
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 24 May 2024
Sec. Molecular Bacterial Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1411333
This article is part of the Research TopicThe Role of Transcriptional Regulation in Mycobacterium PhysiologyView all 5 articles
Mycobacterium abscessus (Mab) is an opportunistic pathogen afflicting individuals with underlying lung disease such as Cystic Fibrosis (CF) or immunodeficiencies. Current treatment strategies for Mab infections are limited by its inherent antibiotic resistance and limited drug access to Mab in its in vivo niches resulting in poor cure rates of 30-50%. Mab’s ability to survive within macrophages, granulomas and the mucus laden airways of the CF lung requires adaptation via transcriptional remodeling to counteract stresses like hypoxia, increased levels of nitrate, nitrite, and reactive nitrogen intermediates. Mycobacterium tuberculosis (Mtb) is known to coordinate hypoxic adaptation via induction of respiratory nitrate assimilation through the nitrate reductase narGHJI. Mab, on the other hand, does not encode a respiratory nitrate reductase. In addition, our recent study of the transcriptional responses of Mab to hypoxia revealed marked down-regulation of a locus containing putative nitrate assimilation genes, including the orphan response regulator nnaR (nitrate/nitrite assimilation regulator). These putative nitrate assimilation genes, narK3 (nitrate/nitrite transporter), nirBD (nitrite reductase), nnaR, and sirB (ferrochelatase) are arranged contiguously while nasN (assimilatory nitrate reductase identified in this work) is encoded in a different locus. Absence of a respiratory nitrate reductase in Mab and down-regulation of nitrogen metabolism genes in hypoxia suggest interplay between hypoxia adaptation and nitrate assimilation are distinct from what was previously documented in Mtb. The mechanisms used by Mab to fine-tune the transcriptional regulation of nitrogen metabolism in the context of stresses e.g. hypoxia, particularly the role of NnaR, remain poorly understood. To evaluate the role of NnaR in nitrate metabolism we constructed a Mab nnaR knockout strain (MabΔnnaR) and complement (MabΔnnaR+C) to investigate transcriptional regulation and phenotypes. qRT-PCR revealed NnaR is necessary for regulating nitrate and nitrite reductases along with a putative nitrate transporter. Loss of NnaR compromised the ability of Mab to assimilate nitrate or nitrite as sole nitrogen sources highlighting its necessity. This work provides the first insights into the role of Mab NnaR setting a foundation for future work investigating NnaR’s contribution to pathogenesis.
The nontuberculous mycobacteria Mycobacterium abscessus (Mab) is an opportunistic pathogen afflicting the immunocompromised and individuals with pre-existing lung disorders such as cystic fibrosis (CF) (Brown-Elliott and Wallace, 2002; Olivier et al., 2003; Lee et al., 2015). CF patients are particularly vulnerable to bacterial infections due to reduced clearance of pathogens caused by viscous mucus buildup in the airways of the lungs and impaired innate immune responses leading to high rates of infections and morbidity (Lyczak et al., 2002; Chmiel and Davis, 2003). Mab is the most prevalent rapidly growing mycobacteria recovered from the lungs of CF patients, often causing deleterious effects such as lung function decline, resulting in extended hospitalization and in some cases exclusion from lung transplants (Olivier et al., 2003; Esther et al., 2010; Lopeman et al., 2019). Treatment options for Mab infections are limited due to inherent antibiotic resistance, resulting in cure rates of less than 50% (Hurst-Hess et al., 2017; Molina-Torres et al., 2018; Story-Roller et al., 2018; Lopeman et al., 2019). Like M. tuberculosis (Mtb), Mab’s ability to reside within macrophages and granulomas physically limits exposure to antibiotics and also promotes drug tolerance by driving metabolic and transcriptional remodeling, further complicating the treatment of these infections (Bernut et al., 2016; Peddireddy et al., 2017; Molina-Torres et al., 2018). Mab is also able to survive within the mucus that accumulates in the CF airway. In addition to creating a hypoxic microenvironment with a pO2 of ~1%, this mucus layer is characterized by increased levels of nitrite, nitrate and nitric oxide (Hassett et al., 2002; Worlitzsch et al., 2002; Robinson et al., 2017). Adaptation to these conditions along with other host-derived stressors requires extensive alterations in gene expression to allow Mab to establish persistent pulmonary infections. In Mtb, it is apparent that hypoxia survival strategies are inextricably linked to nitrogen metabolism via a shift to nitrate respiration as a way for bacilli to cope with low-oxygen microenvironments (Wayne and Hayes, 1998; Sohaskey and Wayne, 2003; Sohaskey, 2008). Our current understanding of how Mab adapts to these stresses in vitro or in vivo remains rudimentary at best. Several recent reports, including one from our group, highlighted the role of the Mab DosRS two-component system in the induction of a species-specific regulon in response to hypoxia (Belardinelli et al., 2022; Simcox et al., 2023). On the other hand, research examining the regulation of nitrogen metabolism in Mab is lacking. Prompted by our recent analysis of the hypoxia-induced transcriptional responses in Mab (Simcox et al., 2023), the current study provides additional evidence that Mab relies on strategies of nitrogen metabolism distinct from Mtb in response to in vivo relevant stresses.
The mechanisms for assimilation of nitrogen from nitrate and nitrite, which are readily accessible to mycobacteria during infection (Gouzy et al., 2014), have been well characterized in Mtb and M. smegmatis (Msm) however little is known about this facet of Mab host-pathogen interactions. Nitrate assimilation not only provides inorganic sources of nitrogen for the production of biomolecules but can also protect microbes against host iNOS (inducible nitric oxide synthase) generated nitric oxide (NO) an important factor used by the host to control infection (Kelm, 1999; Lundberg et al., 2004; Cunningham-Bussel et al., 2013; Gouzy et al., 2014). Host derived NO, after conversion to nitrate through auto-oxidation, can be exploited by Mycobacterium via the nitrate assimilation pathway resulting in the production of ammonium which can be stored as glutamate (Gouzy et al., 2014). Mtb constitutively expresses NarGHJI which serves as both an assimilatory nitrate reductase (NR), which converts nitrate to nitrite, and a respiratory NR allowing utilization of nitrate as a terminal electron acceptor (Sohaskey, 2008; Sohaskey and Modesti, 2009; Khan and Sarkar, 2012). The transcriptional and post-translational activation of NarK2, a putative H+/nitrate symporter (Giffin et al., 2012), under conditions that induced DosR (hypoxia, NO, CO), serves to induce nitrogen assimilation and respiration under hypoxic conditions (Voskuil et al., 2003; Khan and Sarkar, 2012). The excess, potentially toxic nitrite generated by NarGHJI is either extruded or further reduced to ammonium by the NirBD nitrite reductase (NiR) en route to storage or assimilation (Sohaskey and Wayne, 2003; Malm et al., 2009; Khan and Sarkar, 2012; Gouzy et al., 2014). Msm also encodes a constitutively expressed NarGHJI respiratory NR, however, unlike Mtb, it was shown to be non-functional for assimilation of nitrogen from nitrate (Sohaskey and Wayne, 2003; Malm et al., 2009; Cardoso et al., 2021). Instead, Msm relies on an unusual NADPH-dependent, diflavin-containing NR called NasN which is essential for nitrate utilization in aerobic conditions (Tan et al., 2020; Cardoso et al., 2021) as well as a NirBD ortholog to catalyze reduction of nitrite (Akhtar et al., 2013; Martin and Liras, 2019). Each of these components – narGHJI, nirBD, and nasN – along with a putative NarK nitrate/nitrite antiporter (MSMEG_0433) represent distinct operons or transcripts (Jenkins et al., 2013; Antczak et al., 2018). In contrast to Mtb and Msm, Mab does not encode a respiratory NarGHJI suggesting an inability to utilize nitrate as an alternative electron acceptor in low oxygen conditions. However, the Mab genome does include orthologs of other key components of nitrate/nitrite metabolism, including narK3 (Mab_3523c), nirBD (Mab_3522c-3521c), nnaR (transcriptional regulator Mab_3520c) and sirB (siroheme ferrochelatase Mab_3519c). These five genes are arranged as a predicted operon that we found to be dramatically repressed in response to hypoxia in a DosR-independent manner (Simcox et al., 2023). Missing from this locus was a candidate NasN-type assimilatory NR. As detailed herein, we have identified a putative NasN ortholog (Mab_2438) that was co-regulated with the narK3-sirB operon (Mab_3523c-3519c) we predict catalyzes the reduction of nitrate to nitrite.
Both Mycobacteria and related Streptomyces rely upon an intricate network of regulators including two-component systems (TCS), serine-threonine protein kinases (STPKs), and orphan response regulators (ORR) to coordinate the integration of multiple cues to control nitrogen metabolism. GlnR, an ORR that can be activated by phosphorylation of Ser/Thr residues (Hong et al., 2007; Lin et al., 2014), is known as the master regulator of nitrogen metabolism in actinobacteria such as Mycobacteria and Streptomyces (Amin et al., 2012; Jenkins et al., 2013; He et al., 2023). Mtb GlnR is required for growth on nitrate and nitrite when these are the only nitrogen sources available and was shown to be a positive regulator of nirBD (Malm et al., 2009). Additionally, GlnR has been shown to crosstalk with the response regulators PhoP and MtrA which act as repressors of nitrogen metabolism through competition with GlnR for binding sites under nitrogen replete conditions (Rodriguez-Garcia et al., 2009; Sola-Landa et al., 2013; Martin and Liras, 2019; Zhu et al., 2019). As discussed in more detail below, an additional ORR, NnaR, with an unusual domain structure comprised of a HemD (uroporphyrinogen-III synthase) domain fused to a DNA-binding domain, is also implicated in nitrate and nitrite assimilation (Amin et al., 2012). Despite extensive studies examining Mtb and Msm transcriptional regulation of nitrogen metabolism and nitrate/nitrite assimilation, this remains a significant knowledge gap for Mab (Malm et al., 2009; Huang et al., 2015; Williams et al., 2015; Fan et al., 2024). Recently, the GlnR ortholog in Mab was shown to regulate narK3, nirBD, glnA (glutamine synthetase), and amt (ammonium transporter) although a role in the regulation of Mab nasN was not reported (Fan et al., 2024). The inactivation of GlnR in Mab led to an inability to form biofilms resulting in drug susceptibility dependent upon glutamine and glutamate availability (Fan et al., 2024). The role of other transcription factors in the regulation of nitrogen metabolism in Mab remains to be investigated. Notably, there have been no reports of the identification of a NnaR ortholog or its role in the use of nitrate/nitrite as nitrogen sources or detoxification of nitrite.
