- 1Division of Infectious Disease, Far Eastern Memorial Hospital, New Taipei City, Taiwan
- 2Department of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan
- 3Department of Medical Laboratory Science and Biotechnology, Asia University, Taichung, Taiwan
- 4Department of Biotechnology and Laboratory Science in Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan
- 5Division of Infectious Diseases, Department of Medicine, Taipei Veterans General Hospital, Taipei, Taiwan
- 6School of Medicine, National Yang Chiao Tung University, Taipei, Taiwan
Introduction: The hemin acquisition system is composed of an outer membrane TonB-dependent transporter that internalizes hemin into the periplasm, periplasmic hemin-binding proteins to shuttle hemin, an inner membrane transporter that transports hemin into the cytoplasm, and cytoplasmic heme oxygenase to release iron. Fur and HemP are two known regulators involved in the regulation of hemin acquisition. The hemin acquisition system of Stenotrophomonas maltophilia is poorly understood, with the exception of HemA as a TonB-dependent transporter for hemin uptake.
Methods: Putative candidates responsible for hemin acquisition were selected via a homolog search and a whole-genome survey of S. maltophilia. Operon verification was performed by reverse transcription-polymerase chain reaction. The involvement of candidate genes in hemin acquisition was assessed using an in-frame deletion mutant construct and iron utilization assays. The transcript levels of candidate genes were determined using quantitative polymerase chain reaction.
Results: Smlt3896-hemU-exbB2-exbD2-tonB2 and tonB1-exbB1-exbD1a-exbD1b operons were selected as candidates for hemin acquisition. Compared with the parental strain, hemU and tonB1 mutants displayed a defect in their ability to use hemin as the sole iron source for growth. However, hemin utilization by the Smlt3896 and tonB2 mutants was comparable to that of the parental strain. HemA expression was repressed by Fur in iron-replete conditions and derepressed in iron-depleted conditions. HemP negatively regulated hemA expression. Like hemA, hemU was repressed by Fur in iron-replete conditions; however, hemU was moderately derepressed in response to iron-depleted stress and fully derepressed when hemin was present. Unlike hemA and hemU, the TonB1-exbB1-exbD1a-exbD1b operon was constitutively expressed, regardless of the iron level or the presence of hemin, and Fur and HemP had no influence on its expression.
Conclusion: HemA, HemU, and TonB1 contribute to hemin acquisition in S. maltophilia. Fur represses the expression of hemA and hemU in iron-replete conditions. HemA expression is regulated by low iron levels, and HemP acts as a negative regulator of this regulatory circuit. HemU expression is regulated by low iron and hemin levels in a hemP-dependent manner.
1 Introduction
Iron is an essential element for almost all bacteria because of its role as a cofactor for metabolic enzymes and components of the electron transport chain (Jordan and Reichard, 1998; Imlay, 2008; Roux et al., 2009). Owing to its low solubility, most ferric iron in mammalian hosts is bound by heme groups and other iron-binding proteins (Stojiljkovic and Perkins-Balding, 2002). Hemin is the most abundant source of iron for bacterial pathogens in their hosts (Choby and Skaar, 2016). To survive in infectious niches, pathogens have evolved several hemin acquisition systems that ensure iron availability. The hemin acquisition system is generally equipped with outer membrane TonB-dependent transporters (TBDTs) to internalize extracellular hemin into the periplasm, periplasmic proteins to shuttle hemin, inner membrane ABC transporters to transport hemin from the periplasm into the cytosol, and cytoplasmic proteins to provide ATP and degrade hemin (Wandersman and Delepelaire, 2004; Runyen-Janecky, 2013). Hemin uptake via TBDT is energy-dependent, and energy is transduced from the inner to the outer membrane using the TonB/ExbB/ExbD system (Krewulak and Vogel, 2011). Known hemin acquisition systems include hemPRST of Yersinia enterocolitica, hmuRSTUV of Yersinia pestis, phuRSTUV of Pseudomonas aeruginosa, shmR-hmuPSTUV of Ensifer meliloti, hmuR-hemP-hmuTUV of Bradyrhizobium japonicum, hemP-hmuRSTUV of Burkholderia multivorans, and hemP-hemA of S. maltophilia. A comparison of these hemin acquisition systems was summarized in a recent study (Shih et al., 2022). Generally, genes encoding TBDTs, periplasmic hemin transporter proteins, inner membrane permease proteins, and ATP-binding proteins are clustered or organized into an operon.
In most bacteria, ferric uptake regulator (Fur) is the master regulator of iron hemostasis (Troxell and Hassan, 2013). As a repressor, the Fur-Fe2+ complex binds to the consensus DNA sequence, designated as a Fur box, to exclude RNA polymerase binding, which blocks transcriptional initiation. When the intracellular ferrous iron level is too low to bind with Fur, Fur dissociates from the Fur box, triggering the expression of Fur-regulated genes. In addition to Fur, HemP/HmuP is another regulator known to be involved in hemin acquisition. The transcription of hemP/hmuP is repressed by Fur under iron-replete conditions. In iron-depleted conditions, HemP/HmuP is upregulated and functions as a transcription factor contributing to TBDT expression in alpha- and beta-proteobacteria (Sato et al., 2017).
Stenotrophomonas maltophilia is an opportunistic environmental bacterium that can cause broad-spectrum infections of the respiratory tract, soft tissue, urinary tract, eyes, and wounds. S. maltophilia infections are difficult to treat because the pathogen exhibits intrinsic and acquired resistance to a broad spectrum of antibiotics (Liu et al., 2023). To adapt to diverse environments, S. maltophilia is equipped with several iron acquisition systems for the utilization of ferric iron, ferrous iron, hemin, ferric citrate, and Pseudomonas aeruginosa pyochelin (Nas and Cianciotto, 2017; Kalidasan et al., 2018; Liao et al., 2022; Pan et al., 2022; Shih et al., 2022). Hemin is the most abundant source of iron for pathogens in the infection niche; hence, hemin acquisition is a critical determinant of pathogen virulence. The involvement of hemP-hemA-smlt0796-smlt0797 and Smlt2357-Smlt2356-Smlt2355 clusters (the homologs of hmuV, hmuU, and hmuT, respectively) in hemin acquisition was evaluated in a recent study (Shih et al., 2022). Of the seven genes, hemP and hemA are involved in hemin acquisition. smlt0796, smlt0797, smlt2357, smlt2356, and smlt2355 are not individually critical for hemin acquisition. HemA is a TBDT for hemin uptake. Distinct from the known HemP/HmuP in alpha- and beta-proteobacteria, HemP of S. maltophilia negatively regulates hemA expression in iron-depleted conditions (Shih et al., 2022). In this study, we aimed to further elucidate the other members, in addition to hemA and hemP, responsible for hemin acquisition in S. maltophilia.
2 Materials and methods
2.1 Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1.
2.2 Construction of in-frame deletion mutants
The double-crossover homologous recombination strategy was used to construct in-frame deletion mutants, as described previously (Yang et al., 2009). Briefly, two DNA fragments, upstream and downstream of the gene intended for deletion, were amplified from the chromosome of S. maltophilia KJ via polymerase chain reaction (PCR). The primer sets are listed in Supplementary Table S2. The two PCR amplicons were cloned into the pEX18Tc vector to generate the recombinant plasmids listed in Supplementary Table S1. The pEX18Tc-derived plasmids were introduced into S. maltophilia KJ or KJΔEnt via conjugation. Transconjugants were selected on LB plates containing 1.5 μg/mL norfloxacin and 30 μg/mL tetracycline. Then, the selected transconjugants were transferred to LB containing 10% sucrose for the selection of deletion mutant. The correctness of deletion mutants was checked by PCR and sequencing. Double mutant and triple mutant were constructed sequentially form the single mutant by the same procedure.
