- 1Veterinary Research Institute, Xinjiang Academy of Animal Sciences (Animal Clinical Medical Research Center, Xinjiang Academy of Animal Sciences), Urumqi, China
- 2Veterinary Department, Animal Husbandry and Veterinary Station of the Xinjiang Production and Construction Corps, Urumqi, China
- 3Animal Disease Monitoring Department, Changji Animal Disease Prevention and Control Center, Changji, China
- 4Inspection and Monitoring Department, Ili Kazak Autonomous Prefecture Center for Animal Disease Control and Diagnosis, Yining, China
- 5Inspection Department, Kekedala Supply and Marketing Cooperative Federation of the Fourth Division of Xinjiang Production and Construction Corps, Kekedala, China
Introduction: Alveolar echinococcosis (AE) is a life-threatening disease in humans caused by the larval stage of Echinococcus multilocularis. Domestic animals, dogs, foxes, and small mammals constitute the circular chain of AE. To evaluate the infection, distribution, and genetic polymorphism of AE in the Ili Prefecture (Nilka, Xinyuan and Zhaosu), we conducted this survey.
Methods: In June and July 2018, 267 small mammals were captured using water-infusion and mousetrap methods. Combined pathogenic and molecular biological methods were used to observe the histopathology of Echinococcus carried by rodents, amplify the mitochondrial nad1 gene of the pathogen, and investigate the genotype and haplotype diversity of Echinococcus in rodents in Ili Prefecture.
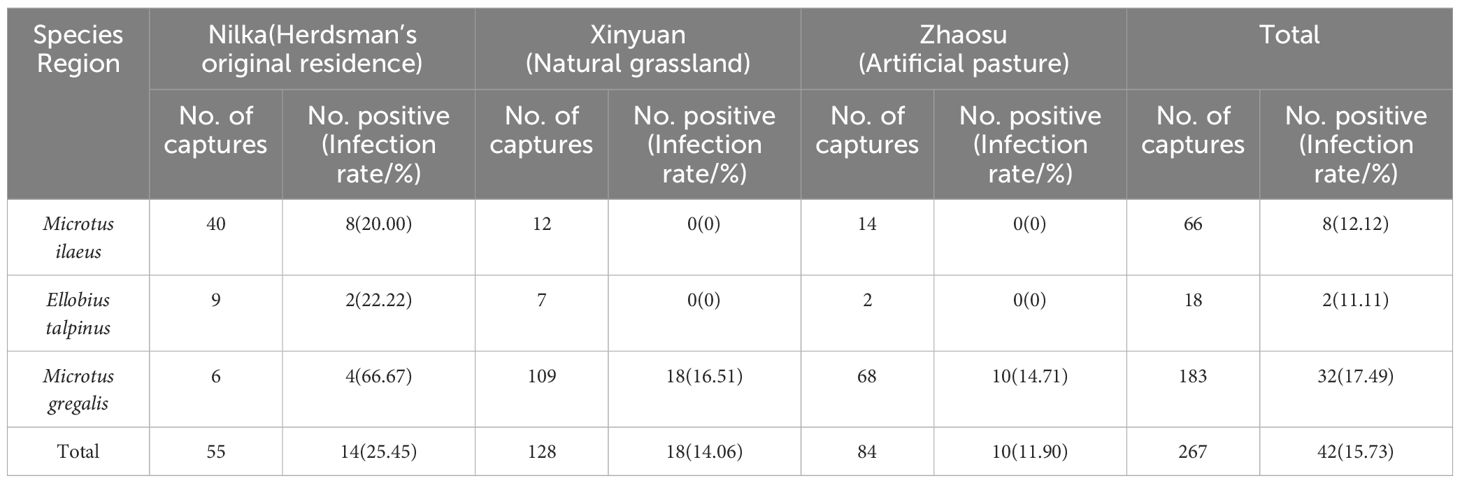
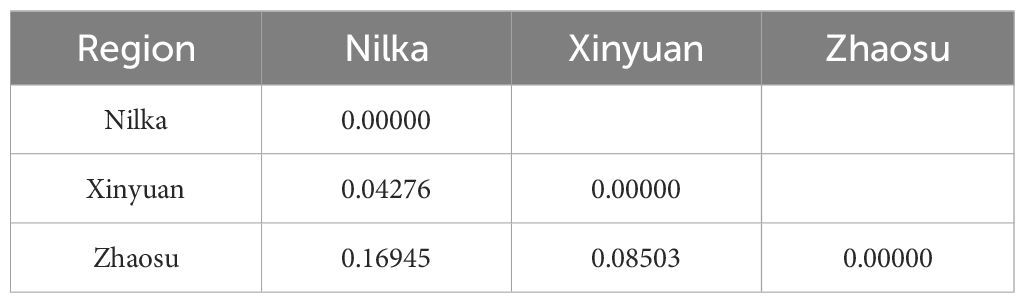
Results: Morphological identification revealed that these captured small mammals belonged to three species, with Microtus gregalis being the dominant species (183/267). Pathological and molecular biological results confirmed that E. multilocularis was the pathogen of echinococcosis in small mammals, with an infection rate of 15.73% (42/267). Among the three areas sampled, the highest infection rate of rodents was 25.45% (14/55) in Nilka County. However, there was no significant difference in the infection rates between regions (χ2 = 5.119, p > 0.05). Of the three captured rodent species, M. gregalis had the highest infection rate of 17.49% (32/183), but there was no significant difference in infection rates between the rodent species (χ2 = 1.364, p > 0.05). Phylogenetic analyses showed that the nad1 gene sequences obtained in this study clustered in the same clade as isolates from China. These isolates contained 21 haplotypes (Hap_1-21); Hap_2 was the most common haplotype (9/42). Furthermore, haplotype diversity (0.925 ± 0.027) and nucleotide diversity (0.01139 ± 0.00119) were higher in the Ili Prefecture than in other regions, indicating that population differentiation was high. Tajima’s D and Fu’s Fs tests were negative (p > 0.10), indicating that the population had expanded. The low fixation index (Fst) ranged from 0.00000 to 0.16945, indicating that the degree of genetic differentiation was different among different populations.
Discussion: In summary, Ili Prefecture is a high incidence area of AE, and Microtus spp. may play an important role in the transmission of AE in this area. The results of this study provide basic data for further study of the molecular epidemiology, genetic differences, and control of E. multilocularis in the Ili Prefecture, Xinjiang.
Introduction
Hydatid disease, also known as Echinococcosis, is a serious zoonotic parasitic disease caused by the larva of Echinococcus in humans and animals, with a global distribution (Xiao et al., 2006; Mandal and Mandal, 2012). More than 1 million people worldwide suffer from this disease (Zeng et al., 2014), which not only affects impoverished populations in developing countries and regions but also poses a barrier to international trade in animals and animal products (McManus et al., 2003; Qi and Wu, 2013). China is one of the countries with the most serious hydatidosis in the world, among which Xinjiang, Qinghai, Ningxia, Gansu, Inner Mongolia, Tibet, Western Sichuan, and other provinces (regions) are the most serious (Li et al., 2009). At present, there are three species of Echinococcus in China, namely Echinococcus multilocularis, Echinococcus granulosus, and Echinococcus shiquicus (Ni et al., 2012). The life cycle of Echinococcus is complex, requiring different intermediate and definitive hosts to complete its life cycle. In addition to ruminants such as cattle and sheep, as well as humans, intermediate hosts also include various wild rodents such as voles and brown rats (Wang, 2003). The connection between wild animals and domestic animals promotes the spread and prevalence of echinococcosis.
