- 1Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, Xi’an, China
- 2Provincial Key Laboratory of Biotechnology of Shaanxi Province, Northwest University, Xi’an, China
- 3Ultrasound Diagnosis Center, Shaanxi Provincial People’s Hospital, Shaanxi, Xi’an, China
The indiscriminate use of antibiotics has resulted in a growing resistance to drugs in Pseudomonas aeruginosa. The identification of antibiotic resistance genes holds considerable clinical significance for prompt diagnosis. In this study, we established and optimized a Recombinase-Aided Amplification (RAA) assay to detect two genes associated with drug resistance, oprD and arr, in 101 clinically collected P. aeruginosa isolates. Through screening for the detection or absence of oprD and arr, the results showed that there were 52 Imipenem-resistant P. aeruginosa (IRPA) strains and 23 Rifampin-resistant P. aeruginosa (RRPA) strains. This method demonstrated excellent detection performance even when the sample concentration is 10 copies/μL at isothermal conditions and the results could be obtained within 20 minutes. The detection results were in accordance with the results of conventional PCR and Real-time PCR. The detection outcomes of the arr gene were consistently with the resistance spectrum. However, the antimicrobial susceptibility results revealed that 65 strains were resistant to imipenem, while 49 strains sensitive to imipenem with oprD were identified. This discrepancy could be attributed to genetic mutations. In summary, the RAA has higher sensitivity, shorter time, and lower-cost instrument requirements than traditional detection methods. In addition, to analyze the epidemiological characteristics of the aforementioned drug-resistant strains, we conducted Multilocus Sequence Typing (MLST), virulence gene, and antimicrobial susceptibility testing. MLST analysis showed a strong correlation between the sequence types ST-1639, ST-639, ST-184 and IRPA, while ST-261 was the main subtype of RRPA. It was observed that these drug-resistant strains all possess five or more virulence genes, among which exoS and exoU do not coexist, and they are all multidrug-resistant strains. The non-coexistence of exoU and exoS in P.aeruginosa is related to various factors including bacterial regulatory mechanisms and pathogenic mechanisms. This indicates that the relationship between the presence of virulence genes and the severity of patient infection is worthy of attention. In conclusion, we have developed a rapid and efficient RAA (Recombinase-Aided Amplification) detection method that offers significant advantages in terms of speed, simplicity, and cost-effectiveness (especially in time and equipment aspect). This novel approach is designed to meet the demands of clinical diagnostics.
1 Introduction
Pseudomonas aeruginosa, a prevalent opportunistic pathogen within the family Pseudomonadaceae, stands as a significant causative agent of hospital-acquired infections (HAIs) in tertiary hospitals (Logan et al., 2017; Curran et al., 2018). Infections caused by P. aeruginosa can manifest in various areas, including the lungs, urinary tract, wounds, blood (such as septicemia), eyes, and other diseases like pneumonia (Lee et al., 2018; Tacconelli et al., 2018). P. aeruginosa is widely distributed, it is typically non-pathogenic under normal circumstances. However, the infection rate of P. aeruginosa increases when the host’s immune system is weakened (Prithiviraj et al., 2005; Bassetti et al., 2017). In the intensive care unit (ICU), P. aeruginosa infections are linked to high incidence and mortality rates across various populations, including individuals with chronic obstructive pulmonary disease and cystic fibrosis (Diekema et al., 2000; Cui et al., 2022). The entire genome size of P. aeruginosa ranges from 5.5 to 7 Mbp, showcasing robust genetic coding capabilities that contribute to metabolic diversity and drug resistance (Montgomery et al., 2018). The spread of multidrug-resistant strains has made treating diseases caused by those pathogens increasingly challenging (Burrows, 2018; Arbune et al., 2021). Mechanisms of antibiotic resistance in P. aeruginosa include natural resistance, acquired resistance, and adaptive resistance (Shen and Fang, 2015; Chevalier et al., 2017). IRPA and RRPA exhibit resistance to various antibiotics and robust survivability in harsh environments such as hospitals, posing significant challenges to clinical treatment (Abniki et al., 2024). The overuse of antibiotics has contributed to the emergence of strains resistant to imipenem and rifampicin. According to Hamid Vaez et al., IRPA prevalence in Iran was reported at 54% (Vaez et al., 2017). Suwantarat et al. reported IRPA prevalence in Southeast Asian countries, noting it was 31.1% in Philippines (Suwantarat and Carroll, 2016). Rifampicin, a derivative of rifamycin, exhibits a broad spectrum of antibacterial activity against Gram-positive bacteria. The lower prevalence of IRPA isolates could be attributed to the limited use of rifampicin antibiotics in these regions (Liu et al., 2020).
Currently, in addition to traditional methods, laboratories and clinical platforms have also established immunological methods based on antigen-antibody specific binding and molecular biological methods (DNA sequencing technology, PCR-based technology, new molecular detection technology) for pathogen detection (Rajapaksha et al., 2019). However, many of these methods are time-consuming and cannot be easily implemented in primary-level laboratories (Bonetta et al., 2016; Yoon et al., 2021).
RAA is based on the recombinase polymerase amplification (RPA) technology. RPA was initially introduced by Niall Armes in 2006 (Cambridge, United Kingdom, founded by the Wellcome Trust Sanger Institute) (Piepenburg et al., 2006; Li et al., 2018). RAA is developed by TwistDx in the UK and further advanced by Jiangsu Qitian Gene Biotechnology (Fan et al., 2020; Li et al., 2021), has become a molecular tool widely used in the identification of genes of various pathogens (Mao et al., 2022; Yan et al., 2023).
RAA employs single-strand binding proteins, DNA polymerase, and recombinases to amplify nucleic acids at an isothermal temperature (38-41°C) (Wu et al., 2021). This technology uses recombinases from Escherichia coli, allowing for tight binding with primer DNA at room temperature, forming an enzyme-primer aggregate (Song et al., 2018).The single-stranded DNA-binding protein aids in unraveling the double-stranded structure of the template DNA, and the DNA polymerase catalyzes the formation of new complementary DNA strands, resulting in exponential growth of the amplification product (Zhao et al., 2020). With the addition of a fluorescence probe, RAA can perform Real-time detection, and the results can be obtained in just 5-20 minutes with high sensitivity (Shelite et al., 2021; Cao et al., 2023).
In this paper, we have established a high-speed platform based on RAA technology to detect the resistance genes oprD/arr in P. aeruginosa. This platform enables efficient and rapid detection of resistance genes, with results attainable in just 10 minutes and a detection sensitivity of 10 copies/μL. The epidemiological analysis revealed that both IRPA and RRPA contain more than five virulence genes, and all strains tested exhibited multidrug resistance. IRPA of ST-1639 and RRPA of ST-261 are the most commonly identified types.
2 Materials and methods
2.1 Bacterial strains and clinical isolates
A total of 101 clinical antibiotics-resistance samples of P. aeruginosa were collected from the Xi’an Children’s Hospital of Shaanxi Province. These samples included 92 sputum samples, 3 urine samples, 3 blood samples, 2 bronchoalveolar lavage fluid samples, and 1 skin secretion sample. Isolates were obtained from the above clinical specimens. The strains tested were indeed cultured as isolates before testing with the RAA method. All isolated strains underwent bacterial identification. Isolates were obtained from various clinical specimen types listed. PAO1 (stored in our laboratory) was used as the wild type strain in this experiment.
2.2 DNA extraction
The total DNA of the bacterial strains was extracted using the Sangon Quick Bacterial Genomic DNA Isolation Kit (Sangon Biotech, China). The DNA was eluted with 100 μL of enzyme-free water and stored at -80°C for future use.
2.3 Preparation of recombinant plasmids
Different data sources of National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) and Pseudomonas Genome DB (https://www.pseudomonas.com) were used to further screen the drug-resistant genes oprD and arr as target genes. The full sequences of oprD (1332 bp) and arr (1578 bp) corresponding to the nucleotide sequences of P. aeruginosa (PAO1, NC_002516) were cloned into the Pme6032 vector (Our laboratory stored). The number of copies of the recombinant plasmid was calculated using plasmid concentration measured with Nanodrop One (Thermo Fisher Scientific, Waltham, MA, United States). The recombinant plasmid is diluted from 107 copies/μL to 100 copies/μL using the following formula, DNA copies (copies/μL) = [6.02×1023×plasmid concentration (ng/μL)×10−9]/[DNA length (bp)×660], stored at -80°C for following detection.
2.4 RAA primer and probe design
The screening of oprD and arr gene sequences was conducted using data retrieved from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). The principles of RAA primer and probe design are as follows: the primers range from 30 to 35 base pairs (bp) in length, while the probes range from 46 to 52 bp. The size of the RAA amplification product falls within the range of 100 to 200 bp. Both the probes and primers are designed to target conserved regions of the gene. The specificity of primers and probes was confirmed using NCBI’s Primer-BLAST program. RAA-oprD-primer1-fw (30 bp), RAA-oprD-primer1-rv (32 bp), RAA-oprD-probe2 (41 bp) were selected as the primers and probes for IRPA detection; RAA-arr-primer3-fw (30 bp), RAA-arr-primer3-rv (32 bp) and RAA- arr-probe2 (44 bp) serves as primer and probe for RRPA detection. Additionally, conserved regions of the 16S rRNA gene were selected for the design of internal positive controls. All primers and probes are synthesized and purified using Biotech (Shanghai, China) by high performance liquid chromatography.
2.5 PCR and real-time PCR
The 20 μL reaction volume contains the following components for all PCR: 10 μL PCR master mix reagents (2×SanTaq Fast PCR Master Mix, Beijing, China), 8 μL of Sterile water, 0.5 μL of 10 μM oprD-fw (or arr-fw) primers and oprD-rv (or arr-rv) primers, and 1 μL DNA templates. The primer sequences are showed in Table 1.
Real-time PCR based on the oprD gene for the detection of oligonucleotide sequences of oprD forward primers (30-50 bp), oprD reverse primers (30-50 bp), and oprD-probe (FAM, BHQ1). Real-time PCR based on the arr gene for the detection of oligonucleotide sequences of arr forward primers (30-50 bp), arr reverse primers (30-50 bp), and arr-probe (FAM, BHQ1). The reactions were prepared as a 25 μL reaction volume containing 12.5 μL TaqMan Universal Master Mix, 0.5 μL forward primers, 0.5 μL reverse primers, 8.5 μL double distilled water, 1 μL probe and 2 μL extracted DNA. The concentration of primer and probe was 10 μM. A positive sample is defined as a sample with a period threshold (CT) value < 30. The primer and probe sequences are listed in Table 1.
