- 1Central Laboratory, Peking University School and Hospital of Stomatology & National Center for Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices, Beijing, China
- 2Fifth Clinical Division, Peking University School and Hospital of Stomatology & National Center for Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices, Beijing, China
- 3Department of Cariology and Endodontology, Peking University School and Hospital of Stomatology & National Center for Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices, Beijing, China
- 4Department of Stomatology, Peking University International Hospital, Beijing, China
- 5Department of Implantology, Peking University School and Hospital of Stomatology & National Center for Stomatology & National Clinical Research Center for Oral Diseases & National Engineering Research Center of Oral Biomaterials and Digital Medical Devices, Beijing, China
- 6Center of Stomatology, Peking University Hospital, Beijing, China
Introduction: Microbial pathogens invade various human organs, including the oral cavity. Candida albicans (C.a) and Streptococcus mutans (S.m) served respectively as representative oral pathogenic fungi and bacteria to stimulate dental pulp stem cells (DPSCs) and to screen the DPSC subcluster that specifically responded to fungal infection.
Methods: DPSCs were obtained from the impacted third molars of six healthy subjects. Then, cells were mixed and divided into three samples, two of which were stimulated with C.a and S.m, respectively; the third sample was exposed to cell medium only (Ctrl). Single-cell mRNA sequencing analysis of treated DPSCs was performed.
Results: DPSCs were composed of four major clusters of which one, DPSC.7, exhibited unique changes compared to those of other subclusters. The DPSC.7 cell percentage of the C.a sample was twice those of the Ctrl and S.m samples. DPSC.7 cells expressed genes associated with the response to reactive oxygen species (ROS) response. DPSC.7 subgroup cells established characteristic aggregation under the stimulation of different pathogens in UMAP. The MAPK/ERK1/2 and NF-κB pathways were up-regulated, DUSP1/5/6 expressions were suppressed, FOS synthesis was activated, the immune-related pathway was induced, and the levels of cytokines, including IL-6 and CCL2, were up-regulated in DPSC.7 cells when stimulated with C.a.
Conclusions: Our study analyzed the cellular and molecular properties of DPSCs infected by oral fungi and bacteria with single-cell RNA sequencing. A subcluster of DPSCs responded specifically to infections with different pathogens, activating the MAPK and NF-κB pathways to induce immune responses via the ROS pathway. This suggests novel treatment strategies for fungal infections.
1 Introduction
Microbial infections often cause organ dysfunctions and immune disorders threatening human health. Infectious diseases impose a substantial economic burden and constitute one of the most severe health issues worldwide (Barreto et al., 2006). Microbial pathogens colonize and invade many human organs, including the oral cavity. Various dental diseases are initiated by excess oral microflora. For instance, oral thrush and Candida-associated denture stomatitis are linked to fungal infection, dental caries, and periodontitis associated with bacterial irritation (Espinosa-Cristóbal et al., 2009; Farges et al., 2009; Gao et al., 2018; Xiao et al., 2018). Periodontal diseases, the putative pathogens of which include Porphyromonas gingivalis and Tannerella forsythensis, are highly prevalent and could affect up to 90% of the worldwide population (Pihlstrom et al., 2005; Patel et al., 2021).
Candida albicans and Streptococcus mutans are representative oral fungal and bacterial pathogens. C. albicans is the most common opportunistic fungal pathogen in the oral cavity. The carriage rates of C. albicans range from 20%-75% in general populations, 65%-88% in those living in acute and long-term care facilities, and 95% in those infected with human immunodeficiency virus (HIV) (Akpan and Morgan, 2002). Oral candidiasis is a common opportunistic infection caused by an overgrowth of Candida species in the oral cavity, the most frequent of which is C. albicans (Akpan and Morgan, 2002; Singh et al., 2014; Matsubara et al., 2017; Sherrington et al., 2017). S. mutans as one species of oral streptococcus, constitutes about 26.9% of all oral microorganisms (Costalonga and Herzberg, 2014) and is associated with dental caries (Hamada and Slade, 1980; Espinosa-Cristóbal et al., 2009; Pitts et al., 2017). Untreated, permanent teeth caries was the most prevalent health condition in 2019, affecting almost 2.0 billion prevalent cases worldwide (Wen et al., 2022).
Both C. albicans and S. mutans trigger inflammation of dental pulp cells. Under certain conditions, such as a compromised immune system or poor oral hygiene, C. albicans may overgrow and invade the pulp tissue. One meta-analysis found that Candida spp. account for 8.20% of root canal infections, approximately two-thirds of which were caused by C. albicans (Mergoni et al., 2018), underscoring the significant role that C. albicans plays in the etiology of endodontic diseases. The presence of this fungus may interfere with the normal functioning of pulp cells, leading to an inflammatory response and tissue damage. Caries-related pathogens may spread and irritate dental pulp cells through dentinal tubules (Hahn and Liewehr, 2007). S. mutans accounted for almost 8.1% of all Streptococcus in young permanent teeth with severe caries (Gross et al., 2010). The bacteria generate acidic byproducts that erode dental enamel, initiating cavity formation. As the cavity deepens, the invasive bacteria and their metabolites penetrate deeper into the tooth’s pulp, triggering inflammation and subsequent infection.
The dental pulp in the tooth center is mainly responsible for maintaining dentin and responding to diverse damages (Pagella et al., 2021; Lee et al., 2022). The dental pulp contains odontoblasts, fibroblasts, dental pulp stem cells, endothelial cells, and T cells (Vainio et al., 1993; Yin et al., 2021). Of these, the dental pulp stem cells (DPSCs) are clonogenic, highly proliferative, mesenchymal stem cells derived from human dental pulp (Gronthos et al., 2000; Miura et al., 2003). DSPCs are heterogeneous, not uniformly expressing many markers and reflecting differences in the developmental stages (Gronthos et al., 2000; Miura et al., 2003; Yamada et al., 2019; Yin et al., 2021). As an essential part of the innate immune response of dental pulp, DPSCs engage in tissue repair and secrete inflammatory cytokines to maintain homeostasis, and have immense potential for tissue regeneration (Liu et al., 2014; Cataldi et al., 2022).
Several issues require attention. Gronthos et al. considered DPSCs homogeneous cells and failed to capture the gene expression of each subcluster. It remains unclear how DPSC subclusters respond to different pathogens (Kawashima et al., 2005; Liu et al., 2014; Macosko et al., 2015; Mao et al., 2024). Also, although the effects of oral bacteria on DPSCs have been studied clearly, there is still the need to investigate the contribution of oral microbes from other species, including fungi and viruses, in dental caries and infected root canals. Furthermore, the specific mechanisms by which DPSCs subclusters respond to fungal infections remain largely unexplored, presenting an opportunity to uncover novel strategies for antifungal treatment.
Thus, we employed single-cell RNA sequencing (scRNA-seq) to capture the responses of DPSC subclusters infected by oral bacteria and fungi, represented by S. mutans and C. albicans, at single-cell resolution. We depict the DPSC transcriptome changes after infection, identify DPSC subclusters countering the fungal pathogen, and reveal the differentiation of DPSCs to suggest future, specific therapeutic approaches toward fungal infections.
2 Methods
2.1 DPSCs collection and cultivation
DPSCs were obtained from six healthy patients aged 18-23 years. All patients have informed written consent. The experimental protocol was approved by the Ethics Committee of Peking University School and Hospital of Stomatology (Approval No. PKUSSIRB-202163047). Immediately after tooth extraction, the pulp tissues were separated and minced into 0.1 × 0.1 × 0.1 cm3 cubes and digested with 3 mg/mL collagenase type I (Worthington). Then, the tissues were seeded into six-well plates containing minimal essential medium-α (α-MEM, Gibco) with 10% fetal bovine serum (FBS, Kang Yuan Biology) and 1% penicillin-streptomycin (Gibco), and incubated at 37°C under 5% CO2. Cells of passage 4 or 5 were applied (Zhang et al., 2023).
2.2 The C. albicans and S. mutans strains and the growth conditions
Candida albicans 90028 (C. albicans) and Streptococcus mutans UA159 (S. mutans) were obtained from the Central Laboratory of Peking University School of Stomatology. C. albicans was cultured in Sabouraud medium (5% CO2, 37°C). S. mutans was grown in brain heart infusion (BHI) medium supplemented with 1% hemin and 0.5% vitamin K in an anaerobic atmosphere.
2.3 Infection of DPSCs by C. albicans and S. mutans
All the DPSCs from six patients were mixed and seeded into 24-well plates at a density of 5 × 104 per well and cultured overnight. The final concentration of the bacterial suspension was measured using the optical density at 630 nm (OD630). An OD630 = 0.1 was equivalent to a bacterial concentration of 1 × 109 colony-forming units (CFU)/mL. The bacterial suspension was diluted to attain the desired multiplicity of infection (MOI). After DPSCs adhered to the plates, the cells were stimulated with C. albicans and S. mutans with a MOI of 1:1 for 4 h at 37°C with 5% CO2. There were no antibiotics in the post-infection medium.
2.4 scRNA-seq using the Singleron GECSCOPE platform
A hemocytometer and an inverted microscope were used to assess the quality and concentration of suspensions. We carried out single-cell preparations with 1 × 105 cells/mL in concentration in PBS (HyClone) and loaded them onto microfluidic devices. scRNA-seq libraries were established by Singleron GEXSCOPE® protocol utilizing GEXSCOPE® Single-Cell RNA Library Kit (Singleron Biotechnologies). Then, individual libraries were diluted to 4 nM, pooled, and subjected to paired-end sequencing on the Illumina Novaseq 6000 platform.
2.5 scRNA-seq alignment and unique molecular identifier calculation
First, raw reads were processed using the CeleScope pipeline (https://github.com/singleron-RD/CeleScope, version 1.9.0) to remove low-quality reads and Cutadapt v1.17 to trim poly-A tail and adapter sequences (Martin, 2011). We extracted the cell barcodes and the UMIs. Then, STAR (version 2.6.1a) was applied to map reads to GRCh38 (ensembl version 92 annotation) (Dobin et al., 2013). UMI counts and gene counts were obtained with featureCounts (version 2.0.1) and used to produce expression matrix files for future analysis (Liao et al., 2014).