The Ili River is located in the north of Tianshan Mountains, Xinjiang Uygur Autonomous Region, with an altitude of about 900–1100 m. It flows through the Kunes grassland and the agricultural and pastoral areas of 8 counties and 1 city in Ili Kazakh Autonomous Prefecture. Echinococcosis spread along the Ili River Valley, and people and animals living in this area may be infected. In particular, farmers and herdsmen in the Kunes grassland have the habit of keeping dogs. Some people kill foxes, wolves, marmots, and other animals for meat, resulting in a high infection rate. For example, the incidence of alveolar echinococcosis in Nalati township, a sheep breeding farm of Xinyuan County, and Ulastai township of Nilka County is far higher than the national average incidence (Gao et al., 2005). Zhaosu County has developed animal husbandry, a large number of dogs, abundant grassland, and severe rodent infestations. In recent years, investigations have found the existence of wild red foxes in the area (Ma, 2014). Domestic animals, dogs, foxes, and small mammals constitute the circular chain of echinococcosis (McManus et al., 2003), which also provides convenience for the spread of echinococcosis. However, currently, there are relatively few reports on the infection and the genetic polymorphism of Echinococcus in rodents in this region.
To prevent the spread of echinococcosis through rodents and trace the origin of echinococcosis, this study combined pathogenic and molecular biological methods to observe the histopathology of Echinococcus carried by rodents, and amplified the mitochondrial nad1 gene of the pathogen to investigate the genotype and haplotype diversity of Echinococcus in rodents in Ili Prefecture. The aim is to understand the basic situation of Echinococcus infection in rodents, clarify the genetic variation characteristics of this area, and provide basic information for the effective prevention and control of rodents and echinococcosis in Ili Prefecture of Xinjiang.
Materials and methods
Ethics statement
The forestry and veterinary departments in the selected grassland area were informed of the study and agreed to the capture of small wild mammals. All procedures were carried out according to the ethical guidelines for the use of animal samples permitted by the experimental animal ethics committee of the Veterinary Research Institute, Xinjiang Academy of Animal Sciences, Urumqi, China (approval No.: 2017–03).
Study area
Ili Prefecture is located to the west of the Tianshan Mountains in Xinjiang Uygur Autonomous Region of the People’s Republic of China, bordering Kazakhstan (Figure 1). The Ili River, formed by the confluence of three major upstream rivers: the Tekes River, Kunse River, and Kashi River, traverses the whole prefecture (Guo et al., 2021). Nilka County (43°74’N, 83°86’E), Xinyuan County (43°25’N, 84°10’E), and Zhaosu County (43°03’N, 80°89’E), where the survey was conducted, are located in the high-altitude mountain pastoral areas upstream of the Ili River. The annual average temperature in this region is 2.9–5.9°C, with an average rainfall of 600 mm. The grassland area is vast and the stock of domestic animals is high. The warm season lasts from mid-June to late August, which is a suitable time to catch small mammals.
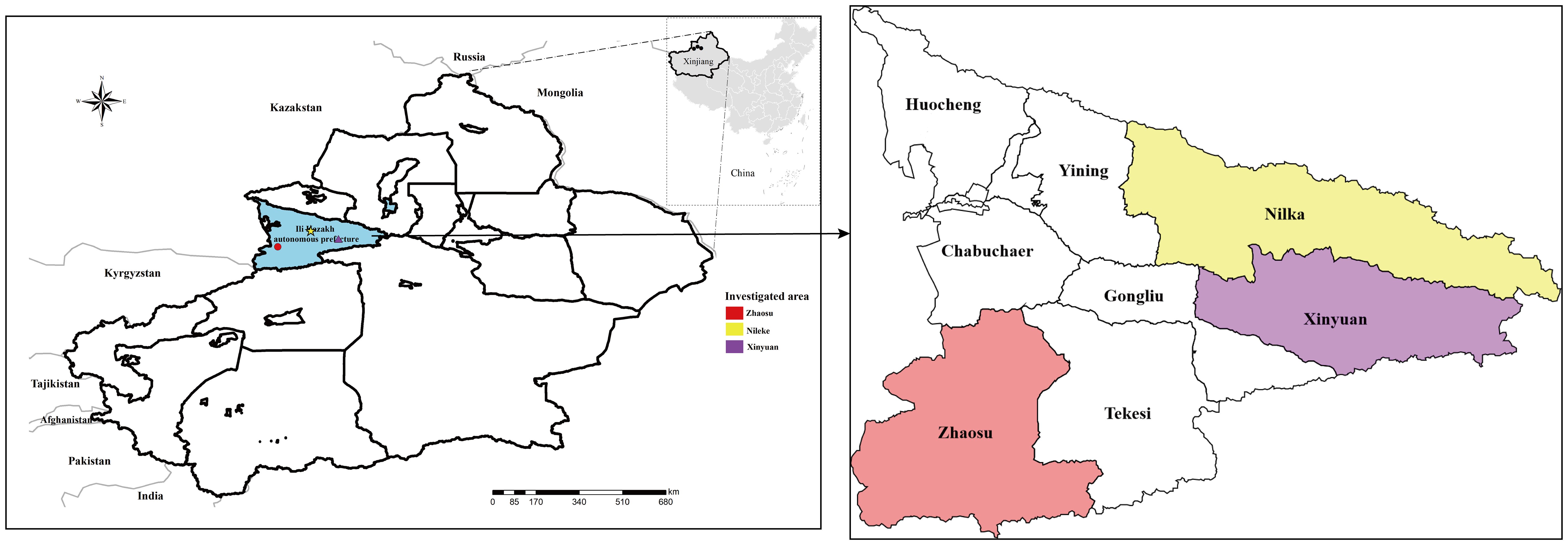
Figure 1 Small mammals were captured from parts of the high-altitude mountainous pasture areas in Ili Kazakh Autonomous Prefecture, Xinjiang, China. In the figure, “different shapes in different colors” indicate the sampling location.
Small mammal collection
Small mammals were collected in June and July 2018. In order to capture small mammals in Nilka and Xinyuan Counties, water was injected into rat holes to drive away small mammals, while collectors caught escaped animals in traps at the entrance of the holes. In Zhaosu County, small mammals were captured using the mousetrap method (Vaniscotte et al., 2009). Break-back traps were set in the rat path and entrance of their dens. Each of the 25 snap traps were arranged in a straight line with a spacing of 5 m and a row spacing of 20 m. The traps were laid each afternoon and collected the next morning. The bait was peanuts.
Each small mammal was dissected, and any lesions of Echinococcus spp. in its organs were carefully checked. Suspected infected livers showing typical (e.g. white dots with a diameter > 1 mm or cyst like lesions) and atypical lesions (e.g. white dots with a diameter < 1 mm, calcified or abnormal surface appearances), were fixed with 75% (v/v) ethanol and stored at 4°C until parasite species identification. The livers without visible lesions were stored under the same conditions as a negative control for subsequent experiments. The rest of the bodies were stored in a 50 ml tube with 75% (v/v) ethanol for further species identification.
Small mammal identification
According to Luo et al (Luo et al., 2000), Smith & Xie (Smith et al., 2010), and the key table in “Rodentology” (Zheng et al., 2008), two rodent identification experts were invited to identify the rodents based on pelt color patterns, skull morphological characteristics, and body measurement data.
Histopathological examination
All rodent liver lesions suspected of Echinococcus infection were excised, fixed in 4% paraformaldehyde, manually dehydrated with gradient ethanol, and then embedded in paraffin after transparent. Four μm thick sections were cut with Leica microtome and placed on glass slides. After hematoxylin and eosin staining (H&E), pathological observation was performed under an Olympus microscope. Ten uninfected liver sections were stained as negative controls.
DNA extraction and PCR amplification
Genomic DNA was extracted from each liver sample fixed with 75% ethanol using a TIANamp Genomic DNA Kit (TIANGEN BIOTECH CO., LTD, CHINA). Extracted DNA was stored at -20°C until further analysis. According to reference (Armua-Fernandez et al., 2011), universal primers E1-ND1 F (5’-GGKTATTCTCARTTTCGTAAGGG-3’) and E1-ND1 R (5’-ATCAAATGGAGTACGATTAGTYTCAC-3’) for the nad1 gene of Echinococcus were synthesized, and the extracted DNA was amplified by polymerase chain reaction (PCR). The final PCR reaction volume was 25 μl, containing 2 μl of each DNA template with a set of primers. Distilled water was used as the negative control for every experiment. The amplification was performed in a T100™ thermal cycler (Applied Bio-Rad, CA, USA) with the following program: initial denaturation at 94°C for 4 min; followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s and extension at 72°C for 1 min, with the last extension at 72˚C for 10 min. The amplification products (5 μl) were visualized on 1% agarose gel electrophoresis detection. PCR products were sent to Sangon Biotech (Beijing, China) for Sanger sequencing.
