- 1Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 2Trauma Center, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
Introduction: The emergence of disinfectant resistance has become a severe threat due to reduced effectiveness. This study was undertaken to determine how bacteria adapt to survive exposure to disinfectants in the busiest section of a tertiary care hospital in Varanasi, India.
Methods: Four isolates (two Klebsiella pneumoniae, Kp1 and Kp2; two Pseudomonas aeruginosa, Pa1 and Pa2) were obtained from chlorhexidine (CHX)–based handwash during microbiological surveillance of “in-use disinfectants” in hospital. Six disinfectants [4% CHX, 2% glutaraldehyde, 7.5% hydrogen peroxide, 1% sodium hypochlorite and 0.1% benzalkonium chloride (BAC), and 70% ethyl alcohol] were tested against these four isolates to determine minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). Antibiotic profile, change in MIC on exposure to disinfectants and biofilm formation in the presence and absence of disinfectants was studied. Whole genome sequencing (WGS) was done to identify the resistance mechanisms.
Result: The isolates showed the highest MBC/MIC ratio (4) against glutaraldehyde. Exposure to supra-inhibitory concentration of BAC for 21 days resulted in doubling of MIC/MBC. The majority (75%) of the isolates were multidrug resistant. All the isolates were strong biofilm producers. The reduction rate of biofilm formation decreased with an increase in the concentration of disinfectants (p = 0.05 for BAC). WGS revealed multiple AMR genes including blaDIM-1, disinfectant-resistant gene and efflux pump genes.
Conclusion: The study emphasized the various adaptation strategies of these isolates for survival in disinfectant environment, thus posing a huge challenge for their control in the hospital environment.
1 Introduction
The rapid emergence of antimicrobial resistance (AMR) is a global threat (Prestinaci et al., 2015). Along with it, there has been a parallel increase in disinfectant resistance resulting in their reduced effectiveness (Rozman et al., 2021). The driving force behind the sudden emergence of disinfectant resistance can be largely attributed to the abuse and misuse of disinfectants as well as lack of understanding of their resistance mechanisms (Carlie et al., 2020). The absence of a widely accepted definition for resistance to disinfectants is a critical hurdle, which might have contributed to the limited attention received from practitioners, administrators, and authorities on this challenging issue (Harbarth et al., 2014). Microorganisms, often dwell on hospital surfaces including patient surroundings and in biofilms where disinfectants cannot reach effortlessly (Tezel and Pavlostathis, 2015). The concentrations of disinfectants utilized in practice are much higher than the minimum inhibitory concentration (MIC) values, which often goes unnoticed (van Dijk and Verbrugh, 2022).
During routine microbiological surveillance in a tertiary care hospital, Gram-negative bacteria were found in “in-use” handwash. Consequently, we hypothesized that these isolates were adapted to survival mechanisms to overcome the disinfectant effect. With this background, the following study was undertaken to study in detail the adaptive strategies employed by the bacterial isolates to survive disinfectant exposures in the hospital environment through a series of phenotypic and genotypic techniques.
2 Materials and methods
2.1 Study site
This descriptive study was conducted in the Department of Microbiology and the associated tertiary care hospital in Varanasi, North India.
2.2 Bacterial isolates identification
During the microbiological surveillance of “in-use disinfectants” from major wards and intensive care units of the tertiary care hospital, four bacterial isolates were obtained from chlorhexidine (CHX)–based handwash. These were isolated from aseptically collected disinfectants from the dispensers in more than one of the busiest areas of the hospital with high patient footfall. Disinfectants were inoculated in blood agar and MacConkey agar (HiMedia Laboratories Pvt Ltd, India) and incubated at 37°C. Organisms were identified by standard biochemical tests (Crichton, 1996). The lot number of the dispensers was checked and noted.
2.3 In-use testing of disinfectant contamination
To check the “in-use” CHX-based disinfectants, “in-use testing” was done for the entire batch from the store with the same lot number as well as from those collected in the surveillance. One milliliter of CHX-based disinfectant was added to 9 mL of the media along with 1mL of 5% (w/v) autoclaved yeast suspension. A volume of 0.02 mL was placed on each of the two nutrient agar plates. One of the plates was incubated at 37°C for 3 days and the other at room temperature for 7 days. Five or more colonies on each plate indicated contamination (Rao S, 2008).
2.4 Determination of MIC against disinfectants
This was done to determine the MIC of the four isolates using the macro broth dilution method against six commonly used disinfectants (Chemical disinfectants from the Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008). Benzalkonium chloride (BAC), sodium hypochlorite, glutaraldehyde, CHX, (all from Sigma-Aldrich Chemicals Pvt. Ltd, India) hydrogen peroxide and ethyl alcohol (both from Thermo Fisher Scientific India Pvt. Ltd.). The appropriate concentration of the disinfectants was prepared by adding distilled water to give the strength of the chemicals as per the effective strength of disinfectant (Rani et al., 2017). Serial twofold dilutions of the disinfectants (4% CHX, 2% Glutaraldehyde, 7.5% hydrogen peroxide, 1% sodium hypochlorite and 0.1% BAC, and 70%–30% ethyl alcohol) were prepared and mixed with the inoculum which was prepared by mixing 1–2 colonies in normal saline to match 0.5 McFarland standard (1.5 × 108 c.f.u. mL−1). Bacterial suspensions at the log growth phase were incubated with serial dilutions of disinfectants in tubes and incubated at 37°C overnight to observe visible growth. For quality control, Escherichia coli ATCC® 25922 and Klebsiella pneumoniae ATCC® 70063 were used.
2.5 Determination of MBC
The MBC analysis was performed using a modified version of the method described in the BSAC Susceptibility Guide Testing (Andrews, 2001). Following the MIC incubation period, MBC was determined by plating an aliquot of 10 μL on nutrient agar plates from each tube demonstrating no visible growth. Resultant colonies were counted after incubation at 37°C overnight. The MBC endpoint was defined as the lowest concentration of the antimicrobial agent showing at least a 99.9% killing of the initial inoculums where no visible growth of the bacteria was observed.
