
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 04 July 2024
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1410997
 Hyun Mi Kang1,2,3
Hyun Mi Kang1,2,3 Kyung Ran Kim4
Kyung Ran Kim4 Gahee Kim5
Gahee Kim5 Dong-gun Lee3,6
Dong-gun Lee3,6 Yae Jean Kim4
Yae Jean Kim4 Eun Hwa Choi1,7
Eun Hwa Choi1,7 Jina Lee5*†
Jina Lee5*† Ki Wook Yun1,7*†
Ki Wook Yun1,7*†Background: Acinetobacter baumannii (AB) has emerged as one of the most challenging pathogens worldwide, causing invasive infections in the critically ill patients due to their ability to rapidly acquire resistance to antibiotics. This study aimed to analyze antibiotic resistance genes harbored in AB and non-baumannii Acinetobacter calcoaceticus-baumannii (NB-ACB) complex causing invasive diseases in Korean children.
Methods: ACB complexes isolated from sterile body fluid of children in three referral hospitals were prospectively collected. Colistin susceptibility was additionally tested via broth microdilution. Whole genome sequencing was performed and antibiotic resistance genes were analyzed.
Results: During January 2015 to December 2020, a total of 67 ACB complexes were isolated from sterile body fluid of children in three referral hospitals. The median age of the patients was 0.6 (interquartile range, 0.1–7.2) years old. Among all the isolates, 73.1% (n=49) were confirmed as AB and others as NB-ACB complex by whole genome sequencing. Among the AB isolates, only 22.4% susceptible to carbapenem. In particular, all clonal complex (CC) 92 AB (n=33) showed multi-drug resistance, whereas 31.3% in non-CC92 AB (n=16) (P<0.001). NB-ACB showed 100% susceptibility to all classes of antibiotics except 3rd generation cephalosporin (72.2%). The main mechanism of carbapenem resistance in AB was the blaoxa23 gene with ISAba1 insertion sequence upstream. Presence of pmr gene and/or mutation of lpxA/C gene were not correlated with the phenotype of colistin resistance of ACB. All AB and NB-ACB isolates carried the abe and ade multidrug efflux pumps.
Conclusions: In conclusion, monitoring and research for resistome in ACB complex is needed to identify and manage drug-resistant AB, particularly CC92 AB carrying the blaoxa23 gene.
Acinetobacter baumannii (AB) and its close relatives—Acinetobacter calcoaceticus, Acinetobacter pittii, Acinetobacter nosocomialis, Acinetobacter seifertii, and Acinetobacter dijkshoorniae—comprise the Acinetobacter calcoaceticus-baumannii (ACB) complex, which mainly causes hospital-acquired pneumonia and bacteremia (Towner, 2009; Chusri et al., 2014). The ACB complex is regarded as a group of phenotypically indistinguishable opportunistic pathogens. Among the ACB complexes, infections with AB are a major public health threat in health care settings worldwide (Doi et al., 2015). Non-baumannii ACB (NB-ACB) species also have more been reported as pathogens causing invasive infections than before (Chuang et al., 2011; Lee et al., 2011; Said et al., 2021).
In the recent decades, outbreaks caused by carbapenem resistant AB (CRAB) have been reported in many countries. Super-infection by carbapenem resistant AB and other gram negative organsims was observed in up to 20% of the patients hospitalized with coronavirus disease-2019 (COVID-19), and studies have shown that CRAB had independent association with 14-day mortality in COVID-19 respiratory sub-intensive care units (Casale et al., 2023; Iacovelli et al., 2023). The production of carbapenemases is the most common mechanism of carbapenem resistance, and so far, the most common carbapenemases found in AB are OXA-23 (Mugnier et al., 2010). In addition, CRAB isolates carrying NDM-1 have also been rapidly increasing worldwide (Chen et al., 2011; Pillonetto et al., 2014; Alcock et al., 2020). To make matters worse, there have been reports of a global emergence of resistant to colistin, which is essentially the last line of defense in treating extensively-drug resistant (XDR) AB (Cai et al., 2012).
In South Korea, expansion of ST191 and ST451 that belong to clonal complex (CC) 208 in adult patients from multiple centers have been observed. Clones that belong to CC208 have been shown to exhibit higher resistance to antimicrobials, therefore their expansion have been a global concern due to difficulties in treatment (Jun et al., 2021; Jun et al., 2023).
Currently most of the data on invasive AB infections are derived from adults, and data on the molecular epidemiology and antimicrobial resistance mechanism of AB and NB-ACB causing invasive infections in the pediatric population is very limited. Therefore, this study aimed to identify AB and NB-ACB genotypes causing invasive diseases in children and analyze their antibiotic resistome by using whole genome sequencing (WGS).
All ACB complexes isolated from sterile body fluids in children who were admitted to three referral hospitals located in Seoul, South Korea, between 2015 and 2020 were collected prospectively. The specimens were collected from three hospitals during the patient’s routine clinical care and were transported to the microbiology unit of the laboratory in the corresponding hospitals for routine microbiological analysis. Identification of the ACB complex and antibiotic susceptibility test (AST) were performed using a VITEK2 automated system (BioMériux, Marcy l’Étoile, France). The results of AST were retrospectively collected and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guideline (CLSI, 2020). Multidrug resistance (MDR) was defined as resistance to at least one antimicrobial drug in three or more antimicrobial categories, and XDR was defined as non-susceptibility to all antimicrobial agents except in two or fewer antimicrobial categories. The isolates were stored at -80°C before the extraction of genomic DNA. Demographic and clinical information were collected from the medical chart review. The protocol for this study was approved by the institutional review board of Seoul National University Hospital (no. H-1812–080-995). The requirement for informed consent was waived due to the retrospective design.
Colistin susceptibility was additionally tested via broth microdilution (BMD) according to the CLSI-EUCAST guidelines. Minimum inhibitory concentrations (MICs) were determined using untreated MicroWell trays (Thermo Fisher Scientific, Inc., Waltham, MA, US) and 5×105 colony-forming units/mL of AB was inoculated. The plates were then incubated for 24 hours and read using the Sensititre Manual Viewer (Thermo Fisher Scientific, Inc., Waltham, MA, US). Colistin resistance was defined as MIC ≥4 μg/mL according to the CLSI criteria, and MIC <4 μg/mL was considered intermediate susceptible (CLSI, 2020).
