
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 05 June 2024
Sec. Bacteria and Host
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1410385
This article is part of the Research Topic Synergistic Approaches to Managing Gram-negative Bacterial Resistance View all 14 articles
Introduction: Stenotrophomonas is a prominent genus owing to its dual nature. Species of this genus have many applications in industry and agriculture as plant growth-promoting rhizobacteria and microbial biological control agents, whereas species such as Stenotrophomonas maltophilia are considered one of the leading gram-negative multi-drug-resistant bacterial pathogens because of their high contribution to the increase in crude mortality and significant clinical challenge. Pathogenic Stenotrophomonas species and most clinical isolates belong to the Stenotrophomonas maltophilia complex (SMc). However, a strain highly homologous to S. terrae was isolated from a patient with pulmonary tuberculosis (TB), which aroused our interest, as S. terrae belongs to a relatively distant clade from SMc and there have been no human association reports.
Methods: The pathogenicity, immunological and biochemical characteristics of 610A2T were systematically evaluated.
Results: 610A2T is a new species of genus Stenotrophomonas, which is named as Stenotrophomonas pigmentata sp. nov. for its obvious brown water-soluble pigment. 610A2T is pathogenic and caused significant weight loss, pulmonary congestion, and blood transmission in mice because it has multiple virulence factors, haemolysis, and strong biofilm formation abilities. In addition, the cytokine response induced by this strain was similar to that observed in patients with TB, and the strain was resistant to half of the anti-TB drugs.
Conclusions: The pathogenicity of 610A2T may not be weaker than that of S. maltophilia. Its isolation extended the opportunistic pathogenic species to all 3 major clades of the genus Stenotrophomonas, indicating that the clinical importance of species of Stenotrophomonas other than S. maltophilia and potential risks to biological safety associated with the use of Stenotrophomonas require more attention.
Stenotrophomonas is a member of the family Xanthomonadaceae. Because of advancements in molecular biology and identification technology, the number of species in this genus has increased rapidly in recent years. Based on our interpretation of the current available NCBI taxonomy, from the first species, Stenotrophomonas maltophilia, isolated in 1885 to 2011, 16 strains have been isolated within 117 years, and 12 species were identified within 9 years from 2016 to 2024. Stenotrophomonas is ubiquitous and associated with a wide range of habitats, including humans, animals, plant hosts, and extreme environments. Most members of Stenotrophomonas are particularly known for producing protective osmotic substances and are considered as plant rhizosphere growth-promoting bacteria. They also have valuable applications in the fields of agriculture and industry as emerging sources of biodegradation and substitutes for synthetic fungicides (Kumar et al., 2023). Stenotrophomonas strains can interact with other microorganisms on plant surfaces and in the soil through biofilms, inhibit plant pathogenic fungi and viruses, and alter the composition of rhizosphere microorganisms through extracellular enzyme decomposition and competition for iron. By inducing the enrichment of plant hormones, such as jasmonic acid, and upregulating the transcription level of jasmonic acid-sensitive genes, Stenotrophomonas strains help plants defend against various crop pests, such as Spodoptera litura. Their function in promoting plant growth is through the production of auxin and hydrogen cyanide; dissolution of phosphorus and potassium salts; nitrogen fixation; enzymatic degradation of soil odour, explosive pollutants, keratin, macrocyclic hydrocarbons, nitrophenols, and other substances; removal of various chemical pesticides, insecticides, and environmental pollutants; and effective bioremediation of agricultural soil without harming the ecosystem (Kumar et al., 2023).
A few species of this genus are pathogenic to humans and are represented by S. maltophilia. As a globally emerging organism, S. maltophilia is recognised for its multi-drug resistance (Gil-Gil et al., 2020), ability to cause various infections in the human body, and high contribution to the increase in crude mortality (Tan et al., 2014). Based on global clinical data, the attributed mortality rates for pulmonary infection and bacteraemia caused by this bacterium are as high as 30% and 65%, respectively (Zhou et al., 2013). S. maltophilia is listed by the World Health Organization as one of the leading gram-negative, multi-drug-resistant bacterial pathogens in hospitals (Gröschel et al., 2020) and poses a great clinical challenge (Mojica et al., 2022; Tamma et al., 2022).
Because of their phenotypic and genotypic diversity, the taxonomic status of the genus Stenotrophomonas and S. maltophilia has changed several times. Besides the species in the Stenotrophomonas maltophilia complex (SMc), S. maltophilia (Chang et al., 2015; Patil et al., 2018; Bansal et al., 2021; Brooke, 2021) and S. sepilia (Gautam et al., 2021), only S. pavanii (Iebba et al., 2018), S. acidaminiphila (Ježek et al., 2020; Zhang et al., 2022) and S. rhizophila (Chao et al., 2022) have been isolated from human samples. The clinically important species S. africana (Coenye et al., 2004) is classified as a synonym of S. maltophilia, whereas S. pavanii has been removed from SMc (Rouf et al., 2011; Patil et al., 2018; Maturana and Cardenas, 2021; Deng et al., 2022). Most of the clinical isolates belong to the 23 monophyletic lineages of SMc (Gröschel et al., 2020), and only S. acidaminiphila and S. rhizophila have no close genetic relationship to S. maltophilia.
In our previous small surveillance conducted in a district tuberculosis (TB) hospital in Beijing, we found that Stenotrophomonas was a major co-occurring species of Mycobacterium tuberculosis, with a crude isolation rate of 6.74%. Stenotrophomonas may be an important opportunistic pathogen for patients with TB, similar to that for patients with cystic fibrosis (Li et al., 2023). On the basis of 16S rDNA sequencing, 2 out of 9 Stenotrophomonas isolates did not belong to S. maltophilia, and one had the highest identity with S. terrae strain AFS037341 (99.24%) and S. humi strain AFS068096 (98.96%). These similarity values were more than 98.65%, which has been previously used as the threshold for differentiating Stenotrophomonas species (Deng et al., 2022). To the best of our knowledge, S. terrae and its closely related species have not been isolated from human resources, which sparked our interest in the pathogenicity of these strains. However, after PacBio whole-genome sequencing, the average nucleotide identity (ANI) between the strain 610A2T, which was highest similar to S. terrae, and all existing species of the genus Stenotrophomonas was less than 88%. Therefore, in addition to animal experiments and pathogenicity assessments, we conducted a systematic evaluation of 610A2T according to the description required for new taxa.
Strain 610A2T was isolated from a 26-year-old male patient with pulmonary TB under treatment at the tuberculosis clinic of the Chaoyang District Center for Disease Control and Prevention (39.89 N, 116.40 E) in our previous surveillance under ethics approval No. ICDC-2022010 (Li et al., 2023). It was first identified as Stenotrophomonas by amplification and sequencing with 16S rDNA universal primers for bacteria (16S-U: 5′AGA GTT TGA TCM TGG CTC AG 3′ and/L: 5′ CCG TCA ATT CMT TTR AGT TT 3′).
Genome of 610A2T was sequenced using the PacBio sequel II and DNBSEQ platform at the Beijing Genomics Institute (BGI, Shenzhen, China). Four SMRT cell zero-mode waveguide arrays for sequencing were used to generate the sub-read set. The PacBio subreads (length < 1 kb) were removed. Canu and GATK (https://www.broadinstitute.org/gatk/) were used for self-correction and single-base corrections, to improve the accuracy of the genome sequences.
Gene prediction was performed using glimmer3 with Hidden Markov models (http://www.cbcb.umd.edu/software/glimmer/). Other genomic elements, such as RNA, tandem repeats, genomic regions, small satellite DNA, and microsatellite DNA, were identified using tRNAscan SE, RNAmmer, Tandem Repeat Finder (http://tandem.bu.edu/trf/trf.html), Genomic Island Suite of Tools (http://www5.esu.edu/cpsc/bioinfo/software/GIST/), PHAge Search Tool (PHAST, http://phast.wishartlab.com), and CRISPRFinder. Eleven databases, namely, KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp), COG (Clusters of Orthologous Groups, http://www.ncbi.nlm.nih.gov), NR (Non-Redundant Protein Database databases, ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz), Swiss-Prot (http://ngdc.cncb.ac.cn), GO (Gene Ontology, http://www.geneontology.org), TrEMBL, EggNOG (http://eggnog.embl.de), VFDB (Virulence Factors of Pathogenic Bacteria, http://www.mgc.ac.cn), ARDB (Antibiotic Resistance Genes Database, http://www.argodb.org/), CAZy (Carbohydrate-Active enZYmes Database, http://www.cazy.org), and T3SS (Type III secretion system effector protein, EggNOG database 1.0.1, http://effectivedb.org), were used for general function annotation and pathogenicity/drug resistance analysis.
