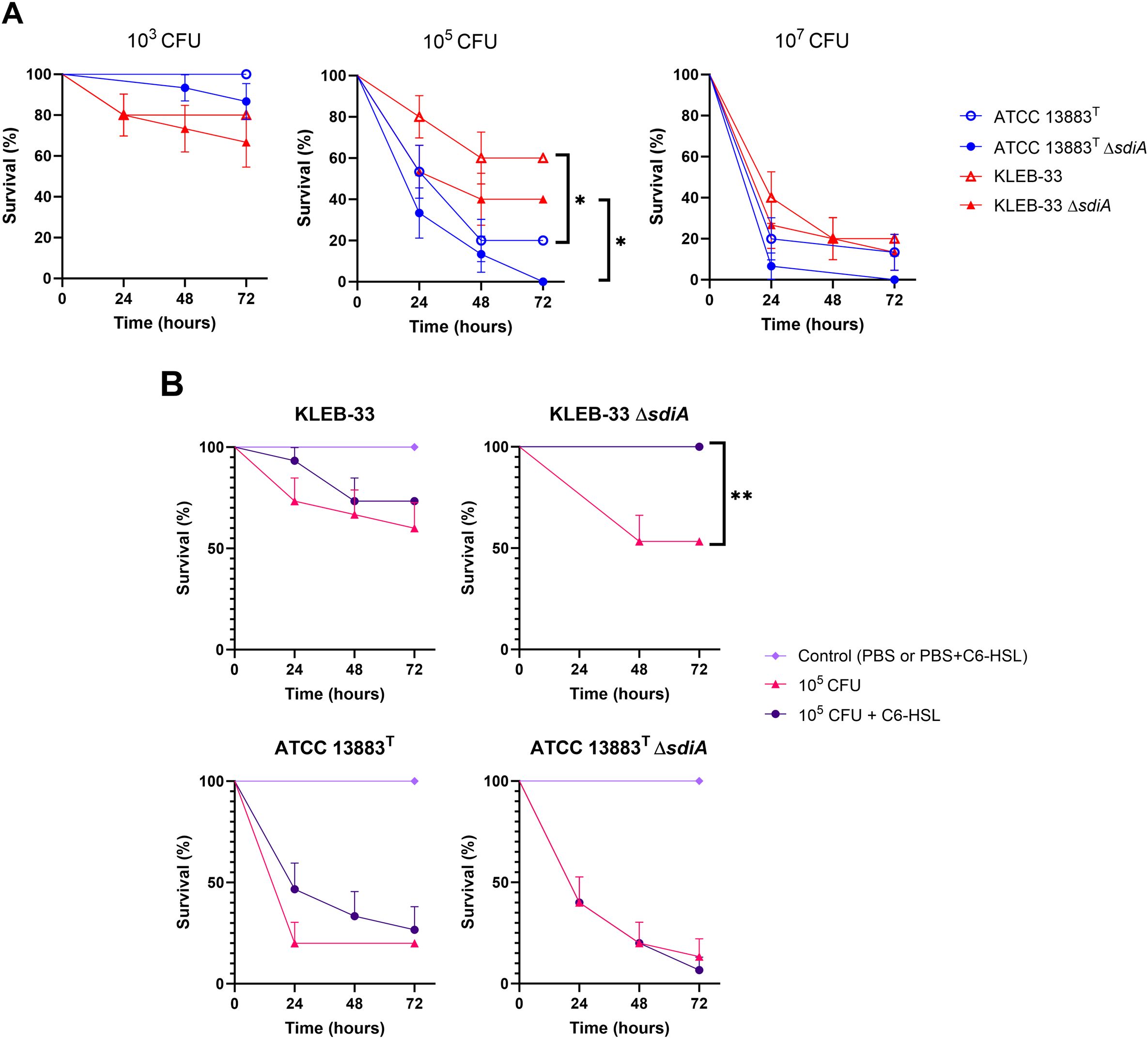
Figure 8. Survival analysis of Galleria mellonella following infection with K. pneumoniae KLEB-33 and ATCC 13883T wild-type and ΔsdiA strains. The survival of the insects was recorded at 24-hour intervals up to 72 hours. The experiments were repeated twice. A total of 5 larvae per condition and sample (N = 3) was used. The results of the comparison between the concentrations of the strains (A) and the effect of C6-HSL addition (5 µM) (B) are presented. An equal amount of solvent was added to the control cultures.
