- 1Department of Pain Management, Affiliated Hospital of Hebei University/School of Clinical Medicine, Hebei University, Baoding, Hebei, China
- 2School of Basic Medicine, Hebei University, Baoding, Hebei, China
- 3Department of Anesthesia, Affiliated Hospital of Hebei University/School of Clinical Medicine, Hebei University, Baoding, Hebei, China
Postherpetic neuralgia (PHN) is a common, severe, and hard-to-treat chronic pain condition in clinics. Although PHN is developed from herpes zoster (HZ), the developing mechanism is unknown. A previous study investigated blood metabolomic and proteomic profiling in patients with PHN and HZ. The current study aims to explore the blood transcriptomic signature of PHN compared to HZ patients. Whole blood from eight PHN and 15 HZ patients was used for RNA-Seq analysis. There were 82 and 1,788 genes detected specifically in the PHN and HZ groups, respectively. PHN-specific genes are involved in viral infection, lipid and carbohydrate metabolism, and immune response. For genes coexpressed in PHN and HZ patients, there were 407 differential expression genes (DEGs), including 205 upregulated (UP DEGs) and 202 downregulated (DOWN DEGs) in PHN compared to HZ groups. DEGs are involved in viral infection, type I interferon (IFN), and hemoglobin and oxygen carrier activity. UP DEGs are associated with regulatory T cells (Tregs), activated NK cells, and neutrophils, while DOWN DEGs are associated with Tregs, resting NK cells, and monocytes. The results suggest that the metabolism of lipid, glycan, and nucleotides, type I IFN signaling, and altered neutrophil activation are associated with and might contribute to the development of PHN in HZ. It is also suggested that persistent or altered activation of nonspecific immunity may contribute to the development of PHN from HZ.
Introduction
Herpes zoster (HZ) develops in about 30% of the world population in their lifetime (without the use of a vaccine), and postherpetic neuralgia (PHN) is the most common complication of HZ (van Oorschot et al., 2021; Curran et al., 2022; Pan et al., 2022). The overall risk of PHN after HZ is about 10%–20%, with the majority of these cases occurring at the age of 50 or older (Delaney et al., 2009; Sampathkumar et al., 2009; Saguil et al., 2017). PHN often refers to neuropathic pain that persists for more than 3 months after the disappearance of the rash and blisters associated with HZ. It is one of the most severe and intractable chronic pain conditions in clinics.
HZ is caused by the reactivation of the Varicella-Zoster virus (VZV). The first VZV infection results in chickenpox, which often self-resolves in 7–10 days. Afterward, VZV becomes latent in dorsal root ganglion (DRG) and trigeminal ganglion (TG) neurons. An increase in age or immunosuppression can trigger the reactivation of VZV, leading to HZ. However, it is unknown how HZ develops into PHN.
Due to the host selectivity (to humans) of VZV, there is a lack of valid animal models for HZ and PHN studies. Previous patient studies involve peripheral blood mononuclear cells, serum molecules associated with inflammation, and postmortem examination of immune cell infiltration in DRG and TG (Gowrishankar et al., 2010; Xing et al., 2013; Mercan et al., 2021; Peng et al., 2022). Particularly, a recent study has investigated blood metabolomic and proteomic profiling in patients with PHN and HZ (Zhou et al., 2022). However, blood transcriptomes have not been studied in these patients. The current study aims to explore the differential blood transcriptomic signatures between PHN and HZ, focusing on the indication for the positive association and development of PHN from HZ.
Experimental procedures
Study population
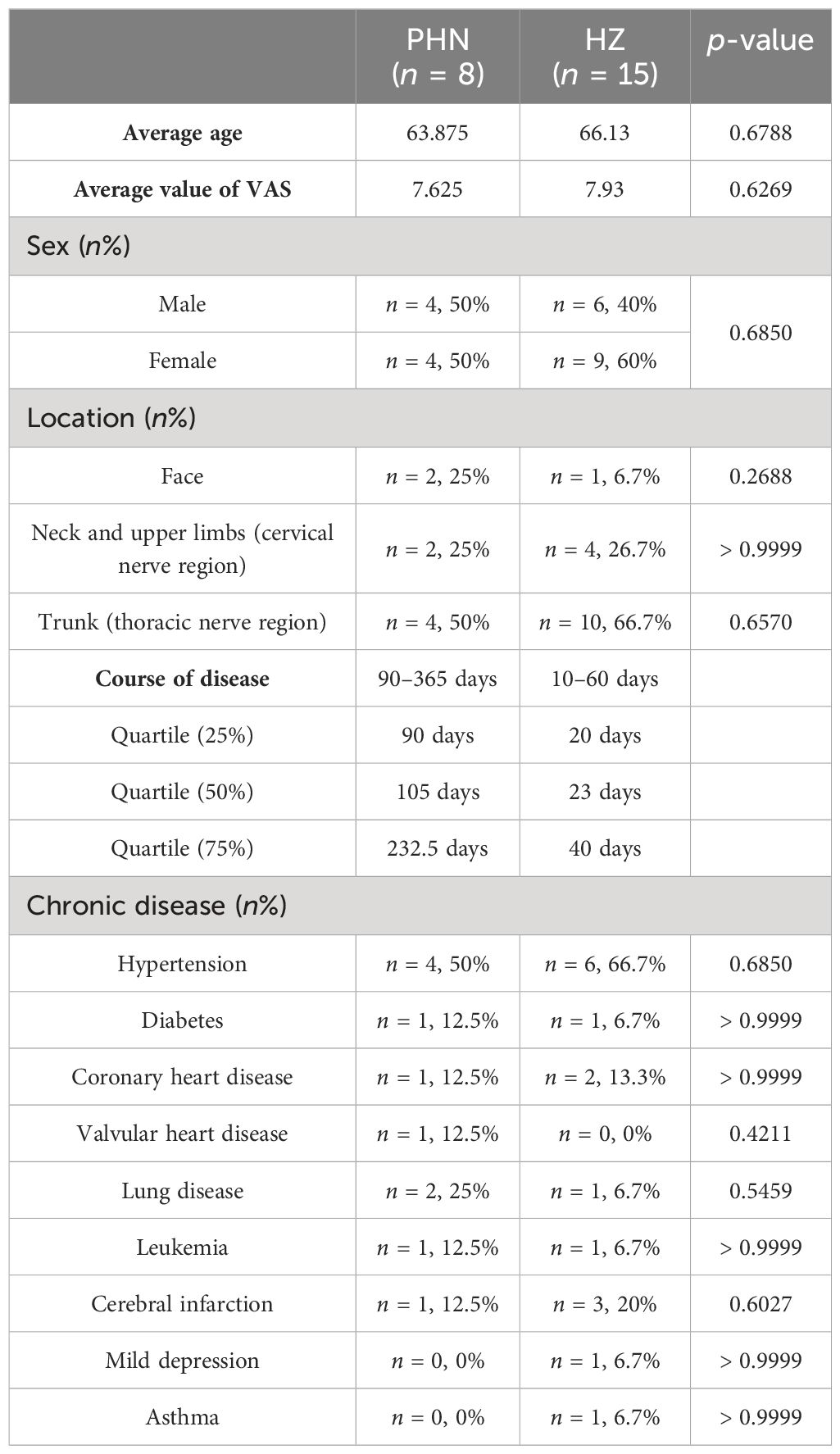
The subjects of the current study were inpatients from October 2022 to May 2023 in the Department of Pain Management at Hebei University Affiliated Hospital. Fifteen HZ and eight PHN patients were included (Table 1). Patients’ information was collected, including gender, age, disease course, location of zoster, Visual Analog Scale (VAS), and medical conditions. A regular blood examination was conducted. The inclusion criteria of HZ and PHN are as follows: (1) meet the diagnosis criteria in the “Chinese consensus on the diagnosis and management of herpes zoster (2018)”; (2) have a VAS > 3 at admission; (3) be > 18 years old; and (4) be able to evaluate self-pain and give a VAS score. Exclusion criteria include the following: conditions (such as mental and nervous diseases) that do not allow cooperative treatment. The study was approved by the ethics committee at Hebei University Affiliated Hospital. Written informed consent was obtained from all participants before the study.
Transcriptome analysis
Blood samples of 1 ml were drawn using EDTA anticoagulation tubes. After adding 3 ml of TRIzol, mixing gently by blowing, and incubating for 5-min, the mixture was frozen and stored in a − 80°C freezer.
RNA purification, reverse transcription, cDNA library construction, and sequencing were conducted by Shanghai Majorbio Bio-Pharm Biotechnology Co. Ltd. (Shanghai, China) according to the relative instructions of the manufacturer (Illumina, San Diego, CA, USA). RNAs were extracted by TRIzol®. RNA quality was detected by a bioanalyzer (Agilent 5300). RNA quantification was determined by a NanoDrop (ND-2000, NanoDrop Technologies, Carlsbad, CA, United States). High-quality RNA samples (OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, > 1 μg) were used for construction of sequencing library.
