
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 11 March 2025
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1534084
This article is part of the Research Topic Synergistic Approaches to Managing Gram-negative Bacterial Resistance View all 14 articles
 Runzhi Zhang1,2†
Runzhi Zhang1,2† Yan Yu2,3†
Yan Yu2,3† Lulu Huang4
Lulu Huang4 Susu Chen4
Susu Chen4 Ruxi Hu4
Ruxi Hu4 Xiuxiu Wang4
Xiuxiu Wang4 Dawei Huang5
Dawei Huang5 Chunhan Song6
Chunhan Song6 Junwan Lu6
Junwan Lu6 Qiyu Bao2,3,4,6
Qiyu Bao2,3,4,6 Yunliang Hu3*
Yunliang Hu3* Pengfei Jiang1*
Pengfei Jiang1* Wei Pan5*
Wei Pan5*Background: M. morganii is a species of the genus Morganella in the family Enterobacteriaceae. This species primarily causes infections of postoperative wounds and the urinary tract. Some isolates of M. morganii exhibit resistance to multiple antibiotics due to multidrug resistance traits, complicating clinical treatment; thus, there is a growing need to elucidate the resistance mechanisms of this pathogen.
Methods: A total of 658 bacterial strains were isolated from anal fecal swabs from poultry and livestock and from the surrounding environment in Wenzhou, China, via plate streaking. The full genome sequences of the bacteria were obtained via next-generation sequencing platforms. The standard agar dilution method was employed to determine the minimum inhibitory concentrations (MICs) of various antimicrobial agents. The resistance gene (fosA13) of the isolate was identified using the Comprehensive Antibiotic Resistance Database (CARD) and confirmed via molecular cloning. The FosA13 protein encoded by the novel resistance gene fosA13 was expressed with the vector pCold I, and its enzyme kinetics parameters were characterized. The genetic background and evolutionary process of the sequence of this novel resistance gene were analyzed by means of bioinformatics methods.
Results: In this study, we identified a new chromosomally encoded fosfomycin resistance gene, designated fosA13, from the M. morganii isolate DW0548, which was isolated from poultry on a farm in Wenzhou, China. Compared with the control strain (pUCP19/DH5α), the recombinant strain carrying fosA13 (pUCP19-fosA13/DH5α) presented a fourfold increase in the MIC value for fosfomycin. The enzyme kinetics data of FosA13 revealed effective inactivation of fosfomycin, with a kcat/Km of (1.50 ± 0.02)×104 M-1·s-1. Among functionally characterized resistance proteins, FosA13 presented the highest amino acid (aa) homology (55.6%) with FosA. FosA13 also contained essential functional residues of FosA proteins. The isolate M. morganii DW0548 presented high MIC values (≥ 8 μg/mL) for 5 classes of antimicrobials, namely, aminoglycosides, β-lactams, quinolones, tetracycline, and chloramphenicol, but only two functionally characteristic antimicrobial resistance genes (ARGs) have been identified in the complete genome: a β-lactam resistance gene (blaDHA-16) and a phenol resistance gene (catII). These findings indicate that in addition to the novel resistance gene identified in this work, other uncharacterized resistance mechanisms might exist in M. morganii DW0548.
Conclusion: A novel chromosomal fosfomycin resistance gene, fosA13, was identified in an animal M. morganii isolate, and its enzymatic parameters were characterized. This protein shares the highest aa identity of 55.6% with the functionally characterized protein FosA and has all the essential functional residues of FosA proteins. Exploring more antimicrobial resistance mechanisms of this pathogen would help clinicians choose effective drugs to treat infectious diseases in animal husbandry and clinical practice and facilitate the development of methods to prevent the spread of resistance between bacteria of different species.
In 1969, a natural antibiotic named fosfomycin was first discovered in the fermentation broth of Streptomyces (Hendlin et al., 1969). Although some species can produce fosfomycin, its concentrations are generally low. Fosfomycin exhibits bactericidal properties against various bacteria, including both Gram-negative and Gram-positive bacteria such as staphylococci (Raz, 2012). Fosfomycin was a decommissioned antibiotic, however, given the increasing prevalence of multidrug-resistant uropathogens, the limited treatment options available, and the lack of new antibiotics, older antibiotics need to be reevaluated. Owing to its unique bactericidal mechanism and physicochemical properties, fosfomycin has the advantages of no cross-resistance, strong antibacterial activity, a wide tissue distribution, and synergistic bactericidal effects when used in combination with other drugs, and was defined by the World Health Organization (WHO) as a “vital” antibiotic (Collignon et al., 2016), which has attracted the interest of many clinicians. With an increase in the frequency of clinical fosfomycin use, resistance to fosfomycin has also increased in some bacteria, such as Acinetobacter, Vibrio fischeri, Chlamydia trachomatis, and so on (Silver, 2017).
The earliest case of fosfomycin resistance dates back to 1977. Since then, there have been epidemics of drug-resistant strains in all countries worldwide (Aghamali et al., 2019). Although the mechanism of action and the structure of fosfomycin are unique, which made the cross-resistance uncommon (Falagas et al., 2020), with the increased use of fosfomycin, bacterial resistance to it has also increased rapidly. Data show that the use of fosfomycin in the treatment of urinary tract infections caused by Escherichia coli, as well as in the treatment of some uropathogen infections, leads to an increase in fosfomycin resistance (Jiang et al., 2015).