Genomic DNA was prepared using the MasterPure™ Complete DNA Purification Kit (Lucigen, Middleton, WI, USA) from colonies grown on sheep blood agar plates according to the manufacturer’s instructions. The DNA was qualified using a NanoDropTM (ThermoFisher Scientific, Waltham, MA, USA) and was quantified using a 1X dsDNA HS Assay kit on a QubitTM 4 fluorometer (ThermoFisher Scientific). Equal amounts of DNA (~400 ng) from each isolate were used for library preparation using the Rapid Barcoding Sequencing Kit (SQK-RBK004) according to the manufacturer’s protocol (Oxford Nanopore Technologies, Oxford, UK). Pooled libraries of six isolates were run on a FLO-MIN106 flow cell with MinION next-generation sequencing (NGS) equipment and MinKNOW software v.4.1.22 (Oxford Nanopore Technologies) for 48 hours. Basecalling was performed on Guppy software v.4.0 (Oxford Nanopore Technologies), which was integrated into MinKNOW after a sequencing run had been completed. The base-called data were packed into FASTQ with a maximum of 4,000 reads per file. The FASTQ files were then uploaded to the EPI2ME cloud platform via EPI2ME Agent v3.1.3 (Oxford Nanopore Technologies) for the post-basecalling analyses.
What’s in my pot (WIMP) is an EPI2ME workflow for the taxonomic classification of basecalled sequences/reads generated by Nanopore sequencing. The WIMP initially filters FASTQ files with a mean Q-score below a minimum threshold of 6. For reads above the quality threshold, the Centrifuge classification engine (RefSeq for bacteria) is executed to assign each read to a taxon in the National Center for Biotechnology Information taxonomy. The centrifuge classification results were then filtered and aggregated to calculate and report the counts of the reads at the species rank. Bacterial species were confirmed when the 1st-ranked species included >90% of the sequences that were tested.
The seven multilocus sequence typing (MLST) gene (cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) sequences were automatically extracted from the draft WGS and were compared with the reference allele sequences in the database (https://pubmlst.org/organisms/acinetobacter-baumannii/) using CLC Genomics Workbench 7.0.4 (Qiagen). The sequence type (ST) was determined based on the Pasteur scheme of Acinetobacter species MLST. The BUST program in the PubMLST site (https://pubmlst.org/) was used to assign the STs to a CC, which was defined as STs differing from the confounding ST at one or two loci. The relationship between the STs was visualized with a phylogenetic tree on ST by the PhyloViz program (https://online.phyloviz.net/).
Antimicrobial resistance (AMR) mapping application (ARMA pathway on EPI2ME program) analysis allows the detection of genes responsible for AMR. The AMR Comprehensive Antibiotic Resistance Database (CARD) component will align input reads with the minmap2 program against all of the reference sequences available in the CARD, which includes protein references, drug classes, and resistance ontologies that describe associated resistance mechanisms (Mugnier et al., 2010). Among the CARD resistance model types, the protein homolog model included most of the CARD genes (92.2%). Because of the relatively high error rate of Nanopore sequencing and the abundance in the AMR mechanism of the ACB complex, only protein homolog models were counted in this study. Additionally, presence and location of ISAba insertion sequences and quinolone resistance gene (gyrA and parC) mutations were manually inspected for all mapped and assembled sequences. For colistin resistance, the pmr and lpxA/C gene sequences were extracted and a comparative analysis was performed with the reference sequence, AB strain A1 (accession no. CP010781.1).
We performed statistical analyses using SPSS version 25.0 software for Windows (IBM SPSS, Chicago, IL). Rates and proportions were compared between the groups using the chi-square test or Fisher’s exact test where appropriate. A P value <0.05 was considered statistically significant.
During January 2015 to December 2020, a total 67 ACB complexes (reported as AB from microbiology laboratory) were isolated from sterile body fluid cultures of pediatric patients from three tertiary university referral hospitals. The median age of the patients was 0.6 (interquartile range, 0.1–7.2) years old, 55.2% were male, all patients had underlying diseases, and the most common underlying diseases were preterm (28.4%) followed by non-malignant chronic disease (26.9%). The seven- and 30-day mortality were 23.9% and 29.8%, respectively (Table 1). Among all the isolates, 73.1% (n=49) were confirmed as AB, and 26.9% (n=18) were confirmed as NB-ACB complex by using Nanopore sequencing. Of the NB-ACB complex, A. nosocomialis was most commonly isolated (n=9), followed by A. pittii (n=7) and A. seifertii (n=2). Compared to patients infected with NB-ACB, patients with AB had a significantly higher seven-day (30.6% vs. 5.6%, P=0.033) and 30-day (38.8% vs. 5.6%, P=0.002) mortality rate (Table 1).
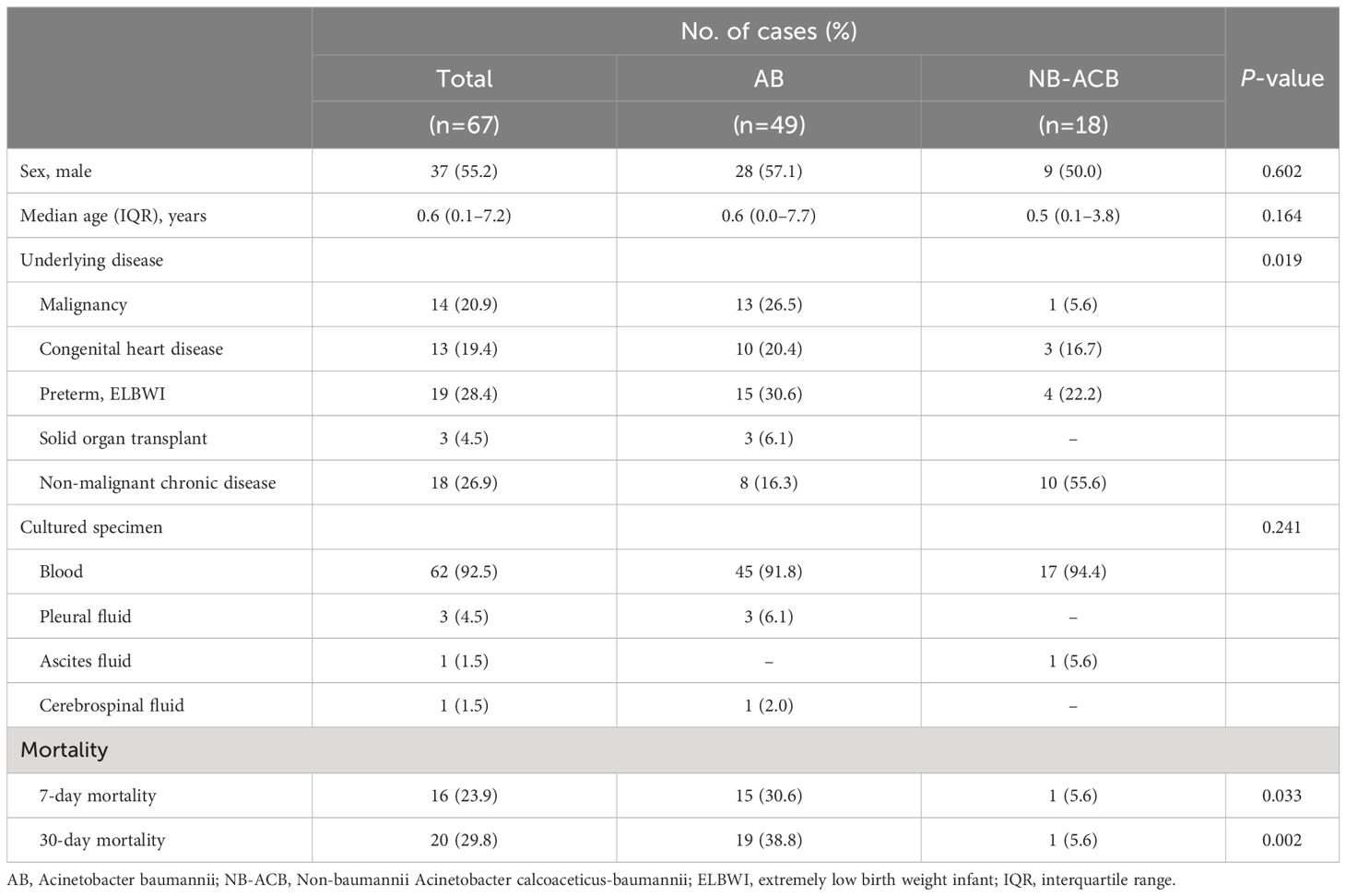
Table 1 Demographics of Korean children infected with Acinetobacter calcoaceticus-baumannii complex in this study.