The transcriptome library of PHN and HZ patients was constructed by Illumina® Stranded mRNA Prep and Ligation from Illumina (San Diego, CA, USA) using 1 μg of total RNA. Briefly, mRNAs were isolated by oligo (dT) beads, and a fragment buffer was used for fragment partitioning. Double-stranded cDNAs were synthesized by SuperScript double-stranded cDNA synthesis Kit (Invitrogen, CA, Wilmington, DE, United States) and random hexamer primers (Illumina, San Diego, CA, USA). DNA end repair, phosphorylation, and “A” base addition were conducted according to the Illumina library construction protocols. cDNA target fragments (300 bp) were chosen at 2% low-range ultra agarose. cDNA amplification was conducted by 15 cycles of PCR using Phusion DNA polymerase (NEB). After quantification by Qubit 4.0, the NovaSeq Xplus sequencer (2 bp × 150 bp read length) was used for sequencing of the paired-end RNA-Seq library.
Pair-end reads were trimmed and controlled for quality using fastp (Chen et al., 2018) with default parameters. Clean reads were compared with the reference genome using HISAT2 (Kim et al., 2015) in the orientation mode. The mapped reads were assembled by StringTie (Pertea et al., 2015) in a reference-based approach for each sample. The expression level of each transcript was calculated by transcript per million (TPM). RSEM (Li and Dewey, 2011) was used to quantify transcript abundances in order to recognize DEGs between two samples. Differential expression analysis was performed by DESeq2 or DEGseq (Wang et al., 2010; Love et al., 2014). DEGs with |log2FC|≥1 and FDR < 0.05 (DESeq2) or FDR < 0.001 (DEGseq) were considered significantly different transcripts. Goatools and Python scipy were used for GO and KEGG and DO analyses, respectively. All data analysis was conducted on the Majorbio Cloud Platform (https://cloud.majorbio.com/).
qPCR
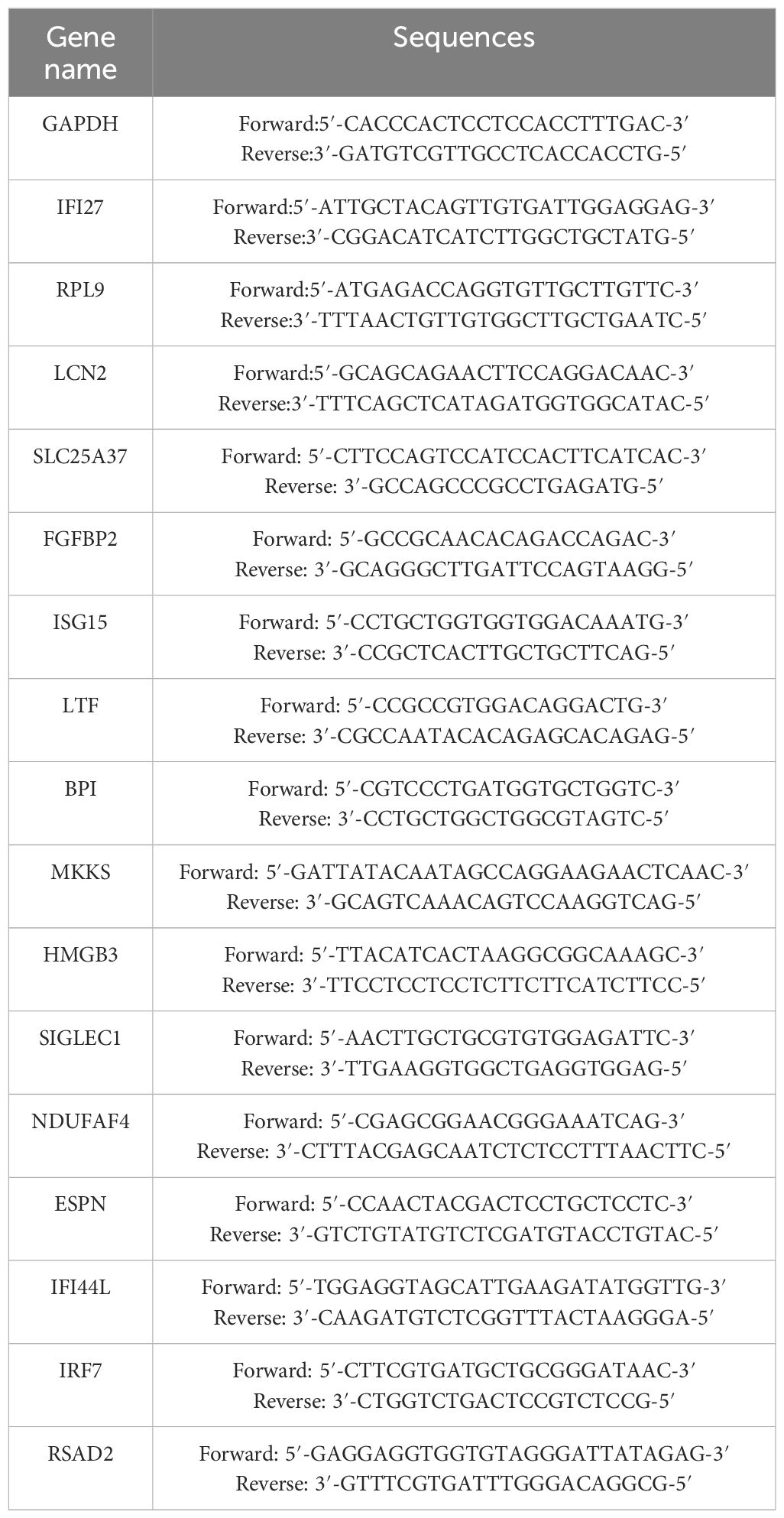
Reverse transcription was conducted using HiScript®III RT SuperMix for the qPCR kit (Vazyme, Nanjing, China). DNA was first degraded by mixing RNA and 4×gDNA wiper Mix. 5×HiScript qRT SuperMix was then used for reverse transcription of mRNA to cDNA. ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme, Nanjing, China) was used for cDNA amplification. Real-time qPCR was conducted by Lightcycler 96 system (Roche, Basel, Switzerland). Primer sequences are listed in Table 2.
Statistical analysis
Averaged data were presented as mean ± SEM. Student’s t-test or Fisher’s exact test was used to examine the statistical significance. The significance level was set at p = 0.05.
Results
Participant demographics
Demographic and disease information were compared between PHN and HZ patients (Table 1). There was no significant difference in gender, age, pain scale, location, or chronic diseases (VAS). The trunk (thoracic nerve region) is the major skin location in both groups. Hypertension is the most frequent chronic disease in both groups. Regular blood examination found that there was a decreasing trend in white blood cells (p = 0.054) and basophils (p = 0.087) count, and an increasing trend in platelet large cell ratio (p = 0.067) (Supplementary Table S1). In addition, the neutrophil-to-lymphocyte ratio (NLR) was insignificantly decreased (from 4.73 to 3.38, p = 0.469). For HZ patients, nine out of 15 were cured and reported no pain 3 months later. Three of them reported a low level of pain (VAS < 3) while the other three reported a medium level of pain (3 < VAS < 6) 3 months later. For all eight PHN patients, a positive treatment had been received before the development of chronic pain.
Overall transcriptomic comparison
The expression of viral transcripts (ORF61, ORF63, and ORF66) was not detected in all the samples. As shown in Figure 1A, the distribution range of transcriptomic expression was similar between PHN and HZ patients. However, it seems the average level of expression was slightly lower in PHN compared to HZ. Moreover, there was less than 1% PHN-specific expression compared to more than 20% HZ-specific expression in the Venn plot (Figure 1B). On the other hand, PCA analysis did not separate the transcriptome of PHN from HZ. All the transcripts detected were included in Supplementary File 1.
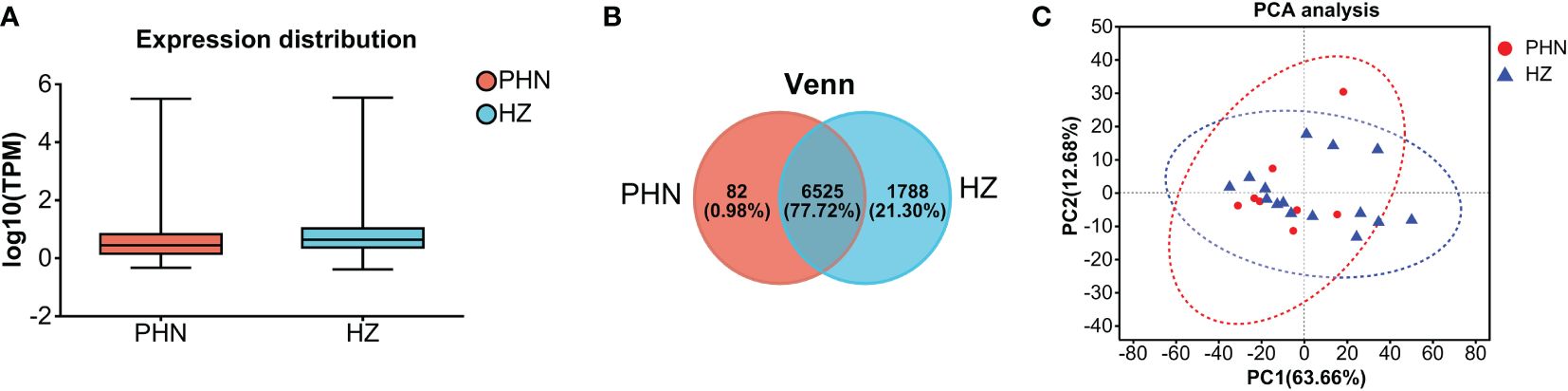
Figure 1. Comparison of overall transcriptomic profiling between PHN and HZ patients. (A) Distribution of transcriptomes detected; (B) Venn plot; (C) PCA analysis.
