
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 September 2024
Sec. Molecular Bacterial Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1454373
Hypervirulent K. pneumoniae (hvKp) have emerged as clinically important pathogens, posing a serious threat to human health. RfaH, a transcriptional elongation factor, has been regarded as implicated in facilitating the transcription of long virulence operons in certain bacterial species. In K. pneumoniae, RfaH plays a vital role in promoting CPS synthesis and hypermucoviscosity, as well as mediating bacterial fitness during lung infection. In this study, we aim to conduct a systematic investigation of the roles of rfaH in the survival, dissemination, and colonization of hvKp through in vitro and in vivo assays. We found that bacterial cells and colonies displayed capsule -deficient phenotypes subsequent to the deletion of rfaH in K. pneumoniae NTUH-K2044. We confirmed that rfaH is required for the synthesis of capsule and lipopolysaccharide (LPS) by positively regulating the expression of CPS and LPS gene clusters. We found that the ΔrfaH mutant led to a significantly decreased mortality of K. pneumoniae in a mouse intraperitoneal infection model. We further demonstrated that the absence of rfaH was associated with slower bacterial growth under conditions of low nutrition or iron limitation. ΔrfaH displayed reduced survival rates in the presence of human serum. Besides, the engulfment of the ΔrfaH mutant was significantly higher than that of NTUH-K2044 by macrophages in vivo, indicating an indispensable role of RfaH in the phagocytosis resistance of hvKp in mice. Both mouse intranasal and intraperitoneal infection models revealed a higher bacterial clearance rate of ΔrfaH in lungs, livers, and spleens of mice compared to its wild type, suggesting an important role of RfaH in the bacterial survival, dissemination, and colonization of hvKp in vivo. Histopathological results supported that RfaH contributes to the pathogenicity of hvKp in mice. In conclusion, our study demonstrates crucial roles of RfaH in the survival, colonization and full virulence of hvKp, which provides several implications for the development of RfaH as an antibacterial target.
Klebsiella pneumoniae is a Gram-negative strain that frequently causes pulmonary infection, urinary tract infections, bacteremia, and liver abscesses (Paczosa and Mecsas, 2016). Hypervirulent K. pneumoniae (hvKP) is associated with severe infections and exhibits an extremely high invasive and transmissible ability, causing higher morbidity and mortality for infected patients (Thomas A. Russo, 2019). hvKp strains may evolve into highly drug-resistant hvKp by acquiring various drug-resistant plasmids (Tian et al., 2022). Especially in recent years, the emergence of multi-resistant or even pan-resistant K. pneumoniae has significantly increased the risk of conversion of hvKp to highly drug-resistant hvKp, such as carbapenems-resistant hvKp isolates (Zhang et al., 2020; Yang et al., 2022), which poses a formidable challenge to human health globally.
RfaH is an antiterminator that directly binds to RNA polymerase (RNAP) and modifies it into a processive, pause-resistant state, thus reducing transcriptional pausing at certain positions and increasing RNAP processivity (Svetlov et al., 2007; Burmann et al., 2012). It specifically promotes the transcription of long pathogenicity operons that encode extracytoplasmic components in bacteria, such as adhesins, capsular polysaccharides, and toxins (Artsimovitch and Landick, 2002; Nagy et al., 2002). RfaH was initially revealed as a regulator of lipopolysaccharide (LPS) synthesis in Salmonella (Lindberg and Hellerqvist, 1980) and Escherichia coli (Creeger et al., 1984). It was also identified to regulate the F plasmid (Sanderson and Stocker, 1981), various types of the capsule (Clarke et al., 1999), hemin receptor (Nagy et al., 2002), and the α-hemolysin in E.coli (Stevens et al., 1994).
Numerous genetic factors contribute to the pathogenicity and virulence of K. pneumoniae strains, such as variable capsular polysaccharide (CPS) and LPS, siderophore, enterobactin, and fimbriae, are basic requirements for establishing opportunistic infections (Wyres et al., 2020). Although a hypermucoviscosity (HMV) colony phenotype is not pathognomonic for hvKp, it has been strongly associated with hvKp strains (Walker and Miller, 2020). Previous studies hinted that RfaH is required for the CPS synthesis and HMV (Mike et al., 2021), and is critical for K. pneumoniae fitness in the lung, by functioning in resisting complement-mediated serum killing, and for maximal growth in serum (Bachman et al., 2015; Short et al., 2020). However, a research gap exists regarding the regulatory roles of RfaH in the pathogenicity and virulence of K. pneumoniae, especially in hvKp.
In this study, we showed that the deletion of rfaH caused significant attenuation of virulence in K. pneumoniae NTUH-K2044, a reference strain of serotype K1 that is highly virulent and hypermucoviscous (Wu et al., 2009). We demonstrate that RfaH contributes to pathogenicity by facilitating bacterial growth in low nutrition and iron-limited conditions, and by regulating bacterial evasion of complement-mediated killing and phagocytosis by macrophages. Especially, we further clarified that RfaH plays critical roles in helping bacterial dissemination, colonization, and full virulence of K. pneumoniae by two different mouse infection models.
Bacterial strains and plasmids used in this study are listed in Table 1. The K. pneumoniae NTUH-K2044, a strain that was isolated from the blood of a patient with community-acquired primary liver abscess and metastatic meningitis, was kindly gifted by professor Jin Town Wang of the National Taiwan University Hospital, Taipei, Taiwan. All the test strains were cultured in Luria-Bertani (LB) broth with 220 rpm shaking or on LB agar plates at 37°C.
The primers for construction of mutant and complementation strains are listed in Supplementary Table S1. To delete rfaH, a ~1kb DNA fragment of the upstream and downstream regions flanking rfaH were PCR amplified from the chromosome of NTUH-K2044, and ligated into the suicide vector pKO3-Km that was cut with NotI (New England Biolabs) using the ClonExpress II One Step Cloning Kit (Vazyme, China). The resulting recombinant plasmid pKO3-km-rfaH L/R was transformed into NTUH-K2044 by electroporation and allowed to recover for 1 h at 30°C. The chromosomal integrates were selected on LB plates containing 50 μg/ml kanamycin after incubation at 43°C for ~16h. After the first round of DNA allelic exchange, the whole recombinant plasmid was expected to have incorporated into the chromosome. To select cells in which the rfaH gene was excised and the plasmid was lost, colonies from the first round DNA exchange event were diluted and simultaneously plated on LB plates containing 10% sucrose and those containing 50 μg/ml kanamycin, respectively. After incubation at 30°C for ~24h, kanamycin-sensitive colonies were picked and the deletion of rfaH was confirmed by PCR and DNA Sanger sequencing using gene specific and genome flanking primers. Comparative analysis of sequencing results was performed by BLASTn. The resulting mutant was denoted as ΔrfaH. For complementation, the intact rfaH gene containing its native promoter was amplified by PCR and cloned into the intergenic region between open reading frames (ORFs), pgpA and yajO, using a pKO3-Km pgpA-yajO recombinant vector (Hsieh et al., 2010). The recombinant plasmid pKO3-km-pgpA-yajO-rfaH was transformed into ΔrfaH by electroporation. After two rounds of homologous recombination, the chromosomal integration of rfaH was selected by LB plates containing 50 μg/ml kanamycin and those containing 10% sucrose, respectively. After PCR analysis and Sanger sequencing, the resulting complemented strain was named ΔrfaH-comp.
NTUH-K2044, ΔrfaH and ΔrfaH-comp were cultured overnight at 37°C. For LB growth experiment, a fresh culture was inoculated at 1:100 from the overnight culture and grown at 37°C for 36 h in 96-well plates. For M9 minimal medium growth experiment, overnight culture of the tested strains were sub-cultured at 1:100 to M9 broth and grown at 37°C for 18 h in 96-well plates. For the iron limitation growth experiment, the tested strains were sub-cultured at 1:100 in LB broth containing 100, 200, 400, or 800 μM 2,2’-dipyridine (Sangon, Shanghai, China), respectively, and grown at 37°C for 12 h in 96-well plates. The bacterial growth status was automatically monitored by Synergy H1 Hybrid Multi-Mode Reader (BioTek, America) at optical density 600 nm (OD600 nm).
In vitro stress assays were performed as described previously with slight modifications (Hsieh et al., 2010; Srinivasan et al., 2012; Bulger et al., 2017). NTUH-K2044, ΔrfaH and ΔrfaH-comp were grown to mid-exponential phase (OD600 nm reaches 0.6-0.8). Cells were pelleted by centrifuging at 10,000 × g for 5 min and resuspended in LB broth. For the oxidative stress assays, cells were treated with or without 0.5 mM H2O2 for 30 min. For the temperature stress assays, cells were incubated at 40°C, 50°C, and 60°C for 20 min, respectively. For the acid stress assays, cells were resuspended in LB broth at pH 5.5 or 7.4 for 1 h, followed by centrifugation and resuspension in LB broth at pH 3.0 for 1 h. Cultures of these stress assays were diluted serially and spread onto LB plates for colony counting. For the osmotic stress assays, pellet cells were diluted serially and spread on LB plates with or without 0.3 M potassium chloride. The results are expressed as the ratio of the number of colony-forming units (CFUs) obtained after stress treatment to the number of CFUs obtained from untreated cultures. All these experiments were independently performed three times.
The string test was carried out by stretching a mucoviscous string from the colony grown on a blood agar plate using a sterile loop as previously described (Fang et al., 2004). The viscous string >5 mm was identified as a positive result. The sedimentation assay was performed to further measure the levels of HMV as described previously (Palacios et al., 2018). Briefly, the tested strains cultured in LB broth (OD600 nm ~1.0) were centrifuged for 5 min at 1,000 × g, respectively. The supernatant was gently removed without disturbing the pellet for OD600 measurement.
NTUH-K2044 and ΔrfaH were cultured overnight, and then sub-cultured at 1:100 to 15 ml fresh LB broth for 6 h, centrifuged at 1,0000 × g for 5 min, and filled with 3% glutaraldehyde fixative. The samples were observed under the transmission electron microscope JEM-1400FLASH (Japan Electronics, Japan).
Capsule was extracted as previously described (Rosen et al., 2016; Walker et al., 2019). NTUH-K2044, ΔrfaH, and ΔrfaH-comp were cultured overnight, and uronic acid (UA) was extracted from 500 μl culture with Zwittergent 3-14 in 100 mM citric acid. The culture was then precipitated with absolute ethanol and resuspended in tetraborate/sulfuric acid. Following the addition of 3-hydroxy diphenol in 0.5% NaOH, the amount of uronic acid was determined by measuring absorbance at 520 nm. A standard curve was generated with glucuronic acid.
