
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 08 May 2024
Sec. Molecular Bacterial Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1375312
 Miao Yao1,2
Miao Yao1,2 Kun Wang1,2
Kun Wang1,2 Guangming Song1,2
Guangming Song1,2 Yumeng Hu1,2
Yumeng Hu1,2 Jiali Chen1,2
Jiali Chen1,2 Tingting Li1,2
Tingting Li1,2 Longying Liang1,2
Longying Liang1,2 Jie Wu2
Jie Wu2 Hongmei Xu3
Hongmei Xu3 Libin Wang3
Libin Wang3 Yuqiang Zheng3
Yuqiang Zheng3 Xuemei Zhang3
Xuemei Zhang3 Yibing Yin3
Yibing Yin3 Shifei Yao1,2*
Shifei Yao1,2* Kaifeng Wu1,2*
Kaifeng Wu1,2*Competence development is essential for bacterial transformation since it enables bacteria to take up free DNA from the surrounding environment. The regulation of teichoic acid biosynthesis is tightly controlled during pneumococcal competence; however, the mechanism governing this regulation and its impact on transformation remains poorly understood. We demonstrated that a defect in lipoteichoic acid ligase (TacL)-mediated lipoteichoic acids (LTAs) biosynthesis was associated with impaired pneumococcal transformation. Using a fragment of tacL regulatory probe as bait in a DNA pulldown assay, we successfully identified several regulatory proteins, including ComE. Electrophoretic mobility shift assays revealed that phosphomimetic ComE, but not wild-type ComE, exhibited specific binding to the probe. DNase I footprinting assays revealed the specific binding sequences encompassing around 30 base pairs located 31 base pairs upstream from the start codon of tacL. Expression of tacL was found to be upregulated in the ΔcomE strain, and the addition of exogenous competence-stimulating peptide repressed the tacL transcription in the wild-type strain but not the ΔcomE mutant, indicating that ComE exerted a negative regulatory effect on the transcription of tacL. Mutation in the JH2 region of tacL upstream regulatory sequence led to increased LTAs abundance and displayed higher transformation efficiency. Collectively, our work identified the regulatory mechanisms that control LTAs biosynthesis during competence and thereby unveiled a repression mechanism underlying pneumococcal transformation.
Streptococcus pneumoniae (S. pneumoniae) continues to pose a significant global health challenge among children and the elderly, especially in the context of epidemic viral lung infections including SARS-CoV-2 (Li et al., 2023; Zhu et al., 2023). Progress in controlling pneumococcal disease has also been greatly impeded by the emergence of multidrug-resistant isolates and serotype replacement within S. pneumoniae strains (Chochua et al., 2017; Yan et al., 2021; Vidal et al., 2022). This issue is closely linked to genetic transformation that is employed by pneumococci for acquiring antibiotic resistance, adapting to their environment, and even generating novel species (Straume et al., 2015; Kim et al., 2019; Lo et al., 2019). Consequently, there has been an increasing interest in investigating the mechanisms underlying gene transfer (Vollmer et al., 2019; Lam et al., 2021; Zhang et al., 2023).
Competence development is crucial for bacterial transformation since it permits the uptake of more efficient exogenous DNA (Claverys et al., 2009; Slager et al., 2019; Lam et al., 2021). In S. pneumoniae, this process is triggered by the presence of ComC-derived competence-stimulating peptide (CSP) (Håvarstein et al., 1995). The CSP signal is sensed via the histidine kinase receptor ComD which then undergoes auto-phosphorylation. This process leads to the phosphorylation of its cognate response regulator ComE that is a central component of the adaptive response network. This results in the activation of other network components including the comX gene that triggers genes essential for S. pneumoniae transformation (competence) (Pestova et al., 1996; Ween et al., 1999). However, it remains to be determined whether there are additional factors regulated by ComE that may also contribute to competence.
Bacterial surface polysaccharides also influence transformation capacity (Weiser and Kapoor, 1999; Li et al., 2019; Minhas et al., 2023). For example, the pneumococcal cell envelope that is primarily composed of capsular polysaccharides (CPS) may impede DNA uptake and hence influence transformation (Weiser and Kapoor, 1999). Non-encapsulated strains demonstrate a higher level of transformability than encapsulated counterparts (Ottolenghi and Macleod, 1963). Teichoic acids (TAs) have also been associated with bacterial transformation, as the transparent phenotype, characterized by elevated TAs and reduced CPS levels and it exhibits enhanced transformation efficiency compared to the opaque phenotype (Weiser et al., 1994; Li et al., 2019; Minhas et al., 2023). Additionally, the deletion of licD2, which leads to a reduction of phosphocholine modification in TAs, significantly decreases pneumococcal transformation (Zhang et al., 1999). These findings indicate that cell wall-associated glycopolymers are associated with pneumococcal transformation.
TAs biosynthesis involves multiple operons that collaborate to synthesize the TA precursor chain (Denapaite et al., 2012; Vollmer et al., 2019) that are subsequently linked to either peptidoglycan through LCP phosphotransferase family proteins (Cps2A, Psr and LytR), forming wall teichoic acids (WTAs), or to membrane glycolipids via TacL, forming lipoteichoic acids (LTAs) (Heß et al., 2017; Ye et al., 2018). Interestingly, previous observations have indicated that tacL (AE007480) is an in vivo-induced gene (Meng et al., 2008). Recently, Minhas et al. revealed that genes involved in TAs biosynthesis are essential and tacL mRNA levels decrease during competence (Aprianto et al., 2018; Minhas et al., 2023). These findings suggest that TacL is one of the crucial enzymes that control the TAs ratios in two cellular locations: membrane versus peptidoglycan, which in turn modulates the location of choline-binding proteins, and the regulation of tacL during competence may be a key factor that determines transformation. Therefore, its regulation during competence is important to understand.
The tacL gene may be transcriptionally regulated during competence so we utilized a tacL DNA promoter fragment to physically screen for regulatory proteins capable of binding to the fragment. DNA affinity chromatography was coupled with mass spectrometry (MS) techniques and resulted in the identification of binding proteins, most notably ComE that has previously been established as a transcription factor essential for competence development. Our findings demonstrated that ComE exerts a negative regulatory effect on LTAs biosynthesis by repressing tacL transcription. Furthermore, our data indicated that repression of tacL by phosphorylated ComE during competence hampers pneumococcal transformation. Overall, these discoveries unveil an innovative mechanism governing pneumococcal transformation through transcriptional regulation of LTAs biosynthesis via phosphorylated ComE.
The bacterial strains, plasmids, and primers utilized in this study are listed in Supplementary Tables S1, S2. The S. pneumoniae D39 strain (serotype 2, NCTC7466), R6 (non-encapsulated strain, ATCC BAA-255), and their derivatives were cultivated in Todd-Hewitt Broth containing 2% Yeast Extract (THY, Hopebio Qingdao, China) medium or blood agar plates (Autobio, Zhengzhou, China) at 37°C in a 5% CO2 atmosphere. Escherichia coli (E. coli) strains used for cloning and plasmid amplification were grown in Luria-Bertani (LB) broth or on agar plates at 37°C. All bacterial growth media were supplemented with appropriate antibiotics (Supplementary Table S1).
Gene deletion of tacL was performed with the long-arm homologous PCR technique (LH-PCR) (Prudhomme et al., 2002; Meng et al., 2008). In brief, we amplified the upstream and downstream regions of tacL (GeneBank SPV_1672) from S. pneumoniae D39 using primers tacL up P1/P2 and tacL dw P3/P4, respectively. We also amplified the erythromycin resistance gene from the S. pneumoniae CPM8 strain using the erm-F/R primer pair. After PCR amplification, these fragments were ligated together by overlapping PCR to create a recombinant fragment. This fragment was then respectively transformed into D39, D39Δcps (SPW3) and R6 strains (The tacL gene exhibits high conservation). Colonies were selected on agar plates containing 0.25 μg/mL erythromycin and confirmed through PCR with primers tacL up P1 and tacL dw P4 to obtain the erythromycin-resistant mutant strain ΔtacL. The plasmid pJWV25 (Eberhardt et al., 2009) and the full-length tacL fragments were digested with SpeI and NotI restriction enzymes for cloning. The resulting recombinant plasmids pJWV25-tacL (pWKF1) were used to transform into D39ΔtacL (SPW1), D39ΔcpsΔtacL (SPW4) and R6ΔtacL (SPW6) strains and were selected on agar plates containing tetracycline to generate the complemented strains D39ΔtacL::PJWtacL (SPW2), D39ΔcpsΔtacL::PJWtacL (SPW5) and R6ΔtacL::PJWtacL (SPW7).
The streptomycin resistance background of strain D39 was utilized for allelic replacement with the counter-selectable Janus cassette (JC) as previously described (Sung et al., 2001). This construct consists of a streptomycin target ribosomal protein S12 and a dominant wild-type rpsL+ allele encoding a kanamycin resistance gene. This resulted in the construction of a series of DNA fragments containing ComE-tacL promoter binding sequence mutation (Sung et al., 2001; Kirkham et al., 2006). Briefly, a K56T codon change was made in rspL to generate the streptomycin-resistant strain D39s (SPW8). Subsequently, upstream (JH up) and downstream (JH dw) sequences of the ComE binding site in tacL were amplified using primers JH up-P1/P2 and JH dw-P3/P4 with D39s as a template. The JC sequence was amplified using primers JC-F/JC-R. A recombinant fragment was generated using an overlapping PCR approach with primers JH up-P1/JH dw-P4 (JH up-JC-JH dw) followed by transformation into D39s. Positive recombinant clones D39ΔJHtacL (SPW9) selected on kanamycin (200 µg/mL) plates that harbored the JH up-JC-JH dw fragment were identified by PCR using primers JH up-P1/JH dw-P4. D39ΔJHtacL indicated that the JHtacL sequence was deleted from the genome of the wild-type D39 strain. The binding sequence of ComE-P to tacL (JHtacL) that exhibited sequence similarity to TATCCTAAATGGT binding sites was identified in the Streptococcus mutans genome (Hung et al., 2011). This enabled the construction of mutant sequences for JH1 (complementary paired base sequences), JH2 and JH3 (near complementary paired base sequences) that were then amplified using D39s genomic DNA as a template. Overlapping PCR with primer pairs at both ends yielded recombinant fragments (JH1 up-JH1 dw, JH2 up-JH2 dw, JH3 up-JH3 dw). These recombinant fragments were transformed into D39ΔJHtacL followed by plating on agar plates containing streptomycin (150 µg/mL). Positive recombinant clones (D39ΔJH1 (SPW10), D39ΔJH2 (SPW 11) and D39ΔJH3 (SPW 12)) carrying mutations were identified using primers JH1 up-P1/JH1 dw -P4, JH2 up-P1/JH2 dw-P4, JH3 up-P1/JH3 dw-P4.