Several categories of fosfomycin drug resistance mechanisms have been characterized (Karageorgopoulos et al., 2012). These mechanisms involve reducing drug absorption, altering drug binding targets, and inactivating fosfomycin. Resistance to fosfomycin is typically associated with the inactivation of fosfomycin by modifying enzymes (such as fosA, fosB, fosC, and fosX) (Yang et al., 2019; Findlay et al., 2023), and kinases (fomA and fomB) (Kobayashi et al., 2000). FosA is a dimeric glutathione S-transferase (GST) that catalyzes the binding of glutathione to fosfomycin with the help of Mn2+ and K+ ions, inactivating fosfomycin, and it can be encoded on either a plasmid or a chromosome (Suárez and Mendoza, 1991). FosA was first found to be present in the plasmid Tn2921 transposon of Serratia marcescens, and it is predominantly present in Enterobacteriaceae, Pseudomonas spp (Biggel et al., 2021). The FosA proteins are a number of metalloenzymes able to disrupt the epoxide ring of fosfomycin. It depends on manganese (II) and potassium as cofactors, and glutathione (GSH) as a nucleophilic molecule (Mattioni Marchetti et al., 2023). The fosA genes are commonly distributed in Providencia stuartii, Providencia rettgeri, Klebsiella pneumoniae, Klebsiella oxytoca, Serratia marcescens, Enterobacter aerogenes and Enterobacter cloacae, however, they are rarely reported in Citrobacter freundii, Proteus mirabilis and Acinetobacter baumannii (Zurfluh et al., 2020). FosB is an L-cysteine thioltransferase that inactivates fosfomycin via the nucleophilic addition of thiols to fosfomycin with the help of Mg2+ (Michalopoulos et al., 2011). FosC is a protease inactivating fosfomycin by adding a phosphate group to it by using ATP as a substrate (García et al., 1995). FosX is a Mn2+-dependent epoxide hydrolase that inactivates fosfomycin by adding a hydroxyl group to the fosfomycin C1 position and opening its epoxide ring, using water as a substrate (Rigsby et al., 2005). FomA and FomB are kinases involved in the degradation of fosfomycin, and the role of these kinases may be to protect fosfomycin producers from fosfomycin (Kobayashi et al., 2000). Chromosomal mutations can affect the transport function of the cell membrane, leading to reduced intracellular levels of fosfomycin. Two transport systems for the uptake of fosfomycin into cells, which involve glycerol-3-phosphate transporter protein (GlpT) and hexose phosphate transporter protein (UhpT), are present in E. coli (Takahata et al., 2010). Altering the drug’s target of action is another mechanism of fosfomycin resistance. Among Gram-positive bacteria, the affinity between the MurA protein and fosfomycin in S. aureus is reduced by mutations in the murA gene (Xin et al., 2022); however, elevated expression levels of this gene can lead to bacterial resistance to fosfomycin (Raina et al., 2021).
M. morganii, a pathogen that was first isolated from pediatric fecal cultures by Morgan et al. in 1906 (Morgan and de, 1906), is a parthenogenetic anaerobic rod-shaped Gram-negative enteric bacterium that produces virulence factors such as hemolysin and causes urinary tract wound infections, and the risk of infection by M. morganii has been highlighted in many epidemiological data. Clinical disease caused by multidrug-resistant (MDR) or even extensively drug-resistant (XDR) M. morganii often results in treatment failure. Studies have shown that the development of intrinsic and acquired multidrug resistance is of concern as the prevalence of M. morganii infections increases, necessitating the global identification of M. morganii as a major pathogen (Liu et al., 2016a; Bandy, 2020a; Li et al., 2023). Rising rates of M. morganii infections are a reminder not only of the need for increased precautions in public areas but also of the need to include this microorganism in the differential diagnosis list in the clinical setting (Bandy, 2020).
In this work, we report a novel chromosomally encoded fosfomycin gene, fosA13, in the isolate DW0548, which was isolated from a farm animal. DW0548 was subjected to whole-genome sequencing for genome-wide characterization. To determine the function of the fosA13 gene, molecular cloning was performed. The protein encoded by fosA13 was expressed, and its enzyme kinetics were also analyzed.
To analyze the drug resistance status of bacteria isolated from animals and the environment of the animal farms, 658 bacterial strains were obtained from the poultry and livestock anal fecal swabs, and sewage and soil of the animal farms in Wenzhou, China. An fosA-like gene, designated fosA13, was found in an isolate from poultry named DW0548. Bacterial species identification was performed via 16S rRNA gene homology and genome-wide average nucleotide identity (ANI) analyses. Table 1 lists the plasmids and strains used in this study.
The MICs of the antimicrobials were determined via the agar dilution method according to Clinical Laboratory Standards Institute (CLSI) guidelines (CLSI, 2024). Medium with glucose-6-phosphate (G6P) at a constant concentration of 25 μg/mL was used when the MIC of fosfomycin was tested. E. coli ATCC 25922 was used as a quality control. Table 2 shows the MIC data for the 25 antibiotics from the six antibacterial categories.
The bacteria were cultured in liquid LB medium and incubated at 37°C for 16 h. Total DNA was extracted from the bacteria with the Generay Genomic DNA Small Volume Preparation Kit (Shanghai Generay Biotech Co., Ltd., Shanghai, China). Whole-genome sequencing was completed on the Illumina NovaSeq and PacBio RS II platforms at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The Illumina short reads and the PacBio long reads were assembled using MEGAHIT v1.2.9 (Li et al., 2016) and Trycycler v0.5.1 (Wick et al., 2021), respectively. Using Pilon v1.24, the quality of the genome sequence from PacBio sequencing was corrected by mapping the Illumina short reads onto the PacBio read assembly (Walker et al., 2014). The open reading frames (ORFs) were predicted with Prokka v1.14.6 (Seemann, 2014). The functions of the predicted proteins were annotated by searching the ORFs against the NCBI nonredundant protein database with DIAMOND v2.0.11 (Buchfink et al., 2021). The promoter region of a gene was predicted using the BPROM tool (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb). Annotation of drug resistance genes was conducted by using Resistance Gene Identifier v5.2.0 (RGI) and the Comprehensive Antibiotic Resistance Database (CARD) database (https://github.com/arpcard/rgi) (McArthur et al., 2013). Homology analysis of the 16S rRNA gene from the target genome was performed by comparison with the 16S ribosomal RNA sequence database in NCBI (McArthur et al., 2013). The ANI was computed using FastANI v1.33 (Jain et al., 2018). Calculation of digital DNA−DNA hybridization (dDDH) values was performed on the basis of the Type strain Genome Server (TYGS) online database (Type Strain Genome Server) (Lian et al., 2021). Multiple sequence alignment of fosA13, fosA, fosA2, and other related genes was performed with MAFFT v7.487 (Katoh and Standley, 2013). A neighbor-joining (N-J) phylogenetic tree including FosA13 and other functionally characterized fosfomycin resistance enzymes was constructed with MEGA11 (Kumar et al., 2018). The phylogenetic tree of FosA13 with other Fos proteins was visualized with the online website iTol (iTOL: Interactive Tree Of Life) (Letunic and Bork, 2021).
Referring to a previous publication (Zhao et al., 2023), primers to clone the predicted resistance gene with its promoter region and the primers to clone the open reading frame (ORF) of the gene were designed. The PCR products were amplified and then inserted into the vectors pUCP19 and pCold I, respectively, via the DNA ligation kit Ver.2.1 (Takara Bio, Inc., Dalian, China). The recombinant plasmids pUCP19-fosA13 and pCold I-fosA13 were transformed into E. coli DH5α and E. coli BL21, respectively. The inserted sequences in the recombinants were verified by Sanger sequencing. Table 3 shows the primer sequences and details.