LPS was extracted by the hot-phenol water method with minor modifications (Lee and Inzana, 2021). Briefly, NTUH-K2044, ΔrfaH and ΔrfaH-comp were cultured overnight, followed by washing with PBS and resuspension in ddH2O. Following ultrasonication, an equal volume of pre-warmed 90% (w/v) phenol solution was added to the bacterial suspension. Stir the mixture vigorously at 68°C for 30 min, and collect the supernatant by centrifugation at 10,000 × g for 10 min. To precipitate the LPS, one-tenth volume of 5 M NaCl, and 5-10 volumes of cold (-20°C) 95% ethanol were added, and the mixture was kept at -20°C overnight. After centrifugation at 2,000 × g for 10 min, suspending the opaque pellet in 1mL of distilled water, followed by digestion with proteinase K at 59°C for 3h. The resulting LPS sample was stored at -20°C. The phenol sulfate method was used for LPS quantification. 1 ml of LPS sample was added with 1 ml ddH2O and 6% phenol, and 5 ml concentrated sulfuric acid. The amount of LPS was detected by measuring absorbance at 490 nm and determined from the standard curve. The LPS was applied to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were stained using the silver staining method.
The biofilm formation assays were performed as previously described (Li et al., 2016). Briefly, overnight culture of the test strains was diluted at 1:100 into fresh LB and inoculated statically in sterile 96-well microtiter plates at 37°C for 12 h. The cultures were removed and the wells were washed by immersing them in water, followed by fixing with methanol and staining with crystal violet (0.1%, w/v) per well. After that, crystal violet was removed from wells and washed with ddH2O. The bound dye was released by adding 100 µl of acetic acid (33%, v/v) and the optical density of dissolved material was determined by measuring absorbance at 595 nm.
The serum bactericidal assay was performed as described previously with minor modifications (Podschun et al., 1993). Briefly, NTUH-K2044, ΔrfaH and ΔrfaH-comp were grown to mid-exponential phase. We mixed 25 μL of each bacterial strain with 75 μL fresh or heat-inactivated (56°C for 30 min) human serum with informed consent from healthy volunteers. The serum-strain mixture was incubated at 37°C for 1h without shaking, and CFUs were enumerated by serial dilution and plating on LB agar plates. The survival percentage in serum was calculated by the ratio of the number of colonies in the fresh serum treatment group to the number of colonies in the heat-inactivated serum treatment group.
2 ml of NTUH-K2044 and ΔrfaH were grown to an OD600 of 0.6-0.8. Total RNA extraction and reverse transcription of RNA were carried out using the MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan, and the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Japan), respectively, according to the manufacturer’s instructions. The primers for qRT-PCR are listed in Supplementary Table S2. qRT-PCR amplification was performed as follows: pre-denaturation (95°C for 30 s), cyclic reactions (40 cycles of 95°C for 10 s, 60°C for 30 s), and melting curve (95°C for 15 s, 60°C for 1 min, 95°C for 15 s). qRT-PCR reactions were conducted using the LineGene 9600 Plus PCR Detection System (BIOER, China). All experiments were repeated for three times, independently, and were performed in triplicate. K. pneumoniae 16s rRNA was used as a reference gene for normalization. Data were analyzed using the 2-ΔΔCt method (Schmittgen and Livak, 2008).
Animal experiments were approved by the Southwest Medical University Institutional Animal Care and Use Committee (Project license: swmu20220089). Four- to six-week-old BALB/c mice (SPF Biotechnology Co., Ltd, Beijing, China) were used in this study. Before and following inoculation, mice had unlimited access to food and water.
To detect whether the virulence of ΔrfaH was attenuated, we used a mouse intraperitoneal infection model as previously described with minor modifications (Russo et al., 2021). Briefly, 10 mice were randomly selected and were intraperitoneally inoculated with 100 μl bacterial suspension (~ 1 × 105 CFU) of NTUH-K2044 or ΔrfaH. Three mice injected with 100 μl sterile PBS alone were used as a negative control. The mental state and death of the mice were monitored at 12-h intervals for 7 days.
For the phagocytosis assays, mice (five per group) were lightly anesthetized with isoflurane and then inhaled with 50 μl of bacterial suspension (~ 1 × 108 CFU) via the intranasal route. At 90 min after inoculation, the trachea of mice were dissected and intubated with the catheter (38mm length, 20 gauge), and lavaged with 1 ml PBS to collect alveolar phagocytes 3 times. Suspensions were treated with 50 μg/ml of gentamicin at 37°C for 1 h, followed by centrifugation at 4000 × g for 5 min and resuspension in 1 ml PBS. The phagocytes were lysed using 1% TritonX-100 solution, vortexed for 15 s, serially diluted, and plated for bacterial enumeration.
In the intranasal infection model, mice were intranasally infected with NTUH-K2044 or ΔrfaH as previously described with minor modifications (Sharma et al., 2017; Palacios et al., 2018). Briefly, mice (twelve per group) were lightly anesthetized with isoflurane, followed by inhalation with 50 μl of bacterial suspension (~ 1 × 105 CFU). The control mice were inhaled with 50 μl of PBS. Inoculated mice were monitored daily for two days, and their activity, breath rate, posture, diarrhea, hair, eyes, nose, and weight were evaluated as health-scoring criteria. For the organ burden assays, four mice were euthanized with pentobarbital sodium at a dose of 150 mg/kg at the indicated time points (6 h, 24 h, and 48 h). After that, the lungs, livers, and spleens of mice in each group were removed, weighed under sterile conditions, and homogenized in 1 ml PBS, followed by serial dilution for the enumeration of CFU on LB agar. The number of CFU detected in the organs was standardized per 0.1 g of wet organ weight. For histopathology, mice (three per group) were inhaled with 50 μl of bacterial suspension (~ 1 × 105 CFU), and the control mice were inhaled with 50 μl of PBS. Inoculated mice were monitored and euthanized by injection of pentobarbital sodium after 24 h post-infection. Lungs, livers, and spleens of mice were removed and fixed with 10 volumes of 10% neutral-buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin-eosin (HE).
For the organ burden assay in the intraperitoneal infection model, mice (twelve per group) were injected with 100 μl of bacterial suspension (~ 1 × 104 CFU), with the control mice being injected with 100 μl of PBS. Inoculated mice were monitored daily for two days, and their activity, breath rate, posture, diarrhea, hair, eyes, nose and weight were evaluated as health-scoring criteria. Four mice were euthanized with pentobarbital sodium at the indicated time points (6 h, 24 h, and 48 h post-infection). For the organ burden assays, the lungs, livers, and spleens of mice in each group were removed, weighed, homogenized, serially diluted, and plated on LB agar for colony counts as described above. For histopathology, mice (three per group) were inhaled with 50 μl of bacterial suspension (~ 1 × 104 CFU). Lungs, livers, and spleens of mice were removed at 24 h post-infection, embedded in paraffin, and stained with HE.
Data were presented as means ± standard deviation (SD) and were analyzed with GraphPad 9.0 software. Two tailed unpaired student’s t-test was employed to determine the difference between the two groups. A two-tailed Mann-Whitney U test was used for data analysis in the organ burden assays. A log-rank (Mantel-Cox) test was used for the analysis of the mouse survival curve. The statistical significance was set to *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
RfaH protein of K. pneumoniae NTUH-K2044 (162aa) encoded by the 489-bp rfaH gene, exhibited 99.38%, 86.5%, 83.95%, and 62.11% identity to RfaH in Klebsiella quasipneumoniae (Accession no. WP_131081528), E. coli (MWO99696), Salmonella enterica (EEO3944103), and Yersinia pestis (WP_016594782), respectively, by BLASTp analysis (Figure 1A). To determine the roles of RfaH, a rfaH deletion mutant was created by homologous recombination using the suicide plasmid pKO3-km in the wild-type K. pneumoniae NTUH-K2044. The genetic complementation of rfaH was done by integrating an intact rfaH gene containing its native promoter into the intergenic region between pgpA and yajO on the chromosome of ΔrfaH. PCR followed by DNA sequencing was performed to confirm the deletion and complementation of rfaH.
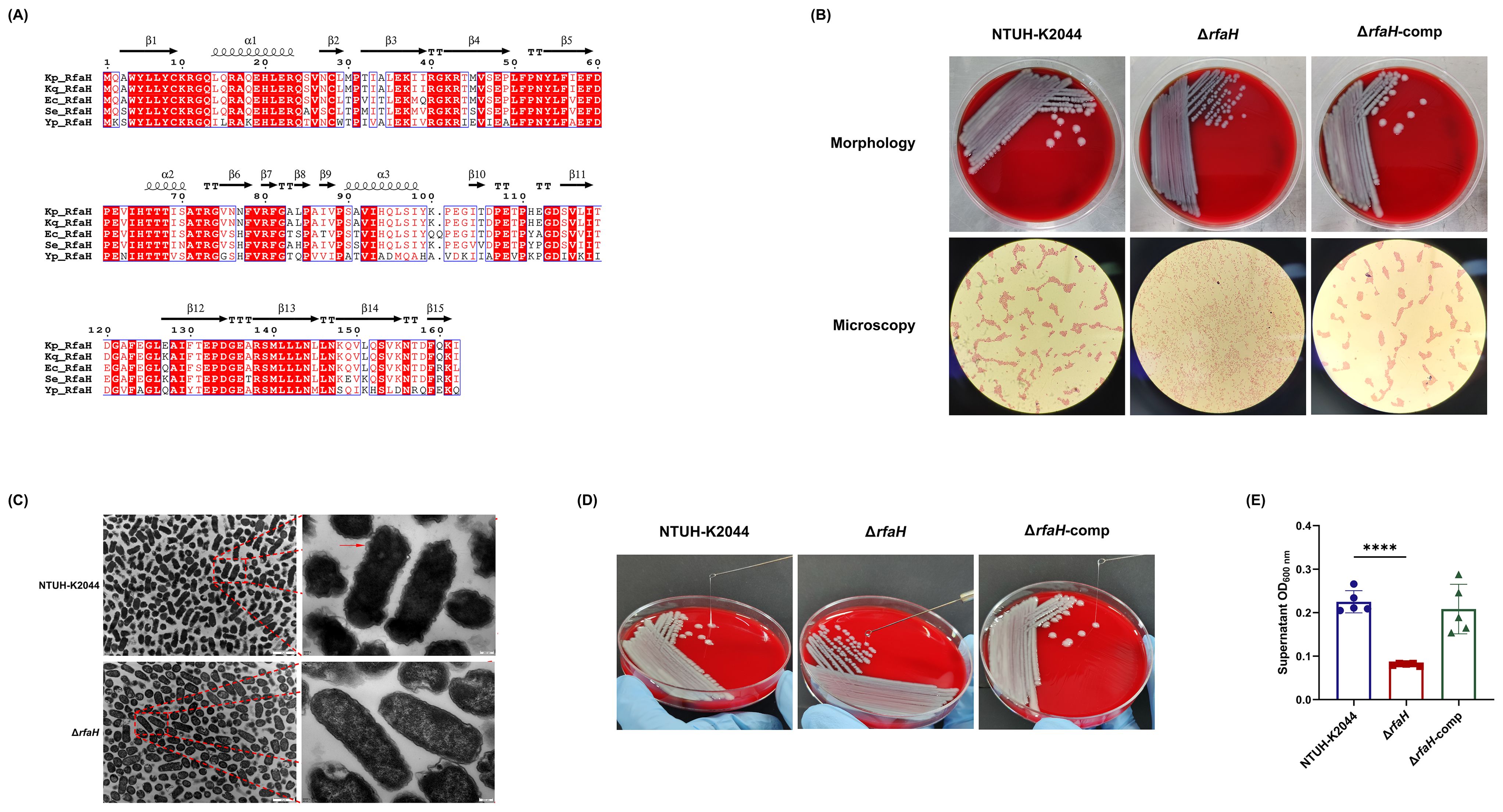
Figure 1. Construction and morphology characteristics of ΔrfaH. (A) Alignment of amino acid sequences of RfaH proteins from Klebsiella quasipneumoniae (Accession no. WP_131081528), Escherichia coli (MWO99696), Salmonella enterica (EEO3944103), and Yersinia pestis (WP_016594782). Analysis was performed using the ESPript 3.x (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Secondary structures of Kp_RfaH predicted by SWISS-MODEL are indicated by coils for -helices and arrows for -strands. The background of identical residues is marked in red, and homologous residues are highlighted in red. (B) Representative colony phenotypes on the blood agar plates and the optical microscope examination (Gram-staining, 100×10) of NTUH-K2044, ΔrfaH, and ΔrfaH-comp. (C) Transmission electron microscope images of NTUH-K2044 and ΔrfaH. The red arrow indicates the capsule. (D) The string test. The viscous string of NTUH-K2044 and ΔrfaH-comp were positive, measuring >5 mm; the viscous string of ΔrfaH was negative, measuring <5 mm. (E) HMV assay by low-speed centrifugation. Data represent the means of at least three independent experiments, and error bars represent the standard error of the mean. Student’s t-test was performed for statistical analyses, ****P < 0.0001.
