- 1School of Traditional Chinese Medicine, Hunan University of Medicine, Huaihua, Hunan, China
- 2Research and Development Center, Kangpu Pharmaceutical Co., Ltd., Changde, Hunan, China
Background: Tongxieyaofang (TXYF) was a traditional Chinese medicine (TCM) formula for the treatment of diarrhea with liver stagnation and spleen deficiency syndrome, but the potential targets and mechanisms have not been fully clarified. This study aims to explore the potential mechanisms of TXYF in alleviating diarrhea using network pharmacology and full-length 16S rRNA gene sequencing.
Methods: Network pharmacology was applied to identify bioactive compounds and potential targets involved in the role of TXYF in alleviating diarrhea. Meanwhile, a model of diarrhea with liver stagnation and spleen deficiency syndrome was constructed by intragastric administration of Folium senna extract combined with restraint and tail pinch stress. The effect of TXYF on intestinal mucosal microbiota of diarrhea mice was analyzed by full-length 16S rRNA gene sequencing.
Results: Network pharmacology analysis showed that kaempferol, wogonin, naringenin, and nobiletin were compounds associated with the efficacy of TXYF. TXYF may alleviate diarrhea via multiple BPs and pathways, including TNF signaling pathway, IL-17 signaling pathway, and Toll-like receptor signaling pathway, which are involved in TCM-gut microbiota-host interactions. Then, we found that TXYF administration reshaped the diversity and composition of the intestinal mucosal microbial community of diarrhea mice. Lactobacillus, primarily Lactobacillus johnsonii, was enriched by the administration of TXYF. After TXYF administration, the abundance of Lactobacillus, particularly Lactobacillus johnsonii, was enriched.
Conclusion: Oral administration of TXYF may alleviate diarrhea through remodeling intestinal mucosal microbiota. Promoting the colonization of beneficial commensal bacteria in the intestinal mucosa through gut microbiota-host interactions may be a potential mechanism of TXYF in the treatment of diarrhea.
1 Introduction
Diarrhea is one of the most common symptoms in digestive. The incidence of diarrhea ranges from 2.2% to 73.8%, with variability depending on the definition applied (Dionne and Mbuagbaw, 2023). Diarrhea remains a common issue in clinical practice, and finding effective administration strategies is still necessary. Tongxieyaofang (TXYF), a traditional Chinese medicine (TCM) formula, consists of Rhizoma Atractylodis Macrocephalae (BaiZhu, dried rootstocks of Atractylodes macrocephala Koidz.), Radix Paeoniae Alba (BaiShao, dried roots of Paeonia lactiflora Pall.), Pericarpium Citri Reticulatae (ChenPi, dried pericarps of Citrus reticulata Blanco and its Cultivars), and Radix Saposhnikoviae (FangFeng, dried roots of Saposhnikovia divaricata (Turcz.) Schischk.) in a ratio of 3:2:1.5:1. TXYF, initially recorded in “Danxi’s Experiential Therapy” during the 14th century, was a treatment with proven effects for diarrhea accompanied by abdominal pain resulting from liver stagnation and spleen deficiency. It was officially recommended as a fundamental prescription for patients afflicted with diarrhea associated with liver stagnation and spleen deficiency syndrome in the Expert Consensus on the TCM Diagnosis and Treatment for Diarrhea (2017). According to TCM theory, TXYF can soothe the liver and regulate Qi, strengthen the spleen, transform dampness, and stop diarrhea. Randomized controlled clinical studies have shown that TXYF, as a primary prescription, has significant efficacy in relieving loose stools, abdominal discomfort, and bloating in IBS-D (Tang et al., 2019; Liang et al., 2022), which provides clinical evidence for its effect on alleviating diarrhea and abdominal pain. Although the clinical efficacy is definite, the potential mechanism of TXYF in stopping diarrhea needs to be further clarified.
Alterations in gut microbiota, also called dysbacteriosis, have been found in many gastrointestinal diseases with diarrhea as the main clinical manifestation (Qiao et al., 2023; Wu et al., 2023; Zhou et al., 2023; Luo et al., 2024). In previous work, we also observed significant differences in the intestinal cavity and intestinal mucosal microbiota between mice with diarrhea due to liver stagnation and spleen deficiency and control mice (Yuan et al., 2020; Zhang et al., 2020). Fecal microbiota transplantation of human fecal samples into germ-free mice can also construct human disease phenotype models (Wang et al., 2022). Targeted manipulation of gut microbiota to treat diarrhea also has shown great potential and attracted wide attention (Qiao et al., 2024; Zhou et al., 2024). Under this background, researchers specializing in TCM uncovered that TCM can modulate the structure and metabolism of the gut microbiota to exert therapeutic effects (Shao et al., 2020; Zhu et al., 2023; Wu et al., 2024). These discoveries provide a new scientific rationale for the application of TCM in modern society, particularly in gastrointestinal health and disease management. Delving into the effects of TCM on the gut microbiota can help reveal the modern scientific basis for its traditional efficacy and pave the way for the development of novel treatment approaches. Therefore, we designed to investigate if the effect of TXYF in alleviating diarrhea with liver stagnation and spleen deficiency syndrome is associated with gut microbiota regulation.
Network pharmacology, a novel model characterized by ‘multi-component and network target’ approaches in TCM research, has been introduced as an encouraging method to study the mechanisms of herbal formulas (Yao et al., 2022; Zhao et al., 2023). In this study, we aimed to identify the potential targets of TXYF alleviating diarrhea through network pharmacology and assess the effects of TXYF on the intestinal mucosa microbiota in mice with diarrhea with liver stagnation and spleen deficiency syndrome using full-length 16S rRNA gene sequencing.
2 Materials and methods
2.1 Animals and ethics statement
Fifteen one-month-old male specific-pathogen-free Kunming mice, weighing (20 ± 2) g, were purchased from the Hunan SJA Laboratory Animal Co., Ltd (License No: SCXK (Xiang) 2016-0002). Mice were housed in the barrier environment of the Center for Experimental Animals of the Hunan University of Chinese Medicine with (23-25) °C, (47-53) % humidity, 12h light/dark cycle, with free access to water and food during adaptive feeding. Animal experiments were performed following the National Guidelines for Experimental Animal Welfare, approved by the Animal Ethics and Welfare Committee of Hunan University of Chinese Medicine.
2.2 Preparation of experimental decoctions
2.2.1 Folium senna extract
All traditional Chinese herbs were purchased from the pharmacy of the First Affiliated Hospital of Hunan University of Chinese Medicine. The authenticated voucher specimens were deposited at School of Chinese Medicine, Hunan University of Chinese Medicine. Folium senna (Fanxieye, dried leaf of Cassia angustifolia Vahl or Cassia acutifolia Delile) samples (500 g) were cut into small pieces and soaked in boiled water (5000 mL) for 10 min. Then, the filtrates were evaporated to 500 mL with a thermostat water bath at 75°C to get the Folium senna extract with a crude drug concentration of 1 g/mL, which was stored at 4°C for model preparation later.
2.2.2 TXYF decoction
The TXYF herbal materials (45 g), consisting of Rhizoma Atractylodis Macrocephalae (18 g), Radix Paeoniae Alba (12 g), Pericarpium Citri Reticulatae (9 g), and Radix Saposhnikoviae (6 g), were soaked in cold water (400 mL) for 30 minutes before boiled for 30 minutes. The filtrates were collected, and the herb residues were decocted again in the same way. Two times’ filtrates were mixed and evaporated to 100 mL to get the TXYF decoction with a crude drug concentration of 0.24 g/mL, which was stored at 4°C for drug administration later.
2.3 Process of animal experiment
After three days of adaptive feeding in a barrier environment, mice were randomly divided into a control group (gtcm), a placebo group (gtmm), and a TXYF group (gttm), with 5 mice in each group. The repeated stress-related diarrhea model was established referring to our previous study (Yuan et al., 2020; Zhang et al., 2020). Briefly, mice in the placebo and TXYF groups were gavage with Folium Senna extract at a dose of 17.5 mL/kg/d after fasting for 12 hours. At the period of Folium Senna administration, each mouse in the placebo and TXYF groups was put in a 50 mL centrifuge tube separately to restrict its activities for 1 hour/day, with its tail nipped at the distal 1/3 using a binder clip at the same time. Mice in the control group were given the same dose of sterile water and free access to water, food, and activities during the period of modeling. After seven days of continuous modeling, mice in the placebo and TXYF groups were successfully induced symptoms of diarrhea with liver stagnation and spleen deficiency syndrome. Then, mice in the TXYF group were gavage with 0.35 mL TXYF decoction twice a day for three days. The control and placebo groups were given the same dose of sterile water at the same time.
At the end of the TXYF treatment period, mice were sacrificed by cervical dislocation. The small intestinal mucosa samples of mice were collected referring to our previous study (Shao et al., 2020; Zhang et al., 2020). Briefly, the abdominal cavity of the mouse was opened at an ultra-clean workbench to take out the small intestine. To sample the mucosa conveniently, the intestinal wall was cut along the long axis. A sterile cover glass was used to scrape the intestinal mucosa tissue. All samples were collected and marked separately and stored at -80°C for further PacBio SMRT sequencing.
2.4 Network pharmacology analysis
All ingredients in the four Chinese herbal medicines of TXYF were retrieved from the traditional Chinese medicine systems pharmacology (TCMSP) database (Ru et al., 2014) (https://tcmspw.com/tcmsp.php). Drug-likeness (DL) and oral bioavailability (OB) were two crucial properties of absorption, distribution, metabolism, and excretion (ADME)-related parameters of ingredients. To obtain the ideal potential bioactive compounds, we set the screening criteria of DL greater than or equal to 0.18 and OB greater than or equal to 30%. Protein targets related to every active ingredient of TXYF were obtained using TCMSP databases and converted to target names in Homo sapiens using UniProtKB (UniProt, 2021) (https://www.uniprot.org/).
