
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 13 November 2024
Sec. Molecular Bacterial Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1465460
This article is part of the Research Topic Controlling Biofilm-Related Infections in Healthcare Settings View all 8 articles
Proteus mirabilis is a Gram-negative bacterium commonly found in urinary tract infections (UTIs) and catheter-associated urinary tract infections (CAUTIs). The pathogenic mechanisms of Proteus mirabilis are complex and diverse, involving various virulence factors, including fimbriae, flagella, urease, polyphosphate kinase, lipopolysaccharides, cyclic AMP receptor protein, Sigma factor RpoE, and RNA chaperone protein Hfq. These factors play crucial roles in bacterial colonization, invasion, evasion of host immune responses, biofilm formation, and urinary stone formation. This paper is the first to comprehensively describe the hydrogenase system, autotransporter proteins, molybdate-binding protein ModA, and two-component systems as virulence factors in Proteus mirabilis, providing new insights into its pathogenic mechanisms in urinary tract infections. This review explores the mechanisms of biofilm formation by Proteus mirabilis and the various virulence factors involved in UTIs, revealing many newly discovered virulence factors from recent studies. These findings may offer new targets for clinical treatment of UTIs and vaccine development, highlighting the importance of understanding these virulence factors.
Proteus mirabilis is widely distributed in natural environments such as water, soil, and the gastrointestinal tracts of humans and animals (Drzewiecka, 2016; Wasfi et al., 2020). In the human gut microbiota, Proteus species constitute less than 0.05% in healthy individuals (Yatsunenko et al., 2012). Among all species of the genus Proteus, Proteus mirabilis is the most common cause of human infections (Jacobsen and Shirtliff, 2011). Proteus mirabilis can also cause osteomyelitis, mastoiditis, rheumatoid arthritis (Salehi et al., 2023; Juárez-Chairez et al., 2024), wound infections, otitis media (Leichtle et al., 2024), keratitis (Mo et al., 2021), Crohn’s disease (Zhang et al., 2021),neonatal meningitis, hemorrhagic meningoencephalitis (Senior and Sweeney, 1984; Chang et al., 2002; Phan and Lehman, 2012; Ebringer and Rashid, 2014), and hepatic encephalopathy in patients with acute liver failure (Samtoy and DeBeukelaer, 1980). Previous studies have reported that the most common type of kidney stone caused by urinary tract infections is struvite, which contains magnesium ammonium phosphate, and the most common microorganisms found in these stones are urease-producing bacteria, particularly Proteus mirabilis (Griffith, 1982; McLean et al., 1985; Miano et al., 2007).Furthermore, Proteus mirabilis has recently been implicated in the pathogenesis of Parkinson’s disease (Li et al., 2017; Choi et al., 2018). Proteus mirabilis is also a common cause of urinary tract infections in patients with certain anatomical or functional abnormalities of the urinary system (Odoki et al., 2019). This is particularly problematic for patients with long-term indwelling catheters (Gambrill et al., 2024; Khorasani et al., 2024), as it adheres to the catheter surface and obstructs urine flow, eventually forming a crystalline biofilm (Pelling et al., 2019). As a result, these patients are at significantly higher risk of catheter-associated urinary tract infections.
During urinary tract infections, Proteus mirabilis initially bypasses the host’s natural defense systems to infect the urethra, where it subsequently colonizes. It can then migrate to the bladder and further to the kidneys. The adherence to the uroepithelium or catheters relies on fimbriae and adhesins. During migration, Proteus mirabilis differentiates from individual short rod-shaped “swimming cells” into multicellular elongated “swarming cells.” This differentiation involves rearrangements of bacterial membrane components (Stolarek et al., 2023), increased expression of urease, ZapA protease, and hemolysin, and an increase in flagella and nucleoid-like structures that form cellular communities (Jones et al., 2004; Rather, 2005; Jacobsen et al., 2008), which plays a crucial role in kidney colonization (Allison et al., 1994). After the adherence process, Proteus mirabilis can form a crystalline biofilm on the surface of the host or catheter: (1) initial colonization by bacteria surrounded by a large amount of glycosylated carbohydrates, (2) formation of sheet-like microcrystalline material, (3) accumulation of dispersed crystalline substances, and (4) formation of a biofilm containing defined crystals and highly motile Proteus mirabilis cells (Wilks et al., 2015). The biofilm is a complex structure where bacterial cells are surrounded by extracellular polymeric substances, primarily composed of polysaccharides, proteins, extracellular DNA, glycoproteins, lipids, teichoic acids, and lipopolysaccharides (Gmiter and Kaca, 2022). The process of urinary tract infection induced by Proteus mirabilis is shown in Figure 1. Concurrently, several recently identified virulence factors also promote the progression of urinary tract infections, which I will introduce in detail in the following sections.
Proteus mirabilis secretes urease, a nickel-containing enzyme located in either the cytoplasm or outer membrane of this bacterium. Urease plays a crucial role in the formation of stones and the onset of acute pyelonephritis (Mobley, 2019). Urease hydrolyzes urea into CO2 and ammonium ions, promoting crystal formation as an initiator (Schaffer and Pearson, 2015; Combaz-Söhnchen and Kuhn, 2017; Manoharan et al., 2023). The hydrolysis process produces high concentrations of ammonia, which raises the pH of the urine. The increased concentrations of ammonium, bicarbonate, and phosphate ions lead to ion precipitation within infection stones (Chakkour et al., 2024; Prywer et al., 2024; Szczerbiec et al., 2024). These stones quickly grow larger, potentially resulting in kidney disease.
The urease gene cluster in Proteus mirabilis consists of ureABC and ureDEFG. ureABC encodes the structural subunits of the apoenzyme, while ureDEFG encodes proteins responsible for facilitating the insertion of essential nickel ions into the catalytic site (Poore and Mobley, 2003). For urease maturation, nickel ions must be transported into the cytoplasm and incorporated into the apoenzyme. UreR is the regulatory factor responsible for activating urease operon expression, and without the ureR gene, urease cannot be produced (Parra and Collins, 2012). UreR is a positive regulator of urease and nickel transport, and in the presence of urea, it activates the expression of the ure gene cluster (Fitzgerald et al., 2024).
Studies using Alizarin Red staining and immunofluorescence to detect calcium and magnesium salts and extracellular clusters compared urease-negative ureC mutants with control groups. As expected, no Alizarin Red staining or extracellular clusters were observed in bladder sections from mice infected with the ureC mutant Proteus mirabilis, whereas strong Alizarin Red staining and extracellular clusters were observed in the control group. These findings underscore the critical role of urease in the formation of extracellular clusters and stone deposition by Proteus mirabilis in vivo. At 24 hours post-infection, the CFU of the ureC mutant was 23 times lower than that of the control group, although the mutant did not exhibit growth defects under various in vitro conditions (Schaffer et al., 2016).
Urease secreted by Proteus mirabilis also exhibits some non-enzymatic properties, such as promoting platelet aggregation, increasing intracellular calcium levels in mouse neuroblastoma cells, and inducing a pro-inflammatory phenotype in HEK293 cells (derived from human embryonic kidney). In the presence of urease, this results in the production of reactive oxygen species and the secretion of IL-1β and TNF-α. In addition to the toxicity caused by ammonia to urinary tract tissues, urease may also exacerbate tissue damage through other non-enzymatic mechanisms (Grahl et al., 2021). The virulence factors of Proteus mirabilis, along with their associated genes, related functions, and references, are summarized in Table 1.
