- 1Neonatology Department, Shenzhen Nanshan Maternity and Child Healthcare Hospital, Shenzhen, China
- 2Neonatology Department, Affiliated Shenzhen Children's Hospital of Shantou University Medical College, Shenzhen, China
- 3Neonatology Department, Guangdong Women and Children Hospital, Guangzhou, China
Background: Women with vulvovaginal candidiasis (VVC) are known to experience vaginal microbial dysbiosis. However, the dynamic alterations of the vaginal microbiome in pregnant women with VVC and its effect on neonatal gut microbiome remain unclear. This study aims to characterize the vaginal microbiome in pregnant women with VVC and its impact on their offspring’s meconium microbiome.
Methods: Forty-four pregnant women, including 17 with VVC (VVC group) and 27 healthy controls (HC group), along with their 44 offspring, were enrolled in this study. Maternal vaginal samples were collected during the pre- and post-delivery phases. Meconium samples from their newborns were also obtained. Microbial communities were characterized using 16S rRNA sequencing.
Results: The vaginal microbiome of healthy pregnant women was predominantly composed of the genus Lactobacillus. The Bray-Curtis dissimilarity index indicated significant alterations in the vaginal microbiome of the VVC group, with a notable decrease in Lactobacillus and significant increases in Delftia, Burkholderia during both the pre- and post-delivery phases compared to the HC group. Additionally, the neonatal meconium microbiome exhibited significant differences between the VVC and HC groups, with L. salivarius and L. helveticus significantly decreased and Delftia significantly increased in the VVC group. Similar trends in microbial variation were observed across maternal and neonatal microbiomes, indicating intergenerational concordance associated with VVC.
Conclusion: VVC alters the microbiota of both pregnant women and their neonates at birth, suggesting a form of microbial inheritance. These findings underscore the distinctive characteristics of the vaginal microbiome associated with VVC and its potential impact on the formation of early-life gut microbiome.
Introduction
The vaginal microbiome during pregnancy is of considerable interest due to its impact on both maternal and neonatal health (Fettweis et al., 2019; Zhao et al., 2021). The maternal vaginal microbiome may serve as an important source of pioneer bacteria for the neonatal gut microbiome (Dominguez-Bello et al., 2010; Kervinen et al., 2019), profoundly affecting host metabolism and immunity (Gaziano et al., 2023) and improving neonatal neurodevelopment (Zhou et al., 2023). During a healthy pregnancy, the vaginal microbiome is characterized by low bacterial species diversity and is typically dominated by one of several different species of Lactobacillus (Ravel et al., 2011; Zhang et al., 2022a). Researchers have categorized the vaginal microbiome into five community state types (CSTs) based on the dominant bacterial species. CST I, CST II, CST III, and CST V are dominated by L. crispatus, L. gasseri, L. iners, and L. jensenii, respectively. In contrast, CST IV is characterized by the absence of L. spp. and comprises a diverse array of strict and facultative anaerobes (Ravel et al., 2011; DiGiulio et al., 2015; MacIntyre et al., 2015). The composition of the vaginal microbiome is influenced by genetic factors, such as ethnicity, as well as environmental, individual and lifestyle factors (Kervinen et al., 2019). Additionally, infectious diseases and their treatments play a significant role in shaping the vaginal microbiome (Greenbaum et al., 2019; Chen et al., 2021b; Zhang et al., 2022b).
Vulvovaginal candidiasis (VVC) is a multifactor infectious disease affecting the lower reproductive tract in women, predominantly caused by Candida albicans, and results in pathological inflammation (Farr et al., 2021). VVC is one of the most common forms of infectious vaginitis, with an estimated 75% of women experiencing at least one episode in their lifetime. Recurrent VVC affects nearly 8% of women worldwide (Denning et al., 2018). Vulvovaginal yeast infections caused by Candida species are more prevalent in pregnant women compared to non-pregnant women, potentially due to hormonal and immunological changes that occur during pregnancy (Aguin and Sobel, 2015; Chatzivasileiou and Vyzantiadis, 2019). These infections during pregnancy are common and can lead to extensive inflammation, which may contribute to adverse perinatal outcomes (Chatzivasileiou and Vyzantiadis, 2019; Schuster et al., 2020).
Accumulating studies have demonstrated alterations in vaginal microbial communities during genital infections, including VVC, bacterial vaginosis infection, and Chlamydia trachomatis infection (Ceccarani et al., 2019; Zhao et al., 2023). These microbial communities are also affected by drug treatments (Xiao et al., 2022). Moreover, the vaginal microbiome is dynamic during and after pregnancy and in the postpartum period (MacIntyre et al., 2015; Li et al., 2022). However, information regarding dynamic changes in maternal vaginal microbiome due to VVC is limited, and its effect on the gut microbiome of offspring remains to be understood. Therefore, this study aimed to compare the alterations in maternal vaginal microbial composition before and right after delivery in pregnant women with VVC versus those without VVC. Additionally, the meconium microbiome was also compared between the offspring of VVC and those without.
Methods
Study subjects
Pregnant women and neonates were recruited at Shenzhen Nanshan Maternity and Child Health Care Hospital. The study protocols were approved by the ethics committee of Shenzhen Nanshan Maternity and Child Health Care Hospital (No. NSFYEC-KY-2022018) and conducted in accordance with all relevant guidelines and regulations. Written informed consent was obtained from all participants, and for neonates, consent was provided by their parents or legal guardians. Participants who did not consent to participation were excluded from the study. Additionally, participants were excluded if they had taken antibiotics or amicrobial therapy during pregnancy, consumed probiotics within three months before delivery, experienced adverse pregnancy outcomes, had a history of smoking and alcohol consumption during pregnancy, had complicated singleton pregnancies, had any underlying medical diseases prior to pregnancy, or if the newborn had physical abnormalities.
A total of forty-four healthy women with a gestational age (GA) greater than 37 weeks who delivered vaginally at maternity centers were included. Specifically, 17 pregnant women were classified into the VVC group, while the remaining 27 pregnant women were designated as the normal controls (HC group). VVC diagnosis was established through microscopic and culture-based identification of Candida, as previously reported (Ceccarani et al., 2019). All pregnant women were tested at 35-36 weeks of gestation at our hospital, and only asymptomatic, without therapeutic intervention VVC cases recruited to minimize the interference of external factors on study outcomes.
Sample collection
Vaginal swab samples were collected from all study participants in two different phases: upon admission to the hospital for delivery (phase BD, before delivery), and immediately after delivery (phase PD, after delivery). To ensure sufficient DNA concentration, three sterile cotton swab sticks were used to collect vaginal swab samples by trained nurses. Following a previously reported method (Li et al., 2023), the sterile swab was placed carefully on the vaginal sidewall about halfway between the introitus and the cervix, and rolled dorsally-ventrally back and forth four times to coat the swab.
Meconium samples were obtained within 24 hours after delivery and collected in a sterile tube under antiseptic handling by trained nurses according to previously reported (Turunen et al., 2021). All samples were stored at -80°C until DNA extraction.