NnaR was first identified in Streptomyces coelicolor (S. coelicolor) as a GlnR target and co-activator with GlnR of nitrite reductase (nirB), nitrate transporter (narK) and nitrate reductase (nasA) (Amin et al., 2012). In the absence of NnaR, S. coelicolor displayed diminished capacity to reduce nitrite to ammonium resulting in a growth defect when nitrate or nitrite were used as sole nitrogen sources. However, nitrate was reduced to nitrite suggesting NnaRSc is not a regulator of nitrate reductase (Amin et al., 2012). Similar to S. coelicolor, Msm NnaR regulates key nitrogen assimilation genes and is required for use of nitrate and nitrite as sole nitrogen sources (Antczak et al., 2018). Subsequent research by Tan et al., determined GlnRMsm is responsible for regulation of the assimilatory nitrate reductase NasN, confirming regulation of nitrate assimilation in Msm involves both GlnR (nitrate>nitrite) and NnaR (nitrite > ammonium) (Tan et al., 2020). The distinct roles for these two transcription factors in regulating different steps of nitrate assimilation were affirmed by EMSA data showing binding of NnaR to the nirBD promoter and GlnR binding upstream of nasN (Antczak et al., 2018; Tan et al., 2020). An ortholog of NnaR is also encoded by Mtb (Rv0260c), but little is known about it beyond its induction by multiple stress conditions including hypoxia, nutrient starvation, acid pH, and stationary phase (Vilcheze et al., 2022). The regulon and physiological role of Mab NnaR remains unknown, highlighting the need for further investigation of NnaR-mediated transcriptional regulation.
The goal of this study was to elucidate the previously unexplored role of Mab NnaR in gene regulation and nitrogen metabolism. We generated a Mab nnaR knockout strain (MabΔnnaR) and complement strain (MabΔnnaR+C) to further study NnaR and its function. Our work revealed that Mab NnaR regulates the nark3-nirBD-nnaR-sirB operon and nasN in a species-specific manner enabling nitrate and nitrite utilization. qRT-PCR analyses demonstrated that both loci were highly upregulated when nitrate or nitrite were the sole nitrogen sources, and with the exception of sirB, this gene induction was dependent on NnaR. The unexpected induction of sirB upon deletion of nnaR, which was restored to baseline levels in the complemented strain, suggests additional layers of regulation on this component of the operon. The inability of MabΔnnaR to regulate nitrogen assimilation genes resulted in growth defects when nitrate, and to a lesser degree nitrite, were used as the sole nitrogen source. This study highlights novel species-specific aspects of nitrogen metabolism in Mab and the critical role of NnaR and its downstream regulon in nitrate/nitrite utilization and detoxification. These findings support the hypothesis that NnaR is important for host adaptation and Mab pathogenesis.
MabΔnnaR was engineered via recombineering as described by van Kessell and Hatfull in the strain Mab 390S obtained from the Thomas Byrd lab (Byrd and Lyons, 1999; van Kessel et al., 2008). In brief, an allelic exchange substrate (AES) was generated containing an apramycin resistance cassette flanked by ~600 nucleotides upstream and downstream of the nnaR gene (MAB_3520c) for homologous recombination. To construct the AES, the nnaR gene plus ~600 nucleotides upstream and downstream were PCR amplified and cloned into pCRBluntII-TOPO vector (Thermo Fisher) followed by round-the-horn PCR to remove the nnaR gene, retaining nucleotide overlap from genes upstream and downstream of nnaR (Li et al., 2011). After removal of nnaR, a phosphorylated apramycin resistance cassette was ligated between the upstream and downstream flanking regions. PCR amplification was then used to generate the AES consisting of the apramycin cassette flanked by upstream and downstream nucleotides. Mab::pJV53 competent cells induced with 0.02% acetamide for 4 hours were electroporated with 100 ng AES, recovered in 7H9 OADC media for 24 hours, and plated on 7H10 agar supplemented with apramycin 50 µg/ml. Complement strain, MabΔnnaR+C containing nnaR and sirB inserted downstream of the constitutive hsp60 promoter (Phsp60) in the episomal vector pVV16 was engineered using round the horn PCR of the vector (Li et al., 2011) and blunt ligation of the phosphorylated insert (Parikh et al., 2013). The sirB gene downstream of nnaR was included due to anticipated polar effects of inactivation of nnaR. This construct was introduced by electroporation into MabΔnnaR, followed by recovery in 7H9 OADC media for 24 hours and plating on 7H10 agar supplemented with kanamycin 50 µg/ml. Mab nnaR knockout was confirmed via PCR, sequencing and qRT-PCR. PCR screening entailed amplification of the apramycin resistance cassette or nnaR plus upstream and downstream flanking regions generating a product of 2,071 bp for ΔnnaR or a 2,287 bp product for WT Mab (refer to primer table for PCR screening, nnaR-F and nnaR-R). Sequencing of positive nnaR knockout clone was performed by Azenta.
Mycobacterial Protein Fragment Complementation (M-PFC) constructs were generated in the background plasmids pUAB100, pUAB200, and pUAB400 (Singh et al., 2006) using round the horn cloning and ligation. Mycobacterium smegmatis competent cells were electroporated with 100ng of each pUAB100:dosS/pUAB400:dosR, pUAB100:dosS/pUAB200:nnaR or pUAB100:dosS/pUAB400:nnaR at the same time and recovered in LB broth for 4 hours before plating on LB Agar supplemented with hygromycin B 50 µg/ml and kanamycin 50 µg/ml. Primers used for cloning are listed in Supplementary Table S1. Strains and plasmids are listed in Supplementary Table S1.
Mab cultures were grown in 7H9 OADC+.05% tyloxapol from glycerol stocks and incubated at 37° C while shaking. Growth kinetic studies under nitrogen limitation were carried out in Sauton’s nitrogen free minimal media (0.05% KH2PO4, 0.05% MgSO4, 0.2% citric acid, 0.005% ferric citrate, 0.2% glycerol, 0.0001% ZnSO4, 0.015% tyloxapol) supplemented with 2.5 mM sodium nitrate, 0.5 mM sodium nitrite or 2.5 mM ammonium sulfate as previously described (Jenkins et al., 2013; Antczak et al., 2018). Briefly, cultures were grown to mid-log phase in 7H9 OADC+0.05% tyloxapol washed twice with Sauton’s minimal media and then diluted to 0.2 OD followed by nitrogen supplementation. On days 0-5 cultures were serially diluted and spot plated on LB Agar for CFU enumeration.
RNA was extracted as described by Rohde et al. (Rohde et al., 2007) in triplicate from cultures grown in 7H9 OADC+.05% tyloxapol or Sauton’s nitrogen free minimal media supplemented with nitrogen sources. Cultures were pelleted at 4,300 rpm for 10 minutes, resuspended in guanidine thiocyanate buffer, pelleted again at 12,000 rpm for 5 min, and stored at -80° C until processing. Thawed pellets were resuspended in 65° C Trizol then lysed using 0.1mM silicon beads in a BeadBeater at max speed for 1 minute 2x followed by cooling on ice for 1 minute between bead beating. Isolation of total RNA from Trizol lysates was performed using chloroform extraction and Qiagen RNeasy column purification. Total RNA was treated with Turbo DNase (Invitrogen) to eliminate DNA contamination. 50 ng/µl of total RNA was used to generate cDNA using iScriptTM cDNA synthesis kit (Bio-Rad) for qRT-PCR and RT-PCR. qRT-PCR reactions were carried out in a QuantStudio7 thermocycler using SYBR green as readout. Primers used for qRT-PCR are listed in Supplementary Table S1. To confirm whether genes in the putative nark3-sirB operon were transcribed as a single transcript, 400ng cDNA generated from RNA as described above was PCR amplified using primers that went across gene junctions (primers are listed in Supplementary Table S1) and analyzed by agarose gel electrophoresis.
Mycobacterium smegmatis containing M-PFC constructs were grown to mid-log phase in LB 0.05% Tween 80 supplemented with hygromycin B 50 µg/ml and kanamycin 50 µg/ml were diluted to 0.0005 OD in same media. Cultures were then incubated in a 96-well plate for 48 hours with 2-fold serial dilutions of trimethoprim ranging from 200-3.125 µg/ml. After 48 hours 0.01% resazurin was added to wells and left to incubate for 4 hours. Data was collected using a Synergy 4 plate reader with fluorescence set to 530 nm excitation and 590 nm emission (Singh et al., 2006).
The ability for Mab to utilize nitrite was analyzed via colorimetric Griess reagent assay as described by Yang et., with minor modifications (Yang et al., 2016). On days 2 and 4, 50 µl of supernatant was added to a 96 well plate with 100 µl Griess Reagent (Thermo Fisher Scientific # 328670500) consisting of 1% sulphanilic acid in 5% phosphoric acid and 0.1% naphthylethylenediamine dihydrochloride. After plate was incubated at 37° C for 10 minutes absorbance was read at 535 nm on a Synergy 4 Plate reader. A standard curve with known nitrite concentrations was used to calculate concentrations of nitrite in supernatant.
