- 1College of Veterinary Medicine, Yunnan Agricultural University, Kunming, China
- 2Faculty of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
Streptococcus agalactiae (Strep. agalactiae) is bovine mastitis pathogen and has thus became a matter of concern to dairy farms worldwide in terms of economic loss. The aims of this study were to (a) determine virulence genes, and (b) characterize the antimicrobial resistance (AMR) profiles and AMR genes and (c) figure out the relationship between AMR phenotypes and genotypes of Strep. agalactiae isolated from dairy cows in north China. A total of 20 virulence genes and 23 AMR genes of 140 isolates collected from 12 farms in six provinces were studied. The antimicrobial susceptibility of 10 veterinary commonly used antimicrobials were tested using the broth microdilution method. Results showed that all the isolates harbored the virulence genes lacIV, gapC, and dltA. The isolates that harbored the genes lacIII, fbsA, hylB, and cfb exhibited the high prevalence (99.29%), followed by isolates that harbored lacI (98.57%), bibA (97.86%), cylE (97.14%), lacII (92.14%), cspA (52.14%), pavA (25%), bca (2.14%), and scpB (0.71%). The fbsB, lmb, spbI, bac, and rib genes were not detected. The virulence patterns of B (fbsA_cfb_cylE_ hylB_bibA_cspA_ gapC_dltA_lacIII/IV) and C (fbsA_cfb_ bibA _ gapC_ dltA_lacIV) were dominant, accounting for 97.86% of the isolates. The following AMR genes were prevalent: pbp1A (97.14%), tet(M) (95.00%), lnu (A) (80.71%), erm (B) (75.00%), tet(O) (72.14%), blaZ (49.29%), tet(S) (29.29%), blaTEM (25.71%), erm (A) (17.14%), erm (C) (13.57%), tet (L) (10.71%), linB (2.86%), and erm (TR) (2.86%). The pbp2b, mecA1, mecC, lnu (D), erm (F/G/Q), and mef (A) genes were not detected. Eighty percent of the isolates harbored AMR genes and were highly resistant to tetracycline, followed by macrolides (10.71%), lincosamides (9.29%) and β-lactams (4.29%). In conclusion, isolates only exhibited well correlation between tetracyclines resistance phenotype and genotype, and almost all isolates harbored intact combination of virulence genes.
Introduction
Bovine mastitis is one of the costliest diseases in the dairy industry due to the discarding of milk, costs of treatments, and even the culling of cows (Shaheen et al., 2016; Krömker and Leimbach, 2017; Gussmann et al., 2019). Streptococcus agalactiae (Strep. agalactiae), as one of the major mastitis pathogens, causing 11%–60% of mastitis cases in Brazil (Reyes et al., 2017). The implementation of the five-point mastitis control program has reduced the prevalence of Strep. agalactiae mastitis to less than 10% in dairy herds in Europe and North America (Jørgensen et al., 2016). However, the herd prevalence of Strep. agalactiae is still high in developing countries, such as Colombia (34.4%), Brazil (60%), and China (92%) (Ramírez et al., 2014; Bi et al., 2016; Carvalho-Castro et al., 2017). Meanwhile, the re-emergence of Strep. agalactiae mastitis in Denmark and Norway has been reported (Katholm et al., 2012; Jørgensen et al., 2016). Moreover, the harm caused by Streptococcus agalactiae to China’s dairy farming industry is still very serious (Yang et al., 2016).
Strep. agalactiae is considered one of contagious pathogens that cause bovine mastitis, which can spread among cows (Thompson-Crispi et al., 2014). Once Strep. agalactiae colonize the bovine mammary gland, it obtains nutrient sources from milk for its proliferation and causes long-term and harmful effects. Therefore, the ability of metabolism and capability for adhesion, invasion, and immune evasion of Strep. agalactiae might play crucial roles in the bovine mastitis (Keefe, 2012). Strep. agalactiae harbors a great range of virulence genes encoding virulence factors, such as fbsA/B and lmb, which are involved in adhesion, cylE and hylB, which are involved in invasion, cspA, which is involved in immune evasion, and LacI/II/III/IV, which play a role in metabolism.
Antimicrobial treatment is major option for treating Strep. agalactiae inducing mastitis (Keefe, 2012). However, the excessive use of antimicrobials increased the risks of antimicrobial resistance (AMR), which is a public health concern worldwide (Flynn and Guarner, 2023). Monitoring the resistance of Strep. agalactiae associated with bovine mastitis is important to the control of AMR of the bacterium.
Antimicrobial resistance genes, pbp1A, lnuA/D, tetO/M/L/S, ermA/B/C/F/G/Q/TR, and mefA, which are involved in resistance to β-lactams, lincosamide, tetracycline, and macrolide have been detected in Strep. agalactiae usually (Poyart et al., 2003; Dogan et al., 2005; Duarte et al., 2005), while AMR gene carrying status of the strains involved in this study is still unclear.
Investigations on virulence genes, and the phenotype and genotype of AMR can contribute to treatment decision and optimization of Strep. agalactiae control programs (Kaczorek et al., 2017). This study aims 1) to determine the antimicrobial resistance and virulence gene profiles of Strep. agalactiae, 2) to detect the AMR profiles of Strep. agalactiae under in vitro conditions, and 3) to determine the correlation between phenotypic and genotypic resistance patterns of Strep. agalactiae isolated in China.
Materials and methods
Statement of ethics
All experiments followed the China Ministry of Science and Technology. Regulations of Experimental Animals (2008) issued by China Ministry of Science and Technology. All animal procedures were approved by the Institutional Animal Care and Use Committee of Yunnan Agricultural University (Approval No: 202403058).
Sample collection and identification of pathogens
Milk samples were collected from cows with clinical mastitis from large dairy farms (>500 cows) in China from 2017 to 2019 (Supplementary Table S1). Milk sampling details were provided by Gao et al. (2017). In brief, udders were disinfected before sample collection, the first three streams of milk were discarded, and 1–2 mL of quarter milk samples were aseptically collected using 50 mL sterile centrifuge tubes. The samples were packed in ice boxes and delivered to the laboratory to be processed within 10 h.
The quarter milk (200 uL) of each sample was coated on Edwards medium (Oxoid, USA) and incubated at 37°C for 24 h, and putative blue colonies without fermentation were enriched in 4 mL of Mueller–Hinton broth containing 5% fetal bovine serum. The putative isolates were identified through PCR using 16S rRNA amplification with the primer (5’-AGAGTTTGATCCTGGCTCAG-3’, 5’-CGGCTACCTTGTTACGACTT-3’) concentration of 5 μmol/L (Frank et al., 2008). The confirmed Strep. agalactiae isolates were stored at −80°C.
