- 1Institute of Bioinformatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 2Medical Molecular Biology Laboratory, School of Medicine, Jinhua Polytechnic, Jinhua, China
- 3Department of Laboratory Sciences, The People’s Hospital of Yuhuan, Yuhuan, China
- 4Department of Laboratory Sciences, Pingyang Hospital of Wenzhou Medical University, Pingyang, China
Background: Aminoglycoside-modifying enzymes (AMEs) play an essential role in bacterial resistance to aminoglycoside antimicrobials. With the development of sequencing techniques, more bacterial genomes have been sequenced, which has aided in the discovery of an increasing number of novel resistance mechanisms.
Methods: The bacterial species was identified by 16S rRNA gene homology and average nucleotide identity (ANI) analyses. The minimum inhibitory concentration (MIC) of each antimicrobial was determined by the agar dilution method. The protein was expressed with the pCold I vector in E. coli BL21, and enzyme kinetic parameters were examined. The whole-genome sequence of the bacterium was obtained via the Illumina and PacBio sequencing platforms. Reconstruction of the phylogenetic tree, identification of conserved functional residues, and gene context analysis were performed using the corresponding bioinformatic techniques.
Results: A novel aminoglycoside resistance gene, designated aph(3’)-Ie, which confers resistance to ribostamycin, kanamycin, sisomicin and paromomycin, was identified in the chromosome of the animal bacterium Citrobacter gillenii DW61, which exhibited a multidrug resistance phenotype. APH(3’)-Ie showed the highest amino acid identity of 74.90% with the functionally characterized enzyme APH(3’)-Ia. Enzyme kinetics analysis demonstrated that it had phosphorylation activity toward four aminoglycoside substrates, exhibiting the highest affinity (Km, 4.22 ± 0.88 µM) and the highest catalytic efficiency [kcat/Km, (32.27 ± 8.14) × 104] for ribomycin. Similar to the other APH(3’) proteins, APH(3’)-Ie contained all the conserved functional sites of the APH family. The aph(3’)-Ie homologous genes were present in C. gillenii isolates from different sources, including some of clinical significance.
Conclusion: In this work, a novel chromosomal aminoglycoside resistance gene, designated aph(3’)-Ie, conferring resistance to aminoglycoside antimicrobials, was identified in a rabbit isolate C. gillenii DW61. The elucidation of the novel resistance mechanism will aid in the effective treatment of infections caused by pathogens carrying such resistance genes.
Introduction
Since the discovery and isolation of the first aminoglycoside antibiotic, streptomycin, from soil bacteria in 1944 (Jones et al., 1944), aminoglycoside antibiotics have been widely used as important anti-infection drugs (Zárate et al., 2018). They usually have good intrinsic activity against gram-negative and some gram-positive bacteria, killing them by binding to bacterial ribosomes and inhibiting normal protein synthesis (Kudo and Eguchi, 2016). Previous studies have shown that bacterial resistance to aminoglycoside antibiotics is usually driven by three main mechanisms: inactivation of the drug by aminoglycoside-modifying enzymes (AMEs), efflux, and targeted modification (e.g., by methylases) (Dozzo and Moser, 2010; Bastian et al., 2022).
Aminoglycoside O-phosphotransferases (APHs) are AMEs that utilize ATP or GTP to mediate the catalytic transfer of a phosphate group to an aminoglycoside molecule and make the antimicrobials inactivated. There are 35 functionally characterized APHs in the database CARD, and they are classified into 7 subfamilies [APH (2”), APH(3’), APH(3”), APH(4), APH(6’), APH(7”) and APH(9)] (Alcock et al., 2023). Located over chromosomes and plasmids, they are sometimes associated with mobile genetic elements (MGEs), such as transposons, integrons and conjugative elements, which made them spread between bacteria of different species or genera by means of horizontal gene transfer (Toth et al., 2010).
Including 21 species, Citrobacter is a genus of the family Enterobacteriaceae. The bacteria of this genus have been clinically reported to cause central nervous system infections and sepsis in neonates and immunocompromised hosts (Doran, 1999). The species Citrobacter gillenii (type strain ATCC 51117 = CCUG 30796 = CIP 106783 = DSM 13694) of the genus was characterized by DNA hybridization and biochemical analysis in 1999 (Brenner et al., 1999). In the recent years, it has been increasingly isolated from aquaculture animals, where it has been found to carry multidrug resistance (MDR) genes which became a challenge for the treatment of farmed animals (Duman et al., 2017; Concha et al., 2021; Türe et al., 2022).
In this study, based on the whole genome sequencing, a novel aminoglycoside O-phosphotransferase gene, designated aph(3’)-Ie, was identified in the chromosome of an animal isolate C. gillenii DW61, and its molecular and functional characteristics were further investigated.
Materials and methods
Bacteria and plasmids
C. gillenii DW61 was isolated from an anal swab of an Oryctolagus cuniculus f. domesticus from a farm in Wenzhou, Zhejiang Province, China. The anal swab sample was streaked onto a standard Luria–Bertani agar plate, and single colonies were then isolated and purified by the same method. Species classification was first performed by 16S rRNA gene homology comparison (Wachino and Arakawa, 2012) and then confirmed by average nucleotide identity (ANI) calculations (Lindsey et al., 2023) and DNA-DNA hybridization (isDDH) analysis (Meier-Kolthoff et al., 2014). The strains and plasmids used in this study are listed in Table 1.
Drug susceptibility testing
The minimum inhibitory concentration (MIC) was tested by the plate dilution method using Mueller–Hinton (MH) agar (Thermo Fisher Scientific Inc. Beijing, China) in accordance with the most recent M100 performance standards for antimicrobial susceptibility testing by the Clinical and Laboratory Standards Institute (CLSI, 2024). After 16-20 h of constant incubation at 37°C, the results were interpreted following the CLSI M100 performance standards and the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2022). Escherichia coli ATCC 25922 was used for a quality control. The experiments were conducted in triplicate.
Genome sequencing, assembly, annotation, and bioinformatics analysis
The bacterial genomic DNA was extracted using the Universal Genomic DNA Purification Mini Spin Kit (Beyotime Biotechnology Co., Ltd., Shanghai, China). DNA sequencing was performed by using the Illumina NovaSeq 6000 and PacBio Sequel II platforms by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The Illumina short reads were assembled by SKESA v2.4.0 (Souvorov et al., 2018). The long reads from PacBio Sequel II were hybrid assembled with short reads in a short-read-first manner using Unicycler (v0.4.8) (Wick et al., 2017) and then polished by Pilon (v 1.23) (Walker et al., 2014). The prediction and annotation of the open reading frames (ORFs) were performed using prokka (Seemann, 2014) against UniProtKB/Swiss-Prot (http://web.expasy.org/docs/swiss-prot_guideline.html), and further annotation was performed by comparison with the NCBI nonredundant (nr) protein database using DIAMOND (v2.0.14) (Buchfink et al., 2021). MAFFT (v7.407) (Rozewicki et al., 2019) was used to perform multiple-sequence alignment. To construct a phylogenetic tree, IQ-TREE v2.2.2.3 (Minh et al., 2020b) was used to select the model that minimized the Bayesian information criterion (BIC) score, employing the log-likelihood method (Minh et al., 2020a). CD-search (Marchler-Bauer and Bryant, 2004) was used to predict the protein domains of APH(3’)-Ie by comparison with the Conserved Domain Database (CDD) (Wang et al., 2023). Clinker (v.0.0.25) (Gilchrist and Chooi, 2021) was used to perform genetic context analyses of aph(3’)-Ie and other related sequences.
