- 1Department of Respiratory and Critical Care Medicine, Senior Department of Infectious Diseases, the Fifth Medical Center of PLA General Hospital, Beijing, China
- 2The Fifth Clinical Medical College, Anhui Medical University, Hefei, Anhui, China
- 3Senior Department of Infectious Diseases, The Fifth Medical Center of PLA General Hospital, National Clinical Research Center for Infectious Diseases, Beijing, China
Mycoplasma pneumoniae is one of the most significant pathogens responsible for respiratory infections in humans. Macrolides are recommended as the first-line treatment for M. pneumoniae infection. The prevalence of macrolide-resistant M. pneumoniae has increased significantly in recent decades, particularly in China. The mechanisms of resistance in M. pneumoniae to macrolides have been extensively studied in pediatric patients. However, a paucity reports regarding the resistance characteristics and mechanisms exhibited in adults. The aim of this study was to elucidate the resistance of M. pneumoniae to macrolides and the underlying mechanisms in adult patients. Pharyngeal swab specimens were collected from adult patients presenting with subacute cough or community-acquired pneumonia at our hospital from January 2011 to June 2017 to identify and isolate M. pneumoniae strains. The antimicrobial susceptibility of these isolates to 3 macrolide antibiotics was assessed using broth microdilution method. The 23S rRNA genes of macrolide-resistant M. pneumoniae strains were sequenced, and the presence of target methylation genes (ermA, ermB, and ermC), efflux pump genes (mefA, mefA/E, msrA, and msrA/B), and the macrolide resistance gene mphC was identified through polymerase chain reaction (PCR) testing. Additionally, MICs were determined with and without the efflux pump inhibitor reserpine. A total of 72 M. pneumoniae strains were isolated from adult patients, with 41.7% (30/72) exhibiting macrolide resistance. Among the 3 macrolides tested, the 16-membered-ring midecamycin exhibited the greatest activity (MIC90: 16 µg/ml) against M. pneumoniae. All macrolide-resistant M. pneumoniae strains harbored mutations at the 2063 site in domain V of the 23S rRNA gene. Two macrolide-resistant M. pneumoniae clinical isolates were found to harbor the efflux pump genes msrA/B and mefA. The efflux pump inhibitor reserpine reduced the MIC for azithromycin in these two strains to a quarter of their original values. In summary, macrolide-resistant M. pneumoniae is commonly observed among adults in Beijing. Point mutations are the primary mechanism responsible for macrolide resistance in adults with M. pneumoniae. Additionally, the efflux pump mechanism may contribute partially to this resistance. Midecamycin presents a promising alternative drug for treating M. pneumoniae infections, particularly in cases of azithromycin-resistant M. pneumoniae infection in young children.
1 Introduction
Mycoplasma pneumoniae is one of the most significant pathogens responsible for respiratory infections in humans. It is estimated to account for 10% to 30% of cases of community-acquired pneumonia (CAP) (Atkinson et al., 2008). Although M. pneumoniae infection is typically self-limiting, severe M. pneumoniae pneumonia has been increasingly reported in recent years (Cillóniz et al., 2016; Waites et al., 2017; Ha et al., 2023; Lai et al., 2024). M. pneumoniae is an atypical pathogen as it lacks a cell wall, rendering it innately resistant to a wide range of antimicrobial drugs that target the cell wall, such as β-lactams (Lee et al., 2018; Waites et al., 2017). Macrolides, fluoroquinolones, and tetracyclines are three major classes of antibiotics effective against M. pneumoniae (Waites et al., 2017; Gautier-Bouchardon, 2018). Macrolides are recommended as the first-line treatment for M. pneumoniae infection in adults and are preferred for children (Principi and Esposito, 2013; Qu et al., 2013). However, as the prescription of macrolide antibiotics for outpatients with CAP has increased, the acquired resistance of M. pneumoniae to macrolide antibiotics has gradually emerged in response to antibiotic selective pressure. Recent studies have demonstrated a significant worldwide increase in the prevalence of macrolide-resistant M. pneumoniae, with a particularly marked rise observed in Asia (Zhou et al., 2015; Waites et al., 2017). Recent reports from China indicate that the macrolide resistance rate of M. pneumoniae can be as high as 80% to 100% in various regions (Zhou et al., 2015; Zhao et al., 2019; Wang et al., 2022b, 2022a). Macrolide-resistant M. pneumoniae infections are more prevalent in children than in adults (Miyashita et al., 2012; Yan et al., 2020; Kim et al., 2022; Jiang et al., 2024), however, the disease burden caused by these infections in adults also warrants attention (Lai et al., 2024). How macrolides should be deployed in adult patients with M. pneumoniae infection, has become a matter of urgent concern in the clinical community.
Bacterial resistance to macrolides is mediated by various mechanisms, including modification of target sites by methylation or mutation in the 23S rRNA or large ribosomal subunit proteins, drug-inactivating and efflux of macrolides from bacterial cell resulting from efflux pump expression (Dinos, 2017). In M. pneumoniae, resistance to macrolides is primarily attributed to mutations in the domains V and/or II of 23S rRNA (Gaynor and Mankin, 2003; Bébéar and Pereyre, 2005). Furthermore, mutations in ribosomal proteins L4 and L22 also play a role in conferring macrolide resistance in M. pneumoniae (Pereyre et al., 2004; Liu et al., 2014; Wang et al., 2023). Nevertheless, the resistance mechanisms of some resistant strains could not be fully explained by target mutations. M. pneumoniae and Streptococcus pneumoniae infections are prevalent causes of CAP in China, and co-infections with these two pathogens are also common (Cao et al., 2010). Target methylation modification represents the primary mechanism of macrolide resistance in S. pneumoniae in China (Zhao et al., 2020). In such cases, it remains unclear whether the target methylation genes are transferred from S. pneumoniae to M. pneumoniae through plasmids or other mobile genetic elements, conferring high levels of resistance to macrolides in M. pneumoniae. Furthermore, there has been no investigation into efflux pump mechanism in drug-resistant clinical isolates of M. pneumoniae in adults, and it remains unclear whether drug inactivation mechanisms are involved in macrolide resistance in M. pneumoniae.
We investigated the resistance of M. pneumoniae clinical isolates to macrolides in adult patients to guide the effective use of currently available macrolides. Furthermore, to elucidate the mechanisms of macrolide resistance in M. pneumoniae in adults, we performed an extensive analysis of target mutations, target methylation modifications, efflux pump activity, and drug-inactivating enzymes in resistant clinical isolates.