Complete 16S rDNA gene sequences were extracted from the sequenced genome and reference genomes of 26 species of the genus Stenotrophomonas, except for S. detusculanense and S. indologenes, which do not have publicly available genome sequences. ClustalX was used for multiple sequence alignments, and a bootstrap neighbour-joining (NJ) phylogenetic tree was constructed using Treeview (1.6.6) with 1000 replicas. ANI was estimated using the ANI calculator (http://enve-omics.ce.gatech.edu/ani/). Syntenies were performed using MUMmer and BLAST. Core/Pan genes were clustered by CD-HIT rapid clustering of similar proteins software with a threshold of 50% pairwise identity and 0.7 length difference cutoff in amino acid. Hcluster_sg software was used to perform gene family clustering, and a phylogenetic tree was constructed using TreeBeST with the NJ method.
The strain’s growth ability was tested on 4 media, tryptone soy (TSA; Difco), Luria-Bertani (LB; Difco), Columbia blood agar (OXOID), and Mueller-Hinton (MH; Difco), at 37 °C for 24–72 h. Growth at different temperatures (4, 15, 20, 25, 28, 35, 37, 40, and 45 °C), pH (pH 4–11, at intervals of 1 pH unit, 37 °C) and NaCl concentrations (0–10%, w/v, in intervals of 1%) was also evaluated using LB broth and agar at 37 °C. Cellular motility was monitored using 0.75% semi solid agar medium, and morphological features were observed using a transmission electron microscope (HT7700; Hitachi, Japan). Utilisation of carbon sources and enzyme production were tested with API 20NE (48 h, 28 °C), API ZYM (4 h, 28 °C), and API 50 CH (inoculated with AUX medium, 48 h, 28 °C) (20050, 50300, 25200; BioMerier France, France), according to the manufacturer’s instructions. Cellular fatty acids and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) were analysed using the Sherlock Microbial Identification System (MIDI) with the MIDI Sherlock software program (version 6.3), RTSBA6 (6.21) library, and Autoflex speed TOF/TOF (Bruker Daltonics GmbH, Germany), according to the manufacturers’ instructions.
Two kinds of AST plates, the customised AST plate for Chinese Pathogen Identification Net (CHNENF, Trek Diagnostic Systems Ltd, West Sussex, United Kingdom) and the Sensititre™ MYCOTB minimum inhibitory concentration (MIC) plate (Trek Diagnostic Systems, Cleveland, OH, USA), were used to determine the strain’s sensitivity to 17 drugs commonly used for gram-negative strains and 12 anti-TB drugs. MICs were determined according to the standards of the Clinical and Laboratory Standard Institute (CLSI, 2018) and manufacturer’s instructions.
Biofilm formation was determined using crystal violet staining. Fresh bacterial suspension was prepared and adjusted to 0.5 McFarland. After 1:30-fold dilution with tryptone soy broth (TSB; Difco), 150 μL/well suspension was added to a 96-well flat-bottom microtitre plate, and the plate was incubated at 37 °C. At each time point, bacterial turbidity (OD630 nm) was measured using an enzyme-linked immunosorbent assay (ELISA) reader (Bio-Rad 680; Bio-Rad Laboratories, Japan). The cultures were discarded, and the wells were washed 3 times with PBS (pH 7.3) to remove planktonic cells. The biofilms were stained with 200 μL/well of 0.1% crystal violet for 15 min. The wells were washed again, and 200 μL of 99% ethanol was added and OD403 nm was measured after 15 min. The biofilm index was calculated as OD403 nm/OD630 nm. Six clinically isolated S. maltophilia strains, including strain 11066 (GDMCC 1.4335), were used as controls.
Animal experiments were conducted with the approval of the Laboratory Animal Welfare & Ethics Committee of the National Institute for Communicable Disease Control and Prevention (Issue number 2023-021).
Using a random number method, female Kunming mice aged 6–7 weeks and weighing 30–35 g were randomly divided into infection and control groups. Five mice in each infection group were intranasally inoculated with 50 μL of 1.5 × 108 CFU/mL of tested strains, and mice in the control groups were inoculated with PBS. At 4 h, 12 h, 1 day, 2 days, 3 days, 5 days, and 7 days post-infection, the mice were sacrificed and the lung and spleen tissues were collected aseptically.
The left lobes of the lungs were fixed in 10% neutral formalin for 24 h, followed by tissue processing and paraffin embedding. The paraffin blocks were sectioned at 2 µm, stained with haematoxylin and eosin, and blindly examined under a microscope by an expert in the field of laboratory animal pathology. Pulmonary clearance and occurrence of disseminated infection were monitored via quantitative bacteriology of the lung and spleen homogenates, respectively. Briefly, tissues were homogenised (1000 rpm) on ice in 1 mL of sterile normal saline by using a homogeniser (RZ-GR96A; Beijing Guoke Rongzhi Biotechnology Co., Ltd., China). Then, 100 μL of homogenates and 10-fold serial dilutions were inoculated on LB agar. The number of colonies was counted 24–48 h after incubation at 37°C. Bacterial colony counts were normalised according to the wet tissue weight and calculated as CFU/g. The levels of murine tumour necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), gamma interferon (IFN-γ), interleukin (IL)-2, IL-4, IL-6, IL-10, IL-12p70, and IL-17A were quantified using Luminex technology (R&D Systems, Minneapolis, MN, USA) and reagents (12002798; BIO-RAD, USA).
For comparison of multiple groups, one-way ANOVA was performed using GraphPad Prism version 9.3.1 for Windows (GraphPad Software, San Diego, California, USA).
The total length of 610A2T complete genome was 4,681,496 bp, and GC content was 63.29% with an N50 length of 11,086 bp. A total of 4,070 CDSs, 70 tRNAs, 6 5S rRNAs, 4 16S rRNAs, 4 23S rRNAs, 37 sRNAs, 353 tandem repeats, 175 minisatellite DNAs, and 90 microsatellite DNAs were annotated.
In the phylogenetic tree based on the complete 16S rDNA gene (1545 bp), 610A2T was clustered in a clade including S. terrae, S. nitritireducens, S. humi, and S. pictorum and formed a subclade between S. terrae and S. humi (Figure 1). Dispensable gene heat maps and phylogenetic trees based on corePan and genefamily1 results showed completely consistent positioning (Figures 2A, B). When compared with the 26 Stenotrophomonas type species, 610A2T displayed lower ANI values ranging from 87.53 to 80.22, less than 95%, the generally accepted threshold for species (Maturana and Cardenas, 2021; Deng et al., 2022). Synteny analyses at the nucleotide and amino acid levels were compared with 8 type strains: S. humi DSM 18929, S. sepilia SM16975, S. geniculata FLMAT1, S. maltophilia NCTC10257, S. terrae DSM 18941, S. nitritireducens DSM 12575, S. rhizophila DSM 14405, and S. acidaminiphila T0-18. S. terrae had the highest homology, with a gene median identity of 91.34% and map length rate of 63.327% (Table 1). These data suggest that 610A2T represents a novel species within the genus Stenotrophomonas, and it is closely related to, but distinct from, S. terrae. The name Stenotrophomonas pigmentata sp. nov. is proposed for the distinct brown water-soluble pigment it produces.
