- 1Xinxiang Key Laboratory of Pathogenic Biology, Department of Pathogenic Biology, School of Basic Medical Sciences, Xinxiang Medical University, Xinxiang, Henan, China
- 2Department of Laboratory, the Third Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan, China
Blastocystis spp. is a ubiquitous protozoon in the intestinal tract of human and many animals. Microscopic examination is the main method of clinical diagnosis for Blastocystis spp., which is prone to false negative. A simple and rapid diagnosis of Blastocystis spp. infection is an important step to prevent and control blastocystosis. Here, a recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay was developed for rapid visual detection of Blastocystis spp. DNA amplification could be performed within 18 min at 37°C. The minimum DNA detection limit was 1 pg/μL, and there was no cross-reactivity with 12 other non-target pathogens, which was consistent with the sensitivity of conventional PCR (cPCR). Furthermore, 56 fecal samples from the Third Affiliated Hospital of Xinxiang Medical University were tested using RPA and cPCR methods respectively, and the results were completely consistent. The results show that RPA-LFD method has high accuracy and visual results, which provides a new choice for the differential diagnosis and rapid field detection of Blastocystis spp.
1 Introduction
Blastocystis spp. is a common intestinal protozoan parasites found in humans and many animals (Mei et al., 2022). The infection rate of Blastocystis spp. is about 0.5%~57% in developed countries and 30%~60% in developing countries (Zhang et al., 2019). More than 12 provinces in China have reported Blastocystis spp. infection, with a total infection rate of 3.37%. Among them, Fujian Province had the lowest prevalence (0.80%) and Guangdong Province had the highest prevalence (100%) (Deng et al., 2019). Several studies have reported that Blastocystis spp. can lead to clinical symptoms such as abdominal pain, diarrhea, vomiting, constipation, and irritable bowel syndrome (IBS) (Légeret et al., 2020; Asghari et al., 2021). In addition, patients with ulcerative colitis develop hematochezia after infection with Blastocystis spp (Unalan-Altintop et al., 2022). Rapid and accurate diagnosis of Blastocystis spp. infection is an important step in the prevention and control of blastocystosis, which plays an essential role in public health (Liu et al., 2023). However, the clinical manifestations of blastocystosis are non-specific and similar to those caused by other gastrointestinal diseases such as irritable bowel syndrome. Microscopic examination is the main method of clinical diagnosis for Blastocystis spp (McHardy et al., 2014)., which is prone to false negative results. Therefore, establishing a rapid, accurate, and convenient detection method for Blastocystis spp. has great significance.
Molecular biological diagnosis has great advantages in specificity and sensitivity, and is one of the commonly used diagnostic methods for blastocystosis. Among them, conventional PCR (cPCR) has been widely used in the diagnosis and genotyping of Blastocystis spp (Yoshikawa et al., 2004; Stensvold et al., 2006; Domínguez-Márquez et al., 2009; Khademvatan et al., 2018). However, specialized instruments and equipment are required for cPCR, including precise thermal cycling instruments, personnel trained in the technology, and maintaining a cold chain to preserve the heat-unstable reagents. The detection is usually completed in specialized testing centers, which are typically located far away from the high-risk population of infection. In contrast, field detection has certain limitations and has not yet been popularized. Therefore, cPCR detection is usually only performed in the laboratory and rarely used in the initial clinical diagnosis. A more rapid, simple, and accurate detection method is required.
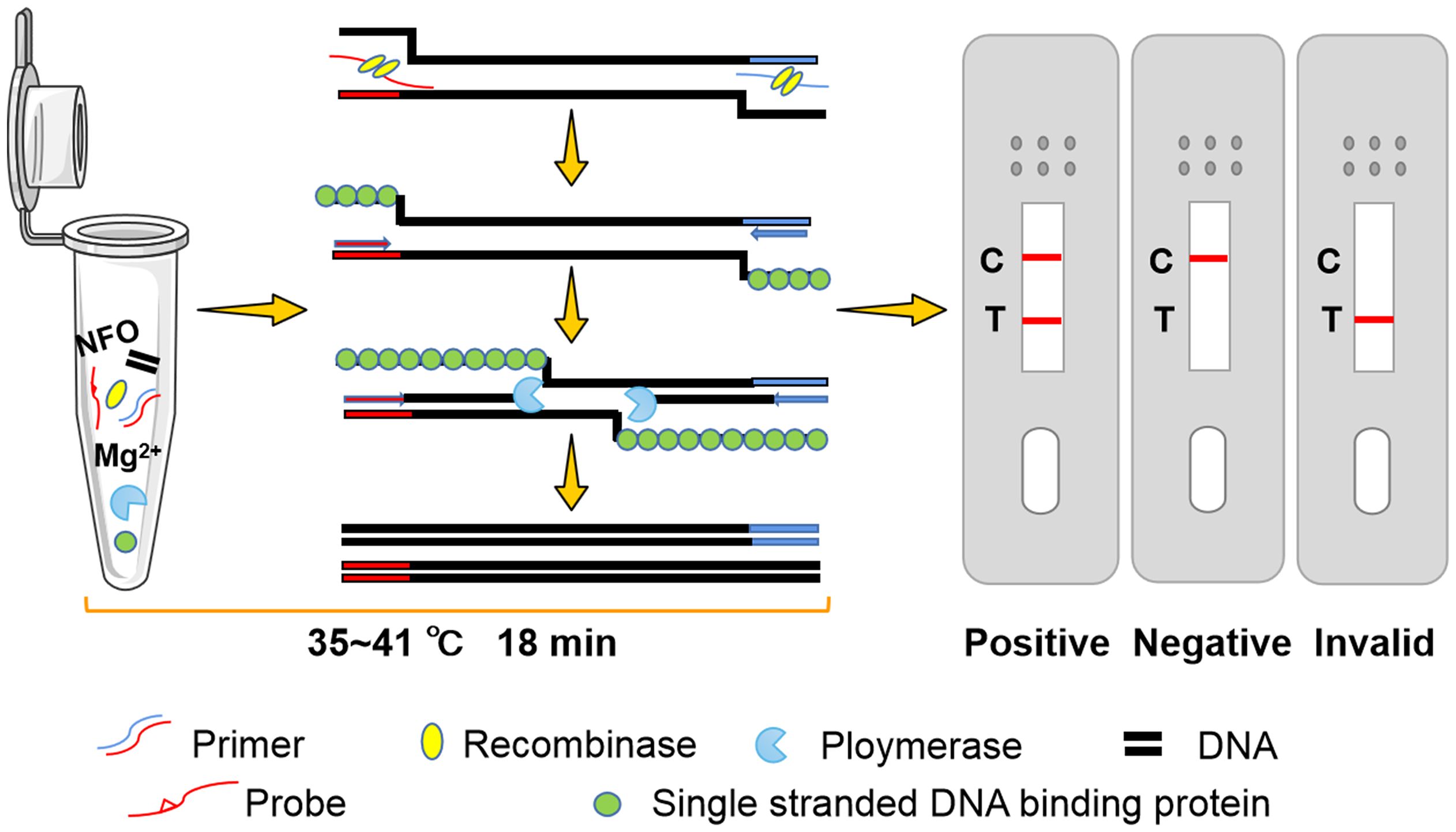
In 2006, recombinase polymerase amplification (RPA) was developed in the UK, providing an isothermal amplification technique for nucleic acids (Piepenburg et al., 2006). The amplification of double-stranded DNA is accomplished through a series of enzyme reactions without thermal or chemical denaturation. Moreover, The results of RPA can be displayed by a lateral flow dipstick (LFD), which is suitable for detection in resource-poor areas or field areas (Wang et al., 2022). RPA-LFD overcomes the need for precision thermal cycling instruments and greatly improves detection efficiency while preserving high sensitivity, thereby allowing rapid visualization of the results (Piepenburg et al., 2006). In recent years, there have been several reports describing the successful application of RPA technology in different parasite detection. including Giardia duodenalis (Crannell et al., 2015), Entamoeba histolytica (Nair et al., 2015) and Leishmania Viannia spp (Saldarriaga et al., 2016).. For Blastocystis spp., only RPA diagnostic methods have been reported (Mei et al., 2023b), it is regrettable that there is no more convenient and visual RPA-LFD diagnostic method at present.