2 Materials and methods
2.1 Clinical M. pneumoniae isolates
All M. pneumoniae clinical strains were isolated from oropharyngeal samples of adult patients presenting with subacute cough and suspected M. pneumoniae infection with CAP. These samples were collected from both respiratory outpatient and inpatient departments of the Fifth Medical Center of PLA General Hospital from January 1, 2011, to June 30, 2017. Screening criteria for patients with subacute cough were similar to those described by Yuan et al., except for age ≥18 years (Yuan et al., 2014). The screening criteria for CAP patients with suspected M. pneumoniae infection were based on the rapid scoring system for M. pneumoniae Pneumonia of the Japanese Respiratory Society (JRS) with modifications (Ishida, et al., 2007). The presence of M. pneumoniae pneumonia is indicated by meeting four of the criteria in this scoring system, or three of the first five criteria. All strains were detected through culture and real-time quantitative polymerase chain reaction (PCR). M. pneumoniae was cultured using an established methodology (Waites et al., 2001). Positive cultures were identified by a color change from red to yellow in the broth medium (CM0403, OXOID, UK) and the presence of characteristic “fried egg” colonies on the agar medium (CM0401, OXOID, UK). A real-time quantitative PCR was used to quantify bacterial load by detecting the 16S rDNA of M. pneumoniae, utilizing primers and probe sequences as described previously (Yuan et al., 2014).
2.2 Antimicrobial susceptibility test
The in vitro susceptibility of the strains to 3 macrolide antibiotics (erythromycin, azithromycin, and midecamycin) was assessed using the broth microdilution method (Matsuoka et al., 2004). All 3 antibiotics were purchased from the National Institute for the Control of Pharmaceutical and Biological Products. A reference strain of M. pneumoniae designated FH (ATCC 15531), was used as the drug-sensitive control in this study. The minimum inhibitory concentration (MIC) for each agent was determined as the lowest concentration of each antimicrobial agent that prevented the color change, observed at the time when the growth controls first showed a color change (Waites et al., 2001). The MIC50 and MIC90 were defined as the MIC required to inhibit the growth of 50% and 90% of the subject bacteria, respectively, in a batch of tests. Each antimicrobial susceptibility test was performed in triplicate. Throughout the study, antimicrobial susceptibility tests were performed on all strains in strict accordance with the methodology described previously by Matsuoka et al (Matsuoka et al., 2004), and all referenced previous resistance breakpoint, the results were defined as resistant with a MIC of ≥32 µg/ml for erythromycin, azithromycin and midecamycin (Xin et al., 2009).
2.3 Polymerase chain reaction amplification and DNA sequencing
Total DNA was manually extracted using the QIAamp DNA Mini kit (QIAGEN, Germany). Primers were designed and synthesized based on GenBank and relevant literature (Matsuoka et al., 2003; Lu et al., 2010) to amplify M. pneumoniae 5S rRNA, 23S rRNA, genes encoding target site-modifying rRNA methylases ermA/B/C, efflux pump genes mefA, mefA/E, msrA, and msrA/B, and the macrolide 2’-phosphotransferase mphC. The primer sequences are listed in Supplementary Table 1. The PCR products of M. pneumoniae 5S rRNA, 23S rRNA genes were sequenced by INVITROGEN using a 3730XL DNA sequencer, and the PCR amplification products of ermA/B/C, mefA, mefA/E, msrA, msrA/B, and mphC genes were subjected to electrophoresis to visualize the target bands.
2.4 Effect of an efflux pump inhibitor (reserpine) on MICs
Reserpine was obtained from the National Institute for the Control of Pharmaceutical and Biological Products. MICs were assessed under 3 conditions: in the presence of each macrolide alone, the efflux pump inhibitor reserpine alone, and a combination of macrolides and reserpine. The microbroth dilution test methodology remains largely unchanged (Matsuoka et al., 2004), apart from a minor modification in the configuration of the 96-well plate. In summary, MICs were determined for each of the resistant strains in three distinct scenarios: exposure to macrolides alone, exposure to efflux pump inhibitor reserpine alone (20 µg/ml), and exposure to macrolides and reserpine (20 µg/ml).
3 Results
3.1 Antimicrobial susceptibility for M. pneumoniae
In this study, 72 strains of M. pneumoniae were isolated, and the MICs of all 72 clinical M. pneumoniae isolates, as well as standard control strain, were tested against 3 different macrolides. A total of 27 males and 45 females were included in the study, with an age range of 18-75 years. 26 specimens were derived from patients with CAP, while 46 were obtained from patients with subacute cough. The MICs of the standard strain FH (ATCC 15531) for the 3 tested agents were consistent with those of the standard strain M129 (ATCC 29342) provided by Dr. Waites KB, all were < 0.5µg/ml. Among the 3 macrolides, 41.7% (30/72) of the strains were resistant to erythromycin, the resistance rate to azithromycin was 38.9% (28/72), and 1.4% (1/72) strains showed resistance to midecamycin, with the following MIC values: Erythromycin (MIC50: < 0.5µg/ml, MIC90: ≥ 128 µg/ml), azithromycin (MIC50: <0.5µg/ml, MIC90: ≥ 128 µg/ml), midecamycin (MIC50: < 0.5µg/ml, MIC90: 16 µg/ml). 54 strains were obtained during the cold season, and 18 during the warm season. No significant difference was observed in resistance rates between strains from the two seasons (P = 0.408) (Supplementary Table 2). Table 1 presents a detailed MIC distribution of the 3 agents tested against 30 resistance M. pneumoniae.
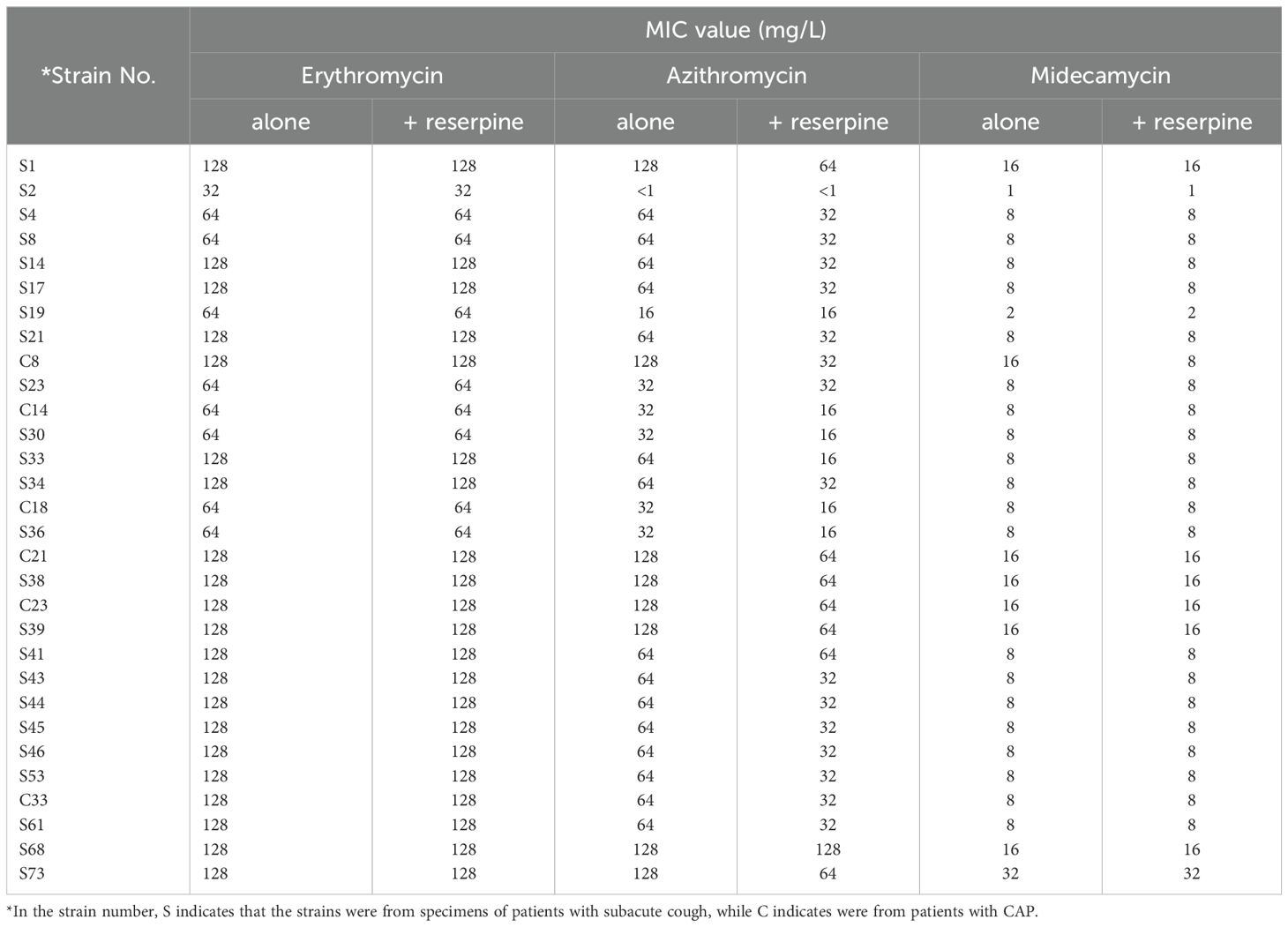
Table 1. Minimal inhibitory concentrations and the effect of efflux pump inhibitor reserpine on 30 resistant clinical isolates of M. pneumonia.
