- 1State Key Laboratory of Pathogenesis, Prevention, Treatment of High Incidence Diseases in Central Asian, Department of Clinical Laboratory, First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang Uygur Autonomous Region, China
- 2State Key Laboratory of Pathogenesis, Prevention, Treatment of High Incidence Diseases in Central Asian, Clinical Medical Research Institute, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang Uygur Autonomous Region, China
Introduction: Mycoplasma pneumoniae (MP) is the major cause of respiratory infections that threaten the health of children and adolescents worldwide. Therefore, an early, simple, and accurate detection approach for MP is critical to prevent outbreaks of MP-induced community-acquired pneumonia.
Methods: Here, we explored a simple and accurate method for MP identification that combines loop-mediated isothermal amplification (LAMP) with the CRISPR/Cas12b assay in a one-pot reaction.
Results: In the current study, the whole reaction was completed within 1 h at a constant temperature of 57°C. The limit of detection of this assay was 33.7 copies per reaction. The specificity of the LAMP-CRISPR/Cas12b method was 100%, without any cross-reactivity with other pathogens. Overall, 272 clinical samples were used to evaluate the clinical performance of LAMP-CRISPR/Cas12b. Compared with the gold standard results from real-time PCR, the present method provided a sensitivity of 88.11% (126/143), specificity of 100% (129/129), and consistency of 93.75% (255/272).
Discussion: Taken together, our preliminary results illustrate that the LAMP-CRISPR/Cas12b method is a simple and reliable tool for MP diagnosis that can be performed in resource-limited regions.
1 Introduction
Mycoplasma pneumoniae (MP)is a small prokaryotic microorganism, which is a major cause of morbidity and mortality in children and adolescents. Notably, MP is commonly associated with community-acquired pneumonia (CAP) (Jain et al., 2015). In clinical practice, the typical symptoms of MP infection include fever, cough, sore throat, muscle pain, and others (Jiang et al., 2021). Additionally, bronchospasms, pneumonia, acute respiratory distress syndrome (ARDS), myocarditis, and meningitis have also been reported in severe cases (Narita, 2010). More than 12% of hospitalized children with MP infection are admitted to the ICU, and over 25% of patients with MP infection can develop severe pneumonia or extrapulmonary complications (Kutty et al., 2019; Rodman Berlot et al., 2023). As the manifestations of MP infection are non-specific and similar to those of other respiratory pathogens, such as the influenza virus, a misdiagnosis or delayed treatment poses a serious health threat to the public (Poddighe, 2018); therefore, it is unreliable to make a definite diagnosis of MP infection simply by clinical presentation. Moreover, the incubation period for MP is approximately 2–4 weeks (Principi and Esposito, 2001), and most patients remain unaware of the invasion of this pathogen for 14 days after infection, which greatly increases the risk of pathogen transmission in crowded places, such as schools. Therefore, MP causes frequent CAP outbreaks in school-aged children aged 5–15 years (Yiş et al., 2008). Therefore, rapid detection methods capable of accurately diagnosing MP early in the incubation period are critical for preventing outbreaks of MP-caused CAP.
Currently, the main diagnostic methods for MP infection include culture, serological tests, and polymerase chain reaction (PCR)-based methods (Waites et al., 2017). Culture-based methods provide a solid foundation for MP infection; however, culturing MP is time-consuming and relatively insensitive owing to its slow growth and fastidious nature (Leal et al., 2020). Regarding serological tests, reliable results are highly dependent on antibodies and exhibit high rates of false negatives (She et al., 2010). The results collected from PCR-based methods serve as the gold standard for MP identification with the requirement for specific equipment and skilled technicians, which are restricted to grassroots laboratories or point-of-care detection (Schmitt et al., 2013). To address this, several isothermal amplification techniques are developed for MP detection, such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) (Cai et al., 2019; Fan et al., 2020). In the LAMP assay, the reaction can be performed at a constant temperature using simple operation and equipment, and the reaction time is shorter than that of PCR-based methods (Hong et al., 2023). However, much more effort is required to overcome the shortcomings of low specificity and false-positive results (Nagai et al., 2016).
Recently, clustered and regularly interspaced short palindromic repeats and their associated protein (CRISPR/Cas) systems have provided a novel method for pathogen diagnostics. Because collateral cleavage was discovered in some Cas proteins (Cas12, Cas13a, and Cas14), the CRISPR/Cas assay was modified for nucleic acid detection. Cas12 has cis-cleavage activity and cleaves targeted double-stranded DNA (dsDNA) by using a specific single CRISPR RNA (crRNA), and produces collateral trans-cleavage and non-specifically degrades single-stranded DNA (ssDNA) (Li et al., 2019), thereby the cleavage results of CRISPR/Cas12 can be visualized by using fluorophore and quencher-labeled ssDNA, indicating the presence of the target gene (Zhu et al., 2021). However, the sensitivity of the CRISPR system is low and it cannot be directly applied in clinical testing. Therefore, isothermal amplification technology was introduced to the CRISPR/Cas system, which yields a rapid, ultrasensitive, and specific detection method for MP (Deng et al., 2022; Li et al., 2022). Moreover, the CRISPR-Cas-Integrated LAMP has been applied for Pathogen-specific point-of-care (PoC) diagnostic tests (Atçeken et al., 2022). In current CRISPR-based methods, pre-amplification of the target gene and CRISPR-mediated detection are the two main steps. Therefore, the entire reaction consists of multiple manual operations that cannot avoid cross-contamination due to complicated diagnostic procedures. To address these drawbacks, a one-step, one-pot CRISPR-based nucleic acid detection platform (CRISPR-top) was developed (Li et al., 2021; Qiu et al., 2022). However, an appropriate platform for the identification of MP remains unknown.
In the present study, we integrated the LAMP assay with the CRISPR/Cas12b method to achieve a rapid, simple, sensitive, and specific detection method for MP detection (MP-LAMP-CRISPR/Cas12b). Our findings not only provide solid data for the reaction conditions, but also demonstrate the potential value of the present platform in resource-limited regions.
2 Materials and methods
2.1 Reagents and instruments
The reagents used in this study were a LAMP nuclear amplification kit (25102, TOLO Biotech, China) and dNTP mix (DN32, Hongene, China)
MgSO4 (B1003S, NEB, USA), Glycine (A610235, Sangon, China), SYTO-9 (S34854ThermoFisher, USA), AapCas12 (32118, TOLO Biotech, China), HOLMES ssDNA reporter (FAM) (31101, TOLO Biotech, China), Cas12b High Yield sgRNA Synthesis and Purification Kit (31904, TOLO Biotech, China), Digital PCR Mixture (MX0108, ZHENZHUN BIO, China). Nuclease-free water (R1600, Solarbio Life Sciences, China).
The instruments used in this study were as follows: Real-Time PCR system (SLAN-96S, HONGSHI, Shanghai), QuantStudio 3 real-time quantitative PCR system (QuantStudio 3, ThermoFisher, USA), QuantStudio 5 real-time quantitative PCR system (QuantStudio 5, ThermoFisher, USA), Qubit Fluorescent spectrophotometer (Qubit4, ThermoFisher, USA), and AccuMini Digital PCR system (AccuMini, ZHENZHUN BIO, China).
2.2 Sample preparation
The MP strains used in this study were isolated from clinically positive samples. A commercial DNA isolation kit (S20025, Sansure, Shanghai, China) was used to extract genomic DNA from MP according to the manufacturer’s instructions. To generate the recombinant plasmid pUC57-MP_PIP, the target gene was inserted into the pUC57 vector. The concentration of the recombinant plasmid was 23.5 ng/μL by using the AccuMini Digital PCR system (AccuMini, ZHENZHUN BIO, China) according to the instructions given by the manufacturers, which corresponds to a copy number of 6.54 ×109 copies/μL. It was subsequently used as a standard product for the LAMP-CRISPR/Cas12b detection system.
2.3 LAMP Primer and sgRNA design
The Online NEB LAMP primer design tool (https://lamp.neb.com/#!/) was used to design a LAMP primer set targeting the MP gene putative intracellular protease (PIP, GenBank ID: CP039761.1). Cas12b sgRNA was designed based on the coding sequence of PIP. The primers and sgRNAs used in this study were synthesized by Sangon Biotech (Shanghai, China). Cas12b sgRNA was purified using the Cas12b High Yield sgRNA Synthesis and Purification Kit (31904, ToloBio, China) according to the manufacturer’s instructions.
2.4 LAMP reaction
The total volume of the LAMP reaction 25 µL, the mixture includes 2.5 µL of 10 x LAMP isothermal reaction buffer, 1 µL dNTP mix (25 mM), 2 µL MgSO4 (100 mM), 2.5 µL 10× MP PIP LAMP primer mix, 6 µL glycine (2M), 1 µL of Bst 2.0 DNA polymerase, 1.25 µL MP PIP sgRNA (10 μM), 2.5 µL template, and nuclease-free water up to 25 µL. LAMP amplification was performed at 57°C for 45 min using pUC57-MP PIP plasmid and nuclease-free DW (NTC, negative control). The products of LAMP amplification were monitored using the Applied Biosystems QuantStudio 5 Real-Time PCR System (QuantStudio 5, ThermoFisher, USA). Each reaction was repeated three times.