The methods used to express and purify the recombinant FosA13 protein were mainly based on a previous publication (Gao et al., 2022). In brief, the recombinant strain (pCold I-fosA13/BL21) was cultured in LB broth. When the OD value of the culture reached between 0.45 and 0.55, the expression of the protein was induced by 1 mM isopropyl-β-dithiogalactopyranoside (IPTG). Bacteria were harvested by centrifugation (4,000 × g, 10 min) at 4°C, resuspended in 5 mL of nondenaturing lysis buffer, and fragmented by ultrasonication for 10 min at 4°C. After centrifugation, the supernatant containing the recombinant protein was collected. The recombinant protein was purified with BeyoGold His-tag purification resin using a nondenaturing elution solution from a His-tag protein purification kit (Beyotime, Shanghai, China). The His-tag of the recombinant protein was removed by enterokinase (EK enzyme). The purity of the protein was determined via SDS−PAGE, and the protein concentration was subsequently determined using a BCA protein concentration assay kit (Beyotime, Shanghai, China).
The kinetic parameters of purified FosA13 with fosfomycin were analyzed via high-performance liquid chromatography (HPLC) using a Thermo Scientific AcceLA HPLC system (Thermo Fisher Scientific, Inc., China) with a 100 μL final reaction volume at 37°C. The 100 μL reaction system consisted of 10 mM GSH, 250 µm MnCl2, 100 mM KCl, and 100 mM Tris-HCl, and gradient concentrations of fosfomycin (0, 25, 50, 100, 200, 400, and 800 µM) were added in a volume of 10 μL. After the reaction system was preheated for 20 minutes, the purified FosA13 protein was added. The reaction was conducted for 5 minutes, and the assay was carried out on the analysis system. The mobile phases were a mixture containing K2HPO4 (200 mM), KH2PO4 (200 mM), acetonitrile and methanol at a percentage volume ratio of 1.68: 78.32: 10: 10 (Arca et al., 1990; Bernat et al., 1997, Bernat et al., 1999). The steady-state kinetic parameters kcat and Km were nonlinearly regressed against the initial reaction rate via the Michaelis−Menten equation in Prism (version 8.0.2) software (GraphPad software, CA, United States). The value is the average of three independent measures.
The GenBank accession numbers for the novel fosfomycin fosA13 gene, the plasmid pMMDW0548 and the chromosome sequences of DW0548 are PQ600006, CP173708 and CP173707, respectively.
To clarify the mechanisms of antimicrobial resistance in bacteria isolated from animals and the environment, we sequenced 658 bacterial isolates isolated from poultry and livestock anal fecal swabs and environmental samples from animal farms in Wenzhou, China. Annotation of genomic data revealed resistance genes against different antibiotics. Notably, among the predicted genes were numerous putative fosfomycin resistance genes, including but not limited to homologs of fosA, fosB, fosC, and fosX. These genes shared less than 80.0% aa sequence identity with functionally characterized fosfomycin resistance genes. Some of these genes, including the fosA2, fosA5, fosB, fosLC2 and fosL1 homologs, were randomly selected and cloned, and their resistance functions were determined. Finally, a gene homologous to fosA2 (designated fosA13 in this work) that conferred resistance to fosfomycin was identified, and it was encoded in the chromosome of an isolate named DW0548.
To confirm the resistance function of this gene, the ORF with its promoter region was cloned into the vector pUCP19, and the recombinant plasmid containing fosA13 was transformed into E. coli DH5α cells. The transformant strain carrying fosA13 (pUCP19-fosA13/E. coli DH5α) showed a 4-fold increase in the MIC value to fosfomycin (2 μg/mL) compared with that of the control strain (pUCP19/DH5α, 0.5 μg/mL) (Table 2). The MICs of the previously cloned fosfomycin resistance genes varied. Compared with the recipients, the cloned fosY (Chen et al., 2022) and fosA6 (Guo et al., 2016a) genes increased the MIC levels by 16- and 32-fold, respectively, whereas fosI (Pelegrino et al., 2016) and fosA7 (Rehman et al., 2017) increased the MIC levels by 128- and >256-fold, respectively (Supplementary Table S1).
16S rRNA gene homology analysis revealed that the 16S rRNA gene of the isolate DW0548 shared the highest similarity (96.0% coverage and 99.0% identity) with that of M. morganii M11 (NR_028938.1). In addition, ANI analysis of all 759 Morganella genomes in the NCBI database revealed that 78 of these genomes had ≥ 95.0% ANI (the threshold value to define a bacterial species) with the isolate DW0548 genome. Seventy-three of these genomes were from the M. morganii genomes. The result of the digital DNA–DNA hybridization (dDDH) analysis revealed that the isolate DW0548 presented the highest dDDH value (75.7%) with M. morganii NBRC 3848, which was greater than the threshold (70.0%) for classifying a bacterial species. Therefore, on the basis of the results above, the isolate DW0548 was ultimately included in the species M. morganii and was thus designated M. morganii DW0548.
The whole genome of M. morganii DW0548 consists of a chromosome and a plasmid named pMM548. The chromosome size was approximately 4.31 Mb, and the average GC content was 50.35%, with 4,380 coding sequences (CDSs). The plasmid was 96,349 bp in length with the average GC content of 50.45%, and it encoded 109 CDSs (Table 4).
The result of the susceptibility test demonstrated that M. morganii DW0548 had high MICs (≥ 8 μg/mL) for 11 of the 25 antimicrobials tested, which included members of 5 classes of antimicrobials, with 4 aminoglycosides (streptomycin, kanamycin, spectinomycin and paromomycin), 4 β-lactams (cefazolin, cefothiophene, cefoxitin and cefuroxime), 1 quinolone (naphridixic acid), tetracycline, and chloramphenicol (Table 2).
In examining the relationship between the drug resistance phenotype and genotype, we found that even though the bacterium presented high MICs for antimicrobials from the 5 classes tested, only two genes (a β-lactam resistance gene, blaDHA-16, and a phenicol resistance gene, catII), which shared ≥ 80% aa similarity with functionally characterized antimicrobial resistance genes (ARGs), were identified. No aminoglycoside, tetracycline or quinolone resistance genes were found in the whole-genome sequence (Supplementary Table S2). Similar to the isolate DW0548, other M. morganii strains have been reported to be resistant to β-lactams, aminoglycosides, tetracyclines, fluoroquinolones, fosfomycin, and other types of antibiotics (Liu et al., 2016).