Human genes associated with diarrhea were identified from the GeneCards database (Safran et al., 2010) (https://www.genecards.org) with the keyword “diarrhea”. Then, the potential targets of TXYF in treating diarrhea were obtained by a comprehensive analysis of the direct related targets of TXYF and the diarrhea targets using the online tool Venny2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
Potential targets for TXYF treat diarrhea were analyzed using the online database STRING11.0 (https://string-db.org/) to construct a functional protein-protein interaction (PPI) network of multiple proteins (Szklarczyk et al., 2019). To obtain appropriate confidence in identified protein interactions in the PPI network, we set the screening criteria of interaction scores greater than 0.9. To evaluate the role of each target in the PPI network, hub targets were identified using four different analysis methods (Degree, Closeness, MCC, and EcCentricity) in “cytoHubba” of cytoscape3.8.0 software (Shannon et al., 2003). Then, active compounds related to hub targets were screened and visualized with a Sankey diagram to introduce the relationship between hub targets and corresponding bioactive ingredients.
The “clusterProfiler” package in R software (v3.6.3) was employed to conduct GO and KEGG enrichment analysis.
2.5 Full-length 16S rRNA gene sequencing on the PacBio SMRT platform
Total bacterial genomic DNA was extracted from intestinal mucosa samples using the E.Z.N.A.® Bacterial DNA Kit (OMEGA, USA). The quality and quantity of the extracted genomic DNA were checked using a 0.8% agarose gel electrophoresis and an ultraviolet spectrophotometer.
For amplification of full-length 16S rRNA gene, the following primers were used: forward primer 27F (5’-AGAGTTTGATCMTGGMCTCAG-3’) and reverse primer 1492R (5’-ACCTTGTTACGACTT-3’). Polymerase chain reaction (PCR) amplification of 16S rRNA genes was conducted using the Q5® High-Fidelity DNA Polymerase (New England BioLabs, USA). Amplified DNA products were checked using a 2% agarose gel electrophoresis and purified using Axygen® AxyPrep DNA Gel Extraction Kit. The quantity of amplified DNA products was determined using Quant-iT PicoGreen dsDNA Assay Kit. Mock community PacBio library was constructed using PacBio SMRTbell Template Prep Kit 1.0. The prepared DNA library was sequenced on the PacBio Sequel platform by Shanghai Personal Biotechnology Co., Ltd.
2.6 Bioinformatics analysis
Quality control of raw sequencing reads was processed to generate high-quality sequences using quantitative insights into microbial ecology (QIIME, v1.8.0) software. The high-quality sequences were clustered into operational taxonomic units (OTUs) at a more than 97% similarity threshold by UCLUST. OTU taxonomic classification was carried out by BLAST searching the representative sequence of each OTU set against the Greengenes database. Bioinformatics analysis was performed based on a filtered OTUs table, in which OTUs containing less than 0.001% of total sequences across all samples were discarded. Alpha and beta diversity were introduced to distinguish the diversity within samples and between samples. For alpha diversity analysis, observed species, Chao1, and Shannon indexes were calculated in the QIIME program and visualized by GraphPad Prism 8. For beta diversity analysis, nonmetric multidimensional scaling (NMDS) analysis was performed based on unweighted and weighted UniFrac distances using R. The analysis of similarities (ANOSIM) based on unweighted and weighted UniFrac distances was carried out to compare different groups by QIIME. Linear discriminant analysis (LDA) effect size (LEfSe) (Segata et al., 2011) was conducted to define microbial biomarkers of the three groups, which provided biological class explanations to establish statistical significance, biological consistency, and effect size estimation of predicted biomarkers. The threshold on the logarithmic LDA score for distinguishing features was set to 4.0. Predictive functional profiling of microbial communities was conducted using 16S rRNA data by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (Langille et al., 2013) in the context of the Kyoto encyclopedia of genes and genomes (KEGG). The raw data will be made available by the authors, without undue reservation, to any qualified researcher.
2.7 Statistical analysis
The data were expressed as mean ± standard deviation (SD) where applicable. SPSS 22.0 and GraphPad Prism 8.0 were used for statistical analysis. Because the data does not conform to the normal distribution, the Kruskal-Wallis H test and Mann-Whitney U test were used to evaluate the statistical significance of the differences between the three groups and the two groups respectively. Differences were considered statistically significant when p < 0.05. The threshold of p-value was determined with the original false discovery rate (FDR) method of Benjamini and Hochberg (BH) in multiple comparisons.
3 Results
3.1 Network pharmacology analysis
3.1.1 Potential targets for TXYF treat diarrhea
After filtered by the ADME threshold (OB ≥ 30% and DL ≥ 0.18) and removed the duplicates, a total of 32 bioactive ingredients in TXYF were screened by the TCMSP database (Supplementary Table 1). Based on identified bioactive compounds in TXYF, a total of 145 potential target genes responding to compounds of TXYF were retrieved using the TCMSP data server. An herb-compound-target (CT) network was constructed by Cytoscape 3.8.0 software, which included 181 nodes (four herb nodes, 32 bioactive compound nodes, and 145 compound-associated target nodes) and 431 compound-target corresponding connections (Figure 1). According to the degree of the nodes in the CT network, kaempferol (degree = 62), wogonin (degree = 46), beta-sitosterol (degree = 39), naringenin (degree = 38), nobiletin (degree = 35), 5-O-Methylvisamminol (degree = 25) were recognized as main compounds of TXYF.
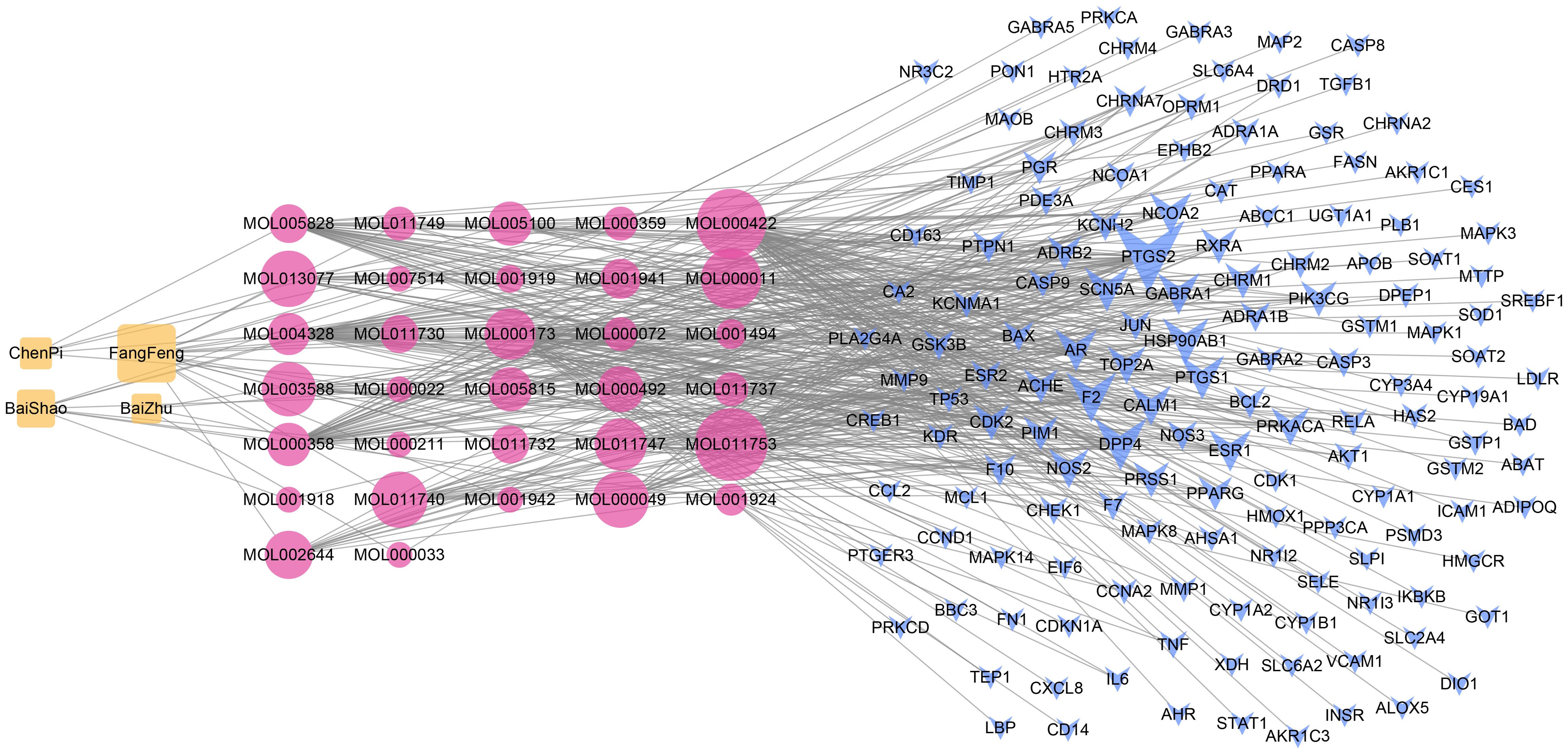
Figure 1. The herb-compound-target (CT) network. The orange nodes stand for herbs in TXYF. The red nodes stand for bioactive compounds in TXYF. The blue nodes stand for targets responding to the efficacy of TXYF. The grey lines stand for corresponding connections between different category nodes. The size of each node indicates the number of connections. The larger the node, the more the connections.