Sequence analysis
The obtained sequence identities and similarities were determined by BLASTn search on the NCBI database, from which the closest sequences were retrieved, and sequences were multiple aligned using Clustal X2 (Larkin et al., 2007) and BioEdit 7.0.9.1 (Alzohairy, 2011). The phylogenetic tree was constructed in the MEGA 7.0 program (Guo et al., 2023), with the neighbor-joining (NJ) method using the Kimura 2-parameter model, which was determined to be the best substitution model. The robustness of phylogenetic trees was tested by bootstrapping using 1000 replicates (Mi et al., 2014). Population diversity indexes (number of haplotypes (Hn), haplotype diversity (Hd), and nucleotide diversity (π) values) and neutrality (Tajima’s D, and Fu’s Fs) were tested using DnaSPv 6.10.04 (Julio et al., 2017). The genetic distances between isolates of E. multilocularis from different regions were analyzed using pairwise fixation index (Fst) by Arlequin 3.5.2.2 (Laurent and Lischer, 2010). Haplotype network was drawn with the PopART 1.7 (Population Analysis with Reticulate Trees) network analysis software (Leigh and Bryant, 2015) using the TCS network inference method (Clement et al., 2000).
Statistical analysis
IBM SPSS Statistics 20.0 software was used for statistical analysis. The difference in infection rates in different regions and different rodent species were compared by Pearson’s Chi-square test (χ2 test). When p < 0.05, the difference was considered significant.
Results
Small mammal species identification
A total of 267 small mammals were captured from the study sites in June and July 2018. All small mammals trapped were voles and identified as three species: Microtus ilaeus 24.72% (66/267), Ellobius talpinus 6.74% (18/267), and Microtus gregalis 68.54% (183/267). In total, 40 M. ilaeus (40/55, 72.73%), 9 E. talpinus (9/55, 16.36%), and 6 M. gregalis (6/55, 10.91%) were captured in Nilka County; 12 M. ilaeus (12/128, 9.38%), 7 E. talpinus (7/128, 5.47%), and 109 M. gregalis (109/128, 85.16%) were captured in Xinyuan County; and 14 M. ilaeus (14/84, 16.67%), 2 E. talpinus (2/84, 2.38%), and 68 M. gregalis (68/84, 80.95%) were captured in Zhaosu County. There were significant differences in the species composition of voles among the three trapping plots (χ2 = 109.603, p < 0.01) (Table 1). The results showed that there were differences in the species composition of voles among different regions, with Nilka County being the habitat of M. ilaeus, and Xinyuan County and Zhaosu County being the habitat of M. gregalis.
Prevalence of Echinococcus in small mammals
Of the 267 rodents were dissected, 37 were found to have suspected Echinococcus lesions (cysts) through visual examination. All lesions were located in the liver. The cysts varied in shape and size, most of which were translucent or opalescent round or oval vesicular nodules. Among them, 31 cysts were embedded in the liver, with a long diameter of 0.717 cm ± 0.275 cm and a short diameter of 0.461 cm ± 0.236 cm; 6 cysts were attached and protruded on the liver surface, with a long diameter of 0.750 cm ± 0.384 cm and a short diameter of 0.400 cm ± 0.255 cm (Figure 2).
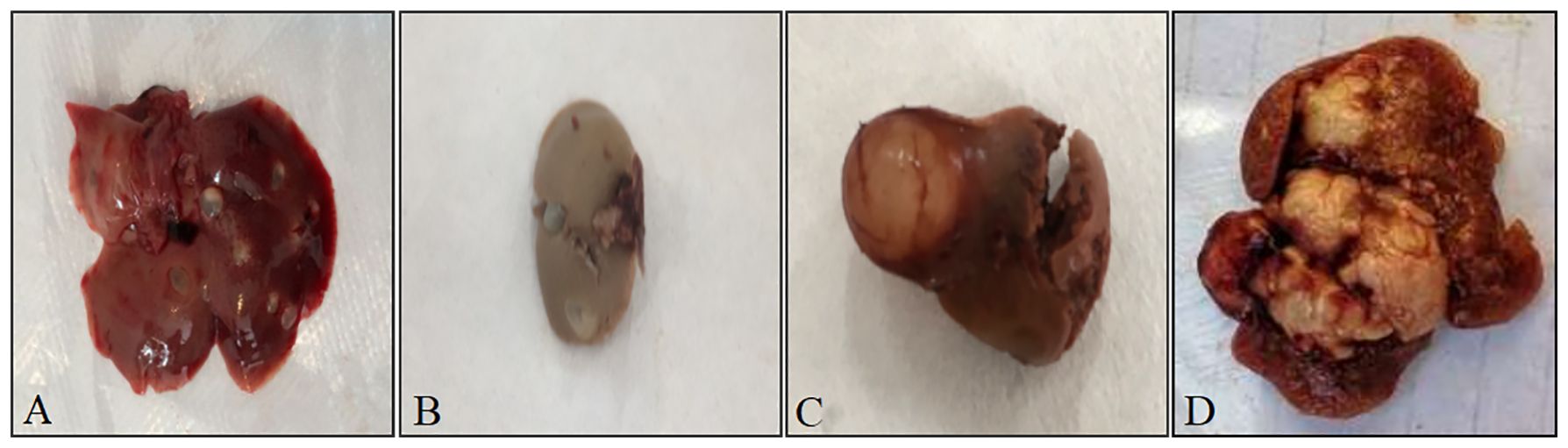
Figure 2 Different appearances of rodent liver cysts. (A) Cysts in the liver of rodents, the reddish-brown is the liver, grayish-white translucent vesicles are cysts; (B) Cysts in the liver of rodents, the grayish-brown is the liver, grayish-white translucent vesicles are cysts; (C) Cysts attached to and protruding from the surface of rodent liver, the reddish-brown is the liver, the grayish-white spheres with blood vessels attached on the surface are cysts; (D) Cysts attached to and protruding from the surface of rodent liver, the reddish-brown is the liver, white tumor-like is the cyst.
The H&E staining results of tissue sections suspected to be infected with Echinococcus showed that the lesion area lost normal liver tissue structure and interstitial connective tissue hyperplasia. There were many vesicles of different sizes in the lesion area. The cyst wall was divided into two layers, the inner layer is the germinal layer, and the outer layer is the cuticle, which were stained in a homogeneous pink color. The vesicles contained many round or oval protoscoleces (PSCs) and round vacuoles of different sizes or calcareous particles dyed dark blue, with inflammatory cell infiltration around the vesicles (Figure 3).