In a similar manner, we generated D39ΔJD3 (SPW22) strains using the Janus cassette. In brief, the upstream (JD3 up) and downstream (JD3 dw) were amplified using primers JD3 up-P1/P2 and JD3 dw-P3/P4 with D39 as a template. An overlapping PCR approach with primers JD3 up-P1 and JD3 dw-P4 resulted in a recombinant fragment (JD3 up-JC-JD3 dw) that was then transformed into D39s to generate the D39ΔJD3. The tacL-up and tacL-dw fragments were then amplified by pairs tacL-up P1/P2 and tacL-dw P3/P4, and this generated the tacL deletion fragment using overlapping PCR. These four distinct phenotyping strains for bacterial wall polysaccharides were created. The tacL deletion fragment was transformed into the D39ΔJD3 and D39Δcps as background strains to construct the D39ΔtacLΔJD3 (SPW23) and D39ΔtacLΔcps (SPW24). We similarly transformed the JD3 and CPS (dexB-cps2A) deletion fragments to D39ΔJH2 to generate D39ΔJH2ΔJD3 (SPW25) and D39ΔJH2Δcps (SPW26).
D39ΔcomE (SPW13) was generated by homologous recombination. Briefly, the upstream and downstream regions of comE were respectively amplified from the D39 strain using comE up P1/P2 and comE dw P3/P4 primers. Additionally, the erythromycin resistance gene was amplified from S. pneumoniae CPM8 using the erm-F/R primer pair. These amplicons were ligated by overlapping PCR to create a recombinant fragment that was transformed into D39. Colonies were selected on agar plates containing erythromycin (0.25 μg/mL) and confirmed through PCR with primers comE up P1 and comE dw P4 to obtain the erythromycin-resistant mutant strain. The mutant strains were complemented using plasmid pJWV25 that contained full-length comE and comED58E genes. The comED58E variant was used to mimic the phosphorylated active state of ComE (Martin et al., 2013). The plasmid pJWV25 and DNA fragment were digested by SpeI and NotI restriction enzymes for subsequent cloning. To obtain the D39::PJWcomEWT (SPW14) and D39::PJWcomED58E (SPW16) strains, the resulting recombinant plasmids pJWV25-comEWT (pWKF2) and pJWV25-comED58E (pWKF3) were used to transform into the D39 strain and selected on agar plates containing tetracycline (1 μg/mL). and then the comE deletion fragment was transformed into the D39::PJWcomEWT and D39::PJWcomED58E constructed the complemented D39ΔcomE::PJWcomEWT (SPW15) and D39ΔcomE::PJWcomED58E (SPW17) strains.
To construct luciferase reporter strains, the upstream regulatory sequence of tacL was amplified from the genomic DNA of the D39 strain using tacL-luc F/R primer. The plasmid pEVP3-luc (pWKF6) and the tacL upstream regulatory sequence were digested with XhoI-BamHI to generate a linearized fragment. The resulting fragment was subsequently inserted into the 3,738-bp long plasmid pEVP3-luc that confers chloramphenicol resistance, yielding pEVP3-PtacL-luc (pWKF7). Similarly, comE-luc transcriptional fusion was generated in plasmid pEVP3-luc by amplifying a DNA fragment of the comE promoter from D39 using primer pairs comE-luc F/R, followed by digestion with XhoI/BamHI and subsequent ligation to XhoI/BamHI-digested pEVP3-luc to generate pEVP3-PcomE-luc (pWKF8). The recombinant plasmid was transformed into D39 to obtain D39::pEVP3-PcomE (SPW18) and D39::pEVP3-PtacL (SPW20) strains. Subsequently, the comE deletion fragment was transformed into the D39::pEVP3-PcomE and D39::pEVP3-PtacL to generate D39ΔcomE::pEVP3-PcomE (SPW19) and D39ΔcomE::pEVP3-PtacL (SPW21) strains.
D39 cell lysates were collected in cell cultures at OD595nm of 0.5 and subjected to repetitive freeze-thawing followed by ultrasonic fragmentation to obtain cell lysates (200~300 µg/mL) (Jutras et al., 2012). A 207-bp DNA fragment (400 µg/mL) was labeled with biotin at 5’-end and amplified from the D39 genomic DNA strain using Pulldown-tacL F/R primers. Subsequently, the purified target and non-specific DNA probe were immobilized separately on streptavidin-coated M-280 immunomagnetic beads and co-incubated with D39 cell lysates. The mixture was subjected to purification using elution buffers containing varying NaCl concentrations. The eluted proteins were separated by SDS-PAGE and stained with Coomassie blue, and compared to the control group for enrichment analysis. Finally, the enriched proteins were excised from the stained gels and identified by mass spectrometry.
The pneumococcal comE and comED58E gene from S. pneumoniae D39 was amplified by PCR, sequenced to ensure accuracy. The pET-28α (+) plasmid and comED58E/comEWT fragments were subjected to restriction enzyme digestion and subsequent ligation to generate recombinant plasmids pET-28a-comEWT (pWKF4) and pET-28a-comED58E (pWKF5). Subsequently, they were transformed into E. coli BL21 (DE3) and protein production were induced by the addition of isopropyl-β-d-thiogalactoside (IPTG) to 1 mM. Following incubation, the cultures were collected and resuspended in PBS prior to sonication. The recombinant protein was purified from the cell lysate using Nickel column affinity chromatography and stored at -80°C.
The protein-DNA binding assays were performed using the Scientific Light-Shift kit (Thermo Fisher, Pittsburg, PA, USA) according to the manufacturer’s protocol. The purified ComED58E and ComEWT proteins were respectively incubated in a reaction buffer (1 × binding buffer, 2.5% glycerol, 5 mM MgCl2, 5 mM ZnCl2, 50 ng/μL Poly (dI-dC), 0.05% NP-40) at 4°C for 5 min, followed by addition of biotin-labelled or un-labelled DNA probes and incubation at 37°C for 30 min. Samples were electro-transferred onto nylon membranes (Bio-Rad, Hercules, CA, USA) in a solution containing 0.5 × Tris-borate-EDTA (TBE) and subjected to UV crosslinking. Finally, band images were captured using a chemiluminescent imaging system (Bio-Rad).
The 400 ng probes for tacL (PtacL) were incubated with varying amounts of ComED58E in a total volume of 40 µL to incubate for 30 min at 25°C. DNase I (Promega, Madison, WI, USA) was further added to the reaction system and incubated for 1 min at 37°C. Subsequently, the samples underwent phenol-chloroform extraction and ethanol precipitation and the resulting DNA was dissolved in 30 µL Mini-Q water. The prepared DNA was analyzed using an automated DNA analyzer (ABI Prism 3100 DNA sequencer; Thermo Fisher).
Bacterial suspensions were collected at 4°C that were washed with PBS and resuspended in PBS to an optical density OD595nm of 0.5 to normalize the sample. Lysis buffer containing 0.5% deoxycholate (Sigma) was then added and the solutions were incubated at 37°C for 15 min. Samples were boiled for 10 min, subjected to SDS-PAGE on a 12% gel, and electro-transferred onto Immunobilon (Millipore, Burlington, MA, USA) and incubated for 2 h at room temperature in Tris-buffered saline with 0.1% Tween 20 (TBST) solution containing 5% skim milk. The membranes were incubated overnight at 4°C with the primary antibody and then washed three times with TBST. The primary antibodies used included rabbit anti-CWPS (Identification of TAs, 10-25kDa defined as LTAs (Wu et al., 2014; Heß et al., 2017; Flores-Kim et al., 2019)) IgG (1: 1000) which targeted TAs as well as mouse anti-ComE (1: 1000). A peroxidase-conjugated secondary antibody goat anti-rabbit IgG antibody was utilized 1: 8000 and the blot was developed using a Clarity Western Enhanced chemiluminescence (ECL) substrate kit (Epizyme, Cambridge, MA, USA) and visualized on a gel imager. Dot blot analysis utilized transfer membranes activated in methanol for 1 min and subsequently washed in PBS to achieve charge equilibrium. Bacterial lysates (2.5 μL) were serially diluted to the indicated concentrations before being applied to the activated transfer membrane and allowed to fully dry. Subsequent steps followed the protocols described in Western blot.
Bacteria were cultured in THY until OD595nm of 0.5 and harvested at 4°C, then was resuspended in PBS. The bacterial suspension was diluted with the antigen coating solution and spread onto 96-well plates overnight at 4°C and then rinsed three times with PBS followed by blocking in PBS containing 2% BSA for 1 h at 37°C. The primary antibody rabbit anti-CWPS (1: 1000) and secondary antibody goat anti-rabbit IgG (1: 8000) were used for processing. After color development, the TAs content was measured by absorption at 450 nm.
S. pneumoniae cultured in THY medium were harvested at 10000 × g for 3 min upon reaching an OD595nm of 0.5 and washed three times with PBS containing 0.05% Tween 20. The resulting pellets were resuspended in 100 µL 1% BSA in PBS and incubated with (rabbit anti-CWPS) 1: 50 for 1 h at 37°C. Subsequently, the pellets were washed as per above and stained with phycoerytherin (PE)-labeled goat anti-rabbit IgG for 1 h at 37°C. The samples were suspended in 200 µL PBS and analyzed using a BD FACS Calibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Bacteria were harvested by centrifugation at 10000 × g for 3 min at 4°C and the cell pellets were resuspended in lysis buffer containing 0.5% deoxycholate (Sigma) and incubated at 37°C. The total RNA was isolated from strains using the Trizol reagent (Takara, Shiga, Japan) and RNA (1 μg) was reverse transcribed to cDNA using an iScript gDNA synthesis kit (Takara). The mRNA levels of the target genes were quantified using real-time PCR using Power SYBR Green PCR Master Mix (TsingKe Biotech, Beijing, China), following the manufacturer’s protocol for relative quantitative analysis. The primers used for PCR are listed in Supplementary Table S2. The gyrB served as internal control.