2.5 One-step LAMP-CRISPR/Cas12b reaction condition
To identify the optimum conditions for the one-step LAMP-CRISPR/Cas12b assay, the total volume of MP LAMP-CRISPR/Cas12b reaction mixture is 25 µL and prepared as follows, 2.5 µL 10×LAMP buffer, 0.8/1.0/1.2/1.4 µL dNTP mix (25 mM), 1.5/1.75/2/2.25/2.5 µL MgSO4 (100 mM), 2.5 µL 10 × MP PIP LAMP primer mix, 6 µL Glycine (2 M), 1.25 µL HOLMES ssDNA reporter (10 μM), 0.625/1.25/1.875/2.5 µL AapCas12b (10 μM), 1 µL Bst (8 U/μL), 0.625/1.25/1.875/2.5 µL MP PIP sgRNA (10 μM), 2.5 µL template and nuclease-free water up to 25 µL. The reaction was performed at 57°C for 45 min and monitored using a fluorescence reader to collect the FAM fluorescent channel signals every 30 s.
2.6 Ethics statement
This study was approved by the Research Ethics Committee of First Affiliated Hospital of Xinjiang Medical University (No. K202403-32). All the experiments were performed according to the relevant regulations.
2.7 Human nasopharyngeal swab samples
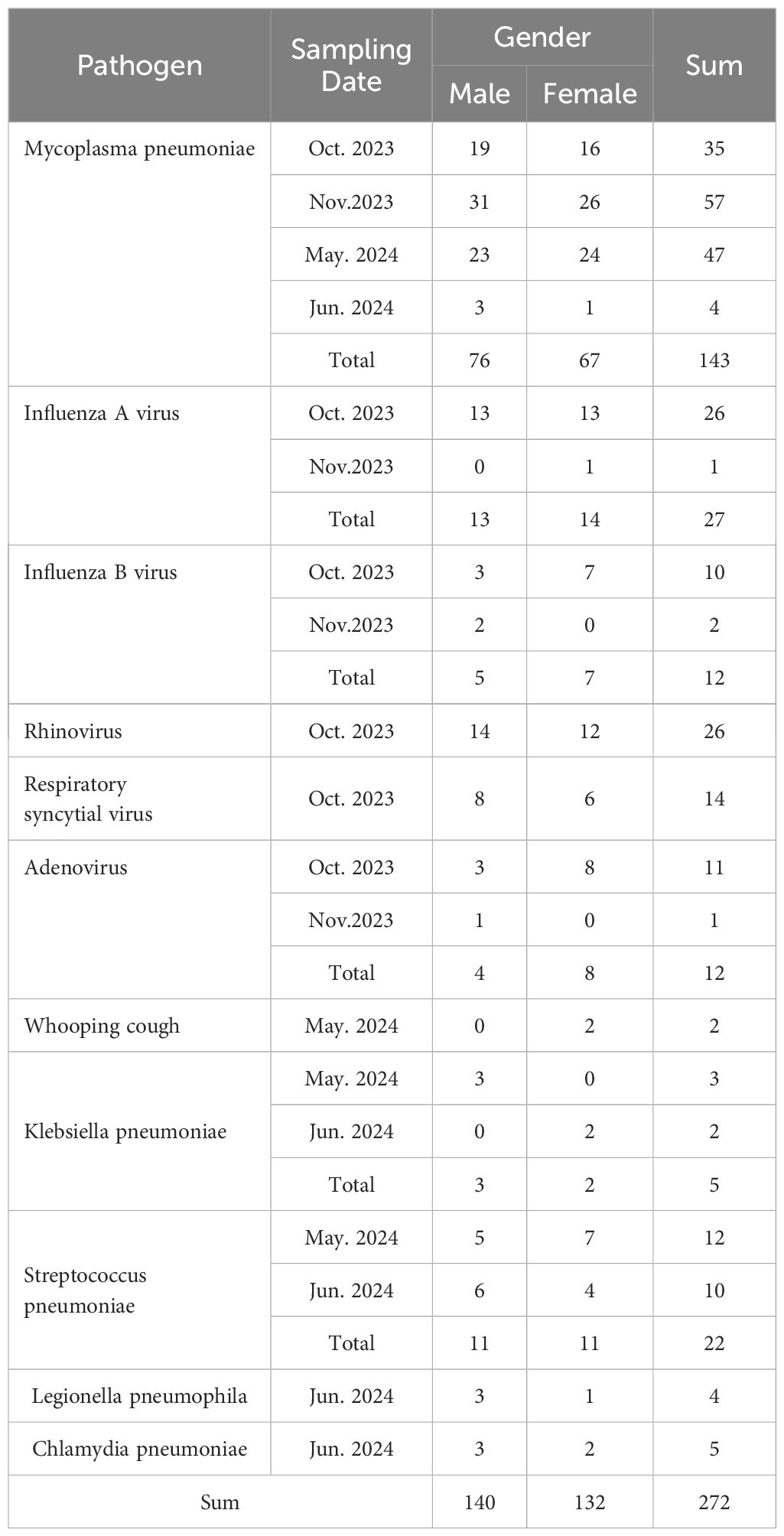
A total of 272 human nasopharyngeal swab samples were collected from The First Affiliated Hospital of Xinjiang Medical University from October 19th, 2023, to June 1st, 2024, from 140 males and 132 females (age range: 1–35 years). There were 143 MP and 129 non-MP samples (27 Influenza A virus, 12 Influenza B virus, 26 rhinovirus, 14 respiratory syncytial virus, 12 adenovirus Adenovirus, 2 Whooping cough, 5 Klebsiella pneumoniae, 22 Streptococcus pneumoniae, 4 Legionella pneumophila, 5 Chlamydia pneumoniae), according to clinical justification. All patients were well informed and signed the consent form. The details are shown in Table 1.
3 Results
3.1 The workflow of MP-LAMP-CRISPR/Cas12b detection system
The workflow of the MP-LAMP-CRISPR/Cas12b assay is shown in Figure 1. The MP-LAMP-CRISPR/Cas12b detection platform combines LAMP with the CRISPR/Cas12b-based detection assay, which can be performed in a single reaction step at a constant temperature for the detection of MP. The crude genomic DNA of MP was extracted using a commercial DNA isolation kit. The LAMP assay was used to amplify a target gene containing a PAM site (TTG) specific to Cas12b. Subsequently, the trans-cleavage activity of Cas12b was activated once the LAMP-amplified products were recognized by the corresponding AapCas12b/sgRNA complex, followed by non-specific cleavage of the reporter DNA that was labeled with the fluorophore 6-FAM and quencher BHQ1, resulting in the release of fluorescence, which could subsequently be detected by real-time PCR fluorescence readout or other small and portable blue light instruments. The entire test, including rapid template preparation (15 min), the LAMP-CRISPR/Cas12b reaction, and result detection (45 min), was completed within 1 h.
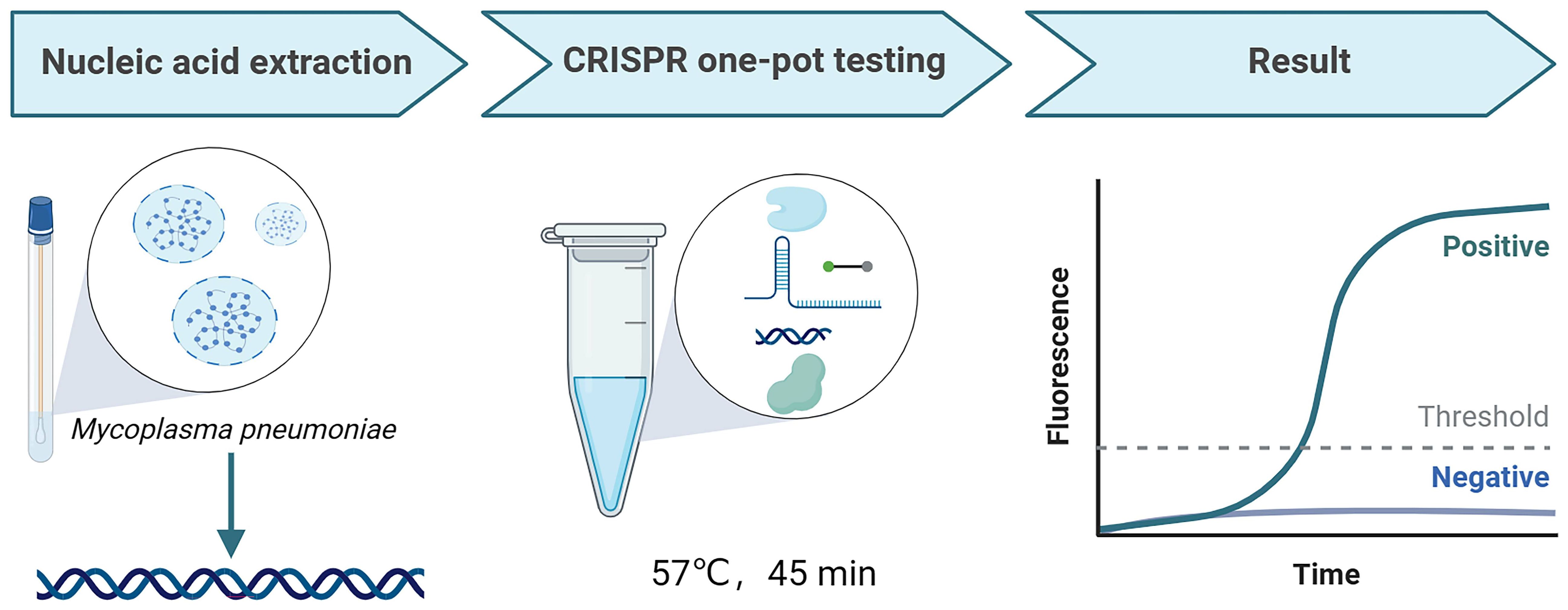
Figure 1 The workflow of MP-LAMP-CRISPR/Cas12b detection system. The whole assay consisted of MP nucleic acid extraction, CRISPR one-pot testing, and result readout. The MP nucleic acid amplification and detection were simultaneously performed in one-pot by one-step at a constant temperature.
