
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 May 2024
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1380678
This article is part of the Research Topic Fighting Antimicrobial Resistance in Developing Countries: Innovative Approaches and Challenges View all 9 articles
Introduction: The increasing incidence of Klebsiella pneumoniae and carbapenem-resistant Klebsiella pneumoniae (CRKP) has posed great challenges for the clinical anti-infective treatment. Here, we describe the molecular epidemiology and antimicrobial resistance profiles of K. pneumoniae and CRKP isolates from hospitalized patients in different regions of China.
Methods: A total of 219 K. pneumoniae isolates from 26 hospitals in 19 provinces of China were collected during 2019–2020. Antimicrobial susceptibility tests, multilocus sequence typing were performed, antimicrobial resistance genes were detected by polymerase chain reaction (PCR). Antimicrobial resistance profiles were compared between different groups.
Results: The resistance rates of K. pneumoniae isolates to imipenem, meropenem, and ertapenem were 20.1%, 20.1%, and 22.4%, respectively. A total of 45 CRKP isolates were identified. There was a significant difference in antimicrobial resistance between 45 CRKP and 174 carbapenem-sensitive Klebsiella pneumoniae (CSKP) strains, and the CRKP isolates were characterized by the multiple-drug resistance phenotype.There were regional differences among antimicrobial resistance rates of K. pneumoniae to cefazolin, chloramphenicol, and sulfamethoxazole,which were lower in the northwest than those in north and south of China.The mostcommon sequence type (ST) was ST11 (66.7% of the strains). In addition, we detected 13 other STs. There were differences between ST11 and non-ST11 isolates in the resistance rate to amikacin, gentamicin, latamoxef, ciprofloxacin, levofloxacin, aztreonam, nitrofurantoin, fosfomycin, and ceftazidime/avibactam. In terms of molecular resistance mechanisms, the majority of the CRKP strains (71.1%, 32/45) harbored blaKPC-2, followed by blaNDM (22.2%, 10/45). Strains harboring blaKPC or blaNDM genes showed different sensitivities to some antibiotics.
Conclusion: Our analysis emphasizes the importance of surveilling carbapenem-resistant determinants and analyzing their molecular characteristics for better management of antimicrobial agents in clinical use.
Klebsiella pneumoniae is a gram-negative bacterium capable of colonizing, invading, and causing infections in different anatomical sites of the human body. It is an opportunistic pathogen that can cause pneumonia, urinary tract infections, and bloodstream infections (Martin and Bachman, 2018). In China, the isolation rate of K. pneumoniae was second only to that of Escherichia coli, which ranks it second among gram-negative bacteria infections (Hu et al., 2019). With the excessive use and misuse of broad-spectrum antimicrobial agents, the increasing incidence of antimicrobial resistance has become a serious threat to public and global health as it complicates the clinical management of infectious diseases (Logan and Weinstein, 2017; van Duin and Doi, 2017; Liu et al., 2022). Thus, the World Health Organization (WHO) lists K. pneumoniae as one of the species of high priority and promotes the research and development of new antibiotics due to the growing global problem of antimicrobial resistance (Tacconelli et al., 2018).
Carbapenem antibiotics are still the “mainstay” in the treatment of K. pneumoniae infection at present. However, many carbapenem-resistant Klebsiella pneumoniae (CRKP) phenotypes have widely emerged in recent years. Yigit et al. (2001) first discovered CRKP in a hospital in North Carolina, USA, and subsequently, CRKP was reported in various countries (Mouloudi et al., 2014; Jin et al., 2015; Yu et al., 2016). The threat of drug-resistant bacterial infection posed by CRKP has garnered increasing attention worldwide. It poses a serious challenge to clinical management due to the limited availability of effective antimicrobial agents. Therefore, strengthening drug resistance monitoring and epidemiological research of CRKP is crucial to control the spread of multi-drug resistant bacteria and guide clinical rational anti-infection treatment.
The resistance mechanisms of CRKP mainly include the production of β-lactamases, overexpression of efflux pump genes, loss of outer membrane porins, and modification of drug targets. The production of carbapenemases is one of the major drivers of carbapenem resistance. The carbapenemase genes in CRKP strains mainly include blaKPC, blaNDM, blaOXA, and blaIMP, of which blaKPC is the most clinically frequent in most countries (Tzouvelekis et al., 2012; Munoz-Price et al., 2013). To better understand CRKP epidemiology, explore the molecular resistance mechanisms, and analyze the characteristics of drug resistance, we initiated a multicenter surveillance study involving 26 hospitals in 19 provinces of China during 2019–2020.
In this study, we tested the resistance profiles of K. pneumoniae against 24 commonly used clinical antibiotics and compared the resistance rates among different regions to analyze the drug resistance status of K. pneumoniae in China. Using multilocus sequence typing (MLST), carbapenemase-resistant genes were detected in 45 CRKP strains and antimicrobial resistance among the different types was characterized, which provides experimental evidence for the effective treatment of CRKP infectious diseases.