Bacteria containing luciferase reporter strains were cultivated in the competence medium (THY containing 20 × BSA+CaCl2) until OD595nm of 0.1. Exogenous transformation inducer (CSP, 100 µg/mL) and D-luciferin (Lablead) at a concentration of 2.7 mg/mL were added as previously described (Liu et al., 2017). Luminescence assays were performed in Polystyrol 96-well plates (Corning) at 37°C. The absorbance at OD595nm and luminescence measured as relative luminescence units (RLU) were determined for each strain. The optical density measurements at OD595nm were taken every 5 min and used to normalize luciferase activity.
The pneumococcal strains were cultured in the competence medium supplemented with 20 × BSA + CaCl2 until OD595nm of 0.1. CSP (EMRLSKFFRDFILQRKK) was then added to 100 µg/mL and the mixture was incubated for 10 min at 37°C. The E. coli-S. pneumoniae shuttle vector pIB166 (Biswas et al., 2008) was incubated for 30 min on ice followed by incubation for 90 min at 37°C. The resulting samples were plated on blood agar plates in the presence and absence of 25 μg/mL chloramphenicol to enumerate total numbers and positive transformants. The transformation efficiency was calculated as the ratio between the number of antibiotic-resistant colony-forming units (CFUs) and the number of CFUs on nonselective blood agar plates.
Statistical significance between groups was compared using variance analysis ANOVA or Student’s t-test. All statistical analyses were performed using GraphPad Prism version 8.00 (San Diego, CA, USA). Data are presented as means ± SD. Statistical significance was defined as a P-value less than 0.05.
To investigate the impact of tacL on pneumococcal transformation, we generated a tacL deletion mutant using an erythromycin for allelic replacement. The complete tacL sequence was cloned into plasmid pJWV25 to construct a complemented strain with tacL (D39ΔtacL::PJWtacL). To eliminate the potential impact of integration processes on exogenous DNA uptake and accurately reflect the influence of competence on transformation, we employed the non-integrating plasmid pIB166 as the donor. We found a significant impairment in the transformation efficiency of D39ΔtacL compared to wild-type D39. The complemented strain exhibited an increased transformation efficiency that did not reach the level observed in the wild-type strain but was still significantly higher than the deletion of tacL strain (Figure 1A).
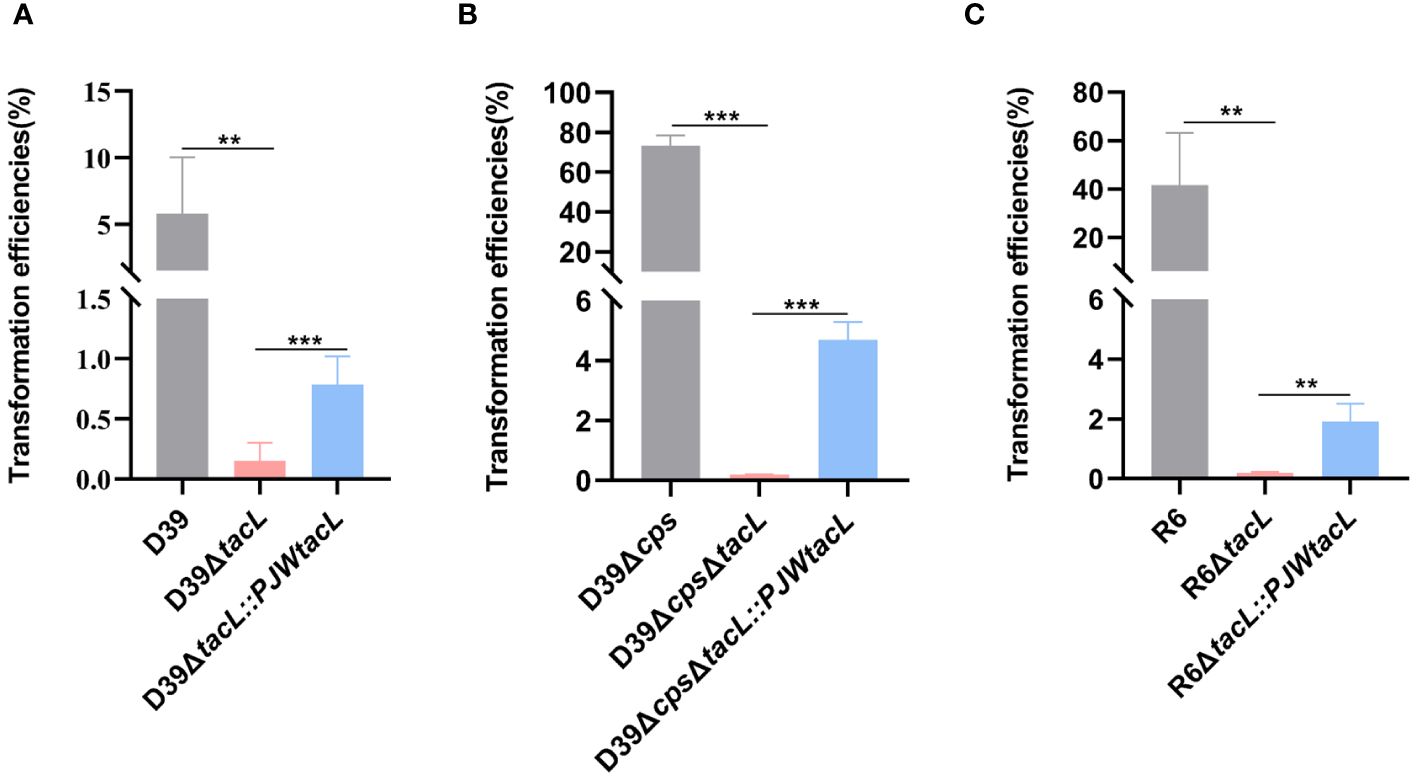
Figure 1 Deletion of tacL impairs pneumococcal transformation. (A) Transformation efficiency of wild-type D39, D39ΔtacL and the tacL complemented strains. (B) Transformation efficiency of non-encapsulated S. pneumoniae D39Δcps, D39ΔcpsΔtacL and complemented strains. (C) Transformation efficiency of non-encapsulated S. pneumoniae R6, R6ΔtacL and complemented strains. Statistical analysis was performed using an unpaired two-tailed Student’s t-test with significance levels denoted as ***P < 0.001, **P < 0.01.
These transformation assays were then conducted using non-encapsulated strains to eliminate any potential interference caused by CPS that is known to affect transformation efficiency. In the non-encapsulated strain D39Δcps, the tacL deletion resulted in a significant decrease in transformation efficiency compared to the cognate parental strain and the complemented strain exhibited higher transformation efficiency than the tacL deletion strain (Figure 1B). Similarly, the deletion of tacL resulted in compromised transformation efficiency in the R6 (non-encapsulated) strain background (Figure 1C). These findings underscore the importance of tacL in pneumococcal transformation.
To establish a correlation between tacL transcription and transformation, we performed DNA pull-down assays coupled with mass spectrometry (MS) analyses. A 207-bp long regulatory DNA probe of tacL was labeled at the 5’-end with biotin. The probe was then incubated with pneumococcal lysates and subjected to DNA affinity chromatography-pulldown. The binding proteins were separated by SDS-PAGE (Figure 2A) and subsequently subjected to MS analysis and this led to the identification of several potential regulatory proteins that included ComE, GntR family SPD_0064, MarR family SPD_0379, GlnR, and RitR25 (Figure 2B). Notably, ComE is a well-established protein that plays a pivotal role in S. pneumoniae transformation (Martin et al., 2010).
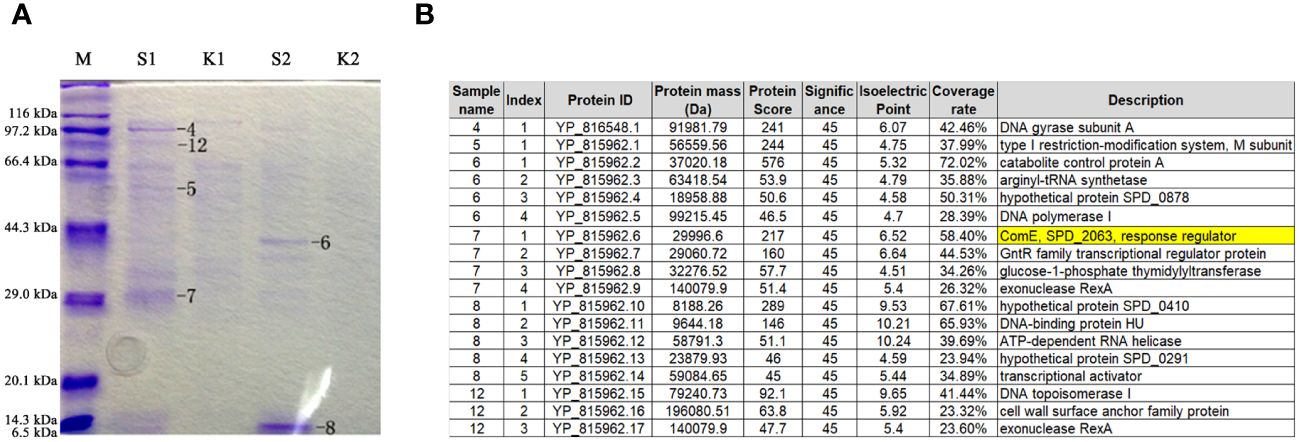
Figure 2 Identification of regulatory proteins capable of binding to the fragment of governing tacL expression. (A) Enriched proteins subjected to SDS-PAGE gel analysis. lane M: Protein molecular mass standards. S1 and S2: elution fractions obtained by binding with the purified specific target probe using 100 mM and 200 mM NaCl, respectively, the numbers 4-8 and 12 represent gel-excised and used for MS analysis. K1 and K2: elution fractions obtained by binding with the non-specific probe using 100 mM and 200 mM NaCl, respectively. (B) Proteins identified by mass spectrometry.
The transcription factor ComE is essential for competence development in S. pneumoniae (Ween et al., 1999; Martin et al., 2010), undergoes phosphorylation at aspartic acid residue 58 (D58) during competence, thereby triggering the expression of early competence genes (Martin et al., 2013). EMSA assays were conducted to validate the binding between ComE or its phosphomimic form and the DNA probe. Distinct shift bands were observed in a dose-dependent manner upon increasing concentrations of ComED58E (Figures 3A, B) while no shift bands were detected for ComEWT (Supplementary Figure S1). DNase I footprinting revealed the presence of a protected region approximately 30 base-pairs in length (Figure 3C) that was identified 31 base-pairs upstream from the start codon (Figure 3D). These findings indicated that phosphomimic ComED58E exhibited the specific binding affinity towards the upstream regulatory sequence of tacL.
