- Institute for Functional Microbial Genomics, Faculty of Mathematics and Natural Sciences, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Attachment and uptake into host cells are pivotal steps in the life cycle of the Chlamydiaceae, a family of obligate intracellular pathogens. Chlamydia trachomatis (Ctr) possesses a family of nine polymorphic membrane proteins (Pmps), which have been shown to be crucial for adhesion and internalization. However, the host-cell molecules involved have so far remained unknown. Here, we show that a fragment of Ctr PmpD, which forms high-molecular-weight oligomers in solution and adheres to epithelial cells, also binds to secreted clusterin (sCLU), a chaperone-like protein that is secreted into the extracellular space by the host cell, and forms part of the chaperone- and receptor-mediated extracellular protein degradation (CRED) pathway. Using in vitro assays, we demonstrate that sCLU interacts directly with soluble rPmpD. In infection experiments, depletion of sCLU from the culture medium leads to a significant decrease in Ctr infection. Thus, sCLU is the first host-cell interaction partner identified for a Ctr Pmp and the first case in which sCLU has been shown to be a vital component for the establishment of a bacterial infection.
Introduction
Chlamydia trachomatis (Ctr) is the most common bacterial cause of sexually transmitted infections worldwide. If untreated, it can lead to infertility or ectopic pregnancies (Rodrigues et al., 2022). In addition, Ctr is the leading cause of trachoma, the world’s primary cause of infectious blindness, which has been targeted for elimination by the end of the decade (Malecela and Ducker, 2021). Ctr belongs to the genus Chlamydia, all of which are obligate intracellular, Gram-negative bacterial pathogens that infect epithelial cells (Elwell et al., 2016). Most chlamydial species are pathogenic to animals. However, two species, Ctr and Chlamydia pneumoniae (Cpn) – which infects the respiratory tract – are pathogenic to humans (Kalman et al., 1999). Ctr can be subdivided into 19 serovars and are grouped into three biovars, each of which is responsible for specific pathological conditions (Murray and McKay, 2021). Serovars A-C lead to trachoma and blindness (Murray and McKay, 2021). Serovars D-K cause infections of the genital tract and can lead to pelvic inflammatory disease (PID), ectopic pregnancy and infertility in women, and urethritis and epididymitis in men (Murray and McKay, 2021). Serovars A-K typically cause local infections only. Serovars L1-L3, also known as the lymphogranuloma venereum biovar, lead to invasive infections of the urogenital and anorectal tract, and cause systemic infections (de Vrieze and de Vries, 2014).
All chlamydial species, including Ctr, utilize a unique type of developmental cycle that involves switching between two morphological forms – the infectious elementary body (EB) and the non-infectious reticulate body (RB) (Moulder, 1991). Chlamydial infections are initiated when EBs adhere to, and are taken up by host cells. After internalization, the EB remains in a membrane-enveloped compartment known as an inclusion, in which it differentiates into an RB. By hijacking nutrients from the host cell, the RB undergoes repeated replication cycles before the pool of RBs asynchronously transitions back into EBs, which are released into the extracellular space either via cell lysis or extrusion of the inclusion (Moulder, 1991; Abdelrahman and Belland, 2005; Elwell et al., 2016; Christensen et al., 2019).
For obligate intracellular pathogens, adhesion to and uptake into the host cell is of the utmost importance. For the successful implementation of these processes, Chlamydiae have evolved a range of adhesins including GroEL1, MOMP, OmcB, Ctad1 and a whole family of polymorphic membrane proteins (Pmps) (Stephens et al., 2001; Mölleken and Hegemann, 2008; Wuppermann et al., 2008; Hegemann and Moelleken, 2012; Fechtner et al., 2013; Becker and Hegemann, 2014; Stallmann and Hegemann, 2016).
Pmps are synthesized by all chlamydial species and can be subdivided into six phylogenetic subtypes (A to H), with 21 members in Cpn and 9 members in Ctr (Grimwood and Stephens, 1999). Pmps share between 19% and 40% sequence identity across species, and are thought to belong to the broader class of type V autotransporters, based on their domain architecture (Henderson and Lam, 2001). These proteins possess an N-terminal secretion signal, a C-terminal β-barrel and a central passenger domain (PD) (Henderson and Lam, 2001). The signal sequence triggers the Sec-dependent transportation of the Pmp(s) across the inner membrane into the periplasmic space, where the Sec sequence is typically split off and the protein folds. The β-barrel is inserted into the outer membrane, supported by the BAM complex, and forms a channel that enables the PD to be exported to the cell surface (Henderson et al., 2004). The available evidence suggests that the extracellular Pmp PDs exist in both membrane-anchored and soluble forms, for which proteolytic processing sites have been identified (Wehrl et al., 2004; Kiselev et al., 2007; Swanson et al., 2009). Furthermore, Pmp PDs are exceptionally rich in FxxN and GGA(I,L,V) motifs, which have been shown to be crucial for the adhesion of EBs to epithelial cells (Mölleken et al., 2010).
Among the nine Ctr Pmps, all of which are known to adhere to epithelial cells and are essential for infection (Becker and Hegemann, 2014), PmpD and its Cpn homologue Pmp21 are the best studied at present. Proteomic studies suggest that PmpD undergoes proteolytic processing, which results in several fragments with or without the β-barrel domain (Kiselev et al., 2009; Swanson et al., 2009). Interestingly, immunoaffinity-purified PmpD from the EB surface is organised into high-molecular-weight complexes, consisting of four to six PmpD fragments (Swanson et al., 2009). In vitro, the formation of such high-molecular-weight oligomers has been demonstrated for Ctr PmpA, PmpD, PmpG and PmpI, and they appear to be important for adhesion to epithelial cells (Favaroni and Hegemann, 2021). Biochemical studies have revealed that the monomeric PmpD PD has a high β-sheet content and probably folds into a triangular β-helical structure (Paes et al., 2018; Cervantes et al., 2024). However, oligomeric forms of the PmpD PD have been suggested to form fibril-like structures, like those formed by the amyloid protein fragment Aβ42 that is associated with Alzheimer’s disease (Gu and Guo, 2013; Paes et al., 2018; Favaroni and Hegemann, 2021). Functional studies have shown that PmpD makes a critical contribution to the infection process. Anti-PmpD antibodies exhibit significant pan-neutralizing activity against a number of different Ctr serovars in cell culture, and a PmpD-based vaccine has shown protective activity in mice (Crane et al., 2006; Paes et al., 2016). In addition, a pmpD null mutant of Ctr serovar D showed significantly reduced adhesion and internalization capacities in human cell lines and non-human primate models, but not in murine cells (Kari et al., 2014). Interestingly, in C. muridarum, transposon-mediated inactivation of PmpD, PmpA or PmpI leads to growth attenuation and reduced the numbers of infectious progeny, suggesting that these proteins may have other functions in addition to adhesion (Wang et al., 2019). For the Ctr PmpD homologue in Cpn, Pmp21, similar structural and functional characteristics have been reported (Mölleken et al., 2010; Mölleken et al., 2013; Luczak et al., 2016). However, the roles of PmpD and Pmp21 in infection are thought to be species-specific, as recombinant Pmp21 reduces Cpn but not Ctr infection, and vice versa (Becker and Hegemann, 2014). Pmp21 binds to and activates the epidermal growth factor receptor (EGFR), which facilitates chlamydial uptake via endocytosis. Thus, Pmp21 functions as both an adhesin and an invasin (Mölleken et al., 2013). Similarly, C. psittaci Pmp17G, which belongs to a different subtype than either Ctr PmpD or Cpn Pmp21, is also suggested to bind the EGFR (Li et al., 2021). Importantly, while several human cell-surface receptors, such as heparan sulfate proteoglycans, CFTR, β1-integrin, ephrin A2, and protein disulphide isomerase have been associated with adhesion and/or internalization of Ctr, no receptor has been identified for Ctr Pmps to date (Romero et al., 2020).
Here, we have used a proteolytically processed form of PmpD from Ctr serovar E, found in both soluble and membrane-bound complexes during infection, to identify its host-cell binding partner(s) (Swanson et al., 2009). Ctr Serovar E is the most commonly identified subtype in clinical cases (Lesiak-Markowicz et al., 2019). Biochemical characterization of this PmpD fragment, called rD72, revealed that it forms high-molecular-weight oligomers, and binds to epithelial cells in a concentration-dependent manner. Incubation of rD72 with human epithelial HEp-2 cells, followed by immunoprecipitation, affinity enrichment and mass spectrometric analysis revealed that rD72 interacts with the secreted form of the host-cell protein clusterin (CLU). Clusterin is a human protein, part of which is secreted into the extracellular space, where it facilitates the in-vivo clearance of misfolded extracellular proteins, with a high preference for amyloid-β-like structures. Secreted CLU (sCLU) is known for its chaperone-like function; it binds to its cargo protein, and induces uptake of the complex into the target cell, where it is eventually degraded. sCLU also acts as a terminal pathway inhibitor of the complement system by interacting with that system’s membrane-attacking complex (MAC) (Choi et al., 1989). Using biochemical and biological approaches, we have confirmed the direct interaction of PmpD with sCLU. Furthermore, we demonstrate that the absence of sCLU in the culture medium significantly reduces Ctr infection in epithelial cells. In summary, we show that Ctr PmpD binds to secreted clusterin, and that the presence of extracellular sCLU is essential for a Ctr infection.
