
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 04 July 2024
Sec. Clinical Microbiology
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1408947
This article is part of the Research TopicRising Stars in Clinical Microbiology: 2023View all 5 articles
Lysine acetylation is an evolutionarily conserved protein modification that changes protein functions and plays an essential role in many cellular processes, such as central metabolism, transcriptional regulation, chemotaxis, and pathogen virulence. It can alter DNA binding, enzymatic activity, protein-protein interactions, protein stability, or protein localization. In prokaryotes, lysine acetylation occurs non-enzymatically and by the action of lysine acetyltransferases (KAT). In enzymatic acetylation, KAT transfers the acetyl group from acetyl-CoA (AcCoA) to the lysine side chain. In contrast, acetyl phosphate (AcP) is the acetyl donor of chemical acetylation. Regardless of the acetylation type, the removal of acetyl groups from acetyl lysines occurs only enzymatically by lysine deacetylases (KDAC). KATs are grouped into three main superfamilies based on their catalytic domain sequences and biochemical characteristics of catalysis. Specifically, members of the GNAT are found in eukaryotes and prokaryotes and have a core structural domain architecture. These enzymes can acetylate small molecules, metabolites, peptides, and proteins. This review presents current knowledge of acetylation mechanisms and functional implications in bacterial metabolism, pathogenicity, stress response, translation, and the emerging topic of protein acetylation in the gut microbiome. Additionally, the methods used to elucidate the biological significance of acetylation in bacteria, such as relative quantification and stoichiometry quantification, and the genetic code expansion tool (CGE), are reviewed.
Post-translational modifications (PTMs) are covalent and generally enzymatic modifications of amino acid residues in a protein that occur after it is synthesized. Amino acid side chains may be modified by adding small chemical groups (e.g., methylation, acetylation, and phosphorylation) or more complex molecules such as oligosaccharides or small peptides (e.g., glycosylation and ubiquitylation) (Mann and Jensen, 2003; Heuts et al., 2009; Macek et al., 2019). The PTMs most widely distributed and frequently reported are phosphorylation, glycosylation, ubiquitination, methylation, and acetylation, among other alkylations (Khoury et al., 2011). Multiple PTMs can occur on a single type of amino acid, or different types of amino acids may be given the same modification, thereby altering the chemical properties of the modified sites. Through protein PTMs, cells regulate their functions and metabolic pathways and increase the variety and complexity of target proteins (Shahin and Javad, 2021).
A wide variety of amino acid residues can be post-translationally acetylated, including:
(1) The irreversible acetylation of the alpha-amino group in the N-terminal amino acid of proteins (Nα-acetylation), a common modification in eukaryotes (50–70% of yeast proteins and 80–90% of human proteins), but rare in prokaryotes.
(2) The reversible acetylation of the hydroxyl side chain of serine or threonine (O-acetylation) is detected only in a few eukaryotic organisms.
(3) The acetylation of serine, threonine, and histidine by a family of bacterial effector molecules from some pathogenic species (Mukherjee et al., 2007; Mittal et al., 2010; Paquette et al., 2012; Christensen et al., 2019b).
(4) The reversible acetylation of the epsilon-amino group of the lysine residue (Nε-acetylation), a modification found in eukaryotes and prokaryotes (Khoury et al., 2011; Diallo et al., 2019; Ramazi and Zahiri, 2021). The lysine epsilon amino group is also the site of other modifications besides acetylation (Gil et al., 2017).
Nε-acetylation occurs either enzymatically by the lysine acetyltransferases (KATs) or non-enzymatically by chemical acetylation. In both cases, the acetyl group comes from a donor molecule, typically acetyl coenzyme A (AcCoA) or acetyl phosphate (AcP). Both types of acetylation are reversible by the action of deacetylase enzymes (Figure 1A) (Hentchel and Escalante-Semerena, 2015).
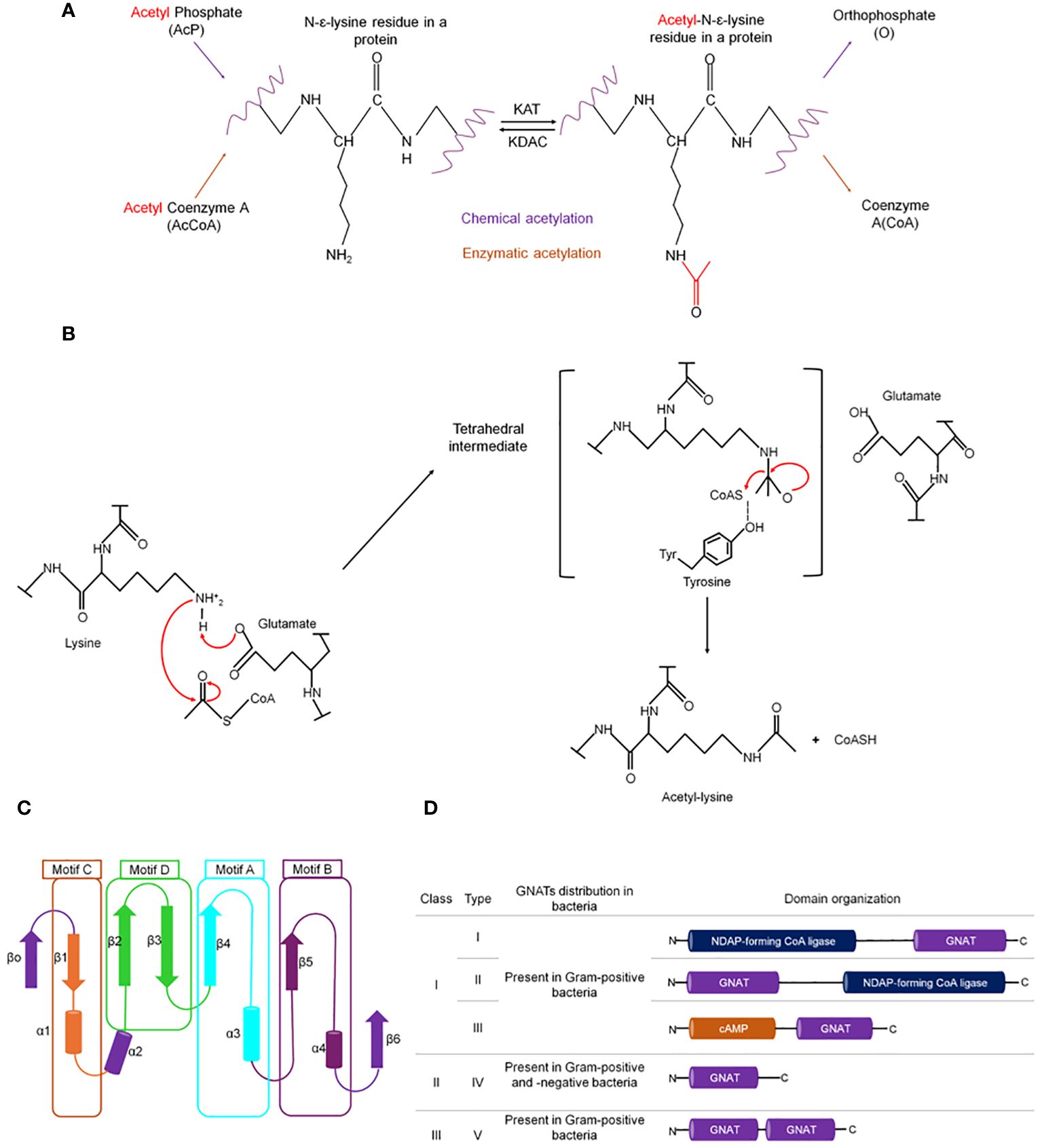
Figure 1 Types of lysine acetylation in prokaryotes. (A) Enzymatic and chemical acetylation and their respective acetyl donor molecule. In bacteria, enzymatic acetylation is catalyzed by Gcn5-related N-acetyltransferase (GNAT). (B) The catalytic mechanism exerted by GNATs is a sequential mechanism where glutamate acts as a general base to deprotonate the amino group of the lysine, enabling nucleophilic attack of the acetyl-CoA carbonyl, followed by the formation of a transient tetrahedral intermediate, that is resolved to yield the acetylated substrate amino group and coenzyme (A) Overall structure of GNATs: (C) Cartoon representation of GNATS topology. The secondary structure elements are colored and represent the different motifs. Motifs C (β1–α1), D (β2–β3), A (β4–α3), and B (β5–α4) are colored orange, green, aquamarine, and blue, respectively. The least conserved secondary structure elements (strands β0 and β6 and helix α2) absent in some GNAT proteins are colored purple. (D) Classification of GNATs in prokaryotes based on their domain organization and the arrangement of the GNAT domain [figure redrawn and modified from VanDrisse and Escalante-Semerena, 2019; Blasl et al., 2021; Lammers, 2021].
Lysine acetylation was discovered 60 years ago in histones and was implicated in transcriptional regulation (Allfrey et al., 1964). Extensive studies in eukaryotic cells have shown that acetylation is an essential protein modification that influences many cellular processes, including protein-protein interaction, protein stability, protein folding, cellular localization, and enzymatic activity. This PTM regulates different biological pathways, such as cell cycle control, cell metabolism, DNA repair, DNA replication, ribosome biogenesis in the nucleus, nuclear transport, translation, and transcription, among others (Berrabah et al., 2011; Oliveira and Sauer, 2012; Gil et al., 2017; Tarazona and Pourquié, 2020). In contrast, research on acetylation in bacteria is relatively new and primarily focused on describing global acetylation through a proteomic approach. The first studies on acetylation as a regulatory mechanism were carried out in Salmonella enterica, where it was demonstrated that the acetyl-CoA synthetase (Acs), an enzyme responsible for the synthesis of acetyl-CoA (Ac-CoA) from acetate, is post-translationally regulated by the protein acetyltransferase (Pat) that acetylates Lys residue 609 of Acs (Starai et al., 2002, Starai et al., 2004). Furthermore, it was found that Acs is activated by CobB, a bacterial protein deacetylase homologous to yeast Sir2, which specifically hydrolyzes the acetyl group of the acetyl-lysine active site of Acs to promote its activity (Starai et al., 2002, Starai et al., 2004). Since then, acetylation in bacteria has been shown to regulate several fundamental pathways, including cellular function, physiology, and metabolism. Proteomics studies have found that in some bacteria, the proteins acetylated on lysine constitute more than 10% of the proteome.
Nε-acetylation refers to adding an acetyl group from a donor molecule, like AcCoA, to an epsilon amino group of lysine side chains. The modification increases the size of the side chain and neutralizes the positive charge of the unmodified amino acid. Nε-acetylation is a reversible process where the acetate added by KATs can be removed by histone deacetylases (HDAC) (Taunton et al., 1996; Haigis and Guarente, 2006; Yang and Seto, 2008; Bürger and Chory, 2018; VanDrisse and Escalante-Semerena, 2019). These opposing enzymatic activities determine the levels of posttranslational modification on a particular protein.
KATs are grouped into three major families based on amino acid sequence homology and biochemical characteristics of catalysis: (i) Gcn5- related N-acetyltransferases (GNATs), (ii) the p300/CBP family, and (iii) MYST family. The MYST and p300/CBP families are present only in eukaryotic cells, while Gcn5-related N-acetyltransferase family members occur in bacteria, eukaryotes, and archaea (Gu and Roeder, 1997; Vetting et al., 2005; Lee and Workman, 2007; Finkel et al., 2009; Favrot et al., 2016).
KAT families differ in sequence similarity, domain organization, substrate specificity, and catalytic mechanism. The GNAT family uses two main types of kinetic mechanisms: a sequential mechanism and a ping-pong mechanism. In the direct transfer mechanism, AcCoA and the protein substrate bind to form a ternary complex in which active site glutamate or aspartate acts as a general base to deprotonate the amino group of the lysine, allowing it to perform a nucleophilic attack on the thioester carbonyl carbon of the acetyl moiety of AcCoA (Figure 1B) (Liu et al., 2008). In the ping-pong mechanism, the acetyl group is covalently attached to the enzyme to form an acyl-enzyme intermediate before being transferred to the substrate. For this type of mechanism, the presence of nucleophilic residues such as cysteine or serine in a proper position in the active site is required (Vetting et al., 2005; Majorek et al., 2013; Baumgartner et al., 2021). Although the MYST family members can also use the sequential or ping-pong mechanisms, this catalysis route occurs via an acetyl-cysteine intermediate (Yan et al., 2002). In contrast, the p300/CBP family does not use a catalytic base to initiate the transfer of the acyl moiety. Instead, this family uses a Theorell–Chance mechanism where a tyrosine in the active site acts as a catalytic acid to increase the nucleophilicity of the lysine side chain (Liu et al., 2008). The protein substrate-AcCoA association binds transiently to the enzyme surface, allowing the lysine residue to receive the acetyl group, followed by rapid protein dissociation (Zhang et al., 2014). The ternary complex formed is kinetically irrelevant for catalysis (Liu et al., 2008).
As mentioned, members of the GNATs family are found in all domains of life and are involved in diverse processes such as synthesis, transcription control, antibiotic resistance, stress regulation, metabolic flux regulation, and virulence regulation (Xie et al., 2014; Hentchel and Escalante-Semerena, 2015; Favrot et al., 2016). The first two reported GNAT members were the aminoglycoside N-acetyltransferase from multidrug-resistant Serratia marcescens and the histone acetyltransferase (HAT1) from Saccharomyces cerevisiae (Dutnall et al., 1998; Wolf et al., 1998). So far, GNATs comprise one of the largest superfamilies, with more than 300,000 GNAT proteins identified (Salah Ud-Din et al., 2016; Burckhardt and Escalante-Semerena, 2020; Krtenic et al., 2020).
GNATs can acetylate a wide variety of substrates, from antibiotics to proteins and peptides (Table 1), so they constitute a highly diverse family of proteins. Interestingly, some acetyltransferases can acetylate multiple substrates. For example, the Eis protein from Mycobacterium tuberculosis was initially described as an aminoglycoside acetyltransferase (Zaunbrecher et al., 2009; Chen et al., 2011; Houghton et al., 2013), but it also acetylates proteins (Kim et al., 2012; Ghosh et al., 2016). A deeper analysis of the GNAT superfamily members’ functions and their target molecules have been discussed in several reviews (Dyda et al., 2000; Vetting et al., 2005; Favrot et al., 2016; Salah Ud-Din et al., 2016; VanDrisse and Escalante-Semerena, 2019; Burckhardt and Escalante-Semerena, 2020).
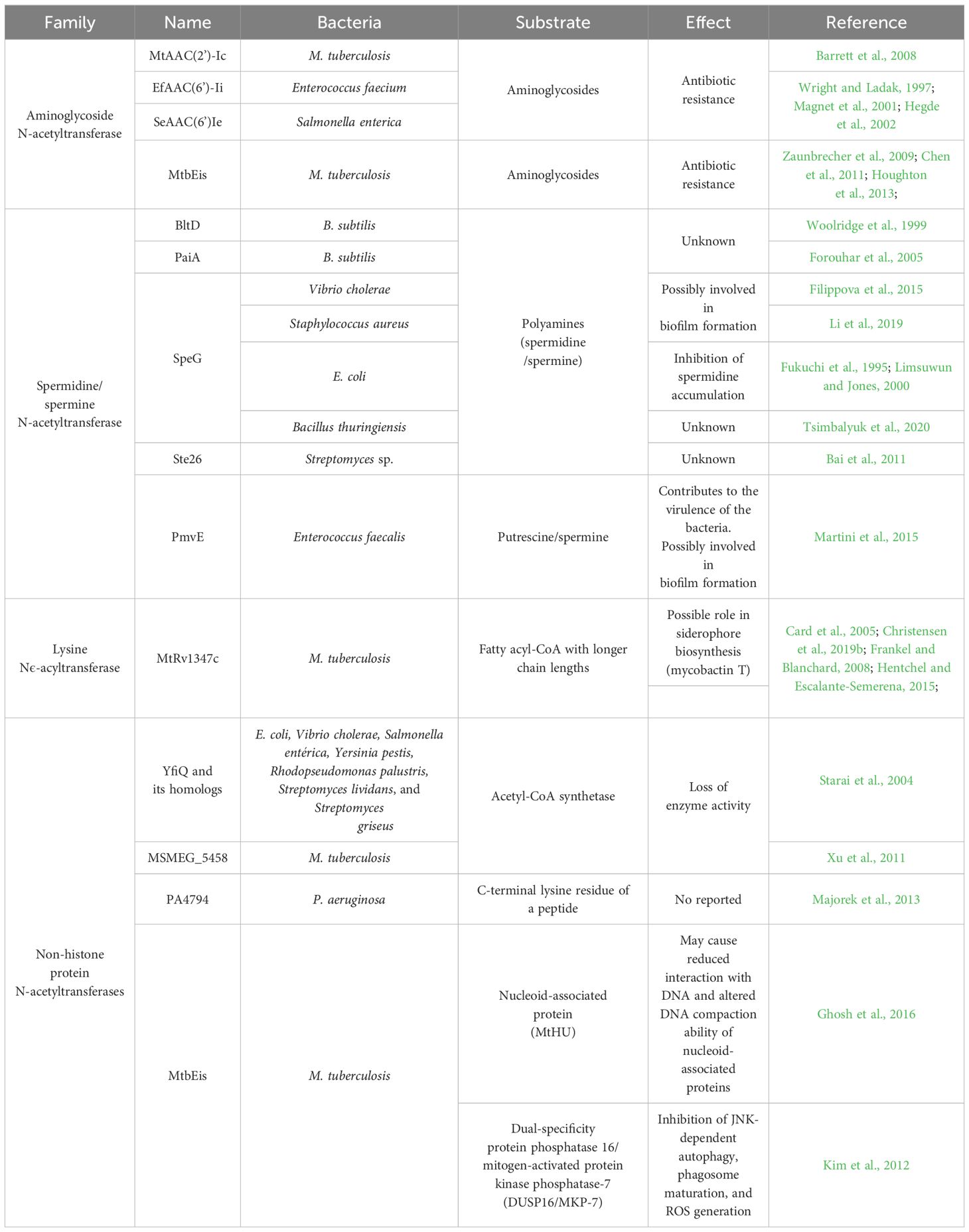
Table 1 Some examples of GNAT enzymes, their targets, and the effect produced by the acetylation are shown.
Although the primary sequence similarity between GNATs is moderate (3%–23%), they are folded into a highly conserved three-dimensional structure. The core secondary elements of the GNAT proteins consist of six or seven β-strands and four α-helices, connected in the order β0-β1-α1-α2-β2-β3-β4-α3-β5-α4-β6. Four conserved motifs, known as the N-acetyltransferase domain, are found in this core arranged in the order C-D-A-B (Figure 1C) (Tercero et al., 1992; Dyda et al., 2000; Hentchel and Escalante-Semerena, 2015).
Motifs C and D are essential in protein stability, while motifs A and B contain the residues involved in acyl-CoA and acceptor substrate binding, respectively (Dyda et al., 2000; Vetting et al., 2005). Motif A contains a six-residue segment (Q/R)-X-X-G-X-(G/A) known as the P-loop (phosphate-binding loop), that serves for substrate recognition and binding and contains glutamic or aspartic acid residues to deprotonate the target lysine (Vetting et al., 2005; Liu et al., 2008; Lu et al., 2017). Across the entire GNAT superfamily, there is structural divergence in the β1β2 loop, the α-4 helix of motif B, and strand β6 at the C-terminal, which together form the binding site for the acceptor substrate. Specifically, the C-terminal region contains a loop of varying length and position, and the residues in the motif B are not well conserved. These structural variants recognize different substrates (Dyda et al., 2000; Salah Ud-Din et al., 2016). For example, in the mycothiol synthase (Rv0819) from M. tuberculosis, the β-strand 1 in the N-terminal domain is missing, the position of helix 2, and a long loop inserted between α3′ and β5′, while aminoglycoside 6´-N-acetyltransferase from Enterococcus faecium has an additional α-helix between the β1 and β2 strands (Wybenga-Groot et al., 1999; Vetting et al., 2003).
The binding of the enzyme to AcCoA/CoA occurs through interactions with the pantetheine and pyrophosphate moieties of CoA (Dutnall et al., 1998; Clements et al., 1999; Lin et al., 1999; Rojas et al., 1999; Wang et al., 2008; Hentchel and Escalante-Semerena, 2015). The pantetheine binding is based on hydrogen bonds with the main chain of β4 and β 5, and the pyrophosphate is coordinated mainly by the main chain nitrogen atoms of the conserved phosphate binding loop between β4 and α3 (Majorek et al., 2017).
Different works have proposed a classification system for bacterial KATs of the GNAT family (Figure 1D) (Hentchel and Escalante-Semerena, 2015; Lu et al., 2017; Christensen et al., 2019a; VanDrisse and Escalante-Semerena, 2019). This classification system recognized that the GNAT family exhibits different sequence lengths, domain architecture, and types. Recently, Christensen et al., 2019a proposed a new system of three main classes of KATs based on sequence length and the present GNAT domains, as well as five different types of KATs based on domain identities and arrangements. Class I consists of a large (>80 KDa) multidomain enzyme, where only the GNAT catalytic domain is conserved. Class II encompasses most bacterial acetyltransferases, smaller enzymes with a single GNAT domain. Class III has a dual arrangement of GNAT domains. Depending on domain position, these classes are further categorized into five types: types I and II contain a domain homologous to nucleotide-diphosphate (NDP)-forming acyl-CoA ligase/synthetase (700–900 aa) but which lack activity and a GNAT domain (~200 aa) at the N- or C-terminal. Type III KATs have a smaller regulatory domain (~300–400 aa) at the N-terminal that binds to an effector (e.g., cAMP, NADP, or amino acids) and a C-terminal GNAT domain. Type IV and V do not contain any regulatory domain and consist only of one GNAT domain (400 aa) and multiple GNAT domains, respectively (Figure 1D) (Hentchel and Escalante-Semerena, 2015; Lu et al., 2017; Christensen et al., 2019a; VanDrisse and Escalante-Semerena, 2019).