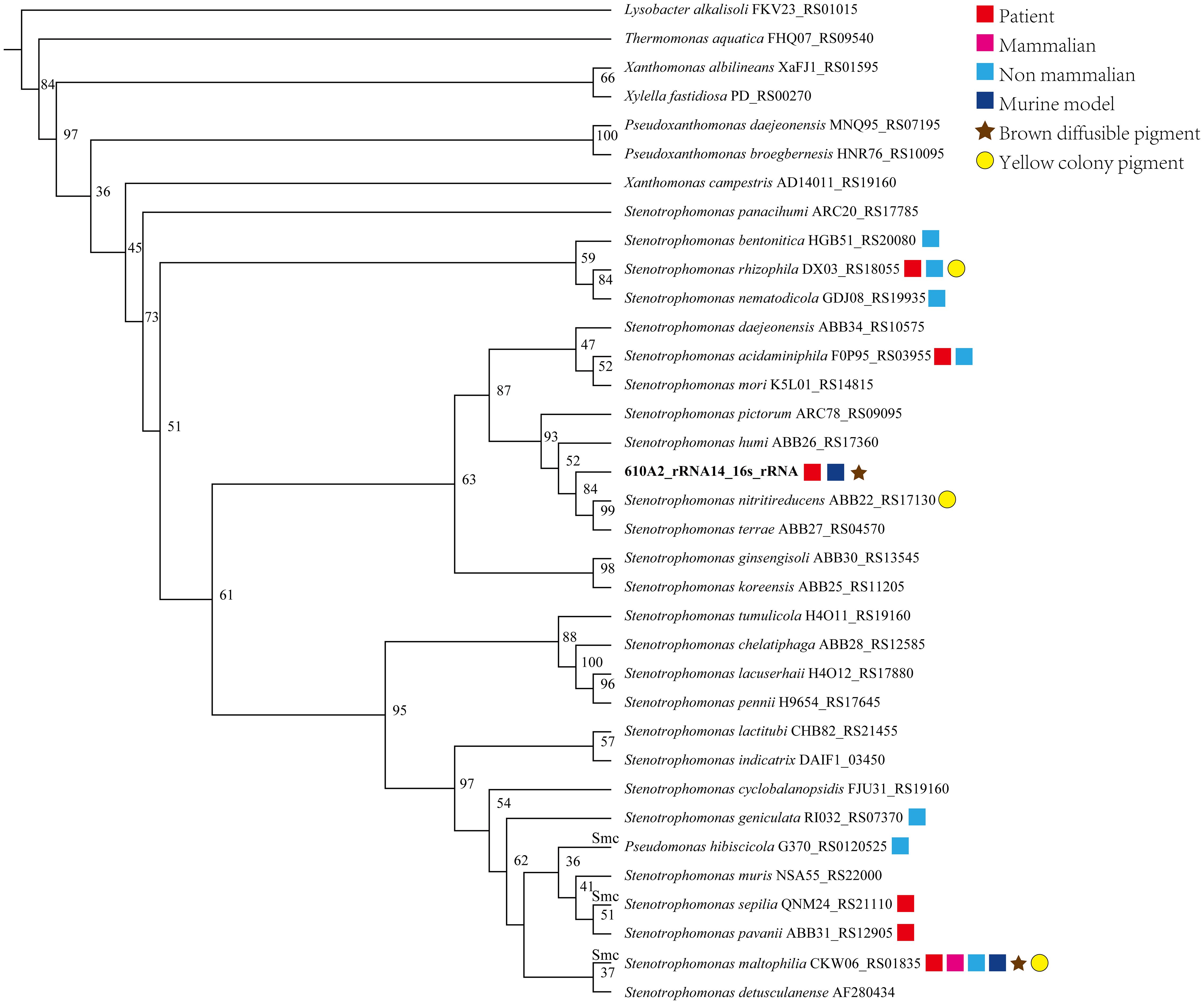
Figure 1 Phylogenetic tree created on the basis of complete 16S rDNA gene sequences of 610A2T and reference genomes of 26 species Stenotrophomonas. ClustalX was used for multiple sequence alignments, and a bootstrap neighbour-joining (NJ) phylogenetic tree was constructed using Treeview (1.6.6) with 1000 replicas. Species isolated from patients or animals or tested on a murine model are marked with symbols following their species name and 16S rDNA locus_tag.
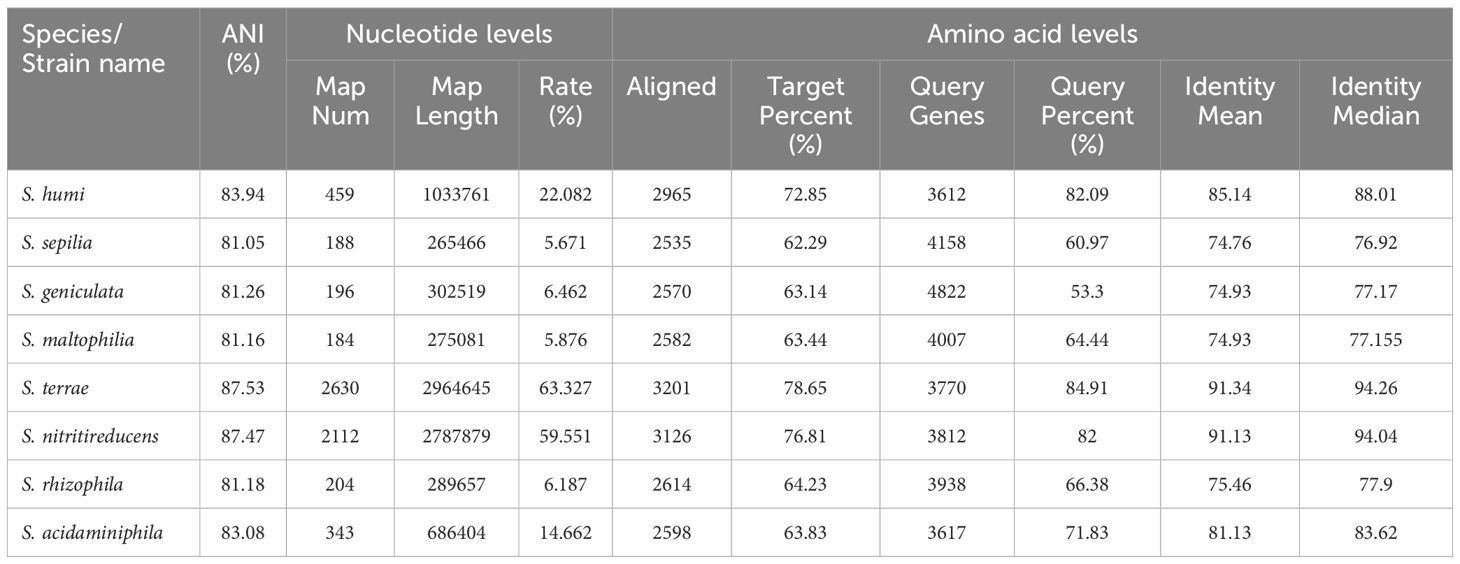
Table 1 ANI values and synteny analyses at the nucleotide and amino acid levels between 610A2 and 8 type strains.
At the infection dose of 50 μL of 1.5 × 108 CFU/mL, no mortality was observed in either the infection or control group. The mice infected with 610A2T showed a weight loss of 3.9 ± 0.9%, 8.0 ± 0.7%, 6.4 ± 1.3%, 2.3 ± 1.2%, 0.8 ± 1.3% at 12 h, 1 day, 2 days, 3 days, and 5 days post-infection, respectively. On the seventh day, weight gain resumed. In addition, no significant differences were observed in the physical signs between the infection and control groups.
The bacterial load per tissue weight (g) in the lung measured via viable count was 1.1 ± 1.5 × 107 CFU/mL·g at 4 h, 1.8 ± 3.6 × 105 CFU/mL·g at 12 h, and 2.0 ± 4.0 × 103 CFU/mL·g at 1 day. Bacteria could not be isolated from lung homogenates 2 days post-infection. 610A2T was isolated only from the spleen homogenate of all infected mice within 4 h, with a viable bacterial count of 1.1 ± 2.2 × 103 CFU/mL g, indicating that 610A2T has strong invasive ability but the host can also quickly clear it.
Histological examination showed that pulmonary damage in the infected group constantly included alveolar septa thickening, congestion, bleeding, inflammatory cell infiltration, compensatory alveolar dilation, mucosal epithelial oedema, and vacuolar degeneration (Figure 3). Although the viable count showed that the bacteria in the lungs had been cleared after 2 days, obvious bloody exudate and a large number of inflammatory cells were detected in the alveoli in 1–3 days, which is consistent with the symptoms for pulmonary congestion. After 7 days, the lung structure of the infected group was restored to normal, whereas that of the control group was normal.