2.6 Antibiotic susceptibility testing profile
Susceptibility toward amoxicillin/clavulanic acid (AMC, 20/10 μg), gentamicin (GEN, 10 μg), ciprofloxacin (CIP, 5 μg), levofloxacin (LE, 5 μg), ceftazidime (CAZ, 30 μg), cefepime (CPM, 30 μg) ceftriaxone (CTR, 30 μg), cotrimoxazole (TMP-SMX 1.25/23.75 μg), piperacillin/tazobactam (PTZ, 100/10 μg), imipenem (IPM, 10 μg), meropenem (MEM, 10 μg), amikacin (AK, 30 μg), ertapenem (ETP 30 μg) (HiMedia Laboratories Pvt. Ltd, India) was tested by Kirby Bauer disc diffusion method. For quality control, E. coli ATCC® 25922 and K. pneumoniae ATCC® 70063 were used. Results were interpreted according to CLSI guidelines 2024 (Clinical Laboratory and Standards Institute, 2024).
2.7 Change in MIC under selective pressure of disinfectants
Determination of MIC under selective pressure of disinfectants was done to note changes in MIC if any. For this, organisms were grown in the presence of the disinfectants. The specific MIC chosen were, half of the MIC value for subinhibitory concentration and twice the MIC for supra-inhibitory concentration. Alcohol (ethyl), Hypochlorite, CHX, and BAC were used for this experiment. MIC was performed in cation-adjusted Mueller Hinton broth (CAMHB) containing tubes with specific concentrations of disinfectants and incubated at 37°C as detailed before. MIC determination was repeated after every 7 days of exposure for a period of 30 days (Merchel Piovesan Pereira et al., 2021).
2.8 Evaluation of formation of the biofilm in the presence and absence of disinfectants
Biofilm forming capacity of the isolates were tested with and without CHX and BAC at different concentrations. Briefly, overnight cultures were diluted to get 1.5 × 108 colony-forming units (CFUs)/mL bacterial suspensions. In a microtiter plate containing 96-well (Tarsons Products Limited, India), the CHX-based disinfectant with concentration ranging from 4% (W/V) to 0.12% (W/V) and BAC ranging from 0.1% (w/v) to 0.02% (w/v) was diluted in freshly prepared Brain Heart infusion broth media supplemented with 0.25% glucose. Ten microliter bacterial suspension was added to each well keeping the final volume at 200 μL. As positive control the disinfectant-free medium containing bacterial suspension was used and wells with only broth medium were considered as negative controls. The activity of the CHX and BAC on biofilm formation was examined on standardized low, moderate, and high biofilm-producing strains of Staphylococcus aureus. After a 48h of incubation period, wells were washed thrice with sterile phosphate buffer saline (PBS) and allowed to air dry before stained with 200 μL of 0.1% crystal violet followed by 20 min of incubation at room temperature and washed twice with PBS. The excess stain was solubilized with 95% ethanol. The plates were again incubated for 20 min at room temperature (Stepanovic et al., 2007). The optical density at 578 nm (OD578) of each well was measured using a microtiter plate reader. (LisaScan® EM Elisa plate reader, Transasia Bio-Medicals Ltd, India). The formation of biofilm was calculated by the following formula (Singh et al., 2017):
ODcut = ODavg of negative control + 3 × standard deviation of ODs of negative control.
OD ≤ ODcut = Non−biofilm−former
ODcut < OD ≤ 2 × ODcut = Weak biofilm former
2 × ODcut < OD ≤ 4 × ODcut = Moderate biofilm former
OD >4 × ODcut = Strong biofilm former.
Percentage reduction in biofilm formation was calculated by (Nunes S de et al., 2021):
2.9 Whole genome sequencing
WGS was performed using Illumina platform (outsourced to Bionivid Technology Private Limited, Bengaluru, India). Fastp 0.23.0 was used for quality control, adapter trimming, and data filtering (Chen et al., 2018). alignment quality was checked by using samtools 1.20 (Kim et al., 2019). Annotation of genomes were done using SnpEff (Cingolani et al., 2012). Variant detection was done using VarScan 2.4.6 (Koboldt et al., 2009).
2.9.1 WGS analysis tools
RGI-CARD (Comprehensive Antibiotic Resistance Database) and ResFinder 4.1 (https://cge.food.dtu.dk/services/ResFinder/) were used to predict resistance genes in assembled bacterial genomes. Multi-locus sequence typing (MLST) patterns were identified using MLST 2.0.9 (https://cge.food.dtu.dk/services/MLST/).
2.10 Statistical analysis
Statistical analysis was performed by ANOVA to compare the biofilm reduction rates, using Microsoft Excel (Version: Microsoft Corporation 2019). Results with a p-value ≤ 0.05 were considered statistically significant.
3 Results
3.1 Isolate details
During the microbiological surveillance, disinfectant samples from 18 separate dispensers distributed in different locations of the hospital were collected. Among these, the four bacterial strains isolated from four sites, namely, Kp1, Kp2, Pa1, and Pa2 were identified as 2 K. pneumoniae and 2 P. aeruginosa. During the same surveillance, gram-negative bacteria were not found on any of the other hospital surfaces.
3.2 In-use testing
The four disinfectants showed the presence of the same bacteria thus implying contamination with colony count >5. However, no contamination was detected in the unused disinfectants of the same batch.