Results
D72, a proteolytically processed fragment of PmpD, forms high-molecular-weight homo-oligomers and binds to epithelial cells
Previous work showed that all nine Ctr Pmps can bind to epithelial cells and block subsequent Ctr infection; this is why Pmps were defined as adhesins (Becker and Hegemann, 2014). The Pmp fragments used in those studies were deliberately designed to focus on regions with high densities of GGA(I,L,V) and FxxN motifs, since these characteristic tetrapeptide motifs had initially been shown to be essential for adhesion (Mölleken et al., 2010). Here, we focus on D72, a fragment of the Ctr PmpD, which was discovered in in-vivo studies (Figure 1A) (Kiselev et al., 2009; Swanson et al., 2009). D72 spans the N-terminal half of the passenger domain (PD) of PmpD, which encompasses amino-acid residues 68 to 761. It therefore includes 12 GGA(I,L,V) and 14 FxxN motifs. Structure prediction tools suggest that the recombinant D72 (rD72) forms a lengthy β-helical structure, as has been suggested for fragments of other Pmp PDs (Supplementary Figure S1) (Debrine et al., 2023).
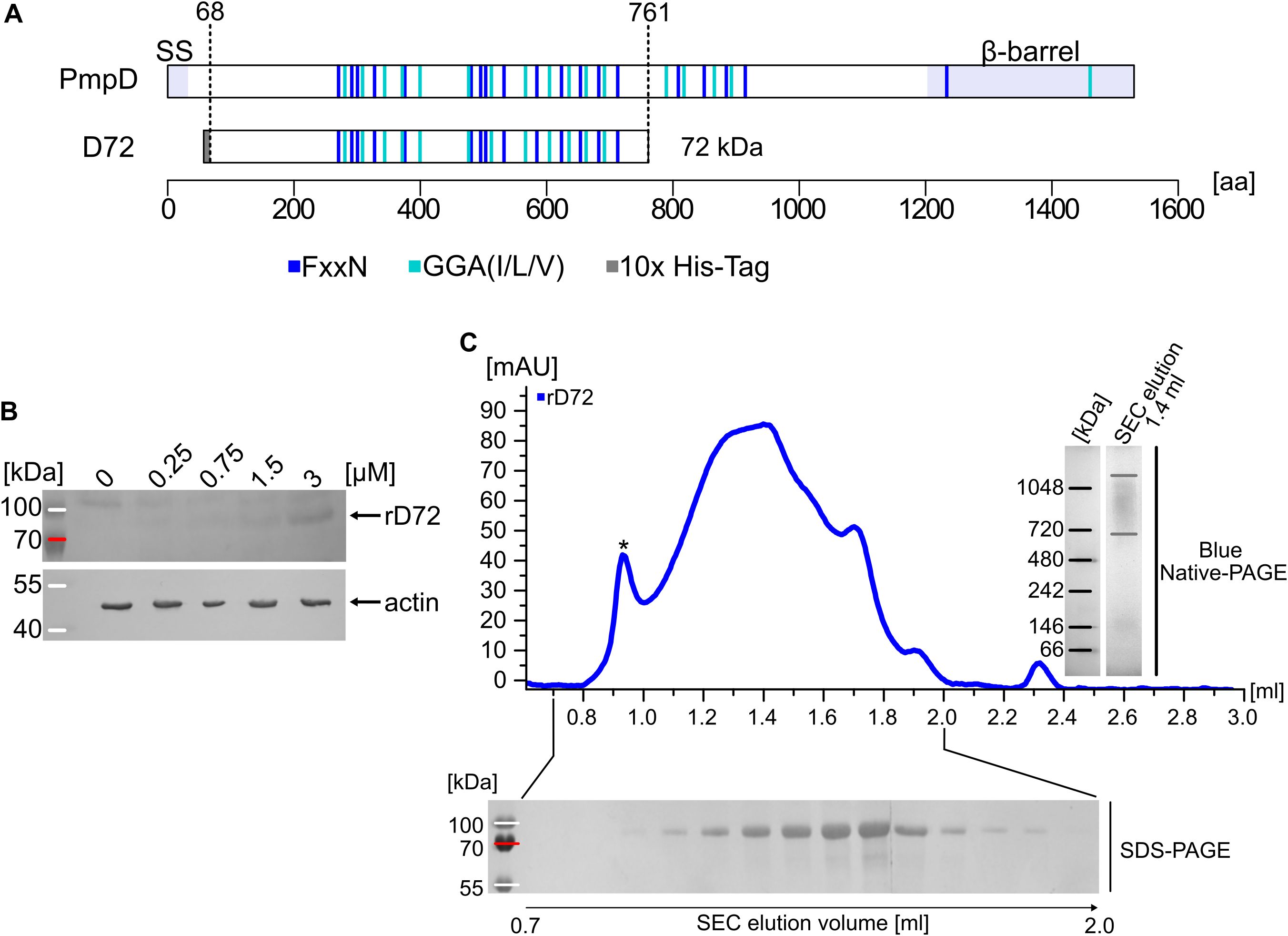
Figure 1. Recombinant D72 is a biologically functional fragment of Ctr PmpD. (A) Schematic representation of Ctr PmpD and its proteolytically processed fragment D72, which spans aa residues 68-761. The N-terminal signal sequence (SS), the passenger domain (PD) and the C-terminal β-barrel are indicated. The positions of FxxN and GGA(I,L,V) motifs are marked in dark and light blue, respectively. The grey box indicates the genetically fused 10x His-Tag. The processed fragment includes 14 FxxN and 12 GGA(I,L,V) motifs. The image was generated in R Studio using a customized code (RStudio Team, 2020; R Core Team, 2024). (B) Increasing concentrations of His-tagged soluble recombinant D72 (rD72) was added to the cell culture medium (ccm) and incubated for 1 h at 4°C with HEp-2 cells, which were grown in this ccm for two days. Bound protein was visualized on a Western blot using an anti-His antibody. Actin was used as an internal loading control. (C) Size-exclusion chromatography (SEC) was used to analyse the oligomeric state of rD72 in solution. The void volume is indicated by an asterisk. Samples of the individual elution fractions were analysed by SDS-PAGE (bottom). A sample of the elution fraction at 1.4 ml was also collected and analysed by Blue Native-PAGE (right).
In order to characterize the biochemical and functional properties of rD72, its ability to (i) adhere to epithelial cells and (ii) form high-molecular-weight oligomers was investigated. To this end, D72 was expressed in E. coli and purified with the aid of an N-terminal 10x His-Tag (rD72, Figure 1A). Adhesion to epithelial cells was tested by adding different concentrations of soluble rD72 to human HEp-2 cells growing in cell culture medium (ccm). Binding of rD72 to HEp-2 cells was detectable even at the lowest concentration used (0.25 µM). With increasing concentration of rD72 in the ccm, binding of rD72 also increased (Figure 1B).
Pmp protein fragments are known to form homo- and hetero-oligomers in vitro (Swanson et al., 2009; Luczak et al., 2016; Paes et al., 2018; Favaroni and Hegemann, 2021). The oligomerisation of rD72 was investigated by size-exclusion chromatography (SEC), Blue Native-PAGE (BN) and SDS-PAGE (Figure 1C). Soluble rD72 was loaded onto a Superose 6 SEC column, and was eluted in a broad peak, which began to rise after the column void volume and eventually spanned one-third of the total volume of the column bed (Figure 1C). SDS-PAGE verified the presence of full-length rD72 (Figure 1C, bottom). BN analysis of the SEC elution fraction at 1.4 ml revealed a high-molecular-weight oligomer of between 720 and 1048 kDa, corresponding to a calculated 10- to 15-mer (Figure 1C, right).
Taken together, these data indicate that rD72 is a biologically functional PmpD fragment that adheres to epithelial cells. In addition, rD72 shows comparable biochemical characteristics to those of other Pmp PD fragments, including a high degree of oligomerisation in solution (Luczak et al., 2016; Paes et al., 2018; Favaroni and Hegemann, 2021). Based on these findings, rD72 was used in further experiments with the aim of identifying host-cell binding partner(s).
Secreted clusterin is a host-cell binding partner for rD72
We carried out immunoprecipitation experiments to identify potential host-cell binding partner(s) for rD72. To this end, confluent HEp-2 cells grown in cell culture medium (ccm) were supplemented with soluble rD72 and incubated for 1 h at 4°C. As the negative control, fresh PBS was added to the ccm. After 1 h, cells were washed three times with HBSS to remove unbound rD72, then cells were lysed, and rD72-containing complexes were isolated by affinity purification (AP) using Ni-NTA agarose. The individual purification steps were monitored via Western-blot analysis, using anti-His antibodies to detect rD72 (Figure 2A; Supplementary Figure 3). The protein composition of the elution fraction, containing rD72, was then subjected to mass spectrometric analysis to identify proteins that co-eluted with rD72.