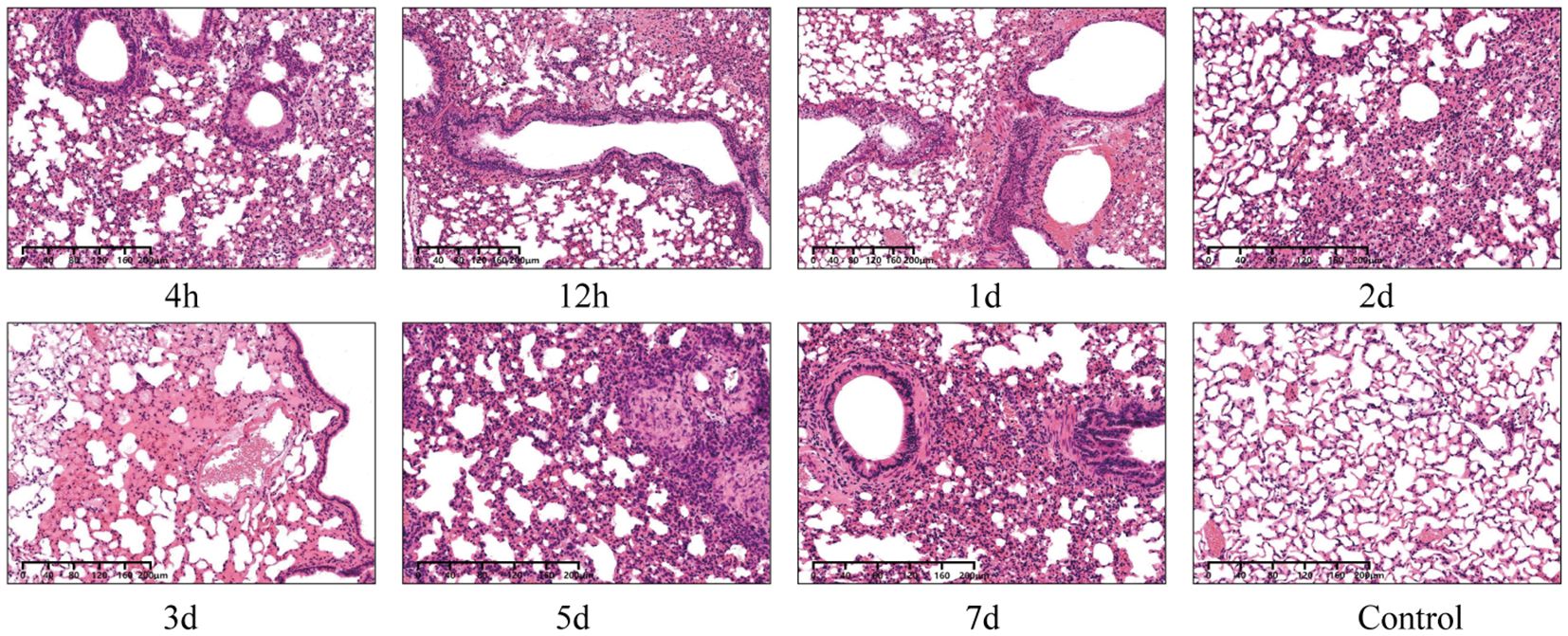
Figure 3 Microscopic images of lung histopathology of KM mice intranasally infected with 610A2T at 7 timepoints (4 h, 12 h, 1 day, 2 days, 3 days, 5 days, and 7 days post-infection). The lung structure of the control group was normal, whereas infiltration of inflammatory cells, compensatory alveolar dilation, mucosal epithelial oedema, and even obvious bloody exudate was observed in the infected lung sections.
Eight of the 9 quantified cytokines showed significant differences between infected and control groups (Figure 4). The means of TNF-α, GM-CSF, IL-4, IL-6, IL-10, and IL-17A at each timepoint were higher than those of the control group, but only the values at 4 h had statistical significance because of the variability caused by individual differences. IFN-γ and IL-2 showed a consistent trend of change, lagging behind other cytokines and showing significant differences from the control group on the 5th day; however, at this time point, TNF-α and IL-6 had recovered to levels no different from the control group.
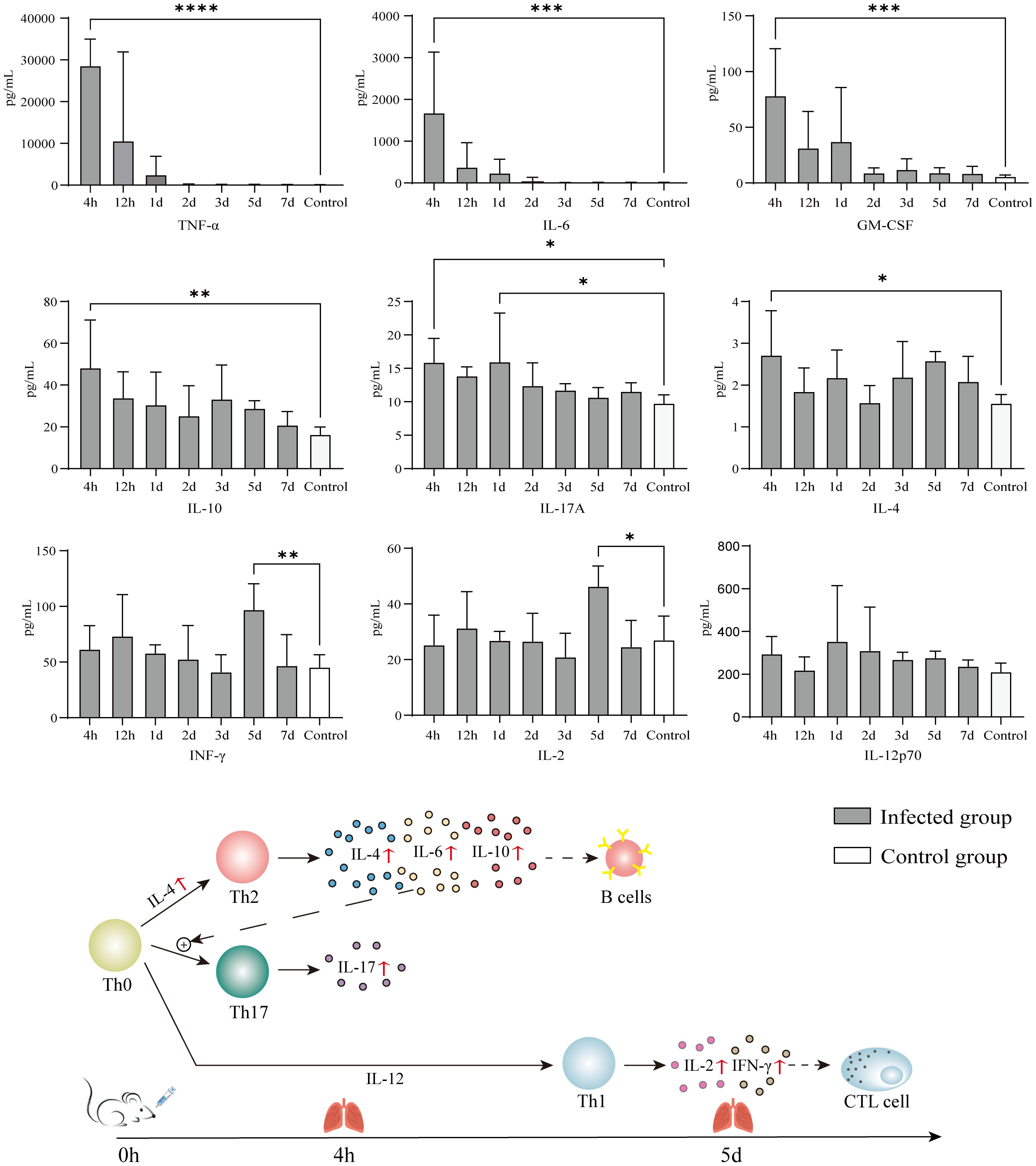
Figure 4 Immune response of KM mice intranasally infected with 610A2T. At 4 h, 12 h, 1 day, 2 days, 3 days, 5 days, and 7 days post-infection, the mice were sacrificed and 9 cytokines, TNF-α, GM-CSF, IFN-γ, IL-2, IL-4, IL-6, IL-10, IL-12p70, and IL-17A, in the lung homogenates were measured using Luminex. Values are mean ± SD (n = 10) obtained from 2 independent experiments. One-way ANOVA followed by Bonferroni’s multiple comparison post-test were used for statistical testing, and the groups with significant differences when compared with the control were marked with asterisks. * P < 0.03, ** P < 0.002, ***P < 0.0002, **** P < 0.0001.