3.3 Determination of MIC and MBC against selected disinfectants
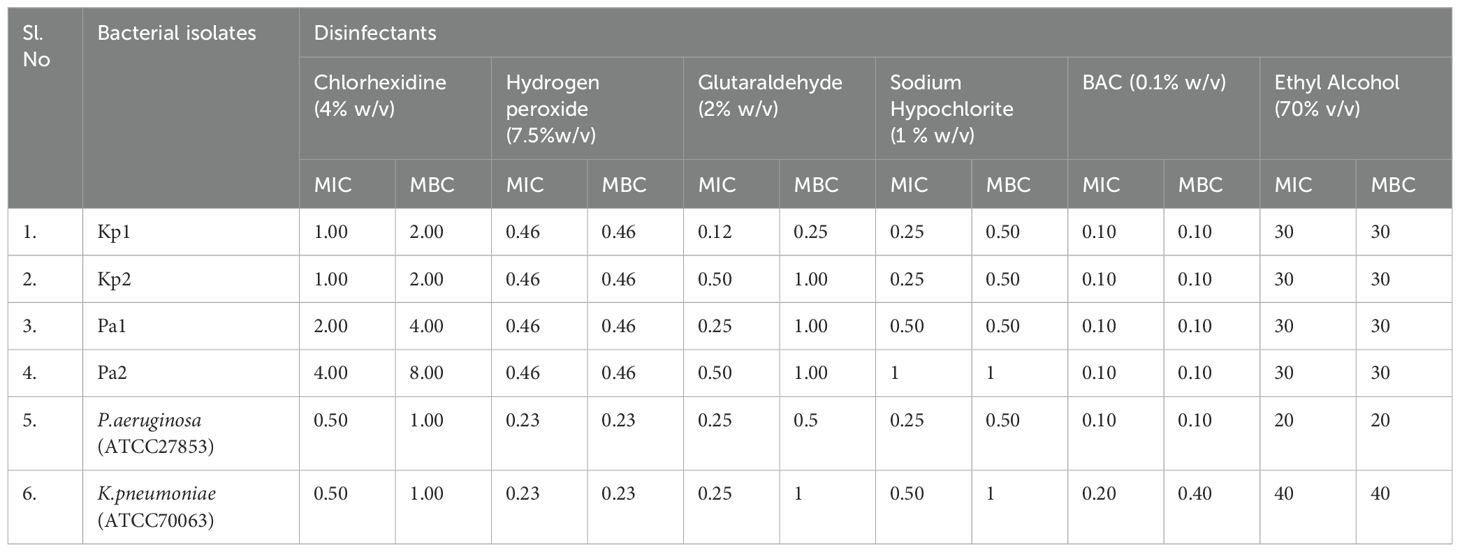
The susceptibility testing demonstrated the MIC values of the CHX ranging from 1% (w/v) to 4% (w/v), for Hydrogen peroxide 0.23% (w/v) to 0.40% (w/v), for sodium hypochlorite 0.25% (w/v) to 1% (w/v), for BAC 0.10% (w/v), for glutaraldehyde 0.12% (w/v) to 0.50% (w/v), and for ethyl alcohol it was 30% against the different test organisms, respectively. The highest MBC/MIC ratio of the CHX, sodium hypochlorite, and BAC against test isolates was 2; for hydrogen peroxide and ethyl alcohol, it was 1; and for glutaraldehyde was 4. The complete data on MIC and MBC values are shown in Table 1.
3.4 AST profile
Among the 2 K. pneumoniae isolates, resistance was noted for AK, LE, IPM, MEM, and TMP-SMX against Kp2 isolate. The other isolate was susceptible to all antibiotics. For the two P. aeruginosa isolates, resistance was noted against PIP, PTZ, LE, MEM, IPM, and CAZ for both the isolates.
3.5 Change in MIC under selective pressure of disinfectants
Changes in MIC and MBC increased to twofolds for Kp2 against BAC after 21 days of exposure. However, no MIC and MBC changes were seen in the other isolates against the tested disinfectants (BAC, CHX, sodium hypochlorite, and ethyl alcohol).
3.6 Evaluation of biofilm formation in the presence and absence of disinfectants
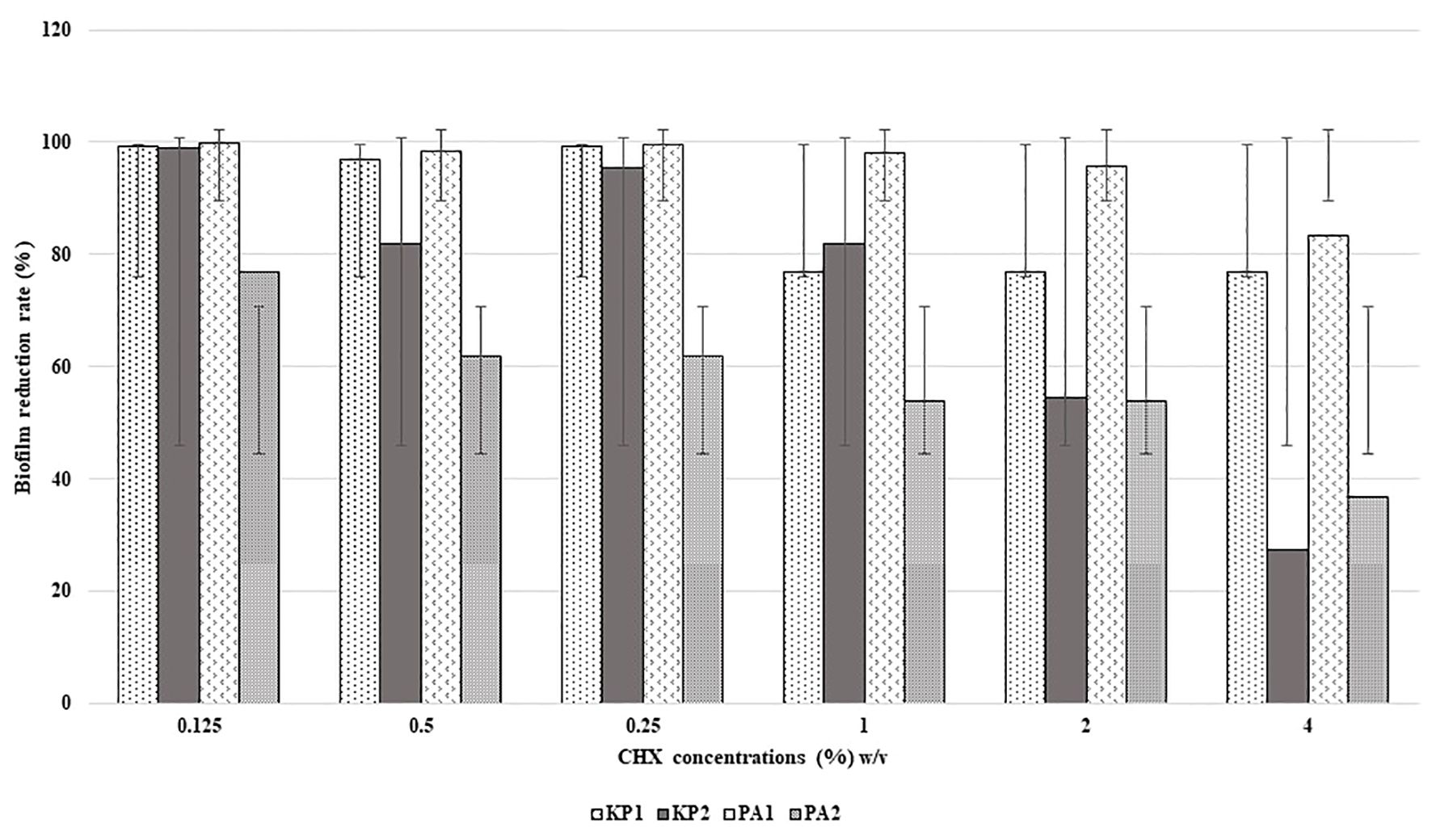
All the isolates were found to be high biofilm producers compared to high and moderate biofilm-producing strains, in the presence and absence of disinfectants. The rate of reduction of biofilm formation decreased with an increase in the concentration of disinfectants as shown in Figures 1 and 2. BAC showed a significant formation (P = 0.05) of biofilm at higher concentrations.
