- Department of Urology II, The First Hospital of Jilin University, Changchun, China
Kidney transplantation (KT) is a life-saving treatment for patients with end-stage renal disease, but post-transplant infections remain one of the most significant challenges. These infections, caused by a variety of pathogens, can lead to prolonged hospitalization, graft dysfunction, and even mortality, particularly in immunocompromised patients. Traditional diagnostic methods often fail to identify the causative organisms in a timely manner, leading to delays in treatment and poorer patient outcomes. This review explores the application of metagenomic next-generation sequencing (mNGS) in the diagnosis of post-KT infections. mNGS allows for the rapid, comprehensive detection of a wide range of pathogens, including bacteria, viruses, fungi, and parasites, without the need for culture-based techniques. We discuss the advantages of mNGS in early and accurate pathogen identification, its role in improving patient management, and the potential challenges in its clinical implementation. Additionally, we consider the future prospects of mNGS in overcoming current diagnostic limitations and its potential for guiding targeted therapies, particularly in detecting antimicrobial resistance and emerging pathogens. This review emphasizes the promise of mNGS as an essential tool in improving the diagnosis and treatment of infections in KT recipients.
1 Introduction
Kidney transplantation (KT) is a surgical procedure that replaces a failed kidney with a functioning one from a donor, offering a life-saving treatment for patients with end-stage renal disease (ESRD) (Reimold et al., 2024). This procedure is especially critical for individuals whose renal function cannot be adequately supported by dialysis or other medical management (Navarrete, 2009; Unruh and Dew, 2014; Viklicky et al., 2020). Over the years, advancements in surgical techniques and immunosuppressive therapies have significantly improved the success rate and accessibility of KT (Lim et al., 2017; Szumilas et al., 2023). However, long-term postoperative management remains a major challenge, with complications such as graft rejection and infection posing significant risks to patient recovery and long-term outcomes (Kim et al., 2023; Voora et al., 2023).
Post-transplant infections are among the most common and severe complications following KT (Agrawal et al., 2022). These infections, caused by bacteria, viruses, fungi, or parasites, are a major clinical concern due to the lifelong use of immunosuppressive therapy, which compromises the immune system’s ability to fight off pathogens (Illesy et al., 2016; Dandamudi et al., 2019; Møller et al., 2021). Such infections can result in prolonged hospitalization, impaired graft function, and in severe cases, mortality. Early and accurate diagnosis of these infections is critical to ensuring timely intervention and effective treatment (Bharati et al., 2023; Pajenda et al., 2023; Grasberger et al., 2024). The selection of appropriate diagnostic methods is a key factor in identifying causative pathogens and guiding targeted therapy, thus improving outcomes for KT recipients (Bharati et al., 2023).
This review aims to summarize the currently available diagnostic methods for post-transplant infections in KT recipients, with a focus on the application of metagenomic next-generation sequencing (mNGS). We highlight the utility of mNGS in diagnosing various types of post-transplant infections, such as pulmonary, urinary tract, and bloodstream infections. Furthermore, we explore the current applications of mNGS in identifying specific pathogens associated with these infections. Lastly, we discuss the limitations and challenges of using mNGS in clinical practice and provide insights into potential improvements that could enhance its application in the management of post-transplant infections, thereby offering valuable guidance for its future use.
2 Diagnostic methods for post-transplant infections in KT
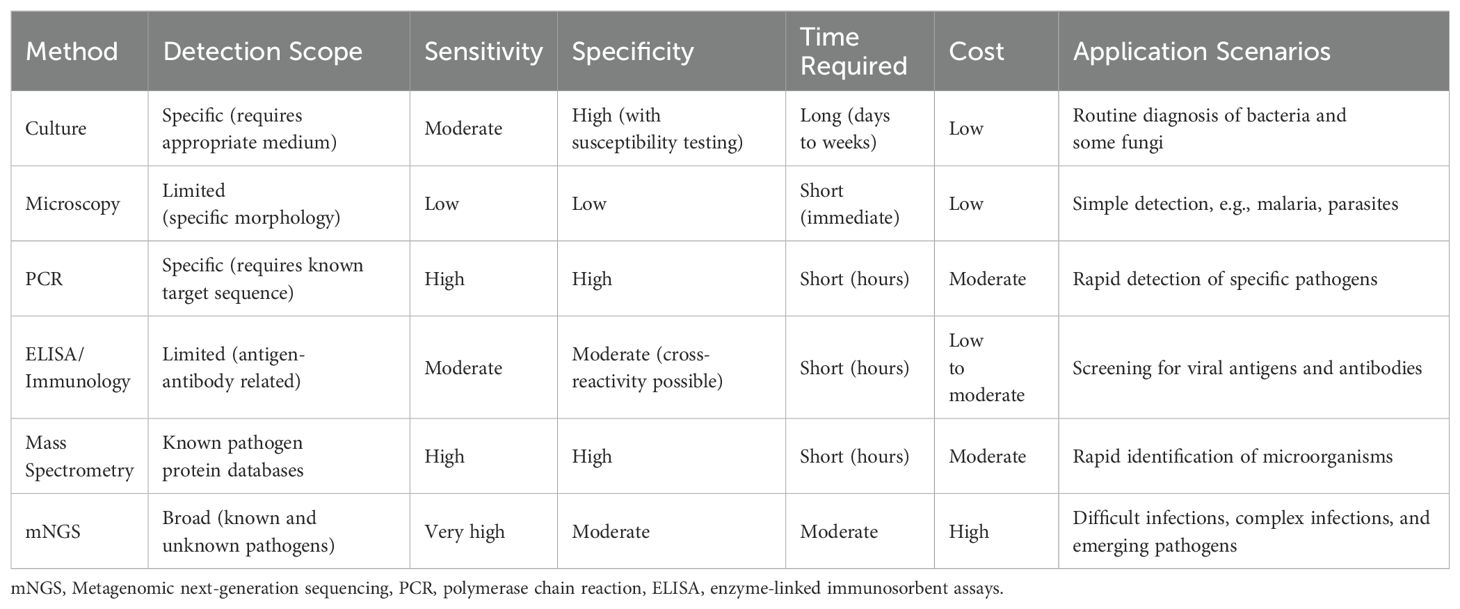
Post-transplant infections are among the most critical complications following KT, particularly in cases of complex infections involving polymicrobial or drug-resistant pathogens (Fishman, 2017; Agrawal et al., 2022). Early and accurate identification of causative pathogens is essential for effective treatment, graft survival, and preventing severe complications (Cippà et al., 2015). Conventional methods for pathogen detection, which remain widely utilized in clinical practice, include microbial culture, microscopy, serological tests, molecular diagnostic techniques, and mass spectrometry analysis (McAteer and Tamma, 2024). Microbial culture, often considered the gold standard, involves inoculating clinical specimens onto selective or differential media to promote the growth of specific microorganisms. It enables the isolation of viable organisms and facilitates antimicrobial susceptibility testing, making it a cornerstone of clinical microbiology. However, its utility is limited in time-sensitive scenarios due to the prolonged incubation period required for certain pathogens and its inability to detect fastidious or non-culturable organisms (Kim et al., 2020; Kobayashi et al., 2021). Microscopy, based on direct visualization of pathogens in stained clinical samples, offers rapid preliminary information and is particularly useful for identifying morphologically distinct pathogens. Despite its simplicity, microscopy often lacks sensitivity and specificity, especially when pathogen loads are low (Lunn et al., 2010; Ernstsen et al., 2017; Laketa, 2018). Molecular diagnostic techniques, such as polymerase chain reaction (PCR), have revolutionized infectious disease diagnostics by enabling the rapid and highly specific detection of pathogens based on their nucleic acid sequences. While PCR-based methods are powerful tools, they are inherently limited by their dependence on prior knowledge of the target sequence, making them less effective for detecting unexpected or unknown pathogens (Tsalik et al., 2018; Liu et al., 2023; Chen et al., 2024). Serological tests, such as enzyme-linked immunosorbent assays (ELISA), are commonly used for detecting pathogen-specific antigens or host antibodies, particularly in viral infections. These tests provide rapid and reliable results but may struggle to distinguish active infections from past exposures, complicating interpretation in certain clinical contexts (Lapošová et al., 2016; Luo et al., 2022; Chen et al., 2023). Mass spectrometry analysis, particularly matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), has emerged as a valuable tool in clinical microbiology for the rapid identification of microorganisms. This technique ionizes microbial proteins to generate mass spectra, which are then compared against reference databases to identify specific pathogens. MALDI-TOF MS offers high-throughput capabilities and rapid turnaround times, significantly enhancing the speed of pathogen identification. However, its effectiveness depends on the quality and comprehensiveness of the reference databases, and it may not reliably identify novel or rare pathogens (Lamy et al., 2020; Ohyama et al., 2020; Kondori et al., 2021). Collectively, these conventional methods have significantly advanced the diagnosis of post-transplant infections, but their limitations underscore the need for innovative diagnostic approaches (Table 1).
mNGS represents a transformative advance in pathogen detection, offering an unbiased and comprehensive approach that is particularly suited to the complex infections often encountered in KT recipients (Hao et al., 2023). Unlike conventional methods, mNGS does not rely on predefined assumptions about the causative pathogen. Instead, it sequences all nucleic acids (DNA or RNA) present in a clinical sample, enabling the simultaneous detection of bacteria, viruses, fungi, and parasites (Han et al., 2019). The typical workflow of mNGS begins with sample preparation, where nucleic acids are extracted from the specimen, and host DNA or RNA is depleted to enhance the detection of microbial sequences. High-throughput sequencing is then performed using advanced platforms such as Illumina or Oxford Nanopore, generating massive volumes of data that are subsequently analyzed using bioinformatics tools to align sequences with reference databases, identify pathogens, and exclude contaminants or background noise (Zhong et al., 2021; Bloemen et al., 2023; Cai et al., 2023; Ying et al., 2024) (Figure 1). The results are interpreted in conjunction with the clinical context to distinguish true pathogens from non-pathogenic or environmental organisms (Cai et al., 2023).
