- 1Microbiology and Clinical Microbiology Unit, Scienze Clinico, Chirurgiche, Diagnostiche, Pediatriche (SCCDP) Department, University of Pavia, Pavia, Italy
- 2Clinical Microbiology, Azienda Unità Sanitaria Locale (AUSL) Modena, Modena, Italy
- 3Infectious Diseases Clinic, Azienda Ospedaliera Universitaria (AOU) Policlinico di Modena, Modena, Italy
- 4I.R.C.C.S. Policlinico S. Matteo, Pavia, Italy
- 5Department of Microbiology, Faculty of Medicine, University Hospital in Pilsen, Charles University, Pilsen, Czechia
- 6Biomedical Center, Faculty of Medicine, Charles University, Pilsen, Czechia
Fosfomycin (FOS) is an effective antibiotic against multidrug-resistant Enterobacterales, but its effectiveness is reducing. Little is known on the current prevalence of FosA enzymes in low-risk pathogens, such as Citrobacter freundii. The aim of the study was the molecular characterization of a carbapenemase- and FosA-producing C. freundii collected in Italy. AK867, collected in 2023, showed an XDR profile, retaining susceptibility only to colistin. AK867 showed a FOS MIC >128 mg/L by ADM. Based on WGS, AK867 belonged to ST116 and owned a wide resistome, including fosA3, blaKPC-2, and blaVIM-1. fosA3 was carried by a conjugative pKPC-CAV1312 plasmid of 320,480 bp, on a novel composite transposon (12,907 bp). FosA3 transposon shared similarities with other fosA3-harboring pKPC-CAV1312 plasmids among Citrobacter spp. We report the first case of FosA3 production in clinical carbapenemase-producing C. freundii ST116. The incidence of FosA3 enzymes is increasing among Enterobacterales, affecting even low-virulence pathogens, as C. freundii.
1 Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) infections, which present a considerable challenge for clinicians, are an increasing global threat. Currently, combination therapy based on carbapenem and fosfomycin is a valid option for the treatment of CRE infections (Bakthavatchalam et al., 2020).
Fosfomycin (FOS) is a phosphoric acid derivate, active against both Gram-negative and Gram-positive bacteria, which regained clinical interest in the last 20 years as a valid candidate in the treatment of multidrug-resistant (MDR) infections (Dijkmans et al., 2017). Currently, FOS is indicated for the treatment of uncomplicated UTIs, whereas the parenteral FOS has been used in case of systemic infections caused by MDR organisms (Falagas et al., 2009). FOS used in combination therapy is usually associated with good clinical outcome and bacteriological cure (Shiju et al., 2020). However, in recent years, an increased rate of resistant bacteria has been reported globally, mainly due to FOS-modifying enzymes (such as FosA).
As of February 2024, 10 fosA variants have been reported in the members of Enterobacterales. Plasmid-mediated dissemination of fosA-like genes is recognized as a worrying new challenge for the public health; fosA3 is the most widespread variant, with endemic cases reported from both veterinary and clinical settings in China (Singkham-In et al., 2020; Zhang et al., 2020; Wang et al., 2021; Zou et al., 2021; Mattioni Marchetti et al., 2023a).
Citrobacter spp. are considered as low-risk pathogens, yet can act as silent reservoirs for relevant resistance genes, especially in case of Citrobacter freundii (Bitar et al., 2019). Recent evidence has suggested that the rate of infections caused by carbapenemase-producing Citrobacter spp. is increasing, with relevant reports among Mediterranean countries (Yao et al., 2021; Nobrega et al., 2023). In this scenario, high FOS MICs may further impair antibiotic effectiveness. The co-occurrence of carbapenemases and FosA in Citrobacter spp. is scarcely reported in the literature, with the sole clinical case from the Czech Republic (Mattioni Marchetti et al., 2023b).
Therefore, the aim of our study is to molecularly characterize an XDR carbapenemase-producing C. freundii isolate with high FOS MIC.
2 Materials and methods
2.1 Identification of the bacterial isolate, susceptibility determination, and detection of enzymes
Identification of the C. freundii strain (AK867) was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with MALDI Biotyper software (Bruker Daltonics, Bremen, Germany). The production of carbapenemases (metallo-β-lactamase, OXA-48, and KPC) was assessed with the ROSCO test and with the NG-Test CARBA 5 immunochromatographic assay (NG Biotech Laboratories) (Chudejova et al., 2021). FOS MICs were evaluated using ADM and interpreted according to EUCAST clinical breakpoints v 13.0 and the new v 14.0, whereas the production of FosA-like and FosC2 enzymes was detected by the PPF test (Nakamura et al., 2014). In accordance with Nakamura in-house protocol, the PPF test requires MH agar plates added with 25 mg/L glucose-6-phosphate (G6P), confluence growth of 0.5 MacFarland solution of the isolate to investigate, one disk of FOS (50 μg), and one of FOS (50 μg) plus PPF (1 mg). The cutoff was set to a 5-mm enlargement in the inhibition zone of FOS plus PPF disk compared with the FOS disk alone (Nakamura et al., 2014; Mattioni Marchetti et al., 2023a).