The succinate dehydrogenase subunit A (SDHA) gene is a novel genetic marker utilized in the typing and evaluation of the genetic diversity of Blastocystis spp (Higuera et al., 2020).. In this study, an RPA-LFD method targeting the SDHA gene was developed to detect Blastocystis spp. rapidly and accurately. This method can be implemented for onsite testing in hospitals or areas with inadequate medical conditions. To our knowledge, this is the first study on the detection of Blastocystis spp. using the RPA-LFD technique.
2 Materials and methods
2.1 DNA samples
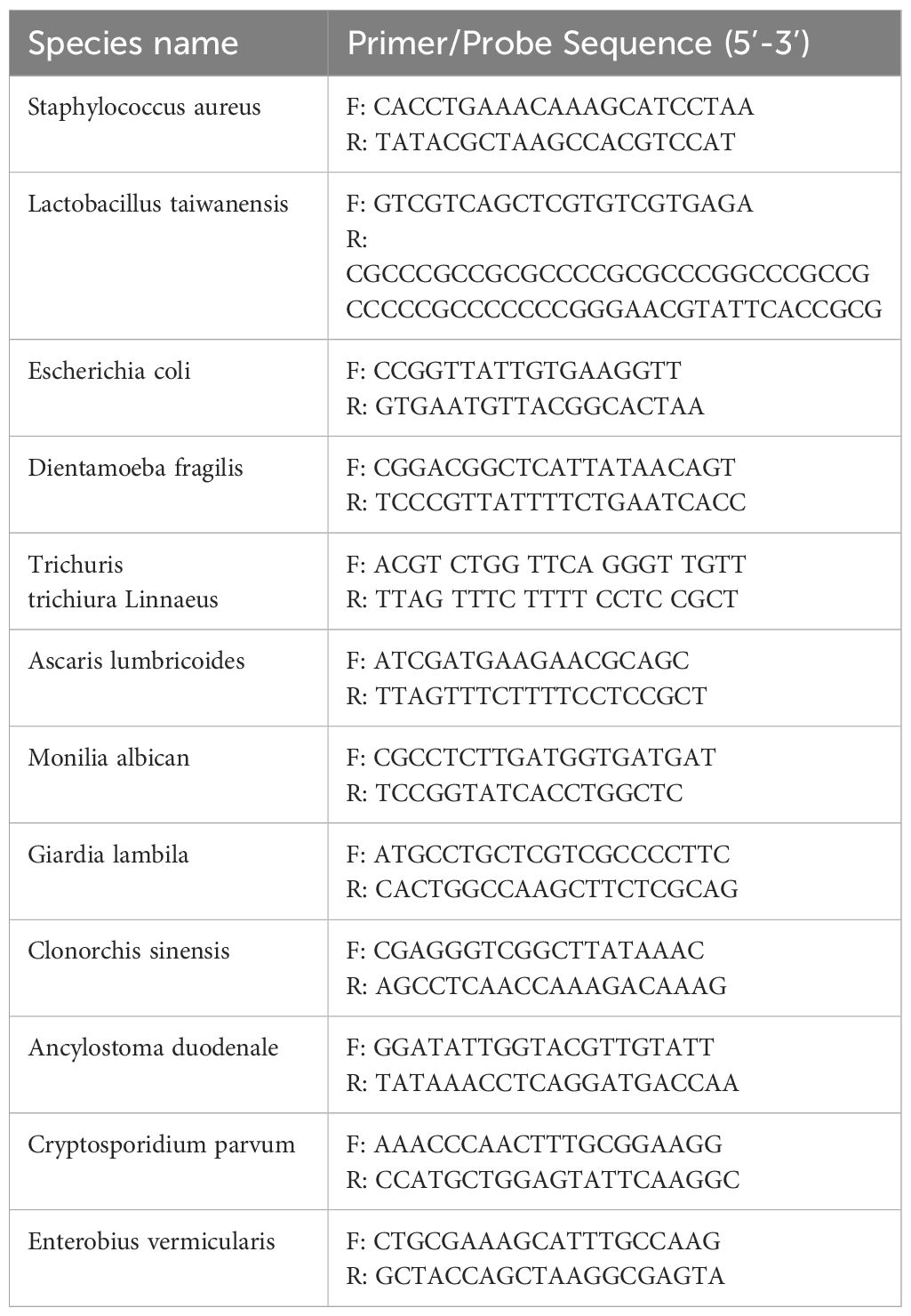
The DNA samples used in this study were obtained from Department of Pathogenic Biology, Xinxiang Medical University, including Blastocystis spp., Dientamoeba fragilis, Giardia lamblia, Cryptosporidium parvum, Ascaris lumbricoides, Enterobius vermicularis, Trichuris trichiura, Clonorchis sinensis, Ancylostoma duodenale, Lactobacillus taiwanensis, Escherichia coli, Candida albican, and Staphylococcus aureus. All DNA samples were amplified by cPCR (the primers for cPCR were shown in Table 1) and sequenced to confirm the species, and stored at -20°C for use.
2.2 Primer and probe design
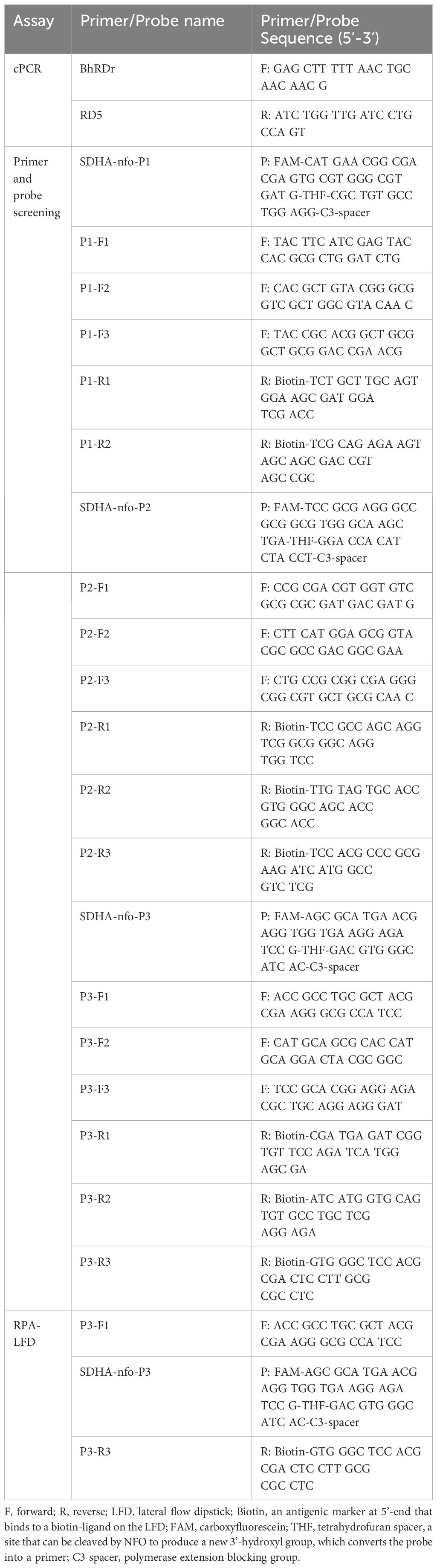
The RPA primers and probes were designed to target the SDHA gene (retrieved from the National Center for Biotechnology Information, Gene ID: 24919714) of Blastocystis spp. according to the requirements of the RPA assay. Here, three probes and corresponding primers were designed, and the cPCR primers used in this study referred to Scicluna SM et al (Scicluna et al., 2006). (see Table 2 for details). Sangon Bioengineering Co., Ltd (Sangon, Shanghai, China) was commissioned to synthesize all primers and probes.