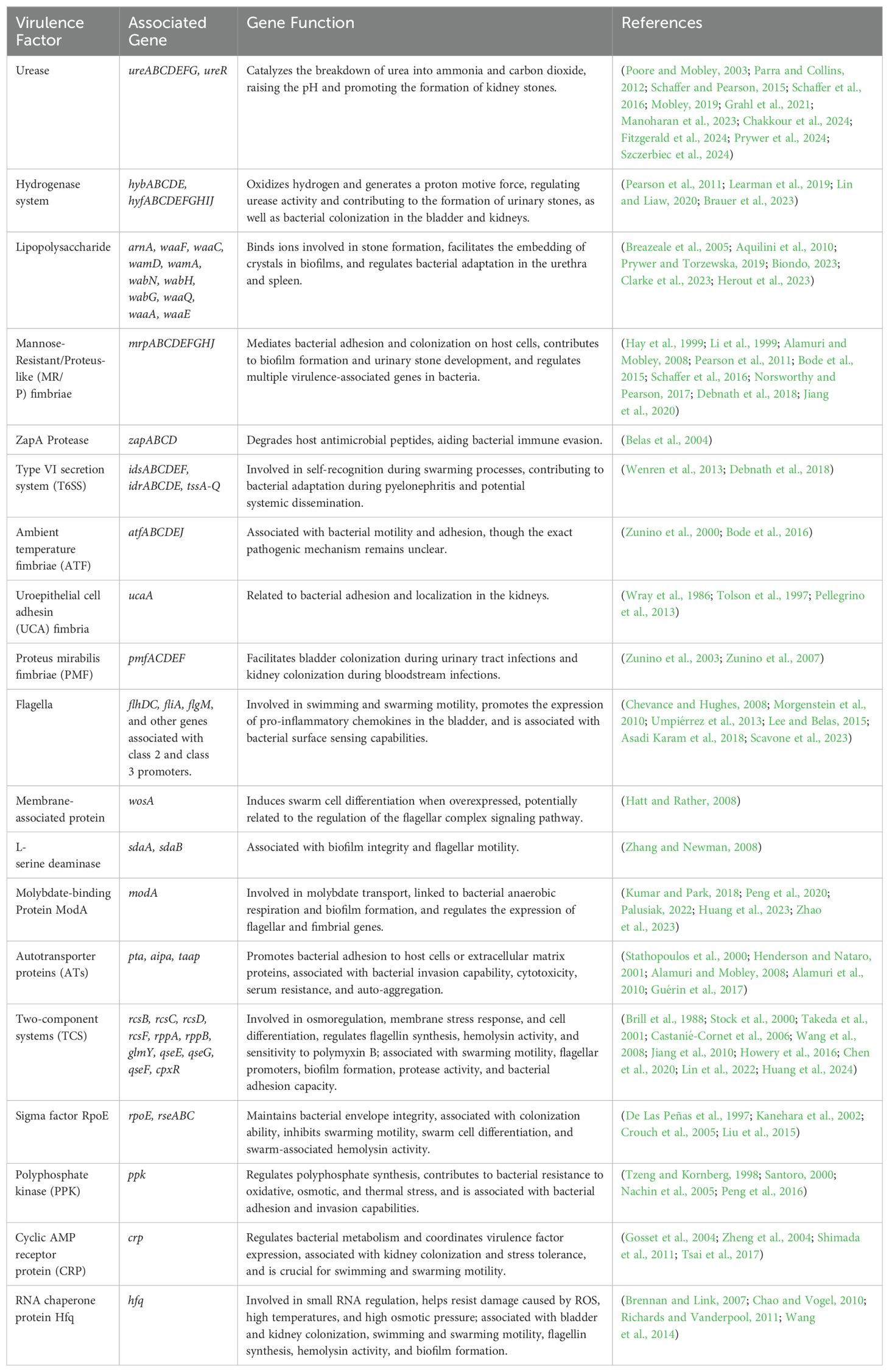
Table 1. Virulence factors, associated genes, related functions, and references of Proteus mirabilis.
Formate hydrogenlyase (FHL), composed of formate dehydrogenase H (FDH) and a [NiFe] hydrogenase complex, links formate oxidation to proton reduction, facilitating H2 production. FhlA, a RpoN-dependent enhancer-binding protein, serves as the transcriptional activator of FHL (Schlensog et al., 1994; Self et al., 2004). The hydrogenase in FHL is a [NiFe] hydrogenase, playing a role in converting protons to hydrogen gas (Schoelmerich and Müller, 2020). In Proteus mirabilis HI4320, two hydrogenases are encoded: the FHL hydrogenase 2 operon (hyb) and hydrogenase 4 operon (hyf). hyfG encodes the [NiFe] hydrogenase large subunit required for hydrogenase activity (Noguchi et al., 2010). The primary role of hyb and hyf is to convert protons into hydrogen gas, deacidifying the environment (Schoelmerich and Müller, 2020). The hyfH gene is significantly upregulated in a mouse model of ascending urinary tract infection (Pearson et al., 2011), and hyfE has been identified as a bladder colonization adaptation factor through transposon sequencing. Schematic representation of hyb,hyf and mrp operon is shown in Figure 2.
In Proteus mirabilis, FhlA regulates the promoter activities of fdhF and hyf, significantly induced by formate and anaerobic conditions (Lin and Liaw, 2020). In the hyfC mutant, hyfA expression is notably higher than in the wild-type or hyfE mutants, suggesting that the hyb operon may influence the hyf operon, although specific mechanisms require further investigation (Brauer et al., 2023).
Deletion of hyfG or fhlA leads to reduced urease activity and urinary stone formation, with significantly lower urease activity observed in these mutants across a pH range of 5 to 8 (Lin and Liaw, 2020). However, hybC and hyfE do not affect urease activity (Brauer et al., 2023).
The structure of hyf resembles respiratory complex I, suggesting involvement in proton gradient formation and energy production (Schoelmerich and Müller, 2020). The rotation of flagella requires the proton motive force (PMF) (Gabel and Berg, 2003). While deletion of hybC, fhlA or hyfG does not impact proton motive force (PMF) phenotypes, the hyf system contributes to PMF under anaerobic conditions (Lin and Liaw, 2020; Brauer et al., 2023).
FhlA and hyfG are essential for Proteus mirabilis colonization. Mutants lacking fhlA or hyfG show significantly reduced colonization in both the bladder and kidneys compared to the wild-type strain (Lin and Liaw, 2020). Deletion of both hybC and hyfE leads to severe colonization defects in the bladder, kidneys, and spleen, while the loss of either gene alone does not affect colonization or adaptation. Studies have shown that co-culturing Proteus mirabilis with Enterococcus faecalis can increase cytotoxicity, and mice with co-infections exhibit more severe disease symptoms compared to single infections. 96% of the co-infected mice developed bacteremia (an 18% increase compared to Proteus mirabilis single infections), and 56% developed urolithiasis (a 30% increase compared to Proteus mirabilis single infections). Additionally, co-infection leads to severe bladder tissue damage and inflammation (Learman et al., 2019). When Proteus mirabilis is co-cultured with Enterococcus faecalis, the loss of both hybC and hyfE operons does not cause adaptation defects. In competitive co-culture, hybC and hyfE offer a slight adaptation advantage during the lag and early log phases, but their high expression inhibits growth in the stationary phase. Direct competition showed that the CFU of the hyfE mutant was higher than the hybC mutant, indicating that increased hyf expression in the hybC mutant leads to adaptation defects (Brauer et al., 2023).
Lipopolysaccharide, present in the outer membrane of Gram-negative bacteria, safeguards bacterial integrity and acts as a critical virulence factor. The LPS of Proteus mirabilis is composed of three parts: lipid A, the core region, and the O-antigen polysaccharide (Palusiak, 2013). The O-antigen exhibits the highest biological activity. Its absence can lead to various changes in the bacteria, such as increased sensitivity to antimicrobial agents, altered hydrophobicity, impaired swarming motility, adhesion to atherosclerotic plaques, and heightened sensitivity to human serum (Xue et al., 2020; Wang et al., 2021; Liu et al., 2022; Clarke et al., 2023). In struvite stones resulting from urinary tract infections involving Proteus mirabilis, LPS can participate in stone formation, mainly by binding ions (Prywer and Torzewska, 2019). The negatively charged polysaccharides can bind free cations in urine (such as calcium and magnesium ions), causing these ions to aggregate around the bacteria, which accelerates stone formation and incorporates them into the stone.
The waa gene cluster in the core region of Proteus mirabilis LPS includes waaC, which is involved in core synthesis (Aquilini et al., 2010). Inactivation of waaC leads to partial loss of the core region and O-antigen, resulting in reduced motility, impaired swarming, increased serum sensitivity, and significantly heightened sensitivity to antimicrobial agents (with MIC reductions up to 64-fold). Crucially, in an in vitro bladder model, the ability of Proteus mirabilis to form crystalline biofilms is significantly reduced, thereby delaying catheter blockage. This may be due to the incomplete LPS structure from waaC gene deletion, inhibiting crystal embedding into biofilms. Further research is warranted to explore waaC as a potential target for inhibiting stone formation (Clarke et al., 2023).
The modification of lipid A in LPS requires the bifunctional polymyxin resistance protein ArnA, allowing bacteria to resist host antimicrobial peptides (Breazeale et al., 2005). ArnA mutants show reduced adaptability in the urethra and spleen, with significantly lower CFU counts adhering to bladder cells (Herout et al., 2023). However, the specific mechanisms and regulatory networks of ArnA in Proteus mirabilis regarding resistance to host immunity and adaptation to environmental stress still require further exploration.