2.2 Long-read sequencing
For genomic characterization, genomic DNA was extracted using the NucleoSpin Microbial DNA kit (Macherey-Nagel, Duren, Germany) and sheared using the Hydropore-long on Megaruptor 2 (Diagenode). Microbial multiplexing library preparation was performed without size selection according to the manufacturer’s instructions. The multiplexed library was sequenced using long-read sequencing technology using the Sequel I platform (Pacific Biosciences, Menlo Park, CA, USA) for a 10-h movie run. Assembly was performed using the “Microbial Assembly” pipeline offered by the SMRT Link v10.0. with the default settings (minimum seed coverage of 30×). Assembled sequences were annotated using the RAST (Rapid Annotation using Subsystems Technology) server (Aziz et al., 2008). In-silico multilocus sequence typing (MLST) of the strains and of the plasmids (pMLST) were performed when applicable. Reconstruction of the resistome, plasmidome, and virulome of the isolates was accomplished using ResFinder, PlasmidFinder, and the Virulence Factors Database (VFDB) via ABRicate (github.com/tseemann/ABRicate). BRIG v.0.95 was used to produce figures of comparison of the circular plasmids’ sequences. A linear map of chromosomal environments was created by using Easyfig (Sullivan et al., 2011) and the graphic editor Procreate (Savage Interactive, Tasmania, Australia).
2.3 Phylogenetic analysis
Phylogenetic relationships between AK867 and 117 global genomes, downloaded from the NCBI assembly database, including complete and draft genomes, were investigated. SNP-based phylogeny was depicted using parsnp v1.2 (Treangen et al., 2014) and using randomly GCF_029840125.1 as the reference genome. A graphic illustration of the trees was built with the Interactive Tree Of Life (iTOL) (https://itol.embl.de/). The clustering of genetic sequence was performed on the total pool of C. freundii ST116 by the FastBaps algorithm (Tonkin-Hill et al., 2019).
2.4 Conjugation/transformation assay
The conjugal transfer of fosA genes was tested in liquid medium using the E. coli J53 strain (RIFr) as a recipient. Transconjugants were selected on MacConkey agar plates (Scharlab, SL, Barcelona, Spain) containing rifampicin (100 mg/L) (Sigma-Aldrich, St. Louis, MO, USA), FOS (64 mg/L) (Sigma-Aldrich), and G6P (25 mg/L) (Roche). The presence of fosA-like genes and the plasmid content in transconjugants were further confirmed by PCR and PCR replicon typing (PBRT 2.0 kit, Diatheva), respectively (Carattoli et al., 2005).
2.5 Data access
The plasmid sequence of pfosA3_CFR867 has been uploaded to GenBank under the accession number CP151860–CP151866.
3 Results
3.1 Isolation and antimicrobial susceptibility profile
On 31/01/2023, a C. freundii (AK867) from rectal swab was collected. The sample was part of ongoing 3-year surveillance on carbapenemase-producing Enterobacterales (CPE) conducted locally at Modena Hospital in Italy. AK867 was isolated from a 41-year-old patient admitted in Modena Hospital, suffering from fever due to inguinal abscess by anaerobic bacteria. AK867 showed an extensively drug-resistant (XDR) phenotype, being susceptible only to colistin. FOS MIC was evaluated by ADM (FOS MIC >128 mg/L). The high FOS MIC was corroborated by the production of FosA-like enzymes, as suggested by a positive phenotypic PPF test.
3.2 WGS
Based on the WGS analysis, AK867 belonged to the sequence type 116 (ST116) and carried three large plasmids: an IncA (pMLST: 12) of 177,013 bp carrying the resistance genes aph(3′)-XV, aadA1, aac(6′)-Ib-cr, blaSHV-12, blaOXA-1, blaVIM-1, mph(A), catB2, catB3, qnrS1, ARR-3, sul1 (x2), and dfrA14; a multireplicon IncFIB-HI1B (pMLST IncF: F-:A-:B-; IncHI1: unknown) plasmid of 252,890 bp harboring blaKPC-2, blaTEM-1A, and blaOXA-9; and a pKPC-CAV1321 of 320,480 bp carrying aac(3)-IIa, aadA2b, aac(6′)-Ib-cr, blaOXA-1, fosA3, ere(A), cmlA1, catB3, ARR-3, sul1 (x2), tet(A), and dfrA19.
