
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 24 October 2024
Sec. Clinical Microbiology
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1462620
Acinetobacter baumannii, a predominant nosocomial pathogen, represents a grave threat to public health due to its multiple antimicrobial resistance. Managing patients afflicted with severe infections caused by multiple drug-resistant A. baumannii is particularly challenging, given the associated high mortality rates and unfavorable prognoses. The diminishing efficacy of antibiotics against this superbug underscores the urgent necessity for novel treatments or strategies to address this formidable issue. Bacteriophage-derived polysaccharide depolymerase enzymes present a potential approach to combating this pathogen. These enzymes target and degrade the bacterial cell’s exopolysaccharide, capsular polysaccharide, and lipopolysaccharide, thereby disrupting biofilm formation and impairing the bacteria’s defense mechanisms. Nonetheless, the narrow host range of phage depolymerases limits their therapeutic efficacy. Despite the benefits of these enzymes, phage-resistant strains have been identified, highlighting the complexity of phage-host interactions and the need for further investigation. While preliminary findings are encouraging, current investigations are limited, and clinical trials are imperative to advance this treatment approach for broader clinical applications. This review explores the potential of phage-derived depolymerase enzymes against A. baumannii infections.
Acinetobacter baumannii (A. baumannii) has emerged as one of the most formidable multidrug-resistant (MDR) nosocomial pathogens. The primary complications associated with infections by various MDR A. baumannii strains include pneumonia, bloodstream infections, urinary tract infections, skin and wound infections (Nguyen and Joshi, 2021). The World Health Organization (WHO, 2024) and the Centers for Disease Control and Prevention (CDC, 2019) have designated this bacterium as a critical priority pathogen due to the scarcity of effective treatment options, emphasizing the urgent need for additional research to address this challenge.
The exopolysaccharide (EPS), capsular polysaccharide (CPS), lipo-oligosaccharide (LOS), outer membrane protein, pili, adhesion, metal ion uptake system etc. of A. baumannii have been identified as the major virulence factors based on numerous studies (Geisinger and Isberg, 2015; Talyansky et al., 2021; Zhao et al., 2021; Karampatakis et al., 2024). Studies involving mutant strains lacking capsular functionality have elucidated their roles in proliferating within soft tissue infection sites, inducing lethality in murine septicemia models, providing defense against serum-mediated killing, and modulating biofilm formation (Russo et al., 2010; Lees-Miller et al., 2013). EPSs play a pivotal role in facilitating bacterial aggregation, leading to the formation of multicellular consortia, wherein the biofilm operates as a highly protected multicellular system. The dense EPS matrix constitutes an effective barrier that impedes the penetration of antibiotics to different layers of the biofilm (Singh et al., 2021). Pathogenic bacteria, including Staphylococcus aureus, A. baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Enterococcus faecalis, have the capacity to form biofilms on medical instrument surfaces, within hospital environments, and on human and animal tissues (Mariani and Galvan, 2023; Weber et al., 2023). Approximately 80% of chronic and recurrent microbial infections in the human body are attributable to bacterial biofilms, which demonstrate antibiotic resistance levels that are 10 to 1000 times higher than those of planktonic cells, thereby substantially contributing to these types of infections (Sharma et al., 2019). In planktonic cells, A. baumannii exhibit higher susceptibility to antibiotics (Wences et al., 2022). However, after the establishment of biofilms on appropriate surfaces, they manifest resistance to antibiotics (Al-Shamiri et al., 2021). These biofilms obstruct the ingress of antibiotics into deeper layers, predominantly characterized by polysaccharide constituents, thereby curtailing antibiotic access to bacterial cells (Singh et al., 2021). Hence, there exists a critical imperative to investigate and innovate novel strategies to address the complexities associated with MDR strains of A. baumannii.
Bacteriophages have resurfaced as an alternative approach for combating drug-resistant A. baumannii, offering a promising avenue for mitigating the challenges posed by antibiotic resistance. Bacteriophages, commonly referred to as phages, are obligate parasitic entities that infects and replicates through either the lytic cycle or integrates its DNA into the host genome in the lysogenic cycle, leading to the production of new phages. These innate bactericidal agents are being revisited to counter bacteria that have developed resistance to multiple antimicrobial agents (Wegrzyn, 2022). Notwithstanding its recognized potential as a substitute for conventional antibiotics, phage therapy encounters efficacy impediments due to the necessity for tailored strategies and the formulation of comprehensive cocktails to address bacterial resistance. The restricted host spectrum and the emergence of phage-resistant bacterial variants are pivotal factors contributing to these challenges (Oechslin, 2018; Pirnay and Kutter, 2021). The coevolution of bacteria and phages drives their selection and infectivity (Wright et al., 2016). Bacteria develop resistance through several mechanisms such as blocking phage receptors, producing extracellular matrices, using CRISPR-Cas and restriction systems to cleave phage DNA, or employing abortive infection systems (Ambroa et al., 2022). Phages, in turn, evolve mutations to counter bacterial defenses. In biofilms, bacteria are shielded from harsh conditions and phage attacks, but phages can evolve enzymes like depolymerases to break down biofilm structures, resulting in a constant arms race between phages and bacteria as they adapt to each other’s defenses (Pires et al., 2021).
A prevalent resistance mechanism observed in Acinetobacter entails the secretion of cell-surface capsules, which serve to obscure the primary receptors, rendering it difficult for phages to adhere to the bacterial host (Oromi-Bosch et al., 2023). Nevertheless, researchers have investigated phage-encoded depolymerases capable of enzymatically degrading bacterial EPS, CPS and lipopolysaccharide (LPS) materials. In numerous animal infection models, phage-derived depolymerases are exhibiting therapeutic promise by selectively targeting bacterial capsules and exhibiting antibiotic efficacy (Lin et al., 2014; Chen et al., 2018, 2020). In phage therapy, the efficacy of phages may be compromised due to the frequent development of bacterial resistance against them. Conversely, free depolymerases, distinct from intact phage particles, do not often engender the emergence of resistant bacterial strains. This phenomenon stems from the fact that depolymerases do not directly induce bacterial cell death; instead, they augment bacterial susceptibility to immune responses (Chen et al., 2022; Guo et al., 2023). Hence, phage-derived depolymerases have emerged as innovative alternative antimicrobials targeting MDR bacteria, particularly those harboring polysaccharide capsules that confer resistance to antibiotics. Numerous phages are equipped with tail-associated depolymerases capable of degrading bacterial CPS, EPS, and LPS, thereby uncovering binding sites for antimicrobial agents. Additionally, they facilitate phage attachment, enhance innate immune responses, and optimize antibiotic effectiveness, including penetration into biofilms (Hsieh et al., 2017; Wang et al., 2024a). The aims of this review are to summarize the state-of-the-art knowledge on the phage derived depolymerase enzymes that can degrade polysaccharide materials and their potential role in treating MDR A. baumannii infections.
The capsule that surrounds the A. baumannii surface is a crucial fitness and virulence factor. Capsules facilitate bacteria to adhere to biotic and abiotic surfaces promoting colonization in diverse niches and thereby biofilm formation (Kaur and Dey, 2023). Poly-N-acetyl glucosamine (PNAG), an important polysaccharide, is well described as a building block of A. baumannii biofilm. In A. baumannii, the pgaABCD locus is found to be associated with the production of PNAG. Deletion of the pgaABCD locus led to a reduced biofilm phenotype, which was restored by complementation, demonstrating the role of polysaccharides in biofilm formation (Choi et al., 2009). Composed of repeating polysaccharide units known as K units, this capsule forms a protective coating around the bacterial cell wall, shielding it from environmental stresses such as desiccation and disinfection (Tipton Kyle et al., 2018). The ability of A. baumannii to survive in the hospital environment is favored by resistance to disinfectants and desiccation. In A. baumannii, polysaccharide materials provide a physical barrier that enhances water retention, allowing the bacteria to survive for long periods of desiccation (Kaur and Dey, 2023). A study showed that the A. baumannii strain AB5075 wild-type virulent opaque cells produced a capsule with a 2-fold increased thickness than avirulent translucent cells. In desiccation and disinfectants, virulent opaque cells showed higher survival on dry surfaces compared to avirulent translucent cells (Chin et al., 2018). Besides, CPS also involves other virulence factors such as motility, confirming its role as a determinant of virulence (Rakovitsky et al., 2021).
Additionally, the A. baumannii capsule plays a vital role in defending against host cell killing and evading activation of the innate immune response (Akoolo et al., 2022). A capsular mutant, designated as wza, of A. baumannii exhibited heightened susceptibility to whole blood, complement, and neutrophil-mediated killing, underscoring the critical role of the polysaccharide capsule in immune evasion (Bjanes et al., 2023). The capsule is also known to play a role in phagocytosis. In hyper mucoid A. baumannii, the capsule assembly gene gtr6 protects it from phagocytosis by inhibiting the deposition of complement component 3 (Talyansky et al., 2021; Gong et al., 2022). In a study, wild-type mice with Toll-like receptor 4 died from A. baumannii-induced septic shock, while TLR4-deficient mice survived despite having a similar bacterial load. The hypervirulent A. baumannii releases more LPS, which activates TLR4, leading to lethal sepsis in the host. The LpxC gene, involved in lipid A biosynthesis, (a component of LPS), is targeted by LpxC inhibitor. Though the inhibitor did not inhibit bacterial growth, it suppressed TLR4 activation by A. baumannii. Mice treated with the LpxC inhibitor showed enhanced opsonophagocytic killing, reduced serum LPS concentrations and inflammation, and were completely protected from lethal infection (Lin et al., 2012). The anti-OmpA monoclonal antibody enhanced macrophages to kill the K1 capsule-negative mutant A. baumannii 307.30, except for those containing thick capsules, particularly extensively drug-resistant A. baumannii. The binding affinity of monoclonal antibodies with drug-resistant clinical isolates of A. baumannii showed weaker affinity than that between mABs and ATCC19606, possibly due to capsular polysaccharide blocking MAb access to OmpA (Wang-Lin Shun et al., 2017).