Functional annotation revealed that the 610A2T genome contained 304 virulence factors of pathogenic bacteria in VFDB and 100 human disease pathway genes in KEGG database, of which 16 genes were related to bacterial infectious diseases. In details, 53 genes were related to flagellar biosynthesis, flagellar basal body protein, flagellar motor switching, and chemotaxis; 32 type IV pili genes; 3 outer membrane usher protein genes; 24 iron/haeme uptake and utilisation relative genes; and 1 biofilm-controlling response regulator. 610A2T harbours 2 haemolysin-encoding genes, hlyB and hlyIII, as well as the haemolysin activation/secretion protein gene fhaC, a transcriptional regulator of haemolysin slyA, and 3 HlyD family secretion proteins, which are consistent with the haemolytic activities on Columbia blood agar containing sheep blood.
The annotation results showed that 610A2T may possess 4 types of secretion systems, consisting of 3 T3SS genes, 5 T4SS effectors, 1 T5SS gene, 19 T6SS genes (tssA-H, tssK, tssL, and tssM), concentrated in 2 regions of the chromosome. In addition, 610A2T harbours 9 lipopolysaccharide biosynthesis genes; 11 haeme biosynthesis and utilisation genes; 4 superoxide dismutase genes (Fe-Mn and Cu-Zn families); 1 superoxide oxidase gene; 3 glutathione peroxidase genes; 2 metalloendopeptidase OMA1; 1 melanin-producing gene cluster containing MarR transcriptional regulator, 4-hydroxyphenylpyruvate dioxygenase, and homogentisate 1,2-dioxygenase; 3 catalase genes; 12 alginate biosynthesis and regulation genes; 2 nitrate reductase genes; 1 isocitrate lyase gene; and 1 succinate dehydrogenase gene.
Owing to the slow growth rate of 610A2T, its AST could not be tested using the BD Phonix-100 equipment (BD, Sparks, MD, USA) with BD Phonix™ NMIC 413 panels. Among the drugs detected with CHNENF and MYCOTB plates, 610A2T was sensitive to 10 drugs: azithromycin (8 μg/mL), chloramphenicol (16 μg/mL), tetracycline (8 μg/mL), streptomycin (16 μg/mL), tigecycline (0.5 μg/mL), ofloxacin (1 μg/mL), moxifloxacin (0.06 μg/mL), and rifampin (0.12 μg/mL), with amikacin (<4 μg/mL) and colistin (<0.25 μg/mL) being the most sensitive. 610A2T was resistant to ertapenem (8 μg/mL) and rifabutin (0.5 μg/mL). Fifteen drugs exceeded the maximum detection concentrations of the assay: cycloserine (>256 μg/mL), kanamycin (>40 μg/mL), para-aminosalicylic acid (>64 μg/mL), ethionamide (>40 μg/mL), isoniazid (>4 μg/mL), ethambutol (>32 μg/mL), cefotaxime (>16 μg/mL), ceftazidime (>32 μg/mL), ceftazidime/avibactam (>16/4 μg/mL), meropenem (>2 μg/mL), ampicillin/sulbactam (>32/16 μg/mL), ciprofloxacin (> 0.12 μg/mL), nalidixic acid (> 32 μg/mL), ampicillin (> 32 μg/mL), and trimethoprim-sulfamethoxazole (TMP-SMX, > 8/152 μg/mL).
Comparison against the ARDB, CARD, and KEGG databases revealed that 610A2T carried 41 antibiotic-resistant relative genes on its genome, including 12 genes, aac6-I, acrb, adeb, ceob, emre, macb, mexc, mexe, oprn, smed, smee, and ykkd, coding for multi-drug resistance efflux pump for chloramphenicol, aminoglycoside, fluoroquinolone, macrolide, and tetracycline resistance; extended-spectrum beta-lactamase (ESBL) gene blaCTX-M, subclass B3 metallo-β-lactamase gene blaB, and pbp1a and pbp2b for cephalosporin, penicillin, and β-lactam resistance; rosA and rosB for fosmidomycin resistance; as well as some special resistant genes, bacA for bacitracin, ksgA for kasugamycin, dfrA-26 for trimethoprim, and vanA for vancomycin and teicoplanin.
610A2T grew in all 4 tested media. The colonies on nutrient agar were translucent, smooth, and moist, with brown water-soluble pigment (Figure 5A); on blood agar, they were greyish white with a stimulating ammonia odour and a haemolytic ring (Figure 5B). The strain grew under conditions of 15–37 °C, pH 6–8, and NaCl concentration of 0–2%, but it did not grow below 4 °C and above 40 °C and did not tolerate 5% salt concentration like S. maltophilia and S. terrae. During semi-solid puncture culture, bacteria grew diffusely along the puncture line; however, no pigment was produced, indicating motility; pigment production was aerobic. 610A2T was rod-shaped gram-negative, and without spores or capsules. Under electron microscopy, the bacterial body was about 1.5–2 × 0.5 μM with a single extreme flagella (Figure 5C).
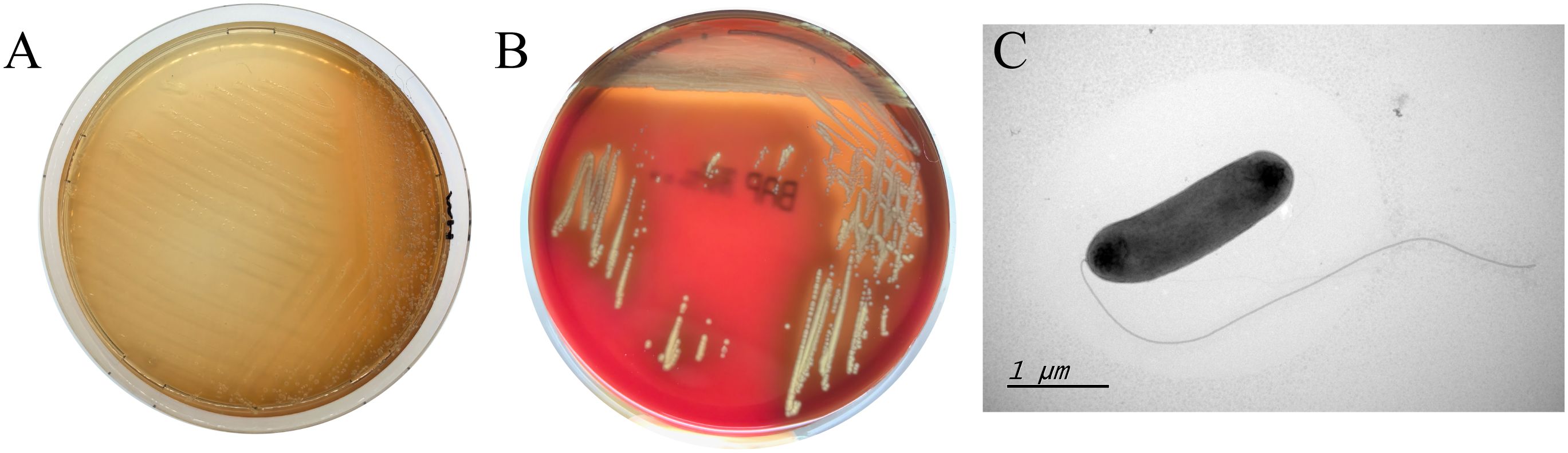
Figure 5 Clones of 610A2T on MH medium (A), columbia blood agar (B) and its transmission electron microscopy image (C). Obvious brown water-soluble pigment and haemolytic ring can be observed on MH medium and blood agar, respectively. The bacterial body of 610A2T is about 1.5–2 × 0.5 μM with a single extreme flagella.
610A2T grew slower than S. maltophilia strains in TSB. After 48 h, the cultures of 610A2T showed turbidity, but the entire growth curve was flat without a logarithmic phase (Figure 6A). Although the growth rate was slow, the biofilm index showed that 610A2T had a strong biofilm-forming ability, which was significantly higher than that of all S. maltophilia strains after 3 h of cultivation (Figure 6B).