Virulence gene identification
Multiplex PCR was conducted three times for the examination of 13 virulence genes (cspA, pavA, cylE, hylB, lmb, fbsB, scpB, bca, pbp1A/ponA, bac, cfb, rib, and fbsA). The final volume of the multiplex PCR mixture was 25 µL, and the mixture contained the template composed of 1 μL (final amount of 20 ng) of bacterial genome, 12.5 μL of premixed 2×PCR master mix (Sangon, Shanghai, China), 1 μL of each primer (final concentration of 5 μmol/L), and ddH2O. The amplification program is provided in Supplementary Table S1, in detail, for SET 1 (cspA, pavA, cylE, hylB, lmb), the amplification program was as follows: 95°C for 5 min; 35 cycles of 95°C for 60 s; annealing temperature for 60 s; and 72°C for 10 min; for SET 2 (fbsB, scpB, bca), the amplification program was as follows: 95°C for 5 min; 35 cycles of 95°C for 60 s; annealing temperature for 60 s; and 72°C for 10 min; for SET 3 (pbp1A/ponA, bac, cfb, rib, and fbsA), the amplification program was as follows: 95°C for 5 min; 35 cycles of 95°C for 60 s; annealing temperature for 60 s; and 72°C for 10 min.
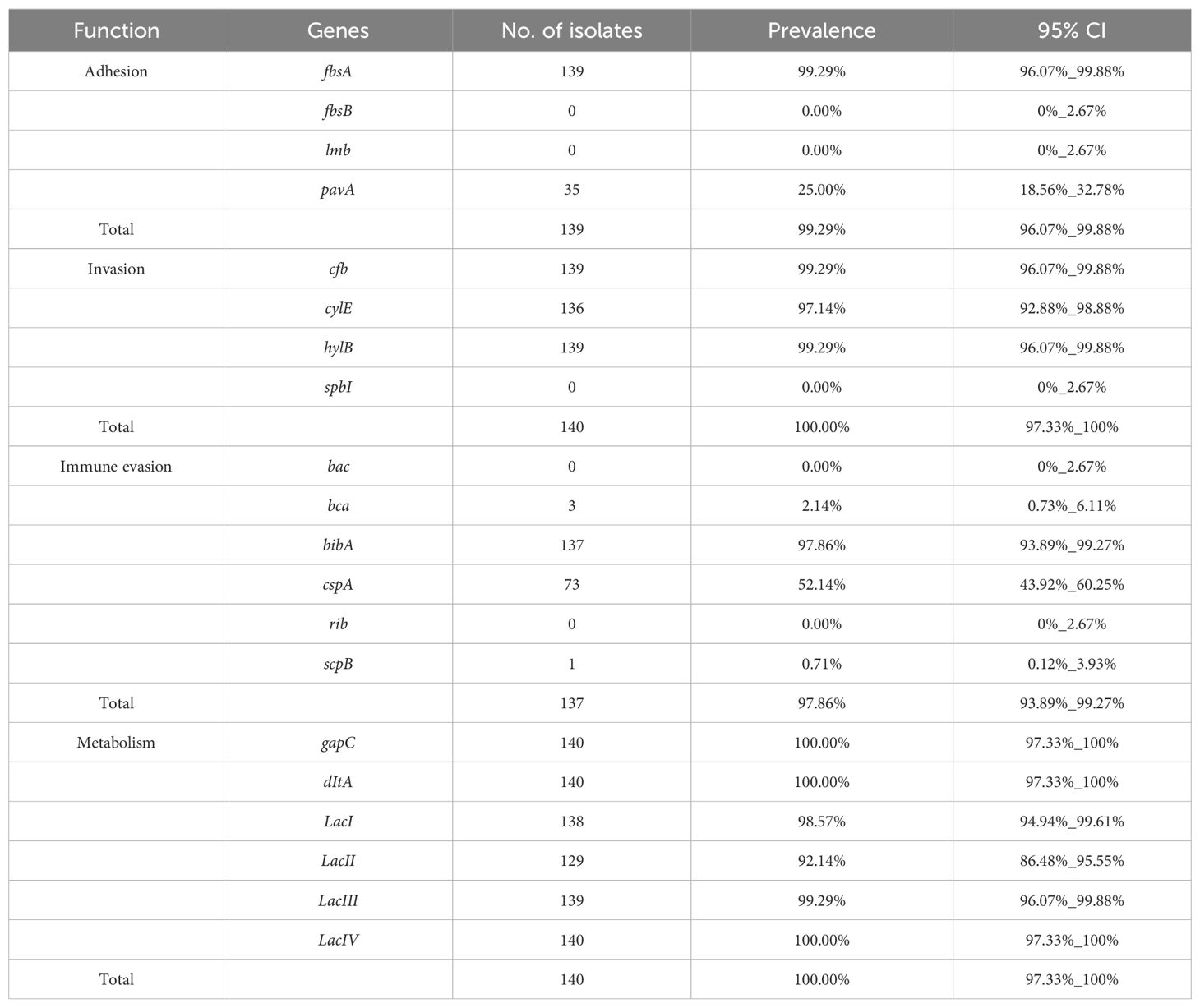
The rest of the virulence genes (spb1, dltA, bibA, gapC, and lacI/II/III/IV) were detected using normal PCR assay. The final volume of the PCR mixture was 25 µL, and the mixture contained the template composed of 1 μL (final amount of 20 ng) of bacterial genome, 12.5 μL of premixed 2×PCR master mix (Sangon, Shanghai, China), 1 μL of primers (final concentration of 5 μmol/L), and 10.5 μL of ddH2O. The amplification program was as follows: 95°C for 5 min; 35 cycles of 95°C for 60 s; annealing temperature for 60 s; and 72°C for 10 min (Supplementary Table S2). Strep. agalactiae ATCC 13813 was used as positive control and PCR mixture without bacterial genome was used as negative control. Agarose gel electrophoresis (AGE) and UV transillumination was conducted to analyze the PCR products (Supplementary Figure S1). The virulence genes were divided into four groups: adhesion(fbsA/B, lmb, pavA), invasion(cfb, cylE, hylB, spbI), immune evasion(bac, bca, bibA, cspA, rib, scpB), and metabolism(gapC, dltA, LacI/II/III/IV).
