- 1The Second Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China
- 2The Third Clinical Medical College, Guangzhou Medical University, Guangzhou, China
- 3Guangdong Provincial People's Hospital, Guangzhou, China
- 4State Key Laboratory of Traditional Chinese Medicine Syndrome, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China
- 5The Second Affiliated Hospital (Guangdong Provincial Hospital of Chinese Medicine), Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China
- 6Guangdong Provincial Key Laboratory of Research on Emergency in TCM, Guangzhou, China
- 7State Key Laboratory of Dampness Syndrome of Chinese Medicine, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
- 8Guangdong Provincial Key Laboratory of Clinical Research on Traditional Chinese Medicine Syndrome, Guangzhou, China
Background: Gut microbiota is closely related to the occurrence and development of sepsis. However, the causal effects between the gut microbiota and sepsis, and whether circulating inflammatory proteins act as mediators, remain unclear.
Methods: Gut microbiota, circulating inflammatory proteins, and four sepsis-related outcomes were identified from large-scale genome wide association studies (GWAS) summary data. Inverse Variance Weighted (IVW) was the primary statistical method. Additionally, we investigated whether circulating inflammatory proteins play a mediating role in the pathway from gut microbiota to the four sepsis-related outcomes.
Results: There were 14 positive and 15 negative causal effects between genetic liability in the gut microbiota and four sepsis-related outcomes. Additionally, eight positive and four negative causal effects were observed between circulating inflammatory proteins and the four sepsis-related outcomes. Circulating inflammatory proteins do not act as mediators.
Conclusions: Gut microbiota and circulating inflammatory proteins were causally associated with the four sepsis-related outcomes. However, circulating inflammatory proteins did not appear to mediate the pathway from gut microbiota to the four sepsis-related outcomes.
1 Introduction
Sepsis is an unusual systemic response to a common infection, representing a mode of immune system response to injury (Faix, 2013). It can cause symptoms in multiple organ systems, such as the heart, lungs, kidneys, and digestive system (Lelubre and Vincent, 2018). According to epidemiological studies, the prevalence of sepsis and the 28-day mortality rate in hospitals range from 25% to 30% (Cohen et al., 2015a). The hyper-inflammatory response is followed by a period of immunosuppression, during which patients develop multiple organ dysfunction and are prone to nosocomial infections (Faix, 2013). Despite its alarming prevalence and severe consequences, treatment options remain limited and have for many years revolve around antibiotics and supportive therapies (Gotts and Matthay, 2016).
Gut microecology, as a unit of interaction between the gut microbiota and the host, is closely related to human health, with gut microbiota being the most important (Alkasir et al., 2017). Intestinal flora refers to the various microbial communities living in the intestinal cavity that adhere to the intestinal mucosa, including Archaea and Eukaryotes, which are predominantly bacterial. They have evolved together with the host in symbiotic relationships and play important roles in substance metabolism, nutrition, and immune protection (O’Hara and Shanahan, 2006; Dang and Marsland, 2019). Previous studies have shown that gut microbiota is linked to sepsis (Zhang et al., 2023). Several well-established risk factors for sepsis such as aging, immunization, and antibiotic resistance are associated with significant changes in the composition and function of the gut microbiome (Takiishi et al., 2017; Ling et al., 2022; Santacroce et al., 2023). However, the extent to which gut microbes are associated with sepsis remains unclear. A better understanding of the causal effects of the gut microbiota and the potential mediators between them will provide evidence for further mechanistic and clinical studies on the management and treatment of sepsis. Additionally, gut microbiota plays an important role in regulating the circulating inflammatory proteins (Xue et al., 2023).
To the best of our knowledge, inflammation plays an important role in promoting sepsis. Previous studies have revealed that sepsis is a common complication of combat injuries and trauma and is defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection (Liu et al., 2022). During the body’s response to infection, an inflammatory response is essential. We hypothesized that circulating inflammatory proteins might mediate the pathway from the gut microbiota to sepsis.
Although randomized controlled trials can help establish a causal relationship between gut microbiota or circulating inflammatory proteins and sepsis, they are difficult to perform in humans due to objective limitations, such as screening the gut microbiota and elevating circulating inflammatory protein levels (Cheng et al., 2023; Combrink et al., 2023). Therefore, most current research conclusions are based on observations of composition and changes in the intestinal flora in the feces of patients with sepsis. Some observational cohort studies have shown that gut microbiota may be associated with sepsis (Deng et al., 2023; Park et al., 2024). Preclinical studies have demonstrated that the gut microbiota plays a key role in the immune response to systemic inflammation, and disruption of this symbiotic relationship increases the susceptibility to sepsis (Haak et al., 2018).
Although probiotic supplementation has positive effects (Vulevic et al., 2015; Panigrahi et al., 2017; Shimizu et al., 2018), its effectiveness and safety remain controversial (Zhang et al., 2023). Therefore, further research is required to determine the specificity and safety of these probiotic supplements. Mendelian randomization (MR) is an epidemiological method that uses genetic variation as an instrumental variable for exposure to estimate the causal effect of exposure on outcomes and strengthens causal inference. It overcomes the limitations of traditional observational study designs by using Mendelian laws, exploiting the random assignment to genotypes at conception, making genotypes independent of potential confounders, and avoiding reverse causality (Smith et al., 2007). Two-sample MR analysis is an extension of the MR approach that allows summary statistics from genome-wide association studies (GWAS) to be used in MR studies without the direct analysis of individual-level data (Davey Smith and Hemani, 2014). In recent years, two-sample MR studies have gradually been recognized by researchers, allowing data between genetic instrument variables and phenotypes, phenotypes, and diseases to come from two different independent populations, thereby improving the efficiency and statistical power of the study (Burgess et al., 2016; Gupta et al., 2017; Davies et al., 2018).
This study employed a comprehensive MR analysis to investigate the causal association between gut microbiota, circulating inflammatory proteins, and sepsis-related outcomes. Additionally, we examined the potential mediating role of circulating inflammatory proteins in the pathway from gut microbiota to sepsis. Furthermore, reverse causality analysis was conducted to assess the impact of genetic susceptibility to sepsis on gut microbiota and circulating inflammatory proteins.
2 Methods
2.1 Study design
This study followed the STROBE-MR guidelines (Skrivankova et al., 2021) and key principles of the Strengthening Epidemiological Observational Research Reporting (STROBE) guidelines (von Elm et al., 2007). Detailed information is provided in Supplementary Table S1. As shown in Figure 1, this study consisted of three main components: analysis of the causal effects of 207 gut microbiota on four sepsis-related outcomes (step 1A), analysis of the causal effects of 91 circulating inflammatory proteins on four sepsis-related outcomes (step 2A), and mediation analysis of circulating inflammatory proteins in the pathway from gut microbiota to sepsis (step 3). In Mendelian randomization, single-nucleotide poly-morphisms (SNPs) are defined as instrumental variables (IVs). This approach is based on three core assumptions: (1) the IVs are closely associated with the exposure factors, (2) IVs are not associated with confounding factors, and (3) IVs do not affect the outcome directly and can only affect outcomes via exposure (Bowden and Holmes, 2019).
