- 1Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 2Agrobiodiversity in Highland Agriculture and Sustainable Utilization Research Group, Chiang Mai University, Chiang Mai, Thailand
- 3Key Laboratory of Phytochemistry and Natural Medicines, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China
- 4Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 5Vishnugupta Vishwavidyapeetam, Gokarna, India
- 6Department of Economic Plants and Biotechnology, Yunnan Key Laboratory for Wild Plant Resources, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China
- 7Honghe Center for Mountain Futures, Kunming Institute of Botany, Chinese Academy of Sciences, Yunnan, China
- 8Centre for Mountain Futures (CMF), Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China
- 9CIFOR-ICRAF China Program, World Agroforestry (ICRAF), Kunming, China
- 10Office of the Research Administration, Chiang Mai University, Chiang Mai, Thailand
During investigations of freshwater fungi in Hunan and Yunnan provinces, China, Chaetopsina yunnanensis sp. nov. (Nectriaceae), Parafuscosporella hunanensis sp. nov. (Fuscosporellaceae), and Pleurotheciella yunnanensis sp. nov. (Pleurotheciaceae) were discovered on submerged decaying wood and branches. Based on phylogenetic analyses, C. yunnanensis formed a separate branch with Chaetopsina pinicola and nested among other Chaetopsina species in Nectriaceae (Hypocreales). Furthermore, hitherto known Chaetopsina beijingensis shared the same branch with Chaetopsina fulva, a type species of the genus, demonstrating their conspecific status. Therefore, C. beijingensis is formally synonymized under C. fulva, with an amended species circumscription. Pa. hunanensis formed a well-separated subclade with the ex-type strain of Parafuscosporella mucosa and clustered with other Parafuscosporella within Fuscosporellaceae (Fuscosporellales). In addition, the genus Parafuscosporella is treated as distinct from Vanakripa due to a lack of phylogenetic evidence in clarifying their congeneric status with the latter. Pl. yunnanensis is found to be sister to Pleurotheciella saprophytica, forming a subclade with Pleurotheciella dimorphospora within the Pleurotheciaceae (Pleurotheciales). Morphologically, C. yunnanensis fits well with the generic concept of Chaetopsina in forming a holomorphic state with hyphomycetous asexual morph producing pigmented, setiform conidiophores, phialidic conidiogenous cells, hyaline conidia, and nectria-like sexual morph. Pa. hunanensis fits well with Parafuscosporella in having acrogenous, apiosporous, versicolored, obovoid to obpyriform conidia. In contrast, Pl. yunnanensis resembles Pl. dimorphospora in forming asexual dimorphism with two types of conidia (Type I, brown, muriform/phragmosporous conidia; Type II, hyaline, amerosporous/didymorsporous conidia). The novelty of taxa is explained with detailed descriptions, photo-micrographic illustrations, polymorphism, and multigene phylogenetic analyses of Bayesian inference and maximum likelihood criteria.
1 Introduction
Freshwater fungi are a diverse and heterogeneous taxonomic group occurring saprobically on partially or fully submerged organic substrates in aquatic habitats. Their life cycle, in whole or part, relies on free freshwater and submerged substrates (Calabon et al., 2020, 2023a). Freshwater environments are categorized into three types: 1) lentic, any natural aquatic environment lacking continuous flow but exhibiting static, low, or slow movement of water as in lakes, ponds, swamps, and pools; 2) lotic, any natural aquatic environment with a continuous flow of water such as rivers, streams, creeks, and brooks; and 3) other habitats, which include artificial water bodies such as in cooling towers and tree holes (Luo et al., 2004). Fungi found in freshwater habitats were grouped into several morphological and ecological entities, viz., freshwater ascomycetes, freshwater hyphomycetes (i.e., Ingoldian fungi, aero-aquatic hyphomycetes or asexual ascomycetes, terrestrial–aquatic hyphomycetes, and submerged aquatic hyphomycetes), freshwater basidiomycetes, coelomycetes, microsporidia, zoosporic fungi, and zygomycetes (Tsui et al., 2016; Schuster et al., 2022). Furthermore, freshwater fungi subsist as saprobes, mutualists, or parasites and have also been isolated as endophytes (Tsui et al., 2016). However, freshwater fungi have an important ecological function in decomposing wood by breaking down complex organic compounds into simpler inorganic materials and facilitating the passage of energy and nutrients across all trophic levels in the food chain (Bärlocher et al., 2011; Sridhar and Sudheep, 2011; Tsui et al., 2016). Two crucial biological characteristics of these fungi include the ability to sporulate underwater and thrive on decaying deciduous leaves and twigs in streams and rivers (Krauss et al., 2011; Wurzbacher et al., 2014; Tsui et al., 2016). In addition, the biotechnological potential of freshwater fungi as producers of bioactive metabolites, with promising values in drug discovery, is becoming evident (Duarte et al., 2013; Shen et al., 2022; El-Elimat et al., 2021; Calabon et al., 2023a). Lignicolous freshwater fungi can degrade indigestible lignocellulose in submerged wood, releasing nutrients into the water. However, their precise ecological role and economic value remain less understood (Bucher et al., 2004; Shen et al., 2022).
Freshwater fungi have been documented since the mid-19th century, and a wealth of information is now available (de Wildeman, 1895; Ingold, 1942, 1955; Calabon et al., 2021). Over the recent past decades, these fungi have been relatively well-studied in Asia, particularly in China, India, and Thailand (Sridhar and Sudheep, 2011; Mehboob et al., 2021; Luo et al., 2019; Dong et al., 2020; Shen et al., 2022; Calabon et al., 2023a, b). In the early stages of studies on freshwater fungi, identification has primarily relied on morphology (Ingold, 1975; Krauss et al., 2011; Calabon et al., 2023a). In the case of yeasts, additional methods such as biochemical, fermentation, and assimilation tests were included. Presently, in addition to morphology, molecular data analyses were applied to taxonomic studies of freshwater fungi (Ranghoo et al., 1999; Nikolcheva and Bärlocher, 2002; Luo et al., 2019; Dong et al., 2020; Calabon et al., 2023a, b). Recent studies revealed that lignicolous freshwater fungi constitute a highly diverse taxonomic group with a substantial population. To date, more than 3,870 species of freshwater fungi have been documented from various substrates and geographical locations (Luo et al., 2019; Dong et al., 2020; Shen et al., 2022; Calabon et al., 2023a, b). Lignicolous freshwater fungi are complex assemblages of mostly filamentous fungi, single-celled chytrids, and yeasts (Tsui et al., 2016). Freshwater fungi belong to Ascomycota, Basidiomycota, Chytridiomycota, and Rozellomycota; in class levels within the Ascomycota, most species are accommodated in Dothideomycetes and Sordariomycetes (Calabon et al., 2022).
Chaetopsina (family Nectriaceae) was established by Rambelli (1956), with the type species Chaetopsina fulva, which was isolated from decaying leaves in northern Italy. Chaetopsina is a dematiaceous hyphomycete genus, the members of which occur on decaying wood, leaves, pine needle litter, and bark, as well as on ascomycetous stromata and soil (Seifert et al., 2011). Most Chaetopsina species are found in tropical and subtropical areas (Lechat and Fournier, 2019). Chaetopsina is initially characterized by reddish-brown setose conidiophores turning yellow in lactic acid, and hyaline, smooth, fusiform ameroconidia, and fertile regions situated terminally or along the axes of the setiform conidiophores (Samuels, 1985; Kirk and Sutton, 1985). Later, some species with dark brown conidiophores or lateral branches on the conidiophores also have been accommodated (Bakhit and Abdel-Aziz, 2021). According to Luo and Zhuang (2010), Chaetopsinectria chaetopsinae was distinct as the sexual morph of C. fulva. Lombard et al. (2015) and Rossman et al. (2016) subsequently recommended using Chaetopsina despite Chaetopsinectria in respect to the 1F = 1N policy, and hitherto, Chaetopsinectria was treated as a synonym of Chaetopsina. The sexual morph of Chaetopsina is seen featuring superficial, non-stromatic, reddish brown to bright red, oval ascomata with an acute ostiolar apex, with walls composed of cells of textura epidermoidea, with paler papilla; unitunicate, clavate, short-stipitate asci containing 8-spored, fusiform, 0–1-septate, hyaline ascospores, and growing in association with upright conidiophores (Lechat and Fournier, 2020). So far, a total of 33 species epithets are listed under Chaetopsina, and four epithets are listed under Chaetopsinectria in Index Fungorum (https://indexfungorum.org/Names/Names.asp; accessed on 21 June 2024). However, eight previously described Chaetopsina epithets have been synonymized under other genera in Nectriaceae or incertae sedis (Index Fungorum, 2024), and the genus now embodies only 25 accepted species wherein molecular data of only 13 species are available (Lechat and Fournier, 2019; Bakhit and Abdel-Aziz, 2021). There are four species in Chaetopsina that have so far been recorded from freshwater habitats, including Chaetopsina aquatica, C. fulva, Chaetopsina hongkongensis, and Chaetopsina polyblastia (Sivichai et al., 2000; Ho et al., 2002; Luo et al., 2019; Bakhit and Abdel-Aziz, 2021). C. fulva and C. polyblastia were found on woody test blocks of Xylia dolabriformis in a freshwater stream of Khao Yai National Park, Thailand (Sivichai et al., 2000). C. hongkongensis was found on decaying submerged wood in Tai Po Kau Forest Stream, Hong Kong, China (Ho et al., 2002). Chaetopsina beijingensis (treated herein as a synonym of C. fulva) was isolated from decaying wood submerged in a freshwater stream in Yunnan, China (Luo et al., 2019). C. aquatica was collected on decaying submerged stems of Phragmites australis (Poaceae) in the River Nile, Sohag, Egypt (Bakhit and Abdel-Aziz, 2021).
Parafuscosporella (family Fuscosporellaceae), typified by Parafuscosporella moniliformis, was introduced by Yang et al. (2016). Yang et al. (2016) also introduced the new order Fuscosporellales to accommodate a new single family Fuscosporellaceae and to which six genera, viz., Bactrodesmiastrum, Fuscosporella, Mucispora, Parafuscosporella, Plagiascoma, and Pseudoascotaiwania were initially accommodated. Parafuscosporella is one of four new genera established by Yang et al. (2016) when the new order Fuscosporellales and the new family Fuscosporellaceae were established. The genus is characterized by spherical to cushion-shaped, black, gelatinous sporodochia, with a jelly-like cover, semi-macronematous, mononematous, compact, flexuous, simple or branched, mostly moniliform, globose to subglobose, ellipsoidal or clavate celled conidiophores, monoblastic, integrated, sometimes discrete, terminal, globose or subglobose, ellipsoidal or clavate conidiogenous cells and acrogenous, ellipsoidal to broadly obpyriform, smooth, dark brown to black conidia with a septum near the base, sometimes with a small protuberance and a pale brown basal cell (Boonyuen et al., 2016; Yang et al., 2016). Ten species are listed in Parafuscosporella, comprising Parafuscosporella aquatica, Parafuscosporella ellipsoconidiogena, Parafuscosporella garethii, Parafuscosporella lignicola, Pa. moniliformis, Parafuscosporella mucosa, Parafuscosporella nilotica, Parafuscosporella obovata, Parafuscosporella pyriformis, and Parafuscosporella xishuangbannaensis (Yang et al., 2016, 2020; Boonyuen et al., 2021; Boonmee et al., 2021; Wang et al., 2023; Li et al., 2023). Species of the genus have been reported from freshwater habitats in China and Thailand (Yang et al., 2016, 2020; Boonmee et al., 2021; Boonyuen et al., 2021). Goh et al. (2023) introduced two novel freshwater fungi from Taiwan, namely, Vanakripa oblonga and Vanakripa taiwanensis. Based on the morphological resemblance between Parafuscosporella and Vanakripa, Goh et al. (2023) treated Parafuscosporella as a synonym of Vanakripa and transferred all Parafuscosporella species to Vanakripa. In contrast, Vanakripa chiangmaiensis and Vanakripa minutiellipsoidea clustered with Conioscypha species were excluded from Vanakripa (Goh et al., 2023). Unfortunately, the type species of Vanakripa, Vanakripa gigaspora, lacks molecular data to clarify the phylogenetic placement. Hence, the congeneric status of Parafuscosporella and Vanakripa is questionable.
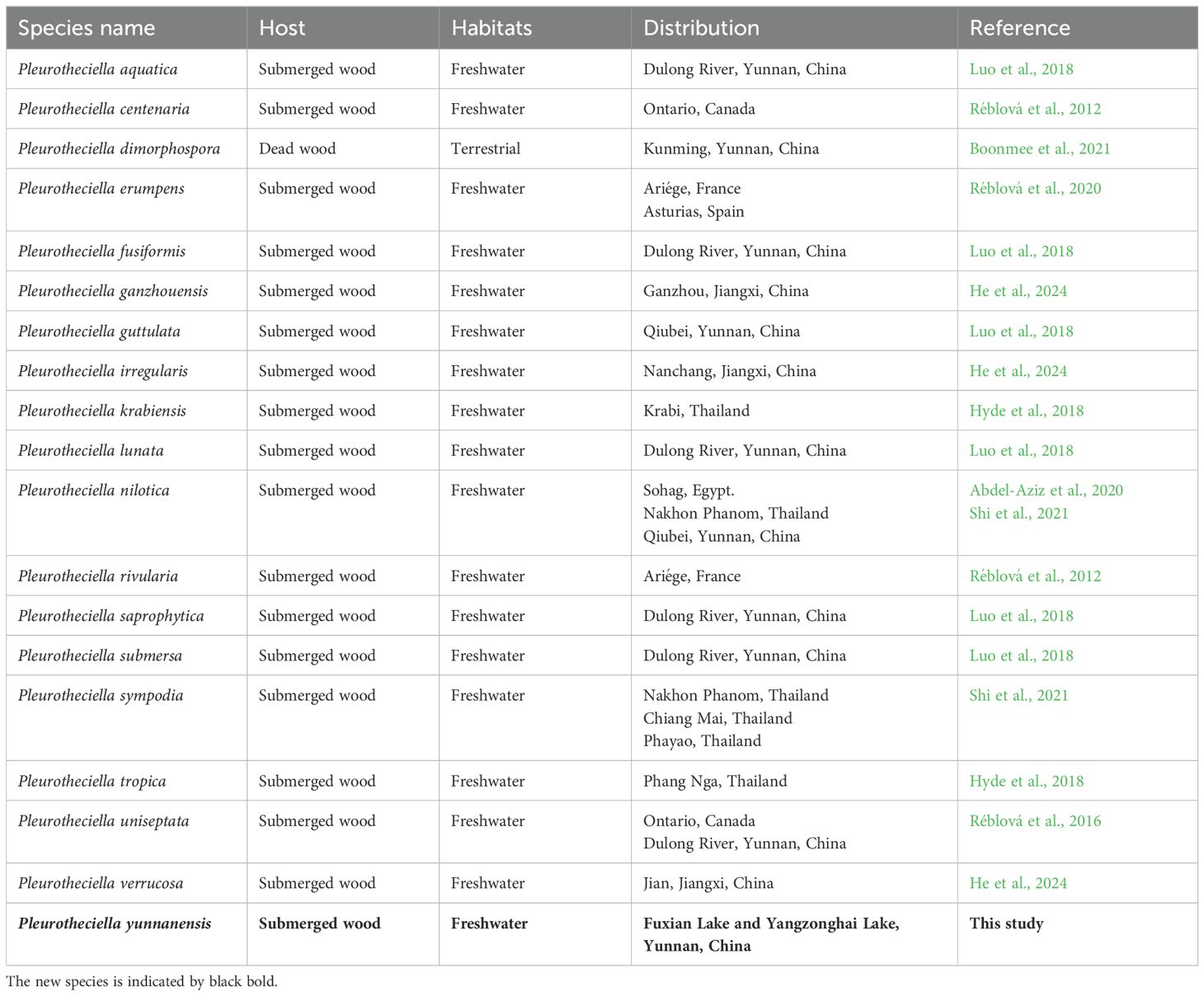
Pleurotheciella (family Pleurotheciaceae), typified by Pleurotheciella rivularia, was introduced by Réblová et al. (2012). To date, within the genus Pleurotheciella, only three species, viz., Pleurotheciella erumpens, Pleurotheciella fusiformis, and Pl. rivularia, are known for their sexual morphs, with non-stromatic perithecia, unitunicate asci, and hyaline to subhyaline, 1- or 3–5-septate ascospores, lacking a mucilaginous sheath. The asexual morph of the genus is a dactylaria-like hyphomycete, which is characterized by polyblastic, denticulate conidiogenesis, subhyaline conidiophores, and hyaline, ellipsoidal to ellipsoidal-fusiform, aseptate to multi-septate conidia (Luo et al., 2018). At present, 18 species are accommodated in Pleurotheciella, viz., Pleurotheciella aquatica, Pleurotheciella centenaria, Pleurotheciella dimorphospora, Pl. erumpens, Pl. fusiformis, Pleurotheciella ganzhouensis, Pleurotheciella guttulata, Pleurotheciella irregularis, Pleurotheciella krabiensis, Pleurotheciella lunata, Pleurotheciella nilotica, Pl. rivularia, Pleurotheciella saprophytica, Pleurotheciella submersa, Pleurotheciella sympodia, Pleurotheciella tropica, Pleurotheciella uniseptata, and Pleurotheciella verrucosa, and all accepted species of the genus have been reported for their asexual morphs (Hyde et al., 2018; Luo et al., 2018; Abdel-Aziz et al., 2020; Réblová et al., 2020; Shi et al., 2021; Boonmee et al., 2021; He et al., 2024). The genus has been so far represented from freshwater habitats in China and Thailand, except for Pl. dimorphospora, which is the only species described from terrestrial habitats in China (Boonmee et al., 2021).