Antimicrobial resistance gene identification
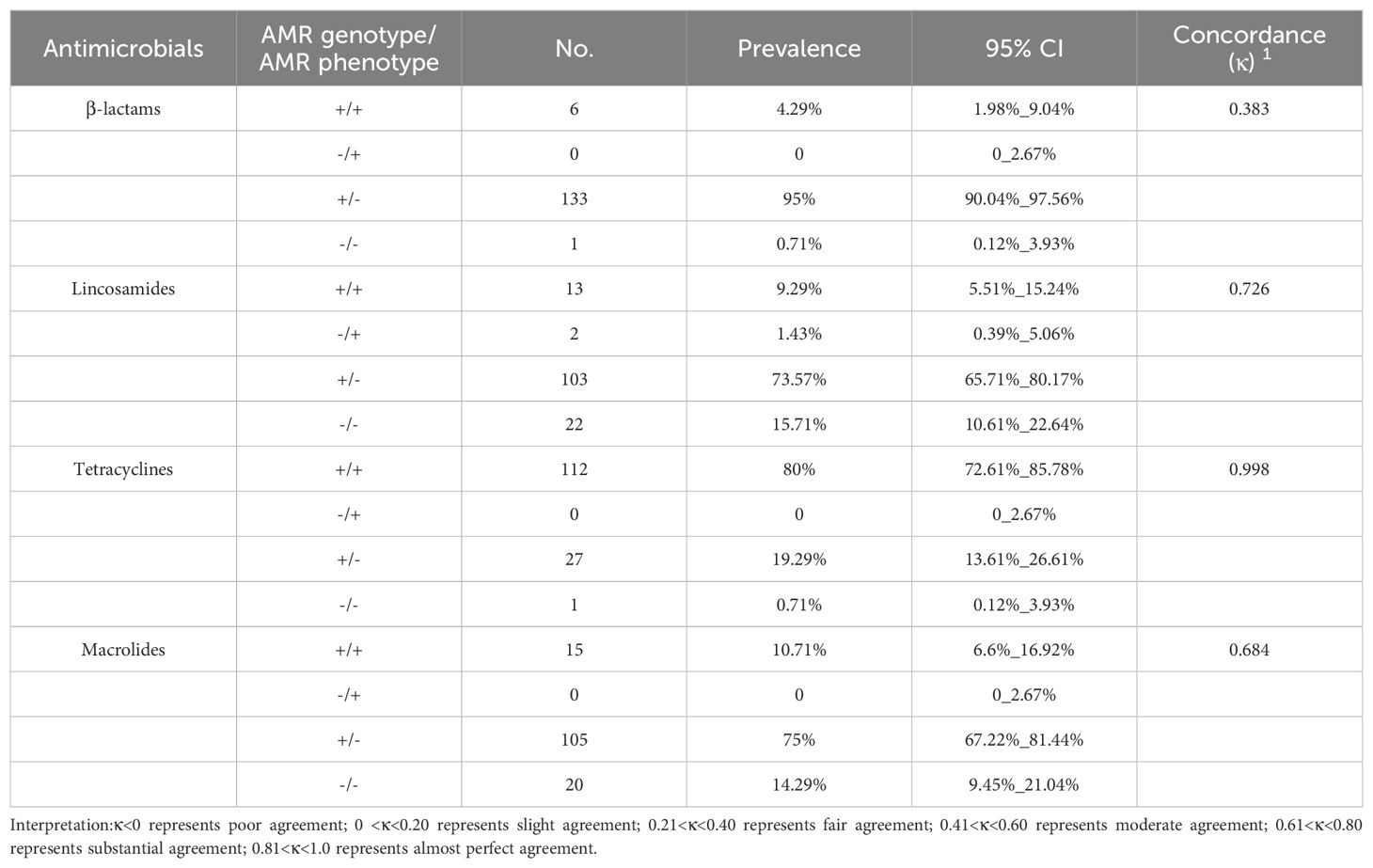
AMR genes associated with resistance to four kinds of antimicrobials: β_lactams (blaTEM, blaZ, pbp2b, mecA1, and mecC), lincosamides (lnuA, lnuD, and linB), tetracyclines (tetO/M/L/S), and macrolides (ermA/B/C/F/G/Q/TR, mefA) were detected using normal PCR assay. The final volume of the PCR mixture was 25 µL containing a template composed of 1 μL (final amount of 20 ng) of bacterial genome, 12.5 μL of premixed 2×PCR master mix (Sangon, Shanghai, China), 1 μL of primers (final concentration of 5 μmol/L), and 10.5 μL of ddH2O. The amplification program was as follows: 95°C for 5 min; 35 cycles of 95°C for 60 s; annealing temperature for 60 s; and 72°C for 10 min (Supplementary Table S3). Strep. agalactiae ATCC 13813 was used as positive control and PCR mixture without bacterial genome was used as negative control. Agarose gel electrophoresis (AGE) and UV transillumination was conducted to analyze the PCR products (Supplementary Figure S2).
Antimicrobial resistance testing
Antimicrobial resistance testing of all the isolates were conducted using the broth microdilution method according to Clinical and Laboratory Standards Institute (CLSI, 2020). Strep. pneumonia ATCC 49619 and Strep. agalactiae ATCC 13813 were used as quality control strains. Antimicrobials commonly used in practice for mastitis treatment and in medicines for humans (penicillin, cefalexin, ceftiofur, cefquinome, oxacillin, clindamycin, tetracycline, enrofloxacin, amoxicillin/clavulanate, and erythromycin) were selected for antimicrobial resistance testing.
Statistical analysis
The online statistical tool VassarStats (http://www.vassarstats.net/) was used in calculating the proportion of genes and its 95% confidence interval (95% CI). Correlation calculation was performed using SPSS 26.0 (IBM Corp, Armonk, NY). The cluster of AMR genes and virulence genes were obtained using R (version 4.0.5) and the package “pheatmap” (the clustering method of “complete” and “ward.D” were used).
Results
Detection and pattern of virulence genes
The virulence genes were divided into four groups: adhesion, invasion, immune evasion, and metabolism. The dominant virulence genes in the adhesion group were fbsA (99.29%; n=139) and pavA (25%, n=35), and cfb, cylE, gapC, and hylB genes were the predominant invasion genes, accounting for 99.29% (n=139), 97.14% (n=136), 100% (n=140), and 99.29% (n=139), respectively. bibA, cspA, bca, and scpB existed in the immune evasion group, with detection rate of which were 97.86% (n=137), 52.14% (n=73), 2.14% (n=3), and 0.71% (n=1), respectively. dltA was exhibited in all the isolates (n=140), and detection rate of Lac I/II/III/IV genes were 98.57% (n=138), 92.14% (n=129), 99.29% (139), and 100% (n=140), respectively. fbsB and lmb were not detected in the adhesion group, spbI was not detected in the invasion group, and bac and rib were not detected in the immune evasion group (Table 1).
Virulence genes can be grouped into subgroups A (cylE_hylB_gapC_dltA_lacI/II/III/IV), B (fbsA_cfb_cylE_hylB_bibA_cspA_gapC_dltA_lacIII/IV), and C (fbsA_cfb_bibA_gapC_dltA_lacIV). Subgroups B and C were the predominant subgroups, accounting for 97.86% of the isolates (Figure 1).