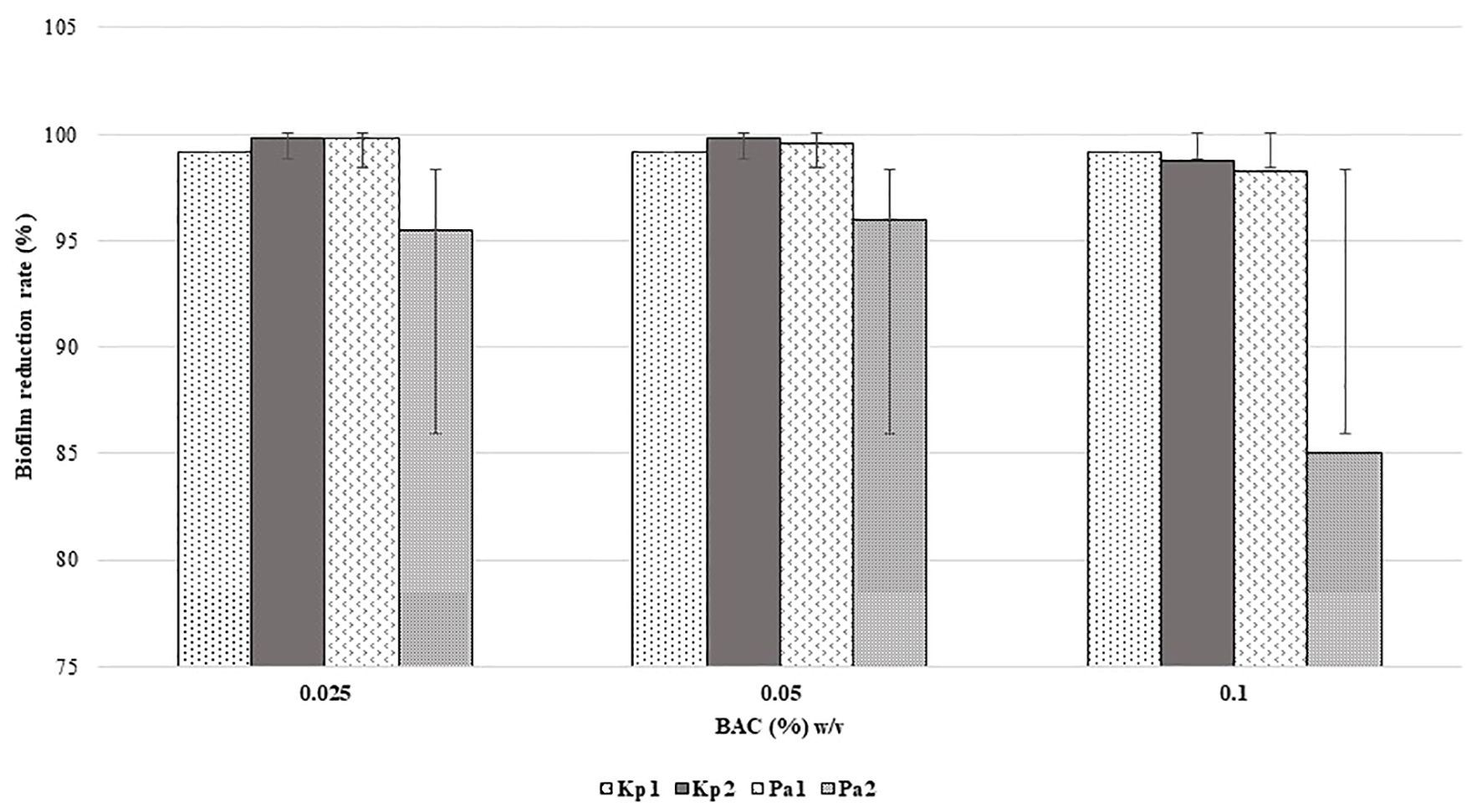
Figure 2. Reduction in biofilm formation according to concentration of BAC. (p = 0.05, two-way ANOVA applied).
3.7 Analysis of WGS results
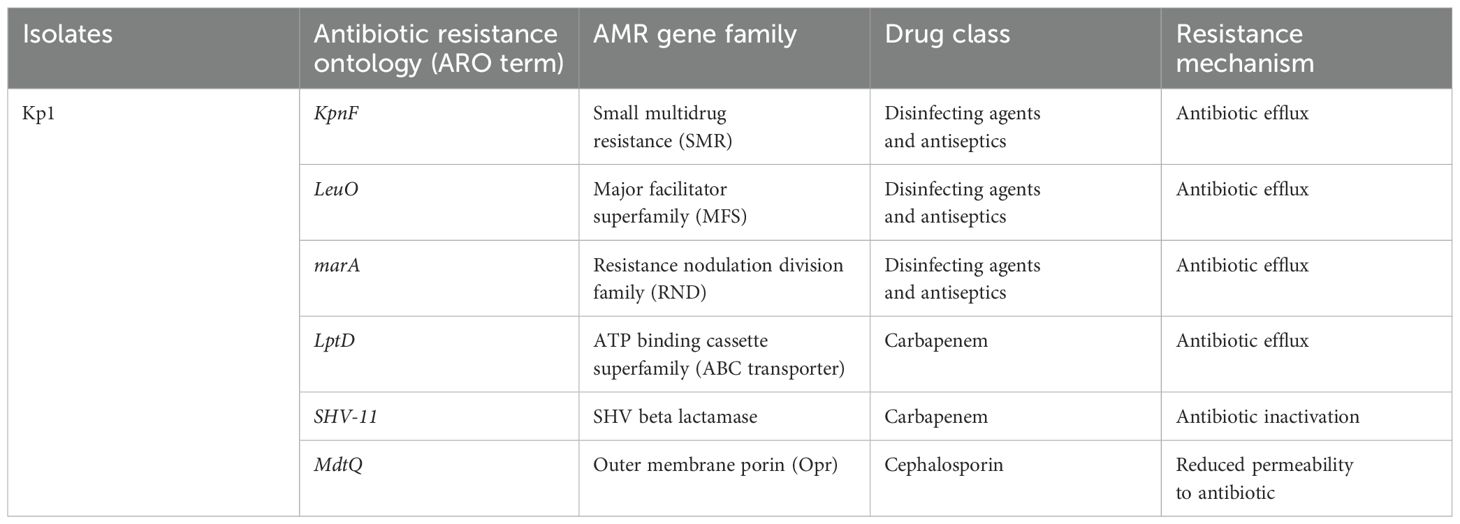
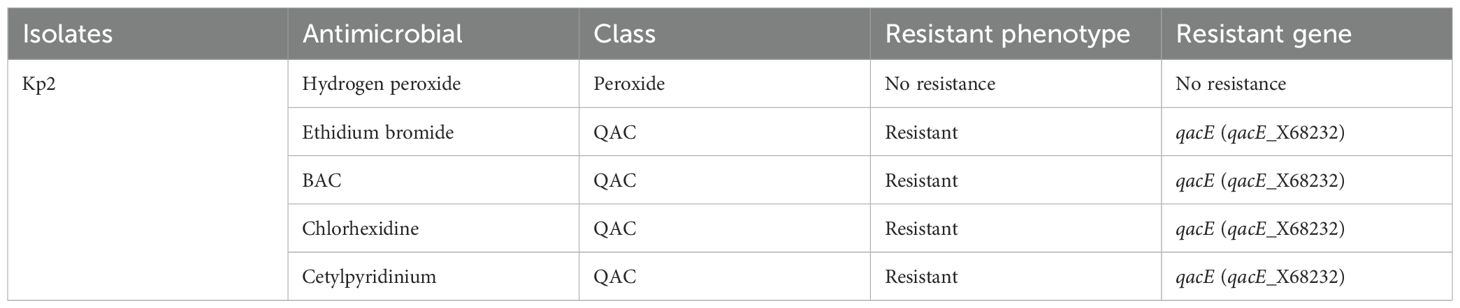
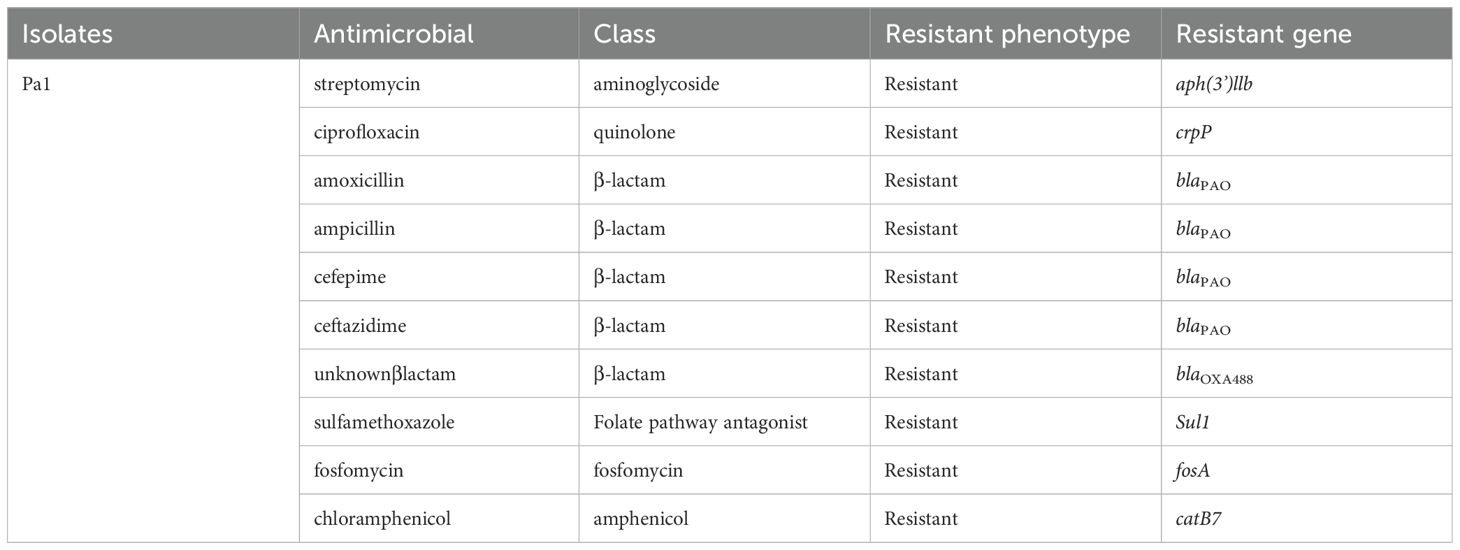
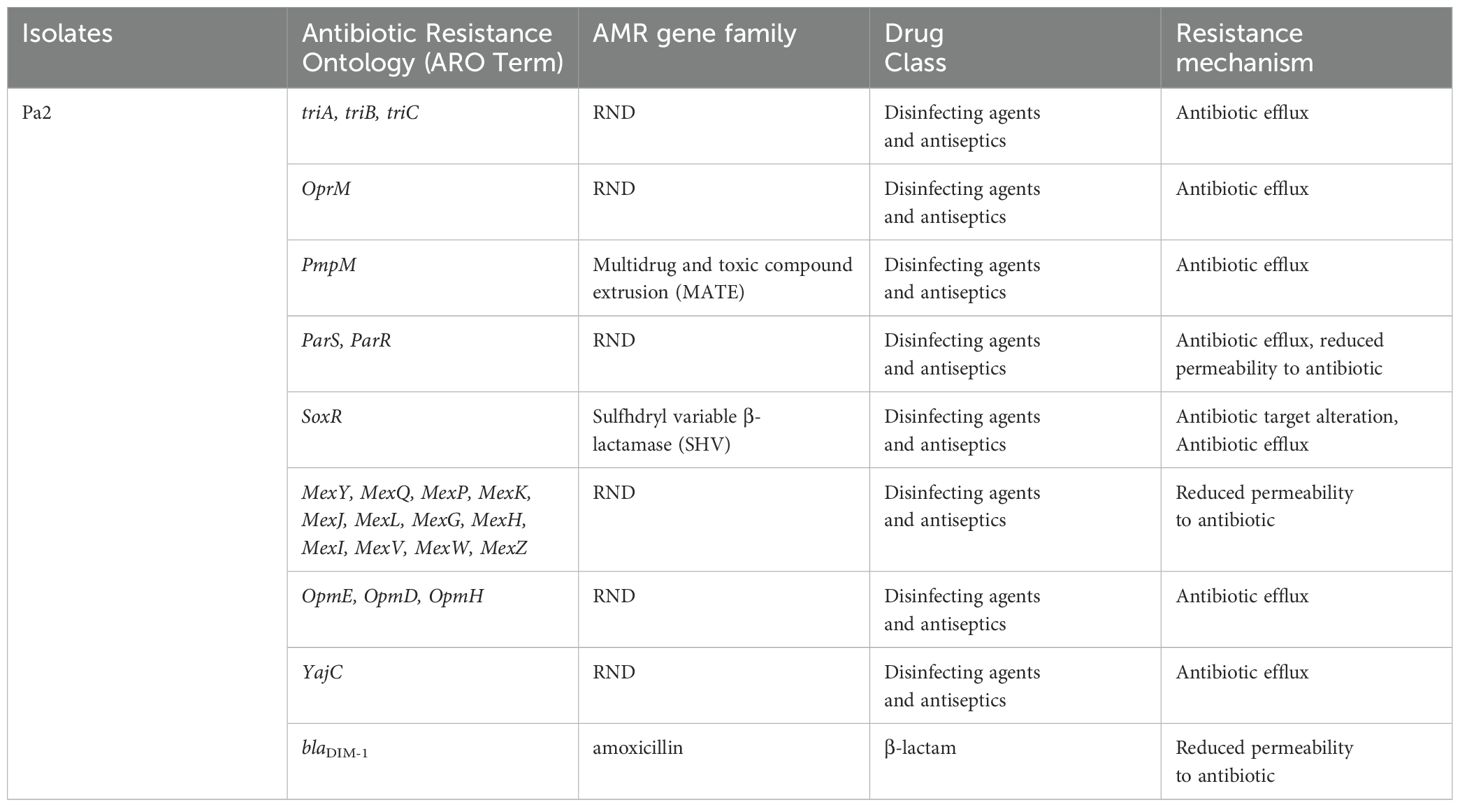
Isolate Kp1 was found to carry KpnE of SMR family, leuO, acrA, and marA of RND family. Sequence type (ST) was unknown of this isolate. MLST for Kp2 was ST 2676 and was found to harbor QAC-resistant gene qacE and antibiotic-resistant genes like β-lactam resistant gene blaOXA-1, aminoglycoside resistant gene aph(3’)-la, blaDHA-1, blaOKP-A, fosfomycin resistant gene fosA, quinolone-resistant gene qnrB1, qnrB4, sulfonamide resistant gene sul-1, trimethoprim resistant gene dfrA, and tetracycline-resistant gene triA. Isolate Pa1 belonged to ST 1047 and showed presence of antibiotic-resistant genes like aph, crpP, blaPAO, blaOXA-488, sul-1, fosA, and catB. Pa2 had tri genes, mex genes, OprM, outer membrane porins, and yajC genes conferring resistance to RND family, soxR conferring resistance to SHV family, PmpM conferring resistance to MATE family. Antibiotic-resistant genes of β-lactam family blaDIM-1, blaPAO, blaOXA488, for aminoglycoside family ant(2’’)-Ia, aph (6)-Id were detected and ST of this isolate was ST 4249. Resistant genes of Kp1, Kp2, Pa1, and Pa2 have been shown in Tables 2–5 respectively and resistance mechanism along with genes for Kp1, Kp2, Pa1, and Pa2 has been shown in representative Supplementary Figures S1–S4 respectively.
4 Discussion