3.3 Genomic characterization of pfosA3_CFR867
The pKPC-CAV1321 plasmid (pfosA3_CFR867) harbored fosA3 as part of a large genomic island (55,446 bp), starting with an HNH endonuclease and ending with an IS66, and composed of five antimicrobial resistance islands (ARI): the first ARI is a IS26-aac(3), the second a IS26-fosA3, the third Int1-aac(6′)-Ib-cr-blaOXA-1-catB3-ARR-3-qacEΔ1-sul2, the fourth a ISCR1-dfrA19-ΔInt1-ant(3ʺ)-Ia-cmlA-ereA-qacEΔ1-sul2, and the fifth is composed of ΔIS110-IS5075-IS4321-tetR-tet(A) (Figure 1). fosA3 is inserted in a large composite transposon (12,907 bp), flanked by IS26 at both sides, organized in IS26-fosA3-fidL-helix-motif-acrR-H1-virG-ABC-transporters-yccJ-NAD(P)H-ymdE-rutR-IS26 (Figure 2). The fosA3 transposon is entirely shared with pTEM-2262, pCFA1707-1, pCF1807-1, and pF321-1. Differently, in pS39_1, the fosA3 transposon appeared to be split, with the presence of additional orfs, enlarging the size of the fosA3 transposon (Figure 1). These results highlighted both the conservative nature of the transposon and its ability to acquire further features.
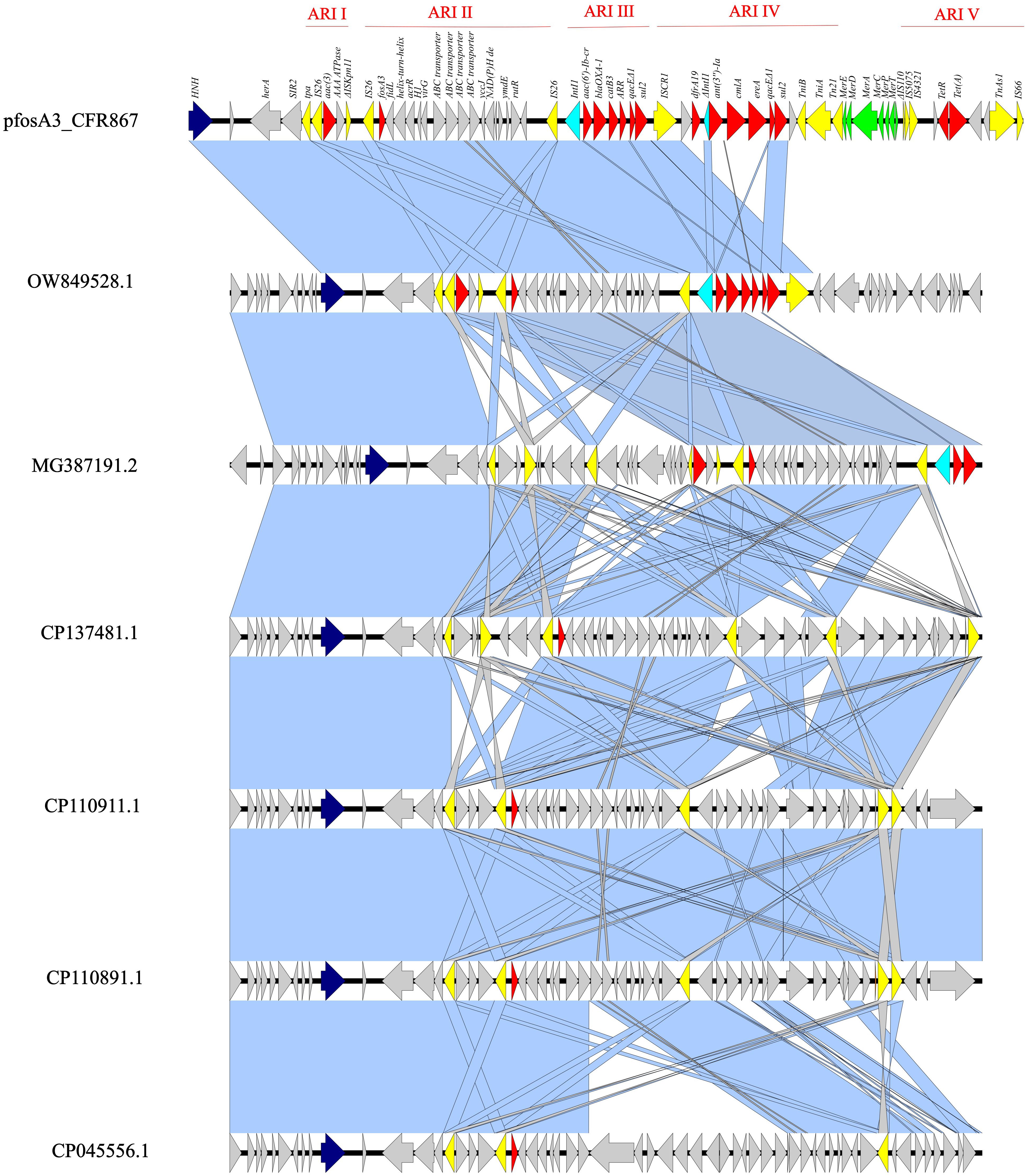
Figure 1 Linear map of pfosA3_CFR867 (from 241,351 bp to 296,797 bp) against OW849528.1, MG387191.2, CP137481.1, CP110911.1, CP110891.1, and СР045556.1. Dark blue: HNH endonuclease; yellow: IS; red: AMR genes; light blue: Int1; green: mercury system locus. Light blue shadows refer to 100% identity; gray shadows refer to identity less than 100%.
