
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 03 February 2025
Sec. Bacteria and Host
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1546991
 Xueyao Fang1†
Xueyao Fang1† Yanhui Chen1†
Yanhui Chen1† Longhua Hu1
Longhua Hu1 Shumin Gu1
Shumin Gu1 Junqi Zhu1
Junqi Zhu1 Yaping Hang1
Yaping Hang1 Xingwei Cao1
Xingwei Cao1 Yanping Xiao1
Yanping Xiao1 Hong Luo1
Hong Luo1 Chuwen Zhao1
Chuwen Zhao1 Lianhua Xiao2
Lianhua Xiao2 Qiaoshi Zhong1*
Qiaoshi Zhong1*Indole, a crucial bacterial signaling molecule, plays a fundamental role in regulating various physiological processes within bacteria, including growth, acid tolerance, biofilm development, motility, and other cellular functions. Its regulatory influence extends beyond indole-producing bacteria, significantly impacting the physiological activities in non-indole-producing species. In this study, we demonstrate that indole enhances the pathogenicity and viability of Klebsiella pneumoniae using the Galleria mellonella infection model and serum killing assay. Concurrently, indole has varying effects on biofilm formation in K. pneumoniae, with some strains showing enhanced biofilm formation ability. To elucidate the underlying molecular mechanisms, transcriptome analysis revealed that indole exposure in K. pneumoniae led to the upregulation of genes associated with pili formation and iron acquisition systems, while simultaneously inducing oxidative stress responses. Additionally, our analysis uncovered extensive metabolic remodeling. Specifically, we observed significant upregulation of genes involved in simple carbohydrate utilization pathways, including those responsible for galactose, mannose, and maltose metabolism, as well as enhanced expression of genes associated with pyrimidine biosynthesis. These findings collectively indicate that indole enhances the intestinal colonization and pathogenicity of K. pneumoniae primarily by modulation of fimbriae expression and metabolic pathway regulation.
Indole, a molecule produced by various bacteria, plays a crucial role in bacterial cell-to-cell communication and behavioral regulation (Vega et al., 2012). This compound is known to regulate multiple physiological processes within bacteria, including growth, acid tolerance, biofilm development, motility, and other cellular functions (Hirakawa et al., 2009; Lee et al., 2015b; Hashidoko and Kim, 2021). Interestingly, the influence of indole extends beyond indole-producing bacteria, impacting various physiological activities in non-indole-producing species as well. For instance, in Pseudomonas aeruginosa, indole and its derivatives suppress motility and virulence factor production while enhancing antimicrobial resistance (Lee et al., 2009). In Agrobacterium tumefaciens, a soil-borne bacterium, indole inhibits growth and motility and upregulates genes involved in biofilm formation, which in turn enhances antibiotic tolerance (Lee et al., 2015a). Similarly, in Klebsiella pneumoniae (K. pneumoniae), indole has been shown to increase antibiotic resistance and promote biofilm formation by modulation of efflux pump activity (Salama et al., 2024). However, despite these observations, the specific molecular mechanisms through which indole regulates virulence factor expression and metabolic pathways in K. pneumoniae remain poorly understood.
K. pneumoniae, a prevalent clinical pathogen, can cause various severe or potentially lethal infections, including pneumonia, liver abscess, and bloodstream infections, particularly in individuals with compromised immunity or other susceptibility factors. Its pathogenicity is primarily attributed to a variety of virulence factors, including capsular polysaccharides, lipopolysaccharides, pili, and iron acquisition systems (Russo and Marr, 2019). Research has established that intestinal colonization is a critical stage in the progression of K. pneumoniae-induced systemic infections (Gorrie et al., 2017). To successfully colonize, K. pneumoniae employs sophisticated mechanisms to overcome host immune defenses and microbial competition, involving the coordinated production of multiple bacterial factors, such as colibactin, microcin E492, and salmochelin (Tan et al., 2024). Additionally, key factors such as iron utilization, purine synthesis, fimbriae production, and capsule formation significantly impact its ability to colonize the intestine (Lasaro et al., 2009; Vogel-Scheel et al., 2010; Hsu et al., 2019; Chen et al., 2023; Hecht et al., 2024).
Studies have revealed that indole concentrations in human and mammalian feces range from 0.30 to 6.64 millimolar (Darkoh et al., 2015), levels sufficient to significantly influence the behavior and physiological functions of gut microbiota (Tennoune et al., 2022). The response to indole signaling molecules, however, varies significantly among different bacterial species. For instance, in a Caenorhabditis elegans gastrointestinal infection model, indole signaling enhances Salmonella typhimurium antibiotic tolerance in the host intestine through the activation of oxidative stress and bacteriophage shock response systems (Vega et al., 2013); however, it exerts inhibitory effects on Staphylococcus aureus by suppressing hemolysin production and reducing its invasiveness (Lee et al., 2013). Furthermore, Aman Kumar et al. demonstrated in a mouse model that lower concentrations of indole enhanced the pathogenesis of enteric pathogens enterohemorrhagic Escherichia coli and Citrobacter rodentium, while higher concentrations suppressed the expression of bacterial virulence genes (Kumar and Sperandio, 2019).
Based on the regulatory functions of indole in intestinal bacteria, we hypothesized that this signaling molecule might modulate the pathogenicity and physiological status of K. pneumoniae by influencing its virulence factors and metabolic pathways. This study aimed to investigate the role of indole signaling in K. pneumoniae intestinal colonization, with the goal of developing novel therapeutic strategies to control K. pneumoniae infections through a better understanding of the molecular mechanisms underlying its effects on bacterial virulence and metabolism.
The clinical K. pneumoniae strain N442 isolated from blood of a patient in the Hepatobiliary Surgery Unit, the Second Affiliated Hospital of Nanchang University, China. K. pneumoniae ATCC 700603 was obtained from the China National Health Inspection Center. NTUH - K2044, a highly virulent strain of K. pneumoniae isolated from patient with liver abscess, was donated by Professor Fangyou Yu. Indole, dimethylformamide (DMF) were of analytical grade, and purchased from Aladdin (Shanghai, China). Crystal Violet was purchased from Solarbio (Beijing, China). Methanol and glacial acetic acid were purchased from Xilong Chemical (China). Galleria mellonella (G. mellonella) larvae were purchased from Huiyude Company (Tianjin, China).
To determine the toxicities of indole, the specific growth rates of the N442 strain, the NTUH-K2044 and the ATCC700603 strain were measured in the different concentrations of indole (0 mM, 0.5 mM, 1.0 mM, 2.0 mM, and 5.0 mM), respectively. Indole was dissolved in dimethylformamide (DMF, ≥99.8%), and 0.5 vol% of DMF was added as a control group at all experiments. All strains were inoculated in LB (Luria–Bertani) broth at 37°C and shaken at 200rpm. Bacterial growth was assessed by monitoring the OD600 values of triplicate samples. Except for the growth rate measurements, a concentration of 0.5 mM indole was selected as the treatment solution in all other experiments.