3.2 Target mutations and target modifications associated with macrolide resistance
No mutations were detected in the 5S rRNA of the 30 macrolide-resistant M. pneumoniae isolates. An A2063G point mutation was observed in the 23S rRNA gene of 29 of these isolates (Figure 1A), and one strain (S19) exhibited an A-to-R transition at point 2063 (A2063R, heterozygote) (Figure 1B). In addition to an A2063G mutation, one strain (S68) harbored a G648R mutation (Figures 1C, D). Additionally, an A1029G mutation was identified in 30 macrolide-resistant clinical strains, the M. pneumoniae reference strain, and 30 macrolide-sensitive strains, indicating that this mutation is not associated with macrolide resistance. Moreover, the amplification of ermA, ermB, and ermC target methylation genes in 30 resistant M. pneumoniae strains failed to yield any gene products.
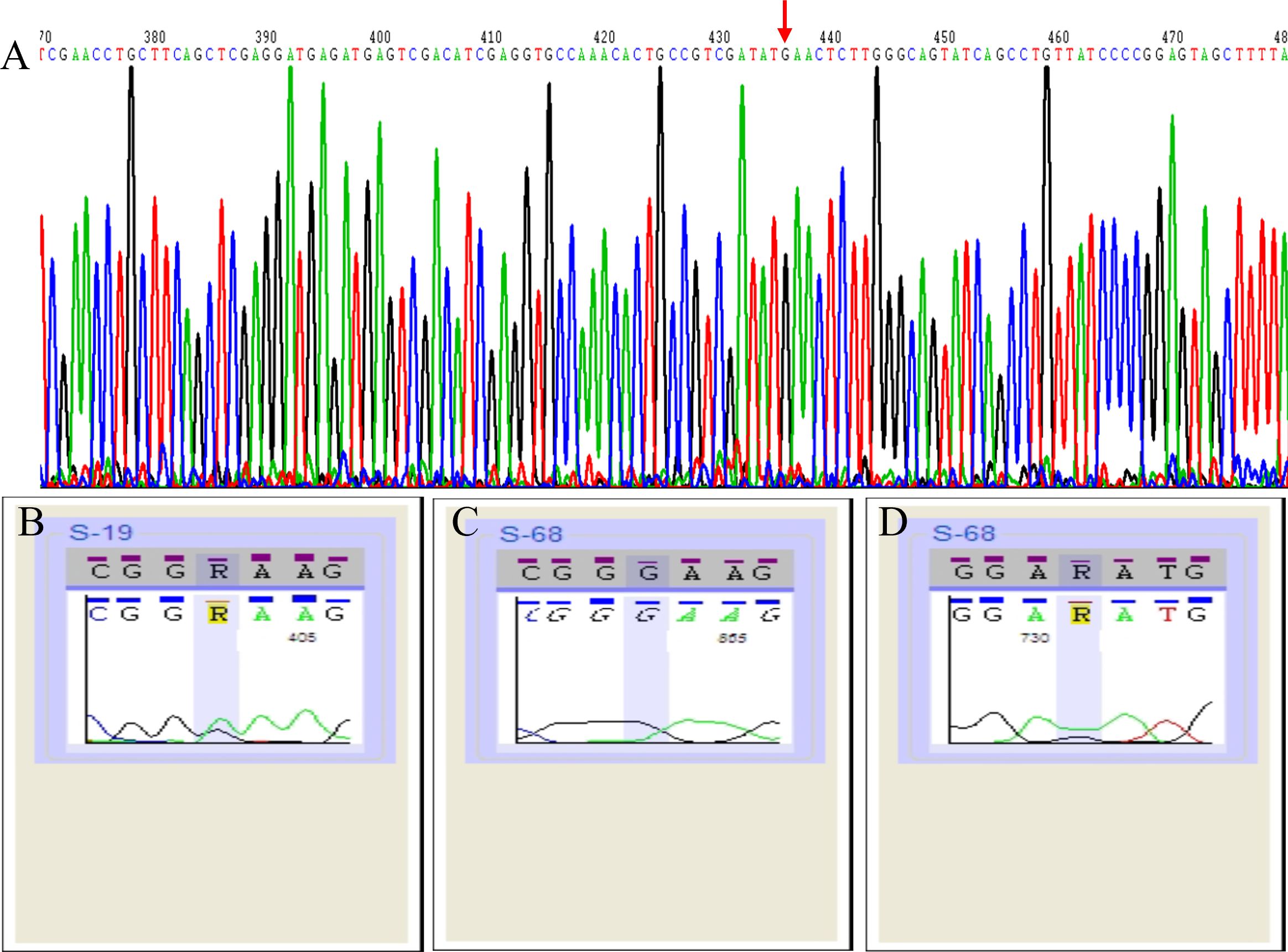
Figure 1. Schematic representation of 23S rRNA sequencing results of 30 macrolide-resistant Mycoplasma pneumoniae isolates. Red arrow indicate the A2063G point mutation in the gene sequence (A). A2063R point mutation (heterozygote) in one strain (S19) (B). A2063G mutation (C) and G648R mutation (D) in one strain (S68).