Targets related to diarrhea were obtained from the GeneCards database. Finally, 2 698 diarrhea-related targets (relevance score > median) were incorporated into the study. There were 94 intersection targets between targets responding to compounds of TXYF and targets related to diarrhea, which were recognized as potential targets for TXYF treat diarrhea.
3.1.2 GO and KEGG enrichment analysis
The results showed that with a post-adjusted p-value threshold of 0.05, 94 targets had enriched 1935 biological processes (BPs), 48 cellular components (CCs), and 131 molecular functions (MFs). The top 10 enriched GO terms from each category were directly shown in Figure 2A. The results showed that targets were enriched in BPs related to antibacterial and inflammation, including the response to lipopolysaccharide, response to molecule of bacterial origin, and response to oxidative stress. The CCs and MFs results indicated that most targets were localized in the membrane and related to steroid hormone receptor activity, nuclear receptor activity, ligand-activated transcription factor activity, and phosphatase binding. Meanwhile, the top 20 enriched KEGG pathways were visualized in Figure 2B. The results showed that majority targets are associated with TNF signaling pathway, IL-17 signaling pathway, and Toll-like receptor signaling pathway. These findings suggest the complex pathological process of diarrhea and the therapeutic potential of TXYF as a treatment for diarrhea.
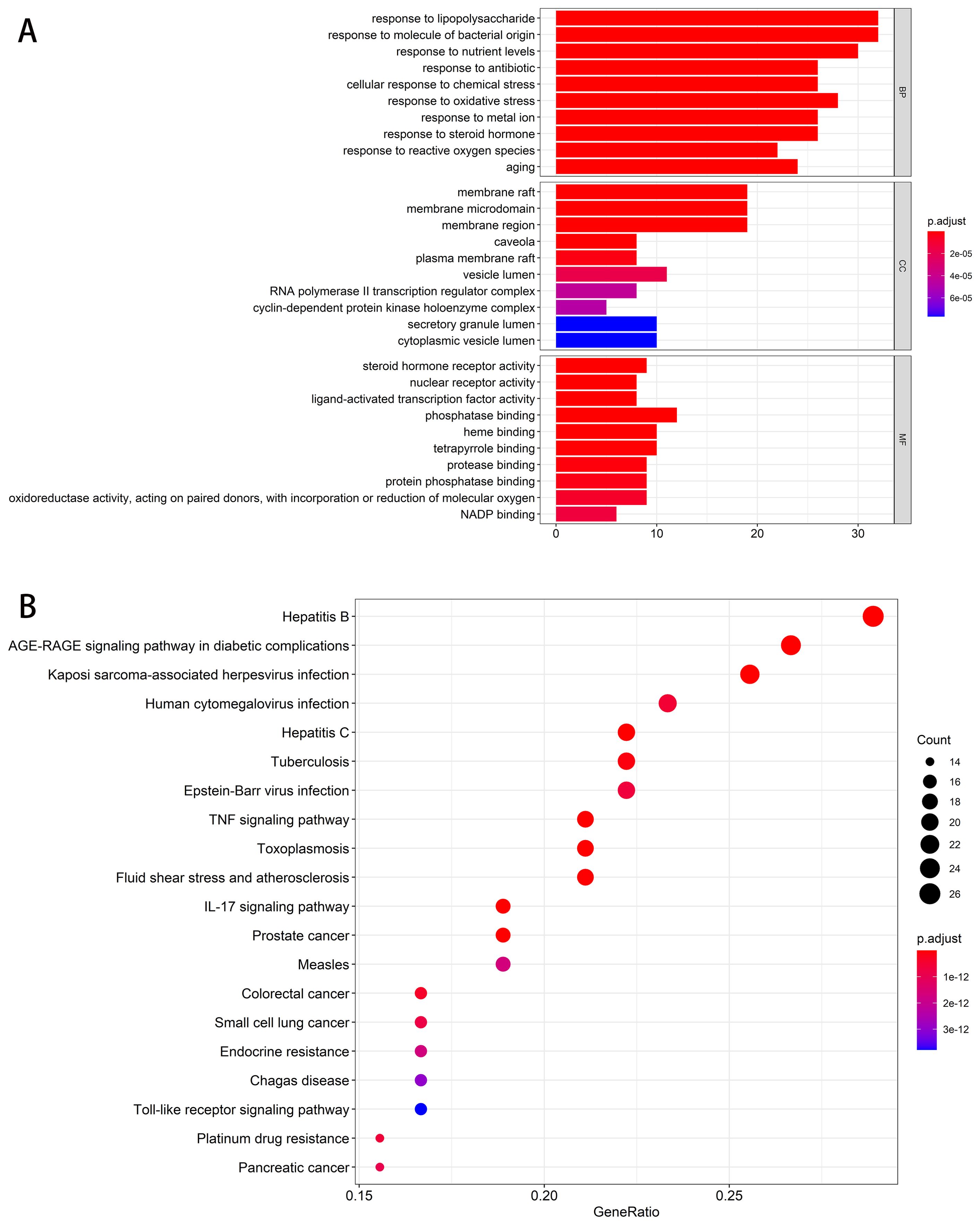
Figure 2. Enrichment analysis. (A) Bar plot for top 10 enriched GO terms of each sub-class. The length of each bar stands for enriched counts, and the color of each bar stands for adjusted p-value. The abscissa represents the enriched gene counts. (B) Dot plot for top 20 enriched KEGG pathway terms. The size of each node stands for enriched counts, and the color of each node stands for adjusted p-value. The abscissa represents the enriched gene ratio.
3.1.3 PPI network analysis and hub targets screening
To investigate the relationships among the 94 intersectant targets, we constructed a PPI network using the STRING database and visualized it with Cytoscape software. After removing disconnected nodes, a PPI network including 82 nodes and 282 interactions were constructed with the highest confidence score of 0.9 as the threshold. Hub targets were identified using four different algorithms, Betweenness, Degree, Maximal clique centrality (MCC), and Closeness, in the cytoHubba app (Figure 3A). Then, core hub targets of the PPI network were obtained by intersecting hub targets (top 10) identified from four algorithms (Figure 3B). Therefore, five targets (AKT1, MAPK14, MAPK3, TP53, and MAPK1) were identified as the core hub targets of the PPI network. Four bioactive compounds (kaempferol, naringin, wogonin, nobiletin) linked to five core hub targets were then selected in reverse. They were also the main compounds in TXYF. The relationships between core hub targets and corresponding bioactive compounds were shown directly using a Sankey diagram (Figure 3C).
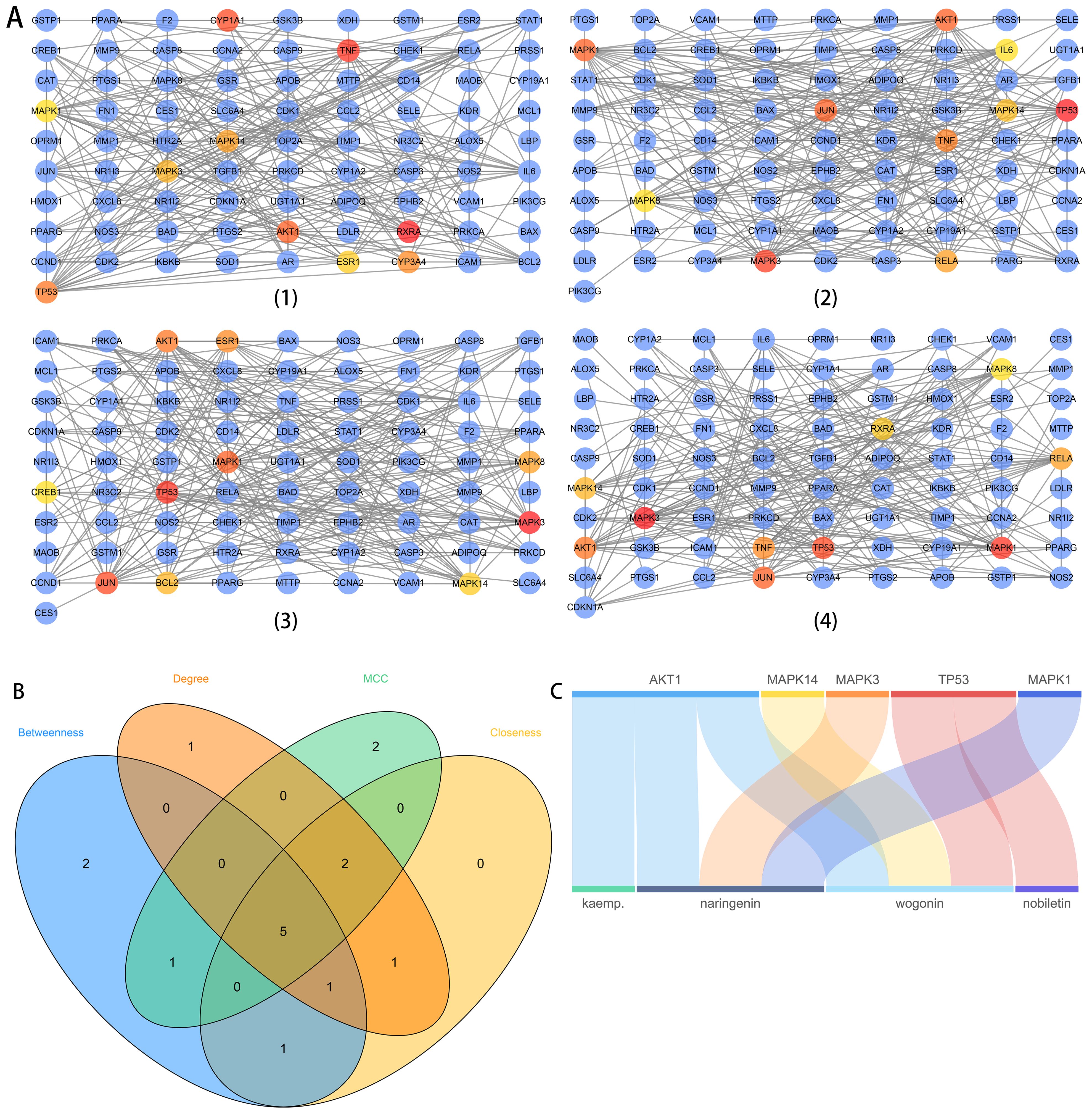
Figure 3. The PPI network analysis (A), the Venn diagram of core hub targets (B), and the Sankey diagram of core hub targets and corresponding compounds (C). In (A), (1) to (4) networks represent the analysis results based on four algorithms, Betweenness, Degree, MCC, and Closeness, respectively. The highlight red-to-yellow nodes are hub targets (top 10). The blue nodes are also protein targets with the highest confidence (0.9) from the PPI network analysis. In (B), hub targets (top 10) identified from four algorithms were intersected to obtain the core hub targets for further analysis. In (C), the relationships between core hub targets obtained from the Venn diagram and corresponding bioactive compounds were shown directly. The up blocks stand for the core hub targets, and the underneath blocks stand for compounds, as follows: kaempferol, naringenin, wogonin, and nobiletin.