Microbiome profiling
Microbial DNA was extracted using the QIAamp DNA Mini kit, and its concentration and purity were measured using the NanoDrop One (Thermo Fisher Scientific, MA, USA). The V3-4 variable region of the 16S rRNA gene was amplified using forward primer 338F (5’-ACTCCTACGGGAGGCAGCAG-3’), and the reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’) (Chen et al., 2021a). PCR reactions, containing 25 μL 2× Premix Taq (Takara Biotechnology, Dalian Co. Ltd., China), 2 μL of each 10 mM primer, and 3 μL DNA template in a volume of 50 μL, were amplified under the following thermal profile: 94 °C for 5 minutes, then 30 cycles of 94 °C for 30 seconds, 52 °C for 30 seconds, 72 °C for 30 seconds, followed by 72 °C for 10 minutes. PCR products were then pooled in equimolar and paired-end sequenced on an Illumina MiSeq platform with V3 chemistry.
The 16S rRNA gene sequences were analyzed using the bioinformatics software package QIIME2 (version 2021.8) (Bolyen et al., 2019). Paired-end reads were denoised by QIIME2 using the command “qiime dada2 denoise-paired”, which aimed to merge paired-end reads, perform quality filtering, and exclude chimeric and phiX sequences. Taxonomic assignment was performed against the Greengenes (13_8 revision) database using command “qiime feature-classifier classify-sklearn”. Alpha- and beta-diversity measures were generated using the commands “qiime phylogeny align-to-tree-mafft-fasttree” and “qiime diversity core-metrics-phylogenetic”.
Statistical analysis
Continuous variables were presented as the mean ± standard deviation (SD), and categorical characteristics were reported as numbers (percentages). All comparisons were performed using R software at a significant level of 0.05, with chi-square and t-tests for categorical and continuous variables, respectively.
Principal coordinate ordination analysis (PCoA) was performed on Bray-Curtis distances and accompanied by permutational multivariate analysis of variance (PERMANOVA, 999 permutations) using the R package “vegan” and MicrobiotaProcess (Xu et al., 2023). Differentially abundant taxa between the two groups were estimated using LEfSe (Linear Discriminate Analysis of Effect size) software, with a significance threshold of P < 0.05 and LDA > 2.0 (Segata et al., 2011).