Cloning of the aph(3’)-Ie gene
PCR was performed using primers to amplify the ORF of aph(3’)-Ie with its upstream promoter region predicted by BPROM (www.softberry.com) (Table 2). The PCR product was subsequently inserted into the T-Vector pMD™19 using T4 ligase (Takara Biomedical Technology Co., Ltd.). The recombinant plasmid in the ligation mixture was then transformed into competent E. coli DH5α cells by chemical transformation (Koppel et al., 2017). After screening colonies using an agar plate containing 100 μg/mL ampicillin, the sequence of the cloned fragment was verified by Sanger DNA sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China).
Expression and purification of APH(3’)-Ie
APH(3’)-Ie was expressed according to previously described methods with minor modifications (Qing et al., 2004; Sha et al., 2023). The ORF of the aph(3’)-Ie gene with the thrombin cleavage site was PCR-amplified (Table 2) and then ligated into the pCold I vector, and the recombinant plasmid pCold I-aph(3’)-Ie was subsequently transformed into E. coli BL21 (Table 1). To obtain APH(3’)-Ie, recombinant BL21(pColdI- aph(3’)-Ie) was grown to an OD600 of 0.5 in 100 mL of LB at 37°C. Then, 1 mM isopropyl-β-D-thiogalactopyranoside was added, and the mixture was incubated at 16°C for 16 h. Bacteria were collected by centrifugation at 10,000 × g for 4 min, resuspended in 4 mL of nondenatured lysate, and immediately sonicated for 10 min. The lysis products were subsequently centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was collected and re-equilibrated with nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Beyotime Biotechnology, Shanghai, China). The mixture was gently agitated for 10 h at 4°C. Standard Ni-NTA affinity chromatography was used to isolate the His-tagged recombinant protein, and the His tag was removed using thrombin for 6 h at 37°C. The purified protein APH(3’)-Ie was validated using SDS-PAGE, and the protein concentration was determined with the BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China).
Enzyme kinetics analysis
The kinetic parameters of APH(3’)-Ie were measured in accordance with the previously reported methodology (Lu et al., 2021; Sha et al., 2023), with minor modifications. In brief, coupling between the production of ADP resulting from aminoglycoside phosphorylation and NADH oxidation was achieved through the use of pyruvate kinase (PK) and lactate dehydrogenase (LDH). The mixing volume for the entire reaction was 250 μL, which included 1 mM phosphoenolpyruvate (Beijing Solarbio Science & Technology Co., Ltd.), 600 μM NADH, 1 mM ATP, 100 mM HEPES (pH 7.0), 1 mM MgCl2, 2 mM KCl, a commercial mixture of PK and LD (Sigma P0294; 18-26 U/mL PK and 25-35 U/mL LDH, final concentrations), 30-40 nM APH(3′)-Ie and an aminoglycoside substrate at various concentrations. The ADP production rate was determined by monitoring the decrease in absorbance at 340 nm using a SpectraMax M5 multifunctional microplate reader (Molecular Devices, CA, United States) at 25°C. The experiments were conducted in triplicate. The velocities at steady state were determined by analyzing the linear phase of the reaction progress curve and then plotted as a function of the substrate concentration. The Michaelis-Menten equation was used to fit the data through nonlinear regression, with velocity as a function of substrate. GraphPad Prism 8.0.2 (GraphPad Software, CA, United States) was used for curve fitting and calculation of the dynamic parameters Km and kcat.
Data availability
The nucleotide sequences of the C. gillenii DW61 genome have been submitted to GenBank under accession numbers CP151088 for the chromosome, CP151089 for the plasmid pDW61, and PP596862 for the aph(3’)-Ie gene.
Results and discussion
Identification of the novel resistance gene and classification and molecular characterization of the isolate DW61
To elucidate novel resistance mechanisms exhibited by bacterial isolates from local animals in response to antimicrobial agents, a total of 576 bacteria isolated from anal fecal swabs of domestic avians and livestock and environment in Wenzhou, China, were subjected to sequencing analysis (Feng et al., 2022; Gao et al., 2022; Zhao et al., 2023). Annotation of the genome data revealed the presence of resistance genes against various classes of antibiotics. Notably, among the predicted genes were some putative aminoglycoside antibiotic resistance genes, including but not limited to aph(3’)-Ia, ant(9)-Ia, aadA5, aph(6)-Ic, aac(3)-IIIb, aac(6’)-Iaa, aph(6)-Id, and aac(2’)-IIb homologues. These genes shared amino acid sequence identities below 80.0% with functionally characterized aminoglycoside resistance genes. Some of the genes were randomly selected and further cloned, and their resistance functions were determined. Ultimately, a novel aph(3’)-Ia homologous gene (designated aph(3’)-Ie in this work) that confers resistance to several aminoglycoside agents was identified from an isolate named DW61.
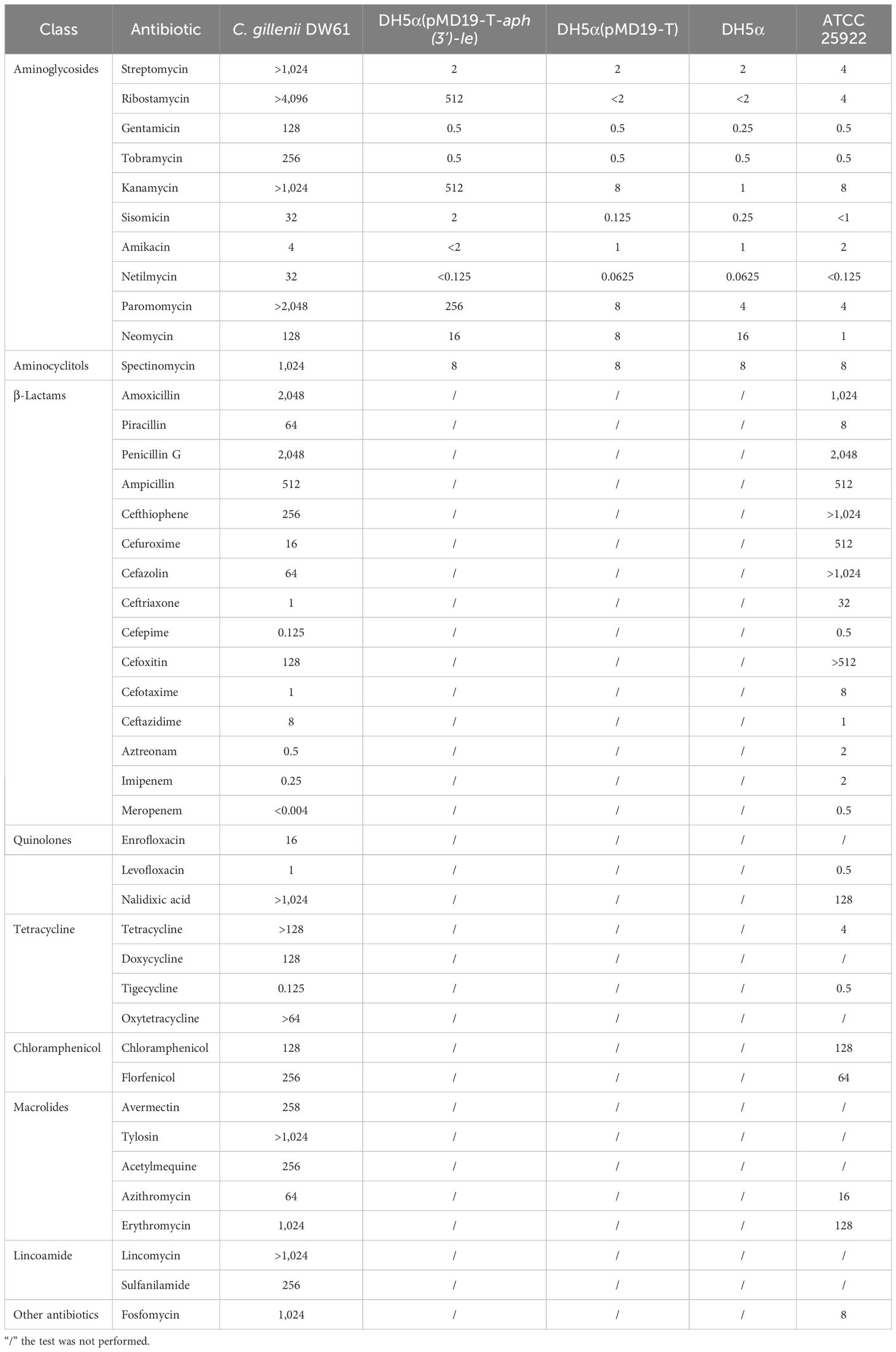
DW61 was isolated from the anal feces of a rabbit, and it showed high MICs of ≥ 16 µg/mL for 76.7% (33/43) of the antibiotics tested, particularly for aminoglycosides (Table 3). For the 10 aminoglycosides tested, except for amikacin, which had an MIC of 4 µg/mL, the MICs were ≥ 32 µg/mL. The MICs of ribostamycin, paromomycin, kanamycin and streptomycin were > 4096, > 2048, > 1024 and > 1024 µg/mL, respectively (Table 3).