In addition to protect from host defense system, capsules also confer resistance to a wide range of antibiotics by limiting the penetration of antibiotics to the outer membrane where the antibiotics can access the inner membrane. Geisinger and Isberg (Geisinger and Isberg, 2015) reported that when A. baumannii is exposed to sub inhibitory concentrations of chloramphenicol or erythromycin, it leads to increased production of capsular polysaccharide. This elevated CPS production is reversible and non-mutational, and occurs alongside with heightened resistance to the antibiotic independent of the K locus. Colony phase variation by A. baumannii responds to differential alterations in tolerance to antibiotics. Mushtaq et al. (Mushtaq et al., 2024) demonstrated that in opaque variants of A. baumannii AB5075, extracellular polysaccharide material plays a role in colistin tolerance at the single-cell level. At the community level, the opaque variant forms a mushroom-shaped biofilm. Thus, demonstrating its fitness advantage over the translucent variants and its ability to tolerate colistin.
Phage depolymerases represent enzymatic proteins designed to dismantle polymers, particularly targeting bacterial outer membrane polysaccharides essential for bacteriophage adsorption. These enzymatic entities play a crucial role in facilitating phage attachment to the bacterial cell surface and subsequent degradation of the bacterial capsule (Knecht et al., 2019). Tail fibers or tail spikes, integral components of bacteriophage structure, serve as anchoring points for depolymerases, which are linked to the base plate of the phage, with a few exceptions found on the neck (Lai et al., 2016; Pires et al., 2016; Blundell-Hunter et al., 2021) (Figure 1A). Biochemically, depolymerases are dichotomized into two distinct types: lyases and hydrolases (Cai et al., 2023; Magill and Skvortsov, 2023). Hydrolases break glycosidic bonds by reacting with water, disrupting the glycosyl-oxygen bond. Lyases, on the other hand, cleave glycosidic bonds through β-elimination, creating a new double bond without the need for water (Li et al., 2022) (Figure 1B). Predominantly harbored in receptor binding proteins (RBPs), depolymerases are crucial for triggering phage infection through the targeting and enzymatic disassembly of bacterial capsules at the onset (Latka et al., 2019). Receptor binding proteins (RBPs) possessing depolymerase activity exhibit a modular architecture comprising three distinct domains: An N-terminal domain responsible for tethering to the phage tail, a central β-helical domain facilitating enzymatic function, and a C-terminal domain associated with receptor binding and potential chaperone activity (Li et al., 2022; Guo et al., 2023; Klumpp et al., 2023; Maciejewska et al., 2023) (Figure 2). The modular configuration of RBPs enables expedited alterations in host specificity, accomplished via horizontal gene transfer such as transduction, which mediates the interchange of host-specific central domains, and vertical gene transfer, a process marked by the accrual of mutations within polysaccharide-depolymerizing domains (Matilla and Salmond, 2014; Latka et al., 2017, 2019; Pas et al., 2023). Two variants of phage depolymerases are discernible: one bound within phage particles and the other existing as soluble proteins following host cell lysis (Drulis-Kawa et al., 2015; Wang et al., 2019). Ultimately, the structural diversity and modular nature of phage depolymerases are fundamental to their ability to degrade bacterial polysaccharides, thereby promoting efficient phage-host interactions and successful infection.
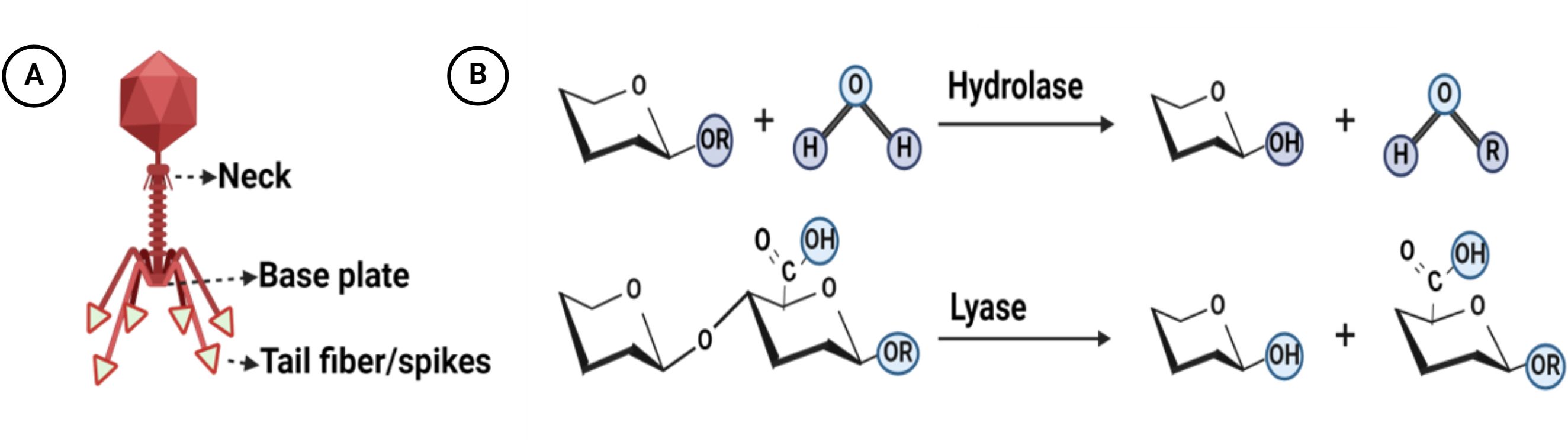
Figure 1. Structure and mechanisms of action of bacteriophage depolymerases: (A) Distribution of depolymerase within the structure of bacteriophages (B) The overarching mechanisms by which depolymerases catalyze polysaccharide degradation involve two fundamental processes: hydrolysis and lysis. During hydrolysis, depolymerases enzymatically cleave the glycosidic bonds between sugar monomers, resulting in the fragmentation of polysaccharides into smaller sugar units. In the lysis process, these enzymes compromise the structural integrity of the polysaccharide matrix. Created with BioRender.com.
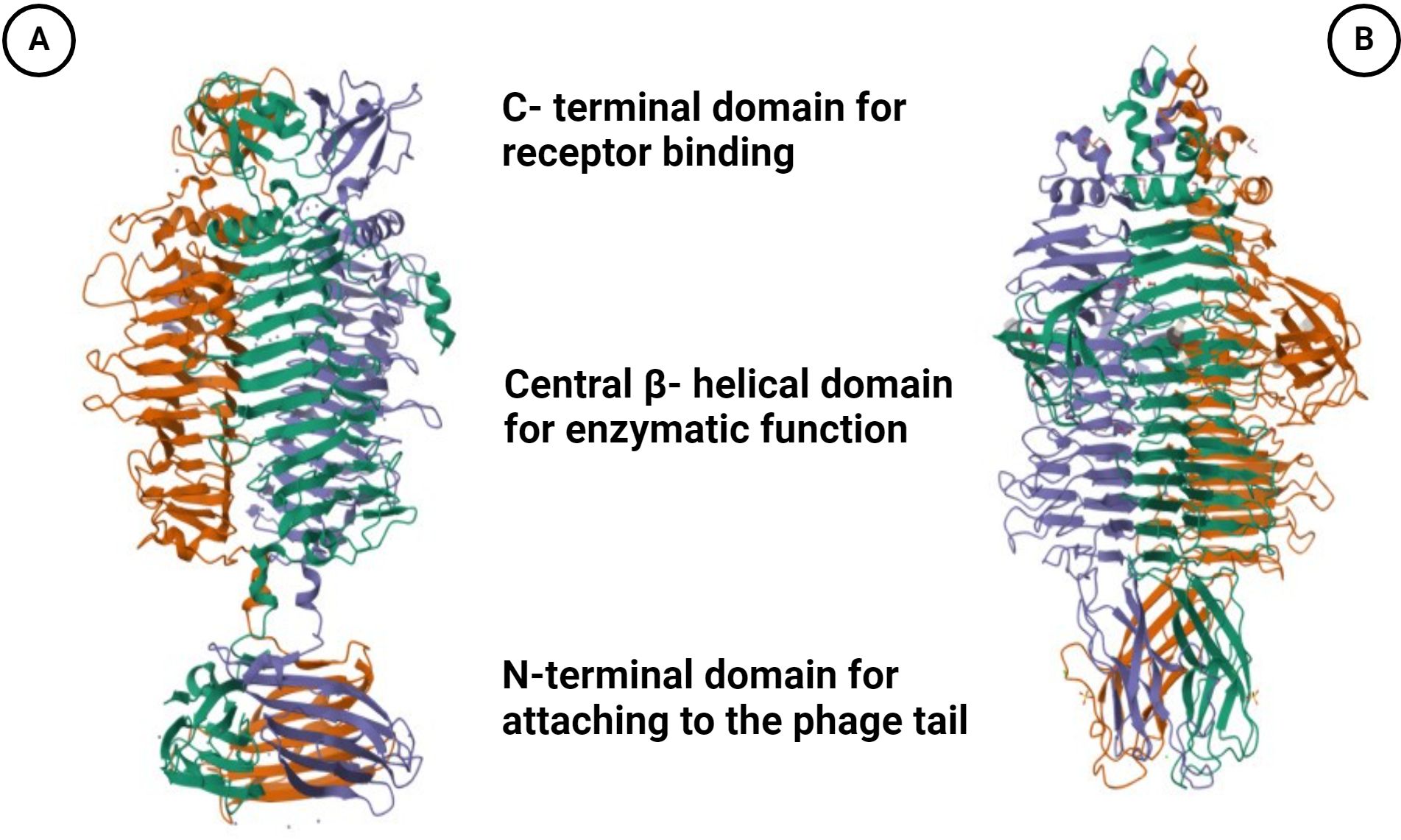
Figure 2. Illustration is the trimeric crystal structure of the tail spike protein. (A) gp49 tail spike protein (PDB ID: 6C72) derived from A. baumannii bacteriophage Fri1, a capsular polysaccharide depolymerase (Hydrolase). (B) Gp54 tail spike (PDB ID: 4Y9V) of A baumannii bacteriophage AP22 polysaccharide degrading lyase. Three monomers are marked by different colors. Created with BioRender.com.