Results
Demographic and clinical data of study subjects
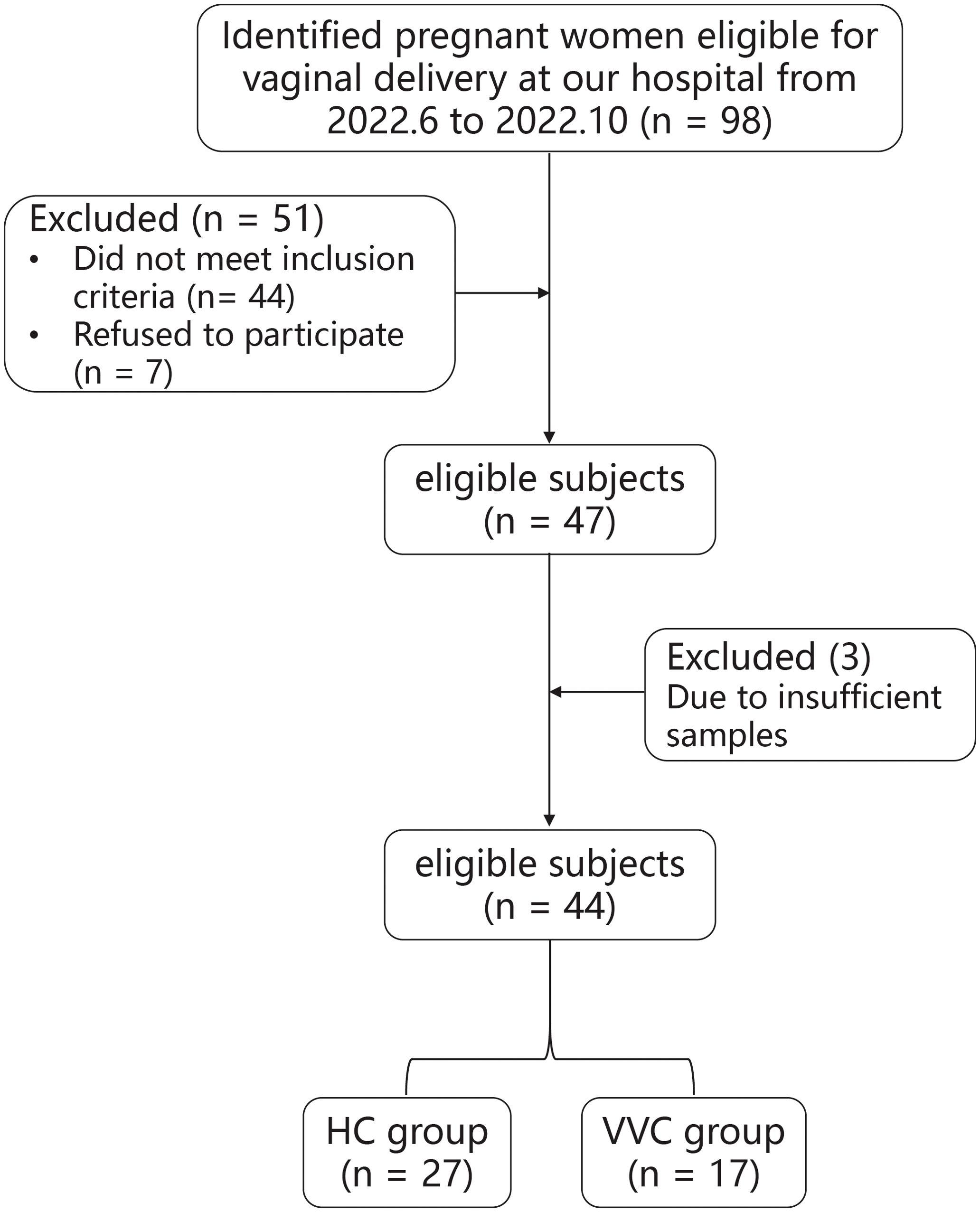
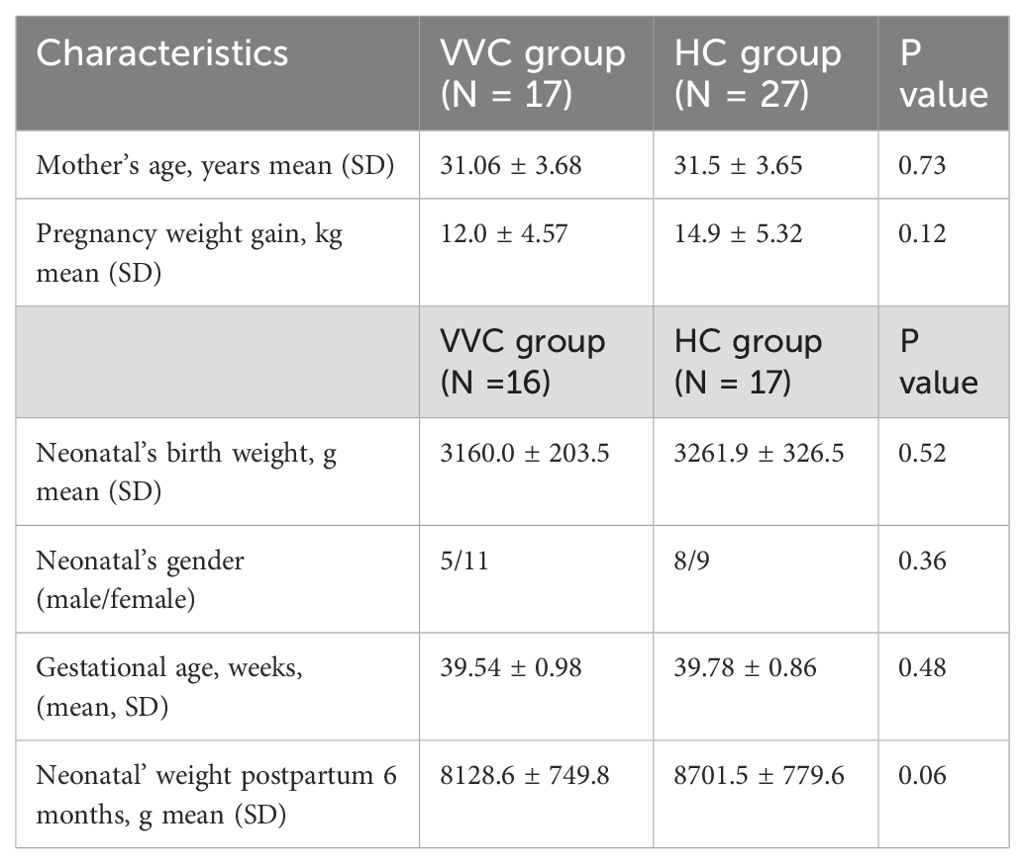
The flowchart illustrating the subject recruitment and exclusion process was detailed in Figure 1. Ultimately, forty-four pregnant women (VVG group, n = 17; HC group, n = 27) were included in this study. All participants were Han Chinese residing in Shenzhen city. The characteristics of all participants were summarized in Table 1, there were no differences in terms of maternal age, gestational age, neonatal birth length and neonatal gender. Due to insufficient DNA in some meconium samples, only 16 neonates in the VVC group and 17 neonates in the HC groups were retained for further analysis. Then in the data analysis process, two steps were undertaken: first, the number of maternal vaginal samples was adjusted to match the number of newborn meconium samples, and second, all maternal vaginal samples were standardized to ensure consistency across groups.
Comparisons of vaginal microbiome before delivery between the VVC and HC groups
Alpha diversity was assessed using the observed feature number, Pielou index, and Shannon index. The results demonstrated that the mean values of the observed feature number significantly increased in the VVC group compared to the HC group (P = 0.043), while no significant differences were observed in the Pielou (P = 0.19) and Shannon indices (P = 0.056) (Figure 2A). Similar findings were observed in the analysis of all 44 study subjects (Supplementary Figure S1A). Furthermore, PCoA with PERMANOVA based on Bray-Curtis distance showed a trend toward significant differences in vaginal microbial communities between the two groups (R2 = 0.061, P = 0.069, Figure 2B). Upon expanding the sample size, a significant difference was confirmed (R2 = 0.065, P = 0.015, Supplementary Figure S1B).
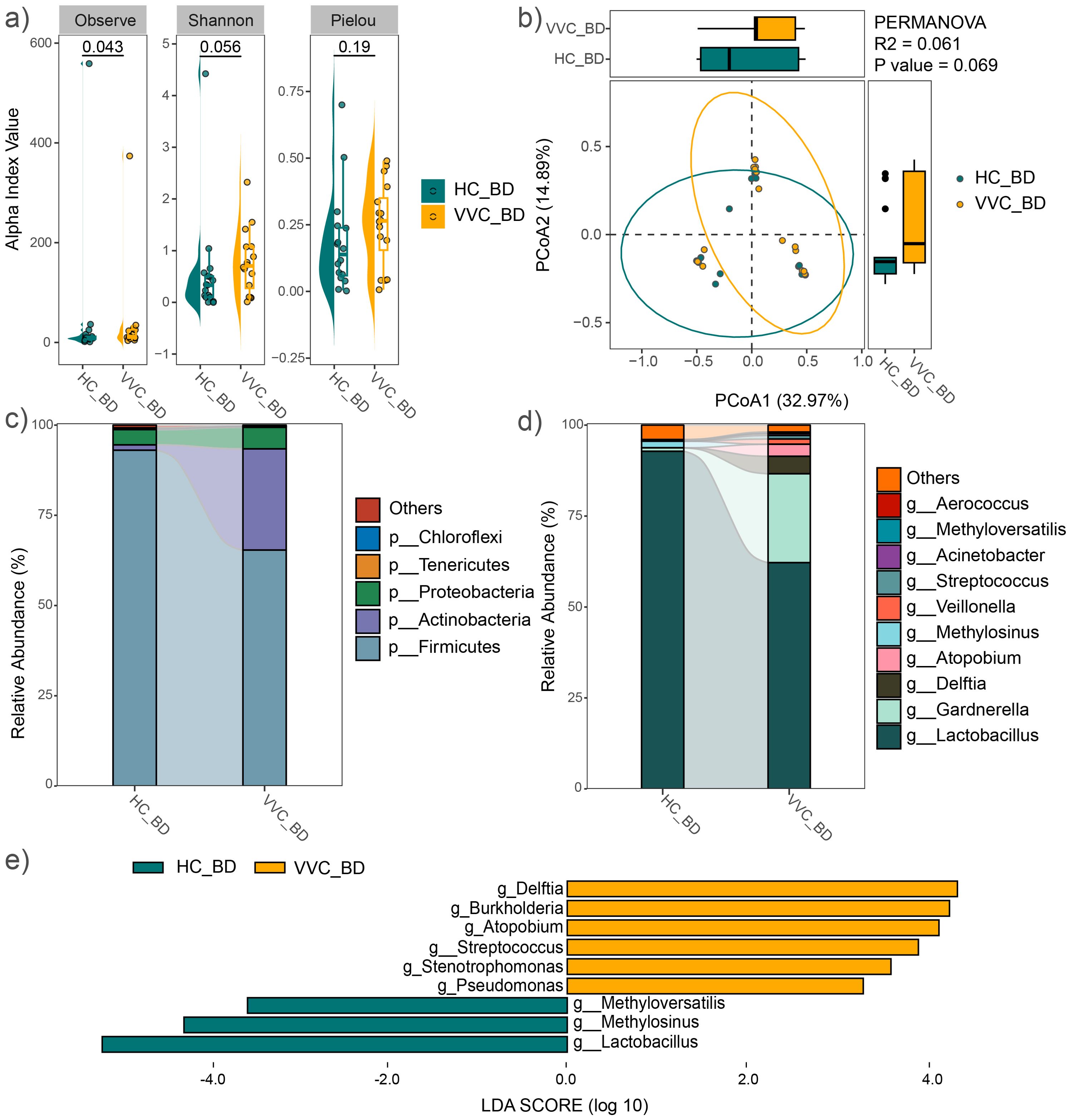
Figure 2. Comparison of vaginal microbial community before delivery between the VVC and HC groups. (A) Comparisons of alpha diversity indices. (B) PCoA plot illustrating the differences in microbial communities between the two groups. (C) Relative abundances of the dominant phyla. (D) Relative abundances of the predominant genus. (E) Significant different genus between the two groups.