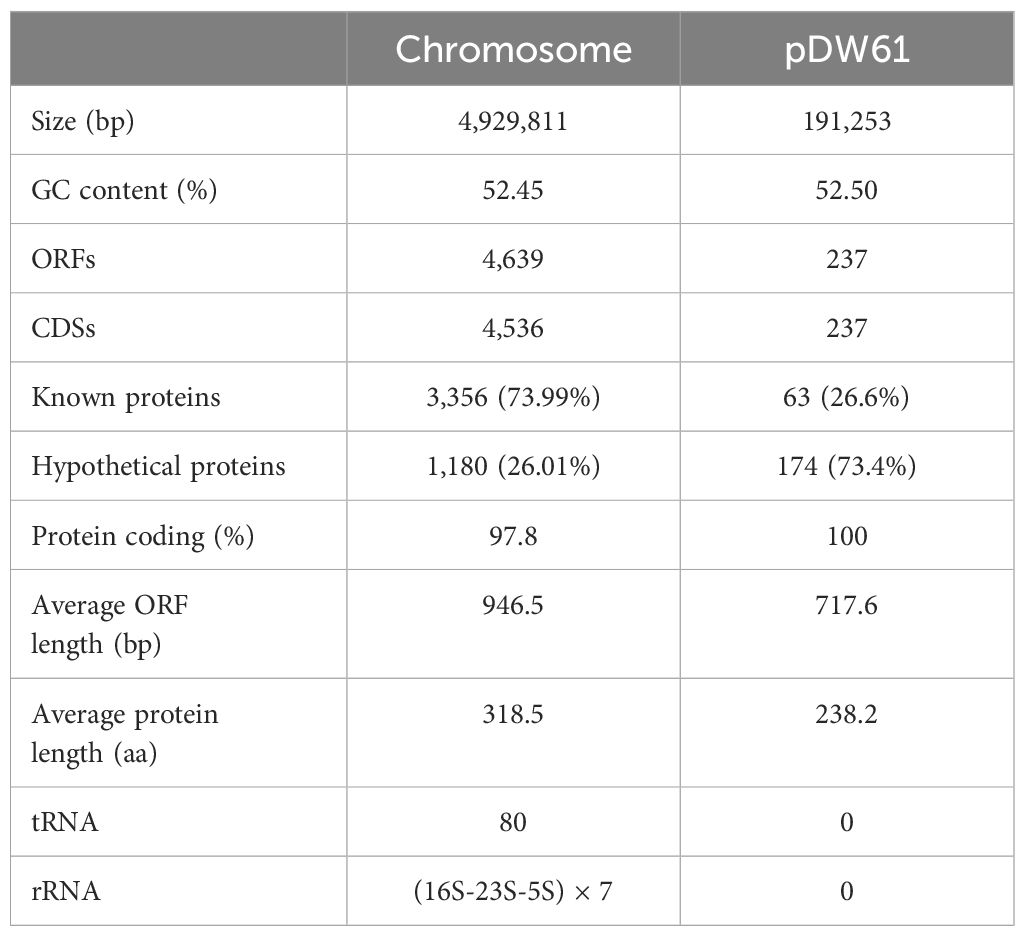
The genome of DW61 consists of a chromosome and a plasmid (designated pDW61-191). The chromosome is approximately 4.93 Mb in length and encodes 4,639 ORFs, and pDW61 is approximately 191.6 kb in length and encodes 237 ORFs (Table 4). 16S rRNA gene homology analysis revealed that the 16S rRNA gene of DW61 showed the highest identity (99.61%) with that of C. gillenii (AF025367.1). The genome-wide ANI analysis showed that the strain shared the highest identity (98.71%) with C. gillenii MBT-C3 (GCF_003429605.1) (Figure 1). Further DNA-DNA hybridization (DDH) indicated 87.80% identity between DW61 and C. gillenii MBT-C3 (GCF_003429605.1). This bacterium was thus classified as C. gillenii and designated C. gillenii DW61. The genome size of C. gillenii DW61 (5.13 Mb) was similar to that of three C. gillenii genomes (C. gillenii MBT-C3, GCA_003429605.1, 4.92 Mb; C. gillenii UMG736, GCA_013337685.1, 4.93 Mb; and C. gillenii AF64-5pH9A, GCA_027681665.1, 5.17 Mb) available in the NCBI genome database.