Poorly crystalline and amorphous precipitate (PCaAP) is a component of infectious urinary stones, characterized by its lack of crystallinity and amorphous nature. It includes amorphous calcium carbonate (ACC), amorphous calcium phosphate (ACP), and/or amorphous carbonated calcium phosphate (ACCP). PCaAP can aggregate with Proteus mirabilis and struvite, accelerating stone formation and resulting in larger stones. LPS is the primary virulence factor in Proteus mirabilis, promoting PCaAP aggregation. The effect of LPS on PCaAP aggregation depends on its concentration. LPS at 12 μg/ml and 50 μg/ml enhances PCaAP aggregation, while 360 μg/ml and 440 μg/ml inhibit it (Prywer and Torzewska, 2019), This inhibitory effect may be due to high concentrations of LPS coating the PCaAP particles, preventing further aggregation. However, this does not imply that increasing LPS levels would generally reduce the formation of poorly crystalline and amorphous precipitates, but rather that it influenced the aggregation process under specific experimental conditions. LPS provides favorable sites for PCaAP nucleation, and they can bind through van der Waals forces or hydrogen bonds. This interaction may lower the energy threshold for PCaAP nucleation, thereby accelerating nucleation (Yang et al., 2003). Future research focusing more on biofilm formation and bacterial surface components related to adhesion and invasion capabilities would greatly enhance our understanding and treatment of Proteus mirabilis-induced urinary tract infections (Biondo, 2023).
Fimbriae enable bacteria to adhere to various abiotic and biotic surfaces. This adhesion capability allows many fimbriae to function as virulence factors, interacting with hosts and materials (Blomfield and van der Woude, 2007). Proteus mirabilis contains 17 potential chaperone-usher fimbrial operons, the highest number recorded in any sequenced bacterial genome to date (Pearson et al., 2008). Four fimbrial operons have been linked to the virulence of Proteus mirabilis: Mannose-Resistant/Proteus-like fimbriae (MR/P), urothelial cell adhesin (UCA, also known as non-agglutinating fimbriae or NAF), Proteus mirabilis fimbria (PMF), and Fimbria 14. The virulence mechanisms of ambient-temperature fimbria (ATF) remain unclear. The following sections will introduce each type of fimbriae.
The formation of extracellular clusters by Proteus mirabilis is an essential early stage in urinary stone development, requiring MR/P fimbriae and urease. Thus, MR/P fimbriae are crucial for urinary stone formation induced by Proteus mirabilis (Schaffer et al., 2016). The mrp operon comprises mrpABCDEFGHJ (Pearson et al., 2011), with mrpA encoding the major structural subunit, mrpC encoding the outer membrane usher, mrpD encoding the periplasmic chaperone, mrpB and mrpE-G encoding four minor subunits, mrpH encoding the tip-located adhesin, and mrpJ encoding the transcriptional regulator (Li et al., 2002). The gene mrpI, separate from the mrp operon and independent of the mrp promoter, encodes a recombinase that can switch the Invertible Element (IE) from “ON” to “OFF” or from “OFF” to “ON” to control MR/P fimbriae expression (Li et al., 2002).
MrpH, located at the tip of mannose-resistant Proteus-like (MR/P) fimbriae, is essential for the assembly of MR/P fimbriae and necessary for their adhesion to surfaces (Li et al., 1999). The adhesive protein domain of MrpH features a unique folding structure with a Zn2+ binding site composed of three conserved histidine residues. MrpH relies on the binding of zinc ions for biofilm formation, and disrupting the zinc binding site can significantly inhibit the biofilm formation of Proteus mirabilis. Additionally, zinc ions can reversibly regulate the biofilm formation process (Jiang et al., 2020). In the future, we hope to see research targeting MrpH to explore whether MrpH can prevent biofilm formation of Proteus mirabilis and to investigate the specific molecular mechanisms involved.
MrpJ is a predicted helix-turn-helix (HTH) protein essential for an ascending UTI mouse model. The study found that the MrpJ mutant showed significantly reduced ability to colonize the urinary tract in mice (Norsworthy and Pearson, 2017). MrpJ can regulate multiple virulence factor genes in Proteus mirabilis, which are closely related to urinary tract infections and the induction of urinary stones caused by Proteus mirabilis. MrpJ can inhibit the flagellar gene flhDC, fimbriae genes pmfA, fim8A, and fim14A, as well as lipopolysaccharide (LPS) modification genes pagP, lpxK, and msbB (which are suspected to be involved in bacterial immune evasion). Additionally, it inhibits the swarming regulators umoA and umoB, the cell differentiation gene lrhA involved in swarming, and the gene ccm that determines cell shape with the strongest inhibitory effect (Hay et al., 1999; Bode et al., 2015). MrpJ can also promote the expression of the Pta toxin and inhibit the expression of the ZapA protease, where Pta is an autotransporter protein that promotes bacterial aggregation and can interact with bladder and renal epithelial cells (Alamuri and Mobley, 2008). The ZapA protease helps bacteria evade the immune system by cleaving antimicrobial peptides in urine (Belas et al., 2004). In addition, MrpJ can promote the expression of the T6SS system (Bode et al., 2015), which is related to self-recognition during the swarming process and contributes to bacterial adaptation during pyelonephritis and potential systemic dissemination (Wenren et al., 2013; Debnath et al., 2018). Recently, genes encoding the T6SS, such as tssA, idsA, hcp, and hcp3, have been identified as direct targets of MrpJ (Debnath et al., 2018).
Proteus mirabilis employs fimbriae-associated transcriptional regulators to balance these opposing processes by upregulating fimbrial adhesion while downregulating flagellar expression.
Research on ambient temperature fimbriae (ATF) is limited, with existing studies indicating that it is not essential for infection (Zunino et al., 2000). Recent research has revealed the role of a site within the atf promoter, atfJ. atfJ is essential for the expression of the atf operon and has a dual regulatory role in motility and adhesion. However, the specific relationship between atfJ and the pathogenic mechanism of Proteus mirabilis remains unclear and requires further research for confirmation (Bode et al., 2016).
The uroepithelial cell adhesin (UCA) fimbria, also known as nonagglutinating fimbriae (NAF), was the first fimbria identified in Proteus mirabilis (Wray et al., 1986), also known as nonagglutinating fimbriae (NAF). NAF can mediate the attachment of Proteus mirabilis to surface receptors of different cells (Tolson et al., 1997). The absence of UCA fimbriae significantly impairs the adhesive ability of Proteus mirabilis, suggesting that UCA may play a role in its localization within the kidneys (Pellegrino et al., 2013).
PMF contributes to bladder colonization during urinary tract infections and kidney colonization during bloodstream infections (Zunino et al., 2003). Both PMF and MR/P fimbriae have specific and cumulative effects in urinary tract infections caused by Proteus mirabilis (Zunino et al., 2007).
For ATF fimbriae, different scholars have demonstrated that it does not play a virulence role in urinary tract infections caused by Proteus mirabilis. However, experiments conducted by Paola Scavone and colleagues confirmed that UCA mutants and PMF mutants produced more extracellular matrix after two days of biofilm assessment compared to the control group. After seven days of cultivation, they produced the highest total bacterial volume (Scavone et al., 2016). Although some studies have addressed this field, the functions of UCA fimbriae and PMF and their roles in Proteus mirabilis biology have not been fully explored. Thus, further research on UCA fimbriae and PMF is essential to uncover their potential mechanisms in bacterial growth, environmental adaptation, and host interactions.
The primary functions of bacterial flagella are motility and pathogenicity. In Proteus mirabilis, flagella influence both swimming and swarming motility, aiding in ascension to the kidneys and migration into the bloodstream (Asadi Karam et al., 2018; Scavone et al., 2023).
Proteus mirabilis mutants lacking flagellin cannot swim or swarm on LB agar plates and cannot migrate along latex and silicone catheters. Their biofilm formation ability in LB broth is significantly impaired (p < 0.0001) (Scavone et al., 2023).
In Proteus mirabilis, flagella expression is regulated through a three-tiered hierarchy (Chevance and Hughes, 2008). The flhDC operon, expressed from class 1 promoters, encodes the master flagellar transcriptional regulator complex FlhD4C2. FlhD4C2 activates class 2 promoters, which transcribe genes including fliA (encoding the flagellar-specific sigma factor σ28), flgM (encoding the anti-σ28 factor), and genes encoding proteins that comprise the HBB complex and the type III flagellar secretion system. FliA is essential for transcribing class 3 promoters, which include genes necessary for flagellum production, stator complexes, and chemotaxis (Morgenstein et al., 2010).
Flagellin promotes the expression of pro-inflammatory chemokines in the bladder, leading to inflammation and recruiting leukocytes to the bladder epithelium (Umpiérrez et al., 2013).
The fliL gene is the first gene in the class 2 operon fliLMNOPQR, which is controlled by FlhD4C2 and FliA. Research has found that bacteria lacking fliL can still swarm. However, the deletion of fliL alters Proteus mirabilis’s surface sensing (i.e., ΔfliL cells are more sensitive to low-viscosity agar), enhances the differentiation of Proteus mirabilis swarm cells, and makes the motility of cells lacking fliL temperature-dependent (Lee and Belas, 2015).