3.3 Efflux pump mechanism in M. pneumoniae isolates
To investigate the presence of an efflux pump, PCR amplification products for the corresponding efflux pump genes mefA, mefA/E, msrA, and msrA/B genes were subjected to electrophoresis to visualize the target bands. The target fragment for the mefA gene was 488 bp. Agarose gel electrophoresis revealed a 488 bp fragment, indicating successful amplification from one sample of macrolide-resistant M. pneumoniae clinical strains (Figure 2A). For the msrA/B gene (target fragment size 399 bp), one sample was found to amplify a 399 bp fragment (Figure 2B). BLAST analysis of the PCR product revealed a 99% similarity to the msrC gene (405 bp), which encodes an ABC (ATP-binding cassette) transporter protein in Enterococcus faecium (GeneBank: AJ243209.1), and a 95% similarity to the macrolide resistance-like protein gene (GeneBank: AY004350.1; 2479 bp) acquired from E. faecium strain TX2465 (Supplementary Table 3). Additionally, the protein sequence corresponding to this gene was found to be homologous to the P-loop_NTPase superfamily (Figure 2C), showing 99% similarity to the ABC transporter protein (ZP_00603470.1) and the acquired macrolide-resistant protein (AAF91071.1) in E. faecium.
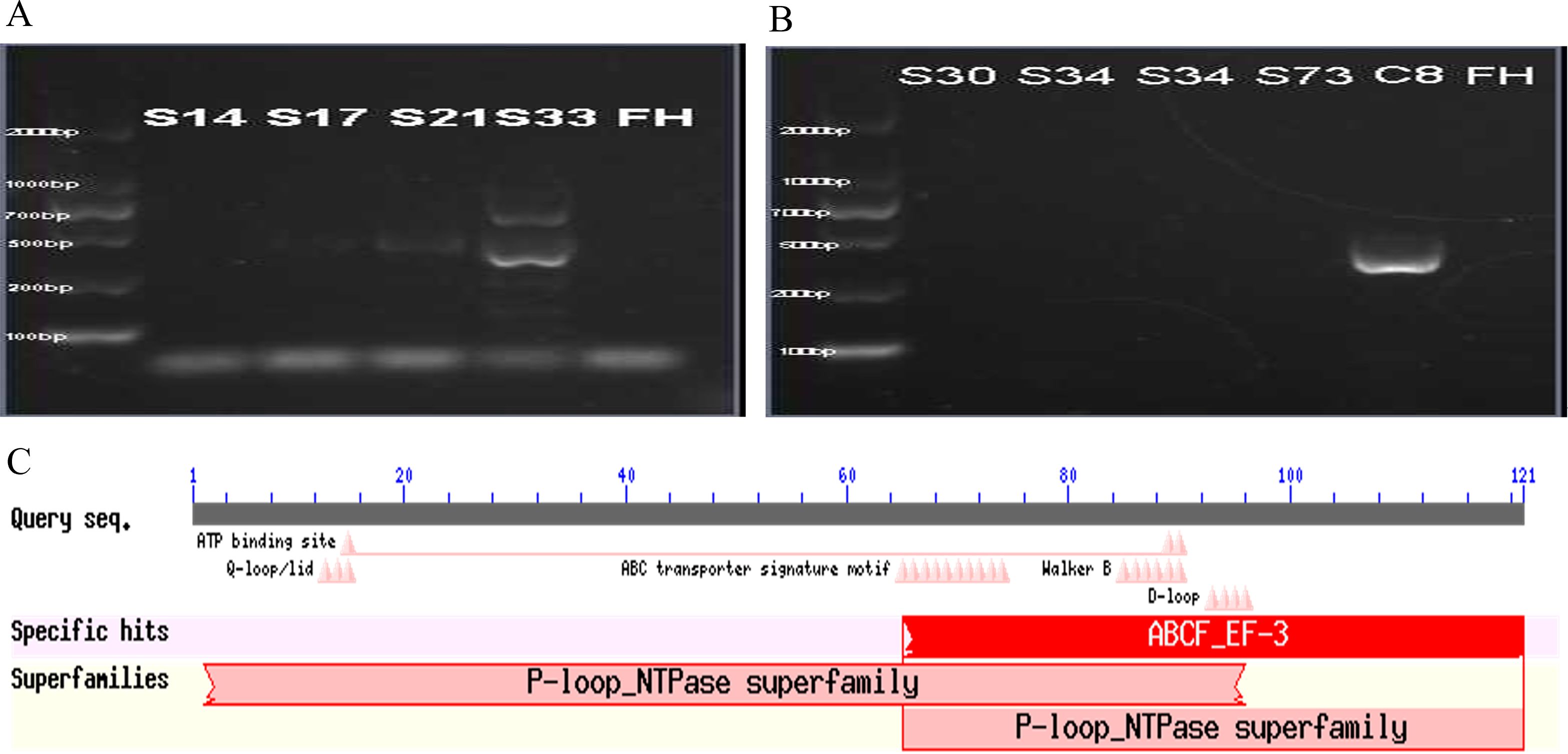
Figure 2. Detection of efflux pump genes. The PCR product (488 bp) of the mefA gene of one clinical macrolides-resistant M. pneumoniae strain (A). The PCR product (399 bp) of msrA/B gene of one clinical macrolides-resistant M. pneumoniae strain (B). The molecular weights labeled on the left side of the figure A and figure B are, from bottom to top, 100bp, 200bp, 600bp, 700bp, 1000bp and 2000bp. Results of conserved protein sequence comparison after efflux pump gene sequencing (C).
In order to evaluate the potential role of the efflux pump in macrolide resistance, MICs were determined with and without the efflux pump inhibitor reserpine. Reserpine alone did not inhibit the growth of M. pneumoniae and did not affect the MICs for the three macrolides in the reference strain FH. Although reserpine did not change the MIC for erythromycin in the 30 macrolide-resistant clinical isolates, it reduced the MIC for azithromycin in 2 strains which harbored efflux pump genes to a quarter, in 23 of these strains to half of their original values, and the remaining 3 strains unchanged. Additionally, in one macrolide-resistant clinical isolate, reserpine decreased the MIC for midecamycin to half of its original value (Tabe 1).
3.4 Macrolide passivating enzymes in M. pneumoniae isolates
The PCR products of the resistant M. pneumoniae strains were subjected to agarose gel electrophoresis, which revealed the absence of macrolide 2’-phosphotransferase mphC.
4 Discussion
In the present study, we measured the resistance of M. pneumoniae to macrolides and observed a relatively high prevalence of macrolide resistance to M. pneumoniae in adult patients with M. pneumoniae infections in Beijing. An A to G mutation at the 2063 site in domain V of the 23S rRNA gene was identified in all resistant isolates. Interestingly, we also identified the presence of the efflux pump gene. Furthermore, midecamycin demonstrated higher susceptibility to M. pneumoniae among the three macrolides.