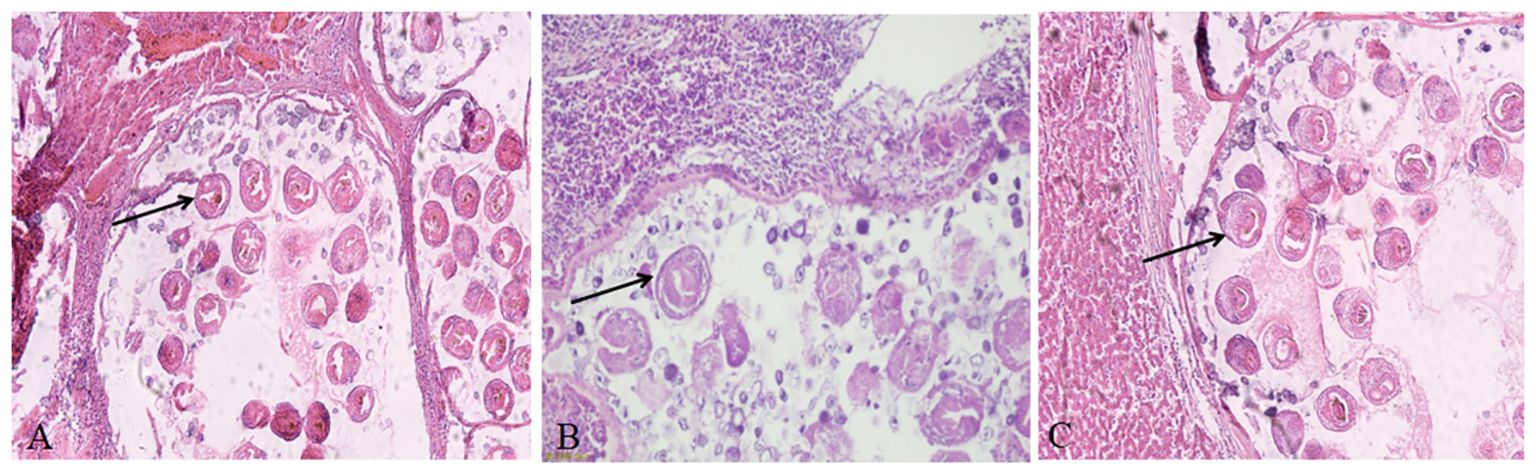
Figure 3 Hematoxylin and eosin staining showing the characteristic pathological response in echinococcosis. (A) Liver cyst sections of rodents from herdsman’s original residence; (B) Liver cyst sections of rodents from artificial pasture; (C) Liver cyst sections of rodents from natural grassland. The original magnification of all tissue sections was 100×; black arrow indicates PSCs.
To further confirm the number of positive samples of Echinococcus infection in rodents, molecular analyses later detected Echinococcus mtDNA. In total, 42 Echinococcus-positive samples were detected from 267 individuals, including 37 samples containing protoscoleces detected by H&E staining and 5 samples from other individuals without lesions. The total infection rate was 15.73% (42/267). Furthermore, 14 infected rodents (including 8 M. ilaeus, 2 E. talpinus, 4 M. gregalis) were from Nilka, with an infection rate of 25.45% (14/55); 18 infected rodents (all M. gregalis) were from Xinyuan, with an infection rate of 14.06% (18/128); and 10 infected rodents (all M. gregalis) were from Zhaosu, with an infection rate of 11.90% (10/84). There was no significant difference in the infection rate among the different regions (χ2 = 5.119, p > 0.05). The infection rates of different rodent species were as follows: the M. ilaeus infection rate was 12.12% (8/66), the E. talpinus infection rate was 11.11% (2/18), and the M. gregalis infection rate was 17.49% (32/183), and there was no significant difference in the infection rates among the different rodent species (χ2 = 1.364, p > 0.05) (Table 1).
Haplotype characteristics and phylogenetic analysis
In total, 42 PCR products were directly sequenced to determine genetic variation among the isolates. The sizes of the amplified DNA fragments were 510 bp. DNA consensus sequences of 453 bp were obtained by trimming low-quality chromatogram data. Compared with the publicly available nad1 sequences, all isolates were identified as E. multilocularis. Blast analysis of the study’s isolates revealed higher than 98.25% similarity with the GenBank sequences of E. multilocularis.
Based on the comparison of the nad1 gene fragment, 21 different haplotypes were identified and named Hap_1-Hap_21 (Table 2). Among all haplotypes, Hap_2 was the major haplotype, accounting for 23.81% (10/42) of the isolates, which were all from M. gregalis. Hap_8 was the second common haplotype, observed in 11.90% (5/42) isolates, two from M. ilaeus and three from M. gregalis. Other haplotypes (Hap_1, Hap_3–7, and Hap_9–21) were found in 64.29% (27/42) isolates. In addition, Hap_1, Hap_10–16, and Hap_18–21 were only identified in M. gregalis (40.48%, 17/42); Hap_3–7 were only found in M. ilaeus (14.29%, 6/42); and Hap_17 was only found in E. talpinus (2.38%, 1/42) (Table 2).
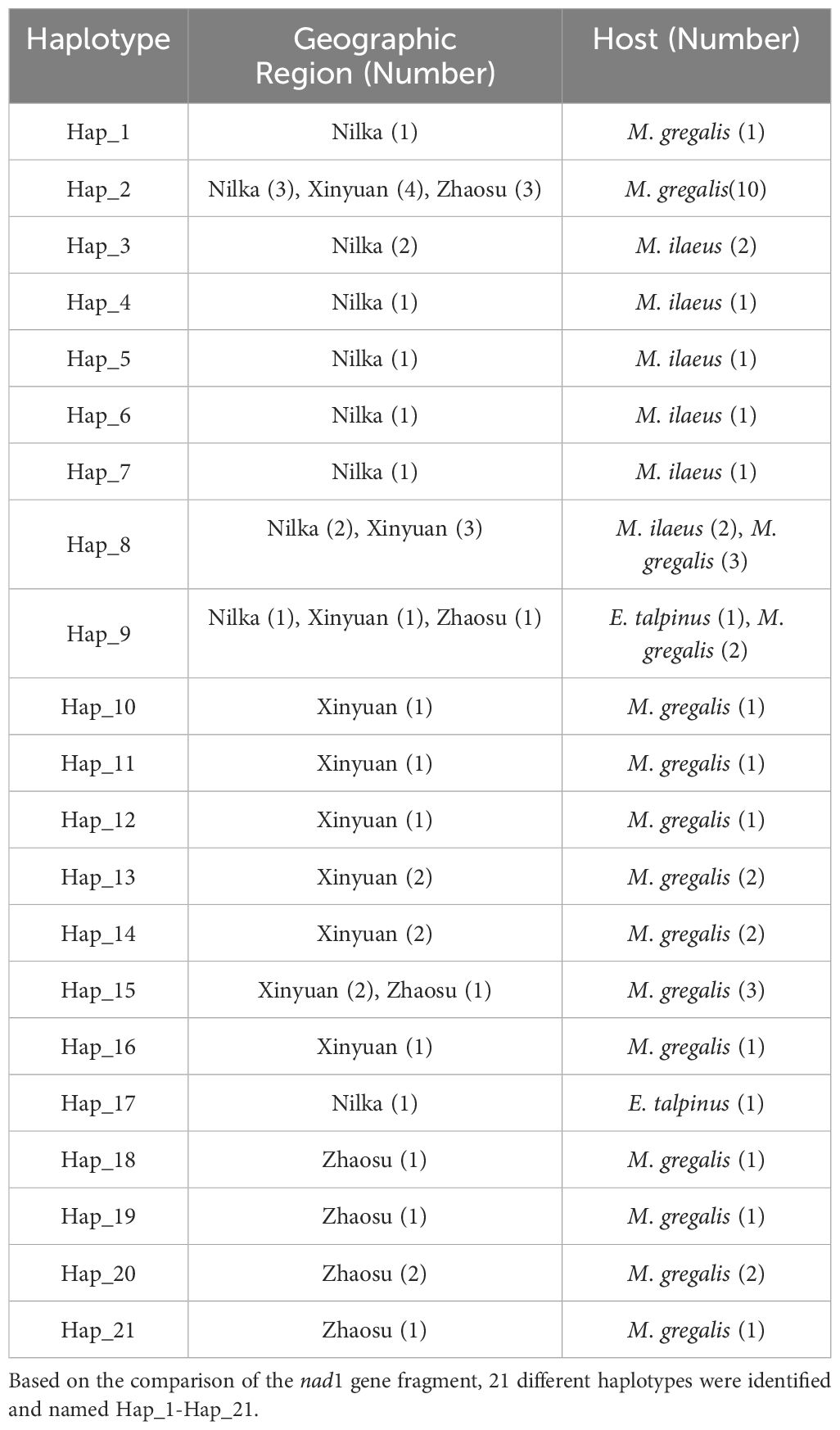
Table 2 E. multilocularis haplotypes characterized by partial nad1 sequence and used for phylogenetic analysis.