2.6 RAA technique
We used the RAA kit (Qitian, China) to conduct the RAA assay. The reaction system consisted of 50 μL, including 2.1 μL of forward primer, 2.1 μL of reverse primer, and 0.6 μL of probe, all at a concentration of 10 μM. Additionally, reaction buffer (25 μL), magnesium acetate (2.5 μL of 280 mM), deionized water (15.7 μL), and 2 μL of template DNA were included. Note that due to the high sensitivity of RAA detection and to avoid contamination, template DNA should be added last. The reaction mixture was briefly centrifuged and mixed in RAA-B6100—an isothermal shaking incubator (QiTian, Wuxi, China) at 39°C for 4 minutes. Subsequently, the mixture was placed in RAA-F1620—a fluorescence detector (QiTian, Wuxi, China) to measure FAM fluorescence signals every 20 seconds.
2.7 Phenotype analysis method of IRPA and RRPA
Typically, this experiment involves cultivating bacteria in media containing varying concentrations of antibiotics, followed by the observation of bacterial growth under different antibiotic conditions (Blair et al., 2015; Denis et al., 2019). The antimicrobial susceptibility testing for the chosen strains were conducted through the disk diffusion method, and the results were interpreted following the guidelines provided by the Clinical and Laboratory Standards Institute. The antibiotics used include ciprofloxacin (CIP), tobramycin (TOB), cefepime (SCF), aztreonam (AZT), polymyxin (PMB), piperacillin (PIP), meropenem (MEM), imipenem (IPM), cefepime (FEP), ceftazidime (CAZ), levofloxacin (LEV), gentamicin (GN), Rifampin (RFP), and amikacin (AMK).
2.8 Determination of MLST and virulence factor
MLST analysis of P. aeruginosa is conducted by analyzing the sequence variations of housekeeping genes (nuoD, mutL, trpE, acsA, aroE, guaA and ppsA). The PCR products are sent to Shanghai Sangon Biotech for sequencing, and the sequencing results are submitted to the P. aeruginosa MLST database (https://pubmlst.org/paeruginosa/) for analysis. Strain types that do not match existing databases will be identified as new sequence typing (ST). Virulence analysis using the genes encompassed plcH, aprA, algD, exoS, exoT, exoU, exoY, toxA and nor.
2.9 Technical route of this study
This experiment primarily utilizes RAA as the detection method, with the technical route shown in 178 Figure 1. From gene screening to RAA detection, we have gone through the following steps: (1) Following the isolation and identification of clinical bacterial strains, collect and culture bacteria and store them in a strain storage center. (2) Antimicrobial susceptibility testing and PCR for drug-resistant isolates. (3) Selection of genes: drug-resistant genes consistent with the resistant phenotype will be selected for RAA detection and optimization. (4) High temperature denaturation extraction of bacterial DNA. (5) RAA detection. RAA is a novel isothermal nucleic acid amplification technology that requires the design of specific primers and probes. Visual analysis can be achieved by a fluorescence detector (QT-RAA-1620; Jiangsu Qitian Bio-Tech Co., Ltd., China) (6).
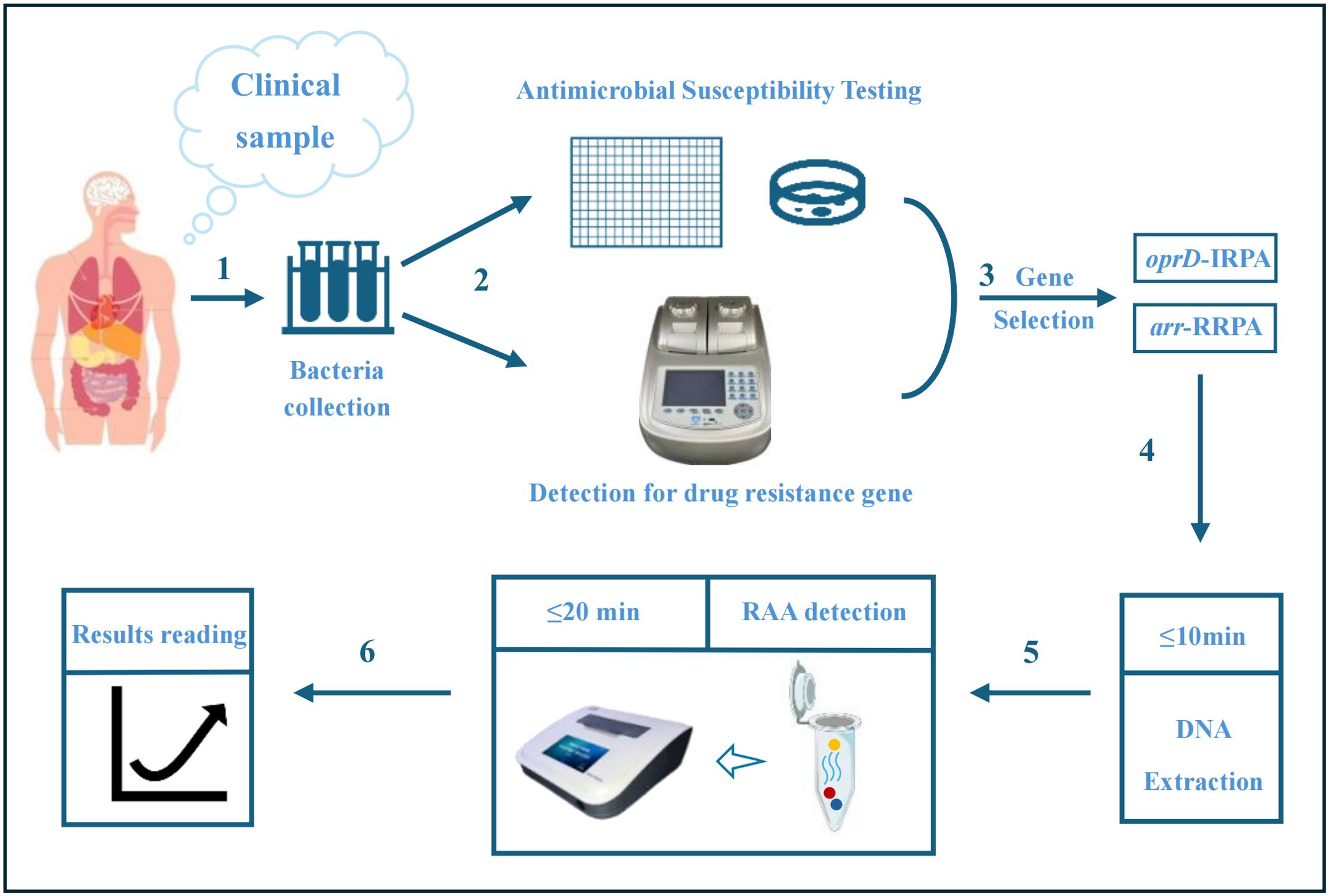
Figure 1. Technical route of this study. (I) Clinical isolates are collected and cultured. (II) Antimicrobial susceptibility testing and PCR for drug-resistant genes. (III) Selection of genes. (IV) High temperature denaturation extraction of bacterial DNA. (V-VI) Detection of oprD/arr by RAA. The blue curve represents the template DNA, the blue circle represents the probe, the red circle represents the primer, and the orange represents the recombinant enzyme medium. The reaction results can be detected with a constant temperature instrument.
2.10 Statistical analysis
Statistical analysis was performed using SPSS 21.0 (IBM, Armonk, NY, USA). Probit analysis for the detection limit of the PCR, Real-time PCR and RAA was performed at a 95% probability level. We performed repeated experiments and calculated p-values and kappa values for RAA, PCR, and Real time PCR.
3 Results
RAA technology performs DNA amplification at isothermal temperature, significantly saving time and cost. Here, we established RAA technology to detect oprD (outer membrane channel protein genes) or arr (aminoglycoside response regulator gene) rapidly and used it to analyze the clinical samples of antibiotic-resistant P. aeruginosa.
3.1 Optimizing the RAA assay-primer, probe and temperature
RAA detection exhibits high sensitivity towards target sequences, enabling reliable detection in complex samples. We designed primers and probes for RAA, utilizing recombinant oprD or arr plasmids as positive templates and sterile water as negative templates to optimize the detection of RAA. Five sets of primers (RAA-oprD-primer1, RAA-oprD-primer2, RAA-oprD-primer3, RAA-oprD-primer4, RAA-oprD-primer5) and two probes (RAA-oprD-probe1, RAA-oprD-probe2) were designed in the conserved region of the gene, and the amplification efficiency was determined based on the change in fluorescence intensity at 492 nm. Finally, we selected RAA-oprD-primer1 as the best primer and RAA-oprD-probe2 as the best probe. The same method was used for the optimization of arr, and RAA-arr-primer3 and RAA-arr-probe2 were finally selected (Table 1; Figure 2).

Figure 2. Selection of specific regions and primer positions for RAA assay. (A) Primer and probe set to amplify the oprD gene. (B) Primer and probe set to amplify the arr gene. Probes are indicated by blue, primer are indicated by orange.
The suitable reaction temperature range for the Recombinase is 38-42°C (Yan et al., 2023). Next, we used primer pairs of RAA-oprD-primer1 or RAA-arr-primer3, adjusted the reaction temperature to compare the amplification efficiency of RAA assay, and chose appropriate reaction temperature within 50 µL system. The results showed that the fluorescence generated by RAA at 39°C was significantly higher than that at other temperatures. Therefore, 39°C was the optimal temperature for our system with the highest amplification efficiency (Figures 3A, B). In addition, the amplification efficiency under different probe concentrations was compared, and the probe concentration selected in our system was 10 µM (Figures 3C, D).
