- 1Center for Marine Environmental Studies (CMES), Ehime University, Matsuyama, Ehime, Japan
- 2Department of Biological Sciences, College of Science, Central Luzon State University, Science City of Muñoz, Nueva Ecija, Philippines
Addressing the global antimicrobial resistance (AMR) crisis requires a multifaceted innovative approach to mitigate impacts on public health, healthcare and economic systems. In the complex evolution of AMR, biofilms and the acquisition of antimicrobial resistance genes (ARGs) play a pivotal role. Aeromonas is a major AMR player that often forms biofilm, harbors ARGs and is frequently detected in wastewater. Existing wastewater treatment plants (WWTPs) do not have the capacity to totally eliminate antimicrobial-resistant bacteria favoring the evolution of ARGs in wastewater. Besides facilitating the emergence of AMR, biofilms contribute significantly to biofouling process within the activated sludge of WWTP bioreactors. This paper presents the inhibition of biofilm formation, the expression of biofilm-linked genes and ARGs by phytochemicals andrographolide, docosanol, lanosterol, quercetin, rutin and thymohydroquinone. Aeromonas species were isolated and purified from activated sludge samples. The ARGs were detected in the isolated Aeromonas species through PCR. Aeromonas biofilms were quantified following the application of biocompounds through the microtiter plate assay. qPCR analyses of related genes were done for confirmation. Findings showed that the natural compounds inhibited the formation of biofilms and reduced the expression of genes linked to biofilm production as well as ARGs in wastewater Aeromonas. This indicates the efficacy of these compounds in targeting and controlling both ARGs and biofilm formation, highlighting their potential as innovative solutions for combating antimicrobial resistance and biofouling.
1 Introduction
Wastewater treatment plants (WWTPs) and wastewater are critical transition points for the emergence of antimicrobial-resistant bacteria (ARB) and antimicrobial resistance genes (ARG) (Skwor et al., 2020) following the release of residual antibiotics not fully metabolized after therapeutic use. Since existing WWTPs do not have the capacity to totally eliminate pollutants specifically ARB (Shrout and Nerenberg, 2012; Uluseker et al., 2021; Perveen et al., 2023), this creates a favorable environment for the expansion of antimicrobial resistance. Among the diverse bacterial species found in wastewater, Aeromonas species are known to harbor a wide range of ARGs and virulence factors, making them ideal model organisms for monitoring AMR and studying the mechanisms of resistance development (Usui et al., 2016; Skwor et al., 2020). Aeromonas genomes harboring ARGs have increased considerably (Roh and Kannimuthu, 2023) and highlights their potential as a global public health risk (Piotrowska et al., 2017; Skwor et al., 2020).
Aside from facilitating the acquisition and exchange of ARGs, Aeromonas is also known for forming biofilms in water systems (Talagrand-Reboul et al., 2017). Biofilm architecture ensures stability and barrier (Hall-Stoodley et al., 2004; Rather et al., 2021) and makes biofilms particularly resistant to antibiotics. Studies have shown a substantial increase in antibiotic resistance in biofilm-associated bacteria compared to their planktonic forms, as well as a high frequency of ARGs (Chen et al., 2020; Michaelis and Grohmann, 2023). Aeromonas, akin to other bacteria, can induce biofouling in WWTP activated sludge bioreactors by forming biofilms on surfaces, filters, pumps, and membranes (Taiswa et al., 2024). This compromises the flow of water through the system, leading to decreased pressure and increased energy consumption, resulting in reduced efficiency of filters, membranes and flux rate and surge in maintenance costs.
ARGs and biofilm play a key part in the emergence of antimicrobial resistance and has been linked to most persistent infections. Strategies and emerging therapeutic options for antipathogenesis in MDR bacteria are limited, leading to higher risks of AMR development, increased morbidity and mortality rates and consequently, significant economic burdens. As antibiotics become less effective while more multi-drug resistant (MDR) pathogens emerge and spread globally, research efforts and new antimicrobials are urgently needed to produce novel, effective strategies not only for the public health but also to mitigate effects on significant economic losses (WHO, 2023). Targeting two mechanisms at the same time, such as ARGs and biofilm is a practical alternative to antibiotics use and display high potential in efficiently managing bacterial pathogenicity and antimicrobial resistance.
Natural products are now being extensively explored for their ability to interfere with ARGs and biofilm formation in bacteria. Plants have evolved intricate defense mechanisms, including the production of a wide array of secondary metabolites, many of which exhibit antimicrobial properties and play an integral part in the plant’s immune system (Algburi et al., 2017). The exploration of natural products for their potential to interfere with ARGs and biofilms is an emerging area of antimicrobial research. Several natural products have been shown to modulate or inhibit biofilm formation and downregulate ARG and biofilm-linked genes. The enormous diversity of natural products provides a vast reservoir of antimicrobial compounds that may contribute to the development of novel antimicrobial drugs to combat resistance mechanisms in bacteria.
The role in pathogenesis and increasing incidences of persistent infections influenced by ARGs and biofilm formation makes these important targets in the development of antimicrobials for Aeromonas sp. while avoiding the development of AMR driven by antibiotic over-use and mis-use. The capacity of natural compounds to influence biofilm formation and the expression of biofilm-linked genes and ARGs offers a plausible approach to addressing AMR in Aeromonas. Hence, this paper explores the action of six plant-derived compounds to inhibit the formation of biofilm and downregulate the expression of biofilm-linked genes and ARGs in 6 wastewater Aeromonas species.
2 Materials and methods
2.1 Isolation and purification of bacteria
Aeromonas species were isolated from wastewater activated sludge collected from bioreactors in 2 wastewater treatment facilities in Japan. A total of 3 liters of water were collected. The water samples were immediately filtered using a 50 µm mesh, stored in ice and processed within 3 hours. Appropriately diluted samples were plated onto Mueller-Hinton Agar plates and incubated at 37°C for 24 h. Colonies were picked on the plates and inoculated to 20 ml Luria-Bertani (LB) Broth and incubated for 24 h at 37 0C. Pure bacterial cultures were obtained after successive subculture of colonies in LB plates.
2.2 Identification of bacteria
Sixty isolates were subjected to DNA extraction and 16srRNA PCR amplification to identify the bacterial species present in the collected wastewater activated sludge. Genomic DNA was extracted using the DNeasy kit (Qiagen) following the manufacturer’s protocol. In 2ml tubes, 1.75 ml of bacterial culture was centrifuged at 20,000 x g for 5 minutes. The resulting supernatant was decanted. 180 µl of lysis buffer was added to the tube, vortexed and incubated at 37°C for 30 minutes. 25 μl of proteinase K and 200 ul of Buffer AL were added to the tube, vortexed and incubated at 56°C for 30 minutes. 200 µl 95% ethanol was added to the sample and vortexed. This was transferred to the DNeasy spin column placed in a 2 ml collection tube centrifuged at ≥6000 x g (8000 rpm) for 1 minute. The flow-through and collection tube was discarded. The spin column was placed in a new 2 ml collection tube. 500 μl Buffer AW1 was added and centrifuged for 1 min at ≥6000 x g. The flow-through and collection tube was discarded. The spin column was placed in a new 2 ml collection tube. 500 μl Buffer AW2 was added and centrifuged for 3 min at 20,000 x g (14,000 rpm). The flow-through and collection tube was discarded. The spin column was transferred to a new 1.5 ml or 2 ml microcentrifuge tube. To elute the DNA, 100 μl Buffer AE was added to the center of the spin column membrane, incubated for 1 min at room temperature (15–25°C) and centrifuge for 1 min at ≥6000 x g. The DNA was stored at -20°C until use. 16S PCR amplification was performed to accurately identify the bacteria. PCR amplification was done in a mix containing molecular-grade water, 10X Taq buffer, DMSO, MgCl2, dNTPs, primers, Taq polymerase and DNA samples. Primers used are the following: 16S 341F 5’-CCTACGGGAGGCAGCAG-3’ and 16S 907R 5’-CCGTCAATTCMTTTRAGTTT-3’ (Lane, 1991). PCR conditions include initial denaturation at 94 °C for 3 minutes followed by 30 cycles of 94 °C, 52 °C and 72 °C; and a final extension of 72°C for 5 minutes. The PCR products were visualized using 1.5% agarose gel and sequenced. Sequences were run in NCBI BLAST for the identification of Aeromonas species.
