- Quorum Sensing Laboratory, Centre for Research in Infectious Diseases (CRID), School of Chemical and Biotechnology, SASTRA Deemed to be University, Thanjavur, India
Infectious diseases represent a significant global health challenge, with bacteria, fungi, viruses, and parasitic protozoa being significant causative agents. The shared symptoms among diseases and the emergence of new pathogen variations make diagnosis and treatment complex. Conventional diagnostic methods are laborious and intricate, underscoring the need for rapid, accurate techniques. Aptamer-based technologies offer a promising solution, as they are cost-effective, sensitive, specific, and convenient for molecular disease diagnosis. Aptamers, which are single-stranded RNA or DNA sequences, serve as nucleotide equivalents of monoclonal antibodies, displaying high specificity and affinity for target molecules. They are structurally robust, allowing for long-term storage without substantial activity loss. Aptamers find applications in diverse fields such as drug screening, material science, and environmental monitoring. In biomedicine, they are extensively studied for biomarker detection, diagnostics, imaging, and targeted therapy. This comprehensive review focuses on the utility of aptamers in managing infectious diseases, particularly in the realms of diagnostics and therapeutics.
1 Introduction
Pathogens, such as bacteria, fungi, viruses, or parasitic protozoa transmitted throughout populations, are typically the source of infectious diseases, some recognized as potentially fatal (Wan et al., 2021; Krüger et al., 2021; Zhang et al., 2021). Infectious diseases continue to be a significant global public health concern, representing the primary causes of morbidity and mortality (Cohen, 2000). Similar signs and symptoms are common among numerous diseases, and the diagnosis, treatment, and management of infectious diseases may face significant difficulties due to the emergence of novel pathogens as well as the reappearance and rise of previously identified pathogen variations (Chen et al., 2022b) (Wan et al., 2021). The rise of antimicrobial resistance can be attributed to the improper or empirical use of antibiotics in the treatment of infections. This underscores the need for careful and evidence-based antibiotic management in addressing infectious diseases (Fair and Tor, 2014; Rahbi et al., 2023). While laboratory testing, imaging scans, and biopsies based on clinical signs and epidemiological data have been successfully used to identify infections, these conventional procedures are either labor-intensive or highly complex (Wan et al., 2021; Zhang et al., 2021).
Therefore, it is imperative to develop new, quick, and precise diagnostic and therapeutic techniques to address the issues of drug resistance and anti-microbial resistance (Krüger et al., 2021). Aptamer-based diagnostic technologies are among the diagnostic approaches that are rapidly being employed for molecular disease diagnosis due to their cost-effectiveness, sensitivity, specificity, and convenience (Wan et al., 2021). The Latin word “aptus”, which means “to fit,” and the Greek word “meros”, which means “region,” are the sources of the word “aptamer” (Ku et al., 2015). Aptamers, single-stranded RNA or DNA oligonucleotide sequences with a length of approximately 25–80 bases, are the nucleotide counterparts of monoclonal antibodies. They may bind target molecules with high affinity and specificity, demonstrating the nucleic acid’s multifunctional nature (Ni et al., 2021). Aptamers offer a range of benefits, such as being cost-effective, exhibiting minimal batch-to-batch variation, demonstrating low immunogenicity, and possessing a small size for improved tissue penetration (Otte et al., 2022). Despite their potential, aptamers are constrained by their rapid clearance through renal filtration and susceptibility to nuclease hydrolysis, leading to a very short half-life in vivo (Kovacevic et al., 2018; Ni et al., 2021). In response to these limitations, several techniques have been developed to extend the half-life. These include PEGylation for sustained action, modification of sugar ring or base, phosphodiester linkage, and 3′ end capping with inverted thymidine (Ni et al., 2017). Due to their structural stability, aptamers can be manufactured in large quantities and stored for extended periods without significant activity loss (Srivastava et al., 2021).
Various aptamers have been developed against various targets such as hormones, viruses, metal ions, proteins, viruses, and bacteria (Zhou and Rossi, 2017; Shraim et al., 2022). These complexes form stable and specific targets with dissociation constants in the nanomolar range. Additionally, aptamers have a greater target range, it is easier to regenerate, substantially smaller, and is neither poisonous nor immunogenic (García-Recio et al., 2016; Zheng et al., 2015; Roxo et al., 2019). New aptamer reports are released nearly daily due to their broad applicability. A specific database has been built (https://sites.utexas.edu/aptamerdatabase) to classify the aptamer-related data and enable access to information about various existent aptamers (Askari et al., 2024). Aptamers have drawn a lot of interest in the biomedical community due to their unique qualities and wide applications in a variety of sectors, including drug screening, material science, and environmental monitoring (Chen et al., 2022a).
Aptamers have been extensively studied and developed over the past 20 years by researchers in several biomedical fields, including biomarker detection, diagnostics, imaging, and targeted therapy. Aptamers that are now utilized in cancer treatment can bind to and block the immunoregulatory components of carcinogenesis, which are particular to molecular targets that are characteristic of various diseases. In December 2004, the US Food and Drug Administration approved pegaptanib (Macugen), the first medication based on aptamer technology, for the treatment of age-related macular degeneration (Adachi and Nakamura, 2019). Despite the lack of new aptamers approved for clinical use, there is promising progress in the development of aptamers for blood disorders, with several of them currently undergoing different stages of clinical trials and proof-of-concept investigations (Aljohani et al., 2022). Aptamers demonstrate a wide range of applications, highlighting their versatile nature in the field of infectious diseases. Thus, the review provides an in-depth insight into the general mechanism of aptamer selection and its applications in the diagnostic and therapeutic fields. Furthermore, it addresses recent advances and challenges in the field of aptamers, aiming to inspire further exploration of aptamer-based approaches in combating infectious diseases.
2 Mechanisms of aptamer selection
The process of aptamer selection includes a range of methodologies designed to identify nucleic acid sequences that can bind specific target molecules with high affinity and specificity (Kinghorn et al., 2017). Both SELEX (Systematic Evolution of Ligands by Exponential Enrichment) and non-SELEX approaches are used to refine methods. SELEX employs iterative rounds of selection, in which a nucleic acid library interacts with the target molecule under controlled conditions, to enhance sequences with optimal binding properties (Uemachi et al., 2021). Contrastingly, Non-SELEX methods steer clear of traditional scaffold-based approaches, opting instead for innovative strategies to bolster aptamer stability, specificity, and interaction dynamics (Kong and Byun, 2013). These diverse methodologies empower researchers to confidently tailor aptamer selection processes according to the specific requirements of their applications, from diagnostics to therapeutic interventions.
2.1 Systematic evolution of ligands by exponential enrichment
SELEX is a method used to derive aptamers from a pool of nucleotide sequences that exhibit high affinity and selectivity (Chen et al., 2016). The process involves several key steps to select aptamers through a repetitive cycle of amplification and enrichment. Initially, a large and diverse library of nucleic acid sequences (DNA or RNA) is synthesized and incubated with the target molecule in an appropriate buffer at a specific temperature (Sun et al., 2014). The partitioning or the eluting steps involves removal of the unbound nucleotide by chromatography, electrophoresis or filtration (Dong et al., 2018). A low ratio of nucleic acid sequences to the target molecule is used, ensuring effective binding. The aptamer-target complexes are then separated from unbound sequences using techniques such as capillary electrophoresis (Hamedani and Müller, 2016), magnetic bead separation (Yüce et al., 2015), and flow cell methodologies (Gopinath, 2007).
The bound sequences are eluted from the target and amplified using PCR for DNA aptamers or reverse transcription followed by PCR for RNA aptamers, creating a new, enriched library. These processes are repeated for several rounds, typically 8-15, to enhance the prevalence of high-affinity species, which eventually dominate the library (Zhou and Rossi, 2017) (Figure 1). After multiple rounds of selection, the enriched library is cloned and sequenced to identify individual aptamer sequences, which are then validated for their binding performance. Through these iterative rounds, SELEX effectively isolates aptamers that can bind to specific target molecules with high affinity and specificity. However, a common drawback of aptamers derived from traditional SELEX methods is poor or nonspecific detection performance in diagnostic applications (Bakhtiari et al., 2021). To overcome these shortfalls, different methodologies are incorporated over conventional SELEX, some of which are discussed below.
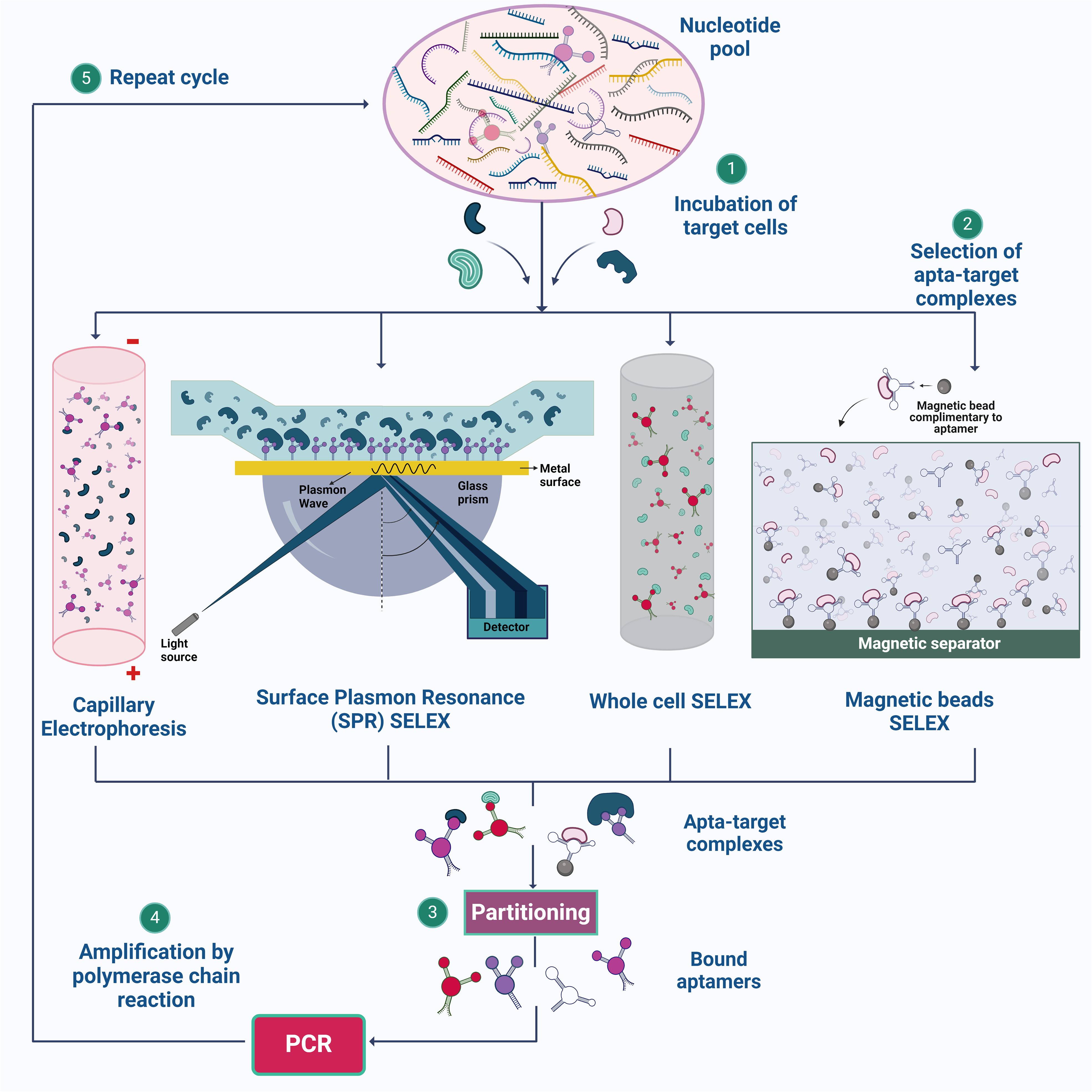
Figure 1. Illustration of SELEX strategies for aptamer synthesis. (1) The process begins with the preparation of a nucleotide pool, which is then incubated with target cells. (2) Various SELEX methods, such as Capillary Electrophoresis SELEX, SPR-SELEX, Whole Cell SELEX, and Magnetic Beads SELEX, are employed to facilitate the selection of aptamer-target complexes. (3) & (4) These complexes are isolated, and the bound aptamers are subsequently amplified by PCR. (5) The cycle is repeated multiple times to enhance the specificity and affinity of the aptamers for their targets. Created using Biorender.com.
2.1.1 Magnetic beads SELEX
Magnetic SELEX is a method that is commonly employed as this method offers ease in the separation of the target and nucleotide sequence easily from the remaining reaction mixture by employing a magnet (Yüce et al., 2015). When the DNA sequence binds with the target molecule, the mixture is now added with magnetic beads coated with a molecule that can selectively bind with the nucleic acid sequence attached to the target molecule. The elution of the bound nucleic acid sequences from the magnetic beads is achieved by altering the buffer’s properties, applying heat, or utilizing other methods that hinder the nucleic acids’ binding to the magnetic beads (Komarova and Kuznetsov, 2019). In previous research, the isolation of Metamitron (MTM) aptamers using magnetic-bead SELEX has been successful. MTM, a widely used herbicide in agriculture, has been the subject of a thorough investigation. It is important to note that even with significant exposure, the negative health effects on humans are minimal. Following ten rounds of screening, high-throughput sequencing successfully identified six outstanding candidate aptamers with remarkable affinity and specificity (Xie et al., 2022).
2.1.2 Capillary electrophoresis
Apart from using traditional gel electrophoresis, capillary electrophoresis (CE) is employed to derive aptamer candidates on the metrics of sizes and charge; under the electric field, capillary electrophoresis can separate molecules as tiny as porphyrin30 (Yüce et al., 2015). When performing CE-SELEX, the target molecules are subjected to incubation with the random library in free solution, and the resulting combination of free target molecules, target-ssDNA complexes, and free ssDNA is then fed into a capillary column, then split apart using a high voltage. Taking a sample of the output fraction at the designated retention time, target-bond ssDNA provides the chance to collect DNA aptamers that bind to a specific target (Hamedani and Müller, 2016). Demonstrating the perspective of CE-SELEX for small-molecule targets in just four rounds. Small-molecule targets are anticipated to alter the mobility of the complex only slightly from the nonbinding sequences, leading to only partial separation of the bound and unbound sequences. However, even if just a tiny amount of the complex can be recovered, adequate enrichment can be accomplished since nucleic acids can be exponentially amplified by polymerase chain reaction (PCR). Additionally, repeated recurrent rounds of enrichment can eventually lead to the evolution of an abundant pool with high quality, even in the situation of separation with poor resolution (Yang and Bowser, 2013).
When CE-SELEX and high throughput sequencing (HTS) gave higher efficiency with faster separation of target-ssDNA complex and free ssDNA in free solution, aptamers can be chosen with relatively fewer rounds of selection thanks to HTS, which offers insight into the sequence evolution during the CE-SELEX process and makes it possible to characterize the entire evolutionary path. This reduces the need for the pool to occupy a consensus sequence and increases selection efficiency (Zhu et al., 2021).
2.1.3 Whole cell SELEX
While the major targets for the other SELEX techniques are highly purified targets, whole cell-SELEX uses a complete cell as the target. The cell-SELEX procedure may aim for extracellular cell surface proteins or unidentified cell structures. This SELEX approach makes it possible to create whole-cell targeting aptamers without much prior information on the cell’s surface proteins, which facilitates the identification of new biomarkers primarily for diagnosis and imaging (Yüce et al., 2015). The whole-cell SELEX method is used to create highly selective aptamers by different rounds of SELEX and counter SELEX. Aptamer can be separated using methods such as flow cytometry, Magnetic-Activated Cell Sorting, Differential Centrifugation, and Label-free methods. Whole-cell SELEX yields aptamers with high affinity and specificity when targeting bacterial surface compounds and live bacterial cells. Flow cytometry is a vital method for identifying target aptamers that bind selectively to cells. The technique overcomes the limitations of whole-cell SELEX by sorting, counting, and detecting fluorescence (Moon et al., 2013). Within flow cytometry techniques, fluorescence-Activated Cell Sorting (FACS) technique offers the ability to simultaneously differentiate and separate cell subpopulations, facilitating the identification of bound and unbound aptamers with specificity along with isolation of functional nucleic acids. By utilizing a sorting device that efficiently separates specific cells based on their fluorescence, FACS streamlines the process of finding aptamers that target different cell types contributing to a better yield of the aptamer candidates. The effectiveness of FACS in SELEX for functional aptamer selection is apparent in its successful separation of E. coli cells that produce RNA mimics (Nishimoto et al., 2007; Mayer et al., 2010; Zou et al., 2015). FACS is an effective method for large-scale aptamer screening because it is a fast and accurate technique that can process thousands of cells per second. It can sort cells based on multiple parameters and select aptamers based on their binding to live cells or complex mixtures, which may be more representative of physiological conditions than selections made in vitro. The possibility of obtaining high-quality aptamers is increased by the capacity to sort and enrich high-affinity binders from a huge library. FACS employs both positive and negative selection strategy, thereby reducing the experimental steps and experimental errors in the cell SELEX process, hence saves times. DNA Aptamers against Burkitt’s lymphoma cells which exhibit a characteristic phenotype was chosen using positive selection methods (Ohuchi, 2012; Raddatz et al., 2008; Sola et al., 2020). Despite the benefits of the cell-SELEX system, the low aptamer enrichment performance of this technique is caused by the co-expression of several off-target surface indicators and compounds on the target cells (Sun et al., 2014).
2.1.4 Surface plasmon resonance or flow cell SELEX
SPR- SELEX utilizes SPR for the selection process, differentiating it from the other methods. A Randomized library is passed over a surface coated (gold surface) with the target molecule (Yüce et al., 2015). In the library, a diverse range of oligonucleotides interact with the target in various ways. Oligonucleotides demonstrating strong binding will firmly adhere to the target-coated surface, while those with weak binding or unbound sequences will be effectively washed away (Jia et al., 2018). In SPR the nucleic acid sequence bound to the target molecule will be monitored in real time by observing the change in the refractive index on the surface leading to the change in surface plasmon signal (Ferhan et al., 2016). With the help of the above-mentioned steps, specific aptamer candidates are carefully selected and amplified using PCR (Jia et al., 2018).
