
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 02 July 2024
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1410921
Objective: The emergence of clinical Klebsiella pneumoniae strains harboring acrAB-tolC genes in the chromosome, along with the presence of two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65 genes on a plasmid, has presented a significant clinical challenge.
Methods: In order to study the detailed genetic features of K. pneumoniae strain SC35, both the bacterial chromosome and plasmids were sequenced using Illumina and nanopore platforms. Furthermore, bioinformatics methods were employed to analyze the mobile genetic elements associated with antibiotic resistance genes.
Results: K. pneumoniae strain SC35 was found to possess a class A beta-lactamase and demonstrated resistance to all tested antibiotics. This resistance was attributed to the presence of efflux pump genes, specifically acrAB-tolC, on the SC35 chromosome. Additionally, the SC35 plasmid p1 carried the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65, as well as blaTEM-1 with rmtB, which shared overlapping structures with mobile genetic elements as In413, Tn3, and TnAs3. Through plasmid transfer assays, it was determined that the SC35 plasmid p1 could be successfully transferred with an average conjugation frequency of 6.85 × 10-4.
Conclusion: The structure of the SC35 plasmid p1 appears to have evolved in correlation with other plasmids such as pKPC2_130119, pDD01754–2, and F4_plasmid pA. The infectious strain SC35 exhibits no susceptibility to tested antibioticst, thus effective measures should be taken to prevent the spread and epidemic of this strain.
The increasing prevalence and global spread of multidrug-resistant bacteria, including in regions such as South/North America, Europe, Africa, the Middle East, South-East Asia, and Oceania, posed a significant threat to human health worldwide (Hamidian and Nigro, 2019). Among these bacteria, carbapenem-resistant Klebsiella pneumoniae had emerged as a major cause of infections globally (World Health Organization (WHO), 2014). Plasmids played a crucial role in the dissemination of drug-resistant genes, such as blaKPC-2 (Zhai et al., 2021). Various types of plasmids had been found to carry blaKPC, with the IncFIIk plasmid being the most frequently detected and widely reported (Villa et al., 2010; Pitout et al., 2015; Li et al., 2021). The production of carbapenemase enzymes, including KPC, NDM, and OXA-48, was one of the major resistance mechanisms in K. pneumoniae (Pitout et al., 2015).
The presence of CTX-M type extended-spectrum beta-lactamases (ESBLs) was another significant factor contributing to bacterial resistance to cephalosporins. These ESBLs had been widely detected in European countries, as well as in Asia and South America (Cantón and Coque, 2006). Mobile genetic factors played an important role in the emergence of multidrug-resistant bacteria (Yu et al., 2024). There were notable variations in the distribution of CTX-M types across different countries and continents (Yu et al., 2024). In Asia, CTX-M-14 and CTX-M-55 were the predominant genotypes, while CTX-M-1 was more prevalent in Europe (Yu et al., 2024), and higher frequency of IncI-I(α) plasmids were found in Eurasian and Asian transmission clusters (Yu et al., 2024). The transfer of CTX-M-14 was mainly mediated by ISEcpl, while IS26 played a significant role in the transfer of CTX-M-65. Additionally, blaTEM and blaCTX-M were frequently found together within the same transposable unit (Yu et al., 2024). Studies had reported concurrent involvement of blaKPC-2, blaCTX-M-65, and blaSHV-12 on the same plasmid (Xiang et al., 2016; Liu et al., 2018), indicating a close relationship and a highly similar genetic backbone among these resistance genes (Liu et al., 2018). Sequencing results had demonstrated that IS26 mediated the duplication of the IS26-fosA3-blaCTX-M-65 plasmid insertion element in certain strains (Wang et al., 2023).
Bacterial resistance to antimicrobial drugs could occur through four main mechanisms: (1) Inactivation of the drugs through degradation or chemical modification, (2) Mutations in the targets of the drugs, resulting in reduced binding or efficacy, (3) Changing in membrane permeability, and (4) Active efflux of drugs, reducing intracellular concentrations (Reygaert, 2018). Mechanisms (1) and (2) generally conferred resistance to specific or similar antibiotics, such as KPC or CTX-M-65, which could be present in the same strain, leading to resistance to multiple drugs within their respective antibiotic families (Yamasaki et al., 2023). On the other hand, mechanisms (3) and (4) could increase resistance to multiple structurally unrelated antibiotics (Reygaert, 2018). For example, a single multidrug efflux pump, such as tmexCD1-toprJ1 (Qu et al., 2023) or acrAB-tolC, had the ability to transport multiple structurally unrelated substrates, and its high expression could result in bacterial resistance to various antibiotic families (Yamasaki et al., 2023). Therefore, the presence or acquisition of resistance genes, along with cellular drug efflux pumps or channels, significantly enhanced bacterial multidrug resistance, making clinical treatment more challenging and complex, and posing a greater risk to infected patients.
This study aims to characterize the multidrug resistance genes present on the chromosome harboring acrAB-tolC and a plasmid carrying two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65. The research will focus on annotating the structure of resistance genes on the plasmid, comparing the structural relationships between similar plasmids, and exploring possible evolutionary processes. The study will provide detailed data that can be used for the treatment of highly multidrug-resistant strains with similar resistance mechanisms.
A 57-year-old man was admitted to the intensive care unit with a closed craniocerebral injury resulting in intracranial and pulmonary infection, but empiric antimicrobial treatment with tigecycline plus polymyxin was ineffective. Subsequently, K. pneumoniae SC35, a carbapenem- insensitive strain, was isolated from the patient’s sputum at Taizhou Municipal Hospital affiliated with Taizhou University in 2021, and for this reason, this selected strain was further investigated in depth in this paper.
For species verification, the almost complete 16S rRNA gene was amplified using PCR. The forward primer used was 5’-AGAGTTTGATYMTGGGGCTCAG’, and the reverse primer used was 5’-TACCTTGTTACGACTT-3’ (where Y represents T or C, and M represents A or C). The length of the amplicon obtained was approximately 1,500 bp (Frank et al., 2008). The PCR amplification was performed using a mixture of Fermentas Taq and Pfu enzymes (in a 3:1 ratio) from ThermoFisher Scientific. The reaction mixture contained 30 ml of the enzyme mixture, and the PCR conditions included an initial denaturation step at 94°C for 3 min, followed by denaturation at 94°C for 40 s, annealing at 50°C for 40 s, extension at 72°C for 1 min, and a final extension step at 72°C for 5 min. A total of 30 cycles were performed. To characterize the PCR products, bidirectional sequencing was carried out. This sequencing method allowed for sequencing in both the forward and reverse directions, providing more accurate and reliable results.