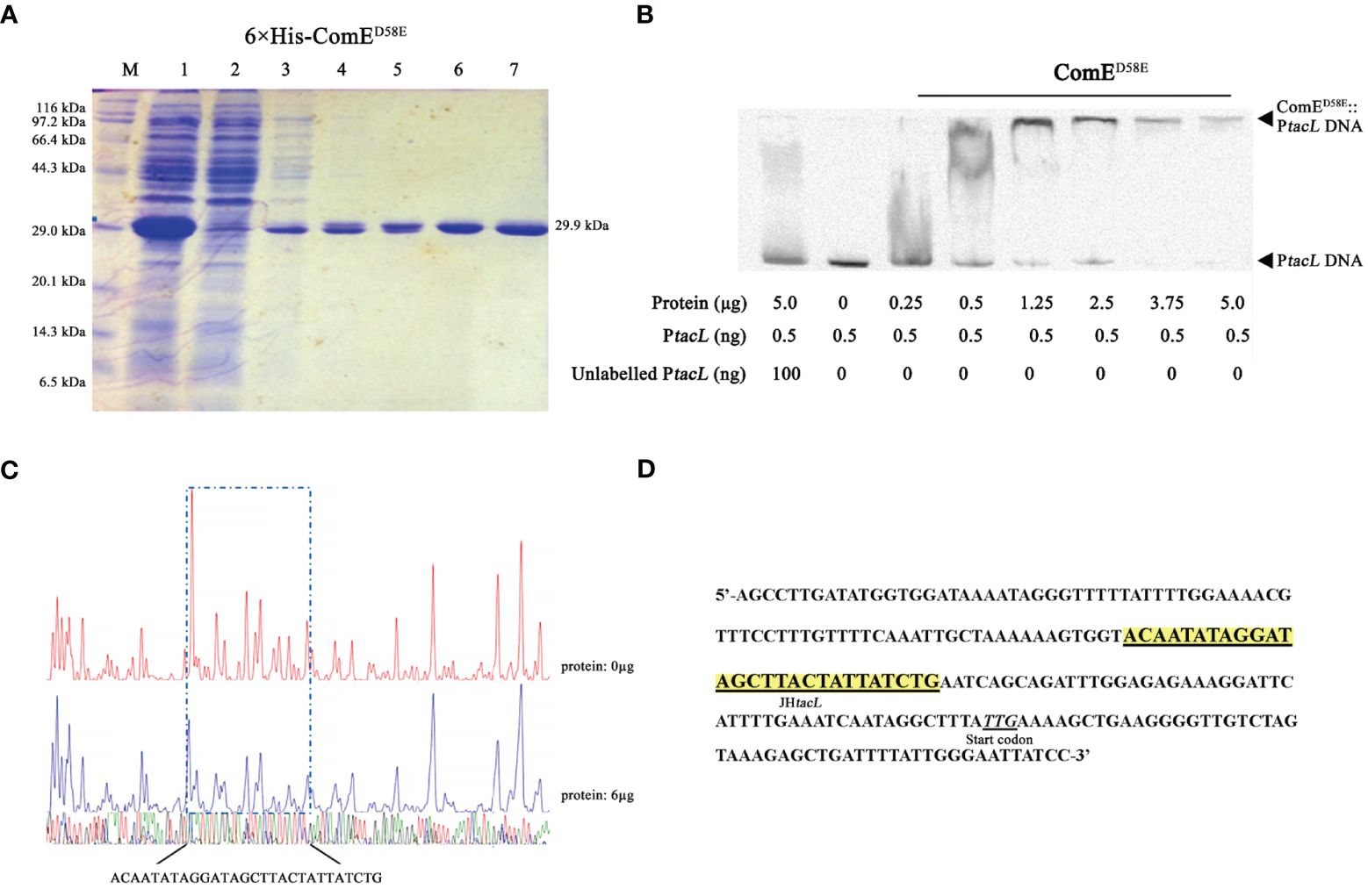
Figure 3 Phosphomimic ComED58E binds to the upstream regulatory region of tacL. (A) The recombinant 6×His-ComED58E protein was analyzed by SDS-PAGE and migrated with an apparent molecular mass of 29.9 kDa, Lane M: Marker; Lane 1, Protein expression induced by IPTG; Lane 2: Ni-NTA flow-through; Lanes 3 – 6: elution from the columns with imidazole at 20, 40, 80 and 500 mM, respectively. Lane 7: the purified form of 6×His-ComED58E. (B) EMSA results of binding between ComED58E and PtacL as indicated. (C) Sequence analysis of DNase I footprinting protection assay between ComED58E and the tacL DNA probe. (D) Structural organization of the tacL promoter-proximal region. The start codon was indicated. Binding sequences for ComED58E are highlighted in yellow.
The impact of the regulatory sequence of tacL on LTAs biosynthesis was demonstrated by generating a D39ΔJHtacL strain, in which the binding region identified by DNase I footprinting assay was deleted (Figure 4A). The mRNA expression level of tacL in D39ΔJHtacL was significantly upregulated compared to that in the wild-type strain (Figure 4B). ELISA, flow cytometry and dot-blot analyses revealed an increased abundance of teichoic acids and an up-regulated LTAs (manifested as smaller molecular-weight bands 10 and 25 kDa) was displayed by western blot in the JHtacL mutant when compared with the wild-type strain (Figures 4C–F).
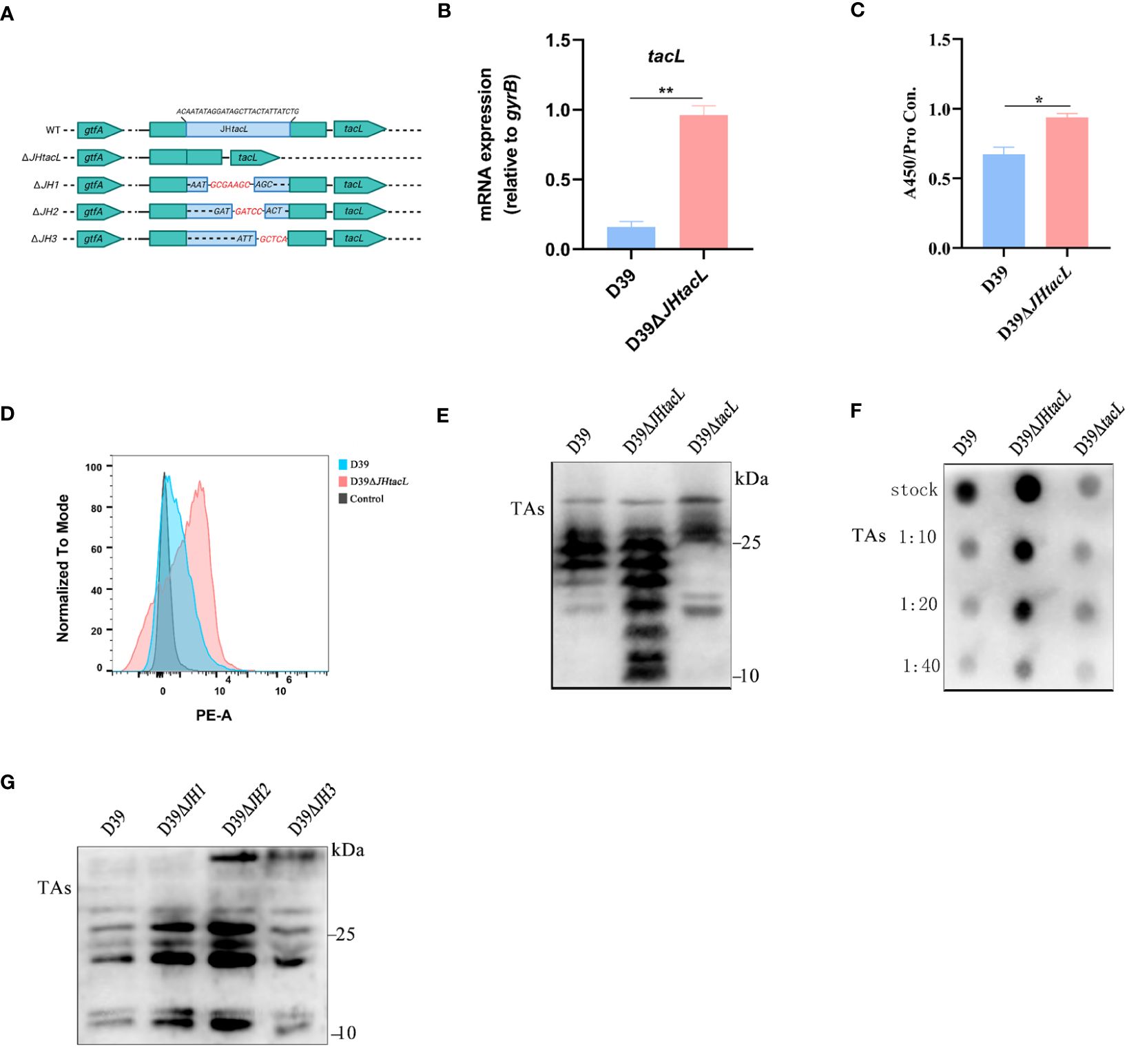
Figure 4 Deletion of JHtacL resulted in increased transcription of tacL and levels of LTAs. (A) Mutation patterns of the binding sites of JHtacL. (B) The transcript levels of tacL for the indicated strains. (C) ELISA analysis for TAs detection. (D) Flow cytometry to determine the surface exposed TAs. (E) Western blot and (F) Dot blot analysis of TAs for the indicated strains. (G) Western blot analysis of TAs of wild-type D39 and the indicated tacL promoter binding sequence mutations for a series of mutants (JH1/JH2/JH3). Bacteria were harvested at OD595nm of 0.5 for tacL transcription and TAs analysis, we used an bacterial density (defined as an OD595nm of 0.5) for normalizing. Statistical analysis was carried out using unpaired two-tailed Student’s t-test. ** P < 0.01 and * P < 0.05.