2.6 Data importation, transformation, and integration
We imported the database derived as described above into Origin Software (2020b) and R software (version 4.0.3). The filtered, feature barcode matrices were inputted using the ‘Read10X’ and ‘CreateSeuratObject’ functions. The Seurat R package (version 4.0.2) was applied to control quality, normalize the database, and facilitate downstream analysis (Stuart et al., 2019). Cells expressing 200 to 4,000 genes and less than 15% mitochondrial genes were reserved to exclude low-quality and artifact cells. We normalized data to transform gene expression matrices. Analysis of integrated data and the correct batch was performed with the ‘FindIntegrationAnchors’ and ‘IntegrateData’ functions.
2.7 Dimensionality reduction, cell clustering, and identification
We used ‘CellCycleScoring’ and ‘ScaleData’ to mitigate the influence of cell cycle heterogeneity (Tirosh et al., 2016). Then, the ‘FindNeighbors’ and ‘FindClusters’ functions (reduction.type = pca, resolution = 1.0, dims = 1:50) were employed to define cell clusters. The ‘FindAllMarkers’ function was used to identify specific marker genes of each cluster.
2.8 Gene ontology categories and gene set variation analysis
Significantly upregulated cluster-defining genes were computed and carried out gene set over-representation with the gsfisher R package (https://github.com/sansomlab/gsfisher, version 0.2) and acquired gene sets from the GO database.
GSVA, a non-parametric and unsupervised analytical method, was employed to evaluate the relative regulation of pathway activity by comparing different samples using the GSVA package (version 1.38.2). Pathway gene sets were downloaded from the GSEA database (https://www.gsea-msigdb.org/gsea/index.jsp, h.all.v7.5.1.symbols and c5.all.v7.5.1.symbols) (Subramanian et al., 2005). Also, we applied the R package AUCell (version 1.12.0) to calculate the area under the curve (AUC) scores of each cell for pathway activities with default parameters.
2.9 Transcription factor module analysis
The R package SCENIC workflow (https://github.com/aertslab/SCENIC, version 1.2.4, RcisTarget version 1.10.0, and AUCell version 1.12.0) was used to analyze active TF modules in DPSC.7 subgroups (Aibar et al., 2017). We inputted the raw UMI counts for each sample in SCENIC. Then, we applied the GENIE3 method to identify potential TF targets according to the standard SCENIC procedure. AUCell was used to compute a score for each TF module of all cells.
2.10 Pseudotime ordering and lineage trajectories
The pseudotime trajectories were identified using the Monocle2 (version 2.18.0) R package to discover potential developmental transitions (Qiu et al., 2017; Cao et al., 2019). 10,000 cells were selected from the database randomly and imported into Monocle2. Then, we followed the standard workflow and preprocessing steps.
3 Results
3.1 A DPSCs subcluster proportion, DPSC.7, displaying a specific characteristic
DPSCs were collected from the impacted third molars of six healthy subjects. Then, we mixed DPSCs and separated them into three samples, one of which was cultured with cell medium without additional microbial stimulation (Ctrl), and two of which were infected with Candida albicans (C.a) and Streptococcus mutans (S.m), respectively, to emulate the early stages of microbial infection (Infected). We performed scRNA-seq to elucidate the cellular and molecular characteristics of DPSCs (Figure 1A).
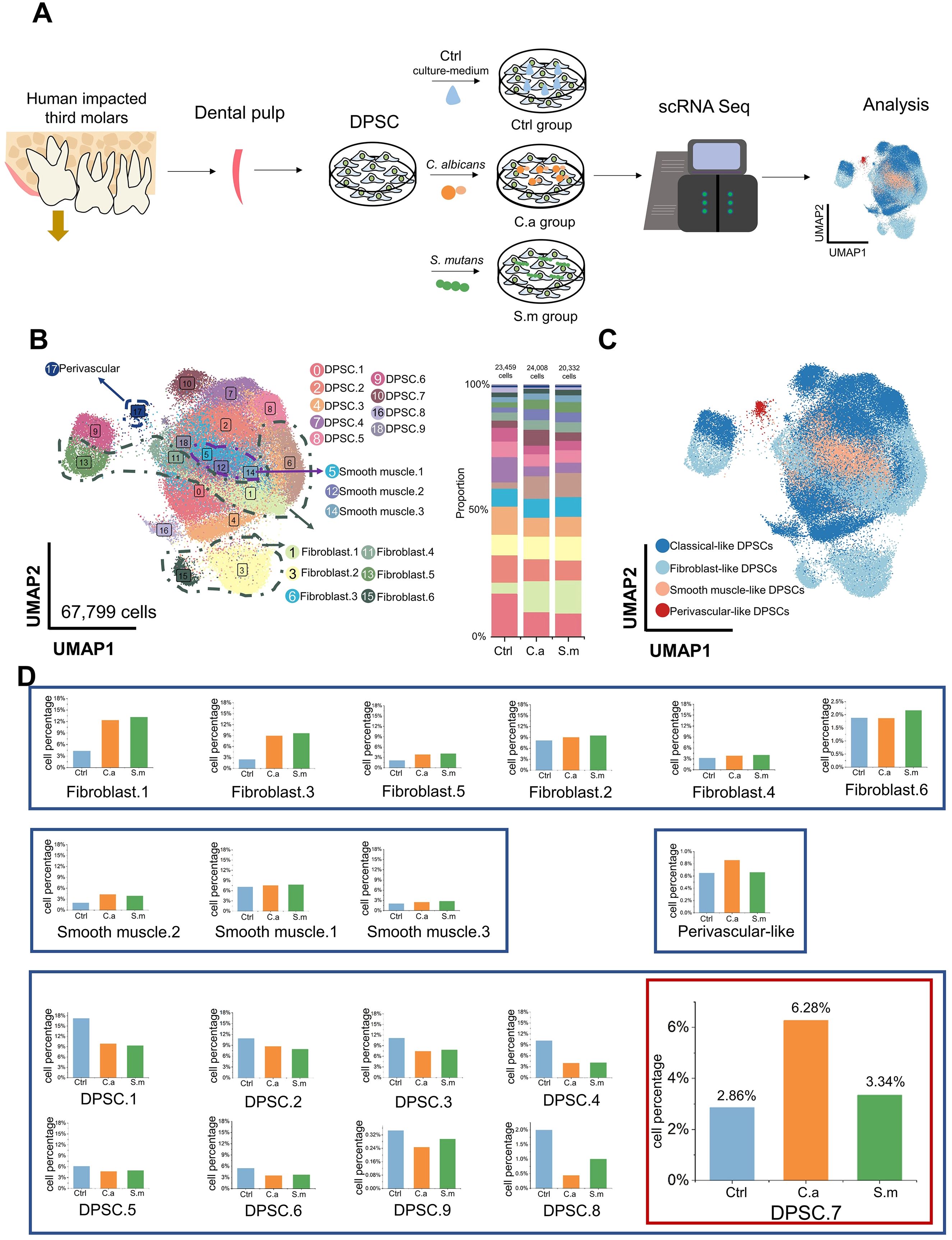
Figure 1. Specific characteristics of DPSC.7 infected by C. albicans and S. mutans. (A) Schematic representation of scRNA-seq experimental design strategy. (B) Distribution of 19 subclusters of DPSCs visualized in UMAP (left) and bar graph of relative cell proportion divided by samples (right). (C) UMAP representation of 4 major cell types, including classical-like DPSCs, fibroblast-like DPSCs, smooth muscle-like DPSCs, and perivascular-like DPSCs. (D) Bar graphs showing the cell percentage of 19 subclusters of DPSCs. DPSC.7 percentage in the C.a sample was approximately twice in the Ctrl and the S.m samples.
After quality control, 67,799 cells were yielded in total, including 23,459 cells in the Ctrl sample, 24,008 cells in the C.a sample, and 20,332 cells in the S.m sample (Supplementary Figure S1). Nineteen subclusters were identified through unsupervised clustering and further annotated (Figure 1B; Supplementary Figures S2, S3). Since several subclusters shared common cellular and functional properties, we integrated cell subclusters, which resulted in four major clusters, including classical-like DPSCs, fibroblastic-like DPSCs, smooth muscle-like DPSCs, and perivascular-like DPSCs (Figure 1C; Supplementary Table S1).
We observed the change in cell percentage under the different conditions of infection. The proportion of several subclusters increased in both C.a and S.m samples, including Fibroblast.1, Fibroblast.3, Fibroblast.5, and Smooth muscle.2, which expressed genes involved in responding to microbial infection, such as toll-like receptor signaling pathway, regulation of chemotaxis, and ossification; these are the common pathway for resistance to pathogens, consistent with previous studies (Supplementary Figure S4) (Kawashima et al., 2005; Liu et al., 2014; Cataldi et al., 2022).
No apparent differences in cell ratio in a few subclusters were shown between the Infected and Ctrl samples, including Fibroblast.2, Fibroblast.4, Fibroblast.6, Smooth muscle.1, and Smooth muscle.3. Also, the percentage of six subclusters in the Infected samples was lower than that in the Ctrl sample (DPSC.1, DPSC.2, DPSC.3, DPSC.4, DPSC.5, and DPSC.6), expressing a gene signature consistent with cell cycle-related pathways. Interestingly, a subcluster, DPSC.7, showed a specific characteristic. Cell proportion in the C.a sample was more than twice higher than that in the Ctrl sample and approximately twice higher than that in the S.m sample (Ctrl [672 cells, 2.86%], C.a [1507 cells, 6.28%], S.m [680 cells, 3.34%], Figure 1D).
3.2 Characteristic aggregation shown in DPSC.7 under different pathogens stimulation
DPSC.7 expressed signature genes, including TXN, MALAT1, MMP3, HIST1H4C, CCND1, and RPS27L, linked to response to reactive oxygen species (ROS), chemical stress, and cell cycle-related pathway (Figures 2A, B; Supplementary Tables S2, S3). To determine whether there were specific subgroups in DPSC. 7 responding to C. albicans infection, we carried out further subgroup clustering.