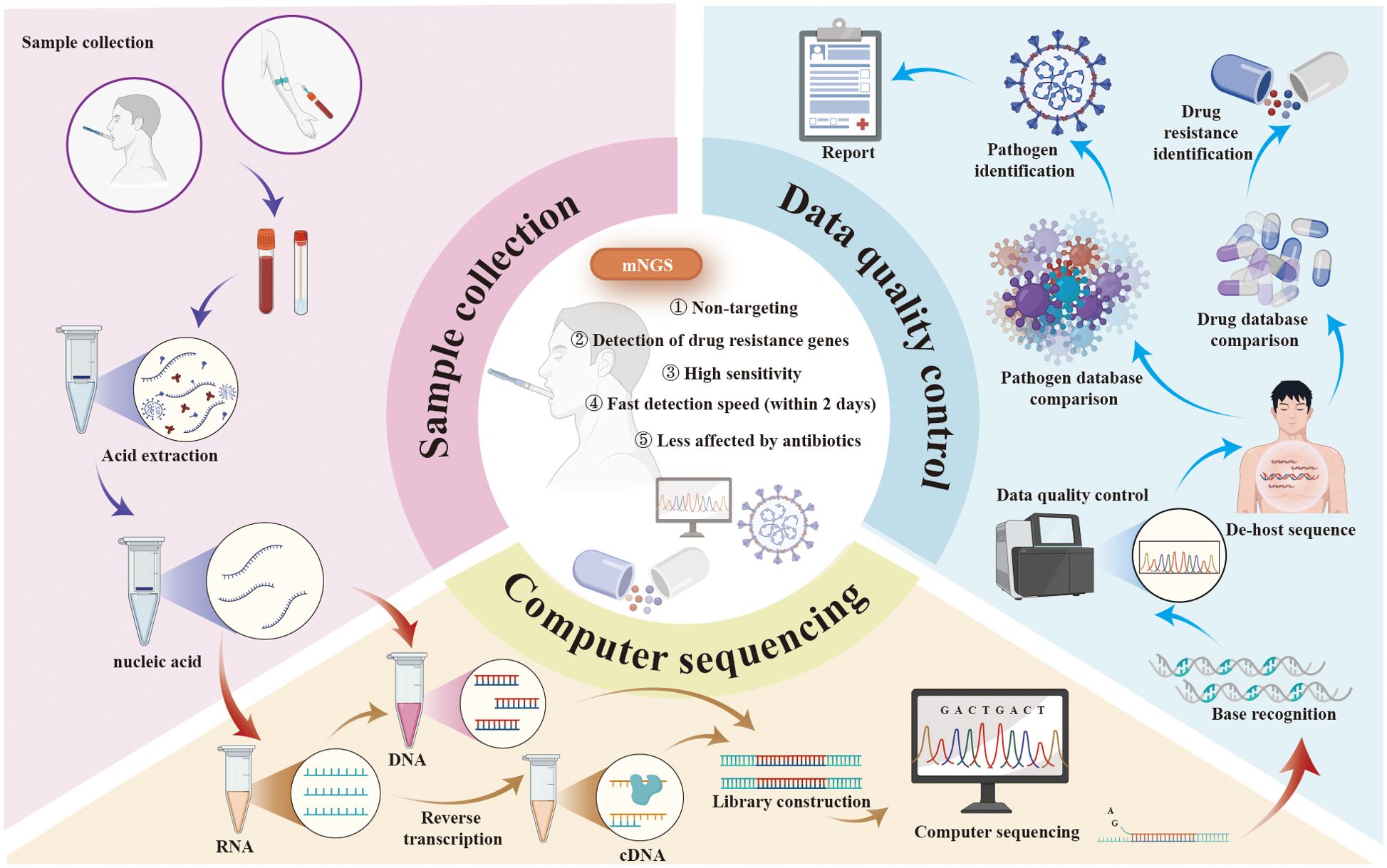
Figure 1. The protocol, advantages, and applications of mNGS. mNGS, metagenomic next-generation sequencing.
mNGS has undergone significant advancements, transitioning from Sanger sequencing to second- and third-generation technologies (Goodwin et al., 2016; Ghosh et al., 2018; Hu et al., 2021). Short-read platforms, such as Illumina, revolutionized genomic research in the mid-2000s by significantly increasing throughput and reducing costs, while long-read platforms like PacBio and Oxford Nanopore addressed limitations of short reads, including difficulties in resolving structural variants and repetitive regions (Goodwin et al., 2016; Ghosh et al., 2018; Hu et al., 2021). These innovations have expanded the applications of mNGS in areas such as diagnostics, pathogen detection, and environmental metagenomics.
Compared to traditional methods such as culture and PCR assays, mNGS offers higher sensitivity and faster detection, often delivering results within 24 hours (Chen et al., 2023; Wang et al., 2023). This rapid turnaround enables timely and targeted antimicrobial therapy, improving treatment efficacy and reducing the adverse consequences of antibiotic misuse. By enhancing the accuracy of pathogen identification, mNGS helps shorten the time to appropriate treatment, lowers healthcare costs, and significantly improves patient outcomes (Rodino and Simner, 2024). Additionally, mNGS provides valuable insights into pathogen resistance, virulence factors, and genomic variations, making it a cornerstone of precision medicine. Studies have highlighted its effectiveness in diagnosing mixed infections, detecting rare or low-frequency pathogens, and assessing host immune responses (Sun et al., 2022; Zhao et al., 2022; Zheng et al., 2022). A notable example of mNGS's impact is its role in the rapid identification of SARS-CoV-2, which expedited the development of targeted treatments and vaccines for COVID-19 (Zhong et al., 2021). In summary, mNGS represents a powerful, comprehensive diagnostic approach, revolutionizing the management of infections through enhanced accuracy, reduced treatment delays, and informed use of antimicrobials. Its contributions to precision medicine and public health underscore its critical role in modern infectious disease diagnostics (Figure 1).
3 Applications of mNGS in post-KT infections
Recent studies have explored the application of mNGS in diagnosing post-transplant infections in KT recipients, highlighting its potential to overcome many of the limitations associated with traditional diagnostics. By enabling early and precise pathogen identification, mNGS has demonstrated significant clinical value in guiding targeted therapies, improving patient outcomes, and optimizing infection management in this vulnerable population. As research continues to refine this technology, mNGS holds promise for transforming the diagnosis and treatment of post-transplant infections, offering a powerful tool for addressing the complex challenges associated with KT.
3.1 Urinary tract infections
UTIs are the most prevalent infections among KT recipients, with incidence rates ranging from 7% to 80% within the first-year post-transplantation (Coussement et al., 2018; Aydın et al., 2020; Coussement et al., 2020; Promsuwan et al., 2023; Szumilas et al., 2023). These infections are associated with significant complications, including sepsis, acute graft dysfunction, rejection, and even graft loss. Risk factors contributing to UTIs in this population include prolonged use of bladder catheters, immunosuppressive therapy, and the development of new-onset diabetes mellitus post-transplantation (Brennan et al., 2006; Cruz et al., 2018; Gerges-Knafl et al., 2020; Hsiao et al., 2021). UTIs in KT recipients can be categorized into asymptomatic bacteriuria, uncomplicated UTIs, complicated UTIs, and recurrent UTIs (Agrawal et al., 2022).
The predominant pathogens responsible for UTIs in KT recipients are Gram-negative bacteria, particularly Escherichia coli, Enterococcus faecalis (Abo Basha et al., 2019). Traditional diagnostic methods, such as urine culture, are considered the gold standard for identifying these pathogens. However, urine cultures can be time-consuming and may yield inaccurate results, especially if patients are undergoing antibiotic treatment (Janes et al., 2022; Kafi et al., 2022). Additionally, the emergence of drug-resistant strains, including extended-spectrum beta-lactamase (ESBL)-producing Gram-negative bacteria and carbapenem-resistant Enterobacteriaceae, poses significant challenges in the management of UTIs in KT recipients (Agrawal et al., 2022). Prophylactic antibiotic use in this population has shown limited efficacy and carries the risk of promoting resistant microorganisms (Wu et al., 2020). Studies have demonstrated that mNGS can detect a rich and diverse array of pathogens, with a significantly higher positive rate compared to traditional urine cultures. For instance, mNGS has shown an extraordinary positive detection rate in certain studies, surpassing the lower rates observed with conventional culture methods. Moreover, mNGS has proven effective in identifying viral, fungal, and mixed infections, which are often missed by standard diagnostic techniques. This comprehensive detection capability facilitates timely and targeted therapeutic interventions, thereby enhancing patient outcomes (Duan et al., 2022).
3.2 Pulmonary infections
Pulmonary infections are a leading cause of infection-related mortality in KT recipients (Huang et al., 2024). The spectrum of pathogens responsible for these infections is diverse and varies by region, including bacteria such as Streptococcus pneumoniae, Escherichia coli, Klebsiella species, Pseudomonas aeruginosa, and Mycobacterium tuberculosis, as well as viruses like cytomegalovirus (CMV) and BK virus (BKV) (Ahmad et al., 2020; Mangalgi et al., 2021; Qian et al., 2021; Bharati et al., 2023; Meira de Faria et al., 2023).