The macrolide resistance rate in M. pneumoniae in adults was found to be 41.7% in our study, which is lower than the rates reported in most other studies conducted in China. Over the past two decades, investigations into macrolide-resistant M. pneumoniae in children have demonstrated the highest rates of resistance in some East Asian regions, with resistance rates reaching 81.6% (493/604 specimens) in Japan and up to 100% (49/49 specimens) in some parts of China (Tanaka et al., 2017; Zhao et al., 2019; Wang et al., 2022b, 2022a). Although macrolide resistance in M. pneumoniae is relatively low in Europe and the United States, with reported rates of 10% (10/114 specimens) in the United States (Rothstein et al., 2022) and 1% to 25% in Europe (Álvaro Varela et al., 2023) in children, resistance rates are on the rise in these regions as well. The disparate rates of macrolide resistance observed across different countries may be attributed to variations in the frequency of macrolide utilization, and there is a well-quantified correlation between antibiotic usage and the emergence of resistance (Klein et al., 2018; Bell et al., 2014). These findings indicate the necessity for heightened surveillance of macrolide-resistant M. pneumoniae, particularly in China. However, there is a paucity of data regarding the prevalence of macrolide-resistant M. pneumoniae in adults. Cao et al. reported a 69% macrolide resistance rate in M. pneumoniae among adults in China in 2010 (Cao et al., 2010); Yin et al. showed an 80% resistance rate to erythromycin in adult CAP isolates from three different Chinese cities between 2010 and 2012 (Yin et al., 2017). Zhou et al. observed a 100% macrolide resistance in M. pneumoniae isolates from adult patients with CAP in Zhejiang Province of China from 2012 to 2014 (Zhou et al., 2015); Jiang et al. investigated 41 M. pneumoniae-positive samples in Beijing, China, and found that only 10.5% exhibited the A2063G resistance mutation (Jiang et al., 2024). Our results contribute further data on macrolide resistance in M. pneumoniae among adults in China, demonstrating a lower rate of macrolide resistance than that reported in the majority of domestic studies. This discrepancy may be attributed to the varied sources of strains. M. pneumoniae has been shown to be highly prevalent among patients with subacute cough, as indicated by our previous study (Yuan et al., 2014). In our study, most isolates were obtained from patients presenting with subacute cough. Some of these patients had only been administered oral β-lactam antibiotics during the course of their illness, which had minimal impact on M. pneumoniae. Consequently, the probability of developing induced resistance was diminished.
The resistance of M. pneumoniae to macrolides does not appear to be reflected in resistance to all macrolide antibiotics. Our data revealed that erythromycin and azithromycin exhibited reduced activity against M. pneumoniae, whereas midecamycin showed good activity. Midecamycin, a 16-membered-ring macrolide, has shown in vitro activity against erythromycin-resistant Streptococcus pyogenes (Schlegel et al., 2001). In in vitro studies, the MIC of acetylmidecamycin (diacetate of midecamycin) was found to be considerably lower than that of other macrolide antibiotics, indicating a more potent activity for M. pneumoniae activity of acetylmidecamycin (Pereyre et al., 2001; Wang et al., 2020). Furthermore, the dosage and safety of midecamycin have been established in pediatric patients in a variety of countries (Wang et al., 2023; Kikuchi et al., 1979; Yoshida et al., 1982; Morikawa et al., 1994). In vitro resistance induction experiments have shown that screening for M. pneumoniae mutant strains of midecamycin is more difficult compared to other macrolide antibiotics (Wang et al., 2023). Additionally, strains resistant to midecamycin remained susceptible to 14- and 15-membered-ring macrolide antibiotics (Wang et al., 2023). Macrolides remain the most commonly utilized antibiotics for the treatment of M. pneumoniae infections in clinical practice. The frequency of antibiotic use is associated with the emergence of drug resistance. In China, erythromycin and azithromycin are more widely used than midecamycin, which may account for the higher MIC values observed for the former antibiotics. Since the MIC of midecamycin against M. pneumoniae is lower than that of other 14- or 15-membered-ring macrolides, midecamycin may serve as a promising treatment option for M. pneumoniae infections, particularly in children.
Macrolide antibiotics bind to specific nucleotides in structural domains II and/or V of 23S rRNA in the 50S bacterial ribosomal subunit, thereby blocking protein synthesis by causing premature dissociation of the peptidyl-tRNA from the ribosome and achieving antimicrobial efficacy. Specific mutations in binding sites result in a reduction in the binding of drugs to M. pneumoniae, ultimately conferring resistance to macrolides (Vázquez-Laslop and Mankin, 2018; Dinos, 2017). In M. pneumoniae, the A2063G mutation, which is the primary mutation responsible for resistance to macrolides, is situated within the peptidyl-transferase center of the 23S rRNA V region (Gaynor and Mankin, 2003; Bébéar and Pereyre, 2005). This structural domain has been demonstrated to serve as a binding site for macrolide antibiotics (Jelić and Antolović, 2016). The A2063 locus mutations were identified in all M. pneumoniae strains exhibiting macrolide resistance in our experiment. These findings are consistent with previous studies that point mutations in the peptidyl transferase loop of the 23S rRNA in M. pneumoniae, such as A2063G/T/C, C2617G, A2064G/C, and A2067G, are key contributors to macrolide resistance (Lucier et al., 1995; Bébéar and Pereyre, 2005; Suzuki et al., 2013; Waites et al., 2017). The 2063 site point mutation is capable of causing high levels of resistance to 14- and 15-membered-ring macrolides in M. pneumoniae. For the 16-membered-ring macrolides, mutations at sites 2063 and 2064 were found to be associated with low to moderate levels of resistance. Whereas, the A2067G mutation resulted in the highest level of resistance due to its ability to form a specific covalent bond with the 16-membered ring (Cardinale et al., 2011; Principi and Esposito, 2013). The differences in sensitivity may be attributed to variations in the binding sites, drug orientation, and binding kinetics between 16-membered-ring macrolide antibiotics and 14- and 15-membered-ring macrolide antibiotics (Starosta et al., 2010; Wang et al., 2023). However, no mutations were observed at locus 2067 in our study. Antimicrobial susceptibility testing results of clinical isolates for midecamycin also demonstrated that mutations at locus A2063 had a minimal effect on susceptibility to the 16-membered-ring macrolides. Most A2063G mutant strains were either susceptible to midecamycin or showed only a low level of resistance. This supports the potential of midecamycin as a promising alternative agent for treating M. pneumoniae infections. However, one strain was highly resistant to midecamycin (MIC >32 mg/L), indicating that mechanisms beyond target mutation may contribute to resistance against 16-membered-ring macrolides.