3.2 TXYF administration altered the community diversity in intestinal mucosal microbiota in diarrhea mice
By high-throughput sequencing, 50005, 44322, and 42813 filtered high-quality sequences were obtained from samples of the control, placebo, and TXYF groups respectively, with the length concentrated at 1500 bp. A Venn diagram of OTU numbers showed that the gut microbiota community was changed with the treatment of TXYF (Figure 4A). The shared OTUs among the three groups were 188 (11.39%). The rarefaction curve demonstrated that the current sequencing depth was adequate to detect the microbial diversity in these community samples (Figure 4B).
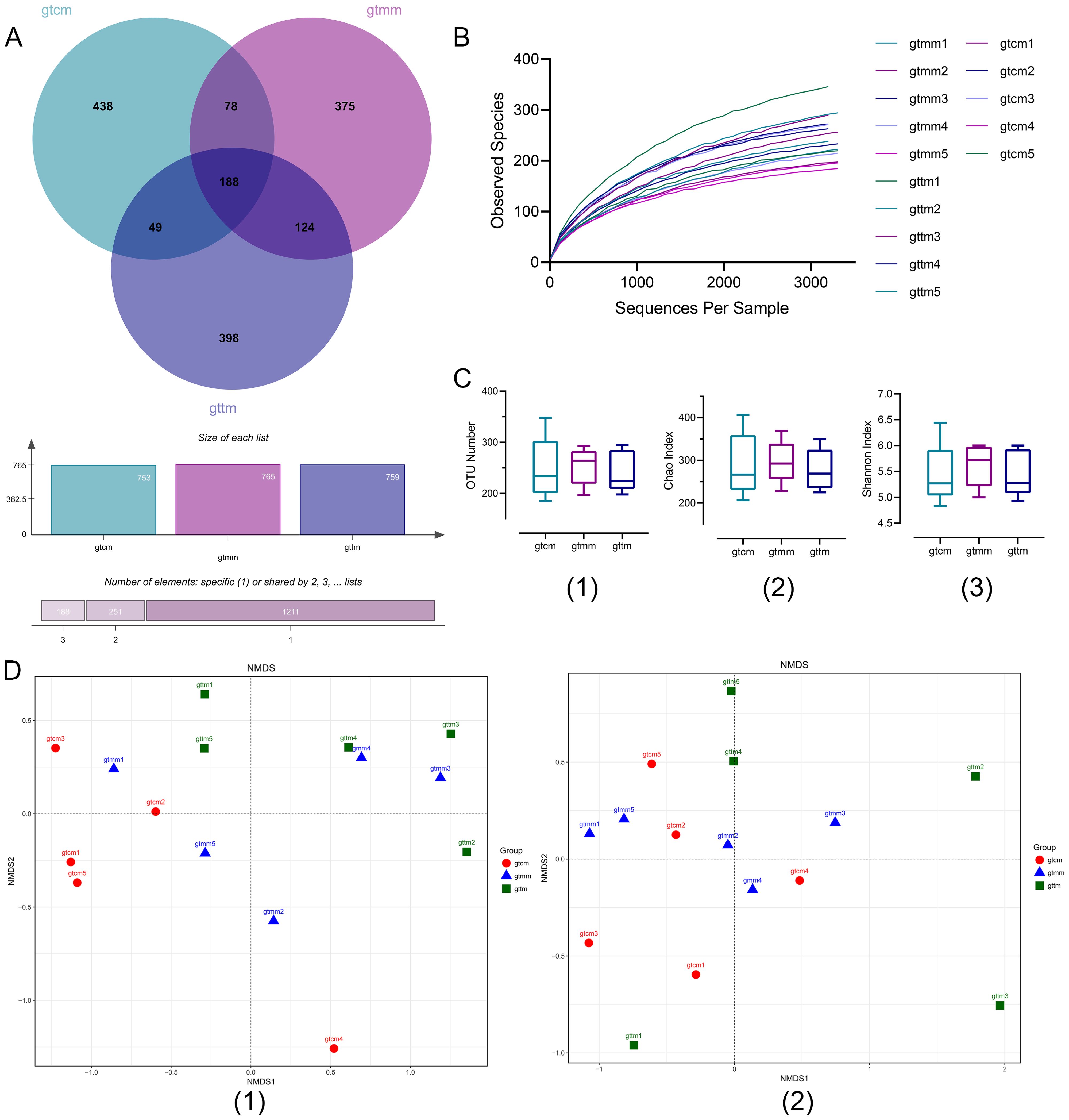
Figure 4. (A) Venn diagram of OTUs (at distance 0.03). (B) Rarefaction curve based on observed species. (C) Box plots of α-diversity index of OTUs number (1), Chao1 (2), and Simpson (3). (D) NMDS of OTU-based unweighted (1) and weighted (2) UniFrac distances. gtcm, the control group; gtmm, the placebo group; gttm, the TXYF group.
Diversity analysis was performed at two levels – within the sample and across samples. Alpha diversity indicators, such as observed species, Chao1 and Shannon indexes, were applied to reflect richness and evenness of community within a single sample (Figure 4C). As the results suggested, the placebo group has the highest community richness among the three groups. Compared with the placebo group, community richness and diversity decreased in the TXYF group and reached the same level as the control group. However, there was no statistical differences among the three groups (p > 0.05). Nonmetric multidimensional scaling (NMDS) of OTU-based weighted and unweighted UniFrac distance was conducted to present the similarities and differences in community composition across different samples and groups (Figure 4D). NMDS highlighted a significant segregation of the gut microbiota according to the unweighted UniFrac distance (ANOSIM, R = 0.1724, p = 0.040; Adonis, R2 = 0.18382, p = 0.033), suggesting that TXYF administration was a potential factor in alterating community composition.
3.3 TXYF administration changed the community composition in intestinal mucosal microbiota in diarrhea mice
To investigate whether there were representative taxa with differences, which may contribute to differences in diversity, we analyzed the community structure and composition of the three groups at the taxonomic level from phylum to species. OTUs were annotated to 17 phyla, 35 classes, 61 orders, 116 families, 199 genus, and 375 species. The proportion of unclassified OTUs was 4.06% (67/1650), suggesting that the full-length 16S sequencing technology has a high resolution in cluster analysis. In Figure 5A, the numbers of taxa at class and family levels in the TXYF group were significantly less than the control group (p < 0.05). In detail, compared with the placebo group, class Acidimicrobiia and 13 family taxa were newly colonized in the TXYF group, while seven class taxa and 21 family taxa were missing from the TXYF group.