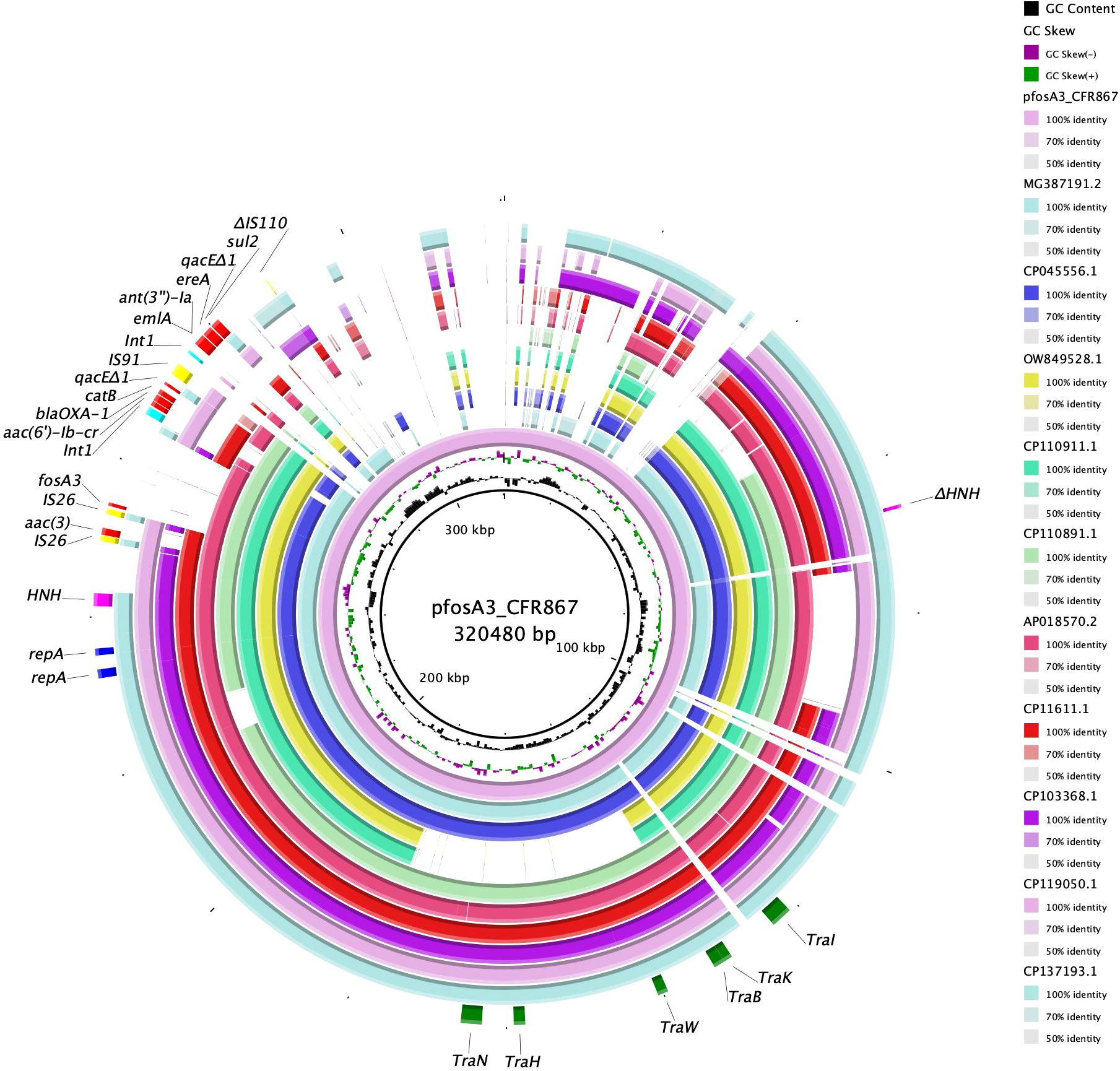
Figure 2 BRIG circular map of pfosA3_CFR867 against MG387191.2 (turquoise), СР045556.1 (violet), OW849528.1 (yellow), CP110911.1 (green), CP110891.1 (light green), AP018570.2 (fuchsia), CP11611.1 (red), CP103368.1 (purple), CP119050.1 (pink), and CP137193.1 (jade green). At the outer curved segments, blue, fuchsia, yellow, red, light blue and green refer to replication (repA), HNH endonuclease, IS, AMR genes, Int1 and Tra system locus.