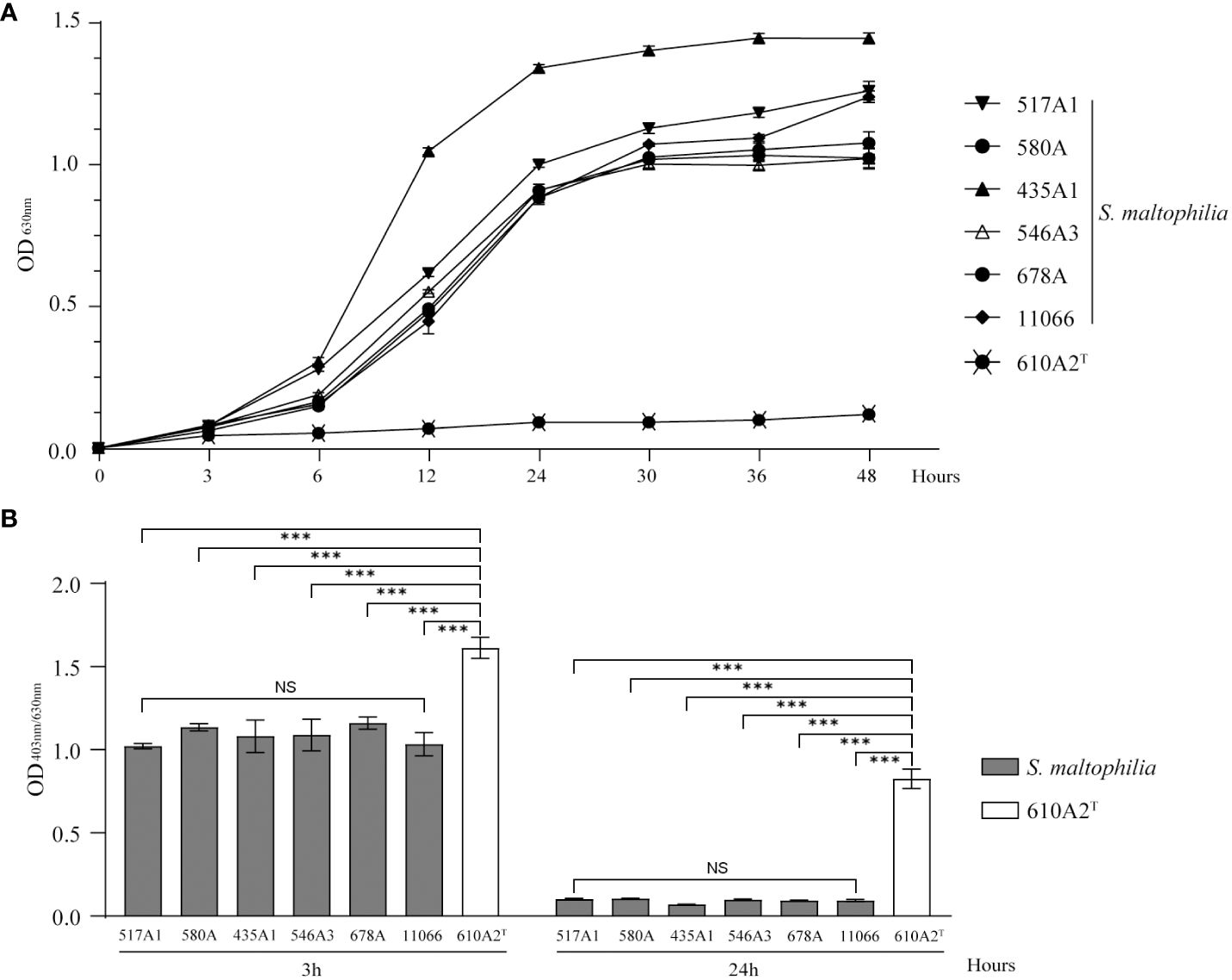
Figure 6 Growth curve (A) and biofilm formation (B) of 610A2T and control strains in TSB medium. Significant differences between groups are marked with asterisks, ***P < 0.0002. NS, without statistical significance.
The biochemical results showed that, in the API 20NE test, 610A2T was positive for nitrate reduction, hydrolysis of quercetin and gelatin, assimilation of N-acetylglucosamine, maltose, and citric acid. In API ZYM, it was positive for alkaline phosphatase, esterase lipase (C8), leucine arylamidase, trypsin, acid phosphatase, and naphthol-AS-BI phosphate hydrolase. In API 50CH, only starch fermentation was detected. The major differentiating feature of 610A2T was that it could not assimilate glucose and mannose, but it could ferment starch.
In the MALDI-TOF spectrum, the 4 main peaks of 610A2T were located at 5254.065 Da, 4857.675 Da, 2770.713 Da, and 6118.187 Da, of which the 4857 Da peak was genus-specific and the 2770.713 Da peak was 610A2T-specific. In the local Bruker_MSP library, only 5 Stenotrophomonas species were found: S. acidaminiphila, S. maltophilia, S. pictorum, S. rhizophila, and S. nitritireducens. The identification scores of 610A2T for these species ranged from 1.625 to 1.370, with the highest similarity to S. pictorum. These data indicate that 610A2T is a distinct species closely associated with these species of Stenotrophomonas.
The predominant fatty acids of 610A2T were iso-C15:0, iso-C14:0, summed Feature 3 (C16:1ω7c/16:1ω6c), iso-C16:0, and C15:0 anteiso and sum in Feature 9 (iso-C17:1ω9c). It contained all 3 fatty acids, iso-C11:0, iso-C11:03 3OH, and iso-C13:0 3OH, characteristic of the genus Stenotrophomonas, but no matches were found in RTSBA6. 610A2T also contained all characteristic fatty acids of S. terrae and S. humi: iso-C14:0 (14.2% and 15.7%), iso-C15:1 (4.6% and 2%), iso-C16:0 (8% and 12.7%), and iso-C17:1ω9c (7.2% and 4.6%, respectively) (Heylen et al., 2007). The contents of these 4 fatty acids in 610A2T were 15.45%, 1.5%, 11.9%, and 5.22%, respectively, indicating that the similarity of 610A2T to S. humi is higher than that to S. terrae.
When evaluating the pathogenicity of strain 610A2T, we used the outbred mouse KM, which has strong disease resistance and adaptability and can better reflect the genetic diversity of the human population. Within the first day post-infection, the bacterial load of 610A2T in the lungs decreased sharply, and 610A2T was cleared completely within 48 h. The lung clearance rate of 610A2T in KM mice was faster than that of S. maltophilia in many inbred mouse strains, which also showed a larger decrease in CFU at 24 h, but could generally still be detected on day 3–7 post-infection and could even replicate in A/J mice with a transient increase in CFU at 4 and 8 h post-inoculation (Di Bonaventura et al., 2010; Rouf et al., 2011; Pompilio et al., 2018). After infection, both S. maltophilia (Di Bonaventura et al., 2010; Pompilio et al., 2018) and 610A2T could be isolated in the spleen, suggesting that, at the same time of lung infection, Stenotrophomonas could disrupt the integrity of the lung epithelial barrier, cause bacteraemia, spread to other tissues, and pose an uncertain risk of assisting other pathogens in spreading and forming extrapulmonary lesions.
Combining with the acute infectious cytokine responses, the animal experiment in this study can confirm the pathogenicity of 610A2T on the basis of the significant weight loss in the mice, tissue reactions due to inflammatory infection, and pulmonary congestion detected in the histopathological sections, although 610A2T can be completely cleared from a healthy host. 610A2T belonged to the S. terrae clade in the phylogenetic tree. In phylogenetic trees, whether constructed based on the concatenation of translated protein sequences (Patil et al., 2018) or the 16S rDNA gene, it is a clade that is relatively far from SMc. All species in this clade have been isolated from the environment and never been isolated from humans (Heylen et al., 2007). The isolation of 610A2T changed the distribution of the pathogenic species. Together with S. acidaminiphila and S. rhizophila, all 3 main clades of the genus Stenotrophomonas have isolates from human sources.