To assess the effect by indole on K. pneumoniae biofilm formation, the assay referred to the previous literature with minor adjustments. The overnight bacterial cultures were adjusted to an OD600 of 1 and subsequently diluted 1:100 in 1 mL of LB medium with or without 0.5 mM indole supplementation, and static incubated at 37°C for 48 hours. After the supernatant was discarded, cells were fixed with 99% methanol for 15 min and then aspirated. Place at room temperature until completely dry, and then stained with 1% crystal violet for 15 min. The 24-well plate was washed with running water until colorless, and then eluted with 33% glacial acetic acid, the absorbance was measured at 570 nm (O’Toole and Kolter, 1998). The experiment was repeated for three times, and the average value was calculated for biofilm quantification.
To observe the effect of indole on the virulence of K. pneumoniae, the survival of G. mellonella larvae were measured, as described earlier with slight modifications (Li et al., 2020). The strains were grown to stationary phase in LB added with or without 0.5mM of indole, then wash the bacterial centrifugation with sterile saline for three times, and diluted to about 1×107 CFU/ml for subsequent injections. The larvae measuring 2 - 2.5 cm in length, weigh 200 - 400 mg, have no black spots on the body surface, show uniform creamy white coloration and display vigorous movement. Each experimental group consists of ten to twelve larvae. A 10-μL bacterial suspension was injected into the body of the larvae via the last left proleg by use of a Hamilton syringe with a 30-gauge needle. The syringes are sequentially flushed with 75% ethanol and sterile saline between strain procedures. After challenge, larvae were placed in petri dish and incubated in dark at 37°C for 3 days. Mortality was observed and recorded every 12 hours. The experiment was repeated for three times in each group.
The experiments were performed in accordance with the method described by Juyoun Shin et al (Shin and Ko, 2015). After incubating the strain to be tested overnight, the bacterial concentration was adjusted to 1×106 CFU/ml. Subsequently, 25-μL of the bacterial solution was mixed with 75-μL of serum, thoroughly shaken, and then placed on a constant temperature shaker at 37°C for 3 hours. The LB broth was serially diluted tenfold and plated onto MH plates. The counts of serum-treated bacterial suspensions were determined and compared with the counts of unsaturated bacterial suspensions to calculate the survival rate.
The strains N442 was cultured in LB supplemented with or without 0.5mM of indole for 16 hours, then transferred to fresh LB and shaken to logarithmic growth phase, and were collected by centrifugation at 3000 × g for 5 min at 4°C. The precipitate was immediately preserved with liquid nitrogen. The bacterial precipitate was sent to Majorbio (Shanghai, China) under dry ice, with three replicates each group. Total RNA extraction, RNA purification, cDNA synthesis, DNA library construction, sequencing and data analyses of the transcriptome were performed by Illumina HiSeq X platform. The database used for alignment was the reference genome of K. pneumoniae HS11286 (GCA_000240185.2) obtained from the National Center for Biotechnology Information (NCBI). DESeq2 software was utilized to conduct differential expression analysis on the raw data, and the Benjamini-Hochberg method was used to correct the p-value for multiple tests to obtain the adjusted p-value (P-adjust). Differentially expressed genes (DEGs) were screened with a P-adjust < 0.05 and an absolute value of log2 fold change (log2FC) ≥ 1. Furthermore, RSEM software was employed to quantitatively analyze gene expression, and the quantitative index was TPM (transcripts per million/kilobases). The transcriptome volcano plot and heatmap plot were performed in Hiplot Pro (https://hiplot.com.cn/). The ClusterProfiler R software package was utilized to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs. The pathway enrichment analysis was assessed using statistical methods based on Fisher’s exact test.
To investigate the effect of indole on K. pneumoniae growth, growth curves were generated in LB broth containing various indole concentrations. While all K. pneumoniae strains demonstrated growth in indole-containing media, their growth was inhibited at elevated indole concentrations, as illustrated in Figure 1. Notably, DMF, used as the indole solvent, showed no effect on K. pneumoniae growth. At lower indole concentrations (0.1 mM and 0.5 mM), K. pneumoniae exhibited growth patterns comparable to those observed in indole-free conditions, with no statistically significant differences, indicating tolerance within this concentration range. However, when indole concentrations were increased to 1 mM, 2 mM, and 5 mM, bacterial growth was significantly inhibited, with the magnitude of inhibition directly proportional to indole concentration. At 5 mM indole, bacterial growth was severely restricted, approaching a bacteriostatic state.
