
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 05 July 2024
Sec. Clinical Microbiology
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1402001
This article is part of the Research Topic Frontiers in Chronic Hepatitis B Clinical Cure View all 4 articles
 Ying Liu1†
Ying Liu1† Di Wu1†
Di Wu1† Kui Zhang2
Kui Zhang2 Rongrong Ren3
Rongrong Ren3 Yuxuan Liu1
Yuxuan Liu1 Shuya Zhang1
Shuya Zhang1 Xuanyu Zhang1
Xuanyu Zhang1 Jilin Cheng4
Jilin Cheng4 Liping Chen5*
Liping Chen5* Jun Huang1*
Jun Huang1*Viral hepatitis, caused by its etiology, hepatitis virus, is a public health problem globally. Among all infections caused by hepatitis-associated viruses, hepatitis B virus (HBV) infection remains the most serious medical concern. HBV infection particularly affects people in East Asia and Africa, the Mediterranean region, and Eastern Europe, with a prevalence rate of > 2%. Currently, approximately 1 billion people worldwide are infected with HBV, and nearly 30% of them experience chronic infection. Chronic HBV infection can lead to chronic hepatitis B (CHB), liver cirrhosis, and hepatocellular carcinoma (HCC), resulting in the related death of approximately 1 million people annually. Although preventative vaccines and antiviral therapies are currently available, there is no cure for this infection. Clinical testing is not only the gateway for diagnosis of HBV infection, but also crucial for judging the timing of medication, evaluating the effect of antiviral therapy, and predicting the risk of relapse after drug withdrawal in the whole follow-up management of hepatitis B infected persons. With advances in detection technology, it is now possible to measure various viral components in the blood to assess the clinical status of HBV infection. Serum viral products of HBV infection, such as HBV DNA, HBV RNA, hepatitis B surface antigen, hepatitis B e-antigen, and hepatitis B core-related antigen, are non-invasive indicators that are critical for the rapid diagnosis and management of related diseases. Improving the sensitivity of monitoring of these products is essential, and the development of corresponding detection technologies is pivotal in achieving this goal. This review aims to offer valuable insights into CHB infection and references for its effective treatment. We provide a comprehensive and systematic overview of classical and novel methods for detecting HBV serum viral products and discusses their clinical applications, along with the latest research progress in this field.
Hepatitis B virus (HBV) is a 3.2 kb partially double-stranded DNA virus that belongs to the Hepadnaviridae family (Barker et al., 1975; Li et al., 2015). The discovery of the “Australia antigen,” now known as the hepatitis B virus surface antigen (HBsAg), by Dr. Baruch Blumberg in the 1960s paved the way for the diagnosis, prevention, and treatment of HBV infection (Blumberg, 1964; Blumberg et al., 1965; Li et al., 2020). The discovery history of serum viral products of HBV was shown in Figure 1. The European Association for the Study of the Liver (EASL) spliced chronic HBV infection into the following categories: hepatitis B e-antigen (HBeAg)-positive chronic infection (formerly known as the immune tolerance period), HBeAg-positive chronic hepatitis (formerly known as the HBeAg-positive immune activity period or immune clearance period), HBeAg-negative chronic infection (formerly known as the inactive carrier phase or low replication phase), and HBeAg-negative chronic hepatitis (formerly known as the HBeAg-negative immune active phase or reactivation phase) based on the evaluation of HBV-related liver disease indicators (Table 1) (European Association for the Study of the Liver, 2017; Chinese Society of Hepatology, 2022a; Zhuang, 2022). However, approximately 40% of patients cannot be categorized under the stages mentioned above (Chinese Society of Hepatology, 2022a); consequently, a new category of infection, referred to as the “uncertain period” of chronic HBV infection, has been defined. Furthermore, the risk of chronic hepatitis B (CHB) progression in patients during the “uncertain period” remains high (Chinese Society of Hepatology, 2022a).
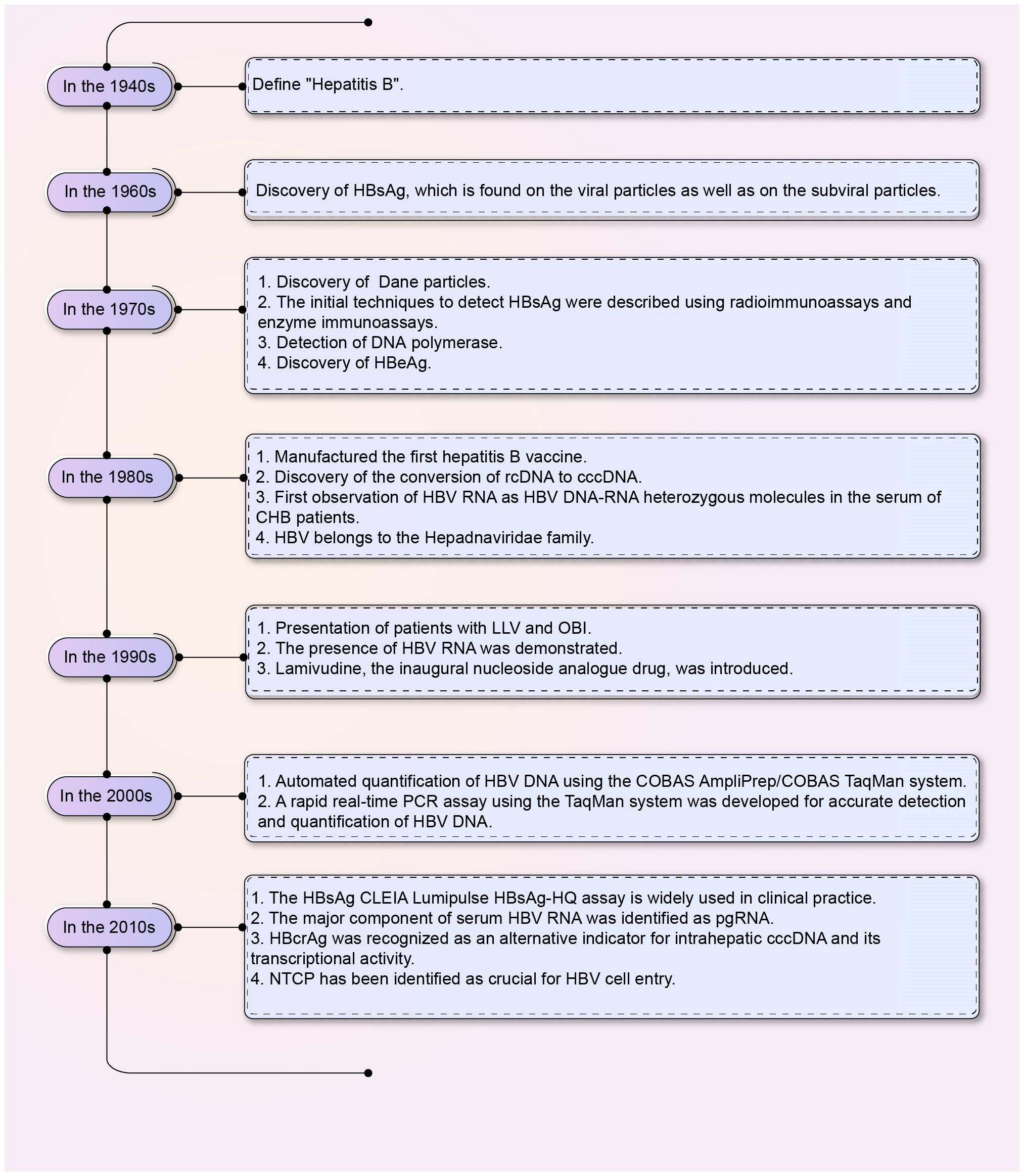
Figure 1 The discovery of HBV serum viral products. The chart shows the discovery of HBV serum viral products from the 1940s to the 2010s. HBV, Hepatitis B virus; HBsAg, HBV surface antigen; cccDNA, Covalently closed circular DNA; rcDNA, Relaxed circular DNA; HBeAg, Hepatitis B e-antigen; CHB, Chronic hepatitis B; CLEIA, Chemiluminescent enzyme immunoassay; pgRNA, Pre-genomic RNA; LLV, Low-level viremia; OBI, Occult hepatitis B virus infection; HBcrAg, Hepatitis B core-related antigen; NTCP, Sodium taurocholate co-transporting polypeptides.
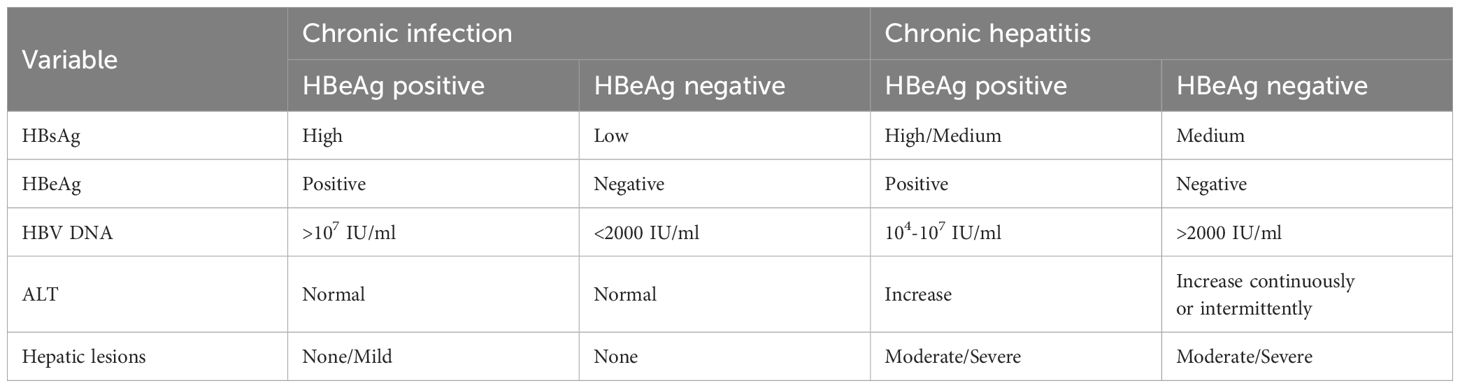
Table 1 Natural history of chronic HBV infection (Chinese Society of Hepatology, 2022a; Jung & Nguyen, 2023; Zhuang, 2022).
Over the past 30 years, there has been extensive research on the life cycle of HBV. The virus binds to liver-specific receptors such as sodium taurocholate co-transporting polypeptides and heparan sulfate proteoglycans, resulting in endocytosis and the release of HBV core particles (Yan et al., 2012; Sureau and Salisse, 2013; Wu and Chang, 2015). These particles are transported to the nucleus, where the HBV genome is released and gets converted from relaxed circular double-stranded DNA (rcDNA) to covalently closed circular DNA (cccDNA), which is highly stable and contributes to the persistent HBV infection and low cure rates (Gerlich and Robinson, 1980; Molnar-Kimber et al., 1983; Tuttleman et al., 1986; Tsukuda and Watashi, 2020). HBV cccDNA interacts with host transcription factors to produce pre-genomic RNA (pgRNA), HBV X mRNA, pre-core mRNA, and preS/S mRNA (Su et al., 1989a; Su et al., 1989b; Nassal, 2015). These HBV RNAs encode viral proteins, including surface (HBs), precore or e (HBe), and core (HBc) antigen, polymerase, and X (HBx) proteins (Moon et al., 2024). The HBV mRNA is primarily transported to the cytoplasm and translated (Wieland et al., 2000; McCoullough et al., 2024). pgRNA is translated into core proteins and viral polymerase in the cytosol. The pgRNA and the viral polymerase are encapsidated. The pgRNA is further reverse-transcribed to rcDNA by the HBV polymerase within the nucelocapsids (core particles) (Bruss, 2007; SChadler and Hildt, 2009; Imam et al., 2018). Mature core particles re-migrate to the nuclear pore complex or are enveloped by viral surface proteins and secreted by multivesicular bodies (MVB) (Babst, 2005; Watanabe et al., 2007; Pan et al., 2023). Early assessment of histological damage and residual levels of de novo infection caused by viral activity during the above HBV life cycle is crucial for preventing hepatitis recurrence and liver disease progression and ultimately achieving a functional cure(Maintain negative Hepatitis B surface antigen, undetectable HBV DNA, and normal liver biochemical indicators)against CHB infection (Author Group of Expert, 2022). The life cycle of HBV and its corresponding detection methods were summarized and shown in Figure 2.