2.3 Reverse transcription-PCR and operon verification
An overnight culture of KJ cells was inoculated into fresh LB containing 50 μg/mL 2, 2’-dipyridyl (DIP) and 150 μM hemin at an initial OD450 of 0.15. After a 15-h incubation at 37°C, DNA-free RNA was prepared and reverse transcribed into cDNA using the primer ExbB2-C (Supplementary Table S2, Figure 1A). cDNA was used as the template for PCR using the primer sets 3896Q-F/R, HemUQ-F/R, ExbB2Q-F/R, and TonB2Q-F/R (Supplementary Table S2, Figure 1A). TonB2Q-F/R primer sets were used as a control for DNA contamination check.

Figure 1 Genetic organization and operon verification of the smlt3896-hemU-exbB2-exbD2-tonB2 cluster of Stenotrophomonas maltophilia. (A) Genetic organization of the hemO-hemU-exbB2-exbD2-tonB2 cluster of S. maltophilia. The orientation of the genes is indicated by arrows. The small black arrow indicates the locations of the ExbB2-C primer used for reverse transcription. The bars indicate the polymerase chain reaction (PCR) amplicons obtained using primer sets 3896Q-F/R (blue), HemUQ-F/R (green), ExbB2Q-F/R (red), and TonB2Q-F/R (purple). The DNA sequences encoding the N-termini of smlt3897 and smlt3896, and the intergenic region of smlt3897 and smlt3896 are placed in a rectangle. smlt3897 and smlt3896 genes are marked in orange and blue, respectively. The putative promoter of the smlt3896-hemU-exbB2-exbD2-tonB2 operon is underlined according to the prediction made using the tool at https://www.fruitfly.org/seq_tools/promoter.html. The putative Fur box is marked in yellow, based on the previously reported Fur box sequence. (B) Operon analysis of the smlt3896-hemU-exbB2-exbD2-tonB2 cluster. DNA-free RNA was prepared from logarithmical phase S. maltophilia KJ cells treated with 50 mg/mL 2, 2’-dipyridyl and 150 μM hemin. cDNAs were obtained by reverse transcription using the primer ExbB2-C and then used as the template for PCR. PCR amplicons were separated by agarose gel electrophoresis and stained with ethidium bromide. Lane 1, PCR amplicon generated by 3896Q-F and 3896Q-R; Lane 2, PCR amplicon generated by HemUQ-F and HemUQ-R; Lane 3, PCR amplicon generated by ExbB2Q-F and ExbB2Q-R; Lane 4, PCR amplicon generated by TonB2Q-F and TonB2Q-R (negative control).
2.4 Iron utilization assay
An overnight culture of bacteria was inoculated into fresh LB at an initial OD450 of 0.15. After a 5-h incubation at 37°C, logarithmic-phase bacterial cells were adjusted to 2 × 105 CFU/μL and then serially 10-fold diluted. A 5-µL bacterial aliquot was spotted onto Luria-Bertani (LB) agar containing different compounds as indicated. Cell viability was observed after 24 h of incubation at 37°C. The experiment was performed in triplicate.
2.5 Construction of complementation plasmids pHemU and pTonB1
The hemU and tonB1 genes were amplified from S. maltophilia KJ using the primer sets HemU-F/R and TonB1-F/R (Supplementary Table S1), and cloned into pRK415 to yield pHemU and pTonB1, respectively.
2.6 Quantitative real-time PCR
An overnight culture of KJ, KJΔFur, and KJΔHemP was inoculated into fresh LB with or without additives as indicated at an initial OD450 of 0.15. After a 5-h incubation at 37°C, DNA-free RNA was prepared and reverse transcribed to cDNA using random hexamers primers. hemA, hemU, and tonB1 transcript levels were then determined. The primers used are listed in Supplementary Table S2. 16S rRNA was used as an internal control for the normalization of gene expression levels. Fold change was calculated using the ΔΔCT method (Livak and Schmittgen, 2001).
2.7 Bioinformatics assay
A protein homolog search was carried out using the BLASTp tool from the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against S. maltophilia K279a genome (Accession no. AM74169.1). Protein sequences comparison was performed using the Global Align tool from the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The putative promoter was predicted using the website https://www.fruitfly.org/seq_tools/promoter.html.
2.8 Statistical analysis
Student’s t test was used for comparison of means between the groups. The Bonferroni correction method was applied to adjust the P values.
3 Results
3.1 smlt3896, hemU, exbB2, exbD2, and tonB2 form an operon
To elucidate the other candidates responsible for hemin acquisition, in silico analysis of the S. maltophilia K279a genome (Crossman et al., 2008) revealed a five-gene cluster, Smlt3896-Smlt3892, which, when combined, was predicted to encode a system for hemin acquisition (Figure 1A). Smlt3896 encodes a 195-aa cytosolic protein that is homologous to Pseudomonas aeruginosa PigA (Ratliff et al., 2001) (53% similarity, 41% identity), Pseudomonas aeruginosa BphO (Wegele et al., 2004) (40% similarity, 31% identity), Neisseria meningitidis HemO (Zhu et al., 2000) (49% similarity, 34% identity), and Corynebacterium diphtheriae HemO (Wilks and Schmitt, 1998) (39% similarity, 23% identity). PigA, BphO, and HemO are heme oxygenases that catalyze the degradation of hemin to produce biliverdin, ferrous iron, and carbon monoxide. The protein encoded by Smlt3895 was predicted to be a 155-aa inner membrane transmembrane protein and displayed no significant similarity with the proteins of known functions. The proteins encoded by Smlt3894, Smlt3893, and Smlt3892 were annotated as ExbB, ExbD, and TonB, respectively. The ExbB-ExbD-TonB complex is an energy transducer embedded in the inner membrane that provides ATP to support the function of TonB-dependent outer membrane proteins (Koebnik, 2005). Recently, a tonB gene (Smlt0009) was identified in S. maltophilia and characterized by Calvopiña et al (Calvopiña et al., 2020). Furthermore, the inner membrane permeases for known hemin acquisition systems in other bacteria are annotated as HemU, HmuU, or PhuU (Shih et al., 2022). Based on the following results and to avoid confusion with Smlt0009, we designated the five genes smlt3896, hemU, exbB2, exbD2, and tonB2. A short intergenic region (31 bp) between smlt3896 and hemU, and a 29-bp overlap between hemU and exbB2 were noticed (Figure 1A). A promoter prediction tool (https://www.fruitfly.org/seq_tools/promoter) demonstrated that a putative promoter located upstream of smlt3896, but not upstream of hemU and exbB2. Furthermore, exbB, exbD, and tonB are generally organized as an operon (Ratliff et al., 2022). Thus, we speculated that smlt3896, hemU, exbB2, exbD2, and tonB2 form an operon. We used revers transcription (RT) to investigate whether smlt3896, hemU, and exbB2 were co-transcribed. Considering that hemin acquisition-associated genes are generally expressed under iron-depleted conditions, we prepared DNA-free RNA from the DIP-treated KJ cells, and RT was carried out using the primer ExbB2-C. No positive results were obtained; therefore, we sought to determine whether hemin is a crucial inducer of expression of the five-gene cluster. DNA-free RNA prepared from KJ cells treated with 50 μg/mL DIP and 150 μM hemin was used as a template for operon verification. The results demonstrated that smlt3896, hemU, and exbB2 locate in a single transcript (Figure 1B) and they can be part of the smlt3896-hemU-exbB2-exbD2-tonB2 operon.