A distinct colony morphology was observed between NTUH-K2044 and ΔrfaH: the colony of NTUH-K2044 was large (3.2 ± 0.17 mm on average), raised, moist, and mucus, while ΔrfaH appeared as a non-mucus, smaller (1.83 ± 0.06 mm on average), colorless, and flat colonies (Figure 1B). ΔrfaH-comp had a similar colony morphology to that of the wild-type strain. Visualization of cells under optical microscope showed that NTUH-K2044 and ΔrfaH-comp cells formed distinct aggregates, while ΔrfaH cells displayed a dispersed arrangement (Figure 1B). Ultrastructural examination by TEM revealed that NTUH-K2044 was rough, with thick extracellular polysaccharides on the surface, whereas the ΔrfaH mutant was smooth, lacking of the capsule on the surface (Figure 1C). CPS production and HMV are closely related (Mike et al., 2021). The viscous string from the colonies of ΔrfaH was 1.33 ± 0.58 mm in length, indicating a negative string test result, while NTUH-K2044 generated 22.0 ± 4.0 mm string (Figure 1D). Sedimentation assay confirmed that ΔrfaH has reduced HMV, as the supernatant of NTUH-K2044 had an OD600 of approximately 0.225 ± 0.023 after centrifugation, while that of the ΔrfaH appeared clearer, measuring around 0.081 ± 0.0024 (ΔrfaH vs NTUH-K2044, P<0.0001) (Figure 1E). Complementation of rfaH restored HMV. These results suggest that RfaH is required for maintaining the hypercapsule morphology of bacterial cells and colonies, and is linked to the HMV phenotype.
The morphology changes of bacterial cells and colonies are most likely attributed to the impaired CPS biosynthesis in ΔrfaH (Mike et al., 2021). In this study, we confirmed that ΔrfaH displayed diminished UA content, measuring at 54.92 ± 0.95 μg/109 CFU, while NTUH-K2044 (88.91 ± 2.81 μg/109 CFU, ΔrfaH vs NTUH-K2044, P=0.0012) and ΔrfaH-comp (91.09 ± 9.02 μg/109 CFU, ΔrfaH vs ΔrfaH-comp, P=0.0102) produced approximately 30-40% more UA (Figure 2A), confirming an essential role of rfaH in the CPS production of K. pneumoniae. We further assessed the transcription levels of capsule synthesis genes wzi and manC (Ares et al., 2016). Among them, the expressions of wzi and manC exhibited 58.7% (P=0.013) and 64.3% (P=0.0053) reduction, respectively, in ΔrfaH when compared to the wild-type strain (Figure 2B). These results suggest that RfaH promotes CPS production by positively regulating the expression of cps gene cluster containing wzi and manC. It has been shown that RcsA could activate cps gene expression in numerous strains (Su et al., 2018; Walker and Miller, 2020). While, we found no significant difference in the expression levels of rcsA between ΔrfaH and its parent strain (P=0.9313, Figure 2B).
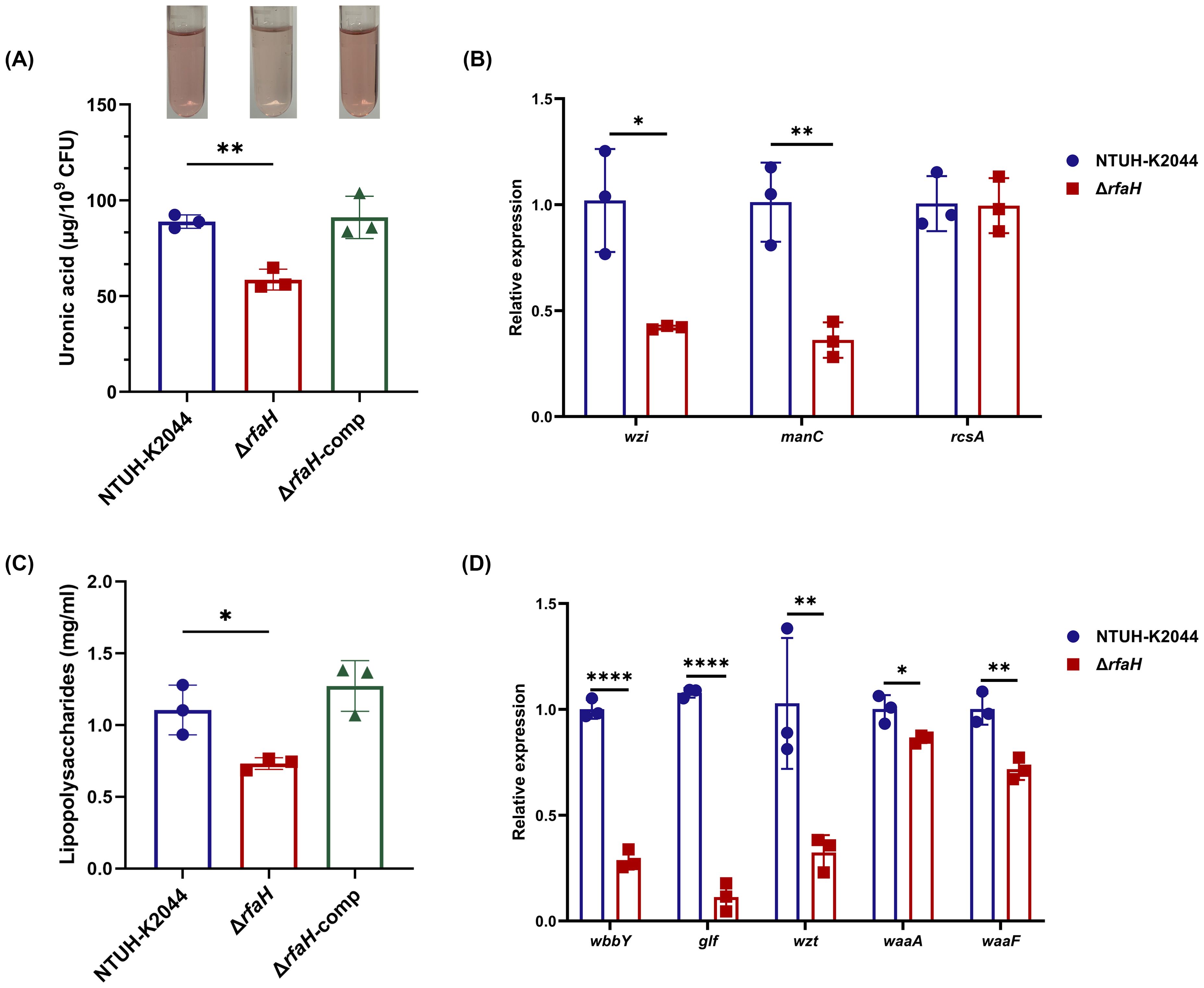
Figure 2. ΔrfaH produces less CPS and LPS. (A) Quantification of the capsule by the uronic acid assay. A representative result of this experiment is shown above the bar chart. (B) The transcription levels of genes associated with capsule production (wzi, manC, and rcsA) in NTUH-K2044 and ΔrfaH. (C) Quantification of LPS with phenol-sulfuric acid method; (D) The transcription levels of genes associated with LPS O-antigen (wbbY, glf, and wzt), and core lipopolysaccharide (waaA, and waaF) in NTUH-K2044 and ΔrfaH. Transcript abundance was measured using the 2-ΔΔCt method with 16s rRNA as a reference gene, and was normalized to the NTUH-K2044. Data represent the mean of at least three independent experiments (in triplicate), and error bars represent the standard error of the mean. P values were calculated using student’s t-tests for statistical analyses, *P < 0.05, **P < 0.01, ****P<0.0001.
It has been shown that RfaH enhances the transcription of a select group of operons controlling bacterial surface features, including LPS. Therefore, we quantified the LPS and found that its content in ΔrfaH (0.732 ± 0.034 mg/ml) was significantly lower than that of NTUH-K2044 (1.105 ± 0.141 mg/ml, P=0.0219) (Figure 2C). Complementation of rfaH restored the LPS to the wild-type level (1.273 ± 0.144 mg/ml, ΔrfaH vs ΔrfaH-comp, P=0.0067). Additionally, SDS-PAGE silver staining also revealed a lower level of LPS in ΔrfaH when compared to NTUH-K2044 and ΔrfaH-comp (Supplementary Figure S1). qRT-PCR showed that the transcription levels of O-antigen genes wbbY, glf, and wzt were reduced by 71.3% (P<0.0001), 89.5% (P<0.0001), and 68.5% (P=0.0029), respectively, in ΔrfaH compared to the wild-type strain. Meanwhile, the expression of core lipopolysaccharide genes waaA and waaF also showed 14.06% and 28.36% reduction (ΔrfaH vs NTUH-K2044, P=0.0239, and P=0.0054, respectively) (Figure 2D), confirming the positive regulation role of rfaH in the expression of O-antigen biosynthesis enzymes and core lipopolysaccharide. These results indicate that RfaH promotes the synthesis of LPS in K. pneumonia, as reported in E. coli and Yersinia sp (Singh et al., 2014; Hoffman et al., 2017).