The average relative abundance of dominant phyla, including Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes, and Tenericutes, was shown in Figure 2C. Firmicutes and Actinobacteria together accounted for over 90% of the microbial composition in both groups. At the genus level, the predominant taxa included Lactobacillus, Gardnerella, Delftia, Atopobium, Methylosinus, Veillonella, Streptococcus, Acinetobacter, Methyloversatilis, and Aerococcus (Figure 2D). Similar microbial distributions were observed across all 44 study subjects (Supplementary Figures S1C, D). LEfSe analysis revealed significant differences at the genus level between the two groups, with Delftia, Burkholderia, Atopobium, Streptococcus, Stenotrophomonas and Pseudomonas significantly enriched in the VVC group, while Methyloversatilis, Methylosinus, and Lactobacillus significantly decreased compared to the HC group (Figure 2E). Expanding sample size confirmed a significant increase in Delftia, Burkholderia and Stenotrophomonas, along with a significant decrease in Methylosinus and Lactobacillus were observed in the VVC group compared to the HC group (Supplementary Figure S1E).
Comparisons of vaginal microbiome after delivery between the VVC and HC groups
After delivery, the mean values of the observed feature number, Pielou and Shannon indices showed no significant differences between the VVC and HC groups (Figure 3A). Consistent findings were observed in the analysis of all 44 study subjects (Supplementary Figure S2A). However, PCoA with PERMANOVA based on Bray-Curtis distance revealed significant differences in the vaginal microbial communities between the two groups (R2 = 0.1394, P = 0.0001, Figure 3B). When the sample size was expanded, this significant difference was further confirmed (Supplementary Figure S2B).
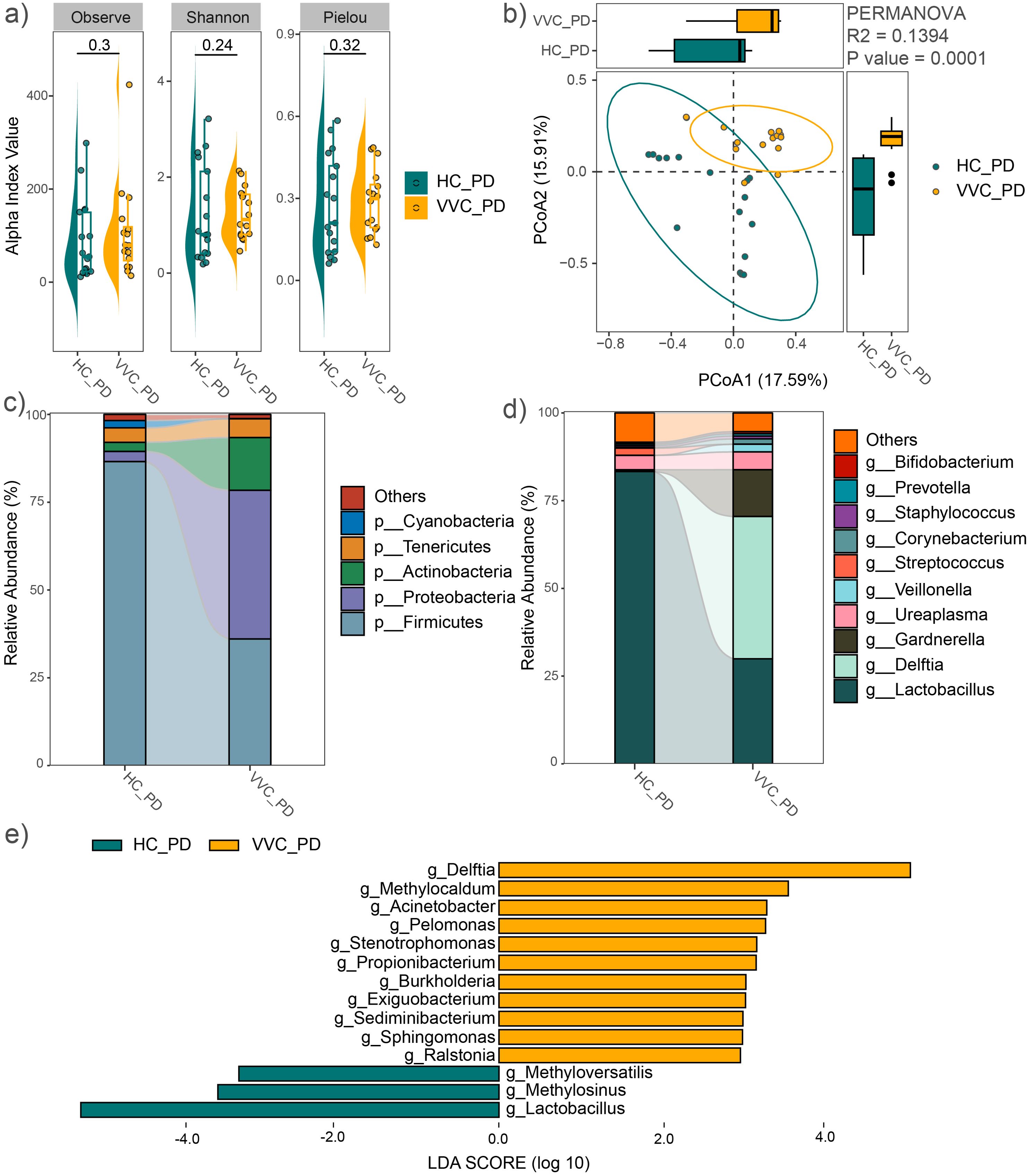
Figure 3. Comparison of vaginal microbial community after delivery between the VVC and HC groups. (A) Comparisons of alpha diversity indices. (B) PCoA plot illustrating the differences in microbial communities between the two groups. (C) Relative abundances of the dominant phyla. (D) Relative abundances of the predominant genus. (E) Significant different genus between the two groups.
The average relative abundance of dominant phyla was shown in Figure 3C, similar to the vaginal microbiome before delivery, including Firmicutes, Proteobacteria, Actinobacteria, Tenericutes, and Bacteroidetes. The predominant genera included Lactobacillus, Delftia, Gardnerella, Ureaplasma, Veillonella, Streptococcus, Corynebacterium, Staphylococcus, Prevotella, Enterococcus, and Bifidobacterium (Figure 3D). These findings were confirmed across all study subjects (Supplementary Figures S2C, D). LEfSe analysis revealed significant genus-level differences between the two groups, with Delftia, Methylocaldum, Acinetobacter, Pelomonas, Stenotrophomonas, Propionibacterium, Burkholderia, Exiguobacterium, Sediminibacterium, Sphingomonas, and Ralstonia significantly enriched, while Methyloversatilis, Methylosinus, and Lactobacillus significantly decreased in the VVC group compared to the HC group (Figure 3E). These differences in genus were also confirmed with the expanded sample size (Supplementary Figure S2E).
Consistently, both before delivery and after delivery, Delftia, Burkholderia, and Stenotrophomonas were significantly enriched in the VVC group. All these genera belong to the phylum Proteobacteria. This was reflected in the differences at the phylum level, with Proteobacteria significantly enriched in the VVC group at both BD (LDA = 4.61, P < 0.001) and PD (LDA = 5.33, P < 0.001) phases.