To perform conjugation, the EC600 receptor strain and SC35 donor strain were mixed together in lysozyme broth (LB), which served as the growth medium. Logarithmic phase cells of both the donor and recipient strains (0.5 mL each) were taken and cultured in 4 mL of fresh LB at 35°C for 18–24 hours without shaking. This incubation period allowed for the transfer of genetic material from the donor to the recipient strain. After incubation, Tryptic Soy Agar (TSA) plate containing selective antibiotics were used to screen for transconjugants. Specifically, the TSA plates were supplemented with 10 μg/L rifampicin and 0.02 μg/L imipenem. These antibiotic concentrations were effective in inhibiting the growth of the donor and recipient strains but allowed the growth of transconjugants that had acquired the resistance genes from the donor strain through conjugation. The transconjugants, which were the result of successful conjugation and acquisition of the resistance genes, could be identified and isolated on the TSA plates supplemented with the selective antibiotics.
For plasmid electroporation, E. coli DH5α cells were used as the recipient cells for transformation experiments. The conjugation frequency was calculated by determining the number of transduced conjugates per initial donor cell. To prepare the electroactive cells, the bacteria were grown to an optical density (OD600) of 0.5–0.6. The cells were then precipitated by centrifugation at 4°C and washed twice with 1 volume of milliQ water, followed by centrifugation at 6,000 rpm at 4°C. Finally, the cells were centrifuged with 1/50 volume of 10% glycerol. This washing process helped in removing any residual media and preparing the cells for electroporation. The prepared electroactive cells were aliquoted and stored at -70°C for backup. For electroporation, aliquots of the electroactive cells were mixed with less than 10 ng of DNA in a 0.2 cm cuvette. The electroporation was performed using a MicroPulser apparatus (Bio-Rad, California, USA) at specific settings, such as 2.5 kV, 25 mOhm, and 200 Ω. This electrical pulse facilitates the uptake of the plasmid DNA by the recipient cells. After electroporation, the cells were immediately transferred into 1 mL of LB medium and incubated at 37°C with shaking. To ensure the expression of the antibiotic resistance encoded by the plasmids, the cells were incubated in a culture medium containing antibiotics. In the case of strain SC35, the plasmids were properly screened using TSA plates supplemented with 10 μg/L rifampicin and 0.02 μg/L imipenem, which allowed for the selection and growth of cells carrying the plasmids.
In the bacterial resistance assay, the BioMerieux VITEK2 system was used to determine the Minimum Inhibitory Concentration (MIC) values of various antibiotics (Table 1). Additionally, the disk diffusion test was performed to measure the diameter (mm) of the zones of inhibition, indicating bacterial resistance. The results obtained were confirmed using the 2022 Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute (CLSI), 2022). A total of 33 antibiotics, including antibiotics combined with enzyme inhibitors, were tested. The MIC values obtained from the VITEK2 system and the zones of inhibition from the disk diffusion test were recorded in Table 1. To ensure the reliability of the assay, E. coli ATCC 25922 was selected as the quality control strain. The MIC values and zone diameter measurements were accurately determined bacterial resistance, and the results obtained from the assay were compared to the breakpoints and interpretive criteria.
According to the CLSI Guidelines 2022, the presence of extended-spectrum beta-lactamases (ESBLs) was determined based on the difference in the diameter of the inhibition zone for specific groups of drugs. Specifically, positive ESBL production was confirmed if the difference in the circle of inhibition diameter was equal to or greater than 5 mm for any of the following drug groups: ceftazidime (CAZ, 30 µg) and ceftazidime/clavulanic acid (30/30 µg), ceftazidime (CTX, 30 µg) and ceftizoxime/clavulanic acid (30/30 µg). This criterion was used to identify bacteria that produced ESBLs, which were enzymes capable of hydrolyzing and inactivating a broad range of beta-lactam antibiotics, including cephalosporins and monobactams.
According to the CLSI Guidelines 2022 (Clinical and Laboratory Standards Institute (CLSI), 2022), the modified carbapenem inactivation method (mCIM) and the modified carbapenem inactivation method + EDTA (eCIM) were used to determine the presence of metal-β-lactamases, specifically class B carbapenemases. In the mCIM, a carbapenem antibiotic was combined with a EDTA to inhibit the activity of metallo-β-lactamases (MBLs). If there was no MBL present, the carbapenem antibiotic was able to inhibit the bacterial growth, resulting in a zone of inhibition. If the addition of EDTA increased the zone of inhibition by 5 mm or more compared to the mCIM, it was considered positive for metal-β-lactamases (class B carbapenemases).
On the other hand, the identification of the class D carbapenemase phenotype, known as class D β- lactamases, currently lacked a definitive method. However, it could be inferred by a measure of exclusion. If the strain did not exhibit inhibition by class A or B inhibitors, it was inferred to possess a class D carbapenemase.
The genome of strain SC35 was sequenced using two different sequencing platforms, PacBio Sequel II and Illumina NovaSeq 6000. The DNA libraries used had an average size of 15 kb for PacBio Sequel II and 400 bp for Illumina NovaSeq 6000. Raw PacBio Sequel II and Illumina reads were trimmed using Canu v2.2 (https://github.com/marbl/canu) and Trimmomatic v0.39 (https://github.com/usadellab/Trimmomatic), respectively. After trimming, 13.7 million clean Illumina reads were obtained with an average length of 148 bp and 100% of reads >=Q20. The paired-end short Illumina reads were then assembled using either Unicycler v0.4.5 (https://github.com/rrwick/Unicycler) or SPAdes v3.15.3 (https://github.com/ablab/spades) to improve the accuracy of data processing. Finally, the post-assembly sequence corrections were performed on the Illumina reads using Pilon v1.24 (https://github.com/broadinstitute/pilon). This comprehensive approach resulted in the generation of accurate DNA sequences for strain SC35.
The prediction of open reading frames (orfs) and pseudogenes was performed using RAST 2.0 (Brettin et al., 2015), BLASTP/BLASTN (Boratyn et al., 2013), UniProtKB/Swiss-Prot (Boutet et al., 2016), and RefSeq databases (O’Leary et al., 2016). Fine annotation of drug-resistance genes, mobile elements, and other features was undertaken through online databases such as CARD 2023 (Alcock et al., 2023), ResFinder 4.0 (Bortolaia et al., 2020), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Tansirichaiya et al., 2019). Cloned MLSTs were identified using MLST 2.0 (https://cge.food.dtu.dk/services/MLST/) and BacWGSTdb 2.0 (Feng et al., 2021). The SC35 plasmid p1 and its related plasmids were compared for multiple and pairwise sequence comparisons using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Circular plots of the chromosome and plasmids were produced by CGView (Stothard et al., 2019). All plasmids comparison figures were created using the R package genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/) and edited using Inkscape v0.48.1 (https://inkscape.org/en).