Among AB, 33 (67.3%) isolates were assigned to CC92 by MLST using the Oxford scheme (Figure 1A). Although majority of the STs in the three hospitals differed, the following four STs were shared between hospitals; ST 191 (center 1, n=1; center B, n=2), ST 208 (center 1, n=1; center C, n=1), ST 369 (center 1, n=1; center 3, n=3), ST 784 (center 1, n=13; center 2, n=1).
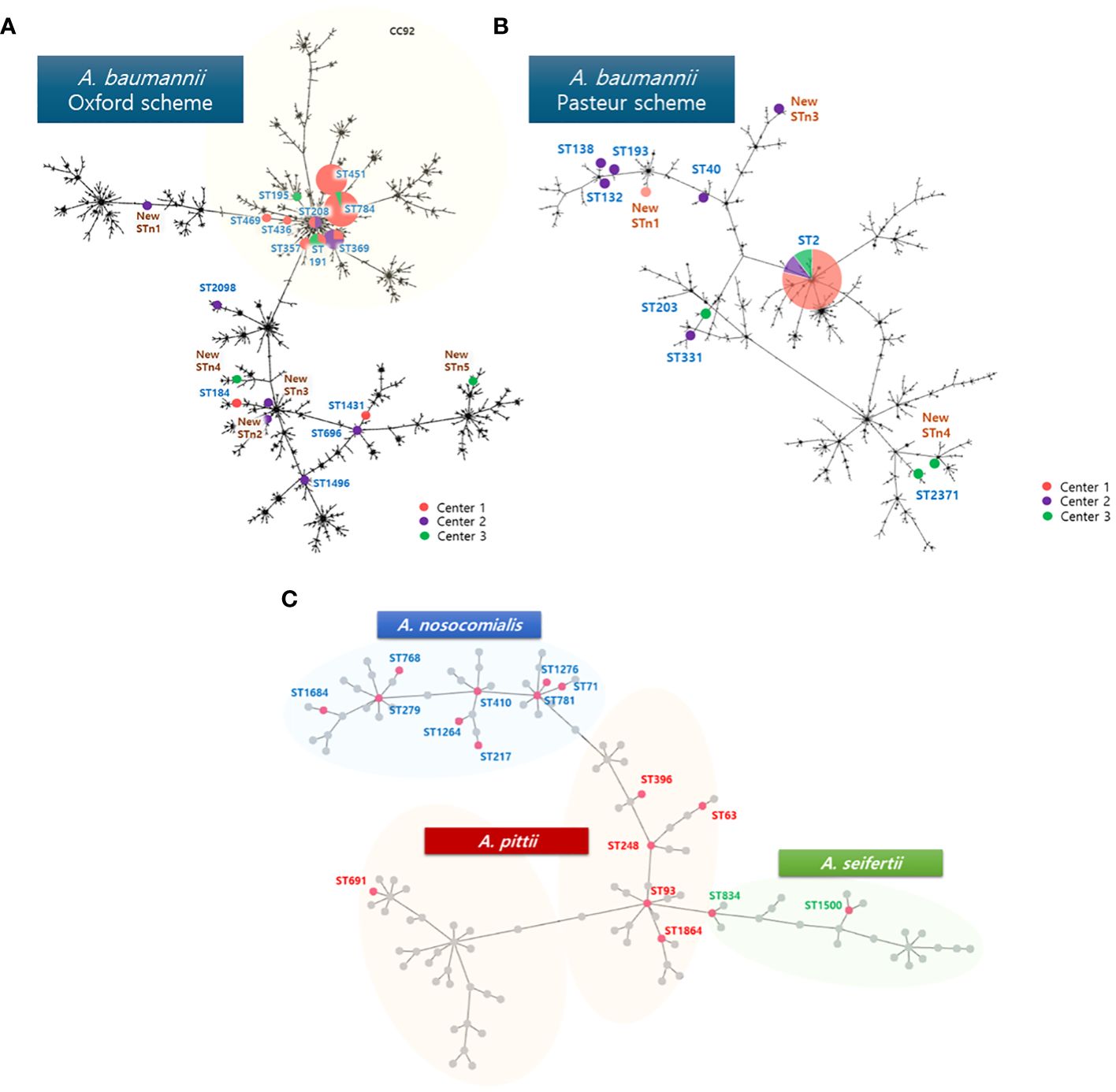
Figure 1 Phylogenetic tree with (A) sequence types of AB analyzed with Oxford scheme, (B) Pasteur scheme, and (C) non-AB ACB complex analyzed with Pasteur scheme in the MLST database. One new ST, STn2, could not be analyzed with the Pasteur scheme. The size of the circles represent the number isolated. Center 1, Seoul National University Children’s Hospital; Center 2, Asan medical center; Center 3, Samsung medical center; AB, A baumannii; CC, clonal complex; ST, sequence type.