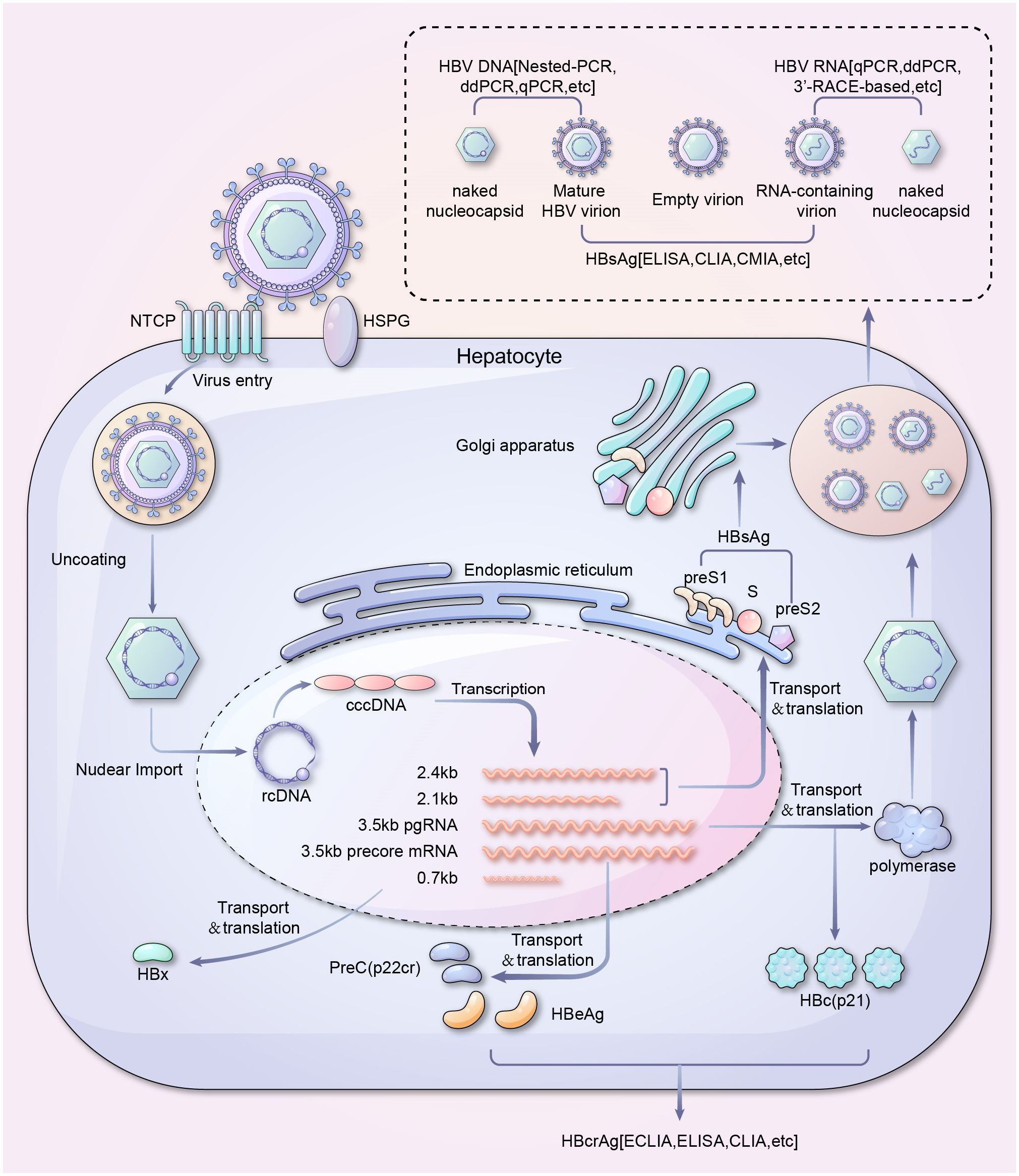
Figure 2 Life cycle and serum viral products detection of HBV. A schematic overview of serum viral products in HBV life cycle and their corresponding detection methods. It should be noted that covalently closed circular DNA (cccDNA) is the template for all the HBV mRNAs, while integrated HBV DNA can transcribe X and S/preS2/preS1 RNAs only. ddPCR, Droplet digital PCR; qPCR, Quantitative polymerase chain reaction; HBsAg, HBV surface antigen; ELISA, Enzyme-linked immunosorbent assay; CLIA, Chemiluminescence immunoassay; CMIA, Chemiluminescent microparticle immunoassay; NTCP, sodium taurocholate co-transporting polypeptide; HSPG, Heparan sulfate proteoglycan; ECLIA, Electrochemiluminescence immunoassay; cccDNA, Covalently closed circular DNA; rcDNA, Relaxed circular DNA; HBeAg, Hepatitis B e-antigen.
In clinical practice, based on the nature of specific viral products, valuable information for evaluating the phase of HBV infection can be obtained by quantifying serum viral products. Table 2 listed the detection methods for HBV serum viral products. Classical detection methods include the following: quantitative polymerase chain reaction (qPCR), enzyme-linked immunosorbent assay (ELISA), radioimmunoassay (RIA), gold immunochromatography assay (GICA), and time-resolved fluoroimmunoassay (TRFIA) (Hu et al., 2012; Maity et al., 2012; Irshad et al., 2016; Jiang et al., 2020; Dera et al., 2023). Novel stable and accurate detection technologies, including chemiluminescence immunoassay (CLIA), chemiluminescent microparticle immunoassay (CMIA), microparticle enzyme immunoassay (MEIA), and automatic chemiluminescent enzyme immunoassay (CLEIA), have also emerged (Ollier et al., 2008; Khadem-Ansari et al., 2014; Liu et al., 2014; Ghosh et al., 2015; Amini et al., 2017; Inoue et al., 2021).
Detection methods for HBV serum viral products can be divided into molecular biological detection and immunological detection methods (Rong et al., 2021).qPCR is a robust molecular biological detection method that involves the addition of a fluorescent gene to the PCR reaction system (Heid et al., 1996). This allows for real-time monitoring of the entire PCR process using fluorescence signal accumulation, thereby facilitating the accurate detection of an unknown template using a standard curve (Caliendo et al., 2011; Siegel et al., 2023). Compared to clinical detection, molecular biological detection and other assays, qPCR-based detection has a relatively low degree of automation and requires high levels of expertise from operators and careful management of experimental conditions (Paraskevis et al., 2010; Caliendo et al., 2011; Irshad et al., 2016; Liu et al., 2019). Using TaqMan probes, HBV RNA in serum samples can be quantified using qPCR (Ji et al., 2020). However, several influencing factors such as homologous and heterologous DNA backgrounds and oligonucleotide hybridization specificity, which may cause quantitative bias in qPCR reactions, need to be considered during qPCR operation. Accordingly, to ensure the accuracy and reliability of the results, it is crucial to optimize the experimental conditions during the operation (Wikstrom et al., 2000; Tay et al., 2001).
RIA is an immunoassay that has been used in clinical practice since the 1960s (Yalow and Berson, 1960; Utiger et al., 1962). It is based on the principle that the labeled and non-labeled antigens (to be tested) bind competitively to a limited number of specific antibodies; the greater the radioactivity of the Ag-Ab complex, the more the labeled antigen binds and the lower is the concentration of non-labeled antigen (to be tested) (von der Waart et al., 1978). This detection method has the characteristic of good cross-reflection. However, clinical trials have shown that RIA can easily lead to radioactivity-related contamination and cannot quickly complete detection operations.
ELISA has become ubiquitous in medical laboratories, in vitro diagnostic product manufacturing industries, regulatory agencies, and external quality assessment and capability verification organizations (Engvall and Perlmann, 1971; Engvall et al., 1971; Lequin, 2005; Maity et al., 2012). It is an immunological detection method commonly used to detect the presence of HBV in patients. ELISA, while valuable and cost-effective for virus detection, is predominantly marketed for qualitative analysis at present. Additionally, it is prone to problems such as false negatives due to hook effects (Lequin, 2005; Maity et al., 2012; Hu et al., 2023a). GICA has the advantages of simple operation, rapidity, and easy preservation (Jiang et al., 2007). However, its sensitivity is relatively low, which may lead to missed detection. TRFIA is a novel ultramicron immunoassay method that integrates the advantages of ELISA and RIA (Halonen et al., 1983). It can quantitatively detect HBV serum viral products with high specificity and sensitivity and samples with low level of replication, thus avoiding missed detection. It should be noted that TRFIA has a significant disadvantage, which is complicated operation (Fu et al., 2020).
CLIA is a widely used technology in clinical medicine for detecting trace amounts of antigens and antibodies (Velan and Halmann, 1978). It is a labeled immunoassay that combines luminescence analysis with immune system reactions (Khadem-Ansari et al., 2014; Shen et al., 2019). CLIA emerged after ELISA and RIA. Due to its simple operation, convenient marking, high stability and sensitivity, high speed, and low environmental impact, it has been well-received by medical examiners and doctors in clinical practice. Electrochemiluminescence immunoassay (ECLIA), a susceptible detection technology that combines electrochemical luminescence with immunoassays, is dependent on chemiluminescence reactions and is a subtype of CLIA. Its significant stability and sensitivity make it ideal for detecting trace substances (Kovac et al., 2020). However, ambiguous results obtained from ECLIA-based detection need to be further confirmed using additional detection methods. CLEIA, a type of CLIA, is a detection method wherein a photon signal is generated via the interaction of an enzyme labeled on an antigen or antibody with a luminescent substrate. Generally, horseradish peroxidase and alkaline phosphatase (ALP) are the two most common enzyme markers used in CLEIA. In practice, its use is limited by the extensive sample quality requirements and high reagent costs. MEIA is an immunoassay technique that uses microparticle endofactors to form a complex with the substance to be tested (Duverlie et al., 1988). The complex then reacts with an ALP conjugate compound to produce a fluorescent product. It has the advantages of high sensitivity, specificity, reproducibility, and simplicity of operation. CMIA is a technique that involves two methods, competitive and double antibody sandwich (Amini et al., 2017). The small solid-phase magnetic particles used in this technique have a diameter of only 1.0 μm. This small size increases the coating surface area, amount of adsorption of the antigen or antibody, and reaction speed and reduces the probability of pollution and cross-contamination (Chen et al., 2005; Li et al., 2022). In CMIA, the antigen or antibody is labeled with ALP, which undergoes luminescence reactions by reacting with its substrate, dioxane phosphate (Loglio et al., 2020). CMIA includes a variety of serum immunoassays with high sensitivity (as low as 0.1 ng/mL) for detecting HBsAg with good repeatability and specificity (Hadziyannis et al., 2008; Motyka et al., 2022). However, the cost of related equipment is high (Motyka et al., 2022).