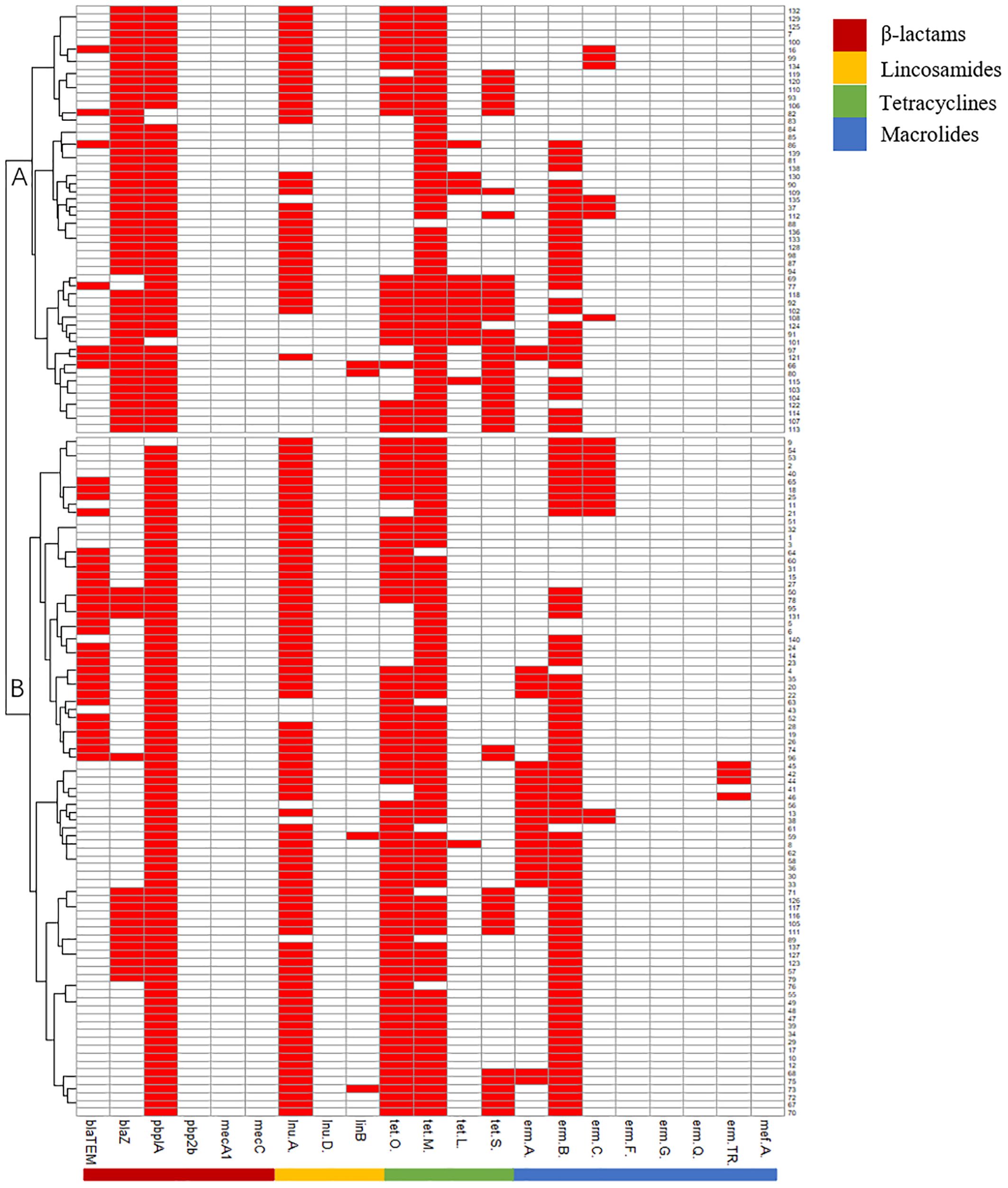
Figure 1. Pattern of virulence genes of 140 Strep.agalactiae. white square means absence of virulence genes, red square means presence of virulence genes.
Antimicrobial resistance testing
The isolates were sensitive to most of the tested antimicrobials: penicillin, ceftiofur, Amoxi/clav, and cefquinome (100%); cefalexin (97.9%); oxacillin (96.4%); enrofloxacin (95.7%); erythromycin (89.3%); and clindamycin (88.6%), but only 19.3% of the isolates were sensitive to tetracycline (Liu et al., 2022).
Detection and patterns of antimicrobial resistance genes
The antimicrobial resistance (AMR) genes were divided into four groups: β-lactam, lincosamide, tetracycline, and macrolide groups according to the type of antimicrobials. The main AMR genes in the β-lactam group were pbp1A (97.14%; n=136), blaZ (49.29%; n=69), and blaTEM (25.71%; n=36). lnu(A) and linB existed in the lincosamide group, with detection rate of 80.71% (n=113) and 2.86% (n=4), respectively. tet(O/M/L/S) were all found in the tetracycline group, with detection rate of 72.14% (n=101), 95.00% (n=133), 10.71% (n=15), and 29.29% (n=41), respectively. In the macrolide group, erm(B) was dominant, with a detection rate of 75.00% in the isolates (n=105), followed by erm(A), erm(C) and erm(TR), with detection rate of 17.14% (n=24), 13.57% (n=19), and 2.86% (n=4), respectively. pbp2b, mecA1, and mecC were not detected in the β-lactam group, lnu(D) gene was not detected in the lincosamide group, and erm(F/G/Q) and mef(A) were not detected in the macrolide group (Table 2).
AMR genes can be divided into subgroups A and B. Subgroup A harbored more blaZ and tetL genes than subgroup B, and subgroup B harbored more blaTEM and ermAgenes than subgroup A (Figure 2).