To determine the contribution of the regions within JHtacL that associate with phosphorylated ComE, a series of mutants were constructed to evaluate LTAs biosynthesis (Figure 4A). Among all tested site mutants, the JH2 mutant exhibited significantly enhanced LTAs production (Figure 4G). Collectively, these findings provide compelling evidence that the JHtacL region is involved in the regulation of tacL transcription, thereby exerting an influence on the biosynthesis of LTAs.
We postulated that ComE may exert regulatory control over the transcription of tacL during competence. By generating the D39ΔcomE mutant and its complementary strain, we observed a significant increase in mRNA levels of tacL in the comE mutant compared to the wild-type D39 strain, while no significant changes were observed in D39ΔcomE::PJWcomEWT (no available antibody for TacL, we resorted to assessing the transcript level of tacL) (Figure 5A). The active form of ComE protein, as assessed using a phosphomimic ComED58E, was previously identified as a regulator of cps locus transcription (Zheng et al., 2017). While, the ComE may be not activated to ComED58E during the bacteria’s logarithmic growth phase. Therefore, we constructed a complemented strain expressing ComED58E (ΔcomE::PJWcomED58E) to evaluate its impact on tacL transcription. We found a decreasing trend was evident in the complementing strain D39ΔcomE::PJWcomED58E (Figure 5B). These findings indicated that phosphorylated ComE is the form of the protein required to repress tacL transcription.
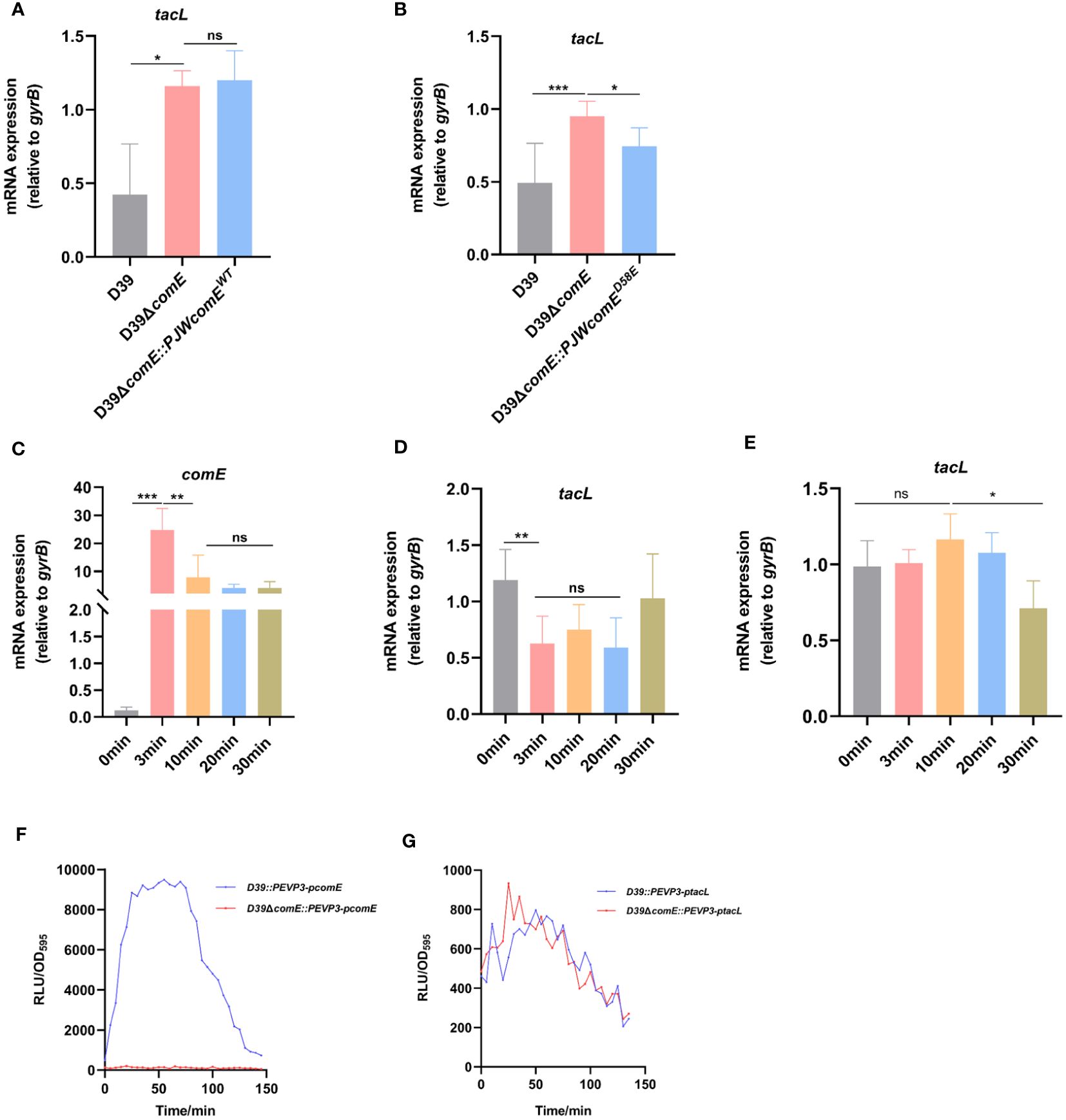
Figure 5 ComE negatively regulates tacL transcription during competence. (A, B) RT-PCR analysis of tacL expression in (A) the wild-type D39, D39ΔcomE, D39ΔcomE::PJWcomEWT and (B) D39ΔcomE:: PJWcomED58E. Bacteria were harvested at OD595nm of 0.5. The mRNA expression levels of tacL were normalized to gyrB expression. (C–E) RT-PCR to determine expression levels of (C) comE and (D) tacL in the wild-type D39 and examined the transcription of (E) tacL in D39ΔcomE. These strains were grown in competence THY medium until OD595nm 0.1 and induced with CSP for different durations (0 min, 3 min, 10 min, 20 min and 30 min). The mRNA expression levels of tacL were normalized to gyrB expression. (F, G) Transcriptional activity of (F) comE and (G) tacL was assessed using luciferase assays for the wild-type D39 and D39ΔcomE (comE-luc, tacL-luc) strains. These strains were grown to OD595nm 0.1 followed by the addition of 100 µg/mL of CSP and 2.7 mg/ml of luciferase. Luciferase activity and OD595nm were measured at 5 min intervals. Statistical analysis was performed using the unpaired two-tailed Student’s t-test or one-way ANOVA with Tukey’s multiple-comparison test. Statistical significance was denoted as *** P < 0.001, ** P < 0.01, * P < 0.05, while ns indicated non-significance.
To further elucidate this regulatory mechanism, we induced competence in the D39 strain by exogenous supplementation with CSP and monitored the expression of comE and tacL. During the initial phase, there was an upregulation in comE mRNA level (Figure 5C) while tacL exhibited a transient downregulation in the wild-type strain (Figure 5D), consistent with the transcriptome data was reported (Aprianto et al., 2018). The transcription of tacL remained unaltered following CSP induction in the D39ΔcomE mutant (Figure 5E). The luciferin reporter assay demonstrated enhanced transcriptional activity of comE (Figure 5F) while tacL experienced a transient decrease in D39 but remained unchanged in the D39ΔcomE strain upon CSP stimulation (Figure 5G). These findings indicated that ComE exerts a negative regulatory effect on tacL transcription during the initial phase of competence.
To demonstrate the regulatory role of ComE in TAs biosynthesis, we constructed the strains D39ΔcomE and D39ΔcomE::PJWcomEWT. The expression of ComE was absent in D39ΔcomE while GFP-ComE could be detected in the complemented strain (Figure 6A). The comE mutant displayed significantly elevated levels of LTAs (manifested as smaller molecular-weight bands 10 and 25 kDa) while the overall level of LTAs in the complemented strain remained unchanged compared to that in the ΔcomE mutant (Figure 6B). S. pneumoniae senses the CSP signal, the histidine kinase receptor ComD that undergoes autophosphorylation causing its cognate response regulator ComE to phosphorylate (Pestova et al., 1996; Martin et al., 2013). Therefore, exogenous CSP was used to induce the phosphorylation of ComE. We found that the complemented strain (D39ΔcomE::PJWcomEWT) induced by CSP displayed inhibition of LTAs content (Figure 6C). Meanwhile, the complemented strain expressing ComED58E (ΔcomE::PJWcomED58E) also exhibited a significant reduction in LTAs synthesis in Figures 6D, E. These findings suggested that phosphorylated ComE is involved in regulating the LTAs biosynthesis.