The sequences of the SC35 chromosome, SC35 plasmid p1, SC35 plasmid p2, SC35 plasmid p3, and SC35 plasmid p4 were deposited on the GenBank database with the accession numbers CP147174.1, CP147175.1, CP147176.1, CP147177.1, and CP147178.1, respectively. The characterization of the related plasmids compared with the SC35 plasmid p1 and F4_plasmid pB could be found in Table 2.
The strain SC35 was identified as K. pneumoniae through BLAST analysis of its 16S rRNA and genome sequences, as well as through nucleotide homology analysis. Table 1 provided the MIC values and circle of inhibition diameters of the strain SC35 in the drug susceptibility testing conducted using BioMerieux VITEK2 and the paper diffusion method. The strain SC35 exhibited resistance to all tested antibiotics (Table 1).
Enzyme characterization revealed that the strain SC35 produced class A beta-lactamase. The MLST sequence was characterized as sequence type (ST) 11 using MLST 2.0 and BacWGSTdb 2.0. Through bacterial conjugation transfer and electroporation experiments, the integrated SC35 plasmid p1 was successfully recovered, with an average conjugation frequency of 6.85 × 10-4.
In the strain SC35, a chromosome and four plasmids were identified, including the SC35 plasmid p1, SC35 plasmid p2, SC35 plasmid p3, and SC35 plasmid p4. On the SC35 chromosome, a total of 40 different genes associated with drug resistance were found. These genes included drug-resistant nodule disintegration (RND) antibiotic efflux pumps (e.g., acrAB-tolC), major facilitator superfamily (MFS) antibiotic efflux pumps (e.g., kpnGH), and ATP-binding cassette (ABC) antibiotic efflux pumps (e.g., msbA) (Figure 1A). For example, it had been documented that upregulation of the expression of the AcrAB-TolC was the major cause of marA-mediated MDR (Li and Nikaido, 2004), whereas AcrAB-TolC overexpression significantly improved the response of marA-mediated MDR to all antibiotics (Ruiz and Levy, 2010). The characterization of drug-resistance genes on the SC35 chromosome could be found in Table 3.
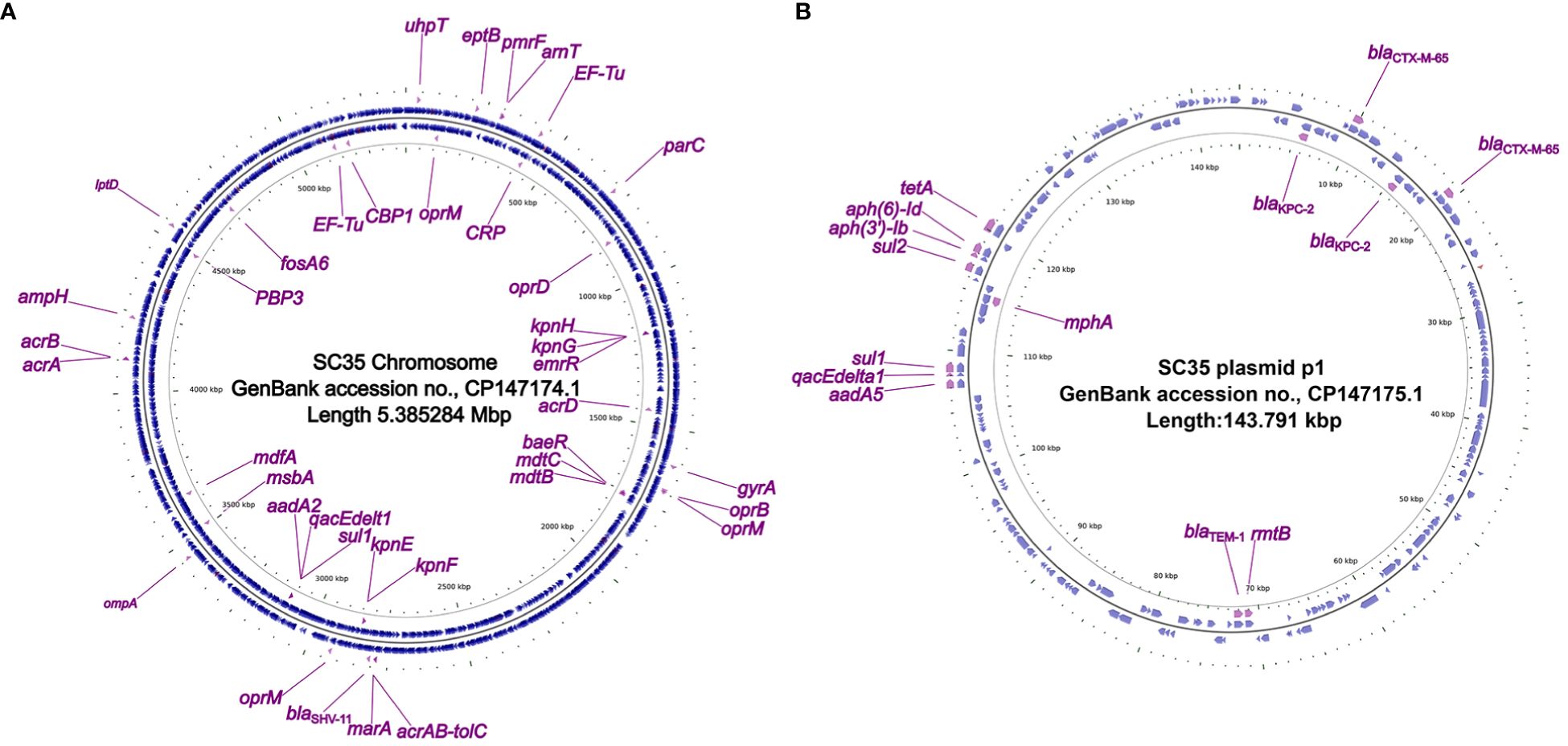
Figure 1 Circos plot of the SC35 chromosome and SC35 plasmid p1 and localization of antibiotic resistance genes. (A) The SC35 chromosome of 5.385284 Mbp in length showing antibiotic resistance genes. (B) The SC35 plasmid p1 of 143.791 kbp in length showing antibiotic resistance genes. The Circos plot of F4_chromosome was established using CGview v2.0.3 (https://github.com/paulstothard/cgview). The locations of the antibiotic resistance genes were drawn manually using Inkscape 0.48.1 (https://inkscape.org/en).