A critical aspect of managing CHB infection involves monitoring the process of HBV replication (Burns and Thompson, 2014). Serological and histological products are typically detected during the diagnosis of HBV infection. Throughout the infection, cccDNA accumulates in the nucleus, persists as a stable inclusion, and serves as a template for the transcription of viral genes (Ding et al., 2023). Several studies have suggested that HBV cccDNA has a relatively short half-life (Lu et al., 2017; Gao et al., 2022); however, its persistence remains a significant challenge in eradicating HBV infection. Another major challenge in HBV cccDNA research is the absence of an efficient method that can directly detect cccDNA in liver biopsy tissues with high sensitivity, significant specificity, and accurate quantification (Tu et al., 2021). To address this issue and facilitate the study of cccDNA, various new methods, including PCR-based methods and in situ hybridization, have recently been applied (Li et al., 2017). PCR-based methods encompass conventional qPCR, competitive qPCR, semi-nested and nested qPCR, and droplet-digital PCR (ddPCR), among others. These methods offer advantages of being simpler, faster, more accurate, cost-effective, sensitive, and capable of higher throughput. In situ hybridization is capable of distinguishing and locating different DNA and RNA. Proteins can be identified in conjunction with the method by utilizing immunohistochemistry (IHC) or immunofluorescence. Serological detection is currently considered an alternative approach; it is non-invasive, easy to operate, and cost-effective. Accordingly, finding an ideal serological biomarker that can reflect the presence of HBV cccDNA and its transcriptional activity is a crucial clinical requirement.
The detection and study of HBV serum viral products have significant value in diagnosing and treating HBV infection and play a crucial role in promoting the development of anti-viral therapies. Currently, common serological products that are used to diagnose HBV infection include HBsAg, anti-HBs, HBeAg, anti-HBe, anti-HBc, and HBV DNA. Among these, HBsAg is a widely used viral product for diagnosing hepatitis B and is considered superior to other viral products in terms of indicating clinical cure after treatment with pegylated interferon and nucleos(t)ide analogs (Chuaypen et al., 2018; Lim et al., 2021; Wang et al., 2023). HBsAg is an antigen that is found on the surface of HBV; it comprises small, medium, and large HBV proteins. In addition, HBsAg is self-assembled in non-infectious subviral particles (SVPs) which are secreted in a large excess compared to the viral particles (Boulon et al., 2020). This antigen plays a crucial role in initiating the infection process of HBV by facilitating the attachment of the virus to the host cell membrane (Marquardt et al., 1987; Pontisso et al., 1989; Lee and Ahn, 2011). HBsAg is the earliest serological product to appear in patients with acute hepatitis B infection. It is also frequently detected in chronic disease carriers, indicating ongoing viral transcriptional activity (Blumberg et al., 1967; Kao, 2008; de Almeida Ponde, 2022). CLEIA, CMIA, CLIA, or standard ELISA is generally performed to detect HBsAg (Takeda et al., 2013; Kim, 2017). Chronic HBV infection is defined as a persistent infection lasting more than six months, during which HBsAg can be detected (Chinese Society of Hepatology, 2022b). Patients with low levels of HBsAg may still experience active replication of the virus and associated liver injury and may also be capable of spreading the infection (Wu et al., 2021; Moini and Fung, 2022). As diagnostic technologies and treatment for HBV continue to advance, researchers are increasingly focusing on individuals with low levels of HBsAg (Wu et al., 2022). It has been shown that a combination of HBsAg quantification and the expression of certain T-cell markers could be a potential predictor of HBsAg clearance in patients with chronic HBV infection within 12 months (Wang et al., 2023). CMIA-based quantitative analysis of HBsAg demonstrates high sensitivity in detecting not only free HBsAg protein but also antigen-antibody complexes and mutant forms of HBsAg.
Hepatitis B e-antigen (HBeAg) is a soluble component of the hepatitis B core antigen. The presence of HBeAg indicates the risk of active replication of HBV and transmission of the infection (Liaw et al., 2010; Cornberg et al., 2017). Clinical laboratories detect HBeAg using procedures such as ELISA, ECLIA, or CLIA. Research studies focusing on these methods have demonstrated that HBeAg quantification is a valuable tool in predicting the antiviral efficacy of HBV reactivation. Additionally, HBeAg levels are higher in patients experiencing HBV reactivation than in those with acute infection and HBeAg-positive chronic infection (Piermatteo et al., 2021). HBeAg seroconversion is a crucial goal encompassing antiviral treatment for patients with CHB. When a patient undergoes HBeAg seroconversion, it indicates that HBV has entered the low-level replication stage, which further indicates a reduction in the likelihood of progression to cirrhosis and liver cancer as well as the risk of infectivity (Hadziyannis and Laras, 2018). Several factors can influence the seroconversion of HBeAg. Numerous cytokines/chemokines and other indicators, including IL-37, IP-10, IL-21, and CLEC18, are correlated with HBeAg seroconversion in patients with CHB who are being treated with nucleos(t)ide analogues (NAs) (Ma et al., 2012; Guo et al., 2016; Zhao et al., 2021). Studies conducted on mice demonstrated that in Kupffer cells, HBeAg inhibited the transcription of NLRP3 and pro-interleukin 1β by reducing the phosphorylation level of NF-κB. Additionally, it inhibited caspase-1 activation and IL-1β maturation by blocking the production of reactive oxygen species (Yu et al., 2017).
High levels of HBV DNA in the serum of a patient indicate active replication of the virus in the liver, which is a critical factor in the progression of liver disease. Recent research suggests that even persistently low levels of HBV DNA can contribute to CHB progression (Sun et al., 2020). The detection of HBV DNA in the serum of a patient is considered the most reliable method for determining hepatitis B viremia; additionally, HBV DNA in the serum is the most reliable product of active viral replication (Robinson et al., 1974; Raimondo et al., 2019). The level of HBV DNA in the serum of a patient is closely associated with the risk of developing liver fibrosis and hepatocellular carcinoma (HCC) in patients with CHB (Kim et al., 2022). Over the past decade, significant advancements have been made in the detection methods used to assess serum HBV DNA levels, resulting in increased sensitivity of detection. Currently, in addition to nested PCR and ddPCR, qPCR is the most widely used method for detecting secreted HBV DNA (Wang et al., 2014; Saitta et al., 2022; Zhang and Tu, 2022). Liver samples in every instance must undergo proper processing to prevent cross-contamination. Moreover, inclusion of suitable negative controls is essential to validate the assay’s specificity.
Recently, there has been a growing trend toward using novel serological viral nucleic acid products, such as serum HBV RNA, for monitoring the clinical status of HBV patients. Previous studies indicated that HBV RNA in serum is pgRNA, which is encapsulated in HBV-like virus particles that can be secreted extracellularly. This suggests that HBV may have an alternative form of virion, in which the nucleic acid is composed of RNA rather than DNA (Wang et al., 2016). HBV RNA has been identified as an indicator of intrahepatic transcriptional activity of cccDNA and was found to be associated with liver histological changes in patients with CHB who have been treated with nucleoside (acid) analogs (Wang et al., 2016; Wang et al., 2017; Jiang et al., 2020). Serum HBV RNA levels have emerged as a useful alternative for assessing the transcriptional activity of cccDNA. Numerous studies have shown that although HBV DNA is below the detection limit, or HBsAg has seroconverted, HBV RNA still exists. Therefore, compared to HBV DNA and other indicators, HBV RNA has higher clinical value in evaluating the efficacy of anti-HBV therapy, selecting the timing for discontinuation of treatment, and predicting the risk of recurrence after cessation of treatment.
A case-control study involving 104 patients receiving entecavir (ETV) treatment revealed that after adjusting for various risk factors such as age, sex, presence or absence of cirrhosis, and duration of antiviral therapy, the level of HBV RNA during treatment was associated with an increased risk of developing HCC within the next two years (Carey et al., 2020; Fan et al., 2020; Dahari et al., 2021; Lok et al., 2022). To date, relatively few studies have been published on the association between HBV RNA levels and the risk of developing HCC. It has been reported that HBV RNA serves as a predictive product not only for HBsAg response during early antiviral therapy but also for the risk of HBsAg reversal after discontinuing the treatment (Wang et al., 2018). Intrahepatic HBV RNA levels approaching those of inactive carriers are also considered a useful viral endpoint for discontinuation of NA therapy. Additionally, HBV RNA should be used as an indicator for discontinuation of testing; this is also included in several guideline consensus (Wang et al., 2017; Berg and Petersen, 2018). At 24 weeks, HBV RNA levels declined more rapidly in patients who received HBsAg serologic conversion than in those who did not (Mak et al., 2022). Several different techniques, including rapid amplification RT-qPCR based on cDNA terminal (RACE), qPCR, and ddPCR, are available for quantifying RNA levels (Limothai et al., 2020; Lok et al., 2022; Yu et al., 2022; Hu et al., 2023b). However, standardization of HBV RNA detection methods is essential. Various experimental methods exist for quantifying intrahepatic and serum HBV RNA, all relying on quantitative RT-qPCR assays. However, consensus remains elusive regarding a singular technical or commercial assay for HBV RNA detection (Hu et al., 2023b).
Hepatitis B core-related antigen (HBcrAg) includes several proteins, including HBV core antigen, HBeAg, and pre-core/core protein (p22cr), and has a molecular weight of 22 kDa (Kimura et al., 2005; Hong et al., 2021; Adraneda et al., 2023). Its quantitative measurement is of great significance for guiding the clinical management of chronic HBV infection (Lee and Ahn, 2011; Ye et al., 2021; Adraneda et al., 2023). HBcrAg has emerged as a novel product of CHB infection and is correlated with the responses to current antiviral therapies for HBeAg-positive CHB; this product should be considered in addition to secreted HBV RNA when evaluating new antiviral therapies that directly or indirectly target hepatic cccDNA, with the goal of achieving functional cure (Chuaypen et al., 2016; Wong et al., 2017; Kramvis et al., 2022). The use of HBcrAg detection methods in patients with undetectable HBV DNA and HBsAg is anticipated to become a beneficial prognostic factor for determining the long-term prognosis of patients with CHB infection (Honda et al., 2016; Watanabe et al., 2021; Sonneveld et al., 2022). The predicted performance of HBcrAg may vary depending on the clinical endpoint being considered for CHB infection (Tseng et al., 2023). The clinically anticipated performance of HBcrAg is inconsistent, and there is a poor correlation between HBsAg loss and antiviral treatment (Wong et al., 2023). Therefore, HBcrAg-related results should be interpreted carefully in clinical practice (Tseng et al., 2022; Adraneda et al., 2023). CLEIA is a method primarily used for detecting HBcrAg in patient serum. CLEIA detects a combination of HBcAg, HBeAg (both free and in the HBeAg–HBe antibody complex), and precore proteins in blood, validated for dried blood spot (Wang et al., 2007).