4 Conclusions
The aim of this study was to elucidate the function of SdiA in the virulence and biofilm formation of K. pneumoniae, which is postulated to be the receptor of AHLs. To this end, we firstly observed that C6-HSL is the most active AHL among 10 different signals. Following, a series of phenotypes were characterised in sdiA-lacking strains and after the addition of the C6-HSL, which was identified as a biofilm-promoting factor in this bacterium. Our findings appear to indicate that SdiA and C6-HSL influence several traits, with some exhibiting a joint effect, but the majority displaying independent regulation.
The results confirm that SdiA plays a role in repressing biofilm formation, and that its absence was linked to a reduction in resistance to human serum and phage infection, as well as a notable promotion of cell filamentation. However, no impact was observed on macroscopic capsule synthesis. Conversely, the exogenous addition of C6-HSL was found to promote capsule production in a sdiA-dependent manner in one of the strains studied. Moreover, C6-HSL was observed to enhance serum resistance independently of SdiA. Nevertheless, no impact on phage sensitivity was noted. Regarding biofilm formation, C6-HSL has a promoting effect when SdiA is present, but is decreased in its absence. This observation is consistent with the virulence data recorded in G. mellonella, which is reduced following the addition of C6-HSL in the absence of SdiA. Additionally, neither SdiA nor C6-HSL affects the composition of the biofilm matrix.
In view of these findings, it seems reasonable to conclude that C6-HSL is not the primary ligand of SdiA and that they act independently in the K. pneumoniae strains under consideration. Our results indicate that SdiA and C6-HSL are involved in different pathways in some cases, and their effects may even be counteracted by hierarchical regulation. This is corroborated by the observation that certain effects of C6-HSL could only be observed in the absence of SdiA. It is important to note that some of the results were dependent on the strain of K. pneumoniae studied, as not all the phenotypes observed were consistent in both strains. Our study provides new insights in QS regulation in this pathogen, even so more experiments are necessary to continue with the characterisation of their respective ways of action.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. PM: Data curation, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. AO: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. MR: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the projects and funds PID2019-104439RB-C21/AEI/10.13039/501100011033 (Agencia Estatal de Investigación, Spain); ED431B 2023/41 (Xunta de Galicia). Sergio Silva-Bea was supported by the “Formación de Profesorado Universitario” programme (FPU21/01147). Manuel Romero was supported by the Maria Zambrano Programme and Research Consolidation Grant (CNS2023-145299).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1562402/full#supplementary-material
Abbreviations
AA, Active Attachment; CV, Crystal Violet; MDR, Multi-Drug Resistance; AHL, N-acyl-homoserine lactone; HSL, homoserine lactone; C6-HSL, N-hexanoyl-L-homoserine lactone; QS, Quorum Sensing; RBB, Rolling-Biofilm Bioreactor; CLSM, Confocal-Laser Scanning Microscope; QQ, Quorum Quenching; CFUs, Colony Forming Units; MOI, Multiplicity of Infection.