Using the Pasteur scheme to analyze AB (Figure 1B), we found that the study isolates had less discriminatory power compared to the Oxford scheme. A total 8 STs were observed, with ST 2 shared by all three centers (center 1, n=30; center 2, n=4; center 3, n=4). Other STs included one isolate each: ST 40, ST 132, ST 138, ST 193, ST 203, ST 331, and ST 2,371.
All NB-ACB isolates had different STs except for two A. pittii isolates, which were both ST63. For the three NB-ACB species identified in the current study, ST phylogeny showed a clearly separated distribution and relationship (Figure 1C).
The AB isolates showed an overall low susceptibility to all tested antibiotics, with only 22.4% (n=11/49) susceptible to carbapenem, 26.9% (n=7/26) to amikacin, and 38.8% (n=19/49) to trimethoprim-sulfamethoxazole. Furthermore, 28.6% (n=14/49) had resistance to colistin. An overall 77.6% (n=38/49) were MDR, and 71.4% (n=35/49) were XDR. Within the AB isolates, a significant difference in antimicrobial susceptibility was observed between CC92 and non-CC92. The percentage of MDR was 100% (n=33/33) in CC92 and 31.3% (n=5/16) in non-CC92 (P<0.001). NB-ACB showed 100% susceptibility to all classes of antibiotics except 3rd generation cephalosporin, where 5 isolates (2 A. nosocomialis and 3 A. pittii) showed intermediate susceptibility (MIC=16 mg/L). One A. seifertii isolate also showed resistance to colistin (MIC>4 mg/L) (Table 2).
All AB isolates contained blaADC, blaOXA51, and blaOXA23 genes, but the ISAba1 insertion sequence was detected at upstream of blaOXA23 gene in all CRAB and no carbapenem-susceptible (CS) AB. Three (9.1%) and none of CRAB carried ISAba1 upstream to blaADC and blaOXA51, respectively. All (n=11) CSAB, which included in non-CC92 AB, carried ISAba1 upstream to blaADC, but none of them showed resistance to 3rd-generation cephalosporin. Among 10 AB harboring blaTEM, all but one isolates also harbored ISAba1-blaOXA23, and thus were MDR as well as CRAB. The other AB isolate was non-CC92 and showed exclusive susceptibility to all antibiotics (Figure 2).
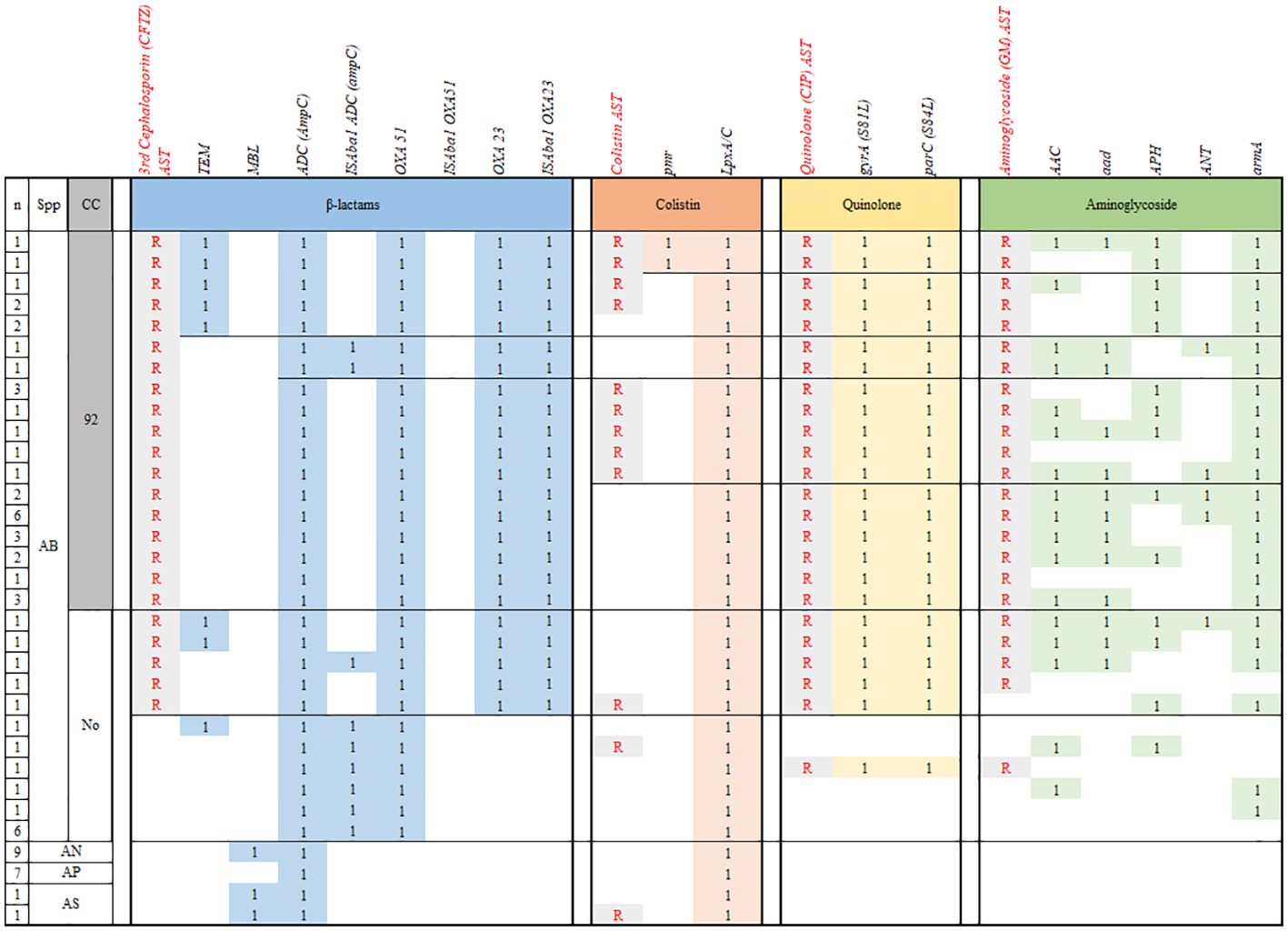
Figure 2 Comparison of the antibiotic resistome profiles of AB isolated from children with invasive infections. n, number; CC, clonal complex; CP, Chloramphenicol; CRAB, carbapenem-resistant A. baumannii; MDR, multidrug resistant; ML, macrolide; R, phenotypic resistance; SA, sulfonamide; SM, streptomycin; SMC, Samsung medical center; TC, tetracycline; 1, presence of resistance gene.