3.2 Selection and confirmation of LAMP primers
putative intracellular protease (PIP) (GenBank: CP039761.1), which is a highly conserved gene in MP, was selected as the target gene for MP (Ratliff et al., 2014; Kanwar et al., 2018). In the present study, a plasmid containing a specific conserved sequence region of PIP was synthesized, and six pairs of LAMP primers (see Table 2) were designed by using the online NEB LAMP primer design tool (https://lamp.neb.com) based on the targeting conserved region. To explore the validity of the primers, LAMP reactions were performed at 60°C for 30 min. The LAMP primer pre-mixture was prepared by using 16 µL each of the forward inner primer (FIP) and backward inner primer (BIP), 4 µL each of the loop forward (LF) and loop backward (LB) primers, and 2 µL each of the forward outer primer (F3) and backward outer primer (B3), and 56 µL nuclease-free water. Taking the take-off time and amplification efficiency into consideration, the MP-PIP-5 primer showed higher effectiveness than the others, and without non-specific amplification product (Figures 2A–F). Therefore, the MP-PIP-5 primer was selected for the following reactions.
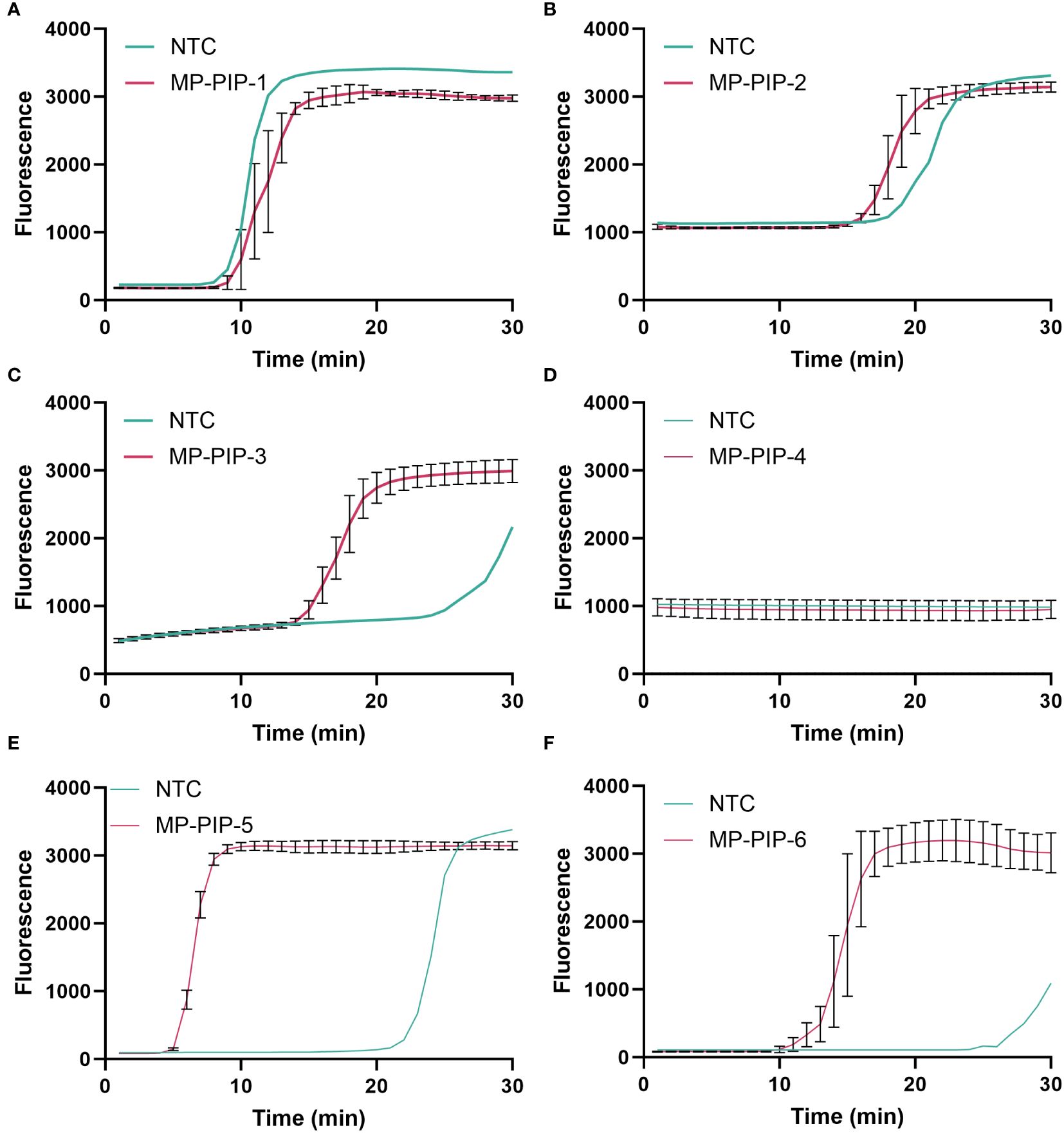
Figure 2 Selection of LAMP primer for PIP amplification. A total of Six groups LAMP primers were designed for selection. The amplification curve was presented from (A–F), corresponding to MP-PIP-1 to MP-PIP-6. NTC, no template control.
3.3 Selection and confirmation of Cas12b sgRNA
Based on the amplification products of primer MP-PIP-5, five Cas12b single guide RNA (Cas12b sgRNA1-5, shown in Table 3) were designed and synthesized. Moreover, a LAMP-based system was used to select the sgRNA. Both the fluorescence and take-off time of sgRNA-1 were better than those of the others, and sgRNA-1 was chosen and used in the following assays (Figures 3A–E).
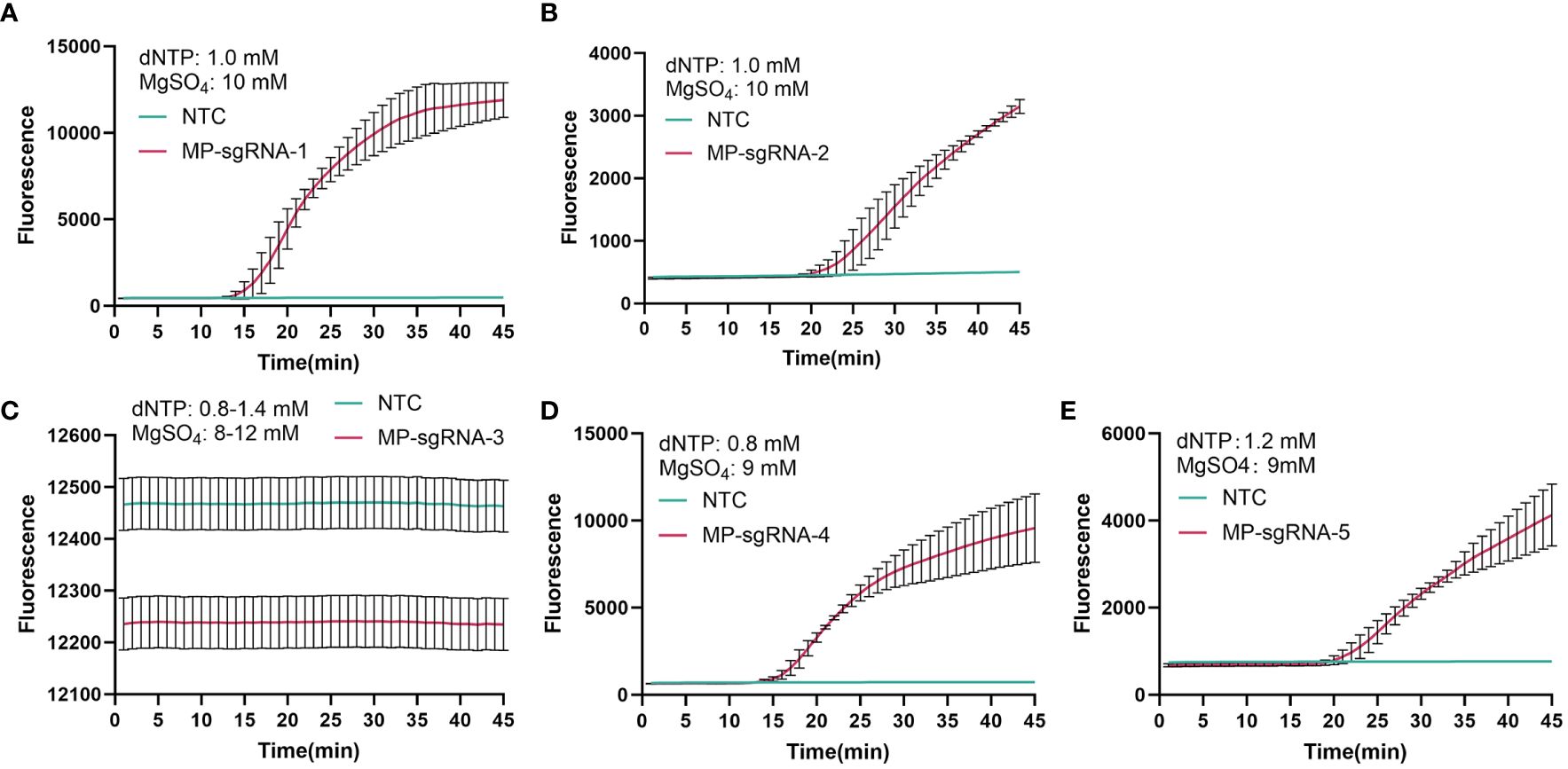
Figure 3 Selection of Cas12b sgRNA for PIP detection. A total of five Cas12b sgRNA that targeting PIP were designed for selection. The detection curve was presented from (A–E), corresponding to MP-sgRNA-1 to MP- sgRNA-5. NTC, no template control.