2.3 Detection of ARGs
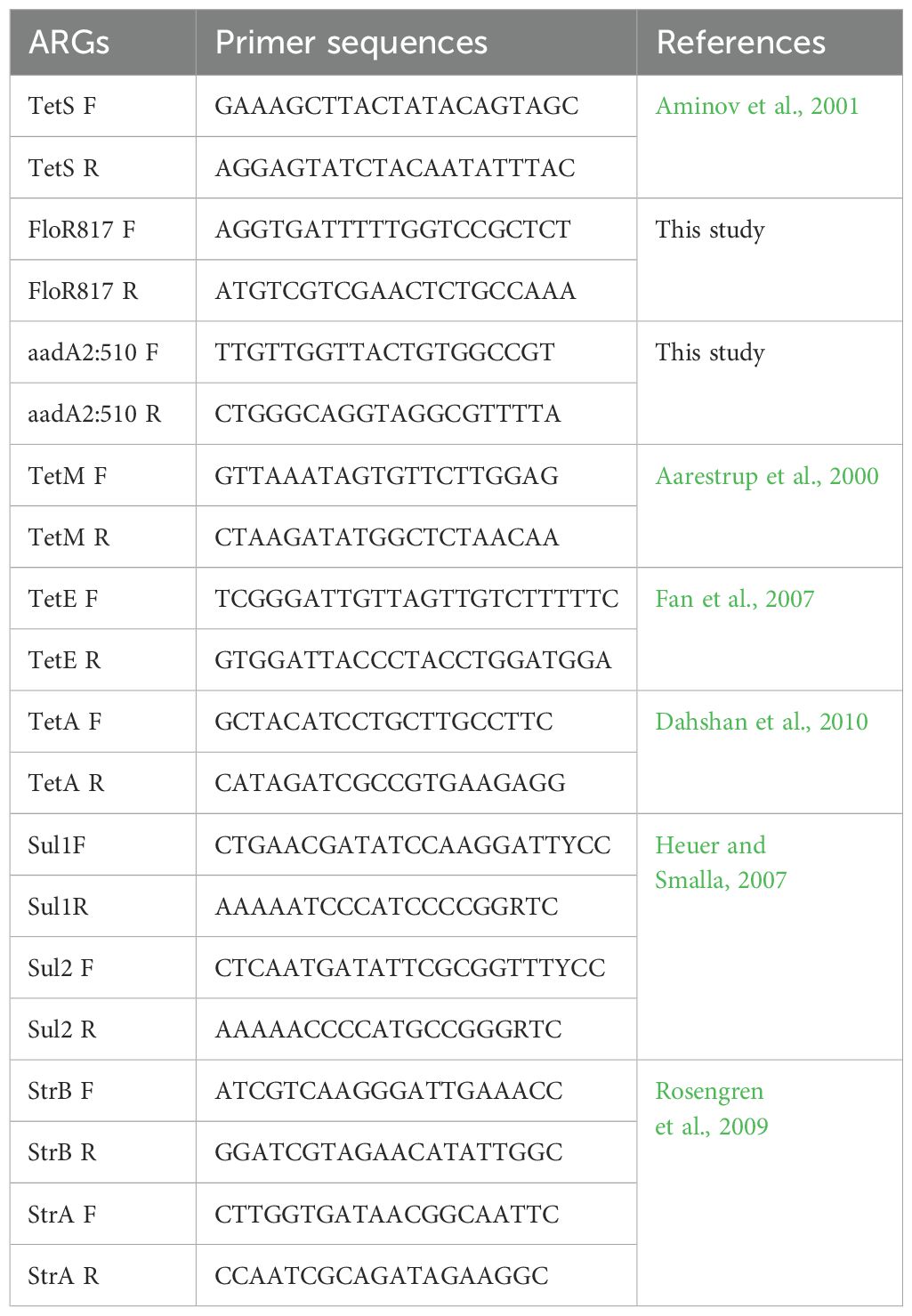
For the detection of ARGs, PCR amplification was done using specific primers (Table 1). Primers generated in this study were designed using the Primer3 program (Untergasser et al., 2012). The reaction mix included molecular-grade water, 10X Taq buffer, DMSO, MgCl2, dNTPs, primers, Taq polymerase and DNA. PCR conditions include initial denaturation at 94 °C for 5 minutes followed by 30 cycles of denaturation 94°C, annealing temperature range of 50 °C – 60 °C and extension of 72 °C; and a final extension of 72 °C for 5 minutes. The PCR products were visualized using 1.5% agarose gel and sequenced. Known DNA samples positive for the ARGs were ran alongside the samples of this study on agarose gel electrophoresis. The ARG sequences were checked on ResFinder 4.5.0 - Center for Genomic Epidemiology (CGE) (Clausen et al., 2018; Bortolaia et al., 2020). The selection of the primer pairs facilitated the identification of prevalent and clinically significant ARGs, allowing an initial assessment of compound effects.
2.4 Biocompounds for bioassays
Biocompounds andrographolide (ANDR), rutin (RUT), quercetin (QUE), docosanol (DOC), lanosterol (LAN), and thymohydroquinone (THQ) were obtained from Sigma-Aldrich (St. Louis, MO, United States) and were prepared as corresponding stock solutions following product information: ANDR - 3mg/ml in DMSO; RUT 2.5 mg/ml using DMSO; QUE- 0.5 mg/ml in DMSO; DOC - 1mg/ml in ethanol; LAN - 1mg/ml in DMSO; THQ - 1mg/ml in DMSO. All stock solutions were stored at -20°C upon preparation and used immediately to ensure freshness and stability.
2.5 Antibacterial assay and determination of minimum inhibitory concentration
Disc diffusion method for antimicrobial susceptibility testing was carried out to assess the presence of antibacterial activities of biocompounds (Rezaei et al., 2011). The 24-hour culture of Aeromonas spp. in nutrient broth was adjusted to 0.5 McFarland standard. Sterile swabs were immersed into the bacterial culture and were aseptically swabbed onto the surface of bacterial medium plates evenly. Twenty μl of each treatment, positive control Enrofloxacin and distilled water for negative control, were pipetted on sterile petri plates containing sterile paper discs which were dipped for 30 minutes. The discs were placed equidistantly on the surface of the agar medium specific for each pathogen using sterile forceps. Treatments were done in triplicates and incubated at 37°C for 24 hours. The zone of inhibition was observed after 12 and 24 hours of incubation. Positive result in the antibacterial assay established the command to proceed on the determination of Minimum Inhibitory Concentration (MIC) determination. The determination of Minimal Inhibitory Concentrations (MICs in mg/ml) of the treatments were determined by performing an adaptation of the method according to Clinical and Laboratory Standards Institute (2015) and with an inoculum of 1 × 105 Colony-Forming Unit/ml. Two-fold dilution method using Nutrient Broth was used to prepare a series of concentrations for the assay. The assay was carried out in a 96-well polystyrene microtiter plate, with 10 wells for each concentration. MIC is defined as the minimum concentration of the biocompounds which inhibit the visible growth of the Aeromonas sp. The lowest concentration that reduced the rezasurin dye was considered the MIC. In the case of THQ with observed antibacterial activity, the sub-MIC was through rezasurin-based assay in microtiter plates with two-fold dilution consisting of 8 concentrations – 1 mg/ml, 0.5 mg/ml, 0.25 mg/ml, 0.125 mg/ml, 0.0625 mg/ml, 0.03125 mg/ml, 0.0156 mg/ml and 0.0078 mg/ml (Gajic et al., 2022).
2.6 Microtiter plate biofilm formation assay
180 µl of overnight cultures of Aeromonas sp. were added to 20 µl biocompounds in each well and incubated at 30°C for 40 h. Untreated bacterial cultures served as negative control. A biological blank (PBS) was also prepared. The setup was run twice, with each run consisting of 6 replicates. After incubation, microtiter plates were rinsed with sterile distilled water five (5) times to remove planktonic cells, air-dried for 45 min, and stained with 150 µl of 1% crystal violet solution. Plates were rinsed five (5) times to remove excess stain. Quantification of biofilm production was done by adding 200 µl of 95% ethanol to destain the wells. Then, 100 μl from each well was transferred to a new microtiter plate and the optical density (OD) values were measured at 595 nm (MultiSkan FC, Thermo Scientific) (Fernando and Judan Cruz, 2020).
2.7 Expression analysis of ARGs and biofilm-linked genes
The total RNA of the bacteria were extracted using the RNeasy Minikit protocol (Qiagen, GmbH, Germany). For each sample, 25-50 mg acid-washed glass beads (150-600 μm diameter) were weighed in 2 ml safe-lock tubes. Bacteria were collected by centrifugation at 5000 x g for five minutes at 4°C. The supernatant was decanted and aspirated to ensure the removal of the remaining media. Buffer RLT was added (350 μl for <5 x 108 and 700 μl for 5 x 108 – 1 x 109 number of bacteria). The suspension was transferred into the 2ml safe-lock tube containing the acid-washed beads. Cells were disrupted in the Tissue Lyser for five minutes at maximum speed. The suspension was centrifuged for 10 seconds at maximum speed. The supernatant was transferred into a new tube and the volume of the sample was determined. An equal volume of 70% ethanol was added and mixed by pipetting. Up to 700 μl lysate was transferred to a spin column placed in a 2 ml collection tube and centrifuged for 15 seconds at ≥8000 x g. The flow-through tube was discarded. 700 μl Buffer RW1 was added to the spin column. With the lid closed gently, the spin column was centrifuged for 15 seconds at ≥8000 x g to wash the spin column membrane. The spin-column was placed in a new 2 ml collection tube with the flow-through. Lids were closed gently and centrifuged at full speed for one minute. The spin-column was placed in a new 1.5 ml collection tube. 30-50 μl RNase-free water was added directly to the spin column membrane and was centrifuged for one minute at ≥8000 x g to elute the RNA (Qiagen, 2023).
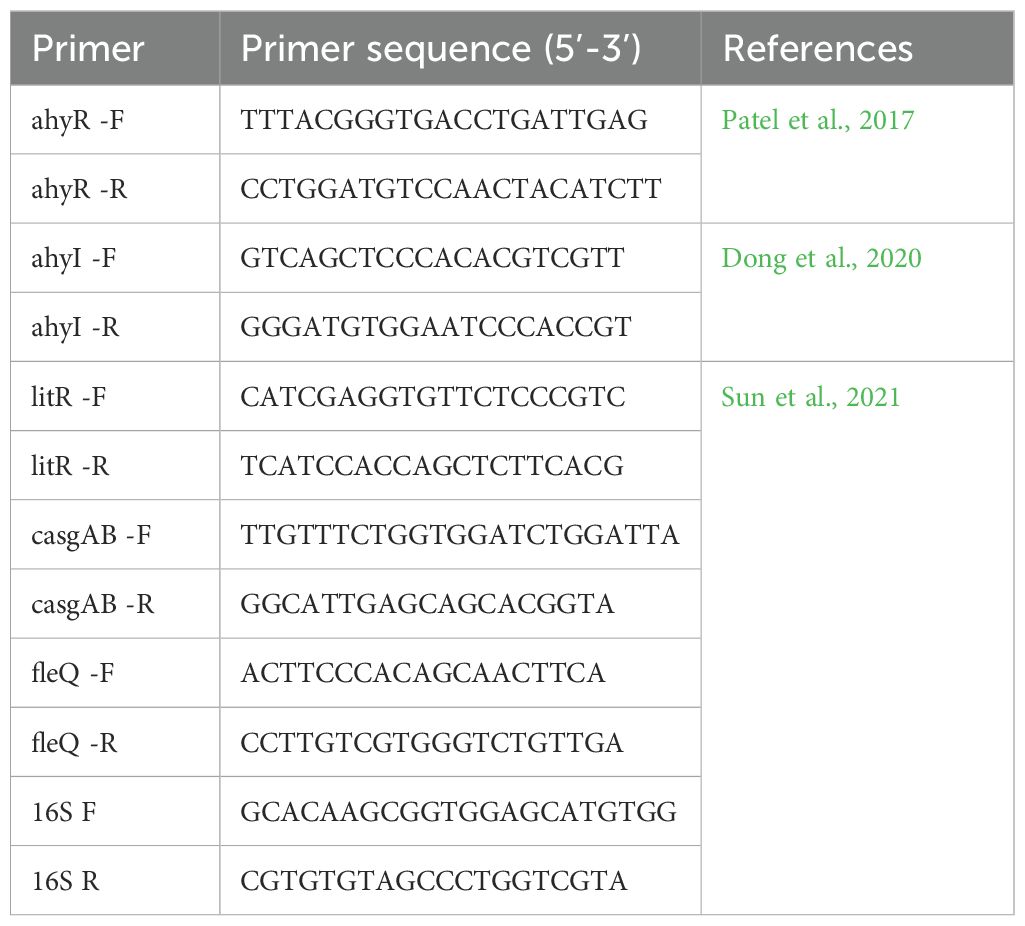
The expression analysis of ARGs and biofilm-linked genes was done using qRT-PCR with specific primers (Tables 1, 2) using Promega GoTaq ® 1-step RT-qPCR kit following the recommended proportions. A 10 μl reaction mix was prepared for each of the samples. qRT-PCR conditions included the following: 1 cycle at 42°C for 5 min and 95°C for 2 min for initial denaturation, followed by 45 cycles at 95°C for 20 s for denaturation, 55°C for 20 s for annealing, and 72°C for 20 s for the extension. For the ARGs expression, annealing temperature ranged from 55°C to 60°C. The relative expression of genes was analyzed using the LIVAK method of 2−ΔΔCt (Livak and Schmittgen, 2001).