3.2 HemU contributes to hemin utilization
To eliminate the growth bias caused by stenobactin-mediated iron acquisition in iron-depleted media, KJΔEnt, a stenobactin-null mutant (Pan et al., 2022), was chosen as the parental strain for the following experiments. An array of in-frame deletion mutants was constructed using KJΔEnt to generate KJΔEntΔ3896, KJΔEntΔHemU, and KJΔEntΔTonB2. The resulting mutants and their parental strain, KJΔEnt, were subjected to an iron utilization assay, using hemin as the sole iron source. Of the three mutants tested, only KJΔEntΔHemU lost viability in DIP- and hemin-containing medium (Figure 2, Supplementary Figure S1A), even though the incubation time was prolonged to 48 h. To assess whether hemU deletion causes a polar effect on the expression of downstream genes, the tonB2 transcript levels in KJΔEnt and KJΔEntΔHemU were determined by qRT-PCR. The results demonstrated that KJΔEnt and KJΔEntΔHemU exhibited comparable tonB2 transcript levels. Furthermore, a hemU complementation strain KJΔEntΔHemU(pHemU) restored the ability to use hemin as an iron source for growth (Figure 2, Supplementary Figure S1B). Collectively, HemU contributes to hemin utilization.
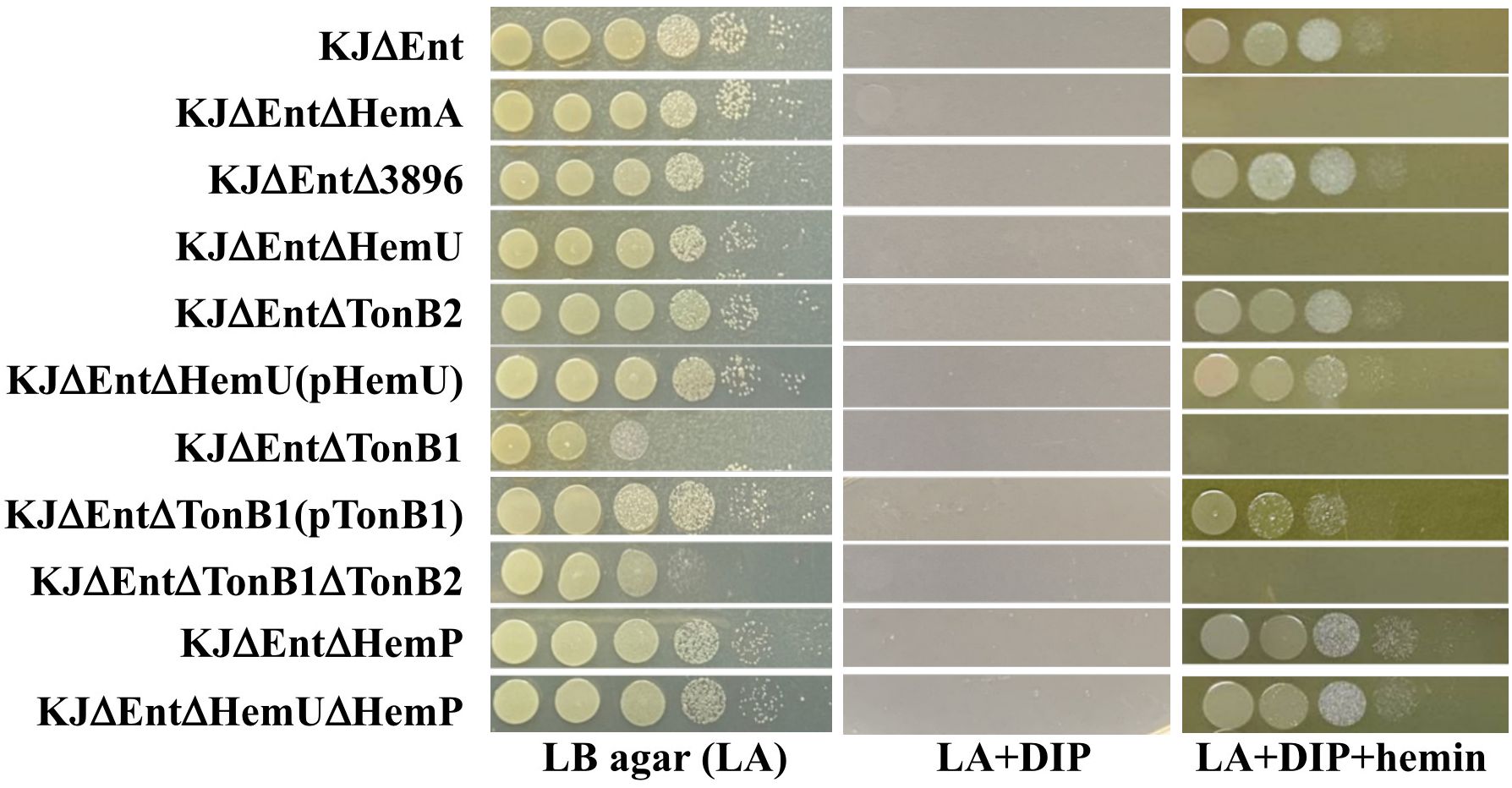
Figure 2 Cell viability of parental strain KJΔEnt, its derived mutants, and complementation strains under iron-replete (LA), iron-depleted (LA + DIP), and iron-depleted with hemin as sole iron source (LA + DIP + hemin). Bacterial cells (2 × 105 CFU/μL) were 10-fold serially diluted and a 5-μL aliquot was spotted onto Luria-Bertani (LB) agar as indicated. After 24 h of incubation at 37°C, the bacteria were imaged to assess their viability. The experiment was performed in triplicate, and one was selected as a representative. 2, 2’-dipyridyl (DIP), 50 μg/mL; hemin, 150 μM.
In addition to hemin, we considered the involvement of the smlt3896-hemU-exbB2-exbD2-tonB2 operon in ferric citrate and ferristenobactin utilization. These results tentatively ruled out the contribution of the smlt3896-hemU-exbB2-exbD2-tonB2 operon to ferric citrate and ferri-stenobactin utilization (Supplementary Figure S2).
3.3 TonB1 (Smlt0009) contributes to hemin utilization
A more interesting finding from the above results (Figure 2) was that exbB2, exbD2, and tonB2 do not contribute to hemin acquisition, although they and hemU are in the same operon (Figure 1). Considering this functional redundancy, we tested whether other TonB systems were involved in hemin acquisition. Another tonB-associated cluster, Smlt0009-Smlt0012, was reported by Calvopina et al (Calvopiña et al., 2020). One peculiar observation was the presence of two exbD genes in the four-gene cluster. We annotated the four genes as tonB1, exbB1, exbD1a, and exbD1b (Supplementary Figure S3). The protein sequence identities and similarities between the two exbB-exbD-tonB clusters are summarized in Supplementary Figure S3. To assess the role of TonB1 in hemin acquisition, tonB1 deletion mutant and tonB1/tonB2 double mutants were generated in KJΔEnt to yield KJΔEntΔTonB1 and KJΔEntΔTonB1ΔTonB2, respectively. KJΔEntΔTonB1 displayed compromised growth on LB agar, which is consistent with previous observations (Calvopiña et al., 2020). Furthermore, KJΔEntΔTonB1 and KJΔEntΔTonB1ΔTonB2 showed no visible viability in DIP- and hemin-containing media (Figure 2, Supplementary Figure S1C). Complementation of KJΔTonB1 with an intact tonB1 gene partially restored the ability to utilize hemin for growth in iron-depleted conditions (Figure 2, Supplementary Figure S1C). Collectively, TonB1 contributes to hemin utilization.