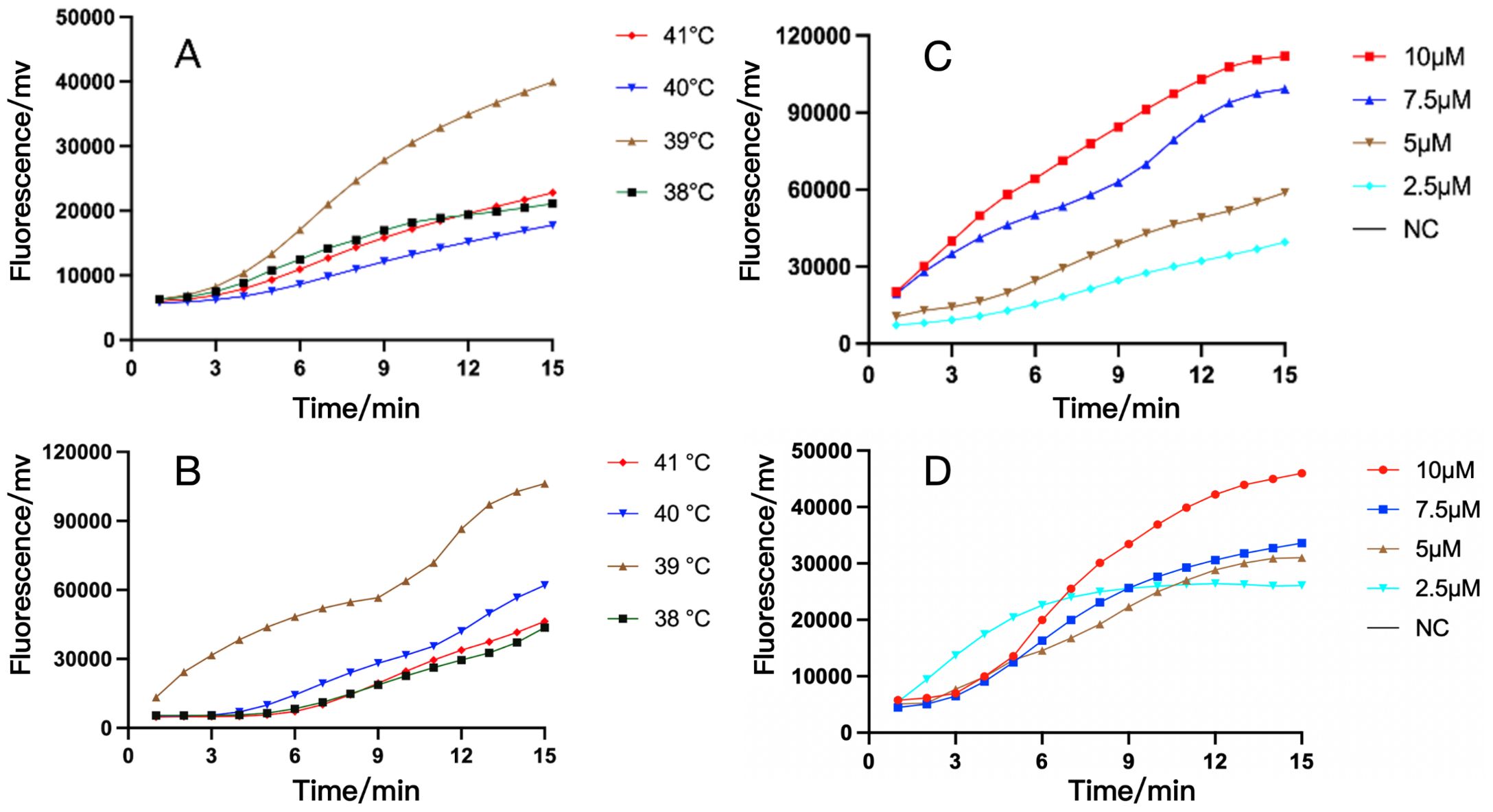
Figure 3. The optimization of temperature and probe concentration of RAA assay for IRPA and RRPA detection. (A, B) The RAA analysis of oprD gene at different temperature (A) and different probe concentration (B). (C, D) The RAA analysis of arr gene at different temperature (C) and different probe concentration (D). The results showed that their optimal temperature is 39°C and the optimal probe amplification concentration is 10 μM. NC, Negative Control.
3.2 Sensitivity for RAA detection
Next, we used RAA technology to detect the oprD or arr to determine the sensitivity of RAA detection. In the experiment, we diluted the constructed recombinant plasmids containing oprD or arr from 107 copies to 100 copies respectively to detect the fluorescence signal (Figure 4). We found that clear signals could be detected using 10 copies/reaction (Figures 4A, B), whereas PCR detection required 103 copies/reaction (Figures 4C, D), which is similar to previous reports (Fu et al., 2022).
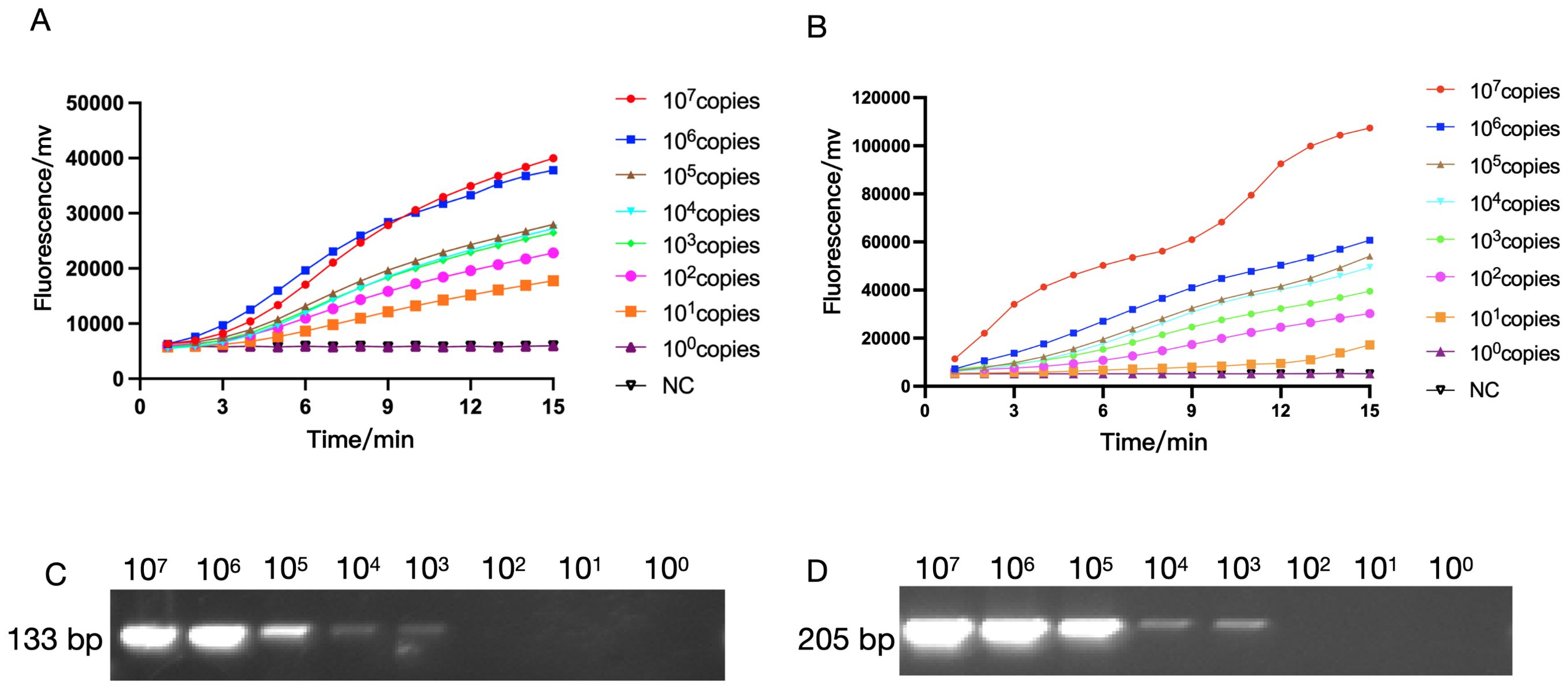
Figure 4. The sensitivity of RAA and PCR assay for IRPA and RRPA detection. The sensitivity of RAA analysis using the primer and probe set RAA-oprD-primer1 (A), RAA-arr-primer2 (B). The plasmid ranged from 1×100 copies/μL to 1 × 107 copies/μL. NC, Negative Control. The sensitivity of PCR for IRPA and RRPA detection. (C, D) The sensitivity of RAA and conventional PCR detection.
3.3 Analytical specificity for RAA detection
Subsequently, we tested the specificity of oprD or arr using the RAA platform, and as a control, we also detected 7 other resistance genes including msr, tetA, mph, etc. As shown in Figure 5, only strains containing oprD or arr were able to exhibit fluorescence signals. Also, we amplified the standard strain with PCR primers and detected it by agarose gel electrophoresis. We found that only strains with oprD or arr showed specific amplification bands, which was consistent with the results of the RAA test. This confirmed the specificity of RAA detection.
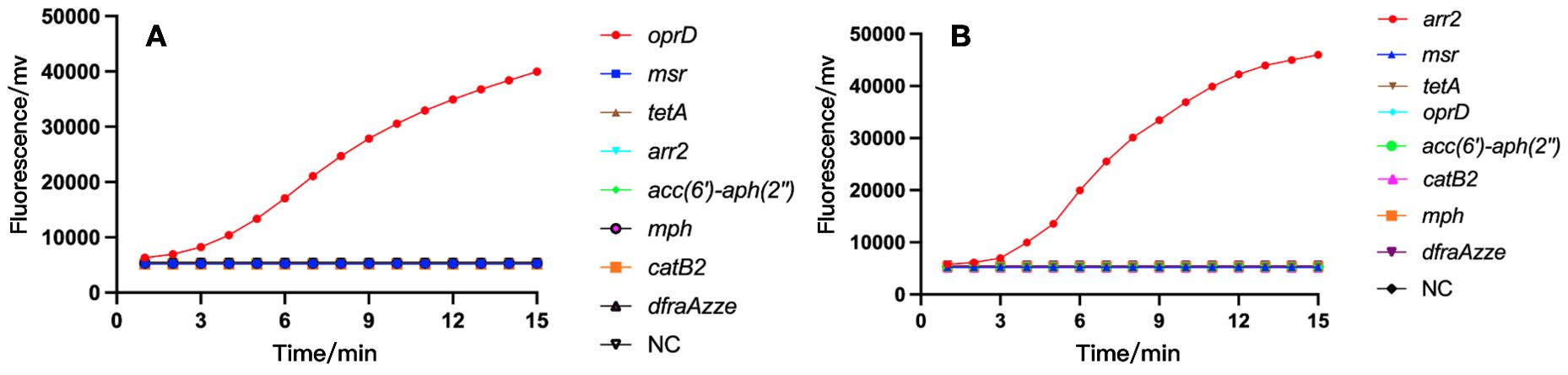
Figure 5. The specificity of RAA assay for IRPA (A) and RRPA (B) detection. Only the recombinant plasmids produced amplification signals, whereas the negative control and control bacterial samples produced negative amplification signals. NC, Negative Control.
3.4 Clinical sample detection by RAA assay
After establishing the RAA detection technology, we simultaneously used conventional PCR, Real-time PCR and RAA technique to detect P. aeruginosa in 101 samples collected from Xi’an Children’s Hospital (Shaanxi, China). Compared with PCR and Real-time PCR methods, RAA technology has comparable detection accuracy for oprD or arr genes, but the time is significantly shortened (Tables 2A, 2B) (Figure 6). PCR and Real-time methods typically take 2 hours, while th RAA method takes less than 20 minutes.