The wosA gene encodes a membrane-associated protein that can induce swarm cell differentiation when overexpressed. The transcription levels of the flhDC operon and flaA are upregulated with wosA overexpression, suggesting that wosA may play a role in the signaling pathway of the flagellar motility switch complex (Hatt and Rather, 2008).
The sdaA and sdaB genes encode enzymes that catalyze the deamination of L-serine (Zhang and Newman, 2008), which serves as a carbon and nitrogen source for metabolism in Proteus mirabilis, supporting its growth and motility (Brauer et al., 2019). When both genes are absent, biofilm integrity is compromised, proton motive force is disrupted, and the bacteria exhibit severe defects in flagellar motility (Brauer et al., 2022).
Molybdenum (Mo) is a transition metal and a vital trace element for nearly all living organisms (Kumar and Park, 2018). The modA gene encodes the molybdate-binding protein ModA, which binds molybdate (MoO42−) with high affinity for its transport. In bacteria, molybdate is transported and incorporated into a molybdopterin precursor before the synthesis of molybdoenzymes, which play a crucial role in anaerobic respiration. ModA influences the transport of molybdate in Proteus mirabilis, affecting the synthesis of molybdoenzymes and, consequently, bacterial anaerobic respiration. The deletion of modA enhances bacterial aggregation and motility (Huang et al., 2023), which is consistent with previous research showing significant upregulation of the modA gene in Proteus mirabilis strains with migration defects (Peng et al., 2020). Additionally, the absence of ModA upregulates several genes in the flagellar assembly pathway, such as flhB, while downregulating multiple genes in the pilus assembly and bacterial adhesion pathways, including mrpA, papA, fimF, fimC, and fimA. Although bacterial aggregation and motility increase under anaerobic conditions, biofilm formation decreases. This paradox is attributed to the downregulation of the mrpA gene, triggered by the deletion of modA, which is known to contribute to in vitro biofilm formation via MR/P fimbriae. The downregulation of fimbrial genes, which hinders bacterial adhesion and invasiveness, is likely a key factor in the reduction of biofilm formation. We propose that biofilm formation is a complex process regulated by various factors, such as environmental conditions, nutrient availability, signaling pathways, and bacterial interactions. Additionally, modA is directly related to bacterial metabolism, and its deletion may disrupt specific metabolic pathways, leading to the inhibition of biofilm formation. Interestingly, transcriptomic profiling and RT-qPCR results from ΔmodA and DP2019 strains showed significant upregulation of the hyfB and fdhF genes, which are part of the hydrogenase system mentioned earlier (Huang et al., 2023). However, no studies have yet clarified the potential link between modA and the regulation of hydrogenase genes. Therefore, we hope that future research will focus on exploring this connection and determining whether other regulatory factors or signaling pathways are involved in mediating this process. We believe that such studies will offer a more comprehensive understanding of ModA’s role in bacterial metabolism.
Understanding the mechanisms of ModA is crucial for developing new therapeutic strategies against harmful pathogens. Current research on ModA is still limited, and many of its mechanisms remain unclear. Studies on Enterococcus ModA (KpModA) have explored high-resolution anaerobic anion and molybdate-binding crystal structures, detailing interactions between ModA and molybdate and tungstate anions. This foundational research provides a pathway for targeting ModA to inhibit Klebsiella pneumoniae virulence (Zhao et al., 2023). Given that both Klebsiella pneumoniae and Proteus mirabilis are uropathogens and share many similarities (Palusiak, 2022), further investigation into ModA in Proteus mirabilis could yield valuable insights. However, such studies have not yet been conducted. Expanding this research could significantly advance our understanding and control of these pathogens.
Autotransporter proteins (ATs) play critical roles in urinary tract infections by promoting bacterial adhesion to host cells or extracellular matrix proteins, host cell invasion, cytotoxicity, serum resistance, and autoaggregation (Stathopoulos et al., 2000; Henderson and Nataro, 2001). Trimeric autotransporter (AT-2) proteins form an AT subfamily with similar structural features, and their passenger domains primarily consist of adhesins, proteases, or esterases (Guérin et al., 2017).The autotransporter protein Pta, identified and named in 2008, has its cytotoxicity associated with its α-domain (Pta-α), which induces actin depolymerization and disrupts nuclear membrane stability. The cytotoxicity and autoaggregation ability of Pta mutants are significantly reduced (Alamuri and Mobley, 2008). Praveen Alamuri et al. first demonstrated two AT-2 proteins in Proteus mirabilis: AipA and TaaP. Like other AT-2 proteins, they exhibit high affinity for extracellular matrix proteins. In their study, AipA demonstrated host cell invasion while TaaP showed autoaggregation (Alamuri et al., 2010). Despite these findings, research on autotransporter proteins in Proteus mirabilis remains limited. Future studies could focus on more virulence-related experiments on these proteins or identify additional ATs and AT-2s. Overall, this field holds significant research potential, and deepening our understanding could pave the way for innovative therapeutic strategies.
Two-component systems (TCS) play essential roles in urinary tract infections caused by Proteus mirabilis (Huang et al., 2024). They regulate bacterial processes like osmoregulation, membrane stress response, and cell differentiation. TCS consists of a sensor histidine kinase (HK) and a response regulator (RR) (Stock et al., 2000), with the HK detecting environmental changes and triggering signal transduction through autophosphorylation. The RR then adjusts downstream gene expression to help the bacteria adapt. Understanding TCS mechanisms in Proteus mirabilis could lead to targeted therapies for urinary tract infections.
The Rcs phosphorelay includes RcsF, RcsC, RcsD, and RcsB (Brill et al., 1988; Howery et al., 2016). RcsF senses membrane stress and activates RcsC, which initiates a phosphate transfer to RcsB through RcsD, activating downstream pathways (Takeda et al., 2001; Castanié-Cornet et al., 2006).
The response regulator RppA in Proteus mirabilis regulates flagellin synthesis, hemolysin activity, and sensitivity to polymyxin B (PB). Knockout of RppA increases sensitivity to PB, alters LPS, and reduces differentiation (Wang et al., 2008). RppA also controls pmrI expression, and pmrI knockout mutants show similar changes in PB sensitivity, biofilm formation, and resistance to imipenem (Jiang et al., 2010).
A regulatory factor of the TCS, QseF, participates in the regulation of swarming motility in Proteus mirabilis through GlmY, an sRNA of Proteus mirabilis. The glmY-qseE-qseG-qseF genes form an operon. Mutants of glmY and qseF exhibit reduced swarming motility and related phenotypes, including shorter cells, fewer flagella, significantly lower flhDC promoter activity, increased rcsB mRNA levels, and significantly reduced cheA mRNA levels (Lin et al., 2022).
CpxR, a key regulator in the TCS, controls biofilm formation via the zapABCD operon. Mutants of zapD and cpxR show reduced biofilm formation, protease activity, and adhesion. CpxR also responds to copper, enhancing biofilm formation through MR/P fimbriae, ZapA, eDNA, and EPS (Chen et al., 2020).
RpoE, as an alternative sigma factor, is crucial for maintaining the cell envelope integrity of Gram-negative bacteria (De Las Peñas et al., 1997). The rpoE gene is co-transcribed with genes encoding the anti-sigma factor rseA and the auxiliary proteins rseB and rseC (Kanehara et al., 2002; Crouch et al., 2005). Liu et al. first introduced the function of RpoE in Proteus mirabilis, revealing its ability to inhibit swarming motility, swarming cell differentiation, and swarming-associated hemolysin activity. RpoE affects mrp expression by regulating mrpI (the mrp operon was previously described). Mutation of rpoE increases the cytotoxicity of Proteus mirabilis, significantly impairing the bacteria’s colonization ability and resulting in more immune cell infiltration in the bladder and kidneys during the early stages of infection. Additionally, RpoE can sense urea in the host urethra and polymyxin B, both of which can induce RpoE expression (Liu et al., 2015).
Polyphosphate kinase (PPK), encoded by the ppk gene, is a key enzyme involved in the synthesis of inorganic polyphosphate (polyP) from ATP and is highly conserved across bacterial species (Tzeng and Kornberg, 1998). The study by Peng revealed the role of PPK in Proteus mirabilis, demonstrating its ability to promote bladder inflammation. PPK mutants lack resistance to oxidative, osmotic, and heat stress, exhibit reduced adhesion and invasion capabilities to urothelial cells, and show significantly decreased colonization in the bladder. Their swarming motility and biofilm formation abilities are also significantly weakened. Additionally, PPK mutants exhibit downregulated expression of universal stress protein G and heat shock proteins (Peng et al., 2016), both of which are important for protecting cells from stress (Santoro, 2000; Nachin et al., 2005).