A comparison of FosA13 with these functionally characterized proteins in the CARD revealed that it had the greatest aa sequence similarities with FosA (55.6%), followed by FosA2 (55.2%), FosA3 (55.2%), FosA4 (55.2%), FosA5 (55.2%), FosA6 (55.2%), FosA7 (52.9%), FosA7.5 (52.9%), and FosA8 (52.9%). Sixteen function and structure essential residues of FosA (Beharry and Palzkill, 2005a) are conserved in FosA13. Half of them act as ligands for Mn2+ (His7, His64 and Glu110) and K+ (Ser94, Ser98 and Glu95) and ligands (Arg119 and Tyr100, within the hydrogen-bonding site of fosfomycin) involved in FosA binding to fosfomycin, and the other half of the residues (W34, Y39, W46, C48, Y65, D103, H107, Y128) were located in the putative fosfomycin/GSH binding channel. Although FosA13 shares only about 50–60% identity with these functionally characterized FosA proteins, the active site residues essential for FosA function remain unchanged (Figure 1). Further analysis of the evolutionary relationship between FosA13 and different glutathione transferases revealed that FosA13 was most similar to FosA2 and formed a new branch (Figure 2).
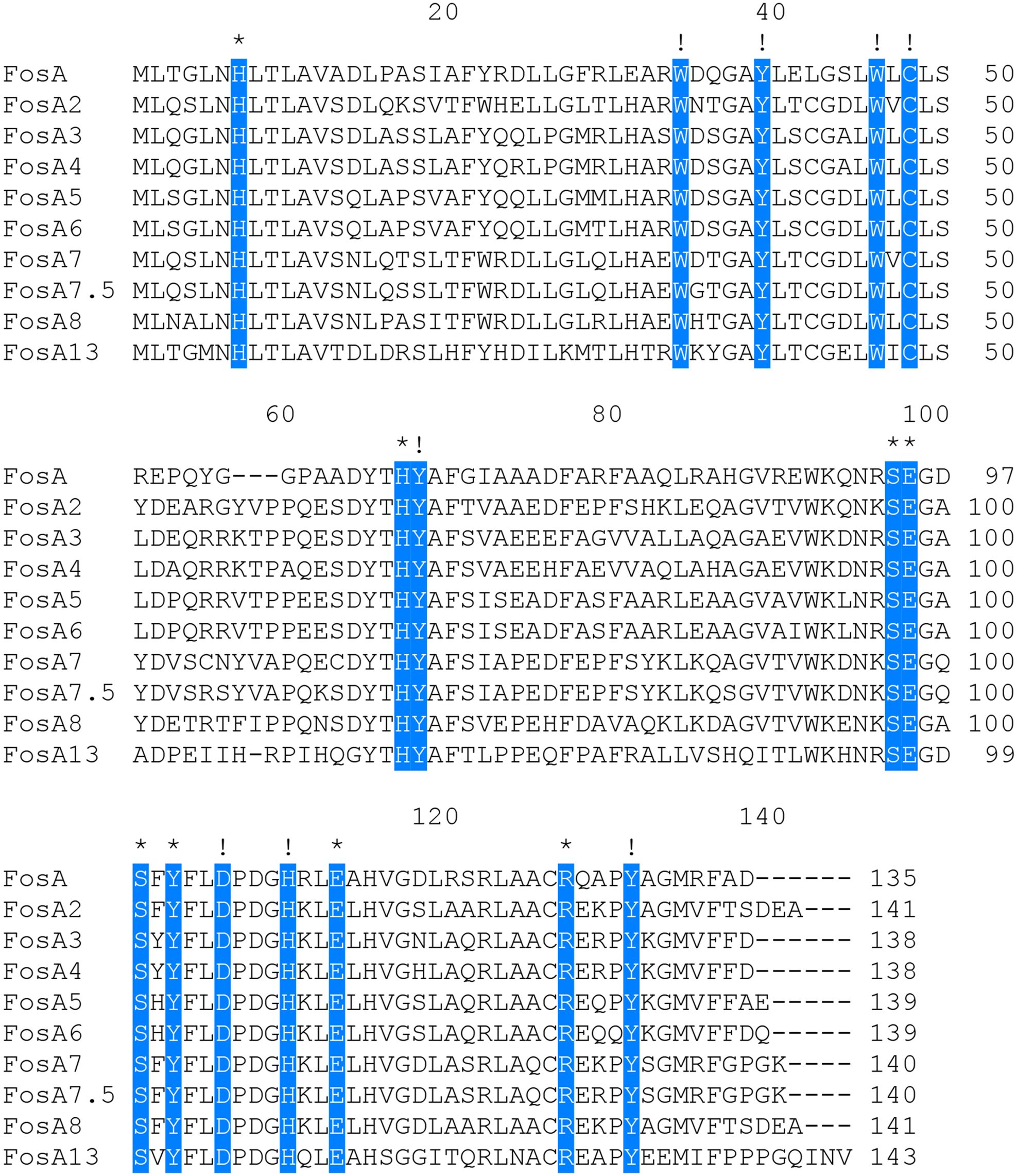
Figure 1. Multiple alignment of the deduced amino acid sequences of FosA13 and its close relatives. The 16 amino acids shaded in blue are function and structure essential residues of FosA proteins, of which those indicated by asterisks are residues that act as ligands for Mn2+, K+ and fosfomycin and those indicated by exclamation points are residues located in the putative fosfomycin/GSH-binding channel. Spaces are indicated by hyphens. The numbers on the right represent the corresponding sequence lengths.