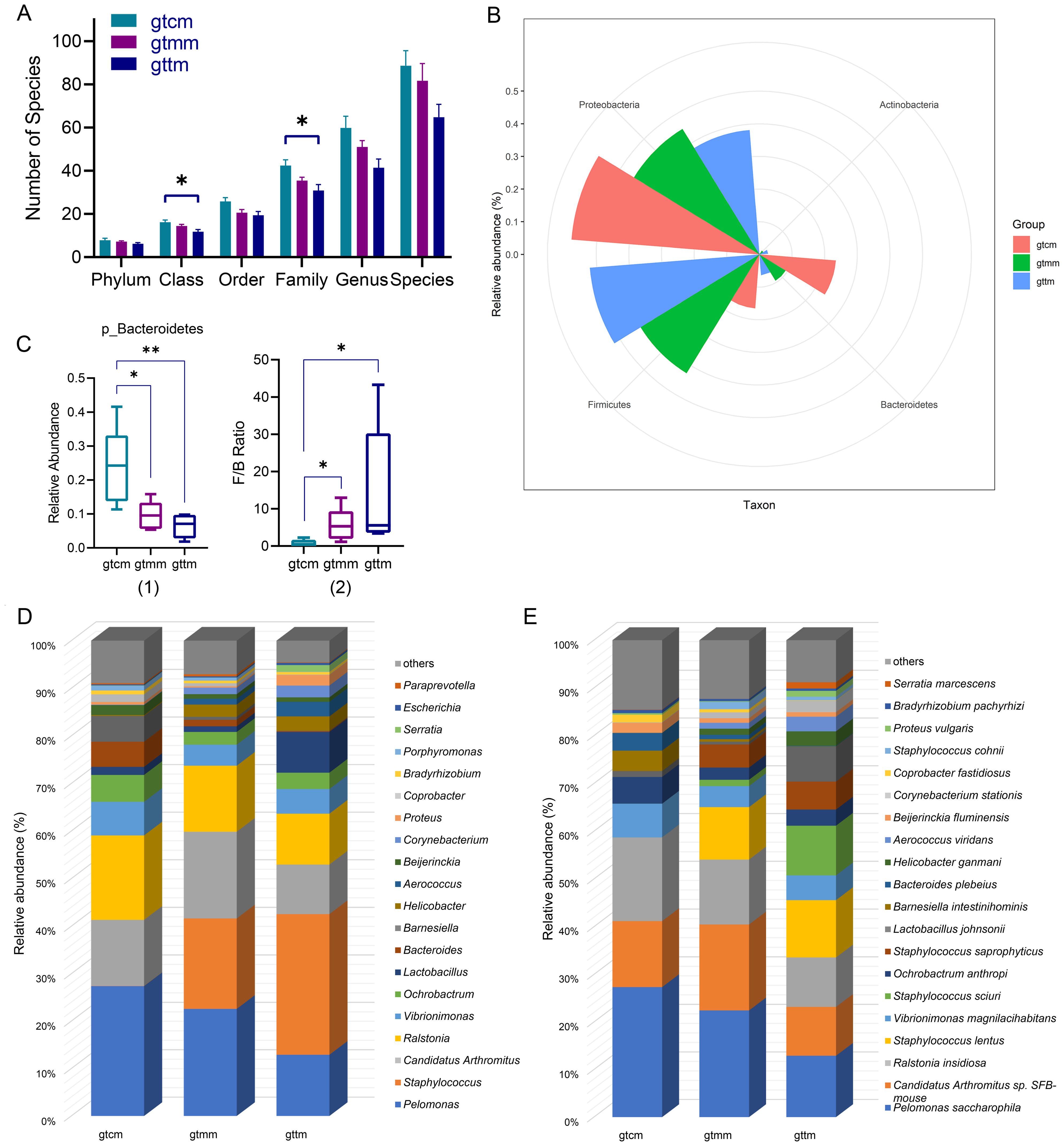
Figure 5. (A) Histogram of the number of microbial species of each sample at different taxonomic levels. (B) Radar graph of the relative abundance of dominant phylum in the intestinal mucosa. (C) Box plots of relative abundance of Bacteroidetes (1) and F/B ratio (2). (D, E) Stack diagram of relative abundance of genus (D) and species (E) in the intestinal mucosa. *p < 0.05, **p < 0.01, Kruskal-Wallis H test. gtcm, the control group; gtmm, the placebo group; gttm, the TXYF group.
Firmicutes (52.09%), Proteobacteria (38.21%), Bacteroidetes (6.41%), and Actinobacteria (2.69%), accounting for more than 98%, were the dominant communities in the intestinal mucosa of mice in the TXYF group (Figure 5B). Moreover, compared with the control group, the relative abundances of Bacteroidetes in both placebo and TXYF groups were significantly decreased, which resulted in a significant increase in the Firmicutes to Bacteroidetes (F/B) ratios in both groups (p < 0.05) (Figure 5C).
At the genus rank, dominant populations varied among groups (Figure 5D). Specifically, Staphylococcus (29.64%), Pelomonas (12.91%), Ralstonia (10.69%), Candidatus Arthromitus (10.41%), Lactobacillus (8.57%), and Vibrionimonas (5.18%), whose relative abundance was greater than 5%, were the six enriched genera in the TXYF group. They accounted for 77.4% of the total in the TXYF group. Only four genera had relative abundance greater than 5% in the placebo group, namely Pelomonas(22.59%), Staphylococcus(19.05%), Candidatus Arthromitus(18.21%), and Ralstonia(13.88%). They accounted for 73.73% of the total in the placebo group. The relative abundances of Pelomonas(27.39%), Ralstonia(17.74%), Candidatus Arthromitus(13.91%), Vibrionimonas(7.06%), Ochrobactrum(5.65%), Barnesiella(5.41%), and Bacteroides(5.28%)were greater than 5%, accounting for 82.44% of the total in the control group.
At the species level, enriched species with a relative abundance greater than 5% in the TXYF group were Pelomonas saccharophila (12.91% vs 22.44% in gtmm), Staphylococcus lentus (12% vs 10.98% in gtmm), Staphylococcus sciuri (10.4% vs 1.31% in gtmm), Ralstonia insidiosa (10.4% vs 13.59% in gtmm), Candidatus Arthromitus sp. SFB-mouse (10.27% vs 18.04% in gtmm), Lactobacillus johnsonii (7.3% vs 0.57% in gtmm), Staphylococcus saprophyticus (5.85% vs 4.84% in gtmm), Vibrionimonas magnilacihabitans (5.18% vs 4.41% in gtmm) (Figure 5E).
3.4 Microbial biomarkers associated with the administration of TXYF
To further identify the biomarkers associated with TXYF administration, we used LEfSe analysis to analyze microbial abundances at taxonomic levels from phylum to species. A total of 23 specific microbial biomarkers were identified across the three groups with linear discriminant analysis (LDA) score greater than 4 and p-value less than 0.05 (Figure 6A). Taxa including Gammaproteobacteria, Bacillales, Lactobacillales, Corynebacteriales, Staphylococcaceae, Aerococcaceae, Staphylococcus, Aerococcus, and Aerococcus_viridans were significantly enriched in the TXYF group, which could be recognized as biomarkers associated with the treatment of TXYF.
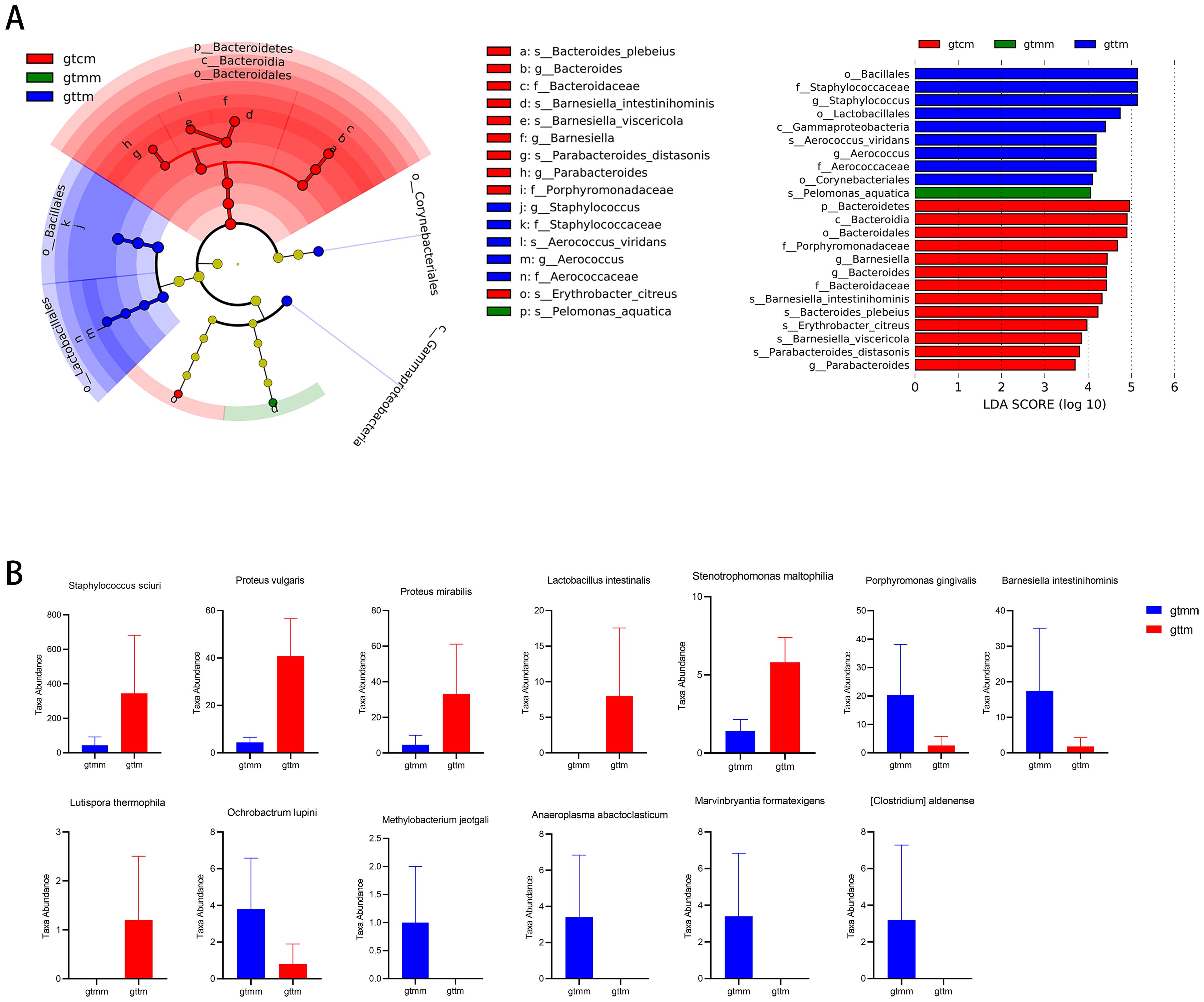
Figure 6. The potential biomarkers associated with the administration of TXYF. (A) Identified taxonomic features of significant differences across three groups by LEfSe (LDA score ≥ 4 and p-value ≤ 0.05). (B) Identified discriminative species between the TXYF (gttm) and placebo (gtmm) groups by Metastats analysis.