Based on the above results, tonB1, but not tonB2, contributed to hemin utilization (Figure 2). We wondered whether the deletion of tonB2 was complemented by an increase in tonB1 expression. To test this notion, the tonB1 transcript levels in KJΔEnt and KJΔEntΔTonB2 were determined by qRT-PCR. A comparable tonB1 transcript level was observed in KJΔEnt and KJΔEntΔTonB2.
3.4 Regulation of smlt3896-hemU-exbB2-exbD2-tonB2 and tonB1-exbB1-exbD1a-exbD1b operons
The contributions of hemA and hemP to hemin acquisition have previously been reported (Shih et al., 2022). The above results support the involvement of hemU and tonB1 in hemin acquisition (Figure 2). Fur and HemP are two known regulators participating in hemin acquisition; thus, we were interested in investigating the regulatory roles of Fur and HemP in the expression of tonB1-exbB1-exbD1a-exbD1b and smlt3896-hemU-exbB2-exbD2-tonB2 operons. For convenience, the expression of the hemA-hemP-Smlt0796-Smlt0797 operon, described in our previous study (Shih et al., 2022), is also shown in Figure 3 for comparison.
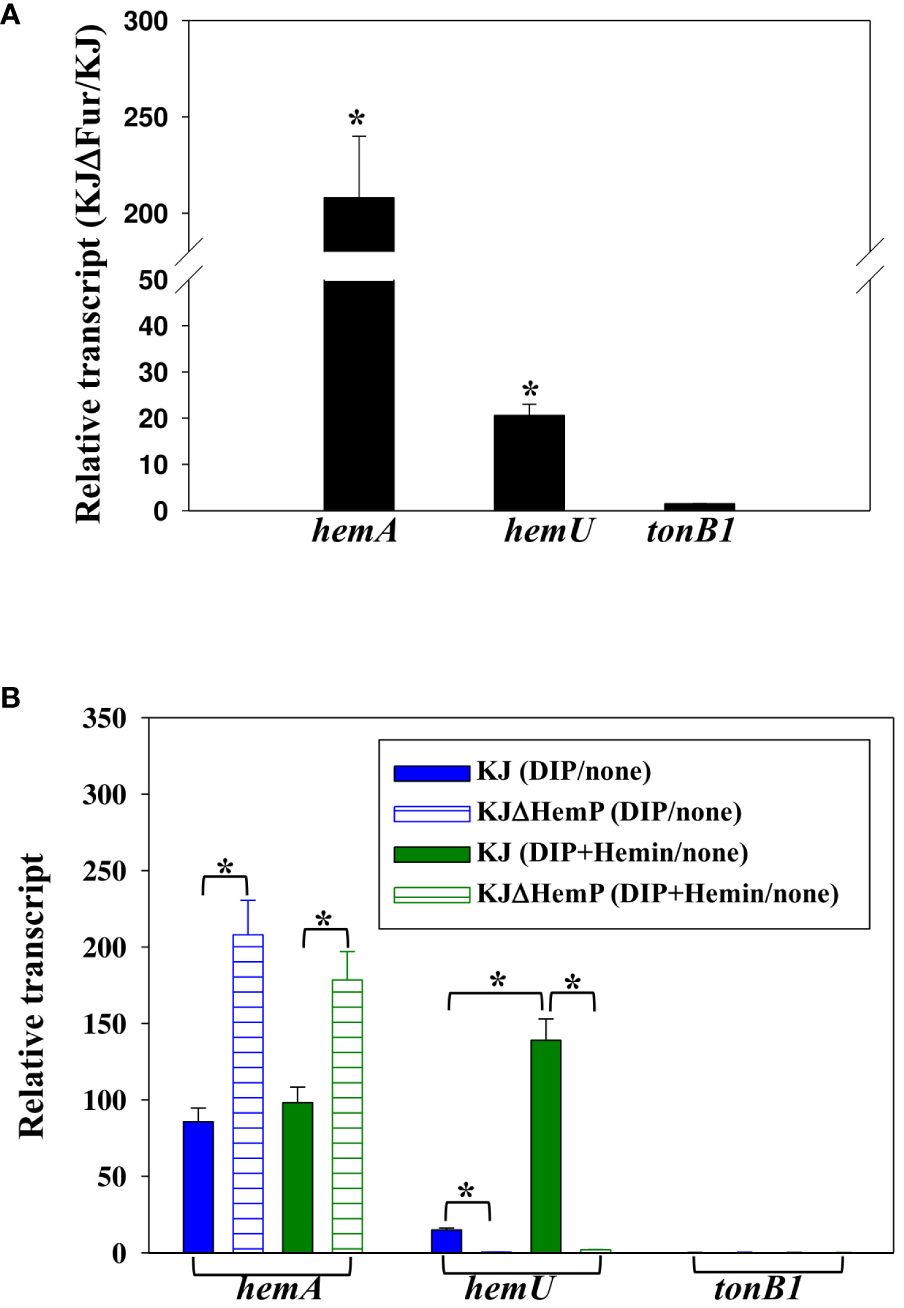
Figure 3 Regulation of hemA, hemU, and tonB1 expression. Data are the means from three independent experiments. The error bars represent the standard deviations for triplicate samples. *, P ≤ 0.05 (significance calculated by Student’s t test). (A) Role of Fur in the expression of hemA, hemU, and tonB1. KJ and KJΔFur were cultured overnight and inoculated into fresh LB medium at an initial OD450 of 0.15. After a 15-h incubation, the hemA, hemU, and tonB1 transcript levels were determined by quantitative RT-PCR. The relative transcript levels were calculated based on the transcript level of KJ cells assigned as 1. (B) Impact of HemP, iron level, and hemin on the expression of hemA, hemU, and tonB1. KJ and KJΔHemP cells were cultured overnight and inoculated into fresh LB medium with or without additives as indicated. After 15 h of incubation, hemA, hemU, and tonB1 transcript levels were determined by quantitative RT-PCR. For DIP treatment, the DIP concentration was 30 μg/mL. For DIP and hemin treatment, the concentrations of DIP and hemin added were 50 μg/mL and 150 μM, respectively. The relative transcript levels were calculated based on the transcript level in the counterpart without additives assigned as 1.
First, we sought to determine whether a putative Fur box is present upstream of the three operons, and positive results were found in the hemA-hemP-Smlt0796-Smlt0797 and smlt3896-hemU-exbB2-exbD2-tonB2 operons (Shih et al., 2022). To further confirm the regulatory role of Fur in the expression of the three operons, the hemA, hemU, and tonB1 transcript levels of KJ and KJΔFur, a fur deletion mutant (Liao et al., 2020), were determined by quantitative RT-PCR. As expected, the levels of hemA and hemU transcripts, but not the tonB1 transcript, were increased in KJΔFur (Figure 3A), verifying the repressor role of Fur in the expression of the hemA-hemP-Smlt0796-Smlt0797 and smlt3896-hemU-exbB2-exbD2-tonB2 operons.
In a recent study, we verified that the transcription factor HemP negatively regulates the expression of hemA (Shih et al., 2022). To investigate whether the three operons were regulated by HemP, iron levels, and hemin, the hemA, hemU, and tonB1 transcripts of KJ and KJΔHemP, with or without DIP and hemin, were determined by quantitative RT-PCR. The hemA transcript of wild-type KJ showed an approximately 90-fold increase in its DIP- and DIP/hemin-additive counterparts (Figure 3B). Furthermore, the upregulation level of the hemA transcript was more significant in the KJΔHemP group (Figure 3B). These observations indicated that the hemA-hemP-Smlt0796-Smlt0797 operon was repressed by Fur in iron-replete conditions and derepressed in response to low iron levels, regardless of the presence of hemin. HemP functions as a negative transcription factor for the expression of the hemA-hemP-Smlt0796-Smlt0797 operon.