A total of 219 K. pneumoniae isolates were collected from 26 hospitals in China through the National Science and Technology Basic Research Program of China (2019FY101200). The participating hospitals are listed in Table 1. In October 2020, a retrospective collection of K. pneumoniae isolates from the participating facilities, collected between January 1 and December 31, 2019, was submitted to the central research team for further analysis. The strains originated from different sites of infection, including the respiratory tract, blood, enterocele, urine, skin and soft-tissue, central nervous system, reproductive tract, and digestive tract. The Qinling Mountains-Huaihe River line was used as the dividing line between the northern and southern regions; the Greater Hinggan Mountains-Yinshan-Helan Mountains line was used as the dividing line between the northern region and the northwestern region. All strains were identified using the VITEK-MS mass spectrometer (bioMerieux, France).
The antimicrobial susceptibility of the 219 K. pneumoniae strains was determined using the automated microorganisms drug sensitivity analyzer SAST60 (Deere, Zhuhai, China). Strains were identified as CRKP based on susceptibility testing against imipenem, meropenem, and ertapenem based on the minimum inhibitory concentrations (MIC) of ≥ 4 μg/mL for imipenem and meropenem, or ≥ 2 μg/mL for ertapenem as per the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI M100-S33, 2023). ATCC 25922 was used as the quality control strain.
MLST was performed as described on the Institute Pasteur database (http://bigsdb.pasteur.fr/klebsiella/). To identify the sequence types (STs) of the 45 CRKP strains, seven housekeeping genes, namely, gapA, infB, mdh, pgi, phoE, rpoB, and tonB, were amplified by polymerase chain reaction (PCR) and sequenced. The FASTA sequences of all these housekeeping genes were submitted to the K. pneumoniae MLST database, and the STs were determined.
The genomic DNA of the 45 CRKP isolates was extracted using a boiling method. Briefly, pure colonies that were cultured overnight on Columbia blood agar plates were suspended in sterile distilled water and the tubes placed in boiling water for 10 min, followed by rapid cooling in ice water. The lysate was then centrifuged at 10000 rpm for 2 min and the supernatant transferred to a sterile tube for further use. Six carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, blaOXA-23, and blaOXA-48) and extended spectrum β-lactamase genes (blaCTX-M) were amplified (Zhang et al., 2022). The detailed primer information and amplification conditions are listed in Table 2. Amplified products were visualized using 1% agarose gel electrophoresis. The amplified gene fragments were sequenced and alignments were performed using the Basic Local Alignment Search Tool (BLAST).