2.3 Primer screening for RPA assay
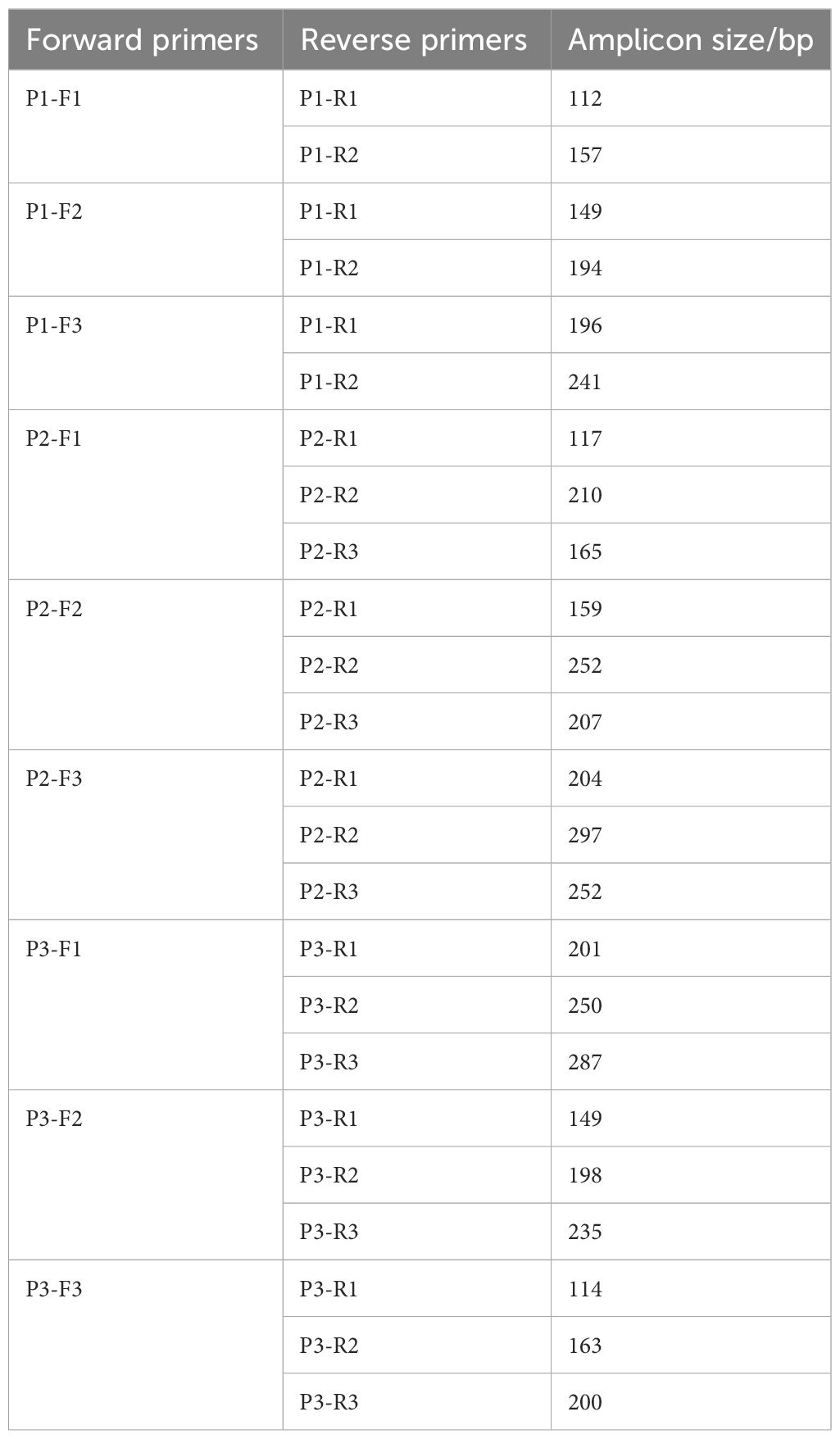
The forward and reverse primers of each probe were randomly combined, as shown in Table 3. The primer screening was performed using commercial basic freeze-dried powder (Amplification future Biotech, Changzhou, China). The process of RPA assay was as follows: 29.4 μL rehydration buffer, 9.1 μL preparation of nuclease-free water, 2 μL forward primer, and 2 μL reverse primer are added into the basic freeze-dried powder and thoroughly mixed. The 5 μL of DNA template was then added to the mixture and 2.5 μL magnesium acetate solution for initiating the reaction was added to the tube cap. The reaction tubes were mixed upside down, centrifuged, and incubated in pre-heated 37°C metal bath for 30 min. Finally, the primer pairs were screened by agarose gel electrophoresis of the amplified products.
2.4 Development of the RPA-LFD assay
The PRA-LFD assay was performed as follows: After the reaction Mix (Rehydration buffer, 29.4 μL; forward primer, 2 μL; reverse primer, 2 μL; probe, 0.6 μL; Nuclease-free water, 8.5 μL; 42.5 μL) was prepared, added it to a NFO freeze-dried powder (Amplification future Biotech, Changzhou, China) and mixed well. The primers and probe were shown in Table 2. Subsequently, 5μL DNA template and 2.5 μL magnesium acetate solution were added to the suspension, The reaction tubes were incubated in a metal bath at 37°C for 15 min. At the end of the reaction, the LFD (Amplification future Biotech, Changzhou, China) was placed into the reaction solution after 10 times dilution. The band on the LFD determines whether the reaction was positive or negative.
2.5 Negative testing of the primers and probes
The primer and probe could easily bind, resulting in false positive in RPA-LFD assay. Therefore, the DNA template of the RPA-LFD assay was replaced with water, and the negative primer and probe was screened through the visualization results of LFD.
2.6 Sensitivity analysis
The DNA templates were diluted to 10 pg/μL, 1 pg/μL, 100 fg/μL, and 10 fg/μL, respectively and amplified by RPA-LFD assay. The cPCR assay were performed simultaneously to compare the sensitivity of RPA-LFD assay. The themal cPCR system consists of 12.5 μL of 2×Taq Plus Master Mix, 8.5 μL nuclease-free water, 2 μL DNA, 1 μL forward primer and 1 μL reverse primer. The thermal cycler program settings for cPCR was as follows: predenaturation at 94°C for 5 min; denaturation at 94°C for 45 s, annealing at 59°C for 1 min, extension at 72°C for 1 min, 30 cycles; final extension at 72°C for 10 min.
2.7 Determination of specificity
For specificity analysis, 12 non-target pathogens were evaluated by RPA-LFD assay and cRCR assay, including Staphylococcus aureus, Lactobacillus taiwanensis, Escherichia coli, Dientamoeba fragilis, Trichuris trichiura Linnaeus, Ascaris lumbricoides, Monilia albican, Giardia lambila, Clonorchis sinensis, Ancylostoma duodenale, Cryptosporidium parvum, and Enterobius vermicularis.
2.8 Optimum reaction time and temperature
The primers and probes screened in the previous experiment were employed for RPA-LFD assay. Different times (3, 6, 9, 12, 15, 18, 21, 24, 27, and 30 min) and different temperatures (25, 30, 35, 37, 39, 41, 43, and 45°C) were set to determine the optimum time and temperature.
2.9 Clinical sample testing
A total of 56 clinical fecal samples were collected from the Third Affiliated Hospital of Xinxiang Medical University for RPA-LFD detection. The cPCR assay was also carried out at the same time, and the application effect of RPA-LFD in clinical sample was analyzed by comparing the results of cPCR.