In the SC35 plasmid p1, a total of 14 drug-resistance genes were identified (Figure 1B). These genes conferred resistance to β-lactams, aminoglycosides, cephalosporins, tetracyclines, macrolides, sulfonamides, disinfectants, and antiseptics (Tables 2, 3). Further characterization of these resistance genes would be addressed later (Figures 2, 3). However, no resistance genes were found in the SC35 plasmids p2, p3, and p4 (Supplementary Figure S1).
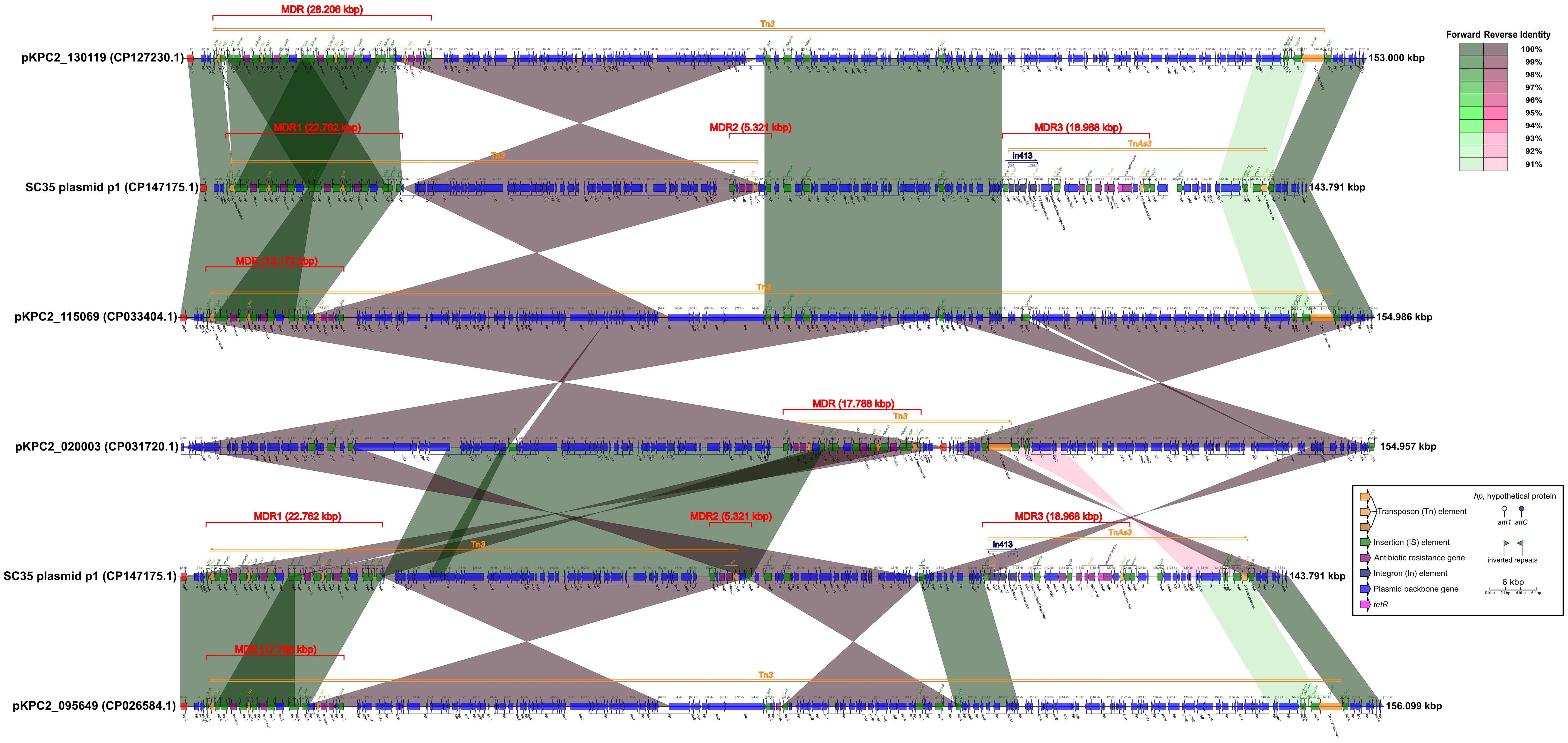
Figure 2 Comparison of structures for SC35 plasmid p1 with closely related plasmids pKPC2_ 130119, pKPC2_115069, pKPC2_020003, and pKPC2_095649. The figure was created by the R package genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/).