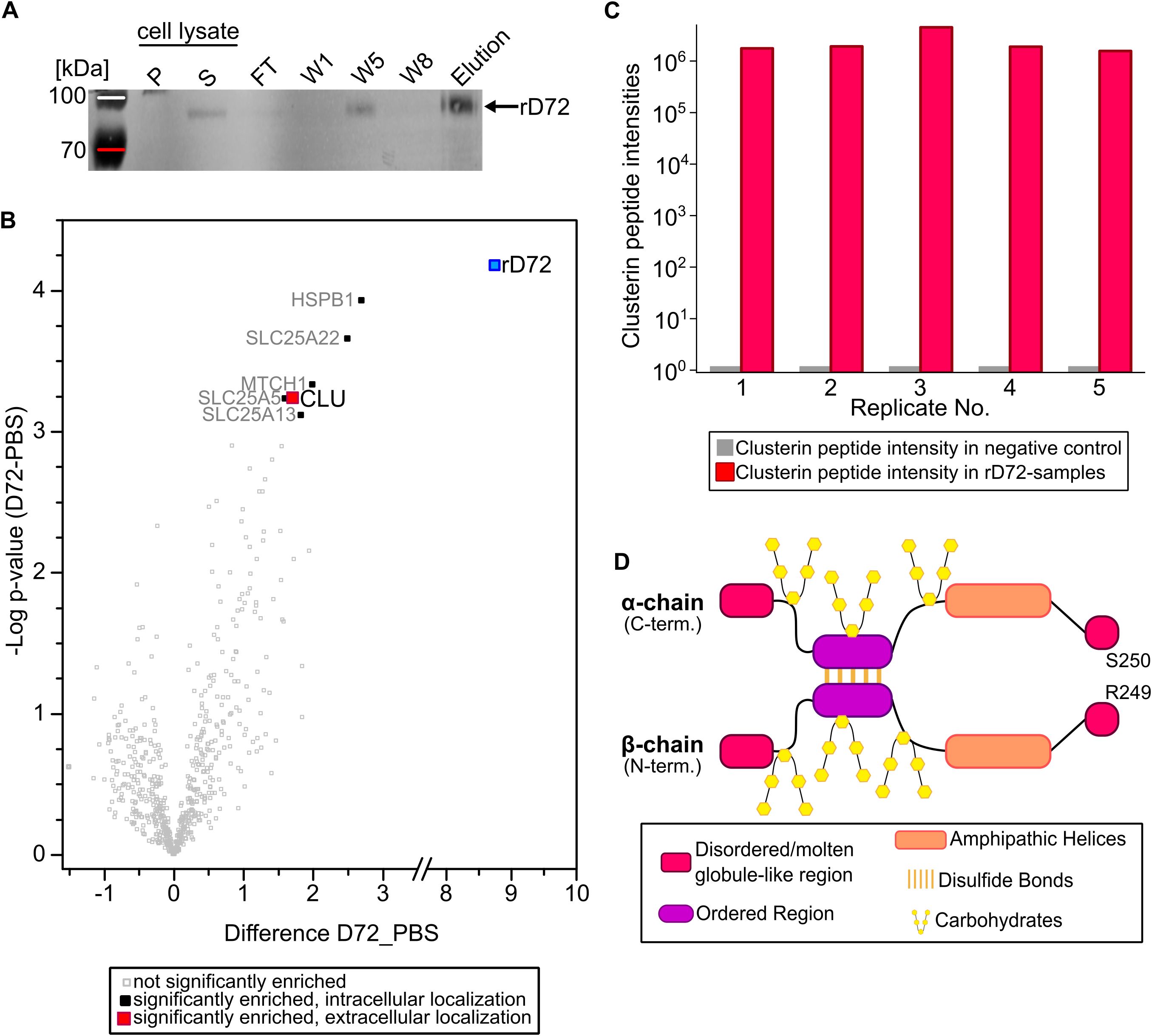
Figure 2. rD72 binds to the secreted host-cell chaperone clusterin. (A) Pulldown experiment in which rD72 was added to HEp-2 cells which had been grown for 2 days in cell culture medium (ccm). Cells were lysed to solubilize protein complexes and the lysate was cleared by centrifugation (P = pellet, S = supernatant). rD72 and any bound host-cell interaction partner(s) were co-purified using Ni-NTA agarose. Fractions of the individual Ni-NTA purification steps were analysed on Western blots, and probed with an anti-His antibody. (FT = flow-through, W1 = wash 1, W5 = wash 5, W8 = wash 8) (B) Volcano plot of human proteins identified by mass spectrometry after affinity purification (n=5). Proteins that were significantly enriched relative to the negative control (PBS) are labelled (Uniprot nomenclature). The fold change (rD72 minus PBS) is plotted against the difference in mean values of log2 label-free quantitation (LFQ) intensities (rD72 minus PBS). (C) The intensities of clusterin peptides in each replicate are compared between the test sample containing rD72 (red columns) and the corresponding negative control (PBS, grey columns). (D) Schematic representation of secreted human clusterin (sCLU). In the cytoplasm, the clusterin precursor is N-glycosylated and is cleaved between residues R249 and S250. The resulting α- and β-chains are then linked in an antiparallel fashion by five intramolecular disulfide bonds, thus generating the secreted isoform (sCLU). Mature sCLU has a molecular weight of ~70 kDa and is secreted into the extracellular space.
Intriguingly, apart from rD72 itself, only five significantly enriched host cell proteins were detected in the AP elution fraction (Figure 2B; Supplementary Figure S2; Supplementary Table S2). All five of these were identified as intracellular proteins, and thus should not be accessible for interactions with extracellular rD72. Indeed, four of them are predicted to be mitochondrial components (Supplementary Table S2). However, the fifth, clusterin (CLU), is also known to be secreted in a soluble extracellular form, designated as sCLU, and is therefore a potential candidate binding partner for rD72 (Bettens et al., 2015). Detailed analysis of the CLU peptide intensities detected by mass-spectrometric analysis confirmed that CLU was enriched in all five replicates of the test samples, and was not found in the negative controls (Figure 2C).
CLU is an ATP-independent chaperone whose main function is to stabilise misfolded proteins and inhibit the formation of amorphous or amyloid-like aggregates, particularly in the extracellular space (Poon et al., 2000; Hatters et al., 2001). The sCLU isoform is derived from a precursor polypeptide of ~50 kDa, which undergoes extensive post-translational modification, including extensive glycosylation (Gross et al., 2023). Furthermore, sCLU forms five intramolecular disulphide bonds, which connect the α- and β-chains in an antiparallel arrangement following cleavage of the precursor molecule (Figure 2D) (Burkey et al., 1991). The fully glycosylated α- and β-chains have molecular weights of 34-36 kDa and 36-39 kDa, respectively (de Silva et al., 1990).
In vitro pulldown assays verify the interaction between rD72 and sCLU
To investigate the respective levels of intra- and extracellular CLU, HEp-2 cells were seeded in sub-confluent amounts and grown for two days at 37°C in ccm. The ccm and the cultured cells were then separately analysed by Western blotting with an antibody raised against the clusterin α-chain (Figure 3A). Fresh ccm served as the negative control, and no non-specific signals were observed. In contrast to this, ccm obtained from HEp-2 cells grown for two days was found to contain a broad protein band ranging in size from approximately 40 kDa to 45 kDa, which corresponded to the secreted CLU α-chain (Figure 3A). Multiple bands were observed in the cell lysate, representing the cleaved and uncleaved clusterin precursor, which had undergone different degrees of post-translational modification (Figure 3A). Thus, growing HEp-2 cells secrete mature clusterin into the extracellular medium.
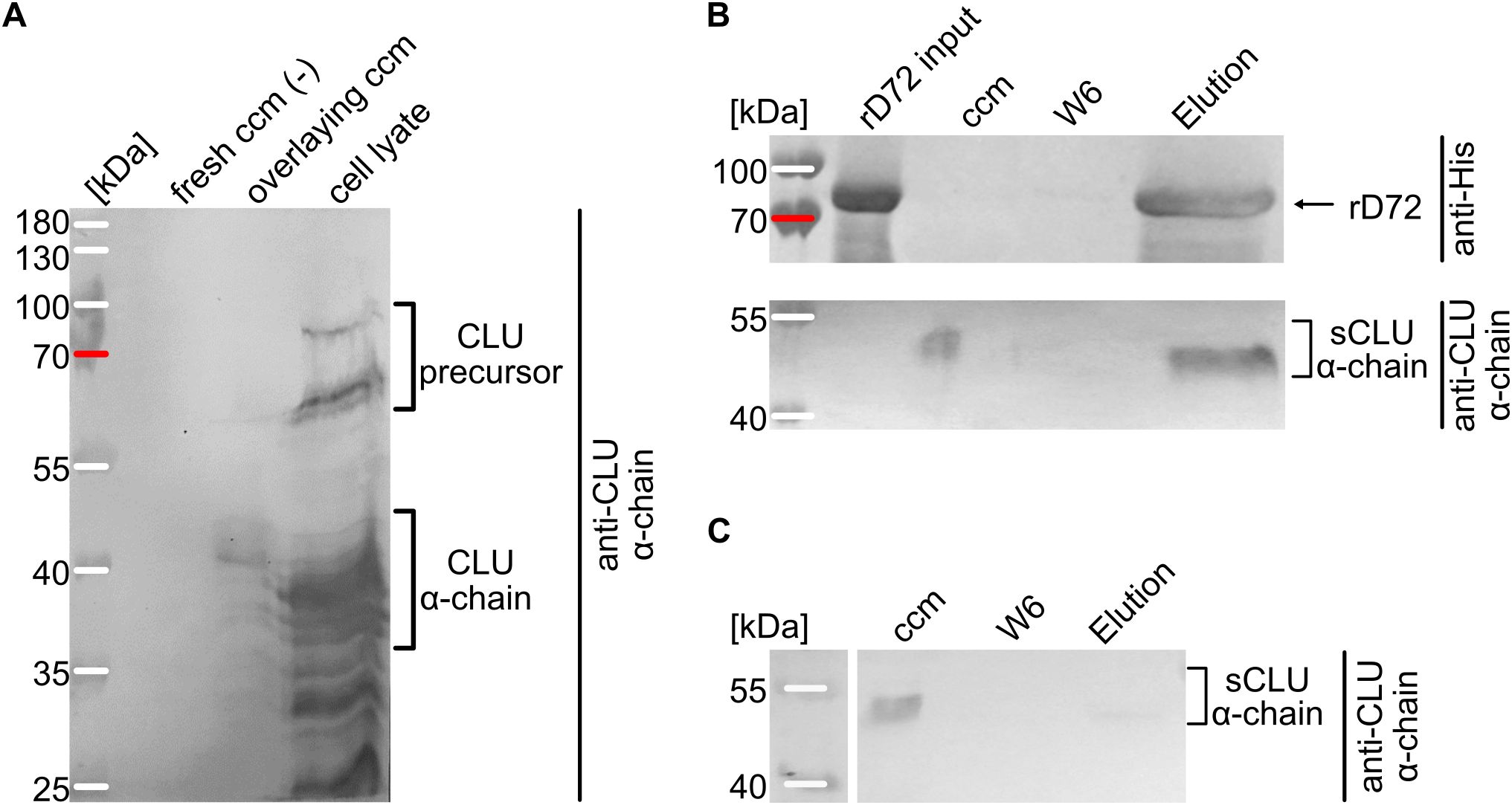
Figure 3. In vitro interaction of rD72 with clusterin. (A) Analysis of clusterin abundance in HEp-2 cells by Western-blot analysis, using an antibody against the clusterin α-chain. Fresh cell culture medium (ccm) was used as the negative control (-). In the ccm isolated from cells that had been grown for 2 days (overlaying ccm), the post-translationally modified clusterin α-chain is detected at ~40-45 kDa (theoretical MW: 36-39 kDa). In the cell lysate, bands appear at 60-80 kDa (corresponding to the clusterin precursor) and the bands at ~40-45 kDa represent the fully processed α-chain of secreted clusterin (sCLU). (B) Ni-NTA pulldown assays. rD72 was incubated with ccm isolated from epithelial cells that had been grown for two days, and used as bait. The protein composition of the individual fractions of the pulldown assay was probed after Western blotting, using anti-His and anti-clusterin α-chain antibodies. (W6 = wash 6). (C) Negative control for the Ni-NTA pulldown assay. The ccm used was obtained from HEp-2 cells that had been grown for two days in the absence of rD72. Fractions were separated by SDS/PAGE and analysed via Western blot, using an anti-clusterin α-chain antibody.