Statistical analysis was performed using the 20.0 version of SPSS software. Categorical variables were described as proportions and associations assessed by Chi-square test or Fisher’s exact test. Results with P-values < 0.05 were considered statistically significant.
A total of 219 K. pneumoniae isolates were included in this study which originated from multiple sites of infection, including 22.4% (49/219) from the respiratory tract, 20.1% (44/219) from the blood, 16.4% (36/219) from enterocele, 13.7% (30/219) from urine, 12.8% (28/219) from skin and soft tissue, 7.8% (17/219) from the central nervous system, 5.0% (11/219) from the reproductive tract, and 1.8% (4/219) from the digestive tract. Of all the enrolled patients, 60.9% (134/219) were male, and the ages of all patients ranged from 1 day to 95 years old (median age: 54 years old). The proportions of K. pneumoniae isolates from patients in the surgical wards, medical wards, and intensive care units (ICUs) were 32.3%, 25.5%, and 15.5%, respectively.
The antimicrobial resistance rates of the K. pneumoniae isolates are shown in Table 3. The resistance rates of K. pneumoniae isolates to imipenem, meropenem, and ertapenem were 20.1%, 20.1%, and 22.4%, respectively. Strains resistant to at least one of the three antimicrobial agents were identified as CRKP, which accounted for 20.5% (45/219) of all strains.
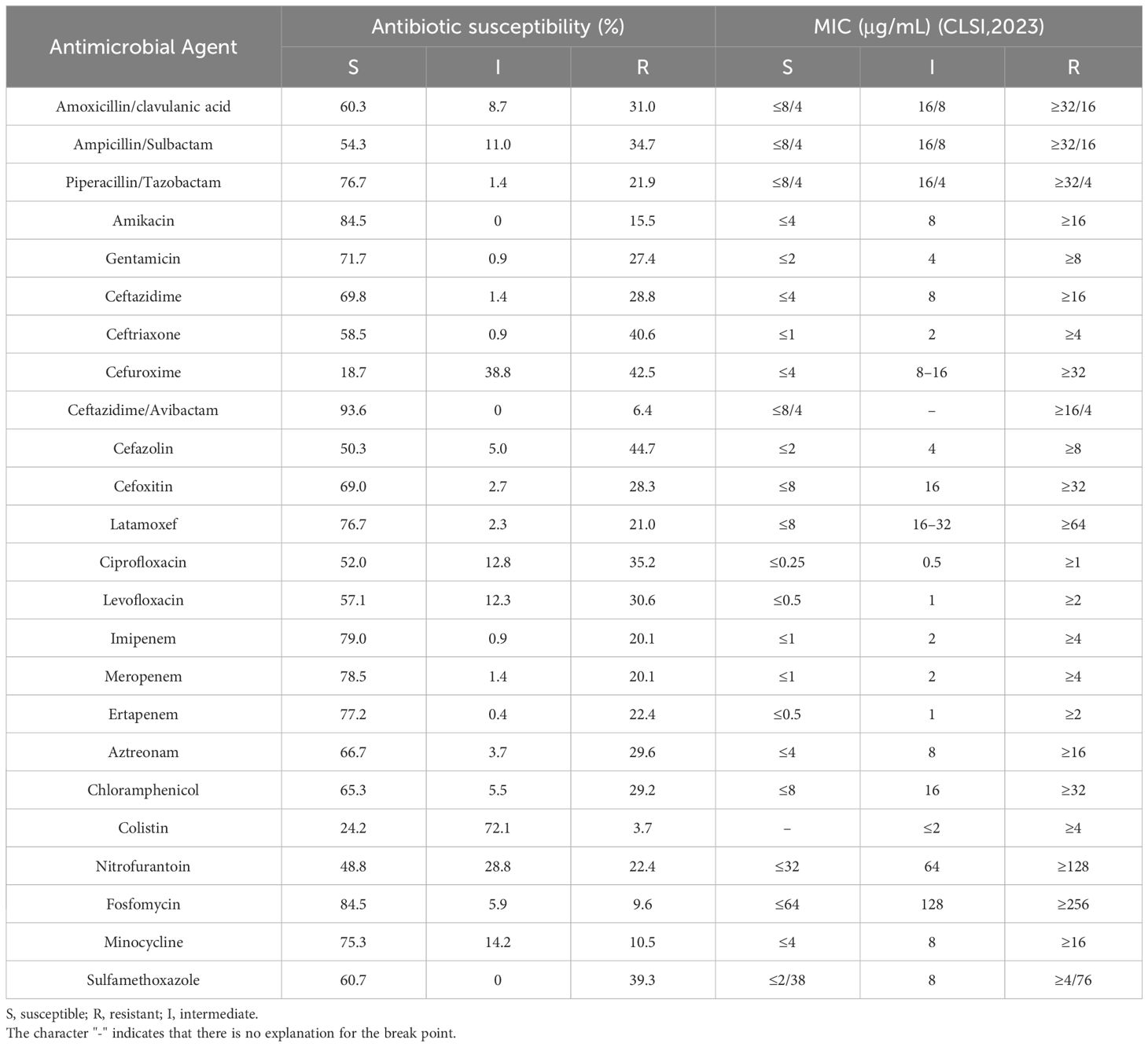
Table 3 Antimicrobial susceptibility of the 219 Klebsiella pneumoniae isolates to 24 antimicrobial agents.
Then, we divided the regional source of strains into three groups: north, south, and northwest according to geographical region, and compared the difference in the antimicrobial resistance rates among these regions by Chi-square test; P < 0.05 indicated that there were differences among the groups. Statistical results showed that some of the antimicrobial resistance rates were different in different regions of China. For example, the resistance rates of cefazolin, chloramphenicol, and sulfamethoxazole in northwest were lower than those in north and south of China (Table 4).
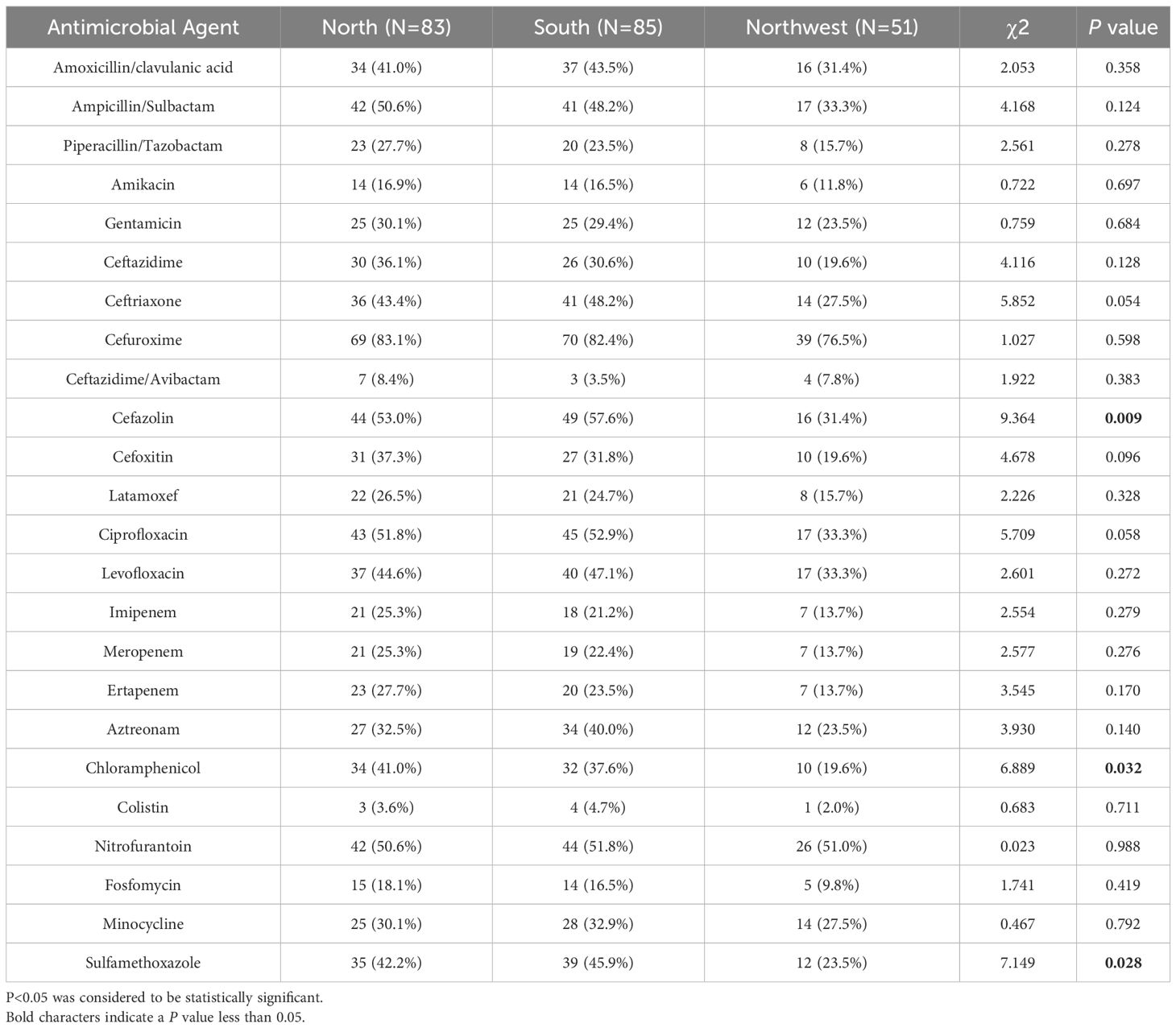
Table 4 Antimicrobial resistance profiles of 219 K. pneumoniae isolates from different regions in China.