Our previous study investigating Mab transcriptional modulation during hypoxic adaptation revealed dramatic down-regulation of a gene locus containing an uncharacterized transcriptional regulator (MAB_3520c) that appeared to be an orphan response regulator (ORR) linked to multiple nitrogen assimilation genes (MAB_3523c, MAB_3522c, MAB_3521c) (Simcox et al., 2023). The down-regulation of these genes in response to hypoxia contrasts with reported transcriptional adaptations Mtb to hypoxia (Wayne and Hayes, 1998; Sohaskey and Wayne, 2003; Sohaskey, 2008) prompting our interest in the role of this ORR and nitrogen assimilation in Mab. Bioinformatic analysis of MAB_3520c using UniProt and Alphafold structural modeling revealed a C-terminal OmpR/PhoB-type winged helix DNA binding domain commonly found in response regulators of two-component systems (Figure 1A). However, lack of an adjacent histidine kinase gene and an unusual N-terminal domain containing a HemD domain rather than a receiver domain with aspartate phosphorylation sites indicated a unique ORR. This atypical ORR is orthologous to the NnaR response regulator of S. coelicolor, Msm and Mtb with 58%, 55%, and 54% sequence identity, respectively. Alignments between these organisms display homology between the C-terminal DNA binding domain of helix 3 and the unique HemD domain residues (Figure 1B). Due to homology within the DNA binding domain between organisms and published data documenting S. coelicolor, Msm, and Mtb NnaR binding motifs, we were able to justify using these orthologous motifs to identify Mab NnaR promoter binding sites (Amin et al., 2012; Antczak et al., 2018).
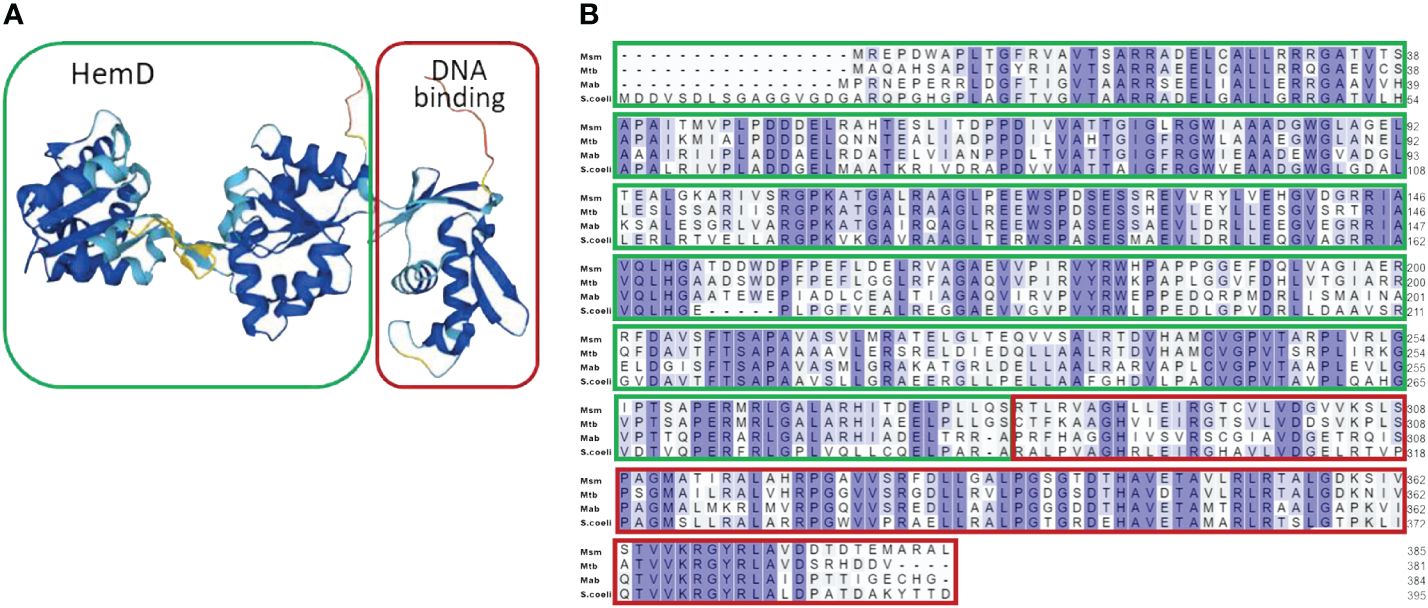
Figure 1 Bioinformatic analysis of NnaR domain structure and sequence alignment. Uniprot Bioanalysis Online Tool was used to generate the (A) Alpha-fold model of 3-D structure of Mab NnaR HemD-ORR protein. and (B) sequence alignment of Mab NnaR (MAB_3520c) compared to Msm NnaR (MSMEG_0432), Mtb (Rv0260c) and S. coelicolor NnaR (SCO2958). The N-terminal HemD domain is outlined in green, and C-terminal DNA-binding domain in red. Sequences for Mab, Msm and Mtb were obtained from the online data base Mycobrowser. S. coelicolor sequence was obtained from the online KEGG data base.
Initially, we sought to leverage previously published NnaR binding sites to define a putative NnaR regulon in Mab via in silico analysis of the Mab ATCC_19977 genome using the DNA Pattern Find Tool (Amin et al., 2012). Using either the motif (CTCAC[A/C][Cg].[13-16bp].GTGAG[CG][GA] based on predicted S. coelicolor and Mtb promoters or (CTCAC[A/C].16bp.[T/GGTGAG) reported by Antczak et al. for Msm (Antczak et al., 2018), only two predicted binding sites for Mab NnaR were identified. These NnaR promoters were positioned 63 bp and 66 bp upstream of the translational start sites of narK3 and MAB_2438, respectively. MAB_2438 encodes a previously uncharacterized protein annotated as a possible oxidoreductase (Figure 2A). Subsequent bioinformatic analysis of MAB_2438 revealed 61% identity (71% similarity) with the recently described NADPH-dependent assimilatory NR Msm NasN encoded by MSMEG_4206, which had been erroneously annotated as a pseudogene due to an unconfirmed frame-shift mutation (Tan et al., 2020). Based on this observation, we will refer to the product of MAB_2438 as NasN, which we noted was significantly shorter than its ortholog in Msm (MSMEG_4206 = 1351aa, MAB_2438 = 1257aa). Primary amino acid sequence alignments of the two NasN orthologs (Supplementary Figure S1) revealed two reasons for this difference: 1) MSMEG_4206 contains a ~75aa longer linker between the N-terminal nitrate reduction catalytic domain and C-terminal diflavin reductase domain (Tan et al., 2020), and 2) the annotated start codon for MAB_2438 is likely incorrect. We predict that NasNMab contains an additional 25 aa on the N-terminus based on homology with NasNMsm and the presence of a critical [4Fe-4S] binding site. NnaR-mediated regulation of nitrate assimilation would be unique to Mab given the absence of NnaR binding sites upstream of Msm nasN as previously mentioned (Tan et al., 2020). We next sought to verify whether genes nark3-nirBD-nnaR-sirB are co-transcribed as an operon via RT-PCR using intergenic primers amplifying the 3’ end of one gene and the 5’ end of the next gene. Amplification of junctions between nark3-nirB, nirB-nirD, nirD-nnaR and nnar-sirB all yielded the expected bands in the DNA (positive control) and cDNA lanes, with no bands present in the RNA (no RT negative controls) lanes (Figure 2B). These data confirm that the five-gene narK3-sirB locus positioned downstream of a predicted NnaR binding site is expressed as a single mRNA transcript. This represents a species-specific organization of genes involved in nitrate/nitrite assimilation, compared to Mtb or Msm in which these genes are not arranged in a single operon (Supplementary Figure S2).
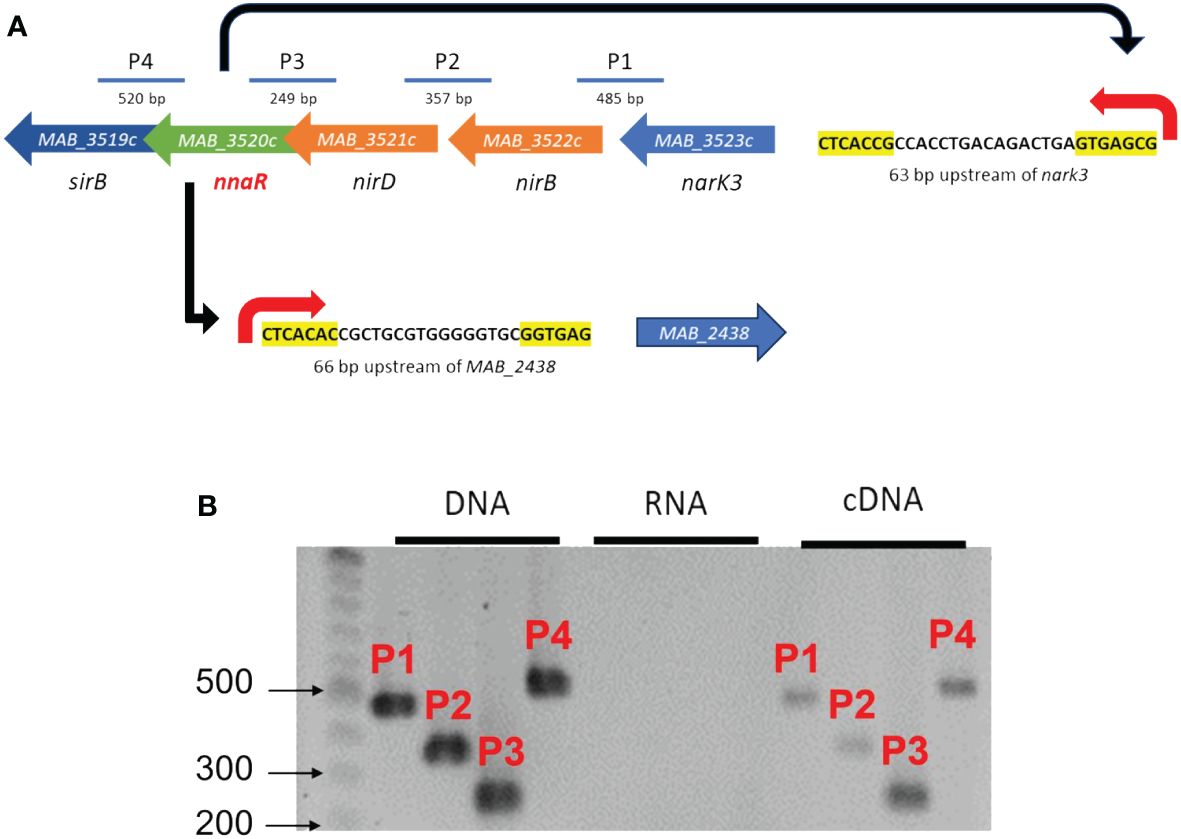
Figure 2 Genetic organization of the NnaR operon and regulon. (A) Schematic depicting nitrogen assimilation gene arrangement and NnaR binding sites discovered using DNA Pattern Find. Yellow highlighted nucleotides coincide with motifs used to find promoter region. P1-P4 indicates intergenic amplicons for RT-PCR. (B) Agarose gel of RT-PCR products P1-P4 amplified from DNA (positive control), RNA (negative control for DNA contamination), or cDNA.