2.2 Non-SELEX methods
SELEX uses a nucleic acid scaffold to develop the aptamer; however, other techniques do not require scaffolds (Reverdatto et al., 2015). For instance, aptamers are produced in the RNase III-deficient E. coli HT115(DE3), and 5′- and 3′ ends of the RNA transcript are protected from the RNase using double stranded spacers. This method only required fewer nucleotides than scaffold-based methods like the other different types of SELEX used to avoid RNase activity on the formed aptamer (Zou et al., 2023). PhotoSELEX, featuring photoreactive nucleic acids, confidently enhances control over the selection process. Upon exposure to light, the photoreactive groups confidently form covalent bonds between the selected aptamers and the target molecule, confidently providing a reliable method for identifying and capturing aptamer-target complexes (Brody et al., 1999). Graphene oxide (GO) is composed of carbon atoms arranged in a hexagonal lattice. Its unique properties allow for the immobilization of arbitrary DNA or RNA sequences on its surface, forming an oligonucleotide library with diverse sequences. During the GO-SELEX process, the target molecule interacts with the library-immobilized sequences. In the presence of the target molecule, the immobilized sequences on the GO surface are released and precisely interact with the target. This stage allows for the selection of aptamers with a high affinity for the target molecule (Nguyen et al., 2014; Ding and Liu, 2023). In the Capture-SELEX process, a DNA library is immobilized onto a substrate. The target of interest is then passed through to extract eluted aptamers. Aptamers are specifically chosen using this strategy for solute targets (Boussebayle et al., 2019). These non-SELEX methods provide versatile alternatives, overcoming challenges such as RNase degradation, and enhancing binding affinity through innovative selection techniques.
The aptamers that are selected can be used in various applications. One groundbreaking application is the use of apta-sensors for detecting infectious diseases. These biosensors use aptamers as recognition elements, and they provide fast, sensitive, and specific detection of pathogens. By incorporating aptamers selected through SELEX or Non-SELEX methods, apta-sensors can accurately detect infectious agents, greatly improving diagnostic capabilities. Their versatility and ability to detect a wide range of pathogens make aptasensors extremely valuable tools in epidemiology, healthcare settings, and biodefense (Brosseau et al., 2023).
3 Aptamers in diagnostics of infectious diseases
Traditional methodologies for detection encompass culture-based techniques and color culture medium approaches. However, these methodologies are encumbered by limitations, necessitating professional expertise, and demanding cumbersome labor and time commitments. The procedural intricacies include pre-enrichment, selective enrichment, and biochemical identification, typically leading to a confirmed outcome after 2-3 days (Bell et al., 2016). Due to the limitations present in these methods, there is a need for more efficient, rapid and accurate diagnostic methods. Immunological assays, such as ELISA and immunosensors, are commonly used for bacterial detection. However, their sensitivity is limited because proteins like immunoglobulins cannot be amplified. Furthermore, nucleic acid-based assays are unable to distinguish between viable and non-viable cells, as DNA can persist in the environment long after cell viability has been lost. This creates a need for a more specific, sensitive, and convenient diagnostic method that can bridge the gap between the detection. Aptamer-based assays are utilized for the detection of pathogens and biomarkers. Aptamers synthesis is rapid compared to the antibody production, and these rapid turnaround time helps in timely diagnosis. Furthermore, they have increased stability and shelf life compared to antibodies and reduced risk of immunogenicity due to ease of modifications that increase the stability, binding affinity and functionality (Ali et al., 2019). These assays enhance detection methods by providing improved specificity and sensitivity even at lower concentrations compared to traditional methods (Aslan et al., 2023). By delivering rapid results, which are ideal for point-of-care settings, this approach enhances diagnostic efficiency across various healthcare applications (Majdinasab et al., 2022). Further, the review delineates a comprehensive analysis of the diverse categories of apta-sensors. (Figure 2, Table 1).
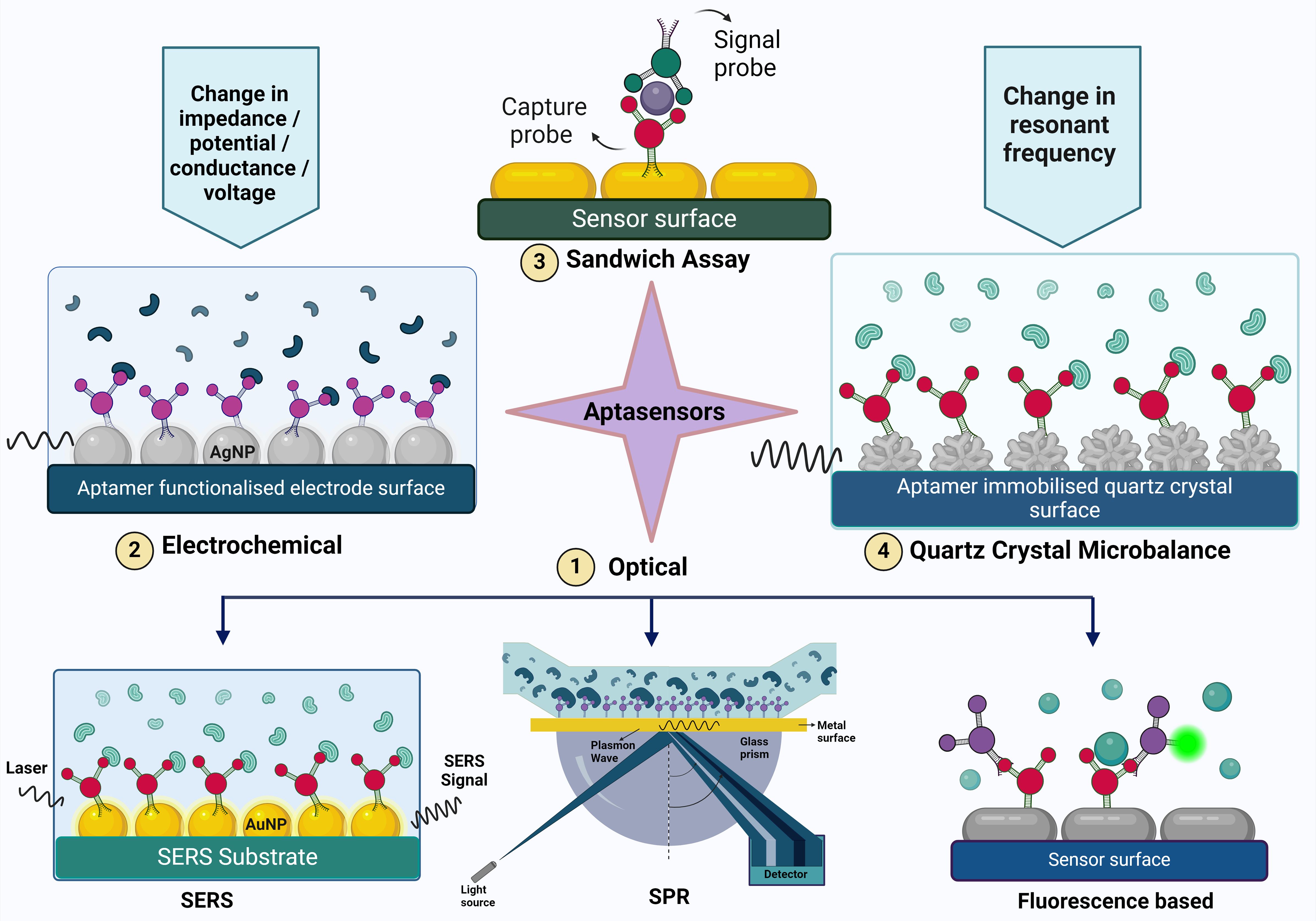
Figure 2. The figure illustrates various aptasensing mechanisms used for detecting target molecules. These mechanisms include (1) Optical sensors (such as Surface-Enhanced Raman Scattering (SERS), Surface Plasmon Resonance (SPR), and fluorescence-based methods), (2) Electrochemical sensors, (3) Sandwich assays, and (4) Quartz Crystal Microbalance (QCM). Each mechanism provides a unique approach to aptamer-based detection, highlighting the versatility and specificity of aptamers in biosensing applications.
3.1 Optical aptasensors
The components of an optical biosensor are an optical transducer system coupled with a biorecognition sensor. Optical biosensors are designed to generate a signal that is directly proportional to the concentration of the analyte (Damborský et al., 2016). Optical aptasensors are biosensors in which the biorecognition sensing element is an aptamer. The transduction method can be SPR, fluorescence, surface enhanced raman scattering (SERS) and chemiluminescence (Uniyal et al., 2023). Optical sensors are frequently used in aptasensors because of their high sensitivity, robustness, reliability, good temporal and spatial control, selectivity, simplicity, versatility, and wide linear range for biomolecule detection (Chen et al., 2021c).
3.1.1 Surface plasmon resonance based aptasensors
When a plane polarized light falls on a thin sheet of metal, plasmons (group of electrons that undergo oscillation due to energy absorption) are formed. In context of aptasensors, aptamer-functionalized metal particles are used. When the analyte binds to the aptamer, it causes changes in the refractive index at the interface, altering the resonance condition of the surface plasmons. These changes can be observed as variations in the angle or intensity of reflected light (Schasfoort, 2017). The sensitivity and selectivity of SPR-based sensors can be significantly improved by utilizing gold nanoparticles linked to ligands that are specific to the target. SPR assays are commonly used in dual-recognition biosensors and sandwich assays to enhance detection capabilities (Kim et al., 2018a).
3.1.2 Fluorescence based aptasensors
In this type of biosensing there are usually two probes involved- the capture probe that binds to the infectious agent and the signaling probe which is usually a nanoparticle that is tagged with a fluorophore. The interaction between the analyte and the aptamers leads to a rise in the fluorescence signal, which is detectable and can be analyzed in both qualitatively and quantitatively. Examples of fluorescent labels are Lanthanide-doped upconversion nanoparticles (UCNPs), silver nanoclusters (AgNCs) (Zhang et al., 2020), carbon quantum dots (CQDs) (Pebdeni et al., 2020), CdTe quantum dots and thiazole orange (Pang et al., 2015).
UCNPs have distinctive optical and chemical characteristics, including excellent photostability, low light scattering, low autofluorescence backgrounds, and low toxicity (Liu et al., 2021). AgNCs have the advantages of high quantum yield, strong photostability, low toxicity, adjustable fluorescence emission, and excellent biocompatibility (Zhang et al., 2020).
An important application of fluorescence spectroscopy is förster resonance energy transfer (FRET). It involves non-radiative transfer of energy from an excited donor fluorophore to an acceptor fluorophore that are in proximity. This phenomenon is also called quenching. Graphene oxide is a commonly used quencher molecule (Verma et al., 2023). For example, Pebdeni et al. discovered that CDQs emit blue-colored fluorescence, which is quenched in the presence of aptamers and gold nanoparticles. With the introduction of specific bacteria, the aptamer-target complex was effectively assembled, leading to the restoration of free CQD emission. The linear range of this aptasensor was 108 to 101 CFU/mL, with a detection limit as low as 10 CFU/mL for S. aureus (Pebdeni et al., 2020). Colorimetric aptasensors work by detecting changes in the color due to the binding of the aptamer to the analyte. This is done with the help of UV-visible spectroscopy. A peak is obtained at a specific wavelength and stokes shift takes place (Weerathunge et al., 2019).
3.1.3 SERS based aptasensors
Surface-enhanced Raman scattering (SERS) is a phenomenon in which the Raman scattering signals are amplified by enhancing the sensor surface. Nanostructured surfaces, usually made of metals such as gold or silver, are shaped into nanoparticles, nanorods, or nanostars. These structures demonstrate strong localized surface plasmon resonance (LSPR), resulting in the enhancement of Raman signals of nearby molecules through electromagnetic and chemical mechanisms. The SERS substrates are aptamers and when the infectious agent binds to the aptamer, there is a change in the raman signal that is detected (Zhou et al., 2020).
3.1.4 Chemiluminescence based aptasensors
Chemiluminescence-based aptasensors rely on the emission of light resulting from a chemical reaction between a luminophore (a molecule capable of emitting light) and a substrate or analyte, often facilitated by enzymatic reactions (Chen et al., 2021b). A DNA aptasensor to detect norovirus GII capsid was developed based on guanine chemiluminescence detection and the principle of intra chemiluminescent resonance transfer. The high-energy intermediates formed from the reaction of extra guanines and TMPG transferred the energy to 6-FAM which caused bright chemiluminescence (Kim et al., 2018a).
3.2 Electrochemical biosensors
A variety of electrochemical transducer systems, including impedimetric, potentiometric, amperometric, voltammetric, conductometric, and FET-based biosensors, can be integrated with aptamers for enhanced functionality.
3.2.1 Impedimetric aptasensor
When the target analyte binds to the aptamer functionalized sensor surface, inducing changes in the electrical properties at the interface such as charge transfer kinetics, dielectric properties, or surface conductivity at the sensor interface. The change in impedance is converted into a measurable electrical signal. Impedance spectroscopy measures the impedance change of the sensor due to exposure to the target analyte and computes how the sensors electrical impedance changes over a range of frequencies. In a study conducted by Roushani et al. (2019) NH2-aptamer was immobilized covalently on the surface of a glassy carbon electrode through electrodeposition modification of AgNPs. The conductivity and the charge transfer resistance before and after the addition of P.aeruginosa to the aptasensor was studied. The impedance increases on going from 102 to 107 CFU/mL concentrations of P. aeruginosa, and the detection limit was found to be 33 CFU/mL (for S/N=3). In a study conducted by Ramanathan et al. (2022) carbon nanodiamond enhanced gold interdigitated electrode was used to detect the nucleocapsid protein of SARS-CoV-2. The aptasensor which was portable, showed a good selectivity with a lower detection limit of 0.389 fM; at a linear detection range from 1 fM to 100 pM; showing 30 & 33% loss with stability & reusability. A rapid (30 mins) label-free aptasensor was constructed by Bagheryan et al., using screen-printed electrodes (SPEs) that were modified with diazonium salt for the detection of Salmonella typhimurium in spiked apple juice samples. The aptasensor had a linear detection range of 1×101 to 1×108 CFU mL−1 (Bagheryan et al., 2016).
3.2.2 Voltammetry based aptasensors
In a recent study, Fathi et al. (2020) developed a novel voltammetric aptasensor for detecting Salmonella enterica serovar. The sensor utilized a pencil graphite electrode modified with chitosan-coated electrospun carbon nanofibers and gold nanoparticles. The presence of the analyte on the electrode surface led to an increase in charge transfer resistance, with the change in current being measured as a function of voltage. Electrochemical detection of Salmonella was achieved using differential pulse voltammetry in a methylene blue solution. The aptasensor demonstrated a linear detection range of 10 to 105 CFU/mL, with a limit of detection (LOD) of 1.223 CFU/mL, outperforming the PCR technique.
3.2.3 Graphene FET based aptasensors
An aptamer with high affinity against HCV (hepatitis C virus) was functionalized on graphene solution-gated field-effect transistors (g-SGFET) and the developed aptasensor was used to amplify and detect the change in conductance caused by the interaction between the aptamer and the HCV core protein (Palacio et al., 2023). Similarly, Almeida et al., fabricated a graphene FET aptasensor to detect Zika virus (ZIKV). The aptamer (termed ZIK60), selected by CE-SELEX was complimentary to the Zika virus non-structural protein 1 (NS1) and counterselection against the NS1 proteins of DENV (serotypes 1, 2, 3, and 4) and YFV (Almeida et al., 2022).
3.2.4 Quartz crystal microbalance based aptasensors
QCM aptasensor is an acoustic (mass-based) piezoelectric biosensor that detect changes in mass on the aptamer immobilized surface of quartz crystal due to its interaction with the analyte molecules by detecting changes in the resonance frequency of the crystal. QCM-based aptasensors are highly sensitive, label free, portable and can be miniaturised and hence are suitable for point-of-care diagnostics. Aptamer selected using whole cell SELEX was utilized to fabricate a QCM sensor to detect E. coli O157:H7. The aptasensor had a LOD that was as low as 1.46 × 103 CFU/mL and outperformed most QCM-based immunosensors for pathogen detection. In addition, the quick response time of 50 min showed the possibility of using this aptamer in various other types of biosensors used for rapid detection and investigation of E. coli O157:H7 outbreaks (Yu et al., 2018). An interesting study conducted by Wang et al., demonstrates the use of QCM based SELEX to effectively select the ssDNA aptamer and subsequent construction of QCM based aptasensor which was able to detect 103 CFU/mL of S. typhimurium within 1 h (Wang et al., 2017a). Another example is a QCM aptasensor in which a nanowell based electrode effectively increased the immobilization capacity of aptamers for the detection of avian influenza virus. The result showed that the binding of target AIV H5N1 onto the immobilized aptamers decreased the sensor’s resonant frequency, and the frequency change correlated to the virus titer. The detection range of 2−4 to 24 hemagglutination units (HAUs)/50 μL was obtained with a detection limit of 2−4 HAU/50 μL for AIV H5N1 with a detection time of 10 mins using a label free assay. (Wang et al., 2017a, 2017b)
3.3 Dual recognition aptasensor
As the name suggests, dual recognition sensors make use of two different recognition principles facilitating a highly specific detection. Li et al., developed an aptasensor for detecting S. typhimurium by combining the methods of immune hybridization chain reaction (HCR) with SERS achieving double amplification and high sensitivity with a limit of detection of 6 CFU/mL in 3.5 h (Li et al., 2021). Bagheri Pebdeni et al., proposed an aptamer and antibiotic-based dual detection sensor that combines copper nanoclusters (CuNCs) as an effective approach for the recognition and quantification of S. aureus. The use of dual receptors enhanced fluorescence signal linearly with S. aureus concentrations between 102 -108 CFU/mL, and the detection limit was 80 CFU/mL after 45 min (Bagheri Pebdeni et al., 2021). Aptasensors like the electrochemiluminescence aptasensors come under both electrochemical and optical sensors. It works by detecting the luminescence that is produced due to the electrochemical interactions between the aptamer and the analyte molecules (Chen et al., 2021c; Chen et al., 2021b).