Conventional methods involve identifying potential pathogens using initial lab tests, imaging results, and exposure history, then conducting a thorough targeted assessment and treatment. Broad-spectrum empiric antibiotic therapy is considered for moderate to severe cases. Research has indicated that the mNGS technique is more sensitive in identifying pathogens in samples from transbronchoscopic lung biopsy (TBLB), bronchoalveolar lavage fluid (BALF), and bronchial needle brush (BB) compared to traditional culture methods (Dong et al., 2023). When diagnosing infectious pneumonia, mNGS of bronchoalveolar lavage samples provide more comprehensive results than transbronchial lung biopsy (Dong et al., 2023). MNGS can be more efficient in searching for pathogens in lung infections after KT, providing precise treatment, reducing costs, and improving cure rates, which is worthy of widespread application (Lian et al., 2024).
3.3 Bloodstream infections
BSIs are a significant concern in KT recipients, particularly within the first-year post-transplantation, with an incidence rate of approximately 10% (He et al., 2024). The primary pathogens involved are Gram-negative bacteria, notably Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa (Eviatar et al., 2022). In kidney transplant recipients, the primary sources of bloodstream infections are the urinary tract and access points for dialysis or central venous catheters. Approximately 35% of Enterobacteriaceae bacteria in this population are known to produce extended beta-lactamases (ESBL), which can hinder accurate diagnosis (Eviatar et al., 2022; Bharati et al., 2023). Traditional blood culture methods, while considered the gold standard, often require extended time to yield results and may fail to detect fastidious or non-culturable organisms, leading to delays in appropriate treatment. mNGS has emerged as a powerful diagnostic tool in this context. By sequencing all nucleic acids present in a blood sample, mNGS can rapidly identify a broad spectrum of pathogens, including bacteria, viruses, fungi, and parasites, without the need for prior knowledge of the causative agent. This comprehensive approach enables timely initiation of targeted antimicrobial therapy, which is crucial for reducing morbidity and mortality associated with BSIs in KT recipients. Moreover, mNGS can detect antimicrobial resistance genes, providing valuable information for optimizing treatment strategies (Tian et al., 2022).
3.4 Tuberculosis
Tuberculosis remains a leading cause of morbidity and mortality worldwide, and KT recipients are at an increased risk due to immunosuppressive therapy. The incidence of TB in KT recipients is significantly higher than in the general population, with rates reported to be 20-50 times greater (Vargas Barahona et al., 2022). TB in this population can result from reactivation of latent infection, donor-derived transmission, or new exposure post-transplantation (Krishnamoorthy et al., 2019). The clinical presentation is often atypical, and there is a higher likelihood of extrapulmonary or disseminated disease, complicating diagnosis and management (Zou et al., 2022).
Traditional diagnostic methods, such as the tuberculin skin test (TST) and interferon-gamma release assays (IGRAs), have limitations in immunocompromised patients, often yielding false-negative results (Li et al., 2022). Microbiological confirmation through culture is time-consuming and may delay treatment initiation. Moreover, certain anti-TB medications, like rifampicin, can interact with immunosuppressive drugs, necessitating careful management to prevent graft rejection (Hamon et al., 2023). mNGS offers a rapid and sensitive alternative for TB diagnosis in KT recipients. By detecting Mycobacterium tuberculosis DNA directly from clinical samples such as blood, sputum, or bronchoalveolar lavage fluid, mNGS facilitates early diagnosis, even in cases with atypical presentations or extrapulmonary involvement. Studies have reported that mNGS can identify TB infections with high sensitivity and specificity, enabling prompt initiation of appropriate therapy (Duan et al., 2021).
In conclusion, mNGS represents a significant advancement in the diagnosis of infections in KT recipients. Its ability to rapidly and accurately identify a wide range of pathogens, including those that are difficult to detect using conventional methods, makes it a valuable tool in the management of post-transplant infections. By facilitating early and precise pathogen identification, mNGS can guide targeted therapies, reduce the emergence of drug-resistant strains, and ultimately improve patient outcomes.
4 The role of mNGS in detecting various pathogens post-KT
KT recipients are particularly susceptible to infections due to immunosuppressive therapy, which can lead to severe complications (Sugi et al., 2019; Aydın et al., 2020; Xin et al., 2020; Dharia et al., 2022; Pilmis et al., 2023). Conventional diagnostic methods often fall short in promptly and accurately identifying the causative pathogens. mNGS has emerged as a powerful tool in this context, offering comprehensive pathogen detection across various infection types (Han et al., 2019; Liu et al., 2022; Chen et al., 2024) (Table 2).
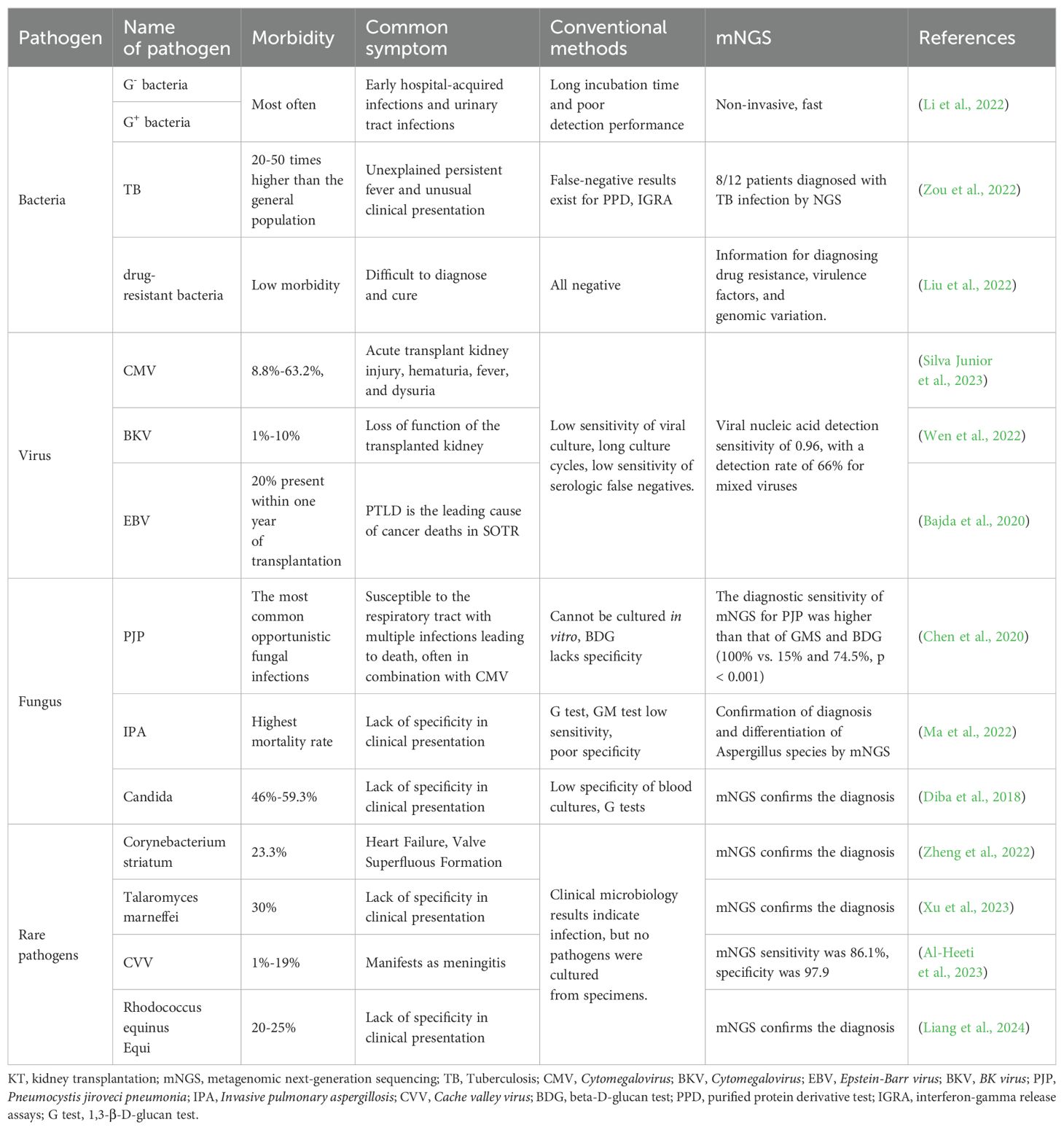
Table 2. The advantages of mNGS in the detection of various types of pathogens in post-KT recipients.
4.1 Frequent bacterial infections following KT
Bacterial infections are prevalent in the early stages following KT, with pathogens such as Escherichia coli, Streptococcus pneumoniae, Klebsiella, Pseudomonas aeruginosa, and Enterococci being common culprits (Bharati et al., 2023; Liu et al., 2024). Mixed bacterial infections are also frequent, and traditional culture methods often struggle to identify them effectively. Studies have demonstrated that mNGS significantly outperforms conventional methods in detecting mixed infections, with detection rates as high as 48.9% compared to 4.3% for traditional techniques (Zheng et al., 2022; Zhang et al., 2023). Additionally, mNGS can rapidly identify drug-resistant genes without the need for isolating resistant strains, thereby guiding the rational use of antimicrobial agents (Hao et al., 2023).