2.8 Statistical analysis
To determine statistical differences in biofilm formation, quantification through OD values were analyzed through an independent sample non-parametric Mann-Whitney U test with 0.05 level of significance using SPSS 13.0 program. The statistical analyses for the gene regulation were done using the Kruskal-Wallis test (non-parametric ANOVA) where means between the control and experimental setup were compared and the significance were determined if the F-values were greater than the F-crit at 0.05 level of significance. For the analysis of the relative mRNA expression in qPCR, the LIVAK method (2−ΔΔCt) was used. The correlation analyses between biofilm formation and biofilm-linked genes, biofilm formation and ARGs as well as biofilm-linked genes and ARGS were done through Spearman correlation coefficient using the function pairs.panel in psychs package in R v. 4.3.1 following the standard procedure for non-parametric correlation analysis.
3 Results
3.1 16S rRNA Identification of Aeromonas wastewater bacteria
Comparison of 16S rRNA sequences of 60 wastewater bacterial isolates with the NCBI database (BLAST) confirmed the presence of known bacterial pathogens, from which five (5) species of Aeromonas were selected for further experiments on the detection and inhibition of ARGs and biofilm formation. These are Aeromonas veronii, Aeromonas jandaei, Aeromonas hydrophila, Aeromonas dhakensis and Aeromonas caviae.
3.2 Detection of ARGs
Major ARGs coding resistance to streptomycin, tetracyclines, sulfonamide and aminoglycosides were evaluated for their occurrence in the identified wastewater Aeromonas species. To identify and analyze the presence of ARGs within the Aeromonas spp., sequences were checked on ResFinder 4.5.0 - Center for Genomic Epidemiology (CGE) (Clausen et al., 2018; Bortolaia et al., 2020). Eleven ARGs were evaluated for their presence: StrA, StrB, TetS, TetM, TetE, TetA, FloR, Sul1, Sul2, aadA1 and aadA2. The results obtained from ResFinder revealed the presence of three ARGs in two Aeromonas spp: A. caviae carries three ARGs namely aadA1, aadA2 and sul1; sul1 was also detected in A. jandaei.
3.3 Biocompounds inhibit the expression of Aeromonas ARGs
The changes in gene expression levels were visualized using a heatmap to present effect of the biocompounds on the expression of ARGs (Figure 1; Supplementary Table 1). The numbers on the right side of the heat map indicate the standardized relative quantification values. The cluster delineating between the effect of the biocompounds versus the untreated sample is clear and distinct. The decrease in aadA1, aadA2 and sul1expression was statistically significant (p <0.05) indicating a downregulating effect of the biocompounds. While RUT’s effects on ARG expression varied slightly from the other biocompounds, still, showed substantial downregulation as compared to the untreated (UN) (p <0.05).
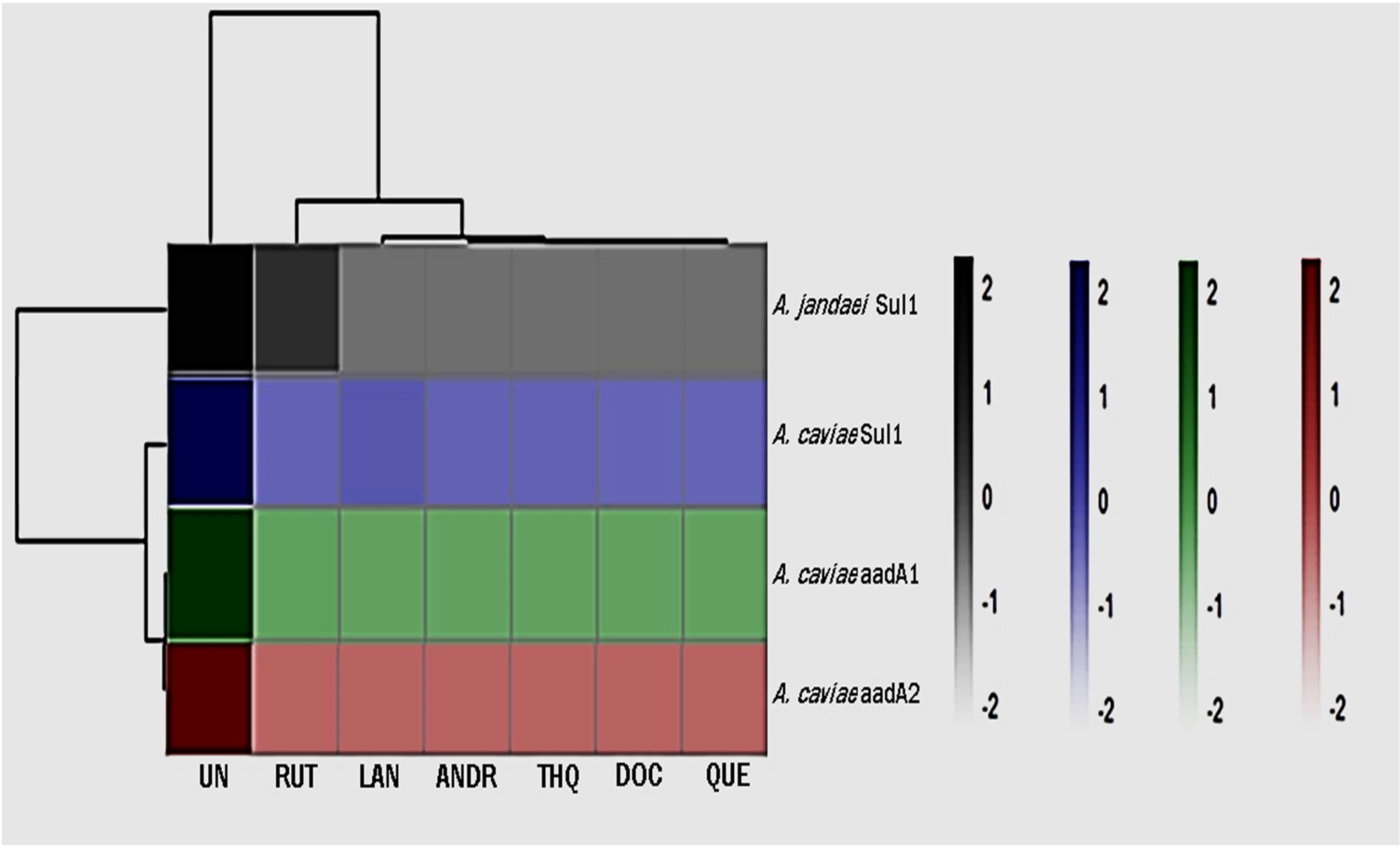
Figure 1. Heatmap of gene expression of ARGs as affected by the treatments. UN (untreated); RUT (rutin); LAN (lanosterol); QUE (quercetin); ANDR (andrographolide); DOC (docosanol); THQ.
3.4 Antibacterial activity of biocompounds against the Aeromonas spp.
Before conducting evaluations on biofilm activity, an antibacterial screening was performed to ensure that any observed reduction in the production of biofilm was not due to antibacterial properties. For biocompounds with confirmed antibacterial activity as noted by a zone of inhibition, the sub MIC was determined through a rezasurin-based assay. The biocompounds ANDR, RUT, QUE, DOC and LAN did not show any antibacterial activity in the disk diffusion assay across all tested Aeromonas spp, hence, their original concentrations were retained for further assessments. THQ demonstrated antibacterial activity against all Aeromonas spp. except A. veronii, hence, the rezasurin-based assay was performed to determine the sub-MIC that resulted to 0.5 mg/ml. This sub-MIC was then used for both assays on ARGs and biofilm since the objective was to see the effects on both mechanisms using the same concentration.