3.4 Sandwich assay based aptasensors
A sandwich assay involves two aptamers – the capture probe and the signal probe. The capture probe is immobilized on the surface of the sensor and after the analyte is added, the signal probe is added forming an aptamer-aptamer sandwich platform. This method is desirable because of the high sensitivity and selectivity that it offers. S. Kim et al., demonstrated a nanorod enhanced SPR with sandwich enzyme-linked immunosorbent assay (ELISA) for the attomolar detection of the norovirus (NoV) capsid protein (Kim et al., 2018b). RNA aptamer-based sandwich assays were used to detect the NS1 protein of dengue virus serotype 2 and a LOD of 2 nM was attained (Thevendran et al., 2023).
Another notable example is an aptamer/antibody sandwich constructed by Ge et al., for the digital detection of SARS-CoV2 nucleocapsid protein using fluorometry. The detection limit of this digital method for N protein was 33.28 pg/mL, which was 300 times lower than traditional double-antibody sandwich-based ELISA (Ge et al., 2022). Even though sandwich ELISA assay offers various advantages, it has a complex workflow, more optimization is required, is labor intensive and the time of detection is a little high.
3.5 Other aptasensors
There are aptasensors based on principles other than the above mentioned, for example, F. Jia et al. developed a low-field magnetic resonance imaging (LF-MRI) aptasensor based on the difference in magnetic behavior of two magnetic nanoparticles covalently immobilized with aptamers for the rapid detection of P.aeruginosa. Under optimum conditions, the LF-MRI platform provides both image analysis and quantitative detection of P. aeruginosa, with a detection limit of 100 CFU/mL (Jia et al., 2021).
Aptamer-based assays represent a significant advancement in the diagnostics of infectious diseases, addressing the limitations of traditional methods. Optical aptasensors, including surface plasmon resonance, fluorescence, and surface-enhanced Raman scattering, excel in sensitivity and specificity, ideal for detailed biomolecule detection. Electrochemical aptasensors, such as impedimetric, voltammetric, and graphene FET-based sensors, offer robust, portable solutions with high sensitivity for point-of-care applications. Meanwhile, dual-recognition and sandwich assay-based aptasensors combine multiple detection principles to enhance accuracy and detection limits. This comprehensive range of aptamer-based technologies demonstrates their potential to revolutionize diagnostic practices by providing versatile, efficient, and precise tools for infectious disease management. The most suitable method can be selected by understanding the strengths and limitations for each approach.
4 Notable aptasensor case studies
4.1 For the detection of methicillin resistant Staphylococcus aureus-contaminated surfaces
Methicillin-resistant Staphylococcus aureus (MRSA) is a well-known pathogen that causes healthcare-associated infections. Hospitals with contaminated environments are important sources for the spread of MRSA and other nosocomial infections. In a study, researchers have developed a new swab called a pathogen aptasensor which can specifically detect MRSA on contaminated non-absorbable surfaces. The visual detection limit of the MRSA aptasensor swab was less than 100 CFU/mL, and theoretically, using a standard curve, it was 2 CFU/mL. The assay has a short turnaround time of 5 minutes, with a linear range of quantitation from 10^2 to 10^5 CFU/mL. The MRSA aptamers bind to the swab’s activated aldehyde group, and when exposed to an MRSA-contaminated surface, the activated nanobeads conjugate with the aptamer, causing the swab to turn blue. The intensity of the color change is proportional to the concentration of MRSA, allowing for both qualitative and quantitative detection (Raji et al., 2021).
4.2 Simultaneous detection of E.coli O157:H7 and S.typhimurium
Simultaneous detection of E.coli and S.typhimurium was achieved using an evanescent wave dual-color fluorescence aptasensor based on time resolved effect. Two fluorescence labeled aptasensors, Cy3-apt-E and Cy5.5-apt-S that were complimentary to E.coli O157:H7 and S.typhimurium were alternatively excited by evanescent waves originated from 520 nm to 635 nm excitation lights, respectively. The fiber nanoprobe with in-situ etched nanopores was used for distinguishing free aptamer and aptamers bound to pathogenic bacteria based on the limited penetrated depth of evanescent wave and the significant size difference of bacteria and nanopore. The E. coli O157:H7 and S. typhimurium were directly and simultaneously quantitated in less than 35 min without the requirement of the complex immobilization of biorecognition molecules and bacteria enrichment/separation processes. The limits of detection of E. coli O157:H7 and S. typhimurium were 340 CFU/mL and 180 CFU/mL, respectively (Fang et al., 2021).
4.3 Colorimetric aptasensor for detecting Salmonella spp., Listeria monocytogenes, and Escherichia coli in meat samples
Aptasensors are revolutionizing infectious disease detection by enhancing the specificity and sensitivity of aptamers. These biosensors provide versatile solutions for streamlining diagnostic processes in healthcare by rapidly and precisely identifying pathogens.
A recent study introduced a quick detection method that can simultaneously identify Salmonella spp., Listeria monocytogenes, and E. coli. This method uses visual colorimetric detection with labeled colloidal gold nanoparticles and UV absorbance determination at optimized wavelengths of 625 nm and 525 nm. The aptasensor has a detection limit as low as 105 CFU/mL. Notably, this colorimetric aptasensor enables one-step detection without the need for pre-culture, DNA extraction, or amplification steps. As a result, it provides a simple, rapid, specific, and qualitative assay suitable for point-of-care testing, allowing for direct detection of multiple foodborne pathogens (Ledlod et al., 2020). Additionally, exploring virulence factors as potential targets for aptamers is helping us understand pathogen behavior and leading to the development of targeted therapeutic interventions.
5 Aptamer applications: targeting virulence factors and recent advances
5.1 Virulence factors and potential aptamer targets
Aptamer is one of the most promising therapeutic candidates because of its selectivity. In the field of therapeutics, they serve various crucial roles, including acting as a drug delivery vehicle (Ninomiya et al., 2014), functioning as a targeting molecule for genes or whole cells, thereby reducing the expression of virulent genes in pathogens and enhancing susceptibility to the immune system (Lai et al., 2014). Furthermore, it serves as a binding agent for toxins and specific proteins that contribute to increased pathogen virulence (Gribanyov et al., 2021). When it comes to treating viral infections that have no known treatment and drug-resistant microorganisms that cause infectious diseases, aptamers may be a useful therapeutic tool (Figure 3).
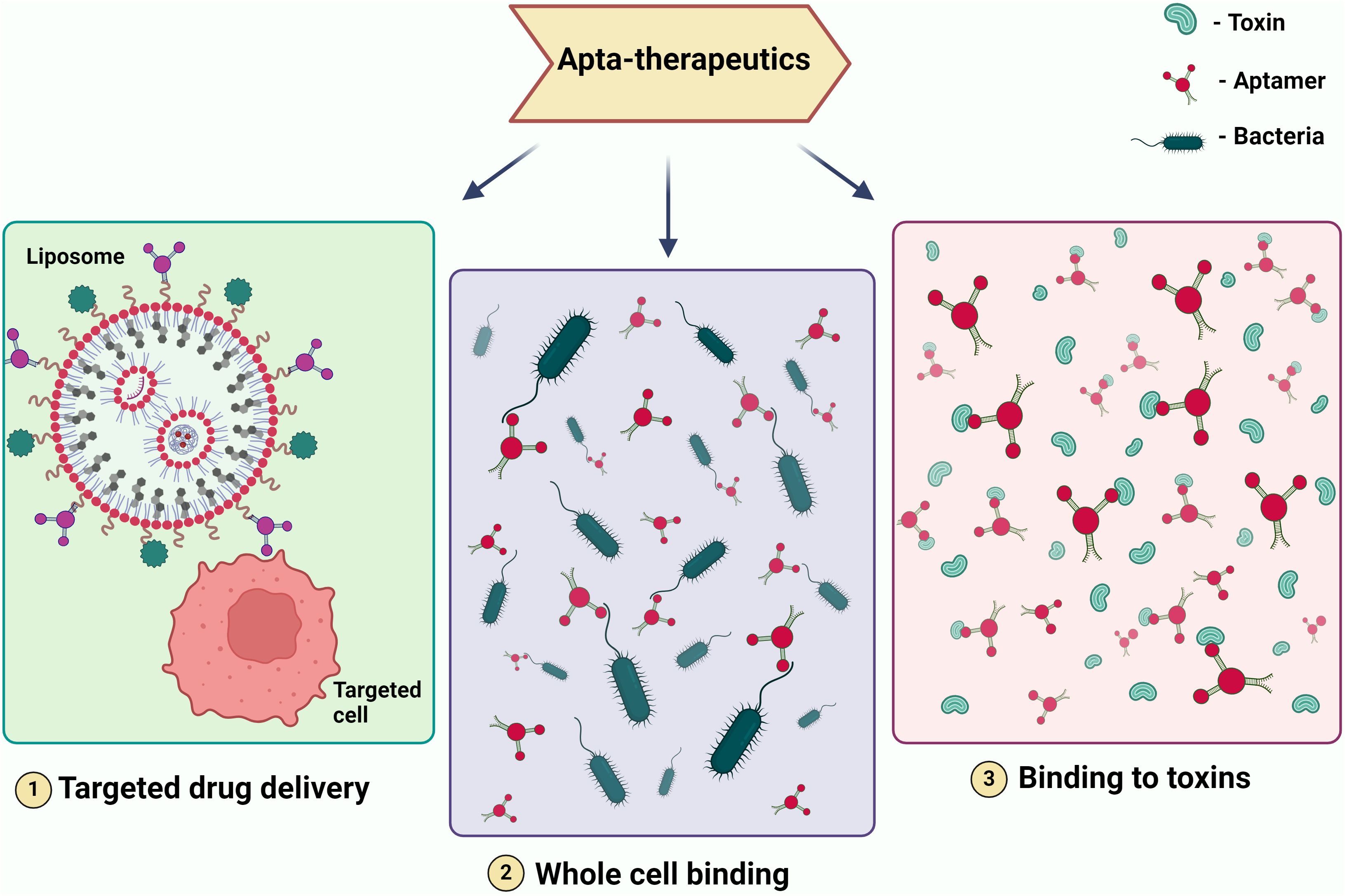
Figure 3. Illustrating Therapeutic Modalities Employing Aptamers. The figure depicts various therapeutic applications of aptamers, including (1) targeted drug delivery, where aptamers are used to direct drugs specifically to diseased cells; (2) whole cell binding, where aptamers bind to specific cells for therapeutic purposes; and (3)binding to toxins, where aptamers neutralize toxins by binding to them. These modalities showcase the potential of aptamers in precision medicine and targeted therapies.
5.1.1 Bacteria
By removing important virulence components from bacteria, aptamers present a viable strategy for treating bacterial illnesses. (Tables 2, 3). The innovative technology enhance the treatment efficacy against pathogens such as Staphylococcus aureus, Mycobacterium tuberculosis, Salmonella typhi, Listeria monocytogenes, Streptococcus pneumoniae, and Escherichia coli. In the fight against S. aureus infections, aptamers AT-33 and AT-36 have been specifically engineered to target and neutralize the α-toxin, a key virulence factor. These aptamers effectively inhibit α-toxin-induced cell death and cytokine upregulation in human cells, offering a promising therapeutic approach (Ommen et al., 2022). Another set of aptamers targets S. aureus biofilms, binding to the biofilm matrix to enhance antibiotic delivery and significantly improve treatment outcomes by overcoming biofilm-associated resistance. This dual approach of targeting both toxins and biofilms represent a significant advancement in therapeutic strategies against S. aureus infections.
To identify and target important molecules connected to M. tuberculosis, aptamers have also been developed. For instance, mannose-capped lipoarabinomannan (ManLAM), a major glycolipid on the bacterial surface, serves as a target for specific aptamers, aiding in both diagnostic and therapeutic applications. Additionally, aptamers targeting the GlcB and HspX antigens disrupt bacterial metabolism and persistence, offering potential therapeutic benefits (Zhou et al., 2021). Furthermore, aptamers targeting ESAT-6, a critical virulence factor secreted by M. tuberculosis, can reduce the bacterium’s virulence and increase its vulnerability to immune system attacks (Sreejit et al., 2014). M. tuberculosis produces a lipid called phthiocerol dimycocerosate (PDIM), which is essential to the pathogenicity and virulence of the bacteria (Augenstreich et al., 2020).
Researchers have developed an aptamer that interacts with and neutralizes the InvA gene of S. typhi, a crucial element in the bacterium’s invasion process. Additionally, the S9 aptamer targets the outer membrane protein of S. typhi, further contributing to the bacterium’s neutralization (Pathania et al., 2017; Yang et al., 2013). SPI1, or Salmonella pathogenicity island 1, is essential for Salmonella’s interaction with host cells, facilitating penetration through the T3SS, also known as the needle complex, which assembles proteins to translocate effector proteins into host cells (Raffatellu et al., 2005; Lerminiaux et al., 2020). The pathogenicity of S. typhi is enhanced by the release of typhoid toxin and the Vi capsular antigen, which has anti-opsonic and antiphagocytic properties (Galán, 2016; Tran et al., 2010; Wain et al., 2005). These harmful factors can be targeted by specifically curated aptamers.
In order to prevent L. monocytogenes from invading host cells, aptamers that target InlB, one of the bacteria’s virulence factors, have been created. By blocking this key infection pathway, these aptamers offer a promising therapeutic strategy for preventing L. monocytogenes infections (Chen et al., 2024). L. monocytogenes produces listeriolysin O (LLO), a pore-forming toxin dependent on cholesterol (Dramsi and Cossart, 2002). LLO damages the vacuolar membrane, facilitating bacterial escape into the cytosol (Petrišič et al., 2021). These vulnerable parts of the pathogen can be exploited by targeting them with protein-specific aptamers.
For S. pneumoniae, aptamers have shown good specificity; the Lyd-3 aptamer in particular has shown promise. Lyd-3 effectively inhibits biofilm formation, a critical factor in the pathogen’s virulence and antibiotic resistance. By significantly reducing biofilm formation, Lyd-3 enhances treatment outcomes, especially when used in combination with antibiotics (Afrasiabi et al., 2020). The pneumococcus’s polysaccharide capsule is a significant virulence component, aiding in immune evasion and colonization (Jonsson et al., 1985). PspK mediates adherence to human epithelial cells, independent of the pneumococcal isolate genetic background (Keller et al., 2013).
Four aptamers have demonstrated high affinity and specificity for E. coli cells, making them valuable tools for both diagnostic and therapeutic applications. These aptamers offer precise detection and effective targeting of E. coli (Marton et al., 2016). E. coli causes various infections, including urinary tract infections, and relies on colonization factors and toxins for virulence. Aptamers can target these virulent factors, disrupting E. coli’s pathogenic mechanisms and enhancing treatment efficacy (Johnson, 1991; Kaper et al., 2004; Terlizzi et al., 2017).
By leveraging the specificity and affinity of aptamers, we can target key virulence factors in various bacterial pathogens, offering innovative and effective therapeutic strategies.
5.1.2 Virus
Aptamers are also increasingly recognized for their potential to combat viral infections by targeting and neutralizing specific viral components (Table 4). It can enhance the efficacy of existing antiviral treatments and provides new therapeutic avenues for diseases like Zika virus, Nipah virus, Ebola virus, and Influenza A virus.
Zika virus, a member of the Flaviviridae family transmitted by Aedes aegypti mosquitoes (Diagne et al., 2015), has been targeted with various antiviral strategies. Ribavirin has shown efficacy in suppressing viremia in ZIKV-infected STAT-1-deficient mice (Kamiyama et al., 2017), while favipiravir and BCX4430 inhibit viral RNA synthesis by targeting viral RNA-dependent RNA polymerase (Furuta et al., 2009; Eyer et al., 2017). The similarity between the envelope proteins of dengue and Zika viruses underscores their close evolutionary relationship (Lunardelli et al., 2023). NS1 protein plays critical roles in ZIKV replication (Valente and Moraes, 2019), and aptamer technology holds promise for enhancing antiviral drug efficacy by targeting specific virulence factors (Feng et al., 2011).
Nipah virus lacks specific antiviral treatments, making aptamer-based therapies a potential breakthrough by targeting its virulence factors, such as the F protein that mediates viral entry through ephrin B2/B3 receptors (Sun et al., 2018; Weis and Maisner, 2015). The Nipah virus V protein inhibits STAT proteins, crucial for interferon signaling, enhancing viral pathogenesis (Shaw et al., 2004). Aptamers designed to bind these proteins could mitigate infection severity.
Ebola virus VP35 and VP24 proteins are key virulence factors that disrupt host immune responses (Leung et al., 2010; Zhang et al., 2012), with aptamers identified to target VP35’s interferon inhibitory domain (Binning et al., 2013). These aptamers offer potential therapeutic avenues against Ebola virus by restoring interferon response pathways.
Influenza A viruses, characterized by their surface proteins HA and NA, play crucial roles in viral entry and replication (Bouvier and Palese, 2008). Aptamers targeting HA have demonstrated significant antiviral effects in animal models, inhibiting viral replication and reducing infection rates across different influenza strains (Nobusawa, 1997; Gopinath et al., 2006; Jeon et al., 2004). Aptamer research continues to explore novel therapeutic strategies, addressing the challenges posed by viral mutation and enhancing treatment efficacy (Musafia et al., 2014; Sanjuán, 2012).
5.2 Recent advancements and case studies
5.2.1 Notable developments in aptamer research for infectious diseases
5.2.1.1 Gold nanoparticle-DNA aptamer conjugate-assisted delivery of antimicrobial peptide (CA2634987A1)
Gold nanoparticles are a durable and widely used delivery technology that offers various benefits over liposomes and PLGA. It was demonstrated that combining antimicrobial peptides with a gold nanoparticle-aptamer complex was effective in eliminating intracellular Salmonella enterica serovar Typhimurium (Yeom et al., 2016).
5.2.1.2 Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer (WO2022016163A2)
An innovative test technique that combines a pre-conjugated aptamer with the enzyme invertase, which is then attached to a magnetic bead. Because the aptamer is highly specific to the antigen found in the corona virus, it goes through a confirmational change that releases the enzyme into the medium, where it is separated by magnetic separation. The medium’s invertase then breaks down sucrose into glucose, and measuring the glucose yields an assay of the antigen present in the sample. This model operates on this concept (Singh et al., 2021).