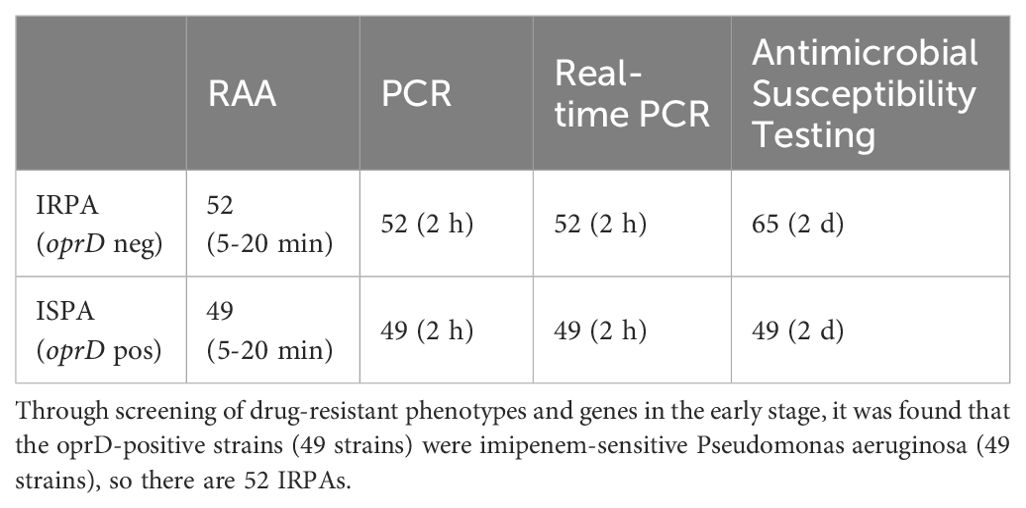
Table 2A. Comparison of recombinant enzyme-assisted amplification methods (RAA) with PCR and real-time PCR for the detection of Imipenem-resistant P. aeruginosa (IRPA).
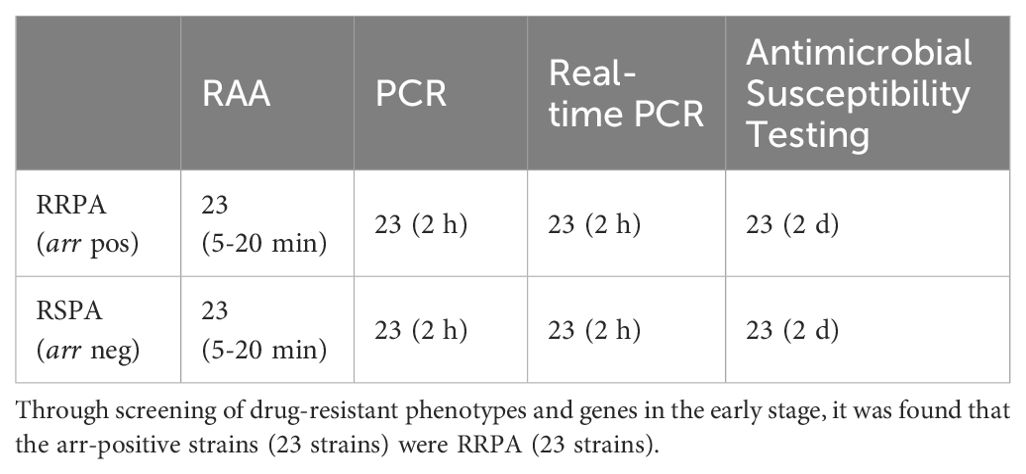
Table 2B. Comparison of recombinant enzyme-assisted amplification methods (RAA) with PCR and real-time PCR for the detection of rifampicin resistant P. aeruginosa (RRPA).
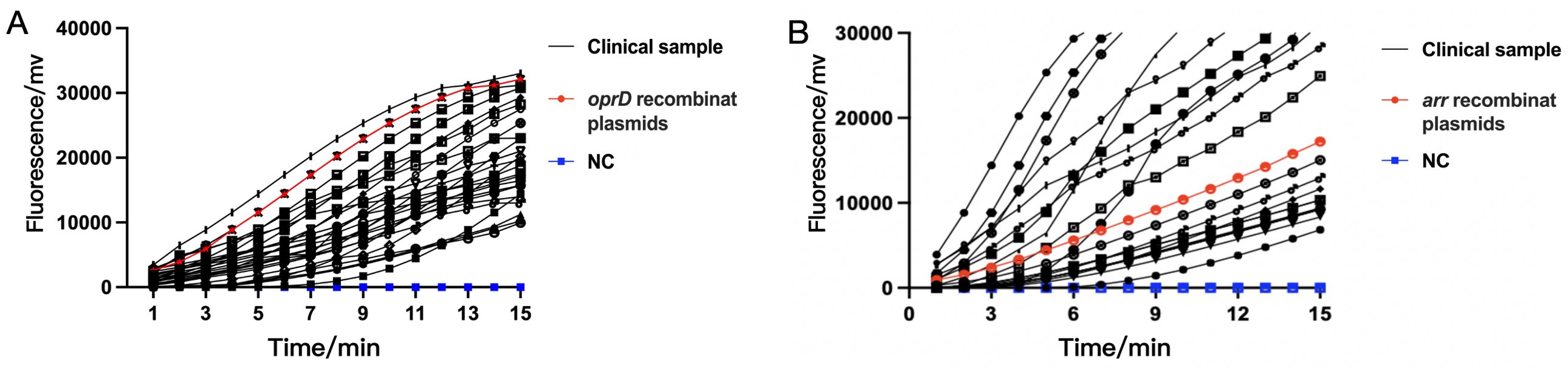
Figure 6. The detection performance of RAA assay in clinical samples for IRPA (A) and RRPA (B). Both IRPA and RRPA are multidrug-resistant strains. NC, Negative Control.
Among the 101 clinical antibiotic-resistant P. aeruginosa isolates collected, we detected 23 strains that contained the arr gene and showed resistant to Rifampin, which was highly consistent with the results obtained by other methods. However, the results for detection of oprD did not match the antimicrobial susceptibility results. There should be 52 strains of P. aeruginosa that are resistant to imipenem based on the lack of detection of the oprD gene, but antimicrobial susceptibility testing results indicated 65 IRPA strains. The previous reports indicate this discrepancy may be associated with mutations or deletions in the oprD gene and other factors. The absence or mutation of oprD leads to the closure of bacterial channels, impeding the entry of imipenem into bacteria and inhibiting its effects, consequently resulting in the development of resistance to imipenem (Suresh et al., 2020; Gonzalez-Vazquez et al., 2021).
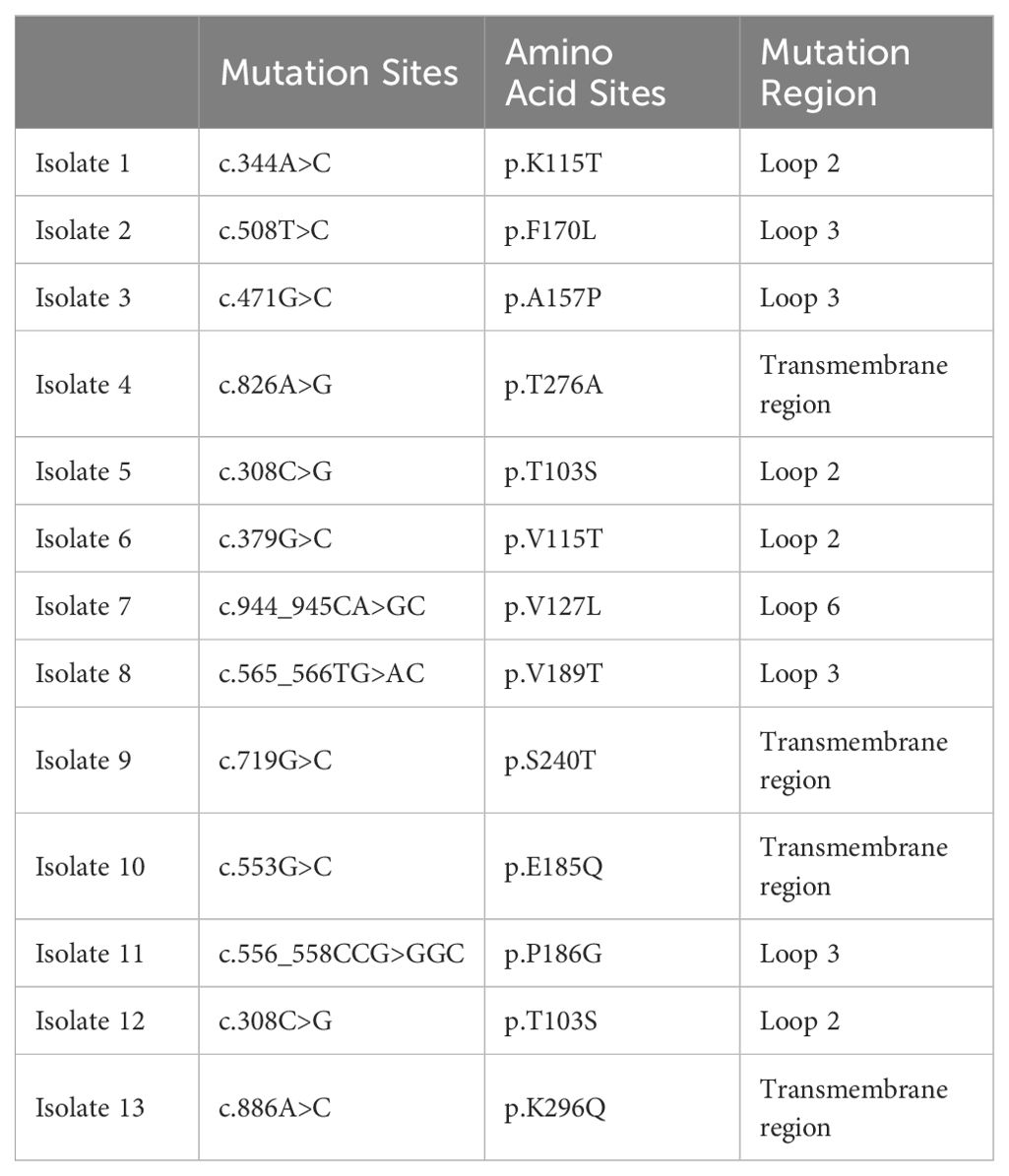
Mutations in the oprD gene are the primary cause of P. aeruginosa’s resistance to imipenem. We also analyzed the types of oprD mutations in this batch of strains. The mutation sites were mainly located the transmembrane region, loop 2, and loop 3 of the protein (Table 3), which is similar to the results of previous reports (Ochs et al., 2000; Li et al., 2012).