3.4 Optimal conditions for the MP one-step LAMP-CRISPR/Cas12b assay
3.4.1 Optimal concentrations of Cas protein and sgRNA
To determine the optimal concentrations of Cas12b protein and Cas12b sgRNA, Cas12b protein and Cas12b sgRNA were diluted to four different concentrations, including 250 nM, 500 nM, 750 nM, and 1000 nM in the reaction. As shown in Figure 2, the threshold time at 750 nM for Cas12b protein and Cas12b sgRNA was the shortest among all concentrations (Figure 4). Therefore, the optimum reaction concentration of the Cas12b protein and Cas12b sgRNA was 750 nM.
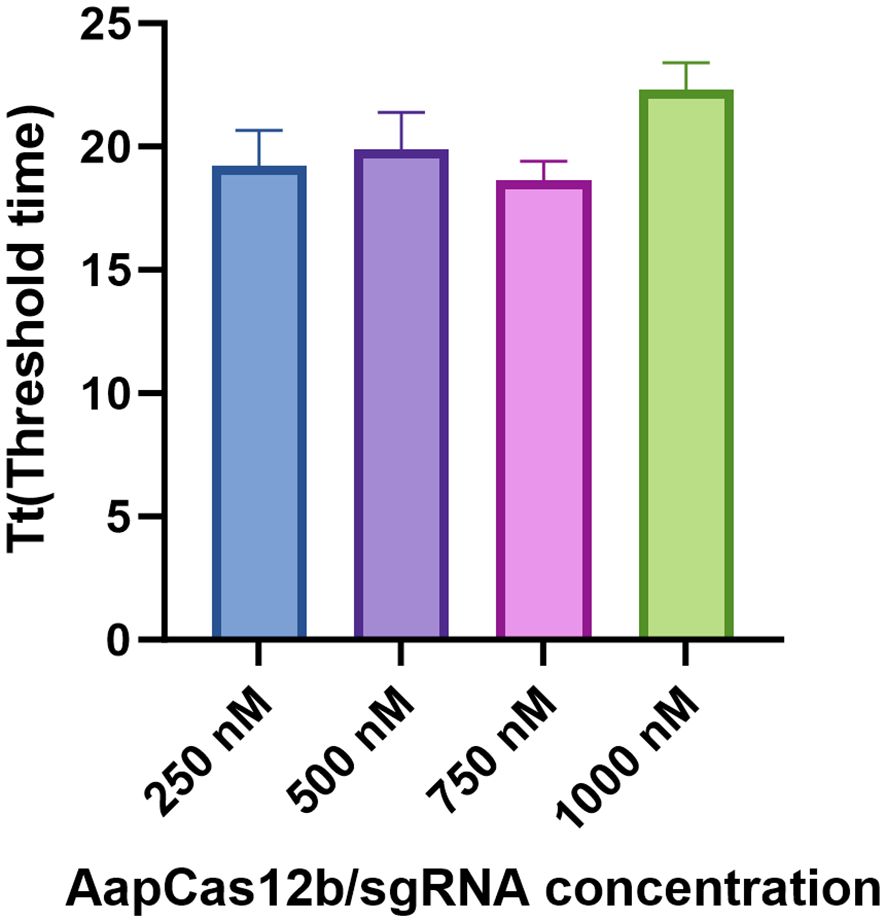
Figure 4 Determination of the optimal concentrations for Cas12b protein and Cas12b sgRNA. The reaction was set by using different concentrations of Cas12b and sgRNA, including 250 nM, 500 nM, 750 nM and 1000 nM respectively. The concentration was optimized according to the threshold time for the reaction. Three replications were established for each reaction.
3.4.2 Optimal reaction temperature
The optimal reaction temperature was determined. The reaction temperature was set from 56°C to 60°C with 1°C increments. Nuclease-free water served as the negative control (NTC). Clearly, the reaction condition in the temperature of 57°C showed the highest fluorescence intensity than that of other temperatures (Figure 5). Hence, 57°C was the optimum reaction temperature of the MP CRISPR-top assay.
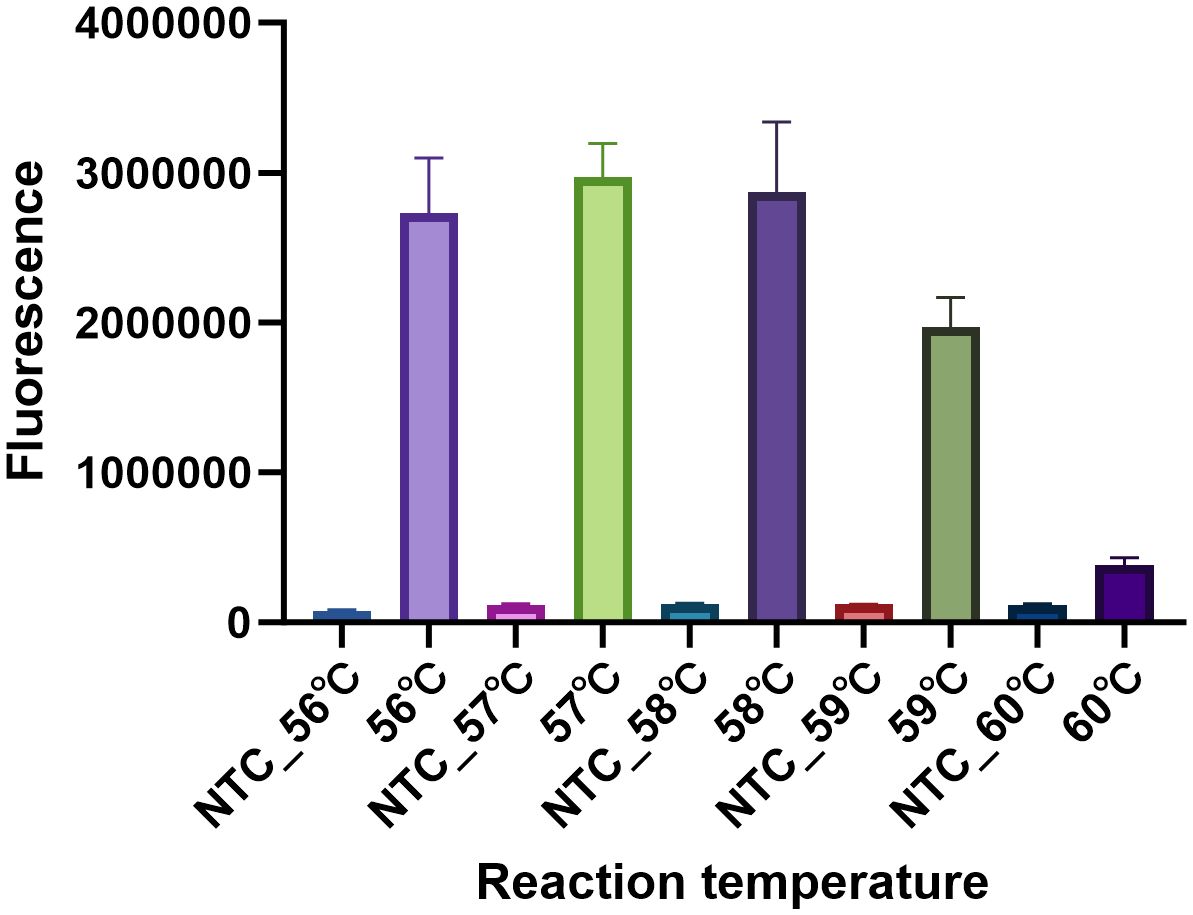
Figure 5 Determination of the optimal reaction temperature. To select the optimum reaction temperature, the assay was performed at 56 °C, 57 °C, 58 °C, 59 °C and 60 °C respectively. The temperature was optimized by the fluorescence intensity, three replications were established for each reaction. The fluorescence intensity of NTC at different temperatures was nearly to background.
3.4.3 Optimal ssDNA reporters
Here, a total of 4 HOLMES ssDNA-FQ reporters were used in the present examination. Based on the sequence, the reporters were named as: 8A-FQ (5’-/6-FAM/AAAAAAAA/BHQ1/-3’), 8T-FQ (5’-/6-FAM/TTTTTTTT/BHQ1/-3’), 8C-FQ (5’-/6-FAM/CCCCCCCC/BHQ1/-3’), and 8G-FQ (5’-/6-FAM/GGGGGGGG/BHQ1/-3’), respectively. Nuclease-free water served as the negative control (NTC). As shown in Figure 6, all the reporters were able to produce a fluorescent signal, especially 8T-FQ. Therefore, the 8T-FQ reporter was used in the SE LAMP-CRISPR-Top assay.