In this study, we introduce three new species, Chaetopsina yunnanensis, Parafuscosporella hunanensis, and Pleurotheciella yunnanensis, based on morphology and phylogenetic studies, from freshwater habitats in China. In addition, C. beijingensis is formally synonymized under C. fulva, and the congeneric status of Parafuscosporella and Vanakripa is discussed.
2 Materials and methods
2.1 Sample collection, morphological studies, and isolation
Submerged decaying wood and branches were collected from a freshwater stream and lakes in Hunan and Yunnan provinces, China, from July to September 2022 (wet season). Detailed environmental parameters (e.g., pH, temperature, and dissolved oxygen) were recorded for each sample collected from lakes. Fresh specimens were brought to the laboratory in Ziploc bags and studied following the methods described by Luo et al. (2018). The samples were incubated in high-density plastic boxes lined with moisturized absorbent paper at room temperature for 1 week. Macro-morphological characters of the fungi on the host surface were observed using an Optec SZ760 compound stereomicroscope. Temporarily prepared microscope slides were placed under a Nikon ECLIPSE 80i compound microscope fitted with a Nikon DS-Ri2 digital camera for observation and micro-morphological photography. The morphologies of colonies on the substrates were photographed using a Nikon SMZ1000 stereo zoom microscope. Microscopic structures were measured using the Tarosoft® Image Frame Work program, and the photographic plates were processed using Adobe Photoshop CS6 version 10.0 software (Adobe Systems, San Jose, CA, USA).
Single spore isolation was performed following the method described by Luo et al. (2018). The germinated conidia were aseptically transferred to fresh potato dextrose agar (PDA) plates and incubated at room temperature. The specimens were dried under natural light, wrapped in absorbent paper, and placed in a Ziploc bag with mothballs. Herbarium specimens were deposited in the Herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (KUN-HKAS), Kunming, China. The cultures were deposited in Kunming Institute of Botany, Chinese Academy of Sciences (KUNCC), Kunming, Yunnan, China. The novel species were registered in the Index Fungorum repository (https://indexfungorum.org/Names/IndexFungorumRegisterName.asp; accessed on 27 September 2024).
2.2 DNA extraction, PCR amplification, and sequencing
Fresh mycelia were scraped from colonies grown on PDA medium. DNA extraction was carried out using a DNA extraction kit (TOLOBIO Plant Genomic DNA Extraction Kit, Tsingke Company, Beijing, China) following the manufacturer’s instructions. PCR amplification was performed using primer pairs LR0R/LR5 (Vilgalys and Hester, 1990) for the nuclear ribosomal large subunit 28S rDNA gene (LSU), NS1/NS4 (White et al., 1990) for the nuclear ribosomal small subunit 18S rDNA gene (SSU), ITS5/ITS4 (White et al., 1990) for the internal transcribed spacer rDNA region (ITS), and fRPB2-5F/fRPB2-7cR (Liu et al., 1999; Rehner and Buckley, 2005) for the RNA polymerase second largest subunit (RPB2). The PCR amplification was carried out in a 25-μL reaction volume containing 12.5 μL of 2× Power Taq PCR Master Mix, 1 μL of each forward and reward primer (10 μM), 1 μL of genomic DNA template (30–50 ng/μL), and 9.5 μL sterilized double-distilled water. Amplifications were carried out using the BioTeke GT9612 thermocycler (Tsingke Company, Beijing, China). The PCR amplification conditions for ITS, LSU, and SSU consisted of initial denaturation at 98°C for 3 minutes, followed by 35 cycles of denaturation at 98°C for 20 seconds, annealing at 53°C for 10 seconds, an extension at 72°C for 20 seconds, and a final extension at 72°C for 5 minutes. The PCR amplification conditions for RPB2 consisted of initial denaturation at 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 1 minute, annealing at 52°C for 2 minutes, an extension at 72°C for 90 seconds, and a final extension at 72°C for 10 minutes. The quality of PCR products was checked using 1% agarose gel electrophoresis, and distinct bands were visualized in the gel documentation system (Compact Desktop UV Transilluminator analyzer GL-3120). The PCR products were purified, and Sanger sequences were obtained by Tsingke Company, Beijing, China.
2.3 Sequence assembly, alignment, and phylogenetic analyses
The newly generated sequences were subjected to the nucleotide BLAST search via the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 1 May 2024) to search the closely related taxa and confirm the correctness of the sequences. The sequence datasets were obtained by compiling the closely related taxa of the novel taxa retrieved from GenBank based on nucleotide BLAST searches and recent publications (Luo et al., 2018; Lechat and Fournier, 2020; Bakhit and Abdel-Aziz, 2021; Boonmee et al., 2021; Shi et al., 2021; Li et al., 2023; He et al., 2024). Outgroups were selected based on recently published data (Bakhit and Abdel-Aziz, 2021; Shi et al., 2021; Li et al., 2023) (Tables 1–3). Multiple sequence alignments were aligned with MAFFT v.7 (http://mafft.cbrc.jp/alignment/server/index.html; accessed on 10 May 2024) (Katoh et al., 2019) and automatically trimmed using TrimAl (http://phylemon.bioinfo.cipf.es/utilities.html; accessed on 10 May 2024) (Capella-Gutiérrez et al., 2009). A combined sequence dataset was obtained using SquenceMatrix v.1.7.8 (Vaidya et al., 2011). Phylogenetic relationships of the new taxa were performed based on maximum likelihood (ML) and Bayesian inference (BI) analyses.
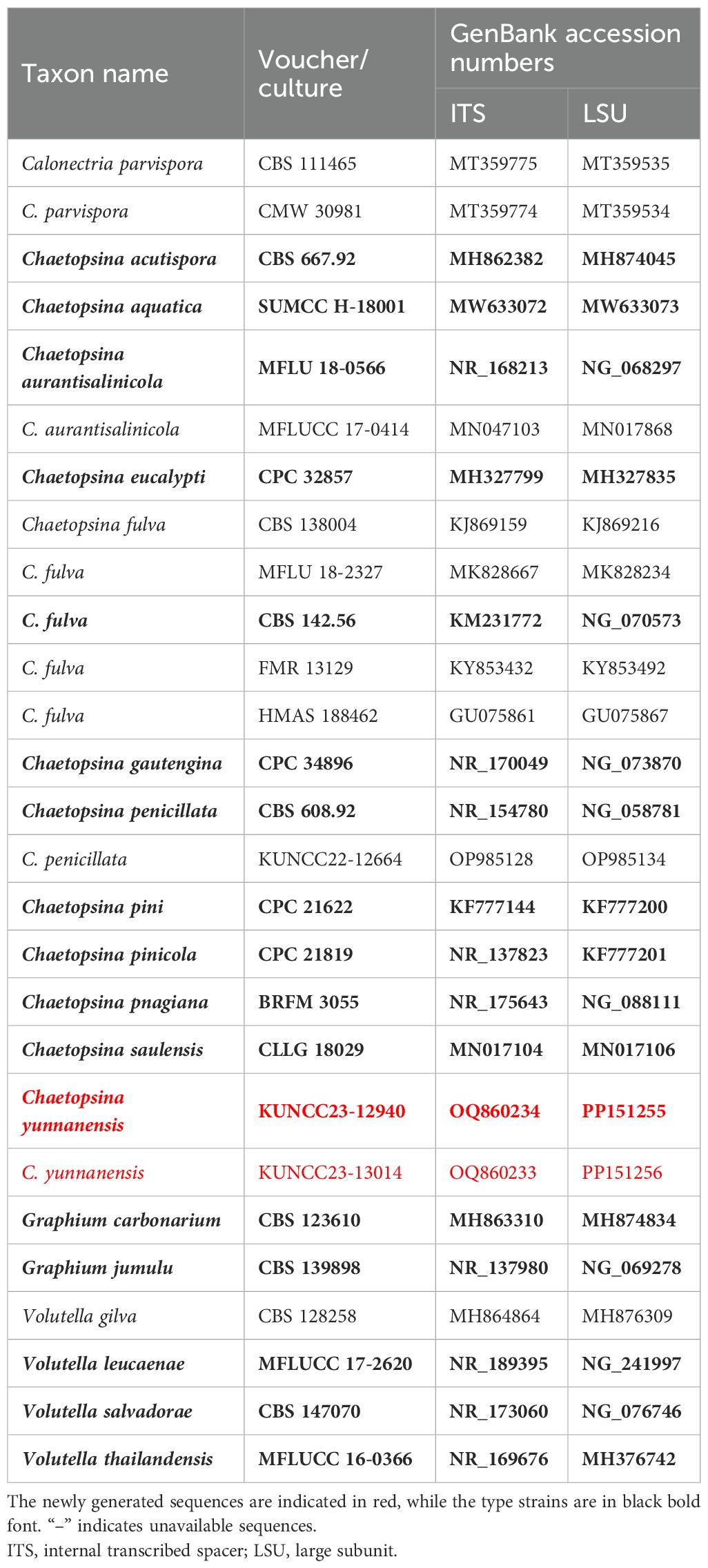
Table 1. Taxon names, strain numbers, and GenBank accession numbers of the ITS and LSU sequences used in the phylogenetic analyses of Chaetopsina.
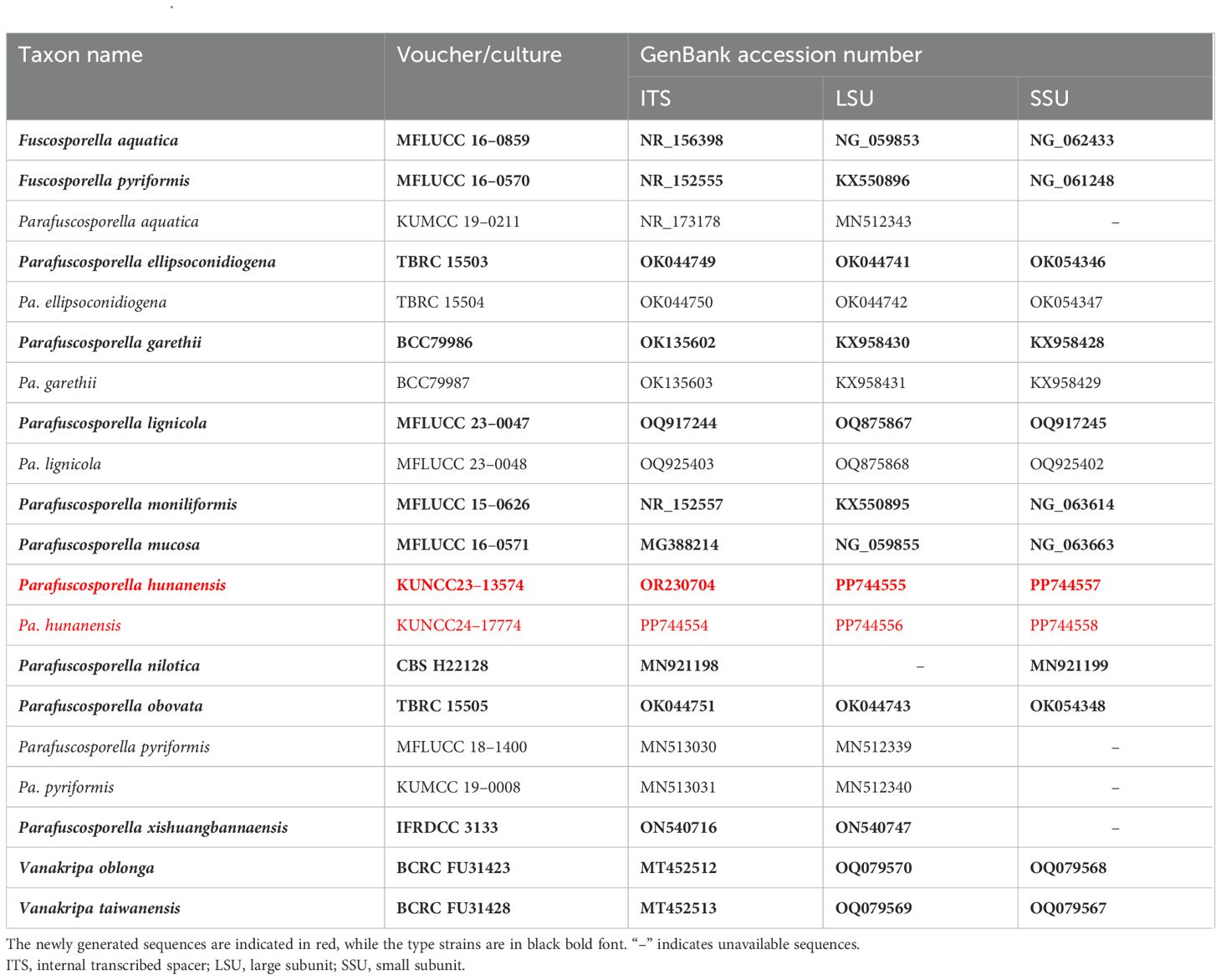
Table 2. Taxon names, strain numbers, and GenBank accession numbers of the ITS, LSU, and SSU and sequences used in the phylogenetic analyses of Parafuscosporella.
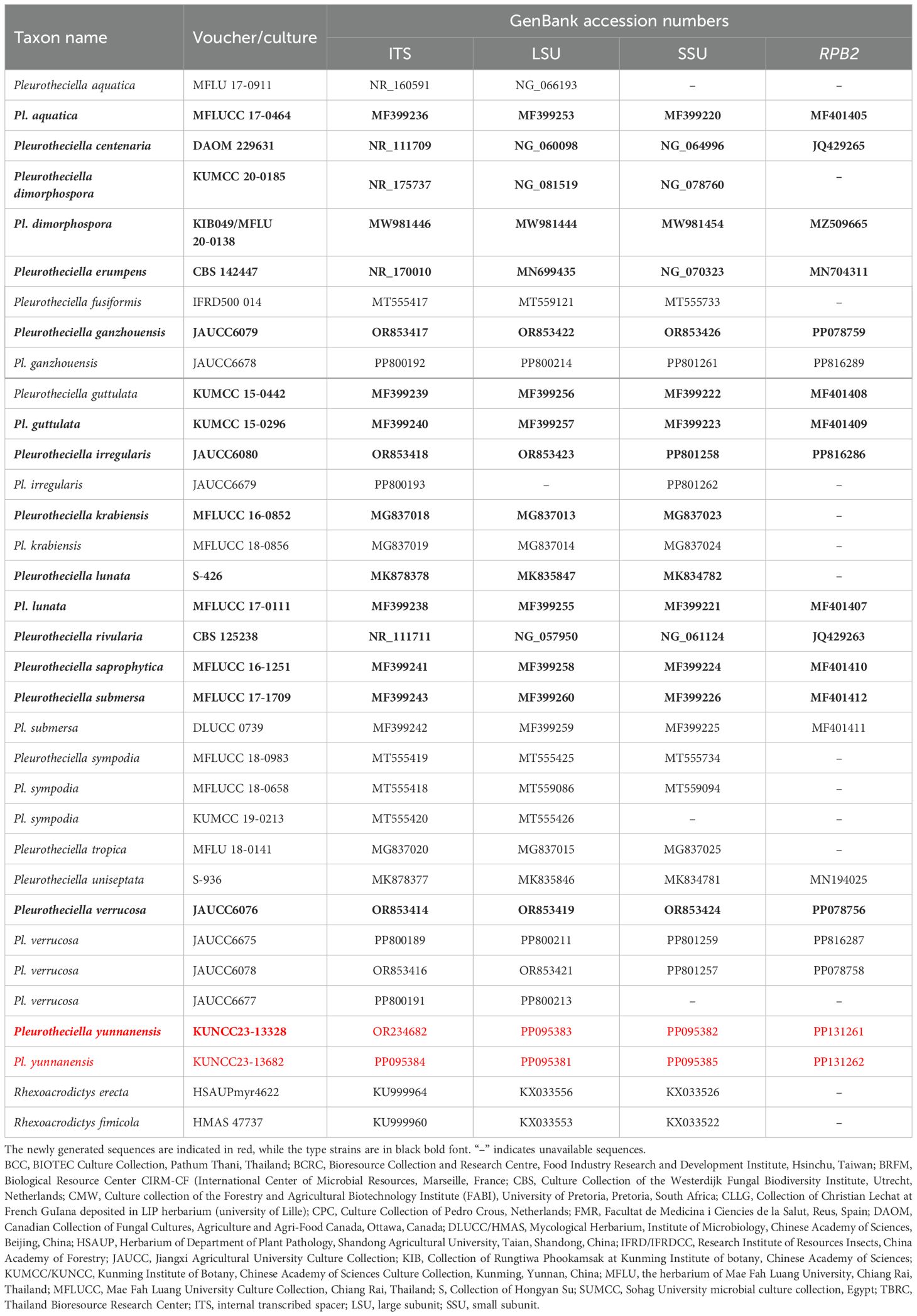
Table 3. Taxon names, strain numbers, and GenBank accession numbers of the ITS, LSU, SSU, and RPB2 sequences used in the phylogenetic analyses of Pleurotheciella.