The diversity of domain architectures and organization of GNATs indicate that in bacteria, lysine acetylation is regulated by diverse metabolic signals in response to physiological conditions and environmental changes. However, more studies are needed to elucidate the substrates and structures of these enzymes and their role in these organisms.
Acetyl-CoA is synthesized by the oxidative decarboxylation of pyruvate during glycolysis, the catabolism of isoleucine, leucine, and threonine, the reverse TCA cycle, and fatty acid β-oxidation. Whereas KATs use AcCoA to acetylate proteins, it can also modify the amino group of the lysine side chains without the assistance of an enzyme. Non-enzymatic acetylation (chemical acetylation) was first mentioned for histone proteins by Phillips, 1963. A few years later, it was demonstrated that purified histones, albumin, and synthetic lysine homopolymer are acetylated in vitro at a pH ~9 in the presence of AcCoA without enzymes (Paik et al., 1970). In eukaryotic cells, specifically in the mitochondria, the absence of acetyltransferases and the unique conditions, such as the high concentration of AcCoA (1.5 mM steady-state) and alkaline pH (7.9–8.0), suggests that lysine acetylation occurs chemically. For example, many mitochondrial proteins are acetylated under conditions mimicking those of the mitochondria matrix in a mechanism that is generally pH- and acyl-CoA concentration-dependent (Wagner and Payne, 2013).
Direct chemical acetylation has also been observed in bacteria. Both AcCoA and AcP can non-enzymatically acetylate lysine residues on many proteins. On the one hand, demonstrating that non-enzymatic AcCoA-dependent acetylation occurs in vivo is difficult since AcCoA is an essential molecule, and deleting it would result in a lack of viability of the organisms. However, it has been shown that AcCoA can acetylate purified bacterial proteins at a concentration comparable to or below physiological levels (Barak et al., 2006; Ramos-Montañez et al., 2010; Sun et al., 2016; Wang et al., 2017). On the other hand, the reactive chemical properties of AcP support the hypothesis that it could function directly as an acetyl donor. In water, AcP hydrolyzes quickly under basic or acidic conditions in the presence of Mg2+ or Ca2+ (Lipmann and Tuttle, 1944; Koshland and Daniel, 1952). Also, acyl group transfer is favored over hydrolysis in the presence of nucleophilic reagents such as thiols, hydroxyl groups, or the amino groups of lysine side chains (Jencks, 1969; Kuhn et al., 2014).
In bacteria, substantial evidence has shown that AcP-dependent non-enzymatic acetylation directly correlates with the levels of this high-energy intermediate. When cells are unable to produce AcP, acetylation levels are reduced, whereas when cells accumulate it, the acetylation levels increase significantly (Weinert et al., 2013; Kuhn et al., 2014; Kosono et al., 2015; Post et al., 2017; Bontemps-Gallo et al., 2018). This PTM seems more global and less specific than enzymatic acetylation (Weinert et al., 2013).
Many studies on protein acetylation in bacteria have focused on describing enzymatic acetylation and its implication in cellular physiology, pathogenesis, and response to environmental conditions. However, non-enzymatic acetylation is also possible (Table 2). The acetylation by AcP has been mainly described by proteomic studies, in which a comparative analysis of the acetylome of different strains allows us to determine if there is any difference in the protein acetylation levels and infer the acetylation mechanism. This has been achieved in Escherichia coli, Neisseria gonorrhoeae and Borrelia burgdorferi (Table 2) (Weinert et al., 2013; Kuhn et al., 2014; Kosono et al., 2015; Schilling et al., 2015; Post et al., 2017; Bontemps-Gallo et al., 2018; Reverdy et al., 2018). These studies have focused on comparing the proteomic data of the acetylated proteins of the wild-type strain with different isogenic mutant strains. The acetylome data using E. coli as a model demonstrated that acetylation depends on AcP formation and occurs at a low level in growth-arrested cells. Mutant cells unable to synthesize (pta ackA mutant) or metabolize (ackA mutant) AcP had the opposite behavior, with the former mutant having significantly reduced acetylation levels and the latter mutant, which accumulated AcP, having substantially higher acetylation levels. It was also shown that AcP acetylates lysine residues in vitro at a concentration comparable to that found in vivo. These data establish AcP as a critical donor for acetylation and suggest that AcP acts nonenzymatically to regulate acetylation levels in response to glucose (Table 2) (Weinert et al., 2013; Kuhn et al., 2014; Schilling et al., 2015).
Since AcCoA and AcP are derived from multiple metabolic pathways in microorganisms, it is difficult to establish whether both or only one of these molecules is the acetyl group donor. The bacterium Borrelia burgdorferi is advantageous in this regard because acetate metabolism is limited to the mevalonate pathway that synthesizes lipid I for cell wall biogenesis. In this pathway, acetate is converted to AcP by acetate kinase (AckA), which is metabolized to AcCoA by phosphotransacetylase (Pta) (Richards et al., 2015). Analysis by immunoblotting demonstrated that AckA and Pta were not produced in ΔackA and Δpta mutants, respectively. In the ackA mutant complemented with a functional ackA gene, AckA was expressed at a level greater than that of the wild-type. Surprisingly, in the pta mutant complemented with pta, Pta was not overexpressed (Richards et al., 2015).
The acetylome analysis of these mutants and their respective complements showed that no acetylation occurred in the ΔackA mutant, which synthesizes neither AcP nor AcCoA. Increased acetylation was observed in the ackA complemented strain, while the Δpta complemented strain had acetylation levels similar to the wild type. Lysine hyper-acetylation was found in the Δpta mutant due to AcP accumulation. Together, these results demonstrate that AcP is the primary source of acetylation (Bontemps-Gallo et al., 2018) (Table 2).
To establish the metabolic processes that acetylation by AcP regulates, the modified proteins can be analyzed with different software (PANTHER, DAVID, ERGO, and KEEG). Functional analysis of the E. coli proteome revealed that elongation factors, most of the ribosomal subunits, and aminoacyl-tRNA ligases are acetylated in an AcP-dependent manner (Kuhn et al., 2014; Post et al., 2017; Christensen et al., 2019b). In B. burgdorferi, the acetylated proteins were involved in genetic information, metabolism and transport, protein folding and degradation, detoxification, motility, and chemotaxis (Bontemps-Gallo et al., 2018). Similarly, some metabolic pathways for carbohydrates (glycolysis/gluconeogenesis, pentose phosphate pathway, pyruvate metabolism, and the TCA cycle), fatty acid, and pantothenate metabolism are sensitive to AcP-dependent acetylation (Kuhn et al., 2014; Schilling et al., 2015).
As mentioned, non-enzymatic acetylation is favored by the physicochemical conditions in mitochondria, where the basic pH causes lysine to be deprotonated, which increases its nucleophilicity (Figure 2). However, the cytoplasmatic pH in bacteria is maintained at near neutrality: the cytoplasmic pH in neutrophils is ~7.5–7.7, alkaliphiles maintain a basic internal pH of 7.5–8.3, and the pH of acidophilic bacteria is close to 6.5 (Slonczewski et al., 2009; Krulwich et al., 2011). Under these conditions, the epsilon amino groups of most lysine side chains are protonated due to their high pKa and are not very reactive.
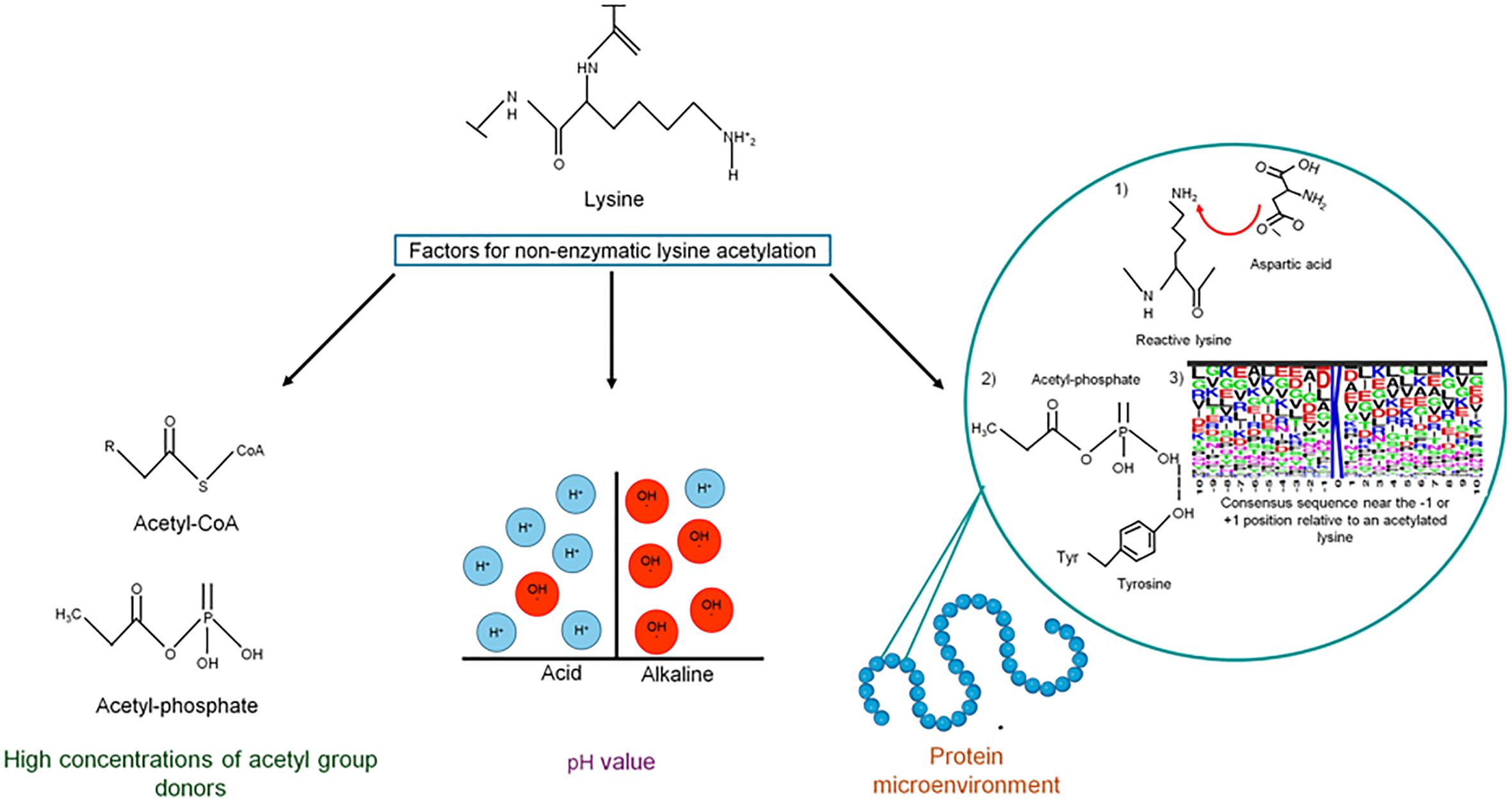
Figure 2 Factors contributing to non-enzymatic acetylation. Acetyl-CoA and acetyl-phosphate are very reactive molecules that, when increasing intracellular concentrations, can non-enzymatically acetylate lysine residues on many proteins. The pH value also plays an essential role in chemical acetylation since, at basic pH, deprotonation of the lysine side chain is favored, increasing its nucleophilicity. In addition to these factors, the efficiency of chemical acetylation also depends on the microenvironment of the protein.
For chemical acetylation, lysine reactivity depends on the protein’s microenvironment. First, the presence of negatively charged amino acids (i.e., Asp or Glu) or water molecules deprotonates the reactive lysine. Second, residues with positive charges (Lys or Arg), hydroxyls (Ser, Thr, or Tyr), or amide groups (Gln or Asn) coordinate AcP binding through ionic and hydrogen bonds, which allows the nucleophilic attack of the active lysine on the phosphoryl group of AcP (Hebert et al., 2013; Wagner and Payne, 2013; Kuhn et al., 2014). A tendency to have negatively charged glutamate (E) and/or aspartate (D) residues near the -1 or +1 position relative to an acetylated lysine has been observed, which reduces the pKa of lysine and enhances its reactivity (Kuhn et al., 2014; Post et al., 2017; Christensen et al., 2019b). If the local pH around a lysine is basic, the epsilon amino group can act as a nucleophile toward AcP (Christensen et al., 2019b).
Enzymatic or non-enzymatic lysine acetylation can be reversed by lysine deacetylase (KDAC), also known as histone deacetylases (HDAC), since histones were the first identified substrate of these enzymes (Inoue and Fujimoto, 1969; Starai et al., 2002; Gardner et al., 2006). There are two types of KDAs: Zn2+-dependent deacetylases and NAD+-dependent sirtuins (Gregoretti et al., 2004; Lombardi et al., 2011; Hentchel and Escalante-Semerena, 2015). Bacteria encode putative homologs of both families, but not all function as true deacetylases (Hildmann et al., 2007; Hentchel and Escalante-Semerena, 2015; Christensen et al., 2019b). The first metal-dependent and NAD-dependent deacetylases were described in Bacillus subtilis and Salmonella enterica serovar Typhimurium, respectively. In both organisms, the acetylation/deacetylation system regulated the activity of the central metabolic enzyme, acetyl-coenzyme A synthetase (Acs) (Tsang and Escalante-Semerena, 1998; Starai et al., 2002, Starai et al., 2004; Gardner et al., 2006).
Sirtuins, also known as Sir2 proteins (silent information regulator 2), have a conserved catalytic core domain characterized by its requirement for nicotine adenine dinucleotide (NAD) to catalyze protein deacetylation by transferring the acetyl group from the lysine to NAD+, resulting in deacetylated lysine, nicotinamide (NAM) and 2′-O-acetyl-ADP- ribose (Figure 3A) (Rine and Herskowitz, 1987). These proteins are evolutionarily conserved within the domains of bacteria, archaea, and eukaryotes (Avalos et al., 2004; Hawse et al., 2008).
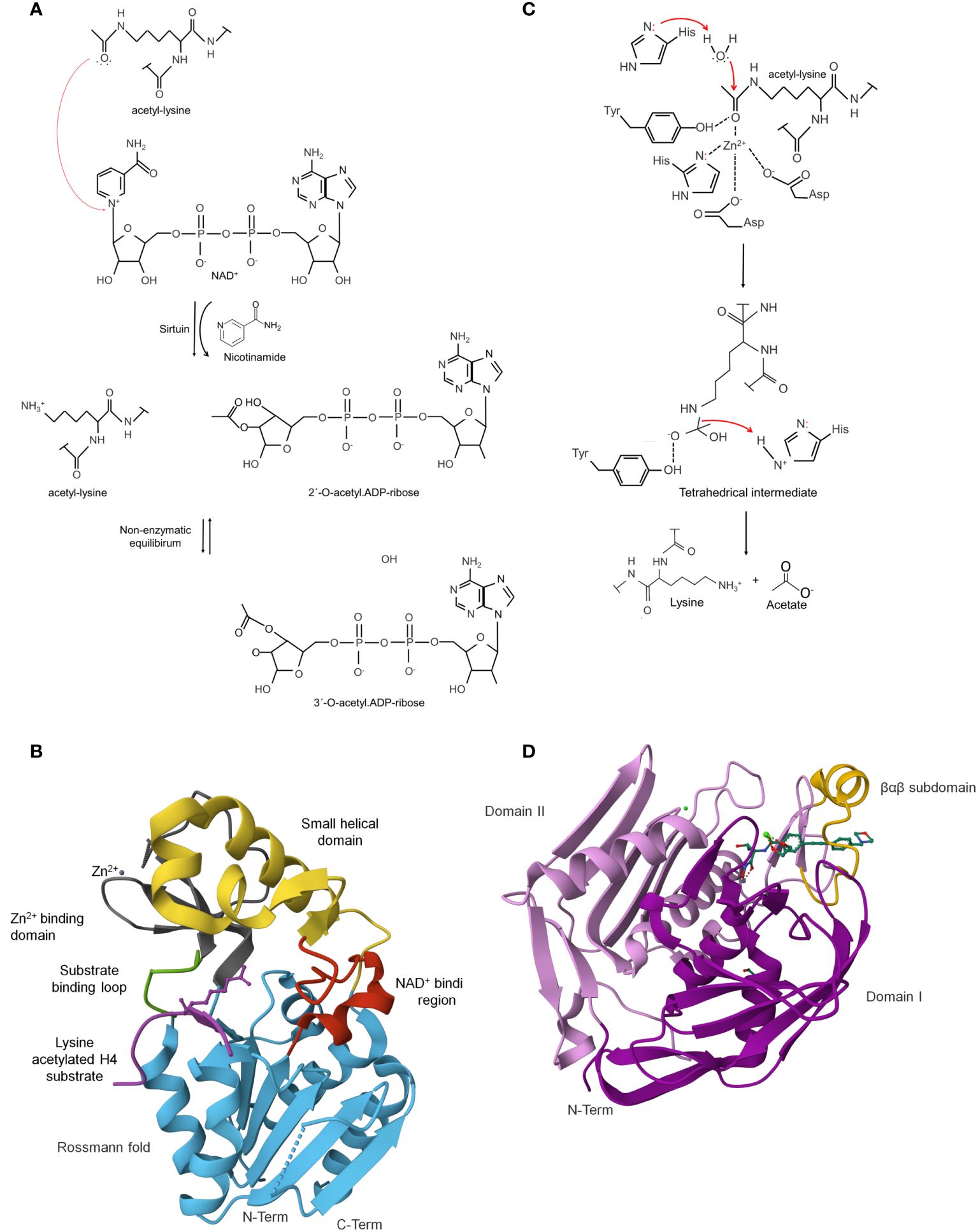
Figure 3 Lysine deacetylases (KDAC) can reverse enzymatic or non-enzymatic lysine acetylation. Catalytic mechanisms used by sirtuins: (A) Sirtuins are NAD+-dependent lysine deacetylases as a co-substrate for catalysis. In the reaction, the carbonyl-oxygen of the acetyl group of lysine performs a nucleophilic attack on the electrophilic C-1’ of the NAD+ ribose, resulting in the fast release of nicotinamide and the formation of a C-1´-O-alkylamidate intermediate. The intermediate is hydrolyzed, forming the deacetylated lysine and 2´-O-acetyl-ADP-ribose in a non-enzymatic equilibrium with 3´-O-acetyl-ADP ribose. Sirtuins are NAD+-dependent lysine-deacetylases (B). Crystal structure ofE. coli sirtuin deacetylase CobB in a complex with a lysine-acetylated substrate (PDB: 1S5P). The characteristic secondary structures of these enzymes are shown in different colors (Zhao et al., 2004) [the figure was generated with PyMOL v.2.3.4 (DeLano, 2002)]. Catalytic mechanisms used by classical deacetylases (C). HDACs use a catalytic water molecule coordinated and polarized by the catalytic Zn2+ ion. The ion, together with a histidine residue, interacts with the carbonyl oxygen of the acetyl group. The histidine is polarized and oriented by an aspartic residue (Asp). It acts as a general base to deprotonate the water molecule, thereby increasing the nucleophilicity for attacking the carbonyl carbon of the acetyl group. A second histidine, again polarized and oriented by another Asp, acts as electrostatic catalyst. A tetrahedral oxyanion intermediate is formed, which is stabilized by a histidine and the Zn2+ ion, to finally release acetate and the deacetylated lysine [figure redrawn and modified from the decomposition of the oxyanionic tetrahedral intermediate [figure redrawn and modified from Ali et al., 2018; Blasl et al., 2021; Lammers, 2021]. Zinc-dependent classical deacetylase (HDAC/KDAC). are metalloenzymes. (D) Crystal structure of Pseudomonas aeruginosa zinc-dependent deacetylase LpxC in a complex with the potent BB-78485 inhibitor (PDB: 2ves). LpxC domain consists of two homologous domains, I (colored in magenta) and II (colored in purple) (Mochalkin et al., 2008) [the figure was generated with PyMOL v.2.3.4 (DeLano, 2002).
Genomes of bacteria and most archaea encode one or two sirtuins, whereas eukaryotes typically contain multiple sirtuins. Lammers (2021) suggests that the low number of sirtuins in bacteria allows them to control specific physiological processes where they can present a very narrow substrate range or may have a high degree of promiscuity in substrate recognition. An enzyme with high substrate promiscuity may also have specificity for certain substrates because they are processed more efficiently due to the region within the cell where the reaction occurs or by transcriptional regulation of their expression levels (Lammers, 2021).
The alignment of sirtuin primary sequences shows a highly conserved catalytic core, while the N- and C-terminal regions are variable in length and sequence (Yuan and Marmorstein, 2012).
The catalytic core adopts an elongated shape containing a conserved large Rossmann-fold domain and a smaller and more structurally diverse domain for acyl peptide and NAD+ binding. These domains are connected by a series of loops that contribute to forming a cleft between the large and small domains (North and Verdin, 2004; Zhao et al., 2004; Sanders et al., 2010).
The large domain comprises an inverted prototypical open α/β Rossmann fold structure, widely distributed in proteins that bind to NAD in its oxidized (NAD+) or reduced (NADH) form. This domain comprises six parallel β strands forming a central β sheet packed between several α helices. The exact number of α helices depends on the protein. For example, E. coli sirtuin CobB contains eight α helices. Also, a conserved Gly-X-Gly sequence important for phosphate binding, a pocket to accommodate an NAD+ molecule, and charged residues responsible for ribose group binding are found (Figure 3B) (Sanders et al., 2010).