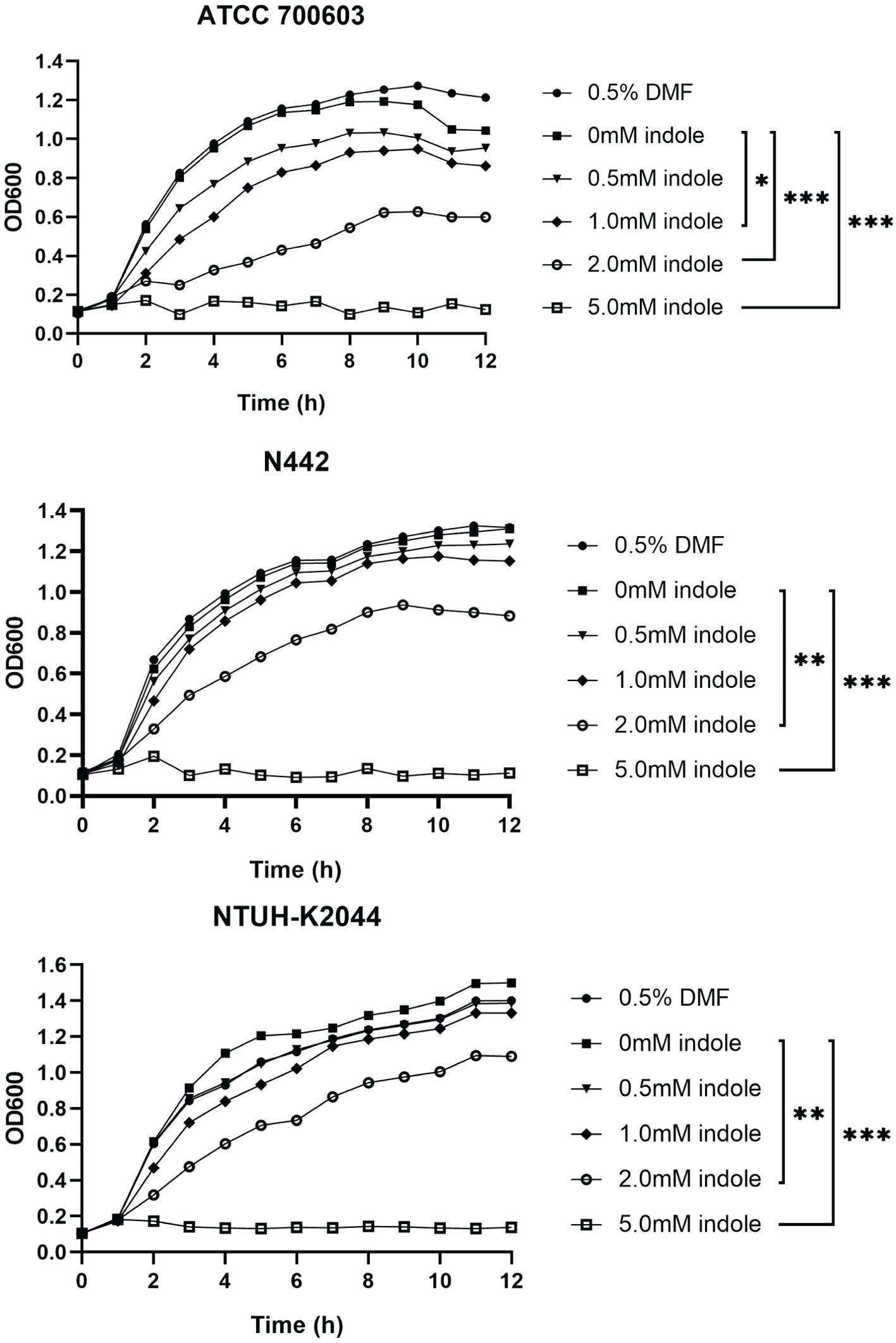
Figure 1. Indole concentration affected the growth of K. pneumoniae. Distinct shapes signify the presence of indole solutions with diverse concentrations in the culture medium. P values of each concentration were calculated by comparing growth curves at other concentrations with that at 0mM. Dunnett’s test was utilized for the differential analysis of the growth curves among strains N442, ATCC700603, and NTUH-K2044. Statistical analysis was performed using Two-way ANOVA with Dunnett’s multiple comparison test. “*” means P<0.05, “**” means P<0.01, “***” means P<0.001.
These findings suggest that K. pneumoniae can tolerate indole concentrations between 0 and 0.5 mM, but concentrations exceeding this threshold become increasingly toxic, inhibiting bacterial growth. Based on these results, 0.5 mM indole was chosen as the treatment concentration for subsequent experiments to observe its effects on toxicity, biofilm formation, and transcriptome metabolism at a tolerable indole concentration for K. pneumoniae.
Crystal violet staining, a well-established assay for biofilm-forming ability (Muzaki et al., 2024), was employed to assess the impact of indole treatment on biofilm formation in K. pneumoniae, as shown in Figure 2. The statistical results indicated that indole treatment significantly enhanced the biofilm formation ability of ATCC700603 (P=0.0067) and N442 (P=0.0105). However, no significant effect was observed on the biofilm formation ability of K2044 (P=0.2036). These results imply that indole facilitates biofilm formation within particular strains of K. pneumoniae.
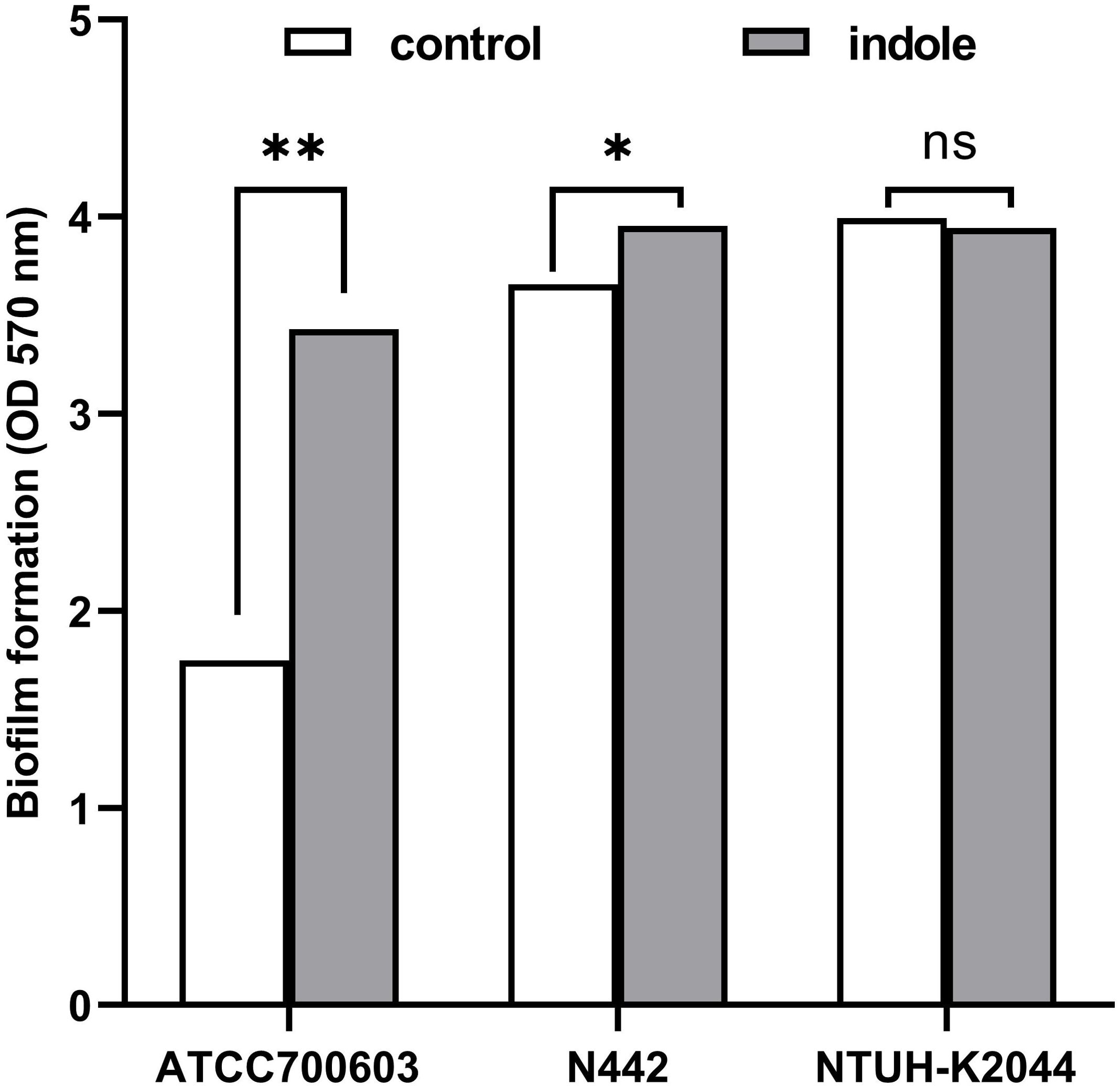
Figure 2. Indole-induced biofilm formation of K. pneumoniae. White squares denote bacteria cultured in standard LB broth, while gray squares signify bacteria cultured in broth with 0.5 mM indole. The statistical analysis was conducted using a two-tailed paired t-test. “ns” means P>0.05, “*” means P<0.05, “**” means P<0.01.