We next investigated transcription of the nark3-sirB operon and nasN in the presence of nitrate or nitrite as sole nitrogen sources via qRT-PCR (Figures 3A, B). WT Mab390S cultures were supplemented with either 2.5 mM ammonium sulfate (AS) (control), 2.5 mM of sodium nitrate, or 0.5 mM of sodium nitrite in Sauton’s minimal media for 4 hours prior to RNA extraction. Sodium nitrite was used at a lower concentration due to toxicity at concentrations equivalent to AS and sodium nitrate (Supplementary Figure S3). qRT-PCR confirmed a 1-2 log induction of all genes within the nark3-sirB operon and nasN in the presence of nitrate (Figure 3A) and nitrite (Figure 3B) compared to the AS control, which bypasses the first two-steps of nitrate assimilation. These studies demonstrated the coordinated induction of the predicted NnaR regulon, including the nark3-sirB operon as well as nasN, specifically when nitrate or nitrite (compared to ammonium sulfate) are utilized as sole nitrogen sources.
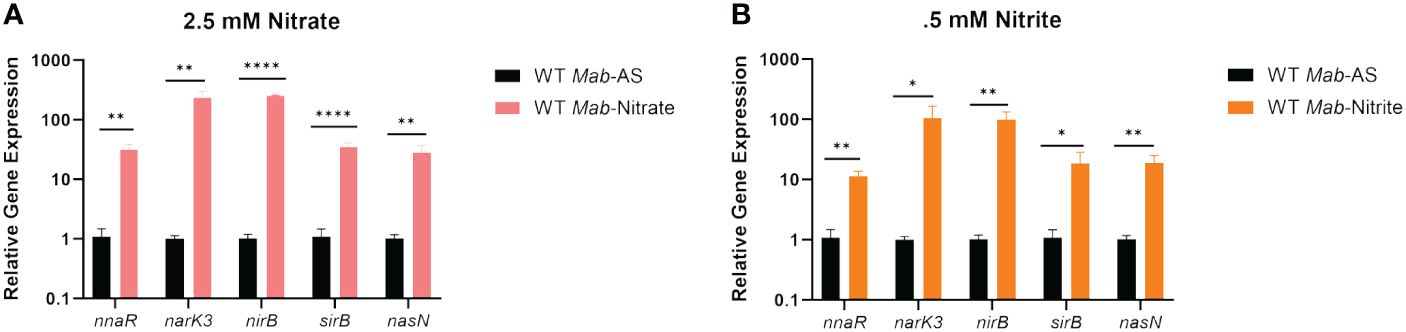
Figure 3 The NnaR regulon is induced when nitrate or nitrite are the sole sources of nitrogen. qRT-PCR was performed to confirm induction of nnaR regulon when nitrate (A) or nitrite (B) are the sole nitrogen sources. Mab390S supplemented with 2.5 mM ammonium sulfate (black bars), 2.5 mM nitrate (pink bars) and 0.5mM nitrite (orange bars). qRT-PCR data is representative of 3 experiments performed in duplicate. P values were calculated via t-test using GraphPad. *P-value <0.05, **P-value <0.01 ****P-value <0.0001.
To further investigate the role of NnaR, a Mab nnaR knockout strain (MabΔnnaR) was generated via recombineering (van Kessel and Hatfull, 2007). Anticipating polar effects, a corresponding complement strain (MabΔnnaR+C) containing both nnaR and sirB constitutively expressed from the hsp60 promoter in the episomal vector pVV16 was constructed (van Kessel et al., 2008; Parikh et al., 2013). In addition to PCR (Supplementary Figure S4) and sequencing, qRT-PCR was used to confirm the knockout and complement via expression levels of nnaR and the effect on genes within the predicted nnaR regulon (Figure 4A). Data from log-phase cultures in 7H9 media confirmed nnaR transcripts were not detectable in MabΔnnaR with restoration to WT levels in MabΔnnaR+C. Consistent with the predicted role for NnaR as an activator of its own operon plus nasN, we expected loss of NnaR to lead to downregulation of its regulon. Surprisingly, gene expression levels for narK3, nirBD and nasN in all three strains grown in 7H9 media were unaltered in the absence of NnaR, despite the presence of a NnaR binding site upstream nark3 and nasN. Closer analysis of qRT-PCR raw data showed high Ct values for all genes, indicating minimal expression of these genes under nitrogen replete conditions. This observation is consistent with Msm NnaR studies which found no difference in the expression levels of narK and nirB in the absence of NnaR when grown in nitrogen replete media (Antczak et al., 2018). In contrast to WT levels of nark3, nirB and nasN, sirB exhibited significant ~3-log up-regulation in the ΔnnaR mutant strain, which was restored to WT levels in the complement strain. Thus, our data indicate that the basal expression of nnaR and associated nitrate/nitrite assimilation genes is low in WT Mab under nitrogen replete conditions such that the absence of NnaR had little effect in gene expression. The unusual expression pattern of sirB, which appears to be cotranscribed with the narK3-sirB operon but suppressed by NnaR, points to the possibility of additional internal promoters and/or additional layers of regulation acting on sirB.
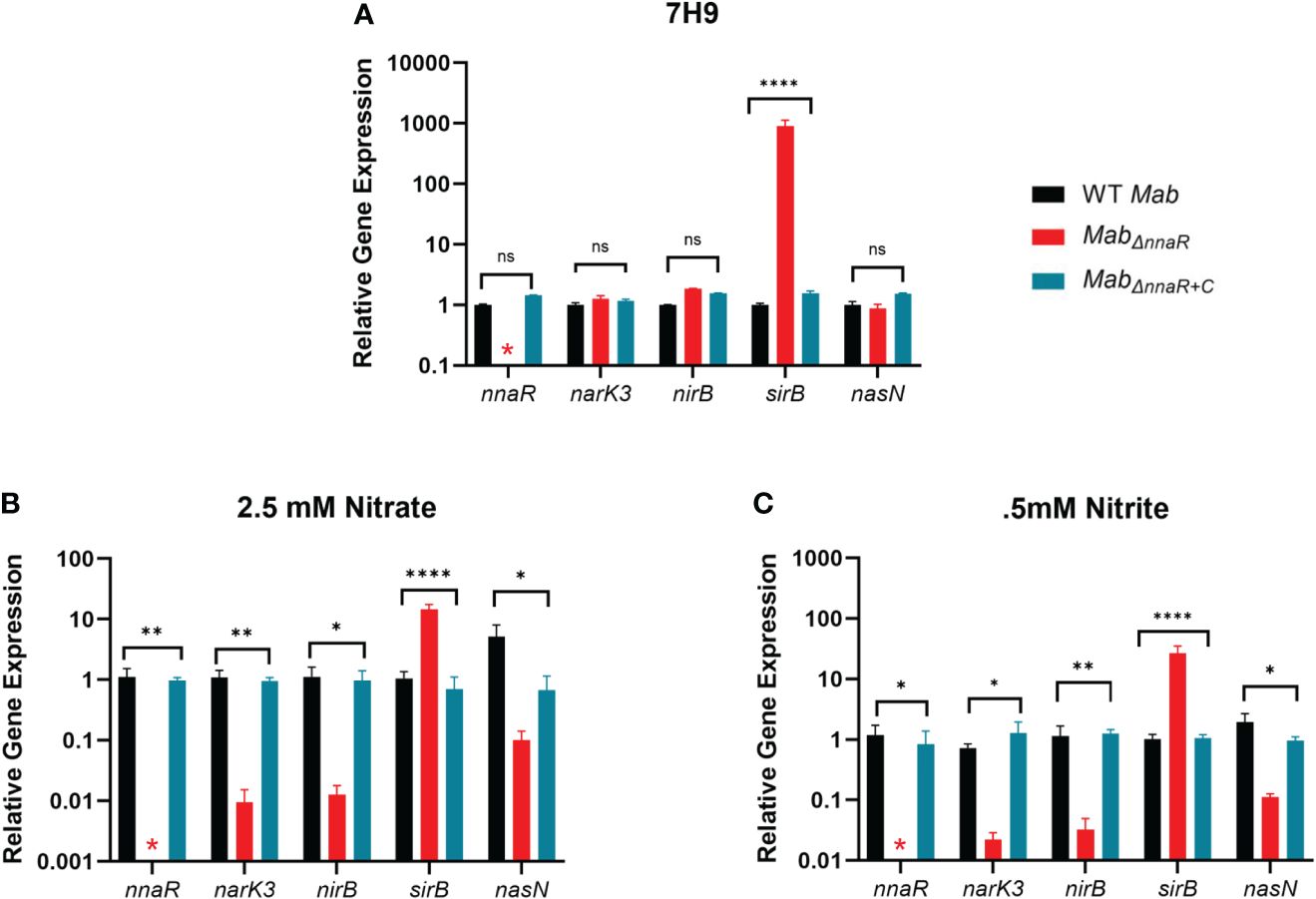
Figure 4 NnaR regulates the nark3-sirB operon and nasN. qRT-PCR was performed to confirm the deletion and restoration of nnaR in MabΔnnaR and MabΔnnaR+C, respectively, and effect on predicted regulon in 7H9 media (A), in minimal media supplemented with nitrate (B) or nitrite (C). WT Mab 390S (black), MabΔnnaR (red) and MabΔnnaR+C (blue). Data is representative of 3 experiments performed in duplicate. P values were calculated via one-way ANOVA using GraphPad. Red stars indicate Ct values were not detected for Mab nnaR in the mutant strain. *P-value <0.05, **P-value <0.01 ****P-value <0.0001. ns denotes samples that were not significantly different than WT.