PHN-specific expression
As shown in Figure 2A, GO analysis found that binding and catalytic activity, cell part and organelle, and cellular process and metabolic process were the top annotations for biological process, cellular components, and molecular function, respectively. KEGG analysis found that PHN-specific expression was more associated with infectious disease (viral), signal transduction, translation, replicate and repair, lipid metabolism, glycan biosynthesis and metabolism (Figure 2B). DO analysis found that cancer was the top disease expressing PHN-specific genes (Figure 2C). Moreover, functional analysis of top 20 genes revealed that a majority of them is involved in biosynthesis and metabolism (TMA16, HPGD, GEMIN6, SRD5A3, MMAB), immunity and inflammation (HMGB3, ELOVL6, TMIGD3, Siglec-1), and mitochondria respiratory chain (NDUFAF4, SMIM4) (Figure 2D). PHN-specific expression was included in Supplementary File 1.
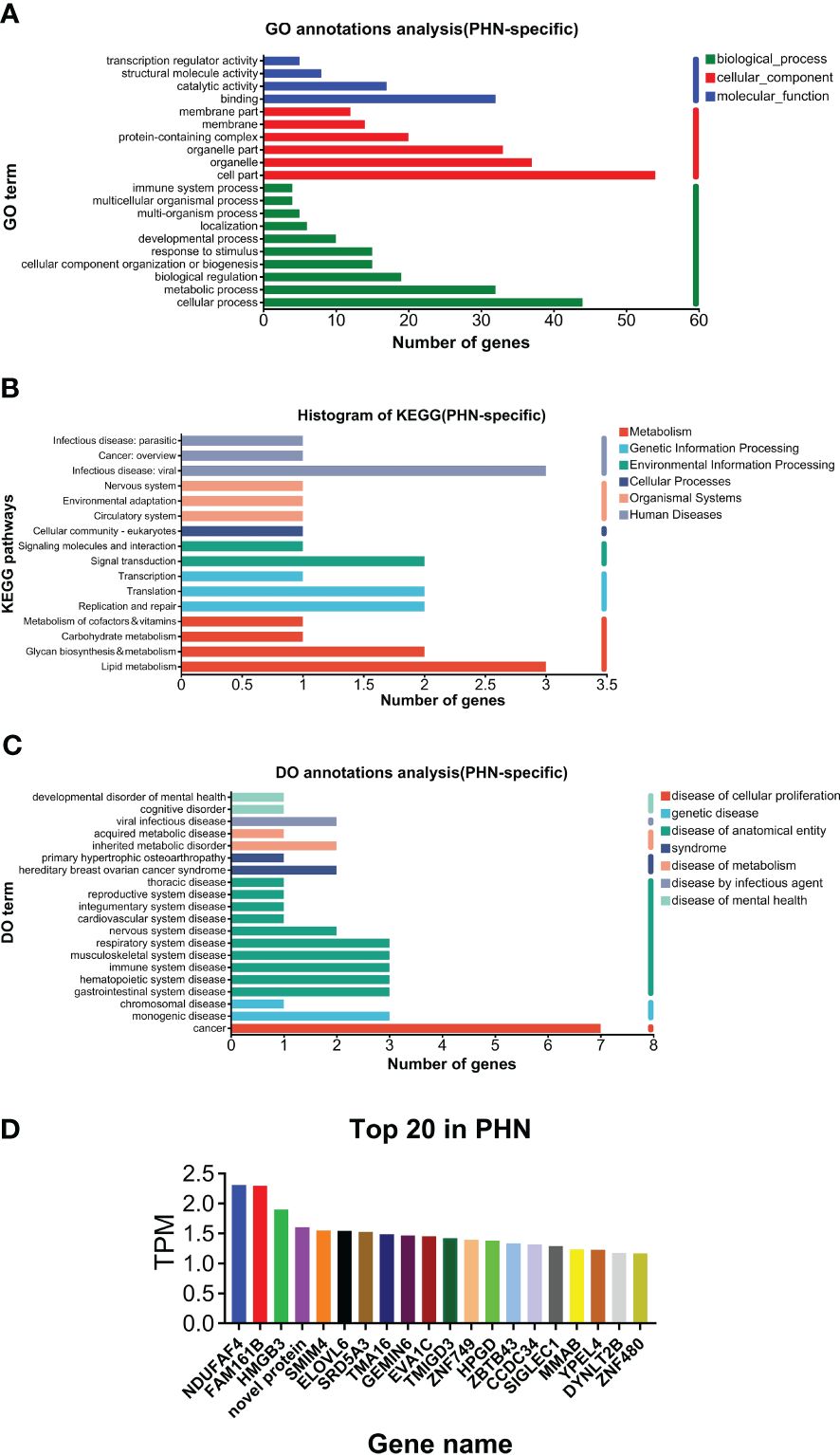
Figure 2. Analysis of PHN-specific expression. (A) GO analysis. (B) KEGG analysis. (C) DO analysis. (D) Top 20 expressed genes.
DEGs
Among the 6,525 gene expressions detected in both PHN and HZ patients, there were 407 DEGs, including 205 upregulated DEGs (UP DEGs) and 202 downregulated DEGs (DOWN DEGs) (Figure 3A). The volcano distribution of UP and DOWN DEGs is shown in Figure 3B. Heatmap analysis found that half of the PHN patients were clustered on the right side, about one-sixth to one-third from the right end (Figure 3C). The other half spread out leftward with increasing distance (Figure 3C). All DEGs are listed in the Supplementary File 1.
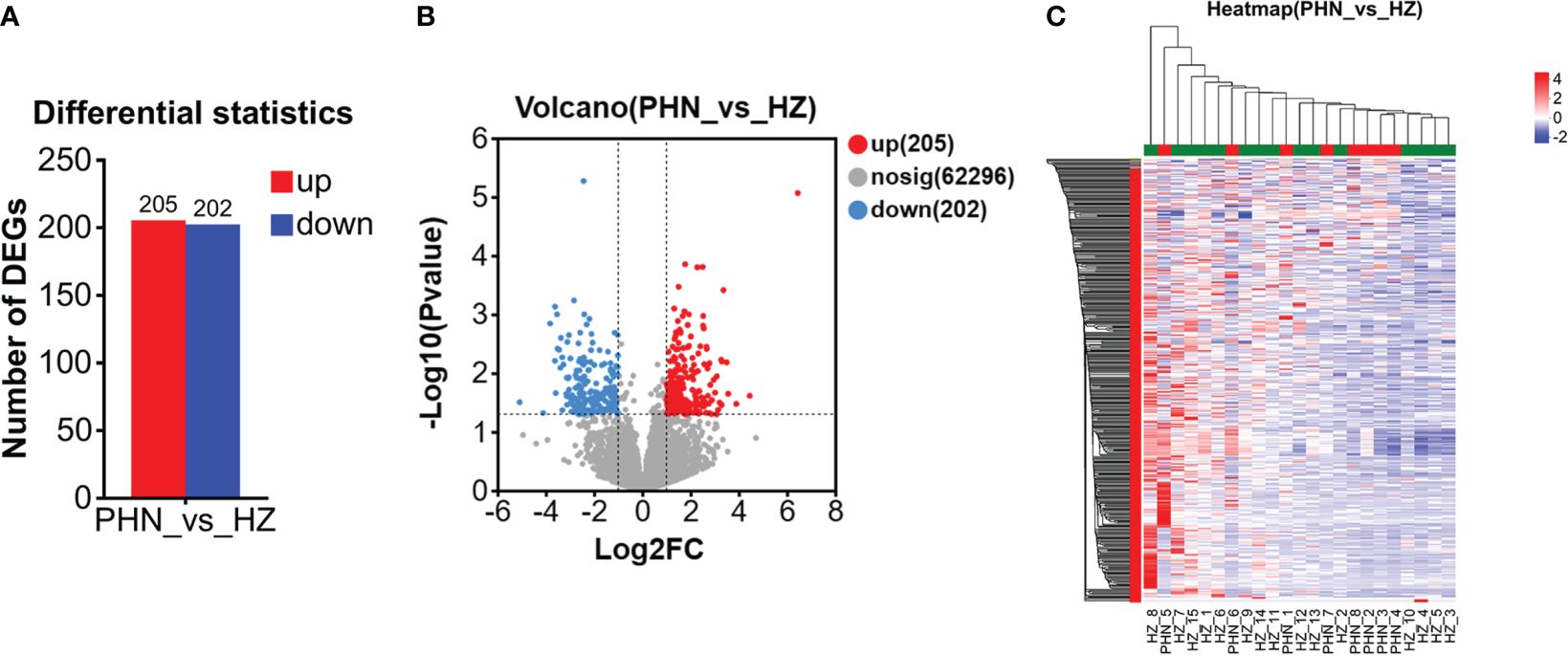
Figure 3. Comparison of overall DEG profiling between PHN and HZ patients. (A) Number of UP and DOWN DEG. (B)Volcano plot. (C) Heat map.