The G. mellonella infection model, which has the capacity to replicate the bacterial infection process within the host organism, was utilized to evaluate the virulence of (D’Apolito et al., 2020). Following exposure to 0.5 mM indole, K. pneumoniae-infected larvae showed significantly decreased survival rates, as shown in Figure 3A. Specifically, larvae infected with indole-treated N442 (P=0.0095) and ATCC700603 (P=0.0473) strains exhibited higher mortality rates compared to those infected with untreated K. pneumoniae. Using K. pneumoniae NTUH-K2044 as a reference strain, the 12-hour mortality rates were significantly elevated in indole-treated strains. The serum killing test is a method of indirectly assessing the resistance of bacteria to the complement system, as shown in Figure 3B. In this assay, strains N442 (P=0.0032) and K0244 (P=0.0293) exhibited significantly higher survival rates compared to the controls lacking indole after 3 hours of incubation in indole-supplemented serum. These results demonstrate that indole treatment significantly enhanced the pathogenicity and viability of K. pneumoniae strain N442 in the host.

Figure 3. (A) Survival rate of G. mellonella post K. pneumoniae infection. Different shapes represent the various bacterial suspensions used for injection. Statistical analysis was performed using Two-way ANOVA with Bonferroni correction for multiple comparisons. (B) Serum killing assay. The survival rate was calculated through comparing the ratio of colony forming units (CFUs) of bacterial suspensions when serum was added to that of bacterial suspensions in the absence of serum. Statistical analysis was performed using a two-tailed paired t-test. “*” means P<0.05, “**” means P<0.01.
To investigate changes in gene expression of K. pneumoniae in response to indole exposure, RNA-seq experiments were conducted using the clinical strain N442. The transcriptome analysis compared two groups: the experimental group treated with 0.5 mM indole (designated as indole-N442) and the untreated control group (designated as N442). Analysis revealed 159 differentially expressed genes (DEGs) in response to indole treatment, comprising 102 up-regulated and 57 down-regulated genes. Among these DEGs, 34 were sRNAs and 29 encoded hypothetical proteins, while the remaining genes were annotated with defined functions, as illustrated in Figures 4A, B.
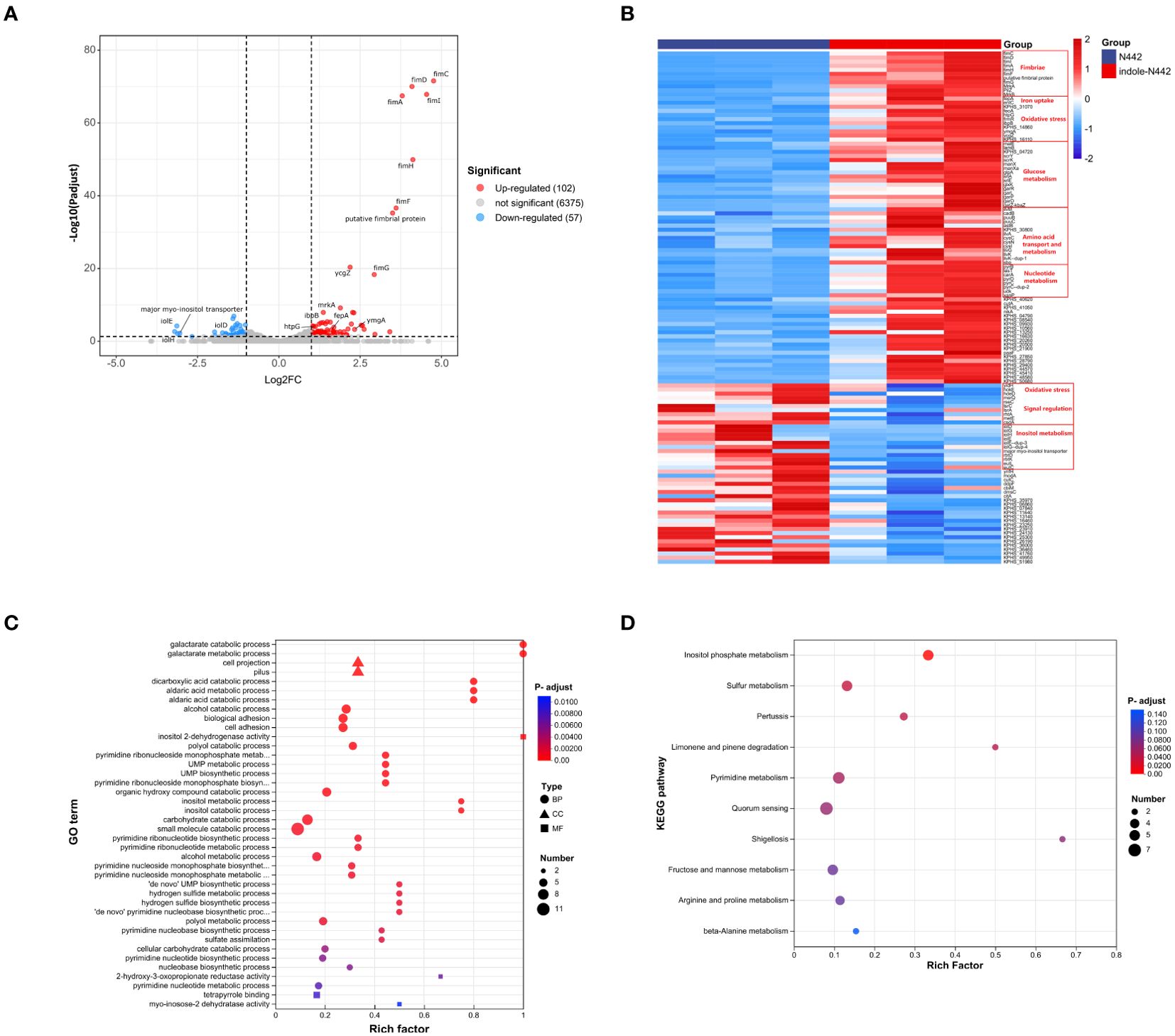
Figure 4. Transcriptome analysis of K. pneumoniae strain N442. (A) Volcano plot depicts up-regulated (red dots) and downregulated (blue dots) DEGs in N442. The thresholds for significant differential expression were set as log2|FC| ≥ 1.0 and P-adjust < 0.05. (B) Heatmap. Post indole treatment, DEGs of N442 strain were screened for heat map analysis. Upregulated genes in red and downregulated in blue. To ensure the accuracy of the analysis, all small RNA (sRNA) sequences have been excluded from the differential gene list. (C) GO enrichment analysis. Different shapes indicate different GO classifications: biological processes (BP) are represented by circles; cellular components (CC) by triangles; and molecular functions (MF) by squares. (D) KEGG enrichment analysis. The X-axis represents the Rich factor, which is the ratio of the number of genes in the set of DEGs annotated to the GO term or KEGG pathway to the number of genes in the set of all genes annotated to the GO term or KEGG pathway. The Y-axis represents the top 40 GO terms or top 10 KEGG pathways, sorted by adjusted p-values (P-adjust), with category names labeled in the figure. The size of the bubbles represents the number of genes enriched in the pathway, while the color of the bubbles indicates the statistical significance of the enrichment, with red representing a small p-value (high significance) and blue representing a large p-value (low significance).