3 Results
3.1 Primer screening
Five pairs of primers were selected according to the results of agarose gel electrophoresis, including P1-F1/P1-R2, P1-F3/P1-R1, P2-F1/P2-R1, P2-F1/P2-R3, and P3-F1/P3-R3 (Figure 1).
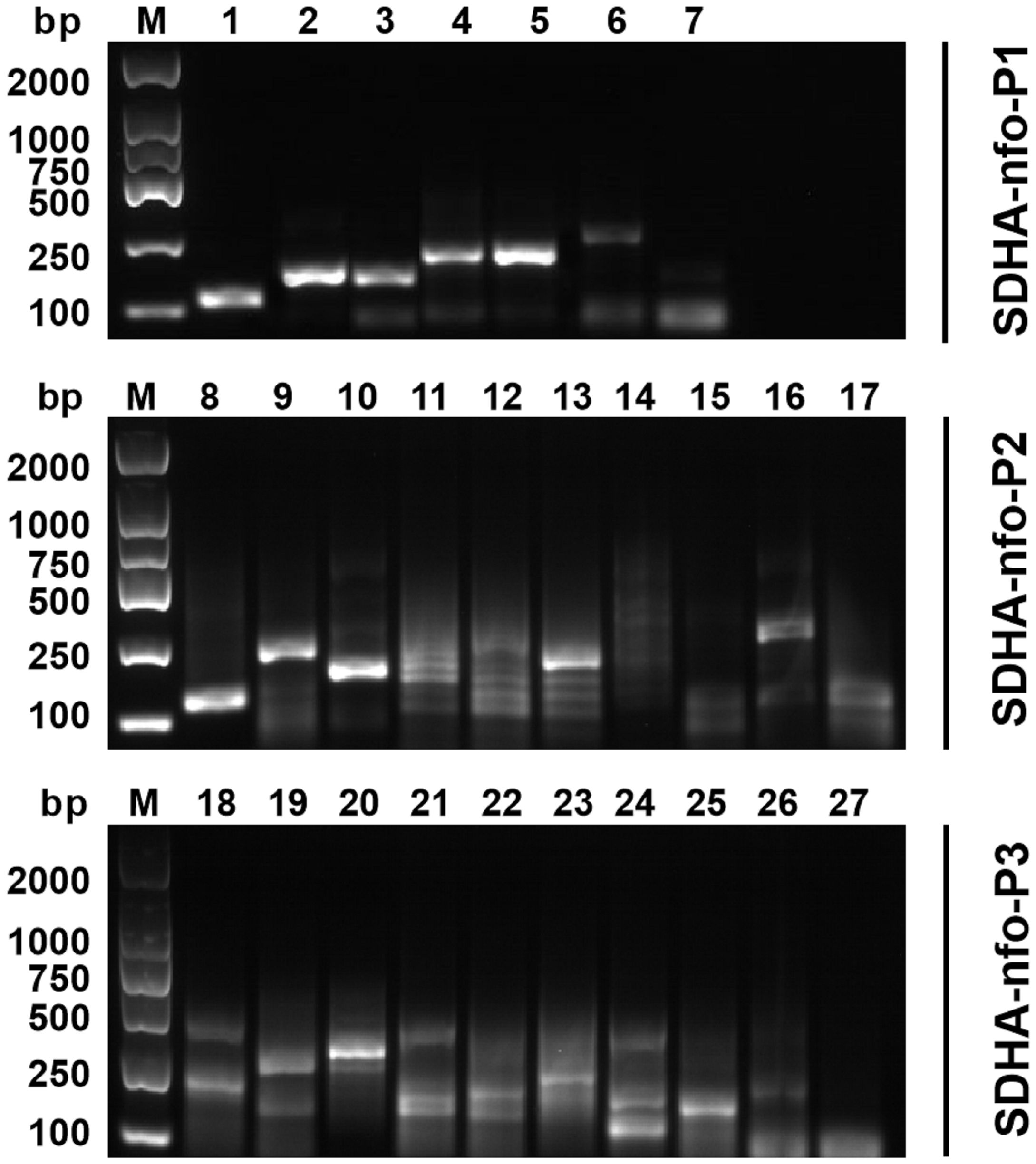
Figure 1 Primer screening for RPA. 1: P1-F1/P1-R1; 2: P1-F1/P1-R2; 3: P1-F2/P1-R1; 4: P1-F2/P1-R2; 5: P1-F3/P1-R1; 6: P1-F3/P1-R2; 7: negative control; 8: P2-F1/P2-R1; 9: P2-F1/P2-R2; 10: P2-F1/P2-R3; 11: P2-F2/P2-R1; 12: P2-F2/P2-R2; 13: P2-F2/P2-R3; 14: P2-F3/P2-R1; 15: P2-F3/P2-R2; 16: P2-F3/P2-R3; 17: negative control; 18: P3-F1/P3-R1; 19: P3-F1/P3-R2; 20: P3-F1/P3-R3; 21: P3-F2/P3-R1; 22: P3-F2/P3-R2; 23: P3-F1/P3-R3; 24: P3-F3/P3-R1; 25: P3-F3/P3-R2; 26: P3-F3/P3-R3; 27: negative control; M, DNA marker.
3.2 Negative testing of primers and probes
Negative testing was performed for the 5 pairs of primers and probes. The results showed false positives with the combinations of SDHA-nfo-P1/P1-F1/P1-R2, SDHA-nfo-P1/P1-F3/P1-R1, and SDHA-nfo-P2/P2-F1/P2-R1. In contrast, the combinations of SDHA-nfo-P2/P2-F1/P2-R3, SDHA-nfo-P3/P3-F1/P3-R3 were negative which could be further screened for sensitivity (Figure 2).

Figure 2 Negative testing of the primers and probes. 1: SDHA-nfo-P1/P1-F1/P1-R2; 2: SDHA-nfo-P1/P1-F3/P1-R1; 3: SDHA-nfo-P2/P2-F1/P2-R1; 4: SDHA-nfo-P2/P2-F1/P2-R3; 5: SDHA-nfo-P3/P3-F1/P3-R3; C, control line; T, test line.
3.3 Sensitivity analysis
The results of the sensitivity test are displayed in Figure 3. Among the 2 pairs of primers and probes, SDHA-nfo-P3/P3-F1/P3-R3 showed the highest sensitivity. The lowest concentration of DNA that could be detected was 1 pg/μL when the RPA-LFD assay was performed using SDHA-nfo-P3/P3-F1/P3-R3, which was consistent with cPCR. The findings demonstrated that the RPA-LFD had high sensitivity, and the samples showed positive results when the concentration of Blastocystis spp. was above 1 pg/μL.