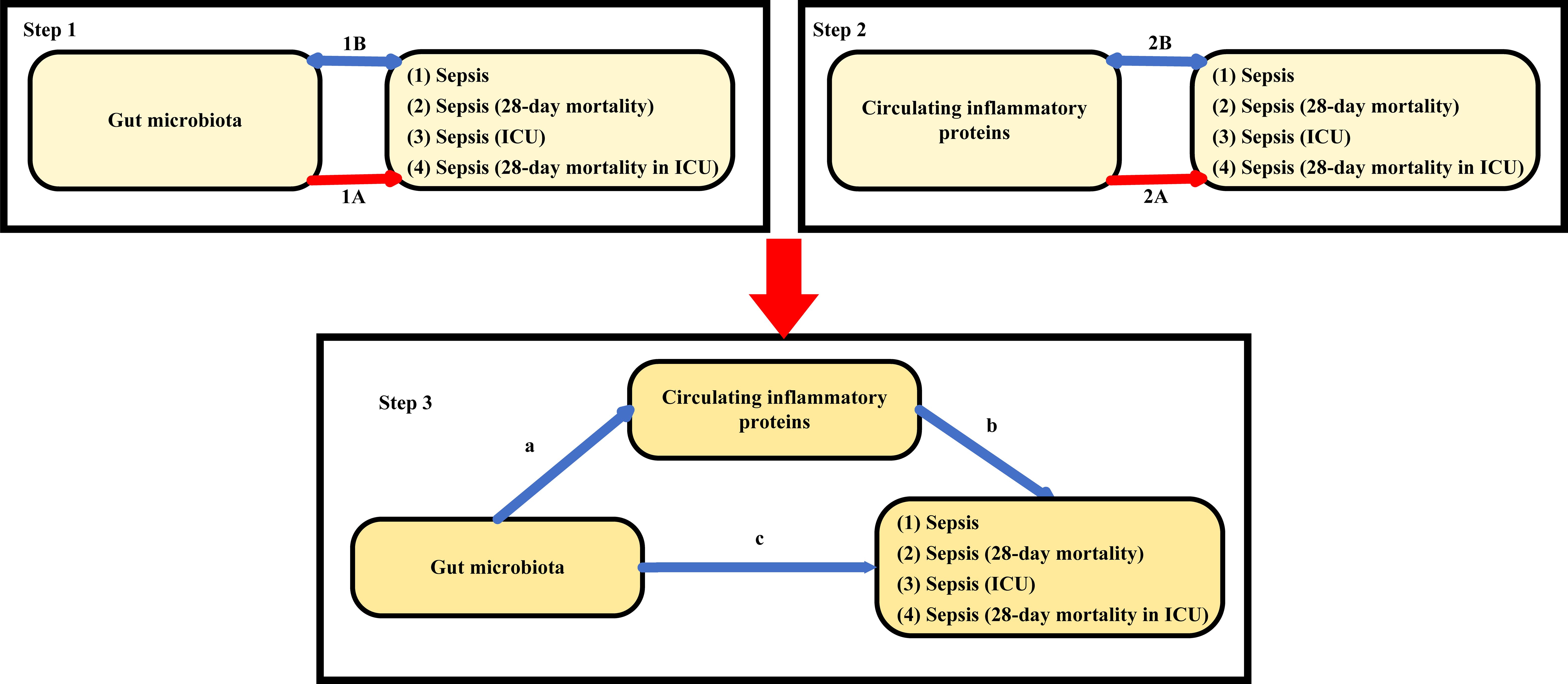
Figure 1 Study overview. Step 1A represents the causal effects of gut microbiota on four sepsis-related outcomes. Step 1B represents the bi−directional causal effects between gut microbiota and four sepsis-related outcomes. Step 2A represents the causal effects of circulating inflammatory proteins on four sepsis-related outcomes. Step 2B represents the bi−directional causal effects between circulating inflammatory proteins and four sepsis-related outcomes. Step 3 represents the mediating analysis of circulating inflammatory proteins in the pathway from the gut microbiota to four sepsis-related outcomes: path c was the total effect of gut microbiota on four sepsis-related outcomes; path b was the causal effect of circulating inflammatory proteins on four sepsis-related outcomes; path a was the causal effect of gut microbiota on circulating inflammatory proteins.
2.2 Data source
The Dutch Microbiome Project (DMP) provides summary data from the GWAS of 7,738 European individuals to obtain species-level data on the gut microbiota and maximize statistical power (Lopera-Maya et al., 2022). The participants in this database were relatively homogeneous, and it is currently the largest database of species-level gut microbiota data. Based on shotgun metagenomic sequencing of the stool samples, we identified 207 taxa (5 phyla, 10 classes, 13 orders, 26 families, 48 genera, and 105 species) in the gut microbiome. Genetic data for circulating inflammatory proteins were obtained from a previous GWAS (14,824 individuals). Circulating inflammatory proteins were measured using an Olink Target-96 Inflammation immunoassay plate, which measures 91 inflammation-related proteins (Zhao et al., 2023).
The main outcomes of this study were four sepsis-associated outcomes: the occurrence of sepsis, sepsis necessitating admission to critical care, mortality within 28 days in the intensive care unit (ICU) after experiencing sepsis, and sepsis 28-day mortality. In the Integrative Epidemiology Unit (IEU) OpenGWAS project database (https://gwas.mrcieu.ac.uk/) (Hemani et al., 2018), these outcomes were identified by the IDs ieu-b-4980, ieu-b-4982, ieu-b-4981 and ieu-b-5086, respectively. The Global Burden of Disease (GBD) study codes were used to define sepsis based on the International Classification of Diseases (ICD)-10 (Rudd et al., 2020).
This study was a secondary analysis of publicly available GWAS summary statistics. Ethical approval was granted to each original GWAS. Moreover, no individual-level data were used in this study. Therefore, no additional ethical review board approval was required.
2.3 Instrumental variables selection
First, we selected the SNPs with significant associations for gut microbiota (P < 1×10–5) (Shi et al., 2024). SNPs with significant associations with circulating inflammatory proteins were selected (P < 5×10–8) (Yan et al., 2024). We excluded the SNPs with linkage disequilibrium (LD) in the analysis. The LD of selected SNPs strongly related to the gut microbiota and circulating inflammatory proteins should meet the conditions of r2 < 0.001 and distance > 10,000 kb (Myers et al., 2020). After matching the outcomes, we removed the palindromic SNPs. SNPs significantly associated with the gut microbiota and circulating inflammatory proteins are listed in Supplementary Tables S2 and S3.
To assess the strength of the relationship between the identified independent variables (IVs) and exposure, we computed the explained variance (R2) and F-statistic parameters. Steiger’s Test was used to detect and avoid reverse causality. The Steiger Test is a new methodology that enhances the robustness of the IV extraction (Hemani et al., 2017). SNPs with F-statistic parameters >10 were considered strong instruments (Burgess et al., 2017). R2 = 2 × EAF × (1-EAF) × β2, F = R2 × (N-2)/(1-R2) (Papadimitriou et al., 2020).
2.4 Primary analysis
To assess the causal impact of the gut microbiota and circulating inflammatory proteins on the four sepsis-related outcomes, we conducted a two-sample Mendelian randomization (MR) analysis for each outcome (step 1A and 2A in Figure 1). The Inverse Variance Weighted (IVW) approach was employed as the primary analytical method, while the Wald ratio test was used for features consisting solely of one independent variable (IV) (Burgess et al., 2013). The outcomes of the MR analysis were presented as odds ratios (ORs) accompanied by their respective 95% confidence intervals (CI). The findings were considered statistically significant when the P-value of the IVW method was below 0.05, and the IVW and MR-Egger estimates exhibited consistent directional effects (Ji et al., 2024).
2.5 Mediation analysis
Using a two-sample analysis (step 1A and 2A in Figure 1), the gut microbiota and circulating inflammatory proteins with significant causal effects on the four sepsis-related outcomes were included in the mediation analysis. As shown in Figure 1, we examined the possible causal impact of the gut microbiota on circulating inflammatory proteins (step 3, path a). If this is the case, we will perform multiple MR analyses to explore whether circulating inflammatory proteins are the mediating factors in the pathway from the gut microbiota to the four sepsis-related outcomes.
2.6 Bi−directional causality analysis
To evaluate bi-directional causation effects between gut microbiota, circulating inflammatory proteins, and four sepsis-related outcomes, we used four sepsis-related outcomes as “exposure” and gut microbiota or circulating inflammatory proteins associated with four sepsis-related outcomes as “outcome” (step 1B and step 2B in Figure 1). We selected SNPs that were significantly associated with four sepsis-related outcomes (P < 5×10–8) as IVs (Chen et al., 2024).