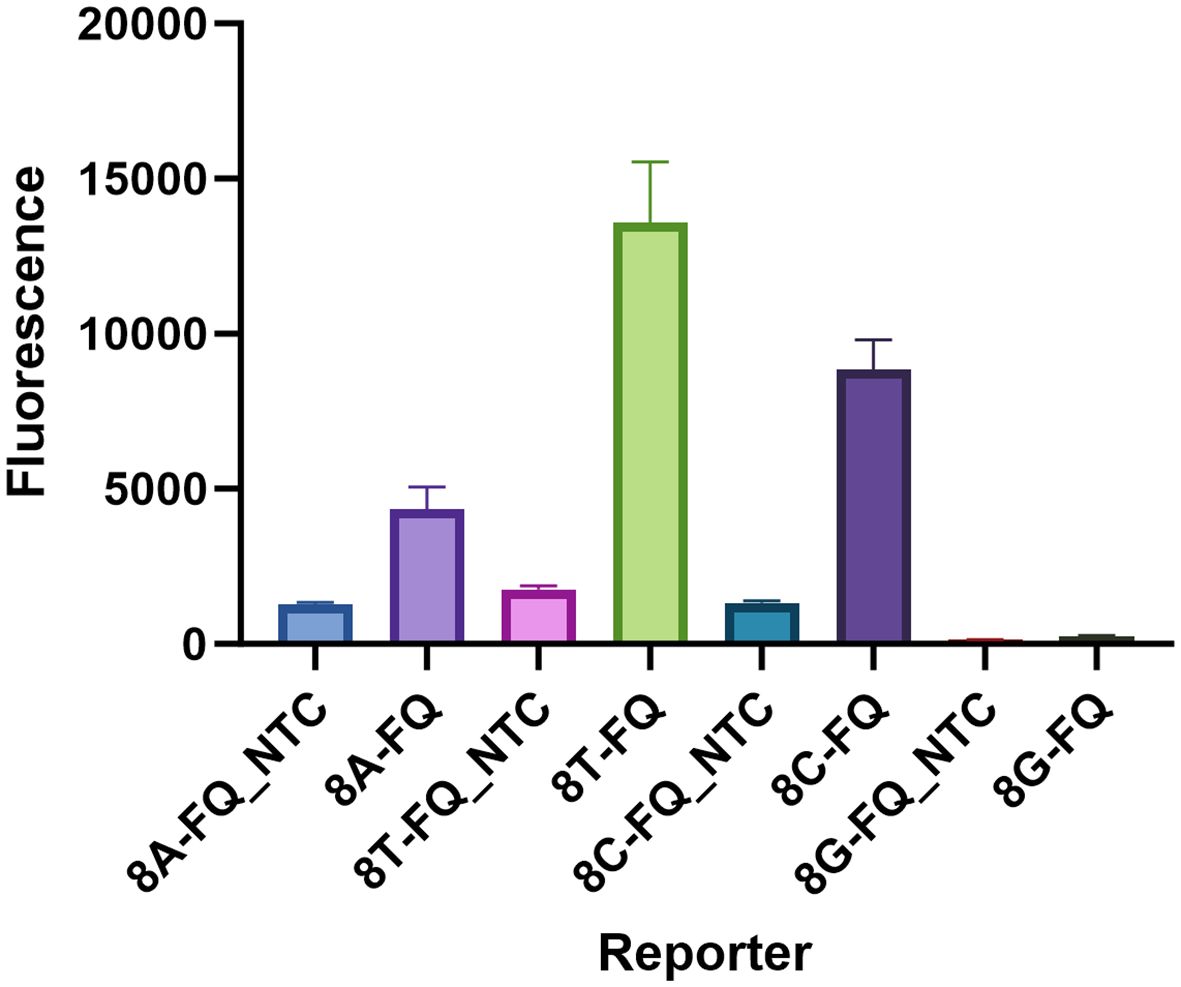
Figure 6 Determination of the optimal ssDNA reporter. Only 8G-FQ reporter was not worked, the left 8A-FQ, 8T-FQ, and 8C-FQ were all well-functioned. The 8T-FQ reporter was chosen based on the fluorescence intensity. Three replications were established for each reaction.
3.4.4 Optimal chemical additives for the reaction
Four chemical additives at different concentrations were chosen to improve the assay performance: including betaine (200 mM, 400 mM, 600 mM, and 800 mM), glycine (120 mM, 240 mM, 360 mM, and 480 mM), glycerol (1%, 2.5%, 5%, and 7.5%), and GuHCl (10 mM, 20 mM, 30 mM, and 40 mM). As shown in Figure 7, the addition of 480 mM glycine significantly increased the reaction efficiency, as it produced the shortest threshold time among the additives.
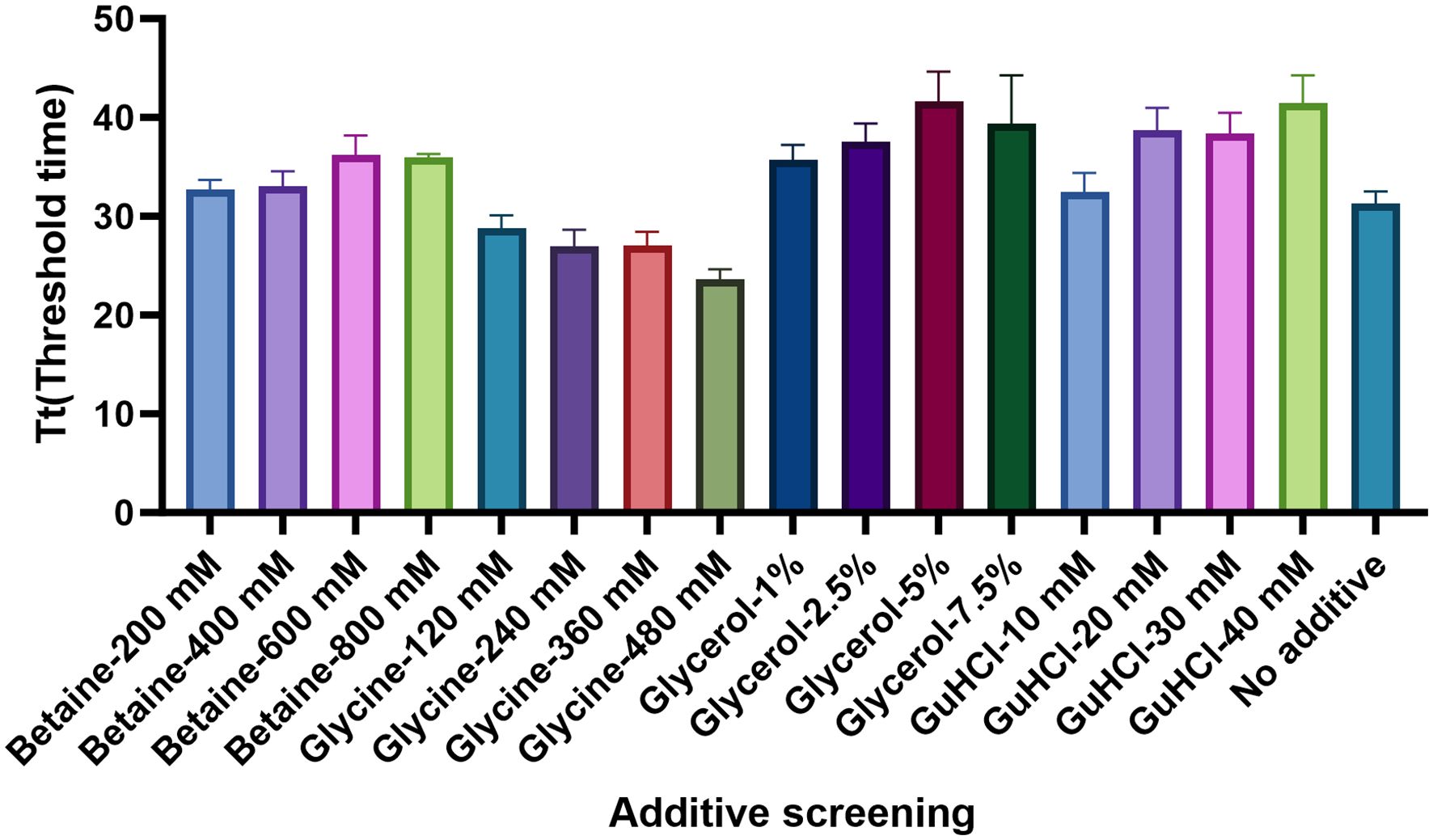
Figure 7 Determination of the optimal chemical additives. The optimum concentrations of Betaine, Glycine, Glycerol, and GuHCl were examined. According to the threshold time, the Glycine in the dose of 480 Mm presented the best performance. There replications were established for each reaction.
3.5 Specificity of the MP one-step LAMP-CRISPR/Cas12b method
The specificity of the MP one-step LAMP-CRISPR/Cas12b assay was confirmed using various templates extracted from different pathogens, including Mycoplasma pneumoniae, Influenza A virus, Influenza B virus, Rhinovirus, respiratory syncytial virus, Adenovirus, Whooping cough, Klebsiella pneumoniae, Streptococcus pneumoniae, Legionella pneumophila and Chlamydia pneumoniae. All pathogens were isolated from the corresponding positive clinical samples. Nuclease-free water served as the negative control (NTC), whereas the recombinant plasmid was used as the positive control (PC). As shown in Figure 8, all the positive results were obtained from the PC and Mycoplasma pneumoniae groups, whereas all the negative results were obtained from the non-MP templates.