3.5 Antimicrobial susceptibility analysis
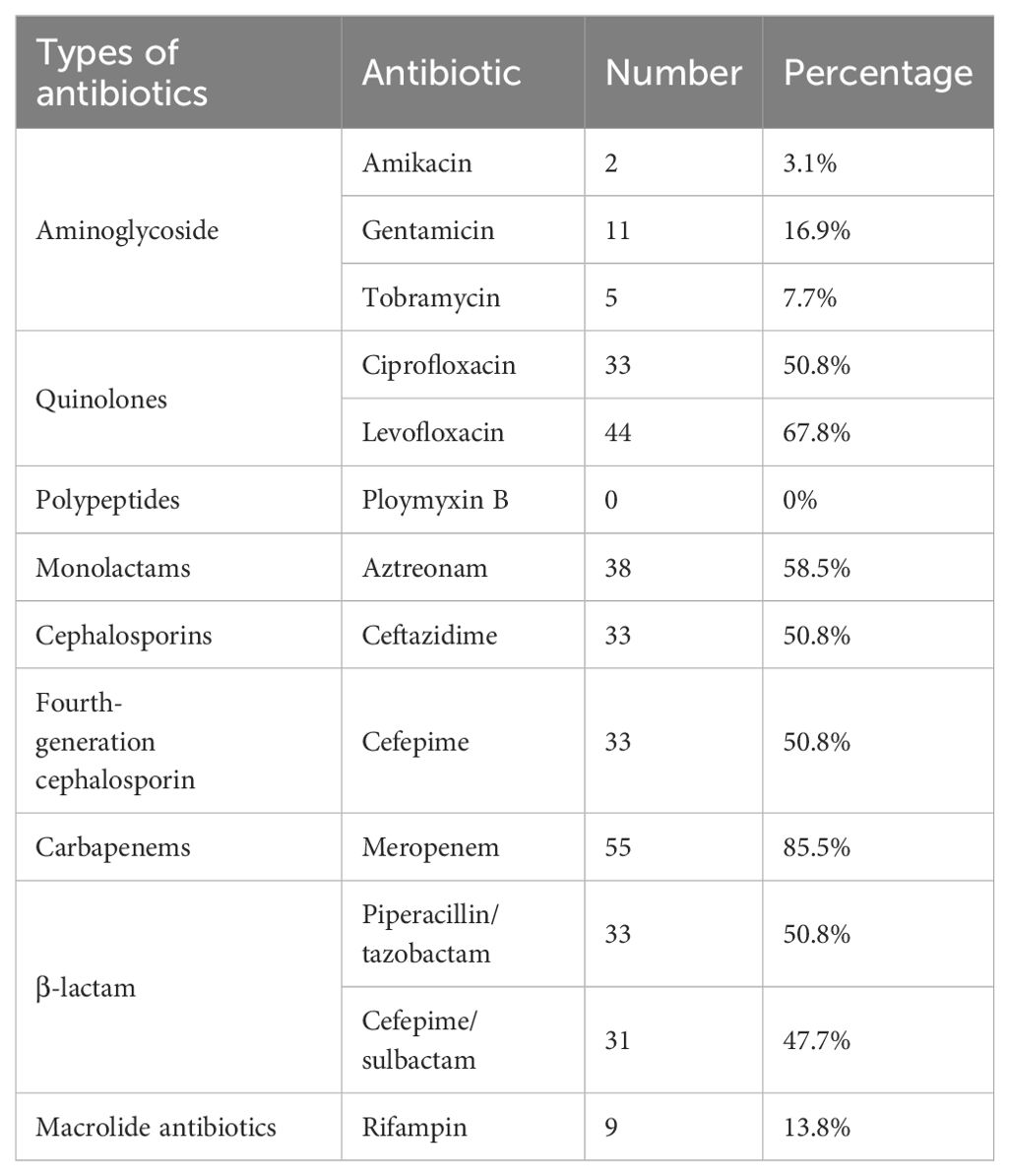
Furthermore, we carried out the antimicrobial susceptibility testing for IRPA and RRPA. Among the 65 IRPA strains, 85.5% (55 strains) were resistant to meropenem, 67.8% (44 strains) were resistant to levofloxacin, 58.5% (38 strains) were resistant to aztreonam, and 50.8% (33 strains) were resistant to ciprofloxacin, ceftazidime, cefepime and piperacillin/tazobactam combination (Table 4).
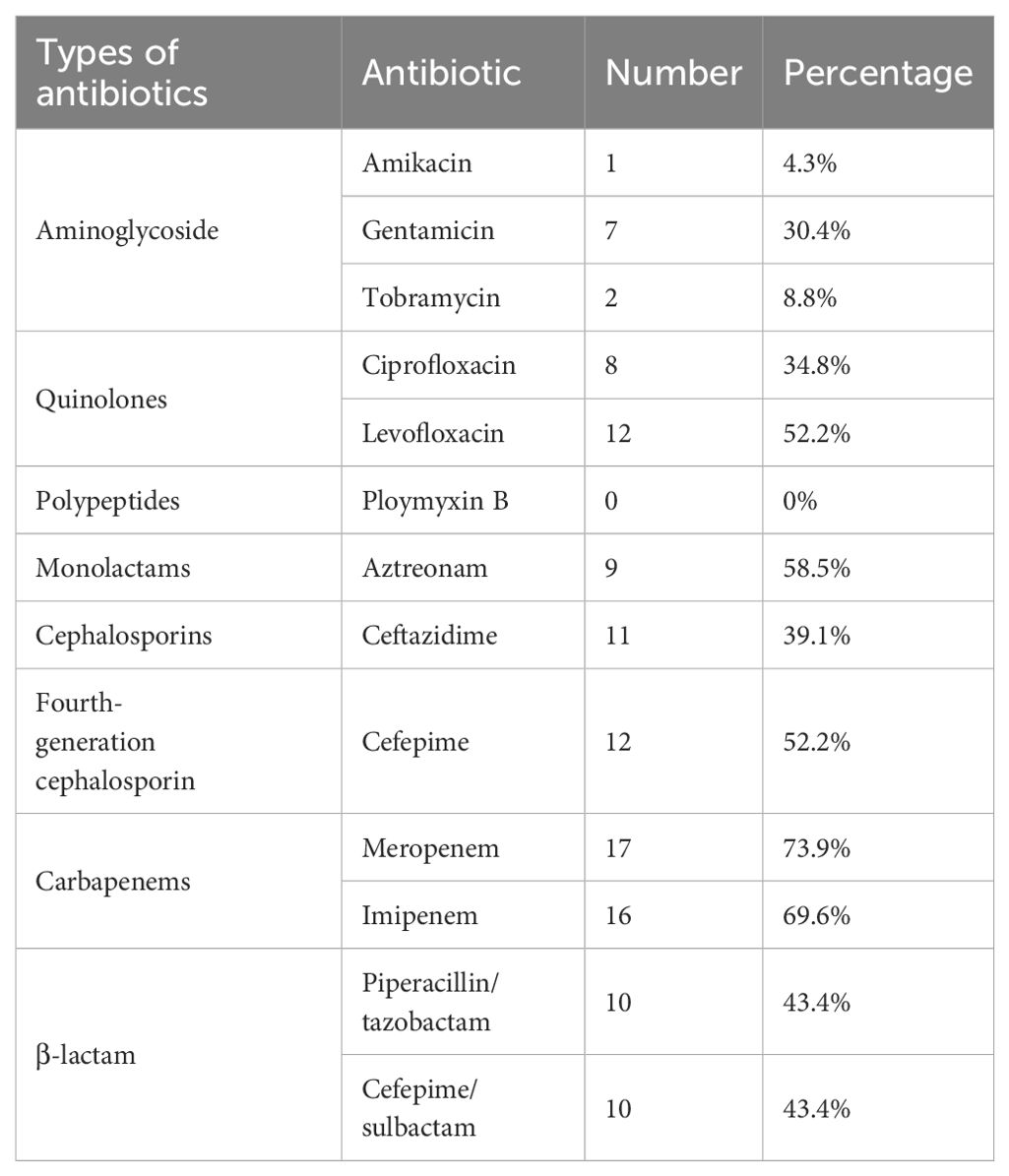
The 101 clinical strains of P. aeruginosa collected include 23 strains of RRPA. The antimicrobial susceptibility tests indicate that among them, 17 strains (73.9%) were resistant to meropenem, 16 strains (69.6%) were resistant to piperacillin/tazobactam combination, and 14 strains (60.9%) were resistant to imipenem (Table 5).
The results showed that most of the clinical antibiotic-resistant P. aeruginosa collected were multidrug-resistant. The most of imipenem, meropenem, and piperacillin-resistant P. aeruginosa also exhibit resistance to levofloxacin.
3.6 Molecular characteristics of the IRPA and RRPA isolates
To understand the distribution and epidemiological characteristics of the pathogen, Multilocus Sequence Typing (MLST) and antimicrobial susceptibility testing was used to classify IRPA and RRPA. MLST analysis was conducted on 65 strains of IRPA and the results revealed that ST-1639 is the predominant genotype, followed by ST-261, ST-485, ST-2375, and ST-2389. Among the 23 isolated strains of RRPA, a total of 12 ST types were identified. Notably, ST-261 was the predominant genotype, accounting for 30.43% of the strains. Particularly interesting is the discovery of one RRPA strain that exhibited resistance to all tested antibiotics, and its ST type was identified as ST-639. The statistical results are shown in the Figure 7, Supplementary Tables 1 and 2.
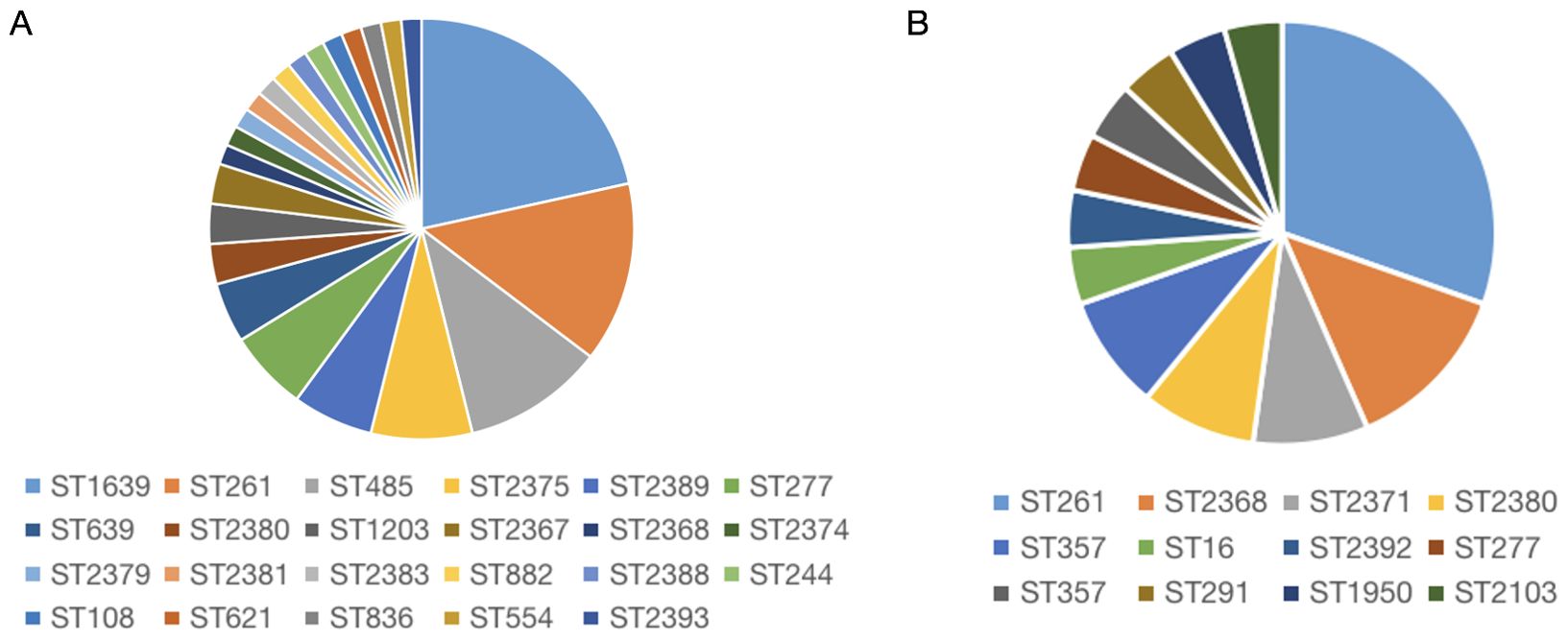
Figure 7. Molecular characteristics of IRPA (A) and RRPA (B) isolates. The result shows that the most common type is ST1639 for IRPA (A), and ST261 for RRPA (B).