2.7 Sensitivity analysis
We performed Cochran’s Q test to evaluate the heterogeneity of each SNP (Cohen et al., 2015b) and generated scatter plots of SNP exposure and outcome associations to visualize the MR results. Leave-one-out analysis was performed to evaluate whether each SNP affected the results (Burgess and Thompson, 2017). In addition, we use MR-PRESSO and MR-Egger regressions to test for the potential horizontal pleiotropy effect. MR-PRESSO was used to detect significant outliers and correct for horizontal plural effects by removing outliers (Verbanck et al., 2018). All analyses were performed using R (v4.3.0) statistical software. MR analysis was performed using the “TwoSampleMR” package. The “MR_PRESSO” package was used for multiplicity tests (Ong and MacGregor, 2019).
3 Results
3.1 Causal effects of gut microbiota and circulating inflammatory proteins on four sepsis-related outcomes
3.1.1 Sepsis
As shown in Figure 2, MR analysis suggested that the genetic prediction of four gut microbiota (species Clostridium asparagiforme, species Bacteroidales bacterium ph8, species Ruminococcus lactaris, and genus Bacteroidale) was associated with an increased risk of sepsis. Genetic prediction of five gut microbiota (genus Adlercreutzia, species Adlercreutzia equolifaciens, genus Pseudoflavonifractor, species Ruminococcus callidus, and species Escherichia unclassified) was associated with a decreased risk of sepsis (Supplementary Table S4).
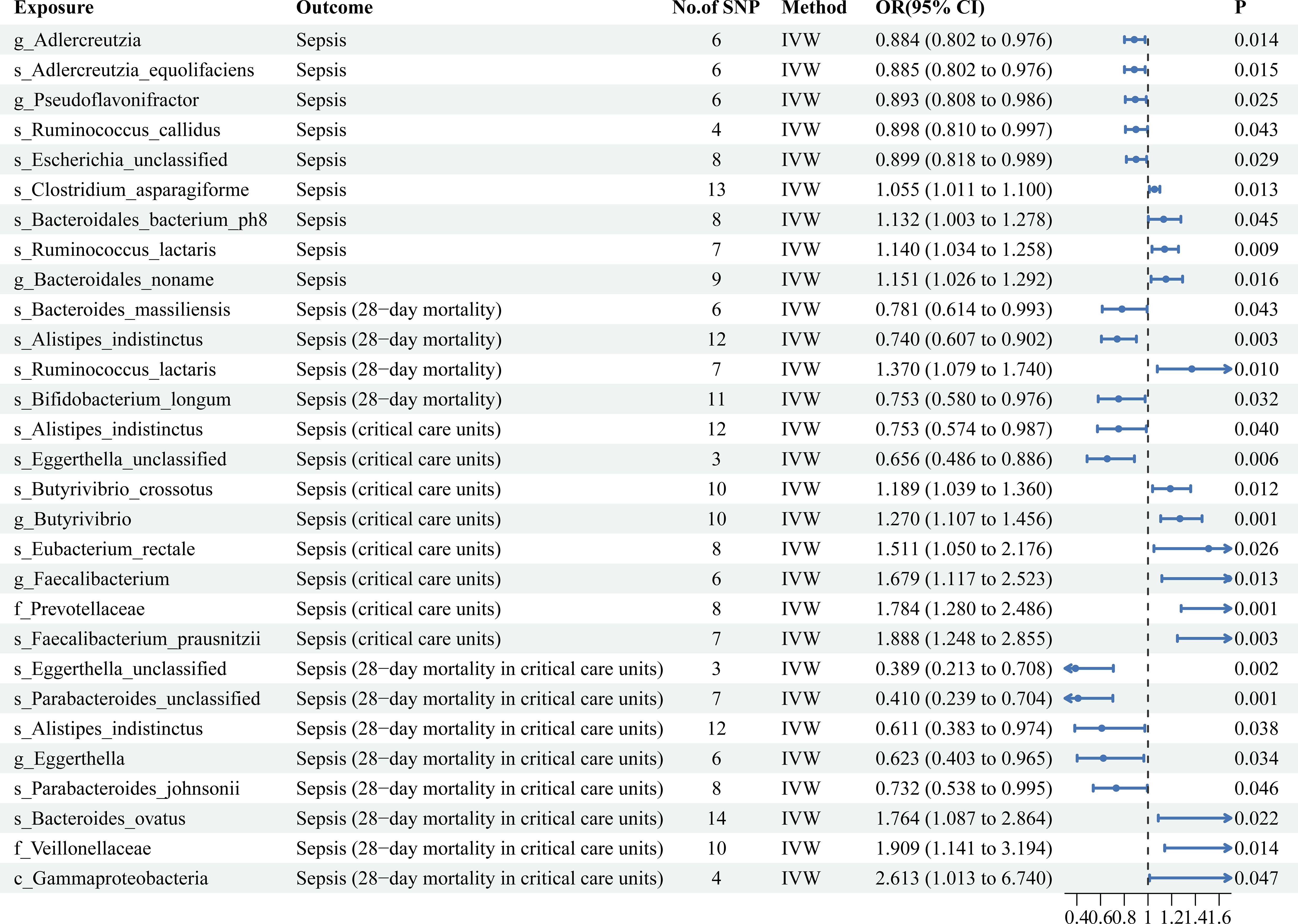
Figure 2 Mendelian randomization results of causal effects between gut microbiotas and four sepsis-related outcomes.
As shown in Figure 3, TNF-related apoptosis-inducing ligand levels (TRAIL) (OR = 1.094, 95%CI = 1.012 ~ 1.183, P = 0.025) and vascular endothelial growth factor A levels (VEGF-A) (OR = 1.182, 95%CI = 1.016 ~ 1.375, P = 0.031) were associated with an increased risk of sepsis. Beta-nerve growth factor levels (β-NGF) (OR = 0.769, 95%CI = 0.599 ~ 0.987, P = 0.039) significantly decreased the incidence of sepsis (Supplementary Table S4).
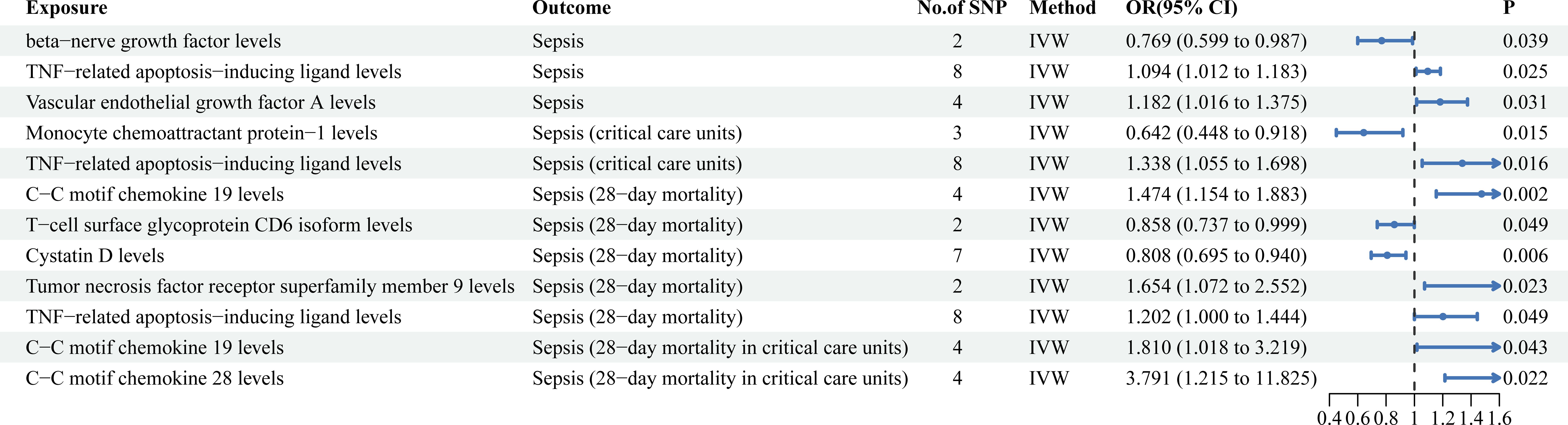
Figure 3 Mendelian randomization results of causal effects between circulating inflammatory proteins and four sepsis-related outcomes.