3.5 Biofilm formation inhibitory effect of the biocompounds against Aeromonas spp.
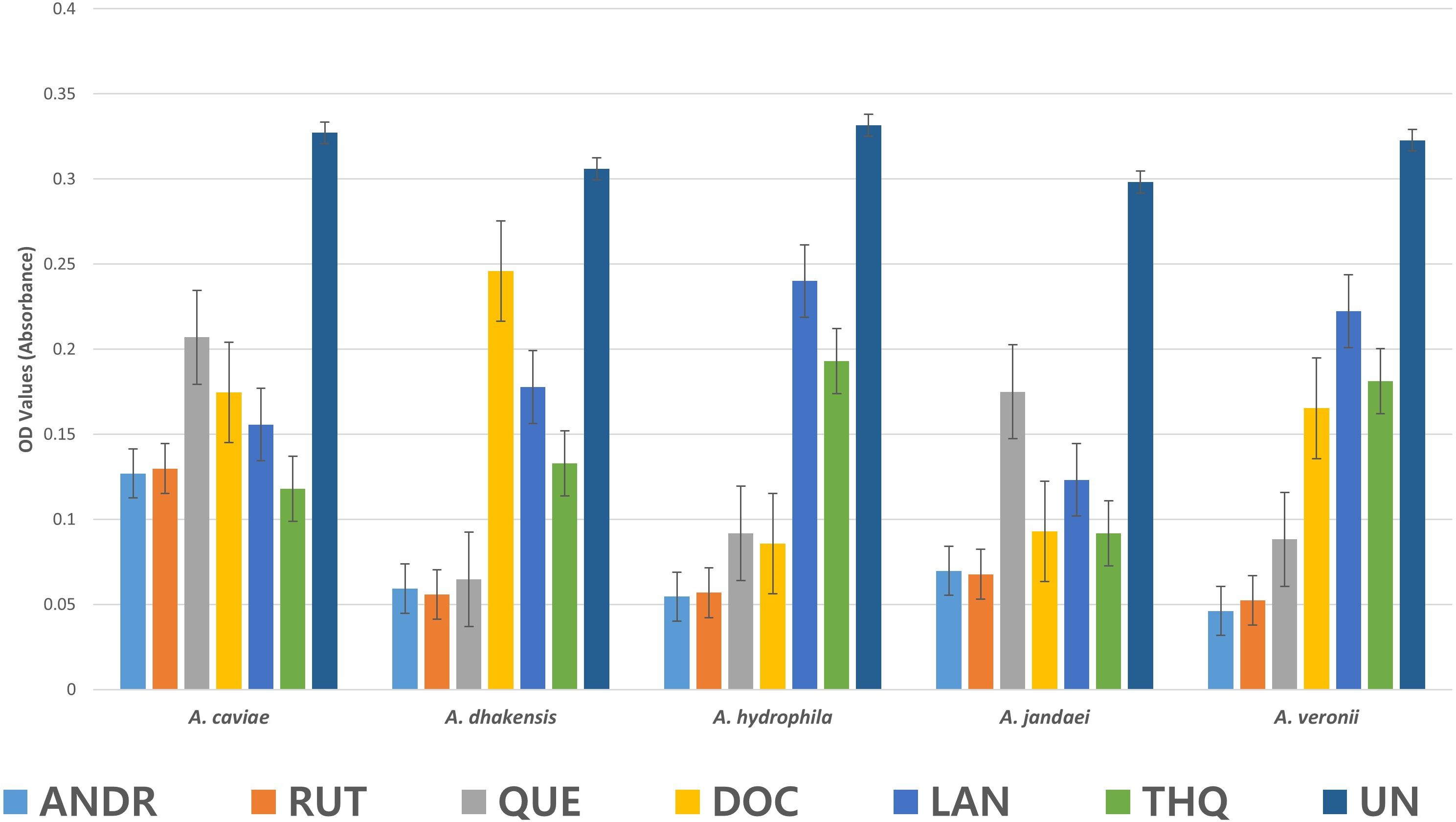
The biofilm formation activity in Aeromonas spp as affected by the biocompounds was lower than the control (UN) (Figure 2). The values presented are OD values for each treatment. In A. caviae strong inhibition by ANDR (0.127), RUT (0.129), DOC (0.174), LAN (0.156) and THQ (0.118) were noted compared to (UN) (0.327) (p < 0.01). A. caviae treated with QUE was not significantly different but showed lower OD values at 0.207 compared to UN (p > 0.05). The OD values in A. dhakensis also significantly decreased (p < 0.01) when treated with AND (0.059), RUT (0.056), QUE (0.65) and LAN (0.178). DOC (0.246) and THQ (0.133) showed lower OD values but was not significantly different from the UN (control) (0.306) (p > 0.05). The same significant observations were seen in A. hydrophila when exposed to ANDR (0.055), RUT (0.057), QUE (0.092) and DOC (0.086) (p < 0.01). Although LAN (0.240) and THQ (0.193) showed decrease in biofilm formation, it showed no significant difference with UN (0.33) (p > 0.05); in A. jandaei, all compounds except QUE (0.175) have strong anti-biofilm activity versus UN (0.298) (p < 0.01) as ANDR (0.069), RUT (0.068), docosanol (0.093), lanosterol (0.123) and THQ (0.092; and in A. veronii, ANDR (0.046), rutin (0.052), quercetin (0.088), docosanol (0.165) substantially suppressed biofilm formation (p < 0.01); OD values of LAN (0.222) and THQ (0.181) was observed lower but not significant compared to UN (0.323) (p > 0.05). The mean OD values in Aeromonas spp. treated with the biocompounds showed significant decrease in biofilm formation indicates efficiency in controlling the growth of biofilm. Overall, all biocompounds showed inhibition in biofilm formation against the wastewater Aeromonas spp.
3.6 Biocompounds inhibit the expression of Aeromonas biofilm-linked genes
The changes in gene expression levels were visualized in a heatmap, indicating the relative expression levels of key biofilm-linked genes across different samples. The heatmap of relative gene expression of biofilm-linked genes as affected by the compounds clearly shows defined clustering (Figure 3; Supplementary Table 2). The biocompounds ANDR, QUE, RUT, DOC, LAN and THQ clustered in one group showing similar downregulation of all genes compared to the untreated culture (UN) which suggests significant effects (p <0.05) of these compounds against the expression of the genes. This can be noted in A. dhakensis and A. jandaei (p <0.05). Different effects were observed in A. caviae, A. hydrophila, and A. veronii. For instance, fleQ as affected by RUT (A. caviae), AhyI, AhyR, fleQ, and casgAB in ANDR (A. hydrophila and A. veronii), DOC (A. hydrophila), RUT (A. veronii) showed higher expression of genes than other compounds, nevertheless, still showed significant downregulation when compared to treatments without the compounds (p <0.05).
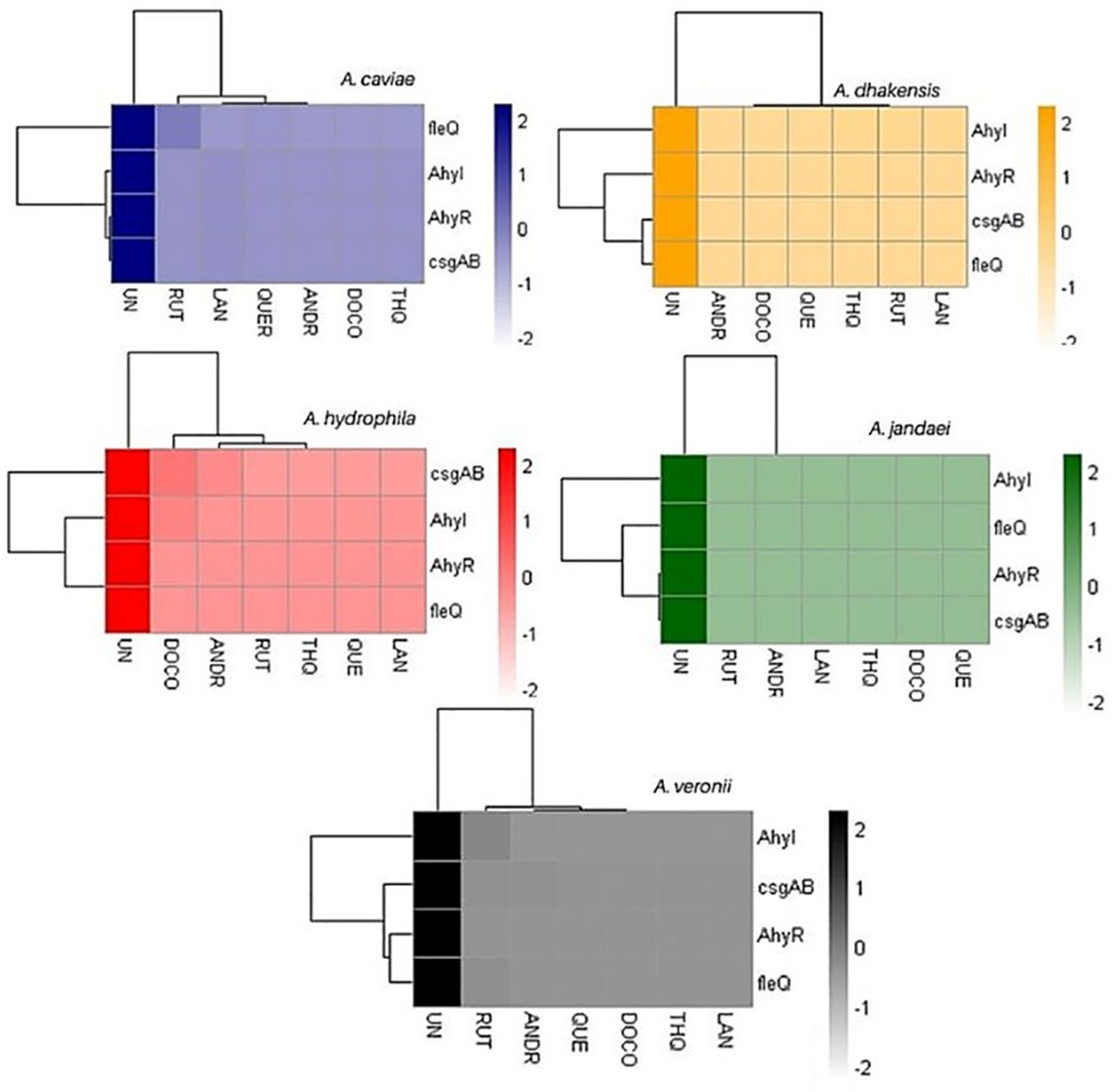
Figure 3. Heatmap of gene expression of biofilm-linked genes as affected by the treatments. UN (untreated); RUT (rutin); LAN (lanosterol); QUE (quercetin); ANDR (andrographolide); DOC (docosanol); THQ (thymohydroquinone).