The proteins encoded by these DEGs significantly influence various vital processes in K. pneumoniae, including pili formation, transcriptional regulation, DNA damage repair, and energy metabolism. Notably, genes encoding both type 1 and type 3 fimbriae showed substantial upregulation. The type 1 fimbrial genes exhibited marked increases in expression, including fimA (fold change: 13.863, P-adjust= 3.45E-68), fimC (fold change: 27.041, P-adjust= 2.71E-72), fimD (fold change: 17.066, P-adjust= 9.57E-71), fimF (fold change: 12.164, P-adjust=2.43E-37), fimG (fold change: 7.634, P-adjust=4.69E-19), fimH (fold change: 17.391, P-adjust=1.22E-50), and fimI (fold change: 23.319, P-adjust=1.38E-68). Similarly, type 3 fimbrial genes showed significant upregulation, including mrkA (fold change: 2.579, P-adjust=1.21E-08), mrkB (fold change: 2.358, P-adjust=0.004), and pilZ (fold change: 4.949, P-adjust=1.51E-08). These fimbriae are known to play crucial roles in host colonization and virulence of K. pneumoniae (Alcántar-Curiel et al., 2013).
Four genes associated with iron acquisition demonstrated significant upregulation. EntC, which encodes isochorismate synthetase, showed a 3.277-fold increase in expression (P-adjust=0.004), while fepA, encoding an outer membrane ligand-gated porin, exhibited a substantial 7.634-fold change (P-adjust=4.69E-19). These genes work synergistically in the enterobactin synthesis pathway. Additionally, two genes involved in ferrous iron transport were upregulated: feoA (fold change: 2.028, P-adjust=0.03) and KPHS_31070 (fold change: 2.076, P-adjust=0.015). The coordinated upregulation of these iron acquisition genes ensures adequate iron availability for essential bacterial metabolic processes and survival (Kumar et al., 2019).
Indole exposure triggered differential regulation of genes encoding oxidative stress-response proteins and chaperones in K. pneumoniae. Notably, two heat shock proteins showed significant upregulation: htpG, encoding heat shock protein 90 (fold change: 2.177, P-adjust=8.01E-05), and ibpB, encoding a small heat shock protein (fold change: 2.454, P-adjust=8.89E-06). These proteins play crucial roles in preventing protein degradation under oxidative stress conditions (Grudniak et al., 2015). Additionally, frmR, which encodes a formaldehyde-responsive transcriptional regulator, exhibited increased expression (fold change: 2.192, P-adjust=0.030). However, several other stress response genes showed downregulation, including hdeD (fold change: 0.455, P-adjust=0.002), hokE (fold change: 0.458, P-adjust=0.014), and msrQ (fold change: 0.494, P-adjust=0.012).
Indole stress also significantly altered the expression of genes involved in quorum sensing and transcriptional regulation in K. pneumoniae. The expression levels of several regulatory genes were upregulated, including ycgZ, ymgA, and two transcriptional regulators: KPHS_14860 (a putative LysR family transcriptional regulator) and KPHS_16110 (a LuxR transcriptional regulator). Conversely, downregulation was observed in rseC (encoding a sigma-E factor negative regulator) and two components of the lsr operon: lsrA and lsrC, which are involved in AI-2-mediated quorum sensing.
To elucidate effects of indole on metabolism, we conducted KEGG pathway and GO enrichment analyses. The GO enrichment analysis encompassed 104 differentially expressed genes (DEGs), comprising 66 upregulated (63.5%) and 38 downregulated genes (36.5%) (Figure 4C). Among the top 30 enriched GO functional categories, 27 were biological processes (BP), 2 were cellular components (CC), and 1 was molecular function (MF). Within the BP category, 19 significantly upregulated processes were identified, primarily involving carbohydrate and acid metabolism, cell adhesion, pyrimidine biosynthesis, and sulfur-containing compound metabolism. Conversely, 8 significantly downregulated BP categories were identified, predominantly associated with alcohol metabolism, particularly inositol metabolism. In the CC category, upregulation was observed in fimbriae-like proteins and cell projections. The sole enriched MF category showed downregulation of inositol 2-dehydrogenase activity.
KEGG metabolic pathway enrichment analysis was performed on 89 DEGs, consisting of 60 upregulated (67.4%) and 29 downregulated genes (32.6%) (Figure 4D). The upregulated DEGs showed significant enrichment in multiple metabolic pathways, including sulfur metabolism, limonene and pinene degradation, pyrimidine metabolism, pertussis, shigellosis, and amino acid metabolism. In contrast, downregulated DEGs were predominantly enriched in inositol phosphate metabolism pathways.
As a signaling molecule, indole is prevalent in the gut and significantly influences the growth and metabolic activity of the gut microbial community (Melander et al., 2014; Lee et al., 2015b). Studies have demonstrated that elevated colonization rates of K. pneumoniae within the gut are highly correlated with increased risks of host infection (Martin et al., 2016; Chen et al., 2023; Cai et al., 2024). However, the specific role of indole in regulating the growth, metabolism, and virulence of K. pneumoniae remains to be fully elucidated. This study investigated the modulatory effects of indole on the virulence and metabolism of K. pneumoniae, as well as its potential to promote intestinal colonization.