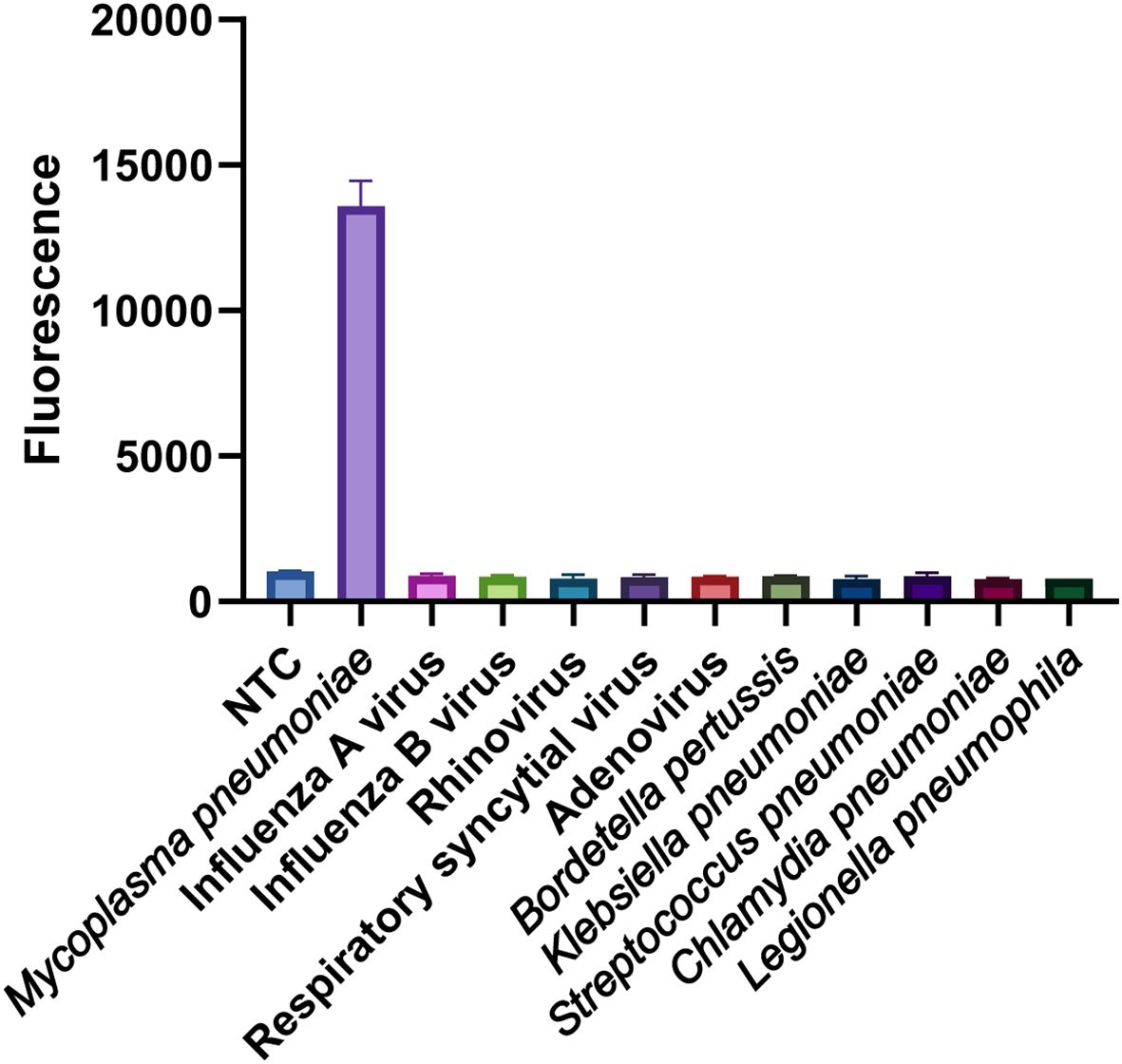
Figure 8 Specificity analysis of the MP one-step LAMP-CRISPR/Cas12b method. Only MP strain showed a remarkable fluorescence intensity, while the value of other non-MP strains was closely to background of NTC. Three replications were established for each reaction.
3.6 Sensitivity of the MP one-step LAMP-CRISPR/Cas12b method
To determine the limit of detection (LOD) of the MP one-step LAMP-CRISPR/Cas12b assay, the recombinant plasmid was diluted ranging from 25 copies/μL to 100 copies/μL at intervals of 25 copies,10 replicates were necessary for per dilution of plasmid. The analytical sensitivity of the assay was 33.7 copies per reaction (Figure 9).
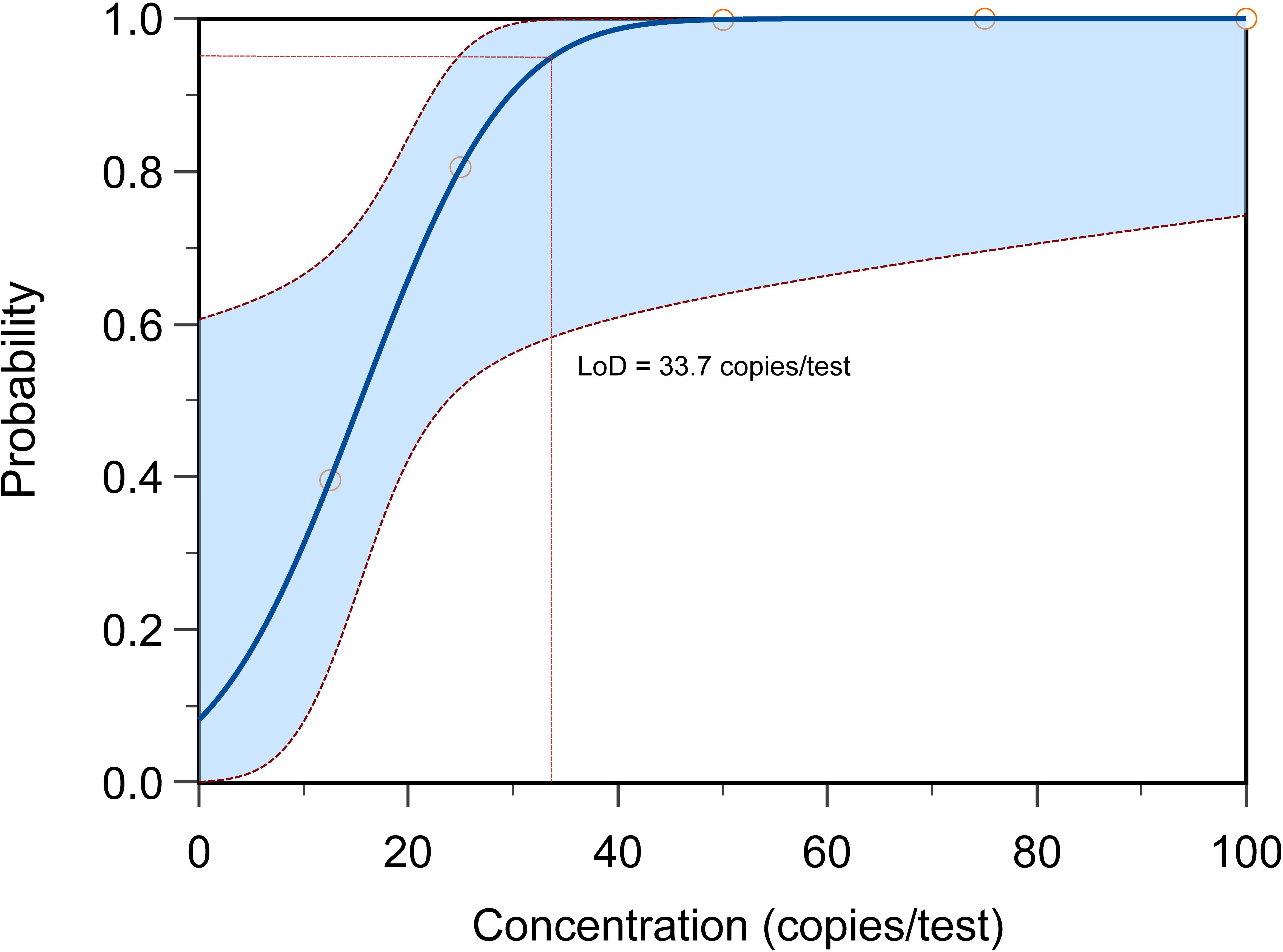
Figure 9 The LOD determination of MP one-step LAMP-CRISPR/Cas12b method. The LoD of present reaction achieved 33.7 copies per test with the probability about 95%, each reaction was performed in 10 replicates.
3.7 Validation of MP one-step LAMP-CRISPR/Cas12b method for clinical samples
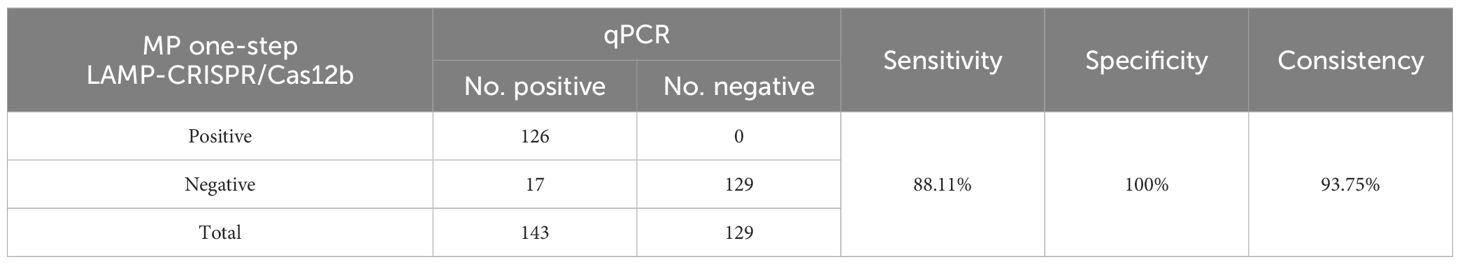
A total of 272 human swab samples were tested using PCR and the MP one-step LAMP-CRISPR/Cas12b method. Taking the results of qRCR as the gold standard, the sensitivity and specificity of the MP one-step LAMP-CRISPR/Cas12b method were 88.11% (126/143), specificity of 100% (129/129), and consistency of 93.75% (255/272), the results have been presented in Table 4. The diagnostic results obtained using the MP one-step LAMP–CRISPR/Cas12b assay were similar to those obtained using qRT-PCR.