Capsular polysaccharide and biofilm of A. baumannii are virulence determinants that play a major role in resistance to antimicrobial agents, evasion of host immune responses and adaptation to other stresses (Geisinger and Isberg, 2015; Mendes et al., 2023). These structural components act as protective barriers, hindering the access of various antimicrobial agents to their respective binding sites (Roy et al., 2022). Biofilms are complex assemblies of bacteria adhering to surfaces and enclosed within a matrix composed of extracellular polymeric substances, comprising proteins, lipids, nucleic acids, polysaccharides, and other constituents. In response to the challenge of biofilm resistance, alternative approaches such as the utilization of phage depolymerases have been explored. Encouragingly, phages carrying depolymerases possess the capacity to degrade the polysaccharide constituents of both capsules and biofilms (Figure 3). The polysaccharide components present on the outer surface of A. baumannii cells are the primary targets for bacteriophages harboring specific structural depolymerases. These depolymerases are crucial for mediating the initial interaction between the phage and the host bacterium (Drobiazko et al., 2022). A solitary phage depolymerase possesses the capacity to address bacterial surface polysaccharides and biofilms. EPSs are crucial for the attachment of bacterial cells together and forming multicellular consortia in biofilms. They are vital for the structural stability, functionality, and virulence of bacterial biofilms (Karygianni et al., 2020). The dense EPS matrix has a high binding affinity to antimicrobial agents, creating a barrier that hinders antibiotic diffusion into biofilm layers (Sharma et al., 2023). Phage-encoded depolymerase enzymes target EPS by recognizing, binding to, and degrading the polysaccharide matrix of bacterial cell walls. This enzymatic action disrupts biofilm structure, reducing bacterial virulence and increasing susceptibility to the host immune system (Tian et al., 2021). Depolymerases degrade polysaccharides like capsular CPSs, LOSs, O-polysaccharides, and EPSs in biofilms. Hydrolase break down α-1,4-glycosidic bonds in polysaccharides to produce oligosaccharides through hydrolysis (Wang et al., 2024b). Lyases, on the other hand, cleave anionic polyuronides by a β-elimination reaction. This process involves removing a proton from the C5 position of the uronide ring and breaking the ether linkage at the C4 position with the help of a general acid-base catalyst, resulting in an unsaturated product (Pandey et al., 2023).
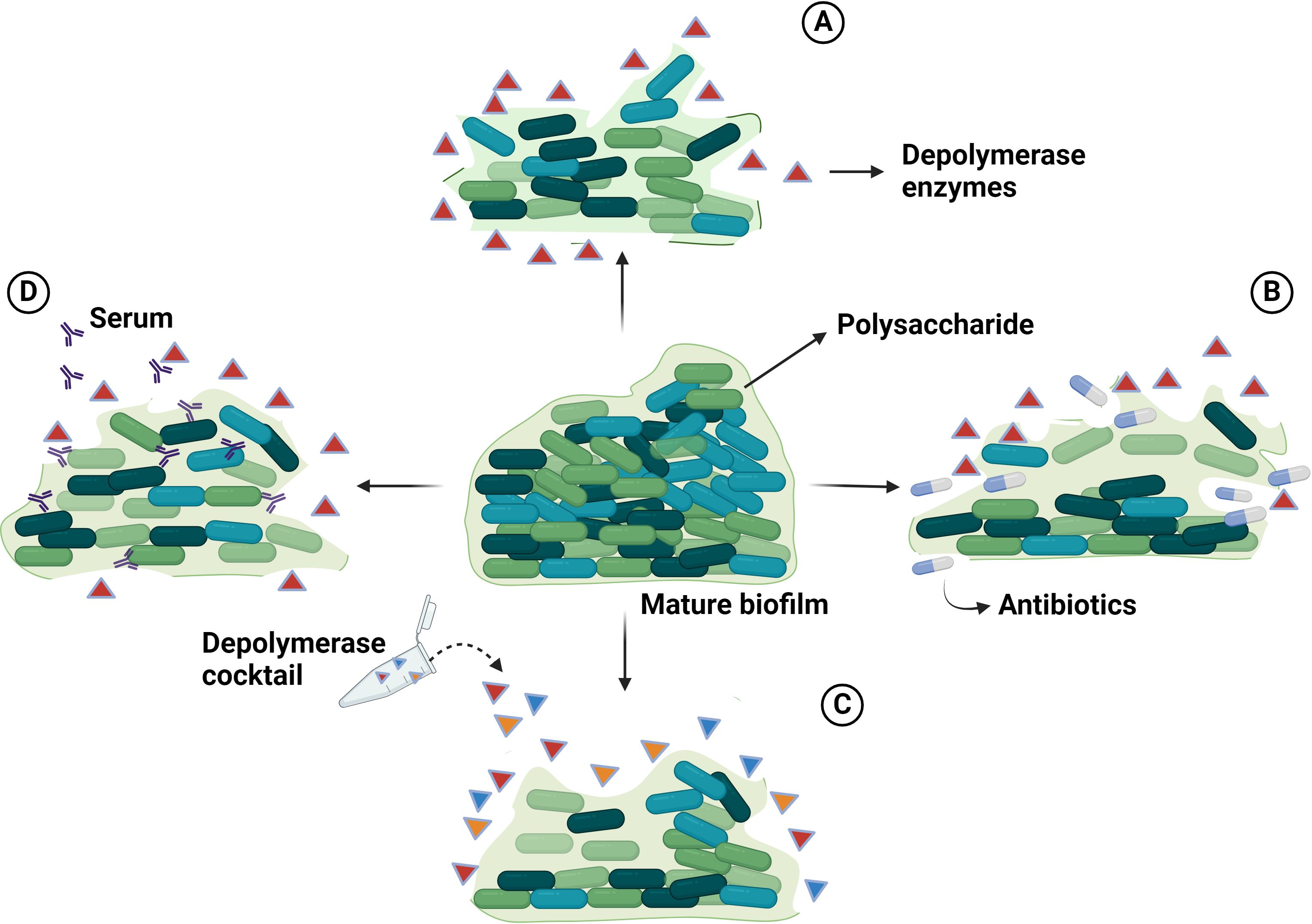
Figure 3. The illustration delineates the anti-biofilm mechanisms of phage-encoded depolymerases: (A) Phage-encoded depolymerases impede biofilm formation by degrading the extracellular polysaccharides that protect the bacterial cells within the biofilm. (B) The synergistic application of phage-encoded depolymerases and antibiotics enhances the bactericidal efficacy against A. baumannii biofilms. The depolymerases degrade the extracellular polysaccharide, rendering the bacteria more susceptible to antibiotic treatment. (C) A consortium of phage-encoded depolymerases can effectively dismantle biofilms by degrading the polysaccharide matrix. (D) Phage-encoded depolymerases sensitize bacteria to the host immune response by degrading the protective polysaccharide matrix of biofilms. Created with BioRender.com.
However, achieving comprehensive eradication of these virulence determinants may necessitate the utilization of combinations of phage depolymerase cocktails or their concurrent administration with antibiotics, or they may serve as adjuncts. Hence, the utilization of phage depolymerases emerges as a prospective tactic for addressing A. baumannii infections, offering potential as standalone interventions or in tandem with antibiotic therapies. This underscores the promising trajectory of phage-derived depolymerases in the realm of antimicrobial strategies against this resilient pathogen.
A single phage depolymerase can efficiently target and disperse the biofilm matrix of Acinetobacter. By degrading the extracellular polymeric substances that shield and reinforce the biofilm, this enzyme disrupts its structure, rendering the bacterial cells more susceptible to treatment (Figure 3A). This enzymatic action aids in facilitating the phage’s DNA injection into the bacterial host. Unlike certain other phage-encoded enzymes, depolymerases do not induce direct bacterial cell lysis. Rather, they degrade bacterial CPS, rendering them vulnerable to host immune responses and antibacterial treatments (Dicks and Vermeulen, 2024). Hydrolases are enzymatic catalysts that cleave glycosidic bonds by disrupting the glycosyl-oxygen bond. Within this category, various subclasses exist, including sialidases, levanases, xylosidases, dextranases and rhamnosidases. Conversely, lyases function by catalyzing the separation of the glycosidic link between monosaccharides and the C4 position in uronic acid molecules. This process involves the creation of a double bond via the β-elimination mechanism between the C4 and C5 positions in nonreducing uronic acid molecules. Examples of lyases include hyaluronidases, alginate lyases, and pectin/pectate lyases (Cai et al., 2023). The utilization of depolymerases for antibacterial purposes can occur through two approaches: as enzymes expressed by bacteriophages and as recombinant protein; and both strategies can be used in combination with antibiotics to broaden their effectiveness against a wider range of hosts (Wang et al., 2024a). These bacteriophages possess inherent mechanisms for recognizing specific bacterial strains, facilitating targeted interventions (Hibstu et al., 2022). Moreover, their capacity to deliver depolymerases directly to infection sites and penetrate biofilms presents advantages over conventional antibiotics (Jo et al., 2023). Recombinant protein synthesis enables standardized, large-scale production of depolymerases with stringent quality control measures, ensuring uniform potency and reliability. Furthermore, genetic engineering methodologies offer avenues for refining recombinant depolymerases to augment their stability, enzymatic activity, or specificity towards various polysaccharide targets (Drulis-Kawa et al., 2015; Hassan et al., 2021; Noreika et al., 2023).