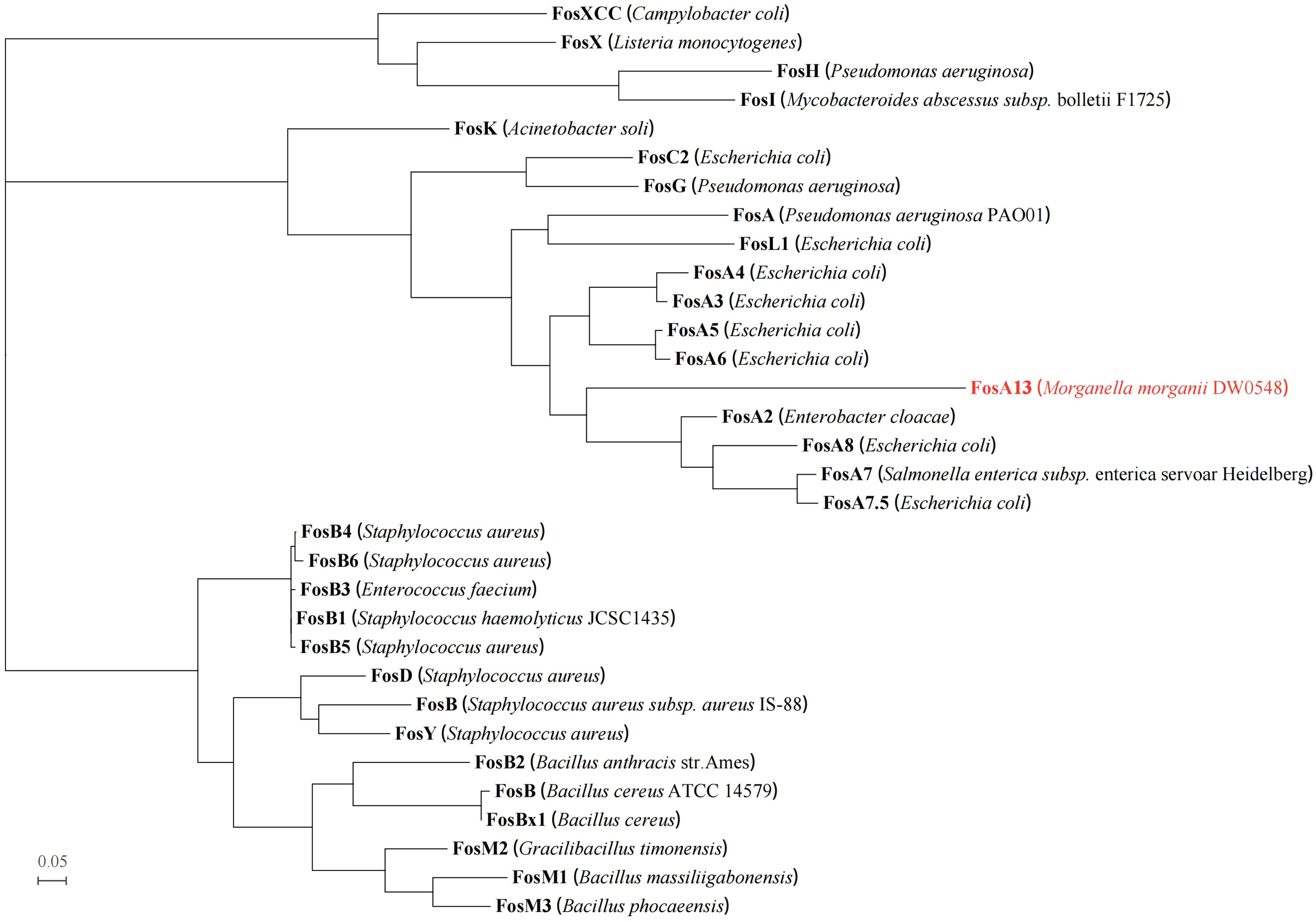
Figure 2. Phylogenetic tree showing the relationship of FosA13 with other functionally characterized proteins (with identities between 20.0% and 60.0%). FosA13 is highlighted in red. The other proteins include FosA (AAG04518.1), FosA2 (ACC85616.1), FosA3 (AEG78825.1), FosA4 (BAP18892.1), FosA5 (AJE60855.1), FosA6 (AMQ12811.1), FosA7 (KKE03230.1), FosA7.5 (ANQ03635.1), FosA8 (QEI22965.1), FosB (EHS19134.1, S. aureus), FosB (AAP08996.1, (B) cereus), FosB1 (BAE05988.1), FosB2 (AAP27834.1), FosB3 (ADX95999.1), FosB4 (ALM24139.1), FosB5 (ALN12426.1), FosB6 (ALM24145.1), FosBx1 (QLF01382.1), FosC2 (BAJ10053.1), FosD (BAG12271.1), FosG (RTB44598.1), FosH (ADF48907.1), FosI (AFJ38137.1), FosK (BAO79518.1), FosL1 (QHR93773.1), FosM1 (DAC85639.1), FosM2 (DAC85640.1), FosM3 (DAC85641.1), FosX (CWV56762.1), FosXCC (AIF29598.1), and FosY (QTE33800.1).
The fosA13 gene is 432 bp in length and encodes a 143 aa protein (FosA13). The predicted molecular weight of the mature glutathione-S-transferase is 16.48 kDa, with a pI of 6.08. The purified FosA13 has the ability to hydrolyze fosfomycin, with a Km of 0.427 ± 0.007 µM, kcat of 6.43 ± 0.04 s-1 and kcat/Km of (1.50 ± 0.02) × 104 M-1. s-1. In terms of enzyme kinetics, compared with the other two FosA proteins, FosA13 showed 6-fold lower hydrolytic activity against fosfomycin than that of FosA (Beharry and Palzkill, 2005b) (kcat/Km of 1.5 × 104 M-1). s-1 vs. 9.0 × 104 M-1. s-1) and approximately 5-fold lower than that of FosA6 (Guo et al., 2016) (kcat/Km of 1.5 × 104 M-1. s-1 vs. 0.3 × 104 M-1. s-1).
To investigate the distribution of fosA13-homologous genes, the nucleotide sequence of fosA13 was used as a query to search for similar genes in the NCBI nucleotide database. As a result, a total of 82 similar genes with similarities between 87.3% and 100.0% were obtained, all of which were derived from the M. morganii genomes (Supplementary Table S3). Among these 82 genes, only one had a similarity of 87.3%, and all the others had similarities of ≥ 92.13%. The gene context of the fosA13-homologous genes was further analyzed. The 20-kb sequences with the fosA13-homologous genes at the center were intercepted and clustered. Finally, these 83 20-kb fragments (including the gene identified in this study) were grouped into 13 clusters with a similarity threshold of 90.0%. The gene context of 13 sequences consisting of one from each of the 13 clusters was compared (Figure 3, Supplementary Table S3).