Growth curve experiments demonstrated that indole at a relatively low concentration (0.5 mM) had no significant effect on K. pneumoniae growth, consistent with previous studies (Yaikhan et al., 2019). This finding suggests that at such low concentrations, indole may not directly interfere with bacterial multiplication. However, at concentrations of 1 mM and above, bacterial growth was significantly inhibited, with the inhibitory effect intensifying as indole concentrations increased. These results indicate that high indole concentrations may suppress bacterial growth by disrupting cell membranes or interfering with metabolic pathways (Chimerel et al., 2013; Zarkan et al., 2020). This implies that elevated indole concentrations within the gut might restrict the growth and colonization of K. pneumoniae, thereby influencing its ecological niche in this environment.
The G. mellonella infection model is an effective tool for evaluating the virulence of K. pneumoniae, as it is capable of simulating the bacterial infection process within the host (Li et al., 2020; Asai et al., 2023). Concurrently, the serum killing assay functions as a methodological framework for assessing the resistance of bacteria to the complement system (Shin and Ko, 2015; Liu et al., 2019). Induction with 0.5 mM indole significantly reduced the survival rate of G. mellonella and enhanced the serum resistance of K. pneumoniae. These findings indicate that indole may play a role in the adaptation of K. pneumoniae to host immune pressure, potentially enhancing its survival and pathogenicity.
Consistent with the findings of Yaikhan et al (Yaikhan et al., 2019; Tan et al., 2024), the effect of indole on K. pneumoniae biofilm formation varied across strains, with strain K2044 showing no significant enhancement. The variations in the responses of distinct strains to biofilm formation may be closely related to factors (Donlan, 2001; Yin et al., 2019) such as their genetic background, metabolic mechanism, surface structure, stress response, and specific response to indole. It is proposed that, as a highly mucoid strain, K2044 inherently synthesizes large amounts of exopolysaccharides and capsule layers (Peng et al., 2018). These surface structures confer upon the strain a strong biofilm-forming capacity, thereby resulting in minimal effects of indole treatment on its biofilm properties. Future studies should explore the interaction between these bacterial traits and environmental factors to uncover additional details of the biofilm formation mechanism.
Transcriptome analysis revealed that indole significantly modulates bacterial gene expression, particularly affecting genes related to virulence and fitness. Specifically, indole upregulated genes associated with type 1 and type 3 fimbriae formation, iron acquisition systems, and oxidative stress responses. These molecular changes suggest that indole enhances the pathogenicity of K. pneumoniae through multiple mechanisms. On one hand, it strengthens the assembly of bacterial surface structures, such as fimbriae and biofilms, thereby enhancing its adhesive and invasive potential in the environment (Struve et al., 2008; Alcántar-Curiel et al., 2013). On the other hand, it boosts iron acquisition and oxidative stress tolerance, promoting bacterial survival and adaptation in the host gut (Mason et al., 1999; Vega et al., 2013; Denby et al., 2016; Genest et al., 2018; Azaharuddin et al., 2023; Kumar et al., 2019).
In particularly, the ycgZ-ymgABC operon-related genes manifested a significant up-regulation in expression, further supports role of indole in promoting K. pneumoniae adaptation and biofilm formation under environmental stress. Previous studies have demonstrated a close association between the ycgZ-ymgABC operon and the regulation of bacterial biofilms under environmental stress (Tschowri et al., 2009, 2012; Kettles et al., 2019). Additionally, pili, especially type 1 and type 3 fimbriae, directly contribute to biofilm formation. Type 1 fimbriae are crucial for bacteria-host cell adhesion and initial biofilm formation, whereas type 3 fimbriae maintain biofilm stability in a fluid shear environment (Alcántar-Curiel et al., 2013).Indole may enhance the adhesion capacity and biofilm-forming propensity of K. pneumoniae via modulating the expression of ycgZ-ymgABC operon and fimbria, thereby enhancing its adaptability to the host environment.
The transcriptome analysis also revealed substantial indole-induced restructuring of metabolic networks of K. pneumoniae. Most notably, genes involved in the utilization of specific carbon sources—including galactose, mannose, and maltose—showed significant upregulation. This metabolic adaptation suggests that indole enhances the ability of bacterium to utilize available host nutrients, thereby supporting growth and survival in the intestinal environment (Chang et al., 2004; Hecht et al., 2024). This conclusion is further supported by previous studies in E. coli, where enhanced pyrimidine metabolism was shown to maintain bacterial growth under indole stress (Vogel-Scheel et al., 2010). The parallel findings in these related species suggest a conserved mechanism whereby metabolic remodeling contributes to bacterial adaptation and survival.
Targeting the indole signaling pathway represents a promising strategy for treating K. pneumoniae infections, given indole’s crucial role in regulating the bacterium’s virulence and metabolism. By modulating indole levels to influence bacterial growth, virulence, and biofilm formation, its pathogenicity in the intestine can be effectively mitigated, thereby reducing the risk of systemic infection. Studies have shown that indole and its derivatives act as modulators of the intestinal microbiota, promoting intestinal health and mitigate the risk of infection, thus offering novel therapeutic approaches that do not rely on traditional antibiotics (Lee and Lee, 2010; Kim and Park, 2015; Qin et al., 2020; Qayed et al., 2021). Future studies should focus on evaluating the safety and efficacy of indole-based therapies and exploring their clinical potential in treating K. pneumoniae infections and other microbiome-related diseases.
However, this study has certain limitations. The molecular mechanisms underlying the regulation of K. pneumoniae by indole remain incompletely understood, and no animal models were used to simulate the human intestinal environment. Future studies should further investigate the associated signaling pathways and molecular targets, and incorporate in vivo models to gain deeper insights into its pathogenesis and therapeutic potential.
Overall, indole concentrations have a significant impact on K. pneumoniae colonization within the intestinal microbiome. We found that indole enhances the adaptability and virulence of K. pneumoniae within the gut milieu by potentiation pili formation, iron uptake, and oxidative stress tolerance. Concurrently, indole refines the bacterial proficiency in host nutrient utilization via metabolic remodeling, thereby facilitating bacterial in the intestinal environment. These findings provide new insights into how microbiota-derived indole modulates pathogenic bacterial colonization in the gut.
The original contributions presented in the study are publicly available. This data can be found here: Sequence Read Archive (SRA) / PRJNA1215061.
XF: Conceptualization, Writing – original draft. YC: Data curation, Writing – review & editing. LH: Resources, Writing – review & editing. SG: Validation, Writing – review & editing. JZ: Methodology, Writing – review & editing. YH: Investigation, Writing – review & editing. XC: Validation, Writing – review & editing. YX: Visualization, Writing – review & editing. HL: Investigation, Writing – review & editing. CZ: Software, Writing – review & editing. LX: Supervision, Writing – review & editing. QZ: Conceptualization, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by The Jiangxi Provincial Health Commission Science and Technology Plan Project (No.202410035).