Active efflux mechanism plays a role in macrolide resistance (Ma et al., 2024). Of particular interest, we further screened for common efflux pump genes and identified the presence of mefA and msrA/B genes. Sequencing of the msrA/B gene in this resistant strain revealed a high degree of similarity between the gene and the msrC gene, which encodes an ABC transporter protein in E. faecium. The protein encoded by this msrA/B gene is also similar to the E. faecium-acquired macrolide-resistant protein, which is absent in the reference strain. Efflux pump inhibitor reserpine was used to assess the role of efflux pumps in macrolide resistance in M. pneumoniae. Our findings revealed that for 2 resistance strains in which the efflux pump gene was identified, the MICs of azithromycin and midecamycin were reduced to half to a quarter of their original values; for 23 resistance strains, the MICs were reduced to half; and for other 3 resistance strains, no change was observed. The disparate effects on the MICs may be attributed to the following factors: Firstly, a not significant multiplicative change in MIC with the addition of an efflux pump inhibitor may not be indicative of the presence of an efflux pump in a strain (Baron and Rolain, 2018); secondly, the expression levels of efflux pump genes in these strains were different (Heijden et al., 2023); thirdly, there may be the potential existence of additional resistance mechanisms beyond the 23S rRNA point mutation and the efflux pump effect. Further studies are necessary to substantiate these hypotheses. Six bacterial drug efflux pump families have been identified as being involved in the efflux pathway (Du et al., 2018; Zhang et al., 2024). The efflux pump msrA/B gene identified in our study may belong to the ATP-binding cassette (ABC) family. Reserpine is known to inhibit multiple drug resistance (MDR) efflux pumps, including ATP-dependent efflux pumps (Frempong-Manso et al., 2009; Huang et al., 2013; Li et al., 2017). Additionally, the small multidrug resistance (SMR) family has been linked to macrolide efflux. However, it remains uncertain whether the 23 strains exhibiting halved MICs possess other efflux pumps like SMR family. These findings suggest the involvement of an efflux pump system mechanism, possibly an ABC transporter, may play an important role in promoting macrolide resistance, thereby providing further insight into the mechanisms of macrolide resistance in M. pneumoniae.
There are several limitations to our study. Firstly, a relatively small sample size affects the generalizability of the findings. Variations in subject populations may have contributed to discrepancies in observed resistance rates. Secondly, our study did not include subgroup analysis to compare differences in resistance between clinical isolates from patients with subacute cough and those with pneumonia. Thirdly, this study did not undertake a more in-depth exploration of resistance mechanisms through the use of whole genome sequencing and bioinformatics tools. Additionally, the resistance criteria were based on the 2006 CLSI standards, which may have led to discrepancies when compared to those of more recent studies. As the present study was initiated prior to the release of the most recent guideline on methods for antimicrobial susceptibility testing for M. pneumoniae and was in strict accordance with the already developed method throughout the study, reference was made to previous resistance breakpoints. Despite these limitations, our study highlights the prevalence of macrolide-resistant M. pneumoniae in adults and supports the hypothesis that efflux pump genes contribute to macrolide resistance in clinical M. pneumoniae isolates.
In conclusion, this study presented valuable insights on M. pneumoniae resistance to macrolides in adults. Macrolide-resistant M. pneumoniae is highly prevalent in adults. Resistance is primarily attributed to mutations in domain V of the 23S rRNA gene, and presence of efflux pump may also bring about the resistance phenotype. The observed reduction in MICs for azithromycin against macrolides-resistant M. pneumoniae in the presence of reserpine suggests that reserpine might be a promising candidate for combination therapy. Additionally, our findings support the potential of midecamycin as an effective alternative for the treatment of macrolide-resistant M. pneumoniae infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the ethics committee of the Fifth Medical Center of PLA General Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
PX: Formal Analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. YZ: Formal Analysis, Investigation, Writing – original draft. YQ: Visualization, Writing – original draft. YF: Data curation, Software, Writing – review & editing. NY: Investigation, Software, Visualization, Writing – review & editing. YB: Investigation, Writing – review & editing. SZ: Investigation, Software, Writing – review & editing. WN: Conceptualization, Methodology, Writing – review & editing. FW: Conceptualization, Writing – review & editing. XY: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China, grant number 81400009.
Acknowledgments
We would like to thank the enrolled subjects of the study and all the staff for their help with the collection of specimens and clinical data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1496521/full#supplementary-material
References
Álvaro Varela, A. I., Aguinaga Pérez, A., Navascués Ortega, A., Castilla Catalán, J., Ezpeleta Baquedano, C. (2023). Macrolide-resistant Mycoplasma pneumoniae: Do we know the situation in Europe]? Rev. Esp Quimioter 36, 259–266. doi: 10.37201/req/118.2022
Atkinson, T. P., Balish, M. F., Waites, K. B. (2008). Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol. Rev. 32, 956–973. doi: 10.1111/j.1574-6976.2008.00129.x
Baron, S. A., Rolain, J.-M. (2018). Efflux pump inhibitor CCCP to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and Gram-negative bacteria. J. Antimicrobial Chemotherapy 73, 1862–1871. doi: 10.1093/jac/dky134
Bébéar, C. M., Pereyre, S. (2005). Mechanisms of drug resistance in Mycoplasma pneumoniae. Curr. Drug Targets Infect. Disord. 5, 263–271. doi: 10.2174/1568005054880109
Bell, B. G., Schellevis, F., Stobberingh, E., Goossens, H., Pringle, M. (2014). A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect. Dis. 14, 13. doi: 10.1186/1471-2334-14-13
Cao, B., Zhao, C.-J., Yin, Y.-D., Zhao, F., Song, S.-F., Bai, L., et al. (2010). High prevalence of macrolide resistance in Mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin. Infect. Diseases: Off. Publ. Infect. Dis. Soc. America 51, 189–194. doi: 10.1086/653535
Cardinale, F., Chironna, M., Dumke, R., Binetti, A., Daleno, C., Sallustio, A., et al. (2011). Macrolide-resistant Mycoplasma pneumoniae in paediatric pneumonia. Eur. Respir. J. 37, 1522–1524. doi: 10.1183/09031936.00172510
Cillóniz, C., Torres, A., Niederman, M., van der Eerden, M., Chalmers, J., Welte, T., et al. (2016). Community-acquired pneumonia related to intracellular pathogens. Intensive Care Med. 42, 1374–1386. doi: 10.1007/s00134-016-4394-4
Dinos, G. P. (2017). The macrolide antibiotic renaissance. Br. J. Pharmacol. 174, 2967–2983. doi: 10.1111/bph.13936
Du, D., Wang-Kan, X., Neuberger, A., van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi: 10.1038/s41579-018-0048-6
Frempong-Manso, E., Raygada, J. L., DeMarco, C. E., Seo, S. M., Kaatz, G. W. (2009). Inability of a reserpine-based screen to identify strains overexpressing efflux pump genes in clinical isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 33, 360–363. doi: 10.1016/j.ijantimicag.2008.10.016
Gautier-Bouchardon, A. V. (2018). Antimicrobial resistance in mycoplasma spp. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.ARBA-0030-2018
Gaynor, M., Mankin, A. S. (2003). Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 3, 949–961. doi: 10.2174/1568026033452159
Ha, E. K., Kim, J. H., Cha, H. R., Han, B. E., Shin, Y. H., Baek, H.-S., et al. (2023). Investigating the occurrence of autoimmune diseases among children and adolescents hospitalized for Mycoplasma pneumoniae infections. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1165586
Heijden, Y. F., Maruri, F., Blackman, A., Morrison, R., Guo, Y., Sterling, T. R. (2023). Mycobacterium tuberculosis gene expression associated with fluoroquinolone resistance and efflux pump inhibition. J. Infect. Dis. 228, 469–478. doi: 10.1093/infdis/jiad112
Huang, T.-S., Kunin, C. M., Wang, H.-M., Yan, B.-S., Huang, S.-P., Chen, Y.-S., et al. (2013). Inhibition of the Mycobacterium tuberculosis reserpine-sensitive efflux pump augments intracellular concentrations of ciprofloxacin and enhances susceptibility of some clinical isolates. J. Formos Med. Assoc. 112, 789–794. doi: 10.1016/j.jfma.2012.03.009
Ishida, T., Miyashita, N., Nakahama, C. (2007). Clinical differentiation of atypical pneumonia using Japanese guidelines. Respirology 12, 104–110. doi: 10.1111/j.1440-1843.2006.00927.x
Jelić, D., Antolović, R. (2016). From erythromycin to azithromycin and new potential ribosome-binding antimicrobials. Antibiotics (Basel) 5, 29. doi: 10.3390/antibiotics5030029
Jiang, Y., Dou, H., Xu, B., Xu, B., Zhou, W., Wang, H., et al. (2024). Macrolide resistance of Mycoplasma pneumoniae in several regions of China from 2013 to 2019. Epidemiol. Infect. 152, e75. doi: 10.1017/S0950268824000323
Kikuchi, N., Kobayashi, A., Kanno, H., Ishihara, K., Kato, S., Kiuchi, N., et al. (1979). Therapeutic effect of midecamycin on Mycoplasma pneumoniae pneumonia in adults (author’s transl). Jpn J. Antibiot 32, 555–561.