4.2 Common viral infections after KT
Viral infections, including those caused by Cytomegalovirus (CMV), BK virus (BKV), and Epstein-Barr virus (EBV), are significant contributors to morbidity and mortality post-transplantation (Savassi-Ribas et al., 2019; Nowak et al., 2021; Agrawal et al., 2022). CMV, a herpesvirus, is a common opportunistic infection that significantly affects kidney transplant outcomes, with prevalence rates among recipients ranging from 8.8% to 63.2% (Nowak et al., 2021; Silva Junior et al., 2023). CMV is strongly linked to complications such as pneumonia, hepatitis, uveitis, and acute or chronic rejection following transplantation. BKV infects 1–10% of kidney transplant recipients (Myint et al., 2021). While most individuals acquire BKV during childhood, with 80–90% of adults carrying the virus latently in renal tubular and urinary tract epithelial cells (Burek Kamenaric et al., 2020), immunosuppression can trigger a progression to BK virus-associated nephropathy (BK-VAN). In the U.S., BK-VAN affects 5–10% of kidney transplant recipients, with 50–80% of these cases resulting in graft failure (Srivastava et al., 2020; Kotla et al., 2021). EBV, another herpesvirus affecting 90% of adults, can lead to severe complications when reactivated, including post-transplant lymphoproliferative disorder (PTLD). PTLD accounts for 21% of cancers in transplant recipients and is a leading cause of cancer-related mortality in this population (Portuguese et al., 2023; Szumilas et al., 2023). Traditional methods for detecting viral infections post-transplant include molecular assays, antigenemia testing, histopathology, viral culture, and serological testing (Shirley et al., 2023). While viral culture is highly specific, its low sensitivity and lengthy turnaround time limit its clinical utility (Cui et al., 2023). Serological tests often fail in early-stage infections due to insufficient antibody levels, resulting in false negatives and low sensitivity (Nagarajah et al., 2020; Roubalová et al., 2020). PCR technology, especially quantitative real-time PCR (qPCR), has become the gold standard for diagnosing and monitoring viral infections due to its sensitivity and reliability (Peinetti et al., 2021). However, as a targeted method, PCR requires prior knowledge of the pathogen and is prone to false negatives, posing limitations in certain clinical scenarios.
Our research demonstrates that mNGS surpasses traditional tests in sensitivity for detecting postoperative lung infections in KT patients and aids in identifying viral infections. For suspected drug-resistant viral infections, mNGS is recommended to assess genotypic drug resistance (Kleiboeker, 2023). During the COVID-19 pandemic, mNGS played a vital role in diagnosing viral infections in kidney transplant recipients, detecting 15 viral nucleic acids with a sensitivity of 0.96 and identifying a wide array of viruses, including rare ones (Tian et al., 2022). Additionally, mNGS demonstrated a 66% detection rate for mixed viral infections (Tian et al., 2022). Prompt antiviral therapy guided by mNGS results can effectively control infections, reduce mortality, and minimize complications in transplant recipients.
4.3 Common fungal infections after KT
Fungal infections are a significant concern in KT recipients, with Pneumocystis jiroveci pneumonia (PJP), invasive pulmonary aspergillosis (IPA), and candidiasis being the most common types. PJP, caused by Pneumocystis jiroveci, often presents acutely or subacutely and is associated with increased graft failure and mortality (Zhang et al., 2019; Chen et al., 2020; Zhu et al., 2023). Traditional diagnostic methods, including microscopic examination, staining, and serum beta-D-glucan (BDG) testing, have low sensitivity and specificity, while obtaining respiratory specimens is challenging (Le Gal et al., 2019). mNGS offers superior sensitivity, enabling early detection of Pneumocystis jiroveci in blood or sputum and identifying mixed infections, with CMV being the most frequent co-pathogen (Zhang et al., 2021). mNGS significantly outperforms conventional tests like GMS and BDG in diagnostic accuracy, improving treatment outcomes (Wang et al., 2022).
Similarly, IPA, caused by Aspergillus species, remains a leading cause of mortality despite advancements in diagnostic techniques such as imaging, BDG, galactomannan assays, and fungal cultures, which often lack precision and are limited by invasive sampling requirements (Ma et al., 2022; Shi et al., 2023). mNGS provides a non-invasive, highly sensitive alternative, detecting various Aspergillus strains, including Aspergillus fumigatus and Aspergillus flavus, even in culture-negative cases (Zhang et al., 2021). It also identifies co-infections to guide antifungal therapy (Ma et al., 2022). For invasive candidiasis, traditionally diagnosed through blood cultures with low sensitivity, mNGS has demonstrated superior diagnostic accuracy, particularly in patients with underlying conditions or severe pneumonia (Huseynov et al., 2021; Atiencia-Carrera et al., 2022; Shi et al., 2023; Thomsen et al., 2024). By identifying Candida species and guiding targeted treatment strategies, mNGS significantly improves clinical outcomes for these high-risk patients.
Overall, mNGS is a transformative diagnostic tool, offering enhanced sensitivity and specificity across a range of fungal infections in KTR, enabling timely and effective interventions (Chen et al., 2020; Zhang et al., 2021; Zhang et al., 2021; Ma et al., 2022; Wang et al., 2022; Huang et al., 2023; Shi et al., 2023; Thomsen et al., 2024).
4.4 Infections by rare pathogens
mNGS demonstrates significant advantages in diagnosing rare infections such as Streptococcus endocarditis, Toxoplasma marneffei, Cache Valley virus (CVV), Rhodococcus equi, and Algeria Bacillus (Zheng et al., 2022; Al-Heeti et al., 2023; Xu et al., 2023; Liang et al., 2024). It is particularly recommended when clinical suspicion of infection exists, but pathogens cannot be cultured from specimens. Unlike traditional methods, mNGS detects a broad range of pathogens simultaneously without requiring prior targeting (Liang et al., 2024). With superior accuracy and sensitivity, mNGS surpasses conventional techniques in pathogen detection, aiding clinicians in making timely and precise diagnoses (Zhang et al., 2023). It effectively identifies pathogens, including those transmitted through unconventional methods, and excels in diagnosing mixed infections (Weiqin et al., 2023). The technology offers faster detection, broader pathogen coverage, and greater clinical utility compared to traditional microbial culture. As sequencing technologies advance and costs decrease, mNGS is becoming increasingly integrated into clinical practice.
In summary, mNGS represents a significant advancement in the detection and management of diverse infections in KT recipients, offering rapid, comprehensive, and accurate pathogen identification that informs targeted therapeutic interventions.
5 Limitations and potential solutions of mNGS in post-KT infection diagnosis
While mNGS has demonstrated significant potential in diagnosing infections following KT, several limitations hinder its widespread clinical application (Zhong et al., 2021). One primary challenge is the high cost associated with mNGS, encompassing expenses for sequencing reagents, extraction, library preparation, and computational analysis. These costs often surpass those of traditional diagnostic methods, making routine use in clinical settings economically unfeasible (Zhong et al., 2021). Another significant limitation is the complexity of data interpretation. mNGS generates vast amounts of sequencing data, which can be challenging to analyze accurately. The presence of host DNA and commensal microorganisms can complicate the identification of pathogenic organisms, leading to potential misinterpretation of results. This complexity necessitates advanced bioinformatics tools and expertise, which may not be readily available in all clinical laboratories (Li et al., 2022; Li et al., 2023; Lu et al., 2023; Chen et al., 2024; Kan et al., 2024). Sensitivity and specificity issues also pose challenges. The detection of low-abundance pathogens can be difficult due to the overwhelming presence of host DNA, potentially leading to false negatives. Conversely, contamination or the presence of non-pathogenic microorganisms can result in false positives, complicating clinical decision-making (Han et al., 2019; Wang et al., 2020; Duan et al., 2022; Liu et al., 2022; Chen et al., 2024). Additionally, the lack of standardized protocols and bioinformatics pipelines hinders reproducibility across studies, and regulatory hurdles impede the integration of mNGS into routine clinical workflows (Ghosh et al., 2018; Hu et al., 2021).
To address these challenges, several strategies can be implemented. Reducing costs through technological advancements and streamlined workflows can improve the feasibility of mNGS for routine diagnostics. Developing standardized protocols and rigorous quality control measures can enhance data accuracy and reliability. Advanced bioinformatics pipelines are essential for effectively filtering out host DNA and distinguishing between pathogenic and non-pathogenic microorganisms, ensuring more precise data interpretation. Integrating mNGS with traditional diagnostic methods may provide a comprehensive approach, leveraging the strengths of both techniques to improve overall diagnostic accuracy. Hybrid sequencing approaches that combine short- and long-read technologies are also emerging as promising solutions, offering a balance between accuracy, cost, and read length.
In conclusion, while mNGS holds promise for diagnosing infections in KT recipients, addressing its current limitations is crucial for its effective integration into clinical practice. Ongoing research and technological advancements are essential to overcome these challenges and fully realize the potential of mNGS in improving patient outcomes.
6 Conclusion and future considerations
In conclusion, the application of mNGS has shown significant promise in revolutionizing the diagnosis and treatment of infections in KT recipients. This advanced technology enables the detection of a wide range of pathogens, including those that are difficult to identify using traditional microbiological methods. The ability to simultaneously identify bacteria, viruses, fungi, and parasites without the need for culture-based techniques provides a crucial advantage in the early diagnosis and management of post-transplant infections. Despite its potential, several challenges remain in the routine clinical adoption of mNGS for post-transplant infection diagnosis. These include the need for standardized protocols, cost-effectiveness considerations, and the integration of mNGS results into clinical decision-making. Furthermore, the interpretation of mNGS data can be complex, and it requires specialized expertise to distinguish between clinically significant pathogens and potential contaminants or colonizers.