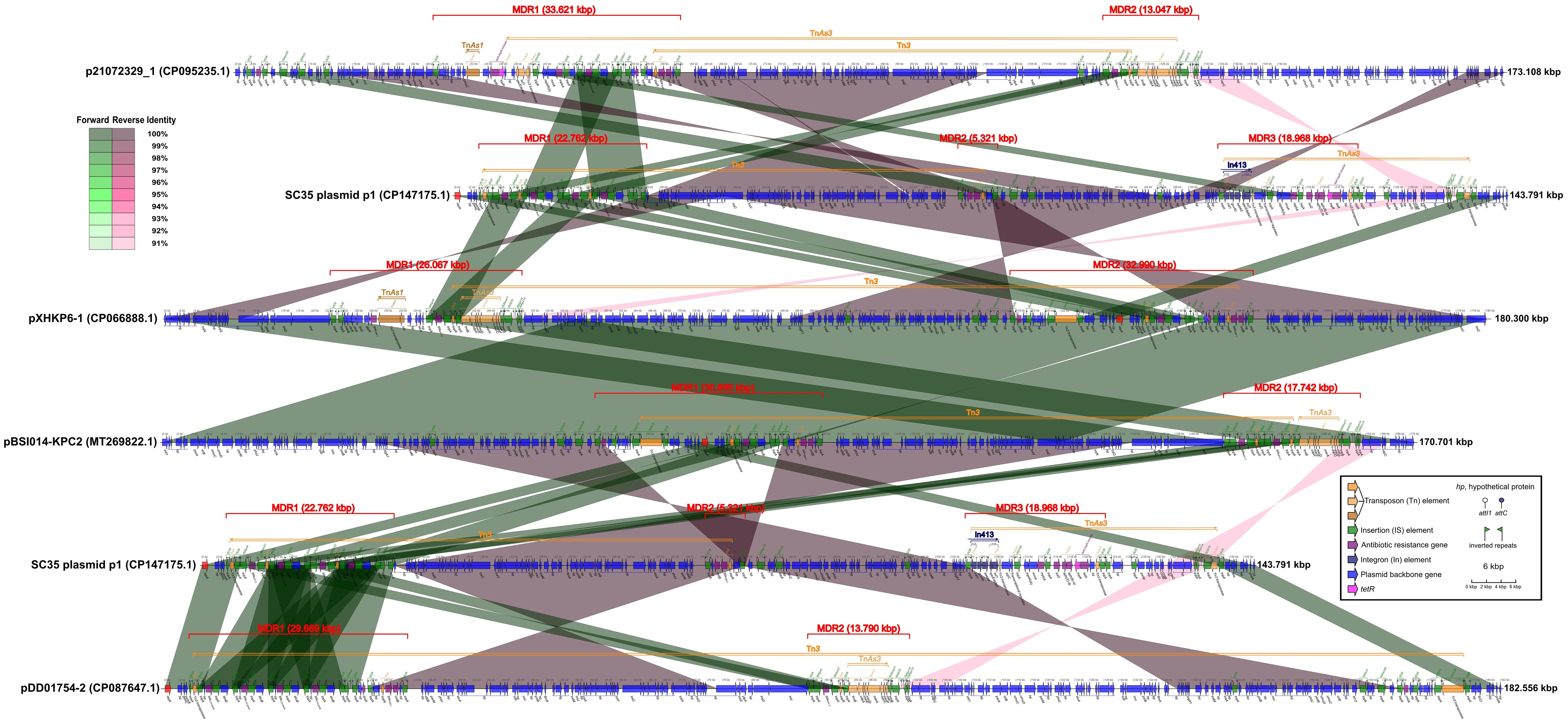
Figure 3 Comparison of structures for SC35 plasmid p1 with other related plasmids p21072329_1, pXHKP6–1, pBSI014-KPC2, and pDD01754–2, China, harboring core structures for blaKPC-2 and blaCTX-M-65. The figure was created by the R package genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/).
The SC35 plasmid p1 contained three multidrug-resistant (MDR) regions, namely MDR1, MDR2, and MDR3. The MDR1 region had a sequence length of 22.762 kbp, ranging from 3.383 kbp to 26.144 kbp. It consisted of the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65, which were double repeats of the genetic array of IS26-ISKpn6-blaKPC-2-ISKpn27-tnpR-IS26-hp- blaCTX-M-65-IS903-btuB-IS26 (Figure 2). The MDR2 region had a sequence length of 5.321 kbp, ranging from 68.642 kbp to 73.962 kbp, and contained the genetic array IS26-hp-rmtB- blaTEM-1-tnpR-hp-hp-IS26. The MDR3 region had a sequence length of 18.968 kbp, ranging from 104.056 kbp to 123.023 kbp, and included In413 as well as the antibiotic resistance genes mphA, sul2, aph(3’)-Ib, aph(6)-Id and tetA (Figure 2). The structure of SC35 plasmid p1 was characterized by the overlap and intersection of mobile genetic elements as Tn3, TnAs3, and In413 (Figure 2). Comparison of the SC35 plasmid p1 with pKPC2_130119 showed that the MDR1 region of SC35 plasmid p1 shared excellent identities with the MDRs of both pKPC2_130119 and pKPC2_115069 (Figure 2). The MDR2 region of SC35 plasmid p1 was inversely identified with the posterior regions of the MDR of pKPC2_130119, ranging from 26.918 kbp to 31.518 kbp (Figure 2). The MDR3 region of SC35 plasmid p1 had no identities with the MDRs of pKPC2_130119 and pKPC2_115069 (Figure 2). Similarly, the MDR1 region of SC35 plasmid p1 was identical to the MDRs of pKPC2_ 115069 and pKPC2_095649, respectively, only in a genetic array of IS26-ISKpn6-blaKPC-2-ISKpn27- tnpR-IS26-hp-blaCTX-M-65-IS903-btuB-IS26 (Figure 2). The MDR2 region of SC35 plasmid p1 was inversely identified with the posterior regions of the MDRs of pKPC2_115069 and pKPC2_095649, ranging from 15.850 kbp to 21.170 kbp (Figure 2). The MDR3 region of SC35 plasmid p1 had no identities with the MDRs of pKPC2_115069 and pKPC2_095649 (Figure 2). The MDR1 region of SC35 plasmid p1 had inversely extreme identity with the MDR of pKPC2_020003, ranging from 82.857 kbp to 96.028 kbp (Figure 2). The MDR2 region of SC35 plasmid p1 was inversely identified with a portion of the MDR of pKPC2_020003, ranging from 78.241 kbp to 83.561 kbp. The MDR3 region of SC35 plasmid p1 also had no identity with the MDR of pKPC2_020003 (Figure 2).
When SC35 plasmid p1 was compared to p21072329_1, pXHKP6–1, and pBSI014- KPC2, one of the two blaCTX-M-65 core structures (IS26-hp-blaCTX-M-65-IS903-btuB- IS26) in the MDR of SC35 plasmid p1 was identical to a blaCTX-M-65 core structure in the MDR regions of p21072329_1, pXHKP6–1, and pBSI014-KPC2 (Figure 3). One of the two blaKPC-2 core structures (IS26-ISKpn6-blaKPC-2-ISKpn27 -tnpR-IS26) in the MDR region of SC35 plasmid p1 was almost identical to a blaKPC-2 core structure in the MDR regions of p21072329_1, pXHKP6–1, and pBSI014-KPC2. However, the arrangement of blaKPC-2 or blaCTX-M-65 core structure differed between the SC35 plasmid p1, p21072329_1, pXHKP6–1, and pBSI014-KPC2 (Figure 3). Additionally, a genetic array of hp-fosA-IS26-hp-hp-tnpR-blaTEM-1-rmtB-hp-IS26 followed the blaCTX-M-65 core structure in p21072329_1, pXHKP6–1, and pBSI014-KPC2, but was absent in the SC35 plasmid p1 (Figure 3). The MDR3 region of SC35 plasmid p1 also had no identity with the MDR regions of p21072329_1, pXHKP6–1, and pBSI014- KPC2 (Figure 3). When the SC35 plasmid p1 was compared to pDD01754–2, one of the two blaCTX-M-65 core structures (IS26-hp-blaCTX-M-65-IS903-btuB-IS26) in the MDR of SC35 plasmid p1 was identical to one of the four repetitive tandem blaCTX-M-65 core structures in the MDR of pDD01754–2. Either of the two blaCTX-M-65 core structures (IS26-hp-blaCTX-M-65-IS903-btuB-IS26) in the MDR region of SC35 plasmid p1 was identical to either of the four repetitive tandem blaCTX-M-65 core structures in the MDR region of pDD01754–2 (Figure 3). One of the two blaKPC-2 core structures (ISKpn6-blaKPC-2-ISKpn27-tnpR-IS26) in the MDR region of SC35 plasmid p1 was identical to a blaKPC-2 core structure in the MDR region of pDD01754–2 (Figure 3). The MDR3 region of SC35 plasmid p1 also had no identity with the MDR region of pDD01754–2 (Figure 3).