To investigate whether the deletion of rfaH would affect the virulence of K. pneumoniae NTUH-K2044, we intraperitoneally inoculated BALB/c mice with ~1×105 CFU of ΔrfaH or NTUH-K2044 and monitored survival. After being infected with NTUH-K2044, 20% mice died within 24 h, 90% died within 48 h, and 100% died at 4.5 days. While all mice survived the injection of ΔrfaH over 7 days (Figure 3). These data show a significant attenuation of virulence after the deletion of rfaH (P < 0.0001), supporting the role for RfaH in contributing to the virulence of K. pneumoniae.
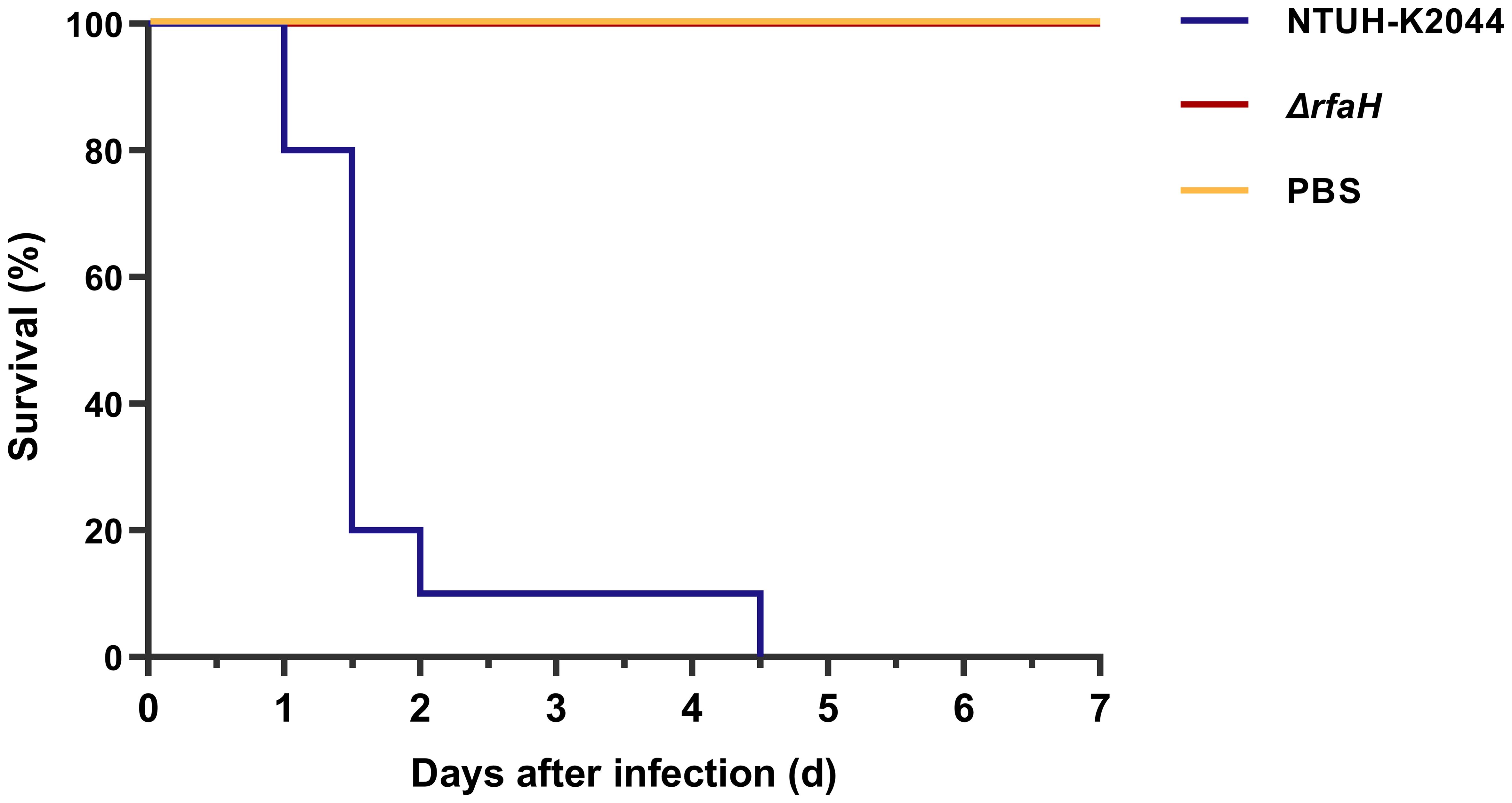
Figure 3. ΔrfaH is significantly attenuated in virulence. Survival curves of mice intraperitoneally inoculated with 1 × 105 CFU of NTUH-K2044 or ΔrfaH show that the survival rate of ΔrfaH-infected mice was significantly enhanced (P < 0.0001, by a log-rank [Mantel-Cox] test).
Despite the obvious difference in colony morphology, ΔrfaH and NTUH-K2044 had similar growth ability in LB broth, a rich laboratory medium (Figure 4A). To investigate the involvement of rfaH in bacterial growth under nutrition-limited conditions, the growth kinetics of NTUH-K2044, ΔrfaH, and ΔrfaH-comp were compared. In the low-nutrition M9 broth, ΔrfaH exhibited significantly slower growth since 4 h when compared to NTUH-K2044 (P<0.0001), and the maximum bacterial concentration in the stationary phase of ΔrfaH was significantly lower than that of NTUH-K2044 (P<0.0001) (Figure 4B). In the iron-chelated LB broth, ΔrfaH and NTUH-K2044 exhibited similar stunted growth in the presence of 100 μM 2,2’-bipyridine (Figure 4C). While, as the concentration of 2,2’-bipyridine increased to 200, 400, or 800 μM, ΔrfaH displayed significantly decreased growth ability compared to the wild-type strain (overall, P<0.05, Figure 4C). Complementation with rfaH restored growth to a level resembling that of the wild-type parent. These results suggest that the deletion of rfaH leads to an impairment of overall growth kinetics under low-nutrient or iron-restricted conditions.
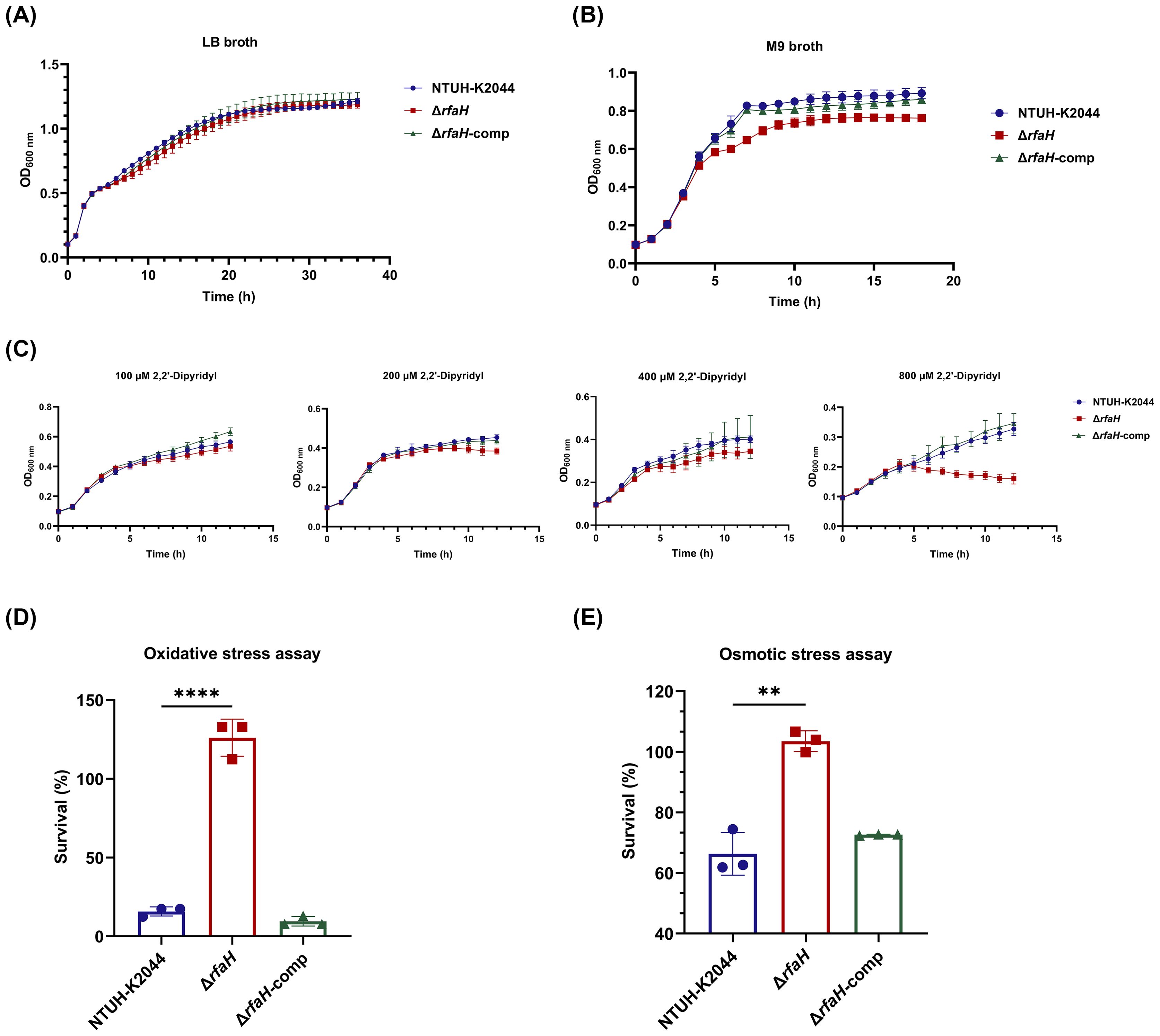
Figure 4. Growth ability and stress resistance assays. (A) Growth curves of NTUH-K2044, ΔrfaH, and ΔrfaH-comp in LB broth. The growth ability of ΔrfaH was comparable to the wild-type strain. (B) Growth curves of NTUH-K2044, ΔrfaH, and ΔrfaH-comp in M9 broth. ΔrfaH exhibited significantly slower growth since 4 h when compared to NTUH-K2044 (P<0.0001), and the maximum bacterial concentration in the stationary phase of ΔrfaH was significantly lower than that of NTUH-K2044 (P<0.0001). (C) Growth curves of NTUH-K2044, ΔrfaH and ΔrfaH-comp in LB broth in the presence of 100 μM, 200 μM, 400 μM or 800 μM 2,2’- dipyridine. ΔrfaH exhibited similar growth ability to NTUH-K2044 in the presence of 100 μM 2,2’-dipyridine, while a significant decreased growth of ΔrfaH was observed in the presence of 200 μM, 400 μM or 800 μM (Overall, P<0.05). (D) The oxidative stress assays. ΔrfaH showed increased resistance to 10 mM H2O2. (E) The osmotic stress assays. ΔrfaH showed increased resistance to 0.3 M potassium chloride. Data represent the mean of at least three independent experiments (in triplicate), and error bars represent the standard error of the mean. P values were calculated using student’s t-tests for statistical analyses, **P<0.01, ****P<0.0001.