Cyclic AMP receptor protein (Crp) is a transcriptional regulator that modulates bacterial metabolism and coordinates the expression of virulence factors through a process known as carbon catabolite repression (CCR) (Gosset et al., 2004; Zheng et al., 2004; Shimada et al., 2011). The study by Yi-Lin Tsai et al. first revealed the role of Crp in Proteus mirabilis, showing that Crp activity is regulated by sugar content within the host and may be associated with urinary tract infections. Compared to the wild-type strain, crp mutants showed increased kidney colonization, decreased flhDC expression, and almost no swimming or swarming motility. The crp mutants also exhibited increased adhesion to renal epithelial cells, higher pmp fimbriae mRNA expression, enhanced survival rates within macrophages, increased stress tolerance, and elevated expression of the sigma factor RpoS (Tsai et al., 2017).
Hfq, a bacterial RNA chaperone, can bind small RNAs (sRNAs) and mRNAs, participating in the riboregulation of different genes (Brennan and Link, 2007). It is also associated with stress responses, iron homeostasis, and other bacterial physiological activities (Chao and Vogel, 2010; Richards and Vanderpool, 2011) The study by Wang et al. first revealed the role of Hfq in Proteus mirabilis. Hfq helps resist damage caused by reactive oxygen species (ROS), high temperatures, and high osmotic pressure. Compared to the wild-type, Hfq mutants have impaired colonization ability in the bladder and kidneys, and promote the production of the key antibacterial cytokines IL-8 and MIF by urothelial cells. They also exhibit reduced swimming and swarming motility, fewer flagella, significantly decreased hemolysin activity, and significantly reduced biofilm formation. Additionally, Hfq mutants show impaired adhesion and invasion capabilities to urothelial cells and increased promoter activity of RpoE (as previously mentioned). Hfq mutants also lock the expression of the mrp operon, which controls MR/P fimbriae expression, in the off state (Wang et al., 2014). Various virulence factors of Proteus mirabilis is shown in Figure 3.
This article provides comprehensive and detailed information on the specific mechanisms of virulence factors in Proteus mirabilis-induced urinary tract infections. However, there is still much to be explored regarding the changes in the expression of virulence factors during the transition from swimming to swarming motility. The interactions between different virulence factors within Proteus mirabilis remain unclear. The latest research indicates that Proteus mirabilis encodes multiple secretion system-related genes, but the specific functions of most of these systems remain largely unexplored (Tatarenkov et al., 2024). Furthermore, the host urinary tract contains multiple bacterial species, and biofilm formation involves interactions among them. Therefore, further investigation is essential to fully understand the pathogenic capacity of Proteus mirabilis in urinary tract infections and its interactions within the host microbiome.
AY: Methodology, Writing – original draft. YT: Visualization, Writing – original draft. XL: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Innovation Team Project of Department of Education of Liaoning Province (Grant No.: 507317).
Figure 3 in this article was drawn by Figdraw, BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relat ionships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alamuri, P., Löwer, M., Hiss, J. A., Himpsl, S. D., Schneider, G., Mobley, H. L. (2010). Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect Immun. 78, 4882–4894. doi: 10.1128/IAI.00718-10
Alamuri, P., Mobley, H. L. (2008). A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol. Microbiol. 68, 997–1017. doi: 10.1111/j.1365-2958.2008.06199.x
Allison, C., Emödy, L., Coleman, N., Hughes, C. (1994). The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J. Infect. Dis. 169, 1155–1158. doi: 10.1093/infdis/169.5.1155
Aquilini, E., Azevedo, J., Jimenez, N., Bouamama, L., Tomás, J. M., Regué, M. (2010). Functional identification of the Proteus mirabilis core lipopolysaccharide biosynthesis genes. J. Bacteriol. 192, 4413–4424. doi: 10.1128/JB.00494-10
Asadi Karam, M. R., Shirzad, A. M., Habibi, M., Bouzari, S. (2018). A heterologous prime-boost route of vaccination based on the truncated MrpH adhesin and adjuvant properties of the flagellin from Proteus mirabilis against urinary tract infections. Int. Immunopharmacol. 58, 40–47. doi: 10.1016/j.intimp.2018.03.009
Belas, R., Manos, J., Suvanasuthi, R. (2004). Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect. Immun. 72, 5159–5167. doi: 10.1128/IAI.72.9.5159-5167.2004
Biondo, C. (2023). New insights into the pathogenesis and treatment of urinary tract infections. Pathogens 12 (10), 1213. doi: 10.3390/pathogens12101213
Blomfield, I., van der Woude, M. (2007). Regulation of fimbrial expression. EcoSal Plus. 2. doi: 10.1128/ecosal.2.4.2.2
Bode, N. J., Chan, K. W., Kong, X. P., Pearson, M. M. (2016). Distinct residues contribute to motility repression and autoregulation in the Proteus mirabilis fimbria-associated transcriptional regulator AtfJ. J. Bacteriol. 198, 2100–2112. doi: 10.1128/JB.00193-16
Bode, N. J., Debnath, I., Kuan, L., Schulfer, A., Ty, M., Pearson, M. M. (2015). Transcriptional analysis of the MrpJ network: modulation of diverse virulence-associated genes and direct regulation of mrp fimbrial and flhDC flagellar operons in Proteus mirabilis. Infect. Immun. 83, 2542–2556. doi: 10.1128/IAI.02978-14
Brauer, A. L., Learman, B. S., Armbruster, C. E. (2023). Differential Contribution of Hydrogen Metabolism to Proteus mirabilis Fitness during Single-Species and Polymicrobial Catheterized Urinary Tract Infection. Pathog. (Basel Switzerland). 12, 1377. doi: 10.3390/pathogens12121377
Brauer, A. L., Learman, B. S., Taddei, S. M., Deka, N., Hunt, B. C., Armbruster, C. E. (2022). Preferential catabolism of l- vs d-serine by Proteus mirabilis contributes to pathogenesis and catheter-associated urinary tract infection. Mol. Microbiol. 118, 125–144. doi: 10.1111/mmi.v118.3
Brauer, A. L., White, A. N., Learman, B. S., Johnson, A. O., Armbruster, C. E. (2019). d-Serine Degradation by Proteus mirabilis Contributes to Fitness during Single-Species and Polymicrobial Catheter-Associated Urinary Tract Infection. mSphere 4. doi: 10.1128/msphere.00020-19
Breazeale, S. D., Ribeiro, A. A., McClerren, A. L., Raetz, C. R. (2005). A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 280, 14154–14167. doi: 10.1074/jbc.M414265200
Brennan, R. G., Link, T. M. (2007). Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10, 125–133. doi: 10.1016/j.mib.2007.03.015
Brill, J. A., Quinlan-Walshe, C., Gottesman, S. (1988). Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170, 2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988
Castanié-Cornet, M. P., Cam, K., Jacq, A. (2006). RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188, 4264–4270. doi: 10.1128/JB.00004-06
Chakkour, M., Hammoud, Z., Farhat, S., El Roz, A., Ezzeddine, Z., Ghssein, G. (2024). Overview of Proteus mirabilis pathogenicity and virulence. Insights into the role of metals. Front. Microbiol. 15, 1383618. doi: 10.3389/fmicb.2024.1383618
Chang, W. N., Tsai, Y. C., Chien, C. C., Huang, C. R., Lu, C. H. (2002). Frequent association with neurosurgical conditions in adult Proteus mirabilis meningitis: report of five cases. Clin. Neurol. Neurosurg. 104, 121–124. doi: 10.1016/S0303-8467(01)00193-7
Chao, Y., Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. doi: 10.1016/j.mib.2010.01.001
Chen, H. H., Chang, C. C., Yuan, Y. H., Liaw, S. J. (2020). A CpxR-Regulated zapD Gene Involved in Biofilm Formation of Uropathogenic Proteus mirabilis. Infect Immun. 88. doi: 10.1128/iai.00207-20