To verify the direct interaction between sCLU and rD72, an in vitro pulldown assay was performed, using rD72 as bait (Figure 3B; Supplementary Figure 4). Supernatant obtained from HEp-2 cells that had been grown for two days was collected and incubated with soluble rD72. rD72 was then affinity-purified using Ni-NTA agarose, and the elution fractions were analysed on a Western blot. As a negative control, 2-day-old ccm was incubated with Ni-NTA agarose in the absence of rD72 (Figure 3C). While the negative control showed no non-specific binding of sCLU to the agarose matrix, the test samples consisting of ccm with the His-tagged rD72 as bait clearly demonstrated co-elution of both proteins in the elution fraction. These findings confirm that sCLU can interact with rD72.
Depletion of secreted clusterin from the culture medium inhibits infection by C. trachomatis
Having verified that sCLU interacts directly with Ctr PmpD, the role of sCLU during the initial stages of a chlamydial infection was investigated. To this end, HEp-2 cells were grown in ccm for two days, and the level of clusterin in the ccm was manipulated, prior to infection, using four different experimental approaches (Figure 4A). We first set up a regular infection experiment that served as a control (i). In this case, Ctr EBs were added to HEp-2 cells, which had been grown in ccm for 2 days, yielding “old medium” enriched in sCLU (Figures 4A, B). In the second approach (ii), ccm in which cells had been grown for two days was removed and replaced by fresh medium that was devoid of sCLU (“new medium”, Figure 4A, B), which was then supplemented with Ctr EBs to initiate infection. In the third approach (iii), after two days of cell growth, soluble rD72 was added to sCLU-rich “old medium” and incubated for 1 h before Ctr EBs were added (“rD72 treatment”, Figure 4A). Western-blot analysis showed no significant change in sCLU abundance after 1 h incubation with rD72, relative to the level of sCLU employed in approach (i) (Figure 4B; Supplementary Figure 5). In the fourth approach (iv), “old medium” in which cells had been grown for two days was isolated, and the sCLU contained in it was removed by affinity purification (AP). The flow-through fraction, depleted in sCLU, was added to the cell monolayer, which was then infected with Ctr EBs (Figures 4A, B).
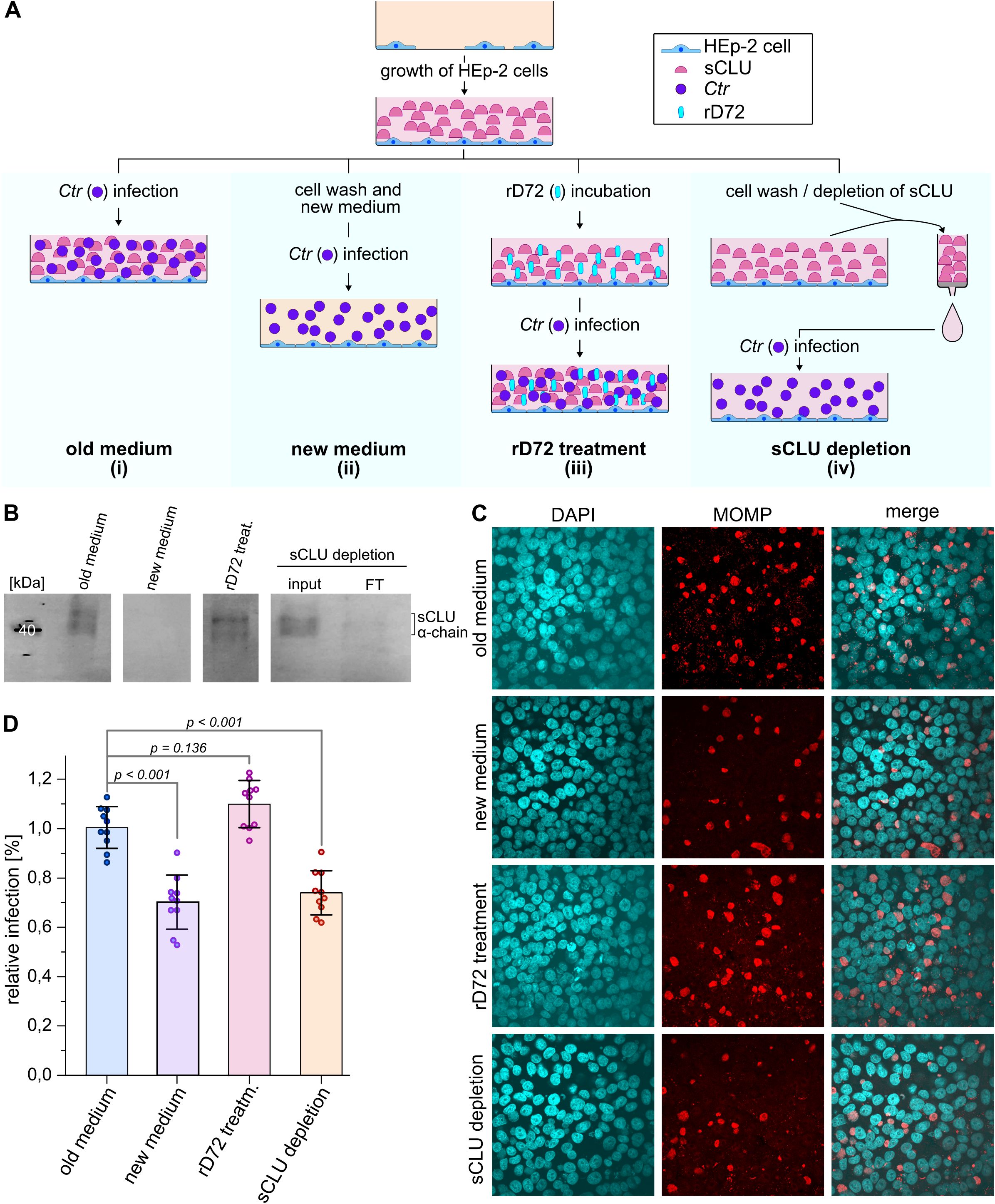
Figure 4. sCLU facilitates uptake of Ctr into host cells. (A) Schematic representation of the different experimental setups used to test the effect of sCLU during the initial steps of a chlamydial infection. In all four experiments (i-iv), confluent cells were grown for two days in ccm. In approach (i), Ctr EBs were added to confluently grown cells. For approach (ii), the cells were washed and fresh ccm supplemented with Ctr EBs was added, in the absence of sCLU. For approach (iii), the ccm was supplemented with soluble rD72 and incubated with HEp-2 cells for 1 h before Ctr EBs were added to the ccm. For approach (iv), ccm that had been used for cell growth was isolated and sCLU was extracted via affinity purification (AP), while the flow-through, depleted of sCLU, was returned to the HEp-2 cells, supplemented with Ctr EBs. For all four approaches, EBs were incubated with HEp-2 cells for 2 h at 37°C before the free EBs were removed. After 24 h, the infection was stopped by fixing and permeabilizing the cells with PFA and methanol. (B) Western blot analysis of ccm, using an anti-clusterin α-chain antibody. For approaches (i) to (iii), samples were taken from the ccm immediately prior to supplementation with Ctr EBs. For approach (iv), the AP input (input) and the AP flow-through (FT) were analysed. (C) Sections of immunofluorescence microscopy pictures of the individual infection approaches as depicted in (A). DNA is visualized with DAPI (cyan) and inclusions are visualized with an anti-MOMP antibody (red). Images are representative of three biologically independent replicates (n=3). (D) Infection rates of the individual approaches (i-iv) were determined by calculating the ratio of inclusions to the number of HEp-2 cells. The infection rate of approach (i) was set to 1 and approaches (ii-iv) were normalized to it. Three biologically independent experiments (n=3) were performed, and for each replicate, 3-4 images on different sections were taken. The means of the individual data points are indicated by the height of the individual bars (± SD) and the p-values are indicated.
In all cases, EBs were allowed to adhere to epithelial cells for 2 h prior to removal of the remaining free EBs, and the infection was allowed to proceed for 24 h (Figure 4C). Interestingly, the level of infection detected in cells grown in the presence of “fresh medium” that initially contained no detectable sCLU (approach (ii)) was found to be significantly reduced (by 30%) relative to the control experiment (i) (Figure 4D). Virtually the same reduction in the infection rate (to 0.72) was observed in approach (iv), in which “old medium” that had been depleted of sClu was used (Figure 4D). In contrast, pre-incubation of “old medium” (abundant in sClu) with rD72 in approach (iii) led to a modest increase in the infection rate (1.1) compared to the control (i) (Figure 4D). Importantly, we did not see any obvious differences in the inclusion size in the infection assays (i) to (iv).
