- 1Division of Infectious Diseases, Weill Department of Medicine, Weill Cornell Medicine, New York, NY, United States
- 2Department of Cell and Developmental Biology, Weill Cornell Medicine, Graduate School of Medical Sciences, New York, NY, United States
- 3Orthopaedic Soft Tissue Research Program, Hospital for Special Surgery, New York, NY, United States
- 4Center of Infectious Diseases and Pathogen Biology, Key Laboratory of Organ Regeneration and Transplantation of The Ministry of Education, State Key Laboratory for Diagnosis and Treatment of Severe Zoonotic Infectious Diseases, The First Hospital of Jilin University, Changchun, China
Editorial on the Research Topic
Metabolomics in bacterial infections
Despite advancements in technology, bacterial infections continue to pose significant global health challenges. Metabolic responses are the first line of changes during infection and represent the best measure to understand the pathogenesis. In this context, metabolomics has emerged as a powerful tool to interpret complex and dynamic biochemical changes. This approach provides valuable insights into bacterial physiology, virulence, and host immune responses, offering new directions for therapeutic development (Figure 1). The aim of this Research Topic, Metabolomics in Bacterial Infections, was to compile various research articles, methods, and reviews on host-pathogen metabolic interactions, including their modulation and regulation, role in virulence, potential as biomarkers, and promise for novel therapeutic development. We have curated four manuscripts that provide novel insights into host responses to Coxiella burnetii infection, metabolic changes during enteric infections and their impact on gut dysbiosis, the genotype-serotype relationship of Group B Streptococcus, and a review of various host-pathogen metabolic changes following Legionella infection.
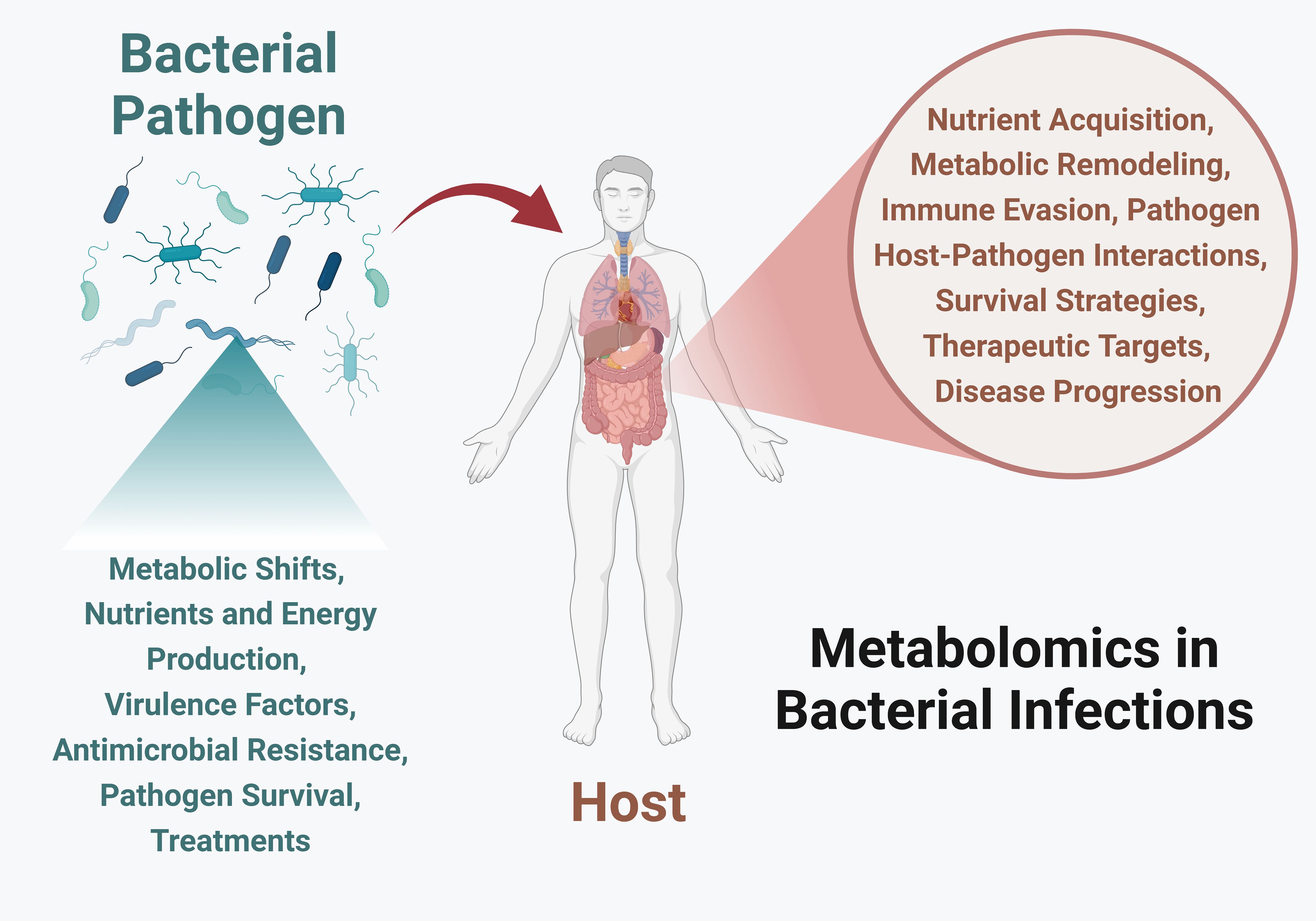
Figure 1. Metabolomics in bacterial infections provides crucial insights into the dynamic interactions between host and pathogens. By studying bacterial metabolic responses, we can understand their nutrient and energy production strategies, and various ways to adapt to host environments, generate virulence factors, evade the immune system, and develop antibiotic resistance. By investigating host metabolism, we can learn about nutrient foraging, changes in metabolic pathways, immune response modulation, pathogen survival strategies, potential therapeutic targets, and the discovery of biomarkers for early detection. Image created using Biorender.