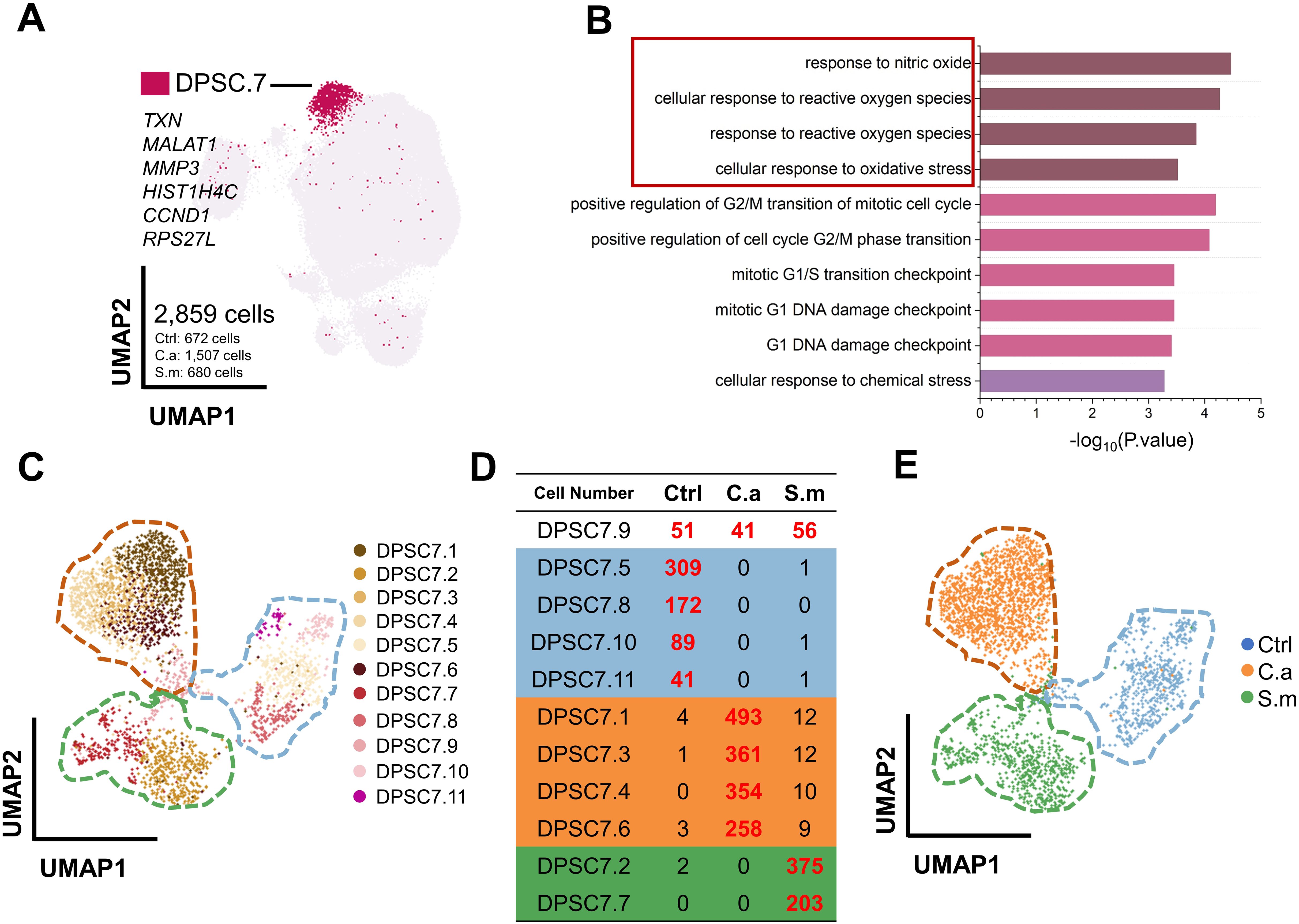
Figure 2. DPSC.7 established specific aggregation under C. albicans and S. mutans infection. (A) UMAP plot showing DPSC.7 cells (2,859 cells, Ctrl [672 cells], C.a [1,507 cells], S.m [680 cells]), whose marker genes were TXN, MALAT1, MMP3, HIST1H4C, CCND1, and RPS27L. (B) Bar graphs depicting GO terms enriched in DPSC.7. (C) Eleven DPSC.7 subclusters were identified and divided by unsupervised clustering shown in UMAP. (D) List showing cell number of eleven DPSC.7 subclusters, which were divided into four categories, including Class One distributed in three samples (DPSC7.9), Class Two primarily from cells in Ctrl sample (DPSC7.5, DPSC7.8, DPSC7.10, and DPSC7.11), Class Three mainly composed of cells in C.a sample (DPSC7.1, DPSC7.3, DPSC7.4, and DPSC7.6), and Class Four mainly consisting of cells in S.m sample (DPSC7.2 and DPSC7.7). (E) UMAP plot visualization of DPSC.7 separated by Ctrl, C.a, and S.m samples distinctly, where cells in the Ctrl sample were distributed on the left side of the plot, cells in the C.a sample were dispersed on the upper right side of the plot, and cells in the S.m sample were occupied in the lower right side of the plot.
This clustering resulted in eleven DPSC.7 subgroups (Figure 2C; Supplementary Table S4). Surprisingly, these subgroups were further divided into four categories (Figure 2D). Cells of the Class One subgroup existed in three samples (DPSC7.9), distributed in the junction center of three samples in a UMAP plot. In subgroups of Class Two, most cells were in the Ctrl sample (DPSC7.5, DPSC7.8, DPSC7.10, and DPSC7.11) occupied on the right side of the UMAP plot. In subgroups of Class Three, cells were mainly in the C.a sample (DPSC7.1, DPSC7.3, DPSC7.4, and DPSC7.6), displayed in the upper left of the UMAP plot. In subgroups of Class Four, most of the cells were in the S.m sample (DPSC.2 and DPSC7.7), exhibiting in the lower left of the UMAP plot. DPSC.7 subgroups exhibited characteristic aggregation in response to stimulation by different pathogens (Figure 2E).
3.3 DPSC.7 subgroups infected by C. albicans expressing genes involved in ERK1/2 signaling pathway
To discover how DPSC.7 might respond to C. albicans infection, we performed differentially expressed genes (DEG) and pathway enrichment analysis on eleven subgroup cells of DPSC.7 (Figures 3A–C; Supplementary Figures S5, S7).
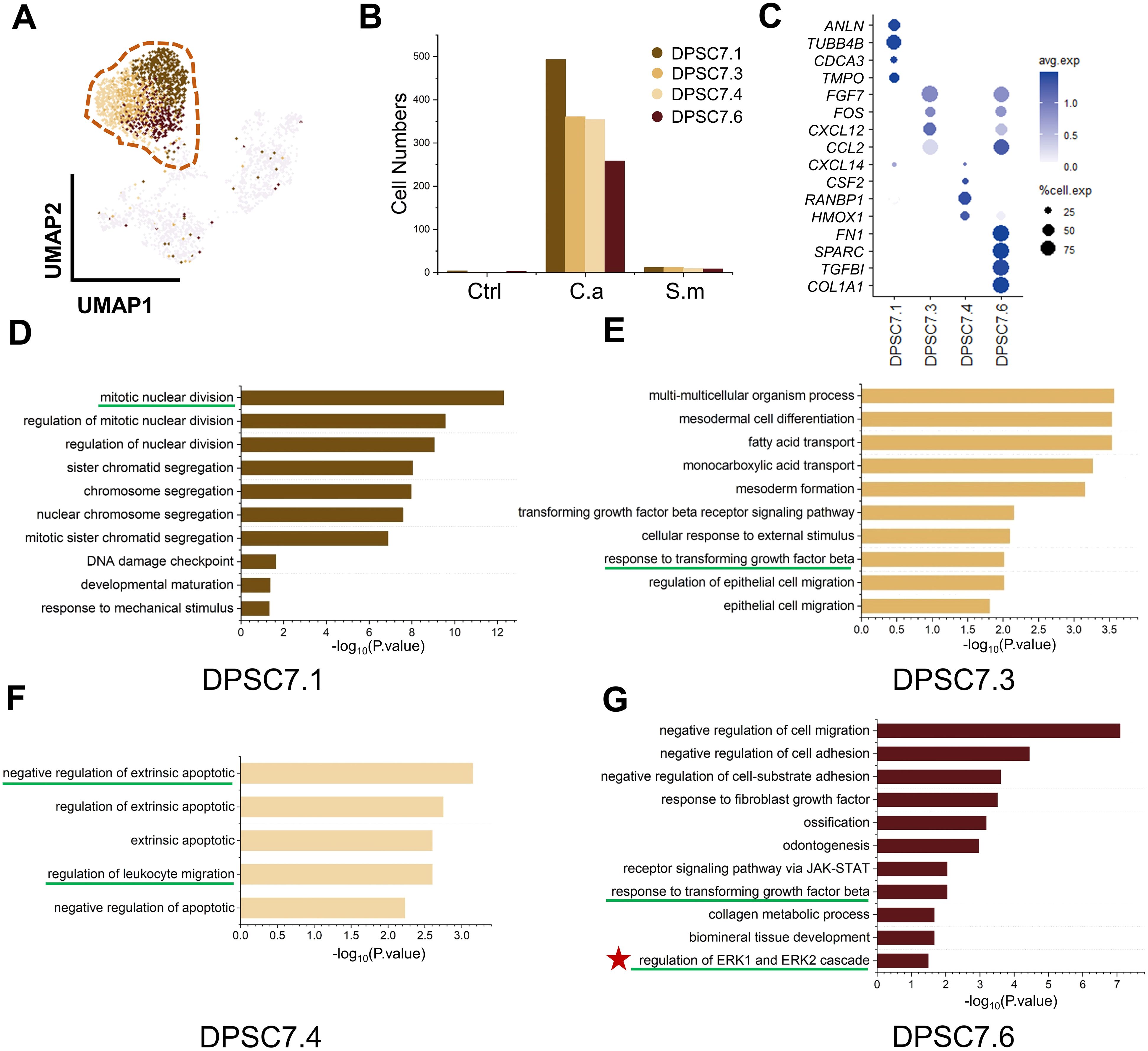
Figure 3. DPSC.7 subgroups cells in C.a sample expressed gene related to ERK1/2 pathway. (A) UMAP plot of DPSC7.1, DPSC7.3, DPSC7.4, and DPSC7.6. (B) Bar graphs showing the cell number of DPSC7.1 (Ctrl [4 cells], C.a [493 cells], and S.m [12 cells]), DPSC7.3 (Ctrl [1 cell], C.a [361 cells], and S.m [12 cells]), DPSC7.4 (C.a [354 cells], and S.m [10 cells]) and DPSC7.6 (Ctrl [3 cells], C.a [258 cells], and S.m [9 cells]) in three groups. (C) Dot plots showing the expression of signature genes and the percentage of cells expressing each gene for four subgroups, including DPSC7.1, DPSC7.3, DPSC7.4, and DPSC7.6. Expression values were normalized and scaled averages. (D–G) Bar graphs showing pathways enriched in DPSC7.1 (D), DPSC7.3 (E), DPSC7.4 (F), and DPSC7.6 (G).