In addition, the Metastats analysis based on the number of sequences (i.e., absolute abundance) was performed to compare the differences in composition at species level between the TXYF and placebo groups. As shown in Figure 6B, there were significant increases in six species, such as Staphylococcus sciuri, Proteus vulgaris, Proteus mirabilis, Lactobacillus intestinalis, Stenotrophomonas maltophilia, and Lutispora thermophila, and decreases in seven species, including Porphyromonas gingivalis, Barnesiella intestinihominis, Ochrobactrum lupini, Methylobacterium jeotgali, Anaeroplasma abactoclasticum, Marvinbryantia formatexigens, and Clostridium aldenense, in the TXYF group compared with the placebo group.
3.5 TXYF administration regulated the microbial function
To predict the potential functions of microbial communities and assess the differences in microbial functions across groups, the BIOM file of OTU abundances was matched with the reference genomes of the KEGG database. The 16S sequencing dataset of 15 samples resulted in a functional dataset containing 5734 KOs. Further statistical analysis showed that 48 KOs were significantly altered between the TXYF and placebo groups (Figure 7A). Administration of TXYF significantly increased relative abundances of KOs in pathways such as two-component system, quorum sensing, biosynthesis, and metabolism of cofactor, butanoate, and tryptophan (Figure 7B). But in the meantime, TXYF reduced the relative abundance of KOs in pathways such as bacterial secretion system, secondary bile acid biosynthesis, and citrate cycle (Figure 7C).
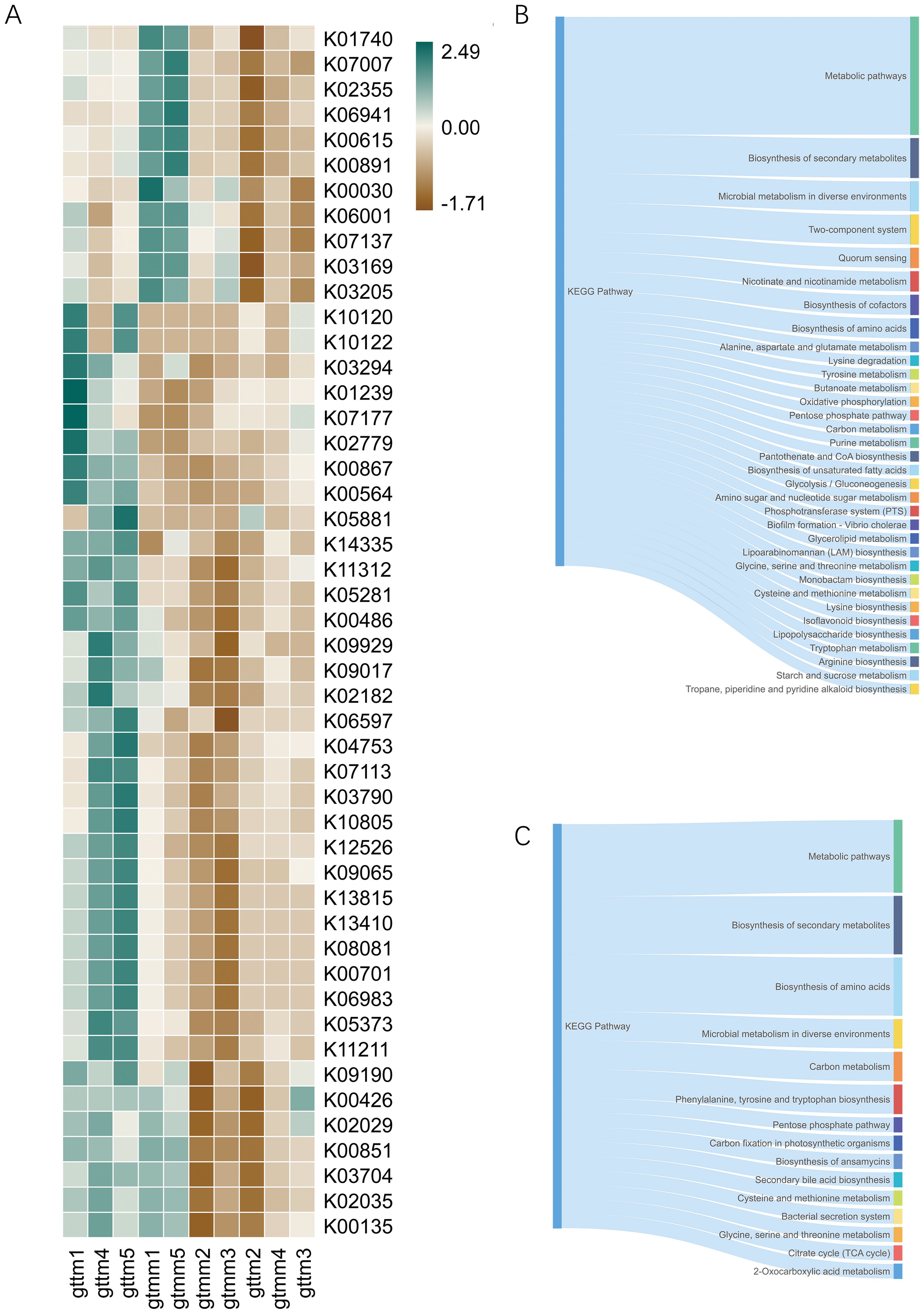
Figure 7. Function prediction based on the KEGG database by PICRUSt. (A) Heatmap for the relative abundance of discriminative KOs between the TXYF (gttm) and placebo (gtmm) groups. (B) The main KEGG pathway of the KOs with significantly increased expression in the TXYF group. (C) The main KEGG pathway of the KOs with significantly decreased expression in the TXYF group.
4 Discussion
According to the theory of TCM, TXYF was widely used to treat diarrhea due to liver stagnation and spleen deficiency syndrome in clinical practices. A meta-analysis of randomized trials also indicated that TXYF as a treatment for IBS-D was more effective and safer in improving the clinical effectiveness rate and decreasing the adverse events than conventional medication such as probiotics, pinaverium bromide, trimebutine, and Oryzanol (Zhou et al., 2019b). The clinical efficacy of TXYF is positive, but the mechanism of TXYF relieving diarrhea is indefinite. Folium senna combined with restraint stress is an effective method of establishing the mice model of liver stagnation and spleen deficiency syndrome (Yuan et al., 2020; Zhang et al., 2020). In this study, we used network pharmacology to identify potential active compounds and targets for TXYF to alleviate diarrhea and focused on the effects of TXYF on the intestinal mucosal microbiota of diarrhea model mice using PacBio SMRT sequencing technology.
Network pharmacology was an efficient approach to elucidate the intrinsic multi-component, multi-target, and multi-pathway features of Chinese medicine prescriptions. In this study, we obtained 32 compounds, including kaempferol, wogonin, naringenin, nobiletin, paeoniflorin, and Atractylenolide IIIt as active fractions of TXYF. The intestinal epithelium barrier consists of a single layer of enterocytes and tight junctions (TJs). A leaky barrier may contribute to hyper-intestinal permeability, which will lead to intestinal dysfunction and systemic diseases (Camilleri et al., 2017). Malfunction of TJ can also trigger the development of infection, inflammation, obesity, and even autoimmune diseases (Wen et al., 2020). Chen et al (Chen et al., 2020). reported that kaempferol supplementation up-regulated the expression of tight junction proteins, butyrate receptors, and transporters in the intestinal mucosa in acute alcoholic liver injury mice. The beneficial action of kaempferol on LPS-induced inflammation and barrier dysfunction in a coculture model of intestinal epithelial cells and intestinal microvascular endothelial cells was associated with inhibiting the NF-κB signaling pathway activation (Bian et al., 2020). In addition, studies suggested that supplementations of TCM bioactive compounds such as wogonin, naringenin, nobiletin, and paeoniflorin could suppress inflammatory response and maintain the intestinal barrier function (Azuma et al., 2013; Wang et al., 2015; Wu et al., 2019; Wen et al., 2020). Atractylodes III was also considered to have an inhibitory effect on mRNA and protein expressions of pro-inflammatory (IL-1β, TNF-α, IL-6) and anti-inflammatory cytokines (Zhou et al., 2019a).
Furthermore, the anti-inflammatory effect of TCM bioactive compounds was related to gut microbiota and its metabolites. It has been reported that the anti-arthritis activities of kaempferol were linked to functions of reversion the perturbation of metabolites involved in energy production and the tryptophan, fatty acid, and secondary bile acid metabolisms in the gut contents (Aa et al., 2020). Paeoniflorin has active impacts in decreasing the serum contents of corticosterone, corticotropin-releasing hormone, and adrenocorticotropic hormone and increasing serotonin and 5-Hydroxyindoleacetic acid in the prefrontal cortex and hippocampus in post-traumatic stress disorder mice (Qiu et al., 2018). Moreover, the anti-inflammatory and immunomodulatory actions of paeoniflorin were microbiota-dependent and related to indole-3-lactate and epithelial autophagy (Fan et al., 2020). Therefore, we speculate that the bioactive compounds of TXYF may interact with gut microbiota after oral administration and play roles in alleviating diarrhea by maintaining the intestinal epithelial barrier and regulating intestinal microbes and their metabolites.