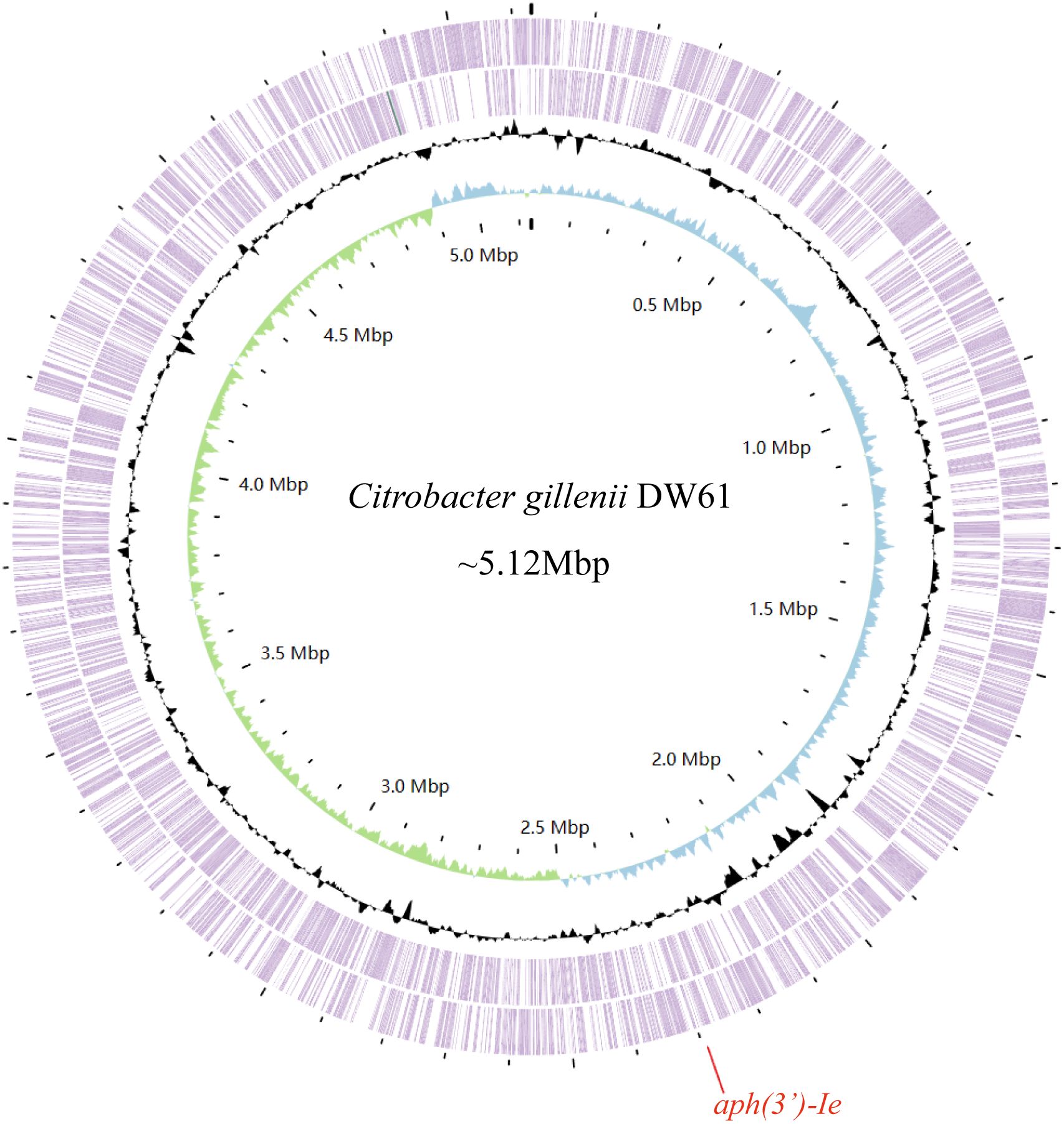
Figure 1 The genome map of C. gillenii DW61. The circles 1-4 from inside to outside represent the GC skew, GC content, and genes encoded in the forward and reverse strands of the chromosome of C. gillenii DW61, respectively. The red line on the bottom indicates the location of the novel resistance gene aph(3’)-Ie.
Functional analysis of the novel aminoglycoside 3’-phosphotransferase gene aph(3’)-Ie
The recombinant strain carrying aph(3’)-Ie showed >256-, 64-, 32-, 8-, and 2-fold greater MICs for ribostamycin, kanamycin, paromomycin, sisomicin, and neomycin, respectively, compared with the control strain (pMD19-T/DH5α). It did not show resistance to streptomycin, gentamicin, tobramycin, amikacin or netilmicin. This resistance phenotype is generally consistent with that associated with the aph(3’)-I class genes. The four close relatives of aph(3’)-Ie [aph(3’)-Id, aph(3’)-Ib, aph(3’)-Ia and aphA15] were previously reported to be resistant to kanamycin, neomycin, ribostamycin and paromomycin, similar to the resistance phenotype of aph(3’)-Ie. Moreover, these four aph genes also conferred susceptibility to gentamicin (except aph(3’)-Ia) or to amikacin (except aphA15) (Siregar et al., 1995; Riccio et al., 2001; Arcari et al., 2023; Sha et al., 2023).
The aph(3’)-Ie gene is 816 bp in length and encodes a 271-amino-acid protein with a theoretical pI of 5.19. Notably, the comparison of APH(3’)-Ie with the functionally characterized proteins in the CARD database revealed that the proteins with the highest identities to the novel aminoglycoside 3’-phosphotransferase APH(3’)-Ie were APH(3’)-Ia (74.90%), APH(3’)-Ib (57.20%), APH(3’)-IIc (40.0%) and APH(3’)-IIa (35.97%). Phylogenetic analysis of APH(3’)-Ie with APH(3’)s revealed that it is also located closest to APH(3’)-Ia (Figure 2; Supplementary Table S1). This finding further confirmed that this protein is an aminoglycoside 3’-phosphotransferase (GenBank accession no. cd05150).
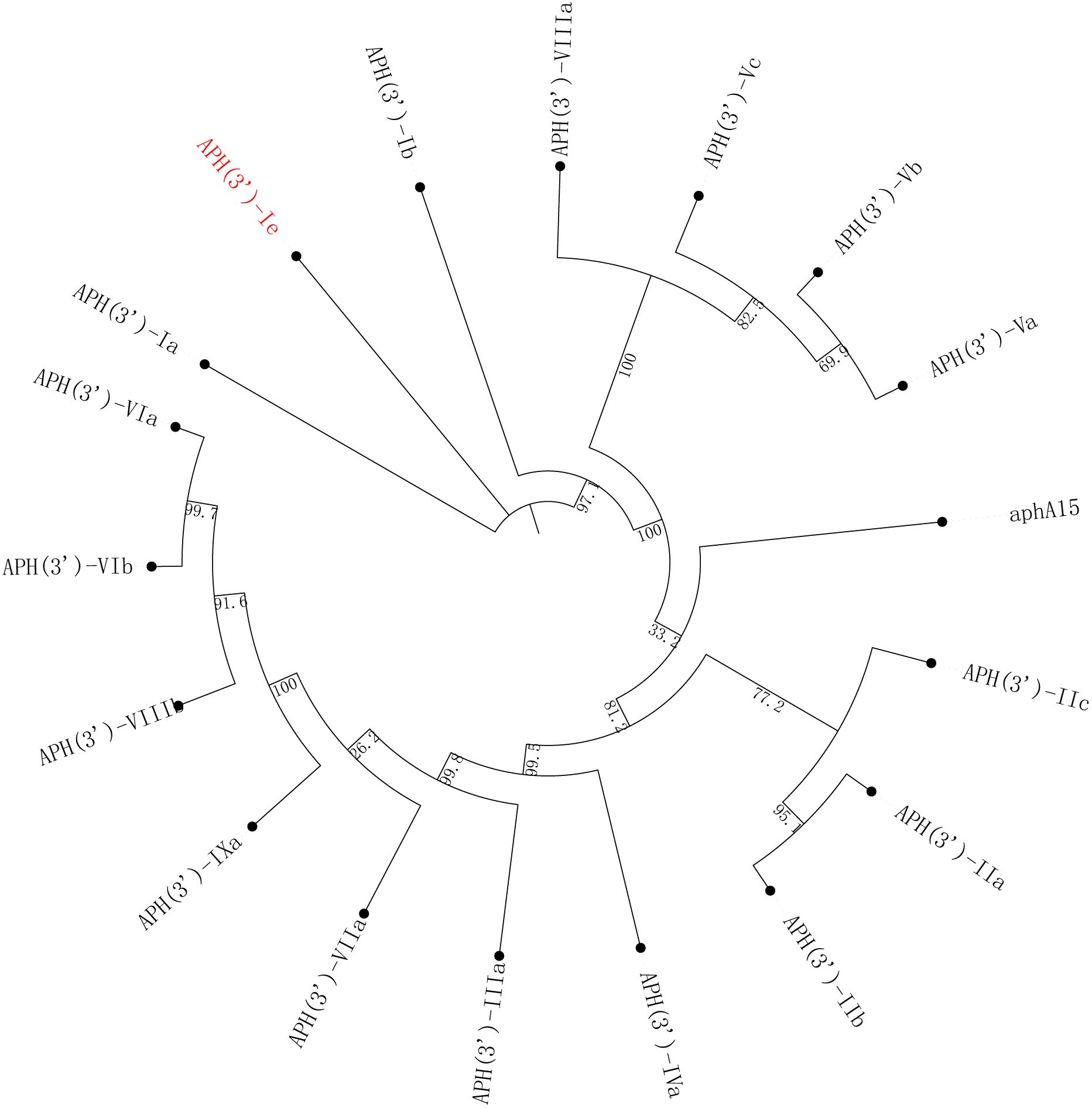
Figure 2 A phylogenetic tree showing the relationships of APH(3’)-Ie with other functionally characterized APH(3’)s. APH(3’)-Ie is highlighted in red. The accession numbers of these proteins are listed in Supplementary Table S1.