3.7 Correlation analysis
The Spearman’s rank correlation coefficient (R) and its significance level (p-value) were calculated to analyze the relationship between the expression levels of genes and biofilm formation (Figure 4). In A. caviae, a strong significant positive correlation between biofilm-linked gene expression and biofilm formation was noted: AhyR (R = 0.89; p < 0.01), AhyI (R = 0.76; p < 0.05) and casgAB (R = 0.89; p < 0.01). This was also observed between biofilm formation and expression of ARGs aadA1 (R = 0.87; p < 0.05) and aadA2 (R = 0.87; p < 0.05). A strong positive correlation was also noted between the aadA1 and biofilm-linked genes AhyR (R = 0.91; p < 0.01), casgAB (R = 0.97; p < 0.001) and fleQ (R = 0.78; p < 0.05), as well as in aadA2: AhyR (R = 0.88; p < 0.01), casgAB (R = 0.98; p < 0.001) and fleQ (R = 0.84; p < 0.05. A strong significant positive correlation between biofilm-linked gene expression and biofilm formation was also seen in A. jandaei: AhyI (R = 0.77; p < 0.05), casgAB (R = 0.84; p < 0.05), fleQ (R = 0.79; p < 0.05). sul1 expression registered high significant correlation values between all biofilm-linked genes AhyR (R = 0.79; p < 0.01), AhyI (R = 0.95; p < 0.01), casgAB (R = 0.91; p < 0.01) and fleQ (R = 0.94; p < 0.01). The R value between biofilm formation and sul1 expression (0.73) was not significant, nevertheless, still showed a strong correlation. No ARGs were detected in A. dhakensis, A. hydrophila and A. veronii, hence, the correlation only presented biofilm and biofilm-linked genes. R values displayed positive correlation between biofilm formation and biofilm-linked genes in A. dhakensis, A. hydrophila and A. veronii through the R values but were not significant. However, strong interactions (p < 0.001) were observed between biofilm-linked genes.
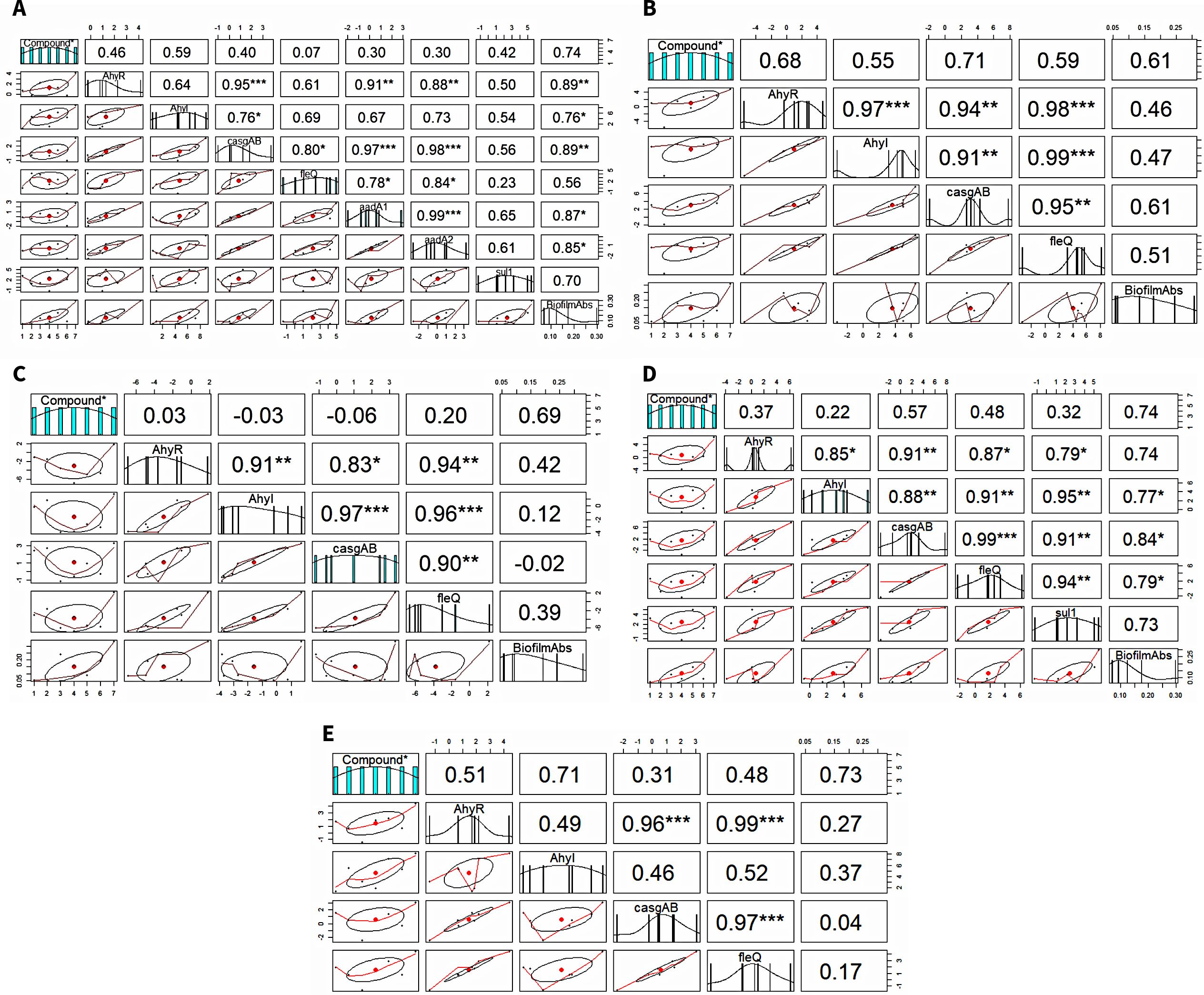
Figure 4. Correlation analysis between biofilm formation, biofilm-linked genes and ARGs. (A) A. caviae, (B) A. dhakensis, (C) A. hydrophila, (D) A. jandaei, (E) A. veronii.
4 Discussion
In this study, three ARGs—aminoglycoside resistance genes aadA1 and aadA2, and sulfonamide resistance gene sul1—were identified in wastewater Aeromonas spp. This work shows effective downregulation of these ARGs when exposed to the biocompounds, suggesting their potential in targeting AMR. These ARGs are involved in enzymatic degradation or modification of respective antibiotics i.e., sul1 encoding the DHPS enzymes with pronounced insensitivity to sulfonamides (Venkatesan et al., 2023); aadA1 and aadA2 modify aminoglycosides by adding an adenyl group rendering them inactive. As such, their efficiency adds up to strategies targeting antimicrobial-resistant enzymes and opens up the possibility of potentially developing these compounds for use in combination therapies as antibiotic adjuvants. Bacteria employ various mechanisms to resist antibiotics, including efflux pumps, antibiotic structural modifications, and in enzyme-catalyzed deactivation (El-Khoury et al., 2022; Murugaiyan et al., 2022). Targeting these mechanisms is crucial for enhancing antibiotic efficacy. Antibiotic adjuvants are used in cases of combination therapies (Kumar and Tudu, 2023) to augment the action of antibiotics by suppressing a mechanism of resistance thereby increasing bacterial susceptibility (Álvarez-Martínez et al., 2020). Natural products, for example, extracts from Rhus coriaria (Adwan et al., 2010) and plant-based compounds like tea catechins (Hu et al., 2002), quercetin and kaempferol (Lin et al., 2008), have shown synergy with antibiotics against resistant pathogens, such as Pseudomonas aeruginosa and MRSA (methicillin-resistant Staphylococcus aureus). While adjuvants are less effective in monotherapy, their combination with traditional antibiotics enhances therapeutic outcomes (Álvarez-Martínez et al., 2020). Efflux pump inhibitors (EPIs), a class of antibiotic adjuvants, have shown promise in preclinical and clinical settings (Van Bambeke et al., 2006; Zhang et al., 2023; Compagne et al., 2023) though challenges remain in their development, including pharmacokinetic profiling and addressing bacterial specificity, particularly their potential side effects on beneficial bacteria. Combination therapy of antibiotics + natural product adjuvants offer new promising strategies in synergistic approaches to manage recurring infections and control the spread of AMR.
The Aeromonas spp. identified in activated sludge in this study aligns with the previous reports of their consistent presence and ubiquity in wastewater samples (Skwor et al., 2020). Aeromonas spp are key indicator bacteria for assessing water quality risks, chiefly in antimicrobial resistance and health issues related to aquatic environments (Usui et al., 2016). The presence of the detected ARGs likely reflects past exposure and selective pressure from antibiotics, notably aminoglycosides and sulfonamides, which are frequently detected in wastewater before and after treatment (Antunes et al., 2005). Sulfonamides are widely disseminated in wastewater, often exceeding sulfonamide levels in aquatic environments linked to wastewater discharge (Proia et al., 2016; Hanna et al., 2018; Kovalakova et al., 2020). In contrast, aminoglycosides, like streptomycin, are typically found at lower concentrations, possibly due to reduced clinical use attributed to toxicity effects and has been in applied only in veterinary practices (Mutuku et al., 2022). Aeromonas spp. exhibits broad antibiotic resistance, notably to β-lactams, quinolones, aminoglycosides, sulfonamides, and tetracyclines, mediated by ARGs on mobile genetic elements (MGEs) such as plasmids, insertion sequences, transposons, and mobile integron gene cassettes (Piotrowska and Popowska, 2015). In Aeromonas, aadA1 and aadA2 are the most predominant ARGs in resistance integrons (Partridge et al., 2009; Piotrowska and Popowska, 2014; Khorshidi et al., 2022) while sul1 is considered a good marker of Class 1 integrons (Arabi et al., 2015; De los Santos et al., 2021). As also seen in this work, sul1 and aadA are found to be colocalized in Aeromonas species (Lee et al., 2023) which provides higher risk of transmission and renders the Aeromonas spp co-resistance to wider spectrum of antibiotics. Although this study only detected three out of 11 target ARGs, their detection in two isolated Aeromonas species in WWTPs already provides evidence not only of their presence but also shows their diversity of AMR mechanisms (Piotrowska and Popowska, 2014).
Biofilm formation in Aeromonas spp was also effectively inhibited by all biocompounds evaluated in this study (Figure 2). When comparing the OD values, the treatment without biocompounds showed considerably higher values, indicating higher biofilm formation. Biofilm is a critical factor in bacterial virulence and the evolution of AMR, directly contributing to approximately 80% of human microbial infections (Shunmugaperumal, 2010; Davies, 2003). Our findings demonstrate that the evaluated biocompounds disrupt biofilm formation, potentially through interference with quorum sensing (QS). QS is a communication mechanism that bacteria use to coordinate biofilm formation by influencing adhesion and exopolysaccharide (EPS) production (Miller and Bassler, 2001; Shrout and Nerenberg, 2012). By targeting QS, the biocompounds in this work may reduce pathogenicity without affecting bacterial growth, thereby lowering selective pressure and the development of antimicrobial resistance. This alternative therapeutic approach of inhibiting QS and biofilm formation offers a promising strategy for managing bacterial infections and combating AMR.