An NJ tree was constructed based on 36 sequences, including 21 E. multilocularis haplotype sequences from this study, 10 reference sequences of E. multilocularis, 3 different types of E. granulosus sequences, and 1 E. shiquicus sequence, with Taenia saginata as outgroups (Supplementary Table 1). Phylogenetic analysis showed that the 21 E. multilocularis haplotype sequences obtained in this study belonged to the same robust group (100% bootstrap value), in which Hap_2 (the major haplotype) was close to the published sequences EU704122 (Sichuan, China) and Hap_8–9, Hap_16, and Hap_18–21 were close to the published sequences KY094609 (Qinghai, China), AJ237639 (China), and MH259778 (China) (Figure 4).
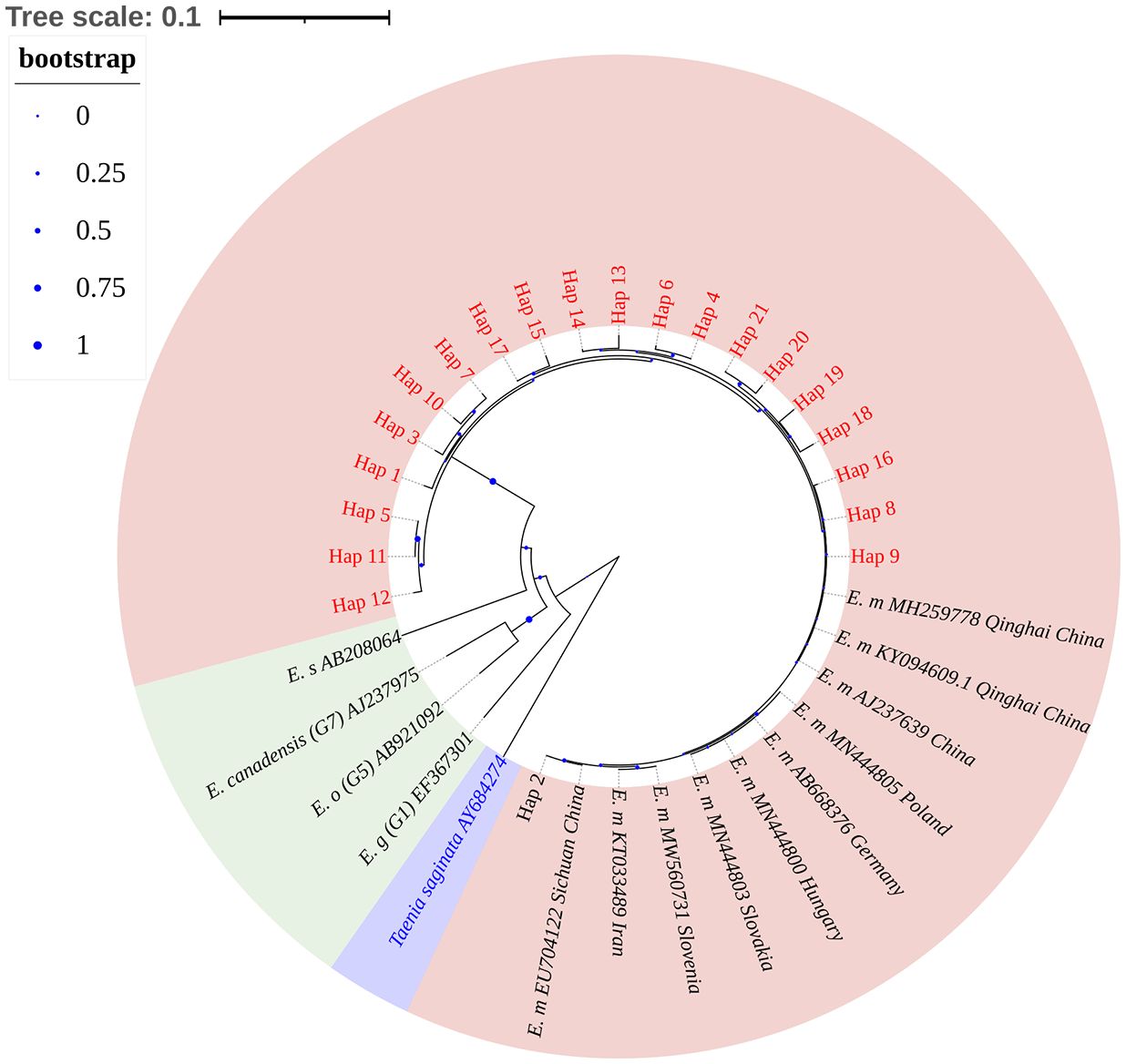
Figure 4 Phylogenetic analysis of Echinococcus nad1 gene sequences by the neighbor-joining method based on the Kimura 2-parameter model. The bootstrap method via 1000 pseudo replicates was used to assess the reliability of the tree. The pink area in the figure indicates sample sequences and sequences of different isolates of E. multilocularis; green area indicates other Echinococcus; purple area indicates outgroup sequences. The blue dots on the branches represent the bootstrap values; the smaller the bootstrap values, the smaller the blue dot, and vice versa. Phylogenetic tree landscaping was performed using an online website: https://itol.embl.de/.
Haplotype networks analysis
The haplotype network of E. multilocularis was constructed with 83 nad1 sequences, as shown in Figure 5. Among these, 42 were acquired from Ili Prefecture in this study (Table 2), and the other 41 sequences were from online data from Xinjiang, Qinghai, Gansu, Sichuan, Poland, Italy, Slovakia, Hungary, Austria, and Germany (Supplementary Table 1). The network has a typical star-like shape connected by 9 haplotypes, of which Hap_2 (39.76%, 33/83) was in the center. Hap_2, Hap_4, Hap_5, Hap_6, and Hap_14 formed an approximate ellipse. This haplotype network included 25 haplotypes. Among these, 20 haplotypes were only prevalent in specific regions, while 5 haplotypes (Hap_2, Hap_5, Hap_12, Hap_19, and Hap_22) were simultaneously prevalent in more than 2 regions. Hap_2, as the dominant haplotype, contains one sequence from Italy and all the other sequences were from various regions of China, indicating that this haplotype is the major haplotype prevalent in China. Hap_19, the second largest haplotype, mainly contains sequences from Poland, Italy, Slovakia, Hungary, and Germany, indicating that it is a relatively common haplotype in Europe.