The results obtained from approaches (i) to (iv) indicate the relevance of the availability of secreted clusterin for successful Ctr infection.
Discussion
The initial adhesion to, and subsequent uptake of EBs into epithelial host cells are critical steps in Ctr´s developmental cycle. The primary contact between pathogen and host is mediated by chlamydial adhesins including GroEL1, MOMP, OmcB, Ctad1 and Pmps that bind to specific host-cell proteins (Mölleken and Hegemann, 2008; Wuppermann et al., 2008; Mölleken et al., 2010; Stallmann and Hegemann, 2016; Romero et al., 2020). The family of Pmps plays a complex but crucial role in the adhesion step and the ensuing internalization processes (Vasilevsky et al., 2016; Romero et al., 2020). All nine Ctr Pmps have been demonstrated to adhere to epithelial cells, and infection-blocking assays using soluble recombinant Pmp fragments have indicated that each of these proteins contributes to Ctr infectivity. However, their host-cell binding partners have remained unknown up to now (Becker and Hegemann, 2014). Here, we show that Ctr PmpD, which forms high-molecular-weight (hMW) oligomers in solution and attaches to epithelial cells, interacts with the secreted host-cell chaperone clusterin (sCLU). Under the experimental conditions used, infectivity is significantly reduced in the absence of sCLU in the cell culture medium (ccm) during the early stages of Ctr infection.
A fragment of the central passenger domain of PmpD, referred to here as rD72, can be found in vivo during infection (Swanson et al., 2009), and forms hMW oligomers in solution, as indicated by SEC analysis and Blue Native-PAGE (Figure 1). The SEC elution profile for rD72 resembles that of the 65-kDa PmpD PD fragment investigated by Paes et al (Paes et al., 2018). Similarly, Favaroni et al., using yet another motif-rich 66-kDa PmpD PD fragment, obtained similar results and further suggested, based on transmission electron microscopy, that these hMW oligomers might form protofibril-like structures (Favaroni and Hegemann, 2021). Furthermore, Luczak et al. working on Pmp21, the Cpn homologue of Ctr PmpD, showed that the hMW oligomers formed by Pmp21 exhibit an amyloid-like character (Luczak et al., 2016) that is very similar to the amyloid fibrils formed by Aβ42, a protein fragment which plays a vital role in Alzheimer’s disease (Gu and Guo, 2013). Taken together, these converging data, in combination with results obtained with structure-prediction tools, suggest that all Pmp PDs, including rD72, form hMW oligomers that are made up of protofibril structures with amyloid-like properties.
Using rD72 bound to HEp-2 cells as bait in pulldown assays, we identified CLU as a direct interaction partner via mass spectrometry (Figures 2, 3). CLU is a ubiquitously expressed protein present in nearly all human tissues, and occurs in multiple isoforms, including a secreted isoform, sCLU, that is found in the extracellular space (Gross et al., 2023). Secretion of CLU generally occurs from vesicles via the secretory pathway (Nizard et al., 2007). Once outside the cell, sCLU acts in chaperone-like fashion to clear the extracellular space of misfolded proteins that might otherwise give rise to unstructured, amorphous or amyloid aggregates (Humphreys et al., 1999; Poon et al., 2000; Wilson and Easterbrook-Smith, 2000). Thus, sCLU is associated with several proteinopathies, including Alzheimer´s disease (Satapathy and Wilson, 2021). In Alzheimer’s disease, Aβ peptides accumulate in the extracellular space of brain cells and eventually oligomerize and aggregate, forming amyloid fibrils. sCLU is known to bind to Aβ oligomers in the brain, thus preventing their aggregation and inducing the clearance of Aβ via transportation across the blood-brain-barrier (Howlett et al., 2013; Madadi et al., 2019). The endocytic mechanisms triggered by sCLU that enable uptake of cargo and its delivery to lysosomes for degradation have not yet been fully identified. However, it was recently demonstrated that sCLU participates in the chaperone- and receptor-mediated extracellular protein degradation (CRED) pathway for aberrant extracellular proteins, and that sCLU-cargo complexes bind to heparan sulfate (HS) receptors on cells via electrostatic interactions (Itakura et al., 2020).
Our data indicate that sCLU binds directly to soluble rD72, which has been shown to form hMW oligomers that probably have amyloid-like characteristics. Hence, we suggest that the sCLU-rD72 interaction is based on rD72’s capacity to form amyloid-like aggregates, which are natural targets of sCLU. Importantly, alteration of the level of sCLU in the ccm either by replacing spent medium with fresh medium shortly before infection or by immunodepletion of sCLU from 2-day-old ccm, prior to infection with Ctr EBs, results in an approximately 30% reduction in the infection rate in the absence of sCLU (Figure 4). Based on these experiments, we cannot formally rule out the possibility that other components of the cell culture medium directly or indirectly might also have an influence on the infection rate. Most likely, however, our data suggest that sCLU promotes Ctr infectivity.
Furthermore, we would also like to postulate that sCLU not only binds to soluble recombinant PmpD, but also to PmpD that is localized on the surface of EBs. All Ctr Pmp proteins act as adhesins and are essential for infection (Becker and Hegemann, 2014). Several Pmp proteins form homo and hetero oligomers with a similar fibrillar structure in vitro (Favaroni and Hegemann, 2021; Luczak et al., 2016). For all Pmp passenger domains, AlphaFold predicts a highly similar beta-helical domain structure, which allows for an oligomerization (Cervantes et al., 2024). Based on this, we suspect that sCLU could also bind other, possibly even all Pmps (Favaroni and Hegemann, 2021; Cervantes et al., 2024). This must be tested in further experiments.
Previous work has shown that pre-incubation with soluble rPmpD (and all other rPmps) blocks subsequent infection. However, in these experiments the 2-day-old ccm (enriched in sCLU) was replaced with fresh ccm (lacking sCLU) prior to infection with Ctr (Becker and Hegemann, 2014). Moreover, the slight increase in infectivity obtained in approach (iii) in Figure 4, confirms published data, which have shown that incubation of infectious EBs with adhesion-competent D72 can functionally replace the naturally exposed adhesive structures, probably because rD72 may be able to bind to the EB cell surface alone as well as in a complex with CLU (Favaroni and Hegemann, 2021). Thus, future experiments will have to clarify how soluble recombinant PmpD mechanistically affects the CLU-dependent Ctr infection.
It is at present unclear but likely, that other Pmp proteins can also interact with sCLU. In vitro, Pmp proteins have the capacity to interact with themselves and with other Pmps forming functional, polyadhesive, homo- and heteromeric autotransporter complexes (Favaroni and Hegemann, 2021). This makes a PmpD analysis by testing a pmpD gene deletion strain challenging. Furthermore, although a PmpD knock-out is available it has been generated in serovar D while our study was performed with the PmpD protein from Ctr serovar E (Kari et al., 2014). There is currently no protocol for the transformation of Ctr serovars of the urogenital tract. To date, all interventions in the Ctr genome have been performed on the L2 serovar strain. The serovar-specific differences make the transfer of data between serovars difficult. We also did not attempt to genetically inactivate the CLU gene because clusterin is a ubiquitously expressed glycoprotein that is involved in a whole range of biological processes including lipid transport, membrane recycling, regulation of apoptosis and inhibiting formation of the complement-mediated membrane attack complex (MAC) (Satapathy and Wilson, 2021). The consequences of CLU inactivation on a chlamydial infection could therefore be direct and/or indirect, which would make their assessment impossible.
Our model is that EBs may be treated as cargo by sCLU, thus facilitating their internalization by the host cell, similar to the mechanism of Aβ clearance from the extracellular space via the CRED pathway. Uptake into host cells may be mediated by heparan sulfate receptors. In addition, the involvement of a co-receptor in facilitating endocytic uptake of the EB by CRED seems likely. Previous studies on various bacterial and viral pathogens that make use of heparan sulfate for host-cell entry have shown that they also rely on co-receptors to promote internalization into the host cell via endocytic processes (Yayon et al., 1991; Liu and Thorp, 2002; Pillay et al., 2016). Indeed, heparan sulfate is already known to be the host receptor for the Ctr adhesin OmcB (Mölleken and Hegemann, 2008). Interestingly, blocking adhesion of Ctr EBs to HEp-2 cells with heparin (a heparan sulfate analogue) reduces binding by approximately 95%, while addition of rOmcB reduced adhesion of EBs by about 70%. These differences may be explained by the fact that heparin blocks binding of both OmcB and sCLU, while blocking by rOmcB only blocks its specific binding sites on heparan sulfate (Mölleken and Hegemann, 2008).
After sCLU-triggered internalization, the endocytic vesicle is typically designated for lysosomal degradation (Kounnas et al., 1995; Itakura et al., 2020). However, chlamydia avoid this fate, most probably through the tight regulation of Rab GTPase recruitment to the early inclusion membrane, which mediates the transition from the early endosome to a slowly recycling endosome, thus providing a protective “inclusion”, within which the chlamydial development cycle can proceed (Mölleken and Hegemann, 2017). While sCLU recruitment and the CRED pathway may facilitate EB uptake, a possible inhibitory effect of CLU on the terminal complement pathway must also be considered, as CLU has been shown to be associated with EBs of Ctr serovar L2 (Partridge et al., 1996; Hallstrom et al., 2015; Fox and Parks, 2022; Lausen et al., 2021).
In conclusion, our data support a unique adhesion and invasion strategy driven by the interaction of Ctr PmpD (and very likely all other Pmps) with sCLU. By binding to sCLU, Ctr may hijack a host component to facilitate its own uptake into the cell. Thus, sCLU may be crucial for the early Ctr infection process.