Over the past decade, there has been significant progress in evaluating the progression of non-invasive liver disease in CHB patients. Although existing antiviral drugs for treating hepatitis B can effectively control the progression of such diseases, they can rarely eliminate the virus or achieve functional cure (Fanning et al., 2019; Chinese Society of Hepatology, 2022a; Watanabe et al., 2022). Additionally, cccDNA may still exist in the nuclei of the livers of patients, increasing their likelihood of HBV reactivation and developing HCC (Wang et al., 2014; Moro et al., 2018; World Health, 2019). The levels of novel HBV products have been demonstrated to correlate with the regression and prognosis of CHB disease. The levels of novel serological viral products, including HBV RNA, HBcrAg, and cccDNA, along with their transcriptional activity, can serve as exploratory endpoints in new drug research (Testoni et al., 2023).. However, the detection of these innovative serum viral products faces methodological challenges. Novel serum viral products can be compared or combined with routine serum viral products (such as HBV DNA, HBeAg, and HBsAg) to assess disease progression clinically. However, the detection should be conducted in centralized labs using thoroughly validated standardized reagents and platforms, accompanied by comprehensive detection protocols (Kuhns et al., 2021). Novel serum viral products aid in analyzing the mechanism of new therapeutic drugs. Anticipated technological advancements and the progress in ultrasensitive assays might potentially redefine the meaning of “functional cure” or even “partial functional cure” in the foreseeable future (Kramvis et al., 2022; Lok et al., 2022).
Overall, the detection of novel serum viral products allows for assessing antiviral effectiveness and predicting the relapse risk after drug withdrawal. This assists clinicians in providing better treatments for patients with CHB. Here, we systematically describe the traditional serum products of HBV (HBsAg, HBeAg, and HBV DNA) and the emerging serum viral products (HBV RNA and HBcrAg) and discuss their detection methods and applications. The principles of these detection methods vary widely, and each has its own unique advantages and disadvantages. CLIA is a non-radioactive immunoassay method that has rapidly advanced in the past 30 years. Owing to the high sensitivity of chemiluminescence and strong specificity of immunoassays, it has attracted wide attention recently. CLIA is widely used in clinical diagnosis and biochemical analysis to detect various tumor markers, cytokines (Yang et al., 2019), and hormones. Its sensitivity and specificity have been improved at both the qualitative to quantitative levels, its procedure has been fully automated, and the detection time has been remarkable reduced. The ECLIA method has ideal clinical application value, with advantages of high detection rates and a wide detection range. The reagents utilized in ECLIA remain long-term stability without the risk of toxicity or contamination. Currently, advanced chemiluminescence systems are being manufactured by Abbott, Siemens, Roche, Beckman, and other international brands as well as domestic brands, such as Mindray and Avron.
Several reports indicate that 27.8% to 59.5% of chronic HBV-infected patients are in “uncertain period” (Chinese Society of Hepatology, 2022b). Patients with chronic HBV infection in the “uncertain period” are at higher risk of progression of liver fibrosis, cirrhosis, and HCC than those without “uncertain period.” The value of noninvasive liver fibrosis assessments (e.g., hepatic transient elastography) in evaluating progression in “uncertain” chronic HBV-infected patients is debatable. Certainly, new serologic assays are being investigated, and better results are expected for clinical guidance. Among patients with CHB infection undergoing antiviral therapy, although potent low-resistance oral antiviral therapy results in potent suppression of HBV replication, low-level viremia (LLV) persists in some patients. Additionally, more LLV is present in some patients in the “uncertain period”. LLV is a term used to describe the detection of HBV DNA in the serum of a patient at levels ranging from 20 to 2000 IU/ML after 48 weeks of antiviral treatment (European Association for the Study of the Liver, 2017; Terrault et al., 2018). According to the guidelines of the American Association for Liver Research and the EASL, potent antiviral drugs such as tenofovir alafenamide fumarate, tenofovir, and ETV are recommended for the management of LLV, after excluding the issues related to compliance and detection errors (Lee et al., 2022). “Uncertain period” LLV refers to the “gray zone” between inactive carriers and HBeAg-negative CHB in patients with first-treatment chronic HBV infection who, after 1 year of follow-up, exhibit a pattern of HBV DNA and ALT that differs from the patterns of the four traditional stages of chronic HBV infection, with levels of either HBV DNA or ALT intermediate between those of inactive carriers and those of HBeAg-negative CHB (Yan and Sun, 2023). Highly sensitive detection methods such as ECLIA and CLIA for HBsAg or HBeAg are needed for patients in this period.
High sensitivity detection of HBsAg and HBV DNA significantly influences the selection and adjustment of antiviral therapy, aiding in predicting efficacy, guiding cessation, and enhancing transfusion safety. High sensitivity method also identifies OBI and shortens the window period for detecting acute HBV infection. Patients with OBI are negative for serum HBsAg, while HBV DNA is present in the liver with detectable/undetectable levels in the serum (Chinese Society of Hepatology, 2022b). Im YR et al. found that the prevalence of OBI reflects the prevalence of Hepatitis B: 0.98% in high prevalence areas, 0.12% in moderately prevalent areas as well as 0.06% in low prevalence areas (Im et al., 2022). OBI has been associated with advanced chronic liver disease, especially HCC, and patients with OBI can transmit HBV (Chen et al., 2019; Saitta et al., 2022; Wu et al., 2024). Combining novel serum viral products with routine serum viral products improves the sensitivity and specificity of identifying OBI and predicting the presence of OBI in the liver. Clinically, HBsAg and HBV DNA are two important serum viral products necessary for HBV diagnosis based on the infection’s natural history. Many treated patients primarily use standard sensitivity detection, considering high sensitivity as supplementary. There is a significant cost difference between the two methods; the high sensitivity detection method has a higher clinical value than the ordinary sensitivity detection; however, considering the economic burden of patients, the ordinary sensitivity detection can be applied to most of the patients.
HBV has high genetic diversity due to different genotypes and even intergenotypic recombinants (Bollyky et al., 1996; Ghosh et al., 2013; Liao et al., 2017; Liu et al., 2018; Feng et al., 2020). Genotypes may influence disease progression, drug resistance, response to antiviral therapy and prognosis. Intergenotype recombination is an important mechanism of virus evolution. Naturally occurring deletions/insertions have been found in the HBV core promoter, which may affect the production of core antigens (Peng et al., 2015). The HBV S gene tends to accumulate immune escape mutations, which may interfere with the use of immunological methods for clinical detection (Ding et al., 2014; Liu et al., 2024). Sun et al. showed that mutations in the pre-S region of the HBV genome may be associated with the development of OBI. The pre-S mutants of genotype B located in the pre-S2/S promoter significantly reduced the production of HBsAg by influencing the promoter activity, thus promoting the occurrence of OBI (Sun et al., 2022). Further, Jiang et al. found that the high frequency mutation of S protein transmembrane domain may be related to the occurrence of OBI (Jiang et al., 2022). It has been shown that mutations in precore, basal core promoter and preS in HBeAg-negative patients are associated with quantitative HBsAg serum levels and HBV DNA levels (Li et al., 2010; Kuhnhenn et al., 2018). Deletion/insertion mutations in the whole genome of HBV are prevalent in HBeAg-positive CHB patients prior to antiviral therapy, and the higher the detection rate of these mutations, the better the response to lamivudine and adefovir dipivoxil combination therapy (Hao et al., 2015). However, more studies are needed to reveal the impact of HBV genetic diversity and genotype on the production and detection of HBV serum markers.
YiL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft. DW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft. KZ: Conceptualization, Investigation, Methodology, Validation, Writing – review & editing. RR: Conceptualization, Formal analysis, Methodology, Resources, Writing – review & editing. YuL: Conceptualization, Data curation, Investigation, Methodology, Resources, Visualization, Writing – original draft. SZ: Conceptualization, Investigation, Methodology, Software, Writing – original draft. XZ: Conceptualization, Data curation, Investigation, Methodology, Software, Validation, Writing – original draft. JC: Conceptualization, Investigation, Methodology, Validation, Visualization, Writing – review & editing. LC: Conceptualization, Investigation, Methodology, Software, Supervision, Validation, Writing – review & editing. JH: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Natural Science Foundation of China (Project Reach Youth, 81903419), the Key Research and Development Project of Henan Province (Science and Technology Research Project, 222102310622) and Zhengzhou University First Class Course Project (2021ZZUKCLX057).
We also thank Figdraw (www.figdraw.com) for the assistance in drawing Figures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.

Adraneda, C., Tan, Y. C., Yeo, E. J., Kew, G. S., Khakpoor, A., Lim, S. G. (2023). A critique and systematic review of the clinical utility of hepatitis B core-related antigen. J. Hepatol. 78, 731–741. doi: 10.1016/j.jhep.2022.12.017
Alves, T. G., Melo, M. C. C., Kasamatsu, T. S., Oliveira, K. C., Souza, J. S., Conceicao, R. R. D., et al. (2017). Novel immunoassay for TSH measurement in rats. Arch. Endocrinol. Metab. 61, 460–463. doi: 10.1590/2359-3997000000293
Amini, A., Varsaneux, O., Kelly, H., Tang, W., Chen, W., Boeras, D. I., et al. (2017). Diagnostic accuracy of tests to detect hepatitis B surface antigen: a systematic review of the literature and meta-analysis. BMC Infect. Dis. 17, 698. doi: 10.1186/s12879-017-2772-3
Author Group of Expert Opinions (2022). [Expert opinions on technical guidance for clinical trials of antiviral drugs for chronic hepatitis B]. Zhonghua Gan Zang Bing Za Zhi 30, 493–504. doi: 10.3760/cma.j.cn501113-20220416-00203
Barker, L. F., Maynard, J. E., Purcell, R. H., Hoofnagle, J. H., Berquist, K. R., London, W. T. (1975). Viral hepatitis, type B, in experimental animals. Am. J. Med. Sci. 270, 189–195. doi: 10.1097/00000441-197507000-00026
Berg, T., Petersen, J. (2018). Nucleos(t)ide analogue interruption: Alternative approach to intrahepatic set point for spontaneous control of HBV replication? J. Hepatol. 68, 611–612. doi: 10.1016/j.jhep.2017.09.025
Blumberg, B. S. (1964). Polymorphisms of the serum proteins and the development of iso-precipitins in transfused patients. Bull. N Y Acad. Med. 40, 377–386.