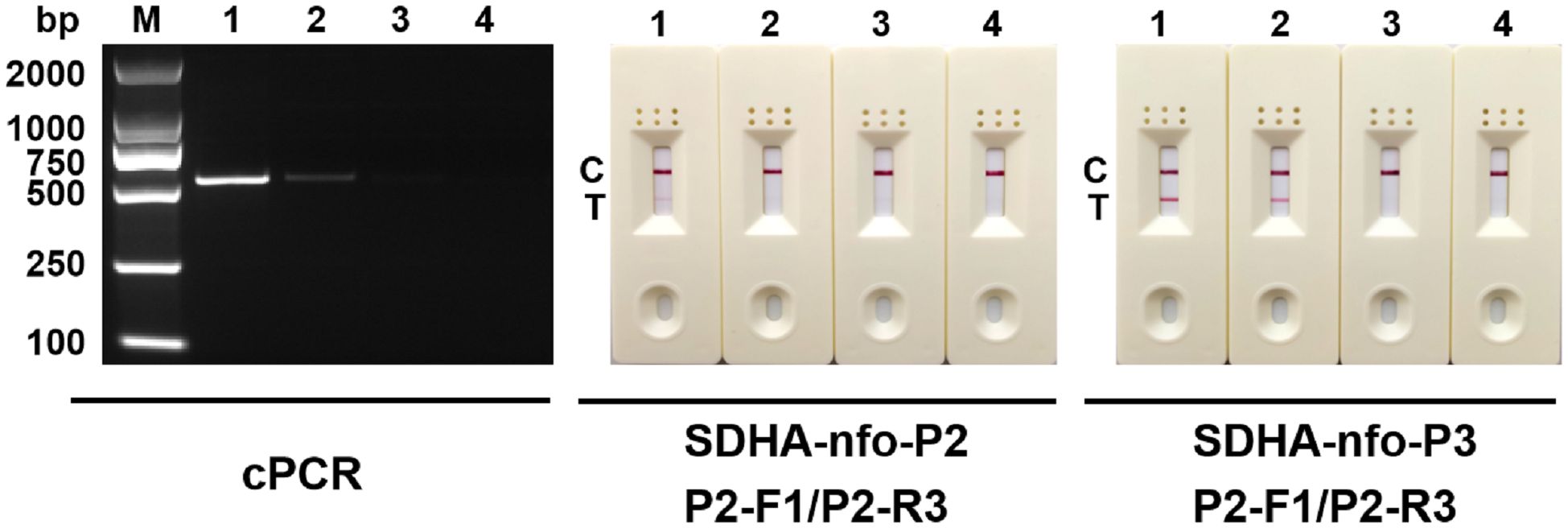
Figure 3 Sensitivity analysis. Different concentrations of DNA were used for cPCR assay and RPA-LFD assay. 1: 10 pg/μL; 2: 1 pg/μL; 3: 100 fg/μL; 4: 10 fg/μL; M, DNA marker; C, control line; T, test line.
3.4 Determination of specificity
The previously screened SDHA-nfo-P3/P3-F1/P3-R3 were further verified by specificity test. As shown in Figure 4, no false positives or false negatives were found in the RPA-LFD and cPCR, suggesting that the RPA-LFD method has high specificity.
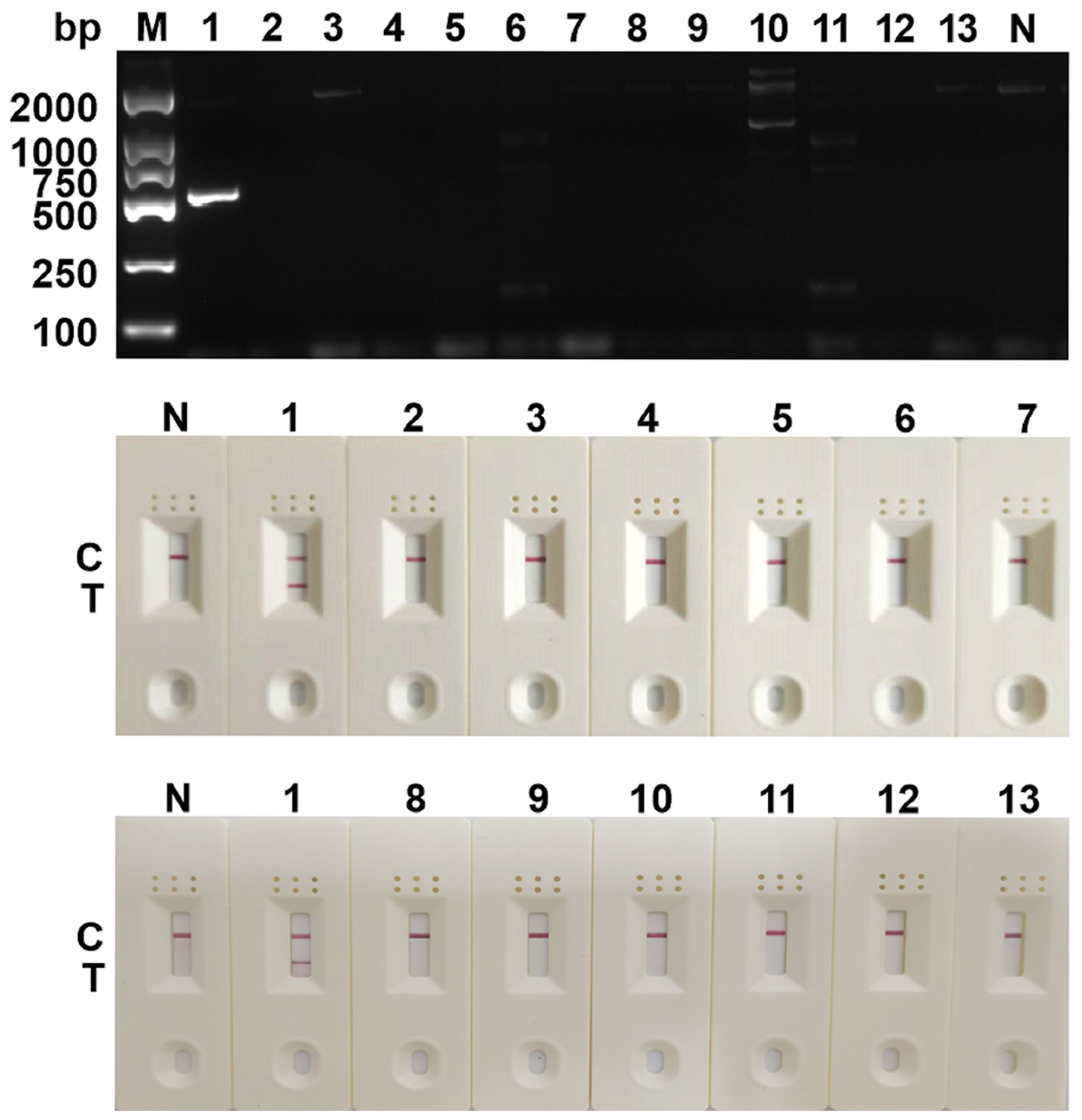
Figure 4 Determination of specificity. The specificity analysis of cPCR and RPA-LFD assay. 1: Blastocystis spp.; 2: Staphylococcus aureus; 3: Lactobacillus taiwanensis; 4: Escherichia coli; 5: Dientamoeba fragilis; 6: Trichuris trichiura Linnaeus; 7: Ascaris lumbricoides; 8: Monilia albican; 9: Giardia lambila; 10: Clonorchis sinensis; 11: Ancylostoma duodenale; 12: Cryptosporidium parvum; 13: Enterobius vermicularis; M, DNA marker; N, negative control; C, control line; T, test line.
3.5 Optimum reaction time and temperature
The RPA-LFD assay was conducted with a combination of primer and probe (SDHA-nfo-P3/P3-F1/P3-R3) at different reaction times (3, 6, 9, 12, 15, 18, 21, 24, 27, and 30 min) and different reaction temperatures (25, 30, 35, 37, 39, 41, 43, and 45°C). The test line of the RPA reaction could be observed in the LFD as from 3 min. The detection line was clearest when the RPA reaction lasted for 18 min (Figure 5), and no significant change was observed when the reaction time was extended. The reaction was successfully completed in the temperature range of 25~45°C, and the bands are similar at 35~41°C (Figure 6). 37°C was selected as the final reaction temperature of this method for clinical sample detection.