Materials and methods
Antibodies and reagents
The following primary antibodies were used: anti-clusterin α-chain (Santa Cruz, sc-5289), anti-penta-His (Qiagen, #34660), anti-β-actin (Thermo Scientific, MA5-15739) and anti-MOMP (Bio-Rad, #1990-0804). The secondary antibodies – anti-mouse, coupled to alkaline phosphatase, and anti-goat, coupled to Alexa Fluor™ 594 – were purchased from Thermo-Fisher Scientific.
Bacterial strains and cell lines
The Saccharomyces cerevisiae strain CEN.PK2 was used for cloning steps. E. coli XL1 blue (Stratagene) and Origami (Novagen) were used for plasmid amplification and protein expression, respectively.
HEp-2 cells (ATCC: CCL-23) were cultured in cell culture medium (ccm), composed of DMEM medium (Thermo Scientific, Waltham, MA, USA) supplemented with 10% heat-inactivated FCS, MEM vitamins and non-essential amino acids (Thermo Scientific, Waltham, MA, USA), and the antibiotics amphotericin B and gentamycin (Life Technologies).
Ctr serovar E (DK-20) (London) (NCBI Accession Number: CP015304.1) was propagated in HEp-2 cells in cell culture medium supplemented with 1.2 μl/ml cycloheximide (Sigma), purified by centrifugation and stored in SPG buffer (220 mM sucrose, 3.8 mM KH2PO4, 10.8 mM Na2HPO4, 4.9 mM L-glutamine), as previously described (Jantos et al., 1997).
Cloning, protein expression and purification
Cloning steps were carried out by in-vivo homologous recombination in S. cerevisiae. The DNA fragment of pmpD encoding the D72 protein variant (residues D68-A761) was amplified via PCR from Ctr serovar E (DK20) genomic DNA, and cloned into the expression vector pKM32 (generating an N-terminal 10xHis-fusion) (Mölleken et al., 2010). Plasmids were amplified in E. coli XL1 blue and the sequence was verified prior to use.
Expression of the His-tagged protein was carried out in the E. coli Origami strain. Proteins were purified using Ni-NTA agarose (Thermo Scientific) and dialysed in phosphate-buffered saline (PBS) (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4).
Immunoblotting and Coomassie Blue staining
SDS-PAGE and immunoblotting were performed according to the standard protocol described by Sambrook and Maniatis et al (Sambrook et al., 1989). His-tagged recombinant proteins were detected with monoclonal anti-His antibodies and clusterin was detected with anti-clusterin α-chain antibodies. Both were visualized with AP-conjugated antibodies. SDS gels were stained with Coomassie Brilliant Blue G250 (Serva).
Blue-Native PAGE
Blue-Native PAGE analysis was performed using the Native PAGE Novex 3-12% Bis-Tris gel system, following the manufacturer’s protocol (Thermo Scientific). Proteins were visualized by staining the gels with Coomassie Brilliant Blue G250 (Serva).
Size-exclusion chromatography
Recombinantly expressed proteins dissolved in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) were analysed on a Superose 6 Increase 3.2/300 column (Cytivia) at a flow rate of 0.05 ml/min, and 0.1-ml fractions were collected. All runs were performed at 4°C. The void volume was estimated as described in the manufacturer’s instructions.
Adhesion assay
Confluent HEp-2 cells were grown at 37°C and specific concentrations of soluble recombinant protein was added to the ccm in a total volume of 250 µl. Binding was allowed to proceed for 1 h at 4°C. After extensive washing, cells were lysed in phospho-lysis buffer (1% NP-40, 1% Triton X-100, 20 mM Tris/HCl, 150 mM NaCl, 1 mM Na2VO4). The lysate subjected to SDS/PAGE and immunoblotting using anti-His antibodies for the quantification of recombinant proteins. Anti-β-actin antibody served as the internal loading control.
Pulldown assays
Confluent HEp-2 cells were grown in 25-cm² flasks, and soluble recombinant His-tagged proteins (2 µM) were mixed with the overlying ccm (without FCS and antibiotics) to a total volume of 3 ml. Binding was allowed to proceed for 1 h at 4°C, then cells were washed three times with HBSS (Gibco™) and lysed with phospho-lysis buffer (1% NP-40, 1% Triton X-100, 20 mM Tris/HCl, 150 mM NaCl, 1 mM Na2VO4). Lysate was centrifuged at 1000 rpm and the soluble fraction was loaded on Ni-NTA agarose. His-tagged proteins and their bound host-cell binding partners were eluted with PBS containing 500 mM imidazole. Proteins enriched in the elution fraction were identified by mass spectrometry.
To verify the direct interaction between sCLU and rD72, an in vitro pulldown assay was performed, using rD72 as bait. HEp-2 cells were seeded subconfluently and grown for 2 days to confluence. This ccm carrying secreted CLU was collected (without HEp-2 cells) and incubated with rD72 for 1 h at 4°C followed by the in vitro pulldown assay. The protein solution was loaded on Ni-NTA agarose and the His-tagged rD72 and its bound host-cell binding partner were eluted with PBS containing 500 mM imidazole. The protein composition of the individual fractions of the pulldown assay was probed by Western blotting, using anti-His and anti-clusterin α-chain antibodies.
Quantitative mass-spectrometric analysis of rD72 host-cell binding partners
Eluates from His-PmpD pulldown assays were prepared for mass spectrometric analysis by in-gel digestion with trypsin, essentially as described earlier (Grube et al., 2018).
Briefly, eluates were fractionated in a polyacrylamide gel, proteins were then reduced with dithiothreitol, alkylated with iodoacetamide and digested overnight with 0.1 µg trypsin. Peptides were extracted from the gel and subsequently purified by solid-phase extraction (HLB µ-elution plate, Waters) according to the manufacturer’s instructions. Finally, peptides were dried in a vacuum concentrator, resuspended in 17 µl 0.1% (v/v) trifluoroacetic acid and analyzed by liquid-chromatography-coupled mass spectrometry as described previously, with the modifications mentioned below (Prescher et al., 2021).
First, peptides were trapped on a 2-cm long pre-column, then separated with the aid of a 1-h gradient on a C18 analytical column (25 cm long) using an Ultimate 3000 rapid separation liquid chromatography system (Thermo Fisher Scientific). Peptides were then injected into an QExactive plus (Thermo Fisher Scientific) mass spectrometer online coupled by a nano-source electrospray interface. The mass spectrometer was operated in data-dependent positive mode. Survey spectra were recorded with following settings: resolution 140000, automatic gain control target 3000000, maximum ion time 50 ms, scan range 200-2000 m/z, profile mode. Up to 20 precursors were selected by the quadrupole of the instrument (4 m/z isolation window), fragmented by higher-energy collisional dissociation (normalized collision energy: 30), and fragment spectra were recorded in the orbitrap analyzer: resolution 17500, automatic gain control target 100000, maximum ion time 50 ms, scan range 200-2000 m/z, centroid mode. Fragmented peptides were excluded for the next 10 seconds.
Peptide and protein identification was carried out with MaxQuant version 2.1.3.0 (MaxPlack Institute for Biochemistry, Planegg, Germany) with standard parameters, if not stated otherwise. Protein sequences from Homo sapiens (79038 entries of UP000005640, UniProt knowledge base, downloaded on 18th January 2022), as well as one entry for PmpD, were used, and the match between runs function and label-free quantification (LFQ) enabled.
Identified proteins were filtered (contaminants, “identified by site”, reverse hits and proteins only identified with one peptide removed) and only proteins identified with at least four valid values in one experimental group were considered for statistical analysis on log2 transformed LFQ-intensities. Missing values were imputed with values drawn from a downshifted (1.8 standard deviations (s.d.)) normal distribution (0.3 s.d.) and differentially abundant proteins between the two groups (PBS, PmpD) determined by the significance analysis of microarray method using an FDR of 5% and So of 0.1 (Tusher et al., 2001).
Infection assay
For all infection assays, cells were seeded at sub-confluent density in 24-well plates and grown in cell culture medium (ccm) to confluence for two days at 37°C. For approach (i), chlamydial EBs (MOI = 10) were added to the 2-day-old ccm covering the confluent cell layer and incubated for 2 h at 37°C. For approach (ii), the confluent cell layer was washed three times with HBSS, and then fresh ccm, supplemented with EBs (MOI = 10) was added and incubated for 2 h at 37°C. For approach (iii), soluble recombinant rD72 was added to the 2-day-old ccm covering the confluent cell layer. After 1 h of incubation at 37°C, chlamydial EBs (MOI = 10) were added and incubated for 2 h at 37°C. For approach (iv), the 2-day-old ccm used for cell growth was isolated and cells were washed three times with HBSS. From the isolated ccm, sCLU was extracted via immunoprecipitation using Protein-G agarose (Merck) and an anti-clusterin α-chain antibody, following the manufacturer’s protocol. The flow-through fraction depleted in sCLU was then added to HEp-2 cells, and chlamydial EBs (MOI = 10) were added and incubated for 2 h at 37°C.
For all four approaches, after the initial 2 h of incubation with EBs without centrifugation, the ccm containing non-bound chlamydial EBs was removed and fresh ccm supplemented with cycloheximide (1.2 µl/ml) was added to the cells. Infection was allowed to proceed for 24 h at 37°C, before the cells were prepared for immunofluorescence microscopy. The presence of sCLU in the ccm used for the different approaches was monitored by immunoblotting using an anti-clusterin α-chain antibody.
Immunofluorescence microscopy
Infected cells were fixed with 3% para-formaldehyde for 10 min at RT, washed three times with PBS, and permeabilized with 96% methanol (-20°C) at room temperature. The primary antibody (anti-MOMP, 1:500) was diluted in bufferX (5% BSA, 0.5% Triton-X 100, 0.2% Tween20) and incubated for 1 h at RT. Cells were washed three times with PBS, and the secondary antibody (anti-goat coupled to Alexa594, 1:1000), diluted in buffer X, was incubated for 60 min at RT. Cells were then washed three times and DAPI was used to visualize DNA. Microscopy was performed using a Nikon Eclipse Ti-E C2 confocal microscope (DS-Qi1MC Camera) and confocal images were generated using NIS Element software (Nikon) and quantified using ImageJ.