By comparing the SC35 plasmid p1 with pDD01754.2 and F4_plasmid pA, it was found that there were good identities between the MDR regions of SC35 plasmid p1, pDD01754.2, and F4_plasmid pA (Figure 4A). The sequence structure of F4_plasmid pA from 68.014 kbp to 90.440 kbp was nearly identical to that of pDD01754.2 from 82.573 kbp to 100.253 kbp, except for an additional blaKPC-2 core structure in F4_plasmid pA (IS26-ISKpn6-blaKPC-2-ISKpn27-tnpR-IS26) (Figure 4A). Either of the two blaKPC-2 core structures on the SC35 plasmid p1 matched exactly with either of the two tandem blaKPC-2 core structures on F4_plasmid pA (Figure 4A).
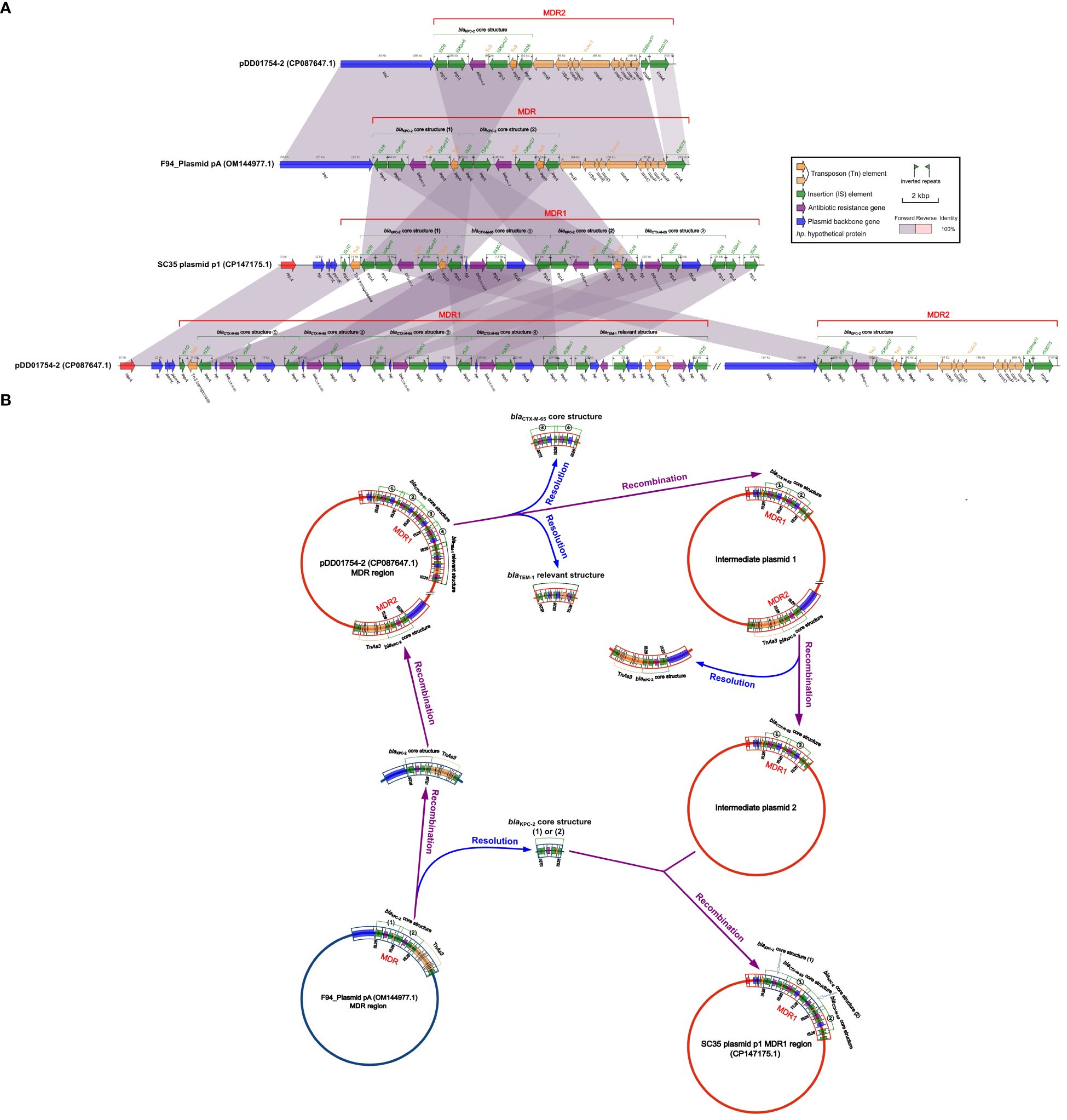
Figure 4 Structural relationship diagram between SC35 plasmid p1, pDD01754.2 and F4_plasmid pA. (A) Comparison of structures btween SC35 plasmid p1, pDD01754.2 and F4_plasmid pA. The figure was created by the R package genoPlotR v0.8.11 software (http://genoplotr.r-forge.r-project.org/). (B) Inference plot of the relationship between SC35 plasmid p1, pDD01754.2 and F4_plasmid pA. The figure was created with hand finishing.