Blumberg, B. S., Alter, H. J., Visnich, S. (1965). A “New” Antigen in leukemia sera. JAMA 191, 541–546. doi: 10.1001/jama.1965.03080070025007
Blumberg, B. S., Gerstley, B. J., Hungerford, D. A., London, W. T., Sutnick, A. I. (1967). A serum antigen (Australia antigen) in Down’s syndrome, leukemia, and hepatitis. Ann. Intern. Med. 66, 924–931. doi: 10.7326/0003-4819-66-5-924
Bollyky, P. L., Rambaut, A., Harvey, P. H., Holmes, E. C. (1996). Recombination between sequences of hepatitis B virus from different genotypes. J. Mol. Evol. 42, 97–102. doi: 10.1007/BF02198834
Boulon, R., Blanchet, M., Lemasson, M., Vaillant, A., Labonte, P. (2020). Characterization of the antiviral effects of REP 2139 on the HBV lifecycle in vitro. Antiviral Res. 183, 104853. doi: 10.1016/j.antiviral.2020.104853
Bruss, V. (2007). Hepatitis B virus morphogenesis. World J. Gastroenterol. 13, 65–73. doi: 10.3748/wjg.v13.i1.65
Burns, G. S., Thompson, A. J. (2014). Viral hepatitis B: clinical and epidemiological characteristics. Cold Spring Harb. Perspect. Med. 4, a024935. doi: 10.1101/cshperspect.a024935
Caliendo, A. M., Valsamakis, A., Bremer, J. W., Ferreira-Gonzalez, A., Granger, S., Sabatini, L., et al. (2011). Multilaboratory evaluation of real-time PCR tests for hepatitis B virus DNA quantification. J. Clin. Microbiol. 49, 2854–2858. doi: 10.1128/JCM.00471-11
Carey, I., Gersch, J., Wang, B., Moigboi, C., Kuhns, M., Cloherty, G., et al. (2020). Pregenomic HBV RNA and hepatitis B core-related antigen predict outcomes in hepatitis B e antigen-negative chronic hepatitis B patients suppressed on nucleos(T)ide analogue therapy. Hepatology 72, 42–57. doi: 10.1002/hep.31026
Chen, D., Kaplan, L., Liu, Q. (2005). Evaluation of two chemiluminescent immunoassays of ADVIA Centaur for hepatitis B serology markers. Clin. Chim. Acta 355, 41–45. doi: 10.1016/j.cccn.2004.11.031
Chen, J., Wang, F., Li, J., Zuo, Q., Wu, D., Xiao, C. (2024). Clinical trial and performance evaluation of the Wantai HBsAg (CMIA) diagnostic kit for screening blood donors in China. Sci. Rep. 14, 1891. doi: 10.1038/s41598-024-51910-1
Chen, X. P., Long, X., Jia, W. L., Wu, H. J., Zhao, J., Liang, H. F., et al. (2019). Viral integration drives multifocal HCC during the occult HBV infection. J. Exp. Clin. Cancer Res. 38, 261. doi: 10.1186/s13046-019-1273-1
Chinese Society of Hepatology, C.M.A. (2022a). [Expert opinion on expanding anti-HBV treatment for chronic hepatitis B]. Zhonghua Gan Zang Bing Za Zhi 30, 131–136. doi: 10.3760/cma.j.cn501113-20220209-00060
Chinese Society of Hepatology, C.M.A.C.S.o.I.D., Chinese Medical Association (2022b). [Guidelines for the prevention and treatment of chronic hepatitis B (version 2022)]. Zhonghua Gan Zang Bing Za Zhi 30, 1309–1331. doi: 10.3760/cma.j.cn501113-20221204-00607
Chuaypen, N., Posuwan, N., Payungporn, S., Tanaka, Y., Shinkai, N., Poovorawan, Y., et al. (2016). Serum hepatitis B core-related antigen as a treatment predictor of pegylated interferon in patients with HBeAg-positive chronic hepatitis B. Liver Int. 36, 827–836. doi: 10.1111/liv.13046
Chuaypen, N., Posuwan, N., Chittmittraprap, S., Hirankarn, N., Treeprasertsuk, S., Tanaka, Y., et al. (2018). Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin. Microbiol. Infect. 24, 306 e7–306 e13. doi: 10.1016/j.cmi.2017.07.016
Cornberg, M., Wong, V. W., Locarnini, S., Brunetto, M., Janssen, H. L.A., Chan, H. L. (2017). The role of quantitative hepatitis B surface antigen revisited. J. Hepatol. 66, 398–411. doi: 10.1016/j.jhep.2016.08.009
Dahari, H., Shlomai, A., Cotler, S. J. (2021). Early HBV RNA kinetics under NA treatment may reveal new insights into HBV RNA dynamics and NA mode of action-more detailed kinetic studies are needed. J. Viral Hepat 28, 687–688. doi: 10.1111/jvh.13463
de Almeida Ponde, R. A. (2022). Detection of the serological markers hepatitis B virus surface antigen (HBsAg) and hepatitis B core IgM antibody (anti-HBcIgM) in the diagnosis of acute hepatitis B virus infection after recent exposure. Microbiol. Immunol. 66, 1–9. doi: 10.1111/1348-0421.12943
Deguchi, M., Yamashita, N., Kagita, M., Asari, S., Iwatani, Y., Tsuchida, T., et al. (2004). Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J. Virol. Methods 115, 217–222. doi: 10.1016/j.jviromet.2003.10.002
Dera, A., Sanou, A. M., Ouattara, M. N.G., Ilboudo, A. K., Lankoande, D. B., Ilboudo, D., et al. (2023). Evaluation of the diagnostic performances of the SD-bioline((R))HBeAg rapid test used routinely for the management of HBV-infected individuals in Burkina Faso. Diagnostics (Basel) 13, 3144. doi: 10.3390/diagnostics13193144
Ding, H., Liu, B., Zhao, C., Yang, J., Yan, C., Yan, L., et al. (2014). Amino acid similarities and divergences in the small surface proteins of genotype C hepatitis B viruses between nucleos(t)ide analogue-naive and lamivudine-treated patients with chronic hepatitis B. Antiviral Res. 102, 29–34. doi: 10.1016/j.antiviral.2013.11.015
Ding, J., Yi, Z., Zai, W., Wu, M., Chen, B., Cai, Q., et al. (2023). Illuminating the live-cell dynamics of hepatitis B virus covalently closed circular DNA using the CRISPR-tag system. mBio, e0355022. doi: 10.1128/mbio.03550-22
Dogbe, E. E., Arthur, F. (2015). Diagnostic accuracy of blood centers in the screening of blood donors for viral markers. Pan Afr Med. J. 20, 119. doi: 10.11604/pamj.2015.20.119.5263
Dong, C., Yu, J., Zhu, Y., Dong, C. (2013). Inhibition of hepatitis B virus gene expression & replication by crude destruxins from Metarhizium anisopliae. Var. dcjhyium. Indian J. Med. Res. 138, 969–976.
Duverlie, G., Roussel, C., Driencourt, M., Orfila, J. (1988). Microparticle enzyme immunoassay for determination of immunoglobulin G antibodies to human cytomegalovirus. J. Clin. Microbiol. 26, 2229–2230. doi: 10.1128/jcm.26.10.2229-2230.1988
Engvall, E., Jonsson, K., Perlmann, P. (1971). Enzyme-linked immunosorbent assay. II. Quantitative assay of protein antigen, immunoglobulin G, by means of enzyme-labelled antigen and antibody-coated tubes. Biochim. Biophys. Acta 251, 427–434. doi: 10.1016/0005-2795(71)90132-2
Engvall, E., Perlmann, P. (1971). Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 8, 871–874. doi: 10.1016/0019-2791(71)90454-X
Fan, R., Zhou, B., Xu, M., Tan, D., Niu, J., Wang, H., et al. (2020). Association between negative results from tests for HBV DNA and RNA and durability of response after discontinuation of nucles(t)ide analogue therapy. Clin. Gastroenterol. Hepatol. 18, 719–727 e7. doi: 10.1016/j.cgh.2019.07.046
Fanning, G. C., Zoulim, F., Hou, J., Bertoletti, A. (2019). Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discovery 18, 827–844. doi: 10.1038/s41573-019-0037-0
Feng, Y., Ran, J., Feng, Y. M., Miao, J., Zhao, Y., Jia, Y., et al. (2020). Genetic diversity of hepatitis B virus in Yunnan, China: identification of novel subgenotype C17, an intergenotypic B/I recombinant, and B/C recombinants. J. Gen. Virol. 101, 972–981. doi: 10.1099/jgv.0.001147
Fu, L., Qian, Y., Zhou, J., Zheng, L., Wang, Y. (2020). Fluorescence-based quantitative platform for ultrasensitive food allergen detection: From immunoassays to DNA sensors. Compr. Rev. Food Sci. Food Saf. 19, 3343–3364. doi: 10.1111/1541-4337.12641
Gao, L., Mao, T. H., Peng, S. W., Wang, J., Chen, X. M., Lu, F. M. (2022). [A short half-life of cccDNA offer or ignite hope for hepatitis B cure under nucleos(t)ide analogues treatment]. Zhonghua Gan Zang Bing Za Zhi 30, 99–102. doi: 10.3760/cma.j.cn501113-20200527-00277
Gerlich, W. H., Robinson, W. S. (1980). Hepatitis B virus contains protein attached to the 5’ terminus of its complete DNA strand. Cell 21, 801–809. doi: 10.1016/0092-8674(80)90443-2
Ghosh, S., Banerjee, P., Deny, P., Mondal, R. K., Nandi, M., Roychoudhury, A., et al. (2013). New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India. J. Viral Hepat 20, 209–218. doi: 10.1111/j.1365-2893.2012.01655.x
Ghosh, M., Nandi, S., Dutta, S., Saha, M. K. (2015). Detection of hepatitis B virus infection: A systematic review. World J. Hepatol. 7, 2482–2491. doi: 10.4254/wjh.v7.i23.2482
Guo, R., Mao, H., Hu, X., Zheng, N., Yan, D., He, J., et al. (2016). Slow reduction of IP-10 Levels predicts HBeAg seroconversion in chronic hepatitis B patients with 5 years of entecavir treatment. Sci. Rep. 6, 37015. doi: 10.1038/srep37015
Hadziyannis, E., Manesis, E., Vassilopoulos, D., Georgiou, A., Archimandritis, A. (2008). Performance characteristics of microparticle enzyme and chemiluminescence immunoassays for measurement of anti-HBc immunoglobulin M in sera of patients with HBeAg-negative chronic hepatitis B virus infection. Clin. Vaccine Immunol. 15, 385–387. doi: 10.1128/CVI.00414-07
Hadziyannis, E., Laras, A. (2018). Viral biomarkers in chronic HBeAg negative HBV infection. Genes (Basel) 9. doi: 10.3390/genes9100469
Halonen, P., Meurman, O., Lovgren, T., Hemmila, I., Soini, E.. (1983). Detection of viral antigens by time-resolved fluoroimmunoassay. Curr. Top. Microbiol. Immunol. 104, 133–146. doi: 10.1007/978-3-642-68949-9_8
Hao, R., Xiang, K., Peng, Y., Hou, J., Sun, J., Li, Y., et al. (2015). Naturally occurring deletion/insertion mutations within HBV whole genome sequences in HBeAg-positive chronic hepatitis B patients are correlated with baseline serum HBsAg and HBeAg levels and might predict a shorter interval to HBeAg loss and seroconversion during antiviral treatment. Infect. Genet. Evol. 33, 261–268. doi: 10.1016/j.meegid.2015.05.013
Heid, C. A., Stevens, J., Livak, K. J., Williams, P. M. (1996). Real time quantitative PCR. Genome Res. 6, 986–994. doi: 10.1101/gr.6.10.986
Honda, M., Shirasaki, T., Terashima, T., Kawaguchi, K., Nakamura, M., Oishi, N., et al. (2016). Hepatitis B virus (HBV) core-related antigen during nucleos(t)ide analog therapy is related to intra-hepatic HBV replication and development of hepatocellular carcinoma. J. Infect. Dis. 213, 1096–1106. doi: 10.1093/infdis/jiv572
Hong, X., Luckenbaugh, L., Mendenhall, M., Walsh, R., Cabuang, L., Soppe, S., et al. (2021). Characterization of hepatitis B precore/core-related antigens. J. Virol. 95, e01695-01620. doi: 10.1128/JVI.01695-20
Hu, Z., Li, M., Huang, B., Liu, J., Yu, L., Chen, G. (2012). Detection of hepatitis B virus PreS1 antigen using a time-resolved fluoroimmunoassay. J. Immunoassay Immunochem 33, 156–165. doi: 10.1080/15321819.2011.609576
Hu, X., Gao, X., Chen, S., Guo, J., Zhang, Y. (2023a). DropLab: an automated magnetic digital microfluidic platform for sample-to-answer point-of-care testing-development and application to quantitative immunodiagnostics. Microsyst Nanoeng 9, 10. doi: 10.1038/s41378-022-00475-y
Hu, X., Zhao, L., Ou, M., Chen, Y., Wei, H., Xia, Y., et al. (2023b). Evaluation of reverse transcription-polymerase chain reaction and simultaneous amplification and testing for quantitative detection of serum hepatitis B virus RNA. Heliyon 9, e18557. doi: 10.1016/j.heliyon.2023.e18557
Huang, P. X., Wang, N., Qian, J. H., Jiang, F., Yang, Y. L., Lin, W. Y., et al. (2017). [A 22-year-follow-up cohort study on primary liver cancer in Haimen city of Jiangsu province]. Zhonghua Liu Xing Bing Xue Za Zhi 38, 1376–1379. doi: 10.3760/cma.j.issn.0254-6450.2017.10.016
Im, Y. R., Jagdish, R., Leith, D., Kim, J. U., Yoshida, K., Majid, A., et al. (2022). Prevalence of occult hepatitis B virus infection in adults: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 7, 932–942. doi: 10.1016/S2468-1253(22)00201-1
Imam, H., Khan, M., Gokhale, N. S., McIntyre, A. B. R., Kim, G. W., Jang, J. Y., et al. (2018). N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. United States America 115, 8829–8834. doi: 10.1073/pnas.1808319115
Inoue, T., Matsui, T., Tanaka, Y. (2021). Novel strategies for the early diagnosis of hepatitis B virus reactivation. Hepatol. Res. 51, 1033–1043. doi: 10.1111/hepr.13699
Irshad, M., Gupta, P., Mankotia, D. S., Ansari, M. A. (2016). Multiplex qPCR for serodetection and serotyping of hepatitis viruses: A brief review. World J. Gastroenterol. 22, 4824–4834. doi: 10.3748/wjg.v22.i20.4824
Ji, X., Xia, M., Zhou, B., Liu, S., Liao, G., Cai, S., et al. (2020). Serum hepatitis B virus RNA levels predict HBeAg seroconversion and virological response in chronic hepatitis B patients with high viral load treated with nucleos(t)ide analog. Infect. Drug Resist. 13, 1881–1888. doi: 10.2147/IDR.S252994
Jiang, T., Liang, Z., Chen, J., He, J. J., Lu, L., Ma, W. M., et al. (2007). [Establishment of a colloid gold-immunochromatography assay for detection of type Asia I foot-and-mouth disease virus]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 23, 1021–1024.