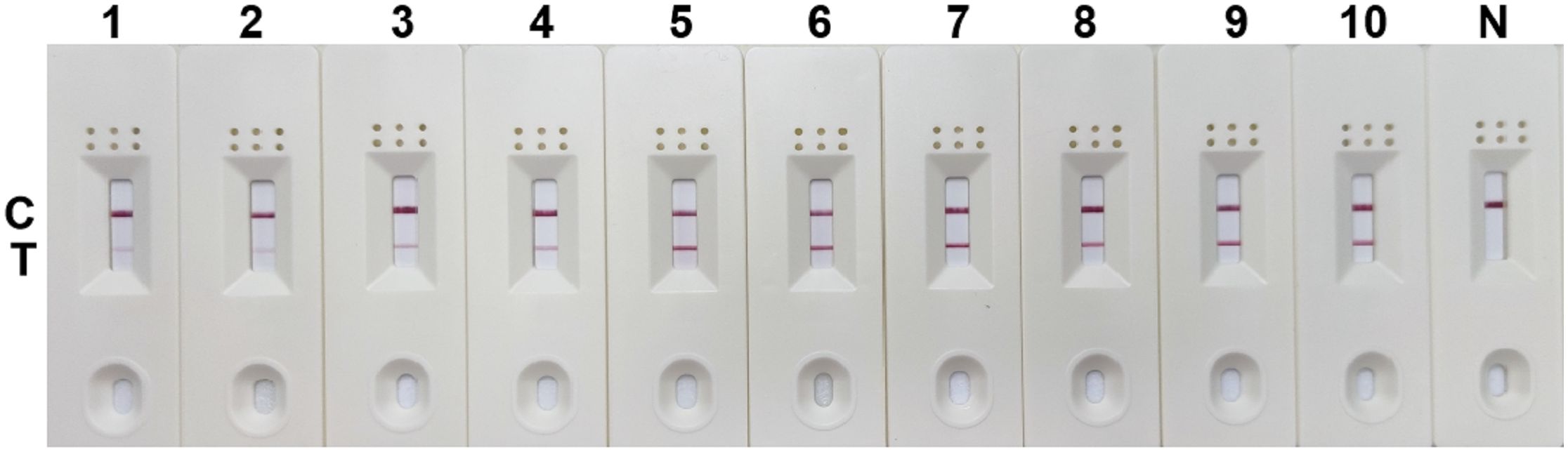
Figure 5 Optimum reaction time for RPA-LFD assay. 1: 3 min; 2: 6 min; 3: 9 min; 4: 15 min; 5: 18 min; 6: 21 min; 7: 24 min; 8: 27min; 9: 30 min; N, negative control; C, control line; T, test line.
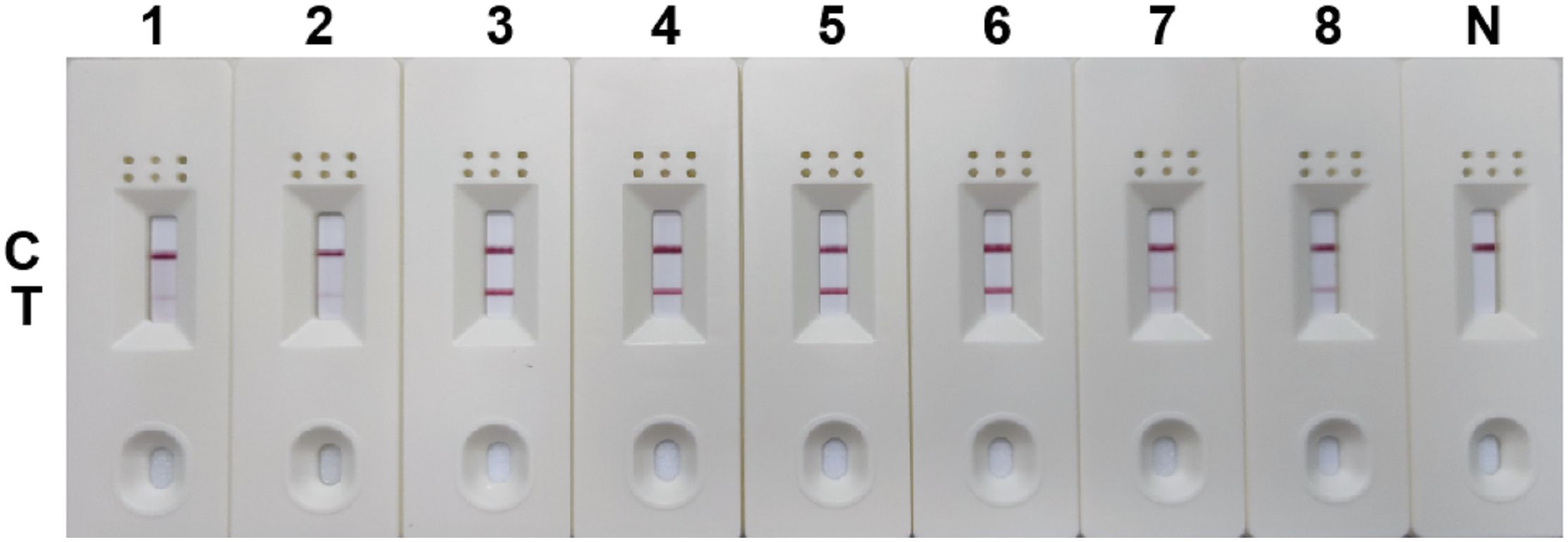
Figure 6 Optimum reaction temperature for RPA-LFD assay. 1: 25°C; 2: 30°C; 3: 35°C; 4: 37°C; 5: 39°C; 6: 41°C; 7: 43°C; 8: 45°C; N, negative control; C, control line; T, test line.
3.6 Application effect of clinical sample detection
The detection results from 56 clinical samples showed that 9 fecal samples were positive for Blastocystis spp. and 47 were negative (Figure 7). The results of RPA-LFD were consistent with those of cPCR. Further sequencing results showed that 9 positive samples contained 3 subtypes, namely ST3 (6/9, 66.7%), ST4 (1/9, 11.1%) and ST7 (2/9, 22.2%).
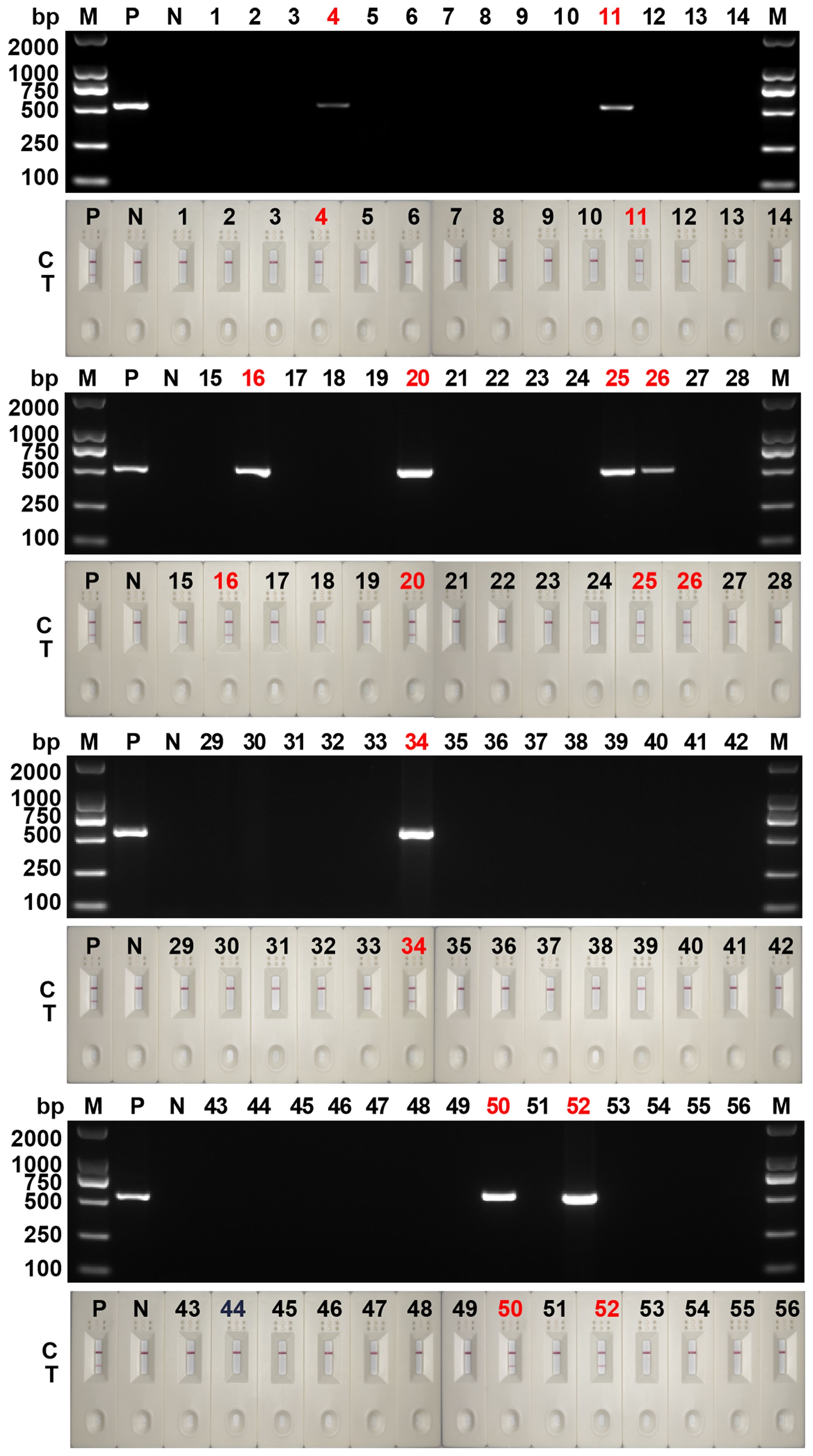
Figure 7 Clinical sample testing for cPCR assay and RPA-LFD assay. 1~56: the numbers of 56 clinical samples; M, DNA marker; P, positive control; N, negative control; C, control line; T, test line.