The structural Zn2+-binding domain is composed of three antiparallel β-strands and a variable α helical region. A zinc ion is generally bound to four conserved lysine residues in the β-sheet module in a tetrahedral conformation, except for CobB, where it is linked to two cysteine residues and contains three of the four expected zinc-coordinating cysteine residues according to sequence alignment (Sanders et al., 2010; Yuan and Marmorstein, 2012). The zinc ion does not participate directly in the deacetylation but has an essential structural role in the integrity of the catalytic core domain (Figure 3B) (Blander and Guarente, 2004; Yuan and Marmorstein, 2012).
A binding site located between the sirtuin large and small domains, linked to each other by two flexible loops (L1 and L2), forms a cleft that acts as the enzyme’s active site. NAD+ and acetyl-lysine substrate bind at this cleft (Yuan and Marmorstein, 2012). The binding region is divided into three spatially distinct sites: site A, for an adenine-ribose moiety of NAD binding; Site B, for the nicotinamide-ribose moiety binding; and Site C site is located deep in the NAD-binding pocket for nicotinamide moiety binding (Figure 3B) (Sanders et al., 2010).
Metal-dependent HDACs are hydrolases that cleave the acetyl group from lysine to yield free lysine and acetate (Figure 3C). They belong to many proteins, including acetylpolyamine amidohydrolases, acetoin utilization proteins, and histone deacetylases (Hernick and Fierke, 2006).
These enzymes are classified in eukaryotic organisms based on their homology, domain organization, and cellular localization with respect to yeast deacetylases. Class I HDACs are closely related to the transcriptional regulator RPD3 of Saccharomyces cerevisiae, with a length of 350–500 amino acids. They are principally localized in the nucleus and have a variable C-terminus with nuclear import and export signals. Class II HDACs are about 1,000 amino acids long, with a catalytic domain containing several conserved sequence motifs. These enzymes are primarily localized in the cytoplasm, have unique binding sites at their N-termini to control translocation of the protein in and out of the nucleus in response to specific cellular signals, and thus are at least in part cytoplasmic and, in some cases, act on non-histone protein substrates (Ruijter et al., 2003; Hildmann et al., 2006; Wagner et al., 2013). Class IV contains only a single enzyme localized to the nucleus.
In bacteria, some proteins have been identified as members of the family of zinc-dependent deacetylases. For example, the LpxC in gram-negative bacteria (E. coli, Aquifex aeolicus, and Pseudomonas aeruginosa) (Jackman et al., 2000; Whittington et al., 2003; Hernick and Fierke, 2006);, PA3774 of Pseudomonas aeruginosa (Majorek et al., 2013), AcuC of Bacillus subtilis and Aeromonas hydrophila (Gardner et al., 2006; Jiang et al., 2017), FB188 HDAH (histone deacetylase-like amidohydrolase) from Bordetella/Alcaligenes strain FB188 (Hildmann et al., 2004; Nielsen et al., 2005) and LdaA of Rhodopseudomonas palustris (Crosby et al., 2010).
The HDAC structure is characterized by an α/β fold topology (Yang and Seto, 2008; Hentchel and Escalante-Semerena, 2015). In some family members, crystallography shows that one or two domains form the tertiary structure. For example, in the LpxC proteins, the two domains have an identical topology of secondary structural elements that includes a five-stranded parallel β-sheet and two principal α-helices connected by a 16-residue linker (Whittington et al., 2003). In contrast, the HDAC8 comprises a single domain consisting of an eight-stranded parallel β-sheet sandwiched between 13 α-helices (Figure 3D) (Somoza et al., 2004). Interestingly, the crystal structure of PA3774 from the human pathogen P. aeruginosa shares a high degree of homology with class IIb HDACs and consists of two dimers that are close to each other, forming a tetramer, which may be essential for substrate recognition and selectivity (Kraümer et al., 2016). Despite these differences, the structural comparison shows that the structural difference is mainly restricted to the loop regions.
The catalytic center contains a zinc ion commonly pentacoordinated by two aspartic acids, a histidine, and a water molecule. In addition to the zinc ligands, two histidine, two aspartic acids, and one tyrosine form hydrogen bonds with bound ligands. Mutation of any of these residues completely abolishes activity (Kraümer et al., 2016). Furthermore, the surface of this site reveals the formation of a narrow pocket that probably serves to accommodate the acetylated lysine during the catalytic reaction (Vannini et al., 2004). The reaction requires that a conserved histidine residue acting as a general base to activate a metal-bound water that attacks the carbonyl of the acetyl group (Figure 3D) (Finnin et al., 1999; Finnin, 2005; Hernick and Fierke, 2006).
To survive and compete in their natural habitats, bacteria must be able to respond to environmental disturbances or nutrient fluctuations by adapting their metabolism. A quick response mechanism is regulated by the activity of metabolic enzymes, which can be controlled by the amount of enzyme, the catalytic activity, and the substrate accessibility (Figure 4) (Xiong and Guan, 2012). Reversible post-translational modification, such as lysine acetylation, may be involved in at least one of these aspects.
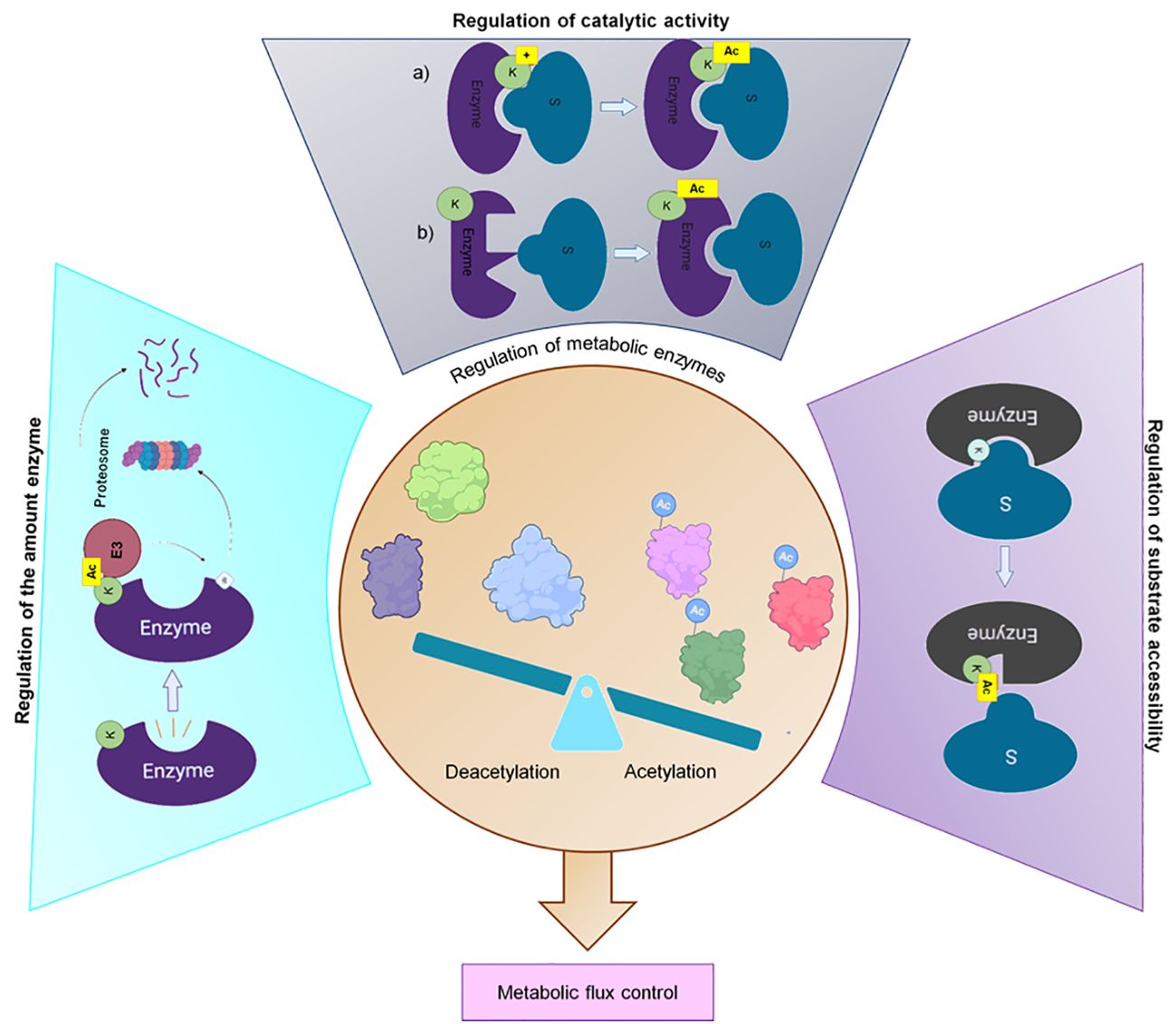
Figure 4 Regulation of the metabolic enzyme activity. Acetylation regulates 1) The number of metabolic enzymes by promoting their degradation through the ubiquitin–proteasomal system, 2) The catalytic activity through a) neutralizing the positive charge of lysine residues in the active site or b) causing allosteric changes and 3) The substrate accessibility to metabolic enzymes by modifying the conserved lysine residues to hinder the entry of substrate (Xiong and Guan, 2012; Liu et al., 2021). Created with BioRender.com.
Acetylation regulates the amount of an enzyme by promoting the assembly of functional multimeric structures that bind to the proteasome for the degradation of proteins or by targeting the substrate for ubiquitylation and proteasome-dependent degradation (Xiong and Guan, 2012; Liu et al., 2021). The enzymatic activity regulation is related to changes in the physical-chemical properties of the catalytic pocket, which could inhibit substrate accessibility (Kim et al., 2006; Nakayasu et al., 2017). In this regard, Nakayasu and co-workers demonstrated that some glycolytic and TCA cycle enzymes are acetylated on highly conserved lysine residues located within the catalytic pocket of the enzyme. This modification dramatically alters the charge and shape of the lysine residue by neutralizing its positive charge and increasing its size, which inhibits enzymatic activity (Nakayasu et al., 2017).
In bacteria, the role of N-epsilon lysine in regulating enzymatic activity was first reported in the Acetyl-CoA synthetase (Acs) of S. enterica (Starai et al., 2002). In vivo and in vitro assays showed that the acetyltransferase Pat interacts with leucine 641 of Acs, leading to acetylation of lysine-609, decreasing Acs’s activity—incubation of the acetylated enzyme with NAD+-dependent deacetylase CobB results in its activation. A Leu-641 Acs mutant showed that position 641 has a structural contribution that allows the interaction of Acs with the protein acetyltransferase (Pat) (Starai et al., 2002; Starai and Escalante-Semerena, 2004b; Starai et al., 2004). In other bacteria (E. coli, B. subtilis, Saccharopolyspora erythraea, Rhodopseudomonas palustris, M. smegmatis and Staphylococcus aureus) similar acetylation-dependent regulatory modes of Acs have been described (Gardner et al., 2006; Xu et al., 2011; Crosby et al., 2012; Kim et al., 2013b; You et al., 2014; Castaño-Cerezo et al., 2015; Burckhardt et al., 2019). Acetylation also negatively regulates the isocitrate dehydrogenase (ICDH) activity from Mycobacterium tuberculosis. The enzyme is acetylated in two lysine residues by Rv2170, reducing the enzymatic activity to around 30% (Lee et al., 2017).
As demonstrated by Venkat et al. (2017a), protein acetylation can also increase enzyme activity. The enzymatic activity of different acetylated variants of malate dehydrogenase (MDH) obtained by an expansion of the genetic code strategy showed that only the acetylation at positions K99 and K140 and the MDH acetylated at both positions increased the enzyme activity. In other variants, no effect was observed. The authors also demonstrated that MDH acetylation can occur either enzymatically or non-enzymatically and that the level of MDH acetylation increases in a glucose-dependent manner (Venkat et al., 2017a). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an essential enzyme in glycolysis and gluconeogenesis, catalyzes the oxidation of D-glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate with NAD+ reduction to NADH. In E. coli, acetylation positively regulates the enzyme; incubation of purified GAPDH with the acetyltransferase PatZ led to a two-fold increase in its enzymatic activity (Plekhanova et al., 2023).
In other proteins, the effect on activity depends on the lysine residue that is acetylated. For example, the enzyme assay and kinetic analysis of different acetylated variants of E. coli citrate synthase (CS) showed that lysine acetylation could decrease the overall CS enzyme activity, mainly due to the acetylation of K295, which impaired the binding of acetyl-coenzyme A. However, acetylation at K283 increased the enzymatic activity since the binding of acetyl-coenzyme A is promoted (Venkat et al., 2019). A similar result was observed in the isocitrate dehydrogenase (ICDH) (Venkat et al., 2018a). Acetylation at K55 or K350 of ICDH significantly increased the activity, while acetylation at K235 caused a loss of about 40% of the activity, and acetylation at K100 or K230 almost eliminated activity (Venkat et al., 2018a). For lactate dehydrogenase (LdhA), acetylation levels for some lysine residues correlate with enzymatic activities. Residues K154 and K248 present low acetylation levels and a decrease of approximately 30% in enzymatic activity. However, residue K9 also showed the same decrease in the acetylation levels, but its enzymatic activity increased 2.5-fold. According to the 3D-modelled structure of LdhA, K154 may be involved in binding the substrate and cofactor, while K248 and K9 are not the catalytic key sites, but both help stabilize the LdhA conformation (Liu et al., 2022).
Reversible lysine acetylation (RLA) has been demonstrated for some of these enzymes. In Salmonella enterica, Acs’s activity can be restored by the NAD+-dependent CobB sirtuin deacetylase, which removes the acetyl group from acetylated Acs (Starai et al., 2002; Starai and Escalante-Semerena, 2004b; Starai et al., 2004, Starai et al., 2005). For B. subtilis, Acs’s activity is restored by two deacetylases, AcuC and SrtN; the first does not require NAD+, while the second is a class III NAD+-dependent sirtuin deacetylase (Gardner et al., 2006; Gardner and Escalante-Semerena, 2009). This suggests that the regulatory mechanisms of Acs activity in these two bacteria differ. For both bacteria, inactivation of the enzyme by acetylation produced a growth defect on acetate, which can be restored by deacetylation of the enzyme. However, B. subtilis could respond to different cell energy charge indicators. On the one hand, the activation by AcuC may be triggered in response to a low AcCoA: CoA ratio. On the other hand, if the NAD+:NAD ratio H becomes high, the SrtN-dependent is activated. In Salmonella enterica, the mechanism seems to be more straightforward. Acs synthesizes AcCoA in two steps, the first in which the acetate is adenylated and a subsequent thioester formation step. In response to high levels of AMP due to the uncontrolled activity of the enzyme, acetylation of K609 blocks the adenylation reaction and stops the synthesis of acyl-AMP intermediates without altering the thioester-forming activity (Starai et al., 2002; Starai and Escalante-Semerena, 2004a; Starai et al., 2004, Starai et al., 2005; VanDrisse and Escalante-Semerena, 2019). The tight regulation of the enzyme by the acetylation/deacetylation mechanism occurs because high levels of AMP in the cell indirectly lead to lower levels of ATP synthesis (VanDrisse and Escalante-Semerena, 2019). In addition, the cell could use this mechanism to maintain the acetate pool and prevent unnecessary ATP hydrolysis (Starai et al., 2003; Chan et al., 2011). We highly recommend reading the article of Hentchel and Escalante-Semerena, which discusses the RLA in gram-negative and gram-positive bacteria in depth.
It has also been shown that CobB sirtuin has site-specificities. Most of the CobB-sensitive acetylated lysine sites are located on the surface of the protein, while CobB-resistant sites are in regions of the protein where the enzyme does not have easy access. For example, some acetylated lysines are located at the dimer interface or are deeply buried near the active site, limiting the access of CobB (Fatema and Fan, 2023).
These studies prove that acetylation modulates the activity of central metabolic enzymes and eventually alters protein function to control competing pathways in response to physiological needs.
Comparative studies with different carbon sources have explored the effect of protein acetylation during growth under glycolytic and oxidative conditions. As Wang and collaborators demonstrated, S. enterica presents changes in cell growth and significant differences in the percentage of acetylated proteins in response to glucose or citrate. A total of 15 enzymes were identified with altered acetylation status in response to a carbon source, and all of them showed higher acetylation levels in cells grown in glucose (Wang et al., 2010). The main targets of lysine acetylation were enzymes involved in central metabolism, and a correlation was observed in the cell growth and the acetylation levels of these proteins. In a medium with glucose, cells grow faster, and an increase in the acetylation of central metabolic enzymes was observed, but when the acetylation decreases, opposite growth properties were observed (Wang et al., 2010). In B. subtilis, different acetylation levels have also been reported in different carbon sources. In decreasing order, protein acetylation occurs in a medium containing only carbon source pyruvate, glucose, glycerol, or citrate (Kosono et al., 2015). The main differences in the acyl modification patterns were observed in glucose and citrate. Under these two conditions, changes in the acetylation sites were determined. Acetylation was upregulated at 13 sites in glucose relative to citrate, and, as in S. enterica, acetylation positively modulated the growth of this bacterium in a glucose medium (Kosono et al., 2015). Western immunoblot analysis found that glucose and lactate produce global acetylation in E. coli K-12, which increased after the cells had entered the stationary phase. Induction of acetylation at this growth point requires continued exposure to carbon sources, specifically glucose (Schilling et al., 2015). The results suggest that protein acetylation may have a physiological role in mediating adaptation to different carbon sources.
Another factor that could determine lysine acetylation levels is the sugar concentration. In E. coli, the increase in acetylation levels occurs during growth at high concentrations of glucose or xylose (4% for both carbon sources), compared to low concentrations of 0.4%. No changes in the profiles associated with the type of carbon source were observed, suggesting that the sensitivity of lysines to acetylation depends on the amount of sugar available in the medium. Furthermore, it was found that significantly more acetate is produced under these conditions, and E. coli cannot consume it. The results support the hypothesis that most acetylation results from acetate overflow metabolism and is independent of the specific catabolic route (Schilling et al., 2015, Schilling et al., 2019).
The concept of metabolic overflow refers to a phenomenon that occurs during growth at a high concentration of carbon source or during the transition from a limited substrate condition to a rich one. Under these conditions, cells exhibit a metabolic shift characterized by the secretion of many primary metabolites, such as acetate, ethanol, or lactate, even in the presence of oxygen. This occurs when the rate of the substrate (e.g., glucose) uptake exceeds the capacity of the cell´s respiratory machinery to fully oxidize it via the TCA cycle and oxidative phosphorylation (Pinu et al., 2018).
Metabolic overflow has been studied in the model microorganisms E. coli and S. cerevisiae. In E. coli, acetate overflow is observed when cells are grown aerobically in high glucose concentrations (Paczia et al., 2012; Pinu et al., 2018). Under these conditions, there is an imbalance between the fluxes of glucose uptake, energy production, and carbon conversion into biomass and products, causing the excretion of significant amounts of acetate (Bernal et al., 2016). The excreted acetate can be used after the depletion of the primary carbon source by the acetyl-CoA synthetase (Acs), which converts acetate to acetyl-coenzyme A (Ac-CoA), which is mainly metabolized by the glyoxylate shunt and the TCA cycle (Starai and Escalante-Semerena, 2004a; Castaño-Cerezo et al., 2015). Acs is tightly controlled at the transcriptional level, and its activity, as mentioned, is posttranslationally regulated by protein acetyltransferase.
The role of the regulation of Acs by the acetylation/deacetylation mechanisms in acetate metabolism was first described in Salmonella enterica, where it was observed that acetyltransferase Pat acetylates Acs inhibiting its activity and the sirtuin deacetylase CobB reactivate the Acs enzyme, restoring growth on acetate (10 mM) as the sole carbon source (Starai et al., 2002, Starai et al., 2003; Starai and Escalante-Semerena, 2004b; Starai et al., 2004, Starai et al., 2005; VanDrisse and Escalante-Semerena, 2019).
In E. coli and Mycobacterium smegmatis, acetate metabolism is also regulated by the acetylation/deacetylation of Acs (Castaño-Cerezo et al., 2011; Hayden et al., 2013; Castaño-Cerezo et al., 2014; Peebo et al., 2014). Studies in E. coli have demonstrated that deleting the NAD+-dependent deacetylase (ΔcobB) strongly affects its physiology when grown in a minimal medium with acetate or glucose as the only carbon source. First, the bacterium is unable to grow at low acetate concentrations (10 mM). Secondly, the mutant strain exhibits a high glucose consumption rate in glucose cultures, leading to more significant acetate accumulation in the medium and slower acetate absorption (Castaño-Cerezo et al., 2011). Under glucose-limited conditions, an increase in acetylation of Acs is observed, with activity decreasing to almost half that of the wild‐type (Castaño-Cerezo et al., 2014). These results support the idea that the inactivation of Acs by acetylation affects the high-affinity Acs pathway for acetate assimilation, which primarily serves to scavenge acetate at low concentrations. Additionally, it has been hypothesized that the regulation of the isocitrate node involves two post‐translational modifications, phosphorylation and acetylation, each operating at distinct levels. AceK is proposed to oversee a broad regulatory mechanism that impedes the flux through the TCA cycle. Meanwhile, acetylation is suggested to contribute to the fine-tuning regulation of the glyoxylate shunt, partially inhibiting its activity. This partially inhibited glyoxylate shunt activity could explain the reduced growth rate observed in the ΔcobB mutant under acetate and glucose-limited conditions (Castaño-Cerezo et al., 2011, Castaño-Cerezo et al., 2014).