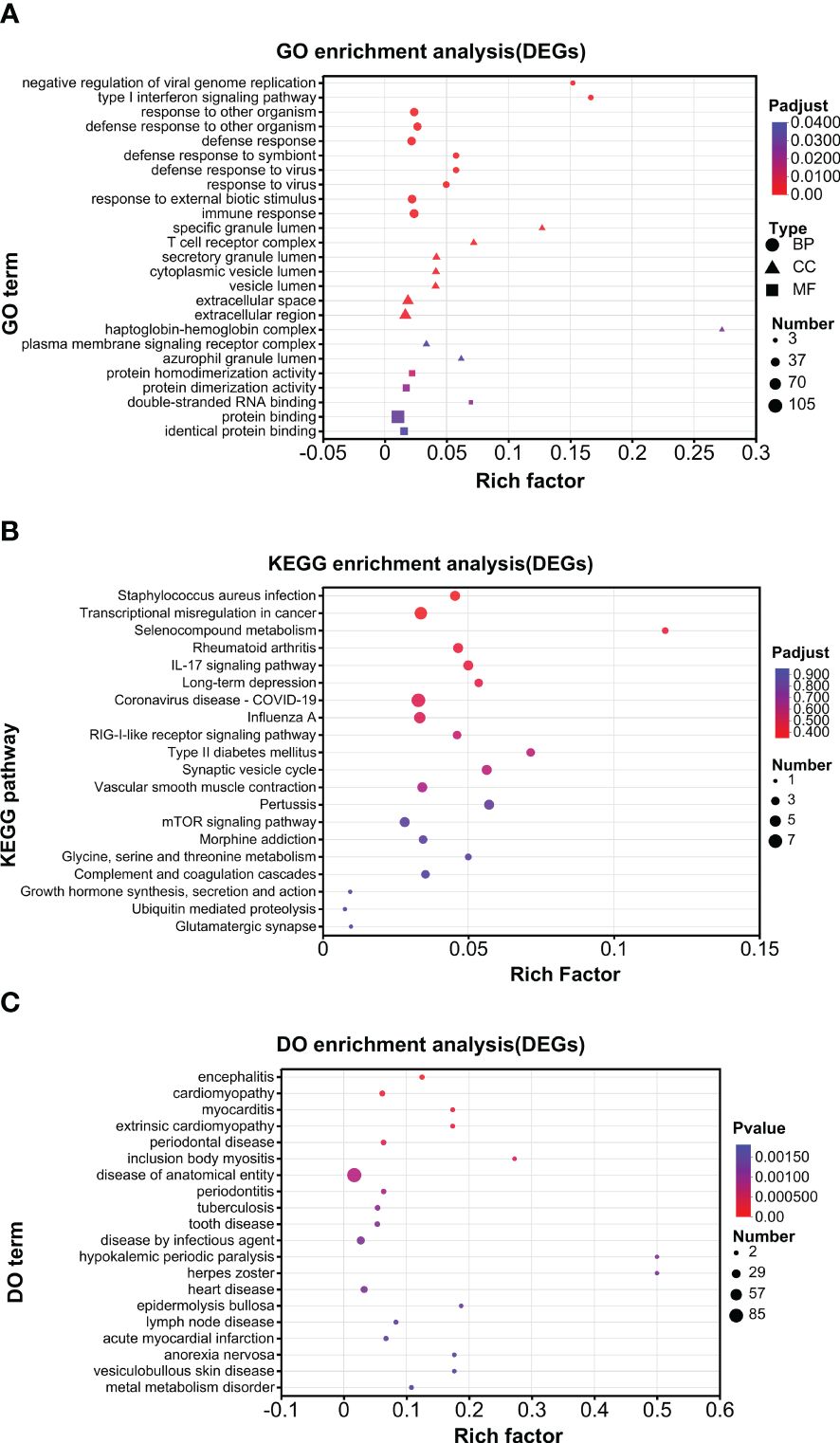
An enrichment analysis was conducted for DEGs. For all the DEGs (UP and DOWN DEGs), GO analysis found that negative regulation of viral genome replication and type I interferon signaling pathway were the top enriched biological processes; that specific granule lumen and haptoglobin–hemoglobin complex were the top enriched cellular components; and that 2′–5′-oligoadenylate synthetase activity and oxygen carrier activity were the top enriched molecular function (Figure 4A). KEGG analysis found that pertussis, synaptic vesicle cycle, type II diabetes mellitus, IL-17 signaling pathway, selenocompound metabolism, and long-term depression were enriched at higher levels (Figure 4B). DO analysis found that herpes zoster and hypokalemic periodic paralysis were the top diseases (Figure 4C).
Further enrichment analysis was conducted for UP DEGs and DOWN DEGs separately. For GO analysis, a similar pattern of annotations was observed between UP and DOWN DEGs (Supplementary Figures S1A, 2A). For example, in both groups, cellular process and biological regulation, cell part and organelle, and binding and catalytic activity were the top 2 annotations for biological process, cellular component, and molecular function, respectively. For KEGG analysis, the overall pattern of annotations was similar between UP and DOWN DEGs (Supplementary Figures S1B, 2B). For example, in both groups, infectious disease (viral.) and cancer (overview), immune system, cellular community (eukaryotes), signal transduction were the top annotations for human diseases, organismal system, cellular process, and environment information processing, respectively. However, endocrine and metabolic diseases and neurodegenerative diseases were selectively marked in the UP but not DOWN DEG groups and were among the top half annotations in the UP DEG group (Supplementary Figure S1B). Moreover, nucleotide metabolism was selectively in the UP DEG group, while metabolism of other amino acids and the metabolism of cofactors and vitamins were only in the DOWN DEG group (Supplementary Figures S1B, S2B). For DO analysis, the top 8 diseases were the same between UP and DOWN DEGs (Supplementary Figures S1C, S2C). However, nervous system disease ranked 2 and 8 in the UP and DOWN DEG groups, respectively (Supplementary Figures S1C, S2C).
The top 20 UP and DOWN DEGs are listed in Figure 5A. The UP DEGs were increased by less than four to more than 100 times. The DOWN DEGs were reduced by about three- to30-fold (Figure 5A). GO analysis found that binding and cell part were the top annotations for molecular function and cellular component, respectively (Figure 5C). In addition, immune system processes, responses to stimulus, and cellular processes were the top 3 biological process (Figure 5C). For KEGG, immune system and infectious disease (viral) were the top 2 annotations, followed by infectious disease (bacteria), immune disease, and cancer (overview) (Figure 5D). DO analysis found that viral infection, acquired metabolic, monogenic, cancer, and multiple anatomical entity diseases were higher enriched (Figure 5E). PPI analysis found that major hub proteins included RTP4, IFI27, IFI44L, IFI44, LCN2, ISG15, MX1, IFIT1, LTF, RSAD2, RETN, MPO, CEACAM8, IFI6, IRF7, BPI, and RNASE3 (Figure 5B). UP and DOWN DEGs are listed in Supplementary File 1.
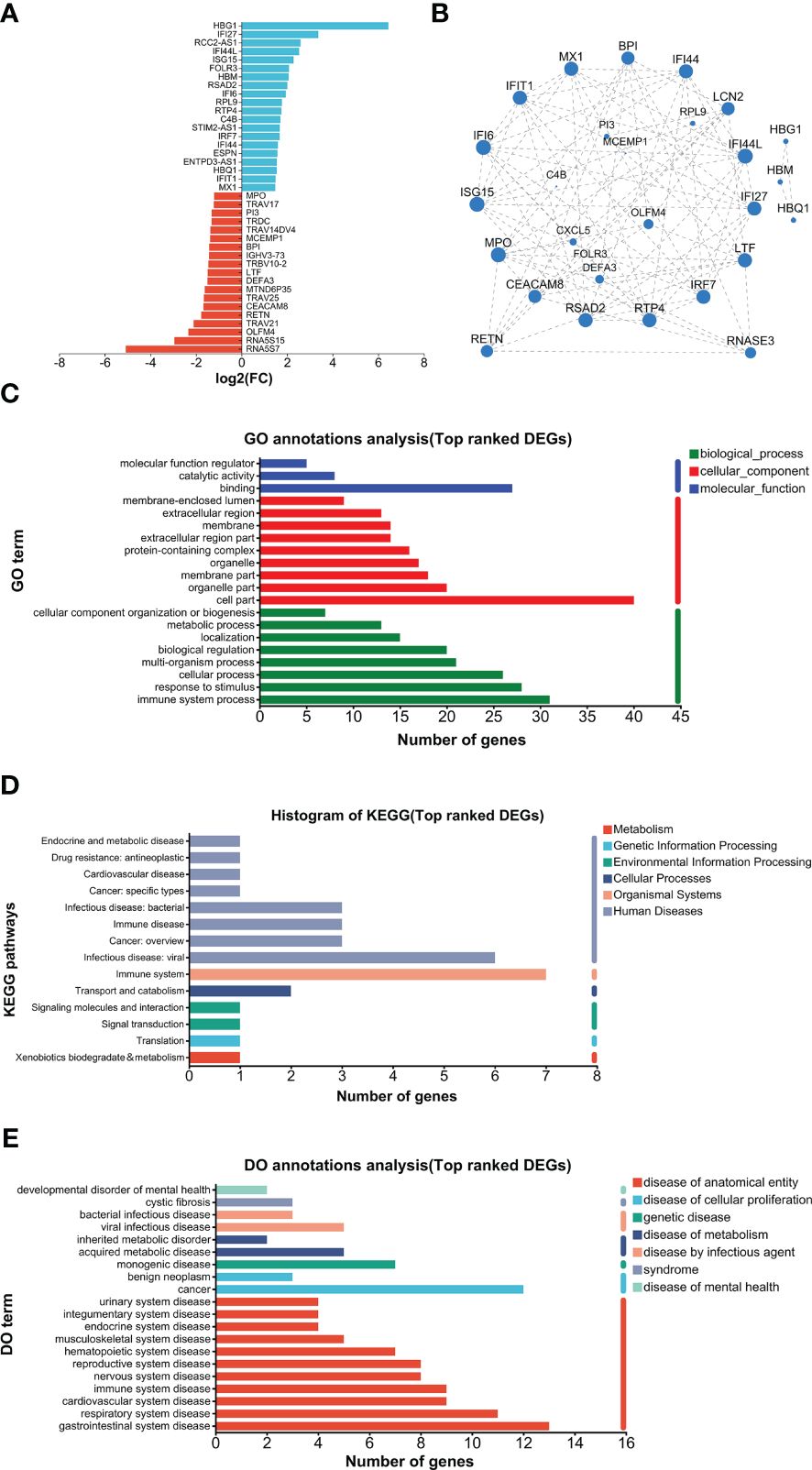
Figure 5. Analysis of top-ranked DEGs. (A) List of top-ranked DEGs. (B) PPI analysis. (C) GO analysis. (D) KEGG analysis. (E) DO analysis.