This study clearly demonstrated the ability of the bacterial contaminants to persist in the presence of the disinfectants, particularly CHX and BAC. Bacterial tolerance to BAC increased on prolonged exposure to the disinfectant and the rate of reduction in bacterial biofilm formation decreased with increasing concentration of disinfectants. On these aspects, the study is important because there is very limited data on bacterial responses towards disinfectants. In this context, the present study highlighted the following points.
Firstly, there is lack of standard definition/criteria of disinfectant resistance in the scientific community, which could be one of the reasons for scarce literature on this aspect (van Dijk and Verbrugh, 2022). Studies have either compared MIC/MBC values of the test isolates with those of the standard isolates or have studied their exact MIC values at varying disinfectant concentrations. In this study, though MIC was not overtly raised in any of the isolates MBC was double the MIC for CHX, glutaraldehyde, and sodium hypochlorite (isolates Kp1, Kp2). This increase in MBC could hint towards the increasing “tolerance” to disinfectants in these isolates (Nordholt et al., 2021). The MIC/MBC values of all the isolates against CHX, hydrogen peroxide, and alcohol were more as compared to the standard strains. There have been reports of emerging disinfectant resistance across the globe (Tong et al., 2021). Varying resistance rates (1–256µg/mL) for P. aeruginosa against CHX has been reported (Mal et al., 2016). Similarly, a study on 126 isolates of K. pneumoniae showed high MIC values and decreased susceptibility in 90% of isolates (Naparstek et al., 2012). A recent Indian study documented higher MIC