The entire genomic island showed level of identity with other fosA3-harboring pKPC-CAV1312 plasmids isolated from different C. freundii: query 75% and ID 99.94% with pTEM-2262 (MG387191.2), from a C. freundii collected in China; query 63% and ID 99.98% with P1 (OW849528.1), from C. freundii ST22 collected in 2018 from a Spanish patient; query 63% and ID 99.96% with pCFA1707-1 (CP110911.1) and pCF1807-1 (CP110891.1), from two C. freundii ST107 and collected in 2017/2018 from China; query 64% and ID 99.91% with pMH17-012_4 (AP018570.2), from a C. freundii collected in Vietnam in 2017; query 65% and ID 99.85% with pS39_1 (CP045556.1), from a C. freundii ST169 collected in China in 2017; query 48% and ID 99.99% with pF321-1 (CP137481.1) from C. portucalensis ST252 that has been collected from urine in China in 2021 (Figure 2). The backbone of pfosA3_CFR867 is enriched with IS sequences, genes involved in transferability (Tra locus), defense system against mercury (Mer locus), two copies of replication genes (rep), and two copies of HNH endonucleases (Figure 2). These data together suggest an initial fitting of fosA3-harboring plasmids among Citrobacter spp. strains of several STs and the rearrangement ease of such plasmids. Moreover, pfosA3_CFR867 showed levels of identity with other pKPC-CAV1312 not-fosA3-harboring plasmids characterized from Chinese and USA C. freundii strains, such as CP011611.1, CP103368.1, CP119050.1, and CP137193, pointing out the conservative nature of pfosA3_CFR867 (Figure 2). PfosA3_CFR867 was transferable by conjugation in E. coli J53, shifting from FOS MIC = 0.5 mg/L to a resistance phenotype (FOS MIC >128 mg/L) in E. coli J53, in accordance with EUCAST clinical breakpoints v 14.0.
3.4 Phylogenetic analysis
The SNP-based phylogeny on the 118 C. freundii ST116 genomes downloaded from the NCBI pointed out the presence of three clades (CL1, CL2, and CL3), confirmed by the FastBaps algorithm (Figure 3). AK867 falls into CL3 and clustered together with GCF_032192735.1, collected in 2017 from a German patient, and GCA_028404165.2, collected in 2023 from an American patient. According to the available metadata, the three clades are circulating worldwide since 2012, with CL3 as the predominant clade. Referring to the antimicrobial resistance content, C. freundii ST116 revealed cluster-related resistomes, with carbapenemase KPC-2 common in the three clades. Moreover, except for AK867, the occurrence of VIM-1 carbapenemase is rare in ST116. Interestingly, the presence of fosA3 already occurred in C. freundii ST116, but in CL1 from sewage sample in China (GCF_032747095.1). Moreover, another fosA-like gene, fosA5, fits in C. freundii ST116 CL1 as reported by three human samples collected in China (GCF_029104415.1, GCF_029104385.1, GCF_029104405.1) (Figure 3). Additionally, the three clusters revealed different plasmidome, with a predominance of pKPC-CAV1312 plasmids in CL2 (Supplementary Figure S1). pKPC-CAV1312 seemed to not easily fit in CL1 and CL3, where there is a higher incidence of IncFIB, IncFII, and IncA/C2 (Supplementary Figure S1). Thus, the entry and stabilization of pKPC-CAV1312 that harbor fosA-like genes in ST116 provide further knowledge on the real incidence of FOS resistance within C. freundii ST116. The presence of multireplicon IncFIB: HI1B was not reported in ST116 except for AK867. Concerning the virulome, all the three clusters shared similar virulence gene content, including adhesions (csg locus), metabolism (chuX, entB, entE, fepC), invasion (ompA), and chemotaxis (fliG) genes (Figure 3). Interestingly, AK867 showed a wider virulome, carrying the adhesion genes fyuA, irp-1 and -2, and the locus ybt for siderophores (Figure 3).

Figure 3 SNP-based phylogeny pictured using iTOL v6. Salmon circle grid: AMR genes. At the end of branches, red dots refer to CL3, light blue dots to CL2, and blue dots to CL1. On the squared grid (virulome), red = toxins; green = adhesion; yellow = metabolism; light green = siderophores; lilac = invasion; blue = chemotaxis.