5.2.1.3 Graphene aptasensor for the detection of hepatitis C virus (EP4124855A1)
Changes in their surroundings, particularly the attachment of bio-receptors to the graphene surface, cause Graphene Field-Effect Transistor Biosensors (gFET) to detect changes in electrical metrics, such as conductivity. Graphene’s sensing potential is increased by chemical modification. As an example of how biological molecules can be sensed, researchers have created chemically functionalized gFETs that can detect negatively charged exosomes when they are bound to the graphene surface. In this particular case, researchers have created sgFETs with aptamer that can detect HCV protein even at lower concentrations making it an ultrasensitive aptasensor. Attomolar detection of the viral protein target is made possible by the enhanced sensitivity brought about by induced polarization at the graphene interface (Kwong Hong Tsang et al., 2019; Palacio et al., 2023).
5.2.1.4 Aptamer binding hemagglutinin of H7N7 subtype influenza virus (JP2014008002A)
An aptamer capable of differentiating between influenza A serotypes and interfering with the HA-glycan interaction was created by researchers. To ensure the aptamer’s stability in the presence of endo-ribonucleases, 2′-fluoro cytidine is employed, which does not interfere with its binding to HA. This aptamer has applications in detecting and diagnosing H5N1 and H7N7 viruses, as well as in synthesizing virucidal drugs that selectively target these viruses, impeding their early interactions with hosts (Suenaga and Kumar, 2014). The glycoprotein known as hemaglutinin (HA), which is present on the influenza virus’s surface, is essential to the virus’s capacity to bind to and penetrate host cells. Aptamers work by specifically targeting HA, which stops the virus from attaching to host cell receptors and preventing it from entering the cells (Zou et al., 2019)
5.2.2 Case studies: aptamers that inhibit biofilm
Quorum sensing, a mechanism that enables signaling and communication within bacteria, plays a key role in the formation of P.aeruginosa biofilms. Three main QS systems in P.aeruginosa: las system, rhl system and Pseudomonas quinolone signal system (PQS) encode for various signaling molecules that act as regulator for the transcription of numerous virulence factor genes (Zhang et al., 2013; Chadha et al., 2022). Zhao et al., conducted a study in which they screened DNA aptamers complimentary to the signal molecule C4-HSL of the rhl system. Depressing the rhl system affects the formation and maintenance of the biofilm. It was observed that the biofilm formation of P.aeruginosa was efficiently reduced to about 1/3 by the aptamers compared with that of the groups without the aptamers in the in vitro biofilm inhibition experiments (Zhao et al., 2019) (Figure 4).
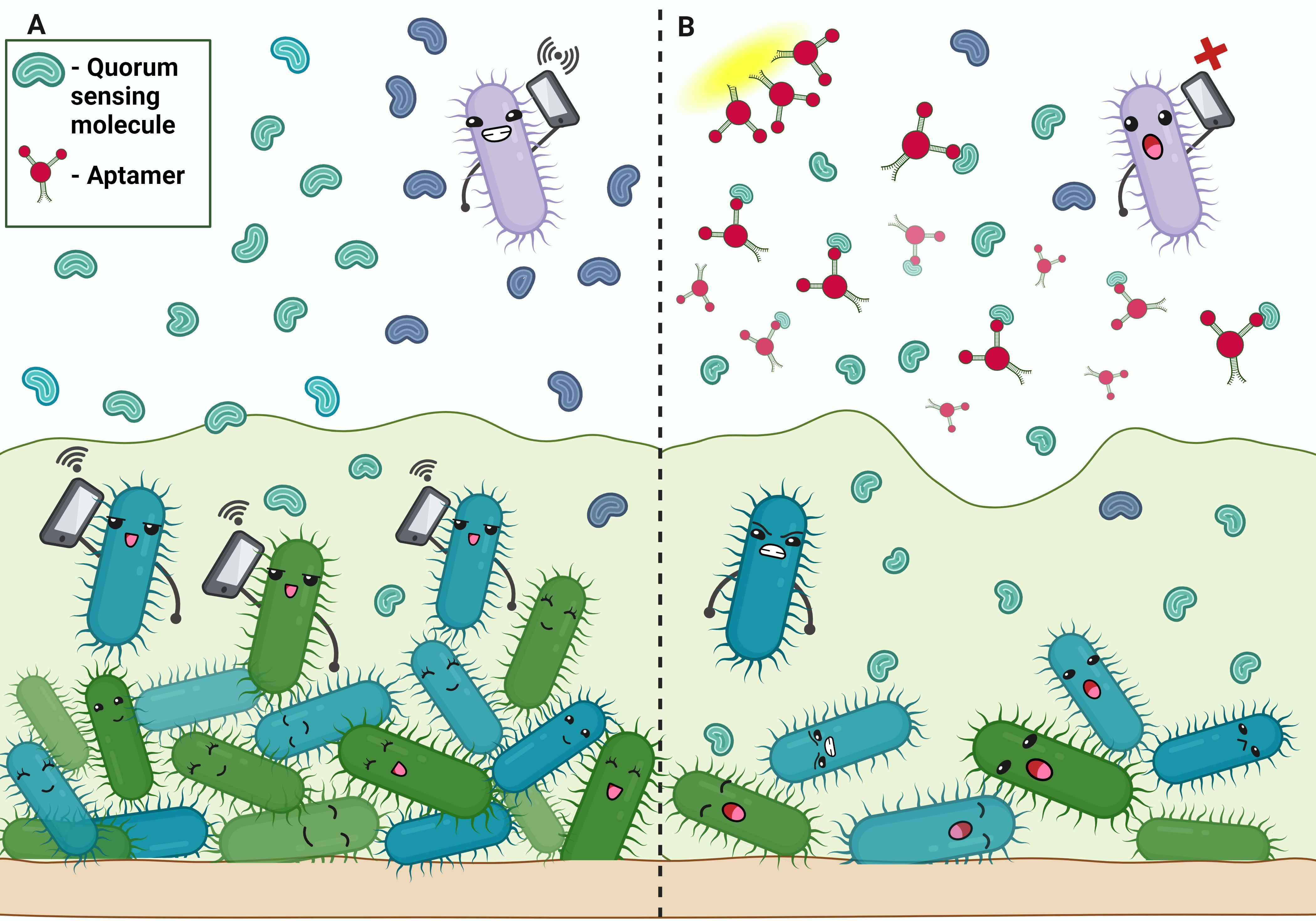
Figure 4. Aptamers disrupting quorum sensing in biofilm. (A) Communication in bacterial biofilm by quorum sensing molecule. (B) Aptamer binds to the quorum-sensing molecules, disrupting signaling.
Matchawong et al., constructed a 2’ -fluoropyrimidine modified nuclease-resistant RNA aptamersusing cell SELEX against Streptococcus suis serotype 2, strain P1/7. The R8-su12 RNA aptamer significantly reduced the S. suis biofilm formation and had the ability to bind to other pathogenic S. suis (serotype ½, 1, 9, and 14) (Matchawong et al., 2022). Candida albicans was grown in the exposure of condensed cigarette smoke (CSC), prepared from clove (CCSC) and non-clove (NCSC) cigarettes, for 48 h (Bachtiar et al., 2021).It was found that the presence of added CCSC or NCSC significantly enhanced C. albicans biofilm development but when C. albicans was precoated with aptamer (Ca-apt1) there was a significant impairment in the biofilm development accelerated by the NCSC and CCSC. This could be attributed to the enhancement of the morphological changes of C. albicans (from yeast to hypha formation) due to CCSC or NCSC was reduced due to precoating the aptamer.
Ning et al.,conducted an interesting study in which a GO-loaded aptamer/berberine bifunctional complex specific to penicillin-binding protein 2a (PBP2a) significantly inhibited MRSA biofilm formation (Ning et al., 2022). The aptamer blocks the function of PBP2a, reducing surface-cell attachment and berberine attenuates the level of the accessory gene regulator (agr) system, which is essential for MRSA biofilm formation. Furthermore, GO also has the potential to disrupt cell membranes, attributing to the antibiofilm activity (Saravanan et al., 2023). Lijuan et al., developed an aptamer-ampicillin bifunctional conjugate that targeted bacterial flagella for treating biofilms (Lijuan et al., 2017).
6 Challenges and limitations
6.1 Stability and delivery concerns of aptamers
The term “steady state” describes the dynamic balance that results from consistent dosage between the total amount of a drug taken and its elimination. For aptamers, the steady-state or nearly steady-state concentration can be attained three to five times the half-life following aptamer administration, which has a steady-state duration of 17 to 29 hours (Lee et al., 2015). This emphasize on the importance of aptamer’s stability i.e.,The longer the aptamer stays in circulation for the treatment of infectious disorders, the greater the likelihood that it may encounter pathogens. The pharmacokinetic profile of the aptamer must be determined to proceed with trials, firstly the aptamer’s half-life in vivo is rather brief, lasting roughly 2 mins. Unmodified ssDNA oligonucleotides have a half-life of less than one minute (Griffin et al., 1993) (Sanjuán, 2012).
In conditioned media, HEK cells infected with M. fermentans exhibit ribonuclease activity that rapidly degrades RNA carrying 2′-fluoro- and 2′-O-methyl-modified pyrimidines. Similar ribonuclease activity was seen in a pure culture of M.fermentans, but not in a culture of uncontaminated HEK cells (Hernandez et al., 2012). RA-36, an aptamer with antithrombin properties, have shown rapid bloodstream elimination with a half life of 1 minute As opposed to 23 minutes in tissues (Zavyalova et al., 2017). The tissue type, aptamer sequence, and their formulation affects the outcomes of aptamer uptake ad distribution. A single bolus oral dose of aptamer was administered to mice, and after tissue contamination was eliminated with perfusion buffer, the aptamer was diffused into the bloodstream from the peritoneum and into multiple organs, including the brain and spinal cord, within minutes of oral administration. The uptake of the aptamer was reduced within a few hours (Perschbacher et al., 2015). When aptamer and nanoparticles are conjugated, the physiochemical properties including the size and distribution of the particles are altered (Ghassami et al., 2018).
6.2 Potential for off-target effects and safety issues
Drugs undergoing clinical trials may have side effects and safety problems of their own, but it is crucial to understand these effects in order to design a therapeutic version with less side effects. Investigations’ findings by Zhao et.al., in the tested settings, the SGC8 aptamer exhibited neither mutagenicity nor genetic toxicity using total body-positron emitting tomography (TB PET) (D. Ding et al., 2023). When an aptamer-tagged radioactive element was injected intravenously, the kidney contained the highest quantities of radioactivity. This indicates that the pharmacokinetics profile of absorption from intravenous aptamer injection results in a relatively low absorption rate. However, giving aptamer was administered in many doses, but this did not cause aptamer to accumulate in plasma. The factor most likely limiting the drug’s rate of disposal is its rate of absorption (Siddiqui and Keating, 2005).
Researchers discovered that the absence of conjugating cholesterol has certain undesirable effects, such as altering the expression of genes associated to innate immunity and cellular survival (Lee et al., 2015). In addition, the aptamer’s overall negative charge causes it to attach to positively charged substances without being specific. As stated previously, the aptamer’s short length and compact size promote bio clearance (Gholikhani et al., 2022).
6.3 Strategies to overcome challenges and ongoing research in the field
The aforementioned problems can be solved in a number of ways, and some of these tactics are (1) substituting sulfur for one of the monothio or dithio groups of the phosphoryl non-bridging oxygen atoms results in a number of benefits, including increased binding to the target, resistance to nuclease action, and faster absorption into the cells. However, there is a small drawback to this the aptamer may become less specific (Thiviyanathan et al., 2007; Keefe et al., 2010). (2) integrating aptamer into a larger molecular framework in the shape of a multivalent circle by offering nucleolytic stabilization that guards against exonucleases (Di Giusto et al., 2006). (3) the body’s nuclease enzymes could not break down aptamers synthesized with an L nucleotide sequence. This type of sequence, called spigelmers, is the mirror image of an oligonucleotide but contains L nucleotide instead of R nucleotide (Maasch et al., 2008; Chen et al., 2017). (4) RNA sequences that have aldehyde derivatives appended to the 5’ end, facilitate affinity purification and coupling with other molecules (Pfander et al., 2007). (5) producing the most stable hybrids by employing nucleoside analogues that have a methylene bond between the ribose ring’s 2′-O and 4′-C in order to create a locked nucleic acid sequence. The sugar moiety is thus locked in a C3′-endo configuration (Lebars et al., 2007). (6) an aptamer was modified by adding PEG linkers to decrease stearic hinderance and 2’-fluoro-pyrimidines (2’F) (Derbyshire et al., 2012; Ni et al., 2021). (7) the highest tissue exposure was achieved by the aptamer, which was prepared as a 3′ biotin derivative coupled with tetrameric streptavidin (Perschbacher et al., 2015). To achieve this, a cholesterol moiety was linked to the 5’ end of a 29-nucleotide RNA aptamer that had been modified with 2’-F against the HCV NS5B protein. This modification was chosen because previous studies have shown that conjugating oligonucleotide molecules with cholesterol can prolong their plasma half-life by associating with plasma lipoproteins and enhance their uptake by hepatic cells through receptor-mediated endocytosis. (8) An improved aptamer half-life results by conjugating cholesterol with the aptamer (Lee et al., 2015). (9) The aptamer attached to MetCyc, facilitating its interaction with other molecules helping evade attacks from nucleases (Borbas et al., 2007; Ni et al., 2021). (10) In some circumstances, we can employ liposomal conjugated aptamers to lengthen the drug’s half-life and make it more covert (Jiang et al., 2020; Alameh et al., 2021; Kim et al., 2001). (11) To shield the RNA from exonuclease degradation, the derivative’s two terminals are capped with an extended stem structure, allowing for the effective in vivo expression of the aptamer (Mori et al., 2012). Creating chimeric aptamers by combining segments from different aptamers or combining with other functional molecules for enhancing specificity (Cheng et al., 2023). In conclusion, these diverse strategies and modifications illustrate ongoing efforts to optimize aptamer technology, enhancing their stability, specificity, and therapeutic efficacy across various biomedical applications.
7 Conclusion and future perspectives
The diverse applications of aptamers in the realm of infectious diseases underscore their immense potential in diagnostics, therapeutics, and biosensing. The ability of aptamers to specifically recognize and bind to a wide range of pathogenic targets, including viruses, bacteria, and fungus, has paved the way for innovative solutions in disease detection and treatment. In diagnostics, aptamers have demonstrated exceptional sensitivity and specificity, enabling the development of rapid and accurate diagnostic assays. Their incorporation into biosensors has facilitated the detection of infectious agents at early stages, contributing to timely interventions and improved patient outcomes. Aptamer-based diagnostic platforms also offer the advantage of portability and cost-effectiveness, making them particularly valuable in resource-limited settings. Aptamers have proven their mettle in therapeutic applications, where they can be engineered to inhibit viral entry, replication, or modulate the host immune response. The versatility of aptamers allows for the design of tailored therapeutic interventions, offering a promising avenue for the development of antiviral and antibacterial agents. Moreover, the potential for aptamers to mitigate the emergence of drug-resistant strains adds another layer of significance to their therapeutic applications. Looking ahead, the future perspectives of aptamer research in infectious diseases are exciting and multifaceted. Advancements in aptamer selection technologies, such as SELEX, will likely enhance the discovery of aptamers with improved binding affinities and specificities. The integration of aptamers with emerging technologies, such as CRISPR-based diagnostics and gene editing, holds promise for the development of next-generation diagnostic and therapeutic tools. Furthermore, the exploration of aptamer-nanoparticle conjugates and other delivery systems may enhance the targeted delivery of aptamers to infected tissues, improving their therapeutic efficacy. Collaborations between academia, industry, and healthcare providers will be crucial in translating aptamer-based technologies from the laboratory to clinical practice.