In NB-ACB isolates, all carried blaADC gene without upstream ISAba1. The metallo-β-lactamase (MBL) gene was detected in all of the A. nosocomialis and A. sefertii isolates, whereas the blaOXA421 gene was detected in all of the A. pittii isolates. None of blaMBL MBL and blaOXA421 genes were accompanied with the upstream ISAba1 and none of NB-ACB isolates with these β-lactamases showed a resistance to β-lactam antibiotics including carbapenem (Table 3).
The pmr gene was detected in two of 14 colistin-resistant (Col-R) AB isolates and none of 25 colistin-susceptible (Col-S) AB and 18 NB-ACB isolates. Plasma-encoded mcr gene was not detected in any of the isolates. Meanwhile, all AB and NB-ACB isolates harbored both the lpxA and lpxC genes. Although several single nucleotide polymorphisms were observed in lpxA and lpxC genes from ACB isolates in this study, all were synonymous mutations (Supplementary Figures 1, 2). Any other AMR genes detected were significantly different in frequency between Col-R and Col-S ACB isolates (Table 3).
For fluoroquinolone resistance, the presence of both mutations in gyrA and parC causing antibiotic target alteration (S81L and S84L, respectively) was exclusively matched to phenotypical quinolone-resistance in AB (n=38, 77.6%; Figure 2). No NB-ACB isolate showed quinolone-resistance and mutations in gyrA and parC genes. A variety of resistance genes to aminoglycoside were carried in AB isolates: antibiotic inactivation genes (aac, aad, APH, ANT), antibiotic target alteration genes (armA), and resistance-nodulation-cell division (RND) antibiotic efflux pumps (smeB and amrB). All AB isolates carried the abe and ade efflux pumps, regardless of resistance to aminoglycoside. The AMR genes most closely correlated with the aminoglycoside resistance were aad and armA genes: 63.2% and 94.7% in aminoglycoside-resistant (n=38) and 0% and 27.3% in aminoglycoside-susceptible AB isolates (n=11), respectively. NB-ACB isolates were exclusively susceptible to aminoglycoside, and only abe and ade efflux genes were detected in all these isolates (Table 3).
AB has emerged as a major global threat, especially due to its exceptional ability to acquire resistance genes to all classes of antibiotics currently available. This study aimed to identify circulating AB and NB-ACB genotypes causing invasive diseases in Korean children, analyze their antibiotic resistomes, and investigate the mechanism of carbapenem and colistin resistance. Compared to NB-ACB isolates, AB had a significantly higher mortality rate with an overall low susceptibility to antibiotics, with carbapenem and colistin resistances in 77.6% and 27.1% of the isolates, respectively. In particular, CC92 AB were exclusively CRAB and MDR, with the main resistance mechanism found to be OXA23 β-lactamase. Colistin resistance was not correlated with the presence of pmr gene and/or mutations in lpxA/C genes.
ACB complex is usually reported as AB from the laboratory using commercial bacterial identification system. However, correctly identifying ACB subspecies is important because of the poorer prognosis of AB than NB-ACB infections (Kuo et al., 2013; Xiao et al., 2017; da Silva et al., 2018). In Japan, between 2001 and 2014, among 155 invasive Acinetobacter species strains, 27.1% was A. pittii and 25.8% was AB, and imipenem nonsusceptibility was detected only in 4 strains (Kiyasu et al., 2020). In the previous study in Mexico, a total of 88 strains were identified as ACB complex by VITEK II during 2015–2017. Among them, AB accounted for 89.8%; A. pittii, 6.8%; and A. nosocomialis, 3.4%. In addition, 44.3% were MDR strains and, 11.4% were XDR (Mancilla-Rojano et al., 2020). In the study from Russia and Kazakhstan in 2016–2022, 234 Acinetobacter isolates were identified as NB-ACB isolates, which comprised 6.2% of Acinetobacter spp. Most NB-ACB isolates were susceptible to all antibiotics; however, sporadic isolates were resistant to carbapenems (Sheck et al., 2023). As shown above, the proportion of NB-ACB strain and its subspecies among Acinetobacter species and their AMR vary according to country, period, specimen type, and isolation method. Also, in the current study, among the 68 invasive isolates reported as AB, one-fourth was NB-ACB, which had much lower antimicrobial resistance and mortality than AB. Among the NB-ACB species, the most abundant were A. nosocomialis and A. pittii as in the previous studies (Mancilla-Rojano et al., 2020; Sheck et al., 2023).
AB emerged as a global pathogen due to the successful expansion of a few epidemic lineages producing acquired OXA-type carbapenemases. Particularly ST2 (Pasteur scheme), known as international clone (IC) II, the most common clone globally, are distributed in Europe, Asia, and Latin America (Levy-Blitchtein et al., 2018; Villalon et al., 2019). Genotype replacement with CC92 (Oxford scheme), which is the same strain to ST2 (Pasteur scheme), has been a global phenomenon, especially in China where CC92 CRAB harboring blaOXA23 has been increasing (Ruan et al., 2013; Chen et al., 2017). The spread of this drug-resistant pathogen should be closely monitored and managed, particularly in critical care. In a previous single center study in Korea, 27 AB were isolated from blood of patients in PICU, and MLST analysis showed that ST138 (Oxford scheme, CC92) was predominant (70%) (Kim et al., 2022). Also in the current study, majority of AB were ST2, which showed XDR characteristics, with compete resistance to 3rd/4th generation cephalosporin, piperacillin-tazobactam, amikacin, ciprofloxacin, and even carbapenem. Furthermore, 36.4% were resistant to colistin.
In our previous study, we identified carbapenem nonsusceptibility as a risk factor for mortality due to AB in children. An early administration of appropriate antibiotics should be enacted, especially in children with neutropenia (Choe et al., 2019). The blaOXA-23 gene is one of the most prevalent β-lactamase genes on the genome of CRAB and is distributed worldwide (Mugnier et al., 2010; Ruan et al., 2013). Among 86 AB isolates obtained during 2008–2015 in Central Illinois, 70.9% were nonsusceptible to carbapenems. Among the CRAB isolates, blaOXA-23 was the most frequently detected carbapenemase gene (52%) (Koirala et al., 2020). However, it is suggested that ISAba1 is providing the promoter for blaOXA-51 and blaOXA-23 genes (Turton et al., 2006). In the current study, the presence of blaOXA-23 genes and ISAba1 upstream of blaOXA-23 were exactly matched to carbapenem resistance in AB. Any other β-lactamase or other AMR mechanism could be differentiated between CRAB and CSAB. We may conclude that the main mechanism of carbapenem resistance in AB isolated in Korean children during 2015–2020 was ISAba1-blaOXA-23. Because the detection of AB with blaOXA-23 and MDR phenotye is associated with increased mortality, prompt identification and management is extremely important in the clinical field. Similar results of high mortality associated with MDR AB carrying blaOXA-23 was observed globally in different STs, and the blaOXA-23 gene found to be present on a conjugative transferable plasmid indicated the advantage in transferability of the genetic element (Lopes et al., 2015; da Silva et al., 2018).