Future research should focus on addressing these challenges. Efforts to streamline the data analysis process, improve the sensitivity and specificity of mNGS, and establish clear clinical guidelines for its use in post-KT infections will be crucial for its broader implementation. Additionally, longitudinal studies are needed to evaluate the long-term impact of mNGS on patient outcomes, including graft survival and overall survival. As technology advances, it is expected that mNGS will become an integral part of the diagnostic arsenal, improving the accuracy and timeliness of infection management in kidney transplant recipients. Moreover, exploring the role of mNGS in detecting emerging pathogens, monitoring antimicrobial resistance patterns, and guiding personalized therapeutic strategies will enhance its value in clinical practice. Collaborative efforts between clinicians, microbiologists, and bioinformaticians will be essential to maximize the full potential of mNGS in improving the care of KT patients.
Author contributions
HW: Conceptualization, Writing – original draft, Writing – review & editing. HC: Conceptualization, Data curation, Formal analysis, Writing – original draft. XG: Conceptualization, Data curation, Writing – original draft. CS: Conceptualization, Data curation, Formal analysis, Writing – original draft. LW: Conceptualization, Data curation, Formal analysis, Writing – original draft. BG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abo Basha, J., Kiel, M., Görlich, D., Schütte-Nütgen, K., Witten, A., Pavenstädt, H., et al. (2019). Phenotypic and genotypic characterization of escherichia coli causing urinary tract infections in kidney-transplanted patients. J. Clin. Med. 8, 988. doi: 10.3390/jcm8070988
Agrawal, A., Ison, M. G., Danziger-Isakov, L. (2022). Long-term infectious complications of kidney transplantation. Clin. J. Am. Soc. Nephrol. 17, 286–295. doi: 10.2215/CJN.15971020
Ahmad, Z., Bagchi, S., Naranje, P., Agarwal, S. K., Das, C. J. (2020). Imaging spectrum of pulmonary infections in renal transplant patients. Indian J. Radiol. Imaging. 30, 273–279.
Al-Heeti, O., Wu, E.-L., Ison, M. G., Saluja, R. K., Ramsey, G., Matkovic, E., et al. (2023). Transfusion-transmitted cache valley virus infection in a kidney transplant recipient with meningoencephalitis. Clin. Infect. Diseases: Off. Publ. Infect. Dis. Soc. America. 76, e1320–e13e7.
Atiencia-Carrera, M. B., Cabezas-Mera, F. S., Tejera, E., MaChado, A. (2022). Prevalence of biofilms in Candida spp. bloodstream infections: A meta-analysis. PloS One 17, e0263522.
Aydın, S., Patil, A., Desai, M., Simforoosh, N. (2020). Five compelling UTI questions after kidney transplant. World J. Urology. 38, 2733–2742. doi: 10.1007/s00345-020-03173-4
Bajda, S., Blazquez-Navarro, A., Samans, B., Wehler, P., Kaliszczyk, S., Amini, L., et al. (2020). The role of soluble mediators in the clinical course of EBV infection and B cell homeostasis after kidney transplantation. Sci. Rep. 10.
Bharati, J., Anandh, U., Kotton, C. N., Mueller, T., Shingada, A. K., Ramachandran, R. (2023). Diagnosis, prevention, and treatment of infections in kidney transplantation. Semin. Nephrol. 43, 151486. doi: 10.1016/j.semnephrol.2023.151486
Bloemen, B., Gand, M., Vanneste, K., Marchal, K., Roosens, N. H. C., De Keersmaecker, S. C. J. (2023). Development of a portble on-site applicable metagenomic data generation workflow for enhanced pathogen and antimicrobial resistance surveillance. Sci. Rep. 13.
Brennan, D. C., Daller, J. A., Lake, K. D., Cibrik, D., Del Castillo, D. (2006). Rabbit antithymocyte globulin versus basiliximab in renal transplantation. New Engl. J. Med. 355, 1967–1977. doi: 10.1056/NEJMoa060068
Burek Kamenaric, M., Ivkovic, V., Kovacevic Vojtusek, I., Zunec, R. (2020). The role of HLA and KIR immunogenetics in BK virus infection after kidney transplantation. Viruses. 12, 1417. doi: 10.3390/v12121417
Cai, Y., Ding, H., Chen, X., Chen, Y., Huang, C., Zhang, C., et al. (2023). Optimization and standardization of mNGS-based procedures for the diagnosis of Mycoplasma periprosthetic joint infection: A novel diagnostic strategy for rare bacterial periprosthetic joint infection. Front. Cell Infect. Microbiol. 13, 1089919. doi: 10.3389/fcimb.2023.1089919
Chen, H., Tang, M., Yao, L., Zhang, D., Zhang, Y., Zhao, Y., et al. (2023). Early application of metagenomics next-generation sequencing may significantly reduce unnecessary consumption of antibiotics in patients with fever of unknown origin. BMC Infect. Dis. 23. doi: 10.1186/s12879-023-08417-3
Chen, J., He, T., Li, X., Wang, X., Peng, L., Ma, L. (2020). Metagenomic next-generation sequencing in diagnosis of a case of pneumocystis jirovecii pneumonia in a kidney transplant recipient and literature review. Infection Drug Resistance. 13, 2829–2836. doi: 10.2147/IDR.S257587
Chen, Q., Yi, J., Liu, Y., Yang, C., Sun, Y., Du, J., et al. (2024). Clinical diagnostic value of targeted next−generation sequencing for infectious diseases (Review). Mol. Med. Rep. 30. doi: 10.3892/mmr.2024.13277
Chen, Y., Wang, J., Niu, T. (2024). Clinical and diagnostic values of metagenomic next-generation sequencing for infection in hematology patients: a systematic review and meta-analysis. BMC Infect. Diseases. 24. doi: 10.1186/s12879-024-09073-x
Chen, Y., Zhang, M., Chen, T., Wang, J., Zhao, Q., Zhou, E. M., et al. (2023). Development and application of a nanobody-based competitive ELISA for detecting antibodies against hepatitis E virus from humans and domestic animals. Microbiol. Spectr. 11, e0360722. doi: 10.1128/spectrum.03607-22
Cippà, P. E., Schiesser, M., Ekberg, H., van Gelder, T., Mueller, N. J., Cao, C. A., et al. (2015). Risk stratification for rejection and infection after kidney transplantation. Clin. J. Am. Soc. Nephrol. 10, 2213–2220. doi: 10.2215/CJN.01790215
Coussement, J., Argudín, M. A., Heinrichs, A., Racapé, J., de Mendonça, R., Nienhaus, L., et al. (2018). Host and microbial factors in kidney transplant recipients with Escherichia coli acute pyelonephritis or asymptomatic bacteriuria: a prospective study using whole-genome sequencing. Nephrol. Dialysis Transplantation. 34, 878–885. doi: 10.1093/ndt/gfy292
Coussement, J., Kaminski, H., Scemla, A., Manuel, O. (2020). Asymptomatic bacteriuria and urinary tract infections in kidney transplant recipients. Curr. Opin. Infect. Diseases. 33, 419–425. doi: 10.1097/QCO.0000000000000678
Cruz, A., Wale, D. J., Wong, K. K., Arnkoff, B. M., Viglianti, B. L. (2018). Unexpected vesicoureteral reflux into a nonfunctioning transplant kidney on renal scintigraphy. Clin. Nucl. Med. 43, 533–534. doi: 10.1097/RLU.0000000000002101
Cui, H., Zhang, C., Tu, F., Zhao, K., Kong, Y., Pu, J., et al. (2023). Rapid detection of influenza A viruses using a real-time reverse transcription recombinase-aided amplification assay. Front. Cell. Infection Microbiol. 12. doi: 10.3389/fcimb.2022.1071288
Dandamudi, R., Smith, J., Dharnidharka, V. R. (2019). Renal transplantation and predisposition to opportunistic infections. Curr. Opin. Pediatr. 31, 226–231. doi: 10.1097/MOP.0000000000000728
Dharia, A. A., Huang, M., Nash, M. M., Dacouris, N., Zaltzman, J. S., Prasad, G. V. R. (2022). Post-transplant outcomes in recipients of living donor kidneys and intended recipients of living donor kidneys. BMC Nephrology. 23. doi: 10.1186/s12882-022-02718-6
Diba, K., Makhdoomi, K., Nasri, E., Vaezi, A., Javidnia, J., Gharabagh, D. J., et al. (2018). Emerging Candida species isolated from renal transplant recipients: Species distribution and susceptibility profiles. Microbial Pathogenesis. 125, 240–245. doi: 10.1016/j.micpath.2018.09.026
Dong, Y., Chen, Q., Tian, B., Li, J., Li, J., Hu, Z. (2023). Advancing microbe detection for lower respiratory tract infection diagnosis and management with metagenomic next-generation sequencing. Infection Drug Resistance. 16, 677–694. doi: 10.2147/IDR.S387134
Duan, H., Li, X., Mei, A., Li, P., Liu, Y., Li, X., et al. (2021). The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases. BMC Infect. Dis. 21. doi: 10.1186/s12879-020-05746-5
Duan, W., Yang, Y., Zhao, J., Yan, T., Tian, X. (2022). Application of metagenomic next-generation sequencing in the diagnosis and treatment of recurrent urinary tract infection in kidney transplant recipients. Front. Public Health 10, 901549. doi: 10.3389/fpubh.2022.901549
Ernstsen, C. L., Login, F. H., Jensen, H. H., Nørregaard, R., Møller-Jensen, J., Nejsum, L. N. (2017). Detection and quantification of intracellular bacterial colonies by automated, high-throughput microscopy. J. Microbiol. Methods 139, 37–44. doi: 10.1016/j.mimet.2017.05.001
Eviatar, N., Dafna, Y., Nadav, M., Tzzipy, S., Eytan, M., Hefziba, G. (2022). The long-term impact of bloodstream infections on patient and graft survival following kidney transplantation. Clin. Transplant. 36, e14694. doi: 10.1111/ctr.14694
Fishman, J. A. (2017). Infection in organ transplantation. Am. J. Transplant. 17, 856–879. doi: 10.1111/ajt.14208
Gerges-Knafl, D., Pichler, P., Zimprich, A., Hotzy, C., Barousch, W., Lang, R. M., et al. (2020). The urinary microbiome shows different bacterial genera in renal transplant recipients and non-transplant patients at time of acute kidney injury – a pilot study. BMC Nephrol. 21.