GO and KEGG pathway enrichment analysis further verified the above inference. The effect of TXYF in alleviating diarrhea may be related to multiple BPs and pathways, such as the inflammatory responses and the immune systems, which are mediated by the TNF signaling pathway, the IL-17 signaling pathway, and the Toll-like receptor (TLR) signaling pathway. Specifically, TNF signaling has an active impact on maintaining epithelial cell tight junctions and intestinal barriers. It was stated that loss of intestinal barrier function mediated by TNF signaling was highly relevant to the inflammatory pathophysiology observed in Crohn’s disease and celiac disease (Kolodziej et al., 2011). TLR signaling pathway was proven to play an essential role in maintaining tolerance to commensal microbiota and regulating specific immune responses against pathogens, which is vital for the homeostasis of the intestine and activation of innate immunity (Takeda and Akira, 2004). For example, Yi et al (Yi et al., 2021). found that dietary anethole supplementation can alleviate intestinal barrier disruption and intestinal inflammation induced by enterotoxigenic Escherichia coli through modification of TLR signaling and intestinal microbiota. However, further studies are needed to confirm the exact role of these pathways in the process of TXYF relieving diarrhea.
Clear evidence from full-length 16S rRNA gene sequencing indicates that TXYF has a direct effect on the intestinal mucosal microbiota of diarrhea model mice. From a macro perspective, although no significant difference was observed in bacterial richness and evenness across groups according to alpha diversity analysis, beta diversity analysis based on unweighted UniFrac distance suggested a statistical difference in microbial composition among the three groups. We also found that the Chao1 and Simpson indices of the TXYF group were in the middle level of the three groups. These findings indicate that Folium senna combined with restraint stress might perturb the biodiversity in the intestinal mucosa by increasing bacterial richness, which is consistent with early studies (Zhang et al., 2020, 2021). TXYF might be a driving force in regulating the intestinal mucosal microbiota of diarrhea mice to a healthy state.
Then, we further concentrated on the changes in specific taxa related to the administration of TXYF. We recognized some positive taxa that were associated with the protective effects of TXYF. In this study, the relative abundance of Lactobacillus increased significantly in the TXYF group. Lactobacillus, a commensal inhabitant of animal and human gastrointestinal tracts, is considered friendly bacteria with a wide range of applications from probiotics to therapeutic bacteria in both humans and animals (O’Flaherty et al., 2020). In our study, Lactobacillus johnsonii and Lactobacillus acidophilus, which are important members of the Lactobacillus acidophilus complex (LAC) (Ramachandran et al., 2013), were enriched with the administration of TXYF. L. johnsonii completely lacked genes encoding biosynthetic pathways to produce essential nutrients, such as amino acids, purine nucleotides, and cofactors. Thus, it has high nutritional requirements for the intestinal environment, and its development and function depend on the host or other intestinal microorganisms to maintain (Pridmore et al., 2004). Co-occurrence network analysis showed that L. johnsonii was positively correlated with L. intestinalis, suggesting a mutualism relationship between L. johnsonii and L. intestinalis. It has been established that L. johnsonii L531 could alleviate Salmonella infantis-associated enteritis and promote intestinal secretory IgA production via regulation of NLRC4 and NLRP3 inflammasomes, NF-κB signaling pathway activation and suppression of mitochondrial damage (Xia et al., 2020). Oral pre-administration of L. acidophilus played a protective role in dextran sodium sulfate-induced colitis by targeting the intestinal TJ barrier that involves the TLR-2 receptor complex (Al-Sadi et al., 2021). Based on the effect of inducing the expression of µ-opioid and cannabinoid receptors in intestinal epithelial cells and mediating analgesic functions in the gut, L. acidophilus was recognized as a potential therapeutic approach for abdominal pain and irritable bowel syndrome (Rousseaux et al., 2007). However, further studies are warranted to investigate the exact beneficial mechanism of L. johnsonii and L. acidophilus in diarrhea mice with liver stagnation and spleen deficiency syndrome.
5 Conclusion
In this study, network pharmacology and full-length 16S rRNA gene sequencing were used to investigate the active compounds and targets of TXYF in alleviating diarrhea and the effect of TXYF on the intestinal mucosal microbiota of diarrhea model mice. Kaempferol, wogonin, naringenin, and nobiletin are compounds associated with the efficacy of TXYF in relieving diarrhea. TXYF may alleviate diarrhea via multiple BPs and pathways, including TNF signaling pathway, IL-17 signaling pathway, and Toll-like receptor signaling pathway, which are involved in TCM-gut microbiota-host interactions. TXYF was directly verified to remodel the diversity and composition of the intestinal mucosal microbiota in mice with diarrhea associated with live stagnation and spleen deficiency syndrome. Lactobacillus, primarily Lactobacillus johnsonii, was enriched by the administration of TXYF and were identified as being involved in the therapeutic role of TXYF in alleviating diarrhea. In conclusion, oral administration of TXYF may alleviate diarrhea through remodeling intestinal mucosal microbiota. Promoting the colonization of beneficial commensal bacteria in the intestinal mucosa through gut microbiota-host interactions may be a potential mechanism of TXYF in the treatment of diarrhea.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA861669.
Ethics statement
The animal study was approved by Animal Ethics and Welfare Committee of Hunan University of Chinese Medicine. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HS: Funding acquisition, Conceptualization, Investigation, Formal analysis, Writing – original draft. LW: Investigation, Visualization, Writing – review & editing. HZ: Supervision, Writing – review & editing.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Scientific Research Foundation of Hunan Provincial Education Department (No. 23B1054) and the Start-up Fund for Doctoral Research of Hunan University of Medicine (No. 202304).
Conflict of interest
Author HZ was employed by Kangpu Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1502373/full#supplementary-material
Supplementary Table 1 | The information of obtained bioactive compounds of TXYF.
References
Aa, L. X., Fei, F., Qi, Q., Sun, R. B., Gu, S. H., Di, Z. Z., et al. (2020). Rebalancing of the gut flora and microbial metabolism is responsible for the anti-arthritis effect of kaempferol. Acta Pharmacol. Sin. 41, 73–81. doi: 10.1038/s41401-019-0279-8
Al-Sadi, R., Nighot, P., Nighot, M., Haque, M., Rawat, M., Ma, T. Y. (2021). Lactobacillus acidophilus induces a strain-specific and toll-like receptor 2-dependent enhancement of intestinal epithelial tight junction barrier and protection against intestinal inflammation. Am. J. Pathol. 191, 872–884. doi: 10.1016/j.ajpath.2021.02.003
Azuma, T., Shigeshiro, M., Kodama, M., Tanabe, S., Suzuki, T. (2013). Supplemental naringenin prevents intestinal barrier defects and inflammation in colitic mice. J. Nutr. 143, 827–834. doi: 10.3945/jn.113.174508
Bian, Y., Dong, Y., Sun, J., Sun, M., Hou, Q., Lai, Y., et al. (2020). Protective effect of kaempferol on LPS-induced inflammation and barrier dysfunction in a coculture model of intestinal epithelial cells and intestinal microvascular endothelial cells. J. Agric. Food Chem. 68, 160–167. doi: 10.1021/acs.jafc.9b06294
Camilleri, M., Sellin, J. H., Barrett, K. E. (2017). Pathophysiology, evaluation, and management of chronic watery diarrhea. Gastroenterology 152, 515–532.e512. doi: 10.1053/j.gastro.2016.10.014
Chen, J., Xuan, Y. H., Luo, M. X., Ni, X. G., Ling, L. Q., Hu, S. J., et al. (2020). Kaempferol alleviates acute alcoholic liver injury in mice by regulating intestinal tight junction proteins and butyrate receptors and transporters. Toxicology 429, 152338. doi: 10.1016/j.tox.2019.152338
Dionne, J. C., Mbuagbaw, L. (2023). Diarrhea in the critically ill: definitions, epidemiology, risk factors and outcomes. Curr. Opin. Crit. Care 29, 138–144. doi: 10.1097/MCC.0000000000001024
Fan, Q., Guan, X., Hou, Y., Liu, Y., Wei, W., Cai, X., et al. (2020). Paeoniflorin modulates gut microbial production of indole-3-lactate and epithelial autophagy to alleviate colitis in mice. Phytomedicine 79, 153345. doi: 10.1016/j.phymed.2020.153345
Kolodziej, L. E., Lodolce, J. P., Chang, J. E., Schneider, J. R., Grimm, W. A., Bartulis, S. J., et al. (2011). TNFAIP3 maintains intestinal barrier function and supports epithelial cell tight junctions. PloS One 6, e26352. doi: 10.1371/journal.pone.0026352