Since the absence of NnaR had no effect on gene expression levels in nitrogen rich media, we evaluated gene expression between Mab390S and MabΔnnaRunder inducing conditions with nitrate or nitrite as the sole nitrogen sources. Briefly, cultures were incubated in minimal media supplemented with 2.5 mM AS, 2.5 mM sodium nitrate or 0.5 mM sodium nitrite for 4hr prior to RNA isolation and qRT-PCR analysis. During growth on defined inorganic nitrogen sources, the absence of NnaR resulted in a significant down-regulation (≥ 1 log) or lack of induction of nark3, nirB and nasN, with restoration to wild-type levels in MabΔnnaR+C (Figures 4B, C). Consistent with our observations in 7H9 media, sirB transcript levels in MabΔnnaRremained elevated in nitrate or nitrite supplemented cultures, with restoration to WT levels in MabΔnnaR+C. As noted above, this lack of correlation between the transcription profiles of sirB and upstream genes appears inconsistent with their co-transcription as part of an operon. Decreased transcription of nark3, nirBD, and nasN in the mutant strain under inducing conditions affirms that NnaR mediates upregulation of its regulon in response to nitrate and nitrite. Induction of a putative assimilatory nitrate reductase nasN represents a species-specific role for NnaRMab since NnaRMsm does not appear to regulate nasN (Tan et al., 2020). This data provides the first insights regarding regulation of nitrogen assimilation for Mab outside of what has been previously published for GlnR (Fan et al., 2024).
Based on our data revealing NnaR-dependent upregulation of genes linked to nitrogen metabolism, we next investigated whether the absence of NnaR affects growth when nitrate or nitrite are the sole sources of nitrogen. Cultures of Mab390S, MabΔnnaR, MabΔnnaR+C were grown in minimal media or minimal media supplemented with 2.5 mM AS, 2.5 mM sodium nitrate or 0.5 mM sodium nitrite, with CFUs enumerated daily through day 4 (Figure 5). The slight initial increase in CFU of Mab390S cultures supplemented with AS which was sustained until day 4, in contrast to the ~4-log decline in CFU observed in minimal media alone (Figure 5A), affirms that Mab can utilize AS as a sole nitrogen source. Notably, the growth of MabΔnnaR in AS was indistinguishable from WT (Figure 5A) indicating that NnaR is not required when the first two-steps of nitrate assimilation (nitrate>nitrite>ammonium) are bypassed by providing ammonium as the sole nitrogen source (Cardoso et al., 2021). When supplemented with nitrate or nitrite, on the other hand, the WT and complement either maintained a steady number of CFU (nitrate) or decreased slightly (nitrite) (Figures 5B, C). We suspect the slight decline in CFU for nitrite supplemented cultures of strains expressing NnaR is due to nitrite toxicity resulting in growth inhibition, a phenomenon also observed in Mtb (Cunningham-Bussel et al., 2013). As predicted, the survival of MabΔnnaR was severely impaired when nitrate was the only nitrogen source, with a final CFU difference of ~3 logs on day 4 (Figure 5B). This verified the importance of NnaR in nitrate assimilation. The dramatic MabΔnnaR phenotype observed on day 4 with nitrate supplementation was less pronounced with nitrite supplementation (Figure 5C). However, a log difference was observed on day 2 and 3 when comparing MabΔnnaR to WT and complement with nitrite as the sole nitrogen source (Figure 5C). Due to decreased CFU from day 3 to day 4 in WT and complement in nitrite and a plateau in CFU of MabΔnnaR during the same period, there was only a difference of 0.5 log in CFU by the end of the assay (Figure 5C). Although the reason why loss of NnaR had a less deleterious effect on growth/survival of MabΔnnaR on nitrite versus nitrate is unclear, it is evident that NnaR plays a critical role in nitrogen assimilation and/or nitrite detoxication when nitrite is the sole nitrogen source available. To biochemically validate the role of NnaR in nitrite utilization, we compared the ability of WT, MabΔnnaR, and MabΔnnaR+C to use nitrite as a nitrogen source via Griess assay. Briefly, nitrite concentrations in supernatants taken from the growth kinetic study were measured on day 2 and day 4 to gauge nitrite utilization by Mab strains. Nitrite concentrations in the supernatant of MabΔnnaR were significantly higher than WT and complement at both day 2 and 4 (Figure 5D), affirming a reduction in the ability to metabolize nitrite in the absence of NnaR. Further studies are required to elucidate the biochemical functions and physiological roles of NnaR regulated enzymes in Mab nitrogen metabolism.
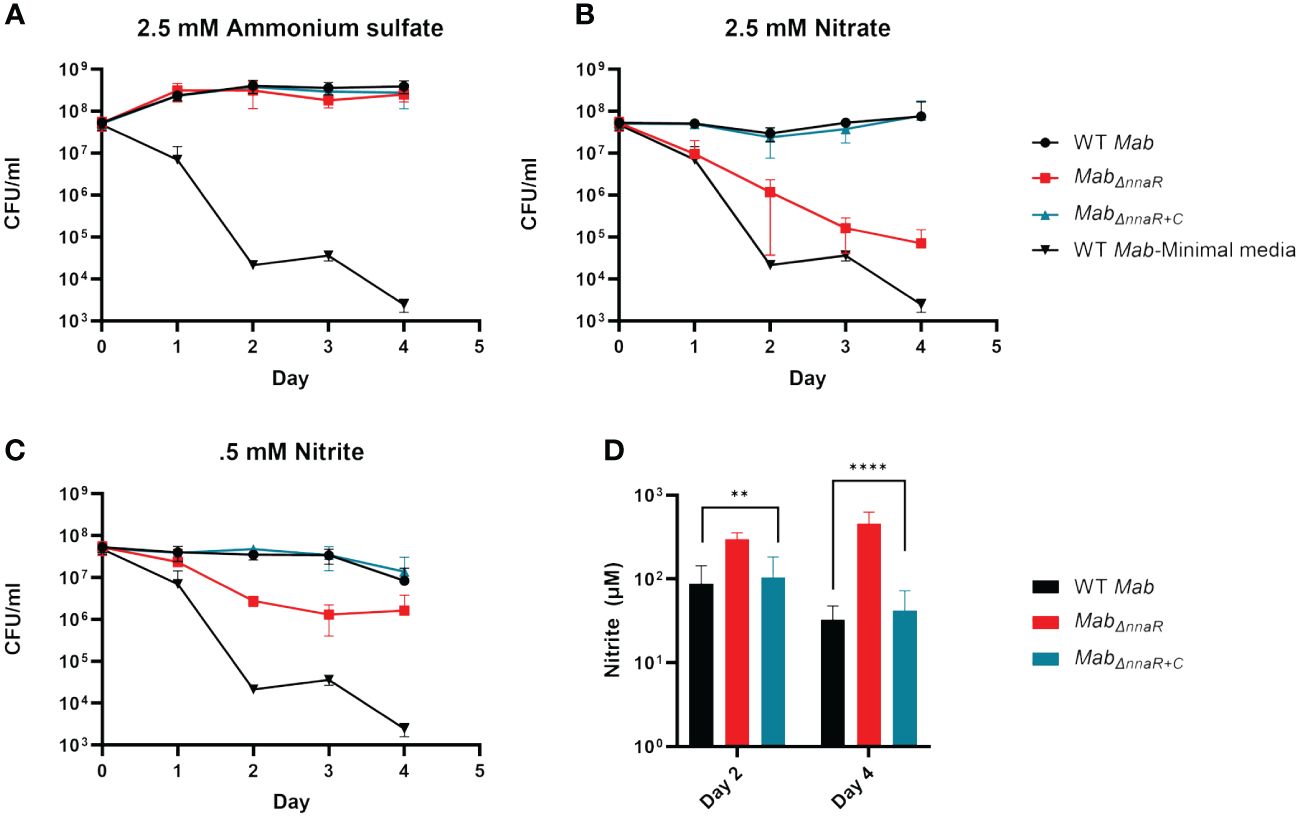
Figure 5 NnaR is responsible for nitrite utilization and growth when nitrate and nitrite are sole sources of nitrogen. Growth kinetics in minimal media supplemented with various nitrogen sources was assessed via serial dilutions, spot plating and enumeration of CFU/ml on days 0-4. Cultures supplemented with (A) ammonium sulfate, (B) nitrate or (C) nitrite. WT Mab (black circle), MabΔnnaR (red square), MabΔnnaR+C (blue triangle), WT Mab-Minimal Media (serving as negative control-no nitrogen supplementation, black upside down triangle). Griess assay (D) was performed to measure nitrite accumulation on day 2 and 4. Mab 390S (black bars), MabΔnnaR (red bars) and MabΔnnaR+C (blue bars). Data reflects 3 independent experiments performed in triplicate. ** P-value <0.01, **** P-value <0.001.