4 Discussion
A variety of intestinal pathogens, including viruses, bacteria and parasitic protozoa, could cause diarrhea, which is the second leading cause of death among children (Kotloff et al., 2013; Caner et al., 2020; dos Santos David et al., 2022; Kotloff, 2022). Among them, the cause of chronic diarrhea is increasingly thought to be caused by parasitic protozoa infection (Lima and Guerrant, 1992), such as Blastocystis spp., Giardia spp., Entamoeba spp. Among them, Blastocystis spp. is the most widely distributed protozoa (Kaewjai et al., 2023). Similar clinical symptoms and different treatment methods highlight the importance of establishing a rapid and accurate diagnostic method for Blastocystis spp.
In this study, a simple, rapid, sensitive, and accurate diagnostic method for Blastocystis spp. was achieved by RPA-LFD assay. This method resolved the low sensitivity limitations of microscopic observation and the requirement for expensive equipment for cPCR in remote areas. Furthermore, this method has high sensitivity, and the samples showed positive results when the DNA concentration of Blastocystis spp. was greater than 1 pg/μL. The species with the same colonization site or close relationship with the target pathogen were prone to false positives in molecular diagnosis. The RPA-LFD method established in this study was proved to have no cross-reaction with these non-target pathogens, indicating that the method has good specificity.
In contrast to PCR, which can be heated to avoid non-specific amplification caused by binding between primers, nucleic acid amplification achieved by RPA under constant temperature easily generates primer dimers. The formation of primer dimers not only affects the amplification efficiency but also may affect the test results in case of low or no template concentration (Özay and McCalla, 2021). To overcome this limitation, several RPA primers and probes were designed with the SDHA gene as the target sequence. Five pairs of primers with higher amplification efficiency were screened by RPA assay. Subsequently, two pairs of primers and probes passed negative testing. In the further sensitivity test, SDHA-nfo-P3/P3-F1/P3-R3 showed the highest sensitivity, which then passed the specificity test. Therefore, SDHA-nfo-P3/P3-F1/P3-R3 was finally determined to be the best combination primer and probe.
The RPA-LFD assay was effectively carried out at 25~45°C, indicating that the RPA-LFD assay had lower requirements for temperature conditions and a wider detection range, which was more convenient for the detection of Blastocystis spp (Abd El Wahed et al., 2015).. In addition, existing RPA methods require agarose gel electrophoresis to read the results (Mei et al., 2023b). In contrast, the RPA-LFD method established in this study can make a diagnosis faster, as the LFD can observe the T-line within 3 min. The accuracy of the method was further confirmed by the detection of clinical samples. However, the distribution of different subtypes of Blastocystis spp. is related to many factors such as geographical location and season. The clinical samples used in this study were from Xinxiang City, China. It was reported that there were 8 subtypes (ST1-ST7, ST12) in the Chinese, and only ST1, ST3, ST4, ST6 and ST7 were reported in Xinxiang City (Deng et al., 2019; Zhang et al., 2019; Mei et al., 2023a). Although only ST3, ST4 and ST7 were included in the 56 clinical samples tested here, the results of RPA-LFD were completely consistent with those of cPCR, indicating that this method has good clinical application potential. In future studies, clinical samples containing more subtypes of Blastocystis spp. can be tested to further ensure the accuracy and stability of the method.
5 Conclusions
In summary, a rapid detection method for Blastocystis spp. was developed using RPA technology, with the detection results being visualized by LFD. This method overcame the requirements of cPCR for thermal cycling instruments and agarose gel electrophoresis instruments. Moreover, amplification was effectively completed in the temperature range of 25~45°C. The detection time was also greatly shortened, and the results were observed by LFD after 3 min of RPA reaction. The lowest detection limit of DNA was 1 pg/μL, which was consistent with the cPCR. In addition, this method showed good specificity. This study provided a simple, rapid, and accurate method for the diagnosis of Blastocystis spp.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Ethical Commission of the Xinxiang Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The manuscript presents research on animals that do not require ethical approval for their study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
XM: Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. CS: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. JX: Formal analysis, Investigation, Writing – original draft. LJ: Data curation, Investigation, Writing – original draft. SZ: Data curation, Investigation, Writing – original draft. ZY: Formal analysis, Writing – original draft. TX: Conceptualization, Formal analysis, Writing – original draft. ZZ: Conceptualization, Writing – original draft. SW: Conceptualization, Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundation of Henan Province (No. 232300421313), the Doctoral Scientific Research Activation Foundation of Xinxiang Medical University (No. XYBSKYZZ202140), the Training Plan for Young Backbone Teachers in Universities of Henan Province (No. 2021GGJS101).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd El Wahed, A., Patel, P., Faye, O., Thaloengsok, S., Heidenreich, D., Matangkasombut, P., et al. (2015). Recombinase polymerase amplification assay for rapid diagnostics of dengue infection. PloS One 10, e0129682. doi: 10.1371/journal.pone.0129682
Asghari, A., Hassanipour, S., Hatam, G. (2021). Comparative molecular prevalence and subtypes distribution of Blastocystis sp. a potentially zoonotic infection isolated from symptomatic and asymptomatic patients in Iran: A systematic review and meta-analysis. Acta Parasitol. 66, 745–759. doi: 10.1007/s11686-021-00360-0
Caner, A., Zorbozan, O., Tunalı, V., Kantar, M., Aydoğdu, S., Aksoylar, S., et al. (2020). Intestinal protozoan parasitic infections in immunocompromised child patients with diarrhea. Jpn. J. Infect. Dis. 73, 187–192. doi: 10.7883/yoken.JJID.2019.054