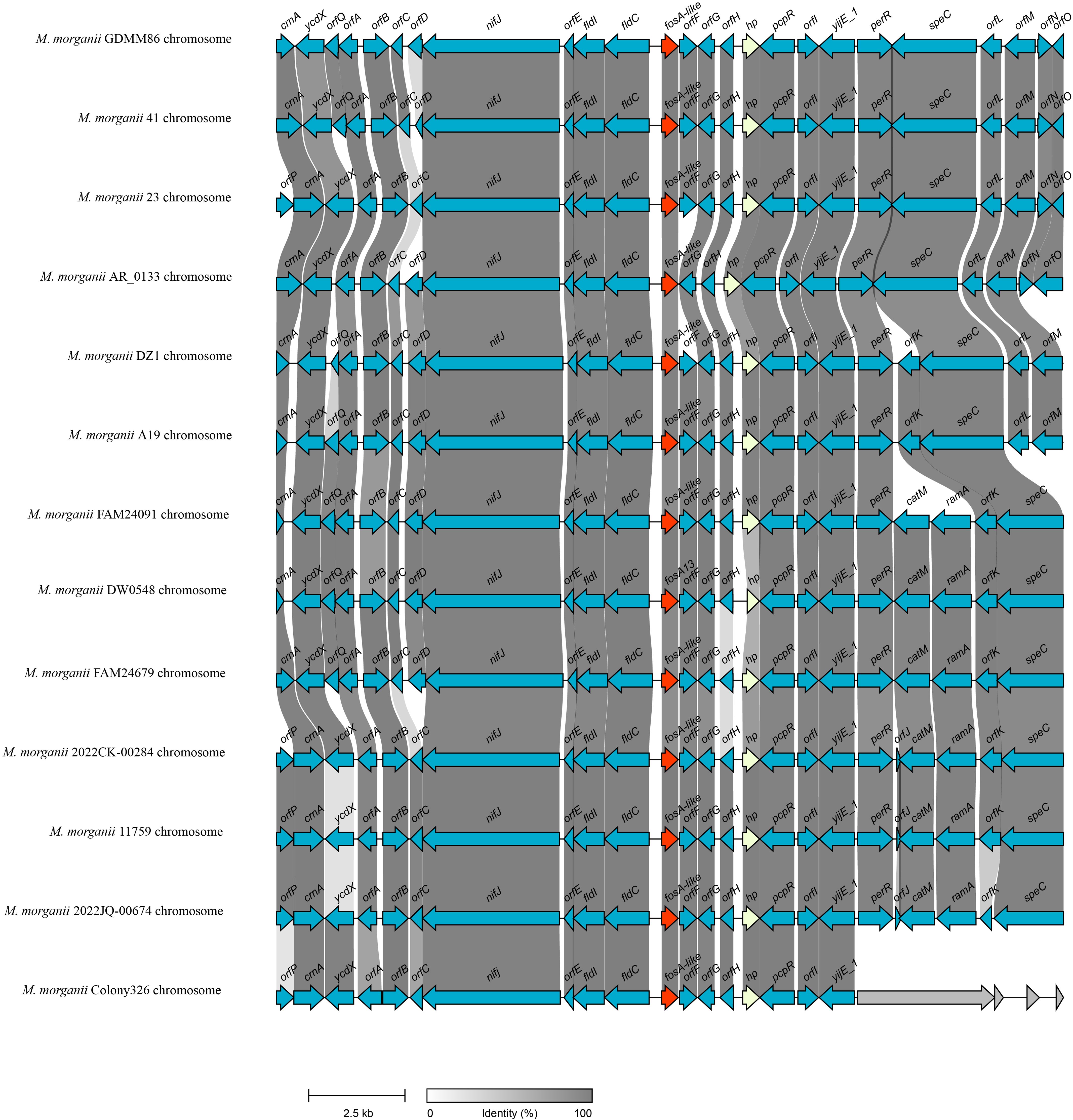
Figure 3. Genetic environment of fosA13 and fosA13-homologous genes. The fosA genes are in red. orfA, DUF523 domain-containing protein; orfB, HAD family hydrolase; orfC, DUF5339 domain-containing protein; orfD, GNAT family N-acetyltransferase; orfE, DUF3343 domain-containing protein; orfF, glyoxalase; orfG, carboxymuconolactone decarboxylase family protein; orfH, DMT family protein; orfI, DMT family transporter; orfJ, aminoglycoside 6’-acetyltransferase; orfK, SMI1/KNR4 family protein; orfL, GNAT family N-acetyltransferase; orfM, SDR family NAD(P)-dependent oxidoreductase; orfN, MerR family transcriptional regulator; orfO, methyl-accepting chemotaxis protein; orfP, cytosine permease; orfQ, glyoxalase/bleomycin resistance/dioxygenase family protein.
As mentioned above and illustrated in Figure 3, the fosA13 and fosA13-homologous genes and their surrounding sequences were relatively conserved in the M. morganii genomes. Further structural analysis revealed that no mobile genetic element (GME) was present in their flanking regions. Upstream of fosA13 are genes encoding proteins related to nifj [ferredoxin (flavodoxin) oxidoreductase], fldI [phenyllactate dehydratase activator] and fld [(R)-phenyllactyl-CoA dehydratase beta subunit], whereas downstream of fosA13 are genes related to pcpR (PCP degradation transcriptional activation protein), yijE_1 (putative cystine transporter YijE_1) and perR (HTH-type transcriptional regulator PerR). However, many other fos genes, such as fosC2 (Wachino et al., 2010), fosA3 (Wachino et al., 2010), fosA5 (Ma et al., 2015), fosA6 (Guo et al., 2016) and fosA8 (Poirel et al., 2019), are related to MGEs and are encoded on plasmids.
This paper presents the discovery of a novel chromosomal fosfomycin resistance gene, designated fosA13, from an animal M. morganii isolate. Although fosA13 encodes a protein that shares less than 80% aa similarity with functionally characterized FosA proteins, the function and structure essential residues of these proteins are conserved within it. Many fosA-type genes are located on plasmids of different bacterial species. These genes are easily captured by mobile genetic elements and transmitted between bacteria of different species by means of horizontal gene transfer, which results in widespread resistance. Identifying more resistance mechanisms would greatly benefit the treatment of bacterial infections in the clinic and the monitoring of resistance transmission.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
This study used strains obtained from poultry, livestock anal fecal swabs, and the environment in animal farms in Wenzhou, China. The owners of the farms were written informed of the study and expressed approval for sampling of animals. The studies involving human participants and animals were reviewed and approved by the Animal Welfare and Ethics Committee of Wenzhou Medical University, Zhejiang Province, China (Protocol number: wydw2021-0323).