AmpC β-lactamase coded in blaADC gene could induce resistance to 3rd-generation cephalosporin of gram-negative bacteria during the use (Tamma et al., 2019). In the current study, all ACB complex isolates harbored blaADC gene, but they did not show resistance to 3rd-genderation cephalosporin unless they also had ISAba1-blaOXA-23, regardless of the presence of upstream ISAba1. Thus, we may conclude that the harboring of blaADC itself is not the requirement of AMR, and upstream ISAba1 is not sufficient to activate blaADC within AB.
Because colistin is usually the only remaining treatment available, the emergence of colistin-resistant CRAB is devastating. Colistin resistance due to the acquisition of plasmid-mediated mobile colistin-resistant (mcr) genes has been described in numerous Enterobacterales, especially Klebsiella pneumonia, however also found in AB. In Pseudomonas aeruginosa, mcr-5 has been identified, however, activation of histidine kinase (PmrB) or the response regulaor (PmrA) following an amino-acid substitution has been found to be a key mechanism for colistin resistance (Liu et al., 2016; Jeannot et al., 2021; Novović and Jovčić, 2023). To date only chromosomally encoded colistin resistance mechanisms have been reported in AB. It is suggested that mutations in the pmr and lpx genes confer colistin resistance via modification and complete loss of lipopolysaccharide, respectively (Adams et al., 2009; Moffatt et al., 2010; Nie et al., 2020). In the current study, two isolates were found to harbor the pmr gene. The significant mutations in pmrA gene related to colistin-resistance (D82G and S119T) were not found, but mutations in pmrB gene (G21V and V227A) were found in both isolates. Furthermore, lpxA and lpxC were found in all the isolates, but the change in the amino acid sequence of lpxA found in one isolate, which were unlikely to be associated with colistin resistance mechanisms in our isolates. This shows that there are other mechanisms that have not yet been uncovered, that are associated with colistin resistance.
The most common mechanism of resistance to fluoroquinolone in AB involves alterations in the genes that encode subunits of the quinolone targets DNA gyrA and parC (Park et al., 2011). Phenotypical quinolone resistance was perfectly matched to both gyrA and parC gene mutations in the current study. All quinolone resistant AB were CRAB, which harbored ISAba1-blaOXA-23. None of CSAB had either gyrA or parC gene mutations.
There are two main mechanisms underlying aminoglycoside resistance in AB: aminoglycoside-modifying enzymes, which were encoded by aac, aad, ant, and aph genes, and antibiotic target alteration by 16S rRNA methylases (encoded by armA) (Bakour et al., 2014). In this study, genes encoding aminoglycoside-modifying enzymes were found in 30.3%~69.7% of CC92 AB, 18.8%~31.3% of non-CC92 AB, and none of NB-ACB. The armA gene was also detected in 100%, 37.5%, and 0%, respectively. In a previous report from 2 hospitals in South Korea, invasive AB isolates from adults during 2004–2005 showed 85.2% of harboring armA (Cho et al., 2009).
The resistance-nodulation-cell division (RND) pumps are the most prevalent efflux pumps in AB and one of the most important determinants of MDR. Typically, efflux pumps (especially those chromosomally encoded) are controlled by negative regulators. Mutations within the negative regulators which releases the ‘block’ on the efflux pump leads to increased expression of the pump causing antibiotics resistance (Fernando and Kumar, 2013; Davin-Regli et al., 2021; Kornelsen and Kumar, 2021). A high prevalence of this pump was observed in up to 97% of MDR AB isolates worldwide (Nowak et al., 2015). In the invasive AB strains from this study, the ade RND pumps were present in 100%. Furthermore, abe, which is a chromosomally encoded drug efflux pump of the small multidrug resistance (SMR) family, was also found in 100% of the isolates in this study. Although just the presence of these efflux pumps did not correlate with MDR in AB, it might show us the potential of AB to be MDR pathogen whenever they get the certain promotor.
Rapid detection of AB and main carbapenemase genes can allow prompt intervention in patients with invasive infections. By identifying whether the identified AB is MDR phenotype, early initiation of adequate antibiotics can improve patient outcome. Commercial molecular rapid testing are available and have already implemented in routine diagnostics in many centers (Dunbar et al., 2022). Improvements have been made in providing rapid phenotypic AST (Briggs et al., 2021), the benefits and limitations of these rapid platforms are currently under study and more studies are needed in critically-ill children.
There were several limitations of this study. First, although the three centers included were representative centers in South Korea with the highest number of beds for treating critically ill children in the pediatric intensive care unit, the resistance profiles of AB causing nosocomial infections may differ at each hospital throughout Korea. Second, the total number of cases included is limited, and because this was a multicenter study, we were unable to obtain all clinical data including information on treatment regimens and duration for all patients. However, this is the first study to investigate, via WGS, the antibiotic resistome profiles of AB dominant in strains isolated from critically ill children, and is deemed to be valuable for the monitoring and treatment of invasive AB infections.
To conclude, we found that CC92 AB have an exceptional ability to acquire a wide variety of antibiotic resistomes which enhance their AMR and survival fitness. Non-CC92 isolates also harbored a variety of resistance genes for which they were phenotypically susceptible to, depicting the necessity for continuous monitoring. Many efforts are needed to prevent their outbreak in critical patients, including methods such as decreasing overall antibiotic use and relieving selective pressure, strict contact precaution and hand hygiene of medical staff to prevent transmission, and environmental disinfection to decrease sources of possible infection. Carbapenem resistance in AB was likely induced by blaOXA-23 carbapenemases promoted by ISAba1. Although only one-third of the isolates were shown to be resistant to colistin, we found that all AB harbored mutations in enzymes that produce lipid A, meaning all AB have the potential to become resistant to colistin. Further studies are necessary to understand resistance mechanisms in order to combat the highly virulent XDR AB, and evaluate susceptibility to novel approved drugs such as cefiderocol and sulbactam/durlobactam.
Original datasets are available in a publicly accessible repository:The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA1127342.