610A2T inherently possesses many pathogenic mechanisms, which can be inferred from the composition of functional genes on the genome, including those for motility (flagella and type IV pili), surface adherence (flagella, fimbriae, and LPS), damage capacity to host (protein secretion systems and extracellular enzymes), iron uptake and utilization, biofilm formation, protection against host defence (superoxide dismutase, hydroperoxidase, catalase, and melanin) as well as multi-antibiotic resistance (Di Bonaventura et al., 2010; Brooke, 2021). In addition, the genomic analysis suggested that 610A2T produces type III haemolysin, for which erythrocyte lysis capacity and virulence have been confirmed in a mouse model infected with Vibrio vulnificus (Baida and Kuzmin, 1996; Chen et al., 2004; Moonah et al., 2014). In the genus Stenotrophomonas, colony pigmentation and brown diffusible pigments have been reported in only S. maltophilia (Romanenko et al., 2008; Liaw et al., 2010), S. nitritireducens (Finkmann et al., 2000), and S. rhizophila (Romanenko et al., 2008) isolates. Pigments and biofilm formation interact, and both are components of bacterial pathogenicity, enabling the colonisation or infection of hosts and promoting the attachment of other pathogens (Tamma et al., 2022). 610A2T showed stronger abilities than S. maltophilia in both aspects, indicating that its pathogenicity may not be weaker than that of S. maltophilia.
However, the strain-specific cytokine responses induced by 610A2T may be difference from those induced by S. maltophilia. In this study, 9 cytokines were selected on the basis of their inflammatory responses in S. maltophilia-infected murine model and patients with TB (Di Bonaventura et al., 2010; Mvubu et al., 2018). Among these cytokines, early inflammatory markers TNF-α and IL-6 showed consistently high expression after 610A2T and S. maltophilia infection, with IFN-γ, IL-4, and IL-10 exhibiting different or even opposite changes (McDaniel et al., 2020) (Table 2). In mice infected with S. maltophilia, characteristic changes in cytokines were the persistent hyperexpression of IFN-γ for more than 3 days and sustained significantly lower level of IL-4 when compared with the control group (Di Bonaventura et al., 2010). IFN-γ was released in the early stage, along with large amounts of TNF-α and IL-2 and was closely associated with the ratio of T cells. Blocking the PD-1/PD-L1 pathway inhibited the apoptosis-inducing effect of S. maltophilia on T cells. The pleotropic cytokine IL-4 is a product of Th2 lymphocytes and inhibits Th1 cell differentiation. Therefore, the immune response to acute infection by S. maltophilia is predominantly a Th1-type response (Rouf et al., 2011). S. maltophilia may participate in the activation of T cells and induce subsequent T-cell exhaustion by activating the PD-1/PD-L1 signalling pathway, as the concentration of cytokines is reduced in the later stages (Wang et al., 2021; Xu et al., 2023). In contrast, in the 610A2T-infected KM mice, cytokines IL-4, IL-6, and IL-10 secreted by Th2 cells showed a rapid and significant increase; there was no difference in IL-12, the main cytokine that induces Th1 cell differentiation and inhibits Th2 cells, indicating that the immune response of 610A2T was Th2 type (Figure 4). In addition, the lagging release of IFN-γ and IL-2 in 610A2T-infection indicated that Th1 type was disinhibition in the later stage and also involved in the host’s defence against the pathogen. In-depth research of the pathogenic Stenotrophomonas species including 610A2T in same mouse strain infection model will further elucidate pathogenic and immunological characteristics.
Moreover, 610A2T is resistant to half of the commonly used anti-TB drugs, including isoniazid, and carries ESBL and metallo-β-lactamase, which can hydrolyse all bicyclic β-lactam antibiotics (Hinchliffe et al., 2023). The co-existence of Stenotrophomonas may affect therapeutic efficacy in patients with TB. The AST results showed that all drugs recommended by the Infectious Diseases Society of America for the treatment of S. maltophilia infection (Tamma et al., 2022) may be applicable to 610A2T, except TMP-SMX, which is recommended as the first-line agent for the treatment of S. maltophilia infection owing to the low isolation rate of resistant strains (Chang et al., 2015; Gröschel et al., 2020). A combination of colistin and rifampicin may be used for patients with mixed 610A2T and TB infections.
Stenotrophomonas pigmentata sp. nov. (pig.men.ta’ta. L. fem. adj. pigmentata, pigmented, coloured).
The type strain 610A2T was isolated from patients with pulmonary TB. It is a Gram-negative bacillus with a single extreme flagella and can produce brown water-soluble pigment. Growth is observed at 15–37 °C, pH 6–8, and 0–2% NaCl concentration, but it does not grow below 4 °C, above 40 °C, and at 5% salt concentration. 610A2T yielded positive results for nitrate reduction, quercetin and gelatin hydrolysis, and starch fermentation. The 4 main peaks in the MALDI-TOF spectrum were located at 5254.065 Da, 4857.675 Da, 2770.713, Da and 6118.187 Da. The predominant fatty acids are iso-C15:0, iso-C14:0, summed Feature 3 (C16:1ω7c/16:1ω6c), iso-C16:0, and C15:0 anteiso and sum in Feature 9 (iso-C17:1ω9c). 610A2T has multi-drug resistance and intrinsic resistance to β-lactams, carbapenems, and trimethoprim-sulfamethoxazole, harbouring several multi-drug resistance efflux pump and antibiotic resistant genes. This strain is pathogenic to mice.
The complete genome of type strain 610A2T comprises 4681496 bp and GC content of 63.29%, and the complete genome and 16S rDNA gene sequence are deposited in GenBank under accession numbers CP130832.1 and OR936313, respectively. The type strain is 610A2T (=GDMCC 1.4134 T =JCM 36488 T).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP130832.1, https://www.ncbi.nlm.nih.gov/genbank/, OR936313.
The animal study was approved by the Laboratory Animal Welfare & Ethics Committee of the National Institute for Communicable Disease Control and Prevention (Issue number 2023-021). The study was conducted in accordance with the local legislation and institutional requirements.
YL: Investigation, Writing – review & editing. ZY: Investigation, Writing – review & editing. XF: Investigation, Writing – review & editing. DX: Investigation, Writing – review & editing. HL: Writing – review & editing. XZ: Investigation, Writing – review & editing. RW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors thank the nomenclature reviewers Dr. Bernhard Schink and Dr. Aharon Oren of International journal of systematic and evolutionary microbiology for their assistance in nomenclature.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baida, G. E., Kuzmin, N. P. (1996). Mechanism of action of hemolysin III from Bacillus cereus. Biochim. Biophys. Acta 1284, 122–124. doi: 10.1016/S0005-2736(96)00168-X
Bansal, K., Kumar, S., Kaur, A., Singh, A., Patil, P. B. (2021). Deep phylo-taxono genomics reveals Xylella as a variant lineage of plant associated Xanthomonas with Stenotrophomonas and Pseudoxanthomonas as misclassified relatives. Genomics. 113(6), 3989–4003113(6):3989-4003. doi: 10.1101/2021.08.22.457248
Brooke, J. S. (2021). Advances in the microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 34, e0003019. doi: 10.1128/CMR.00030-19
Chang, Y. T., Lin, C. Y., Chen, Y. H., Hsueh, P. R. (2015). Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00893
Chao, A., Chao, A. S., Lin, C. Y., Weng, C. H., Wu, R. C., Yeh, Y. M., et al. (2022). Analysis of endometrial lavage microbiota reveals an increased relative abundance of the plastic-degrading bacteria Bacillus pseudofirmus and Stenotrophomonas rhizophila in women with endometrial cancer/endometrial hyperplasia. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1031967
Chen, Y. C., Chang, M. C., Chuang, Y. C., Jeang, C. L. (2004). Characterization and virulence of hemolysin III from Vibrio vulnificus. Curr. Microbiol. 49, 175–179. doi: 10.1007/s00284-004-4288-5
CLSI. (2018). Performance standards for susceptibility tesing of Mycobacteria, Nocardia spp., and other aerobic actinomycetes, M64 (Wayne, PA19089 USA: The Clinical and Laboratory Standard Institute).