Moreover, CRKP exhibited 100% resistance rate to ampicillin/sulbactam, ceftazidime, ceftriaxone, cefuroxime, cefazolin, cefoxitin, and ertapenem. The antimicrobial resistance rates of CRKP isolates to all tested antibiotics except for polymyxin were significantly different to the CSKP (P <0.05) (Table 5).
To further understand the genetic diversity of CRKP, we performed MLST analysis (Table 6). The results showed that 14 distinct STs were identified within the 45 isolates, and the diversity index was 31.1% (14/45). Among them, the most frequently represented ST was ST11 (66.7%,30/45). Other STs included ST48 (4.4%,2/45) and ST299 (4.4%,2/45), and ST23, ST2807, ST15, ST147, ST3410, ST4926, ST661, ST367, ST638, ST345, and ST1540 accounted for 2.2% of the isolates.
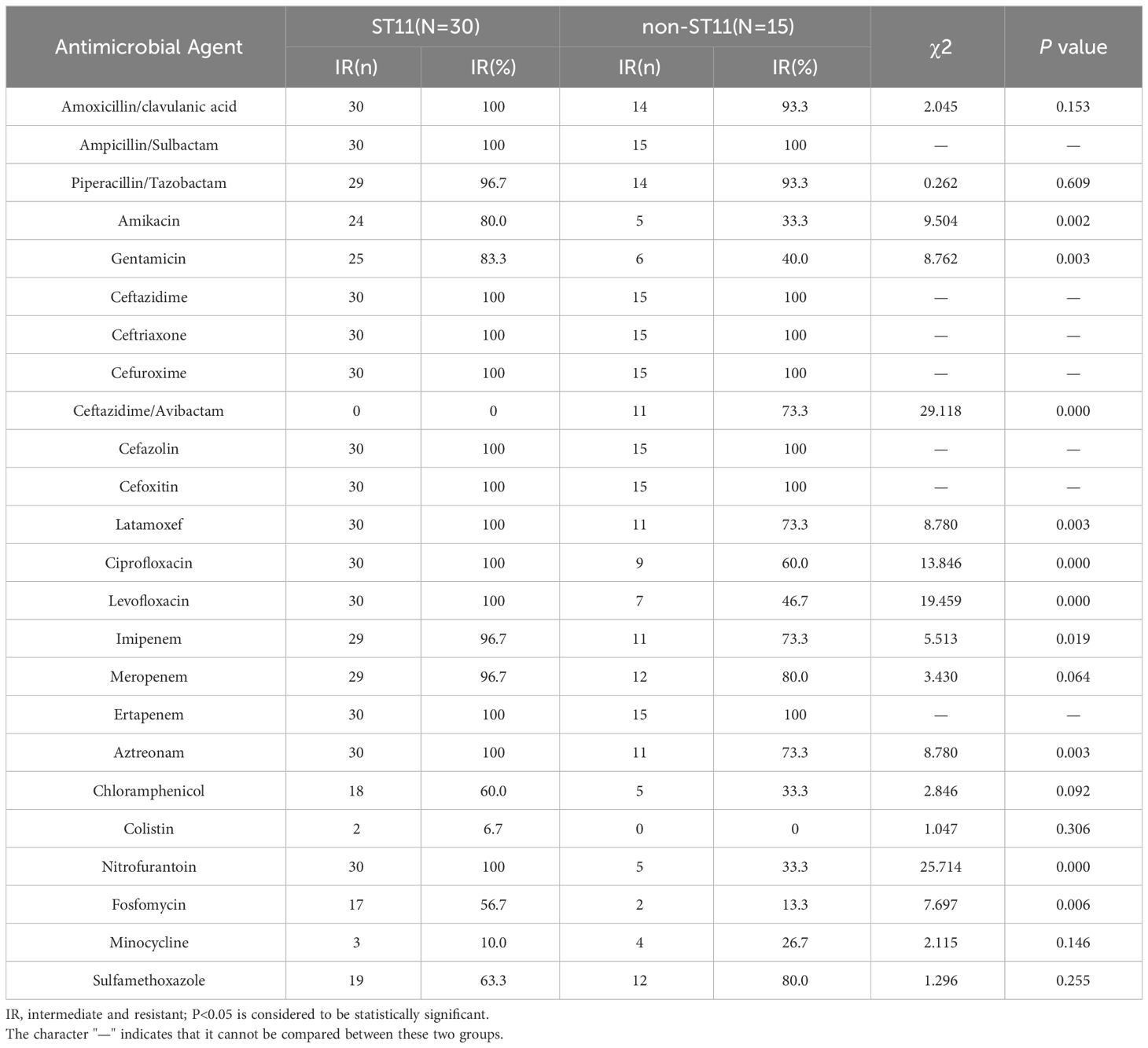
Table 6 Antimicrobial resistance of 45 CRKP isolates against antimicrobial agents among different STs.