Kinetic parameters and structural characterization of APH(3’)-Ie
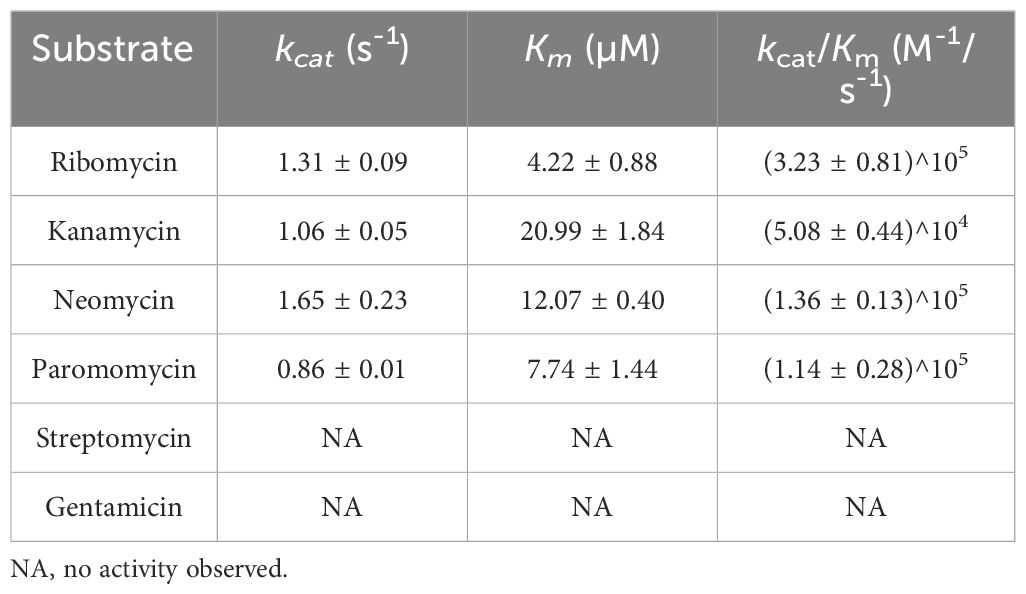
The phosphotransferase activities of APH(3’)-Ie were generally consistent with the MIC results (Table 5). Among the six aminoglycoside substrates tested, APH(3’)-Ie exhibited phosphorylation activity toward ribomycin, kanamycin, neomycin and paromomycin but not gentamicin or streptomycin. It exhibited the highest affinity (Km, 4.22 ± 0.88 µM) and the highest catalytic efficiency [kcat/Km, (32.27 ± 8.14)^104] for ribomycin.
APH(3’)-Ia showed a similar affinity for kanamycin with APH(3’)-Ie (Km, 19.7 ± 2.5 vs. 20.99 ± 1.84 µM), but the catalytic efficiency of APH(3’)-Ia was much greater than that of APH(3’)-Ie [kcat/Km, 8.5 × 107 vs. (5.08 ± 0.44) × 104 M -1/s-1], which was attributed to the much greater substrate transfer rate of APH(3’)-Ia for kanamycin than that of APH(3’)-Ie (kcat, 102 ± 14 vs. 1.06 ± 0.05 s-1) (Siregar et al., 1995). In contrast to APH(3’)-Ia, APH(3’)-IIa showed lower affinities for kanamycin and neomycin than did APH(3’)-Ie, with Km values of 3 vs. 20.99 ± 1.84 µM for kanamycin and 6 vs. 12.07 ± 0.40 µM for neomycin, respectively, but its catalytic efficiencies for both substrates were greater than those of APH(3’)-Ie [with kcat/Km of 1.3 × 106 vs. (5.08 ± 0.44) × 104 M -1/s-1 for kanamycin and 1.8 × 106 vs. (1.36 ± 0.13) × 105 M -1/s-1 for neomycin, respectively]. This was also because the substrate transfer rates of APH(3’)-IIa for these two substrates were greater than those of APH(3’)-Ie (with kcat of 4 vs. 1.06 ± 0.05 s-1 for kanamycin and 11 vs. 1.65 ± 0.23 s-1 for neomycin, respectively) (Siregar et al., 1994).
To analyze the structural conservation of the protein, the amino acid sequences of 5 APH(3’) proteins (including the one from this study) with close evolutionary relationships were compared. APH(3’)-Ie contained all 30 active residues of the conserved APH protein domain family (GenBank accession no. cd05150), including 15 residues of the ATP-binding site and 12 residues of the antibiotic-binding site (Fong and Berghuis, 2002; Nurizzo et al., 2003; Stogios et al., 2013) (Figure 3).