ML analysis was performed by RAxML-HPC2 v.8.2.12 on the XSEDE (8.2.12) tool via the CIPRES Science Gateway (http://www.phylo.org/portal2; accessed on 25 May 2024) (Stamatakis, 2006; Miller et al., 2015) following the default setting but adjusted by setting 1,000 bootstrap replications and GTRGAMMA model of nucleotide substitution. The evolutionary model of nucleotide substitution for the BI analyses was performed independently for each locus using MrModeltest v 2.3 (Nylander, 2008). GTR+I+G was selected as the best-fit model for ITS, LSU, SSU, and RPB2 datasets in all analyses under the Akaike information criterion (AIC). Markov Chain Monte Carlo (MCMC) sampling was computed to estimate Bayesian posterior probabilities (BYPP) in MrBayes v.3.2.7 (Ronquist et al., 2012). Two parallel runs with six simultaneous Markov chains were run for 1,000,000 generations but stopped automatically when the critical value for the topological convergence diagnostic reached 0.01. Trees were sampled every 200th generation. The first 10% of the total trees were set as burn-in and were discarded. The remaining trees were used to calculate posterior probabilities in the majority rule consensus tree.
Phylograms were visualized using FigTree v1.4.4 (Rambaut, 2018) and rearranged in Adobe Photoshop CS6 software (Adobe Systems, USA). The new sequences were deposited in GenBank (Tables 1–3), and the final alignment and phylogenetic tree were registered in TreeBASE under the submission IDs: 31916 (C. yunnanensis), 31910 (Pa. hunanensis) and 31135 (Pl. yunnanensis) (http://www.treebase.org/ accessed on 25 December 2024).
3 Results
3.1 Phylogenetic analyses
Analysis 1
The concatenated LSU and ITS sequence dataset comprises 27 representative taxa in Calonectria, Chaetopsina, and Volutella with Graphium carbonarium (CBS 123610) and Graphium jumulu (CBS 139898) as the outgroup taxa (Table 1). The concatenated sequence matrix comprises 1,420 characters, including gaps (LSU, 828 bp; ITS, 588 bp). BI and ML analyses of the combined dataset were performed to determine the placement of our new taxon and infer relationships at the intrageneric level as well as resolve the phylogenetic relationships of the core genera in Nectriaceae. The phylogenetic trees obtained from BI and ML analyses resulted in trees with largely similar topologies. A phylogenetic investigation based on ML analysis was carried out with the best RAxML tree with a final likelihood value of −5,889.450811. The matrix had 385 distinct alignment patterns, with 6.19% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.238371, C = 0.250160, G = 0.286306, T = 0.225163, with substitution rates AC = 1.768750, AG = 2.223229, AT = 2.277765, CG = 1.698772, CT = 7.040906, GT = 1.000000; gamma distribution shape parameter α = 0.526092. The final average standard deviation of split frequencies at the end of total MCMC generations was calculated as 0.009709 in BI analysis.
Molecular analysis of a concatenated LSU and ITS sequence dataset (Figure 1) demonstrated that the phylogenetic relationship of the representative genera, Calonectria, Chaetopsina, and Volutella, is not well-resolved. The genera Calonectria and Volutella formed distinct clades with well-supported value in ML analyses but low-supported value in BI analysis. Chaetopsina formed a distinct clade with Calonectria and Volutella with low support. The interspecific status of many Chaetopsina species is not well-resolved in the present study, including Chaetopsina acutispora, Chaetopsina pinicola, Chaetopsina pnagiana, and Chaetopsina saulensis. Two new strains (KUNCC23-12940 and KUNCC23-13014) formed a robust subclade [100% maximum likelihood bootstrap support (MLBS) and 1.00 Bayesian posterior probabilities (BYPP)] and clustered with C. pinicola (CPC 21819) with low-supported values. C. beijingensis (CBS 138004) shared the same branch with the type (CBS 142.56) and representative strains of C. fulva (FMR 13129 and MFLU 18-2327). In contrast, C. fulva (HMAS 188462) remained distinct from other strains.
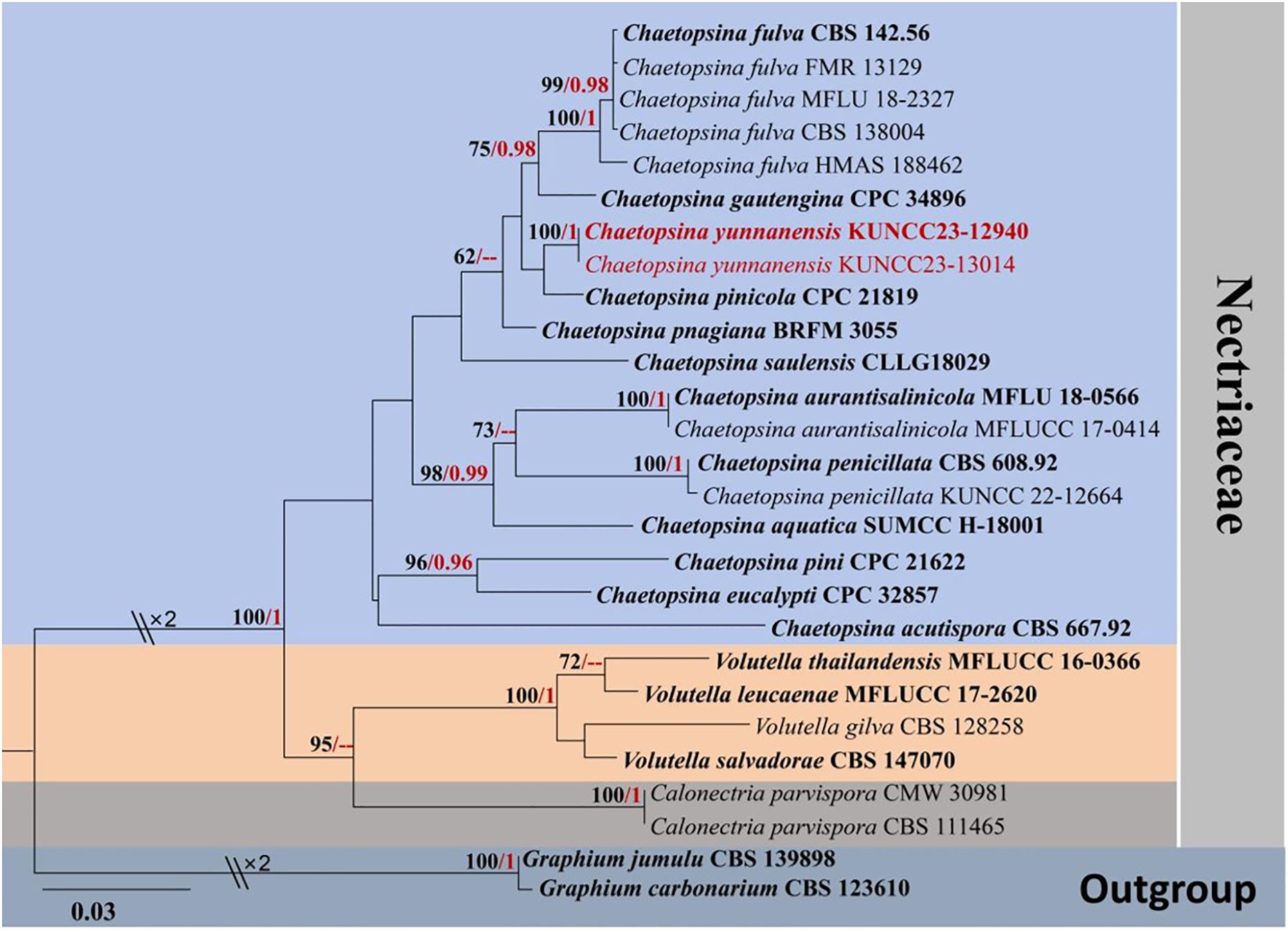
Figure 1. RAxML phylogenetic tree of a concatenated LSU and ITS sequence dataset. The BI and ML support values equal to or greater than 0.90 BYPP and 60% MLBS are shown as “MLBS/BYPP” at the nodes. The tree is rooted to Graphium carbonarium (CBS 123610) and Graphium jumulu (CBS 139898). Type strains are in bold, and newly generated strains are in red. LSU, large subunit; ITS, internal transcribed spacer; BI, Bayesian inference; ML, maximum likelihood.
Analysis 2
The phylogenetic analyses of concatenated ITS, LSU, and SSU sequence data were conducted to demonstrate the phylogenetic relationships of the new isolates with other species in Parafuscosporella. Twenty strains were included in the combined dataset (Table 2), which comprised 2,340 characters (ITS, 535 bp; LSU, 829 bp; SSU, 976 bp) after alignment (including gaps). The best RAxML tree with a final likelihood value of −6,818.339932 is presented (Figure 2). RAxML analysis yielded 415 distinct alignment patterns and 16.25% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.231018, C = 0.253572, G = 0.287376, T = 0.228033, with substitution rates AC = 1.005383, AG = 1.978020, AT = 1.365847, CG = 0.802886, CT = 4.380984, GT = 1.000000; gamma distribution shape parameter alpha = 0.765977. The final average standard deviation of split frequencies at the end of total MCMC generations for BI analysis was 0.009867.
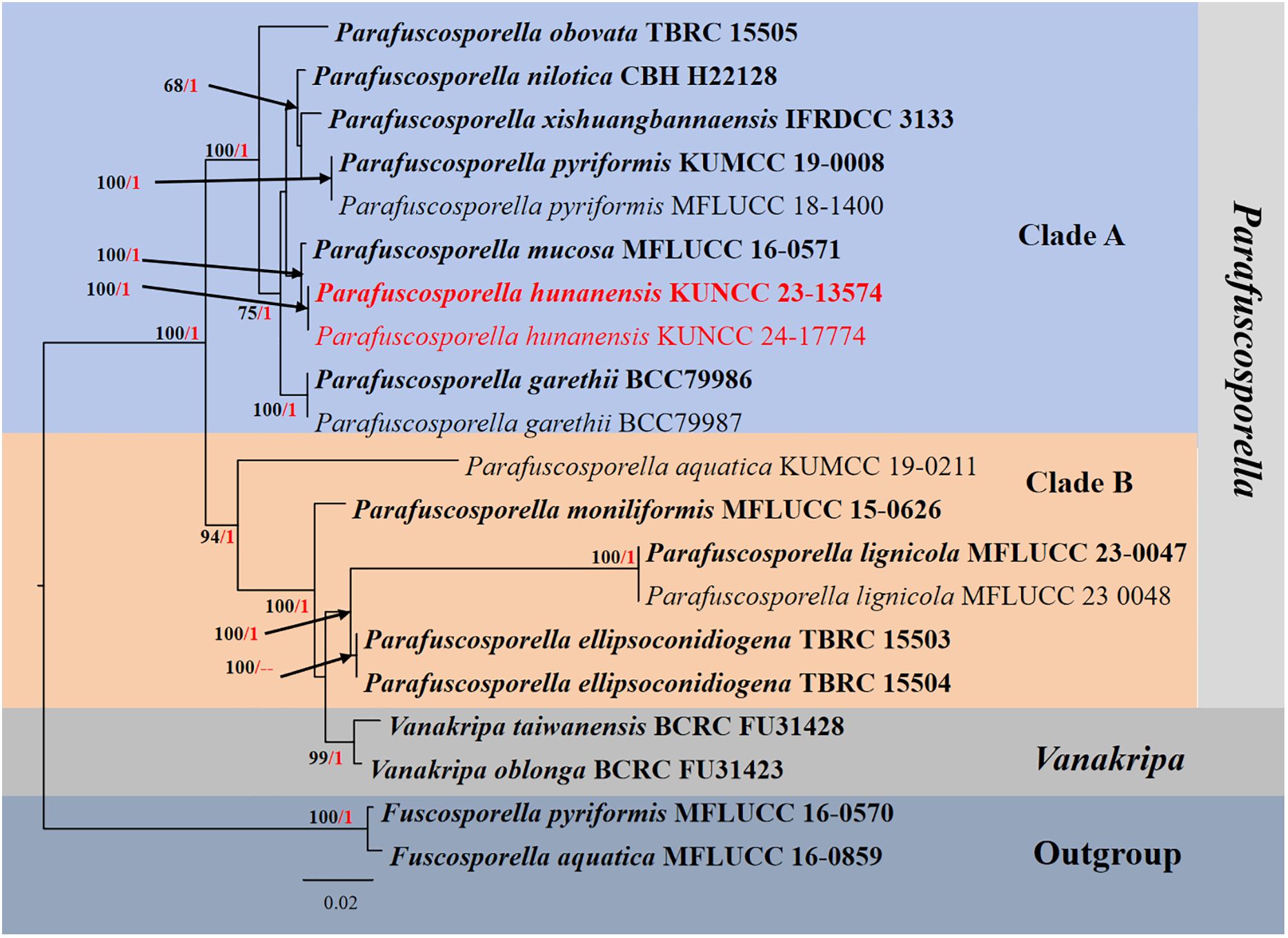
Figure 2. RAxML phylogenetic tree of a concatenated ITS, LSU, and SSU sequence datasets. The BI and ML support values equal to or greater than 0.90 BYPP and 60% MLBS are shown as “MLBS/BYPP” at the nodes. The tree is rooted to Fuscosporella aquatica (MFLUCC 16–0859) and Fuscosporella pyriformis (MFLUCC 16–0570). Type strains are in bold, and newly generated strains are in red. ITS, internal transcribed spacer; LSU, large subunit; SSU, small subunit; BI, Bayesian inference; ML, maximum likelihood.
Phylogenetic analyses retrieved from ML and BI analyses were not significantly different and showed similar topologies. Phylogenetic analyses demonstrated that Parafuscosporella formed the well-resolved clade and separated into two subclades: clade A comprises Pa. garethii, Pa. hunanensis, Pa. mucosa, Pa. nilotica, Pa. obovata, Pa. pyriformis, and Pa. xishuangbannaensis, whereas clade B comprises Pa. aquatica, Pa. ellipsoconidiogena, Pa. lignicola, and Pa. moniliformis. Two Vanakripa species, V. oblonga and V. taiwanensis, also formed a high-support subclade closely related to Pa. ellipsoconidiogena and Pa. lignicola. The interspecific status of most Parafuscosporella is well-clarified, except for Pa. xishuangbannaensis, in the present study. The two strains (KUNCC23-13574 and KUNCC24-17774) of Pa. hunanensis sp. nov. clustered with Pa. mucosa in subclade A with 100% MLBS and 1.00 BYPP support values.
Analysis 3
The phylogenetic analyses of concatenated ITS, LSU, SSU, and RPB2 sequence data were conducted to demonstrate the relationship between the new species and other known species in Pleurotheciella. Thirty-four strains were included in the combined dataset (Table 3), which comprised 2,919 characters (ITS, 564 bp; LSU, 825 bp; SSU, 682 bp; RRB2, 848 bp) after alignment (including gaps). Rhexoacrodictys erecta (HSAUPmyr4622) and Rhexoacrodictys fimicola (HMAS 47737) were selected as the outgroup taxa. The best RAxML tree with a final likelihood value of −10,754.565198 was selected to represent the phylogenetic affinity of the novel species with closely related species (Figure 3). RAxML analysis yielded 677 distinct alignment patterns and 18.84% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.234132, C = 0.258625, G = 0.287873, T = 0.219371, with substitution rates AC = 1.285686, AG = 3.496823, AT = 1.635783, CG = 0.882608, CT = 9.634626, GT = 1.000000; gamma distribution shape parameter alpha = 0.461774. The final average standard deviation of split frequencies at the end of total MCMC generations for BI analysis was 0.009778.