The majority of CRKP (71.1%, 32/45) were found to harbor blaKPC-2, while the rest carried blaNDM(22.2%, 10/45), blaCTX-M(4.4%, 2/45), and blaOXA-23 (2.2%, 1/45). Other carbapenemase genes, including blaOXA-48, blaVIM, and blaIMP were not detected in these strains. Both blaKPC and blaNDM were the most commonly identified in this study. Strains carrying different carbapenemases showed different sensitivity to antibiotics (Table 7). Most CRKP strains with blaKPC were resistant to ciprofloxacin, levofloxacin, nitrofurantoin, fosfomycin, chloramphenicol, and aztreonam, whereas CRKP strains with blaNDM were susceptible to these antibiotics. There were significant differences in resistance to ceftazidime/avibactam between blaKPC and blaNDM. Only one CRKP strain with blaKPC was resistant to ceftazidime/avibactam, while almost all CRKP strains with blaNDM were resistant to the antibiotic.
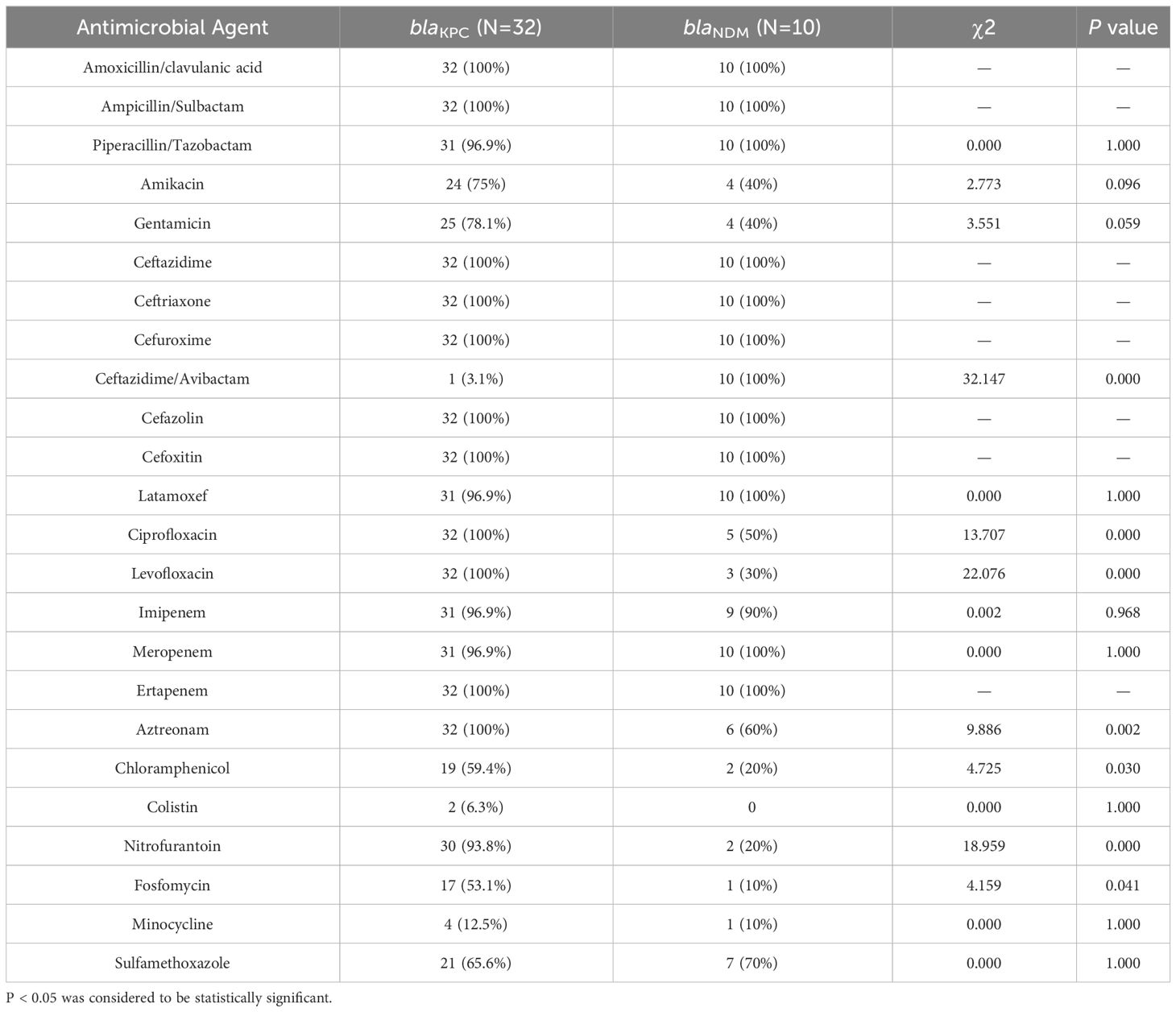
Table 7 Comparison of antibiotic resistance profiles of 45 CRKP isolates harboring blaKPC and blaNDM.