Ghosh, M., Sharma, N., Singh, A. K., Gera, M., Pulicherla, K. K., Jeong, D. K. (2018). Transformation of animal genomics by next-generation sequencing technologies: a decade of challenges and their impact on genetic architecture. Crit. Rev. Biotechnol. 38, 1157–1175. doi: 10.1080/07388551.2018.1451819
Goodwin, S., McPherson, J. D., McCombie, W. R. (2016). Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351. doi: 10.1038/nrg.2016.49
Grasberger, J., Ortiz, F., Ekstrand, A., Sallinen, V., Ahopelto, K., Finne, P., et al. (2024). Infection-related hospitalizations after simultaneous pancreas-kidney transplantation compared to kidney transplantation alone. Transpl Int. 37, 12235. doi: 10.3389/ti.2024.12235
Hamon, A., Liegeon, G., Louis, K., Cambau, E., De Castro, N. (2023). Atypical presentation of Mycobacterium xenopi pulmonary infection in a kidney transplant recipient: A case report and literature review. IDCases. 31, e01675. doi: 10.1016/j.idcr.2022.e01675
Han, D., Li, Z., Li, R., Tan, P., Zhang, R., Li, J. (2019). mNGS in clinical microbiology laboratories: on the road to maturity. Crit. Rev. Microbiol. 45, 668–685. doi: 10.1080/1040841X.2019.1681933
Hao, L., Wen, P., Song, W., Zhang, B., Wu, Y., Zhang, Y., et al. (2023). Direct detection and identification of periprosthetic joint infection pathogens by metagenomic next-generation sequencing. Sci. Rep. 13.
He, K. D., Naqvi, S. S., Cowan, V. L., Stack, C. M., Alonso, C. D., Blair, B. M. (2024). Epidemiology and outcomes associated with enterococcal blood stream infection among liver and kidney transplant recipients. Clin. Transplantation. 38. doi: 10.1111/ctr.15285
Hsiao, C.-Y., Chen, T.-H., Lee, Y.-C., Wang, M.-C. (2021). Ureteral stone with hydronephrosis and urolithiasis alone are risk factors for acute kidney injury in patients with urinary tract infection. Sci. Rep. 11.
Hu, T., Chitnis, N., Monos, D., Dinh, A. (2021). Next-generation sequencing technologies: An overview. Hum. Immunol. 82, 801–811. doi: 10.1016/j.humimm.2021.02.012
Huang, L., Xu, S., Huang, Z., Chen, Y., Xu, N., Xie, B. (2023). Risk factors associated with Pneumocystis jirovecii pneumonia in non-HIV immunocompromised patients and co-pathogens analysis by metagenomic next-generation sequencing. BMC Pulmonary Med. 23. doi: 10.1186/s12890-022-02300-8
Huang, Z., Zou, S., Liu, Q., Qi, W., Sharma, A., Wang, Y., et al. (2024). Inferring the diagnostic potential of 18F-FDG-PET/CT in post-renal transplantation from a unique case harboring multiple rare complications. Front. Med. 11. doi: 10.3389/fmed.2024.1353466
Huseynov, R. M., Javadov, S. S., Osmanov, A., Khasiyev, S., Valiyeva, S. R., Almammadova, E., et al. (2021). The burden of serious fungal infections in Azerbaijan. Ther. Adv. Infect. Dis. 8. doi: 10.1177/20499361211043969
Illesy, L., Szabo-Pap, M., Toth, F., Zadori, G., Zsom, L., Asztalos, L., et al. (2016). Bacterial infections after kidney transplantation: A single-center experience. Transplant. Proc. 48, 2540–2543. doi: 10.1016/j.transproceed.2016.07.011
Janes, V. A., Matamoros, S., Munk, P., Clausen, P. T. L. C., Koekkoek, S. M., Koster, L. A. M., et al. (2022). Metagenomic DNA sequencing for semi-quantitative pathogen detection from urine: a prospective, laboratory-based, proof-of-concept study. Lancet Microbe 3, e588–ee97. doi: 10.1016/S2666-5247(22)00088-X
Kafi, H., Emaneini, M., Halimi, S., Rahdar, H. A., Jabalameli, F., Beigverdi, R. (2022). Multiplex high-resolution melting assay for simultaneous detection of five key bacterial pathogens in urinary tract infections: A pilot study. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1049178
Kan, C.-M., Tsang, H. F., Pei, X. M., Ng, S. S. M., Yim, A. K.-Y., Yu, A. C.-S., et al. (2024). Enhancing clinical utility: utilization of international standards and guidelines for metagenomic sequencing in infectious disease diagnosis. Int. J. Mol. Sci. 25, 3333. doi: 10.3390/ijms25063333
Kim, M. A., Rosa, V., Min, K. S. (2020). Characterization of Enterococcus faecalis in different culture conditions. Sci. Rep. 10, 21867. doi: 10.1038/s41598-020-78998-5
Kim, P. Y., Shoghi, A., Fananapazir, G. (2023). Renal transplantation: immediate and late complications. Radiol. Clin. North Am. 61, 809–820. doi: 10.1016/j.rcl.2023.04.004
Kleiboeker, S. B. (2023). Prevalence of cytomegalovirus antiviral drug resistance in transplant recipients. Antiviral Res. 215, 105623. doi: 10.1016/j.antiviral.2023.105623
Kobayashi, T., Ikeda, M., Okada, Y., Higurashi, Y., Okugawa, S., Moriya, K. (2021). Clinical and microbiological characteristics of recurrent escherichia coli bacteremia. Microbiol. Spectr. 9, e0139921. doi: 10.1128/Spectrum.01399-21
Kondori, N., Kurtovic, A., Piñeiro-Iglesias, B., Salvà-Serra, F., Jaén-Luchoro, D., Andersson, B., et al. (2021). Mass Spectrometry Proteotyping-Based Detection and Identification of Staphylococcus aureus, Escherichia coli, and Candida albicans in Blood. Front. Cell Infect. Microbiol. 11, 634215. doi: 10.3389/fcimb.2021.634215
Kotla, S. K., Kadambi, P. V., Hendricks, A. R., Rojas, R. (2021). BK polyomavirus-pathogen, paradigm and puzzle. Nephrol. Dial Transplant. 36, 587–593. doi: 10.1093/ndt/gfz273
Krishnamoorthy, S., Kumaresan, N., Zumla, A. (2019). Latent tuberculosis infection and renal transplantation – Diagnosis and management. Int. J. Infect. Diseases. 80, S73–SS6. doi: 10.1016/j.ijid.2019.01.049
Laketa, V. (2018). Microscopy in infectious disease research-imaging across scales. J. Mol. Biol. 430, 2612–2625. doi: 10.1016/j.jmb.2018.06.018
Lamy, B., Sundqvist, M., Idelevich, E. A. (2020). Bloodstream infections - Standard and progress in pathogen diagnostics. Clin. Microbiol. Infect. 26, 142–150. doi: 10.1016/j.cmi.2019.11.017
Lapošová, K., Lukáčiková, Ľ, Ovečková, I., Pastoreková, S., Rosocha, J., Kuba, D., et al. (2016). Development and application of ELISA for the detection of IgG antibodies to lymphocytic choriomeningitis virus. Acta Virol. 60, 143–150. doi: 10.4149/av_2016_02_143
Le Gal, S., Toubas, D., Totet, A., Dalle, F., Abou Bacar, A., Le Meur, Y., et al. (2019). Pneumocystis infection outbreaks in organ transplantation units in France: A nation-wide survey. Clin. Infect. Diseases. 70, 2216–2220. doi: 10.1093/cid/ciz901
Li, D., Gai, W., Zhang, J., Cheng, W., Cui, N., Wang, H. (2022). Metagenomic next-generation sequencing for the microbiological diagnosis of abdominal sepsis patients. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.816631
Li, X.-X., Niu, C.-Z., Zhao, Y.-C., Fu, G.-W., Zhao, H., Huang, M.-J., et al. (2023). Clinical application of metagenomic next-generation sequencing in non-immunocompromised patients with severe pneumonia supported by veno-venous extracorporeal membrane oxygenation. Front. Cell. Infection Microbiol. 13. doi: 10.3389/fcimb.2023.1269853
Li, Q., Ren, W., Yuan, J., Guo, H., Shang, Y., Wang, W., et al. (2022). Significant difference in Th1/Th2 paradigm induced by tuberculosis-specific antigens between IGRA-positive and IGRA-negative patients. Front. Immunol. 13. doi: 10.3389/fimmu.2022.904308
Li, Y., X-w, Y., Tang, L., W-j, D., T-l, L., Fan, J., et al. (2022). Diagnostic efficiency of metagenomic next-generation sequencing for suspected spinal tuberculosis in China: A multicenter prospective study. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1018938
Lian, Q., Song, X., Yang, J., Wang, L., Xu, P., Wang, X., et al. (2024). Alterations of lung microbiota in lung transplant recipients with pneumocystis jirovecii pneumonia. Respir. Res. 25, 125. doi: 10.1186/s12931-024-02755-9
Liang, G.-F., Chao, S., Sun, Z., Zhu, K.-J., Chen, Q., Jia, L., et al. (2024). Pleural empyema with endobronchial mass due to Rhodococcus equi infection after renal transplantation: A case report and review of literature. World J. Clin. Cases. 12, 224–231. doi: 10.12998/wjcc.v12.i1.224