values for 12.5% and 50% of K. pneumoniae, 80% and 20% of P. aeruginosa against 70% ethanol and sodium hypochlorite respectively (Gupta and Bhatia, 2018). It has been seen that the concentrations at which disinfectants are used in the hospital are considerably higher than their MIC values. Yet, they do not assure successful disinfection procedure (van Dijk and Verbrugh, 2022).
For BAC, comparable MIC/MBC values were noted against the standard isolates. Both lower (0.02–0.2) and higher MIC (30–120) values have been noted in studies and higher MIC (30–120) has been noted in studies in India and the USA (Rajamohan et al., 2010; Gupta and Bhatia, 2018). However, it was interesting to note that one K. pneumoniae (Kp2) isolate showed double increase in the MIC and MBC of BAC following 21 days of exposure to supra-inhibitory concentration of BAC. It has been advocated that exposure to subinhibitory concentration of disinfectants, which is mostly seen at the periphery of the disinfected area, can initiate several stress responses in the bacteria (Koboldt et al., 2009). On WGS, this isolate Kp2 had shown the presence of qac genes the expression of which could have accounted for this variation in MIC/MBC. In this context, study had shown 53% presence of qacE gene responsible for biocide resistance in K. pneumoniae isolates (Abuzaid et al., 2012). The qac genes have been reported widely for causing disinfection tolerance. This plasmid-mediated group of genes are also responsible for dissemination of disinfection resistance. The qacE gene has been located often in association with integron, thus increasing the risk of horizontal transmission (Tong et al., 2021).