Kim, K., Jung, S., Kim, M., Park, S., Yang, H.-J., Lee, E. (2022). Global trends in the proportion of macrolide-resistant mycoplasma pneumoniae infections: A systematic review and meta-analysis. JAMA Netw. Open 5, e2220949. doi: 10.1001/jamanetworkopen.2022.20949
Klein, E. Y., Van Boeckel, T. P., Martinez, E. M., Pant, S., Gandra, S., Levin, S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U.S.A. 115, E3463–E3470. doi: 10.1073/pnas.1717295115
Lai, C.-C., Hsueh, C.-C., Hsu, C.-K., Tsai, Y.-W., Hsueh, P.-R. (2024). Disease burden and macrolide resistance of Mycoplasma pneumoniae infection in adults in Asia-Pacific region. Int. J. Antimicrob. Agents 64, 107205. doi: 10.1016/j.ijantimicag.2024.107205
Lee, H., Yun, K. W., Lee, H. J., Choi, E. H. (2018). Antimicrobial therapy of macrolide-resistant Mycoplasma pneumoniae pneumonia in children. Expert Rev. Anti Infect. Ther. 16, 23–34. doi: 10.1080/14787210.2018.1414599
Li, S. L., Sun, H. M., Zhu, B. L., Liu, F., Zhao, H. Q. (2017). Whole Genome Analysis Reveals New Insights into Macrolide Resistance in Mycoplasma pneumoniae. Biomed. Environ. sciences: BES 30, 343–350. doi: 10.3967/bes2017.045
Liu, X., Jiang, Y., Chen, X., Li, J., Shi, D., Xin, D. (2014). Drug resistance mechanisms of Mycoplasma pneumoniae to macrolide antibiotics. BioMed. Res. Int. 2014, 320801. doi: 10.1155/2014/320801
Lu, R., Lu, C., Ma, H., Lai, W., Ye, T., Zhu, G., et al. (2010). Distributional difference of erythromycin resistance genes in different Ureaplasma Urealyticum biovars. Chin. J. Derm Venereol 24, 699–701.
Lucier, T. S., Heitzman, K., Liu, S. K., Hu, P. C. (1995). Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 39, 2770–2773. doi: 10.1128/AAC.39.12.2770
Ma, Y., Pirolo, M., Jana, B., Mebus, V. H., Guardabassi, L. (2024). The intrinsic macrolide resistome of Escherichia coli. Antimicrob. Agents Chemother. 68, e0045224. doi: 10.1128/aac.00452-24
Matsuoka, M., Inoue, M., Endo, Y., Nakajima, Y. (2003). Characteristic expression of three genes, msr(A), mph(C) and erm(Y), that confer resistance to macrolide antibiotics on Staphylococcus aureus. FEMS Microbiol. Lett. 220, 287–293. doi: 10.1016/S0378-1097(03)00134-4
Matsuoka, M., Narita, M., Okazaki, N., Ohya, H., Yamazaki, T., Ouchi, K., et al. (2004). Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob. Agents Chemother. 48, 4624–4630. doi: 10.1128/AAC.48.12.4624-4630.2004
Miyashita, N., Kawai, Y., Akaike, H., Ouchi, K., Hayashi, T., Kurihara, T., et al. (2012). Macrolide-resistant Mycoplasma pneumoniae in adolescents with community-acquired pneumonia. BMC Infect. Dis. 12, 126. doi: 10.1186/1471-2334-12-126
Morikawa, K., Oseko, F., Morikawa, S., Iwamoto, K. (1994). Immunomodulatory effects of three macrolides, midecamycin acetate, josamycin, and clarithromycin, on human T-lymphocyte function in vitro. Antimicrob. Agents Chemother. 38, 2643–2647. doi: 10.1128/AAC.38.11.2643
Pereyre, S., de Barbeyrac, B., Renaudin, H., Poutiers, F., Bébéar, C., Bébéar, C. M. (2001). In vitro activity of midecamycin diacetate against Mycoplasma pneumoniae and Chlamydia pneumoniae. J. Antimicrob. Chemother. 47, 240–241. doi: 10.1093/jac/47.2.240
Pereyre, S., Guyot, C., Renaudin, H., Charron, A., Bébéar, C., Bébéar, C. M. (2004). In vitro selection and characterization of resistance to macrolides and related antibiotics in Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 48, 460–465. doi: 10.1128/AAC.48.2.460-465.2004
Principi, N., Esposito, S. (2013). Macrolide-resistant Mycoplasma pneumoniae: its role in respiratory infection. J. Antimicrob. Chemother. 68, 506–511. doi: 10.1093/jac/dks457
Qu, J., Gu, L., Wu, J., Dong, J., Pu, Z., Gao, Y., et al. (2013). Accuracy of IgM antibody testing, FQ-PCR and culture in laboratory diagnosis of acute infection by Mycoplasma pneumoniae in adults and adolescents with community-acquired pneumonia. BMC Infect. Dis. 13, 172. doi: 10.1186/1471-2334-13-172
Rothstein, T. E., Cunningham, S. A., Rieke, R. A., Mainella, J. M., Mutchler, M. M., Patel, R. (2022). Macrolide resistance in mycoplasma pneumoniae, midwestern United States 2014 to 2021. Antimicrob. Agents Chemother. 66, e0243221. doi: 10.1128/aac.02432-21
Schlegel, L., Merad, B., Rostane, H., Broc, V., Bouvet, A. (2001). In vitro activity of midecamycin diacetate, a 16-membered macrolide, against Streptococcus pyogenes isolated in France 1995-1999. Clin. Microbiol. Infect. 7, 362–366. doi: 10.1046/j.1198-743x.2001.00280.x
Starosta, A. L., Karpenko, V. V., Shishkina, A. V., Mikolajka, A., Sumbatyan, N. V., Schluenzen, F., et al. (2010). Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem. Biol. 17, 504–514. doi: 10.1016/j.chembiol.2010.04.008
Suzuki, Y., Itagaki, T., Seto, J., Kaneko, A., Abiko, C., Mizuta, K., et al. (2013). Community outbreak of macrolide-resistant Mycoplasma pneumoniae in Yamagata, Japan in 2009. Pediatr. Infect. Dis. J. 32, 237–240. doi: 10.1097/INF.0b013e31827aa7bd
Tanaka, T., Oishi, T., Miyata, I., Wakabayashi, S., Kono, M., Ono, S., et al. (2017). Macrolide-resistant mycoplasma pneumoniae infection, Japan 2008-2015. Emerg. Infect. Dis. 23, 1703–1706. doi: 10.3201/eid2310.170106
Vázquez-Laslop, N., Mankin, A. S. (2018). How macrolide antibiotics work. Trends Biochem. Sci. 43, 668–684. doi: 10.1016/j.tibs.2018.06.011
Waites, K. B., Bébéar, C. M., Robertson, J. A., Talkington, D. F., Kenny, G. E. (2001). Laboratory diagnosis of Mycoplasma Infections (Washington DC: ASM Press).