Unlike hemA expression, hemU transcript levels in wild-type KJ showed an approximately 15-fold increase upon DIP challenge and a 139-fold increase in the presence of DIP and hemin (Figure 3B), indicating that hemin is a crucial determinant of the full induction of the smlt3896-hemU-exbB2-exbD2-tonB2 operon. This observation provides a reasonable explanation for the detection of the smlt3896-hemU-exbB2-exbD2-tonB2 transcript in DIP- and hemin-treated KJ cells, but not in DIP-treated KJ cells (Figure 1B). In addition, we observed that the DIP- and hemin-mediated upregulation of hemU was not observed in KJΔHemP cells (Figure 3B), indicating that the expression of the smlt3896-hemU-exbB2-exbD2-tonB2 is dependent on HemP.
A similar strategy was used to study the expression of the tonB1-exbB1-exbD1a-exbD1b operon. Of note, the tonB1-exbB1-exbD1a-exbD1b operon was intrinsically highly expressed and its expression level was not affected by Fur, HemP, iron levels, or hemin.
An interesting observation attracted our attention. Based on the findings in this study, HemU contributed to hemin utilization (Figure 2) and its expression was HemP-dependent (Figure 3B). Expectedly, a hemP mutant should be defective in hemin utilization in iron-depleted conditions. However, in our previous study, we showed that hemP deletion has no impact on hemin utilization (Shih et al., 2022, Figure 2, Supplementary Figure S1D). To elucidate the contradiction, we constructed a hemU and hemP mutant in a stenobactin-null background, KJΔEntΔHemUΔHemP, for hemin utilization evaluation. Interestingly, KJΔEntΔHemU lost the ability of hemin utilization in iron-depleted condition, but KJΔEntΔHemUΔHemP reverted this ability (Figure 2, Supplementary Figure S1D), indicating that a hemU-independent mechanism responsible for hemin utilization in hemP mutant.
4 Discussion
Hemin is an important source of iron in infection niches, and it is utilized by most pathogens. Extracellular hemin can be directly captured by pathogens and transported into the cytoplasm via the outer membrane TBDT, periplasmic transport proteins, and inner membrane transporters. Once hemin is located in the cytoplasm, it must be released for further utilization by the pathogen. Heme oxygenase (HemO) is an enzyme involved in this process. Heme oxygenase catalyzes the NADPH-reductase-dependent cleavage of hemin into biliverdin, carbon monoxide, and free iron (Frankenberg-Dinkel, 2004; Wilks and Burkhard, 2007). Heme oxygenases have been identified in several pathogenic bacteria, including Corynebacterium spp. HmuO (Schmitt, 1997; Wilks and Schmitt, 1998), Neisseria meningitidis HemO (Zhu et al., 2000), P. aeruginosa PigA and BphO (Ratliff et al., 2001; Wegele et al., 2004), E. coli ChuS (Suits et al., 2005), Helicobacter pylori HugZ (Guo et al., 2008), Campylobacter jejuni ChuZ (Zhang et al., 2011), and Mycobacterium tuberculosis MhuD (Nambu et al., 2013). However, the protein sequence identities among these heme oxygenases are not high. Phylogenetic analysis of Smlt3896 and the known HemO proteins of other bacteria revealed that Smlt3896 phylogenetically clustered with N. meningitidis HemO and P. aeruginosa PigA (Figure 4). We found that KJΔEntΔ3896 was able to grow using hemin as an iron source (Figure 2), even though Smlt3896 was annotated as hemO in the sequenced K279a genome and was phylogenetically close to known HemO proteins (Figure 4). This observation led us to speculate that heme oxygenase redundancy may exist in S. maltophilia. P. aeruginosa was the first organism known to encode two functional heme oxygenases, PigA and BphO, each capable of oxidizing heme to a different biliverdin isomer. PigA oxidizes heme into a mixture of β- and δ-biliverdin (Caignan et al., 2002), whereas BphO favors the formation of α-biliverdin (Wegele et al., 2004), similarly to the heme oxygenases of Neisseria meningitidis (HemO) (Zhu et al., 2000). The protein identity between PigA and BphO is as low as 24%. Of the two heme oxygenases, PigA plays a major role in iron supply (Ratliff et al., 2001). Based on this rationale, we attempted to survey the putative heme oxygenase in S. maltophilia using the known heme oxygenases of other bacteria, listed in Figure 4, as queries; however, no positive results were obtained. Thus, the exact mechanism responsible for the release of iron from hemin in S. maltophilia requires further investigation.
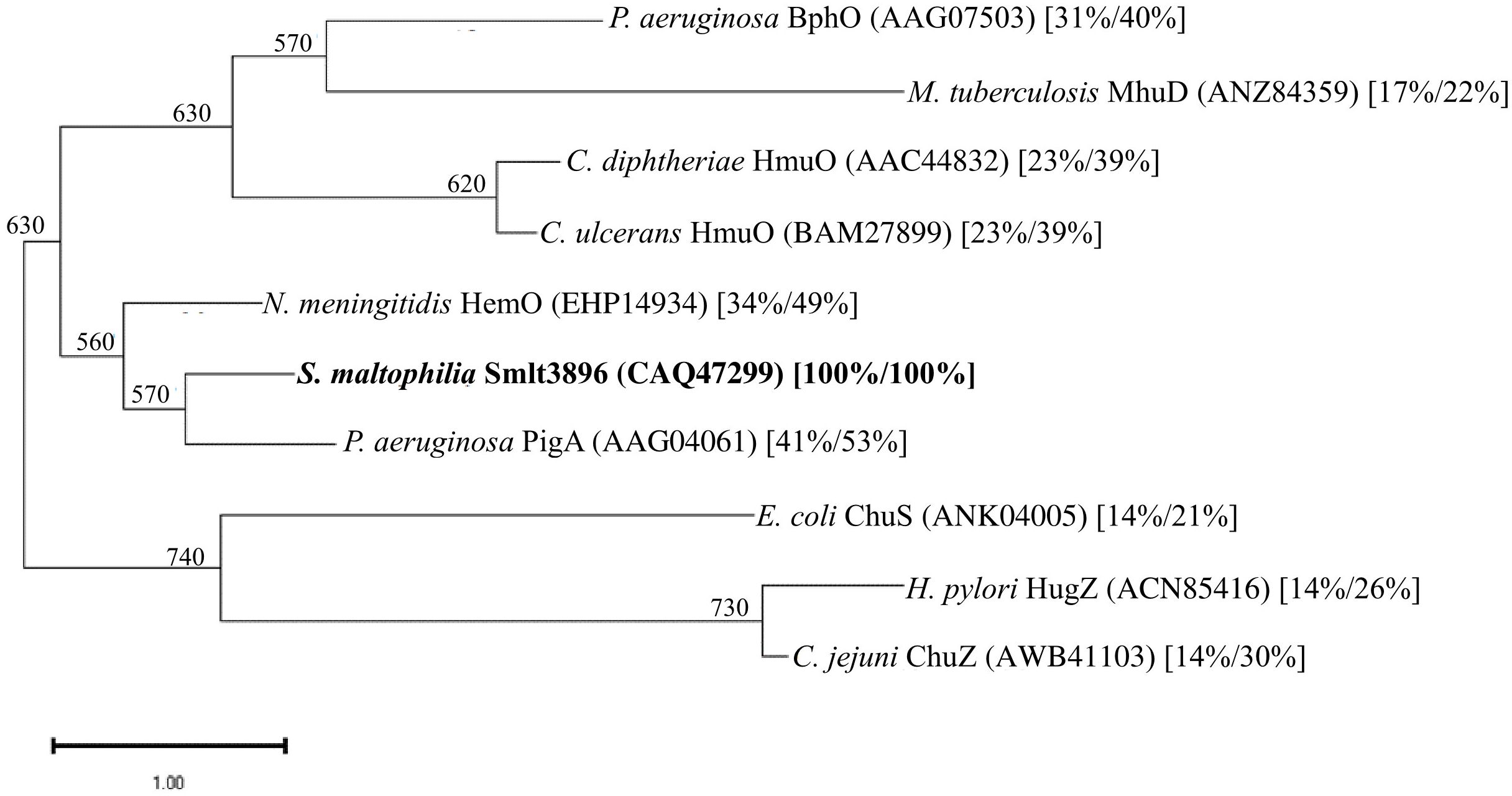
Figure 4 Phylogenetic analysis of S. maltophilia Smlt3896 and known HemO proteins of other bacteria. A dendrogram was constructed with the amino acid sequences of the proteins using the neighbor-joining method. The numbers below the branches indicate the number of bootstraps, which were calculated from 1,000 replicates. The protein accession numbers are included in brackets. The numbers in square brackets are the protein identity and similarity values compared with Smlt3896.