4 Discussion
MP is one of the most common causative pathogens of CAP, accounting for 30–50% of CAP during the peak years. Early and rapid diagnostic methods are the key to guiding clinicians in their choice of antibiotics. However, the common methods for the identification of MP infection are culture separation, serological tests, and PCR, which are underutilized owing to shortcomings, such as time-intensiveness, insensitivity, high cost, requirement for precise instruments, and skilled technicians (Ratliff et al., 2014; Qin et al., 2023; Zhou et al., 2023). Consequently, the early, rapid, and accurate detection of MP is crucial for the diagnosis and rational selection of antibiotics.
Currently, isothermal amplification of nucleic acid technology is widely used for rapid pathogen detection of infectious diseases owing to its high efficiency and simplicity and plays an increasingly important role in promoting the on-site detection and control of pathogen diseases. LAMP was developed by Notomi et al. in 2000 (Notomi et al., 2000). In 2019, Arfaatabar et al. investigated a LAMP technique for the rapid detection of MP in clinical specimens, showing ‘substantial’ (κ=0.77) and ‘almost perfect’ (κ=0.86) congruence between the LAMP assay, culture, and PCR methods, respectively (Arfaatabar et al., 2019). However, the interpretation of the results of the LAMP assays was subjective. Thus, Wang et al. developed a LAMP coupled with a nanoparticle-based lateral flow biosensor (LFB) assay for the objective identification of MP (Wang et al., 2019a). In addition to LAMP, other isothermal amplification techniques have also been applied for the rapid detection of MP, such as multiple cross displacement amplification (MCDA) (Wang et al., 2019b), recombinase-aided amplification (RAA) (Xue et al., 2020), RPA (Jiang et al., 2022), and strand exchange amplification (SEA) (Yang et al., 2020). However, the amplified production of isothermal amplification methods often generates false-positive results; therefore, it is essential to control undesired non-specific amplification.
The specific merit of CRISPR-Cas systems is that they can be combined with isothermal amplification, which effectively solves the problem of false-positives. In 2022, Li et al. and Deng et al. developed the RPA-CRISPR/Cas12a and ERA/CRISPR–Cas12a assays for Mycoplasma pneumoniae detection, respectively (Deng et al., 2022; Li et al., 2022). In 2023, Jia et al. coupled the MCDA technique with a CRISPRCas12a-based biosensing system to design a novel detection platform for MP infection diagnosis and clinical application (Jia et al., 2023). Compared to Cas12a, Cas12b is ultrasensitive to mismatches between the guide RNA (gRNA) and target DNA, resulting in fewer off-target or false-positive effects and presenting a more accurate system in clinical applications (Strecker et al., 2019; Yang et al., 2023). Recently, Zhou et al. developed a CRISPR-Cas12b-based detection platform combined with RPA for MP infection that accurately distinguished MP strains from non-MP strains without cross-reactivity (Zhou et al., 2023). However, the contamination from aerosol cannot be totally avoided during the transferring of RPA products to CRISPR detection system.
The existing CRISPR-based tests for MP infection diagnosis require two separate reaction steps: (1) pre-amplification of the target nucleic acid using isothermal amplification assays and (2) detection of the resulting amplification products through CRISPR-based collateral reporter decoding. These two reaction steps require multiple manual operations and liquid handling, thus complicating the diagnostic procedures, increasing the risk of cross-contamination, and hampering their wider application in various fields.
However, application of the CRISPR system to detect MP with a one-step process has not yet been established. In this study, we established a one-step LAMP-CRISPR/Cas12b platform for MP detection. The working temperatures of Cas12b (37°C to 60°C) and LAMP (55°C to 60°C) reaction were observed to overlap (Dehghan Esmatabadi et al., 2015; Yang et al., 2016), which laid the foundation to integrate CRISPR/Cas12b with LAMP in one-pot. In the current study, the whole reaction was performed in one pot at 57°C and can be finished within 1 h. Moreover, there was no need to transfer the LAMP product to the CRISPR/Cas12b system. Therefore, our method achieved rapid detection of MP and prevented cross-contamination from multiple manual operations. As the entire assay was conducted at a constant temperature, an isothermal hot block and portable blue light equipment were sufficient for the MP-LAMP-CRISPR/Cas12b detection reaction. Moreover, the total cost of per reaction was about 10 RMB and most of the component can be pre-prepared via lyophilization. Hence, the proposed method performed efficiently in resource-limited settings. In addition, The LOD of the MP-LAMP-CRISPR/Cas12b system was 33.7 copies per reaction without any cross-reactivity with other non-MP pathogens. A preliminary evaluation using 272 clinical samples showed a sensitivity of 88.11% (126/143), specificity of 100% (129/129), and consistency of 93.75% (255/272) compared to the real-time PCR method, demonstrating that the developed method is reliable for the rapid diagnosis of MP.
5 Conclusion
In summary, we successfully established a rapid, simple, and accurate one-step method for MP detection by combining the LAMP assay and CRISPR/Cas12b methods in one-pot (MP-LAMP-CRISPR/Cas12b). These data indicate that the developed assay exhibits specificity and efficiency. Importantly, the MP-LAMP-CRISPR/Cas12b method has contributed to the extended detection of MP infections in various respiratory specimens, especially for point-of-care detection in resource-limited areas.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Approval Letter of Ethics Committee of First Affiliated Hospital of Xinjiang Medical University(K202403-32). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
TL: Formal analysis, Funding acquisition, Project administration, Resources, Software, Writing – original draft. QL: Conceptualization, Funding acquisition, Investigation, Methodology, Validation, Writing – original draft. FC: Methodology, Software, Writing – review & editing. YS: Data curation, Writing – review & editing. GM: Validation, Writing – original draft. ZY: Data curation, Formal analysis, Writing – review & editing. SZ: Writing – original draft. XL: Writing – review & editing. HL: Writing – review & editing. ZC: Project administration, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Science and Technology Assistance Project of Xinjiang Uygur Autonomous Region(2021E02072), Natural Science Foundation of Xinjiang Autonomous Region(2021D01E31), State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia Fund(SKL-HIDCA-2020-SG2).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arfaatabar, M., Noori Goodarzi, N., Afshar, D., Memariani, H., Azimi, G., Masoorian, E., et al. (2019). Rapid detection of mycoplasma pneumoniae by loop-mediated isothermal amplification (LAMP) in clinical respiratory specimens. Iran J. Public Health 48, 917–924. doi: 10.18502/ijph.v48i5.1809
Atçeken, N., Yigci, D., Ozdalgic, B., Tasoglu, S. (2022). CRISPR-cas-integrated LAMP. Biosensors 12 (11), 1035. doi: 10.3390/bios12111035
Cai, Z. H., Dai, Y. Y., Huang, L. Y., Zhang, W. S., Guo, X. G. (2019). Diagnosis of mycoplasma pneumoniae by loop-mediated isothermal amplification: systematic review and meta-analysis. BMC Infect. Dis. 19, 173. doi: 10.1186/s12879-019-3799-4
Dehghan Esmatabadi, M. J., Bozorgmehr, A., Motalebzadeh, H., Bodaghabadi, N., Farhangi, B., Babashah, S., et al. (2015). Techniques for evaluation of LAMP amplicons and their applications in molecular biology. Asian Pac J. Cancer Prev. 16, 7409–7414. doi: 10.7314/APJCP.2015.16.17.7409
Deng, Z., Hu, H., Tang, D., Liang, J., Su, X., Jiang, T., et al. (2022). Ultrasensitive, specific, and rapid detection of mycoplasma pneumoniae using the ERA/CRISPR-Cas12a dual system. Front. Microbiol. 13, 811768. doi: 10.3389/fmicb.2022.811768
Fan, X., Li, L., Zhao, Y., Liu, Y., Liu, C., Wang, Q., et al. (2020). Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 11, 1696. doi: 10.3389/fmicb.2020.01696
Hong, S., Park, J., Bok, J., Cho, E., Rhee, J. (2023). Assessment of measurement accuracy of amplified DNA using a colorimetric loop-mediated isothermal amplification assay. Biotechniques. 76, 114–118. doi: 10.2144/btn-2023-0104
Jain, S., Williams, D. J., Arnold, S. R., Ampofo, K., Bramley, A. M., Reed, C., et al. (2015). Community-acquired pneumonia requiring hospitalization among U. S. children. N Engl. J. Med. 372, 835–845. doi: 10.1056/NEJMoa1405870
Jia, N., Zhou, J., Xiao, F., Zheng, B., Huang, X., Sun, C., et al. (2023). A CRISPR-Cas12a-Based platform for ultrasensitive, rapid, and highly specific detection of Mycoplasma pneumonia in clinical application. Front. Bioeng Biotechnol. 11, 1022066. doi: 10.3389/fbioe.2023.1022066
Jiang, T., Wang, Y., Jiao, W., Song, Y., Zhao, Q., Wang, T., et al. (2022). Recombinase polymerase amplification combined with real-time fluorescent probe for mycoplasma pneumoniae detection. J. Clin. Med. 11 (7), 1780. doi: 10.3390/jcm11071780
Jiang, Z., Li, S., Zhu, C., Zhou, R., Leung, P. H. M. (2021). Mycoplasma pneumoniae infections: pathogenesis and vaccine development. Pathogens (Basel, Switzerland) 10 (2), 119. doi: 10.3390/pathogens10020119
Kanwar, N., Pence, M. A., Mayne, D., Michael, J., Selvarangan, R. (2018). Evaluation of the illumigene mycoplasma direct DNA amplification assay. J. Clin. Microbiol. 56 (7), e01930-17. doi: 10.1128/JCM.01930-17
Kutty, P. K., Jain, S., Taylor, T. H., Bramley, A. M., Diaz, M. H., Ampofo, K., et al. (2019). Mycoplasma pneumoniae among children hospitalized with community-acquired pneumonia. Clin. Infect. Dis. 68, 5–12.