In summary, we have identified an orphan response regulator designated NnaR based on homology with recently characterized ortholog in Msm, its unusual domain structure (fusion of HemD domain with DNA binding domain), and critical role in Nitrate/Nitrite Assimilation Regulation (NnaR). It is encoded in a species-specific operon that is downregulated by hypoxia (Simcox et al., 2023) but highly induced by inorganic nitrogen sources, specifically nitrate and nitrite. Our data indicated that NnaRMab activates transcription of a six gene regulon responsible for reduction of nitrate to nitrite (NasN) and nitrite to ammonia (NirBD), transport of nitrate and/or nitrite (NarK3), and synthesis of the siroheme cofactor for nitrite reductase. Finally, NnaR is required for the utilization of nitrate and nitrite as sole nitrogen sources and detoxification of nitrite, suggesting an important role for this transcription factor in Mab host adaptation and pathogenesis.
Mab persistence within the hostile niches of macrophage phagosomes, granulomas, and mucus-laden CF lung requires intricate gene regulation to combat low oxygen tension, reactive nitrogen intermediates (RNI), nutrient depletion, and exposure to excess nitrate and nitrite. Given the capacity of Mtb to adapt to similar niches and presence of many orthologous virulence factors and transcriptional regulators, it is tempting to extrapolate from available Mtb data to understand Mab gene regulation and function. However, emerging data from our lab and others highlights significant species-specific differences in transcriptional regulatory networks and strategies of adapting to in vivo conditions that urge caution when doing this. Unique strategies of Mab host adaptation and persistence stem from its distinct repertoire of genes compared to Mtb (Wee et al., 2017), variable genomic arrangements of the same genes, as well as shared orthologous transcription factors that control species-specific regulons (Gerasimova et al., 2011; Miranda-CasoLuengo et al., 2016; Bryant et al., 2021; Simcox et al., 2023).
In Mtb, the overlap and integration of genes involved in adaptation to hypoxia and nitrate metabolism, not only as a means of alternative respiration but also to defend against host generated RNI, are well-documented (Sohaskey, 2008; Malm et al., 2009; Akhtar et al., 2013; Gouzy et al., 2014; Vilcheze et al., 2022). On the other hand, relatively little is known about mechanisms used by Mab to coordinate responses to these two intertwined host cues. Our interest in nitrate assimilation in Mab and the interplay with hypoxia was sparked by our recent observation that genes implicated in nitrate assimilation are downregulated under hypoxic conditions (Simcox et al., 2023). This data contrasts with the response of Mtb to hypoxia, including upregulation of narK2 and nirBD (Sohaskey, 2008; Malm et al., 2009; Akhtar et al., 2013; Gouzy et al., 2014), which facilitates utilization of nitrate as a terminal electron acceptor via reduction by the constitutively expressed narGHIJ, which acts as both a respiratory and assimilatory nitrate reductase (NR) (Sohaskey and Wayne, 2003; Malm et al., 2009; Gouzy et al., 2014). The resulting nitrite produced by NR could then be extruded by the putative nitrite transporter NarK2 to avoid toxicity or be further reduced to ammonium via the nitrite reductase nirBD under nitrogen-limited conditions (Malm et al., 2009; Gouzy et al., 2014). This strategy is not an option for Mab, due to the conspicuous lack of a NarGHJI-like respiratory NR. Our recently reported RNAseq analysis of Mab hypoxia adaption showed down-regulation of two response regulators implicated in nitrate assimilation, MAB_0744 (glnR) and MAB_3520c (nnaR), in addition to the five additional genes of the NnaR regulon (this study), which may serve to reduce protein synthesis and aid transition to slowed growth under stressful conditions (Simcox et al., 2023).
In addition to species-specific regulation of the nitrate metabolism apparatus under hypoxic conditions, the genomic organization of genes predicted to be involved in this process is unique in Mab. The genes encoding proteins required for nitrate/nitrite transport (NarK3), reduction of nitrite to ammonia (NirBD), an ORR associated with nitrate assimilation (NnaR), and synthesis of the siroheme cofactor for NirBD (SirB) are clustered in the MAB_3523c-MAB_3519c locus, with the nasN NiR gene located elsewhere. The overlap between open reading frames of nirB-nnaR and nnaR-sirB suggest close translational coupling of these components (Huber et al., 2019). All five genes appear to be transcribed as a single unit, as evidenced by our RT-PCR analysis and tight co-regulation under hypoxia and nitrate/nitrite supported growth (Figures 3A, B). However, we cannot rule out the possibility of additional transcripts of subsets of genes from internal promoters. While narK3, nnaR, and sirB are arranged contiguously in Mtb and Msm, the nirBD operon is upstream and oriented divergently and nitrate reductases (narGHJI-Mtb and nasN-Msm) are located elsewhere (Supplementary Figure S2). Following identification of an uncharacterized orphan response regulator designated NnaR embedded within this operon, our focal point became elucidating its role in Mab gene regulation and nitrate assimilation.
In silico analyses identified only 2 putative NnaR binding sites in the Mab genome - upstream of nark3-sirB operon and nasN (Figure 2A) – suggesting a small regulon of genes with focused roles on nitrate and nitrite metabolism. This is comparable to the small repertoire of genes reportedly controlled by the S. coelicolor ortholog shown to regulate narK, nirAB (NiR), and nasA (assimilatory NR) (Amin et al., 2012). In Msm, NnaR controls a slightly larger geneset comprised of seven mRNA transcripts (four single genes, 3 operons), however only 3 target loci (narK3, nirBD, nasN) have corresponding orthologs in Mab (Antczak et al., 2018). The coordinated induction of the nark3-sirB operon and nasN during growth on inorganic nitrogen sources is consistent with regulation by the same transcription factor, NnaR. It is also worth noting that the dramatic ~20- to 200-fold induction of the NnaR regulon (Figures 4B, C) is in reference to equimolar concentrations of nitrogen in the form of ammonium sulfate. This would suggest that NnaR-mediated transcriptional activation is not in response to low nitrogen levels overall but is somehow specific to nitrate and nitrite. Based on these observations, the lack of induction of nasN, nark3, and nirBD in MabΔnnaR during growth on nitrate or nitrite was not surprising. However, the upregulation of sirB in the absence of nnaR in both nitrogen replete and deplete conditions was unexpected (Figure 4). This is inconsistent with sirB being expressed only by NnaR-activated co-transcription with the four upstream genes. An initial hypothesis we considered was upregulation due to read-through from the promoter of the apramycinR cassette used to inactivate nnaR. However, restoration of sirB to wild-type levels when both nnaR and sirB were constitutively expressed in trans argues against this. If anything, an additional copy of sirB on the complement construct (included because we anticipated a polar negative effect) was expected to boost sirB expression in the complement, not restore levels to normal. It is possible that deletion of nnaR disrupts the proposed translational coupling with sirB (despite care taken not to delete the overlapped 5’ end of sirB) such that diminished SirB activity triggers compensatory upregulation of sirB. SirB is a ferrochelatase responsible for catalyzing the last step of siroheme synthesis, a cofactor required for the activity of nitrite reductase (NirBD) and sulfite reductases (Tripathy et al., 2010; Dailey et al., 2017; Pennington et al., 2020). This would require a second internal promoter able to drive expression of sirB alone (or with additional upstream genes since putative promoter cannot be within nnaR) activated by a transcription factor that is induced by NnaR. More research will be needed to understand this unexpected regulation of sirB and the role of NnaR.
The mechanism by which NnaR senses nitrate and nitrate, either directly or indirectly, and becomes activated to promote transcription of its regulon remains a mystery. No studies of the better characterized orthologs in S.coelicolor, Msm, or Mtb have addressed this question. We initially hypothesized that NnaR activation may be mediated by DosS, the sensor histidine kinase (HK) partner of the response regulator (RR) DosR, known to sense hypoxia, NO, and CO in Mtb (Voskuil et al., 2003, 2004; Shiloh et al., 2008; Sivaramakrishnan and de Montellano, 2013; Zheng et al., 2017). This was based on a report by Gautam et al. of potential protein-protein interactions (PPI) between DosS and the Mtb NnaR ortholog Rv0260 (Gautam et al., 2019). Using the same M-PFC (Mycobacterial Protein Fragment Complementation) method (Singh et al., 2006), we were able to detect Mab DosS-DosR PPI, but were unable to detect an interaction between Mab DosS and NnaR (Supplementary Figure S5). This is perhaps not surprising given the other species-specific differences we have noted between Mtb and Mab in their responses to hypoxia and nitrogen limitation. Further in silico analysis also failed to identify a receiver domain with aspartate residues likely to serve as phosphorylation sites (data not shown) suggesting NnaR is not likely to be activated by a non-cognate HK. Alternative hypotheses include NnaR activation by serine/threonine kinases as observed with the ORR GlnR (Hong et al., 2007; Lin et al., 2014; Amin et al., 2016), or self-activation via the unique HemD domain. The domain architecture of NnaR consisting of an N-terminal HemD domain (also known as a uroporphyrinogen III synthase) fused to a C-terminal OmpR DNA binding domain is restricted to Actinomycetes (Amin et al., 2012). Uroporphyrinogen III synthase is responsible for the cyclization of hydroxymethylbilane (HMB) which forms uroporphyrinogen III (UroPIII), a precursor for vitamin B12 and siroheme. It is worth noting that the HemD domain of NnaR in S.coelicolor is catalytically inactive (Schubert et al., 2008; Tripathy et al., 2010; Amin et al., 2012; Dailey et al., 2017). The role of the NnaR HemD domain remains unclear but it is plausible it acts as a sensing domain for HMB or UroPIII and self-activates resulting in conformational changes in the DNA-binding domain as evidenced by the self-modulating RRs, JadR1 and RedZ of streptomycetes (Gao et al., 2019).