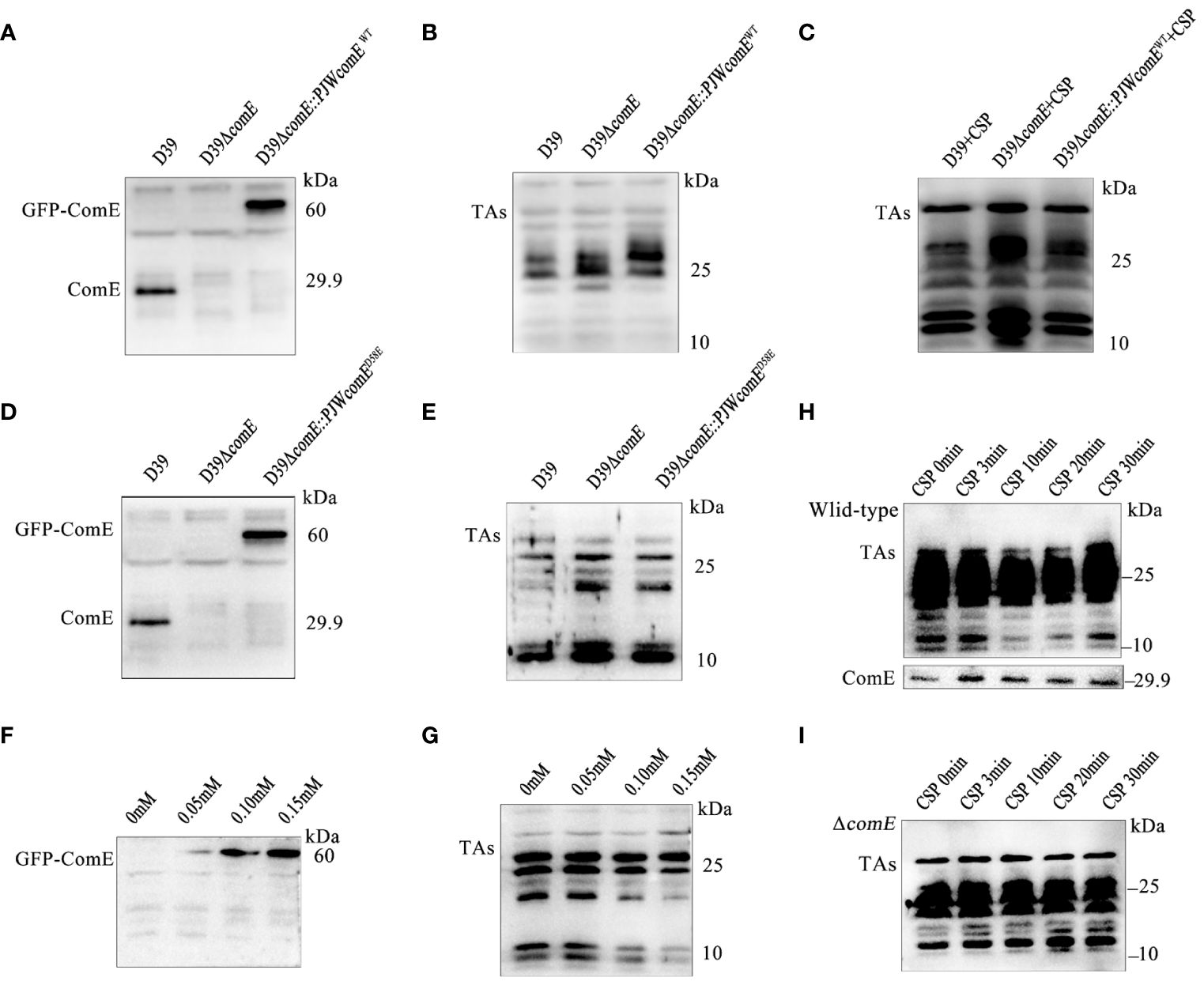
Figure 6 ComE negatively regulates LTAs production. (A, B) Western blot analysis levels of (A) ComE and (B) TAs in the indicated strains (wild type D39, D39ΔcomE and D39ΔcomE::PJWcomEWT), cultured to OD595nm of 0.5. (C) Western blot analysis of TAs levels for the indicated strains (wild type D39, D39ΔcomE and D39ΔcomE::PJWcomEWT). These strains cultured to OD595nm 0.1 followed by CSP induction for 15 min. Bacterial suspensions were harvested and adjusted for bacterial density (defined as an OD595nm of 0.5) for normalization. (D, E) Western blot analysis to determine levels of (D) ComE and analyze (E) TAs for the wild-type D39, D39ΔcomE, and D39ΔcomE:: PJWcomED58E strains, cultured to OD595nm of 0.5. (F, G) Western blot to determine levels of (F) ComE and (G) TAs for the D39ΔcomE:: PJWcomED58E strain in the presence of differing levels of ZnCl2 in the culture medium (0 mM, 0.05 mM, 0.10 mM and 0.15 mM) cultured to OD595nm of 0.5. (H, I) Western blot to assess the levels of TAs and ComE for the (H) wild-type D39 and (I) D39ΔcomE. The strains were cultured to OD595nm 0.1 followed by CSP induction for different durations (0 min, 3 min, 10 min, 20 min and 30 min). Bacterial suspensions were harvested and adjusted for bacterial density (defined as an OD595nm of 0.5) for normalization.
To validate the effect of phosphorylated ComE on LTAs biosynthesis, the D39ΔcomE-PJWcomED58E strain was grown in THY medium supplemented with varying concentrations of ZnCl2 to fine-tune the expression of ComED58E. ZnCl2 exposure led to a concentration-dependent up-regulation of GFP-ComED58E expression (Figure 6F) resulting in decreased levels of LTAs (Figure 6G). While a clear suppression of LTAs biosynthesis during competence was evident for the wild-type D39 (Figure 6H), no corresponding inhibitory effects were observed in D39ΔcomE during competence (Figure 6I). These findings strongly indicated that phosphorylated ComE plays a crucial role in inhibiting LTAs biosynthesis during competence.
To investigate the impact of ComE-regulated LTAs biosynthesis on pneumococcal transformation, we compared the tacL transcript levels and transformation efficiency among wild-type D39, D39ΔJH2, and D39ΔtacL strains. The tacL mRNA could not be detected in D39ΔtacL strain. In contrast to the wild-type strain, elevated levels of tacL transcript were observed in D39ΔJH2 (Figure 7A) and this correlated with a significantly enhanced transformation efficiency (Figure 7B).
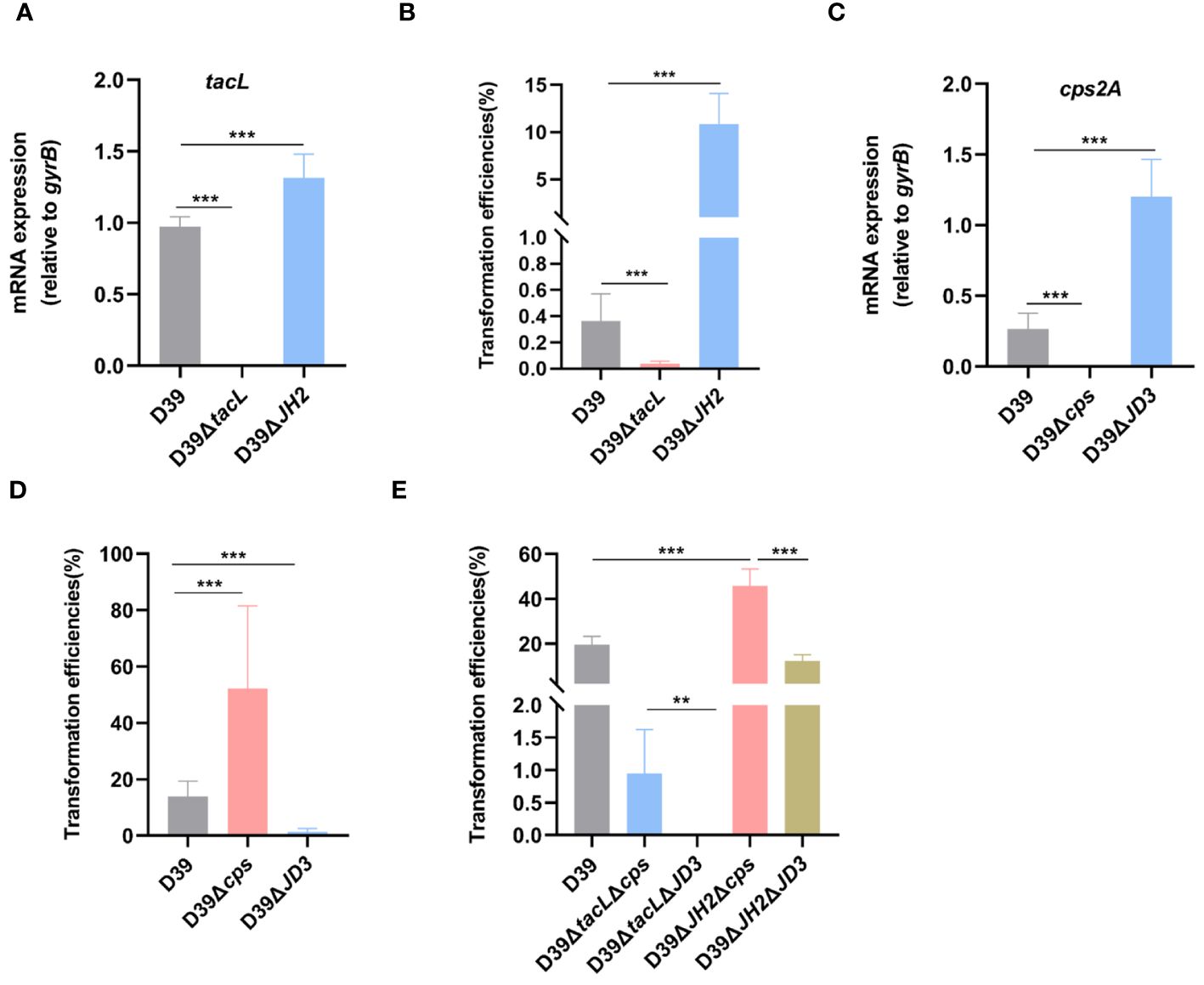
Figure 7 TacL-mediated LTAs biosynthesis affects pneumococcal transformation. (A) RT-PCR analysis of mRNA levels of tacL for the indicated strains (wild-type D39, D39ΔtacL and D39ΔJH2) harvested at OD595nm 0.5. The mRNA levels were expressed relative to gyrB. (B) Transformation efficiency of the indicated strains (wild-type D39, D39ΔtacL and D39ΔJH2) that expressed varying LTAs levels. (C) RT-PCR analysis of mRNA levels of cps2A for the indicated strains (wild-type D39, D39Δcps and D39ΔJD3). (D) Transformation efficiency of the indicated strains (wild-type D39, D39Δcps and D39ΔJD3) that expressed varying CPS contents. (E) Transformation efficiency of different bacterial phenotypes as indicated (D39, ΔtacLΔcps, ΔtacLΔJD3, ΔJH2Δcps and ΔJH2ΔJD3). Statistical analysis was performed using unpaired two-tailed Student’s t-test or one-way ANOVA with Tukey’s multiple-comparison test. Significance differences were denoted as *** P < 0.001; ** P < 0.01.
A previous study reported that ComE functions as a transcriptional repressor of CPS biosynthesis and thereby exerts a negative regulatory effect via binding to the promoter region JD3 of the cps gene (Zheng et al., 2017). We therefore confirmed that the D39ΔJD3 strain exhibited high expression levels of capsular polysaccharides (Supplementary Figure S2). We further validated the impact of ComE-regulated CPS biosynthesis on pneumococcal transformation and evaluated the transformation efficiency of the JD3 mutant. In comparison to the wild-type D39, D39Δcps exhibited a significant enhancement in transformation efficiency, whereas it was suppressed in the JD3 mutant (Figures 7C, D).
Furthermore, transformation efficiencies of mutants generated by combining the ComE-binding sites of tacL with cps regulatory sequences revealed that strains with higher LTAs levels (ΔJH2Δcps and ΔJH2ΔJD3) exhibited significantly enhanced transformation abilities compared to the other strains with lower levels in ΔtacLΔcps and ΔtacLΔJD3. Additionally, the pneumococcal transformation of the strains with higher CPS (ΔtacLΔJD3 and ΔJH2ΔJD3) was found to be suppressed in comparison to the D39ΔtacLΔcps and D39ΔJH2Δcps (Figure 7E). These findings indicated that ComE regulates both CPS and LTAs biosynthesis and this subsequently influences pneumococcal transformation capacity.