References
Abell-King, C., Costas, A., Duggin, I. G., Söderström, B. (2022). Bacterial filamentation during urinary tract infections. PLoS Pathog. 18, e1010950. doi: 10.1371/journal.ppat.1010950
Ahmer, B. M. M. (2004). Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52, 933–945. doi: 10.1111/j.1365-2958.2004.04054.x
Ahmer, B. M. M., Van Reeuwijk, J., Timmers, C. D., Valentine, P. J., Heffron, F. (1998). Salmonella typhimurium encodes an sdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180, 1185–1193. doi: 10.1128/JB.180.5.1185-1193.1998
Askoura, M., Almalki, A. J., Lila, A. S. A., Almansour, K., Alshammari, F., Khafagy, E. S., et al. (2021). Alteration of Salmonella enterica Virulence and Host Pathogenesis through Targeting sdiA by Using the CRISPR-Cas9 System. Microorganisms 9, 2564. doi: 10.3390/microorganisms9122564
Campoccia, D., Montanaro, L., Arciola, C. R. (2021). Extracellular DNA (eDNA). A major ubiquitous element of the bacterial biofilm architecture. Int. J. Mol. Sci. 22, 9100. doi: 10.3390/ijms22169100
Cao, Y., Li, L., Zhang, Y., Liu, F., Xiao, X., Li, X., et al. (2022). Evaluation of Cronobacter sakazakii biofilm formation after sdiA knockout in different osmotic pressure conditions. Food Res. Int. 151, 110886. doi: 10.1016/j.foodres.2021.110886
Chan, K. G. (2013). Expression of Klebsiella sp. lactonase ahlK gene is growth-phase, cell-population density and N -acylhomoserine lactone independent. Front. Life Sci. 7, 132–139. doi: 10.1080/21553769.2013.833141
Das, T., Manefield, M. (2012). Pyocyanin promotes extracellular DNA release in pseudomonas aeruginosa. PLoS One 7, e46718. doi: 10.1371/journal.pone.0046718
Davis, B. M., Jensen, R., Williams, P., O’Shea, P. (2010). The interaction of N-acylhomoserine lactone quorum sensing signaling molecules with biological membranes: implications for inter-kingdom signaling. PLoS One 5, e13522. doi: 10.1371/journal.pone.0013522
Dorman, M. J., Feltwell, T., Goulding, D. A., Parkhill, J., Short, F. L. (2018). The Capsule Regulatory Network of Klebsiella pneumoniae Defined by density-TraDISort. mBio 9, e01863–e01818. doi: 10.1128/mBio.01863-18
Dyszel, J. L., Smith, J. N., Lucas, D. E., Soares, J. A., Swearingen, M. C., Vross, M. A., et al. (2010a). Salmonella enterica Serovar Typhimurium Can Detect Acyl Homoserine Lactone Production by Yersinia enterocolitica in Mice. J. Bacteriol. 192, 29–37. doi: 10.1128/JB.01139-09
Dyszel, J. L., Soares, J. A., Swearingen, M. C., Lindsay, A., Smith, J. N., Ahmer, B. M. M. (2010b). E. coli K-12 and EHEC Genes Regulated by sdiA. PLoS One 5, e8946. doi: 10.1371/journal.pone.0008946
Exterkate, R. A. M., Crielaard, W., Ten Cate, J. M. (2010). Different response to amine fluoride by Streptococcus mutans and polymicrobial biofilms in a novel high-throughput active attachment model. Caries Res. 44, 372–379. doi: 10.1159/000316541
Gahan, C. G., Patel, S. J., Chen, L. M., Manson, D. E., Ehmer, Z. J., Blackwell, H. E., et al. (2021). Bacterial quorum sensing signals promote large-scale remodeling of lipid membranes. Langmuir 37, 9120–9136. doi: 10.1021/acs.langmuir.1c01204