HZ-specific expression
As shown in Figure 6A, GO analysis found that RNA polymerase binding and T-cell receptor complex were the top annotations for molecular function and cellular components, respectively. For biological processes, snRNA transcription by RNA polymerase II, snRNA transcription, snRNA metabolic process, ncRNA transcription, and regulation of cilium assembly were the top annotations (Figure 6A). KEGG analysis found that base excision repair, antifolate resistance, pantothenate and CoA biosynthesis, and apoptosis (multiple species) were highly enriched (Figure 6B). DO analysis found that hereditary multiple exostoses were the top enriched disease (Figure 6C). The top 20 genes are listed in Figure 6D. HZ-specific expression is listed in Supplementary File 1.
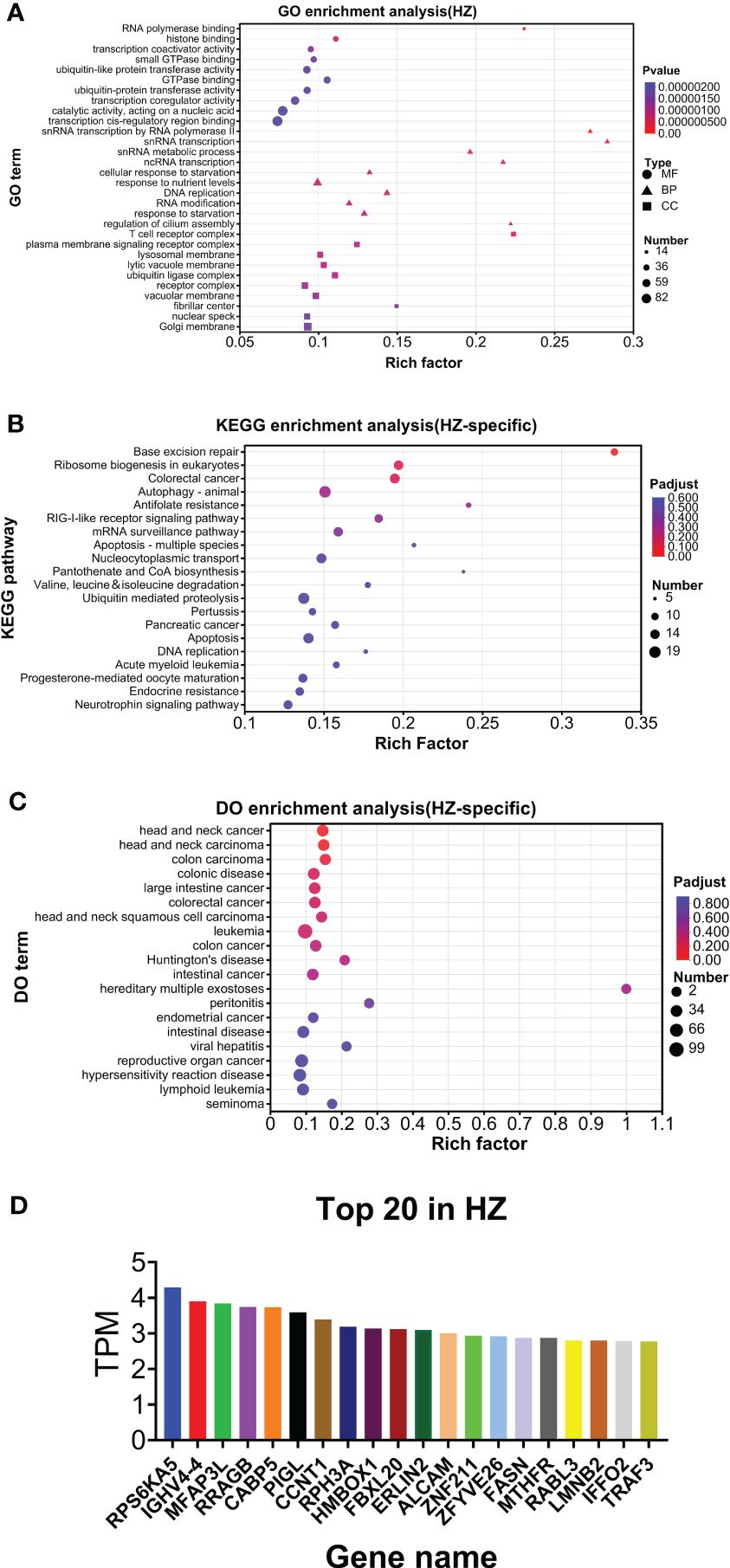
Figure 6. Analysis of HZ-specific expression. (A) GO analysis. (B) KEGG analysis. (C) DO analysis. (D) Top 20 expressed genes.
CIBERSORT
CIBERSORT analysis was conducted for DEGs and HZ-specific expression (Figure 7). Due to the small number of expressed genes, CIBERSORT was not conducted for PHN-specific expression.
CIBERSORT analysis for the UP DEG group was compared to the DOWN DEG group (Figures 7A, B). Among the 7 top-ranked cell types of the UP DEG group, neutrophils and naive CD4+ T cells were selective for the UP but not the DOWN group. The other 5 types of cells were in both UP and DOWN DEG groups including Treg, activated NK cell, monocytes, resting NK cell, and T cells gamma delta (Tγδ). In addition, M0 macrophages and resting memory CD4+ T cells were selectively ranked in the top 7 cell types of the DOWN but not the UP DEG group.
CIBERSORT analysis for HZ-specific expression found that the top 7-ranked cell types were Treg, NK resting cells, activated mast cells, monocytes, resting mast cells, M0 macrophages, and M2 macrophages (Figure 7C). Compared to the top 7-ranked cells in UP and DOWN DEG groups, Treg, resting NK cells, and monocytes were listed in all three groups, M0 macrophages were shared by HZ-specific expression and DOWN DEG groups; activated mast cells, resting mast cells and M2 macrophages were selectively seen in the HZ-specific expression group.
qPCR validation
qPCR was conducted for 16 genes. Most of the PHN-specific genes and UP DEGs were significantly or insignificantly increased in PHN compared to HZ groups (Figure 8). On the other hand, DOWN DEGs and HZ-specific genes were insignificantly decreased or increased in PHN compared to HZ patients (Figure 8).
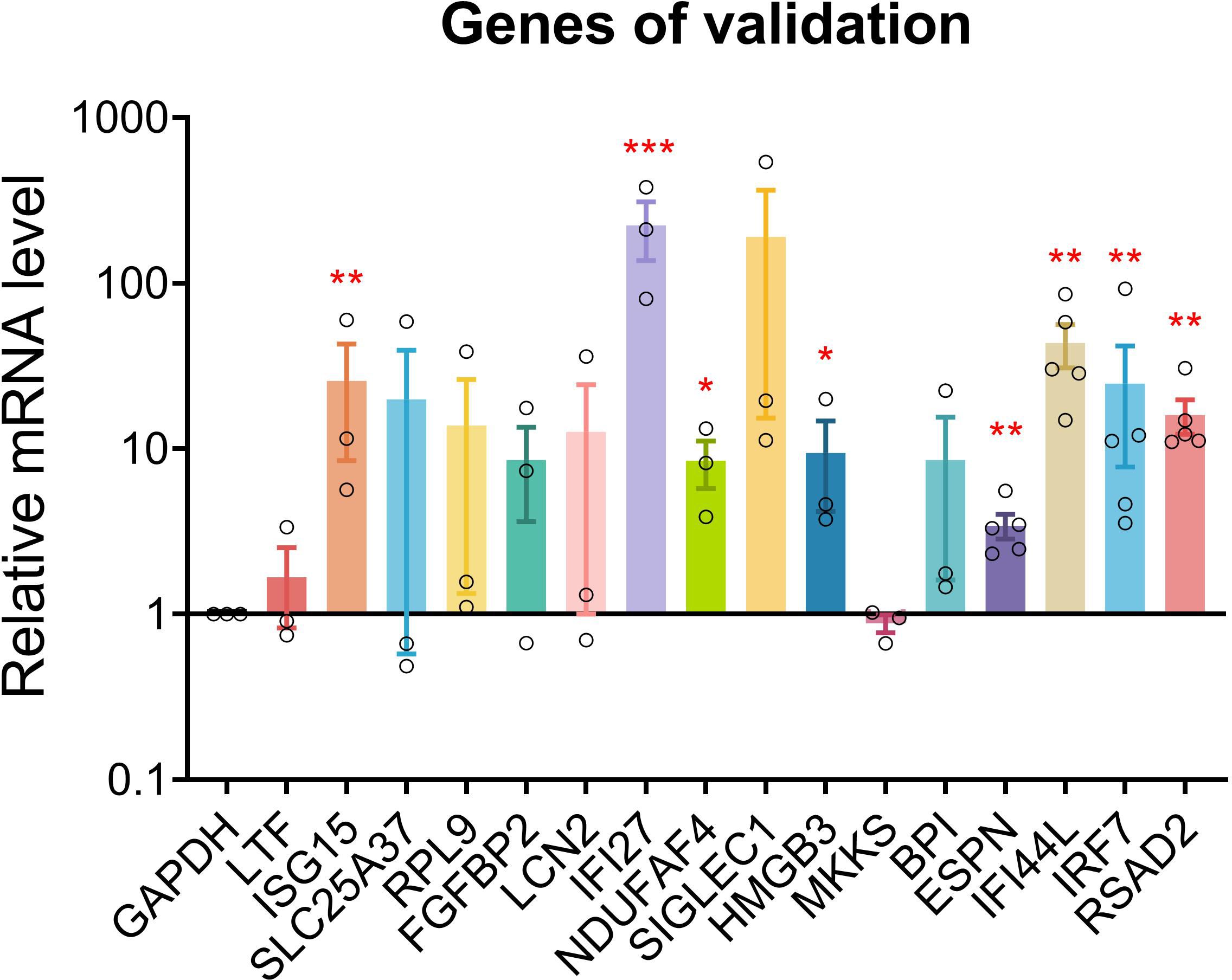
Figure 8. qPCR validation. qPCR validation was conducted for 17 genes between PHN and HZ groups. Data were expressed as mean ± SEM. *p < 0.05; **p < 0.01; ***, p < 0.001; Student’s t-test, PHN vs. HZ (n = 3–5).