Author contributions
RN: Data curation, Visualization, Writing – original draft. BS: Visualization, Writing – original draft, Conceptualization. SS: Data curation, Visualization, Writing – original draft. AS: Data curation, Visualization, Writing – review & editing. SR: Data curation, Visualization, Writing – review & editing. HD: Conceptualization, Data curation, Supervision, Visualization, Writing – review & editing. AS: Conceptualization, Data curation, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are grateful to SASTRA University for providing us with an excellent infrastructure and for providing the opportunity to be a part of the Quorum Sensing Lab (QSL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adachi, T., Nakamura, Y. (2019). Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules 24, 4229. doi: 10.3390/molecules24234229
Afrasiabi, S., Pourhajibagher, M., Raoofian, R., Tabarzad, M., Bahador, A. (2020). Therapeutic applications of nucleic acid aptamers in microbial infections. J. Biomed. Sci. 27, 6. doi: 10.1186/s12929-019-0611-0
Agrelli, A., de Moura, R. R., Crovella, S., Brandão, L. A. C. (2019). ZIKA virus entry mechanisms in human cells. Infection Genet. Evol. 69, 22–29. doi: 10.1016/j.meegid.2019.01.018
Aguilar, H. C., Matreyek, K. A., Filone, C. M., Hashimi, S. T., Levroney, E. L., Negrete, O. A., et al. (2006). N-Glycans on Nipah Virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 80, 4878–4889. doi: 10.1128/jvi.80.10.4878-4889.2006
Alameh, M.-G., Tombácz, I., Bettini, E., Lederer, K., Ndeupen, S., Sittplangkoon, C., et al. (2021). Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54, 2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001
Ali, M. H., Elsherbiny, M. E., Emara, M. (2019). Updates on aptamer research. Int. J. Mol. Sci. 20, 2511. doi: 10.3390/ijms20102511
Aljohani, M. M., Cialla-May, D., Popp, J., Chinnappan, R., Al-Kattan, K., Zourob, M. (2022). Aptamers: potential diagnostic and therapeutic agents for blood diseases. Molecules 27, 383. doi: 10.3390/molecules27020383
Almeida, N. B. F., Sousa, T. A. S. L., Santos, V. C. F., Lacerda, C. M. S., Silva, T. G., Grenfell, R. F. Q., et al. (2022). DNA aptamer selection and construction of an aptasensor based on graphene FETs for Zika virus NS1 protein detection. Beilstein J. Nanotechnology 13, 873–881. doi: 10.3762/bjnano.13.78
Amouzadeh Tabrizi, M., Acedo, P. (2022). Highly sensitive aptasensor for the detection of SARS-CoV-2-RBD using aptamer-gated methylene blue@mesoporous silica film/laser engraved graphene electrode. Biosensors Bioelectronics 215, 114556. doi: 10.1016/j.bios.2022.114556
A Ocsoy, M., Yusufbeyoglu, S., Ildiz, N., Ulgen, A., Ocsoy, I. (2021). DNA aptamer-conjugated magnetic graphene oxide for pathogenic bacteria aggregation: selective and enhanced photothermal therapy for effective and rapid killing. ACS Omega 6, 20637–20643. doi: 10.1021/acsomega.1c02832
Askari, A., Kota, S., Ferrell, H., Swamy, S., Goodman, K. S., Okoro, C. C., et al. (2024). UTexas Aptamer Database: the collection and long-term preservation of aptamer sequence information. Nucleic Acids Res. 52, D351–D359. doi: 10.1093/nar/gkad959
Aslan, Y., Atabay, M., Chowdhury, H. K., Göktürk, I., Saylan, Y., Inci, F. (2023). Aptamer-based point-of-care devices: emerging technologies and integration of computational methods. Biosensors 13, 569. doi: 10.3390/bios13050569
Augenstreich, J., Haanappel, E., Sayes, F., Simeone, R., Guillet, V., Mazeres, S., et al. (2020). Phthiocerol dimycocerosates from Mycobacterium tuberculosis increase the membrane activity of bacterial effectors and host receptors. Front. Cell. Infection Microbiol. 10. doi: 10.3389/fcimb.2020.00420
Bachour Junior, B., Batistuti, M. R., Pereira, A. S., de Sousa Russo, E. M., Mulato, M. (2021). Electrochemical aptasensor for NS1 detection: Towards a fast dengue biosensor. Talanta 233, 122527. doi: 10.1016/j.talanta.2021.122527
Bachtiar, B. M., Gani, B. A., Deviana, A., Utami, N. R., Andriyani, A. D., Bachtiar, E. W. (2021). The discrepancy between clove and non-clove cigarette smoke-promoted Candida albicans biofilm formation with precoating RNA-aptamer. F1000Research 10, 372. doi: 10.12688/f1000research.52266.3
Bagheri Pebdeni, A., Mousavizadegan, M., Hosseini, M. (2021). Sensitive detection of S. aureus using aptamer- and vancomycin -copper nanoclusters as dual recognition strategy. Food Chem. 361, 130137. doi: 10.1016/j.foodchem.2021.130137
Bagheryan, Z., Raoof, J.-B., Golabi, M., Turner, A. P. F., Beni, V. (2016). Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosensors Bioelectronics 80, 566–573. doi: 10.1016/j.bios.2016.02.024
Bai, X., Nakatsu, C. H., Bhunia, A. K. (2021). Bacterial biofilms and their implications in pathogenesis and food safety. Foods 10, 2117. doi: 10.3390/foods10092117
Bakhtiari, H., Palizban, A. A., Khanahmad, H., Mofid, M. R. (2021). Novel approach to overcome defects of Cell-SELEX in developing aptamers against aspartate β-hydroxylase. ACS Omega 6, 11005–11014. doi: 10.1021/acsomega.1c00876
Bayraç, A. T., Donmez, S. I. (2018). Selection of DNA aptamers to Streptococcus pneumonia and fabrication of graphene oxide based fluorescent assay. Analytical Biochem. 556, 91–98. doi: 10.1016/j.ab.2018.06.024
Baz-Martnez, M., El Motiam, A., Ruibal, P., Condezo, G. N., de la Cruz-Herrera, C. F., Lang, V., et al. (2016). Regulation of Ebola virus VP40 matrix protein by SUMO. Sci. Rep. 6. doi: 10.1038/srep37258
Bell, R. L., Jarvis, K. G., Ottesen, A. R., McFarland, M. A., Brown, E. W. (2016). Recent and emerging innovations in Salmonella detection: a food and environmental perspective. Microbial Biotechnol. 9, 279–292. doi: 10.1111/1751-7915.12359
Bharaj, P., Wang, Y. E., Dawes, B. E., Yun, T. E., Park, A., Yen, B., et al. (2016). The matrix protein of nipah virus targets the E3-ubiquitin ligase TRIM6 to inhibit the IKKϵ kinase-mediated type-I IFN antiviral response. PLoS Pathog. 12. doi: 10.1371/journal.ppat.1005880
Binh, N. T., Wakai, C., Kawaguchi, A., Nagata, K. (2013). The N-terminal region of influenza virus polymerase PB1 adjacent to the PA binding site is involved in replication but not transcription of the viral genome. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00398
Binning, J. M., Wang, T., Luthra, P., Shabman, R. S., Borek, D. M., Liu, G., et al. (2013). Development of RNA aptamers targeting ebola virus VP35. Biochemistry 52, 8406–8419. doi: 10.1021/bi400704d
Borbas, K. E., Ferreira, C. S. M., Perkins, A., Bruce, J. I., Missailidis, S. (2007). design and synthesis of mono- and multimeric targeted radiopharmaceuticals based on novel cyclen ligands coupled to Anti-MUC1 aptamers for the diagnostic imaging and targeted radiotherapy of cancer. Bioconjugate Chem. 18, 1205–1212. doi: 10.1021/bc0700741
Boussebayle, A., Groher, F., Suess, B. (2019). RNA-based Capture-SELEX for the selection of small molecule-binding aptamers. Methods 161, 10–15. doi: 10.1016/j.ymeth.2019.04.004
Bouvier, N. M., Palese, P. (2008). The biology of influenza viruses. Vaccine 26, D49–D53. doi: 10.1016/j.vaccine.2008.07.039
Brandenburg, B., Koudstaal, W., Goudsmit, J., Klaren, V., Tang, C., Bujny, M. V., et al. (2013). Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One 8 (12), e80034. doi: 10.1371/journal.pone.0080034
Brody, E., Willis, M., Smith, J., Jayasena, S., Zichi, D., Gold, L. (1999). The use of aptamers in large arrays for molecular diagnostics. Mol. Diagnosis 4, 381–388. doi: 10.1016/S1084-8592(99)80014-9
Brooks, L. R. K., Mias, G. I. (2018). Streptococcus pneumoniae’s virulence and host immunity: Aging, diagnostics, and prevention. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01366
Brosel-Oliu, S., Ferreira, R., Uria, N., Abramova, N., Gargallo, R., Muñoz-Pascual, F.-X., et al. (2018). Novel impedimetric aptasensor for label-free detection of Escherichia coli O157:H7. Sensors Actuators B: Chem. 255, 2988–2995. doi: 10.1016/j.snb.2017.09.121
Brosseau, N. E., Vallée, I., Mayer-Scholl, A., Ndao, M., Karadjian, G. (2023). Aptamer-based technologies for parasite detection. Sensors 23, 562. doi: 10.3390/s23020562
Canova, M. J., Molle, V. (2014). Bacterial serine/threonine protein kinases in host-pathogen interactions. J. Biol. Chem. 289, 9473–9479). doi: 10.1074/jbc.R113.529917
Capatina, D., Lupoi, T., Feier, B., Blidar, A., Hosu, O., Tertis, M., et al. (2022). Label-free electrochemical aptasensor for the detection of the 3-O-C12-HSL quorum-sensing molecule in Pseudomonas aeruginosa. Biosensors 12, 440. doi: 10.3390/bios12070440
Chadha, J., Harjai, K., Chhibber, S. (2022). Revisiting the virulence hallmarks of Pseudomonas aeruginosa : a chronicle through the perspective of quorum sensing. Environ. Microbiol. 24, 2630–2656. doi: 10.1111/1462-2920.15784
Chauhan, P., Datta, I., Dhiman, A., Shankar, U., Kumar, A., Vashist, A., et al. (2022). DNA aptamer targets Mycobacterium tuberculosis DevR/DosR response regulator function by inhibiting its dimerization and DNA binding activity. ACS Infect. Dis. 8, 2540–2551. doi: 10.1021/acsinfecdis.2c00414
Chen, X., Chang, Y., Ye, M., Wang, Z., Wu, S., Duan, N. (2024). Rational design of a robust g-quadruplex aptamer as an inhibitor to alleviate Listeria monocytogenes infection. ACS Appl. Materials Interfaces 16, 15946–15958. doi: 10.1021/acsami.4c00496
Chen, W., Cui, L., Song, Y., Chen, W., Su, Y., Chang, W., et al. (2021b). Detection of Listeria monocytogenes using luminol-functionalized AuNF-Labeled aptamer recognition and magnetic separation. ACS Omega 6, 26338–26344. doi: 10.1021/acsomega.1c03527
Chen, Z., Luo, H., Gubu, A., Yu, S., Zhang, H., Dai, H., et al. (2023). Chemically modified aptamers for improving binding affinity to the target proteins via enhanced non-covalent bonding. Front. Cell Dev. Biol. 11, 1091809. doi: 10.3389/fcell.2023.1091809
Chen, X., Ma, Y., Xie, Y., Pu, J. (2022a). Aptamer-based applications for cardiovascular disease. Front. Bioengineering Biotechnol. 10. doi: 10.3389/fbioe.2022.1002285
Chen, H., Park, S.-G., Choi, N., Moon, J.-I., Dang, H., Das, A., et al. (2020). SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosensors Bioelectronics 167, 112496. doi: 10.1016/j.bios.2020.112496
Chen, W., Wu, J., Li, S., Zhang, H., Cui, L., Liu, J., et al. (2021c). Ultrasensitive detection of Listeria monocytogenes using solid-state electrochemiluminescence biosensing based on the quenching effect of ferrocene on ruthenium pyridine. J. Food Saf. 41. doi: 10.1111/jfs.12868
Chen, H., Xie, S., Liang, H., Wu, C., Cui, L., Huan, S.-Y., et al. (2017). Generation of biostable L-aptamers against achiral targets by chiral inversion of existing D-aptamers. Talanta 164, 662–667. doi: 10.1016/j.talanta.2016.11.001
Chen, M., Yu, Y., Jiang, F., Zhou, J., Li, Y., Liang, C., et al. (2016). Development of Cell-SELEX technology and its application in cancer diagnosis and therapy. Int. J. Mol. Sci. 17, 2079. doi: 10.3390/ijms17122079
Chen, X.-F., Zhao, X., Yang, Z. (2022b). Aptasensors for the detection of infectious pathogens: design strategies and point-of-care testing. Microchimica Acta 189, 443. doi: 10.1007/s00604-022-05533-w
Chen, S., Zong, X., Zheng, J., Zhang, J., Zhou, M., Chen, Q., et al. (2021a). A colorimetric strategy based on aptamer-catalyzed hairpin assembly for the on-site detection of salmonella typhimurium in milk. Foods 10 (11), 2539. doi: 10.3390/foods10112539
Chessa, D., Spiga, L., de Riu, N., Delaconi, P., Mazzarello, V., Ganau, G., et al. (2014). Lipopolysaccharides belonging to different Salmonella serovars are differentially capable of activating toll-like receptor 4. Infection Immun. 82, 4553–4562. doi: 10.1128/IAI.02297-14
Cohen, M. L. (2000). Changing patterns of infectious disease. Nature 406, 762–767. doi: 10.1038/35021206
Dai, G., Li, Y., Li, Z., Zhang, J., Geng, X., Zhang, F., et al. (2022). Zirconium-based metal–organic framework and Ti3C2Tx nanosheet-based faraday cage-type electrochemical aptasensor for Escherichia coli detection. ACS Appl. Nano Materials 5, 9201–9208. doi: 10.1021/acsanm.2c01548
Damborský, P., Švitel, J., Katrlík, J. (2016). Optical biosensors. Essays Biochem. 60, 91–100. doi: 10.1042/EBC20150010
Derbyshire, N., White, S. J., Bunka, D. H. J., Song, L., Stead, S., Tarbin, J., et al. (2012). Toggled RNA aptamers against aminoglycosides allowing facile detection of antibiotics using gold nanoparticle assays. Analytical Chem. 84, 6595–6602. doi: 10.1021/ac300815c
Dhiman, A., Kumar, C., Mishra, S. K., Sikri, K., Datta, I., Sharma, P., et al. (2019). Theranostic application of a novel G-quadruplex-forming DNA aptamer targeting malate synthase of Mycobacterium tuberculosis. Mol. Ther. Nucleic Acids 18, 661–672. doi: 10.1016/j.omtn.2019.09.026
Diagne, C. T., Diallo, D., Faye, O., Ba, Y., Faye, O., Gaye, A., et al. (2015). Potential of selected Senegalese Aedes spp. mosquitoes (Diptera: Culicidae) to transmit Zika virus. BMC Infect. Dis. 15, 492. doi: 10.1186/s12879-015-1231-2
Di Giusto, D. A., Knox, S. M., Lai, Y., Tyrelle, G. D., Aung, M. T., King, G. C. (2006). Multitasking by multivalent circular DNA aptamers. ChemBioChem 7, 535–544. doi: 10.1002/cbic.200500316
Ding, Y., Liu, J. (2023). Quantitative comparison of Capture-SELEX, GO-SELEX, and Gold-SELEX for enrichment of aptamers. Analytical Chem. 95, 14651–14658. doi: 10.1021/acs.analchem.3c02477
Ding, D., Zhao, H., Wei, D., Yang, Q., Yang, C., Wang, R., et al. (2023). The first-in-human whole-body dynamic pharmacokinetics study of aptamer. Research 6. doi: 10.34133/research.0126
Doherty, M. K., Shaw, C., Woods, L., Weimer, B. C. (2023). Alpha-gal bound aptamer and vancomycin synergistically reduce Staphylococcus aureus infection In vivo. Microorganisms 11 (7), 1776. doi: 10.3390/microorganisms11071776
Dong, Y., Wang, Z., Wang, S., Wu, Y., Ma, Y., Liu, J. (2018). “Introduction of SELEX and important SELEX variants,” in Aptamers for Analytical Applications: Affinity Acquisition and Method Design, pp.1–pp25.
Dramsi, S., Cossart, P. (2002). Listeriolysin O. J. Cell Biol. 156, 943–946. doi: 10.1083/jcb.200202121
Du, J., Chen, X., Liu, K., Zhao, D., Bai, Y. (2022). Dual recognition and highly sensitive detection of Listeria monocytogenes in food by fluorescence enhancement effect based on Fe3O4@ZIF-8-aptamer. Sensors Actuators B: Chem. 360, 131654. doi: 10.1016/j.snb.2022.131654
Duan, N., Chang, B., Zhang, H., Wang, Z., Wu, S. (2016). Salmonella typhimurium detection using a surface-enhanced Raman scattering-based aptasensor. Int. J. Food Microbiol. 218, 38–43. doi: 10.1016/j.ijfoodmicro.2015.11.006
Duan, N., Wu, S., Zhu, C., Ma, X., Wang, Z., Yu, Y., et al. (2012). Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella Typhimurium and Staphylococcus aureus. Analytica Chimica Acta 723, 1–6. doi: 10.1016/j.aca.2012.02.011
El-Wekil, M. M., Halby, H. M., Darweesh, M., Ali, M. E., Ali, R. (2022). An innovative dual recognition aptasensor for specific detection of Staphylococcus aureus based on Au/Fe3O4 binary hybrid. Sci. Rep. 12, 12502. doi: 10.1038/s41598-022-15637-1
Escolano, J. M., Díaz-Durán, B., DeMiguel-Ramos, M., Olivares, J., Geday, M. A., Iborra, E. (2017). Selection of aptamers to Neisseria meningitidis and Streptococcus pneumoniae surface specific proteins and affinity assay using thin film AlN resonators. Sensors Actuators B: Chem. 246, 591–596. doi: 10.1016/j.snb.2017.02.098
Eyer, L., Zouharová, D., Širmarová, J., Fojtíková, M., Štefánik, M., Haviernik, J., et al. (2017). Antiviral activity of the adenosine analogue BCX4430 against West Nile virus and tick-borne flaviviruses. Antiviral Res. 142, 63–67. doi: 10.1016/j.antiviral.2017.03.012
Fair, R. J., Tor, Y. (2014). Antibiotics and bacterial resistance in the 21st century. Perspect. Medicinal Chem. 6, PMC.S14459. doi: 10.4137/PMC.S14459
Fang, S., Song, D., Zhuo, Y., Chen, Y., Zhu, A., Long, F. (2021). Simultaneous and sensitive determination of Escherichia coli O157:H7 and Salmonella typhimurium using evanescent wave dual-color fluorescence aptasensor based on micro/nano size effect. Biosensors Bioelectronics 185, 113288. doi: 10.1016/j.bios.2021.113288
Fathi, S., Saber, R., Adabi, M., Rasouli, R., Douraghi, M., Morshedi, M., et al. (2020). Novel competitive voltammetric aptasensor based on electrospun carbon nanofibers-gold nanoparticles modified graphite electrode for salmonella enterica serovar detection. Biointerface Res. Appl. Chem. 11, 8702–8715. doi: 10.33263/BRIAC112.87028715
Faucher, S. P., Viau, C., Gros, P. P., Daigle, F., Le Moual, H. (2008). The prpZ gene cluster encoding eukaryotic-type Ser/Thr protein kinases and phosphatases is repressed by oxidative stress and involved in Salmonella enterica serovar Typhi survival in human macrophages. FEMS Microbiol. Lett. 281, 160–166. doi: 10.1111/j.1574-6968.2008.01094.x
Feng, H., Beck, J., Nassal, M., Hu, K. (2011). A SELEX-Screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS One 6, e27862. doi: 10.1371/journal.pone.0027862
Ferhan, A. R., Jackman, J. A., Cho, N.-J. (2016). Integration of quartz crystal microbalance-dissipation and reflection-mode localized surface plasmon resonance sensors for biomacromolecular interaction analysis. Analytical Chem. 88, 12524–12531. doi: 10.1021/acs.analchem.6b04303
Fowler, C. C., Galán, J. E. (2018). Decoding a Salmonella typhi regulatory network that controls typhoid toxin expression within human cells. Cell Host Microbe 23, 65–76.e6. doi: 10.1016/j.chom.2017.12.001
Furuta, Y., Takahashi, K., Shiraki, K., Sakamoto, K., Smee, D. F., Barnard, D. L., et al. (2009). T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 82, 95–102. doi: 10.1016/j.antiviral.2009.02.198
Galán, J. E. (2016). Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc. Natl. Acad. Sci. 113, 6338–6344. doi: 10.1073/pnas.1606335113
Gao, R., Zhong, Z., Gao, X., Jia, L. (2018). Graphene oxide quantum dots assisted construction of fluorescent aptasensor for rapid detection of Pseudomonas aeruginosa in food samples. J. Agric. Food Chem. 66, 10898–10905. doi: 10.1021/acs.jafc.8b02164
García-Recio, E. M., Pinto-Díez, C., Pérez-Morgado, M. I., García-Hernández, M., Fernández, G., Martín, M. E., et al. (2016). Characterization of MNK1b DNA Aptamers that inhibit proliferation in MDA-MB231 breast cancer cells. Mol. Ther. - Nucleic Acids 5, e275. doi: 10.1038/mtna.2015.50
Ge, C., Feng, J., Zhang, J., Hu, K., Wang, D., Zha, L., et al. (2022). Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein. Talanta 236, 122847. doi: 10.1016/j.talanta.2021.122847
Gerlach, R. G., Jäckel, D., Stecher, B., Wagner, C., Lupas, A., Hardt, W. D., et al. (2007). Salmonella Pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9, 1834–1850. doi: 10.1111/j.1462-5822.2007.00919.x
Ghassami, E., Varshosaz, J., Jahanian-Najafabadi, A., Minaiyan, M., Rajabi, P., Hayati, E. (2018). Pharmacokinetics and in vitro/in vivo antitumor efficacy of aptamer-targeted Ecoflex® nanoparticles for docetaxel delivery in ovarian cancer. Int. J. Nanomedicine Volume 13, 493–504. doi: 10.2147/IJN.S152474
Gholikhani, T., Kumar, S., Valizadeh, H., Mahdinloo, S., Adibkia, K., Zakeri-Milani, P., et al. (2022). Advances in aptamers-based applications in breast cancer: drug delivery, therapeutics, and diagnostics. Int. J. Mol. Sci. 23, 14475. doi: 10.3390/ijms232214475
Gopinath, S. C. B. (2007). Methods developed for SELEX. Analytical Bioanalytical Chem. 387, 171–182. doi: 10.1007/s00216-006-0826-2
Gopinath, S. C. B., Misono, T. S., Kawasaki, K., Mizuno, T., Imai, M., Odagiri, T., et al. (2006). An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 87, 479–487. doi: 10.1099/vir.0.81508-0
Gribanyov, D., Zhdanov, G., Olenin, A., Lisichkin, G., Gambaryan, A., Kukushkin, V., et al. (2021). SERS-Based colloidal aptasensors for quantitative determination of influenza virus. Int. J. Mol. Sci. 22, 1842. doi: 10.3390/ijms22041842
Griffin, L., Tidmarsh, G., Bock, L., Toole, J., Leung, L. (1993). In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood 81, 3271–3276. doi: 10.1182/blood.V81.12.3271.3271
Gründling, A., Burrack, L. S., Bouwer, H. A., Higgins, D. E. (2004). Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci. 101 (33), 12318–12323.