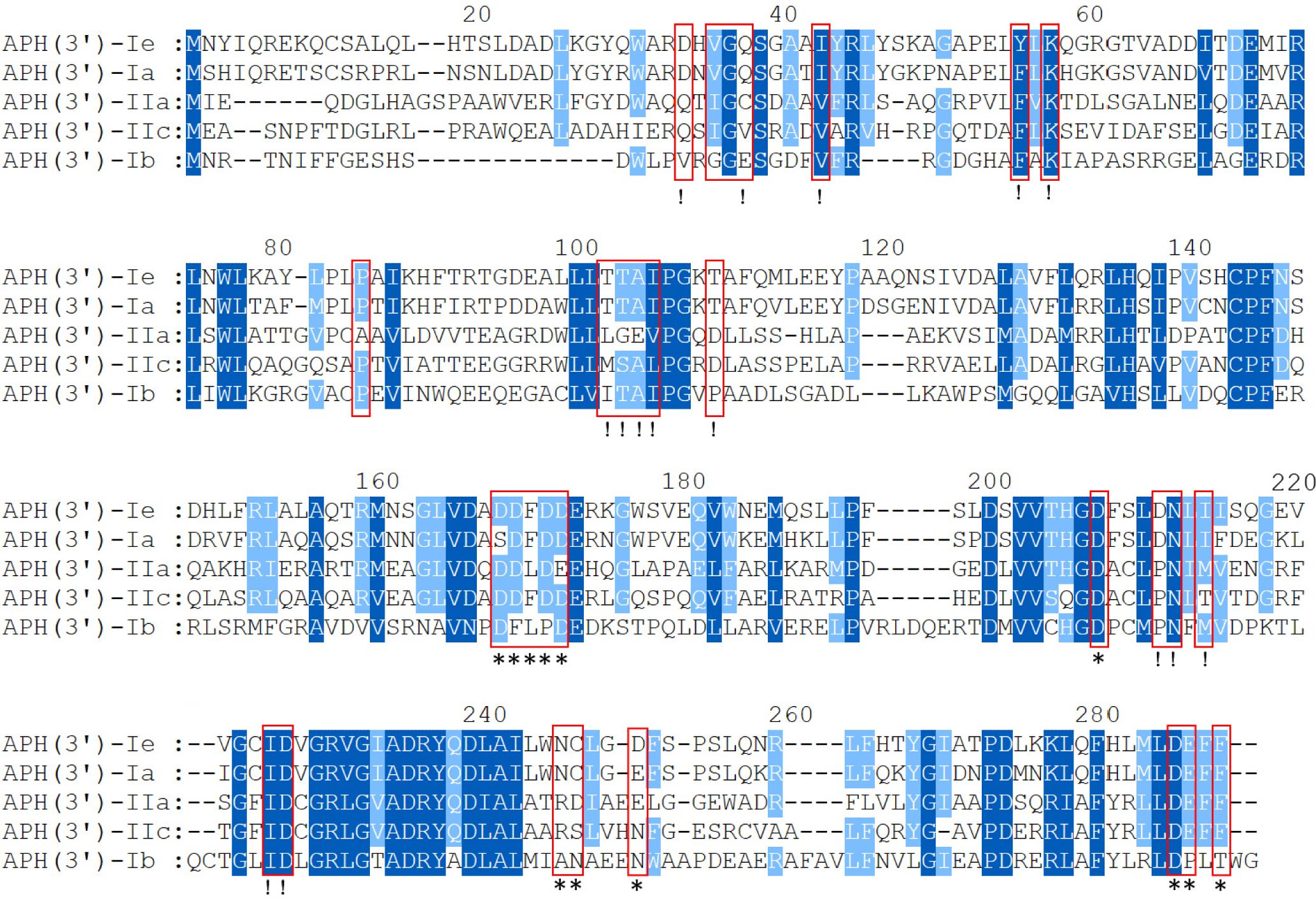
Figure 3 Multiple-sequence alignment of APH(3’)-Ie with its close relatives. The numbers on the right represent the corresponding amino acid sequence lengths. The red frames represent the residues of the conserved APH protein domain family. The exclamation marks indicate the residues of the ATP-binding sites, and the asterisks indicate the 12 residues of the antibiotic-binding site. The conserved residues are shown in dark blue, while the relatively conserved residues are shown in light blue. Gaps are represented using hyphens.
Distribution of the aph(3’)-Ie genes and structural characterization of the aph(3’)-Ie-related sequences
To analyze the distribution of the aph(3’)-Ie gene and its close relatives, the amino acid sequence of APH(3’)-Ie was used as a query to search for homologous protein-encoding nucleotide sequences using the tBLASTn program against the NCBI nucleotide collection (nt) database (Figure 4), and a total of 28 sequences showing ≥ 98.15% amino acid sequence identity with APH(3’)-Ie were found in the database. The three strains with the highest identities (100.0%) were all C. freundii strains (including C. freundii RHBSTW-00714, CP056333.1; C. freundii RHBSTW-00006, CP056910.1; and C. freundii RHBSTW-00334, CP056597.1). Among the remaining 25 sequences, one was from the uncultured bacterium pJM6 from a metagenomic library of an environmental sample (FJ537710.1, identity 98.89%) and one was from Citrobacter werkmanii NBRC 105721 (LR699014.1, identity 98.15%), while the other 23 genes were also from C. freundii. Of the 28 genes, one was from a metagenomic library of an environmental sample, while the other 27 were from strains isolated from different sources, including animals (92.59%, 25/27), the environment (3.70%, 1/27) and humans (3.70%, 1/27) (Supplementary Table S2).
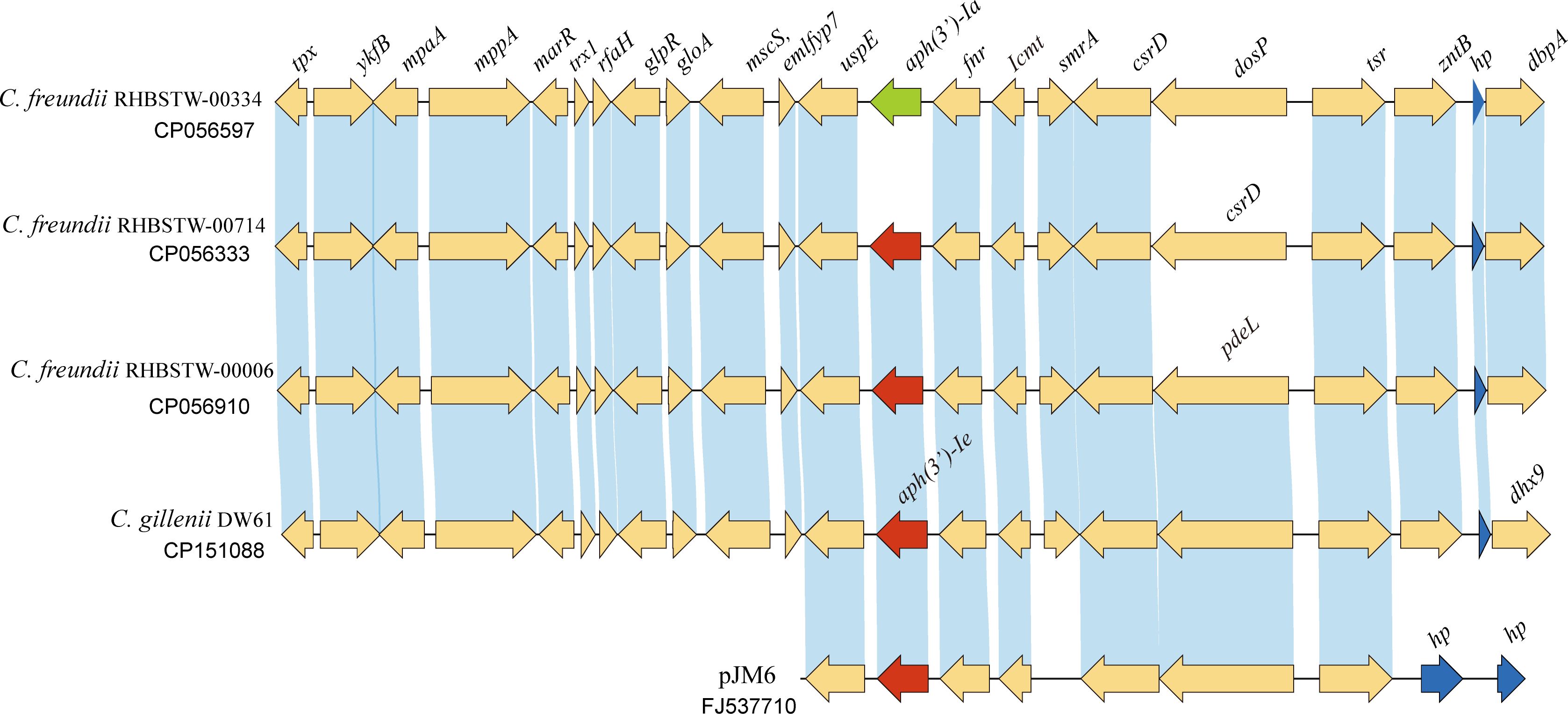
Figure 4 Genetic background of the aph(3’)-Ie gene and the aph(3’)-Ie homologous genes. Regions with an amino acid identity of ≥ 80% are in blue. hp, hypothetical protein. The aph(3’)-Ie and aph(3’)-Ie-like genes (similarity > 98.15%) are shown in red. The aph(3’)-Ia gene is shown in green, and the hypothetical protein-encoding genes are shown in blue.