Four subgroups mainly existed in the C.a sample (Supplementary Table S5). DPSC7.1 subgroup cells were linked to the cell-cycle pathway (Figure 3D). DPSC7.3 subgroup cells characterized by the expression of FGF7, FOS, CXCL12, and CCL2 were associated with the TGF-β receptor signaling pathway and epithelial cell migration (Figure 3E). DPSC7.4 subgroup cells displayed gene signatures related to negative regulation of extrinsic apoptotic and regulation of leukocyte migration (CXCL14, CSF2, RANBP1, HMOX1) (Figure 3F). DPSC7.6 subgroup cells expressed genes involved in response to TGF-β, collagen metabolic process, and regulation of ERK1 and ERK2 cascade (Figure 3G). Marker genes and enrichment pathways in other subgroups are described in Supplementary Figures S6, S7 (Supplementary Tables S10–S13).
3.4 Activation of MAPK/P38, MAPK/ERK1/2 signaling pathways in DPSC.7 subgroups
Our results showed that DPSC7.6 expressed genes linked to ERK1 and ERK2 pathways (Figure 3G). Previous studies have described human cells’ response to C. albicans via NF-κB and a biphasic MAPK pathway. Activation of NF-κB and the first MAPK phase – c-Jun might be related to fungal cell wall recognition. And activation of the second MAPK phase – MKP1 and c-Fos, relying on hypha formation and fungal burdens, was associated with proinflammatory responses (Moyes et al., 2010; Naglik et al., 2014; Böhringer et al., 2016; Rast et al., 2016). Therefore, we used GSVA and AUcell to calculate the activity of MAPK and related pathways in DPSC.7 subgroups. We found the activity of MAPK, P38, ERK1/2, and NF-κB pathway in the Infected group was higher than that in the Ctrl group, but there was no apparent difference between C.a and S.m groups (Figures 4A–C).
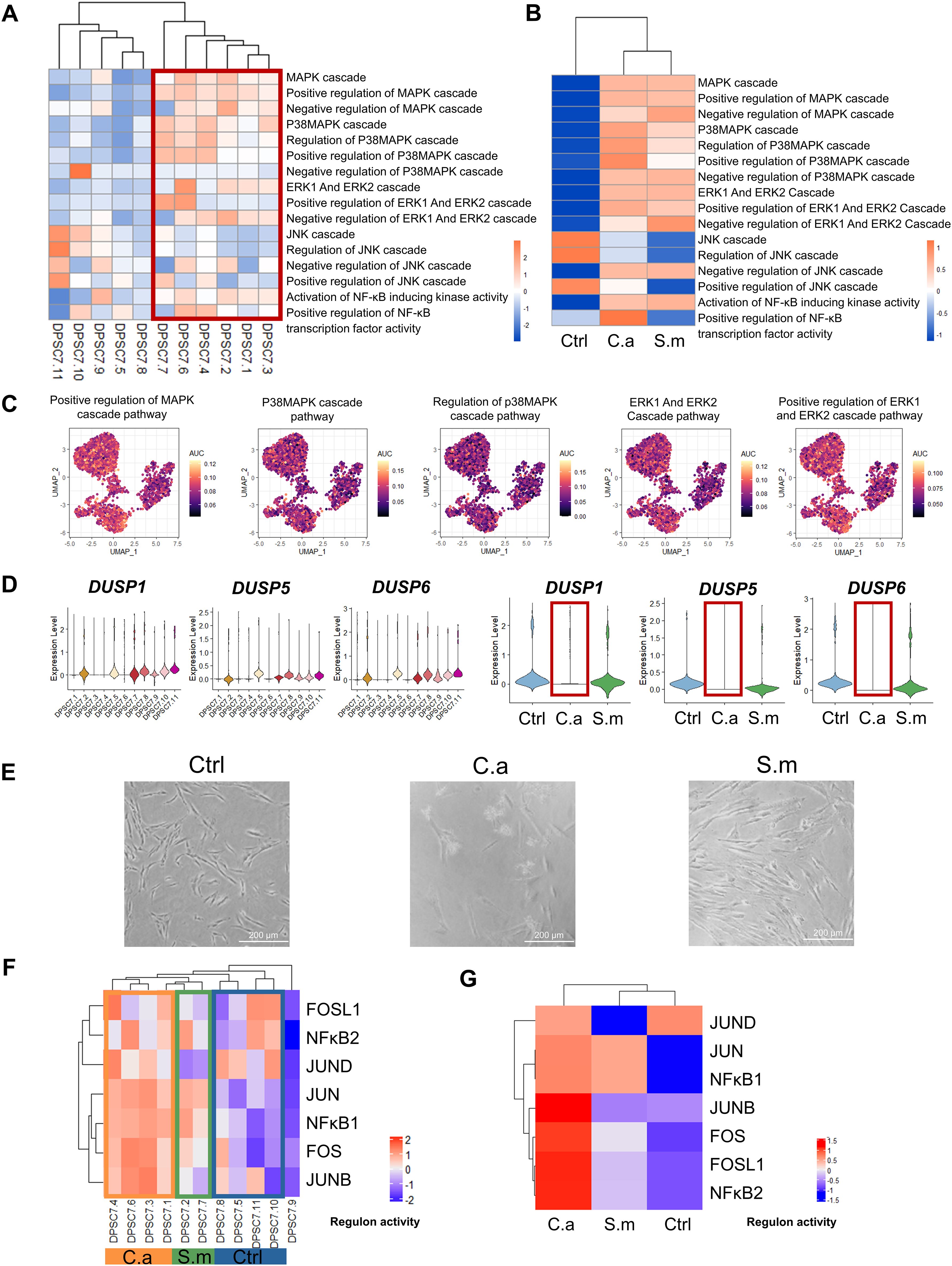
Figure 4. The activity of pathways in DPSC.7 subgroups cells infected by C. albicans. The activity of MAPK/P38, MAPK/ERK1/2 signaling pathways, and transcription factors, including FOS and FOSL1, was up-regulated, and expression of DUSP1/5/6 decreased in the DPSC.7 subgroup under C.albicans infection. (A, B) Heatmap showing the differences of pathway activity associated with MAPK and NF-κB signaling pathway by GSVA enrichment scores among eleven DPSC.7 clusters (A) and three samples (B). (C) UMAP of AUC scores of pathways related to MAPK signaling pathway in DPSC.7. (D) Violin plots of DUSP1, DUSP5, and DUSP6 expression among eleven DPSC.7 clusters and three samples. (E) Imaging of DPSCs in Ctrl, C.a, and S.m samples under the microscope (Scale = 200 μm). (F, G) Heatmap showing the scaled regulon activity by the subclusters of DPSC.7 (F) and three samples (G).
MKP1(DUSP1) and c-FOS activation are related to response specifically to damage-inducing hyphae (Moyes et al., 2010). Thus, we detected the expression of dual specificity phosphatase (DUSP) family genes. DUSP is the key phosphatase acting as the negative regulator of MAPK activity, which could recognize and inactivate ERK1/2, P38, and JNK (Owens and Keyse, 2007; Keyse, 2008). Of interest, the expression of DUSP1/5/6 in the C.a sample was lower than that in the Ctrl and S.m sample (Figure 4D). However, it was not the result of the lack of existence of hyphae. Our microscopic results showed that C. albicans hypha had formed and DPSCs were in contact with hyphae before sequencing (Figure 4E). Besides, our TF activity analysis demonstrated that the activity level of FOS in the C.a sample, other key factors response to C. albicans, was higher than that in the Ctrl and S.m sample with SCENIC. At the same time, the activities of FOS, FOSL1, and JUNB in the C.a group were higher than those in Ctrl and S.m samples (Figures 4F, G; Supplementary Figure S8, Supplementary Table S6).
3.5 The same trend of MAPK/ERK1/2 and ROS pathway activity with the pseudotime
DPSC.7 displayed a gene signature corresponding to the ROS pathway (Figure 2B). The activity of the ROS pathway in the C.a sample was higher than that in the other two samples. Previous studies indicated that NF-kB and MAPK have been the best-characterized oxidation-reduction-sensitive pathways (Baas and Berk, 1995; Ushio-Fukai et al., 1998; Sano et al., 2001; Fukawa et al., 2012). Moreover, MAP kinase phosphatase has been sensitive to ROS, and ROS promotes sustained MAPK pathway activation (Kamata et al., 2005; Levinthal and Defranco, 2005; Cagnol and Chambard, 2010).
Therefore, we used Monocle2 to explore the relationship between ROS and MAPK pathways. We found that cells in several subgroups, including DPSC7.1, DPSC7.5, DPSC7.7, DPSC7.10, and DPSC7.11, were mainly distributed in the early phase of pseudotime, associated with expressed genes related to cell cycle regulation. Cells in other subgroups were mostly in the late phase of pseudotime, related to expressed genes involved in immune response and mineralization (Figures 5A–C). Along the pseudotime, the activity of the ROS biosynthetic process was raised, accompanied by the increased activity of MAPK/ERK1/2 and immune-related pathways, whose related genes were COL1A1, LOX, IL6, and CXCL12 (Figures 5D, E; Supplementary Table S7). Besides, we constructed pseudotime analysis in DPSC7.1, 7.3, 7.4, and 7.6. The activity of ROS and MAPK pathways was the highest at the early phase of pseudotime. Along the pseudotime, the activity of ERK1/2 and immune-related pathways was up-regulated, whose related genes were HMGB1, FOSL1, DCN, and CCL2 (Figures 5F–J; Supplementary Table S8).