Chevance, F. F., Hughes, K. T. (2008). Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465. doi: 10.1038/nrmicro1887
Choi, J. G., Kim, N., Ju, I. G., Eo, H., Lim, S. M., Jang, S. E., et al. (2018). Oral administration of Proteus mirabilis damages dopaminergic neurons and motor functions in mice. Sci. Rep. 8, 1275. doi: 10.1038/s41598-018-19646-x
Clarke, O. E., Pelling, H., Bennett, V., Matsumoto, T., Gregory, G. E., Nzakizwanayo, J., et al. (2023). Lipopolysaccharide structure modulates cationic biocide susceptibility and crystalline biofilm formation in Proteus mirabilis. Front. Microbiol. 14, 1150625. doi: 10.3389/fmicb.2023.1150625
Combaz-Söhnchen, N., Kuhn, A. (2017). A systematic review of mycoplasma and ureaplasma in urogynaecology. Geburtshilfe und Frauenheilkunde. 77, 1299–1303. doi: 10.1055/s-0043-119687
Crouch, M. L., Becker, L. A., Bang, I. S., Tanabe, H., Ouellette, A. J., Fang, F. C. (2005). The alternative sigma factor sigma is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol. Microbiol. 56, 789–799. doi: 10.1111/j.1365-2958.2005.04578.x
Debnath, I., Stringer, A. M., Smith, S. N., Bae, E., Mobley, H. L. T., Wade, J. T., et al. (2018). MrpJ Directly Regulates Proteus mirabilis Virulence Factors, Including Fimbriae and Type VI Secretion, during Urinary Tract Infection. Infect Immun. 86 (10). doi: 10.1128/iai.00388-18
De Las Peñas, A., Connolly, L., Gross, C. A. (1997). The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24, 373–385. doi: 10.1046/j.1365-2958.1997.3611718.x
Drzewiecka, D. (2016). Significance and roles of Proteus spp. Bacteria in natural environments. Microbial Ecol. 72, 741–758. doi: 10.1007/s00248-015-0720-6
Ebringer, A., Rashid, T. (2014). Rheumatoid arthritis is caused by a Proteus urinary tract infection. APMIS: Acta pathologica microbiologica immunologica Scandinavica. 122, 363–368. doi: 10.1111/apm.2014.122.issue-5
Fitzgerald, M. J., Pearson, M. M., Mobley, H. L. T. (2024). Proteus mirabilis UreR coordinates cellular functions required for urease activity. J. Bacteriol. 206 (4), e00031–e00024. doi: 10.1128/jb.00031-24
Gabel, C. V., Berg, H. C. (2003). The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc. Natl. Acad. Sci. U.S.A 100, 8748–8751. doi: 10.1073/pnas.1533395100
Gambrill, B., Pertusati, F., Hughes, S. F., Shergill, I., Prokopovich, P. (2024). Materials-based incidence of urinary catheter associated urinary tract infections and the causative micro-organisms: systematic review and meta-analysis. BMC Urology. 24, 186. doi: 10.1186/s12894-024-01565-x
Gmiter, D., Kaca, W. (2022). Into the understanding the multicellular lifestyle of Proteus mirabilis on solid surfaces. Front. Cell. Infect. Microbiol. 12, 864305. doi: 10.3389/fcimb.2022.864305
Gosset, G., Zhang, Z., Nayyar, S., Cuevas, W. A., Saier, M. H., Jr. (2004). Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186, 3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004
Grahl, M. V. C., Uberti, A. F., Broll, V., Bacaicoa-Caruso, P., Meirelles, E. F., Carlini, C. R. (2021). Proteus mirabilis urease: unsuspected non-enzymatic properties relevant to pathogenicity. Int. J. Mol. Sci. 22, 7205. doi: 10.3390/ijms22137205
Griffith, D. P. (1982). Infection-induced renal calculi. Kidney Int. 21, 422–430. doi: 10.1038/ki.1982.40
Guérin, J., Bigot, S., Schneider, R., Buchanan, S. K., Jacob-Dubuisson, F. (2017). Two-partner secretion: combining efficiency and simplicity in the secretion of large proteins for bacteria-host and bacteria-bacteria interactions. Front. Cell. Infect. Microbiol. 7, 148. doi: 10.3389/fcimb.2017.00148
Hatt, J. K., Rather, P. N. (2008). Characterization of a novel gene, wosA, regulating FlhDC expression in Proteus mirabilis. J. Bacteriol. 190, 1946–1955. doi: 10.1128/JB.01010-07
Hay, N. A., Tipper, D. J., Gygi, D., Hughes, C. (1999). A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J. Bacteriol. 181, 2008–2016. doi: 10.1128/JB.181.7.2008-2016.1999
Henderson, I. R., Nataro, J. P. (2001). Virulence functions of autotransporter proteins. Infect. Immun. 69, 1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001
Herout, R., Khoddami, S., Moskalev, I., Reicherz, A., Chew, B. H., Armbruster, C. E., et al. (2023). Role of bacterial surface components in the pathogenicity of Proteus mirabilis in a murine model of catheter-associated urinary tract infection. Pathog. (Basel Switzerland). 12, 509. doi: 10.3390/pathogens12040509
Howery, K. E., Clemmer, K. M., Rather, P. N. (2016). The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr. Genet. 62, 775–789. doi: 10.1007/s00294-016-0579-1
Huang, Y., Chen, J., Jiang, Q., Huang, N., Ding, X., Peng, L., et al. (2023). The molybdate-binding protein ModA is required for Proteus mirabilis-induced UTI. Front. Microbiol. 14, 1156273. doi: 10.3389/fmicb.2023.1156273
Huang, W. S., Lee, Y. J., Wang, L., Chen, H. H., Chao, Y. J., Cheng, V., et al. (2024). Copper affects virulence and diverse phenotypes of uropathogenic Proteus mirabilis. J. microbiology immunology infection = Wei mian yu gan ran za zhi. 57, 385–395. doi: 10.1016/j.jmii.2024.02.007
Jacobsen, S. M., Shirtliff, M. E. (2011). Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence 2, 460–465. doi: 10.4161/viru.2.5.17783
Jacobsen, S. M., Stickler, D. J., Mobley, H. L., Shirtliff, M. E. (2008). Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 21, 26–59. doi: 10.1128/CMR.00019-07
Jiang, S. S., Liu, M. C., Teng, L. J., Wang, W. B., Hsueh, P. R., Liaw, S. J. (2010). Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrobial Agents Chemother. 54, 1564–1571. doi: 10.1128/AAC.01219-09
Jiang, W., Ubhayasekera, W., Breed, M. C., Norsworthy, A. N., Serr, N., Mobley, H. L. T., et al. (2020). MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation. PloS Pathogens. 16, e1008707. doi: 10.1371/journal.ppat.1008707
Jones, B. V., Young, R., Mahenthiralingam, E., Stickler, D. J. (2004). Ultrastructure of Proteus mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect. Immun. 72, 3941–3950. doi: 10.1128/IAI.72.7.3941-3950.2004
Juárez-Chairez, M. F., Cid-Gallegos, M. S., Jiménez-Martínez, C., Prieto-Contreras, L. F., Bollain, Y.-l. (2024). The role of microbiota on rheumatoid arthritis onset. Int. J. Rheum. Dis. 27, e15122. doi: 10.1111/1756-185X.15122
Kanehara, K., Ito, K., Akiyama, Y. (2002). YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 16, 2147–2155. doi: 10.1101/gad.1002302
Khorasani, A., Moghim, S., Wagemans, J., Lavigne, R., Mirzaei, A. (2024). Antibiotic profile classification of Proteus mirabilis using machine learning: An investigation into multidimensional radiomics features. Comput. Biol. Med. 182, 109131. doi: 10.1016/j.compbiomed.2024.109131
Kumar, V., Park, S. (2018). Potential and limitations of Klebsiella pneumoniae as a microbial cell factory utilizing glycerol as the carbon source. Biotechnol. Adv. 36, 150–167. doi: 10.1016/j.biotechadv.2017.10.004
Learman, B. S., Brauer, A. L., Eaton, K. A., Armbruster, C. E. (2019). A rare opportunist, Morganella morganii, decreases severity of polymicrobial catheter-associated urinary tract infection. Infect Immun 88 (1). doi: 10.1128/iai.00691-19
Lee, Y. Y., Belas, R. (2015). Loss of FliL alters Proteus mirabilis surface sensing and temperature-dependent swarming. J. Bacteriol. 197, 159–173. doi: 10.1128/JB.02235-14
Leichtle, A., Lupatsii, M., Graspeuntner, S., Jeschke, S., Penxová, Z., Kurabi, A., et al. (2024). Anti-inflammatory response to 1,8-Cineol and associated microbial communities in Otitis media patients. Sci. Rep. 14, 16362. doi: 10.1038/s41598-024-67498-5
Li, X., Johnson, D. E., Mobley, H. L. (1999). Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect. Immun. 67, 2822–2833. doi: 10.1128/IAI.67.6.2822-2833.1999