PghP is a bacteriophage-encoded peptidase that specifically hydrolyzes γ-polyglutamic acid (γ-PGA), a polymeric extracellular barrier produced by certain bacterial species. This enzymatic activity is critical for bacteriophage entry, as PghP degrades the γ-PGA matrix surrounding the bacteria. PghP catalyzes the cleavage of γ-PGA into smaller oligo-γ-glutamates, further processing these into tri-, tetra-, and penta-γ-glutamate units (Kimura and Itoh, 2003). Sialidases, also referred to as neuraminidases, are enzymes that catalyze the cleavage of the α-linkage of terminal sialic acid residues in glycoconjugates (Keil et al., 2022). Pathogenic bacteria, including Escherichia coli K1, Acinetobacter baumannii, Haemophilus influenzae, and Neisseria spp., exploit sialic acid to evade host immune defenses by incorporating it into their surface structures or utilizing it as a metabolic nutrient source (Ko et al., 2018; Jennings et al., 2022). Furthermore, numerous bacterial species produce sialidases to enhance their survival within mucosal environments and facilitate interactions with other microorganisms, thereby contributing to their pathogenic potential (Lewis and Lewis, 2012; Tailford et al., 2015). Bacteriophages encode endosialidases that degrade capsular polysaccharides, rendering bacteria more vulnerable to host immune responses. Studies have demonstrated that phage-derived endosialidases can sensitize Escherichia coli to immune defenses, promoting phagocytosis and offering protection against systemic infection in murine models (Mushtaq et al., 2004). Levanases (EC 3.2.1.65) are enzymes present in microorganisms such as Bacillus and Pseudomonas, which catalyze the hydrolysis of the β-2,6-linked chains of levan, a fructan polymer (Murakami et al., 1992). Bacteriophage ϕNIT1 possesses the pghP gene, encoding a γ-PGA hydrolase that enables the phage to overcome the host cell’s defense mechanisms. Additionally, ϕNIT1 contains a gene encoding LevP, an active levan hydrolase classified as an endo-levanase, which cleaves levan, an exopolysaccharide produced by Bacillus subtilis (Ozaki et al., 2017). Xylosidases (EC 3.2.1.37), dextranases (EC 3.2.1.11), lipases (EC 3.1.1.3), and rhamnosidases (EC 3.2.1.40) represent enzyme classes that are among the least frequently identified depolymerase domains in bacteriophages. These enzymes are involved in the hydrolytic degradation of specific substrates: xylan, dextran, triacylglycerols, and rhamnogalacturonan, respectively. Hyaluronate lyases (EC 4.2.99.1) and hyaluronidases (EC 4.2.2.1) are enzymes responsible for the degradation of hyaluronate. Phage-encoded hyaluronidases, such as those identified in Streptococcus pyogenes bacteriophages, specifically target hyaluronan, aiding in the degradation of the bacterial hyaluronan capsule (Baker et al., 2002). Alginate lyases (EC 4.2.2) are enzymes that catalyze the degradation of alginic acids, found in organisms such as Pseudomonas aeruginosa and Azotobacter vinelandii phages. These enzymes, including mannuronate lyase (EC 4.2.2.3) and guluronate lyase (EC 4.2.2.11), specifically target alginates, which serve as structural components in brown algae and bacterial extracellular polysaccharides (EPS) (Kim et al., 2011).
Pectin, a polysaccharide with an α-1,4-linked D-galacturonic acid backbone and α-1,2-l-rhamnose units, is catalyzed by pectin/pectate lyase, which cleaves the α-1,4 bonds of polygalacturonic acid is catalyzed by pectate lyase, which cleaves the α-1,4 bonds of polygalacturonic acid (Latka et al., 2017; Minzanova et al., 2018), are typically present in the cell wall or capsule of A. baumannii. DpoMK34, a pectate lyase, derived from the A. baumannii MK34 phage, demonstrates considerable efficacy even at low concentrations, falling within sub-micromolar ranges. Its activity is not limited to A. baumannii MK34 but also extends to strain CIP110467, which possesses a K2 capsular serotype, suggesting its potential for broad-spectrum effectiveness (Abdelkader et al., 2022). A study revealed the presence of 12 podoviruses featuring depolymerases equipped with a pectate lyase domain, facilitating the breakdown of capsules within the Acinetobacter baumannii-Acinetobacter calcoaceticus complex. This observation implies that these podoviruses may have undergone evolutionary refinement through the acquisition of specialized pectate lyase coding regions, thereby enhancing their infectivity and viability within this bacterial cohort (Oliveira et al., 2017). Dpo48, recognized as a capsule depolymerase, displays a broad activity spectrum, and maintains effectiveness across varied pH and temperature conditions. Its capacity to efficiently strip capsules from A. baumannii, even at elevated concentrations, facilitates bacterial eradication by serum complement in vitro (Liu et al., 2019). The Acinetobacter baumannii phage Bϕ-R2096, sourced from sewage water, exhibits robust bacteriolytic activity and generates distinct circular plaques surrounded by halos, indicative of the potential presence of phage-derived depolymerases. In vivo investigations utilizing Galleria mellonella larvae and mice infected with carbapenem-resistant A. baumannii (CRAB) reveal that Bϕ-R2096 significantly enhances survival rates and mitigates histologic lung damage in infected hosts, with no reported mortality or adverse effects (Jeon et al., 2019).
A study by Hernandez-Morales et al. (Hernandez-Morales et al., 2018) investigated the bacteriophage Petty, which harbors the depolymerase gene Dpo1, capable of degrading exopolysaccharides and infecting both A. nosocomialis and A. baumannii. In vitro assessments revealed that Dpo1 effectively reduced the viscosity of EPS and partially disrupted biofilms formed by the tested Acinetobacter strains, resulting in a modest reduction of approximately 20%. Although complete eradication of the biofilms was not achieved, Dpo1 likely attenuated the virulence of the examined strains by degrading EPS components. Notably, plaques formed by Petty exhibited halos, indicative of depolymerase activity targeting capsular exopolysaccharides. Additionally, gene 39, encoding a putative tail fiber, displayed depolymerase activity, suggesting its potential application in diagnostics and therapeutics against drug-resistant Acinetobacter strains through biofilm disruption. Another investigation revealed that depolymerases sourced from bacteriophages AM24, BS46, and APK09 possess the capability to degrade the K9 CPS of the antibiotic-resistant A. baumannii GC1(ST1IP) isolate SGH0807. Upon exposure to the K70 CPS from SGH0807, these depolymerases induced the cleavage of monomers and dimers at the K-unit linkage, resulting in the generation of oligosaccharide fragments. This finding underscores the potential of these depolymerases as a targeted strategy against the antibiotic-resistant strain by degrading its K70 CPS. Moreover, the observed specificity of these phages’ depolymerases for distinct CPS types suggests their potential efficacy in infecting and eradicating the SGH0807 K70 isolate (Kasimova et al., 2023).
Timoshina et al. (Timoshina et al., 2023) made an intriguing discovery of seven novel Friunaviruses and proceeded to dissect their functionalities and mechanisms in the degradation of CPS. Recombinant depolymerases were expressed in Escherichia coli B834 (APK09_gp48, APK14_gp49, APK16_gp47, APK86_gp49, APK127v_gp47, APK128_gp45, and APK37.1_gp49) generated, demonstrating effective degradation of the CPS of A. baumannii under laboratory conditions. Remarkably, recombinant depolymerase APK09_gp48 exhibited significant efficacy in reducing mortality rates among Galleria mellonella larvae infected with A. baumannii of K9 capsular type. Moreover, when administered independently, TSD APK09_gp48 did not result in larval mortality, underscoring its safety. These outcomes highlight the potential therapeutic utility of depolymerase APK09_gp48 in combating A. baumannii infections. Eight bacteriophages were identified, each carrying depolymerase genes. These genes were cloned, expressed, and the resulting enzymes were purified. The enzymes broke down the capsular polysaccharides (CPSs) of their A. baumannii hosts into monomers or dimers of CPS repeats (K units) through a hydrolytic process. Specific depolymerases, such as APK2_gp43, APK32_gp46, APK37_gp44, APK44_gp44, APK48_gp43, APK87_gp48, APK89_gp46, and APK116_gp43, were found to be glycosidases. Notably, APK2_gp43 could interact with both K2 and K93 CPSs because of their similar K unit structures and identical linkages, enabling the enzyme to break down both types effectively (Popova et al., 2021). The recombinant depolymerase derived from the tail spike protein (TSP) of the bacteriophage φAB6 exhibits proficient degradation of A. baumannii biofilms in vitro. It effectively impedes biofilm formation and dismantles pre-existing biofilms. Even at minimal concentrations, TSP significantly hampers biofilm formation and adhesion on Foley catheter surfaces. Furthermore, it induces bacterial cell death and augments survival rates in zebrafish, suggesting its potential as a novel antibiotic against MDR A. baumannii and as a biocontrol agent for preventing biofilm formation on medical devices (Shahed-Al-Mahmud et al., 2021).
Depolymerases used in therapeutic applications are typically prepared as liquid formulations for parenteral injections, aerosols for inhalations and topical delivery methods also in use (Maciejewska et al., 2018). The enzyme production requires a comprehensive protein purification process to remove bacterial endotoxins. The delivery of enzymes through aerosolization and topical application allows for targeted drug accumulation at the infection site while minimizing systemic exposure (Ryan et al., 2011). Specific pH and buffers are required for long-term storage or to ensure delivery to the reaction site. Chen et al. (Chen et al., 2022) found that storing Dpo71 in phosphate-buffered saline at 4°C maintained its stability for up to six months without significant loss of activity. Another study showed that KP32gp37 enzyme was resistant to SDS detergent and trypsin protease, while KP32gp38 was not (Majkowska-Skrobek et al., 2018). These findings provide safe formulation strategies for depolymerases for therapeutic use to prevent denaturation by body fluids.