4 Discussion
FosA3 is a subtype of the FosA enzyme family that, since its first report in 2010, is currently worldwide disseminated, especially in China (Hou et al., 2012; Mattioni Marchetti et al., 2023a). The major vehicles involved in dissemination are IncFII (Hou et al., 2012), followed by IncI1 (Sato et al., 2013), IncN (Liu et al., 2022), IncHI2 (Chen et al., 2021), and IncP (Hameed et al., 2022). Furthermore, it has been documented that IS26 plays a fundamental role in antimicrobial resistance genes (AMR) transposition and dissemination among Enterobacterales (Lv et al., 2020). Here, we depicted the genomic organization of a novel fosA3-harboring transposon on a pKPC-CAV1312 plasmids. As suggested by the available genomes on NCBI, pKPC-CAV1312 plasmids are likely to promote the entry and the subsequent fitting of fosA3 in Citrobacter spp. Interestingly, pfosA3_CFR867 did not show a perfect identity with others fosA3-mediated pKPC-CAV1312 plasmids, suggesting consistent rearrangements in the plasmid backbone structure. The plasmid-mediated fosA3 is generally organized in a composite transposon of 4 kb in size, consisting in two IS26 elements with the same orientation, flanking the cassette fosA3-orf1-orf2-Δorf3 (Wachino et al., 2010). In the present study, fosA3 was included in a large composite transposon of >12 kb that contained several orfs. The occurrence of fosA3 in large transposon, with the ability to carry different genes, and combined with the transposition potential of IS26, poses a further challenge in containing the spread of such emergent antibiotic-resistant strains at a global level.
The HNH endonucleases are a group of homing endonucleases that can act as selfish genetic elements, like transposons, breaking double-strand DNA and allowing the acquisition of functional attributes to the host cell, such as AMR genes (Edgell, 2009). The association between fosA-like genes and HNH endonucleases has already been pointed out in literature, speculating an undefined role of HNH in the dissemination of fosA genes, in absence of any surrounding insertion elements, within Citrobacter spp (Li et al., 2020; Mattioni Marchetti et al., 2023b). The pfosA3_CFR867 contained an HNH endonuclease, at a 6,530-bp distance from the fosA3 transposon.
In the present study, AK867 showed an XDR profile due to the co-presence of clinically relevant carbapenemases KPC-2 and VIM-1. In this prospective, the resistance to carbapenems and high FOS MIC in C. freundii could represent a novel menace and reduce the current antimicrobial armamentarium. This possibility is also strengthened by the easy transfer of such gene into the E. coli recipient, inducing an increase in MIC values beyond the current EUCAST breakpoints (resistance category with MIC >8 mg/L).
EUCAST clinical breakpoints for FOS underwent on a recent revision, with FOS cutoffs applicable on E. coli only and not recommending the use of FOS for other Enterobacterales than E. coli. However, FOS still represents a valid option in the treatment of urinary tract infection by ESBL-producing E. coli and C. freundii (Bielen and Likic, 2019). Moreover, FOS is also recognized as a valid option in combination therapy against several MDR Gram-negative infections, due to its relevant synergistic effect (Antonello et al., 2020). Based on the previous EUCAST breakpoints, C. freundii strains maintain high susceptibility levels to FOS (average MIC = 4 mg/L), but an eventual stabilization of transferable FosA enzymes may mark a turning point in the evolution of antibiotic resistance in C. freundii (Bielen et al., 2018).
In fact, cases of FosA-like enzymes in uncommon pathogens, as C. freundii, are slowly emerging in the literature (Chen et al., 2021; Mattioni Marchetti et al., 2023a; Mattioni Marchetti et al., 2023b). For this reason, despite the EUCAST revision, FosA enzyme detection and related transferability should be assessed even in low-risk pathogens, in order to track the transmission routes from these pathogens to clinically relevant clones, such as E. coli ST131 (Chudejova et al., 2024). Doubtless, large-scale surveillance on FOS-resistance profiles among Enterobacterales are demanding due to the lack of rapid kit as reliable as the reference ADM method. However, the coexistence of carbapenemases and FosA-like enzymes requires additional effort in clinical surveillance programs (Nakamura et al., 2014).
In conclusion, the incidence of FOS resistance is increasing globally among Enterobacterales, reaching and fitting even in low-risk pathogens, such as C. freundii. Together with carbapenem resistance, FOS resistance strains pose clinical challenges that, to be addressed, required dedicated surveillance programs and alternative rapid detection methods.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The study was designed and conducted in accordance with the Helsinki Declaration. The work described herein is molecular study performed on bacterial isolate from human sample that were obtained as part of routine hospital care and used anonymously. Consent to participate was not required, as samples were collected as part of standard patient care. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by-product of routine care or industry. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
VM: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. IV: Formal analysis, Writing – review & editing. TC: Data curation, Formal analysis, Writing – review & editing. MM: Data curation, Writing – review & editing. RM: Validation, Writing – review & editing. IB: Funding acquisition, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was supported by the research project grant NU23J-09-00067 provided by the Czech Health Research Council and by the project National Institute of Virology and Bacteriology (Programme EXCELES, ID Project No. LX22NPO5103)—funded by the European Union—Next Generation EU and by the EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT).
Acknowledgments
We thank Dr. Mario Sarti for the support provided to the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1447933/full#supplementary-material
Supplementary Figure 1 | Heatmap representation of plasmids content among 118 ST116 C. freundii. Light blue dots= absence; red dots= presence. Blue labels=CL1, light blue labels=CL2, red labels=CL3.