Jiang, S., Zhu, Y., Cheng, C., Li, Y., Ma, T., Peng, Z., et al. (2020). NK cells contribute to hepatic CD8(+) T cell failure in hepatitis B virus-carrier mice after alcohol consumption. Virus Res. 286, 198085. doi: 10.1016/j.virusres.2020.198085
Jiang, B., Su, R., Ren, D., Zheng, X., Cao, Y., Mi, Y., et al. (2020). Evaluation of HBV serological markers in treatment-naive HBV mono-infected patients and HBV-HIV co-infected patients. Virus Res. 290, 198117. doi: 10.1016/j.virusres.2020.198117
Jiang, X., Chang, L., Yan, Y., Ji, H., Sun, H., Xiao, Y., et al. (2022). Role of S protein transmembrane domain mutations in the development of occult hepatitis B virus infection. Emerg. Microbes Infect. 11, 2184–2196. doi: 10.1080/22221751.2022.2114849
Jung, J., Nguyen, M. H. (2023). Liver-Related Mortality in Hepatitis B Virus Core Antibody+/Hepatitis B Virus Surface Antigen− Patients: Occult Hepatitis B Virus, Hepatitis B Virus Reactivation, and Hepatocellular Carcinoma Development. American Journal of Gastroenterology 118 (1), 24–25. doi: 10.14309/ajg.0000000000002030
Kalita, D., Deka, S., Chamuah, K., Ahmed, G. (2022). Laboratory evaluation of hepatitis C virus infection in patients undergoing hemodialysis from north east India. J. Clin. Exp. Hepatol. 12, 475–482. doi: 10.1016/j.jceh.2021.05.011
Kao, J. H. (2008). Diagnosis of hepatitis B virus infection through serological and virological markers. Expert Rev. Gastroenterol. Hepatol. 2, 553–562. doi: 10.1586/17474124.2.4.553
Khadem-Ansari, M. H., Omrani, M. D., Rasmi, Y., Ghavam, A. (2014). Diagnostic validity of the chemiluminescent method compared to polymerase chain reaction for hepatitis B virus detection in the routine clinical diagnostic laboratory. Adv. BioMed. Res. 3, 116. doi: 10.4103/2277-9175.133178
Kim, S. H. (2017). ELISA for quantitative determination of hepatitis B virus surface antigen. Immune Netw. 17, 451–459. doi: 10.4110/in.2017.17.6.451
Kim, S. K., Fujii, T., Kim, S. R., Nakai, A., Lim, Y. S., Hagiwara, S., et al. (2022). Hepatitis B virus treatment and hepatocellular carcinoma: controversies and approaches to consensus. Liver Cancer 11, 497–510. doi: 10.1159/000525518
Kim, J. H., Lee, S. Y., Lee, S. K. (2021). Development of novel lab-on-a-chip platform for high-throughput radioimmunoassay. Appl. Radiat. Isot 168, 109526. doi: 10.1016/j.apradiso.2020.109526
Kimura, T., et al. (2002). Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 40, 439–445. doi: 10.1128/JCM.40.2.439-445.2002
Kimura, T., Ohno, N., Terada, N., Rokuhara, A., Matsumoto, A., Yagi, S., et al. (2005). Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 280, 21713–21719. doi: 10.1074/jbc.M501564200
Kovac, M., Risch, L., Thiel, S., Weber, M., Grossmann, K., Wohlwend, N., et al. (2020). EDTA-anticoagulated whole blood for SARS-coV-2 antibody testing by electrochemiluminescence immunoassay (ECLIA) and enzyme-linked immunosorbent assay (ELISA). Diagnostics (Basel) 10, 593. doi: 10.3390/diagnostics10080593
Kramvis, A., Chang, K. M., Dandri, M., Farci, P., Glebe, D., Hu, J., et al. (2022). A roadmap for serum biomarkers for hepatitis B virus: current status and future outlook. Nat. Rev. Gastroenterol. Hepatol. 19, 727–745. doi: 10.1038/s41575-022-00649-z
Kroes, A. C., Quint, W. G., Heijtink, R. A. (1991). Significance of isolated hepatitis B core antibodies detected by enzyme immunoassay in a high risk population. J. Med. Virol. 35, 96–100. doi: 10.1002/jmv.1890350205
Kuhnhenn, L., Jiang, B., Kubesch, A., Vermehren, J., Knop, V., Susser, S., et al. (2018). Impact of HBV genotype and mutations on HBV DNA and qHBsAg levels in patients with HBeAg-negative chronic HBV infection. Aliment Pharmacol. Ther. 47, 1523–1535. doi: 10.1111/apt.14636
Kuhns, M. C., Holzmayer, V., McNamara, A. L., Anderson, M., Cloherty, G. A.. (2021). Hepatitis B seroconversion revisited: new insights into the natural history of acute hepatitis B virus (HBV) infection from quantitative and highly sensitive assays and novel biomarkers . Virol. J. 18, 235. doi: 10.1186/s12985-021-01706-w
Lee, J. M., Ahn, S. H. (2011). Quantification of HBsAg: basic virology for clinical practice. World J. Gastroenterol. 17, 283–289. doi: 10.3748/wjg.v17.i3.283
Lee, H., Jang, S., Ahn, S. H., Kim, B. K. (2022). Cost-effectiveness of antiviral therapy in untreated compensated cirrhosis patient with serum HBV-DNA level < 2000 IU/mL. Hepatol. Int. 16, 294–305. doi: 10.1007/s12072-022-10310-1
Lequin, R. M. (2005). Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 51, 2415–2418. doi: 10.1373/clinchem.2005.051532
Li, X., Wang, L., Zhong, Y., Wong, V. W., Xu, Z., Liu, Y., et al. (2010). Hepatitis B virus (HBV) subgenotypes C2 and B2 differ in lamivudine- and adefovir-resistance-associated mutational patterns in HBV-infected Chinese patients. J. Clin. Microbiol. 48, 4363–4369. doi: 10.1128/JCM.01518-10
Li, X., Zhu, J., Lai, G., Yan, L., Hu, J., Chen, J., et al. (2015). The infection efficiency and replication ability of circularized HBV DNA optimized the linear HBV DNA in vitro and in vivo. Int. J. Mol. Sci. 16, 5141–5160. doi: 10.3390/ijms16035141
Li, X., Zhao, J., Yuan, Q., Xia, N. (2017). Detection of HBV covalently closed circular DNA. Viruses 9, 139. doi: 10.3390/v9060139
Li, H., Yan, L., Shi, Y., Lv, D., Shang, J., Bai, L., et al. (2020). Hepatitis B virus infection: overview. Adv. Exp. Med. Biol. 1179, 1–16. doi: 10.1007/978-981-13-9151-4_1
Li, F., Deng, Y., Zhang, S., Zhu, B., Wang, J., Wang, J., et al. (2022). Human hepatocyte-enriched miRNA-192–3p promotes HBV replication through inhibiting Akt/mTOR signalling by targeting ZNF143 in hepatic cell lines. Emerg. Microbes Infect. 11, 616–628. doi: 10.1080/22221751.2022.2037393
Liao, H., Li, X., Liu, Y., Xu, Z., Huang, P., Nian, X., et al. (2017). Intergenotype recombinant analysis of full-length hepatitis B virus genomes from 516 Chinese patients with different illness categories. J. Med. Virol. 89, 139–145. doi: 10.1002/jmv.24609
Liaw, Y. F., Lau, G. K., Kao, J. H., Gane, E. (2010). Hepatitis B e antigen seroconversion: a critical event in chronic hepatitis B virus infection. Dig Dis. Sci. 55, 2727–2734. doi: 10.1007/s10620-010-1179-4
Lim, S. G., Phyo, W. W., Ling, J. Z.J., Cloherty, G., Butler, E. K., Kuhns, M. C., et al. (2021). Comparative biomarkers for HBsAg loss with antiviral therapy shows dominant influence of quantitative HBsAg (qHBsAg). Aliment Pharmacol. Ther. 53, 172–182. doi: 10.1111/apt.16149
Limothai, U., Chuaypen, N., Poovorawan, K., Chotiyaputta, W., Tanwandee, T., Poovorawan, Y., et al. (2020). Reverse transcriptase droplet digital PCR vs reverse transcriptase quantitative real-time PCR for serum HBV RNA quantification. J. Med. Virol. 92, 3365–3372. doi: 10.1002/jmv.25792
Liu, C., Chen, T., Lin, J., Chen, H., Chen, J., Lin, S., et al. (2014). Evaluation of the performance of four methods for detection of hepatitis B surface antigen and their application for testing 116,455 specimens. J. Virol. Methods 196, 174–178. doi: 10.1016/j.jviromet.2013.10.039
Liu, B., Yang, J. X., Yan, L., Zhuang, H., Li, T. (2018). Novel HBV recombinants between genotypes B and C in 3’-terminal reverse transcriptase (RT) sequences are associated with enhanced viral DNA load, higher RT point mutation rates and place of birth among Chinese patients. Infect. Genet. Evol. 57, 26–35. doi: 10.1016/j.meegid.2017.10.023
Liu, J., Liang, W., Jing, W., Liu, M. (2019). Countdown to 2030: eliminating hepatitis B disease, China. Bull. World Health Organ 97, 230–238. doi: 10.2471/BLT.18.219469
Liu, H., Chen, S., Liu, X., Lou, J. (2024). Effect of S-region mutations on HBsAg in HBsAg-negative HBV-infected patients . Virol. J. 21, 92. doi: 10.1186/s12985-024-02366-2
Liubavina, I. A., Zinchenko, A. A., Lebedin Iu, S., Chukanov, S. V. (2007). [Development of a rapid method for the detection of prostate-specific antigen by immunochromatography]. Bioorg Khim 33, 550–554. doi: 10.1134/S1068162007050081
Liver, E.A.f.t.S.o.t. (2017). EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 67, 370–398. doi: 10.1016/j.jhep.2017.03.021
Loglio, A., Iavarone, M., Facchetti, F., Di Paolo, D., Perbellini, R., Lunghi, G., et al. (2020). The combination of PIVKA-II and AFP improves the detection accuracy for HCC in HBV caucasian cirrhotics on long-term oral therapy. Liver Int. 40, 1987–1996. doi: 10.1111/liv.14475
Lok, J., Dusheiko, G., Carey, I., Agarwal, K.. (2022). Review article: novel biomarkers in hepatitis B infection. Aliment Pharmacol. Ther. 56, 760–776. doi: 10.1111/apt.17105
Lu, F. M., Wang, J., Chen, X. M., Jiang, J. N., Zhang, W. H., Zhao, J. M., et al. (2017). [The potential use of serum HBV RNA to guide the functional cure of chronic hepatitis B]. Zhonghua Gan Zang Bing Za Zhi 25, 105–110. doi: 10.3760/cma.j.issn.1007-3418.2017.02.005