To assess the impact of rfaH deletion on bacterial responses to host stresses, NTUH-K2044, ΔrfaH, and ΔrfaH-comp underwent heat shock, acidic, osmotic, and oxidative challenges. As a result, ΔrfaH showed survival ability comparable to the wild-type strain after heat (ΔrfaH vs NTUH-K2044, P=0.9942 for 40°C, P=0.6154 for 50°C, and P=0.171 for 60°C, respectively) or acid (ΔrfaH vs NTUH-K2044, P=0.1141) treatments (Supplementary Figures S2, S3), indicating that the deletion of the rfaH does not affect bacterial response to heat-shock and acid adaptation. Unexpectedly, upon exposure to 10 mM H2O2 for 30 min, the survival rate of ΔrfaH was 126.1 ± 9.64%, notably higher than that of NTUH-K2044 (15.75 ± 2.35%, ΔrfaH vs NTUH-K2044, P<0.0001) and ΔrfaH-comp (9.52 ± 2.45%, ΔrfaH vs ΔrfaH-comp, P<0.0001) (Figure 4D). Similarly, ΔrfaH exhibited higher survival rates 103.5 ± 2.79% compared to the parental strain (66.33 ± 5.76%, ΔrfaH vs NTUH-K2044, P=0.0012) and ΔrfaH-comp (72.62 ± 0.17%, ΔrfaH vs ΔrfaH-comp, P<0.0001) in the presence of 0.3 M potassium chloride (Figure 4E). These results suggest that RfaH of K. pneumoniae is implicated in regulating bacterial growth in oxidative and osmotic conditions, while the underlying mechanism remains unknown.
Biofilm formation facilitates bacterial colonization and has been associated with reduced susceptibility to host immune responses (Zhu et al., 2021). To determine whether RfaH plays a role in the biofilm formation of K. pneumoniae, biofilm biomass formed by ΔrfaH and NTUH-K2044 was quantified using a crystal violet staining. Unexpectedly, the amount of biofilm in ΔrfaH displays a 29.8% increase compared to that of NTUH-K2044 (ΔrfaH vs NTUH-K2044,1.40 ± 0.137 vs 0.983 ± 0.062, P=0.0171, Figure 5A). Continuous monitoring from 0 to 36 hours further confirmed this result, with the biofilm production in ΔrfaH surpassing that of NTUH-K2044 from the 2nd hour and lasting till the 36th hour (P=0.0025, Figure 5B). The biofilm formation returned to the wild-type level in the complemented strain.
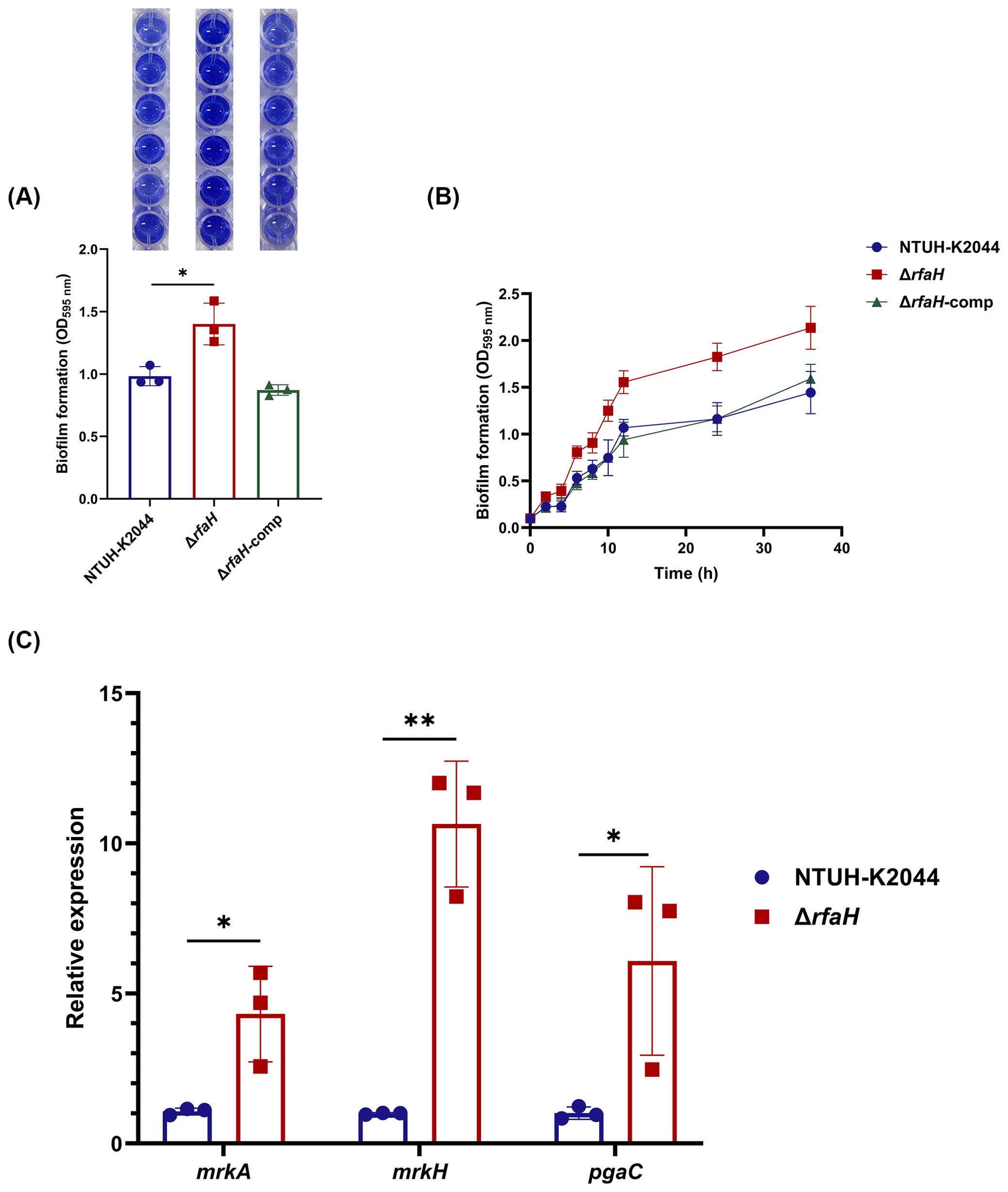
Figure 5. rfaH deletion improves biofilm formation. (A) Quantification of biofilm formation by crystal violet staining in 96-wells plate. This experiment was performed for three independent times, with six repetitions. A representative result is shown above the bar chart. (B) Continuous monitoring of the biofilm formation for 36 h. (C) The transcription level of genes associated with biofilm formation (mrkA, mrkH, and pgaC). Transcript abundance was measured using the 2-ΔΔCt method with 16s rRNA as a reference gene, and was normalized to the NTUH-K2044. Data represent the mean of at least three independent experiments (in triplicate), and error bars represent the standard error of the mean. P values were calculated using student’s t-tests for statistical analyses, *P < 0.05, **P<0.01.
It was shown that type 3 fimbriae strongly promote biofilm formation in K. pneumoniae (Schroll et al., 2010). We found that levels of mrkA (encoding major pilus subunit MrkA) and mrkH (a transcriptional regulator of the mrk gene cluster) were 4.03-fold (P=0.0245) and 10.64-fold (P=0.0013) higher in ΔrfaH than in NTUH-K2044 (Figure 5C). In addition, analysis of the transcription level of pgaC, a gene encoding for Poly β-1,6-N-acetyl-D-glucosamine (PNAG) that is required for biofilm formation in K. pneumoniae (Chen et al., 2014), revealed a 6.08-fold (P=0.0492) increase in ΔrfaH (Figure 5C). These results suggest that RfaH may play an inhibitory role in the biofilm formation of K. pneumoniae by down-regulating the expression of some related factors, including type 3 fimbriae and PNAG production.
To examine the involvement of RfaH in bacterial survival under serum exposure, serum bactericidal assays were performed. We found that the survival rate of ΔrfaH (2.82 ± 0.64%) was significantly lower than that of NTUH-K2044 (103.67 ± 24.9%, ΔrfaH vs NTUH-K2044, P=0.0046). The complementation of rfaH restored the survival rate to 106.42 ± 11.07% (ΔrfaH vs ΔrfaH-comp, P=0.0062). This result confirms the essential role of RfaH in resisting the complement-mediated killing of K. pneumoniae (Figure 6A). To assess directly the roles of RfaH in resisting early immune clearance in vivo, we performed alveolar lavage after the mice were infected with ΔrfaH or NTUH-K2044 by the intranasal route and counted the viable bacteria engulfed by alveolar macrophages. As a result, a significantly higher number of engulfed bacteria were obtained from mice infected by ΔrfaH (1.24 ± 0.59×105 CFU/ml) when compared to those infected by NTUH-K2044 (3.51 ± 0.95×105 CFU/ml, P=0.0079), yielding 1.67- to 3.99-fold more engulfment of ΔrfaH than the parental strain (Figure 6B), indicating that the absence of rfaH leads to significantly increased susceptibility to phagocytosis by macrophages. This result reveals a critical role of RfaH of K. pneumoniae in resisting phagocytosis in mice.