By interfering with QS cell-cell signaling, biofilm synthesis genetic pathways, adhesins, efflux pumps, and motility by repressing flagellar genes, phytochemicals function as natural antibiofilm agents that can target all stages of development (Mishra et al., 2020). Corollary to the inhibition of biofilm formation, biofilm-linked genes ahyR, ahyI, fleQ and casgAB were subsequently downregulated confirming the effects of the biocompounds and likewise points out that the compounds were able to target these genetic pathways. AhyR and AhyI are homologues of the Lux QS proteins (Kirke et al., 2004). Biofilm development in Aeromonas is dependent on the production of many QS signaling molecules, N-acylhomoserine lactone (AHL), via the AhyRI pathways (Lynch et al., 2002; Jin et al., 2020) wherein ahyR encodes AhyR which acts as a transcription regulator of ahyI for the synthesis of AHLs. fleQ is involved in the regulation of flagellar biogenesis and motility which is closely intertwined with the processes leading to biofilm formation (Du et al., 2020; Laganenka et al., 2020; Benyoussef et al., 2022). csgAB operon is responsible for the production of curli fimbriae which facilitates cell adhesion and biofilm formation (Barnhart and Chapman, 2006; Kozlova et al., 2011; Boya et al., 2022).
Our study evaluated the efficacy of six biocompounds in inhibiting biofilm formation and ARG expression in Aeromonas spp. The results demonstrated significant antibiofilm activity for all tested compounds, with andrographolide consistently showing substantial inhibition. Quercetin also showed significant inhibition in the majority of cases but was not as effective in all Aeromonas spp. tested. Andrographolide, a diterpenoid from A. paniculata, effectively reduced biofilm formation in Aeromonas, corroborating its known antimicrobial properties against high-risk pathogens such as S. aureus, Escherichia coli, and P. aeruginosa (Guo et al., 2014; Banerjee et al., 2017; Zhang et al., 2020). Andrographolide has also been reported to inhibit QS in Listeria monocytogenes by targeting the Agr system (Yu et al., 2022). Quercetin, a flavonoid, suppressed QS molecules and biofilm-related genes in Aeromonas, aligning with its previously reported effects on P. aeruginosa and Klebsiella pneumoniae, where it inhibited QS molecules lasI, lasR, rhlI, and rhlR, reducing pyocyanin production and swarming (Gopu et al., 2015; Ouyang et al., 2016; Paczkowski et al., 2017; Wang et al., 2021). It also affected gene expression linked to flagellar motility, biofilm formation, and virulence in Vibrio parahaemolyticus (Roy et al., 2022). Docosanol, primarily recognized for its antiviral properties (Katz et al., 1991; Pope et al., 1998; Sadowski et al., 2021), also exhibited antibiofilm activity in our study. This finding is consistent with limited reports on its bacterial effects, such as the suppression of biofilm and virulence factors in MRSA and the downregulation of stress response proteins (Lakshmi et al., 2020, 2022), and in K. pneumoniae (Umaru et al., 2019). Similarly, rutin has shown effectiveness against QS, biofilm formation, and expression of virulence genes in E. coli and S. aureus (Peng et al., 2018; Matilla-Cuenca et al., 2020). lanosterol and thymohydroquinone (THQ), despite their limited prior research on antimicrobial properties, showed promising results in our assays. Lanosterol has documented anticancer (Ivankovic et al., 2006) and antioxidant attributes (Tesarova et al., 2011), while THQ has shown potential but remains largely unexplored in terms of antimicrobial properties. Natural products display a rich repertoire of chemically and structurally diverse molecules with novel antimicrobial activities. This diversity can target different aspects of bacterial mechanisms and ensures that bacteria cannot easily predict or develop resistance to phytochemicals simultaneously. The dynamic nature of these compounds allows them to interfere with various bacterial processes, making them promising candidates for combating biofilm-related infections and reducing the evolution of antimicrobial resistance. Our findings provide new insights into the antibiofilm and QS-inhibiting properties of these natural biocompounds in Aeromonas spp., suggesting their potential as alternative therapeutic agents to combat biofilm-related infections and reduce the evolution of antimicrobial resistance. This study contributes to the limited research on the effects of these compounds on biofilm formation and ARG expression, highlighting their promise as antibiotic adjuvants.
The association between the ARGs, biofilm-linked genes and biofilm formation within the Aeromonas species as affected by the compounds was explored. The strong positive correlations identified in our study suggest a clear interaction between biofilm formation and the expression of biofilm-linked genes along with ARGs. Our findings align with previous studies suggesting a link between antibiotic resistance and biofilm formation, reinforcing that biofilms play a crucial role in the persistence and spread of antibiotic-resistant strains. The correlation between ARGs and biofilm-linked genes somehow suggests the coordinated regulation of these genes in the development of biofilm-associated antibiotic resistance. To strengthen these findings, future studies could consider increasing the sample size or conducting relevant experiments to further elucidate these correlations. This association has significant implications for simultaneously managing two resistance mechanisms, as well as understanding bacterial resistance mechanisms in biofilms. ARGs, such as in efflux pumps and antibiotic-modifying enzymes, have been linked to AMR in bacteria in biofilms (Kvist et al., 2008; Soto, 2013) wherein their absence can lead to diminished biofilm formation due to compromised adherence of microorganisms and the establishment of mature biofilm structures (Ren et al., 2024), thus, ARGs are now an emerging target for anti-biofilm applications and in enhancing the efficacy of AMR strategies. The limitations of this study confined the number of ARGs that could be detected to only 11, and given that only three ARGs were found and none were efflux pumps, it is plausible that other efflux pumps associated with biofilm formation in Aeromonas spp. were also affected. Hence, given this limitation, it is evident that further detection of other ARGs should be done, particularly those associated with efflux pumps. This will enhance the evaluations on the full spectrum of AMR mechanisms influenced by the biocompounds.
Suppression of both ARGs and biofilm formation by the biocompounds point to their significant role in the future of wastewater treatment. Since WWTPs are recognized as hotspots for horizontal gene transfer (HGT) of ARGs between bacterial populations, targeting these mechanisms in wastewater can prevent the transmission of antibiotic resistance, a critical issue concerning wastewater treatment. By reducing the transmission of ARGs in wastewater, there is a decreased risk of antibiotic resistance spreading into the environment, which could affect human health and ecosystems (Che et al., 2019). This also addresses reduction of biofouling in bioreactors. Reduction in biofouling, is a constant concern in wastewater treatment, and this justifies the application of biocompounds substances as a cost-effective and environmentally friendly solution paving the way for sustainable solutions to wastewater treatments and its implication to global health.
5 Conclusion
To combat the increasing multidrug resistance in Aeromonas, a comprehensive, multifaceted approach is essential, targeting both ARGs and biofilm formation. This work demonstrates that naturally-derived compounds effectively address these issues. Specifically, our results revealed that these compounds significantly reduced ARG expression levels and markedly decreased biofilm formation in Aeromonas.
The study highlights the effectiveness of these natural products in mitigating the development and transmission of AMR in wastewater bacteria. By disrupting biofilm formation and inhibiting ARG expression, these compounds present a promising strategy for enhancing sustainable wastewater treatment and preventing biofouling in treatment facilities. Furthermore, the ability of these biocompounds to act as adjuvants to antibiotics — by inhibiting ARGs and biofilms — offers a significant advancement in improving the efficacy of antibiotic therapies for biofilm-forming pathogens.
Overall, the findings underscore the potential of these natural products to provide a dual-action approach against AMR, both in molecular and structural aspects of bacterial resistance. This work not only addresses the challenge of AMR in wastewater bacteria but also contributes to reducing the broader burden of AMR. The study emphasizes the need for continued exploration of these bioactive compounds to fully understand their mechanisms and maximize their potential in addressing the global AMR crisis.
Data availability statement
All relevant data is contained within the article: The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
KJ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. OT: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft. KB: Formal analysis, Methodology, Software, Validation, Visualization, Writing – review & editing. EG: Investigation, Methodology, Writing – review & editing. NK: Methodology, Validation, Writing – review & editing. KW: Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Japan Society for the Promotion of Science (JSPS) and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1456700/full#supplementary-material
References
Aarestrup, F. M., Agerso, Y., Gerner-Smidt, P., Madsen, M., Jensen, L. B. (2000). Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37, 127–137. doi: 10.1016/s0732-8893(00)00130-9
Adwan, G., Abu-Shanab, B., Adwan, K. (2010). Antibacterial activities of some plant extracts alone and in combination with different antimicrobials against multidrug–resistant Pseudomonas aeruginosa strains. Asia Pac J. Trop. Med. 3 (4), 266–269. doi: 10.1016/S1995-7645(10)60064-8
Algburi, A., Comito, N., Kashtanov, D., Dicks, L. M. T., Chikindas, M. L. (2017). Control of biofilm formation: antibiotics and beyond. Appl. Environ. Microbiol. 83, e02508–e02516. doi: 10.1128/AEM.02508-16
Álvarez-Martínez, F. J., Barrajón-Catalán, E., Micol, V. (2020). Tackling antibiotic resistance with compounds of natural origin: A comprehensive review. Biomedicines 8, 405. doi: 10.3390/biomedicines8100405
Aminov, R. I., Garrigues-Jeanjean, N., Mackie, R. I. (2001). Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67, 2–32. doi: 10.1128/AEM.67.1.22-32.2001
Antunes, P., Machado, J., Sousa, J. C., Peixe, L. (2005). Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49 (2), 836–839. doi: 10.1128/AAC.49.2.836-839.2005
Arabi, H., Pakzad, I., Nasrollahi, A., Hosainzadegan, H., Azizi Jalilian, F., Taherikalani, M., et al. (2015). Sulfonamide resistance genes (sul) M in Extended Spectrum Beta Lactamase (ESBL) and Non-ESBL producing Escherichia coli isolated from Iranian hospitals. Jundishapur J. Microbiol. 8, e19961. doi: 10.5812/jjm.19961v2
Banerjee, M., Parai, D., Chattopadhyay, S., Mukherjee, S. K. (2017). Andrographolide: antibacterial activity against common bacteria of human health concern and possible mechanism of action. Folia Microbiol. (Praha). 62 (3), 237–244. doi: 10.1007/s12223-017-0496-9
Barnhart, M. M., Chapman, M. R. (2006). Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147. doi: 10.1146/annurev.micro.60.080805.142106
Benyoussef, W., Deforet, M., Monmeyran, A., Henry, N. (2022). Flagellar motility during E. coli biofilm formation provides a competitive disadvantage which recedes in the presence of co-colonizers. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.896898
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Boya, B. R., Lee, J. H., Lee, J. (2022). Antibiofilm and antimicrobial activities of chloroindoles against uropathogenic. Escherichia coli. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.872943
Che, Y., Xia, Y., Liu, L., Li, A.-D., Yang, Y., Zhang, T. (2019). Mobile antibiotic resistome in wastewater treatment plants revealed by nanopore metagenomic sequencing. Microbiome 7, 44. doi: 10.1186/s40168-019-0663-0
Chen, J., Li, W., Zhang, J., Qi, W., Li, Y., Chen, S., et al. (2020). Prevalence of antibiotic resistance genes in drinking water and biofilms: The correlation with the microbial community and opportunistic pathogens. Chemosphere 259, 127483. doi: 10.1016/j.chemosphere.2020.127483
Clausen, P. T. L. C., Aarestrup, F. M., Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinf. 19, 307. doi: 10.1186/s12859-018-2336-6
Clinical and Laboratory Standards Institute (2015). Performance Standards for Antimicrobial Susceptibility Testing; 25th Informational Supplement (Wayne, PA: Clinical and Laboratory Standards Institute). CLSI Document M100-S25.