Statistical analysis
Graphics were created in OriginPro v.2021b (OriginLab). The data represent the means ± s.d. A 1-way ANOVA (with multiple comparisons) was used to compare all four groups shown in Figure 4D. Images were prepared using the open-source software Inkscape (www.inkscape.org).
Data availability statement
All relevant data is contained within the article. The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
JHH: Writing – original draft, Writing – review & editing, Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Validation. FK: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by a scholarship to FK by the Graduate School “Molecules of Infection IV (MOI IV)” founded by the Jürgen Manchot Foundation. We acknowledge grant support from the DFG to JHH (Project-ID 267205415) of CRC 1208.
Acknowledgments
We thank Dr. Reiners for the help with SEC analysis. We thank BMFZ HHU, specifically Dr. Poschmann and Prof. Dr. Stühler for processing the mass-spectrometric analysis of the rD72 pulldown experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1519883/full#supplementary-material
References
Abdelrahman, Y. M., Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959. doi: 10.1016/j.femsre.2005.03.002
Becker, E., Hegemann, J. H. (2014). All subtypes of the Pmp adhesin family are implicated in chlamydial virulence and show species-specific function. Microbiologyopen 3, 544–556. doi: 10.1002/mbo3.2014.3.issue-4
Bettens, K., Vermeulen, S., Van Cauwenberghe, C., Heeman, B., Asselbergh, B., Robberecht, C., et al. (2015). Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol. Neurodegener. 10, 30. doi: 10.1186/s13024-015-0024-9
Burkey, B. F., deSilva, H. V., Harmony, J. A. (1991). Intracellular processing of apolipoprotein J precursor to the mature heterodimer. J. Lipid Res. 32, 1039–1048. doi: 10.1016/S0022-2275(20)42000-0
Cervantes, P. W., Segelke, B. W., Lau, E. Y., Robinson, B. V., Abisoye-Ogunniyan, A., Pal, S., et al. (2024). Sequence, structure prediction, and epitope analysis of the polymorphic membrane protein family in Chlamydia trachomatis. PloS One 19, e0304525. doi: 10.1371/journal.pone.0304525
Choi, N. H., Mazda, T., Tomita, M. (1989). A serum protein SP40,40 modulates the formation of membrane attack complex of complement on erythrocytes. Mol. Immunol. 26, 835–840. doi: 10.1016/0161-5890(89)90139-9
Christensen, S., McMahon, R. M., Martin, J. L., Huston, W. M. (2019). Life inside and out: making and breaking protein disulfide bonds in Chlamydia. Crit. Rev. Microbiol. 45, 33–50. doi: 10.1080/1040841X.2018.1538933
Crane, D. D., Carlson, J. H., Fischer, E. R., Bavoil, P., Hsia, R. C., Tan, C., et al. (2006). Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc. Natl. Acad. Sci. U. S. A. 103, 1894–1899. doi: 10.1073/pnas.0508983103
Debrine, A. M., Karplus, P. A., Rockey, D. D. (2023). A structural foundation for studying chlamydial polymorphic membrane proteins. Microbiol. Spectr. 11, e0324223. doi: 10.1128/spectrum.03242-23
de Silva, H. V., Harmony, J. A., Stuart, W. D., Gil, C. M., Robbins, J. (1990). Apolipoprotein J: structure and tissue distribution. Biochemistry 29, 5380–5389. doi: 10.1021/bi00474a025
de Vrieze, N. H., de Vries, H. J. (2014). Lymphogranuloma venereum among men who have sex with men. An epidemiological and clinical review. Expert Rev. Anti Infect. Ther. 12, 697–704. doi: 10.1586/14787210.2014.901169
Elwell, C., Mirrashidi, K., Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400. doi: 10.1038/nrmicro.2016.30
Favaroni, A., Hegemann, J. H. (2021). Chlamydia trachomatis polymorphic membrane proteins (Pmps) form functional homomeric and heteromeric oligomers. Front. Microbiol. 12, 709724. doi: 10.3389/fmicb.2021.709724
Fechtner, T., Stallmann, S., Moelleken, K., Meyer, K. L., Hegemann, J. H. (2013). Characterization of the interaction between the chlamydial adhesin OmcB and the human host cell. J. Bacteriol. 195, 5323–5333. doi: 10.1128/JB.00780-13
Fox, C. R., Parks, G. D. (2022). Complement inhibitors vitronectin and clusterin are recruited from human serum to the surface of coronavirus OC43-infected lung cells through antibody-dependent mechanisms. Viruses 14, 29. doi: 10.3390/v14010029
Grimwood, J., Stephens, R. S. (1999). Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics 4, 187–201. doi: 10.1089/omi.1.1999.4.187
Gross, C., Guerin, L. P., Socol, B. G., Germain, L., Guerin, S. L. (2023). The ins and outs of clusterin: its role in cancer, eye diseases and wound healing. Int. J. Mol. Sci. 24, 13182. doi: 10.3390/ijms241713182
Grube, L., Dellen, R., Kruse, F., Schwender, H., Stuhler, K., Poschmann, G. (2018). Mining the secretome of C2C12 muscle cells: data dependent experimental approach to analyze protein secretion using label-free quantification and peptide based analysis. J. Proteome Res. 17, 879–890. doi: 10.1021/acs.jproteome.7b00684
Gu, L., Guo, Z. (2013). Alzheimer’s Abeta42 and Abeta40 peptides form interlaced amyloid fibrils. J. Neurochem. 126, 305–311. doi: 10.1111/jnc.2013.126.issue-3
Hallstrom, T., Uhde, M., Singh, B., Skerka, C., Riesbeck, K., Zipfel, P. F. (2015). Pseudomonas aeruginosa uses dihydrolipoamide dehydrogenase (Lpd) to bind to the human terminal pathway regulators vitronectin and clusterin to inhibit terminal pathway complement attack. PloS One 10, e0137630. doi: 10.1371/journal.pone.0137630
Hatters, D. M., Lindner, R. A., Carver, J. A., Howlett, G. J. (2001). The molecular chaperone, alpha-crystallin, inhibits amyloid formation by apolipoprotein C-II. J. Biol. Chem. 276, 33755–33761. doi: 10.1074/jbc.M105285200
Hegemann, J. H., Moelleken, K. (2012). “Chlamydial adhesion and adhesins,” in Intracellular Pathogens I: Chlamydiales. Eds. Tan, M., Bavoil, P. (ASM Press, Washington, DC).
Henderson, I. R., Lam, A. C. (2001). Polymorphic proteins of Chlamydia spp.–autotransporters beyond the Proteobacteria. Trends Microbiol. 9, 573–578. doi: 10.1016/S0966-842X(01)02234-X
Henderson, I. R., Navarro-Garcia, F., Desvaux, M., Fernandez, R. C., Ala’Aldeen, D. (2004). Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68, 692–744. doi: 10.1128/MMBR.68.4.692-744.2004
Howlett, D. R., Hortobagyi, T., Francis, P. T. (2013). Clusterin associates specifically with Abeta40 in Alzheimer’s disease brain tissue. Brain Pathol. 23, 623–632. doi: 10.1111/bpa.2013.23.issue-6
Humphreys, D. T., Carver, J. A., Easterbrook-Smith, S. B., Wilson, M. R. (1999). Clusterin has chaperone-like activity similar to that of small heat shock proteins. J. Biol. Chem. 274, 6875–6881. doi: 10.1074/jbc.274.11.6875
Itakura, E., Chiba, M., Murata, T., Matsuura, A. (2020). Heparan sulfate is a clearance receptor for aberrant extracellular proteins. J. Cell Biol. 219 (3), e201911126. doi: 10.1083/jcb.201911126
Jantos, C. A., Heck, S., Roggendorf, R., Sen-Gupta, M., Hegemann, J. H. (1997). Antigenic and molecular analyses of different Chlamydia pneumoniae strains. J. Clin. Microbiol. 35, 620–623. doi: 10.1128/jcm.35.3.620-623.1997
Kalman, S., Mitchell, W., Marathe, R., Lammel, C., Fan, J., Hyman, R. W., et al. (1999). Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21, 385–389. doi: 10.1038/7716
Kari, L., Southern, T. R., Downey, C. J., Watkins, H. S., Randall, L. B., Taylor, L. D., et al. (2014). Chlamydia trachomatis polymorphic membrane protein D is a virulence factor involved in early host-cell interactions. Infect. Immun. 82, 2756–2762. doi: 10.1128/IAI.01686-14
Kiselev, A. O., Skinner, M. C., Lampe, M. F. (2009). Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2. PloS One 4, e5191. doi: 10.1371/journal.pone.0005191
Kiselev, A. O., Stamm, W. E., Yates, J. R., Lampe, M. F. (2007). Expression, processing, and localization of PmpD of Chlamydia trachomatis serovar L2 during the chlamydial developmental cycle. PloS One 2, e568. doi: 10.1371/journal.pone.0000568
Kounnas, M. Z., Loukinova, E. B., Stefansson, S., Harmony, J. A., Brewer, B. H., Strickland, D. K., et al. (1995). Identification of glycoprotein 330 as an endocytic receptor for apolipoprotein J/clusterin. J. Biol. Chem. 270, 13070–13075. doi: 10.1074/jbc.270.22.13070