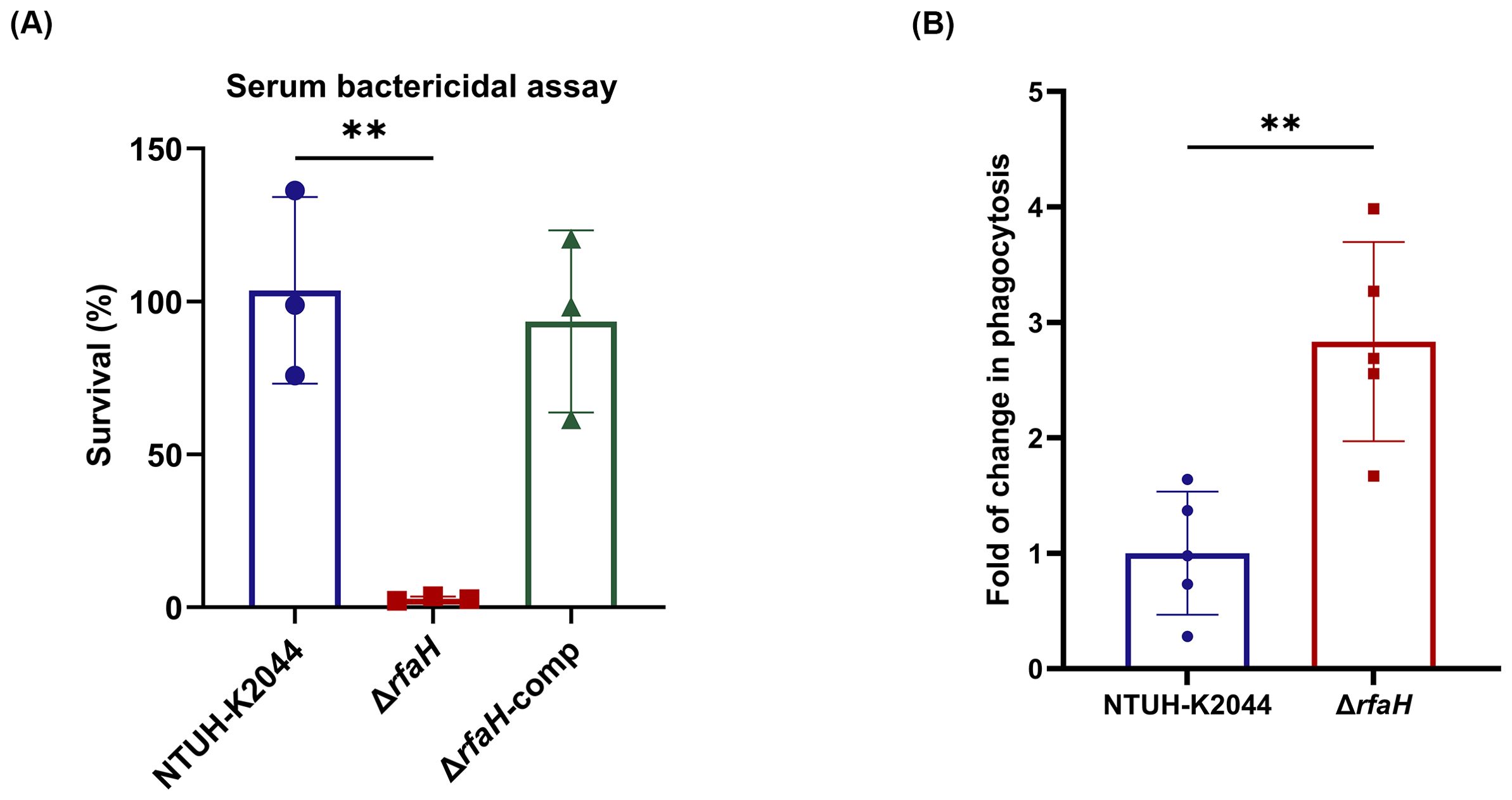
Figure 6. rfaH deletion reduces serum and phagocytosis resistance. (A) The serum bactericidal assays. ΔrfaH showed survival defects in 75% serum. Data represent the mean of at least three independent experiments (in triplicate), and error bars represent the standard error of the mean. (B) in vivo phagocytosis assays. Mice (five per group) were intranasally inoculated with 1 × 108 CFU of NTUH-K2044 or ΔrfaH, respectively. A significantly increased engulfment of ΔrfaH by mouse macrophages was observed (P=0.0079). Each symbol represents 1 animal and error bars represent the standard error of the mean. P values were calculated using student’s t-tests for statistical analyses, **P<0.01.
To assess the role of RfaH in contributing to the pathogenesis of K. pneumoniae, BALB/c mice were intranasally inoculated with 1×105 CFU of NTUH-K2044 or ΔrfaH, respectively. The bacterial loads in the lungs of mice infected by ΔrfaH were 1 log lower at 6 hours post-infection (hpi, NTUH-K2044 vs ΔrfaH, 8.6 (± 4.0) × 103 vs 6.9 (± 2.6) × 102 CFU/0.1g, P<0.05), 4 logs lower at 24 hpi (NTUH-K2044 vs ΔrfaH, 1.2 (± 0.2) × 105 vs 6.4 (± 4.3) × 101 CFU/0.1g, P<0.05), and 6 logs lower at 48 hpi than those in mice infected by NTUH-K2044 (NTUH-K2044 vs ΔrfaH, 2.0 (± 0.7) × 106 vs 2.0 (± 1.0) × 100 CFU/0.1g, P<0.05, Figure 7A). ΔrfaH was almost eliminated in the lung at 48 hpi. In spleens and livers, ΔrfaH was under the detection limit throughout the experiment. While the spleen titers in NTUH-K2044-infected mice were under detection limit at 6 hpi, but increased to 6.7 (± 5.6) × 102 CFU/0.1g at 24 hpi, and 1.0 (± 1.0) × 102 CFU/0.1g at 48 hpi (Figure 7A). Similar results were also observed in the livers, where the bacterial loads of NTUH-K2044 were undetected at 6 hpi, but increased to 1.0 (± 0.5) × 102 CFU/0.1g at 24 hpi, and 8.6 (± 8.4) × 101 CFU/0.1g at 48 hpi (Figure 7A). Altogether, these results showed that the absence of rfaH enhanced clearance of bacteria in vivo, indicating a critical role of RfaH in helping bacterial survival, dissemination, and maximal colonization in mice. Histopathology examination of the mice after 24 h of intranasal infection showed that NTUH-K2044 infection caused mild infiltration of inflammatory cells in the bronchi and adjacent tissues, along with dilated alveoli and congested blood vessels in the lung. In comparison, ΔrfaH infection caused scattered inflammatory cells in the alveolus, with no significant vascular congestion. In the spleen, the NTUH-K2044-infected mice exhibited dilated splenic blood sinusoids and an increase in the number of megakaryocytes, while ΔrfaH-infected mice showed slightly dilated splenic blood sinusoid with a minor megakaryocyte proliferation. Histopathology of the liver displayed no significant difference between two groups (Figure 7B). Considering the relatively short infection duration, the liver’s histopathological changes may not be evident, which is consistent with the bacterial load data.
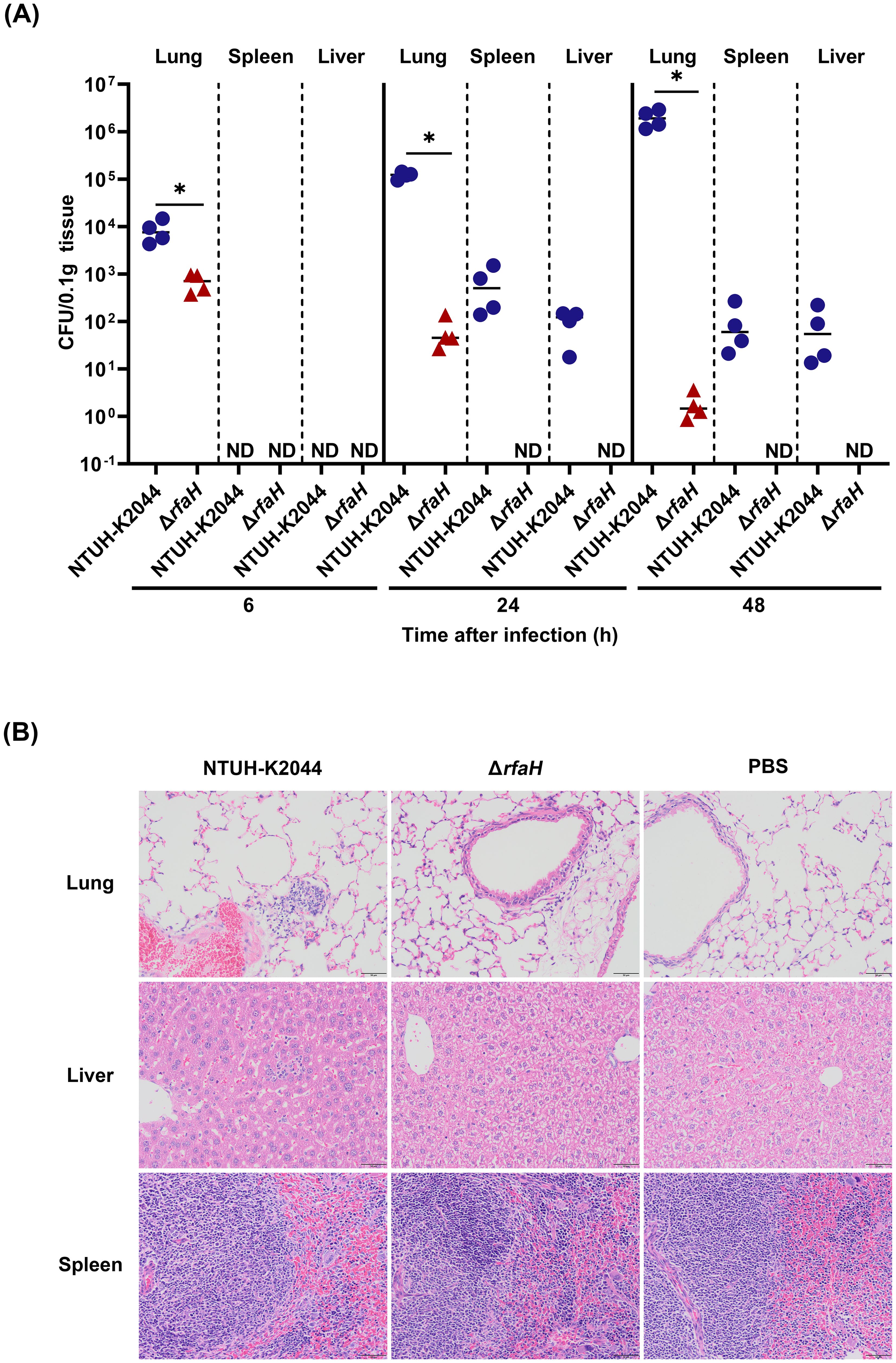
Figure 7. rfaH deletion decreases bacterial dissemination and colonization in mice by the intranasal route. Mice were intranasally inoculated with 1 × 105 CFU of NTUH-K2044 or ΔrfaH, respectively. At the indicated times, mice were euthanized, and the lungs, spleens and livers were homogenized and plated for bacterial enumeration. (A) The organ burden assays after intranasal infection for 6 h, 24 h, and 48 h. Each symbol represents 1 animal, and short bars represent geometric means of each group. The Mann-Whitney test was used for statistical analyses, *P<0.05. ND, not detected. (B) Histopathology of lungs, spleens, and livers in mice after intranasal infection for 24 h.
To further decipher the basis for the attenuation of mutant ΔrfaH, we analyzed bacterial colonization and pathogenicity by intraperitoneally inoculated BALB/c mice with 1 × 104 CFU of NTUH-K2044 or ΔrfaH. Mice infected with ΔrfaH bore significantly (P<0.05) lower bacterial burdens in the lungs (NTUH-K2044 vs ΔrfaH, 1.5 (± 1.0) × 102 vs 1.3 (± 0.1) × 100 CFU/0.1g), liver (NTUH-K2044 vs ΔrfaH, 6.8 (± 2.3) × 101 vs 1.6 (± 0.1) × 10-1 CFU/0.1g), and spleens (NTUH-K2044 vs ΔrfaH, 1.2 (± 0.4)× 103 vs 2.3 (± 0.1) × 100 CFU/0.1g) at 6 hpi. Similarly, after intraperitoneal infection for 24 h, the lungs, livers and spleens of mice infected with ΔrfaH also harbored decreased bacterial loads relative to those infected with NTUH-K2044 (Figure 8A). During this period, an increased number of NTUH-K2044 was observed in the lungs, spleens, and livers at 6 and 24 hpi, indicating bacterial multiplication of NTUH-K2044 through utilization of limited nutrients in mice, while ΔrfaH was rarely detected as a result of elimination by the immune system. All mice infected with NTUH-K2044 died during 24 to 48 h, so we were unable to compare the differences in bacterial loads between NTUH-K2044- and ΔrfaH- infected mice at 48 hpi. These results collectively suggest that RfaH contributes to bacterial fitness and colonization of K. pneumoniae in mice.