Ma, S. W., Huang, X., Li, Y. Y., Tang, L. B., Sun, X. F., Jiang, X. T., et al. (2012). High serum IL-21 levels after 12 weeks of antiviral therapy predict HBeAg seroconversion in chronic hepatitis B. J. Hepatol. 56, 775–781. doi: 10.1016/j.jhep.2011.10.020
Maity, S., Nandi, S., Biswas, S., Sadhukhan, S. K., Saha, M. K. (2012). Performance and diagnostic usefulness of commercially available enzyme linked immunosorbent assay and rapid kits for detection of HIV, HBV and HCV in India . Virol. J. 9, 290. doi: 10.1186/1743-422X-9-290
Mak, L. Y., Wong, D., Kuchta, A., Hilfiker, M., Hamilton, A., Chow, N., et al. (2022). HBV pgRNA and HBcrAg reductions at week 4 predict favourable HBsAg response upon long-term nucleos(t)ide analogue in CHB. Clin Mol Hepatol 29, 146–162. doi: 10.3350/cmh.2022.0172
Marquardt, O., Heermann, K. H., Seifer, M., Gerlich, W. H. (1987). Cell type specific expression of pre S 1 antigen and secretion of hepatitis B virus surface antigen. Brief Report. Arch. Virol. 96, 249–256. doi: 10.1007/BF01320964
McCoullough, L. C., Sadauskas, T., Sozzi, V., Mak, K. Y., Mason, H., Littlejohn, M., et al. (2024). The in vitro replication phenotype of hepatitis B virus (HBV) splice variants Sp3 and Sp9 and their impact on wild-type HBV replication. J. Virol. 98, e0153823. doi: 10.1128/jvi.01538-23
Moini, M., Fung, S. (2022). HBsAg loss as a treatment endpoint for chronic HBV infection: HBV cure. Viruses 14, 657. doi: 10.3390/v14040657
Molnar-Kimber, K. L., Summers, J., Taylor, J. M., Mason, W. S. (1983). Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J. Virol. 45, 165–172. doi: 10.1128/jvi.45.1.165-172.1983
Moon, J. S., Lee, W., Cho, Y. H., Kim, Y., Kim, G. W. (2024). The significance of N6-methyladenosine RNA methylation in regulating the hepatitis B virus life cycle. J. Microbiol. Biotechnol. 34, 233–239. doi: 10.4014/jmb.2309.09013
Moro, P. L., Zheteyeva, Y., Barash, F., Lewis, P., Cano, M. (2018). Assessing the safety of hepatitis B vaccination during pregnancy in the Vaccine Adverse Event Reporting System (VAERS), 1990–2016. Vaccine 36, 50–54. doi: 10.1016/j.vaccine.2017.11.039
Motyka, J., Gacuta, E., Kicman, A., Kulesza, M., Lawicki, P., Lawicki, S. (2022). Plasma levels of CXC motif chemokine 1 (CXCL1) and chemokine 8 (CXCL8) as diagnostic biomarkers in luminal A and B breast cancer. J. Clin. Med. 11, 6694. doi: 10.3390/jcm11226694
Nassal, M. (2015). HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984. doi: 10.1136/gutjnl-2015-309809
Ollier, L., Laffont, C., Kechkekian, A., Doglio, A., Giordanengo, V. (2008). Detection of antibodies to hepatitis B core antigen using the Abbott ARCHITECT anti-HBc assay: analysis of borderline reactive sera. J. Virol. Methods 154, 206–209. doi: 10.1016/j.jviromet.2008.09.006
Pan, Y., Xia, H., He, Y., Zeng, S., Shen, Z., Huang, W. (2023). The progress of molecules and strategies for the treatment of HBV infection. Front. Cell Infect. Microbiol. 13, 1128807. doi: 10.3389/fcimb.2023.1128807
Paraskevis, D., Beloukas, A., Haida, C., Katsoulidou, A., Moschidis, Z., Hatzitheodorou, H., et al. (2010). Development of a new ultra sensitive real-time PCR assay (ultra sensitive RTQ-PCR) for the quantification of HBV-DNA . Virol. J. 7, 57. doi: 10.1186/1743-422X-7-57
Peng, Y., Liu, B., Hou, J., Sun, J., Hao, R., Xiang, K., et al. (2015). Naturally occurring deletions/insertions in HBV core promoter tend to decrease in hepatitis B e antigen-positive chronic hepatitis B patients during antiviral therapy. Antivir Ther. 20, 623–632. doi: 10.3851/IMP2955
Piermatteo, L., Alkhatib, M., D'Anna, S., Malagnino, V., Bertoli, A., Andreassi, E., et al. (2021). HBeAg levels vary across the different stages of HBV infection according to the extent of immunological pressure and are associated with therapeutic outcome in the setting of immunosuppression-driven HBV reactivation. Biomedicines 9, 1352. doi: 10.3390/biomedicines9101352
Pontisso, P., Ruvoletto, M. G., Gerlich, W. H., Heermann, K. H., Bardini, R., Alberti, A. (1989). Identification of an attachment site for human liver plasma membranes on hepatitis B virus particles. Virology 173, 522–530. doi: 10.1016/0042-6822(89)90564-3
Raimondo, G., Locarnini, S., Pollicino, T., Levrero, M., Zoulim, F., Lok, A. S., et al. (2019). Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J. Hepatol. 71, 397–408. doi: 10.1016/j.jhep.2019.03.034
Robinson, W. S., Clayton, D. A., Greenman, R. L. (1974). DNA of a human hepatitis B virus candidate. J. Virol. 14, 384–391. doi: 10.1128/jvi.14.2.384-391.1974
Rong, X., Ailing, F., Xiaodong, L., Jie, H., Min, L. (2021). Monitoring hepatitis B by using point-of-care testing: biomarkers, current technologies, and perspectives. Expert Rev. Mol. Diagn. 21, 195–211. doi: 10.1080/14737159.2021.1876565
Saitta, C., Pollicino, T., Raimondo, G. (2022). Occult hepatitis B virus infection: an update. Viruses 14, 1504. doi: 10.3390/v14071504
SChadler, S., Hildt, E. (2009). HBV life cycle: entry and morphogenesis. Viruses 1, 185–209. doi: 10.3390/v1020185
Shen, Y., Prinyawiwatkul, W., Xu, Z. (2019). Insulin: a review of analytical methods. Analyst 144, 4139–4148. doi: 10.1039/C9AN00112C
Siegel, S. R., Ulrich, M., Logue, S. F. (2023). Comparison qPCR study for selecting a valid single copy gene for measuring absolute telomere length. Gene 860, 147192. doi: 10.1016/j.gene.2023.147192
Sonneveld, M. J., Park, J. Y., Kaewdech, A., Seto, W. K., Tanaka, Y., Carey, I., et al. (2022). Prediction of sustained response after nucleo(s)tide analogue cessation using HBsAg and HBcrAg levels: A multicenter study (CREATE). Clin. Gastroenterol. Hepatol. 20, e784–e793. doi: 10.1016/j.cgh.2020.12.005
Su, T. S., Lai, C. J., Huang, J. L., Lin, L. H., Yauk, Y. K., Chang, C. M., et al. (1989a). Hepatitis B virus transcript produced by RNA splicing. J. Virol. 63, 4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989
Su, T. S., Lui, W. Y., Lin, L. H., Han, S. H., P'Eng, F, K. (1989b). Analysis of hepatitis B virus transcripts in infected human livers. Hepatology 9, 180–185. doi: 10.1002/(ISSN)1527-3350
Sun, Y., Wu, X., Zhou, J., Meng, T., Wang, B., Chen, S., et al. (2020). Persistent low level of hepatitis B virus promotes fibrosis progression during therapy. Clin. Gastroenterol. Hepatol. 18, 2582–2591.e6. doi: 10.1016/j.cgh.2020.03.001
Sun, H., Chang, L., Yan, Y., Ji, H., Jiang, X., Song, S., et al. (2022). Naturally occurring pre-S mutations promote occult HBV infection by affecting pre-S2/S promoter activity. Antiviral Res. 208, 105448. doi: 10.1016/j.antiviral.2022.105448
Sureau, C., Salisse, J. (2013). A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology 57, 985–994. doi: 10.1002/hep.26125
Takeda, K., Maruki, M., Yamagaito, T., Muramatsu, M., Sakai, Y., Tobimatsu, H., et al. (2013). Highly sensitive detection of hepatitis B virus surface antigen by use of a semiautomated immune complex transfer chemiluminescence enzyme immunoassay. J. Clin. Microbiol. 51, 2238–2244. doi: 10.1128/JCM.00324-13
Tay, S. T., Hemond, F. H., Krumholz, L. R., Cavanaugh, C. M., Polz, M. F. (2001). Population dynamics of two toluene degrading bacterial species in a contaminated stream. Microb. Ecol. 41, 124–131. doi: 10.1007/s002480000089
Terrault, N. A., Lok, A. S.F., McMahon, B. J., Chang, K. M., Hwang, J. P., Jonas, M. M., et al. (2018). Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 67, 1560–1599. doi: 10.1002/hep.29800
Testoni, B., Scholtes, C., Plissonnier, M. L., Paturel, A., Berby, F., Facchetti, F., et al. (2023). Quantification of circulating HBV RNA expressed from intrahepatic cccDNA in untreated and NUC treated patients with chronic hepatitis B. Gut. 73, 659–667. doi: 10.1136/gutjnl-2023-330644
Ti, Y. N., Han, B., Liu, T. F., Yuan, Y. J., Zhang, L. Y. (2022). [Efficacy and safety of inactivated novel coronavirus vaccine inoculation in patients with chronic hepatitis B]. Zhonghua Gan Zang Bing Za Zhi 30, 1370–1374. doi: 10.3760/cma.j.cn501113-20220825-00437
Tseng, T. C., Hosaka, T., Liu, C. J., Suzuki, F., Hong, C. M., Kumada, H., et al. (2022). Hepatitis B core-related antigen stratifies the risk of liver cancer in HBeAg-negative patients with indeterminate phase. Am. J. Gastroenterol. 117, 748–757. doi: 10.14309/ajg.0000000000001691
Tseng, T. C., Chiang, C., Liu, C. J., Hong, C. M., Su, T. H., Yang, H. C., et al. (2023). Low hepatitis B core-related antigen levels correlate higher spontaneous seroclearance of hepatitis B surface antigen in chronic hepatitis B patients with high hepatitis B surface antigen levels. Gastroenterology 164, 669–679. doi: 10.1053/j.gastro.2023.01.005
Tsukuda, S., Watashi, K. (2020). Hepatitis B virus biology and life cycle. Antiviral Res. 182, 104925. doi: 10.1016/j.antiviral.2020.104925