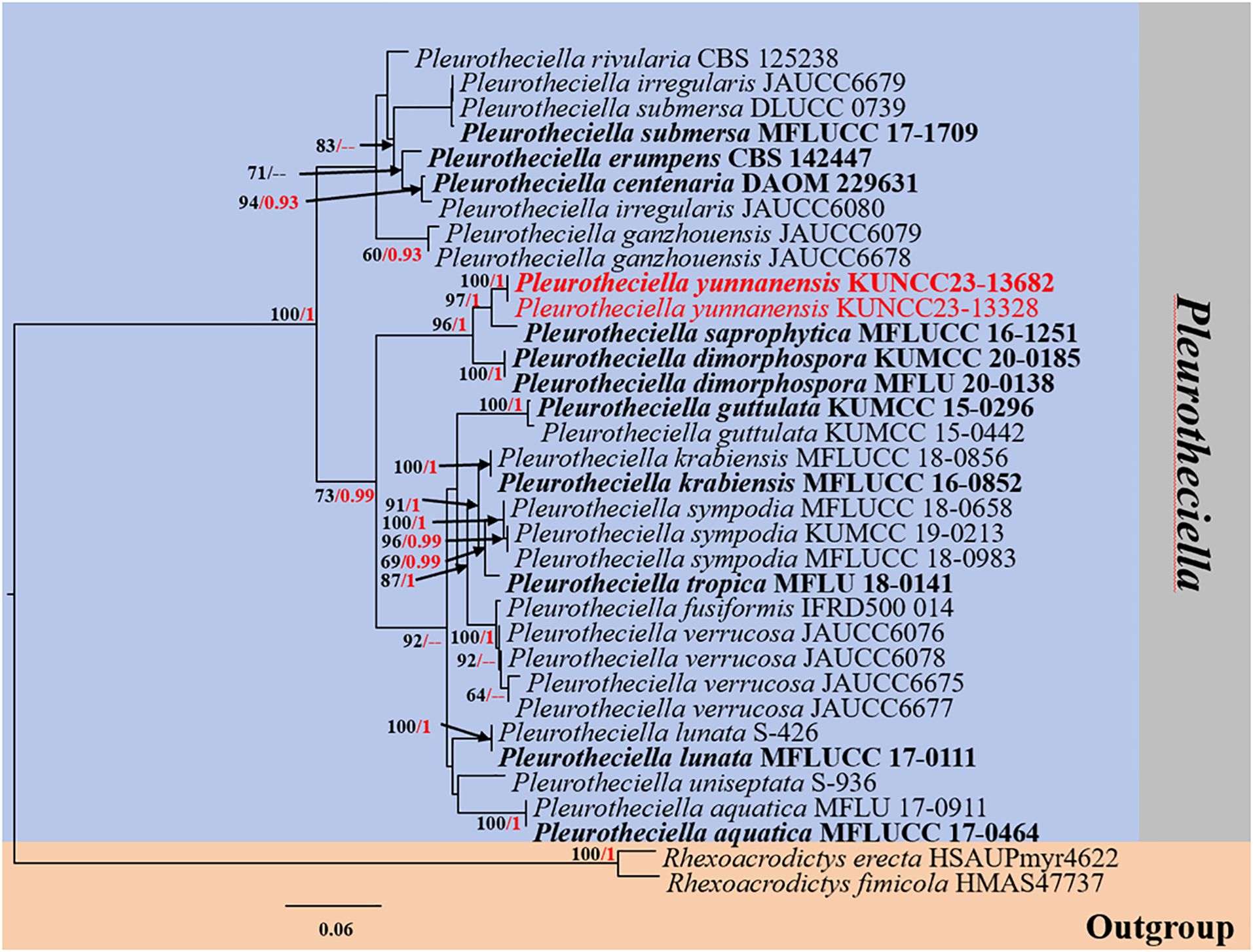
Figure 3. RAxML phylogenetic tree of a concatenated ITS, LSU, SSU, and RPB2 sequence dataset. The BI and ML support values equal to or greater than 0.90 BYPP and 60% MLBS are shown as “MLBS/BYPP” at the nodes. The tree is rooted to Rhexoacrodictys erecta (HSAUPmyr4622) and Rhexoacrodictys fimicola (HMAS 47737). Type strains are in bold, and newly generated strains are in red. ITS, internal transcribed spacer; LSU, large subunit; SSU, small subunit; BI, Bayesian inference; ML, maximum likelihood.
Phylogenetic analyses retrieved from ML and BI analyses were not significantly different and showed similar topologies. Phylogenetic affinities of most Pleurotheciella species are well-resolved (up to 70% MLBS and 0.90 BYPP support values) in the present study, except for Pl. uniseptata. However, the species always formed a stable subclade with Pl. aquatica. Two strains (KUNCC23-13328 and KUNCC23-13682) of the new species, Pl. yunnanensis, formed a distinct subclade (100% MLBS/1.00 BYPP), sister to Pl. saprophytica (MFLUCC 16-1251) with significant support (96% MLBS/1.00 BYPP) and clustered with Pl. dimorphospora (KUMCC 20-0185) with significant support (96% MLBS/1.00 BYPP).
3.2 Taxonomy
Class Sordariomycetes O.E. Erikss. & Winka
Subclass Hypocreomycetidae O.E. Erikss. & Winka
Order Hypocreales Lindau
Family Nectriaceae Tul. & C. Tul.
Genus Chaetopsina Rambelli
Chaetopsina fulva Atti Accad. Sci. Ist. Bologna, Cl. Sci. Fis., Rendiconti 11: 5 (1956). Amend. L. Li, Phookamsak & Bhat
Index Fungorum number: IF 294735
= C. beijingensis Crous & Y. Zhang ter, in Crous et al., Persoonia 32: 267 (2014)
Typification: Italy, on fallen leaves (needle) of Cedrus deodara (Pinaceae), Feb 1956, A. Rambelli, IMI 62199 (type of C. fulva), ex-type culture, CBS 142.56; China, Beijing, Fragrant Hill, N39°59′18.4″ E116°11′25″, on needles of Pinus tabulaeformis (Pinaceae), 1 Sep 2013, P.W. Crous & Y. Zhang, CBS H-21718 (holotype of C. beijingensis), CPC 23629 = CBS 138004.
Saprobic on Pinaceae and various plant hosts as well as soil. Sexual morph: Referred to Chaetopsinectria chaetopsinae (≡ Nectria chaetopsinae Samuels; Lombard et al., 2015; Rossman et al., 2016). Asexual morph: Conidiophores 180–300 × 7.5–10 µm, macronematous, simple, sparse, erect, setiform, often swollen, bulbous at the base (up to 15–20 µm), tapering toward acutely rounded apex, straight or slightly curved, mostly flexuous, yellow-brown to red-brown, less pigmented at the base, turning red-brown in 3% KOH and yellow in lactic acid, unbranched, smooth to verruculose, up to 13-septate (12–16-septate as in C. beijingensis), thick-walled (2 µm diam.), sterile or occasionally fertile; fertile region situated below the middle of the main axis or higher, occasionally terminal, comprising an irregularly branched, densely aggregated, hyaline, conidiogenous apparatus. Conidiogenous cells 6–12(–20) × 3.5–5 µm monophialidic, discrete, hyaline, ampulliform to lageniform, smooth, irregularly branched, periclinal thickening visible, with minute collarettes. Conidia 8–12 × 2 µm, solitary, hyaline, subcylindrical to cylindrical, with rounded ends, aseptate, smooth, with guttules, rarely with flattened hilum (adopted from Ellis, 1971; Kirk and Sutton, 1985; Samuels, 1985; Luo and Zhuang, 2010; Crous et al., 2014; Perera et al., 2023).
Known habitats and host: On needle of C. deodara and fallen leaves of Laurus nobilis, Quercus, and Carpinus, needles of P. tabulaeformis (as C. beijingensis), woody test blocks of X. dolabriformis, dead leaves, decaying wood and soil (Rambelli, 1956; Ellis, 1971; Sivichai et al., 2000; Luo and Zhuang, 2010; Crous et al., 2014; Arias et al., 2015; Hernández-Restrepo et al., 2017; Luo et al., 2019; Vu et al., 2019; Perera et al., 2023).
Known distribution: Canada, China, Italy, Spain, and Thailand (Rambelli, 1956; Ellis, 1971; Sivichai et al., 2000; Luo and Zhuang, 2010; Crous et al., 2014; Arias et al., 2015; Hernández-Restrepo et al., 2017; Luo et al., 2019; Vu et al., 2019; Perera et al., 2023).
Notes: C. fulva was designated as the type species of Chaetopsina when Rambelli (1956) introduced the genus. The species was poorly studied to begin with, and molecular data of the type specimen were obtained by Vu et al. (2019), who also confirmed its phylogenetic affinity in the Nectriaceae. The species, originally isolated from a needle of C. deodara and fallen leaves of L. nobilis, Quercus, and Carpinus in northern Italy, produces pigmented, erect, septate, setiform conidiophores that are apically sterile (Rambelli, 1956; Kirk and Sutton, 1985; Bakhit and Abdel-Aziz, 2021; Index Fungorum, 2024). Ellis (1971) provided a detailed morphological description of C. fulva, which produces macronematous, up to 280 × 5–8 µm conidiophores, with bases often swollen to 15–20 µm. The conidiogenous cells (phialides) are 7–15 µm long, with a swollen base 3–4 µm wide and a phialidic neck approximately 1 µm thick. Conidia are 7–11 × 1 µm. The species were reported from dead fallen leaves and soil, with distribution noted in Canada and Italy. Kirk and Sutton (1985) re-circumscribed the species based on type studies and emended the generic circumscription incorporating only five described species, viz., Chaetopsina catenulata, C. fulva, Chaetopsina penicillata, C. polyblastia, and Chaetopsina splendida. Kirk and Sutton (1985) treated Chaetopsina romantica (= Chaetopsis romantica) as a synonym of C. fulva on the insistence of Rambelli (1987).
Luo and Zhuang (2010) introduced a sexual genus Chaetopsinectria to accommodate sexual morphs of Chaetopsina. Based on ITS and 28S rDNA sequence analyses, Luo and Zhuang (2010) mentioned that C. fulva, C. chaetopsinae, and C. chaetopsinae-penicillatae have a close relationship. Regarding the generic type, C. fulva, the common features include red-brown and setose conidiophores that turn yellow in lactic acid, hyaline phialides, and sienna-colored colonies on PDA; these morphological traits are shared by other Chaetopsina asexual morph of nectriaceous fungi and provide phenotypic information in defining this asexual genus (Samuels, 1985; Okada et al., 1997; Luo and Zhuang, 2010). However, Rossman et al. (2016) mentioned that C. fulva is the asexual morph of C. chaetopsinae (≡ N. chaetopsinae), and hence, these generic names were synonyms and further recommended to use Chaetopsina rather than Chaetopsinectria due to its prior establishment, following the 1F = 1N policy (McNeill et al., 2012).
Crous et al. (2014) introduced C. beijingensis, collected from decaying wood submerged in a freshwater stream, in Beijing, China, and further demonstrated the similarity of nucleotide pairwise (ITS and LSU) between C. beijingensis and C. fulva (HMAS 188462) with 98% similarity of ITS and 100% similarity of LSU. Morphologically, C. beijingensis resembles C. fulva but can be distinguished from the latter in having subcylindrical and slightly larger conidia [(11–)12–13(–14) × 2(–2.5) μm; Crous et al., 2014]. In contrast, C. fulva (described from the type’s slide) has cylindrical, 8–12 × 1.5 μm conidia (Kirk and Sutton, 1985). Vu et al. (2019) provided the molecular data from the type of C. fulva (CBS 142.56). Subsequently, Lechat and Fournier (2019) informally treated C. beijingensis as a synonym of C. fulva, in view of their morphological resemblance, phylogenetic evidence, and nucleotide pairwise similarities—99.6% similarity of ITS and 100% similarity of LSU sequences based on the nucleotide pairwise comparison of the type strains (CBS 142.56 vs. CBS 138004)—but did not taxonomically synonymize these taxa. Phylogenetically, C. beijingensis (CBS 138004) shares the same branch with the type strain of C. fulva and other representative strains with high support (100% MLBS/1.00 BYPP; Figure 1) in the present study. In accordance with Lechat and Fournier (2019) and our own observations, we formally synonymized C. beijingensis under C. fulva based on morphological indistinctiveness and phylogenetic support coupled with the conspecific in nucleotide polymorphism. Detailed description is amended to incorporate the morphological features of C. beijingensis and C. fulva.
Chaetopsina yunnanensis L. Li, Bhat & Phookamsak, sp. nov.
Index Fungorum number: IF901629, Figure 4

Figure 4. Chaetopsina yunnanensis (KUN-HKAS 126987, holotype). (A) Hyphomycetous colonies associated with ascomata (in group) on pseudostomata on submerged branch. (B) Pyriform ascoma with a conical apex. (C) Upper view of ascoma with pore-like opening ostiole. (D) Vertical section of peridium. (E, F) Conidiophores and conidiogenous cells bearing conidia. (G, H) Conidia. (I, J) Culture characteristics on PDA (I = from up-front, J = down-reverse). Scale bars: (B) = 100 μm, (C–E) = 50 μm, (F) = 20 μm, and (G, H) = 10 μm. PDA, potato dextrose agar.
Etymology: The name reflects the location of Yunnan where the holotype was collected.
Holotype: KUN-HKAS 126987
Saprobic on an unidentified branch, submerged in a freshwater habitat. Sexual morph: Ascomata perithecial, raised from pseudostomatic, visible as orangish to reddish brown, solitary or in groups (2–3 ascomata), superficial, scattered, associated with Chaetopsina-like asexual morph, ovoid to obpyriform, with an acutely conical apex, ostiolate with pore-like opening, soft and freshly, shiny, collapsing when dry. Peridium 20–50 µm ( = 38 µm, n = 20) thick in vertical section, composed of thick-walled, orangish cells, of textura angularis to textura prismatica or epidermoidea, becoming paler and slightly elongated toward interior. Asci and ascospores were not observed. Asexual morph: Conidiophores 200–310 µm long, 5–11 µm wide, broader at bulbose base with 8–10 µm wide, erect, macronematous, mononematous, straight to slightly curved, 5–8-septate, thick-walled, smooth, orangish brown to reddish brown, paler toward apex, fertile at above half. Fertile region yellow to orangish brown comprising loosely and regularly arranged penicillate, smooth-walled, 2-septate branches bearing densely aggregated conidiogenous cells, arranged in several whorls. Conidiogenous cells 6–8 × 4–6 µm ( = 7 × 4.5 µm, n = 20), discrete, ampulliform, phialidic, hyaline. Conidia 8–11 × 5–7 µm ( = 9.5 × 6 µm, n = 20), cylindrical to cylindric-fusoid or ellipsoidal, hyaline, solitary, unicellular, guttulate, smooth.
Culture characteristics: Conidia germinating on PDA within 48 h. Colonies on PDA reaching 20 mm diam. at room temperature in natural light after 1 month. Colonies from above medium dense, circular, dull, flat to slightly raised, effuse to fairly fluffy, white to cream, edge entire, with a rough surface, slightly radiating with pale yellowish concentric ring outward colonies, sometimes with raised floccose mycelial turfs; reverse: dark reddish brown, paler at margins, with black reddish pigments produced in PDA. Sporulation not observed.
Material examined: China, Yunnan Province, Xishuangbanna (99°56′–101°50′E, 21°08′–22°36′N, 470–2,429.5 msl), on a branch submerged in a freshwater stream, 9 September 2022, L. Li, LILU-133 (KUN-HKAS 126987, holotype), ex-type living culture KUNCC23-12940; ibid., Kunming, Yang Zonghai Lake (102°5′–103°02′E, 24°51′–24°58′N, 1,770 msl; pH of water = 8.92, temperature = 26°C, dissolved oxygen = 7.6 mg/L, purification level 3), on submerged wood, 12 July 2022, L. Li, LILU-133-1 (KUN-HKAS 126988), living culture KUNCC23-13014.
Notes: The nucleotide BLAST searches of ITS and LSU sequences indicated that C. yunnanensis sp. nov. (KUNCC23-12940) is close to Chaetopsina species, with LSU of 99% similarity to C. pinicola (CBS 136444), Chaetopsina gautengina (CPC 34896), and C. pnagiana (BRFM 3055); ITS of 97% similarity to C. pinicola (CBS 136444); and 96% similarity to C. fulva (CBS 138004) and C. gautengina (CPC 34896). In addition, the morphological characteristics of C. yunnanensis well match the concept of Chaetopsina, with red-pigmented upright conidiophores bearing discrete, phialidic conidiogenous cells and unicellular, cylindrical to cylindric-fusoid or ellipsoidal conidia (Kirk and Sutton, 1985; Samuels, 1985; Lechat and Fournier, 2020). The concatenated ITS and LSU phylogenetic analyses also demonstrated that the new strains KUNCC23-12940 and KUNCC23-13014 clustered within the species group of Chaetopsina but constituted an independent branch and sister to C. pinicola (CPC 21819) (Figure 1). The nucleotide base pair comparison between the type strain of the new species, KUNCC23-12940, and C. pinicola (CPC 21819) revealed 18/586 bp (3.07%) of ITS and 4/800 bp (0.5%) of LSU differences. Morphologically, C. yunnanensis differs from C. pinicola in having darker reddish-pigmented, 5–8-septate conidiophores, with penicillate fertile region at the apex and cylindric-fusoid or ellipsoidal conidia. In contrast, C. pinicola has medium brown, turning red-brown in 3% KOH, unbranched, verruculose, 11–15-septate conidiophores, with fertile mid region and subcylindrical, larger conidia [(11–)13–15(–17) × 2(–2.5) µm; Crous et al., 2013].
Class Sordariomycetes O.E. Erikss. & Winka
Subclass Savoryellomycetidae O.E. Erikss. & Winka
Order Fuscosporellales Jing Yang, Bhat & K.D. Hyde
Family Fuscosporellaceae Jing Yang, Bhat & K.D. Hyde
Genus Parafuscosporella Jing Yang & K.D. Hyde
Parafuscosporella hunanensis L. Li, Bhat & Phookamsak, sp. nov.