3.7 Virulence factors of the P. aeruginosa isolates
We utilized PCR to ascertain the presence of virulence factors (exoY, exoS, exoT, exoU, plcH, aprA, pys, toxA) in clinical isolates. The aprA gene is associated with T1SS, while toxA and plcH are associated with T2SS, and exoS, exoT, exoY, and exoU are associated with T3SS.
Positive IRPA virulence gene detections encompass 11 types, with all multidrug-resistant P. aeruginosa isolates carrying at least 5 virulence genes. The detection rate of the toxA gene associated with Type II Secretion System is 100%. Its virulence is modulated by three secretion systems, particularly exerting significant influence on IRPA virulence, especially those associated with the T3SS secretion system (exoY, exoS, exoT) (Supplementary Table 1). All virulence factors in RRPA showed relatively high detection rates among the 23 strains, with all containing toxA. The toxA gene is secreted via T2SS, resulting in modification and loss of intracellular proteins. In order of detection rates, virulence genes are arranged as follows: toxA, algD, aprA, exoY, norC (Supplementary Table 1).
4 Discussion
P. aeruginosa causes various diseases and localized infections resulting from surgical procedures or burns often lead to fatal injury (Dunn and Wunderink, 1995). Multidrug-resistant P. aeruginosa (MDR-PA) is commonly identified in major hospitals worldwide and the Centers for Disease Control and Prevention (CDC) has categorized MDR-PA as a significant menace (Kang et al., 2003; Karruli et al., 2023). Antibiotic resistance, especially multidrug resistance, is one of the most critical factors in patients acquiring infections with MDR-PA (Denis et al., 2019; Naik et al., 2021). The rapid diagnosis of antibiotic resistance genes aims to detect whether patients carry or possess specific drug-resistant gene characteristics related to certain diseases, and timely diagnosis can avoid unnecessary antibiotic use and treatment delays, thereby reducing healthcare cost and conserving medical resources.
RAA demonstrates excellent detection capability and this method not only saves time but also does not require complex instrumentation. Due to the extremely high sensitivity of the RAA detection method we developed, it can directly test the sputum or alveolar lavage fluid of infected patients without the need for complex and time-consuming cultivation. This process can be completed within 20 minutes, ensuring rapid turnaround times. Furthermore, the visualization of results significantly enhances the efficiency of doctors’ diagnoses and minimizes patient waiting times to the greatest extent possible. As an initial attempt, we selected the oprD and arr genes of P. aeruginosa to test the characteristics of RAA in rapidly identifying resistance genes. OprD protein plays a role as a specific channel protein for the rapid entry of imipenem into P. aeruginosa and decreased expression or loss of the oprD lead to imipenem resistance (Quinn et al., 1991; Sun et al., 2016), and the aminoglycoside response regulator gene arr can deactivate rifampicin through ribosylation, which is a crucial factor in bacterial resistance to rifampicin (Tribuddharat and Fennewald, 1999; Arlet et al., 2001; Naas et al., 2001; Alexander et al., 2003).
In clinical practice, the detection of drug-resistant genes traditionally relies on comprehensive antimicrobial susceptibility testing, which is time-consuming and costly (Shen and Fang, 2015; Holbrook and Garneau-Tsodikova, 2018). We conducted a comparative analysis of RAA, conventional PCR, and Real-time PCR, examining factors such as cost, sensitivity, scope of application, and their respective strengths and weaknesses (Table 6). The results showed that RAA has obvious advantages. When we apply RAA assay to clinical gene diagnosis and commercialize it, it can replace the time-consuming and labor-intensive traditional detection methods, which can greatly improve the accuracy of gene diagnosis, and help clinicians make quick decisions. In this study, we use RAA method to detected oprD and arr genes. The results indicate that the minimum detection limit of the RAA assay is 10 copies/reaction, which approaches the highest sensitivity of most Real-time PCR methods. Furthermore, the RAA demonstrated specificity in detecting IRPA and RRPA without cross-reactivity with other antibiotic resistance genes in P. aeruginosa, such as ges and msr. In order to assess the clinical feasibility of this method, 101 clinical isolates of P. aeruginosa were tested using the RAA assay, conventional PCR, and TaqMan probe-based Real-time PCR. The result of gene detection for oprD and arr are consistent with those obtained from conventional PCR and Real-time PCR. This indicates that the RAA method exhibits good sensitivity comparable to PCR and Real-time PCR, but in a much shorter time. RAA can be completed in 20 minutes, while PCR and Real-time PCR require about 2 hours. Therefore, the RAA assay established in our study has the potential to be developed into a portable on-site kit for rapid screening of IRPA and RRPA in basic clinical laboratories.
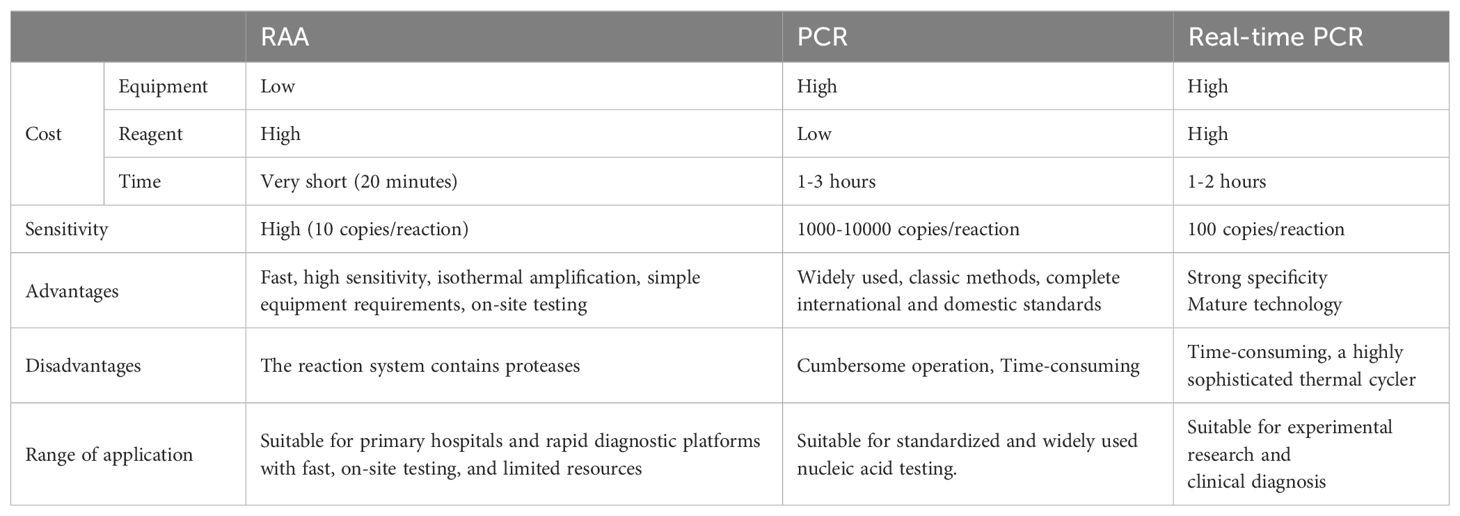
Table 6. Comparison of costs, advantages, and disadvantages of RAA, conventional PCR, and real-time PCR.
The genetic testing of RRPA is consistent with the results of drug susceptibility testing, indicating a higher correlation between arr and drug-resistant phenotypes. As has been previously reported, arr genes are located on the integron (Da Fonseca et al., 2008; Firoozeh et al., 2023). However, there is a certain discrepancy between the RAA detection results of oprD and the drug sensitivity results. Upon comparing the results with drug sensitivity tests, the concordance rate between RAA-IRPA detection results and drug resistance is 80%, surpassing the detection rate of the drug sensitivity tests. This is attributed to the tendency of oprD gene to undergo mutations (Quale et al., 2006; Kao et al., 2016; Do Rego and Timsit, 2023). It was discovered that this is caused by mutations, such as membrane topology and site-specific mutagenesis of P. aeruginosa porin OprD (Huang et al., 1995; Kim et al., 2016). In the deduced topological structure of the OprD protein, Li et al. identified external loops 2 and 3 as the entrance for basic amino acids and the binding site for imipenem. Furthermore, any substitution or deletion within loops 2 and 3 that results in a conformational change can cause imipenem resistance (Li et al., 2012). Ochs et al. found that deletions of amino acids 74-81, 84-91, 80-87, or 94-101 in loop L2, or amino acids 156-163 in loop L3, resulted in reduced sensitivity to imipenem compared to the wild type. This suggests that these amino acids play an important role in imipenem binding or transmembrane transfer (Ochs et al., 2000). Overall, OprD is a well-characterized imipenem influx channel, and mutations in oprD render P. aeruginosa resistant to imipenem, posing significant challenges in clinical practice. These show that for some antibiotic resistances, detecting only one gene may be inaccurate and that all factors need to be considered to get more accurate results.
The establishment of RAA assay significantly saves time, cost, and manpower. However, rapid detection of resistance genes is still limited, and although RAA maintains its sensitivity and specificity, it may be impacted by the propensity for mutation in resistance genes, changes in drug resistance caused by mutated genes require careful consideration. Antimicrobial susceptibility testing, whole-genome sequencing, and other technologies can also serve as alternative methods. Additionally, the results of our experiments need validation from larger clinical samples to enhance the reliability of the findings.