Compagne, N., Vieira Da Cruz, A., Müller, R. T., Hartkoorn, R. C., Flipo, M., Pos, K. M. (2023). Update on the discovery of efflux pump inhibitors against critical priority gram-negative bacteria. Antibiotics 12, 180. doi: 10.3390/antibiotics12010180
Dahshan, H., Chuma, T., Shahada, F., Akiba, M., Fujimoto, H., Akasaka, K., et al. (2010). Characterization of antibiotic resistance and the emergence of AmpC-producing Salmonella Infantis from pigs. J. Vet. Med. Sci. 72, 1437–1442. doi: 10.1292/jvms.10-0186
Davies, D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discovery. 2, 114–122.
De los Santos, E., Laviña, M., Poey, M. E., Part, A. (2021). Strict relationship between class 1 integrons and resistance to sulfamethoxazole in Escherichia coli. Microb. Pathog. 161, 105206. doi: 10.1016/j.micpath.2021.105206
Dong, J., Zhang, L., Liu, Y., Xu, N., Zhou, S., Yang, Q., et al. (2020). Thymol protects channel catfish from Aeromonas hydrophila infection by inhibiting aerolysin expression and biofilm formation. Microorganisms 8, 636. doi: 10.3390/microorganisms8050636
Du, B., Gu, Y., Chen, G., Wang, G., Liu, L. (2020). Flagellar motility mediates early-stage biofilm formation in oligotrophic aquatic environment. Ecotoxicol Environ. Saf. 194, 110340. doi: 10.1016/j.ecoenv.2020.110340
El-Khoury, C., Mansour, E., Yuliandra, Y., Lai, F., Hawkins, B. A., Du, J. J., et al. (2022). The role of adjuvants in overcoming antibacterial resistance due to enzymatic drug modification. RSC Med. Chem. 13, 1276–1299. doi: 10.1039/d2md00263a
Fan, W., Hamilton Webster-Sesay, T. S., Nikolich, M. P., Lindler, L. E. (2007). Multiplex real-time SYBR Green I PCR assay for detection of tetracycline efflux genes of gram-negative bacteria. Mol. Cell Probes. 21, 245–256. doi: 10.1016/j.mcp.2006.12.005
Fernando, S. I. D., Judan Cruz, K. G. (2020). Ethnobotanical biosynthesis of gold nanoparticles and its downregulation of quorum sensing-linked AhyR gene in Aeromonas hydrophila. SN Appl. Sci. 2, 570. doi: 10.1007/s42452-020-2368-1
Gajic, I., Kabic, J., Kekic, D., Jovicevic, M., Milenkovic, M., Mitic Culafic, D., et al. (2022). Antimicrobial susceptibility testing: A comprehensive review of currently used methods. Antibiot. (Basel) 11, 427. doi: 10.3390/antibiotics11040427
Gopu, V., Kothandapani, S., Shetty, P. H. (2015). Quorum quenching activity of Syzygium cumini (L.) skeels and its anthocyanin malvidin against Klebsiella pneumoniae. Microb. Pathog. 79, 61–69. doi: 10.1016/j.micpath.2015.01.010
Guo, X., Zhang, L. Y., Wu, S. C., Xia, F., Fu, Y. X., Wu, Y. L., et al. (2014). Andrographolide interferes quorum sensing to reduce cell damage caused by avian pathogenic Escherichia coli. Vet. Microbiol. 174, 496–503. doi: 10.1016/j.vetmic.2014.09.021
Hall-Stoodley, L., Costerton, J. W., Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Hanna, N., Sun, P., Sun, Q., Li, X., Yang, X., Ji, X., et al. (2018). Presence of antibiotic residues in various environmental compartments of Shandong province in eastern China: its potential for resistance development and ecological and human risk. Environ. Int. 114, 131–142. doi: 10.1016/j.envint.2018.02.003
Heuer, H., Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Hu, Z. Q., Zhao, W. H., Yoda, Y., Asano, N., Hara, Y., Shimamura, T. (2002). Additive, indifferent and antagonistic effects in combinations of epigallocatechin gallate with 12 non-beta-lactam antibiotics against methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 50, 1051–1054. doi: 10.1093/jac/dkf250
Ivankovic, S., Stojkovic, R., Jukic, M., Milos, M., Milos, M., Jurin, M. (2006). The antitumor activity of thymoquinone and thymohydroquinone in vitro and in vivo. Exp. Oncol. 28, 220–224.
Jin, L., Zhang, X., Shi, H., Wang, W., Qiao, Z., Yang, W., et al. (2020). Identification of a novel N-acyl homoserine lactone synthase, AhyI, in Aeromonas hydrophila and structural basis for its substrate specificity. J. Agri Food Chem. 68 (8), 2516–2527. doi: 10.1021/acs.jafc.9b07833
Katz, D. H., Marcelletti, J. F., Khalil, M. H., Pope, L. E., Katz, L. R. (1991). Antiviral activity of 1-docosanol, an inhibitor of lipid-enveloped viruses including herpes simplex. Proc. Natl. Acad. Sci. U.S.A. 88, 10825–10829. doi: 10.1073/pnas.88.23.10825
Khorshidi, A., Zadeh, N. M., Khaledi, A., Moosavi, G. A., Shakerimoghaddam, A., Matinpur, A. (2022). Investigation of class 1 integrons and biofilm formation in multi-drug resistance uropathogenic Escherichia coli isolated from patients with urinary tract infection in Shohadaye Qom Hospital, Iran. IntArch Heal Sci. 9, 47–52. doi: 10.4103/iahs.iahs_163_21
Kirke, D. F., Swift, S., Lynch, M. J., Williams, P. (2004). The Aeromonas hydrophila LuxR homologue AhyR regulates the N-acyl homoserine lactone synthase, AhyI positively and negatively in a growth phase-dependent manner. FEMS Microbiol. Lett. 241, 109–117. doi: 10.1016/j.femsle.2004.10.011
Kovalakova, P., Cizmas, L., McDonald, T. J., Marsalek, B., Feng, M., Sharma, V. K. (2020). Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere. 251, 126351. doi: 10.1016/j.chemosphere.2020.126351
Kozlova, E. V., Khajanchi, B. K., Sha, J., Chopra, A. K. (2011). Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 50, 213–223. doi: 10.1016/j.micpath.2011.01.007
Kumar, G., Tudu, A. K. (2023). Tackling multidrug-resistant Staphylococcus aureus by natural products and their analogues acting as NorA efflux pump inhibitors. Bioorg. Med. Chem. 80, 117187. doi: 10.1016/j.bmc.2023.117187
Kvist, M., Hancock, V., Klemm, P. (2008). Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74, 7376–7382. doi: 10.1128/AEM.01310-08
Laganenka, L., López, M. E., Colin, R., Sourjik, V. (2020). Flagellum-mediated mechanosensing and RflP control motility state of pathogenic Escherichia coli. mBio. 11 (2), 3e02269–19. doi: 10.1128/mbio.02269-19
Lakshmi, S. A., Bhaskar, J. P., Krishnan, V., Sethupathy, S., Pandipriya, S., Aruni, W., et al. (2020). Inhibition of biofilm and biofilm-associated virulence factor production in methicillin-resistant Staphylococcus aureus by docosanol. J. Biotechnol. 317, 59–69. doi: 10.1016/j.jbiotec.2020.04.014
Lakshmi, S. A., Prasath, K. G., Tamilmuhilan, K., Srivathsan, A., Shafreen, R. M. B., Kasthuri, T., et al. (2022). Suppression of thiol-dependent antioxidant system and stress response in methicillin-resistant Staphylococcus aureus by docosanol: explication through proteome investigation. Mol. Biotechnol. 64, 575–589. doi: 10.1007/s12033-021-00434-4
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics. Eds. Stackebraundt, E., Goodfellow, M. (John Wiley & Sons Ltd, New York), 115–175.
Lee, H. J., Storesund, J. E., Lunestad, B. T., Hoel, S., Lerfall, J., Jakobsen, A. N. (2023). Whole genome sequence analysis of Aeromonas spp. isolated from ready-to-eat seafood: antimicrobial resistance and virulence factors. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1175304
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lynch, M. J., Swift, S., Kirke, D. F., Keevil, C. W., Dodd, C. E., Williams, P. (2002). The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4, 18–28.