In general, the mechanism of action for phage depolymerases involves the cleavage of glycosidic bonds present within bacterial polysaccharides, leading to the formation of oligosaccharide products either through hydrolysis or lysis. The EPS found in A. baumannii biofilms exhibit three distinct bond types: β-1,6-glycosidic, β-1,3-glycosidic and β-2,6-glycosidic bond (Lee et al., 2017). Each phage depolymerase possesses the ability to hydrolyze at least one of these bond types (Guo et al., 2023). Hence, employing a combination of phage depolymerases in a cocktail may present a promising approach for the comprehensive eradication of A. baumannii biofilms.
Integrating antibiotics with phage-derived depolymerases offers a potent approach to addressing MDR bacterial infections. Depolymerases alone might be less effective due to biofilm variability; however, their combination with antibiotics increases bacterial susceptibility by breaking down protective polysaccharide matrices. This combined mechanism enhances antibiotic penetration, making the treatment more effective in eradicating infections, particularly those caused by resistant bacteria (Topka-Bielecka et al., 2021) (Figure 3B).
Colistin is considered a last-resort antibiotic used to treat infections caused by MDR A. baumannii. However, there have been reports of A. baumannii strains developing resistance to colistin (Elham and Fawzia, 2019; Seleim et al., 2022). Combining phage depolymerases with colistin could help to address this issue. The synergistic use of colistin and phage depolymerases, such as Dpo71, could effectively counter MDR A. baumannii. Dpo71, a phage-derived enzyme, boosts bacterial susceptibility to immune system attacks and enhances colistin’s antimicrobial activity (Chen et al., 2022). It helps to clears bacteria by human serum, degrades biofilms, and improves survival outcomes in infection models. The strain-specific action of Dpo71 demonstrates its potential as a complementary therapy to antibiotics against resistant bacterial infections (Chen et al., 2022). Phage vWU2001, which was isolated from treated wastewater, produced large plaques with halo zones, suggesting the presence of a phage-encoded depolymerase. When combined with colistin, phage vWU2001 significantly inhibited the growth and viability of carbapenem-resistant A. baumannii more effectively than either treatment alone. This combination also improved the survival of G. mellonella and enhanced bacterial clearance, indicating a synergistic effect (Wintachai et al., 2022). Here, phage depolymerases might act as an adjuvant that could help prevent the emergence of resistant cells through synergistic interactions.
Single-phage depolymerase therapy exhibits dose-dependent repression of bacterial proliferation, albeit encountering impediments such as the emergence of phage-resistant bacterial strains, particularly a problem in the case of A. baumannii. To mitigate this challenge, the utilization of cocktails comprising multiple phages encoding depolymerases exhibiting diverse host specificities could demonstrate heightened effectiveness by broadening the host range and mitigating resistance occurrences (Figure 3C). Additionally, the incorporation of multiple depolymerases within these cocktails addresses the inherent limitations in the specificity of individual depolymerases for eradicating bacterial biofilms. In sum, the employment of phage depolymerase cocktails augments therapeutic efficacy by targeting a wider array of bacterial strains, potentially delaying the onset of phage resistance, bolstering antimicrobial effectiveness (Chang et al., 2022; Tan et al., 2022). For instance, the co-administration of Ab105-2phiΔCI404ad with phage vB_AbaP_B3, known for their depolymerase expression, synergistically enhanced the antimicrobial efficacy against diverse strains of A. baumannii, encompassing both clinical and reference strains (Blasco et al., 2022).
Phage cocktails are recognized for their ability to enhance antibacterial activity against biofilms. Yet, there is a lack of research focusing on the potential use of various phage depolymerases within these cocktails to target bacterial capsules, especially in the context of A. baumannii infections. Bacteriophage cocktails have been tested to treat septicemia (Patel et al., 2021), wound infections (Ilomuanya et al., 2022), nasal infections (Cha et al., 2018), and pneumonia (Li et al., 2024) caused by A. baumannii in animal models. In addition, phage cocktails have been effectively used in clinical settings to treat human urinary tract infections (Grygorcewicz et al., 2021), necrotizing pancreatitis (Schooley et al., 2017), and post-operative cerebritis (Lavergne et al., 2018) caused by MDR A. baumannii. Further research is needed to investigate the efficacy and safety of phage depolymerase cocktails in treating A. baumannii infections.
Polysaccharide components are crucial for shielding bacteria from immune assaults and adverse environmental conditions, including phage infection. Notably, the application of depolymerases in conjunction with human serum or immune cells has significantly augmented antibacterial efficacy (Figure 3D). Phage depolymerases render bacteria more susceptible to human serum, resulting in heightened bacterial eradication. This approach represents a promising strategy for combating infections and surmounting bacterial defenses (Maciejewska et al., 2018). Hugo Oliveira et al. (Oliveira et al., 2019) reported that the recently discovered myovirus vB_AbaM_B9, along with its depolymerase B9gp69, effectively dismantles exopolysaccharides sourced from various strains of A. baumannii. This functionality increases susceptibility to serum-mediated bacterial elimination while simultaneously reducing the likelihood of bacterial resistance development. Specifically engineered to target the virulent K45 capsule type, this recombinant depolymerase demonstrates potential as an antimicrobial supplement, fortifying the host immune defense against A. baumannii infections. The analysis of A. baumannii phage IME285 resulted in the detection of a depolymerase gene, ORF49, following genomic scrutiny. Subsequent investigation led to the cloning and expression of the recombinant enzyme, Dp49, originating from this gene, showcasing substantial depolymerase activity both in murine experimental models, and in vitro, as evidenced by the formation of translucent halos surrounding bacterial plaques. Dp49 displayed efficacy against 25 of the 49 A. baumannii strains tested, amplifying the inhibitory impact of serum on bacterial proliferation in vitro and augmenting survival rates in A. baumannii-infected mice. These observations indicate the potential utility of Dp49 as a promising intervention for managing infections instigated by MDR A. baumannii (Wang et al., 2020). Therefore, there is a good reason to delve into further exploration of depolymerases that can effectively target various bacterial types and their corresponding strains.
Phage-encoded enzymes, such as polysaccharide depolymerases raised attention as novel antibacterial agents due to their potential to eradicate biofilms and capsular polysaccharides that hinder phage adsorption to host bacteria (Bleriot et al., 2024). In contrast to whole phage particles, bacteria hardly develop resistance against free depolymerases, making them a promising antibacterial agent (Chen et al., 2022). Genetically engineering phage depolymerases to modify host ranges holds considerable promise for phage applications (Anyaegbunam et al., 2022). In Gram-negative E. coli, genetically engineered phage enzymes significantly prevent biofilm formation (Lu and Collins, 2007). Following this, engineered depolymerase could be effective in A. baumannii. As our comprehension of bacteriophage therapy advances, the engineered phage-depolymerases could become a viable solution for biofilm management across various settings. By synthesizing biofilm-degrading enzymes by genetic engineering during the infection process, these phages can simultaneously target bacterial cells and the biofilm matrix, resulting in more efficient biofilm eradication compared to traditional bacteriophage treatments. This underscores the promise of employing engineered enzymatic bacteriophages in combating bacterial biofilms, showcasing the benefits of synthetic biology in addressing both medical and industrial challenges (Lu and Collins, 2007). Advancements in DNA sequencing and synthetic biology present promising avenues for the enhancement of phage-derived enzymes-based antimicrobials, providing critical solutions to address the antibiotic crisis. Comprehensive analysis of the uncharacterized functional proteins in A. baumannii phages, alongside the development of phage-derived enzyme formulations, can significantly improve the efficacy and expand the scope of phage applications.
Despite the promising in vitro capabilities of phage depolymerases in polysaccharide degradation, clinical trials assessing their therapeutic effectiveness remain scarce. Further investigation is essential to thoroughly understand the clinical utility of phage depolymerases. This includes the development of robust delivery systems to guarantee the stability of depolymerases during storage and transport and their precise delivery to targeted sites while ensuring safety.
Phages represent a naturalistic solution in combating MDR strains, offering a novel approach to addressing antibiotic resistance. Depolymerases, derived from phages, present a distinct mechanism for combating antibiotic-resistant pathogens, mitigating the risk of resistant strain development. These phage-derived depolymerases exhibit potent antimicrobial properties as alternative antibiotic, in combination with antibiotic, in phage cocktails and as adjuvants by targeting bacterial capsular polysaccharide, particularly effective against biofilm-related infections. This strategy signifies a significant avenue in the ongoing battle against antibiotic resistance. A number of depolymerases from several studies have been identified as promising alternatives to antibiotics against A.baumannii, including TSP from the φAB6 phage, Dp49, and Dpo71. However, further research is warranted to identify and characterize additional depolymerases capable of targeting a diverse range of bacterial species and strains comprehensively. Such endeavors hold the potential to significantly contribute to the development of novel antimicrobial strategies and enhance our ability to combat infectious diseases caused by antibiotic-resistant pathogens.