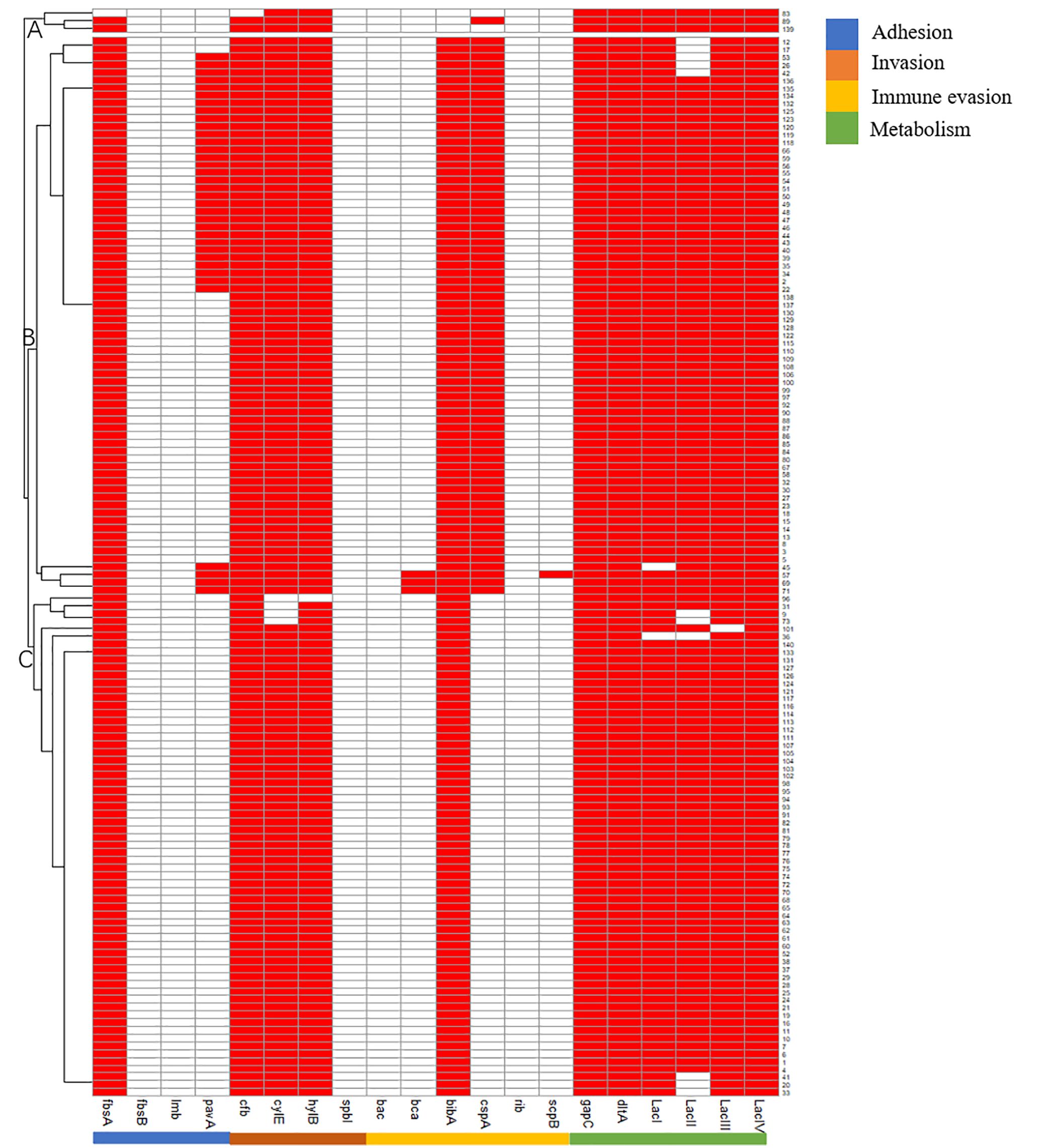
Figure 2. Pattern of antimicrobials resistant genes of 140 Strep.agalactiae. Note: white square means absence of antimicrobials resistant genes, red square means presence of antimicrobials resistant genes.
Relationship between AMR genotype and AMR phenotype
Four types of relationships between the phenotypes and genotypes of the five classes of antimicrobials were examined: resistant phenotype vs presence of ARG (+/+), sensitive phenotype vs presence of ARG (-/+), resistant phenotype vs absence of ARG (+/-), and sensitive phenotype vs absence of ARG (-/-). The pattern in which resistant phenotype vs absence of ARG (+/-) was dominant in the β-lactam, lincosamide, tetracycline, and macrolide groups, accounting for 95% (n=133), 73.57% (n=103), 19.29% (n=27), and 75% (n=105), respectively. In the tetracycline group, 80% of the isolates (n=112) were positive in AMR genes harboring AMR. Only 4.29% (n=6), 9.29% (n=13), and 10.71% (n=15) of the isolates were resistant phenotype and harboring AMR genes in the β-lactam, lincosamide, and macrolide groups. 0.71% (n=1), 15.71% (n=22), 0.71% (n=1), and 14.29% (n=20) of the isolates exhibited sensitive phenotype and absence of ARG in the β-lactam, lincosamide, tetracycline, and macrolide groups. In the lincosamides group, two isolates, accounting for 1.43%, did not harbor any AMR genes but exhibited resistance to clindamycin (Table 3).
Discussion
A total of 140 Strep. agalactiae isolates were collected from 12 large dairy farms in north China. The percentages of isolates harboring the virulence genes of adhesion(fbsA/B, lmb, pavA), invasion(cfb, cylE, hylB, spbI), immune evasion(bac, bca, bibA, cspA, rib, scpB), and metabolism(gapC, dltA, LacI/II/III/IV). were 99.29%, 100%, 47.86%, and 100%, respectively. The percentages of the isolates harboring antimicrobial resistance genes of β-lactams, lincosamides, tetracyclines, and macrolides were 99.29%, 82.86%, 99.29%, and 85.71%, respectively. 95%, 73.57%, and 75% of the isolates harbored the antimicrobial resistance genes of β-lactams, lincosamides, but macrolides, and they did not show resistance to the corresponding antimicrobials.
Bovine mastitis induced by Streptococcus can be divided into four steps: adhesion and colonization on bovine mammary epithelium cells (bMECs), invasion across or into bMECs, immune evasion, and metabolism (Keefe, 2012). The virulence genes of Strep. agalactiae can be categorized into four clusters: adhesion, invasion, immune evasion, and metabolism. In this study, the prevalence of adhesion, invasion, and metabolism clusters were relatively high, and the virulence gene of immune evasion accounted for 47.86% of the isolates. The adhesion genes fbsA and bibA accounted for 99.29% and 97.86% of the isolates, respectively. The main invasion genes were gapC (100%), hylB (99.29%), cfb (99.29%), and cylE (97.14%). The metabolism genes were conservative, and their detection rate were relatively high (dltA, 100%; LacI, 98.57%; LacII, 92.14%; LacIII, 99.29%; and LacIV, 100%). The results were consistent with those in previous research (Keefe, 2012; Morach et al., 2018). The high detection rate of these genes indicated that these genes are essential for the development of bovine mastitis.
The virulence genes fbsA/B encode fibrinogen-binding proteins, allowing Strep. agalactiae to bind to bMECs and extracellular proteins (Gutekunst and Eikmanns, 2004; Tenenbaum et al., 2005; Pietrocola et al., 2006; Buscetta et al., 2014). In a previous study, lmb was found to be associated with the adherence of Strep. agalactiae, but it was rarely harbored by bovine mastitis isolates (Duarte et al., 2005; Wu et al., 2016). Our results showed that the major virulence gene in charge of adhesion was fbsA, accounting for 99.29% (139/140). The low frequency or absence of pavA, fbsB, and lmb indicated that these genes are not essential to the pathogenesis of bovine mastitis. cfb encodes the CAMP factor involved in hemolytic activation (Lasagno et al., 2011). The cspA gene encoding serine protease and hemolysin encoded by cylE play crucial roles in the virulence of Strep. agalactiae (Chou et al., 2019). Hyaluronidase encoded by hylB promotes Strep. agalactiae invasion in host cells and promotes its host tissue-spreading ability (Oviedo et al., 2013; Coleman et al., 2023). Our study was consistent with the studies of Whist and Osterås (2007) and Pang et al. (2017), who indicated that cfb, cylE, and hylB were the main virulence genes of Strep. agalactiae (Whist and Osterås, 2007; Keefe, 2012). The high frequencies of virulence genes associated with invasion indicated that these genes were essential to induce clinical bovine mastitis for Strep. agalactiae (Keefe, 2012).