Comparisons of neonatal meconium microbiome between the VVC and HC groups
For the neonatal meconium microbiome, the mean values of the observed feature number, Pielou and Shannon indices showed no differences between the VVC and HC groups (Figure 4A). However, PCoA with PERMANOVA revealed significant differences in the neonatal meconium microbial communities between the two groups (R2 = 0.0518, P = 0.0001). The VVC group exhibited more closely clustered microbial communities, indicating a more homogeneous community structure among the neonates in the VVC group (Figure 4B).
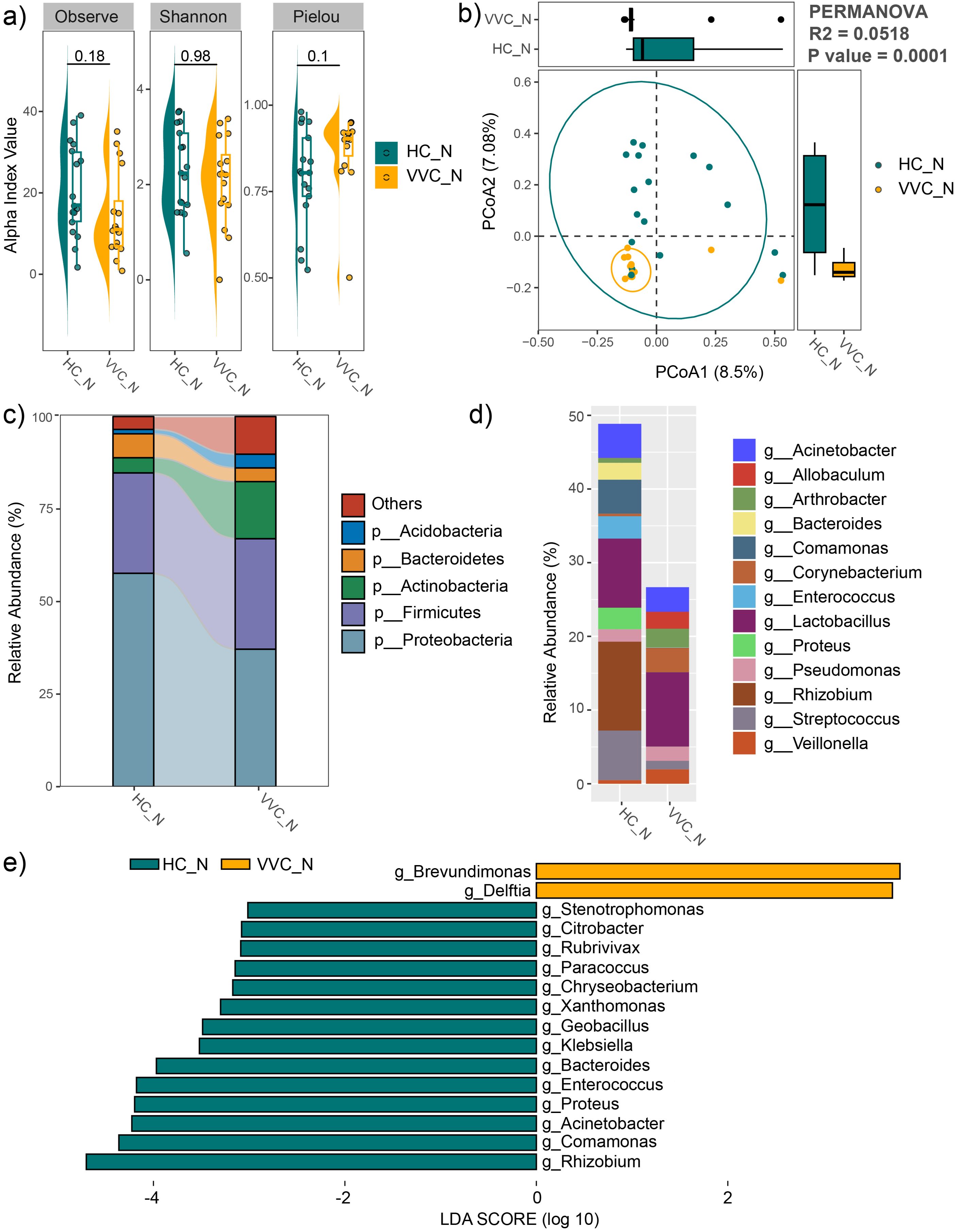
Figure 4. Comparison of neonatal meconium microbiome between the VVC and HC groups. (A) Comparisons of alpha diversity indices. (B) PCoA plot illustrating the differences in microbial communities between the two groups. (C) Relative abundances of the dominant phyla. (D) Relative abundances of the predominant genus. (E) Significant different genus between the two groups.
The average relative abundance of dominant phyla was shown in Figure 4C, including Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Acidobacteria. The predominant genus included Veillonella, Streptococcus, Rhizobium, Proteus, Lactobacillus, Enterococcus, Corynebacterium, Comamonas, Bacteroides, Arthrobacter, Allobaculum and Acinetobacter (Figure 4D). LEfSe analysis revealed significant differences at the genus level between the two groups, with Brevundimonas, and Delftia significantly enriched, while Stenotrophomonas, Citrobacter, Rubrivivax, Paracoccus, Chryseobacterium, Xanthomonas, Geobacillus, Klebsiella, Bacteroides, Enterococcus, Proteus, Acinetobacter, Comamonas and Rhizobium significantly decreased in the VVC group compared to the HC group (Figure 4E).
Lactobacillus species’ differences between the VVC and HC groups
It is well-established that the female genital tract is predominantly inhabited by Lactobacillus species, which protect against infections through the production of L-lactic acid and H2O2. Pathological changes in their profile may render the vagina susceptible to infections. In this study, we identified various Lactobacillus species, including L. iners, L. helveticus, L. reuteri, L. zeae, L. coleohominis, L. mucosae, L. manihotivorans, L. salivarius and L. acidipiscis (Supplementary Figure S3). Among these, L. iners and L. helveticus were the most abundant species in both vaginal and meconium samples (Supplementary Figure S3). Differences in these Lactobacillus species were further investigated using LEfSe analysis. At the BD phase, no significant differences in the relative abundance of these Lactobacillus species were observed. However, the overall abundance of the genus Lactobacillus was significantly decreased in the VVC group compared to the HC group (LDA = 5.24, P = 0.029), a result confirmed across all 44 study subjects (LDA = 5.14, P = 0.027). A similar pattern was observed at the PD phase, with a significant decrease in the genus Lactobacillus in the VVC group (LDA = 5.44, P < 0.01), particularly in species L. helveticus (LDA = 5.20, P = 0.014). Only the significant difference in genus Lactobacillus was confirmed with the expanded sample size (LDA = 5.32, P = 0.001). In contrast, neonatal meconium samples showed no significant differences of the genus Lactobacillus, though L. unclassified (LDA = 2.67, P = 0.021), L. salivarius (LDA = 2.85, P = 0.042), L. helveticus (LDA = 4.47, P = 0.023) were significantly more abundant in the HC group.