RZ: Writing – original draft, Writing – review & editing, Data curation, Investigation, Supervision, Validation, Visualization. YY: Writing – original draft, Writing – review & editing, Data curation, Investigation, Supervision, Validation, Visualization. LH: Writing – review & editing, Investigation, Supervision, Validation. SC: Writing – review & editing, Investigation, Supervision, Validation. RH: Writing – review & editing, Investigation, Supervision, Validation. XW: Writing – review & editing, Investigation, Supervision, Validation. DH: Writing – review & editing, Investigation, Supervision, Validation. CS: Writing – review & editing, Data curation, Visualization. JL: Writing – review & editing, Data curation, Visualization. QB: Writing – original draft, Writing – review & editing, Conceptualization, Methodology. YH: Writing – review & editing, Conceptualization, Methodology. PJ: Writing – review & editing, Conceptualization, Methodology. WP: Writing – original draft, Writing – review & editing, Conceptualization, Methodology.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Science & Technology Project of Jinhua City, China (2023-3-159, 2022-2-013), the Science and Technology Plan Project of Taizhou (21ywb128), Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2023KY1350) and the Science & Technology Project of Wenzhou City, China (N20210001).
The authors would like to acknowledge all study participants and individuals who contributed to this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1534084/full#supplementary-material
Aghamali, M., Sedighi, M., Zahedi Bialvaei, A., Mohammadzadeh, N., Abbasian, S., Ghafouri, Z., et al. (2019). Fosfomycin: mechanisms and the increasing prevalence of resistance. J. Med. Microbiol. 68, 11–25. doi: 10.1099/jmm.0.000874
Arca, P., Hardisson, C., Suárez, J. E. (1990). Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob. Agents Chemother. 34, 844–848. doi: 10.1128/AAC.34.5.844
Bandy, A. (2020). Ringing bells: Morganella morganii fights for recognition. Public Health 182, 45–50. doi: 10.1016/j.puhe.2020.01.016
Beharry, Z., Palzkill, T. (2005a). Functional analysis of active site residues of the fosfomycin resistance enzyme fosA from pseudomonas aeruginosa*. J. Biol. Chem. 280, 17786–17791. doi: 10.1074/jbc.M501052200
Beharry, Z., Palzkill, T. (2005b). Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J. Biol. Chem. 280, 17786–17791. doi: 10.1074/jbc.M501052200
Bernat, B. A., Laughlin, L. T., Armstrong, R. N. (1997). Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry 36, 3050–3055. doi: 10.1021/bi963172a
Bernat, B. A., Laughlin, L. T., Armstrong, R. N. (1999). Elucidation of a monovalent cation dependence and characterization of the divalent cation binding site of the fosfomycin resistance protein (FosA). Biochemistry 38, 7462–7469. doi: 10.1021/bi990391y
Biggel, M., Zurfluh, K., Treier, A., Nüesch-Inderbinen, M., Stephan, R. (2021). Characteristics of fosA-carrying plasmids in E. coli and Klebsiella spp. isolates originating from food and environmental samples. J. Antimicrob. Chemother. 76, 2004–2011. doi: 10.1093/jac/dkab119
Buchfink, B., Reuter, K., Drost, H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18 (4), 366–368. doi: 10.1038/s41592-021-01101-x
Chen, Y., Ji, S., Sun, L., Wang, H., Zhu, F., Chen, M., et al. (2022). The novel fosfomycin resistance gene fosY is present on a genomic island in CC1 methicillin-resistant Staphylococcus aureus. Emerg. Microbes Infect. 11, 1166–1173. doi: 10.1080/22221751.2022.2058421
Collignon, P. C., Conly, J. M., Andremont, A., McEwen, S. A., Aidara-Kane, A., for the World Health Organization Advisory Group, B. M. @ on I. S. @ of A. R. (WHO-A, et al. (2016). World health organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 63, 1087–1093. doi: 10.1093/cid/ciw475
Falagas, M. E., Vouloumanou, E. K., Samonis, G., Vardakas, K. Z. (2020). Fosfomycin. Clin. Microbiol. Rev. 29, 321–347. doi: 10.1128/cmr.00068-15
Findlay, J., Sierra, R., Raro, O. H. F., Aires-de-Sousa, M., Andrey, D. O., Nordmann, P. (2023). Plasmid-mediated fosfomycin resistance in Escherichia coli isolates of worldwide origin. J. Glob. Antimicrob. Resist. 35, 137–142. doi: 10.1016/j.jgar.2023.09.003
Gao, M., Feng, C., Ji, Y., Shi, Y., Shi, W., Zhang, L., et al. (2022). AadA36, a novel chromosomal aminoglycoside nucleotidyltransferase from a clinical isolate of Providencia stuartii. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1035651
García, P., Arca, P., Evaristo Suárez, J. (1995). Product of fosC, a gene from Pseudomonas syringae, mediates fosfomycin resistance by using ATP as cosubstrate. Antimicrob. Agents Chemother. 39, 1569–1573. doi: 10.1128/AAC.39.7.1569
Guo, Q., Tomich, A. D., McElheny, C. L., Cooper, V. S., Stoesser, N., Wang, M., et al. (2016). Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J. Antimicrob. Chemother. 71, 2460–2465. doi: 10.1093/jac/dkw177
Hendlin, D., Stapley, E. O., Jackson, M., Wallick, H., Miller, A. K., Wolf, F. J., et al. (1969). Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science 166, 122–123. doi: 10.1126/science.166.3901.122
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9 (1), 5114 doi: 10.1038/s41467-018-07641-9
Jiang, Y., Shen, P., Wei, Z., Liu, L., He, F., Shi, K., et al. (2015). Dissemination of a clone carrying a fosA3-harbouring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. Int. J. Antimicrob. Agents 45, 66–70. doi: 10.1016/j.ijantimicag.2014.08.010
Karageorgopoulos, D. E., Wang, R., Yu, X., Falagas, M. E. (2012). Fosfomycin: evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J. Antimicrob. Chemother. 67, 255–268. doi: 10.1093/jac/dkr466