This study provides novel and significant insights into the regulation of teichoic acids biosynthesis during pneumococcal competence. Our findings demonstrate that phosphomimic ComE specifically binds to regulatory sequences spanning approximately 30 base pairs located 31 base pairs upstream from the start codon of the tacL gene, resulting in repression of tacL transcription. Moreover, our results emphasize the importance of regulation of TAs biosynthesis during competence and establish the involvement of tacL-mediated LTAs biosynthesis in pneumococcal transformation.
A previous CRISPRi-seq analysis indicated that tacL expression is suppressed during competence (Aprianto et al., 2018). We sought to identify the precise mechanism that underlies this inhibition. We screened the potential transcriptional regulators involved in linking competence with tacL and identified several potential regulators, including ComE. In S. pneumoniae, ComE undergoes phosphorylation in the presence of phosphorylated ComD, a transmembrane protein histidine kinase responsible for sensing CSP (Yang and Tal-Gan, 2019). Phosphorylated ComE is a central component of the cellular adaptive-response network, demonstrating a pivotal role in gene regulation, including comX gene that triggers genes essential for S. pneumoniae transformation (Kowalko and Sebert, 2008; Martin et al., 2013). Given its well-established function as a transcription factor and its involvement in competence (Martin et al., 2010; Zheng et al., 2017), we focused on elucidating the transcriptional regulatory mechanism used by ComE to regulate tacL expression. Since ComE phosphorylation at D58 is associated with competence, we expressed a recombinant functional phosphomimic ComED58E and conducted EMSA assays to unveil their interaction. We observed that ComED58E specifically bound to the tacL regulatory region. In vitro footprinting experiments also indicated that ComED58E bound to the regulatory region of tacL, and there was a specific binding sequence. We also observed that LTAs expression patterns were altered in the strain following deletion of the JHtacL sequence from the tacL promoter. This phenomenon may be caused by unpolymerized RU of TAs or other factors that we did not identify. Moreover, the JHtacL sequence exhibits sequence similarity with the TATCCTAAATGGT binding sites identified in S. mutans (Hung et al., 2011). The transcription of tacL is repressed by ComE during competence, resulting in the inhibition of LTAs biosynthesis. The repression is primarily attributed to the binding of ComE to the upstream regulatory region of tacL. Notably, CSP stimulation no longer reduces LTAs biosynthesis in comE mutants. Collectively, these findings indicate that ComE regulates the transcription of tacL by binding to its upstream regulatory region, thereby linking competence and TAs remodeling.
Upon CSP stimulation, we also found an immediate up-regulation of ComE protein levels, resulting in a reduction in tacL transcription (Figure 5D). As the response progressed, comE mRNA levels remained up-regulated and the tacL transcripts increased (Figures 5C, D). These findings suggested the presence of a complex regulatory network governing tacL transcription.
The tacL gene deletion strongly reduced CSP-induced transformation and agreed with a previous observation emphasizing the importance of phosphocholine modifications in TAs for pneumococcal transformation (Zhang et al., 1999; Minhas et al., 2023). Therefore, we hypothesized that tacL might be upregulated during competence. However, our findings indicated the contrary; the phosphomimic ComE suppressed the transcription of tacL during competence, and mutation in the binding sites resulted in increased tacL levels and LTAs amounts that promoted transformation. These results suggested a complex role of tacL-mediated LTAs biosynthesis during competence that requires further clarification.
Bacterial cell wall polysaccharides play numerous roles in transformation (Mirouze et al., 2018; Minhas et al., 2023) and in particular, CPS impedes pneumococcal transformation (Weiser and Kapoor, 1999; Li et al., 2019). Furthermore, phosphorylated ComE exerts a repressive effect on cps expression by binding to the cps promoter region, thereby inhibiting CPS biosynthesis (Zheng et al., 2017). However, it remains unclear whether this regulatory mechanism contributes to the process of pneumococcal transformation. Our findings underscore the significant involvement of ComE-mediated cps transcription in pneumococcal transformation, as evidenced by the impaired transformation resulting from mutations in the regulatory sequences of the cps locus This also partially reveals why competent bacteria possess lower levels of CPS (Weiser and Kapoor, 1999; Minhas et al., 2023). Collectively, we propose that cell wall remodeling is an event driven by ComE at the transcriptional level.
The competence response has been demonstrated to potentially induce fatal bacterial injury (Claverys et al., 2007; Straume et al., 2015). Therefore, stringent regulation of competence is imperative in order to prevent bystander damage. LTAs are glycopolymers that play crucial roles in cell division, surface adhesion and biofilm formation (Moscoso et al., 2006; Heß et al., 2017; Ye et al., 2018). The precise reasons underlying the simultaneous production of both LTAs and WTAs by S. pneumoniae remain incompletely understood. Activation of choline-binding proteins on the bacterial wall occurs during competence, resulting in lysis of non-competent pneumococci (Eldholm et al., 2010; Wei and Håvarstein, 2012). Simultaneously, immunity proteins within the competent pneumococcal cell walls are activated to provide protection against cleavage (Håvarstein et al., 2006; Straume et al., 2017). S. pneumoniae exhibits a phenomenon known as “fratricide” where competent cells engage in natural genetic transformation lyse and thereby eliminate noncompetent siblings present within their environment (Claverys et al., 2007). This phenomenon relies on the activation of the late competence protein CbpD. Competent pneumococci are protected from fratricide by producing the immunity proteins ComM and LytR that are regulated by the transcription factor ComE (Eldholm et al., 2009; Straume et al., 2015; Minhas et al., 2023). This subsequently results in an elevation of surface WTAs levels that function as a protective mechanism against lysis (Minhas et al., 2023). Although we did not provide evidence demonstrating the influence of ComE-regulated LTAs biosynthesis on autolysin activity, based on previous findings (Flores-Kim et al., 2019; Minhas et al., 2023), it is plausible to propose that the involvement of tacL-mediated LTAs biosynthesis regulated by ComE in competence is accountable for the regulation of cell wall lysis. The ComE-tacL-LTAs pathway may represent a novel molecular mechanism contributing to pneumococcal transformation. The regulation of fratricide activity partially elucidates the necessity for S. pneumoniae to produce both LTAs and WTAs.
In conclusion, our study provides compelling evidence that transcriptional repression of tacL-mediated LTAs biosynthesis by ComE constitutes a pivotal mechanism underlying the remodeling of the cell wall during competence, thereby exerting an impact on pneumococcal transformation.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
KFW, SY, MY and KW designed experiments. MY, GS, YH, JC, TL and LL carried out experiments. KW, JW, YZ, LW and HX analyzed experimental results. MY, KW and GS wrote the manuscript. KFW, SY, MY, YY and XZ reviewed and edited the manuscript. KFW, SY, and MY: Writing – review & editing and Writing – original draft. GS: Writing – original draft. KW, YH, JC, TL, LL, JW, HX, LW, YZ, XZ, and YY: Writing –review & editing. All authors contributed to the article and approved the submitted version.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support for this study was provided by the Guizhou High-level (BAI) Innovative Talents Project (QIANKehe Platform & Talents-GCC [2022]042-1), the Graduate Education Innovation Program Project of Zunyi Medical University (ZYK144), the Key Discipline Project of Clinical Laboratory Diagnostics funded by Guizhou Provincial Health Commission (QianWeijianhan [2021]160), Scientific project provided by Shanghai Municipal Health Commission (20215024), and the Key Discipline Project of Clinical Laboratory Diagnostics funded by Zunyi Municipal Health Bureau (2022-1444).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1375312/full#supplementary-material.