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Liang, S. B., Han, M., Cheng, H. J., Zhang, Q. Y., Zhang, N. W., Jia, B. Y., et al. (2022). Chinese herbal formula Tongxie Yaofang for diarrhea-predominant irritable bowel syndrome: study protocol for a randomized, multiple-blind, placebo-controlled trial. Trials 23, 226. doi: 10.1186/s13063-022-06142-x
Luo, W., Zhao, M., Dwidar, M., Gao, Y., Xiang, L., Wu, X., et al. (2024). Microbial assimilatory sulfate reduction-mediated H(2)S: an overlooked role in Crohn’s disease development. Microbiome 12, 152. doi: 10.1186/s40168-024-01873-2
O’Flaherty, S., Foley, M. H., Rivera, A. J., Theriot, C. M., Barrangou, R. (2020). Complete genome sequence of lactobacillus johnsonii NCK2677, isolated from mice. Microbiol. Resour. Announc. 9 (43), e00918–20. doi: 10.1128/MRA.00918-20
Pridmore, R. D., Berger, B., Desiere, F., Vilanova, D., Barretto, C., Pittet, A. C., et al. (2004). The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. U.S.A. 101, 2512–2517. doi: 10.1073/pnas.0307327101
Qiao, B., Liu, J., Deng, N., Cai, Y., Bian, Y., Wu, Y., et al. (2023). Gut content microbiota dysbiosis and dysregulated lipid metabolism in diarrhea caused by high-fat diet in a fatigued state. Food Funct. 14, 3880–3892. doi: 10.1039/d3fo00378g
Qiao, B., Xiao, N., Deng, N., Tan, Z. (2024). Shenling Baizhu powder attenuates lard diet in a fatigued state-induced diarrhea via targeting microbial metabolites short chain fatty acids-mediated lipid metabolism. 3 Biotech. 14, 203. doi: 10.1007/s13205-024-04045-z
Qiu, Z. K., He, J. L., Liu, X., Zeng, J., Xiao, W., Fan, Q. H., et al. (2018). Anxiolytic-like effects of paeoniflorin in an animal model of post traumatic stress disorder. Metab. Brain Dis. 33, 1175–1185. doi: 10.1007/s11011-018-0216-4
Ramachandran, P., Lacher, D. W., Pfeiler, E. A., Elkins, C. A. (2013). Development of a tiered multilocus sequence typing scheme for members of the Lactobacillus acidophilus complex. Appl. Environ. Microbiol. 79, 7220–7228. doi: 10.1128/AEM.02257-13
Rousseaux, C., Thuru, X., Gelot, A., Barnich, N., Neut, C., Dubuquoy, L., et al. (2007). Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat. Med. 13, 35–37. doi: 10.1038/nm1521
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform 6, 13. doi: 10.1186/1758-2946-6-13
Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., et al. (2010). GeneCards Version 3: the human gene integrator. Database (Oxford) 2010, baq020. doi: 10.1093/database/baq020
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shao, H., Zhang, C., Wang, C., Tan, Z. (2020). Intestinal mucosal bacterial diversity of antibiotic-associated diarrhea (AAD) mice treated with Debaryomyces hansenii and Qiweibaizhu powder. 3 Biotech. 10, 392. doi: 10.1007/s13205-020-02383-2
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Takeda, K., Akira, S. (2004). TLR signaling pathways. Semin. Immunol. 16, 3–9. doi: 10.1016/j.smim.2003.10.003
Tang, X. D., Zhang, S. S., Hou, X. H., Li, Z. H., Chen, S. N., Feng, P. M., et al. (2019). Post-marketing re-evaluation of tongxiening granules () in treatment of diarrhea-predominant irritable bowel syndrome: A multi-center, randomized, double-blind, double-dummy and positive control trial. Chin. J. Integr. Med. 25, 887–894. doi: 10.1007/s11655-019-3030-x
UniProt, C. (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi: 10.1093/nar/gkaa1100
Wang, W., Xia, T., Yu, X. (2015). Wogonin suppresses inflammatory response and maintains intestinal barrier function via TLR4-MyD88-TAK1-mediated NF-kappaB pathway in vitro. Inflammation Res. 64, 423–431. doi: 10.1007/s00011-015-0822-0
Wang, Y., Zhang, Z., Liu, B., Zhang, C., Zhao, J., Li, X., et al. (2022). A study on the method and effect of the construction of a humanized mouse model of fecal microbiota transplantation. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1031758
Wen, X., Zhao, H., Wang, L., Wang, L., Du, G., Guan, W., et al. (2020). Nobiletin attenuates DSS-induced intestinal barrier damage through the HNF4alpha-claudin-7 signaling pathway. J. Agric. Food Chem. 68, 4641–4649. doi: 10.1021/acs.jafc.0c01217
Wu, Y., Deng, N., Liu, J., Cai, Y., Yi, X., Tan, Z. (2024). Unlocking the therapeutic potential of Huoxiang Zhengqi San in cold and high humidity-induced diarrhea: Insights into intestinal microbiota modulation and digestive enzyme activity. Heliyon 10, e32789. doi: 10.1016/j.heliyon.2024.e32789
Wu, Y., Deng, N., Liu, J., Jiang, P., Tan, Z. (2023). Alterations in intestinal microbiota and enzyme activities under cold-humid stress: implications for diarrhea in cold-dampness trapped spleen syndrome. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1288430
Wu, X. X., Huang, X. L., Chen, R. R., Li, T., Ye, H. J., Xie, W., et al. (2019). Paeoniflorin prevents intestinal barrier disruption and inhibits lipopolysaccharide (LPS)-induced inflammation in caco-2 cell monolayers. Inflammation 42, 2215–2225. doi: 10.1007/s10753-019-01085-z
Xia, B., Yu, J., He, T., Liu, X., Su, J., Wang, M., et al. (2020). Lactobacillus johnsonii L531 ameliorates enteritis via elimination of damaged mitochondria and suppression of SQSTM1-dependent mitophagy in a Salmonella infantis model of piglet diarrhea. FASEB J. 34, 2821–2839. doi: 10.1096/fj.201901445RRR
Yao, Z., Fu, S., Ren, B., Ma, L., Sun, D. (2022). Based on network pharmacology and gut microbiota analysis to investigate the mechanism of the laxative effect of pterostilbene on loperamide-induced slow transit constipation in mice. Front. Pharmacol. 13. doi: 10.3389/fphar.2022.913420
Yi, Q., Liu, J., Zhang, Y., Qiao, H., Chen, F., Zhang, S., et al. (2021). Anethole Attenuates Enterotoxigenic Escherichia coli-Induced Intestinal Barrier Disruption and Intestinal Inflammation via Modification of TLR Signaling and Intestinal Microbiota. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.647242
Yuan, Z., Zhang, C., Peng, X., Shu, L., Long, C., Tan, Z. (2020). Intestinal microbiota characteristics of mice treated with Folium senna decoction gavage combined with restraint and tail pinch stress. 3 Biotech. 10, 180. doi: 10.1007/s13205-020-02172-x
Zhang, C. Y., Peng, X. X., Shao, H. Q., Li, X. Y., Wu, Y., Tan, Z. J. (2021). Gut microbiota comparison between intestinal contents and mucosa in mice with repeated stress-related diarrhea provides novel insight. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.626691
Zhang, C., Shao, H., Peng, X., Liu, T., Tan, Z. (2020). Microbiotal characteristics colonized in intestinal mucosa of mice with diarrhoea and repeated stress. 3 Biotech. 10, 372. doi: 10.1007/s13205-020-02368-1
Zhao, L., Zhang, H., Li, N., Chen, J., Xu, H., Wang, Y., et al. (2023). Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J. Ethnopharmacol 309, 116306. doi: 10.1016/j.jep.2023.116306
Zhou, K., Chen, J., Wu, J., Wu, Q., Jia, C., Xu, Y. X. Z., et al. (2019a). Atractylenolide III ameliorates cerebral ischemic injury and neuroinflammation associated with inhibiting JAK2/STAT3/Drp1-dependent mitochondrial fission in microglia. Phytomedicine 59, 152922. doi: 10.1016/j.phymed.2019.152922
Zhou, Y., Han, S., He, Y. (2019b). Clinical effects and safety of tongxieyaofang on diarrhea predominant irritable bowel syndrome: A meta-analysis of randomized trails. Evid Based Complement Alternat Med. 2019, 4893876. doi: 10.1155/2019/4893876
Zhou, M., Li, X., Wang, X., Deng, N., Cai, Y., Tan, Z. (2023). The dysfunction in intestinal microorganisms and enzyme activity as significant contributors to diarrhea with kidney-yang deficiency syndrome. Front. Microbiol. 14, 88. doi: 10.3389/fmicb.2023.1324938
Zhou, M., Li, X., Liu, J., Wu, Y., Tan, Z., Deng, N. (2024). Adenine's impact on mice's gut and kidney varies with the dosage administered and relates to intestinal microorganisms and enzyme activities. 3 Biotech. 14, 88. doi: 10.1007/s13205-024-03959-y
Zhu, J., Li, X., Deng, N., Zhou, K., Qiao, B., Li, D., et al. (2023). Intestinal mucosal flora of the intestine-kidney remediation process of diarrhea with deficiency kidney-yang syndrome in Sishen Pill treatment: Association with interactions between Lactobacillus johnsonii, Ca(2+)-Mg(2+)-ATP-ase, and Na(+)-K(+)-ATP-ase. Heliyon 9, e16166. doi: 10.1016/j.heliyon.2023.e16166
Keywords: Chinese medicine, intestinal mucosal microbiota, diarrhea, Tongxieyaofang (TXYF), full-length 16S rRNA gene sequencing
Citation: Shao H, Wang L and Zhang H (2024) Chinese herbal medicine, Tongxieyaofang, alleviates diarrhea via gut microbiota remodeling: evidence from network pharmacology and full-length 16S rRNA gene sequencing. Front. Cell. Infect. Microbiol. 14:1502373. doi: 10.3389/fcimb.2024.1502373
Received: 26 September 2024; Accepted: 18 October 2024;
Published: 15 November 2024.
Edited by:
Xinhua Shu, Glasgow Caledonian University, United KingdomReviewed by:
Huaying Hui, Hunan University of Chinese Medicine, ChinaYunting Wang, University of Houston, United States
Copyright © 2024 Shao, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hualing Zhang, NDc1NzQxNjM0QHFxLmNvbQ==
 Haoqing Shao
Haoqing Shao Liping Wang
Liping Wang Hualing Zhang2*
Hualing Zhang2*