Comparison between five PHN and HZ patients
The expression profiles of five PHN patients (located in the lower left corner of PCA in Figure 1) and 15 HZ patients were compared preliminarily. As shown in Supplementary Figure S3A, the distribution range of transcriptomic expression was similar between PHN and HZ patients. However, the average level of expression was slightly lower in PHN compared to HZ. Moreover, there were only 13 PHN-specific expressions compared to 3,276 HZ-specific expressions in the Venn plot (Supplementary Figure S3B). PCA analysis is shown in Supplementary Figure S3C. PHN-specific and HZ-specific transcripts were included in Supplementary File 2.
Among the 5,037 gene expressions detected in both PHN and HZ patients, there were 487 DEGs, including 224 upregulated DEGs (UP DEGs) and 263 downregulated DEGs (DOWN DEGs) (Supplementary Figure S4A). The volcano distribution of UP and DOWN DEGs is shown in Supplementary Figure S4B. GO, KEGG, and DO analysis is shown in Supplementary Figures S4C–E. All DEGs were listed in Supplementary File 2. The top 20 UP and DOWN DEGs are listed in Supplementary Figure S5A. PPI analysis is shown in Supplementary Figure S5B. GO, KEGG, and DO analyses of the top 20 UP and DOWN DEGs are shown in Supplementary Figures S5C–E. All DEGs are included in Supplementary File 2.
Due to the limited number of PHN-specific genes. Enrichment analysis of these genes was not conducted. For the HZ-specific genes, GO, KEGG, and DO analyses were conducted (Supplementary Figures S6A–C). The top 20 genes of HZ-specific group are shown in Supplementary Figure S6D. CIBERSORT analysis for UP and DOWN DEGs and for the HZ-specific genes is shown in Supplementary Figure S7.
Discussion
The overall transcriptomic data suggests that there is a reduced expression level in PHN compared to HZ in general (Figures 1A, B). Considering that HZ is a virus-induced inflammatory skin disease while PHN is free of zoster for months, it is not surprising that HZ would result in an upregulation of transcription related to systemic immunity and inflammation. In contrast, there are only 287 transcripts, including 82 PHN-specific expressions and 205 UP DEGs, compared to the 1,990 transcripts, which included 1,788 HZ-specific expressions and 202 DOWN DEGs (Figures 1B, 3A, B). As the 287 transcripts display an opposite way of change (increase in PHN) compared to the 1,990 transcripts (decrease in PHN), it would be more likely to associate these increased transcripts with the development of PHN from HZ. On the other hand, the decreased transcripts could largely result from the disappearance of rash and the resolution of acute inflammation. Moreover, the evolution of HZ to PHN could be due to the progression of the diseases or to certain genetic characteristics that predispose some individuals to PHN. Considering the HZ patients develop much less severe pain 3 months later compared to the PHN patients in the current study, our analysis of genes increased in PHN may reflect the influence of a combination of both disease progression and genetic disposition on the development of PHN from HZ.
The PCA analysis does not separate the two patient groups in the current study (Figure 1C). It suggests that the major components of transcriptomic are not different between the PHN and HZ groups. This mixed distribution pattern could be due to the fact that both PHN and HZ involve the same ganglion neurons, similar dermatomes, and relatively small skin areas. However, in the heatmap of DEGs, half of the PHN patients are located side by side (Figure 3C). Moreover, these four clustered PHN patients, plus another PHN patient who is nearest to them in the heatmap, are all located in the same quarter of the PCA plot (down and left quarter) (Figures 1C, 3C). These results suggest that there is an intrinsic homogeneity in transcriptome among PHN patients that enables them to be partially separated from the transcription pattern of HZ patients in the heatmap. On the other hand, the other three PHN patients are located in the other three quarters of the PCA plot, respectively. It may be related to the disease and demographic characteristics of PHN patients (Figure 1C; Table 1).
Enrichment analysis data suggest that PHN-specific expression participates in the lipid and glycan metabolism processes (Figures 2B, C). In addition, nucleotide metabolism and metabolic diseases are associated with UP DEGs (Supplementary Figure S1B). These results suggest that increased expression of genes in lipid, glycan, and nucleotide metabolism might be associated with and/or contribute to the development of PHN from HZ. A recent plasma metabolomic study suggests that there is an increase in amino acid metabolites in PHN compared to HZ patients (Zhou et al., 2022). Moreover, lipid metabolites are increased in PHN men, while amino acid metabolites are more pronouncedly increased in PHN women compared to their respective HZ counterparts. Therefore, increased metabolism overall, or in some category, might be associated with and/or contribute to the development of PHN from HZ overall or in some subpopulations of patients.
Several top-ranked PHN-specific genes encode proinflammatory molecules, including HMGB3, ELOVL6, TMIGD3, and Siglec-1 (Figure 2D). For example, Siglec-1 is a member of sialic acid-binding lectins of the Ig superfamily (Siglec) expressed in a subset of macrophages. On the one hand, Siglec-1 is involved in the virus infection (Mikulak et al., 2017; Perez-Zsolt et al., 2022). On the other hand, Siglec-1 mediates crosstalk between macrophages and other immune cells such as neutrophils, dendritic cells, and Tregs (Crocker et al., 1995 (Wu et al., 2009; Perez-Zsolt et al., 2022). Particularly, the Siglec-1-mediated interaction between macrophage and Tregs promotes inflammation through suppressing Tregs (Wu et al., 2021). This mechanism is associated with autoimmune diseases of the nervous system such as experimental autoimmune uveitis, experimental autoimmune encephalomyelitis, and neuronal ceroid lipofuscinoses (Jiang et al., 2006; Wu et al., 2009; Groh et al., 2016). These results might suggest that increased activation of Siglec-1 is associated with or contributes to the development of PHN from HZ.
A majority of the TOP UP DEGs are related to type I IFN signaling, including IFI27, IFI44L, ISG15, RSAD2, IFI6, IRF7, IFI44, and IFIT1. It has been suggested that type I IFN induces headache and flu-like pain symptoms in multiple sclerosis patients (Goldschmidt and Hua, 2020). Moreover, type I IFN therapy increases somatic pain scores in a subpopulation of chronic hepatitis C virus (HCV) patients who also develop depression following IFN treatment (Lin et al., 2020). A number of animal studies have been conducted to investigate type I IFN signaling of DRG neurons in chronic pain conditions (Donnelly et al., 2021; Wang et al., 2021). The majority of these studies suggest that upregulation of type I IFN signaling results in the sensitization of DRG neurons and chronic pain (Fitzgibbon et al., 2019; Sun et al., 2022; Wong et al., 2023; Zhang et al., 2023). Particularly, a recent study suggests that type I IFN directly acts on nociceptors to induce pain sensitization (Barragan-Iglesias et al., 2020). Moreover, the chemotherapy drug vinorelbine may induce pain by sequential activation of STING, pIRF3, type I IFN, mitogen-activated protein kinase interacting kinase (MNK), and eIF4E phosphorylation in DRG neuron (Franco-Enzastiga et al., 2024). Overall, our results suggest that activation of type I IFN signaling might play an important role in the development of PHN from HZ, possibly through effects on VZV-reactivated DRG neurons.