Liang, W., Zhang, Q., Qian, Q., Wang, M., Ding, Y., Zhou, J., et al. (2024). Diagnostic strategy of metagenomic next-generation sequencing for gram negative bacteria in respiratory infections. Ann. Clin. Microbiol. Antimicrobials 23. doi: 10.1186/s12941-024-00670-x
Lim, M. A., Kohli, J., Bloom, R. D. (2017). Immunosuppression for kidney transplantation: Where are we now and where are we going? Transplant. Rev. (Orlando) 31, 10–17. doi: 10.1016/j.trre.2016.10.006
Liu, Q., Jin, X., Cheng, J., Zhou, H., Zhang, Y., Dai, Y. (2023). Advances in the application of molecular diagnostic techniques for the detection of infectious disease pathogens (Review). Mol. Med. Rep. 27. doi: 10.3892/mmr.2023.12991
Liu, H., Wei, X., Wang, Z., Huang, X., Li, M., Hu, Z., et al. (2024). LysSYL: a broad-spectrum phage endolysin targeting Staphylococcus species and eradicating S. aureus biofilms. Microbial Cell Factories 23. doi: 10.1186/s12934-024-02359-4
Liu, H., Zhang, Y., Yang, J., Liu, Y., Chen, J. (2022). Application of mNGS in the etiological analysis of lower respiratory tract infections and the prediction of drug resistance. Microbiol. Spectr. 10, e0250221. doi: 10.1128/spectrum.02502-21
Lu, D., Abudouaini, M., Kerimu, M., Leng, Q., Wu, H., Aynazar, A., et al. (2023). Clinical evaluation of metagenomic next-generation sequencing and identification of risk factors in patients with severe community-acquired pneumonia. Infection Drug Resistance 16, 5135–5147. doi: 10.2147/IDR.S421721
Lunn, A., Holden, S., Boswell, T., Watson, A. R. (2010). Automated microscopy, dipsticks and the diagnosis of urinary tract infection. Arch. Dis. Child. 95, 193–197. doi: 10.1136/adc.2009.166835
Luo, J., Brakel, A., Krizsan, A., Ludwig, T., Mötzing, M., Volke, D., et al. (2022). Sensitive and specific serological ELISA for the detection of SARS-CoV-2 infections. Virol. J. 19, 50. doi: 10.1186/s12985-022-01768-4
Ma, X., Zhang, S., Xing, H., Li, H., Chen, J., Li, H., et al. (2022). Invasive pulmonary aspergillosis diagnosis via peripheral blood metagenomic next-generation sequencing. Front. Med. 9, 751617. doi: 10.3389/fmed.2022.751617
Mangalgi, S., Madan, K., Das, C. J., Singh, G., Sati, H., Yadav, R., et al. (2021). Pulmonary infections after renal transplantation: a prospective study from a tropical country. Transplant. Int. 34, 525–534. doi: 10.1111/tri.13817
McAteer, J., Tamma, P. D. (2024). Diagnosing and managing urinary tract infections in kidney transplant recipients. Infect. Dis. Clin. North Am. 38, 361–380. doi: 10.1016/j.idc.2024.03.008
Meira de Faria, L., Nobre, V., Ribeiro de Oliveira Guardão, L., Magalhães Souza, C., Damasceno de Souza, A., dos Reis Estrella, D., et al. (2023). Factors associated with pulmonary infection in kidney and kidney-pancreas transplant recipients: a case-control study. Jornal Brasileiro Pneumologia., e20220419.
Møller, D. L., Sørensen, S. S., Wareham, N. E., Rezahosseini, O., Knudsen, A. D., Knudsen, J. D., et al. (2021). Bacterial and fungal bloodstream infections in pediatric liver and kidney transplant recipients. BMC Infect. Dis. 21, 541. doi: 10.1186/s12879-021-06224-2
Myint, T. M., Chong, C. H. Y., Wyld, M., Nankivell, B., Kable, K., Wong, G. (2021). Polyoma BK virus in kidney transplant recipients: screening, monitoring, and management. Transplantation. 106, e76–e89.
Nagarajah, S., Rasmussen, M., Hoegh, S. V., Tepel, M. (2020). Prospective study of MGAT3-AS1, and viremia of BK polyomavirus and cytomegalovirus in living donor renal transplant recipients. Kidney Int. Rep. 5, 2218–2227. doi: 10.1016/j.ekir.2020.09.005
Navarrete, R. V. (2009). Renal transplantation with living-donor surgical strategies: a view with 40 years of experience. Eur. Urol. 56, 38–39. doi: 10.1016/j.eururo.2009.02.018
Nowak, H., Vornweg, S., Rump, K., Rahmel, T., Unterberg, M., Koos, B., et al. (2021). The NFKB1 promoter polymorphism (-94ins/delATTG) is associated with susceptibility to cytomegalovirus infection after kidney transplantation and should have implications on CMV prophylaxis regimens. Cells. 10, 380. doi: 10.3390/cells10020380
Ohyama, Y., Nakajima, K., Renfrow, M. B., Novak, J., Takahashi, K. (2020). Mass spectrometry for the identification and analysis of highly complex glycosylation of therapeutic or pathogenic proteins. Expert Rev. Proteomics. 17, 275–296. doi: 10.1080/14789450.2020.1769479
Pajenda, S., Gerges, D. A., Freire, R., Wagner, L., Hevesi, Z., Aiad, M., et al. (2023). Acute kidney injury and BK polyomavirus in urine sediment cells. Int. J. Mol. Sci. 24. doi: 10.3390/ijms242417511
Peinetti, A. S., Lake, R. J., Cong, W., Cooper, L., Wu, Y., Ma, Y., et al. (2021). Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci. Adv. 7.
Pilmis, B., Weiss, E., Scemla, A., Le Monnier, A., Grossi, P. A., Slavin, M. A., et al. (2023). Multidrug-resistant Enterobacterales infections in abdominal solid organ transplantation. Clin. Microbiol. Infection. 29, 38–43. doi: 10.1016/j.cmi.2022.06.005
Portuguese, A. J., Gauthier, J., Tykodi, S. S., Hall, E. T., Hirayama, A. V., Yeung, C. C. S., et al. (2023). CD19 CAR-T therapy in solid organ transplant recipients: case report and systematic review. Bone Marrow Transplant. 58, 353–359. doi: 10.1038/s41409-022-01907-z
Promsuwan, O., Malathum, K., Ingsathit, A. (2023). Epidemiology of extended-spectrum β-lactamase–producing Enterobacterales infection in kidney transplant recipients. Antimicrobial Resistance Infection Control. 12. doi: 10.1186/s13756-023-01308-x
Qian, Y., Zhu, Y., Li, Y., Li, B. (2021). Legend of the sentinels: development of lung resident memory T cells and their roles in diseases. Front. Immunol. 11. doi: 10.3389/fimmu.2020.624411
Reimold, P., Aksoy, C., Beckmann, J., Zacharis, A., Groeben, C., Karschuck, P., et al. (2024). Development and outcomes of surgical and urological kidney transplantation programs in Germany: a total population analysis from 2006 to 2021. World J. Urol. 42. doi: 10.1007/s00345-023-04740-1
Rodino, K. G., Simner, P. J. (2024). Status check: next-generation sequencing for infectious-disease diagnostics. J. Clin. Invest. 134. doi: 10.1172/JCI178003
Roubalová, K., Němečková, Š, Kryštofová, J., Hainz, P., Pumannová, M., Hamšíková, E. (2020). Antigenic competition in the generation of multi-virus-specific cell lines for immunotherapy of human cytomegalovirus, polyomavirus BK, Epstein-Barr virus and adenovirus infection in haematopoietic stem cell transplant recipients. Immunol. Letters. 228, 64–69. doi: 10.1016/j.imlet.2020.09.009
Savassi-Ribas, F., Gomes dos Santos de Almeida, S., Baez, C. F., Magalhães de Souza, L., Wagner, T. C. S., Matuck, T. A., et al. (2019). Impact assessment and investigation of factors associated with herpesviruses viremia in the first year of renal transplantation. J. Med. Virology. 92, 107–112.
Shi, Y., Peng, J.-M., Hu, X.-Y., Yang, Q.-W., Wang, Y. (2023). Metagenomic next-generation sequencing for detecting Aspergillosis pneumonia in immunocompromised patients: a retrospective study. Front. Cell. Infection Microbiol. 13. doi: 10.3389/fcimb.2023.1209724
Shirley, J. D., Bennett, S. A., Binnicker, M. J. (2023). Current regulatory landscape for viral point-of-care testing in the United States. J. Clin. Virology. 164, 105492. doi: 10.1016/j.jcv.2023.105492
Silva Junior, H. T., Tokat, Y., Cai, J., Singh, I., Sandhu, A., Demuth, D., et al. (2023). Epidemiology, management, and burden of cytomegalovirus in solid organ transplant recipients in selected countries outside of Europe and North America: A systematic review. Transplant. Infect. Dis. 25.