Thirdly, differences in disinfectant susceptibility might be accompanied by antibiotic resistance by a phenomenon of cross-resistance. As shown by AST and WGS, these isolates harbored various AMR genes of importance. In this study, three of the four isolates were resistant to multiple antibiotics. WGS revealed several genes that could account for this resistance. The presence of multidrug-resistant (MDR) genes in clinical isolates of K. pneumoniae and P. aeruginosa had been a known fact. One of the isolate (Kp1) carried multiple genes for antibiotic efflux pump like LptD, SHV-11, MdtQ, which could explain the resistance to beta lactam agents like cephalosporins and carbapenems in this isolate. The Pseudomonas isolates were widely resistant to beta lactam antibiotics, aminoglycosides, fosfomycin and also possessed genes like blaPAO, aminoglycoside phosphotransferases corresponding to their phenotypic resistance. In this context, the study also revealed the presence of blaDIM-1 gene in Pa2, a less studied carbapenem-encoding gene. In India, blaDIM-1 was first identified in a P. stutzeri strain collected in 2000 and reported in 2014 (Deshpande et al., 2014). As carbapenems are considered to be one of the last resorts of antibiotics for P. aeruginosa infections, the identified blaDIM-1 gene in this study is quite concerning.
Besides, several disinfectant resistance genes conferring resistant to the efflux pump families were also found in these isolates. They carried genes from all the five efflux pump membrane protein families (RND, MFS, MATE, SMR, and ABC). These efflux pumps have been noted to affect disinfection activity of the biocides against the organisms harboring them (Tong et al., 2021). As efflux pumps are one of the most effective resistance mechanisms that bacteria employ against stress, it could be possible that the diversity in these pumps was an adaptation in these isolates to survive within the disinfectant environment. Additionally, Pa1 belonged to ST 1047, which is a globally circulating clone of MDR strains especially in the Southeast Asian region (Tada et al., 2019; Takahashi et al., 2021).
Last, the study reinstated the importance of biofilm formation as an adaptation strategy for survival in the hospital environment. All the four bacterial isolates were high biofilm producers both in the presence and absence of disinfectants namely CHX and BAC. When biofilm production was assessed at increasing concentration of these disinfectants, it was very interesting to note that the rate of reduction of biofilm formation decreased as the concentration of disinfectants increased. This phenotypic presentation could be one of the most important strategy of these bacterial isolates to survive in the disinfectant environment. It could be possible that the possession of several types of efflux pumps as detected by WGS, which are implicated in biofilm production, could have been activated at subinhibitory concentration of disinfectants thus promoting more biofilm formation (Sekiya et al., 2003). A study on expression profile of these pumps under these circumstances, which was not within the scope of the present study, could confirm the prediction. This finding also raises concern as use of disinfection in appropriate dilutions is an existing issue. If bacteria like P. aeruginosa, K. pneumoniae can survive the working concentrations of disinfectants through biofilm formation, rapid emergence of resistance is inevitable. However, unless there is considerable baseline data on disinfectant resistance/tolerance that is widely prevalent, it would be very difficult to comment on whether disinfectant resistance is emerging or already existing.
The study was not without limitations. While we could not locate the exact area where the bacteria were residing in the disinfectant container which could have translated the expected finding into the real scenario. We could only access the biofilm formation ability of the organisms, while visualization of the real biofilm structure would have added more value. Moreover, experiments of efflux pumps could not be done which could have confirmed the phenotypic presentations. Nevertheless, the study is of utmost importance because to the best of the knowledge, it is the first study on adaptation of bacterial isolates in the disinfectant environment of the hospital, highlighting the different strategies adopted by the bacteria to persist.
5 Conclusion
Bacterial isolates, 2 K. pneumoniae, and 2 P. aeruginosa were found to survive in CHX-based disinfectants in the hospital environment. All the four isolates had increased MIC/MBC against glutaraldehyde and sodium hypochlorite. Exposure to supra-inhibitory concentration of BAC resulted in doubling of MIC/MBC. All the isolates were strong biofilm producers both in the presence and absence of disinfectants with reduced rate of reduction in biofilm formation at increasing concentrations. WGS revealed multiple AMR genes including blaDIM-1, disinfectant-resistant gene and efflux pump genes in these isolates. The study emphasized the various adaptation strategies of these isolates for survival in disinfectant environment, thus posing a huge challenge for their control in the hospital environment.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The study was approved by the Institute ethical committee via letter No. Dean/2023/EC/6834.
Author contributions
PR: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. AS: Formal analysis, Writing – original draft, Software. RS: Writing – original draft, Investigation, Resources. TB: Conceptualization, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are grateful to Banaras Hindu University, India for providing the facility to conduct this study. We also acknowledge the IoE phase II grant for consumables.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The AE DK declared a shared parent affiliation with the authors at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1442914/full#supplementary-material
References
Abuzaid, A., Hamouda, A., Amyes, S. G. B. (2012). Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE effluxpump genes and antibiotic resistance. J. Hosp. Infect. 81, 2. doi: 10.1016/j.jhin.2012.03.003
Andrews, J. (2001). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48, 5–16. doi: 10.1093/jac/48.suppl_1.5
(2008). Chemical disinfectants from the Guideline for Disinfection and Sterilization in Healthcare Facilities. Available online at: https://www.cdc.gov/infection-control/hcp/disinfection-sterilization/chemical-disinfectants.html (Accessed May 1st, 2024).
Carlie, S. M., Boucher, C. E., Bragg, R. R. (2020). Molecular basis of bacterial disinfectant resistance. Drug Resistance Updates. 48, 100672. doi: 10.1016/j.drup.2019.100672
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, 17. doi: 10.1093/bioinformatics/bty560
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain. Fly Austin. 6, 2. doi: 10.4161/fly.19695
Clinical Laboratory and Standards Institute (2024). “Performance standard for antimicrobial susceptibility testing,” in CLSI supplement M100, 34th (Clinical and Laboratory Standards Institute, Wayne, PA), 58.
Crichton, P. B. (1996). “Enterobacteriaceae: Escherichia, Klebsiella, Proteus and other genera,” in Mackie and McCartney practical medical microbiology, fourteenth. Eds. Collee, J. G., Mackie, T. J., McCartney, J. E. (Churchill Livingstone, London, UK), 361–364.