Based on the consistency of the structures of SC35 plasmid p1 with pDD01754.2 and F4_plasmid pA (Figure 4A), genetic inferences were made as shown in Figure 4B. In the evolutionary processes, under suitable conditions, two of the four repetitive tandem blaCTX-M-65 core structures (③ and ④) and the blaTEM-1 related structures (hp-fosA-IS26-hp-hp-tnpR-blaTEM-1-rmtB-hp-IS26) were detached from the MDR1 region of pDD01754.2, forming an intermediate plasmid 1 (Figure 4B). Next, the MDR2 region of intermediate plasmid 1, which contained the blaKPC-2 core structure, was delinked, resulting in the development of an intermediate plasmid 2 that only had two repetitive tandem blaCTX-M-65 core structures (① and ②) (Figure 4B). Subsequently, F4_plasmid pA, which contained the two repetitive tandem blaKPC-2 core structures, underwent disassociation into one blaKPC-2 core structure (1) or (2), or two blaKPC-2 core structures (1) and (2), along with a blaKPC-2 core structure + TnAs3 (Figure 4B). The resolved blaKPC-2 core structures (1) and (2) were then recombined with intermediate plasmid 2, which contained the two repetitive tandem blaCTX-M-65 core structures (① and ②). A blaKPC-2 core structure (1) was integrated in front of blaCTX-M-65 ①, and another blaKPC-2 core structure (2) was inserted in front of blaCTX-M-65 ②, resulting in the formation of a novel MDR1 region. This novel MDR1 region corresponds to the MDR1 region of SC35 plasmid p1 (Figure 4B). Simultaneously, the resolved blaKPC-2 core structure + TnAs3 was recombined into pDD01754.2, and the MDR2 region of pDD01754.2 was reorganized (Figure 4B).
The acrAB-tolC resistance-nodulation-cell division (RND) antibiotic efflux pump, composed of the extracellular cytoplasmic fusion protein AcrA, the drug proton transporter AcrB, and the outer membrane channel protein TolC, was responsible for bacterial resistance to various agents including fluoroquinolones, tetracyclines, cephalosporins, glycylcyclines, penams, rifamycins, phenicol antibiotics, disinfectants, and antiseptics (Li et al., 2015) (Table 3). The expression of the marA gene, which regulated the transcription of the acrAB system, significantly increased the activity of the efflux pump by more than 2.8-fold and induced resistance to a wide range of antibiotics (Shi et al., 2022). In the SC35 strain, the presence of acrAB-tolC on the chromosome led to a high level of resistance to various antibiotics including fluoroquinolones, tetracyclines, cephalosporins, cephalomycins, meropenem, and glycylcyclines (tigecycline) (Table 1).
In addition to the RND efflux pump, the SC35 chromosome also contained MFS antibiotic efflux pump and ABC antibiotic efflux pump (Table 3). These efflux pumps contributed to the resistance phenotype of the SC35 strain. Furthermore, the presence of various non-efflux pump resistance genes on both the SC35 chromosome and SC35 plasmid p1, along with the overlapping and interaction of mobile genetic elements as In413, Tn3, and TnAs3 on the SC35 plasmid p1 (Figures 1–3), further contributed to the emergence of a highly multidrug-resistant strain SC35 (Table 1). The combination of these resistance mechanisms resulted in the almost complete resistance observed in the strain SC35.
In China, ST11 had been the most prevalent carbapenem-resistant strain of K. pneumoniae, and the majority of these ST11-KPC-2 strains were considered highly threatening clones. Infections caused by these prevalent strains were often difficult to treat, as they exhibited limited response to available treatment options (Zhang et al., 2017; Wang et al., 2018; Zhang et al., 2018). The SC35 strain, belonging to ST11, produced carbapenemase and possessed antibiotic resistance genes solely on the SC35 plasmid p1. Through sequencing analysis, it was discovered that the SC35 plasmid p1 carried the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65 in the MDR1 region, as well as blaTEM-1 with rmtB in the MDR2 region, and other resistance genes that overlapped with mobile genetic elements as In413 and TnAs3 in the MDR3 region (Figures 2, 3). The combination of resistance genes with various functions on both the chromosome and plasmids was what contributed to the challenging situation of having limited treatment options for the SC35 strain.
By comparing the SC35 plasmid p1 with closely related plasmids from Chengdu, China (pKPC2_130119, pKPC2_115069, pKPC2_020003, and pKPC2_095649), it was found that the SC35 plasmid p1 shared the highest similarity with pKPC2_130119 in the MDR1 and MDR2 regions. Both plasmids contained the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65, and blaTEM-1 with rmtB (forward or reversed). In contrast, the other plasmids only had one tandem core structure for blaKPC-2 and blaCTX-M-65, and blaTEM-1 with rmtB (forward or reversed). None of these plasmids, except the SC35 plasmid p1, had an MDR3 region (Figure 2). Therefore, The SC35 plasmid p1 showed a high level of similarity to the plasmid from Chengdu, China, with the only difference being the absence of an MDR3 region. Similarly, when comparing the SC35 plasmid p1 with other related plasmids (p21072329_1, pXHKP6–1, pBSI014-KPC2, and pDD01754–2) that had the blaKPC-2 core structure arranged separately from the blaCTX-M-65 core structure, it was observed that the blaCTX-M-65 core structure of SC35 plasmid p1 was more similar to that of pDD01754–2. On the other hand, the arrangement of the blaKPC-2 core structure in pDD01754–2 was identical to that of the SC35 plasmid p1 (Figure 4A). This finding was consistent with the previous discovery of F4_plasmid pA, which shared perfect identity with the SC35 plasmid p1 (Ma et al., 2023). Based on these observations, it was hypothesized that pDD01754–2, F4_plasmid pA, and SC35 plasmid p1 might be evolutionarily linked (Figure 4B).
The presence of similarities in the structure of SC35 plasmid p1 with plasmids such as pKPC2_130119, pDD01754–2, and F4_plasmid pA suggests that these plasmids are interconnected in their evolutionary processes. The strain SC35, which carries multiple efflux pumps on its chromosome (such as acrAB-tolC) and harbors the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65, and blaTEM-1 with rmtB on a plasmid with overlapping structures of mobile genetic elements as In413, Tn3, and TnAs3, poses a significant threat. Effective preventive measures should be taken to stop the spread and prevalence of such antibiotic-resistant bacteria.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
This study was approved by the Ethics Committee of Taizhou Municipal Hospital, Zhejiang, China, and written informed consent was obtained from each of the participants in accordance with the Declaration of Helsinki. The rights of the research subjects were protected throughout, and we confirm that this study was conducted in our hospital. The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of the indicated institutions, and the protocols were carried out in accordance with approved guidelines.