Iron complex uptake via the outer membrane TBDT is an energy-dependent process that relies on the inner membrane TonB-ExbB-ExbD complex. TonB-ExbB-ExbD uses a proton gradient across the inner membrane as an energy source to transport the iron complex from the extracellular environment to the periplasm (Torres and Payne, 1997). TonB is a well-structured protein with an N-terminal transmembrane helix anchored to the inner membrane, a proline-rich periplasmic domain, and a C-terminal domain that interacts with the TonB box of TBDTs (Kohler et al., 2010). The number of tonB genes varies significantly among different species, being as low as one in E. coli and up to 15 homologs in Bacteroides thetaiotaomicron (Chu et al., 2007). High sequence diversity has been observed across the characterized TonB proteins, even within the same species, suggesting that each TonB protein functions differently. Nevertheless, functional redundancy has also been reported in TonBs (Liao et al., 2015). Thus, multiple TonB proteins in an organism can function in redundant or unique manners. A genome-wide survey of the S. maltophilia K279a genome revealed that S. maltophilia harbors 10 annotated tonB genes (Crossman et al., 2008). Among the 10 tonB homologs, only tonB1 (Smlt0009) and tonB2 (Smlt3892) were linked to and in the same transcriptional orientation as exbB and exbD (Supplementary Figure S2). The only previous study of S. maltophilia TonBs showed linkage between TonB1 and β-lactam susceptibility (Calvopiña et al., 2020). In this study, we investigated the involvement of TonB1 and TonB2 in hemin acquisition and found that TonB1 predominantly contributed to hemin acquisition. Although tonB2 and hemU are organized into an operon, our results do not support the involvement of TonB2 in hemin acquisition.
Of note, the 185-aa TonB1 protein has a relatively short N-terminus. Protein alignment between E. coli TonB and S. maltophilia TonB1 revealed that the putative hydrophobic transmembrane region of E. coli TonB (amino acid residues 12–32) (Jaskula et al., 1994), is absent in S. maltotphilia TonB1 (Figure 5). The N-terminus 1–39 residues of E. coli TonB are essential for its ability to transduce energy (Jaskula et al., 1994); however, this feature has not been observed in S. maltophilia TonB1. Thus, the N-terminal membrane anchor of TonB may not be the only mechanism by which TonB associates with the cytoplasmic membrane. In contrast, the YP motif is the most conserved feature among TonB proteins (Chu et al., 2007) and is conserved in E. coli TonB and S. maltophilia TonB1 (Figure 5).

Figure 5 Protein sequence alignment of Esherichia coli TonB and S. maltophilia TonB1. Protein sequence alignment was carried out using Needleman-Wunsch global alignment using sequences in the National Center for Biotechnology Information database. The putative hydrophobic transmembrane region of E. coli TonB, amino acid residues 12–32, is marked in yellow. The most conserved YP motif among the TonB proteins is marked in blue. The protein accession numbers for E. coli TonB and S. maltophilia TonB1 are EOW16097 and CAQ43627, respectively.
Distinct from the known hemin permeases reported in other bacteria, such as HemU of Y. enterocolitica, PhuU of P. aeruginosa, and HmuUs of B. japonicum, E. meliloti, and B. multivorans (Shih et al., 2022), HemU of S. maltophilia owns some peculiar features. (i) Compared to the other hemin permeases of 326-372 amino acids, HemU of S. maltophilia is relative shorter (155 amino acids) and displays no significant protein similarity with the known hemin permease. S. maltophilia HemU can be a novel type of hemin permease. (ii) The role of HemU in hemin utilization is HemP dependent. In a hemP mutant, there is an unidentified HemU-independent iron utilization mechanism.
The hemin acquisition and regulatory mechanisms in S. maltophilia are shown in Figure 6. Under iron-depleted conditions, hemin is loaded onto the outer membrane of TBDT HemA, followed by subsequent delivery into the periplasmic space in an energy-dependent manner. This energy for this process is provided by the TonB1-ExbB1-ExbD1a-ExbD1b complex. Hemin is then transported into the cytoplasm via the inner membrane transporter HemU. The genes involved in hemin acquisition are in three operons, hemP-hemA-Smlt0796-Smlt0797, Smlt3896-hemU-exbB2-exbD2-tonB2, and tonB1-exbB1-exbD1a-exbD1b. There is precedence for the coordination of the expression of these three operons. The tonB1-exbB1-exbD1a-exbD1b operon was constitutively expressed regardless of iron levels (Figures 6A–C), indicating that this operon may be involved in other physiological functions in addition to hemin acquisition. The involvement of TonB proteins in distinct functions other than iron acquisition has been reported, such as the involvement of P. aeruginosa TonB3 in twitching motility and type IV pili assembly (Huang et al., 2004), S. maltophilia TonB1 in β-lactam susceptibility (Calvopiña et al., 2020), and Anabaena sp. TonB2 in outer membrane integrity (Schätzle et al., 2021). The hemP-hemA-Smlt0796-Smlt0797 operon is repressed by Fur in iron-replete conditions (Figure 6A). This operon was highly upregulated in response to iron-depletion stress, and HemP played a negative role in this regulatory circuit (Figure 6B). However, hemin did not significantly affect the expression of the hemP-hemA-Smlt0796-Smlt0797 operon (Figure 6C). The expression of the Smlt3896-hemU-exbB2-exbD2-tonB2 operon was regulated in a stepwise manner by Fur and HemP. In iron-rich situations, the Smlt3896-hemU-exbB2-exbD2-tonB2 operon was repressed by Fur (Figure 6A). In response to iron-depleted stress, the Smlt3896-hemU-exbB2-exbD2-tonB2 operon displays a moderate level of upregulation due to the loss-of-function of Fur and can be further fully expressed when hemin is present (Figures 6B, C). Compared with other known hemin acquisition systems (Shih et al., 2022), the periplasmic-binding proteins and HemO-like iron-releasing enzymes in S. maltophilia require further elucidation.