Immune evasion enables Strep. agalactiae to escape from host immunity killing. The α/β-C protein, as a surface protein, facilitates the invasion of Strep. agalactiae in cells and resistance to the clearance of phagocyte; the protein is encoded by bac and bca (Oviedo et al., 2013; Pulido-Colina et al., 2021). bac and bca usually appear together (Delannoy et al., 2013). The detection rate of bac is low in bovine isolates (Duarte et al., 2005). Our results showed the low detection rate of bac and bca, indicating they were not essential to bovine mastitis pathogenicity. C5a peptidase cleaving human C5a and BibA known as the C4-binding protein are encoded by scpB and bibA, respectively. Both proteins hamper the complement system, thereby reducing immune killing (Manne et al., 2020; Cullen et al., 2024). Our result indicated that bibA is the main virulence gene involved in the immune evasion of Strep. agalactiae. However, Duarte et al. (2005) revealed that 66% of Strep. agalactiae isolates from bovine harbor scpB (Duarte et al., 2005). Rib encoded by rib confers the ability of immune evasion and has been found in most isolates that caused invasive infections (Pulido-Colina et al., 2021). Consistent with our study, previous research indicated that only a small part of Strep. agalactiae isolated from bovine (20% and 26%) harbors the rib gene (B. Jain et al., 2012).
Rohmer et al. (2011) assumed that bacteria evolved to access specific nutrients that hosts provided and develop pathogenicity (Rohmer et al., 2011). Lac encodes lactose operon, and dItA encodes D-alanylation of lipoteichoic acid, which is involved in the completion of the cell wall of Gram-positive bacteria. The genes were conserved in all Strep. agalactiae isolates. Glyceraldehyde-3-phosphate dehydrogenase encoded by gapC is involved in carbohydrate metabolism. Our results indicated the ability of Strep. agalactiae to use milk as a nutrient resource due to these metabolism genes (Keefe, 2012). Overall, the results of our research indicated that the integrity of the parts of the virulence genes (adhesion, invasion, immune evasion, and metabolism) mediates the pathogenesis of Strep. agalactiae.
Isolates were sensitive to most of the tested antimicrobials: penicillin, ceftiofur, amoxi/clav, cefquinome, and vancomycin (100%); cefalexin (97.9%); oxacillin (96.4%); enrofloxacin (95.7%); erythromycin (89.3%); and clindamycin (88.6%). However, only 19.3% of the isolates were sensitive to tetracycline, and 0.7% were sensitive to daptomycin (Liu et al., 2022).
The percentage of the isolates resistant to tetracycline was 80%, in line with the results of Gao et al. (2012) and Tomazi et al. (2018), who reported that the percentages of resistance were 72.5% in China and 68.6% in Brazil, respectively (Gao et al., 2012; Tomazi et al., 2018). The low efficacy of tetracycline in treating mastitis has been reported worldwide, and one of the reasons is its excessive use in treatment and growth promotion (Kaczorek et al., 2017). This antimicrobial should be used prudently in the treatment of mastitis.
In addition to the AMR profiles of Strep. agalactiae under in vitro conditions, genotypic AMR detection was performed for the selected AMR genes encoding different resistance mechanisms.
The results of our research were consistent with those of Kannika et al. (2017) (Kannika et al., 2017). pbplA/ponA (penicillin-binding protein 1A) was the dominant gene encoding resistance to β-lactams and accounted for 97.14%, followed by blaZ (49.29%).
The lincosamides resistant genes we detected are the lnu A/D and linB genes, nucleotidyl transferases are encoded by lnu genes, resulting in enzymatic inactivation of lincosamides. The lnu gene was first identified in Enterococcus faecium and then observed in Strep. agalactiae (Arana et al., 2014; Kaczorek et al., 2017). Our results indicated that the detection rate of lnu(A) was 80.71%, which may raise concerns about the spreading of AMR genes among bacteria.
We detected four genes responsible for resistance to tetracyclines: tet(M), tet(O), tet(S). and tet(L), which encodes resistance through ribosomal protection and efflux pump (Poyart et al., 2003; Dogan et al., 2005; Gao et al., 2012). In our research, the tet(M) and tet(O) genes were predominant, which is consistent with previous study (Gao et al., 2012; Rato et al., 2013). The high detection rate of these genes can be attributed to horizontal gene transfer in the same genus of bacteria (Gao et al., 2012; Ruegg et al., 2015).
Eight genes encoding macrolide resistance were detected. erm(B) was predominant, consistent with previous reports (Loch et al., 2005; Gao et al., 2012; Rato et al., 2013). erm(B) can encode methylase, reducing the number of macrolides binding to Strep. agalactiae (Denamiel et al., 2005). erm(B) can transfer among bacteria in the same genus (Loch et al., 2005), and this feature explains the high detection rate of the gene. mef(A), harbored by only 2.86% of isolates, was examined as well. The results were consistent with those of previous research (Rato et al., 2013).
There were two isolates that exhibited resistance to lincosamides but did not harbor the examined AMR genes, the possible reason for which is that we failed to detect other resistance genes that encoding lincosamides resistance. Nevertheless, some isolates exhibited AMR genes carrying but negative in AMR phenotype. The reasons were as follows: (1) AMR genes may not transcribe nor translate because the corresponding antimicrobials were not used in bovine mastitis treatment, so they were far from a promoter or associated with a weak promoter; (2) mutations or lack of promoters induce the silencing of the AMR genes of the isolates. Advanced research is essential to discover the mechanisms of the insufficient correlation between the genotype and phenotype of AMR (Gao et al., 2012).
For the reason of insufficient controls (positive controls for each AMR or virulence genes) were used in our research, the genes that did not be detected may due to the following situations: (1) the isolates did not harbor corresponding genes, (2) the primers failed to combine to template on account of gene mutation.
Conclusion
Some isolates resistant to lincosamides did not necessarily carry any tested gene. Conversely, a large part of β-lactams, lincosamides, and macrolides sensitive isolates contained corresponding AMR genes, which may not be expressed in these isolates. Furthermore, based on almost all isolates harbored virulence genes encoded the ability of adhesion, invasion, immune evasion and metabolism, we inferred that intact combination of virulence genes is essential to the pathogenesis of Strep. agalactiae inducing bovine mastitis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
All animal procedures were approved by the Institutional Animal Care and Use Committee of Yunnan Agricultural University (Approval No: 202403058). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
KL: Methodology, Writing – original draft. XL: Investigation, Writing – review & editing. JY: Data curation, Software, Writing – review & editing. XG: Data curation, Formal analysis, Validation, Writing – review & editing. LZ: Project administration, Resources, Visualization, Writing – review & editing. WQ: Data curation, Funding acquisition, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by Yunnan Fundamental Research Projects (grant no. 202301AS070081), Rural Vitalization Science and technology Project -Gejiu Cattle Industry Science and Technology Mission in Yunnan Province (grant no. 202304BI090007).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1417299/full#supplementary-material
References
Arana, D. M., Rojo-Bezares, B., Torres, C., Alós, J. I. (2014). First clinical isolate in Europe of clindamycin-resistant group B Streptococcus mediated by the lnu(B) gene. Rev. Esp Quimioter. 27, 106–109.