Furthermore, acetylation of Acs may be involved in controlling the co-utilization of fermentable substrates. A mutation in Leu-641 makes Acs insensitive to acetylation even at high glucose concentrations (Starai et al., 2005). The overexpression of this protein in E. coli W allows the efficient co-utilization of glucose and acetate. In a batch process containing glucose and high acetate concentrations, a 2.7-fold increase in acetate uptake was observed in comparison to a control strain (Novak et al., 2018).
Most microorganisms have developed different strategies to sequentially metabolize a mixture of simple carbohydrates, favoring the utilization of glucose as a carbon and energy source to sustain a higher growth rate. The metabolic plasticity allows them to obtain specific carbon sources efficiently and survive in competitive environments (Vinuselvi et al., 2012). Most bacteria regulate their metabolism via carbon catabolite repression (CCR), which involves a complex interplay between metabolism, signaling by proteins and metabolites, and the regulation of gene expression (Kremling et al., 2015). Alternative splicing, mRNA stability, translation, and protein degradation control of enzymes are other mechanisms that can be used to modulate carbon flux. These coarse regulations usually respond to long-term environmental changes (Wegner et al., 2015).
Post-translational modifications affect flux distribution between important metabolic branches, such as glycolysis and gluconeogenesis, TCA cycle and glyoxylate shunt, and glycolysis and TCA cycle. Wang et al. (2010) demonstrated that carbon source-associated acetylation modulates metabolic flux profiles in S. enterica. In the presence of glucose, acetylation increases the glycolysis/gluconeogenesis flux ratio 2.07-fold, while acetylation reduces the glyoxylate bypass/TCA flux ratio under a citrate-based carbon source.
The isocitrate node is a vital regulation point of carbon flux between the TCA cycle and the glyoxylate shunt. Isocitrate, the substrate of isocitrate dehydrogenase (ICDH) and isocitrate lyase (AceA), is converted to α-ketoglutarate by ICDH or is cleaved to succinate and glyoxylate by AceA, directing the carbon source flow to TCA cycle or to glyoxylate shunt, respectively. In the 70s, Holms and Bennett, (1971) were the first to report the inactivation of ICDH at the end of the growth of E. coli on glucose, and later, this same behavior was reported when E. coli was grown on acetate as the only carbon source (Bennett and Holms, 1975). The authors also demonstrated that when pyruvate, malate, or succinate were added to cultures growing on acetate, the ICDH activity increased (Bennett and Holms, 1975). The inhibition of ICDH enzymatic activity was attributed to phosphorylation, which decreases isocitrate flux to the TCA cycle and forces it through the glyoxylate bypass (Garnak and Reeves, 1979). The phosphorylated serine residue at position 113 completely inactivates the enzyme, while dephosphorylation restores the impaired activity (Dean et al., 1989). Although historically, it was considered that phosphorylation was the only way to regulate the enzyme, proteomic studies have demonstrated that metabolic enzymes, including ICDHs, are favorable targets for lysine acetylation (Xu et al., 2017). In E. coli, proteomic studies have identified more than 20 acetylated lysine residues in ICDH (Yu et al., 2008; Zhang et al., 2009; Weinert et al., 2013) and, as mentioned in the regulation of enzymatic activity section, ICDH activity in in vitro assays can be regulated by lysine acetylation. However, it would have to be demonstrated that this phenomenon also occurs in vivo. In Salmonella enterica serovar Typhimurium ICDH is also regulated by phosphorylation (Wang and Koshland, 1982).
In M. tuberculosis, this metabolic node is regulated by the acetylation of the ICDH. Acetylation suppresses enzyme activity in the presence of fatty acids, reducing carbon flow into the TCA cycle (Lee et al., 2017). The activation of glyoxylate bypass allows the conversion of AcCoA to the metabolic intermediate succinate to support the growth in the presence of non-carbohydrate substrates such as fatty acids or acetate (Cronan and Laporte, 2005; Lee et al., 2017).
Recently, it was demonstrated that glyceraldehyde 3-phosphate dehydrogenase (GapA) and 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (GpmA) were sensitive to non-enzymatic acetylation in vitro at physiological AcP concentrations. In both enzymes, acetylation reduced their activity, which could be reflected in reduced glycolytic/gluconeogenic flux in conditions with higher concentrations of AcP (Schastnaya et al., 2023).
The flux from glucose to glutamate is increased when the cell excretes glutamate. Factors like the depletion of biotin and the addition of detergents or antibiotics trigger glutamate overproduction and, therefore, a change in the flux of central carbon metabolism to favor glutamate production (Shirai et al., 2007). It has been proposed that in addition to the decrease in 2-oxoglutarate dehydrogenase complex (ODHC) activity, the regulation of phosphoenolpyruvate carboxylase (PEPC) activity by acetylation may be a mechanism involved in the change in metabolic flux during overproduction of glutamate. PEPC catalyzes the irreversible carboxylation of phosphoenolpyruvate to generate oxaloacetate, ensuring that the carbon flow is directed toward glutamate production via the TCA cycle. Acetylation at the K653 site regulates enzyme activity and is, therefore, the mechanism that maintains metabolic flux under glutamate-producing conditions (Mizuno et al., 2016; Nagano-Shoji et al., 2017).
Hence, acetylation may provide a new strategy for regulating protein activity and improving the utilization of different carbon sources.
Although the use of antibiotics to treat infectious diseases has improved quality of life and reduced mortality, their excessive, unnecessary, and incorrect usage has caused the rapid emergence of antibiotic-resistant strains. It is estimated that antibiotic resistance is a growing problem that accounts for approximately 1.27 million global deaths (Tacconelli et al., 2018; WHO, 2023). Furthermore, this number is expected to increase in the coming years, leading cause of 10 million deaths worldwide by 2050 (Zalewska-Piątek, 2023). Understanding the antibiotic resistance mechanisms in bacteria will allow the development of new antimicrobial agents with novel mechanisms of action or new strategies for innovative non-antibiotic therapeutic methods.
Bacteria have developed sophisticated molecular strategies to counteract the effect of antibiotics; for example, antibiotic sequestration by specific proteins can prevent them from reaching their targets. Increased efflux pump activity to extrude the antibiotic or decreased influx to avoid antibiotic entry reduces the effective concentration of antibiotics and, therefore, decreases their efficiency (Luther et al., 2018). Finally, bacteria can use enzymatic strategies to inactivate antibiotics. In this mechanism, the antibiotic can be degraded by the action of β-lactamases, macrolide esterases, and epoxidases or can be covalently modified. These chemical modifications include O-phosphorylation, O-nucleotidylation, O-ribosylation, O-glycosylation, O-acetylation, and N-acetylation (Wright, 2005).
Aminoglycosides are a large family of antibiotics with broad-spectrum used to treat Gram-negative and Gram-positive bacterial infections (Magalhaes and Blanchard, 2005). Aminoglycosides inhibit bacterial protein synthesis by binding to the aminoacyl-tRNA site (A-site) of 16S rRNA, resulting in an erroneous reading and/or hindering the translocation step (Magnet and Blanchard, 2005; Barrett et al., 2008; Vetting et al., 2008). The emergence of a growing number of resistant strains has limited its use. One of the principal causes of aminoglycoside resistance is related to its enzymatic modification by aminoglycoside N-acetyltransferases (AACs), proteins members of the GNAT superfamily.
Genes encoding aminoglycoside N-acetyltransferases have been described in pathogenic bacteria. AACs catalyze the acetyl-CoA-dependent N-acetylation of the amino group of aminoglycosides, inhibiting their interaction with the 16S rRNA, resulting in microbial resistance and complications in the clinical treatment (Teran et al., 1991; Shaw et al., 1992; Costa et al., 1993; Magnet et al., 1999; Vetting et al., 2004; Magnet and Blanchard, 2005). ACCs selectively transfer the acetyl group to one of the four amino groups of the typical aminoglycoside structure. According to this regiospecificity, these enzymes are grouped into four major classes: AAC(1), AAC(3), AAC(2′), and AAC(6′) (Draker and Wright, 2004; Magalhaes and Blanchard, 2005; Magnet and Blanchard, 2005; Wright, 2005). However, not all the AACs are related to aminoglycoside resistance; the decreased binding of the aminoglycoside to the A-site is only associated with the acetylation at the position three′ of the 2-deoxystreptamine ring by AAC(3) or the acetylation at position 6′of the amino hexose by AAC(6′) of the aminoglycoside (Magnet and Blanchard, 2005; Barrett et al., 2008).
Interestingly, some clinical isolates of Gram-negative bacteria contain a variant of an aminoglycoside-modifying enzyme, the aminoglycoside acetyltransferase AAC(6′)-Ib, which has two amino acid substitutions, Trp102Arg and Asp179Tyr, that are not found in other variants (Park et al., 2006; Robicsek et al., 2006; Jung et al., 2009; Kim et al., 2013a). In particular, the Asp179Tyr mutation allows the enzyme to acetylate fluoroquinolones, representing an alternative resistance mechanism for this synthetic drug (Vetting et al., 2008).
Recently, it has been found that lysine acetylation of proteins involved in essential metabolic processes allows the bacteria to respond to antibiotic stress, which can significantly affect the bactericidal effect of antibiotics. An acetylome analysis of E. coli strains resistant to three types of antibiotics (ampicillin, kanamycin, or polymyxin B) showed a common regulatory mechanism. Acetylation positively regulates bacterial motility by reducing the acetylation of flagellar proteins. It negatively regulates energy metabolism by increasing the acetylation levels of central metabolic proteins. This reduces the metabolic level of bacteria to regulate antibiotic resistance. Lower levels of acetylation cause downregulation of the ribosome, RNA degradation, signal transduction, and protein export, which could inhibit signal transduction and prevent the antibiotic from binding to the target protein (Fang et al., 2022). Also, the KEGG analysis of lysine acetylation shows enrichment of this post-translational modification in different metabolic processes such as the TCA cycle, pyruvate metabolism, and glycolysis (Fang et al., 2022). Similar results have been found in other pathogenic organisms, demonstrating that central metabolic pathways regulate bacterial antibiotic susceptibility (Peng et al., 2015; Xie et al., 2016; Pang et al., 2020a). However, more studies are necessary to elucidate the correlation between acetylation and antibiotic resistance.
The other way acetylation controls antibiotic response is related to virulence factors. In Salmonella Typhimurium, deleting the acetyltransferase pat reduced the pathogenicity island 1 (SPI-1) expression, affecting its ability to invade HeLa cells and intramacrophage replication. This mutant has attenuated virulence compared to the mouse model’s wild type. It has been proposed that HilD, a critical transcriptional regulator of SPI-1, could mediate the acetylation-related virulence in Salmonella. The acetylation stabilizes this transcriptional regulator and promotes the transcription of HilA, leading to the activation of SPI-1 (Sang et al., 2016). The first Salmonella acetylome study identified HilD as an acetylated protein (Wang et al., 2010). In contrast, when HilD is deacetylated, it tends to be degraded and does not activate HilA transcription, lowering SPI-1 expression and leading to attenuated virulence of the bacteria (Sang et al., 2016; Koo et al., 2020).
Another vital virulence factor in Salmonella is the PhoP/PhoQ two-component system, which regulates virulence in response to various signals within the mammalian host. PhoP is acetylated and deacetylated in vitro and in vivo by Pat and CobB, respectively. Specifically, when the conserved lysine residue 201(K201) located in the C-terminal DNA-binding domain of PhoP is acetylated, its ability to bind to DNA is inhibited, altering the transcription of phoP and PhoP-regulated genes. This enzymatic acetylation impairs bacterial intracellular replication and inflammation response in macrophages and significantly attenuates the bacteria virulence, suggesting that non-acetylated PhoP is essential for Salmonella pathogenesis. With these results, the authors propose that regulating transcriptional factors by acetylation of the DNA binding domain may be a new regulatory mechanism of gene expression involved in bacterial virulence (Ren et al., 2016, Ren et al., 2017).
The global acetylome analysis of different bacteria has allowed the identification of other acetylated virulence factors. In M. tuberculosis, the heat-shock protein X (HspX) linked to the latent infection and the isocitrate lyase involved in the persistence, virulence, and antibiotic resistance are acetylated (Liu et al., 2014; Xie et al., 2015). In Salmonella, two virulence factors (inositol phosphate and the pathogenicity island one effector protein SipC) are acetylated after the bacteria acquire drug resistance (Li et al., 2018). The virulence factors exotoxin A, chitin-binding protein, serine protease, and hemolysin have been characterized as lysine-acetylated proteins in P. aeruginosa (Ouidir et al., 2015). In Vibrio vulnificus and Vibrio alginolyticus, several virulence factors are found to be acetylated (Pang et al., 2020a, Pang et al., 2020b). For E. amylovora, the proteins exopolysaccharide amylovoran biosynthesis- and type III secretion-associated proteins involved in virulence are lysine-acetylated (Zhao et al., 2009; Wu et al., 2013). In P. aeruginosa, some virulence factors (exotoxin A, chitin-binding protein, serine protease, and hemolysin) have been characterized as lysine-acetylated proteins (Ouidir et al., 2015). These data suggest that acetylation may play a regulatory role in the virulence of these bacteria.
Biofilms are bacterial multicellular communities composed of polysaccharides and proteins in which individual cells stick together and are encapsulated in a beneficial and protective environment for the microbial community (Joo and Otto, 2012). Biofilm formation is often triggered in response to environmental stresses, representing an important adaptation strategy. However, when the associated microorganisms are pathogenic, this ability becomes a significant virulence factor, protecting the bacteria from host immune responses and antibiotics (Donlan, 2000; Høiby et al., 2011).
Dental caries is a disease that occurs due to a combination of factors, such as diet, poor dental cleaning, and biofilm formation. Cariogenic and commensal microorganisms form biofilm on tooth surfaces. The factor lays the foundation for the pathogenesis of dental caries, as they provide a platform for pathogenic bacteria to colonize and accumulate (Ma et al., 2021). Among the more than 700 different bacterial species identified in dental biofilm, Streptococcus mutans is considered the principal etiologic agent of dental caries (Kang et al., 2011), with essential virulence factors, such as acidogenicity, aciduricity, and biofilm formation (Daboor et al., 2015; Ma et al., 2021).
The extracellular biofilm matrix is mainly managed by three kinds of glucosyltransferases (Gtfs), which synthesize extracellular polysaccharides (EPS) from sucrose and promote the adhesion of S. mutans to tooth surfaces (Selwitz et al., 2007). Compared with S. mutans in planktonic conditions, the quantitative proteome analysis revealed that the lysine acetylation levels of three Gtfs (glucosyltransferase-SI, glucosyltransferase-I, and glucosyltransferase-S) decreased in S. mutans in biofilm conditions (Lei et al., 2021). To determine the role of this modification, Ma and coworkers (2011) induced the overexpression of 15 genes whose products are annotated as GNATs in S. mutans. Only one GNATs, ActG, could enzymatically acetylate two Gtfs, reducing their enzymatic activity and water-insoluble EPS production, biofilm formation, and caries formation in a rat caries model (Ma et al., 2021). The results suggest acetylation could function as a switch-off for regulating glucosyltransferase activities (Lei et al., 2021).
In a study performed to characterize the acetylome of M. tuberculosis, deleting the sirtuin-like gene (rv1151c) results in the hyperacetylation of different proteins. Particularly, reversible acetylation of several fatty-acid-CoA ligases can modulate their activity. The acetylation inhibits their activity and results in a deficient biofilm formation, possibly due to the inhibition of fatty acid metabolic enzymes (Liu et al., 2014). In Neisseria gonorrhoeae, the AcP accumulation probably produces a nonenzymatic acetylation of the essential biofilm regulatory proteins, which leads to a marked defect in the maintenance of the biofilm structure over time. However, further analysis is required to determine which essential biofilm regulatory proteins are acetylated in N. gonorrhoeae (Post et al., 2017).
On the contrary, in Yersinia pestis, the acetylation of Lys 73 of a transcriptional regulator for the MarR-family (SlyA protein), which can regulate the expression of many genes associated with virulence and biofilm formation, inhibits its binding to the promoter of target genes and significantly enhances biofilm formation (Tan et al., 2023). In Aeromonas hydrophila, the S-ribosyl homocysteine lyase (LuxS) involved in the production of the quorum-sensing (QS) auto-inducer 1 (AI-1) type signal molecule, which plays an important role in biofilm formation, is acetylated at K165, and, interestingly, this site is also succinylated. Acetylation of K165 inhibits enzymatic activity, while succinylation has the opposite effect (Sun et al., 2019; Li et al., 2020a). In a previous study, the luxS isogenic mutant of A. hydrophila exhibited enhanced biofilm production and virulence in the septicemic mouse model. Therefore, the acetylation of this protein may enhance biofilm formation and virulence (Kozlova et al., 2008; Sun et al., 2019).
Bacteria can colonize different environments since they respond quickly to fluctuating environmental stresses, including changes in temperature, pH, osmolarity, radiation, and the concentration of nutrients and toxins (Aertsen and Michiels, 2004). To ensure their survival from these adversities, bacteria coordinate various mechanisms, such as changes in the expression of the genes for combating harmful stresses, DNA repair mechanisms, elimination of molecules that cause damage (for example, reactive oxygen species), post-translational modification systems, among others (Hecker and Völker, 2001; Aertsen and Michiels, 2004).
For foodborne pathogens, the ability to survive in an extremely acid stomach environment is necessary to invade the intestinal epithelium further. Various regulatory mechanisms can be used to sense and respond to the acidic environment. S. Typhimurium can respond to acid stress by a reversible protein acetylation system. The S. Typhimurium transcriptome analysis under acid stress showed that the acetylation level may decrease through the regulation of pat transcription and the NAD+/NADH ratio of the cell in response to the acid signal. Furthermore, the ability to tolerate acidic conditions in the pat or cobB mutant compared to the WT strain showed that the ΔcobB mutant has a significantly lower survival rate than the other strains, both in the log and stationary phases (Ren et al., 2015). For other pathogens, acetylation is involved in response to oxygen limitation stress. In Mycobacterium tuberculosis (Mtb), hypoxia induces the deacetylation at K182 of DosR (dormancy survival regulon). This regulon may be a critical factor in latency adaptation, abolishing the affinity of DosR for DNA in vitro and further altering the transcription of DosR-regulated genes, leading to the rapid response of Mtb to hypoxia (Bi et al., 2018; Yang et al., 2018).
Lysine acetylation is also involved in oxidative stress protection. When reactive oxygen species exceed the antioxidant and reducing capacities within the cell, it causes damage to biomolecules such as proteins, lipids, and nucleotides (Cabiscol et al., 2000). Sodium hypochlorite (NaOCl) in solution forms hypochlorous acid (HOCl), which is a portent oxidant that causes irreversible protein aggregation by disrupting their structure (McMillan et al., 2018). The exposure to hypochlorite increases the abundance of lysine-acetylated peptides in the H. volcanii and Δsir2 mutant strains. The protein enrichment of the proteins showed significant enrichment of DNA topological change networks, AcCoA biosynthesis, and enzymes involved in translation. In the Δsir2 mutant, a dramatic downregulation of sulfur metabolism, cobalamin synthesis, division septum assembly, and DNA topological change proteins occurred after hypochlorite exposure (Couto-Rodríguez et al., 2023). For E. coli, protein lysine acetylation allows the bacterium to reach higher cell densities and become more resistant to oxidative stress. To survive this stress, some two-component system proteins may need to be acetylated for catalase activity. Interestingly, this study demonstrated that other genes are repressed by deacetylation, including genes for heat shock, osmotic stress, acid resistance, cold shock, carbon starvation, and general stress, indicating that protein acetylation is involved in various bacterial stress response systems (Ma and Wood, 2011).
Under other types of stress, it has been observed that acetylated proteins are related to metabolic enzymes, critical proteins involved in DNA damage repair and cellular processes such as translation, ribosomal structure and biogenesis, amino acid transport and metabolism, and energy conservation and conversion (Zhang et al., 2022; Xiao et al., 2023; Yao et al., 2023). These studies show a global view of the response to different types of stress and the PTM of lysine acetylation.
Protein synthesis (translation) is carried out by a translational machinery composed of ribosomes, translation factors, and aminoacyl-tRNA synthetases. Ribosomes are involved in a highly dynamic and complex process of translating mRNA into proteins. They have a basic structure highly conserved in all cellular organisms, composed of two major subunits: the large ribosomal subunit (LSU) and the small ribosomal subunit (SSU). Each subunit is constituted of ribosomal RNAs (rRNAs) and ribosomal proteins (r-proteins) (Selmer et al., 2006; Ban et al., 2014; Ni et al., 2023). For example, in E. coli, the small ribosomal subunit comprises 16S ribosomal RNA and 24 ribosomal proteins. The large ribosomal subunit comprises a 23S ribosomal RNA and 34 ribosomal proteins (Jeffares et al., 1998).
Although different PTMs have been identified in a growing number of r-proteins and rRNAs, which regulated the fidelity of protein synthesis, including methylation, phosphorylation, and ubiquitylation, among others, the physiological role of translation-related protein acetylation, remains poorly studied (Martin et al., 2011; Sung et al., 2016; Natchiar et al., 2017). Mass spectrometry analysis of different bacterial proteomes has identified many acetylation sites on proteins. The functional annotation showed that some of the acetylated proteins are involved in translation and transcription (Weinert et al., 2013; Nakayasu et al., 2017; Christensen et al., 2019b; Marakasova et al., 2020; Liu et al., 2023).
Recently, it was found that AcP significantly reduced the relative translation rate, and deacetylation partially restored the translation activity. Using the cell-free translation system of E. coli, where the yellow fluorescent protein (YFP) is used as a signal output of translation activity, the YFP synthesis rate was examined after the E. coli extracts were incubated with AcP at different times. The analysis of the extracts by western blot to detect the acetylation level and the YFP synthesis rates of translation machinery showed a time-dependent acetylation pattern of the translation machinery in the presence of AcP and inhibition of translation by reducing the rate of elongation, while deacetylation partially restored it. Both results implied acetylation inhibits protein synthesis in vitro (Zhang et al., 2020a).