In this study, we collected 219 K. pneumoniae isolates from different regions of China in 2020. The main sources of specimens were surgical wards (32.4%), medical wards (25.4%), and ICUs (15.5%). This may be related to more invasive procedures such as ventilator placement and indwelling devices in surgical wards and ICUs, as well as longer hospital stay and long-term use of antibiotics (Karakonstantis et al., 2020). Tian et al. (2016) also reported that morbidity and mortality rates for CRKP-infected patients in ICUs were much higher than those for non-ICU patients. This suggests that environmental hygiene and disinfection of medical devices should be strengthened in clinical settings, and attention should be paid to aseptic practices of medical staff as well as the rational use of antibiotics.
We surveyed the molecular characteristics and antibiotic resistance profiles of our K. pneumoniae isolates. The results showed that K. pneumoniae is inherently resistant to ampicillin, and the resistance rates of the first, second, and third generation cephalosporins exceeded 40%. This implied that cephalosporins in China may be excessively used. Carbapenem antibiotics are the last line of defense against K. pneumoniae; however, with the widespread use of these antibiotics, the detection rates of CRKP strains had been increasing gradually. In our study, among the 219 K. pneumoniae isolates, 45 (20.5%) were identified as CRKP. This is generally consistent with other research. For example, Hu et al. (2019) reported the resistance rates of K. pneumoniae to imipenem and meropenem as 25% and 26.3%, respectively, in 2018. In addition, a retrospective observational study from Zhejiang, China suggested that imipenem and meropenem resistance rates in 2018 were 15.8% and 20.9%, respectively (Hu et al., 2020), which were lower than the rates observed in our study. Notably, most CRKP strains showed multiple drug resistance; thus, we compared the difference of antibiotics resistance between CRKP and CSKP. Our findings confirmed that CRKP strains were resistant to most of clinically common antibiotics, which included penicillins, cephalosporins, aminoglycosides, quinolones, carbapenems, and so on, but not for colistin and minocycline.
Antimicrobial Resistance Collaborators (2022) estimated that Australasia had the lowest antimicrobial resistance burden among the 21 Global Burden of Diseases, Injuries, and Risk Factors Study regions in 2019, while the highest rates of antimicrobial resistance burden were in sub-Saharan Africa. This confirmed that bacterial antimicrobial resistance rates differed by geographic location. China as a vast nation exhibited significant variations in the levels of antibiotic resistance across different provinces and regions. In our study, there were regional differences among antimicrobial resistance rates of K. pneumoniae to cefazolin, chloramphenicol, and sulfamethoxazole. Resistance to these antibiotics was lower in Northwest China, but there was no difference in resistance to other antibiotics. This difference may hint at differences in medication habits in different regions. Because of these regional differences, the epidemiological precautions and medication policies that guide them also differ geographically. In recent years, China has issued the “National Action Plans for Combating Bacterial Resistance (2022–2025)”, implementing effective antimicrobial stewardship and developing a rational antibiotic use policies (Ding and Hu, 2023). China’s efforts to combat antimicrobial resistance have shown significant results. According to CHINET, the resistance rates of K. pneumoniae to imipenem and meropenem have straightly climbed from 3.0% and 2.9% in 2005, to 25.0% and 26.3% in 2018 (Hu et al., 2019). However, a stabilizing trend is observed from 2019 onwards, with rates of 22.6% and 24.2% in 2022 (Qin et al., 2024). Therefore, drug use in various regions should be adapted to local conditions through the use of corresponding drugs according to the regional resistance rate to replace other antibiotics. For example, in areas with high rates of chloramphenicol resistance, use of chloramphenicol should be reduced. Therefore, it is necessary to continuously monitor the molecular epidemiological characteristics of the same strain isolate in a specific region, so as to develop appropriate antibiotic therapy and reduce unnecessary antibiotic treatment.
MLST of 45 CRKP isolates was conducted, and a total of 14 ST types were identified, of which, ST11 accounted for the largest proportion (about 66.7%). There are 13 other ST types, accounting for a smaller proportion, including ST48, ST299, ST23, ST2807, ST15, ST147, ST3410, ST4926, ST661, ST367, ST638, ST345, and ST1540. Although the number of the strains included in this study was insufficient, it still showed that the distribution of ST had strong regional characteristics. At present, there are more than 2000 K. pneumoniae STs identified globally. The predominant ST among CRKP strains in European countries, such as Norway and Sweden, and in the United States, is ST258 (Deleo et al., 2014), whereas ST11 predominates in China (Zhang et al., 2017; Zhou et al., 2020). Liu et al. (2022) reported that ST11 accounted for 64.2% of all CRKP strains in China in 2016–2020, which was consistent with our results, showing that ST11 was the most common type of CRKP strains. The number of virulence plasmids and mobile elements carried by ST11 were higher than those of other ST strains, which resulted in the dissemination of virulence plasmids in hospitals and different areas (Mathers et al., 2011; Martin et al., 2017). Horizontal gene transfer within and between bacterial species ultimately led to the high prevalence of ST11 in China.