LH: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. WW: Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing, Software. LM: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing, Project administration. DW: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing, Conceptualization, Funding acquisition, Writing – original draft. SL: Data curation, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Programs of Zhejiang Health Department of China (2021KY405 and 2023KY1319), Zhejiang Provincial Natural Science Foundation of China (LTGD24H160008), and the Science and Technology Program of Wenling City (No.2020S0180089), Zhejiang Province, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1410921/full#supplementary-material
Supplementary Figure 1 | Circos plots of SC35 plasmid p2, SC35 plasmid p3, and SC35 plasmid p4. The figure was established using CGview v2.0.3 (https://github.com/paulstothard/cgview).
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST; a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978–1-4939–3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Cantón, R., Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Clinical and Laboratory Standards Institute (CLSI) (2022). Performance Standards for Antimicrobial Susceptibility Testing. Thirty-two Informational Supplement. M100–S32. Wayne, PA: Clinical and Laboratory Standards. https://clsi.org
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, Y., Zou, S., Chen, H., Yu, Y., Ruan, Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49, D644–DD650. doi: 10.1093/nar/gkaa821
Frank, J. A., Reichm, C. I., Sharma, S., Weisbaumm, J. S., Wilson, B. A., Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16 S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Hamidian, M., Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 5, e000306. doi: 10.1099/mgen.0.000306
Li, X. Z., Nikaido, H. (2004). Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204. doi: 10.2165/00003495-200464020-00004
Li, X. Z., Plesiat, P., Nikaido, H. (2015). The challenge of efux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Li, X., Quan, J., Ke, H., Wu, W., Feng, Y., Yu, Y., et al. (2021). Emergence of a KPC variant conferring resistance to ceftazidime-avibactam in a widespread ST11 carbapenem-resistant Klebsiella pneumoniae clone in China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.724272
Liu, J., Xie, J., Yang, L., Chen, D., Peters, B. M., Xu, Z., et al. (2018). Identification of the KPC plasmid pCY-KPC334: new insights on the evolution pathway of the epidemic plasmids harboring fosA3–blaKPC-2 genes. Int. J. Antimicrob. Agents. 52, 510–511. doi: 10.1016/j.ijantimicag.2018.04.013
Ma, L., Wang, W., Qu, Y., Wang, D. (2023). Characterization of the two repetitivetandem repeats for the KPC-2 core structures on a plasmid from hospital-derived Klebsiella pneumoniae. Sci. Rep. 13, 12049. doi: 10.1038/s41598-023-38647-z
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, 733–745. doi: 10.1093/nar/gkv1189
Pitout, J. D., Nordmann, P., Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/AAC.01019-15
Qu, Y., Wang, W., Lu, Q., Qiu, J., Wang, D., Ma, L. (2023). Occurrence and characterization of plasmids carrying tmexCD1-toprJ1, blaDHA-1 and blaCTX-M-127, in Klebsiella pneumoniae clincal strains. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1260066
Reygaert, W. C. (2018). An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 4, 482–501. doi: 10.3934/microbiol.2018.3.482
Ruiz, C., Levy, S. B. (2010). Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob. Agents Chemother. 54, 2125–2134. doi: 10.1128/AAC.01420-09
Shi, D., Hao, H., Wei, Z., Yang, D., Yin, J., Li, H., et al. (2022). Combined exposure to non-antibiotic pharmaceutics and antibiotics in the gut synergistically promote the development of multi-drug-resistance in Escherichia coli. Gut Microbes 14, 2018901. doi: 10.1080/19490976.2021.2018901
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stothard, P., Grant, J. R., Van Domselaar, G. (2019). Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 20, 1576–1582. doi: 10.1093/bib/bbx081
Tansirichaiya, S., Rahman, M. A., Roberts, A. P. (2019). The transposon registry. Mob. DNA. 10, 40. doi: 10.1186/s13100-019-0182-3
Villa, L., García-Fernández, A., Fortini, D., Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wang, Y., He, J., Sun, L., Jiang, Y., Hu, L., Leptihn, S., et al. (2023). IS26 mediated blaCTX-M-65 amplification in Escherichia coli increase the antibiotic resistance to cephalosporin in vivo. J. Glob. Antimicrob. Resist. 35, 202–209. doi: 10.1016/j.jgar.2023.09.018
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China, (2012–2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
World Health Organization (WHO) (2014). Antimicrobial resistance: global report on surveillance (Geneva, Switzerland: WHO).
Xiang, D. R., Li, J. J., Sheng, Z. K., Yu, H. Y., Deng, M., Bi, S., et al. (2016). Complete sequence of a novel IncR-F33:A-:B-plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob. Agents Chemother. 60, 1343–1348. doi: 10.1128/AAC.01488-15
Yamasaki, S., Zwama, M., Yoneda, T., Hayashi-Nishino, M., Nishino, K. (2023). Drug resistance and physiological roles of RND multidrug efflux pumps in Salmonella enterica, Escherichia coli and. Pseudomonas aeruginosa. Microbiol. (Reading). 169, 1322. doi: 10.1099/mic.0.001322
Yu, K., Huang, Z., Xiao, Y., Gao, H., Bai, X., Wang, D. (2024). Global spread characteristics of CTX-M-type extended-spectrum β-lactamases: A genomic epidemiology analysis. Drug Resist. Updat. 73, 101036. doi: 10.1016/j.drup.2023.101036
Zhai, Y., Li, D., Du, P., Zhang, Z., He, Z., Guo, Y., et al. (2021). Complete sequences of two new KPC-harbouring plasmids in Klebsiella pneumoniae ST11 strains in China. J. Glob. Antimicrob. Resist. 24, 114–120. doi: 10.1016/j.jgar.2020.11.023
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBio Med. 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Keywords: K. pneumoniae, SC35 chromosome, SC35 plasmid p1, acrAB-tolC, the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65
Citation: He L, Wang W, Ma L, Wang D and Long S (2024) Emergence of a clinical Klebsiella pneumoniae harboring an acrAB-tolC in chromosome and carrying the two repetitive tandem core structures for blaKPC-2 and blaCTX-M-65 in a plasmid. Front. Cell. Infect. Microbiol. 14:1410921. doi: 10.3389/fcimb.2024.1410921
Received: 04 April 2024; Accepted: 14 June 2024;
Published: 02 July 2024.
Edited by:
Maria De Lurdes Enes Dapkevicius, University of the Azores, PortugalReviewed by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyCopyright © 2024 He, Wang, Ma, Wang and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongguo Wang, d2RndHpzQDE2My5jb20=; Shanshan Long, bG9uZ3NzNDIzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.