Further analysis of the sequences with lower identities revealed that of the 100 sequences with identities ≥ 70.94%, which included the 28 sequences mentioned above, 72 sequences had identities between 74.91% and 70.94%, and no sequence with an identity between > 74.91% and < 98.15% was present. Notably, of these 72 sequences with identities ranging from 70.94% to 74.91%, three sequences were from Klebsiella pneumoniae, including two encoded on plasmids (pBWH77 of K. pneumoniae 5214773, NG_047431.1, and pBWH77 of K. pneumoniae, X62115.1) and one in the chromosome (K. pneumoniae GDKA1, NG_047441.1), while the others were from other orders of bacteria, cloning vectors or synthetic constructs (Supplementary Table S2).
When analyzing the structure of the aph(3’)-Ie gene-related sequences, a nucleotide sequence approximately 20 kb in length with the aph(3’)-Ie gene at the center from the C. gillenii DW61 chromosome was used as a query to search for homologous sequences in the NCBI nucleotide database. A total of 28 sequences with identities ≥ 80.0% were retrieved, and they were all from the genus Citrobacter. Of these 28 sequences, 27 carried one aph(3’)-Ie(-like) gene each and were from C. freundii, except one from C. werkmanii, MGYG-HGUT-02535 (LR699014.1, identity 98.41%). One sequence free of an aph(3’)-Ie-like gene was from another species, Citrobacter ructae SNUWT2 (CP038469.1), and it shared a lower identity (81.62%) with that of C. gillenii DW61. No mobile genetic elements (MGEs) were detected within these 20 kb fragments.
Conclusion
In this study, a novel aminoglycoside phosphotransferase gene, designated aph(3’)-Ie, was identified in the chromosome of the MDR Citrobacter gillenii isolate DW61, which showed high MICs (≥ 16 μg/mL) for 76.7% of the 43 antibiotics tested. This novel resistance gene conferred resistance to some aminoglycosides, such as ribostamycin and kanamycin, and was present in C. gillenii from different sources, including some of clinical significance. The discovery of new antimicrobial resistance mechanisms is helpful for effective clinical treatment of bacterial infectious diseases.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
This study used strains isolated from animals in animal farms in Wenzhou, China. The owners of the farms were informed in writing of the study and provided approval for the sampling of animals. The studies involving human participants and animals were reviewed and approved by the Animal Welfare and Ethics Committee of Wenzhou Medical University, Zhejiang Province, China (protocol number: wydw2021-0323).
Author contributions
NL: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft. YS: Writing – original draft. GZ: Writing – original draft. CS: Writing – original draft. YZ: Writing – original draft. JZ: Writing – original draft. DH: Writing – original draft. JL: Writing – original draft. QB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. WP: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Science & Technology Project of Jinhua City, China (2023-3-159, 2022-2-013), the Science & Technology Project of Taizhou City, China (21ywb128), Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2023KY1350) and the Science & Technology Project of Wenzhou City, China (N20210001).
Acknowledgments
The authors would like to acknowledge the teachers and scientists of the Science and Technology Platforms of Wenzhou Medical University who helped with the analysis of the enzyme kinetic parameters.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1435123/full#supplementary-material
Supplementary Table 1 | Proteins used for phylogenetic tree construction.
Supplementary Table 2 | Distribution of aph(3’)-Ie(-like) genes.
References
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–d699. doi: 10.1093/nar/gkac920
Arcari, G., Hennart, M., Badell, E., Brisse, S. (2023). Multidrug-resistant toxigenic Corynebacterium diphtheriae sublineage 453 with two novel resistance genomic islands. Microbial Genomics 9(1). doi: 10.1099/mgen.0.000923
Bastian, A. A., Bastian, M., Jäger, M., Loznik, M., Warszawik, E. M., Yang, X., et al. (2022). Late-stage modification of aminoglycoside antibiotics overcomes bacterial resistance mediated by APH(3') kinases. Chem. (Weinheim an Der Bergstrasse Germany) 28, e202200883. doi: 10.1002/chem.202200883
Brenner, D. J., O'Hara, C. M., Grimont, P. A., Janda, J. M., Falsen, E., Aldova, E., et al. (1999). Biochemical identification of Citrobacter species defined by DNA hybridization and description of Citrobacter gillenii sp. nov. (formerly Citrobacter genomospecies 10) and Citrobacter murliniae sp. nov. (formerly Citrobacter genomospecies 11). J. Clin. Microbiol. 37, 2619–2624. doi: 10.1128/JCM.37.8.2619-2624.1999
Buchfink, B., Reuter, K., Drost, H.-G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
CLSI (2024). Performance standards for antimicrobial susceptibility testing. 34th Edition. (Berwyn, PA 19312: Clinical and Laboratory Standards Institute).
Concha, C., Miranda, C. D., Rojas, R., Godoy, F. A., Romero, J. (2021). Characterization of a Novel Variant of the Quinolone-Resistance Gene qnrB (qnrB89) Carried by a Multi-Drug Resistant Citrobacter gillenii Strain Isolated from Farmed Salmon in Chile. Antibiotics (Basel Switzerland) 10 (236). doi: 10.3390/antibiotics10030236
Doran, T. I. (1999). The role of Citrobacter in clinical disease of children: review. Clin. Infect. Dis. an Off. Publ. Infect. Dis. Soc. America 28, 384–394. doi: 10.1086/515106
Dozzo, P., Moser, H. E. (2010). New aminoglycoside antibiotics. Expert Opin. On Ther. Patents 20, 1321–1341. doi: 10.1517/13543776.2010.506189
Duman, M., Saticioglu, I. B., Buyukekiz, A. G., Balta, F., Altun, S. (2017). Molecular characterization and antimicrobial resistance profile of atypical Citrobacter gillenii and Citrobacter sp. isolated from diseased rainbow trout (Oncorhynchus mykiss). J. Global Antimicrobial Resistance 10, 136–142. doi: 10.1016/j.jgar.2017.05.014
EUCAST (2022). Breakpoint tables for interpretation of MICs and zone diameters. Available online at: http://www.eucast.org.