Srivastava, A., Bodnar, J., Osman, F., Jorgenson, M. R., Astor, B. C., Mandelbrot, D. A., et al. (2020). Serum albumin level before kidney transplant predicts post-transplant BK and possibly cytomegalovirus infection. Kidney Int. Rep. 5, 2228–2237. doi: 10.1016/j.ekir.2020.09.012
Sugi, M. D., Joshi, G., Maddu, K. K., Dahiya, N., Menias, C. O. (2019). Imaging of renal transplant complications throughout the life of the allograft: comprehensive multimodality review. RadioGraphics. 39, 1327–1355. doi: 10.1148/rg.2019190096
Sun, H., Wang, F., Zhang, M., Xu, X., Li, M., Gao, W., et al. (2022). Diagnostic value of bronchoalveolar lavage fluid metagenomic next-generation sequencing in pneumocystis jirovecii pneumonia in non-HIV immunosuppressed patients. Front. Cell. Infection Microbiol. 12. doi: 10.3389/fcimb.2022.872813
Szumilas, K., Wilk, A., Wiśniewski, P., Gimpel, A., Dziedziejko, V., Kipp, M., et al. (2023). Current status regarding immunosuppressive treatment in patients after renal transplantation. Int. J. Mol. Sci. 24. doi: 10.3390/ijms241210301
Thomsen, J., Abdulrazzaq, N. M., Oulhaj, A., Nyasulu, P. S., Alatoom, A., Denning, D. W., et al. (2024). Emergence of highly resistant Candida auris in the United Arab Emirates: a retrospective analysis of evolving national trends. Front. Public Health 11. doi: 10.3389/fpubh.2023.1244358
Tian, X., Duan, W., Zhang, X., Wu, X., Zhang, C., Wang, Z., et al. (2022). Metagenomic next-generation sequencing reveals the profile of viral infections in kidney transplant recipients during the COVID-19 pandemic. Front. Public Health 10, 888064. doi: 10.3389/fpubh.2022.888064
Tsalik, E. L., Bonomo, R. A., Fowler, V. G., Jr. (2018). New molecular diagnostic approaches to bacterial infections and antibacterial resistance. Annu. Rev. Med. 69, 379–394. doi: 10.1146/annurev-med-052716-030320
Unruh, M., Dew, M. A. (2014). Asking dialysis patients about what they were told: a new strategy for improving access to kidney transplantation? J. Am. Soc. Nephrol. 25, 2683–2685. doi: 10.1681/ASN.2014060571
Vargas Barahona, L., Henao-Cordero, J., Smith, J., Gray, A., Marshall, C. B., Scherger, S., et al. (2022). Disseminated tuberculosis in a lung transplant recipient presenting as tenosynovitis, subcutaneous nodules, and liver abscesses. Ther. Adv. Infect. Dis. 9.
Viklicky, O., Novotny, M., Hruba, P. (2020). Future developments in kidney transplantation. Curr. Opin. Organ Transplant. 25, 92–98. doi: 10.1097/MOT.0000000000000722
Voora, S., Shah, S., Nadim, M. K. (2023). Management of the kidney transplant recipient in the intensive care unit. Curr. Opin. Crit. Care 29, 587–594. doi: 10.1097/MCC.0000000000001098
Wang, D., Fang, S., Hu, X., Xu, Q., Chu, X., Mei, X., et al. (2022). Metagenomic next-generation sequencing is highly efficient in diagnosing pneumocystis jirovecii pneumonia in the immunocompromised patients. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.913405
Wang, L., Liu, J., Peng, L. (2023). High-dose tigecycline for the treatment of progressive pneumonia caused by chlamydia psittaci: case series and literature review. Infection Drug Resistance. 16, 115–124. doi: 10.2147/IDR.S393647
Wang, Q., Wu, B., Yang, D., Yang, C., Jin, Z., Cao, J., et al. (2020). Optimal specimen type for accurate diagnosis of infectious peripheral pulmonary lesions by mNGS. BMC Pulmonary Med. 20. doi: 10.1186/s12890-020-01298-1
Weiqin, W., Xiang, H., Hongmei, Z., Hong, S., Qingsong, S. (2023). Application value of next-generation sequencing of bronchial alveolar lavage fluid in emergency patients with infection. Cell. Mol. Biol. 69, 45–49. doi: 10.14715/cmb/2023.69.8.7
Wen, J., Sun, R., Yang, H., Ran, Q., Hou, Y. (2022). Detection of BK polyomavirus-associated nephropathy using plasma graft-derived cell-free DNA: Development of a novel algorithm from programmed monitoring. Front. Immunol. 13. doi: 10.3389/fimmu.2022.1006970
Wu, D., Chen, C., Liu, T., Wan, Q. (2020). Risk factors for acquisition of carbapenem-resistant klebsiella pneumoniae and mortality among abdominal solid organ transplant recipients with K. pneumoniae Infections. Med. Sci. Monitor. 26. doi: 10.12659/MSM.922996
Xin, Z., Wu, L., Zhou, J., Zhuang, J., Peng, W., Song, T., et al. (2020). Analysis of factors influencing kidney function of recipients after renal transplantation in southwestern China: A retrospective study. Front. Med. 7. doi: 10.3389/fmed.2020.519582
Xu, L., Chen, X., Yang, X., Jiang, H., Wang, J., Chen, S., et al. (2023). Disseminated Talaromyces marneffei infection after renal transplantation: A case report and literature review. Front. Cell. Infection Microbiol. 13, 1115268. doi: 10.3389/fcimb.2023.1115268
Ying, L., Hu, Z., Lu, Y., Tao, Q., Xiong, F., Shu, Y., et al. (2024). An oncogene regulating chromatin favors response to immunotherapy: Oncogene CHAF1A and immunotherapy outcomes. Oncoimmunology. 13, 2303195. doi: 10.1080/2162402X.2024.2303195
Zhang, Y., Ai, J.-W., Cui, P., Zhang, W.-H., Wu, H.-L., Ye, M.-Z. (2019). A cluster of cases of pneumocystis pneumonia identified by shotgun metagenomics approach. J. Infection. 78, 158–169. doi: 10.1016/j.jinf.2018.08.013
Zhang, W. W., Ai, C., Mao, C. T., Liu, D. K., Guo, Y. (2023). Prevotella oris-caused meningitis and spinal canal infection: A case report. World J. Clin. Cases. 11, 3830–3836. doi: 10.12998/wjcc.v11.i16.3830
Zhang, F., Chen, J., Huang, H., Deng, X., Zhang, W., Zeng, M., et al. (2021). Application of metagenomic next-generation sequencing in the diagnosis and treatment guidance of Pneumocystis jirovecii pneumonia in renal transplant recipients. Eur. J. Clin. Microbiol. Infect. Diseases: Off. Publ. Eur. Soc. Clin. Microbiol. 40, 1933–1942. doi: 10.1007/s10096-021-04254-x
Zhang, H., Liu, G., He, L., Zhu, Y., Tu, H., Zhuang, S. (2023). Life-threatening pulmonary coinfection with Mycobacterium tuberculosis and Aspergillus lentulus in a diabetic patient diagnosed by metagenome next-generation sequencing. BMC Infect. Diseases. 23. doi: 10.1186/s12879-023-08052-y
Zhang, B., Zhou, J., Gui, R., Li, Z., Zu, Y., Wang, J., et al. (2021). Metagenomic next generation sequencing in the detection of pathogens in cerebrospinal fluid of patients after alternative donor transplantation: A feasibility analysis. Front. Cell. Infection Microbiol. 11. doi: 10.3389/fcimb.2021.720132
Zhao, D., Guo, L., Lian, D., Gu, Y., Yan, X., Hu, H., et al. (2022). Diagnostic value and clinical application of mNGS for post-liver transplantation infection: A cross-sectional study with case reports. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.919363
Zheng, Y.-R., Lin, S.-H., Chen, Y.-K., Cao, H., Chen, Q. (2022). Application of metagenomic next-generation sequencing in the detection of pathogens in bronchoalveolar lavage fluid of infants with severe pneumonia after congenital heart surgery. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.954538
Zheng, M.-M., Shang, L.-M., Du, C.-K., Zhang, L., Sun, W., Wang, Z.-P., et al. (2022). Corynebacterium striatum endocarditis after renal transplantation confirmed by metagenomic next-generation sequencing: case report and literature review. Infection Drug Resistance. 15, 4899–4906. doi: 10.2147/IDR.S376985
Zhong, Y., Xu, F., Wu, J., Schubert, J., Li, M. M. (2021). Application of next generation sequencing in laboratory medicine. Ann. Lab. Med. 41, 25–43. doi: 10.3343/alm.2021.41.1.25
Zhu, L., Xu, H., Pu, Y., Fu, C., Pan, Q., Zhao, H. (2023). Case Report: Comprehensive Management of Pneumocystis Jiroveci Pneumonia (PJP) and Secondary Infections of Multiple-Drug Resistant Enterobacter cloacae complex and Pseudomonas aeruginosa in a Kidney Transplant Recipient with Sulfonamide Allergies. Infection Drug Resistance. 16, 6185–6193. doi: 10.2147/IDR.S428890
Keywords: kidney transplantation, metagenomic next-generation sequencing, infection, pathogens, diagnosis
Citation: Wu H, Cao H, Gao X, Shi C, Wang L and Gao B (2025) The role of metagenomic next-generation sequencing in diagnosing and managing post-kidney transplantation infections. Front. Cell. Infect. Microbiol. 14:1473068. doi: 10.3389/fcimb.2024.1473068
Received: 30 July 2024; Accepted: 16 December 2024;
Published: 07 January 2025.
Edited by:
Friedrich Thaiss, University of Hamburg, GermanyCopyright © 2025 Wu, Cao, Gao, Shi, Wang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoshan Gao, Z2FvYnNAamx1LmVkdS5jbg==
†These authors have contributed equally to this work
 Hao Wu
Hao Wu Hongliang Cao
Hongliang Cao Xin Gao
Xin Gao Chengdong Shi
Chengdong Shi Lei Wang
Lei Wang Baoshan Gao
Baoshan Gao