Comparisons of vaginal microbiome between the pre- and post- delivery phases
In both the VVC and HC groups, the observed feature number and Shannon index significantly increased in the PD phase compared to the BD phase (Figures 5A, B), indicating an increase of microbial diversity after delivery. Moreover, PCoA with PERMANOVA based on Bray-Curtis distance revealed significant differences in the vaginal microbial communities between pre- and post- delivery (R2 = 0.0429, P = 0.0254 in the HC group, Figure 5C; R2 = 0.0810, P = 0.001 in the VVC group, Figure 5D). Additionally, after delivery, the relative abundance of Lactobacillus significantly decreased in both the HC and VVC groups (LDA = 4.98, P < 0.001 in the HC group, Figure 5E; LDA = 5.18, P = 0.026 in the VVC group; Figure 5F).
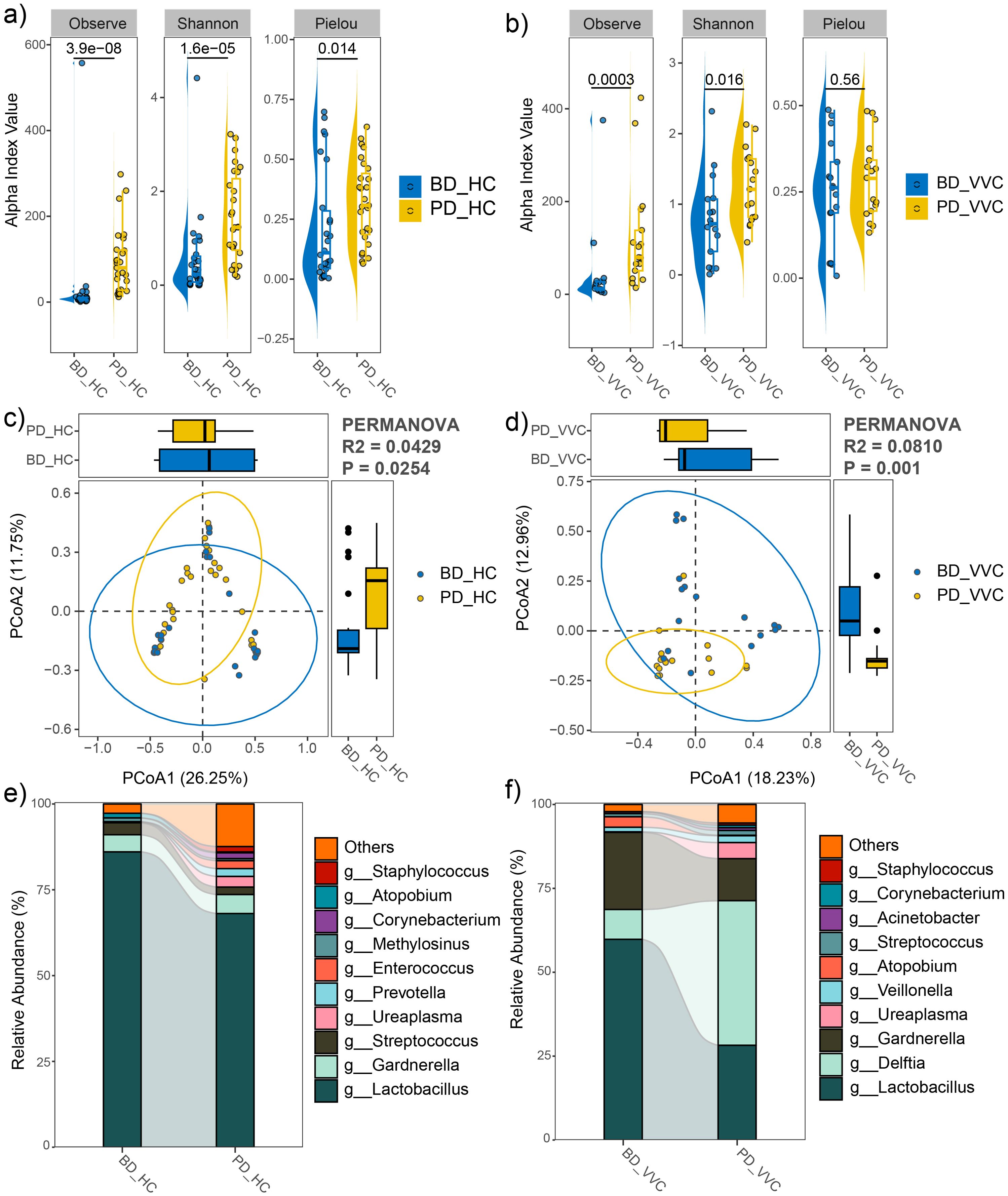
Figure 5. Comparison of vaginal microbial community between pre- and post- delivery phases in both the HC and VVC groups. (A) Alpha diversity indices between two phases in the HC group. (B) Alpha diversity indices between two phases in the VVC group. (C) PCoA plot illustrating the differences in microbial communities between the two phases in the HC group. (D) PCoA plot illustrating the differences in microbial communities between the two phases in the VVC group. (E) Relative abundances of the predominant genus in the HC group, a decrease of Lactobacillus was observed. (F) Relative abundances of the predominant genus in the VVC group, a decrease of Lactobacillus was observed.
Discussion
This study aimed to characterize the vaginal microbial profile in pregnant women affected by vulvovaginal candidiasis and its effect on their offspring. The results revealed significant differences in the vaginal microbiome associated with VVC, as evidenced by PCoA based on the Bray-Curtis dissimilarity index at both pre- and post-delivery phases. Additionally, this study showed that the vaginal microbiome of most women in HC group was dominated by Lactobacillus, consistent with the typical vaginal microbiota composition in healthy women as previously reported (DiGiulio et al., 2015; MacIntyre et al., 2015; Zhang et al., 2022a). The significant decrease in the relative abundance of Lactobacillus observed in the VVC group is consistent with previous studies (Ceccarani et al., 2019). Moreover, the meconium microbiome of the offspring of women with VVC also presented significant differences compared to the healthy control group, with L. salivarius and L. helveticus significantly decreased. Lactobacillus species help maintain an acidic environment (low pH) in the vagina, which is essential for preventing the overgrowth of harmful bacteria, yeast, and viruses (Wu et al., 2024). These bacteria produce substances such as lactic acid, hydrogen peroxide, and bacteriocins that inhibit the growth of pathogenic microorganisms (Pendharkar et al., 2023). Lactobacillus can also form biofilms that act as a physical barrier against pathogens, providing an additional layer of protection (Chee et al., 2020). L. salivarius and L. helveticus are types of lactic acid bacteria commonly used as probiotic for infants (Manzano et al., 2017; Chen et al., 2023). These Lactobacillus species aid digestion and nutrient absorption, enhance the production of secretory immunoglobulin A, which are essential for neonatal growth and development, and help reduce the risk of infections and inflammation (Gritz and Bhandari, 2015). Additionally, they support the maturation of the neonatal immune system. The significant decrease in these bacteria in both maternal vaginal and neonatal gut microbiome may be closely related to their health status.