Wan et al. demonstrated that infection with Coxiella burnetii upregulates the transcription of the lyst gene, which regulates lysosome size and is associated with the pathogen’s effector secretion. LYST limits the expansion of Coxiella-containing vacuoles (CCVs) and enhances macrophage immunity by promoting nitric oxide production through the upregulation of nos2. These mechanisms help restrict C. burnetii replication. Therefore, targeting LYST, by developing small molecule inhibitors, can advance therapeutic strategies against C. burnetii and Q fever. However, further investigation into the precise molecular mechanisms involved is needed.
In another interesting study, Hansen et al. employed shotgun metagenomics and untargeted meta-metabolomics to investigate metabolome changes in the human gut microbiome during and after acute enteric infections and recovery. Analyzing data from 60 patients, their findings revealed an increased capacity of microbial metabolic pathways during infection, despite a reduction in taxonomic diversity. Pathways associated with inflammation, such as LPS production, nitrogen metabolism, and glycolysis, were enriched, highlighting the role of Enterobacteriaceae under stress. Post-recovery, a greater diversity of metabolites and the presence of compounds like bile acids indicated a return to homeostasis. Also, these metabolites can distinguish between infected and recovered samples, indicating their potential as biomarkers. The genes involved in these pathways can be further investigated to understand their roles in infection and to aid in the discovery of new drugs.
Further, to understand host-pathogen relation, Zeng et al. used strain-level genomic analysis and assessed the distributions of serotypes and genotypes and their association with virulence genes and host factors (interacting with Srr1 and Srr2) in Group B Streptococcus (GBS). This study highlights the impact of host immunity, strain-specific virulence, and colonization dynamics. Virulence genes such as srr2, hygA, and hvgA are pivotal in GBS hypervirulence, promoting colonization and invasion, while surface proteins like Fbs and Alp help in evading the immune system. The research emphasizes the significance of virulence gene combinations in pathogenicity and suggests that detecting these genes could enhance GBS surveillance, prevention, and vaccine development. However, the study notes the need for broader epidemiological research.
Finally, in a concise mini-review, Wang and Song. explored the metabolic alterations happening in both host and pathogen during Legionella infections. They highlighted the role of the bacterial Type IV Secretion System (T4SS) in infection and detailed the roles of key effectors such as MavQ, SidP, and Lgt1-3. These effectors interrupt carbohydrate, lipid, and protein metabolism, manipulate phosphoinositide pathways, and alter membrane properties to inhibit protein biosynthesis, escape immune defenses, and support bacterial growth. The review underscores how understanding Legionella’s metabolic interactions with its host can delineate novel drug targets and diagnostic approaches. Despite new discoveries, further studies are required to elucidate these mechanisms across different host systems, paving the way for improved disease management and reduced public health risks.
In conclusion, the studies presented highlight the novel functionality of the lyst gene in C. burnetii replication, the critical role of the gut microbiome and metabolome during enteric infections, and the intricate relationship between virulence genes, host factors, and metabolic aspects in host-pathogen interactions. These insights can guide novel drug discoveries and serve as biomarkers for disease diagnosis or surveillance. Metabolomics, as a growing field, holds significant promise, and the diverse research contributions in this Research Topic offer new perspectives to combat infectious diseases in a systemic manner.
Author contributions
VS: Conceptualization, Investigation, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. AU: Writing – original draft, Writing – review & editing. YZ: Project administration, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: metabolomics, host pathogen interaction, metabolism, bacterial metabolomis, system biology
Citation: Soni V, Upadhyay A and Zhang Y (2025) Editorial: Metabolomics in bacterial infections. Front. Cell. Infect. Microbiol. 15:1557057. doi: 10.3389/fcimb.2025.1557057
Received: 07 January 2025; Accepted: 22 January 2025;
Published: 04 February 2025.
Edited and Reviewed by:
Jon Skare, Texas A&M Health Science Center, United StatesCopyright © 2025 Soni, Upadhyay and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vijay Soni, dmlzMjAzMkBtZWQuY29ybmVsbC5lZHU=
 Vijay Soni
Vijay Soni Aditya Upadhyay
Aditya Upadhyay Yong Zhang
Yong Zhang