Feng, C., Gao, M., Jiang, W., Shi, W., Li, A., Liu, S., et al. (2022). Identification of a novel aminoglycoside O-nucleotidyltransferase AadA33 in Providencia vermicola. Front. In Microbiol. 13. doi: 10.3389/fmicb.2022.990739
Fong, D. H., Berghuis, A. M. (2002). Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 21, 2323–2331. doi: 10.1093/emboj/21.10.2323
Gao, M., Feng, C., Ji, Y., Shi, Y., Shi, W., Zhang, L., et al. (2022). AadA36, a novel chromosomal aminoglycoside nucleotidyltransferase from a clinical isolate of Providencia stuartii. Front. In Microbiol. 13. doi: 10.3389/fmicb.2022.1035651
Gilchrist, C. L. M., Chooi, Y.-H. (2021). clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinf. (Oxford England) 37, 2473–2475. doi: 10.1093/bioinformatics/btab007
Jones, D., Metzger, H. J., Schatz, A., Waksman, S. A. (1944). Control of gram-negative bacteria in experimental animals by streptomycin. Sci. (New York N.Y.) 100, 103–105. doi: 10.1126/science.100.2588.103.b
Koppel, N., Maini Rekdal, V., Balskus, E. P. (2017). Chemical transformation of xenobiotics by the human gut microbiota. Sci. (New York N.Y.) 356, eaag2770. doi: 10.1126/science.aag2770
Kudo, F., Eguchi, T. (2016). Aminoglycoside antibiotics: new insights into the biosynthetic machinery of old drugs. Chem. Rec. (New York N.Y.) 16, 4–18. doi: 10.1002/tcr.v16.1
Lindsey, R. L., Gladney, L. M., Huang, A. D., Griswold, T., Katz, L. S., Dinsmore, B. A., et al. (2023). Rapid identification of enteric bacteria from whole genome sequences using average nucleotide identity metrics. Front. In Microbiol. 14. doi: 10.3389/fmicb.2023.1225207
Lu, W., Li, K., Huang, J., Sun, Z., Li, A., Liu, H., et al. (2021). Identification and characteristics of a novel aminoglycoside phosphotransferase, APH(3')-IId, from an MDR clinical isolate of Brucella intermedia. J. Antimicrobial Chemotherapy 76, 2787–2794. doi: 10.1093/jac/dkab272
Marchler-Bauer, A., Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Meier-Kolthoff, J. P., Klenk, H.-P., Göker, M. (2014). Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Systematic Evolutionary Microbiol. 64, 352–356. doi: 10.1099/ijs.0.056994-0
Minh, B. Q., Hahn, M. W., Lanfear, R. (2020a). New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 37, 2727–2733. doi: 10.1093/molbev/msaa106
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020b). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Nurizzo, D., Shewry, S. C., Perlin, M. H., Brown, S. A., Dholakia, J. N., Fuchs, R. L., et al. (2003). The crystal structure of aminoglycoside-3'-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327, 491–506. doi: 10.1016/S0022-2836(03)00121-9
Qing, G., Ma, L.-C., Khorchid, A., Swapna, G. V. T., Mal, T. K., Takayama, M. M., et al. (2004). Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882. doi: 10.1038/nbt984
Riccio, M. L., Pallecchi, L., Fontana, R., Rossolini, G. M. (2001). In70 of plasmid pAX22, a bla(VIM-1)-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrobial Agents Chemotherapy 45, 1249–1253. doi: 10.1128/AAC.45.4.1249-1253.2001
Rozewicki, J., Li, S., Amada, K. M., Standley, D. M., Katoh, K. (2019). MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47, W5–W10. doi: 10.1093/nar/gkz342
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinf. (Oxford England) 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sha, Y., Lin, N., Zhang, G., Zhang, Y., Zhao, J., Lu, J., et al. (2023). Identification and characterization of a novel chromosomal aminoglycoside 3'-O-phosphotransferase, APH(3')-Id, from Kluyvera intermedia DW18 isolated from the sewage of an animal farm. Front. In Microbiol. 14. doi: 10.3389/fmicb.2023.1224464
Siregar, J. J., Lerner, S. A., Mobashery, S. (1994). Purification and characterization of aminoglycoside 3'-phosphotransferase type IIa and kinetic comparison with a new mutant enzyme. Antimicrobial Agents Chemotherapy 38, 641–647. doi: 10.1128/AAC.38.4.641
Siregar, J. J., Miroshnikov, K., Mobashery, S. (1995). Purification, characterization, and investigation of the mechanism of aminoglycoside 3'-phosphotransferase type Ia. Biochemistry 34, 12681–12688. doi: 10.1021/bi00039a026
Souvorov, A., Agarwala, R., Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19, 153. doi: 10.1186/s13059-018-1540-z
Stogios, P. J., Spanogiannopoulos, P., Evdokimova, E., Egorova, O., Shakya, T., Todorovic, N., et al. (2013). Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance. Biochem. J. 454, 191–200. doi: 10.1042/BJ20130317
Toth, M., Frase, H., Chow, J. W., Smith, C., Vakulenko, S. B. (2010). Mutant APH(2'')-IIa enzymes with increased activity against amikacin and isepamicin. Antimicrobial Agents Chemotherapy 54, 1590–1595. doi: 10.1128/AAC.01444-09
Türe, M., Cebeci, A., Özcelep, T. (2022). The first outbreak of citrobacteriosis caused by Citrobacter gillenii in reared Russian sturgeon (Acipenser gueldenstaedtii) in Turkiye. Veterinary Res. Forum an Int. Q. J. 13, 323–329. doi: 10.30466/vrf.2021.137808.3076
Wachino, J.-i., Arakawa, Y. (2012). Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resistance Updates Rev. Commentaries In Antimicrobial Anticancer Chemotherapy 15, 133–148. doi: 10.1016/j.drup.2012.05.001
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Zárate, S. G., de la Cruz Claure, M. L., Benito-Arenas, R., Revuelta, J., Santana, A. G., Bastida, A. (2018). Overcoming aminoglycoside enzymatic resistance: design of novel antibiotics and inhibitors. Molecules (Basel Switzerland) 23, 284–302. doi: 10.3390/molecules23020284
Keywords: Citrobacter gillenii, resistance mechanism, aminoglycoside 3’phosphotransferase, aph(3')-Ie, kinetic parameter
Citation: Lin N, Sha Y, Zhang G, Song C, Zhang Y, Zhao J, Huang D, Lu J, Bao Q and Pan W (2024) APH(3’)-Ie, an aminoglycoside-modifying enzyme discovered in a rabbit-derived Citrobacter gillenii isolate. Front. Cell. Infect. Microbiol. 14:1435123. doi: 10.3389/fcimb.2024.1435123
Received: 19 May 2024; Accepted: 15 July 2024;
Published: 30 July 2024.
Edited by:
Ronni Mol Joji, Arabian Gulf University, BahrainReviewed by:
Maritza Alvarez-Ainza, Universidad de Sonora, MexicoCemal Sandalli, Recep Tayyip Erdoğan University, Türkiye
Mohd W. Azam, Albert Einstein College of Medicine, United States
Copyright © 2024 Lin, Sha, Zhang, Song, Zhang, Zhao, Huang, Lu, Bao and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=; Wei Pan, MjU2NTg1MDdAcXEuY29t
 Naru Lin
Naru Lin Yuning Sha
Yuning Sha Guozhi Zhang
Guozhi Zhang Chunhan Song2
Chunhan Song2 Yuan Zhang
Yuan Zhang Jingxuan Zhao
Jingxuan Zhao Junwan Lu
Junwan Lu Qiyu Bao
Qiyu Bao