Li, X., Lockatell, C. V., Johnson, D. E., Mobley, H. L. (2002). Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol. Microbiol. 45, 865–874. doi: 10.1046/j.1365-2958.2002.03067.x
Li, W., Wu, X., Hu, X., Wang, T., Liang, S., Duan, Y., et al. (2017). Structural changes of gut microbiota in Parkinson's disease and its correlation with clinical features. Sci. China Life Sci. 60, 1223–1233. doi: 10.1007/s11427-016-9001-4
Lin, W. Y., Lee, Y. J., Yu, P. H., Tsai, Y. L., She, P. Y., Li, T. S., et al. (2022). The QseEF two-component system-GlmY small RNA regulatory pathway controls swarming in uropathogenic Proteus mirabilis. Int. J. Mol. Sci. 23, 487. doi: 10.3390/ijms23010487
Lin, W. Y., Liaw, S. J. (2020). Deacidification by FhlA-dependent hydrogenase is involved in urease activity and urinary stone formation in uropathogenic Proteus mirabilis. Sci. Rep. 10, 19546. doi: 10.1038/s41598-020-76561-w
Liu, M. C., Kuo, K. T., Chien, H. F., Tsai, Y. L., Liaw, S. J. (2015). New aspects of RpoE in uropathogenic Proteus mirabilis. Infect. Immun. 83, 966–977. doi: 10.1128/IAI.02232-14
Liu, A. N., Teng, K. W., Chew, Y., Wang, P. C., Nguyen, T. T. H., Kao, M. C. (2022). The effects of HP0044 and HP1275 knockout mutations on the structure and function of lipopolysaccharide in helicobacter pylori strain 26695. Biomedicines 10, 145. doi: 10.3390/biomedicines10010145
Manoharan, A., Farrell, J., Aldilla, V. R., Whiteley, G., Kriel, E., Glasbey, T., et al. (2023). N-acetylcysteine prevents catheter occlusion and inflammation in catheter associated-urinary tract infections by suppressing urease activity. Front. Cell. Infect. Microbiol. 13, 1216798. doi: 10.3389/fcimb.2023.1216798
McLean, R. J., Nickel, J. C., Noakes, V. C., Costerton, J. W. (1985). An in vitro ultrastructural study of infectious kidney stone genesis. Infect. Immun. 49, 805–811. doi: 10.1128/iai.49.3.805-811.1985
Miano, R., Germani, S., Vespasiani, G. (2007). Stones and urinary tract infections. Urologia Internationalis. 79 Suppl 1, 32–36. doi: 10.1159/000104439
Mo, S., Durrani, A. F., Safiullah, Z., Kowalski, R. P., Jhanji, V. (2021). Proteus mirabilis keratitis: risk factors, clinical features, treatment outcomes, and microbiological characteristics. Cornea 40, 704–709. doi: 10.1097/ICO.0000000000002469
Mobley, H. L. T. (2019). Proteus mirabilis overview. Methods Mol. Biol. (Clifton NJ). 2021, 1–4. doi: 10.1007/978-1-4939-9601-8_1
Morgenstein, R. M., Szostek, B., Rather, P. N. (2010). Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis. FEMS Microbiol. Rev. 34, 753–763. doi: 10.1111/j.1574-6976.2010.00229.x
Nachin, L., Nannmark, U., Nyström, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005
Noguchi, K., Riggins, D. P., Eldahan, K. C., Kitko, R. D., Slonczewski, J. L. (2010). Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli. PLoS One 5, e10132. doi: 10.1371/journal.pone.0010132
Norsworthy, A. N., Pearson, M. M. (2017). From catheter to kidney stone: the uropathogenic lifestyle of Proteus mirabilis. Trends Microbiol. 25, 304–315. doi: 10.1016/j.tim.2016.11.015
Odoki, M., Almustapha Aliero, A., Tibyangye, J., Nyabayo Maniga, J., Wampande, E., Drago Kato, C., et al. (2019). Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi district, Uganda. Int. J. Microbiol. 2019, 4246780. doi: 10.1155/2019/4246780
Palusiak, A. (2013). Immunochemical properties of Proteus penneri lipopolysaccharides–one of the major Proteus sp. virulence factors. Carbohydr. Res. 380, 16–22. doi: 10.1016/j.carres.2013.06.025
Palusiak, A. (2022). Proteus mirabilis and Klebsiella pneumoniae as pathogens capable of causing co-infections and exhibiting similarities in their virulence factors. Front. Cell. Infection Microbiol. 12, 991657. doi: 10.3389/fcimb.2022.991657
Parra, M. C., Collins, C. M. (2012). Mutational analysis of the N-terminal domain of UreR, the positive transcriptional regulator of urease gene expression. Microbiological Res. 167, 433–444. doi: 10.1016/j.micres.2012.03.005
Pearson, M. M., Sebaihia, M., Churcher, C., Quail, M. A., Seshasayee, A. S., Luscombe, N. M., et al. (2008). Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190, 4027–4037. doi: 10.1128/JB.01981-07
Pearson, M. M., Yep, A., Smith, S. N., Mobley, H. L. (2011). Transcriptome of Proteus mirabilis in the murine urinary tract: virulence and nitrogen assimilation gene expression. Infect. Immun. 79, 2619–2631. doi: 10.1128/IAI.05152-11
Pellegrino, R., Scavone, P., Umpiérrez, A., Maskell, D. J., Zunino, P. (2013). Proteus mirabilis uroepithelial cell adhesin (UCA) fimbria plays a role in the colonization of the urinary tract. Pathog. Dis. 67, 104–107. doi: 10.1111/2049-632X.12027
Pelling, H., Nzakizwanayo, J., Milo, S., Denham, E. L., MacFarlane, W. M., Bock, L. J., et al. (2019). Bacterial biofilm formation on indwelling urethral catheters. Lett. Appl. Microbiol. 68, 277–293. doi: 10.1111/lam.2019.68.issue-4
Peng, L., Chen, D. Q., Jiang, G. M., Ou, J. Y., Jiang, Q., Zeng, L. T., et al. (2020). Transcriptome analysis of two strains of Proteus mirabilis with swarming migration deficiency isolated from patients with urinary tract infection. Curr. Microbiol. 77, 1381–1389. doi: 10.1007/s00284-020-01931-6
Peng, L., Jiang, Q., Pan, J. Y., Deng, C., Yu, J. Y., Wu, X. M., et al. (2016). Involvement of polyphosphate kinase in virulence and stress tolerance of uropathogenic Proteus mirabilis. Med. Microbiol. Immunol. 205, 97–109. doi: 10.1007/s00430-015-0430-1
Phan, H., Lehman, D. (2012). Cerebral abscess complicating Proteus mirabilis meningitis in a newborn infant. J. Child Neurol. 27, 405–407. doi: 10.1177/0883073811422271
Poore, C. A., Mobley, H. L. T. (2003). Differential regulation of the Proteus mirabilis urease gene cluster by UreR and H-NS. Microbiol. (Reading England). 149, 3383–3394. doi: 10.1099/mic.0.26624-0
Prywer, J., Torzewska, A. (2019). Aggregation of poorly crystalline and amorphous components of infectious urinary stones is mediated by bacterial lipopolysaccharide. Sci. Rep. 9, 17061. doi: 10.1038/s41598-019-53359-z
Prywer, J., Torzewska, A., Mielniczek-Brzóska, E. (2024). Understanding the role of zinc ions on struvite nucleation and growth in the context of infection urinary stones. Metallomics: integrated biometal science. 16. doi: 10.1093/mtomcs/mfae017
Rather, P. N. (2005). Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7, 1065–1073. doi: 10.1111/j.1462-2920.2005.00806.x
Richards, G. R., Vanderpool, C. K. (2011). Molecular call and response: the physiology of bacterial small RNAs. Biochim. Biophys. Acta 1809, 525–531. doi: 10.1016/j.bbagrm.2011.07.013
Salehi, S., Mahmoudinezhad Dezfouli, S. M., Azadeh, H., Khosravi, S. (2023). Immune dysregulation and pathogenic pathways mediated by common infections in rheumatoid arthritis. Folia Microbiologica. 68, 325–335. doi: 10.1007/s12223-023-01036-0
Samtoy, B., DeBeukelaer, M. M. (1980). Ammonia encephalopathy secondary to urinary tract infection with Proteus mirabilis. Pediatrics 65, 294–297. doi: 10.1542/peds.65.2.294
Santoro, M. G. (2000). Heat shock factors and the control of the stress response. Biochem. Pharmacol. 59, 55–63. doi: 10.1016/S0006-2952(99)00299-3
Scavone, P., Iribarnegaray, V., Caetano, A. L., Schlapp, G., Härtel, S., Zunino, P. (2016). Fimbriae have distinguishable roles in Proteus mirabilis biofilm formation. Pathog. Dis. 74. doi: 10.1093/femspd/ftw033
Scavone, P., Iribarnegaray, V., González, M. J., Navarro, N., Caneles-Huerta, N., Jara-Wilde, J., et al. (2023). Role of Proteus mirabilis flagella in biofilm formation. Rev. Argent. Microbiologia. 55, 226–234. doi: 10.1016/j.ram.2023.01.005