Bi, Y., Wang, Y. J., Qin, Y., Guix Vallverdú, R., Maldonado García, J., Sun, W., et al. (2016). Prevalence of bovine mastitis pathogens in bulk tank milk in China. PloS One 11, e0155621. doi: 10.1371/journal.pone.0155621
Buscetta, M., Papasergi, S., Firon, A., Pietrocola, G., Biondo, C., Mancuso, G., et al. (2014). FbsC, a novel fibrinogen-binding protein, promotes Streptococcus agalactiae-host cell interactions. J. Biol. Chem. 289, 21003–21015. doi: 10.1074/jbc.M114.553073
Carvalho-Castro, G. A., Silva, J. R., Paiva, L. V., Custódio, D. A. C., Moreira, R. O., Mian, G. F., et al. (2017). Molecular epidemiology of Streptococcus agalactiae isolated from mastitis in Brazilian dairy herds. BrazJ Microbiol. 48, 551–559. doi: 10.1016/j.bjm.2017.02.004
Chou, C. C., Lin, M. C., Su, F. J., Chen, M. M. (2019). Mutation in cyl operon alters hemolytic phenotypes of Streptococcus agalactiae. Infect. Genet. Evol. 67, 234–243. doi: 10.1016/j.meegid.2018.11.003
CLSI. (2020). Performance Standards for Antimicrobial Susceptibility Testing, CLSI Document (Wayne, PA: M100. Clinical Laborratory Standards Institute (CLSI).
Coleman, M., Orvis, A., Brokaw, A., Furuta, A., Sharma, K., Quach, P., et al. (2023). GBS hyaluronidase mediates immune suppression in a TLR2/4- and IL-10-dependent manner during pregnancy-associated infection. mBio 14, e0204923. doi: 10.1128/mbio.02049-23
Cullen, R., Teçza, M., Miclot, T., Behan, S., Jain, M., Avink, M. K., et al. (2024). The 1.7 Å crystal structure of the C5a peptidase from Streptococcus agalactiae (ScpB) reveals an active site competent for catalysis. Proteins 92, 427–431. doi: 10.1002/prot.26625
Delannoy, C. M., Crumlish, M., Fontaine, M. C., Pollock, J., Foster, G., Dagleish, M. P., et al. (2013). Human Streptococcus agalactiae strains in aquatic mammals and fish. BMC Microbiol. 13, 41. doi: 10.1186/1471-2180-13-41
Denamiel, G., Llorente, P., Carabella, M., Rebuelto, M., Gentilini, E. (2005). Anti-microbial susceptibility of Streptococcus spp. isolated from bovine mastitis in Argentina. J. Vet. Med. B-Infect Dis. Vet. Public Health 52, 125–128. doi: 10.1111/j.1439-0450.2005.00830.x
Dogan, B., Schukken, Y. H., Santisteban, C., Boor, K. J. (2005). Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J. Clin. Microbiol. 43, 5899–5906. doi: 10.1128/JCM.43.12.5899-5906.2005
Duarte, R. S., Bellei, B. C., Miranda, O. P., Brito, M. A., Teixeira, L. M. (2005). Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 49, 97–103. doi: 10.1128/AAC.49.1.97-103.2005
Flynn, C. E., Guarner, J. (2023). Emerging antimicrobial resistance. Mod Pathol. 36, 100249. doi: 10.1016/j.modpat.2023.100249
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Gao, J., Barkema, H. W., Zhang, L., Liu, G., Deng, Z., Cai, L., et al. (2017). Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J. Dairy Sci. 100, 4797–4806. doi: 10.3168/jds.2016-12334
Gao, J., Yu, F. Q., Luo, L. P., He, J. Z., Hou, R. G., Zhang, H. Q., et al. (2012). Antibiotic resistance of Streptococcus agalactiae from cows with mastitis. Vet. J. 194, 423–424. doi: 10.1016/j.tvjl.2012.04.020
Gussmann, M., Denwood, M., Kirkeby, C., Farre, M., Halasa, T. (2019). Associations between udder health and culling in dairy cows. Prev. Vet. Med. 171, 104751. doi: 10.1016/j.prevetmed.2019.104751
Gutekunst, H., Eikmanns, B. J. (2004). Reinscheid DJ The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72, 3495–3504. doi: 10.1128/IAI.72.6.3495-3504.2004
Jain, B., Tewari, A., Bhandari, B. B., Jhala, M. (2012). Antibiotic resistance and virulence genes in Streptococcus agalactiae isolated from cases of bovine subclinical mastitis. Vet. Arch. 82, 423–432. doi: 10.3389/fcimb.2022.1049167
Jørgensen, H. J., Nordstoga, A. B., Sviland, S., Zadoks, R. N., Sølverød, L., Kvitle, B., et al. (2016). Streptococcus agalactiae in the environment of bovine dairy herds–rewriting the textbooks? Vet. Microbiol. 184, 64–72. doi: 10.1016/j.vetmic.2015.12.014
Kaczorek, E., Małaczewska, J., Wójcik, R., Rękawek, W., Siwicki, A. K. (2017). Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J. Dairy Sci. 100, 6442–6453. doi: 10.3168/jds.2017-12660
Kannika, K., Pisuttharachai, D., Srisapoome, P., Wongtavatchai, J., Kondo, H., Hirono, I., et al. (2017). Molecular serotyping, virulence gene profiling and pathogenicity of Streptococcus agalactiae isolated from tilapia farms in Thailand by multiplex PCR. J. Appl. Microbiol. 122, 1497–1507. doi: 10.1111/jam.13447
Katholm, J., Bennedsgaard, T. W., Koskinen, M. T., Rattenborg, E. (2012). Quality of bulk tank milk samples from Danish dairy herds based on real-time polymerase chain reaction identification of mastitis pathogens. J. Dairy Sci. 95, 5702–5708. doi: 10.3168/jds.2011-5307
Keefe, G. (2012). Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Veterinary Clinics North America: Food Anim. Pract. 28, 203–216. doi: 10.1016/j.cvfa.2012.03.010
Krömker, V., Leimbach, S. (2017). Mastitis Treatment-Reduction in antibiotic usage in dairy cows. Reprod. Domest Anim. 52, 21–29. doi: 10.1111/rda.13032
Lasagno, M. C., Reinoso, E. B., Dieser, S. A., Calvinho, L. F., Buzzola, F., Vissio, C., et al. (2011). Phenotypic and genotypic characterization of Streptococcus uberis isolated from bovine subclinical mastitis in Argentinean dairy farms. Rev. Argent Microbiol. 43, 212–217. doi: 10.1590/S0325-75412011000300009
Liu, K., Zhang, L., Gu, X., Liu, G., Liu, Y., Chen, P., et al. (2022). The prevalence, molecular characterization and antimicrobial resistance profiling of Streptococcus agalactiae isolated from clinical mastitis cases on large dairy farms in China. J. Dairy Res. 28, 1–5. doi: 10.1017/S0022029922000152