Index Fungorum number: IF 902698, Figure 5
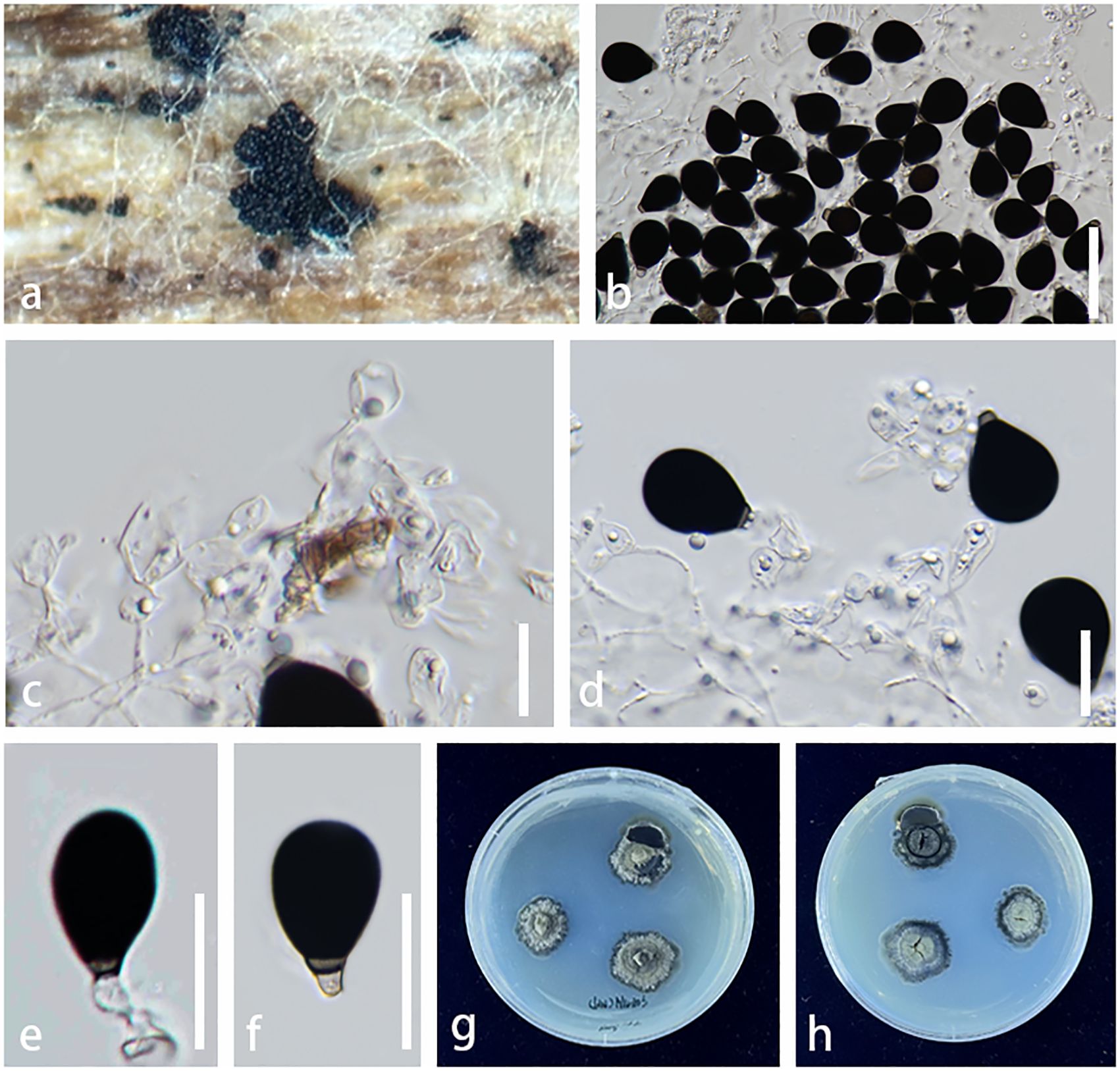
Figure 5. Parafuscosporella hunanensis (KUNCC23–13574, holotype). (A) Sporodochia on host surface. (B) Conidial mass with dense, inconspicuous conidiophores, embedded in gelatinous matrix. (C) Micronematous, branched septate conidiophores with ampulliform to doliiform conidiogenous cells. (D) Conidiophores and conidiogenous cells bearing conidia. (E, F) Conidia with a distinct basal frill derived from the distal end of the conidiogenous cell or with pale brown to hyaline protuberance. (G, H) Colony on PDA (G = from above, H = from below). Scale bars: (B) = 50 μm, (C–F) = 20 μm. PDA, potato dextrose agar.
Etymology: The name reflects the location Hunan Province of China where the holotype was collected.
Holotype: KUN-HKAS 136260
Saprobic on decaying submerged wood in a freshwater stream. Sexual morph: Undetermined. Asexual morph Colonies on natural substrate sporodochial, scattered, sparse, visible as black, dull, soft, granular on the host surface. Mycelium semi-immersed to superficial, composed of septate, hyaline, and smooth hyphae. Conidiophores 10–35 × 3–10 µm ( = 23 × 7 µm, n = 20), micronematous to semi-macronematous, mononematous, inconspicuous, hyaline, cylindrical, dense, erect or flexuous, branched, septate, smooth-walled. Conidiogenous cells 7–15 × 4–12 µm ( = 11 × 8 µm, n = 20), holoblastic, monoblastic, integrated, terminal, determinate, hyaline to pale brown, ampulliform to doliiform, smooth-walled. Conidia 20–29 × 15–22 µm ( = 26 × 19 µm, n = 20), acrogenous, obovoid to pyriform, with base truncate, dark brown to black, versicolored, paler brown at basal cell, 1-septate near the base, sometimes with pale brown to hyaline protuberance, or with a distinct basal frill derived from the distal end of the conidiogenous cell on cessation, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 24 h. Germ tubes produced from both ends. Colonies on PDA reaching 1 mm diam. at room temperature in natural light after 3 months. Colonies from above, dense, irregular in shape, with undulate margin, dull, flat to slightly raised, velvety to felted, dark greenish gray at the margin, pale greenish gray toward the center, slightly radiated with convex ring, surface rough, with winkled folded aspect; reverse, dark greenish at the margin, pale greenish gray toward the center, radiated with black and white ring, radially furrowed at the edge; not produced pigmentation. Sporulation not observed.
Material examined: China, Hunan Province (108°47′–114°15′E, 24°38′–30°08′N, 500–1,500 msl), saprobic on decaying wood submerged in a freshwater stream, 26 August 2022, L. Li, LILU-203 (KUN-HKAS 136260, holotype), ex-type living culture, KUNCC23–13574; ibid., LILU-203-2 (KUN-HKAS 136261), living culture KUNCC24-17774.
Notes: The nucleotide BLAST searches of ITS, LSU, and SSU sequences indicated that Pa. hunanensis sp. nov. (KUNCC23-13574) is similar to Pa. mucosa (MFLUCC 16-0571, type strain) with 97.73% similarity of ITS, 99.88% similarity of LSU, and 100% similarity of SSU. Multigene phylogenetic analyses demonstrated that Pa. hunanensis (KUNCC23-13574 and KUNCC24-17774) clustered with the type strain of Pa. mucosa (MFLUCC 16-0571) with high support (100% MLBS/1.00 BYPP; Figure 2). Pa. hunanensis morphologically resembles Pa. mucosa (MFLU 16-1980) in having obovoid to pyriform, versicolored conidia, with brown to dark brown distal cell, paler basal cell, truncate base, and septate near the basal cell. However, Pa. hunanensis can be distinguished from Pa. mucosa by the absence of jelly-like cover on sporodochia and with inconspicuous, branched, septate, cylindrical conidiophores, hyaline to pale brown, doliiform conidiogenous cells and obovoid to pyriform conidia. In contrast, Pa. mucosa produced sporodochia with distinct jelly-like covering, macronematous, conspicuous, cylindrical conidiophores with globose, subglobose, ellipsoidal, or clavate conidiogenous cells, and obovoid to pyriform, or ellipsoidal conidia (Yang et al., 2016). The nucleotide pairwise difference comparison between Pa. hunanensis (KUNCC23-13574) and Pa. mucosa (MFLUCC 16-0571) revealed 14/602 bp (2.32%) for ITS, 1/839 bp (0.11%) for LSU, and 0/777 bp (0%) for SSU sequences. Based on the morphological distinctions, and phylogenetic evidence, coupled with the significant nucleotide pairwise differences of ITS region, the new species Pa. hunanensis is introduced herein.
Class Sordariomycetes O.E. Erikss. & Winka
Subclass Savoryellomycetidae O.E. Erikss. & Winka
Order Pleurotheciales Réblová & Seifert
Family Pleurotheciaceae Réblová & Seifert
Genus Pleurotheciella Réblová
Pleurotheciella yunnanensis L. Li, Bhat & Phookamsak, sp. nov.
Index Fungorum number: IF901610, Figure 6
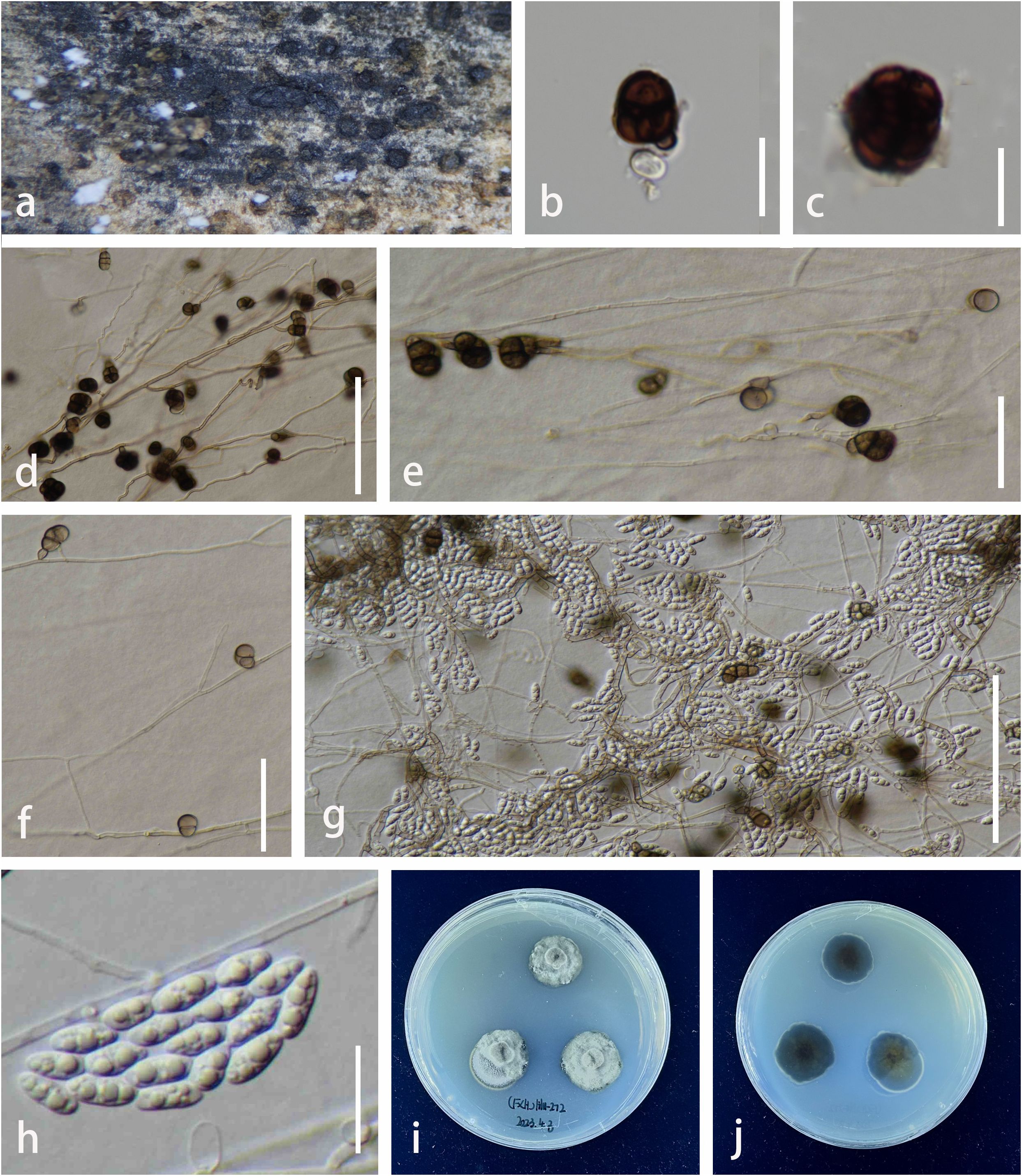
Figure 6. Pleurotheciella yunnanensis (KUN-HKAS 132016, holotype). (A) Colonies on surface of submerged wood. (B, C) Dark brown, muriform conidia on natural substrate. (D–F) Conidiogenous cells bearing muriform conidia in vitro. (G) Conidiogenous cells bearing brown, muriform conidia (Type I) and hyaline, ellipsoidal conidia (Type II) on PDA. (H) Hyaline conidia (Type II). (I, J) Colonies on PDA (I = from above, J = from below). Scale bars: (B–E, G, H) = 20 µm; (F) = 10 µm. PDA, potato dextrose agar.
Etymology: In reference to Yunnan Province of China, where the holotype was collected.
Holotype: KUN-HKAS 132016
Saprobic on decaying, submerged wood from freshwater habitats. Sexual morph: Undetermined. Asexual morph: Colonies on natural substrates visible as small, black, scattered, dots on the host surface. Mycelium immersed to partially superficial, brown to dark brown, composed of branched, septate, smooth, 2–3 µm wide, thin-walled hyphae. Conidiophores difficult to distinguish on the host, semi-macronematous, mononematous, sub-hyaline to brown, erect, or bent on the host surface. Conidiogenous cells 3–5 × 3–6 µm ( = 4 × 4.5 µm, n = 20), holoblastic, hyaline, raised from hyphae, terminal or intermediate. Conidia 18–25 × 22–30 µm ( = 21.5 × 26 µm, n = 20), varied in shape, ellipsoidal to subglobose, dark brown to black, initially forming phragmoconidia, later becoming muriform, chairoid at maturity. In vitro: Dimorphic, with two conidial types. Type I: Conidiophores 30–50 × 3–5 µm ( = 36 × 4.5 µm, n = 20), semi-macronematous or macronematous, mononematous, hyaline to dark brown, cylindrical, septate, unbranched. Conidiogenous cells holo- to polyblastic, terminal, lateral, or intercalary, brown. Conidia 15–22 × 12–15 µm ( = 19 × 13.5 µm, n = 20), phragmosporous to muriform, variedly shaped, brown to dark brown, subglobose to cordiform, or irregular in shape, with a protuberant hilum, initially 2–3-phragmoseptate, at maturity becoming 1–2 transverse and longitudinally dictyoseptate, sectored, leaf clover-like, brown to dark brown. Type II: Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–8 × 2–6 µm ( = 5.5 × 5 µm, n = 20), phialidic, terminal, integrated, with minute denticles, subhyaline to pale brown, 1–2-septate, unbranched, arising in pseudo-chains. Conidia 7–12 × 3–5 µm ( = 9.5 × 4 µm, n = 20), hyaline, ellipsoidal, 0–1-septate, guttulate, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 48 h. Germ tubes produced from the basal cell. Colonies reaching 2.3 mm diam at room temperature in normal day and night light after 1 month. Colonies from above, white to pale gray, dense, slightly circular to irregular in shape, slightly raised to umbonate, rough at the surface, with wrinkled folded aspect, undulate at the edge, slightly radiating, velvety to felted; reverse, white at the margin, dark green to black in the middle, olive in the center, wrinkled folded; not producing pigmentation. Mycelium superficial to immersed in media, brown, composed of septate, branched, smooth hyphae. Sporulation on PDA after 2 weeks.
Material examined: China, Yunnan Province, Fuxian Lake (102°43′–102°59′E, 24°31′–24°51′N, 1,720 msl; pH of water = 8.593333, temperature = 23°C, dissolved oxygen = 7.39 mg/L, purification level 1), on decaying wood submerged in a freshwater lake, 14 August 2022, L. Li, LILU-272 (KUN-HKAS 132016, holotype), ex-type living culture KUNCC23-13328; ibid., Yang Zonghai Lake (102°5′–103°02′E, 24°51′–24°58′N, 1,770 msl; pH of water = 8.92, temperature = 26°C, dissolved oxygen = 7.6 mg/L, purification level 3), 12 July 2022, L. Li, LILU-136 (KUN-HKAS 132017), living culture KUNCC23-13682.
Notes: Based on the nucleotide BLAST search, Pl. yunnanensis (KUNCC23-13328) is similar to Pl. dimorphospora (KUMCC 20-0185) with 96.21% similarity of the ITS, 98.88% similarity of LSU, 99.44% similarity of SSU, and 92.29% similarity of RPB2. The multigene phylogenetic analyses confirmed the phylogenetic affinity of the new species (KUNCC23-13328 and KUNCC23-13682) as a member of Pleurotheciella by nested with Pl. dimorphospora and Pl. saprophytica forming a well-resolved subclade among other species within the Pleurotheciella (Figure 3). The nucleotide pairwise comparison between Pl. yunnanensis (KUNCC23-13328) and Pl. saprophytica (MFLU 17-0915) revealed 16/464 bp (3.4%) of ITS, 7/804 bp of LSU (0.9%), 0/813 bp of SSU (0%), and 40/781 bp of RPB2 (5%) differences. Pl. yunnanensis shares similar morphology with Pl. dimorphospora in having two types of conidial morphology in vitro. However, they differ by phragmoconidia comprising 2–3-septate and leaf clover-like, brown to dark brown dictyoconidia. In contrast, Pl. dimorphospora mainly produced multi-septate, sectored, ellipsoidal to subglobose dictyoconidia (Boonmee et al., 2021). Furthermore, Pl. dimorphospora was found in terrestrial habitats, whereas Pl. yunnanensis has been found in aquatic habitats. Pl. yunnanensis differs from Pl. saprophytica by the lack of cylindrical or apically tapering sympodial, conspicuously denticulate conidiogenous cells and subcylindrical to obovoidal, obtuse conidia with a rounded apex and tapering base (Luo et al., 2018). However, both species were isolated from submerged wood in a freshwater environment in Yunnan Province, China. Interestingly, most Pleurotheciella have been found in aquatic environments, except for Pl. dimorphospora. Details on habitats, hosts, and distributions of Pleurotheciella are provided in Table 4. Furthermore, Pl. yunnanensis fits well with the generic concept of Pleurotheciella by sharing similar morphological characteristics as polyblastic, denticulate conidiogenesis, subhyaline conidiophores, and hyaline, ellipsoidal to ellipsoidal-fusiform, and aseptate to multi-septate conidia. Detailed morphological characteristics of all known Pleurotheciella species are also provided in Table 5.