MI: Formal analysis, Investigation, Methodology, Project administration, Software, Visualization, Writing – original draft. NM: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. WS: Conceptualization, Funding acquisition, Writing – review & editing. MO: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant Nos. 2022R1F1A1071415 and NRF-RS-2023-00275307) and Basic Science Research Capacity Enhancement Project through Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (Grant No. 2019R1A6C1010033).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelkader, K., Gutierrez, D., Latka, A., Boeckaerts, D., Drulis-Kawa, Z., Criel, B., et al. (2022). The specific capsule depolymerase of phage PMK34 sensitizes acinetobacter baumannii to serum killing. Antibiot. (Basel) 11. doi: 10.3390/antibiotics11050677
Akoolo, L., Pires, S., Kim, J., Parker, D. (2022). The Capsule of Acinetobacter baumannii Protects against the Innate Immune Response. J. Innate Immun. 14, 543–554. doi: 10.1159/000522232
Al-Shamiri, M. M., Zhang, S., Mi, P., Liu, Y., Xun, M., Yang, E., et al. (2021). Phenotypic and genotypic characteristics of Acinetobacter baumannii enrolled in the relationship among antibiotic resistance, biofilm formation and motility. Microb. Pathogen. 155, 104922. doi: 10.1016/j.micpath.2021.104922
Ambroa, A., Blasco, L., López, M., Pacios, O., Bleriot, I., Fernández-García, L., et al. (2022). Genomic analysis of molecular bacterial mechanisms of resistance to phage infection. Front. microbiol. 12. doi: 10.3389/fmicb.2021.784949
Anyaegbunam, N. J., Anekpo, C. C., Anyaegbunam, Z. K. G., Doowuese, Y., Chinaka, C. B., Odo, O. J., et al. (2022). The resurgence of phage-based therapy in the era of increasing antibiotic resistance: From research progress to challenges and prospects. Microbiol. Res. 264, 127155. doi: 10.1016/j.micres.2022.127155
Baker, J. R., Dong, S., Pritchard, D. G. (2002). The hyaluronan lyase of Streptococcus pyogenes bacteriophage H4489A. Biochem. J. 365, 317–322. doi: 10.1042/bj20020149
Bjanes, E., Koh, T., Qayum, T., Zurich, R., Mccabe, S., Hampel, K., et al. (2023). Exploring roles of the polysaccharide capsule in pathogenesis of hypervirulent acinetobacter baumannii clinical isolate lac-4. Antibiot. (Basel) 13. doi: 10.3390/antibiotics13010010
Blasco, L., Bleriot, I., González De Aledo, M., Fernández-García, L., Pacios, O., Oliveira, H., et al. (2022). Development of an anti-acinetobacter baumannii biofilm phage cocktail: genomic adaptation to the host. Antimicrob. Agents Chemother. 66, e0192321. doi: 10.1128/aac.01923-21
Bleriot, I., Pacios, O., Blasco, L., Fernández-García, L., López, M., Ortiz-Cartagena, C., et al. (2024). Improving phage therapy by evasion of phage resistance mechanisms. JAC-Antimicrob. Resist. 6. doi: 10.1093/jacamr/dlae017
Blundell-Hunter, G., Enright, M. C., Negus, D., Dorman, M. J., Beecham, G. E., Pickard, D. J., et al. (2021). Characterisation of bacteriophage-encoded depolymerases selective for key klebsiella pneumoniae capsular exopolysaccharides. Front. Cell Infect. Microbiol. 11, 686090. doi: 10.3389/fcimb.2021.686090
Cai, R., Ren, Z., Zhao, R., Lu, Y., Wang, X., Guo, Z., et al. (2023). Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides. Microbiol. Spectr. 11, e0530422. doi: 10.1128/spectrum.05304-22
CDC (2019). Antibiotic Resistance Threats in The United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC.
Cha, K., Oh, H. K., Jang, J. Y., Jo, Y., Kim, W. K., Ha, G. U., et al. (2018). Characterization of two novel bacteriophages infecting multidrug-resistant (MDR) acinetobacter baumannii and evaluation of their therapeutic efficacy in vivo. Front. microbiol. 9 doi: 10.3389/fmicb.2018.00696
Chang, C., Yu, X., Guo, W., Guo, C., Guo, X., Li, Q., et al. (2022). Bacteriophage-mediated control of biofilm: A promising new dawn for the future. Front. Microbiol. 13, 825828. doi: 10.3389/fmicb.2022.825828
Chen, Y., Li, X., Wang, S., Guan, L., Li, X., Hu, D., et al. (2020). A novel tail-associated O91-specific polysaccharide depolymerase from a podophage reveals lytic efficacy of shiga toxin-producing escherichia coli. Appl. Environ. Microbiol. 86. doi: 10.1128/AEM.00145-20
Chen, X., Liu, M., Zhang, P., Xu, M., Yuan, W., Bian, L., et al. (2022). Phage-derived depolymerase as an antibiotic adjuvant against multidrug-resistant acinetobacter baumannii. Front. Microbiol. 13, 845500. doi: 10.3389/fmicb.2022.845500
Chen, Y., Sun, E., Yang, L., Song, J., Wu, B. (2018). Therapeutic application of bacteriophage PHB02 and its putative depolymerase against pasteurella multocida capsular type A in mice. Front. microbiol. 9. doi: 10.3389/fmicb.2018.01678
Chin, C. Y., Tipton, K. A., Farokhyfar, M., Burd, E. M., Weiss, D. S., Rather, P. N. (2018). A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat. Microbiol. 3, 563–569. doi: 10.1038/s41564-018-0151-5
Choi, A. H., Slamti, L., Avci, F. Y., Pier, G. B., Maira-Litrán, T. (2009). The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963. doi: 10.1128/JB.00647-09
Dicks, L. M. T., Vermeulen, W. (2024). Bacteriophage-host interactions and the therapeutic potential of bacteriophages. Viruses 16. doi: 10.3390/v16030478
Drobiazko, A. Y., Kasimova, A. A., Evseev, P. V., Shneider, M. M., Klimuk, E. I., Shashkov, A. S., et al. (2022). Capsule-targeting depolymerases derived from acinetobacter baumannii prophage regions. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23094971
Drulis-Kawa, Z., Majkowska-Skrobek, G., Maciejewska, B. (2015). Bacteriophages and phage-derived proteins–application approaches. Curr. Med. Chem. 22, 1757–1773. doi: 10.2174/0929867322666150209152851
Elham, B., Fawzia, A. (2019). Colistin resistance in Acinetobacter baumannii isolated from critically ill patients: clinical characteristics, antimicrobial susceptibility and outcome. Afr Health Sci. 19, 2400–2406. doi: 10.4314/ahs.v19i3.13
Geisinger, E., Isberg, R. R. (2015). Antibiotic modulation of capsular exopolysaccharide and virulence in acinetobacter baumannii. PloS Pathog. 11, e1004691. doi: 10.1371/journal.ppat.1004691
Gong, X., Zhao, Q., Wu, Y., Zhou, H., Ding, S., Zhu, K. (2022). Mucoid Acinetobacter baumannii enhances anti-phagocytosis through reducing C3b deposition. Front. Med. (Lausanne) 9, 879361. doi: 10.3389/fmed.2022.879361
Grygorcewicz, B., Wojciuk, B., Roszak, M., Łubowska, N., Błażejczak, P., Jursa-Kulesza, J., et al. (2021). Environmental phage-based cocktail and antibiotic combination effects on acinetobacter baumannii biofilm in a human urine model. Microb. Drug Resist. 27, 25–35. doi: 10.1089/mdr.2020.0083
Guo, Z., Liu, M., Zhang, D. (2023). Potential of phage depolymerase for the treatment of bacterial biofilms. Virulence 14, 2273567. doi: 10.1080/21505594.2023.2273567
Hassan, A. Y., Lin, J. T., Ricker, N., Anany, H. (2021). The age of phage: friend or foe in the new dawn of therapeutic and biocontrol applications? Pharmaceuticals 14, 199. doi: 10.3390/ph14030199
Hernandez-Morales, A. C., Lessor, L. L., Wood, T. L., Migl, D., Mijalis, E. M., Cahill, J., et al. (2018). Genomic and biochemical characterization of acinetobacter podophage petty reveals a novel lysis mechanism and tail-associated depolymerase activity. J. Virol. 92. doi: 10.1128/JVI.01064-17
Hibstu, Z., Belew, H., Akelew, Y., Mengist, H. M. (2022). Phage therapy: A different approach to fight bacterial infections. Biologics 16, 173–186. doi: 10.2147/BTT.S381237
Hsieh, P. F., Lin, H. H., Lin, T. L., Chen, Y. Y., Wang, J. T. (2017). Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci. Rep. 7, 4624. doi: 10.1038/s41598-017-04644-2
Ilomuanya, M. O., Enwuru, N. V., Adenokun, E., Fatunmbi, A., Adeluola, A., Igwilo, C. I. (2022). Chitosan-based microparticle encapsulated acinetobacter baumannii phage cocktail in hydrogel matrix for the management of multidrug resistant chronic wound infection. Turk J. Pharm. Sci. 19, 187–195. doi: 10.4274/tjps
Jennings, M. P., Day, C. J., Atack, J. M. (2022). How bacteria utilize sialic acid during interactions with the host: snip, snatch, dispatch, match and attach. Microbiol. (Reading) 168. doi: 10.1099/mic.0.001157
Jeon, J., Park, J. H., Yong, D. (2019). Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 19, 70. doi: 10.1186/s12866-019-1443-5
Jo, S. J., Kwon, J., Kim, S. G., Lee, S. J. (2023). The biotechnological application of bacteriophages: what to do and where to go in the middle of the post-antibiotic era. Microorganisms 11. doi: 10.3390/microorganisms11092311
Karampatakis, T., Tsergouli, K., Behzadi, P. (2024). Pan-genome plasticity and virulence factors: A natural treasure trove for acinetobacter baumannii. Antibiot. (Basel) 13. doi: 10.3390/antibiotics13030257