We thank Prof. Fangyou Yu for donating the highly virulent Klebsiella pneumoniae strain NTUH- K2044.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alcántar-Curiel, M. D., Blackburn, D., Saldaña, Z., Gayosso-Vázquez, C., Iovine, N., de la Cruz, M. A., et al. (2013). Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 4, 129–138. doi: 10.4161/viru.22974
Asai, M., Li, Y., Newton, S. M., Robertson, B. D., Langford, P. R. (2023). Galleria mellonella–intracellular bacteria pathogen infection models: the ins and outs. FEMS Microbiol. Rev. 47, fuad011. doi: 10.1093/femsre/fuad011
Azaharuddin, M., Pal, A., Mitra, S., Dasgupta, R., Basu, T. (2023). A review on oligomeric polydispersity and oligomers-dependent holding chaperone activity of the small heat-shock protein IbpB of Escherichia coli. Cell Stress Chaperones 28, 689. doi: 10.1007/s12192-023-01392-3
Cai, S., Wang, Z., Han, X., Hu, H., Quan, J., Jiang, Y., et al. (2024). The correlation between intestinal colonization and infection of carbapenem-resistant Klebsiella pneumoniae: A systematic review. J. Global Antimicrobial Resistance 38, 187–193. doi: 10.1016/j.jgar.2024.04.013
Chang, D.-E., Smalley, D. J., Tucker, D. L., Leatham, M. P., Norris, W. E., Stevenson, S. J., et al. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. United States America 101, 7427. doi: 10.1073/pnas.0307888101
Chen, Q., Wang, M., Han, M., Xu, L., Zhang, H. (2023). Molecular basis of Klebsiella pneumoniae colonization in host. Microbial Pathogenesis 177, 106026. doi: 10.1016/j.micpath.2023.106026
Chimerel, C., Murray, A. J., Oldewurtel, E. R., Summers, D. K., Keyser, U. F. (2013). The effect of bacterial signal indole on the electrical properties of lipid membranes. Chemphyschem 14, 417–423. doi: 10.1002/cphc.201200793
D’Apolito, D., Arena, F., Conte, V., De Angelis, L. H., Di Mento, G., Carreca, A. P., et al. (2020). Phenotypical and molecular assessment of the virulence potential of KPC-3-producing Klebsiella pneumoniae ST392 clinical isolates. Microbiological Res. 240, 126551. doi: 10.1016/j.micres.2020.126551
Darkoh, C., Chappell, C., Gonzales, C., Okhuysen, P. (2015). A rapid and specific method for the detection of indole in complex biological samples. Appl. Environ. Microbiol. 81, 8093. doi: 10.1128/AEM.02787-15
Denby, K. J., Iwig, J., Bisson, C., Westwood, J., Rolfe, M. D., Sedelnikova, S. E., et al. (2016). The mechanism of a formaldehyde-sensing transcriptional regulator. Sci. Rep. 6, 38879. doi: 10.1038/srep38879
Donlan, R. M. (2001). Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 33, 1387–1392. doi: 10.1086/322972
Genest, O., Wickner, S., Doyle, S. M. (2018). Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 294, 2109. doi: 10.1074/jbc.REV118.002806
Gorrie, C. L., Mirceta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Grudniak, A. M., Markowska, K., Wolska, K. I. (2015). Interactions of Escherichia coli molecular chaperone HtpG with DnaA replication initiator DNA. Cell Stress Chaperones 20, 951–957. doi: 10.1007/s12192-015-0623-y
Hashidoko, Y., Kim, D. (2021). Bidirectional cell-cell communication via indole and cyclo(Pro-tyr) modulates interspecies biofilm formation. Appl. Environ. Microbiol. 87, e01277. doi: 10.1128/AEM.01277-21
Hecht, A. L., Harling, L. C., Friedman, E. S., Tanes, C., Lee, J., Firrman, J., et al. (2024). Dietary carbohydrates regulate intestinal colonization and dissemination of Klebsiella pneumoniae. J. Clin. Invest. 134, e174726. doi: 10.1172/JCI174726
Hirakawa, H., Kodama, T., Takumi-Kobayashi, A., Honda, T., Yamaguchi, A. (2009). Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157 : H7. Microbiology 155, 541–550. doi: 10.1099/mic.0.020420-0
Hsu, C.-R., Chang, I.-W., Hsieh, P.-F., Lin, T.-L., Liu, P.-Y., Huang, C.-H., et al. (2019). A novel role for the Klebsiella pneumoniae sap (Sensitivity to antimicrobial peptides) transporter in intestinal cell interactions, innate immune responses, liver abscess, and virulence. J. Infect. Dis. 219, 1294–1306. doi: 10.1093/infdis/jiy615
Kettles, R. A., Tschowri, N., Lyons, K. J., Sharma, P., Hengge, R., Webber, M. A., et al. (2019). The Escherichia coli MarA protein regulates the ycgZ-ymgABC operon to inhibit biofilm formation. Mol. Microbiol. 112, 1609. doi: 10.1111/mmi.14386
Kim, J., Park, W. (2015). Indole: a signaling molecule or a mere metabolic byproduct that alters bacterial physiology at a high concentration? J. Microbiol. 53, 421–428. doi: 10.1007/s12275-015-5273-3
Kumar, A., Chakravorty, S., Yang, T., Russo, T. A., Newton, S. M., Klebba, P. E. (2019). Siderophore-mediated iron acquisition by Klebsiella pneumoniae. J. Bacteriol 206, e00024-24. doi: 10.1128/jb.00024-24
Kumar, A., Sperandio, V. (2019). Indole signaling at the host-microbiota-pathogen interface. mBio 10, e01031–e01019. doi: 10.1128/mBio.01031-19
Lasaro, M. A., Salinger, N., Zhang, J., Wang, Y., Zhong, Z., Goulian, M., et al. (2009). F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 75, 246–251. doi: 10.1128/AEM.01144-08
Lee, J., Attila, C., Cirillo, S. L. G., Cirillo, J. D., Wood, T. K. (2009). Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microbial Biotechnol. 2, 75–90. doi: 10.1111/j.1751-7915.2008.00061.x
Lee, J.-H., Cho, H. S., Kim, Y., Kim, J.-A., Banskota, S., Cho, M. H., et al. (2013). Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl. Microbiol. Biotechnol. 97, 4543–4552. doi: 10.1007/s00253-012-4674-z
Lee, J.-H., Kim, Y.-G., Baek, K.-H., Cho, M. H., Lee, J. (2015a). The multifaceted roles of the interspecies signalling molecule indole in Agrobacterium tumefaciens. Environ. Microbiol. 17, 1234–1244. doi: 10.1111/1462-2920.12560
Lee, J.-H., Lee, J. (2010). Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34, 426–444. doi: 10.1111/j.1574-6976.2009.00204.x