4 Discussion
Freshwater fungi exhibit remarkable ecological diversity, playing essential roles in aquatic ecosystems. They act as decomposers, breaking down organic matter like wood and leaf litter, which contributes to nutrient cycling and energy flow. These fungi also form symbiotic relationships with aquatic plants and algae, aiding in nutrient uptake and survival. Some freshwater fungi are pathogens, impacting aquatic plants and animals, while others help control populations within ecosystems. Adapted to both flowing (lotic) and still (lentic) water, they show a variety of morphological and physiological adaptations that allow them to thrive in diverse aquatic environments. Freshwater fungi are also important as bioindicators, providing insights into water quality and ecosystem health. Overall, they are key contributors to ecosystem function, biodiversity, and the balance of freshwater habitats (Gessner et al., 2007; Krauss et al., 2011; Jones et al., 2014; Kuehn, 2016).
Sordariomycetes is the largest class of lignicolous freshwater fungi in Ascomycota, containing approximately 823 species and 298 genera (Luo et al., 2019; Calabon et al., 2022; Shen et al., 2022). In this study, three new freshwater species belonging to the subclasses Hypocreomycetidae and Savoryellomycetidae (Sordariomycetes) are introduced from Hunan and Yunnan provinces, China. The three new species, C. yunnanensis, Pa. hunanensis, and Pl. yunnanensis, are found inhabiting the freshwater habitats, both lotic and lentic. C. yunnanensis was collected in a lake (lentic) and a freshwater stream (lotic), Pa. hunanensis in a freshwater stream (lotic), and Pl. yunnanensis from a lake (lentic). These three species were isolated from submerged decaying branch and wood, indicating their roles as decomposers degrading organic matter in nutrient cycling. It is notable that species of Chaetopsina, Parafuscosporella, and Pleurotheciella are commonly known from aquatic habitats. However, many species such as Chaetopsina guyanensis, C. saulensis, and Pl. dimorphospora have also been found in terrestrial habitats (Lechat and Fournier, 2019; Boonmee et al., 2021). Hence, we speculate the species in genera Chaetopsina and Pleurotheciella tend to inhabit a wide range of environments, including submerged freshwater or wet terrestrial environments. However, all species of Parafuscosporella have so far been found in aquatic environments.
Similar to Tsui et al. (2016), the three new species—C. yunnanensis, Pa. hunanensis, and Pl. yunnanensis—may belong to the group of aquatic–terrestrial hyphomycetes (mitosporic ascomycetes) that are initially found growing on decaying plant material and capable of sporulation underwater. The freshwater fungi in this group such as Canalisporium, Dactylaria, Dictyochaeta, and Sporoschisma, were typically distinguished based on the features of their conidia, conidiophores, and the process of conidiogenesis (Tsui et al., 2016). Since C. yunnanensis, Pa. hunanensis, and Pl. yunnanensis exhibit characteristics similar to those described in aquatic–terrestrial hyphomycetes, they should fit well into this group. Their ability to grow in aquatic environments, along with their possible role in decaying plant material (e.g., branches, leaves, and wood), aligns with the ecological characteristics described for this group of fungi. However, further studies would be required to confirm their precise classification within the group, especially considering conidial morphology and conidiogeneses, as noted by Tsui et al. (2016).
Some freshwater fungi exhibit morphological and ecological specificity (e.g., aquatic spore morphology, attachment structures, growth form adaptation, and spore release strategies), which may display different morphological traits depending on their growth environment, and these traits are often linked to factors such as habitat type, nutrient sources, and interactions with other organisms (Suberkropp, 2011; Naranjo-Ortiz and Gabaldón, 2019). In this study, the three new species, C. yunnanensis, Pa. hunanensis, and Pl. yunnanensis, do not exhibit specific morphological and ecological adaptations to lotic and lentic environments, although detailed studies on their precise adaptations in these habitats are limited. The special morphological characteristics may inconspicuously occur in these three new species, which allow them to adapt to an aquatic environment. However, these three new three species were explored from decaying plant material submerged in freshwater lakes and streams. For instance, most species in Parafuscosporella possess a jelly-like cover on sporodochia or hyaline appendages at the base of conidia. Pa. hunanensis may produce a jelly-like cover; however, this characteristic may disappear during the shifting from the collection site to the laboratory, or this species may generally occur as saprobe in terrestrial and produce some ecological functions that allow its adaptation during its submerged in aquatic environments.
Nonetheless, we can infer potential adaptations based on their general ecological characteristics and roles in aquatic ecosystems. C. yunnanensis, Pa. hunanensis, and Pl. yunnanensis may exhibit morphological traits suited to their respective aquatic environments. In lotic systems, where water flow is continuous, these species may develop stronger attachment structures on their spores or hyphae, which help them remain anchored to substrates such as decaying wood or aquatic plants. In contrast, species in lentic environments, where the water is still, may not require such robust attachment features but may have adaptations for better spore dispersal in stagnant water. Additionally, species in flowing water (lotic systems) may possess conidia with characteristics that allow them to float or travel further, optimizing dispersal in currents. Conversely, in still water (lentic systems), these fungi may produce denser spores or develop morphology that enhances their survival and growth on submerged wood and organic matter, as they would be more reliant on localized nutrient sources (Kuehn, 2016).
The three new species may play crucial roles in nutrient cycling, particularly in wood degradation. In both lotic and lentic environments, they contribute to the breakdown of organic matter such as decomposing wood, which is an important source of nutrients for other organisms in the ecosystem. Their activity in wood degradation releases essential nutrients like nitrogen, carbon, and phosphorus back into the ecosystem, influencing nutrient cycling and supporting a wide range of aquatic life (Gulis et al., 2006, 2008). Moreover, the three new species may interact with other aquatic organisms, including bacteria and invertebrates, through their decomposition activities. The release of small organic molecules during wood degradation can attract microbial communities, facilitating a rich nutrient web. These interactions are crucial for maintaining the health and balance of aquatic ecosystems. The ecological relevance of these fungi in nutrient cycling and wood degradation in lotic and lentic systems is profound. In lotic environments, where constant water movement aids the dispersal of spores, the fungi can contribute to nutrient cycling over large areas, potentially impacting the entire aquatic food web. In lentic environments, they may play a more localized role but still significantly contribute to the recycling of nutrients trapped in stagnant water and organic matter. While studies on these species-specific interactions with aquatic organisms are limited, it is likely that their roles in wood degradation and nutrient cycling are shared across many freshwater fungi.
The species of Chaetopsina are distributed in different climate zones in various countries such as China (C. fulva as C. beijingensis and C. hongkongensis) (Goh and Hyde, 1997; Crous et al., 2014) Egypt (C. aquatica) (Bakhit and Abdel-Aziz, 2021), France (C. guyanensis, C. pnagiana, and C. saulensis) (Lechat and Fournier, 2019, 2020), Thailand (C. penicillata) (Bao et al., 2023), and South Africa (C. gautengina) (Crous et al., 2020). This genus has been reported from both freshwater and terrestrial habitats. For instance, C. penicillata was introduced by Samuels (1985) from terrestrial habitats in Ecuador, Jamaica, and New Zealand. Bao et al. (2023) reported C. penicillata from freshwater habitats in China. Through our study on C. yunnanensis, we noticed the Chaetopsina species can be segregated by their morphological characteristics, such as ascomata dimensions and perithecial wall anatomy, size, shape, and ornamentation of ascospores and conidia of the asexual morph. Furthermore, the morphological traits of our isolates closely resemble those attributed to Chaetopsina. These include red-pigmented erect conidiophores producing unicellular, cylindrical to cylindro-fusoid or ellipsoidal conidia. Phylogenetic analyses of a combined ITS and LSU sequence data indicated that our isolates C. yunnanensis (KUNCC23-12940 and KUNCC23-13014) are grouped with the species of Chaetopsina, forming a distinct lineage that is basal to C. pinicola (CPC 21819). Therefore, considering both the morphological comparisons and the phylogenetic analyses, we proposed the establishment of a new species, C. yunnanensis, in this study.
Meanwhile, we noticed that C. beijingensis has not been formally synonymized under C. fulva so far. While describing a new aquatic species, C. aquatica, from River Nile in Egypt, Bakhit and Abdel-Aziz (2021), quoting Lechat and Fournier (2019), referred that C. beijingensis is a synonym of C. fulva, the type species of the genus. Lechat and Fournier (2019) described two new species of Chaetopsina from Saül (French Guiana) and noted that C. beijingensis is similar to C. fulva, both morphologically and phylogenetically but did not taxonomically synonymize these taxa. In the present study, the phylogenetic analyses coupled with the comparison of their polymorphism demonstrated that C. beijingensis is conspecific with C. fulva, concurring with previous studies (Lechat and Fournier, 2019; Bakhit and Abdel-Aziz, 2021). Hence, C. beijingensis is formally synonymized under C. fulva herein.
Only ITS and LSU sequence data are available for most species of Chaetopsina, of which the ITS gene is currently remarkable as the sufficient phylogenetic marker in delineating the interspecific status of Chaetopsina (Lechat and Fournier, 2019, 2020; Bakhit and Abdel-Aziz, 2021). However, many sufficient phylogenetic markers derived from the protein-coding genes were recommended for resolving a better phylogenetic resolution of the family Nectriaceae where Chaetopsina does belong as well as the class Sordariomycetes [e.g., the ATP citrate lyase (ACL1), α-actin (ACT), β-tubulin (TUB2), calmodulin (CMDA), histone H3 (HIS3), the RNA polymerase II largest subunit (RPB1), the RNA polymerase II second largest subunit (RPB2), and translation elongation factor 1 alpha (TEF1-α)] (Lombard et al., 2015; Perera et al., 2023). Of these, sequences of the protein-coding genes are available for few Chaetopsina species such as C. acutispora, C. fulva (type species), and C. penicillata (Lombard et al., 2015). The limitation on molecular data of Chaetopsina can cause taxonomic ambiguities. We, therefore, recommend utilizing multigene phylogeny for resolving taxonomic ambiguities and also providing a better taxonomic resolution on Chaetopsina, expanding to Nectriaceae as well as the class Sordariomycetes, of which the sufficient phylogenetic markers based on the protein-coding genes of Chaetopsina should be derived.
In our studies on Pa. hunanensis, we noticed that the species of Parafuscosporella are morphologically indistinguishable. Hence, it was found necessary to understand their taxonomic boundaries using multigene phylogenetic markers. Phylogenetic affinities of species in Parafuscosporella have been delineated by ITS, LSU, and SSU phylogenetic markers, of which ITS region is currently recommended as a measurable gene for resolving phylogenetic relationships among species in Parafuscosporella (Boonmee et al., 2021; Boonyuen et al., 2021; Li et al., 2023). Nevertheless, the RPB2 gene has always been utilized for delineating taxa in Fuscosporellaceae (Fuscosporellales) where Parafuscosporella accommodated, with other related orders such as Conioscyphales, Pleurotheciales, and Savoryellales in Savoryellomycetidae (Boonyuen et al., 2016; Yang et al., 2016, 2020; Wang et al., 2023). Unfortunately, the RPB2 gene is only available for Pa. garethii, Pa. pyriformis, and Pa. xishuangbannaensis in the total of the known species. This may cause the insufficient phylogenetic resolution on Parafuscosporella correlated with the closely related genera in the family Fuscosporellaceae, extending to the correlation with taxa among the closely related orders in Savoryellomycetidae. Thus, the RPB2 gene is recommended in further clarifying species levels of Parafuscosporella corresponding with other closely related genera in Fuscosporellaceae.
The confusion between Parafuscosporella and Fuscosporella was also discussed in a previous study (Yang et al., 2016). Fuscosporella is morphologically similar to Parafuscosporella but differs in the structure of conidia, which are produced in culture. Fuscosporella produces multi-celled, filamentous to helicoid conidia, while Parafuscosporella produces globose to obpyriform, uni-septate conidia in culture. The same situation exists for Conioscypha in that the species of Conioscypha are largely indistinguishable in morphology. Li et al. (2024) proposed to use the potential of phylogenetic markers to clarify their phylogenetic relationships. The single gene trees of Conioscypha (ITS, LSU, SSU, and RPB2) and combined sequence datasets (LSU-ITS, LSU-ITS-SSU, and LSU-ITS-RPB2) were previously obtained to compare the reliable phylogenetic markers. The results of these prior analyses demonstrated that analysis of the RPB2 gene could provide a better phylogenetic resolution of Conioscypha. Therefore, the RPB2 gene is recommended as a genetic marker for resolving phylogenetic relationships among species in Conioscypha (Li et al., 2024). Simultaneously, the protein-coding genes such as TEF1-α and RPB2 have been recommended for clarifying the taxonomic ambiguities of various genera in Sordariomycetes due to their high utility in resolving phylogenetic relationships at multiple taxonomic levels. TEF1-α and RPB2 are relatively conserved within Sordariomycetes but also exhibit enough variability to distinguish closely related species or genera. Their nucleotide sequences evolve at rates suitable for both deep and shallow phylogenetic analyses, making them useful for resolving both inter- and intra-generic relationships (Matheny et al., 2007; Lücking et al., 2020, 2021). Therefore, it is concluded that the protein-coding genes, especially the RPB2 gene region, are necessary for resolving the taxonomic ambiguities of some genera in Savoryellomycetidae as well as Sordariomycetes, and hence, TEF1-α and RPB2 should be derived for novel taxa in further study.
It is worth noting that Goh et al. (2023) synonymized Parafuscosporella under Vanakripa due to the morphological features of V. gigaspora, the type of Vanakripa, resembling Parafuscosporella. Based on the principle of nomenclatural priority, all nine Parafuscosporella species were transferred to Vanakripa (Goh et al., 2023). However, V. gigaspora as the type species of Vanakripa lacks molecular data to confirm their phylogenetic placement. Hence, this leads the taxonomic confusion on the phylogenetic placement of Vanakripa. Vanakripa was introduced by Bhat and Kendrick (1993), with V. gigaspora as the type species. The genus was considered as genus incertae sedis in Pezizomycotina (Wijayawardene et al., 2021), while a key to species of the genus was provided by Arias et al. (2008). Vanakripa has a special clavate to vermiform, hyaline, separating cells attached to the conidia (Bhat and Kendrick, 1993). The genus currently accommodated a few species lacking genetic sequences. In Goh et al. (2023), phylogenetic analyses demonstrated that Vanakripa formed separated clades relating to Conioscypha and Parafuscosporella. Two new species that were identified as V. oblonga and V. taiwanensis by Goh et al. (2023) formed a clade with Parafuscosporella in Fuscosporellaceae, whereas V. chiangmaiensis and V. minutiellipsoidea are related to Conioscypha in Conioscyphaceae. Goh et al. (2023) re-circumscribed morphological features of Vanakripa compared to Parafuscosporella. Based on conidial ontogeny resemblance, Goh et al. (2023) demonstrated that Parafuscosporella is congeneric with Vanakripa. Therefore, Goh et al. (2023) transferred all Parafuscosporella to Vanakripa without providing phylogenetic evidence from type studies.
In accordance with the morphological trait, many genera in Fuscosporellaceae and Conioscyphaceae are morphologically somewhat similar; however, these genera can be distinguished based on multigene phylogeny of sufficient genes. Unfortunately, Vanakripa formed polyphyletic clades in Conioscyphales and Fuscosporellales (Goh et al. (2023). Considering the morphology of Vanakripa species, Goh et al. (2023) excluded V. chiangmaiensis and V. minutiellipsoidea (clade in Conioscyphales) and some other described species from Vanakripa (viz., Vanakripa chinensis, Vanakripa ellipsoidea, Vanakripa fasciata, Vanakripa inflata, Vanakripa menglaensis, Vanakripa parva, and Vanakripa rhizophorae) due to their conidial morphology being different from V. gigaspora, the type species of Vanakripa, in producing ellipsoidal or broadly obovoid, one-celled conidia. In contrast, V. oblonga, V. taiwanensis (clade as basal of Parafuscosporella in Fuscosporellales in the present study), and other synonymized Parafuscosporella species were treated as Vanakripa sensu stricto due to the conidial morphology resemblance in producing septate, apiosporous, versicolored conidia, with or without the presence of a hyaline appendage at the conidial base (Goh et al., 2023). With this point of view, it is reasonable to segregate Vanakripa into two morphological groups. However, molecular data of V. gigaspora, the type species of Vanakripa, have not yet been derived, and/or the epitype has not yet been designated, leading to the phylogenetic uncertainty for Vanakripa. Hence, the treatment of Vanakripa in different orders may cause taxonomic confusion in Vanakripa, corresponding with Parafuscosporella. In the present study, we, therefore, tentatively place Parafuscosporella as a distinct genus from Vanakripa to avoid taxonomic confusion until the type strain of V. gigaspora is derived from molecular data and/or the epitype is designated for clarifying its phylogenetic affinity. We also believe that caution of synonymization should be maintained at this stage. However, if future studies could confirm that Parafuscosporella and Vanakripa are congeneric based on morphological and phylogenetic evidence, this would greatly advance the taxonomic discussion of the Fuscosporellaceae. We recognize that any changes in classification should take into account the importance of nomenclatural stability, ensuring that the naming and classification system of the family remains consistent and carefully considered within the scientific community.