References
Antonello, R. M., Principe, L., Maraolo, A. E., Viaggi, V., Pol, R., Fabbiani, M., et al. (2020). Fosfomycin as partner drug for systemic infection management. A systematic review of its synergistic properties from in vitro and in vivo studies. Antibiotics (Basel) 9, 500. doi: 10.3390/antibiotics9080500
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Bakthavatchalam, Y. D., Shankar, A., Muthuirulandi Sethuvel, D. P., Asokan, K., Kanthan, K., Veeraraghavan, B. (2020). Synergistic activity of fosfomycin-meropenem and fosfomycin-colistin against carbapenem resistant Klebsiella pneumoniae: an in vitro evidence. Future Sci. OA 6, FSO461. doi: 10.2144/fsoa-2019-0074
Bielen, L., Likic, R. (2019). Experience with fosfomycin in the treatment of complicated urinary tract infections caused by extended-spectrum beta-lactamase-producing Enterobacteriaceae. Ther. Adv. Infect. Dis. 6, 2049936119858883. doi: 10.1177/2049936119858883
Bielen, L., Likić, R., Erdeljić, V., Mareković, I., Firis, N., Grgić-Medić, M., et al. (2018). Activity of fosfomycin against nosocomial multiresistant bacterial pathogens from Croatia: a multicentric study. Croat Med. J. 59, 56–64. doi: 10.3325/cmj.2018.59.56
Bitar, I., Caltagirone, M., Villa, L., Mattioni Marchetti, V., Nucleo, E., Sarti, M., et al. (2019). Interplay among IncA and blaKPC-carrying plasmids in Citrobacter freundii. Antimicrob. Agents Chemother. 63, e02609–e02618. doi: 10.1128/AAC.02609-18
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chen, L., Ou, B., Zhang, M., Chou, C. H., Chang, S. K., Zhu, G. (2021). Coexistence of fosfomycin resistance determinant fosA and fosA3 in Enterobacter cloacae isolated from pets with urinary tract infection in Taiwan. Microb. Drug Resist. 27, 415–423. doi: 10.1089/mdr.2020.0077
Chudejova, K., Caltagirone, M. S., Mattioni Marchetti, V., Rezzani, A., Navarra, A., Bitar, I. (2024). FosA8-producing E. coliST131: clinical cases in Italy, February 2023. Euro Surveill 29, 2400276. doi: 10.2807/1560-7917.ES.2024.29.21.2400276
Chudejova, K., Kraftova, L., Mattioni Marchetti, V., Hrabak, J., Papagiannitsis, C. C., Bitar, I. (2021). Genetic plurality of OXA/NDM-encoding features characterized from Enterobacterales recovered from Czech hospitals. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.641415
Dijkmans, A. C., Zacarías, N. V. O., Burggraaf, J., Mouton, J. W., Wilms, E. B., van Nieuwkoop, C., et al. (2017). Fosfomycin: pharmacological, clinical and future perspectives. Antibiotics 6, 24. doi: 10.3390/antibiotics6040024
Edgell, D. R. (2009). Selfish DNA: homing endonucleases find a home. Curr. Biol. 19, R115–R117. doi: 10.1016/j.cub.2008.12.019
Falagas, M. E., Roussos, N., Gkegkes, I. D., Rafailidis, P. I., Karageorgopoulos, D. E. (2009). Fosfomycin for the treatment of infections caused by Gram-positive cocci with advanced antimicrobial drug resistance: a review of microbiological, animal and clinical studies. Expert Opin. Investig. Drugs 18, 921–944. doi: 10.1517/13543780902967624
Hameed, M. F., Chen, Y., Bilal, H., Khan, S., Ge, H., Xiaofang, C., et al. (2022). The Co-occurrence of mcr-3 and fosA3 in IncP plasmid in ST131 Escherichia coli: a novel case. J. Infect. Dev. Ctries 16, 622–629. doi: 10.3855/jidc.15943
Hou, J., Huang, X., Deng, Y., He, L., Yang, T., Zeng, Z., et al. (2012). Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 56, 2135–2138. doi: 10.1128/AAC.05104-11
Li, Z., Lin, Y., Lu, L., Wang, K., Yang, L., Li, P., et al. (2020). Genetic characterisation of a complex class 1 integron in an NDM-1-producing Citrobacter freundii ST396 clinical strain isolated from a urine sample. J. Glob Antimicrob. Resist. 23, 64–66. doi: 10.1016/j.jgar.2020.08.002
Liu, F., Tian, A., Wang, J., Zhu, Y., Xie, Z., Zhang, R., et al. (2022). Occurrence and molecular epidemiology of fosA3-bearing Escherichia coli from ducks in Shandong province of China. Poult Sci. 101, 101620. doi: 10.1016/j.psj.2021.101620
Lv, L., Huang, X., Wang, J., Huang, Y., Gao, X., Liu, Y., et al. (2020). Multiple plasmid vectors mediate the spread of fosA3 in extended-spectrum-β-lactamase-producing enterobacterales isolates from retail vegetables in China. mSphere 5, e00507-20. doi: 10.1128/mSphere.00507-20