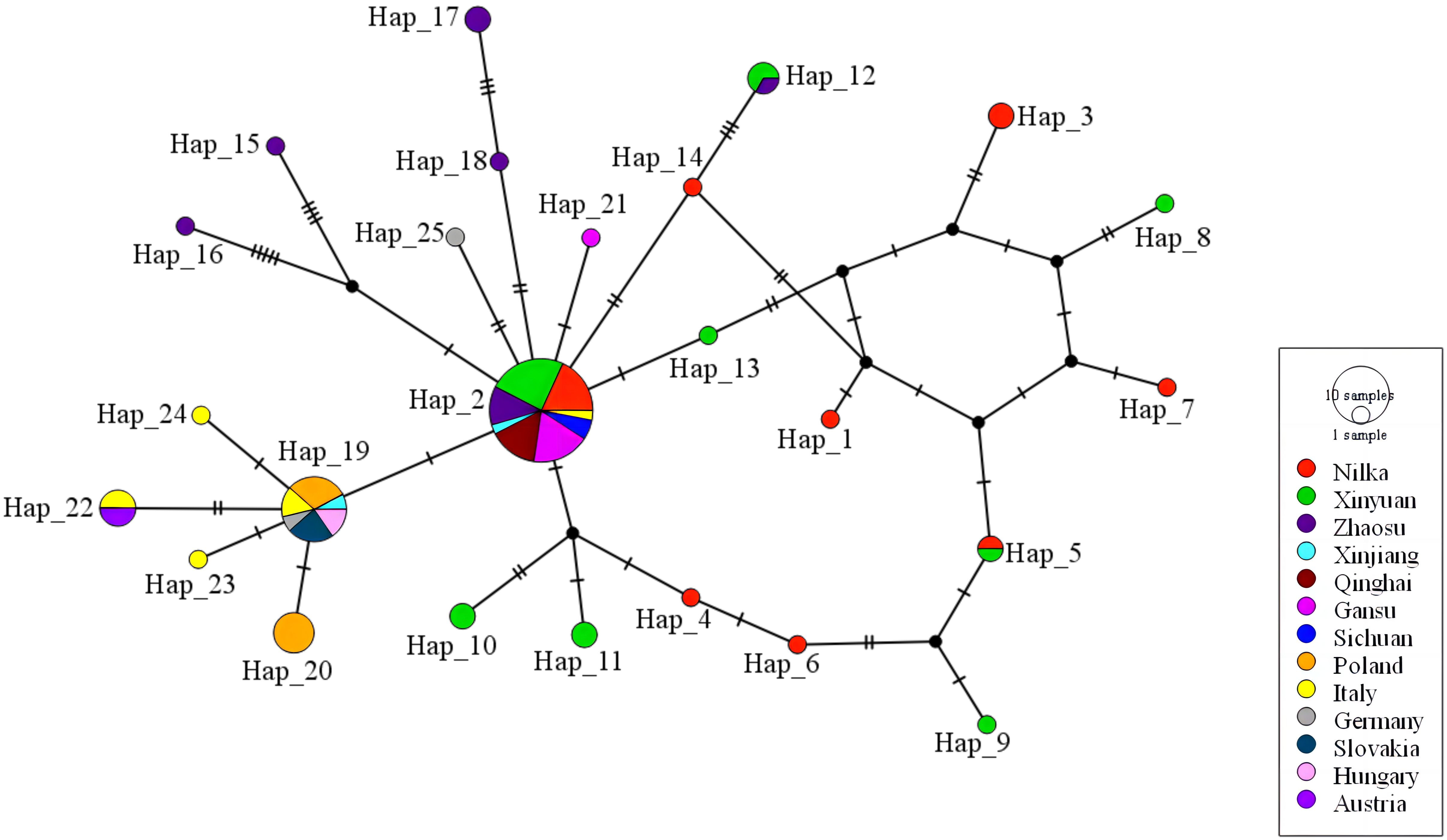
Figure 5 The haplotype network for the nad1 gene of E. multilocularis. Geographical distribution of the haplotypes is indicated by different colors. Size of circles is proportional to the frequency of each haplotype. Each cross-sectional line represents a gene mutation site. Black circles: hypothetical intermediate haplotypes. Haplotype network was drawn with the PopART 1.7 software using the TCS network inference method.
Population genetics analysis
The haplotype and nucleotide diversities for the E. multilocularis nad1 gene in this study were 0.925 ± 0.027 and 0.01139 ± 0.00119, respectively, indicating that the population diversity in Ili Prefecture was high. The negative values of both Tajima’s D (D = -1.07256, p > 0.10) and Fu’s Fs (Fs = -7.172, p > 0.10) tests indicated that the E. multilocularis population in small mammals of Ili Prefecture is currently expanding. The haplotype diversity ranged from 0.797 to 0.844, and the nucleotide diversity ranged from 0.00876 to 0.01063 in the three regions of Ili Prefecture (Table 3). These results showed that the E. multilocularis population in Ili Prefecture exhibits high levels of haplotype diversity and relatively low levels of nucleotide diversity, suggesting minimal population genetic differentiation in this region. In contrast, previously uploaded haplotype sequences of E. multilocularis from Xinjiang showed higher haplotype diversity (Hd =1.000) and lower nucleotide diversity (0.00226) than those from other regions (Supplementary Table 2).
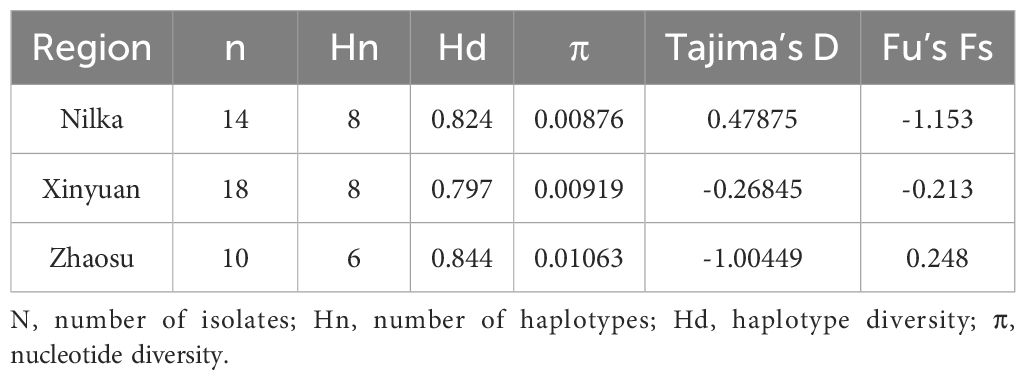
Table 3 Population diversity indices of E. multilocularis isolates from Ili Prefecture, calculated from the nad1 gene fragment.
The Fst estimated the population differentiation between different regions, ranging from 0.00000 to 0.16945 (Table 4), and indicated low genetic differentiation between Nilka and Xinyuan, moderate genetic differentiation between Xinyuan and Zhaosu, and greater genetic differentiation between Nilka and Zhaosu.
Discussion
E. multilocularis is one of the most pathogenic and widespread species of the genus Echinococcus. Small rodents serve as the primary intermediate hosts for E. multilocularis, with their habitats and activity ranges directly influencing the presence and transmission of the parasite. Changes in the species composition and distribution of small rodents also affect the epidemiology of E. multilocularis (Shang et al., 2020). Therefore, we conducted an investigation and analysis of the epidemiology, geographic distribution, and phylogeny of small rodents infected with alveolar echinococcosis (AE).
This epidemiological survey of E. multilocularis in Ili Prefecture included 3 regions (Nilka, Xinyuan, dmg Zhaosu), the sampling points located in the high-altitude mountainous pasture, with an altitude of 1980–2500 meters. The captured rodents were all voles. The possible reason is that, geographically, altitude and rainfall impact the landscape and habitats, and grasslands are better suitable for the survival of voles (Giraudoux et al., 2003; Giraudoux et al., 2013a; Giraudoux et al., 2013b; Cadavid et al., 2016). However, the dominant species of small rodents captured varied in different regions. In Nilka County, the captured rodents were mainly M. ilaeus (40/55, 72.73%), while in Xinyuan and Zhaosu County, they were mainly M. gregalis (109/128, 85.16%; 68/84, 80.95%). M. gregalis is mainly distributed in Xinjiang, Inner Mongolia, Hebei, and northern Shanxi in China. In foreign countries, it is mainly distributed in Mongolia and Russia (Chen et al., 2014). Previous reports have indicated that M. gregalis is a dominant species of artificial grassland in Zhaosu, and can be infected with E. multilocularis (Ma, 2014). M. ilaeus, ranking second in quantity in this survey, was initially found to be infected with E. multilocularis during a wild animal disease investigation in Nilka County, northwest Xinjiang, in 1999 (Jiang et al., 2000). These findings suggest that rodents are widely distributed in the Ili region and may play an important role in the epidemic transmission of AE.
Whether the intermediate host is infected with protoscoleces is mainly confirmed by characteristic lesions, microscopic observation, tissue sectioning, immunohistochemistry, and PCR analysis. In this study, captured rodents were dissected first, and the characteristic lesions were identified through visual examination. The results found that all lesions were in the livers of rodents, but not in other organs such as lungs and kidneys, which was consistent with the fact that alveolar echinococcus was nearly 100% primary in the liver (Brehm and Koziol, 2017). The morphology of the infected cysts in rodents was similar to that observed in the investigation on echinococcosis in wild rodents at Alashankou port (Xiao et al., 2015), but most of them were single translucent gray-white round cysts, which may be due to the different sampling times, so the development status of cysts was different. In our study, cysts were detected in only 13.86% (37/267) of the liver samples by H&E staining of tissue sections, but this increased to 15.73% (42/267) positive for E. multilocularis infection with PCR in the livers of all sampled specimens. A possible reason was that in this study only positive samples with obvious cysts found by visual inspection were subjected to tissue sectioning, and visual inspection has certain limitations and subjectivity.