Coenye, T., Vanlaere, E., Falsen, E., Vandamme, P. (2004). Stenotrophomonas africana Drancourt et al. 1997 is a later synonym of Stenotrophomonas maltophilia (Hugh 1981) Palleroni and Bradbury 1993. Int. J. Syst. Evol. Microbiol. 54, 1235–1237. doi: 10.1099/ijs.0.63093-0
Deng, Y., Han, X. F., Jiang, Z. M., Yu, L. Y., Li, Y., Zhang, Y. Q. (2022). Characterization of three Stenotrophomonas strains isolated from different ecosystems and proposal of Stenotrophomonas mori sp. nov. and Stenotrophomonas lacuserhaii sp. nov. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1056762
Di Bonaventura, G., Pompilio, A., Zappacosta, R., Petrucci, F., Fiscarelli, E., Rossi, C., et al. (2010). Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect. Immun. 78, 2466–2476. doi: 10.1128/IAI.01391-09
Finkmann, W., Altendorf, K., Stackebrandt, E., Lipski, A. (2000). Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 50 Pt 1, 273–282. doi: 10.1099/00207713-50-1-273
Gautam, V., Patil, P. P., Bansal, K., Kumar, S., Kaur, A., Singh, A., et al. (2021). Description of Stenotrophomonas sepilia sp. nov., isolated from blood culture of a hospitalized patient as a new member of Stenotrophomonas maltophilia complex. New Microbes New Infect. 43, 100920. doi: 10.1016/j.nmni.2021.100920
Gil-Gil, T., Martínez, J. L., Blanco, P. (2020). Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: a review of current knowledge. Expert Rev. Anti Infect. Ther. 18, 335–347. doi: 10.1080/14787210.2020.1730178
Gröschel, M. I., Meehan, C. J., Barilar, I., Diricks, M., Gonzaga, A., Steglich, M., et al. (2020). The phylogenetic landscape and nosocomial spread of the multidrug-resistant opportunist Stenotrophomonas maltophilia. Nat. Commun. 11, 2044. doi: 10.1038/s41467-020-15123-0
Heylen, K., Vanparys, B., Peirsegaele, F., Lebbe, L., De Vos, P. (2007). Stenotrophomonas terrae sp. nov. and Stenotrophomonas humi sp. nov., two nitrate-reducing bacteria isolated from soil. Int. J. Syst. Evol. Microbiol. 57, 2056–2061. doi: 10.1099/ijs.0.65044-0
Hinchliffe, P., Calvopiña, K., Rabe, P., Mojica, M. F., Schofield, C. J., Dmitrienko, G. I., et al. (2023). Interactions of hydrolyzed β-lactams with the L1 metallo-β-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products. J. Biol. Chem. 299, 104606. doi: 10.1016/j.jbc.2023.104606
Iebba, V., Guerrieri, F., Di Gregorio, V., Levrero, M., Gagliardi, A., Santangelo, F., et al. (2018). Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci. Rep. 8, 8210. doi: 10.1038/s41598-018-26509-y
Ježek, P., Šafránková, R., Mališová, L. (2020). Unusual microbiological findings in bacteremia cases - three case reports. Klin Mikrobiol Infekc Lek 26, 106–110.
Kumar, A., Rithesh, L., Kumar, V., Raghuvanshi, N., Chaudhary, K., Abhineet, et al. (2023). Stenotrophomonas in diversified cropping systems: friend or foe? Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1214680
Li, Y., Zhao, A., Yu, Q., Yu, N., Cui, Y., Ma, X., et al. (2023). Effect of Stenotrophomonas maltophilia on tuberculosis. Microbiol. Spectr. 11(4), e0094423. doi: 10.1128/spectrum.00944-23
Liaw, S. J., Lee, Y. L., Hsueh, P. R. (2010). Multidrug resistance in clinical isolates of Stenotrophomonas maltophilia: roles of integrons, efflux pumps, phosphoglucomutase (SpgM), and melanin and biofilm formation. Int. J. Antimicrob. Agents 35, 126–130. doi: 10.1016/j.ijantimicag.2009.09.015
Maturana, J. L., Cardenas, J. P. (2021). Insights on the evolutionary genomics of the Blautia genus: potential new species and genetic content among lineages. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.660920
McDaniel, M. S., Schoeb, T., Swords, W. E. (2020). Cooperativity between Stenotrophomonas maltophilia and Pseudomonas aeruginosa during polymicrobial airway infections. Infect. Immun. 88(4), e00855-19. doi: 10.1128/IAI.00855-19
Mojica, M. F., Humphries, R., Lipuma, J. J., Mathers, A. J., Rao, G. G., Shelburne, S. A., et al. (2022). Clinical challenges treating Stenotrophomonas maltophilia infections: an update. JAC Antimicrob. Resist. 4, dlac040. doi: 10.1093/jacamr/dlac040
Moonah, S., Sanders, N. G., Persichetti, J. K., Sullivan, D. J., Jr. (2014). Erythrocyte lysis and Xenopus laevis oocyte rupture by recombinant Plasmodium falciparum hemolysin III. Eukaryot Cell 13, 1337–1345. doi: 10.1128/EC.00088-14
Mvubu, N. E., Pillay, B., Mckinnon, L. R., Pillay, M. (2018). Mycobacterium tuberculosis strains induce strain-specific cytokine and chemokine response in pulmonary epithelial cells. Cytokine 104, 53–64. doi: 10.1016/j.cyto.2017.09.027
Patil, P. P., Kumar, S., Midha, S., Gautam, V., Patil, P. B. (2018). Taxonogenomics reveal multiple novel genomospecies associated with clinical isolates of Stenotrophomonas maltophilia. Microb. Genom 4. doi: 10.1099/mgen.0.000207
Pompilio, A., Crocetta, V., Di Bonaventura, G. (2018). Stenotrophomonas maltophilia mutant lacking flagella remains virulent in DBA/2N mice but is less efficient in stimulating TNF-alpha expression. FEMS Microbiol. Lett. 365. doi: 10.1093/femsle/fny205
Romanenko, L. A., Uchino, M., Tanaka, N., Frolova, G. M., Slinkina, N. N., Mikhailov, V. V. (2008). Occurrence and antagonistic potential of Stenotrophomonas strains isolated from deep-sea invertebrates. Arch. Microbiol. 189, 337–344. doi: 10.1007/s00203-007-0324-8
Rouf, R., Karaba, S. M., Dao, J., Cianciotto, N. P. (2011). Stenotrophomonas maltophilia strains replicate and persist in the murine lung, but to significantly different degrees. Microbiol. (Reading) 157, 2133–2142. doi: 10.1099/mic.0.048157-0
Tamma, P. D., Aitken, S. L., Bonomo, R. A., Mathers, A. J., Van Duin, D., Clancy, C. J. (2022). Infectious Diseases Society of America guidance on the treatment of AmpC β-lactamase-producing Enterobacterales, carbapenem-resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia infections. Clin. Infect. Dis. 74, 2089–2114. doi: 10.1093/cid/ciab1013
Tan, R., Liu, J., Li, M., Huang, J., Sun, J., Qu, H. (2014). Epidemiology and antimicrobial resistance among commonly encountered bacteria associated with infections and colonization in intensive care units in a university-affiliated hospital in Shanghai. J. Microbiol. Immunol. Infect. 47, 87–94. doi: 10.1016/j.jmii.2012.11.006
Wang, M., Yin, S., Qin, Q., Peng, Y., Hu, Z., Zhu, X., et al. (2021). Stenotrophomonas-maltophilia inhibits host cellular immunity by activating PD-1/PD-L1 signaling pathway to induce T-cell exhaustion. Mol. Immunol. 130, 37–48. doi: 10.1016/j.molimm.2020.12.019
Xu, G., Liu, H., Xia, D., Zhao, Y., Qian, Y., Han, H., et al. (2023). Time-course transcriptome analysis of lungs from mice infected with inhaled aerosolized Stenotrophomonas maltophilia. J. Thorac. Dis. 15, 4987–5005. doi: 10.21037/jtd-23-1138
Zhang, Y., Li, D., Yan, Q., Xu, P., Chen, W., Xin, H., et al. (2022). Genome-wide analysis reveals the emergence of multidrug resistant Stenotrophomonas acidaminiphila strain SINDOREI isolated from a patient with sepsis. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.989259
Keywords: Stenotrophomonas, tuberculosis, pathogen, multiple-drug resistance, pigment
Citation: Li Y, Yu Z, Fan X, Xu D, Liu H, Zhao X and Wang R (2024) A novel pathogenic species of genus Stenotrophomonas: Stenotrophomonas pigmentata sp. nov. Front. Cell. Infect. Microbiol. 14:1410385. doi: 10.3389/fcimb.2024.1410385
Received: 01 April 2024; Accepted: 17 May 2024;
Published: 05 June 2024.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Alexey V. Rakov, Central Research Institute of Epidemiology (CRIE), RussiaCopyright © 2024 Li, Yu, Fan, Xu, Liu, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruibai Wang, d2FuZ3J1aWJhaUBpY2RjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.