Figure 8. rfaH deletion decreases bacterial colonization and pathogenicity in mice by the intraperitoneal route. Mice were intraperitoneally inoculated with 1 × 104 CFU of NTUH-K2044 and ΔrfaH, respectively. At the indicated times, mice were euthanized, and the lungs, spleens and livers were homogenized and plated for bacterial enumeration. (A) The organ burden assays after intraperitoneal infection for 6 h, 24 h, and 48 h. Each symbol represents 1 animal, and short bars represent geometric means of each group. The Mann-Whitney test was used for statistical analyses, *P<0.05. ND, not detected. (B) Histopathology of the lungs, spleens, and livers in mice after intraperitoneal infection for 24 h.
Histopathology examination showed that NTUH-K2044 infection caused moderate infiltration of inflammatory cells in bronchi and adjacent tissues, along with dilated alveoli and sporadic signs of hemorrhage in the lung of mice. In contrast, mice infected by ΔrfaH showed mild infiltration of inflammatory cells in the alveolus, with no significant sign of hemorrhage (Figure 8B). In the spleen, mice from both groups displayed dilated splenic blood sinusoids, accompanied by an increase in the number of megakaryocytes. There were no significant histopathological changes in the liver of both NTUH-K2044- and ΔrfaH-infected mice. The histopathology results are consistent with survival data and mouse pneumonia model data, which collectively highlights a requirement of RfaH for the full virulence of K. pneumoniae in mice.
RfaH is well studied as an important pathogen virulence factor in E. coli by promoting LPS synthesis, K15 capsule and α-hemolysin (Nagy et al., 2002) and S. enterica by impacting the production of LPS (Nagy et al., 2006). In Y. pestis, RfaH contributes to the expression of LPS but has little effect on bacterial acute virulence (Hoffman et al., 2017). Previous studies suggested an essential role of RfaH in the CPS production in K. pneumoniae, and its involvement in bacterial fitness in mice (Bachman et al., 2015; Mike et al., 2021). In our work, we demonstrate that RfaH plays a critical role in the CPS synthesis of K. pneumoniae NTUH-K2044, a hvKp characterized by hyper-capsule. The loss of RfaH results in reduced bacterial survival, dissemination, colonization, and full virulence in mice, which is likely an indirect consequence of CPS deficiency. Collectively, this makes RfaH a promising drug target, especially for treating infections by hvKp.
In this study, we confirmed that RfaH contributes to CPS biosynthesis by directly promoting the transcription of cps operon in K. pneumoniae, as found in E. coli (Bailey et al., 1997). It is known that RcsAB directly activates the transcription of cps operon by binding to a DNA sequence called the RcsAB box located upstream of the cps gene cluster in several bacterial organisms (Walker and Miller, 2020). We found here that the significantly reduced expression of cps operon as a result of rfaH deletion did not bring changes to the expression of rcsA. This finding indicates that RcsAB and RfaH regulate the transcription of cps operon independently, and the decreased expression of cps operon did not lead to upregulation of rcsA as a response in our case.
A previous study revealed that the transcription of rfaH in Salmonella enterica serovar Typhi shows a growth-phase dependent regulation, with maximal expression at the late exponential and stationary phase (Rojas et al., 2001) and that nitrogen limitation increases the transcription of the rfaH (Bittner et al., 2002). That is, RfaH may be involved in regulating the utilization of nitrogen sources, particularly during the nutrient deficiency phase. In this study, the growth rate of ΔrfaH significantly decreased when compared to its parental strain during the logarithmic and stationary phases in the M9 medium. We reasoned that the absence of RfaH may lead to dysregulation or suboptimal utilization of nitrogen sources in M9 medium, causing bacterial growth retardation. The specific mechanism of RfaH in regulating energy metabolism remains to be further studied. In the iron-limited condition, the lower growth kinetics of ΔrfaH was most likely result from its CPS deficiency, due to the close correlation between the CPS biosynthesis and environmental iron availability in hvKp (Thomas A. Russo, 2019).
In stress-response assays, ΔrfaH showing higher survival rates under oxidative and osmotic stress conditions were unexpected. In response to oxidative stress, bacteria can deploy the OxyR and SoxRS systems to transcriptionally activate genes either to protect or repair damage caused by intracellular ROS accumulation in K. pneumoniae (Anes et al., 2021). In our work, the transcription levels of soxS and oxyR were unchanged after the deletion of rfaH (Data not shown), suggesting that rfaH may respond to oxidative stress in a soxS- or oxyR-independent way. It has been shown that outer membrane porin OmpK36 contributes to high osmotic resistance by regulating membrane permeability (Wang et al., 2022). While the absence of rfaH did not change the expression of ompK-36 under high osmotic stress in this work (Data not shown). We speculate that the defects in LPS caused by rfaH deletion might disturb the assembly of outer membrane proteins, resulting in a decrease in membrane permeability (Tian et al., 2022), which may become a growth advantage in oxidative or osmotic stress conditions.
Biofilm formation is a critical factor in the pathogenesis of K. pneumoniae, influenced by various aspects such as environmental conditions, flagella, fimbriae, capsule, and quorum sensing systems (Clegg and Murphy, 2016; Wang et al., 2020). Our study found that ΔrfaH significantly increased biofilm formation, which would be most likely caused by unmasking of proteinaceous adhesins as a result of CPS loss (Sahly et al., 2000). Type 3 fimbriae encoded by the mrk operon (mrkABCDFJ) is known to strongly promote biofilm formation in K. pneumoniae (Schroll et al., 2010). mrkH acts as a regulatory factor facilitating the transcription of the mrk operon (Johnson et al., 2011). The simultaneous increased expression of mrkA and mrkH in ΔrfaH indicates that RfaH is likely to participate in the repression of the mrk operon in a mrkH-dependent way. PNAG encoded by the pgaABCD operon is a bacterial surface component required for biofilm formation (Wang et al., 2005). The deletion of pgaC results in a significantly reduction in the biofilm formation in K. pneumoniae (Chen et al., 2014; Huang et al., 2014). In our study, we observed an up-regulated expression of pgaC after the deletion of rfaH (data not shown), suggesting an inhibitory role of RfaH in the expression of pgaC. The upregulated expression of type 3 fimbriae and PNAG gives alternative explanations for the increased biofilm formation in ΔrfaH. As a transcriptional elongation factor, RfaH displays inhibitory functions in the mrk and pga operons is unexpected. Efforts are underway to further investigate the underlying mechanisms.
Complement proteins are important components of host innate immunity, and both CPS and LPS play crucial roles in bacterial resistance to complement-mediated killing in human serum (Short et al., 2020). The abolished CPS production and truncated LPS in ΔrfaH could explain its impaired survival capabilities in human serum. Meanwhile, ΔrfaH showed significant susceptibility to macrophage phagocytosis during lung infection in mice. It was reported that HMV blocks adherence and internalization by macrophages (Mike et al., 2021). Therefore, the decreased HMV and the increased complement-mediated opsonization as a result of diminished CPS in ΔrfaH may jointly lead to an enhanced macrophage phagocytosis in vivo. These results echo the requirement of RfaH in serum resistance and demonstrate its essential role in bacterial defense against phagocytosis by macrophages. The organ burden assays indicated a higher bacterial clearance rate of ΔrfaH in the lungs in the intranasal infection model, and that ΔrfaH almost could not spread to the livers and spleens of infected mice. Similarly, ΔrfaH was quickly cleared by livers, spleens, and lungs in the intraperitoneal infection model. The decreased bacterial colonization, dissemination and pathogenicity explained the attenuation in virulence of ΔrfaH in mice. These results collectively revealed that RfaH helps bacterial survival and maximal colonization in the host, and ultimately contribute to the full virulence of hvKp. In conclusion, our work demonstrates that RfaH positively regulates the pathogenicity of hvKp, by promoting the CPS production to mediate successful colonization and full virulence. Therefore, developing small molecule inhibitors targeting RfaH could have great potential to treat K. pneumoniae infections. This is of particular significance for hvKp, as increased capsule production is an established hvKp-specific virulence factor.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was approved by Southwest Medical University Institutional Animal Care and Use Committee (Project license: swmu20220089). The study was conducted in accordance with the local legislation and institutional requirements.