Gato, E., Vázquez-Ucha, J. C., Rumbo-Feal, S., Álvarez-Fraga, L., Vallejo, J. A., Martínez-Guitián, M., et al. (2020). Kpi, a chaperone-usher pili system associated with the worldwide-disseminated high-risk clone Klebsiella pneumoniae ST-15. Proc. Natl. Acad. Sci. 117, 17249–17259. doi: 10.1073/pnas.1921393117
Ghosh, D., Roy, K., Williamson, K. E., Srinivasiah, S., Wommack, K. E., Radosevich, M. (2009). Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75, 7142–7152. doi: 10.1128/AEM.00950-09
González--Ferrer, Peñaloza, H. F., Budnick, J. A., Bain, W. G., Nordstrom, H. R., Lee, J. S., et al. (2021). Finding order in the chaos: outstanding questions in Klebsiella pneumoniae pathogenesis. Infect. Immun. 89, e00693–e00620. doi: 10.1128/IAI.00693-20
Gopu, V., Meena, C. K., Murali, A., Shetty, P. H. (2016). Petunidin as a competitive inhibitor of acylated homoserine lactones in Klebsiella pneumoniae. RSC Adv. 6, 2592–2601. doi: 10.1039/C5RA20677D
Habyarimana, F., Sabag-Daigle, A., Ahmer, B. M. M. (2014). The sdiA-regulated gene srgE encodes a type III secreted effector. J. Bacteriol. 196, 2301–2312. doi: 10.1128/JB.01602-14
Hosny, R. A., Fadel, M. A. (2021). Detection of quorum sensing N-acyl-homoserine lactone molecules produced by different resistant Klebsiella pneumoniae isolates recovered from poultry and different environmental niches. Appl. Biochem. Biotechnol. 193, 3351–3370. doi: 10.1007/s12010-021-03605-w
Høyland-Kroghsbo, N. M., Mærkedahl, R. B., Svenningsen, S. L. A. (2013). Quorum-sensing-induced bacteriophage defense mechanism. mBio 4, e00362–e00312. doi: 10.1128/mBio.00362-12
Hughes, R. B., Smith, A. C. (2007). Capsule-Stain-Protocols (American Society for Microbiology). Available at: https://asm.org/ASM/media/Protocol-Images/Capsule-Stain-Protocols.pdf?ext=.pdf (Accessed February, 2024).
Huja, S., Oren, Y., Biran, D., Meyer, S., Dobrindt, U., Bernhard, J., et al. (2014). Fur is the master regulator of the extraintestinal pathogenic Escherichia coli response to serum. mBio 5, e01460–e01414. doi: 10.1128/mBio.01460-14
Insua, J. L., Llobet, E., Moranta, D., Pérez-Gutiérrez, C., Tomás, A., Garmendia, J., et al. (2013). Modeling Klebsiella pneumoniae Pathogenesis by Infection of the Wax Moth Galleria mellonella. Infect. Immun. 81, 3552–3565. doi: 10.1128/IAI.00391-13
Janssens, J. C. A., Metzger, K., Daniels, R., Ptacek, D., Verhoeven, T., Habel, L. W., et al. (2007). Synthesis of N -Acyl homoserine lactone analogues reveals strong activators of sdiA, the Salmonella enterica serovar typhimurium LuxR homologue. Appl. Environ. Microbiol. 73, 535–544. doi: 10.1128/AEM.01451-06
Kochan, T. J., Nozick, S. H., Valdes, A., Mitra, S. D., Cheung, B. H., Lebrun-Corbin, M., et al. (2023). Klebsiella pneumoniae clinical isolates with features of both multidrug-resistance and hypervirulence have unexpectedly low virulence. Nat. Commun. 14, 7962. doi: 10.1038/s41467-023-43802-1
Król, J. E., Hall, D. C., Balashov, S., Pastor, S., Sibert, J., McCaffrey, J., et al. (2019). Genome rearrangements induce biofilm formation in Escherichia coli C – an old model organism with a new application in biofilm research. BMC Genomics 20, 767. doi: 10.1186/s12864-019-6165-4
Lee, J., Maeda, T., Hong, S. H., Wood, T. K. (2009). Reconfiguring the Quorum-Sensing Regulator SdiA of Escherichia coli To Control Biofilm Formation via Indole and N -Acylhomoserine Lactones. Appl. Environ. Microbiol. 75, 1703–1716. doi: 10.1128/AEM.02081-08