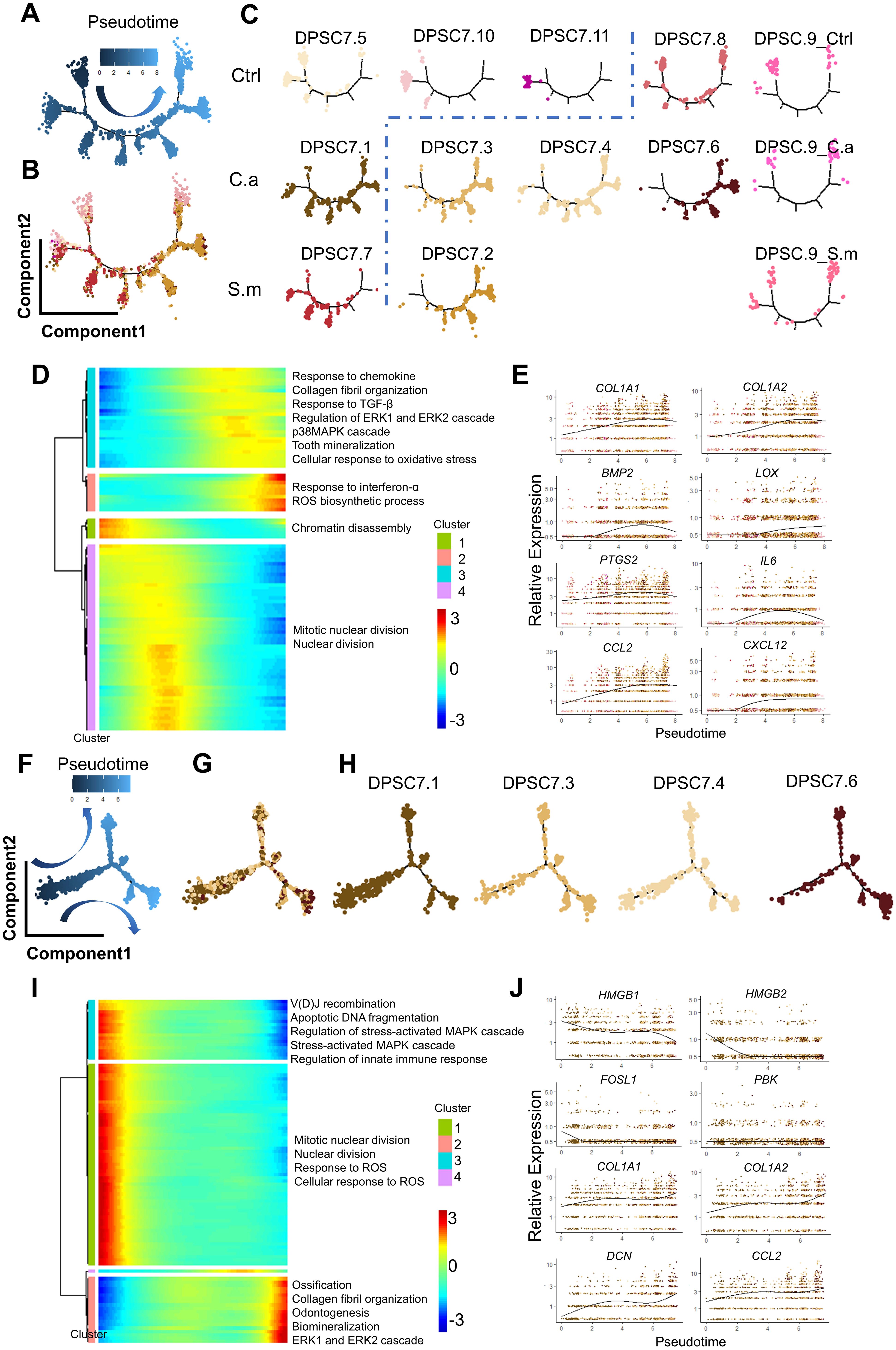
Figure 5. Pseudotime trajectory of DPSC.7. The pseudotime trajectory of DPSC.7 suggests that the activity of MAPK/ERK1/2 and immune-related pathways increased with the upregulation of the ROS pathway. (A–C) Monocle 2 trajectory plot showing the dynamics of DPSC.7 and the pseudotime curve (A) and DPSC.7 subclusters together (B) and respectively (C). (D) The differentially expressed genes were hierarchically clustered into four subclusters along with the pseudotime of DPSC.7. The annotated GO terms of each cluster were selected and provided. (E) Specific genes associated with pathways chosen in (D), including COL1A1, COL1A2, BMP2, LOX, PTGS2, IL6, CCL2, and CXCL12, are listed to describe expression kinetics. (F–H). Pseudotime trajectory analysis shows the relationships of nine subclusters of mesenchymal cells, colored by pseudotime (F) and by major clusters together (G) and separately (H). (I) The differentially expressed genes were hierarchically divided into four subgroups through pseudotime in DPSC7.1, DPSC7.3, DPSC7.4, and DPSC7.6. The annotated GO terms of each cluster were selected and provided. (J) Several genes related to pathways chosen in (I), including HMGB1, HMGB2, FOSL1, PBK, COL1A1, COL1A2, DCN, and CCL2, are listed to show expression kinetics.
3.6 Activation of immune-related pathway and cytokine expression in DPSC.7 subgroups
Finally, we detected the activity of the immune-related pathway in each subgroup. The activity of the immune-related pathway in the C.a sample, including inflammatory response, complement, and IFN-α response, was higher than that in the Ctrl and the S.m samples (Figures 6A–C). In addition, the average expression of IL-6, CCL2, and CXCL2 and the percentage of IL-6, CCL2, and CXCL2 positive cells in the C.a group were also higher than that in the Ctrl and the S.m sample (Figures 6D, E; Supplementary Figure S9).
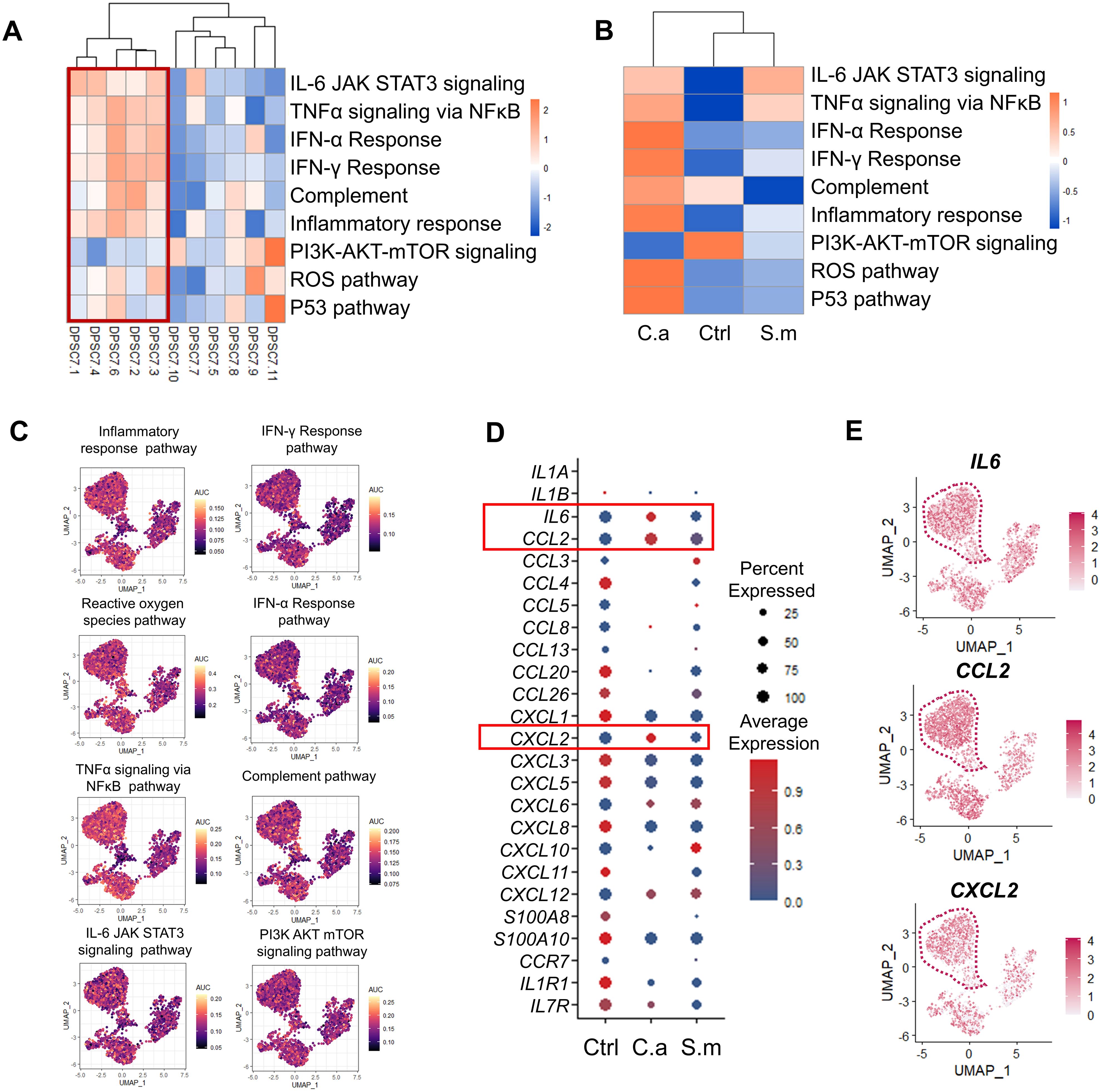
Figure 6. The activity of immune-related pathways and genes was up-regulated in the C.a sample. (A, B) Heatmap representing the GSVA enrichment score for the pathway related to inflammation-related responses among 11 subclusters (A) and three samples (B). (C) UMAP visualization of individual cell AUC score for the selected immune-related pathway. (D) Dot plot indicating chemokine receptor and chemokine expression in DPSC.7 among three samples, whose expression values were normalized and scaled averages. (E) UMAP plots reveal the expression of IL6, CCL2, and CXCL2 in each cell of DPSC.7.
At the same time, we observed that the activity of TGF-β/SMAD pathways increased in the Infected sample. Transforming growth factor–β (TGF-β) was reported to induce collagen deposit around newly recruited polymorphonuclear cells to avoid microbial diffusing (Santus et al., 2017). Similarly, we also found that the activity of odontogenic, ossification, and biomineralization pathways in the Infected sample was higher than in the Ctrl sample (Supplementary Figure S9, Supplementary Table S9).
4 Discussion
Our study analyzed the cellular and molecular characteristics of DPSCs stimulated by oral fungi and bacteria infections represented by Candida albicans and Streptococcus mutans by scRNA-seq. DPSCs were divided into four major clusters: classical-like DPSCs, fibroblastic-like DPSCs, smooth muscle-like DPSCs, and perivascular-like DPSCs. These clusters exhibited both convergent and divergent responses to the various pathogenic stimuli. We found a specific subcluster, DPSC.7, expressed genes linked to the ROS pathway. DPSC.7 subgroups showed characteristic aggregation under different pathogen infections. The activity of MAPK/P38, MAPK/ERK1/2, and NF-κB signaling pathways were up-regulated, transcription factors (FOS, FOSL1, JUN, JUNB) were activated, DUSP1/5/6 expression decreased, and immune-related pathway and cytokine expression were driven in DPSC.7 subgroups infected by C. albicans. Also, the activity of MAPK/ERK1/2 and ROS pathways had the same trend with pseudotime analysis. This finding might be instrumental in searching for new treatment strategies for antifungal infections.