Six of the top 20 DOWN DEGs belong to T-cell receptors, including TRAV21, TRAV25, TRBV10-2, TRAV14DV4, TRDC, and TRAV17 (Figure 5A). Moreover, the T-cell receptor complex is a top annotation in GO analysis of HZ-specific expression. Multiple studies have investigated T cells in HZ patients. An earlier study has found no change in CD8+ T cells and a decrease in CD4+ T cells in HZ compared to control patients (Xing et al., 2013). In contrast, a recent study has found a transient increase of CD4+ and CD8+ T cells and B cells in the HZ stage (Peng et al., 2022). In addition, there is a significant decrease in CD4+ T cells in PHN compared to non-PHN groups (Peng et al., 2022). Partially similar to and different from each of the above two studies, another recent study has reported an increase in CD8+ T cells and a decrease in CD4+ T cells in HZ compared to control patients (Chen et al., 2023). These results might suggest that CD8+ T cells may be activated in the HZ stage compared to healthy controls. On the other hand, CD4+ T cells could be decreased in the HZ (vs. healthy control) and PHN (vs. non-PHN) stages. Moreover, our results suggest that there is an overall decrease in T cell activation in PHN compared to the HZ stage.
In addition to T-cell receptors, five of the top 20 DOWN DEGs are related to neutrophils, including DEFA3, LTF, BPI, MPO, and LCN2 (Figure 5A). In contrast, CIBERSORT analysis suggests that neutrophils are selectively associated with UP but not DOWN DEGs (Figures 7A–C). These results might suggest that the expression profile of neutrophils is altered in PHN compared to HZ stages. A recent study suggests that transient activation of neutrophils during the acute inflammation stage protects against the development of chronic low back pain (Parisien et al., 2022). Considering neutrophil infiltration is a common mechanism of chronic pain (Yang et al., 2022; Zhao et al., 2023), our study might suggest that persisted but altered activation of neutrophils contributes to the development of PHN from HZ.
On top of neutrophils, M1 and M2 macrophages are selectively associated with UP, while M0 macrophages are selectively associated with DOWN DEGs (Figures 7A, B). These results appear to suggest a transition from resting to activated states of macrophages from HZ to PHN. However, all M0, M1, and M2 macrophages are associated with HZ-specific genes (Figure 7C). Therefore, it might be suggested that there is a disappearance of M0 macrophages from HZ to PHN. Interestingly, the ratio of M1/M2 is higher in UP DEGs compared to the PHN-specific group (Figures 7A, C). As M1 macrophages are proinflammatory and cause pain, while M2 macrophages are anti-inflammatory and cause pain in general, our results might suggest that a higher M1/M2 ratio of macrophages contributes to the development of PHN in HZ.
Ramsay Hunt syndrome (RHS) is an acute peripheral facial neuropathy that occurs as a complication of shingles. It has been reported that patients with RHS who have an elevated NLR have poor outcomes, including a higher degree of paralysis and a lower probability of complete recovery (Soh et al., 2019). NLR is an independent risk factor for herpes zoster infection in patients with rheumatic diseases (Mok et al., 2023). Preoperative NLR is significantly associated with HZ and PHN in living donor liver transplantation recipients (Sim et al., 2021). In patients with HZ and malignancy, a higher NLR is associated with fatal outcomes (Yamamoto and Aoyama, 2023). These results suggest a higher NLR is associated with a higher frequency and worse outcomes of HZ or HZ-associated diseases. Our results (insignificant decrease of NLR in PHN) do not suggest whether or not a higher NLR may be critical in the development of PHN from HZ.
In addition to neutrophils, CIBERSORT analysis suggests that UP DEGs are associated with higher activated NK cells and a higher ratio of activated/resting NK cells compared to DOWN DEGs or HZ-specific groups (Figures 7A–C). Increased NK cells have been reported in HZ patients compared to healthy controls (Xing et al., 2013; Peng et al., 2022). However, NK cells are not different between PHN and non-PHN patients (Peng et al., 2022). Our results suggest that activated NK cells are increased in PHN compared to HZ stages. It has been reported that NK cells are negatively associated with mechanical pain sensitivity in chronic peripheral neuropathic pain conditions (Lassen et al., 2021). NK cells may help restore normal sensory function and reduce pain by eliminating miswired nerve endings in peripheral sensory neurons (Kim et al., 2023). Therefore, our results might suggest that abnormal axon degeneration/regeneration of DRG neurons contributes to the development of PHN in HZ.
There are several limitations to the current study. First, due to the lack of healthy controls and non-PHN patients (who had HZ previously), the current study cannot address how HZ develops from a healthy control or what differentiates PHN from non-PHN. Second, given the lack of a valid animal model for PHN (VZV selectively infects human cells), no animal experiments on effects and mechanisms were conducted. Finally, the study is limited by the small sample size and the presence of multiple chronic complications in the patients.
By comparing the blood transcriptomic signature of PHN to that of HZ patients, the current study suggests metabolism of lipid, glycan, and nucleotides, type I IFN signaling, and altered activation of neutrophils are associated with and potentially contribute to the development of PHN from HZ.
Data availability statement
The sequence data presented in the study are deposited in the NCBI Sequence Read Archive database, accession number PRJNA1142765.
Ethics statement
The studies involving humans were approved by Ethics Committee of Affiliated Hospital at Hebei University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
CW: Conceptualization, Data curation, Investigation, Methodology, Writing – review & editing. KZ: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. YB: Data curation, Investigation, Methodology, Validation, Writing – review & editing. YL: Formal analysis, Writing – review & editing. YZ: Conceptualization, Writing – review & editing. Y-HJ: Conceptualization, Writing – review & editing. HW: Conceptualization, Investigation, Supervision, Writing – review & editing. Z-YT: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Z-YT was supported by Natural Science Foundation of Hebei Province (C2023201035).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1425393/full#supplementary-material
Abbreviations
DEG, differential expression gene; DO, disease ontology; DOWN DEG, downregulated DEG; GO, gene ontology; HZ, herpes zoster; IFN, type I interferon; NLR, neutrophil-to-lymphocyte ratio; ORF, open reading frame; PCA, principal component analysis; PHN, postherpetic neuralgia; PPI, protein-protein interaction; Treg, regulatory T cell; Tγδ, T-cell gamma delta; UP DEG, upregulated DEG; VAS, Visual Analog Scale.
References
Barragan-Iglesias, P., Franco-Enzastiga, U., Jeevakumar, V., Shiers, S., Wangzhou, A., Granados-Soto, V., et al. (2020). Type I interferons act directly on nociceptors to produce pain sensitization: Implications for viral infection-induced pain. J. Neurosci. 40, 3517–3532. doi: 10.1523/JNEUROSCI.3055-19.2020
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–ii90. doi: 10.1093/bioinformatics/bty560
Chen, W., Zhu, L., Shen, L. L., Si, S. Y., Liu, J. L. (2023). T lymphocyte subsets profile and toll-like receptors responses in patients with herpes zoster. J. Pain Res. 16, 1581–1594. doi: 10.2147/JPR.S405157
Crocker, P. R., Freeman, S., Gordon, S., Kelm, S. (1995). Sialoadhesin binds preferentially to cells of the granulocytic lineage. J Clin Invest. 95, 645–643. doi: 10.1172/JCI117708
Curran, D., Callegaro, A., Fahrbach, K., Neupane, B., Vroling, H., van Oorschot, D., et al. (2022). Meta-regression of herpes zoster incidence worldwide. Infect. Dis. Ther. 11, 389–403. doi: 10.1007/s40121-021-00567-8
Delaney, A., Colvin, L. A., Fallon, M. T., Dalziel, R. G., Mitchell, R., Fleetwood-Walker, S. M. (2009). Postherpetic neuralgia: from preclinical models to the clinic. Neurotherapeutics 6, 630–637. doi: 10.1016/j.nurt.2009.07.005
Donnelly, C. R., Jiang, C., Andriessen, A. S., Wang, K., Wang, Z., Ding, H., et al. (2021). STING controls nociception via type I interferon signalling in sensory neurons. Nature 591, 275–280. doi: 10.1038/s41586-020-03151-1
Fitzgibbon, M., Kerr, D. M., Henry, R. J., Finn, D. P., Roche, M. (2019). Endocannabinoid modulation of inflammatory hyperalgesia in the IFN-alpha mouse model of depression. Brain Behav. Immun. 82, 372–381. doi: 10.1016/j.bbi.2019.09.006