QY: Formal analysis, Methodology, Resources, Writing – original draft. LX: Formal analysis, Methodology, Writing – review & editing. MY: Methodology, Writing – review & editing. CF: Methodology, Writing – review & editing. XD: Writing – review & editing, Software. LZ: Writing – review & editing, Conceptualization, Supervision. YL: Conceptualization, Writing – review & editing, Formal analysis.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by National Natural Science Foundation of China (82302576). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1454373/full#supplementary-material
Anes, J., Dever, K., Eshwar, A., Nguyen, S., Cao, Y., Sivasankaran, S. K., et al. (2021). Analysis of the Oxidative Stress Regulon Identifies soxS as a Genetic Target for Resistance Reversal in Multidrug-Resistant Klebsiella pneumoniae. mBio 12, e0086721. doi: 10.1128/mBio.00867-21
Ares, M. A., Fernandez-Vazquez, J. L., Rosales-Reyes, R., Jarillo-Quijada, M. D., von Bargen, K., Torres, J., et al. (2016). H-NS nucleoid protein controls virulence features of klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front. Cell Infect. Microbiol. 6. doi: 10.3389/fcimb.2016.00013
Artsimovitch, I., Landick, R. (2002). The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203. doi: 10.1016/s0092-8674(02)00724-9
Bachman, M. A., Breen, P., Deornellas, V., Mu, Q., Zhao, L., Wu, W., et al. (2015). Genome-Wide Identification of Klebsiella pneumoniae Fitness Genes during Lung Infection. MBio 6, e00775. doi: 10.1128/mBio.00775-15
Bailey, M. J., Hughes, C., Koronakis, V. (1997). RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26, 845–851. doi: 10.1046/j.1365-2958.1997.6432014.x
Bittner, M., Saldı As, S., Estévez, C., Zaldı Var, M., Marolda, C. L., Valvano, M. A., et al. (2002). O-antigen expression in Salmonella enterica serovar Typhi is regulated by nitrogen availability through RpoN-mediated transcriptional control of the rfaH gene. Microbiol. (Reading) 148, 3789–3799. doi: 10.1099/00221287-148-12-3789
Bulger, J., MacDonald, U., Olson, R., Beanan, J., Russo, T. A. (2017). Metabolite Transporter PEG344 Is Required for Full Virulence of Hypervirulent Klebsiella pneumoniae Strain hvKP1 after Pulmonary but Not Subcutaneous Challenge. Infect. Immun. 85, e00093-17. doi: 10.1128/IAI.00093-17
Burmann, B. M., Knauer, S. H., Sevostyanova, A., Schweimer, K., Mooney, R. A., Landick, R., et al. (2012). An α Helix to β Barrel domain switch transforms the transcription factor rfaH into a translation factor. Cell 150, 291–303. doi: 10.1016/j.cell.2012.05.042
Chen, K. M., Chiang, M. K., Wang, M., Ho, H. C., Lu, M. C., Lai, Y. C. (2014). The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb. Pathog. 77, 89–99. doi: 10.1016/j.micpath.2014.11.005
Clarke, B. R., Pearce, R., Roberts, I. S. (1999). Genetic organization of the escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol 181, 2279–2285. doi: 10.1128/JB.181.7.2279-2285.1999
Clegg, S., Murphy, C. N. (2016). Epidemiology and virulence of klebsiella pneumoniae. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.UTI-0005-2012
Creeger, E. S., Schulte, T., Rothfield, L. I. (1984). Regulation of membrane glycosyltransferases by the sfrB and rfaH genes of Escherichia coli and Salmonella typhimurium. J. Biol. Chem. 259, 3064–3069. doi: 10.1016/s0021-9258(17)43260-1
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Hoffman, J. M., Sullivan, S., Wu, E., Wilson, E., Erickson, D. L. (2017). Differential impact of lipopolysaccharide defects caused by loss of RfaH in Yersinia pseudotuberculosis and Yersinia pestis. Sci. Rep. 7, 10915. doi: 10.1038/s41598-017-11334-6
Hsieh, P. F., Lin, H. H., Lin, T. L., Wang, J. T. (2010). CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae. J. Infect. Dis. 202, 52–64. doi: 10.1086/653079
Huang, T.-W., Lam, I., Chang, H.-Y., Tsai, S.-F., Palsson, B. O., Charusanti, P. (2014). Capsule deletion via a λ-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res. Notes 7, 13. doi: 10.1186/1756-0500-7-13
Johnson, J. G., Murphy, C. N., Sippy, J., Johnson, T. J., Clegg, S. (2011). Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol 193, 3453–3460. doi: 10.1128/JB.00286-11
Lee, Y.-J., Inzana, T. (2021). Extraction and electrophoretic analysis of bacterial lipopolysaccharides and outer membrane proteins. Bio-Protocol 11, e4263. doi: 10.21769/BioProtoc.4263
Li, Y., Cao, S., Zhang, L., Lau, G. W., Wen, Y., Wu, R., et al. (2016). A tolC-like protein of actinobacillus pleuropneumoniae is involved in antibiotic resistance and biofilm formation. Front. Microbiol. 07. doi: 10.3389/fmicb.2016.01618
Lindberg, A., Hellerqvist, C. G. (1980). Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J. Gen. Microbiol. 116, 25–32. doi: 10.1099/00221287-116-1-25
Mike, L. A., Stark, A. J., Forsyth, V. S., Vornhagen, J., Smith, S. N., Bachman, M. A., et al. (2021). A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog. 17, e1009376. doi: 10.1371/journal.ppat.1009376
Nagy, G., Danino, V., Dobrindt, U., Pallen, M., Chaudhuri, R., Emödy, L., et al. (2006). Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 74, 5914–5925. doi: 10.1128/iai.00619-06
Nagy, G., Dobrindt, U., Schneider, G., Khan, A. S., Hacker, J., Emoüdy, L. (2002). Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70, 4406–4413. doi: 10.1128/IAI.70.8.4406-4413.2002
Paczosa, M. K., Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/mmbr.00078-15
Palacios, M., Miner, T. A., Frederick, D. R., Sepulveda, V. E., Quinn, J. D., Walker, K. A., et al. (2018). Identification of two regulators of virulence that are conserved in klebsiella pneumoniae classical and hypervirulent strains. mBio 9, e01443-18. doi: 10.1128/mBio.01443-18
Pan, Y.-J., Fang, H.-C., Yang, H.-C., Lin, T.-L., Hsieh, P.-F., Tsai, F.-C., et al. (2008). Capsular polysaccharide synthesis regions in klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46, 2231–2240. doi: 10.1128/jcm.01716-07
Podschun, R., Sievers, D., Fischer, A., Ullmann, U. (1993). Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of klebsiella isolates causing human urinary tract infections. J. Infect. Dis. 168, 1415–1421. doi: 10.1093/infdis/168.6.1415
Rojas, G., Saldías, S., Bittner, M., Zaldívar, M., Contreras, I. (2001). The rfaH gene, which affects lipopolysaccharide synthesis in Salmonella enterica serovar Typhi, is differentially expressed during the bacterial growth phase. FEMS Microbiol. Lett. 204, 123–128. doi: 10.1111/j.1574-6968.2001.tb10874.x
Rosen, D. A., Hilliard, J. K., Tiemann, K. M., Todd, E. M., Morley, S. C., Hunstad, D. A. (2016). Klebsiella pneumoniae fimK promotes virulence in murine pneumonia. J. Infect. Dis. 213, 649–658. doi: 10.1093/infdis/jiv440
Russo, T. A., MacDonald, U., Hassan, S., Camanzo, E., LeBreton, F., Corey, B., et al. (2021). An assessment of siderophore production, mucoviscosity, and mouse infection models for defining the virulence spectrum of hypervirulent klebsiella pneumoniae. mSphere 6, e00045-21. doi: 10.1128/mSphere.00045-21
Sahly, H., Podschun, R., Oelschlaeger, T. A., Greiwe, M., Parolis, H., Hasty, D., et al. (2000). Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect. Immun. 68, 6744–6749. doi: 10.1128/iai.68.12.6744-6749.2000
Sanderson, K. E., Stocker, B. A. (1981). Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J. Bacteriol 146, 535–541. doi: 10.1128/jb.146.2.535-541.1981
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schroll, C., Barken, K. B., Krogfelt, K. A., Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10, 179. doi: 10.1186/1471-2180-10-179
Sharma, R., Patel, S., Abboud, C., Diep, J., Ly, N. S., Pogue, J. M., et al. (2017). Polymyxin B in combination with meropenem against carbapenemase-producing Klebsiella pneumoniae: pharmacodynamics and morphological changes. Int. J. Antimicrob. Agents 49, 224–232. doi: 10.1016/j.ijantimicag.2016.10.025
Short, F. L., Di Sario, G., Reichmann, N. T., Kleanthous, C., Parkhill, J., Taylor, P. W., et al. (2020). Genomic profiling reveals distinct routes to complement resistance in klebsiella pneumoniae. Infect. Immun. 88, e00043-20. doi: 10.1128/iai.00043-20
Singh, S. S., Singh, N., Bonocora, R. P., Fitzgerald, D. M., Wade, J. T., Grainger, D. C. (2014). Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 28, 214–219. doi: 10.1101/gad.234336.113
Srinivasan, V. B., Vaidyanathan, V., Mondal, A., Rajamohan, G. (2012). Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7, e33777. doi: 10.1371/journal.pone.0033777
Stevens, M. P., Hänfling, P., Jann, B., Jann, K., Roberts, I. S. (1994). Regulation of Escherichia coli K5 capsular polysaccharide expression: Evidence for involvement of RfaH in the expression of group II capsules. FEMS Microbiol. Lett. 124, 93–98. doi: 10.1111/j.1574-6968.1994.tb07267.x
Su, K., Zhou, X., Luo, M., Xu, X., Liu, P., Li, X., et al. (2018). Genome-wide identification of genes regulated by RcsA, RcsB, and RcsAB phosphorelay regulators in Klebsiella pneumoniae NTUH-K2044. Microb. Pathog. 123, 36–41. doi: 10.1016/j.micpath.2018.06.036
Svetlov, V., Belogurov, G. A., Shabrova, E., Vassylyev, D. G., Artsimovitch, I. (2007). Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 35, 5694–5705. doi: 10.1093/nar/gkm600
Thomas A. Russo, C. M. M. (2019). Hypervirulent klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–e00019. doi: 10.1128/MMBR.00078-15
Tian, D., Liu, X., Chen, W., Zhou, Y., Hu, D., Wang, W., et al. (2022). Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg. Microbes Infect. 11, 1936–1949. doi: 10.1080/22221751.2022.2103454
Walker, K. A., Miller, V. L. (2020). The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol. 54, 95–102. doi: 10.1016/j.mib.2020.01.006
Walker, K. A., Miner, T. A., Palacios, M., Trzilova, D., Frederick, D. R., Broberg, C. A., et al. (2019). A Klebsiella pneumoniae Regulatory Mutant Has Reduced Capsule Expression but Retains Hypermucoviscosity. mBio 10, e00089-19. doi: 10.1128/mBio.00089-19
Wang, G., Zhao, G., Chao, X., Xie, L., Wang, H. (2020). The characteristic of virulence, biofilm and antibiotic resistance of klebsiella pneumoniae. Int. J. Environ. Res. Public Health 17, 6278. doi: 10.3390/ijerph17176278
Wang, M., Tian, Y., Xu, L., Zhang, F., Lu, H., Li, M., et al. (2022). High Osmotic Stress Increases OmpK36 Expression through the Regulation of KbvR to Decrease the Antimicrobial Resistance of Klebsiella pneumoniae. Microbiol. Spectr. 10, e0050722. doi: 10.1128/spectrum.00507-22
Wang, X., Dubey, A. K., Suzuki, K., Baker, C. S., Babitzke, P., Romeo, T. (2005). CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56, 1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x
Wu, K. M., Li, L. H., Yan, J. J., Tsao, N., Liao, T. L., Tsai, H. C., et al. (2009). Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol 191, 4492–4501. doi: 10.1128/JB.00315-09
Wyres, K. L., Lam, M. M. C., Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1
Yang, X., Sun, Q., Li, J., Jiang, Y., Li, Y., Lin, J., et al. (2022). Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg. Microbes Infect. 11, 841–849. doi: 10.1080/22221751.2022.2049458
Zhang, Y., Jin, L., Ouyang, P., Wang, Q., Wang, R., Wang, J., et al. (2020). Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 75, 327–336. doi: 10.1093/jac/dkz446
Keywords: hypervirulent Klebsiella pneumoniae, rfaH, capsule, anti-phagocytosis, virulence
Citation: Qiu Y, Xiang L, Yin M, Fang C, Dai X, Zhang L and Li Y (2024) RfaH contributes to maximal colonization and full virulence of hypervirulent Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 14:1454373. doi: 10.3389/fcimb.2024.1454373
Received: 25 June 2024; Accepted: 19 August 2024;
Published: 17 September 2024.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Zheng Fan, Capital Institute of Pediatrics, ChinaCopyright © 2024 Qiu, Xiang, Yin, Fang, Dai, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Li, THlpbmcxMDE5QHN3bXUuZWR1LmNu; Luhua Zhang, emhsdWh1YUBzd211LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.