Crannell, Z. A., Cabada, M. M., Castellanos-Gonzalez, A., Irani, A., White, A. C., Richards-Kortum, R. (2015). Recombinase polymerase amplification-based assay to diagnose Giardia in stool samples. Am. J. Trop. Med. Hyg. 92, 583–587. doi: 10.4269/ajtmh.14-0593
Deng, L., Chai, Y., Zhou, Z., Liu, H., Zhong, Z., Hu, Y., et al. (2019). Epidemiology of Blastocystis sp. infection in China: a systematic review. Parasite. 26, 41. doi: 10.1051/parasite/2019042
Domínguez-Márquez, M. V., Guna, R., Muñoz, C., Gómez-Muñoz, M. T., Borrás, R. (2009). High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain). Parasitol. Res. 105, 949–955. doi: 10.1007/s00436-009-1485-y
dos Santos David, E., da Costa Fonseca, E., de Carvalho, J. F. N., Marinho, R. D. S. S., Duro, R. L. S., Komninakis, S. V., et al. (2022). Metagenomics applied to the detection of diarrhea viruses in humans: Systematic Review. Acta Trop. 227, 106287. doi: 10.1016/j.actatropica.2021.106287
Higuera, A., Muñoz, M., López, M. C., Reyes, P., Urbano, P., Villalobos, O., et al. (2020). Succinate dehydrogenase gene as a marker for studying Blastocystis genetic diversity. Heliyon. 6, e05387. doi: 10.1016/j.heliyon.2020.e05387
Kaewjai, C., Tonsomboon, A., Pawiwongchai, J., Prommano, O. (2023). Antiprotozoal activity of Boesenbergia rotunda (L.) Mansf and Ganoderma lucidum (Fr.) Kart extracts against Blastocystis hominis. Vet. World 16, 187–193. doi: 10.14202/vetworld.2023.187-193
Khademvatan, S., Masjedizadeh, R., Yousefi-Razin, E., Mahbodfar, H., Rahim, F., Yousefi, E., et al. (2018). PCR-based molecular characterization of Blastocystis hominis subtypes in southwest of Iran. J. Infect. Public Health 11, 43–47. doi: 10.1016/j.jiph.2017.03.009
Kotloff, K. L. (2022). Bacterial diarrhoea. Curr. Opin. Pediatr. 34, 147–155. doi: 10.1097/MOP.0000000000001107
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Légeret, C., Rüttimann, C., Furlano, R. I., Ruf, T., Poppert, S., Fankhauser, H., et al. (2020). Blastocystis in Swiss children: a practical approach. Eur. J. Pediatr. 179, 979–984. doi: 10.1007/s00431-020-03599-3
Lima, A. A., Guerrant, R. L. (1992). Persistent diarrhea in children: epidemiology, risk factors, pathophysiology, nutritional impact, and management. Epidemiol. Rev. 14, 222–242. doi: 10.1093/oxfordjournals.epirev.a036088
Liu, H., Ni, H., Zhu, N., Liu, S., Wang, R., Cao, J., et al. (2023). Blastocystis infection among diarrhea outpatients in Ningbo, Southeast China: A potential zoonotic health threat. Microb. Pathog. 181, 106219. doi: 10.1016/j.micpath.2023.106219
McHardy, I. H., Wu, M., Shimizu-Cohen, R., Couturier, M. R., Humphries, R. M. (2014). Detection of intestinal protozoa in the clinical laboratory. J. Clin. Microbiol. 52, 712–720. doi: 10.1128/JCM.02877-13
Mei, X. F., Su, C. W., Wang, W. J., Zhang, B., Wei, L., Zhang, Z. C., et al. (2022). Molecular prevalence and subtypes distribution of Blastocystis sp. amongst outpatients and inpatients in north and south areas of Henan Province, China. J. Eukaryot. Microbiol. 70, e12960. doi: 10.1111/jeu.12960
Mei, X. F., Su, C. W., Wang, W. J., Zhang, B., Wei, L., Zhang, Z. C., et al. (2023a). Molecular prevalence and subtypes distribution of Blastocystis sp. among outpatients and inpatients in north and south areas of Henan Province, China. J. Eukaryot. Microbiol. 70, e12960. doi: 10.1111/jeu.12960
Mei, X. F., Su, C. W., Zhang, S. R., Jia, L. W., Yang, Z. K., Tian, X. W., et al. (2023b). Development and application of recombinase polymerase amplification assay for rapid detection of Blastocystis sp. Parasitology 150, 1221–1225. doi: 10.1017/S0031182023000975
Nair, G., Rebolledo, M., White, A. C., Jr., Crannell, Z., Richards-Kortum, R. R., Pinilla, A. E., et al. (2015). Detection of Entamoeba histolytica by recombinase polymerase amplification. Am. J. Trop. Med. Hyg. 93, 591–595. doi: 10.4269/ajtmh.15-0276
Özay, B., McCalla, S. E. (2021). A review of reaction enhancement strategies for isothermal nucleic acid amplification reactions. Sensors Actuators Rep. 3, 100033. doi: 10.1016/j.snr.2021.100033
Piepenburg, O., Williams, C. H., Stemple, D. L., Armes, N. A. (2006). DNA detection using recombination proteins. PloS Biol. 4, e204. doi: 10.1371/journal.pbio.0040204
Saldarriaga, O. A., Castellanos-Gonzalez, A., Porrozzi, R., Baldeviano, G. C., Lescano, A. G., de Los Santos, M. B., et al. (2016). An innovative field-applicable molecular test to diagnose cutaneous Leishmania Viannia spp. infections. PloS Negl. Trop. Dis. 10, e0004638. doi: 10.1371/journal.pntd.0004638
Scicluna, S. M., Tawari, B., Clark, C. G. (2006). DNA barcoding of blastocystis. Protist. 157, 77–85. doi: 10.1016/j.protis.2005.12.001
Stensvold, R., Brillowska-Dabrowska, A., Nielsen, H. V., Arendrup, M. C. (2006). Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J. Parasitol. 92, 1081–1087. doi: 10.1645/GE-840R.1
Unalan-Altintop, T., Vahabov, C., Ergunay, K., Kurt, O., Kav, T., Akyon, Y., et al. (2022). Investigation of Dientamoeba fragilis and Blastocystis in patients from Turkey with ulcerative colitis and irritable bowel syndrome: Any relation with genotypes? Acta Trop. 231, 106451. doi: 10.1016/j.actatropica.2022.106451
Wang, H., Yang, X., Tuo, D., Liu, Y., Zhou, P., Shen, W., et al. (2022). Rapid detection of sweepoviruses through lateral flow dipstick-based recombinase polymerase amplification. Acta Virol. 66, 186–191. doi: 10.4149/av_2022_208
Yoshikawa, H., Wu, Z., Kimata, I., Iseki, M., Ali, I. K. M., Hossain, M. B., et al. (2004). Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol. Res. 92, 22–29. doi: 10.1007/s00436-003-0995-2
Keywords: isothermal amplification, Blastocystis spp., lateral flow dipstick, visual detection, rapid detection
Citation: Mei X, Su C, Xin J, Jia L, Zhang S, Yang Z, Xiaowei T, Zhang Z and Wang S (2024) Recombinase polymerase amplification - lateral flow dipstick for rapid and visual detection of Blastocystis spp.. Front. Cell. Infect. Microbiol. 14:1391943. doi: 10.3389/fcimb.2024.1391943
Received: 26 February 2024; Accepted: 29 April 2024;
Published: 14 May 2024.
Edited by:
Stanislas Tomavo, Délégation Ile-de-France Sud (CNRS), FranceReviewed by:
Iraj Sharifi, Kerman University of Medical Sciences, IranJuan David Ospina-Villa, Colombian Institute of Tropical Medicine (ICMT), Colombia
Copyright © 2024 Mei, Su, Xin, Jia, Zhang, Yang, Xiaowei, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Wang, dG9uZ2JhaXdzMTAwM0AxNjMuY29t
 Xuefang Mei
Xuefang Mei Changwei Su1
Changwei Su1 Zhenke Yang
Zhenke Yang Tian Xiaowei
Tian Xiaowei Zhenchao Zhang
Zhenchao Zhang Shuai Wang
Shuai Wang