Accompanying with the decrease of Lactobacillus in the vaginal microbiome of the VVC group was a significant increase in genera such as Delftia and Burkholderia from the phylum Proteobacteria. Delftia species, which have been isolated from vaginal discharge, are found related to cervical intra-epithelial neoplasia and unexplained recurrent pregnancy loss (Wu et al., 2020; Mori et al., 2023). Some strains of Delftia carry multiple antibiotic resistance genes, complicating treatment if infections occur (Zhang et al., 2024). An increase in Burkholderia is associated with negative reproductive outcomes, such as recurrent implantation failure in assisted reproduction technology procedures (Chen et al., 2024). Interestingly, a significant increase in Delftia was observed in the meconium microbiome of the offspring of women with VVC. Some species of Delftia are pathogenic to infants and have been associated with respiratory infection (Ranc et al., 2018; Agarwal et al., 2023). Unfortunately, this study was unable to identify the specific Delftia species based on 16S sequencing data, which limited further analysis. Collectively, their increase may be harmful to maternal and neonatal health.
An interesting finding in this study was the significant alterations of vaginal microbiome observed between the pre- and post- delivery phases in both the HC and VVC groups, with a notable decrease in Lactobacillus abundance after delivery. This is consistent with previous reports by Li et al (Li et al., 2022, 2023), which suggest associations with povidone-iodine use during delivery. Our results provide additional evidence supporting these findings.
Furthermore, the significant decrease in Lactobacillus species and the concomitant increase in Delftia species observed in the VVC group were consistent across both the maternal vaginal microbiome and neonatal meconium microbiome. This parallel in microbial shifts between mothers and their neonates suggests a potential intergenerational transmission associated with VVC. However, to validate these findings, it is essential to enlarge the sample size and recruit participants from multiple hospitals to account for potential confounding factors such as diet and demographic distances. Additionally, conducting metagenomic sequencing would provide deeper insights into the observed alterations in the maternal vaginal and neonatal meconium microbiome with VVC. Collecting comprehensive health data for both mothers and their offspring in future studies will also be crucial. These steps would strengthen the robustness and applicability of the study’s findings, paving the way for more comprehensive and insightful research in this critical area.
In conclusion, a notable distinction was observed in the maternal vaginal microbiome and neonatal meconium microbiome between VVC and healthy control groups. The persistence of these differences and their potential impact on growth and development will be the focus of forthcoming studies. This ongoing investigation will shed further light on the intricate relationship between vaginal microbial composition in pregnant women with VVC.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://db.cngb.org/search/project/CNP0006080.
Ethics statement
The studies involving humans were approved by ethical committee of Shenzhen Nanshan Maternity and Child Healthcare Hospital (No. NSFYEC-KY-2022018). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
HZ: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Writing – original draft, Writing – review & editing. HL: Formal analysis, Funding acquisition, Methodology, Visualization, Writing – original draft. RZ: Funding acquisition, Investigation, Resources, Writing – review & editing. LJ: Data curation, Investigation, Resources, Writing – review & editing. JC: Data curation, Visualization, Writing – review & editing. CN: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. WH: Conceptualization, Formal analysis, Funding acquisition, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 82101811), Science and Technology Planning Project of Shenzhen Nanshan District (No. NS2022060 and NS2022128), Shenzhen Science and Technology Program (No. RCJC20231211085923029), Shenzhen Science and Technology Innovation Commission Fund (No. JCYJ20230807152302005), Guangdong Basic and Applied Basic Foundation (2023A1515010674), Guangzhou Science and Technology Key Research and Development Program Project (2024B03J1297), Sanming Project of Medicine in Shenzhen (No. SZSM202311027) and Guangdong High-level Hospital Construction Fund.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1480200/full#supplementary-material
References
Agarwal, N., Jindal, A., Bhargava, A. (2023). Delftia acidovorans: rarely a pathogen: A case report. Pediatr. Infect. Dis. J. 42, e130–e131. doi: 10.1097/INF.0000000000003818
Aguin, T. J., Sobel, J. D. (2015). Vulvovaginal candidiasis in pregnancy. Curr. Infect. Dis. Rep. 17, 30. doi: 10.1007/s11908-015-0462-0
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Ceccarani, C., Foschi, C., Parolin, C., D’Antuono, A., Gaspari, V., Consolandi, C., et al. (2019). Diversity of vaginal microbiome and metabolome during genital infections. Sci. Rep. 9, 14095. doi: 10.1038/s41598-019-50410-x
Chatzivasileiou, P., Vyzantiadis, T.-A. (2019). Vaginal yeast colonisation: From a potential harmless condition to clinical implications and management approaches-A literature review. Mycoses 62, 638–650. doi: 10.1111/myc.12920
Chee, W. J. Y., Chew, S. Y., Than, L. T. L. (2020). Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb. Cell Factories 19, 203. doi: 10.1186/s12934-020-01464-4
Chen, D.-L., Dai, Y.-C., Zheng, L., Chen, Y.-L., Zhang, Y.-L., Tang, Z.-P. (2021a). Features of the gut microbiota in ulcerative colitis patients with depression: A pilot study. Med. (Baltimore) 100, e24845. doi: 10.1097/MD.0000000000024845
Chen, X., Lu, Y., Chen, T., Li, R. (2021b). The female vaginal microbiome in health and bacterial vaginosis. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.631972
Chen, J.-F., Ou-Yang, M.-C., Hsia, K.-C., Neonatal Probiotic Use and Safety Research Group, Li, C.-M., Yeh, Y.-T., et al. (2023). A Three-Arm, Randomized, Double-Blind, Placebo-Controlled Study to Evaluate the Safety of Lactobacillus salivarius AP-32 and Bifidobacterium animalis CP-9 Used Individually in Healthy Infants. Nutrients 15, 3426. doi: 10.3390/nu15153426
Chen, X., Sui, Y., Gu, J., Wang, L., Sun, N. (2024). The implication of the vaginal microbiome in female infertility and assisted conception outcomes. doi: 10.21203/rs.3.rs-4194198/v1
Denning, D. W., Kneale, M., Sobel, J. D., Rautemaa-Richardson, R. (2018). Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect. Dis. 18, e339–e347. doi: 10.1016/S1473-3099(18)30103-8