Waites, K. B., Xiao, L., Liu, Y., Balish, M. F., Atkinson, T. P. (2017). Mycoplasma pneumoniae from the respiratory tract and beyond. Clin. Microbiol. Rev. 30, 747–809. doi: 10.1128/CMR.00114-16
Wang, X., Li, M., Luo, M., Luo, Q., Kang, L., Xie, H., et al. (2022b). Mycoplasma pneumoniae triggers pneumonia epidemic in autumn and winter in Beijing: a multicentre, population-based epidemiological study between 2015 and 2020. Emerging Microbes Infections 11, 1508–1517. doi: 10.1080/22221751.2022.2078228
Wang, G., Wu, P., Tang, R., Zhang, W. (2022a). Global prevalence of resistance to macrolides in Mycoplasma pneumoniae: a systematic review and meta-analysis. J. Antimicrob. Chemother. 77, 2353–2363. doi: 10.1093/jac/dkac170
Wang, N., Xu, X., Xiao, L., Liu, Y. (2023). Novel mechanisms of macrolide resistance revealed by in vitro selection and genome analysis in Mycoplasma pneumoniae. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1186017
Wang, N., Zhou, Y., Zhang, H., Liu, Y. (2020). In vitro activities of acetylmidecamycin and other antimicrobials against human macrolide-resistant Mycoplasma pneumoniae isolates. J. Antimicrob. Chemother. 75, 1513–1517. doi: 10.1093/jac/dkaa027
Xin, D., Mi, Z., Han, X., Qin, L., Li, J., Wei, T., et al. (2009). Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob. Agents Chemother. 53, 2158–2159. doi: 10.1128/AAC.01563-08
Yan, C., Yang, H., Sun, H., Zhao, H., Feng, Y., Xue, G., et al. (2020). Diversity in genotype distribution of mycoplasma pneumoniae obtained from children and adults. Jpn J. Infect. Dis. 73, 14–18. doi: 10.7883/yoken.JJID.2019.037
Yin, Y.-D., Wang, R., Zhuo, C., Wang, H., Wang, M.-G., Xie, C.-M., et al. (2017). Macrolide-resistant Mycoplasma pneumoniae prevalence and clinical aspects in adult patients with community-acquired pneumonia in China: a prospective multicenter surveillance study. J. Thorac. Dis. 9, 3774–3781. doi: 10.21037/jtd.2017.09.75
Yoshida, T., Watanabe, T., Shomura, T., Someya, S., Okamoto, R., Ishihara, S., et al. (1982). Bacteriological evaluation of midecamycin acetate and its metabolites. Jpn J. Antibiot 35, 1462–1474.
Yuan, X., Liu, Y., Bai, C., Luo, Y., Wang, R., Wang, R., et al. (2014). Mycoplasma pneumoniae infection is associated with subacute cough. Eur. Respir. J. 43, 1178–1181. doi: 10.1183/09031936.00067213
Zhang, L., Tian, X., Sun, L., Mi, K., Wang, R., Gong, F., et al. (2024). Bacterial efflux pump inhibitors reduce antibiotic resistance. Pharmaceutics 16, 170. doi: 10.3390/pharmaceutics16020170
Zhao, F., Li, J., Liu, J., Guan, X., Gong, J., Liu, L., et al. (2019). Antimicrobial susceptibility and molecular characteristics of Mycoplasma pneumoniae isolates across different regions of China. Antimicrobial Resistance Infection Control 8, 143. doi: 10.1186/s13756-019-0576-5
Zhao, C., Xie, Y., Zhang, F., Wang, Z., Yang, S., Wang, Q., et al. (2020). Investigation of antibiotic resistance, serotype distribution, and genetic characteristics of 164 invasive streptococcus pneumoniae from north China between april 2016 and october 2017. Infect. Drug Resist. 13, 2117–2128. doi: 10.2147/IDR.S256663
Keywords: Mycoplasma pneumoniae, macrolide resistance, resistant mechanism, point mutations, efflux pump
Citation: Xie P, Zhang Y, Qin Y, Fang Y, Yang N, Bai Y, Zhi S, Niu W, Wang F and Yuan X (2025) Macrolide resistance in Mycoplasma pneumoniae in adult patients. Front. Cell. Infect. Microbiol. 15:1496521. doi: 10.3389/fcimb.2025.1496521
Received: 14 September 2024; Accepted: 12 February 2025;
Published: 04 March 2025.
Edited by:
Dongsheng Zhou, Academy of Military Medical Science, ChinaReviewed by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyMilena Milakovic Obradovic, Ludwig Maximilian University of Munich, Germany
Horacio Reyes-Vivas, National Institute of Pediatrics, Mexico
Özgen Köseoglu Eser, Hacettepe University, Türkiye
Copyright © 2025 Xie, Zhang, Qin, Fang, Yang, Bai, Zhi, Niu, Wang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Yuan, eGlueXVhbm5hdGlsZUBzaW5hLmNvbQ==; Wenkai Niu, bml1d2s4OEBzaW5hLmNvbQ==; Fusheng Wang, ZnN3YW5nMzAyQDE2My5jb20=
†These authors have contributed equally to this work
 Panpan Xie
Panpan Xie Yue Zhang1,3†
Yue Zhang1,3† Wenkai Niu
Wenkai Niu Fusheng Wang
Fusheng Wang Xin Yuan
Xin Yuan