Lindsay, A., Ahmer, B. M. M. (2005). Effect of sdiA on biosensors of N -acylhomoserine lactones. J. Bacteriol. 187, 5054–5058. doi: 10.1128/JB.187.14.5054-5058.2005
Lv, J., Zhu, J., Wang, T., Xie, X., Wang, T., Zhu, Z., et al. (2022). The role of the two-component QseBC signaling system in biofilm formation and virulence of hypervirulent Klebsiella pneumoniae ATCC43816. Front. Microbiol. 13, 817494. doi: 10.3389/fmicb.2022.817494
Majkowska-Skrobek, G., Markwitz, P., Sosnowska, E., Lood, C., Lavigne, R., Drulis-Kawa, Z. (2021). The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 23, 7723–7740. doi: 10.1111/1462-2920.15476
Mambu, J., Virlogeux-Payant, I., Holbert, S., Grépinet, O., Velge, P., Wiedemann, A. (2017). An updated view on the Rck invasin of Salmonella: still much to discover. Front. Cell Infect. Microbiol. 7, 500. doi: 10.3389/fcimb.2017.00500
Mann, E. E., Rice, K. C., Boles, B. R., Endres, J. L., Ranjit, D., Chandramohan, L., et al. (2009). Modulation of eDNA Release and Degradation Affects Staphylococcus aureus Biofilm Maturation. PLoS One 4, e5822. doi: 10.1371/journal.pone.0005822
Mayer, C., Borges, A., Flament-Simon, S. C., Simões, M. (2023). Quorum sensing architecture network in Escherichia coli virulence and pathogenesis. FEMS Microbiol. Rev. 47, fuad031. doi: 10.1093/femsre/fuad031
Michael, B., Smith, J. N., Swift, S., Heffron, F., Ahmer, B. M. M. (2001). SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001
Miles, A. A., Misra, S. S., Irwin, J. O. (1938). The estimation of the bactericidal power of the blood. Epidemiol. Infect. 38, 732–749. doi: 10.1017/S002217240001158X
Milton, D. L., Chalker, V. J., Kirke, D., Hardman, A., Cámara, M., Williams, P. (2001). The luxM homologue VanM from Vibrio Anguillarum directs the synthesis of N -(3-hydroxyhexanoyl)homoserine lactone and N -hexanoylhomoserine lactone. J. Bacteriol. 183, 3537–3547. doi: 10.1128/JB.183.12.3537-3547.2001
Ngeow, Y., Cheng, H., Chen, J., Yin, W. F., Chan, K. G. (2013). Short chain N-acylhomoserine lactone production by clinical multidrug resistant Klebsiella pneumoniae strain CSG20. Sensors 13, 15242–15251. doi: 10.3390/s131115242
Nunez, C., Kostoulias, X., Peleg, A. Y., Short, F., Qu, Y. (2023). A comprehensive comparison of biofilm formation and capsule production for bacterial survival on hospital surfaces. Biofilm 5, 100105. doi: 10.1016/j.bioflm.2023.100105
Pacheco, T., Gomes, A. É. I., Siqueira, N. M. G., Assoni, L., Darrieux, M., Venter, H., et al. (2021). SdiA, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front. Microbiol. 12, 597735. doi: 10.3389/fmicb.2021.597735
Panchal, J., Prajapati, J., Dabhi, M., Patel, A., Patel, S., Rawal, R., et al. (2024). Comprehensive computational investigation for ligand recognition and binding dynamics of sdiA: a degenerate LuxR -type receptor in Klebsiella pneumoniae. Mol. Divers. 28, 3897–3918. doi: 10.1007/s11030-023-10785-6
Papenfort, K., Bassler, B. L. (2016). Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Parga, A., Muras, A., Otero-Casal, P., Arredondo, A., Soler-Ollé, A., Àlvarez, G., et al. (2023). The quorum quenching enzyme Aii20J modifies in vitro periodontal biofilm formation. Front. Cell Infect. Microbiol. 13, 1118630. doi: 10.3389/fcimb.2023.1118630