3.1.2 Sepsis (28-day mortality)
As shown in Figure 2, the MR analysis suggested that the genetic prediction of one gut microbiota (species Ruminococcus lactaris) was associated with an increased risk of sepsis (28-day mortality). Genetic prediction of three gut microbiota (species Bacteroides massiliensis, species Alistipes indistinctus, and species Bifidobacterium longum) was associated with a decreased risk of sepsis (28-day mortality) (Supplementary Table S4).
As shown in Figure 3, C-C motif chemokine 19 levels (CCL19) (OR = 1.474, 95% CI = 1.154 ~ 1.883, P = 0.002), Tumor necrosis factor receptor superfamily member 9 levels (TNFRSF9) (OR = 1.654, 95%CI = 1.071 ~ 2.552, P = 0.023) and TRAIL (OR = 1.202, 95% CI = 1.000 ~ 1.444, P = 0.049) were associated with an increased risk of sepsis (28-day mortality). T-cell surface glycoprotein CD6 isoform levels (CD6) (OR = 0.858, 95% CI = 0.737 ~ 0.999, P = 0.049) and Cystatin D levels (OR = 0.808, 95% CI = 0.695 ~ 0.940, P = 0.006) were associated with a decreased risk of sepsis (28-day mortality) (Supplementary Table S4).
3.1.3 Sepsis (critical care units)
As shown in Figure 2, MR analysis suggested that the genetic prediction of six gut microbiota (species Butyrivibrio crossotus, family Prevotellaceae, species Eubacterium rectale, species Faecalibacterium prausnitzii, genus Butyrivibrio, and genus Faecalibacterium) was associated with an increased risk of sepsis (critical care units). Genetic prediction of two gut microbiota (species Alistipes indistinctus and species Eggerthella unclassified) was associated with a decreased risk of sepsis (critical care units) (Supplementary Table S4).
As shown in Figure 3, TRAIL (OR = 1.338, 95% CI = 1. 055 ~ 1.698, P = 0.016) were associated with an increased risk of sepsis (critical care units). Monocyte chemoattractant protein-1 levels (MCP-1) (OR = 0.642, 95% CI = 0. 448 ~ 0.918, P = 0.015) were associated with a decreased risk of sepsis (critical care units) (Supplementary Table S4).
3.1.4 Sepsis (28-day mortality in critical care units)
As shown in Figure 2, MR analysis suggested that the genetic prediction of the three gut microbiota (species Bacteroides ovatus, family Veillonellaceae, and class Gammaproteobacteria) was associated with an increased risk of sepsis (28-day mortality in critical care units). Moreover, all three factors significantly increased the risk of sepsis (28-day mortality in critical care units). Genetic prediction of five gut microbiota (species Eggerthella unclassified, species Parabacteroides unclassified, species Alistipes indistinctus, genus Eggerthella, and species Parabacteroides johnsonii) was associated with a decreased risk of sepsis (28-day mortality in critical care units). Moreover, all five significantly reduce the risk of sepsis development (28-day mortality in critical care units) (Supplementary Table S4).
As shown in Figure 3, CCL19 (OR = 1.810, 95% CI = 1.018 ~ 3.219, P = 0.043) and C-C motif chemokine 28 levels (CCL28) (OR = 3.791, 95% CI = 1.215 ~ 11.825, P = 0.022) significantly increased risk of sepsis (28-day mortality in critical care units) (Supplementary Table S4).
By comparing the results horizontally and vertically, interesting and surprising findings were obtained. In the gut microbiota, species Ruminococcus lactaris not only increased the risk of sepsis incidence but was also strongly associated with sepsis 28-day mortality. Although species Alistipes inditinctus had no significant effect on the incidence of sepsis, it had significant causal effects on sepsis (28-day mortality), sepsis (critical care units), and sepsis (28-day mortality in critical care units). Additionally, species Eggerthella had significant causal effects on the risk reduction of both sepsis (critical care units) and sepsis (28-day mortality in critical care units). Therefore, regulating species Alistipes indistintus and Eggerthella may be an important measure to prevent and treat critical sepsis. Among circulating inflammatory proteins, CCL19 not only increased the risk of sepsis (28-day mortality), but was also strongly associated with sepsis (28-day mortality in critical care units). TRAIL had significant effects on multiple sepsis-related outcomes.
3.2 Sensitivity analyses
Based on the MR-Egger regression intercept method, the outcomes were not influenced by genetic pleiotropy, and further analysis using MR-PRESSO demonstrated the absence of horizontal pleiotropy in the MR study (Supplementary Tables S5, S6). Cochran’s Q test revealed no significant heterogeneity (Supplementary Table S7). The findings from the “leave-one-out” analysis provide evidence supporting the reliability of MR analysis (Supplementary Figures S1-S5).
3.3 Bi−directional causal effects of four sepsis-related outcomes on gut microbiota and circulating inflammatory proteins
A P-value threshold of less than 5×10-8 was employed as the criterion to identify SNPs that exhibit significant associations with the three types of sepsis. However, no statistically significant SNPs were identified, indicating that the presence of a reverse causal effect was not substantial.
3.4 Mediation analysis
This study elucidated the causal relationships between gut microbiota, circulating inflammatory proteins, and four sepsis-related outcomes. Circulating inflammatory proteins appear to play a mediating role in the pathway from the gut microbiota to the four sepsis-related outcomes. One of the prerequisites for establishing a mediating effect is a significant association between the gut microbiota and circulating inflammatory proteins. Class Gammaproteobacteria (OR = 1.166, 95% CI = 1.010 ~ 1.345, P = 0.035) significantly increased beta-nerve growth factor levels. Species Faecalibacterium prausnitzii (OR = 0.861, 95% CI = 0.765 ~ 0.968, P = 0.012) significantly decreased C-C motif chemokine 19 levels (step 3a in Figure 1; Supplementary Table S8). However, the mediation analysis results showed that beta-nerve growth factor and C-C motif chemokine 19 could not mediate the influence of gut microbiota on sepsis-related outcomes, indicating that circulating inflammatory proteins did not act as mediators in the pathway from gut microbiota and four sepsis-related outcomes.
4 Discussion
In this study, we evaluated the bidirectional association between the gut microbiota and sepsis and investigated whether circulating inflammatory proteins act as mediators. We found 14 positive and 15 negative causal effects between genetic liability in the gut microbiota and four sepsis-related outcomes. Additionally, eight positive and four negative causal effects were observed between circulating inflammatory proteins and four sepsis-related outcomes. Circulating inflammatory proteins do not act as mediators. Notably, despite these findings, our reverse transcription analysis did not indicate any influence of sepsis on the gut microbiota or circulating inflammatory proteins.
There is a close relationship between the gut microbiota and sepsis. First, before the onset of sepsis, alterations in the gut microbiome increase the susceptibility to sepsis through multiple mechanisms, including (1) allowing the expansion of pathogenic gut bacteria, (2) providing a robust proinflammatory response to the immune system, and (3) reducing the production of beneficial microbial products, such as short-chain fatty acids (Adelman et al., 2020). Pathogenic bacteria present in the gut of healthy hosts may fail to proliferate and cause diseases in the presence of protective commensals (Chow et al., 2011). However, when protective bacterial taxa are lost, pathogens can proliferate and cause diseases (Kim et al., 2019; Woodworth et al., 2019). Previous studies have demonstrated the role of the gut microbiome in the initiation of the immune system in response to sepsis. After an episode of sepsis, changes in the gut microbiome can affect the inflammatory responses. Clinical studies on sepsis have shown a link between alterations in the gut microbiome, characterised by an increase in pathogenic bacteria and a robust immune response (Li et al., 2015; Cernada et al., 2016; Singer et al., 2018). In this study, we identified several gut microbiota that showed protective effects against sepsis, including species Alistipes indistintus and species Eggerthella. However, some taxa show potentially harmful effects, such as species Eubacterium rectale and genus Bacteroidales. Additionally, species Ruminococcus lactaris not only increased the risk of sepsis but was also strongly associated with sepsis mortality at 28 days.