Loch, I. M., Glenn, K., Zadoks, R. N. (2005). Macrolide and lincosamide resistance genes of environmental streptococci from bovine milk. Vet. Microbiol. 111, 133–138. doi: 10.1016/j.vetmic.2005.09.001
Manne, K., Chattopadhyay, D., Agarwal, V., Blom, A. M., Khare, B., Chakravarthy, S., et al. (2020). Novel structure of the N-terminal helical domain of BibA, a group B streptococcus immunogenic bacterial adhesin. Acta Crystallogr. D Struct. Biol. 76, 759–770. doi: 10.1107/S2059798320008116
Morach, M., Stephan, R., Schmitt, S., Ewers, C., Zschöck, M., Reyes-Velez, J., et al. (2018). Population structure and virulence gene profiles of Streptococcus agalactiae collected from different hosts worldwide. Eur. J. Clin. Microbiol. Infect. Dis. 37, 527–536. doi: 10.1007/s10096-017-3146-x
Oviedo, P., Pegels, E., Laczeski, M., Quiroga, M., Vergara, M. (2013). Phenotypic and genotypic characterization of Streptococcus agalactiae in pregnant women. First study in a province of Argentina. Braz. J. Microbiol. 44, 253–258. doi: 10.1590/S1517-83822013005000030
Pietrocola, G., Visai, L., Valtulina, V., Vignati, E., Rindi, S., Arciola, C. R., et al. (2006). Multiple interactions of FbsA, a surface protein from Streptococcus agalactiae, with fibrinogen: affinity, stoichiometry, and structural characterization. Biochemistry 45, 12840–12852. doi: 10.1021/bi060696u
Poyart, C., Jardy, L., Quesne, G., Berche, P., Trieu-Cuot, P. (2003). Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob. Agents Chemother. 47, 794–797. doi: 10.1128/AAC.47.2.794-797.2003
Pulido-Colina, A., Pastrana, J. S., Valencia-Bazalar, E., Apestegui, M. Z. (2021). Caracterización molecular de genes de virulencia (lmb, bca y rib) y de resistencia a macrólidos (ermB, ermTR y mefA) en aislamientos clínicos de Streptococcus agalactiae [Molecular characterization of virulence (lmb, bca and rib) and macrolid resistance genes (ermB, ermTR and mefA) in clinical isolates of Streptococcus agalactiae]. Rev. Peru Med. Exp. Salud Publica. 38, 615–620. doi: 10.17843/rpmesp.2021.384.8726
Ramírez, N. F., Keefe, G., Dohoo, I., Sánchez, J., Arroyave, O., Cerón, J., et al. (2014). Herd- and cow-level risk factors associated with subclinical mastitis in dairy farms from the High Plains of the northern Antioquia, Colombia. J. Dairy Sci. 97, 4141–4150. doi: 10.3168/jds.2013-6815
Rato, M. G., Bexiga, R., Florindo, C., Cavaco, L. M., Vilela, C. L., Santos-Sanches, I. (2013). Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Vet. Microbiol. 161. doi: 10.1016/j.vetmic.2012.07.043
Reyes, J., Chaffer, M., Rodriguez-Lecompte, J. C., Sánchez, J., Zadoks, R. N., Robinson, N., et al. (2017). Short communication: Molecular epidemiology of Streptococcus agalactiae differs between countries. J. Dairy Sci. 100, 9294–9297. doi: 10.3168/jds.2017-13363
Rohmer, L., Hocquet, D., Miller, S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. doi: 10.1016/j.tim.2011.04.003
Ruegg, P. L., Oliveira, L., Jin, W., Okwumabua, O. (2015). Phenotypic antimicrobial susceptibility and occurrence of selected resistance genes in gram-positive mastitis pathogens isolated from Wisconsin dairy cows. J. Dairy Sci. 98, 4521–4534. doi: 10.3168/jds.2014-9137
Shaheen, M., Tantary, H. A., Nabi, S. U. (2016). A treatise on bovine mastitis: Disease and disease economics, etiological basis, risk factors, impact on human health, therapeutic management, prevention and control strategy. J. Adv. Dairy Res. 4, 150.
Tenenbaum, T., Bloier, C., Adam, R., Reinscheid, D. J., Schroten, H. (2005). Adherence to and invasion of human brain microvascular endothelial cells are promoted by fibrinogen-binding protein FbsA of Streptococcus agalactiae. Infect. Immun. 73, 4404–4409. doi: 10.1128/IAI.73.7.4404-4409.2005
Thompson-Crispi, K., Atalla, H., Miglior, F., Mallard, B. A. (2014). Bovine mastitis: frontiers in immunogenetics. Front. Immunol. 5, 493. doi: 10.3389/fimmu.2014.00493
Tomazi, T., de Souza Filho, A. F., Heinemann, M. B., Dos Santos, M. V. (2018). Molecular characterization and antimicrobial susceptibility pattern of Streptococcus agalactiae isolated from clinical mastitis in dairy cattle. PloS One 13, e0199561. doi: 10.1371/journal.pone.0199561
Whist, A. C., Osterås, O. (2007). Associations between somatic cell counts at calving or prior to drying-off and clinical mastitis in the remaining or subsequent lactation. J. Dairy Res. 74, 66–73. doi: 10.1017/S0022029906002172
Wu, X., Hou, S., Zhang, Q., Ma, Y., Zhang, Y., Kan, W. (2016). Prevalence of virulence and resistance to antibiotics in pathogenic enterococci isolated from mastitic cows. J. Vet. Med. Sci. 78, 1663–1668. doi: 10.1292/jvms.15-0718
Keywords: bovine mastitis, Streptococcus agalactiae, antimicrobial resistance, virulence gene, AMR genes detection
Citation: Liu K, Liu X, Yang J, Gu X, Zhang L and Qu W (2024) Streptococcus agalactiae isolated from clinical mastitis cases on large dairy farms in north China: phenotype, genotype of antimicrobial resistance and virulence genes. Front. Cell. Infect. Microbiol. 14:1417299. doi: 10.3389/fcimb.2024.1417299
Received: 14 April 2024; Accepted: 27 June 2024;
Published: 04 September 2024.
Edited by:
Ilke Pala-Ozkok, University of Stavanger, NorwayReviewed by:
Zhanbo Zhu, Heilongjiang Bayi Agricultural University, ChinaTariq Ali, Veterinary Research Institute, Peshawar, Pakistan
Copyright © 2024 Liu, Liu, Yang, Gu, Zhang and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijie Qu, MTQwMTA3NTAxMEBxcS5jb20=; Limei Zhang, MjAwNzMxNzVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Kai Liu1,2†
Kai Liu1,2† Xiaolong Gu
Xiaolong Gu Limei Zhang
Limei Zhang Weijie Qu
Weijie Qu