Hamedani, N. S., Müller, J. (2016). Capillary electrophoresis for the selection of DNA aptamers recognizing activated protein C. Nucleic acid aptamers: selection, characterization, and application. 1380, 61–75. doi: 10.1007/978-1-4939-3197-2_5
Hameed, S. S., Al-Ogaili, A. S., Noori, N. (2022). Single-stranded DNA aptamer-based rolling circle amplification as anti-chicken Salmonella bacteriostatic. Veterinary World 15, 1171–1176. doi: 10.14202/vetworld.2022.1171-1176
Hernandez, F. J., Stockdale, K. R., Huang, L., Horswill, A. R., Behlke, M. A., McNamara, J. O. (2012). Degradation of nuclease-stabilized RNA oligonucleotides in mycoplasma-contaminated cell culture media. Nucleic Acid Ther. 22, 58–68. doi: 10.1089/nat.2011.0316
Hong, X., Xue, L., Gao, J., Jiang, Y., Kou, X. (2022). Epochal coevolution of minor capsid protein in norovirus GII.4 variants with major capsid protein based on their interactions over the last five decades. Virus Res. 319, 198860. doi: 10.1016/j.virusres.2022.198860
Huang, X., Zheng, M., Wang, P., Mok, B. W. Y., Liu, S., Lau, S. Y., et al. (2017). An NS-segment exonic splicing enhancer regulates influenza A virus replication in mammalian cells. Nat. Commun. 8 (1), 14751. doi: 10.1038/ncomms14751
Hui, Y., Peng, H., Zhang, F., Zhang, L., Liu, Y., Jia, R., et al. (2022). An ultrasensitive sandwich-type electrochemical aptasensor using silver nanoparticle/titanium carbide nanocomposites for the determination of Staphylococcus aureus in milk. Microchimica Acta 189, 276. doi: 10.1007/s00604-022-05349-8
Jeon, S. H., Kayhan, B., Ben-Yedidia, T., Arnon, R. (2004). A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 279, 48410–48419. doi: 10.1074/jbc.M409059200
Jia, F., Bai, X., Zhang, X., Fu, Y., Li, Y., Li, X., et al. (2021). A low-field magnetic resonance imaging aptasensor for the rapid and visual sensing of Pseudomonas aeruginosa in food, juice, and water. Analytical Chem. 93, 8631–8637. doi: 10.1021/acs.analchem.1c01669
Jia, F., Duan, N., Wu, S., Dai, R., Wang, Z., Li, X. (2016). Impedimetric Salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchimica Acta 183, 337–344. doi: 10.1007/s00604-015-1649-7
Jia, W., Li, H., Wilkop, T., Liu, X., Yu, X., Cheng, Q., et al. (2018). Silver decahedral nanoparticles empowered SPR imaging-SELEX for high throughput screening of aptamers with real-time assessment. Biosensors Bioelectronics 109, 206–213. doi: 10.1016/j.bios.2018.02.029
Jia, F., Xu, L., Yan, W., Wu, W., Yu, Q., Tian, X., et al. (2017). A magnetic relaxation switch aptasensor for the rapid detection of Pseudomonas aeruginosa using superparamagnetic nanoparticles. Microchimica Acta 184, 1539–1545. doi: 10.1007/s00604-017-2142-2
Jiang, L., Wang, H., Chen, S. (2020). Aptamer (AS1411)-conjugated liposome for enhanced therapeutic efficacy of mirna-29b in ovarian cancer. J. Nanoscience Nanotechnology 20, 2025–2031. doi: 10.1166/jnn.2020.17301
Johnson, J. R. (1991). Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4, 80–128. doi: 10.1128/CMR.4.1.80
Jonsson, S., Musher, D. M., Chapman, A., Goree, A., Lawrence, E. C. (1985). Phagocytosis and killing of common bacterial pathogens of the lung by human alveolar macrophages. J. Infect. Dis. 152, 4–13. doi: 10.1093/infdis/152.1.4
Kadurugamuwa, J. L., Beveridge, T. J. (1995). Virulence factors are released from pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177 (14), 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Kalra, P., Mishra, S. K., Kaur, S., Kumar, A., Prasad, H. K., Sharma, T. K., et al. (2018). G-Quadruplex-forming DNA aptamers inhibit the dna-binding function of HupB and Mycobacterium tuberculosis entry into host cells. Mol. Ther. Nucleic Acids 13, 99–109. doi: 10.1016/j.omtn.2018.08.011
Kamiyama, N., Soma, R., Hidano, S., Watanabe, K., Umekita, H., Fukuda, C., et al. (2017). Ribavirin inhibits Zika virus (ZIKV) replication in vitro and suppresses viremia in ZIKV-infected STAT1-deficient mice. Antiviral Res. 146, 1–11. doi: 10.1016/j.antiviral.2017.08.007
Kaper, J. B., Nataro, J. P., Mobley, H. L. T. (2004). Pathogenic escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Kaur, H., Shorie, M., Sabherwal, P. (2020). Electrochemical aptasensor using boron-carbon nanorods decorated by nickel nanoparticles for detection of E. coli O157:H7. Microchimica Acta 187, 461. doi: 10.1007/s00604-020-04444-y
Kaur, H., Shorie, M., Sharma, M., Ganguli, A. K., Sabherwal, P. (2017). Bridged rebar graphene functionalized aptasensor for pathogenic E. coli O78:K80:H11 detection. Biosensors Bioelectronics 98, 486–493. doi: 10.1016/j.bios.2017.07.004
Keefe, A. D., Pai, S., Ellington, A. (2010). Aptamers as therapeutics. Nat. Rev. Drug Discovery 9, 537–550. doi: 10.1038/nrd3141
Keller, L. E., Jones, C. V., Thornton, J. A., Sanders, M. E., Swiatlo, E., Nahm, M. H., et al. (2013). PspK of Streptococcus pneumoniae increases adherence to epithelial cells and enhances nasopharyngeal colonization. Infection Immun. 81, 173–181. doi: 10.1128/IAI.00755-12
Ker, D. S., Jenkins, H. T., Greive, S. J., Antson, A. A. (2021). CryoEM structure of the Nipah virus nucleocapsid assembly. PLoS Pathog. 17 (7), e1009740. doi: 10.1371/journal.ppat.1009740
Kim, B., Chung, K. W., Lee, J. H. (2018a). Non-stop aptasensor capable of rapidly monitoring norovirus in a sample. J. Pharm. Biomed. Anal. 152, 315–321. doi: 10.1016/j.jpba.2018.02.022
Kim, S., Lee, S., Lee, H. J. (2018b). An aptamer-aptamer sandwich assay with nanorod-enhanced surface plasmon resonance for attomolar concentration of norovirus capsid protein. Sensors Actuators B: Chem. 273, 1029–1036. doi: 10.1016/j.snb.2018.06.108
Kim, E. S., Lu, C., Khuri, F. R., Tonda, M., Glisson, B. S., Liu, D., et al. (2001). A phase II study of STEALTH cisplatin (SPI-77) in patients with advanced non-small cell lung cancer. Lung Cancer 34, 427–432. doi: 10.1016/S0169-5002(01)00278-1
Kinghorn, A., Fraser, L., Liang, S., Shiu, S., Tanner, J. (2017). Aptamer bioinformatics. Int. J. Mol. Sci. 18, 2516. doi: 10.3390/ijms18122516
Komarova, N., Kuznetsov, A. (2019). Inside the black box: what makes SELEX better? Molecules 24, 3598. doi: 10.3390/molecules24193598
Kong, H. Y., Byun, J. (2013). Nucleic acid aptamers: new methods for selection, stabilization, and application in biomedical science. Biomolecules Ther. 21, 423–434. doi: 10.4062/biomolther.2013.085
Kovacevic, K. D., Gilbert, J. C., Jilma, B. (2018). Pharmacokinetics, pharmacodynamics and safety of aptamers. Advanced Drug Delivery Rev. 134, 36–50. doi: 10.1016/j.addr.2018.10.008
Kristian, S. A., Hwang, J. H., Hall, B., Leire, E., Iacomini, J., Old, R., et al. (2015). Retargeting pre-existing human antibodies to a bacterial pathogen with an alpha-Gal conjugated aptamer. J. Mol. Med. 93, 619–631. doi: 10.1007/s00109-015-1280-4
Krüger, A., de Jesus Santos, A. P., de Sá, V., Ulrich, H., Wrenger, C. (2021). Aptamer applications in emerging viral diseases. Pharmaceuticals 14, 622. doi: 10.3390/ph14070622
Ku, T.-H., Zhang, T., Luo, H., Yen, T., Chen, P.-W., Han, Y., et al. (2015). Nucleic acid aptamers: an emerging tool for biotechnology and biomedical sensing. Sensors 15, 16281–16313. doi: 10.3390/s150716281
Kumar De, S., Ray, S., Rawat, Y., Mondal, S., Nandy, A., Verma, P., et al. (2021). Porous Au-seeded Ag nanorod networks conjugated with DNA aptamers for impedimetric sensing of DENV-2. Sensors Actuators B: Chem. 348, 130709. doi: 10.1016/j.snb.2021.130709
Kusumawati, A., Mustopa, A. Z., Umami, R. N., Santoso, A., Wibawan, I. W. T., Setiyono, A., et al. (2022). Antibiofilm activity and binding specificity of polyclonal dna aptamers on Staphylococcus aureus and Escherichia coli. Microbiol. Biotechnol. Lett. 50, 328–336. doi: 10.48022/mbl.2206.06001
Kwong Hong Tsang, D., Lieberthal, T. J., Watts, C., Dunlop, I. E., Ramadan, S., del Rio Hernandez, A. E., et al. (2019). Chemically functionalised graphene fet biosensor for the label-free sensing of exosomes. Sci. Rep. 9, 13946. doi: 10.1038/s41598-019-50412-9
Lai, H.-C., Wang, C.-H., Liou, T.-M., Lee, G.-B. (2014). Influenza A virus-specific aptamers screened by using an integrated microfluidic system. Lab. Chip 14, 2002–2013. doi: 10.1039/C4LC00187G
Lebars, I., Richard, T., Di Primo, C., Toulmé, J.-J. (2007). LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood Cells Molecules Dis. 38, 204–209. doi: 10.1016/j.bcmd.2006.11.008
Ledlod, S., Areekit, S., Santiwatanakul, S., Chansiri, K. (2020). Colorimetric aptasensor for detecting Salmonella spp., Listeria monocytogenes, and Escherichia coli in meat samples. Food Sci. Technol. Int. 26, 430–443. doi: 10.1177/1082013219899593
Lee, C. H., Lee, S.-H., Kim, J. H., Noh, Y.-H., Noh, G.-J., Lee, S.-W. (2015). Pharmacokinetics of a cholesterol-conjugated aptamer against the Hepatitis C Virus (HCV) NS5B protein. Mol. Ther. - Nucleic Acids 4, e254. doi: 10.1038/mtna.2015.30
Lee, J. Y., Nguyen, T. T. N., Myoung, J. (2020). Zika virus-encoded NS2A and NS4A strongly downregulate NF-κB promoter activity. J. Microbiol. Biotechnol. 30, 1651–1658. doi: 10.4014/JMB.2011.11003
Lee, B., Park, J., Ryu, M., Kim, S., Joo, M., Yeom, J. H., et al. (2017). Antimicrobial peptide-loaded gold nanoparticle-DNA aptamer conjugates as highly effective antibacterial therapeutics against Vibrio vulnificus. Sci. Rep. 7 (1), 13572. doi: 10.1038/s41598-017-14127-z
Lerminiaux, N. A., MacKenzie, K. D., Cameron, A. D. S. (2020). Salmonella pathogenicity island 1 (spi-1): the evolution and stabilization of a core genomic type three secretion system. Microorganisms 8, 576. doi: 10.3390/microorganisms8040576
Leung, D. W., Prins, K. C., Basler, C. F., Amarasinghe, G. K. (2010). Ebolavirus VP35 is a multifunctional virulence factor. Virulence 1, 526–531. doi: 10.4161/viru.1.6.12984
Li, Y., Chen, M., Fan, X., Peng, J., Pan, L., Tu, K., et al. (2022). Sandwich fluorometric method for dual-role recognition of Listeria monocytogenes based on antibiotic-affinity strategy and fluorescence quenching effect. Analytica Chimica Acta 1221, 340085. doi: 10.1016/j.aca.2022.340085
Li, T., Ou, G., Chen, X., Li, Z., Hu, R., Li, Y., et al. (2020). Naked-eye based point-of-care detection of E.coli O157: H7 by a signal-amplified microfluidic aptasensor. Analytica Chimica Acta 1130, 20–28. doi: 10.1016/j.aca.2020.07.031
Li, J., Yun, W., Zhang, H., Chen, L., Ho, H.-P., Pu, X., et al. (2023). MoS2 nanosheets based label-free colorimetric aptasensor for Escherichia coli O157: H7 detection. Colloids Surfaces A: Physicochemical Eng. Aspects 674, 131955. doi: 10.1016/j.colsurfa.2023.131955
Li, A., Zuo, P., Ye, B.-C. (2021). An aptamer biosensor based dual signal amplification system for the detection of Salmonella typhimurium. Analytical Biochem. 615, 114050. doi: 10.1016/j.ab.2020.114050
Lier, C., Becker, S., Biedenkopf, N. (2017). Dynamic phosphorylation of Ebola virus VP30 in NP-induced inclusion bodies. Virology 512, 39–47. doi: 10.1016/j.virol.2017.09.006
Lijuan, C., Xing, Y., Minxi, W., Wenkai, L., Le, D. (2017). Development of an aptamer-ampicillin conjugate for treating biofilms. Biochem. Biophys. Res. Commun. 483, 847–854. doi: 10.1016/j.bbrc.2017.01.016
Liu, R., Zhang, Y., Ali, S., Haruna, S. A., He, P., Li, H., et al. (2021). Development of a fluorescence aptasensor for rapid and sensitive detection of Listeria monocytogenes in food. Food Control 122, 107808. doi: 10.1016/j.foodcont.2020.107808
Lum, J., Wang, R., Hargis, B., Tung, S., Bottje, W., Lu, H., et al. (2015). An impedance aptasensor with microfluidic chips for specific detection of H5N1 avian influenza virus. Sensors 15, 18565–18578. doi: 10.3390/s150818565
Lunardelli, V. A. S., Almeida, B. S., Apostolico, J. S., Rezende, T., Yamamoto, M. M., Pereira, S. S., et al. (2023). Diagnostic and vaccine potential of Zika virus envelope protein (E) derivates produced in bacterial and insect cells. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1071041
Ma, X., Xie, L., Wartchow, C., Warne, R., Xu, Y., Rivkin, A., et al. (2017). Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit. Sci. Rep. 7 (1), 9385. doi: 10.1038/s41598-017-09538-x
Ma, X., Xu, X., Xia, Y., Wang, Z. (2018). SERS aptasensor for Salmonella typhimurium detection based on spiny gold nanoparticles. Food Control 84, 232–237. doi: 10.1016/j.foodcont.2017.07.016
Maasch, C., Buchner, K., Eulberg, D., Vonhoff, S., Klussmann, S. (2008). Physicochemical stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for therapeutic applications. Nucleic Acids Symposium Ser. 52, 61–62. doi: 10.1093/nass/nrn031
Majdinasab, M., Badea, M., Marty, J. L. (2022). Aptamer-based lateral flow assays: current trends in clinical diagnostic rapid tests. Pharmaceuticals 15, 90. doi: 10.3390/ph15010090
Marton, S., Cleto, F., Krieger, M. A., Cardoso, J. (2016). Isolation of an aptamer that binds specifically to E. coli. PLoS One 11, e0153637. doi: 10.1371/journal.pone.0153637
Matchawong, A., Srisawat, C., Sangboonruang, S., Tharinjaroen, C. S. (2022). The ability of nuclease-resistant RNA aptamer against Streptococcus suis Serotype 2, Strain P1/7 to reduce biofilm formation in vitro. Molecules 27, 3894. doi: 10.3390/molecules27123894
Mayer, G., Ahmed, M.-S. L., Dolf, A., Endl, E., Knolle, P. A., Famulok, M. (2010). Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 5, 1993–2004. doi: 10.1038/nprot.2010.163
McAuley, J. L., Gilbertson, B. P., Trifkovic, S., Brown, L. E., McKimm-Breschkin, J. L. (2019). Influenza virus neuraminidase structure and functions. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00039
Mishra, A., Pilloton, R., Jain, S., Roy, S., Khanuja, M., Mathur, A., et al. (2022). Paper-based electrodes conjugated with tungsten disulfide nanostructure and aptamer for impedimetric detection of Listeria monocytogenes. Biosensors 12, 88. doi: 10.3390/bios12020088
Mok, J., Jeon, J., Jo, J., Kim, E., Ban, C. (2021). Novel one-shot fluorescent aptasensor for dengue fever diagnosis using NS1-induced structural change of G-quadruplex aptamer. Sensors Actuators B: Chem. 343, 130077. doi: 10.1016/j.snb.2021.130077
Moon, J., Kim, G., Lee, S., Park, S. (2013). Identification of Salmonella Typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J. Microbiological Methods 95, 162–166. doi: 10.1016/j.mimet.2013.08.005
Mori, Y., Nakamura, Y., Ohuchi, S. (2012). Inhibitory RNA aptamer against SP6 RNA polymerase. Biochem. Biophys. Res. Commun. 420, 440–443. doi: 10.1016/j.bbrc.2012.03.014
Muniandy, S., Teh, S. J., Appaturi, J. N., Thong, K. L., Lai, C. W., Ibrahim, F., et al. (2019). A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 127, 136–144. doi: 10.1016/j.bioelechem.2019.02.005
Musafia, B., Oren-Banaroya, R., Noiman, S. (2014). Designing anti-influenza aptamers: Novel quantitative structure activity relationship approach gives insights into aptamer – virus interaction. PLoS One 9, e97696. doi: 10.1371/journal.pone.0097696
Nguyen, V.-T., Kwon, Y. S., Kim, J. H., Gu, M. B. (2014). Multiple GO-SELEX for efficient screening of flexible aptamers. Chem. Commun. 50, 10513–10516. doi: 10.1039/C4CC03953J
Ni, S., Yao, H., Wang, L., Lu, J., Jiang, F., Lu, A., et al. (2017). Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 18, 1683. doi: 10.3390/ijms18081683
Ni, S., Zhuo, Z., Pan, Y., Yu, Y., Li, F., Liu, J., et al. (2021). Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Materials Interfaces 13, 9500–9519. doi: 10.1021/acsami.0c05750
Ning, Y., Wang, X., Chen, P., Liu, S., Hu, J., Xiao, R., et al. (2022). Targeted inhibition of methicillin-resistant Staphylococcus aureus biofilm formation by a graphene oxide-loaded aptamer/berberine bifunctional complex. Drug Delivery 29, 1675–1683. doi: 10.1080/10717544.2022.2079768
Ninomiya, K., Yamashita, T., Kawabata, S., Shimizu, N. (2014). Targeted and ultrasound-triggered drug delivery using liposomes co-modified with cancer cell-targeting aptamers and a thermosensitive polymer. Ultrasonics Sonochemistry 21, 1482–1488. doi: 10.1016/j.ultsonch.2013.12.023
Nishimoto, K. P., Newkirk, D., Hou, S., Fruehauf, J., Nelson, E. L. (2007). Fluorescence activated cell sorting (FACS) using RNAlater to minimize RNA degradation and perturbation of mRNA expression from cells involved in initial host microbe interactions. J. Microbiological Methods 70, 205–208. doi: 10.1016/j.mimet.2007.03.022
Nishimoto, A. T., Rosch, J. W., Tuomanen, E. I. (2020). Pneumolysin: pathogenesis and therapeutic target. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01543
Nobusawa, E. (1997). Structure and function of the hemagglutinin of influenza viruses. Nihon Rinsho 55, 2562–2569. Japanese Journal of Clinical Medicine.