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kobayashi, S., Kuzuyama, T., Seto, H. (2000). Characterization of the fomA and fomB gene products from Streptomyces wedmorensis, which confer fosfomycin resistance on Escherichia coli. Antimicrob. Agents Chemother. 44, 647–650. doi: 10.1128/AAC.44.3.647-650.2000
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, D., Luo, R., Liu, C.-M., Leung, C.-M., Ting, H.-F., Sadakane, K., et al. (2016). MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods San Diego Calif 102, 3–11. doi: 10.1016/j.ymeth.2016.02.020
Li, C., Wang, H., Zhang, J., Wang, Z., Wei, Y., Zhu, Y. (2023). Endocarditis induced by M. morganii in an immunocompetent patient without underlying valvular abnormalities. Heliyon 9, e17069. doi: 10.1016/j.heliyon.2023.e17069
Lian, F.-B., Jiang, S., Zhu, K.-L., Shang, D.-D., Zhang, J., Du, Z.-J. (2021). Salegentibacter maritimus sp. nov., isolated from marine coastal sediment. Syst. Appl. Microbiol. 44, 126209. doi: 10.1016/j.syapm.2021.126209
Liu, H., Zhu, J., Hu, Q., Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
Ma, Y., Xu, X., Guo, Q., Wang, P., Wang, W., Wang, M. (2015). Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett. Appl. Microbiol. 60, 259–264. doi: 10.1111/lam.12366
Mattioni Marchetti, V., Hrabak, J., Bitar, I. (2023). Fosfomycin resistance mechanisms in Enterobacterales: an increasing threat. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1178547
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Michalopoulos, A. S., Livaditis, I. G., Gougoutas, V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15, e732–e739. doi: 10.1016/j.ijid.2011.07.007
Morgan, H., de, R. (1906). Report xcv. Upon the bacteriology of the summer diarrhoea of infants. Br. Med. J. 1, 908–912. doi: 10.1136/bmj.1.2364.908
Pelegrino, K., de, O., Campos, J. C., Sampaio, S. C. F., Lezirovitz, K., Seco, B. M., et al. (2016). fosI is a new integron-associated gene cassette encoding reduced susceptibility to fosfomycin. Antimicrob. Agents Chemother. 60, 686–688. doi: 10.1128/AAC.02437-15
Poirel, L., Vuillemin, X., Kieffer, N., Mueller, L., Descombes, M.-C., Nordmann, P. (2019). Identification of FosA8, a Plasmid-Encoded Fosfomycin Resistance Determinant from Escherichia coli, and Its Origin in Leclercia adecarboxylata. Antimicrob. Agents Chemother. 63, e01403–e01419. doi: 10.1128/AAC.01403-19
Raina, D., Tiwari, H., Sharma, S., Deepika, N., Chinthakindi, P. K., Nargotra, A., et al. (2021). Screening of compound library identifies novel inhibitors against the MurA enzyme of Escherichia coli. Appl. Microbiol. Biotechnol. 105, 3611–3623. doi: 10.1007/s00253-021-11272-4
Raz, R. (2012). Fosfomycin: an old–new antibiotic. Clin. Microbiol. Infect. Off. Publ. Eur. Soc Clin. Microbiol. Infect. Dis. 18, 4–7. doi: 10.1111/j.1469-0691.2011.03636.x
Rehman, M. A., Yin, X., Persaud-Lachhman, M. G., Diarra, M. S. (2017). First Detection of a Fosfomycin Resistance Gene, fosA7, in Salmonella enterica Serovar Heidelberg Isolated from Broiler Chickens. Antimicrob. Agents Chemother. 61, e00410–e00417. doi: 10.1128/AAC.00410-17
Rigsby, R. E., Fillgrove, K. L., Beihoffer, L. A., Armstrong, R. N. (2005). Fosfomycin resistance proteins: a nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily. Methods Enzymol. 401, 367–379. doi: 10.1016/S0076-6879(05)01023-2
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silver, L. L. (2017). Fosfomycin: mechanism and resistance. Cold Spring Harb. Perspect. Med. 7, a025262. doi: 10.1101/cshperspect.a025262
Suárez, J. E., Mendoza, M. C. (1991). Plasmid-encoded fosfomycin resistance. Antimicrob. Agents Chemother. 35, 791–795. doi: 10.1128/AAC.35.5.791
Takahata, S., Ida, T., Hiraishi, T., Sakakibara, S., Maebashi, K., Terada, S., et al. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 35, 333–337. doi: 10.1016/j.ijantimicag.2009.11.011
Wachino, J., Yamane, K., Suzuki, S., Kimura, K., Arakawa, Y. (2010). Prevalence of Fosfomycin Resistance among CTX-M-Producing Escherichia coli Clinical Isolates in Japan and Identification of Novel Plasmid-Mediated Fosfomycin-Modifying Enzymes. Antimicrob. Agents Chemother. 54, 3061–3064. doi: 10.1128/AAC.01834-09
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Judd, L. M., Cerdeira, L. T., Hawkey, J., Méric, G., Vezina, B., et al. (2021). Trycycler: consensus long-read assemblies for bacterial genomes. Genome Biol. 22, 266. doi: 10.1186/s13059-021-02483-z
Xin, L., Hu, Z., Han, R., Xu, X., Wang, C., Li, D., et al. (2022). Asp50Glu mutation in MurA results in fosfomycin resistance in Enterococcus faecium. J. Glob. Antimicrob. Resist. 30, 50–55. doi: 10.1016/j.jgar.2022.05.026
Yang, T.-Y., Lu, P.-L., Tseng, S.-P. (2019). Update on fosfomycin-modified genes in Enterobacteriaceae. J. Microbiol. Immunol. Infect. 52, 9–21. doi: 10.1016/j.jmii.2017.10.006
Zhao, J., Zhang, Y., Sha, Y., Lin, N., Zhang, G., Lu, J., et al. (2023). BlaPSZ-1, a novel AmpC gene identified from a Pantoea isolate. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1222703
Keywords: Morganella morganii, resistance gene, fosA13, fosfomycin glutathione transferase, kinetic parameter
Citation: Zhang R, Yu Y, Huang L, Chen S, Hu R, Wang X, Huang D, Song C, Lu J, Bao Q, Hu Y, Jiang P and Pan W (2025) Characterization of FosA13, a novel fosfomycin glutathione transferase identified in a Morganella morganii isolate from poultry. Front. Cell. Infect. Microbiol. 15:1534084. doi: 10.3389/fcimb.2025.1534084
Received: 25 November 2024; Accepted: 21 February 2025;
Published: 11 March 2025.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyCopyright © 2025 Zhang, Yu, Huang, Chen, Hu, Wang, Huang, Song, Lu, Bao, Hu, Jiang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunliang Hu, aHV5dW5saWFuZzY2QDE2My5jb20=; Pengfei Jiang, cGVuZy1mZWlqaWFuZ0Bob3RtYWlsLmNvbQ==; Wei Pan, MjU2NTg1MDdAcXEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.