Karygianni, L., Ren, Z., Koo, H., Thurnheer, T. (2020). Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/j.tim.2020.03.016
Kasimova, A. A., Sharar, N. S., Ambrose, S. J., Knirel, Y. A., Shneider, M. M., Timoshina, O. Y., et al. (2023). The Acinetobacter baumannii K70 and K9 capsular polysaccharides consist of related K-units linked by the same Wzy polymerase and cleaved by the same phage depolymerases. Microbiol. Spectr. 11, e0302523. doi: 10.1128/spectrum.03025-23
Kaur, N., Dey, P. (2023). Bacterial exopolysaccharides as emerging bioactive macromolecules: from fundamentals to applications. Res. Microbiol. 174, 104024. doi: 10.1016/j.resmic.2022.104024
Keil, J. M., Rafn, G. R., Turan, I. M., Aljohani, M. A., Sahebjam-Atabaki, R., Sun, X. L. (2022). Sialidase inhibitors with different mechanisms. J. Med. Chem. 65, 13574–13593. doi: 10.1021/acs.jmedchem.2c01258
Kim, H. S., Lee, C.-G., Lee, E. Y. (2011). Alginate lyase: Structure, property, and application. Biotechnol. Bioprocess Eng. 16, 843–851. doi: 10.1007/s12257-011-0352-8
Kimura, K., Itoh, Y. (2003). Characterization of poly-gamma-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of Bacillus subtilis encapsulated with poly-gamma-glutamate. Appl. Environ. Microbiol. 69, 2491–2497. doi: 10.1128/AEM.69.5.2491-2497.2003
Klumpp, J., Dunne, M., Loessner, M. J. (2023). A perfect fit: Bacteriophage receptor-binding proteins for diagnostic and therapeutic applications. Curr. Opin. Microbiol. 71, 102240. doi: 10.1016/j.mib.2022.102240
Knecht, L. E., Veljkovic, M., Fieseler, L. (2019). Diversity and function of phage encoded depolymerases. Front. Microbiol. 10, 2949. doi: 10.3389/fmicb.2019.02949
Ko, T. P., Lai, S. J., Hsieh, T. J., Yang, C. S., Chen, Y. (2018). The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase from Acinetobacter baumannii: A comparative study with human GNE. J. Biol. Chem. 293, 10119–10127. doi: 10.1074/jbc.RA118.001971
Lai, M. J., Chang, K. C., Huang, S. W., Luo, C. H., Chiou, P. Y., Wu, C. C., et al. (2016). The tail associated protein of acinetobacter baumannii phage phiAB6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PloS One 11, e0153361. doi: 10.1371/journal.pone.0153361
Latka, A., Leiman, P. G., Drulis-Kawa, Z., Briers, Y. (2019). Modeling the architecture of depolymerase-containing receptor binding proteins in klebsiella phages. Front. microbiol. 10. doi: 10.3389/fmicb.2019.02649
Latka, A., Maciejewska, B., Majkowska-Skrobek, G., Briers, Y., Drulis-Kawa, Z. (2017). Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 101, 3103–3119. doi: 10.1007/s00253-017-8224-6
Lavergne, S., Hamilton, T., Biswas, B., Kumaraswamy, M., Schooley, R. T., Wooten, D. (2018). Phage therapy for a multidrug-resistant acinetobacter baumannii craniectomy site infection. Open Forum Infect. Dis. 5, ofy064. doi: 10.1093/ofid/ofy064
Lee, I. M., Tu, I. F., Yang, F.-L., Ko, T.-P., Liao, J.-H., Lin, N.-T., et al. (2017). Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage ΦAB6 tailspike protein. Sci. Rep. 7, 42711. doi: 10.1038/srep42711
Lees-Miller, R. G., Iwashkiw, J. A., Scott, N. E., Seper, A., Vinogradov, E., Schild, S., et al. (2013). A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89, 816–830. doi: 10.1111/mmi.12300
Lewis, A. L., Lewis, W. G. (2012). Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol. 14, 1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x
Li, P., Ma, W., Shen, J., Zhou, X. (2022). Characterization of Novel Bacteriophage vB_KpnP_ZX1 and Its Depolymerases with Therapeutic Potential for K57 Klebsiella pneumoniae Infection. Pharmaceutics 14. doi: 10.3390/pharmaceutics14091916
Li, S., Wei, B., Xu, L., Cong, C., Murtaza, B., Wang, L., et al. (2024). In vivo efficacy of phage cocktails against carbapenem resistance Acinetobacter baumannii in the rat pneumonia model. Virol. J. 0, e00467–e00424. doi: 10.1128/jvi.00467-24
Lin, T. L., Hsieh, P. F., Huang, Y. T., Lee, W. C., Tsai, Y. T., Su, P. A., et al. (2014). Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210, 1734–1744. doi: 10.1093/infdis/jiu332
Lin, L., Tan, B., Pantapalangkoor, P., Ho, T., Baquir, B., Tomaras, A., et al. (2012). Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. mBio 3. doi: 10.1128/mBio.00312-12
Liu, Y., Mi, Z., Mi, L., Huang, Y., Li, P., Liu, H., et al. (2019). Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 7, e6173. doi: 10.7717/peerj.6173
Lu, T. K., Collins, J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. PNAS 104, 11197–11202. doi: 10.1073/pnas.0704624104
Maciejewska, B., Olszak, T., Drulis-Kawa, Z. (2018). Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 102, 2563–2581. doi: 10.1007/s00253-018-8811-1
Maciejewska, B., Squeglia, F., Latka, A., Privitera, M., Olejniczak, S., Switala, P., et al. (2023). Klebsiella phage KP34gp57 capsular depolymerase structure and function: from a serendipitous finding to the design of active mini-enzymes against K. pneumoniae. mBio 14, e0132923. doi: 10.1128/mbio.01329-23
Magill, D. J., Skvortsov, T. A. (2023). DePolymerase Predictor (DePP): a machine learning tool for the targeted identification of phage depolymerases. BMC Bioinf. 24, 208. doi: 10.1186/s12859-023-05341-w
Majkowska-Skrobek, G., Latka, A., Berisio, R., Squeglia, F., Maciejewska, B., Briers, Y., et al. (2018). Phage-borne depolymerases decrease klebsiella pneumoniae resistance to innate defense mechanisms. Front. Microbiol. 9, 2517. doi: 10.3389/fmicb.2018.02517
Mariani, F., Galvan, E. M. (2023). Staphylococcus aureus in polymicrobial skinand soft tissue infections: impact of inter-species interactionsin disease outcome. Antibiot. (Basel) 12. doi: 10.3390/antibiotics12071164
Matilla, M. A., Salmond, G. P. C. (2014). Bacteriophage ϕMAM1, a viunalikevirus, is a broad-host-range, high-efficiency generalized transducer that infects environmental and clinical isolates of the enterobacterial genera Serratia and Kluyvera. Appl. Environ. Microbiol. 80, 6446–6457. doi: 10.1128/AEM.01546-14
Mendes, S. G., Combo, S. I., Allain, T., Domingues, S., Buret, A. G., Da Silva, G. J. (2023). Co-regulation of biofilm formation and antimicrobial resistance in Acinetobacter baumannii: from mechanisms to therapeutic strategies. Eur. J. Clin. Microbiol. Infect. Dis. 42, 1405–1423. doi: 10.1007/s10096-023-04677-8
Minzanova, S. T., Mironov, V. F., Arkhipova, D. M., Khabibullina, A. V., Mironova, L. G., Zakirova, Y. M., et al. (2018). Biological activity and pharmacological application of pectic polysaccharides: A review. Polymers 10, 1407. doi: 10.3390/polym10121407
Murakami, H., Kuramoto, T., Mizutani, K., Nakano, H., Kitahata, S. (1992). Purification and some properties of a new levanase from bacillus sp. No. 71. Biosci. Biotechnol. Biochem. 56, 608–613. doi: 10.1271/bbb.56.608
Mushtaq, F., Nadeem, A., Yabrag, A., Bala, A., Karah, N., Zlatkov, N., et al. (2024). Colony phase variation switch modulates antimicrobial tolerance and biofilm formation in Acinetobacter baumannii. Microbiol. Spectr. 12, e02956–e02923. doi: 10.1128/spectrum.02956-23
Mushtaq, N., Redpath, M. B., Luzio, J. P., Taylor, P. W. (2004). Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob. Agents Chemother. 48, 1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004
Nguyen, M., Joshi, S. G. (2021). Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J. Appl. Microbiol. 131, 2715–2738. doi: 10.1111/jam.15130
Noreika, A., Stankeviciute, J., Rutkiene, R., Meskys, R., Kaliniene, L. (2023). Exploring the enzymatic activity of depolymerase gp531 from Klebsiella pneumoniae jumbo phage RaK2. Virus Res. 336, 199225. doi: 10.1016/j.virusres.2023.199225
Oechslin, F. (2018). Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10. doi: 10.3390/v10070351
Oliveira, H., Costa, A. R., Ferreira, A., Konstantinides, N., Santos, S. B., Boon, M., et al. (2019). Functional analysis and antivirulence properties of a new depolymerase from a myovirus that infects acinetobacter baumannii capsule K45. Virol. J. 93. doi: 10.1128/jvi.01163-01118
Oliveira, H., Costa, A. R., Konstantinides, N., Ferreira, A., Akturk, E., Sillankorva, S., et al. (2017). Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ. Microbiol. 19, 5060–5077. doi: 10.1111/1462-2920.13970
Oromi-Bosch, A., Antani, J. D., Turner, P. E. (2023). Developing phage therapy that overcomes the evolution of bacterial resistance. Annu. Rev. Virol. 10, 503–524. doi: 10.1146/annurev-virology-012423-110530
Ozaki, T., Abe, N., Kimura, K., Suzuki, A., Kaneko, J. (2017). Genomic analysis of Bacillus subtilis lytic bacteriophage ϕNIT1 capable of obstructing natto fermentation carrying genes for the capsule-lytic soluble enzymes poly-γ-glutamate hydrolase and levanase. Biosci. Biotechnol. Biochem. 81, 135–146. doi: 10.1080/09168451.2016.1232153