In order to analyze the molecular characteristics of these drug-resistant strains, we performed MLST typing, virulence gene analysis, and antimicrobial susceptibility testing. Molecular epidemiological methods were employed to analyze virulence genes, antibiotic resistance genes, and MLST (Curran et al., 2004; Huang et al., 2019; Blanc et al., 2020). MLST holds profound significance in preventing and controlling drug resistance (Maiden et al., 1998; Waters et al., 2012; Castaneda-Montes et al., 2018). Recently, this method has been applied in molecular studies of environmental microbiota and eukaryotic organisms (Chan et al., 2001; Cooper and Feil, 2004). Our results indicate that ST-1639 is the predominant subtype in IRPA (presumably a specific context or population), but there is no significant association between the ST type and virulence genes. ST-261 is the predominant sequence type of RRPA, and no significant association between ST typing and virulence genes was observed.
P. aeruginosa adapts to adverse host environments by secreting multiple virulence factors, aiding in successful host infection and disease onset. Detection of these virulence factors is crucial for understanding bacterial pathogenic mechanisms and guiding clinical treatments. The T3SS is a crucial secretion system that can inject various virulence factors into host cells, facilitating bacterial infection and pathogenesis (Soscia et al., 2007). Our results show a remarkably high detection rate for exoS, exoT, and exoY, indicating that these virulence factors play a driving role in the multidrug resistance of clinical P. aeruginosa strains. It is noteworthy that the detection rate of the exoU gene, a phospholipase with strong cytotoxicity, is extremely low in all clinical strains. Consistent with previous reports, this gene is almost always mutually exclusive to exoS (Juan et al., 2017; Zhao et al., 2023). The exoU and exoS genes encode distinct exotoxins, which have different pathogenic mechanisms and host effects. The selective expression of these exotoxins helps avoid competition for resources, allowing P. aeruginosa to efficiently utilize resources in specific environments. This mechanism also increases its ability to survive and reproduce in its host (Shaver and Hauser, 2004; Horna et al., 2019).
In summary, we established an RAA assay method for detecting IRPA and RRPA and evaluated its performance through sensitivity detection, specificity detection, and clinical sample detection. The results demonstrated that this method has high sensitivity and excellent detection performance. RAA assay does not rely on expensive equipment or specialized technicians, it is suitable for diagnostic laboratories with limited resources. This method is beneficial for future clinical treatment and disease control in primary hospitals. Therefore, RAA is an excellent tool for epidemiological surveillance.
For epidemiological analysis, we employed MLST, virulence gene identification, and Antimicrobial Susceptibility Testing. MLST analysis revealed that ST-1639 is the most common type of IRPA, while ST-261 is the most common type of RRPA. All tested strains possessed five or more virulence genes, indicating that the presence of these genes and their relationship to the severity of infections in patients warrants further attention. These analyses provide valuable guidance for clinical diagnosis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed from the corresponding author upon reasonable request.
Ethics statement
The studies involving humans were approved by Northwest University Experimental Animal Care and Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
YZ: Formal analysis, Writing – original draft, Writing – review & editing. RS: Writing – review & editing. LM: Resources, Conceptualization, Writing – review & editing. LT: Methodology, Software, Supervision, Writing – review & editing. MZ: Project administration, Validation, Writing – review & editing. WL: Methodology, Visualization, Writing – review & editing. YC: Conceptualization, Data curation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (grant no. 31970050) to YC and Opening Foundation of Key Laboratory of Resource Biology and Biotechnology in Western China (Northwest University), Ministry of Education (grant no. ZSK2019004) to YC.
Acknowledgments
This is a short text to acknowledge the contributions of Chen laboratory that aided the efforts of this program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1428827/full#supplementary-material
References
Abniki, R., Tashakor, A., Masoudi, M., Mansury, D. (2024). Global resistance of imipenem/relebactam against gram-negative bacilli: systematic review and meta-analysis. Curr. Ther. Res. Clin. Exp. 100, 100723. doi: 10.1016/j.curtheres.2023.100723
Alexander, D. C., Jones, J. R., Liu, J. (2003). A rifampin-hypersensitive mutant reveals differences between strains of Mycobacterium smegmatis and presence of a novel transposon, IS1623. Antimicrob. Agents Chemother. 47, 3208–3213. doi: 10.1128/AAC.47.10.3208-3213.2003
Arbune, M., Gurau, G., Niculet, E., Iancu, A. V., Lupasteanu, G., Fotea, S., et al. (2021). Prevalence of antibiotic resistance of ESKAPE pathogens over five years in an infectious diseases hospital from south-east of Romania. Infect. Drug Resist. 14, 2369–2378. doi: 10.2147/IDR.S312231
Arlet, G., Nadjar, D., Herrmann, J. L., Donay, J. L., Lagrange, P. H., Philippon, A. (2001). Plasmid-mediated rifampin resistance encoded by an arr-2-like gene cassette in Klebsiella pneumoniae producing an ACC-1 class C beta-lactamase. Antimicrob. Agents Chemother. 45, 2971–2972. doi: 10.1128/AAC.45.10.2971-2972.2001
Bassetti, M., Poulakou, G., Ruppe, E., Bouza, E., Van Hal, S. J., Brink, A. (2017). Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: a visionary approach. Intensive Care Med. 43, 1464–1475. doi: 10.1007/s00134-017-4878-x
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Blanc, D. S., Magalhaes, B., Koenig, I., Senn, L., Grandbastien, B. (2020). Comparison of whole genome (wg-) and core genome (cg-) MLST (BioNumerics(TM)) versus SNP variant calling for epidemiological investigation of pseudomonas aeruginosa. Front. Microbiol. 11, 1729. doi: 10.3389/fmicb.2020.01729
Bonetta, S., Pignata, C., Lorenzi, E., De Ceglia, M., Meucci, L., Bonetta, S., et al. (2016). Detection of pathogenic Campylobacter, E. coli O157:H7 and Salmonella spp. in wastewater by PCR assay. Environ. Sci. pollut. Res. Int. 23, 15302–15309. doi: 10.1007/s11356-016-6682-5
Burrows, L. L. (2018). The therapeutic pipeline for pseudomonas aeruginosa infections. ACS Infect. Dis. 4, 1041–1047. doi: 10.1021/acsinfecdis.8b00112
Cao, Y., Fang, T., Shen, J., Zhang, G., Guo, D., Zhao, L., et al. (2023). Development of recombinase aided amplification (RAA)-exo-probe assay for the rapid detection of shiga toxin-producing escherichia coli. J. AOAC Int. 106, 1246–1253. doi: 10.1093/jaoacint/qsad063
Castaneda-Montes, F. J., Avitia, M., Sepulveda-Robles, O., Cruz-Sanchez, V., Kameyama, L., Guarneros, G., et al. (2018). Population structure of Pseudomonas aeruginosa through a MLST approach and antibiotic resistance profiling of a Mexican clinical collection. Infect. Genet. Evol. 65, 43–54. doi: 10.1016/j.meegid.2018.06.009
Chan, M. S., Maiden, M. C., Spratt, B. G. (2001). Database-driven multi locus sequence typing (MLST) of bacterial pathogens. Bioinformatics 17, 1077–1083. doi: 10.1093/bioinformatics/17.11.1077
Chevalier, S., Bouffartigues, E., Bodilis, J., Maillot, O., LesouHaitier, O., Feuilloley, M. G. J., et al. (2017). Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 41, 698–722. doi: 10.1093/femsre/fux020
Cooper, J. E., Feil, E. J. (2004). Multilocus sequence typing–what is resolved? Trends Microbiol. 12, 373–377. doi: 10.1016/j.tim.2004.06.003
Cui, G., Zhang, Y., Xu, X., Liu, Y., Li, Z., Wu, M., et al. (2022). PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa. Sci. Adv. 8, eadd4220. doi: 10.1126/sciadv.add4220
Curran, C. S., Bolig, T., Torabi-Parizi, P. (2018). Mechanisms and targeted therapies for pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 197, 708–727. doi: 10.1164/rccm.201705-1043SO
Curran, B., Jonas, D., Grundmann, H., Pitt, T., Dowson, C. G. (2004). Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42, 5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004
Da Fonseca, E. L., Freitas Fdos, S., De Amorim, J. C., Vicente, A. C. (2008). Detection of new arr-4 and arr-5 gene cassettes in clinical Pseudomonas aeruginosa and Klebsiella pneumoniae strains from Brazil. Antimicrob. Agents Chemother. 52, 1865–1867. doi: 10.1128/AAC.00017-08
Denis, J. B., Lehingue, S., Pauly, V., Cassir, N., Gainnier, M., Leone, M., et al. (2019). Multidrug-resistant Pseudomonas aeruginosa and mortality in mechanically ventilated ICU patients. Am. J. Infect. Control 47, 1059–1064. doi: 10.1016/j.ajic.2019.02.030
Diekema, D. J., Pfaller, M. A., Jones, R. N., Doern, G. V., Kugler, K. C., Beach, M. L., et al. (2000). Trends in antimicrobial susceptibility of bacterial pathogens isolated from patients with bloodstream infections in the USA, Canada and Latin America. SENTRY Participants Group. Int. J. Antimicrob. Agents 13, 257–271. doi: 10.1016/S0924-8579(99)00131-4
Do Rego, H., Timsit, J. F. (2023). Management strategies for severe Pseudomonas aeruginosa infections. Curr. Opin. Infect. Dis. 36, 585–595. doi: 10.1097/QCO.0000000000000981
Dunn, M., Wunderink, R. G. (1995). Ventilator-associated pneumonia caused by Pseudomonas infection. Clin. Chest Med. 16, 95–109. doi: 10.1016/S0272-5231(21)00980-1
Fan, X., Li, L., Zhao, Y., Liu, Y., Liu, C., Wang, Q., et al. (2020). Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 11, 1696. doi: 10.3389/fmicb.2020.01696
Firoozeh, F., Ghorbani, M., Zibaei, M., Badmasti, F., Farid, M., Omidinia, N., et al. (2023). Characterization of class 1 integrons in metallo-beta-lactamase-producing Acinetobacter baumannii isolates from hospital environment. BMC Res. Notes 16, 365.