Carbapenem antibiotics are a general term for a class of broad-acting antibiotics that are commonly used as last-resort antibiotics to treat serious bacterial infections. Therefore, we focused on the resistance of carbapenem antibiotics and deeply researched carbapenemase as it proved to be the main mechanism of drug resistance in CRKP. In our study, 71.1% (32/45) of CRKP isolates were found to harbor blaKPC-2, followed by blaNDM (22.2%, 10/45), blaCTX-M (4.4%, 2/45), and blaOXA-23 (2.2%, 1/45). Among 32 strains harboring blaKPC, 93.8% (30/32) belonged to ST11, whereas the STs of the strains carrying blaNDM were more dispersed. K. pneumoniae carbapenemase (KPC)-producing CRKP was reported first in 2007 (Wei et al., 2007), and blaKPC-2 subsequently became the dominant genotype in China (Zhang et al., 2018; Li et al., 2022), which was most commonly associated with the epidemic clone ST11 (Liu et al., 2018). KPC enzymes account for more than 70% of CRKP, followed by New Delhi metallo-beta-lactamase (NDM)-producing strains, while the prevalence of impenemase (IMP), verona integron-encoded metallo-beta-lactamase (VIM), and oxacillinase-48 (OXA-48) enzymes is relatively low in China (Hu et al., 2020), which was similar to our results.
We also discussed the relationship between antimicrobial resistance and different STs or carbapenemase genotypes. The results showed that antibiotic resistance is also different in different STs. The resistance rates of ST11 to amikacin, gentamicin, latamoxef, ciprofloxacin, levofloxacin, aztreonam, nitrofurantoin, and fosfomycin were higher than those of non-ST11. In contrast, ST11 was sensitive to ceftazidme/avibactam, while non-ST11 showed resistance to it. Of all the 45 CRKP isolates, there were 32 strains harboring blaKPC and 10 strains carrying blaNDM. Antibiotic resistance analysis found that the resistance rates of the two strains to some antibiotics differed, and the difference was statistically significant. CRKP strains with blaKPC showed higher rates of resistance to ciprofloxacin, levofloxacin, nitrofurantoin, fosfomycin, chloramphenicol, and aztreonam, while CRKP strains with blaNDM were almost completely resistant to ceftazidime/avibactam. Similar to previous findings, blaNDM-carrying CRKP strains were resistant to ceftazidime/avibactam, but blaKPC-containing CRKP strains were sensitive to it. This indicated that CRKP resistance is closely related to its different resistance genotypes and STs. CRKP carrying blaKPC and blaNDM differed in antibiotic resistance, with different resistance to different antibiotics. The detection of drug resistance genotypes would be helpful to adjust the antibiotic application strategy in a timely manner.
This study has a few limitations. First, the number of strains included in this study may be insufficient to provide a comprehensive representation of the molecular epidemiological characteristics and antimicrobial resistance profiles of K. pneumoniae and CRKP in China. Second, whole genome sequencing of the CRKP was not performed, which would be included in our further research priorities to provide deeper insights into the pathogens.
This study described the prevalence and molecular epidemiological characteristics of K. pneumoniae and CRKP in China during 2019–2020. The resistance rates of K. pneumoniae to different antibiotics differed by region, which could be attributed to regional variations in antibiotic use. Compared with CSKP, CRKP had more severe drug resistance and showed more multiple drug resistance rates. Although CRKP exhibited a wide range of strain types, ST11 was still the major strain type. Other strain types were relatively rare and more sporadic. In terms of molecular resistance mechanisms, the majority of the CRKP strains harbored blaKPC-2, followed by blaNDM. Moreover, there was an association between antibiotic resistance profiles of CRKP strains with different STs and different carbapenemase genes. Hence, understanding the resistance phenotype and molecular epidemiology of CRKP isolates is essential to control the transmission of these drug-resistant bacteria and to promote the rational use of antibiotics.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
In accordance with the local legislation and institutional requirements, ethical review and approval were waived for this study as the samples were collected during routine care. Patient consent was waived in accordance with the national legislation and the institutional requirements as the patient data were completely anonymized and study findings were not used for the clinical management of the patients.