Sixteen acetylated sites on 12 proteins were identified by mass spectrometry. In particular, the acetylation of K411 and K464 in domains D5 and D6 of the ribosomal protein S1 reduced the chaperone activity of the protein, suggesting that acetylation may affect S1-mediated recruitment of mRNAs into the ribosome (Zhang et al., 2020a).
However, other studies showed that acetylation plays a more complex role than translational inhibition. Feid and coworkers (2022) reported that adding AcP and AcCoA inhibits translation but not transcription. The elongation rates using a LacZ induction assay were measured to confirm this observation. The results indicated that acetylation does not significantly change the elongation rate but increases the proportions of dissociated 30S and 50S ribosomes. In addition, when the 30S and 50S ribosomal subunits are disassociated, the ribosomal proteins are more acetylated than when the 70S complex is fully assembled. The authors concluded that lysine acetylation interferes with the association of ribosomal subunits, inhibiting translation (Feid et al., 2022).
The same observation was made by analyzing the acetylation of r-proteins of Salmonella enterica serovar Typhimurium (S. Typhimurium). By mass spectrometry, 289 AcK sites were identified in 52 of 54 r-proteins; of these, the small subunit protein S1 contained 19 Kac sites. Most of the acetylated lysine residues of r-proteins were found to be regulated by both the acetyltransferase Pat and the metabolic intermediate AcP. Also, it was found that acetylation plays a critical role in the assembly of the mature 70S ribosome complex by modulating r-proteins binding to rRNA and that appropriate acetylation is necessary for the interaction between elongation factors and polysomes, as well as regulating ribosome translation efficiency and fidelity (Ni et al., 2023). These data show that ribosome acetylation is crucial for their correct assembly and function.
The human gut microbiota is a complex ecosystem composed of bacteria, archaea, viruses, and eukarya. The microbiota interacts with one another and the host in a mutually beneficial relationship, significantly impacting human health and physiology (Clemente et al., 2012). Compositional and functional alterations of the gut microbiome result in dysbiosis, which has been associated with different diseases, including obesity, diabetes, Crohn’s disease (CD), cancer, and cardiovascular diseases (Myers and Hawrelak, 2004; Clemente et al., 2012). Several studies have been carried out to determine which factors (diet, age, pregnancy, antibiotic use, among others) modulate the composition and functionality of the microbiota and how this can induce dysbiosis (Myers and Hawrelak, 2004; Brown et al., 2012; O’Toole and Jeffery, 2015; Edwards et al., 2017; Ramos and Martín, 2021), little is known about whether PTMs modifications have a regulatory effect on enzymatic activities of the microbiota and its impact.
To establish a methodology that allows studying protein acetylation in the microbiome, Zhang et al. (2020) found an experimental and bioinformatic workflow for its characterization. The proteolytic peptides were generated and aliquoted from human fecal samples for metaproteomics and acetylomics analysis. An enrichment process was carried out using a seven-plex anti-Kac peptide antibody cocktail for the lysine acetylome analysis. The metaproteomics/lysine acetylomics data analysis showed that a higher number of acetylated peptides were identified with the enrichment step. In this sample, 31,821 acetylated sites (35,200 acetylated peptides) were identified, of which 31,307 were from microbes, and 497 were of human origin. A deeper analysis revealed that protein acetylation level is much higher in Prokaryotes than human proteins in the microbiome samples and that lysine acetylation is widely distributed in gut microbial metabolic pathways, including anaerobic fermentation to generate short-chain fatty acids.
The methodology was applied to the analyses of the microbiomes of patients with CD. A total of 52 hosts and 136 microbial acetylated sites differentially abundant in disease versus controls were identified. Also, the authors demonstrated that the lysine acetylation levels in gut microbial metabolic pathways to produce short-chain fatty acids (SCFAs) were down-regulated in CD patients (Zhang et al., 2020b).
Considering that a significant challenge in metaproteomics studies is to control the discovery rate, the same group reanalyzed the previous Kac data set using an open search workflow. The open search (extensive precursor ion tolerant search) approach determines the identity of unassigned peptides using an ultra-tolerant Sequest database search that allows peptide matching even with modifications of unknown masses up to ± 500 Da (Chick et al., 2015). With this approach, they identified 57,406 Kac peptides from six microbiome samples; this is 1.6 times more. The taxonomy analysis showed that most lysine-acylated peptides were from the Firmicutes and Bacteroidetes phylum (Zhang et al., 2021). Interestingly, the study of untreated rheumatoid arthritis patients’ feces showed that lysine acetylation was downregulated and significantly positively correlated with the low relative abundance of Fusicatenibacter (a genus of Firmicutes) (Yu et al., 2022).
The results demonstrate that new methodologies for studying PTMs in complex samples such as the microbiome are necessary. Furthermore, these may provide more information about how PTMs regulate microbiota functions and the effect this has.
Host-microbiota interactions depend partly on metabolites produced by the microbiota that can function as receptor ligands, enzyme inhibitors, or metabolic precursors (Levy et al., 2016). Some of these metabolites can alter metabolic activities in the host through covalent modification. For example, the comparison of germ-free mice (GF) vs. colonized adult mice (CONV-D) showed that in both the colon and the liver samples, there is an increase in lysine ϵ-acetylation on CONV-D vs. GF. In addition, it was demonstrated that colonization also affected the acetylation of lysines in metabolic enzymes expressed in the liver, including the glycolytic enzyme phosphoglycerate mutase and the melanin-biosynthetic enzyme dopachrome decarboxylase. While in the colon, the most dramatically elevated site of lysine ϵ-acetylation was found in α-1-antitrypsin (Simon et al., 2012).
To study the biological role of protein acetylation, appropriate methods must be applied to identify and quantify acetylated proteins and assign acetylation sites. Different techniques can be used in the same study, depending on the aim of the investigation, sample complexity, and type (Table 3).
As a first step in the investigation, detection, and semi-quantification of the targeted acetylated protein can be made by western blot analysis using specific antibodies against acetylated proteins (Yu et al., 2008; Hirano, 2012; Diallo et al., 2019). This type of analysis gives a global view of the acetylation profiles under different conditions (temperature, growth phase, stressors, among others) and in different strains to determine the differences in acetylation levels qualitatively (Kosono et al., 2015; Schilling et al., 2015; Christensen et al., 2018; Xu et al., 2018; Dale et al., 2023). However, this approach usually presents an insensitive and non-specific signal, so it is necessary to introduce negative controls to demonstrate the specificity of the anti-acetyl lysine antibody. Furthermore, it is not possible to know the number/assignment of specific acetylation sites, and protein identification requires additional steps (Diallo et al., 2019).
Liquid chromatography coupled to mass spectrometry (LC-MS/MS) systems has become the method for sequencing and identifying thousands of proteins. Due to its sensitivity, precision, and accuracy, it is also necessary to study PTMs.
Mass spectrometry has been widely used for global acetylome analysis in several bacterial species. The data generated are commonly used to determine the number of proteins and acetylated sites and the metabolic processes that this modification regulates through the functional annotation of the acetylated proteins and identify the potential consensus motifs of the acetylated sites (Table 3). One of the first published acetylomes in bacteria was from E. coli using an anti-acetyl lysine antibody with nano-HPLC/MS/MS. This study identified 125 lysine-acetylated sites in 85 proteins involved in diverse physiological functions, including carbon metabolism, protein synthesis, and amino acid metabolism (Yu et al., 2008). In the following years, many groups conducted global screening and characterization of lysine acetylation in other organisms. In Streptococcus pneumoniae, 653 lysine-acetylated sites on 392 proteins were identified. The gene ontology enrichment analysis demonstrated that in the metabolic process, the major targets of lysine acetylation are involved in the cellular amide metabolic process, gene expression, and macromolecule biosynthetic process. The acetylated proteins identified mainly possess structural constituents of the ribosome, ion binding, organic cyclic compound binding, and ligase activity for molecular function. The acetylated proteins were significantly enriched in intracellular, non-membrane-bound organelle ribonucleoprotein complexes and intracellular organelles for cellular components GO enrichment analysis. The motif analysis of the identified acetylated peptides showed that all the amino acids in these motifs are hydrophilic (Liu et al., 2018). Moreover, the KEGG pathways enrichment analysis allows further insights into the functional correlation of lysine acetylation. In V. alginolyticus, the functional correlation of acetylation and bile salt stress through KEGG pathways enrichment analysis revealed that the largest group of acetylated proteins was involved in metabolic pathways (20.5%), biosynthesis of antibiotics (13.3%), ribosome (5.8%), purine metabolism (3.3%), carbon metabolism (2%), beta-lactam resistance (2.0%), and RNA degradation (1.7%). In general, these data indicated that the enzymes in the metabolic pathway play an important role in enabling bacteria to respond to stress (Xiao et al., 2023).
To better understand how lysine acetylation regulates protein function and if this mechanism is evolutionarily conserved, Nakayasu and coworkers investigated by LC-MS/MS whether acetylation occurs on lysine residues conserved in different bacteria. For this, the proteomic analysis of 48 phylogenetically diverse bacteria from six phyla: Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Cyanobacteria, and Fibrobacteres were analyzed. The data showed that acetylated proteins were significantly enriched in glycolysis, pyruvate metabolism, ribosomes, and the TCA cycle. Some enzymes involved in glycolysis and the TCA cycle were further examined to analyze whether these enzymes contained catalytically critical lysine residues involved in substrate or cofactor binding. The results showed that lysine residues are localized in the active site that binds to the substrate or cofactor, demonstrating that lysine acetylation occurs in evolutionarily conserved residues of the enzyme catalytic sites in multiple organisms (Nakayasu et al., 2017). Moreover, the in-silico modeling analysis adding acetylation to the enzyme structure evidence that the modification alters the charge and shape of the lysine residue by neutralizing its positive charge and increasing its size; therefore, the catalytic activity is inhibited (Nakayasu et al., 2017).
Mass spectrometry is not an inherently quantitative method because proteolytic peptides have different physiochemical properties (size, charge, hydrophobicity, and more), which produce variations in the MS/MS spectra. Therefore, for accurate quantification, it is generally required to compare each peptide between experiments (Bantscheff et al., 2007). For this reason, different labeling techniques coupled to LC-MS have been developed: stable-isotope labeling with amino acids in cell culture (SILAC), isotope-coded affinity tags (ICAT), tandem mass tag (TMT), iTRAQ, multiplex isobaric tags, and heavy peptide AQUA are some examples (Gingras et al., 2007; Lindemann et al., 2017; Zhang and Elias, 2017).
As shown in Table 3, using different labeling strategies, it has been possible to quantify acetylated sites and proteins robustly and precisely, which has allowed us to elucidate the role of N-acetylation in processes such as biofilm formation and pathogenesis, determine how the dynamics of acetylation during bacterial growth and if the carbon source influences the PTM rate. For the pathogenic bacterium Bacillus nematocida B16 (B16), an antagonistic against plant-parasitic nematodes, the lysine acetylome quantification using TMT-(Tandem Mass Tag) labeling during its interaction with worms allowed determining that the acetylation levels of 18 sites were up-regulated and those of 19 sites were down-regulated during pathogenesis. Acetylated peptides were further characterized based on the Gene Ontology annotation (COG). The analysis suggested that acetylated proteins participate in diverse biological processes, mainly in metabolism and protein synthesis. Furthermore, analysis of the acetylated proteins revealed that many identified proteins are involved in B16 infection, suggesting that lysine acetylation may play a role in regulating B16-C. elegans interaction (Sun et al., 2018). In E. coli, label-free quantification allowed to determine the substrate specificity of four new Nϵ-lysine acetyltransferases. Using an E. coli strain that lacked known acetylation mechanisms (YfiQ and AcP), 818 acetylation sites on 434 proteins whose acetylation increased with overexpression of at least one KAT were identified. Of the proteins identified as acetylated by the newly identified KATs, several are central metabolic proteins, and many are components of the translational machinery. These new Kats tend to acetylate enzymes that regulate the branch points of central metabolism, while AcP seems to modify many of the central metabolic enzymes. These suggest that enzymatic acetylation has evolved to regulate key flux points in metabolism specifically and that non-enzymatic acetylation is a mechanism global to respond to the carbon flux (Christensen et al., 2018).
Although relative quantification is a powerful technique that detects variations in the acetylation levels in different conditions, it is limited by the fact that the changes are relative to the total protein abundance, which can vary from one condition to another, giving misinterpretations about the physiological significance of this PTM. Analysis of acetylation stoichiometry or occupancy can allow us to identify the critical acetylation sites whose changes in abundance are important.
However, determining the acetylation stoichiometry is complex, mainly because the ionization efficiency of modified and unmodified peptides in a mass spectrometer is different and artifact-prone. For this reason, various working groups have reported methods and workflows for the precise quantification of site-specific protein acetylation occupancy, which are based on the comparison of the proportion of endogenously acetylated lysine versus chemically labeled lysine that is not endogenously acetylated (Baeza et al., 2014; Weinert et al., 2014; Gil et al., 2017; Miyagi, 2017; Weinert et al., 2017; Wei et al., 2018).
The first protocol developed for directly quantifying stoichiometry of site-specific acetylation in bacteria was based on the chemical acetylation of free lysine residues with isotopic acetic anhydride, followed by trypsin cleavage and MS analysis. The method was applied to analyze the entire proteome of E. coli precisely to determine the role of deacetylases, CobB, on both site-specific and global acetylation (Baeza et al., 2014). In a similar study, Weinert et al. (2017) determined the absolute acetylation stoichiometry but used a serial dilution of SILAC-labeled peptides (SDSILAC). Although the methodologies differ in the way of labeling the peptides, with both approaches, it was shown that sirtuin deacetylase deficiency affects central metabolism and leads to both site-specific and global changes in protein acetylation stoichiometry (Baeza et al., 2014; Weinert et al., 2017).
Exploring the relationship of chemical acetylation and how it affects the enzymatic activity of glycolytic proteins, it was found that possibly a maximum of 10% of non-enzymatically acetylated proteins reach a stoichiometry that could inhibit their activity and that enzymes such as GapA and GpmA are acetylated at high stoichiometry (Schastnaya et al., 2023). The authors suggest that AcP-acetylation is specific and may exert control over metabolism.
Knowing the stoichiometry of acetylation can help to establish how it changes and whether it exerts a regulatory effect or only has a constitutive function necessary for protein folding or stable interaction.
The advances in mass spectrometry techniques enabled researchers to identify thousands of acetylation sites in various proteins. As mentioned throughout the review, analyzing the acetylome of multiple organisms using either stable isotope labeling workflows or label-free protocols or analyzing the acetylation through stoichiometry measurements has shown that acetylation regulates gene expression and physiological processes. However, it should be considered that lysine acetylation is highly dynamic and mostly reversible in cells. Therefore, the extraction of acetylated proteins with identical acetylation stoichiometry is difficult. Also, chemical lysine acetylation usually occurs at multiple sites simultaneously in one protein, and other modifications can compete with acetylation for the same lysine residue (Fatema and Fan, 2023). Although different methods have been developed to overcome these limitations, such as direct mutagenesis to mimic the acetylated lysine residue (substitution of lysine for glutamine), the study of acetylation remains challenging mainly because the substitution only imitates the electronic effects of the modifications but not their steric effects.
The genetic code expansion technique (GCE) has been applied to generate site-specifically acetylated proteins. Some reviews summarize the use of this technique for the analysis of different PTMs and the strategies to produce site-specifically modification (Neumann et al., 2008; Schmidt and Summerer, 2014; Krauskopf and Lang, 2020; Neumann-Staubitz et al., 2021; Chen and Tsai, 2022; Fatema and Fan, 2023). Specifically, proteins with the actual modifications at the desired positions can be generated recombinantly, facilitating biochemical studies (Chen and Tsai, 2022).
The GCE strategy to site-specifically incorporate Nϵ-thioacetyl-l-lysine (TAcK) as an analog of Nϵ-acetyl-l-lysine (AcK) into green fluorescent protein and malate dehydrogenase (MDH) in Escherichia coli, showed that TAcK could serve as an ideal functional mimic for AcK. The replacement of lysine residue 140 in MDH showed that the MDH-140TAcK variant could increase the enzyme activity by 3-fold, like that of the MDH-140AcK variant (Venkat et al., 2017b). Another enzyme studied with this approach is the aconitase of E. coli. Aconitase catalyzes the second reaction of the tricarboxylic acid cycle, the reversible conversion of citrate and isocitrate. With GCE 14, site-specifically acetylated aconitase variants were produced, and their enzymatic analysis and kinetic analyses showed that acetylation of AcnA K684 decreased the enzyme activity. In contrast, acetylation of AcnB K567 increased the enzyme activity. Moreover, in vitro acetylation and deacetylation assays were performed, and the results showed that the enzyme could be acetylated by acetyl-phosphate chemically and be deacetylated by the CobB deacetylase at most lysine sites (Araujo et al., 2022).
GCE has been used to study the effect of coexisting multiple distinct PTMs in one protein. Malate dehydrogenase was used to validate the method, and phosphorylation and acetylation were studied simultaneously in E. coli. The simultaneous incorporation of phosphoserine (Sep) and acetyl lysine (AcK) was confirmed by both Western blotting, and then the enzyme activities of wild-type, AcK only containing, Sep-only containing, and AcK/Sep-containing MDHs were measured. Lysine acetylation at position 140 increased the enzyme activity by 3.5-fold, while serine phosphorylation at position 280 decreased the enzyme activity to only 30% of that of the wild-type enzyme. Interestingly, the combination of 140-AcK and 280-Sep had a similar activity with the wild-type enzyme, indicating that two PTMs work separately to affect MDH activity (Venkat et al., 2018b).
These studies demonstrated practical applications of genetic code expansion in acetylation studies.
In recent years, the field of research on acetylation in bacteria has increased considerably, demonstrating its relevance in various cellular processes. Protein acetylation is a highly dynamic modification that occurs on short time scales (minutes to hours), where the acetylation levels are dictated by two closely related mechanisms: the addition and removal of the acyl group (Baeza et al., 2020). This property allows the cell to respond quickly to different environmental changes by modifying the functionality of the protein to adjust metabolic processes.
The development of protein labeling and enrichment methods and technological advances in mass spectrometry for identifying and quantifying acetylated sites and proteins have allowed the study of lysine acetylation in various organisms under different conditions. Thus, it has been shown that acetyl-lysines must result from one of the recognized acetylation mechanisms, whether enzymatic or non-enzymatic. Furthermore, it has been identified that the target acetylation proteins participate in various cellular and metabolic processes, highlighting the acetylation of conserved lysines in central metabolism proteins. However, for >99% of proteins, the biological relevance of this modification remains to be determined.
Another critical aspect yet to be investigated profoundly is the determination of the acetylation stoichiometry. Quantification of fold changes only sometimes has crucial biological significance. For example, if acetylation has functional consequences, such as loss of function, the modification should have a high stoichiometry. In comparison, a low percentage of acetylation may suggest a modification related to protein folding or the formation of a stable interaction (Carabetta et al., 2016). So far, the techniques available for determining the stoichiometry have revealed that most acetylated proteins occur at low stoichiometry (0–10%), and in approximately 4% of acetylated sites, a >20% stoichiometry is observed (Baeza et al., 2014; Weinert et al., 2017). This indicates that only a tiny fraction of the acetylome is of physiological importance. Also, it is essential to consider that proteomic analysis is based on experiments under a particular condition and in isolated organisms, so it would be convenient to consider performing a comparative analysis under different conditions (temperature, carbon sources, among others) and their behavior in co-culture or symbiosis for a correct interpretation of the data.
Several functional monitoring studies are still being carried out using in vitro assays. Although they allow us to know the effect of acetylation on the protein, they only sometimes reflect the natural consequence of this post-translational regulation process. Therefore, it is necessary to generate hybrid approaches that help us identify and characterize not only the modified proteins but also which metabolic pathways are involved and how this impacts the cell’s metabolism. Such approaches can integrate the analysis of proteomic, transcriptomic, and metabolomics data sets.
Incorporating synthetic biology can expand our understanding of how lysine acetylation regulates protein function. In this regard, genetic code expansion (GCE) has become essential for studying biological processes such as post-translational modifications. Rather than adding acetyl groups after protein translation, this approach relies on heterologous pairs of aminoacyl-tRNA-synthetases (aaRS) and its cognate tRNA from Methanosarcinaceae species that enable the co-translational incorporation of Nϵ-acetyllysine (AcK) in response to stop codon (Schmidt and Summerer, 2014). This approach has been used for some groups to evaluate the role of lysine acetylation using recombinantly expressed proteins, finding that the enzymatic activity of several proteins is positively or negatively regulated. Once again, we believe this type of study should be applied to studies of bacteria in vivo.
Non-enzymatic acetylation is an inevitable consequence of the cell’s metabolic states. At high concentrations of AcP this mechanism is favored. However, it should be considered that this molecule can also be used as an acyl group donor for acetyltransferases catalyzing protein acetylation. Therefore, it cannot be ruled out that these mechanisms coexist. The roles of global acetylation and other acyl donors (succinyl-CoA, propionyl-CoA, and malonyl-CoA) for enzymatic or non-enzymatic protein acylation remain to be elucidated. Finally, it is essential to understand how lysine acetylation interferes with and cross-links with other post-translational modifications.
JR: Writing – review & editing, Writing – original draft. SE-G: Writing – review & editing, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Programa “Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM)” supported part of this work, granting IN-213522 to SE-G.