Nutho, B., Mulholland, A. J., Rungrotmongkol, T. (2019). The reaction mechanism of Zika virus NS2B/NS3 serine protease inhibition by dipeptidyl aldehyde: A QM/MM study. Phys. Chem. Chem. Phys. 21, 14945–14956. doi: 10.1039/c9cp02377a
O’Neill, R. E., Talon, J., Palese, P. (1998). The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17 (1), 288–296. doi: 10.1093/emboj/17.1.288
Ohuchi, S. (2012). Cell-SELEX technology. Biores. Open Access 1, 265–272. doi: 10.1089/biores.2012.0253
Ommen, P., Hansen, L., Hansen, B. K., Vu-Quang, H., Kjems, J., Meyer, R. L. (2022). Aptamer-targeted drug delivery for Staphylococcus aureus biofilm. Front. Cell. Infection Microbiol. 12. doi: 10.3389/fcimb.2022.814340
Oravczová, V., Tatarko, M., Süle, J., Hun, M., Kerényi, Z., Hucker, A., et al. (2020). Detection of Listeria innocua by acoustic aptasensor. In Proceedings. MDPI 60 (1), 8. doi: 10.3390/IECB2020-07079
Oroh, S. B., Mustopa, A. Z., Budiarti, S., Budiarto, B. R. (2020). Inhibition of enteropathogenic Escherichia coli biofilm formation by DNA aptamer. Mol. Biol. Rep. 47, 7567–7573. doi: 10.1007/s11033-020-05822-8
Otte, D.-M., Choukeife, M., Patwari, T., Mayer, G. (2022). “Nucleic acid aptamers: from basic research to clinical applications,” in Handbook of chemical Biology of Nucleic Acids (Springer Nature, Singapore), 1–25. doi: 10.1007/978-981-16-1313-5_25-1
Otto, M. (2014). Staphylococcus aureus toxins. Curr. Opin. In Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Palacio, I., Moreno, M., Náñez, A., Purwidyantri, A., Domingues, T., Cabral, P. D., et al. (2023). Attomolar detection of hepatitis C virus core protein powered by molecular antenna-like effect in a graphene field-effect aptasensor. Biosensors Bioelectronics 222, 115006. doi: 10.1016/j.bios.2022.115006
Palmqvist, N., Foster, T., Tarkowski, A., Josefsson, E. (2002). Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microbial Pathogenesis 33, 239–249. doi: 10.1006/mpat.2002.0533
Pan, Q., Zhang, X. L., Wu, H. Y., He, P. W., Wang, F., Zhang, M. S., et al. (2005). Aptamers that preferentially bind type IVB pili and inhibit human monocytic-cell invasion by Salmonella enterica serovar typhi. Antimicrobial Agents Chemotherapy 49, 4052–4060. doi: 10.1128/AAC.49.10.4052-4060.2005
Pang, Y., Rong, Z., Wang, J., Xiao, R., Wang, S. (2015). A fluorescent aptasensor for H5N1 influenza virus detection based-on the core–shell nanoparticles metal-enhanced fluorescence (MEF). Biosensors Bioelectronics 66, 527–532. doi: 10.1016/j.bios.2014.10.052
Park, G., Lee, M., Kang, J., Park, C., Min, J., Lee, T. (2022). Selection of DNA aptamer and its application as an electrical biosensor for Zika virus detection in human serum. Nano Convergence 9, 41. doi: 10.1186/s40580-022-00332-8
Park, H., Lee, H., Lee, M., Baek, C., Park, J. A., Jang, M., et al. (2023). Synthesis of isolated DNA aptamer and its application of AC-electrothermal flow-based rapid biosensor for the detection of dengue virus in a spiked sample. Bioconjugate Chem. 34, 1486–1497. doi: 10.1021/acs.bioconjchem.3c00249
Pathania, P., Sharma, A., Kumar, B., Rishi, P., Raman Suri, C. (2017). Selective identification of specific aptamers for the detection of non-typhoidal salmonellosis in an apta-impedimetric sensing format. Microchimica Acta 184, 1499–1508. doi: 10.1007/s00604-017-2098-2
Pebdeni, A. B., Hosseini, M., Ganjali, M. R. (2020). Fluorescent turn-on aptasensor of Staphylococcus aureus based on the FRET between green carbon quantum dot and gold nanoparticle. Food Analytical Methods 13, 2070–2079. doi: 10.1007/s12161-020-01821-4
Perschbacher, K., Smestad, J. A., Peters, J. P., Standiford, M. M., Denic, A., Wootla, B., et al. (2015). Quantitative PCR Analysis of DNA aptamer pharmacokinetics in mice. Nucleic Acid Ther. 25, 11–19. doi: 10.1089/nat.2014.0515
Petrišič, N., Kozorog, M., Aden, S., Podobnik, M., Anderluh, G. (2021). The molecular mechanisms of listeriolysin O-induced lipid membrane damage. Biochim. Biophys. Acta (BBA) - Biomembranes 1863, 183604. doi: 10.1016/j.bbamem.2021.183604
Pfander, S., Fiammengo, R., Kirin, S. I., Metzler-Nolte, N., Jäschke, A. (2007). Reversible site-specific tagging of enzymatically synthesized RNAs using aldehyde–hydrazine chemistry and protease-cleavable linkers. Nucleic Acids Res. 35, e25. doi: 10.1093/nar/gkl1110
Pistor, S., Chakraborty, T., Niebuhr, K., Domann, E., Wehland, J. (1994). The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 13, 758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x
Portnoy, D. A., Jacks, P. S., Hinrichs, D. J. (1988). Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 167 (4), 1459–1471. doi: 10.1084/jem.167.4.1459
Qi, X., Ye, Y., Wang, H., Zhao, B., Xu, L., Zhang, Y., et al. (2022). An ultrasensitive and dual-recognition SERS biosensor based on Fe3O4@Au-Teicoplanin and aptamer functionalized Au@Ag nanoparticles for detection of Staphylococcus aureus. Talanta 250, 123648. doi: 10.1016/j.talanta.2022.123648
Raddatz, M. L., Dolf, A., Endl, E., Knolle, P., Famulok, M., Mayer, G. (2008). Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angewandte Chemie Int. Edition 47, 5190–5193. doi: 10.1002/anie.200800216
Raffatellu, M., Wilson, R. P., Chessa, D., Andrews-Polymenis, H., Tran, Q. T., Lawhon, S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 Contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infection Immun. 73, 146–154. doi: 10.1128/IAI.73.1.146-154.2005
Rahbi, F. A., Salmi, I. A., Khamis, F., Balushi, Z. A., Pandak, N., Petersen, E., et al. (2023). Physicians’ attitudes, knowledge, and practices regarding antibiotic prescriptions. J. Global Antimicrobial Resistance 32, 58–65. doi: 10.1016/j.jgar.2022.12.005
Raji, M. A., Suaifan, G., Shibl, A., Weber, K., Cialla-May, D., Popp, J., et al. (2021). Aptasensor for the detection of Methicillin resistant Staphylococcus aureus on contaminated surfaces. Biosensors Bioelectronics 176, 112910. doi: 10.1016/j.bios.2020.112910
Ramanathan, S., Gopinath, S. C. B., Ismail, Z. H., Md Arshad, M. K., Poopalan, P. (2022). Aptasensing nucleocapsid protein on nanodiamond assembled gold interdigitated electrodes for impedimetric SARS-CoV-2 infectious disease assessment. Biosensors Bioelectronics 197, 113735. doi: 10.1016/j.bios.2021.113735
Ranadheera, C., Proulx, R., Chaiyakul, M., Jones, S., Grolla, A., Leung, A., et al. (2018). The interaction between the Nipah virus nucleocapsid protein and phosphoprotein regulates virus replication. Sci. Rep. 8 (1), 15994. doi: 10.1038/s41598-018-34484-7
Ranjbar, S., Shahrokhian, S. (2018). Design and fabrication of an electrochemical aptasensor using Au nanoparticles/carbon nanoparticles/cellulose nanofibers nanocomposite for rapid and sensitive detection of Staphylococcus aureus. Bioelectrochemistry 123, 70–76. doi: 10.1016/j.bioelechem.2018.04.018
Rastogi, M., Singh, S. K. (2020). Zika virus NS1 affects the junctional integrity of human brain microvascular endothelial cells. Biochimie 176, 52–61. doi: 10.1016/j.biochi.2020.06.011
Reich, P., Stoltenburg, R., Strehlitz, B., Frense, D., Beckmann, D. (2017). Development of an impedimetric aptasensor for the detection of Staphylococcus aureus. Int. J. Mol. Sci. 18, 2484. doi: 10.3390/ijms18112484
Ren, J., Liang, G., Man, Y., Li, A., Jin, X., Liu, Q., et al. (2019). Aptamer-based fluorometric determination of Salmonella typhimurium using Fe3O4 magnetic separation and CdTe quantum dots. PLoS One 14, e0218325. doi: 10.1371/journal.pone.0218325
Reverdatto, S., Burz, D., Shekhtman, A. (2015). Peptide aptamers: Development and applications. Curr. Topics Medicinal Chem. 15, 1082–1101. doi: 10.2174/1568026615666150413153143
RM, R., Maroli, N., J, A., Ponmalai, K., K, K. (2020). Highly adaptable and sensitive FRET-based aptamer assay for the detection of Salmonella paratyphi A. Spectrochimica Acta Part A: Mol. Biomolecular Spectrosc. 243, 118662. doi: 10.1016/j.saa.2020.118662
Roberts, P. C., Lamb, R. A., Compans, R. W. (1998). The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240 (1), 127–137. doi: 10.1006/viro.1997.8916
Roushani, M., Sarabaegi, M., Pourahmad, F. (2019). Impedimetric aptasensor for Pseudomonas aeruginosa by using a glassy carbon electrode modified with silver nanoparticles. Microchimica Acta 186, 725. doi: 10.1007/s00604-019-3858-y
Roxo, C., Kotkowiak, W., Pasternak, A. (2019). G-Quadruplex-forming aptamers—characteristics, applications, and perspectives. Molecules 24, 3781. doi: 10.3390/molecules24203781
Sanjuán, R. (2012). From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog. 8, e1002685. doi: 10.1371/journal.ppat.1002685
Saravanan, H., Subramani, T., Rajaramon, S., David, H., Sajeevan, A., Sujith, S., et al. (2023). Exploring nanocomposites for controlling infectious microorganisms: charting the path forward in antimicrobial strategies. Front. Pharmacol. 14. doi: 10.3389/fphar.2023.1282073
Schasfoort, R. B. M. (2017). “Introduction to surface plasmon resonance,” in handbook of surface plasmon resonance (The Royal Society of Chemistry), 1–26. doi: 10.1039/9781788010283-00001
Schmitz, F. R. W., Cesca, K., Valério, A., de Oliveira, D., Hotza, D. (2023). Colorimetric detection of Pseudomonas aeruginosa by aptamer-functionalized gold nanoparticles. Appl. Microbiol. Biotechnol. 107, 71–80. doi: 10.1007/s00253-022-12283-5
Shang, Z., Song, H., Shi, Y., Qi, J., Gao, G. F. (2018). Crystal structure of the capsid protein from zika virus. J. Mol. Biol. 430, 948–962. doi: 10.1016/j.jmb.2018.02.006
Shatila, F., Yalçın, H. T., Özyurt, C., Evran, S., Çakır, B., Yaşa, İ., et al. (2020a). Single-stranded DNA (ssDNA) Aptamer targeting SipA protein inhibits Salmonella Enteritidis invasion of intestinal epithelial cells. Int. J. Biol. Macromolecules 148, 518–524. doi: 10.1016/j.ijbiomac.2020.01.132
Shatila, F., Yaşa, İ., Yalçın, H. T. (2020b). Inhibition of Salmonella enteritidis biofilms by Salmonella invasion protein-targeting aptamer. Biotechnol. Lett. 42, 1963–1974. doi: 10.1007/s10529-020-02920-2
Shaw, M. L., Garciía-Sastre, A., Palese, P., Basler, C. F. (2004). Nipah Virus V and W proteins have a common stat1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78, 5633–5641. doi: 10.1128/JVI.78.11.5633-5641.2004
Shraim, A. S., Abdel Majeed, B. A., Al-Binni, M. A., Hunaiti, A. (2022). Therapeutic potential of aptamer–protein interactions. ACS Pharmacol. Trans. Sci. 5, 1211–1227. doi: 10.1021/acsptsci.2c00156
Siddiqui, M. A. A., Keating, G. M. (2005). Pegaptanib. Drugs 65, 1571–1577. doi: 10.2165/00003495-200565110-00010
Singh, N. K., Ray, P., Carlin, A. F., Magallanes, C., Morgan, S. C., Laurent, L. C., et al. (2021). Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosensors Bioelectronics 180, 113111. doi: 10.1016/j.bios.2021.113111
Sohouli, E., Ghalkhani, M., Zargar, T., Joseph, Y., Rahimi-Nasrabadi, M., Ahmadi, F., et al. (2022). A new electrochemical aptasensor based on gold/nitrogen-doped carbon nano-onions for the detection of Staphylococcus aureus. Electrochimica Acta 403, 139633. doi: 10.1016/j.electacta.2021.139633
Sola, M., Menon, A. P., Moreno, B., Meraviglia-Crivelli, D., Soldevilla, M. M., Cartón-García, F., et al. (2020). Aptamers against live targets: is in vivo SELEX finally coming to the edge? Mol. Ther. - Nucleic Acids 21, 192–204. doi: 10.1016/j.omtn.2020.05.025
Somerville, J. E., Jr., Cassiano, L., Darveau, R. P. (1999). Escherichia coli msbB Gene as a virulence factor and a therapeutic target. Infection Immun. 67 (12), 6583–6590. doi: 10.1128/IAI.67.12.6583-6590.1999
Sreejit, G., Ahmed, A., Parveen, N., Jha, V., Valluri, V. L., Ghosh, S., et al. (2014). The ESAT-6 Protein of Mycobacterium tuberculosis interacts with Beta-2-Microglobulin (β2M) affecting antigen presentation function of macrophage. PLoS Pathog. 10, e1004446. doi: 10.1371/journal.ppat.1004446
Srivastava, S., Abraham, P. R., Mukhopadhyay, S. (2021). Aptamers: An emerging tool for diagnosis and therapeutics in tuberculosis. Front. Cell. Infection Microbiol. 11. doi: 10.3389/fcimb.2021.656421
Sudagidan, M., Yildiz, G., Onen, S., Al, R., Temiz, Ş.N., Yurt, M. N. Z., et al. (2021). Targeted mesoporous silica nanoparticles for improved inhibition of disinfectant resistant Listeria monocytogenes and lower environmental pollution. J. Hazardous Materials 418, 126364. doi: 10.1016/j.jhazmat.2021.126364
Suenaga, E., Kumar, P. K. R. (2014). An aptamer that binds efficiently to the hemagglutinins of highly pathogenic avian influenza viruses (H5N1 and H7N7) and inhibits hemagglutinin–glycan interactions. Acta Biomaterialia 10, 1314–1323. doi: 10.1016/j.actbio.2013.12.034
Sun, B., Jia, L., Liang, B., Chen, Q., Liu, D. (2018). Phylogeography, transmission, and viral proteins of Nipah Virus. Virologica Sin. 33, 385–393. doi: 10.1007/s12250-018-0050-1
Sun, J., Siroy, A., Lokareddy, R. K., Speer, A., Doornbos, K. S., Cingolani, G., et al. (2015). The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat. Struct. Mol. Biol. 22, 672–678. doi: 10.1038/nsmb.3064
Sun, H., Zhu, X., Lu, P. Y., Rosato, R. R., Tan, W., Zu, Y. (2014). Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. - Nucleic Acids 3, e182. doi: 10.1038/mtna.2014.32
Terlizzi, M. E., Gribaudo, G., Maffei, M. E. (2017). UroPathogenic Escherichia coli (UPEC) infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01566
Thevendran, R., Rogini, S., Leighton, G., Mutombwera, A., Shigdar, S., Tang, T.-H., et al. (2023). The diagnostic potential of RNA aptamers against the NS1 protein of dengue virus serotype 2. Biology 12, 722. doi: 10.3390/biology12050722
Thiviyanathan, V., Somasunderam, A. D., Gorenstein, D. G. (2007). Combinatorial selection and delivery of thioaptamers. Biochem. Soc. Trans. 35, 50–52. doi: 10.1042/BST0350050
Tran, Q. T., Gomez, G., Khare, S., Lawhon, S. D., Raffatellu, M., Baöumler, A. J., et al. (2010). The Salmonella enterica Serotype typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infection Immun. 78, 527–535. doi: 10.1128/IAI.00972-09
Turgis, M., Khanh, D. V., Majid, J., Behnoush, M., Monique, L. (2016). Synergistic antimicrobial effect of combined bacteriocins against food pathogens and spoilage bacteria. Microb. Res. Inter. 4 (1), 1–5.