Fu, H., Gan, L., Tian, Z., Han, J., Du, B., Xue, G., et al. (2022). Rapid detection of Burkholderia cepacia complex carrying the 16S rRNA gene in clinical specimens by recombinase-aided amplification. Front. Cell Infect. Microbiol. 12, 984140. doi: 10.3389/fcimb.2022.984140
Gonzalez-Vazquez, M. C., Rocha-Gracia, R. D. C., Carabarin-Lima, A., Bello-Lopez, E., Huerta-Romano, F., Martinez-Laguna, Y., et al. (2021). Location of OprD porin in Pseudomonas aeruginosa clinical isolates. APMIS 129, 213–224. doi: 10.1111/apm.13118
Holbrook, S. Y. L., Garneau-Tsodikova, S. (2018). Evaluation of aminoglycoside and carbapenem resistance in a collection of drug-resistant pseudomonas aeruginosa clinical isolates. Microb. Drug Resist. 24, 1020–1030. doi: 10.1089/mdr.2017.0101
Horna, G., Amaro, C., Palacios, A., Guerra, H., Ruiz, J. (2019). High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant "high-risk clones" of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci. Rep. 9, 10874. doi: 10.1038/s41598-019-47303-4
Huang, H., Jeanteur, D., Pattus, F., Hancock, R. E. (1995). Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol. Microbiol. 16, 931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x
Huang, H., Shao, X., Xie, Y., Wang, T., Zhang, Y., Wang, X., et al. (2019). An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 10, 2931. doi: 10.1038/s41467-019-10778-w
Juan, C., Pena, C., Oliver, A. (2017). Host and pathogen biomarkers for severe pseudomonas aeruginosa infections. J. Infect. Dis. 215, S44–S51. doi: 10.1093/infdis/jiw299
Kang, C. I., Kim, S. H., Kim, H. B., Park, S. W., Choe, Y. J., Oh, M. D., et al. (2003). Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin. Infect. Dis. 37, 745–751. doi: 10.1086/377200
Kao, C. Y., Chen, S. S., Hung, K. H., Wu, H. M., Hsueh, P. R., Yan, J. J., et al. (2016). Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol. 16, 107. doi: 10.1186/s12866-016-0719-2
Karruli, A., Catalini, C., D'amore, C., Foglia, F., Mari, F., Harxhi, A., et al. (2023). Evidence-based treatment of pseudomonas aeruginosa infections: A critical reappraisal. Antibiotics (Basel) 12, 399. doi: 10.3390/antibiotics12020399
Kim, C. H., Kang, H. Y., Kim, B. R., Jeon, H., Lee, Y. C., Lee, S. H., et al. (2016). Mutational inactivation of OprD in carbapenem-resistant Pseudomonas aeruginosa isolates from Korean hospitals. J. Microbiol. 54, 44–49. doi: 10.1007/s12275-016-5562-5
Lee, M. S., Oh, J. Y., Kang, C. I., Kim, E. S., Park, S., Rhee, C. K., et al. (2018). Guideline for antibiotic use in adults with community-acquired pneumonia. Infect. Chemother. 50, 160–198. doi: 10.3947/ic.2018.50.2.160
Li, H., Luo, Y. F., Williams, B. J., Blackwell, T. S., Xie, C. M. (2012). Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int. J. Med. Microbiol. 302, 63–68. doi: 10.1016/j.ijmm.2011.10.001
Li, J., Macdonald, J., Von Stetten, F. (2018). Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 144, 31–67. doi: 10.1039/C8AN01621F
Li, F., Ye, Q., Chen, M., Zhou, B., Zhang, J., Pang, R., et al. (2021). An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens Bioelectron 179, 113073. doi: 10.1016/j.bios.2021.113073
Liu, J., Chen, F., Wang, X., Peng, H., Zhang, H., Wang, K. J. (2020). The synergistic effect of mud crab antimicrobial peptides sphistin and sph(12-38) with antibiotics azithromycin and rifampicin enhances bactericidal activity against pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 10, 572849. doi: 10.3389/fcimb.2020.572849
Logan, L. K., Gandra, S., Mandal, S., Klein, E. Y., Levinson, J., Weinstein, R. A., et al. (2017). Multidrug- and carbapenem-resistant pseudomonas aeruginosa in children, United States 1999-2012. J. Pediatr. Infect. Dis. Soc. 6, 352–359.
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Mao, L., Ying, J., Selekon, B., Gonofio, E., Wang, X., Nakoune, E., et al. (2022). Development and characterization of recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of monkeypox virus. Viruses 14, 2112. doi: 10.3390/v14102112
Montgomery, R., Boswell, T., Mahida, N. (2018). Pseudomonas aeruginosa control in healthcare settings: outpatient dialysis units are not augmented care units. J. Hosp Infect. 98, 65–66. doi: 10.1016/j.jhin.2017.09.027
Naas, T., Mikami, Y., Imai, T., Poirel, L., Nordmann, P. (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol 183, 235–249. doi: 10.1128/JB.183.1.235-249.2001
Naik, P., Singh, S., Rudraprasad, D., Dave, V. P., Kumar, A., Joseph, J. (2021). Multidrug-resistant pseudomonas aeruginosa triggers differential inflammatory response in patients with endophthalmitis. Transl. Vis. Sci. Technol. 10, 26. doi: 10.1167/tvst.10.9.26
Ochs, M. M., Bains, M., Hancock, R. E. (2000). Role of putative loops 2 and 3 in imipenem passage through the specific porin OprD of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 1983–1985. doi: 10.1128/AAC.44.7.1983-1985.2000
Piepenburg, O., Williams, C. H., Stemple, D. L., Armes, N. A. (2006). DNA detection using recombination proteins. PloS Biol. 4, e204. doi: 10.1371/journal.pbio.0040204
Prithiviraj, B., Bais, H. P., Weir, T., Suresh, B., Najarro, E. H., Dayakar, B. V., et al. (2005). Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect. Immun. 73, 5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005
Quale, J., Bratu, S., Gupta, J., Landman, D. (2006). Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 50, 1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006
Quinn, J. P., Darzins, A., Miyashiro, D., Ripp, S., Miller, R. V. (1991). Imipenem resistance in pseudomonas aeruginosa PAO: mapping of the OprD2 gene. Antimicrob. Agents Chemother. 35, 753–755. doi: 10.1128/AAC.35.4.753
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., Chapman, J. (2019). A review of methods for the detection of pathogenic microorganisms. Analyst 144, 396–411. doi: 10.1039/C8AN01488D
Shaver, C. M., Hauser, A. R. (2004). Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72, 6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004
Shelite, T. R., Uscanga-Palomeque, A. C., Castellanos-Gonzalez, A., Melby, P. C., Travi, B. L. (2021). Isothermal recombinase polymerase amplification-lateral flow detection of SARS-CoV-2, the etiological agent of COVID-19. J. Virol. Methods 296, 114227. doi: 10.1016/j.jviromet.2021.114227
Shen, J. L., Fang, Y. P. (2015). Detection of drug-resistance mechanism of Pseudomonas aeruginosa developing from a sensitive strain to a persister during carbapenem treatment. Genet. Mol. Res. 14, 6723–6732. doi: 10.4238/2015.June.18.16
Song, Z., Ting, L., Kun, Y., Wei, L., Jian-Feng, Z., Li-Chuan, G., et al. (2018). Establishment of a recombinase-aided isothermal amplification technique to detect Schistosoma japonicum specific gene fragments. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 30, 273–277.
Soscia, C., Hachani, A., Bernadac, A., Filloux, A., Bleves, S. (2007). Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J. Bacteriol 189, 3124–3132. doi: 10.1128/JB.01677-06
Sun, Q., Ba, Z., Wu, G., Wang, W., Lin, S., Yang, H. (2016). Insertion sequence ISRP10 inactivation of the oprD gene in imipenem-resistant Pseudomonas aeruginosa clinical isolates. Int. J. Antimicrob. Agents 47, 375–379. doi: 10.1016/j.ijantimicag.2016.02.008
Suresh, M., Skariyachan, S., Narayanan, N., Pullampara Rajamma, J., Panickassery Ramakrishnan, M. K. (2020). Mutational Variation Analysis of oprD Porin Gene in Multidrug-Resistant Clinical Isolates of Pseudomonas aeruginosa. Microb. Drug Resist. 26, 869–879. doi: 10.1089/mdr.2019.0147
Suwantarat, N., Carroll, K. C. (2016). Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob. Resist. Infect. Control 5, 15. doi: 10.1186/s13756-016-0115-6
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tribuddharat, C., Fennewald, M. (1999). Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 960–962. doi: 10.1128/AAC.43.4.960
Vaez, H., Salehi-Abargouei, A., Khademi, F. (2017). Systematic review and meta-analysis of imipenem-resistant Pseudomonas aeruginosa prevalence in Iran. Germs 7, 86–97. doi: 10.18683/germs.2017.1113
Waters, V., Zlosnik, J. E., Yau, Y. C., Speert, D. P., Aaron, S. D., Guttman, D. S. (2012). Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 31, 3341–3350. doi: 10.1007/s10096-012-1701-z
Wu, K., Zhang, Y., Zeng, S., Liu, X., Li, Y., Li, X., et al. (2021). Development and application of RAA nucleic acid test strip assay and double RAA gel electrophoresis detection methods for ASFV and CSFV. Front. Mol. Biosci. 8, 811824. doi: 10.3389/fmolb.2021.811824
Yan, C., Zhou, Y., Du, S., Du, B., Zhao, H., Feng, Y., et al. (2023). Recombinase-Aided Amplification Assay for Rapid Detection of Hypervirulent Klebsiella pneumoniae (hvKp) and Characterization of the hvKp Pathotype. Microbiol. Spectr. 11, e0398422. doi: 10.1128/spectrum.03984-22
Yoon, S. A., Park, S. Y., Cha, Y., Gopala, L., Lee, M. H. (2021). Strategies of detecting bacteria using fluorescence-based dyes. Front. Chem. 9, 743923. doi: 10.3389/fchem.2021.743923
Zhao, Y., Chen, D., Ji, B., Zhang, X., Anbo, M., Jelsbak, L. (2023). Whole-genome sequencing reveals high-risk clones of Pseudomonas aeruginosa in Guangdong, China. Front. Microbiol. 14, 1117017. doi: 10.3389/fmicb.2023.1117017
Keywords: Pseudomonas aeruginosa, recombinase-aided amplification, rapid detection, antimicrobial susceptibility testing, ARR, OprD
Citation: Zhou Y, Shi R, Mu L, Tian L, Zhou M, Lyu W and Chen Y (2024) Recombinase-aided amplification assay for rapid detection of imipenem-resistant Pseudomonas aeruginosa and rifampin-resistant Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 14:1428827. doi: 10.3389/fcimb.2024.1428827
Received: 07 May 2024; Accepted: 24 July 2024;
Published: 10 September 2024.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Michelle M. Tabb, DiaSorin Molecular LLC, United StatesElena Niculet, de Jos University, Romania
Copyright © 2024 Zhou, Shi, Mu, Tian, Zhou, Lyu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaodong Chen, eWRjaGVuQG53dS5lZHUuY24=
 Yao Zhou
Yao Zhou Ruiqing Shi
Ruiqing Shi Liang Mu3
Liang Mu3 Yaodong Chen
Yaodong Chen