Deshpande, L. M., Jones, R. N., Woosley, L. N., Castanheira, M. (2014). Retrospective molecular analysis of DIM-1 metallo-β-lactamase discovered in pseudomonas stutzeri from India in 2000. Antimicrob. Agents Chemother. 58, 1. doi: 10.1128/AAC.01541-13
Gupta, P., Bhatia, M. (2018). Emerging biocide resistance among multidrug-resistant bacteria: myth or reality?A pilot study. J. Pharm. Bioallied Sci. 10, 2. doi: 10.4103/JPBS.JPBS_24_18
Harbarth, S., Tuan, S. S., Horner, C., Wilcox, M. H. (2014). Is reduced susceptibility to disinfectants and antiseptics a risk in healthcare settings? A point/counterpoint review. J. Hosp. Infect. 87, 4. doi: 10.1016/j.jhin.2014.04.012
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 8. doi: 10.1038/s41587-019-0201-4
Koboldt, D. C., Chen, K., Wylie, T., Larson, D. E., McLellan, M. D., Mardis, E. R., et al. (2009). VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 17. doi: 10.1093/bioinformatics/btp373
Mal, P., Farooqi, J., Irfan, S., Hughes, M., Khan, E. (2016). Reduced susceptibility to chlorhexidine disinfectant among New Delhi metallo-beta-lactamase-1 positive Enterobacteriaceae and other multidrug-resistant organisms: Report from a tertiary care hospital in Karachi, Pakistan. Indian J. Med. Microbiol. 34, 3. doi: 10.4103/0255-0857.188338
Merchel Piovesan Pereira, B., Wang, X., Tagkopoulos, I. (2021). Biocide-induced emergence of antibiotic resistance in escherichia coli. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.640923
Naparstek, L., Carmeli, Y., Chmelnitsky, I., Banin, E., Navon-Venezia, S. (2012). Reduced susceptibility to chlorhexidine among extremely-drug-resistant strains of Klebsiella pneumoniae. J. Hosp. Infect. 81, 1. doi: 10.1016/j.jhin.2012.02.007
Nordholt, N., Kanaris, O., Schmidt, S. B. I., Schreiber, F. (2021). Persistence against benzalkonium chloride promotes rapid evolution of tolerance during periodic disinfection. Nat. Commun. 12, 1. doi: 10.1038/s41467-021-27019-8
Nunes S de, O., Rosa H da, S., Canellas, A. L. B., Romanos, M. T. V., dos Santos, K. R. N., Muricy, G., et al. (2021). High reduction of staphylococcal biofilm by aqueous extract from marine sponge-isolated Enterobacter sp. Res. Microbiol. 172, 1. doi: 10.1016/j.resmic.2020.10.002
Prestinaci, F., Pezzotti, P., Pantosti, A. (2015). Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob Health 109, 7. doi: 10.1179/2047773215Y.0000000030
Rajamohan, G., Srinivasan, V. B., Gebrey, W. A. (2010). Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J. Antimicrob. Chemother. 65, 2. doi: 10.1093/jac/dkp427
Rani, R., Sharma, D., Chaturvedi, M., Yadav, J. P. (2017). Antibacterial activity of twenty different endophytic fungi isolated from Calotropis procera and time kill assay. Clin. Microbiol. 6, 3. doi: 10.4172/2327-5073.1000280
Rao S, N. P. (2008). Testing of disinfectants. Available online at: https://www.microrao.com/micronotes/pg/testing_of_disinfectants.pdf (Accessed January 15,2024).
Rozman, U., Pušnik, M., Kmetec, S., Duh, D., Šostar Turk, S. (2021). Reduced susceptibility and increased resistance of bacteria against disinfectants: A systematic review. Microorganisms. 10, 12. doi: 10.3390/microorganisms9122550
Sekiya, H., Mima, T., Morita, Y., Kuroda, T., Mizushima, T., Tsuchiya, T. (2003). Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a pseudomonas aeruginosa mutant. Antimicrob. Agents Chemothe. 47, 2990–2. doi: 10.1128/AAC.47.9.2990-2992.2003
Singh, A. K., Prakash, P., Achra, A., Singh, G. P., Das, A., Singh, R. K. (2017). Standardization and classification of In vitro biofilm formation by clinical isolates of Staphylococcus aureus. J. Global Infect. Dis. 9, 3. doi: 10.4103/jgid.jgid_91_16
Stepanovic, S., Vukovic, D., Hola, V., Di Bonaventura, G., Djukic, S., Cirkovic, I., et al. (2007). Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. APMIS 115, 8. doi: 10.1111/j.1600-0463.2007.apm_630.x
Tada, T., Hishinuma, T., Watanabe, S., Uchida, H., Tohya, M., Kuwahara-Arai, K., et al. (2019). Molecular characterization of multidrug-resistant pseudomonas aeruginosa isolates in hospitals in Myanmar. Antimicrob. Agents Chemother. 63, 5. doi: 10.1128/AAC.02397-18
Takahashi, T., Tada, T., Srestha, S., Hishinuma, T., Sherchan, J. B., Tohya, M., et al. (2021). Molecular characterisation of carbapenem-resistant pseudomonas aeruginosa clinical isolates in Nepal. J. Global Antimicrobial Resistance. 26, 279–84. doi: 10.1016/j.jgar.2021.07.003
Tezel, U., Pavlostathis, S. G. (2015). Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 33, 296–304. doi: 10.1016/j.copbio.2015.03.018
Tong, C., Hu, H., Chen, G., Li, Z., Li, A., Zhang, J. (2021). Disinfectant resistance in bacteria: Mechanisms, spread, and Resolution Strategies. Environ. Res. 195. doi: 10.1016/j.envres.2021.110897
Keywords: disinfectant resistance, MBC, WGS, chlorhexidine, benzalkonium chloride, biofilm
Citation: Rakshit P, Singh A, Singh R and Banerjee T (2024) An in-depth study on survival mechanism of bacterial isolates in disinfectants within the hospital environment. Front. Cell. Infect. Microbiol. 14:1442914. doi: 10.3389/fcimb.2024.1442914
Received: 03 June 2024; Accepted: 23 July 2024;
Published: 15 August 2024.
Edited by:
Divya Kushwaha, Banaras Hindu University, IndiaReviewed by:
Mujde Eryilmaz, Acıbadem University, TürkiyeSahana Vasudevan, Institute for Stem Cell Science and Regenerative Medicine (inStem), India
Copyright © 2024 Rakshit, Singh, Singh and Banerjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tuhina Banerjee, ZHJ0dWhpbmFAeWFob28uY29t; ZHJ0dWhpbmExMDVAYmh1LmFjLmlu
 Pue Rakshit
Pue Rakshit Aradhana Singh1
Aradhana Singh1 Tuhina Banerjee
Tuhina Banerjee