Lausen, M., Thomsen, M. E., Christiansen, G., Karred, N., Stensballe, A., Bennike, T. B., et al. (2021). Analysis of complement deposition and processing on Chlamydia trachomatis. Med. Microbiol. Immunol. 210, 13–32. doi: 10.1007/s00430-020-00695-x
Lesiak-Markowicz, I., Schötta, A. M., Stockinger, H., Stanek, G., Markowicz, M. (2019). Chlamydia trachomatis serovars in urogenital and ocular samples collected 2014-2017 from Austrian patients. Sci. Rep. 9, 18327. doi: 10.1038/s41598-019-54886-5
Li, X., Zuo, Z., Wang, Y., Hegemann, J. H., He, C. (2021). Polymorphic membrane protein 17G of chlamydia psittaci mediated the binding and invasion of bacteria to host cells by interacting and activating EGFR of the host. Front. Immunol. 12, 818487. doi: 10.3389/fimmu.2021.818487
Liu, J., Thorp, S. C. (2002). Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 22, 1–25. doi: 10.1002/med.v22:1
Luczak, S. E., Smits, S. H., Decker, C., Nagel-Steger, L., Schmitt, L., Hegemann, J. H. (2016). The chlamydia pneumoniae adhesin pmp21 forms oligomers with adhesive properties. J. Biol. Chem. 291, 22806–22818. doi: 10.1074/jbc.M116.728915
Madadi, S., Schwarzenbach, H., Saidijam, M., Mahjub, R., Soleimani, M. (2019). Potential microRNA-related targets in clearance pathways of amyloid-beta: novel therapeutic approach for the treatment of Alzheimer’s disease. Cell Biosci. 9, 91. doi: 10.1186/s13578-019-0354-3
Malecela, M. N., Ducker, C. (2021). A road map for neglected tropical diseases 2021-2030. Trans. R. Soc. Trop. Med. Hyg. 115, 121–123. doi: 10.1093/trstmh/trab002
Mölleken, K., Becker, E., Hegemann, J. H. (2013). The Chlamydia pneumoniae invasin protein Pmp21 recruits the EGF receptor for host cell entry. PloS Pathog. 9, e1003325. doi: 10.1371/journal.ppat.1003325
Mölleken, K., Hegemann, J. H. (2008). The Chlamydia outer membrane protein OmcB is required for adhesion and exhibits biovar-specific differences in glycosaminoglycan binding. Mol. Microbiol. 67, 403–419. doi: 10.1111/j.1365-2958.2007.06050.x
Mölleken, K., Hegemann, J. H. (2017). Acquisition of Rab11 and Rab11-Fip2-A novel strategy for Chlamydia pneumoniae early survival. PloS Pathog. 13 (8), e1006556. doi: 10.1371/journal.ppat.1006556
Mölleken, K., Schmidt, E., Hegemann, J. H. (2010). Members of the Pmp protein family of Chlamydia pneumoniae mediate adhesion to human cells via short repetitive peptide motifs. Mol. Microbiol. 78, 1004–1017. doi: 10.1111/j.1365-2958.2010.07386.x
Moulder, J. W. (1991). Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55, 143–190. doi: 10.1128/mr.55.1.143-190.1991
Murray, S. M., McKay, P. F. (2021). Chlamydia trachomatis: Cell biology, immunology and vaccination. Vaccine 39, 2965–2975. doi: 10.1016/j.vaccine.2021.03.043
Nizard, P., Tetley, S., Le Drean, Y., Watrin, T., Le Goff, P., Wilson, M. R., et al. (2007). Stress-induced retrotranslocation of clusterin/ApoJ into the cytosol. Traffic 8, 554–565. doi: 10.1111/j.1600-0854.2007.00549.x
Paes, W., Brown, N., Brzozowski, A. M., Coler, R., Reed, S., Carter, D., et al. (2016). Recombinant polymorphic membrane protein D in combination with a novel, second-generation lipid adjuvant protects against intra-vaginal Chlamydia trachomatis infection in mice. Vaccine 34, 4123–4131. doi: 10.1016/j.vaccine.2016.06.081
Paes, W., Dowle, A., Coldwell, J., Leech, A., Ganderton, T., Brzozowski, A. (2018). The Chlamydia trachomatis PmpD adhesin forms higher order structures through disulphide-mediated covalent interactions. PloS One 13, e0198662. doi: 10.1371/journal.pone.0198662
Partridge, S. R., Baker, M. S., Walker, M. J., Wilson, M. R. (1996). Clusterin, a putative complement regulator, binds to the cell surface of Staphylococcus aureus clinical isolates. Infect. Immun. 64, 4324–4329. doi: 10.1128/iai.64.10.4324-4329.1996
Pillay, S., Meyer, N. L., Puschnik, A. S., Davulcu, O., Diep, J., Ishikawa, Y., et al. (2016). An essential receptor for adeno-associated virus infection. Nature 530, 108–112. doi: 10.1038/nature16465
Poon, S., Easterbrook-Smith, S. B., Rybchyn, M. S., Carver, J. A., Wilson, M. R. (2000). Clusterin is an ATP-independent chaperone with very broad substrate specificity that stabilizes stressed proteins in a folding-competent state. Biochemistry 39, 15953–15960. doi: 10.1021/bi002189x
Prescher, N., Hansch, S., Knobbe-Thomsen, C. B., Stuhler, K., Poschmann, G. (2021). The migration behavior of human glioblastoma cells is influenced by the redox-sensitive human macrophage capping protein CAPG. Free Radic. Biol. Med. 167, 81–93. doi: 10.1016/j.freeradbiomed.2021.02.038
R Core Team. (2024). R: A Language and Environment for Statistical Computing (Vienna, Austria). The R Foundation. Available online at: https://www.R-project.org/ (Accessed January 09, 2025).
Rodrigues, R., Sousa, C., Vale, N. (2022). Chlamydia trachomatis as a current health problem: challenges and opportunities. Diagnostics (Basel) 12, 1795. doi: 10.3390/diagnostics12081795
Romero, M. D., Moelleken, K., Hegemann, J. H., Carabeo, R. A. (2020). “Chlamydia adhesion and invasion,” in Chlamydia Biology: From Genome to Disease. (Norfolk, UK: Caister Academic Press), 59–84.
RStudio Team. (2020). RStudio: Integrated Development Environment for R (Boston, MA: RStudio). Available at: http://www.rstudio.com/ (Accessed January 09, 2025).
Sambrook, J., Maniatis, T., Fritsch, E. (1989). Molecular cloning: A laboratory manual (Cold Spring Harbor, N. Y: Cold Spring Harbor laboratory).
Satapathy, S., Wilson, M. R. (2021). The dual roles of clusterin in extracellular and intracellular proteostasis. Trends Biochem. Sci. 46, 652–660. doi: 10.1016/j.tibs.2021.01.005
Stallmann, S., Hegemann, J. H. (2016). The Chlamydia trachomatis Ctad1 invasin exploits the human integrin beta1 receptor for host cell entry. Cell Microbiol. 18, 761–775. doi: 10.1111/cmi.12549
Stephens, R. S., Koshiyama, K., Lewis, E., Kubo, A. (2001). Heparin-binding outer membrane protein of chlamydiae. Mol. Microbiol. 40, 691–699. doi: 10.1046/j.1365-2958.2001.02418.x
Swanson, K. A., Taylor, L. D., Frank, S. D., Sturdevant, G. L., Fischer, E. R., Carlson, J. H., et al. (2009). Chlamydia trachomatis polymorphic membrane protein D is an oligomeric autotransporter with a higher-order structure. Infect. Immun. 77, 508–516. doi: 10.1128/IAI.01173-08
Tusher, V. G., Tibshirani, R., Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98, 5116–5121. doi: 10.1073/pnas.091062498
Vasilevsky, S., Stojanov, M., Greub, G., Baud, D. (2016). Chlamydial polymorphic membrane proteins: regulation, function and potential vaccine candidates. Virulence 7, 11–22. doi: 10.1080/21505594.2015.1111509
Wang, Y., LaBrie, S. D., Carrell, S. J., Suchland, R. J., Dimond, Z. E., Kwong, F., et al. (2019). Development of transposon mutagenesis for chlamydia muridarum. J. Bacteriol. 201 (23), e00366-19. doi: 10.1128/JB.00366-19
Wehrl, W., Brinkmann, V., Jungblut, P. R., Meyer, T. F., Szczepek, A. J. (2004). From the inside out–processing of the Chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51, 319–334. doi: 10.1046/j.1365-2958.2003.03838.x
Wilson, M. R., Easterbrook-Smith, S. B. (2000). Clusterin is a secreted mammalian chaperone. Trends Biochem. Sci. 25, 95–98. doi: 10.1016/S0968-0004(99)01534-0
Wuppermann, F. N., Mölleken, K., Julien, M., Jantos, C. A., Hegemann, J. H. (2008). Chlamydia pneumoniae GroEL1 protein is cell surface associated and required for infection of HEp-2 cells. J. Bacteriol. 190, 3757–3767. doi: 10.1128/JB.01638-07
Keywords: PmpD adhesin, secreted human clusterin, interaction, Chlamydia trachomatis, infection
Citation: Kocher F and Hegemann JH (2025) The secreted host-cell protein clusterin interacts with PmpD and promotes Chlamydia trachomatis infection. Front. Cell. Infect. Microbiol. 14:1519883. doi: 10.3389/fcimb.2024.1519883
Received: 30 October 2024; Accepted: 30 December 2024;
Published: 27 January 2025.
Edited by:
Nicolas Jacquier, Centre Hospitalier Universitaire Vaudois (CHUV), SwitzerlandReviewed by:
George W. Liechti, Uniformed Services University of the Health Sciences, United StatesLuís Jaime Mota, NOVA School of Science and Technology, Portugal
Copyright © 2025 Kocher and Hegemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes H. Hegemann, am9oYW5uZXMuaGVnZW1hbm5AaGh1LmRl
 Fabienne Kocher
Fabienne Kocher Johannes H. Hegemann
Johannes H. Hegemann