Pandey, S., Berger, B. W., Acharya, R. (2023). Structural analyses of substrate–pH activity pairing observed across diverse polysaccharide lyases. Biochemistry 62, 2775–2790. doi: 10.1021/acs.biochem.3c00321
Pas, C., Latka, A., Fieseler, L., Briers, Y. (2023). Phage tailspike modularity and horizontal gene transfer reveals specificity towards E. coli O-antigen serogroups. Virol. J. 20, 174. doi: 10.1186/s12985-023-02138-4
Patel, S. R., Pratap, C. B., Nath, G. (2021). Evaluation of bacteriophage cocktail on septicaemia caused by colistin-resistant Acinetobacter baumannii in immunocompromised mice model. Indian J. Med. Res. 154, 141–149. doi: 10.4103/ijmr.IJMR_2271_18
Pires, D. P., Melo, L. D. R., Azeredo, J. (2021). Understanding the complex phage-host interactions in biofilm communities. Annu. Rev. Virol. 8, 73–94. doi: 10.1146/annurev-virology-091919-074222
Pires, D. P., Oliveira, H., Melo, L. D., Sillankorva, S., Azeredo, J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151. doi: 10.1007/s00253-015-7247-0
Pirnay, J. P., Kutter, E. (2021). Bacteriophages: it's a medicine, Jim, but not as we know it. Lancet Infect. Dis. 21, 309–311. doi: 10.1016/S1473-3099(20)30464-3
Popova, A. V., Shneider, M. M., Arbatsky, N. P., Kasimova, A. A., Senchenkova, S. N., Shashkov, A. S., et al. (2021). Specific interaction of novel friunavirus phages encoding tailspike depolymerases with corresponding acinetobacter baumannii capsular types. J. Virol. 95. doi: 10.1128/JVI.01714-20
Rakovitsky, N., Lellouche, J., Ben David, D., Frenk, S., Elmalih, P., Weber, G., et al. (2021). Increased capsule thickness and hypermotility are traits of carbapenem-resistant acinetobacter baumannii ST3 strains causing fulminant infection. Open Forum Infect. Dis. 8, ofab386. doi: 10.1093/ofid/ofab386
Roy, S., Chowdhury, G., Mukhopadhyay, A. K., Dutta, S., Basu, S. (2022). Convergence of biofilm formation and antibiotic resistance in acinetobacter baumannii infection. Front. Med. (Lausanne) 9, 793615. doi: 10.3389/fmed.2022.793615
Russo, T. A., Luke, N. R., Beanan, J. M., Olson, R., Sauberan, S. L., Macdonald, U., et al. (2010). The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 78, 3993–4000. doi: 10.1128/IAI.00366-10
Ryan, E. M., Gorman, S. P., Donnelly, R. F., Gilmore, B. F. (2011). Recent advances in bacteriophage therapy: how delivery routes, formulation, concentration and timing influence the success of phage therapy. J. Pharm. Pharmacol. 63, 1253–1264. doi: 10.1111/j.2042-7158.2011.01324.x
Schooley, R. T., Biswas, B., Gill, J. J., Hernandez-Morales, A., Lancaster, J., Lessor, L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.00954-17
Seleim, S. M., Mostafa, M. S., Ouda, N. H., Shash, R. Y. (2022). The role of pmrCAB genes in colistin-resistant Acinetobacter baumannii. Sci. Rep. 12, 20951. doi: 10.1038/s41598-022-25226-x
Shahed-Al-Mahmud, M., Roy, R., Sugiokto, F. G., Islam, M. N., Lin, M. D., Lin, L. C., et al. (2021). Phage phiAB6-Borne Depolymerase Combats Acinetobacter baumannii Biofilm Formation and Infection. Antibiot. (Basel) 10. doi: 10.3390/antibiotics10030279
Sharma, D., Misba, L., Khan, A. U. (2019). Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8, 76. doi: 10.1186/s13756-019-0533-3
Sharma, S., Mohler, J., Mahajan, S. D., Schwartz, S. A., Bruggemann, L., Aalinkeel, R. (2023). Microbial biofilm: A review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganisms 11. doi: 10.3390/microorganisms11061614
Singh, S., Datta, S., Narayanan, K. B., Rajnish, K. N. (2021). Bacterial exo-polysaccharides in biofilms: role in antimicrobial resistance and treatments. J. Genet. Eng. Biotechnol. 19, 140. doi: 10.1186/s43141-021-00242-y
Tailford, L. E., Owen, C. D., Walshaw, J., Crost, E. H., Hardy-Goddard, J., Le Gall, G., et al. (2015). Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat. Commun. 6, 7624. doi: 10.1038/ncomms8624
Talyansky, Y., Nielsen, T. B., Yan, J., Carlino-Macdonald, U., Di Venanzio, G., Chakravorty, S., et al. (2021). Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PloS Pathog. 17, e1009291. doi: 10.1371/journal.ppat.1009291
Tan, Y., Su, J., Fu, M., Zhang, H., Zeng, H. (2022). Recent advances in phage-based therapeutics for multi-drug resistant acinetobacter baumannii. Bioeng. (Basel) 10. doi: 10.3390/bioengineering10010035
Tian, F., Li, J., Nazir, A., Tong, Y. (2021). Bacteriophage - A promising alternative measure for bacterial biofilm control. Infect. Drug Resist. 14, 205–217. doi: 10.2147/IDR.S290093
Timoshina, O. Y., Kasimova, A. A., Shneider, M. M., Matyuta, I. O., Nikolaeva, A. Y., Evseev, P. V., et al. (2023). Friunavirus phage-encoded depolymerases specific to different capsular types of acinetobacter baumannii. Int. J. Mol. Sci. 24. doi: 10.3390/ijms24109100
Tipton Kyle, A., Chin, C.-Y., Farokhyfar, M., Weiss David, S., Rather Philip, N. (2018). Role of capsule in resistance to disinfectants, host antimicrobials, and desiccation in acinetobacter baumannii. Antimicrob. Agents Chemother. 62. doi: 10.1128/aac.01188-01118
Topka-Bielecka, G., Dydecka, A., Necel, A., Bloch, S., Nejman-Falenczyk, B., Wegrzyn, G., et al. (2021). Bacteriophage-derived depolymerases against bacterial biofilm. Antibiot. (Basel) 10. doi: 10.3390/antibiotics10020175
Wang, C., Li, P., Niu, W., Yuan, X., Liu, H., Huang, Y., et al. (2019). Protective and therapeutic application of the depolymerase derived from a novel KN1 genotype of Klebsiella pneumoniae bacteriophage in mice. Res. Microbiol. 170, 156–164. doi: 10.1016/j.resmic.2019.01.003
Wang, C., Li, P., Zhu, Y., Huang, Y., Gao, M., Yuan, X., et al. (2020). Identification of a novel acinetobacter baumannii phage-derived depolymerase and its therapeutic application in mice. Front. Microbiol. 11, 1407. doi: 10.3389/fmicb.2020.01407
Wang, H., Liu, Y., Bai, C., Leung, S. S. Y. (2024a). Translating bacteriophage-derived depolymerases into antibacterial therapeutics: Challenges and prospects. Acta Pharm. Sin. B 14, 155–169. doi: 10.1016/j.apsb.2023.08.017
Wang, M., Xian, Y., Lu, Z., Wu, P., Zhang, G. (2024b). Engineering polysaccharide hydrolases in the product-releasing cleft to alter their product profiles. Int. J. Biol. Macromol. 256, 128416. doi: 10.1016/j.ijbiomac.2023.128416
Wang-Lin Shun, X., Olson, R., Beanan Janet, M., Macdonald, U., Balthasar Joseph, P., Russo Thomas, A. (2017). The capsular polysaccharide of acinetobacter baumannii is an obstacle for therapeutic passive immunization strategies. Infect. Immun. 85. doi: 10.1128/iai.00591-00517
Weber, D. J., Rutala, W. A., Anderson, D. J., Sickbert-Bennett, E. E. (2023). Biofilms on medical instruments and surfaces: Do they interfere with instrument reprocessing and surface disinfection. Am. J. Infect. Control 51, A114–A119. doi: 10.1016/j.ajic.2023.04.158
Wegrzyn, G. (2022). Should bacteriophages be classified as parasites or predators? Pol. J. Microbiol. 71, 3–9. doi: 10.33073/pjm-2022-005
Wences, M., Wolf, E. R., Li, C., Singh, N., Bah, N., Tan, X., et al. (2022). Combatting planktonic and biofilm populations of carbapenem-resistant acinetobacter baumannii with polymyxin-based combinations. Antibiot. (Basel) 11. doi: 10.3390/antibiotics11070959
WHO (2024). WHO Bacterial Priority Pathogens List 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization.
Wintachai, P., Phaonakrop, N., Roytrakul, S., Naknaen, A., Pomwised, R., Voravuthikunchai, S. P., et al. (2022). Enhanced antibacterial effect of a novel Friunavirus phage vWU2001 in combination with colistin against carbapenem-resistant Acinetobacter baumannii. Sci. Rep. 12, 2633. doi: 10.1038/s41598-022-06582-0
Wright, R. C. T., Brockhurst, M. A., Harrison, E. (2016). Ecological conditions determine extinction risk in co-evolving bacteria-phage populations. BMC Evolution. Biol. 16, 227. doi: 10.1186/s12862-016-0808-8
Keywords: bacteriophage, depolymerase, polysaccharide, biofilm, drug resistance
Citation: Islam MM, Mahbub NU, Shin WS and Oh MH (2024) Phage-encoded depolymerases as a strategy for combating multidrug-resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 14:1462620. doi: 10.3389/fcimb.2024.1462620
Received: 10 July 2024; Accepted: 26 September 2024;
Published: 24 October 2024.
Edited by:
Dhammika Leshan Wannigama, Yamagata Prefectural Central Hospital, JapanReviewed by:
Puey Ounjai, Mahidol University, ThailandCopyright © 2024 Islam, Mahbub, Shin and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo Shik Shin, d3NoaW5AbmVvbWVkLmVkdQ==; Man Hwan Oh, eXkxMDkxQGRhbmtvb2suYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.