Lee, J.-H., Wood, T. K., Lee, J. (2015b). Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718. doi: 10.1016/j.tim.2015.08.001
Li, G., Shi, J., Zhao, Y., Xie, Y., Tang, Y., Jiang, X., et al. (2020). Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1673–1679. doi: 10.1007/s10096-020-03890-z
Liu, Y., Long, D., Xiang, T.-X., Du, F.-L., Wei, D. D., Wan, L.-G., et al. (2019). Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J. Antimicrob. Chemother. 74, 1233–1240. doi: 10.1093/jac/dkz023
Martin, R. M., Cao, J., Brisse, S., Passet, V., Wu, W., Zhao, L., et al. (2016). Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 1, e00261. doi: 10.1128/mSphere.00261-16
Mason, C. A., Dünner, J., Indra, P., Colangelo, T. (1999). Heat-induced expression and chemically induced expression of the escherichia coli stress protein htpG are affected by the growth environment. Appl. Environ. Microbiol. 65, 3433. doi: 10.1128/aem.65.8.3433-3440.1999
Melander, R. J., Minvielle, M. J., Melander, C. (2014). Controlling bacterial behavior with indole-containing natural products and derivatives. Tetrahedron 70, 6363–6372. doi: 10.1016/j.tet.2014.05.089
Muzaki, M. Z. B. M., Subramoni, S., Summers, S., Kjelleberg, S., Rice, S. A. (2024). Klebsiella pneumoniae AI-2 transporters mediate interspecies interactions and composition in a three-species biofilm community. NPJ Biofilms Microbiomes 10, 91. doi: 10.1038/s41522-024-00546-0
O’Toole, G. A., Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Peng, D., Li, X., Liu, P., Zhou, X., Luo, M., Su, K., et al. (2018). Transcriptional regulation of galF by RcsAB affects capsular polysaccharide formation in Klebsiella pneumoniae NTUH-K2044. Microbiological Res. 216, 70–78. doi: 10.1016/j.micres.2018.08.010
Qayed, M., Michonneau, D., Socié, G., Waller, E. K. (2021). Indole derivatives, microbiome and graft versus host disease. Curr. Opin. Immunol. 70, 40–47. doi: 10.1016/j.coi.2021.02.006
Qin, H.-L., Liu, J., Fang, W.-Y., Ravindar, L., Rakesh, K. P. (2020). Indole-based derivatives as potential antibacterial activity against methicillin-resistance Staphylococcus aureus (MRSA). Eur. J. Medicinal Chem. 194, 112245. doi: 10.1016/j.ejmech.2020.112245
Russo, T. A., Marr, C. M. (2019). Hypervirulent klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001. doi: 10.1128/CMR.00001-19
Salama, G. G., El-Mahdy, T. S., Moustafa, W. H., Emara, M. (2024). Downregulation of Klebsiella pneumoniae RND efflux pump genes following indole signal produced by Escherichia coli. BMC Microbiol. 24, 312. doi: 10.1186/s12866-024-03443-w
Shin, J., Ko, K. S. (2015). Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int. J. Antimicrobial Agents 46, 214–218. doi: 10.1016/j.ijantimicag.2015.04.012
Struve, C., Bojer, M., Krogfelt, K. A. (2008). Characterization of Klebsiella pneumoniae Type 1 Fimbriae by Detection of Phase Variation during Colonization and Infection and Impact on Virulence. Infection Immun. 76, 4055. doi: 10.1128/IAI.00494-08
Tan, Y. H., Arros, P., Berríos-Pastén, C., Wijaya, I., Chu, W. H. W., Chen, Y., et al. (2024). Hypervirulent Klebsiella pneumoniae employs genomic island encoded toxins against bacterial competitors in the gut. ISME J. 18, wrae054. doi: 10.1093/ismejo/wrae054
Tennoune, N., Andriamihaja, M., Blachier, F. (2022). Production of indole and indole-related compounds by the intestinal microbiota and consequences for the host: the good, the bad, and the ugly. Microorganisms 10, 930. doi: 10.3390/microorganisms10050930
Tschowri, N., Busse, S., Hengge, R. (2009). The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23, 522. doi: 10.1101/gad.499409
Tschowri, N., Lindenberg, S., Hengge, R. (2012). Molecular function and potential evolution of the biofilm-modulating blue light-signalling pathway of Escherichia coli. Mol. Microbiol. 85, 893. doi: 10.1111/j.1365-2958.2012.08147.x
Vega, N. M., Allison, K. R., Khalil, A. S., Collins, J. J. (2012). Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8, 431–433. doi: 10.1038/nchembio.915
Vega, N. M., Allison, K. R., Samuels, A. N., Klempner, M. S., Collins, J. J. (2013). Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. U.S.A. 110, 14420–14425. doi: 10.1073/pnas.1308085110
Vogel-Scheel, J., Alpert, C., Engst, W., Loh, G., Blaut, M. (2010). Requirement of purine and pyrimidine synthesis for colonization of the mouse intestine by escherichia coli. Appl. Environ. Microbiol. 76, 5181. doi: 10.1128/AEM.00242-10
Yaikhan, T., Chuerboon, M., Tippayatham, N., Atimuttikul, N., Nuidate, T., Yingkajorn, M., et al. (2019). Indole and derivatives modulate biofilm formation and antibiotic tolerance of Klebsiella pneumoniae. Indian J. Microbiol. 59, 460–467. doi: 10.1007/s12088-019-00830-0
Yin, W., Wang, Y., Liu, L., He, J. (2019). Biofilms: the microbial “Protective clothing” in extreme environments. Int. J. Mol. Sci. 20, 3423. doi: 10.3390/ijms20143423
Keywords: K. pneumoniae, indole, fimbriae, metabolism, virulence
Citation: Fang X, Chen Y, Hu L, Gu S, Zhu J, Hang Y, Cao X, Xiao Y, Luo H, Zhao C, Xiao L and Zhong Q (2025) Modulation of virulence and metabolic profiles in Klebsiella pneumoniae under indole-mediated stress response. Front. Cell. Infect. Microbiol. 15:1546991. doi: 10.3389/fcimb.2025.1546991
Received: 17 December 2024; Accepted: 13 January 2025;
Published: 03 February 2025.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Sudharshini Jayaraman, Bharathidasan University, IndiaCopyright © 2025 Fang, Chen, Hu, Gu, Zhu, Hang, Cao, Xiao, Luo, Zhao, Xiao and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiaoshi Zhong, emhvbmcyMDAwMDk0N0BzaW5hLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.