This study provides clear evidence that Mab utilizes species-specific strategies of nitrogen assimilation when nitrate or nitrite are the only sources of nitrogen and employs the ORR NnaR in distinct ways compared to Msm and Mtb. This is illustrated by confirmation of a unique operon containing genes narK3-nirBD-nnaR-sirB, regulation of this operon and nasN by NnaR, and the importance of NnaR for survival when nitrate or nitrite are the sole sources of nitrogen. There are still unanswered questions regarding the regulation of nitrogen metabolism in Mab, and the role of NnaR in particular, that remain to be addressed. One key knowledge gap is the degree of regulatory overlap and interaction with GlnR, which typically controls larger regulons involved in nitrogen metabolism and many other processes. Although Fan et al. did report that Mab GlnR regulates 8 genes associated with nitrogen metabolism, they only queried 9 promoters by ChIP-qPCR (Fan et al., 2024). The full extent of the GlnR regulon in Mab remains to be determined. Other relevant questions include how hypoxia triggers down-regulation of the NnaR regulon, what is the function of the HemD domain, what underlies the unusual regulation of sirB, and what signaling mechanisms contribute to NnaR activation. These data revealing a species-specific role for Mab NnaR and its significance in nitrate assimilation establishes a premise for future studies exploring its role in persistence within host environments such macrophage phagosomes, granulomas, and mucus-laden airways affected by CF.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
BS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. KR: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. BS acknowledges funding from a Doctoral Support Award from the University of Central Florida. Article processing charges were provided in part by the UCF College of Graduate Studies Open Access Publishing Fund.
We would like to thank Dr. Rashmi Gupta, Dr. Mollie Jewett, and Dr. Hubert Salvail for technical support and guidance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1411333/full#supplementary-material
Supplementary Figure 1 | NasN sequence alignment. Uniprot sequence amino acid sequence alignment of NasN orthologs MSMEG_4206 and MAB_2438. Conserved amino acids are highlighted in blue. The annotated versus predicted translational start sites for MAB_2438 NasN are indicated by red arrows.
Supplementary Figure 2 | Nitrate assimilation gene organization in Mab, Mtb, and Msm.
Supplementary Figure 3 | Growth kinetics of Mab390S at different concentrations of inorganic nitrogen sources. Growth kinetics were measured by OD600nm on day 4 of growth in minimal media supplemented with (A) 2.5 mM, 5 mM, and 10 mM ammonium sulfate (B) 0.5 mM, 1 mM, 2.5 mM sodium nitrite (C) 2.5 mM, 5 mM, and 10 mM sodium nitrate.
Supplementary Figure 4 | PCR confirmation of nnaR mutant. PCR was used to screen for nnaR mutants via amplification of NnaR or Apramycin resistance cassette plus upstream and downstream flanking region. L1) DNA ladder; L2) WT Mab, 2287 bp amplicon; L3) negative clone; L4) ΔnnaR mutant, 2071 bp amplicon.
Supplementary Figure 5 | MPFC assay displays lack of interaction between NnaR and DosS. Minimum inhibitory concentration of trimethoprim (TRIM) was analyzed to measure interaction of DHFR1,2 and DHFR3 indicative of PPI. Black circle) pUAB100-dosS-(DHFR1,2 on C-term):pUAB200-dosR-(DHFR3 on C-term) Red square) pUAB100-dosS-(DHFR1,2 on C-term):pUAB200-nnaR-(DHFR3 on C-term) Turquoise triangle) pUAB100-dosS-(DHFR1,2 on C-term):pUAB400-nnaR-(DHFR3 on N-term) Blue triangle) pUAB300-GCN4-(DHFR1,2 on C-term):pUAB400-GCN4-(DHFR3 on N-term) Gray Diamond) empty vector.
Akhtar, S., Khan, A., Sohaskey, C. D., Jagannath, C., Sarkar, D. (2013). Nitrite reductase NirBD is induced and plays an important role during in vitro dormancy of Mycobacterium tuberculosis. J. Bacteriol 195, 4592–4599. doi: 10.1128/JB.00698-13
Amin, R., Franz-Wachtel, M., Tiffert, Y., Heberer, M., Meky, M., Ahmed, Y., et al. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: A novel regulatory aspect of the global nitrogen response regulator glnR in S. coelicolor M145. Front. Mol. Biosci. 3, 38. doi: 10.3389/fmolb.2016.00038
Amin, R., Reuther, J., Bera, A., Wohlleben, W., Mast, Y. (2012). A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiol. (Reading) 158, 1172–1182. doi: 10.1099/mic.0.054817-0
Antczak, M., Plocinska, R., Plocinski, P., Rumijowska-Galewicz, A., Zaczek, A., Strapagiel, D., et al. (2018). The NnaR orphan response regulator is essential for the utilization of nitrate and nitrite as sole nitrogen sources in mycobacteria. Sci. Rep. 8, 17552. doi: 10.1038/s41598-018-35844-z
Belardinelli, J. M., Verma, D., Li, W., Avanzi, C., Wiersma, C. J., Williams, J. T., et al. (2022). Therapeutic efficacy of antimalarial drugs targeting DosRS signaling in Mycobacterium abscessus. Sci. Transl. Med. 14, eabj3860. doi: 10.1126/scitranslmed.abj3860
Bernut, A., Nguyen-Chi, M., Halloum, I., Herrmann, J. L., Lutfalla, G., Kremer, L. (2016). Mycobacterium abscessus-induced granuloma formation is strictly dependent on TNF signaling and neutrophil trafficking. PloS Pathog. 12, e1005986. doi: 10.1371/journal.ppat.1005986
Brown-Elliott, B. A., Wallace, R. J., Jr. (2002). Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15, 716–746. doi: 10.1128/CMR.15.4.716-746.2002
Bryant, J. M., Brown, K. P., Burbaud, S., Everall, I., Belardinelli, J. M., Rodriguez-Rincon, D., et al. (2021). Stepwise pathogenic evolution of Mycobacterium abscessus. Science 372. doi: 10.1126/science.abb8699
Byrd, T. F., Lyons, C. R. (1999). Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 67, 4700–4707. doi: 10.1128/IAI.67.9.4700-4707.1999
Cardoso, N. C., Papadopoulos, A. O., Kana, B. D. (2021). Mycobacterium smegmatis does not display functional redundancy in nitrate reductase enzymes. PloS One 16, e0245745. doi: 10.1371/journal.pone.0245745
Chmiel, J. F., Davis, P. B. (2003). State of the art: why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir. Res. 4, 8. doi: 10.1186/1465-9921-4-8
Cunningham-Bussel, A., Zhang, T., Nathan, C. F. (2013). Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, E4256–E4265. doi: 10.1073/pnas.1316894110
Dailey, H. A., Dailey, T. A., Gerdes, S., Jahn, D., Jahn, M., O'Brian, M. R., et al. (2017). Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 81. doi: 10.1128/MMBR.00048-16
Esther, C. R., Jr., Esserman, D. A., Gilligan, P., Kerr, A., Noone, P. G. (2010). Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J. Cyst Fibros 9, 117–123. doi: 10.1016/j.jcf.2009.12.001
Fan, J., Jia, Y., He, S., Tan, Z., Li, A., Li, J., et al. (2024). GlnR activated transcription of nitrogen metabolic pathway genes facilitates biofilm formation by mycobacterium abscessus. Int. J. Antimicrob. Agents 63, 107025. doi: 10.1016/j.ijantimicag.2023.107025
Gao, R., Bouillet, S., Stock, A. M. (2019). Structural basis of response regulator function. Annu. Rev. Microbiol. 73, 175–197. doi: 10.1146/annurev-micro-020518-115931
Gautam, U. S., Mehra, S., Kumari, P., Alvarez, X., Niu, T., Tyagi, J. S., et al. (2019). Mycobacterium tuberculosis sensor kinase DosS modulates the autophagosome in a DosR-independent manner. Commun. Biol. 2, 349. doi: 10.1038/s42003-019-0594-0
Gerasimova, A., Kazakov, A. E., Arkin, A. P., Dubchak, I., Gelfand, M. S. (2011). Comparative genomics of the dormancy regulons in mycobacteria. J. Bacteriol 193, 3446–3452. doi: 10.1128/JB.00179-11
Giffin, M. M., Raab, R. W., Morganstern, M., Sohaskey, C. D. (2012). Mutational analysis of the respiratory nitrate transporter NarK2 of Mycobacterium tuberculosis. PloS One 7, e45459. doi: 10.1371/journal.pone.0045459
Gouzy, A., Poquet, Y., Neyrolles, O. (2014). Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 12, 729–737. doi: 10.1038/nrmicro3349
Hassett, D. J., Cuppoletti, J., Trapnell, B., Lymar, S. V., Rowe, J. J., Yoon, S. S., et al. (2002). Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Delivery Rev. 54, 1425–1443. doi: 10.1016/S0169-409X(02)00152-7
He, J., Kang, X., Wu, J., Shao, Z., Zhang, Z., Wu, Y., et al. (2023). Transcriptional self-regulation of the master nitrogen regulator glnR in mycobacteria. J. Bacteriol 205, e0047922. doi: 10.1128/jb.00479-22
Hong, E., Lee, H. M., Ko, H., Kim, D. U., Jeon, B. Y., Jung, J., et al. (2007). Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J. Biol. Chem. 282, 20667–20675. doi: 10.1074/jbc.M609104200
Huang, Q., Abdalla, A. E., Xie, J. (2015). Phylogenomics of mycobacterium nitrate reductase operon. Curr. Microbiol. 71, 121–128. doi: 10.1007/s00284-015-0838-2
Huber, M., Faure, G., Laass, S., Kolbe, E., Seitz, K., Wehrheim, C., et al. (2019). Translational coupling via termination-reinitiation in archaea and bacteria. Nat. Commun. 10, 4006. doi: 10.1038/s41467-019-11999-9