It is notable that 17 of the total 18 species of Pleurotheciella were collected from freshwater habitats, and only Pl. dimorphospora is from the terrestrial environment. Furthermore, most species have been found in China and Thailand, as well as in Canada, Egypt, France, and Spain (Table 4). In addition, Pl. dimorphospora is dimorphic (with two types of conidial morphology), which can be well distinguished from other species in Pleurotheciella (Boonmee et al., 2021). Initially, the original authors felt that the two types of conidial morphology in vitro were caused by contamination in the culture. However, through single spore isolation and molecular work on both types of conidia, they confirmed that these two types of conidia are of the same species (Boonmee et al., 2021). It is interesting that our study also meets the same situation with the new species, Pl. yunnanensis, showcasing two-type conidial morphology in vitro.
Although in recent times freshwater fungi have been continuously discovered, compared to soil ecosystems, these fungi still lacked sufficient spatial and temporal resolution, especially from those environments falling in various latitudinal zones, ecosystems (such as the water column and sediments), snowclad mountains, and extreme environments like deep-sea vents (Grossart et al., 2019). Yunnan has emerged as a hotspot for lignicolous freshwater fungal research since 2015, resulting in the discovery of a number of new species and new records in some extremely varied genera such as Acrogenospora, Dictyosporium, Distoseptispora, Pleurotheciella, Sporidesmium, and Sporoschisma (Bao et al., 2020; Wan et al., 2021; Shen et al., 2022). In contrast, the lignicolous freshwater fungi in Hunan Province still remained understudied. Hyde et al. (2016) discussed the impacts of riparian vegetation, water pollution, sampling methods, and global warming on the diversity of lignicolous freshwater fungi. However, the diversity, quantitative abundance, and ecological functions of freshwater fungi, particularly their interactions with other microorganisms, remain largely speculative, unexplored, and overlooked in the current understanding of aquatic ecology and biogeochemistry (Haraldsson et al., 2018; Grossart et al., 2019). The three new species explored in the present study from freshwater lakes and streams in Hunan and Yunnan provinces will add to the species number of lignicolous freshwater fungi in China and upgrade the global species numbers of freshwater fungi.
Additionally, genomic studies have revealed that freshwater fungi contain a vast diversity of secondary metabolite pathways (El-Elimat et al., 2021). However, the genes and gene clusters involved in these metabolic processes, as well as the secondary metabolite products, remain mostly unknown (Chiang et al., 2009; El-Elimat et al., 2021). Over the last 30 years, several freshwater fungi have been subjected to chemical investigations, resulting in the isolation of 283 secondary metabolites of wide chemical diversity and a broad range of biological activities (Wang et al., 2008; Hernández-Carlos and Gamboa-Angulo, 2011; Canto et al., 2022; El-Elimat et al., 2021). El-Elimat et al. (2021) reviewed the secondary metabolites of freshwater fungi and summarized those compounds as mainly belonging to alkaloids, terpenes, polyketides, phenylpropanoids and peptides, and some unclassified secondary metabolites. Dong et al. (2009) isolated two novel naphthalene-containing compounds, colelomycerones A and B, and three known metabolites from the culture broth of an unidentified freshwater fungus YMF 1.01029, and those metabolites showed noticeable antifungal and antibacterial activities. Prabhu et al. (2018) found Greensporone C, a secondary metabolite from freshwater fungi inducing mitochondrial-mediated apoptotic cell death in leukemic cell lines. de Souza et al. (2023) investigated communities of 154 culturable freshwater fungi from Antarctic lakes and the capabilities of all cultured fungi to produce various extracellular enzymes at low temperatures and found that the most widely produced enzymes were proteases and pectinases. These active enzymes produced by freshwater fungi have various applications in biotechnological processes in industries including textile, pharmaceutical, food, detergent, and paper, as well as in bioremediation of environmental pollutants (Raghukumar, 2008; Fathima et al., 2022; de Souza et al., 2023). Unfortunately, the study on secondary metabolites of Chaetopsina, Parafuscosporella, and Pleurotheciella is unexplored. Therefore, further studies on secondary metabolites of freshwater fungi in Chaetopsina, Parafuscosporella, and Pleurotheciella will be most rewarding.
The limitations of the current study are essential for providing a balanced perspective and guiding future research. One of the key challenges encountered in this study was the difficulty in morphological differentiation of some freshwater fungal species, particularly given the high morphological plasticity within certain genera. Additionally, our sampling efforts were constrained by limited geographic coverage and sampling time, which may have affected the comprehensiveness of the data. Furthermore, genetic sequencing efforts were limited by the availability of high-quality DNA samples from all species, which constrained the depth of phylogenetic analyses. These limitations underscore the need for more extensive and targeted genetic studies, such as the use of multigene sequencing or the application of environmental DNA (eDNA) sampling techniques, which could help capture a broader diversity of fungi in various aquatic ecosystems. Future research should prioritize these approaches to better understand the full scope of freshwater fungal biodiversity and address the unresolved taxonomic and ecological questions raised in this study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/ ITS: OQ860234, OQ860233, OR230704, PP744554, OR234682, PP095384; LSU: PP151255, PP151256, PP744555, PP744556, PP095383, PP095381; SSU: PP744557, PP744558, PP095382, PP095385; RPB2: PP131261, PP131262. The final alignment and phylogenetic tree were registered in TreeBASE under the submission IDs: 31916 (C. yunnanensis), 31910 (Pa. hunanensis) and 31135 (Pl. yunnanensis) (http://www.treebase.org/ accessed on 25 December 2024).
Author contributions
LL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. DB: Investigation, Methodology, Writing – review & editing. H-BJ: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. J-FL: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. TD: Funding acquisition, Writing – review & editing. FS: Project administration, Writing – review & editing. SH: Project administration, Writing – review & editing. RC: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. RP: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (Grant No. 2019QZKK0503) and partially supported by Chiang Mai University (Fundamental Fund 2023 and grant FF2024) the financial publication of this article is supported by Yunnan Revitalization Talent Support Program “Young Talent” Project (grant no. YNWR-QNBJ-2020-120).
Acknowledgments
The authors would like to thank Professor Qi Zhao for his generosity in providing the experimental platform and all the costs of the experiment. RP sincerely acknowledges Introducing Talents Start-up Fund of Kunming Institute of Botany, Chinese Academy of Sciences, Yunnan Revitalization Talent Support Program “Young Talent” Project (grant no. YNWR-QNBJ-2020-120), Yunnan Revitalization Talent Support Program: High-end Foreign Expert Project), Independent research of Department of Economic Plants and Biotechnology, Yunnan Key Laboratory for Wild Plant Resources, Kunming Institute of Botany, Chinese Academy of Sciences (grant no. Y537731261), and Yunnan Department of Sciences and Technology of China (grant nos. 202302AE090023 and 202303AP140001). H-BJ appreciates the Postdoctoral Directional Training Foundation of Yunnan Province (grant no. E33O38E261) under the Yunnan Provincial Department of Human Resources and Social Security, Yunnan, China, and the 1st batch of the National Postdoctoral Overseas Talent Introduction Program, China. J-FL appreciates the Postdoctoral Directional Training Foundation of Yunnan Province (grant no. E33O386261) under the Yunnan Provincial Department of Human Resources and Social Security, Yunnan, China. DB and TD gratefully acknowledge the financial support provided under the Distinguished Scientist Fellowship Programme (DSFP), at King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Aziz, F. A., Bahkali, A. H., Elgorban, A. M., Abdel-Wahab, M. A. (2020). Pleurotheciella nilotica sp. nov. (Pleurotheciales, Ascomycota) from freshwater habitats in Egypt. Nova Hedwigia 110, 91–98. doi: 10.1127/nova_hedwigia/2020/0570
Arias, R. M. A., Abarca, G. H., Ruíz, R. F. C., Hernández, C. I. B. J. M. (2008). Two new species of Polyschema and Vanakripa and other microfungi recorded from mangrove in Veracruz, Mexico. Mycotaxon 106, 29–40.
Arias, R. M. A., Heredia, G., Castañeda-Ruiz, R. F. (2015). Adiciones al conocimiento de la diversidad de los hongos conidiales saprobios del bosque mesófilo de montaña del estado de Veracruz IV. Acta Bot. Mex 113, 87–101. doi: 10.21829/abm113.2015.1097
Bakhit, M. S., Abdel-Aziz, A. E. (2021). Chaetopsina aquatica sp. nov. (Hypocreales, Nectriaceae) from the River Nile, Egypt. Phytotaxa 511, 289–295. doi: 10.11646/phytotaxa.511.3.8
Bao, D. F., Hyde, K. D., Maharachchikumbura, S. S. N., Rekhani, H. P., Vinodhini, T., Sinang, H., et al. (2023). Taxonomy, phylogeny and evolution of freshwater Hypocreomycetidae (Sordariomycetes). Fungal Divers. 121, 1–94. doi: 10.1007/s13225-023-00521-8
Bao, D. F., McKenzie, E. H., Bhat, D. J., Hyde, K. D., Luo, Z. L., Shen, H. W., et al. (2020). Acrogenospora (Acrogenosporaceae, Minutisphaerales) appears to be a very diverse genus. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01606
Bärlocher, F., Stewart, M., Ryder, D. S. (2011). Analyzing aquatic fungal communities in Australia: impacts of sample incubation and geographic distance of streams. Czech Mycology 63, 113–132. doi: 10.33585/cmy.63202
Bhat, D. J., Kendrick, B. (1993). Twenty-five new conidial fungi from the Western Ghats and the Andaman Islands (India). Mycotaxon 49, 19–90.
Boonmee, S., Wanasinghe, D. N., Calabon, M. S., Huanraluek, N., Chandrasiri, S. K. U., Jones, E. B. G., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3
Boonyuen, N., Chuaseeharonnachai, C., Nuankaew, S., Kwantong, P., Pornputtapong, N., Suwannarach, N., et al. (2021). Novelties in Fuscosporellaceae (Fuscosporellales): Two new Parafuscosporella from Thailand revealed by morphology and phylogenetic analyses. Diversity 13, 517. doi: 10.3390/d13110517
Boonyuen, N., Chuaseeharonnachai, C., Suetrong, S., Sujinda, S., Somrithipol, S. (2016). Parafuscosporella garethii sp. nov. (Fuscosporellales) from a rivulet in a community-based northern forest, in Thailand. Mycosphere 7, 1265–1272. doi: 10.5943/mycosphere/7/9/2
Bucher, V. V. C., Hyde, K. D., Pointing, S. B., Reddy, C. A. (2004). Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Divers. 15, 1–14. doi: 10.1007/s00248-003-0132-x
Calabon, M. S., Hyde, K. D., Jones, E. B. G., Bao, D. F., Bhunjun, C. S., Phukhamsakda, C., et al. (2023a). Freshwater fungal biology. Mycosphere 14, 195–413. doi: 10.5943/mycosphere/14/1/4
Calabon, M. S., Hyde, K. D., Jones, E. B. G., Chandrasiri, S., Dong, W., Fryar, S. C., et al. (2020). www.freshwaterfungi.org, an online platform for the taxonomic classification of freshwater fungi. AJOM 3, 419–445. doi: 10.5943/ajom/3/1/14
Calabon, M. S., Hyde, K. D., Jones, E. B. G., Luo, Z.-L., Dong, W., Hurdeal, V. G., et al. (2022). Freshwater fungal numbers. Fungal Divers. 114, 3–235. doi: 10.1007/s13225-022-00503-2
Calabon, M. S., Jones, E. B. G., Boonmee, S., Doilom, M., Lumyong, S., Hyde, K. D. (2021). Five novel freshwater ascomycetes indicate high undiscovered diversity in lotic habitats in Thailand. J. Fungi 7, 1–27. doi: 10.3390/jof7020117
Calabon, M. S., Jones, E. B. G., Boonmee, S., Li, W. J., Xiao, Y. P., Hyde, K. D. (2023b). Acrocalymmaceae (Pleosporales) from freshwater habitats in Thailand with the introduction of Acrocalymma bilobatum sp. nov. Stud. Fungi. 8, 11. doi: 10.48130/SIF-2023-0011
Canto, E. S. M., Segundo, W. O. P. F., Cortez, A. C. A., Barbosa, F. R., Monteiro, J. S., de Souza, J. V. B. (2022). “Chapter 15 Freshwater fungi in the Amazon as a potential source of antimicrobials,” in Freshwater Mycology: Perspectives of Fungal Dynamics in Freshwater Ecosystems (Elsevier, Amsterdam, Netherlands), 261–275. doi: 10.1016/B978-0-323-91232-7.00009-X
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chiang, Y. M., Lee, K. H., Sanchez, J. F., Keller, N. P., Wang, C. C. (2009). Unlocking fungal cryptic natural products. Nat. Prod. Commun. 4, 1505–1510. doi: 10.1177/1934578X0900401113
Crous, P. W., Shivas, R. G., Quaedvlieg, W., van der Bank, M., Zhang, Y., Summerell, B. A., et al. (2014). Fungal Planet description sheets: 214–280. Persoonia 23, 184–306. doi: 10.3767/003158514X682395
Crous, P. W., Wingfield, M. J., Guarro, J., Cheewangkoon, R., van der Bank, M., Swart, W. J., et al. (2013). Fungal Planet description sheets: 154–213. Persoonia 31, 188–296. doi: 10.3767/003158513X675925
Crous, P. W., Wingfield, M. J., Schumacher, R. K., Akulov, A., Bulgakov, T. S., Carnegie, A. J., et al. (2020). New and interesting fungi. 3. Fungal Syst. Evol. 6, 157–231. doi: 10.3114/fuse.2020.06.09
de Souza, L. M. D., Ogaki, M. B., Teixeira, E. A. A., De Menezes, G. C. A., Convey, P., Rosa, C. A., et al. (2023). Communities of culturable freshwater fungi present in Antarctic lakes and detection of their low-temperature-active enzymes. Braz. J. Microbiol. 54, 1923–1933. doi: 10.1007/s42770-022-00834-x
de Wildeman, E. (1895). Notes mycologiques. XV. Chytridinées. Annales la Societé Belge Microscopie 19, 88–117.
Dong, J. Y., Song, H. C., Li, J. H., Wang, C. R., Sun, R., Tang, Y. S., et al. (2009). Two unusual naphthalene-containing compounds from a freshwater fungus YMF 1.01029. Chem. Biodivers. 6, 569–577. doi: 10.1002/cbdv.200800028
Dong, W., Wang, B., Hyde, K. D., McKenzie, E. H. C., Raja, H. A., Tanaka, K., et al. (2020). Freshwater dothideomycetes. Fungal Divers. 105, 319–575. doi: 10.1007/s13225-020-00463-5
Duarte, S., Fernandes, I., Nogueira, M. J., Cássio, F., Pascoal, C. (2013). Temperature alters interspecific relationships among aquatic fungi. Fungal Ecol. 6, 187–191. doi: 10.1016/j.funeco.2013.02.001
El-Elimat, T., Raja, H. A., Figueroa, M., Al Sharie, A. H., Bunch, R. L., Oberlies, N. H. (2021). Freshwater fungi as a source of chemical diversity: a review. J. Nat. Prod. 84, 898–916. doi: 10.1021/acs.jnatprod.0c01340
Fathima, A., Arafath, Y., Sadasivam, V., Hassan, S., Kiran, G. S., Selvin, J. (2022). “Chapter 8-Biochemical and industrial potential of aquatic fungi,” in Freshwater mycology (Elsevier), p 135–149. doi: 10.1016/B978-0-323-91232-7.00011-8
Gessner, M. O., Gulis, V., Kuehn, K. A., Chauvet, E., Suberkropp, K. (2007). “). Fungal decomposers of Plant Litter in Aquatic Ecosystems,” in Environmental and Microbial Relationships, vol. 4 . Eds. Kubicek, C., Druzhinina, I. (Springer, Berlin, Heidelberg). doi: 10.1007/978-3-540-71840-6_17
Goh, T.-K., Hsieh, S.-Y., Kuo, C.-H. (2023). Synonymy of Parafuscosporella with Vanakripa and descriptions of two new species from Taiwan. Mycol Prog. 22, 61. doi: 10.1007/s11557-023-01907-3
Goh, T., Hyde, K. D. (1997). The generic distinction between Chaetopsina and Kionochaeta, with descriptions of two new species. Mycol. Res. 101, 1517–1523. doi: 10.1017/S0953756297004292
Grossart, H. P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., Rojas-Jimenez, K. (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 17, 339–354. doi: 10.1038/s41579-019-0175-8
Gulis, V., Kuehn, K., Suberkropp, K. (2006). “The role of fungi in carbon and nitrogen cycles in freshwater ecosystems,” in Fungi in biogeochemical cycles. Ed. Gadd, G. M. (Cambridge University Press), 1–15. doi: 10.1017/CBO9780511550522.018
Gulis, V., Suberkropp, K., Rosemond, A. D. (2008). Comparison of fungal activities on wood and leaf litter in unaltered and nutrient-enriched headwater streams. Appl. Environ. Microbiol. 74, 1094–1101. doi: 10.1128/AEM.01903-07
Haraldsson, M., Gerphagnon, M., Bazin, P., Colombet, J., Tecchio, S., Sime-Ngando, T., et al. (2018). Microbial parasites make cyanobacteria blooms less of a trophic dead end than commonly assumed. ISME J. 12, 1008–1020. doi: 10.1038/s41396-018-0045-9
He, W. M., Zhang, J. B., Zhai, Z. J., Tennakoon, D. S., Cui, C. Y., Zhou, J. P., et al. (2024). Four novel species of Pleurotheciaceae collected from freshwater habitats in Jiangxi Province, China. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1452499
Hernández-Carlos, B., Gamboa-Angulo, M. M. (2011). Metabolites from freshwater aquatic microalgae and fungi as potential natural pesticides. Phytochem. Rev. 10, 261–286. doi: 10.1007/s11101-010-9192-y
Hernández-Restrepo, M., Gené, J., Castañeda-Ruiz, R., Mena-Portales, J., Crous, P. W., Guarro, J. (2017). Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/j.simyco.2017.05.002
Ho, W. H., Hyde, K. D., Hodgkiss, I. J. (2002). Seasonality and sequential occurrence of fungi on wood submerged in Tai Po Kau Forest Stream, Hong Kong. Fungal Divers. 10, 21–43.