Alistipes is a Gram-negative bacterium belonging to the phylum Bacteroides (Parker et al., 2020). Recent studies suggest that Alistipes may have protective effects against certain diseases, including colitis (Dziarski et al., 2016), liver fibrosis (Rau et al., 2018) and cardiovascular diseases (Kim et al., 2018; Zuo et al., 2019). The co-organisms of the intestinal microbiome produce short-chain fatty acids (SCFAs), which have the immune function of regulating the intestinal microenvironment. SCFAs play important roles in epithelial cell function. Alistipes is an acetate producer, and since previous studies have shown that short-chain fatty acids have an anti-inflammatory mechanism, it can be shown that Alistipes reduction promotes short-chain fatty acid reduction (Parker et al., 2020). Eggerthella is a bacterium that colonizes the human intestinal tract, female genital tract, oral cavity, and prostate (Lau et al., 2004). Recently, a growing body of evidence has collectively emphasized the association of Eggerthella with various diseases, such as asthma (Wang et al., 2018), multiple sclerosis (Cekanaviciute et al., 2017), systemic lupus erythematosus (Xiang et al., 2021), rheumatoid arthritis (Chen et al., 2016), and sepsis (Priputnevich et al., 2021). Eggerthella is likely to be implicated in sepsis; however, the exact role of Eggerthella in sepsis needs to be further explored. Eubacterium plays important roles in various processes, including the conversion of bile acids and cholesterol, metabolizing oxalate, producing anti-inflammatory molecules, reducing allergic airway inflammation, regulating insulin secretion, and controlling lipid metabolism (Mukherjee et al., 2020; Kamel et al., 2021). Ruminococcus plays an important role in the digestive process, especially in the decomposition and fermentation of plant fibers, such as cellulose and hemicellulose (Crost et al., 2023). Ruminococcus has been associated with various gastrointestinal, immune-related, and neurological disorders (Crost et al., 2023). Some studies have found that the number and activity of Ruminococcus in the guts of patients with these diseases may change, thereby affecting gut health and inflammation levels (Hall et al., 2017). Bacteroides are present in the human gut and have a symbiotic relationship with humans. They help break down food and produce the nutrients and energy required by the body. Specific taxa and their relative abundance in this phylum are associated with various diseases, including metabolic syndrome (Turnbaugh et al., 2009; Pedersen et al., 2016; Vieira-Silva et al., 2020), viral infections (Stefan et al., 2020). These conditions can cause or exacerbate sepsis. An experimental study indicated that in sepsis models, the dominant bacterial population in the lungs was Bacteroides (Dickson et al., 2016). And the relative abundance of Bacteroides in the lung microbiome was significantly correlated with serum TNF-α concentrations, a key mediator of the septic stress response that predicts sepsis patient mortality (Osuchowski et al., 2006). In some cases, Bacteroides may cause sepsis (Arnold et al., 2020). Perforations or ruptures of the intestine, dysbiosis of the intestinal microbiota, and impaired immune function allow intestinal bacteria such as Bacteroides to enter the abdominal cavity or bloodstream, causing infection and sepsis. More research is needed to fully understand the connection between gut microbiota and diseases, especially infection-related diseases such as sepsis.
This study aimed to determine whether the gut microbiota has a positive or negative impact on sepsis outcomes. However, the specific mechanism by which gut microbiota contribute to or worsen sepsis remains unclear. We hypothesized that inflammatory proteins in the bloodstream may play a role in the relationship between the gut microbiota and sepsis.
According to the MR analysis, we found that C-C motif chemokine 19 (CCL19), C-C motif chemokine 28 (CCL28), TNF-related apoptosis-inducing ligand (TRAIL), and vascular endothelial growth factor A (VEGF-A) levels were associated with an increased risk of sepsis-related outcomes. CCL19 and CCL28 are chemokines found in immune cells that play important immune-regulatory roles in the human body. Studies have shown a significant increase in plasma CCL19 levels in patients with sepsis, which may be related to sepsis severity (Tuerxun et al., 2023). However, research on the relationship between CCL19 and CCL28 and sepsis is currently limited. We look forward to a more comprehensive and in-depth research in the future. TRAIL, a biomarker of circulating cell death, is a potent inducer of apoptosis. Previous studies have shown that TRAIL has therapeutic effects in sepsis, peritonitis, and pulmonary inflammation (McGrath et al., 2011). Lower versus higher levels of TRAIL were associated with increased organ dysfunction and septic shock (Schenck et al., 2019). TRAIL is known to exert pluripotent effects in sepsis, leading to an improved survival rate in the early stages of abdominal sepsis and a decreased survival rate in the late phase of hypoinflammation (Boomer et al., 2014; Berg et al., 2023). It is worth noting that in recent years, animal experiments have found that TRAIL mediates immune suppression in sepsis, and that anti-TRAIL antibodies have a protective effect on sepsis mice (Gurung et al., 2011). VEGFA belongs to the ligand-tyrosine kinase receptor system, is widely expressed in the vascular system, and participates in the development of normal vascular barriers. VEGFA belongs to the ligand-tyrosine kinase receptor system, is widely expressed in the vascular system, and participates in the development of normal vascular barriers. During sepsis, the expression of VEGF-A increases, leading to damage to the vascular endothelial barrier, tissue inflammation, vascular leakage, insufficient blood volume, tissue oedema, and changes in microcirculation flow, ultimately leading to MODS (Hauschildt et al., 2020).
The development and management of sepsis are multifactorial, and this study specifically examined microecological differences to understand this condition. It is essential to acknowledge the complexity of sepsis and the need for a comprehensive evaluation of its various factors to gain a thorough understanding. This study is the first to reveal a potential causal relationship between the species levels of gut microbiota and sepsis-related outcomes, with profound implications for public health and the management of individual patients with sepsis. These findings may help diagnose sepsis more accurately. Based on the presence of multiple gut microbiota, patients with sepsis can be screened and assessed for the risk of taking preventive measures to reduce the incidence of sepsis (Peng et al., 2024). In addition, this study contributes to a deeper understanding of the pathogenesis and pathological processes of sepsis, providing a scientific basis for formulating targeted public health policies and prevention and control measures. Alleviates symptoms of sepsis or facilitates recovery by adjusting the gut microbiota balance. Interventions targeting the gut microbiota are needed for more effective treatment of sepsis (Cai et al., 2022).
This study represents a comprehensive initial MR analysis that examined the causal association between the gut microbiome, circulating inflammatory proteins, and sepsis-related outcomes. This study had several strengths. First, we highlighted certain species that showed more significant associations with sepsis than with other microbial classes. Although these associations were not statistically significant after adjusting for multiple testing, they nevertheless constitute important preliminary observations and may indicate underlying biological phenomena. In addition, this study used multiple sensitivity analyses to enhance the reliability of the findings. The consistency of the MR-Egger method with the IVW method demonstrates the robustness of the results. Despite the wide confidence intervals in some results, the overall patterns of the associations remained consistent. Furthermore, we employed the MR-PRESSO technique to identify and exclude potential outliers that may have biased our findings, thereby improving reliability. Finally, given that both the study population and the population surveyed were of European origin, the possibility of bias due to population stratification was reduced.