The studies involving humans were approved by institutional review board of Seoul National University Hospital (no. H-1812-080-995). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
HK: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. KK: Data curation, Investigation, Methodology, Writing – review & editing. GK: Data curation, Investigation, Methodology, Writing – original draft. D-GL: Data curation, Investigation, Methodology, Writing – review & editing. YK: Data curation, Formal Analysis, Investigation, Supervision, Writing – review & editing. EC: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Supervision, Writing – review & editing. JL: Data curation, Formal Analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. KW: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1C1B5085781).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1410997/full#supplementary-material
Adams, M. D., Nickel, G. C., Bajaksouzian, S., Lavender, H., Murthy, A. R., Jacobs, M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53, 3628–3634. doi: 10.1128/AAC.00284-09
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–DD25. doi: 10.1093/nar/gkz935
Bakour, S., Touati, A., Bachiri, T., Sahli, F., Tiouit, D., Naim, M., et al. (2014). First report of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and rapid spread of metallo-β-lactamase NDM-1 in Algerian hospitals. J. infection chemother. 20, 696–701. doi: 10.1016/j.jiac.2014.07.010
Briggs, N., Campbell, S., Gupta, S. (2021). Advances in rapid diagnostics for bloodstream infections. Diagn. Microbiol. Infect. disease 99, 115219. doi: 10.1016/j.diagmicrobio.2020.115219
Cai, Y., Chai, D., Wang, R., Liang, B., Bai, N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrobial. Chemother. 67, 1607–1615. doi: 10.1093/jac/dks084
Casale, R., Bianco, G., Bastos, P., Comini, S., Corcione, S., Boattini, M., et al. (2023). Prevalence and impact on mortality of colonization and super-infection by carbapenem-resistant gram-negative organisms in COVID-19 hospitalized patients. Viruses 15 (9), 1934. doi: 10.3390/v15091934
Chen, Y., Gao, J., Zhang, H., Ying, C. (2017). Spread of the bla(OXA-23)-Containing Tn2008 in Carbapenem-Resistant Acinetobacter baumannii Isolates Grouped in CC92 from China. Front. Microbiol. 8, 163.
Chen, Y., Zhou, Z., Jiang, Y., Yu, Y. (2011). Emergence of NDM-1-producing acinetobacter baumannii in China. J. Antimicrobial. Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
Cho, Y. J., Moon, D. C., Jin, J. S., Choi, C. H., Lee, Y. C., Lee, J. C. (2009). Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn. Microbiol. Infect. disease 64, 185–190. doi: 10.1016/j.diagmicrobio.2009.02.010
Choe, Y. J., Lee, H. J., Choi, E. H. (2019). Risk factors for mortality in children with acinetobacter baumannii bacteremia in South Korea: the role of carbapenem resistance. Microb. Drug Resist. 25, 1210–1218. doi: 10.1089/mdr.2018.0465
Chuang, Y. C., Sheng, W. H., Li, S. Y., Lin, Y. C., Wang, J. T., Chen, Y. C., et al. (2011). Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with Acinetobacter bacteremia. Clin. Infect. Dis. 52, 352–360. doi: 10.1093/cid/ciq154
Chusri, S., Chongsuvivatwong, V., Rivera, J. I., Silpapojakul, K., Singkhamanan, K., McNeil, E., et al. (2014). Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob. Agents Chemother. 58, 4172–4179. doi: 10.1128/AAC.02992-14
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed (Wayne, Pennsylvania: CLSI).
da Silva, K. E., Maciel, W. G., Croda, J., Cayô, R., Ramos, A. C., de Sales, R. O., et al. (2018). A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit. PloS One 13, e0209367. doi: 10.1371/journal.pone.0209367
Davin-Regli, A., Pages, J. M., Ferrand, A. (2021). Clinical status of efflux resistance mechanisms in gram-negative bacteria. Antibiotics (Basel Switzerland) 10. doi: 10.3390/antibiotics10091117
Doi, Y., Murray, G. L., Peleg, A. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 36, 85–98.
Dunbar, S. A., Gardner, C., Das, S. (2022). Diagnosis and management of bloodstream infections with rapid, multiplexed molecular assays. Front. Cell. infection Microbiol. 12, 859935. doi: 10.3389/fcimb.2022.859935
Fernando, D. M., Kumar, A. (2013). Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: role in virulence. Antibiotics (Basel Switzerland) 2, 163–181. doi: 10.3390/antibiotics2010163
Iacovelli, A., Oliva, A., Siccardi, G., Tramontano, A., Pellegrino, D., Mastroianni, C. M., et al. (2023). Risk factors and effect on mortality of superinfections in a newly established COVID-19 respiratory sub-intensive care unit at University Hospital in Rome. BMC pulmonary Med. 23, 30. doi: 10.1186/s12890-023-02315-9
Jeannot, K., Hagart, K., Dortet, L., Kostrzewa, M., Filloux, A., Plesiat, P., et al. (2021). Detection of colistin resistance in pseudomonas aeruginosa using the MALDIxin test on the routine MALDI biotyper sirius mass spectrometer. Front. Microbiol. 12, 725383. doi: 10.3389/fmicb.2021.725383
Jun, S. H., Lee, D. E., Hwang, H. R., Kim, N., Kim, H. J., Lee, Y. C., et al. (2021). Clonal change of carbapenem-resistant Acinetobacter baumannii isolates in a Korean hospital. Infection Genet. evolution: J. Mol. Epidemiol. evolutionary Genet. Infect. diseases 93, 104935.
Jun, S. H., Lee, D. E., Hwang, H. R., Kim, N., Kwon, K. T., Kim, Y. K., et al. (2023). Clonal evolution and antimicrobial resistance of Acinetobacter baumannii isolates from Korean hospitals over the last decade. Infection Genet. evolution: J. Mol. Epidemiol. evolutionary Genet. Infect. diseases 108, 105404.