Turner, J., Torrelles, J. B. (2018). Mannose-capped lipoarabinomannan in Mycobacterium tuberculosis pathogenesis. Pathog Dis. 76 (4), fty026. doi: 10.1093/femspd/fty026
Turrell, L., Lyall, J. W., Tiley, L. S., Fodor, E., Vreede, F. T. (2013). The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat. Commun. 4 (1), 1591. doi: 10.1038/ncomms2589
Ucak, S., Sudagidan, M., Borsa, B. A., Mansuroglu, B., Ozalp, V. C. (2020). Inhibitory effects of aptamer targeted teicoplanin encapsulated PLGA nanoparticles for Staphylococcus aureus strains. World J. Microbiol. Biotechnol. 36 (5), 69. doi: 10.1007/s11274-020-02845-y
Uemachi, H., Kasahara, Y., Tanaka, K., Okuda, T., Yoneda, Y., Obika, S. (2021). Hybrid-Type SELEX for the selection of artificial nucleic acid aptamers exhibiting cell internalization activity. Pharmaceutics 13, 888. doi: 10.3390/pharmaceutics13060888
Uniyal, A., Srivastava, G., Pal, A., Taya, S., Muduli, A. (2023). Recent advances in optical biosensors for sensing applications: a review. Plasmonics 18, 735–750. doi: 10.1007/s11468-023-01803-2
Valente, A. P., Moraes, A. H. (2019). Zika virus proteins at an atomic scale: how does structural biology help us to understand and develop vaccines and drugs against Zika virus infection? J. Venomous Anim. Toxins Including Trop. Dis. 25. doi: 10.1590/1678-9199-jvatitd-2019-0013
Verma, A. K., Noumani, A., Yadav, A. K., Solanki, P. R. (2023). FRET Based Biosensor: Principle applications recent advances and challenges. Diagnostics 13, 1375. doi: 10.3390/diagnostics13081375
Vivekananda, J., Salgado, C., Millenbaugh, N. J. (2014). DNA aptamers as a novel approach to neutralize Staphylococcus aureus α-toxin. Biochem. Biophys. Res. Commun. 444, 433–438. doi: 10.1016/j.bbrc.2014.01.076
Wain, J., House, D., Zafar, A., Baker, S., Nair, S., Kidgell, C., et al. (2005). Vi Antigen Expression in Salmonella enterica Serovar typhi Clinical Isolates from Pakistan. J. Clin. Microbiol. 43, 1158–1165. doi: 10.1128/JCM.43.3.1158-1165.2005
Wan, Q., Liu, X., Zu, Y. (2021). Oligonucleotide aptamers for pathogen detection and infectious disease control. Theranostics 11, 9133–9161. doi: 10.7150/thno.61804
Wang, J. Y., Koshland, D. E., Jr. (1978). Evidence for protein kinase activities in the prokaryote Salmonella typhimurium. J. Biol. Chem. 253 (21), 7605–7608.
Wang, L., Wang, R., Chen, F., Jiang, T., Wang, H., Slavik, M., et al. (2017a). QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 221, 776–782. doi: 10.1016/j.foodchem.2016.11.104
Wang, R., Wang, L., Callaway, Z. T., Lu, H., Huang, T. J., Li, Y. (2017b). A nanowell-based QCM aptasensor for rapid and sensitive detection of avian influenza virus. Sensors Actuators B: Chem 240, 934–940. doi: 10.1038/nature13027
Weerathunge, P., Ramanathan, R., Torok, V. A., Hodgson, K., Xu, Y., Goodacre, R., et al. (2019). Ultrasensitive colorimetric detection of murine norovirus using nanozyme aptasensor. Analytical Chem. 91, 3270–3276. doi: 10.1021/acs.analchem.8b03300
Weis, M., Maisner, A. (2015). Nipah virus fusion protein: Importance of the cytoplasmic tail for endosomal trafficking and bioactivity. Eur. J. Cell Biol. 94, 316–322. doi: 10.1016/j.ejcb.2015.05.005
Wei-Wen Hsiao, W., Sharma, N., Le, T.-N., Cheng, Y.-Y., Lee, C.-C., Vo, D.-T., et al. (2022). Fluorescent nanodiamond-based spin-enhanced lateral flow immunoassay for detection of SARS-CoV-2 nucleocapsid protein and spike protein from different variants. Analytica Chimica Acta 1230, 340389. doi: 10.1016/j.aca.2022.340389
Weng, X., Neethirajan, S. (2017). Aptamer-based fluorometric determination of norovirus using a paper-based microfluidic device. Microchimica Acta 184, 4545–4552. doi: 10.1007/s00604-017-2467-x
Winter, S. E., Winter, M. G., Thiennimitr, P., Gerriets, V. A., Nuccio, S. P., Rüssmann, H., et al. (2009). The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol. Microbiol. 74, 175–193. doi: 10.1111/j.1365-2958.2009.06859.x
Xi, Z., Gong, Q., Wang, C., Zheng, B. (2018). Highly sensitive chemiluminescent aptasensor for detecting HBV infection based on rapid magnetic separation and double-functionalized gold nanoparticles. Sci. Rep. 8, 9444. doi: 10.1038/s41598-018-27792-5
Xie, X., Huang, W., Shen, G., Yu, H., Wang, L. (2022). Selection and colorimetric application of ssDNA aptamers against metamitron based on magnetic bead-SELEX. Analytical Methods 14, 3021–3032. doi: 10.1039/D2AY00566B
Yang, J., Bowser, M. T. (2013). Capillary Electrophoresis–SELEX selection of catalytic dna aptamers for a small-molecule porphyrin target. Analytical Chem. 85, 1525–1530. doi: 10.1021/ac302721j
Yang, M., Peng, Z., Ning, Y., Chen, Y., Zhou, Q., Deng, L. (2013). Highly specific and cost-efficient detection of salmonella paratyphi A combining aptamers with single-walled carbon nanotubes. Sensors 13, 6865–6881. doi: 10.3390/s130506865
Ye, Y., Qi, X., Wang, H., Zhao, B., Xu, L., Zhang, Y., et al. (2022). A surface-enhanced Raman scattering aptasensor for Escherichia coli detection based on high-performance 3D substrate and hot spot effect. Analytica Chimica Acta 1221, 340141. doi: 10.1016/j.aca.2022.340141
Yeom, J. H., Lee, B., Kim, D., Lee, J. K., Kim, S., Bae, J., et al. (2016). Gold nanoparticle-DNA aptamer conjugate-assisted delivery of antimicrobial peptide effectively eliminates intracellular Salmonella enterica serovar typhimurium. Biomaterials 104, 43–51. doi: 10.1016/j.biomaterials.2016.07.009
Yu, X., Chen, F., Wang, R., Li, Y. (2018). Whole-bacterium SELEX of DNA aptamers for rapid detection of E.coli O157:H7 using a QCM sensor. J. Biotechnol. 266, 39–49. doi: 10.1016/j.jbiotec.2017.12.011
Yuan, B., Peng, Q., Cheng, J., Wang, M., Zhong, J., Qi, J., et al. (2022). Structure of the Ebola virus polymerase complex. Nature 610, 394–401. doi: 10.1038/s41586-022-05271-2
Yüce, M., Ullah, N., Budak, H. (2015). Trends in aptamer selection methods and applications. Analyst 140, 5379–5399. doi: 10.1039/C5AN00954E
Zavyalova, E., Samoylenkova, N., Revishchin, A., Turashev, A., Gordeychuk, I., Golovin, A., et al. (2017). The evaluation of pharmacodynamics and pharmacokinetics of anti-thrombin DNA aptamer RA-36. Front. Pharmacol. 8. doi: 10.3389/fphar.2017.00922
Zhan, Z., Li, H., Liu, J., Xie, G., Xiao, F., Wu, X., et al. (2020). A competitive enzyme linked aptasensor with rolling circle amplification (ELARCA) assay for colorimetric detection of Listeria monocytogenes. Food Control 107, 106806. doi: 10.1016/j.foodcont.2019.106806
Zhang, A. P. P., Abelson, D. M., Bornholdt, Z. A., Liu, T., Woods, J. V. L., Saphire, E. O. (2012). The ebolavirus VP24 interferon antagonist. Virulence 3, 440–445. doi: 10.4161/viru.21302
Zhang, X., Khan, I. M., Ji, H., Wang, Z., Tian, H., Cao, W., et al. (2020). A label-free fluorescent aptasensor for detection of staphylococcal enterotoxin a based on aptamer-functionalized silver nanoclusters. Polymers 12, 152. doi: 10.3390/polym12010152
Zhang, L. F., Lepenies, B., Nakamae, S., Young, B. M., Santos, R. L., Raffatellu, M., et al. (2022). The Vi Capsular polysaccharide of Salmonella typhi promotes macrophage phagocytosis by binding the human C-type lectin DC-SIGN. MBio 13 (6). doi: 10.1128/mbio.02733-22
Zhang, T., Lu, Y., Deng, S., Deng, R. (2021). “Aptamers for the diagnosis of infectious diseases,” in Aptamers for Medical Applications (Springer, Singapore), 207–238. doi: 10.1007/978-981-33-4838-7_8
Zhang, Y., Zhang, Y., Yang, Y., Wang, L., Weng, L. (2013). Identification of a Pseudomonas sp. that inhibits rhl system of quorum sensing. Indian J. Microbiol. 53, 28–35. doi: 10.1007/s12088-012-0340-5
Zhao, Q., Du, P., Wang, X., Huang, M., Sun, L.-D., Wang, T., et al. (2021). Upconversion fluorescence resonance energy transfer aptasensors for H5N1 influenza virus detection. ACS Omega 6, 15236–15245. doi: 10.1021/acsomega.1c01491
Zhao, M., Li, W., Liu, K., Li, H., Lan, X. (2019). C4-HSL aptamers for blocking qurom sensing and inhibiting biofilm formation in Pseudomonas aeruginosa and its structure prediction and analysis. PLoS One 14, e0212041. doi: 10.1371/journal.pone.0212041
Zheng, J., Tang, X., Wu, R., Yan, Q., Tang, H., Luo, J., et al. (2015). Identification and characteristics of aptamers against inactivated Vibrio alginolyticus. LWT - Food Sci. Technol. 64, 1138–1142. doi: 10.1016/j.lwt.2015.07.021
Zhou, S., Lu, C., Li, Y., Xue, L., Zhao, C., Tian, G., et al. (2020). Gold nanobones enhanced ultrasensitive surface-enhanced raman scattering aptasensor for detecting Escherichia coli o157:h7. ACS Sensors 5, 588–596. doi: 10.1021/acssensors.9b02600
Zhou, J., Rossi, J. (2017). Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discovery 16, 181–202. doi: 10.1038/nrd.2016.199
Zhou, Y., Xiong, H., Chen, R., Wan, L., Kong, Y., Rao, J., et al. (2021). Aptamer detection of Mycobaterium tuberculosis mannose-capped lipoarabinomannan in lesion tissues for tuberculosis diagnosis. Front. Cell. Infection Microbiol. 11, 634915. doi: 10.3389/fcimb.2021.634915
Zhu, L., Gao, T., Huang, Y., Jin, J., Wang, D., Zhang, L., et al. (2022). Ebola virus VP35 hijacks the PKA-CREB1 pathway for replication and pathogenesis by AKIP1 association. Nat. Commun. 13 (1), 2256. doi: 10.1038/s41467-022-29948-4
Zhu, C., Li, L., Yang, G., Qu, F. (2021). Investigating the influences of random-region length on aptamer selection efficiency based on capillary electrophoresis–SELEX and high-throughput sequencing. Analytical Chem. 93, 17030–17035. doi: 10.1021/acs.analchem.1c03661
Zhu, S., Watanabe, M., Kirkpatrick, E., Murray, A. B., Sok, R., Karst, S. M. (2016). Regulation of norovirus virulence by the VP1 protruding domain correlates with B Cell infection efficiency. J. Virol. 90, 2858–2867. doi: 10.1128/jvi.02880-15
Zou, Y., Duan, N., Wu, S., Shen, M., & Wang, Z. (2018). Selection, identification, and binding mechanism studies of an ssdna aptamer targeted to different stages of E. coli O157:H7. J. Agric. Food Chem. 66, 5677–5682. doi: 10.1021/acs.jafc.8b01006
Zou, J., Huang, X., Wu, L., Chen, G., Dong, J., Cui, X., et al. (2015). Selection of intracellularly functional RNA mimics of green fluorescent protein using fluorescence-activated cell sorting. J. Mol. Evol. 81, 172–178. doi: 10.1007/s00239-015-9718-4
Zou, X., Wu, J., Gu, J., Shen, L., Mao, L. (2019). Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 10, 1462. doi: 10.3389/fmicb.2019.01462
Keywords: aptamers, SELEX, biosensor, therapeutics, diagnosis, bacteria, virus, infectious disease
Citation: Sujith S, Naresh R, Srivisanth BU, Sajeevan A, Rajaramon S, David H and Solomon AP (2024) Aptamers: precision tools for diagnosing and treating infectious diseases. Front. Cell. Infect. Microbiol. 14:1402932. doi: 10.3389/fcimb.2024.1402932
Received: 18 March 2024; Accepted: 03 September 2024;
Published: 25 September 2024.
Edited by:
Andrea Marino, University of Catania, ItalyReviewed by:
Nejat Duzgunes, University of the Pacific, United StatesYingwang Ye, Hefei University of Technology, China
Ricardo Oliveira, National Institute for Agrarian and Veterinariay Research (INIAV), Portugal
Copyright © 2024 Sujith, Naresh, Srivisanth, Sajeevan, Rajaramon, David and Solomon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adline Princy Solomon, YWRsaW5lcHJpbnp5QHNhc3RyYS5hYy5pbg==; Helma David, aGVsbWFkYXZpZEBzY2J0LnNhc3RyYS5hYy5pbg==
†These authors have contributed equally to this work
 Swathi Sujith
Swathi Sujith Rajalakshmi Naresh
Rajalakshmi Naresh B. U. Srivisanth
B. U. Srivisanth Anusree Sajeevan
Anusree Sajeevan Shobana Rajaramon
Shobana Rajaramon Helma David
Helma David Adline Princy Solomon
Adline Princy Solomon