Additionally, it may be assumed that this finding reflected the complexity of distinguishing small immature cysts (Al-Sabi and Kapel, 2011), especially in animals less than 3 months old (Burlet et al., 2011), or in atypical or calcified liver lesions less than 5 mm in diameter (Gottstein et al., 2001; Torgerson and Deplazes, 2009) by microscopic examination, while PCR is particularly suitable for calcified or infertile cysts.
In this study, PCR detection revealed that the total infection rate of Echinococcus in rodents in Ili Prefecture was 15.73% (42/267). This result was similar to previous reports in Ili Prefecture (Guo et al., 2021), but significantly higher than the infection rate of rodent Echinococcus in endemic provinces (autonomous regions) in China in 2022 (Kui et al., 2024). A possible reason is that the sample size was too low to prove the true extent of local infection with E. multilocularis and needs to be confirmed by repeat surveys. Phylogenetic analyses revealed that all 21 nad1 gene haplotypes from Ili Prefecture were closely related and clustered with haplotypes from Qinghai and Sichuan (Figure 4). Shang et al. found that Xinjiang is separated from Qinghai and Sichuan in the Tibetan Plateau by the Kunlun Mountains and Taklimakan Desert, with E. multilocularis subgroups sharing the same primitive haplotype (Shang et al., 2021). Wu et al. also observed the coexistence of E. multilocularis haplotypes in Qinghai and Xinjiang (Wu et al., 2017), indicating that E. multilocularis may have the same origin in this region.
The study of gene polymorphism is an important basis for understanding the biological characteristics and epidemiological patterns of E. multilocularis, evaluating the risk of transmission of E. multilocularis, and formulating and optimizing relevant control strategies (Shang et al., 2021). This study analyzed the genetic polymorphism of the nad1 gene of E. multilocularis in Ili Prefecture of western Xinjiang, and explored the relationship between geographical subgroups of E. multilocularis based on GenBank data. The analysis showed high genetic diversity in E. multilocularis in this region, and the detected haplotypes were distributed in a star shape with one dominant haplotype as the center, suggesting a founder effect in the population. This is similar to the findings of Nakao et al. on the genetic polymorphism of the cox1 gene in Echinococcus (Nakao et al., 2009a). The main haplotype was identical to the previous isolates found in Sichuan (Nakao et al., 2009b) and Qinghai (Li et al., 2018), belonging to the Asian branch of E. multilocularis, but it has also been detected in Italy (Figure 5). Indeed, the atypical cross-state distribution of Asian groups of E. multilocularis was not an exception, the prevalence of Asian strains of E. multilocularis has also been identified in Russia (Konyaev et al., 2013) and Alaska (Nakao et al., 2009b). Nakao et al (Nakao et al., 2009a; Nakao et al., 2009b). suggested that red foxes, which are highly adapted to different environments, may play an important role in the long-distance transmission of the Asian group of E. multilocularis.
Conclusion
In this study, the type of Echinococcus infection in small mammals was E. multilocularis. Among the analyzed sequences, the highly prevalent haplotype was Hap_2. AE is highly prevalent in Ili Prefecture, and M. gregalis, as the dominant species collected, plays an important role in its transmission. Due to herdsmen raising sheep in the summer season in high-altitude pasture areas where there are significant numbers of small mammals infected with E. multilocularis, shepherd dogs come into contact with herdsmen after consuming infected animals, which increases the spread of echinococcosis. Therefore, it is necessary to strengthen the monitoring of E. multilocularis in small rodents and the management of domestic dogs, and develop scientific prevention and control measures to prevent the spread of AE.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Veterinary Research Institute, Xinjiang Academy of Animal Sciences, Urumqi, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BW: Writing – original draft, Data curation, Formal analysis, Investigation, Methodology, Conceptualization. LZ: Writing – original draft, Conceptualization, Data curation, Formal analysis, Methodology. WB: Data curation, Investigation, Writing – review & editing. XZ: Data curation, Investigation, Writing – review & editing. CQ: Data curation, Investigation, Writing – review & editing. MT: Data curation, Investigation, Writing – review & editing. LH: Data curation, Investigation, Writing – review & editing. ZC: Data curation, Investigation, Writing – review & editing. ZZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by National Key Research and Development Program of China (2022YFE0114600). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank Ili Prefecture Animal Husbandry and Veterinary Bureau and Animal Husbandry and Veterinary Stations in Xinyuan County, Nilka County, and Zhaosu County for their assistance in capturing rodents at sampling points.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1433359/full#supplementary-material
References
Al-Sabi, M. N., Kapel, C. M. (2011). Multiplex PCR identification of Taenia spp. in rodents and carnivores. Parasitol. Res. 109, 1293–1298. doi: 10.1007/s00436-011-2373-9
Alzohairy, M. A. (2011). BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2, 60–61.
Armua-Fernandez, M. T., Nonaka, N., Sakurai, T., Nakamura, S., Gottstein, B., Deplazes, P., et al. (2011). Development of PCR/dot blot assay for specific detection and differentiation of taeniid cestode eggs in canids. Parasitol. Int. 60, 84–89. doi: 10.1016/j.parint.2010.11.005
Brehm, K., Koziol, U. (2017). Echinococcus-host interactions at cellular and molecular levels. Adv. Parasitol. 95, 147–212. doi: 10.1016/bs.apar.2016.09.001
Burlet, P., Deplazes, P., Hegglin, D. (2011). Age, season and spatio-temporal factors affecting the prevalence of Echinococcus multilocularis and Taenia taeniaeformis in Arvicola terrestris. Parasit Vectors. 4, 6. doi: 10.1186/1756-3305-4-6
Cadavid, R. A. M., Yang, Y. R., McManus, D. P., Gray, D. J., Giraudoux, P., Barnes, T. S., et al. (2016). The landscape epidemiology of echinococcoses. Infect. Dis. Poverty. 5, 13–25. doi: 10.1186/s40249-016-0109-x
Chen, X. H., Zhang, Z. Z., Ma, Z. G., Xia, H., Shi, Z. S., Wubulihashengmu, S. L. K., et al. (2014). Investigation on rodent pests of artificial grassland in zhaosu regimental farm 77th. Grass-feeding Livestock. 166, 45–48. doi: 10.16863/j.cnki 1003-6377.2014.03.012
Clement, M., Posada, D., Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Gao, Y. S., Zhu, M. B., Guo, Y. Z., DIL, M. R. T., Ar, X., Wang, Y., et al. (2005). Clinical analysis on hepatic hydatid disease in Yili River Valley. Chin. J. Parasitol. Parasit Dis. 23, 12–15.