YaL: Formal analysis, Investigation, Methodology, Writing – original draft, Data curation. CX: Writing – review & editing, Formal analysis, Investigation, Methodology, Validation. ZZ: Writing – review & editing. JL: Writing – review & editing. HC: Data curation, Writing – review & editing. YoL: Writing – review & editing, Funding acquisition, Project administration, Resources. XQ: Conceptualization, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Special Foundation for National Science and Technology Basic Research Program of China, Grant/Award Number: 2019 FY101200. This work was also supported by Medical Masters Project of "Xingliao Talent Plan" in Liaoning Province, Grant/Award Number: No.YXMJ-LJ-09.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Antimicrobial Resistance Collaborators. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140–6736(21)02724–0
Deleo, F. R., Chen, L., Porcella, S. F., Martens, C. A., Kobayashi, S. D., Porter, A. R., et al. (2014). Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 111, 4988–4993. doi: 10.1073/pnas.1321364111
Ding, L., Hu, F. (2023). China's new national action plan to combat antimicrobial resistance, (2022–25). J. Antimicrob. Chemother. 78, 558–560. doi: 10.1093/jac/dkac435
Hu, F., Guo, Y., Yang, Y., Zheng, Y., Wu, S., Jiang, X., et al. (2019). Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur. J. Clin. Microbiol. Infect. Dis. 38, 2275–2281. doi: 10.1007/s10096-019-03673-1
Hu, Y., Liu, C., Shen, Z., Zhou, H., Cao, J., Chen, S., et al. (2020). Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China 2008–2018. Emerg. Microbes Infect. 9, 1771–1779. doi: 10.1080/22221751.2020.1799721
Jin, Y., Shao, C., Li, J., Fan, H., Bai, Y., Wang, Y. (2015). Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PloS One 10, e0119571. doi: 10.1371/journal.pone.0119571
Karakonstantis, S., Kritsotakis, E. I., Gikas, A. (2020). Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: an approach based on the mechanisms of resistance to carbapenems. Infection 48, 835–851. doi: 10.1007/s15010-020-01520-6
Li, C., Jiang, X., Yang, T., Ju, Y., Yin, Z., Yue, L., et al. (2022). Genomic epidemiology of carbapenemase-producing klebsiella pneumoniae in China. Genomics Proteomics Bioinf. 20, 1154–1167. doi: 10.1016/j.gpb.2022.02.005
Liu, C., Dong, N., Chan, E. W. C., Chen, S., Zhang, R. (2022). Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in China 2016–20. Lancet Infect. Dis. 22, 167–168. doi: 10.1016/S1473-3099(22)00009-3
Liu, J., Yu, J., Chen, F., Yu, J., Simner, P., Tamma, P., et al. (2018). Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur. J. Clin. Microbiol. Infect. Dis. 37, 293–299. doi: 10.1007/s10096-017-3131-4
Logan, L. K., Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Martin, J., Phan, H. T. T., Findlay, J., Stoesser, N., Pankhurst, L., Navickaite, I., et al. (2017). Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J. Antimicrob. Chemother. 72, 3025–3034. doi: 10.1093/jac/dkx264
Martin, R. M., Bachman, M. A. (2018). Colonization, infection, and the accessory genome of klebsiella pneumoniae. Front. Cell Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00004
Mathers, A. J., Cox, H. L., Kitchel, B., Bonatti, H., Brassinga, A. K., Carroll, J., et al. (2011). Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2, e00204–e0e211. doi: 10.1128/mBio.00204-11
Mouloudi, E., Massa, E., Papadopoulos, S., Iosifidis, E., Roilides, I., Theodoridou, T., et al. (2014). Bloodstream infections caused by carbapenemase-producing Klebsiella pneumoniae among intensive care unit patients after orthotopic liver transplantation: risk factors for infection and impact of resistance on outcomes. Transplant. Proc. 46, 3216–3218. doi: 10.1016/j.transproceed.2014.09.159
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Qin, X., Ding, L., Hao, M., Li, P., Hu, F., Wang, M., et al. (2024). Antimicrobial resistance of clinical bacterial isolates in China: current status and trends. JAC Antimicrob. Resist. 6, dlae052. doi: 10.1093/jacamr/dlae052
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tian, L., Tan, R., Chen, Y., Sun, J., Liu, J., Qu, H., et al. (2016). Epidemiology of Klebsiella pneumoniae bloodstream infections in a teaching hospital: factors related to the carbapenem resistance and patient mortality. Antimicrob. Resist. Infect. Control 5, 48. doi: 10.1186/s13756-016-0145-0
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
van Duin, D., Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Wei, Z. Q., Du, X. X., Yu, Y. S., Shen, P., Chen, Y. G., Li, L. J. (2007). Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob. Agents Chemother. 51, 763–765. doi: 10.1128/AAC.01053-06
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Yu, J., Tan, K., Rong, Z., Wang, Y., Chen, Z., Zhu, X., et al. (2016). Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect. Dis. 16, 563. doi: 10.1186/s12879-016-1870-y
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang, Z., Wang, D., Li, Y., Liu, Y., Qin, X. (2022). Comparison of the performance of phenotypic methods for the detection of carbapenem-resistant enterobacteriaceae (CRE) in clinical practice. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.849564
Zhang, Y., Wang, Q., Yin, Y., Chen, H., Jin, L., Gu, B., et al. (2018). Epidemiology of carbapenem-resistant enterobacteriaceae infections: report from the China CRE network. Antimicrob. Agents Chemother. 62, e01882–e01817. doi: 10.1128/AAC.01882-17
Keywords: Klebsiella pneumoniae, antimicrobial resistance, carbapenem-resistant, epidemiology, sequence type
Citation: Li Y, Xie C, Zhang Z, Liu J, Chang H, Liu Y and Qin X (2024) Molecular epidemiology and antimicrobial resistance profiles of Klebsiella pneumoniae isolates from hospitalized patients in different regions of China. Front. Cell. Infect. Microbiol. 14:1380678. doi: 10.3389/fcimb.2024.1380678
Received: 02 February 2024; Accepted: 30 April 2024;
Published: 16 May 2024.
Edited by:
Ilke Pala-Ozkok, University of Stavanger, NorwayReviewed by:
Pedro Teixeira, National Institute of Health Dr Ricardo Jorge, PortugalCopyright © 2024 Li, Xie, Zhang, Liu, Chang, Liu and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaosong Qin, cWlueHNAc2otaG9zcGl0YWwub3Jn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.