JR was a Postdoctoral fellow supported by the DGAPA-UNAM Postdoctoral Program. We also thank Michael F. Dunn for critical reading and language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aertsen, A., Michiels, C. W. (2004). Stress and how bacteria cope with death and survival. Crit. Rev. Microbiol. 30, 263–273. doi: 10.1080/10408410490884757
Allfrey, V. G., Faulkner, R., Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. PNAS. 51, 786–794. doi: 10.1073/pnas.51.5.786
Araujo, J., Ottinger, S., Venkat, S., Gan, Q., Fan, C. (2022). Studying acetylation of aconitase isozymes by genetic code expansion. Front. Chem. 10. doi: 10.3389/fchem.2022.862483
Avalos, J. L., Boeke, J. D., Wolberger, C. (2004). Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell. 13, 639–648. doi: 10.1016/S1097-2765(04)00082-6
Baeza, J., Dowell, J. A., Smallegan, M. J., Fan, J., Amador-Noguez, D., Khan, Z., et al. (2014). Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 289, 21326–21338. doi: 10.1074/jbc.M114.581843
Baeza, J., Lawton, A. J., Fan, J., Smallegan, M. J., Lienert, I., Gandhi, T., et al. (2020). Revealing dynamic protein acetylation across subcellular compartments. J. Proteome Res. 19, 2404–2418. doi: 10.1021/acs.jproteome.0c00088
Bai, L., Chang, M., Shan, J., Jiang, R., Zhang, Y., Zhang, R., et al. (2011). Identification and characterization of a novel spermidine/spermine acetyltransferase encoded by gene ste26 from Streptomyces sp. 139. Biochimie. 93, 1401–1407. doi: 10.1016/j.biochi.2011.04.014
Ban, N., Beckmann, R., Cate, J. H., Dinman, J. D., Dragon, F., Ellis, S. R., et al. (2014). A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24, 165–169. doi: 10.1016/j.sbi.2014.01.002
Bantscheff, M., Schirle, M., Sweetman, G., Rick, J., Kuster, B. (2007). Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031. doi: 10.1007/s00216-007-1486-6
Barak, R., Yan, J., Shainskaya, A., Eisenbach, M. (2006). The chemotaxis response regulator CheY can catalyze its own acetylation. J. Mol. Biol. 359, 251–265. doi: 10.1016/j.jmb.2006.03.033
Barrett, O. J., Pushechnikov, A., Wu, M., Disney, M. D. (2008). Studying aminoglycoside modification by the acetyltransferase class of resistance-causing enzymes via microarray. Carbohydr. Res. 343, 2924–2931. doi: 10.1016/j.carres.2008.08.018
Baumgartner, J. T., Mohammad, T. S. H., Czub, M. P., Majorek, K. A., Arolli, X., Variot, C., et al. (2021). Gcn5-related N-acetyltransferases (GNATs) with a catalytic serine residue can play ping-pong too. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.646046
Bennett, P. M., Holms, W. H. (1975). Reversible inactivation of the isocitrate dehydrogenase of Escherichia coli ML308 during growth on acetate. J. Gen. Microbiol. 87, 37–51. doi: 10.1099/00221287-87-1-37
Bernal, V., Castaño-Cerezo, S., Cánovas, M. (2016). Acetate metabolism regulation in Escherichia coli: carbon overflow, pathogenicity, and beyond. Appl. Microbiol. Biotechnol. 100, 8985–9001. doi: 10.1007/s00253-016-7832-x
Berrabah, W., Aumercier, P., Lefebvre, P., Staels, B. (2011). Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett. 585, 1640–1650. doi: 10.1016/j.febslet.2011.03.066
Bi, J., Gou, Z., Zhou, F., Chen, Y., Gan, J., Liu, J., et al. (2018). Acetylation of lysine 182 inhibits the ability of Mycobacterium tuberculosis DosR to bind DNA and regulate gene expression during hypoxia. Emerg. Microbes Infect. 7, 1–12. doi: 10.1038/s41426-018-0112-3
Blander, G., Guarente, L. (2004). The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. doi: 10.1146/annurev.biochem.73.011303.073651
Blasl, A. T., Schulze, S., Qin, C., Graf, L. G., Vogt, R., Lammers, M. (2021). Post-translational lysine ac (et) ylation in health, ageing and disease. Biol. Chem. 403, 151–194. doi: 10.1515/hsz-2021–0139
Bontemps-Gallo, S., Gaviard, C., Richards, C. L., Kentache, T., Raffel, S. J., Lawrence, K. A., et al. (2018). Global profiling of lysine acetylation in Borrelia burgdorferi B31 reveals its role in central metabolism. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02036
Brown, K., DeCoffe, D., Molcan, E., Gibson, D. L. (2012). Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 4, 1095–1119. doi: 10.3390/nu4081095
Burckhardt, R. M., Buckner, B. A., Escalante-Semerena, J. C. (2019). Staphylococcus aureus modulates the activity of acetyl-Coenzyme A synthetase (Acs) by sirtuin-dependent reversible lysine acetylation. Mol. Microbiol. 112, 588–604. doi: 10.1111/mmi.14276
Burckhardt, R. M., Escalante-Semerena, J. C. (2020). Small-molecule acetylation by GCN5-related N-acetyltransferases in bacteria. MMBR. 84, 10–1128. doi: 10.1128/MMBR.00090-19
Bürger, M., Chory, J. (2018). Structural and chemical biology of deacetylases for carbohydrates, proteins, small molecules and histones. Commun. Biol. 1, 217. doi: 10.1038/s42003-018-0214-4
Cabiscol, E., Tamarit Sumalla, J., Ros Salvador, J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Internatl. Microbiol. 3, 3–8.
Carabetta, V. J., Greco, T. M., Tanner, A. W., Cristea, I. M., Dubnau, D. (2016). Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. Msystems. 1, e00005–e00016. doi: 10.1128/mSystems.00005-16
Card, G. L., Peterson, N. A., Smith, C. A., Rupp, B., Schick, B. M., Baker, E. N. (2005). The crystal structure of rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis. J. Biol. Chem. 280, 13978–13986. doi: 10.1074/jbc.M413904200
Castaño-Cerezo, S., Bernal, V., Blanco-Catalá, J., Iborra, J. L., Cánovas, M. (2011). cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 82, 1110–1128. doi: 10.1111/j.1365–2958.2011.07873.x
Castaño-Cerezo, S., Bernal, V., Post, H., Fuhrer, T., Cappadona, S., Sánchez-Díaz, N. C., et al. (2014). Protein acetylation affects acetate metabolism, motility and acid stress response in. Escherichia coli. Mol. Syst. Biol. 10, 762. doi: 10.15252/msb.20145227
Castaño-Cerezo, S., Bernal, V., Röhrig, T., Termeer, S., Cánovas, M. (2015). Regulation of acetate metabolism in Escherichia coli BL21 by protein Nϵ-lysine acetylation. Appl. Microbiol. Biotechnol. 99, 3533–3545. doi: 10.1007/s00253–014-6280–8
Chan, C. H., Garrity, J., Crosby, H. A., Escalante-Semerena, J. C. (2011). In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Mol. Microbiol. 80, 168–183. doi: 10.1111/j.1365-2958.2011.07566.x
Chen, J., Tsai, Y. H. (2022). Applications of genetic code expansion in studying protein post-translational modification. J. Mol. Biol. 434, 167424. doi: 10.1016/j.jmb.2021.167424
Chen, W., Biswas, T., Porter, V. R., Tsodikov, O. V., Garneau-Tsodikova, S. (2011). Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. PNAS. 108, 9804–9808. doi: 10.1073/pnas.1105379108
Chick, J. M., Kolippakkam, D., Nusinow, D. P., Zhai, B., Rad, R., Huttlin, E. L., et al. (2015). A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat. Biotechnol. 33, 743–749. doi: 10.1038/nbt.3267
Christensen, D. G., Baumgartner, J. T., Xie, X., Jew, K. M., Basisty, N., Schilling, B., et al. (2019a). Mechanisms, detection, and relevance of protein acetylation in prokaryotes. MBio. 10, e02708–e02718. doi: 10.1128/mBio.02708-18
Christensen, D. G., Meyer, J. G., Baumgartner, J. T., D’Souza, A. K., Nelson, W. C., Payne, S. H., et al. (2018). Identification of novel protein lysine acetyltransferases in Escherichia coli. MBio. 9, e01905–e01918. doi: 10.1128/mBio.01905-18
Christensen, D. G., Xie, X., Basisty, N., Byrnes, J., McSweeney, S., Schilling, B., et al. (2019b). Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01604
Clemente, J. C., Ursell, L. K., Parfrey, L. W., Knight, R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell. 148, 1258–1270. doi: 10.1016/j.cell.2012.01.035
Clements, A., Rojas, J. R., Trievel, R. C., Wang, L., Berger, S. L., Marmorstein, R. (1999). Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 18, 3521–3532. doi: 10.1093/emboj/18.13.3521
Costa, Y., Galimand, M., Leclercq, R., Duval, J., Courvalin, P. (1993). Characterization of the chromosomal aac (6')-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37, 1896–1903. doi: 10.1128/AAC.37.9.1896
Couto-Rodríguez, R. L., Koh, J., Chen, S., Maupin-Furlow, J. A. (2023). Insights into the lysine acetylome of the Haloarchaeon Haloferax volcanii during oxidative stress by quantitative SILAC-based proteomics. Antioxidants. 12, 1203. doi: 10.3390/antiox12061203
Cronan, J. E., Jr, Laporte, D. (2005). Tricarboxylic acid cycle and glyoxylate bypass. EcoSal Plus. 1. doi: 10.1128/ecosalplus.3.5.2
Crosby, H. A., Heiniger, E. K., Harwood, C. S., Escalante-Semerena, J. C. (2010). Reversible Nϵ-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol. Microbiol. 76, 874–888. doi: 10.1111/j.1365-2958.2010.07127.x
Crosby, H. A., Pelletier, D. A., Hurst, G. B., Escalante-Semerena, J. C. (2012). System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases. J. Biol. Chem. 287, 15590–15601. doi: 10.1074/jbc.M112.352104
Daboor, S. M., Masood, F. S. S., Al-Azab, M. S., Nori, E. E. (2015). A review on Streptococcus mutans with its diseases dental caries, dental plaque and endocarditis. Indian J. Microbiol. Res. 2, 76–82.
Dale, A. L., Man, L., Cordwell, S. J. (2023). Global acetylomics of Campylobacter jejuni shows lysine acetylation regulates CadF adhesin processing and human fibronectin binding. J. Proteome Res. 22, 3519–3533. doi: 10.1021/acs.jproteome.3c00391
Dean, A. M., Lee, M. H., Koshland, D. E. (1989). Phosphorylation inactivates Escherichia coli isocitrate dehydrogenase by preventing isocitrate binding. J. Biol. Chem. 264, 20482–20486. doi: 10.1016/S0021-9258(19)47087-7
DeLano, W. L. (2002) The PyMOL molecular graphics system. Available online at: http://www.pymol.org/.
Diallo, I., Seve, M., Cunin, V., Minassian, F., Poisson, J. F., Michelland, S., et al. (2019). Current trends in protein acetylation analysis. Expert. Rev. Proteom. 16, 139–159. doi: 10.1080/14789450.2019.1559061
Donlan, R. M. (2000). Role of biofilms in antimicrobial resistance. ASAIO J. 46, S47–S52. doi: 10.1097/00002480-200011000-00037
Draker, K. A., Wright, G. D. (2004). Molecular mechanism of the enterococcal aminoglycoside 6 ‘-N-acetyltransferase ‘: Role of GNAT-conserved residues in the chemistry of antibiotic inactivation. Biochemistry. 43, 446–454. doi: 10.1021/bi035667n
Dutnall, R. N., Tafrov, S. T., Sternglanz, R., Ramakrishnan, V. (1998). Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell. 94, 427–438. doi: 10.1016/S0092-8674(00)81584-6
Dyda, F., Klein, D. C., Hickman, A. B. (2000). GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29, 81–103. doi: 10.1146/annurev.biophys.29.1.81
Edwards, S. M., Cunningham, S. A., Dunlop, A. L., Corwin, E. J. (2017). The maternal gut microbiome during pregnancy. MCN Am. J. Matern Child Nurs. 42, 310. doi: 10.1097/NMC.0000000000000372
Fang, Z., Lai, F., Cao, K., Zhang, Z., Cao, L., Liu, S., et al. (2022). Potential role of lysine acetylation in antibiotic resistance of Escherichia coli. Msystems. 7, e00649–e00622. doi: 10.1128/msystems.00649-22
Fatema, N., Fan, C. (2023). Studying lysine acetylation of citric acid cycle enzymes by genetic code expansion. Mol. Microbiol. 119, 551–559. doi: 10.1111/mmi.15052
Favrot, L., Blanchard, J. S., Vergnolle, O. (2016). Bacterial GCN5-related N-acetyltransferases: from resistance to regulation. Biochem. 55, 989–1002. doi: 10.1021/acs.biochem.5b01269
Feid, S. C., Walukiewicz, H. E., Wang, X., Nakayasu, E. S., Rao, C. V., Wolfe, A. J. (2022). Regulation of translation by lysine acetylation in Escherichia coli. Mbio. 13, e01224–e01222. doi: 10.1128/mbio.01224-22
Filippova, E. V., Kuhn, M. L., Osipiuk, J., Kiryukhina, O., Joachimiak, A., Ballicora, M. A., et al. (2015). A novel polyamine allosteric site of SpeG from Vibrio cholerae is revealed by its dodecameric structure. J. Mol. Biol. 427, 1316–1334. doi: 10.1016/j.jmb.2015.01.009
Finkel, T., Deng, C. X., Mostoslavsky, R. (2009). Recent progress in the biology and physiology of sirtuins. Nature. 460, 587–591. doi: 10.1038/nature08197
Finnin, C. A. (2005). Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 433, 71–84. doi: 10.1016/j.abb.2004.08.006
Finnin, M. S., Donigian, J. R., Cohen, A., Richon, V. M., Rifkind, R. A., Marks, P. A., et al. (1999). Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 401, 188–193. doi: 10.1038/43710
Forouhar, F., Lee, I. S., Vujcic, J., Vujcic, S., Shen, J., Vorobiev, S. M., et al. (2005). Structural and functional evidence for Bacillus subtilis PaiA as a novel N1 -spermidine/spermine acetyltransferase. J. Biol. Chem. 280, 40328–40336. doi: 10.1074/jbc.M505332200
Frankel, B. A., Blanchard, J. S. (2008). Mechanistic analysis of Mycobacterium tuberculosis Rv1347c, a lysine Nϵ-acyltransferase involved in mycobactin biosynthesis. Arch. Biochem. Biophys. 477, 259–266. doi: 10.1016/j.abb.2008.05.013
Fukuchi, J. I., Kashiwagi, K., Yamagishi, M., Ishihama, A., Igarashi, K. (1995). Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J. Biol. Chem. 270, 18831–18835. doi: 10.1074/jbc.270.32.18831
Gardner, J. G., Escalante-Semerena, J. C. (2009). In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J. Bacteriol. 191, 1749–1755. doi: 10.1128/JB.01674-08
Gardner, J. G., Grundy, F. J., Henkin, T. M., Escalante-Semerena, J. C. (2006). Control of acetyl-coenzyme a synthetase (AcsA) activity by acetylation/deacetylation without NAD+ involvement in Bacillus subtilis. J. Bacteriol. 188, 5460–5468. doi: 10.1128/JB.00215-06
Garnak, M., Reeves, H. C. (1979). Phosphorylation of isocitrate dehydrogenase of Escherichia coli. Science. 203, 1111–1112. doi: 10.1126/science.34215
Gaviard, C., Broutin, I., Cosette, P., Dé, E., Jouenne, T., Hardouin, J. (2018). Lysine succinylation and acetylation in Pseudomonas aeruginosa. J. Proteome Res. 17, 2449–2459. doi: 10.1021/acs.jproteome.8b00210
Ghosh, S., Padmanabhan, B., Anand, C., Nagaraja, V. (2016). Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization. Mol. Microbiol. 100, 577–588. doi: 10.1111/mmi.13339
Gil, J., Ramírez-Torres, A., Chiappe, D., Luna-Peñaloza, J., Fernandez-Reyes, F. C., Arcos-Encarnación, B., et al. (2017). Lysine acetylation stoichiometry and proteomics analyses reveal pathways regulated by sirtuin 1 in human cells. JBC. 292, 18129–18144. doi: 10.1074/jbc.M117.784546
Gingras, A. C., Gstaiger, M., Raught, B., Aebersold, R. (2007). Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Bio. 8, 645–654. doi: 10.1038/nrm2208
Gregoretti, I., Lee, Y. M., Goodson, H. V. (2004). Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. K. Mol. Biol. 338, 17–31. doi: 10.1016/j.jmb.2004.02.006
Gu, W., Roeder, R. G. (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 90, 595–606. doi: 10.1016/S0092-8674(00)80521-8
Haigis, M. C., Guarente, L. P. (2006). Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes. Dev. 20, 2913–2921. doi: 10.1101/gad.1467506
Hawse, W. F., Hoff, K. G., Fatkins, D. G., Daines, A., Zubkova, O. V., Schramm, V. L., et al. (2008). Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 16, 1368–1377. doi: 10.1016/j.str.2008.05.015
Hayden, J. D., Brown, L. R., Gunawardena, H. P., Perkowski, E. F., Chen, X., Braunstein, M. (2013). Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiol. 159, 1986–1999. doi: 10.1099/mic.0.068585-0
Hebert, A. S., Dittenhafer-Reed, K. E., Yu, W., Bailey, D. J., Selen, E. S., Boersma, M. D., et al. (2013). Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell. 49, 186–199. doi: 10.1016/j.molcel.2012.10.024
Hecker, M., Völker, U. (2001). General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44, 35–91. doi: 10.1016/S0065-2911(01)44011-2
Hegde, S. S., Dam, T. K., Brewer, C. F., Blanchard, J. S. (2002). Thermodynamics of aminoglycoside and acyl-coenzyme A binding to the Salmonella enterica AAC(6‘)-Iy aminoglycoside N-acetyltransferase. Biochemistry. 41, 7519–7527. doi: 10.1021/bi020190l
Hentchel, K. L., Escalante-Semerena, J. C. (2015). Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. MMBR. 79, 321–346. doi: 10.1128/MMBR.00020-15
Hernick, M., Fierke, C. A. (2006). Catalytic mechanism and molecular recognition of E. coli UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase probed by mutagenesis. Biochemistry. 45, 15240–15248. doi: 10.1021/bi061405k
Heuts, D. P., Scrutton, N. S., McIntire, W. S., Fraaije, M. W. (2009). What's in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS. J. 276, 3405–3427. doi: 10.1111/j.1742-4658.2009.07053.x
Hildmann, C., Ninkovic, M., Dietrich, R., Wegener, D., Riester, D., Zimmermann, T., et al. (2004). A new amidohydrolase from Bordetella or Alcaligenes strain FB188 with similarities to histone deacetylases. J. Bacteriol. 186, 2328–2339. doi: 10.1128/JB.186.8.2328-2339.2004
Hildmann, C., Riester, D., Schwienhorst, A. (2007). Histone deacetylases—an important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 75, 487–497. doi: 10.1007/s00253-007-0911-2
Hildmann, C., Wegener, D., Riester, D., Hempel, R., Schober, A., Merana, J., et al. (2006). Substrate and inhibitor specificity of class 1 and class 2 histone deacetylases. J. Biotechnol. 124, 258–270. doi: 10.1016/j.jbiotec.2006.01.030
Hirano, S. (2012). Western blot analysis. Nanotoxicity: Methods Protoc. 926, 87–97. doi: 10.1007/978–1-62703–002-1_6
Høiby, N., Ciofu, O., Johansen, H. K., Song, Z. J., Moser, C., Jensen, P.Ø., et al. (2011). The clinical impact of bacterial biofilms. Int. J. Oral. Sci. 3, 55–65. doi: 10.4248/IJOS11026
Holms, W. H., Bennett, P. M. (1971). Regulation of isocitrate dehydrogenase activity in Escherichia coli on adaptation to acetate. J. Gen. Microbiol. 65, 57–68. doi: 10.1099/00221287-65-1-57
Houghton, J. L., Green, K. D., Pricer, R. E., Mayhoub, A. S., Garneau-Tsodikova, S. (2013). Unexpected N-acetylation of capreomycin by Mycobacterial Eis enzymes. J. Antimicrob. Chemother. 68, 800–805. doi: 10.1093/jac/dks497
Inoue, A., Fujimoto, D. (1969). Enzymatic deacetylation of histone. Biochem. Biophys. Res. Com. 36, 146–150. doi: 10.1016/0006-291X(69)90661-5
Jackman, J. E., Fierke, C. A., Tumey, L. N., Pirrung, M., Uchiyama, T., Tahir, S. H., et al. (2000). Antibacterial agents that target lipid a biosynthesis in gram-negative bacteria: inhibition of diverse UDP-3-O-(r-3-hydroxymyristoyl)-N-acetylglucosamine deacetylases by substrate analogs containing zinc binding motifs. J. Biol. Chem. 275, 11002–11009. doi: 10.1074/jbc.275.15.11002
Jeffares, D. C., Poole, A. M., Penny, D. (1998). Relics from the RNA world. J. Mol. Evol. 46, 18–36. doi: 10.1007/PL00006280
Jiang, Q., Chen, W., Qin, Y., Huang, L., Xu, X., Zhao, L., et al. (2017). AcuC, a histone deacetylase, contributes to the pathogenicity of Aeromonas hydrophila. Microbiologyopen. 6, e00468. doi: 10.1002/mbo3.468
Joo, H. S., Otto, M. (2012). Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem. Biol. 19, 1503–1513. doi: 10.1016/j.chembiol.2012.10.022
Jung, C. M., Heinze, T. M., Strakosha, R., Elkins, C. A., Sutherland, J. B. (2009). Acetylation of fluoroquinolone antimicrobial agents by an Escherichia coli strain isolated from a municipal wastewater treatment plant. J. Appl. Microbiol. 106, 564–571. doi: 10.1111/jam.2009.106.issue-2