Schaffer, J. N., Norsworthy, A. N., Sun, T. T., Pearson, M. M. (2016). Proteus mirabilis fimbriae- and urease-dependent clusters assemble in an extracellular niche to initiate bladder stone formation. Proc. Natl. Acad. Sci. U.S.A 113, 4494–4499. doi: 10.1073/pnas.1601720113
Schaffer, J. N., Pearson, M. M. (2015). Proteus mirabilis and urinary tract infections. Microbiol Spectr 3 (5). doi: 10.1128/microbiolspec.uti-0017-2013
Schlensog, V., Lutz, S., Böck, A. (1994). Purification and DNA-binding properties of FHLA, the transcriptional activator of the formate hydrogenlyase system from Escherichia coli. J. Biol. Chem. 269, 19590–19596. doi: 10.1016/S0021-9258(17)32210-X
Schoelmerich, M. C., Müller, V. (2020). Energy-converting hydrogenases: the link between H(2) metabolism and energy conservation. Cell. Mol. Life Sci: CMLS. 77, 1461–1481. doi: 10.1007/s00018-019-03329-5
Self, W. T., Hasona, A., Shanmugam, K. T. (2004). Expression and regulation of a silent operon, hyf, coding for hydrogenase 4 isoenzyme in Escherichia coli. J. Bacteriol. 186, 580–587. doi: 10.1128/JB.186.2.580-587.2004
Senior, B. W., Sweeney, G. (1984). The association of particular types of Proteus with chronic suppurative otitis media. J. Med. Microbiol. 17, 201–205. doi: 10.1099/00222615-17-2-201
Shimada, T., Fujita, N., Yamamoto, K., Ishihama, A. (2011). Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PloS One 6, e20081. doi: 10.1371/journal.pone.0020081
Stathopoulos, C., Hendrixson, D. R., Thanassi, D. G., Hultgren, S. J., St Geme, J. W., 3rd, Curtiss, R., 3rd (2000). Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2, 1061–1072. doi: 10.1016/S1286-4579(00)01260-0
Stock, A. M., Robinson, V. L., Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Stolarek, P., Bernat, P., Różalski, A. (2023). Adjustment in the Composition and Organization of Proteus mirabilis Lipids during the Swarming Process. Int. J. Mol. Sci. 24, 16461. doi: 10.3390/ijms242216461
Szczerbiec, D., Bednarska-Szczepaniak, K., Torzewska, A. (2024). Antibacterial properties and urease suppression ability of Lactobacillus inhibit the development of infectious urinary stones caused by Proteus mirabilis. Sci. Rep. 14, 943. doi: 10.1038/s41598-024-51323-0
Takeda, S., Fujisawa, Y., Matsubara, M., Aiba, H., Mizuno, T. (2001). A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC –> YojN –> RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40, 440–450. doi: 10.1046/j.1365-2958.2001.02393.x
Tatarenkov, A., Muñoz-Gutiérrez, I., Vargas, I., Behnsen, J., Mota-Bravo, L. (2024). Pangenome analysis reveals novel contact-dependent growth inhibition system and phenazine biosynthesis operons in Proteus mirabilis BL95 that are located in an integrative and conjugative element. Microorganisms 12 (7), 1321. doi: 10.3390/microorganisms12071321
Tolson, D. L., Harrison, B. A., Latta, R. K., Lee, K. K., Altman, E. (1997). The expression of nonagglutinating fimbriae and its role in Proteus mirabilis adherence to epithelial cells. Can. J. Microbiol. 43, 709–717. doi: 10.1139/m97-102
Tsai, Y. L., Chien, H. F., Huang, K. T., Lin, W. Y., Liaw, S. J. (2017). cAMP receptor protein regulates mouse colonization, motility, fimbria-mediated adhesion, and stress tolerance in uropathogenic Proteus mirabilis. Sci. Rep. 7, 7282. doi: 10.1038/s41598-017-07304-7
Tzeng, C. M., Kornberg, A. (1998). Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol. Microbiol. 29, 381–382. doi: 10.1046/j.1365-2958.1998.00887.x
Umpiérrez, A., Scavone, P., Romanin, D., Marqués, J. M., Chabalgoity, J. A., Rumbo, M., et al. (2013). Innate immune responses to Proteus mirabilis flagellin in the urinary tract. Microbes Infect. 15, 688–696. doi: 10.1016/j.micinf.2013.06.007
Wang, W. B., Chen, I. C., Jiang, S. S., Chen, H. R., Hsu, C. Y., Hsueh, P. R., et al. (2008). Role of RppA in the regulation of polymyxin b susceptibility, swarming, and virulence factor expression in Proteus mirabilis. Infect. Immun. 76, 2051–2062. doi: 10.1128/IAI.01557-07
Wang, M. C., Chien, H. F., Tsai, Y. L., Liu, M. C., Liaw, S. J. (2014). The RNA chaperone Hfq is involved in stress tolerance and virulence in uropathogenic Proteus mirabilis. PloS One 9, e85626. doi: 10.1371/journal.pone.0085626
Wang, J., Ma, W., Fang, Y., Liang, H., Yang, H., Wang, Y., et al. (2021). Core oligosaccharide portion of lipopolysaccharide plays important roles in multiple antibiotic resistance in Escherichia coli. Antimicrobial Agents Chemother. 65, e0034121. doi: 10.1128/AAC.00341-21
Wasfi, R., Hamed, S. M., Amer, M. A., Fahmy, L. I. (2020). Proteus mirabilis biofilm: development and therapeutic strategies. Front. Cell. Infect. Microbiol. 10, 414. doi: 10.3389/fcimb.2020.00414
Wenren, L. M., Sullivan, N. L., Cardarelli, L., Septer, A. N., Gibbs, K. A. (2013). Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. Mbio 4 (4). doi: 10.1128/mbio.00374-13
Wilks, S. A., Fader, M. J., Keevil, C. W. (2015). Novel Insights into the Proteus mirabilis Crystalline Biofilm Using Real-Time Imaging. PLoS One 10, e0141711. doi: 10.1371/journal.pone.0141711
Wray, S. K., Hull, S. I., Cook, R. G., Barrish, J., Hull, R. A. (1986). Identification and characterization of a uroepithelial cell adhesin from a uropathogenic isolate of Proteus mirabilis. Infect. Immun. 54, 43–49. doi: 10.1128/iai.54.1.43-49.1986
Xue, Y., Li, Q., Park, C. G., Klena, J. D., Anisimov, A. P., Sun, Z., et al. (2020). Proteus mirabilis Targets Atherosclerosis Plaques in Human Coronary Arteries via DC-SIGN (CD209). Front. Immunol. 11, 579010. doi: 10.3389/fimmu.2020.579010
Yang, L., Zhang, X., Liao, Z., Guo, Y., Hu, Z., Cao, Y. (2003). Interfacial molecular recognition between polysaccharides and calcium carbonate during crystallization. J. Inorganic Biochem. 97, 377–383. doi: 10.1016/S0162-0134(03)00311-8
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Zhang, J., Hoedt, E. C., Liu, Q., Berendsen, E., Teh, J. J., Hamilton, A., et al. (2021). Elucidation of proteus mirabilis as a key bacterium in Crohn's disease inflammation. Gastroenterology 160, 317–30.e11. doi: 10.1053/j.gastro.2020.09.036
Zhang, X., Newman, E. (2008). Deficiency in l-serine deaminase results in abnormal growth and cell division of Escherichia coli K-12. Mol. Microbiol. 69, 870–881. doi: 10.1111/j.1365-2958.2008.06315.x
Zhao, Q., Su, X., Wang, Y., Liu, R., Bartlam, M. (2023). Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae. Biochem. Biophys. Res. Commun. 681, 41–46. doi: 10.1016/j.bbrc.2023.09.055
Zheng, D., Constantinidou, C., Hobman, J. L., Minchin, S. D. (2004). Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32, 5874–5893. doi: 10.1093/nar/gkh908
Zunino, P., Geymonat, L., Allen, A. G., Legnani-Fajardo, C., Maskell, D. J. (2000). Virulence of a Proteus mirabilis ATF isogenic mutant is not impaired in a mouse model of ascending urinary tract infection. FEMS Immunol. Med. Microbiol. 29, 137–143. doi: 10.1111/j.1574-695X.2000.tb01516.x
Zunino, P., Sosa, V., Allen, A. G., Preston, A., Schlapp, G., Maskell, D. J. (2003). Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiol. (Reading England). 149, 3231–3237. doi: 10.1099/mic.0.26534-0
Keywords: urinary tract infections, catheter-associated urinary tract infections, virulence factors, biofilm formation, urinary stone formation, Proteus mirabilis
Citation: Yang A, Tian Y and Li X (2024) Unveiling the hidden arsenal: new insights into Proteus mirabilis virulence in UTIs. Front. Cell. Infect. Microbiol. 14:1465460. doi: 10.3389/fcimb.2024.1465460
Received: 16 July 2024; Accepted: 18 October 2024;
Published: 13 November 2024.
Edited by:
Reham Wasfi, MSA University, EgyptCopyright © 2024 Yang, Tian and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiancheng Li, bGl4aWFuY2hlbmdAZG11LmVkdS5jbg==
†ORCID: Xiancheng Li, orcid.org/0000-0002-8301-2589
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.