Matilla-Cuenca, L., Gil, C., Cuesta, S., Rapún-Araiz, B., Žiemytė, M., Mira, A., et al. (2020). Antibiofilm activity of flavonoids on staphylococcal biofilms through targeting BAP amyloids. Sci. Rep. 10 (1), 18968. doi: 10.1038/s41598-020-75929-2
Michaelis, C., Grohmann, E. (2023). Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiot. (Basel). 12, 328. doi: 10.3390/antibiotics12020328
Miller, M. B., Bassler, B. L. (2001). Quorum sensing in bacteria. Ann. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Mishra, R., Panda, A. K., De Mandal, S., Shakeel, M., Bisht, S. S., Khan, J. (2020). Natural anti-biofilm agents: strategies to control biofilm-forming pathogens. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.566325
Murugaiyan, J., Kumar, P. A., Rao, G. S., Iskandar, K., Hawser, S., Hays, J. P., et al. (2022). Progress in alternative strategies to combat antimicrobial resistance: Focus on antibiotics. Antibiotics. 11 (2), 200. doi: 10.3390/antibiotics11020200
Mutuku, C., Gazdag, Z., Melegh, S. (2022). Occurrence of antibiotics and bacterial resistance genes in wastewater: resistance mechanisms and antimicrobial resistance control approaches. World J. Microbiol. Biotechnol. 38, 152. doi: 10.1007/s11274-022-03334-0
Ouyang, J., Sun, F., Feng, W., Sun, Y., Qiu, X., Xiong, L., et al. (2016). Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J. Appl. Microbiol. 120, 966–974. doi: 10.1111/jam.13073
Paczkowski, J. E., Mukherjee, S., McCready, A. R., Cong, J. P., Aquino, C. J., Kim, H., et al. (2017). Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J. Biol. Chem. 292, 4064–4076. doi: 10.1074/jbc.M116.770552
Partridge, S. R., Tsafnat, G., Coiera, E., Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Patel, B., Kumari, S., Banerjee, R., Samanta, M., Das, S. (2017). Disruption of the quorum sensing regulated pathogenic traits of the biofilm-forming fish pathogen Aeromonas hydrophila by tannic acid, a potent quorum quencher. Biofouling 33, 580–590. doi: 10.1080/08927014.2017.1336619
Peng, L. Y., Yuan, M., Cui, Z. Q., Wu, Z. M., Yu, Z. J., Song, K., et al. (2018). Rutin inhibits quorum sensing, biofilm formation and virulence genes in avian pathogenic Escherichia coli. Microb. Pathog. 119, 54–59. doi: 10.1016/j.micpath.2018.04.007
Perveen, S., Pablos, C., Reynolds, K., Stanley, S., Marugán, J. (2023). Growth and prevalence of antibiotic-resistant bacteria in microplastic biofilm from wastewater treatment plant effluents. Sci. Total Environ. 856, 159024. doi: 10.1016/j.scitotenv.2022.159024
Piotrowska, M., Popowska, M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64, 921–934. doi: 10.1007/s13213-014-0911-2
Piotrowska, M., Popowska, M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00494
Piotrowska, M., Przygodzińska, D., Matyjewicz, K., Popowska, M. (2017). Occurrence and variety of β-lactamase genes among Aeromonas spp. isolated from urban wastewater treatment plant. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00863
Pope, L. E., Marcelletti, J. F., Katz, L. R., Lin, J. Y., Katz, D. H., Parish, M. L., et al. (1998). The anti-herpes simplex virus activity of n-docosanol includes inhibition of the viral entry process. Antiviral Res. 40, 85–94. doi: 10.1016/s0166-3542(98)00048-5
Proia, L., von Schiller, D., Sànchez-Melsió, A., Sabater, S., Borrego, C. M., Rodríguez-Mozaz, S., et al. (2016). Occurrence and persistence of antibiotic resistance genes in river biofilms after wastewater inputs in small rivers. Environ. pollut. 210, 121–128. doi: 10.1016/j.envpol.2015.11.035
Qiagen (2023). RNeasy Mini Kit Handbook. Available online at: https://www.qiagen.com/us/resources (Accessed November 15, 2023).
Rather, M., Gupta, K., Mandal, M. (2021). Microbial biofilm: formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 52, 1701–1718. doi: 10.1007/s42770-021-00624-x
Ren, J., Wang, M., Zhou, W., Liu, Z. (2024). Efflux pumps as potential targets for biofilm inhibition. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1315238
Rezaei, A., Oyong, G. G., Borja, V. B., Inoue, M., Abe, T., Tamamura, R., et al. (2011). Molecular screening of anti-quorum sensing capability of Salvadora persica on Enterococcus faecalis. J. Hard Tis Bio. 20 (2), 115–124. doi: 10.2485/jhtb.20.115
Roh, H., Kannimuthu, D. (2023). Comparative resistome analysis of Aeromonas species in aquaculture reveals antibiotic resistance patterns and phylogeographic distribution. Env. Res. 239, 117273. doi: 10.1016/j.envres.2023.117273
Rosengren, L. B., Waldner, C. L., Reid-Smith, R. J. (2009). Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes of fecal Escherichia coli isolates from healthy grow-finish pigs. Appl. Environ. Microbiol. 75 (5), 1373–1380. doi: 10.1128/AEM.01253-08
Roy, P. K., Song, M. G., Jeon, E. B., Kim, S. H., Park, S. Y. (2022). Antibiofilm efficacy of quercetin against Vibrio parahaemolyticus biofilm on food-contact surfaces in the food industry. Microorganisms 10, 1902. doi: 10.3390/microorganisms10101902
Sadowski, L. A., Upadhyay, R., Greeley, Z. W., Margulies, B. J. (2021). Current drugs to treat infections with herpes simplex viruses-1 and -2. Viruses 13, 1228. doi: 10.3390/v13071228
Shrout, J. D., Nerenberg, R. (2012). Monitoring bacterial twitter: does quorum sensing determine the behavior of water and wastewater treatment biofilms? Environ. Sci. Technol. 46, 1995–2005. doi: 10.1021/es203933h
Shunmugaperumal, T. (2010). Biofilm eradication and prevention: a pharmaceutical approach to medical device infections (Hoboken NJ: John Wiley & Sons).
Skwor, T., Stringer, S., Haggerty, J., Johnson, J., Duhr, S., Johnson, M., et al. (2020). Prevalence of potentially pathogenic antibiotic-resistant Aeromonas spp. in treated urban wastewater effluents versus recipient riverine populations: a 3-year comparative study. Appl. Environ. Microbiol. 86, e02053–e02019. doi: 10.1128/AEM.02053-19
Soto, S. M. (2013). Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4, 223–229. doi: 10.4161/viru.23724
Sun, B., Luo, H., Jiang, H., Wang, Z., Jia, A. (2021). Inhibition of quorum sensing and biofilm formation of esculetin on Aeromonas Hydrophila. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.737626
Taiswa, A., Andriolo, J. M., Hailer, M. K., Skinner, J. K. (2024). Electrospun controlled release anti-quorum sensing filter for biofouling prevention in MCE membranes. Sep Pur Tech. 332, 125874. doi: 10.1016/j.seppur.2023.125874
Talagrand-Reboul, E., Jumas-Bilak, E., Lamy, B. (2017). The social life of Aeromonas through biofilm and quorum sensing systems. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00037
Tesarova, H., Svobodova, B., Kokoska, L., Marsik, P., Pribylova, M., Landa, P., et al. (2011). Determination of oxygen radical absorbance capacity of black cumin (Nigella sativa) seed quinone compounds. Nat. ProdComm 6 (2), 213–216. doi: 10.1177/1934578X1100600214
Uluseker, C., Kaster, K. M., Thorsen, K., Basiry, D., Shobana, S., Jain, M., et al. (2021). A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: mechanisms and perspectives. Front. Microbiol. 12, 717809. doi: 10.3389/fmicb.2021.717809
Umaru, I. J., Ahmad, F. B., Aduwamai, U. H. (2019). Extraction, elucidation, characterization and evaluation of antibacterial activity of four pure compounds from Barringtonia racemosa leaf extract. World Jour Pharm. Pharma Sci. 8, 184–223. doi: 10.20959/wjpps20198-14476
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
Usui, M., Tagaki, C., Fukuda, A., Okubo, T., Boonla, C., Suzuki, S., et al. (2016). Use of Aeromonas spp. as general indicators of antimicrobial susceptibility among bacteria in aquatic environments in Thailand. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00710
Van Bambeke, F., Pagès, J. M., Lee, V. J. (2006). Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent Pat. Antiinfect. Drug Discovery 1, 157–175. doi: 10.2174/157489106777452692
Venkatesan, M., Fruci, M., Verellen, L. A., Skarina, T., Mesa, N., Flick, R., et al. (2023). Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics. Nat. Commun. 4, 4031. doi: 10.1038/s41467-023-39778-7
Wang, Z., Ding, Z., Li, Z., Ding, Y., Jiang, F., Liu, J. (2021). Antioxidant and antibacterial study of 10 flavonoids revealed rutin as a potential antibiofilm agent in Klebsiella pneumoniae strains isolated from hospitalized patients. Microb. Pathog. 159, 105121. doi: 10.1016/j.micpath.2021.105121
World Health Organization (2023). Antimicrobial resistance. Available online at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed February 15, 2024).
Zhang, L., Bao, M., Liu, B., Zhao, H., Zhang, Y., Ji, X., et al. (2020). Effect of andrographolide and its analogs on bacterial infection: a review. Pharmacology 105, 123–134. doi: 10.1159/000503410
Yu, T., Jiang, X., Xu, X., Jiang, C., Kang, R., Jiang, X. (2022). Andrographolide inhibits biofilm and virulence in Listeria monocytogenes as a quorum-sensing inhibitor. Molecules. 27 (10), 3234. doi: 10.3390/molecules27103234
Keywords: ARGS, antimicrobial resistance, biofilm, natural compounds, Aeromonas, wastewater
Citation: Judan Cruz KG, Takumi O, Bongulto KA, Gandalera EE, Kagia N and Watanabe K (2024) Natural compound-induced downregulation of antimicrobial resistance and biofilm-linked genes in wastewater Aeromonas species. Front. Cell. Infect. Microbiol. 14:1456700. doi: 10.3389/fcimb.2024.1456700
Received: 29 June 2024; Accepted: 18 September 2024;
Published: 14 October 2024.
Edited by:
Maria Gabriela Paraje, National University of Cordoba, ArgentinaReviewed by:
Luisa Jordao, National Health Institute Doutor Ricardo Jorge (INSA), PortugalMauro Nicolas Gallucci, National Scientific and Technical Research Council (CONICET), Argentina
Graciela Pucci, National University of Patagonia San Juan Bosco, Argentina
Copyright © 2024 Judan Cruz, Takumi, Bongulto, Gandalera, Kagia and Watanabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khristina G. Judan Cruz, a2pjcnV6QGNsc3UuZWR1LnBo; Kozo Watanabe, d2F0YW5hYmUua296by5takBlaGltZS11LmFjLmpw
 Khristina G. Judan Cruz
Khristina G. Judan Cruz Okamoto Takumi1
Okamoto Takumi1 Emmanuel E. Gandalera
Emmanuel E. Gandalera Kozo Watanabe
Kozo Watanabe