Franco-Enzastiga, U., Natarajan, K., David, E. T., Patel, K., Ravirala, A., Price, T. J. (2024). Vinorelbine causes a neuropathic pain-like state in mice via STING and MNK1 signaling associated with type I interferon induction. iScience 27, 108808. doi: 10.1016/j.isci.2024.108808
Goldschmidt, C. H., Hua, L. H. (2020). Re-evaluating the use of IFN-beta and relapsing multiple sclerosis: safety, efficacy and place in therapy. Degener Neurol. Neuromuscul Dis. 10, 29–38. doi: 10.2147/DNND.S224912
Gowrishankar, K., Steain, M., Cunningham, A. L., Rodriguez, M., Blumbergs, P., Slobedman, B., et al. (2010). Characterization of the host immune response in human Ganglia after herpes zoster. J. Virol. 84, 8861–8870. doi: 10.1128/JVI.01020-10
Groh, J., Ribechini, E., Stadler, D., Schilling, T., Lutz, M. B., Martini, R. (2016). Sialoadhesin promotes neuroinflammation-related disease progression in two mouse models of CLN disease. Glia 64, 792–809. doi: 10.1002/glia.22962
Jiang, H. R., Hwenda, L., Makinen, K., Oetke, C., Crocker, P. R., Forrester, J. V. (2006). Sialoadhesin promotes the inflammatory response in experimental autoimmune uveoretinitis. J. Immunol. 177, 2258–2264. doi: 10.4049/jimmunol.177.4.2258
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, H. W., Wang, S., Davies, A. J., Oh, S. B. (2023). The therapeutic potential of natural killer cells in neuropathic pain. Trends Neurosci. 46, 617–627. doi: 10.1016/j.tins.2023.05.008
Lassen, J., Sturner, K. H., Gierthmuhlen, J., Dargvainiene, J., Kixmuller, D., Leypoldt, F., et al. (2021). Protective role of natural killer cells in neuropathic pain conditions. Pain 162, 2366–2375. doi: 10.1097/j.pain.0000000000002274
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Lin, C. Y., Guu, T. W., Lai, H. C., Peng, C. Y., Chiang, J. Y., Chen, H.T., et al. (2020). Somatic pain associated with initiation of interferon-alpha (IFN-alpha) plus ribavirin (RBV) therapy in chronic HCV patients: A prospective study. Brain Behav. Immun. Health 2, 100035. doi: 10.1016/j.bbih.2019.100035
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mercan, A., Uzun, S. T., Keles, S., Hacibeyoglu, G., Yilmaz, R., Reisli, R. (2021). Immunological mechanism of postherpetic neuralgia and effect of pregabalin treatment on the mechanism: a prospective single-arm observational study. Korean J. Pain 34, 463–470. doi: 10.3344/kjp.2021.34.4.463
Mikulak, J., Di Vito, C., Zaghi, E., Mavilio, D. (2017). Host immune Responses in Hiv-1 infection: The emerging Pathogenic Role of Siglecs and Their Clinical Correlates. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00314
Mok, C. C., Ho, L. Y., Tse, S. M., Chan, K. L., To, C. H. (2023). Prevalence and risk factors of herpes zoster infection in patients with rheumatic diseases not receiving biologic or targeted therapies. Clin. Rheumatol 42, 1019–1026. doi: 10.1007/s10067-022-06450-2
Pan, C. X., Lee, M. S., Nambudiri, V. E. (2022). Global herpes zoster incidence, burden of disease, and vaccine availability: a narrative review. Ther. Adv. Vaccines Immunother. 10, 25151355221084535. doi: 10.1177/25151355221084535
Parisien, M., Lima, L. V., Dagostino, C., El-Hachem, N., Drury, G. L., Grant, A.V., et al. (2022). Acute inflammatory response via neutrophil activation protects against the development of chronic pain. Sci. Transl. Med. 14, eabj9954. doi: 10.1126/scitranslmed.abj9954
Peng, Q., Guo, X., Luo, Y., Wang, G., Zhong, L., Zhu, J., et al. (2022). Dynamic immune landscape and VZV-specific T cell responses in patients with herpes zoster and postherpetic neuralgia. Front. Immunol. 13, 887892. doi: 10.3389/fimmu.2022.887892
Perez-Zsolt, D., Muñoz-Basagoiti, J., Rodon, J., Elosua-Bayes, M., Raïch-Regué, D., Risco, C., et al. (2022). SARS-CoV-2 interaction with Siglec-1 mediates -infection by dendritic cells (Nov, 10.1038/s41423-022-00893-y 2022). Cell. Mol. Immunol. 19, 965–966. doi: 10.1038/s41423-022-00893-y
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Saguil, A., Kane, S., Mercado, M., Lauters, R. (2017). Herpes zoster and postherpetic neuralgia: Prevention and management. Am. Fam Physician 96, 656–663.
Sampathkumar, P., Drage, L. A., Martin, D. P. (2009). Herpes zoster (shingles) and postherpetic neuralgia. Mayo Clin. Proc. 84, 274–280. doi: 10.4065/84.3.274
Sim, J. H., Moon, Y. J., Kim, S. H., Kim, K. S., Lee, J. S., Song, J. G., et al. (2021). Association between neutrophil-lymphocyte ratio and herpes zoster infection in 1688 living donor liver transplantation recipients at a large single center. Biomedicines 9, 963. doi: 10.3390/biomedicines9080963
Soh, Y., Lee, J. H., Jung, J., Dong, S. H., Byun, J. Y., Park, M. S., et al. (2019). A high neutrophil-to-lymphocyte ratio is associated with recovery from ramsay hunt syndrome. ORL J. Otorhinolaryngol Relat. Spec 81, 130–137. doi: 10.1159/000499471
Sun, C., Wu, G., Zhang, Z., Cao, R., Cui, S. (2022). Protein tyrosine phosphatase receptor type D regulates neuropathic pain after nerve injury via the STING-IFN-I pathway. Front. Mol. Neurosci. 15, 859166. doi: 10.3389/fnmol.2022.859166
van Oorschot, D., Vroling, H., Bunge, E., Diaz-Decaro, J., Curran, D., Yawn, B. (2021). A systematic literature review of herpes zoster incidence worldwide. Hum. Vaccin Immunother. 17, 1714–1732. doi: 10.1080/21645515.2020.1847582
Wang, K., Donnelly, C. R., Jiang, C., Liao, Y., Luo, X., Tao, X., et al. (2021). STING suppresses bone cancer pain via immune and neuronal modulation. Nat. Commun. 12, 4558. doi: 10.1038/s41467-021-24867-2
Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wong, C., Tavares-Ferreira, D., Thorn Perez, C., Sharif, B., Uttam, S., Amiri, M., et al. (2023). 4E-BP1-dependent translation in nociceptors controls mechanical hypersensitivity via TRIM32/type I interferon signaling. Sci. Adv. 9, eadh9603. doi: 10.1126/sciadv.adh9603
Wu, C., Rauch, U., Korpos, E., Song, J., Loser, K., Crocker, P. R., et al. (2009). Sialoadhesin-positive macrophages bind regulatory T cells, negatively controlling their expansion and autoimmune disease progression. J. Immunol. 182, 6508–6516. doi: 10.4049/jimmunol.0804247
Wu, G., Murugesan, G., Nagala, M., McCraw, A., Haslam, S. M., Dell, A., et al (1995). Activation of regulatory T cells triggers specific changes in glycosylation associated with Siglec-1-dependent inflammatory responses. Wellcome Open Res. 6, 134. doi: 10.12688/wellcomeopenres
Xing, Q., Hu, D., Shi, F., Chen, F. (2013). Role of regulatory T cells in patients with acute herpes zoster and relationship to postherpetic neuralgia. Arch. Dermatol. Res. 305, 715–722. doi: 10.1007/s00403-013-1367-0
Yamamoto, T., Aoyama, Y. (2023). Risk factor-specific mortality in herpes zoster in a retrospective cohort study: The neutrophil-to-lymphocyte ratio is a useful biomarker for predicting late-onset complications resulting in death. J. Eur. Acad. Dermatol. Venereol 37, e191–ee93. doi: 10.1111/jdv.18602
Yang, J. X., Wang, H. F., Chen, J. Z., Li, H. Y., Hu, J. C., Yu, A. A., et al. (2022). Potential neuroimmune interaction in chronic pain: A review on immune cells in peripheral and central sensitization. Front. Pain Res. (Lausanne) 3, 946846. doi: 10.3389/fpain.2022.946846
Zhang, Y., Wang, W., Gong, Z., Peng, Y., Li, X., Zhang, Z., et al. (2023). Activation of the STING pathway induces peripheral sensitization via neuroinflammation in a rat model of bone cancer pain. Inflammation Res. 72, 117–132. doi: 10.1007/s00011-022-01663-2
Zhao, J., Huh, Y., Bortsov, A., Diatchenko, L., Ji, R. R. (2023). Immunotherapies in chronic pain through modulation of neuroimmune interactions. Pharmacol. Ther. 248, 108476. doi: 10.1016/j.pharmthera.2023.108476
Zhou, R., Li, J., Zhang, Y., Xiao, H., Zuo, Y., Ye, L. (2022). Characterization of plasma metabolites and proteins in patients with herpetic neuralgia and development of machine learning predictive models based on metabolomic profiling. Front. Mol. Neurosci. 15, 1009677. doi: 10.3389/fnmol.2022.1009677
Keywords: postherpetic neuralgia, herpes zoster, RNA-Seq, blood transcriptome, patient, differential expression gene
Citation: Wang C, Zhang K, Bao Y, Liu Y, Zhou Y, Ji Y-H, Wang H and Tan Z-Y (2024) Exploring blood transcriptomic signatures in patients with herpes zoster and postherpetic neuralgia. Front. Cell. Infect. Microbiol. 14:1425393. doi: 10.3389/fcimb.2024.1425393
Received: 29 April 2024; Accepted: 19 July 2024;
Published: 15 August 2024.
Edited by:
Cui Hao, The Affiliated Hospital of Qingdao University, ChinaReviewed by:
Xin Zhang, Duke University, United StatesQian Bai, Second Affiliated Hospital of Zhengzhou University, China
Copyright © 2024 Wang, Zhang, Bao, Liu, Zhou, Ji, Wang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Yong Tan, emhpeW9uZ3Rhbl9oYnVAMTYzLmNvbQ==; Hongjie Wang, aG9uZ2ppZXc2OEAxMjYuY29t
†These authors have contributed equally to this work
 Chunliang Wang1†
Chunliang Wang1† Kaiyi Zhang
Kaiyi Zhang You Zhou
You Zhou Yong-Hua Ji
Yong-Hua Ji Hongjie Wang
Hongjie Wang Zhi-Yong Tan
Zhi-Yong Tan