Rahmati, S., Yang, S., Davidson, A. L., Zechiedrich, E. L. (2002). Control of the AcrAB multidrug efflux pump by quorum-sensing regulator sdiA. Mol. Microbiol. 43, 677–685. doi: 10.1046/j.1365-2958.2002.02773.x
Romero, M., Mayer, C., Heeb, S., Wattanavaekin, K., Cámara, M., Otero, A., et al. (2022). Mushroom-shaped structures formed in Acinetobacter baumannii biofilms grown in a roller bioreactor are associated with quorum sensing–dependent Csu-pilus assembly. Environ. Microbiol. 24, 4329–4339. doi: 10.1111/1462-2920.15985
Russo, T. A., MacDonald, U. (2020). The Galleria mellonella Infection Model Does Not Accurately Differentiate between Hypervirulent and Classical Klebsiella pneumoniae. mSphere 5, e00850–e00819. doi: 10.1128/mSphere.00850-19
Russo, T. A., Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32. doi: 10.1128/CMR.00001-19
Sabag-Daigle, A., Ahmer, B. M. M. (2012). ExpI and PhzI are descendants of the long lost cognate signal synthase for sdiA. PLoS One 7, e47720. doi: 10.1371/journal.pone.0047720
Sabag-Daigle, A., Dyszel, J. L., Gonzalez, J. F., Ali, M. M., Ahmer, B. M. M. (2015). Identification of sdiA-regulated genes in a mouse commensal strain of Enterobacter cloacae. Front. Cell Infect. Microbiol. 5. doi: 10.3389/fcimb.2015.00047
Sabag-Daigle, A., Soares, J. A., Smith, J. N., Elmasry, M. E., Ahmer, B. M. M. (2012). The Acyl Homoserine Lactone Receptor, sdiA, of Escherichia coli and Salmonella enterica Serovar Typhimurium Does Not Respond to Indole. Appl. Environ. Microbiol. 78, 5424–5431. doi: 10.1128/AEM.00046-12
Schwieters, A., Ahmer, B. M. M. (2024). Identification of new sdiA regulon members of Escherichia coli, Enterobacter cloacae, and Salmonella enterica serovars Typhimurium and Typhi. Microbiol. Spectr. 12, e01929–e01924. doi: 10.1128/spectrum.01929-24
Shankar, M., Ponraj, P., Illakkiam, D., Rajendhran, J., Gunasekaran, P. (2013). Inactivation of the Transcriptional Regulator-Encoding Gene sdiA Enhances Rice Root Colonization and Biofilm Formation in Enterobacter cloacae GS1. J. Bacteriol. 195, 39–45. doi: 10.1128/JB.01236-12
Silva-Bea, S., García-Meniño, I., Rey, S., Romero, M., Fernández, J., Hammerl, J. A., et al. (2024a). Draft genome sequence of Klebsiella pneumoniae KLEB-33: a convergent biofilm hyperforming multiresistant strain belonging to the emerging ST16 lineage harboring multiple hypervirulence genes. Microbiol. Resour Announc. 13, e01192–e01123. doi: 10.1128/mra.01192-23
Silva-Bea, S., Romero, M., Parga, A., Fernández, J., Mora, A., Otero, A. (2024b). Comparative analysis of multidrug-resistant Klebsiella pneumoniae strains of food and human origin reveals overlapping populations. Int. J. Food Microbiol. 413, 110605. doi: 10.1016/j.ijfoodmicro.2024.110605
Smith, J. N., Ahmer, B. M. M. (2003). Detection of other microbial species by Salmonella : expression of the sdiA regulon. J. Bacteriol. 185, 1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003