Hurst-Hess, K., Rudra, P., Ghosh, P. (2017). Mycobacterium abscessus whiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.01347-17
Jenkins, V. A., Barton, G. R., Robertson, B. D., Williams, K. J. (2013). Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genomics 14, 301. doi: 10.1186/1471-2164-14-301
Kelm, M. (1999). Nitric oxide metabolism and breakdown. Biochim. Biophys. Acta 1411, 273–289. doi: 10.1016/S0005-2728(99)00020-1
Khan, A., Sarkar, D. (2012). Nitrate reduction pathways in mycobacteria and their implications during latency. Microbiol. (Reading) 158, 301–307. doi: 10.1099/mic.0.054759-0
Lee, M. R., Sheng, W. H., Hung, C. C., Yu, C. J., Lee, L. N., Hsueh, P. R. (2015). Mycobacterium abscessus complex infections in humans. Emerg. Infect. Dis. 21, 1638–1646. doi: 10.3201/2109.141634
Li, C., Wen, A., Shen, B., Lu, J., Huang, Y., Chang, Y. (2011). FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11, 92. doi: 10.1186/1472-6750-11-92
Lin, W., Wang, Y., Han, X., Zhang, Z., Wang, C., Wang, J., et al. (2014). Atypical OmpR/PhoB subfamily response regulator GlnR of actinomycetes functions as a homodimer, stabilized by the unphosphorylated conserved Asp-focused charge interactions. J. Biol. Chem. 289, 15413–15425. doi: 10.1074/jbc.M113.543504
Lopeman, R. C., Harrison, J., Desai, M., Cox, J. A. G. (2019). Mycobacterium abscessus: environmental bacterium turned clinical nightmare. Microorganisms 7. doi: 10.3390/microorganisms7030090
Lundberg, J. O., Weitzberg, E., Cole, J. A., Benjamin, N. (2004). Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2, 593–602. doi: 10.1038/nrmicro929
Lyczak, J. B., Cannon, C. L., Pier, G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222. doi: 10.1128/CMR.15.2.194-222.2002
Malm, S., Tiffert, Y., Micklinghoff, J., Schultze, S., Joost, I., Weber, I., et al. (2009). The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiol. (Reading) 155, 1332–1339. doi: 10.1099/mic.0.023275-0
Martin, J. F., Liras, P. (2019). The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in streptomyces and related actinobacteria. Front. Microbiol. 10, 3120. doi: 10.3389/fmicb.2019.03120
Miranda-CasoLuengo, A. A., Staunton, P. M., Dinan, A. M., Lohan, A. J., Loftus, B. J. (2016). Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics 17, 553. doi: 10.1186/s12864-016-2868-y
Molina-Torres, C. A., Tamez-Pena, L., Castro-Garza, J., Ocampo-Candiani, J., Vera-Cabrera, L. (2018). Evaluation of the intracellular activity of drugs against Mycobacterium abscessus using a THP-1 macrophage model. J. Microbiol. Methods 148, 29–32. doi: 10.1016/j.mimet.2018.03.020
Olivier, K. N., Weber, D. J., Wallace, R. J., Jr., Faiz, A. R., Lee, J. H., Zhang, Y., et al. (2003). Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167, 828–834. doi: 10.1164/rccm.200207-678OC
Parikh, A., Kumar, D., Chawla, Y., Kurthkoti, K., Khan, S., Varshney, U., et al. (2013). Development of a new generation of vectors for gene expression, gene replacement, and protein-protein interaction studies in mycobacteria. Appl. Environ. Microbiol. 79, 1718–1729. doi: 10.1128/AEM.03695-12
Peddireddy, V., Doddam, S. N., Ahmed, N. (2017). Mycobacterial dormancy systems and host responses in tuberculosis. Front. Immunol. 8, 84. doi: 10.3389/fimmu.2017.00084
Pennington, J. M., Kemp, M., McGarry, L., Chen, Y., Stroupe, M. E. (2020). Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site. Nat. Commun. 11, 864. doi: 10.1038/s41467-020-14722-1
Robinson, J. L., Jaslove, J. M., Murawski, A. M., Fazen, C. H., Brynildsen, M. P. (2017). An integrated network analysis reveals that nitric oxide reductase prevents metabolic cycling of nitric oxide by Pseudomonas aeruginosa. Metab. Eng. 41, 67–81. doi: 10.1016/j.ymben.2017.03.006
Rodriguez-Garcia, A., Sola-Landa, A., Apel, K., Santos-Beneit, F., Martin, J. F. (2009). Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 37, 3230–3242. doi: 10.1093/nar/gkp162
Rohde, K. H., Abramovitch, R. B., Russell, D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2, 352–364. doi: 10.1016/j.chom.2007.09.006
Schubert, H. L., Phillips, J. D., Heroux, A., Hill, C. P. (2008). Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex. Biochemistry 47, 8648–8655. doi: 10.1021/bi800635y
Shiloh, M. U., Manzanillo, P., Cox, J. S. (2008). Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3, 323–330. doi: 10.1016/j.chom.2008.03.007
Simcox, B. S., Tomlinson, B. R., Shaw, L. N., Rohde, K. H. (2023). Mycobacterium abscessus DosRS two-component system controls a species-specific regulon required for adaptation to hypoxia. Front. Cell Infect. Microbiol. 13, 1144210. doi: 10.3389/fcimb.2023.1144210
Singh, A., Mai, D., Kumar, A., Steyn, A. J. (2006). Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U.S.A. 103, 11346–11351. doi: 10.1073/pnas.0602817103
Sivaramakrishnan, S., de Montellano, P. R. (2013). The dosS-dosT/dosR mycobacterial sensor system. Biosensors (Basel) 3, 259–282. doi: 10.3390/bios3030259
Sohaskey, C. D. (2008). Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol 190, 2981–2986. doi: 10.1128/JB.01857-07
Sohaskey, C. D., Modesti, L. (2009). Differences in nitrate reduction between Mycobacterium tuberculosis and Mycobacterium bovis are due to differential expression of both narGHJI and narK2. FEMS Microbiol. Lett. 290, 129–134. doi: 10.1111/fml.2008.290.issue-2
Sohaskey, C. D., Wayne, L. G. (2003). Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol 185, 7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003
Sola-Landa, A., Rodriguez-Garcia, A., Amin, R., Wohlleben, W., Martin, J. F. (2013). Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res. 41, 1767–1782. doi: 10.1093/nar/gks1203
Story-Roller, E., Maggioncalda, E. C., Cohen, K. A., Lamichhane, G. (2018). Mycobacterium abscessus and beta-Lactams: Emerging Insights and Potential Opportunities. Front. Microbiol. 9, 2273. doi: 10.3389/fmicb.2018.02273
Tan, W., Liao, T. H., Wang, J., Ye, Y., Wei, Y. C., Zhou, H. K., et al. (2020). A recently evolved diflavin-containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J. Biol. Chem. 295, 5051–5066. doi: 10.1074/jbc.RA120.012859
Tripathy, B. C., Sherameti, I., Oelmuller, R. (2010). Siroheme: an essential component for life on earth. Plant Signal Behav. 5, 14–20. doi: 10.4161/psb.5.1.10173
van Kessel, J. C., Hatfull, G. F. (2007). Recombineering in mycobacterium tuberculosis. Nat. Methods 4, 147–152. doi: 10.1038/nmeth996
van Kessel, J. C., Marinelli, L. J., Hatfull, G. F. (2008). Recombineering mycobacteria and their phages. Nat. Rev. Microbiol. 6, 851–857. doi: 10.1038/nrmicro2014
Vilcheze, C., Yan, B., Casey, R., Hingley-Wilson, S., Ettwiller, L., Jacobs, W. R., Jr. (2022). Commonalities of mycobacterium tuberculosis transcriptomes in response to defined persisting macrophage stresses. Front. Immunol. 13, 909904. doi: 10.3389/fimmu.2022.909904
Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713. doi: 10.1084/jem.20030205
Voskuil, M. I., Visconti, K. C., Schoolnik, G. K. (2004). Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 84, 218–227. doi: 10.1016/j.tube.2004.02.003
Wayne, L. G., Hayes, L. G. (1998). Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber Lung Dis. 79, 127–132. doi: 10.1054/tuld.1998.0015
Wee, W. Y., Dutta, A., Choo, S. W. (2017). Comparative genome analyses of mycobacteria give better insights into their evolution. PloS One 12, e0172831. doi: 10.1371/journal.pone.0172831
Williams, K. J., Jenkins, V. A., Barton, G. R., Bryant, W. A., Krishnan, N., Robertson, B. D. (2015). Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress. Mol. Microbiol. 97, 1142–1157. doi: 10.1111/mmi.13091
Worlitzsch, D., Tarran, R., Ulrich, M., Schwab, U., Cekici, A., Meyer, K. C., et al. (2002). Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325. doi: 10.1172/JCI0213870
Yang, K., Wu, Y., Xie, H., Li, M., Ming, S., Li, L., et al. (2016). Macrophage-mediated inflammatory response decreases mycobacterial survival in mouse MSCs by augmenting NO production. Sci. Rep. 6, 27326. doi: 10.1038/srep27326
Zheng, H., Colvin, C. J., Johnson, B. K., Kirchhoff, P. D., Wilson, M., Jorgensen-Muga, K., et al. (2017). Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 13, 218–225. doi: 10.1038/nchembio.2259
Keywords: Mycobacterium abscessus, nitrogen metabolism, nitrate, nitrite, orphan response regulator, NnaR
Citation: Simcox BS and Rohde KH (2024) Orphan response regulator NnaR is critical for nitrate and nitrite assimilation in Mycobacterium abscessus. Front. Cell. Infect. Microbiol. 14:1411333. doi: 10.3389/fcimb.2024.1411333
Received: 02 April 2024; Accepted: 10 May 2024;
Published: 24 May 2024.
Edited by:
Selvakumar Subbian, The State University of New Jersey, United StatesReviewed by:
Arshad Rizvi, Emory University, United StatesCopyright © 2024 Simcox and Rohde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle H. Rohde, S3lsZS5yb2hkZUB1Y2YuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.