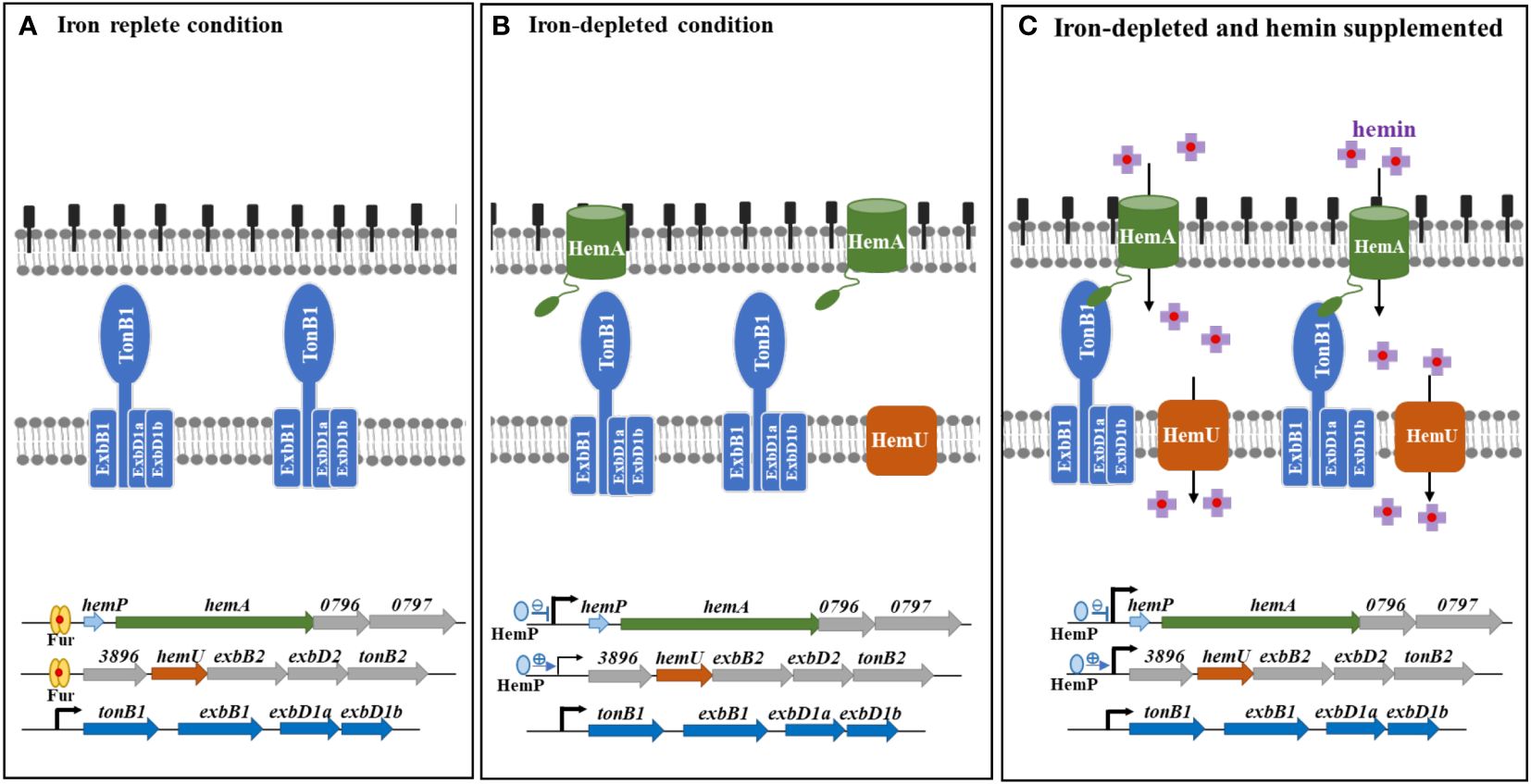
Figure 6 The proposed model for hemin acquisition by S. maltophilia. (A) In iron-replete conditions, hemP-hemA-Smlt0796-Smlt0797 and Smlt3896-hemU-exbB2-exbD2-tonB2 operons are repressed by Fur. Nevertheless, tonB1-exbB1-exbD1a-exbD1b operon is constitutively expressed. (B) In iron-depleted conditions, hemP-hemA-Smlt0796-Smlt0797 and Smlt3896-hemU-exbB2-exbD2-tonB2 operons are derepressed due to the loss-of-function of Fur. Meanwhile, HemP has negative and positive roles in the regulation of hemP-hemA-Smlt0796-Smlt0797 and Smlt3896-hemU-exbB2-exbD2-tonB2 operons, respectively. (C) In iron-depleted and hemin-supplemented condition, Smlt3896-hemU-exbB2-exbD2-tonB2 operon is further upregulated. Hemin is taken up via the outer membrane HemA receptor and TonB1-ExbB1-ExbD1a-ExbD1b provides the energy. Hemin is then transported into the cytoplasm via the inner membrane transporter HemU.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author contributions
C-HL: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – original draft, Writing – review & editing. H-FL: Data curation, Formal analysis, Methodology, Validation, Writing – review & editing. C-WY: Data curation, Formal analysis, Methodology, Writing – review & editing. T-YY: Data curation, Formal analysis, Methodology, Writing – review & editing. Y-TL: Data curation, Formal analysis, Methodology, Writing – review & editing. T-CY: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Science and Technology Council of Taiwan (grant numbers MOST 111-2320-B-A49-025-MY3 and NSTC 112-2320-B-A49-043-MY3) and the National Yang Ming Chiao Tung University Far Eastern Memorial Hospital Joint Research Program (grant number 113DN23).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1380976/full#supplementary-material
References
Caignan, G. A., Deshmukh, R., Wilks, A., Zeng, Y., Huang, H. W., Moënne-Loccoz, P., et al. (2002). Oxidation of heme to beta- and delta-biliverdin by Pseudomonas aeruginosa heme oxygenase as a consequence of an unusual seating of the heme. J. Am. Chem. Soc 124, 14879–14892. doi: 10.1021/ja0274960
Calvopiña, K., Dulyayangkul, P., Heesom, K. J., Avison, M. B. (2020). TonB-dependent uptake of β-lactam antibiotics in the opportunistic human pathogen Stenotrophomonas maltophilia. Mol. Microbiol. 113, 492–503. doi: 10.1111/mmi.14434.
Choby, J. E., Skaar, E. P. (2016). Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 428, 3408–3428. doi: 10.1016/j.jmb.2016.03.018
Chu, B. C. H., Peacock, R. S., Vogel, H. J. (2007). Bioinformatic analysis of the TonB protein family. Biometals 20, 467–483. doi: 10.1007/s10534-006-9049-4
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9, R74. doi: 10.1186/gb-2008-9-4-r74
Frankenberg-Dinkel, N. (2004). Bacterial heme oxygenases. Antioxid. Redox Signal. 6, 825–834. doi: 10.1089/ars.2004.6.825.
Guo, Y., Guo, G., Mao, X., Zhang, W., Xiao, J., Tong, W., et al. (2008). Functional identification of HugZ, a heme oxygenase from Helicobacter pylori. BMC Microbiol. 8, 226. doi: 10.1186/1471-2180-8-226
Huang, B., Ru, K., Yuan, Z., Whitchurch, C. B., Mattick, J. S. (2004). TonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J. Bacteriol. 186, 4387–4389. doi: 10.1128/JB.186.13.4387-4389.2004
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Ann. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
Jaskula, J. C., Letain, T. E., Roof, S. K., Skare, J. T., Postle, K. (1994). Role of TonB amino terminus in energy transduction between membranes. J. Bacteriol. 176, 2326–2338. doi: 10.1128/jb.176.8.2326-2338.1994
Jordan, A., Reichard, P. (1998). Ribonucleotide reductases. Ann. Rev. Biochem. 67, 71–98. doi: 10.1146/annurev.biochem.67.1.71
Kalidasan, V., Azman, A., Joseph, N., Kumar, S., Awang Hamat, R., Neela, V. K. (2018). Putative iron acquisition systems in. Molecules 23, 2048. doi: 10.3390/molecules23082048.