Smith, J. N., Dyszel, J. L., Soares, J. A., Ellermeier, C. D., Altier, C., Lawhon, S. D., et al. (2008). SdiA, an N-Acylhomoserine Lactone Receptor, Becomes Active during the Transit of Salmonella enterica through the Gastrointestinal Tract of Turtles. PLoS One 3, e2826. doi: 10.1371/journal.pone.0002826
Styles, M. J., Early, S. A., Tucholski, T., West, K. H. J., Ge, Y., Blackwell, H. E. (2020). Chemical control of quorum sensing in E. coli : identification of small molecule modulators of sdiA and mechanistic characterization of a covalent inhibitor. ACS Infect. Dis. 6, 3092–3103. doi: 10.1021/acsinfecdis.0c00654
Subramoni, S., Venturi, V. (2009). LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385. doi: 10.1099/mic.0.026849-0
Suzuki, K., Wang, X., Weilbacher, T., Pernestig, A. K., Melefors, Ö., Georgellis, D., et al. (2002). Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol 184, 5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tang, M., Huang, Z., Zhang, X., Kong, J., Zhou, B., Han, Y., et al. (2023). Phage resistance formation and fitness costs of hypervirulent Klebsiella pneumoniae mediated by K2 capsule-specific phage and the corresponding mechanisms. Front. Microbiol. 14, 1156292. doi: 10.3389/fmicb.2023.1156292
Wang, H., Cai, T., Weng, M., Zhou, J., Cao, H., Zhong, Z., et al. (2006). Conditional production of acyl-homoserine lactone-type quorum-sensing signals in clinical isolates of enterobacteria. J. Med. Microbiol. 55, 1751–1753. doi: 10.1099/jmm.0.46756-0
Wang, H., Dong, Y., Wang, G., Xu, X., Zhou, G. (2016). Effect of growth media on gene expression levels in Salmonella Typhimurium biofilm formed on stainless steel surface. Food Control. 59, 546–552. doi: 10.1016/j.foodcont.2015.06.026
Wang, S., Payne, G. F., Bentley, W. E. (2020). Quorum sensing communication: molecularly connecting cells, their neighbors, and even devices. Annu. Rev. Chem. Biomol Eng. 11, 447–468. doi: 10.1146/annurev-chembioeng-101519-124728
Wang, Y., Wang, S., Chen, W., Song, L., Zhang, Y., Shen, Z., et al. (2018). CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 84, e01834–e01818. doi: 10.1128/AEM.01834-18
WHO (2024). WHO Bacterial Priority Pathogens List 2024 Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance (Geneva: World Health Organization).
Xie, Y., Wahab, L., Gill, J. (2018). Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses 10, 189. doi: 10.3390/v10040189
Keywords: quorum sensing, biofilm, AHL (N-acyl-homoserine lactone), serum resistance, bacteriophage, Galleria mellonella, SdiA
Citation: Silva-Bea S, Maseda P, Otero A and Romero M (2025) Regulatory effects on virulence and phage susceptibility revealed by sdiA mutation in Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 15:1562402. doi: 10.3389/fcimb.2025.1562402
Received: 17 January 2025; Accepted: 25 February 2025;
Published: 13 March 2025.
Edited by:
Roberto Rusconi, Humanitas University, ItalyReviewed by:
Brian Ahmer, The Ohio State University, United StatesXiudong Xia, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2025 Silva-Bea, Maseda, Otero and Romero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuel Romero, bWFudWVscm9tZXJvLmJlcm5hcmRlekB1c2MuZXM=; Ana Otero, YW5hbWFyaWEub3Rlcm9AdXNjLmVz
 Sergio Silva-Bea
Sergio Silva-Bea Pablo Maseda
Pablo Maseda Ana Otero
Ana Otero Manuel Romero
Manuel Romero