Hyde, K. D., Chaiwan, N., Norphanphoun, C., Boonmee, S., Camporesi, E., Chethana, K. W. T., et al. (2018). Mycosphere notes 169–224. Mycosphere 9, 271–430. doi: 10.5943/mycosphere/9/2/8
Hyde, K. D., Fryar, S., Tian, Q., Bahkali, A. H., Xu, J.-C. (2016). Lignicolous freshwater fungi along a north–south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. doi: 10.1016/j.funeco.2015.07.002
Index Fungorum (2024). Available online at: http://www.indexfungorum.org/names/names.asp (Accessed 5 January 2024).
Ingold, C. T. (1942). Aquatic hyphomycetes of decaying alder leaves. Trans. Br. Mycol. Soc 25, 339–417. doi: 10.1016/S0007-1536(42)80001-7
Ingold, C. T. (1955). Aquatic ascomycetes: Further species from the English Lake Distrit. Trans. Br. Mycol. Soc 58, 43–56. doi: 10.1016/S0007-1536(55)80026-5
Ingold, C. T. (1975). Guide to Aquatic Hyphomycetes. Scientific Publication (Freshwater Biological Association), 96. No.30.
Jones, E. B. G., Hyde, K. D., Pang, K. L. (2014). Freshwater Fungi and Fungi-like Organisms (Germany: De Gruyter).
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioniform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kirk, P. M., Sutton, B. C. (1985). A reassessment of the anamorph genus Chaetopsina (Hyphomycetes). Trans. Br. Mycol. Soc 85, 709–717. doi: 10.1016/S0007-1536(85)80267-9
Krauss, G. J., Sole, M., Krauss, G., Schlosser, D., Wesenberg, D., Baerlocher, F. (2011). Fungi in freshwaters: ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 35, 620–651. doi: 10.1111/j.1574-6976.2011.00266.x
Kuehn, K. A. (2016). Lentic and lotic habitats as templets for fungal communities: traits, adaptations, and their significance to litter decomposition within freshwater ecosystems. Fungal Ecology. 19, 135–154. doi: 10.1016/j.funeco.2015.09.009
Lechat, C., Fournier, J. (2019). Two new species of Chaetopsina (Nectriaceae) from Saül (French Guiana). Ascomycete.org 11, 127–134. doi: 10.25664/art-0265
Lechat, C., Fournier, J. (2020). Chaetopsina pnagiana (Nectriaceae, Hypocreales), a new holomorphic species from Saül (French Guiana). Ascomycete.org 12, 1–5. doi: 10.25664/ART-0289
Li, L., Du, H. Z., Thiyagaraja, V., Bhat, D. J., Phookamsak, R., Cheewangkoon, R. (2024). “Two novel freshwater hyphomycetes, in Acrogenospora (Minutisphaerales, Dothideomycetes) and Conioscypha (Conioscyphales, Sordariomycetes) from Southwestern China. MycoKeys, vol. 101, 249–273. doi: 10.3897/mycokeys.101.115209
Li, L., Xu, R. J., Luo, Z. L., Zhao, Q., Cheewangkoon, R. (2023). Parafuscosporella lignicola sp. nov. (Fuscosporellaceae), from a freshwater habitat in northern Thailand. Phytotaxa 597, 141–152. doi: 10.11646/phytotaxa.597.2.4
Liu, Y. J., Whelen, S., Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lombard, L., van der Merwe, N. A., Groenewald, J. Z., Crous, P. W. (2015). Generic concepts in nectriaceae. Stud. Mycol. 80, 189–245. doi: 10.1016/j.simyco.2014.12.002
Lücking, R., Aime, M. C., Robbertse, B., Miller, A. N., Aoki, T., Ariyawansa, H. A., et al. (2021). Fungal taxonomy and sequence-based nomenclature. Nat. Microbiol. . 6, 540–548. doi: 10.1038/s41564-021-00888-x
Lücking, R., Aime, M. C., Robbertse, B., Miller, A. N., Ariyawansa, H. A., Aoki, T., et al. (2020). Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11, 14. doi: 10.1186/s43008-020-00033-z
Luo, Z.-L., Hyde, K. D., Liu, J. K., Bhat, D. J., Bao, D. F., Li, W. L., et al. (2018). Lignicolous freshwater fungi from China II: novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9, 444–461. doi: 10.5943/mycosphere/9/3/2
Luo, Z.-L., Hyde, K. D., Liu, J. K., Maharachchikumbura, S. S. N., Jeewon, R., Bao, D. F., et al. (2019). Freshwater sordariomycetes. Fungal Divers. 99, 451–660. doi: 10.1007/s13225-019-00438-1
Luo, J., Yin, J., Cai, L., Zhang, K., Hyde, K. D. (2004). Freshwater fungi in Lake Dianchi, a heavily polluted lake in Yunnan, China. Fungal Divers. 16, 93–112. doi: 10.1023/B:MYCO.0000041833.41085.6f
Luo, J., Zhuang, W.-Y. (2010). Chaetopsinectria (Nectriaceae, Hypocreales), a new genus with Chaetopsina anamorphs. Mycologia 102, 976–984. doi: 10.3852/09-263
Matheny, P. B., Wang, Z., Binder, M., Curtis, J. M., Lim, Y. W., Nilsson, R. H., et al. (2007). Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 43, 430–451. doi: 10.1016/j.ympev.2006.08.024
McNeill, J., Barrie, F. R., Buck, W. R., Demoulin, V., Greuter., W., Hawksworth, D. L., et al. (Eds.) (2012). International Code of Nomenclature for algae, fungi, and plants (Melbourne Code), Adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011, (electronic ed.).
Mehboob, A., Ganie, A., Dar, A. R., Soni, K. K. (2021). Aquatic fungal diversity in two freshwater ecosystems of Madhya Pradesh, India. Stud. Fungi. 6, 116–137. doi: 10.5943/sif/6/1/6
Miller, M. A., Schwartz, T., Pickett, B. E., He, S., Klem, E. B., Scheuermann, R. H., et al. (2015). Restful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinf. 11, 43–48. doi: 10.4137/EBO.S21501
Naranjo-Ortiz, M. A., Gabaldón, T. (2019). Fungal evolution: Major ecological adaptations and evolutionary transitions. Biol. Rev. Cambridge Philos. Soc. 94, 1443–1476. doi: 10.1111/brv.12510
Nikolcheva, L. G., Bärlocher, F. (2002). Phylogeny of Tetracladium based on 18S rDNA. Czech Mycology 53, 285–295. doi: 10.33585/cmy.53404
Nylander, J. (2008). MrModeltest2 v. 2.3 (Program for selecting DNA substitution models using PAUP*) (Uppsala: Evolutionary Biology Centre).
Okada, G., Takematsu, A., Takamura, Y. J. M. (1997). Phylogenetic relationships of the hyphomycete genera Chaetopsina and Kionochaeta based on 18S rDNA sequences. Mycoscience 38, 409–420. doi: 10.1007/BF02461681
Perera, R., Hyde, K. D., Jones, E. B. G., Maharachchikumbura, S., Bundhun, D., Camporesi, E., et al. (2023). Profile of Bionectriaceae, Calcarisporiaceae, Hypocreaceae, Nectriaceae, Tilachlidiaceae, Ijuhyaceae fam. nov., Stromatonectriaceae fam. nov., and Xanthonectriaceae fam. nov. Fungal Diversity 118, 1–29. doi: 10.1007/s13225-022-00512-1
Prabhu, K. S., Siveen, K. S., Kuttikrishnan, S., Iskandarani, A. N., Khan, A. Q., Merhi, M., et al. (2018). Greensporone C, a freshwater fungal secondary metabolite induces mitochondrial-mediated apoptotic cell death in Leukemic cell lines. Front. Pharmacol. 9. doi: 10.3389/fphar.2018.00720
Raghukumar, C. (2008). Marine fungal biotechnology: an ecological perspective. Fungal Divers. 31, 19–35.
Rambaut, A. (2018). “FigTree v1.4.4: Tree figure drawing tool,” in FigTree v1.4.4: Tree figure drawing tool(Edinburgh. UK). Available at: http://tree.bio.ed.ac.uk/software/Figtree/ (Accessed May 1, 2024).
Rambelli, A. (1956). Chaetopsina nuovo genere di ifali Demaziacei. Atti della Accademia delle Sci. dell’Istituto di Bologna 15, 1–6.
Rambelli, A. (1987). A bibliographic reassessment of the genus Chaestopsina. Micologia Italiana 16 (1), 7–13.
Ranghoo, V. M., Hyde, K. D., Liew, E. C. Y., Spatafora, J. W. (1999). Family placement of Ascotaiwania and Ascotacicola based on DNA sequences from the large subunit rRNA gene. Fungal Divers. 2, 159–168.
Réblová, M., Hernández-Restrepo, M., Fournier, J., Nekvindová, J. (2020). New insights into the systematics of Bactrodesmium and its allies and introducing new genera, species and morphological patterns in the Pleurotheciales and Savoryellales (Sordariomycetes). Stud. Mycol. 95, 415–466. doi: 10.1016/j.simyco.2020.02.002
Réblová, M., Seifert, K. A., Fournier, J., Stepanek, V. (2012). Phylogenetic classification of Pleurothecium and Pleurotheciella gen. nov. and its dactylaria-like anamorph (Sordariomycetes) based on nuclear ribosomal and protein-coding genes. Mycologia 104, 1299–1314. doi: 10.3852/12-035
Réblová, M., Seifert, K. A., Fournier, J., Štěpánek, V. (2016). Newly recognised lineages of perithecial ascomycetes: the new orders Conioscyphales and Pleurotheciales. Persoonia 37 (1), 57–81. doi: 10.3767/003158516X689819
Rehner, S. A., Buckley, E. A. (2005). Phylogeny inferred from nuclear ITS and EF1-a sequences evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.1080/15572536.2006.11832842
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rossman, A. Y., Allen, W. C., Braun, U., Castlebury, L. A., Chaverri, P., Crous, P. W., et al. (2016). Overlooked competing asexual and sexually typified generic names of Ascomycota with recommendations for their use or protection. IMA Fungus 7, 289–308. doi: 10.5598/imafungus.2016.07.02.09
Samuels, G. J. (1985). Four new species of Nectria and their Chaetopsina anamorphs. Mycotaxon 22, 13–32.
Schuster, C. J., Sanders, J. L., Couch, C., Kent, M. L. (2022). “Recent advances with fish microsporidia,” in Microsporidia. Experientia Supplementum, vol. 114 . Eds. Weiss, L. M., Reinke, A. W. (Springer, Cham). doi: 10.1007/978-3-030-93306-7_11
Seifert, K. A., Morgan-Jones, G., Gams, W., Kendrick, B. (2011). The Genera of Hyphomycetes Vol. 9 (Utrech: CBS KNAW Fungal Biodiversity centre), 997.
Shen, H. W., Bao, D. F., Bhat, D. J., Su, H. Y., Luo, Z. L. (2022). Lignicolous freshwater fungi in Yunnan Province, China: an overview. Mycology 13, 119–132. doi: 10.1080/21501203.2022.2058638
Shi, L., Yang, H., Hyde, K. D., Wijayawardene, N. N., Wang, G. N., Yu, X. D., et al. (2021). Freshwater Sordariomycetes: new species and new records in Pleurotheciaceae, Pleurotheciales. Phytotaxa 518, 143–166. doi: 10.11646/phytotaxa.518.2.4
Sivichai, S., Jones, E. B. G., Hywel-Jones, N. L. (2000). Fungal colonisation of wood in a freshwater stream at Khao Yai National Park, Thailand. Fungal Divers. 5, 71–88.
Sridhar, K. R., Sudheep, N. M. (2011). The spatial distribution of fungi on decomposing woody litter in a freshwater stream, Western Ghats, India. Microb. Ecol. 61, 635–645. doi: 10.1007/s00248-011-9803-1
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Suberkropp, K. (2011). The influence of nutrients on fungal growth, productivity, and sporulation during leaf breakdown in streams. Can. J. Bot. 73, 1361–1369. doi: 10.1139/b95-398
Tsui, C. K. M., Baschien, C., Goh, T. K. (2016). “Biology and ecology of freshwater fungi,” in Biology of Microfungi. Ed. Li, D. W. (Springer, Cham), 285–313. doi: 10.1007/978-3-319-29137-6_13
Vaidya, G., Lohman, D. J., Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
Wan, Y. L., Bao, D. F., Luo, Z. L., Bhat, D. J., Xu, Y. X., Su, H. Y., et al. (2021). Two new species of Minimelanolocus (Herpotrichiellaceae, Chaetothyriales) from submerged wood in Yunnan, China. Phytotaxa 480, 45–56. doi: 10.11646/phytotaxa.480.1.4
Wang, L., Dong, J. Y., Song, H. C., Shen, K. Z., Wang, L. M., Sun, R., et al. (2008). Screening and isolation of antibacterial activities of the fermentative extracts of freshwater fungi from Yunnan Province, China. Ann. Microbiol. 58, 579–584. doi: 10.1007/BF03175561
Wang, L., Dong, W., Yang, J. X., Zhu, R., Zhang, H., Yu, Z. F. (2023). Parafuscosporella xishuangbannaensis sp. nov. (Fuscosporellales, Sordariomycetes) from submerged wood in China. Phytotaxa 587, 218–228. doi: 10.5281/zenodo.7752908
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (Academic Press, New York), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Dissanayake, L. S., Li, Q. R., Dai, D. Q., Xiao, Y., Wen, T. C., et al. (2021). Yunnan-guizhou plateau: A mycological hotspot. Phytotaxa 523, 1–31. doi: 10.11646/phytotaxa.523.1.1
Wurzbacher, C., Rösel, S., Rychła, A., Grossart, H. P. (2014). Importance of saprotrophic freshwater fungi for pollen degradation. PloS One 9, e94643. doi: 10.1371/journal.pone.0094643
Yang, H., Dong, W., Yu, X. D., Bhat, D. J., Boonmee, S., Zhang, H. (2020). Four freshwater dematiaceous hyphomycetes in Sordariomycetes with two new species of. Parafuscosporella. Phytotaxa 441, 19–34. doi: 10.11646/phytotaxa.441.1.2
Keywords: Fuscosporellaceae, hyphomycetes, morpho-molecular-based taxonomy, Nectriaceae, novel taxa, Pleurotheciaceae
Citation: Li L, Bhat DJ, Jiang H-B, Li J-F, Dawoud TM, Sun F, Haituk S, Cheewangkoon R and Phookamsak R (2025) New insights into freshwater ascomycetes: discovery of novel species in diverse aquatic habitats. Front. Cell. Infect. Microbiol. 14:1515972. doi: 10.3389/fcimb.2024.1515972
Received: 23 October 2024; Accepted: 02 December 2024;
Published: 13 January 2025.
Edited by:
Sinang Hongsanan, Shenzhen University, ChinaReviewed by:
Huang Zhang, Kunming University of Science and Technology, ChinaNattawut Boonyuen, National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand
Dian-Ming Hu, Jiangxi Agricultural University, China
Copyright © 2025 Li, Bhat, Jiang, Li, Dawoud, Sun, Haituk, Cheewangkoon and Phookamsak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ratchadawan Cheewangkoon, cmF0Y2hhZGF3YW4uY0BjbXUuYWMudGg=; Rungtiwa Phookamsak, cGhvb2thbXNha0BtYWlsLmtpYi5hYy5jbg==
 Lu Li
Lu Li Darbhe Jayarama Bhat4,5
Darbhe Jayarama Bhat4,5 Hong-Bo Jiang
Hong-Bo Jiang Jun-Fu Li
Jun-Fu Li Sukanya Haituk
Sukanya Haituk Ratchadawan Cheewangkoon
Ratchadawan Cheewangkoon Rungtiwa Phookamsak
Rungtiwa Phookamsak