Giraudoux, P., Craig, P. S., Delattre, P., Bao, G., Bartholomot, B., Harraga, S., et al. (2003). Interactions between landscape changes and host communities can regulate Echinococcus multilocularis transmission. Parasitology. 127, S121–S131. doi: 10.1017/S0031182003003512
Giraudoux, P., Raoul, F., Afonso, E., Ziadinov, I., Yang, Y. R., Li, L., et al. (2013a). Transmission ecosystems of Echinococcus multilocularis in China and Central Asia. Parasitology. 140, 1655–1666. doi: 10.1017/S0031182013000644
Giraudoux, P., Raoul, F., Pleydell, D., Li, T., Han, X., Qiu, J., et al. (2013b). Drivers of Echinococcus multilocularis transmission in China: small mammal diversity, landscape or climate? PloS Negl. Trop. Dis. 7, e2045. doi: 10.1371/journal.pntd.0002045
Gottstein, B., Saucy, F., Deplazes, P., Reichen, J., Demierre, G., Busato, A., et al. (2001). Is high prevalence of Echinococcus multilocularis in wild and domestic animals associated with disease incidence in humans? Emerg. Infect. Dis. 7, 408–412. doi: 10.3201/eid0703.017307
Guo, B., Zhao, L., Zhao, L., Mi, R. S., Zhang, X., Wang, B. J., et al. (2023). Survey and Molecular Characterization of Echinococcus granulosus sensu stricto from Livestock and Humans in the Altai Region of Xinjiang, China. Pathogens. 12, 134–146. doi: 10.3390/pathogens12010134
Guo, B. P., Zhang, Z. Z., Guo, Y. Z., Guo, G., Wang, H. Y., Ma, J. J., et al. (2021). High endemicity of alveolar echinococcosis in Yili Prefecture, Xinjiang Autonomous Region, the People's Republic of China: Infection status in different ethnic communities and in small mammals. PloS Negl. Trop. Dis. 15, e0008891. doi: 10.1371/journal.pntd.0008891
Jiang, W., Zheng, Q., Istraine, O. S. M. A., Xu, Q. Y., Chai, J. J. (2000). First discovery of natural infection of metacestode of Echinococcus multilocularis in Microtus ilaeus in Nilka County Xinjiang, China. Bull. Dis. Control Prev. (China) 15, 36–37+96. doi: 10.13215/j.cnki.jbyfkztb.2000.01.016
Julio, R., Albert, F. M., Carlos, S. D. J., Sara, G. R., Pablo, L., Sebastián E, R. O., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Konyaev, V. S., Yanagida, T., Nakao, M., Ingovatova, M. G., Shoykhet, N. Y., Bondarev, Y. A., et al. (2013). Genetic diversity of Echinococcus spp. in Russia. Parasitology. 140, 1637–1647. doi: 10.1017/S0031182013001340
Kui, Y., Xue, C. Z., Wang, X., Liu, B. X., Wang, Y., Wang, L. Y., et al. (2024). Progress of echinococcosis control in China, 2022. Chin. J. Parasitol. Parasit Dis. Feb. 42, 8–16.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 21, 2947–2948. doi: 10.1093/bioinformatics/btm404
Laurent, E., Lischer, E. L. H. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Leigh, J. W., Bryant, D. (2015). PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Li, B. J., Guo, S. X., Peng, X. Y. (2009). Current status of epidemiological investigation for hydatidosis. Med. Recapitulate. 15, 1195–1198.
Li, J. Q., Li, L., Fan, Y. L., Fu, B. Q., Zhu, X. Q., Yan, H. B., et al. (2018). Genetic diversity in echinococcus multilocularis from the plateau vole and plateau pika in jiuzhi county, Qinghai Province, China. Front. Microbiol. 9, 2632–2640. doi: 10.3389/fmicb.2018.02632
Luo, Z. X., Chen, W., Gao, W. (2000). “Fauna Sinica Mammalia,” in Rodentia Part III: Cricetidae (Science Press, Beijing), 522.
Ma, Z. (2014). Epidemiological investigation for wild host of alveolar echinococcosis at Zhaosu basin, Xinjiang. Xinjiang Agricultural University, Urumqi.
Mandal, S., Mandal, M. D. (2012). Human cystic echinococcosis: epidemiologic, zoonotic, clinical, diagnostic and therapeutic aspects. Asian Pac J. Trop. Med. 5, 253–260. doi: 10.1016/S1995-7645(12)60035-2
McManus, D. P., Zhang, W. B., Li, J., Bartley, P. B. (2003). Echinococcosis. Lancet. 362, 1295–1304. doi: 10.1016/S0140-6736(03)14573-4
Mi, R., Wang, X. J., Huang, Y., Zhou, P., Liu, Y. X., Chen, Y. J., et al. (2014). Prevalence and molecular characterization of Cryptosporidium in goats across four provincial level areas in China. PloS One 9, e111164. doi: 10.1371/journal.pone.0111164
Nakao, M., Li, T., Han, X., Ma, X., Xiao, N., Qiu, J., et al. (2009a). Genetic polymorphisms of Echinococcus tapeworms in China as determined by mitochondrial and nuclear DNA sequences. Int. J. Parasitol. 40, 379–385. doi: 10.1016/j.ijpara.2009.09.006
Nakao, M., Xiao, N., Okamoto, M., Yanagida, T., Sako, Y., Ito, A. (2009b). Geographic pattern of genetic variation in the fox tapeworm Echinococcus multilocularis. Parasitol. Int. 58, 384–389. doi: 10.1016/j.parint.2009.07.010
Ni, X. W., Yan, H. B., Lou, Z. Z., Jia, W. Z. (2012). Progress in the detection methods of wild animals infected with Echinococcus species. Chin. Vet. Sci. 42, 100–106. doi: 10.16656/j.issn.1673-4696.2012.01.020
Qi, Y. F., Wu, W. P. (2013). Progress on the epidemiology of echinococcosis. Chin. J. Parasitol. Parasit Dis. 31, 143–148.
Shang, J. Y., Zhang, G. J., Yu, W. J., He, W., Liao, S., Li, R. R., et al. (2020). Advances in researches on the genetic diversity of Echinococcus multilocularis. Chin. J. Parasitol. Parasit Dis. 38, 637–642. doi: 10.13350/j.cjpb.210203
Shang, J. Y., Zhang, G. J., Yu, W. J., He, W., Wang, Q., Liao, S., et al. (2021). Genetic polymorphism of Echinococcus multilocularis in northwestern China inferred from cox1 gene sequences. J. Pathogen Biol. 16, 137–142.
Smith, A. T., Gemma, F., Xie, Y., Hoffmann, R. S., Lunde, D., MacKinnon, J. (2010). A guide to the mammals of China (Princeton: Princeton University Press), 55–184. doi: 10.1515/9781400834112
Torgerson, P. R., Deplazes, P. (2009). Echinococcosis: diagnosis and diagnostic interpretation in population studies. Trends Parasitol. 25, 164–170. doi: 10.1016/j.pt.2008.12.008
Vaniscotte, A., Pleydell, D., Raoul, F., Quere, J. P., Qiu, J. M., Wang, Q., et al. (2009). Modelling and spatial discrimination of small mammal assemblages: an example from Western Sichuan (China). Ecol. Modell 220, 1218–1231. doi: 10.1016/j.ecolmodel.2009.02.019
Wu, C. C., Zhang, W. B., Ran, B., Fan, H., Wang, H., Guo, B., et al. (2017). Genetic variation of mitochondrial genes among Echinococcus multilocularis isolates collected in western China. Parasit Vectors. 10, 265–271. doi: 10.1186/s13071-017-2172-y
Xiao, N., Qiu, J. M., Nakao, M., Li, T. Y., Yang, W., Chen, X. W., et al. (2006). Echinococcus shiquicus, a new species from the Qinghai-Tibet plateau region of China: discovery and epidemiological implications. Parasitol. Int. 55, 233–236. doi: 10.1016/j.parint.2005.11.035
Xiao, Y. X., Xu, X. L., Yin, X. P., Wang, A. D., Yan, Z. X., Duan, C. R., et al. (2015). Investigation on Echinococcosis in wild rodents at Ala-shankou port. Acta Vet. Zootech Sin. 46, 1004–1010.
Zeng, X. M., Guan, Y. Y., Wu, W. P. (2014). Epidemiological distribution characteristics of echinococcosis. Chin. J. Zoonoses. 30, 413–417.
Keywords: Echinococcus multilocularis, nad1 gene, genetic polymorphism, rodent, Ili
Citation: Wang B, Zhao L, Ban W, Zhang X, Quan C, Teliewuhan M, He L, Chen Z and Zhang Z (2024) Investigation and genetic polymorphism analysis of rodents infected with Echinococcus in Ili Prefecture, Xinjiang Uygur Autonomous Region, China. Front. Cell. Infect. Microbiol. 14:1433359. doi: 10.3389/fcimb.2024.1433359
Received: 15 May 2024; Accepted: 11 July 2024;
Published: 09 August 2024.
Edited by:
Omar Hamarsheh, Al-Quds University, PalestineReviewed by:
Isabel Mauricio, New University of Lisbon, PortugalVladimir M. Jovanovic, Free University of Berlin, Germany
Copyright © 2024 Wang, Zhao, Ban, Zhang, Quan, Teliewuhan, He, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuangzhi Zhang, enpodWFuZ3poaUBhbGl5dW4uY29t
†These authors have contributed equally to this work and share first authorship
 Bingjie Wang
Bingjie Wang Li Zhao1†
Li Zhao1†