Kang, M. S., Oh, J. S., Lee, H. C., Lim, H. S., Lee, S. W., Yang, K. H., et al. (2011). Inhibitory effect of Lactobacillus reuteri on periodontopathic and cariogenic bacteria. J. Microbiol. 49, 193–199. doi: 10.1007/s12275-011-0252-9
Khoury, G., Baliban, R., Floudas, C. (2011). Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90. doi: 10.1038/srep00090
Kim, K. H., An, D. R., Song, J., Yoon, J. Y., Kim, H. S., Yoon, H. J., et al. (2012). Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. PNAS. 109, 7729–7734. doi: 10.1073/pnas.1120251109
Kim, D. W., Feng, J., Chen, H., Kweon, O., Gao, Y., Yu, L. R., et al. (2013a). Identification of the enzyme responsible for N-acetylation of norfloxacin by Microbacterium sp. strain 4N2–2. Appl. Environ. Microbiol. 79, 314–321. doi: 10.1128/AEM.02347-12
Kim, S. C., Sprung, R., Chen, Y., Xu, Y., Ball, H., Pei, J., et al. (2006). Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 23, 607–618. doi: 10.1016/j.molcel.2006.06.026
Kim, D., Yu, B. J., Kim, J. A., Lee, Y. J., Choi, S. G., Kang, S., et al. (2013b). The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics. 13, 1726–1736. doi: 10.1002/pmic.201200001
Koo, H., Park, S., Kwak, M. K., Lee, J. S. (2020). Regulation of gene expression by protein lysine acetylation in Salmonella. J. Microbiol. 58, 979–987. doi: 10.1007/s12275-020-0483-8
Koshland, J., Daniel, E. (1952). Effect of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymatic reactions. J. Am. Chem. Soc 74, 2286–2292. doi: 10.1021/ja01129a035
Kosono, S., Tamura, M., Suzuki, S., Kawamura, Y., Yoshida, A., Nishiyama, M., et al. (2015). Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 10, e0131169. doi: 10.1371/journal.pone.0131169
Kozlova, E. V., Popov, V. L., Sha, J., Foltz, S. M., Erova, T. E., Agar, S. L., et al. (2008). Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 45, 343–354. doi: 10.1016/j.micpath.2008.08.007
Kraümer, A., Wagner, T., Yildiz, O., Meyer-Almes, F. J. (2016). Crystal structure of a histone deacetylase homologue from Pseudomonas aeruginosa. Biochemistry. 55, 6858–6868. doi: 10.1021/acs.biochem.6b00613
Krauskopf, K., Lang, K. (2020). Increasing the chemical space of proteins in living cells via genetic code expansion. Curr. Opin. Chem. Biol. 58, 112–120. doi: 10.1016/j.cbpa.2020.07.012
Kremling, A., Geiselmann, J., Ropers, D., de Jong, H. (2015). Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbiol. 23, 99–109. doi: 10.1016/j.tim.2014.11.002
Krtenic, B., Drazic, A., Arnesen, T., Reuter, N. (2020). Classification and phylogeny for the annotation of novel eukaryotic GNAT acetyltransferases. PLoS Comput. Biol. 16, e1007988. doi: 10.1371/journal.pcbi.1007988
Krulwich, T. A., Sachs, G., Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Kuhn, M. L., Zemaitaitis, B., Hu, L. I., Sahu, A., Sorensen, D., Minasov, G., et al. (2014). Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9, e94816. doi: 10.1371/journal.pone.0094816
Lammers, M. (2021). Post-translational lysine ac(et)ylation in bacteria: a biochemical, structural, and synthetic biological perspective. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.757179
Lee, W., VanderVen, B. C., Walker, S., Russell, D. G. (2017). Novel protein acetyltransferase, Rv2170, modulates carbon and energy metabolism in Mycobacterium tuberculosis. Sci. Rep. 7, 72. doi: 10.1038/s41598–017-00067–1
Lee, K. K., Workman, J. L. (2007). Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295. doi: 10.1038/nrm2145
Lei, L., Zeng, J., Wang, L., Gong, T., Zheng, X., Qiu, W., et al. (2021). Quantitative acetylome analysis reveals involvement of glucosyltransferase acetylation in Streptococcus mutans biofilm formation. Environ. Microbiol. Rep. 13, 86–97. doi: 10.1111/1758-2229.12907
Levy, M., Thaiss, C. A., Elinav, E. (2016). Metabolites: messengers between the microbiota and the immune system. Genes Dev. 30, 1589–1597. doi: 10.1101/gad.284091.116
Li, B., Maezato, Y., Kim, S. H., Kurihara, S., Liang, J., Michael, A. J. (2019). Polyamine-independent growth and biofilm formation, and functional spermidine/spermine N-acetyltransferases in Staphylococcus aureus and Enterococcus faecalis. Mol. Microbiol. 111, 159–175. doi: 10.1111/mmi.14145
Li, D., Ramanathan, S., Wang, G., Wu, Y., Tang, Q., Li, G. (2020a). Acetylation of lysine 7 of AhyI affects the biological function in Aeromonas hydrophila. Microb. Pathog. 140, 103952. doi: 10.1016/j.micpath.2019.103952
Li, L., Wang, W., Zhang, R., Xu, J., Wang, R., Wang, L., et al. (2018). First acetyl-proteome profiling of Salmonella Typhimurium revealed involvement of lysine acetylation in drug resistance. Vet. Microbiol. 226, 1–8. doi: 10.1016/j.vetmic.2018.09.024
Li, Y., Xue, H., Bian, D. R., Xu, G., Piao, C. (2020b). Acetylome analysis of lysine acetylation in the plant pathogenic bacterium Brenneria nigrifluens. MicrobiologyOpen. 9, e00952. doi: 10.1002/mbo3.952
Limsuwun, K., Jones, P. G. (2000). Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol. 182, 5373–5380. doi: 10.1128/JB.182.19.5373-5380.2000
Lin, Y., Fletcher, C. M., Zhou, J., Allis, C. D., Wagner, G. (1999). Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature. 400, 86–89. doi: 10.1038/21922
Lindemann, C., Thomanek, N., Hundt, F., Lerari, T., Meyer, H. E., Wolters, D., et al. (2017). Strategies in relative and absolute quantitative mass spectrometry based proteomics. Biol. Chem. 398, 687–699. doi: 10.1515/hsz-2017-0104
Lipmann, F., Tuttle, L. C. (1944). Acetyl phosphate: chemistry, determination, and synthesis. J. Biol. Chem. 153, 571–582. doi: 10.1016/S0021-9258(18)72001-2
Liu, M., Guo, L., Fu, Y., Huo, M., Qi, Q., Zhao, G. (2021). Bacterial protein acetylation and its role in cellular physiology and metabolic regulation. Biotechnol. Adv. 53, 107842. doi: 10.1016/j.biotechadv.2021.107842
Liu, M., Huo, M., Liu, C., Guo, L., Ding, Y., Ma, Q., et al. (2022). Lysine acetylation of Escherichia coli lactate dehydrogenase regulates enzyme activity and lactate synthesis. Front. Bioeng. Biotechnol. 10. doi: 10.3389/fbioe.2022.966062
Liu, Y., Liu, X., Dong, X., Yin, Z., Xie, Z., Luo, Y. (2023). Systematic analysis of lysine acetylation reveals diverse functions in Azorhizobium caulinodans Strain ORS571. Microbiol. Spectr. 11, e03539–e03522. doi: 10.1128/spectrum.03539-22
Liu, Y. T., Pan, Y., Lai, F., Yin, X. F., Ge, R., He, Q. Y., et al. (2018). Comprehensive analysis of the lysine acetylome and its potential regulatory roles in the virulence of Streptococcus pneumoniae. J. Proteomics. 176, 46–55. doi: 10.1016/j.jprot.2018.01.014
Liu, X., Wang, L., Zhao, K., Thompson, P. R., Hwang, Y., Marmorstein, R., et al. (2008). The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 451, 846–850. doi: 10.1038/nature06546
Liu, F., Yang, M., Wang, X., Yang, S., Gu, J., Zhou, J., et al. (2014). Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell Proteomics 13, 3352–3366. doi: 10.1074/mcp.M114.041962
Lombardi, P. M., Cole, K. E., Dowling, D. P., Christianson, D. W. (2011). Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 21, 735–743. doi: 10.1016/j.sbi.2011.08.004
Lu, Y. X., Liu, X. X., Liu, W. B., Ye, B. C. (2017). Identification and characterization of two types of amino acid-regulated acetyltransferases in actinobacteria. Biosci. Rep. 37, BSR20170157. doi: 10.1042/BSR20170157
Luther, A., Bisang, C., Obrecht, D. (2018). Advances in macrocyclic peptide-based antibiotics. Bioorg. Med. Chem. 26, 2850–2858. doi: 10.1016/j.bmc.2017.08.006
Ma, Q., Pan, Y., Chen, Y., Yu, S., Huang, J., Liu, Y., et al. (2021). Acetylation of glucosyltransferases regulates Streptococcus mutans biofilm formation and virulence. PLoS Pathogens. 17, e1010134. doi: 10.1371/journal.ppat.1010134
Ma, Q., Wood, T. K. (2011). Protein acetylation in prokaryotes increases stress resistance. Biochem. Biophys. Res. Commun. 410, 846–851. doi: 10.1016/j.bbrc.2011.06.076
Macek, B., Forchhammer, K., Hardouin, J., Weber-Ban, E., Grangeasse, C., Mijakovic, I. (2019). Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 17, 651–664. doi: 10.1038/s41579-019-0243-0
Magalhaes, M. L., Blanchard, J. S. (2005). The kinetic mechanism of AAC (3)-IV aminoglycoside acetyltransferase from Escherichia coli. Biochemistry. 44, 16275–16283. doi: 10.1021/bi051777d
Magnet, S., Blanchard, J. S. (2005). Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105, 477–498. doi: 10.1021/cr0301088
Magnet, S., Courvalin, P., Lambert, T. (1999). Activation of the cryptic aac (6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J. Bacteriol. 181, 6650–6655. doi: 10.1128/JB.181.21.6650-6655.1999
Magnet, S., Lambert, T., Courvalin, P., Blanchard, J. S. (2001). Kinetic and mutagenic characterization of the chromosomally encoded Salmonella enterica AAC (6 ‘)-Iy aminoglycoside N-acetyltransferase. Biochemistry. 40, 3700–3709. doi: 10.1021/bi002736e
Majorek, K. A., Kuhn, M. L., Chruszczm, M., Anderson, W. F., Minor, W. (2013). Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from Pseudomonas aeruginosa. J. Biol. Chem. 288, 30223–30235. doi: 10.1074/jbc.M113.501353
Majorek, K. A., Osinski, T., Tran, D. T., Revilla, A., Anderson, W. F., Minor, W., et al. (2017). Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria. Biochim. Biophys. Acta 1865, 55–64. doi: 10.1016/j.bbapap.2016.10.011
Mann, M., Jensen, O. N. (2003). Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261. doi: 10.1038/nbt0303-255
Marakasova, E., Ii, A., Nelson, K. T., van Hoek, M. L. (2020). Proteome wide profiling of n-ϵ-lysine acetylation reveals a novel mechanism of regulation of the chitinase activity in Francisella novicida. J. Proteome Res. 19, 1409–1422. doi: 10.1021/acs.jproteome.9b00512
Martin, I., Dawson, V. L., Dawson, T. M. (2011). Recent advances in the genetics of Parkinson's disease. Annu. Rev. Genom. Hum. Genet. 12, 301–325. doi: 10.1146/annurev-genom-082410-101440
Martini, C., Michaux, C., Bugli, F., Arcovito, A., Iavarone, F., Cacaci, M., et al. (2015). The polyamine N-acetyltransferase-like enzyme PmvE plays a role in the virulence of Enterococcus faecalis. Infect. Immun. 83, 364–371. doi: 10.1128/IAI.02585-14
McMillan, L. J., Hwang, S., Farah, R. E., Koh, J., Chen, S., Maupin-Furlow, J. A. (2018). Multiplex quantitative SILAC for analysis of archaeal proteomes: a case study of oxidative stress responses. Environ. Microbiol. 20, 385–401. doi: 10.1111/1462-2920.14014
Mittal, R., Peak-Chew., S. Y., Sade, R. S., Vallis, Y., McMahon, H. T. (2010). The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J. Biol. Chem. 285, 19927–19934. doi: 10.1074/jbc.M110.126581
Miyagi, M. (2017). Site-specific quantification of lysine acetylation using isotopic labeling. In Method. Enzymol. 586, 85–95. doi: 10.1016/bs.mie.2016.09.029
Mizuno, Y., Nagano-Shoji, M., Kubo, S., Kawamura, Y., Yoshida, A., Kawasaki, H., et al. (2016). Altered acetylation and succinylation profiles in Corynebacterium glutamicum in response to conditions inducing glutamate overproduction. Microbiologyope. 5, 152–173. doi: 10.1002/mbo3.320
Mochalkin, I., Knafels, J. D., Lightle, S. (2008). Crystal structure of LpxC from Pseudomonas aeruginosa complexed with the potent BB-78485 inhibitor. Prot. Sci. 17, 450–457. doi: 10.1110/ps.073324108
Mukherjee, S., Hao, Y. H., Orth, K. (2007). A newly discovered post-translational modification–the acetylation of serine and threonine residues. TIBS. 32, 210–216. doi: 10.1016/j.tibs.2007.03.007
Myers, S. P., Hawrelak, J. (2004). The causes of intestinal dysbiosis: a review. Altern. Med. Rev. 9, 180–197.
Nagano-Shoji, M., Hamamoto, Y., Mizuno, Y., Yamada, A., Kikuchi, M., Shirouzu, M., et al. (2017). Characterization of lysine acetylation of a phosphoenolpyruvate carboxylase involved in glutamate overproduction in Corynebacterium glutamicum. Mol. Microbiol. 104, 677–689. doi: 10.1111/mmi.13658
Nakayasu, E. S., Burnet, M. C., Walukiewicz, H. E., Wilkins, C. S., Shukla, A. K., Brooks, S., et al. (2017). Ancient regulatory role of lysine acetylation in central metabolism. mBio. 8 (6), e01894-17. doi: 10.1128/mbio.01894-17
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I., Klaholz, B. P. (2017). Visualization of chemical modifications in the human 80S ribosome structure. Nature. 551, 472–477. doi: 10.1038/nature24482
Neumann, H., Peak-Chew, S. Y., Chin, J. W. (2008). Genetically encoding N ϵ-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234. doi: 10.1038/nchembio.73
Neumann-Staubitz, P., Lammers, M., Neumann, H. (2021). Genetic code expansion tools to study lysine acylation. Adv. Biol. 5, 2100926. doi: 10.1002/adbi.202100926
Ni, J., Li, S., Lai, Y., Wang, Z., Wang, D., Tan, Y., et al. (2023). Global profiling of ribosomal protein acetylation reveals essentiality of acetylation homeostasis in maintaining ribosome assembly and function. Nucleic Acids Res. 51, 10411–10427. doi: 10.1093/nar/gkad768
Nielsen, T. K., Hildmann, C., Dickmanns, A., Schwienhorst, A., Ficner, R. (2005). Crystal structure of a bacterial class 2 histone deacetylase homologue. J. Mol. Biol. 354, 107–120. doi: 10.1016/j.jmb.2005.09.065
North, B. J., Verdin, E. (2004). Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5, 1–12. doi: 10.1186/gb-2004-5-5-224
Novak, K., Flöckner, L., Erian, A. M., Freitag, P., Herwig, C., Pflügl, S. (2018). Characterizing the effect of expression of an acetyl-CoA synthetase insensitive to acetylation on co-utilization of glucose and acetate in batch and continuous cultures of E. coli W. Microb. Cell Fact. 17, 1–15. doi: 10.1186/s12934-018-0955-2
Oliveira, A. P., Sauer, U. (2012). The importance of post-translational modifications in regulating Saccharomyces cerevisiae metabolism. FEMS Yeast. Res. 12, 104–117. doi: 10.1111/j.1567-1364.2011.00765.x
O’Toole, P. W., Jeffery, I. B. (2015). Gut microbiota and aging. Science. 350, 1214–1215. doi: 10.1126/science.aac8469
Ouidir, T., Cosette, P., Jouenne, T., Hardouin, J. (2015). Proteomic profiling of lysine acetylation in Pseudomonas aeruginosa reveals the diversity of acetylated proteins. Proteomics. 15, 2152–2157. doi: 10.1002/pmic.201500056
Paczia, N., Nilgen, A., Lehmann, T., Gätgens, J., Wiechert, W., Noack, S. (2012). Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 11, 122. doi: 10.1186/1475-2859-11-122
Paik, W. K., Pearson, D., Lee, H. W., Kim, S. (1970). Nonenzymatic acetylation of histones with acetyl-CoA. Biochim. Biophys. Acta 213, 513–522. doi: 10.1016/0005-2787(70)90058-4
Pang, R., Li, Y., Liao, K., Guo, P., Li, Y., Yang, X., et al. (2020b). Genome-and proteome-wide analysis of lysine acetylation in Vibrio vulnificus Vv180806 reveals its regulatory roles in virulence and antibiotic resistance. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.591287
Pang, H., Li, W., Zhang, W., Zhou, S., Hoare, R., Monaghan, S. J., et al. (2020a). Acetylome profiling of Vibrio alginolyticus reveals its role in bacterial virulence. J. Proteomics. 211, 103543. doi: 10.1016/j.jprot.2019.103543
Paquette, N., Conlon, J., Sweet, C., Rus, F., Wilson, L., Pereira, A., et al. (2012). Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 12710–12715. doi: 10.1073/pnas.1008203109
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., Hooper, D. C. (2006). Prevalence in the United States of aac (6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06
Peebo, S. K., Valgepea, K., Nahku, R., Riis, G., Oun, M., Adamberg, K., et al. (2014). Coordinated activation of PTA-ACS and TCA cycles strongly reduces overflow metabolism of acetate in. Escherichia coli. Appl. Microbiol. Biotechnol. 98, 5131–5143. doi: 10.1007/s00253-014-5613-y
Peng, B. O., Su, Y. B., Li, H., Han, Y. I., Guo, C., Tian, Y. M., et al. (2015). Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262. doi: 10.1016/j.cmet.2015.01.008
Phillips, D. M. P. (1963). The presence of acetyl groups in histones. J. Biochem. 87, 258. doi: 10.1042/bj0870258
Pinu, F. R., Granucci, N., Daniell, J., Han, T. L., Carneiro, S., Rocha, I., et al. (2018). Metabolite secretion in microorganisms: the theory of metabolic overflow put to the test. Metabolomics 14, 1–16. doi: 10.1007/s11306-018-1339-7
Plekhanova, N. S., Altman, I. B., Yurkova, M. S., Fedorov, A. N. (2023). The Effects of Nϵ-acetylation on the enzymatic activity of Escherichia coli glyceraldehyde-3-phosphate dehydrogenase. Appl. Biochem. Microbiol. 59, 778–785. doi: 10.1134/S000368382306011X
Post, D. M., Schilling, B., Reinders, L. M., D’Souza, A. K., Ketterer, M. R., Kiel, S. J., et al. (2017). Identification and characterization of AckA-dependent protein acetylation in Neisseria gonorrhoeae. PLoS One 12, e0179621. doi: 10.1371/journal.pone.0179621
Ramazi, S., Zahiri, J. (2021). Post-translational modifications in proteins: resources, tools and prediction methods. DATABASE-OXFORD 2021, 1–20. doi: 10.1093/database/baab012
Ramos, S., Martín, M.Á. (2021). Impact of diet on gut microbiota. Curr. Opin. Food Sci. 37, 83–90. doi: 10.1016/j.cofs.2020.09.006
Ramos-Montañez, S., Kazmierczak, K. M., Hentchel, K. L., Winkler, M. E. (2010). Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 192, 6390–6400. doi: 10.1128/JB.00995-10
Ren, J., Sang, Y., Lu, J., Yao, Y. F. (2017). Protein acetylation and its role in bacterial virulence. Trends Microbiol. 25, 768–779. doi: 10.1016/j.tim.2017.04.001
Ren, J., Sang, Y., Ni, J., Tao, J., Lu, J., Zhao, M., et al. (2015). Acetylation regulates survival of Salmonella enterica serovar Typhimurium under acid stress. Appl. Environ. Microbiol. 81, 5675–5682. doi: 10.1128/AEM.01009-15
Ren, J., Sang, Y., Tan, Y., Tao, J., Ni, J., Liu, S., et al. (2016). Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS pathogens. 12, e1005458. doi: 10.1371/journal.ppat.1005458
Reverdy, A., Chen, Y., Hunter, E., Gozzi, K., Chai, Y. (2018). Protein lysine acetylation plays a regulatory role in Bacillus subtilis multicellularity. PLoS One 13, e0204687. doi: 10.1371/journal.pone.0204687
Richards, C. L., Lawrence, K. A., Su, H., Yang, Y., Yang, X. F., Dulebohn, D. P., et al. (2015). Acetyl-phosphate is not a global regulatory bridge between virulence and central metabolism in Borrelia burgdorferi. PLoS One 10, e0144472. doi: 10.1371/journal.pone.0144472
Rine, J., Herskowitz, I. (1987). Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 116, 9–22. doi: 10.1093/genetics/116.1.9
Robicsek, A., Strahilevitz, J., Jacoby, G. A., Macielag, M., Abbanat, D., Hye Park, C., et al. (2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. doi: 10.1038/nm1347
Rojas, J. R., Trievel, R. C., Zhou, J., Mo, Y., Li, X., Berger, S. L., et al. (1999). Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 401, 93–98. doi: 10.1038/43487
Ruijter, A. J. D., Gennip, A. H. V., Caron, H. N., Kemp, S., Kuilenburg, A. B. V. (2003). Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749. doi: 10.1042/bj20021321