In our study, the activity of MAPK/P38, MAPK/ERK1/2, and NF-κB pathway of DPSC.7 in the Infected sample was higher than that in the Ctrl sample. Our findings corroborate the established knowledge that C. albicans can elicit NF-κB and biphasic MAPK signaling pathways across diverse cell types, leading to the activation of immune-related pathways (Moyes et al., 2010, Moyes et al., 2011, Moyes et al., 2014; Naglik et al., 2014; Böhringer et al., 2016; Nikou et al., 2022). In particular, oral epithelial cells could orchestrate an innate response against C. albicans infection through NF-κB and biphasic MAPK signaling pathways. Activation of NF-κB, the first MAPK phase, and transcription factor c-Jun was independent of morphology due to the recognition of fungal cell walls. The second MAPK phase was likely associated with C. albicans hyphal invasion and induction of inflammatory mediators (Moyes et al., 2010, Moyes et al., 2011).
Moyes et al. have identified two pivotal factors in the secondary mitogen-activated protein kinase (MAPK) phase: MKP-1 and c-Fos. These factors potentially form a danger response mechanism, enabling cells to remain quiescent in the presence of “commensal” fungal levels, while eliciting a specific and robust host response to “dangerous” fungal burdens (Moyes et al., 2010, Moyes et al., 2011). In our study, we noted an upregulation of FOS and a down-regulation of DUSP1/5/6 in the C.a group compared to the Ctrl and S.m samples, corroborating previous findings that c-Fos activation is strongly associated with mycelial growth and fungal burden (Moyes et al., 2010, Moyes et al., 2011). Microscopic imaging further substantiated the hyphae formation and the intimate contact between DPSCs and hyphae (Figure 4E). The c-Fos pathway was also found to be closely associated with the induction of inflammatory mediators, while DUSP1 activation ensured a potent yet tightly regulated immune response of DPSCs to C. albicans hyphae (Moyes et al., 2010, Moyes et al., 2011). Our findings lead us to hypothesize that DPSC.7 infected by C. albicans mycelia may partially activate immune-related cellular functions rather than informing or protecting. Prior research has indicated that the yeast form of C. albicans triggers c-Jun but fails to initiate a sufficient immune response, whereas its mycelial form activates c-Fos, prompting a vigorous immune reaction in oral epithelial cells (Moyes et al., 2010, Moyes et al., 2011; Naglik et al., 2014). Our study extends this by demonstrating that DPSC.7 infected by C. albicans hyphae activates FOS and immune-related pathways, in contrast to the stimulation by S. mutans, which induces JUN and a less robust immune response, aligning with previous studies (Moyes et al., 2010; Warnatsch et al., 2017).
Then, we explored why DUSP was suppressed in DPSC.7 in the C.a sample. Interestingly, we found that the activity of the ROS pathway was up-regulated in the C.a sample. DUSP was sensitive to ROS, and ROS could further prolong the activity of MAPK-related pathways by inhibiting DUSP (Kamata et al., 2005; Levinthal and Defranco, 2005; Cagnol and Chambard, 2010). These results suggested that DPSC.7 infected by C. albicans via ROS pathway inhibits DUSP expression and activates MAPK/ERK1/2 and MAPK/p38 pathway to provoke an immune-related response. However, although ROS might drive the activation of immune-related responses, the decrease of DUSP expression might lead to deleterious MAPK-related inflammatory responses or even apoptosis (Kamata et al., 2005; Cagnol and Chambard, 2010; Moyes et al., 2010).
ROS might also have other roles during the process of C. albicans infection. NF-κB and MAPK were the typical ROS-sensitive signaling pathways, and the increase of ROS would lead to the activation of MAPK and NF-kB pathways (Baas and Berk, 1995; Ushio-Fukai et al., 1998; Sano et al., 2001; Fukawa et al., 2012). ROS could drive inflammatory and immune responses (Hervera et al., 2018; Liu et al., 2022). Additionally, Warnatsch et al. revealed ROS’s function as microbe size sensors. Tiny microbes, such as C. albicans yeast, triggered ROS intracellularly and selectively suppressed IL-1β expression to restrict neutrophil recruitment. Giant microbes, such as C. albicans hypha, impelled ROS extracellularly and enlarged IL-1β expression to recruit a large number of neutrophils (Warnatsch et al., 2017). In our study, the size of S. mutans was relatively smaller, and ROS and immune-related pathways were not obviously activated; in comparison, the size of C. albicans hypha was larger, and ROS and immune-related pathways were provoked. Thus, a possible mechanism and hypothesis might be that ROS, as sensors of microbe size, amplified immune-related pathway activity by C. albicans hyphae and selectively inhibited the expression of cytokines by S. mutans in DPSCs, which was an essential part of the innate immune response.
We also found that the activity of TGF/SMAD was up-regulated in DPSC.7 infected by C. albicans and S. mutans, as well as activation of the fibrosis and mineralization pathways. TGF-β was recognized as a well-known profibrotic factor (Meng et al., 2016). Also, it was demonstrated that the fibrosis process emerged after pathogen infection in various kinds of cells (Meng et al., 2016; Chen et al., 2019; Lai et al., 2022). Santus et al. described the possible mechanism of TGF-β in C. albicans skin infections. Two major phases of the innate response to C. albicans infections included protective containment (abscess) and elimination (expulsion) phases. TGF-β could lead to collagen deposition and impede microbial diffusion to minimize tissue damage and optimize pathogen elimination during the early containment phases (Santus et al., 2017). Besides, the TGF-β/SMAD pathway was also related to tooth regeneration and biomineralization to help resist microbial irritation (Kaufman and Skrtic, 2016; Widbiller et al., 2018; Hu et al., 2019). Our study showed similar results. TGF-β/SMAD and related pathways, including collagen metabolic process and biomineralization, were activated in DPSC.7 infected by microflora. Moreover, MALAT1, a marker gene of DPSC.7, was demonstrated to regulate various factors, such as TGFβ-1 and BMP2, participating in the mineralization of dental pulp cells (Li et al., 2022). Besides, ROS were indicated to modulate TGF-β signaling through different pathways. ROS would activate TGF-β1 and regulate the fibrogenic effect of TGF-β (Jain et al., 2013; Beshay et al., 2020). Thus, TGF-β might be involved in limiting DPSC.7 migration to control microbial damage, and ROS might promote the TGF-β pathway.
Therefore, we propose a potential mechanism model illustrating the response of DPSC.7 to oral pathogen infection. C. albicans initiates a cascade via the ROS pathway, activating the MAPK/p38, MAPK/ERK1/2, and NF-κB pathways, coupled with the suppression of DUSP1/5/6 expression and the induction of key transcription factors. It also propels an immune response, characterized by an upregulation in the expression of cytokines, including IL6 and CCL2. Meanwhile, the engagement of the TGF-β/SMAD signaling pathway stimulates the expression of genes associated with fibrosis and mineralization, thereby potentially curtailing microbial spread. In parallel, S. mutans instigates a similar response by triggering the MAPK/P38, MAPK/ERK1/2, NF-κB, and TGF-β/SMAD pathways. However, its distinctive effect is the activation of the JUN transcription factor, thereby driving both immune and fibrotic responses (Figure 7). Taken together, our research uncovers the multifaceted immune responses of DPSCs to different microbial infections, shedding new light on the intricate dynamics of oral microbe-host cell interactions and offering novel avenues for investigation.
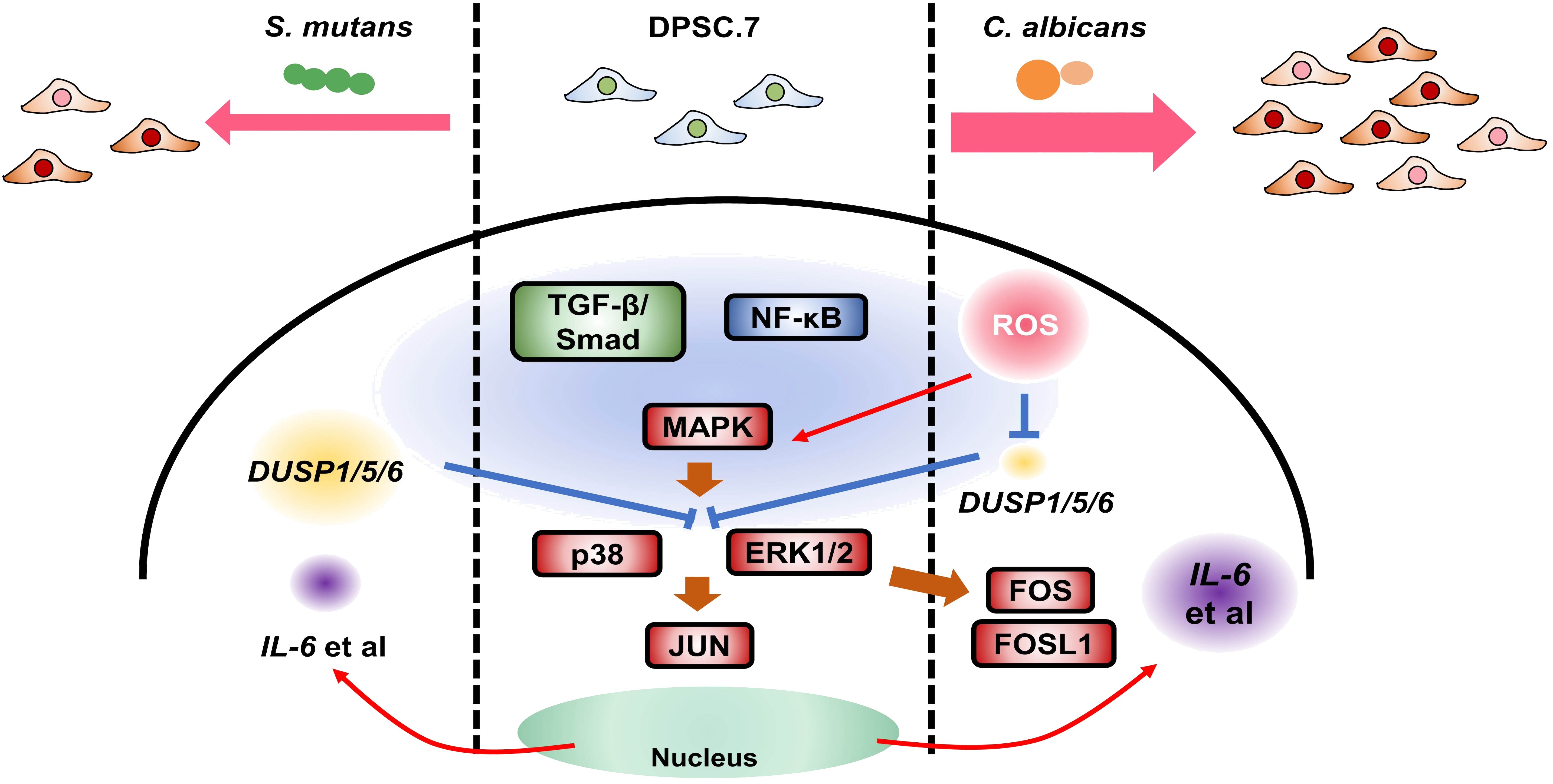
Figure 7. The possible mechanism model responding to oral pathogen infection in DPSC.7. Candida albicans, an oral opportunistic fungus, activated MAPK/p38, ERK1/2, and NF-κB pathway through ROS pathway, suppressed DUSP1/5/6 expression, up-regulated transcription factors (FOS, FOSL1), drove immune response and increased cytokines expression (IL6, CCL2). Meanwhile, the activation of the TGF-β/SMAD signaling pathway induced the expression of genes related to fibrosis and mineralization to limit microbial diffusing. Also, Streptococcus mutans, an oral pathogenic bacterium, could trigger MAPK/P38, ERK1/2, NF-κB, and TGF-β/SMAD pathway, but activated JUN, to induce and weaken immune response and fibrosis response. This study provided new possibilities for antifungal infection with DPSCs in the dental pulp and other kinds of human diseases.