Kim, D., Lee, H., Choi, J. S., Croney, C. M., Park, K. S., Park, H. J., et al. (2022). The changes in epidemiology of imipenem-resistant acinetobacter baumannii bacteremia in a pediatric intensive care unit for 17 years. J. Korean Med. Sci. 37, e196. doi: 10.3346/jkms.2022.37.e196
Kiyasu, Y., Hitomi, S., Funayama, Y., Saito, K., Ishikawa, H. (2020). Characteristics of invasive Acinetobacter infection: A multicenter investigation with molecular identification of causative organisms. J. Infect. Chemother. 26, 475–482. doi: 10.1016/j.jiac.2019.12.010
Koirala, J., Tyagi, I., Guntupalli, L., Koirala, S., Chapagain, U., Quarshie, C., et al. (2020). OXA-23 and OXA-40 producing carbapenem-resistant Acinetobacter baumannii in Central Illinois. Diagn. Microbiol. Infect. Dis. 97, 114999. doi: 10.1016/j.diagmicrobio.2020.114999
Kornelsen, V., Kumar, A. (2021). Update on multidrug resistance efflux pumps in acinetobacter spp. Antimicrobial. Agents chemother. 65, e0051421. doi: 10.1128/AAC.00514-21
Kuo, S. C., Lee, Y. T., Yang, S. P., Chiang, M. C., Lin, Y. T., Tseng, F. C., et al. (2013). Evaluation of the effect of appropriate antimicrobial therapy on mortality associated with Acinetobacter nosocomialis bacteraemia. Clin. Microbiol. infection: Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Diseases 19, 634–639.
Lee, Y. C., Huang, Y. T., Tan, C. K., Kuo, Y. W., Liao, C. H., Lee, P. I., et al. (2011). Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J. Antimicrob. Chemother. 66, 1839–1846. doi: 10.1093/jac/dkr200
Levy-Blitchtein, S., Roca, I., Plasencia-Rebata, S., Vicente-Taboada, W., Velasquez-Pomar, J., Munoz, L., et al. (2018). Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg. Microbes Infect. 7, 119. doi: 10.1038/s41426-018-0127-9
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lopes, B. S., Al-Agamy, M. H., Ismail, M. A., Shibl, A. M., Al-Qahtani, A. A., Al-Ahdal, M. N., et al. (2015). The transferability of blaOXA-23 gene in multidrug-resistant Acinetobacter baumannii isolates from Saudi Arabia and Egypt. Int. J. Med. Microbiol. 305, 581–588. doi: 10.1016/j.ijmm.2015.07.007
Mancilla-Rojano, J., Ochoa, S. A., Reyes-Grajeda, J. P., Flores, V., Medina-Contreras, O., Espinosa-Mazariego, K., et al. (2020). Molecular Epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex Isolated From Children at the Hospital Infantil de Mexico Federico Gomez. Front. Microbiol. 11, 576673. doi: 10.3389/fmicb.2020.576673
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/AAC.00834-10
Mugnier, P. D., Poirel, L., Naas, T., Nordmann, P. (2010). Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerging Infect. diseases 16, 35–40.
Nie, D., Hu, Y., Chen, Z., Li, M., Hou, Z., Luo, X., et al. (2020). Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. sci. 27, 26. doi: 10.1186/s12929-020-0617-7
Novović, K., Jovčić, B. (2023). Colistin resistance in acinetobacter baumannii: molecular mechanisms and epidemiology. Antibiotics (Basel). 12, 3, 516. doi: 10.3390/antibiotics12030516
Nowak, J., Seifert, H., Higgins, P. G. (2015). Prevalence of eight resistance-nodulation-division efflux pump genes in epidemiologically characterized Acinetobacter baumannii of worldwide origin. J. Med. Microbiol. 64, 630–635. doi: 10.1099/jmm.0.000069
Park, S., Lee, K. M., Yoo, Y. S., Yoo, J. S., Yoo, J. I., Kim, H. S., et al. (2011). Alterations of gyrA, gyrB, and parC and Activity of Efflux Pump in Fluoroquinolone-resistant Acinetobacter baumannii. Osong Public Health Res. Perspect. 2, 164–170. doi: 10.1016/j.phrp.2011.11.040
Pillonetto, M., Arend, L., Vespero, E. C., Pelisson, M., Chagas, T. P., Carvalho-Assef, A. P., et al. (2014). First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrobial. Agents chemother. 58, 7592–7594. doi: 10.1128/AAC.03444-14
Ruan, Z., Chen, Y., Jiang, Y., Zhou, H., Zhou, Z., Fu, Y., et al. (2013). Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int. J. antimicrobial. agents 42, 322–328. doi: 10.1016/j.ijantimicag.2013.06.019
Said, D., Willrich, N., Ayobami, O., Noll, I., Eckmanns, T., Markwart, R. (2021). The epidemiology of carbapenem resistance in Acinetobacter baumannii complex in Germany (2014–2018): an analysis of data from the national antimicrobial resistance surveillance system. Antimicrob. Resist. Infect. Control 10, 45. doi: 10.1186/s13756-021-00909-8
Sheck, E., Romanov, A., Shapovalova, V., Shaidullina, E., Martinovich, A., Ivanchik, N., et al. (2023). Acinetobacter non-baumannii species: occurrence in infections in hospitalized patients, identification, and antibiotic resistance. Antibiotics (Basel) 12. doi: 10.3390/antibiotics12081301
Tamma, P. D., Doi, Y., Bonomo, R. A., Johnson, J. K., Simner, P. J. (2019). Antibacterial resistance leadership G. A primer on ampC beta-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin. Infect. Dis. 69, 1446–1455. doi: 10.1093/cid/ciz173
Towner, K. J. (2009). Acinetobacter: an old friend, but a new enemy. J. Hosp. Infect. 73, 355–363. doi: 10.1016/j.jhin.2009.03.032
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/fml.2006.258.issue-1
Villalon, P., Ortega, M., Saez-Nieto, J. A., Carrasco, G., Medina-Pascual, M. J., Garrido, N., et al. (2019). Dynamics of a Sporadic Nosocomial Acinetobacter calcoaceticus - Acinetobacter baumannii Complex Population. Front. Microbiol. 10, 593.
Keywords: Acinetobacter baumannii, genotype, resistome, colistin resistance, children
Citation: Kang HM, Kim KR, Kim G, Lee D-g, Kim YJ, Choi EH, Lee J and Yun KW (2024) Antimicrobial resistance genes harbored in invasive Acinetobacter calcoaceticus-baumannii complex isolated from Korean children during the pre-COVID-19 pandemic periods, 2015–2020. Front. Cell. Infect. Microbiol. 14:1410997. doi: 10.3389/fcimb.2024.1410997
Received: 02 April 2024; Accepted: 15 May 2024;
Published: 04 July 2024.
Edited by:
Juan Carlos Rodriguez Diaz, Hospital General Universitario de Alicante, SpainReviewed by:
Gabriele Bianco, University Hospital Città della Salute e della Scienza di Torino, ItalyCopyright © 2024 Kang, Kim, Kim, Lee, Kim, Choi, Lee and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ki Wook Yun, cGVkd2lsbHlAc251LmFjLmty; Jina Lee, ZW50aWVyQGFtYy5zZW91bC5rcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.