Salah Ud-Din, A. I. M., Tikhomirova, A., Roujeinikova, A. (2016). Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). Int. J. Mol. Sci. 17, 1018. doi: 10.3390/ijms17071018
Sanders, B. D., Jackson, B., Marmorstein, R. (2010). Structural basis for sirtuin function: what we know and what we don't. Biochim. Biophys. Acta 1804, 1604–1616. doi: 10.1016/j.bbapap.2009.09.009
Sang, Y., Ren, J., Ni, J., Tao, J., Lu, J., Yao, Y. F. (2016). Protein acetylation is involved in Salmonella enterica serovar Typhimurium virulence. J. Infect. Dis. 213, 1836–1845. doi: 10.1093/infdis/jiw028
Schastnaya, E., Doubleday, P. F., Maurer, L., Sauer, U. (2023). Non-enzymatic acetylation inhibits glycolytic enzymes in Escherichia coli. Cell Rep. 42, 111950. doi: 10.1016/j.celrep.2022.111950
Schilling, B., Basisty, N., Christensen, D. G., Sorensen, D., Orr, J. S., Wolfe, A. J., et al. (2019). Global lysine acetylation in Escherichia coli results from growth conditions that favor acetate fermentation. J. Bacteriol. 201, e00768–e00718. doi: 10.1128/JB.00768-18
Schilling, B., Christensen, D., Davis, R., Sahu, A. K., Hu, L. I., Walker-Peddakotla, A., et al. (2015). Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 98, 847–863. doi: 10.1111/mmi.13161
Schmidt, M. J., Summerer, D. (2014). Genetic code expansion as a tool to study regulatory processes of transcription. Front. Chem. 2. doi: 10.3389/fchem.2014.00007
Selmer, M., Dunham, C. M., Murphy, F. V., IV, Weixlbaumer, A., Petry, S., Kelley, A. C., et al. (2006). Structure of the 70 S ribosome complexed with mRNA and tRNA. Science. 313, 1935–1942. doi: 10.1126/science.1131127
Selwitz, R. H., Ismail, A. I., Pitts, N. B. (2007). Dental caries. Lancet. 369, 51–59. doi: 10.1016/S0140-6736(07)60031-2
Shahin, R., Javad, Z.. (2021). Post-translational modifications in proteins: resources, tools and prediction methods. Database 2021, baab012. doi: 10.1093/database/baab012
Shaw, K. J., Rather, P. N., Sabatelli, F. J., Mann, P., Munayyer, H., Mierzwa, R., et al. (1992). Characterization of the chromosomal aac (6')-Ic gene from Serratia marcescens. Antimicrob. Agents Chemother. 36, 1447–1455. doi: 10.1128/AAC.36.7.1447
Shirai, T., Fujimura, K., Furusawa, C., Nagahisa, K., Shioya, S., Shimizu, H. (2007). Study on roles of anaplerotic pathways in glutamate overproduction of Corynebacterium glutamicum by metabolic flux analysis. Microb. Cell Fact. 6, 1–11. doi: 10.1186/1475-2859-6-19
Simon, G. M., Cheng, J., Gordon, J. I. (2012). Quantitative assessment of the impact of the gut microbiota on lysine ϵ-acetylation of host proteins using gnotobiotic mice. PNAS. 109, 11133–11138. doi: 10.1073/pnas.1208669109
Slonczewski, J. L., Fujisawa, M., Dopson, M., Krulwich, T. A. (2009). Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 55, 1–317. doi: 10.1016/S0065-2911(09)05501-5
Somoza, J. R., Skene, R. J., Katz, B. A., Mol, C., Ho, J. D., Jennings, A. J., et al. (2004). Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 12, 1325–1334. doi: 10.1016/j.str.2004.04.012
Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., Escalante-Semerena, J. C. (2002). Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Sci. 298, 2390–2392. doi: 10.1126/science.1077650
Starai, V. J., Escalante-Semerena, J. C. (2004a). Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. CMLS. 61, 2020–2030. doi: 10.1007/s00018-004-3448-x
Starai, V. J., Escalante-Semerena, J. C. (2004b). Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012. doi: 10.1016/j.jmb.2004.05.010
Starai, V. J., Gardner, J. G., Escalante-Semerena, J. C. (2005). Residue Leu-641 of acetyl-CoA synthetase is critical for the acetylation of residue Lys-609 by the protein acetyltransferase enzyme of Salmonella enterica. J. Biol. Chem. 280, 26200–26205. doi: 10.1074/jbc.M504863200
Starai, V. J., Takahashi, H., Boeke, J. D., Escalante-Semerena, J. C. (2003). Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics. 163, 545–555. doi: 10.1093/genetics/163.2.545
Starai, V. J., Takahashi, H., Boeke, J. D., Escalante-Semerena, J. C. (2004). A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. COMICR. 7, 115–119. doi: 10.1016/j.mib.2004.02.005
Sun, M., Guo, H., Lu, G., Gu, J., Wang, X., Zhang, X. E., et al. (2016). Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase. Acta Biochim. Biophys. Sin. (Shanghai). 48, 723–731. doi: 10.1093/abbs/gmw066
Sun, X. L., Yang, Y. H., Zhu, L., Liu, F. Y., Xu, J. P., Huang, X. W., et al. (2018). The lysine acetylome of the nematocidal bacterium Bacillus nematocida and impact of nematode on the acetylome. J. Proteomics. 177, 31–39. doi: 10.1016/j.jprot.2018.02.005
Sun, L., Yao, Z., Guo, Z., Zhang, L., Wang, Y., Mao, R., et al. (2019). Comprehensive analysis of the lysine acetylome in Aeromonas hydrophila reveals cross-talk between lysine acetylation and succinylation in LuxS. Emerg. Microbes Infect. 8, 1229–1239. doi: 10.1080/22221751.2019.1656549
Sung, M. K., Porras-Yakushi, T. R., Reitsma, J. M., Huber, F. M., Sweredoski, M. J., Hoelz, A., et al. (2016). A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. 5, e19105. doi: 10.7554/eLife.19105.026
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tan, Y., Liu, W., Chen, Y., Zhou, Y., Song, K., Cao, S., et al. (2023). Comparative lysine acetylome analysis of Y. pestis YfiQ/CobB mutants reveals that acetylation of SlyA Lys73 significantly promotes biofilm formation of Y. pestis. Microbiol. Spectr. 11, e00460–e00423. doi: 10.1128/spectrum.00460–23
Tarazona, O. A., Pourquié, O. (2020). Exploring the influence of cell metabolism on cell fate through protein post-translational modifications. Dev. Cell. 54, 282–292. doi: 10.1016/j.devcel.2020.06.035
Taunton, J., Hassig, C. A., Schreiber, S. L. (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Sci. 272, 408–411. doi: 10.1126/science.272.5260.408
Teran, F. J., Alvarez, M., Suárez, J. E., Mendoza, M. C. (1991). Characterization of two aminoglycoside-(3)-N-acetyltransferase genes and assay as epidemiological probes. J. Antimicrob. Chemother. 28, 333–346. doi: 10.1093/jac/28.3.333
Tercero, J. C., Riles, L. E., Wickner, R. B. (1992). Localized mutagenesis and evidence for post-transcriptional regulation of MAK3. A putative N-acetyltransferase required for double-stranded RNA virus propagation in Saccharomyces cerevisiae. JBC. 267, 20270–20276. doi: 10.1016/S0021-9258(19)88696-9
Tsang, A. W., Escalante-Semerena, J. C. (1998). CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide: 5, 6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 273, 31788–31794. doi: 10.1074/jbc.273.48.31788
Tsimbalyuk, S., Shornikov, A., Kuhn, M. L., Forwood, J. K. (2020). SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency. J. Struct. Biol. 210, 107506. doi: 10.1016/j.jsb.2020.107506
VanDrisse, C. M., Escalante-Semerena, J. C. (2019). Protein acetylation in bacteria. Annu. Rev. Microbiol. 73, 111–132. doi: 10.1146/annurev-micro-020518-115526
Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., et al. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069. doi: 10.1073/pnas.0404603101
Venkat, S., Chen, H., McGuire, P., Stahman, A., Gan, Q., Fan, C. (2019). Characterizing lysine acetylation of Escherichia coli type II citrate synthase. FEBS J. 286, 2799–2808. doi: 10.1111/febs.14845
Venkat, S., Chen, H., Stahman, A., Hudson, D., McGuire, P., Gan, Q., et al. (2018a). Characterizing lysine acetylation of isocitrate dehydrogenase in Escherichia coli. J. Mol. Biol. 430, 1901–1911. doi: 10.1016/j.jmb.2018.04.031
Venkat, S., Gregory, C., Sturges, J., Gan, Q., Fan, C. (2017a). Studying the lysine acetylation of malate dehydrogenase. J. Mol. Biol. 429, 1396–1405. doi: 10.1016/j.jmb.2017.03.027
Venkat, S., Nannapaneni, D. T., Gregory, C., Gan, Q., McIntosh, M., Fan, C. (2017b). Genetically encoding thioacetyl-lysine as a non-deacetylatable analog of lysine acetylation in Escherichia coli. FEBS Open Bio. 7, 1805–1814. doi: 10.1002/2211-5463.12320
Venkat, S., Sturges, J., Stahman, A., Gregory, C., Gan, Q., Fan, C. (2018b). Genetically incorporating two distinct post-translational modifications into one protein simultaneously. ACS synthetic Biol. 7, 689–695. doi: 10.1021/acssynbio.7b00408
Vetting, M. W., de Carvalho, L. P. S., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L., et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226. doi: 10.1016/j.abb.2004.09.003
Vetting, M. W., Magnet, S., Nieves, E., Roderick, S. L., Blanchard, J. S. (2004). A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol. 11, 565–573. doi: 10.1016/j.chembiol.2004.03.017
Vetting, M. W., Park, C. H., Hegde, S. S., Jacoby, G. A., Hooper, D. C., Blanchard, J. S. (2008). Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC (6′)-Ib and its bifunctional, fluoroquinolone-active AAC (6′)-Ib-cr variant. Biochemistry. 47, 9825–9835. doi: 10.1021/bi800664x
Vetting, M. W., Roderick, S. L., Yu, M., Blanchard, J. S. (2003). Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases. Protein Sci. 12, 1954–1959. doi: 10.1110/ps.03153703
Vinuselvi, P., Kim, M. K., Lee, S. K., Ghim, C. M. (2012). Rewiring carbon catabolite repression for microbial cell factory. BMB Rep. 45, 59–70. doi: 10.5483/BMBRep.2012.45.2.59
Wagner, G. R., Payne, R. M. (2013). Widespread and enzyme-independent nϵ-acetylation and nϵ-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045. doi: 10.1074/jbc.M113.486753
Wagner, F. F., Weїwer, M., Lewis, M. C., Holson, E. B. (2013). Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics. 10, 589–604. doi: 10.1007/s13311-013-0226-1
Wang, J. Y. J., Koshland, D. E., Jr (1982). The reversible phosphorylation of isocitrate dehydrogenase of Salmonella typhimurium. Arch. Biochem. Biophys. 218, 59–67. doi: 10.1016/0003-9861(82)90321-6
Wang, L., Tang, Y., Cole, P. A., Marmorstein, R. (2008). Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18, 741–747. doi: 10.1016/j.sbi.2008.09.004
Wang, Y., Wang, F., Bao, X., Fu, L. (2019). Systematic analysis of lysine acetylome reveals potential functions of lysine acetylation in Shewanella baltica, the specific spoilage organism of aquatic products. J. Proteom. 205, 103419. doi: 10.1016/j.jprot.2019.103419
Wang, M. M., You, D., Ye, B. C. (2017). Site-specific and kinetic characterization of enzymatic and nonenzymatic protein acetylation in bacteria. Sci. Rep. 7, 14790. doi: 10.1038/s41598–017-13897-w
Wang, Q., Zhang, Y., Yang, C., Xiong, H., Lin, Y., Yao, J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 327, 1004–1007. doi: 10.1126/science.1179687
Wegner, A., Meiser, J., Weindl, D., Hiller, K. (2015). How metabolites modulate metabolic flux. Curr. Opin. Biotechnol. 34, 16–22. doi: 10.1016/j.copbio.2014.11.008
Wei, L., Meyer, J. G., Schilling, B. (2018). Quantification of site-specific protein lysine acetylation and succinylation stoichiometry using data-independent acquisition mass spectrometry. J. Vis. Exp. 4, 57209. doi: 10.3791/57209
Weinert, B. T., Iesmantavicius, V., Moustafa, T., Scholz, C., Wagner, S. A., Magnes, C., et al. (2014). Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. doi: 10.1002/msb.134766
Weinert, B. T., Iesmantavicius, V., Wagner, S. A., Schölz, C., Gummesson, B., Beli, P., et al. (2013). Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 51, 265–272. doi: 10.1016/j.molcel.2013.06.003
Weinert, B. T., Satpathy, S., Hansen, B. K., Lyon, D., Jensen, L. J., Choudhary, C. (2017). Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol. Cell Proteomics. 16, 759–769. doi: 10.1074/mcp.M117.067587
Whittington, D. A., Rusche, K. M., Shin, H., Fierke, C. A., Christianson, D. W. (2003). Crystal structure of LpxC, a zinc-dependent deacetylase essential for endotoxin biosynthesis. PNAS. 100, 8146–8150. doi: 10.1073/pnas.1432990100
WHO. (2023). Antimicrobial resistance. Available online at: http://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed 28 November 2023).
Wolf, E., Vassilev, A., Makino, Y., Sali, A., Nakatani, Y., Burley, S. K. (1998). Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 94, 439–449. doi: 10.1016/S0092-8674(00)81585-8
Woolridge, D. P., Martinez, J. D., Stringer, D. E., Gerner, E. W. (1999). Characterization of a novel spermidine/spermine acetyltransferase, BltD, from Bacillus subtilis. Biochem. J. 340, 753–758. doi: 10.1042/bj3400753
Wright, G. D. (2005). Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57, 1451–1470. doi: 10.1016/j.addr.2005.04.002
Wright, G. D., Ladak, P. (1997). Overexpression and characterization of the chromosomal aminoglycoside 6'-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 41, 956–960. doi: 10.1128/AAC.41.5.956
Wu, X., Vellaichamy, A., Wang, D., Zamdborg, L., Kelleher, N. L., Huber, S. C., et al. (2013). Differential lysine acetylation profiles of Erwinia amylovora strains revealed by proteomics. J. Proteomics. 79, 60–71. doi: 10.1016/j.jprot.2012.12.001
Wybenga-Groot, L. E., Draker, K., Wright, G. D., Berghuis, A. M. (1999). Crystal structure of an aminoglycoside 6´-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure. 7, 497–507. doi: 10.1016/S0969-2126(99)80066-5
Xiao, X., Li, W., Pan, Y., Wang, J., Wei, Z., Wang, S., et al. (2023). Holistic analysis of lysine acetylation in aquaculture pathogenic bacteria Vibrio alginolyticus under bile salt stress. Front. Vet. Sci. 10. doi: 10.3389/fvets.2023.1099255
Xie, L., Fang, W., Deng, W., Yu, Z., Li, J., Chen, M., et al. (2016). Global profiling of lysine acetylation in human histoplasmosis pathogen Histoplasma capsulatum. Int. J. Biochem. Cell Biol. 73, 1–10. doi: 10.1016/j.biocel.2016.01.008
Xie, L., Wang, X., Zeng, J., Xie, L., Wang, X., Zeng, J., et al. (2015). Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol. 59, 193–202. doi: 10.1016/j.biocel.2014.11.010
Xie, L., Zeng, J., Luo, H., Pan, W., Xie, J. (2014). The roles of bacterial GCN5-related N-acetyltransferases. Crit. Rev. Eukaryot. Gene Expr. 24, 77–87. doi: 10.1615/CritRevEukaryotGeneExpr.v24.i1
Xiong, Y., Guan, K. L. (2012). Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell. Biol. 198, 155–164. doi: 10.1083/jcb.201202056
Xu, H., Hegde, S. S., Blanchard, J. S. (2011). Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry. 50, 5883–5892. doi: 10.1021/bi200156t
Xu, J. Y., Xu, Z., Liu, X., Tan, M., Ye, B. C. (2018). Protein acetylation and butyrylation regulate the phenotype and metabolic shifts of the endospore-forming Clostridium acetobutylicum. Mol. Cell Proteomics. 17, 1156–1169. doi: 10.1074/mcp.RA117.000372
Xu, H., Zhou, J., Lin, S., Deng, W., Zhang, Y., Xue, Y. (2017). PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics 44, 243–250. doi: 10.1016/j.jgg.2017.03.007
Yan, Y., Harper, S., Speicher, D. W., Marmorstein, R. (2002). The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9, 862–869. doi: 10.1038/nsb849
Yang, X. J., Seto, E. (2008). The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 9, 206–218. doi: 10.1038/nrm2346
Yang, H., Sha, W., Liu, Z., Tang, T., Liu, H., Qin, L., et al. (2018). Lysine acetylation of DosR regulates the hypoxia response of Mycobacterium tuberculosis. Emerg. Microbes Infect. 7, 1–14. doi: 10.1038/s41426-018-0032-2
Yao, J., Wang, Z. N., Liu, H., Jin, H., Zhang, Y. (2023). Survey of acetylation for Thermoanaerobacter tengcongensis. Appl. Biochem. Biotechnol. 195, 6081–6097. doi: 10.1007/s12010-023-04361-9
You, D., Yao, L. L., Huang, D., Escalante-Semerena, J. C., Ye, B. C. (2014). Acetyl coenzyme A synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with a single Gcn5-type N-acetyltransferase (GNAT) domain in Saccharopolyspora erythraea. J. Bacteriol. 196, 3169–3178. doi: 10.1128/JB.01961-14
Yu, D., Du, J., Pu, X., Zheng, L., Chen, S., Wang, N., et al. (2022). The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.763507
Yu, B. J., Kim, J., Moon, J. H., Ryu, S. E., Pan, J. G. (2008). The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18, 1529–1536.
Yuan, H., Marmorstein, R. (2012). Structural basis for sirtuin activity and inhibition. J. Biol. Chem. 287, 42428–42435. doi: 10.1074/jbc.R112.372300
Zalewska-Piątek, B. (2023). Phage therapy—Challenges, opportunities and future prospects. Pharmaceuticals. 16, 1638. doi: 10.3390/ph16121638
Zaunbrecher, M. A., Sikes, R. D., Metchock, B., Shinnick, T. M., Posey, J. E. (2009). Overexpression of the chromosomally encoded aminoglycoside acetyltransferase is confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 106, 20004–20009. doi: 10.1073/pnas.0907925106
Zhang, B. Q., Bu, H. L., You, D., Ye, B. C. (2020a). Acetylation of translation machinery affected protein translation in E. coli. Appl. Microbiol. Biotechnol. 104, 10697–10709. doi: 10.1007/s00253-020-10985-2
Zhang, X., Cheng, K., Ning, Z., Mayne, J., Walker, K., Chi, H., et al. (2021). Exploring the microbiome-wide lysine acetylation, succinylation, and propionylation in human gut microbiota. Anal. Chem. 93, 6594–6598. doi: 10.1021/acs.analchem.1c00962
Zhang, L., Elias, J. E. (2017). Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol. Biol. 1550, 185-198. doi: 10.1007/978-1-4939-7108-4
Zhang, Y., Li, N., Wei, Q., Min, R., Liu, F., Wang, F., et al. (2022). Lysine acetylome profiling reveals diverse functions of acetylation in Deinococcus radiodurans. Microbiol. Spectr. 10, e01016–e01021. doi: 10.1128/spectrum.01016-21
Zhang, M., Liu, T., Wang, L., Huang, Y., Fan, R., Ma, K., et al. (2023). Global landscape of lysine acylomes in Bacillus subtilis. J. Proteomics. 271, 104767. doi: 10.1016/j.jprot.2022.104767
Zhang, X., Ning, Z., Mayne, J., Yang, Y., Deeke, S. A., Walker, K., et al. (2020b). Widespread protein lysine acetylation in gut microbiome and its alterations in patients with Crohn’s disease. Nat. Commun. 11, 4120. doi: 10.1038/s41467-020-17916-9
Zhang, X., Ouyang, S., Kong, X., Liang, Z., Lu, J., Zhu, K., et al. (2014). Catalytic mechanism of histone acetyltransferase p300: from the proton transfer to acetylation reaction. J. Phys. Chem. B. 118, 2009–2019. doi: 10.1021/jp409778e
Zhang, J., Sprung, R., Pei, J., Tan, X., Kim, S., Zhu, H., et al. (2009). Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 8, 215–225. doi: 10.1074/mcp.M800187-MCP200
Zhao, K., Chai, X., Marmorstein, R. (2004). Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 337, 731–741. doi: 10.1016/j.jmb.2004.01.060
Keywords: bacteria, protein, acetylation, functions, methods
Citation: Rizo J and Encarnación-Guevara S (2024) Bacterial protein acetylation: mechanisms, functions, and methods for study. Front. Cell. Infect. Microbiol. 14:1408947. doi: 10.3389/fcimb.2024.1408947
Received: 29 March 2024; Accepted: 03 June 2024;
Published: 04 July 2024.
Edited by:
Alessandro Russo, Magna Græcia University, ItalyReviewed by:
Marat R. Sadykov, University of Nebraska Medical Center, United StatesCopyright © 2024 Rizo and Encarnación-Guevara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergio Encarnación-Guevara, ZW5jYXJuYWNAY2NnLnVuYW0ubXg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.