There are a few limitations here, the first being that we carried out the co-culture of DPSCs and microbials for four hours. Various time points were required to explore the mechanism of DPSCs against microbial infection. Besides, we will isolate and culture DPSC.7 to verify gene expression and pathway activity and confirm the process of response to C. albicans infection through animal and cytological experiments.
5 Conclusions
In conclusion, leveraged single-cell RNA sequencing to delve into the cellular and molecular properties of DPSCs in response to oral fungal and bacterial infections. The identification of the DPSC.7 subcluster, which exhibits a specific reactivity to various pathogens via the ROS pathway, marks a significant discovery. Uncovering this connection might be the first step to opening the door to new possibilities for treating fungal infections with DPSCs.
Data availability statement
The datasets presented in this study are deposited in the National Microbiology Data Center (NMDC) repository, accession number NMDC10018945. The specific names of the repository/repositories and accession number(s) can be found in the Supplementary Material.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Peking University School and Hospital of Stomatology (PKUSSIRB-202163047). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
TX: Conceptualization, Formal analysis, Methodology, Software, Visualization, Writing – original draft. YL: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. WZ: Investigation, Methodology, Resources, Writing – review & editing. ML: Data curation, Writing – review & editing. LZ: Data curation, Writing – review & editing. XL: Resources, Writing – review & editing. YZ: Resources, Writing – review & editing. LY: Conceptualization, Writing – review & editing. SL: Data curation, Writing – review & editing. YeL: Conceptualization, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. FC: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was supported by grants-in-aid from the National Science Foundation of China (No. 81991501), the National Key R&D Program of China (2022YFE0118300), the Research Foundation of Peking University School and Hospital of Stomatology (PKUSS20230109), State Key Laboratory of Oral Diseases Open Fund (SKLOD2021OF03, to FC), and National Key R&D Program of China (2022YFA1206100).
Acknowledgments
We thank all the subjects who participated in the study possible and the Singleron GECSCOPE platform.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1452124/full#supplementary-material
References
Aibar, S., González-Blas, C. B., Moerman, T., Huynh-Thu, V. A., Imrichova, H., Hulselmans, G., et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. doi: 10.1038/nmeth.4463
Akpan, A., Morgan, R. (2002). Oral candidiasis. Postgrad Med. J. 78, 455–459. doi: 10.1136/pmj.78.922.455
Baas, A. S., Berk, B. C. (1995). Differential activation of mitogen-activated protein kinases by H2O2 and O2- in vascular smooth muscle cells. Circ. Res. 77, 29–36. doi: 10.1161/01.res.77.1.29
Barreto, M. L., Teixeira, M. G., Carmo, E. H. (2006). Infectious diseases epidemiology. J. Epidemiol. Community Health 60, 192–195. doi: 10.1136/jech.2003.011593
Beshay, O. N., Ewees, M. G., Abdel-Bakky, M. S., Hafez, S., Abdelrehim, A. B., Bayoumi, A. M. A. (2020). Resveratrol reduces gentamicin-induced EMT in the kidney via inhibition of reactive oxygen species and involving TGF-β/Smad pathway. Life Sci. 258, 118178. doi: 10.1016/j.lfs.2020.118178
Böhringer, M., Pohlers, S., Schulze, S., Albrecht-Eckardt, D., Piegsa, J., Weber, M., et al. (2016). Candida albicans infection leads to barrier breakdown and a MAPK/NF-κB mediated stress response in the intestinal epithelial cell line C2BBe1. Cell Microbiol. 18, 889–904. doi: 10.1111/cmi.12566
Cagnol, S., Chambard, J. C. (2010). ERK and cell death: mechanisms of ERK-induced cell death–apoptosis, autophagy and senescence. FEBS J. 277, 2–21. doi: 10.1111/j.1742-4658.2009.07366.x
Cao, J., Spielmann, M., Qiu, X., Huang, X., Ibrahim, D. M., Hill, A. J., et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. doi: 10.1038/s41586-019-0969-x
Cataldi, A., Amoroso, R., di Giacomo, V., Zara, S., Maccallini, C., Gallorini, M. (2022). The inhibition of the inducible nitric oxide synthase enhances the DPSC mineralization under LPS-induced inflammation. Int. J. Mol. Sci. 23, 14560. doi: 10.3390/ijms232314560
Chen, W. Y., Wu, S. Y., Lin, T. C., Lin, S. L., Wu-Hsieh, B. A. (2019). Human dendritic cell-specific ICAM-3-grabbing non-integrin downstream signaling alleviates renal fibrosis via Raf-1 activation in systemic candidiasis. Cell Mol. Immunol. 16, 288–301. doi: 10.1038/s41423-018-0161-5
Costalonga, M., Herzberg, M. C. (2014). The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162, 22–38. doi: 10.1016/j.imlet.2014.08.017
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Espinosa-Cristóbal, L. F., Martínez-Castañón, G. A., Martínez-Martínez, R. E., Loyola-Rodríguez, J. P., Patiño-Marín, N., Reyes-Macias, J. F., et al. (2009). Antibacterial effect of silver nanoparticles against Streptococcus mutans. Materials Lett. 63, 2603–2606. doi: 10.1016/j.matlet.2009.09.018
Farges, J. C., Keller, J. F., Carrouel, F., Durand, S. H., Romeas, A., Bleicher, F., et al. (2009). Odontoblasts in the dental pulp immune response. J. Exp. Zool B Mol. Dev. Evol. 312b, 425–436. doi: 10.1002/jez.b.21259
Fukawa, T., Kajiya, H., Ozeki, S., Ikebe, T., Okabe, K. (2012). Reactive oxygen species stimulates epithelial mesenchymal transition in normal human epidermal keratinocytes via TGF-beta secretion. Exp. Cell Res. 318, 1926–1932. doi: 10.1016/j.yexcr.2012.05.023
Gao, L., Xu, T., Huang, G., Jiang, S., Gu, Y., Chen, F. (2018). Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell 9, 488–500. doi: 10.1007/s13238-018-0548-1
Gronthos, S., Mankani, M., Brahim, J., Robey, P. G., Shi, S. (2000). Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 97, 13625–13630. doi: 10.1073/pnas.240309797
Gross, E. L., Leys, E. J., Gasparovich, S. R., Firestone, N. D., Schwartzbaum, J. A., Janies, D. A., et al. (2010). Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48, 4121–4128. doi: 10.1128/jcm.01232-10
Hahn, C. L., Liewehr, F. R. (2007). Innate immune responses of the dental pulp to caries. J. Endod. 33, 643–651. doi: 10.1016/j.joen.2007.01.001
Hamada, S., Slade, H. D. (1980). Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44, 331–384. doi: 10.1128/mr.44.2.331-384.1980
Hervera, A., De Virgiliis, F., Palmisano, I., Zhou, L., Tantardini, E., Kong, G., et al. (2018). Reactive oxygen species regulate axonal regeneration through the release of exosomal NADPH oxidase 2 complexes into injured axons. Nat. Cell Biol. 20, 307–319. doi: 10.1038/s41556-018-0039-x
Hu, X., Zhong, Y., Kong, Y., Chen, Y., Feng, J., Zheng, J. (2019). Lineage-specific exosomes promote the odontogenic differentiation of human dental pulp stem cells (DPSCs) through TGFβ1/smads signaling pathway via transfer of microRNAs. Stem Cell Res. Ther. 10, 170. doi: 10.1186/s13287-019-1278-x
Jain, M., Rivera, S., Monclus, E. A., Synenki, L., Zirk, A., Eisenbart, J., et al. (2013). Mitochondrial reactive oxygen species regulate transforming growth factor-β signaling. J. Biol. Chem. 288, 770–777. doi: 10.1074/jbc.M112.431973
Kamata, H., Honda, S., Maeda, S., Chang, L., Hirata, H., Karin, M. (2005). Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120, 649–661. doi: 10.1016/j.cell.2004.12.041
Kaufman, G., Skrtic, D. (2016). Structural and recovery mechanisms of 3D dental pulp cell microtissues challenged with Streptococcusmutans in extracellular matrix environment. J. Med. Microbiol. 65, 1332–1340. doi: 10.1099/jmm.0.000353
Kawashima, N., Nakano-Kawanishi, H., Suzuki, N., Takagi, M., Suda, H. (2005). Effect of NOS inhibitor on cytokine and COX2 expression in rat pulpitis. J. Dent. Res. 84, 762–767. doi: 10.1177/154405910508400815
Keyse, S. M. (2008). Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 27, 253–261. doi: 10.1007/s10555-008-9123-1
Lai, L. Y. S., Gracie, N. P., Gowripalan, A., Howell, L. M., Newsome, T. P. (2022). SMAD proteins: Mediators of diverse outcomes during infection. Eur. J. Cell Biol. 101, 151204. doi: 10.1016/j.ejcb.2022.151204
Lee, S., Chen, D., Park, M., Kim, S., Choi, Y. J., Moon, S. J., et al. (2022). Single-cell RNA sequencing analysis of human dental pulp stem cell and human periodontal ligament stem cell. J. Endod. 48, 240–248. doi: 10.1016/j.joen.2021.11.005
Levinthal, D. J., Defranco, D. B. (2005). Reversible oxidation of ERK-directed protein phosphatases drives oxidative toxicity in neurons. J. Biol. Chem. 280, 5875–5883. doi: 10.1074/jbc.M410771200