Leal, S. M., Jr., Totten, A. H., Xiao, L., Crabb, D. M., Ratliff, A., Duffy, L. B., et al. (2020). Evaluation of commercial molecular diagnostic methods for detection and determination of macrolide resistance in mycoplasma pneumoniae. J. Clin. Microbiol. 58 (6), e00242-20. doi: 10.1128/JCM.00242-20
Li, F., Xiao, J., Yang, H., Yao, Y., Li, J., Zheng, H., et al. (2022). Development of a rapid and efficient RPA-CRISPR/Cas12a assay for mycoplasma pneumoniae detection. Front. Microbiol. 13, 858806. doi: 10.3389/fmicb.2022.858806
Li, S., Huang, J., Ren, L., Jiang, W., Wang, M., Zhuang, L., et al. (2021). A one-step, one-pot CRISPR nucleic acid detection platform (CRISPR-top): Application for the diagnosis of COVID-19. Talanta 233, 122591. doi: 10.1016/j.talanta.2021.122591
Li, Y., Li, S., Wang, J., Liu, G. (2019). CRISPR/cas systems towards next-generation biosensing. Trends Biotechnol. 37, 730–743. doi: 10.1016/j.tibtech.2018.12.005
Nagai, K., Horita, N., Yamamoto, M., Tsukahara, T., Nagakura, H., Tashiro, K., et al. (2016). Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: systematic review and meta-analysis. Sci. Rep. 6, 39090. doi: 10.1038/srep39090
Narita, M. (2010). Pathogenesis of extrapulmonary manifestations of Mycoplasma pneumoniae infection with special reference to pneumonia. J. Infect. Chemother. 16, 162–169. doi: 10.1007/s10156-010-0044-X
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, E63. doi: 10.1093/nar/28.12.e63
Poddighe, D. (2018). Extra-pulmonary diseases related to Mycoplasma pneumoniae in children: recent insights into the pathogenesis. Curr. Opin. Rheumatol 30, 380–387. doi: 10.1097/BOR.0000000000000494
Principi, N., Esposito, S. (2001). Emerging role of Mycoplasma pneumoniae and Chlamydia pneumoniae in paediatric respiratory-tract infections. Lancet Infect. Dis. 1, 334–344. doi: 10.1016/S1473-3099(01)00147-5
Qin, D., Gong, Q., Li, X., Gao, Y., Gopinath, S. C. B., Chen, Y., et al. (2023). Identification of Mycoplasma pneumoniae by DNA-modified gold nanomaterials in a colorimetric assay. Biotechnol. Appl. Biochem. 70, 553–559. doi: 10.1002/bab.2377
Qiu, X., Liu, X., Ma, X., Wang, R., Chen, S., Li, F., et al. (2022). One-pot isothermal LAMP-CRISPR-based assay for klebsiella pneumoniae detection. Microbiol. Spectr. 10, e0154522. doi: 10.1128/spectrum.01545-22
Ratliff, A. E., Duffy, L. B., Waites, K. B. (2014). Comparison of the illumigene Mycoplasma DNA amplification assay and culture for detection of Mycoplasma pneumoniae. J. Clin. Microbiol. 52, 1060–1063. doi: 10.1128/JCM.02913-13
Rodman Berlot, J., Mrvič, T., Košnik, M., Keše, D. (2023). The association between mycoplasma pneumoniae genotype and cutaneous disease. Microorganisms 11 (1), 205. doi: 10.3390/microorganisms11010205
Schmitt, B. H., Sloan, L. M., Patel, R. (2013). Real-time PCR detection of Mycoplasma pneumoniae in respiratory specimens. Diagn. Microbiol. Infect. Dis. 77, 202–205. doi: 10.1016/j.diagmicrobio.2013.07.016
She, R. C., Thurber, A., Hymas, W. C., Stevenson, J., Langer, J., Litwin, C. M., et al. (2010). Limited utility of culture for Mycoplasma pneumoniae and Chlamydophila pneumoniae for diagnosis of respiratory tract infections. J. Clin. Microbiol. 48, 3380–3382. doi: 10.1128/JCM.00321-10
Strecker, J., Jones, S., Koopal, B., Schmid-Burgk, J., Zetsche, B., Gao, L., et al. (2019). Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 10, 212. doi: 10.1038/s41467-018-08224-4
Waites, K. B., Xiao, L., Liu, Y., Balish, M. F., Atkinson, T. P. (2017). Mycoplasma pneumoniae from the respiratory tract and beyond. Clin. Microbiol. Rev. 30, 747–809. doi: 10.1128/CMR.00114-16
Wang, Y., Wang, Y., Jiao, W., Li, J., Quan, S., Sun, L., et al. (2019a). Development of loop-mediated isothermal amplification coupled with nanoparticle-based lateral flow biosensor assay for Mycoplasma pneumoniae detection. AMB Express 9, 196. doi: 10.1186/s13568-019-0921-3
Wang, Y., Wang, Y., Quan, S., Jiao, W., Li, J., Sun, L., et al. (2019b). Establishment and application of a multiple cross displacement amplification coupled with nanoparticle-based lateral flow biosensor assay for detection of mycoplasma pneumoniae. Front. Cell Infect. Microbiol. 9, 325. doi: 10.3389/fcimb.2019.00325
Xue, G., Li, S., Zhao, H., Yan, C., Feng, Y., Cui, J., et al. (2020). Use of a rapid recombinase-aided amplification assay for Mycoplasma pneumoniae detection. BMC Infect. Dis. 20, 79. doi: 10.1186/s12879-019-4750-4
Yang, C., Li, Y., Deng, J., Li, M., Ma, C., Shi, C. (2020). Accurate, rapid and low-cost diagnosis of Mycoplasma pneumoniae via fast narrow-thermal-cycling denaturation bubble-mediated strand exchange amplification. Anal. Bioanal Chem. 412, 8391–8399. doi: 10.1007/s00216-020-02977-y
Yang, H., Gao, P., Rajashankar, K. R., Patel, D. J. (2016). PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167, 1814–28.e12. doi: 10.1016/j.cell.2016.11.053
Yang, Y., Wang, D., Lü, P., Ma, S., Chen, K. (2023). Research progress on nucleic acid detection and genome editing of CRISPR/Cas12 system. Mol. Biol. Rep. 50, 3723–3738. doi: 10.1007/s11033-023-08240-8
Yiş, U., Kurul, S. H., Cakmakçi, H., Dirik, E. (2008). Mycoplasma pneumoniae: nervous system complications in childhood and review of the literature. Eur. J. Pediatr. 167, 973–978. doi: 10.1007/s00431-008-0714-1
Zhou, J., Xiao, F., Fu, J., Jia, N., Huang, X., Sun, C., et al. (2023). Rapid, ultrasensitive and highly specific diagnosis of Mycoplasma pneumoniae by a CRISPR-based detection platform. Front. Cell Infect. Microbiol. 13, 1147142. doi: 10.3389/fcimb.2023.1147142
Keywords: Mycoplasma pneumoniae, LAMP, CRISPR, Cas12b, One-step, one-pot
Citation: Liu T, Liu Q, Chen F, Shi Y, Maimaiti G, Yang Z, Zheng S, Lu X, Li H and Chen Z (2024) An accurate and convenient method for Mycoplasma pneumoniae via one-step LAMP-CRISPR/Cas12b detection platform. Front. Cell. Infect. Microbiol. 14:1409078. doi: 10.3389/fcimb.2024.1409078
Received: 29 March 2024; Accepted: 22 July 2024;
Published: 08 August 2024.
Edited by:
Huan Chen, Zhejiang Chinese Medical University, ChinaReviewed by:
Thomas Dandekar, Julius Maximilian University of Würzburg, GermanyRoger Dumke, Technical University Dresden, Germany
Copyright © 2024 Liu, Liu, Chen, Shi, Maimaiti, Yang, Zheng, Lu, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, MjQ3ODA2NTU4MEBxcS5jb20=; Zhaoyun Chen, Y3p5NTIwMTA3MTBAMTYzLmNvbQ==
†These authors contributed equally to this work and share first authorship
 Tao Liu
Tao Liu Qing Liu
Qing Liu Fuqun Chen1
Fuqun Chen1 Shutao Zheng
Shutao Zheng Xiaomei Lu
Xiaomei Lu