However, our study has several limitations. First, the limited European population data on gut microbiota and circulating inflammatory proteins may have biased our results. Second, the number of loci associated with circulating inflammatory proteins was relatively small compared to those associated with sepsis and gut microbiota. Third, our Mendelian randomization study did not have access to individual-level data, such as sex, age, and disease severity, which limited the depth of our analysis.
5 Conclusion
In summary, our bidirectional Mendelian randomization study clearly demonstrated 29 causal effects between genetic liability in the gut microbiota and sepsis-related outcomes, whereas the reverse causality hypothesis did not hold. Notably, our findings indicated that circulating inflammatory proteins do not act as mediators. To gain a more nuanced understanding of the observed association between the gut microbiota and sepsis, future studies should focus on potential mechanistic pathways while also attempting to adjust for potential confounders, such as diet, lifestyle, and medications, given that these factors may have a greater impact on sepsis. Our work is an important step forward in explaining the relationship between gut microbiota and sepsis, but more microbiological and clinical studies are needed to validate and expand our findings. We hope that this research will contribute to the diagnosis, treatment, and development of drugs for sepsis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors. The summary data of genome-wide association studies (GWAS) used in this study can be obtained from the Integrative Epidemiology Unit (IEU) OpenGWAS project database (https://gwas.mrcieu.ac.uk/) and the GWAS catalog (https://www.ebi.ac.uk/gwas/).
Author contributions
ZL: Software, Writing – original draft, Writing – review & editing, Formal analysis, Funding acquisition, Supervision. LL: Data curation, Formal analysis, Investigation, Writing – original draft. YK: Validation, Formal analysis, Methodology, Writing – review & editing. JF: Data curation, Investigation, Writing – original draft, Validation. XR: Data curation, Writing – review & editing, Methodology. YW: Data curation, Writing – review & editing, Validation. XC: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. SW: Writing – review & editing, Conceptualization, Software, Validation. RY: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing. JL: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing, Validation. YtL: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing. YeL: Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – original draft. JC: Funding acquisition, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (82374392), Traditional Chinese Medicine Innovation Team Project of China Administration of Traditional Chinese Medicine (ZYYCXTD-D-202203), National Administration of Traditional Chinese Medicine project (2023ZYLCYJ02-21), Key Technologies Research and Development Program of Guangdong Province (2023B1111020003), Science and Technology Planning Project of Guangdong Province (No. 2023B1212060062), Basic and Applied Basic Research of Guangzhou City-University Joint Funding Project (202201020382), Open project of Guangdong Provincial Key Laboratory of Research on Emergency in TCM (KF2023JZ06), Research Fund for Zhaoyang Talents of Guangdong Provincial Hospital of Chinese Medicine (ZY2022KY10, ZY2022YL04).
Acknowledgments
The authors thank the investigators of the original studies for sharing the GWAS summary statistics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1398756/full#supplementary-material
References
Adelman, M. W., Woodworth, M. H., Langelier, C., Busch, L. M., Kempker, J. A., Kraft, C. S., et al. (2020). The gut microbiome’s role in the development, maintenance, and outcomes of sepsis. Crit. Care. 24, 278. doi: 10.1186/s13054-020-02989-1
Alkasir, R., Li, J., Li, X., Jin, M., Zhu, B. (2017). Human gut microbiota: the links with dementia development. Protein Cell. 8, 90–102. doi: 10.1007/s13238-016-0338-6
Arnold, Z., Wiggins, A., Santos, R. L., Breighner, C. (2020). Bacteroides bundle of joy: sepsis from a degenerating/necrotic fibroid. BMJ Case Rep. 13, e237811. doi: 10.1136/bcr-2020-237811
Berg, A. K., Hahn, E. M., Speichinger-Hillenberg, F., Grube, A. S., Hering, N. A., Stoyanova, A. K., et al. (2023). The impact of TRAIL on the immunological milieu during the early stage of abdominal sepsis. Cancers. 15, 1773. doi: 10.3390/cancers15061773
Boomer, J. S., Green, J. M., Hotchkiss, R. S. (2014). The changing immune system in sepsis: is individualized immuno-modulatory therapy the answer? Virulence. 5, 45–56. doi: 10.4161/viru.26516
Bowden, J., Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: A review. Res. Synthesis Methods. 10, 486–496. doi: 10.1002/jrsm.1346
Burgess, S., Butterworth, A., Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. doi: 10.1002/gepi.21758
Burgess, S., Davies, N. M., Thompson, S. G. (2016). Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 40, 597–608. doi: 10.1002/gepi.21998
Burgess, S., Small, D. S., Thompson, S. G. (2017). A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 26, 2333–2355. doi: 10.1177/0962280215597579
Burgess, S., Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389. doi: 10.1007/s10654-017-0255-x
Cai, Y., Zhang, Y., Wang, W., Geng, J. (2022). Prospect of research hotspots in prevention and treatment of diseases based on intestinal microbiome. Front. Immunol. 13. doi: 10.3389/fimmu.2022.971457
Cekanaviciute, E., Yoo, B. B., Runia, T. F., Debelius, J. W., Singh, S., Nelson, C. A., et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114, 10713–10718. doi: 10.1073/pnas.1711235114
Cernada, M., Bäuerl, C., Serna, E., Collado, M. C., Martínez, G. P., Vento, M. (2016). Sepsis in preterm infants causes alterations in mucosal gene expression and microbiota profiles compared to non-septic twins. Sci. Rep. 6, 25497. doi: 10.1038/srep25497
Chen, J., Wright, K., Davis, J. M., Jeraldo, P., Marietta, E. V., Murray, J., et al. (2016). An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 8, 43. doi: 10.1186/s13073-016-0299-7
Chen, Y., Xie, Y., Ci, H., Cheng, Z., Kuang, Y., Li, S., et al. (2024). Plasma metabolites and risk of seven cancers: a two-sample Mendelian randomization study among European descendants. BMC Med. 22, 90. doi: 10.1186/s12916-024-03272-8
Cheng, Z. X., Wu, Y. X., Jie, Z. J., Li, X. J., Zhang, J. (2023). Genetic evidence on the causality between gut microbiota and various asthma phenotypes: a two-sample Mendelian randomization study. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1270067
Chow, J., Tang, H., Mazmanian, S. K. (2011). Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 23, 473–480. doi: 10.1016/j.coi.2011.07.010
Cohen, J., Vincent, J. L., Adhikari, N. K., MaChado, F. R., Angus, D. C., Calandra, T., et al. (2015a). Sepsis: a roadmap for future research. Lancet Infect. Dis. 15, 581–614. doi: 10.1016/S1473-3099(15)70112-X
Cohen, J. F., Chalumeau, M., Cohen, R., Korevaar, D. A., Khoshnood, B., Bossuyt, P. M. (2015b). Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 68, 299–306. doi: 10.1016/j.jclinepi.2014.09.005