DiGiulio, D. B., Callahan, B. J., McMurdie, P. J., Costello, E. K., Lyell, D. J., Robaczewska, A., et al. (2015). Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. 112, 11060–11065. doi: 10.1073/pnas.1502875112
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Farr, A., Effendy, I., Frey Tirri, B., Hof, H., Mayser, P., Petricevic, L., et al. (2021). Guideline: Vulvovaginal candidosis (AWMF 015/072, level S2k). Mycoses 64, 583–602. doi: 10.1111/myc.13248
Fettweis, J. M., Serrano, M. G., Brooks, J. P., Edwards, D. J., Girerd, P. H., Parikh, H. I., et al. (2019). The vaginal microbiome and preterm birth. Nat. Med. 25, 1012–1021. doi: 10.1038/s41591-019-0450-2
Gaziano, R., Sabbatini, S., Monari, C. (2023). The interplay between Candida albicans, vaginal mucosa, host immunity and resident microbiota in health and disease: an overview and future perspectives. Microorganisms 11, 1211. doi: 10.3390/microorganisms11051211
Greenbaum, S., Greenbaum, G., Moran-Gilad, J., Weintraub, A. Y. (2019). Ecological dynamics of the vaginal microbiome in relation to health and disease. Am. J. Obstet. Gynecol. 220, 324–335. doi: 10.1016/j.ajog.2018.11.1089
Gritz, E. C., Bhandari, V. (2015). The human neonatal gut microbiome: a brief review. Front. Pediatr. 3. doi: 10.3389/fped.2015.00017
Kervinen, K., Kalliala, I., Glazer-Livson, S., Virtanen, S., Nieminen, P., Salonen, A. (2019). Vaginal microbiota in pregnancy: Role in induction of labor and seeding the neonate’s microbiota? J. Biosci. 44, 116. doi: 10.1007/s12038-019-9925-z
Li, H., Jiang, J., Nie, C., Xiao, B., Li, Q., Yu, J. (2022). Community structure and ecological network’s changes of vaginal microbiome in women right after delivery. Front. Pediatr. 10. doi: 10.3389/fped.2022.750860
Li, H., Zhang, H., Geng, L., Huang, H., Nie, C., Zhu, Y. (2023). Association between vaginal microbiome alteration and povidone iodine use during delivery. BMC Microbiol. 23, 348. doi: 10.1186/s12866-023-03014-5
MacIntyre, D. A., Chandiramani, M., Lee, Y. S., Kindinger, L., Smith, A., Angelopoulos, N., et al. (2015). The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 5, 8988. doi: 10.1038/srep08988
Manzano, S., De Andrés, J., Castro, I., Rodríguez, J. M., Jiménez, E., Espinosa-Martos, I. (2017). Safety and tolerance of three probiotic strains in healthy infants: a multi-centre randomized, double-blind, placebo-controlled trial. Benef. Microbes 8, 569–578. doi: 10.3920/BM2017.0009
Mori, R., Hayakawa, T., Hirayama, M., Ozawa, F., Yoshihara, H., Goto, S., et al. (2023). Cervicovaginal microbiome in patients with recurrent pregnancy loss. J. Reprod. Immunol. 157, 103944. doi: 10.1016/j.jri.2023.103944
Pendharkar, S., Skafte-Holm, A., Simsek, G., Haahr, T. (2023). Lactobacilli and their probiotic effects in the vagina of reproductive age women. Microorganisms 11, 636. doi: 10.3390/microorganisms11030636
Ranc, A., Dubourg, G., Fournier, P. E., Raoult, D., Fenollar, F. (2018). Delftia tsuruhatensis, an emergent opportunistic healthcare-associated pathogen. Emerg. Infect. Dis. 24, 594–596. doi: 10.3201/eid2403.160939
Ravel, J., Gajer, P., Abdo, Z., Schneider, G. M., Koenig, S. S. K., McCulle, S. L., et al. (2011). Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. 108, 4680–4687. doi: 10.1073/pnas.1002611107
Schuster, H. J., De Jonghe, B. A., Limpens, J., Budding, A. E., Painter, R. C. (2020). Asymptomatic vaginal Candida colonization and adverse pregnancy outcomes including preterm birth: a systematic review and meta-analysis. Am. J. Obstet. Gynecol. MFM 2, 100163. doi: 10.1016/j.ajogmf.2020.100163
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Turunen, J., Tejesvi, M. V., Paalanne, N., Hekkala, J., Lindgren, O., Kaakinen, M., et al. (2021). Presence of distinctive microbiome in the first-pass meconium of newborn infants. Sci. Rep. 11, 19449. doi: 10.1038/s41598-021-98951-4
Wu, M., Gao, J., Wu, Y., Li, Y., Chen, Y., Zhao, F., et al. (2020). Characterization of vaginal microbiota in Chinese women with cervical squamous intra-epithelial neoplasia. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc 30, 1500–1504. doi: 10.1136/ijgc-2020-001341
Wu, L.-Y., Yang, T.-H., Ou, Y.-C., Lin, H. (2024). The role of probiotics in women’s health: An update narrative review. Taiwan. J. Obstet. Gynecol. 63, 29–36. doi: 10.1016/j.tjog.2023.09.018
Xiao, B., D., A., Qin, H., Mi, L., Zhang, D. (2022). Correlation analysis of vaginal microbiome changes and bacterial vaginosis plus vulvovaginal candidiasis mixed vaginitis prognosis. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.860589
Xu, S., Zhan, L., Tang, W., Wang, Q., Dai, Z., Zhou, L., et al. (2023). MicrobiotaProcess: A comprehensive R package for deep mining microbiome. Innovation 4, 100388. doi: 10.1016/j.xinn.2023.100388
Zhang, Y., Xu, X., Yu, L., Shi, X., Min, M., Xiong, L., et al. (2022b). Vaginal microbiota changes caused by HPV infection in Chinese women. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.814668
Zhang, X., Zhai, Q., Wang, J., Ma, X., Xing, B., Fan, H., et al. (2022a). Variation of the vaginal microbiome during and after pregnancy in Chinese women. Genomics Proteomics Bioinf. 20, 322–333. doi: 10.1016/j.gpb.2021.08.013
Zhang, L., Zhang, X., Bai, H., Li, T., Zhang, Z., Zong, X., et al. (2024). Characterization and genome analysis of the Delftia lacustris strain LzhVag01 isolated from vaginal discharge. Curr. Microbiol. 81, 232. doi: 10.1007/s00284-024-03758-x
Zhao, F., Chen, Y., Gao, J., Wu, M., Li, C., Wang, Z., et al. (2021). Characterization of vaginal microbiota in women with recurrent spontaneous abortion that can be modified by drug treatment. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.680643
Zhao, C., Li, Y., Chen, B., Yue, K., Su, Z., Xu, J., et al. (2023). Mycobiome study reveals different pathogens of vulvovaginal candidiasis shape characteristic vaginal bacteriome. Microbiol. Spectr. 11, e03152–e03122. doi: 10.1128/spectrum.03152-22
Keywords: vulvovaginal candidiasis, vaginal microbiome, meconium microbiome, microbial community, genital infection
Citation: Zhang H, Li H, Zhang R, Ji L, Chen J, Nie C and Huang W (2024) Alterations of the paired maternal vaginal microbiome and neonatal meconium microbiome in vulvovaginal candidiasis positive pregnant women. Front. Cell. Infect. Microbiol. 14:1480200. doi: 10.3389/fcimb.2024.1480200
Received: 13 August 2024; Accepted: 25 November 2024;
Published: 13 December 2024.
Edited by:
António Machado, University of the Azores, PortugalReviewed by:
Daniel C. Stein, University of Maryland, College Park, United StatesVonetta Edwards, University of Maryland, United States
Copyright © 2024 Zhang, Li, Zhang, Ji, Chen, Nie and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weimin Huang, aHdtbmV0QDIxY24uY29t; Chuan Nie, Y2h1YW5uaWVAc2luYS5jb20=
†These authors have contributed equally to this work
 Hongqin Zhang
Hongqin Zhang Hongping Li
Hongping Li Ruolin Zhang
Ruolin Zhang Lingxia Ji1
Lingxia Ji1 Chuan Nie
Chuan Nie Weimin Huang
Weimin Huang