Mattioni Marchetti, V., Hrabak, J., Bitar, I. (2023a). Fosfomycin resistance mechanisms in Enterobacterales: an increasing threat. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1178547
Mattioni Marchetti, V., Kraftova, L., Finianos, M., Sourenian, T., Hrabak, J., Bitar, I. (2023b). Polyclonal spread of fosfomycin resistance among carbapenemase-producing members of the enterobacterales in the Czech Republic. Microbiol. Spectr. 11, e0009523. doi: 10.1128/spectrum.00095-23
Nakamura, G., Wachino, J., Sato, N., Kimura, K., Yamada, K., Jin, W., et al. (2014). Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J. Clin. Microbiol. 52, 3175–3179. doi: 10.1128/JCM.01094-14
Nobrega, D., Peirano, G., Matsumura, Y., Pitout, J. D. D. (2023). Molecular epidemiology of global carbapenemase-producing citrobacter spp. (2015-2017). Microbiol. Spectr. 11, e0414422. doi: 10.1128/spectrum.04144-22
Sato, N., Kawamura, K., Nakane, K., Wachino, J., Arakawa, Y. (2013). First detection of fosfomycin resistance gene fosA3 in CTX-m-producing Escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resist. 19, 477–482. doi: 10.1089/mdr.2013.0061
Shiju, K. S., Pallam, G., Mandal, J., Jindal, B., S, K. (2020). Use of fosfomycin combination therapy to treat multidrug-resistant urinary tract infection among paediatric surgical patients - a tertiary care center experience. Access Microbiol. 2, acmi000163. doi: 10.1099/acmi.0.000163
Singkham-In, U., Muhummudaree, N., Chatsuwan, T. (2020). fosA3 overexpression with transporter mutations mediates high-level of fosfomycin resistance and silence of fosA3 in fosfomycin-susceptible Klebsiella pneumoniae producing carbapenemase clinical isolates. PLoS One 15, e0237474. doi: 10.1371/journal.pone.0237474
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tonkin-Hill, G., Lees, J. A., Bentley, S. D., Frost, S. D. W., Corander, J. (2019). Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 47, 5539–5549. doi: 10.1093/nar/gkz361
Treangen, T. J., Ondov, B. D., Koren, S., Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524. doi: 10.1186/s13059-014-0524-x
Wachino, J., Yamane, K., Suzuki, S., Kimura, K., Arakawa, Y. (2010). Prevalence of fosfomycin resistance among CTX-m-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob. Agents Chemother. 54, 3061–3064. doi: 10.1128/AAC.01834-09
Wang, H., Min, C., Li, J., Yu, T., Hu, Y., Dou, Q., et al. (2021). Characterization of fosfomycin resistance and molecular epidemiology among carbapenem-resistant Klebsiella pneumoniae strains from two tertiary hospitals in China. BMC Microbiol. 21, 109. doi: 10.1186/s12866-021-02165-7
Yao, Y., Falgenhauer, L., Falgenhauer, J., Hauri, A. M., Heinmuller, P., Domann, E., et al. (2021). Carbapenem-resistant Citrobacter spp. as an emerging concern in the hospital-setting: results from a genome-based regional surveillance study. Front. Cell Infect. Microbiol., 11, 744431. doi: 10.3389/fcimb.2021.744431
Zhang, L. J., Gu, X. X., Zhang, J., Yang, L., Lu, Y. W., Fang, L. X., et al. (2020). Characterization of a fosA3 carrying IncC-IncN plasmid from a multidrug-resistant ST17 Salmonella Indiana isolate. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01582
Keywords: fosfomycin, Citrobacter freundii, carbapenemases, fosfomycin resistance, fosA3 gene
Citation: Mattioni Marchetti V, Venturelli I, Cassetti T, Meschiari M, Migliavacca R and Bitar I (2024) FosA3 emerging in clinical carbapenemase-producing C. freundii. Front. Cell. Infect. Microbiol. 14:1447933. doi: 10.3389/fcimb.2024.1447933
Received: 12 June 2024; Accepted: 12 July 2024;
Published: 06 August 2024.
Edited by:
Irena Maliszewska, Wrocław University of Science and Technology, PolandReviewed by:
Özgen Köseoglu Eser, Hacettepe University, TürkiyeMarie Louise Guadalupe Attwood, North Bristol NHS Trust, United Kingdom
Copyright © 2024 Mattioni Marchetti, Venturelli, Cassetti, Meschiari, Migliavacca and Bitar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vittoria Mattioni Marchetti, dml0dG9yaWEubWF0dGlvbmltYXJjaGV0dGlAdW5pcHYuaXQ=
 Vittoria Mattioni Marchetti
Vittoria Mattioni Marchetti Irene Venturelli
Irene Venturelli Tiziana Cassetti2
Tiziana Cassetti2 Roberta Migliavacca
Roberta Migliavacca Ibrahim Bitar
Ibrahim Bitar