Combrink, L., Humphreys, I. R., Washburn, Q., Arnold, H. K., Stagaman, K., Kasschau, K. D., et al. (2023). Best practice for wildlife gut microbiome research: A comprehensive review of methodology for 16S rRNA gene investigations. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1092216
Crost, E. H., Coletto, E., Bell, A., Juge, N. (2023). Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol. Rev. 47, fuad014. doi: 10.1093/femsre/fuad014
Dang, A. T., Marsland, B. J. (2019). Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6
Davey Smith, G., Hemani, G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–R98. doi: 10.1093/hmg/ddu328
Davies, N. M., Holmes, M. V., Davey Smith, G. (2018). Reading Mendelian randomization studies: a guide, glossary, and checklist for clinicians. Bmj. 362, k601. doi: 10.1136/bmj.k601
Deng, F., Chen, Y., Sun, Q. S., Lin, Z. B., Min, Y., Zhao, B. C., et al. (2023). Gut microbiota dysbiosis is associated with sepsis-induced cardiomyopathy in patients: A case-control study. J. Med. Virol. 95, e28267. doi: 10.1002/jmv.28267
Dickson, R. P., Singer, B. H., Newstead, M. W., Falkowski, N. R., Erb-Downward, J. R., Standiford, T. J., et al. (2016). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1, 16113. doi: 10.1038/nmicrobiol.2016.113
Dziarski, R., Park, S. Y., Kashyap, D. R., Dowd, S. E., Gupta, D. (2016). Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PloS One. 11, e0146162. doi: 10.1371/journal.pone.0146162
Faix, J. D. (2013). Biomarkers of sepsis. Crit. Rev. Clin. Lab. Sci. 50, 23–36. doi: 10.3109/10408363.2013.764490
Gotts, J. E., Matthay, M. A. (2016). Sepsis: pathophysiology and clinical management. BMJ. 353, i1585. doi: 10.1136/bmj.i1585
Gupta, V., Walia, G. K., Sachdeva, M. P. (2017). ‘Mendelian randomization’: an approach for exploring causal relations in epidemiology. Public Health. 145, 113–119. doi: 10.1016/j.puhe.2016.12.033
Gurung, P., Rai, D., Condotta, S. A., Babcock, J. C., Badovinac, V. P., Griffith, T. S. (2011). Immune unresponsiveness to secondary heterologous bacterial infection after sepsis induction is TRAIL dependent. J. Immunol. 187, 2148–2154. doi: 10.4049/jimmunol.1101180
Haak, B. W., Prescott, H. C., Wiersinga, W. J. (2018). Therapeutic potential of the gut microbiota in the prevention and treatment of sepsis. Front. Immunol. 9. doi: 10.3389/fimmu.2018.02042
Hall, A. B., Yassour, M., Sauk, J., Garner, A., Jiang, X., Arthur, T., et al. (2017). A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103. doi: 10.1186/s13073-017-0490-5
Hauschildt, J., Schrimpf, C., Thamm, K., Retzlaff, J., Idowu, T. O., von Kaisenberg, C., et al. (2020). Dual pharmacological inhibition of angiopoietin-2 and VEGF-A in murine experimental sepsis. J. Vasc. Res. 57, 34–45. doi: 10.1159/000503787
Hemani, G., Tilling, K., Davey Smith, G. (2017). Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PloS Genet. 13, e1007081. doi: 10.1371/journal.pgen.1007081
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. Elife. 7, e34408. doi: 10.7554/eLife.34408
Ji, D., Chen, W. Z., Zhang, L., Zhang, Z. H., Chen, L. J. (2024). Gut microbiota, circulating cytokines and dementia: a Mendelian randomization study. J. Neuroinflamm. 21, 2. doi: 10.1186/s12974-023-02999-0
Kamel, O., Van Noten, H., Argudín, M. A., Martiny, D. (2021). Butyricimonas faecihominis and Eubacterium callanderi mixed bloodstream infection after appendicular peritonitis. Anaerobe. 71, 102419. doi: 10.1016/j.anaerobe.2021.102419
Kim, S. G., Becattini, S., Moody, T. U., Shliaha, P. V., Littmann, E. R., Seok, R., et al. (2019). Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature. 572, 665–669. doi: 10.1038/s41586-019-1501-z
Kim, S., Goel, R., Kumar, A., Qi, Y., Lobaton, G., Hosaka, K., et al. (2018). Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin. Sci. 132, 701–718. doi: 10.1042/CS20180087
Lau, S. K., Woo, P. C., Woo, G. K., Fung, A. M., Wong, M. K., Chan, K. M., et al. (2004). Eggerthella hongkongensis sp. nov. and eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteremia. Diagn. Microbiol. Infect. Dis. 49, 255–263. doi: 10.1016/j.diagmicrobio.2004.04.012
Lelubre, C., Vincent, J. L. (2018). Mechanisms and treatment of organ failure in sepsis. Nat. Rev. Nephrol. 14, 417–427. doi: 10.1038/s41581-018-0005-7
Li, Q., Wang, C., Tang, C., He, Q., Zhao, X., Li, N., et al. (2015). Successful treatment of severe sepsis and diarrhea after vagotomy utilizing fecal microbiota transplantation: a case report. Crit. Care. 19, 37. doi: 10.1186/s13054-015-0738-7
Ling, Z., Liu, X., Cheng, Y., Yan, X., Wu, S. (2022). Gut microbiota and aging. Crit. Rev. Food Sci. Nutr. 62, 3509–3534. doi: 10.1080/10408398.2020.1867054
Liu, D., Huang, S. Y., Sun, J. H., Zhang, H. C., Cai, Q. L., Gao, C., et al. (2022). Sepsis-induced immunosuppression: mechanisms, diagnosis and current treatment options. Mil Med. Res. 9, 56. doi: 10.1186/s40779-022-00422-y
Lopera-Maya, E. A., Kurilshikov, A., van der Graaf, A., Hu, S., Andreu-Sánchez, S., Chen, L., et al. (2022). Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat. Genet. 54, 143–151. doi: 10.1038/s41588-021-00992-y
McGrath, E. E., Marriott, H. M., Lawrie, A., Francis, S. E., Sabroe, I., Renshaw, S. A., et al. (2011). TNF-related apoptosis-inducing ligand (TRAIL) regulates inflammatory neutrophil apoptosis and enhances resolution of inflammation. J. Leukocyte Biol. 90, 855–865. doi: 10.1189/jlb.0211062
Mukherjee, A., Lordan, C., Ross, R. P., Cotter, P. D. (2020). Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 12, 1802866. doi: 10.1080/19490976.2020.1802866
Myers, T. A., Chanock, S. J., Machiela, M. J. (2020). LDlinkR: an R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front. Genet. 11. doi: 10.3389/fgene.2020.00157
O’Hara, A. M., Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. doi: 10.1038/sj.embor.7400731
Ong, J. S., MacGregor, S. (2019). Implementing MR-PRESSO and GCTA-GSMR for pleiotropy assessment in Mendelian randomization studies from a practitioner’s perspective. Genet. Epidemiol. 43, 609–616. doi: 10.1002/gepi.22207
Osuchowski, M. F., Welch, K., Siddiqui, J., Remick, D. G. (2006). Circulating cytokine/inhibitor profiles reshape the understanding of the SIRS/CARS continuum in sepsis and predict mortality. J. Immunol. 177, 1967–1974. doi: 10.4049/jimmunol.177.3.1967
Panigrahi, P., Parida, S., Nanda, N. C., Satpathy, R., Pradhan, L., Chandel, D. S., et al. (2017). A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 548, 407–412. doi: 10.1038/nature23480
Papadimitriou, N., Dimou, N., Tsilidis, K. K., Banbury, B., Martin, R. M., Lewis, S. J., et al. (2020). Physical activity and risks of breast and colorectal cancer: a Mendelian randomization analysis. Nat. Commun. 11, 597. doi: 10.1038/s41467-020-14389-8
Park, G., Munley, J. A., Kelly, L. S., Kannan, K. B., Mankowski, R. T., Sharma, A., et al. (2024). Gut mycobiome dysbiosis after sepsis and trauma. Crit. Care. 28, 18. doi: 10.1186/s13054-023-04780-4
Parker, B. J., Wearsch, P. A., Veloo, A. C. M., Rodriguez-Palacios, A. (2020). The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 11. doi: 10.3389/fimmu.2020.00906