Aprianto, R., Slager, J., Holsappel, S., Veening, J.-W. (2018). High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res. 46, 9990–10006. doi: 10.1093/nar/gky750
Biswas, I., Jha, J. K., Fromm, N. (2008). Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiol. (Reading England) 154, 2275–2282. doi: 10.1099/mic.0.2008/019265-0
Chochua, S., Metcalf, B. J., Li, Z., Walker, H., Tran, T., McGee, L., et al. (2017). Invasive serotype 35B Pneumococci including an expanding serotype switch lineage, United States 2015-2016. Emerging Infect. Dis. 23, 922–930. doi: 10.3201/eid2306.170071
Claverys, J.-P., Martin, B., Håvarstein, L. S. (2007). Competence-induced fratricide in streptococci. Mol. Microbiol. 64, 1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x
Claverys, J.-P., Martin, B., Polard, P. (2009). The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33, 643–656. doi: 10.1111/j.1574-6976.2009.00164.x
Denapaite, D., Brückner, R., Hakenbeck, R., Vollmer, W. (2012). Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microbial Drug Resistance 18, 344–358. doi: 10.1089/mdr.2012.0026
Eberhardt, A., Wu, L. J., Errington, J., Vollmer, W., Veening, J.-W. (2009). Cellular localization of choline-utilization proteins in Streptococcus pneumoniae using novel fluorescent reporter systems. Mol. Microbiol. 74, 395–408. doi: 10.1111/j.1365-2958.2009.06872.x
Eldholm, V., Johnsborg, O., Haugen, K., Ohnstad, H. S., Håvarstein, L. S. (2009). Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiol. (Reading England) 155, 2223–2234. doi: 10.1099/mic.0.026328-0
Eldholm, V., Johnsborg, O., Straume, D., Ohnstad, H. S., Berg, K. H., Hermoso, J. A., et al. (2010). Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol. Microbiol. 76, 905–917. doi: 10.1111/j.1365-2958.2010.07143.x
Flores-Kim, J., Dobihal, G. S., Fenton, A., Rudner, D. Z., Bernhardt, T. G. (2019). A switch in surface polymer biogenesis triggers growth-phase-dependent and antibiotic-induced bacteriolysis. eLife 8, e44912. doi: 10.7554/eLife.44912.018
Håvarstein, L. S., Coomaraswamy, G., Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Håvarstein, L. S., Martin, B., Johnsborg, O., Granadel, C., Claverys, J.-P. (2006). New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59, 1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x
Heß, N., Waldow, F., Kohler, T. P., Rohde, M., Kreikemeyer, B., Gómez-Mejia, A., et al. (2017). Lipoteichoic acid deficiency permits normal growth but impairs virulence of Streptococcus pneumoniae. Nat. Commun. 8, 2093. doi: 10.1038/s41467-017-01720-z
Hung, D. C. I., Downey, J. S., Ayala, E. A., Kreth, J., Mair, R., Senadheera, D. B., et al. (2011). Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J. Bacteriol. 193, 3642–3652. doi: 10.1128/JB.00155-11
Jutras, B. L., Verma, A., Stevenson, B. (2012). Identification of novel DNA-binding proteins using DNA-affinity chromatography/pull down. Curr. Protoc. In Microbiol. Chapter 1, Unit1F.1. doi: 10.1002/9780471729259.mc01f01s24
Kim, S.-J., Seon, S. H., Luong, T. T., Ghosh, P., Pyo, S., Rhee, D.-K. (2019). Immunization with attenuated non-transformable pneumococcal pep27 and comD mutant provides serotype-independent protection against pneumococcal infection. Vaccine 37, 90–98. doi: 10.1016/j.vaccine.2018.11.027
Kirkham, L.-A. S., Kerr, A. R., Douce, G. R., Paterson, G. K., Dilts, D. A., Liu, D.-F., et al. (2006). Construction and immunological characterization of a novel nontoxic protective pneumolysin mutant for use in future pneumococcal vaccines. Infect. Immun. 74, 586–593. doi: 10.1128/IAI.74.1.586-593.2006
Kowalko, J. E., Sebert, M. E. (2008). The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect. Immun. 76, 3131–3140. doi: 10.1128/IAI.01696-07
Lam, T., Ellison, C. K., Eddington, D. T., Brun, Y. V., Dalia, A. B., Morrison, D. A. (2021). Competence pili in Streptococcus pneumoniae are highly dynamic structures that retract to promote DNA uptake. Mol. Microbiol. 116, 381–396. doi: 10.1111/mmi.14718
Li, X., Liu, Y., Li, M., Bian, J., Song, D., Liu, C. (2023). Epidemiological investigation of lower respiratory tract infections during influenza A (H1N1) pdm09 virus pandemic based on targeted next-generation sequencing. Front. Cell. Infect. Microbiol. 13, 1303456. doi: 10.3389/fcimb.2023.1303456
Li, J., Zhang, J.-R., Fischetti, V. A., Novick, R. P., Ferretti, J. J., Portnoy, D. A., et al. (2019). Phase variation of streptococcus pneumoniae. Microbiol. Spectr. 7, GPP3-005-2018. doi: 10.1128/microbiolspec.GPP3-005-2018
Liu, X., Gallay, C., Kjos, M., Domenech, A., Slager, J., van Kessel, S. P., et al. (2017). High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 13, 931. doi: 10.15252/msb.20167449
Lo, S. W., Gladstone, R. A., van Tonder, A. J., Lees, J. A., du Plessis, M., Benisty, R., et al. (2019). Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect. Dis. 19, 759–769. doi: 10.1016/S1473-3099(19)30297-X
Martin, B., Granadel, C., Campo, N., Hénard, V., Prudhomme, M., Claverys, J.-P. (2010). Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol. Microbiol. 75, 1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x
Martin, B., Soulet, A.-L., Mirouze, N., Prudhomme, M., Mortier-Barrière, I., Granadel, C., et al. (2013). ComE/ComE~P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87, 394–411. doi: 10.1111/mmi.12104
Meng, J.-P., Yin, Y.-B., Zhang, X.-M., Huang, Y.-S., Lan, K., Cui, F., et al. (2008). Identification of Streptococcus pneumoniae genes specifically induced in mouse lung tissues. Can. J. Microbiol. 54, 58–65. doi: 10.1139/W07-117
Minhas, V., Domenech, A., Synefiaridou, D., Straume, D., Brendel, M., Cebrero, G., et al. (2023). Competence remodels the pneumococcal cell wall exposing key surface virulence factors that mediate increased host adherence. PloS Biol. 21, e3001990. doi: 10.1371/journal.pbio.3001990
Mirouze, N., Ferret, C., Cornilleau, C., Carballido-Lopez, R. (2018). Antibiotic sensitivity reveals that wall teichoic acids mediate DNA binding during competence in Bacillus subtilis. Nat. Commun. 9, 5072. doi: 10.1038/s41467-018-07553-8
Moscoso, M., García, E., López, R. (2006). Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188, 7785–7795. doi: 10.1128/JB.00673-06
Ottolenghi, E., Macleod, C. M. (1963). Genetic transformation among living pneumococci in the mouse. Proc. Natl. Acad. Sci. U.S.A. 50, 417–419. doi: 10.1073/pnas.50.3.417
Pestova, E. V., Håvarstein, L. S., Morrison, D. A. (1996). Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21, 853–862. doi: 10.1046/j.1365-2958.1996.501417.x
Prudhomme, M., Libante, V., Claverys, J.-P. (2002). Homologous recombination at the border: insertion-deletions and the trapping of foreign DNA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 99, 2100–2105. doi: 10.1073/pnas.032262999
Slager, J., Aprianto, R., Veening, J.-W. (2019). Refining the pneumococcal competence regulon by RNA sequencing. J. Bacteriol. 2019, 201. doi: 10.1128/JB.00780-18
Straume, D., Stamsås, G. A., Håvarstein, L. S. (2015). Natural transformation and genome evolution in Streptococcus pneumoniae. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 33, 371–380. doi: 10.1016/j.meegid.2014.10.020
Straume, D., Stamsås, G. A., Salehian, Z., Håvarstein, L. S. (2017). Overexpression of the fratricide immunity protein ComM leads to growth inhibition and morphological abnormalities in Streptococcus pneumoniae. Microbiol. (Reading England) 163 (1), 9–21. doi: 10.1099/mic.0.000402
Sung, C. K., Li, H., Claverys, J. P., Morrison, D. A. (2001). An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67, 5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001
Vidal, J. E., Bou Ghanem, E. N., Wu, X., Wu, K., Bai, G., Hammerschmidt, S. (2022). Editorial: Transmission, colonization, and molecular pathogenesis of pneumococcus. Front. Cell. Infect. Microbiol. 12, 1028047. doi: 10.3389/fcimb.2022.1028047
Vollmer, W., Massidda, O., Tomasz, A. (2019). The cell wall of Streptococcus pneumoniae. Microbiol. Spectr. 7 (3), GPP3-0018-2018. doi: 10.1128/microbiolspec.GPP3-0018-2018
Ween, O., Gaustad, P., Håvarstein, L. S. (1999). Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33, 817–827. doi: 10.1046/j.1365-2958.1999.01528.x
Wei, H., Håvarstein, L. S. (2012). Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl. Environ. Microbiol. 78, 5897–5905. doi: 10.1128/AEM.01343-12
Weiser, J. N., Austrian, R., Sreenivasan, P. K., Masure, H. R. (1994). Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62, 2582–2589. doi: 10.1128/iai.62.6.2582-2589.1994
Weiser, J. N., Kapoor, M. (1999). Effect of intrastrain variation in the amount of capsular polysaccharide on genetic transformation of Streptococcus pneumoniae: implications for virulence studies of encapsulated strains. Infect. Immun. 67, 3690–3692. doi: 10.1128/IAI.67.7.3690-3692.1999
Wu, K., Huang, J., Zhang, Y., Xu, W., Xu, H., Wang, L., et al. (2014). A novel protein, RafX, is important for common cell wall polysaccharide biosynthesis in Streptococcus pneumoniae: implications for bacterial virulence. J. Bacteriol. 196, 3324–3334. doi: 10.1128/JB.01696-14
Yan, Z., Cui, Y., Huang, X., Lei, S., Zhou, W., Tong, W., et al. (2021). Molecular characterization based on whole-genome sequencing of streptococcus pneumoniae in children living in Southwest China during 2017-2019. Front. Cell. Infect. Microbiol. 11, 726740. doi: 10.3389/fcimb.2021.726740
Yang, Y., Tal-Gan, Y. (2019). Exploring the competence stimulating peptide (CSP) N-terminal requirements for effective ComD receptor activation in group1 Streptococcus pneumoniae. Bioorganic Chem. 89, 102987. doi: 10.1016/j.bioorg.2019.102987
Ye, W., Zhang, J., Shu, Z., Yin, Y., Zhang, X., Wu, K. (2018). Pneumococcal LytR protein is required for the surface attachment of both capsular polysaccharide and teichoic acids: essential for Pneumococcal virulence. Front. Microbiol. 9, 1199. doi: 10.3389/fmicb.2018.01199
Zhang, J. R., Idanpaan-Heikkila, I., Fischer, W., Tuomanen, E. I. (1999). Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31, 1477–1488. doi: 10.1046/j.1365-2958.1999.01291.x
Zhang, Y., Zhang, J., Xiao, J., Wang, H., Yang, R., Guo, X., et al. (2023). comCDE (Competence) operon is regulated by CcpA in Streptococcus pneumoniae D39. Microbiol. Spectr. 11, e0001223. doi: 10.1128/spectrum.00012-23
Zheng, Y., Zhang, X., Wang, X., Wang, L., Zhang, J., Yin, Y. (2017). ComE, an essential response regulator, negatively regulates the expression of the capsular polysaccharide locus and attenuates the bacterial virulence in Streptococcus pneumoniae. Front. Microbiol. 8, 277. doi: 10.3389/fmicb.2017.00277
Keywords: Streptococcus pneumoniae, transformation, competence, teichoic acids, ComE, transcriptional regulation
Citation: Yao M, Wang K, Song G, Hu Y, Chen J, Li T, Liang L, Wu J, Xu H, Wang L, Zheng Y, Zhang X, Yin Y, Yao S and Wu K (2024) Transcriptional regulation of TacL-mediated lipoteichoic acids biosynthesis by ComE during competence impacts pneumococcal transformation. Front. Cell. Infect. Microbiol. 14:1375312. doi: 10.3389/fcimb.2024.1375312
Received: 23 January 2024; Accepted: 08 April 2024;
Published: 08 May 2024.
Edited by:
Jing-Ren Zhang, Tsinghua University, ChinaReviewed by:
Donald A. Morrison, University of Illinois Chicago, United StatesCopyright © 2024 Yao, Wang, Song, Hu, Chen, Li, Liang, Wu, Xu, Wang, Zheng, Zhang, Yin, Yao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shifei Yao, c2hpZmVpeWFvMTAwNUAxNjMuY29t; Kaifeng Wu, a2lwaG9vbnd1QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.