Koebnik, R. (2005). TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13, 343–347. doi: 10.1016/j.tim.2005.06.005.
Kohler, S. D., Weber, A., Howard, S. P., Welte, W., Drescher, M. (2010). The proline-rich domain of TonB possesses an extended polyproline II-like conformation of sufficient length to span the periplasm of gram-negative bacteria. Protein Sci. 19, 625–630. doi: 10.1002/pro.345.
Krewulak, K. D., Vogel, H. J. (2011). TonB or not TonB: is that the question. Biochem. Cell Biol. 89, 87–97. doi: 10.1139/O10-141
Liao, C. H., Chen, W. C., Li, L. H., Lin, Y. T., Pan, S. Y., Yang, T. C. (2020). AmpR of Stenotrophomonas maltophilia is involved in stenobactin synthesis and enhanced β-lactam resistance in an iron-depleted condition. J. Antimicrob. Chemother. 75, 3544–3551. doi: 10.1093/jac/dkaa358
Liao, H., Cheng, X., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2015). TonB energy transduction systems of Riemerella anatipestifer are required for iron and hemin utilization. PloS One 10, e0127506. doi: 10.1371/journal.pone.0127506
Liao, C. H., Lu, H. F., Huang, H. H., Chen, Y., Li, L. H., Lin, Y. T., et al. (2022). The fciTABC and feoABI systems contribute to ferric citrate acquisition in Stenotrophomonas maltophilia. J. Biomed. Sci. 29, 26. doi: 10.1186/s12929-022-00809-y
Liu, J., Xiang, Y., Zhang, Y. (2023). Stenotrophomonas maltophilia: an urgent threat with increasing antibiotic resistance. Curr. Microbiol. 81, 6. doi: 10.1007/s00284-023-03524-5
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Nambu, S., Matsui, T., Goulding, C. W., Takahashi, S., Ikeda-Saito, M. (2013). A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J. Biol. Chem. 288, 10101–10109. doi: 10.1074/jbc.M112.448399
Nas, M. Y., Cianciotto, N. P. (2017). Stenotrophomonas maltophilia produces an EntC-dependent catecholate siderophore that is distinct from enterobactin. Microbiol. (Reading). 163, 1590–1603. doi: 10.1099/mic.0.000545
Pan, S. Y., Shih, Y. L., Huang, H. H., Li, L. H., Lin, Y. T., Yang, T. C. (2022). The involvement of PacIRA system of Stenotrophomonas maltophilia in the uptake of Pseudomonas aeruginosa pyochelin and intraspecies competition for iron acquisition. J. Microbiol. Immunol. Infect. 55, 273–281. doi: 10.1016/j.jmii.2021.03.001
Ratliff, A. C., Buchanan, S. K., Celia, H. (2022). The ton motor. Front. Microbiol. 13, 852955. doi: 10.3389/fmicb.2022.852955
Ratliff, M., Zhu, W., Deshmukh, R., Wilks, A., Stojiljkovic, I. (2001). Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001
Roux, A., Payne, S. M., Gilmore, M. S. (2009). Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe 6, 115–124. doi: 10.1016/j.chom.2009.07.004
Runyen-Janecky, L. J. (2013). Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol. 3, 55. doi: 10.3389/fcimb.2013.00055
Sato, T., Nonoyama, S., Kimura, A., Nagata, Y., Ohtsubo, Y., Tsuda, M. (2017). The small protein HemP is a transcriptional activator for the hemin uptake operon in Burkholderia multivorans ATCC 17616. Appl. Environ. Microbiol. 83, e00479–e00417. doi: 10.1128/AEM.00479-17
Schätzle, H., Arévalo, S., Fresenborg, L., Seitz, H. M., Flores, E., Schleiff, E. (2021). Functional diversity of TonB-like proteins in the heterocyst-forming Cyanobacterium Anabaena sp. PCC 7120. mSphere 6, e0021421. doi: 10.1128/mSphere.00214-21.
Schmitt, M. P. (1997). Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179, 838–845. doi: 10.1128/jb.179.3.838-845.1997
Shih, Y. L., Wu, C. M., Lu, H. F., Li, L. H., Lin, Y. T., Yang, T. C. (2022). Involvement of the hemP-hemA-smlt0796-smlt0797 operon in hemin acquisition by Stenotrophomonas maltophilia. Microbiol. Spectr. 10, e0032122. doi: 10.1128/spectrum.00321-22
Stojiljkovic, I., Perkins-Balding, D. (2002). Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21, 281–295. doi: 10.1089/104454902753759708
Suits, M. D., Pal, G. P., Nakatsu, K., Matte, A., Cygler, M., Jia, Z. (2005). Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc. Natl. Acad. Sci. U.S.A. 102, 16955–16960. doi: 10.1073/pnas.0504289102
Torres, A. G., Payne, S. M. (1997). Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23, 825–833. doi: 10.1046/j.1365-2958.1997.2641628.x
Troxell, B., Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3, 59. doi: 10.3389/fcimb.2013.00059
Wandersman, C., Delepelaire, P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58, 611–647. doi: 10.1146/annurev.micro.58.030603.123811
Wegele, R., Tasler, R., Zeng, Y., Rivera, M., Frankenberg-Dinkel, N. (2004). The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802. doi: 10.1074/jbc.M408303200
Wilks, A., Burkhard, K. A. (2007). Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat. Prod. Rep. 24, 511–522. doi: 10.1039/b604193k
Wilks, A., Schmitt, M. P. (1998). Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae: iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273, 837–841. doi: 10.1074/jbc.273.2.837
Yang, T. C., Huang, Y. W., Hu, R. M., Huang, S. C., Lin, Y. T. (2009). AmpDI is involved in expression of the chromosomal L1 and L2 beta-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 53, 2902–2907. doi: 10.1128/AAC.01513-08
Zhang, R., Zhang, J., Ding, H., Lu, D., Hu, Y., Wang, D. C., et al. (2011). Crystallization and preliminary crystallographic studies of Campylobacter jejuni ChuZ, a member of a novel haem oxygenase family. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 67, 1228–1230. doi: 10.1107/S1744309111026194
Keywords: Stenotrophomonas maltophilia, hemin acquisition, ExbB-ExbD-TonB complex, HemU, Fur, HemP
Citation: Liao C-H, Lu H-F, Yang C-W, Yeh T-Y, Lin Y-T and Yang T-C (2024) HemU and TonB1 contribute to hemin acquisition in Stenotrophomonas maltophilia. Front. Cell. Infect. Microbiol. 14:1380976. doi: 10.3389/fcimb.2024.1380976
Received: 02 February 2024; Accepted: 12 March 2024;
Published: 26 March 2024.
Edited by:
Yinduo Ji, University of Minnesota Twin Cities, United StatesReviewed by:
Michał Śmiga, University of Wrocław, PolandAvishek Mitra, Oklahoma State University, United States
Copyright © 2024 Liao, Lu, Yang, Yeh, Lin and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsuey-Ching Yang, dGN5YW5nQG55Y3UuZWR1LnR3
†These authors have contributed equally to this work
 Chun-Hsing Liao
Chun-Hsing Liao Hsu-Feng Lu3†
Hsu-Feng Lu3† Ching-Wei Yang
Ching-Wei Yang Yi-Tsung Lin
Yi-Tsung Lin Tsuey-Ching Yang
Tsuey-Ching Yang