Pedersen, H. K., Gudmundsdottir, V., Nielsen, H. B., Hyotylainen, T., Nielsen, T., Jensen, B. A., et al. (2016). Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 535, 376–381. doi: 10.1038/nature18646
Peng, J., Cai, K., Chen, G., Liu, L., Peng, L. (2024). Genetic evidence strengthens the bidirectional connection between gut microbiota and Shigella infection: insights from a two-sample Mendelian randomization study. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1361927
Priputnevich, T., Lyubasovskaya, L., Muravieva, V., Kondrakhin, A., Ignateva, A., Gordeev, A., et al. (2021). Postpartum endometritis and obstetrical sepsis associated with Eggerthella lenta. Case report and review of the literature. J. Matern Fetal Neonatal Med. 34, 313–317. doi: 10.1080/14767058.2019.1602602
Rau, M., Rehman, A., Dittrich, M., Groen, A. K., Hermanns, H. M., Seyfried, F., et al. (2018). Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur. Gastroenterol. J. 6, 1496–1507. doi: 10.1177/2050640618804444
Rudd, K. E., Johnson, S. C., Agesa, K. M., Shackelford, K. A., Tsoi, D., Kievlan, D. R., et al. (2020). Global, regional, and national sepsis incidence and mortality 1990-2017: analysis for the global burden of disease study. Lancet. 395, 200–211. doi: 10.1016/S0140-6736(19)32989-7
Santacroce, L., Di Domenico, M., Montagnani, M., Jirillo, E. (2023). Antibiotic resistance and microbiota response. Curr. Pharm. Des. 29, 356–364. doi: 10.2174/1381612829666221219093450
Schenck, E. J., Ma, K. C., Price, D. R., Nicholson, T., Oromendia, C., Gentzler, E. R., et al. (2019). Circulating cell death biomarker TRAIL is associated with increased organ dysfunction in sepsis. JCI Insight. 4, e127143. doi: 10.1172/jci.insight.127143
Shi, L., Liu, X., Zhang, S., Zhou, A. (2024). Association of gut microbiota with cerebral cortical thickness: A Mendelian randomization study. J. Affect. Disord. 352, 312–320. doi: 10.1016/j.jad.2024.02.063
Shimizu, K., Yamada, T., Ogura, H., Mohri, T., Kiguchi, T., Fujimi, S., et al. (2018). Synbiotics modulate gut microbiota and reduce enteritis and ventilator-associated pneumonia in patients with sepsis: a randomized controlled trial. Crit. Care. 22, 239. doi: 10.1186/s13054-018-2167-x
Singer, B. H., Dickson, R. P., Denstaedt, S. J., Newstead, M. W., Kim, K., Falkowski, N. R., et al. (2018). Bacterial dissemination to the brain in sepsis. Am. J. Respir. Crit. Care Med. 197, 747–756. doi: 10.1164/rccm.201708-1559OC
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Yarmolinsky, J., Davies, N. M., Swanson, S. A., et al. (2021). Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. Jama. 326, 1614–1621. doi: 10.1001/jama.2021.18236
Smith, G. D., Lawlor, D. A., Harbord, R., Timpson, N., Day, I., Ebrahim, S. (2007). Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PloS Med. 4, e352. doi: 10.1371/journal.pmed.0040352
Stefan, K. L., Kim, M. V., Iwasaki, A., Kasper, D. L. (2020). Commensal microbiota modulation of natural resistance to virus infection. Cell. 183, 1312–1324.e1310. doi: 10.1016/j.cell.2020.10.047
Takiishi, T., Fenero, C. I. M., Câmara, N. O. S. (2017). Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. 5, e1373208. doi: 10.1080/21688370.2017.1373208
Tuerxun, K., Eklund, D., Wallgren, U., Dannenberg, K., Repsilber, D., Kruse, R., et al. (2023). Predicting sepsis using a combination of clinical information and molecular immune markers sampled in the ambulance. Sci. Rep. 13, 14917. doi: 10.1038/s41598-023-42081-6
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature. 457, 480–484. doi: 10.1038/nature07540
Verbanck, M., Chen, C. Y., Neale, B., Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Vieira-Silva, S., Falony, G., Belda, E., Nielsen, T., Aron-Wisnewsky, J., Chakaroun, R., et al. (2020). Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature. 581, 310–315. doi: 10.1038/s41586-020-2269-x
von Elm, E., Altman, D. G., Egger, M., Pocock, S. J., Gøtzsche, P. C., Vandenbroucke, J. P. (2007). Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ. 335, 806–808. doi: 10.1136/bmj.39335.541782.AD
Vulevic, J., Juric, A., Walton, G. E., Claus, S. P., Tzortzis, G., Toward, R. E., et al. (2015). Influence of galacto-oligosaccharide mixture (B-GOS) on gut microbiota, immune parameters and metabonomics in elderly persons. Br. J. Nutr. 114, 586–595. doi: 10.1017/S0007114515001889
Wang, Q., Li, F., Liang, B., Liang, Y., Chen, S., Mo, X., et al. (2018). A metagenome-wide association study of gut microbiota in asthma in UK adults. BMC Microbiol. 18, 114. doi: 10.1186/s12866-018-1257-x
Woodworth, M. H., Hayden, M. K., Young, V. B., Kwon, J. H. (2019). The role of fecal microbiota transplantation in reducing intestinal colonization with antibiotic-resistant organisms: the current landscape and future directions. Open Forum Infect. Dis. 6, ofz288. doi: 10.1093/ofid/ofz288
Xiang, K., Wang, P., Xu, Z., Hu, Y. Q., He, Y. S., Chen, Y., et al. (2021). Causal effects of gut microbiome on systemic lupus erythematosus: A two-sample mendelian randomization study. Front. Immunol. 12. doi: 10.3389/fimmu.2021.667097
Xue, F., He, Z., Zhuang, D. Z., Lin, F. (2023). The influence of gut microbiota on circulating inflammatory cytokines and host: A Mendelian randomization study with meta-analysis. Life Sci. 332, 122105. doi: 10.1016/j.lfs.2023.122105
Yan, C., Bao, J., Jin, J. (2024). Exploring the interplay of gut microbiota, inflammation, and LDL-cholesterol: a multiomics Mendelian randomization analysis of their causal relationship in acute pancreatitis and non-alcoholic fatty liver disease. J. Trans. Med. 22, 179. doi: 10.1186/s12967-024-04996-0
Zhang, Z., Cheng, L., Ning, D. (2023). Gut microbiota and sepsis: bidirectional Mendelian study and mediation analysis. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1234924
Zhao, J. H., Stacey, D., Eriksson, N., Macdonald-Dunlop, E., Hedman Å, K., Kalnapenkis, A., et al. (2023). Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat. Immunol. 24, 1540–1551. doi: 10.1038/s41590-023-01588-w
Keywords: circulating inflammatory proteins, genome-wide association study, gut microbiota, Mendelian randomization, sepsis
Citation: Li Z, Lin L, Kong Y, Feng J, Ren X, Wang Y, Chen X, Wu S, Yang R, Li J, Liu Y, Lu Y and Chen J (2024) Gut microbiota, circulating inflammatory proteins and sepsis: a bi-directional Mendelian randomization study. Front. Cell. Infect. Microbiol. 14:1398756. doi: 10.3389/fcimb.2024.1398756
Received: 10 March 2024; Accepted: 08 July 2024;
Published: 08 August 2024.
Edited by:
Yongbo Kang, Shanxi Medical University, ChinaReviewed by:
Georgia Damoraki, National and Kapodistrian University of Athens, GreeceYubao Wang, Tianjin Medical University General Hospital, China
Keke Liu, Shandong Provincial Hospital, China
Copyright © 2024 Li, Lin, Kong, Feng, Ren, Wang, Chen, Wu, Yang, Li, Liu, Lu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiankun Chen, Y2hlbmppYW5rdW5kb2N0b3JAMTI2LmNvbQ==; Yue Lu, Z3p5bHV5dWVAMTI2LmNvbQ==; Yuntao Liu, aWFtbGl1eXVudGFvQDE2My5jb20=; Jiqiang Li, bGlqaXFpYW5naml6aGVuQDE2My5jb20=; Rongyuan Yang, eWFuZ3Jvbmd5dWFuQDE2My5jb20=
 Zuming Li1
Zuming Li1 Liangcai Lin
Liangcai Lin Xueru Chen
Xueru Chen Siyi Wu
Siyi Wu Yue Lu
Yue Lu Jiankun Chen
Jiankun Chen