Tu, T., Zehnder, B., Qu, B., Urban, S. (2021). D e novo synthesis of hepatitis B virus nucleocapsids is dispensable for the maintenance and transcriptional regulation of cccDNA. JHEP Rep. 3, 100195. doi: 10.1016/j.jhepr.2020.100195
Tuttleman, J. S., Pugh, J. C., Summers, J. W. (1986). In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J. Virol. 58, 17–25. doi: 10.1128/jvi.58.1.17-25.1986
Utiger, R. D., Parker, M. L., Daughaday, W. H. (1962). Studies on human growth hormone. I. A radio-immunoassay for human growth hormone. J. Clin. Invest. 41, 254–261. doi: 10.1172/JCI104478
Velan, B., Halmann, M. (1978). Chemiluminescence immunoassay: a new sensitive method for determination of antigens. Immunochemistry 15, 331–333. doi: 10.1016/0161-5890(78)90094-9
von der Waart, M., Snelting, A., Cichy, J., Wolters, G., Schuurs, A. (1978). Enzyme-immunoassay in diagnosis of hepatitis with emphasis on the detection of “e” antigen (HBeAg). J. Med. Virol. 3, 43–49. doi: 10.1002/jmv.1890030111
Wang, L., Tsai, T. H., Huang, C. F., Ho, M. S., Lin, D. B., Ho, Y. C., et al. (2007). Utilizing self-prepared ELISA plates for a cross-population study of different anti-HBe IgG subclass profiles. J. Med. Virol. 79, 495–502. doi: 10.1002/jmv.20852
Wang, W., Liang, H., Zeng, Y., Lin, J., Liu, C., Jiang, L., et al. (2014b). Establishment of a novel two-probe real-time PCR for simultaneously quantification of hepatitis B virus DNA and distinguishing genotype B from non-B genotypes. Clin. Chim. Acta 437, 168–174. doi: 10.1016/j.cca.2014.07.021
Wang, H., Fang, M., Gu, X., Ji, Q., Li, D., Cheng, S. Q., et al. (2014a). The intracellular HBV DNAs as novel and sensitive biomarkers for the clinical diagnosis of occult HBV infection in HBeAg negative hepatocellular carcinoma in China. PloS One 9, e107162. doi: 10.1371/journal.pone.0107162
Wang, X., Zhang, Q., Hao, F., Gao, X., Wu, W., Liang, M., et al. (2014c). Development of a colloidal gold kit for the diagnosis of severe fever with thrombocytopenia syndrome virus infection. BioMed. Res. Int 2014 p, 530621. doi: 10.1155/2014/530621
Wang, J., Shen, T., Huang, X., Kumar, G. R., Chen, X., Zeng, Z., et al. (2016). Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J. Hepatol. 65, 700–710. doi: 10.1016/j.jhep.2016.05.029
Wang, J., Yu, Y., Li, G., Shen, C., Meng, Z., Zheng, J., et al. (2017). Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J. Hepatol. S0168-8278(17), 32261–32264. doi: 10.1016/j.jhep.2017.08.021
Wang, J., Chen, X., Wu, Y., Cao, Z., Wang, L., Huang, H., et al. (2018). Serum HBV RNA is a potential predictor of hepatitis B surface antigen reversion. Hepatol. Commun. 2, 1168–1171. doi: 10.1002/hep4.1249
Wang, Z. L., Zheng, J. R., Yang, R. F., Huang, L. X., Chen, H. S., Feng, B. (2023). An ideal hallmark closest to complete cure of chronic hepatitis B patients: high-sensitivity quantitative HBsAg loss. J. Clin. Transl. Hepatol. 11, 197–206. doi: 10.14218/JCTH.2022.00289
Watanabe, T., Sorensen, E. M., Naito, A., Schott, M., Kim, S., Ahlquist, P. (2007). Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. U.S.A. 104, 10205–10210. doi: 10.1073/pnas.0704000104
Watanabe, T., Hayashi, S., Tanaka, Y. (2022). Drug discovery study aimed at a functional cure for HBV. Viruses 14, 1393. doi: 10.3390/v14071393
Watanabe, T., Inoue, T., Tanaka, Y. (2021). Hepatitis B core-related antigen and new therapies for hepatitis B. Microorganisms 9, 2083. doi: 10.3390/microorganisms9102083
Wieland, S. F., Guidotti, L. G., Chisari, F. V. (2000). Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74, 4165–4173. doi: 10.1128/JVI.74.9.4165-4173.2000
Wikstrom, P., Hagglund, L., Forsman, M. (2000). Structure of a Natural Microbial Community in a Nitroaromatic Contaminated Groundwater Is Altered during Biodegradation of Extrinsic, but Not Intrinsic Substrates. Microb. Ecol. 39, 203–210. doi: 10.1007/s002480000001
Wong, D. K., Seto, W. K., Cheung, K. S., Chong, C. K., Huang, F. Y., Fung, J., et al. (2017). Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver Int. 37, 995–1001. doi: 10.1111/liv.13346
Wong, D. K., Inoue, T., Mak, L. Y., Hui, R. W., Fung, J., Cheung, K. S., et al. (2023). A longitudinal study to detect hepatitis B surface and core-related antigens in chronic hepatitis B patients with hepatitis B surface antigen seroclearance using highly sensitive assays. J. Clin. Virol. 160, 105375. doi: 10.1016/j.jcv.2022.105375
World Health, O. (2019). Hepatitis B vaccines: WHO position paper, July 2017 - Recommendations. Vaccine 37, 223–225. doi: 10.1016/j.vaccine.2017.07.046
Wu, J. F., Chang, M. H. (2015). Natural history of chronic hepatitis B virus infection from infancy to adult life - the mechanism of inflammation triggering and long-term impacts. J. Biomed. Sci. 22, 92. doi: 10.1186/s12929-015-0199-y
Wu, Y., Zhu, Z., Wu, J., Bi, W., Xu, W., Xia, X., et al. (2021). Evolutionary analysis of pre-S/S mutations in HBeAg-negative chronic hepatitis B with HBsAg < 100 IU/ml. Front. Public Health 9, 633792. doi: 10.3389/fpubh.2021.633792
Wu, J., Yu, Y., Dai, Y., Zhang, Y., Cheng, J. (2022). Research progress on the mechanism of persistent low-level HBsAg expression in the serum of patients with chronic HBV infection. J. Immunol. Res 2022 p, 1372705. doi: 10.1155/2022/1372705
Wu, J., He, J., Xu, H. (2024). Global prevalence of occult HBV infection in children and adolescents: A systematic review and meta-analysis. Ann. Hepatol. 29, 101158. doi: 10.1016/j.aohep.2023.101158
Yalow, R. S., Berson, S. A. (1960). Immunoassay of endogenous plasma insulin in man. J. Clin. Invest. 39, 1157–1175. doi: 10.1172/JCI104130
Yan, H., Zhong, G., Xu, G., He, W., Jing, Z., Gao, Z., et al. (2012). Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1, e00049. doi: 10.7554/eLife.00049.027
Yan, M. M., Sun, L. H. (2023). [Natural history of chronic hepatitis B virus infection under low-level viremia]. Zhonghua Gan Zang Bing Za Zhi 31, 322–326. doi: 10.3760/cma.j.cn501113-20220907-00461
Yang, N., Huang, Y., Ding, G., Fan, A. (2019). In situ generation of pRussian blue with potassium ferrocyanide to improve the sensitivity of chemiluminescence immunoassay using magnetic nanoparticles as label. Anal. Chem. 91, 4906–4912. doi: 10.1021/acs.analchem.9b01091
Ye, X., Zhao, Y., Li, R., Li, T., Zheng, X., Xiong, W., et al. (2021). High frequency occult hepatitis B virus infection detected in non-resolved donations suggests the requirement of anti-HBc test in blood donors in southern China. Front. Immunol. 12, 699217. doi: 10.3389/fimmu.2021.699217
Yin, W., Xu, L., Sun, R., Wei, H., Tian, Z. (2012). Interleukin-15 suppresses hepatitis B virus replication via IFN-beta production in a C57BL/6 mouse model. Liver Int. 32, 1306–1314. doi: 10.1111/j.1478-3231.2012.02773.x
Yu, X., Lan, P., Hou, X., Han, Q., Lu, N., Li, T., et al. (2017). HBV inhibits LPS-induced NLRP3 inflammasome activation and IL-1beta production via suppressing the NF-kappaB pathway and ROS production. J. Hepatol. 66, 693–702. doi: 10.1016/j.jhep.2016.12.018
Yu, G., Chen, R., Zheng, S., Liu, Y., Zou, J., Gu, Z., et al. (2022). A standardized assay for the quantitative detection of serum HBV RNA in chronic hepatitis B patients. Emerg. Microbes Infect. 11, 775–785. doi: 10.1080/22221751.2022.2045874
Zhang, H., Tu, T. (2022). Approaches to quantifying hepatitis B virus covalently closed circular DNA. Clin. Mol. Hepatol. 28, 135–149. doi: 10.3350/cmh.2021.0283
Zhao, X. A., Wang, J., Liu, J., Chen, G., Yan, X., Jia, B., et al. (2021). Baseline serum hepatitis B core antibody level predicts HBeAg seroconversion in patients with HBeAg-positive chronic hepatitis B after antiviral treatment. Antiviral Res. 193, 105146. doi: 10.1016/j.antiviral.2021.105146
Zhao, X., Meng, C., Xu, Y., Cao, W. (2022). Correlations between breast milk HBsAg, serum HBsAg, and serum HBV DNA in women with chronic HBV infection. Clin. Lab. 68. doi: 10.7754/Clin.Lab.2021.210641
Keywords: HBV, serum viral products, detection technology, diagnosis, treatment
Citation: Liu Y, Wu D, Zhang K, Ren R, Liu Y, Zhang S, Zhang X, Cheng J, Chen L and Huang J (2024) Detection technology and clinical applications of serum viral products of hepatitis B virus infection. Front. Cell. Infect. Microbiol. 14:1402001. doi: 10.3389/fcimb.2024.1402001
Received: 16 March 2024; Accepted: 12 June 2024;
Published: 05 July 2024.
Edited by:
Haitao Wen, The Ohio State University, United StatesReviewed by:
Mihaela Olivia Dobrica, Institute of Biochemistry of the Romanian Academy, RomaniaCopyright © 2024 Liu, Wu, Zhang, Ren, Liu, Zhang, Zhang, Cheng, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Huang, aHVhbmdqdW5Aenp1LmVkdS5jbg==; Liping Chen, bHBjaGVuMTRAZnVkYW4uZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.