Li, X., Xu, W., Lin, X., Wu, J., Wu, B. (2022). Effect of LncRNA-MALAT1 on mineralization of dental pulp cells in a high-glucose microenvironment. Front. Cell Dev. Biol. 10. doi: 10.3389/fcell.2022.921364
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, C., Hu, F., Jiao, G., Guo, Y., Zhou, P., Zhang, Y., et al. (2022). Dental pulp stem cell-derived exosomes suppress M1 macrophage polarization through the ROS-MAPK-NFκB P65 signaling pathway after spinal cord injury. J. Nanobiotechnology 20, 65. doi: 10.1186/s12951-022-01273-4
Liu, Y., Gao, Y., Zhan, X., Cui, L., Xu, S., Ma, D., et al. (2014). TLR4 activation by lipopolysaccharide and Streptococcus mutans induces differential regulation of proliferation and migration in human dental pulp stem cells. J. Endod. 40, 1375–1381. doi: 10.1016/j.joen.2014.03.015
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. doi: 10.1016/j.cell.2015.05.002
Mao, X., Xu, M., Yan, X., Chen, L., Lin, N., Zhang, Y. (2024). Single cell omics in tracing cellular heterogeneity of drug-induced liver injury: technological landscape and prospective application. hLife. 2, 325–341. doi: 10.1016/j.hlife.2024.01.004
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1
Matsubara, V. H., Ishikawa, K. H., Ando-Suguimoto, E. S., Bueno-Silva, B., Nakamae, A. E. M., Mayer, M. P. A. (2017). Probiotic bacteria alter pattern-recognition receptor expression and cytokine profile in a human macrophage model challenged with candida albicans and lipopolysaccharide. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02280
Meng, X. M., Nikolic-Paterson, D. J., Lan, H. Y. (2016). TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338. doi: 10.1038/nrneph.2016.48
Mergoni, G., Percudani, D., Lodi, G., Bertani, P., Manfredi, M. (2018). Prevalence of candida species in endodontic infections: systematic review and meta-analysis. J. Endod. 44, 1616–1625.e1619. doi: 10.1016/j.joen.2018.07.016
Miura, M., Gronthos, S., Zhao, M., Lu, B., Fisher, L. W., Robey, P. G., et al. (2003). SHED: stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. U.S.A. 100, 5807–5812. doi: 10.1073/pnas.0937635100
Moyes, D. L., Murciano, C., Runglall, M., Islam, A., Thavaraj, S., Naglik, J. R. (2011). Candida albicans yeast and hyphae are discriminated by MAPK signaling in vaginal epithelial cells. PLoS One 6, e26580. doi: 10.1371/journal.pone.0026580
Moyes, D. L., Runglall, M., Murciano, C., Shen, C., Nayar, D., Thavaraj, S., et al. (2010). A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 8, 225–235. doi: 10.1016/j.chom.2010.08.002
Moyes, D. L., Shen, C., Murciano, C., Runglall, M., Richardson, J. P., Arno, M., et al. (2014). Protection against epithelial damage during Candida albicans infection is mediated by PI3K/Akt and mammalian target of rapamycin signaling. J. Infect. Dis. 209, 1816–1826. doi: 10.1093/infdis/jit824
Naglik, J. R., Richardson, J. P., Moyes, D. L. (2014). Candida albicans pathogenicity and epithelial immunity. PLoS Pathog. 10, e1004257. doi: 10.1371/journal.ppat.1004257
Nikou, S. A., Zhou, C., Griffiths, J. S., Kotowicz, N. K., Coleman, B. M., Green, M. J., et al. (2022). The Candida albicans toxin candidalysin mediates distinct epithelial inflammatory responses through p38 and EGFR-ERK pathways. Sci. Signal 15, eabj6915. doi: 10.1126/scisignal.abj6915
Owens, D. M., Keyse, S. M. (2007). Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213. doi: 10.1038/sj.onc.1210412
Pagella, P., de Vargas Roditi, L., Stadlinger, B., Moor, A. E., Mitsiadis, T. A. (2021). A single-cell atlas of human teeth. iScience 24, 102405. doi: 10.1016/j.isci.2021.102405
Patel, J., Wallace, J., Doshi, M., Gadanya, M., Ben Yahya, I., Roseman, J., et al. (2021). Oral health for healthy ageing. Lancet Healthy Longev 2, e521–e527. doi: 10.1016/s2666-7568(21)00142-2
Pihlstrom, B. L., Michalowicz, B. S., Johnson, N. W. (2005). Periodontal diseases. Lancet 366, 1809–1820. doi: 10.1016/s0140-6736(05)67728-8
Pitts, N. B., Zero, D. T., Marsh, P. D., Ekstrand, K., Weintraub, J. A., Ramos-Gomez, F., et al. (2017). Dental caries. Nat. Rev. Dis. Primers 3, 17030. doi: 10.1038/nrdp.2017.30
Qiu, X., Mao, Q., Tang, Y., Wang, L., Chawla, R., Pliner, H. A., et al. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. doi: 10.1038/nmeth.4402
Rast, T. J., Kullas, A. L., Southern, P. J., Davis, D. A. (2016). Human epithelial cells discriminate between commensal and pathogenic interactions with candida albicans. PLoS One 11, e0153165. doi: 10.1371/journal.pone.0153165
Sano, M., Fukuda, K., Sato, T., Kawaguchi, H., Suematsu, M., Matsuda, S., et al. (2001). ERK and p38 MAPK, but not NF-kappaB, are critically involved in reactive oxygen species-mediated induction of IL-6 by angiotensin II in cardiac fibroblasts. Circ. Res. 89, 661–669. doi: 10.1161/hh2001.098873
Santus, W., Barresi, S., Mingozzi, F., Broggi, A., Orlandi, I., Stamerra, G., et al. (2017). Skin infections are eliminated by cooperation of the fibrinolytic and innate immune systems. Sci. Immunol. 2, eaan2725. doi: 10.1126/sciimmunol.aan2725
Sherrington, S. L., Sorsby, E., Mahtey, N., Kumwenda, P., Lenardon, M. D., Brown, I., et al. (2017). Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 13, e1006403. doi: 10.1371/journal.ppat.1006403
Singh, A., Verma, R., Murari, A., Agrawal, A. (2014). Oral candidiasis: An overview. J. Oral. Maxillofac. Pathol. 18, S81–S85. doi: 10.4103/0973-029x.141325
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., et al. (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Tirosh, I., Izar, B., Prakadan, S. M., Wadsworth, M. H., 2nd, Treacy, D., Trombetta, J. J., et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. doi: 10.1126/science.aad0501
Ushio-Fukai, M., Alexander, R. W., Akers, M., Griendling, K. K. (1998). p38 Mitogen-activated protein kinase is a critical component of the redox-sensitive signaling pathways activated by angiotensin II. Role in vascular smooth muscle cell hypertrophy. J. Biol. Chem. 273, 15022–15029. doi: 10.1074/jbc.273.24.15022
Vainio, S., Karavanova, I., Jowett, A., Thesleff, I. (1993). Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell 75, 45–58. doi: 10.1016/S0092-8674(05)80083-2
Warnatsch, A., Tsourouktsoglou, T. D., Branzk, N., Wang, Q., Reincke, S., Herbst, S., et al. (2017). Reactive oxygen species localization programs inflammation to clear microbes of different size. Immunity 46, 421–432. doi: 10.1016/j.immuni.2017.02.013
Wen, P. Y. F., Chen, M. X., Zhong, Y. J., Dong, Q. Q., Wong, H. M. (2022). Global burden and inequality of dental caries 1990 to 2019. J. Dent. Res. 101, 392–399. doi: 10.1177/00220345211056247
Widbiller, M., Eidt, A., Lindner, S. R., Hiller, K. A., Schweikl, H., Buchalla, W., et al. (2018). Dentine matrix proteins: isolation and effects on human pulp cells. Int. Endod. J. 51 Suppl 4, e278–e290. doi: 10.1111/iej.12754
Xiao, J., Huang, X., Alkhers, N., Alzamil, H., Alzoubi, S., Wu, T. T., et al. (2018). Candida albicans and early childhood caries: A systematic review and meta-analysis. Caries Res. 52, 102–112. doi: 10.1159/000481833
Yamada, Y., Nakamura-Yamada, S., Kusano, K., Baba, S. (2019). Clinical potential and current progress of dental pulp stem cells for various systemic diseases in regenerative medicine: A concise review. Int. J. Mol. Sci. 20, 1132. doi: 10.3390/ijms20051132
Yin, W., Liu, G., Li, J., Bian, Z. (2021). Landscape of cell communication in human dental pulp. Small Methods 5, e2100747. doi: 10.1002/smtd.202100747
Keywords: dental pulp stem cells, Candida albicans, MAPK/ERK1/2, FOS, immune response
Citation: Xu T, Liu Y, Zhang W, Li M, Zhang L, Li X, Zhang Y, Yue L, Li S, Lin Y, Zou X and Chen F (2024) Specific cell subclusters of dental pulp stem cells respond to distinct pathogens through the ROS pathway. Front. Cell. Infect. Microbiol. 14:1452124. doi: 10.3389/fcimb.2024.1452124
Received: 20 June 2024; Accepted: 12 August 2024;
Published: 12 September 2024.
Edited by:
J. Christopher Fenno, University of Michigan, United StatesReviewed by:
Daqiang Wu, Anhui University of Chinese Medicine, ChinaAna Paula Piovezan Fugolin, Oregon Health and Science University, United States
Copyright © 2024 Xu, Liu, Zhang, Li, Zhang, Li, Zhang, Yue, Li, Lin, Zou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Chen, Y2hlbmZlbmcyMDExQGhzYy5wa3UuZWR1LmNu; Xiaoying Zou, em91eGlhb3lpbmcxMTI1QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
 Tiansong Xu
Tiansong Xu Yangjia Liu
Yangjia Liu Wen Zhang
Wen Zhang Murong Li
Murong Li Liqi Zhang1
Liqi Zhang1 Yifei Zhang
Yifei Zhang Feng Chen
Feng Chen